Abstract
Motivation: Microbial metagenomic profiling software and databases are advancing rapidly for development of novel disease biomarkers and therapeutics yet three problems impede analyses: 1) the conflation of “genome assembly” and “strain” in reference databases; 2) difficulty connecting DNA biomarkers to a procurable strain for laboratory experimentation; and 3) absence of a comprehensive and unified strain-resolved reference database for integrating both shotgun metagenomics and 16S rRNA gene data.
Results: We demarcated 681,087 strains, the largest collection of its kind, by filtering public data into a knowledge graph of vertices representing contiguous DNA sequences, genome assemblies, strain monikers and bio-resource center (BRC) catalog numbers then adding inter-vertex edges only for synonyms or direct derivatives. Surprisingly, for 10,043 important strains, we found replicate RefSeq genome assemblies obstructing interpretation of database searches. We organized each strain into eight taxonomic ranks with bootstrap confidence inversely correlated with genome assembly contamination. The StrainSelect database is suited for applications where a taxonomic, functional or procurement reference is needed for shotgun or amplicon metagenomics since 636,568 strains have at least one 16S rRNA gene, 245,005 have at least one annotated genome assembly, and 36,671 are procurable from at least one BRC. The database overcomes all three aforementioned problems since it disambiguates strains from assemblies, locates strains at BRCs, and unifies a taxonomic reference for both 16S rRNA and shotgun metagenomics.
Availability: The StrainSelect database is available in igraph and tabular vertex-edge formats compatible with Neo4J. Dereplicated MinHash and fasta databases are distributed for sourmash and usearch pipelines at http://strainselect.secondgenome.com.
Contact:todd.desantis@gmail.com.
Supplementary information: Supplementary data are available online.
Graphical abstract
1. Statement of significance
Problem: Although clinical microbiome data is being evaluated for both precision biomedical decision support and therapeutic discovery, three problems impede translation of data into beneficial products and services: 1) the conflation of “genome assembly” and “strain” in reference databases; 2) difficulty mapping microbiome DNA biomarkers to an extant strain for purchase and experimentation; and 3) absence of a unified comprehensive strain-resolved reference database for integrating both shotgun data and 16S rRNA gene data.
What is Already Known: Reference databases, such as RefSeq, are currently available for organizing microbiome data at the strain-level resolution. Unfortunately, novices are unaware these databases contain multiple genome records generated from a single strain but deposited as separate strains. For instance, sequence data labeled as Mesorhizobium loti HAMBI 1129, M. loti DSM 2626, M. jarvisii ATCC 700743, and M. jarvisii ATCC 33669 are all from the same source strain isolate. As another example, four different RefSeq genome assemblies, GCF_001571425, GCF_001652705, GCF_001678855, and GCF_003628755, are all derived from the same source strain isolate. Most reference databases improperly assume that each name bestowed to an organism and each genome assembly equates to a unique strain.
What This Paper Adds: We describe a method to identify 681,087 unique strains that are represented by over 8 million synonymous monikers in public records. We constructed a database that overcomes all three aforementioned problems since it disambiguates strains from assemblies, maps which strains are available for procurement and experimentation from a culture collection, even if those strains are named differently in the respective catalogs, and allows integration of both shotgun and 16S rRNA gene data against a single organized taxonomy which is a key utility for comprehensive meta-analyses and robust biomarker applications.
2. Introduction
Both shotgun metagenomics and 16S rRNA gene amplicon marker gene publications exhibit year-over-year growth (Fig. 1) due to broad applications in clinical, agricultural, and environmental data sciences. Depending on the experimental design, molecular microbiologists process the raw data to determine, as examples, which genera are significantly elevated in the colons of one group of patients relative to another [16], which combination of bacterial species predicts a beneficial response to a pharmaceutical agent [32], or which novel chromosomes from yet-to-be-cultured bacteria can be reconstructed from 0.1 to 8.0 kilo-base sequencing reads in silico into metagenomic assembled genomes (MAGs) [1], [39], [52] to discover novel CRISPR-Cas systems [6]. But other investigators will go beyond descriptive analytics and will conduct follow-up experiments to establish causation linking certain strains or their products to a particular outcome in an animal model of disease [49], [53] or an agricultural field trial [35]. To accomplish this, microbiome data would need to be interpreted with methods to reveal the individual strains associated with the outcome of interest. Then in an efficient manner those strains would need to be grown in the lab and tested against controls in experiments structured to prove/disprove the causal hypothesis. A data engineer tasked to determine the set of strains within a metagenomics data set that significantly associate with an experimental variable and then to map those strains against worldwide bio-resource centers (BRCs) from which individual strains can be purchased, will need to first settle on a definition of a “strain” that fits this endeavor. Then, the engineer must overcome three challenges which motivated this work: 1) ambiguity between a “genome assembly” and a “strain” in reference databases; 2) difficulty connecting observations in the metagenomic analysis to a procurable strain for laboratory experimentation; and, if confirmation of findings among different library techniques was desired as in Tessler et al. [54], 3) integration of both shotgun and 16S rRNA data against a single reference.
Figure 1.
Quantity of Pubmed indexed publications found with search terms 16S or metagenom* (where * represents a wildcard) have increased throughout the last two decades. Publications leveraging metagenomics are less frequent than those leveraging 16S rRNA gene amplicons. A reference database that enables integration of both technologies is ideal.
An investigator will need to be clear about their operational definition of “strain” for the investigation and they may favor the MAG definition, where each unique MAG is one strain, or the isolate-and-propagate definition where an isolate and its descendants are one strain. If the investigator adopts the definition that any chromosomal variant among any MAG from any biospecimen is a distinct strain, then a reference database is not required nor is a BRC connection valued. Instead, a multitude of isolates would need to be directly cultured from the biospecimens, each sequenced and assembled until the desired MAGs were matched exactly before proceeding to the causative experiments. On the other hand, to accelerate procurement of a live strain for a causative experiment, we suggest the second more traditional and tractable strain definition. In this definition, a single strain encapsulates all the descendants of a single colony isolation in pure (axenic) culture and is disseminated among microbiologists by a succession of cultures [4], [19]. It is appreciated that the initial process of isolation from a living community is itself a selection event which captures one point-in-time of a mutable genome [13]. Nonetheless, these isolated and propagated strains are important tools for experimental microbiology and provide necessary points of reference for scientific communication and intellectual property delineations.
Heterogeneity exists among the methods of naming and bio-banking the descendants of a single isolate among microbiologists and this has led to downstream confusion for the bioinformatician. Oftentimes microbiologists, after isolating and naming a single strain from clinical or environmental material, will send replicate sub-cultures to multiple BRCs, such as ATCC (http://www.atcc.org), DSMZ (http://www.dsmz.de) and JCM (http://jcm.brc.riken.jp) or dozens of other worldwide centers. These BRCs then assign their own catalog numbers. DNA sequencing institutes throughout the international scientific community procure strains from various BRCs, extract and sequence the DNA then upload single genes or whole genome assemblies to public databases, such as GenBank [2], who assigns an identifier for each assembly received. Because this is a decentralized international activity, there has been persistent uncertainty about what data belongs to each strain [3], [21]. A prime example of the need for unification can be seen in a strain isolated from a healthy Japanese male in 2011 [37]. The research team bestowed novel genus and species level nomenclature for the isolate which they publicized as Christensenella minuta YIT 12065. Two independent BRCs (DSM and JCM) also propagated sub-cultures of this strain with their own unique catalog numbers, DSM 22607 and JCM 16072. The University of California at Davis, Beijing Genome Institute, Washington University, and South China University of Technology each procured the strain from one of the BRCs then separately sequenced the extracted DNA and submitted their optimal assembly to public databases which are now downloadable from RefSeq under four different assembly identifiers: GCF_001571425, GCF_001652705, GCF_001678855, and GCF_003628755. Novice users of these public databases could easily misinterpret these four assembles as four different genomes from four different strains. In contrast, we see these as technical replicates. In building the StrainSelect database, we sought to overcome confusion by tracing through the synonymous identifiers for sub-cultures and genomic data records and assign a consistently formatted identifier for the strain, which in this example is “StrainSelectID:t__520”, and connect all the genomic records together.
Now if the investigator decides to match metagenomic data to strains according to the isolate-and-propagate aforementioned definition, the bioinformatician will need to build or acquire a reference database with three properties to overcome three challenges. First, the database will appropriately label each gene and genome assembly by the strain of origin carefully avoiding conflation of genome assemblies as strains. Unfortunately, NCBI, a central foundational database, has announced cessation in efforts to organize data in this fashion [20]. What is needed in a reference database is reliable linkage of clandestine technical replicates, those genome assemblies from the same strain published from two or more institutes using dissimilar monikers. A recent study on of the deleterious effects of duplicate sequence records in bioinformatics reference databases demonstrated inefficiency, obviously in computational search load, and less obvious but more severe, in the manual or scripted assessment of the results of a search [8]. As a simple example of the problem, consider a single query DNA sequence matching the set of database subjects Mesorhizobium loti HAMBI 1129, M. loti DSM 2626, M. jarvisii ATCC 700743, as well as M. jarvisii ATCC 33669, with zero matches outside this set. The inexperienced bioinformatician would likely interpret these match results as a non-strain-specific “hit” since the names share only the genus. But since these are all synonyms for the same strain it would be accurate to conclude that the hit was in fact strain-specific. Second, the database will need a schema to relate each genomic record to zero or one extant procurable strain cultures distributed by one or more BRCs. In other words, users should know if a genomic record is not only linked to a strain but if that strain is available in a BRC. Third, since microbiome meta-analysis provides opportunity to find concordant observations among cohorts often profiled with differing lab technologies [51], a single reference database should enable integration of metagenomic shotgun and the more popular 16S rRNA gene amplicon data (Fig. 1) into a single taxonomic ontology. StrainSelect was built to overcome all three challenges and is available as a reference database (http://strainselect.secondgenome.com) describing 681,087 strains for use in standalone pipelines. The R code to reproducibly generate all tables, figures and text for this manuscript is provided, as well.
2.1. Other notable resources
Over the last decade, several data curators have attempted to solve these problems however each effort has either been abandoned or lacks key features to support current data analysis needs. StrainInfo [55], the early inspiration for StrainSelect, endeavored to build a database that would include both genome assemblies and 16S rRNA genes apart from assemblies, but is no longer maintained. BacDive [48] organizes genome assemblies, 16S rRNA genes and functional attributes via an informative interactive web tool. It contains a small number of the known strains (89,545 strains) and does not provide a downloadable database for high-throughput data pipelines. GOLD [38] appeared more comprehensive representing 395,286 bacterial and archaeal “organisms” but in some cases one strain has multiple organism identifiers as exemplified in Sup. Fig. 1 so the actual strain count is likely less. The Genome Taxonomy Database (GTDB) [43] contains 258,406 genome assemblies taxonomically organized from domain to species but does not attempt to categorize the assemblies by strain and only includes 16S rRNA genes if they are embedded into genome assemblies of pure cultures or connected to a MAG. GTDB has fully disclosed its methods for placing assemblies into species and distributes useful files and software for species-level classification. StrainSelect expands on the esteemed work from StrainInfo, BacDive, GOLD, and GTDB by including more than double the number of strains than previous resources, resolving synonymous organism names for the same strain, and building a unified taxonomy for use with both shotgun or amplicon techniques.
3. Approach
Various known monikers of the isolated and published strains as well as the identifiers for the public genomic records attached to each were collected from relevant sources. Genomic records gathered were either full genome assemblies or 16S rRNA gene assemblies covering eight of the nine hyper-variable regions and both types were filtered by standardized procedures. All monikers and sequence identifiers were placed as vertices (nodes) of a network knowledge graph and inter-vertex edges (connections) were created to represent direct material derivatives. The graph was decomposed into components, where one component is a connected sub-graph of vertices that is disjointed from any other sub-graph. Each component defined exactly one archaeal or bacterial strain and each strain was assigned a StrainSelectID identifier.
Where possible, taxonomic nomenclature for seven levels from domain to species was adapted from GTDB with the additional and relevant constraint that one strain can belong to only one species. For strains with 16S rRNA genes available but without a genome assembly, taxonomic placement was estimated by k-mer similarity. Where formal taxonomic names were not yet coined for demarcated genera-level and species-level groups, provisional identifiers were assigned. The stability of both formally-named and provisionally-named species-level groupings was measured by bootstrapping prompting a subset of provisionally-named species to be merged into formally-named siblings.
Because all data was organized by species and by strain, intrastrain versus intraspecies genomic similarity was contrasted. We present a new estimate of variation among related but distinct strains as well as an estimate of technical variation of genome assemblies from the same strain sequenced and assembled at different institutes.
4. Methods and results
4.1. Software
R, https://www.R-project.org, [47] was used for the majority of the graph construction pipeline with Python, http://www.python.org, used to download and filter NCBI data. The R libraries, data.table, https://CRAN.R-project.org/package=data.table [14] and kableExtra, https://CRAN.R-project.org/package=kableExtra [62] were used for tabular operations and ggplot2 [60], ggnetwork, https://CRAN.R-project.org/package=ggnetwork, [5], and ggbreak [61] for data visualizations. Additional software packages for specific steps are cited in subsequent sections.
4.2. Input data
Monikers (i.e. published names, abbreviated names, machine readable identifiers and synonyms) for strains and their associated genomic data were collected from PATRIC [56] on 2021-11-23, GOLD [38] on 2021-11-23, GTDB [42] on 2021-12-26, BioCyc [30] on 2021-08-05, KEGG [29] on 2021-10-31, RefSeq [22] on 2021-11-28 with the NCBI Type-Strain Report, https://ftp.ncbi.nlm.nih.gov/genomes/ASSEMBLY_REPORTS/ on 2021-11-29, WGS, https://www.ncbi.nlm.nih.gov/genbank/wgs/ on 2021-11-23, and StrainInfo [55] on 2013-10-20. Custom parsers are maintained for each data source and require adjustments as source formats evolve.
Input data was categorized into 13 vertex types as shown in Table 1 and denoted in fixed width font in this description. The StrainSelect data model follows the common data types created as information is generated. An institute generates and assembles sequencing reads from an isolated strain into contiguous DNA sequences (contig) identified by GenBank accession numbers and can encode a single gene, as in the case of the 16S rRNA gene (g16), or encode many genes when assembled from of a whole genome shotgun read library. These shotgun projects are registered at NCBI and assigned a 4 or 6 letter string that becomes the master prefix (wgs_master_pre) for all the project's contigs. A set of one or more contigs representing a genome assembly effort is distributed from GenBank (gb_assembly) and if the set meets certain quality thresholds for completeness and purity will additionally be distributed from RefSeq (rs_assembly). When KEGG or PATRIC annotate an assembly, they re-distribute the data and StrainSelect includes those vertices as kegg_genome or patric_genome, respectively. If BioCyc creates a specially formatted database from an assembly for interactive pathway analyses, then a biocyc_pgdb vertex was included. StrainInfo recognized that one strain can exist as cultures at multiple institutes and established separate culture identifiers for each (si_culture_id) and a list of the disseminated cultures from the same strain defines the si_grouping_id. Bio-resource centers (BRCs), sometimes known as culture collections, will receive live strains then store, propagate and ship the strains under their own catalog numbers (brc_cat_id). The GOLD organism identifier was captured as gold_org. The gss vertex type was established for both human- and machine-readable processes and encodes the genus-species-strain concatenation, as described below.
Table 1.
Vertex types in the graph schema.
| Vertex type | Description | Examples |
|---|---|---|
| contig | Contiguous DNA sequence | JF079054, NZ_FJOC01000002, NC_013353 |
| g16 | 16S rRNA gene | g16_4602054 |
| wgs_master_pre | NCBI WGS master record prefix | wgs_AADD, wgs_FJOC, wgs_CAADNE |
| gb_assembly | Genbank genome assembly | GCA_000155415 |
| rs_assembly | RefSeq genome assembly | GCF_000001635 |
| kegg_genome | KEGG genome | gn_ebw, gn_ecok |
| patric_genome | Patric genome | pat_1131286.3, pat_1123738.3 |
| biocyc_pgdb | BioCyc PGDB | bc_LLAC1295826, bc_GCF_000001635 |
| si_culture_id | Culture recorded by StrainInfo | ci_119674 |
| si_grouping_id | Group of replicate cultures recorded by StrainInfo | gr_2, gr_171641 |
| brc_cat_id | Bioresource center catalog identifier | ATCC 700598, DSM 2281, CCUG 38580 |
| gold_org | GOLD organism | Go0516098, Go0000004 |
| gss | genus species strain string | escherichia.coli.k.12.dh10b |
4.3. Genus-species-strain vertices
Due to differing database entry conventions, strains have been dubbed with slight variations in the formatting of character strings for genus, species and strain names. For example, one strain classified within the species Comamonas terrigena can be found as “R. Hugh 247”, “R.Hugh 247”, and “R Hugh 247”. To prevent the creation of multiple vertices that are only slight deviations in string content, all alphabetical characters are converted to lowercase and each series of non-alpha-numeric characters are converted to a single period. Thus, the genus-species-strain (gss) vertex in each of these cases would be unified to “comamonas.terrigena.r.hugh.247”. Since this same strain has also been referenced as “Vron 31”, a distinct vertex of “comamonas.terrigena.vron.31” is also included. To avoid insufficient vertex name complexity resulting from this process, a gss vertex was not formed when less than three words were available for the concatenation or when the gss would be less than 10 characters thereafter.
4.4. Genome assembly quality control
Genome assemblies in RefSeq are assumed to be more reliable than those only in GenBank since, as the documentation at https://www.ncbi.nlm.nih.gov/assembly/help/anomnotrefseq attests, each has at least one copy of a 16S rRNA gene and are not contaminated with DNA sequence from multiple strains. StrainSelect further scans the set of contigs of each RefSeq assembly using profile hidden Markov models (HMMs) with nhmmer [58]) to obtain the count, lengths, coordinates and taxonomic domains of origin for the 16S and 23S rRNA genes. In total, 250,511 RefSeq assemblies were processed and 4,882 (1.9%) were rejected due to rRNA genes found from more than one domain within the same assembly, suggesting contamination. In other words, after discarding potentially problematic genomes StrainSelect provided 98.1% coverage of RefSeq. Surprisingly, in 5,723 (2.3%) RefSeq assemblies nhmmer failed to find any archaeal nor bacterial rRNA and in 25,938 (10.4%) when a 16S rRNA gene was encountered it was incomplete (under 1,250 nt where ~1,500 nt is expected) or it contained over 1% non-ACGT characters. These assemblies were still retained as vertices but their 16S rRNA genes were not.
4.5. 16S rRNA gene assembly quality control
An NCBI search for 16S rRNA genes ≥1250 nt that were derived from isolated strains and not from clones, unculturable materials, nor PCR libraries resulted in 511,858 records. These records are of the type contig in the StrainSelect graph schema since they are contiguous DNA sequences. Contigs can exist independent from or belonging to one genome assembly. All contigs were processed by nhmmer (described above) to reject those with regions from more than one taxonomic domain or containing under 1,250 nt matching the 16S rRNA gene model or if that span contained over 1% non-ACGT characters. In total 11,279 were rejected, which resulted in 500,579 contigs remaining. A separate vertex was formed from each 16S rRNA gene instance within each contig totaling 917,079 g16 type vertices. Although many 16S rRNA gene sequences are identical across genome assemblies [44], no sequence dereplication was applied at this step.
4.6. Graph composition, component discovery, and component filtering
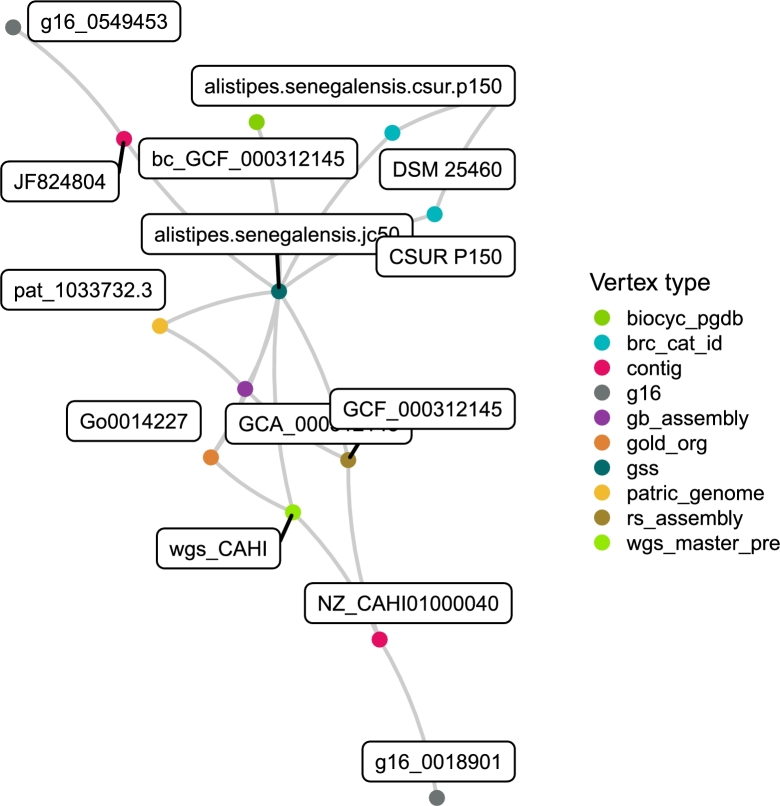
All vertices resulting from parsing input records and filtering sequence data were loaded into the network-based data management software package, iGraph, https://igraph.org [10], with each having exactly one vertex “type” attribute from Table 1. Graph edges, where each edge is a link between exactly two vertices were defined by an equality represented by an input data source or from the HMM analysis. As an example of vertices connected by edges consider the case in Fig. 2. Parsing data from GOLD equated Alistipes senegalensis JC50 [36] (vertex(id=alistipes.senegalensis.jc50, type=gss)) to the organism identifier Go0014227 (vertex(id=Go0014227, type=gold_org)). Identifying a 16S rRNA gene (vertex(id=g16_0018901, type=g16)) spanning positions 4 to 1528 within the sequence NZ_CAHI01000040 (vertex(id=NZ_CAHI01000040, type=contig)) established an edge. Since this contig was from the set of contigs defining one RefSeq genome assembly (vertex(id=GCF_000312145, type=rs_assembly)) submitted in 2012, an edge was established for this relationship, as well. Fig. 2 displays how these edges and others connect all the monikers for this strain. The graph integration of information reveals that RefSeq genome assembly GCF_000312145 was derived from alistipes.senegalensis.jc50 which is available for procurement at two different BRCs and under another synonym, alistipes.senegalensis.csur.p150. The GOLD organism identifier is attached to the gb_assembly and wgs_master_pre. Also notice that two contigs carry high quality 16S rRNA genes, one as described above from a multigene contig and the other from a single gene contig, (vertex(id=JF824804, type=contig)), submitted to NCBI in 2011. For a more complex example see Sup. Fig. 1.
Figure 2.
An exemplary subgraph of the vertices comprising one strain, StrainSelectID t__117676. This subgraph is a disjointed component within the entire knowledge graph and connects vertices of various types shown by color. All data, although distributed from different sources such as NCBI, KEGG, BioCyc. Patric, and GOLD, was derived from an isolate originally named Alistipes senegalensis str. jc50 which is available for procurement at two different BRCs (blue). The subgraph conveys that two high quality 16S rRNA genes (grey) are available for this strain, one from a RefSeq genome assembly (brown) containing a contig (pink) encoding a 16S rRNA gene (grey) and the other is independent of a genome assembly project but was deposited as contig JF824804 (pink).
The entire graph was initialized with 8,412,126 vertices connected by 7,603,203 edges. Vertices with degree=0, in other words vertices with zero edges, were dropped leaving 8,339,151. These vertices are not useful for our purpose since they represent an assembly without a name for the isolate nor a BRC entry or these vertices are cultures in a BRC with no public sequence data available. The remaining graph was decomposed into components, where one component was a connected sub-graph of vertices that is disjointed from any other sub-graph.
Components removed were those encompassing zero gss vertices or zero g16 and rs_assembly vertices, a condition formalized in Eq. (1).
| (1) |
Each of the 681,087 remaining components defined exactly one StrainSelect strain and each strain was assigned an integer identifier prefixed with a t__. For example, the single component in Fig. 2 is StrainSelect strain t__117676 and a vertex-rich component in Sup. Fig. 1 is StrainSelect strain t__47740. The single lowercase letter plus two underscores prefix format [34] was popularized when integrated into the Greengenes database [12] to disambiguate which taxonomic rank was referenced by a term (for example, p__Firmicutes, c__Bacilli, o__Staphylococcales, as the phylum, class and order names, respectively). Since s__ is already the prefix for species rank, t__ was used for the strain rank prefix. This final graph of 4,002,309 vertices and 4,380,302 edges in 681,087 strain components can be obtained in a single R iGraph formatted file as StrainSelect_iGraph.rds or in two tsv files StrainSelect_vertices.tab.txt and StrainSelect_edges.tab.txt.
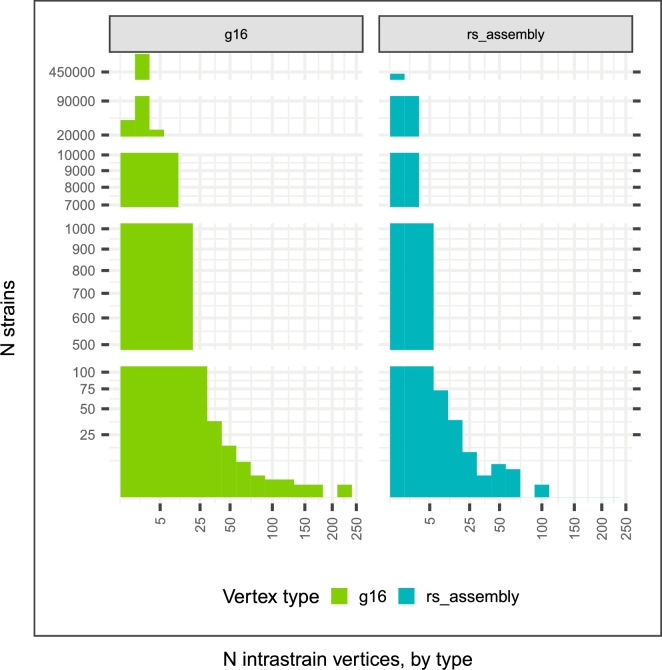
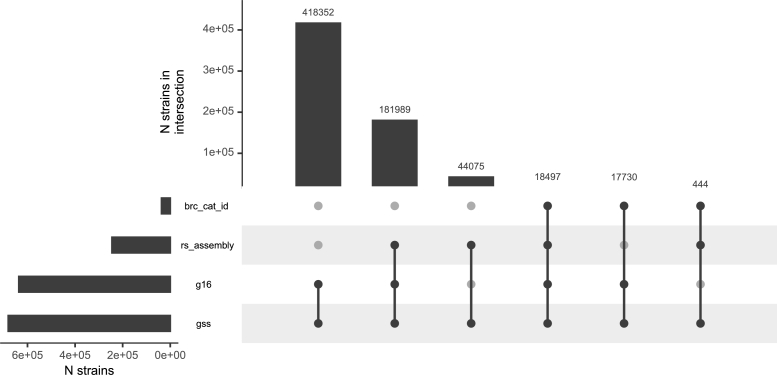
The component-producing procedure did not presume nor constrain that each rs_assembly should belong to a different strain and therefore revealed that 10,043 strains have more than one high-quality assembly (Fig. 3) and, surprisingly, 19 strains have over 25. The general membership of vertex types among components was examined with multiple intersection analysis [9] in Fig. 4. Components containing brc_cat_id, rs_assembly, g16 and a gss vertices represent a large proportion of all components. Components containing g16, gss and brc_cat_id vertices without rs_assembly vertices were the most common. Overall, RefSeq only covers 36% of the strains in StrainSelect since most strains do not yet have high quality assemblies publicly available.
Figure 3.
Distribution of strains binned by count of available 16S rRNA gene records (g16) or RefSeq genome assembly (rs_assembly) records derived from the strain. Most strains have less than 25 of either type but a minority of strains have over 100 of these vertex types suggesting many technical replicates exist in the public databases.
Figure 4.
Component counts based on presence or absence of four types of vertices. For the majority of strains, names and 16S rRNA genes are known but they have not been deposited in a BRC nor has a genome assembly been entered into RefSeq.
4.7. Taxonomy adaptation
The opportunity to apply a single taxonomic ontology to all the sequence records in StrainSelect to create a single ontology to encompass both the 16S rRNA genes and the genome assemblies was challenging. Some strains are missing either a high-quality 16S rRNA gene or a genome assembly, while others have multiples of each. In the StrainSelect schema, a contig belongs to exactly one strain and a strain can belong to only one species, therefore the StrainSelect taxonomy is the first, to our knowledge, to ensure that contigs from the same strain do not end up in different taxonomic lineages. GTDB was conscripted as the base taxonomy because it has balanced traditional microbiological nomenclature with modern tree construction based on similarity across multiple genes [42] and has placed the majority of the RefSeq assemblies into categories from domain to species. The adaptation of GTDB taxonomy to satisfy the schema constraints of StrainSelect was accomplished for 236,992 strains as described below.
In a first step, taxon names in GTDB that are not in Latin form but instead take a variety of formats as placeholder strings used until agreement in the nomenclatural literature emerges, were identified. To these, StrainSelect assigned a consistently formatted provisional identifier using the characters “PROV” for reliable machine reading/parsing. For instance, the name “s__PROV_95247” indicates a species level taxon without a formal Latin name.
A GTDB taxonomic placement was available for at least one genome assembly from 185,872 strains. Because the StrainSelect data model recognizes that some strains have replicate genome assemblies, we had to examine if GTDB had placed replicates in different lineages. We found, for small percentage (429 strains, 0.2%), the replicate assemblies were spread into more than one GTDB species. The discordance was minor. For example, two assemblies from one strain, t__104183, were placed by GTDB in distinct but sister species, GCF_001490875 in Listeria monocytogenes and GCF_001711055 in Listeria monocytogenes_B. Of the 185,443 strains without this discrepancy, 165,904 have one or more 16S rRNA genes, useful for anchor points for taxonomic estimation where only a 16S rRNA gene is available without a RefSeq genome assembly.
To classify all the BRC deposited strains not yet placed into a single GTDB lineage but with available 16S rRNA genes, the kmer-based sintax algorithm of usearch [18] was applied to each 16S sequence to make an initial placement for each gene. For strains with multiple 16S rRNA genes split to multiple species placements due to dissimilarities, preference was given for the Latin-named, non-provisional species placement with the greatest sintax confidence score and that preferred species was applied to all 16S rRNA genes of that strain. To test the stability of these initial strain-to-species memberships, 100 bootstrap cycles were performed where each 16S gene was compared against up to 200 randomly chosen intrafamily 16S genes and one randomly chosen 16S gene from a near-neighbor taxon outside the family (out-group). A multiple sequence alignment was solved by muscle [17], the hamming symmetric distance matrix [23] was calculated then partitioned by pamk, https://CRAN.R-project.org/package=fpc [24] as visualized with t-distributed stochastic neighbor embedding (TSNE) in Fig. 5. Partitions were created purely from the distance matrix without any added parameters for mutational rates nor tree-constructions since phylogeny was not the objective. Each gene in each bootstrap was affixed with the species name that comprised the majority of its partition. The percentage of bootstrap cycles where a gene was affixed to the same species was the gene-to-species bootstrap support score.
Figure 5.
Example of species placements of 16S rRNA genes from one bootstrap cycle within one family. In the Anaerotignaceae family, the DNA sequence distance matrix between the known high-quality 16S rRNA genes is partitioned around medoids (PAM) and visualized with t-distributed stochastic neighbor embedding (TSNE). Each point represents one 16S rRNA gene and the symbol represents their membership to one of six species before the initiation of the bootstrapping process. In this family, four formally and two provisionally (PROV) named species were available. After partitioning, the majority species within each partition is determined as represented by the color. The points highlighted with white circles are 16S rRNA genes that were affixed with new species names in this cycle. One gene within s__PROV_231201 (inverted triangle) was affixed with the species name s__Anaerotignum__lactatifermentans (blue) and two genes within s__Anaerotignum__neopropionicum (plus symbol) were affixed with the species name s__Anaerotignum__propionicum. After 100 cycles, only PROV assignments were adjusted in the final taxonomy when >50% of the bootstrap cycles were concordant. Also noteworthy are the three genes from s__PROV_231201 (inverted triangles) separated on both axes indicating, at least in one bootstrap cycle comparing these 16S rRNA genes, the instability of this taxon group.
To then summarize the support from all genes from a strain, the strain-to-species bootstrap support score was the average observed among its 16S genes. Bootstrap support varied among strains and was compared against attributes of genome assemblies reported by GTDB. An inverse correlation (p < 1e-90) between bootstrap support and various metrics of genome size, G+C percentage and contamination was observed (Fig. 6, Sup. Fig. 3).
Figure 6.
Bootstrap support for species assignments from 16S rRNA analysis inversely correlates with assembly contamination. For 161,094 strains, all three of the following were available: RefSeq assemblies, GTDB-reported assembly contamination and 16S rRNA genes. Where multiple assemblies for a strain were available, the mean assembly contamination was calculated. A significant inverse relationship (Spearman correlation coefficient = -0.16, p<1e-90) between the magnitude of a strain's genome assembly contamination and the likelihood that the strain's 16S rRNA genes come from the same species was observed.
All strains originally placed in provisionally-named species but whose bootstrap support was >50% for an alternate species were re-assigned. Of the 429 strains with GTDB discrepancies described above, 211 had 16S rRNA genes available and were placed into a single species using this same method. In total, 236,992 strains were placed into a structured taxonomy with specific ranks from domain to strain.
4.8. Knowledge graph quality control
To verify the reliability of the final information linkage within the StrainSelect graph it was compared to pre-existing knowledge. Two highly dissimilar sources of pre-existing knowledge were used in the comparisons: NCBI's Prokaryote Type Strain Report (PTSR) and previously reported sequence similarity within taxonomic boundaries. The PTSR contains a map between brc_cat_ids that are replicate cultures of the same strain and one or more of the synonymous gss names. In this file was 9,267 edges between brc_cat_id and gss vertices and 8,953 of those edges have vertices that met all criteria for StrainSelect inclusion (96.6% coverage). If the construction of the StrainSelect graph introduced errant linkages, we should find cases where the two vertices connected by these 8,953 PTSR edges ended-up in different StrainSelectIDs as different strains. We observed zero of these errors. Thus, based on the PTSR comparison, the StrainSelect database includes nearly all type strains and, when passing all filters, reliably connects synonymous information into the same component.
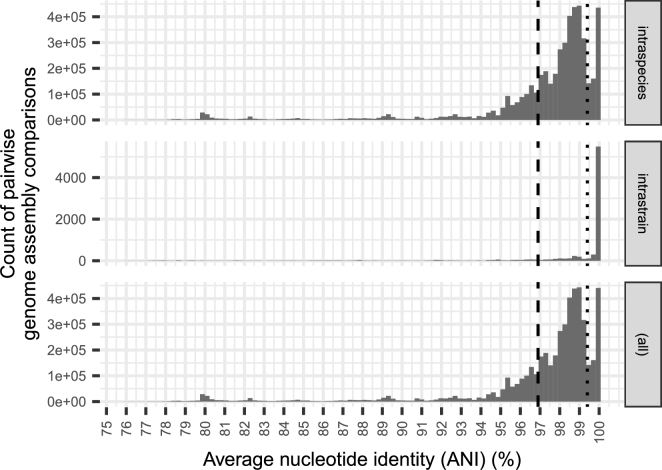
The second comparison of the final graph to pre-existing knowledge was based on DNA sequence comparisons. Since the vertices of the knowledge graph are not connected by edges defined by genome sequence identity, the validity of components was evaluated by this metric as a post hoc analysis. If a meaningful demarcation among strains existed in the graph, we would expect the majority of components with more than one assembly to have low DNA divergence between those assemblies explained by technical variation expected when independent institutes sequence the same strain. Conversely, if the graph-building methods resulted in a poor-quality over-connected graph generating components that unintentionally merged assemblies derived from different strains, we would expect high divergence among intracomponent assemblies. To test this, a sampling of 80,741 assemblies within 75,894 strains from 5,436 species where >2 and <700 intraspecies genome assemblies were available were compared with FastANI [26] to determine the average nucleotide identity (ANI). Limiting the sampling to species with under 700 assemblies held-out species within Escherichia and Shigella which contain large numbers of assemblies with species boundaries under debate for likely reorganization [25] but included well-studied species such as Yersinia pestis, Haemophilus influenzae, and Bacillus subtilis. In previously published observations, intraspecies ANI among genome assembles is typically >95% [26], [31], [43]. Since strains are a finer taxonomy rank than species, we expected that most intrastrain ANIs should be at least this high. We observed that of the 3,621 strains investigated, only 264 (7.3%) contained a pair of assemblies <95% ANI, indicating that nearly all components had avoided over-merging vertices belonging to different species. This analysis also allowed a systematic estimate, for the first time to our knowledge, of the technical variation observed among assemblies from the same strain to be approximately 0.6% (Fig. 7). Consequently, we determined the distributions of intraspecies identities without the bias of repetitive intrastrain comparisons in Fig. 7 and observed an intraspecies ANI of >96.9%.
Figure 7.
Distribution of pairwise identities between genome assemblies within the same species (intraspecies) and within the same strain (intrastrain). Displayed are the observations from a set 80,741 assemblies within 75,894 strains from 5,436 species. Assemblies were compared pairwise for their average nucleotide identity (ANI). 75% of ANIs between assemblies from the same species but from different strains were greater than 96.9% (dashed line) while 75% of intrastrain ANIs was greater than 99.4% (dotted line).
To identify hub vertices potentially over-connecting genome assemblies that do not belong to the same strain, the betweenness centrality for each vertex () was calculated to find the vertices acting as frequent bridges between divergent assemblies <95% ANI within the same component. Eq. (2) defines where is the number of possible shortest paths from one rs_assembly to another and are the count of those paths passing through vertex, V.
| (2) |
The vertex types accumulating the greatest were gss and brc_cat_id indicating that genome assemblies submitted to NCBI that share a genus-species-strain name and/or a BRC catalog identifier can, in rare cases, have divergent DNA sequence contigs. As a case study, we investigated the vertex with the greatest , the gss vertex, serratia.marcescens.cdc.813.60 from strain t__19847. This hub is perhaps reflective of the experimental design (NCBI BioProject: PRJEB40306) to produce many assemblies from isolates generated by thermal mutagenesis of a culture grown from ATCC 13880. In this case, it is not obvious that all these assemblies are still representative of a single strain even though the annotations attached to the assemblies asserted that they were. For all 264 strains containing divergent genome assemblies (<95% ANI), the strain was removed from the set of strains with taxonomy placements although it remains in the graph. This results in 236,992 strains with taxonomy placements of which 219,349 strains (92.6%) have ≥1 genome assembly and 217,454 strains (91.8%) have ≥1 16S rRNA gene.
4.9. Reduced MinHash and fasta files
Although all the DNA sequence data encompassed by StrainSelect can be downloaded from NCBI, we have provided users with reduced dimensionality reference files for taxonomic classification of shotgun metagenomic reads in sourmash's MinHash sketch format [45] with parameters -k 51 –scaled 5000. Sketches were attempted for 219,349 strains but 166 of these strains were omitted as only deprecated RefSeq assemblies were available. Of the remaining 219,183 strains, the single assembly with the lowest GTDB-reported contamination was included to represent the strain. The sum of contigs from all these assemblies is 873 Gb but after sketch formatting, aggregation, and compression all signatures fit into one 5.3 Gb file. The StrainSelect21_README.txt file accompanying the downloadable sourmash reference file contains example commands to assist informatitians in building computational pipelines.
A 16S rRNA gene reference fasta file was prepared using sintax-formatted taxonomy headers containing only intrastrain dereplicated sequences meaning that g16 sequences which are an exact sub-sequence within another from the same strain were not included in the file. After compression the final file is 61 Mb in size and contains 333,204 16S rRNA gene sequences from 217,454 strains preserving intrastrain diversity helpful for training classifiers.
5. Discussion
The conceptual approach of connecting synonymous monikers for each strain sourced from a variety of data sources was overall successful and produced a knowledge graph with a variety of utility. It allowed us to run component discovery to find the boundaries around the data records pertaining to each of 681,087 strains. It facilitated calculations of betweenness centrality to prioritize, for manual inspection, the hub vertices potentially over-connecting identifiers, such as in Serratia marcescens. With the graph we could connect known 16S rRNA records and genome assemblies for each strain and discover cases where technical replicates are available. This empowered dissimilarity analysis among replicate genome assemblies and bootstrap support scoring for the taxonomic placement of species using 16S rRNA genes. Overall, we found the graph methodology to be appropriate for this application and able to cover a large portion of the graph with a structured taxonomy. Surprises that were encountered during the database build are worth consideration as they have implications on the future of microbial genomics and the adept usage of StrainSelect.
5.1. On graph methodological validation
There is a valid concern that bioinformatic creation of mega-graphs from public resources can over-connect information that domain experts would find disagreeable. For examples of problematic false or spurious edges in the domain of protein-protein interaction graphs see López et al. [33]. Since we combined large quantities of relationships from multiple public sources we benefited from a emphpost hoc test to measure the frequency of improbable connections, namely genome assemblies connected within the same strain but with divergence beyond what is likely from technical variation. We demonstrated that the StrainSelect graph building method defined reasonable boundaries between strains by component decomposition and revealed intracomponent (intrastrain) ANI was over 99.6% for 75% of the comparisons. In a second test of component integrity, we verified that 96.6% of type strain synonyms published by NCBI were included in StrainSelect and of those none were improperly separated into different components by any methodological step in the graph construction method. These observations, one using sequence comparison method independent of how StrainSelect constructed and the other using a knowledge preservation test reveals minor limitations of StrainSelect but provides evidence that the components, which are simply groups of data and monikers from a single strain, are generally reliable.
5.2. On comprehensive taxonomy
In building the taxonomic ontology for StrainSelect, we valued the work of Greengenes which implemented consistent data filtering, DNA similarity based taxa and consistent machine-friendly taxonomic ranks for all tree leaves and, even more so, GTDB which has carried the burden of balancing traditional microbiological taxonomic nomenclature with hierarchical incongruities revealed in multi-gene tree construction. Therefore, the basic ontology for StrainSelect will be familiar to users of either. Only 429 strains had multiple assemblies split between different GTDB species and those were either resolved to a single species based on the 16S rRNA genes (211 strains) or withheld from the structured taxonomy. Thus, 99.8% agreement exists between GTDB taxonomy and StrainSelect. The larger future endeavor will be to incorporate into the taxonomy the over 400,000 strains known only by a gss name and a 16S rRNA gene (Fig. 4). In the current version, these strains were left out of the taxonomy but with the steady reduction in DNA sequencing costs many of these strains' genomes are likely to become publicly available. The group of over 17,000 strains with 16S rRNA genes as well as cultures deposited at a BRC but without a RefSeq assembly (Fig. 4) are possibly queued for laboratory or bioinformatics progression for eventual broadcast via RefSeq. It's likely that at any point in time there will be a set of strains at this stage and StrainSelect includes them to build a more comprehensive taxonomy based on available 16S rRNA genes. Overall, the taxonomy includes 236,992 strains of which 219,349 (92.6%) have a genome assembly and 217,454 (91.8%) have a 16S rRNA gene. We expect both of these percentages to increase in future versions.
5.3. On implications for the field of microbial genomics
As a by-product of constructing this database and overcoming challenges in DNA sequence contamination, clandestine technical replicate records, and incorrect metadata, we formed some remarks on the general state of the field.
In this work 1.9% of RefSeq assemblies were eliminated from entering the knowledge graph due to interdomain contamination which means we were more permissive compared to EMBL's estimate that 5.2% of RefSeq genomes are impure [41]. EMBL may be correct because even after our RefSeq filter, we observed that even minor genome assembly contamination levels (Fig. 6) were inversely correlated to the bootstrap confidence of a strain's placement into a species. These observations are unsettling to the assumption that RefSeq is a pristine reference database for any genomics inquiry. It holds valuable data and has been a dependable resource with consistent availability for international collaborative research. But, until sequencing facilities or RefSeq editors can optimize the identification and elimination of contaminant contig regions, users should be aware that taxonomic placement, and more broadly, phylogenetic conclusions are subject to improvements.
Since duplicates and redundant information exist in biological databases, any database maintainer should assist their users by documenting how these cases are identified and handled. The presence of duplicate sequence records in bioinformatics reference databases creates inefficiency in computational search load, and in the assessment of the results of a search [8]. Most users would agree that clear duplicates, for instance an assembly from the same strain sequenced once at one institute but deposited at NCBI twice under different accession identifiers ought to be removed. But these types of duplicates don't appear to be the problem. Instead, we counted that for 10,043 strains submitters created genome assemblies from the same strain in different sequencing projects usually at different institutes. Whether these repeats produced slightly different results or identical results, these data observations should be made public to enable measurements of technical variation, for instance, but should be clearly labeled as such. Since NCBI does not attempt this after genome submissions but instead allows rich metadata to accompany a submission [20], it is up to the user to either determine from the metadata which assemblies are technical replicates and which are from distinct strains or to use a resource such as StrainSelect. In the StrainSelect graph, our findings of technical replicates derived from the same strain are documented and all are labeled with the same StrainSelectID.
Despite the capacity for NCBI data contributors to include metadata to describe one or a collection of assemblies, we uncovered a problem in naming isolates created from a mutagenesis experiment (NCBI BioProject: PRJEB40306) by re-applying the same name as the origin (parental) strain. This led to a graph component connecting a set of assemblies that were <95% ANI. Thus, it is recommended in these cases if a mutagen was applied and the genome content changed then the new isolate should be given a separate name from the parent strain. Otherwise the mutant genome assemblies would be assumed to be taken from a single strain and the casual data consumer would attribute divergence to technical artifacts/errors instead of the intentional experimental design.
We observed a genetic discontinuity between strains (Fig. 7) at 99.4% ANI. We contemplated an interassembly ANI exceeding this threshold as an edge in the graph construction process in future versions of StrainSelect. To add these ANI-based edges would result in fewer overall components but would merge genetic information where subject matter experts would keep them discriminated due to critical genes. For example, strains within the pathogenic species Corynebacterium bovis such as t__915 (synonyms: str. DSM 20582, str. Evans, str. CIP 54-80T, and 14 others) and t__254639 (synonym: str. MI 82-1021) have >99.7% ANI but have dissimilar virulence genes [7] warranting their distinction. Future research could involve weighting edges more when direct culture sharing is known and less if ANI is the only connection, then implementing a more sophisticated component boundary definition that would resolve a set of training cases such as within C. bovis, but in the current version no ANI edges were created in the knowledge graph.
5.4. On future directions for StrainSelect
In addition to potential future improvements in leveraging ANI, we also foresee opportunities in leveraging MAGs and consensus assemblies. It's conceivable that identical MAGs will be observed in multiple biospecimens as is suggested by a clinical study where >50% of a MAG can be >99.999% similar in two different stool samples [40]. Once metagenomic technology advances to enable entire MAGs to be found as nearly identical in among biospecimens, and the recurring MAGs are dissimilar to known isolates then StrainSelect should recognize them as yet-to-be-isolated strains. In the meantime, if a research team has ample resources to culture isolates matching MAGs from their metagenomic sequencing, then the isolates and corresponding genome assemblies should be submitted to NCBI and BRCs, respectively, to increase the diversity available.
As the number of MAGs and assemblies grow, the number of strains with technical replicate assemblies will also grow. In our current build, when technical replicates were found, we selected the least contaminated for inclusion in the MinHash (sourmash) database. Alternatively, one could create a single consensus assembly before the MinHash is derived. A potential tool to implement this process would be Trycycler [59] although in its current implementation requires subjective steps in post-processing, or polishing, which would introduce a non-reproducible step in the StrainSelect build. Once a validated automated process is available, StrainSelect will likely focus MinHashes to regions harmonious across technical replicates.
5.5. On usage of StrainSelect
The first published usage of StrainSelect was described for organizing raw fecal 16S rRNA gene sequencing data to identify a composite biomarker for colorectal cancer [51]. In the manuscript, binning the reads by unique matches to one strain, where possible, was compared to binning the reads by a popular operational taxonomic unit (OTU) method. The StrainSelect method produced biomarkers that outperformed the OTU method in accurately classifying patients. Other notable examples are the use of StrainSelect to pinpoint the strains phagocytized by specific macrophage types in Crohn's Disease patients [50], and to enable a strain-level meta-analysis across 21 Inflammatory Disease datasets [46] and across 10 Autism Spectrum Disorder data sets [57].
With the description of StrainSelect herein, biologists can now choose to organize microbiome profiling data from shotgun or amplicon techniques for strain-level analyses. Compared to shotgun metagenomic laboratory techniques, it is expected that a fewer number of individual strains will be uniquely detected from short 16S rRNA gene amplicons covering only 1 or 2 of the 9 hyper-variable regions of the 16S rRNA gene [28]. The short NGS reads covering 1 or 2 hyper-variable regions of the gene often align equally well to sequences from multiple taxa [27], limiting the ability to pinpoint specific strains. Longer amplicons that span all 9 regions [15] are preferable and can be assessed by Sanger sequencing, probe arrays such as the PhyloChip [11] and now also possible with long read high-throughput sequencing [28]. As with all bioinformatic sequence reference databases, as knowledge of new strains expands, we expect two changes to previously published shotgun metagenomic and 16S rRNA amplicon findings. First, additional reads from the raw data will match to the newly isolated and sequenced strains and, second, some reads that were perceived as evidence of unique strain hits in the past will be determined to be non-unique in the future. Data platforms will need to be developed that can easily remap all public raw data into strain bins in a cost-effective manner with each update of StrainSelect. These platforms will need stable funding and resources since the growth of this data is not exhibiting deceleration (Fig. 1).
6. Conclusion
StrainSelect is a reference database of archaeal and bacterial genomic identifiers organized by strain (see Graphical abstract for a visual summary). StrainSelect assigns a consistently formatted identifier for known strains that have been isolated and have had their genome assembled or at least their 16S rRNA gene assembled and shared publicly. StrainSelect has three important properties. First, the database appropriately labels each contig and genome assembly by the strain of origin carefully avoiding conflation of genome assemblies as strains and in doing so identified over 10,000 strains with at least two technical replicate assemblies. Second, each strain is mapped to the bio-resource centers where the live strain can be procured if extant. Third, a single comprehensive domain to strain taxonomic ontology is included integrating both 16S rRNA genes and genome assemblies as points of reference so meta-analysis sourced from both technologies are possible. StrainSelect, with 681,087 strains demarcated, is the largest collection of its kind. The database can be inspected in graph or tabular formats in its entirety allowing mapping between StrainSelect strain identifiers, genome assemblies, 16S rRNA genes, international bio-resource center catalog identifiers for strain procurement, and genome function-focused databases.
With the StrainSelect foundation, research teams can annotate microbial community data into strain-level biomarkers, accelerate translational research after biomarker discovery into in vivo laboratory experiments with those strains to establish causality and confirm findings with meta-analyses across a growing public data warehouse containing 16S rRNA and shotgun metagenomics data.
StrainSelect database is available for download at http://strainselect.secondgenome.com.
Funding
This work was supported in part by the National Institutes of Health's (NIH) National Institute on Drug Abuse (NIDA) [R44DA043954].
CRediT authorship contribution statement
Todd Z. DeSantis: Conceptualization, Software, Formal analysis, Data Curation, Writing, Visualization. Cesar Cardona: Software, Investigation. Nicole R. Narayan: Methodology, Investigation. Satish Viswanatham: Resources, Supervision. Divya Ravichandar: Methodology. Brendan Wee: Software. Cheryl-Emiliane Chow: Methodology. Shoko Iwai: Methodology.
Declaration of Competing Interest
All authors were employed by Second Genome, Inc. during the course of the work. Second Genome, Inc. is a biomarkers and therapeutics company with products in development derived from bacterial polypeptides to treat cancer, ulcerative colitis and other human diseases. A publication announcing the availability of StrainSelect for academic use will allow greater transparency on the contents of our reference database but will not affect the value of our therapeutic products.
Acknowledgements
The authors thank Dr. Peter Karp at SRI International, Dr. Phil Hugenholtz at the University of Queensland, Dr. Nikos Krypides at the Joint Genome Institute, and Drs. Wim De Smet and Peter Dawyndt at Ghent University for their helpful discussions regarding the databases they initiated and curated. We also appreciate the guidance of Dr. Alex Probst in forming the discussion on MAGs as future entities in StrainSelect.
Footnotes
Supplementary material related to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e13314.
Appendix A. Supplementary material
The following is the Supplementary material related to this article.
Supplemental figures.
References
- 1.Almeida A., Mitchell A.L., Boland M., Forster S.C., Gloor G.B., Tarkowska A., Lawley T.D., Finn R.D. A new genomic blueprint of the human gut microbiota. Nature. 2019;568(7753):499–504. doi: 10.1038/s41586-019-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W. GenBank. Nucleic Acids Res. 2013;41(Database issue) doi: 10.1093/nar/gks1195. D36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bidartondo M.I. Preserving accuracy in GenBank. Science. 2008;319(5870):1616. doi: 10.1126/science.319.5870.1616a. [DOI] [PubMed] [Google Scholar]
- 4.Boone D.R., Castenholz R.W. 2nd edition. Springer; New York: 2001. Bergey's Manual of Systematic Bacteriology. [Google Scholar]
- 5.Briatte F. 2021. ggnetwork: Geometries to Plot Networks with ggplot2. [Google Scholar]
- 6.Burstein D., Harrington L.B., Strutt S.C., Probst A.J., Anantharaman K., Thomas B.C., Doudna J.A., Banfield J.F. New CRISPR-Cas systems from uncultivated microbes. Nature. 2017;542(7640):237–241. doi: 10.1038/nature21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheleuitte-Nieves C., Gulvik C.A., McQuiston J.R., Humrighouse B.W., Bell M.E., Villarma A., Fischetti V.A., Westblade L.F., Lipman N.S. Genotypic differences between strains of the opportunistic pathogen corynebacterium bovis isolated from humans, cows, and rodents. PLoS ONE. 2018;13(12) doi: 10.1371/journal.pone.0209231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Q., Zobel J., Verspoor K. Duplicates, redundancies and inconsistencies in the primary nucleotide databases: a descriptive study. Database (Oxford) 2017;2017 doi: 10.1093/database/baw163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conway J.R., Lex A., Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33(18):2938–2940. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Csardi G., Nepusz T. 2006. The Igraph Software Package for Complex Network Research. [Google Scholar]
- 11.DeSantis T.Z., Dubosarskiy I., Murray S.R., Andersen G.L. Comprehensive aligned sequence construction for automated design of effective probes (CASCADE-P) using 16S rDNA. Bioinformatics. 2003;19(12):1461–1468. doi: 10.1093/bioinformatics/btg200. [DOI] [PubMed] [Google Scholar]
- 12.DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dijkshoorn L., Ursing B.M., Ursing J.B. Strain, clone and species: comments on three basic concepts of bacteriology. J. Med. Microbiol. 2000;49(5):397–401. doi: 10.1099/0022-1317-49-5-397. [DOI] [PubMed] [Google Scholar]
- 14.Dowle M., Srinivasan A. 2021. data.table: Extension of data.frame. [Google Scholar]
- 15.Durso L.M., Harhay G.P., Smith T.P.L., Bono J.L., Desantis T.Z., Harhay D.M., Andersen G.L., Keen J.E., Laegreid W.W., Clawson M.L. Animal to animal variation in fecal microbial diversity among beef cattle. Appl. Environ. Microbiol. 2010;76(14):4858–4862. doi: 10.1128/AEM.00207-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duvallet C., Gibbons S.M., Gurry T., Irizarry R.A., Alm E.J. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat. Commun. 2017;8(1):1784. doi: 10.1038/s41467-017-01973-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 19.Federhen S. Type material in the NCBI taxonomy database. Nucleic Acids Res. 2015;43(Database issue) doi: 10.1093/nar/gku1127. D1086–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Federhen S., Clark K., Barrett T., Parkinson H., Ostell J., Kodama Y., Mashima J., Nakamura Y., Cochrane G., Karsch-Mizrachi I. Toward richer metadata for microbial sequences: replacing strain-level NCBI taxonomy taxids with BioProject, BioSample and Assembly records. Stand. Genom. Sci. 2014;9(3):1275–1277. doi: 10.4056/sigs.4851102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Field D., Garrity G., Gray T., Morrison N., Selengut J., Sterk P., Tatusova T., Thomson N., Allen M.J., Angiuoli S.V., Ashburner M., Axelrod N., Baldauf S., Ballard S., Boore J., Cochrane G., Cole J., Dawyndt P., De Vos P., DePamphilis C., Edwards R., Faruque N., Feldman R., Gilbert J., Gilna P., Glöckner F.O., Goldstein P., Guralnick R., Haft D., Hancock D., Hermjakob H., Hertz-Fowler C., Hugenholtz P., Joint I., Kagan L., Kane M., Kennedy J., Kowalchuk G., Kottmann R., Kolker E., Kravitz S., Kyrpides N., Leebens-Mack J., Lewis S.E., Li K., Lister A.L., Lord P., Maltsev N., Markowitz V., Martiny J., Methe B., Mizrachi I., Moxon R., Nelson K., Parkhill J., Proctor L., White O., Sansone S.-A., Spiers A., Stevens R., Swift P., Taylor C., Tateno Y., Tett A., Turner S., Ussery D., Vaughan B., Ward N., Whetzel T., San Gil I., Wilson G., Wipat A. The minimum information about a genome sequence (MIGS) specification. Nat. Biotechnol. 2008;26(5):541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haft D.H., DiCuccio M., Badretdin A., Brover V., Chetvernin V., O'Neill K., Li W., Chitsaz F., Derbyshire M.K., Gonzales N.R., Gwadz M., Lu F., Marchler G.H., Song J.S., Thanki N., Yamashita R.A., Zheng C., Thibaud-Nissen F., Geer L.Y., Marchler-Bauer A., Pruitt K.D. RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res. 2018;46(D1):D851–D860. doi: 10.1093/nar/gkx1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamming R.W. Error detecting and error correcting codes. Bell Syst. Tech. J. 1950;29(2):147–160. [Google Scholar]
- 24.Hennig C. 2020. fpc: Flexible Procedures for Clustering. [Google Scholar]
- 25.Hu D., Liu B., Wang L., Reeves P.R. Living trees: high-quality reproducible and reusable construction of bacterial phylogenetic trees. Mol. Biol. Evol. 2020;37(2):563–575. doi: 10.1093/molbev/msz241. [DOI] [PubMed] [Google Scholar]
- 26.Jain C., Rodriguez-R L.M., Phillippy A.M., Konstantinidis K.T., Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018;9(1):5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeong J., Yun K., Mun S., Chung W.-H., Choi S.-Y., Nam Y.-d., Lim M.Y., Hong C.P., Park C., Ahn Y.J., Han K. The effect of taxonomic classification by full-length 16S rRNA sequencing with a synthetic long-read technology. Sci. Rep. 2021;11(1):1727. doi: 10.1038/s41598-020-80826-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson J.S., Spakowicz D.J., Hong B.-Y., Petersen L.M., Demkowicz P., Chen L., Leopold S.R., Hanson B.M., Agresta H.O., Gerstein M., Sodergren E., Weinstock G.M. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019;10(1):5029. doi: 10.1038/s41467-019-13036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karp P.D., Billington R., Caspi R., Fulcher C.A., Latendresse M., Kothari A., Keseler I.M., Krummenacker M., Midford P.E., Ong Q., Ong W.K., Paley S.M., Subhraveti P. The BioCyc collection of microbial genomes and metabolic pathways. Brief. Bioinform. 2019;20(4):1085–1093. doi: 10.1093/bib/bbx085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim M., Oh H.-S., Park S.-C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014;64(Pt 2):346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 32.Lee K.A., Thomas A.M., Bolte L.A., Björk J.R., de Ruijter L.K., Armanini F., Asnicar F., Blanco-Miguez A., Board R., Calbet-Llopart N., Derosa L., Dhomen N., Brooks K., Harland M., Harries M., Leeming E.R., Lorigan P., Manghi P., Marais R., Newton-Bishop J., Nezi L., Pinto F., Potrony M., Puig S., Serra-Bellver P., Shaw H.M., Tamburini S., Valpione S., Vijay A., Waldron L., Zitvogel L., Zolfo M., de Vries E.G.E., Nathan P., Fehrmann R.S.N., Bataille V., Hospers G.A.P., Spector T.D., Weersma R.K., Segata N. Cross-cohort gut microbiome associations with immune checkpoint inhibitor response in advanced melanoma. Nat. Med. 2022;28(3):535–544. doi: 10.1038/s41591-022-01695-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.López Y., Nakai K., Patil A. HitPredict version 4: comprehensive reliability scoring of physical protein-protein interactions from more than 100 species. Database (Oxford) 2015;2015 doi: 10.1093/database/bav117. bav117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDonald D., Price M.N., Goodrich J., Nawrocki E.P., DeSantis T.Z., Probst A., Andersen G.L., Knight R., Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6(3):610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendes R., Kruijt M., de Bruijn I., Dekkers E., van der Voort M., Schneider J.H.M., Piceno Y.M., DeSantis T.Z., Andersen G.L., Bakker P.A.H.M., Raaijmakers J.M. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332(6033):1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 36.Mishra A.K., Gimenez G., Lagier J.-C., Robert C., Raoult D., Fournier P.-E. Genome sequence and description of Alistipes senegalensis sp. nov. Stand. Genom. Sci. 2012;6(3):1–16. doi: 10.4056/sigs.2625821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morotomi M., Nagai F., Watanabe Y. Description of Christensenella minuta gen. nov., sp. nov., isolated from human faeces, which forms a distinct branch in the order Clostridiales, and proposal of Christensenellaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2012;62(Pt 1):144–149. doi: 10.1099/ijs.0.026989-0. [DOI] [PubMed] [Google Scholar]
- 38.Mukherjee S., Stamatis D., Bertsch J., Ovchinnikova G., Katta H.Y., Mojica A., Chen I.-M.A., Kyrpides N.C., Reddy T. Genomes OnLine database (GOLD) v. 7: updates and new features. Nucleic Acids Res. 2019;47(D1):D649–D659. doi: 10.1093/nar/gky977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nayfach S., Shi Z.J., Seshadri R., Pollard K.S., Kyrpides N.C. New insights from uncultivated genomes of the global human gut microbiome. Nature. 2019;568(7753):505–510. doi: 10.1038/s41586-019-1058-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olm M.R., Crits-Christoph A., Bouma-Gregson K., Firek B.A., Morowitz M.J., Banfield J.F. inStrain profiles population microdiversity from metagenomic data and sensitively detects shared microbial strains. Nat. Biotechnol. 2021;39(6):727–736. doi: 10.1038/s41587-020-00797-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orakov A., Fullam A., Coelho L.P., Khedkar S., Szklarczyk D., Mende D.R., Schmidt T.S.B., Bork P. GUNC: detection of chimerism and contamination in prokaryotic genomes. Genome Biol. 2021;22(1):178. doi: 10.1186/s13059-021-02393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parks D.H., Chuvochina M., Waite D.W., Rinke C., Skarshewski A., Chaumeil P.-A., Hugenholtz P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018;36(10):996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 43.Parks D.H., Chuvochina M., Rinke C., Mussig A.J., Chaumeil P.-A., Hugenholtz P. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 2022;50(D1):D785–D794. doi: 10.1093/nar/gkab776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pei A.Y., Oberdorf W.E., Nossa C.W., Agarwal A., Chokshi P., Gerz E.A., Jin Z., Lee P., Yang L., Poles M., Brown S.M., Sotero S., DeSantis T.Z., Brodie E., Nelson K., Pei Z. Diversity of 16S rRNA genes within individual prokaryotic genomes. Appl. Environ. Microbiol. 2010;76(12):3886–3897. doi: 10.1128/AEM.02953-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pierce N.T., Irber L., Reiter T., Brooks P., Brown C.T. Large-scale sequence comparisons with sourmash. F1000Res. 2019;8:1006. doi: 10.12688/f1000research.19675.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ravichandar J.D., Rutherford E., Chow C.-E.T., Han A., Yamamoto M.L., Narayan N., Kaplan G.G., Beck P.L., Claesson M.J., Dabbagh K., Iwai S., DeSantis T.Z. Strain level and comprehensive microbiome analysis in inflammatory bowel disease via multi-technology meta-analysis identifies key bacterial influencers of disease. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.961020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.R_Core_Team . 2021. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 48.Reimer L.C., Sardà Carbasse J., Koblitz J., Ebeling C., Podstawka A., Overmann J. BacDive in 2022: the knowledge base for standardized bacterial and archaeal data. Nucleic Acids Res. 2022;50(D1):D741–D746. doi: 10.1093/nar/gkab961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rooks M.G., Veiga P., Reeves A.Z., Lavoie S., Yasuda K., Asano Y., Yoshihara K., Michaud M., Wardwell-Scott L., Gallini C.A., Glickman J.N., Sudo N., Huttenhower C., Lesser C.F., Garrett W.S. QseC inhibition as an antivirulence approach for colitis-associated bacteria. Proc. Natl. Acad. Sci. USA. 2017;114(1):142–147. doi: 10.1073/pnas.1612836114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sekido Y., Nishimura J., Nakano K., Osu T., Chow C.-E.T., Matsuno H., Ogino T., Fujino S., Miyoshi N., Takahashi H., Uemura M., Matsuda C., Kayama H., Mori M., Doki Y., Takeda K., Uchino M., Ikeuchi H., Mizushima T. Some Gammaproteobacteria are enriched within CD14+ macrophages from intestinal lamina propria of Crohn's disease patients versus mucus. Sci. Rep. 2020;10(1):2988. doi: 10.1038/s41598-020-59937-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shah M.S., DeSantis T.Z., Weinmaier T., McMurdie P.J., Cope J.L., Altrichter A., Yamal J.-M., Hollister E.B. Leveraging sequence-based faecal microbial community survey data to identify a composite biomarker for colorectal cancer. Gut. 2018;67(5):882–891. doi: 10.1136/gutjnl-2016-313189. [DOI] [PubMed] [Google Scholar]
- 52.Sharon I., Kertesz M., Hug L.A., Pushkarev D., Blauwkamp T.A., Castelle C.J., Amirebrahimi M., Thomas B.C., Burstein D., Tringe S.G., Williams K.H., Banfield J.F. Accurate, multi-kb reads resolve complex populations and detect rare microorganisms. Genome Res. 2015;25(4):534–543. doi: 10.1101/gr.183012.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sivan A., Corrales L., Hubert N., Williams J.B., Aquino-Michaels K., Earley Z.M., Benyamin F.W., Lei Y.M., Jabri B., Alegre M.-L., Chang E.B., Gajewski T.F. Commensal bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350(6264):1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tessler M., Neumann J.S., Afshinnekoo E., Pineda M., Hersch R., Velho L.F.M., Segovia B.T., Lansac-Toha F.A., Lemke M., DeSalle R., Mason C.E., Brugler M.R. Large-scale differences in microbial biodiversity discovery between 16S amplicon and shotgun sequencing. Sci. Rep. 2017;7(1):6589. doi: 10.1038/s41598-017-06665-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verslyppe B., De Smet W., De Baets B., De Vos P., Dawyndt P. StrainInfo introduces electronic passports for microorganisms. Syst. Appl. Microbiol. 2014;37(1):42–50. doi: 10.1016/j.syapm.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 56.Wattam A.R., Brettin T., Davis J.J., Gerdes S., Kenyon R., Machi D., Mao C., Olson R., Overbeek R., Pusch G.D., Shukla M.P., Stevens R., Vonstein V., Warren A., Xia F., Yoo H. Assembly, annotation, and comparative genomics in PATRIC, the all bacterial bioinformatics resource center. Methods Mol. Biol. 2018;1704:79–101. doi: 10.1007/978-1-4939-7463-4_4. [DOI] [PubMed] [Google Scholar]
- 57.West K.A., Yin X., Rutherford E.M., Wee B., Choi J., Chrisman B.S., Dunlap K.L., Hannibal R.L., Hartono W., Lin M., Raack E., Sabino K., Wu Y., Wall D.P., David M.M., Dabbagh K., DeSantis T.Z., Iwai S. Multi-angle meta-analysis of the gut microbiome in Autism Spectrum Disorder: a step toward understanding patient subgroups. Sci. Rep. 2022;12(1) doi: 10.1038/s41598-022-21327-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wheeler T.J., Eddy S.R. nhmmer: DNA homology search with profile HMMs. Bioinformatics. 2013;29(19):2487–2489. doi: 10.1093/bioinformatics/btt403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wick R.R., Judd L.M., Cerdeira L.T., Hawkey J., Méric G., Vezina B., Wyres K.L., Holt K.E. Trycycler: consensus long-read assemblies for bacterial genomes. Genome Biol. 2021;22(1):266. doi: 10.1186/s13059-021-02483-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wickham H. 2nd ed. 2016 edition. Springer International Publishing: Imprint: Springer; Cham: 2016. ggplot2: Elegant Graphics for Data Analysis. Use R! [Google Scholar]
- 61.Xu S., Chen M., Feng T., Zhan L., Zhou L., Yu G. Use ggbreak to effectively utilize plotting space to deal with large datasets and outliers. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.774846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu H. 2021. kableExtra: Construct Complex Table with kable and Pipe Syntax. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figures.