Abstract
Pain is often debilitating, and current treatments are neither universally efficacious nor without risks. Transient receptor potential (TRP) ion channels offer alternative targets for pain relief, but little is known about the regulation or identities of endogenous TRP ligands that affect inflammation and pain. Here, transcriptomic and targeted lipidomic analysis of damaged tissue from the mouse spinal nerve ligation (SNL)-induced chronic pain model revealed a time-dependent increase in Cyp1b1 mRNA and a concurrent accumulation of 8,9-epoxyeicosatrienoic acid (EET) and 19,20-EpDPA post injury. Production of 8,9-EET and 19,20-EpDPA by human/mouse CYP1B1 was confirmed in vitro, and 8,9-EET and 19,20-EpDPA selectively and dose-dependently sensitized and activated TRPA1 in overexpressing HEK-293 cells and Trpa1-expressing/AITC-responsive cultured mouse peptidergic dorsal root ganglia (DRG) neurons. TRPA1 activation by 8,9-EET and 19,20-EpDPA was attenuated by the antagonist A967079, and mouse TRPA1 was more responsive to 8,9-EET and 19,20-EpDPA than human TRPA1. This latter effect mapped to residues Y933, G939, and S921 of TRPA1. Intra-plantar injection of 19,20-EpDPA induced acute mechanical, but not thermal hypersensitivity in mice, which was also blocked by A967079. Similarly, Cyp1b1-knockout mice displayed a reduced chronic pain phenotype following SNL injury. These data suggest that manipulation of the CYP1B1–oxylipin–TRPA1 axis might have therapeutic benefit.
Key words: CYP1B1, Spinal nerve ligation, Epoxides, TRPA1, Calcium imaging, Hyperalgesia, Chronic pain, Inflammation
Graphical abstract
Targeting CYP1B1–Oxylipin–TRPA1 network may represent a novel treatment option for preventing chronic pain in patients.
1. Introduction
Pain is elicited by injury and inflammation. Acute pain serves as a defense mechanism and pain-sensing neurons also play a key role in the initiation of repair processes1,2. In comparison, chronic pain has many different origins, and can be maladaptive and pathologic in nature1. Chronic pain emerges because of unresolved sensitization of neural circuitry, as well as synaptic reorganization2. In general, sensitization is characterized by an enhancement in excitatory transmission and a suppression of inhibitory signaling within pain circuits peripherally and in the central nervous system3. These phenomena can be both biochemically and transcriptionally driven.
Chronic pain is a major health concern as it is estimated to affect more than 1.5 billion people worldwide. In the United States, 30% of the population suffers from some form of chronic pain, with an annual estimated cost of $635 billion3,4. Conventional therapeutics such as opioids, non-steroidal anti-inflammatory drugs, and tumor necrosis factor alpha inhibitors are only marginally effective and are associated with the potential for adverse side effects. Thus, there is a clear and urgent need for the discovery of safer, non-addictive and efficacious treatments for chronic pain.
Transient receptor potential (TRP) ankyrin-1 (TRPA1), vanilloid-1 (TRPV1), and vanilloid-4 (TRPV4), among other TRPs, continue to emerge as key mediators of inflammation and acute and persistent pain5, 6, 7. Peripheral pain-sensing neurons originating from dorsal root ganglia (DRG) express many TRPs, and these neurons contain an array of bioactive lipids such as 9,10-EpOME, 9-HODE, and 5,6-EET. These lipids, and other “oxylipins” (i.e., products of cyclooxygenase, lipoxygenase, or CYP450 enzymes) are generally transient species that function as local signaling molecules. Some oxylipins have been shown to sensitize and/or directly activate TRPA1 or TRPV1 in neurons, promoting substance P and calcitonin gene-related peptide (CGRP) release, as well as to further inflammation by engaging adjacent non-neuronal cells in the inflammatory process8, 9, 10. Some oxylipins may also be inhibitory. However, the degree to which these endogenous TRP ligands are involved in the activation, sensitization, or inhibition of TRP channels under various conditions is not fully understood. In particular, there is much to learn about how such dynamic interactions alter the trajectory of inflammation and affect both acute pain and the development of chronic pain.
The purpose of this study was to identify endogenous mediators of inflammation and pain, focusing on the involvement of TRP channels. Transcriptomic analysis of surgically damaged neurons (spinal nerve ligation; SNL model) revealed time-dependent increases in mRNA for both cytochrome P450 (Cyp) 1b1 and Cyp4b1. These specific CYPs oxidize ω-6/ω-3 fatty acids, including arachidonic acid (AA) and linoleic acid to produce biologically active epoxides and hydroxides including the epoxyeicosatrienoic acids (EETs), hydroxyeicosatetraenoic acids (HETEs), epoxyoctadecenoic acids (EpOMEs), and epoxyeicosatetraenoic acids (EEQs)11,12. 9,10-EpOME, a linoleic acid metabolite produced by CYP2J2, has been shown to sensitize TRPV1, increase CGRP release from sciatic and DRG neurons, and promote mechanical and thermal hypersensitivity in mice11. Because lipids play a key role in the initiation, propagation and chronicity of inflammation and pain, it was hypothesized that CYP1B1-derived lipids would interact with TRP channels, thereby affecting acute and chronic pain. Results show a role for CYP1B1, CYP1B1-derived 8,9-EET and 19,20-EpDPA, and TRPA1 in modulating both acute and chronic pain in mice.
2. Materials and methods
2.1. Reagents and chemicals
Mouse CYP1B1, low reductase bactosomes from Escherichia coli, was obtained from Sekisui XenoTech (Kansas City, KS, USA). Recombinant human CYP1B1 Supersomes derived from insect cells, and an NADPH regenerating system consisting of solution A (26 mmol/L NADP+, 66 mmol/L glucose-6-phosphate and 66 mmol/L magnesium chloride in aqueous solution) and solution B (40 U/mL glucose-6-phosphate dehydrogenase in 5 mmol/L sodium citrate) were obtained from Corning (Glendale, AZ, USA). H89-dihydrochloride, Go6983 (pan-protein kinase C/PKC inhibitor) and GF 109203X (potent PKCα, β1, δ, ε, ζ, and weak PKA inhibitor) were obtained from Tocris (Minneapolis, MN, USA). Fetal bovine serum (FBS), geneticin, trypsin, and penicillin/streptomycin (pen/strep) were purchased from Thermo Fisher Scientific/Life Technologies (Carlsbad, CA, USA). All other chemicals were purchased from Sigma–Aldrich (St Louis, MO, USA) or Cayman Chemical (Ann Arbor, MI, USA). High purity water (Milli-Q water) was prepared using a Barnstead GenPure™ Water Purification system (ThermoFisher, Waltham, MA, USA). All reagents were prepared and stored according to manufacturer's recommendations.
2.2. Animals
Experimental protocols were approved by the University of Utah Institutional Animal Care and Use Committee. C57BL/6J mice (20–25 g; 6–8 weeks) were obtained from Jackson Laboratories. Cyp1b1−/− mice (C57BL/6J background) were obtained from Dr. Bhagavatula Moorthy of the Baylor College of Medicine (Houston, TX, USA), under a material transfer agreement with Dr. Frank J. Gonzalez, of the National Cancer Institute (Bathesda, MD, USA). Cyp1b1−/− mice were maintained as a breeding colony on site. All mice were housed in an AAALAC-approved vivarium under controlled environmental conditions consisting of 12 h light/12 h dark cycles, a temperature of 23–26 °C, and 40%–50% relative humidity. Mice were fed standard lab chow and water ad libitum.
2.3. Spinal nerve ligation (SNL) injury model
Mouse spinal nerve ligation (SNL) was performed according to the procedure described by Kim and Chung13 and Rigaud et al14. Briefly, male C57BL/6J mice were anesthetized with 4% isoflourane and maintained at 2.5% with 0.8 L/min oxygen. Following a 1-cm incision in the midline of the dorsal skin and removal of the left L5 transverse process, the left L4 spinal nerve was exposed and tightly ligated with a 5-0 silk suture (AD Surgical, USA). The incision was closed with 5–0 silk and Vetbond tissue adhesive (3M, USA). Animals were injected intramuscular with a 50/50 mixture of 2% lidocaine and 0.5% bupivacaine after surgery and returned to their cages. Sham controls received the same surgical procedure except that the nerves were not ligated. At the end of each experiment, each mouse was inspected for ligation quality. All mice with unsuccessful ligation and/or conspicuous left paw flexor weakness were removed from data analysis.
2.4. Mouse behavioral/pain tests
Mice were habituated on the Von Frey, hot plate and running wheel apparatuses for 30 min, respectively, for 4 consecutive workdays immediately before the surgery day. The same apparatuses were used by each mouse throughout the experiment. On the surgery day (day 0), baseline Von Frey, hot plate and running wheel activities were assayed in series before surgical operation. After the surgery, on days 1, 3, or 7, mice were evaluated for pain responses and euthanized (with overdose of isoflurane) for tissue collection. Behavior tests were performed by an experimenter blinded to the treatments, as described previously15,16.
For Von Frey tests, animals were acclimated in individual Plexiglas chambers on a wire mesh grid (Bioseb) for 30 min. A total of three sets of stimuli (10 applications/set) were then applied to the plantar surface of hind paws, with at least a 5 s interval between two consecutive applications, and 5 min between two consecutive sets of stimuli. The total number of nocifensive responses (fast withdrawal or flinching/licking/biting of the paw) were counted and compared.
Mice were then placed on a metal hotplate surface maintained at 52 °C to evaluate responses to noxious heat. Response latency for hind paw withdrawal, licking, or jumping were measured as the average from three trials carried out over 5 min intervals. A cut-off of 30 s was used to prevent tissue damage.
Finally, mice were tested on voluntary running wheels (diameter 11 cm, width 5.5 cm) in polycarbonate cages for 30 min. The number of revolutions were recorded by a custom multi-channel digital counter connected to magnetic switches placed adjacent to the wheels. Travel distances were calculated and compared.
2.5. RNA sequencing
Lumbar 3–5 DRGs were harvested on days 0, 1, 3, and 7 following SNL surgery. The ipsilateral (SNL-injured) and contralateral (uninjured) DRGs were separately collected into RLT buffer (RNeasy kit Qiagen), homogenized and stored at −80 °C. DRGs from three mice per time point were pooled and RNA was isolated using the RNeasy Kit (Qiagen) with on column DNaseI digestion. RNA sequencing was performed by the High Throughput Genomics Core Facility at the Huntsman Cancer Institute, University of Utah, as described previously17. Briefly RNA quality was assessed with an Agilent Technologies RNA ScreenTape Assay and library construction was performed using the Illumina TruSeq Stranded mRNA Sample Preparation kit using established protocols. The sequencing libraries (18 pmol/L) were then subject to RNA sequencing on an Illumina HiSeq instrument (HCS v2.0.12 and RTA v1.17.21.3), with a 50-cycle single read sequence run. Data were processed by the University of Utah Bioinformatics core. The data presented in this publication have been deposited in NCBI's Gene Expression Omnibus18 and are accessible through GEO Series accession number (GSE194174).
2.6. Quantitative real-time PCR
T12 to L1 spinal cord, corresponding to the L3–L5 DRG injury, was collected. The ipsilateral side was isolated and frozen at −80 °C before being homogenized and processed for RNA isolation using the PureLink® RNA Mini Kit (ThermoFisher). Total RNA (1 μg), was then converted to cDNA using the Applied Biosystems High-capacity cDNA Reverse Transcription kit (ThermoFisher) and analyzed by quantitative real-time PCR (qPCR) performed on a Life Technologies QuantStudio 6 Flex instrument. TaqMan probes for mouse Cyp1b1 (Mm00487229_m1), mouse Cyp4b1 (Mm00484138_m1), mouse Cyp1a1 (Mm00487218_m1), mouse Cyp1a2 (Mm00487224_m1) and mouse Cyp2j6 (Mm01268197_m1) were used to measure gene expression. Ct values were normalized to Gapdh (Mm99999915_g1), and quantification of mRNA utilized the ΔΔC(T) method. The sequences of the probes are listed in Supporting Information Table S1.
2.7. Preparation of tissue-derived lipids
As above, ipsilateral spinal cord from SNL-injured and sham-operated mice were isolated and frozen at −80 °C. Non-esterified oxylipins, endocannabinoids, and polyunsaturated fatty acids in spinal cord were isolated after protein precipitation using a 1:1 (v/v) methanol/acetonitrile mixture and quantified by ultra-performance liquid chromatography–tandem mass spectrometry (UPLC–MS/MS) by the West Coast Metabolomics Center at UC-Davis, as previously described19. Briefly, 15 μg of spinal cord was homogenized using a GenoGrider 2000 sample homogenizer (SPEX Sample Prep; Metuchen, NJ, USA), together with 5 μL BHT/EDTA (in 1:1 MeOH:water), 5 μL of 1250 nmol/L deuterated oxylipin stock and endocannabinoid surrogates in methanol, 5 μL 1-cyclohexyl ureido, 3-dodecanoic acid (CUDA) and 1-phenyl ureido 3-hexanoic acid (PUHA; 5 μmol/L in 1:1 methanol:acetonitrile) and 185 μL of 1:1 methanol:acetonitrile. The homogenate mixture was then placed at −20 °C for 30 min and centrifuged at 15,000×g for 10 min to remove the protein precipitates. The supernatant was further filtered through 0.2 μm PVDF using 96-well filter plates and collected for subsequent analysis.
Oxylipin/analyte analysis was carried out on an API 6500 QTRAP (Sciex; Redwood City, CA, USA) equipped with a 2.1 mm × 150 mm, 1.7 μm BEH C18 column (Waters, Milford, MA, USA). Analyte detection utilized negative electrospray ionization (–ESI) with multi-reaction monitoring. Quantification was achieved using 7–9 point calibration curves generated using the analyte to internal standard ratio and authentic standards20.
2.8. In vitro metabolism assays
Incubations were carried out in accordance with the enzyme supplier protocols. Briefly, incubation mixtures contained 100 mmol/L potassium phosphate buffer (pH 7.4), 50 μmol/L AA or DHA, and 50 pmol/mL human/mouse CYP1B1 enzyme. The mixture was warmed to 37 °C for 5 min, and the reaction was initiated by the sequential addition of the NADPH regenerating system solutions A and B. Aliquots of 300 μL were withdrawn at 0, 30, 60, 90, 120, 150 and 180 min. Reactions were terminated by mixing with an equal volume of ice-cold acetonitrile containing 0.1 μmol/L internal standard (8,9-EET-d11). The mixture was centrifuged at 12,000 × g at 4 °C for 5 min, and the supernatant was extracted 3 times using 1:1 (v/v) ethyl acetate. The extracts were pooled, dried under nitrogen, and analyzed immediately. Control samples (without enzyme) and blank samples (without substrate) were included for each assay, using phosphate buffer as the substitute.
2.9. Quantification of AA and DHA metabolites
A Thermo LTQ-VELOS Pro interfaced with a Vanquish UPLC–MS/MS system was used. Sample residues were dissolved in 50 μL methanol, and 5 μL was injected onto an Acquity UPLC BEH C18 (1.7 μm, 2.1 mm × 150 mm) column. Analytes were eluted using a gradient of (A) 0.1% acetic acid and (B) 9:1 (v/v) acetonitrile–isopropanol. The gradient was as follows: 0 → 10 min (65% B → 75% B); 10 → 15 min (75% B → 95% B); 15 → 18 min (95% B); 18 → 18.1 min (95% B → 65% B); and 18.1 → 22 min (65% B). The flow rate was 0.4 mL/min, and the auto-sampler compartment and column temperatures were 4 and 35 °C, respectively. MS detection utilized –ESI with selected reaction monitoring (SRM) using the following precursor-to-product transitions: 8,9-EET (319.2 → 155.2), 11,12-EET (319.2 → 167.0), 14,15-EET (319.2 → 219.1), 16,17-EpDPA (343.2 → 274.1), 19,20-EpDPA (343.2 → 285.2), AA (303.2 → 259.2), DHA (327.2 → 283.3). Authentic standards were used as reference materials and deuterated analogues of each analyte were used to normalize peak area and to create calibration curves, as described above.
2.10. Cell culture
Human embryonic kidney 293 cells (HEK-293; ATCC; Rockville, MD, USA) were cultured in DMEM/F12 (Life Technologies; Carlsbad, CA, USA) media, supplemented with 5% FBS and 1 × penicillin–streptomycin. HEK-293 cells stably overexpressing various human TRP channels, or the ultra-sensitive fluorescent protein calcium sensor GCaMP6s were generated as described previously21,22, and cultured with 0.3 mg/mL geneticin in culture media. Cells were maintained in a humidified cell culture incubator with 5% CO2 at 37 °C. In general, all cell assays were performed 24–48 h after plating at 80%–90% confluence. Cells were sub-cultured using trypsin, and 96-well assay plates were pre-coated with 1% gelatin.
2.11. Site-directed mutagenesis and transient transfection
Site-directed mutagenesis and transient transfection were performed as previously described22. Mutation of TRPA1 was carried out using the QuickChange II XL kit (Stratagene, La Jolla, CA, USA), according to manufacturer's instructions. Sequences of all plasmids were verified before use. HEK-GCaMP6s-overexpressing cells were plated at 35,000 cells/well in gelatin-coated 96-well plates at 37 °C. After 24 h the cells were transfected with sequence-verified plasmid DNA (100 ng/well) using Lipofectamine 2000 (Life Technologies) at a 2:1 lipid/DNA in 50 μL Opti-MEM media (Life Technologies) for 4 h, followed by addition of 50 μL recovery media (DMEM/F12 + 5% FBS)/well. Cells were used for calcium assays after overnight incubation at 37 °C.
2.12. Calcium imaging in HEK-293 cells
Calcium flux was determined on an EVOS FL Auto microscope (ThermoFisher, Waltham, MA, USA), as previously described19,21. Briefly, calcium flux in TRP-overexpressing HEK-293 was measured using the Fluo-4 Direct assay kit (Invitrogen, Carlsbad, CA, USA). Cells were plated in gelatin-coated 96-well plates at a density of 30,000 cells/cm2 and incubated at 37 °C. Culture media was removed after 48 h, and each well was replenished with 50 μL loading buffer (1 × Fluo-4 Direct reagent prepared in LHC-9 media). The cells were incubated at 37 °C for 1 h. After removal of the loading buffer, 50 μL calcium buffer (HBSS containing 20 mmol/L HEPES, pH 7.3) containing 1 mmol/L probenecid and 0.75 mmol/L trypan red, with or without pre-treatments, was added to each well. For experiments involving transiently transfected HEK-293 cells, HEK-GCaMP6s cells were treated with 50 μL calcium buffer (HBSS containing 20 mmol/L HEPES, pH 7.3) containing 0.75 mmol/L trypan red, with or without pre-treatments. All agonist treatment solutions were prepared in calcium buffer at 3-fold concentration and added to cells at 37 °C maintained in 5% CO2 with humidification. Images were captured prior to agonist treatment and every 9 s for 72 s following the application of agonists. Changes in fluorescence were quantified using an Image-J-based program created in Matlab. Data were normalized by subtracting baseline fluorescence and then to the maximum attainable change in fluorescence elicited by ionomycin (10 μmol/L) post-treatment.
2.13. Calcium imaging in mouse DRG neurons
For dissociated DRG cells, calcium flux was quantified using constellation pharmacology, originally described by Teichert et al. and applied by others16,23, 24, 25. Briefly, isolated L1–L6 lumbar DRG neurons from CGRP-GFP mice were dissociated with 0.25% trypsin at 37 °C for 20 min and triturated with fire-polished Pasteur pipets of decreasing diameter. Cells/neurons were then plated into the center of a silicone ring (10 mm outer diameter, 4.5 mm inner diameter) pre-attached in a 24-well poly-d-lysine coated plate followed by incubation at 37 °C for 1 h to allow the cells to adhere. After adding 0.7 mL minimal essential medium containing 10% fetal bovine serum, 1 × penicillin/streptomycin, 10 mmol/L HEPES, and 0.4% (w/v) glucose into each well, the plated cells were incubated in a 5% CO2 incubator at 37 °C overnight. Media was removed and the cells were loaded with 4 μmol/L Fura2-AM dye in 0.7 mL MEM at 37 °C for 1 h and equilibrated at room temperature for 0.5 h prior to assay. The cells were then washed three times (0.7 mL each) with observation solution (145 mmol/L NaCl, 5 mmol/L KCl, 2 mmol/L CaCl2, 1 mmol/L MgCl2, 1 mmol/L sodium citrate, 10 mmol/L HEPES, and 10 mmol/L glucose) after removing the MEM solution. The observation solution was removed from each well via a peristaltic pump controlled by a microfluidic system. Then, cells were exposed to each treatment. After 15 s of application, the solution was removed. The cells were then washed 5 times with observation solution and equilibrated for 5 min with observation solution with or without antagonists (e.g., A967079) prior to the next treatment.
The fluorescence ratio (340 nm/380 nm excitation; 510 nm emission) was determined using a fluorescence microscope to reveal the relative level of intracellular calcium in each cell while applying a pre-determined series of pharmacological agents at room temperature. Pharmacological agents were used to differentiate cell types in each experiment included KCl (20, 30, and 40 mmol/L; all excitable cells), allyl-isothiocyanate (100 μmol/L AITC; TRPA1-expressing cells), menthol (400 μmol/L; TRPM8-expressing cells), capsaicin (300 nmol/L; TRPV1-expressing cells), and conotoxin κM-RIIIJ (1 μmol/L; large neuronal cells). Images were acquired in real time at 2 s intervals for the duration of the experiment. In each experiment, more than 1000 cells were imaged with individual cell responses simultaneously recorded. For activation and inhibition assays, cells were treated in series with 19,20-EpDPA (20 μmol/L, 15 s) or A967079 (20 μmol/L, 5 min). For sensitization assays, cells were treated in series with AITC (100 μmol/L, 15 s) or 19,20-EpDPA (0.5 μmol/L, 5 min). At the end of the calcium imaging experiments, cells were incubated with Alexa-Fluor 568 isolectin B4 (IB4, 2.5 μg/mL) for 5 min at room temperature followed by washing (3 times), incubation for an additional 5 min, and 3 additional washes. IB4 staining was captured using a rhodamine filter set. Calcium flux trace data were analyzed using an in-house software program developed in R, with stratification of neurons by sub-class (i.e., CGRP-GFP expression, IB4 staining, responses to AITC, capsaicin, etc.), as described previously.
2.14. Statistical analysis
Results are presented as the mean ± standard deviation (SD) or SEM from a minimum of 3 biological replicates, as specified in the figure legends. A difference between two treatment groups was analyzed using the unpaired, two-tailed Student's t-test. One-way ANOVA or two-way ANOVA with a Bonferroni post hoc test was used for comparisons of >2 treatment groups/treatment conditions. All statistical analyses were performed using GraphPad Prism 8.0.2 (GraphPad Software Inc., La Jolla, CA, USA). A P value < 0.05 was set as the threshold for significance.
3. Results
3.1. SNL-induced chronic pain promoted neuronal Cyp1b1 mRNA expression
Mice subjected to SNL were monitored for the development of mechanical hypersensitivity using the Von Frey test. Responses to stimulation of both hind paws was compared between sham-operated and SNL-injured mice for 7-days post-surgery. The left (ipsilateral) paw of SNL-injured mice was ∼3.6-fold more sensitive to stimulation than the right (contralateral) paw, confirming an SNL-induced chronic pain phenotype (Fig. 1A and B).
Figure 1.
SNL-evoked chronic pain enhanced Cyp1b1 mRNA expression in both DRG neurons and spinal cord. (A, B) Von Frey test confirming the development of mechanical hyperalgesia in SNL-injured versus sham operated control mice. (A) left (ipsilateral) paw (B) right (contralateral) paw. Data were normalized to the threshold at baseline and presented as the mean ± Standard Error of mean (SEM) (n = 12–13). ∗∗∗∗P < 0.0001 versus the sham group, using a two-tailed Student's t-test. (C) MA plot of RNA sequencing results comparing mRNA expression in DRG neurons isolated from sham versus SNL-injured mice 7-days post-injury. Elevated Cyp1b1 and Cyp4b1 mRNA in injured DRG of SNL mice are highlighted. (D) mRNA expression of Cyp1b1 and (E) Cyp4b1 in control and SNL-injured DRG as a function of time. Data are representative of mean transcript levels from pooled DRGs from 3 mice/group. (F, G) quantification of mRNA expression (qPCR) of Cyp1b1 and Cyp4b1 in adjacent (to injury site) spinal cord tissue of sham and SNL-injured mice. Data are presented as the mean ± SD (n = 3). ∗P < 0.05, versus the sham group, using a two-tailed Student's t-test.
DRG mRNA transcripts were compared at days 0, 1, 3, and 7 post-SNL injury using transcriptome analysis. Fig. 1C shows an MA plot of the day 7 transcriptome dataset comparing sham-operated and SNL-injured DRGs, while Fig. 1D and E shows the relative abundance of the Cyp1b1 and 4b1 transcripts as a function of time post-sham and SNL injury. Analysis of the datasets revealed time-dependent increases in the expression of Cyp1b1 and Cyp4b1 mRNA. The expression of Cyp1b1 and Cyp4b1 mRNA in adjacent spinal cord tissue was also quantified using qPCR. Cyp1b1, but not Cyp4b1 was increased ∼2-fold compared to tissue from sham-operated animals (Fig. 1F and G). Analysis of other Cyp epoxygenases demonstrated a reduction in Cyp1a1, but no changes in Cyp1a2 and Cyp2j6 as a function of proximity to injury site (Supporting Information Fig. S1).
3.2. SNL-induced chronic pain paralleled the accumulation of selected oxylipins in neuronal tissue
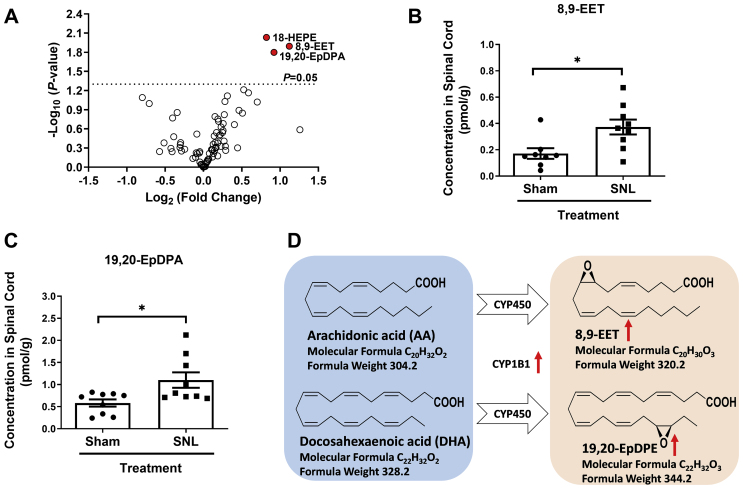
CYP epoxygenases metabolize polyunsaturated fatty acids to epoxides that have a range of biological activities11,12. Accordingly, differences in the abundance of 81 oxylipins and related analytes were compared in spinal cord tissue from control and SNL-injured mice using UPLC–MS/MS. A volcano plot was used to visualize differences in oxylipin abundance between sham-operated control and SNL-injured tissue (Fig. 2A). Elevated concentrations of the oxidized arachidonic acid (AA) and docosahexaenoic acid (DHA) metabolites 8,9-EET and 19,20-EpDPA (Fig. 2B and C) and the non-enzymatic oxidized eicosapentaenoic acid (EPA) metabolite 18-HEPE (Fig. 2A) were detected in spinal cord tissue of SNL-injured mice. There were no significant differences in any of the other analytes/lipids assayed, including those immediately up (i.e., AA and DHA) or downstream of 8,9-EET (i.e., 8,9-DiHET) and 19,20-EpDPA (i.e.,19,20-DiHDPA) (Supporting Information Fig. S2A) and oxylipins derived from the LOX and COX pathways. Significant increases in both the ratio of 8,9-EET/AA and 19,20-EpDPA/DHA were also observed within the spinal cords of SNL-injured mice compared to sham operated mice (Figs. S2B and S2C). These results suggest a possible mechanism in which CYP1B1 produced 8,9-EET and 19,20-EpDPA via epoxidation of AA and DHA, respectively (Fig. 2D). Further, the transcriptomic and targeted lipidomic results indicated a potentially vital role for the CYP1B1-8,9-EET and 19,20-EpDPA network in SNL-induced chronic pain.
Figure 2.
Lipidomics analysis revealed synthesis of 8,9-EET and 19,20-EpDPA during SNL-induced chronic pain. (A) Volcano plot comparing the concentrations of 81 oxylipins in spinal cord tissue collected adjacent to DRGs of sham and SNL-injured mice. (B) Concentration of 8,9-EET and (C) 19,20-EpDPA in spinal cord. (D) Scheme showing CYP450-derived metabolites of AA and DHA, as revealed by oxylipin analysis. Data are presented as mean ± SD (n = 9). ∗P < 0.05 versus the sham group, using a two-tailed Student's t-test.
3.3. Human and mouse CYP1B1 produced 8,9-EET and 19,20-EpDPA in vitro
Next, we sought to validate whether CYP1B1 is implicated in the synthesis of 8,9-EET and 19,20-EpDPA. Using recombinant CYP1B1 enzymes, it was confirmed that 8,9-EET and 19,20-EpDPA, among other related oxylipins, are produced from AA and DHA. For mouse CYP1B1, the primary epoxide metabolites of AA were 14,15-EET, 11,12-EET and 8,9-EET, whereas the primary metabolites of DHA were 19,20-EpDPA and 16,17-EpDPA (Supporting Information Fig. S3A and S3B). Similarly, 14,15-EET, 11,12-EET and 8,9-EET were the metabolites of AA generated by human CYP1B1, and 19,20-EpDPA from DHA (Fig. S3C and S3D). Notably, 14,15-EET, 11,12-EET and 8,9-EET increased rapidly in the human CYP1B1 incubations, peaked after 1 h, and gradually decreased over 3 h. Further analysis showed that these epoxides were further hydrolyzed into the corresponding diol metabolites in the human CYP1B1 incubations which was partially attenuated by a potent inhibitor of soluble epoxide hydrolase (i.e., AUDA), whereas this did not occur with mouse CYP1B1 (Supporting Information Fig. S4). Thus, this phenomenon appears to be due to differences in epoxide hydrolase content between the CYP1B1 insect cell-derived supersomes and CYP1B1 bactosomes from E. coli, as opposed to a species-specific difference in CYP1B1 activity.
3.4. 8,9-EET and 19,20-EpDPA activated human TRPA1 in HEK-293 overexpressing cells
The effects of 8,9-EET and 19,20-EpDPA on the activity of human TRPA1 was evaluated using TRPA1-overexpressing HEK-293 cells and a calcium flux assay. For TRPA1, both 8,9-EET and 19,20-EpDPA stimulated calcium flux in a concentration-dependent manner, which was greater in TRPA1-overexpressing HEK cells than normal HEK-293 cells (Fig. 3A and B). Further, calcium flux caused by 8,9-EET and 19,20-EpDPA treatment at 50 and 100 μmol/L in TRPA1 over-expressing cells was largely attenuated by A967079, a TRPA1 antagonist (Fig. 3A and B). Videos of the representative calcium imaging are included as Movie 1. The EC50 for 8,9-EET and 19,20-EpDPA were 32.7 and 31.4 μmol/L, respectively (Supporting Information Fig. S5A). For comparison, the EC50 for 2,4-DTBP and AITC were 225.3 and 4.1 μmol/L, respectively (Fig. S5B). The selectivity of 8,9-EET and 19,20-EpDPA for TRPA1 activation was also evaluated using multiple human TRP channel over-expressing cells. Neither 8,9-EET nor 19,20-EpDPA, at concentrations up to 50 μmol/L, activated human TRPV1, V3, V4 or M8 in HEK-293 cells (Supporting Information Fig. S6).
Figure 3.
8,9-EET and 19,20-EpDPA activate and sensitize TRPA1 in TRPA1-overexpressing HEK-293 cells. (A) 8,9-EET and (B) 19,20-EpDPA cause calcium flux in a dose-dependent manner, which was inhibited by the TRPA1 antagonist A-967079 (20 μmol/L). ∗P < 0.05, ∗∗P < 0.001, ∗∗∗P < 0.0001, ###P < 0.001, using two-way ANOVA with Bonferroni correction for multiple comparisons. (C, D) Graphs of maximum 2,4-DTBP-evoked calcium flux following sensitization with non-activating concentrations of 8,9-EET and 19,20-EpDPA. TRPA1 overexpressing cells were pretreated with 0.5, 1, 2, or 4 μmol/L 8,9-EET and 19,20-EpDPA for 5 min. Data are normalized to the response of ionomycin (10 μmol/L) and are represented as the mean ± SD (n = 4–6). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ###P < 0.001 using one-way ANOVA with Bonferroni correction for multiple comparisons.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.apsb.2022.09.007
The following is/are the supplementary data related to this article:
Videos showing that the calcium flux induced by 8,9-EET and 19,20-EpDPA was inhibited by the TRPA1 antagonist A-967079 in TRPA1-overexpressing HEK-293 cells.
3.5. Lipids up and downstream of 8,9-EET and 19,20-EpDPA also weakly activated human TRPA1 in HEK-293 overexpressing cells
Lipids up- and downstream of 8,9-EET and 19,20-EpDPA were also tested as TRPA1 agonists. AA, DHA, and the diol metabolites 8,9-DiHET and 19,20-DiHDPA all induced calcium flux in a concentration-dependent manner, albeit to a lesser degree and with slightly lower potency than either 8,9-EET or 19,20-EpDPA (Supporting Information Fig. S7). With all lipids, there was some degree of non-specificity at 100 μmol/L, in that calcium flux was observed in normal HEK-293 cells and inhibition using A967079 was incomplete. Accordingly, all subsequent experiments utilized 50 μmol/L as the maximum concentration.
3.6. 8,9-EET and 19,20-EpDPA also sensitized human TRPA1 in HEK-293 overexpressing cells
Sensitization of TRPA1 by low concentrations of 8,9-EET and 19,20-EpDPA, as opposed to direct activation was also evaluated. At 0.5, 1, 2 and 4 μmol/L, both 8,9-EET and 19,20-EpDPA enhanced 2,4-DTBP-induced calcium flux (Fig. 3C and D, and Supporting Information Fig. S8A and S8B). Interestingly, the lipids did not increase AITC-evoked calcium flux (Fig. S8C and S8D). Of note, neither 8,9-EET nor 19,20-EpDPA at 2 or 4 μmol/L sensitized the TRPV1 response to capsaicin, TRPV3 response to drofenine, TRPV4 response to GSK1016790A, or TRPM8 response to menthol (Supporting Information Fig. S9).
3.7. 8,9-EET and 19,20-EpDPA also selectively activated and sensitized TRPA1 in DRG neurons
TRPA1 is expressed by a sub-population of pain-sensing DRG neurons, and 8,9-EET has been shown to promote and sensitize AITC-induced calcium flux26,27. Application of 19,20-EpDPA (20 μmol/L) was also found to trigger robust calcium flux in AITC-responsive DRG neurons (Fig. 4A), which was inhibited by the antagonist A967079 (20 μmol/L; Fig. 4B). Like human TRPA1, application of 19,20-EpDPA to DRGs at 0.5 μmol/L also enhanced the amplitude of TPRA1-dependent calcium flux (Fig. 4C).
Figure 4.
Effects of 19,20-EpDPA on calcium flux in cultured mouse DRG neurons. (A) Representative neuronal response trace to 19,20-EpDPA treatment. DRG neurons were stimulated 2X with 19,20-EpDPA (20 μmol/L, 15 s), in series following an initial KCl (30 mmol/L) pulse and prior to AITC (100 μmol/L). (B) Representative neuronal response trace to 19,20-EpDPA prior to and immediately after A-967079 treatment (20 μmol/L; 5 min). (C) Representative neuronal response trace illustrating sensitization of TRPA1. DRG neurons were stimulated with AITC 2X (100 μmol/L, 15 s) over a 10-min interval. 19,20-EpDPA (0.5 μmol/L) was applied for 5 min prior to the second application of AITC. (D) Distribution of peptidergic and nonpeptidergic neurons activated by 19,20-EpDPA (n = 659). For panel D, the test agents were KCl (30 mmol/L), AITC (100 μmol/L), menthol (400 μmol/L), and capsaicin (300 nmol/L). Peptidergic and non-peptidergic neurons were differentiated by CGRP-GFP expression vs. IB4 staining, and further classified by responses to AITC, capsaicin, and menthol.
3.8. 19,20-EpDPA preferentially activated TRPA1-expressing peptidergic DRG neurons
Quantification of the specific neuronal sub-types activated by 19,20-EpDPA was also evaluated by relating response data with CGRP-GFP expression and IB4 staining. Based on CGRP expression and IB4 staining, 41.2% of 19,20-EpDPA-responsive neurons were CGRP+, of which the majority (92.9%) were AITC-sensitive. An additional 5.9% of 19,20-EpDPA-responsive neurons were IB4+ or co-labeled for both CGRP+ and IB4+, of which all were AITC-sensitive. Regardless, most 19,20-EpDPA-responsive neurons were AITC-sensitive (Fig. 4D), indicating that 19,20-EpDPA activates peptidergic (CGRP+) and to a lesser degree, nonpeptidergic (IB4+) neurons that express TRPA1.
3.9. Trpa1 and Cyp1b1 are co-expressed in pain-sensing neurons
Results from the calcium imaging-based constellation pharmacology assays and single cell RNA sequencing data were used to classify different neuronal cell types. This approach is an expansion of that described by Giacobassi et al.28. In our dataset and in a large external single cell RNA sequencing dataset29, co-expression of Cyp1b1 and Trpa1 was primarily observed in peptidergic neurons and to a lesser extent in nonpeptidergic neurons (Supporting Information Fig. S10), supporting the idea that CYP1B1-derived oxylipins have the capacity to directly affect TRPA1 in selected neurons, in addition to paracrine mechanisms.
3.10. 8,9-EET and 19,20-EpDPA activated TRPA1 independent of the typical electrophile and non-electrophile agonist sites
Electrophilic chemicals (e.g., AITC) covalently modify cysteine and lysine residues localized on the intracellular N-terminal domain of TRPA130, whereas non-electrophilic compounds such as menthol and 2,4-DTBP interact with S873 and T874 (abbreviated as TRPA1-ST) located in transmembrane 5 of TRPA131. HEK293-GCaMP6s-overexpressing cells were transiently transfected with either human TRPA1 or a TRPA1-S873 A/T874A (TRPA1-ST) mutant and treated with 8,9-EET and 19,20-EpDPA, with or without pre-incubation of the lipids with the electrophile scavenger, glutathione (GSH). As expected, TRPA1 activation by AITC was abolished by GSH (Fig. 5A). In contrast, 8,9-EET and 19,20-EpDPA-induced calcium flux was not affected by GSH. Similarly, cells transiently transfected with the TRPA1-ST mutant exhibited reduced responses to 2,4-DTBP, while the responses to 8,9-EET and 19,20-EpDPA were not affected (Fig. 5B). Videos showing calcium flux are included as Movie 2A and 2B. These data imply that 8,9-EET and 19,20-EpDPA do not appear to activate TRPA1 through either the electrophile or non-electrophile binding sites. However, one cannot fully rule out that binding of highly reactive cysteine residues in TRPA1 occurs, even in the presence of GSH pre-treatment32.
Figure 5.
8,9-EET and 19,20-EpDPA active TRPA1 independent of the traditional electrophilic and non-electrophilic agonist binding sites, potentially by residues in the extracellular pore forming/loop domains. (A, B) Calcium flux in HEK-293 GCaMP6s-overexpressing cells transiently transfected with human TRPA1, a human TRPA1-ST mutant, or a control vector (CV; pcDNA3.1/V5-His/lacZ). Treatments were AITC (150 μmol/L), 2,4-DTBP (250 μmol/L), 19,20-EpDPA (50 μmol/L) and 8,9-EET (50 μmol/L), with or without pre-incubation of the agonist stock solutions in 6.7 mmol/L GSH for 10 min. (C) Activation of TRPA1 by 8,9-EET and 19,20-EpDPA is attenuated by PKA inhibitors. HEK-293 cells over-expressing TRPA1 were pre-treated with the PKA inhibitor H89 (20 μmol/L) or the PKC inhibitors GF 109203X (10 μmol/L) or Go6983 (10 μmol/L) for 24 h. Data are normalized to the response of ionomycin (10 μmol/L) and represented as mean ± SD (n = 6). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, using two-way ANOVA with a Bonferroni post hoc test. (D) Responses of human TRPA1 and mouse Trpa1 elicited by equal concentrations (as indicated) of AITC (150 μmol/L), 2,4-DTBP (250 μmol/L), 8,9-EET (50 μmol/L) and 19,20-EpDPA (50 μmol/L) (n = 6). (E) Calcium flux elicited by 8,9-EET (50 μmol/L) and (F) 19,20-EpDPA (50 μmol/L) in GCaMP6-overexpressing cells transiently transfected with either human TRPA1, mouse Trpa1, or mutant TRPA1 channels wherein the human sequence was mutated to the corresponding residue of mouse Trpa1. Calcium flux responses were normalized by subtracting responses observed for a control vector and then further normalized to the response of human TRPA1-WT. Data are represented as mean ± SEM (n ≥ 5). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, using two-way ANOVA with the Bonferroni post hoc test or one-way ANOVA.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.apsb.2022.09.007
The following is/are the supplementary data related to this article:
Videos revealing that 8,9-EET and 19,20-EpDPA activated TRPA1 independent of the traditional electrophilic agonist binding sites using GSH pretreatment to scavenge/inactivate electrophiles.
Videos demonstrating that 8,9-EET and 19,20-EpDPA activated TRPA1 independent of the non-electrophilic agonist binding site using HEK-293 GCaMP6s-overexpressing cells transiently transfected with human TRPA1 and the human TRPA1-ST mutant.
3.11. Activation of TRPA1 by 8,9-EET and 19,20-EpDPA is modulated by PKA and PKC
Regulation of TRPA1 activity by PKA and PKC has been reported33, but modulation of responses to oxylipins is not well understood. TRPA1 activation by 8,9-EET and 19,20-EpDPA was, to varying degrees, attenuated by pre-treatment of cells with selective PKA and PKC inhibitors, but only the PKA inhibitor H89 significantly reduced calcium flux (Fig. 5C). Videos showing calcium flux are included as Movie 2C. Partial inhibition by GF 109203X likely reflects the low-level inhibition of PKA reported for this agent34.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.apsb.2022.09.007
The following is/are the supplementary data related to this article:
Videos showing that activation of TRPA1 by 8,9-EET and 19,20-EpDPA was attenuated by the PKA inhibitor H89.
3.12. Mouse and human TRPA1 exhibited differential sensitivity to 8,9-EET and 19,20-EpDPA
Examples of species-specific responses to TRPA1 modulators have been reported30,35, and such differences are important to understand with respect to translation of rodent studies to humans. As shown in Fig. 5D, mouse Trpa1 exhibited ∼2–4-fold greater response to AITC, 2,4-DTBP, 8,9-EET and 19,20-EpDPA compared to human TRPA1. Videos showing calcium flux are included as Movie 2D. Sequence alignment of mouse and human TRPA1 revealed differences in amino acids that map to the putative pore-loop region. Mouse Trpa1 residues were subsequently substituted into human TRPA1 to verify which residues may be involved in the species-specific effects of 8,9-EET and 19,20-EpDPA. The mutants were transiently transfected into GCaMP6-over-expressing HEK-293 cells and the effects on 8,9-EET and 19,20-EpDPA were tested in the calcium flux assay. The substitutions H933Y and A939G enhanced TRPA1 activation by 8,9-EET and S921A caused a significant reduction in response to 8,9-EET (Fig. 5E). H933Y also increased responses to 19,20-EpDPA, whereas L894F and S921A significantly reduced activation by 19,20-EpDPA (Fig. 5F). Videos showing calcium flux are included as Movie 2E and 2F. A939G was also implicated in the response to 2,4-DTBP, while L894F, S921A and H933Y mutations failed to trigger alterations in response to AITC and 2,4-DTBP (Supporting Information Fig. S11A and S11B). These results revealed the following insights: 1) S921 is critical for the activation of human TRPA1 by lipids, whereas P897 may limit responses to AITC and 2,4-DTBP; 2) H933 and A939 appear to limit human TRPA1 responses to the lipids, particularly 8,9-EET; and 3) L894 is important for the activation of TRPA1 activation by 19,20-EpDPA, but not 8,9-EET. Finally, S900 appears to be important for the activation of human TRPA1 by 2,4-DTBP, presumably due to its proximity to the ST binding site. Of note, there was limited overlap between residues that affected the responses of TRPA1 to lipids and either AITC or 2,4-DTBP, with the following exception: The A939G mutation enhanced both 8,9-EET and 2,4-DTBP responses, suggesting perhaps a degree of overlap in substrate binding sites or agonist-induced structural dynamics between the lipids and non-electrophilic TRPA1 agonists.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.apsb.2022.09.007
The following is/are the supplementary data related to this article:
Videos highlighting different responses of human and mouse TRPA1 evoked by equal concentrations of AITC, 2,4-DTBP, 8,9-EET and 19,20-EpDPA.
Videos illustrating differences in calcium flux induced by 8,9-EET in GCaMP6-overexpressing cells transiently transfected with either human TRPA1, mouse Trpa1, or mutant channels.
Videos illustrating differences of calcium flux induced by 19,20-EpDPA in GCaMP6-overexpressing cells transiently transfected with either human TRPA1, mouse Trpa1, or mutant channels.
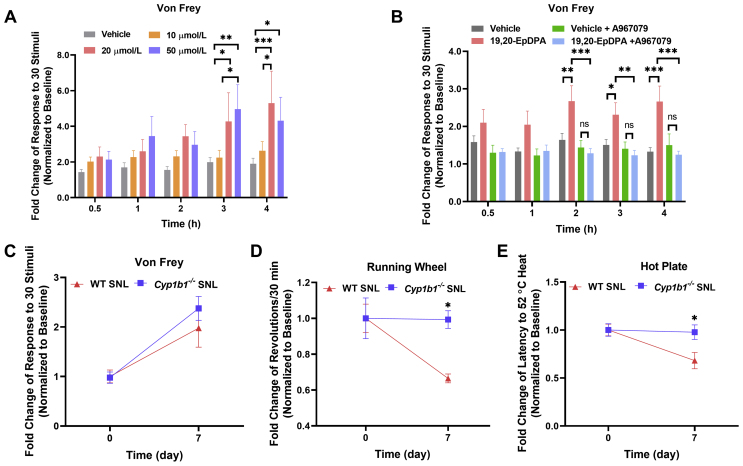
3.13. 19,20-EpDPA evoked mechanical, but not thermal hyperalgesia in mice
Injection of 8,9-EET has been previously shown to cause mechanical hyperalgesia, but not thermal hyperalgesia27. 19,20-EpDPA was injected into the left hind paw of C57BL/6J mice and pain was assessed using the Von Frey and hot plate tests to measure mechanical and thermal responses up to 4 h post-injection. Like 8,9-EET, 19,20-EpDPA dose-dependently increased sensitivity to mechanical stimulation, but not thermal stimulation, over time (Fig. 6A and Supporting Information Fig. S12A). Consistent with a role for TRPA1 in mediating these effects, pretreatment with the TRPA1 antagonist A967079 (dosed i.p. at 40 mg/kg, using an 8 mg/mL stock in MCT oil) prevented mechanical hypersensitivity (Fig. 6B and Fig. S12B).
Figure 6.
19,20-EpDPA caused mechanical pain in vivo and Cyp1b1 knockout prevented the development of pain hypersensitivity in SNL-injured mice. (A) Hind paw injection of 19,20-EpDPA triggered time- and dose-dependent mechanical. Male C57BL/6J mice received an intraplantar injection of 15 μL vehicle (DMSO/saline 0.5%, v/v) or 19,20-EpDPA (10, 20, or 50 μmol/L). (B) Attenuation of 19,20-EpDPA-induced mechanical pain by the TRPA1 antagonist A967079. Male C57BL/6J mice received an intraperitoneal injection of 150 μL vehicle (DMSO/MCT oil 1%, v/v) or A967079 (40 mg/kg, 8 mg/mL in MCT oil) 30 min prior to intraplantar injection of 15 μL vehicle (DMSO/saline 0.2%, v/v) or 20 μmol/L 19,20-EpDPA. Data were normalized to the threshold at baseline. Data represents the mean ± SEM (n = 9–12). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, using two-way ANOVA with the Bonferroni post hoc test. (C) Mechanical hypersensitivity (Von Frey), (D) voluntary activity (running wheel) and (E) thermal (hot plate) hypersensitivity in wild-type (red) versusCyp1b1-deficient mice (blue) before and 7 days after SNL-injury. Data were normalized to the threshold at baseline. Data represents the mean ± SEM (n = 4–6). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ns = not significant, using two-way ANOVA with the Bonferroni post hoc test.
3.14. Cyp1b1-defecient mice exhibited reduced hyperalgesia following SNL injury
The role of CYP1B1, and by extension CYP1B1-derived 8,9-EET and 19,20-EpDPA in SNL-induced chronic pain was assessed. SNL-injured Cyp1b1−/− mice exhibited similar responses to Von Frey (mechanical) stimulation (Fig. 6C) compared to C57BL/6J mice 7-days post-surgery. However, Cyp1b1-deficient mice exhibited significantly greater voluntary activity and fewer responses to thermal stimulation compared to SNL-injured C57BL/6J mice 7-days post-surgery (Fig. 6D and E). Together, these data suggest a role for CYP1B1-derived 8,9-EET and 19,20-EpDPA in promoting acute and chronic pain.
4. Discussion
Acute and chronic pain are complex and distinct phenotypes with variable etiology. Many forms of injury cause pain, and depending upon the extent of the injury, varying levels of inflammation and injury repair occur over time. In some instances, improper or inadequate resolution and repair can result in dysfunction at the tissue and/or cellular levels, leading to loss of function. Additionally, neuronal changes can develop which can manifest as chronic and occasionally debilitating pain. Many factors influence the outcome of injury, and here we have evaluated the contributions of a selected CYP epoxygenase, CYP1B1 and endogenous agonists of the TRPA1 ion channel produced by this enzyme as mediators of pain. The roles of CYP enzymes and TRP channels in inflammation are widely studied, but precisely how they integrate to shape the development of chronic pain phenotypes as a function of time is not fully understood.
CYP1B1 is an aryl-hydrocarbon receptor (AhR)-regulated gene and is commonly induced by cell stressors, pro-inflammatory and damaging agents. An example is when lung epithelial cells are exposed to AhR ligands in pneumotoxic environmental pollution or cigarette smoke36. Previous reports have also documented a role for CYP1B1 in estrogen metabolism and increased risk for breast cancer due to the production of reactive quinones that cause DNA damage37. However, there have been limited studies regarding the roles of CYP1B1 and CYP1B1-derived lipid epoxides in pain.
CYP1B1 is expressed in mouse brain, human neurons, and astrocytes38, which suggests that CYP1B1 may have crucial functions in neuronal tissues, in addition to its canonical roles in xenobiotic metabolism elsewhere in the body (e.g., liver and lung). Moreover, AA is a substrate for CYP1B1 and several other CYP enzymes39.
In this study, increased expression of Cyp1b1 mRNA was observed in both SNL-injured DRG and adjacent spinal cord tissue (Fig. 1). Expression was time-dependent and paralleled the development of pain hypersensitivity, quantified using the Von Frey test. Concomitantly, increased concentrations of the CYP1B1-generated and AA- and DHA-derived epoxide metabolites 8,9-EET and 19,20-EpDPA were enriched in the spinal cords of SNL-injured mice 7-days post-surgery (Fig. 2B and C). Interestingly, substantive differences in the concentrations of other oxylipins that are produced by other CYP epoxygenases, such as 9,10- and 12,13-EpOME (CYP2C or CYP2J enzymes), which have been reported to be TRPV1 agonists and to promote inflammatory hyperalgesia40, were not observed. Additionally, oxylipins derived from the LOX and COX pathways, including prostaglandins, thromboxanes and leukotrienes, which have previously been identified as modulators of pain signaling41, were seemingly not affected in the spinal cords of SNL-injured mice. These results suggest a potentially important and specific role for CYP1B1, 8,9-EET and 19,20-EpDPA in SNL-injury and associated pain.
TRPA1 is a polymodal receptor predominantly expressed by peptidergic pain-sensing neurons. TRPA1 is an established pain sensor and mediator of inflammation triggered by an array of irritating and noxious stimuli including electrophilic agents, and for example, urushiol, and propofol34,42. TRPA1 activation downstream of metabotropic receptors is also known to elicit pain (nociceptive, inflammatory, and neuropathic), itch, and airway hypersensitivity43, 44, 45, making it an attractive therapeutic target. However, analgesics targeting TRPA1 have not been advanced to market.
Here, it was found that 8,9-EET and 19,20-EpDPA were capable of sensitizing and directly activating TRPA1 in both human TRPA1 over-expressing HEK-293 cells and cultured mouse DRG neurons (Figure 3, Figure 4). Electrophiles activate TRPA1 through covalent modification of cysteine (C621, C641, and C665) and lysine (K710) residues (C622, C642, C666, K712 for mouse Trpa1) localized within the N-terminal domain of human TRPA130,46. Non-electrophilic TRPA1 agonists involve residues on transmembrane segment 5 (S873 and T874), among others (S876 and T877 for mouse Trpa1)42,47,48. In contrast, 8,9-EET and 19,20-EpDPA appear to activate TRPA1 via a mechanism that involves residues within the pore-loop and adjacent extracellular region of transmembrane helices 5 and 6 (Fig. 5E and F). Specifically, based on species-specific responses to the oxylipins, it appears that L894 at the extracellular termination of transmembrane helix 5, S921 on the intramembrane pore-forming domain, as well as S933 and A939 in the extracellular domain between the pore and S6, are involved in TRPA1 activation by 8,9-EET and 19,20-EpDPA. Interestingly, AITC, 2,4-DTBP and 8,9-EET and 19,20-EpDPA have distinct characteristics with respect to the effects of mutations to residues that vary across species suggesting unique interaction/binding sites for these agonists and species-specific responses of TRPA1 in general (Fig. 5E, F and Fig. S11). Accordingly, the potential of targeting these alternative ligand binding sites for oxylipins with novel TRPA1 inhibitors may exist, while species-specific differences in TRPA1 responses to TRPA1 agonists in general emphasizes the need to better understand the role of TRPA1 in pain through clinical studies.
The identification of 8,9-EET and 19,20-EpDPA as TRPA1 sensitizers and agonists specifically provides new insight into mechanisms that may influence the development of chronic pain phenotypes. In behavioral experiments, the nocifensive responses to AITC began immediately upon injection, and subsided by 30 min, indicating a fast onset of transient pain behavior30. In contrast, 19,20-EpDPA evoked a protracted mechanical pain response with a slower onset, occurring 2–4 h after injection (Fig. 6A and B). As per a prior report, 8,9-EET also significantly reduced the threshold for mechanical pain 1–2 h after injection27. This discrepancy may be due to differences in activation kinetics, agonist stability, and/or tissue penetration among other factors. AITC is reactive and likely saturates TRPA1 in short time, while also being scavenged by GSH and other nucleophiles. However, the 8,9-EET and 19,20-EpDPA may be more selective for TRPA1 in that they do not indiscriminately react with cellular nucleophiles, promote concentration-dependent effects on TRPA1 (sensitization in addition to direct activation), and are part of a family of related lipids that also modulate TRPA1. Hence, activation of TRPA1 by lipids can be transient, reversing with time as a function of concentration. Further, the ability of AA, DHA, and the diol metabolites of 8,9-EET and 19,20-EpDPA to activate TRPA1 may have specific effects in the context of TRPA1 activity and the dynamics of inflammation and pain that are unique to the proposed CYP1B1–oxylipin–TRPA1 paradigm, and relevant to perhaps modulation of inflammation and pain proximal to injury to prevent chronic pain from developing.
With respect to pain causation, 8,9-EET and 19,20-EpDPA elicited pharmacological effects that would be predicted for a TRPA1 agonist; they promoted pain when injected, which was attenuated by a TRPA1 antagonist. However, a bolus dose of lipid is not necessarily relevant to pain caused by injury. The fact that Cyp1b1 knockout prevented the development of pain hypersensitivity in SNL-injured mice raises the distinct possibility that low-level TRPA1 activation, elicited in part by sensitization of TRPA1 to other endogenous agonists produced following injury (e.g., ROS, lipid-derived α,β-unsaturated aldehydes, etc.), is how these molecules affect chronic pain phenotypes. Accordingly, and as above, inhibition of the CYP1B1–oxylipin–TRPA1 axis may have therapeutic potential, particularly proximal to injury and in people suffering from chronic pain that is resistant to other mainstream therapies.
Finally, CYP1B1 is a polymorphic enzyme. There are multiple gain and loss-of-function variants that have been associated with susceptibility to different types of cancer, as well as glaucoma49, 50, 51, 52. In general, the hypothesis is that altered catalytic processing of exogenous chemicals and endogenous signaling processes underlie these associations. Accordingly, it is possible that differences in CYP1B1 as well as TRPA1 genetics may have an impact on the susceptibility of some people to inflammation and pain following injury, which may influence the frequency at which certain injuries produce chronic pain. This latter idea is particularly intriguing and clinical genotype:phenotype association studies could help reveal the potential (or lack thereof) of inhibiting the CYP1B1–oxylipin–TRPA1nexus for therapeutic purposes.
5. Conclusions
This study shows an enrichment of Cyp1b1 mRNA and the CYP1B1-generated oxylipins 8,9-EET and 19,20-EpDPA in mice subjected to SNL injury. In vitro, 8,9-EET and 19,20-EpDPA preferentially activated TRPA1 independent of known TRPA1 agonist binding sites, raising the possibility of developing selective TRPA1 antagonists for lipid activators. Injection of mice with 19,20-EpDPA support a role for this lipid in triggering acute pain, and a reduction in pain-related outcomes in SNL-injured Cyp1b1-deficient mice indicates a role for CYP1B1-derived 8,9-EET and 19,20-EpDPA in establishing chronic pain phenotypes following nerve injury. Accordingly, targeting the CYP1B1–oxylipin–TRPA1 network may represent a novel option for preventing pain arising from certain types of injury.
Acknowledgments
The authors acknowledge the West Coast Metabolomics Center and Drs. John Newman, PhD and Kamil Borkowski, PhD for carrying out the oxylipin analyses. This work was funded by the Department of Defense [W81XWH-17-1-0413, USA].
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2022.09.007.
Author contributions
Lili Sun and Christopher A. Reilly designed the research; Lili Sun, Jie Zhang, Changshan Niu, Ronald W. Hughen, and Markel Walter performed animal behavior tests; Changshan Niu performed calcium imaging in mouse DRG neurons; John G. Lamb performed site-directed mutagenesis; Lili Sun, Changshan Niu, Cassandra E. Deering–Rice, Katherine Rose, Kevin M. Chase, and Marysol Almestica-Roberts analyzed data. Lili Sun, Jie Zhang, Changshan Niu, Cassandra E. Deering–Rice, Eric W. Schmidt, Alan R. Light, Baldomero M. Olivera, and Christopher A. Reilly wrote the manuscript.
Conflicts of interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Scholz J., Woolf C.J. Can we conquer pain?. Nat Neurosci. 2002;5:1062–1067. doi: 10.1038/nn942. [DOI] [PubMed] [Google Scholar]
- 2.Ji R.R., Nackley A., Huh Y., Terrando N., Maixner W. Neuroinflammation and central sensitization in chronic and widespread pain. Anesthesiology. 2018;129:343–366. doi: 10.1097/ALN.0000000000002130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ji R.R., Donnelly C.R., Nedergaard M. Astrocytes in chronic pain and itch. Nat Rev Neurosci. 2019;20:667–685. doi: 10.1038/s41583-019-0218-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rwt Gereau, Sluka K.A., Maixner W., Savage S.R., Price T.J., Murinson B.B., et al. A pain research agenda for the 21st century. J Pain. 2014;15:1203–1214. doi: 10.1016/j.jpain.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.da Costa D.S.M., Meotti F.C., Andrade E.L., Leal P.C., Motta E.M., Calixto J.B. The involvement of the transient receptor potential A1 (TRPA1) in the maintenance of mechanical and cold hyperalgesia in persistent inflammation. Pain. 2010;148:431–437. doi: 10.1016/j.pain.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Brandt M.R., Beyer C.E., Stahl S.M. TRPV1 Antagonists and chronic pain: beyond thermal perception. Pharmaceuticals (Basel) 2012;5:114–132. doi: 10.3390/ph5020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alessandri-Haber N., Dina O.A., Joseph E.K., Reichling D.B., Levine J.D. Interaction of transient receptor potential vanilloid 4, integrin, and SRC tyrosine kinase in mechanical hyperalgesia. J Neurosci. 2008;28:1046–1057. doi: 10.1523/JNEUROSCI.4497-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Premkumar L.S. Targeting TRPV1 as an alternative approach to narcotic analgesics to treat chronic pain conditions. AAPS J. 2010;12:361–370. doi: 10.1208/s12248-010-9196-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Julius D. TRP channels and pain. Annu Rev Cell Dev Biol. 2013;29:355–384. doi: 10.1146/annurev-cellbio-101011-155833. [DOI] [PubMed] [Google Scholar]
- 10.Chiu I.M., von Hehn C.A., Woolf C.J. Neurogenic inflammation and the peripheral nervous system in host defense and immunopathology. Nat Neurosci. 2012;15:1063–1067. doi: 10.1038/nn.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sisignano M., Angioni C., Park C.K., Meyer Dos Santos S., Jordan H., Kuzikov M., et al. Targeting CYP2J to reduce paclitaxel-induced peripheral neuropathic pain. Proc Natl Acad Sci U S A. 2016;113:12544–12549. doi: 10.1073/pnas.1613246113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panigrahy D., Kaipainen A., Greene E.R., Huang S. Cytochrome P450-derived eicosanoids: the neglected pathway in cancer. Cancer Metastasis Rev. 2010;29:723–735. doi: 10.1007/s10555-010-9264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho Kim S., Mo Chung J. An experimental model for peripheral neuropathy produced by segmental spinal nerve ligation in the rat. Pain. 1992;50:355–363. doi: 10.1016/0304-3959(92)90041-9. [DOI] [PubMed] [Google Scholar]
- 14.Rigaud M., Gemes G., Barabas M.E., Chernoff D.I., Abram S.E., Stucky C.L., et al. Species and strain differences in rodent sciatic nerve anatomy: implications for studies of neuropathic pain. Pain. 2008;136:188–201. doi: 10.1016/j.pain.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deuis J.R., Dvorakova L.S., Vetter I. Methods used to evaluate pain behaviors in rodents. Front Mol Neurosci. 2017;10:284. doi: 10.3389/fnmol.2017.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niu C., Leavitt L.S., Lin Z., Paguigan N.D., Sun L., Zhang J., et al. Neuroactive type-A γ-aminobutyric acid receptor allosteric modulator steroids from the hypobranchial gland of marine mollusk. Conus geographus. J Med Chem. 2021;64:7033–7043. doi: 10.1021/acs.jmedchem.1c00562. [DOI] [PubMed] [Google Scholar]
- 17.Deering-Rice C.E., Stockmann C., Romero E.G., Lu Z., Shapiro D., Stone B.L., et al. Characterization of transient receptor potential vanilloid-1 (TRPV1) variant activation by coal fly ash particles and associations with altered transient receptor potential ankyrin-1 (TRPA1) expression and asthma. J Biol Chem. 2016;291:24866–24879. doi: 10.1074/jbc.M116.746156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ron E., Michael D., Alex E.L. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen N.D., Memon T.A., Burrell K.L., Almestica-Roberts M., Rapp E., Sun L., et al. Transient receptor potential ankyrin-1 and vanilloid-3 differentially regulate endoplasmic reticulum stress and cytotoxicity in human lung epithelial cells after pneumotoxic wood smoke particle exposure. Mol Pharmacol. 2020;98:586–597. doi: 10.1124/molpharm.120.000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pedersen T.L., Newman J.W. In: Clinical metabolomics: methods and protocols. Giera M., editor. Springer New York; New York, NY: 2018. Establishing and performing targeted multi-residue analysis for lipid mediators and fatty acids in small clinical plasma samples; pp. 175–212. [DOI] [PubMed] [Google Scholar]
- 21.Deering-Rice C.E., Johansen M.E., Roberts J.K., Thomas K.C., Romero E.G., Lee J., et al. Transient receptor potential vanilloid-1 (TRPV1) is a mediator of lung toxicity for coal fly ash particulate material. Mol Pharmacol. 2012;81:411–419. doi: 10.1124/mol.111.076067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamb J.G., Romero E.G., Lu Z., Marcus S.K., Peterson H.C., Veranth J.M., et al. Activation of human transient receptor potential melastatin-8 (TRPM8) by calcium-rich particulate materials and effects on human lung cells. Mol Pharmacol. 2017;92:653–664. doi: 10.1124/mol.117.109959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teichert R.W., Schmidt E.W., Olivera B.M. Constellation pharmacology: a new paradigm for drug discovery. Annu Rev Pharmacol Toxicol. 2015;55:573–589. doi: 10.1146/annurev-pharmtox-010814-124551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teichert R.W., Memon T., Aman J.W., Olivera B.M. Using constellation pharmacology to define comprehensively a somatosensory neuronal subclass. Proc Natl Acad Sci U S A. 2014;111:2319–2324. doi: 10.1073/pnas.1324019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Curtice K.J., Leavitt L.S., Chase K., Raghuraman S., Horvath M.P., Olivera B.M., et al. Classifying neuronal subclasses of the cerebellum through constellation pharmacology. J Neurophysiol. 2016;115:1031–1042. doi: 10.1152/jn.00894.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Logu F., Nassini R., Materazzi S., Carvalho Goncalves M., Nosi D., Rossi Degl'Innocenti D., et al. Schwann cell TRPA1 mediates neuroinflammation that sustains macrophage-dependent neuropathic pain in mice. Nat Commun. 2017;8:1887. doi: 10.1038/s41467-017-01739-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brenneis C., Sisignano M., Coste O., Altenrath K., Fischer M.J., Angioni C., et al. Soluble epoxide hydrolase limits mechanical hyperalgesia during inflammation. Mol Pain. 2011;7:78. doi: 10.1186/1744-8069-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giacobassi M.J., Leavitt L.S., Raghuraman S., Alluri R., Chase K., Finol-Urdaneta R.K., et al. An integrative approach to the facile functional classification of dorsal root ganglion neuronal subclasses. Proc Natl Acad Sci U S A. 2020;117:5494–5501. doi: 10.1073/pnas.1911382117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma N., Flaherty K., Lezgiyeva K., Wagner D.E., Klein A.M., Ginty D.D. The emergence of transcriptional identity in somatosensory neurons. Nature. 2020;577:392–398. doi: 10.1038/s41586-019-1900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu C., Reese R., Vu S., Rouge L., Shields S.D., Kakiuchi-Kiyota S., et al. A non-covalent ligand reveals biased agonism of the TRPA1 ion channel. Neuron. 2021;109:273–284.e4. doi: 10.1016/j.neuron.2020.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Memon T., Yarishkin O., Reilly C.A., Krizaj D., Olivera B.M., Teichert R.W. trans-Anethole of fennel oil is a selective and nonelectrophilic agonist of the trpa1 ion channel. Mol Pharmacol. 2019;95:433–441. doi: 10.1124/mol.118.114561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bahia P.K., Parks T.A., Stanford K.R., Mitchell D.A., Varma S., Stevens SM Jr, et al. The exceptionally high reactivity of Cys 621 is critical for electrophilic activation of the sensory nerve ion channel TRPA1. J Gen Physiol. 2016;147:451–465. doi: 10.1085/jgp.201611581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Talavera K., Startek J.B., Alvarez-Collazo J., Boonen B., Alpizar Y.A., Sanchez A., et al. Mammalian transient receptor potential TRPA1 channels: from structure to disease. Physiol Rev. 2020;100:725–803. doi: 10.1152/physrev.00005.2019. [DOI] [PubMed] [Google Scholar]
- 34.Liu B., Escalera J., Balakrishna S., Fan L., Caceres A.I., Robinson E., et al. TRPA1 controls inflammation and pruritogen responses in allergic contact dermatitis. FASEB J. 2013;27:3549–3563. doi: 10.1096/fj.13-229948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chernov-Rogan T., Gianti E., Liu C., Villemure E., Cridland A.P., Hu X., et al. TRPA1 modulation by piperidine carboxamides suggests an evolutionarily conserved binding site and gating mechanism. Proc Natl Acad Sci U S A. 2019;116:26008–26019. doi: 10.1073/pnas.1913929116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaligama S., Patel V.S., Wang P., Sallam A., Harding J., Kelley M., et al. Radical containing combustion derived particulate matter enhance pulmonary Th17 inflammation via the aryl hydrocarbon receptor. Part Fibre Toxicol. 2018;15:20. doi: 10.1186/s12989-018-0255-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu C., Miyajima T., Melangath G., Miyai T., Vasanth S., Deshpande N., et al. Ultraviolet A light induces DNA damage and estrogen–DNA adducts in Fuchs endothelial corneal dystrophy causing females to be more affected. Proc Natl Acad Sci U S A. 2020;117:573–583. doi: 10.1073/pnas.1912546116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh P., Dutta S.R., Song C.Y., Oh S., Gonzalez F.J., Malik K.U. Brain testosterone-CYP1B1 (cytochrome P450 1B1) generated metabolite 6β-hydroxytestosterone promotes neurogenic hypertension and inflammation. Hypertension. 2020;76:1006–1018. doi: 10.1161/HYPERTENSIONAHA.120.15567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amirmokhtari N., Foresi B.D., Dewan S.S., Bouhenni R.A., Smith M.A. Absence of cytochrome P450-1b1 increases susceptibility of pressure-induced axonopathy in the murine retinal projection. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.636321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patwardhan A.M., Akopian A.N., Ruparel N.B., Diogenes A., Weintraub S.T., Uhlson C., et al. Heat generates oxidized linoleic acid metabolites that activate TRPV1 and produce pain in rodents. J Clin Invest. 2010;120:1617–1626. doi: 10.1172/JCI41678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baral P., Udit S., Chiu I.M. Pain and immunity: implications for host defence. Nat Rev Immunol. 2019;19:433–447. doi: 10.1038/s41577-019-0147-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ton H.T., Phan T.X., Abramyan A.M., Shi L., Ahern G.P. Identification of a putative binding site critical for general anesthetic activation of TRPA1. Proc Natl Acad Sci U S A. 2017;114:3762–3767. doi: 10.1073/pnas.1618144114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caceres A.I., Brackmann M., Elia M.D., Bessac B.F., del Camino D., D'Amours M., et al. A sensory neuronal ion channel essential for airway inflammation and hyperreactivity in asthma. Proc Natl Acad Sci U S A. 2009;106:9099–9104. doi: 10.1073/pnas.0900591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J., Joshi S.K., DiDomenico S., Perner R.J., Mikusa J.P., Gauvin D.M., et al. Selective blockade of TRPA1 channel attenuates pathological pain without altering noxious cold sensation or body temperature regulation. Pain. 2011;152:1165–1172. doi: 10.1016/j.pain.2011.01.049. [DOI] [PubMed] [Google Scholar]
- 45.Wilson S.R., Nelson A.M., Batia L., Morita T., Estandian D., Owens D.M., et al. The ion channel TRPA1 is required for chronic itch. J Neurosci. 2013;33:9283–9294. doi: 10.1523/JNEUROSCI.5318-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao J., Lin King J.V., Paulsen C.E., Cheng Y., Julius D. Irritant-evoked activation and calcium modulation of the TRPA1 receptor. Nature. 2020;585:141–145. doi: 10.1038/s41586-020-2480-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ton H.T., Phan T.X., Ahern G.P. Inhibition of ligand-gated TRPA1 by general anesthetics. Mol Pharmacol. 2020;98:185–191. doi: 10.1124/mol.119.118851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woll K.A., Skinner K.A., Gianti E., Bhanu N.V., Garcia B.A., Carnevale V., et al. Sites contributing to TRPA1 activation by the anesthetic propofol identified by photoaffinity labeling. Biophys J. 2017;113:2168–2172. doi: 10.1016/j.bpj.2017.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aklillu E., Oscarson M., Hidestrand M., Leidvik B., Otter C., Ingelman-Sundberg M. Functional analysis of six different polymorphic CYP1B1 enzyme variants found in an Ethiopian population. Mol Pharmacol. 2002;61:586–594. doi: 10.1124/mol.61.3.586. [DOI] [PubMed] [Google Scholar]
- 50.Banerjee A., Chakraborty S., Chakraborty A., Chakrabarti S., Ray K. Functional and structural analyses of CYP1B1 variants linked to congenital and adult-onset glaucoma to investigate the molecular basis of these diseases. PLoS One. 2016;11 doi: 10.1371/journal.pone.0156252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lao X., Qin X., Peng Q., Chen Z., Lu Y., Liu Y., et al. Association of CYP1B1 Leu432Val polymorphism and lung cancer risk: an updated meta-analysis. Lung. 2014;192:739–748. doi: 10.1007/s00408-014-9618-1. [DOI] [PubMed] [Google Scholar]
- 52.Zhang H., Li L., Xu Y. CYP1B1 polymorphisms and susceptibility to prostate cancer: a meta-analysis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0068634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Videos showing that the calcium flux induced by 8,9-EET and 19,20-EpDPA was inhibited by the TRPA1 antagonist A-967079 in TRPA1-overexpressing HEK-293 cells.
Videos revealing that 8,9-EET and 19,20-EpDPA activated TRPA1 independent of the traditional electrophilic agonist binding sites using GSH pretreatment to scavenge/inactivate electrophiles.
Videos demonstrating that 8,9-EET and 19,20-EpDPA activated TRPA1 independent of the non-electrophilic agonist binding site using HEK-293 GCaMP6s-overexpressing cells transiently transfected with human TRPA1 and the human TRPA1-ST mutant.
Videos showing that activation of TRPA1 by 8,9-EET and 19,20-EpDPA was attenuated by the PKA inhibitor H89.
Videos highlighting different responses of human and mouse TRPA1 evoked by equal concentrations of AITC, 2,4-DTBP, 8,9-EET and 19,20-EpDPA.
Videos illustrating differences in calcium flux induced by 8,9-EET in GCaMP6-overexpressing cells transiently transfected with either human TRPA1, mouse Trpa1, or mutant channels.
Videos illustrating differences of calcium flux induced by 19,20-EpDPA in GCaMP6-overexpressing cells transiently transfected with either human TRPA1, mouse Trpa1, or mutant channels.