Abstract
Immunohistochemistry (IHC) highlights specific cell types in tissues and traditionally involves antibody staining together with a hematoxylin counterstain. The intensity and pattern of hematoxylin staining differs between cell types and reveals morphological characteristics of cells. Here, we propose that features in the hematoxylin stain can be used to predict IHC labels, such as Neurofibromin (encoded by the gene NF1). The dataset consists of 7.2 million cells from benign and kidney cancer cores in a tissue microarray. Morphology and hematoxylin (H&M) features defined within QuPath are subjected to a clustering analysis in CytoMap. H&M features are also used to train 4 different XGBoost models to predict high, low, and negative NF1 stain classes in benign renal tubules, clear cell (ccRCC), papillary (PRCC), and chromophobe (ChRCC) renal carcinoma. The prediction accuracies of NF1 staining classes in benign, ccRCC, ChRCC, and PRCC range between 70% and 90% with areas under the precision recall curve PRAUCNF1-high = 0.82+0.12, PRAUCNF1-low = 0.62+0.25, and PRAUCNF1-negative = 0.83+0.16. The most important feature for predicting the NF1 class involves the minimum cellular hematoxylin staining intensity. Together, these results demonstrate the feasibility to predict NF1 expression solely from features in hematoxylin staining using open source software. Since the hematoxylin features can be obtained from regular H&E and IHC slides, the proposed workflow has broad applicability.
Keywords: QuPath, CytoMap, Kidney cancer, Prediction
Highlights
-
•
Hematoxylin and morphology (H&M) features are different between cell types.
-
•
H&M features at single cell level can be defined in open source software QuPath.
-
•
H&M features can predict immunohistochemistry labels with high accuracy.
Introduction
Immunohistochemistry (IHC) has emerged as a powerful approach to interrogate cellular mechanisms in normal and diseased tissues, as well as to assist with diagnostic and prognostic questions.1,2 Using IHC protocols, tissue sections on glass slides are stained with antibodies to visualize protein expression in single cells and subcellular compartments. Further, tissues are counterstained with hematoxylin (H) to identify cells and observe tissue organization. IHC staining can be examined using a regular brightfield microscope and is widely used in clinical pathology practice. Slides stained by IHC can be digitized on regular slide scanners and analyzed by image analysis. Evaluating the staining of specific cells types within heterogenous cell populations can be challenging because pathologists have to rely sole on H without Eosin (E) for cellular characterization. However, IHC staining only uses H without E. Machine-learning algorithms have the potential to help with cellular classifications since they can be trained to distinguish different morphologies in H&E stained tissues.3, 4, 5, 6 However, it is unclear whether algorithms can utilize H staining by itself to separate cell types and thus improve the interpretation of slides stained by IHC.
Neurofibromin 1 (NF-1) is a tumor suppressor protein that negatively regulates highly oncogenic RAS proteins.7 NF-1 contains an intrinsic guanine nucleotide hydrolysis (GAP) function to convert active RAS-GTP to RAS-GDP.8 In addition, NF-1 functions as a RAS effector, regulating RAS signals for pathway activation. NF1 mRNA is expressed at varying levels in adult tissues and is developmentally regulated during embryogenesis.9 To increase the activity of the tumor promoting RAS pathway, NF1 expression is decreased in multiple cancer types.10,11
Neurofibromin is a large (∼280 kDa) multifunctional protein and is one of the largest genes in the human genome.12 Its best-known function is as a GTPase-activating protein for Ras (Ras-GAP), thereby acting as an “off” signal for the RAS-GTPase.13,14 However, neurofibromin has multiple additional interacting partners such as FAK, tubulin, and Spred (Sprouty-Related EVH1 domain-containing protein, another negative regulator of the Ras-MAPK pathway), and is involved in several cell signaling pathways, including the Ras/MAPK, Akt/mTOR, ROCK/LIMK/cofilin, and cAMP/PKA pathways, thereby regulating many fundamental cellular processes including proliferation and migration.7 NF1 loss-of-function mutation(s) within the germline results in NF1 disease, a syndrome associated with an increased predisposition for benign peripheral nerve sheath tumors, neurofibromas, as well as for malignant sarcomas, gliomas, pheochromocytomas, gastrointestinal stromal tumors, and myeloid leukemia.15,16 Somatic NF1 mutations have been observed in many sporadic tumor types particularly in melanoma, lung adenocarcinoma, and glioblastoma.17
QuPath is a multifunctional open-source software package for analysis of whole-slide images stained with single or multiple stains. Using QuPath commands, the user can co-register images, perform segmentation of anatomical structures using pretrained convolutional neural networks, draw the outlines of nuclei by using feature-based or deep learning-based models and obtain size, shape, and texture feature values from segmented objects.18 As such, QuPath has broad applicability without the need of coding expertise. Recently, another open-source software called Histo-Cytometric Multidimensional Analysis Pipeline (CytoMAP) became available.19 CytoMap provides tools for data clustering, positional correlation, dimensionality reduction, and 2D/3D region reconstruction to identify localized cellular networks and reveal both cellular- and tissue-level relationships. We applied the unsupervised clustering function of CytoMap to QuPath features. Cells in clusters can be visualized within the context of the tissue architecture revealing interactions between clusters and allowing for interpretation of cell morphology.19 Together, QuPath and CytoMap provide a powerful toolkit based on multiple machine learning algorithms for comprehensive analysis of cellular phenotypes and spatial relationships in tissues stained by IHC.
In this study, we apply QuPath functionality to segment renal tubules and single cells in kidney with and without cancer. We extract hematoxylin and morphology (H&M) feature values from single cells and demonstrate that H&M features can be used to separate cell populations in tissues stained by IHC using clustering in CytoMap and to predict NF1 protein expression.
Methods
QuPath software
We analyzed immunohistochemistry images from tissue sections in QuPath (version 0.3.0), which is an open-source software for digital pathology and whole-slide image analysis.20 Briefly, the software was developed using Java 8, with a JavaFX interface for object annotation and visualization. QuPath has built-in algorithms for general tasks, such as cell and tissue detection, and interactive machine learning for object and pixel classification. The software supports several image formats through Bio-Formats and OpenSlide, including whole-slide images of IHC antibody stains.
CytoMap
CytoMap is an MatLab-based Histo-Cytometric Multidimensional Analysis Pipeline (CytoMap) for spatial analysis of segmented cell objects,19 which utilizes diverse statistical approaches to extract and quantify information about cellular spatial positioning, preferential cell–cell associations, and global tissue structure. CytoMAP is capable of simplifying spatial analysis by grouping cells into local neighborhoods and revealing complex patterns of cellular composition and regional tissue structures.19
Cases, IHC staining, and TMA construction and imaging
We obtained tissue blocks from the pathology archive at the University of Utah of patients who underwent nephrectomy for kidney cancer. The study was approved by the Institutional Review Board under protocol #00067518. Excess formalin-fixed and paraffin-embedded tissue was used to generate tissue microarrays (TMA). The cases were deidentified before blocks were used for construction of TMAs. The digital slides from these TMAs were also fully deidentified. For TMA construction, cylindrical 2 mm cores from benign and cancer regions were sampled from donor blocks and placed into columns of recipient tissue array blocks. Each column consists of 6 cores from 1 patient, divided in 3 cores entirely composed of benign renal parenchyma and 3 cores of cancer. Cases include 50 clear cell renal cell carcinomas (ccRCC) in 5 TMA blocks, 30 chromophobe renal carcinomas (ChRCC) in 3 TMA blocks, and 17 papillary renal carcinomas (PRCC) in 2 TMA blocks. TMA blocks were sectioned at 4-microns. Slides were stained with the NF1 antibody on the Leica Bond Rx. NF1 antibody was purchased from Sigma–Aldrich (catalog #HPA045502), and was validated in the Human Protein Atlas. The protocol on the Leica Bond Rx consists of antigen retrieval #2 for 30 min and a 15-min incubation with the antibody at a dilution of 1:200.
The NF1 antibody binding was visualized using 3, 3'-diaminobenzidine (DAB). Slides were scanned on the Aperio AT2 slide scanner. Digital slides were opened in QuPath and images of individual cores were saved with a label of the row, column, and case ID number.
Renal tubule segmentation
We used a random tree-based algorithm available in QuPath to train a pixel-level classification model to outline renal tubules. An algorithm was trained in QuPath to outline renal tubules and consists of a random tree-based pixel-wise classification model. The model was trained on manual outlines of tubules from 10 tissue regions. To access the software module, we applied the following commands: Classify>>Pixel classification>>Train pixel classifier and used the detailed settings in Supplementary Table 1. After training, the model was applied to all normal tissue cores to obtain outlines of individual tubules with the minimum object size and minimum hole size set at 50 and 20 μm2, respectively. The step-by-step process for model training is illustrated in Supplementary Fig. 1.
Nuclear segmentation and generation of cell outlines
After applying the tubule segmentation algorithm, we segment nuclei within the outlines of renal tubules in benign cores or across the entire tissue of cancer cores. For segmentation of nuclei, we apply the STARDIST nucleus segmentation algorithm21 to images at ×20 magnification. This model is available within QuPath without further training. We do not assess its performance in our case because it was previously validated.21 To obtain cell borders, we expanded the nuclear outlines by 5 pixels. The parameter settings of the STARDIST code for nuclear segmentation are available in the Supplementary Table 1.
Determining NF1 expression levels and calculation of H-scores
Determining cell-wise NF1 levels
Throughout the study, we use 3 ways to determine the expression of NF1 protein in the cytoplasm in tissue sections stained by IHC.
-
1.
Pixel level NF1 staining intensity: determined as the average intensity of brown color in cytoplasmic pixels, named: “NF1 staining intensity.”
-
2.
NF1 class: determined by an algorithm trained in QuPath that classifies cells into negative, low, and high NF1 staining groups based on NF1 staining intensity level, named: “NF1 class.”
-
3.
Predicted NF1 class: determined by an XGBoost model trained on H&M features and named “predicted NF1 class.”
Calculation of NF1 staining intensity
The cell segmentation approach described above leads to a division of cells into nuclear, cytoplasmic, and membrane compartments. The NF1 protein is expressed in the cytoplasm. Therefore, for each cell, the NF1 staining intensity is calculated as the average intensity of cytoplasmic pixels. Each pixel receives a value for the DAB channel proportional to the darkness of brown staining in the pixel. The average of DAB pixel values in each cell amounts to the cellular NF1 staining intensity. To determine the average NF1 staining intensity of cells in the low or high NF1 classes, we averaged the NF1 staining intensity across all cells in the class within each core.
Determining cell-wise NF1 classes
We propose 3 NF1 classes, NF1-negative, -low, and -high. To determine the class to each cell, we trained an algorithm to perform an automated classification based on cytoplasmic NF1 staining. The algorithm was trained using the object classification model in QuPath. Areas of tubules containing high or low NF1 stained cells as well as negative cells outside tubules were manually annotated in 10 regions and used for training of a random tree-based algorithm. Briefly, the training was performed using the Classify>>Object classification>>Train object classifier module with the detailed settings in Supplementary Table 1. The step-by-step training process of the model and the settings used in QuPath is available in Supplementary Fig. 2. Performance of the algorithm is assessed by visual evaluation of each core. If the performance of tree-based NF1 classification model is deemed unsatisfactory for a core, the algorithm is further trained with examples from that specific core.
The trained NF1 classification algorithm is applied to segmented cells in renal tubules and to cancer cores. The algorithm assigns each cell to either the NF1-high, NF1-low, or to the NF1-negative class. Cell numbers in each class are determined in Python 3.7.
Digital H-score
Digital H-scores are calculated using NF1 classes to summarize the NF1 expression in each core.22 The H-score is composed of percentages of cells within high-, low-, or negative NF1 classes on a scale from 0 to 2 and calculated using the equation:
In the benign cores, we only include cells within renal tubules to calculate the H-scores. In the cancer cores, we included all the cells.
Extraction of features from the hematoxylin channel, and cell clustering
Color deconvolution happens automatically, separating hematoxylin, DAB and residual channels when images are uploaded to QuPath. We obtained features from nuclear, cytoplasmic, and membrane compartments using the hematoxylin channel. The features are divided into hematoxylin (H) features capturing parameters from the distribution hematoxylin staining intensities and morphology (M) features capturing nuclear and cellular shapes. Altogether, QuPath provides values of 33 H&M features from the hematoxylin channel.
After nuclear segmentation in renal tubules and cancer regions, we exported the coordinates and H&M features of each cell from QuPath. We import feature values into CytoMap channels, normalize the features to the maximum of each channel, and perform an unsupervised cell clustering. Self-organizing map (SOM),23 an unsupervised clustering approach, was used to assign each cell to a cluster designation. The Davies-Bouldin index provides a score to determine the optimal number of clusters in the data by calculating the ratio of similarity of feature values between clusters to within a cluster. We retrieve the assignment of cell to cluster in QuPath for visualization. We describe the code to provide cluster labels of cells in tissue images in detail in the Supplementary Methods section.
Interrogation of biases in H&M feature values
Preanalytical variables during tissue collection and processing can generate abnormal hematoxylin staining, which in turn can introduce a bias in hematoxylin-derived features. To identify a potential bias across TMA cores, we used principal component analysis (PCA) on the hematoxylin features from benign cores of all TMAs and plot the first 2 components. This analysis showed the overlap of cases from 3 kidney cancer subtypes (ccRCC, ChRCC, and PRCC) and no separation of cases into clouds, indicating the homogeneity of hematoxylin-derived feature values across all TMAs.
In addition to a potential preanalytical bias, we examined a bias that might have arisen through incomplete unmixing between the hematoxylin and DAB channel. This issue might lead to a contribution of brown NF1 staining intensity to the intensity of staining by blue hematoxylin. We calculated the Spearman correlation (R2 = 0.0017, Supplementary Fig. 3) between the average staining intensity of DAB and hematoxylin in the cytoplasm of each cell, indicating the absence of a bias caused by insufficient unmixing of colors.
Training of models to predict the NF1 class
To train NF1 classification models using H&M features from each cell, we divided cores into training, validation, and testing groups at the patient level according to a ratio of 0.6:0.2:0.2. We trained separate models for renal tubules, ccRCC, PRCC, and CRCC using the NF1 class as the prediction target value. We evaluated the model performance using the classification accuracy, which is the percentage of correctly classified cells. We performed 5-fold cross validation on the training set in Python 3.7.
We tested the following models: DT – decision tree,24 RF – random forest,25 LDA – linear discriminant analysis,26 XGBoost – regularization gradient boosting framework,27 kNN – k nearest neighbor,28 MLP – multilayer perceptron,29 and NB – naïve Bayesian.30 Model training parameters in QuPath and Python are listed in Supplementary Table 2. We used the published,31 default hyperparameters for each of the models.
To obtain variable importance of each of 33 H&M features, we applied the XGBoost classification model on all cases of renal tubules, ccRCC, PRCC, or ChRCC cores. The XGBoost model outputs the variable importance score of each feature in terms of its contribution to the overall accuracy.
Statistical analysis
Box plots were used to visualize differences between groups. Within each box, the mean value is indicated by a black line and the whiskers of the box include the 25th and 75th percentiles of data points.
We used OriginLab Pro 2022b to generate all visualizations. We used One-way ANOVA to compare NF1 scores for 3 group (negative, low, and high) comparisons and t-tests for 2 group comparison. We use a P-value threshold of .05 to indicate significance.
We use accuracy, the area under the precision-recall receiver operating characteristic curve (PRAUC), and the F1 score as metrics of performance for the classification models, comparing both model performance with all features and for models trained on the top 5 features. We calculate accuracy as the ratio between the number of correct predictions to the total number of predictions, incorporating all 3 classes. We calculate the PRAUC and the F1 score (the harmonic mean between precision and recall), for each class versus the other 2 classes. The PRAUC and F1 score are particularly useful for imbalanced data, i.e., when one class is rare compared to the others.
Results
Overview of workflow
The overall workflow in QuPath and CytoMap is depicted in Fig. 1. 10 TMA slides with clear cell renal cell carcinoma (ccRCC), papillary renal carcinoma (PRCC), and chromophobe renal carcinoma (ChRCC) were stained for NF1, digitized, and analyzed in QuPath. We trained a pixel-wise classification algorithm to segment benign renal tubules and applied the STARDIST nuclear segmentation algorithm for nuclear segmentation. We applied a tool in Qupath to generate cell outlines by expanding nuclear contours. The analysis of benign tissues involved only cells within tubules (Supplementary Fig. 1).
Fig. 1.
Workflow of data generation and visualization. (A) Data structure. 10 TMA slides include 5 ccRCC TMAs (50 patients), 3 ChRCC TMAs (30 patients), and 2 PRCC TMAs (17 patients) displaying 3 benign and 3 cancer cores from each of 97 patients. TMA slides are stained by IHC with an anti-NF1 antibody. (B) Digital H-score. A digital H-score is generated in QuPath for each core or for individual tubules. (C) Targeted feature extraction. After color deconvolution into hematoxylin (H channel) and DAB (NF1 channel) channels in QuPath, 33 targeted feature values of morphology and hematoxylin (H&M features) are exported from each cell for further analysis. (D) Unsupervised cell clustering based on H&M features using CytoMap. (E) Training of XGBost prediction model. H&M feature values are used as the input into prediction models that predict the NF1 staining intensity class.
Next, we trained an algorithm in QuPath to automatically classify cells into NF1-high (class label 2), NF1-low (class label 1), and NF1-negative (class label 0) classes. To summarize the NF1 staining intensity in each core, a digital H-score was calculated as the sum of products from each class label times the percentage of cells in the class. The class designation was also used for the training of models that predict the NF1 class from features in the hematoxylin stain.
QuPath automatically unmixes the brown NF1 stain from the blue hematoxylin stain and calculates 33 feature values in the hematoxylin channel. The features pertain to cell and nuclear morphology and hematoxylin intensity (H&M features) (Supplementary Table 3). We exported cell-wise H&M features for unsupervised clustering of cells in CytoMap. In addition, we examined feature values for potential biases using principle component analysis, before using the features to train machine learning models predicting the NF1 class of each cell.
Digital H-score calculation in QuPath
In benign renal parenchyma, we observe three types of tubules composed of cells within NF1-high, -low and -negative classes. Because proportions of tubules vary amongst benign cores, we calculated a digital H-score to determine the overall NF1 staining level of each core. Digital H-scores were computed as described in the Methods section using the cell-wise NF1 classes (Fig. 2A). We did not observe a significant difference (P > .05) when comparing H-scores of benign cores associated with ccRCC, PRCC, or ChRCC (Fig. 2B). For comparison with core-wise H-scores, H-scores were calculated for populations of individual tubules that contain a majority of either NF1-high or NF1-low cells. The same NF1 classification used for benign tubules was applied to calculate the H-scores of cancer cores (Fig. 2C). Hence, QuPath is capable of computing H-scores of tissue sections with heterogenous IHC staining.
Fig. 2.
Digital H score. (A) Benign tissue. Tubules are outlined as described in Fig. 1. Cells within tubules are classified based on NF1 staining intensity. NF1-high – red, NF1-low – purple, and NF1-negative – blue. (B) H-scores in benign tubules. H-scores from entire cores are shown for benign cores associated with ccRCC, PRCC, and ChRCC cases. Each dot in the box plot represents the H-score from 1 benign tissue core. For comparison of core-wise H-scores to H-scores of NF1-high and NF1-low tubules shown in the cell segmentation panel in (A), the H-scores for high- and low-NF1 expressing tubules are shown in orange and light blue, respectively. H-scores of NF1 in tumor cores. All cells in the core are analyzed. (C) H-scores of NF1 in tumor cores. All cells in the core are analyzed.
Cell cluster analysis in CytoMap
We generated cell clusters based on H&M features using the internal functionality of CytoMap. The list of the 33 H&M features that are generated by QuPath is shown in Supplementary Table 2. The 20 hematoxylin features include the mean, median, minimum, and maximum H staining intensity and standard deviation in whole cell, nuclear, cytoplasmic, and cell surface compartments. The 13 morphology features that we obtained from the H channel include the area, length, circularity, solidity (area of nucleus/convex hull area), minimum, and maximum diameter of the nuclear or cell outline and the nuclear to whole cell ratio. We excluded potential biases in feature values caused preanalytical variables or inadequate unmixing of hematoxylin and DAP channels.
We transferred the 33 feature values together with the coordinates of each cell from QuPath to CytoMap. An unsupervised cell clustering analysis was performed in CytoMap using all H&M features (Fig. 3A). The Davies-Bouldin index reveals that 4 cell clusters are the optimal number for the data (Fig. 3B). For each cluster, we determined the average NF1 expression level in the cytoplasm and observed an average NF1 intensity of clusters 1 and 4 similar to high-NF1-expressing renal tubules, while clusters 2 and 3 displayed NF1 expression levels in the range of negative- and low-NF1 tubules. Finally, using the cellular coordinates of each cell, we returned the cell-wise cluster designations to QuPath. This allows visualizing the locations of cells within each cluster using a function called measurement map in QuPath. Cells belonging to each cluster are labeled by a separate color in a tissue overlay (Fig. 3C).
Fig. 3.
Cell clustering using CytoMap. (A) Schematic workflow using CytoMap. Cell-wise H&M feature values together with cell coordinates are entered into CytoMap. (B) CytoMap output. CytoMap identifies the optimal number of clusters in the data and provides each cell with a cluster label. The pink shaded areas represent the range of NF1 staining intensity in high-NF1 expressing tubules, while the gray shaded area indicates NF1 staining levels in low-NF1 tubules. (C) Cluster visualization. Cluster labels are retuned to QuPath for overlay with the original image.
Tumor cores contain heterogeneous cell populations comprising cancer, immune, and stromal cells. To determine if clustering in CytoMap can separate cell types, we analyzed cells from 1 core of ccRCC, PRCC, and ChRCC (Fig. 4A). We obtained H&M features using QuPath from all the cells in the core. While ccRCC and PRCC cores revealed 3 cell clusters, cells from ChRCC were divided into 2 clusters (Fig. 4B). In ccRCC, the proportions of cells in the three clusters are similar in NF1-negative, -low, and -high classes, while in ChRCC and PRCC, NF1-negative cells are primarily associated with cluster 2 and cluster 1, respectively (Fig. 4C).
Fig. 4.
Cluster analysis of ccRCC, ChRCC, and PRCC. (A) Representative region of interest from cores analyzed in CytoMap. Columns 1 and 2 show corresponding H&E and IHC images after co-registration. The distance between the H&E and IHC tissue sections hinders a direct comparison at the cell level. The IHC image is analyzed in QuPath as described in Fig. 2 to identify the NF1 class of each cell (third column). Fourth column shows the CytoMap cluster designation of each cell in the tissue context. (B) NF1 intensity comparison among clusters. Box plots show the NF1 staining intensity in each cluster. (C) Cluster representation inside NF1 classes. The percentage of cells within each cluster is plotted on the y-axis for NF1-negative, NF1-low, and NF1-high classes (X-axis). Tissue images are screenshots at 40X magnification in QuPath.
Next, we overlaid the cluster designation on an NF1 stained tissue image to allow for visual interpretation of cell types (Fig. 4A, column 4). In ccRCC, we observed NF1 staining of stromal cells and a few cancer cells, while in PRCC, immune cells, and stromal cells revealed greatest NF1 staining. Cluster 1 in ChRCC contains NF1-high and -low cancer cells, while cluster 2 contains mostly cells from the tumor microenvironment. Altogether, the data demonstrate that the cluster analysis can reveal distinct cell types solely based on H&M features. It is notable that the high NF1 expression in PRCC are immune and not cancer cells. More importantly, CytoMap in conjunction with QuPath helps to resolve cell types within tissues and improve the interpretation of positive cell types.
Prediction of NF1 expression levels using H&M features
We find that H&M features can be used to predict NF1 protein expression levels in renal tubules and subtypes of kidney cancers. This is supported by the associations between NF1 classes and cell clusters that are generated based on H&M features (Fig. 4). We trained 7 different machine learning models using cells from benign cores to determine which model is best suited to predict the NF1 class of a cell using the parameters for each model described in SCIKIT-learn package.31 Cells were divided at the patient level into training, validation, and testing groups. The training set consists of 764 498, 752 750, and 376 243 cells in the NF1-negative, -low, and -high classes, respectively. We performed a 5-fold cross validation using the cells in the training set to determine how the selection of training data effects the performance of the model. We assessed the performance of each model by the percentage of accurately classified cells. Fig. 5A shows that the Random Forrest (RF) model, the regularization gradient boosting framework (XGBoost) model, and a multi-layer perceptron (MLP) outperform the other 4 models. Based on its fast processing time, we selected XGBoost to address additional classification questions.
Fig. 5.
Prediction of NF1 expression from H&M. (A) Model selection. Each model is trained on single cells using 33 H&M features per cell to predict the NF1 class (high, low, and negative NF1 class). Data are divided at the patient level into training (60%), validation (20%), and testing (20%) sets. The accuracy (percent correctly classified cells) of each model is determined and repeated in a 5-fold cross-validation approach. The resulting data points in the held-out test set are plotted on the y-axis. The types of models are indicated below the x-axis (DT - decision tree, RF - random forest, LDA - linear discriminant analysis, XGBoost – regularization gradient boosting framework, kNN – k nearest neighbor, MLP – multilayer perceptron, and NB – naïve Bayesian). (B) Comparison of “true class” and “predicted class” number of cells in NF1 classes. Cell numbers in NF1-high, -low and -negative classes are depicted in the left bar, while cell numbers predicted by XGBoost are shown in the right bar. (C) Precision recall ROC curves of model trained on 33 H&M features. Gray – NF1 negative class, blue – NF1 low class, green – NF1 high class.
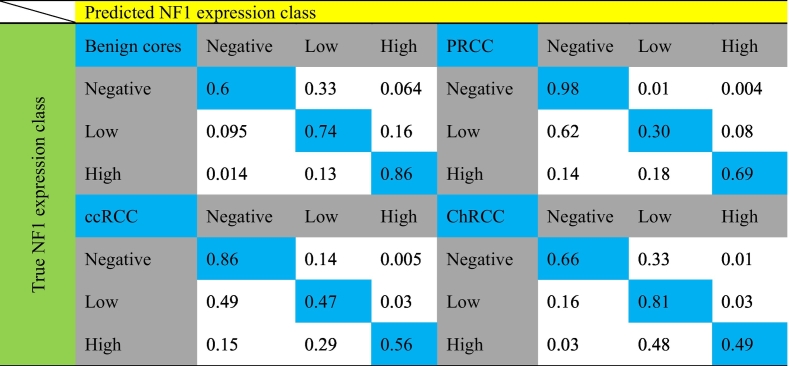
As a next step, we used XGBoost to predict the NF1 class of cancer cells. We trained separate models to predict NF1 classes in ccRCC, PRCC, and ChRCC. Results shown in Fig. 5B demonstrate that numbers in the predicted NF1 classes are comparable to ground-truth NF1 classes and that we have imbalanced number of classes. Misclassifications occurred primarily between negative and weak NF1 expressing classes in ccRCC and PRCC, while in ChRCC, misclassification was primarily observed between low and high NF1 classes (Table 1).
Table 1.
Predicted versus ground-truth NF1 classes for benign tubules, clear cell renal cell carcinoma (ccRCC), papillary renal cell carcinoma (PRCC), and chromophobe renal cell carcinoma (ChRCC).
Because of the class imbalance of NF1 classes, we used precision and recall to evaluate the classification performance of each class (Fig. 5C). We generated receiver operator curves (ROC) for 1 predicted NF1 class against the other 2 classes and used the area under the curve (AUC) to evaluate the performance of each model. The XGBoost models possess a good performance for predicting NF1-high and -negative cells. However, the performance of predicting the NF1-low class was reduced in ccRCC and PRCC with PRAUC = 0.4 and PRAUC = 0.41, respectively. Using the harmonic mean of precision and recall, we calculated F1 scores for all models and classes (Supplementary Table 4), confirming the PRAUC results. We noticed poor performance of an XGBoost model trained on benign tubules and applied to cancer cores (data not shown). Altogether, the XGBoost models trained separately on benign tubules and 3 subtypes of kidney cancer possess adequate performance to predict the NF1 classes, demonstrating feasibility of predicting an IHC label from hematoxylin features.
While separate training of models was required for benign and cancer, models may rely on the same H&M features to predict the NF1 class in the respective tissue entity. To determine whether the 4 XGBoost models prioritize the same or different features for NF1 class prediction, we obtained a rank list of feature importance for each model (Supplementary Fig. 5A). We also performed an ablation study, comparing models that used decreasing numbers of features in the prediction, starting with a model trained on only the top-ranked feature (Supplementary Fig. 5B). Next, we compared the top 5 features that each model selected and noticed considerable redundancy across models (Table 2). Altogether, there are only 10 distinct features in the top 20 from all 4 models. The top selected feature in all 4 models constitutes the minimum hematoxylin intensity in the cell and encompasses the minimum hematoxylin intensities in the nucleus and cytoplasm, which were also amongst the top features in the 3 cancer models. In addition, the standard deviation of hematoxylin pixel intensity in nucleus was selected by 3 models, which can loosely be related to chromatin compaction. Altogether, analysis of IHC stained tissues with QuPath together with CytoMap is broadly applicable and reveals novel associations of hematoxylin features and IHC-stained cell types. Together, the open source software packages can be used for quantitative analysis of tissue staining and interpretation of staining patterns in tissues.
Table 2.
Feature ranking in XGboost models trained on benign or individual cancer cells.
Discussion
Manual scoring of IHC images using H-scores, which is the current standard of practice, is known to have several shortcomings in terms of reproducibility and scalability to large studies. The introduction of automated digital image analysis has brought a new technique that may help to standardize and objectify pathological analysis for assessment of biomarkers. In addition, the separation of hematoxylin and DAB channels in IHC-stained slides allows us to determine whether features in the hematoxylin channel can predict IHC labels.
We performed a comprehensive image analysis of tissues stained by IHC to determine the expression of NF1 by leveraging the functionalities of the interactive QuPath and CytoMap software packages. Compared to commercial digital image analysis software (Halo and VisioPharm), QuPath is an open source software for analysis of tissues stained by H&E, IHC, and multiplexed immunofluorescence. Researchers have a chance to examine and modify the code in order to optimize the performance for specific use cases, reproduce analysis results through publishing the settings selected for training of models, and share trained models with collaborating teams, thereby benefitting the scientific community around the world through transparency and rigor of image analysis approaches.
The features measured by QuPath belong to the category of targeted, handcrafted (HC) features that can easily be interpreted. Several other studies highlight the ability of HC features and in particular nuclear features, to predict patient outcomes.32 However, the currently published nuclear features are related to shape, density, and orientation of nuclei and not related to the staining intensity of hematoxylin. In contrast to models using morphology features, our models, which predict NF1 IHC staining intensity, mostly select hematoxylin features. As of December 2022, we did not find a study in PubMed aiming to develop a model that predicts IHC labels solely based on multiple features in the hematoxylin channel. The closest to our approach is a report that uses QuPath to identify features in tumor cells that predict Ki67 positivity in the nucleus. The mean optical density of hematoxylin in the nucleus emerged as the best feature to distinguish Ki67 positive from negative cells, providing the pathologist with a fast method of identifying the proliferating compartment of the tumor through a quantitative assessment of only 1 nuclear feature in the hematoxylin channel.5 Thus, it appears that our approach using multiple features in the hematoxylin channel has not been used previously. A potentially broad applicability of using hematoxylin-derived features for prediction relies on the color unmixing functions in QuPath that can generate hematoxylin channels from slides stained with H&E, IHC, or Mason trichrome. Therefore, as a follow-up study it would be interesting to compare the prediction of NF1 staining from unmixed H&E images or from virtual hematoxylin stains.
Because IHC staining is an expensive, time-consuming process which can introduce discordant results due to variability in sample preparation and pathologist subjectivity, multiple groups attempted to predict IHC labels from H&E stained digital slides. In contrast to IHC staining, H&E staining—which highlights cellular morphology—is quick and less expensive. In 2016, a group at the University of Helsinki identified immune cell-rich and immune cell-poor regions in H&E stained cancer tissues guided by the pan-leukocyte marker, CD45. Using a pretrained convolutional neural network (CNN) to quantify immune cell infiltration reached a high agreement with the assessment by a pathologist.3 A different group trained a CNN to detect Ki67+ and Ki67- cells in an H&E image. The group reported a correlation coefficient of 0.8 between the percentage of Ki67 positive nuclei predicted by the model and the percentage of nuclei stained by Ki67 in a parallel tissue section.4 A similar strategy was applied to another use case, predicting the estrogen receptor status in H&E images of breast cancer. The authors reported an AUC = 0.92 in a heterogenous, multi-country dataset of 3474 patients.6 Most recently, the HEROHE grand challenge was organized to identify high performing machine learning algorithms for prediction of HER2 expression in breast cancer from H&E images.33 The top-performing teams achieved AUCs of 0.71–0.74, with 1 team reaching an AUC of 0.84. The H&E images and ground-truth IHC and ISH HER2 annotations for each case are publicly available. The above studies represent a few examples of a large effort within the digital pathology community to develop algorithms that can be used to predict IHC labels in cells from H&E stained tissues. It is unclear whether both hematoxylin and eosin are needed for the prediction or if one stain would be sufficient.
While we believe that our study bears considerable novelty, it also has several limitations. The cases used for TMA construction represent a single institution study, were processed by the same pathology laboratory over a 10-year period with minimal changes in protocols, were stained with the same hematoxylin formulation and stained as 1 batch on the autostainer for NF1. Neurofibromin was selected as a marker to provide proof-of-concept of this analytical approach because of its broad relevance to cancer and to the availability of validated antibodies. The slides were scanned altogether on the same slide scanner. Therefore, the technical variability of the study is smaller than what we would expect to see in a multi-institutional cohort study and a larger, independent study set would be desirable to confirm our results. The hematoxylin features provided by QuPath do not include features that capture patterns of hematoxylin staining. It would be interesting to determine whether the model performance could be improved by including texture features in the hematoxylin channel, which exist in Matlab.34 Finally, we experienced imbalances in the data. The ccRCC and PRCC kidney cancer subtypes contain 70–80% of NF1 negative cells, while the ChRCC mostly consists of low-expressing NF1 cells. Even though, we used precision and recall to evaluate the performance of the models, this does not overcome the imbalance effecting the training data. Furthermore, the PRAUC analysis compares 1 group versus the other 2 groups. This does take into consideration the ordinal structure of the data. The data structure could be simplified by using a 2-tiered system instead of our arbitrary choice of a 3-tiered system. The ultimate use of NF1 results is needed to decide the appropriate number of NF1 classes. Altogether, there is a need to further optimize and validate the approach we propose and to determine how it might work for other IHC labels that are different from NF1.
Conclusion
In conclusion, the open source digital image analysis software package, QuPath, provides a well-designed and multi-functional platform for data generation and visualization in clinical studies and research that does not require coding skills by the users. The compatibility of QuPath with other software packages, for example, CytoMap, provides additional analytical opportunities. Our study shows that morphology and hematoxylin features can predict NF1 expression levels in single cells from renal tubules and kidney cancer, indicating the potential of machine learning approaches to improve cancer diagnosis, prognosis, and treatment decisions.
Funding Statement
This project is supported by NIH/NCI grant R01CA217905 to MYK. Research reported in this publication utilized the Biorepository and Molecular Pathology Shared Resource at Huntsman Cancer Institute at the University of Utah and was supported by the National Cancer Institute of the National Institutes of Health under Award Number P30CA042014. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author Contributions
WZ, MYK, DS and BK designed the study. WZ collected and analyzed the imaging data. JY and BB carried out the statistical analysis. WZ and BK drafted the manuscript. All authors have read and approved the final manuscript.
Conflict of interest
The author(s) declare they have no competing interests
Acknowledgments
We acknowledge the generous support by the Department of Pathology and the Huntsman Cancer Institute. We thank Jeff Stanley and the Biospecimen and Molecular Pathology Shared Resource for the NF1 staining. We acknowledge the direct financial support for the research reported in this publication provided by the Huntsman Cancer Foundation and the Experimental Therapeutics Program at Huntsman Cancer Institute.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpi.2023.100196.
Contributor Information
Wei Zhang, Email: Wei.zhang@hci.utah.edu.
Beatrice S. Knudsen, Email: beatrice.knudsen@path.utah.edu.
Appendix A. Supplementary data
Supplementary material
References
- 1.Deng Y., Bartosovic M., Ma S., et al. Spatial profiling of chromatin accessibility in mouse and human tissues. Nature. 2022;609:375–383. doi: 10.1038/s41586-022-05094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ram S., Vizcarra P., Whalen P., et al. Pixelwise H-score: a novel digital image analysis-based metric to quantify membrane biomarker expression from immunohistochemistry images. PLoS One. 2021;16 doi: 10.1371/journal.pone.0245638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turkki R., Linder N., Kovanen P.E., Pellinen T., Lundin J. Antibody-supervised deep learning for quantification of tumor-infiltrating immune cells in hematoxylin and eosin stained breast cancer samples. J Pathol Inform. 2016;7:38. doi: 10.4103/2153-3539.189703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y., Li X., Zheng A., et al. Predict Ki-67 positive cells in H&E-stained images using deep learning independently from IHC-stained images. Front Mol Biosci. 2020:7. doi: 10.3389/fmolb.2020.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martino F., Varricchio S., Russo D., et al. A machine-learning approach for the assessment of the proliferative compartment of solid tumors on hematoxylin-eosin-stained sections. Cancers (Basel) 2020:12. doi: 10.3390/cancers12051344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naik N., Madani A., Esteva A., et al. Deep learning-enabled breast cancer hormonal receptor status determination from base-level H&E stains. Nat Commun. 2020;11:5727. doi: 10.1038/s41467-020-19334-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergoug M., Doudeau M., Godin F., Mosrin C., Vallee B., Benedetti H. Neurofibromin structure, functions and regulation. Cells. 2020:9. doi: 10.3390/cells9112365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takacs T., Kudlik G., Kurilla A., Szeder B., Buday L., Vas V. The effects of mutant Ras proteins on the cell signalome. Cancer Metastasis Rev. 2020;39:1051–1065. doi: 10.1007/s10555-020-09912-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutmann D.H., Geist R.T., Wright D.E., Snider W.D. Expression of the neurofibromatosis 1 (NF1) isoforms in developing and adult rat tissues. Cell Growth Differ. 1995;6:315–323. [PubMed] [Google Scholar]
- 10.Ratner N., Miller S.J. A RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor. Nat Rev Cancer. 2015;15:290–301. doi: 10.1038/nrc3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tao J., Sun D., Dong L., Zhu H., Hou H. Advancement in research and therapy of NF1 mutant malignant tumors. Cancer Cell Int. 2020;20:492. doi: 10.1186/s12935-020-01570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cawthon R.M., Weiss R., Xu G.F., et al. A major segment of the neurofibromatosis type 1 gene: cDNA sequence, genomic structure, and point mutations. Cell. 1990;62:193–201. doi: 10.1016/0092-8674(90)90253-b. [DOI] [PubMed] [Google Scholar]
- 13.Xu G., O’Connell P., Viskochil D., et al. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell. 1990;62:599–608. doi: 10.1016/0092-8674(90)90024-9. [DOI] [PubMed] [Google Scholar]
- 14.Bollag G., Clapp D.W., Shih S., et al. Loss of NF1 results in activation of the Ras signaling pathway and leads to aberrant growth in haematopoietic cells. Nat Genet. 1996;12:144–148. doi: 10.1038/ng0296-144. [DOI] [PubMed] [Google Scholar]
- 15.Yap Y.S., McPherson J.R., Ong C.K., et al. The NF1 gene revisited - from bench to bedside. Oncotarget. 2014;5:5873–5892. doi: 10.18632/oncotarget.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jett K., Friedman J.M. Clinical and genetic aspects of neurofibromatosis 1. Genet Med. 2010;12:1–11. doi: 10.1097/GIM.0b013e3181bf15e3. [DOI] [PubMed] [Google Scholar]
- 17.Philpott C., Tovell H., Frayling I.M., Cooper D.N., Upadhyaya M. The NF1 somatic mutational landscape in sporadic human cancers. Hum Genomics. 2017;11:13. doi: 10.1186/s40246-017-0109-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Humphries M.P., Maxwell P., Salto-Tellez M. QuPath: the global impact of an open source digital pathology system. Comput Struct Biotechnol J. 2021;19:852–859. doi: 10.1016/j.csbj.2021.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoltzfus C.R., Filipek J., Gern B.H., et al. CytoMAP: a spatial analysis toolbox reveals features of myeloid cell organization in lymphoid tissues. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bankhead P., Loughrey M.B., Fernandez J.A., et al. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7:16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt U., Weigert M., Broaddus C., Myers G. In: Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. Frangi A.F., Schnabel J.A., Davatzikos C., Alberola-López C., Fichtinger G., editors. Springer International Publishing; Cham: 2018. Cell detection with star-convex polygons; pp. 265–273. [Google Scholar]
- 22.Fedchenko N., Reifenrath J. Different approaches for interpretation and reporting of immunohistochemistry analysis results in the bone tissue - a review. Diagn Pathol. 2014;9:221. doi: 10.1186/s13000-014-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schizas C.N., Pattichis C.S. Learning systems in biosignal analysis. Biosystems. 1997;41:105–125. doi: 10.1016/s0303-2647(96)01668-1. [DOI] [PubMed] [Google Scholar]
- 24.Kingsford C., Salzberg S.L. What are decision trees? Nat Biotechnol. 2008;26:1011–1013. doi: 10.1038/nbt0908-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Breiman L. Random forests. Mach Learn. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 26.Tharwat A., Gaber T., Ibrahim A., Hassanien A.E. Linear discriminant analysis: a detailed tutorial. Ai Commun. 2017;30:169–190. doi: 10.3233/AIC-170729. [DOI] [Google Scholar]
- 27.Chen T., Guestrin C. 2016. XGBoost: A Scalable Tree Boosting System. [Google Scholar]
- 28.Zhang Z. Introduction to machine learning: k-nearest neighbors. Ann Transl Med. 2016;4:218. doi: 10.21037/atm.2016.03.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pal S.K., Mitra S. Multilayer perceptron, fuzzy sets, and classification. IEEE Trans Neural Netw. 1992;3:683–697. doi: 10.1109/72.159058. [DOI] [PubMed] [Google Scholar]
- 30.Webb G.I. In: Naïve Bayes. Encyclopedia of Machine Learning. Sammut C., Webb G.I., editors. Springer US; Boston, MA: 2010. pp. 713–714. [Google Scholar]
- 31.Pedregosa F., Varoquaux G., Gramfort A., et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 32.Lu C., Romo-Bucheli D., Wang X., et al. Nuclear shape and orientation features from H&E images predict survival in early-stage estrogen receptor-positive breast cancers. Lab Invest. 2018;98:1438–1448. doi: 10.1038/s41374-018-0095-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conde-Sousa E., Vale J., Feng M., et al. HEROHE challenge: predicting HER2 status in breast cancer from hematoxylin-eosin whole-slide imaging. J Imaging. 2022:8. doi: 10.3390/jimaging8080213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gertych A., Ing N., Ma Z., et al. Machine learning approaches to analyze histological images of tissues from radical prostatectomies. Comput Med Imaging Graph. 2015;46(Pt 2):197–208. doi: 10.1016/j.compmedimag.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material