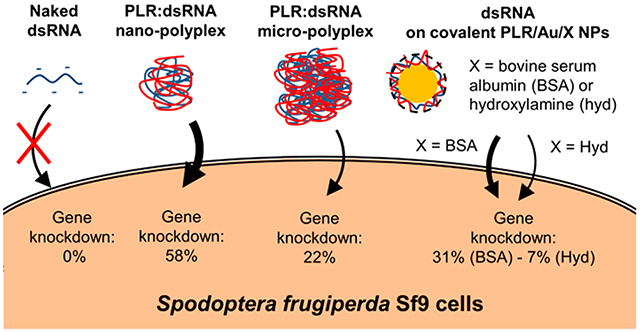
Abstract
This work focused on the delivery of dsRNA either complexed with poly-l-arginine (PLR-polyplex) or loaded onto poly-L-arginine functionalized 20 nm Au nanoparticles (PLR/Au NPs). RNA interference (RNAi) of a luciferase gene expressed in Spodopteria frugiperda Sf9 stable cell line (Sf9_LUC) was used as a model system. The binding affinity of dsRNA with two modes of functionalization of Au NPs was investigated: the electrostatic binding of PLR on citrate stabilized Au NPs (e-PLR/Au NPs) via the layer-by-layer method or the covalent-linking reaction of the polymer with NHS groups on the Au NPs surface (c-PLR/Au NPs) with excess groups quenched with either hydroxylamine (c-PLR/Au/Hyd NPs) or bovine serum albumin (c-PLR/Au/BSA NPs). The formation of PLR-polyplex particles resulting from the complexation of PLR and dsRNA were revealed by transmission electron microscope (TEM), scanning transmission electron microscope (STEM), and elemental mapping by energy dispersive X-ray spectroscopy (EDS). Luciferase gene knockdown was evaluated after exposure of Sf9 cells for 72 h to 600 ng of dsRNA. Gene knockdown (GK) was found to be more efficient by exposing Sf9 cells to nanoscaled (size <100 nm) PLR-polyplex (58% GK), in contrast to microscaled (size >1 μm) PLR-polyplex (20% GK) or covalent PLR/Au/Hyd (7% GK) particles. The replacement of hydroxylamine by bovine serum albumin as ligand has significantly enhanced the efficiency of GK (31% GK). Investigation of endosomal escape, a key physiological barrier for dsRNA delivery, with CypHer5E labeled dsRNA showed the good ability for the dsRNA conjugated to the different nanosystems to enter the cells compared to the unconjugated one. This study provides valuable information concerning the required properties of materials used for dsRNA delivery in lepidopteran cells such as nanoparticle size, stability in the cell culture media, the polymer molecular weight and binding strength to the nanoparticle, and the nature of ligands on the surface.
Keywords: RNA interference, Spodoptera frugiperda, dsRNA, delivery, Sf9 cells, poly-l-arginine, polyplexes, Au nanoparticles
Graphical Abstract
INTRODUCTION
Insect pests are responsible for the destruction of 10–16% of the world’s annual crop production, leading to billions of dollars of losses every year to the agriculture industry.1 Chemical pesticides are subject to insecticide resistance, public health issues, and potential environmental damage.2 There is need for new specific, affordable, efficient, and environmentally friendly alternatives. Among them, RNA interference technology (RNAi) has been considered as a potential strategy for pest control.3 RNAi, initially reported in plants,4 then later in the nematode Caenorhabditis elegans5 and mammalian cells,6 is a post-transcriptional cellular mechanism inducing gene silencing initiated by the introduction of double-stranded RNA (dsRNA) into cells.7 Considerable effort has been made on crop protection through triggering an RNAi response in insects.8 Although many studies have reported the silencing of a specific target gene leading to arrested development or death in insects,9,10 the RNAi efficiency differs among insect taxa (dsRNA triggers a highly efficient RNAi response in coleopterans but is ineffective in lepidopterans for example) because of different dsRNA pathways, processing mechanisms, stability, or physiological factors.11
Another key technological problem to solve for the widespread development of RNAi-based crop protection resides in the ability for dsRNAs to gain entry into cells, given that lipid bilayers allow only small (<1000 Da), neutral, and hydrophobic molecules to diffuse across them passively.12 Double-stranded RNA is a hydrophilic and highly negatively charged macromolecule that can be taken up by endocytosis but remains trapped inside of endosomes.13 Several strategies have been considered to overcome this barrier, such as the chemical modification of the oligonucleotide backbone,14,15 supramolecular assembly into polymer-based nanosystems (polyplexes),16 or covalent conjugation with transport vehicles.17 Since dsRNA’s charged phosphodiester linkage has been recalcitrant to chemical manipulation while maintaining proper biological activity and stability,18 noncovalent complexation with cationic polymers or adsorption into nanoparticles (liposomes, dendrimers, and inorganic nanoparticles) were found to be interesting alternatives for the delivery of medium-sized and large nucleic acid such as mRNA.19 Cationic polymers possess similar size and opposite charge for electrostatic interaction with nucleic acids but also bind to cell surfaces and trigger intracellular uptake.20 Among them, poly-l-arginine (PLR) has been used in several systems for the delivery of siRNA21,22 and was shown to enter cells efficiently.23 Further, poly-l-arginine is relatively inexpensive to produce at scale and is biodegradable making it suitable for agricultural applications. Inorganic particles, and in particular Au nanoparticles (Au NPs), are also promising vectors for drug and nucleic acid delivery applications due to their tunable size and surface properties, along with multifunctional capabilities.24 Another interesting feature relies on the possibility of the functionalization of Au NP surface and conjugation with nucleic acids in a noncovalent manner25,26 via a supramolecular assembly with positively charged Au surface or covalently via COOH-, S-, or NHS- groups grafted on the surface.27 While the application of Au NPs may be limited in agricultural settings, they provide an ideal model system by which particle properties can be systematically varied to determine the relationship between particle properties and efficacy.
This work aims to trigger RNAi activity in Sf9 cell line of the fall armyworm, Spodoptera frugiperda, a major pest species responsible for many billions of dollars of crop damage across North America, South America, Africa, and Asia.28 Sf9 cells are derived from IPLBSf21-AE, a well-established cell line isolated from Spodoptera frugiperda ovary tissue29 and were proved to be useful laboratory model for the development of materials for use in other insect pest species.30 The Sf9 cells were transfected with the luciferase gene construct (pIZT/V5-His-OpIE2:Luciferase),31 and stable cell line Sf9_LUC was selected and maintained in SF900 II serum-free medium (SF900 II SFM). We used dsRNA targeted to the luciferase gene (dsLUC), which is capable of inhibiting the luciferase protein expression in Sf9 cells and dsRNA targeted to a green fluorescent protein (GFP) gene were used as a negative control. Herein, we analyzed the data obtained with two delivery modes of dsRNA (PLR-polyplex versus dsRNA conjugated on PLR-functionalized inorganic Au particles) in terms of (i) their capacity to bind dsRNA, (ii) stability in SF900 II SFM cell culture medium, (iii) endosomal escape, and (iv) knockdown of LUC expression. The delivery of dsRNA on PLR functionalized 20 nm Au NPs was investigated with PLR either electrostatically (e-PLR/Au NPs) or covalently (c-PLR/Au NPs) bound to the Au surface.
The results presented here are of high significance toward the development of an effective and low-toxicity poly-l-arginine based delivery system able to bind dsRNA, deliver it to the cytoplasm of insect cells, and affect gene knockdown.
MATERIALS AND METHODS
Au NP Functionalization and Characterization.
Chemicals and Au NPs.
Monodisperse poly-l-arginine having molar masses of 9.6 kDa (PLR10) or 38.5 kDa (PLR40) were purchased from Alamanda polymers (Huntsville, AL, U.S.A.). PLR stock solution (5 mg mL−1) was prepared by dissolving 5 mg of poly-l-arginine (10 and 40 kDa) in 1 mL of DI water and then sonicating for 15 min in a bath sonicator before use. Commercially available 20 nm spherical citrate (aqueous suspension) and NHS ester functionalized Au NPs (lyophilized powder) were purchased from Nanopartz Inc. (Loveland, CO, U.S.A.). All chemicals were used without any further purification.
Au NP Concentration.
The concentration of Au NP was determined by measuring the optical density at 520 nm (OD520). An OD520 of 1 corresponds to a concentration of 56.6 μg mL−1 as verified by ICP-MS and indicated on the certificate of analysis provided by the manufacturer. The UV–vis spectra were collected on a Varian Carry Bio 50 (Agilent Technologies, Santa Clarita, CA, U.S.A.), by dropping 1 μL of sample on a tray cell (Hellma, Plainville, NY, USA; optical path = 1 mm). Samples were always diluted to an OD < 1 prior to analysis. The linearity of the calibration curve over OD520 = 0 to 1 had an r2 of 0.99. The mean relative percent difference (RPD) between replicates was 7 ± 8% (n = 5) and a method detection limit of 7 μg mL−1
Electrostatic Functionalization of 20 nm Citrate-Au NPs with Poly-l-arginine (e-PLR/Au NPs).
This approach used layer-by-layer assembly to functionalize citrate-coated Au NPs.32 A 200 μL of Citrate-Au NPs suspension (OD520 = 10) was added vigorously to 4 mL of a 100 μg mL−1 PLR10 aqueous solution. The suspension was then mixed 2 h at room temperature with an end-to-end sample rotator. After incubation, the reaction mixture was centrifuged at 5510g for 15 min. The supernatant was discarded and the pellet (e-PLR10/Au NPs) washed once (1×) or twice (2×) with deionized water (DI). The purified pellet was resuspended in DI water for further study.
Covalent Binding of Poly-l-arginine on NHS-Au NPs Surface (c-PLR/Au NPs).
The protocol for the conjugation of NHS-Au NPs with PLR10 was adapted from the Nanopartz, Inc. technical instructions for handling and use. Nanopartz NHS-Au NPs were redispersed in 5 mM potassium phosphate buffer (pH 7.4) then sonicated for 5 min at 50% amplitude with a Misonix S-4000 cup-horn sonicator (Newton, CT, U.S.A.) to ensure good dispersion of the powder. Then, 200 μL of an OD520 = 10 suspension was added to 4 mL potassium phosphate buffer (pH 7.4) containing 80 μL of PLR10 stock solution (PLR10 concentration: 100 μg mL−1). The solution was then mixed for 1 h on an end-to-end sample rotator. Ten microliters of 0.1% BSA protein (c-PLR10/Au/BSA NPs) or 5 μL of 5% w/v hydroxylamine (c-PLR10/Au/Hyd NPs) were then added to quench the unreacted NHS groups. The suspension of NPs was then centrifuged at 5510g for 1 h. The pellet was collected and resuspended in DI water.
Hydrodynamic Diameter and Charge of the Nanoparticles.
Mean hydrodynamic diameters (intensity-weighted average) and zeta potentials of the NP suspensions were measured in DI water or SF900 II SFM medium with a Zetasizer NanoZS (Malvern Instruments, Malvern, U.K.) using dynamic light scattering (DLS) or phase analysis light scattering (PALS), respectively. The Hückel approximation was used to estimate zeta potential from electrophoretic mobility. The nanoparticle suspensions were diluted down to a concentration of 10 μg mL−1 in DI water or in SF900 II SFM medium (Thermo Fisher Scientific, U.S.A.). We periodically analyze a polystyrene/latex standard reference material provided by Malvern to ensure the accuracy of PALS/DLS measurements. Additionally, we followed manufacturers recommended quality control included in the software package which evaluates goodness of fit of correlograms. The software also automatically adjusts the number of replicates to achieve the required precision.
Electron Microscopy.
Nanoparticle size and morphology were examined by transmission electron microscopy (TEM) and scanning transmission electron microscopy (STEM) using a Talos F200X analytical electron microscope (Thermo Scientific, Waltham, MA, U.S.A.), operated at 200 keV and equipped with a four-element silicon drift detector (SDD)-based EDS system for quantitative chemical composition analysis and elemental distribution mapping. Sample preparation was performed by dipping lacey carbon grids (Ted Pella, Redding, CA, U.S.A.) in 50 μg mL−1 suspensions of nanoparticles.
dsRNA Binding and Luciferase Assay.
Synthesis of dsRNA.
The 348 bp fragment of the gene coding for green fluorescence protein (GFP, negative control) and 355 bp fragment of the luciferase were PCR amplified using L4440-GFP RNAi vector DNA and pIZT/V5:Luciferase gene construct and gene-specific primers containing T7 promoter sequence on the 5′ end of each primer. The PCR products were purified by QIAquick PCR Purification kit (Qiagen, U.S.A.) and analyzed by NanoDrop 2000 Spectrophotometer (ThermoFisher Scientific, Waltham, MA, U.S.A.) and 1% agarose gel electrophoresis. The MEGAscript T7 Transcription Kit (Invitrogen, ThermoFisher Scientific, U.S.A.) was used to synthesize the dsRNA. A total of 2 μL of each 10X Buffer, ATP, GTP, CTP, UTP, and 500 ng of each T7-GFP and T7-Luciferase PCR product and 2.0 μL of T7 enzyme were mixed in 1.5 mL microcentrifuge tube and incubated at 37 °C for 16 h. The input DNA was digested using 1.0 μL of TURBO DNase (Invitrogen, ThermoFisher Scientific, USA) for 30 min at 37 °C followed by heat inactivation at 70 °C for 10 min. The dsRNA products were precipitated with 1/10th volume of 0.3 M sodium acetate solution (pH 5.2) and 3 volumes of ice-cold 100% ethanol and kept at −20 °C for 1 h and then centrifuged at 13 450g for 15 min at 4 °C followed by washing with 750 μL of 70% ethanol. The washed dsRNA pellets were dried in the Vacufuge plus (Eppendorf, Germany) at 30 °C for 10 min. The pellet was then resuspended in 25 μL of nuclease free water and the quantity and quality of the product were measured using NanoDrop 2000 Spectrophotometer and 1% agarose gel electrophoresis, respectively as described previously.26,11
Poly-l-arginine:dsRNA Complexation Assay.
Isotherms for complexation of poly-l-arginine with dsRNA (dsGFP or dsLUC) were determined by adding dropwise 100 μL of a 50 μg mL−1 solution of dsRNA (5 μg) to 100 μL solutions containing various amount of PLR (PLR:dsRNA mass ratios: 1:50; 1:5; 1:2; 1:1, and 2:1). After incubation for 30 min at 25 °C, solutions were centrifuged at 16 873g for 1 h. The supernatant was separated from the pellet, which contained the PLR-polyplex. The dsRNA concentration in the supernatant was measured using a NanoDrop 2000 Spectrophotometer to determine the amount of unbound dsRNA, while the hydrodynamic diameter and zeta potential of the resuspended pellets were measured by DLS and PALS, respectively. The linearity of the calibration curve of this instrument of the dsRNA concentration of 0–50 μg mL−1 was excellent with an r2 of 1.00. The RPD was 11 ± 11% and the detection limit was 1.35 μg mL−1.
dsRNA Binding Affinity Assays with e- or c-PLR/Au NPs.
Binding of dsRNA with the different PLR10/Au NPs was investigated by adding 1 μg of dsRNA (50 μg mL−1 solution) dropwise to solutions containing various amount of PLR10/Au NPs (NP:dsRNA mass ratios: 1:1, 3:1, 5:1, and 7:1). The solutions were incubated for 30 min at room temperature then centrifuged at 5510g for 30 min. The supernatants were collected and the pellets washed then resuspended in DI water or SF900 II SFM medium for characterization.
Agarose Gel Electrophoresis.
Agarose gel electrophoresis (1% agarose) was performed on the collected supernatants and resuspended pellets in water to verify the binding of dsRNA. TBE buffer was used with electrolyte and the gels were run for 30 min at constant 100 V with variable current. The dsRNA was visualized under UV light after staining with ethidium bromide.
Luciferase Protein Assay.
A stable Sf9_LUC cell line was used for RNAi investigation by estimating the luciferase protein level. A dsRNA sequence targeting the genes coding for luciferase (dsLUC) and green fluorescent protein (dsGFP) were used as target and control, respectively. A 3 × 104 Sf9_LUC stable cells/well were seeded in a 96-well plate and incubated overnight at 27 °C. A total of 600 ng of naked dsRNA (dsGFP and dsLUC), complexed with PLR-polyplex or conjugated onto e-PLR10/Au, c-PLR10/Au/BSA and c-PLR10/Au/Hyd NPs were dispersed in 100 μL of SF900 II SFM medium before exposure to cells. After 72 h incubation at 27 °C, the cells were washed with 100 μL of 1X PBS and lysed with 100 μL of lysis buffer for 10 min. Then 10 μL of cell lysate were used for measuring the luciferase activity using a SpectraMax i3x multimode plate reader (Molecular Devices, Sunnyvale, CA, U.S.A.). Luciferase activity was normalized to total protein concentration determined using Bradford’s assay33 and expressed as relative luminescence units (RLU/mg protein).
Endosomal Escape.
Labeling of dsRNA with CypHer5E Fluorescent Dye.
The labeling of dsRNA with a pH-sensitive fluorescent cyanine dye (CypHer5E) was carried out as previously described.34 Aminoallyl dsRNA was synthesized by in vitro transcription using MEGAscript T7 kit (Ambion, U.S.A.) according to the manufacturer’s instructions except that UTP was replaced with amino-allyl UTP (Thermo Fisher Scientific, U.S.A.). A 348 bp T7-GFP PCR product purified using Qiagen PCR purification columns followed by synthesized dsRNA with amino-allyl UTP. The purified dsRNA was conjugated with a pH-sensitive cyanine dye, CypHer5E mono-NHS ester (GE Healthcare, U.K.) with an excitation and emission wavelength of 644 and 663 nm, respectively. One vial of CypHer5E dye (1 mg) was dissolved in 40 mL of DMSO and divided into aliquots of 2 μL in Eppendorf tubes. The dye was dried using a freeze-dryer (Labconco Freezone 6) and stored at 40 °C until use. For conjugation of dye to dsRNA, one vial of dye (~50 mg) was resuspended in a mixture containing 2 μg of aminoallyl UTP-labeled dsRNA (in 3.33 μL H2O), 5 μL of DMSO, and 1.66 μL of 0.3 M sodium bicarbonate buffer, pH 9.0. The conjugation reaction was carried in the dark for 1 h at room temperature. The conjugated dsRNA was repurified using Qiagen PCR purification columns and eluted in DEPC-treated water.
Internalization and Endosomal Escape Study of CypHer5E Labeled dsRNA in Sf9 Cells.
Since CypHer5E has a fluorescence maximum around pH 5 and almost no fluorescence around pH 7, it can indicate localization within the late endosomes.35 One day prior to the experiment, 1.2 × 105 cells/well were seeded in eight well chamber microscope slides (NuncTM Lab-TekTM II, ThermoFisher Scientific, Waltham, MA). The next day, 25 ng of CypHer5E-dsGFP, either naked or conjugated with PLR10 (PLR:dsRNA mass ratio of 1:1), c-PLR10/Au/BSA and c-PLR10/Au/Hyd NPs (NP:dsRNA mass ratio of 5:1) and washed one time with DI water to remove free CypHer5E-dsGFP were mixed with 100 μL of SF900 II SFM medium then added to cells. At 4 h after exposure, the cells were washed twice with 1X phosphate-buffered saline (PBS), fixed with 100 μL of 4% paraformaldehyde for 20 min at room temperature and stained with EverBrite mounting medium (Biotium, Fremont, CA) containing 4′,6-diamidino-2-phenylindole (DAPI). The cells were visualized with Leica SP8 confocal microscope at 63× magnification by overlapping DAPI (Blue; for nuclei), Alexa 633 (Red; for CypHer5E labeled dsRNA), and bright-field (BF) filters images.
Statistical Analysis.
The data from the in vitro study were analyzed using one-way ANOVA with Tukey’s posthoc multiple comparisons at a significance level of α = 0.05 (GraphPad Prism v.5 Software, Inc., U.S.A.). The data are presented as mean ± standard error (n = 6). Error bars represent the standard error of the mean of six biological replicates. Luciferase activity in dsLUC treatments was compared to control cells treated with dsGFP.
RESULTS AND DISCUSSION
RNAi in Sf9 Cells Using Poly-l-arginine Polyplexes.
Particle Characterization.
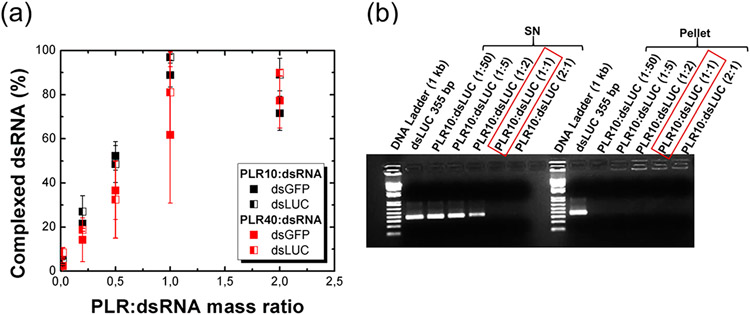
The percentage of complexed dsRNA with poly-l-arginine as a function of the PLR:dsRNA mass ratio (Figure 1a) showed the progressive increase of complexed dsRNA with poly-l-arginine when increasing the amount of polymer. In the case of PLR10, the absorbance begins to increase again after reaching the maximum of complexation (respectively 88.7 ± 5.4% and 96.9 ± 1.3% of complexed dsRNA, mean ± standard deviation, for dsGFP and dsLUC at a PLR10:dsRNA mass ratio of 1:1). The decrease of the amount of complexed dsRNA (71.5 ± 7.8% and 89.0 ± 7.4% of complexed dsRNA for dsGFP and dsLUC at ratio 2:1) indicates that the complexation reaction is highly sensitive to the stoichiometry between the biomolecule and the polymer. Although the complexation rate appeared lower in the case of PLR40 with a maximum complexation obtained for a PLR40:dsRNA mass ratio of 2:1 (77.2 ± 12.4% and 89.9 ± 1.4% complexed dsRNA for dsGFP and dsLUC), the conversion into molar ratio gives an equilibrium point reached for a PLR:dsRNA molar ratio of 1:24 and 1:3 for PLR10 and PLR40, respectively. The dsLUC molecule possesses a slightly higher affinity for PLR than dsGFP, although the experimental slopes remain very close to each other. Around 96% of dsLUC is complexed with PLR10 compared to 88% of dsGFP at a mass ratio of 1:1. As GFP and LUC double-stranded RNAs possess almost the same number of base pairs (around 350 bp), this slight difference in reactivity could be explained by their sequences. To confirm the absorbance based method, 1% agarose gel electrophoresis was performed on both supernatants (SN) and the particles pellets issued from centrifugation and redispersed in DI water at different mass ratios for PLR10:dsLUC polyplex (Figure 1b). As observed for the absorbance quantification method, the amount of dsRNA found in the supernatant progressively decreases when increasing the PLR:dsRNA mass ratio (dsRNA totally absent in the supernatant at a mass ratio of 1) while no free dsRNA is observed in the pellets.
Figure 1.
Complexation of dsRNA (5 μg) of similar base pair numbers (dsGFP: 348 bp and dsLUC: 355 bp) with poly-l-arginine (PLR) of different molecular weights (PLR10:10 kDa and PLR40:40 kDa). The complexation reaction titration as a function of the PLR:dsRNA mass ratio was determined according to (a) the presence of free dsRNA in the supernatant determined by the absorbance at 260 nm and (b) 1% agarose gels performed on supernatants (SN) and pellets in the case of PLR10:dsLUC polyplexes. Error bars were determined from the standard deviation of duplicate experiments.
The pellets resulting from two duplicate experiments (noted as Exp 1 and Exp 2) formed at a PLR:dsRNA mass ratio of 1 were redispersed in DI water or in SF900 II SFM medium. Table 1 presents the intensity-weighted hydrodynamic diameter and zeta potential value of the so-formed precipitate obtained in the case of PLR10 and PLR40 polymers complexed with dsGFP and dsLUC. DLS measurements for Exp 1 showed the presence of particles with hydrodynamic diameters in the hundreds of nanometers for both dsRNAs and PLR molecular weights, whereas particles found in Exp 2 are in the micrometer range. PLR-polyplexes from Exp 1 and Exp 2 present net positive zeta potentials value (in the range 5–30 mV) in DI water. After dispersion in SF900 II SFM medium, however, the zeta potentials became negative and reach a value of around −20 mV for each PLR-polyplex. The SF900 II SFM medium is a serum-free medium, but it contains a proprietary protein substitute. It is likely that this protein substitute formed a corona on the particles conferring a net negative charge. It has been observed previously that when particles with a wide range of surface chemistries are dispersed in serum, they have a consistent negative zeta potential.36 In this medium, the particles possess a narrower hydrodynamic diameter distribution than in DI water, showing good stability and dispersion of the so-formed PLR-polyplex nanoparticles in the culture cell medium.
Table 1.
Intensity-Weighted Hydrodynamic Diameter (Z-Average Diameter) and Zeta Potential in DI Water and in SF900 II SFM Media of the Polyplex Particles Formed by the Reaction of dsRNA with 10 kDa (PLR10) and 40 kDa (PLR40) Poly-l-arginine during Two Duplicate Experiments Labelled Exp 1 and Exp 2 with a PLR:dsRNA Mass Ratio of 1:1
| DI water (pH 5.9) |
SF900 II SFM(pH 7.4) |
||||
|---|---|---|---|---|---|
| duplicate experiment | samples | Z-average diameter (nm) | zeta potential (mV) | Z-average diameter (nm) | zeta potential (mV) |
| Exp 1 | PLR10:dsGFP | 480 ± 60 | +10.5 ± 4.6 | 201 ± 84 | −22.5 |
| PLR10:dsLUC | 270 ± 36 | +30.8 ± 5.2 | 213 ± 95 | −19.5 | |
| PLR40:dsGFP | 188 ± 65 | +34 ± 9.2 | 189 ± 65 | −22.8 | |
| PLR40:dsLUC | 420 ± 120 | +6 ± 7 | 205 ± 40 | −22.9 | |
| Exp 2 | PLR10:dsGFP | 1022 ± 260 | +5.9 ± 7.0 | - | - |
| PLR10:dsLUC | 1077 ± 224 | +21.8 ± 4.8 | - | - | |
| PLR40:dsGFP | 2228 ± 530 | +31.3 ± 6.4 | - | - | |
| PLR40:dsLUC | 1935 ± 350 | +75 ± 8.8 | - | - | |
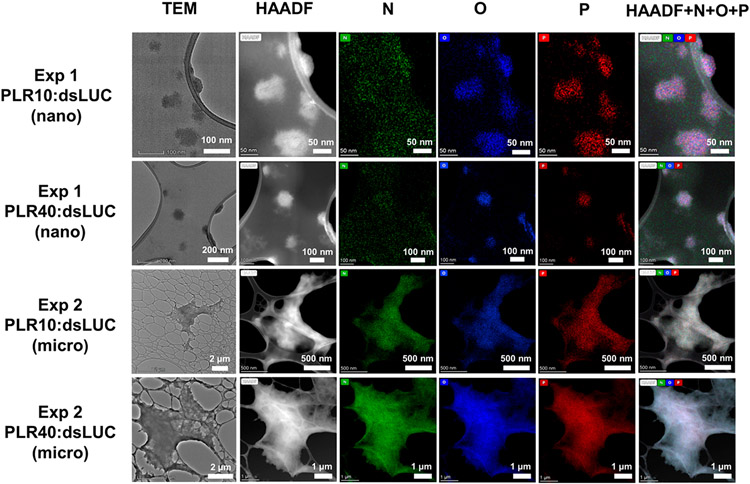
Transmission electron microscopy (TEM) and high angle annular dark-field (HAADF) images of the resuspended pellets in the case of PLR-polyplexes are presented in Figure 2. Exp 1 particles had spherical shapes with sizes ranging from 50 to 100 nm for PLR10 and PLR40 polymer, respectively. Conversely, particles from Exp 2 had an irregular morphology and diameters of several micrometers. HAADF imaging and elemental mapping by energy dispersive X-ray spectroscopy (EDS) were carried out to look at the elements composing the polyplexes. The presence of elements in the targeted particle such as O, N and particularly P, marker of the presence of dsRNA, nicely confirmed that these particles are the products of complexation between poly-l-arginine and dsRNA.
Figure 2.
Transmission electron microscopy (TEM), high angle annular dark-field (HAADF) images, and elemental mapping (N, O, P) by energy dispersive X-ray spectroscopy (EDS) of polyplexes formed by the reaction of dsLUC with poly-l-arginine (PLR10:10 kDa and PLR40:40 kDa) during experiments one (Exp 1, nanoparticles) and two (Exp 2, microparticles) with a PLR:dsRNA mass ratio of 1:1.
RNAi Activity of the Particles.
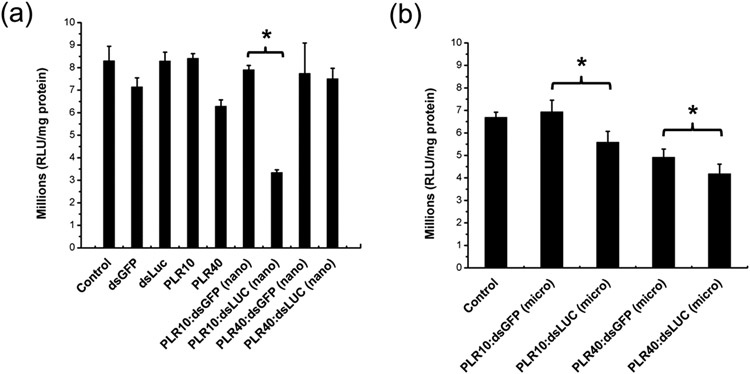
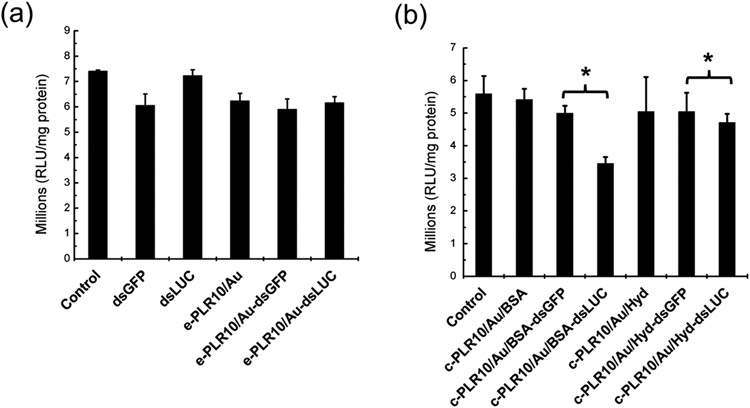
A statistically significant reduction of the luciferase activity (up to 58%) was observed in the case of nanometer-sized PLR10:dsLUC particles obtained during Exp 1 (Figure 3a) as compared with the PLR10:dsGFP used as a negative control. However, only a 20% reduction of luciferase activity was observed in the case of PLR10:dsLUC in Exp 2, where the particles were of micrometer dimensions (Figure 3b). In an attempt to improve the knockdown of the luciferase gene, we increased the dose of micropolyplexes up to 3 μg, but contrary to our expectation, a reduction in the decrease in luciferase activity from 22% down to 15% was observed (see Figure S1). Analysis of the cell phenotype (Figure S2) showed the formation of dark spots at a higher concentration that could be possibly attributed to aggregated particles settling onto the cells. However, the PLR10:dsRNA polyplex presents no visual abnormality in the cells contrary to free PLR10 polymer showing abnormal cells at a concentration of 30 μg mL−1. The PLR40:dsLUC sample (Exp 1) had a decrease in LUC activity of only 3.1% compared to the PLR40:dsGFP control (Figure 3a) and 15% in the case of Exp 2 (Figure 3b) with less healthy cells, indicating a lack of efficacy for higher molecular weight PLR.
Figure 3.
Luciferase assay in Sf9_LUC stable cells without exposure (control) or by exposing the cells to 600 ng of naked dsRNA (dsLUC, dsGFP), free poly-l-arginine (PLR10:10 kDa and PLR40:40 kDa) or dsRNA complexed with poly-l-arginine (1:1 PLR:dsRNA mass ratio) in the case of (a) size-controlled (nano) and (b) aggregated (micro) polyplexes particles issued from Exp 1 and Exp 2. Data analyzed using one-way ANOVA with Tukey’s posthoc multiple comparisons. (*) Significantly different at α = 0.05. Error bars determined from the standard error of the mean of six biological replicates.
This result suggests the importance of the careful control of the nanoparticle size to obtain an effective gene knockdown. Complexation can lead to the formation of nanoparticles if the precipitation is done in a controlled fashion. Duplicate experiments (Exp 1 and Exp 2) were performed under the same conditions (dsRNA and PLR concentrations, incubation time, centrifugation speed) but led to the formation of two different particle sizes, highlighting the sensitivity to a minor variable that was not controlled for, such as room temperature or mixing rate. The polyplex formation process is an electrostatic interaction between the polyanionic nucleic acid and the multivalent cationic polymers.37 This reaction is able to produce nanoparticles containing the nucleic acid in compacted and protected form as observed with PLR-polyplex particles such as observed in Exp 1. It appears that the polyplex size is critical. It is possible that the larger micron-sized particles have difficulty entering the cells. We also found that higher molecular weight poly-l-arginine caused more toxicity in cells than the lower molecular weight poly-l-arginine. It is also important to note that we saw a decrease in LUC activity for PLR40:dsGFP particles, perhaps indicating cellular toxicity, which decreased LUC expression. Condensation of the dsRNA is difficult because of the high rigidity of the short double-helical structures38 resulting in the lack of cooperativity in the polyplex structure.39 This cooperativity can be enhanced by the use of a cross-linker. Shen et al.40 have successively complexed low molecular weight poly-l-lysine with siRNA forming nanopolyplexes by using epigallo-catechin-3-O-gallate (EGCG) as cross-linker precomplexing the siRNA. An alternative is to prefunctionalize the polymer on a size-controlled inorganic core as Au NPs.
RNAi in Sf9 Cells Using PLR10 Functionalized Au NP.
Particle Characterization.
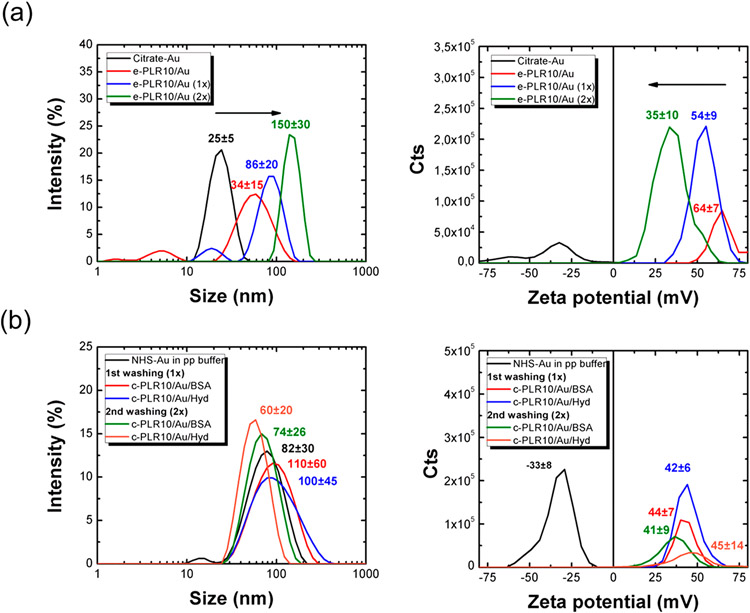
Figure 4 shows the evolution of the Z-average hydrodynamic diameter and zeta potential (Figure 4a) of the e-PLR10/Au NPs before and after incubation with PLR10 and washing steps with DI water. The hydrodynamic diameter of the nanoparticles increases during the process, starting from 25 ± 5 nm before functionalization to 35 ± 15 nm after the addition of the polymer then 86 ± 18 and 150 ± 30 nm after the first and second washing step with water, respectively. A negative zeta potential (ξ = −27 ± 10 mV) was observed for the citrate stabilized Au NPs. These negatively charged particles became highly positively charged (ξ = 64 ± 7 mV) after the addition of PLR10, suggesting the good grafting of the polycationic polymer on the surface. A progressive decrease of the zeta potential is noticed over the washings steps, finally stabilizing at a ξ value of 35 ± 10 mV, suggesting that the PLR on the surface of the particles is labile. The adsorption of PLR10 on the surface was confirmed by UV–Vis spectroscopy (Figure S3). A characteristic ligand-to-ligand charge transfer (LLCT) band is observed at 220 nm in the UV–Vis spectrum of the PLR10 functionalized Au NPs (washed 1× and 2×) in addition to the surface plasmon resonance band centered at 525 nm. The surface plasmon band of the Au NPs is progressively enlarged and shifted toward higher wavelengths after PLR10 addition and washing steps. This can be due to particle aggregation.
Figure 4.
Size and zeta potential before/after incubation with poly-l-arginine 10 kDa (PLR10) then washing with DI water (once (1×) or twice (2×)) of (a) the electrostatically stabilized PLR10 on citrate-Au NPs (e-PLR10/Au) and (b) the covalently stabilized PLR10 on n-hydroxysuccinimide NHS-Au NPs (c-PLR10/Au). For the c-PLR10/Au NPs, the unreacted NHS groups were quenched with 0.1% bovine serum albumin (c-PLR10/Au/BSA NPs) or 5% w/v hydroxylamine (c-PLR10/Au/Hyd NPs) after 30 min of incubation.
To decrease the lability of PLR10 on the Au NPs surface, we covalently bound the PLR10 using an n-hydroxysuccinimide (NHS) ester linkage. Figure S4 shows the evolution of the hydrodynamic diameter and zeta potential of the particles as a function of the incubation time. Before quenching, the size of the particles remains stable over time, with a hydrodynamic diameter between 40 and 50 nm. The initial negatively charged NHS-Au NPs (ξ = −35 mV) became positively charged right after the addition of PLR10 (ξ = +40 mV) before stabilizing at a zeta potential of around +50 mV after 30 min of reaction. The aggregation of the particles was observed after 2 h of incubation. Thus, the reaction was quenched after 30 min of reaction with hydroxylamine (Hyd) or bovine serum albumin (BSA) and the particles were washed once or twice with DI water to remove unreacted or unwanted components (polymer, quencher, or phosphate buffer). The hydrodynamic diameter and zeta potential after the different washing steps are presented in Figure 4b. Contrary to the e-PLR10/Au NPs, the hydrodynamic diameter and zeta potential values of the different c-PLR/Au NPs (quenched with Hyd or BSA) were found to be highly stable over the DI water washing step with sizes of around 60 and 100 nm for c-PLR10/Au/Hyd and c-PLR10/Au/BSA NPs, respectively. The hydrodynamic diameter is higher in the case of BSA quenched particles and can be easily explained by the intrinsic hydrodynamic diameter of the quencher. The zeta potential of the particles remains positive and constant during the washing step, with narrowing-ranged values between +40 and +45 mV.
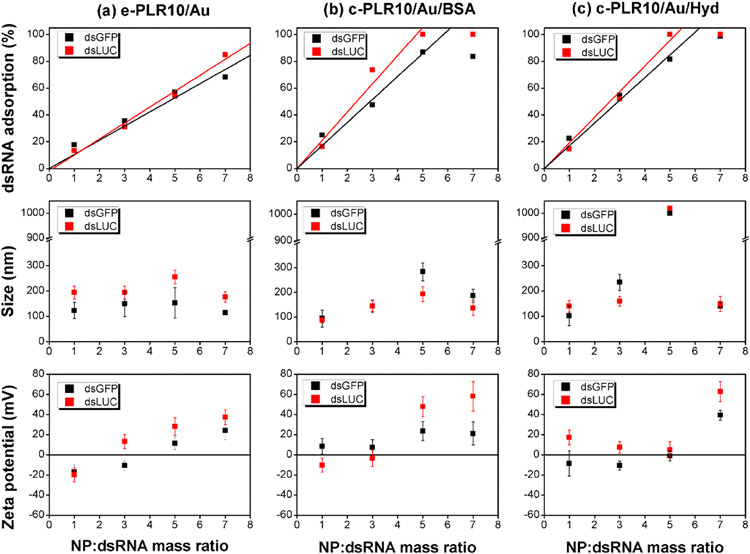
Figure 5 summarizes the results obtained for the different Au NPs functionalized with PLR10. The adsorption of the dsRNA on the PLR10/Au NPs as a function of the NP:dsRNA mass ratio follows a linear increase for the three types of nanoparticles investigated before reaching a plateau at a higher mass ratio when the maximum of adsorption is reached. For all PLR10/Au NPs, we can observe better binding for dsLUC compared to dsGFP as observed with PLR-polyplexes. By comparison of the different slopes, c-PLR10/Au/BSA (1×) NPs possess a slightly better dsLUC affinity than c-PLR10/ Au/Hyd (1×) NPs with 100% of dsLUC (1 μg) adsorbed at a NP:dsRNA mass ratio of 4.8 (0.21 μg adsorbed dsLUC/μg Au NP) versus a NP:dsRNA mass ratio of 5.2 for c-PLR10/Au/Hyd (1×; 0.19 μg adsorbed dsLUC/μg NP). The adsorption observed for the c-PLR10/Au was better than those observed for the e-PLR10/Au NPs where 100% of dsLUC was found to be absorbed at a NP:dsRNA mass ratio of 8.4 (equivalent to 0.12 μg adsorbed dsLUC/μg Au NP) by linear regression. These results were confirmed by agarose gel electrophoresis performed on the supernatants and the resuspended pellets (Figure S5 in the Supporting Information). In these experiments, the presence of free dsRNA progressively vanished when increasing the NP:dsRNA ratio. In DI water, the evolution of the hydrodynamic diameter and zeta potential as a function of the NP:dsRNA mass ratio is directly correlated to the adsorption of dsRNA on the nanoparticles. After addition of dsRNA, the hydrodynamic diameter of the nanoparticles increased up to 200 nm and the zeta potential turned negative then became progressively positive again when increasing the NP:dsRNA ratio until reaching their initial value above the maximum of adsorption. Aggregation of the c-PLR10/Au/Hyd NPs was observed at a ratio of 5:1 that was directly associated with the 0 zeta potential value and the neutralization of the charges at this ratio.
Figure 5.
dsRNA adsorption (1 μg) on the different PLR10/Au NPs as a function of the NP:dsRNA mass ratio, and resulting nanoparticles size and zeta potential after redispersion in DI water for: (a) e-PLR10/Au, (b) c-PLR10/Au/BSA, and (c) c-PLR10/Au/Hyd NPs (e: electrostatic PLR10/Au binding, c: covalent binding).
Because of the loose immobilization of the polymer with the layer-by-layer method, the e-PLR10/Au NPs were not stable in SF900 II SFM media contrary to the c-PLR10/Au-dsRNA NPs, with hydrodynamic diameters very close to or smaller than those observed in DI water.
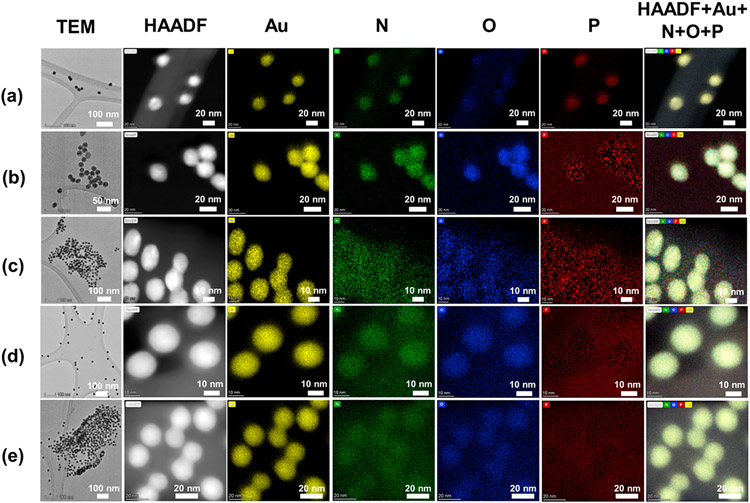
The morphology and elemental composition of the different Au NPs were analyzed by electron microscopy. Figure 6 presents TEM and HAADF images as well as elemental mapping (Au, N, O, and P) of the citrate- and NHS-Au NP before and after functionalization with PLR10 and loading with dsLUC (NP:dsRNA mass ratio of 5:1). All citrate- and NHS-Au NPs suspensions showed the presence of spherical shaped nanoparticles of 15–20 nm of diameter as expected. Before functionalization (Figure 6a), the citrate-Au are mainly composed of Au but some amount of O and N can be found on the surface. Presence of S and P is also found in the case of the citrate-Au NPs before functionalization and loading with dsRNA due to the strong overlapping between the peaks of S and P with the one of Au (see Figures S6 and S7 for EDS spectra of citrate-Au and NHS-Au NPs, respectively). The contrast of the O and N is, however, more enhanced after functionalization (Figure 6b) and could be attributed to the presence of PLR10 surrounding the Au core. This observation confirms the good and homogeneous grafting via the layer-by-layer method. After loading with dsLUC (NP:dsRNA mass ratio of 5:1), a halo of P surrounds the different Au NPs suggesting the presence of dsRNA anchored to the Au NPs surface.
Figure 6.
Transmission electron microscopy (TEM), high angle annular dark-field (HAADF) images and elemental mapping (N, O, and P) by energy dispersive X-ray spectroscopy (EDS) of the citrate Au NPs before (a) and after (b) the electrostatic functionalization with poly-l-arginine 10 kDa then (c) loaded with dsLUC (NP:dsRNA mass ratio of 5:1). TEM, HAADF, and elemental mapping of the covalently functionalized Au NPs: (d) c-PLR10/Au/BSA and (e) c-PLR10/Au/Hyd NPs loaded with dsLUC (NP:dsRNA mass ratio of 5:1).
RNAi Activity of the Particles.
A NP:dsRNA ratio of 5:1 (or 5.5 for the c-PLR10/Au/Hyd NPs to avoid their aggregation due to neutral zeta potential) was chosen for all NPs to have a maximum adsorption while sustaining a good nanoparticle size distribution and positive zeta potential to maximize their entrance in the cells. Due to their aggregation and instability in SF900 II SFM medium, no knockdown of the e-PLR10/Au-dsLUC sample was observed compared with e-PLR10/Au-dsGFP exposed cells (Figure 7a). Stability of the particles in extracellular media is thus an important feature as the interaction with electrolytes, proteins, or cellular surfaces can cause partial or complete dissociation and, as observed for citrate-Au NPs, loss of delivery efficacy and irreversible aggregations.41 This stability issue was overcome with covalently binding PLR10 to the Au NPs. As for the c-PLR10/Au NPs (Figure 7b), c-PLR10/Au/BSA-dsLUC and c-PLR10/Au/Hyd-dsLUC exposed cells had 31% and 6.7% knockdown of the luciferase gene, respectively. At comparable particle size after dsRNA loading, the gene knockdown for dsRNA complexed with PLR covalently functionalized on Au NPs (GK < 31%) is less effective than in the case of the condensed form of PLR-polyplexes nanoparticles (GK 58%). According to the literature, the successful gene delivery required the high compaction and condensation of the DNA/RNA with the nonviral vector.42 ,43 PLR on Au-NPs possess a lower degree of liberty and higher structure rigidity for dsRNA compaction and condensation as compared to the more important number of binding sites available with free PLR.
Figure 7.
Luciferase activity in Sf9_LUC stable cell line without (control) and after exposure to 600 ng of naked dsGFP and dsLUC then conjugated to poly-l-arginine 10 kDa (PLR10) functionalized Au NPs at a NP:dsRNA mass ratio of 5:1 in the case of (a) e-PLR10/Au NPs (e: electrostatic binding) and (b) c-PLR10/Au NPs (c: covalent binding) quenched with BSA (c-PLR10/Au/BSA) or hydroxylamine (c-PLR10/Au/Hyd). Cells were exposed to the pellets redispersed in SF900 II SFM media prior to exposure. Data analyzed using one-way ANOVA with Tukey’s posthoc multiple comparisons. (*) Significantly different at α = 0.05. Error bars were determined from the standard error of the mean of six biological replicates.
The particle uptake and release of dsRNA to the cytoplasm is also an important consideration. We found that when we quenched the unreacted NHS groups with BSA, they caused greater knockdown than particles quenched with hydroxylamine. It is also possible that the BSA-quenched particles had greater uptake due the presence of BSA molecules on the surface. A previous study showed that BSA-coated Au NPs had greater uptake in human cells than citrate coated particles for example.44-46
Endosomal Escape.
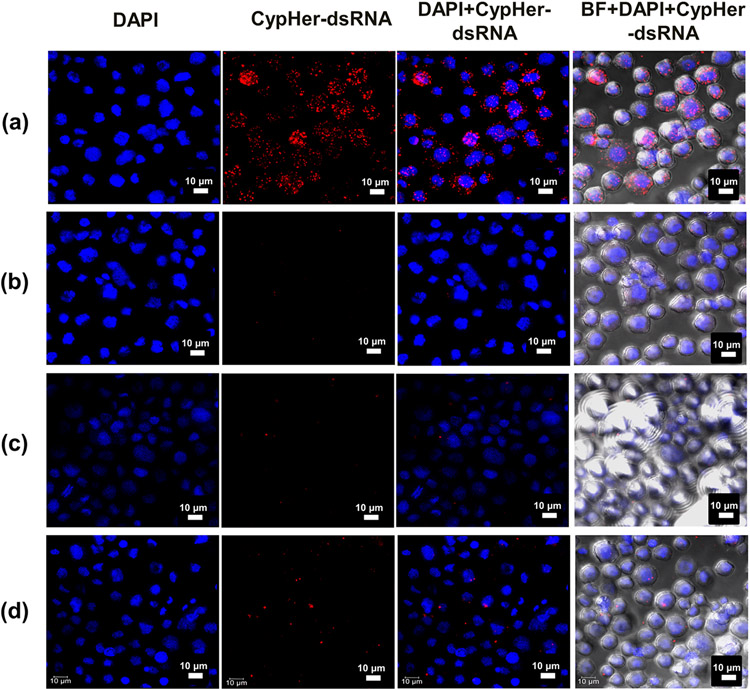
CypHer5E dye is designed to fluoresce only in the acidic environment when fully protonated at around pH 5.4 (pKa = 6.4).47 Fluorescence of the dye indicates the presence of dsRNA in the endosomes, where the pH is estimated to be in the 5.0–5.5 range.48 Conjugation of CypHer5E-dsGFP with PLR10 (PLR:dsRNA mass ratio of 1:1), c-PLR10/Au/BSA and c-PLR10/Au/Hyd NPs (NP:dsRNA mass ratio of 5:1) along with the removal of unbound dsRNA during washing was confirmed by agarose gel electrophoresis (Figure S8). Incubation of cells with naked CypHer5E-dsGFP (Figure 8a) resulted in red fluorescent bodies within the cytoplasm. This signal corresponds to the fluorescence of the dye in acidic conditions and previous studies showed that these acidic bodies accumulating dsRNA are early and late endosomes.34 No red bodies were observed in the case of the CypHer5E dye when complexed with PLR10 (Figure 8b) whereas only a few red dots were observed with CypHer5E conjugated to c-PLR10/Au/BSA (Figure 8c) and c-PLR10/Au/Hyd (Figure 8d) NPs. This presence of red bodies is correlated to the gene expression knockdown and can be attributed to the endosomal escape capacity of the different particles, or alternatively uptake into the endosomes. Indeed, a significant decrease of the fluorescence, e.g., a more important escape from endosomes, is observed for PLR-polyplexes that present the highest luciferase gene knockdown (58%). c-PLR10/Au/Hyd NPs exhibits less knockdown than c-PLR10/Au/BSA (7% versus 31% for Hyd and BSA quenched NPs, respectively) but presents a higher CypHer5E fluorescence signal, e.g., more dsRNA trapped in endosomes than the c-PLR10/Au/BSA NPs. As CypHer5E dye only highlights dsRNA trapped in the endosomes, experiments involving anti- EEA-1(labeling of early endosome) or lysotracker Green (labeling of later endosome/lysosome) colocalization would need to be conducted for a more in-depth study of the endosomal escape. Polyplexes possess positive surface potential, which can efficiently induce internalization by electrostatic interaction with the negatively charged cell membranes.49 BSA and hydroxylamine (Hyd) ligands were used as quenchers for the covalent functionalization of PLR on NHS-Au NPs to stop the reaction but also to provide multifunctionality feature to the delivery system. The attachment of targeting ligands to the surface is commonly used to enhance the cell binding via the affinity of ligands for the receptor, facilitate endosome disruption and trigger the release of the delivered therapeutic molecules from the endosome.19 Methods for assisting endosomal escape include the use of acid-responsive chemical functional groups, such as acid cleavage linkers like acetate, hydrazine, and maleic amides; acid protonating groups such as amino esters, imidazole, and sulfon-amide;50 or ligand able to interact with receptors of the cell membrane.51 Another approach is using cell-penetrating peptides or amphipathic peptides such as BSA in our case that lead to protonation of histidine residues to disrupt the endosome via proton sponge effect.52 Experimentally, the BSA quenched c-PLR10/Au particles show a better gene knockdown, which can be associated with a more favorable endosomal escape and cell penetration than the Hyd-quenched particles. Finally, dsRNA should be able to dissociate from the PLR-polyplex or Au-PLR NPs surface and be released in the cytoplasm. Contrary to PLR-polyplex, the strong covalent attachment to the Au NPs surface could lead to a lower transfection efficiency as previously reported53 or a slower release rate.
Figure 8.
Confocal microscopy images showing the internalization in Sf9 cells of CypHer5E labeled dsGFP unconjugated (a) or conjugated to (b) PLR10 (PLR10:CypHer5E-dsGFP mass ratio of 1:1), (c) c-PLR10/Au/BSA, and (d) c-PLR10/Au/Hyd NPs (NP:CypHer5E-dsGFP mass ratio of 5:1). Sf9 stable cells were exposed for 4 h, washed twice with 1X PBS buffer, fixed with 4% paraformaldehyde, and stained with DAPI.
CONCLUSION
This work demonstrated that nanomaterial delivery systems could be used to deliver dsRNA to lepidopteran cells and trigger an RNAi response, while naked dsRNA is completely ineffective. We also showed that material properties (size, stability in the extracellular medium, binding strength) must be tuned in order to achieve a successful knockdown. Polycations such as PLR have the ability to bind more efficiently to dsRNA and form PLR:dsRNA polyplexes stable enough in extracellular media for effective delivery in Sf9 compared to polymers functionalized on the inorganic core. However, the precipitation of the polyplex remains challenging to control and the formation of microparticles led to a drastic decrease of the luciferase gene knockdown efficiency (58% GK for nanopolyplex versus 20% GK for the micropolyplex). Inorganic cores may be used to control the size of the nanoparticles. Noncovalent electrostatic interaction on the Au NP surface resulted in poor attachment of PLR and aggregation in SF900 II SFM media. The immobilization of the PLR and stability in extracellular media were dramatically improved by covalently binding the PLR using an NHS ester linkage. Although both systems (polyplexes and covalently functionalized NPs) significantly favor endosomal escape compared to naked dsRNA, the polyplexes were more efficient at gene knockdown. The presence and nature of additional surface ligands on Au NPs were shown to play an important role for the capacity of the dsRNA loaded particles to induce RNAi (respectively 31% and 6.7% GK only was observed for c-PLR10/Au/BSA and c-PLR10/Au/Hyd NPs).
Toward the formulation/application of PLR-polyplexes or PLR/Au NPs as pest control agents by RNAi, some issues still need to be addressed such as the scaling-up without altering the properties (especially the size of PLR-polyplexes). Cell viability (MTT assays), intracellular uptake (flow cytometry, TEM), or the solution stability inside different concentrations of serum buffer medium of the different systems would also need to be evaluated.
Supplementary Material
ACKNOWLEDGMENTS
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award No. R21AI131427. The authors gratefully acknowledge S. Shrestha.
ABBREVIATIONS
- RNAi
RNA interference
- dsRNA
double-stranded RNA
- PLR
poly-l-arginine
- Au NPs
gold nanoparticles
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsami.0c06234.
Dose–response luciferase expression assay for PLR10:dsRNA (1:1) micropolyplexes, the phenotype of the cells, additional characterizations of the delivery systems (UV–Vis spectroscopy, gel electrophoresis, TEM, STEM-EDS), and complexation of the NPs with CyphEr5E-dsGFP (PDF)
The authors declare no competing financial interest.
Contributor Information
Jérôme Laisney, Department of Plant and Soil Sciences, University of Kentucky, Lexington, Kentucky 40546, United States.
Dhandapani Gurusamy, Department of Entomology, University of Kentucky, Lexington, Kentucky 40546, United States.
Zeinah Elhaj Baddar, Department of Entomology, University of Kentucky, Lexington, Kentucky 40546, United States.
Subba Reddy Palli, Department of Entomology, University of Kentucky, Lexington, Kentucky 40546, United States.
Jason M. Unrine, Department of Plant and Soil Sciences, University of Kentucky, Lexington, Kentucky 40546, United States
REFERENCES
- (1).Bradshaw CJA; Leroy B; Bellard C; Roiz D; Albert C; Fournier A; Barbet-Massin M; Salles J-M; Simard F; Courchamp F Massive Yet Grossly Underestimated Global Costs of Invasive Insects. Nat. Commun 2016, 7 (1), 12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Aktar MW; Sengupta D; Chowdhury A Impact of Pesticides Use in Agriculture: their Benefits and Hazards. Interdiscip. Toxicol 2009, 2 (1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Baum JA; Roberts JK Progress Towards RNAi-Mediated Insect Pest Management. In Advances in Insect Physiology; Dhadialla TS, Gill SS, Eds.; Academic Press: New York, 2014; Chapter 5, pp 249–295. [Google Scholar]
- (4).Napoli C; Lemieux C; Jorgensen R Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans. Plant Cell 1990, 2 (4), 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Fire A; Xu S; Montgomery MK; Kostas SA; Driver SE; Mello CC Potent and Specific Genetic Interference by Double-stranded RNA in Caenorhabditis Elegans. Nature 1998, 391 (6669), 806–811. [DOI] [PubMed] [Google Scholar]
- (6).Elbashir SM; Harborth J; Lendeckel W; Yalcin A; Weber K; Tuschl T Duplexes of 21-nucleotide RNAs Mediate RNA Interference in Cultured Mammalian Cells. Nature 2001, 411 (6836), 494–498. [DOI] [PubMed] [Google Scholar]
- (7).Hannon GJ RNA Interference. Nature 2002, 418 (6894), 244–251. [DOI] [PubMed] [Google Scholar]
- (8).Katoch R; Sethi A; Thakur N; Murdock LL RNAi for Insect Control: Current Perspective and Future Challenges. Appl. Biochem. Biotechnol 2013, 171 (4), 847–873. [DOI] [PubMed] [Google Scholar]
- (9).Zhang H; Li H-C; Miao X-X Feasibility, Limitation and Possible Solutions of RNAi-based Technology for Insect Pest Control. Insect Science 2013, 20 (1), 15–30. [DOI] [PubMed] [Google Scholar]
- (10).Huvenne H; Smagghe G Mechanisms of dsRNA Uptake in Insects and Potential of RNAi for Pest Control: A review. J. Insect Physiol 2010, 56 (3), 227–235. [DOI] [PubMed] [Google Scholar]
- (11).Zhu KY; Palli SR Mechanisms, Applications, and Challenges of Insect RNA Interference. Annu. Rev. Entomol 2020, 65 (1), 293–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Yang NJ; Hinner MJ Getting Across the Cell Membrane: An Overview for Small Molecules, Peptides, and Proteins. In Site-Specific Protein Labeling: Methods and Protocols; Gautier A, Hinner MJ, Eds.; Springer New York: New York, NY, 2015; pp 29–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Dominska M; Dykxhoorn DM Breaking Down the Barriers: siRNA Delivery and Endosome Escape. J. Cell Sci 2010, 123 (8), 1183. [DOI] [PubMed] [Google Scholar]
- (14).Behlke MA Chemical Modification of siRNAs for In Vivo Use. Oligonucleotides 2008, 18 (4), 305–320. [DOI] [PubMed] [Google Scholar]
- (15).Shukla S; Sumaria CS; Pradeepkumar PI Exploring Chemical Modifications for siRNA Therapeutics: A Structural and Functional Outlook. ChemMedChem 2010, 5 (3), 328–349. [DOI] [PubMed] [Google Scholar]
- (16).Gary DJ; Puri N; Won Y-Y Polymer-based siRNA Delivery: Perspectives on the Fundamental and Phenomenological Distinctions from Polymer-based DNA Delivery. J. Controlled Release 2007, 121 (1), 64–73. [DOI] [PubMed] [Google Scholar]
- (17).Jeong JH; Mok H; Oh Y-K; Park TG siRNA Conjugate Delivery Systems. Bioconjugate Chem. 2009, 20 (1), 5–14. [DOI] [PubMed] [Google Scholar]
- (18).Dowdy SF Overcoming Cellular Barriers for RNA Therapeutics. Nat. Biotechnol 2017, 35, 222–229. [DOI] [PubMed] [Google Scholar]
- (19).Shu Y; Pi F; Sharma A; Rajabi M; Haque F; Shu D; Leggas M; Evers BM; Guo P Stable RNA Nanoparticles as Potential New Generation Drugs for Cancer Therapy. Adv. Drug Delivery Rev 2014, 66, 74–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Lächelt U; Wagner E Nucleic Acid Therapeutics Using Polyplexes: A Journey of 50 Years (and Beyond). Chem. Rev 2015, 115 (19), 11043–11078. [DOI] [PubMed] [Google Scholar]
- (21).Zhang C; Tang N; Liu X; Liang W; Xu W; Torchilin VP siRNA-containing Liposomes Modified with Polyarginine Effectively Silence the Targeted Gene. J. Controlled Release 2006, 112 (2), 229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Noh SM; Park MO; Shim G; Han SE; Lee HY; Huh JH; Kim MS; Choi JJ; Kim K; Kwon IC; Kim J-S; Baek K-H; Oh Y-K Pegylated Poly-l-arginine Derivatives of Chitosan for Effective Delivery of siRNA. J. Controlled Release 2010, 145 (2), 159–164. [DOI] [PubMed] [Google Scholar]
- (23).Alhakamy NA; Berkland CJ Polyarginine Molecular Weight Determines Transfection Efficiency of Calcium Condensed Complexes. Mol. Pharmaceutics 2013, 10 (5), 1940–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Ding Y; Jiang Z; Saha K; Kim CS; Kim ST; Landis RF; Rotello VM Gold Nanoparticles for Nucleic Acid Delivery. Mol. Ther 2014, 22 (6), 1075–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Zhao E; Zhao Z; Wang J; Yang C; Chen C; Gao L; Feng Q; Hou W; Gao M; Zhang Q Surface Engineering of Gold Nanoparticles for In Vitro siRNA Delivery. Nanoscale 2012, 4 (16), 5102–5109. [DOI] [PubMed] [Google Scholar]
- (26).Guo S; Huang Y; Jiang Q; Sun Y; Deng L; Liang Z; Du Q; Xing J; Zhao Y; Wang PC; Dong A; Liang X-J Enhanced Gene Delivery and siRNA Silencing by Gold Nanoparticles Coated with Charge-Reversal Polyelectrolyte. ACS Nano 2010, 4 (9), 5505–5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Kim HJ; Takemoto H; Yi Y; Zheng M; Maeda Y; Chaya H; Hayashi K; Mi P; Pittella F; Christie RJ; Toh K; Matsumoto Y; Nishiyama N; Miyata K; Kataoka K Precise Engineering of siRNA Delivery Vehicles to Tumors Using Polyion Complexes and Gold Nanoparticles. ACS Nano 2014, 8 (9), 8979–8991. [DOI] [PubMed] [Google Scholar]
- (28).Goergen G; Kumar PL; Sankung SB; Togola A; Tamò M First Report of Outbreaks of the Fall Armyworm Spodoptera Frugiperda (JE Smith)(Lepidoptera, Noctuidae), a new alien invasive pest in West and Central Africa. PLoS One 2016, 11 (10), No. e0165632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Vaughn JL; Goodwin RH; Tompkins GJ; McCawley P The Establishment of two Cell Lines from the Insect Spodoptera Frugiperda (lepidoptera; noctuidae). In Vitro 1977, 13 (4), 213–217. [DOI] [PubMed] [Google Scholar]
- (30).Shu B; Zhang J; Sethuraman V; Cui G; Yi X; Zhong G Transcriptome Analysis of Spodoptera Frugiperda Sf9 Cells Reveals Putative Apoptosis-related Genes and a Preliminary Apoptosis Mechanism Induced by Azadirachtin. Sci. Rep 2017, 7 (1), 13231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Gurusamy D; Mogilicherla K; Shukla JN; Palli SR Lipids Help Double-stranded RNA in Endosomal Escape and Improve RNA Interference in the Fall Armyworm, Spodoptera frugiperda. Arch. Insect Biochem. Physiol 2020, No. e21678. [DOI] [PubMed] [Google Scholar]
- (32).Elbakry A; Zaky A; Liebl R; Rachel R; Goepferich A; Breunig M Layer-by-Layer Assembled Gold Nanoparticles for siRNA Delivery. Nano Lett. 2009, 9 (5), 2059–2064. [DOI] [PubMed] [Google Scholar]
- (33).Bradford MM A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem 1976, 72 (1), 248–254. [DOI] [PubMed] [Google Scholar]
- (34).Yoon J-S; Gurusamy D; Palli SR Accumulation of dsRNA in Endosomes Contributes to Inefficient RNA Interference in the Fall Armyworm, Spodoptera Frugiperda. Insect Biochem. Mol. Biol 2017, 90, 53–60. [DOI] [PubMed] [Google Scholar]
- (35).Shukla JN; Kalsi M; Sethi A; Narva KE; Fishilevich E; Singh S; Mogilicherla K; Palli SR Reduced Stability and Intracellular Transport of dsRNA Contribute to Poor RNAi Response in Lepidopteran Insects. RNA Biol. 2016, 13 (7), 656–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Walkey CD; Olsen JB; Song F; Liu R; Guo H; Olsen DWH; Cohen Y; Emili A; Chan WCW Protein Corona Fingerprinting Predicts the Cellular Interaction of Gold and Silver Nanoparticles. ACS Nano 2014, 8 (3), 2439–2455. [DOI] [PubMed] [Google Scholar]
- (37).Bloomfield VA Condensation of DNA by Multivalent Cations: Considerations on Mechanism. Biopolymers 1991, 31 (13), 1471–1481. [DOI] [PubMed] [Google Scholar]
- (38).Thi EP; Mire CE; Ursic-Bedoya R; Geisbert JB; Lee AC; Agans KN; Robbins M; Deer DJ; Fenton KA; MacLachlan I; Geisbert TW Marburg Virus Infection in Nonhuman Primates: Therapeutic Treatment by lipid-encapsulated siRNA. Sci. Transl. Med 2014, 6 (250), 250ra116–250ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Bolcato-Bellemin A-L; Bonnet M-E; Creusat G; Erbacher P; Behr J-P Sticky Overhangs Enhance siRNA-Mediated Gene Silencing. Proc. Natl. Acad. Sci. U. S. A 2007, 104 (41), 16050–16055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Shen W; Wang Q; Shen Y; Gao X; Li L; Yan Y; Wang H; Cheng Y Green Tea Catechin Dramatically Promotes RNAi Mediated by Low-Molecular-Weight Polymers. ACS Cent. Sci 2018, 4 (10), 1326–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Burke RS; Pun SH Extracellular Barriers to in Vivo PEI and PEGylated PEI Polyplex-Mediated Gene Delivery to the Liver. Bioconjugate Chem. 2008, 19 (3), 693–704. [DOI] [PubMed] [Google Scholar]
- (42).Mintzer MA; Simanek EE Nonviral Vectors for Gene Delivery. Chem. Rev 2009, 109 (2), 259–302. [DOI] [PubMed] [Google Scholar]
- (43).Shan Y; Luo T; Peng C; Sheng R; Cao A; Cao X; Shen M; Guo R; Tomás H; Shi X Gene Delivery Using Dendrimer-Entrapped Gold Nanoparticles as Nonviral Vectors. Biomaterials 2012, 33 (10), 3025–3035. [DOI] [PubMed] [Google Scholar]
- (44).Chithrani BD; Ghazani AA; Chan WCW Determining the Size and Shape Dependence of Gold Nanoparticle Uptake into Mammalian Cells. Nano Lett. 2006, 6 (4), 662–668. [DOI] [PubMed] [Google Scholar]
- (45).Kummitha CM; Malamas AS; Lu Z-R Albumin Pre-Coating Enhances Intracellular siRNA Delivery of Multifunctional Amphiphile/siRNA nanoparticles. Int. J. Nanomed 2012, 7, 5205–5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Mariam J; Sivakami S; Dongre PM Albumin Corona on Nanoparticles – a Strategic Approach in Drug Delivery. Drug Delivery 2016, 23 (8), 2668–2676. [DOI] [PubMed] [Google Scholar]
- (47).Michael NP; et al. In CypHer 5 - A Novel pH Sensitive Fluor For Measuring Cell Surface Receptor Activation In Live Cell Assays. Poster presented during the 8th annual Conference of the Society of Biomolecular Screening, The Hague, Netherlands, 2002. [Google Scholar]
- (48).Geisow MJ; Evans WH pH in the endosome: Measurements During Pinocytosis and Receptor-Mediated Endocytosis. Exp. Cell Res 1984, 150 (1), 36–46. [DOI] [PubMed] [Google Scholar]
- (49).Varkouhi AK; Scholte M; Storm G; Haisma HJ Endosomal Escape Pathways for Delivery of Biologicals. J. Controlled Release 2011, 151 (3), 220–228. [DOI] [PubMed] [Google Scholar]
- (50).Kwon YJ Before and after Endosomal Escape: Roles of Stimuli-Converting siRNA/Polymer Interactions in Determining Gene Silencing Efficiency. Acc. Chem. Res 2012, 45 (7), 1077–1088. [DOI] [PubMed] [Google Scholar]
- (51).Zu Y; Huang S; Liao W-C; Lu Y; Wang S Gold Nanoparticles Enhanced Electroporation for Mammalian Cell Transfection. J. Biomed. Nanotechnol 2014, 10 (6), 982–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Langlet-Bertin B; Leborgne C; Scherman D; Bechinger B; Mason AJ; Kichler A Design and Evaluation of Histidine-Rich Amphipathic Peptides for siRNA Delivery. Pharm. Res 2010, 27 (7), 1426–1436. [DOI] [PubMed] [Google Scholar]
- (53).Kawano T; Yamagata M; Takahashi H; Niidome Y; Yamada S; Katayama Y; Niidome T Stabilizing of Plasmid DNA In Vivo by PEG-Modified Cationic Gold Nanoparticles and the Gene Expression assisted with Electrical Pulses. J. Controlled Release 2006, 111 (3), 382–389. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.