Abstract
Cardiac-resident macrophages (CRMs) play important roles in homeostasis, cardiac function, and remodeling. Although CRMs play critical roles in cardiac regeneration of neonatal mice, their roles are yet to be fully elucidated. Therefore, this study aimed to investigate the dynamic changes of CRMs during cardiac ontogeny and analyze the phenotypic and functional properties of CRMs in the promotion of cardiac regeneration. During mouse cardiac ontogeny, four CRM subsets exist successively: CX3CR1+CCR2–Ly6C–MHCII– (MP1), CX3CR1lowCCR2lowLy6C–MHCII– (MP2), CX3CR1–CCR2+Ly6C+MHCII– (MP3), and CX3CR1+CCR2–Ly6C–MHCII+ (MP4). MP1 cluster has different derivations (yolk sac, fetal liver, and bone marrow) and multiple functions population. Embryonic and neonatal-derived-MP1 directly promoted cardiomyocyte proliferation through Jagged-1–Notch1 axis and significantly ameliorated cardiac injury following myocardial infarction. MP2/3 subsets could survive throughout adulthood. MP4, the main population in adult mouse hearts, contributed to inflammation. During ontogeny, MP1 can convert into MP4 triggered by changes in the cellular redox state. These findings delineate the evolutionary dynamics of CRMs under physiological conditions and found direct evidence that embryonic and neonatal-derived CRMs regulate cardiomyocyte proliferation. Our findings also shed light on cardiac repair following injury.
Key words: Cardiac resident macrophages, Cardiac regeneration, Cardiac injury, Cardiomyocyte proliferation, Myocardial infarction
Graphical abstract
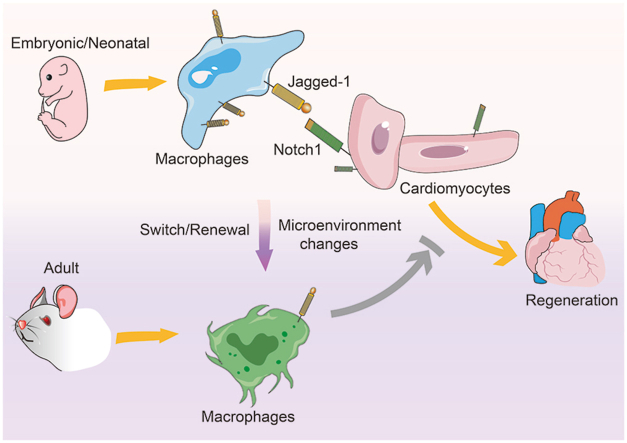
The embryonic and neonatal-derived cardiac-resident macrophages (CRMs) facilitated cardiomyocyte proliferation through Jagged-1–Notch1 axis. After birth, microenvironment changes caused switch or renewal of CRMs, which resulted in the loss of their proliferative potential.
1. Introduction
Resident macrophages are found in all tissues, where they maintain tissue integrity and local homeostasis by defending the organism against pathogens, phagocytizing senescent or dead cells, and clearing up the matrix debris. It was believed that tissue macrophages originate from bone marrow monocytes. Recent evidence, however, suggests that tissue-resident macrophages are established prenatally and originate from the yolk sac and fetal liver progenitors1, 2, 3. In some tissues, these cells can massively expand through local proliferation in response to challenge4 and they can extensively self-renewal without loss of differentiated function. At a steady-state, embryonic-derived macrophage in some organs persist throughout life, and they are exclusively replenished through in situ proliferation, independent of monocyte input. However, some tissue macrophages are notable exceptions, such as intestinal and dermal macrophages, which rely on monocyte input to renew1,5,6.
As an essential component of the heart, cardiac-resident macrophages (CRMs) almost account for 5%–10% of the non-cardiomyocytes7 and are critical for cardiac development, homeostasis, and cardiac healing. For example, yolk sac-derived CRMs facilitate the remodeling of the primitive coronary plexus and are important for the maturation of coronary8; CRMs contribute to efficient conduction of electrical impulses in cardiomyocytes through connexin-43 containing gap junctions9,10. Recently it was established that CRMs expand in neonatal cardiac tissue, and depletion of CRMs abolishes the scarless repair of the heart following cardiac injury. However, similar phenomena are not observed in adult mice11, 12, 13, which suggests that CRMs are associated with the transient capacity for cardiac regeneration. CRMs represent a heterogeneous population with diverse phenotypes, functions, and distinct renewal processes14,15. Although it has been indicated that there are four CRM subsets in the heart of adult mice5,16, it is not clear how the CRMs evolve during the ontogeny in mice. Several other questions, such as the role of CRMs in promoting cardiomyocyte proliferation, the specific phenotype and function of CRMs contributing to cardiac regeneration, and the driving force behind such processes remain unanswered. Therefore, this study aimed: (i) to analyze the constituent characteristics of CRMs at different developmental stages of mouse hearts; (ii) to explore the evidence and mechanism that CRMs directly promote cardiomyocyte proliferation; and (iii) to delineate the evolutionary dynamics of the specific phenotypic and functional CRMs during mouse ontogeny. Our results indicate that four CRMs subsets existed successively (MP1–4) during ontogeny and changed dynamically. MP1 cluster was a different derivation and multiple functions population, embryonic and neonatal derived-MP1 can directly promote cardiomyocytes proliferation through Jagged-1–Notch1 axis and significantly ameliorates cardiac injury following myocardial infarction (MI). MP2/3 subsets can survive throughout adulthood. MP4, the main population in adult mouse hearts, contributes to inflammation. Changes in the redox state drove the replacement of CRMs from MP1 to MP4. Such changes caused the loss of proliferative ability in cardiomyocytes. Our results delineate the evolutionary dynamics of CRMs under physiological conditions and found direct evidence that embryonic and neonatal-derived CRMs regulate cardiomyocyte proliferation. Of course, our results also shed light on cardiac repair following injury.
2. Materials and methods
2.1. Animal and cell co-culture
Wide-type BALB/c mice were purchased from the Animal Center of Jiangsu University and housed in the same center. Animals were given food and water ad libitum. For all surgeries, the mice were anesthetized with 2% isoflurane via breathing using a surgical respiratory anesthesia system from RWD Life Science. All the process of surgeries kept giving 2% isoflurane via breathing. At the end of the procedure, the mice were euthanized by exsanguination. Heart tissues from embryonic Day 18 (E18D) and post-natal 7 days, 4 and 8 weeks, and 1-year-old (P7D, P4W, P8W, P1Y) mice were removed and immediately used in the experiments. All animal procedures performed to conform to the guidelines from Directive 2010/63/EU of the European Parliament on the protection of animals used for scientific, and approved by the Animal Care and Use Committee of Jiangsu University (Zhenjiang, China).
Cardiomyocytes and CRMs were isolated as described previously17, 18, 19. Briefly, hearts were carefully cut into small pieces and digested using 2.25 ng/mL Liberase TH Research Grade (47419000, Roche) (700 μg/mg heart tissue) at 37 °C for 20 min to get single-cell suspensions, and tissue suspensions were passed through a 40-μm cell strainer. Cells were cultured in DMEM medium (11965092, Gibco) to obtain cardiomyocytes containing 10% fetal bovine serum (FBS) (Gibco) for 2 h on cell plates. Cardiomyocytes that did not adhere to the plate were harvested. The purity of cardiomyocytes was detected through immunofluorescence. The cardiomyocytes were labeled with anti-cardiac troponin T (cTNT), and cardiac fibroblasts were labeled with Vimentin. For CRMs, CD45+ cells were purified from cardiac suspensions using CD45 microBeads kit (130-052-301, Miltenyi-Biotec) and enriched over MACS® Column. Cells were stained with anti-CD45 antibody conjugated to BV510, anti-CD64 antibody conjugated to APC (103138 and 139306, Biolegend). CRMs were isolated by fluorescence-activated cell sorting using Aria II (BD Bioscience). CD45+CD64+ CRMs (4 × 103) were directly co-cultured with 2 × 105 neonatal mouse cardiomyocytes in DMEM supplemented with 10% FBS. Indirectly coculture assay were performed using Transwell 24-well plate inserts 0.4 μm pore PC membrane (Transwell, Corning Incorporated), 4 × 103 CD45+CD64+ CRMs were seeded on Transwell inserts, and 2 × 105 cardiomyocytes were cultured in the lower compartment. After 48 h, cardiomyocytes were stained with cTNT, anti-Ki67 (ab8295 and ab15580, Abcam) and anti-Aurora B (RM278, Invitrogen) according to the manufacturer's instructions. All cells were counterstained with DAPI (D1306, Invitrogen, 0.1 mg/mL for 10 min). Fluorescence images were captured using a fluorescence microscope (Thermo Fisher Scientific) with 390-, 475-nm lasers.
2.2. Single cell RNA-Seq (scRNA-seq)
According to the manufacturer's instructions, single live cells, defined by propidium iodide and Hoechst staining, were captured using a 10 × Chromium controller (10 × Genomics). cDNA libraries were generated using Chromium Single Cell 3′-Library & Single Cell 3′-v3 Gel Beads (PN-1000075, Chromium). These libraries were sequenced using the Illumina NovaSeq 6000. The Cell Ranger software pipeline (version 3.1.0) was used to de-multiplex the sequencing data and generate normalized aggregate data across samples, producing a gene expression matrix of cells. The unique molecular identifier count matrix was further processed using the R toolkit Seurat20. Cells with unique molecular identifier/gene numbers outside the limit of mean value ± 2 folds of standard deviations (SD) were excluded, and low-quality cells in which >10% of genes belonging to the mitochondrial genome were discarded. 44,604 cells passed the stringent quality control and used for downstream analysis.
Highly variable genes across single cells were identified using the Seurat Find variable gene function (mean function = ExpMean, dispersion function = the variance to mean ratios of logged values) described previously21. The top variable genes were selected, and principal component analysis was performed to reduce the dimensionality in Seurat. According to their gene expression profiles, graph-based clustering was performed for cell clusters using the Find Clusters function in Seurat20. Cells were visualized using a t-distributed stochastic neighbor embedding (t-SNE) algorithm in Seurat. To find all marker functions in Seurat were used to identify marker genes of each cluster. The R package SingleR method22 and mouse transcriptomic datasets23,24 were used to infer the origin of each of the cells independently and to identify the cell type. Cell types were further annotated based on known marker genes.
Differentially expressed genes (DEGs) were identified in Seurat using the method described previously20. Gene ontology (GO) enrichment analysis of DEGs was performed using Metascape software. Heatmap visualization was obtained using the Heatmapper software. These heatmaps were generated based on the average-linkage clustering and Pearson distance measurement methods.
A single-cell trajectory was constructed using the Monocle package25. We used the differential Gene Test function of the Monocle2 package to select ordering genes (qval <0.01), which were used for a building trace. Cells ordered in pseudotime created seven distinct states.
2.3. Flow cytometry analysis
Cardiac cells were prepared as single-cell suspensions and then stained for 30 min at 4 °C with fixable viability dye (65-0865-18, eBioscience) before labeling them with antibodies from BioLegend: anti-CD45-Brilliant Violet 510, anti-CD64-APC, anti-Ly6C-Alexa Fluor® 488 (128022), anti-CX3CR1 Brilliant Violet 421 (149023), I-A/I-E Percp/cy5.5 (107626) and Jagged-1 (2191034, Invitrogen). Cell fluorescence was measured with a CytoFLEX S (BECKMAN COULTER), and data analysis was performed using FlowJo software.
2.4. Immunofluorescence analysis
Cardiomyocytes from neonatal mice were co-cultured with CRMs isolated from the heart of the embryonic or neonatal mice in a complete medium containing anti-Jagged-1 neutralizing Ab (10 μg/mL, MAB1277-SP, R&D Systems) or PBS for 2 days, or before coculture, the cardiomyocytes were pretreated by 10 μm DAPT, an inhibitor. Immunofluorescence assessed the expression cTNT, F4/80, Ki67, and Aurora B. The positive ratio of Ki67 or Aurora B was Ki67+cTNT+/cTNT+ or Aurora B+cTNT+/cTNT+ in each field of views. The average of randomly selected five fields was taken as the result of one experiment. The same experiment was repeated 3–5 times and then the average was taken as the final positive ratio.
2.5. Oxidant and antioxidant treatment
CD45+CD64+ CRMs of the desired ages were isolated by flow sorting. E18D and P7D-derived CRMs were treated with 5 μmol/L H2O2 and P4W, P8W, and P1Y-derived CRMs were treated with 5 μmol/L glutathione (GSH). After 48 h of culture in DMEM containing 10% FBS, some cells were used to analyze the expression of MHCII and CX3CR1 by flow cytometry. The murine cardiomyocyte cell line (MCMs) was treated with 300 μmol/L H2O2 for 0, 0.5, 1 and 2 h, and after 48 h of culture in DMEM containing 10% FBS, cells were collected for ROS quantify via DCFDA cellular ROS detection assay. For antioxidant treatment, MCMs were pretreated with 300 μmol/L H2O2 for 1 h and then treated with 5 μmol/L GSH for 48 h in DMEM containing 10% FBS. Then treated MCMs were co-cultured with CRMs derived from embryonic or neonatal mice for 48 h, the shift of co-cultured CRMs was detected by flow cytometry.
2.6. Adoptive transfer experiments
For adoptive transfer experiments, six-week-old male BALB/c mice were used to establish a MI model by ligating the left coronary artery as previously described26. Then CRMs derived from E18D or neonatal mice and labeled with 10 μmol/L lipophilic carbocyanine cell tracker dye Dil (1,1′-dioctadecyl-3,3,3′,3′-tetramethyl-indocarbocyanine perchlorate) at 37 °C for 10 min. Dil labeled CRMs (2 × 104) were immediately intra-myocardial injection in the ischemic left anterior descending coronary artery territory, and the control group was treated with an equal volume of PBS. On the 3rd day after MI, mouse hearts were harvested and dissociated into single-cell suspensions, and the Dil-labeled CRMs were detected through flow cytometric to confirm the CRMs entered into the heart. The shift of adoptive transferred CRMs was detected through flow cytometry. After that, CRMs derived from E18D or neonatal and P1Y (2 × 104) mice were intra-myocardial injection in the ischemic left anterior descending coronary artery territory of MI mice immediately and respectively, and on the 7th day after MI, heart tissue was carefully harvested. Mouse hearts were fixed in 4% formalin, embedded in paraffin, and sectioned serially at 5-μm thickness. Hematoxylin and eosin staining was performed to evaluate inflammation. The data were repeated three times, and at least five mice were included every time.
2.7. RNA isolation and RT-qPCR
Total RNA from CRMs or cardiomyocytes was extracted using TRIzol reagent (15596026, Invitrogen), and 500 ng RNA was reverse-transcribed transcribed into first-strand cDNA using a Reverse Transcription and cDNA Synthesis Kit (6130, TaKaRa) following the manufacturer's protocol. For RT-qPCR, cDNA was amplified using primers for genes tested. Gene expression was quantified relative to β-actin. The primers were purchased from iGene Biotechnology, detailed sequences were as following: Notch1 (F: 5′-GATGGCCTCAATGGGTACAAG-3′, R: 5′-TCGTTG TTGTTGATGTCACAGT-3′), Notch2 (F: 5′-GACTGCCAATACTCCACCTCT-3′, R: 5′-CCATTTTCGCAGGGATGAGAT-3′), Notch3 (F: 5′-TGCCAGAGTTCAGTG GTGG-3′, R: 5′-CACAGGCAAATCGGCCATC-3′), Notch4 (F: 5′-CTCTTGC CACTCAATTTCCCT-3′, R: 5′-TTGCAGAGTTGGGTATCCCTG-3′), Hes1 (F: 5′- GGAAATGACTGTGAAGCACCT-3′, R: 5′-GTCACCTCGTTCATGCACTC-3′), Hey1 (F: 5′-CACCTGAAAATGCTGCACAC-3′, R: 5′-ATGCTCAGATAACGGGC AAC-3′) and Actb (F: 5′-GAAGTCCCTCACCCTCCCAA-3′, R: 5′-GGCAT GGACGCGACCA-3′). Results were analyzed with the accompanying software according to the manufacturers' instructions.
2.8. Statistical analysis
Data were expressed as mean ± standard deviation (SD). Statistical analysis was performed by one-way analysis of variance or a two-tailed unpaired Student's t-test. P < 0.05 was considered statistically significant. All experiments were performed as triplicates.
3. Results
3.1. The phenotype and distribution of non-cardiomyocytes dynamically change
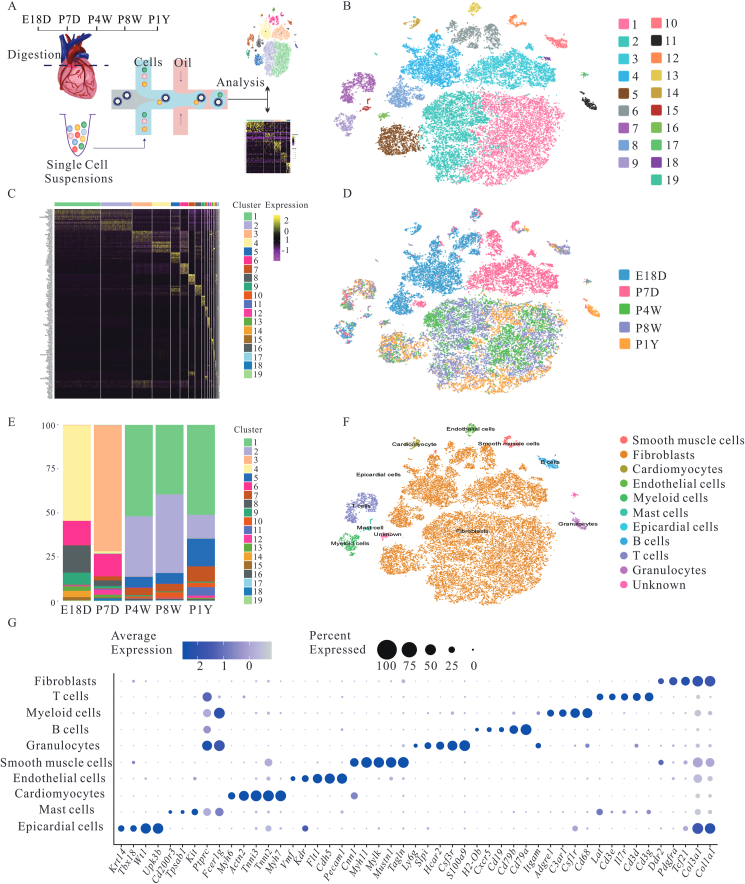
Since non-cardiomyocytes, namely stromal cells, play an indispensable role in physiological and pathological cardiac conditions, the dynamic changes of cardiac stromal cells during ontogeny were analyzed. The RNA from 44,604 stromal cells was sequenced from mice of different ages using scRNA-seq. Cells passed strict quality control and achieved a mean depth of 61,283 reads per cell and 2866 median genes per cell (Fig. 1A, and Supporting Information Fig. S1A–S1C). To acquire insights into the overall stromal cellular composition of the whole heart, unsupervised cell clustering divided all cells into 19 major clusters, consisting of as few as 50 cells to as many as 11,057 cells per cluster, according to their gene expression profiles (Fig. 1B). Cells from various clusters exhibited transcriptomic heterogeneity, suggesting dimensionality reduction. Graph-based clustering was carried out (Fig. 1C). Ontogeny was accompanied by the variation in the shape, structure, and function of somatic cells27. As expected, stromal cells exhibited dynamic alterations in cell clusters at various points, and their distribution was almost identical in adult mice (P4W, P8W, P1Y) (Fig. 1D and E). However, these components from embryonic and neonatal mice were completely different from those in adult mice. The cell types were annotated using the correlation values between the scRNA sequencing data and reference data sets provided by SingleR (Fig. S2B)22. To reliably assign a specific cell type to each cell cluster, cell types were redefined based on the expression of established cell-type markers. T-SNE plots of cells and dot plots of marker genes were used for visualization (Fig. 1F and G). All cells were divided into 19 clusters and they contained the following cell types: fibroblast (cluster 1, 2, 3, 4, 5, 6, 8 and 18; Tcf21, Pdgfra, Col1a1, Col3a1 and Ddr2)28,29, T cells (cluster 7; Cd3g, Cd3d, IL7r, Cd3e and Lat)30, myeloid cells (cluster 9; Cd68, Csf1r, C3ar1, Adgre1 and Itgam)31, B cells (cluster 10; Cd79a, Cd79b, Cd19, Cxcr5 and H2-ob)32, granulocytes (cluster 11; S100a9, Csf3r, Hcar2, Slpi and Ly6g)33,34, smooth muscle cells (cluster 12; Tagln, Mustn1, Mylk, Myh11 and Cnn1)35, endothelial cells (cluster 13; Pecam1, Cdh5, Vwf, Flt1 and Kdr)36, cardiomyocytes (cluster 14; Myh7, Tnnt2, Tnni3, Actn2 and Myh6)37, mast cells (cluster 15; Fcer1g, Ptprc, Kit, Tpsab1 and Cd200r3)38,39 and epicardial cells (cluster 19, Upk3b, Wt1, Tbx18 and Krt14)40. Clusters 16 and 17 contained a mixture of both fibroblasts and T cells or B cells markers, respectively, and they were designated as “unknown” (Supporting Information Fig. S2A). Furthermore, cardiomyocytes were detected only in E18D mouse hearts but not in postnatal mouse hearts. The cardiomyocytes from E18D mouse hearts were small and could pass through the mesh used (10 × Genomics). These results suggest that the composition of cardiac stromal cells changes dynamically with the development of the mouse hearts.
Figure 1.
Stromal cell composition of the mouse heart under steady states during ontogeny. (A) Scheme of the study design and workflow. n = 2 (one male and one female mouse were included at every point. More than 10,000 cells were collected at indicated points). (B) Cells were reduced dimensionally by principal component analysis, clustered based on a graph-based clustering approach, spectral t-SNE was used to reduce to 2-dimension for visualization. Each point depicts a single cell, and different color represents each cluster. (C) Gene-expression Heatmap of top 10 marker genes for each cell cluster. (D) t-SNE plot of single cells isolated from the heart of varying ages. Cells were colored according to their time point of origin. (E) Relative fraction of each cluster was given by the number of cells at a different time point and scaled to 100%. (F) t-SNE plot of cardiac stromal cells composition after unsupervised clustering. (G) Dot plot of marker genes identifying cell type. Individual dots are sized to reflect the proportion of cells and color to reflect the average expression levels of each marker gene across all cells. E18D, P7D, P4W and P1Y mean embryonic Day 18, post-natal 7 days, 4 and 8 weeks, and 1-year-old, respectively.
3.2. There are four successive CRMs subsets
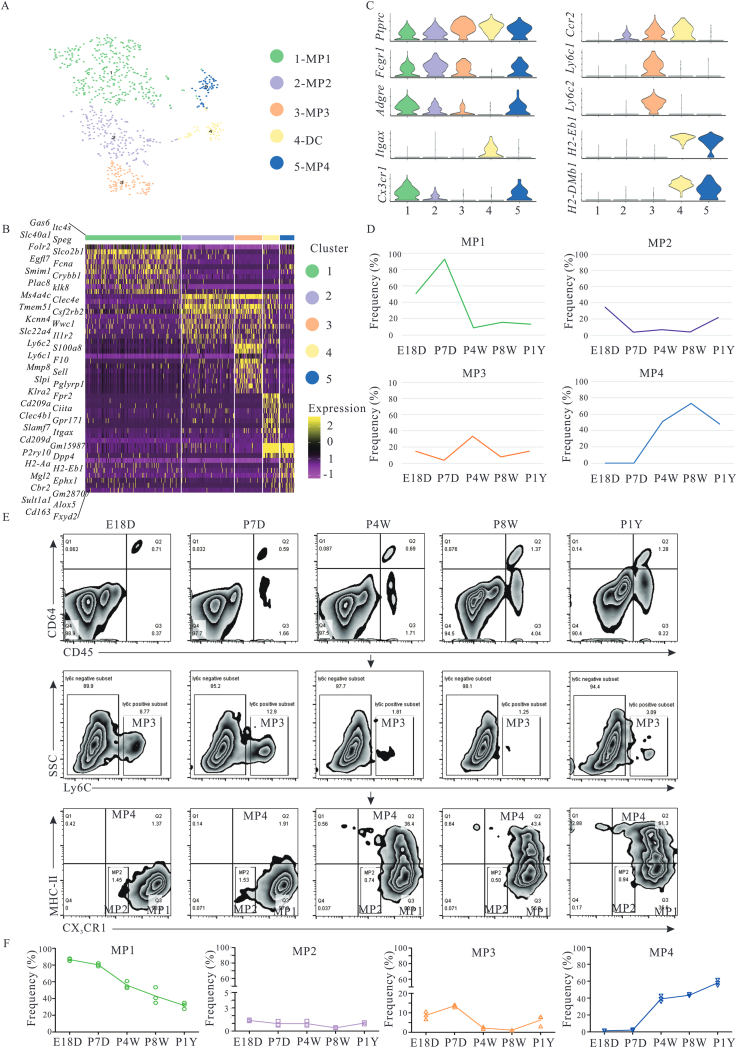
Highly heterogeneous CRMs populations play important roles in maintaining cardiac homeostasis, removing senescent or dying cells, and modulating cardiac development41. However, these functions of CRMs are closely related to specific phenotypes and stages of cardiac development. To reveal the evolution and phenotypic properties of CRMs during cardiac development, 766 myeloid cells were first divided into macrophages and dendritic cells based on common cell surface markers (Fig. 2A). Dendritic cells were separated from macrophages based on Cd11c expression. CRMs were defined by Ptprc, Fcgr1 (Cd64), and Adgre (F4/80) and were classified into four subsets with distinct transcriptomes (referred to as “MP1–4”). The four subsets included CX3CR1+CCR2–Ly6C–MHCII– (MP1), CX3CR1lowCCR2lowLy6C–MHCII– (MP2), CX3CR1–CCR-2+Ly6C+MHCII– (MP3), and CX3CR1+CCR2–Ly6C–MHCII+ (MP4) (Fig. 2B and C). MP1 was the dominant subset during E18D and P7D. However, they abruptly decreased after postnatal Day 7. Conversely, MP4 cells emerged at P4W (may emerge after postnatal Day 7) and persisted throughout adulthood. MP2 and MP3 existed at low frequency during mouse cardiac development. Additionally, MP3, expressing Ly6C, Ccr2, Adgrelow, might be a monocytes population (Fig. 2D). These observations indicated that the heterogeneous CRMs population is changed dynamically. Some CRMs established during embryonic development are long-lived cells independent of their replenishment by bone marrow-derived monocytes under physiological conditions, consistent with published data that embryonic development and neonatal stage, CRMs mainly originate from the yolk sac and fetal liver. After birth, CRMs from the yolk sac and fetal liver are gradually replaced by bone marrow-derived monocytes42,43. To further validate CRMs, the existence and frequency of CRMs subsets (MP1–4 subcluster) in the heart were examined at various development stages using flow cytometry and the results are shown in Fig. 2E. During mouse development, the alterations of CRM subclusters were identical to the scRNA-Seq analysis (Fig. 2D and F).
Figure 2.
CRMs properties at different time points during ontogeny. (A) Unsupervised sub-cluster showed four distinct cell subsets in a t-SNE map. (B) Heatmap of top 10 marker genes in down-sampling of more than 100 cells for each cluster. (C) Violin plots of the marker genes expression across five subsets. (D) Distribution characteristics of different subsets at different time during ontogeny based on scRNA-seq analysis. (E) Four CRM subsets existed successively. Macrophages were identified by the expression of CD45 and CD64 in hearts, and separated into four subsets based on Ly6C, CX3CR1, and MHCII, as determined by flow cytometry. At every point, six to eight mice were included. (F) The dynamic changes of MP1–4 frequencies at different points. E18D, P7D, P4W and P1Y mean embryonic Day 18, post-natal 7 days, 4 and 8 weeks, and 1-year-old, respectively.
3.3. MP1 subset derived from embryonic and neonatal mouse directly promoted cardiomyocytes proliferation via Jagged-1–Notch1 axis
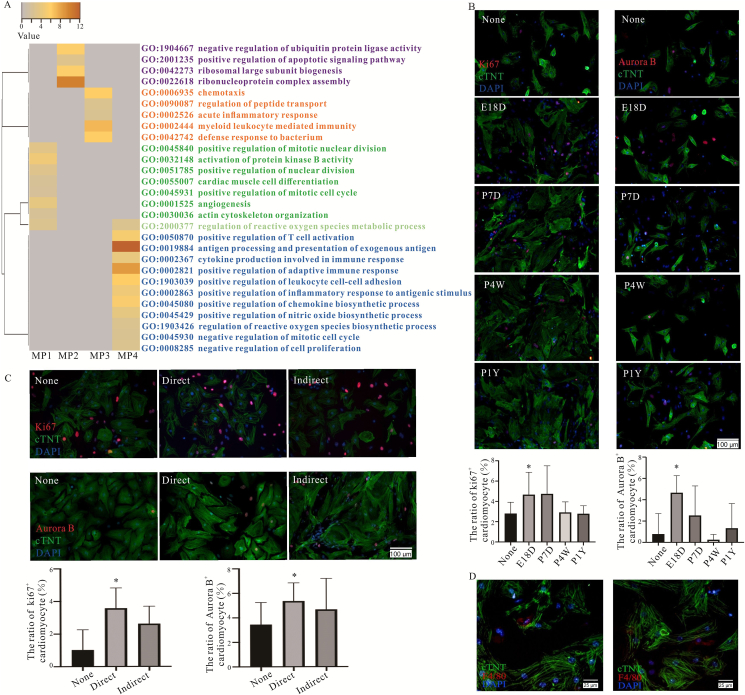
Based on GO enrichment, the MP1 subcluster focused on development, homeostatic and regenerative functions, including positive regulation of mitotic nuclear division, nuclear division and mitotic cell cycle, cell differentiation, actin cytoskeleton organization and angiogenesis. Conversely, the MP4 subcluster was involved in inflammatory pathway (positive regulation of T cell activation, antigen processing and presentation, cytokine and chemokine production) and negative regulation of proliferative pathway (negative regulation of mitotic nuclear division and mitotic cell cycle). It is to be noted that MP1 and MP4 subclusters regulated ROS metabolic process, whereas ROS biosynthetic process pathways were only enriched in the MP4 subcluster (Fig. 3A). Firstly, to confirm the effects of different CRMs subsets on cardiomyocytes proliferation, the purity of isolated cardiomyocytes from neonatal mouse was identified, and up to 90% through immunofluorescence staining (Supporting Information Fig. S3A), CRMs derived from the various ages of mice were co-cultured with neonatal mouse cardiomyocytes. Interestingly, CRMs derived from embryonic and neonatal mice had more pronounced effects in promoting cardiomyocyte proliferation than the cells from adult mice (Fig. 3B), which is also supported by the heatmap of the correlation coefficient of various samples (Fig. S1D). Furthermore, the phenotype of isolated CRMs from embryonic and neonatal mice was maintained throughout culturing and had the proliferative potential in vitro (Fig. S3B–S3D). However, their proliferative ability was weakened in co-culture system compared with cardiomyocytes (Fig. S3E and S3F). CRMs derived from embryonic and neonatal mouse hearts were directly or indirectly co-cultured with neonatal mouse cardiomyocytes to further confirm the effect through cells to cells contact or indirect. The results show that direct co-culturing significantly promoted cardiomyocyte proliferation (Fig. 3C), super-resolution data further approved the conclusion (Fig. 3D), which suggests that cell–cell contact contributed to cardiomyocytes proliferation.
Figure 3.
Four CRM subsets existed successively and exhibited different functions during ontogeny. (A) Selected categories from GO enrichment analysis of DEGs are expressed as –log10P value adjusted for multiple comparisons. (B) Immunofluorescence staining of Ki67 (left) and Aurora B (right) in cTNT positive neonatal mouse cardiomyocytes directly co-cultured with CRMs from mouse hearts of different ages (None, E18D, P7D, P4W, P1Y). The representative images are shown above and statistical analysis shown below. X axis presents different CRMs treatment. (C) Immunofluorescence staining of Ki67 and Aurora B in neonatal mouse cardiomyocytes co-cultured with CRMs from embryonic or neonatal mice directly or indirectly. The representative images are shown left and statistical analysis shown right. (D) Neonatal mouse cardiomyocytes were directly co-cultured with CRMs from embryonic and neonatal mouse hearts by cell–cell contact, CRMs were stained by F4/80, and the cardiomyocytes were stained by cTNT. For B and C, the data were repeated 3–5 times, similar data were obtained, the representative images are shown. ∗P < 0.05. E18D, P7D, P4W and P1Y mean embryonic Day 18, post-natal 7 days, 4 and 8 weeks, and 1-year-old, respectively.
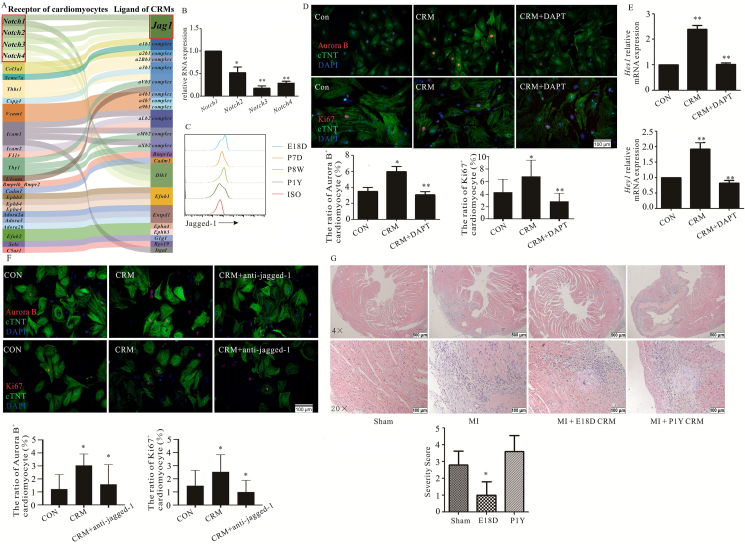
Next, the analysis of biologically relevant ligand-receptor interacting pairs suggested that Jagged-1–Notch axis was a potential cell–cell contact through CellPhoneDB (v2.0) (Fig. 4A). RT-qPCR further showed that Notch1 was predominantly expressed in neonatal mouse hearts (Fig. 4B); furthermore, flow cytometry also demonstrated that Jagged-1 expression was higher on CRMs at the end of embryonic and neonatal CRMs than that on the adult heart (Fig. 4C). To directly assess the role of Jagged-1–Notch1 axis on cardiomyocyte proliferation, DAPT, a novel γ-secretase inhibitor, was used, and obvious changes were observed between CRM-cardiomyocytes with DAPT and those without (Fig. 4D). We also observed a similar pattern of Notch-dependent Hes1 and Hey1 mRNA expression in cardiomyocytes (Fig. 4E). The positive role of CRMs on cardiomyocyte proliferation was abolished upon supplementation with a neutralizing anti-murine Jagged-1 antibody (Fig. 4F), which suggested that Jagged-1–Notch1 axis truly played a key role in cardiomyocyte proliferation. The adoptive transfer experiment was employed to further confirm similar phenomena in vivo. The results show that Dil-labeled CRMs from embryonic and neonatal mice truly entered into the heart, ameliorated the cardiomyocytes necrosis and inflammatory cells infiltration robustly following MI (Fig. 4G and Supporting Information Fig. S4). To further confirm the positive role of CRMs on cardiomyocyte proliferation in vivo, CD11b-DTR mice and liposome-encapsulated clodronate were employed to deplete the resident macrophages in neonatal mouse, respectively. However, all the mice were died (data not shown), which indirectly indicated that resident macrophages are critical for cardiac development and physiological function.
Figure 4.
CRMs derived from embryonic or neonatal mice facilitated the cardiomyocytes proliferation via Jagged-1–Notch. (A) The analysis of biologically relevant ligand–receptor interacting between CRMs from embryonic or neonatal mice and cardiomyocytes. (B) Notch1-4 expression was analyzed. RNA was extracted from purified neonatal mouse cardiomyocytes, and the gene expression of Notch1-4 was measured by RT-qPCR and normalized relative to β-actin (n = 4). (C) Jagged-1 expressions on CRMs of different ages mice (n = 4). (D) Immunofluorescence staining of Aurora B and Ki67 in cTNT positively neonatal mouse cardiomyocytes directly co-cultured with CRMs from embryonic or neonatal mice in the presence or absence of DAPT (n = 3). (E) qPCR analysis of Hes1 and Hey1 mRNA in neonatal mouse cardiomyocytes, results normalized to β-actin (n = 3). (F) Aurora B and Ki67 expression in neonatal mouse cardiomyocytes co-cultured with CRMs from embryonic or neonatal mice with or without pre-exposure to Jagged-1 antibody for 20 min (n = 4). (G) CRMs from embryonic or neonatal mice and P1Y mouse hearts were immediately transferred into the appropriate recipient mice by intra-myocardial injection on Day 1 after MI. 15–18 mice were included in every group. All mice were sacrificed on Day 7 and heart sections were examined by hematoxylin and eosin staining. For D, F and G, the representative images are shown above and statistical analysis shown below. ∗P < 0.05, ∗∗P < 0.01. MI, ISO, E18D, P7D, P4W and P1Y mean myocardial infarction, isotype, embryonic Day 18, post-natal 7 days, 4 and 8 weeks, and 1-year-old, respectively.
3.4. MP1 subset, a powerful and diverse function subset, had different derivations
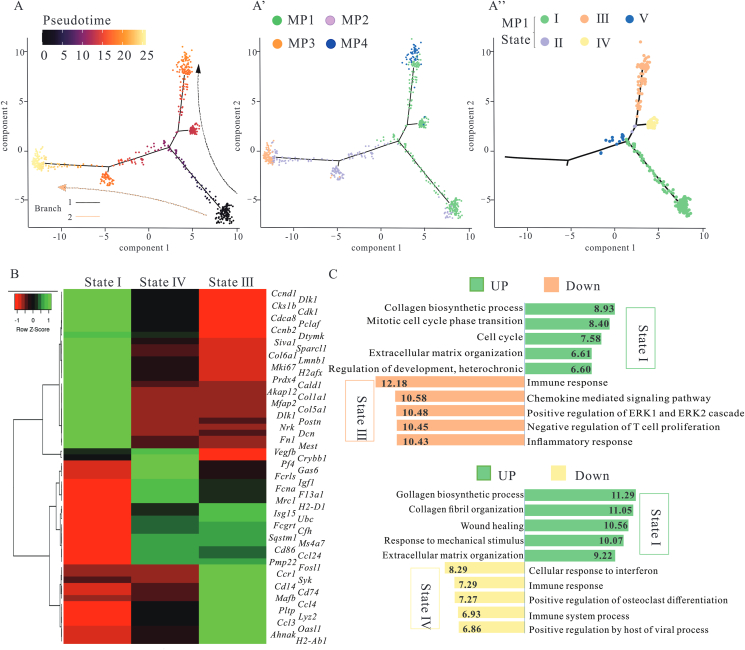
Next, to clarify the characteristics of MP1, firstly, CRM subsets were aligned in pseudotime to model cardiac developmental trajectories based on changes in gene expression. Cells were partitioned based on their branch location, and each region was defined as a state. MP1 mainly straddled the two-branch trajectory (states I–IV), MP4 occupied branch 1 (state III), MP2 and MP3 occupied branch 2 (state V, VII and state VI, respectively) (Fig. 5A and Supporting Information Fig. S5A). Fig. 5A shows that embryonic and neonatal-derived CRMs (MP1) contained a variety of subpopulations with distinct expression patterns, which suggested MP1 had multi-functions, whereas in adults, the function of CRMs was more monotonous. According to differential gene expression analysis, CRMs obtained from state III in MP1 were transcriptionally closer to state IV, and distinct from the state I (Fig. 5B and Fig. S5B). Compared to the other two states, upregulated genes in state I were significantly enriched in biological behaviors related to ECM organization, cell proliferation, and development, whereas MP1 in state III and state IV exhibited alterations related to immune response (Fig. 5C and Fig. S5C). Genes involved in the cell cycle (including Cdk1, Ccnb2, Cald1, and Birc5) were also upregulated in the state I in the MP1. These data indicate that MP1 cluster had different functions and derivations (from yolk sac, fetal liver and bone marrow).
Figure 5.
MP1 cluster, a powerful and diverse function subset, had different derivations. (A) Ordering CRMs along a cell conversion trajectory using Monocle analysis. CRMs were color-labeled by pseudotime or sub-cluster. Cell clustering of MP1 based on the state along pseudotime trajectory showed five distinct trajectory states. (B) Heatmap of marker genes expression in the indicated states of MP1. (C) Selected top GO enrichment categories of the DEGs between different states. 30–50 cells per category were included.
3.5. The shift of MP1 and MP4 might result from cellular redox state
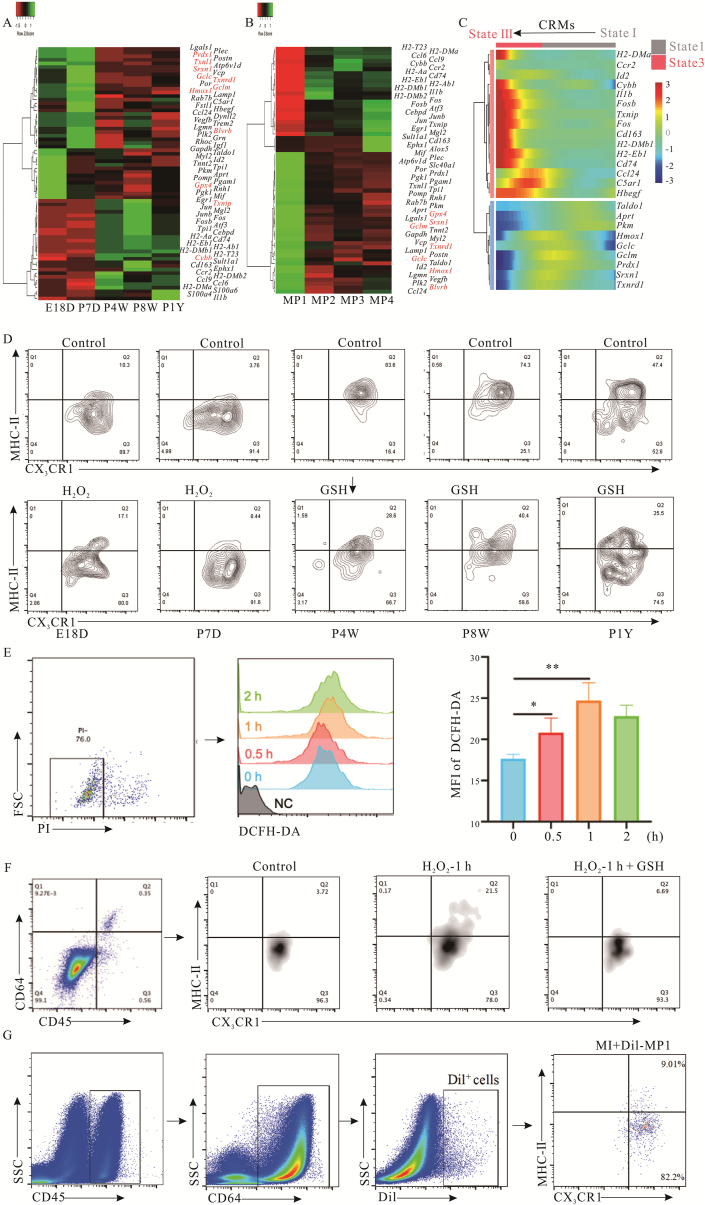
A differential gene expression analysis was performed to elucidate the mechanism of the rapid replacement of the MP1 subset after birth. The results indicated that inflammation (Il1b, Ccr2, and S100a4) and antigen presentation (H2-Aa and H2-Dmb1) associated genes were specifically expressed in adult-derived CRMs, and this might contribute to cardiomyocyte senescence. However, development-associated (Id2, Myl2 and Tnnt2) and proliferation-related genes (Igf1, Hbegf, Vegfb, Fstl1 and C5ar1) were enriched only in the embryonic and neonatal-derived CRMs (Fig. 6A). MP1 was the major subset of embryonic and neonatal-derived CRMs (E18D and <P7D, regenerative stage). In contrast, MP4 was present in greater numbers than other clusters in adult mouse hearts (non-regenerative stage). The different gene expression patterns of MP1 and MP4 also supported the division of regenerative and non-regenerative phases (Fig. 6B). The expression of genes related to antioxidant activity (such as Txnrd1, Gclm, Gclc, Gpx4, Srxn1 and Hmox1) was downregulated during pseudotime progression in MP4, whereas Cybb, which codes NOX2, a well-established ROS-producing enzyme and inflammation-associated genes were enriched (Fig. 6C). ROS is an important byproduct of the electron transport chain of the mitochondrial oxidative phosphorylation system44. During ontogeny, the cardiac microenvironment changes from a hypoxic state to an oxygen-rich state, and the metabolism of cardiomyocytes undergoes a transition from glycolysis to oxidative phosphorylation. However, it is unclear whether the replacement of MP1 by MP4 is related to changes in the redox state in the microenvironment caused by ROS production. Therefore, CRMs were isolated from mice of different ages and treated with H2O2 or GSH, natural redox agents in vivo. The results showed that MP1 subsets from embryonic and neonatal mice converted to MP4 phenotype following H2O2 stimulation. However, MP4 subsets are only partially converted into MP1 phenotype in response to GSH (Fig. 6D). To further confirm the shift of CRMs from MP1 to MP4, MCMs were pretreated with H2O2 with/without GSH and then cultured for 48 h. The pretreated MCMs were co-cultured with CRMs from embryonic or neonatal mice for 48 h, and the results demonstrate that pretreated MCMs up-regulated the ROS and promoted the phenotype shift of CRMs from MP1 to MP4. Conversely, GSH postponed this process (Fig. 6E and F). The shift of CRMs from MP1 to MP4 was further confirmed in vivo (Fig. 6G).
Figure 6.
The replacement of MP1 by MP4 was driven by the cellular redox state. (A, B) Heatmap display DEGs expression level in each time point (A) or MP1–4 (B). (C) Heatmap of gene expression along the pseudotime trajectory of CRMs. For A–C, 30–50 cells per category were included. (D) Frequencies of MP1 and MP4 were determined by flow cytometry. CRMs derived from different age mice were treated by GSH or H2O2 for 48 h and then analyzed using flow cytometry (n = 4). (E) Intracellular ROS levels were measured by flow cytometry using DCFDA dye fluorescence in cells treated with or without H2O2. (F) The phenotypes of CRMs from embryonic or neonatal mice were detected after co-culturing with H2O2 or H2O2+GSH pretreated MCMs by flow cytometry. (G) Dil-labeled MP1s were detected on the 3rd after MI, the phenotype was identified by the expression of CD45, CD64, CX3CR1 and MHCII (n = 6). ∗P < 0.05, ∗∗P < 0.01. E18D, P7D, P4W and P1Y mean embryonic Day 18, post-natal 7 days, 4 and 8 weeks, and 1-year-old, respectively. MI and MCMs mean myocardial infarction and murine cardiomyocyte cell line, respectively.
4. Discussion
Cardiovascular disease, especially devastating heart failure, is becoming a growing health problem. It represents a major reason for morbidity and mortality and the significantly increasing burden on healthcare45,46. The key reason for the high mortality in heart disease is the rapid loss of cardiac regenerative potential after birth in humans, compared to other species such as teleost fish and urodeles47,48. Central to this regenerative potential is CRMs49,50. CRMs, a highly heterogeneous population, play critical roles in maintaining cardiac homeostasis, functions, and protecting the heart from injury and pathogenic infection through dynamic phenotypic changes in response to external stimuli. Following cardiac injury, CRMs, predominant in neonatal cardiac tissue, contribute to scarless repair and coronary angiogenesis11, 12, 13, CRMs depletion abrogates cardiac regeneration51. It is known that yolk sac- and fetal liver-derived CRMs are beneficial for cardiac regeneration. However, there are no data on the underlying mechanism and show the dynamic changes in CRMs during cardiac ontogeny. Hence, it is important to address these above questions.
The composition of cardiac stromal cells of mice during ontogeny (from E18D to P7D, P4W, P8W, and P1Y) was analyzed by scRNA-seq. Four CRM subsets existed successively: MP1–4. Similarly, four subsets have been reported in adult mouse heart: MerTK+Ly6C–MHCIIhiCX3CR1hiCD206intCCR2–, MerTK+Ly-6C–MHCIIloCX3CR1intCD206hi CD11cloCCR2–, MerTK+Ly6-C+MHCIIhi/loCX3CR1hi CD206hi/intCD11chi/loCCR2–, and Mer-TK–Ly6C+MHCII–CX3CR1–CD11cloCD206–CCR2+ 42 or TIMD4+LYVE1+ MHCIIloCCR2–, TIMD4–LYVE1–MHCI-IhiCCR2–, TIMD4–LYVE1– MHCIIhiCCR2+, and CCR-2+MHCIIlo1,49,52. Such differences may be caused by the variations in the germlines and age of the selected mice and cardiac region-related sampling variations. MP2 and MP3 subsets persisted throughout adulthood, which indicated that some embryonic CRMs might be partially renewed during cardiac development. MP1 subset was the dominant CRM subtype during embryonic and neonatal stages and abruptly decreased after 7 days. GO enrichment analysis showed that this subset was associated with development, homeostasis, and regeneration. To further confirm the positive roles of MP1 on cardiac regeneration in vivo, the resident macrophage depletion models in neonatal mice were trying to establish through liposome-encapsulated clodronate and CD11b-DTR mice but all failed, which may be attributed rather to resident macrophage depletion caused cardiac developmental arrest and physiological dysfunction. The published data also suggested the critical roles of CRMs on cardiac development and physiological function8,10. All the data partially illustrated that resident macrophages are critical for cardiac development and function especially at neonatal stage. In this sense, the conditional cardiac-specific CRMs depletion mice should be considered in the future research. However, MP4 subset emerged at P4W and persisted throughout adulthood. Compared with MP1, the function of MP4 was relatively monotonous, and it was enriched majorly during inflammatory pathway activation. Flow cytometry analysis revealed that MP1 sub-cluster also existed after birth, which indicated that they may have multi-sources (derived from yolk sac, fetal liver and bone marrow) or persisted survival. Computational cell trajectory analysis revealed that MP1 subset could be converted into MP4 subset. In vitro data further approved the phenotype shift of MP1 and MP4.
The embryonic and neonatal-derived MP1 directly activated Notch signaling to promote cardiomyocyte proliferation via Jagged-1, which was consistent with previous data that bone marrow-derived mesenchymal stromal cells through Notch1–Jagged-1 pathway promoted the cardiomyocytes proliferation18,53,54. However, during the individual growth after birth, MP1 subsets from embryonic and neonatal stages were replaced by bone marrow monocytes, making them lose their inherent functions and develop new properties. Similar phenomena occur in inflammatory conditions, where the CRMs are replenished following cardiac injury55. The replacement not only directly affected cardiomyocyte proliferation, but also reprogrammed fibroblasts and indirectly regulated cardiomyocyte proliferation. As shown in Fig. 1, the composition, phenotype, and functional characteristics of mouse cardiac fibroblasts are significantly changed during ontogeny. Cardiac fibroblasts, an essential cell type in the heart, deposit ECM and collagen, and provide a structural scaffold for cardiomyocytes. They are involved in cardiac development, mechanical forces distribution, and electrical conduction mediation through the expression of cellular adhesion molecules, such as connexins and cadherins56,57. The switch of fibroblast subsets from neonatal to adult states might also drive cardiomyocyte maturation and cause the cardiomyocytes to lose their proliferative ability58. Therefore, we speculate that replacing yolk sac-or fetal liver-derived MP1 subsets, is the main cause of rapid loss of cardiac regenerative potential after birth. However, further experiments are needed to establish this notion and identify the driving force for this switch.
The difference in physiological function among macrophages is dependent on environmental stimulus and functional needs. Macrophages can rapidly exhibit functional adaption in response to novel stimuli59. Recent data indicate that cellular metabolism is tightly linked to cell proliferation. Glucose metabolism can facilitate cardiomyocyte proliferation and inhibit cardiomyocyte maturation or hypertrophy through the pentose phosphate pathway60. Cardiomyocytes’ glucose uptake declined by more than 99.9%, starting from P7D to E18D. At this point, the cardiomyocytes mature and cell cycle arrest happens60. The adoption of oxidative metabolism is another key determinant of cardiac regeneration or non-regenerative scar formation after injury61. Cardiomyocytes in the heart of developing mice are exposed to the relatively hypoxic condition compared to adult heart62. In a hypoxic environment, energy is produced through glycolysis; and in an oxygen-rich environment after birth, energy is provided through oxidative phosphorylation. The mitochondrial oxidative phosphorylation system generates a large amount of ROS as a byproduct of the electron transport chain44. ROS can influence the phenotype and function of macrophages63. This notion is further supported by the reports that, when exposed to chronic hypoxia, adult mouse hearts show regenerative potential64,65. Furthermore, following birth, changes in mammalian cardiomyocyte metabolism influence the cardiac microenvironment. The metabolic changes of cardiomyocytes are one of the main dynamics of CRMs replacement and function alteration. Our results also approved the speculation, which may be an important reason why adult mammalian hearts suffer from cardiomyopathy following infarction.
5. Conclusions
During mice ontogeny, four CRMs subsets existed successively and changed dynamically. The MP1 subset from embryonic and neonatal stages directly facilitated cardiomyocyte proliferation through Jagged-1–Notch1 axis, significantly ameliorating cardiac injury following MI. However, after birth, the microenvironment changes led to the replacement of MP1 by MP4. Such a switch of CRMs resulted in the loss of proliferative potential. This study is the first to delineate this evolutionary process and dynamics of CRMs under physiological conditions, and demonstrated that yolk sac and fetal-derived MP1 subset could directly contribute to cardiomyocyte proliferation and shed light on cardiac repair following injury.
Acknowledgments
This work was supported by National Natural Science Foundation of China (Grant No. 81871244), Primary Research & Development Plan of Jiangsu Province (BE2019700, China), Jiangsu Province “333” project (BRA2018016, China), six talent peaks project in Jiangsu Province (2019-WSN-122, China), Projects of International Cooperation from Jiangsu (BX2019100, China), and international cooperation and exchange from Zhenjiang (GJ2020010, China), key funds from health commission of jiangsu (ZD2021009, China). We thank Wiley Editing Service and Prof. Chunfu Zheng for editing the manuscript. We thank OE Biotech Co., Ltd. (Shanghai, China) for providing single-cell RNA-seq, Dr. Yongbing Ba for assistance with bioinformatics analysis.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2022.08.016.
Contributor Information
Tianqing Peng, Email: tpeng2@uwo.ca.
Zhaoliang Su, Email: szl30@ujs.edu.cn.
Author contributions
Rong Chen, Shiqing Zhang and Fan Liu contributed equally in the design and preparing the data; Lin Xia, Chong Wang, Siamak Sandoghchian Shotorbani and Huaxi Xu obtained the in vivo data and statistical analysis, Subrata Chakrabarti edited the manuscript, Tianqing Peng and Zhaoliang Su provided the ideas, funds and prepared the manuscript.
Conflicts of interest
The authors have declared that no conflict of interest exists.
Appendix A. Supporting information
The following is the Supplementary data to this article:
References
- 1.Epelman S., Lavine K.J., Randolph G.J. Origin and functions of tissue macrophages. Immunity. 2014;41:21–35. doi: 10.1016/j.immuni.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yona S., Kim K.W., Wolf Y., Mildner A., Varol D., Breker M., et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoeffel G., Wang Y., Greter M., See P., Teo P., Malleret B., et al. Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac-derived macrophages. J Exp Med. 2012;209:1167–1181. doi: 10.1084/jem.20120340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hashimoto D., Chow A., Noizat C., Teo P., Beasley M.B., Leboeuf M., et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tamoutounour S., Guilliams M., Montanana Sanchis F., Liu H., Terhorst D., Malosse C., et al. Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity. 2013;39:925–938. doi: 10.1016/j.immuni.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 6.Zigmond E., Varol C., Farache J., Elmaliah E., Satpathy A.T., Friedlander G., et al. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity. 2012;37:1076–1090. doi: 10.1016/j.immuni.2012.08.026. [DOI] [PubMed] [Google Scholar]
- 7.Heidt T., Courties G., Dutta P., Sager H.B., Sebas M., Iwamoto Y., et al. Differential contribution of monocytes to heart macrophages in steady-state and after myocardial infarction. Circ Res. 2014;115:284–295. doi: 10.1161/CIRCRESAHA.115.303567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leid J., Carrelha J., Boukarabila H., Epelman S., Jacobsen S.E., Lavine K.J. Primitive embryonic macrophages are required for coronary development and maturation. Circ Res. 2016;118:1498–1511. doi: 10.1161/CIRCRESAHA.115.308270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harari E., Guo L., Smith S.L., Braumann R.E., Virmani R., Finn A.V. Heart-resident macrophages: are they involved in the rhythm of every beat?. J Thorac Dis. 2017;9:2264–2267. doi: 10.21037/jtd.2017.07.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hulsmans M., Clauss S., Xiao L., Aguirre A.D., King K.R., Hanley A., et al. Macrophages facilitate electrical conduction in the heart. Cell. 2017;169:510–522.e20. doi: 10.1016/j.cell.2017.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lavine K.J., Epelman S., Uchida K., Weber K.J., Nichols C.G., Schilling J.D., et al. Distinct macrophage lineages contribute to disparate patterns of cardiac recovery and remodeling in the neonatal and adult heart. Proc Natl Acad Sci U S A. 2014;111:16029–16034. doi: 10.1073/pnas.1406508111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez I., Duval V., Silvestre J.S. Cardiomyocytes and macrophages discourse on the method to govern cardiac repair. Front Cardiovasc Med. 2018;5:134. doi: 10.3389/fcvm.2018.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frangogiannis N.G. Emerging roles for macrophages in cardiac injury: cytoprotection, repair, and regeneration. J Clin Invest. 2015;125:2927–2930. doi: 10.1172/JCI83191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mass E., Ballesteros I., Farlik M., Halbritter F., Günther P., Crozet L., et al. Specification of tissue-resident macrophages during organogenesis. Science. 2016;353:aaf4238. doi: 10.1126/science.aaf4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van de Laar L., Saelens W., De Prijck S., Martens L., Scott C.L., Van Isterdael G., et al. Yolk sac macrophages, fetal liver, and adult monocytes can colonize an empty niche and develop into functional tissue-resident macrophages. Immunity. 2016;44:755–768. doi: 10.1016/j.immuni.2016.02.017. [DOI] [PubMed] [Google Scholar]
- 16.Gautier E.L., Shay T., Miller J., Greter M., Jakubzick C., Ivanov S., et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng D., Su Z., Zhang Y., Ni R., Fan G.C., Robbins J., et al. Calpain-2 promotes MKP-1 expression protecting cardiomyocytes in both in vitro and in vivo mouse models of doxorubicin-induced cardiotoxicity. Arch Toxicol. 2019;93:1051–1065. doi: 10.1007/s00204-019-02405-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S.H., Hadipour-Lakmehsari S., Murthy H.R., Gibb N., Miyake T., Teng A.C.T., et al. REEP5 depletion causes sarco-endoplasmic reticulum vacuolization and cardiac functional defects. Nat Commun. 2020;11:965. doi: 10.1038/s41467-019-14143-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nazir S., Gadi I., Al-Dabet M.M., Elwakiel A., Kohli S., Ghosh S., et al. Cytoprotective activated protein C averts Nlrp3 inflammasome-induced ischemia–reperfusion injury via mTORC1 inhibition. Blood. 2017;130:2664–2677. doi: 10.1182/blood-2017-05-782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aran D., Looney A.P., Liu L., Wu E., Fong V., Hsu A., et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han X., Wang R., Zhou Y., Fei L., Sun H., Lai S., et al. Mapping the mouse cell atlas by microwell-seq. Cell. 2018;172:1091–1107.e17. doi: 10.1016/j.cell.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Benayoun B.A., Pollina E.A., Singh P.P., Mahmoudi S., Harel I., Casey K.M., et al. Remodeling of epigenome and transcriptome landscapes with aging in mice reveals widespread induction of inflammatory responses. Genome Res. 2019;29:697–709. doi: 10.1101/gr.240093.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao E., Lei Y.H., Shang X., Huang Z.M., Zuo L., Boucher M., et al. A novel and efficient model of coronary artery ligation and myocardial infarction in the mouse. Circ Res. 2010;107:1445–1453. doi: 10.1161/CIRCRESAHA.110.223925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katano W., Moriyama Y., Takeuchi J.K., Koshiba-Takeuchi K. Cardiac septation in heart development and evolution. Dev Growth Differ. 2019;61:114–123. doi: 10.1111/dgd.12580. [DOI] [PubMed] [Google Scholar]
- 28.Lighthouse J.K., Small E.M. Transcriptional control of cardiac fibroblast plasticity. J Mol Cell Cardiol. 2016;91:52–60. doi: 10.1016/j.yjmcc.2015.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burstein B., Libby E., Calderone A., Nattel S. Differential behaviors of atrial versus ventricular fibroblasts: a potential role for platelet-derived growth factor in atrial-ventricular remodeling differences. Circulation. 2008;117:1630–1641. doi: 10.1161/CIRCULATIONAHA.107.748053. [DOI] [PubMed] [Google Scholar]
- 30.Dulken B.W., Buckley M.T., Navarro Negredo P., Saligrama N., Cayrol R., Leeman D.S., et al. Single-cell analysis reveals T cell infiltration in old neurogenic niches. Nature. 2019;571:205–210. doi: 10.1038/s41586-019-1362-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skelly D.A., Squiers G.T., McLellan M.A., Bolisetty M.T., Robson P., Rosenthal N.A., et al. Single-cell transcriptional profiling reveals cellular diversity and intercommunication in the mouse heart. Cell Rep. 2018;22:600–610. doi: 10.1016/j.celrep.2017.12.072. [DOI] [PubMed] [Google Scholar]
- 32.Tokunaga R., Naseem M., Lo J.H., Battaglin F., Soni S., Puccini A., et al. B cell and B cell-related pathways for novel cancer treatments. Cancer Treat Rev. 2019;73:10–19. doi: 10.1016/j.ctrv.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen H., Assmann J.C., Krenz A., Rahman M., Grimm M., Karsten C.M., et al. Hydroxycarboxylic acid receptor 2 mediates dimethyl fumarate's protective effect in EAE. J Clin Invest. 2014;124:2188–2192. doi: 10.1172/JCI72151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobsen L.C., Sorensen O.E., Cowland J.B., Borregaard N., Theilgaard-Monch K. The secretory leukocyte protease inhibitor (SLPI) and the secondary granule protein lactoferrin are synthesized in myelocytes, colocalize in subcellular fractions of neutrophils, and are coreleased by activated neutrophils. J Leukoc Biol. 2008;83:1155–1164. doi: 10.1189/jlb.0706442. [DOI] [PubMed] [Google Scholar]
- 35.Wirka R.C., Wagh D., Paik D.T., Pjanic M., Nguyen T., Miller C.L., et al. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat Med. 2019;25:1280–1289. doi: 10.1038/s41591-019-0512-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalucka J., de Rooij L., Goveia J., Rohlenova K., Dumas S.J., Meta E., et al. Single-cell transcriptome atlas of murine endothelial cells. Cell. 2020;180:764–779.e20. doi: 10.1016/j.cell.2020.01.015. [DOI] [PubMed] [Google Scholar]
- 37.Wang L., Yu P., Zhou B., Song J., Li Z., Zhang M., et al. Single-cell reconstruction of the adult human heart during heart failure and recovery reveals the cellular landscape underlying cardiac function. Nat Cell Biol. 2020;22:108–119. doi: 10.1038/s41556-019-0446-7. [DOI] [PubMed] [Google Scholar]
- 38.Gilreath J.A., Tchertanov L., Deininger M.W. Novel approaches to treating advanced systemic mastocytosis. Clin Pharmacol. 2019;11:77–92. doi: 10.2147/CPAA.S206615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Metz M., Torene R., Kaiser S., Beste M.T., Staubach P., Bauer A., et al. Omalizumab normalizes the gene expression signature of lesional skin in patients with chronic spontaneous urticaria: a randomized, double-blind, placebo-controlled study. Allergy. 2019;74:141–151. doi: 10.1111/all.13547. [DOI] [PubMed] [Google Scholar]
- 40.Huang G.N., Thatcher J.E., McAnally J., Kong Y., Qi X., Tan W., et al. C/EBP transcription factors mediate epicardial activation during heart development and injury. Science. 2012;338:1599–1603. doi: 10.1126/science.1229765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang S., Chen R., Chakrabarti S., Su Z. Resident macrophages as potential therapeutic targets for cardiac ageing and injury. Clin Transl Immunol. 2020;9:e1167. doi: 10.1002/cti2.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Epelman S., Lavine K.J., Beaudin A.E., Sojka D.K., Carrero J.A., Calderon B., et al. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity. 2014;40:91–104. doi: 10.1016/j.immuni.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ginhoux F., Guilliams M. Tissue-resident macrophage ontogeny and homeostasis. Immunity. 2016;44:439–449. doi: 10.1016/j.immuni.2016.02.024. [DOI] [PubMed] [Google Scholar]
- 44.Li G., Qin Y. Mitochondrial translation factor EF4 regulates oxidative phosphorylation complexes and the production of ROS. Free Radic Res. 2018;52:1250–1255. doi: 10.1080/10715762.2018.1479063. [DOI] [PubMed] [Google Scholar]
- 45.Song Y., Song F., Wu C., Hong Y.X., Li G. The roles of epicardial adipose tissue in heart failure. Heart Fail Rev. 2022;27:369–377. doi: 10.1007/s10741-020-09997-x. [DOI] [PubMed] [Google Scholar]
- 46.Biglu M.H., Ghavami M., Biglu S. Cardiovascular diseases in the mirror of science. J Cardiovasc Thorac Res. 2016;8:158–163. doi: 10.15171/jcvtr.2016.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vadakke-Madathil S., Chaudhry H.W. Cardiac regeneration: time to revisit nature. Circ Res. 2018;123:24–26. doi: 10.1161/CIRCRESAHA.118.313246. [DOI] [PubMed] [Google Scholar]
- 48.Tzahor E., Poss K.D. Cardiac regeneration strategies: staying young at heart. Science. 2017;356:1035–1039. doi: 10.1126/science.aam5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dick S.A., Macklin J.A., Nejat S., Momen A., Clemente-Casares X., Althagafi M.G., et al. Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat Immunol. 2019;20:29–39. doi: 10.1038/s41590-018-0272-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Theret M., Mounier R., Rossi F. The origins and non-canonical functions of macrophages in development and regeneration. Development. 2019;146:dev156000. doi: 10.1242/dev.156000. [DOI] [PubMed] [Google Scholar]
- 51.Aurora A.B., Porrello E.R., Tan W., Mahmoud A.I., Hill J.A., Bassel-Duby R., et al. Macrophages are required for neonatal heart regeneration. J Clin Invest. 2014;124:1382–1392. doi: 10.1172/JCI72181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stevens S.M., von Gise A., VanDusen N., Zhou B., Pu W.T. Epicardium is required for cardiac seeding by yolk sac macrophages, precursors of resident macrophages of the adult heart. Dev Biol. 2016;413:153–159. doi: 10.1016/j.ydbio.2016.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takeshita K., Satoh M., Ii M., Silver M., Limbourg F.P., Mukai Y., et al. Critical role of endothelial Notch1 signaling in postnatal angiogenesis. Circ Res. 2007;100:70–78. doi: 10.1161/01.RES.0000254788.47304.6e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kratsios P., Catela C., Salimova E., Huth M., Berno V., Rosenthal N., et al. Distinct roles for cell-autonomous Notch signaling in cardiomyocytes of the embryonic and adult heart. Circ Res. 2010;106:559–572. doi: 10.1161/CIRCRESAHA.109.203034. [DOI] [PubMed] [Google Scholar]
- 55.Lu H., Chen R., Barnie P.A., Tian Y., Zhang S., Xu H., et al. Fibroblast transdifferentiation promotes conversion of M1 macrophages and replenishment of cardiac resident macrophages following cardiac injury in mice. Eur J Immunol. 2020;50:795–808. doi: 10.1002/eji.201948414. [DOI] [PubMed] [Google Scholar]
- 56.Souders C.A., Bowers S.L., Baudino T.A. Cardiac fibroblast: the renaissance cell. Circ Res. 2009;105:1164–1176. doi: 10.1161/CIRCRESAHA.109.209809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Travers J.G., Kamal F.A., Robbins J., Yutzey K.E., Blaxall B.C. Cardiac fibrosis: the fibroblast awakens. Circ Res. 2016;118:1021–1040. doi: 10.1161/CIRCRESAHA.115.306565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y., Yao F., Wang L., Li Z., Ren Z., Li D., et al. Single-cell analysis of murine fibroblasts identifies neonatal to adult switching that regulates cardiomyocyte maturation. Nat Commun. 2020;11:2585. doi: 10.1038/s41467-020-16204-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bailey J.D., Shaw A., McNeill E., Nicol T., Diotallevi M., Chuaiphichai S., et al. Isolation and culture of murine bone marrow-derived macrophages for nitric oxide and redox biology. Nitric Oxide. 2020;100–101:17–29. doi: 10.1016/j.niox.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakano H., Minami I., Braas D., Pappoe H., Wu X., Sagadevan A., et al. Glucose inhibits cardiac muscle maturation through nucleotide biosynthesis. eLife. 2017;6 doi: 10.7554/eLife.29330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sakaguchi A., Nishiyama C., Kimura W. Cardiac regeneration as an environmental adaptation. Biochim Biophys Acta Mol Cell Res. 2020;1867 doi: 10.1016/j.bbamcr.2019.118623. [DOI] [PubMed] [Google Scholar]
- 62.Dunwoodie S.L. The role of hypoxia in development of the mammalian embryo. Dev Cell. 2009;17:755–773. doi: 10.1016/j.devcel.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 63.O'Neill L.A., Pearce E.J. Immunometabolism governs dendritic cell and macrophage function. J Exp Med. 2016;213:15–23. doi: 10.1084/jem.20151570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakada Y., Canseco D.C., Thet S., Abdisalaam S., Asaithamby A., Santos C.X., et al. Hypoxia induces heart regeneration in adult mice. Nature. 2017;541:222–227. doi: 10.1038/nature20173. [DOI] [PubMed] [Google Scholar]
- 65.Puente B.N., Kimura W., Muralidhar S.A., Moon J., Amatruda J.F., Phelps K.L., et al. The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response. Cell. 2014;157:565–579. doi: 10.1016/j.cell.2014.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.