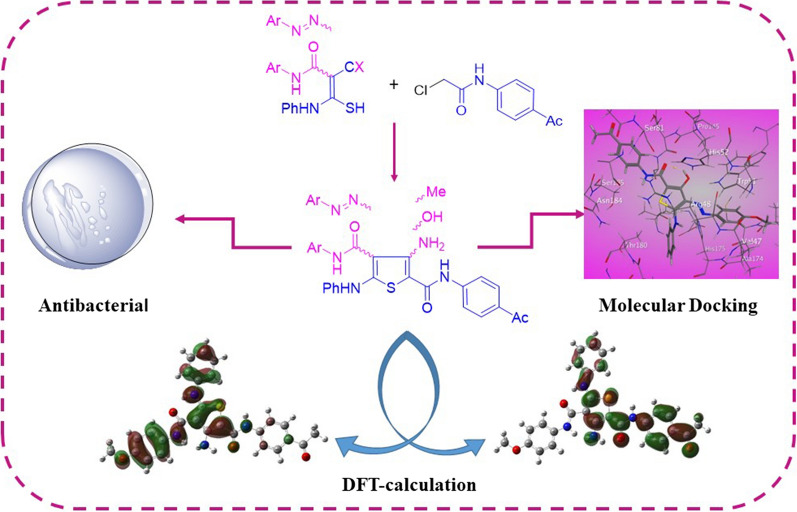
Abstract
Synthetic strategy for the synthesis of thiophene 2-carboxamide derivatives substituted with hydroxyl, methyl and amino groups at position-3 was proposed. The strategy includes the cyclization of the precursor ethyl 2-arylazo-3-mercapto-3-(phenylamino)acrylate derivatives, 2-acetyl-2-arylazo-thioacetanilide derivatives and N-aryl-2-cyano-3-mercapto-3-(phenylamino)acrylamide derivatives with N-(4-acetylphenyl)-2-chloroacetamide in alcoholic sodium ethoxide. IR, 1H NMR, and mass spectroscopic analyses were used to characterize the synthesized derivatives. In addition, molecular, electronic properties of the synthesized products were studied by the density functional theory (DFT) where they exhibited close HOMO–LUMO energy gap (ΔEH-L) in which the amino derivatives 7a-c have the highest while the methyl derivatives 5a-c were the lowest. Using the ABTS method, the antioxidant properties of the produced compounds were evaluated, where amino thiophene-2-carboxamide 7a exhibit significant inhibition activity 62.0% compared to ascorbic acid The antibacterial activity against two pathogenic Gram-positive bacteria (Staphylococcus aureus and Bacillus subtilis) and two of pathogenic Gram-negative bacteria (Escherichia coli and Pseudomonas aeruginosa) revealed that 7b records the highest activity index compared to ampicillin 83.3, 82.6, 64.0, 86.9%, respectively. Furthermore, the thiophene-2-carboxamide derivatives were docked with five different proteins with the use molecular docking tools and the results explained interactions between amino acid residue of enzyme and compounds. Compounds 3b and 3c showed the highest binding score with 2AS1 protein.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13065-023-00917-2.
Keywords: 2-Chloroacetamide, Thiophene-2-carboxamide, Antioxidant, DFT calculations, Molecular docking
Introduction
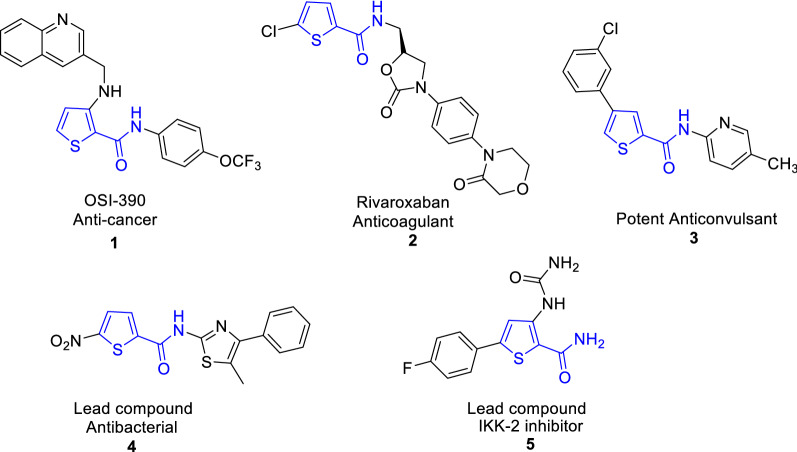
The antibacterial resistance threatens the human health around the world. It appeared due to the human abuse of antibiotics and it reached to danger limit. In 2019, 1.27 out of 4.95 million deaths attributed to antimicrobial resistance, with three leading pathogens for deaths (E. coli, S. aureus, P. aeuroginosa) [1]. The researchers continue their efforts for exploration for more powerful drugs with some requirements that involves, the molecular size of a drug's affinity for its target, drug bio-activation and metabolization. They must also address the design of medications with fewer adverse effects and more desirable small-molecular drug characteristics than existing drugs [2]. Recently, thiosemicarbazone and thio/carbohydrazone grabbed attention due to their potential biological activities [3, 4]. Many five-membered heterocyclic rings were extensively reported due to their chemical properties and versatile biological activities. It is known that heterocyclic compounds containing sulfur in their structures are used widely to eliminate free radicals and stop the antimicrobial resistance in addition to other pharmaceutical applications [5–7]. Generally, thiophenes have wide applications in different fields such as solid-state electrochromic devices [8], industry and medicinal chemistry. They possess biological properties such as antioxidant [9–16], antibacterial [17–19], antifungal [20–22], inflammatory [23] and antitumor [24–26]. Some examples of thiophene 2-carboxamide are marketed drugs, for example OSI-390 is used as anticancer drug (Fig. 1) [27, 28] and Rivaroxaban is used as an antithrombotic agent (Fig. 1) [29]. Moreover, compound 3 was found to have potent anticonvulsive effects in BALB-C mice [30]. Additionally, thiophene-2-carboxamide was considered to be a lead compound for drug discovery [31, 32]; for example; nitro thiophene-2-carboxamide 4 was used as a narrow spectrum antibacterial lead compound [33] and thiophene-2-carboxamide 5 was used as IKK-2 potent lead inhibitor [34]. Previous DFT-study for thiophene-2-carboxamide derivatives showed that N-(thiophen-2-ylmethyl)thiophene-2-carboxamide displays close experimental and theoretical structure parameters with (ΔEH-L) 5.031 eV [35]. While thiophene-thiadiazole hybrid derivatives (FMOs) shows close values between 3.83 and 4.18 eV [36]. As a result, many methods for the preparation of thiophene derivatives have been published either by incorporating thiophene moiety or by construction of the ring. One of the most efficient synthetic strategy includes cyclization of valuable thiocarbamoyl derivatives with α-halogenated reagents. A variety of advantages of this unique synthetic strategy includes the synthesis of a variety polysubstituted thiophenes, easy workup and cleaner products. As a result of previous advantages, we are attracted to explore this method. In light of the aforementioned bacterial resistance findings, DFT- studies and in accordance with the current research focus on preparing bioactive substituted thiophenes, the main goal of this work is to synthesize 3-substituted thiopene 2-carboxamide derivatives decorated with different substituents Cl, OMe, Me, NH2, OH groups aiming at increasing anti-oxidant and antibacterial inhibition power.
Fig. 1.
Versatile thiophene 2-carboxamide derivatives
Result and discussion
Chemistry
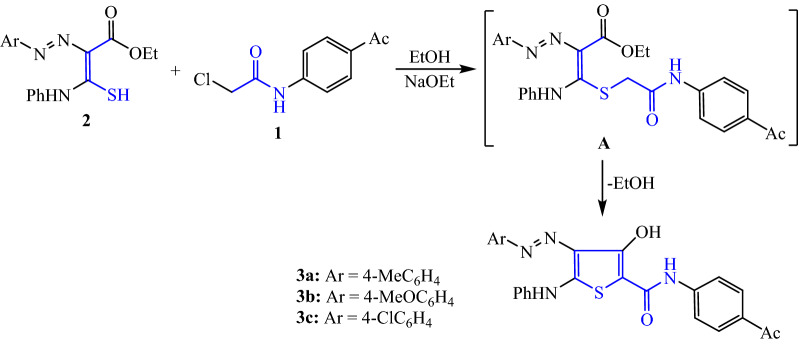
The synthesis of our target thiophene 2-carboxamide derivatives 3, 5 and 7 was aimed as one-step method by condensation of N-(4-acetylphenyl)-2-chloroacetamide (1) and various functionalized thiocarbamoyl compounds 2, 4, and 6, respectively. The highly versatile chloroacetamide compound, N-(4-acetylphenyl)-2-chloroacetamide (1) has been prepared by the reported chloroacetylation reaction of 4-aminoacetopheone [37]. Initially, 2-chloroacetamide derivative 1 was reacted with ethyl 2-arylazo-3-mercapto-3-(phenylamino)acrylate derivatives (2) [38] in ethanolic sodium ethoxide which resulted in the formation of sulfide intermediate A (Scheme 1). Substitution of the chlorine atom from the chloroacetamide derivative 1 by the 3-mercaptoacrylate reagent 2 led to the formation of the intermediate A. Subsequently, ethanol is removed intramolecularly, resulting in intermediate A, which is then used to yield the 3-hydroxythiophene 3a-c. Using IR data and 1H NMR, the structures of all of the newly synthesized compounds were analyzed, and the results were completely consistent with the assigned molecular structures.
Scheme 1.
Synthesis of 3-hydroxy thiophene-2-carboxamides 3a–c
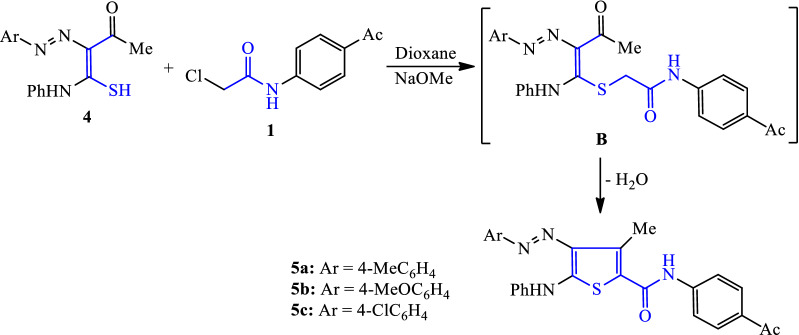
Based upon the above successful one step method for preparation of 4-arylazo-3-hydroxy thiophene derivatives, synthesis of our targeted thiophene derivatives 5a-c was performed. Thus, reaction of N-(4-acetylphenyl)-2-chloroacetamide (1) with 2-acetyl-2-arylazo-thioacetanilide derivatives (4) [39] in boiling dioxane containing sodium methoxide gave the corresponding 4-arylazo-3-methyl-thiophene derivatives 5a–c (Scheme 2). The mechanistic reaction scenario proceeded via the formation of intermediate B, which was followed by heterocyclization of the compound via the nucleophilic addition of the methylene group to the carbonyl function, and terminated with the removal of the water molecule (Scheme 2). The structures of the entire newly synthesized compound were validated using IR and 1H NMR data, which confirmed the molecular structures assigned.
Scheme 2.
Synthesis of 3-methylthiophene-2-carboxamide derivatives 5a–c
Similarly, the reaction of 2-chloroacetamide reagent 1 with N-aryl-2-cyano-3-mercapto-3-(phenylamino)acrylamide derivatives 6 [40–42] was carried out in boiling dioxane containing sodium methoxide to furnish the corresponding 3-aminothiophene derivatives 7a-c. The reaction proceeded via the formation of alkylated sulphide intermediate C, which was then subjected to intramolecular methylene group addition on the nitrile function to yield the corresponding 3-aminothiophene derivatives 7a-c (Scheme 3).
Scheme 3.
Synthesis of 3-aminothiophene-2-carboxamides derivatives 7a–c
Biological activity
Antioxidant assay
In this study, the antioxidant activity of the synthesized thiophene-2-carboxamide derivatives 3a-c, 5a-c and 7a-c were determined using ABTS antioxidant assay [43]. The decrease of absorbance for green ABTS˙+ radical cation at 734 nm was measured by using a UV–Visible spectrophotometer. ABTS antioxidant activity was measured by using L-Ascorbic acid as standard and the obtained results of the ABTS antioxidant assay was reported in (Table 1). The results showed that the 3-amino thiophene-2-carboxamide derivative 7a increased antioxidant activity by 62.0%, which is comparable to the reference antioxidant ascorbic acid (88.44%). Furthermore, 3-amino thiophene-2-carboxamide derivatives 7a-c promoted the highest antioxidant activity with percent inhibition 62.0–46.9%. While, 3-hydroxy thiophene-2-carboxamide derivatives 3a-c shows moderate inhibition percentage 54.9–28.4% and 3-methyl thiophene-2-carboxamide derivatives 5a-c the lowest 22.9–12.0%.
Table 1.
Antioxidant activity of the newly synthesized thiophene-2-carboxamide derivatives
| Compounds | Absorbance of samples | Inhibition (%) |
|---|---|---|
| 3a | 0.374 | 26.7 |
| 3b | 0.230 | 54.9 |
| 3c | 0.365 | 28.4 |
| 5a | 0.428 | 16.1 |
| 5b | 0.393 | 22.9 |
| 5c | 0.449 | 12.0 |
| 7a | 0.194 | 62.0 |
| 7b | 0.271 | 46.9 |
| 7c | 0.248 | 51.4 |
| Control of ABTS | 0.510 | 0 |
| Ascorbic acid | 0.061 | 88.0 |
Antibacterial assay
The newly synthesized compounds 3, 5 and 7 were evaluated for their antibacterial activity against a panel of two pathogenic Gram-positive bacteria (S. aureus and B. subtilis) and two of pathogenic Gram-negative bacteria (E. coli and P. aeruginosa), as shown in (Table 2). The antibacterial activity of the tested compounds was estimated in comparison with Ampicillin. Overall, the investigated compounds were more active against Gram-positive bacterial strains. 3-Amino thiophene-2-carboxamide compounds 7a-c displayed higher antibacterial activity (ranged from 40.0 to 86.9%) than their corresponding 3-hydroxy thiophene-2-carboxamide compounds 3a-c (from 20.0 to 78.3%) and the lowest inhibition values displayed by 3-methyl thiophene-2-carboxamide compounds 5a-c (from no activity to 47.8%). The thiophene-2-carboxamide derivatives 3, 5, 7b (substituted with methoxy group at aryl Th4) showed the best inhibition activity against Gram-positive bacteria (S. aureus and B. subtilis) and against Gram-negative bacterium (P. aeruginosa) rather than their corresponding derivatives substituted with methyl group or chlorine atom.
Table 2.
Antimicrobial activity of the newly synthesized thiophene-2-carboxamide derivatives using various bacterial strains
| Compound | E. coli | P.aeruginosa | S. aureus | B. subtilis |
|---|---|---|---|---|
| 3a | 5 (20.0) | 11 (47.8) | 10 (41.7) | 13 (56.5) |
| 3b | 13 (52.0) | 18 (78.3) | 17 (70.8) | 18 (78.3) |
| 3c | 9 (36.0) | 12 (52.2) | 13 (54.2) | 16 (69.6) |
| 5a | NA (––) | 7 (30.4) | 6 (25.0) | 8 (34.8) |
| 5b | 4 (16.0) | 10 (43.5) | 9 (37.5) | 11 (47.8) |
| 5c | NA(––) | 3 (13.0) | 4 (16.7) | 6 (26.1) |
| 7a | 12 (48.0) | 15 (65.2) | 16 (66.7) | 17 (73.9) |
| 7b | 16 (64.0) | 20 (86.9) | 20 (83.3) | 19 (82.6) |
| 7c | 10 (40.0) | 17 (73.9) | 16 (66.7) | 17 (73.9) |
| Ampicillin | 25 | 23 | 24 | 23 |
NA No activity; results of the antibacterial activity expressed as a mean on inhibition zone diameter (mm) and between brackets activity index (%) for different compounds; E. coli, P. aeruginosa, S. aureus and B. subtilis
Amino thiophene-2-carboxamide compound 7b containing methoxy group showed excellent activity against P. aeruginosa (86.9%), S. aureus (83.3%) and B. subtilis (82.6%) with inhibition zones 20, 20 and 19 mm, respectively. Hydroxy thiophene-2-carboxamide compound 3b having methoxy group showed very good effect against B. subtilis and P. aeruginosa 78.3% (inhibition zone 18 mm) and inhibition activity against S. aureus 70.8% (inhibition zone 17 mm).
Structure activity relationship (SAR) studies
The following structure–activity relationship of newly synthesized thiophene 2-carboxamide derivatives 3a-c, 5a-c, and 7a-c can be derived from antioxidant and antibacterial testing results: (i) Antioxidant and antibacterial activity of the amino thiophene-2-carboxamide derivatives 7a-c are more potent than hydroxyl or methyl thiophene-2-carboxamide 3a-c; which may be attributed to the absence of azo moiety and presence of amino group. Increasing the antioxidant power in 7a-c is caused by presence of electron donating amino group which increases the resonating electron on the thiophene ring, and faceplate the electron trapping for peroxide radical [44, 45]. Moreover, presence of hydroxyl group in derivatives 3a-c showing higher activity than 5a-c, perhaps it increase the solubility of this class of compound [46, 47]. (ii) Antioxidant activity of compound 7a possess potent activity among them; which may be due to absence of substituents on benzene ring Th4. While, compounds 3b and 5b is more potent than their derivatives which may be attributable to the presence of (-OMe group Th4). (iii) Antibacterial activity of amino thiophene-2-carboxamide derivatives 7b was boosted due to presence of (4-Me on carboxamide Th2) against E. coli, P. aeruginosa, S. aureus and B. subtilis. The structural variations such as (4-OMe on carboxamide Th4) and replacement of carboxamide with azo moieties 3b and 5b favors the activity in positive manner [48]. The highest antibacterial values for 3b, 5b, 7b may be attributed to the increase in the hydrophilicity power of the antibacterial drug agent caused by methoxy group [49, 50].
Computational study
Molecular modeling
DFT calculations were used to study how the thiophene 2-carboxamide derivatives 3a-c, 5a-c, and 7a-c differ in their shapes and electronic properties. DFT-optimized structures, atomic numbers, and geometrical parameters, including bond length, angle, and dihedral angle, for 3a-c, 5a-c and 7a-c obtained at B3LYP/6-31G (d,p) are shown in (Additional file 1: Fig. S1) and (Additional file 1: Tables S1-S3). The data of thiophene 2-carboxamide derivatives indicated that the H-atom of NHPh(Th5) is involved in intramolecular hydrogen bond with N1 of azo group(Th4), in 3a-c and 5a-c, while with oxygen of the carboxamide(Th4) in 7a-c. For 3a-c and 7a-c, the H-atoms of thiophene-OH and thiophene-NH2 formed hydrogen bonds with the O-carboxamide (Th2) and not the azo group’s nitrogen atom, (Additional file 1: Fig. S1). The distance between O-carbonyl and H-hydroxyl or H-amino in 3a-c and 7a-c derivatives was 1.730–1.732 Å, which was within the H-bond range [51–54].
Using the average values of the dihedral angles Nazo(1)-Th4-Th5-S and Th4-Nazo(1)-Nazo(2)-CPh(Azo), we can see that the thienyl is flat and coplanar with the phenylazo moiety in compounds 3a-c and 5a-c. Despite, the NH group was in the same plane as the thienyl ring (NH-Th5-S-Th2 = 176.56–177.90°), the phenyl ring was angled on the aminothienyl plane between 21 and 24° in compounds 3a-c and 5a-c and at an angle of 29° in compounds 7a-c. Furthermore, the 2-carboxamide group was skewed for compounds 7a-c = 152° and lied in plane with the thienyl ring as S-Th2-Th3-OH(th) in range 179–176° for compounds 3a-c and 7a-c.
Frontier molecular orbitals (FMOs)
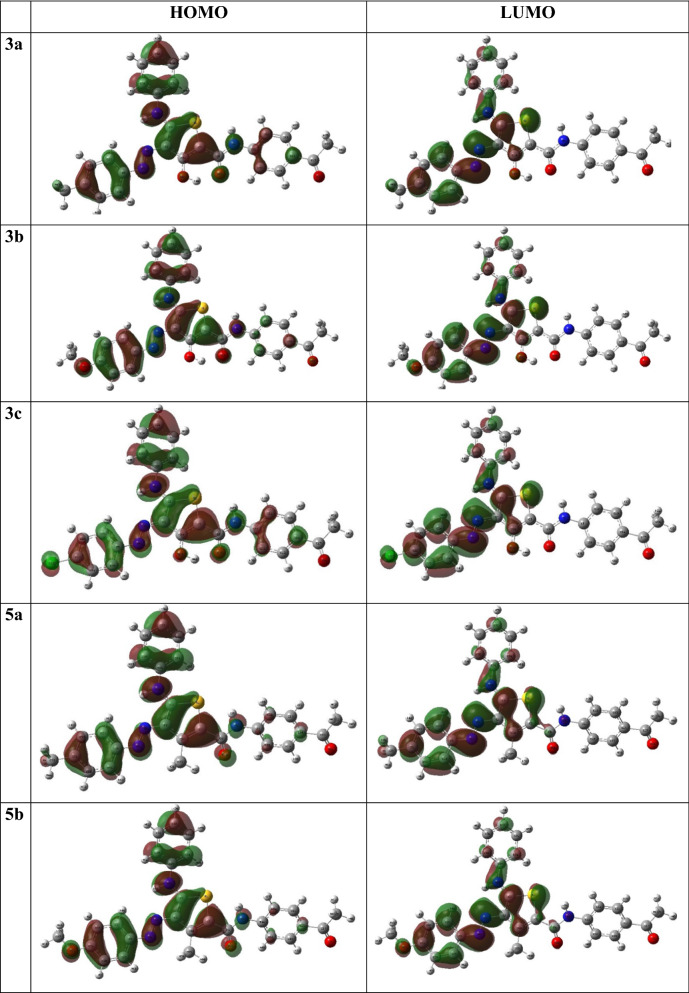
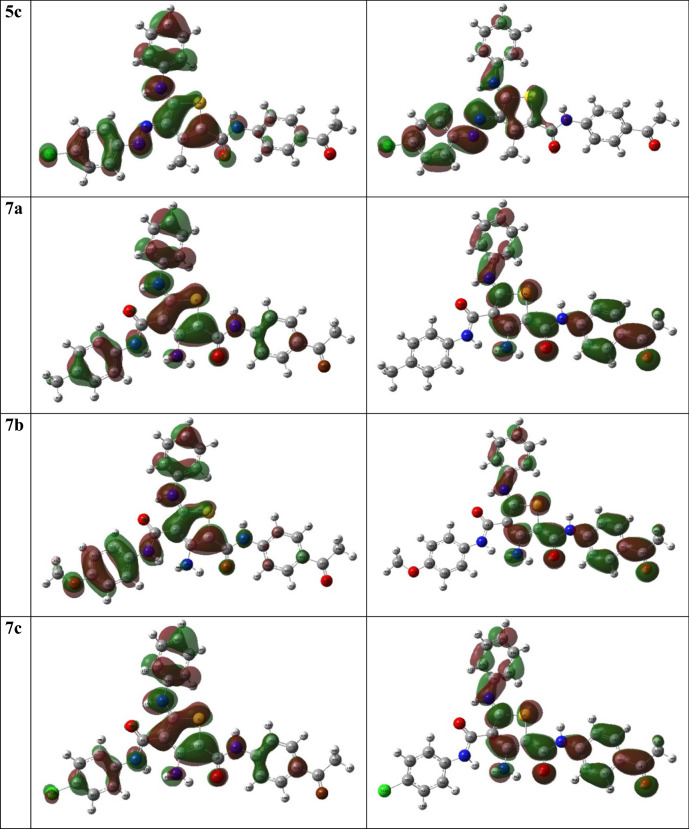
Electronic properties and the chemical reactivity of molecules are chiefly determined by Frontier molecular orbitals (FMOs) [55, 56]. The distribution of highest occupied and lowest unoccupied molecular orbitals, HOMO and LUMO, respectively, of the investigated molecules are presented in (Fig. 2). Figure 2 shows that the 3a-c and 5a-c derivatives HOMO plots are composed primarily of the π-orbitals of the thienyl, phenyl, and azo groups, as well as S, N, and O, non-bonding lone pairs except OH(Th3), which has a slight corporation. LUMO orbitals consist of π∗-orbitals of thienyl, phenyl, and azo groups with minimal heteroatom contributions. For 7a-c derivatives, the HOMO orbitals were distributed over the π-orbitals of the thienyl, phenyl, and amide groups, while their LUMO showed completely different composition, where it is localized only on the 2-carboxamide phenyl thiophene substituent. As shown in (Table 3), the above findings were demonstrated in the EHOMO and ELUMO values. The values of EHOMO were close to each other and ranged from 5.58 to 5.91 eV, while the ELUMO values ranged from 1.99 to 2.73 eV. Furthermore, the p-chlorophenyl derivatives demonstrated the highest EHOMO where 7c ≈ 3c > 5c. The aminothiophene derivatives 7a-c had the highest ELUMO, which might be caused by the presence of the carboxamide group at Th4. The values of HOMO–LUMO energy gap (ΔEH-L) ranged between 3.11–3.83 eV. It reveals that 7a-c derivatives have the maximum value. Also, the hydroxyl derivatives 3a-c were higher than corresponding methyl derivatives 5a-c. Finally, the amino derivatives, 7a-c showed higher ΔEH-L than other derivatives.
Fig. 2.
The HOMO and LUMO distribution pattern of 3a-c, 5a-c and 7a-c
Table 3.
The HOMO Energy (EHOMO), LUMO Energy (ELUMO), HOMO–LUMO Energy Gap (ΔEH-L) in eV, electronegativity (χ), global hardness (η), softness (δ) and electrophilicity (ω) at B3LYP/6-31G* Level of Theory
| Molecules | EHOMO | ELUMO | ΔEH-L | χ | η | δ | ω |
|---|---|---|---|---|---|---|---|
| 3a | −5.74 | −2.46 | 3.29 | 4.10 | 1.64 | 0.61 | 5.11 |
| 3b | −5.64 | −2.39 | 3.25 | 4.02 | 1.63 | 0.61 | 4.96 |
| 3c | −5.90 | −2.68 | 3.22 | 4.29 | 1.61 | 0.62 | 5.72 |
| 5a | −5.68 | −2.50 | 3.18 | 4.09 | 1.59 | 0.63 | 5.26 |
| 5b | −5.58 | −2.43 | 3.15 | 4.01 | 1.57 | 0.64 | 5.10 |
| 5c | −5.84 | −2.73 | 3.11 | 4.28 | 1.56 | 0.64 | 5.89 |
| 7a | −5.77 | −1.99 | 3.79 | 3.88 | 1.89 | 0.53 | 3.97 |
| 7b | −5.69 | −1.99 | 3.71 | 3.84 | 1.85 | 0.54 | 3.98 |
| 7c | −5.91 | −2.08 | 3.83 | 3.99 | 1.91 | 0.52 | 4.16 |
Using the above calculated EHOMO and ELUMO we managed to determine some chemical descriptors such as electronegativity (χ), global hardness (η) and softness (δ) and electrophilicity (ω), respectively (Table 3) [57], where:
The results showed that 3c derivative possessed the highest Lewis acid character, while 7b derivative exhibited the lowest. Although 5c was the softest derivative, 7c was the hardest. Based on the electrophilicity (ω) results, the HOMO–LUMO electron flow in the 5c derivative resulted in a greater decrease in energy compared to the 3c and 7c derivatives.
Mulliken’s charges, Fukui’s and relative indices
Describing the electronegativity and charge transfer processes was offered as an output of quantum chemical calculations. This calculation process is described as the Mulliken’s atomic charges [58] (Additional file 1: Table S4). As expected, the results showed that all heteroatoms had negative charges owing to their high electronegativity, with the exception of Nazo(2), NHCO and NHTh5, each of which has positive charges, showing that its lone pair is involved in the resonance of phenyl ring. Much more, in derivatives 3a-c, the carbon atom of thienyl (Th3) has close negative charges, but in derivatives 5a-c, they have close positive charges. This could be due to the OH group electron donating effect. On parallel, the thienyl carbon atoms Th3 in derivatives 7a-c have close negative charges and lower than 3a-c and the reason behind this decrease is replacing azo group with amide group in position 4. Moreover, determination of the Fukui indices ( and ) was also used to investigate the reactivity of atoms toward nucleophilic and electrophilic attacks were explored using the following equations [59–61], where q k (N), q k (N + 1) and q k (N − 1) are the systems atomic charges with N, N + 1 and N-1 electrons, respectively [62].
It is clear for all derivatives values are higher than for all heteroatoms. Except NHamideTh2, NHTh5, OH (3a-c), Nazo2 (7a-c) and S (7a-c), have lower values than proving lone pair participation in ring resonating structure. In some cases values equals values; Nazo1(7a-c), Oacetyl(3a-c, 5a-c). The data showed Th3 has higher index in all derivatives, reached about 2 folds in comparison to (Additional file 1: Table S4).
Molecular docking
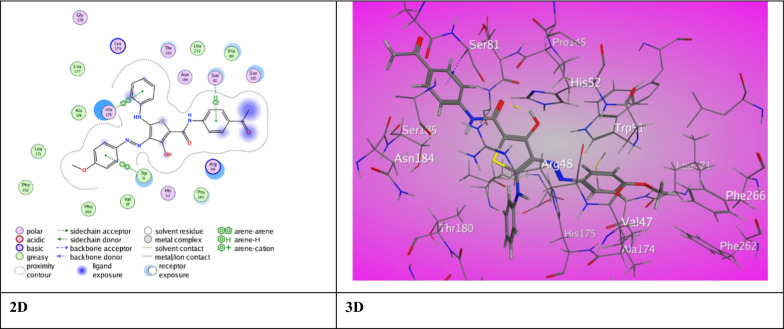
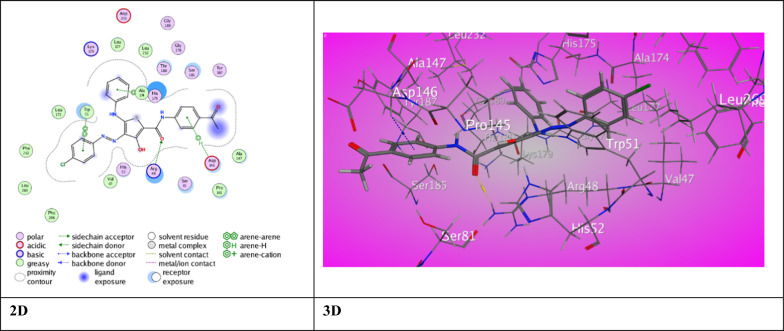
To get a promising approach of synthesized ligands interactions with antioxidant and bacterial protein receptors, docking studies were performed. A molecular docking investigation was conducted on the newly synthesized thiophene-2-carboxamide derivatives in order to investigate their interaction with the crystallographic coordinates available in the RCSB Protein Data Bank. Downloaded from the Protein Data Bank [63] and delivered via the operating MOE “v10.2015.10 software,” (PDB ID 2AS1) was chosen as the antioxidant target for the derivatives so that its activity could be tested and specified (Table 4). It is noteworthy that all of the compounds showed excellent inhibition activities against target proteins and higher than previously prepared compounds [17] perhaps because of the presence of carboxamide and thienyl groups in all the compounds which can develop a variation of associations in the active site of proteins. The 3-hydroxy thiophene carboxamide derivative 3a displayed one intermolecular H-bond between N-aniline (Th5) with Pro 145 (2.99 ˚A) offering a score for binding energy, S = −8.1675 kcal/mol. Derivative 3b showed π- H interaction between aniline (Th5) with Ser 8 (3.84 ˚A) and π- π stick interactions between aryl azo (Th4) and Ph (Th2) with Trp 51 (3.83 ˚A) and His 175 (3.84 ˚A), respectively. An eminent binding energy score, S = −9.3283 kcal/mol was offered for derivative 3b (Fig. 3). But, derivative 3c displayed two distinct binding modes promoting a distinguished score of binding energy, S = −9.1141 kcal/mol (Fig. 4). The first is an H-bond between O-carboxamide (Th2) with Arg 84 (3.02 ˚A). The second binding type was π-H and π- π stick between Ph (Th2), aryl azo (Th4) and aniline (Th5) with Asp 146, Trp 51 and His 175 (4.69 ˚A, 3.85 ˚A, 3.88 ˚A), respectively.
Table 4.
The molecular docking data of the synthesized thiophene 2-carboxamide derivatives with 2AS1 (Anti-oxidant)
| Ligand | S (energy score) (Kcal/mol) | Rmsd (refine unit) | Interaction with ligand | Types of interactions | Distance (A) | rseq | E_conf |
|---|---|---|---|---|---|---|---|
| 3a | −8.1675 | 1.2616 | N-aniline(Th5) with Pro 145 | H-donor | 2.99 | 1 | 11.2872 |
| 3b | −9.3283 | 1.4389 |
Aniline (Th5) with Ser 81 Aryl azo (Th4) with Trp 51 Ph (Th2) with His 175 |
π-H interaction π-π interaction π-π interaction |
3.84 3.83 3.84 |
1 | 16.5201 |
| 3c | −9.1141 | 1.6664 |
O-carboxamide (Th2) Arg 48 Ph (Th2) with Asp 146 Aryl azo (Th4) with Trp 51 Aniline (Th5) with His 175 |
H-acceptor π-H interaction π-π interaction π-π interaction |
3.02 4.69 3.85 3.88 |
1 | 25.7050 |
| 5a | −8.7347 | 1.5232 |
Aniline (Th5) Arg 48 Aryl azo (Th4) Trp 51 |
π-H interaction π-π interaction |
4.14 3.70 |
1 | 23.4919 |
| 5b | −6.4382 | 1.5862 |
O-carboxamide (Th2) Arg 72 O-acetyl group with Glu 135 Aniline (Th5) His 96 |
H-acceptor H-acceptor π-π interaction |
3.04 3.12 3.65 |
1 | 21.0089 |
| 5c | −7.4535 | 1.5972 |
N1-azo group with Ser 81 Thiophene ring with Asp 146 |
H-acceptor π-H interaction |
3.24 4.37 |
1 | 24.2779 |
| 7a | −8.2395 | 1.5308 |
O-carboxamide (Th2) with Trp 51 O-carboxamide (Th2) with His 52 |
H-acceptor H-acceptor |
3.59 3.23 |
1 | −50.4239 |
| 7b | −6.0187 | 1.3779 |
N-carboxamide (Th4) with Asp 148 N-amino group (Th3) with Asp 146 O-acetyl group with Arg 48 Thiophene ring with Ala 147 Ph (Th4) with Lys 149 |
H-donor H-donor H-acceptor π-H interaction π-cation interaction |
3.49 2.90 3.10 4.32 3.44 |
1 | −53.7450 |
| 7c | −7.3131 | 1.7642 |
S-thiophene with Ser 81 Cl-benzamide (Th4) with Leu 177 |
H-donor H-donor |
3.91 3.27 |
1 | −52.8699 |
| Ascorbic acid | −4.7248 | 1.0693 |
O (Lac3) with His 181 O (Eth1) Leu 177 |
H-donor H-donor |
3.16 2.88 |
1 | 87.5356 |
Fig. 3.
The binding interaction of 3-hydroxythiophene 3b with (PDB ID: 2AS1)
Fig. 4.
The binding interaction of 3-hydroxythiophene 3c with (PDB ID: 2AS1)
Meanwhile, 3-methyl thiophene carboxamide derivative 5a exhibited notable binding score (S = −8.7347 kcal/mol). The π-H and π- π stick forces between aniline (Th5) and aryl azo(Th4) with Arg 48 (4.14 ˚A) and Trp 51 (3.70 ˚A) was the reason for binding. A weak score of binding (S = −6.4382 kcal/mol) showed by derivative 5b. This is due to two H-bonds exhibited between O-carboxamide (Th2) and O-acetyl with Arg 72 (3.04 ˚A) Glu 135 (3.12 ˚A) as well π- π stick forces between aniline (Th5) and His 96 (3.65˚A). Moreover, derivative 5c demonstrated a respectable binding score, (S = −7.4535 kcal/mol) due to H-bond between N1-azo group with Ser 81(3.65˚A) and π-H interaction between thiophene ring with Asp 146 (4.37 ˚A). However, 3-amino thiophene carboxamide derivative 7a revealed two intermolecular H-bond between Trp 51 and His 52 with the O-carboxamide (Th2) (3.59 ˚A), (3.23 ˚A) respectively. Derivative 7a exhibited a highly regarded binding score with the amino acids of 2AS (S = −8.2395 kcal/mol). Derivative 7b exhibited three intermolecular hydrogen bonds between N-carboxamide (Th4) with Asp 148 (3.49˚A), and N-amino group (Th3) with Asp 146 (2.90˚A) besides, O-acetyl group with Arg 48 (3.10 ˚A). It also showed π-H interaction between thiophene ring with Ala 147 (4.32˚A), and π-cation interaction between Ph (Th4) with Lys 149 (3.44˚A) recording a weak score (S = −6.0187 kcal/mol). Moreover, derivative 7c presented two intermolecular H-bond between Ser 81 and Leu 177 with the S-atom and Cl atom (3.91 ˚A), (3.27 ˚A) through very good binding score (S = −7.4535 kcal/mol). The ascorbic acid was subjected to amino acid 2AS1 and shows low score (S = −4.7248 kcal/mol) resulted from two H-bonds between O(Lac3) and O(Et1) with His 181 and Leu 177 (3.16 ˚A), (2.88 ˚A) respectively.
For antibacterial target, IMP-1 metallo beta-lactamase (PDB ID 1DD6), from P. aeruginosa and sortase A (PDB ID 2MLM) from S. aureus [64] and rhomboid protease (PDB ID 3ZMI) from E. coli [64] and Bacillus subtilis nitric oxide synthase I218V (PDB ID 4D3V) from B. subtilis [65] was selected and downloaded from the Protein Data Bank. For anti-bacterial protein IMP-1 metallo beta-lactamase (PDB ID 1DD6), the ligands inhibited the target by developing various associations with amino acid residues of active site of P. aeruginosa (Table 5). The 3-hydroxy thiophene carboxamide derivative 3a recorded outstanding score of binding (S = −7.5022 kcal/mol) as a result of π-H interaction between anilide (Th2) with Trp 28 (4.17 ˚A). Moreover, derivative 3b demonstrated intermolecular H-bonds between N-aniline with Leu 39 (2.89 ˚A) and one π- H interaction between aniline (Th5) with Glu 150 (4.30 ˚A) through binding score (S = −6.6965 kcal/mol). Additionally, derivative 3c showed a weak binding score (S = −6.1669 kcal/mol). It has two H-bond interaction between O-carboxamide (Th2) with Tyr 45 (3.08 ˚A) and Lys 71 (2.84˚A) plus π- H interaction between aniline (Th4) with Tyr 97 (3.83 ˚A) and aryl azo (Th5) with Tyr 97(3.85 ˚A).
Table 5.
The molecular docking data of the synthesized thiophene 2-carboxamide derivatives with 1DD6 (P. aeruginosa)
| Code | S (energy score) (Kcal/mol) | Rmsd (refine unit) | Interaction with ligand | Types of interactions | Distance (A) | rseq | E_conf |
|---|---|---|---|---|---|---|---|
| 3a | −7.5553 | 1.3950 | Anilide (Th2) with Trp 28 | H-π interaction | 4.17 | 1 | 19.4438 |
| 3b | −6.6965 | 1.4268 |
N-aniline with Leu 39 Aniline (Th5) with Glu 150 |
H-donor π-H interaction |
2.89 4.30 |
1 | 14.8238 |
| 3c | −6.1669 | 1.1349 |
O-carboxamide (Th2) with Tyr 45 O-carboxamide (Th2) with Lys 71 aniline (Th4) with Tyr 97 aryl azo (Th5) with Tyr 97 |
H-acceptor H-acceptor π-H interaction π-H interaction |
3.08 2.84 3.83 3.85 |
1 | 18.9813 |
| 5a | −7.6609 | 1.4058 |
O-acetyl group with Ser 80 H-acetyl group with His 79 |
H-acceptor H-π interaction |
2.95 3.90 |
1 | 26.8327 |
| 5b | −7.4306 | 1.4020 |
O-carboxamide(Th2) with Asn 167 Aniline (Th5) with Val 25 Aryl azo (Th4) with Ser 80 |
H-acceptor π-H interaction π-H interaction |
3.32 4.11 4.24 |
1 | 25.9591 |
| 5c | −6.0538 | 1.1783 |
O-acetyl with Lys 215 Aniline (Th5) with Tyr 163 |
H-acceptor π-H interaction |
3.02 4.51 |
1 | 27.1141 |
| 7a | −7.4729 | 1.0919 |
N-aniline (Th5) with Leu 39 O-carboxamide (Th4) with Asn 41 Anilide (Th4) with Ala 42 |
H-donor H-acceptor π-H interaction |
3.07 3.05 4.65 |
1 | −50.3092 |
| 7b | −6.2489 | 0.9195 |
O-carboxamide (Th2) with Lys 127 O-carboxamide (Th2) with Lys 127 O-carboxamide (Th4) with Tyr 97 Anilide (Th4) with Tyr 97 Aniline (Th5) with Trp 124 |
H-acceptor H-acceptor H-acceptor π-H interaction π-H interaction |
3.42 2.88 2.92 4.37 4.43 |
1 | −54.4244 |
| 7c | −6.5478 | 1.5580 |
N-amino (Th3) with Asp 170 Thiophene ring with Ser 80 Thiophene ring with His 79 |
H-donor π-H interaction π-π interaction |
3.29 4.95 3.96 |
1 | −47.9835 |
| Ampicillin | −6.8228 | 1.2780 |
O-cyclic amide with His 197 O-carboxyl group with Lys 161 |
H-acceptor H-acceptor |
3.06 2.88 |
1 | 76.1998 |
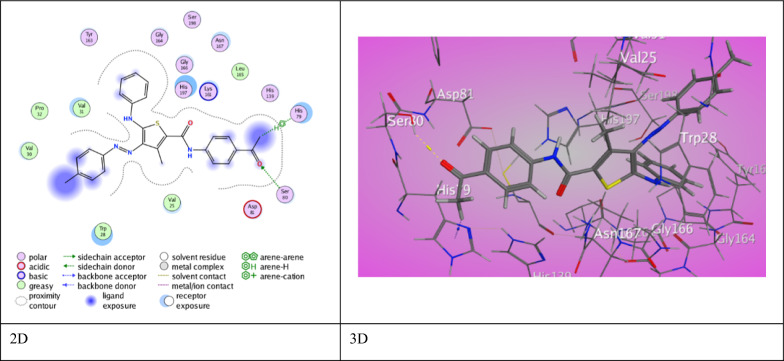
Meanwhile, a good score of binding (S = −7.6609 kcal/mol) offered by 3-methyl thiophene carboxamide derivative 5a through two interaction modes (Fig. 5). An H-bond was presented between O-acetyl group with Ser 80 (2.95 ˚A), and H-π force was exhibited between H-acetyl group with His 79 (3.90 ˚A). Derivative 5b showed decent score (S = −7.4306 kcal/mol) outcomes from one H-bond interaction between O-carboxamide with Asn 167 (3.32 ˚A), and two π-H interaction between aniline with Val 25 (4.11 ˚A) and aryl azo with Ser 80 (4.24 ˚A). Furthermore, derivative 5c shows weak score (S = −6.0538 kcal/mol) as a consequence of one H-bond interaction between O-acetyl group with Lys 215 (3.02 ˚A), and π-H interaction between aniline with Tyr 163 (4.51 ˚A).
Fig. 5.
The binding interaction of 3-methylthiophene 5a with (PDB ID: 1DD6)
However, 3-amino thiophene carboxamide derivative 7a revealed two intermolecular H-bond between N-aniline (Th5) and O-carboxamide (Th4) with Leu 39 (3.07 ˚A) and Asn 41 (3.05 ˚A) besides, π-H interaction between anilide with Ala 42 (4.65 ˚A). Derivative 7a was displayed a highly regarded binding score (S = −7.4729 kcal/mol). Furthermore, derivative 7b exhibited three intermolecular hydrogen bonds between O-carboxamide (Th2) with Lys 127 (3.42˚A), (2.88˚A), respectively. In addition to, O-carboxamide (Th4) with Tyr 97 (2.92 ˚A). It also showed two π-H interaction between anilide(Th4) with Tyr 97 (4.37˚A), and π-H interaction between aniline (Th5) with Try 124 (4.43˚A) offering a weak score (S = −6.2489 kcal/mol). Moreover, derivative 7c presented one H-bond between Asp 170 with N-amino (Th3) (3.29 ˚A). It also displayed π-H and π-π interaction between thiophene ring with Ser 80 (4.95 ˚A) and His 79 (3.96 ˚A) showing a good score (S = −6.5478 kcal/mol). Ampicillin was also docked with 1DD6 protein and showed good binding score (S = −6.8228 kcal/mol) resulted from intermolecular H-bond between O-cyclic amide and O-carboxyl group with His 179 (3.06 ˚A) and Lys 161 (2.88 ˚A).
For anti-bacterial protein sortase A (PDB ID 2MLM), the ligands inhibited the target by developing various associations with amino acid residues of active site of Staphyllococcus aureus (Table 6). The 3-hydroxy thiophene carboxamide derivative 3a exhibited two intermolecular H-bonds between S-thiophene ring with Asn 56 (3.69 ˚A) and O-carboxamide (Th2) with Lys 117 (3.03 ˚A) through a binding score (S = −6.9492 kcal/mol). Also, derivative 3b demonstrated two intermolecular H-bonds between S-thiophene ring with Asn 56 (3.08 ˚A) and N-aniline (Th5) with Thr 122 (3.17 ˚A) through binding score (S = −6.6975 kcal/mol). Moreover, derivative 3c demonstrated two types of interactions. Two intermolecular H-bonds between S-thiophene ring with Gln 114 (3.67 ˚A) and O-carboxamide (Th2) with Lys 117 (3.06 ˚A) and one π- H interaction between thiophene ring with Gln 120 (3.95 ˚A).
Table 6.
The molecular docking data of the synthesized thiophene 2-carboxamide derivatives with 2MLM (S. aureus)
| Code | S (energy score) (Kcal/mol) | Rmsd (refine unit) | Interaction with ligand | Types of interactions | Distance (A) | rseq | E_conf |
|---|---|---|---|---|---|---|---|
| 3a | −6.9492 | 1.4683 |
S-thiophene ring with Asn 56 O-carboxamide (Th2) with Lys 117 |
H-donor H-acceptor |
3.69 3.03 |
1 | 26.3509 |
| 3b | −6.6975 | 1.1433 |
S-thiophene ring with Asn 56 N-aniline (Th5) with Thr 122 |
H-donor H-donor |
3.08 3.17 |
1 | 17.1511 |
| 3c | −6.5149 | 1.0726 |
S-thiophene ring with Gln 114 O-carboxamide (Th2) with Lys 117 Thiophene ring with Gln 120 |
H-donor H-acceptor π-H interaction |
3.67 3.06 3.95 |
1 | 17.7282 |
| 5a | −6.2959 | 0.9967 |
S-thiophene ring with Asn 56 N-aniline (Th5) with Glu 47 N-aniline (Th5) with Glu 47 |
H-donor H-donor H-donor |
3.63 3.02 3.12 |
1 | 24.9926 |
| 5b | −6.3089 | 0.8325 |
S-thiophene ring with Val 108 N-aniline (Th5) with Val 108 O-carboxamide (Th2) with Arg 139 O-carboxamide (Th5) with Arg 139 |
H-donor H-donor H-acceptor H-acceptor |
4.38 3.17 3.20 3.14 |
1 | 23.4584 |
| 5c | −7.0550 | 1.0874 |
O-acetyl with Gln 120 Aryl azo (Th4) with Ala 46 |
H-acceptor π-H interaction |
3.14 4.06 |
1 | 26.0452 |
| 7a | −7.0842 | 1.1622 |
S-thiophene ring with Glu 113 O-acetyl group with Gln 120 Aniline (Th5) with Arg 139 |
H-donor H-acceptor π-cation interaction |
3.68 3.00 3.93 |
1 | −57.4937 |
| 7b | −6.8518 | 0.9057 |
S-thiophene ring with Asn 56 O-carboxamide (Th2) with Lys 117 |
H- donor H-acceptor |
4.09 2.97 |
1 | −54.0559 |
| 7c | −6.7376 | 1.3201 |
N-amide (Th2) with Gln 120 Aniline (Th5) Ser 99 Anilide (Th4) Thr 122 |
H-donor π-H interaction π-H interaction |
3.16 3.68 4.12 |
1 | −50.5308 |
| Ampicillin | −5.0480 | 1.3765 |
C-benzyl with Glu 47 N-amino group with Asn 56 S-thiazole with Glu 47 O-cyclic ketone with Lys 117 O-β-lactam with Lys 117 O-carboxylic ketone with Gln 55 |
H-donor H-donor H-donor H-acceptor H-acceptor H-acceptor |
3.35 3.23 3.63 3.00 3.07 2.93 |
1 | 72.4682 |
Meanwhile, 3-methyl thiophene carboxamide derivative 5a exhibited a binding score (S = −6.2959 kcal/mol). The binding interactions are three H-bonds between N-aniline (Th5) with Glu 47 (3.02 ˚A) (3.12 ˚A) and S-thiophene ring with Asn 56 (3.63 ˚A). A weak binding score was assigned to derivative 5b (S = −6.3089 kcal/mol). Derivative 5b displayed four H-bond between S-thiophene ring and N-aniline (Th5) with Val 108 in addition to O-carboxamide (Th2) and (Th5) with Arg 139 (4.38 ˚A), (3.17 ˚A), (3.20 ˚A), (3.14 ˚A), respectively. Derivative 5c demonstrated a proper binding score, (S = −7.0550 kcal/mol). The interactions in derivative 5c involves H-bond between O-acetyl group with Gln 120 (3.14˚A) and π-H interaction between arylazo (Th4) with Ala 46 (4.06 ˚A).
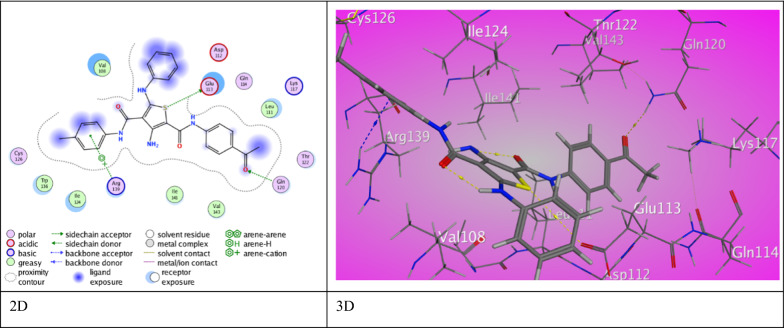
However, 3-amino thiophene carboxamide derivative 7a displayed a highly regarded score of binding with the amino acids of 2MLM (S = −7.0842 kcal/mol) rose from two H-bond between O-acetyl group with Gln 120 (3.00 ˚A) and S-thiophene ring with Glu 113 (3.68 ˚A). Also, a π-cation interaction between aniline (Th5) with Arg 139 (3.93 ˚A) was noticed (Fig. 6). Furthermore, derivative 7b exhibited two H-bonds between S-thiophene ring with Asn 56 (4.09˚A) along with O-carboxamide (Th2) with Lys 117 (2.97 ˚A) through abundant binding score (S = −6.8518 kcal/mol). Additionally, derivative 7c presented H-bond between Gln 120 with N-amide (Th2) (3.16 ˚A). It also exhibited two π-H interactions between aniline (Th5) and (Th4) with Ser 99 and Thr 122 (3.68 ˚A), (4.12 ˚A) respectively through good binding score (S = −6.7376 kcal/mol). Ampicillin showed with 2MLM protein good binding score (S = −5.0480 kcal/mol) resulted from six H-bonds between C-benzyl and S-thiazole with Glu 47 (3.35 ˚A), (3.63 ˚A), respectively. As well, N-amino group with Asn 56 (3.23 ˚A) and O-carboxylic ketone with Gln 55 (2.93 ˚A) was also appeared. Lys 117 was engaged in two H-bonds with O-cyclic ketone and O-β-lactam (3.00 ˚A and 3.07 ˚A).
Fig. 6.
The binding interaction of 3-aminothiophene 7a with (PDB ID: 2MLM)
For anti-bacterial protein rhomboid protease (PDB ID 3ZMI), the ligands inhibited the target by developing various associations with amino acid residues of active site of E. coli (Table 7).
Table 7.
The molecular docking data of the synthesized thiophene 2-carboxamide derivatives with 3ZMI (E. Coli)
| Code | S (energy score) (Kcal/mol) | Rmsd (refine unit) | Interaction with ligand | Types of interactions | Distance (A) | rseq | E_conf |
|---|---|---|---|---|---|---|---|
| 3a | −7.5672 | 1.4487 |
O-acetyl group with Met 208 O-carboxamide (Th2) with Ser 201 Thiophene ring with His 150 |
H-donor H-acceptor π-H interaction |
3.40 2.96 4.12 |
1 | 12.7966 |
| 3b | −6.8908 | 1.3841 | O-carboxamide (Th2) with Arg 92 | H-acceptor | 2.92 | 1 | 25.5604 |
| 3c | −6.8945 | 1.5074 |
N-aniline (Th5) with Ser 201 N1-azo group with Ser 201 Thiophene ring with His 150 |
H-donor H-acceptor π- π interaction |
3.17 3.35 3.58 |
1 | 17.9663 |
| 5a | −6.1292 | 0.8206 | S-thiophene ring with Leu 152 | H-donor | 4.14 | 1 | 21.0952 |
| 5b | −7.3344 | 1.5308 |
N2-azo group with Ser 201 Thiophene ring with His 150 Ph (Th2)with Phe 146 |
H-acceptor π- π interaction π- π interaction |
3.00 3.88 3.58 |
1 | 24.7280 |
| 5c | −7.2949 | 1.5666 |
Thiophene ring with His 150 Ph(Th2) with Phe 146 |
π- π interaction π- π interaction |
3.78 3.95 |
1 | 23.9127 |
| 7a | −7.1277 | 1.2547 |
O-carboxamide (Th4) with Ser 201 O-carboxamide (Th4) with His 254 Thiophene ring with Gly 198 Ph (Th2)with Phe 146 |
H-acceptor H-acceptor π-H interaction π- π interaction |
3.27 3.00 4.78 3.68 |
1 | −49.7131 |
| 7b | −7.0293 | 1.3327 | O-carboxamide with Ala 250 | H-acceptor | 2.96 | 1 | −51.8681 |
| 7c | −7.8889 | 1.0667 | Ph (Th4)with His 150 | π- π interaction | 3.95 | 1 | −51.8923 |
| Ampicillin | −5.5469 | 1.2420 |
O-carboxylic ketone with Arg 168 O-β-lactam with Arg 168 Benzene ring with Arg 92 |
H-acceptor H-acceptor π-cation interaction |
3.51 3.01 3.38 |
1 | 71.6361 |
The 3-hydroxy thiophene carboxamide derivative 3a displayed two H-bonds between O-carboxamide (Th2) with Ser 201 (2.96 ˚A), O-acetyl group with Met 208 (3.40 ˚A) and π- H interaction between thiophene ring with His 150 (4.12 ˚A). These interactions have a strong binding score (S = −7.5672 kcal/mol). Derivative 3b demonstrated binding score (S = −6.8908 kcal/mol) presented in a H-bond between O-carboxamide (Th2) with Arg 92 (2.92˚A). Moreover, derivative 3c shows two mode of interactions via a binding score (S = −6.8945 kcal/mol). Two H-bonds between N-aniline (Th5) (3.17 ˚A) and N1-azo group with Ser 201 (3.35˚A) and π- π interaction between thiophene ring with His 150 (3.58 ˚A).
Meanwhile, 3-methyl thiophene carboxamide derivative 5a exhibited binding score (S = −6.1292 kcal/mol) resulted from H-bonds between S-thiophene ring with Leu 152 (4.14 ˚A). But, derivative 5b showed a rising binding score (S = −7.3344 kcal/mol). Binding modes are, one H-bond displayed between N2-azo group with Ser 201 (3.00 ˚A) as well as π- π stick forces between Ph(Th2) with Phe 146 (3.58 ˚A) and thiophene ring with His 150 (3.88 ˚A). Derivative 5c have a relatively high binding score, (S = −7.2949 kcal/mol). It also forms a π- π stick forces between thiophene ring with His 150 (3.78 ˚A) and Ph (Th2) with Phe 146 (3.95 ˚A).
However, 3-amino thiophene carboxamide derivative 7a established two interaction modes with a binding score (S = −7.1277 kcal/mol). Derivative 7a shows two H-bond between O-carboxamide (Th4) with Ser 201 and His 254 (3.27˚A), (3.00˚A), respectively. A π-H type of interaction between thiophene ring with Gly 198 (4.78 ˚A) and π- π stick forces between Ph (Th2) Phe 146 (3.68 ˚A). Derivative 7b demonstrated one H-bond between O-carboxamide (Th2) with Ala 250 (2.96 ˚A) through a binding score (S = −7.0293 kcal/mol). Derivative 7c presented one π-π stick interaction between His 150 and Ph (Th4) (3.95 ˚A) via a unique binding score (S = −7.8889 kcal/mol) (Fig. 7). Ampicillin established a binding score (S = −5.5469 kcal/mol) with 3ZMI protein resulted from two H-bonds between Arg 168 with O-carboxylic ketone and O-β-lactam (3.51 ˚A and 3.01 ˚A). Besides π-cation interaction between benzene ring with Arg 92 (3.38 ˚A).
Fig. 7.
The binding interaction of 3-aminothiophene 7c with (PDB ID: 3ZMI)
For anti-bacterial protein Bacillus subtilis nitric oxide synthase I218V (PDB ID 4D3V), the ligands inhibited the target by developing various associations with amino acid residues of active site of B. subtilis (Table 8).
Table 8.
The molecular docking data of the synthesized thiophene 2-carboxamide derivatives with 4D3V (B. subtilis)
| Code | S (energy score) (Kcal/mol) | Rmsd (refine unit) | Interaction with ligand | Types of interactions | Distance (A) | rseq | E_conf |
|---|---|---|---|---|---|---|---|
| 3a | −8.2175 | 1.5172 |
N-aniline (Th5) with Trp 329 Ph (Th2) with Gly 68 |
H-donor π-H interaction |
3.17 3.98 |
1 | 24.3131 |
| 3b | −7.9612 | 0.9335 |
N-carboxamide (Th2) with Met 221 O-acetyl group with Lys 360 Aniline (Th5) with Arg 65 Ph (Th2) with His 128 |
H-donor H-acceptor π-H interaction π- π interaction |
4.23 2.92 4.11 3.90 |
1 | 18.0041 |
| 3c | −6.5618 | 0.9875 |
N atom of azo group (N2) with Lys 211 Benzene ring (Th2) with Glu 194 |
H-acceptor π-H interaction |
3.14 3.78 |
1 | 19.0186 |
| 5a | −7.2325 | 1.5263 | Benzene ring (Th2) with Arg 65 | π-H interaction | 4.19 | 1 | 20.4180 |
| 5b | −6.0805 | 1.1811 | Thiophene ring with Pro 196 | π-H interaction | 3.78 | 1 | 20.9885 |
| 5c | −7.3792 | 1.3171 |
S-thiophene ring with Tyr 357 O-carboxamide (Th2) with Arg 65 O-acetyl group with Asn 64 Ph (Th2) with Arg 65 |
H-donor H-acceptor H-acceptor π-H interaction |
3.37 2.89 3.08 4.83 |
1 | 33.2914 |
| 7a | −7.2786 | 1.3658 |
H-acetyl group with Trp 238 Ph (Th5) with Trp 329 |
H-π interaction π-H interaction |
3.87 4.45 |
1 | −51.9118 |
| 7b | −7.7337 | 1.2784 |
N-NH2 (Th3) with Cys 66 N-aniline (Th5) with Met 221 Ph (Th4) with Arg 65 Thiophene ring with Phe 235 |
H- donor H- donor π-H interaction π-H interaction |
3.68 3.70 3.77 4.65 |
1 | −53.0105 |
| 7c | −6.4323 | 1.2726 |
N-carboxamide (Th4) with Asp 256 N-carboxamide (Th2) with Asp 256 O-acetyl group with Lys 257 O-acetyl group with Lys 260 Ph (Th4) with Lys 259 |
H-donor H-donor H-acceptor H-acceptor π-H interaction |
3.29 2.96 2.93 2.98 4.34 |
1 | −45.5377 |
| Ampicillin | −6.5350 | 1.3533 | O-carboxyl group with Glu 243 | H-donor | 2.86 | 1 | 78.6567 |
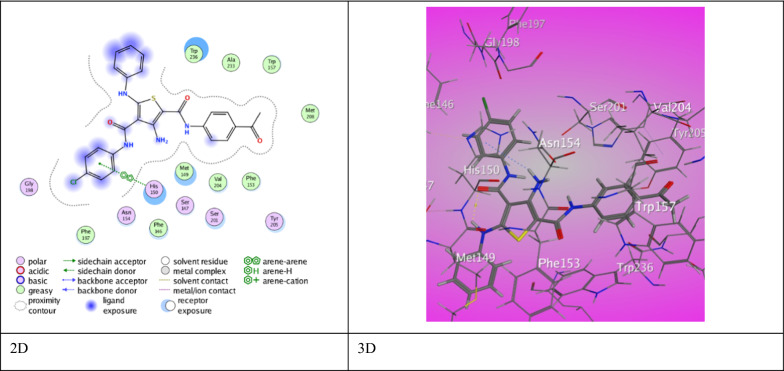
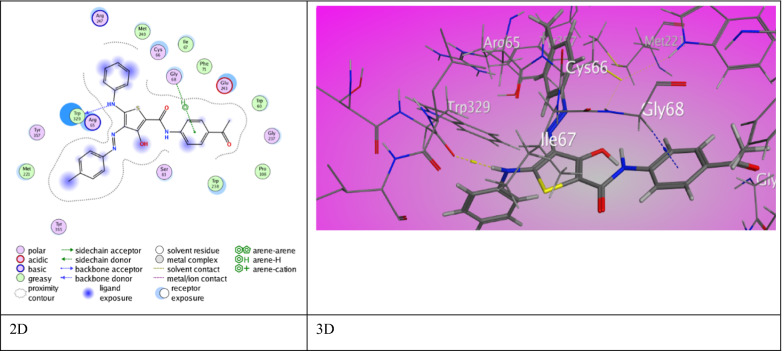
The 3-hydroxy thiophene carboxamide derivative 3a exhibited two types of interactions offering an eminent binding score (S = −8.2175 kcal/mol). A H-bond between N-aniline (Th5) with Trp 329 (3.17 ˚A) and π- H interaction between Ph (Th2) with Gly 68 (3.98 ˚A) (Fig. 8). Likewise, derivative 3b proved pre-eminent binding score (S = −7.9612 kcal/mol) through two modes of interactions. Two H-bonds between N-carboxamide (Th2) with Met 221 (4.23˚A) and O-acetyl group with Lys 360 (2.92˚A). Besides π- H interaction and π- π stick forces between aniline (Th5) and Ph (Th2) with Arg 65 (4.11 ˚A) and His 128 (3.90 ˚A). Derivative 3c showed two modes of interactions and a score of binding (S = −6.5618 kcal/mol). A H-bond between N2-azo group with Lys 211 (3.14 ˚A) and π- H interaction between Ph (Th2) with Glu 194 (3.78 ˚A). Furthermore, 3-methyl thiophene carboxamide derivative 5a revealed binding score (S = −7.2325 kcal/mol) via π-H interaction between Ph (Th2) with Arg 65 (4.19 ˚A). Derivative 5b have a fallen binding score (S = −6.0805 kcal/mol) through π-H interaction between thiophene ring with Pro 196 (3.78 ˚A). Derivative 5c demonstrated a relatively high binding score, (S = −7.3792 kcal/mol) through three H-bonds between S-thiophene ring with Tyr 357 (3.37 ˚A) and O-carboxamide (Th2) with Arg 65 (2.89 ˚A) and O-acetyl group with Asn 64 (3.08 ˚A). Along with π- H interaction between Ph (Th2) with Arg 65 (4.83 ˚A).
Fig. 8.
The binding interaction of 3-hydroxythiophene 3a with (PDB ID: 4d3v)
However, 3-amino thiophene carboxamide derivative 7a presented two interactions through a binding score (S = −7.2786 kcal/mol). It shows H-π interaction between H-acetyl group with Trp 238 (3.87 ˚A) and π-H interaction between Ph (Th5) with Trp 329 (4.45 ˚A). Derivative 7b established excellent binding score (S = −7.7337 kcal/mol) caused by two H-bonds between N-amino (Th3) and N-aniline (Th5) with Cys 66 (3.68 ˚A) and Met 221 (3.70 ˚A) besides two π-H interaction between Ph (Th4) and thiophene ring with Arg 65 (3.77 ˚A) and Phe 235 (4.65 ˚A). Furthermore, derivative 7c presented π-H interaction between Lys 259 with Ph (Th4) (4.34 ˚A) in addition to four H-bonds through binding score (S = −6.4323 kcal/mol). Asp 256 established H-bonds with N-carboxamide (Th4) (3.29 ˚A) and (Th2) (2.96 ˚A). Lys 257 and Lys 260 showed H- bonds with O-acetyl group (2.93 ˚A) (2.98 ˚A). Ampicillin showed with 4D3V protein binding score (S = −6.5350 kcal/mol) resulted from H-bond between Glu 243 with O-carboxyl group (2.86 ˚A).
Conclusion
A series of 3-substituted thiophene 2-carboxamide with functional groups (-OH, -Me and -NH2) were synthesized. In addition to intramolecular H-bonds, the DFT modelling data suggested a comparable Mulliken's charge and HOMO–LUMO contribution. The newly synthesized 3-substituted thiophene 2-carboxamide 3a-c, 5a-c, 7a-c antioxidant activity was examined through ABTS method, while the antibacterial activity was assessed against 4 types of bacteria. Derivatives 7a-c inhibited better than derivatives 3a-c and 4a-c. SAR-study of the obtained compounds shows the enhancement of OMe substituent on the activity and absence of substituent was also a positive impact. In addition, the synthesized compounds were subjected to a molecular docking analysis using (PDB ID 2AS1) (PDB ID 1DD6) (PDB ID 2MLM) (PDB ID 3ZMI) (PDB ID 4D3V). Thiophene derivatives 3b, 5a, 7a, 7c and 3a had better score of binding (S = −9.3283, −7.6609, −7.0842, −7.8889 and −8.2175 kcal/mol) toward the selected proteins.
Experimental
Synthesis of 3-hydroxythiophene compounds 3a-c
A suspension of each 2-(arylhydrazono)-2-ethoxycarbonyl-thioacetanilide derivative 2a, 2b or 2c (0.002 mol) in sodium ethoxide solution (which was prepared from 0.05 g Na and 20 mL absolute ethanol) was stirred for 10 min and then N-(4-acetylphenyl)-2-chloroacetamide (1) (0.42 g, 0.002 mol) was added. The mixture was refluxed for 2 h then was permitted to be cool, and poured onto ice-water. The obtained product was dried out and crystallized from ethanol to give the targeted hydroxythiophenes 3a-c.
N-(4-Acetylphenyl)-3-hydroxy-5-phenylamino-4-(p-tolylazo)-thiophene-2-carboxamide (3a)
Red solid, yield = 56%, m.p. = 270–271 °C. IR (ν/cm−1): 3363 (N–H stretch), 1654 (C = O stretch). 1H NMR (δ/ppm): 2.34 (s, 3H, CH3), 2.52 (s, 3H, COCH3), 7.29 (d, J = 8.0 Hz, 5H, Ar–H), 7.44–7.52 (m, 4H, Ar–H and NH), 7.75 (t, J = 9.0 Hz, 3H, Ar–H), 7.93 (d, J = 8.5 Hz, 2H, Ar–H), 9.86 (s, 1H, NH), 12.38 (s, 1H, OH). 13C NMR (δ/ppm): 20.80, 26.46, 118.55 (3C), 118.92 (2C), 121.36 (4C), 125.93, 129.64 (4C), 129.72 (5C), 129.93 (3C), 160.83, 196.51. MS: m/z (%) = 470 (M+, 22.63), 430.94 (52.51), 405.13 (78.21), 400.79 (100.00), 284.79 (51.30), 284.28 (46.29), 254.68 (44.82), 244.49 (51.91), 237.61 (50.33), 236.67 (45.29), 175.37 (51.74), 172.90 (44.75), 90.64 (45.29), 40.61 (52.01). Analysis calculated for C26H22N4O3S (470.14): C, 66.37; H, 4.71; N, 11.91%. Found: C, 66.07; H, 4.63; N, 12.07%.
N-(4-Acetylphenyl)-4-(p-anisylazo)-3-hydroxy-5-phenylamino-thiophene-2-carboxamide (3b)
Dark red solid, yield = 48%, m.p. = 234–235 °C. IR (ν/cm−1): 3366 (N–H stretch), 1654 (C = O stretch). 1H NMR (δ/ppm): 2.52 (s, 3H, COCH3), 3.84 (s, 3H, OCH3), 7.07 (d, J = 9.0 Hz, 3H, Ar–H), 7.26 (t, J = 7.5, 1H, Ar–H), 7.46–7.52 (m, 3H, Ar–H and NH), 7.74 (d, J = 8.5 Hz, 2H, Ar–H), 7.93 (d, J = 9.0 Hz, 5H, Ar–H), 9.73 (s, 1H, NH), 12.40 (s, 1H, OH). 13C NMR (δ/ppm): 26.48, 55.58, 114.62 (2C), 114.76, 118.55 (2C), 118.99, 121.13 (3C), 122.13 (2C), 125.63, 129.67 (4C), 129.79 (4C), 131.51, 153.91, 159.95, 196.50. Analysis calculated for C26H22N4O4S (486.14): C, 64.18; H, 4.56; N, 11.52%. Found: C, 63.96; H, 4.48; N, 11.68%.
N-(4-Acetylphenyl)-4-((4-chlorophenyl)azo)-3-hydroxy-5-phenylamino-thiophene-2-carboxamide (3c)
Red crystals, yield = 52%, m.p. = 252–253 °C. IR (ν/cm−1): 3366 (N–H stretch), 1653 (C = O stretch). 1H NMR (δ/ppm): 2.52 (s, 3H, COCH3), 7.29 (t, J = 7.5 Hz, 1H, Ar–H), 7.42 (d, J = 7.0 Hz, 2H), 7.49–7.58 (m, 4H, Ar–H and NH), 7.74 (d, J = 8.0 Hz, 3H, Ar–H), 7.87 (d, J = 9.0 Hz, 2H, Ar–H), 7.93 (d, J = 9.0 Hz, 2H, Ar–H), 9.88 (s, 1H, NH), 12.47 (s, 1H, OH). Analysis calculated for C25H19ClN4O3S (490.09): C, 61.16; H, 3.90; N, 11.41%. Found: C, 61.32; H, 3.85; N, 11.50%.
Synthesis of 3-methylthiophene compounds 5a-c
To a solution of each 2-acetyl-2-arylazo-thioacetanilide derivative 4a, 4b or 4c (0.002 mol) in 20 mL dioxane, sodium methoxide (0.05 g, 0.001 mol) and N-(4-acetylphenyl)-2-chloroacetamide (1) (0.42 g, 0.002 mol) were added. The mixture was refluxed for an hour and then sodium methoxide (0.05 g, 0.001 mol) was added again. Refluxing was continued for additional 2 h and then the reaction mixture was permitted to cool. The formed solid product was collected to give the targeted methylthiophenes 5a-c.
N-(4-Acetylphenyl)-3-methyl-5-phenylamino-4-(p-tolylazo)thiophene-2-carboxamide (5a)
Pale red crystals, yield = 70%, m.p. = 290–291 °C. IR (ν/cm−1): 3447, 3277 (N–H stretch), 1675, 1640 (C = O). 1H NMR (δ/ppm): 2.32 (s, 3H, CH3), 2.49 (s, 3H, CH3), 2.51 (s, 3H, COCH3), 7.25 (d, J = 8.0 Hz, 2H, Ar–H), 7.30 (t, J = 8.5 Hz, 1H, Ar–H), 7.38 (d, J = 8.0 Hz, 2H, Ar–H), 7.48–7.55 (m, 4H, Ar–H), 7.75 (d, J = 8.5 Hz, 2H, Ar–H), 7.90 (d, J = 9.0 Hz, 2H, Ar–H). 13C NMR (δ/ppm): 12.94, 20.65, 26.47, 115.09, 116.81 (2C), 119.86 (2C), 120.86 (3C), 121.05 (2C), 125.23, 125.82, 129.26 (2C), 129.36 (2C), 129.66 (3C), 129.98, 130.07 (2C), 170.10, 196.56. Analysis calculated for C27H24N4O2S (468.16): C, 69.21; H, 5.16; N, 11.96%. Found: C, 69.43; H, 5.07; N, 12.11%.
N-(4-Acetylphenyl)-4-(p-anisylazo)-3-methyl-5-phenylamino-thiophene-2-carboxamide (5b)
Red crystals, yield = 63%, m.p. = 319–320 °C. IR (ν/cm−1): 3447, 3289 (N–H stretch), 1676, 1639 (C = O). 1H NMR (δ/ppm): 2.52 (s, 3H, COCH3), 2.57 (s, 3H, CH3), 3.80 (s, 3H, OCH3), 7.05 (d, J = 8.5 Hz, 2H), 7.24 (t, J = 7.5 Hz, 1H, Ar–H), 7.42–7.51 (m, 4H, Ar–H), 7.70 (d, J = 9.0 Hz, 2H, Ar–H), 7.79 (d, J = 9.0 Hz, 2H, Ar–H), 7.91 (d, J = 9.0 Hz, 2H, Ar–H), 8.49 (s, 1H, NH), 10.28 (s, 1H, NH). 13C NMR (δ/ppm): 13.32, 26.50, 55.51, 114.78 (2C), 119.66 (2C), 120.14 (2C), 120.51 (3C), 125.33, 129.29 (3C), 129.76 (3C), 134.35, 141.41, 143.91, 158.76, 161.61, 166.67, 175.08, 196.59. MS: m/z (%) = 484 (M+, 5.10), 475.20 (41.81), 408.21 (33.89), 380.32( 31.14), 370.21 (76.02), 352.11 (30.03), 328.12 (34.06), 327.54 (59.69), 313.37 (55.63), 298.16 (48.14), 297.22 (52.83), 253.13 (44.02), 233.13 (30.90), 206.22 (30.80), 165.38 (85.57), 86.55 (63.81), 75.22 (100.00), 66.44 (83.76). Analysis calculated for C27H24N4O3S (484.16): C, 66.92; H, 4.99; N, 11.56%. Found: C, 66.75; H, 5.06; N, 11.67%.
N-(4-Acetylphenyl)-4-((4-chlorophenyl)azo)-3-methyl-5-phenylamino-thiophene-2-carboxamide (5c)
Orange crystals, yield = 81%, m.p. = 270–271 °C. IR (ν/cm−1): 3448, 3277 (N–H stretch), 1674, 1638 (C = O). 1H NMR (δ/ppm): 2.45 (s, 3H, CH3), 2.52 (s, 3H, COCH3), 7.28 (t, J = 7.5 Hz, 1H, Ar–H), 7.39 (d, J = 8.0 Hz, 2H, Ar–H), 7.47–7.52 (m, 4H, Ar–H), 7.63 (d, J = 8.5 Hz, 2H, Ar–H), 7.78 (d, J = 8.5 Hz, 2H, Ar–H), 7.92 (d, J = 9.0 Hz, 2H, Ar–H), 8.50 (s, 1H, NH), 10.27 (s, 1H, NH). Analysis calculated for C26H21ClN4O2S (488.11): C, 63.86; H, 4.33; N, 11.46%. Found: C, 63.64; H, 4.28; N, 11.61%.
Synthesis of 3-aminothiophene compounds 7a-c
A mixture of each 2-cyano-3-mercapto-N-phenyl-3-(phenylamino)acrylamide derivative 6a, 6b or 6c (0.002 mol), N-(4-acetylphenyl)-2-chloroacetamide (0.44 g, 0.002 mol) and sodium methoxide (0.11 g, 0.002 mol) was refluxed in 20 mL dioxane for 2 h. The mixture was poured into ice-water. The formed solid was collected and recrystallized from ethanol to produce the corresponding 3-aminothiophene compounds 7a-c.
N2-(4-Acetylphenyl)-3-amino-N4-phenyl-5-phenylaminothiophene-2,4-dicarboxamide (7a)
White crystals, yield = 47%, m.p. = 190–191 °C. IR (ν/cm−1): 3346, 3276, 3192 (NH2 & NH), 1654 (broad, C = O stretch). 1H NMR (δ/ppm): 2.51 (s, 3H, COCH3), 5.12 (s, 1H, NH), 7.03–7.08 (m, 4H, Ar–H), 7.29–7.38 (m, 4H, Ar–H), 7.57 (d, J = 7.5 Hz, 2H, Ar–H), 7.68 (d, J = 8.5 Hz, 2H, Ar–H), 7.94 (d, J = 8.5 Hz, 2H, Ar–H), 8.80 (s, 1H, NH), 9.31 (s, 1H, NH), 10.71 (s, 1H, NH), 11.85 (s, 1H, NH). 13C NMR (δ/ppm): 26.50, 118.54 (2C), 119.44 (2C), 119.94, 120.29, 121.21 (2C), 123.21, 124.50, 128.49, 129.01 (2C), 129.28 (2C), 129.63 (2C), 132.20, 138.58, 142.88, 149.95, 163.38, 165.85, 167.48, 196.57. MS: m/z (%) = 470 (M+, 25.44), 453.06 (58.69), 449 (63.16), 44.27 (52.26), 429.94 (52.39), 354.88 (52.44), 174.09 (62.15), 147.41 (75.00), 117.02 (57.18), 104.56 (100), 43.95 (72.07). Analysis calculated for C26H22N4O3S (470.14): C, 66.37; H, 4.71; N, 11.91%. Found: C, 66.48; H, 4.65; N, 11.82%.
N2-(4-Acetylphenyl)-3-amino-5-phenylamino-N4-(p-tolyl)thiophene-2,4-dicarboxamide (7b)
Yellow solid, yield = 54%, m.p. = 230–231 °C. IR (ν/cm−1): 3390, 3252 (NH2 & NH), 1670, 1649 (C = O stretch). 1H NMR (δ/ppm): 2.24 (s, 3H, CH3), 2.51 (s, 3H, COCH3), 5.10 (s, 1H, NH), 7.06–7.14 (m, 3H, Ar–H), 7.36–7.52 (m, 6H, Ar–H), 7.67 (d, J = 9.0 Hz, 2H, Ar–H), 7.94 (d, J = 9.0 Hz, 2H, Ar–H), 8.76 (s, 1H, NH), 9.33 (s, 1H, NH), 10.71 (s, 1H, NH), 11.75 (s, 1H, NH). Analysis calculated for C27H24N4O3S (484.16): C, 66.92; H, 4.99; N, 11.56%. Found: C, 67.08; H, 5.06; N, 11.45%.
N2-(4-Acetylphenyl)-3-amino-N4-(p-anisyl)-5-phenylamino-thiophene-2,4-dicarboxamide (7c)
Orange solid, yield = 62%, m.p. = 244–245 °C. IR (ν/cm−1): 3348, 3234 (NH2 & NH), 1646 (broad, C = O stretch). 1H NMR (δ/ppm): 2.52 (s, 3H, COCH3), 3.77 (s, 3H, OCH3), 5.12 (s, 1H, NH), 6.94 (d, J = 9.0 Hz, 2H, Ar–H), 7.38–7.43 (m, 5H, Ar–H), 7.68 (d, J = 9.0 Hz, 2H, Ar–H), 7.74 (d, J = 9.0 Hz, 2H, Ar–H), 7.98 (d, J = 9.0 Hz, 2H, Ar–H), 8.78 (s, 1H, NH), 9.25 (s, 1H, NH), 10.70 (s, 1H, NH), 11.81 (s, 1H, NH). Analysis calculated for C27H24N4O4S (500.15): C, 64.79; H, 4.83; N, 11.19%. Found: C, 64.84; H, 4.89; N, 11.31%.
Computational details
Quantum chemical calculations for the synthesized compounds were used to optimize the geometry by Gaussian 09W suite program [66] using the Becke3–Lee–Yang–Parr (B3LYP) exchange–correlation functional [67–69] with standard 6–311 + + G (d,p) basis set. The HOMO–LUMO plots and Mulliken’s atomic charges data were obtained using the GaussView program [70]. The Fukui indices were determined by Materials studio package DMol3 module [71] utilizing the GGA and B3LYP functional with DNP (version 3.5) [72].
Docking method
All the molecular modeling studies were carried out using Molecular Operating Environment (MOE, 2015.01) software.The three-dimensional structure (3D) of the selected proteins (PDB ID 2AS1), (PDB ID 1DD6), (PDB ID 2MLM), (PDB ID 3ZMI), (PDB ID 4D3V) were downloaded from the PDB website. The water molecules and repeated chains were removed. Protons were added and the energy of the protein was minimized. The preparation of thiophene 2-carboxamide derivatives 3a-c, 5a-c and 7a-c for docking were carried out by energy minimization and potential energy calculation inside MOE propgram. MOE conducted the docking of the newly synthesized compounds, calculated the binding energies, and provided their binding modes.
Supplementary Information
Additional file 1: Experimental general remarks. Table S1. Bond length, bond angle, dihedral angle for compounds 3a-c. Table S2. Bond length, bond angle, dihedral angle for compounds 5a-c. Table S3. Bond length, bond angle, dihedral angle for compounds 7a-c. Table S4. The atomic Mulliken’s charges and Fukui’s indices of investigated compounds. Fig. S1. DFT optimized structures for compounds 3a-c, 5a-c, and 7a-c. Fig. S2. The binding interaction of 3-hydroxythiophene 3a with (PDB ID: 2AS1). Fig. S3. The binding interaction of 3-hydroxythiophene 5a with (PDB ID: 2AS1). Fig. S4. The binding interaction of 3-hydroxythiophene 5b with (PDB ID: 2AS1). Fig. S5. The binding interaction of 3-hydroxythiophene 5c with (PDB ID: 2AS1). Fig. S6. The binding interaction of 3-hydroxythiophene 7a with (PDB ID: 2AS1). Fig. S7. The binding interaction of 3-hydroxythiophene 7b with (PDB ID: 2AS1). Fig. S8. The binding interaction of 3-hydroxythiophene 7c with (PDB ID: 2AS1). Fig. S9. The binding interaction ascorbic acid with (PDB ID: 2AS1). Fig. S10. The binding interaction of 3-hydroxythiophene 3a with (PDB ID: 1DD6). Fig. S11. The binding interaction of 3-hydroxythiophene 3b with (PDB ID: 1DD6). Fig. S12. The binding interaction of 3-hydroxythiophene 3c with (PDB ID: 1DD6). Fig. S13. The binding interaction of 3-methylthiophene 5b with (PDB ID: 1DD6). Fig. S14. The binding interaction of 3-methylthiophene 5c with (PDB ID: 1DD6). Fig. S15. The binding interaction of 3-aminothiophene 7a with (PDB ID: 1DD6). Fig. S16. The binding interaction of 3-aminothiophene 7b with (PDB ID: 1DD6). Fig. S17. The binding interaction of 3-aminothiophene 7c with (PDB ID: 1DD6). Fig. S18. The binding interaction of with ampicillin (PDB ID: 1DD6). Fig. S19. The binding interaction of 3-hydroxythiophene 3a with (PDB ID: 2MLM). Fig. S20. The binding interaction of 3-hydroxythiophene 3b with (PDB ID: 2MLM). Fig. S21. The binding interaction of 3-hydroxythiophene 3c with (PDB ID: 2MLM). Fig. S22. The binding interaction of 3-methylthiophene 5a with (PDB ID: 2MLM). Fig. S23. The binding interaction of 3-methylthiophene 5b with (PDB ID: 2MLM). Fig. S24. The binding interaction of 3-methylthiophene 5c with (PDB ID: 2MLM). Fig. S25. The binding interaction of 3-methylthiophene 7b with (PDB ID: 2MLM). Fig. S26. The binding interaction of 3-methylthiophene 7c with (PDB ID: 2MLM). Fig. S27. The binding interaction of ampicillin with (PDB ID: 2MLM). Fig. S28. The binding interaction of 3-hydroxythiophene 3a with (PDB ID: 3ZMI). Fig. S29. The binding interaction of 3-hydroxythiophene 3b with (PDB ID: 3ZMI). Fig. S30. The binding interaction of 3-hydroxythiophene 3c with (PDB ID: 3ZMI). Fig. S31. The binding interaction of 3-methylthiophene 5a with (PDB ID: 3ZMI). Fig. S32. The binding interaction of 3-methylthiophene 5b with (PDB ID: 3ZMI). Fig. S33. The binding interaction of 3-methylthiophene 5c with (PDB ID: 3ZMI). Fig. S34. The binding interaction of 3-aminothiophene 7a with (PDB ID: 3ZMI). Fig. S35. The binding interaction of 3-aminothiophene 7b with (PDB ID: 3ZMI). Fig. S36. The binding interaction of with ampicillin (PDB ID: 3ZMI). Fig. S37. The binding interaction of 3-hydroxythiophene 3b with (PDB ID: 4d3v). Fig. S38. The binding interaction of 3-hydroxythiophene 3c with (PDB ID: 4d3v). Fig. S39. The binding interaction of 3-methylthiophene 5a with (PDB ID: 4d3v). Fig. S40. The binding interaction of 3-methylthiophene 5b with (PDB ID: 4d3v). Fig. S41. The binding interaction of 3-methylthiophene 5c with (PDB ID: 4d3v). Fig. S42. The binding interaction of 3-aminothiophene 7a with (PDB ID: 4d3v). Fig. S43. The binding interaction of 3-aminothiophene 7b with (PDB ID: 4d3v). Fig. S44. The binding interaction of 3-aminothiophene 7c with (PDB ID: 4d3v). Fig. S45. The binding interaction of with ampicillin (PDB ID: 4d3v). Fig. S46. IR spectrum of compound 3a. Fig. S47. 1H NMR spectrum of compound 3a. Fig. S48. 13C NMR spectrum of compound 3a. Fig. S49. IR spectrum of compound 3b. Fig. S50. 1H NMR spectrum of compound 3b. Fig. S51. 13C NMR spectrum of compound 3b. Fig. S52. IR spectrum of compound 3c. Fig. S53. 1H NMR spectrum of compound 3c. Fig. S54. IR spectrum of compound 5a. Fig. S55. 1H NMR spectrum of compound 5a. Fig. S56. 13C NMR spectrum of compound 5a. Fig. S57. IR spectrum of compound 5b. Fig. S58. 1H NMR spectrum of compound 5b. Fig. S59. 13C NMR spectrum of compound 5b. Fig. S60. IR spectrum of compound 5c. Fig. S61. 1H NMR spectrum of compound 5c. Fig. S62. IR spectrum of compound 7a. Fig. S63. 1H NMR spectrum of compound 7a. Fig. S64. 13C NMR spectrum of compound 7a. Fig. S65. IR spectrum of compound 7b. Fig. S66. 1H NMR spectrum of compound 7b. Fig. S67. IR spectrum of compound 7c. Fig. S68. 1H NMR spectrum of compound 7c.
Acknowledgements
The authors express their gratitude to University of Mansoura – Faculty of Science for lab facilities for antioxidant and antibacterial activity.
Author contributions
HMM; conceptualization, modelling and docking studies, writing original draft, data analysis, proofreading, manuscript handling. NAK; synthesis, methodology, characterization of compounds. EA-L and MAI; supervision, initial corrections, biological evaluation, data curation. All the authors read and approved the final manuscript.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Availability of data and materials
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Antimicrobial Resistance Collaborators Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–655. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gramec D, Peterlin Mašič L, Sollner Dolenc M. Bioactivation potential of thiophene-containing drugs. Chem Res Toxicol. 2014;27(8):1344–1358. doi: 10.1021/tx500134g. [DOI] [PubMed] [Google Scholar]
- 3.Koçyiğit ÜM, et al. Determination of biological studies and molecular docking calculations of isatin-thiosemicarbazone hybrid compounds. J Mol Struct. 2022;1264:133249. doi: 10.1016/j.molstruc.2022.133249. [DOI] [Google Scholar]
- 4.Muğlu H, et al. Preparation, antioxidant activity, and theoretical studies on the relationship between antioxidant and electronic properties of bis(thio/carbohydrazone) derivatives. J Phys Chem Solids. 2022;164:110618. doi: 10.1016/j.jpcs.2022.110618. [DOI] [Google Scholar]
- 5.Lamberth C, Dinges J. Bioactive heterocyclic compound classes: pharmaceuticals. Hoboken: John Wiley & Sons; 2012. [Google Scholar]
- 6.Isloor AM, Kalluraya B, Pai KS. Synthesis, characterization and biological activities of some new benzo [b] thiophene derivatives. Eur J Med Chem. 2010;45(2):825–830. doi: 10.1016/j.ejmech.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 7.Khan E, et al. Synthesis characterization and DFT calculations of 2, 5-substituted thiophene derivatives. J Chem Crystallogr. 2015;45(5):238–243. doi: 10.1007/s10870-015-0588-9. [DOI] [Google Scholar]
- 8.Chua MH, et al. Towards modulating the colour hues of isoindigo-based electrochromic polymers through variation of thiophene-based donor groups. Polym Chem. 2022;13(7):967–981. doi: 10.1039/D1PY01531A. [DOI] [Google Scholar]
- 9.Vairalakshmi M, et al. Synthesis, structural elucidation, catalytic, antibacterial and antioxidant activity of thiophene derived mixed ligand metal complexes. J Chil Chem Soc. 2018;63(1):3844–3849. doi: 10.4067/s0717-97072018000103844. [DOI] [Google Scholar]
- 10.Harinath Y, et al. Synthesis, spectral characterization and antioxidant activity studies of a bidentate Schiff base, 5-methyl thiophene-2-carboxaldehyde-carbohydrazone and its Cd (II), Cu (II), Ni (II) and Zn (II) complexes. Spectrochim Acta Part A Mol Biomol Spectrosc. 2013;101:264–272. doi: 10.1016/j.saa.2012.09.085. [DOI] [PubMed] [Google Scholar]
- 11.Yanagimoto K, et al. Antioxidative activity of heterocyclic compounds found in coffee volatiles produced by Maillard reaction. J Agric Food Chem. 2002;50(19):5480–5484. doi: 10.1021/jf025616h. [DOI] [PubMed] [Google Scholar]
- 12.Sonmez F, et al. Synthesis, antioxidant activity and SAR study of novel spiro-isatin-based Schiff bases. Mol Diversity. 2019;23(4):829–844. doi: 10.1007/s11030-018-09910-7. [DOI] [PubMed] [Google Scholar]
- 13.Yakan H, et al. A new series of asymmetric bis-isatin derivatives containing urea/thiourea moiety: preparation, spectroscopic elucidation, antioxidant properties and theoretical calculations. J Mol Struct. 2021;1239:130495. doi: 10.1016/j.molstruc.2021.130495. [DOI] [Google Scholar]
- 14.Kurt BZ, et al. Synthesis, anticholinesterase, antioxidant, and anti-aflatoxigenic activity of novel coumarin carbamate derivatives. ChemistrySelect. 2018;3(14):3978–3983. doi: 10.1002/slct.201800142. [DOI] [Google Scholar]
- 15.Kurt BZ, et al. Synthesis, antioxidant and carbonic anhydrase I and II inhibitory activities of novel sulphonamide-substituted coumarylthiazole derivatives. J Enzyme Inhib Med Chem. 2016;31(6):991–998. doi: 10.3109/14756366.2015.1077823. [DOI] [PubMed] [Google Scholar]
- 16.Çakmak Ş, et al. Synthesis, structural investigation, hirshfeld surface analysis, and biological evaluation of N-(3-Cyanothiophen-2-yl)-2-(thiophen-2-yl)acetamide. ACS Omega. 2022;7(13):11320–11329. doi: 10.1021/acsomega.2c00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abu-Melha S. Molecular modeling and antioxidant activity of newly synthesized 3-hydroxy-2-substituted-thiophene derivatives. J Mol Struct. 2022;1250:131821. doi: 10.1016/j.molstruc.2021.131821. [DOI] [Google Scholar]
- 18.Rauf A, et al. Synthesis, structure and antibacterial activity of a copper (II) coordination polymer based on thiophene-2, 5-dicarboxylate ligand. Polyhedron. 2019;166:130–136. doi: 10.1016/j.poly.2019.03.039. [DOI] [Google Scholar]
- 19.Rani M, Mohamad Y. Synthesis, studies and in vitro antibacterial activity of some 5-(thiophene-2-yl)-phenyl pyrazoline derivatives. J Saudi Chem Soc. 2014;18(5):411–417. doi: 10.1016/j.jscs.2011.09.002. [DOI] [Google Scholar]
- 20.Alomar K, et al. Synthesis, structure and antifungal activity of thiophene-2, 3-dicarboxaldehyde bis (thiosemicarbazone) and nickel (II), copper (II) and cadmium (II) complexes: Unsymmetrical coordination mode of nickel complex. J Inorg Biochem. 2013;126:76–83. doi: 10.1016/j.jinorgbio.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 21.Fokialakis N, et al. Antifungal activity of thiophenes from Echinops ritro. J Agric Food Chem. 2006;54(5):1651–1655. doi: 10.1021/jf052702j. [DOI] [PubMed] [Google Scholar]
- 22.Al-Azmi A, John E. Synthesis of 4-arylazo-2-(N-pyrazolylcarboxamido) thiophene disperse dyes for dyeing of polyester and their antibacterial evaluation. Text Res J. 2020;90(23–24):2795–2805. doi: 10.1177/0040517520931476. [DOI] [Google Scholar]
- 23.Molvi KI, et al. Synthesis, anti-inflammatory, analgesic and antioxidant activities of some tetrasubstituted thiophenes. J Enzyme Inhib Med Chem. 2008;23(6):829–838. doi: 10.1080/14756360701626082. [DOI] [PubMed] [Google Scholar]
- 24.Lisboa T, et al. Toxicity and antitumor activity of a thiophene–acridine hybrid. Molecules. 2019;25(1):64. doi: 10.3390/molecules25010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghorab MM, Al-Said MS. Antitumor activity of novel pyridine, thiophene and thiazole derivatives. Arch Pharmacal Res. 2012;35(6):965–973. doi: 10.1007/s12272-012-0603-z. [DOI] [PubMed] [Google Scholar]
- 26.Liu KK-C, et al. Highly selective and potent thiophenes as PI3K inhibitors with oral antitumor activity. ACS Med Chem Lett. 2011;2(11):809–813. doi: 10.1021/ml200126j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pathania S, Chawla PA. Thiophene-based derivatives as anticancer agents: an overview on decade’s work. Bioorg Chem. 2020;101:104026. doi: 10.1016/j.bioorg.2020.104026. [DOI] [PubMed] [Google Scholar]
- 28.Pathania S, Narang RK, Rawal RK. Role of sulphur-heterocycles in medicinal chemistry: an update. Eur J Med Chem. 2019;180:486–508. doi: 10.1016/j.ejmech.2019.07.043. [DOI] [PubMed] [Google Scholar]
- 29.Seecheran R, et al. Rivaroxaban as an antithrombotic agent in a patient with ST-segment elevation myocardial infarction and left ventricular thrombus: a case report. J Investig Med High Impact Case Rep. 2017;5(1):2324709617697991. doi: 10.1177/2324709617697991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahmad G, et al. Synthesis, in-vitro cholinesterase inhibition, in-vivo anticonvulsant activity and in-silico exploration of N-(4-methylpyridin-2-yl) thiophene-2-carboxamide analogs. Bioorg Chem. 2019;92:103216. doi: 10.1016/j.bioorg.2019.103216. [DOI] [PubMed] [Google Scholar]
- 31.Janetka JW, et al. Discovery of a novel class of 2-ureido thiophene carboxamide checkpoint kinase inhibitors. Bioorg Med Chem Lett. 2008;18(14):4242–4248. doi: 10.1016/j.bmcl.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 32.Kojima N, et al. Thiophene-3-carboxamide analogue of annonaceous acetogenins as antitumor drug lead. Eur J Med Chem. 2014;86:684–689. doi: 10.1016/j.ejmech.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 33.Hameed P, S., , et al. Nitrothiophene carboxamides, a novel narrow spectrum antibacterial series: mechanism of action and efficacy. Sci Rep. 2018;8(1):7263. doi: 10.1038/s41598-018-25407-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baxter A, et al. Hit-to-lead studies: the discovery of potent, orally active, thiophenecarboxamide IKK-2 inhibitors. Bioorg Med Chem Lett. 2004;14(11):2817–2822. doi: 10.1016/j.bmcl.2004.03.058. [DOI] [PubMed] [Google Scholar]
- 35.Cakmak S, et al. Synthesis, X-ray structure, antimicrobial activity, DFT and molecular docking studies of N-(thiophen-2-ylmethyl)thiophene-2-carboxamide. Acta Crystallogr C. 2022;78(7):390–397. doi: 10.1107/S2053229622006283. [DOI] [PubMed] [Google Scholar]
- 36.Muğlu H, et al. Exploring of antioxidant and antibacterial properties of novel 1,3,4-thiadiazole derivatives: facile synthesis, structural elucidation and DFT approach to antioxidant characteristics. Comput Biol Chem. 2022;96:107618. doi: 10.1016/j.compbiolchem.2021.107618. [DOI] [PubMed] [Google Scholar]
- 37.Anthwal A, Thakur BK, Rawat M. Synthesis, antibacterial screening and theoretical molecular properties prediction of hydantoin-chalcone conjugates. J Indian Chem Soc. 2014;91:1525–1531. [Google Scholar]
- 38.Harhash AH, et al. Synthesis of Ethyl α-phenylthiocarbamylglyoxalate arylhydrazones and their reactions with hydrazines. Zeitschrift für Naturforschung B. 1976;31(6):846–849. doi: 10.1515/znb-1976-0624. [DOI] [Google Scholar]
- 39.Amer FA, Harhash AH, Awad ML. Synthesis of 2-Methyl-l-phenylthiocarbamoylglyoxal Arylhydrazones and Diethyl (arylazo)(phenylthiocarbamoyl) malonate and their Reactions with Hydrazines. Zeitschrift für Naturforschung B. 1978;33(6):660–662. doi: 10.1515/znb-1978-0620. [DOI] [Google Scholar]
- 40.George RF. Stereoselective synthesis and QSAR study of cytotoxic 2-(4-oxo-thiazolidin-2-ylidene)-2-cyano-N-arylacetamides. Eur J Med Chem. 2012;47:377–386. doi: 10.1016/j.ejmech.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 41.Farag AB, Ewida HA, Ahmed MS. Design, synthesis, and biological evaluation of novel amide and hydrazide based thioether analogs targeting Histone deacteylase (HDAC) enzymes. Eur J Med Chem. 2018;148:73–85. doi: 10.1016/j.ejmech.2018.02.011. [DOI] [PubMed] [Google Scholar]
- 42.Morad M, et al. Copper–acetanilide complexes: synthesis, characterization, crystal structure, computational analysis and their application as heterogeneous catalysts for biodiesel synthesis from frying waste oils. Res Chem Intermed. 2020;46(10):4543–4562. doi: 10.1007/s11164-020-04220-w. [DOI] [Google Scholar]
- 43.Lissi EA, et al. Total antioxidant potential of resinous exudates from Heliotropium species, and a comparison of the ABTS and DPPH methods. Free Radical Res. 1999;30(6):471–477. doi: 10.1080/10715769900300511. [DOI] [PubMed] [Google Scholar]
- 44.Tavadyan LA, et al. Antioxidant properties of selenophene, thiophene and their aminocarbonitrile derivatives. Antioxidants. 2017;6(2):22. doi: 10.3390/antiox6020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abumelha HM, et al. Molecular modeling of new thiazolyl-thiophene based compounds as antioxidant agents. J Mol Struct. 2022;1262:133112. doi: 10.1016/j.molstruc.2022.133112. [DOI] [Google Scholar]
- 46.Keri RS, et al. An overview of benzo[b]thiophene-based medicinal chemistry. Eur J Med Chem. 2017;138:1002–1033. doi: 10.1016/j.ejmech.2017.07.038. [DOI] [PubMed] [Google Scholar]
- 47.Flynn BL, Verdier-Pinard P, Hamel E. A novel palladium-mediated coupling approach to 2,3-disubstituted benzo[b]thiophenes and its application to the synthesis of tubulin binding agents. Org Lett. 2001;3(5):651–654. doi: 10.1021/ol0067179. [DOI] [PubMed] [Google Scholar]
- 48.Shah R, Verma PK. Synthesis of thiophene derivatives and their anti-microbial, antioxidant, anticorrosion and anticancer activity. BMC Chem. 2019;13(1):13–54. doi: 10.1186/s13065-019-0569-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Queiroz MJ, et al. Synthesis and antimicrobial activity studies of ortho-chlorodiarylamines and heteroaromatic tetracyclic systems in the benzo[b]thiophene series. Bioorg Med Chem. 2006;14(20):6827–6831. doi: 10.1016/j.bmc.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 50.Sable PN, Ganguly S, Chaudhari PD. An efficient one-pot three-component synthesis and antimicrobial evaluation of tetra substituted thiophene derivatives. Chin Chem Lett. 2014;25(7):1099–1103. doi: 10.1016/j.cclet.2014.03.044. [DOI] [Google Scholar]
- 51.Ramulu BJ, Koley S, Singh MS. Metal-free Brønsted acid mediated synthesis of fully substituted thiophenes via chemo-and regioselective intramolecular cyclization of α, α′-bis (β-oxodithioesters) at room temperature. Org Biomol Chem. 2016;14(2):434–439. doi: 10.1039/C5OB02081F. [DOI] [PubMed] [Google Scholar]
- 52.Pouzet P, et al. Thiophene 1-oxides. V. comparison of the crystal structures and thiophene ring aromaticity of 2, 5-diphenylthiophene, its sulfoxide and sulfone. J Heterocycl Chem. 1997;34(5):1567–1574. doi: 10.1002/jhet.5570340530. [DOI] [Google Scholar]
- 53.Nohara I, et al. [Cu (POP)(N^ S)][PF 6] and [Cu (xantphos)(N^ S)][PF 6] compounds with 2-(thiophen-2-yl) pyridines. RSC Adv. 2019;9(24):13646–13657. doi: 10.1039/C9RA02617G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Portalone G. Benzamidinium 2-meth-oxy-benzoate. Acta Crystallogr Sect E Struct Rep Online. 2013;69:o1114–o1115. doi: 10.1107/s1600536813016395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sajan D, et al. Natural bond orbital analysis, electronic structure, non-linear properties and vibrational spectral analysis of L-histidinium bromide monohydrate: a density functional theory. Spectrochim Acta Part A Mol Biomol Spectrosc. 2011;81(1):85–98. doi: 10.1016/j.saa.2011.05.052. [DOI] [PubMed] [Google Scholar]
- 56.Bulat FA, et al. Condensation of frontier molecular orbital Fukui functions. J Phys Chem A. 2004;108(2):342–349. doi: 10.1021/jp036416r. [DOI] [Google Scholar]
- 57.Xavier S, Periandy S, Ramalingam S. NBO, conformational, NLO, HOMO–LUMO, NMR and electronic spectral study on 1-phenyl-1-propanol by quantum computational methods. Spectrochim Acta Part A Mol Biomol Spectrosc. 2015;137:306–320. doi: 10.1016/j.saa.2014.08.039. [DOI] [PubMed] [Google Scholar]
- 58.Bhagyasree J, et al. Vibrational spectroscopic (FT-IR, FT-Raman, 1H NMR and UV) investigations and computational study of 5-nitro-2-(4-nitrobenzyl) benzoxazole. Spectrochim Acta Part A Mol Biomol Spectrosc. 2013;102:99–113. doi: 10.1016/j.saa.2012.09.032. [DOI] [PubMed] [Google Scholar]
- 59.Olasunkanmi LO, Obot IB, Ebenso EE. Adsorption and corrosion inhibition properties of N-{n-[1-R-5-(quinoxalin-6-yl)-4, 5-dihydropyrazol-3-yl] phenyl} methanesulfonamides on mild steel in 1 M HCl: experimental and theoretical studies. RSC Adv. 2016;6(90):86782–86797. doi: 10.1039/C6RA11373G. [DOI] [Google Scholar]
- 60.El Adnani Z, et al. DFT theoretical study of 7-R-3methylquinoxalin-2 (1H)-thiones (RH; CH3; Cl) as corrosion inhibitors in hydrochloric acid. Corros Sci. 2013;68:223–230. doi: 10.1016/j.corsci.2012.11.020. [DOI] [Google Scholar]
- 61.Mi H, Xiao G, Chen X. Theoretical evaluation of corrosion inhibition performance of three antipyrine compounds. Comput Theor Chem. 2015;1072:7–14. doi: 10.1016/j.comptc.2015.08.023. [DOI] [Google Scholar]
- 62.Messali M, et al. A new schiff base derivative as an effective corrosion inhibitor for mild steel in acidic media: Experimental and computer simulations studies. J Mol Struct. 2018;1168:39–48. doi: 10.1016/j.molstruc.2018.05.018. [DOI] [Google Scholar]
- 63.Brenk R, et al. Probing molecular docking in a charged model binding site. J Mol Biol. 2006;357(5):1449–1470. doi: 10.1016/j.jmb.2006.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mabkhot YN, et al. Antimicrobial activity of some novel armed thiophene derivatives and petra/osiris/molinspiration (POM) analyses. Molecules. 2016;21(2):222. doi: 10.3390/molecules21020222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Holden JK, et al. Structure-based design of bacterial nitric oxide synthase inhibitors. J Med Chem. 2015;58(2):994–1004. doi: 10.1021/jm501723p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frisch M, et al. Gaussian 09W. Wallingford, CT, USA: Gaussian. Inc.; 2009. [Google Scholar]
- 67.Becke AD. Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys. 1993;98(7):5648–5652. doi: 10.1063/1.464913. [DOI] [Google Scholar]
- 68.Lee C, Yang W, Parr RG. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B. 1988;37(2):785–789. doi: 10.1103/PhysRevB.37.785. [DOI] [PubMed] [Google Scholar]
- 69.Perdew JP, Wang Y. Pair-distribution function and its coupling-constant average for the spin-polarized electron gas. Phys Rev B. 1992;46(20):12947–12954. doi: 10.1103/PhysRevB.46.12947. [DOI] [PubMed] [Google Scholar]
- 70.Dennington R, Keith T, Millam J. Gaussview, version 5. Shawnee Mission, KS: Semichem Inc; 2009. [Google Scholar]
- 71.BIOVIA DS. Materials studio. San Diego: Dassault Systèmes; 2017. [Google Scholar]
- 72.Delley B. Ground-state enthalpies: evaluation of electronic structure approaches with emphasis on the density functional method. J Phys Chem A. 2006;110(50):13632–13639. doi: 10.1021/jp0653611. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Experimental general remarks. Table S1. Bond length, bond angle, dihedral angle for compounds 3a-c. Table S2. Bond length, bond angle, dihedral angle for compounds 5a-c. Table S3. Bond length, bond angle, dihedral angle for compounds 7a-c. Table S4. The atomic Mulliken’s charges and Fukui’s indices of investigated compounds. Fig. S1. DFT optimized structures for compounds 3a-c, 5a-c, and 7a-c. Fig. S2. The binding interaction of 3-hydroxythiophene 3a with (PDB ID: 2AS1). Fig. S3. The binding interaction of 3-hydroxythiophene 5a with (PDB ID: 2AS1). Fig. S4. The binding interaction of 3-hydroxythiophene 5b with (PDB ID: 2AS1). Fig. S5. The binding interaction of 3-hydroxythiophene 5c with (PDB ID: 2AS1). Fig. S6. The binding interaction of 3-hydroxythiophene 7a with (PDB ID: 2AS1). Fig. S7. The binding interaction of 3-hydroxythiophene 7b with (PDB ID: 2AS1). Fig. S8. The binding interaction of 3-hydroxythiophene 7c with (PDB ID: 2AS1). Fig. S9. The binding interaction ascorbic acid with (PDB ID: 2AS1). Fig. S10. The binding interaction of 3-hydroxythiophene 3a with (PDB ID: 1DD6). Fig. S11. The binding interaction of 3-hydroxythiophene 3b with (PDB ID: 1DD6). Fig. S12. The binding interaction of 3-hydroxythiophene 3c with (PDB ID: 1DD6). Fig. S13. The binding interaction of 3-methylthiophene 5b with (PDB ID: 1DD6). Fig. S14. The binding interaction of 3-methylthiophene 5c with (PDB ID: 1DD6). Fig. S15. The binding interaction of 3-aminothiophene 7a with (PDB ID: 1DD6). Fig. S16. The binding interaction of 3-aminothiophene 7b with (PDB ID: 1DD6). Fig. S17. The binding interaction of 3-aminothiophene 7c with (PDB ID: 1DD6). Fig. S18. The binding interaction of with ampicillin (PDB ID: 1DD6). Fig. S19. The binding interaction of 3-hydroxythiophene 3a with (PDB ID: 2MLM). Fig. S20. The binding interaction of 3-hydroxythiophene 3b with (PDB ID: 2MLM). Fig. S21. The binding interaction of 3-hydroxythiophene 3c with (PDB ID: 2MLM). Fig. S22. The binding interaction of 3-methylthiophene 5a with (PDB ID: 2MLM). Fig. S23. The binding interaction of 3-methylthiophene 5b with (PDB ID: 2MLM). Fig. S24. The binding interaction of 3-methylthiophene 5c with (PDB ID: 2MLM). Fig. S25. The binding interaction of 3-methylthiophene 7b with (PDB ID: 2MLM). Fig. S26. The binding interaction of 3-methylthiophene 7c with (PDB ID: 2MLM). Fig. S27. The binding interaction of ampicillin with (PDB ID: 2MLM). Fig. S28. The binding interaction of 3-hydroxythiophene 3a with (PDB ID: 3ZMI). Fig. S29. The binding interaction of 3-hydroxythiophene 3b with (PDB ID: 3ZMI). Fig. S30. The binding interaction of 3-hydroxythiophene 3c with (PDB ID: 3ZMI). Fig. S31. The binding interaction of 3-methylthiophene 5a with (PDB ID: 3ZMI). Fig. S32. The binding interaction of 3-methylthiophene 5b with (PDB ID: 3ZMI). Fig. S33. The binding interaction of 3-methylthiophene 5c with (PDB ID: 3ZMI). Fig. S34. The binding interaction of 3-aminothiophene 7a with (PDB ID: 3ZMI). Fig. S35. The binding interaction of 3-aminothiophene 7b with (PDB ID: 3ZMI). Fig. S36. The binding interaction of with ampicillin (PDB ID: 3ZMI). Fig. S37. The binding interaction of 3-hydroxythiophene 3b with (PDB ID: 4d3v). Fig. S38. The binding interaction of 3-hydroxythiophene 3c with (PDB ID: 4d3v). Fig. S39. The binding interaction of 3-methylthiophene 5a with (PDB ID: 4d3v). Fig. S40. The binding interaction of 3-methylthiophene 5b with (PDB ID: 4d3v). Fig. S41. The binding interaction of 3-methylthiophene 5c with (PDB ID: 4d3v). Fig. S42. The binding interaction of 3-aminothiophene 7a with (PDB ID: 4d3v). Fig. S43. The binding interaction of 3-aminothiophene 7b with (PDB ID: 4d3v). Fig. S44. The binding interaction of 3-aminothiophene 7c with (PDB ID: 4d3v). Fig. S45. The binding interaction of with ampicillin (PDB ID: 4d3v). Fig. S46. IR spectrum of compound 3a. Fig. S47. 1H NMR spectrum of compound 3a. Fig. S48. 13C NMR spectrum of compound 3a. Fig. S49. IR spectrum of compound 3b. Fig. S50. 1H NMR spectrum of compound 3b. Fig. S51. 13C NMR spectrum of compound 3b. Fig. S52. IR spectrum of compound 3c. Fig. S53. 1H NMR spectrum of compound 3c. Fig. S54. IR spectrum of compound 5a. Fig. S55. 1H NMR spectrum of compound 5a. Fig. S56. 13C NMR spectrum of compound 5a. Fig. S57. IR spectrum of compound 5b. Fig. S58. 1H NMR spectrum of compound 5b. Fig. S59. 13C NMR spectrum of compound 5b. Fig. S60. IR spectrum of compound 5c. Fig. S61. 1H NMR spectrum of compound 5c. Fig. S62. IR spectrum of compound 7a. Fig. S63. 1H NMR spectrum of compound 7a. Fig. S64. 13C NMR spectrum of compound 7a. Fig. S65. IR spectrum of compound 7b. Fig. S66. 1H NMR spectrum of compound 7b. Fig. S67. IR spectrum of compound 7c. Fig. S68. 1H NMR spectrum of compound 7c.
Data Availability Statement
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.