Abstract
Purpose:
Covalent inhibitors of KRASG12C specifically target tumors driven by this form of mutant KRAS, yet early studies show that bypass signaling drives adaptive resistance. While several combination strategies have been shown to improve efficacy of KRASG12C inhibitors, underlying mechanisms and predictive strategies for patient enrichment are less clear.
Experimental Design:
We performed mass spectrometry based phosphoproteomics analysis in KRASG12C cell lines after short term treatment with ARS-1620. To understand signaling diversity and cell-type specific markers, we compared proteome and phosphoproteomes of KRASG12C cells. Gene expression patterns of KRASG12C cell lines and lung tumor tissues were examined.
Results:
Our analysis suggests cell-type specific perturbation to ERBB2/3 signaling compensate for repressed ERK and AKT signaling following ARS-1620 treatment in epithelial cell type, and this subtype was also more responsive to co-inhibition of SHP2 and SOS1. Conversely, both high basal and feedback activation of FGFR or AXL signaling was identified in mesenchymal cells. Inhibition of FGFR signaling suppress feedback activation of ERK and mTOR, while AXL inhibition suppress PI3K pathway. In both cell lines and human lung cancer tissues with KRASG12C we observed high basal ERBB2/3 associated with epithelial gene signatures while higher basal FGFR1 and AXL was observed in cells/tumors with mesenchymal gene signatures.
Conclusions:
Our phosphoproteomic study identified cell-type adaptive responses to KRASG12C inhibitors. Markers and targets associated with ERBB2/3 signaling in epithelial subtype and FGFR1/AXL signaling in mesenchymal subtype should be considered in patient enrichment schemes with KRASG12C inhibitors.
Keywords: Lung Cancer, KRAS, mass spectrometry, targeted therapy, drug resistance
INTRODUCTION
Oncogenic KRAS mutations occur in nearly 25% of lung adenocarcinomas, are often associated with poor prognosis, and are notoriously refractory to conventional cytotoxic chemotherapies as well as agents that target receptor tyrosine kinases (RTK) (1,2). Use of MEK inhibitors with cytotoxic agents or other pathway inhibitors (PI3K/Akt) also proved to be either ineffective or toxic (3). Therefore, it is an unmet medical need to identify effective therapies for patients with KRAS mutant lung cancer. The advancements in medicinal chemistry has produced high potency KRASG12C inhibitors (KRASi) such as ARS-1620, rapid binding to the GDP-bound inactive form, and optimized pharmacokinetics to maximize exposure and duration (4,5). Since KRASG12C mutations occur in ~11–16% of lung adenocarcinoma, this represents a large group of patients who can potentially benefit. Subsets of KRASG12C NSCLC cells are highly sensitive to these inhibitors, and AMG-510 and MRTX-849 have demonstrated early activity in KRASG12C mutant lung cancer patients (6–8).
Since nucleotide exchange of RAS signaling can be mediated by upstream RTKs, recent results demonstrated that RTK inhibitors can impede the exchange reaction and sensitize cells to KRASi (9,10). Recent studies identified several combination strategies with KRASi which includes inhibitors of HER kinases, FGFR1, AXL, SHP2, mTOR, MEK, PI3K and CDK4/6, among others (11–16). However, it remains unclear why one co-targeting strategy may be more effective in particular cell types, and what mechanisms underlie the observed effectiveness of some combinations. The exact RTK that mediates this effect may vary from cell line to cell line, despite the common KRAS driver mutation, thus making it difficult to determine how to precisely predict combinations that would be effective in the clinic. In one case, higher basal expression of EGFR signaling in KRASG12C colorectal cancer cell line compared to NSCLC predicted effectiveness of adding anti-EGFR therapy to KRASi (17). However, these results are somewhat perplexing given the high levels of EGFR signaling in NSCLC (18). Since KRASG12C bound to GDP competes with the small molecule inhibitors and exchange factors (i.e., SOS), a better understanding of signaling events that control RAS signaling will be needed to optimize the use of KRASi and design precision medicine with KRASi in the clinic.
Given the diverse responses observed in both preclinical models and human patients, it is critical to understand how cells escape from targeted inhibition, which pathways contribute to resistance, and how to predict pathway utilization for escape to enable precision medicine in the form of combination therapy. Given the diversity of kinases and other signaling molecules known for adaptive resistance, a system-level analysis is the most suitable approach to uncover mechanisms of therapeutic escape to KRASi. To address these challenges, we carried out mass spectrometry-based phosphoproteomics following KRASG12C inhibition in three different lung cancer cell lines. Our data suggest a model in which inhibition of KRASG12C is overcome by adaptive signaling changes. There is growing appreciation for the role of adaptive resistance to targeted agents mediated by changes in feedback programs (19). Our phosphoproteomics data suggest different processes of adaptive rewiring and therapeutic escape to KRASG12C inhibitors and indicate the possibility to predict co-targeting strategies that interfere with compensatory pathways.
MATERIALS AND METHODS
Cell lines
NCI-H358, Calu1, NCI-H1373, NCI-H1792, NCI-H23 and NCI-H2122 were purchased from the American Type Culture Collection (ATCC). HCC-44 and HOP-62 were acquired from MD Anderson Cancer Center (MDACC) and National Cancer Institute, respectively. LU-99 was generous gift from Dr. Sheri Moores (The Janssen Pharmaceutical Companies of Johnson & Johnson).
Drugs
ARS-1620 and BI-3406 was purchased from MedChemExpress. AMG-510, Erlotinib, Afatinib, AZD-4547, Linsitinib, Imatinib, Cabozantinib, Ceritinib, GDC-0941 and RXDX-106 were purchased from Selleckchem. Drugs were reconstituted in DMSO to 10 mM stock concentrations and stored at −80oC. The combination index (CI) was calculated using CompuSyn software (20). The coefficient of drug interaction (CDI) was calculated using the following formula; CDI = AB/(A × B) (21).
Sample preparation for phosphoproteomics and LC-MS/MS
Sample preparation for proteomics was carried out according to Cell Signaling Technology protocol (#8803). Briefly the cells were lysed in denaturing buffer, followed by reduction/alkylation, trypsin digestion and peptide desalting. For phosphotyrosine enrichment, anti-pY-1000 antibodies was used, followed by TMT labeling and IMAC enrichment. For global phosphorylation enrichment, the peptides were labeled with TMT first and fractionated with bRPLC, followed by IMAC enrichment. A nanoflow ultra-high-performance liquid chromatography interfaced with an electrospray quadrupole-orbitrap mass spectrometer (RSLCnano and Q Exactive HF-X, Thermo, CA) was used for LC-MS/MS peptide sequencing and TMT quantification.
Data analysis
MaxQuant software (version 1.6.2.10) was used for peptide identification and reporter ion quantification (22). Data were normalized and analyzed for differential expression between treatment and control time points. Data available via the PRIDE (23) for pY in ARS-1620 treated cells (PXD021607), for global phosphoproteomics after ARS-1620 treatment in H358 (PXD021611), H1792 (PXD021609) and Calu1 (PXD021608), and for pY (PXD021604) and protein expression (PXD021603) in 8 KRASG12C cell lines.
LC-MRM and data analysis
LC-MRM analysis was performed in triplicate on a nanoUHPLC interfaced with an electrospray triple quadrupole mass spectrometer (DionexRSLCnano and TSQ Altis, Thermo). For each sample, peptide mixture (5 μL) was loaded onto the trap column and the trapped peptides were eluted onto a C18 analytical column and separated using a 62.5-minute gradient. Collision energy (CE) values were optimized for this instrument in Skyline version 20.1.0.76 (24) using CE optimization equations empirically derived from data previously collected on the instrument. Skyline (v. 20.1.0.76) was used to evaluate the data (24).
TGFβ-EMT scores
The 105 probeset TGFβ-EMT signature (25) was used to assess the levels of TGFβ-driven EMT in cell lines and tumor samples. For each dataset, rows corresponding to the signature genes were selected and PCA models generated from these TGFβ-EMT rows. Low/High directionality of PC1 was established from the PCA loadings and their signs from the original TGFβ-EMT signature, flipping the sign of PC1 as appropriate so that increasing PC1 values corresponds to increasing levels of TGFβ-driven EMT.
For detailed experimental procedures, see the Supplemental Methods.
RESULTS
Quantitative phosphoproteomics identifies patterns of signaling responses following KRASG12C inhibition
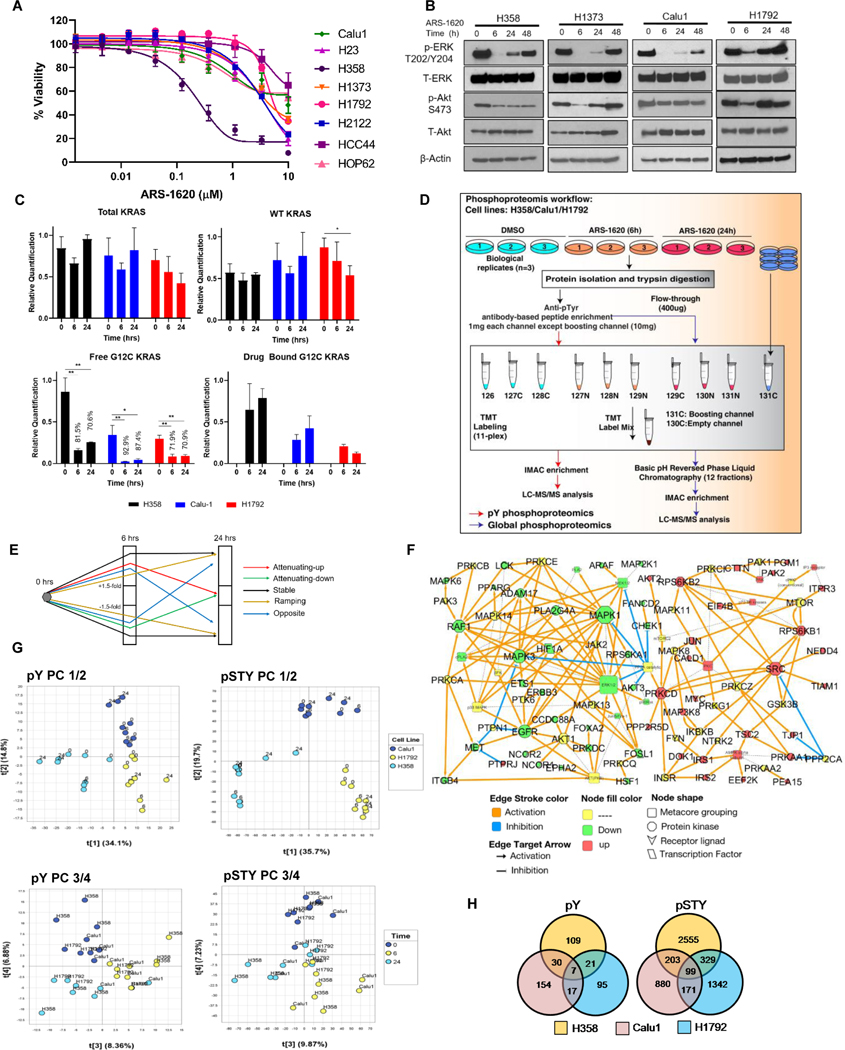
To understand mechanism(s) of short-term signaling adaptation to KRASi using phosphoproteomics, we selected three cell lines with differential sensitivity to ARS-1620. A heterogeneous response to ARS-1620 was observed in a panel of 8 KRASG12C NSCLC cell lines in both 2D and 3D cultures (Fig. 1A; Supplementary Fig. S1A), with H358 exhibited highest sensitivity with half maximal inhibitory concentration (IC50) of 0.4 μM (in 2D culture). Similar heterogeneity of response was also observed with AMG-510 (Supplementary Fig. S1B-C). Using 1 μM doses, 3 cell line models, H358, Calu1 and H1792 cells, were selected and designated as sensitive, moderate and resistant lines, respectively (Supplementary Fig. S1D). In order to select time points for phosphoproteomics analysis, we carried out western blotting of key signaling molecules, pERK (T202/Y204) and pAkt (S473), in a panel KRASG12C cell lines at different treatment time points. Signaling analysis after ARS-1620 and AMG-510 treatment reveals inhibition of p-ERK with evidence of rebound starting at 24 hrs (Fig. 1B; Supplementary Fig. S1E). Similarly, suppression in pAkt was observed only at 6 hrs in H1373 and H1792 following rebound starting at 24 hrs. However, moderate to less impact on Akt phosphorylation was observed in H358 and Calu1 at all treatment time points (Fig. 1B).
Figure 1. Signaling rebound in KRASG12C lung cancer cells following ARS-1620 treatment and phosphoproteomics analysis indicates cell-type specific perturbations.
A) Panel of KRASG12C mutant cell lines were analyzed by CellTiter-Glo after 96 hrs of ARS-1620 treatment. Data represent means of three biological replicates; error bars denote SD. B) The indicated cell lines were treated with ARS-1620 (1μM) and rebound in p-ERK and p-Akt were monitored for 6, 24 and 48 hrs by immunoblotting with the indicated antibodies. C) LC-MRM peptide quantification of WT, total, G12C free and drug bound peptide before and after ARS-1620 treatment in KRASG12C mutant lung cancer cell lines. Target engagement (%) determined by the loss of LC-MRM ion signal for the free KRAS G12C protein (LVVVGAcamCGVGK). Unpaired Student’s t-tests were used to calculate p values: * p < 0.05 and ** p < 0.01. D) Schematic workflow for TMT-based quantitative phosphoproteomics to identify perturbation in phosphoproteome upon 6 and 24 hrs of ARS-1620 treatment in H358, H1792 and Calu1. The experiment was performed in biological triplicates with 2 mass spec injections per sample (a multiplexed TMT set). E) Categorization of differentially expressed phosphosites: “attenuating” (maximum perturbation at 6 hrs, returning towards baseline at 24 hrs), “ramping” (increasingly perturbed over time), “stable” (6 hrs and 24 hrs similarly perturbed), and “opposite” (perturbed in opposite directions at 6 hrs and 24 hrs). The “attenuating” proteins were categorized in to “attenuating-down” (hypo-phosphorylated at 6 hrs, returning towards baseline at 24 hrs) and “attenuating-up” (hyper-phosphorylated at 6 hrs, returning towards baseline at 24 hrs). F) Experimentally consistent literature network was generated for “attenuating” proteins using the MetaCore (Clarivate Analytics) pathway analysis. G) Principal component analysis (PCA) plots generated on pY and pSTY data sets using “Nonlinear Iterative Partial Least Squares” (NIPALS) algorithm. PC1/2 and PC3/4 indicate differences between cell lines and treatment time points, respectively. H) Venn diagram representing distribution of differentially expressed phosphosites among 3 cell lines (H358, Calu1 and H1792).
In parallel, we evaluated KRASG12C target engagement at different times after ARS-1620 treatment using LC-MRM relative quantification of tryptic peptides representing total KRAS, wild-type (WT) KRAS, free G12C KRAS, and drug-bound KRAS G12C (Supplementary Table S1). Total KRAS expression before treatment differs between cell lines: H358 > Calu-1 > H1792. H358 and Calu-1 cell lines show a rebound in total KRAS expression at 24 hrs, while H1792 shows a decreasing trend (p = 0.054). Baseline expression of WT KRAS also differs between the cell lines: H1792 > Calu-1 > H358, while mutant protein is highly expressed in H358 compared with the other cell lines. Target engagement (%) was determined by the loss of ion signal for the free KRAS G12C protein (LVVVGAcamCGVGK); values are listed on the plot (Fig. 1C). Signal for this peptide is significantly decreased in all cell lines at both time points after ARS-1620 treatment, but a minor rebound in free KRAS G12C protein expression is observed in H358 cells. The G12C peptide covalently modified with ARS1620 was also quantified and increased at both time points after drug treatment.
The signaling analysis and LC-MRM peptide quantification guided selection of ARS-1620 treatment time points (6 and 24 hrs) for a multiplexed isobaric-tag-based (TMT) quantitative phosphoproteomics experiment in H358, Calu1 and H1792 cells done in biological triplicates (Fig. 1D). With phosphotyrosine (pY) enrichment, we identified 2,275 human phosphosites in total, corresponding to 1,153 proteins in 987 protein groups, out of which 433 phosphosites were differentially expressed (selected based on ±1.5-fold change and p-value < 0.05) between any treatment time point and control (Supplementary Table S2). Analysis of phosphoproteomics data (pS/T/Y) identified 26,735 human phosphosites, corresponding to 5,577 proteins in 5,527 protein groups, out of which 5,579 phosphosites were differentially expressed (based on ±1.5-fold change and p-value < 0.05) between any treatment time point and control (Supplementary Table S3).
Differentially expressed phosphosites were then categorized into 4 broad patterns of behaviours: “attenuating”, “ramping”, “stable”, and “opposite”. Further, “attenuating” proteins were categorized in to “attenuating-down” and “attenuating-up” (Fig. 1E). We reasoned that “attenuating” proteins indicates direct impact of ARS-1620 treatment on signaling and generated a literature network for these proteins (Fig. 1F) using MetaCore (Clarivate Analytics) pathway analysis. This analysis clustered “attenuating-down” proteins indicating inhibition of ERK signaling. The network on “attenuating-up” proteins indicates activation of Insulin signaling, where IRS1, IRS2, DOK1 and PRKCD are major activation nodes connected to Insulin receptor (INSR). In addition, a network connecting RPS6KB1, RPS6KB2, EIF4B and RPS6 nodes indicates activation of mTOR signaling. Thus, IGFR, PI3K and mTOR serve as adaptive signaling hubs in response to ARS-1620.
We next compared alterations in the phosphoproteomes of these cells following KRASG12C inhibition. Interestingly, principal-component (PC) analysis of the pY and pSTY data set (Fig. 1G) showed separation of the 3 cell lines between PC1 and PC2, which indicates that three cell lines have distinct phosphoproteomic alterations following ARS-1620 treatment. Indeed, the PC plot between PC3 and PC4 indicates more changes at 6 hrs than 24 hrs when compared to DMSO treatment (0 hrs) and lesser change in magnitude between 0 and 24 hrs. This analysis indicates major phosphoproteomics alterations at 6 hrs followed by signaling rebound by 24 hrs, consistent with our western blot observations.
The distribution of differentially expressed phosphosites indicate more of cell line specific response, where only 7 pY and 99 pSTY phosphosites were differentially affected across all 3 cell lines (Fig. 1H, Supplementary Table S2 & S3). The decreased phosphorylation of MAPK3 (pY204) and MAPK6 (pS189) in all cells indicated inhibition of ERK signaling. Among the other proteins with decreased phosphorylation, we identified proteins associated with (1) cell-cycle regulation and DNA replication check point (RB1 pS788, WEE1 pT190, RBL2 pT401, and EMD pS120), (2) transcription and RNA splicing (BCLAF1 pS196, SRSF4 pS431, NCBP1 pS7, RANBP2 pS1509, and SRRM1 pS452) and (3) receptor signaling (EPHA2 pY594, pS901 and pS897, as well as CD44 pS697). The phosphosites with increased phosphorylation associated with (1) mTOR signaling (RPS6 pS247, pS244 and pS240, EIF4G1 pS1147 and RPS6KB1 pS394), (2) cell-motility and survival (SDC4 pY197, IRS2 pS620 and PRKCD pS302), (3) negative regulation of apoptosis (HIPK3 pY359) (4) and regulation of RAS-ABL signaling (RIN1 pY36). Despite this commonality among cell lines, these data suggest that perturbations result in unique phosphosignatures across various cell lines despite all harboring KRASG12C. It also suggested that each cell type required individual data analysis to uncover cell type specific adaptive resistance mechanisms.
HER2/HER3 signaling drives adaptive resistance to KRASG12C inhibitors
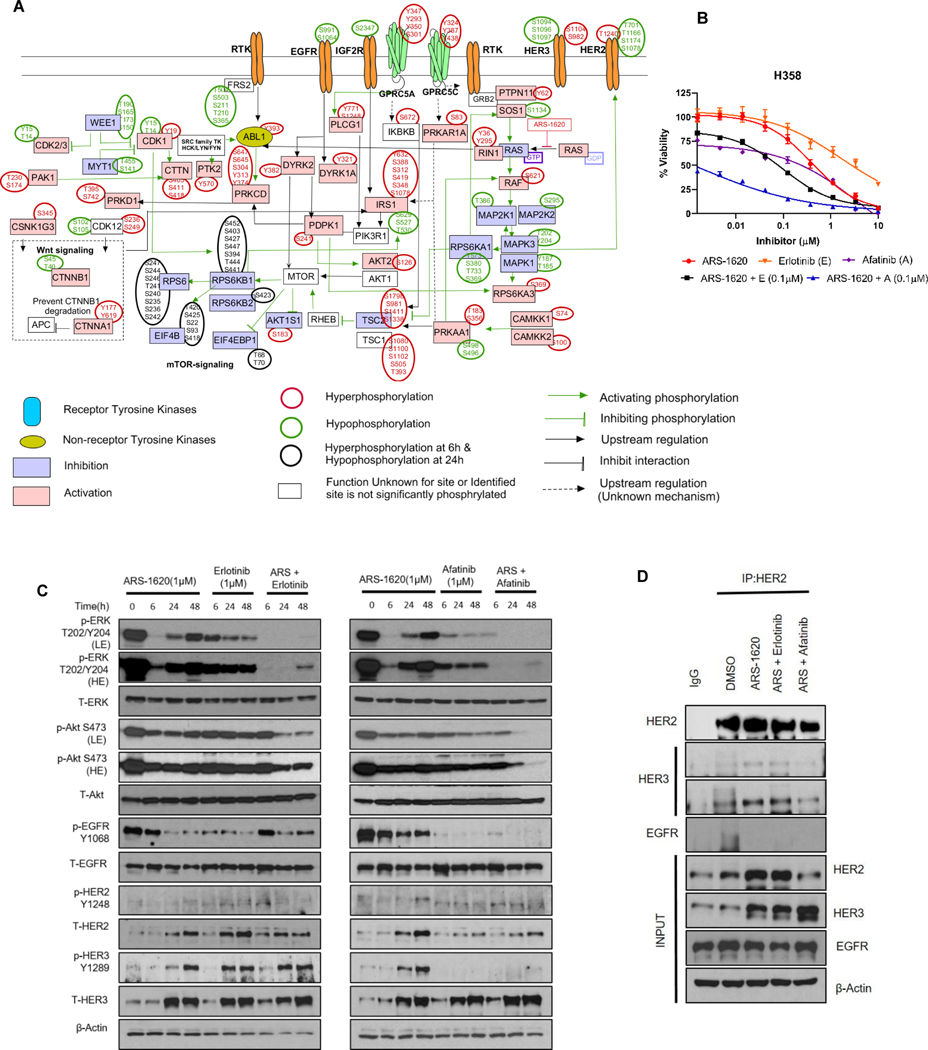
We observed the highest number of perturbed phosphosites in H358 cells, where 167 and 3186 phosphosites were differentially expressed in the pY and pSTY data, respectively. We carried out MetaCore pathway analysis on phosphosites perturbed at 6 hrs to understand an immediate impact of ARS-1620 on signaling. We observed many nodes densely connected in several sub-networks (Supplementary Fig. S2A). The inhibition of network surrounding MAPK1/MAPK3 indicate inhibition of ERK signaling. Among the various activating sub-networks, we observed a signaling hub surrounding RAF1, presented as a direct substrate of several kinases including CAMKK2, CAMK2A, PRKCA, PRKCB, PRKD1 and PAK4.
Having observed the complexity of the Metacore network, we next sought to generate a simplified network by manually curating the pathway based on information retrieved from PhosphoSitePlus (26) and experimentally affirmed phosphosites information in the literature (Fig. 2A). We observed decreased phosphorylation of MAPK1/3 and RPS6KA1 indicating inhibition of MAPK signaling. However, other signaling events such as decreased phosphorylation of RPS6KA1-specific SOS1 site (pS1134) and increased phosphorylation of PRKAA1-specific RAF1 site (pS621) indicate feedback activation of MAPK signaling following KRASG12C inhibition (27,28).
Figure 2. Combination of pan-HER Inhibitor and ARS-1620 overcomes signaling rebound in H358 cells.
A) Depiction of manually curated signaling rebound network in H358 cells based on experimentally affirmed signaling information available in literature for phosphosites differentially perturbed after KRASG12C inhibition. B) H358 - dose response to ARS-1620, erlotinib and afatinib alone or ARS-1620 combination with erlotinib (0.1μM) or afatinib (0.1μM) after 96 hrs of treatment and analysis using CellTiter-Glo. Data represent average of three biological replicates; error bars denote SD. C) H358 cells were treated with ARS-1620 (1μM), erlotinib (1μM), afatinib (1μM), or the indicated combinations, and p-ERK and p-Akt rebound was monitored after 6, 24 and 48 hrs by immunoblotting with the indicated antibodies. Lower exposure (LE) and higher exposures (HE). D) H358 cells were treated for 24 hrs with ARS-1620 (1μM) alone and in combination with erlotinib (1 μM) or afatinib (1μM). Cell lysates were subjected to immunoprecipitation (CoIP) using HER2 antibody and immunoblotted for HER3 (lower and higher exposure) and EGFR. Rabbit normal IgG was used as negative control.
We next checked alterations in EGFR phosphorylation as previous studies reports synergistic combination effects of EGFR/HER family kinase inhibitors with ARS-1620 (14). ARS-1620 treatment decreases phosphorylation levels of known inhibitory phosphorylation sites of EGFR at S1064 and S991 (29). Next, we sought to look in signaling perturbation in other members of ERBB family. Notably, we identified the phospho-peptide for tyrosine activation site (Y1289) of HER3 only in ARS-treated cells. Enhanced phosphorylation on multiple phosphosites of HER2 (T1240) and HER3 (S1104 and S982) was also observed. Conversely, decreased phosphorylation of many phosphosites on HER2 (T701, T1166, S1078, S1174, S1107 and S998) and HER3 (S1094, S1096 and S1097) were observed. The ERK-mediated phosphorylation of T701 (old annotation T677) on HER2 mediates a negative feedback loop to control ERBB receptor dimer formation (30) and MEK inhibition induces ERBB/Akt signaling activation by suppressing HER2 pT701 phosphorylation (31). These alterations in multiple phosphosites of HER2 and HER3 indicate a significant role of HER2/HER3 signaling, beyond EGFR signaling, and could explain recent studies suggesting a lesser role of EGFR in lung cancer.
To address this, we evaluated in vitro efficacy of the ARS-1620 plus ERBB inhibitor combinations in H358 cells. When compared to more specific EGFR inhibition (erlotinib), the ARS-1620 combination with the pan-ERBB inhibitor afatinib had greater impact on reducing cell viability (Fig. 2B) and presented higher synergistic effects even at lower dose (Supplementary Fig. S2 B-C). Western blot-based signaling analysis also indicated longer suppression of pERK and pAKT with the afatinib combination (Fig. 2C). Despite higher total levels of HER2/3 following KRASG12C inhibition (Fig. 2C), decreased phosphorylation on multiple serine and threonine residues suggest these sites as inhibitory phosphorylation sites, thereby driving resistance to KRASi via feedback ERBB signaling activation. ARS-1620 treatment alone increased the phosphorylation of HER3 (Y1289), although a weak and variable signal for HER2 phosphorylation (Y1248) was observed. Conversely, decreased phosphorylation of EGFR (Y1068) was observed in cells treated with ARS-1620 for longer duration. Co-treatment with afatinib suppressed HER2 expression and HER3 phosphorylation while also suppressing EGFR phosphorylation. Since higher expression of HER2 and HER3 was observed following ARS-1620 treatment, we next checked whether ARS-1620 enhances HER2-HER3 dimerization. We observed higher HER2 and HER3 binding following KRASG12C inhibition, which was suppressed by the ARS-1620 plus afatinib combination (Fig. 2D). These findings indicate that activation of HER2/HER3 signaling limits the efficacy of KRASG12C inhibition and provides the mechanistic insight of responsiveness to afatinib combination with KRASG12C inhibition in xenograft models, as well as why lung cancer may be less reliant on EGFR than colon cancer (12).
HER2/HER3 signaling within an Epithelial subtype associates with effectiveness of dual KRASi/pan-HERi inhibition
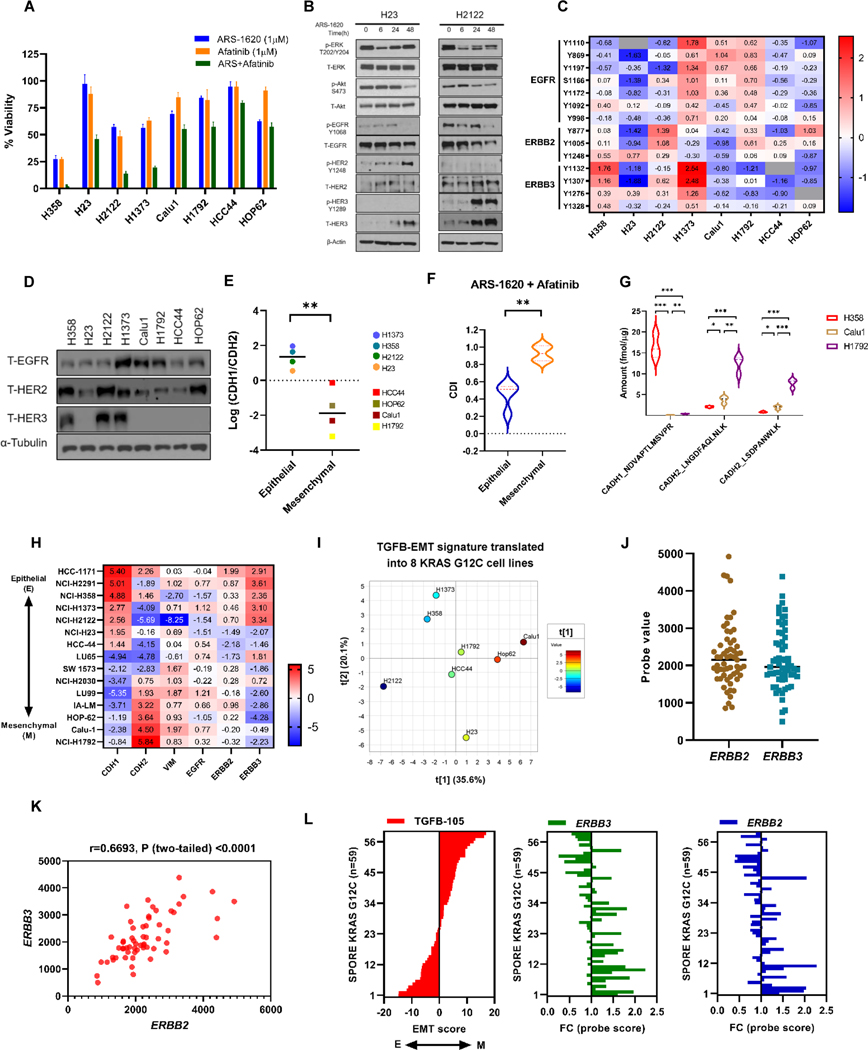
We and others have observed that afatinib treatment as single agent had effects on cell viability in H358 cells (32). This observation indicated that some KRASG12C mutants have active ERBB signaling aiding KRASG12C signaling for tumor progression. Moreover, our results indicated that KRASi further amplifies ERBB signaling as an adaptive mechanism to sustain cell survival. Next, we determined the efficacy of the KRASi plus panHER inhibitor combination on cell viability and then evaluated ERBB signaling in the panel of KRASG12C cell lines. The afatinib combination with ARS-1620 had higher effect on cell viability in H2122, H1373 and H23, along with H358 cells (Fig. 3A). Consistent with our previous observation, enhanced expression and phosphorylation (Y1289) of HER3 and increase in HER2 expression was observed in H2122 following ARS1620 treatment. On the other hand, a higher expression and phosphorylation (Y1248) of HER2 and enhanced expression of HER3 was observed in H23 (Fig. 3B).
Figure 3. Epithelial subtype predicts pan-HER combination strategy in KRASG12C lung cancer cell lines.
A) Panel of KRASG12C mutant lung cancer cell lines were analyzed by CellTiter-Glo after 96 hrs of ARS-1620 (1μM) treatment alone and in combination with Afatinib (1μM). Data represent means of three biological replicates and was normalized to DMSO control; error bars denote SD. B) H23 and H2122 cells were treated with ARS-1620 (1μM) and rebound in signaling was monitored after 6, 24 and 48 hrs by immunoblotting with the indicated antibodies. C) Heatmap showing mean-centered log abundance expression values for various phosphosites belonging to HER family of proteins in KRASG12C mutant lung cancer cell lines. The expression is scaled from lowest (blue) to highest (red); grey indicates peptides that were not detected in particular cell line. D) Cell lysates from KRASG12C mutant NSCLC cell lines were probed with indicated antibodies. E) Log2 difference between E-cadherin (CDH1) and N-cadherin (CDH2) protein expression was plotted for different KRASG12C mutant cell lines. F) The coefficient of drug interaction (CDI) was calculated for ARS-1620 + afatinib combination in KRASG12C mutant NSCLC cell lines (using biological triplicate data presented in Figure 3A) indicating relative survival rate in presence of both drugs when compared with either drug alone. The Student’s t-tests were performed to calculate the significant difference in CDI between epithelial and mesenchymal KRASG12C mutant NSCLC cell lines. The values represent means of three biological replicates for each cell lines. Antagonism, additivity or synergy is represented by values >1, = 1 or <1, respectively. A CDI below 0.7 indicates significant synergism. G) Peptides from CADH1 (CDH1) and CAHD2 (CDH2) proteins were quantified using LC-MRM method. The violin plot indicates the difference in baseline protein expression among 3 KRASG12C mutant NSCLC cell lines. Each cell line was analyzed in biological triplicates. H) Mean-centered CCLE normalized log2 RNA-seq counts for ERBB and EMT genes in KRASG12C mutant NSCLC cell lines. Raw counts were normalized with IRON (iron_generic--rnaseq) against the whole-dataset median sample (RERFLCAI_LUNG). I) TGFβ-EMT analysis of the 8 KRASG12C cell lines using our proteomics data. Positive score = more mesenchymal and negative score = more epithelial. J-K) Scatter plot of probe values representing ERBB2 and ERBB3 gene expression in KRASG12C mutants (n=59) in SPORE442 dataset. Multiple probesets exist for ERBB3, the probeset with the highest average intensity was chosen as the representative for that gene. The Pearson correlation coefficient between ERBB2 and ERBB3 gene expression was calculated along with the p-value. L) Median-centered probe value for ERBB2 and ERBB3 gene were plotted for KRASG12C mutants (n=59) in SPORE442 dataset. TGFβ-EMT scores were calculated for each KRASG12C mutant and samples were sorted based on their EMT score. After sorting on the TGFB-105 score, the p-value for Pearson correlation was calculated for the individual genes using the TGFB-105 High vs. Low median cut, tertitle, and quantiles (Supplementary Table S9). Unpaired Student’s t-tests were used to calculate p values: * p < 0.05, ** p < 0.01. *** p < 0.001.
Next, to ascertain if cell lines responsive to the afatinib/ARS-1620 combination have enhanced ERBB signaling, we carried out TMT-based quantitative phosphoproteomics (with pY enrichment) and expression proteomics of all 8 KRASG12C cell lines (Supplementary Fig. S3A, and Supplementary Table S4 & S5). We observed higher phosphorylation of multiple phosphosites on HER3 in cell lines responsive to afatinib combination (Fig. 3C). Western blot analysis of ERBB proteins indicates a varied basal expression of EGFR among KRASG12C cell lines but higher basal expression of HER2 and HER3 was observed in H358, H2122 and H1373 (Fig. 3D).
We hypothesized that cell states, specifically epithelial or mesenchymal phenotypes, could contribute to the response of combining pan-HER inhibitor with KRASG12C inhibition. To determine EMT status, we assessed the protein ratio of epithelial marker E-cadherin (CADH1/CDH1) to mesenchymal markers N-cadherin (CADH2/CDH2). We observed significantly higher CDH1 to CDH2 ratio in 4 cell lines (H358, H2122, H1373 and H23), which were then designated as Epithelial (E) type (Fig. 3E), and other models as Mesenchymal (M) type. When we compared the coefficient of drug interaction (CDI) for afatinib plus ARS-1620 combination between epithelial and mesenchymal models, we observed higher synergistic effects in E-type KRASG12C models (Fig. 3F). The E-type KRASG12C models also showed significant lower intensities of many phosphosite(s) associated with Vimentin (Y53, Y11, Y38, Y61, Y117 and Y276) and higher intensity for CDH1 Y797 when compared with M-type (Supplementary Fig. S3B). We did not see significant difference in ERBB phosphosites in 2 group (E vs M) comparison. However, a significant difference in ERBB3 phosphosite Y1328 was observed (Supplementary Fig. S3B) when we compared 2 most epithelial (H358 and H1373) with 2 most mesenchymal (H1792 and Calu1) models (Fig. 3E).
The LC-MRM based quantification of EMT makers also confirmed that E-cadherin is highly expressed in H358, while N-cadherin is expressed at higher levels in Calu-1 and H1792 (Fig. 3G; Supplementary Table S6). We next compared gene expression (using RNAseq data) of ERBB genes and EMT markers in KRASG12C cell lines using data from CCLE (33) (Fig. 3H; Supplementary Table S7). In concordance with our proteomics results, we identified KRASG12C models (H1171, H2291, H358, H2122 and H1373) with higher E-cadherin and/or lower N-cadherin expression also has higher ERBB3 gene expression and a significant difference was observed when we performed 2 group (E vs M) comparison (Supplementary Fig. S4A).
As an alternative way to score epithelial versus mesenchymal phenotypes, TGF-β induced epithelial-to-mesenchymal transition (EMT) signature (25) were translated into the protein expression data of KRASG12C cell lines to differentiate (Supplementary Fig. S4B). The PCA plot of TGFβ-EMT score among 8 KRAS G12C cell lines predicted similar phenotypes where H358, H2122 and H1373 appeared to be more epithelial (Low EMT score) than other cell lines (Fig. 3I, Supplementary Table S8).
We next examined if KRASG12C human lung cancer tumor tissues also had a similar distribution of ERBB2 and ERBB3 expression and included both epithelial and mesenchymal phenotypes. Gene expression data of KRASG12C mutants (n=59) was extracted from microarray gene expression data set from 442 lung adenocarcinoma patients (34). A divergent expression of ERBB2 and ERBB3 was observed in KRASG12C mutants with a sub-population of mutants having higher expression of both ERBB2 and ERBB3 (Fig. 3J) (Supplementary Table S9). Moreover, a positive correlation was also observed between ERBB2 and ERBB3 gene expression (Fig. 3K). Next, we examined EMT activity based on the TGF-β EMT gene signature in KRASG12C mutants (25). The samples were ranked by TGFβ −105 score and divided into two groups at the median (ERBB3: p = 1.62E-04 & ERBB2: p = 1.97E-03). We observed a significant negative correlation of EMT score with ERBB2 (r= −0.50, p=5.63E-05) and ERBB3 (r= −0.57, p=2.95E-06) (Fig. 3L & Supplementary Table S9). Therefore, KRASG12C human lung cancer exists in a ERBB2/ERBB3 overexpressing state associated with epithelial subtype.
SHP2, SOS1 and IRS1 contribute to adaptive resistance to KRASG12C inhibition with an epithelial subtype
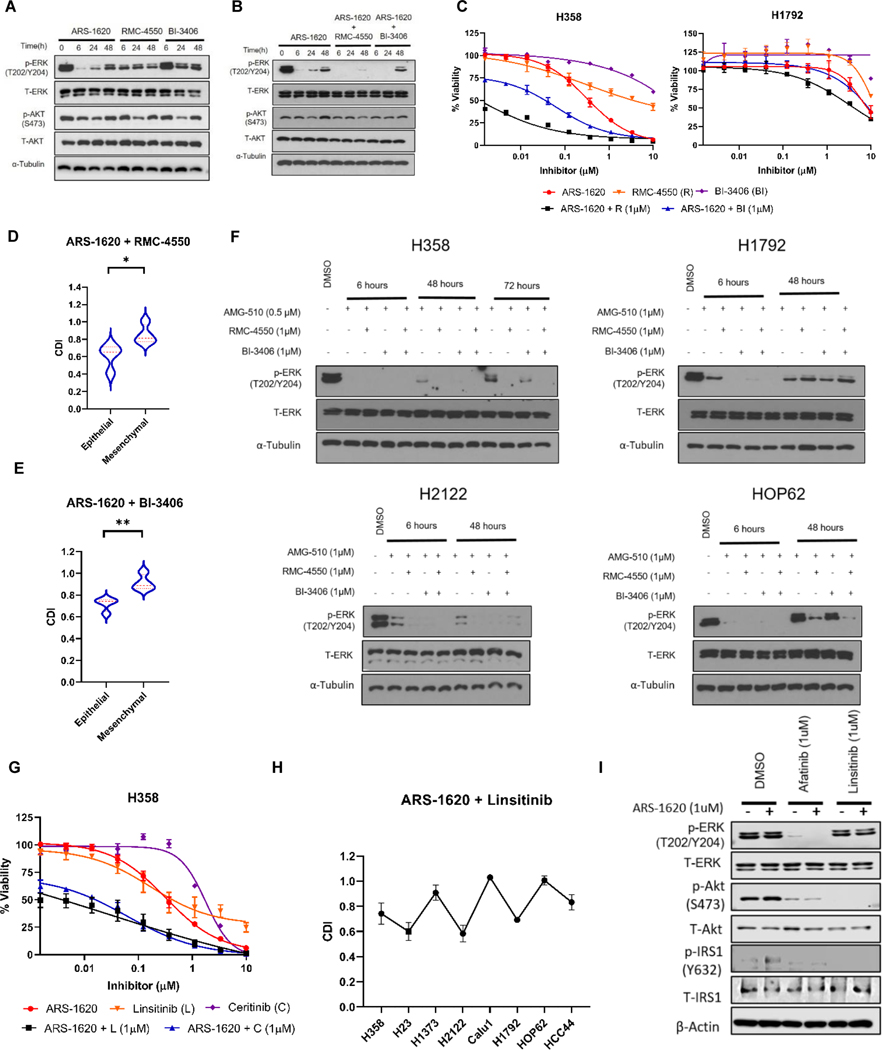
Our data identified rebound of MAPK signaling as one of the underlying mechanisms of resistance in the epithelial subtype (exemplified by H358 cells). In one of such signaling event, we saw decreased phosphorylation of SOS1 (on S1134) only in H358 cells (phospho-peptide not detected in H1792 and Calu1), which relaxes allosteric inhibition of SOS1 (27). We hypothesized that the epithelial phenotype could also be important in affecting cell sensitivity to SHP2 and SOS1 inhibition in combination with KRASG12C inhibition and recapitulate effects observed with afatinib. SHP2 inhibition using RMC-4550 (10) and SOS1 inhibition using BI-3406 (35) as single agents incompletely suppress ERK phosphorylation at any treatment time point when compared to KRASi in H358 cells (Fig. 4A). Individual inhibition of SHP2 or SOS1 is not effective to induce reduction in cell viability in KRASG12C cell lines (Supplementary Fig. S5A-C). Longer suppression of pERK with ARS-1620 was observed when combined with SHP2 and SOS1 inhibitors (Fig. 4B), though pERK rebound was observed with SOS1i at 48 hrs. We observed no effects on pAKT expression which is consistent with previous studies indicating that Akt signaling is independent of SHP2 (36). Cell viability analysis indicates that combining SHP2 or SOS1 inhibitors sensitizes H358 cells to KRASG12C inhibition, but not H1792 cells (Fig. 4C; Supplementary Fig. S5D-E). The cell viability analysis in other KRASG12C NSCLC cells indicates that epithelial subtypes showed greater synergistic effects to SHP2 and SOS1 inhibitor when combined with ARS-1620 (Fig. 4D-E; Supplementary Fig. S5F-G). Of note, complete inhibition of ERK signaling using three-drug combination (KRASi, SHP2i and SOS1i) suppressed pERK expression only in epithelial subtype, while pERK rebound was still observed in mesenchymal cells (Fig. 4F). These results suggest targeting SHP2 and SOS1, downstream of HER signaling, also has combination efficacy within the epithelial subtype.
Figure 4. SHP2 and SOS1 inhibitor(s) recapitulates efficacy observed with combining pan-HER inhibitor with ARS-1620 in epithelial subtypes. IRS1, a common signaling node mediating HER/IGF1R mediated cell survival.
A) Immunoblot analysis of p-ERK and p-AKT in H358 cells after 6, 24 and 48 hrs of treatment with ARS-1620 (1μM), RMC4550 (1μM) and BI-3406 (1μM) treatment. B) H358 cells were treated with ARS-1620 (1μM) and in the combination with RMC4550 (1μM) or BI-3406 (1μM); and expression of p-ERK and p-AKT was monitored after 6, 24 and 48 hrs by immunoblotting. C) H358 and H1792 - dose response to ARS-1620, RMC4550 and BI-3406 alone or ARS-1620 combination with RMC4550 (1μM) or BI-3406 (1μM) after 96 hrs of treatment and analysis using CellTiter-Glo. D-E) NSCLCG12C mutant cell lines were analyzed by CellTiter-Glo after 96 hrs of ARS-1620 (1μM) treatment alone and in combination with RMC4550 (1μM) or BI-3406 (1μM). The data was normalized to DMSO control. The CDI was calculated for both the combination treatments: ARS-1620 + RMC-4550 and ARS-1620 + BI-3406. The Student’s t-tests were used to calculate the significant difference in CDI between epithelial and mesenchymal KRASG12C mutant NSCLC cell lines. F) H358, H1792, H2122 and HOP62 cells were treated with AMG-510 alone and in combination with RMC4550 and/or BI-3406 (1μM). Immunoblot analysis of p-ERK was performed after different treatment time points. G) H358 dose response to ARS-1620, linsitinib and ceritinib alone or ARS-1620 combination with linsitinib (1μM) or ceritinib (1μM) after 96 hrs of treatment and analysis using CellTiter-Glo. H) Lung cancer KRASG12C mutant cell lines were analyzed by CellTiter-Glo after 96 hrs of ARS-1620 (1μM) treatment alone and in combination with linsitinib (1 μM). The data was normalized to DMSO control and CDI for ARS-1620 + linsitinib combination was plotted for all cell lines. I) H2122 cells were treated with ARS-1620, afatinib, linsitinib or the indicated combinations, and expression of pIRS1 (Y632), pAkt (473) and pERK (T202/Y204) was monitored after 24 hrs by immunoblotting. The values in figure panels (C), (D), (E), (G) and (H) represent average of three biological replicates. Unpaired Student’s t-tests were used to calculate p values: * p < 0.05, ** p < 0.01. *** p < 0.001.
We also observed that KRASG12C inhibition in H358 also increased phosphorylation on IRS1, an adaptor protein for IGF1R, on multiple sites. Inhibition of IGF1R-mediated signaling with linsitinib was previously reported to sensitize KRASG12C cell lines to ARS-1620 (13). We sought to extend these findings using ceritinib, a FDA approved inhibitor of ALK which also has activity against IGF1R (37). Both ceritinib and linsitinib combinations with ARS-1620 demonstrated reduced cell viability (Fig. 4G). Next, we assessed the effect of the linsitinib plus ARS-1620 combination on cell viability in other KRASG12C models (Supplementary Fig. S5H). Our data suggest no significant difference in CDI between epithelial and mesenchymal models to linsitinib combination with ARS-1620, where 3 epithelial (H358, H23, H2122) and 1 mesenchymal model (H1792) showed synergistic effects to linsitinib combination (Fig. 4H).
IRS1 is also known to interact with ERBB family kinases further activating the ERBB-PI3K-Akt axis (38,39). KRASG12C inhibition in combination with either afatinib or linsitinib effectively suppressed cell growth in some KRASG12C lung cancer models, we hypothesized that IRS1 was common signaling node downstream of ERBB and IGF receptor signaling. Our proteomics data identified increased phosphorylation of IRS1 (on Y632) following ARS-1620 treatment; hence, we assessed IRS1 signaling after combining KRASi with pan-HER or IGF1R inhibitor (Fig. 4I). Afatinib and linsitinib alone or in combination with ARS-1620 were able to reduce phosphorylation of IRS1. We observed that KRASi/afatinib combination decreased phosphorylation of Akt and ERK, while KRASi/linsitinib combination reduced only Akt phosphorylation. These observations suggest that IRS1 mediates signaling downstream of activated ERBB and IGF receptors regulate Akt signaling to sustain cell survival following KRASG12C inhibition.
FGFR1 activation drives resistance to ARS-1620 in H1792 cells
We next focused on H1792 cells since these cells presented with a more mesenchymal phenotype and showed modest response to dual pan-HER/KRASG12C or SHP2/KRASG12C inhibition. Phosphoproteomics analysis identified 140 pY and 1,941 pSTY modulated by ARS-1620. The MetaCore pathway analysis (Supplementary Fig. S6A) indicate inhibition of ERK signaling, but a well-connected activation network surrounding ABL1, CDK5, PRKCD, EGFR, RPS6KB1, YY1 and MYC was identified. While many proteins appeared to be inter-connected, identification of the upstream source of the signaling was quite challenging.
To gain more in depth understanding of the signaling cascade, we manually curated the network solely on perturbed phosphosites (Fig. 5A). We identified increased phosphorylation of FGFR1 (Y653/Y654) and FGFRL1 (Y502). We also identified perturbation in other components of the FGFR signaling axis, which include increased phosphorylation of PLCG1 (Y977), ABL1 (Y393), and CDK5 (Y15). Increased phosphorylation of multiple phosphosites on RPS6, RPS6KB1 and RPS6KB2 and EIF4EBP1 indicated activation of mTOR signaling. These results are consistent with other observations that MEK inhibition can enhance phosphorylation of the FGFR adaptor protein FRS2, serving a role in adaptive resistance in KRAS mutants (40,41). We also observed increased phosphorylation of FRS2 at Y436 in H1792 following ARS-1620 treatment (Fig. 5B).
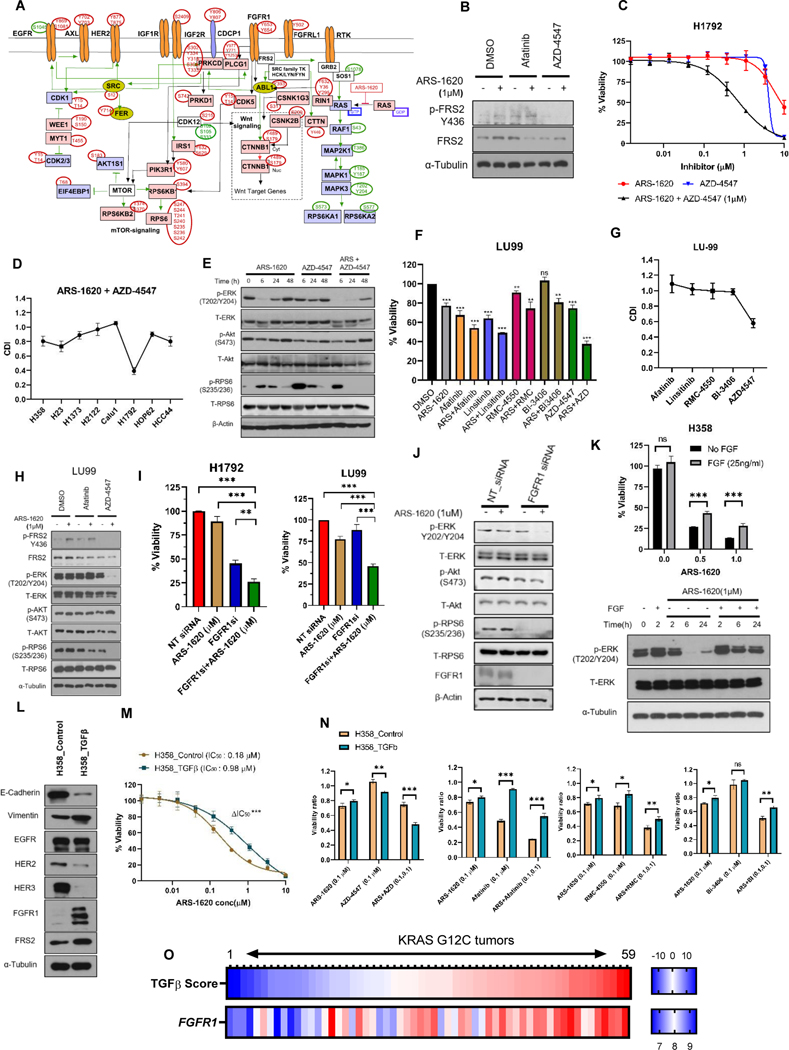
Figure 5. The rebound activation of FGFR signaling cause resistance to KRASG12C specific inhibitors in KRASG12C lung cancer models with mesenchymal sub-type.
A) Depiction of manually curated signaling rebound network in H1792 cells based on experimentally affirmed signaling information available in literature for phosphosites differentially perturbed after KRASG12C inhibition. B) H1792 cells were treated with ARS-1620, afatinib, AZD-4547 or the indicated combinations, and expression of pFRS2 (Y436) was monitored after 24 hrs by immunoblotting. C) H1792 - dose response to ARS-1620 and AZD-4547 alone or ARS-1620 combination with AZD-4547 (1μM) after 96 hrs of treatment and analysis using CellTiter-Glo. D) KRASG12C mutant cell lines were analyzed by CellTiter-Glo after 96 hrs of ARS-1620 (1μM) treatment alone and in combination with AZD-4547 (1μM). The data was normalized to DMSO control and the CDI was plotted for all KRASG12C mutant NSCLC cell lines. E) H1792 cells were treated with ARS-1620 (1μM), AZD-4547 (1μM), or in the combination, and rebound in signaling was monitored after 6, 24 and 48 hrs by immunoblotting with the indicated antibodies. F-G) LU-99 cells treated with ARS-1620 (1μM), afatinib (1μM), linsitinib (1μM), RMC-4550 (1μM), BI-3406 (1μM) and AZD-4547 (1μM) or the indicated combinations. The cell viability was assessed after 96 hrs of treatment using CellTiter-Glo and data was normalized to DMSO control and statistical analysis performed using Unpaired Student’s t-tests. CDI was plotted for each combination treatment. H) LU-99 cells were treated with ARS-1620, afatinib, AZD-4547 or the indicated combinations, and rebound in signaling was monitored after 24 hrs by immunoblotting with the indicated antibodies. I-J) H1792 and/or LU-99 cells were transfected with either non-targeting (NT) control or siRNAs targeting FGFR1 for 24 hrs and then treated with ASR-1620 (1uM). The cell viability was checked after 96 hrs of treatment using CellTiter-Glo. Signaling analysis was performed after 24 hrs of ARS-1620 treatment using antibodies against indicated proteins. FGFR1 protein expression was checked to confirm siRNA-based gene silencing. K) H358 cells were serum starved (0.5% FBS) overnight and then stimulated with FGF (25 ng/ml) for 2 hrs. The cells (stimulated and un-stimulated) were then treated with ARS-1620 at two different concentrations and cell viability was assessed after 96 hrs of treatment using CellTiter-Glo. For signaling analysis, following FGF stimulation cells were treated with ARS-1620 (1μM) and p-ERK rebound was monitored after 24 hrs by immunoblotting. L) H358 cells were chronically treated for 14 days with TGFβ (4ng/ml). H358-TGFβ and untreated (H358-control) cells were checked for expression of EMT markers, RTKs, FRS2 by immunoblotting. M) H358-TGFβ and H358-control: dose response to ARS-1620. Assay readout after 96 hrs using CellTiter-Glo. Unpaired Student’s t-tests was performed to show statistically significant difference (ΔIC50) in IC50s of H358-TGFβ and H358-control. N) H358-TGFβ and H358-control cells treated with ARS-1620, AZD-4547, afatinib, RMC-4550, BI-3406 alone and in indicated combinations. The cell viability was assessed after 96 hrs using CellTiter-Glo. Data was normalized to DMSO control. O) The Heatmap representing TGFβ-EMT score and FGFR1 gene expression (probe value) in KRASG12C mutants (n=59) in SPORE442 dataset. The values in figure panels (C), (D), (F), (G), (I), (K), (M) and (N) represent average of three biological replicates; error bars denote SD. Unpaired Student’s t-tests were used to calculate p values: * p < 0.05, ** p < 0.01. *** p < 0.001.
We next checked the efficacy of AZD-4547 and ARS-1620 combination on cell viability in KRASG12C lung cancer cell lines. Inhibition of FGFR1 signaling using FGFR kinase inhibitor AZD-4547 inhibited tyrosine phosphorylation of FRS2 as expected (Fig. 5B). AZD-4547 effectively enhances the sensitivity of ARS-1620 in H1792 (Fig. 5C-D, Supplementary Fig. S6B). Western blotting indicated that inhibition of FGFR1 delayed pERK rebound; however, decrease in pAKT was not observed. Further, combining AZD-4547 with ARS-1620 repressed mTOR signaling indicated by decrease in RPS6 phosphorylation (Fig. 5E). Notably, we observed similar observations with AZD-4547 combination on cell viability and signaling in another mesenchymal model LU99 (Fig. 5F-H). siRNA-based knockdown of FGFR1 in combination with KRASi also suppressed cell viability; and decreases ERK and RPS6 phosphorylation (Fig. 5I-J). Next, we checked impact of exogenous FGF2 stimulation on KRASi sensitivity and signaling. FGF2 stimulation reduced the sensitivity of H358 cells to ARS-1620 and only moderate suppression in pERK was observed with ARS-1620 (Fig. 5K).
To more directly determine the impact of EMT on sensitivity to KRASG12C inhibition and different combination strategies, H358 cells were chronically treated with TGF-β (41). The induction of EMT, as indicated by enhanced expressed of VIM and suppressed expression of CDH1 (Fig. 5L) reduced the sensitivity to ARS-1620 (Fig. 5M). Our results indicate substantial rewiring of RTKs during epithelial to mesenchymal transition in H358 cells, where we observed reduced expression of HER2 and HER3; and enhanced expression of FGFR1 and FRS2 in the mesenchymal cells. Of note, this observation was consistent with the cell viability analysis where EMT-induced H358 cells become sensitive to both – signal-agent treatment with FGFR1 inhibitor and in combination with ARS-1620 (Fig. 5N). Consistently, we observed decreased sensitivity to pan-HER inhibitor when treated standalone and with ARS-1620 combination. Consistent with our earlier results, sensitivity to dual SHP2i/KRASi and SOSi/KRASi combination treatments also reduced in the mesenchymal H358 cells (Fig. 5N). However, sensitivity to dual pan-HERi/KRASi and SHP2i/KRASi combinations remained unchanged when treated at higher doses of drugs combination (Supplementary Fig. S7 A-D).
Next, we analyzed our proteomics and CCLE RNAseq data (33) of KRASG12C cell lines for FGFR signaling signatures. We observed highest expression of FGF2 protein/gene expression in H1792 and LU99 cells (Supplementary Fig. S8 & Supplementary Table S7). We also observed higher phosphorylation (Y502) and expression of FGFRL1 in H1792 in our proteomics data (Supplementary Fig. S8A). Next, we analyzed CCLE RNAseq data which enabled inclusion of more KRASG12C models where a positive correlation was observed between FGFR1 and TGF-β EMT score (r = 0.72 and p = 0.002) (Supplementary Table S7). Also, we observed significant difference in FGFR1 gene in 2 group (E vs M) comparison (Supplementary Fig. S8B). Finally, we asked if FGFR1 expression in KRASG12C human lung cancer tumor tissues co-relate with the mesenchymal sub-type. We observed a positive correlation between FGFR1 and TGF-β EMT score (r = 0.707 and p = 4.04E-10) (Fig. 5O & Supplementary Table S9). Our results show significant role of FGFR1 signaling in mesenchymal type KRASG12C tumors and we show synergistic effects with dual FGFRi/KRASi combination treatment in a sub-group of KRASG12C models with mesenchymal type.
Phosphoproteomics identified AXL receptor-mediated adaptive rewiring to KRASG12C inhibition
Our results showed that inhibition of IGF1R and FGFR1 does not sensitize Calu1 to ARS-1620 treatment. In addition, combination with a pan-HER or SHP2 inhibitor had only moderate response (Figure 6A). Immunoblot-based signaling analysis also indicated that SHP099 alone or in combination with ARS-1620 does not effectively suppress p-ERK expression (Figure 6B). Our phosphoproteomics data identified 208 pY and 1,353 pSTY phosphosites that were modulated by ARS-1620. The MetaCore pathway analysis on phosphosites perturbed at 6 hrs (Supplementary Fig. S9A) indicate inhibition of ERK signaling, yet other signaling were up-regulated. A well-connected signaling network of PI3K-AKT-mTOR was identified which involves IRS1, IRS2, AKT1, AKT2, RPS6KB1, RPS6, and EIF4B among other nodes.
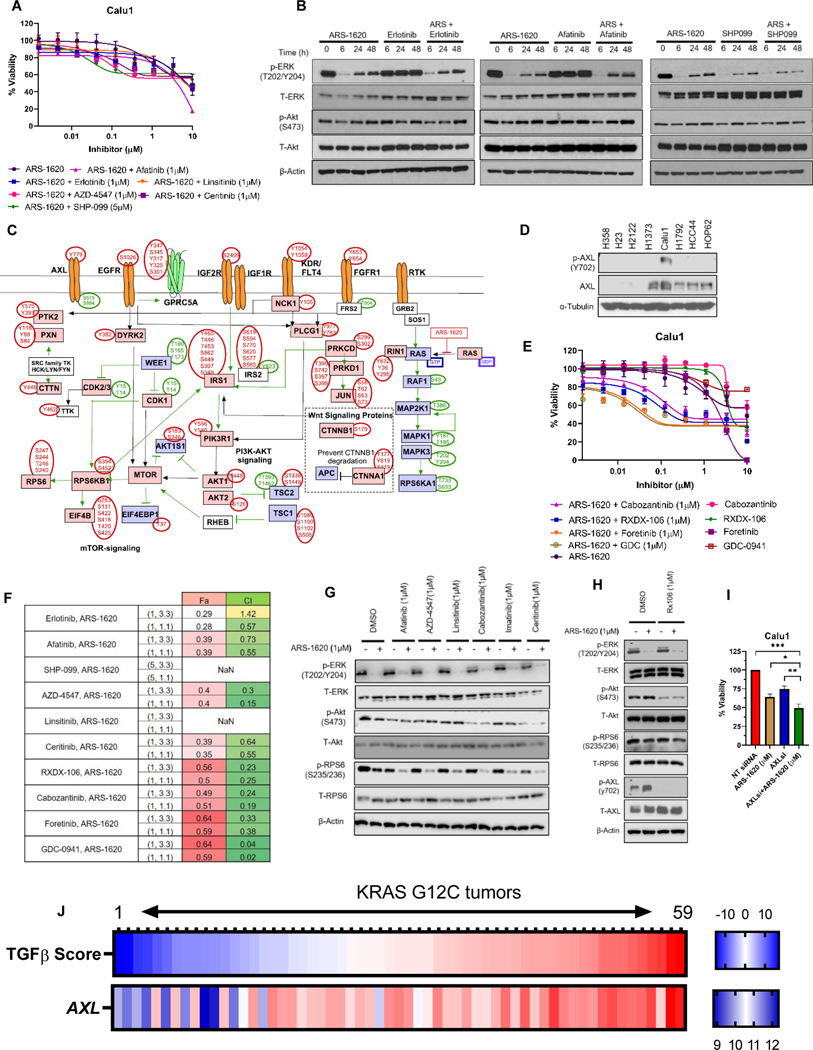
Figure 6. Phosphoproteomics identified activation AXL receptors following KRASG12C inhibition in Calu1.
A) Calu1 cells were analyzed by CellTiter-Glo after 96 hrs of ARS-1620 treatment alone and in combination with indicated drugs. B) Calu1 cells were treated with ARS-1620 (1μM), erlotinib (1μM), afatinib (1μM), SHP-099 (5μM) or the indicated combinations and rebound in p-ERK and p-Akt was monitored after 6, 24 and 48 hrs by immunoblotting with the indicated antibodies. C) Depiction of manually curated signaling rebound network in Calu1 based on phosphosites-based literature evidences. D) Cell lysates from KRASG12C mutant cell lines were probed with indicated antibodies. E-F) Calu1 - dose response to ARS-1620, AXL inhibitors (RXDX-106, foretinib and cabozantinib) and PI3K-AKT inhibitor (GDC-0941) alone or ARS-1620 combination with AXL inhibitors (1μM) or PI3K-AKT inhibitor (1μM) after 96 hrs of treatment and analysis using CellTiter-Glo. CI and Fa was calculated using CompuSyn software. G) Signaling analysis in Calu1 after 24 hrs of ARS-1620 (1μM), afatinib (1μM), AZD-4547(1μM), linsitinib (1μM), cabozantinib (1μM), imatinib (1μM), ceritinib (1μM) treatment alone and ARS-1620 combination with indicated drug. H) Calu1 cells were treated with ARS-1620 and RXDX-106 alone and in combination ARS + RXDX-106 (1μM) combination for 24 hrs and immunoblotted with indicated antibodies. I) Calu1 cells were transfected with either non-targeting (NT) control or siRNAs targeting AXL for 24 hrs and then treated with ASR-1620 (1uM). The cell viability was checked after 96 hrs of treatment using CellTiter-Glo. J) The Heatmap representing TGFβ-EMT score and AXL gene expression (probe value) in KRASG12C mutants (n=59) in SPORE442 dataset. The values in figure panels (A), (E), (F) and (I) represents average of three biological replicates; error bars denote SD. Unpaired Student’s t-tests were used to calculate p values: * p < 0.05, ** p < 0.01. *** p < 0.001.
Resistance to combinations of ARS-1620 with drugs targeting HER, IGF1R, or FGFR in Calu1 led to more comprehensive re-evaluation of perturbed phosphoproteome with manual curation of the signaling network based on differentially expressed phosphosites (Fig. 6C). This strategy also indicated activation of PI3K-AKT pathway. Interestingly, we identified increased phosphorylation of AXL (Y779). We also observed higher phosphorylation and expression (both protein & gene) of AXL in Calu1 when compared with other cell lines (Fig. 6D & Supplementary Table S7). For this reason, we sought to check whether inhibition of AXL can sensitize Calu1 to ARS-1620 treatment. The drug response curves indicated that combination treatment with AXL inhibitor(s) enhances sensitivity to ARS-1620 in Calu1 cells. In addition, our data was in concordance with previous study where PI3K inhibitor showed synergistic effects with KRASi in Calu1 and suppress p-Akt expression (Fig. 6E-F & Supplementary Fig. S9B).
Next, we sought to identify if AXL is the upstream signaling source to PI3K-AKT pathway in Calu1. Signaling analysis indicates that cabozantinib and Foretinib alone or in combination with ARS-1620 was able to reduce p-AXL and p-Akt (Fig. 6G & Supplementary Fig. S9C). AXL inhibition using other small molecule inhibitor RXDX-106 alone or in combination with ARS-1620 was also able to reduce Akt phosphorylation in Calu1 cells (Fig. 6H), but not in H358 and H1792 cells (Supplementary Fig. S9D). The siRNA-based knockdown of AXL in combination with KRASi also suppressed cell viability (Fig. 6I); and decreases pAKT expression (Supplementary Fig. S9E-F). Interestingly, ceritinib showed similar results, attributing to its potential of simultaneously inhibit multiple pathways, including insulin, mTOR, adherens junction, and focal adhesion signaling (37). Combination with other RTK inhibitors (afatinib, AZD-4547, linsitinib and imatinib) did not alter phosphorylation of Akt.
Next, we also determined if AXL expression in KRASG12C human lung cancer tumor tissues associate with the mesenchymal sub-type, similar to what we observed with positive association with FGFR1 and mesenchymal subtype. We observed a positive correlation (r = 0.75 and p = 9.04E-12) between AXL and TGF-β EMT score (Fig. 6J & Supplementary Table S9). Our data suggest another subtype within mesenchymal models where AXL inhibitors-mediated combination strategies with KRASi merits further investigation, and these results suggest the general utility of unbiased phosphoproteomics to elucidate adaptive resistance mechanisms to KRASi.
DISCUSSION
While several combination regimens with KRASi have been suggested, our study describes the various mechanisms of signaling rebound and the mechanistic insight of these combination strategies (11–16). Our phosphoproteomics analysis indicated that each KRASG12C NSCLC model (e.g. H358, H1792, and Calu1 cells) underwent unique adaptive signaling events to KRASG12C inhibition. These results, along with signaling studies of tissues from KRAS mutant GEMM models, argues strongly that cell context will dictate responses to KRASG12C inhibition (42). Further, comparison of proteomes and phosphoproteomes of 8 different KRASG12C lung cancer models indicate that pre-existing transcription states are likely to influence response to KRASG12C inhibitors. As depicted in model (Fig. 7) describing signaling adaptations to ARS-1620 in NSCLC, the epithelial subtype typically adapted to KRASG12C targeted agents by activating ERBB signaling. Conversely, the mesenchymal subtype showed activation of the FGFR or AXL signaling cascade. This analysis suggests a personalized combination strategy with HER2/3 targeting agents along with KRASG12C inhibitors for epithelial subtypes, while FGFR or AXL inhibition should be considered for more mesenchymal tumors. We also observed similar differences in the transcriptome of other cell lines using CCLE data (33) and in KRASG12C tumor tissues taken directly from patients (34). Our results also raise the possibility to analyze the proteomes in re-biopsies of recurrent tumors following targeted therapy to capture adaptive resistance signaling and design personalized combination therapy.
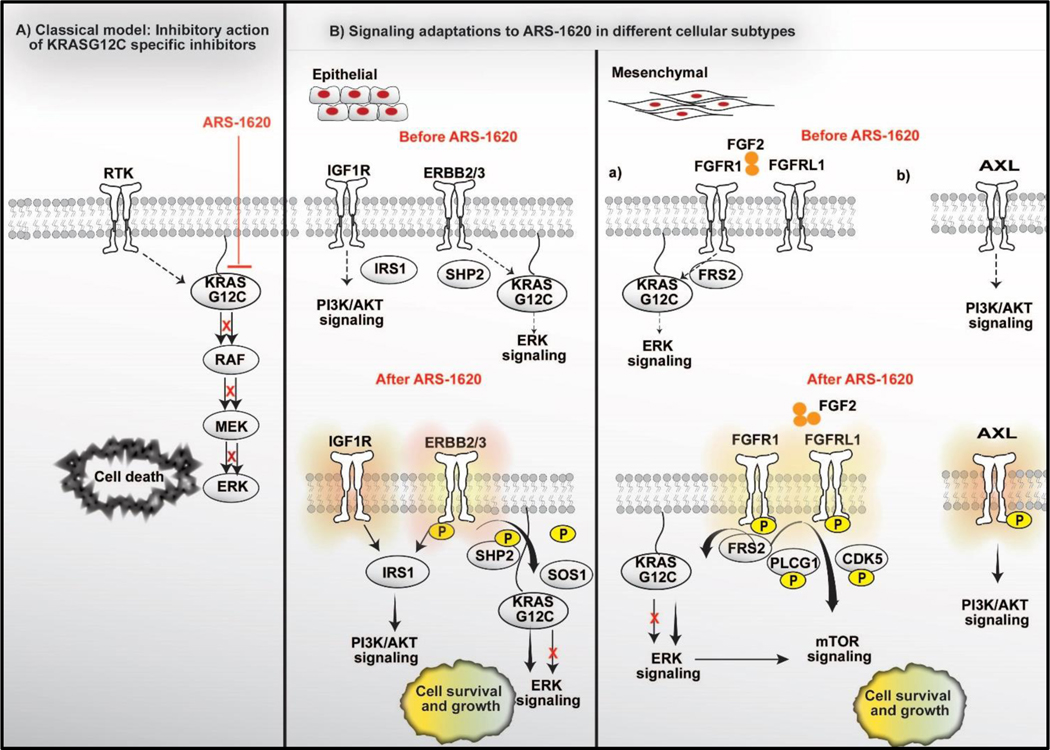
Figure 7. Schematics of the short-term adaptive resistance to KRAS specific inhibitor ARS-1620 via signaling network modulation.
A) A classical model depicting suppression of ERK Signaling by KRASG12C specific covalent inhibitors in tumors with KRASG12C mutation. B) The mechanism of adaptive signaling differs in tumor cells in epithelial vs. mesenchymal states. Epithelial subtypes do have support from HER signaling prior treatment with KRASG12C inhibitor. KRASG12C inhibition further enhances HER signaling support for survival and growth. IRS1 acts a major signaling hub downstream of HER and IGF receptors. Thus, Epithelial subtypes benefit from dual pan-HER/KRASi combination. In contrast, we speculate possibilities of multiple subtypes within mesenchymal type of KRASG12C tumors, where each sub-type might have experienced specific rewiring in signaling during epithelial to mesenchymal transformation. Specifically, within mesenchymal type, our analysis identified activation of two major signaling pathways: (a) FGFR and (b) AXL signaling, likely representing two different sub-group in mesenchymal type. The rebound activation of FGFR signaling cause therapy resistance by regulating MAPK and mTOR signaling, while rebound in AXL signaling cause activation of PI3K-AKT signaling. These specific subtypes identified within mesenchymal models also benefit from FGFR or AXL inhibitor-based combination strategy with KRASG12C inhibitor.
The altered phosphorylation of SOS1, SHP2 and RAF1 show rebound of MAPK signaling as one of the underlying mechanisms of resistance in the epithelial subtype. SHP2 inhibitors have been suggested as an effective combination strategy with KRASi to disrupt RAS activation by RTKs (16). Our results suggest that dual SHP2/KRASG12Ci and SOS1/KRASG12Ci strategies will provide more benefit to patients with epithelial subtype tumors and this should be examined in ongoing trials. We also observed increased phosphorylation of RAF1 (S621) following KRAS inhibition. Phosphorylation of RAF1 at S621, a site for PRKAA1, enhances its interaction with the cross-linker protein, 14-3-3, which stabilizes RAF1/BRAF heterodimers (28,43). In the context to RAS-driven cancer, concomitant suppression of RAF1 and MEK leads to persistent ERK suppression and enhance apoptosis (44). Thus, development of specific RAF1 inhibitors and determination of their synergistic effects with KRASG12C inhibitors on growth inhibition will be important.
The genetic ablation of IRS1 and IRS2 in KRAS-driven genetically engineered mouse models (GEMM) of lung cancer suppresses Akt signaling and delays lung tumorigenesis (45). As compared to xenograft models, GEMM models reliably recapitulate the tumor pathology in epithelial cancers and allow better exploitation of signaling mechanisms to understand process of tumorigenesis (46,47). Given the importance of IRS signaling downstream of ERBB and IGF receptors following KRAS inhibition, our data explain the mechanistic insight of effective PI3K/AKT inhibitor combination strategies and provides a rationale for co-targeting HER2/3 or IGF1 receptors along with KRASi in epithelial subtype of KRASG12C mutants.
Our analysis identified activation of FGFR1 and FGFRL1 signaling as one of the mechanism of therapy resistance in mesenchymal subtype (exemplified by H1792 cells), which provides mechanistic basis to co-target FGFR and KRASG12C. FGFRL1 lacks an intracellular kinase domain and consists of a SH2 domain to facilitate binding to signaling adaptors. FGFRL1 was identified to interact with RASG12V (48) and can enhance ERK phosphorylation either by association with SHP-1 or by enhancing FGFR1 activation to induce MEK-independent activation of ERK1/2 (49). Because, our analysis identified cell-type specific activation of FGFR1 signaling, future interrogations of tumors for FGFR pathway activation, possibly through FGFR proximity ligation assays (PLA) (18) or MRM-based proteomics methods (50) may help predicting personalized dual FGFR/KRASG12Ci treatment strategy. Based on phosphoproteomic signatures, we identified activation of AXL signaling, as an alternative mechanism of resistance in mesenchymal models. We identified higher FGF2 expression and FGFR1 phosphorylation on other NSCLC KRASG12C models such as HCC44 and HOP62, but they did not respond to the FGFR/KRASG12Ci combination. Thus, a more comprehensive phosphoproteomic analysis is required in these models to identify other mechanisms of resistance to KRASG12C inhibitors. Our data set is also likely to harbor other mechanisms and targets of adaptive resistance to KRASG12C inhibitors. For example, we observed many perturbations on phosphosites associated with Wnt signaling proteins (CTNNB1, CTNNA1, CSNK1G3 and CSNK2B) and G Protein-Coupled Receptors (GPCRs). Thus, phenotypic consequences of these perturbations in KRASG12C models remain important to investigate in further detail.
Our data highlighted the importance of mass spectrometry-based proteomics to read out system-wide changes in signaling events to understand the diversity of drug responses based on protein expression and post-translational modifications. Our study provides a more comprehensive view of phosphoproteome alterations to KRASG12C inhibition in various lung cancer models with high clinical relevance to consider biomarker development strategies to suggest clinically effective combinations in diverse group of KRASG12C mutant lung cancer. Further, our analysis raises the possibilities to utilize applications of phosphoproteomics in biopsy/re-biopsy samples, organoids and PDX models to aid personalized approach to cancer care.
Supplementary Material
Translational Relevance:
KRASG12C inhibitors (AMG-510 and MRTX849) have demonstrated efficacy in KRASG12C mutant lung cancer however variability in tumor response and activation of bypass signaling are limiting efficacy. While combination strategies (MEK inhibitor, SHP2 inhibitor, pan-HER inhibitor, and EGFR inhibitor) with KRASG12C specific inhibitors are being investigated in clinical trials, how to best identify responsive tumors for each combination remains a clinical challenge. Our data suggest that depicting epithelial vs mesenchymal state of KRASG12C along with basal expression of ERBB family, FGFR1, and AXL may associate with combination efficacy secondary to mechanisms of adaptive resistance. We identify that ERBB2/3 signaling cause resistance to KRASG12C inhibitors in epithelial type, while FGFR1 or AXL signaling is more prominent in mesenchymal type. Our data highlight the importance of phosphoproteomics-based approach to identify tumor specific adaptive rewiring, which can be utilized to aid personalized patient care in KRASG12C mutants.
ACKNOWLEDGEMENTS
We thank AC Tan for useful discussions in designing TMT experiments. We thank Daniel Chen for his assistance in manuscript revision.
FINANCIAL SUPPORT
The work was supported by State of Florida Bankhead Coley Grant (5BC07) and the Moffitt Lung Cancer Center of Excellence. This work has been supported in part by the Proteomics & Metabolomics Core Facility and the Biostatistics and Bioinformatics Shared Resource at the H. Lee Moffitt Cancer Center& Research Institute, an NCI designated Comprehensive Cancer Center (P30-CA076292). LC-MRM assays for cancer signaling proteins were developed with funding from the NCI CPTAC and RAS Initiative through Leidos (14×270 to J.M.K.).
E.B. Haura reports receiving honorarium from Janssen Pharmaceuticals, and Amgen for serving on advisory boards.
Footnotes
DECLARATION OF INTEREST
No potential conflict of interest was disclosed by other authors.
REFERENCES:
- 1.Singh A, Settleman J. Oncogenic K-ras “addiction” and synthetic lethality. Cell Cycle 2009;8(17):2676–7 doi 9336 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba II, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014;311(19):1998–2006 doi 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedard PL, Tabernero J, Janku F, Wainberg ZA, Paz-Ares L, Vansteenkiste J, et al. A phase Ib dose-escalation study of the oral pan-PI3K inhibitor buparlisib (BKM120) in combination with the oral MEK1/2 inhibitor trametinib (GSK1120212) in patients with selected advanced solid tumors. Clin Cancer Res 2015;21(4):730–8 doi 10.1158/1078-0432.CCR-14-1814. [DOI] [PubMed] [Google Scholar]
- 4.Janes MR, Zhang J, Li LS, Hansen R, Peters U, Guo X, et al. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 2018;172(3):578–89 e17 doi 10.1016/j.cell.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Patricelli MP, Janes MR, Li LS, Hansen R, Peters U, Kessler LV, et al. Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State. Cancer Discov 2016;6(3):316–29 doi 10.1158/2159-8290.CD-15-1105. [DOI] [PubMed] [Google Scholar]
- 6.Hong DS, Fakih MG, Strickler JH, Desai J, Durm GA, Shapiro GI, et al. KRASG12C Inhibition with Sotorasib in Advanced Solid Tumors. New England Journal of Medicine 2020. doi 10.1056/NEJMoa1917239. [DOI] [PMC free article] [PubMed]
- 7.Jänne PA. A phase 1 clinical trial evaluating the pharmacokinetics (PK), safety, and clinical activity of MRTX849, a mutant-selective small molecule KRAS G12C inhibitor, in advanced solid tumors. Data presented at: AACR-NCI-EORTC International Conference on Molecular Targets; 2019 October 28; San Diego, California. [Google Scholar]
- 8.Nagasaka M, Li Y, Sukari A, Ou SI, Al-Hallak MN, Azmi AS. KRAS G12C Game of Thrones, which direct KRAS inhibitor will claim the iron throne? Cancer treatment reviews 2020;84:101974 doi 10.1016/j.ctrv.2020.101974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bivona TG. Dampening oncogenic RAS signaling. Science 2019;363(6433):1280–1 doi 10.1126/science.aav6703. [DOI] [PubMed] [Google Scholar]
- 10.Nichols RJ, Haderk F, Stahlhut C, Schulze CJ, Hemmati G, Wildes D, et al. RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nature cell biology 2018;20(9):1064–73 doi 10.1038/s41556-018-0169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019;575(7781):217–23 doi 10.1038/s41586-019-1694-1. [DOI] [PubMed] [Google Scholar]
- 12.Hallin J, Engstrom LD, Hargis L, Calinisan A, Aranda R, Briere DM, et al. The KRAS(G12C) Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov 2020;10(1):54–71 doi 10.1158/2159-8290.CD-19-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molina-Arcas M, Moore C, Rana S, van Maldegem F, Mugarza E, Romero-Clavijo P, et al. Development of combination therapies to maximize the impact of KRAS-G12C inhibitors in lung cancer. Science translational medicine 2019;11(510) doi 10.1126/scitranslmed.aaw7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lou K, Steri V, Ge AY, Hwang YC, Yogodzinski CH, Shkedi AR, et al. KRAS(G12C) inhibition produces a driver-limited state revealing collateral dependencies. Science signaling 2019;12(583) doi 10.1126/scisignal.aaw9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Misale S, Fatherree JP, Cortez E, Li C, Bilton S, Timonina D, et al. KRAS G12C NSCLC Models Are Sensitive to Direct Targeting of KRAS in Combination with PI3K Inhibition. Clin Cancer Res 2019;25(2):796–807 doi 10.1158/1078-0432.CCR-18-0368. [DOI] [PubMed] [Google Scholar]
- 16.Ryan MB, Fece de la Cruz F, Phat S, Myers DT, Wong E, Shahzade HA, et al. Vertical Pathway Inhibition Overcomes Adaptive Feedback Resistance to KRAS(G12C) Inhibition. Clin Cancer Res 2020;26(7):1633–43 doi 10.1158/1078-0432.CCR-19-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amodio V, Yaeger R, Arcella P, Cancelliere C, Lamba S, Lorenzato A, et al. EGFR Blockade Reverts Resistance to KRAS(G12C) Inhibition in Colorectal Cancer. Cancer Discov 2020;10(8):1129–39 doi 10.1158/2159-8290.CD-20-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith MA, Hall R, Fisher K, Haake SM, Khalil F, Schabath MB, et al. Annotation of human cancers with EGFR signaling-associated protein complexes using proximity ligation assays. Science signaling 2015;8(359):ra4 doi 10.1126/scisignal.2005906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandarlapaty S. Negative feedback and adaptive resistance to the targeted therapy of cancer. Cancer Discovery 2012;2(4):311–9 doi 10.1158/2159-8290.CD-12-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang N, Fu JN, Chou TC. Synergistic combination of microtubule targeting anticancer fludelone with cytoprotective panaxytriol derived from panax ginseng against MX-1 cells in vitro: experimental design and data analysis using the combination index method. Am J Cancer Res 2016;6(1):97–104. [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez-Acevedo M, Grace L, Teoh D, Whitaker R, Adams DJ, Jia J, et al. Dasatinib (BMS-35482) potentiates the activity of gemcitabine and docetaxel in uterine leiomyosarcoma cell lines. Gynecol Oncol Res Pract 2014;1:2 doi 10.1186/2053-6844-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology 2008;26(12):1367–72 doi 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 23.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic acids research 2019;47(D1):D442–D50 doi 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maclean B, Tomazela DM, Abbatiello SE, Zhang S, Whiteaker JR, Paulovich AG, et al. Effect of collision energy optimization on the measurement of peptides by selected reaction monitoring (SRM) mass spectrometry. Analytical chemistry 2010;82(24):10116–24 doi 10.1021/ac102179j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordian E, Welsh EA, Gimbrone N, Siegel EM, Shibata D, Creelan BC, et al. Transforming growth factor beta-induced epithelial-to-mesenchymal signature predicts metastasis-free survival in non-small cell lung cancer. Oncotarget 2019;10(8):810–24 doi 10.18632/oncotarget.26574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic acids research 2015;43(Database issue):D512–20 doi 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saha M, Carriere A, Cheerathodi M, Zhang X, Lavoie G, Rush J, et al. RSK phosphorylates SOS1 creating 14-3-3-docking sites and negatively regulating MAPK activation. The Biochemical journal 2012;447(1):159–66 doi 10.1042/BJ20120938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dhillon AS, Yip YY, Grindlay GJ, Pakay JL, Dangers M, Hillmann M, et al. The C-terminus of Raf-1 acts as a 14-3-3-dependent activation switch. Cellular signalling 2009;21(11):1645–51 doi 10.1016/j.cellsig.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 29.Tong J, Taylor P, Peterman SM, Prakash A, Moran MF. Epidermal growth factor receptor phosphorylation sites Ser991 and Tyr998 are implicated in the regulation of receptor endocytosis and phosphorylations at Ser1039 and Thr1041. Molecular & cellular proteomics : MCP 2009;8(9):2131–44 doi 10.1074/mcp.M900148-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawasaki Y, Sakimura A, Park CM, Tomaru R, Tanaka T, Ozawa T, et al. Feedback control of ErbB2 via ERK-mediated phosphorylation of a conserved threonine in the juxtamembrane domain. Scientific reports 2016;6:31502 doi 10.1038/srep31502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen CH, Hsia TC, Yeh MH, Chen TW, Chen YJ, Chen JT, et al. MEK inhibitors induce Akt activation and drug resistance by suppressing negative feedback ERK-mediated HER2 phosphorylation at Thr701. Mol Oncol 2017;11(9):1273–87 doi 10.1002/1878-0261.12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kruspig B, Monteverde T, Neidler S, Hock A, Kerr E, Nixon C, et al. The ERBB network facilitates KRAS-driven lung tumorigenesis. Science translational medicine 2018;10(446) doi 10.1126/scitranslmed.aao2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghandi M, Huang FW, Jane-Valbuena J, Kryukov GV, Lo CC, McDonald ER, 3rd, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019;569(7757):503–8 doi 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schabath MB, Welsh EA, Fulp WJ, Chen L, Teer JK, Thompson ZJ, et al. Differential association of STK11 and TP53 with KRAS mutation-associated gene expression, proliferation and immune surveillance in lung adenocarcinoma. Oncogene 2016;35(24):3209–16 doi 10.1038/onc.2015.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marco H Hofmann MG, Ramharter Jürgen, Savarese Fabio, Gerlach Daniel, Marszalek Joseph R et al. Discovery of BI-3406: A potent and selective SOS1::KRAS inhibitor opens a new approach for treating KRAS-driven tumors [abstract]. In: Proceedings of the AACR-NCI-EORTC International Conference on Molecular Targets and Cancer Therapeutics; 2019 Oct 26–30; Boston, MA Philadelphia (PA): AACR; Mol Cancer Ther 2019;18(12 Suppl):Abstract nr PL06–01 2019 doi 10.1158/1535-7163.TARG-19-PL06-01. [DOI] [Google Scholar]
- 36.Adachi Y, Ito K, Hayashi Y, Kimura R, Tan TZ, Yamaguchi R, et al. Epithelial-to-Mesenchymal Transition is a Cause of Both Intrinsic and Acquired Resistance to KRAS G12C Inhibitor in KRAS G12C-Mutant Non-Small Cell Lung Cancer. Clin Cancer Res 2020;26(22):5962–73 doi 10.1158/1078-0432.CCR-20-2077. [DOI] [PubMed] [Google Scholar]
- 37.Kuenzi BM, Remsing Rix LL, Stewart PA, Fang B, Kinose F, Bryant AT, et al. Polypharmacology-based ceritinib repurposing using integrated functional proteomics. Nature chemical biology 2017;13(12):1222–31 doi 10.1038/nchembio.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi HJ, Jin S, Cho H, Won HY, An HW, Jeong GY, et al. CDK12 drives breast tumor initiation and trastuzumab resistance via WNT and IRS1-ErbB-PI3K signaling. EMBO reports 2019;20(10):e48058 doi 10.15252/embr.201948058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knowlden JM, Gee JM, Barrow D, Robertson JF, Ellis IO, Nicholson RI, et al. erbB3 recruitment of insulin receptor substrate 1 modulates insulin-like growth factor receptor signalling in oestrogen receptor-positive breast cancer cell lines. Breast cancer research : BCR 2011;13(5):R93 doi 10.1186/bcr3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manchado E, Weissmueller S, Morris JPt, Chen CC, Wullenkord R, Lujambio A, et al. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature 2016;534(7609):647–51 doi 10.1038/nature18600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kitai H, Ebi H, Tomida S, Floros KV, Kotani H, Adachi Y, et al. Epithelial-to-Mesenchymal Transition Defines Feedback Activation of Receptor Tyrosine Kinase Signaling Induced by MEK Inhibition in KRAS-Mutant Lung Cancer. Cancer Discov 2016;6(7):754–69 doi 10.1158/2159-8290.CD-15-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brubaker DK, Paulo JA, Sheth S, Poulin EJ, Popow O, Joughin BA, et al. Proteogenomic Network Analysis of Context-Specific KRAS Signaling in Mouse-to-Human Cross-Species Translation. Cell Syst 2019;9(3):258–70 e6 doi 10.1016/j.cels.2019.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varga A, Ehrenreiter K, Aschenbrenner B, Kocieniewski P, Kochanczyk M, Lipniacki T, et al. RAF1/BRAF dimerization integrates the signal from RAS to ERK and ROKalpha. Science signaling 2017;10(469) doi 10.1126/scisignal.aai8482. [DOI] [PubMed] [Google Scholar]
- 44.Lamba S, Russo M, Sun C, Lazzari L, Cancelliere C, Grernrum W, et al. RAF suppression synergizes with MEK inhibition in KRAS mutant cancer cells. Cell reports 2014;8(5):1475–83 doi 10.1016/j.celrep.2014.07.033. [DOI] [PubMed] [Google Scholar]
- 45.Xu H, Lee MS, Tsai PY, Adler AS, Curry NL, Challa S, et al. Ablation of insulin receptor substrates 1 and 2 suppresses Kras-driven lung tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America 2018;115(16):4228–33 doi 10.1073/pnas.1718414115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kersten K, de Visser KE, van Miltenburg MH, Jonkers J. Genetically engineered mouse models in oncology research and cancer medicine. EMBO molecular medicine 2017;9(2):137–53 doi 10.15252/emmm.201606857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Hagan RC, Heyer J. KRAS Mouse Models: Modeling Cancer Harboring KRAS Mutations. Genes & cancer 2011;2(3):335–43 doi 10.1177/1947601911408080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adhikari H, Counter CM. Interrogating the protein interactomes of RAS isoforms identifies PIP5K1A as a KRAS-specific vulnerability. Nature communications 2018;9(1):3646 doi 10.1038/s41467-018-05692-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silva PN, Altamentova SM, Kilkenny DM, Rocheleau JV. Fibroblast growth factor receptor like-1 (FGFRL1) interacts with SHP-1 phosphatase at insulin secretory granules and induces beta-cell ERK1/2 protein activation. The Journal of biological chemistry 2013;288(24):17859–70 doi 10.1074/jbc.M112.440677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Y, Fisher KJ, Lloyd M, Wood ER, Coppola D, Siegel E, et al. Multiplexed Liquid Chromatography-Multiple Reaction Monitoring Mass Spectrometry Quantification of Cancer Signaling Proteins. Methods Mol Biol 2017;1647:19–45 doi 10.1007/978-1-4939-7201-2_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.