Abstract
Astragalosides are the main active constituents of traditional Chinese medicine Huang-Qi, of which cycloastragenol-type glycosides are the most typical and major bioactive compounds. This kind of compounds exhibit various biological functions including cardiovascular protective, neuroprotective, etc. Owing to the limitations of natural sources and the difficulties encountered in chemical synthesis, re-engineering of biosynthetic machinery will offer an alternative and promising approach to producing astragalosides. However, the biosynthetic pathway for astragalosides remains elusive due to their complex structures and numerous reaction types and steps. Herein, guided by transcriptome and phylogenetic analyses, a cycloartenol synthase and four glycosyltransferases catalyzing the committed steps in the biosynthesis of such bioactive astragalosides were functionally characterized from Astragalus membranaceus. AmCAS1, the first reported cycloartenol synthase from Astragalus genus, is capable of catalyzing the formation of cycloartenol; AmUGT15, AmUGT14, AmUGT13, and AmUGT7 are four glycosyltransferases biochemically characterized to catalyze 3-O-xylosylation, 3-O-glucosylation, 25-O-glucosylation/O-xylosylation and 2ʹ-O-glucosylation of cycloastragenol glycosides, respectively. These findings not only clarified the crucial enzymes for the biosynthesis and the molecular basis for the structural diversity of astragalosides in Astragalus plants, also paved the way for further completely deciphering the biosynthetic pathway and constructing an artificial pathway for their efficient production.
Key words: Cycloastragenol-type astragalosides, Cycloartenol synthase, Glycosyltransferase, Biosynthesis, Astragalus membranaceus
Graphical abstract
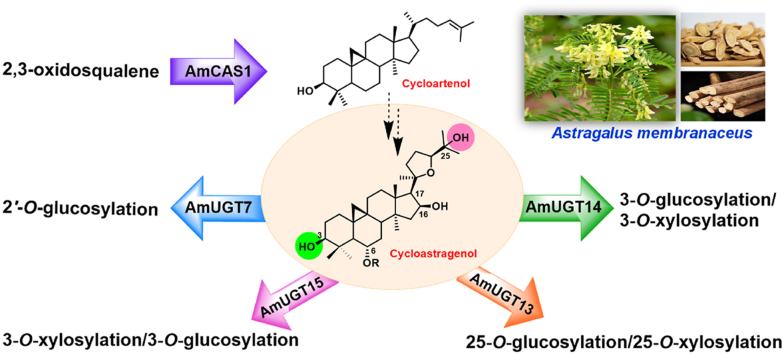
A cycloartenol synthase and four glycosyltransferases with different glycosylation patterns in the biosynthesis of cycloastragenol-type astragalosides were identified from Astragalus membranaceus.
1. Introduction
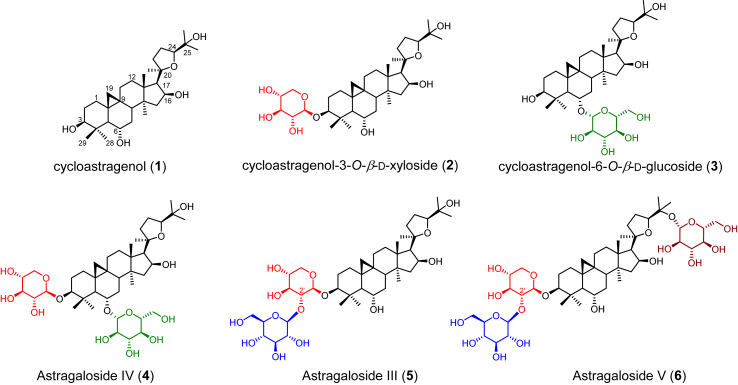
Astragalus is the largest genus of Leguminosae family, and Astragalus membranaceus (Fisch.) Bge. var. mongholicus (Bge.) Hisao and A. membranaceus (Fisch.) Bge are the two most well-known medicinal species in this genus. The dried roots of the two plants (Astragali Radix) are named as “Huang-Qi” in traditional Chinese medicine (TCM), which has long been used as an important “Qi” tonifying adaptogenic herb and prescribed in combination with other Chinese herbal medicines in traditional medicinal preparations1, 2, 3. The chemical constituents of “Huang-Qi” mainly include triterpenoids4, 5, 6, 7, 8, polysaccharides9, flavonoids10, phenylpropanoids11, alkaloids12, and steroids13, and show a variety of biological activities. Astragalosides (triterpenoid saponins) have been commonly considered as the main active constituents, of which cycloastragenol-type glycosides are the most typically important and bioactive compounds of Huang-Qi (1–6, Fig. 1). For example, astragaloside IV (4, Fig. 1) has attracted tremendous concern due to its neuroprotective14, hepatoprotective15, anticancer16, and antidiabetic activity17; astragaloside III (5, Fig. 1) could be a potential drug candidate for its anti-inflammatory18 and substantial antiviral19 activity. However, the low yield and intermittent supply of astragalosides from natural or cultivated sources of A. membranaceus severely limit the drug research and development of this type of bioactive compounds20,21. Moreover, the complex stereochemical rings with multiple chiral centers and various post modifications (oxidation, glycosylation, etc.) make the chemical synthesis of astragalosides very difficult and not readily and commercially accessible22. In comparison, biosynthesis and synthetic biology might be a promising approach to producing such structurally complex but biologically active triterpenoid glycosides23, 24, 25, 26.
Figure 1.
Representative cycloastragenol-type astragalosides in Astragalus species. Diverse glycosylation could occur at the 3-OH (red), 6-OH (green), 25-OH (brown) or 2′-OH (blue).
Cycloastragenol-type glycosides, a kind of rare triterpenoid glycosides in nature, presumably were derived from the initial cyclization of 2,3-oxidosqualene to cycloartenol (7) bearing a typical 9,19-cyclopropane moiety inherent in the cyclization by cycloartenol synthase (CAS). After that cycloastragenol (1, Fig. 1), featuring a 20,24-tetrahydrofuran ring (20,24-epoxy) in the side chain and various hydroxyl groups at different sites (mainly C6, C16, and C25) was hypothesized to be generated through a series of oxidations and furan ring formation. Subsequently, structurally diverse glycosides are biosynthesized via a variety of glycosylation patterns including at different sites (3-OH, 6-OH, 25-OH, and 2′-OH), with different sugars (glucose and xylose) and indeterminate numbers of glycosyl moieties (mono-, di-, tri-, or branched sugar chains). Whereas the genes and their encoding enzymes responsible for these oxygenation, glycosylation, and other reactions are rarely reported. Until recently, only a glycosyltransferase responsible for cycloastragenol glycosides was identified from A. membranaceus27. Therefore, identification of their biosynthetic genes/enzymes is highly necessary for deciphering and further constructing the biosynthetic pathway of bioactive astragalosides.
Herein, guided by the transcriptome and phylogenetic analysis, we described the in vivo (in yeast) and in vitro functional identification of AmCAS1, the first reported CAS catalyzing 2,3-oxidosqualene into cycloartenol in A. membranaceus. Moreover, we biochemically screened 24 candidate genes in vitro and functionally characterized four glycosyltransferases (AmUGT15, AmUGT14, AmUGT13, and AmUGT7) responsible for 3-O-xylosylation, 3-O-glucosylation, 25-O-glucosylation/25-O-xylosylation, and 2′-O-glucosylation in the biosynthesis of cycloastragenol-type glycosides, respectively.
2. Materials and methods
2.1. Chemical reagents
Cycloastragenol (1), astragaloside IV (4), astragaloside III (5), and cycloartenol (7) were purchased from Sigma–Aldrich (St. Louis, MO, USA). Cycloastragenol-3-O-β-d-xyloside (2) and cycloastragenol-6-O-β-d-glucoside (3) were prepared by acid hydrolysis of 428. UDP-xylose (UDP-Xyl) and UDP-glucose (UDP-Glc) were purchased from Beijing BG Biotech Co. Ltd. (Beijing, China). KOD-Plus DNA Polymerase was purchased from Toyobo Biotech Co., Ltd. (Shanghai, China). Primer synthesis was conducted at Generay Company (Shanghai, China), and DNA sequencing was performed at Tsingke Biotech Company (Beijing, China). Restriction enzymes and DNA ligase were purchased from Takara Biotechnology Co. Ltd. (Dalian, China).
2.2. Analytical procedures
The extract of engineered yeast with CASs was analyzed on a Shimadzu GCMS-QP2010 Plus system (Shimadzu, Japan). Enzymatic products were detected on a Shimadzu LC-20AD HPLC system with an evaporative light scattering detector (ELSD) and the molecular weights of products were measured on an Agilent 1260 series HPLC system (Agilent Technologies, Germany) coupled with an LCQ Fleet ion trap mass spectrometer (Thermo Electron Corp., USA) equipped with an electrospray ionization (ESI) source. Optical rotations were measured on a Perkin–Elmer Model-343 digital polarimeter (PerkinElmer Inc., Waltham, MA, USA). 1H and 13C NMR, 1H–1H COSY, HSQC, NOE and HMBC spectra were obtained on Bruker AVIIIHD-400, 500, and 600 spectrometers. Chemical shifts (δ) were referenced to internal solvent resonances and given in parts per million (ppm), and coupling constants (J) were given in hertz (Hz).
2.3. Plant materials and transcriptome sequencing
Young roots of biennial A. membranaceus (Fisch) Bge. var. mongholicus (Bge) were collected from Medicinal Botanical Garden of the Temple of Heaven (Beijing, China) and immediately frozen with liquid nitrogen. Transcriptome sequencing and data was acquired from BGISEQ-500 platform with a total length of 10.29 Gb (The Beijing Genomics Institute, BGI). After assembly and redundancy removal, 74,090 unigenes were obtained, with total length, average length, N50 and GC content of 61, 594, 720 bp, 830 bp, 1297 bp and 39.87%, respectively. Unigenes were compared to seven functional databases, and 44,709 (NR: 60.34%), 41,288 (NT: 55.73%), 27,914 (SwissProt: 37.68%), 33,722 (KOG: 45.51%), 31,902 (KEGG: 43.06%), 27,009 (GO: 36.45%), and 35,888 (InterPro: 48.44%) unigenes were annotated. 36,929 CDS were detected using Transdecoder. In addition, 8211 SSR markers were detected in 7021 unigenes and 1408 unigenes encoding transcription factors were predicted. The RNA-seq data were utilized to mine the biosynthetic genes of astragalosides.
2.4. RNA isolation, cDNA synthesis and gene clone
Total RNA of A. membranaceus (Fisch) Bge. var. mongholicus (Bge) Hsiao biennial root was extracted following manufacture's protocols of E.Z.N.A.™ Plant RNA kit (Omega Bio-Tek, USA). The RNA concentration and the ratios of A260/A280 and A260/A230 were measured with Biospectrometer (Eppendorf, USA) and the quality of RNA was examined by 0.8% gel electrophoresis, followed by the first strand cDNA synthesis instructed by the SMARTer™ RACE cDNA Amplification Kit protocols. The full-length of candidate genes were amplified from the cDNA by PCR using the primer pairs (Supporting Information Table S1) and the missing sequences of the tentative partial cDNAs were obtained by RACE (Rapid Amplification of cDNA End). The PCR process was performed using KOD-Plus DNA polymerase, and the PCR products were cloned into suitable expression vectors and verified by sequencing. The codon-optimized genes were conducted the same procedures as aforementioned.
2.5. Sequence alignment and phylogenetic analysis of cycloartenol synthases and glycosyltransferases
The encoding amino acid sequences of candidate cycloartenol synthases and glycosyltransferases were exported using online NCBI ORF finder and multiple sequence alignment was implemented using DNAMAN software, respectively. The neighbor-joining phylogenetic tree was constructed using MEGA 7.0 software based on ClustalW multiple alignments (bootstrap = 1000). The phylogenetic tree was further annotated by online iTOL website (https://itol.embl.de/).
2.6. Construction of recombinant expression systems for cycloartenol synthase
The genes encoding cycloartenol synthase were amplified from the cDNA of A. membranaceus and inserted into the BamH I restriction enzyme site of pESC-His vector, resulting in the pESC-His-AmCASs for heterologous expression in Saccharomyces cerevisiae INVSc1. Additionally, the gene encoding truncated HMG-CoA reductase29 (tHMG1) was cloned into pESC-Ura vector, and the gene encoding antisense lanosterol synthase29 (ERG7−) was cloned into pESC-Leu vector. The three positive recombinant plasmids were transformed into the competent cells of S. cerevisiae INVSc1 simultaneously instructed by the protocols of Frozen-EZ Yeast Transformation II Kit (Zymo Research, Irvine, CA, USA). As controls, the engineered yeast harboring the empty pESC-His vector and the other two recombinant plasmids with tHMG1 and ERG7–, and the engineered yeast with three empty vectors (pESC-His, pESC-Leu and pESC-Ura) were also individually constructed. All of the engineered strains were verified by PCR analysis of the genomic DNA following the instruction of Rapid Yeast Genomic DNA Isolation Kit (Sangon, China).
2.7. Functional identification of cycloartenol synthase in engineered yeast
The positive colonies were activated on the solid synthetic complete medium without histidine, leucine and uracil (SD-HLU) for 2 days and inoculated into 10 mL SD medium lacking the corresponding nutrition with 20 g/L glucose at 30 °C and 220 rpm for 24 h. The cells were collected by centrifugation at 1000×g for 5 min and washed with sterile water twice, then induced in 200 mL SG-HLU medium with 2% galactose at an initial OD600 of 0.6 to grow at 30 °C and 220 rpm for another 5 days. The cells were collected by centrifugation, weighted and lysed with adding 40 mL 20% KOH and 60 mL 50% ethanol per gram of cells by ultrasonication for 90 min. The resulting lysate was extracted with n-hexane for 3 times and the organic layer was evaporated under reduced pressure to afford a residue, which was redissolved with n-hexane for GC–MS analysis. GC–MS analysis was performed on Shimadzu gas chromatograph (split, 15:1; injector temperature, 250 °C) with a Rtx-5MS (30 mm × 0.25 mm × 0.25 μm) capillary column. 1 μL of the concentrated organic phase was injected under a He flow rate of 1 mL/min with a temperature program of 0 min at 40 °C, followed by a gradient from 40 to 200 °C at 20 °C/min with a 5 min hold time, then to 300 °C at 25 °C/min with a 15 min hold time. The ion trap temperature was 200 °C, and the electron energy was 70 eV. Spectra were recorded in the range of m/z 45–492. To prepare the products, a 10 L scale of fermentation was performed as described above. The resultant n-hexane extracts were chromatographed over normal-phase semi-preparative HPLC with n-hexane: EtOAc (v/v, 10:1) as mobile phase at 4 mL/min to obtain the product-enriched fractions. The samples were further purified by HPLC–DAD at 203 nm with ACN or 95% ACN as mobile phase at 1 mL/min to give the cycloartenol (7) and the two metabolites (7a and 7b). The compounds were redissolved in chloroform-d for NMR analysis.
2.8. In vitro assay for cycloartenol synthase
The constructed plasmid pESC-His-AmCAS1 and the empty pESC-His vector was individually transformed into S. cerevisiae INVSc1 for heterologous expression. The resultant positive colonies were cultured and induced with 2% galactose for 24 h as above described. Yeast cells were harvested and washed by centrifugation, then resuspended in 0.1 mol/L potassium phosphate buffer (pH 7.4, containing 0.45 mol/L sucrose, 1 mmol/L EDTA, 1 mmol/L DTT and 1 mmol/L phenylmethylsulfonyl fluoride (PMSF)) and homogenized with zirconia beads by JXFSTPRP-64 L (Shanghai Jingxin Industrial Development Co., Ltd., China). The homogenate was centrifuged at 12,000×g for 20 min, and the supernatant was further ultracentrifuged at 160,000×g for 90 min. The pellet was resuspended in 300 μL of 0.1 mol/L potassium phosphate buffer to afford microsomal fraction. For enzyme reaction, microsomal fraction (98 μL) was incubated with 10 μg of 2,3-oxidosqualene and 0.1% Triton X-100 in a total volume of 100 μL at 30 °C for 12 h. The reaction was terminated by the addition of 40 μL 20% KOH and 60 μL 50% ethanol and a 30 min sonication step. The resultant mixture was extracted with two volumes of n-hexane twice. The organic layer was evaporated until dryness, then was dissolved in 50 μL n-hexane for GC–MS analysis.
2.9. Expression and purification of candidate glycosyltransferases
The recombinant plasmids with candidate glycosyltransferase genes and empty pET-28a (+) were transformed into Escherichia coli Transetta (DE3) (TransGen Biotech, China) for heterologous expression. The described strains were cultivated overnight in 10 mL LB media containing 50 μg/mL kanamycin and 34 μg/mL chloromycetin at 37 °C and 200 rpm. The cultures were inoculated in fresh LB media at a ratio of 1:100 and shaken at 37 °C and 200 rpm until the optical density of the culture at 600 nm (OD600) reached 0.4–0.6, then induced with 0.2 mmol/L isopropyl β-d-thiogalactoside (IPTG) for 18 h at 16 °C and 200 rpm. The cells were harvested by centrifugation at 8000×g for 6 min and washed twice with deionized water, then re-suspended in binding buffer (20 mmol/L phosphate buffer, 0.5 mol/L NaCl, 20 mmol/L imidazole, pH 7.4) containing 1 mmol/L PMSF for sonication disruption in an ice bath. Afterwards, the cell debris was removed by centrifugation at 10,000×g for 1 h at 4 °C, then the soluble fraction was filtered by a 0.45 μm syringe filter unit and the supernatant protein was immediately applied to a 1 mL column of Ni-NTA resin (GE, USA) that was pre-equilibrated with binding buffer. The resin was subsequently eluted with 5 mL washing buffer (20 mmol/L phosphate buffer, 0.5 mol/L NaCl, 50 mmol/L imidazole, pH 7.4). The protein purification was carried out with 5 mL different elution buffer (20 mmol/L phosphate buffer, 0.5 mol/L NaCl, 75–250 mmol/L imidazole, pH 7.4) at a flow rate of 1 mL/min at 4 °C. The purified proteins were concentrated and buffer exchanged to a desalting buffer (50 mmol/L Tris-HCl buffer, 50 mmol/L NaCl, 1 mmol/L DTT, 1.5% glycerol, pH 7.4) using an Amicon Ultra-30 K centrifugal concentrator (Millipore, USA). The protein expression analysis was conducted by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the protein concentration was determined using the Protein Quantitative Kit.
2.10. Enzymatic activity assays of glycosyltransferases in vitro
The reaction mixture contained 0.4 mmol/L acceptor substrates (1–7) individually, 0.8 mmol/L donor substrates (UDP-Glc or UDP-Xyl), 50 μg purified recombinant enzymes, and 50 mmol/L Tris-HCl buffer (pH 7.4) in a final volume of 100 μL at 30 °C for 12 h. Control experiments were performed with the boiled enzymes under the same condition. The reactions were terminated by the addition of 200 μL ice cold methanol, and centrifuged at 15,000×g for 30 min. The supernatants were performed on a Shiseido Capcell Pak C18 MG III column (250 mm × 4.6 mm id, 5 μm, Shiseido Co., Ltd., Tokyo, Japan) at 30 °C with a flow rate of 1 mL/min. The mobile phase was a gradient elution of solvents A (0.1% formic acid aqueous solution) and B (MeOH). Gradient program was as follows: (a) 65%–84% B 30 min; 84%–100% B 3 min; 100% B 5 min; and the adjusted gradient elution method (b) 65%–100% B 15 min; 100% B 5 min. The ELSD parameters were as follows: nitrogen pressure, 350 kPa; the drift tube temperature, 40 °C; gain value, 9. The ESI source was used in the positive mode with the following setting: spray voltage, 5.0 kV; capillary voltage, 35 V; capillary temperature, 275 °C; tube lens offset voltage, 110 V; sheath gas flow rate, 20 arbitrary units (abr); aux gas flow rate, 5 arbitrary units (abr). Spectra were recorded in the range of m/z 100–1300. Data acquisition was under the control of the Xcalibur software (Thermo Finnigan, San Jose, CA, USA). The conversion rates of the enzyme reactions were calculated from peak areas of glycosylated products and substrates. For quantification, three parallel assays were routinely carried out.
2.11. Effects of pH, temperature and divalent metal ions on glycosyltransferase activity
For pH, the enzymatic reactions were performed in various reaction buffers with pH values in the range of 5.0–6.0 (citric acid-sodium citrate buffer), 6.0–8.0 (Na2HPO4–NaH2PO4 buffer), 7.0–9.0 (Tris-HCl buffer) and 9.0–11.0 (Na2CO3–NaHCO3 buffer). For temperature, the reaction mixtures were incubated at different temperatures (25–60 °C). For divalent metal ions, BaCl2, CaCl2, CoCl2, FeSO4, CuCl2, MgCl2, SnCl2, MnCl2, ZnCl2, SrCl2, and NiSO4 were used individually at final concentrations of 5 mmol/L under optimal pH and temperature, and the group without divalent metal ions and EDTA being added group were setup at the same time. The reactions were analyzed by HPLC-ELSD as described above and three independent experiments were performed.
2.12. Kinetic parameters of glycosyltransferases
To determine the kinetic values of AmUGT15, reactions were performed with acceptor 1 from 10 to 400 μmol/L with the saturating UDP-Xyl (800 μmol/L) or UDP-Xyl from 100 to 1400 μmol/L with 1 (400 μmol/L) at pH 8.0 and 35 °C in a total volume of 50 μL. When using 3 as acceptor, the assay containing 50 mmol/L Na2HPO4–NaH2PO4 (pH 8.0), saturating UDP-Xyl (800 μmol/L) and varying concentration of 3 (10–500 μmol/L) or 3 (400 μmol/L) and varying concentration of UDP-Xyl (50–800 μmol/L) was conducted at 35 °C in a total volume of 50 μL. Aliquots were quenched with 100 μL of ice cold MeOH and centrifuged at 15,000×g for 30 min. Supernatants were analyzed by HPLC-ELSD as mentioned above. All experiments were performed in triplicate. Data fitting was performed using GraphPad Prism 8, and Km, kcat and kcat/Km values represented the mean ± SD of three independent replicates. Likewise, the kinetic parameters of AmUGT14, AmUGT13 and AmUGT7 were measured as described above and the details were listed in Supporting Information Table S2.
2.13. Preparation of glycosylated products
To prepare the glycosylated products, a 10 mL scale of reaction containing 0.4 mmol/L acceptors (1–6 or 1a), 0.8 mmol/L sugar donor (UDP-Glc or UDP-Xyl) and 10 mg purified enzymes was conducted at 35 °C for 12 h. The enzymatic reaction was terminated by extraction with water-saturated n-butanol for 3 times. The organic extract was evaporated under reduced pressure to dryness, and the resultant residue was redissolved with MeOH, which was isolated by reversed-phase semi-preparative HPLC equipped with differential refractive index detector with MeOHH2O (75:25, v/v) as mobile phase at 4 mL/min to afford the glycosylated products. All the obtained glycosylated products were subjected to NMR analysis.
2.14. GenBank accession numbers
The sequence data in this article are deposited in the GenBank under the following accession numbers: AmCAS1 (OM913798), AmUGT15 (OM913794), AmUGT14 (OM913795), AmUGT13 (OM913796), and AmUGT7 (OM913797).
3. Results
3.1. Mining and phylogenetic analysis of cycloartenol synthases
The formation of cycloartane-type skeleton by cycloartenol synthase (CAS) is the first committed step in the proposed biosynthesis of diverse astragalosides. Therefore, cycloartenol synthase or triterpene cyclase was used as keywords to search for the transcriptome annotation files. Using an identified cycloartenol synthase AtCAS from Arabidopsis thaliana as the query sequence30, local BLASTp searching was then performed against the transcriptome, ultimately resulting in three AmCASs, designated as AmCAS1, AmCAS2, and AmCAS3 with high transcript levels as candidate genes.
Subsequently, the neighbor-joining tree of candidate genes and characterized oxidosqualene cyclases from other plants was constructed to search for the desired cycloartenol synthase (Supporting Information Fig. S1). The phylogenetic analysis demonstrated that AmCAS1 was grouped into the clade of cycloartenol synthase and showed highest 92.6% sequence identity with GgCAS from Glycyrrhiza glabra31, while AmCAS2 and AmCAS3 were grouped with GgLUS132 in the clade of lupeol synthase. More detailed alignment (Supporting Information Fig. S2) revealed that AmCAS1 contained a conserved MWCHCR motif, which was commonly existed in cycloartenol synthase; moreover, AmCAS1 had one DCTAE motif for the initiation of the cyclization reaction and four QW (QXXXGXW and QXXXGXXXW) motifs for substrate binding and stabilization of the carbocation intermediates33,34. Thus, AmCAS1 might be the most potential candidate cycloartenol synthase.
3.2. Functional characterization of cycloartenol synthase
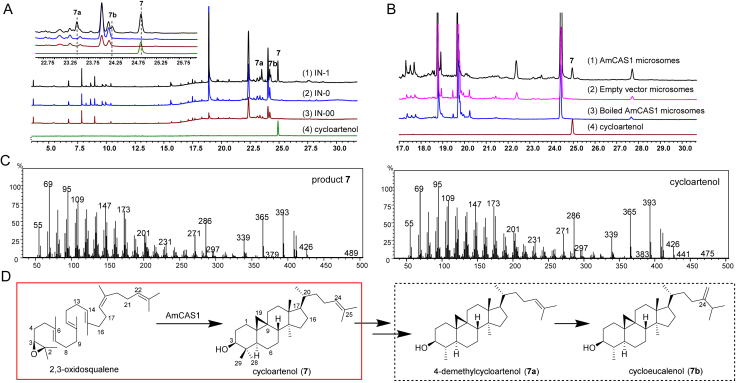
To accumulate sufficient concentrations of 2,3-oxidosqualene for enzyme identification, the precursor of cycloartenol biosynthesis, we optimized its pathway by introducing tHMG1 and ERG7– genes into S. cerevisiae INVSc129,35 (Supporting Information Fig. S3). Three engineered yeast strains harboring the recombinant plasmids of pESC-His-AmCASs with both pESC-Ura-tHMG1 and pESC-Leu-ERG7– were then constructed and designated as IN-1, IN-2, and IN-3, respectively. Transformant IN-0 with the empty pESC-His vector, pESC-Ura-tHMG1, and pESC-Leu-ERG7–, and transformant IN-00 harboring pESC-His, pESC-Leu and pESC-Ura were designated as the two negative controls. GC–MS analysis showed that the IN-1 strain expressing AmCAS1 produced an evident product (7) at tR 24.9 min, and two minor products (7a and 7b) at tR 23.5 min and tR 24.3 min, respectively, of which product 7 exhibited identical retention time and mass spectral characteristics to those of the authentic standard of cycloartenol (Fig. 2A and C, Supporting Information Fig. S4), meanwhile, the two minor products (7a and 7b) were predicted to be cycloartanes based on MS database (Supporting Information Fig. S5). While no corresponding products were generated in the IN-0, IN-00, IN-2, and IN-3 strains (Supporting Information Fig. S6).
Figure 2.
Functional identification of AmCAS1. (A) Functional identification in yeast. (1) IN-1, harboring pESC-His-AmCAS1, pESC-Ura-tHMG1 and pESC-Leu-ERG7−; (2) control yeast IN-0, transformed with the empty pESC-His vector, pESC-Ura-tHMG1 and pESC-Leu-ERG7−; (3) control yeast IN-00, transformed with the empty pESC-His, pESC-Ura and pESC-Leu vector; and (4) the authentic standard of cycloartenol. (B) In vitro functional identification using 2,3-oxidosqualene as substrate. (1) enzymatic reaction with AmCAS1 microsomes; (2) enzymatic reaction with empty vector microsomes; (3) enzymatic reaction with boiled AmCAS1 microsomes and (4) the authentic standard of cycloartenol. (C) MS spectra of product 7 and the authentic standard of cycloartenol. (D) Production and metabolism of 7 in yeast.
A 10 L scale of yeast fermentation was performed to give AmCAS1-derived 7, 7a and 7b, and their structures were determined by extensive spectroscopic analysis (Supporting Information Table S8, Figs. S22–S38). All the spectroscopic data of 7, including 1D NMR and 2D NMR, NOEs, and optical rotation, were in good agreement with those of cycloartenol, thus determining 7 as cycloartenol. Cyclopropane ring protons were found in the 1H NMR spectra of 7a and 7b [δH 0.38 (1H, J = 4.0 Hz), 0.14 (1H, J = 4.0 Hz)] (Table S8, Figs. S29 and S34) and further assignment of signals allowed the determination of 7a to be 4-dimethylcycloartenol and 7b to be cycloeucalenol, as potential metabolites of cycloartenol by S. cerevisiae36,37 (Fig. 2D). In vitro assays with 2,3-oxidosqualene as substrate revealed that cycloartenol (7) was produced by AmCAS1 microsomes (Fig. 2B). All of the results indicated the gene AmCAS1 encodes a cycloartenol synthase to catalyze 2,3-oxidosqualene to form cycloartenol. To our best knowledge, AmCAS1 is the first identified cycloartenol synthase from A. membranaceus.
3.3. Functional identification of glycosyltransferases in the biosynthesis of cycloastragenol-type astragalosides
A variety of bioactive astragalosides with diverse glycosylation patterns have been isolated from A. membranaceus, which implies the existence of corresponding glycosyltransferases. However, little is known about the enzymes that are responsible for such glycosylation. Therefore, searching for glycosyltransferases with specific catalytic properties will facilitate to elucidate the biosynthetic pathway of these cycloastragenol-type glycosides and provide molecular insight into the structural diversity of astragalosides.
3.3.1. Mining of glycosyltransferase candidate genes and phylogenic analysis
Glucosyltransferase or glycosyltransferase was used as the entry to search against the transcriptome to mine the desired glycosyltransferases and 517 unigenes were retrieved, 154 unigenes of which were selected for further investigation through deduplication and exclusion of some primary metabolism related glycosyltransferases. A neighbor-joining phylogenetic tree was constructed with 154 unigenes and known triterpenoid glycosyltransferases from different plant species (Supporting Information Table S3, Fig. S7). The sugar donors of these triterpenoid glycosyltransferases involved UDP-Glc, UDP-Xyl, and UDP-galactose and the glycosylation sites covered 3-OH, 6-OH, -OHs in sugar, and –COOH leading to the formation of glycosides, branched glycosides and sugar esters.
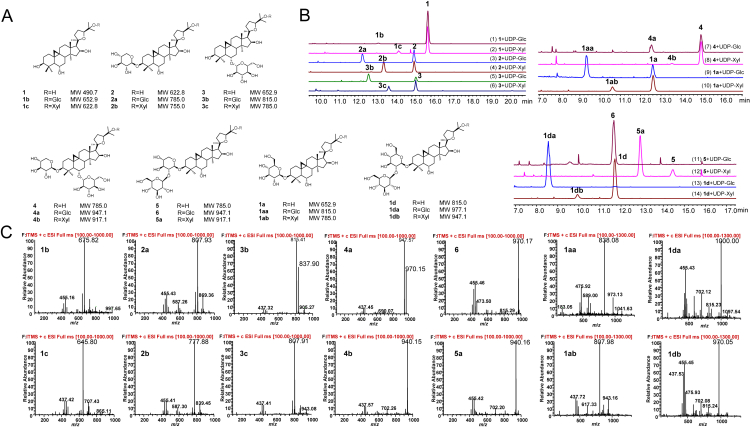
Phylogenetic analysis revealed that 49 unigenes were fell into the clade with characterized triterpenoid UGTs, whereas another 105 unigenes exhibited too phylogenetically distant to be considered. Of the 49 candidate unigenes, 24 unigenes with high transcript levels were successfully amplified in full length and named as AmUGT1–AmUGT24. Subsequently, the catalytic functions of these 24 candidate glycosyltransferases were individually determined by assaying in vitro with acceptors 1–7 and UDP-Xyl or UDP-Glc (Figure 1, Figure 2). Of the 24 recombinant AmUGTs, AmUGT15, AmUGT14, AmUGT13, and AmUGT7 exhibited high activities (Supporting Information Tables S4–S6), and were selected for further characterizing the functions in the biosynthesis of bioactive astragalosides.
3.3.2. AmUGT15 as a 3-O-xylosyltransferase
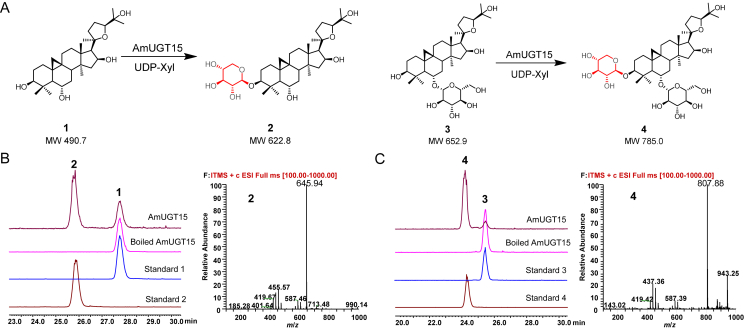
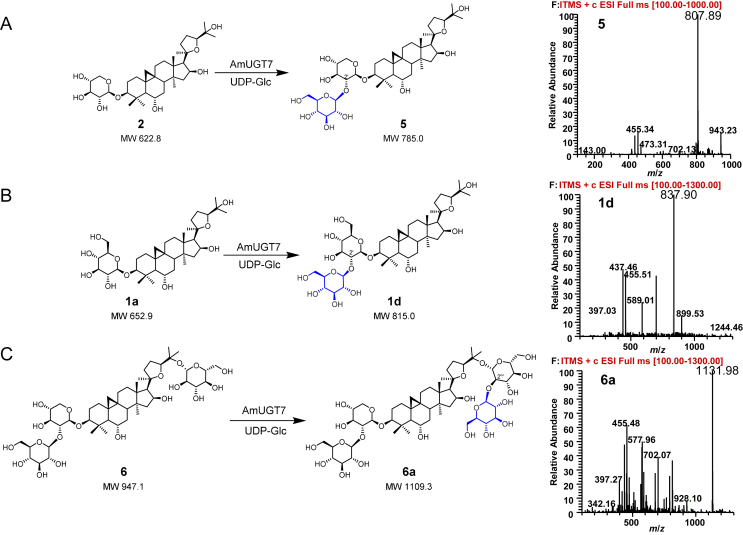
AmUGT15 displayed an open reading frame of 1452 bp, putatively encoding a polypeptide with 483 amino acids and a predicted molecular weight of 58.0 kDa (Supporting Information Fig. S8). In the phylogenetic tree, AmUGT15 was grouped into the UGT73 family clade showing 63.3% sequence identity with UGT73F17, a pentacyclic triterpenoid 30/29-O-glycosyltransferase from Glycyrrhiza uralensis38. HPLC–ELSD and HPLC–MS analysis revealed that recombinant AmUGT15 could catalyze 3 to form a xylosylated product with 95% conversion rate, which was assigned as astragaloside IV (4) by comparing with the authentic standard. Moreover, AmUGT15 also displayed 68% conversion rate for xylosylation of 1, and the product was identified as cycloastragenol-3-O-β-d-xyloside (2) by comparing with the standard sample (Fig. 3, Table S4). In addition to UDP-Xyl, AmUGT15 was also able to recognize UDP-Glc with a low activity (<35%) when 1 was used as acceptor (Table S4, Supporting Information Fig. S9). However, AmUGT15 had no catalytic activity towards cycloartenol (7), indicating that 3-O-glycosylation reaction may occur after oxidative modification (Supporting Information Fig. S10). Therefore, AmUGT15 can be validated as a 3-O-xylosyltransferase for cycloastragenol glycosides.
Figure 3.
3-O-Xylosylation of 1 and 3 catalyzed by recombinant AmUGT15. (A) 3-O-Xylosylated reactions catalyzed by AmUGT15 with 1 and 3. (B) HPLC–ELSD chromatograms of 1 and the xylosylated product 2 and MS spectrum of the xylosylated product 2 (also see Fig. S9). (C) HPLC–ELSD chromatograms of 3 and the xylosylated product 4 and MS spectrum of the xylosylated product 4 (also see Fig. S9). The peaks of molecular ion were displayed in the form of sodium adducts, and the molecular weights of xylosylated products are 132 amu more than those of their substrates.
Biochemical characterization of recombinant AmUGT15 showed that the optimal temperature was 35 °C, and the optimal pH values were ranged from pH 8.0–9.0. AmUGT15 was independent of the divalent cations, but the addition of Cu2+, Zn2+ and Ni2+ completely abolished its activity (Supporting Information Fig. S11). The kinetic parameters of AmUGT15 were then further investigated by using 1 or 3 and UDP-Xyl as acceptor and donor substrates. The Km values were 23.8 μmol/L and 109.3 μmol/L for 1 and 3, respectively, and the corresponding kcat/Km values were 527.9 L/mol·s and 283.6 L/mol·s. Whereas AmUGT15 exhibited a kcat/Km value of 231.2 L/mol·s to UDP-Xyl when using 3 as acceptor, which was nearly 6-fold to that when using 1 (42.8 L/mol·s, Table S2, Fig. S11). These results may explain why AmUGT15 had a higher affinity for 1, but exhibited lower glycosylation conversion rate than that of 3. Cumulatively, both cycloastragenol (1) and its 6-O-glucoside (3) might be the substrates for 3-O-xylosylation in the biosynthesis of bioactive cycloastragenol-type glycosides.
3.3.3. AmUGT14 as a 3-O-glucosyltransferase
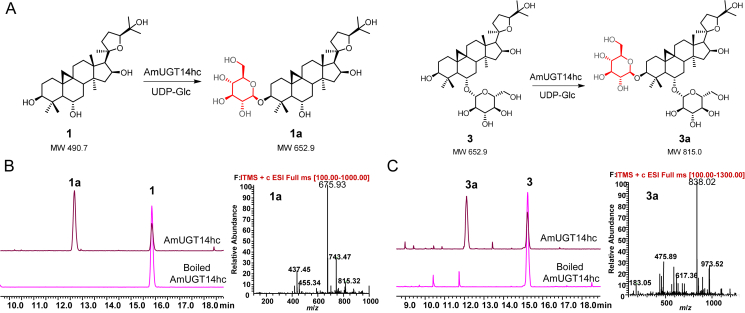
AmUGT14 had an open reading frame of 1449 bp and putatively encoded a polypeptide with 482 amino acids. In the phylogenetic tree, AmUGT14 was also grouped into the UGT73 family clade and in 66.7% identical with AmUGT15. Upon incubation of the crude enzymes of recombinant AmUGT14 with 1 and UDP-Glc, a glucosylated product (Supporting Information Fig. S12) was detected by HPLC–ELSD and HPLC–MS. The further enzymatically preparative synthesis of the product 1a allowed the assignment of the structure to be cycloastragenol-3-O-β-d-glucoside39,40 on the basis of the HMBC correlations of H-1ʹ/C-3 (δH 5.04/δC 89.5), in which the glucosyl group is attached at 3-OH of 1 with a β-configuration and adopting an inverting mechanism according to the anomeric proton at δH 5.04 (1H, d, J = 7.6 Hz, H-1′) in the 1H NMR spectrum (Supporting Information Figs. S39–S43). To obtain the purified recombinant protein and further investigate its catalytic property, the codon-optimized AmUGT14 (designated as AmUGT14hc) was synthesized (Supporting Information Fig. S13) and its glycosylation activity against other substrates was further investigated. The results showed that the recombinant AmUGT14hc could not only convert 1 to 1a with a conversion rate of 80%, but also catalyze 3a from 3 with 55% conversion rate when UDP-Glc was used as sugar donor. In addition to UDP-Glc, recombinant AmUGT14hc was also able to recognize UDP-Xyl to yield cycloastragenol 3-O-β-d-xylosides (2) with a low conversion rate (<10%) with 1 as the acceptor by comparing with the authentic standard (Table S4, Fig. 4 and Supporting Information Fig. S14). The structure of 3a was determined as 3-O-β-d-glucopyranosyl-6-O-β-d-glucopyranosyl cycloastragenol, as a 3-O-glucosylated product of 3 (Supporting Information Table S7, Figs. S60–S64). Thus, AmUGT14 is a 3-O-glucosyltransferase to exert 3-O-glucosylation activity of 1 and 3, but also exhibits tolerance for UDP-Xyl.
Figure 4.
3-O-Glucosylation of 1 and 3 catalyzed by recombinant AmUGT14hc. (A) 3-O-Glucosylated reactions catalyzed by AmUGT14hc with 1 and 3. (B) HPLC-ELSD chromatograms of 1 and the glucosylated product 1a and MS spectrum of the glucosylated product 1a (also see Fig. S14). (C) HPLC-ELSD chromatograms of 3 and the glucosylated product 3a and MS spectrum of the glucosylated product 3a (also see Fig. S14). The peaks of molecular ion were displayed in the form of sodium adducts, and the molecular weights of glucosylated products are 162 amu more than those of their substrates.
Recombinant AmUGT14hc exhibited the maximum activity at pH 8.0 and 40 °C and was independent of divalent cations, but its glucosylation activity was greatly inhibited in the presence of Cu2+ and Zn2+ (Supporting Information Fig. S15). AmUGT14hc displayed quite a dissimilar affinity for 1 (Km = 55.5 μmol/L) and 3 (Km = 86.0 μmol/L). Moreover, kcat value for 1 (0.024 s−1) was significantly higher than 3 (0.0003 s−1). Consequently, AmUGT14hc exhibited about 140-fold higher catalytic efficiency (kcat/Km) using 1 (437.7 L/mol·s) than using 3 (3.1 L/mol·s, Fig. S15). Taken together, cycloastragenol (1) is more likely to be the substrate for the 3-O-glucosylation in the biosynthesis of cycloastragenol-type glycosides.
3.3.4. AmUGT13 as a 25-O-glycosyltransferase
AmUGT13 had an open reading frame of 1422 bp and predictively encoded 473 amino acids polypeptide. In the phylogenetic tree, AmUGT13 was also grouped into the UGT73 family clade and exhibited 54.6% and 59.7% identities with AmUGT15 and AmUGT14, respectively. HPLC–ELSD and HPLC–MS analysis revealed that recombinant AmUGT13 (Supporting Information Fig. S16) could catalyze most of the tested acceptors (1–5, 1a and 1d) with sugar donors (UDP-Glc or UDP-Xyl) to yield the corresponding glycosylated products (Fig. 5), where the molecular weights of the glucosylated products (1b, 2a, 3b, 4a, 1aa, 1da and 6) were increased by 162 amu and the molecular weights of the xylosylated products (1c, 2b, 3c, 4b, 1ab, 1db and 5a) were increased by 132 amu based on MS analysis. Of the 14 glycosylation reactions, high yields (>98%) were achieved with astragaloside III (5) or 3-(2′-O-β-d-glucopyranosyl)-O-β-d-glucopyranosyl cycloastragenol (1d) as acceptor and UDP-Glc as sugar donor, while the conversion rates of 1–4 and 1a with UDP-Glc or UDP-Xyl were lower than 65% (Table S5, Fig. 5). These results indicated that both 5 and 1d, with a sugar chain at C3–OH, might be the natural substrates of AmUGT13; if anything, the conversion rate of 5 with UDP-Xyl was higher than that of 1d.
Figure 5.
25-O-Glycosylation of acceptors 1–5, 1a and 1d catalyzed by recombinant AmUGT13. (A) Structures of 25-O-glucosides and xylosides; (B) HPLC–ELSD chromatograms of 1−5, 1a, 1d and their glycosylated products; (C) The MS spectra of the glycosylated products. The peaks of molecular ion were displayed in the form of sodium adducts, and the molecular weights of glucosylated products are 162 amu more than those of their substrates and the molecular weights of xylosylated products are 132 amu more than those of their substrates.
To characterize the catalytic property of AmUGT13, the glucosylated products 2a, 4a, and 6 as well as the xylosylated product 5a were prepared from scaled-up enzymatic reactions. The structure of 2a was determined as isoastragaloside IV, a 25-O-β-d-glucosylated product41 of 2 based on the 1D and 2D NMR spectroscopic data analysis (Table S7, Supporting Information Figs. S52–S56). The HMBC correlations of H-1′′′ (δH 5.09, Glc)/C-25 (δC 79.05) for 4a (Table S7, Supporting Information Figs. S68–S72), and H-1′′′ (δH 5.08, Glc)/C-25 (δC 79.04) for 6 (Table S7, Supporting Information Figs. S83–S87) were also indicative of the occurrence of glucosylation at 25-OH. Accordingly, the structures of 4a and 6 were identified as astragaloside VII42 and astragaloside V43, respectively. Likewise, the product 5a was characterized to be 3-(2′-O-β-d-glucopyranosyl)-O-β-d-xylopyranosyl-25-O-β-d-xylopyranosyl cycloastragenol according to the HMBC correlations from H-1′′′ (δH 4.96, Xyl) to C-25 (δC 79.0), a new 25-O-β-d-xylosylated derivative of 5 (Table S7, Supporting Information Figs. S78–S82). Therefore, AmUGT13 is a 25-O-glycosyltransferase and exhibits flexibility towards both acceptors and sugar donors.
The biochemical characteristics of recombinant AmUGT13 were further investigated by using 5 and UDP-Glc or UDP-Xyl as acceptor and donor substrates. AmUGT13 displayed the maximum activity at pH 8.0 and 30 °C and was divalent cation-independent. The Km values for 5, UDP-Glc, and UDP-Xyl were 84.6, 414.7 and 479.6 μmol/L, respectively (Supporting Information Fig. S17). AmUGT13 exhibited a kcat/Km value 36.5 L/mol·s to UDP-Glc, which was sevenfold to that of UDP-Xyl (5.3 L/mol·s). Moreover, the conversion rates of 3–5, 1a and 1d with UDP-Glc were higher than that of UDP-Xyl (Table S5). Cumulatively, AmUGT13 might be mainly responsible for the 25-O-glucosylation of astragaloside III (5) or 3-(2′-O-β-d-glucopyranosyl)-O-β-d-glucopyranosyl cycloastragenol (1d), and also contributed to the diversity of cycloastragenol-type glycosides due to its enzyme promiscuity.
3.3.5. AmUGT7 as a 2′-O-glucosyltransferase
AmUGT7 displayed an open reading frame of 1503 bp, putatively encoding a polypeptide with 500 amino acids. Phylogenetic analysis showed that AmUGT7 was also grouped into the UGT73 family clade and had 54.5% identity with GmSGT2 from Glycine max, which was found to transfer a galactosyl group to the 2′-OH of the sugar moiety of GA-3-O-monoglucuronide and GA-3-O-monoglucose44. HPLC–ELSD and HPLC–MS analysis revealed that recombinant AmUGT7 (Supporting Information Fig. S18) could efficiently catalyze 2 and 1a with UDP-Glc to afford 5 and 1d in high conversion rates (>96%), respectively, while the conversion rate of 6 with UDP-Glc was 45% (Table S6, Fig. 6, Supporting Information S19 and S20). However, AmUGT7 was not able to recognize UDP-Xyl. To further confirm the catalytic properties of AmUGT7, scale-up reactions of 1a and 6 were performed to yield the corresponding glucosylated products (1d and 6a). According to the 1D and 2D NMR spectra, and the HMBC correlations of H-1′′ (δH 5.43, Glc)/C2′ (δC 83.9, Glc), the structure of 1d was identified as 3-(2′-O-β-d-glucopyranosyl)-O-β-d-glucopyranosyl cycloastragenol (Table S7, Fig. 6, Supporting Information Figs. S44–S48), and the structure of 6a was 3-(2′-O-β-d-glucopyranosyl)-O-β-d-xylopyranosyl-25-(2′′′-O-β-d-glucopyranosyl)-O-β-d-glucopyranosyl cycloastragenol (Table S7, Fig. 6, Supporting Information Figs. S88–S92). Interestingly, 6a was a new cycloastragenol tetraglycoside with two sugar chains linked at 3-OH and 25-OH branched via a 1,2-glycosidic bond linkage, respectively. These results suggested that AmUGT7 was a 2′-O-glucosyltransferase and could transfer a glucosyl group to the xylosyl or glucosyl moiety at C3 or C25 position, respectively.
Figure 6.
2′-O-Glucosylated reactions catalyzed by AmUGT7 and MS spectra of glycosylated products. The peaks of molecular ion were displayed in the form of sodium adducts, and the molecular weights of glucosylated products are 162 amu more than those of their substrates (also see Figs. S19 and S20).
Recombinant AmUGT7 exhibited optimal pH at 9.0 and optimal temperature in the range of 35–55 °C, and was independent of divalent ions, of which Fe2+ could enhance the catalytic activity. The Km values for 2 and UDP-Glc were 54.2 μmol/L and 74.8 μmol/L, respectively (Supporting Information Fig. S21). Although AmUGT7 could catalyze the first 3-O-glucosylation and successive second 2′-O-glucosylation of 1 to afford 1d as reported in AmGT827, the conversion rate was drastically lower than 7% (Table S6, Fig. S19). Therefore, AmUGT7 can be definitively validated as a 2′-O-glucosyltransferase to carry out the linkage of branched sugar chains in the biosynthesis of cycloastragenol-type glycosides.
4. Discussion
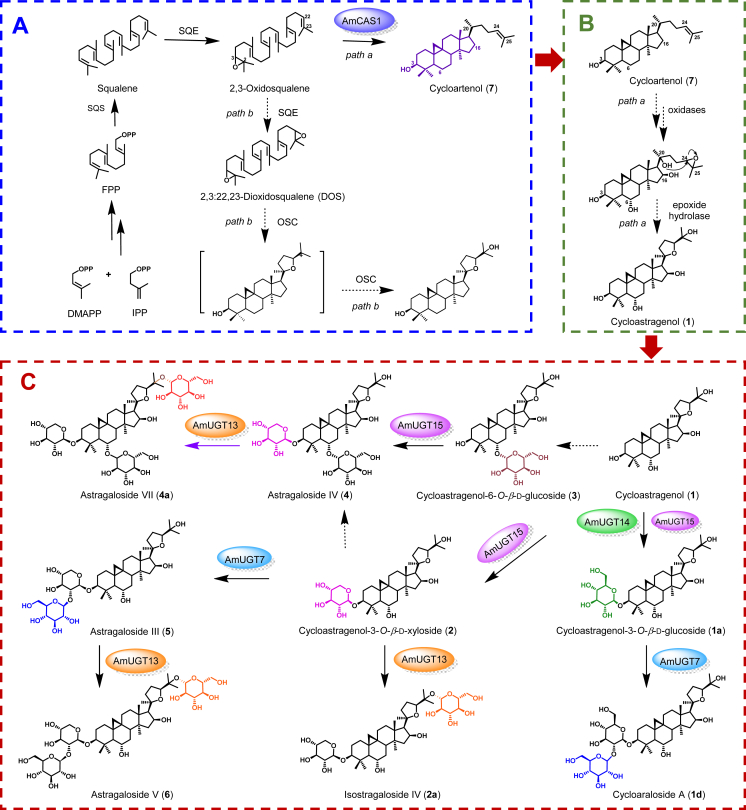
Cycloastragenol-type astragalosides are uniformly distributed in A. membranaceus and structurally belong to the family of cycloartane-type saponins, the group of which with a 20,24-tetrahydrofuran ring are more common representatives of bioactive astragalosides. In the biosynthesis of this type of astragalosides, cycloartenol is an important precursor subjected to diverse modifications, including oxygenation, glycosylation even acylation, resulting in the vast structural diversity. This has been clearly shown in our study with regard to cycloastragenol-type glycosides biosynthesis in which 2,3-oxidosqualene is actually the preferred substrate for cycloartenol synthase AmCAS1 in the biosynthesis of cycloartenol and the resultant cycloartenol will undergo oxygenation modifications at different sites, for example, C6, C16, C20, C24, and C25 positions, which mainly involve regio- and stereo-specific hydroxylation reactions catalyzed by oxidases family especially by P450 monooxygenases. Two possible mechanisms for the formation of the tetrahydrofuran ring, a typical moiety in these compounds, are proposed in Fig. 7. In the pathway a, an epoxidation may occur in the terminal olefinic bond of cycloartenol instead of the single oxygenation at C24 and C25 positions, and the hydroxyl group at the C20 position initiates a nucleophilic attack on the epoxide to form the 5-member cyclic ether (tetrahydrofuran ring) in 5-exo-tet mechanism by an epoxide hydrolase45. By contrast, the pathway b with oxidation before cyclization using 2,3:22,23-dioxidosqualene (DOS) as the substrate received less attention for the considerable difficulties of producing both cyclopropane and heterocyclic structures only by DOS cyclization reaction46. The above-mentioned hydroxylated sites are described to be glycosylated by diverse glycosyltransferases, which will significantly contribute to the chemical complexity of astragalosides. Combined with the identified glycosyltransferases in our study, it seems reasonable that a series of glycosyltransferases showing substrate promiscuity have infused great vitality to the diversification of cycloastragenol glycosides (Tables S4–S6). AmUGT15 not only transferred UDP-Xyl to cycloastragenol, an aglycone acceptor, but also performed xylosylation towards cycloastragenol-6-O-β-d-glucoside, a mono-glycoside, to generate both mono- and di-glycosides. Moreover, AmUGT15 also exhibited flexible selectivity for UDP-Glc to enrich the structural diversity of astragalosides. Similarly, AmUGT13 could catalyze both 25-O-glucosylation and xylosylation of various cycloastragenol glycosides with different types or numbers of glycosyl substitutes, fulfilling the varied structures of bioactive astragalosides. These results provided us a better understanding for the divergent biosynthesis of such diverse triterpenoid glycosides.
Figure 7.
Proposed biosynthetic pathways of cycloastragenol glycosides in A. membranaceus. (A) Cycloartenol skeleton formation; (B) Cycloastragenol formation from cycloartenol; (C) Diverse cycloastragenol glycosides generation.
5. Conclusions
In this work, we identified a triterpene cyclase AmCAS1 involved in the formation of triterpene skeleton, and in vitro biochemically and functionally characterized four glycosyltransferases contributing to the diverse glycosylation patterns of cycloastragenol-type astragalosides. To the best of our knowledge, AmCAS1 was the first identified cycloartenol synthase from A. membranaceus to catalyze the cyclization of 2,3-oxidosqualene into cycloartenol (7). AmUGT15, a novel promiscuous xylosyltransferase catalyzing the 3-O-xylosylation of cycloastragenol (1) and cycloastragenol-6-O-β-d-glucoside (3), is able to synthesize astragaloside IV (4) in vitro. Different preference for sugar donor, AmUGT14 exerts 3-O-glucosylation of cycloastragenol (1) and cycloastragenol-6-O-β-d-glucoside (3). AmUGT13 is a high strict regio-specific 25-O-glycosyltransferase with UDP-Glc as the preferred sugar donor. While, 25-O-xylosides of this type of triterpenoids are rare in nature, thus the utility of AmUGT13 even its engineered variants will facilitate the discovery of unprecedented bioactive derivatives. Additionally, the identification of AmUGT7 allows the new member joining into the glycosidic glycosyltransferase family and AmUGT7 is determined as a 2′-O-glucosyltransferase of triterpenoid glycoside for its regio-selectivity towards 2-OH of glucosyl or xylosyl moiety at C3 or C25 positions. Accordingly, the discovery of cycloartenol synthase and these functionally diverse glycosyltransferases here demonstrates the crucial enzymes for the biosynthesis of astragalosides and the molecular insights into their structurally diversity, and paves the way for elucidating the complete biosynthetic pathway, even constructing the synthetic biology platform for the production of such bioactive astragalosides in an environmentally friendly way.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2020YFA0908000), CAMS Innovation fund for Medical Sciences (CIFMS, No. 2021-I2M-1-029, China), and Beijing Key Laboratory of non-Clinical Drug Metabolism and PK/PD Study (Z141102004414062). We thank Prof. Jinling Yang (Institute of Materia Medica, Chinese Academy of Medical Sciences & Peking Union Medical College, China) for providing plasmids to amplify tHMG1 and ERG7– genes.
Author contributions
Jungui Dai and Dawei Chen designed and guided the experiments; Yangyang Duan and Wenyu Du conducted experiments; Yangyang Duan and Zhijun Song contributed to the preparation and structural characterization of the products; Yangyang Duan, Ridao Chen, Kebo Xie, and Jimei Liu performed bioinformatics analysis and structural elucidation of the products; All the authors analyzed and discussed the experimental data; Yangyang Duan, Dawei Chen, and Jungui Dai wrote the manuscript. All authors approved the final manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2022.05.015.
Contributor Information
Dawei Chen, Email: chendawei@imm.ac.cn.
Jungui Dai, Email: jgdai@imm.ac.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Chinese Pharmacopoeia Commission . China Medical Science and Technology Press (CFDA); Beijing: 2015. Pharmacopoeia of the People's Republic of China. [Google Scholar]
- 2.Chen X., Wang H., Jiang M., Zhao J., Fan C., Wang Y., et al. Huangqi (astragalus) decoction ameliorates diabetic nephropathy via IRS1-PI3K-GLUT signaling pathway. Am J Transl Res. 2018;10:2491–2501. [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou D., Wei W.B., Yang C.X., Ding N., Liu Y., He M.L., et al. Treatment of retinal vein occlusion in rabbits with traditional Chinese medicine Fufang XueShuan Tong. Chin Med J. 2010;123:3293–3298. [PubMed] [Google Scholar]
- 4.Li X., Qu L., Dong Y., Han L., Liu E., Fang S., et al. A review of recent research progress on the Astragalus genus. Molecules. 2014;19:18850–18880. doi: 10.3390/molecules191118850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao L.M., Cao P.H., Luo Z.H., Bao X.F., Zhou Z.Q., Li S., et al. Cycloartane-type triterpenoids from the root of Astragalus membranaceus var. mongholicus. J Asian Nat Prod Res. 2020;22:905–913. doi: 10.1080/10286020.2020.1787994. [DOI] [PubMed] [Google Scholar]
- 6.Bedir E., Calis I., Aquino R., Piacente S., Pizza C. Cycloartane triterpene glycosides from the roots of Astragalus brachypterus and Astragalus microcephalus. J Nat Prod. 1998;61:1469–1472. doi: 10.1021/np9801763. [DOI] [PubMed] [Google Scholar]
- 7.Gülcemal D., Masullo M., Bedir E., Festa M., Karayıldırım T., Alankus-Caliskan O., et al. Triterpene glycosides from Astragalus angustifolius. Planta Med. 2012;78:720–729. doi: 10.1055/s-0031-1298337. [DOI] [PubMed] [Google Scholar]
- 8.Gülcemal D., Masullo M., Napolitano A., Karayıldırım T., Bedir E., Alankus-Caliskan O., et al. Oleanane glycosides from Astragalus tauricolus: isolation and structural elucidation based on a preliminary liquid chromatography-electrospray ionization tandem mass spectrometry profiling. Phytochemistry. 2013;86:184–194. doi: 10.1016/j.phytochem.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Wang J., Jia J., Song L., Gong X., Xu J., Yang M., et al. Extraction, structure, and pharmacological activities of Astragalus polysaccharides. Appl Sci. 2019;9:122. [Google Scholar]
- 10.Lin L.Z., He X.G., Lindenmaier M., Nolan G., Yang J., Cleary M., et al. Liquid chromatography–electrospray ionization mass spectrometry study of the flavonoids of the roots of Astragalus mongholicus and A. membranaceus. J Chromatogr A. 2000;876:87–95. doi: 10.1016/s0021-9673(00)00149-7. [DOI] [PubMed] [Google Scholar]
- 11.Xiao C.J., Zhang Y., Qiu L., Xu W., Zhao M.Z., Dong X., et al. A new schistosomicidal and antioxidative phenylpropanoid from Astragalus englerianus. J Asian Nat Prod Res. 2015;17:772–777. doi: 10.1080/10286020.2014.977787. [DOI] [PubMed] [Google Scholar]
- 12.Molyneux R.J., James L.F. Loco intoxication: indolizidine alkaloids of spotted locoweed (Astragalus lentiginosus) Science. 1982;216:190–191. doi: 10.1126/science.6801763. [DOI] [PubMed] [Google Scholar]
- 13.Agzamova M.A., Isaev I.M., Rakhmatov K.A. Steroids and glycosides from Astragalus turczaninowii. Chem Nat Compd. 2017;53:398–399. [Google Scholar]
- 14.Zhang J., Wu C., Gao L., Du G., Qin X. Astragaloside IV derived from Astragalus membranaceus: a research review on the pharmacological effects. Adv Pharmacol. 2020;87:89–112. doi: 10.1016/bs.apha.2019.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Wang S., Li J., Huang H., Gao W., Zhuang C., Li B., et al. Anti-hepatitis B virus activities of astragaloside IV isolated from Radix Astragali. Biol Pharm Bull. 2009;32:132–135. doi: 10.1248/bpb.32.132. [DOI] [PubMed] [Google Scholar]
- 16.Zhang A., Zheng Y., Que Z., Zhang L., Lin S., Le V., et al. Astragaloside IV inhibits progression of lung cancer by mediating immune function of Tregs and CTLs by interfering with IDO. J Cancer Res Clin Oncol. 2014;140:1883–1890. doi: 10.1007/s00432-014-1744-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu W.S., Li S., Guo W.W., Chen L.L., Li Y.S. Effects of astragaloside IV on diabetic nephropathy in rats. Genet Mol Res. 2015;14:5427–5434. doi: 10.4238/2015.May.22.12. [DOI] [PubMed] [Google Scholar]
- 18.Indu P., Arunagirinathan N., Rameshkumar M.R., Sangeetha K., Divyadarshini A., Rajarajan S. Antiviral activity of astragaloside II, astragaloside III and astragaloside IV compounds against dengue virus: computational docking and in vitro studies. Microb Pathog. 2020;152:104563. doi: 10.1016/j.micpath.2020.104563. [DOI] [PubMed] [Google Scholar]
- 19.Wang H., Yuan R., Cao Q., Wang M., Ren D., Huang X., et al. Astragaloside III activates TACE/ADAM17-dependent anti-inflammatory and growth factor signaling in endothelial cells in a p38-dependent fashion. Phytother Res. 2020;34:1096–1107. doi: 10.1002/ptr.6603. [DOI] [PubMed] [Google Scholar]
- 20.Ma X.Q., Shi Q., Duan J.A., Dong T.T., Tsim K.W. Chemical analysis of Radix Astragali (Huangqi) in China: a comparison with its adulterants and seasonal variations. J Agric Food Chem. 2002;50:4861–4866. doi: 10.1021/jf0202279. [DOI] [PubMed] [Google Scholar]
- 21.Ionkova I., Shkondrov A., Krasteva I., Ionkov T. Recent progress in phytochemistry, pharmacology and biotechnology of Astragalus saponins. Phytochemistry Rev. 2014;13:343–374. [Google Scholar]
- 22.Jiao J., Gai Q.Y., Wang W., Luo M., Zu Y.G., Fu Y.J., et al. Enhanced astragaloside production and transcriptional responses of biosynthetic genes in Astragalus membranaceus hairy root cultures by elicitation with methyl jasmonate. Biochem Eng J. 2016;105:339–346. [Google Scholar]
- 23.Itkin M., Davidovich-Rikanati R., Cohen S., Portnoy V., Doron-Faigenboim A., Oren E., et al. The biosynthetic pathway of the nonsugar, high-intensity sweetener mogroside V from Siraitia grosvenorii. Proc Natl Acad Sci U S A. 2016;113:E7619–E7628. doi: 10.1073/pnas.1604828113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang D., Wang J., Shi Y., Li R., Fan F., Huang Y., et al. Elucidation of the complete biosynthetic pathway of the main triterpene glycosylation products of Panax notoginseng using a synthetic biology platform. Metab Eng. 2020;61:131–140. doi: 10.1016/j.ymben.2020.05.007. [DOI] [PubMed] [Google Scholar]
- 25.Yin Y., Li Y.P., Jiang D., Zhang X.N., Gao W., Liu C.S. De novo biosynthesis of liquiritin in Saccharomyces cerevisiae. Acta Pharm Sin B. 2020;10:711–721. doi: 10.1016/j.apsb.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou M.Q., Wang R.F., Zhao S.J., Wang Z.T. Ginsenosides in Panax genus and their biosynthesis. Acta Pharm Sin B. 2021;11:1813–1834. doi: 10.1016/j.apsb.2020.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang M., Yi Y., Gao B.H., Su H.F., Bao Y.O., Shi X.M., et al. Functional characterization and protein engineering of a triterpene 3-/6-/2′-O-glycosyltransferase reveal a conserved residue critical for the regiospecificity. Angew Chem Int Ed. 2022;61 doi: 10.1002/anie.202113587. [DOI] [PubMed] [Google Scholar]
- 28.Feng L.M., Lin X.H., Huang F.X., Cao J., Qiao X., Guo D.A., et al. Smith degradation, an efficient method for the preparation of cycloastragenol from astragaloside IV. Fitoterapia. 2014;95:42–50. doi: 10.1016/j.fitote.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 29.Liang H., Hu Z., Zhang T., Gong T., Chen J., Zhu P., et al. Production of a bioactive unnatural ginsenoside by metabolically engineered yeasts based on a new UDP-glycosyltransferase from Bacillus subtilis. Metab Eng. 2017;44:60–69. doi: 10.1016/j.ymben.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 30.Corey E.J., Matsuda S.P., Bartel B. Isolation of an Arabidopsis thaliana gene encoding cycloartenol synthase by functional expression in a yeast mutant lacking lanosterol synthase by the use of a chromatographic screen. Proc Natl Acad Sci U S A. 1993;90:11628–11632. doi: 10.1073/pnas.90.24.11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayashi H., Hiraoka N., Ikeshiro Y., Kushiro T., Morita M., Shibuya M., et al. Molecular cloning and characterization of a cDNA for Glycyrrhiza glabra cycloartenol synthase. Biol Pharm Bull. 2000;23:231–234. doi: 10.1248/bpb.23.231. [DOI] [PubMed] [Google Scholar]
- 32.Hayashi H., Huang P., Takada S., Obinata M., Inoue K., Shibuya M., et al. Differential expression of three oxidosqualene cyclase mRNAs in Glycyrrhiza glabra. Biol Pharm Bull. 2004;27:1086–1092. doi: 10.1248/bpb.27.1086. [DOI] [PubMed] [Google Scholar]
- 33.Abe I., Rohmer M., Prestwich G.D. Enzymatic cyclization of squalene and oxidosqualene to sterols and triterpenes. Chem Rev. 1993;93:2189–2206. [Google Scholar]
- 34.Poralla K., Hewelt A., Prestwich G.D., Abe I., Reipen I., Sprenger G. A specific amino acid repeat in squalene and oxidosqualene cyclases. Trends Biochem Sci. 1994;19:157–158. doi: 10.1016/0968-0004(94)90276-3. [DOI] [PubMed] [Google Scholar]
- 35.Wang Q.H., Gao L.L., Liang H.C., Du G.H., Gong T., Yang J.L., et al. Downregulation of lanosterol synthase gene expression by antisense RNA technology in Saccharomyces cerevisiae. Yao Xue Xue Bao. 2015;50:118–122. [PubMed] [Google Scholar]
- 36.Ito R., Mori K., Hashimoto I., Nakano C., Sato T., Hoshino T. Triterpene cyclases from Oryza sativa L.: cycloartenol, parkeol and achilleol B synthases. Org Lett. 2011;13:2678–2681. doi: 10.1021/ol200777d. [DOI] [PubMed] [Google Scholar]
- 37.Venkatramesh M., Nes W.D. Novel sterol transformations promoted by Saccharomyces cerevisiae strain GL7: evidence for 9,19-cyclopropyl to 9(11)-isomerization and for 14-demethylation to 8(14)-sterols. Arch Biochem Biophys. 1995;324:189–199. doi: 10.1006/abbi.1995.9912. [DOI] [PubMed] [Google Scholar]
- 38.He J., Chen K., Hu Z.M., Li K., Song W., Yu L.Y., et al. UGT73F17, a new glycosyltransferase from Glycyrrhiza uralensis, catalyzes the regiospecific glycosylation of pentacyclic triterpenoids. Chem Commun. 2018;54:8594–8597. doi: 10.1039/c8cc04215b. [DOI] [PubMed] [Google Scholar]
- 39.Isaev M.I., Gorovits M.B., Abubakirov N.K. Triterpene glycosides of Astragalus and their genins XXX. cycloaraloside A from Astragalus amarus. Chem Nat Compd. 1989;25:684–687. [Google Scholar]
- 40.Kim J.S., Yean M.H., Lee E.J., Kang S.S. Phytochemical studies on Astragalus root (1) –Saponins. Nat Prod Sci. 2008;14:37–46. [Google Scholar]
- 41.He Z.Q., Findlay J.A. Constituents of Astragalus membranaceus. J Nat Prod. 1991;54:810–815. [Google Scholar]
- 42.Kitagawa I., Wang H., Yoshikawa M. Saponin and sapogenol. XXXVII. Chemical constituents of Astragali Radix, the root of Astragalus membranaceus Bunge. (4). astragalosides VII and VIII. Chem Pharm Bull. 1983;31:716–722. [Google Scholar]
- 43.Kitagawa I., Wang H., Saito M., Yoshikawa M. Saponin and sapogenol. XXXVI. Chemical constituents of Astragali Radix, the root of Astragalus membranaceus Bunge. (3). astragalosides III, V, and VI. Chem Pharm Bull. 1983;31:709–715. [Google Scholar]
- 44.Gao Y., Zhang L., Feng X., Liu X., Guo F., Lv B., et al. The galactosylation of monosaccharide derivatives of glycyrrhetinic acid by UDP-glycosyltransferase GmSGT2 from Glycine max. J Agric Food Chem. 2020;68:8580–8588. doi: 10.1021/acs.jafc.0c03842. [DOI] [PubMed] [Google Scholar]
- 45.Van Wagoner R.M., Satake M., Wright J.L.C. Polyketide biosynthesis in dinoflagellates: what makes it different? Nat Prod Rep. 2014;31:1101–1137. doi: 10.1039/c4np00016a. [DOI] [PubMed] [Google Scholar]
- 46.Shan H., Segura M.J., Wilson W.K., Lodeiro S., Matsuda S.P. Enzymatic cyclization of dioxidosqualene to heterocyclic triterpenes. J Am Chem Soc. 2005;127:18008–18009. doi: 10.1021/ja055822g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.