Abstract
Chemicals possessing reactive electrophiles can denature innate proteins leading to undesired toxicity, and the overdose-induced liver injury by drugs containing electrophiles has been one of the major causes of non-approval and withdraw by the US Food and Drug Administration (FDA). Elucidating the associated proteins could guide the future development of therapeutics to circumvent these drugs' toxicities, but was largely limited by the current probing tools due to the steric hindrance of chemical tags including the common “click chemistry” labels. Taking the widely used non-steroidal anti-inflammatory drug acetaminophen (APAP) as an example, we hereby designed and synthesized an APAP analogue using fluorine as a steric-free label. Cell toxicity studies indicated our analogue has similar activity to the parent drug. This analogue was applied to the mouse hepatocellular proteome together with the corresponding desthiobiotin-SH probe for subsequent fluorine-thiol displacement reactions (FTDRs). This set of probes has enabled the labeling and pull-down of hepatocellular target proteins of the APAP metabolite as validated by Western blotting. Our preliminary validation results supported the interaction of APAP with the thioredoxin protein, which is an important redox protein for normal liver function. These results demonstrated that our probes confer minimal steric perturbation and mimic the compounds of interest, allowing for global profiling of interacting proteins. The fluorine-thiol displacement probing system could emerge as a powerful tool to enable the investigation of drug–protein interactions in complex biological environments.
KEY WORDS: Acetaminophen (APAP), Liver, Hepatotoxicity, Bioorthogonal, Click chemistry, Fluorine displacement, Fluorine thiol displacement reaction, FTDR
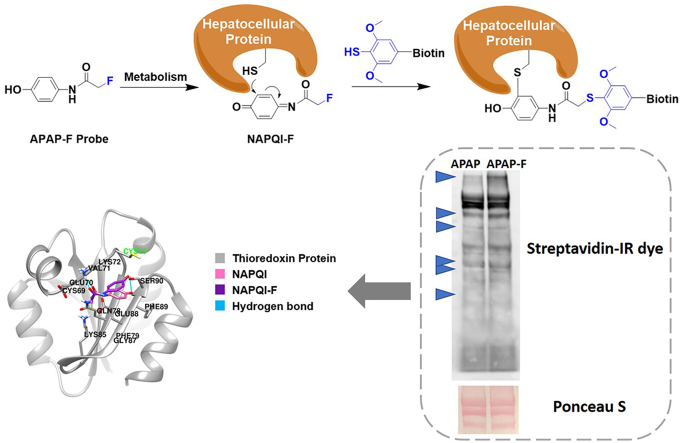
Graphical abstract
Fluorine tagging coupled with aryl-thiol probes enable steric-free profiling of the hepatocellular protein targets of the drug acetaminophen and the validation of thioredoxin as one protein underlying its inherent toxicity.
1. Introduction
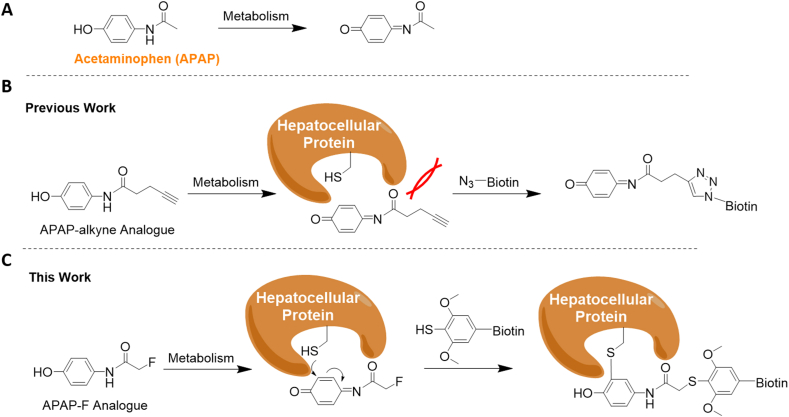
Reactive electrophiles are a common type of toxic metabolites that can react with nucleophilic amino acids1. Such modifications disturb native protein functions and have been implicated in many diseases1. One representative example is the compound acetaminophen (APAP, Tylenol), which is one of the world's most commonly used drug ingredients2, 3, 4. However, APAP is also the world's leading cause of drug overdose and liver injury/failure2,3,5. APAP is primarily metabolized in vivo by the enzyme CYP2E1 which is highly expressed in the liver (Fig. 1A)5, 6, 7. The resulting intermediate N-acetyl-p-benzo-quinone imine (NAPQI) depletes hepatic glutathione (GSH) and reacts exclusively with functional cysteine or selenocysteine residues throughout the liver proteome6, 7, 8, leading to hepatocellular necrosis6,7. Nevertheless, the global interacting proteomes of APAP are poorly understood because of the lack of an efficient and broadly applicable experimental tool. Currently, the most common method entails covalent labeling of the drugs with an alkyne or azide tag (Fig. 1B) that can later be appended via a classical bio-orthogonal reaction, copper-catalyzed azide-alkyne cycloaddition (CuAAC “click” chemistry). The final in situ appending with a biotin affinity probe allows for enrichment by streptavidin-coated beads/resins, target protein pull-down, and chemical proteomics-based protein identification9,10. Yet the alkyne/azide-based tags are still relatively bulky in terms of size and their incorporation into sites may perturb metabolism or interactions with potential protein targets (Fig. 1B)8,11, 12, 13. Moreover, the alkynes are known to serve as inhibitors for the liver enzyme CYP2E18. As a result, the acetaminophen alkyne analogue (APAP-alkyne) has exhibited limited in vitro reactivity in mouse liver homogenates8, and only thioredoxin reductase-1 (TrxR1) has been identified to date10.
Figure 1.
Design of the acetaminophen (APAP) analogues. (A) The in vivo metabolism of APAP parent drug to N-acetyl-p-benzoquinone imine (NAPQI). (B) Previous work on APAP-alkyne analogue for “click chemistry” based appendence with biotin and target protein labeling. (C) Current work on the steric-free APAP-F analogue for “fluorine-thiol displacement reaction (FTDR)” based appendence with biotin and target protein labeling.
We recently demonstrated that fluorine substitution onto natural small molecules such as post-translational modification cofactors/precursors present minimal steric hindrance14, and the fluorine tag alpha to the carbonyl groups can be further converted in situ to other functional probes such as TAMRA fluorophore or biotin through the bioorthogonal fluorine-thiol displacement reaction (FTDR)14. The drug acetaminophen happens to possess an acetamide group that can potentially allow for the derivatization by fluorine and the subsequent FTDR reaction. Herein, we report the synthetic preparation of the fluorine-modified APAP analogue (APAP-F) and the utilization of it to test the hypothesis that fluorinated acetaminophen does not exhibit the steric issues as observed by the alkyne-tagged acetaminophen and thereby can be utilized as a more efficient probe. Along with the benzenethiol containing desthiobiotin probe, our APAP-F compound has served as a better analogue, empowering us to explore the interacting proteome of the drug (Fig. 1C).
2. Results and discussions
2.1. Design and synthesis of APAP analogues
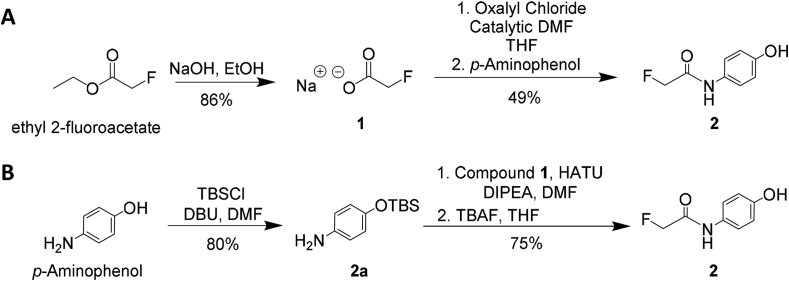
We first prepared the APAP-alkyne derivative by reacting 4-pentynoic acid with oxalyl chloride in DCM followed by addition of the commercially available p-aminophenol (Supporting Information Scheme S2), which affords a 39% yield of the purified derivative with NMR spectra consistent with the reported data8,10. Implementing the similar approach, we treated the freshly hydrolyzed fluoroacetate with oxalyl chloride in THF and reacted the mixture with p-aminophenol to generate the desired APAP-F analogue 2 (Scheme 1A). This route consists of two steps with an overall 42% yield. To obtain a better yield of the analogue, we also attempted the previously reported TBS protection strategy10 by reacting the phenol part with tert-butyldimethylsilyl chloride in the presence of catalytic amount of DBU to afford the intermediate 2a (Scheme 1B). HATU-mediated coupling of 2a with sodium fluoroacetate followed by silyl deprotection with TBAF finally led to APAP-F, which took a total of three steps with an overall 52% yield. Both analogues appeared stable as a solid but also in solution, as demonstrated by LC‒MS.
Scheme 1.
Synthesis of the APAP-F analogue 2 with two separate routes (A) and (B).
2.2. Design and synthesis of biotin probes
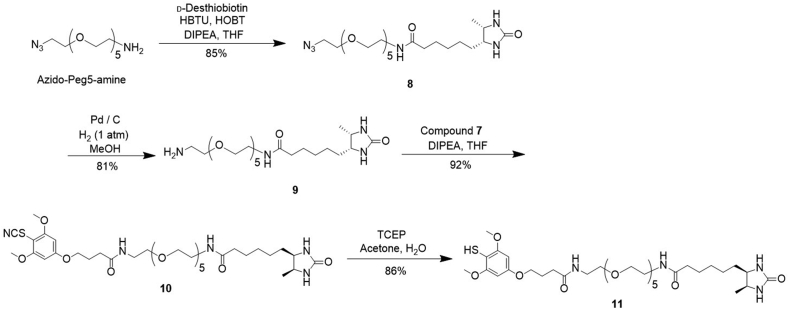
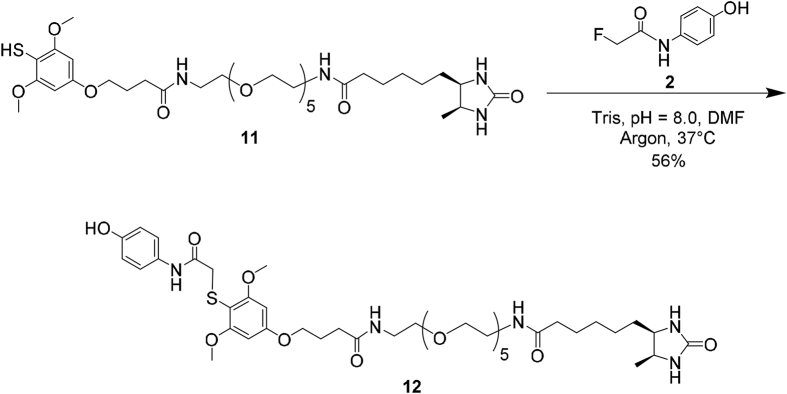
To pair up with the alkyne analogue, we have synthesized the desthiobiotin-azide probe by first preparing the 3-azidopropylamine linker from the commercially available bromopropylamine (Supporting Information Scheme S2)15. Coupling between d-desthiobiotin and 3-azidopropylamine with the aid of 1,1′-carbonyldiimidazole resulted in the desired probe 15 with a 65% yield. In order to convert the fluorine tag on APAP to a biotin functional moiety, we have designed the desthiobiotin-SH probe (11, Scheme 2) by incorporating the previously reported tri-methoxy thiophenol as the warhead and connecting it with desthiobiotin through a polyethylene glycol linker that greatly increases solubility. After preparing the thiocyanate dimethoxy phenol 5 based on the reported procedure14, we performed O-alkylation with t-butyl 4-bromobutanoate to generate intermediate 6 (Supporting Information Scheme S1). Adapting Sammakia's approach16, we treated the tert-butyl ester 6 directly with thionyl chloride, and subsequently with N-hydroxysuccinimide to render the important intermediate 7 with a 43% yield. Concurrently, we carried out amide coupling between d-desthiobiotin and the commercially available azido-PEG5-amine linker (Scheme 2). The resulting azide intermediate 8 was reduced by Pd/C-mediated hydrogenation to generate derivative 9 with an active terminal amine. We further conjugated 9 with the previously prepared NHS-thiocyanate intermediate 7, and consecutively reduced compound 10 with TCEP to finally afford the desired desthiobiotin-SH probe 11. Altogether, the construction of probe 11 took four linear steps with a cumulative yield of 54%.
Scheme 2.
Synthetic route of the desthiobiotin-SH probe 11.
2.3. Cytotoxicity evaluation of APAP analogues
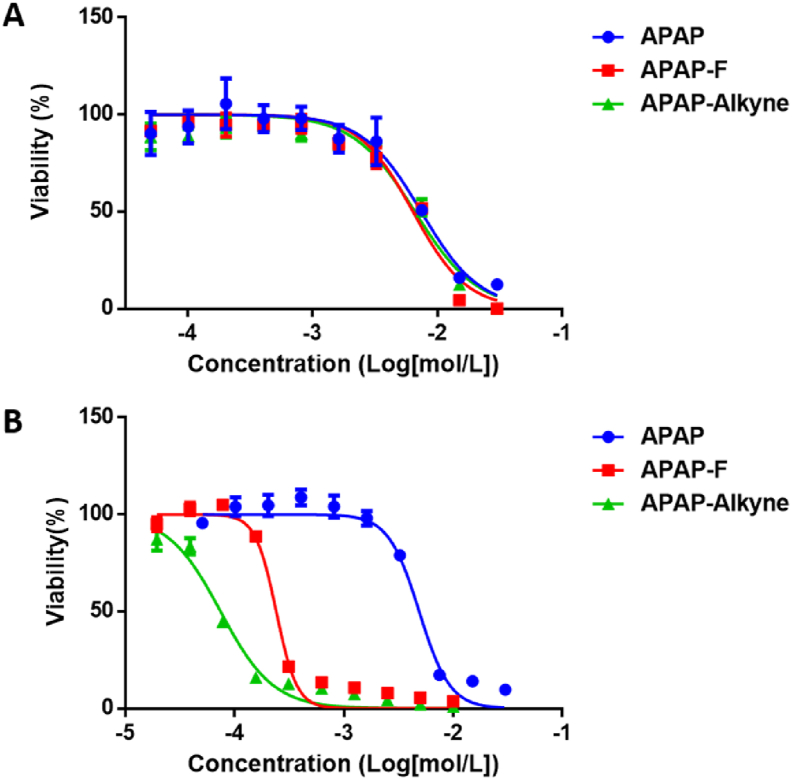
To test the hypothesis that fluorine modification on APAP does not perturb the parent drug's activity, we started by evaluating the cell cytotoxicity of APAP analogues in human liver cell line HepG2 (Fig. 2A). The HepG2 cell line has been commonly used in research related to drug metabolic activities and hepatotoxicity studies17. Treatment with HepG2 cells for 48 h suggested that all compounds displayed similar cytotoxicity, with EC50 values of APAP, APAP-F, and APAP-alkyne equaling to 7.4 ± 1.4, 6.4 ± 1.5, and 6.6 ± 1.8 mmol/L, respectively. Since HepG2 cells express lower level of CYP2E1 enzymes than the primary human hepatocytes18, we then turned our attention to primary mouse hepatocytes. Fresh liver cells were harvested from 12-week-old female mice following the reported procedure19 and were immediately treated with APAP and APAP analogues. As shown in Fig. 2B, the EC50 of APAP was 4.9 ± 0.9 mmol/L, while the EC50s of APAP-F and APAP-alkyne were 0.6 ± 0.04 and 0.8 ± 0.07 mmol/L, respectively. While the toxicities from the derivatives were more significant, the fluorine label did not result in different toxicity from the click chemistry alkyne tag. Taken together, the F-modified analogue inherits similar biological profiles to the parent APAP and the alkyne analogue APAP-alkyne.
Figure 2.
Cell toxicity evaluation of APAP and APAP analogues. (A) Viability of human HepG2 cell lines after treatment with APAP or tag-derivatized APAP analogues. (B) Viability of mouse liver cells after treatment with APAP or APAP analogues.
2.4. Model protein labeling by biotin probes
We then assessed the reactivity of the desthiobiotin-SH probe 11 by first testing its reaction with the APAP-F analogue at the small molecule level (Scheme 3), which successfully proceeded at 37 °C and afforded the expected product 12 with a 56% yield after isolation and purification.
Scheme 3.
Model fluorine-thiol displacement reaction between the APAP-F derivative and the desthiobiotin-SH probe.
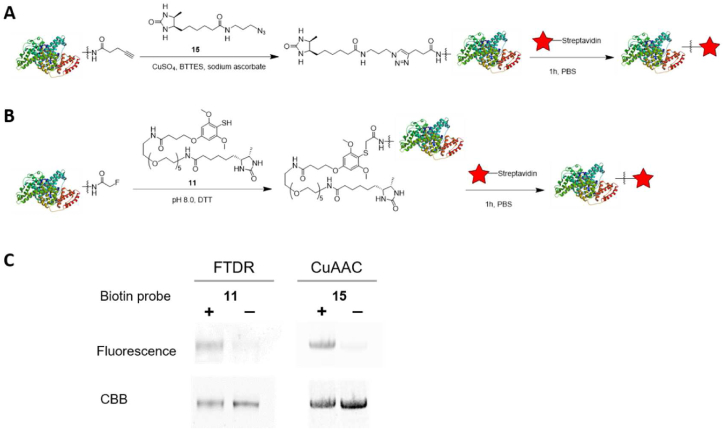
Moving forward, we performed model protein labeling using BSA (Fig. 3). The protein was first modified with alkyne or fluorine acetamide tags based on the random lysine-NHS coupling chemistry20. Subsequent incubation with the corresponding desthiobiotin probes was followed with capturing by infrared (IR) dye-conjugated streptavidin (Fig. 3A and B). This procedure allowed us to readily monitor the two-step labeling process mediated by either “click chemistry” (Fig. 3A) or FTDR (Fig. 3B) in a straightforward manner through IR fluorescence detection. As shown through PAGE gel analysis (Fig. 3C), both biotin probes have resulted in satisfactory labeling of model proteins as long as the first step modification occurred.
Figure 3.
In vitro model protein BSA labeling with desthiobiotin probes. (A) Illustration of “click chemistry” based labeling using desthiobiotin-azido probe 15. (B) Illustration of “FTDR” based labeling using desthiobiotin-SH probe 11. (C) PAGE gel results of the protein labeling. “CBB” is Coomassie brilliant blue staining. The first-step installation of the alkyne (A) or acetyl fluoride (B) tags were based on the random conjugation of lysine-NHS chemistry.
2.5. Labeling of hepatocellular protein targets of APAP
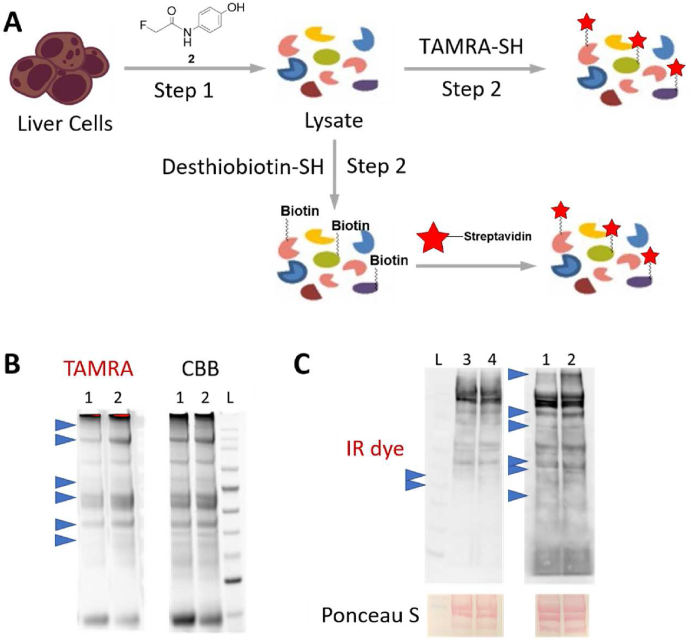
Moving forward, we attempted labeling of APAP-interacting hepatocellular proteins by treating mouse hepatocytes with APAP analogues, which was followed by lysis and immediate conversion of protein tags to functional probes with desthiobiotin-SH or the previously reported TAMRA-SH (Fig. 4A)14. Using the TAMRA-SH probe, we were able to directly observe the hepatocellular proteins that were labeled by fluorine due to the covalent interactions with NAPQI-F metabolic intermediate (Fig. 4B). Compared to the control lane 1, more protein bands (pinpointed by blue arrows) displayed TAMRA fluorescence in lane 2, while both lanes showed equal protein loading by CBB staining. We also converted the tags to desthiobiotin and observed significant protein band differences as a result of FTDR-mediated labeling by APAP-F/Biotin-SH (lanes 1 and 2, Fig. 4C). Contrary to this, the “click chemistry”-based labeling resulted in fewer distinct protein bands (lane 4 versus lane 3, Fig. 4C), which is presumably due to the steric hindrance and the unwanted enzyme reactivity of the alkyne tag8,11, 12, 13. These results demonstrated the unique compatibility of APAP-F analogue with the hepatocellular proteome.
Figure 4.
In situ mouse hepatocellular protein labeling by APAP analogues followed by TAMRA-SH or desthiobiotin-SH/desthiobiotin-azide probes. (A) Illustration of the two-step hepatocellular protein labeling and detection processes. (B) The FTDR labeling results using the parent APAP (lane 1) or APAP-F analogue (lane 2), along with the TAMRA-SH probe for the 2nd step. “CBB”: Coomassie brilliant blue. (C) Comparison of the FTDR labeling (lane 1: APAP, lane 2: APAP-F) and the click chemistry labeling (lane 3: APAP, lane 4: APAP-alkyne) using desthiobiotin probes (desthiobiotin-SH for lanes 1–2, desthiobiotin-azide for lanes 3–4).
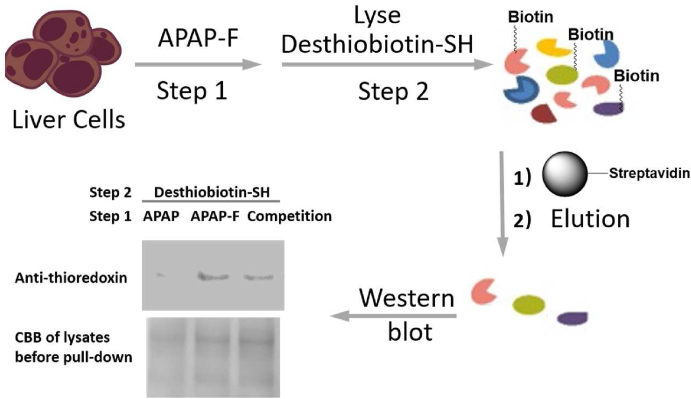
2.6. Pull-down of hepatocellular protein targets of APAP
Finally, we combined the biotin labeling approach with streptavidin-mediated protein pull-down to enrich the target proteins of APAP (Fig. 5). To confirm the enrichment results, we pursued western blotting using antibodies against potential targets of APAP. Previous LC‒MS/MS based proteomics research has identified thioredoxin as a potential APAP interacting protein, although there is no further experimental validation21. We hereby observed the selective presence of mouse thioredoxin in the proteins enriched by our FTDR-based APAP-F/desthiobiotin-SH probing strategy. Notably, the pull-down of thioredoxin can be weakened after competition of the APAP-F incorporation by 10-fold excess of APAP parent drug during the 1st step incubation with liver cells (Fig. 5), which further confirmed that thioredoxin is a target of APAP.
Figure 5.
FTDR-mediated pull-down of mouse hepatocellular proteins that interact with APAP-F, followed by validations with Western blot. “CBB”: Coomassie brilliant blue. Competition has been done by treating mouse liver cells with APAP-F in the presence of 10 equivalents of the APAP parent compound.
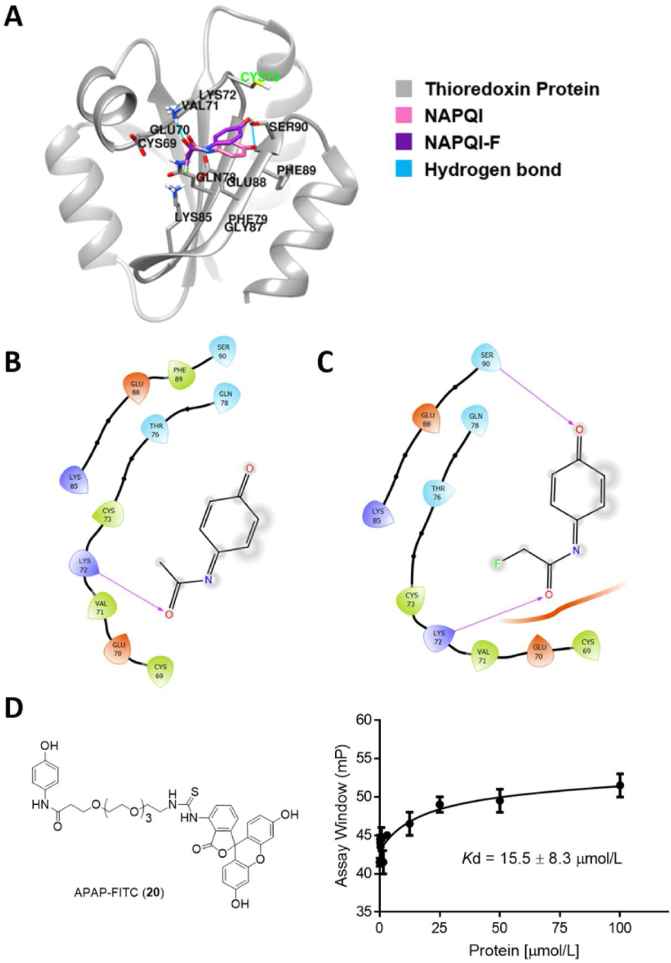
To understand how APAP/NAPQI interacts with the thioredoxin protein at the molecular level, we performed the Schrodinger molecular docking studies (Fig. 6A‒C). Both the parent APAP and APAP-F analogue bound within the same site of thioredoxin and are in close proximity to the nucleophilic cysteine 73 (Fig. 6A), which is consistent with the previous mass spec analysis results21 that cysteine 73 could be the active site of thioredoxin for the NAPQI metabolic intermediate of APAP. Further analysis of the active site (Fig. 6B and C) showed that NAPQI binds via hydrogen bonding with lysine 72 while NAPQI-F interacts with lysine 72 and serine 90. Correspondingly, the resulting docking scores for NAPQI and NAPQI-F were −3.4 and −4.2, respectively, suggesting that NAPQI-F may have a higher affinity for the thioredoxin protein. This mechanistic finding could explain why the APAP-F analogue displayed an improved EC50 during the cytotoxicity evaluations in mouse liver cells (Fig. 2B).
Figure 6.
Molecular docking and experimental validation of the APAP metabolites within the pocket of the identified protein thioredoxin. (A) 3D interaction diagram between the thioredoxin protein (PDB: 1ERU) and the metabolite intermediate NAPQI (pink)/NAPQI-F (purple). (B) 2D interaction diagram between NAPQI and the amino acids within thioredoxin. (C) 2D interaction diagram between NAPQI-F and the amino acids within thioredoxin. (D) Fluorescence polarization studies to confirm the binding of the oxidized APAP-FITC probe with human thioredoxin.
To experimentally confirm the binding of APAP with the thioredoxin protein, we synthesized the FITC conjugate version of the APAP drug. Adopting a convergent synthetic strategy (Supporting Information Scheme S3), we prepared the azido-derivatized PEG4 linker 18 and the TIPS protected APAP drug, respectively. With the aid of thionyl chloride, the coupling of the linker with the amino group of APAP-TIPS proceeded smoothly to yield intermediate 19. A final three-step deprotection and coupling with the isothiocyanate group of FITC rendered the desired APAP-FITC (20) with a yield of ∼20% and a >95% purity as indicated by LC‒MS analysis (Supporting Information). The probe was then oxidized by silver oxide to generate the NAPQI-FITC intermediate22, and subsequently incubated with human thioredoxin. As shown by the fluorescence anisotropy assay result (Fig. 6D)23, significant binding was observed between thioredoxin and the probe, with the Kd estimated to be 15.5 ± 8.3 μmol/L. Future studies may explore the effect on binding with the mutation of Cys73 in thioredoxin, which could further corroborate the molecular mechanisms revealed by our docking studies.
3. Conclusions
In summary, we have applied the steric-free fluorine thiol probing strategy to a classic drug, acetaminophen, to further investigate its underlying hepatotoxicity. The designed analogue, APAP-F, has been synthesized and assessed to demonstrate comparable hepatotoxicity of tagged APAP analogues. We also designed and synthesized a powerful desthiobiotin probe and utilized it in tandem with the APAP-F analogue to successfully carry out the labeling and pull-down assays of the APAP targeting proteins from primary mouse hepatocytes. Our FTDR-based probes have resulted in more explicit protein labeling than the reported probes designed by traditional “click chemistry”, corroborating the initial hypothesis that small molecule drugs modified by fluorine possess minimal steric hindrance and may inherit similar biological profiles of the parent drugs. Further western blotting analysis of the FTDR-enriched hepatocellular proteins has led to the subsequent validation of thioredoxin as one of the protein targets of APAP. Given thioredoxin's ubiquitous antioxidant function and its role in redox signaling24, its contribution towards APAP's hepatotoxicity warrants further investigations including potential development of therapeutic mitigations. Future work would focus on proteomics studies of the enriched hepatocellular proteins in order to systematically elucidate and to obtain a complete set of APAP targets. Nevertheless, this proof-of-concept research has provided strong evidence that the FTDR-based labeling strategy can be potentially applied to many other small molecule compounds and drugs, thereby empowering the interrogation of their pharmacology.
4. Experimental
4.1. Chemistry
The synthetic routes, synthesis and characterization data of compounds have been provided in Supporting Information.
4.2. Biological assays
4.2.1. Probe labeling on model proteins
Bovine serum albumin (BSA) was randomly conjugated by fluoroacetamide or alkyne-derivatized acetamide through lysine-NHS ester chemistry1. For FTDR labeling, the fluorine-tagged BSA (1.5 mg/mL final concentration) was mixed with desthiobiotin-SH probe (2 mmol/L final concentration) along with DTT (5 mmol/L final concentration) in DPBS buffer (pH 8.0). The mixture was incubated at 37 °C in the dark for 6–8 h before SDS-PAGE analysis. For the control “click chemistry” labeling, 4 μL of CuSO4 (5 mmol/L) was pre-incubated with 4 μL of BTTES (5 mmol/L) for 5 min and then mixed with 2 μL of sodium ascorbate (50 mmol/L), 2 μL of desthiobiotin-azide probe (4 mmol/L), and 3 μL of the alkyne-tagged BSA (7.5 mg/mL). The mixture was incubated at 37 °C in the dark for 3 h before SDS-PAGE analysis.
4.2.2. Isolation of primary mouse liver cells
The experiments were exactly pursued following the approved protocols by the Institutional Animal Care and Use Committee (IACUC). About 12-week-old female mice were euthanized by carbon dioxide (CO2) under IACUC guidelines. The fresh liver tissues were harvested, washed with warm Hank's buffer to remove red blood cells, and sliced into small pieces as suspensions in Hank's buffer. After filtrating through a 40 μm cell strainer on ice, the cell suspension was purified by Percoll gradient. Briefly, 20 mL of the suspension was slowly dispensed on top of a 20 mL solution of 45% Percoll/Hank's buffer inside a 50 mL conical tube. The conical tube was then centrifuged at 500×g for 15 min at a minimum acceleration and deceleration speed. The primary mouse liver cells were collected as the pellet at the bottom of the tube, resuspended in cold Hank's buffer and washed twice (repeatedly dissolved in cold Hank's buffer and spined down at 200×g for 7 min). The cells were finally resuspended in warm William E media, and cultured as described below.
4.2.3. Cell culturing
The primary mouse liver cells were incubated at 37 °C under an atmosphere of 5% CO2, in William E medium without phenol red but supplemented with 10% of fetal bovine serum (FBS) and 1% of antibiotic antimycotic solution (AAS). The human HepG2 cell line was incubated at 37 °C under an atmosphere of 5% CO2, in low glucose DMEM medium (final concentration of glucose: 4.5 g/L) supplemented with 1% of AAS.
4.2.4. In vitro hepatotoxicity assays
HepG2 cells or primary mouse liver cells were seeded in 96-well plates at a density of 3.3 × 106 cells per well (90 μL) and incubated for 6 h at 37 °C under an atmosphere of 5% CO2. Serially diluted (1/2 fold) compound stocks (10×) for either the parent compound APAP, the probe APAP-F, or APAP-alkyne were prepared in DPBS buffer that was premixed with 30% of DMSO (v/v). Exactly 10 μL of each stock solution was added into the cells pre-plated within 90 μL of cell medium per sample well, to make final compound concentrations spanning from 0.059 to 30 mmol/L. Treatment with HepG2 cells lasted for 48 h before work-up, while the compounds were incubated with the primary mouse liver cells for only 16 h. After incubation, CellTiter-Glo reagents were added at a scale of 50 μL per well and the luminescence signals were recorded at Synergy H1 plate reader (Biotek) using the Gen5 software. The data were processed and plotted using GraphPad Prism V.11.1.
4.2.5. Hepatocellular protein extraction after live cell treatment with APAP analogues
Approximately 33.3 × 106 primary mouse liver cells were cultured in 9 mL of Hank's buffer (without serum) in a 10 cm cell dish. APAP, APAP-F, or APAP-alkyne probe was each prepared in 30% DMSO/DPBS buffer (v/v), and 1 mL of the probe solution (2.5 mmol/L) was added to the cell culture dish under appropriate mixing by pipetting up and down. The cells were treated with probes for 3 h at 37 °C under an atmosphere of 5% CO2 and were washed twice by repeatedly spinning down at 300×g for 10 min followed by resuspension in DPBS buffer. Lysis was performed in a mild manner by incubating the adherent cells with 1 mL of lysis buffer (CelLytic M, Sigma–Aldrich) on ice for 10 min, followed by scratching them down from the plate surface. The collected cell suspension was then shaken vigorously at room temperature for 15 min. The debris were removed by centrifuging the lysate at 15,000×g for 15 min at 4 °C. Hepatocellular proteins were purified from the remaining supernatant by methanol precipitation. Briefly, methanol was added to the supernatant at 10:1 v/v ratio. The resulting mixture was cooled down at −80 °C for 1–2 h, and spun down at 10,000×g for 5 min at 4 °C. The protein pellets were resuspended with 100% cold methanol and spun down again at 10,000×g for 5 min at 4 °C to wash away the unlabeled probes and salts. Finally, DPBS with 0.1% SDS was added to make a protein solution with a final concentration of 1.5 mg/mL for follow-up characterizations.
4.2.6. TAMRA-SH probe labeling and analysis
Hepatocellular proteins (40 μL, 1.5 mg/mL) harvested after cellular treatment with APAP or APAP-F analogue were mixed with TCEP (5 mmol/L), and the final pH value was adjusted to 8.0. Approximately 4 μL of TAMRA-SH probe (20 mmol/L in DMSO) was added to the mixture and the labeling reaction was allowed to take place at 37 °C in the dark for 6 h. This type of labeling reaction was attempted in either the DPBS buffer or the Hank's buffer. After the 6 h incubation, all samples were added to the loading buffer, heated for 5 min at 90 °C, and directly loaded onto 4%–12% Bis-Tris SDS-PAGE gels for protein separation (Genscript). The resulting PAGE gels were fixed and scanned by the ChemiDoc MP Imaging System (Bio-Rad) for protein bands labeled by TAMRA dye. The gel was finally stained by Coomassie brilliant blue (CBB) as a loading control.
4.2.7. Desthiobiotin probe labeling and analysis
The hepatocellular proteins were harvested after treatment with APAP and APAP-F analogue, respectively, and were incubated with desthiobiotin-SH probe in a similar manner to the TAMRA-SH probe labeling as mentioned above. For lysate proteins harvested from treatment with APAP and APAP-alkyne analogue respectively, the specific protein samples were dissolved in DPBS buffer, pH 7.2. About 40 μL of each lysate protein sample (1.5 mg/mL) was mixed with 2 μL of sodium ascorbate (50 mmol/L). The coupling catalyst (4 μL of CuSO4 (5 mmol/L) and 4 μL of BTTES (5 mmol/L)) was preincubated for 5 min at room temperature and then added to the protein mixture, along with 2 μL of the desthiobiotin-azide probe (4 mmol/L). The reaction mixture was incubated at room temperature for 3 h in the dark. Then all the reaction samples were loaded onto SDS-PAGE. The protein bands on the gel were further transferred to the PVDF membrane through semi-dry Trans-Blot (Bio-Rad). The membrane was subsequently blocked by TBS-Tween buffer/3% BSA overnight at 4 °C, and finally probed with streptavidin-IRdye 630. The biotin-labeled proteins were finally visualized and recorded on ChemiDoc MP Imaging System (Bio-Rad).
4.2.8. Hepatocellular protein pull-down and Western blot analysis
After biotin labeling, unreacted probes and other reagents were removed from lysate proteins by methanol precipitation. Then, lysate proteins were dissolved in DPBS with 0.5% SDS. For each pull-down experiment, 100 μL slurry of streptavidin magnetic beads (NEB S1420S) were mixed with 30 μg of lysate proteins in DPBS buffer with 0.05% SDS and incubated at room temperature for 3 h. After discarding unbound lysate proteins in the supernatant, the beads were washed twice in 0.5% SDS DPBS buffer and twice in DPBS buffer. Enriched proteins were eluted by boiling beads in 1 × LDS loading buffer with 100 mmol/L DTT at 95 °C for 5 min and were loaded onto SDS-PAGE. The Western blot against thioredoxin was detected by thioredoxin rabbit polyclonal antibody (Proteintech 14999-1-AP) and IRDye 680RD secondary antibody (Li-COR). As the loading control, the PAGE gel of the lysates of each experimental group right before pull-down were stained by Coomassie brilliant blue.
4.2.9. Fluorescence polarization assays
APAP-FITC probe (compound 20) was oxidized by silver oxide to NAPQI-FITC intermediate, and roughly purified according to the published procedure22. The intermediate was dissolved in protein buffer (300 mmol/L NaCl, 25 mmol/L Tris, pH 7.2, 2 mmol/L DTT, 2% glycerol, 0.01% Tween) at a final concentration of 10 μmol/L, and was mixed with recombinant human thioredoxin-1 protein (R&D Systems™, MN) that was serially diluted to a final concentration spanning from 0.2 to 100 μmol/L. The final mixtures were plated in a black opaque 384-well plate (Nunc, Sigma–Aldrich) at 10 μL/well and incubated at 37 °C for 1 h. Wells containing buffer only were used as a blank control. The FP signals were measured on a BioTek microplate reader (Synergy H1, USA), with the excitation and emission wavelengths set at 485 and 528 nm, respectively.
4.3. In silico simulations
The crystal structure of the oxidized thioredoxin was derived from the Protein Data Bank with accession code 1ERU. Molecular docking was performed using the Schrodinger molecular docking toolkit (New York, NY, USA). The 3D structures of the NAPQI and NAPQI-F analogue were generated using the construction panel in Maestro 11.8 and optimized using the LigPrep module with default parameters. Molecular docking was carried out using Glide module in its SP mode. The obtained docked poses were analyzed with Chimera. The docking scores were analyzed to determine which compounds have a higher affinity.
Acknowledgments
This work has been supported by NIH grant 5R35GM133468-03, Temple University Startup Funding, and Temple University the Center for Substance Abuse Research (CSAR) under the Pilot P30 grant by National Institute on Drug Abuse. Rongsheng E. Wang is a Cottrell Scholar of Research Corporation for Science Advancement. Support for the NMR facility at Temple University by a CURE grant from the Pennsylvania Department of Health is gratefully acknowledged.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2022.08.003.
Author contributions
Benjamin L. Prather and Shuyue Ji contributed equally to this work, lead the research, data analysis, and writing of the manuscript. Yue Zhao, Robert Maloney, and Rui Zhang performed the biological assays. Femil Joseph Shajan and Zakey Yusuf Buuh carried out the chemical synthesis. Robert Maloney and Carson Cohen performed mass spectrometry analysis. Mi Zhao performed the molecular docking studies. Rongsheng E. Wang supervised the entire research with conceptualization, analysis, and resources.
Conflicts of interest
The authors declare no competing financial interests.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Sibbersen C., Palmfeldt J., Hansen J., Gregersen N., Jorgensen K.A., Johannsen M. Development of a chemical probe for identifying protein targets of alpha-oxoaldehydes. Chem Commun (Camb) 2013;49:4012–4014. doi: 10.1039/c3cc41099d. [DOI] [PubMed] [Google Scholar]
- 2.Nguyen N.T., Akakpo J.Y., Weemhoff J.L., Ramachandran A., Ding W.X., Jaeschke H. Impaired protein adduct removal following repeat administration of subtoxic doses of acetaminophen enhances liver injury in fed mice. Arch Toxicol. 2021;95:1463–1473. doi: 10.1007/s00204-021-02985-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behrends V., Giskeodegard G.F., Bravo-Santano N., Letek M., Keun H.C. Acetaminophen cytotoxicity in HepG2 cells is associated with a decoupling of glycolysis from the TCA cycle, loss of NADPH production, and suppression of anabolism. Arch Toxicol. 2019;93:341–353. doi: 10.1007/s00204-018-2371-0. [DOI] [PubMed] [Google Scholar]
- 4.Ndetan H., Evans M.W., Tanue T., Osuagwu C.C., Elueze E., Singh K.P., et al. Therapeutic use of acetaminophen and light to moderate alcohol: are there early disparate risks for kidney disease?. Health Equity. 2020;4:518–524. doi: 10.1089/heq.2020.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bunchorntavakul C., Reddy K.R. Acetaminophen-related hepatotoxicity. Clin Liver Dis. 2013;17:587–607. doi: 10.1016/j.cld.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Larson A.M. Acetaminophen hepatotoxicity. Clin Liver Dis. 2007;11:525–548. doi: 10.1016/j.cld.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Bartolone J.B., Birge R.B., Sparks K., Cohen S.D., Khairallah E.A. Immunochemical analysis of acetaminophen covalent binding to proteins. Partial characterization of the major acetaminophen-binding liver proteins. Biochem Pharmacol. 1988;37:4763–4774. doi: 10.1016/0006-2952(88)90350-4. [DOI] [PubMed] [Google Scholar]
- 8.Whitby L.R., Obach R.S., Simon G.M., Hayward M.M., Cravatt B.F. Quantitative chemical proteomic profiling of the in vivo targets of reactive drug metabolites. ACS Chem Biol. 2017;12:2040–2050. doi: 10.1021/acschembio.7b00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emnett C., Li H., Jiang X., Benz A., Boggiano J., Conyers S., et al. A clickable analogue of ketamine retains NMDA receptor activity, psychoactivity, and accumulates in neurons. Sci Rep. 2016;6 doi: 10.1038/srep38808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang S., Tian Y., Lu S., Wang R., Shang H., Zhang X., et al. Design and synthesis of acetaminophen probe APAP-P1 for identification of the toxicity targets thioredoxin reductase-1 in HepaRG cells. RSC Adv. 2019;9:15224–15228. doi: 10.1039/c9ra00483a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grammel M., Hang H.C. Chemical reporters for biological discovery. Nat Chem Biol. 2013;9:475–484. doi: 10.1038/nchembio.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang C., Mi J., Feng Y., Ngo L., Gao T., Yan L., et al. Labeling lysine acetyltransferase substrates with engineered enzymes and functionalized cofactor surrogates. J Am Chem Soc. 2013;135:7791–7794. doi: 10.1021/ja311636b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y.Y., Ascano J.M., Hang H.C. Bioorthogonal chemical reporters for monitoring protein acetylation. J Am Chem Soc. 2010;132:3640–3641. doi: 10.1021/ja908871t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyu Z., Zhao Y., Buuh Z.Y., Gorman N., Goldman A.R., Islam M.S., et al. Steric-free bioorthogonal labeling of acetylation substrates based on a fluorine-thiol displacement reaction. J Am Chem Soc. 2021;143:1341–1347. doi: 10.1021/jacs.0c05605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi Y., Pierce J.G. Synthesis of the 5,6-dihydroxymorpholin-3-one fragment of monanchocidin A. Org Lett. 2015;17:968–971. doi: 10.1021/acs.orglett.5b00069. [DOI] [PubMed] [Google Scholar]
- 16.Greenberg J.A., Sammakia T. The conversion of tert-butyl esters to acid chlorides using thionyl chloride. J Org Chem. 2017;82:3245–3251. doi: 10.1021/acs.joc.6b02931. [DOI] [PubMed] [Google Scholar]
- 17.Donato M.T., Tolosa L., Gomez-Lechon M.J. Culture and functional characterization of human hepatoma HepG2 cells. Methods Mol Biol. 2015;1250:77–93. doi: 10.1007/978-1-4939-2074-7_5. [DOI] [PubMed] [Google Scholar]
- 18.Westerink W.M., Schoonen W.G. Cytochrome P450 enzyme levels in HepG2 cells and cryopreserved primary human hepatocytes and their induction in HepG2 cells. Toxicol In Vitro. 2007;21:1581–1591. doi: 10.1016/j.tiv.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 19.Charni-Natan M., Goldstein I. Protocol for primary mouse hepatocyte isolation. STAR Protoc. 2020;1 doi: 10.1016/j.xpro.2020.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nanda J.S., Lorsch J.R. Labeling a protein with fluorophores using NHS ester derivitization. Methods Enzymol. 2014;536:87–94. doi: 10.1016/B978-0-12-420070-8.00008-8. [DOI] [PubMed] [Google Scholar]
- 21.Geib T., Moghaddam G., Supinski A., Golizeh M., Sleno L. Protein targets of acetaminophen covalent binding in rat and mouse liver studied by LC‒MS/MS. Front Chem. 2021;9 doi: 10.3389/fchem.2021.736788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dahlin D.C., Nelson S.D. Synthesis, decomposition kinetics, and preliminary toxicological studies of pure N-acetyl-p-benzoquinone imine, a proposed toxic metabolite of acetaminophen. J Med Chem. 1982;25:885–886. doi: 10.1021/jm00350a001. [DOI] [PubMed] [Google Scholar]
- 23.Du Y. Fluorescence polarization assay to quantify protein‒protein interactions in an HTS format. Methods Mol Biol. 2015;1278:529–544. doi: 10.1007/978-1-4939-2425-7_35. [DOI] [PubMed] [Google Scholar]
- 24.Collet J.F., Messens J. Structure, function, and mechanism of thioredoxin proteins. Antioxid Redox Signal. 2010;13:1205–1216. doi: 10.1089/ars.2010.3114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.