SUMMARY
Palmoplantar skin is structurally and functionally unique, but the transcriptional programs driving this specialization are unclear. Here, we use bulk and single-cell RNA sequencing of human palm, sole, and hip skin to describe the distinguishing characteristics of palmoplantar and non-palmoplantar skin while also uncovering differences between palmar and plantar sites. Our approach reveals an altered immune environment in palmoplantar skin, with downregulation of diverse immunological processes and decreased immune cell populations. Further, we identify specific fibroblast populations that appear to orchestrate key differences in cell-cell communication in palm, sole, and hip. Dedicated keratinocyte analysis highlights major differences in basal cell fraction among the three sites and demonstrates the existence of two spinous keratinocyte populations constituting parallel, site-selective epidermal differentiation trajectories. In summary, this deep characterization of highly adapted palmoplantar skin contributes key insights into the fundamental biology of human skin and provides a valuable data resource for further investigation.
Graphical Abstract
In brief
Wiedemann et al. present a comprehensive characterization of human palmoplantar skin. They find a dampened palmoplantar immune environment, describe differences between palmar and plantar skin, characterize a distinct palmoplantar fibroblast population marked by differential cell-cell signaling, and identify a site-selective dual epidermal differentiation trajectory.
INTRODUCTION
The palmar skin of the hand and the plantar skin of the foot are similar in that the epidermis, and especially the cornified layer, is thicker than in other regions of the body, and there is increased interdigitation between the dermis and the epidermis.1 These factorsare thought to contributeto palmoplantar skin’s increased mechanical strength, as the plantar skin is regularly able to withstand pressures 50 times that of other load-bearing regions of the body.2 Previous studies established that the expression of keratin 9 in the suprabasal layers of the palmoplantar epidermis is important to maintaining its structural integrity and correct terminal differentiation.3,4 In addition, other stress response keratins such as keratins 6 and 16 have been shown to be particularly important in palmoplantar skin differentiation, with mutations of these genes leading to palmoplantar keratodermas.5,6 These and other studies have added to the impression of sameness between palmar and plantar skin. Much less, however, is known about the differences between palmar and plantar skin.
In addition, site-specific manifestations of a variety of skin disorders have been noted. Palmoplantar manifestations, particularly in the case of inflammatory skin diseases such as psoriasis and atopic dermatitis, respond poorly to treatment compared with other body sites and typically require a different treatment regimen.7,8 Yet, the specific modifications of immune response as well as differences in immune cell types found in these sites are not fully understood.
Understanding the control of site-specific epidermal differentiation is also important for understanding skin diseases with altered epidermal differentiation. The distinct patterning of palmoplantar skin is partially due to the influence of a site-dependent fibroblast HOX code on epidermal differentiation.9,10 The induction of the characteristic thickened epidermal phenotype of palmoplantar skin has been achieved via co-culture of keratinocytes with palmoplantar fibroblasts.11 Although these facets of epidermal-mesenchymal signaling have been well established, these interactions have yet to be studied at the single-cell transcriptomic level. Having information at a single-cell level allows for an unbiased, inclusive analysis of signaling pathways at these different sites that was previously impossible. Recent human single-cell studies in healthy skin12–14 have established that fibroblast heterogeneity is important for maintaining dermal structure and enabling immune surveillance. However, these studies have not compared body site-specific differences.
The conventional model of epidermal differentiation is one of stepwise progression, corresponding to the distinct layers of the skin, with transit into each layer marked by a sharp transcriptional activation of a well-characterized series of markers.15 More recent single-cell studies in the mouse indicate that epidermal differentiation is instead gradualistic,16 while human studies have highlighted the heterogeneity within what were previously thought to be discrete epidermal populations.17,18 So far, the site-specific differences in human epidermal populations and their differentiation programs have not been studied.
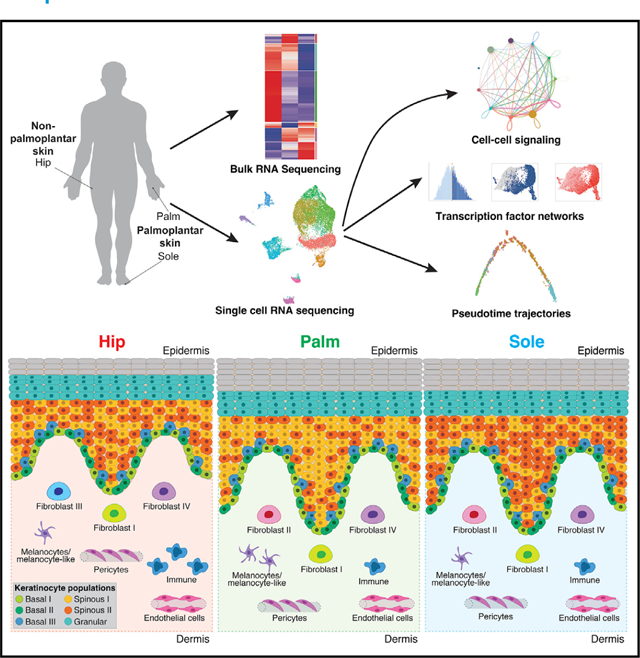
To better understand human epidermal differentiation in different sites of the body, we leveraged both bulk and single-cell transcriptional data from healthy adult donors. Samples were collected from three sites: palm and sole, representing palmoplantar skin, and hip, representing non-palmoplantar skin. Bulk transcriptionalanalysisrevealedhighlydivergent immunological states in palmoplantar and non-palmoplantar skin and, unexpectedly, suggested the presence of specific transcriptional programs in palmar versus plantar skin. To determine the contributions of individual cell types to these site-specific features, we performed single-cell RNA sequencing. We found a marked increase in immune cells in non-palmoplantar skin, a fibroblast population specific to palmoplantar skin, and distinct epithelial-mesenchymal cross-talk patterns in palmoplantar versus non-palmoplantar skin. Focusing on keratinocytes, we observed that the fraction of basal keratinocytes in the palm was markedly decreased, and the fraction of differentiated keratinocytes increased, compared with the sole. Keratinocyte subclustering also uncovered the presence of two discrete spinous cell populations, each marked with distinct transcriptional networks, which show differential enrichment in palm versus sole. Pseudotime trajectory analysis revealed these populations to be concurrent, alternate branches in epidermal differentiation. These spinous populations are conserved across all three sites, albeit in different proportions, and culminate in the same terminally differentiated end state. Altogether, our data support clinical observations of altered inflamma-tory involvement of palmoplantar skin and reveal the specific features of palmar, plantar, and non-palmoplantar skin.
RESULTS
Non-palmoplantar, palm, and sole skin are distinct at a transcriptional level
To identify differences in gene expression between palmoplantar and non-palmoplantar (hip region) skin, we took skin biopsies from 15 subjects, obtaining 30 samples, 10 of which were matched for hip and palm and 5 of which were matched for hip and sole (Figure 1A and S1A). Principal-component analysis (PCA) indicates that the majority of the variability in these samples was due to skin site, with the palmoplantar and non-palmoplantar skin separating into distinct clusters in the first principal component (Figure 1B). But it also indicates subtler differences between palmar and plantar skin, as the palm and sole form discrete clusters in the second principal component; the sole is most distinct from hip skin.
Figure 1. Bulk RNA sequencing reveals differences between palmoplantar and non-palmoplantar skin.
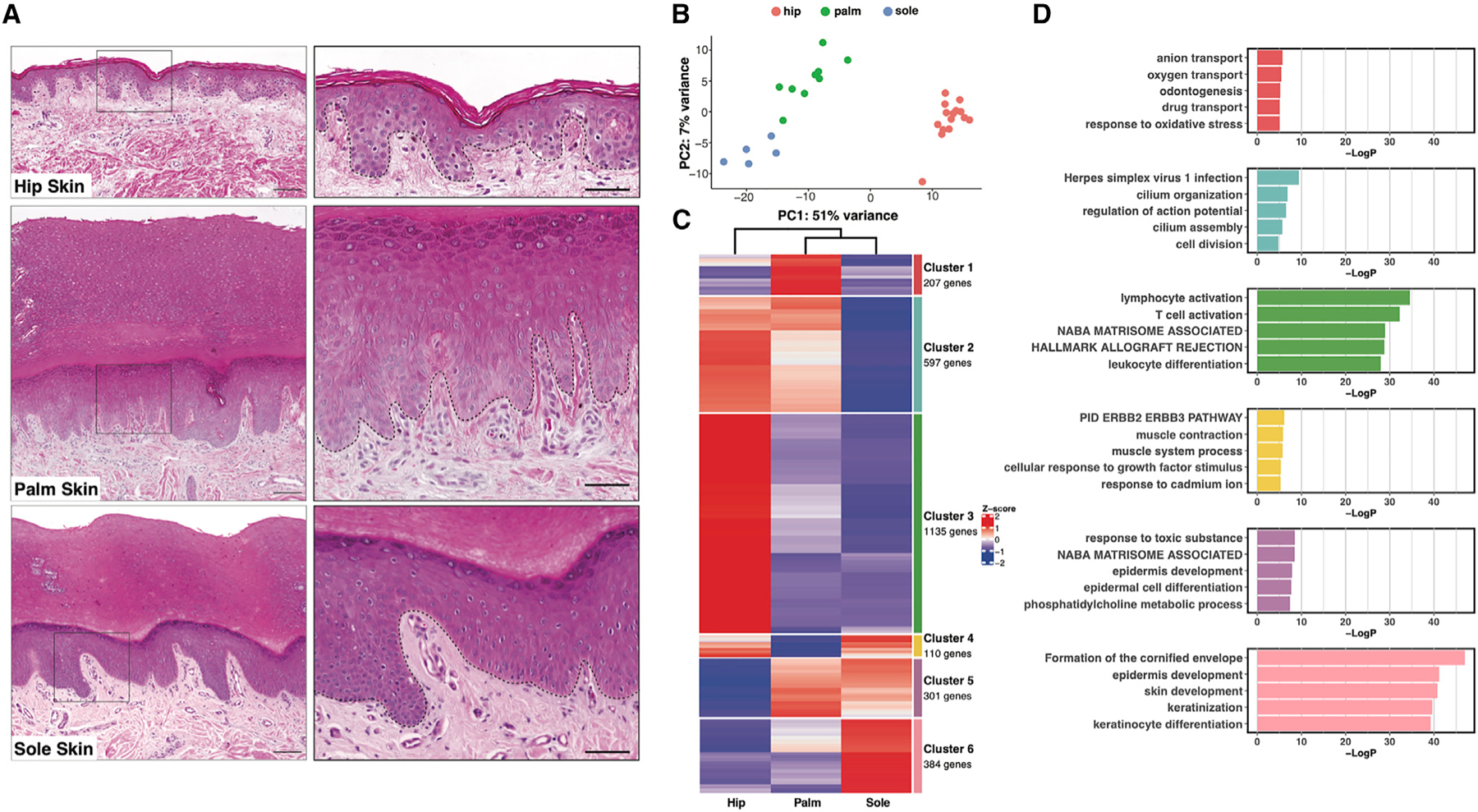
(A) H&E staining of hip, palm, and sole skin. Scale bars: 180 mm (left), 80 mm (right). Black dotted line, dermal-epidermal junction.
(B) Principal-component analysis (PCA) plot for 30 samples (15 from hip, 10 from palm, 5 from sole).
(C) Heatmap visualizing average expression by site of six gene clusters generated by k-means clustering of the 2,734 differentially expressed genes.
(D) Bar plot showing the top five gene ontology (GO) terms for each cluster of genes.
To further interrogate the gene expression differences between these three sites, we performed pairwise comparisons between the hip and the palm, the hip and the sole, and the palm and the sole using DESeq2. We identified a total of 2,734 significantly differentially regulated genes (Figures S1A). To detect distinct and shared patterns of gene expression across the three sites, we performed k-means clustering followed by gene ontology (GO) analysis to define gene pathways enriched or depleted at each site. The genes grouped into six different clusters, each with enrichment of certain functional categories (Figures 1C, 1D, and S1B–D).
Notably, GO terms emerging from this analysis are most significant for clusters 3 (enriched in hip) and 6 (enriched in sole), suggesting that the transcriptional divergence at these sites translates to functional distinctions. Intriguingly, GO terms for cluster 3 (hip), the largest cluster at 1,135 genes, primarily comprise inflammatory and immune response-related terms. This striking enrichment of immune-related genes in hip skin likely accounts for the propensity for many inflammatory skin diseases in non-palmoplantar skin compared with palmoplantar skin. Cluster 3 genes include immune cell-specific markers such as CD3D and CD79A, indicating a higher proportion of immune cells in hip skin, as well as immune-related genes that are expressed by skin tissue cells; these include factors such as TLR2, involved in innate immune responses,19 and CCL27, which drives immune cell recruitment.20 Cluster 3 also includes genes such as hair keratins KRT31, KRT74, and KRT85 related to differences in adnexal structures across the sites, as well as melanocyte markers MLANA, TYR, and TYRP1 and Langerhans cell markers CD1A and CD207 (langerin), corresponding to cell types that are more abundant in non-palmoplantar skin.21 Cluster 6 (sole) is a far smaller cluster, at only 384 genes, but nonetheless shows highly significant GO terms, most of which relate to cornification and keratinization. This corresponds to the greater degree of terminal differentiation in the epidermis of the sole compared with the palm, consistent with the more intense stress and pressure regularly encountered by plantar skin. Accordingly, stress keratins KRT6A/B/C, KRT16, and KRT17 all emerge in cluster 6.
In contrast, despite the transcriptional similarity of palm and sole shown by PCA (Figure 1B), GO terms associated with cluster 5 (representing genes enriched in both palm and sole relative to hip) show comparatively low significance, further highlighting the prominent functional distinctions between palm and sole. The top GO term, response to toxic substance, is driven by antioxidants such as thioredoxin and peroxiredoxins 1 and 6, as well as numerous other factors involved in responses to reactive oxygen species, hinting at possible metabolic differences in palmoplantar skin. Cluster 5 further includes anticipated genes such as FLG (filaggrin), LOR (loricrin), and HRNR (hornerin). However, additional interesting structural genes, such as COL17A1, a critical hemidesmosome component anchoring down the basal epidermis, and desmosomal components DSC1 (desmocollin 1) and DSG1 (desmoglein 1), emerge, as well as a number of immune genes such as IL36G and IL36RN (IL-36 receptor antagonist) that likely further contribute to differences in palmoplantar presentations of immune-mediated skin disease. In addition, cluster 1 (palm) GO terms showed only marginal significance, with many terms being driven by hemoglobin gene transcripts likely derived from residual erythrocytes inadvertently captured in the microvasculature. These transcriptional signals suggest greater functional specialization of non-palmoplantar and plantar than of palmar skin. While our results agree with previous work3,22–24 showing that palm and sole have common characteristics that are distinct from non-palmoplantar skin, we now show important differences between palm and sole.
scRNA-seq reveals differences in cell states and cell types between hip, palm, and sole skin
We next used single-cell RNA sequencing (scRNA-seq) to define cell-type and cell-state differences between palmoplantar and non-palmoplantar skin and to map gene expression differences between palmoplantar and non-palmoplantar skin to specific cell types. Skin biopsies were taken from four subjects; from two subjects, we obtained samples from hip and palm, and from the other two subjects, from hip and sole. In each case, the epidermis and dermis were enzymatically separated and then either recombined or sequenced separately, resulting in a total of 12 datasets (Figure 2A). After quality control, normalization, and integration into a single object in Seurat,25,26 the dataset consisted of 15,243 cells and 17,261 genes (Figure S1E).
Figure 2. Hip, palm, and sole show major differences in cell-type composition.
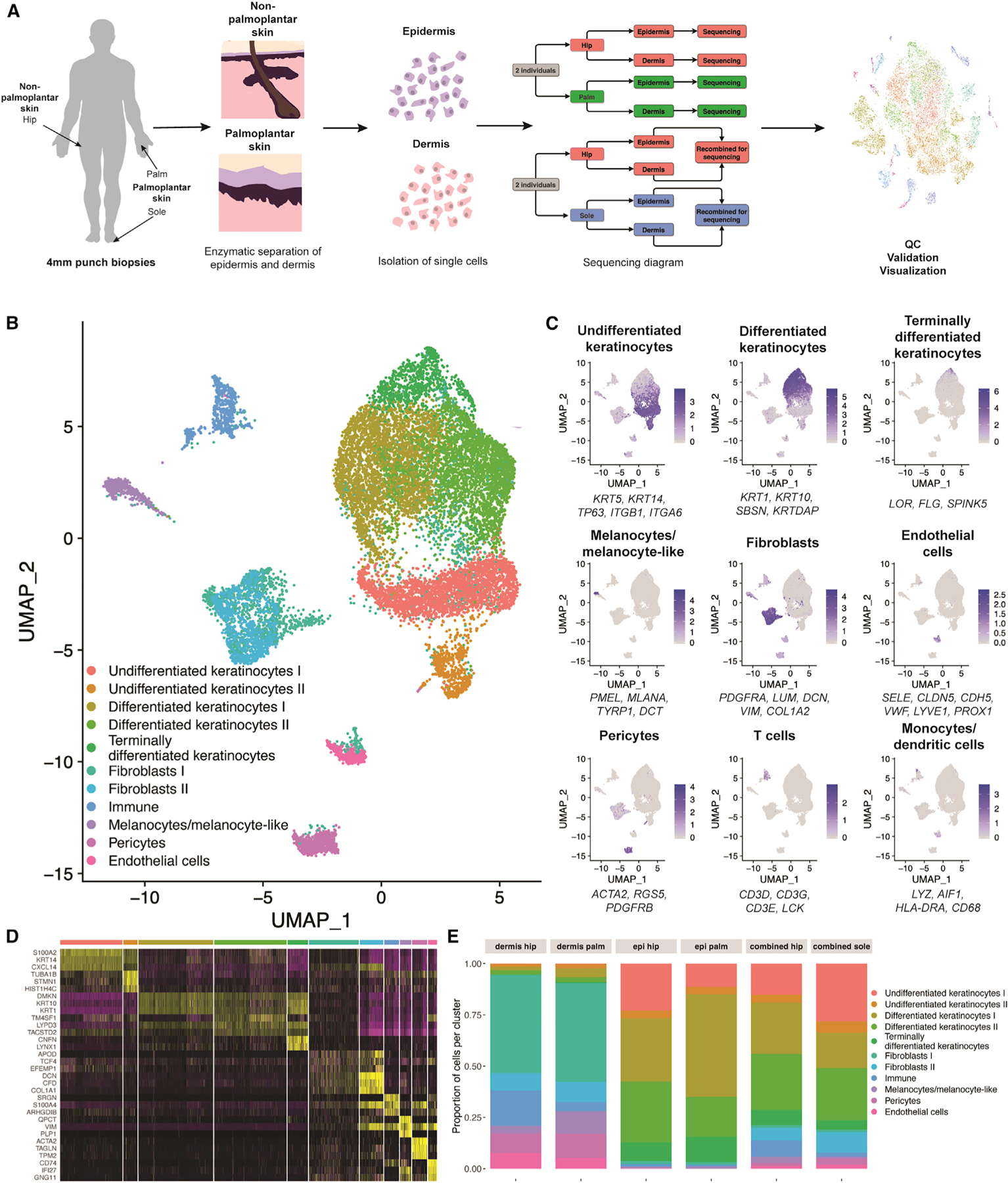
(A) Schematic of sequencing method. Punch biopsies were taken from n = 4 research subjects, with half donating hip and palm and half donating hip and sole.
(B) Uniform manifold approximation and projection (UMAP) visualization of all 12 datasets. Each dot represents a single cell (n = 15,243). Colors were determined by unsupervised clustering performed by Seurat.
(C) Average expression of three to six well-established gene markers projected onto the UMAP plot to annotate cell types.
(D) Heatmap showing the top three most differentially expressed genes for each cluster, as determined by Seurat. Each row represents expression of the marker gene, while each column is an individual cell.
(E) Proportion of cell types normalized by the total number of cells in that sequencing type and site. Because of differences in sequencing, proportions of cell types are best compared within conditions.
Unsupervised clustering of the integrated dataset identified 11 clusters, including keratinocytes, fibroblasts, immune cells, melanocytes/melanocyte-like cells, pericytes, and endothelial cells (Figures 2B and S1F).26,27 These annotations were determined by the projection of established marker genes for each cell type, as well as the top marker genes for each cluster (Figures 2C and 2D).14 To avoid bias introduced by differences in sample preparation, we performed pairwise comparisons (Figures 2E and S1G). As predicted by the bulk RNA-seq data (Figures 1C and 1D), there was a consistent decrease in the number of immune cells, including T cells and mast cells in the sole and T cells, mast cells, Langerhans cells, and other myeloid populations in the palm, compared with the non-palmoplantar skin (Figures 2E and S1H). In addition, we identified downregulation of ITGB28 and TIMP,29 associated with leukocyte migration, and SL2A3,30 associated with Th17 and other immune cell activation, in multiple palmoplantar immune populations (Figure S1I).
The melanocytes/melanocyte-like cluster showed expansion in the palm dermis (11%) compared with hip dermis (3.6%) (Figure 2E), in disagreement with the enrichment of melanocyte markers in bulk RNA-seq cluster 3 (Figure 1C). This cluster showed the opposite trend in the hip and sole matched samples, with a greater fraction (3%) of melanocytes/melanocyte-like cells in the hip and a lesser fraction (0.9%) in the sole. Although this cluster was annotated based on high expression of traditional melanocyte markers, such as KIT and MLANA, these markers were expressed only in a subset of the cluster. In the palm specifically, there was an expansion of the melanocyte-like population that lacked expression of pigment genes (Figure 2C). This agrees with a recent study that reported detection of a melanocyte sub-population with diminished expression of pigment genes and other melanocyte markers in acral skin.31 There were also more differentiated keratinocytes in the palm than in the hip. In comparison, the sole showed greater enrichment in undifferentiated keratinocytes than the hip (Figure 2E). These results support the two major findings of the bulk RNA-seq analysis, corroborating an expanded immune role in the non-palmoplantar skin, as well as differences between palm and sole epidermis.
Palmoplantar skin contains a distinct fibroblast population and features specific cell-cell signaling
Subclustering of 2,303 fibroblasts from the three sites resulted in the formation of four fibroblast clusters (Figures 3A and S2A). Fibroblasts II and III show non-overlapping localization, with fibro-blast II found nearly exclusively in the palmoplantar skin and fibroblast III found only in non-palmoplantar skin (Figures 3B and S2B). However, these two clusters share expression of markers such as WIF1, MMP2, SFRP2, and DPP4, suggesting a shared role in maintaining dermal and extracellular matrix homeostasis (Figures 3C, S2C, S2D, and S2G).12,14,32 The palmo plantar-specific fibroblast II also expressed ANGPTL7 and PRG4, markers of two previously identified minor fibroblast populations (Figures S2G and S2H).12
Figure 3. Palmoplantar skin contains a distinct fibroblast population.
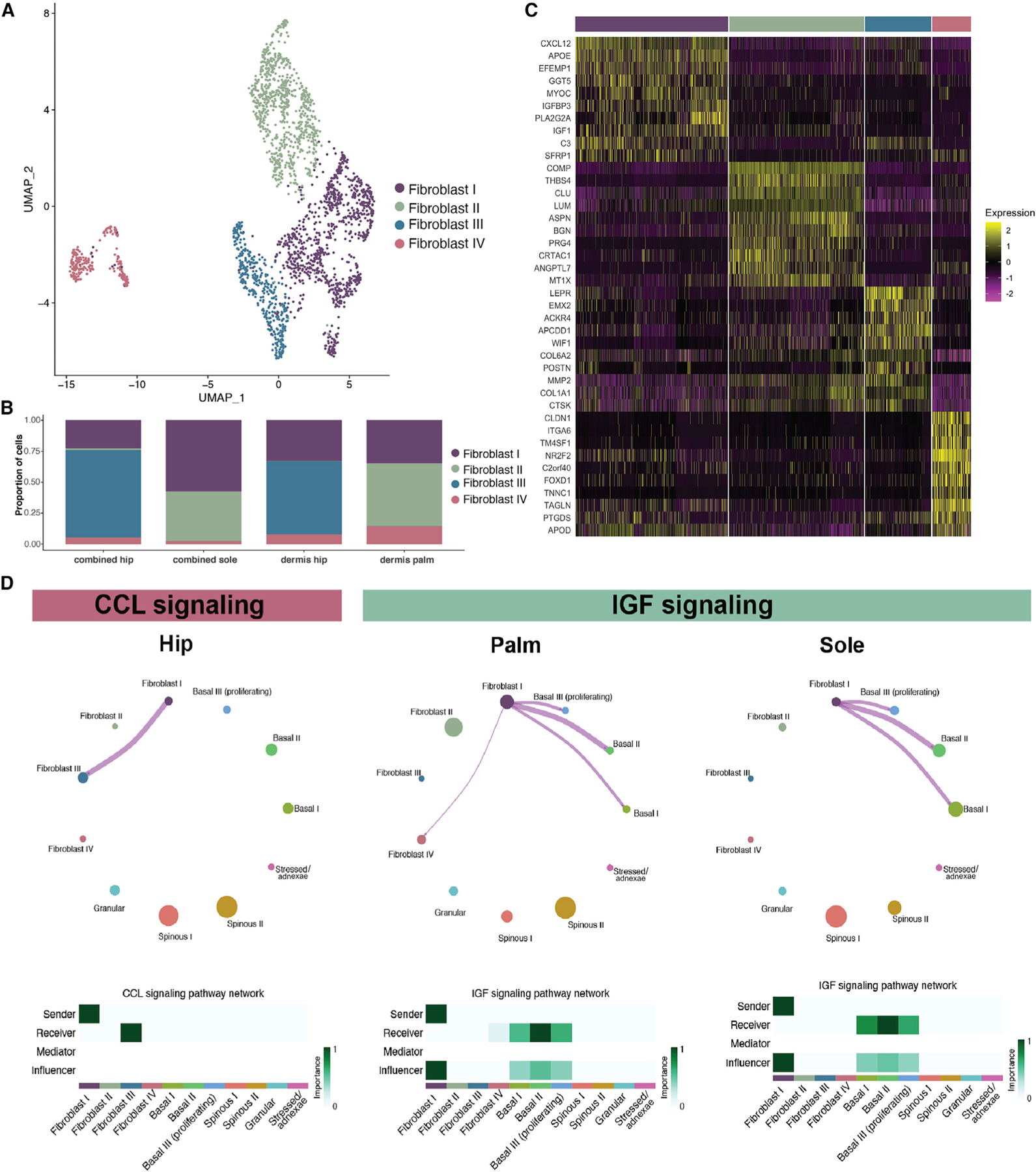
(A) UMAP visualization of fibroblast subclusters. Each dot represents a single cell (n = 2,303).
(B) Proportion of fibroblast subclusters normalized by the total number of cells in that sequencing type.
(C) Heatmap of top 10 marker genes for each of the four fibroblast subclusters.
(D) CellChat circle plots showing cell-cell communication related to CCL and IGF signaling in the indicated sites.
Fibroblast I is marked by upregulation of CXCL12, APOE, FMO1, and LSP1, pointing to a role in immune surveillance and promotion of inflammation (Figures 3C, S2C, S2D, and S2G).12,14,32 Fibroblast IV was marked by APOD, C2orf40, ITGA6, CLDN1, and TM4SF1, overlapping with a specialized fibroblast population previously identified in single-cell analysis of healthy human skin (Figures 3C and S2I).12,32
We calculated a reticular and papillary score32–37 and found that, although the clustering was not driven by localization in the dermis, fibroblast II is marked by a higher papillary score, while fibroblast III has a higher reticular score (Figures S2E and S2F). GO terms for these clusters further support fibroblasts II and III sharing a role in structural maintenance, as both are marked by a number of terms related to extracellular matrix organization (Figure S2J). MetaNeighbor analysis38 supported similarity between the site-specific fibroblast II and fibroblast III and indicated that more subtle differences exist between the hip and the palmoplantar skin in the immunologically active fibro-blast I (Figure S2K). Pairwise comparison of these two populations further found that the palmoplantar-specific cluster (II) shows upregulation of GO terms associated with cell migration, while the non-palmoplantar cluster (III) shows upregulation of WNT signaling terms (Figures S2L–S2N).
We next utilized CellChat39 to infer cell-state-specific signaling communications for the fibroblast and keratinocyte populations for the three sites independently (Figure S3A). This unbiased analysis of signaling pathways revealed key differences between the non-palmoplantar and the palmoplantar skin. First, fibroblast cluster II, found in the palmoplantar skin, and fibroblast cluster III, in the non-palmoplantar skin, play comparable roles in overall signaling, supporting that these two fibroblast populations have similar functional roles in the different sites (Figure S3B). Second, in all three sites, fibroblast cluster I is consistently the fibroblast subtype most involved in signaling. Overall, information flow and signaling strength are higher in the hip and palm than in the sole (Figure S3C). Unbiased signaling analysis revealed specific signaling communications in each of the three sites (Figure S3D).
Notably, CCL signaling within the fibroblasts is exclusively identified in the hip dataset, showing that the increased inflammatory role of non-palmoplantar skin extends to the fibroblast populations (Figures 3D, S3D, and S3H). In contrast, IGF signaling between the fibroblasts and the basal keratinocytes was found only in the palm and sole, indicating that this proliferative signal to the keratinocytes is more prominent in the palmoplantar skin, where it likely contributes to increased epidermal thickness (Figures 3D, S3D, S3I, and S3J). To ensure that these results were not driven by differences in sequencing, we independently analyzed hip datasets from the epidermis and dermis sequenced separately and those that were recombined for sequencing. This analysis recapitulated our findings from the aggregated analysis (Figures S3E–S3G). Having established these clear differences in dermal signaling pathways between the palmoplantar and the non-palmoplantar skin samples, we moved to further investigate the differences in the epidermal populations between sites.
The human epidermis contains multiple clusters of cell states that both divide and span epidermal layers
To understand body-site differences in epidermal differentiation, we focused our studies on the 9,471 keratinocytes from hip, palm, and sole. Subclustering the keratinocytes resulted in seven clusters: three basal clusters (basal I, II, III), expressing KRT5 and KRT14; two spinous clusters (spinous I, II), expressing KRT1 and KRT10; a single granular cluster (granular), expressing FLG and LOR; and one cluster (stressed/adnexae) marked by higher expression of stress-response genes such as KRT6B, as well as genes expressed in the pilosebaceous units and eccrine glands, i.e., MGST1 (Figures 4A and S4A). We also projected onto the uniform manifold approximation and projection (UMAP) an average expression score for canonical differentiation markers to validate the cluster assignments (Figure 4B). These clusters are consistent regardless of dataset and Seurat integration (Figures S4B and S4C). The three basal clusters show similarities to those previously identified in neonatal foreskin,17 with both basal I and basal II being most similar to the stem cell population found at the bottom of rete ridges and basal III to the two proliferative clusters located closer to the suprabasal layer. The fourth basal cluster identified in the neonatal keratinocytes, which localized to the top of the rete ridges, appears to be absent in adult epidermis (Figures S4D and S4E). Although the keratinocyte subclusters are clear and match the expression of canonical markers, we observed transition cells expressing both basal and spinous markers, as has been described in mouse epidermal differentiation,16 indicating gradualistic features of human epidermal differentiation (Figures S4F and S4G).
Figure 4. Consistent keratinocyte populations are detected in adult skin.
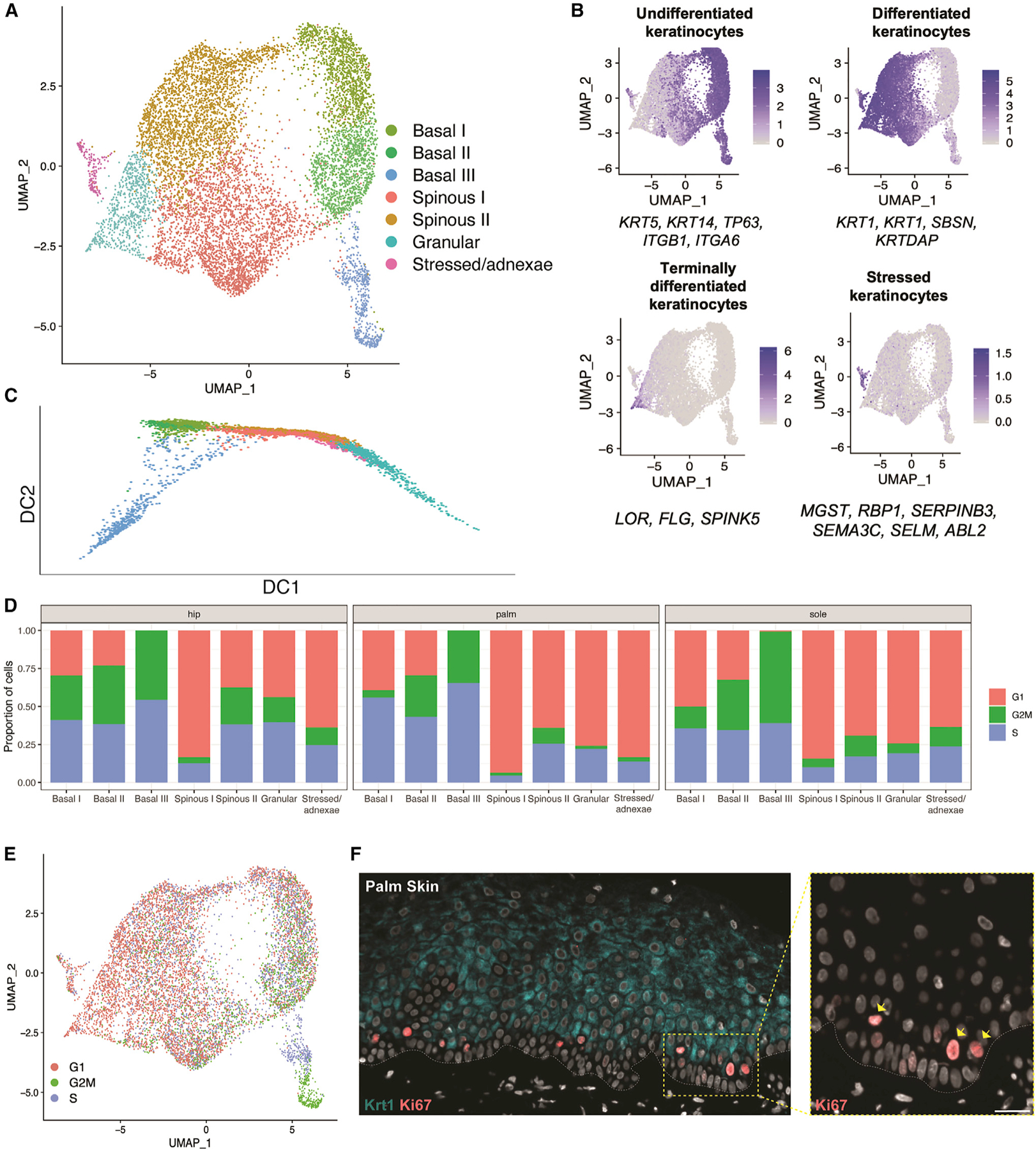
(A) UMAP visualization of keratinocyte subclusters. Each dot represents a single cell (n = 9,801).
(B) Average expression of three to six established keratinocyte marker genes projected onto UMAP plots to identify keratinocyte subclusters.
(C) Diffusion map of keratinocyte subpopulations.
(D) Proportion of cells in each cell-cycle phase (as assigned by the CellCycleScoring function in Seurat) split by site and keratinocyte subcluster.
(E) Keratinocyte UMAP colored by cell-cycle phase.
(F) Co-immunostaining in the palm skin of proliferation marker Ki67 and differentiation marker KRT1. Inset: arrows indicating suprabasal and basal Ki67-positive cells. White dotted line, dermal-epidermal junction. Scale bar: 60 μm.
Proliferative epidermal stem cells are mostly located in the first suprabasal layer and show increased differentiation
Using Scanpy40 to generate a diffusion map of all keratinocytes, we identified a clear trajectory in the first diffusion component, starting with the three basal clusters, moving in parallel through either spinous I or spinous II, and culminating in the terminal granular state, with stressed/adnexae keratinocytes found throughout (Figure 4C). The second diffusion component shows a clear separation of basal III and the granular cluster, which appears to be driven by proliferation and terminal differentiation, respectively. Cell-cycle scoring highlights a consistent decrease in proliferation throughout differentiation, as shown by the increase in G1-phase cells compared with basal cells (Figure 4D). Interestingly, the two spinous populations appear as parallel tracks in the diffusion trajectory. We also found consistent differential proliferation across all three sites, with spinous I keratinocytes largely marked by G1-phase genes, whereas spinous II has a greater number of keratinocytes marked by G2M- and S-phase genes.
Basal III, unlike the other two basal clusters, is entirely composed of cycling cells, with one-half of the cluster in G2M and the other half in S phase (Figure 4E). Immunostaining of proliferation marker Ki67 in palm, sole, and hip skin shows that basal III cells are in fact localized above the first layer of basal cells (Figures 4F and S4H). This is consistent with the proliferating basal III cells being more differentiated than basals I and II, as supported by basal III having the largest fraction of cells co-expressing basal and suprabasal markers across the three sites (Figure S4G).
Palmoplantar keratinocytes exhibit increased expression of oxidative phosphorylation-related genes but decreased expression of immune-related genes
Based on conserved cell state definitions across the three sites, we next investigated differential gene expression among hip, palm, and sole keratinocytes. As expected, KRT9, a well-known marker of suprabasal keratinocytes in palmoplantar skin,4,22,23 is strongly expressed in the differentiated keratinocytes of the palm and sole (Figure S4I). In contrast, KRT16, known to be associated with palmoplantar epidermal differentiation,24,41 is particularly highly enriched in the sole, but also expressed in palmar keratinocytes. CCL27, encoding a T cell activating protein and one of the top upregulated genes in non-palmoplantar skin in the bulk RNA-seq analysis, is selectively expressed in all hip keratinocytes. Pairwise comparisons of the palm versus hip and sole versus hip (between keratinocytes sequenced in the same manner), showed high correspondence with genes identified in the bulk analysis, including palmoplantar upregulation of transcripts such as KRT9, KRT6, KRT16, GJB2, and GJB6 (Figure S4J).
Next, we examined differences in gene expression between palmoplantar and non-palmoplantar skin in each keratinocyte subcluster, performing pairwise comparisons between matched samples. To explore the significance of these differences, we performed gene set enrichment analysis (GSEA)42,43 to identify GO terms enriched in each cell state, as well as those enriched in the bulk data (Figures S4K and S4L). In both the palm and the sole, pathways associated with epidermal development and cornification were upregulated in multiple populations. In addition, there was a strong enrichment for terms associated with oxidative phosphorylation and the electron transport chain, which may indicate divergent metabolic states in palmoplantar and non-palmoplantar epidermis. This altered metabolism in palmoplantar keratinocytes is further supported by similar results in the bulk RNA-seq from the sole (Figure S4K), hearkening back to the emergence of response to toxic substances as the top significant GO term for palm- and sole-enriched gene cluster 5 (Figure 1D).
Non-palmoplantar keratinocytes are markedly enriched in a number of immunological GO terms, including TNF-a, leukocyte differentiation, lymphocyte and T cell activation, and cytokine signaling (Figure S4L). These results, paired with the increased proportion of immune cells in the non-palmoplantar skin (Figure 2E) and the enrichment of similar immune GO terms in the non-palmoplantar skin from the bulk data (Figures 1D, S1C, and S1D), reiterate the existence of distinct immune environments in palmoplantar and non-palmoplantar skin.
Stem cell and spinous differentiation properties are distinct between palm and sole epidermis
Having established key differences between palmoplantar and non-palmoplantar keratinocytes, we then looked for differences in the proportion of cell states in the keratinocyte populations, which could reflect differences in epidermal differentiation among hip, palm, and sole skin. As expected, the palmoplantar epidermis differs from non-palmoplantar epidermis in the distribution of cell states (Figures 5A and 5B). But more surprising are striking differences between palmar and plantar skin types. Plantar epidermis is enriched 1.5-fold in basal keratinocytes, including from proliferating basal III, compared with the hip epidermis. In contrast, the proportion of basal cells in palmar epidermis is decreased nearly 2-fold compared with hip. In addition, the relative ratio of spinous I and spinous II varies between sites, with palm favoring spinous I and sole favoring spinous II, whereas spinous I and spinous II keratinocytes are approximately equal in the hip skin. These differences in proportion are consistent between samples from different individuals (Figure S4B). In conclusion, these results indicate (1) that plantar epidermis is specific in its enrichment of basal keratinocytes, including proliferating cells, and (2) that the epidermal differentiation pathways in palm and sole are different, with the palm favoring spinous I differentiation and the sole favoring spinous II differentiation.
Figure 5. Notch signaling intensity in keratinocytes differs across hip, palm, and sole.
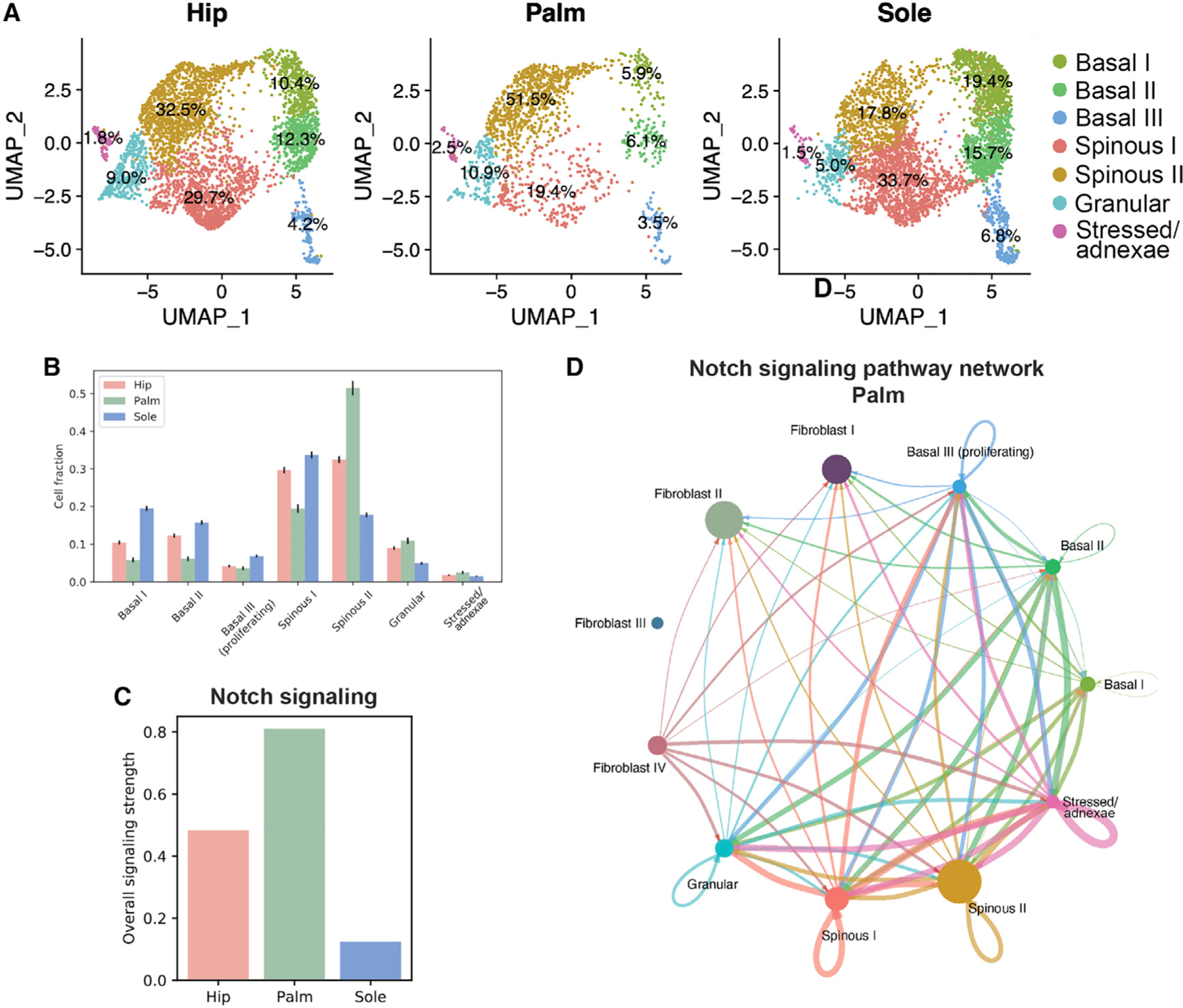
(A) UMAP visualization split up by the three sites (hip, n = 3,845; palm, n = 1,431; sole, n = 4,195), with respective proportions of each cluster.
(B) Bar plot showing cell fraction for each site by cluster. Error bars indicate standard deviation.
(C) Bar plot showing signaling intensity of Notch signaling in the three sites.
(D) Circle plot showing CellChat results for overall Notch signaling in the palm.
CellChat39 inference shows major differences in signaling in keratinocyte populations across the three sites. Strikingly, the strength of Notch signaling, which occurs largely between the keratinocyte populations, is inversely correlated with the size of the basal cell fraction. The palm, which has the smallest fraction of basal cells, has the strongest Notch signal, while the sole has both the largest fraction of basal cells and the weakest Notch signal (Figures 5B–5D and S5A–S5E). This, paired with the correlation between intensity of Notch signaling and granular and total spinous (I and II) cell fractions (Figures 5B–5D), supports the induction of epidermal differentiation via Notch that has been reported in the literature.44,45
Human epidermis uses two parallel differentiation pathways in a site-selective manner
The differences in proportion of the two spinous clusters in hip, palm, and sole point to two distinct differentiation trajectories in which keratinocytes move from the basal state through either the spinous I or the spinous II state but reconverge into a shared terminal differentiation state in the granular cluster. Comparing the two spinous clusters reveals a number of differentially expressed genes and transcription factors (Figure 6A). Analysis with MetaNeighbor38 validated the Seurat clustering, confirming that these populations are present across all sites and demonstrating that the two spinous clusters differed considerably, exhibiting greater similarity to the stressed/adnexae and granular clusters than to each other (Figure 6B). This contrasts with the three basal clusters, which demonstrate distinct markers, but show far greater intergroup similarity.
Figure 6. Adult epidermis contains two distinct spinous populations.
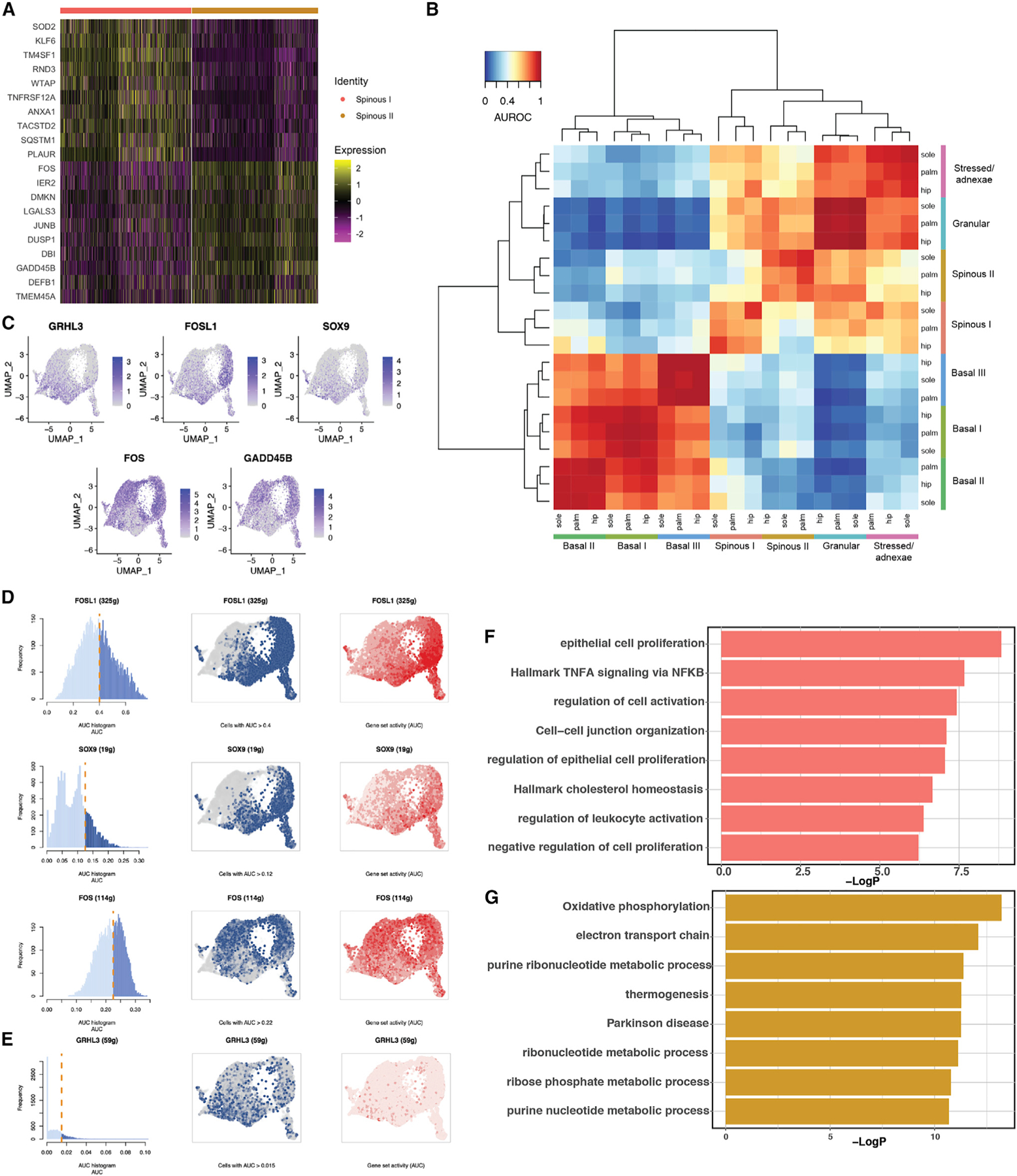
(A) Heatmap of top differentially expressed genes between spinous I and spinous II.
(B) Heatmap of AUROC scores between keratinocyte clusters based on the highly variable gene set using MetaNeighbor.
(C) Feature plot showing the most distinct markers for spinous I and II populations.
(D) SCENIC result, with cutoff for AUC score for each transcription factor network (left), UMAP with cells above the given AUC score threshold colored blue (middle), and UMAP with cells colored by the AUC score in red (right).
(E) AUCell scoring of GRHL3 network based on Klein et al.46 Panels as in (D).
(F) Bar plot showing specific enrichment of GO terms for spinous I.
(G) Bar plot showing specific enrichment of GO terms for spinous II.
As reflected in the differentially expressed genes (Figure 6A), multiple transcription factors with well-known roles in epidermal differentiation vary substantially in expression between the two spinous populations across all skin sites (Figure S5F). Spinous II, which is enriched in the palm, is marked by high expression of FOS and GADD45B. In contrast, spinous I, enriched in the sole, lacks FOS expression but shows expression of GRHL3, FOSL1, and SOX9 (Figure 6C). Next, using SCENIC,47 we found that the transcription factor networks validate these results, with increased activity of the FOS network in spinous II and the FOSL1 and SOX9 networks in spinous I (Figure 6D). Although the GRHL3 network from SCENIC did not show a significant difference between clusters, AUCell scoring of a gene list of previously identified direct GRHL3 targets is consistent with increased activity in spinous I.46,47
We then performed GO term enrichment on the two spinous clusters. Although both clusters share terms associated with epidermal development and keratinocyte differentiation, there are key differences (Figures 6F, 6G, and S5G). While the GRHL3+ cluster (spinous I) is marked by terms such as regulation of epithelial cell proliferation, cell-cell-junction organization, and regulation of immune activation, the FOS+ cluster (spinous II) is marked by oxidative phosphorylation, electron transport, and a number of metabolic process terms, indicating these cells may bemarked bydistinctmetabolic modes. These different pathways, paired with the specific expression of key transcription factors in the two spinous populations, support the concept that these two paths play separate roles in epidermal differentiation.
A recent study also identified concurrent differentiation trajectories in healthy adult keratinocytes from the breast driven by high or low lamellar body (LB) formation in differentiated keratinocytes.18 Although our spinous I population is marked by expression of SOX9, the marker of the LB branch in the breast keratinocytes, the expression of LB- and congenital ichthyosis-related genes was slightly upregulated in the spinous II population (Figure S5H). Integration of the breast skin dataset with ours48 revealed that the LB and non-LB branches in the breast are driven by site in our dataset, with sole associated with LB and hip associated with non-LB, rather than overlapping with our two spinous clusters (Figures S5I–S5K). Projection of spinous I and II markers onto the integrated dataset revealed that GRHL3 expression was higher in the LB branch, while FOS expression was higher in the non-LB branch in both datasets (Figures S5L and S5M). These results indicate that the differences in the two spinous populations, and thus the two differentiation trajectories, are not driven by LB formation.
Spatial localization of two distinct spinous populations
To gain greater insight into the differentiation process across the three body sites, we performed pseudotemporal analysis using Monocle to order the keratinocytes based on differentiation status. Ordering of the Seurat clusters over pseudotime is consistent across all three sites (Figure 7A) and agrees with the earlier diffusion trajectory (Figure 4C), as well as the partition-based graph abstraction (PAGA) trajectory that included keratinocytes from all three sites (Figure S6A).49,50 Notably, although the differentiation trajectories for the hip and the palm are linear, there is a bifurcation in the basal part of the sole trajectory. Since one branch is dominated by cells from the basal III cluster, and the other comprises basal I and basal II clusters, this split is likely driven by the increased number of proliferative basal cells compared with that of the hip or palm (Figure S6B). In addition, the trajectories are consistent across the four research subjects (Figure S6C).
Figure 7. Spinous populations represent two differentiation trajectories with a shared end state.
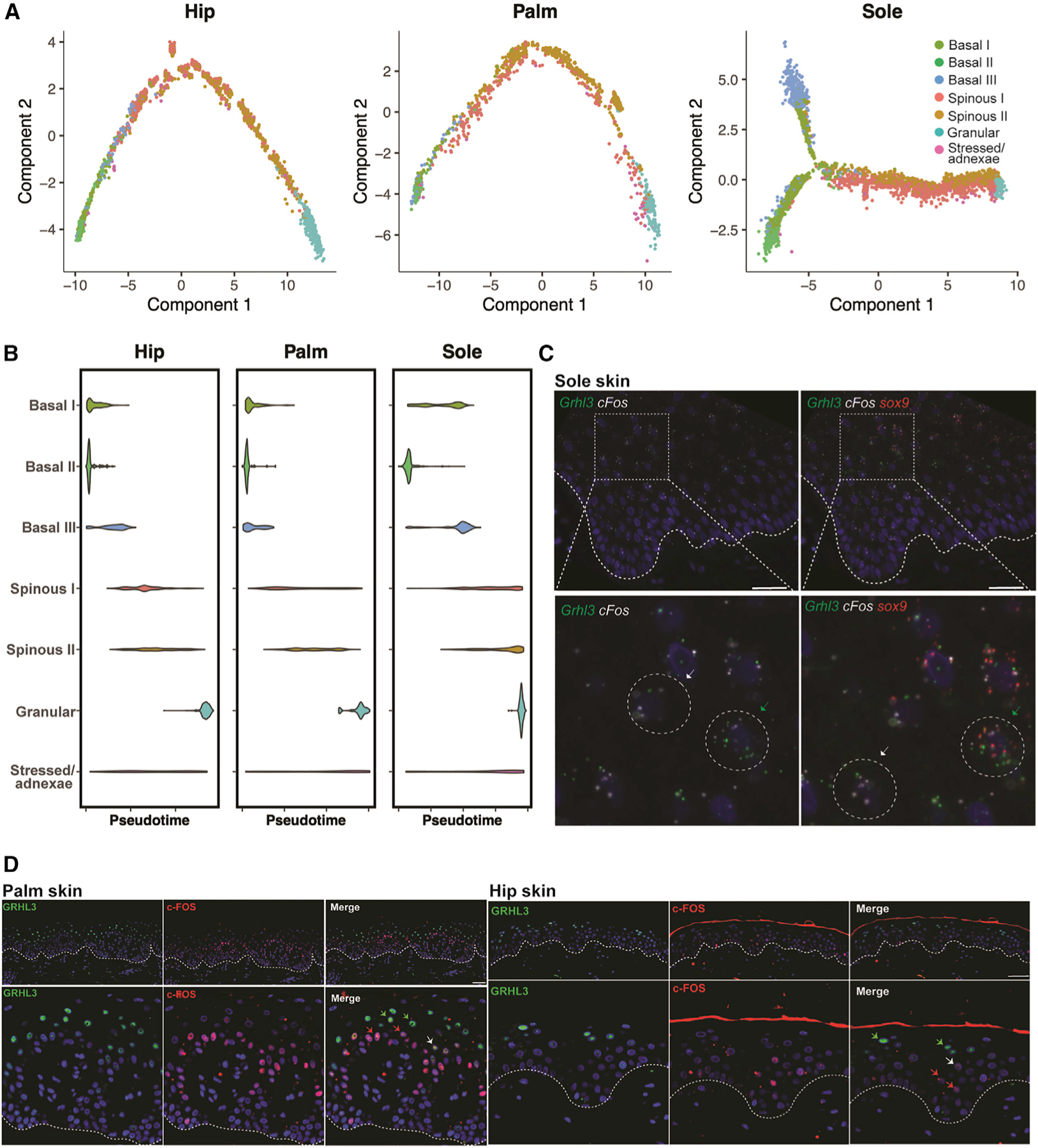
(A) Monocle pseudotime trajectories showing epidermal differentiation for each site.
(B) Violin plots showing distribution of keratinocytes in each cluster over pseudotime for the three sites.
(C) Immunostaining of spinous cluster markers GRHL3 and c-FOS in the palm. White dotted line, dermal-epidermal junction. Scale bars: 80 μm.
(D) Validation of the presence of two distinct spinous populations by co-immunostaining of GRHL3 and c-FOS in hip and palm skin. White dotted line, dermalepidermal junction. Scale bars: 150 mm.
Regardless of site, in pseudotime the two distinct spinous clusters are distributed contemporaneously rather than sequentially (Figure 7B). Thus, the pseudotime trajectory supports the concept of two independent spinous clusters that culminate in a shared terminal differentiation state. RNA fluorescence in situ hybridization (FISH) results reiterated the presence of two populations, with quantification of the spinous layer validating co-localization of GRHL3 and SOX9 and separate expression of FOS in the palm (Figures 7C and S6D–S6F). Differences in patterns of expression of GRHL3 and FOS (Figure 6C) account for the apparent stratification of expression outside of the spinous layer. No statistically significant differences in expression were seen between regions over the rete ridge (Figure S6G). Co-immunostaining further validated that the GRHL3+ and FOS+ cells are two distinct populations, with limited overlap in both palmoplantar (palm) and non-palmoplantar (hip) skin (Figure 7D). In addition, co-immunostaining of GRHL3 and SOX9 corroborated the existence of a population of double-positive cells identified in the single-cell data (Figure S6H). Ultimately, these data support the concept of two transcriptionally distinct differentiation trajectories in human keratinocytes that culminate in a common granular end state.
DISCUSSION
We have conducted comprehensive transcriptomic analysis of healthy adult human skin from three sites—hip, palm, and sole—at both the bulk and the single-cell level and with spatial validation. The study reveals a number of key insights about site-specific differences in skin structure and function. First, increased immune cell populations and heightened inflamma-tory environment characterize non-palmoplantar compared with palmoplantar skin. Second, fibroblast populations that confer key differences in cell-cell signaling selectively mark palmoplantar and non-palmoplantar skin. Third, palmar and plantar skin exhibit differences in keratinocyte subpopulations and gene expression. Fourth, there are two concurrent keratinocyte differentiation trajectories marked by distinct spinous cell populations that operate in a site-biased manner.
At a gene expression level, we demonstrate the greatest differences between non-palmoplantar and palmoplantar skin, with the largest gene cluster (cluster 3), containing two-fifths of the total differentially expressed genes, specifically upregulated in hip skin (Figure 1). Cluster 3 is marked by specific immune cell markers, such as CD3D, CD79A, and CD207, suggesting that hip skin contains a higher fraction of immune cells than palmoplantar skin. In addition, cluster 3 contains a number of genes expressed by skin cells, including innate immune-response genes and genes involved in immune cell recruitment. Together, these results support the notion of a more robust immune environment in hip skin compared with palmoplantar skin. Yet, we find evidence that palmoplantar skin may have distinct pro-inflamma-tory features as well. Cluster 5, which is enriched in palmoplantar skin, also contains distinct immune-related genes, including IL36RN, a key susceptibility gene for both generalized pustular psoriasis and certain palmoplantar pustulosis patients51,52; this feature could further contribute to palmoplantar manifestations of inflammatory skin diseases.
These bulk RNA-seq results are corroborated by the scRNA-seq data. We find a consistent increase in the number of immune cells in non-palmoplantar skin compared with palmoplantar skin, with an ~4-fold increase in both the matched hip versus palm dermis samples and the matched hip versus sole whole-skin samples and an ~1.5-fold increase in matched hip versus palm epidermis samples (Figure 2). In addition, specific CCL signaling between the fibroblasts and the keratinocytes in the hip (Figure 3) and consistent upregulation of immune-response terms in the hip in both keratinocyte populations and the bulk data (Figure S4) further support an increased inflammatory environment in the hip. Taken as a whole, our findings demonstrate an enhanced immune environment in the non-palmoplantar skin, which likely promotes the differential susceptibility to inflammatory disease, such as psoriasis and atopic dermatitis, between non-palmoplantar and palmoplantar sites.7,8
Our analysis also shows that specific fibroblast subtypes, as well as gene expression within subtypes, contribute to differences between non-palmoplantar and palmoplantar skin (Figure 3). Fibroblast I, which is found in both hip and palmoplantar skin, matches a previously described major fibroblast population defined by FMO1/LSP1 expression.12 This population is implicated in immune surveillance and promotion of inflammation.12,14,32 Regardless of location, fibroblast I is the population most involved in signaling, being a major sender of immune signaling via CCL in the hip and a major sender of growth signaling via IGF in palmoplantar skin. Fibroblasts II and III are both marked by SFRP2 and DPP4, matching a second major fibroblast population in human skin that is implicated in matrix deposition and extracellular matrix homeostasis.12 Intriguingly, fibroblasts II and III are, respectively, specific to palmoplantar and hip skin, indicating that this major fibroblast population marked by SFRP2 and DPP4 is specialized in a site-specific manner. We note that the palmoplantar fibroblast II shares certain key markers, namely ANGPTL7 and PRG4, with two other previously identified minor fibroblast populations,12 indicating potential specialization of this cluster. In addition, markers for these two clusters differ in enrichment of GO terms, including cell migration terms in the palmoplantar-specific fibroblast II and WNT signaling terms in the hip-specific fibroblast III.
The final cluster, fibroblast IV, is marked by expression of ANGPTL7 and C2orf40, matching a minor, specialized population of fibroblasts,14 referred to as C4,32 the function of which has not been previously determined. This population is also defined by exclusive expression of ITGA6 and ITGB4, which have been shown to be expressed specifically in Schwann cells and perineural fibroblasts in the dermis,53 as well as CLDN1 and SLC2A, both of which are currently used as markers for perineural cells.54,55 Considering the increased innervation in the palmar skin compared with the rest of the body,56 this perineural role would match with the increased proportion of fibroblast IV cells seen in the palm, where it makes up 14% rather than <8% in the other two sites (Figure 3). In sum, we have discovered specialized palmoplantar fibroblasts (fibroblast II) and identified the location and function of a minor fibroblast population (fibroblast IV). We also demonstrate the impact of site-specific differences in fibroblast signaling in shaping immunological properties of palmoplantar and non-palmoplantar skin.
Through bulk and single-cell transcriptomics we consistently identify upregulation of epidermis development and cornification GO terms in the palmoplantar compared with hip keratinocytes, consistent with the thicker cornified layer of the palmoplantar epidermis. The palmoplantar-specific suprabasal marker KRT9 and stress-response genes KRT16 and KRT6A/B/C, whose mutations are associated with palmoplantar keratoderma,5,6,57 are also upregulated in both the palm and the sole. More unexpect edly, we identify enrichment of GO terms relating to oxidative phosphorylation and metabolism in palmoplantar skin, suggesting metabolic differences between the non-palmoplantar and the palmoplantar epidermis (Figure S4).
Notably, though, our analysis reveals previously unappreciated differences between palmar and plantar skin. These differences are evident in the bulk transcriptomes, but most strikingly in the keratinocyte populations as revealed by scRNA-seq. These differences are likely driven by the contrast in pressure regularly experienced between the palm and the sole.58,59 Contrary to expectations, while the sole has the largest proportion of basal cells (~42%), the palm has the smallest (~16%). In addition, marked differences occur in proportions of the spinous populations I and II between the palm and the sole, whereas the hip skin exhibits an even split (Figure 5). Finally, the palm contains the highest population of granular cells and the sole the lowest. Cell-cell signaling analysis reveals that the basal fraction in each site is inversely correlated to intensity of Notch signaling (Figure 5). This may partially explain how the palmar skin achieves its increased thickness, as this result aligns with the literature, which shows that Notch signaling induces differentiation of the epidermis.
In accordance with recent publications in murine skin, we find a surprising amount of co-expression of basal and differentiated markers within basal and suprabasal clusters (Figure S4).16 This further supports the concept of a gradualistic, rather than stepwise, differentiation process in the human epidermis, conserved across the sites. In addition, we characterize a conserved population of more differentiated, proliferating basal keratinocytes, in agreement with previous studies.17,60–64 Although the proportion of cells varies by site, this population is present and localized similarly in both non-palmoplantar and palmoplantar skin.
Finally, we identify two distinct spinous keratinocyte populations that form two concurrent trajectories in pseudotime and terminate in a shared end state (Figures 4, 6, and 7). Although these populations exist in markedly different proportions, this trajectory is conserved across the three sites. The presence of a dual epidermal differentiation trajectory in healthy adult breast keratinocytes, driven by either the formation or the absence of LBs, lipid-enriched secretory organelles proven important for barrier formation, was recently reported.18,65 Our populations share some characteristics with the breast, including expression of important epidermal transcription factors, such as SOX9, GRHL3, and FOS. However, rather than being driven by LB formation, our differentiation branches are instead marked by differences in GO terms related to cellular metabolism (spinous I) and epithelial cell proliferation and regulation of immune activation (spinous II) (Figures 6 and S5). We hypothesize that the specialization of the differentiated keratinocytes into these two discrete populations may be necessary to carry out two incompatible functions. In our data, we see increased oxidative phosphorylation and metabolism in the spinous I population, which may be incompatible with other processes necessary to maintain epidermal homeostasis. Alternatively, cell-cell communication signaling in the epidermis may necessitate the formation of separate keratinocyte populations to communicate via mutually exclusive signaling pathways.
In addition to these findings, the palmoplantar dataset provides a valuable resource for future studies concerning human epidermal differentiation as well as skin disorders. Applying our dataset to previously published work on GRHL3 knockdown in differentiated human keratinocytes66 confirms that siGRHL3 knockdown is marked by downregulation of terms associated with keratinocyte differentiation and upregulation of more basal-associated terms (Figure S6I). These findings provide in vivo support for GRHL3’s role in terminal epidermal differentiation in humans. By applying markers of our identified keratinocyte populations to human psoriasis and atopic dermatitis data-sets, we are not able to identify any major differences between the keratinocyte populations in non-lesional skin. However, in both lesional atopic dermatitis and psoriatic skin, we show the largest effect size in proliferative cluster basal III, indicating that this population is most associated with disease (Figure S6J).
In summary, we comprehensively categorize the altered immune environment in the palmoplantar skin, including a decrease in immune cells, as well as downregulation of immuno-logical terms in a variety of additional cell types. We also identify site-specific fibroblast populations, characterize a perineural fibroblast population, and categorize differences in cell-cell signaling across the three sites that may drive palmoplantar skin specialization. Finally, in the epidermis, we demonstrate key differences between the palm and the sole, in terms of both cell-cell signaling and basal cell fraction, including the more differentiated, proliferative basal population that we localized to higher in the epidermis. Strikingly, we demonstrate the existence of two separate spinous populations across the three sites, likely with mutually exclusive functions, which establish two parallel, site-selective epidermal differentiation trajectories. Taken as a whole, these results reveal the dampened immune environment of palmoplantar skin, uncover unexpected differences between the palmar and the plantar skin, and offer key insights about human epidermal differentiation.
Limitations of the study
Our study has several limitations. First, the sequencing approach included samples in which dermis and epidermis were sequenced individually or in combination, which complicated some direct comparisons of palmar and plantar samples, as acknowledged in the results. However, the inclusion of the combined sequencing approach provided additional information on the relative proportions of cell types captured through unbiased scRNA-seq. Relatedly, while the study included 15,243 cells, a majority (9,471) of these were keratinocytes. As such, some rarer cell types, particularly immune cells, were low in abundance in our dataset, limiting the conclusions we were able to draw through direct examination of immune cell subsets. This limitation was offset by the inclusion of bulk RNA-seq, which showed strong immunological transcriptional shifts across body sites, as well as the robust immune signals detected by scRNA-seq in fibroblasts and keratinocyte subpopulations that varied in representation across body sites. Third, the lower sequencing depth of scRNA-seq data precluded direct examination of low-abundance transcripts, such as many cytokines and transcription factors that may influence the distinct features of palmoplantar skin. Finally, the data presented herein are intended to serve as a resource, and thus further functional studies are needed to explore the consequences of the transcriptional differences uncovered by these data.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Bogi Andersen (bogi@hs.uci.edu).
Materials availability
All unique reagents generated in this study will be available from the lead contact upon reasonable request.
Data and code availability
The bulk RNA-seq data are available in GEO under accession number GEO: GSE193101 and the scRNA-seq data under accession number GEO: GSE202352. Analyzed scRNA-seq is accessible at UCI interactive portal, with the complete dataset at http://www.skingenes.org/palmoplantar_final/, the fibroblasts at http://www.skingenes.org/palmoplantar_fibroblasts_final/, and keratinocytes at http://www.skingenes.org/palmoplantar_keratinocytes_final/. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human subjects
15 healthy research subjects (7 male, 8 female, between the ages of 18 and 67) were recruited for 4 mm punch biopsies for bulk RNA-seq; 10 contributed hip and palm biopsies and 5 hip and sole. 4 healthy research subjects (3 male, 1 female, between the ages of 39 and 65) were recruited for 4 mm punch biopsies for scRNA-seq; 2 contributed hip and palm biopsies and 2 hip and sole. The study was approved by the University of Michigan Institutional Review Board (IRB), and all patients were consented. The study was conducted according to the Declaration of Helsinki Principles.
METHOD DETAILS
Bulk RNA-seq and gene expression analysis
Skin biopsies were snap-frozen in liquid nitrogen and stored at −80°C until use. Samples were then pulverized and dissolved in complete RLT buffer (Qiagen, Valencia, CA) for RNA extraction. RNA was isolated using the RNeasy Mini kit (Qiagen). Stranded mRNA libraries were prepared using the TruSeq RNA library prep kit (Illumina) and sequenced on the Illumina HiSeq 4000 sequencer by the University of Michigan Advanced Genomics Core. FastQ file parameters and sequence quality metrics were calculated using the FASTX-Toolkit67 and FastQC.68 Removal of the Illumina adaptor sequence and trimming of reads was performed using TrimGalore (version 0.4.5) with a default phred threshold of 20. Ribosomal RNA was filtered using the bbduk.sh script from the BBMap software71 with rRNA sequences from the silva database.79 Read mapping was performed using Hisat2 (version 2.1.0)72,69 with maximum mismatch penalty of 1, minimum mismatch penalty of 0, maximum soft-clipping penalty of 3, minimum soft-clipping penalty of 1, and non-canonical splice site penalty of 20. Resulting alignment bam files were sorted using samtools.73 Quantification of gene abundance was then performed by running stringtie (version 1.3.3)70 in expression estimation mode with the human genome sequence (GRCh38/hg38) used as a reference. Read counts for each gene were tabulated using the prepDE python script included in the stringtie software.70 Post-mapping quality metrics such as percentage of reads mapped to intronic regions were calculated using RSeQC75 and RNA-SeQC.74 Raw gene counts were normalized using the trimmed mean of M-values method.76 Differential expression testing was then performed using negative binomial generalized log-linear models with likelihood ratio testing as implemented in edgeR.80 Raw p-values from differential expression analyses were corrected to control the false discovery rate using the benjamini-hochberg method.81
Single cell RNA-seq and analysis
Generation of single-cell suspensions for scRNA-seq was performed as previously described.82 Dermal and epidermal fractions were either combined or sequenced independently as depicted in Figure 2A. Libraries were constructed by the University of Michigan Advanced Genomics Core on the 103 Genomics Chromium system and sequenced on the Illumina NovaSeq 6000 sequencer to generate 150 bp paired-end reads.
Raw sequencing data were demultiplexed and processed using Cellranger (10× Genomics version 3.0.2) using hg19 reference provided by 10× Genomics. The transcriptomic data from one of the acral skin sites is included in a paper on lipidomic changes in acral skin,83 but all other data is new. Preliminary analysis and visualization of the single cell datasets were performed using Seurat26 in R. For all datasets, cells with <300 and >4500 genes, and >20% mitochondrial genes detected were removed. We performed integrated analysis of the 12 datasets from 3 sites SCTransform normalization separately for each dataset, selected 2000 informative features, and performed integration using the FindIntegrationAnchors function. Cells were visualized using UMAP and tSNE (t-distributed stochastic neighbor embedding) algorithm. Gene Ontology enrichment analysis of the enriched genes was performed using Metascape77 and visualized in R. Gene Set Enrichment Analysis for the keratinocyte populations was carried out using GSEA.42
Gene and probability scores
We calculated the scores for a group of genes associated with different cell cycle phases using CellCycleScoring() in Seurat. The gene lists for the G2M and S phase were the Seurat default. Custom gene scores were calculated using AddModuleScore() in Seurat for reticular and papillary signature and fibroblast subtype, and basal cluster identity.
Trajectory analysis
Scanpy40 was used to calculate the diffusion trajectory of the keratinocytes, using the scanpy.tl.diffmap function. The PAGA50 keratinocyte trajectory was also performed in Scanpy, using the scanpy.tl.paga function. Monocle trajectories were calculated using Monocle2.78
Further analysis
Receptor-ligand analysis for the fibroblasts and keratinocytes for three sites, as well as the hip combined and hip dermis datasets, was performed via CellChat.39 Cell-cell similarity between fibroblast clusters and keratinocyte clusters for the three sites was computed on the integrated matrix using the MetaNeighbor.38 Transcription factor analysis for the keratinocytes was conducted using SCENIC.47
IHC and immunofluorescence
For histology, samples from the three sites were fixed with 10% formalin for 48 h at 4°C, followed by ethanol dehydration. Paraffin-embedded samples were sectioned (10 μm thick) with a microtome. H&E staining was performed as previously described.84
For immunofluorescence, formalin fixed samples were embedded in paraffin and sectioned (10 mm thick) with a cryostat. Slides were fixed in cold acetone for 10–13 min at room temperature (RT). After 3 washes with PBS, slides were fixed with 4% PFA for 10 min. Slides were then permeabilized with permeabilization buffer (Triton X in PBS) for 10–15 min and blocked with blocking buffer (1× PBS+ 2% BSA) for 1 h. For primary antibody, slides were incubated with anti-KI67 and anti- KRT14 or anti-KRT1; or anti–SOX9, anti-GRHL3, and anti-cFOS overnight at 4°C. After several washes with PBS, slides were incubated with secondary antibody (Alexa Fluor 488/ Alexa Fluor 594/ Alexa Fluor 647) for 1 h at RT in the dark. Stained slides were mounted with a mounting DAPI solution for nuclear staining. Slides were imaged with a Keyence BZ-X710 all-in-one fluorescence microscope.
RNA fluorescence in situ hybridization (RNA FISH)
RNA FISH was performed using the RNAscope Multiplex Fluorescent Detection Kit v2 according to manufacturer’s instructions on 10 μM thick formalin fixed paraffin embedded sections. All sections were counterstained with ProLong Gold antifade reagent with DAPI. Images were acquired on a Leica SP8 confocal microscope. To ensure that images were comparable, they were all processed the same maximum intensity projection and brightness.
QUANTIFICATION AND STATISTICAL ANALYSIS
Three biological replicates were analyzed for the palm and the hip RNA staining experiments using ImageJ. Sample sizes, statistical tests and p values are indicated in figure legends. All the quantitative data are presented in mean ± SEM.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Ki67 | Cell Signaling | RRID:AB_2687446 |
| Mouse monoclonal anti-KRT14 | Abcam | RRID:AB_306091 |
| Purified rabbit anti-KRT1 | Gift: Julie Segre, NIH | N/A |
| Mouse monoclonal anti-SOX9 | Abcam | RRID:AB_2194156 |
| Rabbit polyclonal anti-GRHL3 | Sigma Aldrich | RRID:AB_2684180 |
| Rabbit monoclonal anti-cFOS | Cell Signaling | RRID:AB_2247211 |
| Biological samples | ||
| Healthy adult hip, palm, and sole tissue | University of Michigan | N/A |
| Critical commercial assays | ||
| RNeasy Mini kit | Qiagen | Cat#: 74104 |
| TruSeq RNA library prep kit | Illumina | RS-122–2001 |
| Chromium Single Cell 3′ v2 reagent kit | 10x Genomics | PN-120237 |
| RNAscope Multiplex Fluorescent | Advanced Cell | Cat#: 323100 |
| Detection Kit v2 | Diagnostics | |
| Deposited data | ||
| Bulk RNA seq data | This paper | GEO: GSE193101 |
| Single cell RNA seq data | This paper | GEO: GSE202352 |
| Analyzed single cell RNA seq data | This paper | http://www.skingenes.org/palmoplantar_final/; http://www.skingenes.org/palmoplantar_fibroblasts_final/; http://www.skingenes.org/palmoplantar_keratinocytes_final/ |
| Oglionucleotides | ||
| RNAscope Probe- Hs-FOS | Advanced Cell Diagnostics | Cat#: 319901 |
| RNAscope Probe- Hs-GRHL3 | Advanced Cell Diagnostics | Cat#: 1072001 |
| RNAscope Probe- Hs-SOX9 | Advanced Cell Diagnostics | Cat#: 404221 |
| Software and algorithms | ||
| FASTX-Toolkit | (Hannon, 2010)67 | http://hannonlab.cshl.edu/fastx_toolkit/ |
| FastQC | (Brown et al., 2017)68 | http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| TrimGalore v0.4.5 | http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ | RRID:SCR_011847 |
| samtools | (Li et al., 2009)69 | http://htslib.org/ |
| stringtie v1.3.3 | (Pertea et al., 2015)70 | https://ccb.jhu.edu/software/stringtie/ |
| BBMap | (Bushnell, 2014)71 | http://sourceforge.net/projects/bbmap |
| Hisat2 v2.1.0 | (Pertea et al., 2016)72;(Kim et al., 2015)73 | http://ccb.jhu.edu/software/hisat2/index.shtml |
| RNA-SeQC | (DeLuca et al., 2012)74 | http://www.broadinstitute.org/cancer/cga/rna-seqc |
| RSeQC | (Wang etal., 2012)75 | http://code.google.com/p/rseqc/ |
| edgeR v3.24.3 | (Robinson et al., 2010)76 | http://bioconductor.org/packages/edgeR/ |
| Cellranger v3.0.2 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/using/count |
| Seurat v3.1.1 | (Stuart et al., 2019)26 | https://satijalab.org/seurat/ |
| Metascape | (Zhou etal., 2019)77 | http://metascape.org/gp/index.html-/main/step1 |
| MetaNeighbor v1.2.1 | (Crow etal., 2018)38 | https://www.bioconductor.org/packages/release/bioc/html/MetaNeighbor.html |
| CellChat v1.1.0 | (Jin etal., 2021)39 | https://github.com/sqjin/CellChat |
| SCENIC v1.1.2 | (Aibaretal., 2017)47 | https://github.com/aertslab/SCENIC |
| Scanpy v1.6.0 | (Wolf etal., 2018)40 | https://github.com/theislab/scanpy |
| Monocle2 v2.10.1 | (Qiuetal., 2017)78 | http://cole-trapnell-lab.github.io/monocle-release/docs/ |
| ImageJ | https://imagej.net/ | RRID:SCR_003070 |
| BBKNN | (Polański et al., 2020)48 | RRID:SCR_022807 |
Highlights.
Find differences between palmoplantar and non-palmoplantar skin and palm and sole skin
Report an altered immune environment in the palmoplantar skin
Characterize a palmoplantar-specific fibroblast population
Identify two parallel site-selective epidermal differentiation trajectories
ACKNOWLEDGMENTS
This work was supported by NIH grants P30AR075047 and R01AR44882 and the Irving Weinstein Foundation (to B.A.); an NSF grant DMS1763272 and a grant from the Simons Foundation (594598) (Q.N.); and NIH grants K08AR078251 (A.C.B.), R01AR069071 (J.E.G.), R01AR073196 (J.E.G.), P30AR075043 (J.E.G.), R01AR071384 (J.M.K.), K24AR076975 (J.M.K.), and K01AR072129 (L.C.T.). This work was additionally supported by Chan Zuckerberg Initiative grant DAF2022-239946 (B.A., A.C.B, J.G., J.M.K., Q.N., and L.C.T.); Taubman Institute via Innovative Program (J.E.G. and J.M.K.); Parfet Emerging Scholar (J.M.K.) and Wexner Emerging Scholar (A.C.B.) funds; the Lupus Research Alliance (J.M.K.); the Dermatology Foundation (J.E.G. via the Sun Pharma Research Award, A.C.B., and L.C.T.); the Arthritis National Research Foundation (L.C.T.); and the National Psoriasis Foundation (L.C.T.). P.W.H. receives support from CA046592 (University of Michigan Rogel Cancer Center Support Grant) and AR075043 (University of Michigan Skin Biology and Diseases Resource-Based Center); E.X. receives support from the Blue Cross Blue Shield Foundation of Michigan (Student Award) and the University of Michigan Rackham Graduate School. J.W. was supported by T32-GM136624 from the National Institute of General Medical Sciences.
DECLARATION OF INTERESTS
J.M.K. has received grant support from Q32 Bio, Celgene/BMS, Ventus Therapeutics, and Janssen. J.M.K. has served on advisory boards for AstraZeneca, Eli Lilly, GlaxoSmithKline, Bristol Myers Squibb, Avion Pharmaceuticals, ProventionBio, Aurinia Pharmaceuticals, Ventus Therapeutics, and Boehringer Ingelheim. J.E.G. has received grant support from Celgene/BMS, Janssen, Eli Lilly, and Almirall. J.E.G. has served on advisory boards for AstraZeneca, Sanofi, Eli Lilly, Boehringer Ingelheim, Novartis, Janssen, Almirall, and BMS.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.111994.
REFERENCES
- 1.Boyle CJ, Plotczyk M, Villalta SF, Patel S, Hettiaratchy S, Masouros SD, Masen MA, and Higgins CA (2019). Morphology and composition play distinct and complementary roles in the tolerance of plantar skin to mechanical load. Sci. Adv 5, eaay0244. 10.1126/sciadv.aay0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gyi DE, and Porter JM (1999). Interface pressure and the prediction of car seat discomfort. Appl. Ergon 30, 99–107. 10.1016/S0003-6870(98)00018-0. [DOI] [PubMed] [Google Scholar]
- 3.Schweizer J, Baust I, and Winter H (1989). Identification of murine type I keratin 9 (73 kDa) and its immunolocalization in neonatal and adult mouse foot sole epidermis. Exp. Cell Res 184, 193–206. 10.1016/0014-4827(89)90377-7. [DOI] [PubMed] [Google Scholar]
- 4.Fu DJ, Thomson C, Lunny DP, Dopping-Hepenstal PJ, McGrath JA, Smith FJD, Irwin McLean WH, and Leslie Pedrioli DM (2014). Keratin 9 is required for the structural integrity and terminal differentiation of the palmoplantar epidermis. J. Invest. Dermatol 134, 754–763. 10.1038/jid.2013.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reis A, Hennies H-C, Langbein L, Digweed M, Mischke D, Drechsler M, Schröck E, Royer-Pokora B, Franke WW, Sperling K, et al. (1994). Keratin 9 gene mutations in epidermolytic palmoplantar keratoderma (EPPK). Nat. Genet 6, 174–179. 10.1038/ng0294-174. [DOI] [PubMed] [Google Scholar]
- 6.Has C, and Technau-Hafsi K (2016). Palmoplantar keratodermas: clinical and genetic aspects. J. Dtsch. Dermatol. Ges 14, 123–139. 10.1111/ddg.12930. [DOI] [PubMed] [Google Scholar]
- 7.Kumar B, Saraswat A, and Kaur I (2002). Palmoplantar lesions in psoriasis: a study of 3065 patients. Acta Derm. Venereol 82, 192–195. [DOI] [PubMed] [Google Scholar]
- 8.Lillis JV, Guo C-S, Lee JJ, and Blauvelt A (2010). Increased IL-23 expression in palmoplantar psoriasis and hyperkeratotic hand dermatitis. Arch. Dermatol 146, 918–919. [DOI] [PubMed] [Google Scholar]
- 9.Kim D, Hossain MZ, Nieves A, Gu L, Ratliff TS, Mi Oh S, Park A, Han S, Yang NB, Qi J, et al. (2016). To control site-specific skin gene expression, autocrine mimics paracrine canonical Wnt signaling and is activated ectopically in skin disease. Am. J. Pathol 186, 1140–1150. 10.1016/j.ajpath.2015.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rinn JL, Wang JK, Liu H, Montgomery K, van de Rijn M, and Chang HY (2008). A systems biology approach to anatomic diversity of skin. J. Invest. Dermatol 128, 776–782. 10.1038/sj.jid.5700986. [DOI] [PubMed] [Google Scholar]
- 11.Yamaguchi Y, Itami S, Tarutani M, Hosokawa K, Miura H, and Yoshikawa K (1999). Regulation of keratin 9 in nonpalmoplantar keratinocytes by palmoplantar fibroblasts through epithelial–mesenchymal interactions. J. Invest. Dermatol 112, 483–488. 10.1046/j.1523-1747.1999.00544.x. [DOI] [PubMed] [Google Scholar]
- 12.Tabib T, Morse C, Wang T, Chen W, and Lafyatis R (2018). SFRP2/DPP4 and FMO1/LSP1 define major fibroblast populations in human skin. J. Invest. Dermatol 138, 802–810. 10.1016/j.jid.2017.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vorstandlechner V, Laggner M, Kalinina P, Haslik W, Radtke C, Shaw L, Lichtenberger BM, Tschachler E, Ankersmit HJ, and Mild-ner M (2020). Deciphering the functional heterogeneity of skin fibroblasts using single-cell RNA sequencing. FASEB J. 34, 3677–3692. 10.1096/fj.201902001RR. [DOI] [PubMed] [Google Scholar]
- 14.Solé-Boldo L, Raddatz G, Schütz S, Mallm J-P, Rippe K, Lonsdorf AS, Rodríguez-Paredes M, and Lyko F (2020). Single-cell transcriptomes of the human skin reveal age-related loss of fibroblast priming. Commun. Biol 3, 188. 10.1038/s42003-020-0922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watt FM (1983). Involucrin and other markers of keratinocyte terminal differentiation. J. Invest. Dermatol 81, 100s–103s. 10.1111/1523-1747.ep12540786. [DOI] [PubMed] [Google Scholar]
- 16.Lin Z, Jin S, Chen J, Li Z, Lin Z, Tang L, Nie Q, and Andersen B (2020). Murine interfollicular epidermal differentiation is gradualistic with GRHL3 controlling progression from stem to transition cell states. Nat. Commun 11, 5434. 10.1038/s41467-020-19234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, Drummond ML, Guerrero-Juarez CF, Tarapore E, Ma-cLean AL, Stabell AR, Wu SC, Gutierrez G, That BT, Benavente CA, et al. (2020). Single cell transcriptomics of human epidermis identifies basal stem cell transition states. Nat. Commun 11, 4239. 10.1038/s41467-020-18075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reynolds G, Vegh P, Fletcher J, Poyner EFM, Stephenson E, Goh I, Botting RA, Huang N, Olabi B, Dubois A, et al. (2021). Developmental cell programs are co-opted in inflammatory skin disease. Science 371, eaba6500. 10.1126/science.aba6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryu S, Johnson A, Park Y, Kim B, Norris D, Armstrong CA, and Song PI (2015). The alpha-melanocyte-stimulating hormone suppresses TLR2-mediated functional responses through IRAK-M in normal human keratinocytes. PLoS One 10, e0136887. 10.1371/journal.pone.0136887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Homey B, Alenius H, Müller A, Soto H, Bowman EP, Yuan W, McE-voy L, Lauerma AI, Assmann T, Bünemann E, et al. (2002). CCL27–CCR10 interactions regulate T cell–mediated skin inflammation. Nat. Med 8, 157–165. 10.1038/nm0202-157. [DOI] [PubMed] [Google Scholar]
- 21.Vela-Romera A, Carriel V, Martín-Piedra MA, Aneiros-Fernández J, Campos F, Chato-Astrain J, Prados-Olleta N, Campos A, Alaminos M, and Garzón I (2019). Characterization of the human ridged and non-ridged skin: a comprehensive histological, histochemical and immunohistochemical analysis. Histochem. Cell Biol 151, 57–73. 10.1007/s00418-018-1701-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knapp AC, Franke WW, Heid H, Hatzfeld M, Jorcano JL, and Moll R (1986). Cytokeratin No. 9, an epidermal type I keratin characteristic of a special program of keratinocyte differentiation displaying body site spec-ificity. J. Cell Biol 103, 657–667. 10.1083/jcb.103.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moll I, Heid H, Franke WW, and Moll R (1987). Distribution of a special subset of keratinocytes characterized by the expression of cytokeratin 9 in adult and fetal human epidermis of various body sites. Differentiation 33, 254–265. 10.1111/j.1432-0436.1987.tb01565.x. [DOI] [PubMed] [Google Scholar]
- 24.Swensson O, Langbein L, Mcmillan JR, Stevens HP, Leigh IM, Mclean WH, Lane EB, and Eady RA (1998). Specialized keratin expression pattern in human ridged skin as an adaptation to high physical stress. Br. J. Dermatol 139, 767–775. 10.1046/j.1365-2133.1998.02499.x. [DOI] [PubMed] [Google Scholar]
- 25.Satija R, Farrell JA, Gennert D, Schier AF, and Regev A (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol 33, 495–502. 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21. 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rose DM, Alon R, and Ginsberg MH (2007). Integrin modulation and signaling in leukocyte adhesion and migration. Immunol. Rev 218, 126–134. 10.1111/j.1600-065X.2007.00536.x. [DOI] [PubMed] [Google Scholar]
- 29.Johnatty RN, Taub DD, Reeder SP, Turcovski-Corrales SM, Cottam DW, Stephenson TJ, and Rees RC (1997). Cytokine and chemokine regulation of proMMP-9 and TIMP-1 production by human peripheral blood lymphocytes. J. Immunol 158, 2327–2333. [PubMed] [Google Scholar]
- 30.Hochrein SM, Wu H, Eckstein M, Arrigoni L, Herman JS, Schumacher F, Gerecke C, Rosenfeldt M, Grün D, Kleuser B, et al. (2022). The glucose transporter GLUT3 controls T helper 17 cell responses through glycolytic-epigenetic reprogramming. Cell Metab. 34, 516–532.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belote RL, Le D, Maynard A, Lang UE, Sinclair A, Lohman BK, Planells-Palop V, Baskin L, Tward AD, Darmanis S, and Judson-Torres RL (2021). Human melanocyte development and melanoma dedifferentiation at single-cell resolution. Nat. Cell Biol 23, 1035–1047. 10.1038/s41556-021-00740-8. [DOI] [PubMed] [Google Scholar]
- 32.Ascensión AM, Fuertes-Álvarez S, Ibañez-Solé O, Izeta A, and Araúzo-Bravo MJ (2021). Human dermal fibroblast subpopulations are conserved across single-cell RNA sequencing studies. J. Invest. Dermatol 141, 1735–1744.e35. 10.1016/j.jid.2020.11.028. [DOI] [PubMed] [Google Scholar]
- 33.Haydont V, Neiveyans V, Zucchi H, Fortunel NO, and Asselineau D (2019). Genome-wide profiling of adult human papillary and reticular fibro-blasts identifies ACAN, Col XI a1, and PSG1 as general biomarkers of dermis ageing, and KANK4 as an exemplary effector of papillary fibroblast ageing, related to contractility. Mech. Ageing Dev 177, 157–181. 10.1016/j.mad.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 34.Janson DG, Saintigny G, van Adrichem A, Mahé C, and El Ghalbzouri A (2012). Different gene expression patterns in human papillary and reticular fibroblasts. J. Invest. Dermatol 132, 2565–2572. 10.1038/jid.2012.192. [DOI] [PubMed] [Google Scholar]
- 35.Janson D, Saintigny G, Mahé C, and El Ghalbzouri A (2013). Papillary fibroblasts differentiate into reticular fibroblasts after prolonged in vitro culture. Exp. Dermatol 22, 48–53. 10.1111/exd.12069. [DOI] [PubMed] [Google Scholar]
- 36.Haydont V, Neiveyans V, Perez P, Busson É , Lataillade J, Asselineau D, and Fortunel NO (2020). Fibroblasts from the human skin dermo-hypodermal junction are distinct from dermal papillary and reticular fibroblasts and from mesenchymal stem cells and exhibit a specific molecular profile related to extracellular matrix organization and modeling. Cells 9, 368. 10.3390/cells9020368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nauroy P, Barruche V, Marchand L, Nindorera-Badara S, Bordes S, Closs B, and Ruggiero F (2017). Human dermal fibroblast subpopulations display distinct gene signatures related to cell behaviors and matri-some. J. Invest. Dermatol 137, 1787–1789. 10.1016/j.jid.2017.03.028. [DOI] [PubMed] [Google Scholar]
- 38.Crow M, Paul A, Ballouz S, Huang ZJ, and Gillis J (2018). Characterizing the replicability of cell types defined by single cell RNA-sequencing data using MetaNeighbor. Nat. Commun 9, 884. 10.1038/s41467-018-03282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan C-H, Myung P, Plikus MV, and Nie Q (2021). Inference and analysis of cell-cell communication using CellChat. Nat. Commun 12, 1088. 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolf FA, Angerer P, and Theis FJ (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15. 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maruthappu T, Chikh A, Fell B, Delaney PJ, Brooke MA, Levet C, Moncada-Pazos A, Ishida-Yamamoto A, Blaydon D, Waseem A, et al. (2017). Rhomboid family member 2 regulates cytoskeletal stress-associated Keratin 16. Nat. Commun 8, 14174. 10.1038/ncomms14174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. (2003). PGC-1a-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet 34, 267–273. 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 44.Rangarajan A, Talora C, Okuyama R, Nicolas M, Mammucari C, Oh H, Aster JC, Krishna S, Metzger D, Chambon P, et al. (2001). Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 20, 3427–3436. 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moriyama M, Durham A-D, Moriyama H, Hasegawa K, Nishikawa S-I, Radtke F, and Osawa M (2008). Multiple roles of Notch signaling in the regulation of epidermal development. Dev. Cell 14, 594–604. 10.1016/j.devcel.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 46.Klein RH, Lin Z, Hopkin AS, Gordon W, Tsoi LC, Liang Y, Gudjonsson JE, and Andersen B (2017). GRHL3 binding and enhancers rearrange as epidermal keratinocytes transition between functional states. PLoS Genet. 13, e1006745. 10.1371/journal.pgen.1006745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine J-C, Geurts P, Aerts J, et al. (2017). SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086. 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Polański K, Young MD, Miao Z, Meyer KB, Teichmann SA, and Park J-E (2020). BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics 36, 964–965. 10.1093/bioinformatics/btz625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol 32, 381–386. 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolf FA, Hamey FK, Plass M, Solana J, Dahlin JS, Göttgens B, Rajewsky N, Simon L, and Theis FJ (2019). PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 20, 59. 10.1186/s13059-019-1663-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Onoufriadis A, Simpson MA, Pink AE, Di Meglio P, Smith CH, Pull-abhatla V, Knight J, Spain SL, Nestle FO, Burden AD, et al. (2011). Mutations in IL36RN/IL1F5 are associated with the severe episodic inflammatory skin disease known as generalized pustular psoriasis. Am. J. Hum. Genet 89, 432–437. 10.1016/j.ajhg.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takahashi T, Fujimoto N, Kabuto M, Nakanishi T, and Tanaka T (2017). Mutation analysis of IL36RN gene in Japanese patients with palmoplantar pustulosis. J. Dermatol 44, 80–83. 10.1111/1346-8138.13551. [DOI] [PubMed] [Google Scholar]
- 53.Niessen CM, Cremona O, Daams H, Ferraresi S, Sonnenberg A, and Marchisio PC (1994). Expression of the integrin alpha 6 beta 4 in peripheral nerves: localization in Schwann and perineural cells and different variants of the beta 4 subunit. J. Cell Sci 107, 543–552. 10.1242/jcs.107.2.543. [DOI] [PubMed] [Google Scholar]
- 54.North PE, Waner M, Mizeracki A, and Mihm MC (2000). GLUT1: a newly discovered immunohistochemical marker for juvenile hemangiomas. Hum. Pathol 31, 11–22. 10.1016/S0046-8177(00)80192-6. [DOI] [PubMed] [Google Scholar]
- 55.Piña-Oviedo S, and Ortiz-Hidalgo C (2008). The normal and neoplastic perineurium. Adv. Anat. Pathol 15, 147–164. 10.1097/PAP.0b013e31816f8519. [DOI] [PubMed] [Google Scholar]
- 56.Corniani G, and Saal HP (2020). Tactile innervation densities across the whole body. J. Neurophysiol 124, 1229–1240. 10.1152/jn.00313.2020. [DOI] [PubMed] [Google Scholar]
- 57.Wilson NJ, Messenger AG, Leachman SA, O’Toole EA, Lane EB, McLean WHI, and Smith FJD (2010). Keratin K6c mutations cause focal palmoplantar keratoderma. J. Invest. Dermatol 130, 425–429. 10.1038/jid.2009.215. [DOI] [PubMed] [Google Scholar]
- 58.Fransson-Hall C, and Kilbom Å (1993). Sensitivity of the hand to surface pressure. Appl. Ergon 24, 181–189. [DOI] [PubMed] [Google Scholar]
- 59.Hennig EM, and Rosenbaum D (1991). Pressure distribution patterns under the feet of children in comparison with adults. Foot Ankle 11, 306–311. 10.1177/107110079101100507. [DOI] [PubMed] [Google Scholar]
- 60.Dunaway S, Rothaus A, Zhang Y, Luisa Kadekaro A, Andl T, and Andl CD (2019). Divide and conquer: two stem cell populations in squamous epithelia, reserves and the active duty forces. Int. J. Oral Sci 11, 26. 10.1038/s41368-019-0061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jensen KB, and Watt FM (2006). Single-cell expression profiling of human epidermal stem and transit-amplifying cells: lrig1 is a regulator of stem cell quiescence. Proc. Natl. Acad. Sci. USA 103, 11958–11963. 10.1073/pnas.0601886103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kotelnikov VM, Coon IV JS, Taylor IV S, Hutchinson J, Panje W, Caldarelli DD, LaFollette S, and Preisler HD (1996). Proliferation of epithelia of noninvolved mucosa in patients with head and neck cancer. Head Neck 18, 522–528. . [DOI] [PubMed] [Google Scholar]
- 63.Penneys NS, Fulton JE, Weinstein GD, and Frost P (1970). Location of proliferating cells in human epidermis. Arch. Dermatol 101, 323–327. [PubMed] [Google Scholar]
- 64.Pinkus H, and Hunter R (1966). The direction of the mitotic axis in human epidermis. Arch. Dermatol 94, 351–354. [PubMed] [Google Scholar]
- 65.Feingold KR (2012). Lamellar bodies: the key to cutaneous barrier function. J. Invest. Dermatol 132, 1951–1953. 10.1038/jid.2012.177. [DOI] [PubMed] [Google Scholar]
- 66.Hopkin AS, Gordon W, Klein RH, Espitia F, Daily K, Zeller M, Baldi P, and Andersen B (2012). GRHL3/GET1 and trithorax group members collaborate to activate the epidermal progenitor differentiation program. PLoS Genet. 8, e1002829. 10.1371/journal.pgen.1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hannon GJ (2010). FASTX-toolkit. http://hannonlab.cshl.edu/fastx_toolkit/.
- 68.Brown J, Pirrung M, and McCue LA (2017). FQC Dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 33, 3137–3139. 10.1093/bioinformatics/btx373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, and Salzberg SL (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol 33, 290–295. 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bushnell B (2014). BBMap: A Fast, Accurate, Splice-Aware Aligner (Lawrence Berkeley National Lab. (LBNL)).
- 72.Pertea M, Kim D, Pertea GM, Leek JT, and Salzberg SL (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie, and Ballgown. Nat. Protoc 11, 1650–1667. 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Subgroup (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire M-D, Williams C, Reich M, Winckler W, and Getz G (2012). RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 28, 1530–1532. 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang L, Wang S, and Li W (2012). RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185. 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 76.Robinson MD, and Oshlack A (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25. 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, and Chanda SK (2019). Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun 10, 1523. 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, and Trapnell C (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Pe-plies J, and Glöckner FO (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bio-conductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Benjamini Y, and Hochberg Y (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol 57, 289–300. 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 82.Gudjonsson JE, Tsoi LC, Ma F, Billi AC, van Straalen KR, Vossen ARJV, van der Zee HH, Harms PW, Wasikowski R, Yee CM, et al. (2020). Contribution of plasma cells and B cells to hidradenitis suppurativa pathogenesis. JCI Insight 5, e139930. 10.1172/jci.insight.139930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Merleev AA, Le ST, Alexanian C, Toussi A, Xie Y, Marusina AI, Watkins SM, Patel F, Billi AC, Wiedemann J, et al. (2022). Biogeo-graphic and disease-specific alterations in epidermal lipid composition and single cell analysis of acral keratinocytes. JCI Insight 7, e159762. 10.1172/jci.insight.159762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu Z, Lin KK, Bhandari A, Spencer JA, Xu X, Wang N, Lu Z, Gill GN, Roop DR, Wertz P, and Andersen B (2006). The Grainyhead-like epithelial transactivator Get-1/Grhl3 regulates epidermal terminal differentiation and interacts functionally with LMO4. Dev. Biol 299, 122–136. 10.1016/j.ydbio.2006.07.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The bulk RNA-seq data are available in GEO under accession number GEO: GSE193101 and the scRNA-seq data under accession number GEO: GSE202352. Analyzed scRNA-seq is accessible at UCI interactive portal, with the complete dataset at http://www.skingenes.org/palmoplantar_final/, the fibroblasts at http://www.skingenes.org/palmoplantar_fibroblasts_final/, and keratinocytes at http://www.skingenes.org/palmoplantar_keratinocytes_final/. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


