Abstract
The activity of a green tissue-specific promoter of the Rubisco small subunit gene from Arabidopsis (AraSSU) was studied using transgenic chickpea lines. We generated transgenic chickpea lines expressing an AraSSU promoter-driven cry2Aa gene through the Agrobacterium-mediated transformation method. Lines with AraSSU expressed the gene in all green tissues at high levels (> 90 ng/mg of fresh weight tissue) compared to lines generated using CaMV35S (< 10 ng/mg FW). We used vertical cross sections of various tissues of homozygous progeny using microtome for immunolocalization. The immunolocalization showed the expression of the cry2Aa gene in the green mesophyll cells of the leaves of both AraSSU and CaMV35 chickpea lines. Moreover, the accumulation of AraSSU-regulated Cry2Aa protein was also observed in vascular tissues, including enucleate sieve elements and their companion cells. However, no expression was observed in the roots of AraSSU lines. In the case of CaMV35 lines, the transgene expression was observed in all the tissues. Since our data indicated that the AraSSU promoter is active in non-green tissues such as vascular bundles. Therefore, we validated this by RT-PCR. We found Cry2Aa RNA transcripts in leaves, stems without epidermis (for vascular tissues), and roots with and without epidermis. Thus, the AraSSU promoter is active in all above-ground tissues of the chickpea plant.
Keywords: AraSSU promoter, CaMV35S promoter cry2Aa, Chickpea, Immuno-localization
Introduction
The promoter is the main switch of gene regulation as it harbors specific sequences and elements that function as binding sites for RNA polymerase and transcription factors. A plant gene promoter consists of a TATA box, initiator element as well a well-defined transcription initiation site about 20–30 bp downstream of the TATA element (Ali and Kim 2019). Promoters are classified into three broad classes based on their activity: constitutive, inducible, and tissue-specific. Several plant promoters have been identified to date and used for regulating transgenes for crop improvement. When the expression of a transgene is required throughout the developmental stages, a constitutive promoter is used. In inducible expression, it is stimulated by different biotic and abiotic environmental factors, for example, the hsp70 gene promoter is induced by heat shock. Tissue-specific promoters direct the expression of a gene in one or more tissues or at a particular developmental stage of the plant (Ali and Kim 2019). In genetic engineering, constitutive promoters are widely used, for example, the cauliflower mosaic virus 35S (CaMV35S) promoter, the rice Actin1 (OsAct1), and the maize polyubiquitin (ZmUbi1), etc. (Lv and Zhang 2016). However, constitutive promoters often cause large consumption of plants' energy and nutrients leading to metabolic load (Bhullar et al. 2003). Tissue-specific promoters are preferred over constitutive promoters to reduce this burden. One of the classic examples of the tissue-specific promoter is the Arabidopsis Rubisco small subunit gene promoter, which is exclusively expressed in green tissues. Reportedly, the Arabidopsis small subunit (AraSSU) promoter appeared to generate 10- to 20-fold more transgene (cry1Ac) mRNA transcript and protein than the CaMV35S promoter with a duplicated enhancer (Wong et al. 1992) and tobacco small subunit (TobSSU) promoter in tobacco (Tabe et al. 1995).
Genes belonging to the ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit (rbcS) family in Arabidopsis thaliana are differentially regulated by different types of light (Terzaghi and Cashmore 1995). Rubisco is a multimeric protein that catalyzes the initial step of carbon fixation through the assimilation of CO2 by carboxylation of ribulose-1, 5-bisphosphate, and is also involved in photorespiration (Schwarte and Tiedemann 2011). In higher plants, it is composed of eight small subunits (rbcS) and eight large subunits (rbcL) of 14 and 55 kD, respectively (Dedonder et al. 1993). The small subunits are encoded by the rbcS multigene family in the nuclear genome, whereas large subunits are encoded by a single rbcL gene of the chloroplast genome (Izumi et al. 2012). In Arabidopsis thaliana, the small subunits of Rubisco are encoded by four genes and divided into two subfamilies based on linkage and sequence similarities. The three genes (rbcS-1b, rbcS-2b, and rbcS-3b) are tightly linked at a single locus, and the fourth gene (rbcS-1a) is at least 10 kb removed from, or completely unlinked to the B subfamily (Dedonder et al. 1993). It has been found that rbcS promoters can confer light-inducible and tissue-specific expression in transgenic plants (Gilmartin and Chua 1990). Furthermore, the expression pattern of rbcS in leaf tissue was studied by fusion of the rbcS promoter with the β-glucuronidase (gus) reporter gene in transgenic cotton (Song et al. 2000). The rbcS promoter was used for driving transgene expression in the photosynthetic tissues. For example, the rice rbcS promoter was used to express a delta-endotoxin gene (novel cry2AX1) of Bacillus thuringiensis in rice leaves to provide resistance against the rice leaf folder (Manikandan et al. 2016). In chickpea, the rbcS promoter was used to drive the expression of other Bt genes (cry2Aa and cry1Ac) against Helicoverpa (Acharjee et al. 2010; Chakraborty et al. 2016; Hazarika et al. 2019).
The generation of transgenic plants depends on several tissue culture factors. However, for appropriate transgene expression, the selection of promoter is important (Wang and Oard 2003). To obtain the desired level of transgene expression, it is important to choose an appropriate promoter. The activity of the promoter may vary from tissue to tissue as well as from plant to plant. Therefore, understanding the activity of the promoter to achieve optimum levels of transgene expression is crucial prior to initiating transformation experiments. One of the approaches to studying the activity of promoters is by developing transgenic plants. The location of a foreign protein in a transgenic plant could facilitate further insights into the precise function of the promoter. Fusing the promoter with a reporter gene such as β-glucuronidase (gus), GFP or luciferase was a commonly used method to study the promoter activity (Ali and Kim 2019). Also, immunolocalization of transgenic protein in stably transformed lines is useful to know the activity of a promoter (Sudhakar et al. 1998; Kiani et al. 2013; Chakraborty et al. 2016).
Chickpea is an important grain legume that serves as an affordable source of protein for humans and livestock (Jukanti et al. 2012; Rachwa-Rosiak et al. 2015). The crop suffers significant yield losses, annually, due to Helicoverpa armigera (Sarmah et al. 2012). We generated several transgenic lines expressing either a cry2Aa gene regulated by AraSSU promoter (Acharjee et al. 2010). Moreover, chickpea is a self-pollinated crop with an extremely narrow genetic base and variability that restricts the scope of heterosis breeding. Therefore, the introduction of a lepidopteran-specific toxin, such as that encoded by the cry genes of Bacillus thuringiensis (Bt), into chickpea by genetic engineering is a promising option for developing insect resistance. This method has been successfully implemented in other important crop plants through the expression of different Bt genes for insecticidal crystal proteins (Manikandan et al. 2016; Ghosh et al. 2017). Since Helicoverpa prefers to feed on green foliage and reproductive parts (fruits), it was considered that a green tissue-specific promoter would efficiently target the Bt protein to the pod borer. To this end, the AraSSU promoter was used to generate transgenic chickpea lines expressing a cry2Aa gene which has been shown to decrease the growth and survival of pod borer (Acharjee et al. 2010). However, the lines with high levels of Cry2Aa protein exhibited slow growth and vigor, as well as significantly lower seed set per plant. As discussed earlier, the expression of a transgene appears to be highly regulated by a promoter driving the gene.
This work was based on the study done by Acharjee et al. (2010) where transgenic chickpea lines harboring the cry2Aa gene driven by the AraSSU promoter. The use of endogenous promoters was avoided as homologous promoters may lead to transgene and/or the resident gene/transgene silencing attributed to methylation of promoters (Vaucheret 1993; Park et al. 1996). However, Sunilkumar et al. (2002) demonstrated that the introduction of a homologous seed-specific promoter does not cause silencing of either the transgenic or resident promoter in transgenic cotton, Arabidopsis, and tobacco. Here we investigated the activity of AraSSU promoter by performing immunolocalization of transgenic Cry2Aa protein-specific antibodies.
Materials and methods
Vector constructs
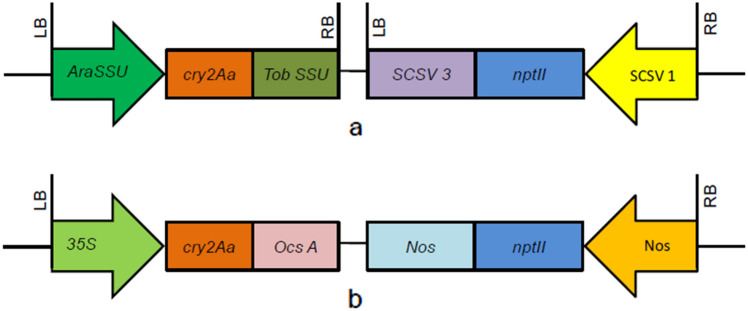
For chickpea transformation, two binary vectors, namely pBK201 and pBin35S2Aa, were used (Fig. 1). The twin binary vector pBK201 (Fig. 1a) was described previously by Acharjee et al. (2010). It harbored a modified synthetic cry2Aa gene controlled by AraSSU promoter and TobSSU terminator. The second construct pBin352Aa (Fig. 1b) was obtained from the ICAR-National Research Centre for Plant Biotechnology, New Delhi, also known as ICAR-NIPB. The pBin352Aa harbored a single T-DNA containing both the cry2Aa and neomycin phosphotransferase (npt-II) expression cassettes. The coding sequence of the cry2Aa gene was driven by a CaMV35S promoter and terminated by a nopaline synthase (nos) gene terminator. Both binary vectors contained the kanamycin resistance gene (npt-II) for the selection of transgenic plants.
Fig. 1.
Schematic representation of plasmid constructs used for genetic transformation of chickpea: a T-DNA of pBK201 harboring cry2Aa gene driven by AraSSU promoter and TobSSU terminator in a twin T-DNA binary vector, b T-DNA of pBin35S2Aa containing cry2Aa gene driven by CaMV35S promoter and nptII gene driven by NOS promoter, LB, RB left and right borders of T-DNA, CaMV35S cauliflower mosaic virus promoter, NOS nopaline synthase promoter and terminator, OCS octopine synthase terminator, AraSSU Arabidopsis thaliana small subunit promoter, TobSSU tobacco small subunit terminator, nptII neomycin phosphotransferase II gene, SCSV1 subclover stunt virus segment 1 promoter, SCSV3 subclover stunt virus segment 3 terminator
Plant material
We used two transgenic chickpea lines for the current study, BS6H (AraSSU-cry2Aa) and 39 J (35S-cry2Aa). The BS6H line contained a single copy of the cry2Aa gene (described in detail in Acharjee et al. 2010) which was also confirmed by its segregation pattern in the T1 generation. For immunolocalization studies, we selected homozygous T8 progeny of transgenic chickpea lines harboring the cry2Aa gene driven by AraSSU promoter (previously generated by Acharjee et al. 2010) and T1 progeny of chickpea lines harboring the cry2Aa gene driven by CaMV35S promoter. Seeds from transgenic and non-transgenic control plants were germinated and grown in the greenhouse. After 26 days, leaves and stems were harvested for immunolocalization studies.
Quantitative estimation of Cry2Aa protein in leaf tissue of transgenic chickpea plants
Quantitative estimation of Cry2Aa expression was determined through enzyme-linked immunosorbent analysis (ELISA) as described by Acharjee et al. (2010). Total leaf protein was extracted after homogenizing 60–100 mg of young open leaves using PBST (Phosphate-Buffered Saline with Tween 20) buffer (Agdia, USA) and subjected to centrifugation (12,000 rpm, 10 min, 4 °C) to collect the supernatant. Protein concentration was determined by Bradford assay (Bradford 1976). The protein samples were added to wells of a primary antibody-coated ELISA plate (Agdia, USA), followed by incubation with a second antibody (AP-conjugated goat anti-rabbit IgG antibody, Promega, Inc., USA) at 1:5000 dilutions at 4 °C overnight in a sealed container and finally washed seven times with PBST buffer. After the addition of the substrate, TMB (3, 3ʹ, 5, 5ʹ-tetramethylbenzidine) color was developed and absorbance was measured at 650 nm by a microplate reader (Bio-Rad, USA) and the amount of expressed Bt protein was calculated using a Cry2Aa toxin standard curve.
Tissue fixation, embedding, and sectioning
Leaves and stem from 26-day-old transgenic chickpea and control plants were fixed in fixation solution (10% formalin, 50% ethanol, and 5% acetic acid + 35% water) for 24 h followed by a rinse in running tap water at room temperature (RT). Tissues were partially dehydrated in a graded ethanol series (70, 90, and 100% (v/v) for 30 min each respectively) followed by ethanol: xylene series (3:1, 1:1, and 1:3 (v/v)) for 1 h each at RT. They were then gradually infiltrated in molten paraffin wax (HiMedia, India). All the infiltration steps were done at room temperature. Finally, the samples were placed in paper boats for about 20 h for the preparation of blocks. The embedded tissues were sliced into 12 µm sections using a microtome (Leica Instruments GmbH, Germany).
Immuno-localization
Transverse sections of leaves and stems from both transgenic and untransformed chickpea plants were used for immunofluorescence studies. Sections were affixed to slides pre-coated with egg albumin. The samples were then deparaffinized with xylene and rehydrated through a series of graded ethanol (100, 90, and 70% (v/v), respectively each for 30 min) at RT. The sections were incubated for 5 min in 10 mM citrate buffer, pH 6.0 at 100 °C to inactivate endogenous alkaline phosphatase activity followed by washing twice in Tris-buffered saline (TBS: 0.1 M Tris, 0.1 M NaCl, pH 7.4), each of 10 min duration. Free binding sites of antibodies were blocked over 1 h with 5% (w/v) bovine serum albumin (BSA, Sigma-Aldrich, USA) in TBS. Anti-Cry2Aa antibody (1:2000 dilutions in TBS with 3% (w/v) BSA) was applied to the sections for 1 h at room temperature (RT). Excess antibody was removed with successive washing in 0.5% (v/v) Tween 20 in TBS. After a second blocking, the secondary antibody (goat anti-rabbit IgG conjugated with alkaline phosphatase at 1:5000 dilutions in TBS) was applied and the preparation was extensively washed as above. Sections were incubated in TBS containing the substrate 5-bromo-4-chloro-3-indolyl-phosphate and 4-nitroblue tetrazolium chloride (BCIP/NBT) tablet to visualize the antigen–antibody complex. Another fluorescently labeled secondary antibody (Alexa Fluor 488 goat anti-rabbit) diluted at 1:200 in TBS was used for immunofluorescence. This preparation was washed as mentioned above. Observations were performed under an Olympus BX60 microscope equipped with fluorescence. Images were collected and assembled by Adobe Photoshop 7.0.
The activity of AraSSU promoter in various tissues by RT-PCR
Total RNA was isolated using 1 ml of TRIZOL reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The homogenate was incubated for 5 min at room temperature. 200 µl of chloroform was added and incubated at RT for another 2–3 min. The mixture was then centrifuged at 12,000 rpm for 15 min at 4 °C. The aqueous phase was transferred into a fresh tube and 500 µl of isopropanol was added and incubated at room temperature for 10 min followed by centrifugation at 12,000 rpm for 10 min at 4 °C. The supernatant was removed and the RNA pellet was washed with 70% ethanol and air-dried. Finally, the RNA pellet was air-dried, dissolved in 20 µl of DEPC-treated water, and stored at − 80 °C. The RNA samples were resolved on 1.2% agarose gel in 1X TAE buffer (40 mM Tris, 20 mM Glacial acetic acid, 1 mM EDTA). The quality and the quantity of total RNA were determined by formaldehyde-denatured agarose gel electrophoresis and an absorption ratio of OD 260/280 in a nanodrop spectrophotometer. RT-PCR analysis of transgenic chickpea lines for semi-quantitative RNA expression of the transgene transcripts was performed using the QIAGEN OneStep RT-PCR Kit according to the protocol provided by the manufacturer and was subsequently amplified by PCR with 5 (TAGCTTCGTACCGACGCAGTC) 3 and 5 (AGCTAGATGTCGCTCCTCACTGC) 3 primers, which were designed based on cry2Aa gene sequence. Amplification of rRNA was done using primers 5 (GTCAGCAACTGGGATGATATGG) 3 and 5 (TCTTCCTTGCTCATCCTGTCAG) 3 which served as an internal control. RT-PCR conditions were reverse transcription at 50 °C for 30 min, 15 min at 95 °C, 28 cycles (1 min at 94 °C, 1 min at 58 °C and 1 min at 72 °C) and 10 min at 72 °C.
Results
Level of expression of Cry2Aa protein in transgenic chickpea lines
A quantitative assessment of Cry2Aa protein accumulated in leaves of transgenic chickpea lines generated with pBK201 and pBin352Aa constructs was performed by ELISA. Among the two promoters (AraSSU and CaMV35S), the concentration of the Cry2Aa protein in the leaves of the AraSSU line was higher (97 ng/mg FW), whereas the CaMV35S lines accumulated less (< 10 ng/mg FW) (Table 1). While no Cry2Aa protein accumulation was found in untransformed chickpea plants, we also looked at the morphology of both the lines and found that CaMV35S lines are similar in growth and architecture compared to the control (data not shown). However, the AraSSU line showed phenotypic aberrations, such as stunting and low seed yield, as reported, previously by Acharjee et al. 2010.
Table 1.
Expression level of Cry2Aa in transgenic chickpea plants based on quantitative ELISA
| Transgenic line | Level of Cry2Aa protein (ng/mg FW) | Mortality (%) | Phenotype |
|---|---|---|---|
| AraSSU-Cry2Aa | 97 | 100 | Reduced yield, stunted growth |
| CaMV-Cry2Aa | 5 | ** | Normal |
**Mortality assay of transgenic plant was not performed due to low levels of expressed Cry2Aa protein
Activity of both AraSSU and CaMV35S promoter
To determine the location of the Cry2Aa protein driven by two different promoters in the leaf sections of transgenic chickpea, immunocytochemical assays were performed using the anti-Cry2Aa antibodies (non-fluorescent and fluorescent) as mentioned in the material and methods section. Since the aim of this study was to establish a putative relationship between the detection of protein and promoter driving the transgene expression in a tissue-specific and constitutive manner, leaf sections were used for immunolocalization studies in the transgenic chickpea lines along with a control. Also, since the AraSSU is a tissue-specific promoter, vertical sections of green tissues (leaves and stem) were used for the study in the high expressing line. Similarly, leaf sections from transgenic chickpea generated using a constitutive promoter, CaMV35S, were used for the study using fluorescent antibodies.
At first, we performed the assay on a transgenic chickpea line generated using the AraSSU promoter. For this experiment, non-fluorescent antibodies produced against the Cry2Aa and alkaline phosphatase-conjugated (anti-rabbit) secondary antibodies were utilized to obtain a histochemical stain for antibody binding. This method was found to be optimal for the detection of Cry2Aa with the reduced background as observed in the control tissues. When leaf sections of transgenic chickpea were immunolabelled, the presence of Cry2Aa protein was visualized as a purple stain (Supplementary Fig. 1). The protein appears located in the cell wall of mesophyll cells, the epidermal layer, and the enucleate sieve elements (xylem and phloem) and their companion cells of the leaf section of the transgenic chickpea. No accumulation of Cry2Aa protein was detected in the leaf sections of the untransformed plant. In stem sections of transgenic chickpea immunolabelled with anti-Cry2Aa, the protein was observed in the enucleate sieve elements (xylem and phloem) and their companion cells. No accumulation of Cry2Aa was detected in the leaf sections of the untransformed plant.
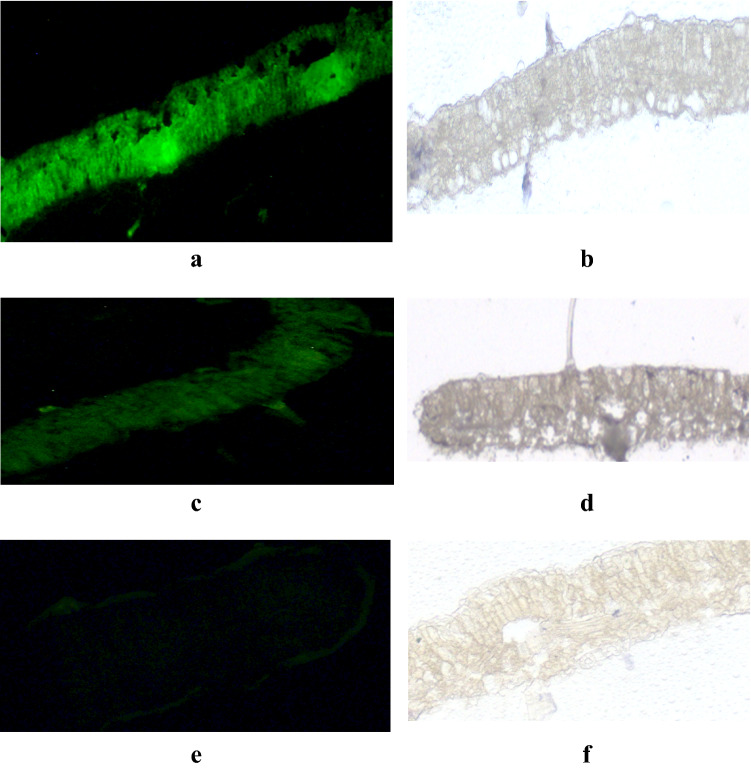
However, this non-fluorescent antibody technique was less conclusive, which was evaluated by using a fluorescently labeled secondary antibody to visualize the accumulation of Cry2Aa protein in the leaf sections of transgenic chickpea lines. Cry2Aa protein was localized in ultrathin leaf sections of 26-day-old transgenic chickpea lines generated using either the AraSSU or CaMV35S promoter. When the fluorescently labeled secondary antibody was used, Cry2Aa protein was detected in the palisade parenchyma cells, mesophyll cells, vascular bundles, and companion cells of the leaf sections of high-expressing AraSSU-2Aa chickpea line (Fig. 2a). This indicates that the AraSSU promoter-driven cry2Aa gene is either expressed by the companion cells of vascular tissues or might be transported into the vascular tissue from the mesophyll cells. The protein was also detected in the trichome (Fig. 2a). Another transgenic chickpea line generated using the CaMV35S promoter-driven cry2Aa gene showed accumulation of Cry2Aa protein over the non-vascular tissues, mesophyll cells, parenchyma cells and trichome (Fig. 2c). However, immunofluorescence of the tracheary elements compared to the vascular tissues of AraSSU line was less intense. Detection of immunofluorescence in the control sections incubated similarly with antibody as in the transgenic sections was found to be negligible (Fig. 2e).
Fig. 2.
Immuno-localization of transgenic and untransformed chickpea leaves. Transverse leaf sections were incubated with anti-Cry2Aa anti-serum as primary antibody and goat anti-rabbit IgG-Alexa Fluor 488-conjugated secondary antibody. The accumulation of Cry2Aa is indicated by the green fluorescence. Leaf sections of transgenic chickpea line AraSSU-Cry2Aa displayed bright green fluorescence (a, b) and CaMV-Cry2Aa demonstrated faint green fluorescence in all cell types (c and d) and untransformed chickpea leaf sections did not show green fluorescence comparatively (e and f). AraSSU-Cry2Aa line showed a deep fluorescence signal compared to the CaMV35S-Cry2Aa line, indicating a higher amount of Bt protein accumulation. Magnification: 10×
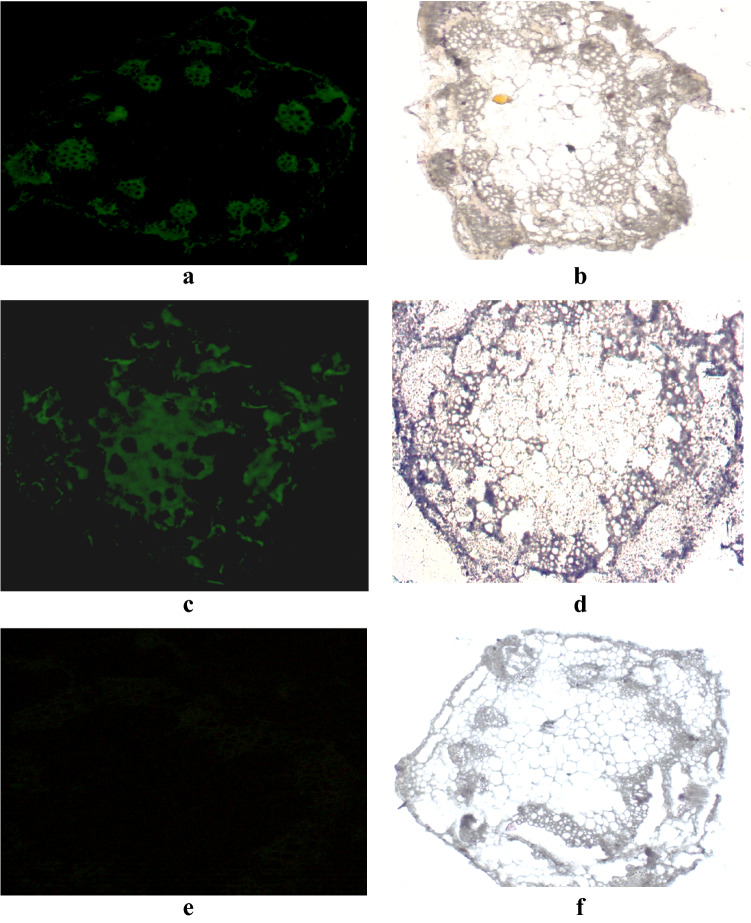
Internodes from the 26-day-old transgenic chickpea plants were dissected, fixed, and embedded for immunolocalization assay to determine the pattern of Cry2Aa expression along the stem. Initially, AraSSU-2Aa plants were used for the assay using anti-Cry2Aa antibodies. In the stem sections of the transgenic chickpea line, where the central pith is surrounded by vascular bundles, immunolabelling was observed in vascular tissues, both xylem, and phloem cells, and the signal was also detected in the sclerenchyma cells (Supplementary Fig. 1). No accumulation of Cry2Aa was detected in the stem sections of the untransformed plant (Supplementary Fig. 1). In stem sections treated with fluorescently labeled secondary antibody, a strong immunolabelling was shown on the epidermis, maturing xylem, phloem, and their companion cells (Fig. 3a). This indicates that the AraSSU-driven cry2Aa gene is either expressed by the companion cells of vascular tissues or might be transported into the vascular tissue from the mesophyll cells. However, no immunolabelling was observed in the pith parenchyma cells (Fig. 3a). A limited degree of immunolabeling also occurred on the cortical parenchyma cells. However, it was attributed to autofluorescence by observing control sections treated with antibody (Fig. 3e).
Fig. 3.
Immuno-localization of Cry2Aa protein in the stem sections of transgenic chickpea line AraSSU-Cry2Aa (a and b) and CaMV-Cry2Aa (c and d) and untransformed chickpea (e and f) using fluorescent antibody (Alexa Fluor 488) Magnification: 10×
In the stem sections of transgenic chickpea line generated using CaMV35S promoter-driven cry2Aa gene, accumulation of Cry2Aa protein was shown in a nonspecific manner with uniform distribution of stain. Cry2Aa was detected over phloem sieve elements (Fig. 3c). Immunofluorescence of the tracheary elements, however, was less intense compared to the vascular tissues of the AraSSU line. Although immunofluorescence was observed in the epidermis and cortical cells, this reactivity was attributed to autofluorescence, since control sections incubated with Cry2Aa antibodies demonstrated epidermal fluorescence. However, no immunofluorescence was detected in the pith parenchyma cells of control sections incubated similarly with antibody as the transgenic sections were found to be negligible (Fig. 3e).
Expression analysis of AraSSU-driven transgene
To analyze the expression of the cry2Aa gene at the transcriptional level, cDNA was synthesized from isolated RNA described in the materials and methods section. The absence of genomic DNA contamination in the isolated total RNA was confirmed by PCR amplification using only RNA samples. The presence of Cry2Aa protein in the non-green tissues such as vascular bundle and root was investigated by semi-quantitative reverse transcriptase PCR analysis (RT-PCR) to know whether the promoter is active in those tissues. We performed RT-PCR of the cry2Aa gene driven by AraSSU promoter in the high-expressing transgenic chickpea line and a control chickpea plant. Total RNA was isolated from leaves, stems without epidermis (for vascular tissues), roots and roots without epidermis (for vascular tissues). The RT-PCR assay revealed that the cry2Aa gene was expressed in the leaves of the transgenic chickpea plant. The removal of the epidermis of stem samples also resulted in the expression of the cry2Aa gene transcript. It may be due to the high expression of Cry2Aa protein in the transgenic chickpea. As previously reported (Amarasinghe et al. 2006), a thin band of staining was also observed in the vasculature of roots of the highest expressing cotton line. No expression was observed in roots (with and without epidermis) in the transgenic line. Untransformed chickpea plants were used as control and total RNA was extracted only from leaves and stems without epidermis. Control sections did not show any expression (Fig. 4). The results are in agreement with the immunolocalization studies which showed the expression of Cry2Aa transcript in the non-green vascular tissues of high-expressing transgenic chickpea plant.
Fig. 4.
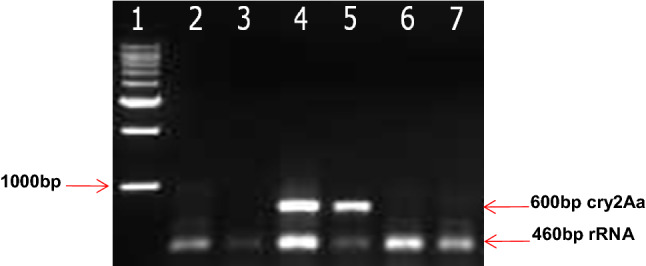
RT-PCR analysis of transgenic line AraSSU-Cry2Aa and control (untransformed plant) with cry2Aa and rRNA gene-specific primers from different tissues Lane 1: Marker; Lane2: Untransformed leaf; Lane 3: Untransformed stem; Lane 4: AraSSU-Cry2Aa leaves; Lane 5: AraSSU-Cry2Aa stem without epidermis; Lane 6: AraSSU-Cry2Aa root with epidermis; Lane 7: AraSSU-Cry2Aa root without epidermis
Discussion
For the first time, we have described immunolocalization of Cry2Aa protein in transgenic chickpea line harboring cry2Aa gene driven by AraSSU promoter and CaMV35S promoter. Two modes of visualization were used for immunodetection of Cry2Aa protein in the transgenic lines, one using an AP-conjugated secondary antibody and another fluorescently labeled secondary antibody. Chickpea is an important source of dietary protein and its production is severely constrained by H. armigera (Sharma et al. 2006) which feeds during vegetative and reproductive stages (Fitt 1989). It is one of the most significant lepidopteran pests attacking more than 180 species of plants (Lone et al. 2017). Among the Bt genes, cry1Ab, cry1Ac, cry2Ab, and cry9C have been used to generate transgenic plants for resistance to lepidopteran pests (Chen et al. 2005; Acharjee et al. 2010; Schünemann et al. 2014; Ghosh et al. 2017; Singh et al. 2018). In case of chickpea, the Bt gene has been previously demonstrated to confer protection against H. armigera (Kumar et al. 2018). The expression of Bt gene in transgenic plants is either tissue-specific or constitutive based on the promoter used. The expression of a transgenic protein depends on various factors, such as codon use, promoter, site of integration, and the number of copy integration into the genome. Generally, the level of expression in a transgenic plant with a single copy appears to be highly regulated by the promoter driving the gene. Transgenic chickpea lines harboring the cry2Aa gene have been generated using the AraSSU promoter. This promoter was shown to confer a tissue-specific expression of cry1Ac in transgenic tobacco plants (Wong et al. 1992). Recently, the CaMV35S promoter was used for constitutive expression of the cry2Aa gene in plant tissues and to evaluate differences in the level of expression between the two promoters. Transgenic chickpea lines harboring the cry2Aa gene driven by both AraSSU and CaMV35S promoters were generated with different levels of Cry2Aa expression. The Cry2Aa line driven by AraSSU promoter was found to be a high expresser as observed in western and northern analysis (Acharjee et al. 2010). On the contrary, the level of expression of the cry2Aa gene driven by the CaMV35S promoter was comparatively lower. The AraSSU promoter was shown to generate 10- to 20-fold more cry1Ac transcript and protein than CaMV35S promoter with duplicated enhancer in tobacco (Wong et al. 1992) and fivefold more SFA8 (sunflower albumin 8) transcript compared to Tobacco SSU and CaMV35S promoter in alfalfa (Tabe et al. 1995). Insect feeding bioassays on transformed chickpea plants showed a different level of toxicity and protection against pod borer (Acharjee et al. 2010). Larval mortality was found to be 100% in the high-expressing transgenic chickpea line compared to a medium-expressing line with 37% larval mortality. The relatively lower expression level of Cry2Aa protein as observed in the transgenic chickpea plants transformed with CaMV35S promoter compared to the AraSSU promoter does not always reflect the lower activity of the promoter. A higher level of Cry1Ac protein was observed in transgenic chickpea plants driven by the CaMV35S promoter (Sanyal et al. 2005; Mehrotra et al. 2011). The lower level of Cry2Aa in the transgenic chickpea line driven by the CaMV35S promoter can be explained based on the genotype of the plant used for transformation. The specificity of a genotype is found to be highly related to physiological conditions of a cell, such as concentration of cell internal hormone, the structure of cell wall, and physiological reaction after wounding. Similar observations on different transformation efficiency of chickpea cultivars were made by previous workers (Indurker et al. 2010). It might be the reason that chickpea cv. semsen showed a better response compared to the cv PBA Hatrick used for genetic transformation with CaMV35S promoter. Higher expression of transgene often exhibits unusual phenotypes. Tissue-specific expression of the cry2Aa gene was used for generating transgenic chickpea to confer protection against pod borer (Acharjee et al. 2010). However, the growth and the vigor of the lines expressing very high levels of Cry2Aa protein were found to be slow and the seed yield per plant reduced significantly (Acharjee et al. 2010). Similarly, transgenic rice expressing higher levels of Bt gene were stunted with poor seed set and matured before non-transgenic control plants (Gahakwa et al. 2000). In plant leaves, RuBisCO represents up to 50% of the total soluble proteins. To achieve higher expression levels of transgenes in green tissues, Rubisco small subunit (rbcS) promoters are most preferred over constitutive promoters (Cai et al. 2007). As stated earlier, expression of cry1Ac driven by Arabidopsis thaliana Rubisco small subunit (ats1A) promoter along with its transit peptide showed 10- to 20-fold mRNA and protein expression than CaMV35S promoter along with a duplication of the enhancer region (CaMV-En35S) (Wong et al. 1992). Similarly, Rubisco promoters from plant families Asteraceae and Fabaceae were also used to drive higher expression of the transgene. In tobacco, higher expression of the gus gene was achieved using a Rubisco small subunit promoter from chrysanthemum with an expression level of up to 10% of total soluble protein (Outchkourov et al. 2003). The expression pattern of rbcS was previously studied by the expression of the gus in transgenic rice, tobacco, and chickpea in green tissues as well as in non-green tissues in high-expressing lines. Histochemical analyses of the gusA gene driven by rbcS promoters from rice and tomato were found in the mesophyll cells of leaves of transgenic rice plants (Kyozuka et al. 1993). Similarly, the heterologous expression of the gus gene driven by a rbcS gene promoters RbsS3CP, from tomato (Lycopersicon esculentum Mill.) and SRS1P from soybean (Glycine max L.) was studied in transgenic apple (Malus pumila Mill.). The gus activity was localized to the mesophyll and palisade cells of the leaf as demonstrated by histochemical analysis (Gittins et al. 2000). Tobacco plants transformed with the gus gene driven by rbcS promoter from coffee (Coffea arabica) showed the gus activity in leaf tissues (Marraccini et al. 2003). Similarly, Chakraborty et al. (2016) reported the expression of a cry1Ac gene driven by chickpea rbcS in the mesophyll cells in transgenic chickpea plants. Further studies on the localization of the gus gene driven by rbcS promoters from cotton (Gossypium hirsutum L. cv. Coker 315) in transgenic Arabidopsis and cotton plants have shown that the protein was confined to green photosynthetic tissues. However, a thin band of staining was observed around the vasculature of high-expressing lines (Amarasinghe et al. 2006) and thus the expression of the cry2Aa gene in non-green tissues of the high-expressing chickpea line as reported in the previous studies (Fig. 2a). In transgenic chickpea plants transformed with the 35S-cry2Aa gene, faint green fluorescence was observed in all cell types including the vascular bundles, and similar results were reported by Chakraborty et al. (2016). It indicates that this promoter is dependent on the genotype of chickpea used for genetic transformation as we described previously.
Conclusion and prospects
In the present study, we have reported the immunochemical analysis of transgenic chickpea plants expressing the Cry2Aa insecticidal protein driven by tissue-specific and constitutive promoters. Simultaneously, we have also demonstrated the stable integration and the expression of the cry2Aa gene in a tissue-specific and constitutive manner using fluorescent and non-fluorescent antibodies. We have recovered transgenic plants expressing high levels of the Bt protein and immunolocalization studies have shown that the AraSSU promoter functions in a predictable manner in terms of driving heterologous gene expression in dicots. As transgenic chickpea plants expressed high levels of Cry2Aa protein, it caused developmental and morphological defects, such as stunting and poor seed set, it would be helpful in future studies to generate transgenic chickpea line harboring cry2Aa gene with the native promoter of Rubisco small subunit gene from chickpea. Thus, a comparative analysis can be made to identify the expression pattern of the cry2Aa gene driven by the rbcS promoter from chickpea and Arabidopsis in transgenic chickpea lines.
Acknowledgements
We would like to express our thanks to Dr. P. Ananda Kumar (Indian Council of Agricultural Research-National Institute for Plant Biotechnology, New Delhi, India) for providing the pBin352A vector and Dr. Montu Bhuyan (Council of Scientific and Industrial Research-North East Institute of Science and Technology, Assam, India) for kindly providing the microtome used in this study. The authors acknowledge the Department of Biotechnology (DBT), Government of India, New Delhi, and the Indian Council of Agriculture Research (ICAR), New Delhi for funding assistance.
Author contributions
SA conceptualized and designed the experiments; BKS and SA contributed all the reagents, materials and analysis tools. RRB carried out transgenic experiments of CaMV35S-2Aa plants, RT-PCR analysis, ELISA, Immuno-histo-fluorescence, and statistical analysis. RRB, TK, SA, and BKS analyzed the data. RRB drafted the manuscript. SA edited the manuscript and supervised the work. BKS reviewed the manuscript. All the authors read and approved the manuscript.
Data availability
Availability of data and materials not applicable.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Rashmi Rekha Boruah, Email: rashmi.r.boruah@gmail.com.
Trishna Konwar, Email: trishna_xyz@yahoo.co.in.
Pranab Kumar Nath, Email: pranabkumar_nath@yahoo.com.
Sumita Acharjee, Email: sumita.acharjee@aau.ac.in, Email: sumita.aus@gmail.com.
Bidyut Kumar Sarmah, Email: bidyutsarmah@aau.ac.in.
References
- Acharjee S, Sarmah BK, Kumar PA, Olsen K, Mahon R, Moar WJ, Moore A, Higgins TJ. Transgenic chickpeas (Cicer arietinum L.) expressing a sequence-modified cry2Aa gene. Plant Sci. 2010;178:333–339. [Google Scholar]
- Ali S, Kim WC. A fruitful decade using synthetic promoters in the improvement of transgenic plants. Front Plant Sci. 2019;10:1433. doi: 10.3389/fpls.2019.01433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasinghe BHRR, Faivre-Nitschke E, Wu Y, Udall JA, Dennis ES, Constable G, Llewellyn DJ. Genomic approaches to the discovery of promoters for sustained expression in cotton (Gossypium hirsutum L.) under field conditions: expression analysis in transgenic cotton and Arabidopsis of a Rubisco small subunit promoter identified using EST sequence analysis and cDNA microarrays. Plant Biotechnol. 2006;23:437–450. [Google Scholar]
- Bhullar S, Chakravarthy S, Advani S, Datta S, Pental D, Burma PK. Strategies for development of functionally equivalent promoters with minimum sequence homology for transgene expression in plants: cis-elements in a novel DNA context versus domain swapping. Plant Physiol. 2003;132:988–998. doi: 10.1104/pp.103.020602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cai M, Wei J, Li X, Xu C, Wang S. A rice promoter containing both novel positive and negative cis-elements for regulation of green tissue-specific gene expression in transgenic plants. Plant Biotechnol J. 2007;5:664–674. doi: 10.1111/j.1467-7652.2007.00271.x. [DOI] [PubMed] [Google Scholar]
- Chakraborty J, Sen S, Ghosh P, Sengupta A, Basu D, Das S. Homologous promoter derived constitutive and chloroplast targeted expression of synthetic cry1Ac in transgenic chickpea confers resistance against Helicoverpa armigera. Plant Cell, Tissue Organ Cult. 2016;125:521–535. [Google Scholar]
- Chen H, Tang W, Xu C, Li X, Lin Y, Zhang Q. Transgenic indica rice plants harboring a synthetic cry2A* gene of Bacillus thuringiensis exhibit enhanced resistance against lepidopteran rice pests. Theor Appl Genet. 2005;111:1330. doi: 10.1007/s00122-005-0062-8. [DOI] [PubMed] [Google Scholar]
- Dedonder A, Rethy R, Fredericq H, Van Montagu M, Krebbers E. Arabidopsis rbcS genes are differentially regulated by light. Plant Physiol. 1993;101:801–808. doi: 10.1104/pp.101.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitt GP. The ecology of Heliothis species in relation to agroecosystems. Annu Rev Entomol. 1989;34:17–53. [Google Scholar]
- Gahakwa D, Maqbool SB, Fu X, Sudhakar D, Christou P, Kohli A. Transgenic rice as a system to study the stability of transgene expression: multiple heterologous transgenes show similar behavior in diverse genetic backgrounds. Theor Appl Genet. 2000;101:388–399. [Google Scholar]
- Ghosh G, Ganguly S, Purohit A, Chaudhuri RK, Das S, Chakraborti D. Transgenic pigeonpea events expressing Cry1Ac and Cry2Aa exhibit resistance to Helicoverpa armigera. Plant Cell Rep. 2017;36:1037–1051. doi: 10.1007/s00299-017-2133-0. [DOI] [PubMed] [Google Scholar]
- Gilmartin PM, Chua NH. Spacing between GT-1 binding sites within a light-responsive element is critical for transcriptional activity. Plant Cell. 1990;2:447–455. doi: 10.1105/tpc.2.5.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittins JR, Pellny TK, Hiles ER, Rosa C, Biricolti S, James DJ. Transgene expression driven by heterologous ribulose-1, 5-bisphosphate carboxylase/oxygenase small-subunit gene promoters in the vegetative tissues of apple (Malus pumila Mill.) Planta. 2000;210:232–240. doi: 10.1007/PL00008130. [DOI] [PubMed] [Google Scholar]
- Hazarika N, Acharjee S, Boruah RR, Babar K, Parimi S, Char B, Armstrong J, Moore A, Higgins TJ, Sarmah BK. Enhanced expression of Arabidopsis rubisco small subunit gene promoter regulated Cry1Ac gene in chickpea conferred complete resistance to Helicoverpa armigera. J Plant Biochem Biotechnol. 2019;1:1–11. [Google Scholar]
- Indurker S, Misra HS, Eapen S. Agrobacterium-mediated transformation in chickpea (Cicer arietinum L.) with an insecticidal protein gene: optimisation of different factors. Physiol Mol Biol Plants. 2010;16:273–284. doi: 10.1007/s12298-010-0030-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M, Tsunoda H, Suzuki Y, Makino A, Ishida H. RBCS1A and RBCS3B, two major members within the Arabidopsis RBCS multigene family, function to yield sufficient Rubisco content for leaf photosynthetic capacity. J Exp Bot. 2012;63:2159–2170. doi: 10.1093/jxb/err434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukanti AK, Gaur PM, Gowda CL, Chibbar RN. Nutritional quality and health benefits of chickpea (Cicer arietinum L.): a review. Br J Nutr. 2012;108:11–26. doi: 10.1017/S0007114512000797. [DOI] [PubMed] [Google Scholar]
- Kiani S, Ali A, Bajwa KS, Muzaffar A, Ashraf MA, Samiullah TR, Shahid AA, Husnain T. Cloning and chloroplast-targeted expression studies of insect-resistant gene with ricin fusion-gene under chloroplast transit peptide in cotton. Electron J Biotechnol. 2013;16:13–13. [Google Scholar]
- Kumar M, Yusuf MA, Nigam M. An update on genetic modification of chickpea for increased yield and stress tolerance. Mol Biotechnol. 2018;60:651–663. doi: 10.1007/s12033-018-0096-1. [DOI] [PubMed] [Google Scholar]
- Kyozuka J, McElroy D, Hayakawa T, Xie Y, Wu R, Shimamoto K. Light-regulated and cell-specific expression of tomato rbcS-gusA and rice rbcS-gusA fusion genes in transgenic rice. Plant Physiol. 1993;102:991–1000. doi: 10.1104/pp.102.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lone SA, Malik A, Padaria JC. Characterization of lepidopteran-specific cry1 and cry2 gene harbouring native Bacillus thuringiensis isolates toxic against Helicoverpa armigera. Biotechnol Rep. 2017;15:27–32. doi: 10.1016/j.btre.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv D, Zhang Y. Isolation and functional analysis of apple MdHMGR1 and MdHMGR4 gene promoters in transgenic Arabidopsis thaliana. Plant Cell, Tissue Organ Cult. 2016;129:133–143. [Google Scholar]
- Manikandan R, Balakrishnan N, Sudhakar D, Udayasuriyan V. Development of leaffolder resistant transgenic rice expressing cry2AX1 gene driven by green tissue-specific rbcS promoter. World J Microbiol Biotechnol. 2016;32:37. doi: 10.1007/s11274-015-2006-z. [DOI] [PubMed] [Google Scholar]
- Marraccini P, Courjault C, Caillet V, Lepage B, Rogers WJ, Tessereau S, Deshayes A. Rubisco small subunit of Coffea arabica: cDNA sequence, gene cloning and promoter analysis in transgenic tobacco plants. Plant Physiol Biochem. 2003;41:17–25. [Google Scholar]
- Mehrotra M, Singh AK, Sanyal I, Altosaar I, Amla DV. Pyramiding of modified cry1Ab and cry1Ac genes of Bacillus thuringiensis in transgenic chickpea (Cicer arietinum L.) for improved resistance to pod borer insect Helicoverpa armigera. Euphytica. 2011;182:87. [Google Scholar]
- Outchkourov NS, Peters J, De Jong J, Rademakers W, Jongsma MA. The promoter–terminator of chrysanthemum rbcS1 directs very high expression levels in plants. Planta. 2003;216:1003–1012. doi: 10.1007/s00425-002-0953-8. [DOI] [PubMed] [Google Scholar]
- Park Y-D, Papp I, Moscone EA, Iglesias VA, Vaucheret H, Matzke AJM, et al. Gene silencing mediated by promoter homology occurs at the level of transcription and results in meiotically heritable alterations in methylation and gene activity. Plant J. 1996;9:183–194. doi: 10.1046/j.1365-313x.1996.09020183.x. [DOI] [PubMed] [Google Scholar]
- Rachwa-Rosiak D, Nebesny E, Budryn G. Chickpeas-composition, nutritional value, health benefits, application to bread and snacks: a review. Crit Rev Food Sci Nutr. 2015;55:1137–1145. doi: 10.1080/10408398.2012.687418. [DOI] [PubMed] [Google Scholar]
- Sanyal I, Singh AK, Kaushik M, Amla DV. Agrobacterium-mediated transformation of chickpea (Cicer arietinum L.) with Bacillus thuringiensis cry1Ac gene for resistance against pod borer insect Helicoverpa armigera. Plant Sci. 2005;168:1135–1146. [Google Scholar]
- Sarmah BK, Acharjee S, Sharma HC. Chickpea: Crop Improvement under Changing Environment Conditions. In: Tuteja N, Gill SS, Tuteja R, editors. Improving Crop Productivity in Sustainable Agriculture. Weinheim, Germany: Wiley-Blackwell; 2012. pp. 361–380. [Google Scholar]
- Schünemann R, Knaak N, Fiuza LM. Mode of action and specificity of Bacillus thuringiensis toxins in the control of caterpillars and stink bugs in soybean culture. ISRN Microbiol. 2014;2014:e135675. doi: 10.1155/2014/135675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarte S, Tiedemann R. A gene duplication/loss event in the ribulose-1, 5-bisphosphate-carboxylase/oxygenase (Rubisco) small subunit gene family among accessions of Arabidopsis thaliana. Mol Biol Evol. 2011;28:1861–1876. doi: 10.1093/molbev/msr008. [DOI] [PubMed] [Google Scholar]
- Sharma KK, Bhatnagar-Mathur P, Jayanand B. Chickpea (Cicer arietinum L.) Agrobacterium Protocols. 2006;343:313–324. doi: 10.1385/1-59745-130-4:313. [DOI] [PubMed] [Google Scholar]
- Singh S, Kumar NR, Maniraj R, Lakshmikanth R, Rao KY, Muralimohan N, Arulprakash T, Karthik K, Shashibhushan NB, Vinutha T, Pattanayak D. Expression of Cry2Aa, a Bacillus thuringiensis insecticidal protein in transgenic pigeon pea confers resistance to gram pod borer, Helicoverpa armigera. Sci Rep. 2018;8:1–12. doi: 10.1038/s41598-018-26358-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song P, Heinen JL, Burns TH, Allen RD. Expression of two tissue-specific promoters in transgenic cotton plants. J Cotton Sci. 2000;4:217–223. [Google Scholar]
- Sudhakar D, Fu X, Stoger E, Williams S, Spence J, Brown DP, Bharathi M, Gatehouse JA, Christou P. Expression and immunolocalization of the snowdrop lectin, GNA in transgenic rice plants. Transgenic Res. 1998;7:371–378. doi: 10.1023/a:1008856703464. [DOI] [PubMed] [Google Scholar]
- Sunilkumar G, Connell JP, Smith CW, Reddy AS, Rathore KS. Cotton α-globulin promoter: isolation and functional characterization in transgenic cotton, Arabidopsis, and tobacco. Transgenic Res. 2002;11(4):347–359. doi: 10.1023/a:1016322428517. [DOI] [PubMed] [Google Scholar]
- Tabe LM, Wardley-Richardson T, Ceriotti A, Aryan A, McNabb W, Moore A, Higgins TJ. A biotechnological approach to improving the nutritive value of alfalfa. J Anim Sci. 1995;73:2752–2759. doi: 10.2527/1995.7392752x. [DOI] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR. Light-regulated transcription. Annu Rev Plant Biol. 1995;46:445–474. [Google Scholar]
- Vaucheret H. Identification of a general silencer for 19S and 35S promoters in transgenic tobacco plant: 90 bp of homology in the promoter sequence is sufficient for trans-inactivation. C R Acad Sci Paris. 1993;316:1471–1483. [Google Scholar]
- Wang J, Oard JH. Rice ubiquitin promoters: deletion analysis and potential usefulness in plant transformation systems. Plant Cell Rep. 2003;22:129–134. doi: 10.1007/s00299-003-0657-y. [DOI] [PubMed] [Google Scholar]
- Wong EY, Hironaka CM, Fischhoff DA. Arabidopsis thaliana small subunit leader and transit peptide enhance the expression of Bacillus thuringiensis proteins in transgenic plants. Plant Mol Biol. 1992;20:81–93. doi: 10.1007/BF00029151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Availability of data and materials not applicable.