Abstract
T cells expressing either alpha-beta or gamma-delta T cell receptors (TCR) are critical sentinels of the adaptive immune system, with receptor diversity being essential for protective immunity against a broad array of pathogens and agents. Programs available to profile TCR clonotypic signatures can be limiting for users with no coding expertise. Current analytical pipelines can be inefficient due to manual processing steps, open to data entry errors and have multiple analytical tools with unique inputs that require coding expertise. Here we present a bespoke webtool designed for users irrespective of coding expertise, coined ‘TCR_Explore’, enabling analysis either derived via Sanger sequencing or next generation sequencing (NGS) platforms. Further, TCR_Explore incorporates automated quality control steps for Sanger sequencing. The creation of flexible and publication ready figures are enabled for different sequencing platforms following universal conversion to the TCR_Explore file format. TCR_Explore will enhance a user’s capacity to undertake in-depth TCR repertoire analysis of both new and pre-existing datasets for identification of T cell clonotypes associated with health and disease. The web application is located at https://tcr-explore.erc.monash.edu for users to interactively explore TCR repertoire datasets.
Keywords: T cells, T cell receptor, TCR repertoire, Shiny R application
Abbreviations: APC, antigen presenting cells; CDR, complementarity determining region; HLA, human leukocyte antigen; NGS, next generation sequencing; QC, quality control; TCR, T cell receptor
Highlights
-
•
Bespoke program for non-specialists in computerised methodologies for deep exploration of TCR repertoire analysis.
-
•
Automated QC and analysis pipelines for Sanger based TCR sequencing coupled with immunophenotyping, with the capacity for integration of other sequencing platform outputs.
-
•
Automated summary processes to aid data visualisation and generation of publication-ready graphical displays.
1. Introduction
Conventional alpha-beta (αβ) T cells and unconventional gamma-delta (γδ) are critical sentinels of the immune system that are equipped with a molecular armory to detect, engage and eliminate abnormal cells 1, 2, 3. Both αβTCR and γδTCR are heterodimeric proteins containing an α- and a β-chain or a γ- and a δ-chain, respectively. The α- and γ-chain, termed TRA or TRG, are encoded by one variable (V), one joining (J) and one constant (C) gene, whilst the β- and δ-chain, termed TRB or TRD, are encoded by one V, variable diversity (D) genes (up to three), one J and a C gene 4, 5. Recombination of various V(D)J genes, including incorporation of non-template encoded N additions, results in distinct TCR complementarity determining region (CDR) 3 protein sequences 4, 6, and leads to a highly diverse TCR repertoire (e.g. 106 to 108 and unique functional αβTCR clonotypes [7]). Determining the composition and diversity of the TCR repertoire associated with different human diseases is an important step in rationalised clinical interventions or development of T cell-based immunotherapies. For example, we have applied single-cell αβTCR [8] and γδTCR [9] gene analysis to decipher MHC-restricted TCR signatures associated with herpesvirus infection 10, 11, autoimmune diseases such as rheumatoid arthritis [12], heterologous immunity [13] and drug hypersensitivity (αβTCR 14, 15). Although, Sanger sequencing collects fewer cells than NGS, a key advantage is a lower sequencing error rate [16]. Due to this lower error rate, Sanger sequencing is the gold standard approach especially for focused analyses. Specifically, Sanger sequencing is particularly useful when smaller numbers of targeted TCRs are being analysed such as antigen-specific responses. Moreover, accurate TCR data with known epitopes is needed for the curation of databases including VDJdb 17, 18. Whilst the acquisition pipeline of single-cell TCR data by Sanger sequencing is relatively standardised (Fig. 1), the downstream quality control (QC) processes including filtering out poor quality sequences and manual pairing of αβ or γδ TCR chains, as well as verification to ensure data transcription accuracy remain labour intensive. Similarly, single chain data aligned utilising specific software or paid services (e.g. ImmunoSEQ [19], MiXCR [20]) can also require post-alignment QC processes that include filtering out poor quality sequences. Moreover, visualisation of TCR repertoire data often requires additional manual reformatting steps to conform to input requirements of figure generation pipelines such as the Circos® online tool [21], downloadable coding-based programs (e.g. TCRdist, VDJtools, VisTCR) 20, 22, 23, 24, 25, 26, 27, 28, 29, or subscribed non-TCR specific statistical programs (e.g. GraphPad Prism 9 [GraphPad, Software, San Diego, CA, USA] or Microsoft®Excel® [Microsoft, Redmond, WA, USA]). Often these applications are restricted to either a single TCR chain analysis 23, 26 or paired TCR chain analysis [22], or specific to NGS 29, 30, 31. Hence, there is an unmet need for an application that requires minimal to no coding expertise, automation of manual processes for Sanger Sequencing data, can facilitate the analysis of both NGS and Sanger Sequencing, elimination of data entry errors, as well as improved flexibility of data analysis and figure generation.
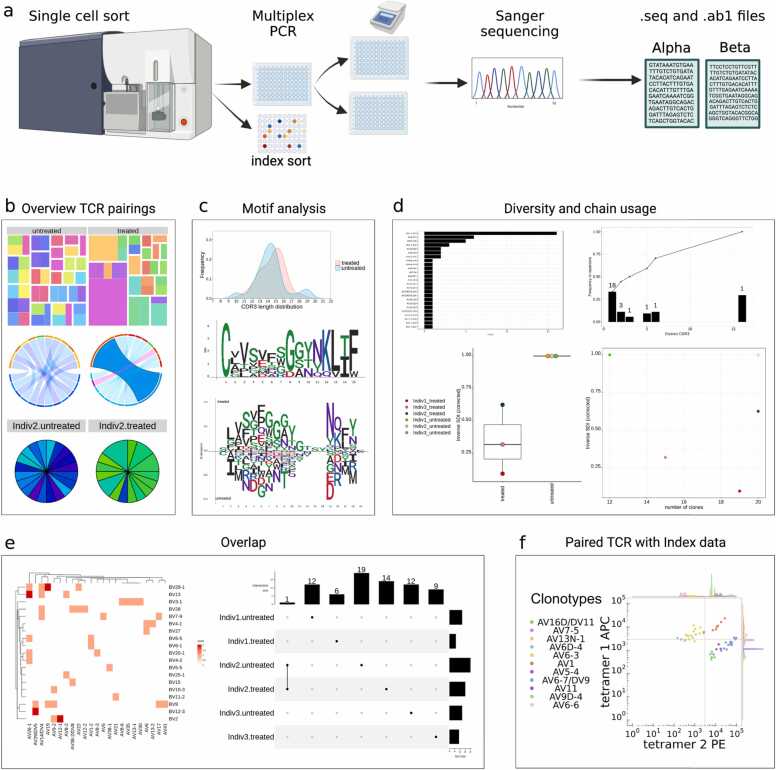
Fig. 1.
Traditional Sanger sequencing pipeline. (A) Targeted T cells are single-cell sorted into 96 well plates by flow cytometry. Single cells undergo reverse transcription to cDNA that is then used as the template for amplification of selected TCR genes by multiplex nested PCR. Following a PCR clean-up step and fluorescent labelling of dNTPs, the amplified DNA undergoes Sanger sequencing, which produces two output files (.seq and .ab1). (B) ‘Overview of TCR pairing’ panel; (top) treemap, (middle) chord diagram, (bottom) pie chart. (C) ‘Motif analysis’ panel; (top) length distribution, (middle), single length motif plot, (bottom) aligned motif plot. (D) ‘Diversity and chain usage’ panel; (top left) chain usage, (top right) frequency of each clonotype, (bottom left) inverse Simpson diversity index (SDI), (bottom right) total number of clones. (E) ‘Overlap’ group comparison panel; (left) heatmap or (right) upset plot. (F) ‘Paired TCR with Index data’ panel; overlaid dot plot with histograms of the functional TCR sequence and two immunophenotyping markers. Figure created using BioRender (BioRender.com).
Here we present TCR_Explore, a Shiny R application available on an open-access webserver (http://tcr-explore.erc.monash.edu) that analyses and visualises TCR repertoire data. TCR_Explore introduces workflows using an automated process that pairing of αβ or γδ TCR chains from Sanger sequencing pipelines and facilitates interrogation of linked flow cytometric index data for immunophenotyping analyses. Additionally, TCR_Explore facilitates conversion and filtering of non-Sanger alignment pipelines (e.g. ImmunoSEQ, MiXCR, 10X Genomics) to the required TCR_Explore format. Moreover, an automated summarisation process from a single input file enables the visualisation of complex data sets and creation of a variety of publication ready figures. Thus, TCR_Explore is a powerful platform for routine analysis of TCR repertoire data of new and reanalysis of pre-existing datasets for greater insight by users irrespective of coding expertise.
2. Methods
2.1. Data and code availability
The demonstration data is from Mifsud et al. (2021) [14] and Lim et al. (2021) [12]. The local version of ‘TCR_Explore’ and all the raw data files and processed datasheets are located on GitHub https://github.com/KerryAM-R/TCR_Explore in the test-data section.
2.2. Single cell sorting and multiplex nested PCR for amplification of TCR chain genes
TCR_Explore was developed for the QC and analysis of Sanger sequencing data generated following multiplex nested PCR (Fig. 1A). Briefly, this included a single cell sort, with and without FACSort index data, into position A1 to H10 of a 96 well plate. Followed by multiplexed PCR of either the TCRα and TCRβ or TCRγ and TCRδ 9, 10, 12, 14. Ideally, sample labelling should follow IndividualID.groupChain-initialwell (e.g. T00020.IFNB-A1). This naming can be either added in the.seq to.fasta conversion step or prior to the pairing process. Sanger sequencing generates two outputs files, .seq and .ab1, that contains the sequencing and chromatogram information, respectively.
2.3. Quality control
2.3.1. Step 1: Alignment of TCR chain sequences using IMGT
Sanger sequencing .seq output files need to be converted into a .fasta file, which can be performed using TCR_Explore (recommend 50 sequences per file), which adds the necessary label (e.g. individual, group, chain or IndividualID.groupChain-initialwell.) used for the merging step (Supplementary data 1). The .fasta file is then uploaded onto the international ImMunoGeneTics information system® (IMGT) 4, 32 website (https://www.imgt.org/IMGT_vquest/input), which aligns a maximum of 50 sequences at a time. To download the Vquest.xls file, the user selects the relevant species (e.g. Homo sapiens), receptor type or locus (e.g. TR), followed by section “C. Excel file” containing only the ‘Summary (1)’ and ‘Junction (6)’ tabs.
2.3.2. Step 2: TCR_Explore quality control
-
(i)
Chromatogram QC files
All the .ab1 files are uploaded to the ‘QC → automated .ab1 QC’ and the same name is added to each file. This process extracts the heterogeneity score (0.33 ratio of the primary/secondary sequence). Lower and negative score indicates poor concordance between the primary/secondary sequences, and suggests multiple sequences are present or poor calling. The score is normalised to the total nucleotide length, which produces values between ‐2 to 2. The user, if needed, can add in the individual, group or chain to name. The user then downloads the .ab1 QC file.
-
(ii)
Uploading the QC file
Upload the Vquest.xls file and .ab1 QC file into the ‘QC→IMGT (Sanger Sequencing)’ tab, select the dataset ‘own_data’ and ‘Select file for IMGT datafile’ using the browse function to upload a file. This will create a downloadable IMGT_onlyQC.date.csv file that contains necessary information for either the TCR_Explore QC process or for compatible files for use in external programs.
-
(iii)
Chromatogram quality and sequence functionality
The program scores both the chromatogram quality and if there are any issues with the sequences based on several IMGT columns (Supplementary Table 1). The 'V.sequence.quality.check’ (column T) flags IMGT outputs that were not aligned, no junction produced, unproductive in ‘V-DOMAIN Functionality’ (column C), if there were either V (<90%) or J (<80%) identity issue from ‘V-REGION identity %’ (column E) and ‘J-REGION identity %’ (column G), respectively or ‘No issue flagged by IMGT’. The ‘chromatogram_check’ (column U), categorises the normalised scores into very high (>1), high (0.9–1), moderate (0.7–0.9), low (0.2–0.7) and poor (<0.2). The ‘clone_quality’ (column V) designated if the sequences ‘pass’ or ‘fail’, based on the following criteria. Those with “No issue flagged by IMGT” and “high” or “very high” chromatogram alignment scores were designated as ‘pass’. Sequences that were ‘low’, ‘poor’, ‘no alignment’, ‘no arrangement’, ‘unproductive’, ’no Junction found’ were designated as ‘fail’. The sequences that ‘fail’ had a reason added to the comment column (column W) including possible need to check moderate quality, either poor sequence quality or no sequence called, unproductive high-quality sequence and possibly resolvable, possible J identity issue, possible V identity issue or other possible issue. The user needs to download the QC file. This process is repeated for all Vquest.xls sequence files, and the data is combined into a single.csv file for downstream TCR chain pairing.
We recommend the user check the reason for a ‘fail’ sequence, as they may be resolvable. The user can create a new file .fasta file with the resolved sequence(s) with ‘man’ added to the end of header (e.g.>T00020. IFNA-A1_A1.seq#1man), as this will not impact the pairing process. The original sequences will ‘fail’, while the manually altered sequences will be designated a ‘pass’. This ensures that all alterations are documented and traceable.
2.3.3. Step 3: Creating the paired TCR file
Upload the completed QC.csv file into the ‘QC → Paired chain file’ tab, select the dataset ‘own_data’ and ‘Completed QC file (.csv)’ using the browse function to upload a file to create the paired_TCR.csv file (Supplementary Table 2). The user can select either alpha-beta or gamma-delta chains as well as the Information included (e.g. Summary+JUNCTION). Only paired chains that have a ‘pass’ assigned will included in the final functional paired TCR repertoire file. The pairing process is based on the IndividualID.groupChain-initialwell. Additionally, the T cell receptor (TR) abbreviation was removed (e.g. TRAV = AV). Moreover, the program adds several columns to the end of the file that shows the genes without the allele for AJ, AV, AVJ, BJ, BV, BD, BVJ, BVDJ, AVJ.BVJ, and AVJ.BVDJ or the γδTCR equivalent (Supplementary Table 2). This will create a downloadable.csv file (e.g. paired_TCR.csv) that contains necessary information for both the ‘TCR analysis’ and ‘Paired TCR with index data’ sections. There is also an option to download the cleaned ‘single chain file’ if pairing is not needed or the Tab space variable (TSV) or.tsv file for use in TCRdist [22].
2.4. Conversion of alternate TCR data outputs to TCR_Explore format
TCR_Explore can convert TCR repertoire data from other alignment programs into a compatible format. For ImmunoSEQ® we utilised data from Heikkila et al. (2021) [33], this process rearranges the file so that the count data is in column A (renamed as cloneCount), keeps the in-frame sequences only, removes empty columns and missing information from either the V or J genes. For MiXCR [20], the program removes sequences with stop codons or frameshifts. For sequencing data not aligned through either ImmunoSEQ®, 10x_scSeq, MiXCR or other, use the ‘Other’ in the ‘Input type’ dropdown menu, as this contains the generic filtering and conversion functions. A video example of the functionality is provided in ‘QC → Convert to TCR_Explore file format → Video of the conversion process’.
2.5. TCR analysis
The ‘TCR analysis’ tab includes features to aid in figure generation and summary statistics. Users can select either test αβTCR data [14] denoted as “ab-test-data2″ or they can upload the single or paired TCR.csv file or from another QC processed TCR dataset (e.g. MiXCR [20]). Interrogation of TCR data is achieved via four distinct analysis platforms (Overview of the TCR, Motif analysis, Diversity and chain usage, Overlap). Each section is further subdivided using tabs or dropdown menus to alter graphing parameters and displays for each figure. In total, there are 14 distinct figures that can be generated in ‘TCR analysis’ section and one in the ‘Paired TCR with Index data’ section.
2.5.1. Overview of TCR pairing
‘Overview of TCR pairing’ tab includes a downloadable summary table for TCRdist3 [27], and three analytical graphs: treemap, chord diagram and pie chart. There are common automated and customisable features of the plots which include: ordering the groups, customisable colours, font type, drop-down menus to change the desired comparison. The drop-down menus enable the user to quickly change their comparison from single chain analysis (e.g. TRAV vs TRAJ) to paired chain analysis (e.g. TRAV-TRAJ vs TRBV-TRBJ) without the need to manually alter the file. Additionally, the chord diagram included options for selective labelling (e.g. Label or colour selected clone/s). A video example of the functionality is available in ‘Tutorials → Video examples → Overview of TCR pairing’.
2.5.2. Motif analysis
There are four sub-tabs in the ‘Motif analysis’ section which presents motif plots based on the unique CDR3 sequences. The first tab is the ‘CDR3 length distribution’, which includes a histogram that can be colour coded by a specific column (e.g. AVJ) or as a density plot that shows the overlap of the groups. The three other sub-tabs show either the ‘Motif (amino acid)’ and ‘Motif (nucleotide sequence)’ for single lengths, while the ‘Motif (AA or NT alignment)’ uses MUSCLE [34] to align the sequences. Both the ‘Motif (amino acid)’ and ‘Motif (AA or NT alignment)’ can compare the differences of two motifs using subtractive analysis. Like the online version of MUSCLE, we restricted sequence alignment to 500 sequences to prevent server timeout issues. A video example of the functionality is provided in ‘Tutorials → Video examples → Motif analysis’.
2.5.3. Diversity and chain usage
There are two tabs in the ‘Diversity and chain usage’ section. The first tab ‘Chain bar graph’ has three distinct graphs of either the chains used per group, the frequency of the repertoire per group and a stacked bar graph. For the frequency graph, the x-axis represents the number of times a clone was observed, the numbers above the bars represent the unique clones, and the line represents the cumulative frequency. The second tab is the ‘Diversity Index’, which is used to calculate the changes in diversity. The program includes access to Shannon Index (how diverse the TCR in a given sample are), Pielou evenness (Shannon index divided by the log of the sample richness [e.g. unique number of clonotypes]), inverse Simpson index (indication of the richness in a TCR repertoire with uniform evenness that would have the same level of diversity), inverse Simpson index corrected (inverse Simpson index divided by unique clonotypes) and Chao1 (is a nonparametric method for estimating the number of TCR’s in a given repertoire). There are two graphs available; (1) shows the index vs the selected group and (2) showcases the index vs either the number of total clones or unique clones. Graph (2) is used to check the total number of sequences is not biasing the results. If two groups are being compared, a standard t-test calculation is available and a one-way ANOVA (groups ∼ index) with a Tukey post-hoc test. For more complex statistical methods, such as a two-way ANOVA or more complex modelling, a third-party program is required, therefore the diversity index table is downloadable for this purpose. Further interrogation of the data can also be conducted using TCRdist [22] or TCRdist3 [27]. A video example of the functionality is in ‘Tutorials → Video examples →Diversity and chain usage’.
2.5.4. Overlap
The ‘Overlap’ section enables users to compare multiple groups using either a heatmap or upset plot. The heatmap compares chain usage from either single or multiple individuals, whilst an upset plot can display the overlap of up to 31 groups, which is a restriction of the package used [35]. These comparisons highlight whether the TCR repertoires are of a public or private nature. The upset plot table data is also downloadable. A video example of this functionality is in ‘Tutorials → Video examples → Overlap’.
2.6. Paired TCR with Index data
This three-step process is showcased as a video in ‘Tutorials → Video examples → Paired TCR with Index data’.
2.6.1. Step 1. Merging the paired TCR with Index data (QC process 1)
The ‘Paired TCR with Index data’ section automates the merging of the paired clone file with the corresponding .fcs file. A background file converts the .fcs xloc and yloc (e.g. 0,0) values to A1 to H10 values to enable merging. This process is limited to one plate at a time as only one .fcs can be uploaded. However, there is no need to reformate the QC paired TCR.csv file. The user needs to select the group, individual and if there were multiple plates. The user can then copy all the samples into one .csv file.
2.6.2. Step 2. Data cleaning steps (QC process 2)
The next QC step occurs in ‘Data cleaning steps’ to covert the negative fluorochrome values to small positive values required for log transformation, and was the method utilised in Lim et al. (2021) [12]. To create the colour scheme for the file, the users can select all necessary columns, but must not select fluorochrome columns, well, cloneCount columns as it will not summarise. This second tab “UMAP reduction”, the user selects the fluorochromes they wish to include in the dimensional reduction as well as decide on cluster range clustering. The pamk() function of the ‘fpc’ package [36] that uses the average silhouette width criterion to decide on the optimal number of clusters with clustering range (can be altered by the user). After downloading, the user can alter names of the fluorochromes, which is restricted to alphabetic characters and numbers.
2.6.3. Step 3. Generation of the analytical plot
Next, in the ‘TCR with index data plot’, the user uploaded the cleaned file from step 2. The user can select any of the fluorochromes to display on the graph. There are over 20 customisable features including the size, colour, and shape of each dot as well as text size and font. These features are either located in the side panel or above the plot, so the user can readily visualise all changes. The figure can be downloaded as either a PNG or PDF.
2.7. R packages
TCR_Explore is an R-based Shiny application constructed using various R packages including: “tidyverse” (version 1.3.1) [37], “ggplot2” (version 3.3.5) [38], “ggrepel”(version 0.9.1) [39], “shiny” (version 1.7.1) [40], “shinyBS” (version 0.61) [41], “gridExtra”(version 2.3) [42], “DT”(version 0.20) [43], “plyr” (version 1.8.6) [44], “dplyr” (version 1.0.7) [45], “reshape2” (version 1.4.4) [46], “treemapify” (version 2.5.5) [47], “circlize” (version 0.4.13) [48], “motifStack” (version 1.36.1) [49], “scales” (version 1.1.1) [50], “flowCore” (version 2.4.0) [51], “readxl” (version 1.3.1) [52], “RcolorBrewer” (version 1.1–2) [53], “randomcoloR” (version 1.1.0.1) [54], “colourpicker” (version 1.1.1) [55], “ComplexHeatmap” (version 2.9.4) [35], “MUSCLE” (version 3.34.0) [34], “DiffLogo” (version 2.16.0) [56], “vegan” (version 2.5–7) [57], “VLF” (version 1.0) [58], “ShinyWidgets” (version 0.7.0) [59], “showtext” (version 0.9–5) [60], “ggseqlogo” (version 0.1) [61], “markdown” (version 1.1) [62], “rmarkdown” (version 2.14) [63], “sangerseqR” (version 1.32.0) [64], “fossil” (version 0.4.0) [65], “umap” (version 0.2.9.0) [66] and “fpc” (version 2.2–9) [36]. All dependent packages used to run the program are in ‘Tutorials → Session info’.
3. Results
3.1. Interrogation of TCR repertoires
TCR_Explore is a web-application for TCR repertoire QC and analysis. The automated QC process was created to aid cleaning and pairing of αβTCRs or γδTCRs from different sequencing alignment pipelines (Fig. 1A) to create a single input file for TCR_Explore. Importantly, both single chain and paired chain TCR repertoires can be interrogated.
The ‘TCR analysis’ tab has four subsections for repertoire analysis. The first section enables data visualisation of TCR repertoire profiles via treemaps, chord diagrams or pie charts (Fig. 1B) and generates a summary table. The second section evaluates CDR3 length distributions and amino acid motifs (both single length and consensus sequences) can be plotted for comparative analyses (Fig. 1C). The third section examines changes in repertoire diversity and chain usage via inverse Simpson diversity index (SDI) values or frequency plots (Fig. 1D). The fourth section facilitates a comparative group overlap analysis using heatmaps or upset plots (Fig. 1E), with a downloadable table output. Collectively, TCR_Explore has enhanced flexibility to perform TCR repertoire analysis and aid in directing the next stages of analysis or experimentation.
3.2. Showcasing TCR immunophenotypes
Unlike existing tools, TCR_Explore enables merging of functional TCR repertoire data with phenotypic expression, which is critical for validation of T cell biomarkers [12]. Previously, manual merging of paired TCR repertoire data with phenotypic markers (i.e. index data) collected during the FACSort was cumbersome and prone to data entry errors (Table 1). In addition, manual conversion of negative expression values for log transformation was also required, as well as the need to select the phenotypic comparison before generating a overlaid dot plot with histograms figure in GraphPad® [12] (Table 1). Due to the time-consuming nature of this workflow, few studies have utilised this validation process 12, 67. Here, the ‘Paired TCR with Index data’ tab automates these processes to generate an overlaid dot plot with histograms (Fig. 1F). Drop-down menus improve flexibility for display of selected phenotypic markers, providing a detailed assessment of T cell specific biomarkers.
Table 1.
Comparison of QC and automation pipelines.
| Step | Process | Current pipeline | TCR_Explore |
|---|---|---|---|
| 1 | Chromatogram visualisation | Manual | Manual |
| 2 | Conversion of .seq to .fasta files | Manual* | Automated |
| 3 | Merging of up to 50 files for IMGT sequence alignment | Manual | Automated |
| Command-line program (e.g. MIXCR) | Requires coding expertise | - | |
| 4 | Classification of productive and non-productive sequences | Manual^ | Automated based on .ab1 quality scoring of Sanger Sequence and IMGT outputs |
| 5 | Pairing of αβ or γδ TCR sequences | Manual* | Automated based on Sequence ID naming |
| 6 | Removal of non-productive sequences | Manual* | Automated |
| If immunophenotyping data available; Index data using.fcs files | |||
| 7 | Pairing of TCR sequence with immunophenotype | Manual* | Automated |
| *8 | Conversion of negative values to allow for log transformation | Manual* | Automated |
Potential for data entry errors, ^ Potential for human interpretation errors
3.3. Improved quality control process for pairing TCR chain genes
TCR_Explore, utilising the R statistical language, automates numerous manual QC processes as depicted in Table 1. This process includes pairing the separate TCRα and TCRβ or TCRγ and TCRδ chains, produced from Sanger sequencing output files (.seq). Importantly, our program also includes automated merging the functional paired TCR file with corresponding phenotype index data (.fcs) file (Table 1; Supplementary Table 3). Both QC merging process, based on the naming convention (IndividualID.groupChain-initialwell), thereby reduce the time needed to create an analysis file, with substantial reduction of data entry errors in the QC process.
3.4. Automated summarisation reduces errors and enables flexible figure generation
Previous workflows for TCR repertoire analysis and visualisation involved the use of multiple tools, each requiring specific file formats 20, 21, 22, 23, 24, 25, 26, 27, 28. This process is time intensive, vulnerable to data entry errors, and inflexible with respect to incorporating data updates or changes to the comparison of interest (e.g. TRAV vs TRAJ to TRAV vs TRBV). Additionally, these programs include limitations in plot customisation (i.e. font choices or colouring of specific chains) 21, 23, inflexible in their export functions (e.g. inability to specify height/width; PNG or PDF only) 21, 23 and restricted to single chain analysis 23, 24. Therefore, there is an unmet need to develop a program to overcome these limitations. To remove these reformatting processes and reduce errors, TCR_Explore was designed to rely on a single integrated input file for the generation of all data figures (Table 2). The program promptly summarises the data based on researcher inputs located in the Shiny R interface, thereby eliminating the need for creating multiple files and removal of data entry errors. The input file structure and user-friendly interface allows flexibility, particularly if samples are added or removed. Overall, TCR_Explore has improved flexibility and breadth of analysis, thereby enhancing opportunity for TCR repertoire discoveries.
Table 2.
Figure generation with available programs or TCR_Explore.
| Figure | Available Programs | TCR_Explore Location |
|---|---|---|
| Chord diagram | Circos® (online) VDJtools# VDJviz# ARResT#^ R ‘circlize’ package# |
Overview of TCR pairing |
| Treemap & pie chart | iRepertoire§ (treemap) GraphPad®§ (pie chart) R ‘treemap’ package# (treemap) R ‘ggplot2′ package# (pie chart) |
Overview of TCR pairing |
| CDR3 length distribution | VDJtools# R ‘ggplot2′ package# VisTCR#^ VDJviz#^ ARResT#^ Immunarch# |
Motif analysis |
| Motif filtering, alignment and plot generation | Microsoft®Excel®§ (filter to one length) MUSCLE (online or R ‘muscle’ package#; Align CDR3 sequences) Motif plot (online or R ‘ggseqlogo’ package#) TCRdist3# Immunarch# |
Motif analysis |
| Stacked bar graph, repertoire frequency | Microsoft®Excel®§ GraphPad®§ R ‘ggplot2′ package# VDJviz#^ ARResT#^ VDJtools# |
Diversity and Chain usage |
| Diversity plot | Microsoft®Excel®§ GraphPad®§ R ‘ggplot2′ package# TCRdist# VDJtools# VisTCR# (Shannon diversity only) Immunarch# |
Diversity* and Chain usage |
| Heatmap & upset plot | R and RStudio# VDJtools# (heatmap only) ARResT#^ (heatmap only) Immunarch# (heatmap only) |
Overlap |
| Overlaid Dot plot | FlowJo§ Microsoft®Excel®§ GraphPad®§ R ‘ggplot2′ package# |
TCR dot plot with Index data |
Inverse Simpson diversity index, Pielou index, Shannon diversity and Choa1, §requires paid subscription, #coding skills required, ^NGS only
3.5. Interface with existing programs for extended data analysis
One statistical program that uses paired TCR data is TCRdist [22], which has many features including identifying the origin of TCR sequences, principal component analysis, αβTCR epitope-specific repertoires, and a robust TCR diversity statistic. These features were beyond the scope of TCR_Explore, therefore we included an interface that serves to provide compatible outputs for TCRdist via the generation of a TSV file (Supplementary data 2). Additionally, we included a.csv output that was compatible with further analysis using TCRdist3 [27] (Supplementary data 3). Moreover, the TCR data outputs from other programs and pipelines including: iRepertoire (deep-sequencing of TCR repertoire [68]), ImmunoSEQ [19], and MiXCR [20]. These external TCR data, once converted (see ‘QC → Convert to TCR_Explore file format’), can be imported into the TCR_Explore ‘TCR analysis’. Overall, TCR_Explore is compatible with external programs and data pipelines.
3.6. Extended analysis of drug-induced human αβTCRs reveals a central residue for TCR activation
In our recent study [14], we examined the carbamazepine-induced αβTCR profile of patients who had previously experienced either Stevens-Johnson syndrome or toxic epidermal necrolysis following prescription of an anti-seizure medication, carbamazepine. These severe allergic responses are classified as T cell-mediated drug hypersensitivity reactions that principally target the skin following carbamazepine exposure via TCR activation 68, 69. To profile the TCR repertoire of our patient cohort, peripheral blood mononuclear cells were in vitro stimulated for 14 days with 25 μg/mL carbamazepine, with 50 U/mL of IL-2 being added from day 4. On day 14, drug-induced T cells were restimulated with HLA allotype matched antigen presenting cells (APC) in the absence or presence of carbamazepine, and single-cell sorted based on the production of the proinflammatory cytokine IFNγ into two subsets: (i) CD8+IFNγ+ (IFN activated group) or (ii) CD8+IFNγ- (CD8 non-activated group). In our initial publication, we reported the drug-induced TCR clonotypes were highly focused to a few TCR clonotypes and this TCR usage was private amongst the cohort. Using TCR_Explore, reanalysis of the pre-existing dataset demonstrated a capacity to not only replicate the published findings but also to interrogate the data to new depths and reveal nuances not previously appreciated.
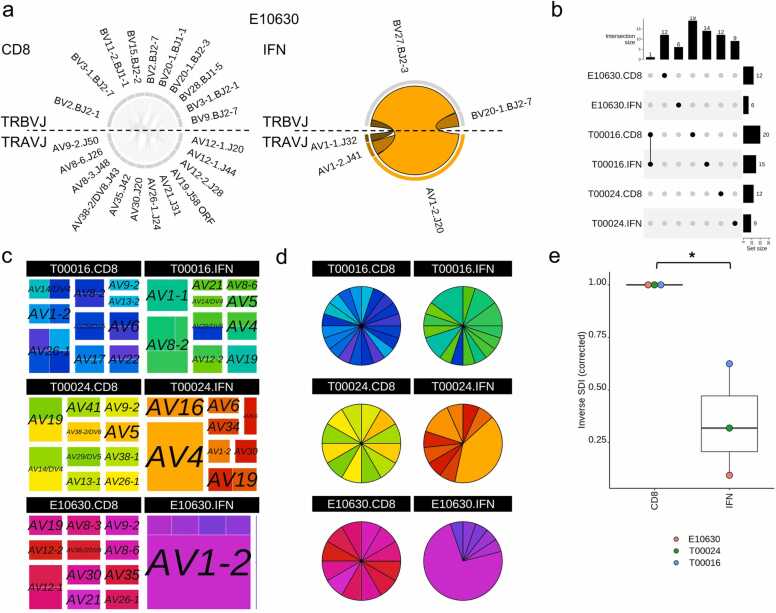
For the reanalysis, we examined three patients E100630, T00016 and T00024 (Supplementary Table 4). Firstly for E10630, drug-specific TCRs (TRAVJ-TRBVJ) were visualised using chord diagrams in TCR_Explore to demonstrate recapitulation of the Circos® plots shown in the publication [14] (Fig. 2a). Next, we confirmed via an upset plot that the paired αβTCR chains were specific to the individual patient (i.e. private TCR repertoire) by comparing multiple individuals TCR repertoires (Fig. 2b). TCR_Explore also enabled greater flexibility for data display using either treemaps (Fig. 2c) or pie charts (Fig. 2d), which represent alternatives to chord diagrams. Additionally, examination of diversity based on unique sequences showed a decrease in TCR clonotypes in the IFN activated group compared to the CD8 non-activated group. This reduced diversity was represented by a significant reduction of the sample size corrected inverse SDI score (p = 0.026; paired t-test; one-tailed), which highlighted that all three individuals expressed carbamazepine-induced TCR clonotypes (Fig. 2e). Here, TCR_Explore provided an opportunity to reveal further nuances within the TCR sequences that warrant further functional validation.
Fig. 2.
TCR_Explore analysis of a pre-existing TCR repertoire dataset. TCR repertoire data derived from in vitro expanded carbamazepine-induced T cells derived from patients with Stevens-Johnson Syndrome (T00016, T00024 and E10630), with CD8 and IFN representing the non-activated and drug-activated subsets, respectively. (a) E10630, chord diagram of drug-induced αβTCR repertoire for CD8 and IFN subsets. No overlapping sequences between the CD8 (grey) and IFN (orange) subsets. (b) Upset plot representing αβTCR CDR3 region overlap. Dots represent the presence of a clonal sequence and lines connect overlapping samples. (c) treemap coloured by AVJ_aCDR3_BVJ_bCDR3 and separated by the TRAV genes (size of the square indicate proportion of each TCR relative to the individual sample; colour represents a unique clone). (d) pie chart coloured by AVJ_aCDR3_BVJ_bCDR3 (size of the segment is proportional to the percentage of each clone; colour represents a unique clone). Same colours were used for both the (c) treemap and (d) pie chart. (e) Inverse Simpson index vs condition to measure change in diversity following drug exposure. Paired Students t-test, *p < 0.05. Dots represent each individual.
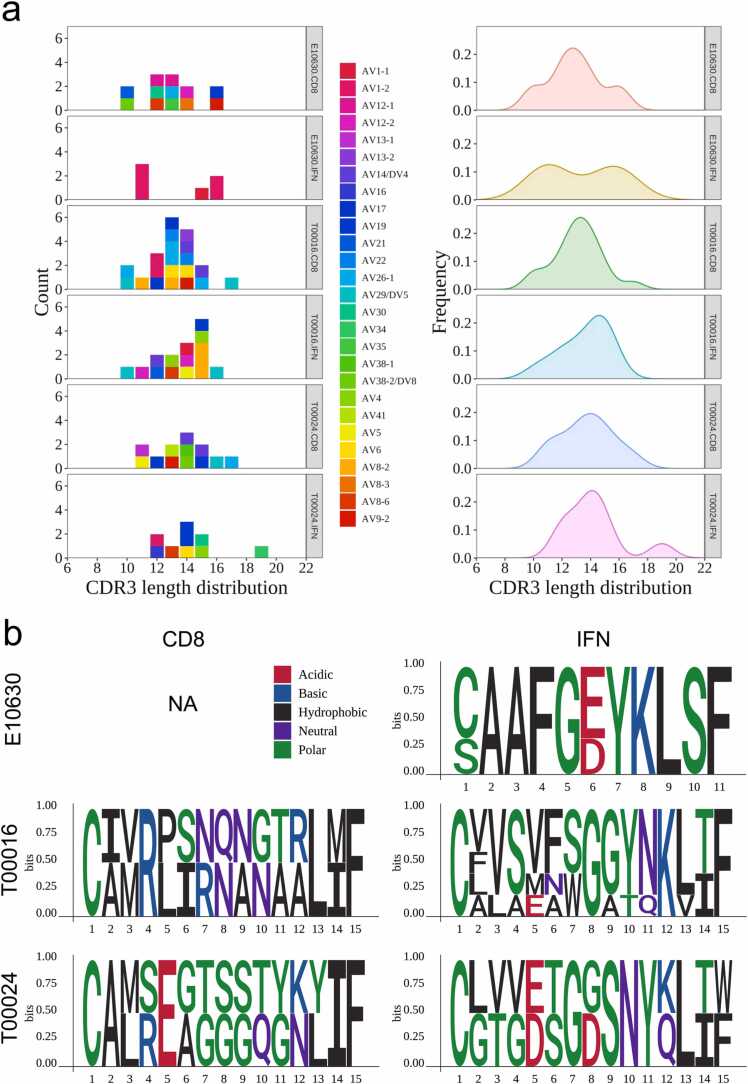
There are three proposed mechanisms for T cell activation by small molecules drugs, with both the hapten/prohapten and altered repertoire mechanisms resulting in alteration to peptides presented by the HLA [15]. In contrast, our paper [14] showed that carbamazepine-induced Stevens-Johnson syndrome or toxic epidermal necrolysis, associated with the pharmacological interaction with immune receptors concept, had minimal impact on the peptide repertoire. Therefore, T cell stimulation is likely to be triggered by direct interactions between the drug and the TCR, bypassing the need for a specific peptide/HLA complex. Using TCR_Explore, interrogation of CDR3α motifs (i.e. unique sequences) highlighted a redistribution of carbamazepine-induced CDR3α lengths towards 11mers (E10630) and 15mers (E10630, T00016), as well as loss of 16 mers (T00024) and 17mers (T00016, T00024) (Fig. 3a). Interestingly, the E10630-derived 11mer (CAAFGDYKLSF) in our original publication was shown to be activated in the presence of both carbamazepine and HLA-B*15:02 (Fig. 3b top). For the 15mers, the IFN activated group was dissimilar from the CD8 non-activated group for both T00016 (Fig. 3b middle) and T00024 (Fig. 3b bottom; major clonotype CDR3α TRAV4-TRAJ33 CLVGETGDSNYQLIW). Interestingly, the E10630 and T00024 CDR3α two clonotypes shown above presented a central aspartate residue (bolded D), that was lacking in the non-activated treatment of the same length sequence (Fig. 3b). A centric aspartic acid residue αCDR3 region was also observed in carbamazepine-induced TCRs of other study participants (E10056, CAAKDGMDSSYKLIF; AP026, CIVRSLRDNYGQNFVF) [14], which were also activated in the presence of carbamazepine and HLA-B*15:02. Moreover, centric aspartic acid residues have been previously reported in carbamazepine-induced Stevens-Johnson syndrome/toxic epidermal necrolysis blister fluid-derived T cells (VFDNTDKLI and AASPPDGNQFY) [68]. In this study, the centric aspartic acid was only observed in different TRAJ genes (underlined section) derived from carbamazepine-induced TCRs of activated IFN T cells but not the non-activated CD8 T cells, suggesting that this feature may be required for TCR activation. Together, TCR_Explore provided an opportunity to further interrogate our dataset that contributed to novel insights and opened further avenues for functional investigation.
Fig. 3.
TCR_Explore reveals CDR3α motif nuances. TCR repertoire data from patients with Stevens-Johnson Syndrome (T00016, T00024 and E10630), with CD8 and IFN representing the non-activated and drug-activated subsets, respectively. (a) CDR3α length distribution coloured by individual or density plot of CD8 non-activated vs IFN activated group. (b) CDR3α motif plot showcasing CD8 non-activated (left) vs IFN activated (right) for (top) E10630 (11mers), (middle) T00016 (15mers) and (bottom) T00024 (15mers). Note, no E10630 CD8 motif shown due to the absence of 11mers for CDR3 length (NA).
3.7. Linking of TCR clonotypes with immunophenotypes in a mouse model of autoimmunity
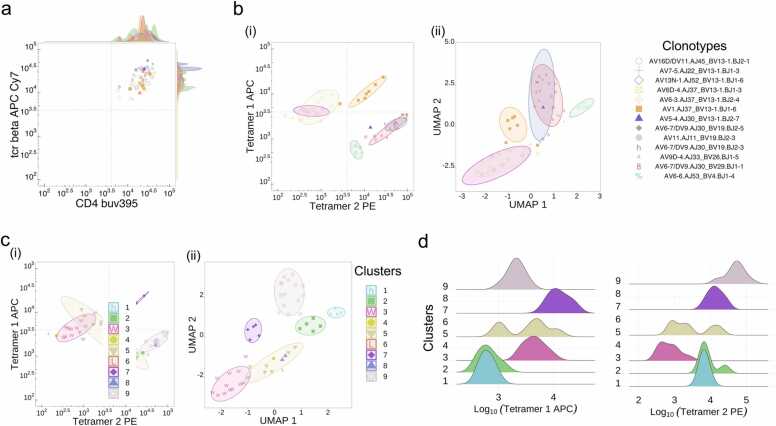
In another recent study [12], we examined the αβTCR repertoire in a mouse model of rheumatoid arthritis that expresses the human susceptibility allele HLA-DRB1*04:01. HLA-DR4 mice were inoculated with a double citrullinated peptide Fibβ-72,74cit69–81, and lymphocytes from draining lymph nodes were collected on day 8 and examined for TCR cross-reactivity to the single- and double-citrullinated epitopes by co-staining with Fibβ-72,74cit69–81 and Fibβ-74cit69–81 tetramers. Individual unique- and cross-reactive CD4+ T cells were index sorted for downstream association of immunophenotype and TCR sequence and single-cell TCRα and TCRβ sequencing. The dot plot can now readily screen through two fluorochrome markers e.g. CD4 vs TCRβ (Fig. 4a). Concordant with the original analysis, TCR_Explore highlighted the cross-reactive αβTCR clone (i.e. TRAV1/J37-TRBV13–1/J1–6; orange square), as depicted by high tetramer expression of both single- and dual-stained citrullinated peptides (Fig. 4bi). To extend the original analysis, the user can perform unsupervised clustering using k-means (Fig. 4ci) as well as perform and visualise the dimensional reduction (UMAP) analysis (Fig. 4bii,cii). Both the dimensional reduction and unsupervised clustering highlighted cluster 7 as the cross-reactive CD4+ T cell population. To aid in understanding the clustering, the user can visualise the markers (log10 normalisation) with a Ridges plot (Fig. 4d) and includes a one-way ANOVA table comparing the distribution of a given fluorochrome. For instance, there was no statistical difference for CD4 expression (data not shown), but cluster 7 had a significantly higher Tetramer 1 APC (adjusted p-value = 8.13e-08; diff=0.79). Overall, TCR_Explore provides a critical platform to examine TCR signatures with immunophenotyping captured via FACSort index data to identify phenotypic markers of interest.
Fig. 4.
Linking TCR clonotype with immunophenotype. CD4+ T cells harvested from draining lymph nodes of an HLA-DR4 mouse immunised with Fibβ-72,74cit69–81 peptide. (a) Dot plot with overlaid histograms depicting CD4 expression of different TCR sequences. Co-staining of CD4+ T cells with HLA-DRB1 * 04:01Fibβ−72,74cit69–81 (tetramer 1 APC) and HLA-DRB1*04:01Fibβ−74cit69–81 (tetramer 2 PE) showing (b) TCR gene analysis and (c) clustering analysis. Visual interrogation can be either the (i) tetramer 1 APC vs tetramer 2 PE and (ii) dimensional reduction of UMAP 1 vs UMAP 2. (d) Ridges plot represents a single fluorochrome marker for each group in the clustering analysis.
4. Discussion
Programs that perform in-depth TCR repertoire analysis (e.g. TCRdist [22], TCRdist3 [27], clusTCR [28], VDJtools [23]) utilise coding languages such as python and R, which effectively limits their usage to individuals with experience in these programming languages or at the very least requires a dedicated time commitment to learn these languages for accessibility. Alternatively, other programs such as Immunarch [26], a coding-based R tool, is aimed to improve access to TCR analysis by minimising the amount of coding needed to a maximum of 5–10 lines [26]. We have further improved the user experience of TCR repertoire analysis by launching our R application TCR_Explore on a website, which does not require coding inputs for program operation.
One of the most critical processes for any dataset analysis is QC of the raw data. Prior to TCR_Explore, our workflow involved the processing of Sanger sequencing information into IMGT to generate the TCR assignments via a vquest.xls file. This file then underwent manual QC to generate a curated file for TCR analysis, which increases the potential for data entry errors to occur. To eliminate both manual QC processing and data entry errors, TCR_Explore was purposefully designed to include an automated QC function, with greatly reduced errors and efficiency that enables users to analyse their data in a shorter timeframe.
Flexibility of data selection is an essential design feature in TCR_Explore to ensure that different variables can be examined without the need to modify the single curated input data file. Previous analyses that required changes to the dataset, such as the addition of individuals and/or treatment groups, would magnify the time required for reanalysis as this would also involve manual reformatting steps required for the various programs being used. TCR_Explore automates reformatting and eliminates the need for the modification of multiple files, enabling the user to interrogate their dataset more thoroughly in the first instance.
The capacity to visualise TCR repertoire data has often been restricted to programs and webtools that employ either coding languages and/or require specific file formats for input data, which also introduces the possibility of data entry errors. Indeed, in some instances more than one webtool is required to visualise different graphical representations of the dataset. Here, TCR_Explore consolidates the generation of 15 different types of figures, with 14 of these able to be created from the same input file in the ‘TCR analysis’ section. To demonstrate the strength of this feature we showcased an increased flexibility and capacity to perform in-depth TCR repertoire analysis by re-examining our previously published human drug-induced TCR dataset [14]. Not only were we able to recapitulate our initial findings but extended our analysis in terms of evaluating both altered diversity of the TCR sequences and CDR3α motif differences. Access to all these figures in the one location enabled us to further interrogate our data with new hypotheses, which led to novel lines of inquiry not previously appreciated.
The capacity to pair the immunophenotype and TCR signature of an antigen-specific T cell provides powerful information for identification of disease biomarkers [67]. TCR_Explore was tailored to readily merge single-cell TCR sequencing and FACS Index sorted information. Re-evaluation of our mouse autoimmune TCR dataset [12] with TCR_Explore improved the robustness of data interrogation and visualisation to showcase immunophenotypic markers of interest (i.e. moderate CD62L expression, data not shown) associated with rheumatoid arthritis. Our program facilitated the examination of two immunophenotypic biomarkers at a time, where the comparisons could be readily changed. Importantly, third party programs involving paid subscriptions or coding-based programs are no longer required to perform this function. Our program has improved capacity to identify T cell biomarkers that could be used in disease diagnoses, as well as identification of immunogenic T cells that have the potential to be developed into T cell-based therapeutics.
5. Conclusion
TCR_Explore is a purpose-designed program to perform automated TCR repertoire analysis and visualisation. TCR_Explore includes a QC pipeline to aid in error-free and proficient TCR repertoire analysis, as well as the generation of a single input file for data analysis and creation of publication ready figures. Use of the Shiny R interface and program maintenance on a webserver ensures that TCR_Explore is accessible to users irrespective of their coding expertise. We anticipate that TCR_Explore will provide a powerful platform for interrogation of TCR repertoires to unravel the complexity of their contribution in human health and disease.
Ethics declaration
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Funding
This work has been financially supported by a National Health and Medicine Research Council of Australia (NHMRC) Project Grant to AWP (1122099). KAM, SJRG, JBZ are supported by an Australian Government Research Training Program (RTP) Scholarship. KAM is also supported by the Monash Biomedical Discovery Institute Departmental Scholarship. CL was supported by an NHMRC CJ Martin Early Career Research Fellowship (1143366). AWP is supported by an NHMRC Principal Research Fellowship (1137739). NLG is supported by funding from NHMRC (1182086) and ARC (DP200102776).
CRediT authorship contribution statement
Kerry A. Mullan and Nicole A. Mifsud: Conceptualization, Methodology, Kerry A. Mullan: Software and Validation, Jerico Revote, Anthony W. Purcell and Chen Li: Resources, Kerry A. Mullan and Nicole A. Mifsud: Writing - original draft, Kerry A. Mullan, Justin B. Zhang, Claerwen M. Jones, Shawn J. R. Goh, Patricia T. Illing, Anthony W. Purcell, Nicole L. La Gruta, Chen Li and Nicole A. Mifsud: Validation, Writing - review & editing.
Competing interests
The authors declare that they have no competing interests.
Acknowledgements
KAM would also like to acknowledge her additional PhD supervisors Prof. Patrick Kwan and Dr Alison Anderson for their support during her candidature.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.01.046.
Contributor Information
Kerry A. Mullan, Email: Kerry.Mullan1@monash.edu.
Nicole A. Mifsud, Email: Nicole.Mifsud@monash.edu.
Appendix A. Supplementary material
Supplementary material.
.
Supplementary material.
.
Supplementary material.
.
Supplementary material.
.
References
- 1.Kumar B.V., Connors T.J., Farber D.L. Human T cell development, localization, and function throughout life. Immunity. 2018;48:202–213. doi: 10.1016/j.immuni.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vantourout P., Hayday A. Six-of-the-best: unique contributions of gammadelta T cells to immunology. Nat Rev Immunol. 2013;13:88–100. doi: 10.1038/nri3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davey M.S., Willcox C.R., Baker A.T., et al. Recasting Human Vdelta1 Lymphocytes in an Adaptive Role. Trends Immunol. 2018;39:446–459. doi: 10.1016/j.it.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lefranc M.P., Giudicelli V., Duroux P., et al. IMGT(R), the international ImMunoGeneTics information system(R) 25 years on. Nucleic Acids Res. 2015;43:D413–D422. doi: 10.1093/nar/gku1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Legut M., Cole D.K., Sewell A.K. The promise of gammadelta T cells and the gammadelta T cell receptor for cancer immunotherapy, Cellular and Molecular Immunology. 2015;12:656–668. doi: 10.1038/cmi.2015.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berland A., Rosain J., Kaltenbach S., et al. PROMIDISalpha: A T-cell receptor alpha signature associated with immunodeficiencies caused by V(D)J recombination defects. J Allergy Clin Immunol. 2019;143:325–334. doi: 10.1016/j.jaci.2018.05.028. e322. [DOI] [PubMed] [Google Scholar]
- 7.Granadier D., Iovino L., Kinsella S., et al. Dynamics of thymus function and T cell receptor repertoire breadth in health and disease. Semin Immunopathol. 2021;43:119–134. doi: 10.1007/s00281-021-00840-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang G.C., Dash P., McCullers J.A., et al. T cell receptor alphabeta diversity inversely correlates with pathogen-specific antibody levels in human cytomegalovirus infection. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3003647. 128ra142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davey M.S., Willcox C.R., Joyce S.P., et al. Clonal selection in the human Vdelta1 T cell repertoire indicates gammadelta TCR-dependent adaptive immune surveillance. Nat Commun. 2017;8:14760. doi: 10.1038/ncomms14760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen T.H., Rowntree L.C., Pellicci D.G., et al. Recognition of distinct cross-reactive virus-specific CD8+ T cells reveals a unique TCR signature in a clinical setting. J Immunol. 2014;192:5039–5049. doi: 10.4049/jimmunol.1303147. [DOI] [PubMed] [Google Scholar]
- 11.Rowntree L.C., Nguyen T.H.O., Farenc C., et al. A Shared TCR Bias toward an Immunogenic EBV Epitope Dominates in HLA-B*07:02-Expressing Individuals. J Immunol. 2020;205:1524–1534. doi: 10.4049/jimmunol.2000249. [DOI] [PubMed] [Google Scholar]
- 12.Lim J.J., Jones C.M., Loh T.J., et al. The shared susceptibility epitope of HLA-DR4 binds citrullinated self-antigens and the TCR. Sci Immunol. 2021:6. doi: 10.1126/sciimmunol.abe0896. [DOI] [PubMed] [Google Scholar]
- 13.Rowntree L.C., van den Heuvel H., Sun J., et al. Preferential HLA-B27 Allorecognition Displayed by Multiple Cross-Reactive Antiviral CD8(+) T Cell Receptors. Front Immunol. 2020;11:248. doi: 10.3389/fimmu.2020.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mifsud N.A., Illing P.T., Lai J.W., et al. Carbamazepine Induces Focused T Cell Responses in Resolved Stevens-Johnson Syndrome and Toxic Epidermal Necrolysis Cases But Does Not Perturb the Immunopeptidome for T Cell Recognition. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.653710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Illing P.T., Vivian J.P., Dudek N.L., et al. Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature. 2012;486:554–558. doi: 10.1038/nature11147. [DOI] [PubMed] [Google Scholar]
- 16.Fozza C., Barraqueddu F., Corda G., et al. Study of the T-cell receptor repertoire by CDR3 spectratyping. J Immunol Methods. 2017;440:1–11. doi: 10.1016/j.jim.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Pogorelyy M.V., Minervina A.A., Shugay M., et al. Detecting T cell receptors involved in immune responses from single repertoire snapshots. PLoS Biol. 2019;17 doi: 10.1371/journal.pbio.3000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goncharov M., Bagaev D., Shcherbinin D., et al. VDJdb in the pandemic era: a compendium of T cell receptors specific for SARS-CoV-2. Nat Methods. 2022;19:1017–1019. doi: 10.1038/s41592-022-01578-0. [DOI] [PubMed] [Google Scholar]
- 19.Morin A., Kwan T., Ge B., et al. Immunoseq: the identification of functionally relevant variants through targeted capture and sequencing of active regulatory regions in human immune cells. BMC Med Genom. 2016;9:59. doi: 10.1186/s12920-016-0220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolotin D.A., Poslavsky S., Mitrophanov I., et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods. 2015;12:380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- 21.Krzywinski M., Schein J., Birol I., et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dash P., Fiore-Gartland A.J., Hertz T., et al. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature. 2017;547:89–93. doi: 10.1038/nature22383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shugay M., Bagaev D.V., Turchaninova M.A., et al. VDJtools: Unifying Post-analysis of T Cell Receptor Repertoires. PLoS Comput Biol. 2015;11 doi: 10.1371/journal.pcbi.1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aouinti S., Giudicelli V., Duroux P., et al. IMGT/StatClonotype for Pairwise Evaluation and Visualization of NGS IG and TR IMGT Clonotype (AA) Diversity or Expression from IMGT/HighV-QUEST. Front Immunol. 2016;7:339. doi: 10.3389/fimmu.2016.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nazarov V.I., Pogorelyy M.V., Komech E.A., et al. tcR: an R package for T cell receptor repertoire advanced data analysis. BMC Bioinforma. 2015;16:175. doi: 10.1186/s12859-015-0613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Team I. immunarch: an R package for painless bioinformatics analysis of T-cell and B-cell immune repertoires. Zenodo10. 2019:5281. [Google Scholar]
- 27.Mayer-Blackwell K., Schattgen S., Cohen-Lavi L., et al. TCR meta-clonotypes for biomarker discovery with tcrdist3: identification of public, HLA-restricted SARS-CoV-2 associated TCR features. BioRxiv. 2021 doi: 10.7554/eLife.68605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valkiers S., Van Houcke M., Laukens K., et al. ClusTCR: a Python interface for rapid clustering of large sets of CDR3 sequences with unknown antigen specificity. Bioinformatics. 2021 doi: 10.1093/bioinformatics/btab446. [DOI] [PubMed] [Google Scholar]
- 29.Ni Q., Zhang J., Zheng Z., et al. VisTCR: An Interactive Software for T Cell Repertoire Sequencing Data Analysis. Front Genet. 2020;11:771. doi: 10.3389/fgene.2020.00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bagaev D.V. Zvyagin IV, Putintseva EV et al. VDJviz: a versatile browser for immunogenomics data. BMC Genom. 2016;17:453. doi: 10.1186/s12864-016-2799-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bystry V., Reigl T., Krejci A., et al. ARResT/Interrogate: an interactive immunoprofiler for IG/TR NGS data. Bioinformatics. 2017;33:435–437. doi: 10.1093/bioinformatics/btw634. [DOI] [PubMed] [Google Scholar]
- 32.Lefranc M.P. Immunoglobulin and T Cell Receptor Genes: IMGT((R)) and the Birth and Rise of Immunoinformatics. Front Immunol. 2014;5:22. doi: 10.3389/fimmu.2014.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heikkila N., Kleino I., Vanhanen R., et al. Characterization of human T cell receptor repertoire data in eight thymus samples and four related blood samples. Data Brief. 2021;35 doi: 10.1016/j.dib.2021.106751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 36.Hennig C. fpc: Flexible procedures for clustering. R Package Version. 2020;2:1–9. [Google Scholar]
- 37.Wickham H., Averick M., Bryan J., et al. Welcome to the Tidyverse. J Open Source Softw. 2019;4:1686. [Google Scholar]
- 38.Wickham H. ggplot2, Wiley Interdiscip Rev-Comput Stat. 2011;3:180–185. [Google Scholar]
- 39.Slowikowski K. ggrepel: Automatically position non-overlapping text labels with ‘ggplot2′. 2018.
- 40.Chang W., Cheng J., Allaire J. et al. Shiny: web application framework for R. 2021.
- 41.Bailey E. shinyBS: twitter bootstrap components for Shiny., 2015.
- 42.Auguie B., Antonov A. gridExtra: miscellaneous functions for “grid” graphics. 2017.
- 43.Xie Y., Cheng J., Tan X. DT: A wrapper of the javascript library “DataTables”. 2018.
- 44.Wickham H. The split-apply-combine strategy for data analysis. J Stat Softw. 2011;40:1–29. [Google Scholar]
- 45.Wickham H., Francois R., Henry L. et al. dplyr: A grammar of data manipulation. 2022.
- 46.Wickham H. Reshaping data with the reshape package. J Stat Softw. 2007;21:1–20. [Google Scholar]
- 47.Wilkins D. treemapify: Draw Treemaps in “ggplot2.”. 2019.
- 48.Gu Z., Gu L., Eils R., et al. circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 49.Ou J., Wolfe S.A., Brodsky M.H., et al. motifStack for the analysis of transcription factor binding site evolution. Nat Methods. 2018;15:8–9. doi: 10.1038/nmeth.4555. [DOI] [PubMed] [Google Scholar]
- 50.Wickham H., Seidel D. Scales: Scale functions for visualization. 2020.
- 51.Ellis B., Haaland P., Hahne F. et al. flowCore: Basic structures for flow cytometry data., 2020.
- 52.Wickham H., Bryan J. readxl: Read Excel Files. 2019.
- 53.Neuwirth E. RColorBrewer: ColorBrewer palettes. 2014.
- 54.Ammar R. randomcoloR: Generate Attractive Random Colors. 2019.
- 55.Attali D. colourpicker: A Colour Picker Tool for Shiny and for Selecting Colours in Plots. 2021.
- 56.Nettling M., Treutler H., Grau J., et al. DiffLogo: a comparative visualization of sequence motifs. BMC Bioinforma. 2015;16:387. doi: 10.1186/s12859-015-0767-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oksanen J., Blanchet F., Friendly M. et al. vegan: community ecology package., 2022.
- 58.Athey T.B.T., McNicholas P.D. VLF: Frequency Matrix Approach for Assessing Very Low Frequency Variants in Sequence Records., 2013.
- 59.Perrier V., Meyer F., Granjon D. shinyWidgets: Custom Inputs Widgets for Shiny., 2022.
- 60.Qiu Y.X. showtext: Using System Fonts in R Graphics. R J. 2015;7:99–108. [Google Scholar]
- 61.Wagih O. ggseqlogo: A ‘ggplot2′extension for drawing publication-ready sequence logos. Bioinformatics. 2017;33:3645–3647. doi: 10.1093/bioinformatics/btx469. [DOI] [PubMed] [Google Scholar]
- 62.Allaire J., Horner J., Xie Y., et al. markdown: Render Markdown with the C Library’Sundown’. R Package Version. 2019:1. [Google Scholar]
- 63.Xie Y., Allaire J.J., Grolemund G.R. Chapman and Hall/CRC; 2018. markdown: The definitive guide. [Google Scholar]
- 64.Hill J.T., Demarest B.L., Bisgrove B.W., et al. Poly peak parser: Method and software for identification of unknown indels using sanger sequencing of polymerase chain reaction products. Dev Dyn. 2014;243:1632–1636. doi: 10.1002/dvdy.24183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vavrek M.J. Fossil: palaeoecological and palaeogeographical analysis tools, Palaeontologia electronica 2011;14:16.
- 66.Konopka T. umap: Uniform Manifold Approximation and Projection.(2020). R package version 0.2. 7.0. 2022.
- 67.Penter L., Dietze K., Bullinger L., et al. FACS single cell index sorting is highly reliable and determines immune phenotypes of clonally expanded T cells. Eur J Immunol. 2018;48:1248–1250. doi: 10.1002/eji.201847507. [DOI] [PubMed] [Google Scholar]
- 68.Pan R.Y., Chu M.T., Wang C.W., et al. Identification of drug-specific public TCR driving severe cutaneous adverse reactions. Nat Commun. 2019;10:3569. doi: 10.1038/s41467-019-11396-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ko T.M., Chung W.H., Wei C.Y., et al. Shared and restricted T-cell receptor use is crucial for carbamazepine-induced Stevens-Johnson syndrome. J Allergy Clin Immunol. 2011;128:1266–U1624. doi: 10.1016/j.jaci.2011.08.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Data Availability Statement
The demonstration data is from Mifsud et al. (2021) [14] and Lim et al. (2021) [12]. The local version of ‘TCR_Explore’ and all the raw data files and processed datasheets are located on GitHub https://github.com/KerryAM-R/TCR_Explore in the test-data section.