Abstract
The epigenome plays an important role in shaping phenotypes. However, whether the environment can alter an organism’s phenotype across several generations through epigenetic remodeling in the germline is still a highly debated topic. In this chapter, we briefly review the mechanisms of epigenetic inheritance and their connection with germline development before highlighting specific developmental windows of susceptibility to environmental cues. We further discuss the evidence of transgenerational inheritance to a range of different environmental cues, both epidemiological in humans and experimental in rodent models. Doing so, we pinpoint the current challenges in demonstrating transgenerational inheritance to environmental cues and offer insight in how recent technological advances may help deciphering the epigenetic mechanisms at play. Together, we draw a detailed picture of how our environment can influence our epigenomes, ultimately reshaping our phenotypes, in an extended theory of inheritance.
1. Introduction
The definition and associated understanding of “epigenetics” have dramatically shifted since the 1940’s coining of the term epigenotype by Waddington (Waddington, 2012). Nearly 60 years later, the currently most accepted definition of epigenetics was put forth by Wu and Morris (Wu & Morris, 2001). They proposed that epigenetics should be defined as: “the study of changes in gene function that are mitotically and/or meiotically heritable and that do not entail a change in DNA sequence.” As evidenced by the extensive correspondence associated with this proposal (Wu & Morris, 2001), the introduction of the concept of heritability in epigenetics was quite remarkable. Indeed, while epigenetic mechanisms operate on a short timescale for gene regulation, the potentially long-lasting nature of some epigenetic modifications has garnered significant attention for its ability to explain biological phenomena that unfold across several generations.
Contrasting with the relative stability of epigenetic marks, the epigenome is remarkably malleable and sensitive to environmental influences. Thus, epigenetics is intimately tied to the research into the Developmental Origins of Health and Disease (DOHaD), which aims to uncover the mechanisms by which early life (pre- or peri-natal) exposures may condition the health of an individual over their life course (Barrett, 2017; Bianco-Miotto, Craig, Gasser, van Dijk, & Ozanne, 2017; Ong, Lin, & Holbrook, 2015). In the context of an in utero (i.e., prenatal) exposure directly altering the health outcomes of the next generation, this phenomenon would call for an intergenerational role for the epigenome. Even more remarkable is the wealth of evidence, in a variety of species, indicating that many different types of environmental cues elicit epigenetic changes and phenotypic alterations over several generations. In many instances, these health outcomes are detected in generations beyond the point at which any directly exposed cells remain. This mechanism of epigenetic inheritance is termed “transgenerational” and is the most intriguing since it implies a transfer of biological information by germ cells independently of DNA sequences, yet, acting on gene expression and functioning in parallel with sequence-based regulation. Perhaps unsurprisingly, transgenerational epigenetic inheritance (TEI) has attracted both significant research interest and controversy when applied to humans (Breton et al., 2021).
In this chapter, we explore the molecular mechanisms underlying TEI. We first review the epigenetic mechanisms of inheritance, then discuss their involvement in germ cells development. We then focus on the evidence for TEI, but also highlight the knowledge gaps that require significant research investment. We end this chapter with forward-thinking approaches that we propose to deploy to address the remaining black boxes in the field.
2. Epigenetic mechanisms of inheritance
Epigenetic mechanisms make use of reversible modifications of chromatin states that impact gene expression (Bird, 2007; Murr, 2010). Such modifications include chemical changes of the DNA itself, in particular DNA methylation, changes in histones post-translational modifications, changes in nucleosomes position and composition, non-coding RNA (ncRNA)-mediated modifications of the chromatin structure and changes in high-order chromatin structure within the nucleus (Fig. 1). Importantly, epigenetic states are heritable and ensure the maintenance and long-term stability of gene expression across cell division and during development (Almouzni & Cedar, 2016). Here, we briefly review known mechanisms of epigenetic inheritance.
Fig. 1.
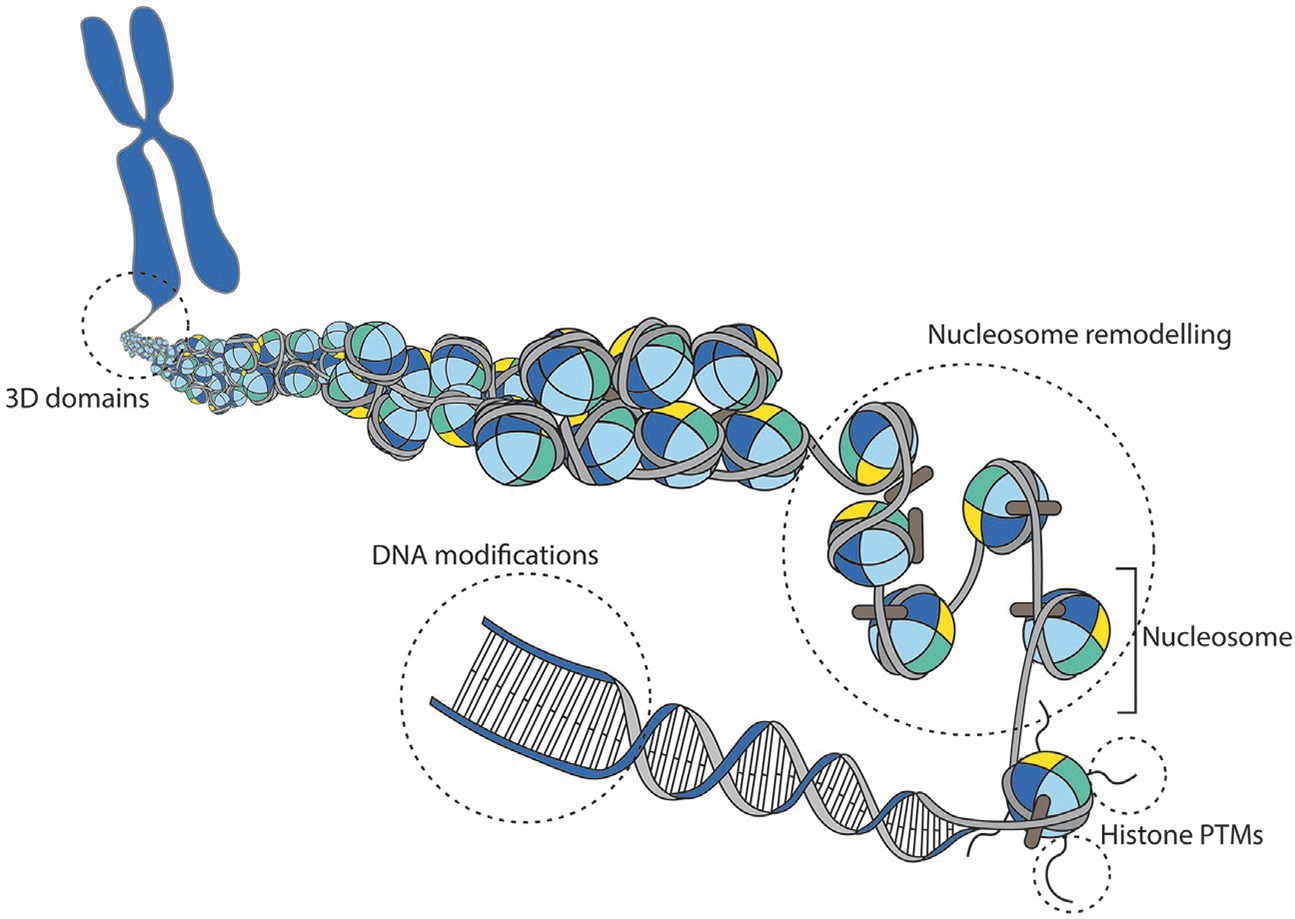
Epigenetic Regulation of Chromatin Structure. The nucleosome is the basic structural unit of chromatin and consists in 146 base pairs of DNA tightly packaged around an octameric protein core of histones. Epigenetic mechanisms include the positioning and remodeling of nucleosomes, post-translational modifications of histones, chemical modifications of DNA itself and higher organization of three-dimensional chromatin domains.
In mammals, DNA methylation mainly consists in the addition of a methyl group onto cytosines (5mC) within the context of CpG dinucleotides (Bird, 2002; Goll & Bestor, 2005). Dense regions of CpGs – termed CpG islands (CGIs) – are associated with the majority of human and mouse genome promoters and their methylation constitutes a major mechanism for stable repression of gene expression (Haberle & Stark, 2018; Zhu, Wang, & Qian, 2016). The DNA methyltransferase DNMT1, recruited by the ubiquitin E3 ligase UHRF1, has been shown to reproduce DNA methylation patterns during DNA replication and repair, ensuring faithful maintenance of methylation profiles (Nishiyama et al., 2020; Sharif et al., 2007). DNMT3A and DNMT3B have long been considered as “de novo” methyltransferases for their capacity to establish new patterns of DNA methylation (Okano, Bell, Haber, & Li, 1999). However, the distinction between maintenance and de novo DNA methyltransferases has been challenged by observations of their cooperation in establishing DNA methylation patterns (Haggerty et al., 2021; Lyko, 2018).
In addition to changes in the DNA sequence itself, changes in nucleosome position and composition, in particular on the amino-terminal tails of histones, have also been shown to participate in epigenetic memory of phenotypes (Bannister & Kouzarides, 2011; Venkatesh & Workman, 2015). Multiple post-translational modifications (PTMs) of histone tails have been identified, including methylation, acetylation, ubiquitination, sumoylation, and this repertoire is in perpetual expansion with the discovery of crotonylation (Fang et al., 2021; Tan et al., 2011), malonylation, succinylation, and glutarylation (Sabari, Zhang, Allis, & Zhao, 2017). Histone PTMs can directly regulate chromatin compaction but can also indirectly regulate gene expression through the recruitment of chromatin-modifying enzymes, which has led to the notion of a histone code (Strahl & Allis, 2000; Tan et al., 2011). Indeed, specific genomic regions are occupied by histones displaying different PTMs and these epigenetic signatures are predictive of transcriptional states. For instance, transcriptionally-active enhancers are enriched in a subset of histone marks, including histone H3 lysine 27 acetylation (H3K27ac) and histone H3 lysine 4 monomethylation (H3K4me1) (Heintzman et al., 2007; Hon, Hawkins, & Ren, 2009). Maintenance of histone marks is regulated by the controlled recruitment of histone modifiers, principally during DNA replication (Shan, Fang, & Jia, 2021).
Non-coding RNAs regulate epigenomes by recruiting chromatin-modifying complexes (Holoch & Moazed, 2015). Regulatory ncRNAs can be classically classified according to their size, with transcripts longer than 200 nt termed long ncRNAs (lncRNAs) and smaller transcripts including microRNAs (miRNAs) or piwi-interacting RNAs (piRNAs) (Kaikkonen, Lam, & Glass, 2011). Small ncRNAs-mediated inheritance of epigenetic states is a hallmark in Schizosaccharomyces pombe, Drosophila and Caenorhabditis elegans (Heyt & Thakur, 2021). In mammals, piRNAs and sperm-borne ncRNAs are the most well-understood small ncRNAs in the inheritance of epigenetic states (Chen, Yan, & Duan, 2016; Yan, 2014). In addition, lncRNAs are important regulators of epigenetic states and serves as hub for the concerted recruitment of multiple types of chromatin modifiers that are relevant for epigenetic inheritance (Legoff, D’Cruz, Tevosian, & Smagulova, 2019).
Finally, the three-dimensional organization of chromatin domains within the nucleus has been shown to be an important epigenetic mechanism (Pombo & Dillon, 2015). Indeed, depending on its transcriptional competence, chromatin dynamically transits between higher-order organization forms within different subnuclear compartments (Pombo & Dillon, 2015). Technical advances in the recent years have provided tools to analyze chromatin conformation but the three-dimensional control of epigenetic inheritance still needs to be better defined (Zheng & Xie, 2019).
3. Substrates for epigenetic inheritance
Germ cells are at the origin of new organisms and ensure a continuous link across generations and species (Lesch & Page, 2012). The germ cell lineage is responsible for the faithful propagation of genetic and epigenetic information to the next generation. Thus, for TEI to occur, the germline information needs to be altered and these changes retained throughout development. In this section, we will briefly review the development of germ cells and their epigenetic dynamics. We highlight key windows of susceptibility to environmental cues. We will further discuss how environmentally-induced epigenetic changes can be sustained during the intense epigenetic remodeling undergone by the germline.
3.1. Epigenetic dynamics during germ cells development
Germ cells are specified early in the developing embryo, in the perigastrulation epiblast, around embryonic day 7.5 (E7.5) in the mouse (Magnúsdóttir & Surani, 2014; Saitou & Yamaji, 2012) (Fig. 2). The founding population of mouse primordial germ cells (mPGCs) consists of about 40 cells and arise following specification cues—WNT3 signaling and bone morphogenetic proteins (BMP4, BMP2 and BMP8b)—from the extraembryonic ectoderm and visceral endoderm. Specification results in the induction of germline transcriptional programs, regulated by the lineage-determining transcription factors PRDM14, BLIMP1/PRDM1 and TFAP2C (Tang, Kobayashi, Irie, Dietmann, & Surani, 2016). From E9.5, mPGCs start migrating through the hindgut endoderm towards the genital ridge. The colonization of the genital ridge is accompanied by cell proliferation and an intense wave of epigenetic remodeling, which creates a naïve epigenome (Tang et al., 2016). In particular, mPGCs undergo a global and extensive loss of DNA methylation to reach a level of 5% of global DNA methylation (Guibert, Forné, & Weber, 2012; Hajkova et al., 2002; Seisenberger et al., 2012) (Fig. 2). Histone reprogramming and changes in histone post-translational modifications have been proposed to be functionally coordinated to preserve genomic stability and safeguard the genome from DNA demethylation and spurious transcription (Ancelin et al., 2006; Kurimoto et al., 2015; Seki et al., 2007). Indeed, mPGCs present a progressive reduction in global levels of H3K9me2 followed by an increase in global levels of H3K27me3, an increase of open domains marked by H3K4me1 and a global decrease in H2A/H4R3me2s (Ancelin et al., 2006; Kurimoto et al., 2015; Nagano et al., 2022; Ohta et al., 2017; Seki et al., 2007). In addition, it has been recently shown that the three-dimensional organization of the chromatin within the nucleus also preserves mPGCs from pervasive transcription, with an augmented chromatin insulation mediated by CTCF (Nagano et al., 2022).
Fig. 2.
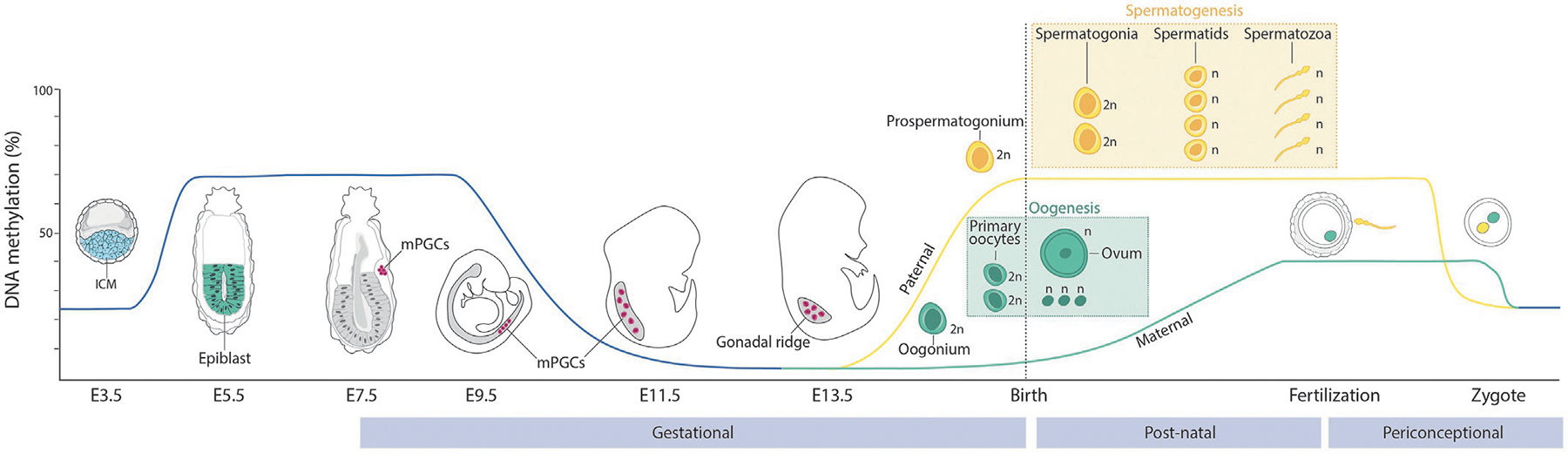
Epigenetic Remodeling during germline development offer unique exposure windows for inheritance. Variations in DNA methylation levels (%) are shown during germline development in the mouse. Spermatogenesis and oogenesis are summarized and depicted in respective frames. Windows of susceptibility for epigenetic memory to environmental exposures are shown for the gestational, post-natal and periconceptional periods, respectively. 2n = diploid, n = haploid.
At their arrival in the developing somatic gonad, by E11.5, mPGCs are on the onset of gametogenesis, during which sex differentiation and meiosis will occur, ultimately generating the functional gametes: oocytes or spermatozoa (Larose et al., 2019). During this time, mPGCs undergo licensing, a developmental transition regulated by the RNA-binding protein DAZL, that enables the cells to acquire the competence to respond to feminizing or masculinizing cues produced by the fetal ovary or testis (Gill, Hu, Lin, & Page, 2011). Subsequently, mPGCs will progress either to spermatogenesis or oogenesis depending on the sex of the embryo and epigenetics marks will be re-established in a sex-specific manner (Fraser & Lin, 2016; Kota & Feil, 2010; Sasaki & Matsui, 2008).
Oogenesis,the process of formation of mature female gametes, begins with differentiation of female PGCs into oogonia around E13.5 (Sánchez & Smitz, 2012) (Fig. 2). Oogonia proliferate by mitosis to form a pool of primary oocytes that arrest at the prophase stage of the first meiotic division until after birth when they are assembled into primordial follicles. Upon puberty, a small subset of oocytes will resume meiosis and undergo meiotic maturation concomitant with the process of ovulation prior to fertilization. During oogenesis, epigenetic marks are distinctively re-established on the genome (Sendžikaitė & Kelsey, 2019; Tomizawa, Nowacka-Woszuk, & Kelsey, 2012). At the later stages of their development, after birth, oocytes undergo de novo DNA methylation (Hiura, Obata, Komiyama, Shirai, & Kono, 2006). Mature oocytes are lowly methylated (40%) and present a unique bimodal DNA methylation pattern, with gene bodies of actively-transcribed genes being mostly hypermethylated (>75%) and large-scale genomic domains being hypomethylated (<25%) termed partially-methylated domains (PMDs) (Kobayashi et al., 2012; Veselovska et al., 2015) (Fig. 2). Mechanistically, de novo DNA methylation is mainly mediated by the DNA methyltransferase DNMT3A, guided by DNMT3L (Bourc’his, Xu, Lin, Bollman, & Bestor, 2001; Kaneda et al., 2004; Lucifero et al., 2007). Regions destined for DNA methylation have been shown to present specific histone signatures (reduced H3K4me2/3 and increased H3K36me3), indicating the importance of crosstalk in epigenetic reprogramming (Stewart et al., 2015; Xu et al., 2019). Furthermore, the oocyte PMDs are also associated with non-canonical distributions of histone modifications. Indeed, H3K4me3, a mark typically enriched at active promoters was found on intergenic domains where it accumulates following the activity of the H3K4 methyltransferases MLL2 and SETD1 and correlates with genome-wide transcriptional silencing (Andreu-Vieyra et al., 2010; Dahl et al., 2016; Hanna et al., 2018; Sha et al., 2020; Yu et al., 2017). Accumulation of the repressive mark H3K27me3 has also been reported on the oocyte PMDs but its functional relevance appears less clear (Inoue, Jiang, Lu, Suzuki, & Zhang, 2017; Zheng et al., 2016). A recent study highlighted that H2AK119ub1 was pervasively distributed in oocyte PMDs and that the co-existence of H3K27me3 and H2AK119ub1 at promoters was associated with gene silencing (Mei et al., 2021), further pointing to the importance of mechanistic crosstalk in oocytes epigenetic reprogramming.
Spermatogenesis, the process of formation of mature male germ cells, begins with differentiation of male PGCs into prospermatogonia/spermatogonial stem cells (SSCs) around E13.5 (Fayomi & Orwig, 2018; Griswold, 2016) (Fig. 2). Prospermatogonia arrest at a mitotic G1/G0 phase until puberty during which mitosis and spermatogenesis resumes. Spermatogenesis takes about 34.5 days to complete in the mouse and is classically divided in three major phases: mitotic divisions of prospermatogonia into spermatogonia, two sets of meiotic divisions of spermatogonia to generate the haploid spermatids and spermiogenesis during which spermatids are transformed into spermatozoa (Fig. 2). Contrary to oocytes, spermatozoa DNA is highly methylated (>80%) and de novo DNA methylation is initiated before birth in the prospermatogonia between E15.5 and E18.5 (Ben Maamar, Beck, Nilsson, McCarrey, & Skinner, 2022; Kubo et al., 2015) (Fig. 2). DNA methylation in spermatozoa is evenly distributed, with the exception of the presence of PMDs in intergenic regions (Kubo et al., 2015; Shirane, Miura, Ito, & Lorincz, 2020). Mechanistically, DNMT3A guided by DNMT3L have also been shown to be important for spermatozoa DNA methylation (Bourc’his et al., 2001; Kato et al., 2007; Kobayashi et al., 2013). As for oocytes, DNA methylation and histone post-translation modifications are functionally linked, with domains destined for DNA methylation marked by H3K36me2 and H3K27me3 (Nagano et al., 2022; Shirane et al., 2020). Importantly, spermatogenesis is characterized by histone-to--protamine transition. While spermatogonia DNA is initially wrapped around canonical histones, these are progressively replaced first by testis-specific histone variants then by protamines during spermiogenesis (Bao & Bedford, 2016). Protamines are small arginine-rich nuclear proteins that allow strong DNA binding and tight packaging into the compact spermatozoa head. About 1% of the mature mouse spermatozoa genome retains canonical histones that present post-translational modifications transmitted to the early zygote and persisting in the early embryo (Brykczynska et al., 2010; Hammoud et al., 2009), suggesting a role for these canonical histones in epigenetic inheritance (Siklenka et al., 2015; Van Der Heijden et al., 2008). Finally, spermatozoa also encode a large proportion of small and long ncRNAs, which roles in sperm epigenetic remodeling remain to be determined (Jodar, 2019), but that have nevertheless been linked to epigenetic inheritance in the offspring (Chen, Yan, & Duan, 2016).
Following fertilization, another extensive epigenetic remodeling occurs asymmetrically to remove parental epigenetic marks in each parental genomes before fusion (Okada & Yamaguchi, 2017). Soon after the entry of the spermatozoon in the oocyte, the paternal genome is demethylated via Tet3-mediated oxidation and protamines are exchanged for the maternally-provided histones that eventually acquire post-translational modifications (Okada & Yamaguchi, 2017). In contrast, the maternal genome remains largely protected from demethylation through the H3K9me2-mediated recruitment of Dppa3/PGC7/STELLA (Nakamura et al., 2012). Certain maternal histone marks, such as H3K36me3, are erased prior zygotic genome activation (ZGA), whereas other marks such as the non-canonically-distributed H3K4me3 are redistributed to canonical promoters upon ZGA (Xia & Xie, 2020). This has led some authors to categorize histone remodeling in the early embryo based on functions: epigenetic marks that are required for gametogenesis, for transcriptional control both in the gametes and in the embryo or “passenger” marks that appear dispensable for the gametes but that are nevertheless maintained and could play a role later in embryogenesis (Xia & Xie, 2020). Of note, although early epigenetic reprogramming erases much of the parental epigenomes, certain loci called imprinting control regions (ICRs) conserve epigenetic features that are parentally-inherited and maintained in the offspring (Tucci et al., 2019). Understanding the mechanisms of epigenetic memory on ICRs is relevant for deciphering the mechanisms of epigenetic inheritance of environmental cues. ZGA marks a key step in early development during which the zygote acquires its transcriptional independency and establishes its own epigenome. As the embryo develops towards the blastocyst stage, active histone marks (H3K4me3 and H3K27ac) are established on promoters and enhancers, respectively, whereas DNA methylation and repressive histone marks are not restored until implantation, which could be functionally relevant for totipotency (Xia & Xie, 2020).
The developmental model for germ cells detailed above corresponds to the one of the mouse and is mostly conserved in mammals. The resolution of the epigenetic remodeling occurring during the germline development in humans has been challenged by technical and ethical constraints. However the development of in vitro germ cell models and low-input epigenetic interrogation techniques has enable to demonstrate that, in general, DNA methylation patterns are relatively conserved between mouse and humans (Hanna, Demond, & Kelsey, 2018). This further supports the use of mouse as a model organism for epigenetic studies in the germline.
3.2. Windows of susceptibility
Germ cell development is uniquely regulated by waves of epigenetic erasures and re-establishment. In particular, two major events of epigenetic reprogramming occur during mammalian development: in PGCs, serving to erase established patterns and restore pluripotency potential, and in the early zygote, serving to erase the gametic epigenomes and regain totipotency. For environmentally-induced changes in the epigenome to be passed across generations, they need to be maintained through these epigenetic events. In particular, three temporal windows of susceptibility to environmental cues can be theoretically defined during: (1) gestation, (2) post-natal development of gametes and (3) around conception (Fig. 2). Here we will further detail these temporal windows of susceptibility and the proposed mechanisms of epigenetic inheritance to environmental cues in the germline. We focus on such mechanisms that directly impact germ cells epigenomes, however, change in somatic cells, particularly in gonads, can indirectly alter the epigenome of germ cells (Bline, Le Goff, & Allard, 2020).
During gestation, maternal environmental exposure could affect PGCs’ epigenome remodeling. DNA methylation, the most studied epigenetic mark, is globally reduced to the lowest-reported levels in mammalian epigenomes and would, therefore, not be a likely medium for transgenerational inheritance. However, certain loci corresponding to transposable elements have been identified as methylation escapees. Indeed, young long-terminal repeat (LTR) retrotransposons, known as endogenous retroviruses, resist programed global demethylation during PGC reprogramming. For instance, the intracisternal A-particle (IAP) in the mouse and Alu and HERVK elements in humans have been shown to display higher DNA methylation that the rest of the epigenome (Gkountela et al., 2015; Guo et al., 2015; Ohta et al., 2017; Seisenberger et al., 2012). Retrotransposon-associated inheritance of epialleles is a well-described phenomenon, particularly in the context of the extensively-studied agouti viable yellow (Avy) epiallele. In agouti mice, an IAP retrotransposon element is inserted upstream of the agouti gene (Duhl, Vrieling, Miller, Wolff, & Barsh, 1994). Changes in the IAP LTR methylation status drive phenotypical changes, with unmethylated LTRs promoting ubiquitous expression of agouti (Duhl et al., 1994). These Avy mice present a phenotype of yellow fur, obesity, glucose intolerance and increased susceptibility to tumors development (Duhl et al., 1994). Hence, epigenetic changes in endogenous retroviruses can drive inheritance of phenotypes in mammals, however, if the endovirome can serve as hotspots for epigenetic memory in the context transgenerational inheritance remains to be determined (Blake & Watson, 2016; Friedli & Trono, 2015; van Otterdijk & Michels, 2016; Zeng et al., 2022). To do so, the epigenetic mechanisms responsible for endogenous retroviruses silencing during normal development need to be better understood before analyzing the effect of epigenetic enzymes recruitment upon altered environmental conditions.
The sex-specific reestablishment of epigenetic marks on gametes occurs in part after birth and during puberty, with differences between oocytes and spermatozoa. Hence, epigenetic changes in gametes postnatally, following exposures to environmental cues, has been proposed as a mechanism of transgenerational inheritance. Multiple studies have, in fact, reported the existence of aberrant differentially-methylated regions (DMRs) in paternal epigenomes in response to environmental cues (see Section 3). However, it is unlikely that phenotypic changes in the offspring are mediated solely by DNA methylation, considering the importance of epigenetic crosstalk in spermatogenesis. Rather, it is proposed that cumulative effects of dysregulation epigenetic patterns in the earlier development are sustained and exacerbated in transcriptional regulatory networks (Kremsky & Corces, 2020). In this sense, changes in sperm small ncRNAs, that has garnered much attention recently as a medium for epigenetic inheritance, could also participate in this global change (Chen, Yan, & Duan, 2016). Compared to the breadth of data on paternal-mediated epigenetic inheritance, much less is known on maternal-mediated inheritance, mainly due to the paucity of biological material for epigenetic studies. The advent of new techniques for chromatin interrogation and for modeling oocytes development in vitro will bring to light mechanisms of maternal-mediated inheritance in response to environmental cues (see Section 4).
Finally, the periconceptional environment could affect early embryo reprogramming and constitutes another window of susceptibility for TEI. In human, this specific period has been much studied for assisted reproductive technologies (ART), with accumulating evidence suggesting that ART may influence developmental trajectories in the offspring (Fleming et al., 2018). Indeed, despite advances in in vitro culture of embryos, developmental conditions remain sub-optimal (Watkins, Lucas, & Fleming, 2010), indicating the importance of the maternal reproductive tract for proper development and its sensitivity to environmental cues. In hypofertile conditions, it remains unclear what can be attributed to the periconceptional environment or to the parental gametes themselves. However, studies in mouse models have shown how variations in culture media components impact the developmental potential of the embryos (Watkins et al., 2010). In fact, the vast majority of data emerging from periconceptional sensitivity to the environment has been made in the context of maternal dietary environment (Fleming et al., 2018). In humans, maternal undernutrition was shown to be associated with subsequent adult cardiometabolic and neurological dysfunction in the offspring (Velazquez, Fleming, & Watkins, 2019). Mechanistically, metabolic dysfunction are linked with epigenetic mechanisms since several metabolites act as cofactors for epigenetic reactions (Boon, Silveira, & Mostoslavsky, 2020; Li, Egervari, Wang, Berger, & Lu, 2018). However, more studies are needed to define how the mammalian embryo responds to its immediate environment and whether epigenetic memory of periconceptional events are maintained throughout adulthood (Novakovic et al., 2019). This is further complexified by the existence of multiple simultaneous mechanisms of inheritance to environmental cues and confounding factors, particularly in social animals.
4. Evidence for environmentally-induced epigenetic inheritance in mammals
Exposure to environmental cues have the capacity to alter epigenomes and phenotypes. However, TEI postulates that an epigenetic memory of exposure is transmitted to the offspring through the germline without continuous exposure. For maternal inheritance, true transgenerational effects are detectable only at the third generation (F3). Indeed, for exposure in pregnant females (P0), the embryo (F1) is also exposed, as well as its fetal germ cells (F2). Thus, effects without continuous exposures do not occur before the F3 generation (Fig. 3). For paternal inheritance, the F2 is the first generation wherein transgenerational effects can be examined (Fig. 3). While TEI is well accepted in plants (Hauser, Aufsatz, Jonak, & Luschnig, 2011) and invertebrate animals, such as Caenorhabditis elegans (Baugh & Day, 2020), the existence or significance of TEI in mammals has been much disputed considering the extensive germline epigenetic reprogramming known to occur. In humans, in particular, evidence for transgenerational epigenetic inheritance to environmental cues is limited (Horsthemke, 2018; Svanes et al., 2021). This is because human studies tend to suffer from heterogeneous populations, reproducibility, recall- and selection- bias among other confounding factors (Marczylo, Jacobs, & Gant, 2016). Despite these conceptual challenges, several studies have provided evidence for at least intergenerational inheritance. Here, we compiled existing evidence of environmentally-induced transgenerational epigenetic inheritance in rodents in response to a range of environmental cues from manufactured chemicals, metals, diet to drugs (Table 1). For each environmental cue, we present the available data in humans and highlight the need for a better understanding of the molecular mechanisms of transgenerational versus intergenerational epigenetic inheritance. Of note, we focus here on environmentally-induced TEI and we do not discuss the epigenetic inheritance of behavioral phenotypes that has been covered elsewhere (Bale, 2014; Jawaid & Mansuy, 2019).
Fig. 3.
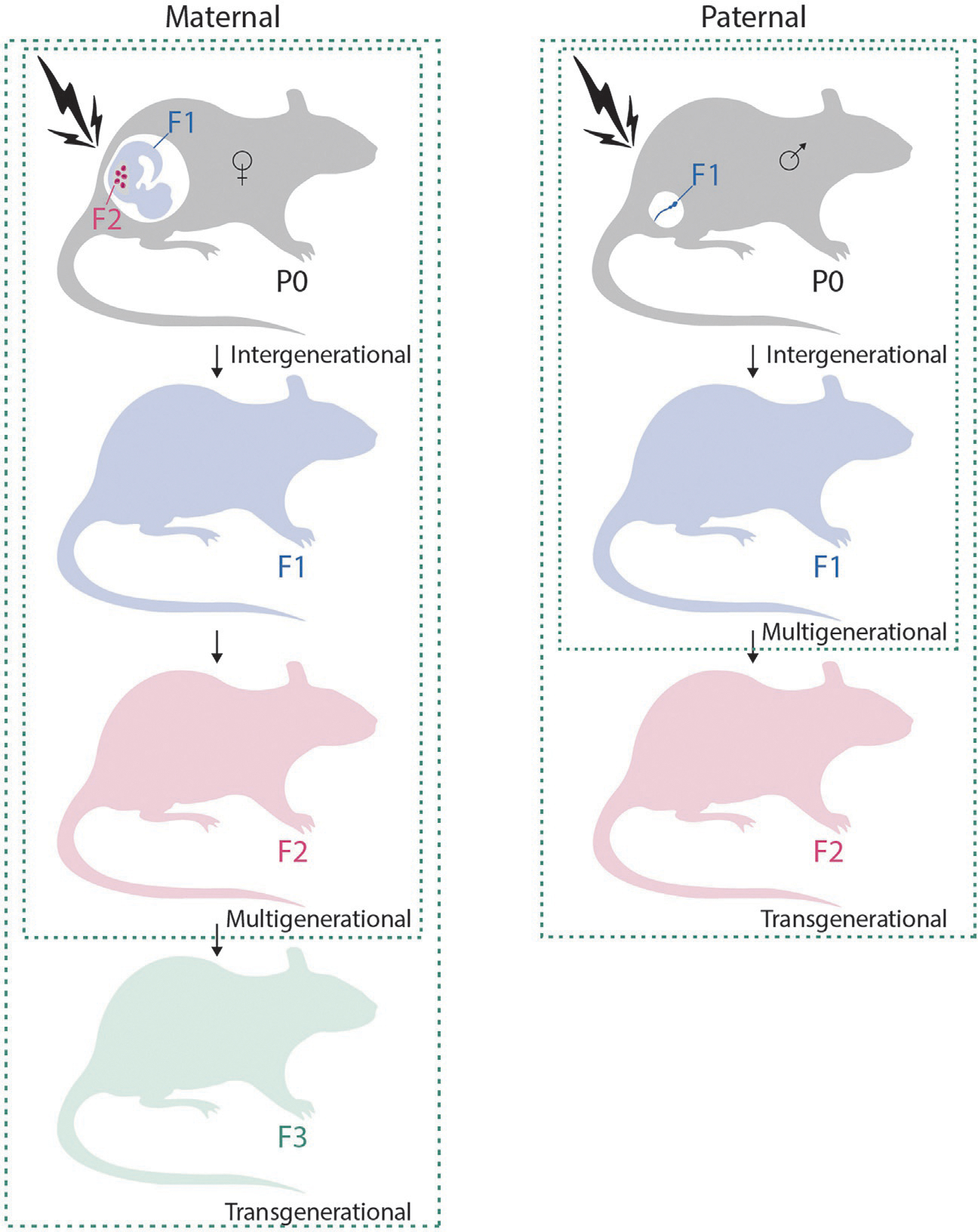
Modes of Inheritance following Environmental Exposure. Exposure in gestational females (P0) also exposes the embryo (F1) and its germ cells (F2). Therefore, only the third generation is the one where the observed phenotype is not due to direct exposure and can truly be transgenerational. For post-natal exposure, as presented under the paternal scheme, exposure affects the F1, hence, the second generation is the one where transgenerational phenotypes can be observed.
Table 1.
Evidence of transgenerational epigenetic inheritance to environmental cues in rodents.
| Animal Model | Environmental cue | Offspring phenotype | Epigenetic mark | Mode of exposure | References |
|---|---|---|---|---|---|
| Rat, Mouse | Manufactured chemical (Vinclozolin) | Impaired male fertility, increased adult onset disease | DNA methylation | Maternal (gestational) | Anway, Cupp, Uzumcu, and Skinner (2005); Anway, Memon, Uzumcu, and Skinner (2006); Anway, Leathers, and Skinner (2006); Guerrero-Bosagna, Settles, Lucker, and Skinner (2010); Guerrero-Bosagna et al. (2012) |
| Rat, Mouse | Manufactured chemical (Dioxin) | Impaired female fertility, increased adult onset disease | DNA methylation | Maternal (gestational) | Bruner-Tran and Osteen (2011); Manikkam, Tracey, Guerrero-Bosagna, and Skinner (2012) |
| Rat, Mouse | Manufactured chemical (BPA) | Impaired female fertility, increased adult onset disease | DNA methylation | Maternal (gestational) Paternal (pre-conceptional) | Manikkam, Tracey, Guerrero-Bosagna, and Skinner (2013); Ziv-Gal, Wang, Zhou, and Flaws (2015); Saidur Rahman, Pang, Ryu, Park, and Pang (2020) |
| Mouse | Manufactured chemical (Atrazine) | Impaired male fertility | H3K4me | Maternal (gestational) | Hao et al. (2016) |
| Mouse | Metal (Arsenic) | Impaired male fertility | DNA methylation | Maternal (gestational) | Yin et al. (2021) |
| Rat | Metal (Arsenic) | Impaired fertility | DNA methylation | Maternal (pre-conceptional) and Paternal (pre-conceptional) | Nava-Rivera et al. (2021) |
| Mouse | Diet (HFD) | Increased male body weight, impaired insulin sensitivity | DNA methylation | Maternal (gestational) | Sarker et al. (2018) |
| Rat | Diet (HFD) | Reduced birth-weight, female increased resistance to HFD-induced weight gain | Small RNAs | Paternal (pre-conceptional) | de Castro et al. (2016) |
| Mouse | Diet (HFD) | Increased obesity, insulin resistance | DNA methylation and small RNAs | Paternal (pre-conceptional) | Fullston et al. (2013); Wei et al. (2014); Chen et al. (2016) |
| Rat | Drugs (Alcohol) | Altered hypothalamic functions | DNA methylation | Maternal (gestational) | Govorko, Bekdash, Zhang, and Sarkar (2012) |
| Mouse | Drugs (Alcohol) | Altered neocortical development and behaviors | DNA methylation | Maternal (gestational) | Abbott, Rohac, Bottom, Patadia, and Huffman (2018); Bottom, Kozanian, Rohac, Erickson, and Huffman (2022) |
| Mouse | Drugs (Nicotine) | Reversal learning deficit | DNA methylation | Paternal (pre-conceptional) | McCarthy et al. (2018) |
4.1. Manufactured chemicals
The first demonstration of transgenerational inheritance of an environmentally-induced phenotype in rodents was made nearly 20 years ago from studying the agricultural fungicide vinclozolin (Anway et al., 2005). Transient gestational exposure in female rats resulted in impaired sperm motility and reduced fertility in the male offspring up to the F4 (Anway et al., 2005; Anway, Memon, et al., 2006). Female descendants also developed transgenerational phenotypes up to the F4, including tumors and kidney disease (Anway, Leathers, & Skinner, 2006). Mechanistically, a methylated DNA immunoprecipitation followed by a promoter tilling microarray (MeDIP-chip) showed the presence of differentially-demethylated regions (DMRs) in F3 spermatozoa (Guerrero-Bosagna et al., 2010, 2012). However, the stability of the generated epimutations, or their relationship with known genetic abnormalities induced by vinclozolin were not assessed. Nevertheless, a following study showed that gestational vinclozolin exposure significantly altered F3 sperm-borne small ncRNAs that correlated with the DMRs and the vinclozolin transgenerational phenotypes (Schuster, Skinner, & Yan, 2016), suggesting a concerted vinclozolin-induced epigenetic transgenerational inheritance phenomenon.
Since these pioneering observations, exposure to multiple other manufactured chemicals, belonging to the large class of endocrine-disrupting chemicals (EDCs), have been shown to be inherited transgenerationally in rodents. However, the epigenetic mechanisms at play have not always been identified. Despite this, the question of EDCs-mediated epigenetic remodeling in the germline and phenotypic inheritance across generations needs to be urgently addressed. Indeed, EDCs are ubiquitous in our environment and because they directly interfere with the endocrine hormonal system, EDCs have been linked with multiple disorders in humans, including pregnancy complications, genital malformations, and cancer (Xin, Susiarjo, & Bartolomei, 2015).
Diethylstilbestrol (DES), a nonsteroidal estrogen, was initially prescribed in the 1950s with the goal of reducing miscarriage (Fénichel, Brucker-Davis, & Chevalier, 2015; Titus et al., 2019). The medication was taken off market in the 1970s after discovering cases of vaginal clear cell adenocarcinoma and breast cancer in the offspring born to women who were gestationally exposed to DES (Fénichel et al., 2015). The cohort of children resulting from DES-exposed women were also found to develop genital tract malformations, including hypospadias and cryptorchidism in males and uterine abnormalities in females (Fénichel et al., 2015). Additionally, both sexes were more likely to develop non-reproductive diseases including diabetes, hypercholesterolemia, hypertension and osteoporosis (Fénichel et al., 2015). Beyond intergenerational effects from mother to offspring, several studies have confirmed that DES has transgenerational effects up to the third generation (Fénichel et al., 2015; Titus et al., 2019). In particular, a study demonstrated that women from the third generation following DES gestational exposure developed higher rates of menstrual irregularity, preterm delivery and vaginal epithelial changes compared to the offspring of unexposed women (Titus et al., 2019). In addition, men from the third generation following DES gestational exposure had a higher likelihood of congenital anomalies, including hypospadias and heart defects (Fénichel et al., 2015). Without any chromosomal or genetic alterations, these offspring of DES-exposed women provide evidence for transmission of epigenetic inheritance across generations. Although the molecular mechanisms of transgenerational inheritance remain unclear, DES exposure may epigenetically alter estrogen receptors and impact the role of sex steroids in differentiating the genital tract during an important fetal exposure window (Fénichel et al., 2015; Titus et al., 2019).
Dioxin/TCDD (2,3,7,8-tetrachlorodibenzo-p-dioxin) is a by-product of industrial combustion processes and gestational exposure of this EDC has been shown to be transgenerationally inherited up to the F3, with an observed phenotype of reduced female fertility (Bruner-Tran & Osteen, 2011). Further studies in rats showed a similar phenotype in the F3 and that the F3 sperm epigenome was altered, with the identification of several DMRs (Manikkam et al., 2012). Again, the functional relevance of these DMRs, and their conservation across rodent models, need to be addressed to properly establish the mechanisms of TEI in response to dioxin. Dioxin exposure in humans is widespread and occurs primarily via food ingestion or contact with contaminated soil, air, and water (Travis & Hattemer-Frey, 1991). Studies in an Italian human cohort, following the explosion of a trichlorophenol manufacturing plant and the release of up to 30 kg of TCDD, showed that men whose mothers were exposed in utero to dioxin had impaired semen quality (reduction in spermatozoa concentration, count and quality) (Mocarelli et al., 2011). In a Russian cohort, whole genome bisulfite sequencing (WGBS) showed the presence of spermatozoa DMRs associated with serum TCDD concentrations (Pilsner et al., 2018). Together, these studies show that dioxin provokes spermatozoa epimutations and intergenerational phenotypes in humans, although impact across a larger number of generations has not been studied.
The plastic monomer bisphenol A (BPA) is another widespread EDC that is readily measured in humans following exposure to food and beverage containers (Vandenberg, Hauser, Marcus, Olea, & Welshons, 2007). BPA directly impacts human metabolic and reproductive organs (Rochester, 2013). However, epidemiological studies tend to support that, although BPA can cause fetal death and growth impairment during infancy at high dose, the risks for low-dose exposure are negligible (Ranjit, Siefert, & Padmanabhan, 2010). Contrarily to the evident paucity of research in humans, several studies in rodents have addressed the consequences of in-utero exposure to BPA across generations. BPA exposure has been associated with transgenerationally-acquired phenotypes of reduced fertility and adult onset disease, correlating with spermatozoa DMRs serving as BPA epimutations (Manikkam et al., 2013; Ziv-Gal et al., 2015). In a recent study focused on paternal inheritance of BPA exposure in mice, P0 males were exposed during the entire period of sexual maturity/puberty and phenotypes were analyzed in subsequent generations (Saidur Rahman et al., 2020). Both F2 and F3 males presented reduced fertility and a global increase in DNA methylation in spermatozoa. This study highlights that EDCs provoke transgenerational phenotypes that are both maternally and paternally inherited. Considering the different role of DNA methylation in oogenesis and spermatogenesis, further studies will need to focus on a better understanding of EDCs-mediated epigenetic remodeling in germ cells from P0 exposed parents.
In this regard, epigenetic crosstalk between histone PTMs and DNA methylation might be relevant in ensuring transgenerational inheritance to EDCs. For instance, gestational exposure to the herbicide atrazine was associated with reduced spermatozoa number in male F3 due to altered spermiogenesis (Hao et al., 2016). This phenotype was associated with a transgenerational global decrease in H3K4me3 level in the F3 (Hao et al., 2016). Occupancy of H3K4me3 was studied by chromatin immunoprecipitation followed by sequencing (ChIP-seq) and integration with transcriptomic data demonstrated that the epigenetic status of promoters of genes associated with germ cell differentiation were transgenerationally altered. Additionally, differential expression of lncRNAs in the F3 was also observed (Hao et al., 2016), further pointing to more potential mechanistic cross-talk in transgenerational inheritance of atrazine.
4.2. Metals
Toxic metals are ubiquitous in our environment and exposure has been associated with serious adverse health outcomes in human populations, such as cancer, cardiovascular diseases, and neurological disorders (Ray, Yosim, & Fry, 2014). With accumulating data showing that environmental toxic metals can impact epigenomes (Ray et al., 2014), their transgenerational inheritance has started to be studied in more depth.
Arsenic is a ubiquitous naturally-occurring toxic substance in the environment (Chung, Do, & Hong, 2014). Arsenic compounds dissolve readily in water; thus, consumption of contaminated water is a global route of exposure (Chung et al., 2014). Arsenic exposure is associated with a constellation of negative health effects impacting all major organs and organ systems and is a major carcinogen (Argos et al., 2010; Smith et al., 1992). For instance, in a Mexican human cohort, elevated arsenic levels in water correlated with hypermethylation in tumor-suppressor genes, pointing towards an epigenetic component in arsenic carcinogenic capacity (Smeester et al., 2011). Transgenerational phenotypes in response to arsenic exposure have been reported in Caenorhabditis elegans (Yu & Liao, 2016), and in zebrafish (Hernández-Sánchez, Valles, & Bardullas, 2022). In humans, a study in a Bengali cohort demonstrated that arsenic exposure in early pregnancy altered the cord blood methylome of the offspring, with more pronounced effects in male than in female newborns (Broberg et al., 2014). The identified DNA methylation changes were associated with cancer-related genes, which would suggest a sex specificity in arsenic carcinogenic capacity (Broberg et al., 2014). In agreement with epidemiological studies in humans, intergenerational effects of parental arsenic exposure have been reported in the mouse (Gong et al., 2021; Yin et al., 2021) and in the rat (Nava-Rivera et al., 2021). The first study on transgenerational inheritance of arsenic exposure in the mouse identified that pre-conceptional paternal exposure was associated with reduced adiposity in the F2 and increased adiposity in the F3 (Gong et al., 2021). Epigenetic mechanisms were not assessed but their adaptative changes were proposed to be responsible for the opposed phenotypes between the F2 and the F3 generations (Gong et al., 2021). More recently, another study in a mouse model of maternal gestational exposure to arsenic showed that the male F3 had impaired fertility associated with a decrease in the methylation levels in two specific imprinted genes (Yin et al., 2021). Finally, a study of pre-conceptional exposure to arsenic in both male and female rats showed impaired reproductive functions in the F2/F3 that were associated with a global increase in DNA methylation in gonads (Nava-Rivera et al., 2021). Thus, while there is evidence of phenotypic transgenerational inheritance of arsenic exposure, the role of DNA methylation in this phenomenon is unclear. This is representative of the unsatisfactory understanding of the effects of arsenic exposure on DNA methylation. Indeed, arsenic exposure provokes both global DNA hypomethylation, due to reduction in the levels of the universal methyl donor S-Adenosyl Methionine SAM, and hypermethylation of specific loci during carcinogenesis (Reichard & Puga, 2010; Ren et al., 2011). More fundamental studies are therefore needed to resolve the opposite effects of arsenic on DNA methylation (genome-wide vs locus-specific). Furthermore, SAM is also the methyl donor for histone methylation; thus, epigenetic crosstalks in arsenic TEI should be investigated.
The number of studies on transgenerational inheritance of metal exposure in rodents is limited, despite their wide distribution in our environment, their reported epigenetic inheritance in other animals (Carvan et al., 2017; Mu et al., 2021; Vandegehuchte, Lemière, Vanhaecke, Vanden Berghe, & Janssen, 2010) and epidemiological studies in humans (Maloney, Bayon, Zawia, & Lahiri, 2018; Sen et al., 2015). Cadmium and mercury co-administration in gestational mice was associated with reduced glucose tolerance up to the male F4 generation (Camsari et al., 2019). Lead gestational exposure in mice provoked growth stunt in the F3 generation (Sobolewski et al., 2020). However, in both studies, potential epigenetic mechanisms for transgenerational inheritance were not studied. Overall, mercury, cadmium, lead, selenium, and aluminum are all prevalent environmental toxic metals and their potential TEI in Mammals needs to be urgently examined.
4.3. Diet
Early long-term epidemiological studies and some empirical observations were the first testimonies that parental nutrition affects the offspring health (Guo, Luo, & Lin, 2020; Soubry, 2015). In recent years, studies in animal models have shed light on how changes in parental diets can modulate germ cells metabolism and epigenomes and induce phenotypic changes in the offspring, particularly the development of metabolic diseases. Here, we discuss how high-fat diet, caloric restriction or diet composition can be transgenerationally-inherited in the offspring upon epigenetic changes in the germline.
Increasing prevalence of obesity has become a major global health concern. A critical question is whether obesity in parents can influence the development of metabolic diseases over several generations in the offspring. Modeling of obesity in rodent models, using high-fat diet (HFD), has revealed the existence of transgenerational epigenetic inheritance of parental diet, both maternally and paternally. Maternal HFD during pregnancy is known to increase the rates of obesity and metabolic syndrome in the first and second generations (Kaspar, Hastreiter, Irmler, Hrabé de Angelis, & Beckers, 2020; Wu & Suzuki, 2006). However, maternal HFD can also be transmitted transgenerationally up to the third generation. Indeed, the F3 female offspring of mice fed with HFD before pregnancy showed increased body size and improved glucose tolerance, that were linked with dynamic patterns in imprinted genes expression (Dunn & Bale, 2011). More recently, the male F3 offspring of mothers perinatally fed with a HFD were shown to present an increased body weight and an impaired sensitivity to insulin (Sarker et al., 2018). These phenotypes were correlated with DNA methylation changes and signature DMRs in F2 spermatozoa (Sarker et al., 2018). Interestingly, observations regarding the phenotypes in the third generation vary between sexes (Sarker et al., 2018) and pre-conceptional versus gestational HFD also provoked different transgenerational phenotypes in the two above-mentioned studies. This indicates that multiple epigenetic mechanisms might be at play in maternal inheritance of HFD and more research is needed (Kaspar et al., 2020). Most studies on transgenerational inheritance of HFD have focused on the paternal route. The first study to address this question showed that female F1 descendent of male rat fed with HFD presented an impaired glucose-insulin homeostasis (Ng et al., 2010). This intergenerational phenotype for paternal inheritance of HFD in the rat was later shown to be transgenerationally transmitted by spermatozoa up to the second generation (de Castro et al., 2016). Indeed, HFD was associated with changes in F1 spermatozoa small RNAs consistent with their metabolic functions and the F2 phenotype (de Castro et al., 2016). Several studies have substantiated these findings in mice and shown that HFD provokes spermatozoa epigenome changes, in terms of DNA methylation and altered small RNA profiles (Chen, Yan, Cao, et al., 2016; Fullston et al., 2013; Wei et al., 2014). Since spermatozoa DNA methylation changes in response to HFD could not completely recapitulate the transcriptional outcomes in the offspring (Wei et al., 2014), attention is now shifting to the understanding of concerted epigenetic reprogramming in spermatozoa following HFD. Another open question is whether dietary changes can restore spermatozoa epigenomes after weight loss.
Inadequate protein intake is one example of malnutrition and can be modeled by feeding rodents with low-protein diet. One study found that the F1 generation of male mice fed with a low-protein diet had reduced DNA methylation levels in the liver (Carone et al., 2010). These epigenetic signatures correlated with increased expression of genes involved in lipid and cholesterol biosynthesis (Carone et al., 2010). However, a later study with the same mice exposure paradigm showed that this intergenerational phenotype was not associated with changes in spermatozoa methylome (Shea et al., 2015), pointing towards the existence of other epigenetic mechanisms such as histone PTMs or small RNAs (Yoshida et al., 2020). In addition, a rat maternal low-protein diet also resulted in altered DNA methylation in the liver of male pups, due to decreased expression of the DNA methylation machinery (Gong, Pan, & Chen, 2010). These studies indicate that low-protein diet results at least in intergenerational phenotypes in rodents. Epidemiological studies in humans show how maternal low protein diet is associated with the lean variant of type 2 diabetes (Vipin, Blesson, & Yallampalli, 2022).Considering the worldwide increase in vegetarian and vegan diets, which are typically associated with low protein consumption (Vipin et al., 2022), more data will be needed to determine whether low protein diet associate with inter- and transgenerational epigenetic inheritance of metabolic disorders.
Undernutrition in early life may affect adult health. The famine period in the Netherlands during World War II, known as the Dutch Hunger Winter of 1944–1945, has allowed to study the intergenerational effect of parental nutrition on the metabolic status of grandchildren in humans (Lumey, Stein, & Susser, 2011). In this historical cohort, famine exposure during pregnancy (daily average of 900 kca/day or less) was associated with an adverse metabolic profile (suboptimal glucose handling, higher body mass index, etc.) and a higher risk for developing schizophrenia in the F1 offspring (Lumey et al., 2011). Adult offspring (F2) of gestationally exposed F1 fathers, but not mothers, were also found to present higher body mass index, suggesting a paternal intergenerational inheritance over two generations (Veenendaal et al., 2013). Subsequently, an methylome-wide approach by reduced representation bisulfite sequencing (RRBS) in the blood of F1 individuals demonstrated the presence of DMRs associated with the phenotypic outcome of early-life exposure to famine (Tobi et al., 2014). Studies in rodents support that gestational undernutrition in mice provokes intergenerational developmental reprogramming, with the apparition of DMRs in the F1 spermatozoa methylome (Radford et al., 2014) (Radford et al., 2014). Male mice undernutrition also resulted in spermatozoa global demethylation and an offspring with reduced postnatal weight (McPherson et al., 2016). However, the only evidence of TEI of undernutrition was found in the rat (Nowacka-Woszuk, Szczerbal, Malinowska, & Chmurzynska, 2018). Indeed, the F3 fetuses of pregnant females fed with a restricted diet presented lower hepatic levels of histone deacetylases (HDAC1 and SIN3A) but the increased H3 acetylation was not statistically significant (Nowacka-Woszuk et al., 2018). Altogether, more studies will be needed to pinpoint the epigenetic mechanisms of inter- and transgenerational inheritance of under- and malnutrition.
The impact of diet onto developmental processes needs to be placed in the larger context of the role of metabolism in early development and of the interplay between metabolism and epigenetics (Dahan, Lu, Nguyen, Kennedy, & Teitell, 2019; Verdikt & Allard, 2021). In this regard, another aspect of nutrition that has been explored is the change in diet composition, in particular, how micro-nutrient deficiency can impact gametes epigenomes and alter the offspring development. Folate is a metabolite that serves as a methyl-donor and its levels influence the availability of SAM (Clare, Brassington, Kwong, & Sinclair, 2019). Female mice carrying mutations in genes involved in the folate metabolism gave rise to an offspring with congenital malformations and tissue-specific loss of DNA methylation (Padmanabhan et al., 2013), demonstrating the importance of folate metabolism in maternal epigenetic inheritance. Folic acid supplementation is often recommended in early pregnancy and studies in humans indicate that changes in maternal plasma folate levels are associated with epigenome-wide DNA methylation variations in newborn cord blood (Caffrey et al., 2018; Joubert et al., 2016). In a recent study in the mouse, paternal inheritance of folate deficiency was shown to occur through dysregulation of H3K4me3 in spermatozoa (Lismer et al., 2021). Importantly, regions with altered H3K4me3 profiles in spermatozoa corresponded to developmental loci, were retained in the zygote and overlapped with differential zygotic gene expression (Lismer et al., 2021). Further research will need to determine whether folate deficiency-induced histone retention interplays with DNA methylation in transgenerational epigenetic inheritance. Another open question is whether the epigenetic modes of inheritance of folate deficiency vary in a sex-specific manner, considering the different role of chromatin methylation in gametogenesis.
4.4. Substances use and abuse
The evidence for epigenetic modifications following exposure to drugs of abuse, and their consequences on rewiring neuronal circuitry to maintain addiction, is well documented (Nestler, 2014). In recent years, several studies have started to address whether drugs of abuse can alter gametes epigenomes and the consequences for transgenerational inheritance (Wanner, Colwell, & Faulk, 2019; Yohn, Bartolomei, & Blendy, 2015; Zeid & Gould, 2020). Inheritance of drug abuse is particularly confounded by genetic and non-genetic (social, cultural, and environmental) factors (Yohn et al., 2015). Hence, modeling parental inheritance to drug abuse in rodent models provides mechanistic bases, i.e., the identification of epigenetic biomarkers, to study the relevance of TEI of drug abuse in human populations.
Ethyl alcohol (ethanol) effects are mediated through interference with several neurotransmitter systems (Chastain, 2006). Alcohol exposure provokes epigenetic and transcriptional changes in the brain, particularly in regions critical to addictive behaviors (Berkel & Pandey, 2017). Mechanistically, alcohol was shown to reduce the production of SAM provoking global hypomethylation in rat liver (Lu & Mato, 2005) as well as cause histone hyperacetylation by directly providing the acetyl groups through ethanol metabolism (Mews et al., 2019), although a tissue-specific understanding of epigenetic remodeling following alcohol exposure is currently lacking. In addition, alcohol exposure provokes genome-wide alteration in DNA methylation patterns in germ cells and several occurrences of intergenerational inheritance of these changes have been found both in rodent models and in human cohorts (Yohn et al., 2015). The risks of maternal alcohol abuse during pregnancy are well-established; however, gestational exposure to alcohol (prenatal ethanol exposure, PrEE) also provokes phenotypic and epigenetic changes across generations. For instance, in a rat model for fetal alcohol exposure, male from the F3 generation presented significant reduced expression of the proopiomelanocortin gene in the hypothalamus, concomitant with increased DNA methylation on its CGIs (Govorko et al., 2012). The fact that F3 male but not female progeny presented these changes support a paternal inheritance, although epigenome-wide studies in the brain are needed to confirm the observation. A similar paternal inheritance of gestational exposure to ethanol was shown in the mouse, where the F3 offspring presented an altered development of the neocortex, associated with a decreased expression of all three DNA methyltransferases and a global neocortical hypomethylation (Abbott et al., 2018). A study in the same transgenerational mouse model for fetal alcohol exposure confirmed that the developmental changes were associated with altered behavioral phenotypes (Bottom et al., 2022). Overall, these studies highlight the importance of paternal inheritance in the transmission of alcohol exposure. Considering recent works showing that chronic ethanol exposure alters spermatozoa small ncRNAs, and their nucleoside modifications (Rompala et al., 2018), further works will need to assess the contribution of pre-conceptional ethanol exposure to TEI of alcohol abuse.
The abuse of opioids, including the illegal drug heroin and pain relievers such as morphine, fentanyl, codeine, and oxycodone, has emerged as a major health threat over the last decade (Gilardi, Augsburger, & Thomas, 2018). Prevalence of opioid medication signifies that exposure in pregnant mothers has increased (Gilardi et al., 2018). While accumulating data evidence that gestational exposure provokes intergenerational phenotypic and epigenetic changes (Gilardi et al., 2018; Zeid & Gould, 2020), there are currently no studies examining epigenetic transgenerational effects of opioids beyond the F1 generation. This is true for another illicit drug: cocaine. Indeed, epigenetic reprogramming in male germ cells has been demonstrated in response to cocaine exposure (González et al., 2020; Swinford-Jackson et al., 2022) and intergenerational inheritance has been reported (He, Lidow, & Lidow, 2006; Swinford-Jackson et al., 2022; Vassoler, White, Schmidt, Sadri-Vakili, & Christopher, 2013). However, future studies will need to address whether pre-conceptional or gestational cocaine exposure can be transmitted through epigenetic remodeling of germ cells across generations.
With the growing use of medical and recreational cannabis, consumption of cannabis products has dramatically increased in recent years, including by expecting mothers (Carlier, Huestis, Zaami, Pichini, & Busardò, 2020; Volkow, Han, Compton, & McCance-Katz, 2019). More than 100 phytocannabinoids have been isolated from Cannabis sativa, with (−)-trans-Δ9-tetrahydrocannabinol (Δ9-THC) being the predominant phytocannabinoid responsible for the psychoactivity of cannabis (Andre, Hausman, & Guerriero, 2016). As a result, the level of Δ9-THC in cannabis has reached up to 20% of total compounds (Chandra et al., 2019). In recent years, several studies have shown that effects of gestational Δ9-THC exposure are intergenerationally inherited in rodents. Indeed, the descendants of rats exposed to Δ9-THC present functional alterations in brain regions controlling compulsive and goal-directed behaviors (Prini et al., 2018; Szutorisz et al., 2014; Vassoler, Johnson, & Byrnes, 2013; Watson et al., 2015). Furthermore, Δ9-THC also has a clearly described impact on the epigenome, especially on DNA methylation and histone PTMs both in the brain (Prini et al., 2018; Prini, Penna, Sciuccati, Alberio, & Rubino, 2017; Szutorisz & Hurd, 2018; Watson et al., 2015) and in spermatozoa (Murphy et al., 2018; Osborne et al., 2020; Schrott & Murphy, 2020). In particular, one study focusing on cross-comparison of spermatozoa methylome in Δ9-THC-exposed rats and human cannabis users showed that similar pathways were altered (Murphy et al., 2018). These DMRs could further serve as biomarkers when investigating the existence of TEI for cannabis in Mammals.
Finally, accumulating data indicate intergenerational epigenetic inheritance following exposure to tobacco smoke (Hammer et al., 2018). In particular, prior evidence has demonstrated that maternal smoking contributes to asthma and poor lung function in the offspring (Magnus et al., 2015; Rehan, Liu, Sakurai, & Torday, 2013). Whether transgenerational inheritance of asthma occurs in humans is more conflicting. Extensive population-based studies have found a significant correlation between a grandmother’s smoking exposure and asthma in grandchildren, independent of the mother’s smoking exposure (Li, Langholz, Salam, & Gilliland, 2005; Magnus et al., 2015). Mechanistically, the transgenerational effects of smoking on asthma susceptibility could arise from differential DNA methylation in fetal oocytes (Li et al., 2005). In this regard, newborn cord blood samples from the Norwegian Mother and Child Cohort Study have confirmed the existence of altered methylation patterns in newborns born to women who smoked (Joubert et al., 2012). Additionally, a Swedish Birth Cohort study observed higher DNA methylation of insulin-like growth factor 2 (IGF2) in newborns whose mothers smoked; this was correlated to the paternally expressed IGF2 with male offspring preferentially affected (Krauss-Etschmann, Meyer, Dehmel, & Hylkema, 2015). Thus, epigenetic mechanisms may play a role in explaining the increasing incidence of asthma in the offspring of smoking mothers. However, whether epigenetic changes in the germline arise from nicotine, the active component of tobacco, or other chemical constituents of tobacco is difficult to assess retroactively (Zeid & Gould, 2020). Studies in rodents have shown that nicotine can remodel the chromatin through DNA methylation and changes in histone PTMs (Levine et al., 2011; Toledo-Rodriguez et al., 2010). Intergenerational epigenetic inheritance of nicotine exposure has been reported in mice (Dai et al., 2017). However, to date, few studies have addressed the mechanisms of epigenetic inheritance to nicotine exposure in rodents further than the second generation. The F2 offspring of male mice exposed to nicotine displayed behavioral changes that might be due to the observed changes in P0 spermatozoa methylome (McCarthy et al., 2018). This warrants further research to understand the epigenetic mechanisms of paternal inheritance to nicotine exposure. Given the rise in the use of smokeless tobacco products, electronic cigarettes in particular (McCarthy et al., 2020), another open question is whether nicotine and other components, such as flavoring agents, synergize in the epigenetic remodeling of germ cells. Data in a mouse intergenerational model for electronic cigarette showed that co-exposure to nicotine and saccharin in the P0 provoked memory deficits in the F1 whereas individual exposure to either substance alone did not (McCarthy et al., 2020). Overall, as for other drugs of abuse, research on TEI of nicotine is still in its early stage. An additional difficulty for nicotine, as well as cannabis and opioids, is to adapt the dose and delivery mode in rodent models as to mimic the best the reported uses in human populations.
5. Harnessing in vitro models for gene-environment interaction studies
Despite multiple lines of evidence for transgenerational inheritance in rodents, our understanding of the epigenetic mechanisms involved in phenotypic inheritance across multiple generations remains limited. Furthermore, the existence of epigenetic inheritance of phenotypes further than the second generation in humans remains controversial. Here we discuss the current limitations in studies on the impact of the environment on germs cells. We further highlight the potential of in vitro germ cell models and in vitro gametogenesis for TEI studies.
5.1. Current limitations to TEI studies in mammals
Beyond epigenetic inheritance, studies on the impact on the environment on subsequent generations must account for other mechanisms that may be interrelated, including genetic changes and non-genetic changes on the individual, ecological, cultural, or social levels (Danchin et al., 2011). Traditional Mendelian inheritance postulates that phenotypes are inherited based on the transmission of DNA across generations and that abnormal DNA sequences lead to abnormal phenotypes (Senaldi & Smith-Raska, 2020). Epigenetic inheritance builds on this genetic model of inheritance and adds that the chromatin structure of gametes, not just their DNA sequence, also contributes to the offspring phenotype. However, an under-appreciated dimension in inheritance studies is how genetic and epigenetic information are interrelated. For instance, DNA sequence variations affect DNA methylation, not just in CpG loci, but genome-wide (Hannon et al., 2018; McRae et al., 2014). In fact, genetic heritability was shown to be the major cause of similarity in DNA methylation levels in a human cohort (McRae et al., 2014), although the mechanisms at play or the contribution of non-genetic inheritance were not assessed. Another dimension is the evolution of epigenomes over time. Indeed, aging is associated with changes in the chromatin structure both globally and locally (Kane & Sinclair, 2019). Hence, aging should be taken into account when studying interactions between the environment and epigenomes (Barrere-Cain & Allard, 2020; Bertozzi et al., 2021). Finally, non-genetic factors also contribute to epigenetic inheritance of phenotypes. In social animals, the inherited environment is shaped by social status, cultural norms and acquired behaviors (Danchin et al., 2011). For instance, in humans, socioeconomic status, political orientation or religious practices may influence a child’s environment, with consequences on their epigenome and phenotype (Danchin et al., 2011). Overall, the epigenome is shaped by its interaction with the genome and the inherited environment. These confounding factors should be considered in the context of an extended theory of inheritance. In addition, studies aiming at proving TEI in animal models must account for genetic biases (by controlling the genetic background) and acknowledge the extent of non-genetic inheritance.
Another major issue in TEI studies is a technical one. Indeed, while epigenetic studies in male germ cells can be easily performed, considering the relatively large amount of material and accessibility, the small number of oocytes in vivo prevent detailed molecular analyses. In fact, the only epigenetic evidence of inheritance of phenotypes across generations following environmental exposure in rodents are paternal, mostly associated with detectable changes in spermatozoa methylome and ncRNAs profiles (Table 1). The development of new low-input chromatin analyses has allowed to further our understanding of the epigenetic mechanisms regulating the homeostatic development of oocytes (Skene & Henikoff, 2017; Xu & Xie, 2018). It is therefore likely that the same techniques will reveal molecular mechanisms of maternal inheritance of environmentally-induced phenotypes.
5.2. In vitro germ cell models
In parallel to the development of low-input chromatin techniques, epigenetic inheritance studies can benefit from in vitro models for germ cells development. Here, we briefly summarize the existing models and highlight how in vitro gametogenesis can be harnessed in studying the impact of the environment on germ cells epigenome.
The generation of functional mature and fertile gametes in vitro requires the reconstitution of the multi-stepped development of germ cells in vivo. Mouse embryonic stem cells (mESCs) are pluripotent stem cells derived from the inner cell mass of blastocysts and thus constitute an immortalized representation of the preimplantation embryo (Evans & Kaufman, 1981; Nichols & Smith, 2009). In early forays into in vitro gametogenesis, direct derivations of mPGCs from mESCs were found to be poorly efficient, with the induced cells not contributing to healthy offspring (Hayashi, Kawaguchi, Durcova-Hills, & Imai, 2018). The first successful reconstitution of the fetal mouse germ cell specification pathway in vitro was made over 10 years ago. In this system, the so-called mouse PGC-like cells (mPGCLCs) are obtained in a two-step differentiation from mESCs to a mouse epiblast-like intermediate (mEpiLCs), therefore mimicking the stepwise differentiation of mPGCs (Hayashi et al., 2018; Hayashi, Ohta, Kurimoto, Aramaki, & Saitou, 2011). Mouse PGCLCs have been found to accurately recapitulate early developmental processes and epigenetic remodeling of mouse PGCs (Hayashi et al., 2011; Ohta et al., 2017; von Meyenn et al., 2016). In particular, using transgenic ESCs harboring fluorescent reporters for germline markers, properly-specified mPGCLCs can be isolated by flow cytometry (Ohinata, Sano, Shigeta, Yamanaka, & Saitou, 2008). The scalability of this PGCLCs differentiation protocol allows, in turn, the rapid generation of a large number of cells, making of it a powerful system for the interrogation of underlying transcriptional and epigenetic mechanisms. In this regard, we have recently explored the developmental trajectory of mPGCLCs following exposure to BPA and showed a dose-dependent effect on mPGCLCs proliferation in XX but not XY cells (Ooi, Jiang, Kang, & Allard, 2021). Furthermore, mPGCLCs transcriptome revealed a dysregulation of X-linked genes and retrotransposons (Ooi et al., 2021). These results represent the first characterization of the effect of a toxicant exposure on fetal germ cells, highlighting their potential for TEI studies.
In the original mPGCLCs protocol, XY cells were shown to properly undergo spermatogenesis following transplantation in the seminiferous tubules of neonatal mice, whereas XX cells aggregated with female gonadal somatic cells in reconstituted ovaries also underwent maturation to oocytes (Hayashi et al., 2011). Since then, multiple studies have attempted to recreate the entire process of gametogenesis in culture. Today, in vitro full reconstitution of mouse spermatogenesis and oogenesis is a reality (Hikabe et al., 2016; Komeya, Sato, & Ogawa, 2018; Luo & Yu, 2022), with the micro-environment of the gonads and communication with somatic cells being replicated in organoids (Yoshino et al., 2021). However, transposition to human gametogenesis has been hindered by ethical and technical limitations (Luo & Yu, 2022). For instance, human PGCLCs (hPGCLCs) can be obtained from human induced-pluripotent stem cells (hiPSCs), however, proper induction cannot be confirmed by the production of fertile offspring as done in the mouse, for obvious ethical reasons (Luo & Yu, 2022). Furthermore, development of germ cells require cocultures of hPGCLCs with human embryonic somatic gonadal cells, which again, cannot be easily obtained for ethical and technical reasons (Luo & Yu, 2022). With the advent of organoid research (Yoshino et al., 2021), reconstituted gonads will provide a platform to study the molecular mechanisms of mammalian germ cells differentiation. These in vitro gametogenesis systems can also be harnessed for TEI studies, by allowing the expansion of initial material for downstream epigenetic interrogation techniques.
6. Conclusion and future challenges
Epigenetic mechanisms represent a formidable tool for organisms to evolve with their environment. Whether transgenerational inheritance of environmental cues across multiple generations occurs in Mammals is still a very much debated topic. Despite the epidemiological evidence in humans and phenotypical evidence in rodent models that we have described, research on the molecular mechanisms of TEI is still in its infancy and multiple open questions remain.
Future studies will need to define what are the epigenetic mechanisms of transgenerational inheritance. We have shown that oftentimes, changes in male germ cells methylome are pointed as a mode of inheritance. However, we have also discussed how DNA methylation and other epigenetic mechanisms, such as histone PTMs, for instance, are coordinated in homeostatic development of germ cells. Hence, understanding how environmental cues alter epigenetic crosstalks in the germline will be critical for TEI studies. Similarly, deciphering the role of sex specificity in epigenetic remodeling in response to the environment will shed light on conflicting results regarding maternal versus paternal modes of inheritance.
Another dimension is an ethical and practical one: if we are to prove epigenetic remodeling in human germ cells and in human embryos, we will need to rely on models. Most studies have been performed in rodent models, which, ultimately do not recapitulate fully the extent of epigenetic remodeling occurring in humans. In addition to these animal models, we have highlighted the relevance of using in vitro gametogenesis to unravel the molecular mechanisms of epigenetic inheritance in response to environmental changes.
Overall, while the study of transgenerational inheritance in response to changes in our environment is still in its early stages, the development of new tools, techniques and models opens the way for exciting future research that may reshape once more our definition of “epigenetics” in an extended theory of inheritance.
References
- Abbott CW, Rohac DJ, Bottom RT, Patadia S, & Huffman KJ (2018). Prenatal ethanol exposure and neocortical development: A transgenerational model of FASD. Cerebral Cortex, 28, 2908–2921. 10.1093/cercor/bhx168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almouzni G, & Cedar H (2016). Maintenance of epigenetic information. Cold Spring Harbor Perspectives in Biology, 8, 1–23. 10.1101/cshperspect.a019372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, et al. (2006). Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nature Cell Biology, 8, 623–630. 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- Andre CM, Hausman J-F, & Guerriero G (2016). Cannabis sativa: The Plant of the Thousand and one Molecules. Frontiers in Plant Science, 7, 19. 10.3389/fpls.2016.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu-Vieyra CV, Chen R, Agno JE, Glaser S, Anastassiadis K, Stewart Francis A, et al. (2010). MLL2 is required in oocytes for bulk histone 3 lysine 4 trimethylation and transcriptional silencing. PLoS Biology, 8, 53–54. 10.1371/journal.pbio.1000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Cupp AS, Uzumcu M, & Skinner MK (2005). Epigenetic transgenerational actions of endocrine disruptors and male fertility. Reproductive Toxicology, 31, 1466–1469. 10.1016/j.reprotox.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Leathers C, & Skinner MK (2006). Endocrine disruptor vinclozolin induced epigenetic transgenerational adult-onset disease. Endocrinology, 147, 5515–5523. 10.1210/en.2006-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Memon MA, Uzumcu M, & Skinner MK (2006). Transgenerational effect of the endocrine disruptor vinclozolin on male spermatogenesis. Journal of Andrology, 27, 868–879. 10.2164/jandrol.106.000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argos M, Kalra T, Rathouz PJ, Chen Y, Pierce B, Parvez F, et al. (2010). Arsenic exposure from drinking water, and all-cause and chronic-disease mortalities in Bangladesh (HEALS): A prospective cohort study. The Lancet, 376, 252–258. 10.1016/S0140-6736(10)60481-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale TL (2014). Lifetime stress experience: Transgenerational epigenetics and germ cell programming. Dialogues in Clinical Neuroscience, 16, 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, & Kouzarides T (2011). Regulation of chromatin by histone modifications. Cell Research, 21, 381–395. 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao J, & Bedford MT (2016). Epigenetic regulation of the histone-to-protamine transition during spermiogenesis. Reproduction, 151, R55–R70. 10.1530/REP-15-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrere-Cain R, & Allard P (2020). An understudied dimension: Why age needs to be considered when studying epigenetic-environment interactions. Epigenet Insights., 13. 10.1177/2516865720947014. 2516865720947014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JR (2017). Programming the future: Epigenetics in the context of DOHaD. Environmental Health Perspectives, A72. 10.1289/ehp.125-A72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, & Day T (2020). Nongenetic inheritance and multigenerational plasticity in the nematode C. elegans. eLife, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Maamar M, Beck D, Nilsson E, McCarrey JR, & Skinner MK (2022). Developmental alterations in DNA methylation during gametogenesis from primordial germ cells to sperm. iScience, 25, 1–29. 10.1016/j.isci.2022.103786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkel TDM, & Pandey SC (2017). Emerging role of epigenetic mechanisms in alcohol addiction. Alcoholism: Clinical and Experimental Research, 41, 666–680. 10.1111/acer.13338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertozzi TM, Becker JL, Blake GET, Bansal A, Nguyen DK, Fernandez-Twinn DS, et al. (2021). Variably methylated retrotransposons are refractory to a range of environmental perturbations. Nature Genetics, 53, 1233–1242. 10.1038/s41588-021-00898-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco-Miotto T, Craig JM, Gasser YP, van Dijk SJ, & Ozanne SE (2017). Epigenetics and DOHaD: From basics to birth and beyond. Journal of Developmental Origins of Health and Disease, 8, 513–519. 10.1017/S2040174417000733. [DOI] [PubMed] [Google Scholar]
- Bird A (2002). DNA methylation patterns and epigenetic memory. Genes & Development, 16, 6–21. 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bird A (2007). Perceptions of epigenetics. Nature, 447, 396–398. 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- Blake GET, & Watson ED (2016). Unravelling the complex mechanisms of transgenerational epigenetic inheritance. Current Opinion in Chemical Biology, 33, 101–107. 10.1016/j.cbpa.2016.06.008. [DOI] [PubMed] [Google Scholar]
- Bline AP, Le Goff A, & Allard P (2020). What is lost in the weismann barrier? Journal of Developmental Biology, 8. 10.3390/jdb8040035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon R, Silveira GG, & Mostoslavsky R (2020). Nuclear metabolism and the regulation of the epigenome. Nature Metabolism, 2, 1190–1203. 10.1038/s42255-020-00285-4. [DOI] [PubMed] [Google Scholar]
- Bottom RT, Kozanian OO, Rohac DJ, Erickson MA, & Huffman KJ (2022). Transgenerational effects of prenatal ethanol exposure in prepubescent mice. Frontiers in Cell and Development Biology, 10, 812429. 10.3389/fcell.2022.812429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc’his D, Xu GL, Lin CS, Bollman B, & Bestor T (2001). Dnmt3L and the establishment of maternal genomic imprints. Science, 294, 2536–2539. 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- Breton CV, Landon R, Kahn LG, et al. (2021). Exploring the evidence for epigenetic regulation of environmental influences on child health across generations. Communications Biology, 4, 769. 10.1038/s42003-021-02316-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broberg K, Ahmed S, Engström K, Hossain MB, Jurkovic Mlakar S, Bottai M, et al. (2014). Arsenic exposure in early pregnancy alters genome-wide DNA methylation in cord blood, particularly in boys. Journal of Developmental Origins of Health and Disease, 5, 288–298. 10.1017/S2040174414000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruner-Tran KL, & Osteen KG (2011). Developmental exposure to TCDD reduces fertility and negatively affects pregnancy outcomes across multiple generations. Reproductive Toxicology, 31, 344–350. 10.1016/j.reprotox.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, et al. (2010). Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nature Structural and Molecular Biology, 17, 679–687. 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- Caffrey A, Irwin RE, McNulty H, Strain JJ, Lees-Murdock DJ, McNulty BA, et al. (2018). Gene-specific DNA methylation in newborns in response to folic acid supplementation during the second and third trimesters of pregnancy: Epigenetic analysis from a randomized controlled trial. American Journal of Clinical Nutrition, 107, 566–575. 10.1093/ajcn/nqx069. [DOI] [PubMed] [Google Scholar]
- Camsari C, Folger JK, Rajput SK, McGee D, Latham KE, & Smith GW (2019). Transgenerational effects of periconception heavy metal administration on adipose weight and glucose homeostasis in mice at maturity. Toxicological Sciences, 168, 610–619. 10.1093/toxsci/kfz008. [DOI] [PubMed] [Google Scholar]
- Carlier J, Huestis MA, Zaami S, Pichini S, & Busardo FP. (2020). Monitoring perinatal exposure to Cannabis and synthetic cannabinoids. Therapeutic Drug Monitoring, 42, 194–204. 10.1097/FTD.0000000000000667. [DOI] [PubMed] [Google Scholar]
- Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, et al. (2010). Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell, 143, 1084–1096. 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvan MJ, Kalluvila TA, Klingler RH, Larson JK, Pickens M, Mora-Zamorano FX, et al. (2017). Mercury-induced epigenetic transgenerational inheritance of abnormal neurobehavior is correlated with sperm epimutations in zebrafish. PLoS One, 12, 1–26. 10.1371/journal.pone.0176155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra S, Radwan MM, Majumdar CG, Church JC, Freeman TP, & ElSohly MA (2019). New trends in cannabis potency in USA and Europe during the last decade (2008–2017). European Archives of Psychiatry and Clinical Neuroscience, 269, 5–15. 10.1007/s00406-019-00983-5. [DOI] [PubMed] [Google Scholar]
- Chastain G (2006). Alcohol, neurotransmitter systems, and behavior. The Journal of General Psychology, 133, 329–335. 10.3200/GENP.133.4.329-335. [DOI] [PubMed] [Google Scholar]
- Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, et al. (2016). Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science, 351, 397–400. 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- Chen Q, Yan W, & Duan E (2016). Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nature Publishing Group, 17, 733–743. 10.1038/nrg.2016.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JY, Do YS, & Hong YS (2014). Environmental source of arsenic exposure. Journal of Preventive Medicine and Public Health, 47, 253–257. 10.3961/jpmph.14.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare CE, Brassington AH, Kwong WY, & Sinclair KD (2019). One-carbon metabolism: Linking nutritional biochemistry to epigenetic programming of long-term development. Annual Review of Animal Biosciences, 7, 263–287. 10.1146/annurev-animal-020518-115206. [DOI] [PubMed] [Google Scholar]
- Dahan P, Lu V, Nguyen RMT, Kennedy SAL, & Teitell MA (2019). Metabolism in pluripotency: Both driver and passenger? The Journal of Biological Chemistry, 294, 5420–5429. 10.1074/jbc.TM117.000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JA, Jung I, Aanes H, Greggains GD, Manaf A, Lerdrup M, et al. (2016). Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature, 537, 548–552. 10.1038/nature19360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Wang Z, Xu W, Zhang M, Zhu Z, Zhao X, et al. (2017). Paternal nicotine exposure defines different behavior in subsequent generation via hyper-methylation of mmu-miR-15b. Scientific Reports, 7, 1–15. 10.1038/s41598-017-07920-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danchin É, Charmantier A, Champagne FA, Mesoudi A, Pujol B, & Blanchet S (2011). Beyond DNA: Integrating inclusive inheritance into an extended theory of evolution. Nature Reviews. Genetics, 12, 475–486. 10.1038/nrg3028. [DOI] [PubMed] [Google Scholar]
- de Castro BT, Ingerslev LR, Alm PS, Versteyhe S, Massart J, Rasmussen M, et al. (2016). High-fat diet reprograms the epigenome of rat spermatozoa and transgenerationally affects metabolism of the offspring. Molecular Metabolism, 5, 184–197. 10.1016/j.molmet.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhl DMJ, Vrieling H, Miller KA, Wolff GL, & Barsh GS (1994). Neomorphic agouti mutations in obese yellow mice. Nature, 8, 59–65. [DOI] [PubMed] [Google Scholar]
- Dunn GA, & Bale TL (2011). Maternal high-fat diet effects on third-generation female body size via the paternal lineage. Endocrinology, 152, 2228–2236. 10.1210/en.2010-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, & Kaufman MH (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature, 292, 154–156. [DOI] [PubMed] [Google Scholar]
- Fang Y, Xu X, Ding J, Yang L, Doan MT, Karmaus PWF, et al. (2021). Histone crotonylation promotes mesoendodermal commitment of human embryonic stem cells. Cell Stem Cell, 1–16. 10.1016/j.stem.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayomi AP, & Orwig KE (2018). Spermatogonial stem cells and spermatogenesis in mice, monkeys and men. Stem Cell Research, 29, 207–214. 10.1016/j.scr.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fénichel P, Brucker-Davis F, & Chevalier N (2015). The history of Distilbène® (diethylstilbestrol) told to grandchildren-the transgenerational effect. Annales d’Endocrinologie, 76, 253–259. 10.1016/j.ando.2015.03.008. [DOI] [PubMed] [Google Scholar]
- Fleming TP, Watkins AJ, Velazquez MA, Mathers JC, Prentice AM, Stephenson J, et al. (2018). Origins of lifetime health around the time of conception: Causes and consequences. The Lancet, 391, 1842–1852. 10.1016/S0140-6736(18)30312-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser R, & Lin CJ (2016). Epigenetic reprogramming of the zygote in mice and men: On your marks, get set, go! Reproduction, 152, R211–R222. 10.1530/REP-16-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedli M, & Trono D (2015). The developmental control of transposable elements and the evolution of higher species. Annual Review of Cell and Developmental Biology, 31, 429–451. 10.1146/annurev-cellbio-100814-125514. [DOI] [PubMed] [Google Scholar]
- Fullston T, Teague EMCO, Palmer NO, Deblasio MJ, Mitchell M, Corbett M, et al. (2013). Paternal obesity initiates metabolic disturbances in two generations of mice with incomplete penetrance to the F2 generation and alters the transcriptional profile of testis and sperm microRNA content. FASEB Journal, 27, 4226–4243. 10.1096/fj.12-224048. [DOI] [PubMed] [Google Scholar]
- Gilardi F, Augsburger M, & Thomas A (2018). Will widespread synthetic opioid consumption induce epigenetic consequences in future generations? Frontiers in Pharmacology, 9, 1–9. 10.3389/fphar.2018.00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill ME, Hu YC, Lin Y, & Page DC (2011). Licensing of gametogenesis, dependent on RNA binding protein DAZL, as a gateway to sexual differentiation of fetal germ cells. Proceedings of the National Academy of Sciences of the United States of America, 108, 7443–7448. 10.1073/pnas.1104501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkountela S, Zhang KX, Shafiq TA, Liao WW, Hargan-Calvopiña J, Chen PY, et al. (2015). DNA demethylation dynamics in the human prenatal germline. Cell, 161, 1425–1436. 10.1016/j.cell.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, & Bestor TH (2005). Eukaryotic cytosine methyltransferases. Annual Review of Biochemistry, 74, 481–514. 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Gong L, Pan YX, & Chen H (2010). Gestational low protein diet in the rat mediates Igf2 gene expression in male offspring via altered hepatic DNA methylation. Epigenetics, 5, 619–626. 10.4161/epi.5.7.12882. [DOI] [PubMed] [Google Scholar]
- Gong Y, Xue Y, Li X, Zhang Z, Zhou W, Marcolongo P, et al. (2021). Inter- and transgenerational effects of paternal exposure to inorganic arsenic.Pdf. Advanced. Science, 8, 1–14. 10.1002/advs.202002715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González B, Gancedo SN, Garazatua SAJ, Roldán E, Vitullo AD, & González CR (2020). Dopamine receptor D1 contributes to cocaine epigenetic reprogramming of histone modifications in male germ cells. Frontiers in Cell and Development Biology, 8, 1–9. 10.3389/fcell.2020.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorko D, Bekdash RA, Zhang C, & Sarkar DK (2012). Male germline transmits fetal alcohol adverse effect on hypothalamic proopiomelanocortin gene across generations. Biological Psychiatry, 72, 378–388. 10.1016/j.biopsych.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold MD (2016). Spermatogenesis: The commitment to meiosis. Physiological Reviews, 96, 1–17. 10.1152/physrev.00013.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Bosagna C, Covert TR, Haque MM, Settles M, Nilsson EE, Anway MD, et al. (2012). Epigenetic transgenerational inheritance of vinclozolin induced mouse adult onset disease and associated sperm epigenome biomarkers. Reproductive Toxicology, 34, 694–707. 10.1016/j.reprotox.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Bosagna C, Settles M, Lucker B, & Skinner MK (2010). Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS One, 5. 10.1371/journal.pone.0013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guibert S, Forné T, & Weber M (2012). Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome Research, 22, 633–641. 10.1101/gr.130997.111.partments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Luo F, & Lin Q (2020). You are affected by what your parents eat: Diet, epigenetics, transgeneration and intergeneration. Trends in Food Science and Technology, 100, 248–261. 10.1016/j.tifs.2020.04.021. [DOI] [Google Scholar]
- Guo F, Yan L, Guo H, Li L, Hu B, Zhao Y, et al. (2015). The transcriptome and DNA methylome landscapes of human primordial germ cells. Cell, 161, 1437–1452. 10.1016/j.cell.2015.05.015. [DOI] [PubMed] [Google Scholar]
- Haberle V, & Stark A (2018). Eukaryotic core promoters and the functional basis of transcription initiation. Nature Reviews. Molecular Cell Biology, 19, 621–637. 10.1038/s41580-018-0028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggerty C, Kretzmer H, Riemenschneider C, Kumar AS, Mattei AL, Bailly N, et al. (2021). Dnmt1 has de novo activity targeted to transposable elements. Nature Structural and Molecular Biology. 10.1038/s41594-021-00603-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, et al. (2002). Epigenetic reprogramming in mouse primordial germ cells. Mechanisms of Development, 117, 15–23. Available: https://www.ncbi.nlm.nih.gov/pubmed/12204247. [DOI] [PubMed] [Google Scholar]
- Hammer B, Wagner C, Divac Rankov A, Reuter S, Bartel S, Hylkema MN, et al. (2018). In utero exposure to cigarette smoke and effects across generations: A conference of animals on asthma. Clinical and Experimental Allergy, 48, 1378–1390. 10.1111/cea.13283. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, & Cairns BR (2009). Distinctive chromatin in human sperm packages genes for embryo development. Nature, 460, 473–478. 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna CW, Demond H, & Kelsey G (2018). Epigenetic regulation in development: Is the mouse a good model for the human? Human Reproduction Update, 24, 556–576. 10.1093/humupd/dmy021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna CW, Taudt A, Huang J, Gahurova L, Kranz A, Andrews S, et al. (2018). MLL2 conveys transcription-independent H3K4 trimethylation in oocytes. Nature Structural and Molecular Biology, 25, 73–82. 10.1038/s41594-017-0013-5. [DOI] [PubMed] [Google Scholar]
- Hannon E, Knox O, Sugden K, Burrage J, Wong CCY, Belsky DW, et al. (2018). Characterizing genetic and environmental influences on variable DNA methylation using monozygotic and dizygotic twins. PLoS Genetics, 14, 1–27. 10.1371/journal.pgen.1007544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao C, Gely-Pernot A, Kervarrec C, Boudjema M, Becker E, Khil P, et al. (2016). Exposure to the widely used herbicide atrazine results in deregulation of global tissue-specific RNA transcription in the third generation and is associated with a global decrease of histone trimethylation in mice. Nucleic Acids Research, 44, 9784–9802. 10.1093/nar/gkw840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser MT, Aufsatz W, Jonak C, & Luschnig C (2011). Transgenerational epigenetic inheritance in plants. Biochimica et Biophysica Acta, Gene Regulatory Mechanisms, 1809, 459–468. 10.1016/j.bbagrm.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Kawaguchi T, Durcova-Hills G, & Imai H (2018). Generation of germ cells from pluripotent stem cells in mammals. Reproductive Medicine and Biology, 17, 107–114. 10.1002/rmb2.12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Ohta H, Kurimoto K, Aramaki S, & Saitou M (2011). Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell, 146, 519–532. 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- He F, Lidow IA, & Lidow MS (2006). Consequences of paternal cocaine exposure in mice. Neurotoxicology and Teratology, 28, 198–209. 10.1016/j.ntt.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Heintzman N, Stuart R, Hon G, Fu Y, Ching C, Hawkins RD, et al. (2007). Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature Genetics, 39, 311–318. 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Hernández-Sánchez J, Valles S, & Bardullas U (2022). Linking arsenic, DNA methylation biomarkers, and transgenerational neurotoxicity: Modeling in zebrafish. Biomarkers. Toxicology, 1–24. 10.1007/978-3-030-87225-0_59-1. [DOI] [Google Scholar]
- Heyt KM, & Thakur J (2021). Regulation of epigenetic processes by non-coding RNAs. Nucleus, 64, 285–301. 10.1007/s13237-021-00372-1. [DOI] [Google Scholar]
- Hikabe O, Hamazaki N, Nagamatsu G, Obata Y, Hirao Y, Hamada N, et al. (2016). Reconstitution in vitro of the entire cycle of the mouse female germ line. Nature, 539, 299–303. 10.1038/nature20104. [DOI] [PubMed] [Google Scholar]
- Hiura H, Obata Y, Komiyama J, Shirai M, & Kono T (2006). Oocyte growth-dependent progression of maternal imprinting in mice. Genes to Cells, 11, 353–361. 10.1111/j.1365-2443.2006.00943.x. [DOI] [PubMed] [Google Scholar]
- Holoch D, & Moazed D (2015). RNA-mediated epigenetic regulation of gene expression. Nature Reviews. Genetics, 16, 71–84. 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon GC, Hawkins RD, & Ren B (2009). Predictive chromatin signatures in the mammalian genome. Human Molecular Genetics, 18, 12–14. 10.1093/hmg/ddp409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsthemke B (2018). A critical view on transgenerational epigenetic inheritance in humans. Nature Communications, 9, 1–4. 10.1038/s41467-018-05445-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Jiang L, Lu F, Suzuki T, & Zhang Y (2017). Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature, 547, 419–424. 10.1038/nature23262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawaid A, & Mansuy IM (2019). Inter- and transgenerational inheritance of behavioral phenotypes. Current Opinion in Behavioral Sciences, 25, 96–101. 10.1016/j.cobeha.2018.12.004. [DOI] [Google Scholar]
- Jodar M (2019). Sperm and seminal plasma RNAs: What roles do they play beyond fertilization? Reproduction, 158, R113–R123. [DOI] [PubMed] [Google Scholar]
- Joubert BR, Den Dekker HT, Felix JF, Bohlin J, Ligthart S, Beckett E, et al. (2016). Maternal plasma folate impacts differential DNA methylation in an epigenome-wide meta-analysis of newborns. Nature. Communications, 7. 10.1038/ncomms10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert BR, Håberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, et al. (2012). 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environmental Health Perspectives, 120, 1425–1431. 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaikkonen MU, Lam MTY, & Glass CK (2011). Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovascular Research, 90, 430–440. 10.1093/cvr/cvr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane AE, & Sinclair DA (2019). Epigenetic changes during aging and their reprogramming potential. Critical Reviews in Biochemistry and Molecular Biology, 54, 61–83. 10.1080/10409238.2019.1570075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda M, Okano M, Hata K, Sado T, Tsujimoto H, Li E, et al. (2004). Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature, 429, 900–903. 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- Kaspar D, Hastreiter S, Irmler M, Hrabé de Angelis M, & Beckers J (2020). Nutrition and its role in epigenetic inheritance of obesity and diabetes across generations. Mammalian Genome, 31, 119–133. 10.1007/s00335-020-09839-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, et al. (2007). Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Human Molecular Genetics, 16, 2272–2280. 10.1093/hmg/ddm179. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Sakurai T, Imai M, Takahashi N, Fukuda A, Yayoi O, et al. (2012). Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genetics, 8. 10.1371/journal.pgen.1002440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Sakurai T, Miura F, Imai M, Mochiduki K, Yanagisawa E, et al. (2013). High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome Research, 23, 616–627. 10.1101/gr.148023.112.Freely. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeya M, Sato T, & Ogawa T (2018). In vitro spermatogenesis: A century-long research journey still half way. Reproductive Medicine and Biology, 17, 407–420. 10.1002/rmb2.12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota SK, & Feil R (2010). Epigenetic transitions in germ cell development and meiosis. Developmental Cell, 19, 675–686. 10.1016/j.devcel.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Krauss-Etschmann S, Meyer KF, Dehmel S, & Hylkema MN (2015). Inter-and transgenerational epigenetic inheritance: Evidence in asthma and COPD? Clinical Epigenetics, 7, 1–13. 10.1186/s13148-015-0085-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremsky I, & Corces VG (2020). Protection from DNA re-methylation by transcription factors in primordial germ cells and pre-implantation embryos can explain trans-generational epigenetic inheritance. Genome Biology, 21, 118. 10.1186/s13059-020-02036-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo N, Toh H, Shirane K, Shirakawa T, Kobayashi H, Sato T, et al. (2015). DNA methylation and gene expression dynamics during spermatogonial stem cell differentiation in the early postnatal mouse testis. BMC Genomics, 16, 1–17. 10.1186/s12864-015-1833-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Hayashi K, Ohta H, Kiyonari H, Mitani T, et al. (2015). Quantitative dynamics of chromatin remodeling during germ cell specification from mouse embryonic stem cells. Cell Stem Cell, 16, 517–532. 10.1016/j.stem.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Larose H, Shami AN, Abbott H, Manske G, Lei L, & Hammoud SS (2019). Gametogenesis: A journey from inception to conception. In Current topics in developmental biology (pp. 257–310). Elsevier Inc. 10.1016/bs.ctdb.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legoff L, D’Cruz SC, Tevosian S, & Smagulova F (2019). Long noncoding RNA and epigenetic inheritance. Transgenerational. Epigenetics, 213–248. 10.1016/b978-0-12-816363-4.00010-9. [DOI] [Google Scholar]
- Lesch BJ, & Page DC (2012). Genetics of germ cell development. Nature Reviews. Genetics. 10.1038/nrg3294. [DOI] [PubMed] [Google Scholar]
- Levine A, Huang YY, Drisaldi B, Griffin EA, Pollak DD, Xu S, et al. (2011). Molecular mechanism for a gateway drug: Epigenetic changes initiated by nicotine prime gene expression by cocaine. Science Translational Medicine, 3, 1–10. 10.1126/scitranslmed.3003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Egervari G, Wang Y, Berger SL, & Lu Z (2018). Regulation of chromatin and gene expression by metabolic enzymes and metabolites. Nature Reviews. Molecular Cell Biology, 19, 563–578. 10.1038/s41580-018-0029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y-F, Langholz B, Salam MT, & Gilliland FD (2005). Maternal and grandmaternal smoking patterns are associated with early childhood asthma. Chest, 127, 1232–1241. 10.1016/s0012-3692(15)34472-x. [DOI] [PubMed] [Google Scholar]
- Lismer A, Dumeaux V, Lafleur C, Lambrot R, Brind’Amour J, Lorincz MC, et al. (2021). Histone H3 lysine 4 trimethylation in sperm is transmitted to the embryo and associated with diet-induced phenotypes in the offspring. Developmental Cell, 56, 671–686.e6. 10.1016/j.devcel.2021.01.014. [DOI] [PubMed] [Google Scholar]
- Lu SC, & Mato JM (2005). Role of methionine adenosyltransferase and S-adenosylmethionine in alcohol-associated liver cancer. Alcohol, 35, 227–234. 10.1016/j.alcohol.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Lucifero D, La Salle S, Bourc’his D, Martel J, Bestor TH, & Trasler JM (2007). Coordinate regulation of DNA methyltransferase expression during oogenesis. BMC Developmental Biology, 7, 1–14. 10.1186/1471-213X-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumey LH, Stein AD, & Susser E (2011). Prenatal famine and adult health L.H. Annual Review of Public Health, 32, 1–26. 10.1146/annurev-publhealth-031210-101230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, & Yu Y (2022). Research advances in gametogenesis and embryogenesis using pluripotent stem cells. Frontiers in Cell and Development Biology, 9, 1–14. 10.3389/fcell.2021.801468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko F (2018). The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nature Reviews. Genetics, 19, 81–92. 10.1038/nrg.2017.80. [DOI] [PubMed] [Google Scholar]
- Magnus MC, Håberg SE, Karlstad Ø, Nafstad P, London SJ, & Nystad W (2015). Grandmother’s smoking when pregnant with the mother and asthma in the grandchild: The Norwegian mother and child cohort study. Thorax, 70, 237–243. 10.1136/thoraxjnl-2014-206438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnúsdóttir E, & Surani MA (2014). How to make a primordial germ cell. Development (Cambridge)., 141, 245–252. 10.1242/dev.098269. [DOI] [PubMed] [Google Scholar]
- Maloney B, Bayon BL, Zawia NH, & Lahiri DK (2018). Latent consequences of early-life lead (pb) exposure and the future: Addressing the pb crisis. Neurotoxicology, 68, 126–132. 10.1016/j.neuro.2018.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manikkam M, Tracey R, Guerrero-Bosagna C, & Skinner MK (2012). Dioxin (TCDD) induces epigenetic transgenerational inheritance of adult onset disease and sperm epimutations. PLoS One, 7, e46249–1–15. 10.1371/journal.pone.0046249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manikkam M, Tracey R, Guerrero-Bosagna C, & Skinner MK (2013). Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS One, 8. 10.1371/journal.pone.0055387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marczylo EL, Jacobs MN, & Gant TW (2016). Environmentally induced epigenetic toxicity: Potential public health concerns. Critical Reviews in Toxicology, 46, 676–700. 10.1080/10408444.2016.1175417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DM, Lowe SE, Morgan TJ, Cannon EN, Biederman J, Spencer TJ, et al. (2020). Transgenerational transmission of behavioral phenotypes produced by exposure of male mice to saccharin and nicotine. Scientific Reports, 10, 1–14. 10.1038/s41598-020-68883-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DM, Morgan TJ Jr., Lowe SE, Williamson MJ, Spencer TJ, Biederman J, et al. (2018). Nicotine exposure of male mice produces behavioral impairment in multiple generations of descendants. PLoS Biology, 16, e2006497. 10.1371/journal.pbio.2006497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson NO, Fullston T, Kang WX, Sandeman LY, Corbett MA, Owens JA, et al. (2016). Paternal under-nutrition programs metabolic syndrome in offspring which can be reversed by antioxidant/vitamin food fortification in fathers. Scientific Reports, 6, 1–14. 10.1038/srep27010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRae AF, Powell JE, Henders AK, Bowdler L, Hemani G, Shah S, et al. (2014). Contribution of genetic variation to transgenerational inheritance of DNA methylation. Genome Biology, 15, R73. 10.1186/gb-2014-15-5-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei H, Kozuka C, Hayashi R, Kumon M, Koseki H, & Inoue A (2021). H2AK119ub1 guides maternal inheritance and zygotic deposition of H3K27me3 in mouse embryos. Nature Genetics, 53, 539–550. 10.1038/s41588-021-00820-3. [DOI] [PubMed] [Google Scholar]
- Mews P, Egervari G, Nativio R, Sidoli S, Donahue G, Lombroso SI, et al. (2019). Alcohol metabolism contributes to brain histone acetylation. Nature, 574, 717–721. 10.1038/s41586-019-1700-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarelli P, Gerthoux PM, Needham LL, Patterson DG, Limonta G, Falbo R, et al. (2011). Perinatal exposure to low doses of dioxin can permanently impair human semen quality. Environmental Health Perspectives, 119, 713–718. 10.1289/ehp.1002134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Y, Hu X, Yang P, Sun L, Gu W, & Zhang M (2021). The effects of cadmium on the development of Drosophila and its transgenerational inheritance effects. Toxicology, 462, 1–10. 10.1016/j.tox.2021.152931. [DOI] [PubMed] [Google Scholar]
- Murphy SK, Itchon-Ramos N, Visco Z, Huang Z, Grenier C, Schrott R, et al. (2018). Cannabinoid exposure and altered DNA methylation in rat and human sperm. Epigenetics, 13, 1208–1221. 10.1080/15592294.2018.1554521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murr R (2010). Interplay between different epigenetic modifications and mechanisms. Advances in Genetics, 70, 101–141. 10.1016/B978-0-12-380866-0.60005-8. [DOI] [PubMed] [Google Scholar]
- Nagano M, Hu B, Yokobayashi S, Yamamura A, Umemura F, Coradin M, et al. (2022). Nucleome programming is required for the foundation of totipotency in mammalian germline development. The EMBO Journal, 41, 1–34. 10.15252/embj.2022110600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Liu YJ, Nakashima H, Umehara H, Inoue K, Matoba S, et al. (2012). PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature, 486, 415–419. 10.1038/nature11093. [DOI] [PubMed] [Google Scholar]
- Nava-Rivera LE, Betancourt-Martínez ND, Lozoya-Martínez R, Carranza-Rosales P, Guzmán-Delgado NE, Carranza-Torres IE, et al. (2021). Transgenerational effects in DNA methylation, genotoxicity and reproductive phenotype by chronic arsenic exposure. Scientific Reports, 11, 8276. 10.1038/s41598-021-87677-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ (2014). Epigenetic mechanisms of drug addiction. Neuropharmacology, 76, 259–268. 10.1016/j.neuropharm.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SF, Lin RCY, Laybutt DR, Barres R, Owens JA, & Morris MJ (2010). Chronic high-fat diet in fathers programs β 2-cell dysfunction in female rat offspring. Nature, 467, 963–966. 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- Nichols J, & Smith A (2009). Naive and primed pluripotent states. Cell Stem Cell, 4, 487–492. 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Nishiyama A, Mulholland CB, Bultmann S, Kori S, Endo A, Saeki Y, et al. (2020). Two distinct modes of DNMT1 recruitment ensure stable maintenance DNA methylation. Nature Communications, 11, 1–7. 10.1038/s41467-020-15006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakovic B, Lewis S, Halliday J, Kennedy J, Burgner DP, Czajko A, et al. (2019). Assisted reproductive technologies are associated with limited epigenetic variation at birth that largely resolves by adulthood. Nature. Communications, 10. 10.1038/s41467-019-11929-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacka-Woszuk J, Szczerbal I, Malinowska AM, & Chmurzynska A (2018). Transgenerational effects of prenatal restricted diet on gene expression and histone modifications in the rat. PLoS One, 13, 1–14. 10.1371/journal.pone.0193464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohinata Y, Sano M, Shigeta M, Yamanaka K, & Saitou M (2008). A comprehensive, non-invasive visualization of primordial germ cell development in mice by the Prdm1-mVenus and Dppa3-ECFP double transgenic reporter. Reproduction, 136, 503–514. 10.1530/REP-08-0053. [DOI] [PubMed] [Google Scholar]
- Ohta H, Kurimoto K, Okamoto I, Nakamura T, Yabuta Y, Miyauchi H, et al. (2017). In vitro expansion of mouse primordial germ cell- like cells recapitulates an epigenetic blank slate. The EMBO Journal, 36, 1888–1907. 10.15252/embj.201695862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, & Yamaguchi K (2017). Epigenetic modifications and reprogramming in paternal pronucleus: Sperm, preimplantation embryo, and beyond. Cellular and Molecular Life Sciences, 74, 1957–1967. 10.1007/s00018-016-2447-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, & Li E (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for De novo methylation and mammalian development. Cell, 99, 247–257. [DOI] [PubMed] [Google Scholar]
- Ong M-L, Lin X, & Holbrook JD (2015). Measuring epigenetics as the mediator of gene/environment interactions in DOHaD. Journal of Developmental Origins of Health and Disease, 6, 10–16. 10.1017/S2040174414000506. [DOI] [PubMed] [Google Scholar]
- Ooi SKT, Jiang H, Kang Y, & Allard P (2021). Examining the developmental trajectory of an in vitro model of mouse primordial germ cells following exposure to environmentally relevant bisphenol a levels. Environmental Health Perspectives, 129, 97013. 10.1289/EHP8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne AJ, Pearson JF, Noble AJ, Gemmell NJ, Horwood LJ, Boden JM, et al. (2020). Genome-wide DNA methylation analysis of heavy cannabis exposure in a New Zealand longitudinal cohort. Translational Psychiatry, 10, 1–10. 10.1038/s41398-020-0800-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan N, Jia D, Geary-Joo C, Wu X, Ferguson-Smith AC, Fung E, et al. (2013). Mutation in folate metabolism causes epigenetic instability and transgenerational effects on development. Cell, 155, 81–93. 10.1016/j.cell.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilsner JR, Shershebnev A, Medvedeva YA, Suvorov A, Wu H, Goltsov A, et al. (2018). Peripubertal serum dioxin concentrations and subsequent sperm methylome profiles of young Russian adults. Reproductive Toxicology, 78, 40–49. 10.1016/j.reprotox.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo A, & Dillon N (2015). Three-dimensional genome architecture: Players and mechanisms. Nature Reviews. Molecular Cell Biology, 16, 245–257. 10.1038/nrm3965. [DOI] [PubMed] [Google Scholar]
- Prini P, Penna F, Sciuccati E, Alberio T, & Rubino T (2017). Chronic Δ9-THC exposure differently affects histone modifications in the adolescent and adult rat brain. International Journal of Molecular Sciences, 18. 10.3390/ijms18102094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prini P, Rusconi F, Zamberletti E, Gabaglio M, Penna F, Fasano M, et al. (2018). Adolescent THC exposure in female rats leads to cognitive deficits through a mechanism involving chromatin modifications in the prefrontal cortex. Journal of Psychiatry & Neuroscience, 43, 87–101. 10.1503/jpn.170082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford EJ, Ito M, Shi H, Corish JA, Yamazawa K, Isganaitis E, et al. (2014). In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science, 345, 1255903–1–8. 10.1126/science.1255903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjit N, Siefert K, & Padmanabhan V (2010). Bisphenol-a and disparities in birth outcomes: A review and directions for future research. Journal of Perinatology, 30, 2–9. 10.1038/jp.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray PD, Yosim A, & Fry RC (2014). Incorporating epigenetic data into the risk assessment process for the toxic metals arsenic, cadmium, chromium, lead, and mercury: Strategies and challenges. Frontiers in Genetics, 5, 1–26. 10.3389/fgene. 2014.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehan VK, Liu J, Sakurai R, & Torday JS (2013). Perinatal nicotine-induced transgenerational asthma. American Journal of Physiology. Lung Cellular and Molecular Physiology, 305, L501–L507. 10.1152/ajplung.00078.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichard JF, & Puga A (2010). Effects of arsenic exposure on DNA methylation and epigenetic gene regulation. Epigenomics, 2, 87–104. 10.2217/epi.09.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, McHale CM, Skibola CF, Smith AH, Smith MT, & Zhang L (2011). An emerging role for epigenetic dysregulation in arsenic toxicity and carcinogenesis. Environmental Health Perspectives, 119, 11–19. 10.1289/ehp.1002114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochester JR (2013). Bisphenol a and human health: A review of the literature. Reproductive Toxicology, 42, 132–155. 10.1016/j.reprotox.2013.08.008. [DOI] [PubMed] [Google Scholar]
- Rompala GR, Mounier A, Wolfe CM, Lin Q, Lefterov I, & Homanics GE (2018). Heavy chronic intermittent ethanol exposure alters small noncoding RNAs in mouse sperm and epididymosomes. Frontiers in Genetics, 9, 1–14. 10.3389/fgene.2018.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Zhang D, Allis CD, & Zhao Y (2017). Metabolic regulation of gene expression through histone acylations. Nature Reviews. Molecular Cell Biology, 18, 90–101. 10.1038/nrm.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saidur Rahman MD, Pang WK, Ryu DY, Park YJ, & Pang MG (2020). Multigenerational and transgenerational impact of paternal bisphenol a exposure on male fertility in a mouse model. Human Reproduction, 35, 1740–1752. 10.1093/humrep/deaa139. [DOI] [PubMed] [Google Scholar]
- Saitou M, & Yamaji M (2012). Primordial germ cells in mice. Cold Spring Harbor Perspectives in Biology, 4. 10.1101/cshperspect.a008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez F, & Smitz J (2012). Molecular control of oogenesis. Biochimica et Biophysica Acta, Molecular Basis of Disease, 1822, 1896–1912. 10.1016/j.bbadis.2012.05.013. [DOI] [PubMed] [Google Scholar]
- Sarker G, Berrens R, von Arx J, Pelczar P, Reik W, Wolfrum C, et al. (2018). Transgenerational transmission of hedonic behaviors and metabolic phenotypes induced by maternal overnutrition. Translational Psychiatry, 8, 1–16. 10.1038/s41398-018-0243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, & Matsui Y (2008). Epigenetic events in mammalian germ-cell development: Reprogramming and beyond. Nature Reviews. Genetics, 9. 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- Schrott R, & Murphy SK (2020). Cannabis use and the sperm epigenome: A budding concern? Environmental Epigenetics, 6, 1–10. 10.1093/eep/dvaa002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster A, Skinner MK, & Yan W (2016). Ancestral vinclozolin exposure alters the epigenetic transgenerational inheritance of sperm small noncoding RNAs. Environmental Epigenetics, 2, 1–10. 10.1093/eep/dvw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, et al. (2012). The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Molecular Cell, 48, 849–862. 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y, Yamaji M, Yabuta Y, Sano M, Shigeta M, Matsui Y, et al. (2007). Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development, 134, 2627–2638. 10.1242/dev.005611. [DOI] [PubMed] [Google Scholar]
- Sen A, Heredia N, Senut MC, Land S, Hollocher K, Lu X, et al. (2015). Multigenerational epigenetic inheritance in humans: DNA methylation changes associated with maternal exposure to lead can be transmitted to the grandchildren. Scientific Reports, 5, 1–10. 10.1038/srep14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senaldi L, & Smith-Raska M (2020). Evidence for germline non-genetic inheritance of human phenotypes and diseases. Clinical Epigenetics, 12, 1–12. 10.1186/s13148-020-00929-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendžikaitė G, & Kelsey G (2019). The role and mechanisms of DNA methylation in the oocyte. Essays in Biochemistry, 63, 691–705. 10.1042/EBC20190043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha QQ, Jiang Y, Yu C, Xiang Y, Dai XX, Jiang JC, et al. (2020). CFP1-dependent histone H3K4 trimethylation in murine oocytes facilitates ovarian follicle recruitment and ovulation in a cell-nonautonomous manner. Cellular and Molecular Life Sciences, 77, 2997–3012. 10.1007/s00018-019-03322-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan CM, Fang Y, & Jia S (2021). Leaving histone unturned for epigenetic inheritance. FEBS Journal, 1–11. 10.1111/febs.16260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S-I, Suetake I, Iwamatsu A, Endo TA, et al. (2007). The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature, 450, 908–912. 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- Shea JM, Serra RW, Carone BR, Shulha HP, Kucukural A, Ziller MJ, et al. (2015). Genetic and epigenetic variation, but not diet, Shape the Sperm Methylome. Developmental Cell, 35, 750–758. 10.1016/j.devcel.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirane K, Miura F, Ito T, & Lorincz MC (2020). NSD1-deposited H3K36me2 directs de novo methylation in the mouse male germline and counteracts Polycomb-associated silencing. Nature Genetics, 52, 1088–1098. 10.1038/s41588-020-0689-z. [DOI] [PubMed] [Google Scholar]
- Siklenka K, Erkek S, Godmann M, Lambrot R, McGraw S, Lafleur C, et al. (2015). Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science, 350, aab2006. 10.1126/science.aab2006. [DOI] [PubMed] [Google Scholar]
- Skene PJ, & Henikoff S (2017). An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife, 6, 1–35. 10.7554/eLife.21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeester L, Rager JE, Bailey KA, Guan X, Smith N, García-Vargas G, et al. (2011). Epigenetic changes in individuals with arsenicosis. Chemical Research in Toxicology, 24, 165–167. 10.1021/tx1004419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AH, Hopenhayn-Rich C, Bates MN, Goeden HM, Hertz-Picciotto I, Duggan HM, et al. (1992). Cancer risks from arsenic in drinking water. Environmental Health Perspectives, 97, 259–267. 10.1289/ehp.9297259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolewski M, Abston K, Conrad K, Marvin E, Harvey K, Susiarjo M, et al. (2020). Lineage-and sex-dependent behavioral and biochemical transgenerational consequences of developmental exposure to lead, prenatal stress, and combined lead and prenatal stress in mice. Environmental Health Perspectives, 128, 1–14. 10.1289/EHP4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubry A (2015). Epigenetic inheritance and evolution: A paternal perspective on dietary influences. Progress in Biophysics and Molecular Biology, 118, 79–85. 10.1016/j.pbiomolbio.2015.02.008. [DOI] [PubMed] [Google Scholar]
- Stewart KR, Veselovska L, Kim J, Huang J, Saadeh H, Tomizawa SI, et al. (2015). Dynamic changes in histone modifications precede de novo DNA methylation in oocytes. Genes and Development, 29, 2449–2462. 10.1101/gad.271353.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, & Allis DC (2000). The language of covalent histone modifications. Nature, 403, 41–45. 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Svanes C, Bertelsen RJ, Accordini S, Holloway JW, Júliússon P., Boateng E, et al. (2021). Exposures during the prepuberty period and future offspring’s health: Evidence from human cohort studies. Biology of Reproduction, 105, 667–680. 10.1093/biolre/ioab158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinford-Jackson SE, Fant B, Wimmer ME, Chan D, Knouse MC, Sarmiento M, et al. (2022). Cocaine-induced changes in sperm Cdkn1a methylation are associated with cocaine resistance in male offspring. The Journal of neuroscience: The Official Journal of the Society for Neuroscience., 42, 2905–2916. 10.1523/JNEUROSCI.3172-20.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szutorisz H, DiNieri JA, Sweet E, Egervari G, Michaelides M, Carter JM, et al. (2014). Parental THC exposure leads to compulsive heroin-seeking and altered striatal synaptic plasticity in the subsequent generation. Neuropsychopharmacology, 39, 1315–1323. 10.1038/npp.2013.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szutorisz H, & Hurd YL (2018). High times for cannabis: Epigenetic imprint and its legacy on brain and behavior. Neuroscience and Biobehavioral Reviews, 85, 93–101. 10.1016/j.neubiorev.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, et al. (2011). Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell, 146, 1016–1028. 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WWC, Kobayashi T, Irie N, Dietmann S, & Surani MA (2016). Specification and epigenetic programming of the human germ line. Nature Reviews. Genetics. 10.1038/nrg.2016.88. [DOI] [PubMed] [Google Scholar]
- Titus L, Hatch EE, Drake KM, Parker SE, Hyer M, Palmer JR, et al. (2019). Reproductive and hormone-related outcomes in women whose mothers were exposed in utero to diethylstilbestrol (DES): A report from the US National Cancer Institute DES third generation study. Reproductive Toxicology, 84, 32–38. 10.1016/j.reprotox.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobi EW, Goeman JJ, Monajemi R, Gu H, Putter H, Zhang Y, et al. (2014). DNA methylation signatures link prenatal famine exposure to growth and metabolism. Nature Communications, 5, 5592. 10.1038/ncomms6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Rodriguez M, Lotfipour S, Leonard G, Perron M, Richer L, Veillette S, et al. (2010). Maternal smoking during pregnancy is associated with epigenetic modifications of the brain-derived neurotrophic Factor-6 exon in adolescent offspring. American Journal of Medical Genetics, 153B, 1350–1354. [DOI] [PubMed] [Google Scholar]
- Tomizawa SI, Nowacka-Woszuk J, & Kelsey G (2012). DNA methylation establishment during oocyte growth: Mechanisms and significance. International Journal of Developmental Biology, 56, 867–875. 10.1387/ijdb.120152gk. [DOI] [PubMed] [Google Scholar]
- Travis CC, & Hattemer-Frey HA (1991). Human exposure to dioxin. The Science of the Total Environment, 104, 97–127. [DOI] [PubMed] [Google Scholar]
- Tucci V, Isles AR, Kelsey G, Ferguson-Smith AC, Bartolomei MS, Benvenisty N, et al. (2019). Genomic imprinting and physiological processes in mammals. Cell, 176, 952–965. 10.1016/j.cell.2019.01.043. [DOI] [PubMed] [Google Scholar]
- Van Der Heijden GW, Ramos L, Baart EB, Van Den Berg IM, Derijck AAHA, Van Der Vlag J, et al. (2008). Sperm-derived histones contribute to zygotic chromatin in humans. BMC Developmental Biology, 8, 6–11. 10.1186/1471-213X-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Otterdijk SD, & Michels KB (2016). Transgenerational epigenetic inheritance in mammals: How good is the evidence? FASEB Journal, 30, 2457–2465. 10.1096/fj.201500083. [DOI] [PubMed] [Google Scholar]
- Vandegehuchte MB, Lemière F, Vanhaecke L, Vanden Berghe W, & Janssen CR (2010). Direct and transgenerational impact on Daphnia magna of chemicals with a known effect on DNA methylation. Comparative Biochemistry and Physiology, Part C: Toxicology & Pharmacology, 151, 278–285. 10.1016/j.cbpc.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Vandenberg LN, Hauser R, Marcus M, Olea N, & Welshons WV (2007). Human exposure to bisphenol a (BPA). Reproductive Toxicology, 24, 139–177. 10.1016/j.reprotox.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Vassoler FM, Johnson NL, & Byrnes EM (2013). Female adolescent exposure to cannabinoids causes transgenerational effects on morphine sensitization in female offspring in the absence of in utero exposure. Journal of Psychopharmacology, 27, 1015–1022. 10.1177/0269881113503504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassoler FM, White SL, Schmidt HD, Sadri-Vakili G, & Christopher PR (2013). Epigenetic inheritance of a cocaine-resistance phenotype. Nature Neuroscience, 16, 42–47. 10.1038/nn.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenendaal MVE, Painter RC, De Rooij SR, Bossuyt PMM, Van Der Post JAM, Gluckman PD, et al. (2013). Transgenerational effects of prenatal exposure to the 1944–45 Dutch famine. BJOG: An International Journal of Obstetrics and Gynaecology, 120, 548–554. 10.1111/1471-0528.12136. [DOI] [PubMed] [Google Scholar]
- Velazquez MA, Fleming TP, & Watkins AJ (2019). Periconceptional environment and the developmental origins of disease. Journal of Endocrinology, 242, T33–T49. 10.1530/JOE-18-0676. [DOI] [PubMed] [Google Scholar]
- Venkatesh S, & Workman JL (2015). Histone exchange, chromatin structure and the regulation of transcription. Nature Reviews. Molecular Cell Biology, 16, 178–189. 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- Verdikt R, & Allard P (2021). Metabolo-epigenetics: The interplay of metabolism and epigenetics during early germ cells development. Biology of Reproduction. 10.1093/biolre/ioab118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veselovska L, Smallwood SA, Saadeh H, Stewart KR, Krueger F, Maupetit Méhouas S, et al. (2015). Deep sequencing and de novo assembly of the mouse oocyte transcriptome define the contribution of transcription to the DNA methylation landscape. Genome Biology, 16, 1–17. 10.1186/s13059-015-0769-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vipin VA, Blesson CS, & Yallampalli C (2022). Maternal low protein diet and fetal programming of lean type 2 diabetes. World Journal of Diabetes, 13, 185–202. 10.4239/wjd.v13.i3.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, Han B, Compton WM, & McCance-Katz EF (2019). Self-reported medical and nonmedical Cannabis use among pregnant women in the United States. JAMA: The Journal of the American Medical Association, 322, 167–169. 10.1001/jama.2018.20391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Meyenn F, Berrens RV, Andrews S, Santos F, Collier AJ, Krueger F, et al. (2016). Comparative principles of DNA methylation reprogramming during human and mouse in vitro primordial germ cell specification. Developmental Cell, 39, 104–115. 10.1016/j.devcel.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH (2012). The epigenotype. 1942. International Journal of Epidemiology, 41, 10–13. 10.1093/ije/dyr184. [DOI] [PubMed] [Google Scholar]
- Wanner NM, Colwell ML, & Faulk C (2019). The epigenetic legacy of illicit drugs: Developmental exposures and late-life phenotypes. Environmental Epigenetics, 5, 1–10. 10.1093/eep/dvz022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins AJ, Lucas ES, & Fleming TP (2010). Impact of the periconceptional environment on the programming of adult disease. Journal of Developmental Origins of Health and Disease, 1, 87–95. 10.1017/S2040174409990195. [DOI] [PubMed] [Google Scholar]
- Watson CT, Szutorisz H, Garg P, Martin Q, Landry JA, Sharp AJ, et al. (2015). Genome-wide DNA methylation profiling reveals epigenetic changes in the rat nucleus Accumbens associated with cross-generational effects of adolescent THC exposure. Neuropsychopharmacology, 40, 2993–3005. 10.1038/npp.2015.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Yang CR, Wei YP, Zhao ZA, Hou Y, Schatten H, et al. (2014). Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proceedings of the National Academy of Sciences of the United States of America, 111, 1873–1878. 10.1073/pnas.1321195111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, & Morris JR (2001). Genes, genetics, and epigenetics: A correspondence. Science, 293, 1103–1105. 10.1126/science.293.5532.1103. [DOI] [PubMed] [Google Scholar]
- Wu Q, & Suzuki M (2006). Parental obesity and overweight affect the body-fat accumulation in the offspring: The possible effect of a high-fat diet through epigenetic inheritance. Obesity Reviews, 7, 201–208. 10.1111/j.1467-789X.2006.00232.x. [DOI] [PubMed] [Google Scholar]
- Xia W, & Xie W (2020). Rebooting the epigenomes during mammalian early embryogenesis. Stem Cell Reports, 15, 1158–1175. 10.1016/j.stemcr.2020.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin F, Susiarjo M, & Bartolomei MS (2015). Multigenerational and transgenerational effects of endocrine disrupting chemicals: A role for altered epigenetic regulation? Seminars in Cell and Developmental Biology, 43, 66–75. 10.1016/j.semcdb.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Xiang Y, Wang Q, Wang L, Brind’Amour J, Bogutz AB, et al. (2019). SETD2 regulates the maternal epigenome, genomic imprinting and embryonic development. Nature Genetics, 51, 844–856. 10.1038/s41588-019-0398-7. [DOI] [PubMed] [Google Scholar]
- Xu Q, & Xie W (2018). Epigenome in early mammalian development: Inheritance, reprogramming and establishment. Trends in Cell Biology, 28, 237–253. 10.1016/j.tcb.2017.10.008. [DOI] [PubMed] [Google Scholar]
- Yan W (2014). Potential roles of noncoding RNAs in environmental epigenetic transgenerational inheritance. Molecular and Cellular Endocrinology, 398, 24–30. 10.1016/j.mce.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin G, Xia L, Hou Y, Li Y, Cao D, Liu Y, et al. (2021). Transgenerational male reproductive effect of prenatal arsenic exposure: Abnormal spermatogenesis with Igf2/H19 epigenetic alteration in CD1 mouse. International Journal of Environmental Health Research, 1–13. 10.1080/09603123.2020.1870668. [DOI] [PubMed] [Google Scholar]
- Yohn NL, Bartolomei MS, & Blendy JA (2015). Multigenerational and transgenerational inheritance of drug exposure: The effects of alcohol, opiates, cocaine, marijuana, andnicotine. Progress in Biophysics and Molecular Biology, 21–33. 10.1016/j.pbiomolbio.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Maekawa T, Ly NH, Fujita SI, Muratani M, Ando M, et al. (2020). ATF7-dependent epigenetic changes are required for the intergenerational effect of a paternal low-protein diet. Molecular Cell, 78, 445–458.e6. 10.1016/j.molcel.2020.02.028. [DOI] [PubMed] [Google Scholar]
- Yoshino T, Suzuki T, Nagamatsu G, Yabukami H, Ikegaya M, Kishima M, et al. (2021). Generation of ovarian follicles from mouse pluripotent stem cells. Science, 373, eabe0237. 10.1126/science.abe0237. [DOI] [PubMed] [Google Scholar]
- Yu C, Fan X, Sha QQ, Wang HH, Li BT, Dai XX, et al. (2017). CFP1 regulates histone H3K4 trimethylation and developmental potential in mouse oocytes. Cell Reports, 20, 1161–1172. 10.1016/j.celrep.2017.07.011. [DOI] [PubMed] [Google Scholar]
- Yu CW, & Liao VHC (2016). Transgenerational reproductive effects of Arsenite are associated with H3K4 Dimethylation and SPR-5 downregulation in Caenorhabditis elegans. Environmental Science & Technology, 50, 10673–10681. 10.1021/acs.est.6b02173. [DOI] [PubMed] [Google Scholar]
- Zeid D, & Gould TJ (2020). Impact of nicotine, alcohol, and cocaine exposure on germline integrity and epigenome. Neuropharmacology, 173, 1–11. 10.1016/j.neuropharm.2020.108127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Wang M, Zhou J, Wang X, Zhang Y, & Su P (2022). A hypothesis: Retrotransposons as a relay of epigenetic marks in intergenerational epigenetic inheritance. Gene, 817, 1–11. 10.1016/j.gene.2022.146229. [DOI] [PubMed] [Google Scholar]
- Zheng H, Huang B, Zhang B, Xiang Y, Du Z, Xu Q, et al. (2016). Resetting epigenetic memory by reprogramming of histone modifications in mammals. Molecular Cell, 63, 1066–1079. 10.1016/j.molcel.2016.08.032. [DOI] [PubMed] [Google Scholar]
- Zheng H, & Xie W (2019). The role of 3D genome organization in development and cell differentiation. Nature Reviews. Molecular Cell Biology, 20, 535–550. 10.1038/s41580-019-0132-4. [DOI] [PubMed] [Google Scholar]
- Zhu H, Wang G, & Qian J (2016). Transcription factors as readers and effectors of DNA methylation. Nature Reviews. Genetics, 17, 551–565. 10.1038/nrg.2016.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv-Gal A, Wang W, Zhou C, & Flaws JA (2015). The effects of in utero bisphenol a exposure on reproductive capacity in several generations of mice. Toxicology and Applied Pharmacology, 284, 354–362. 10.1016/j.taap.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]


