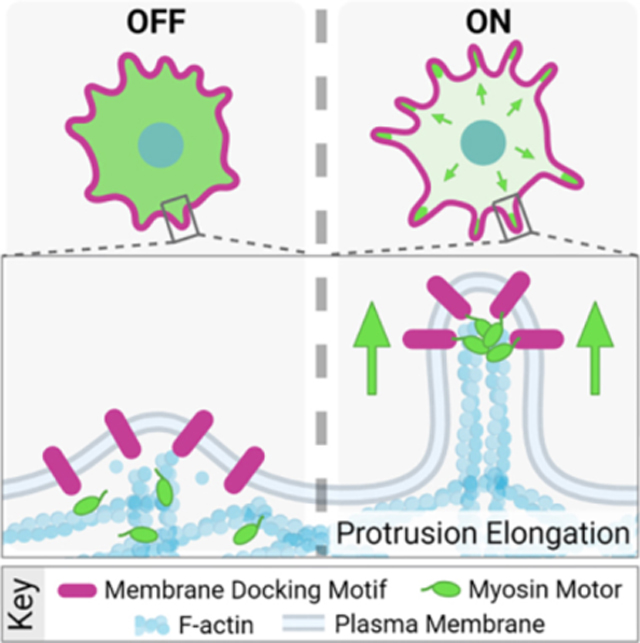
SUMMARY
Actin-based protrusions extend from the surface of all eukaryotic cells, where they support diverse activities essential for life. Models of protrusion growth hypothesize that actin filament assembly exerts force for pushing the plasma membrane outward. However, membrane-associated myosin motors are also abundant in protrusions, though their potential for contributing growth-promoting force remains unexplored. Using an inducible system that docks myosin motor domains to membrane binding modules with temporal control, we found that application of myosin-generated force to the membrane is sufficient for driving robust protrusion elongation in human, mouse, and pig cell culture models. Protrusion growth scaled with motor accumulation, required barbed end-directed force, and was independent of cargo delivery or recruitment of canonical elongation factors. Application of growth-promoting force was also supported by structurally distinct myosin motors and membrane binding modules. Thus, myosin-generated force can drive protrusion growth and this mechanism is likely active in diverse biological contexts.
Keywords: plasma membrane, actin, filopodia, cytoskeleton, microvilli, stereocilia, motor
eTOC blurb
Using an engineered, inducible cell culture model, Fitz et al. show that membrane-bound myosin motors can drive robust protrusion growth. This mechanism requires active, barbed end-directed motor domains, and is supported by structurally distinct myosins and membrane binding motifs, suggesting this elongation mechanism is active in diverse biological contexts.
Graphical Abstract
INTRODUCTION
Actin-based membrane protrusions are ancient cell surface features that have evolved to perform a vast array of biological roles needed for cell function and survival. Microvilli, stereocilia, and filopodia represent a group of structurally related protrusions supported by a core bundle of actin-filaments 1–3. Despite a common morphology, these structures serve distinct physiological functions: microvilli drive nutrient absorption in the gut, stereocilia regulate hearing and balance in the ear, and filopodia contribute to a range of functions including cell migration, cell-cell adhesion, and neurite outgrowth 4. While the function, structure and composition of actin-based protrusions have been studied extensively for decades, the physical mechanisms that control their assembly are still emerging.
A key step in protrusion growth is deformation of the plasma membrane. Previous theoretical and experimental work suggested that positive (i.e. outward) membrane curvature is driven by the mechanical force generated during actin monomer incorporation at the membrane-proximal barbed ends of growing filaments 5–13. Elongation factors such as formin family proteins and enabled/vasodilator-stimulated phosphoprotein (Ena/VASP) have also been implicated in accelerating protrusion elongation, potentially by promoting rapid and processive incorporation of monomers at the membrane interface 5,14,15. However, a recent study found that protrusion length is not tightly coupled to the levels of these canonical elongation factors 16, highlighting the ambiguity of mechanisms that drive growth.
Although actin filament polymerization can drive protrusion growth, the potential for other force generators to contribute to this process has remained unclear. Indeed, myosin superfamily members are some of the most abundant residents of actin-based protrusions 17–19 and may hold the potential to generate significant growth-promoting force. All myosins consist of three structural domains: a conserved N-terminal motor domain that binds actin and hydrolyzes ATP, a central neck that serves as a lever arm to amplify motion produced during the power stroke 20–22, and a class-specific C-terminal tail domain 23. Myosin tail domains mediate cargo binding and association with the plasma membrane (either directly or indirectly). Of all myosin classes, protrusions contain primarily barbed end directed-myosins: microvilli contain myosin-1a, -1d, and -7b 24–26, stereocilia contain myosin-7a, -3a, and -15a 27–29, and filopodia contain myosin-7 30 and myosin-10 31. Several of these motors accumulate in the distal tip compartment, which presumably reflects their robust barbed end-directed movement in these systems. Consistent with this view, previous studies implicate myosins in the delivery of critical regulatory and structural cargoes to the distal tips, suggesting that these motors are important regulators of growth 4. Indeed, loss-of-function studies with tip-targeting myosins generally result in a loss of distal tip components and shorter protrusions in all three systems 28,32,33.
Beyond interacting directly with cargo proteins, the tail domains of myosins also interact with the plasma membrane, either directly or indirectly. Some myosins bind directly to the inner leaflet of the membrane through non-specific electrostatic interaction mediated by their highly basic C-terminal tail 34,35. Others interact with the plasma membrane using motifs that bind to specific membrane lipid head groups 36,37, or through interactions with membrane-associated peripheral or transmembrane proteins 38–40. From this perspective, myosin motors are well-positioned to exert significant force on the membrane, and consequently, impact the kinetics of protrusion growth. However, this hypothesis remains untested due to the technical challenge of uncoupling the contributions of tipward cargo transport from myosin tail membrane interactions.
To address this challenge and determine if membrane-bound myosin-generated force promotes protrusion growth, we developed a synthetic cell-based system that allows for temporally controlled docking of myosin motor domains (lacking cargo binding motifs) to the membrane. Our studies initially leverage the motor domain from the well-studied filopodia resident motor, myosin-10 (Myo10). Myo10 walks towards the barbed ends of polarized actin filament bundles, enriches at the distal tips of filopodia, and is sufficient for the initiation of filopodia formation and subsequent elongation 41. Moreover, the Myo10 tail binds to cargoes 42 that are critical for filopodial function, but also interacts with the plasma membrane using a several distinct mechanisms 37,38. We found that membrane-bound Myo10 motor domains drive robust cell surface protrusion elongation independently of cargo binding and delivery activities. Live-cell and super-resolution imaging studies indicate that while the induced protrusions share many common features with canonical filopodia, they elongate independent of the formin, mammalian diaphanous-related formin 1 (mDia1), and VASP, suggesting myosin-generated force circumnavigates the usual requirement for these factors. Motor domain-driven filopodial elongation was also supported by a variety of membrane binding motifs, although integral membrane motifs produced the most robust response. Elongation activity was not specifically dependent on the mechanical properties of Myo10, as the motor domains from stereocilia motors, myosin-15a and -3a, also promoted filopodia growth. These data reveal that the application of myosin-generated force to the plasma membrane can directly drive protrusion growth.
RESULTS
Membrane-bound Myo10 motor domains drive robust protrusion elongation, independent of cargo binding.
To investigate the role of myosin-generated force in protrusion growth, we used a rapalog inducible system that offers switchable control over the oligomerization of proteins tagged with FRB and FKBP 43,44. These modules were used to engineer constructs that enabled us to rapidly induce interaction between myosin motor domains (tagged with FRB) and plasma membrane binding motifs (tagged with FKBP) upon the addition of rapalog (Fig 1A). We first examined the effect of docking the motor domain from the filopodial motor, Myo10, to the plasma membrane in live HeLa cells. To eliminate cargo binding and specifically probe how the application of force to the membrane affects protrusion growth, we replaced the entire C-terminal tail of Myo10 with FRB (a.a. 1–855, EGFP-Myo10MD-FRB, herein referred to as Myo10MD) (Fig 1B). For a membrane-binding motif, we used the transmembrane domain (TM; a.a. 1155–1310) from cadherin related family member 2 (CDHR2), a single-spanning membrane protein not endogenously found in HeLa cells (CDHR2TM-mCherry-FKBP, herein referred to as CDHR2TM) (Fig 1B). Using live-cell spinning-disc confocal (SDC) microscopy, we visualized cells expressing both Myo10MD and CDHR2TM, imaging every 5 minutes before and after the addition of rapalog (Fig 1C). Upon rapalog treatment, we noticed clear relocalization of the Myo10MD signal from the cytoplasm to the cell periphery, as expected based on the plasma membrane localization of CDHR2TM. Strikingly, cells displayed a robust elongation of finger-like protrusions within 10 minutes of rapalog treatment compared to untreated control cells (Fig 1D, 1E - compare Fig 1Dii to 1Eii–Fig 1F, Video S1, compare top and bottom cell). Additionally, cells expressing the Myo10MD construct alone (in the absence of the membrane binding construct) failed to elongate filopodia in response to rapalog treatment (Fig S1, Video S2). While we were able to directly visualize de novo protrusion growth events in our time-lapse data, and the number of protrusions did trend upward, this increase was not significant (Fig 1G). However, protrusion length increased significantly when Myo10MD was docked to the membrane (Fig 1H, 1I), suggesting that myosin-generated forces contribute primarily to elongating existing structures rather than initiating new ones.
Figure 1: Membrane bound myosin-10 motor domains drive protrusion elongation.
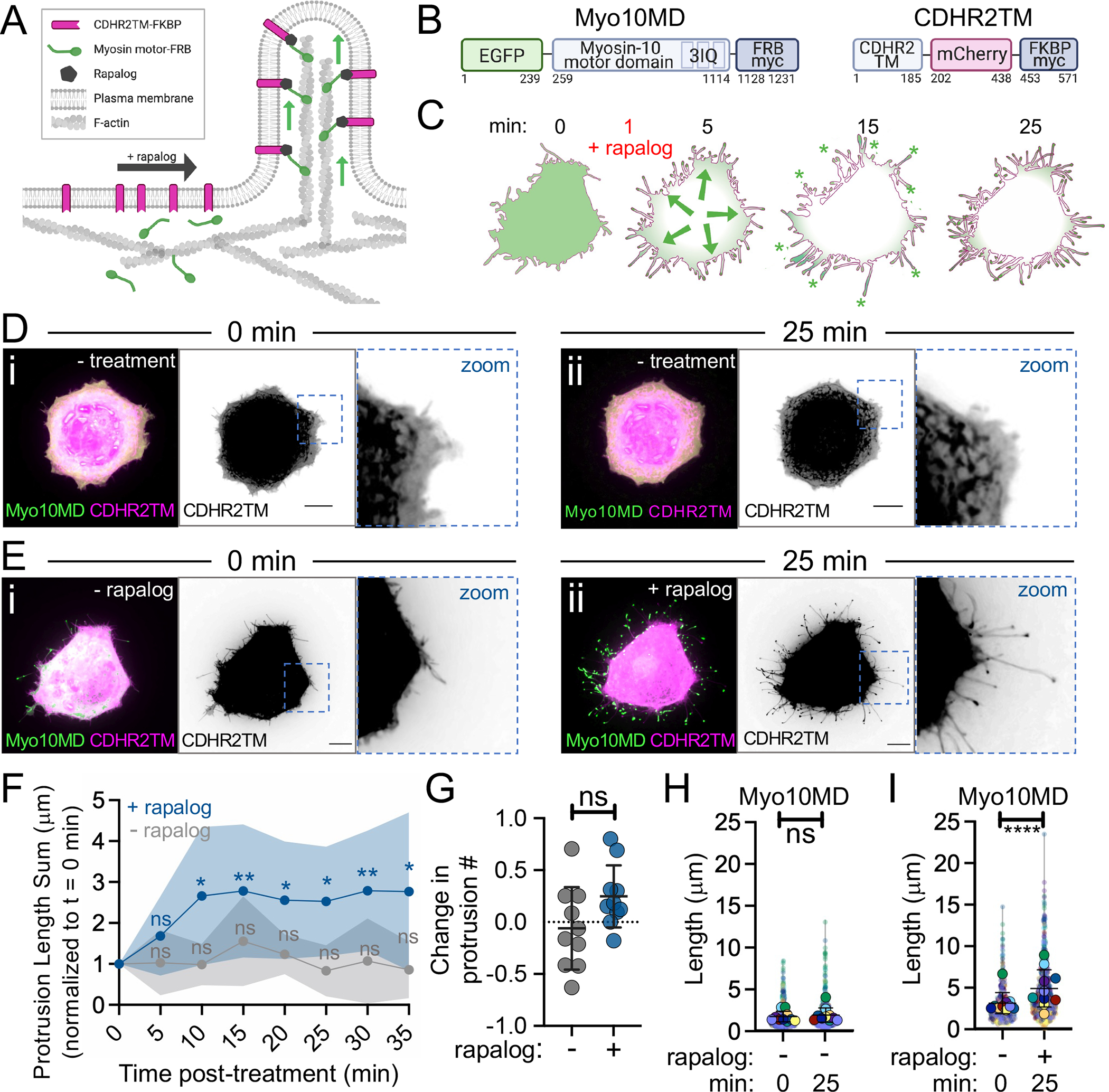
(A) Cartoon of the rapalog inducible system used to dock the myosin motor domains onto the plasma membrane. (B) Cartoons depicting the CDHR2TM and Myo10MD constructs for these experiments; numbers represent amino acids. (C) Cartoon depicting the timeline of rapalog addition and the resultant relocalization of the Myo10MD (green) construct to the CDHR2TM-labeled plasma membrane (magenta). Green asterisks mark Myo10MD enrichment at the tips of protrusions. (D) Confocal maximum intensity projection images of control HeLa cells expressing Myo10MD (green) and CDHR2TM (magenta) at 0 min (i) and 25 min (ii) with no rapalog treatment. (E) Confocal maximum intensity projection of HeLa cells expressing Myo10MD (green) and CDHR2TM (magenta) at 0 min (i) and 25 min after (ii) rapalog treatment. (F) Length sum of protrusions over time in untreated (gray) or rapalog treated (blue) HeLa cells expressing Myo10MD and CDHR2TM; n = 10 individual cells for each treatment. (G) Fractional change in protrusion number from 0 min to 25 min in untreated (gray) or rapalog treated (blue) HeLa cells expressing Myo10MD and CDHR2TM; n = 10 individual cells for each condition. (H) Length of individual protrusions measured at 0 min and 25 min in untreated HeLa cells expressing Myo10MD and CDHR2TM; n = 10 individual cells, n = > 650 individual protrusions. (I) Length of individual protrusions measured at 0 min and 25 min post rapalog treatment in HeLa cells expressing Myo10MD and CDHR2TM; n = 10 individual cells, n = > 550 individual protrusions. Data in (H) and (I) are represented as a SuperPlot, where transparent circles represent the length of individual protrusions, opaque circles represent the average length of protrusions of individual cells, violin plots show the distribution of the data, and all color matched circles represent measurements from the same cell. All graph error bars represent the mean ± SD; ns, p-value > 0.05, * p-value ≤ 0.05, **** p-value ≤ 0.0001. Scale bars = 10 μm.
To determine if protrusion growth induced by membrane-bound Myo10MD was unique to HeLa cells, we co-expressed our system components in B16F1 Mus musculus melanoma cells, which also form fascin-bundled filopodia, and LLC-PK1-CL4 (CL4) Sus scrofa kidney cells, which form microvilli on their apical surface. Induction of Myo10MD membrane binding in both cell lines drove robust protrusion elongation. Thus, the application of myosin-generated force to the plasma membrane is sufficient for driving protrusion growth in distinct cell types derived from a range of tissues (Fig S1).
Protrusions induced by membrane-bound Myo10MD exhibit features of filopodia.
We next sought to determine if the protrusions induced by membrane-bound Myo10MD represent canonical filopodia. To this end, HeLa cells were fixed 25 minutes after the addition of rapalog, and then stained for F-actin (phalloidin)45 and the filopodia-specific parallel filament bundler, fascin 46. Super-resolution structured illumination microscopy (SIM) of these samples revealed that protrusions induced by membrane-bound Myo10MD contained both F-actin and fascin (Fig 2A, 2B, S2). We also analyzed SDC time-lapse data and determined that individual protrusions induced by membrane bound Myo10MD elongated at a similar rate to canonical filopodia. Individual protrusions induced by membrane-bound Myo10MD elongated at 2.16 μm/min, a rate similar to that reported for filopodia in previous studies (~2.2 μm/min)47(Fig 2C, 2Ci zoom, 2D). Based on current models of filopodial growth, we also expected that protrusion growth induced by membrane-bound Myo10MD would require actin polymerization. Indeed, HeLa cells treated with the actin poisons cytochalasin D (caps filament barbed ends) and latrunculin A (sequesters actin monomers), exhibited significantly reduced protrusion elongation in response to rapalog induction (Fig S2C, S2D). Thus, protrusions induced by membrane-bound Myo10MD are similar to canonical filopodia in that they are supported by parallel, fascin-bundled actin filaments, and require both uncapped barbed ends and a pool of free actin monomers to elongate.
Figure 2: Protrusions induced by membrane bound Myo10MD exhibit features of filopodia.
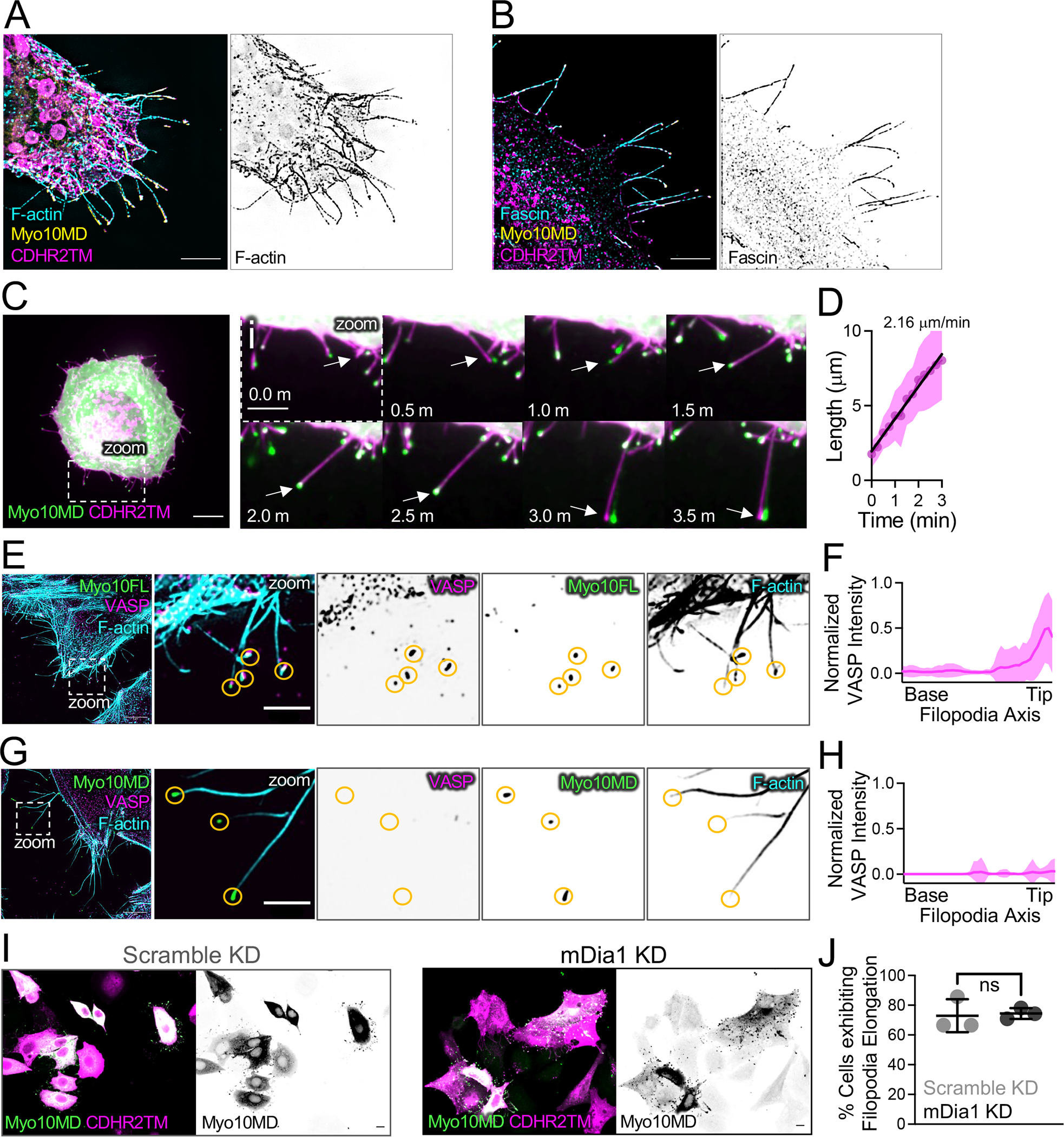
(A) SIM maximum intensity projection image of a HeLa cell expressing Myo10MD (yellow) and CDHR2TM (magenta) and stained for F-actin with phalloidin (cyan). Panel to the right shows inverted single channel image of F-actin (phalloidin). Scale bar = 5 μm. (B) SIM maximum intensity projection image of HeLa cells expressing Myo10MD (yellow) and CDHR2TM (magenta) and stained for endogenous fascin (cyan). Panel to the right shows inverted single channel image of fascin. Scale bar = 5 μm. (C) Confocal maximum intensity projection of HeLa cell expressing Myo10MD (green) and CDHR2TM (magenta) 2 min after rapalog addition. Scale bar = 10 μm. (Ci) Montage of zoomed in region (C) highlighting the growth of a single filopodium (white arrow). (D) Elongation rate of single filopodium induced by rapalog treatment. Rate was calculated via the slope using a simple linear regression; n = 14 individual filopodia from n = 3 separate cells. (E) SIM maximum intensity projection image of a HeLa cell expressing full-length Myosin-10 (green) and stained for endogenous VASP (magenta) and F-actin with phalloidin (cyan). Zoom images to the right show merge and inverted single channel images. Yellow circles denote the tips of individual filopodia. (F) Line scans parallel to the filopodial axis show the intensity distribution of VASP in filopodia generated from over-expressing EGFP-myosin-10 full length; n = 30 protrusions, line represents the average intensity. (G) SIM maximum intensity projection image of a HeLa cell expressing Myo10MD (green) and CDHR2TM (not shown) 25 min after rapalog treatment and stained for endogenous VASP (magenta) and F-actin (cyan). Zoom images to the right show merge and inverted single channel images. Yellow circles denote the tips of individual filopodia. For E and G, scale bar = 5 μm, zoom scale bar = 2 μm. (H) Line scans parallel to the filopodial axis show the intensity distribution of VASP in filopodia; n = 39 filopodia, line represents the average intensity. (I) Confocal maximum intensity projection of HeLa cells transfected with either non-targeting siRNA (scramble KD) or siRNA targeting mDia1 (mDia1 KD) and expressing Myo10MD (green) and CDHR2TM (magenta), imaged 25 min after rapalog treatment. Scale bars = 10 μm. (J) Percent of cells displaying filopodial elongation with or without KD of mDia1; n = 3 replicates with 87–93% knockdown efficiency. Double transfected cells/condition scramble KD (n = 184) and mDia1 KD (n = 63). Ns, p-value > 0.05 calculated using the unpaired Student’s t-test; all graph error bars represent the mean ± SD.
Membrane-bound Myo10MD promotes filopodial growth independent of barbed end elongation factors.
Previous studies identified barbed end elongation factors as proteins that enrich at the distal ends of filopodia and in turn promote their growth. Ena/VASP and formin family proteins are well-studied examples 14,15 and we sought to determine if membrane-bound Myo10MD promoted filopodial growth by recruiting these factors. We first examined HeLa cells for the enrichment of Ena/VASP at the tips of induced filopodia. Ena/VASP is a tetrameric protein that promotes actin monomer incorporation into filaments and protects barbed ends from capping protein, which would otherwise stall core bundle elongation. VASP binds to full-length Myo10 and may be transported to the distal ends of filopodia via this interaction 42, although the possibility of such direct transport is eliminated in our experimental system as Myo10MD constructs lack the C-terminal tail. Using SIM, we visualized cells transfected with either full-length Myo10 or the combination of Myo10MD and CDHR2TM that drives filopodial induction. As expected, filopodia generated by over-expressing full length Myo10 displayed prominent VASP enrichment at their distal tips (Fig 2E, 2F). Remarkably, VASP was undetectable at the tips of filopodia induced by membrane-bound Myo10MD (Fig 2G, 2H). We also asked if formin family proteins were involved in the rapalog-induced response. Based on RNAseq data 48, mDia1 is the most highly expressed formin in HeLa cells. Moreover, previous studies implicate mDia1 as a processive elongator of actin filaments in filopodial core bundles 14. To examine potential involvement of mDia1, we knocked down its expression in HeLa cells using siRNA (Fig S2E), transfected cells with Myo10MD and CDHR2TM, and induced filopodial growth with rapalog treatment. Interestingly, we observed no change in the fraction of cells that were able to elongate filopodia in mDia1 KD cells relative to controls, and no change in filopodial length (Fig 2I, 2J, S2F). Although these results do not rule out the possibility that Myo10MD cooperates with some actin regulatory machinery to drive filopodial elongation, this activity appears to be independent of the canonical barbed end elongation factors VASP and mDia1.
Filopodial elongation temporally correlates with Myo10MD accumulation at the distal tips.
We next sought to determine how membrane-bound Myo10MD elongates filopodia in the absence of barbed end elongation factors. One possibility is that mechanical force applied by Myo10MD directly to the plasma membrane promotes growth by reducing the physical barrier to protrusion elongation. A testable prediction that emerges from this hypothesis is that active, barbed end-directed force generation by membrane-bound Myo10MD is required to induce filopodial growth in response to rapalog treatment. To test this, we first sought to determine if higher levels of Myo10MD, and therefore, higher levels of force, might drive faster elongation rates and/or longer protrusions. While the absolute amount of force generated by Myo10MD in cells is experimentally inaccessible, we reasoned that we could obtain a relative measure of force by monitoring the EGFP intensity from the Myo10MD construct; this signal is proportional to the number of Myo10MD motors and thus local force-generating potential. Using live-cell SDC imaging, we monitored Myo10MD tip intensity during individual filopodial growth events after inducing HeLa cells with rapalog. Because filopodia generated in these experiments are highly dynamic and poorly anchored to the coverslip substrate, culture media was supplemented with 0.5% methylcellulose; this prevented lateral waving of protrusions and improved our ability to monitor the details of elongation with high temporal resolution. Importantly, the addition of methylcellulose did not alter the growth rates of filopodia induced by membrane-bound Myo10MD (Fig S3). Using this approach, we measured the distal tip intensity of Myo10MD in parallel with filopodial length from time-lapse data collected at 2 sec intervals (Fig 3A, Video S3). This analysis revealed a striking and direct temporal correlation between the length of a growing filopodium and the accumulation of Myo10MD at its distal tip (Fig 3B). Thus, during growth, filopodial length is well correlated to the force-generating potential of tip-enriched myosin motors.
Figure 3: Barbed end-directed force is required for filopodial induction by Myo10MD.
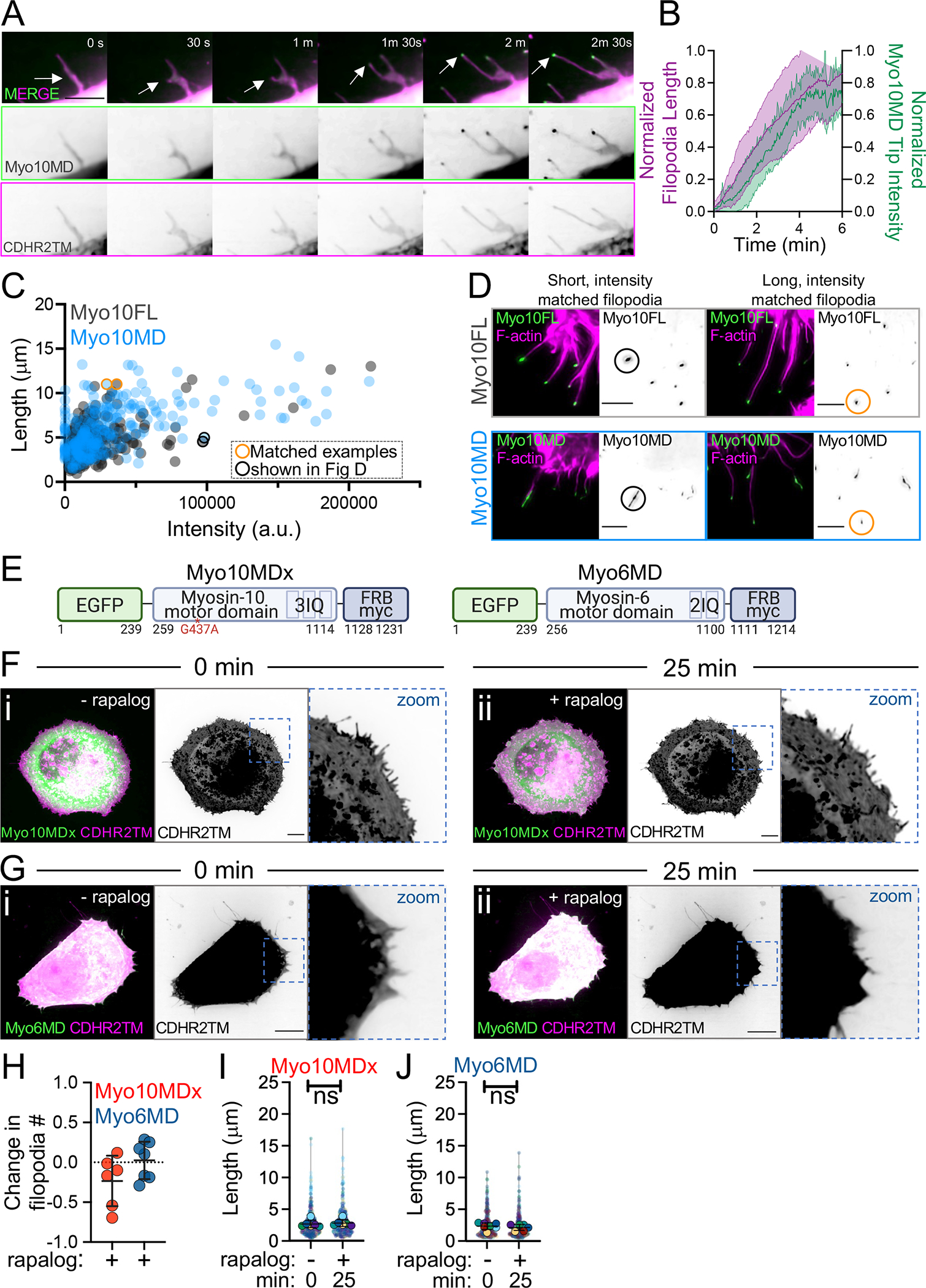
(A) Montage of a filopodial growth event from a HeLa cell expressing Myo10MD (green) and CDHR2TM (magenta) imaged in 0.5% methylcellulose. Arrows denote the tip of a single, growing filopodium. Scale bar = 5 μm. (B) Normalized length (magenta) and tip intensity (green) vs. time curves for individual filopodia on cells expressing Myo10MD and CDHR2TM; n = 10 filopodia. (C) Filopodial length and myosin distal tip-intensity from individual protrusions in cells expressing Myo10FL (gray) or Myo10MD (blue); each circle represents an individual filopodium. Orange and black circled data points represent filopodia shown in D; n = 304 filopodia for Myo10FL and 273 for Myo10MD. (D) Confocal maximum intensity projections of length and tip intensity-matched filopodia in cells expressing Myo10FL or Myo10MD (green) and stained for F-actin with phalloidin (magenta); single inverted channel images are shown to the right. Scale bar = 5 μm. (E) Cartoons depicting the myosin-10 motor domain dead G437A mutant (Myo10MDx) and pointed end-directed myosin-6 motor domain (Myo6MD) constructs; numbers below represent amino acids. (F) Confocal maximum intensity projection of HeLa cells expressing Myo10MDx (green) and CDHR2TM (magenta) before (i) and 25 min after (ii) rapalog treatment. (G) Confocal maximum intensity projection of HeLa cells expressing Myo6MD (green) and CDHR2TM (magenta) before (i) and 25 min after (ii) rapalog treatment. For F and G, scale bar = 10 μm. (H) Fractional change in filopodia number in HeLa cells expressing CDHR2TM and Myo10MDx (red) or Myo6MD (blue) 0 min and 25 min after rapalog treatment; n = 6–7 individual cells for each condition. (I) Filopodial length measured at 0 min and 25 min post rapalog treatment in HeLa cells expressing Myo10MDx and CDHR2TM; n = 6 individual cells, n = > 420 individual protrusions. (J) Filopodial length measured at 0 min and 25 min post rapalog treatment in HeLa cells expressing Myo6MD and CDHR2TM; n = 7 individual cells, n = > 365 individual protrusions. Data in (I) and (J) are represented as a SuperPlot, where transparent circles represent the length of individual filopodia, opaque circles represent the average length of filopodia from individual cells, violin plots show the distribution of the data, and all color-matched circles represent measurements from the same cell. All graph error bars represent the mean ± SD; ns, p-value > 0.05.
To place these findings into context and determine how the distal tip enrichment of Myo10MD compares to wild type full-length Myo10 (Myo10FL), we repeated these experiments in HeLa cells expressing EGFP-Myo10FL and scored tip intensities and lengths from filopodia. This analysis revealed that the tip intensity vs. length distributions obtained for Myo10MD and Myo10FL largely overlapped (Fig 3C). Moreover, filopodia generated by both constructs had similar morphology when matched in length and intensity (Fig 3D - circled data points in 3C). Together these data indicate that, during filopodial elongation, the tip enrichment and thus force-generating potential of membrane-bound Myo10MD is comparable to Myo10FL.
Active, barbed end-directed force is required for filopodial induction.
The observation that filopodial length increases in parallel with distal tip accumulation of membrane-bound Myo10MD is consistent with the proposal that force generated by this motor directly promotes protrusion growth. From this perspective, we next examined whether the mechanical activity of Myo10MD is required for filopodia elongation. We generated two additional Myo10MD constructs containing mutations in switch I (R220A) or switch II (G437A) that are expected to disrupt force generation 49. Docking the motor-dead G437A construct (EGFP-Myo10MDx-FRB, herein referred to as Myo10MDx) (Fig 3E) to the plasma membrane with rapalog treatment failed to elongate filopodia (Fig 3Fi–ii, 3H, 3I, Video S4). A motor-dead R220A Myo10MD mutant also failed to accumulate in filopodia, most likely because of its impaired actin binding (Fig S3). Based on these data, we conclude that active, force-generating motors are needed to drive filopodial elongation in response to rapalog treatment.
To determine if the induction of filopodial elongation specifically requires the application of barbed end-directed force to the plasma membrane, we generated a construct using the motor domain from myosin-6, the sole pointed end-directed member of the myosin superfamily (a.a. 1–844, EGFP-Myo6MD-FRB, herein referred to as Myo6MD) (Fig 3E). Membrane-bound Myo6MD also failed to promote filopodial elongation in response to rapalog treatment (3Gi–ii, 3H, 3J, Video S5). Taken together, these data indicate that the application of barbed end-directed force by membrane-bound motors is needed for robust filopodia elongation.
Filopodial growth induced by membrane-bound Myo10MD is limited by the availability of actin monomers and plasma membrane.
Why the robust filopodial growth induced by membrane-bound Myo10MD stalls at long lengths remains unclear. One possibility is that protrusions become so long that the pool of free actin monomers available for incorporation at the distal barbed ends falls below the level needed to continue elongation. To test this possibility, HeLa cells were subjected to long-term G-actin sequestration with a low concentration of Latrunculin A (Lat A). Based on the reversible nature of Lat A treatment 50, our rationale was that washing out Lat A post-rapalog treatment would rapidly and acutely increase the pool of free actin monomers; any “reactivation” of filopodial growth at that point would indicate that the availability of free G-actin normally limits elongation. For these experiments, HeLa cells were incubated in 100 nM Lat A for 7 days, to allow for long-term adaption prior to transfection with Myo10MD and CDHR2TM (Fig 4A). When we examined how individual cells responded to Lat A washout, we noted a significant increase in filopodia length sum following rapalog treatment (Fig 4B–D, compare blue and gray lines at 75 min, Video S6, S7). These results suggest that the actin monomer pool does become limiting, and that the sustained elongation of protrusions induced by membrane-bound Myo10MD may be at least partially dependent on available monomers.
Figure 4: Myo10MD induced filopodial growth depends on membrane and G-actin availability.
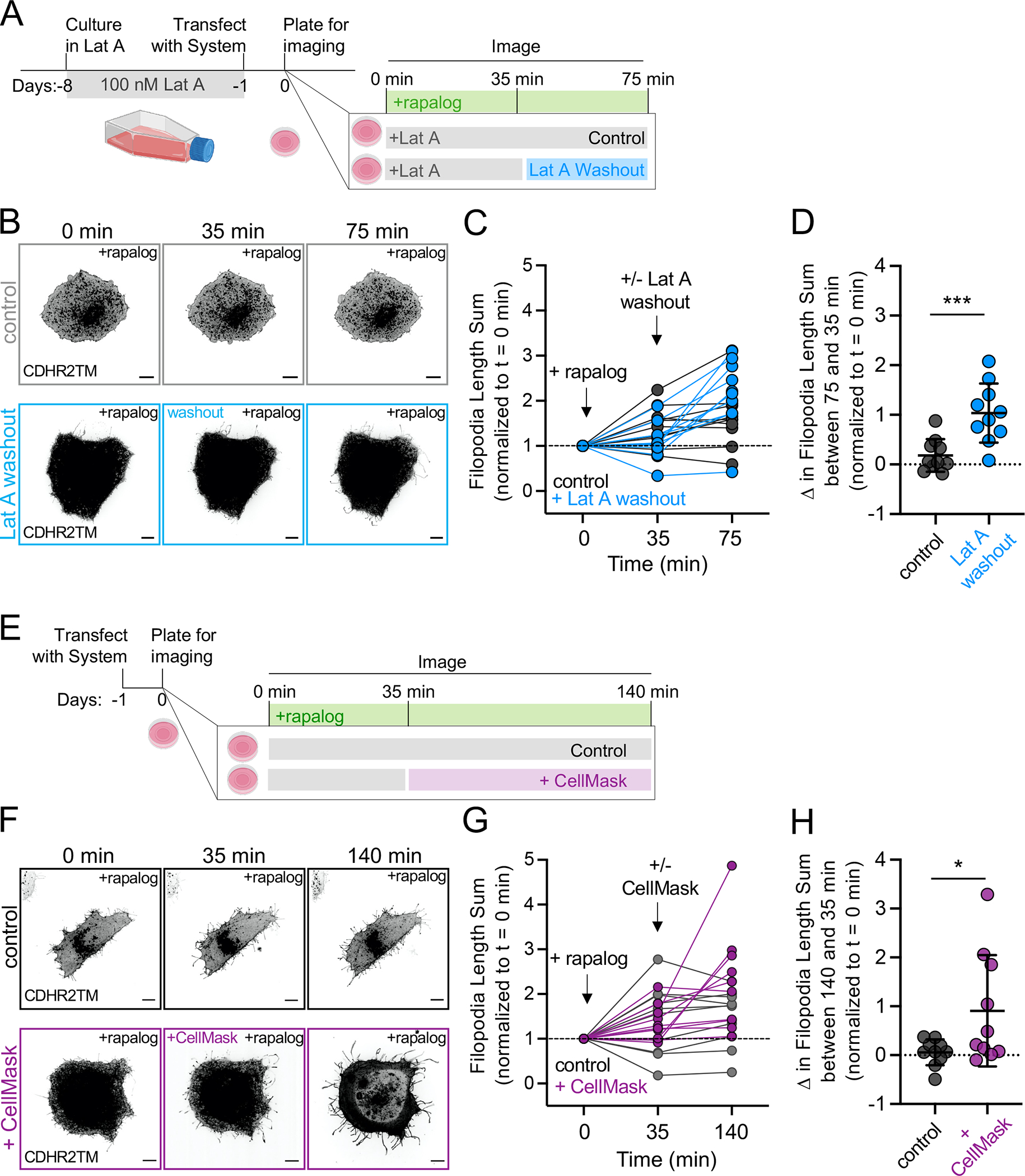
(A) Cartoon of the Latrunculin A (Lat A) washout experimental timeline. (B) Confocal maximum intensity projection of representative HeLa cells expressing Myo10MD (not shown) and CDHR2TM (gray scale), treated with 100 nM Lat A for one week (top; control) or Lat A for one week followed by washout (bottom). (C) Normalized length sum of filopodia generated over time in cells treated with Lat A (gray) or treated with Lat A followed by washout (blue). (D) Change in filopodial length sum between the 75 and 35 min time points for cells treated with Lat A (gray) or treated with Lat A followed by washout (blue). (E) Cartoon of membrane expansion experimental timeline. (F) Confocal maximum intensity projection of representative HeLa cells expressing Myo10MD (not shown) and CDHR2TM (gray scale). Untreated control cell (top) and cell treated with membrane intercalating CellMask for 35 min (bottom). (G) Normalized length sum of filopodia generated over time in control HeLa cells (gray) or cells treated with CellMask (magenta). (H) Change in filopodial length sum between the 140 and 35 min time points for cells treated without (gray) or treated with CellMask (magenta). All graph error bars represent the mean ± SD; * p-value ≤ 0.05, **** p-value ≤ 0.0001. P-values were calculated using unpaired Student’s t-tests comparing treatments at the indicated time point, n = 10 individual cells for each of the four treatments. Scale bars = 10 μm.
Filopodial growth after rapalog induction might also be limited by the availability of encapsulating plasma membrane lipids. To test this idea, we added the fluorescent bilayer intercalating compound CellMask-Orange (herein referred to as CellMask) to cells 35 min after filopodial elongation was initiated with rapalog treatment (Fig 4E). Based on biophysical studies, such compounds are expected to increase membrane surface area while reducing membrane tension 51–53. Interestingly, CellMask addition induced a significant second stage of elongation beyond the initial response driven by membrane-bound Myo10MD (Fig 4F and G, compare magenta and gray lines at 140 min, Videos S8, S9), which resulted in a significant increase in filopodia length sum (Fig 4H). Thus, in addition to being limited by the availability of actin monomers, sustained filopodial growth driven by membrane-bound Myo10MD may also be limited by the availability of encapsulating plasma membrane.
Myosin motor domains from structurally diverse superfamily classes also support rapalog-induced filopodial growth.
Myosin superfamily members exhibit a range of mechanochemical properties, which enable these motors to perform specific subcellular tasks that hold unique kinetic requirements (e.g., cargo anchoring vs. transport). Therefore, we next sought to determine if the robust filopodial elongation driven by Myo10MD is unique to this motor, or could instead be supported by other myosins typically found in other actin-based protrusions, such as microvilli and stereocilia. To test this, we generated additional constructs based on protrusion-resident barbed end-directed myosins that exhibit a range of mechanochemical properties including myosin-1a, myosin-5b (lever arm with 6 IQ motifs (-wildtype) or 3 IQ motifs (-5b3IQ)), myosin-7b, myosin-3a, and myosin-15a. By surveying fixed and stained samples after induction, we noted that the motor domains of myosin-1a, -5b-wildtype, -5b3IQ, and -7b were unable to elongate filopodia when docked onto the plasma membrane (Fig S4). In contrast, motor domains from the stereocilia tip-targeting motors, myosin-3a (a.a. 407–1432, EGFP-Myo3aΔKMD-FRB, herein referred to as Myo3aΔKMD; Fig 5A) and myosin-15a (a.a. 1–750, EGFP-Myo15aMD-FRB, herein referred to as Myo15aMD; Fig 5F) drove significant elongation following rapalog induction (Fig 5Bi, 5Bii, 5Gi, 5Gii, Video S10). Quantification of the length sum of filopodia over time showed that there was a significant increase generated by both motors after 20 and 10 minutes of rapalog treatment, respectively (Fig 5C, 5H). Like the Myo10MD construct, docking either the Myo3aΔKMD or Myo15aMD constructs to the membrane did not significantly change the number of filopodia generated post-rapalog treatment (Fig 5D, 5I), but did increase filopodial length (Fig 5E, 5J). These results indicate that the robust filopodial elongation observed following rapalog induction is not unique to Myo10MD and can also driven by other structurally diverse myosins that reside in distinct protrusion environments.
Figure 5: Motor domains from Myo3a and Myo15a also promote filopodia growth.
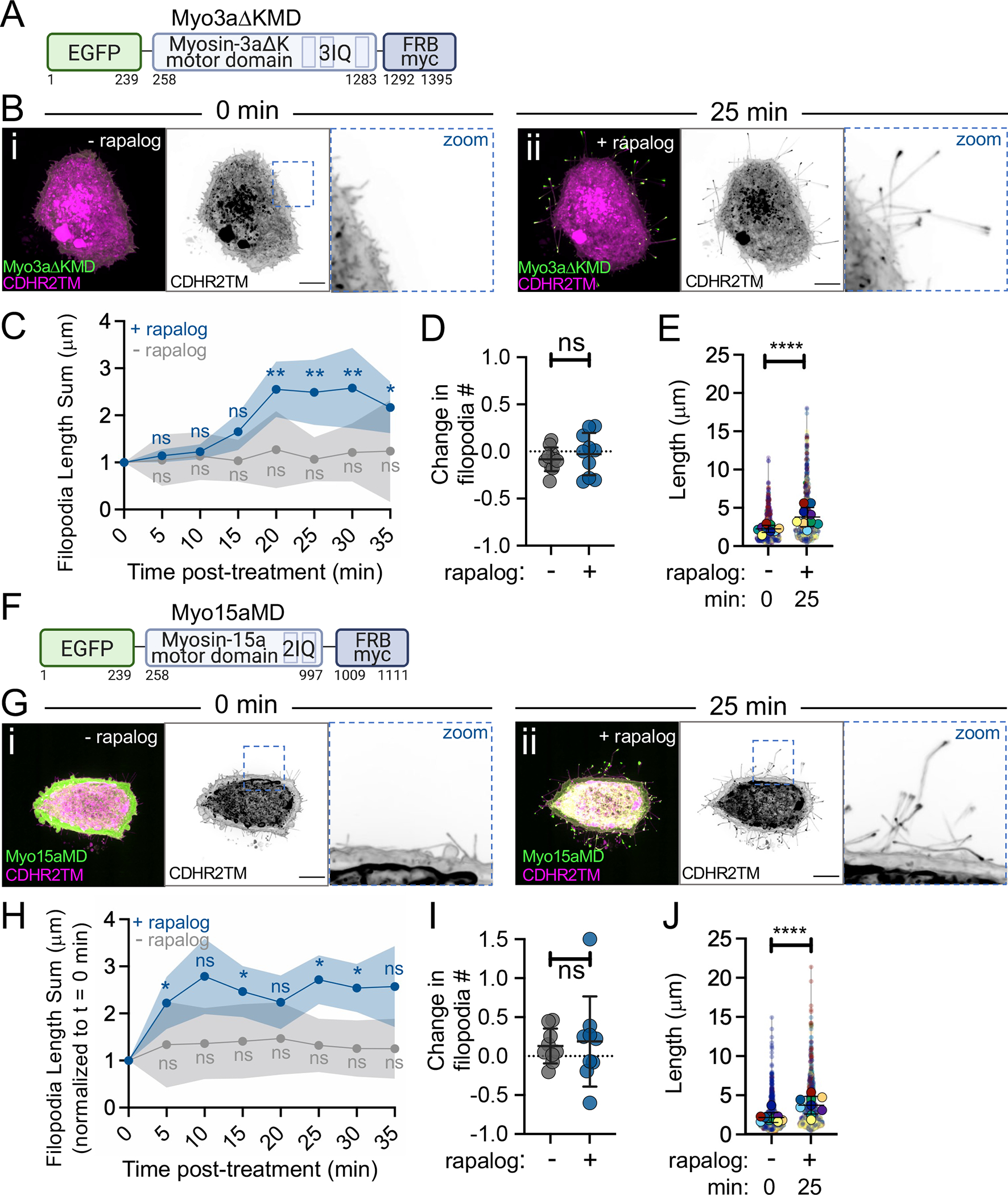
(A) Cartoon depicting the EGFP-Myo3aΔK motor domain-FRB-myc construct (Myo3aΔKMD); numbers below represent amino acids. (B) Confocal maximum intensity projection image of control HeLa cells expressing the Myo3aΔKMD (green) and CDHR2TM (magenta) at 0 min (i) and 25 min after (ii) rapalog treatment. (C) Length sum of filopodia over time in untreated (gray line) or rapalog-treated (blue line) HeLa cells expressing Myo3aΔKMD and CDHR2TM; n = 14–16 cells per treatment. (D) Fractional change in filopodia number from 0 min to 25 min in untreated (gray) or rapalog treated (blue) HeLa cells expressing Myo3aΔKMD and CDHR2TM; n = 10 cells per condition. (E) Filopodia length at 0 and 25 min post rapalog treatment in HeLa cells expressing Myo3aΔKMD and CDHR2TM; n = 9 cells, n = > 670 protrusions. (F) Cartoon depicting the EGFP-Myosin-15a motor domain-FRB-myc construct (Myo15aMD); numbers below represent amino acids. (G) Confocal maximum intensity projection images of control HeLa cells expressing Myo15aMD (green) and CDHR2TM (magenta) before (i) and 25 min after (ii) rapalog treatment. (H) Length sum of filopodia over time in untreated (gray line) or rapalog treated (blue line) HeLa cells expressing Myo15aMD and CDHR2TM; n = 13–16 individual cells per treatment. (I) Fractional change in filopodial number from 0 to 25 min in untreated (gray) or rapalog treated (blue) HeLa cells expressing Myo15aMD and CDHR2TM; n = 9 cells per condition. (J) Filopodia length measured at 0 and 25 min post rapalog treatment in HeLa cells expressing Myo15aMD and CDHR2TM; n = 9 individual cells, n = > 730 protrusions. Data in (E) and (J) are represented as a SuperPlot, where transparent circles represent the length of individual filopodia, opaque circles represent the average length of protrusions of individual cells, violin plots show the distribution of the data, and all color matched circles represent measurements from the same cell. All graph error bars represent the mean ± SD; ns p-value > 0.05, * p-value ≤ 0.05, ** p-value ≤ 0.01, **** p-value ≤ 0.0001. Scale bars = 10 μm.
Myo10MD driven filopodial elongation is supported by diverse membrane binding motifs
Our data show that docking the Myo10MD on the plasma membrane using the single-spanning CDHR2TM leads to robust filopodial elongation. In addition to binding transmembrane proteins, some myosin tail domains interact directly with the plasma membrane. For example, the second PH domain in the myosin-10 tail interacts with the phosphoinositol species, PI(3,4,5)P3, which is found in the plasma membrane along the filopodia shaft, and the TH1 domain of myosin-1a interacts with the acidic phospholipids found in the inner leaflet of the plasma membrane. To determine if these peripheral modes of membrane binding support filopodial elongation, we generated two additional membrane docking constructs by fusing FKBP to: (1) the PH domain of Bruton’s tyrosine kinase (BTK), which binds PI(3,4,5)P3 54,55(BTK-PH-mCherry-FKBP, herein referred to as BTK-PH, Fig 6D), and (2) the TH1 domain of myosin-1a, which binds electrostatically to the inner leaflet of the plasma membrane 35(FKBP-mCherry-TH1, herein referred to as TH1; Fig 6G). Both constructs showed membrane enrichment as expected (Fig 6B, 6E, 6H, pre-bleach). Fluorescence recovery after photobleaching (FRAP) quantification of the turnover kinetics in each case also revealed distinct immobile fractions (I.F.) for each construct (Fig 6C, 6F, 6I). As predicted, the integral, transmembrane CDHR2TM construct (Fig 6A) had the largest I.F. compared to the two peripheral membrane-interacting constructs (compare 6C to 6F and 6I). To determine if these different membrane binding motifs could support filopodial elongation, we turned to a fixed-cell approach, where HeLa cells were transfected with either of the three membrane constructs and the Myo10MD construct and induced to produce filopodia with the addition of rapalog. This analysis showed that all three modes of membrane interactions were able to support filopodia elongation (Fig 6J, 6K, 6L), with the CDHR2TM producing the most robust response (Fig 6M). These results further suggest that the ability of myosin motor domains to drive filopodia elongation is independent of the structural details of membrane association.
Figure 6: Myo10MD driven filopodial elongation is supported by diverse membrane binding motifs.
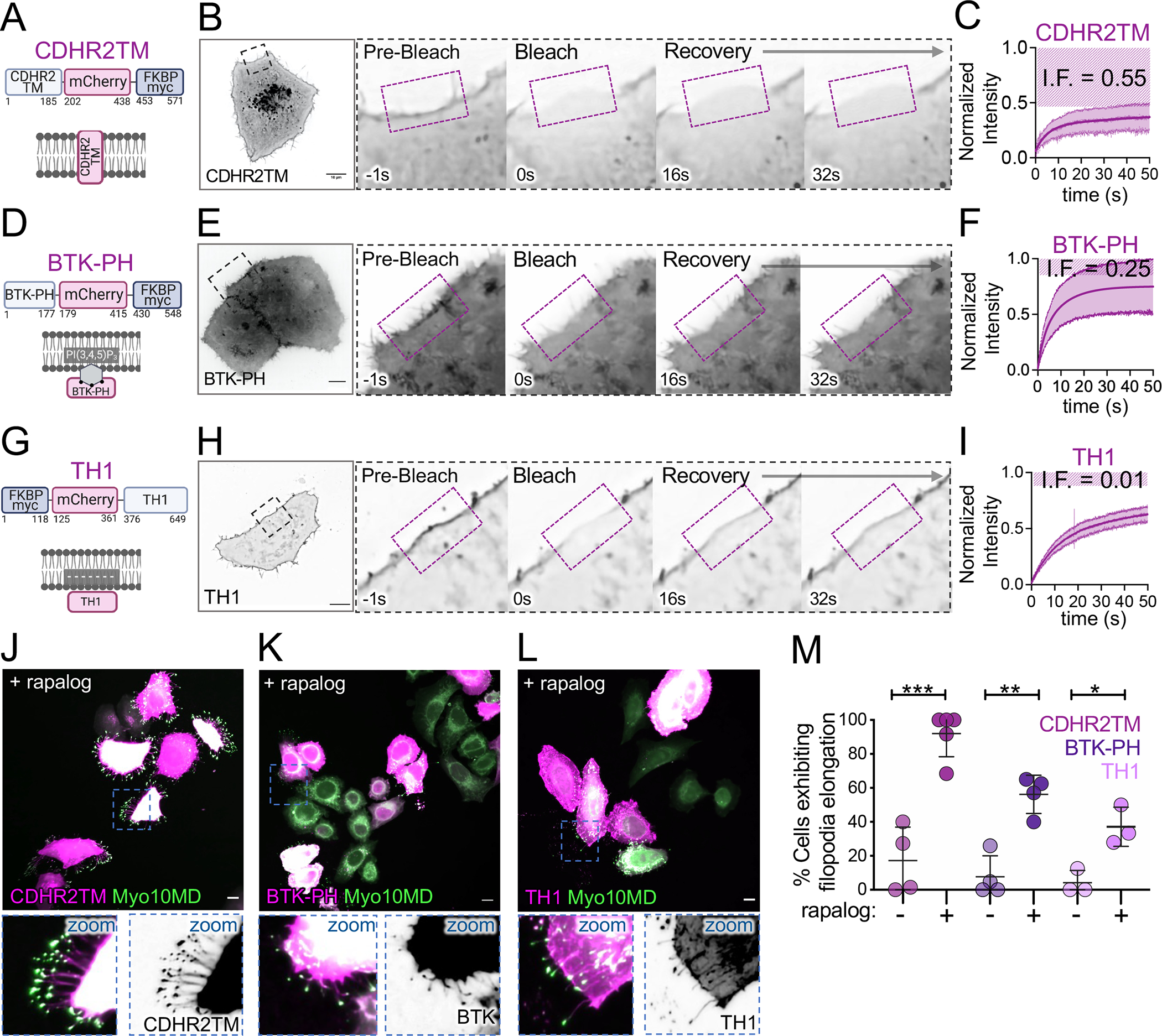
(A) Cartoon depicting the integral transmembrane construct CDHR2TM-mCherry-FKBP construct. (B) Confocal maximum intensity projection timelapse montage showing fluorescence recovery after photobleaching (FRAP) analysis of a HeLa cell expressing CDHR2TM. (C) FRAP recovery curve; measurements were taken from ROI shown in B; n = 6 cells; magenta striped region indicates the immobile fraction. (D) Cartoon showing organization of the peripheral membrane binding construct BTK-PH-mCherry-FKBP (BTK-PH). (E) Confocal maximum intensity projection timelapse montage showing FRAP analysis of a HeLa cell expressing BTK-PH. (F) FRAP recovery curve; measurements were taken from the ROI shown in E; n = 9 cells; magenta striped region indicates the immobile fraction. (G) Cartoon depicting the peripheral membrane binding construct FKBP-mCherry-TH1 (TH1). (H) Confocal maximum intensity projection timelapse montage showing FRAP analysis of a HeLa cell expressing TH1. (I) FRAP recovery curve; measurements were taken from the ROI shown in H; n = 8 cells; magenta striped region indicates the immobile fraction. (J) Confocal maximum intensity projection image of HeLa cells expressing Myo10MD (green) and CDHR2TM (magenta) 25 min after rapalog treatment. (K) Confocal maximum intensity projection image of HeLa cells expressing Myo10MD (green) and BTK-PH (magenta) 25 min after rapalog treatment. (L) Confocal maximum intensity projection image of HeLa cells expressing Myo10MD (green) and TH1 (magenta) 25 min after rapalog treatment. (M) Percentage of untreated or rapalog-treated HeLa cells expressing Myo10MD and CDHR2TM, BTK-PH, or TH1 exhibiting filopodia elongation; n > 170 cells for CDHR2TM, n >140 cells for BTK-PH, and n > 220 cells for TH1. All graph error bars represent the mean ± SD; * p-value ≤ 0.05, ** p-value ≤ 0.01, *** p-value ≤ 0.001. Scale bars = 10 μm.
DISCUSSION
Cell surface protrusions are essential for mediating physical and biochemical interactions with the external environment. Throughout evolution, μm-scale finger-like protrusions have been adapted to fill a range of functional niches, including mechanosensation and solute transport 56. Fundamental to the formation of a surface protrusion is the outward deformation of the fluid plasma membrane. Estimates of the forces a cell must apply to form a protrusion come from optical trap-based measurements of the “tether force”: the force required to pull and hold a thin tubule of membrane at a steady-state length 57,58. Although tether forces are measured by externally imposed mechanical perturbations, they offer an approximation of the physical challenge that the cytoskeletal machinery faces during protrusion formation and suggest that pN-scale pushing force is needed. A long history of previous theoretical and experimental studies have argued that actin polymerization generates this force during protrusion growth 12,59,60. Indeed, the incorporation of new actin subunits into the growing barbed ends of actin filaments generates force in the required pN-scale range 5. Because protrusions are supported by 10s to 100s of parallel filament bundles, these structures should be able to generate outward pushing force that exceeds the threshold suggested by tether force estimates. However, the ability of a growing actin filament to generate force also depends on the availability of free actin monomers 10. This poses a mechanistic conundrum given the tight confines of the intrafilopodial cytoplasm, where the diffusive availability of new monomers at growing barbed ends could become limiting as a protrusion elongates. From this perspective, we set out to test the hypothesis that myosin motors, which are abundant residents of varying actin-based protrusions, provide an additional source of mechanical force for driving protrusion growth.
Based on their mechanical potential and localization at the interface between polymerizing actin filaments and cellular membranes, myosins are well-positioned to drive membrane deformation in diverse biological contexts. Driving the growth of surface membrane protrusions is a long-standing functional hypothesis proposed for several unconventional members of the myosin superfamily, including motors from classes 1, 3, 5, 7, 10, and 15 4. Previous cell biological studies have emphasized cargo transport models, which propose that protrusion myosins deliver specific factors to the distal tip compartment, and these in turn promote growth via a range of molecular mechanisms. We now know that myosins in microvilli, filopodia, and stereocilia are critical for the distal tip enrichment of scaffolding proteins 40,61–63, adhesion molecules 33,39, and proteins that directly impact actin dynamics and organization 42,64–66. However, understanding a given myosin’s contributions to protrusion growth, in terms of tip-directed cargo delivery vs. more direct mechanical effects, has proved challenging largely because an experimental approach for isolating the impact of motor domain force generation in cells has remained elusive.
In this report, we describe an engineered cell-based system that enabled us to directly test whether myosin generated force contributes to protrusion elongation. Using Myo10 as a model myosin, we found that—independent of any cargo binding potential—Myo10 motor domains drive robust filopodial growth when docked onto the plasma membrane using the rapalog-based system described here. Leveraging this approach to develop further insight into the properties of induced filopodial growth, we learned that elongation requires active motor domains that exert barbed end-directed force. Moreover, the elongation of individual protrusions was paralleled by an accumulation of Myo10 motor domains at filopodial tips, strongly suggesting that the local level of force application to the membrane controls filopodial growth kinetics. Importantly, induced filopodial elongation was supported by structurally distinct myosin motors, and membrane binding modules that associate with the plasma membrane using different mechanisms (transmembrane vs. peripheral). We, therefore, propose that the motor-driven filopodial growth revealed in our studies likely reflects a general activity of different unconventional myosins in diverse actin-based protrusions.
Why does docking myosin motor domains onto the plasma membrane drive such robust filopodial growth? We propose that motor domains impart barbed end-directed force directly onto the membrane that encapsulates a growing filopodium, and this, in turn, reduces the physical barrier to elongation. One possibility is that myosin-generated force “softens” the membrane (i.e. lowers membrane tension) in the distal tip compartment, allowing actin monomers to incorporate into the barbed ends of core bundle filaments more readily. Consistent with this idea, we found that addition of a lipid intercalating probe, which is expected to expand bilayer surface area and reduce membrane tension 67, immediately led to additional filopodial elongation. This concept is also supported by theoretical work 68 and biophysical studies showing that application of external force to the tip of a filopodium accelerates actin incorporation at the distal end 69. A second possibility is that force generated by membrane-bound motor domains contributes directly to the retrograde displacement of the actin core bundle; this again would be expected to reduce the barrier to actin monomer incorporation at the distal ends. This latter idea finds support in previous studies on actin treadmilling dynamics in neuronal growth cone filopodia and epithelial microvilli 70,71. A third possibility emerges from recent studies on MYO15A, which revealed a potential for the motor domain to bridge actin monomers and in turn, enhance actin polymerization 72,73.
One open question that re-emerges here is whether a myosin motor can impart mechanical force to a fluid lipid bilayer. In vitro studies have shown that membrane lipids are generally free to diffuse through the plane of the bilayer, as indicated by relatively high diffusion coefficients 74. Based on this, one might expect that mechanical force imparted to proteins in the bilayer (e.g. CDHR2TM) or attached to the surface of the bilayer (e.g. TH1, BTK) would dissipate quickly. However, experimental evidence from more complex lipid mixtures that include cholesterol, indicate much lower rates of diffusion with significant frictional coefficients 75, leading to the possibility that external force applied by myosin motors may persist for a period of time. To be matched to the kinetics of motor domain attachment to actin, this duration only needs to be in the range of milliseconds (e.g. Myo10 exhibits a strongly bound duration of ~80 ms)76. Moreover, filopodia contain 100s to 1000s of Myo10 molecules and because individual motors cycle asynchronously, the population will apply force continuously to the overlying plasma membrane. We, therefore, expect that protrusion resident myosins can exert biologically significant (pN scale) levels of force on the membrane for prolonged periods of time.
Of the numerous unconventional myosin motor domains tested for their ability to support filopodial elongation in this work, only representatives from classes 3, 10, and 15 were able to drive growth. Do these myosins share common features that explain this selectivity? One possible explanation might be linked to the biochemical composition of the core actin filament bundles that support filopodia. Fascin-1 is the dominant bundling protein in these structures and in vitro studies have revealed that the filopodial myosin, Myo10, preferentially interacts with and moves processively along actin filaments that are crosslinked in parallel by this bundler 76,77. Interestingly, as residents of stereocilia, Myo3 and Myo15 are adapted for interacting with core bundles organized by fascin-2 in addition to espin and plastin 78–80. Microvilli are structurally distinct from both filopodia and stereocilia in that their filaments are bundled by plastin, villin, espin and MISP 81–84, but not fascin. Thus, a preference for the close filament spacing that is characteristic of fascin bundling might explain the robust filopodial elongation response demonstrated by the motor domains of Myo3, Myo10, and Myo15, relative to other myosins tested in our assay.
Another possible explanation for why only a subset of myosins can induce filopodial elongation may be related to the natural diversity of kinetic properties exhibited by different motors. Myosins from different superfamily classes exhibit a broad range of actin filament sliding speeds and ATP hydrolysis kinetics 85. It follows that certain kinetic properties might be needed to support robust filopodial elongation in the assay we describe here. One might expect that, at a minimum, any motor capable of driving filopodial growth would need to demonstrate a barbed end-directed mechanical velocity faster than the actin retrograde flow rate. Velocities measured from sliding filaments assays or direct single-molecule observations indicate speeds for Myo3a, Myo10, and Myo15 in the range of 100s of nm/sec (110 nm/s, 310–660 nm/s, 278–429 nm/s, respectively)76,77,86,87, approximately 10-fold higher than typical retrograde flow rates 88. Thus, at least in the case of these three motors, this requirement appears to be met. Another kinetic property relevant here is the duty ratio: the fraction of the total ATPase cycle time that a motor domain will remain bound to actin. A high duty ratio myosin will be able to exert higher time-average force, which will increase its mechanical impact when present in a multi-motor ensemble 87,89,90.
Unconventional myosin isoforms have long been recognized as abundant components of actin-based protrusions. Whether these motors contribute growth-promoting force has remained unclear, primarily because of the technical challenges associated with uncoupling the roles of cargo transport from more direct mechanical effects. The data we report here indicates that myosins are also able to contribute significant growth-promoting force to drive robust protrusion elongation. As one of the main limitations in studying surface protrusions is the stochastic nature of their assembly, we expect that the synthetic, inducible system introduced in this study will prove useful for biologists seeking to uncover a deeper understanding of the biochemical and biophysical nature of protrusion formation.
LIMITATIONS OF THE STUDY
The primary limitation of the current investigation is technical in nature. For all experiments, we took advantage of cell culture models including the use of HeLa, HEK-293FT, B16-F1, and LLC-PK1-CL4 lines. While this approach offers clear experimental advantages (e.g. facile manipulation through transfection, robustness for live imaging, ease of presentation on the light microscope), cell culture models are inherently limited in the extent to which they reflect cell behavior in vivo. Additional studies in organoids or model organisms will be needed to examine the implications of our findings for protrusion formation in a native context.
STAR*METHODS
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Matthew J. Tyska (matthew.tyska@vanderbilt.edu).
Materials availability
Plasmids generated in this study will be made available from the lead contact on request.
Data and code availability
Microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Cell culture
HeLa, HEK293FT, B16-F1, and LLC-PK1-CL4 (CL4), cells were cultured at 37°C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) (Corning #10-013-CV) with high glucose and 2 mM L-Glutamine supplemented with 10% fetal bovine serum (FBS).
Method details
Cloning and constructs
All PCR amplification was completed using Q5 High-Fidelity DNA Polymerase (NEB #M0491S) unless site-mutagenesis was completed, in which case PfuUltra High-Fidelity DNA Polymerase (in Agilent #200523 kit) was used.
Membrane interacting motifs:
To generate the membrane binding motif constructs, we first generated a (1) mCherry-FKBP-myc plasmid by inserting mCherry (a.a. 1329–1564) from CDHR2-mCherry-FKBP-myc using the primers 5’-taagcagaattccatggtgagcaagggcga-3’ and 5’-taagcagcggccgccactgtg-3’ to amplify a PCR product of mCherry flanked by the restriction digest sites for EcoRI (NEB #R3101) and NotI (NEB #R3189). The gel purified mCherry PCR product and pcDNA3.1 Zeo+FKBP-myc (Addgene plasmid #20211) were digested with EcoRI and NotI, gel purified (Macherey-Nagel #740609.50), and ligated together (NEB M2200S). (3) CDHR2TM-mCherry-FKBP-myc was made by amplifying the single-spanning transmembrane domain of a previously generated full-length CDHR2-mCherry-FKBP-myc plasmid. The transmembrane portion of the protein (a.a. 1–185) was amplified using the primers 5’-taagcagctagcaccatggcccagctatgg-3’ and 5’-taagcacttaagcaggtccgtggtgtccagg-3’ to generate a PCR product flanked by the restriction sites NheI (NEB #R0131) and AflII (NEB #R0520). Using traditional cloning, this fragment was then inserted into the mCherry-FKBP-myc plasmid. To generate the (2) mCherry-BTK-PH-FKBP-myc PI(3,4,5)P3 membrane binding motif plasmid, we amplified the BTK-PH domain of hs Bruton tyrosine kinase (NM_000061.2) from the Addgene plasmid #51463 (a.a. 1–177) using the primers 5’-tgcttagaattcggttttaagcttccattcctgttctcca-3’ and 5’-taagcaggtaccaccatggccgcagtgattctggag-3’ to generate a PCR product containing BTK-PH flanked by the restriction digest sites KpnI and EcoRI. The mCherry-FKBP-myc plasmid and gel purified BTK-PH PCR product were both digested with KpnI (NEB #R3142) and EcoRI, gel purified and ligated together. To generate the (3) FKBP-myc-mCherry-TH1 electrostatic membrane binding motif plasmid, Gibson assembly (NEB #E2621) was used to assemble three fragments into pcDNA3.1 Zeo+FKBP-myc: Fragment 1: FKBP-myc (a.a. 1–117) from pcDNA3.1 Zeo FKBP-myc; Fragment 2: mCherry (a.a. 1–236) from the previously generated full length CDHR2-mcherry-FKBP-myc; Fragment 3: TH1 domain (a.a. 772–1043) of hs unconventional myosin-1a (GeneBank™ AF009961) was amplified using the primers 5’-ggcggccgctcgagtctagagccctcaccttggcaga-3’ and 5’-gaaggcacagtcgaggctgatcagcgggtttaaacgggcctcactgcacagtcacctccaag-3’.
Myosin motor domains:
We first generated the (1) EGFP-Myo10MD-FRB-myc construct by inserting the motor domain of Hs myosin-10 (a.a. 1–855) into the pcDNA3.1 Zeo+FRB-myc (Addgene plasmid #20228) plasmid by Gibson assembly. The motor domain was amplified was using the primers 5’-tatagggagacccaagctgggccaccatggtgagcaag-3’ and 5’-actgtgctggatatctgcagcctgcttcctccgttgggc-3’ and the FRB-myc plasmid was linearized by restriction enzyme incubation with NheI and EcoRI. For the additional motor domain constructs, we generated an (2) EGFP-FRB-myc plasmid to insert various motor domains into by amplifying EGFP (a.a. 1–239) from EGFP-Myo10MD-FRB using the primers 5’-taagcaaagcttaccatggtgagcaagggcg-3’ and 5’-taagcagaattcagatctgagtccggacttgtacagc-3’ to generate a PCR product containing EGFP flanked by HindIII and EcoRI. The FRB-myc plasmid and gel purified EGFP PCR product were both digested with HindIII (NEB #R3104) and EcoRI, gel purified and ligated together. To generate the two motor domain dead constructs (G437A, switch II and R220A, switch I) we utilized site-directed mutagenesis (Agilent #200523) of EGFP-Myo10MD-FRB to induce the two, separate amino acid changes using the primers 5’-cacctcaaagttttcaaatgcaaagatgtccaagatgcc-3’ and 5’-ggcatcttggacatctttgcatttgaaaactttgaggtg-3’ for (3) EGFP-Myo10MDxG437A-FRB-myc and 5’-ctgaacaaacttcccgaaggcactggagttattgttgtac-3’ and 5’-gtacaacaataactccagtgccttcgggaagtttgttcag-3’ for (4) EGFP-Myo10MDxR220A-FRB-myc. To generate the (5) EGFP-Myo6MD-FRB-myc plasmid, Gibson assembly was used to assemble three fragments of hs unconventional myosin-6 (GenBank #NM_001300899.2) to replace the Myo10MD in EGFP-Myo10MD-FRB-myc: Fragment 1: a.a. 1–294 was amplified using the primers 5’-agatctcgaatcacaagtttgtacaaaaaagcaggctccgaaatggaggatggaaagcccgt-3’ and 5’-cggttctgtaaaatctgtttgtcagtttcttt-3’; Fragment 2: a.a. 295–576 was amplified using the primers 5’-ttgctaacaaagaaactgacaaacagattttacag-3’ and 5’-gccttcgtcgtctctgatattcctatgaactg-3’; Fragment 3: a.a. 577–844 was amplified using the primers 5’-taagctggcagttcataggaatatcagagacg-3’ and 5’-cagaggattctagactcgagcggccgccactgtgctggattgcccaccttaaccagacca-3’. The backbone (EGFP-Myo10MD-FRB-myc) was linearized by restriction digest via EcoRV (NEB #R3195) and EcoRI. The myosin-3a lacking the N-terminal kinase (ΔK) plasmid was kindly gifted to us by Dr. Christopher Yengo. (6) EGFP-Myo3aΔK-FRB-myc was generated using Gibson assembly to insert the Myo3aΔKMD (a.a. 407–1432) into the EGFP-FRB-myc plasmid. The Myo3aΔMD was amplified using the primers 5’-cacagtggcggccgctcgaggtagatgatttagcaaccctagaagttttgga-3’ and 5’-cacatctcatgccagaggataaaagaagacatcttgtccttcctcaact-3’, and the EGFP-FRB-myc plasmid was linearized using the primers 5’-aggacaagatgtcttcttttatcctctggcatgagatgtgg-3’ and 5’-agggttgctaaatcatctacctcgagcggccgcc-3’. The Mm myosin-15a (NM_182698) plasmid was kindly gifted to us by Dr. Uri Manor. The (7) EGFP-Myo15aMD-FRB-myc plasmid was generated using Gibson assembly to insert the motor domain of myo15a (a.a. 1–750) into EGFP-FRB-myc. The Myo15aMD was amplified using the primers 5’-cacagtggcggccgctcgagatgcactccatacgcaacctgcc-3’ and 5’-atctcatgccagaggattctcagcagactctgcctcatct-3’, and the EGFP-FRB-myc backbone was linearized by PCR using the primers 5’-agatgaggcagagtctgctgagaatcctctggcatgagatgtggc-3’ and 5’-aggttgcgtatggagtgcatctcgagcggccgcc-3’. The (8) EGFP-Myo7bMD-FRB-myc plasmid was generated using Gibson assembly to insert EGFP and the motor domain (a.a. 1 – 968) of Hs myosin-7b (NM001393586.1) into FRB-myc. EGFP and the myosin-7b motor domain were amplified by PCR using the primers 5’-tatagggagacccaagctgggccaccatggtgagcaag-3’ and 5’-actgtgctggatatctgcagcccaggccatccacatcctc-3’ and the FRB-myc backbone was linearized by restriction digest with the enzymes NheI and EcoRI. The Hs myosin-5b plasmid was kindly gifted to us by Dr. James R. Goldenring. The (9) EGFP-Myo5bMD-FRB-myc plasmid was generated using Gibson assembly to insert the motor domain of myosin-5b (a.a. 1–910) into EGFP-FRB-myc. The myosin-5b motor domain fragment was amplified using the primers 5’-acaagtccggactcagatctatgagcgtgggcgagc-3’ and 5’-aggattctagactcgagcggggcctcgattctcagggc-3’, and the EGFP-FRB-myc plasmid was linearized by PCR using the primers 5’-aagccctgagaatcgaggccccgctcgagtctagaatcctctgg-3’ and 5’-tacagctcgcccacgctcatagatctgagtccggacttgtacag-3’. The (10) EGFP-Myo5bΔ3IQ-FRB-myc was generated by amplifying a fragment containing the motor domain and first three IQ motifs (a.a. 1–831) from EGFP-Myo5b-FRB-myc, flanked by EcoRI sites using the primers 5’-taagcagaattctccggactcagatctatgagcgtgg-3’ and 5’-taagcagaattcggcctgtctggcccg-3’. Using traditional cloning, this fragment was then inserted into EGFP-FRB-myc. The (11) EGFP-Myo1aMD-FRB-myc plasmid was generated by amplifying the motor domain (a.a. 1–771) of Hs myosin-1a (NM001256041.2) using the primers 5’-agtccggactcagatctgaaatgcctctcctggaaggt-3’ and 5’-attctagactcgagcggccgagcctctgaccggaaatatttgc-3’. Using Gibson assembly, this fragment was then inserted into a PCR linearized EGFP-FRB-myc backbone using the primers 5’-aatatttccggtcagaggctcggccgctcgagtctaga-3’ and 5’gaaccttccaggagaggcatttcagatctgagtccggacttgtaca-3’. For all plasmids, colonies were first screen by restriction digest to confirm the correct digested band pattern and then verified by sequencing (GENEWIZ, South Plainfield, NJ).
Transfections
Transfections were performed using Lipofectamine 2000 (Thermo Fischer #11668019) according to the manufacturer’s protocol. Cells were incubated in Lipofectamine for 4–6 hrs, after which they were replated onto 35mm glass-bottom dishes (Cellvis #D35-20-1.5-N) and/or coverslips coated with 25 ug/mL laminin (Corning #354232) and allowed to adhere and recover overnight before live imaging or fixing/staining. For DIAPH1 (mDia1) knock down (KD) experiments, HeLa cells were transfected twice with either 10 μM of Accell human DIAPH1 siRNA SMARTPool (Horizon Discovery #E-010347-00-0005) or Accell non-targeting control pool (Horizon Discovery #D-001910-10-05). Cells were allowed to recover overnight before being transfected a second time with their respective siRNA pools. On the third day, HeLa cells were transfected with the EGFP-Myo10MD-FRB and CDHR2TM-mCherry-FKBP constructs and replated as described above.
Immunofluorescence
For Structured Illumination Microscopy (SIM) and A1 confocal imaging, cells were washed with pre-warmed phosphate-buffered saline (PBS) before being fixed in 4% paraformaldehyde (Electron Microscopy Sciences #15710) in PBS for 15 mins at 37°C. After fixation, cells were rinsed three times in PBS and permeabilized with 0.1% Triton X-100 in PBS for 15 mins at 37°C. If cells were stained with anti-Fascin, cells were fixed and permeabilized with ice cold methanol on ice for 15 mins instead of 4% paraformaldehyde. After permeabilization, cells were rinsed three times in PBS and blocked with 5% BSA (Research Products International #9048-46-8) in PBS for 1 hr at 37°C. Immunostaining was performed using the primary antibodies: anti-VASP (1:50; Cell Signaling Technologies #3132S), anti-Fascin (1:100; Agilent Technologies #M356701-8), anti-GFP (1:200; Aves Labs #GFP-1020), anti-mCherry (1:500; Invitrogen #M11217), diluted in 1% BSA in PBS at 37°C for 1 hr. After incubation with primary antibody, coverslips were rinsed three times in PBS and incubated for 1 hr with Alexa Fluor 568-phalloidin or Alexa Fluor 647-phalloidin (1:200; Invitrogen #A12380 and #A22287) at room temperature. Coverslips were then washed five times in PBS and mounted on glass slides in ProLong Gold (Invitrogen #P36930).
Drug treatments
To oligomerize FRB and FKBP constructs, transfected cells were treated with 500 nM of A/C Heterodimerizer (Takara #635057) for 25 mins before being fixed for immunofluorescence or were treated with 500 nM of A/C Heterodimerizer at the onset of live imagining. To inhibit actin polymerization, HeLa cells were treated with either 2.5 uM Cytochalasin D (Sigma #C2618) or 100 nM Latrunculin A (Lat A; Molecular Probes #12370) in addition to 500 nM of the A/C Heterodimerizer and fixed after 25 mins. For the Lat A washout experiments, cells were treated with 100 nM Lat A. For the live-cell imaging membrane addition experiments, cells were treated with CellMask™ (Thermo #C10045) at 1:1000. For live-cell imaging of individual filopodia, 0.5% methylcellulose diluted in DMEM with high glucose and 2 mM L-Glutamine supplemented with 10% FBS was added prior to imaging.
Western blot analysis
To generate lysates for western blotting to confirm DIAPH1 (mDia1) KD, HeLa cells were cultured in 6-well plates, and cell lysates were prepared on ice using CelLytic M lysis buffer (Sigma #C2978) supplemented with 4 mM pefabloc (sc-202041), a pellet protease inhibitor (Roche #04906845001) and 2 mM ATP (Sigma #A1852). Samples were centrifuged at 12,000 rpm for 10 min at 4°C to remove cell debris. The resulting supernatant was normalized and boiled in Laemmli Sample Buffer (Bio-Rad #1610737) with 5% beta-mercaptoethanol (Sigma #M3148) for 5 min. Samples were then loaded on a 4%–12% NuPAGE gradient gel (Invitrogen #NP0233BOX). Gels were transferred onto a nitrocellulose membrane at 30 V for 18 hrs. Membranes were blocked with 5% dry milk diluted in PBS containing 0.1% Tween 20 (PBS-T) for 1 hr at room temperature. Membranes were incubated in primary antibodies against DIAPH1 (Bethyl Laboratories A300-078A) and GAPDH (1:1000; Cell Signaling Technologies #2118L) diluted in 1X PBS-T containing 5% dry milk overnight at 4°C. Membranes were washed 3X for 5 min with 1X PBS-T and incubated in the secondary antibody IRdye 800 donkey anti-rabbit (LI-COR #926-32213) for 1 hr at room temperature. Membranes were then washed 3X for 5 min in 1X PBS-T and imaged using the Odyssey CLx infrared scanner (LI-COR). Images were processed using the FIJI software (NIH) and protein expression levels were normalized to GAPDH.
Light microscopy and image processing
Laser scanning confocal imaging was conducted using a Nikon A1 Microscope equipped with 405, 488, 561, and 645 nm LASERs, Plan Apo 60X/1.4NA, and Plan Apo 25X/1.05 NA silicon (SIL) immersion objectives. Live-cell imaging was performed on a Nikon Ti2 inverted light microscope equipped with a Yokogawa CSU-X1 spinning disk head, equipped with 488 nm, 561 nm, and 647 nm excitation LASERs, a 405 nm photo-stimulation LASER directed by a Bruker mini-scanner to enable targeted photobleaching, a 100X Apo TIRF 100x/1.45 NA objective, and either a Hamamatsu Fusion BT or Photo-metrics Prime 95B sCMOS camera. Cells were maintained in a stage top incubator at 37°C with 5% CO2 (Tokai Hit). Super-resolution imaging was performed using a Nikon Structured Illumination Microscope (N-SIM) equipped with 405, 488, 561 and 640 nm LASERs, an SR Apo TIRF 100X/1.49 NA objective, and an Andor iXon Ultra DU-897 EMCCD camera. Images were reconstructed using Nikon Elements software. For imaging in all microscope modalities, imaging acquisition parameters were matched between samples during image acquisition. All images were denoised and deconvolved in Nikon Elements. As filopodia extremely thin structures, LUTs were optimized to facilitate visualization in figures.
Quantification and statistical analysis
All images were processed and analyzed using Nikon Elements software or FIJI software (https://fiji.sc/).
Analysis of length sum of filopodia over time.
A filopodia Weka macro 91 was used to classify the CDHR2TM channel of the motor domain movies into filopodia, cell body, or background through the Trainable WEKA Segmentation plugin (https://imagej.net/plugins/tws/). The subsequent generated probability maps for classified filopodia were then run through a length measurement macro, which completes an auto threshold, conversion to mask, dilation and skeletonization on the image before being run through the Ridge Detection plugin (https://imagej.net/plugins/ridge-detection) to quantify the length of detected filopodia for the filopodia probability map for each time point.
Analysis of fraction change of filopodia number.
The number of filopodia per cell at 0 min and 25 min either with or without rapalog treatment was counted, and the fraction change was quantified as the number of filopodia at 25 min over 0 min.
Measuring the length and rate of filopodia.
Lengths of individual filopodia were measured in Fiji using the segmented line tool from the edge of the cell body to the end of the myosin-10 EGFP signal. Rate was calculated as the slope of the line fit via simple linear regression to the length over time.
Analysis of VASP tip enrichment in filopodia.
VASP intensity along filopodia was measured using Nikon Elements software. A segmented line was drawn on filopodia from the tip of filopodia to the cell body, with either Myo10MD or Myo10FL at the tips. As all filopodia were different lengths or sometimes overlapped with other filopodia, measurements from the tip to ~1.3 μm towards the cell body were taken and normalized from 0 to 1, where 1 represented the filopodia tip. These normalized average intensity values were plotted, and the line connects the average data points.
Quantification of the percent of cells exhibiting filopodia elongation.
The percentage of cells exhibiting filopodia elongation in HeLa cells was calculated from HeLa cells expressing both the Myo10MD and either of the three membrane docking motifs. A HeLa cell was considered positive for filopodial elongation if the cell expressed both constructs and had at least four filopodia with the motor domain localized to the tip. For the quantification in Fig. S4, cells were considered positive if the cell expressed both constructs and had at least four filopodia where the motor domain was localized into the filopodia. There is no significance bar over Fig. S4C as all values were 0%.
Length and either Myo10MD or Myo10-FL intensity measurements.
ROIs of similar size were drawn over the tips of filopodia induced by either over expression of the Myo10MD-CDHR2TM system or full-length myosin-10 (Myo10-FL). Intensity measurements for either Myo10MD or Myo10-FL were taken, and the length of the associated filopodia was measured using the CDHR2TM channel in Fiji.
Analysis of filopodia length sum for Lat A washout and CellMask addition experiments.
For these experiments, the lengths of individual filopodia were taken manually for the indicated time points. Unpaired t-tests were used to calculate the significance between filopodia length sum of treatment and control cells at 35 min and 75 min (Lat A washout) and 105 min (CellMask); a second unpaired t-test was used to calculate the significance of the change of filopodia length sum between the last and 35 min time point for each treatment.
FRAP analysis.
ROIs of similar area were drawn over the edge of HeLa cells expressing either of the three membrane binding motifs (CDHR2TM-FKBP, BTK-PH-FKBP, or FKBP-TH1) and bleached using a 405 nm LASER steered with a Bruker mini-scanner. Cells were imaged for 1 min prior to bleach and then imaged with no delay for 5 min to capture signal recovery dynamics. The first 2 min of recovery were used for analysis, and the first 50 sec of recovery are displayed on the graphs. All intensity values for each condition were normalized to the cell body intensity and background intensity as a ratio of (stimulated – background)/(cell body – background). These FRAP intensities from multiple cells were then normalized from 0 to 1 and plotted together. Average values for each condition were fit using two-phase association curves. Immobile fractions were calculated as 1 minus the plateau of the curve.
Statistical analysis.
For the purpose of generating SuperPlots 92 we considered the number of individual cells equivalent to the number of biological replicates; this approach allowed us to build plots that communicate the variability between cells and between individual filopodia on a given cell in response to rapalog induction. All experiments were completed at least in triplicate and the number of biological replicates (n) in each case is defined in each figure legend. Statistical significance was performed using the unpaired Student’s t-test for comparisons and the paired Student’s t-test for comparisons of the fraction change of the number of filopodia. All length filopodia sum over time statistical significance was performed using paired Student’s t-tests. All statistical analysis were computed using PRISM v.9.0. (GraphPad).
Supplementary Material
Video S1, related to Figure 1. Membrane bound Myo10MD drives robust elongation of filopodia. (Top) Myo10MD and CDHR2TM: no rapalog. Live cell movie of maximum intensity projections of HeLa cell expressing Myo10MD-FRB (green) and CDHR2TM-FKBP (magenta) not treated with rapalog. 3 μm maximum intensity projections are composed of 16 × 0.2 μm confocal slices. (Bottom) Myo10MD and CDHR2TM: rapalog treatment. Live cell movie of maximum intensity projections of HeLa cell expressing Myo10MD-FRB (green) and CDHR2TM-FKBP (magenta) treated with rapalog. 3 μm maximum intensity projections are composed of 21 × 0.15 μm confocal slices. Images were captured at 1 frame per 15 sec. Time min:sec.
Video S2, related to Figure 1. HeLa cells expressing Myo10MD, without CDHR2TM, and treated with rapalog do not elongate filopodia. Live cell movie of maximum intensity projections of HeLa cell expressing Myo10Md-FRB (green) treated with rapalog. 3 μm maximum intensity projections are composed of 16 × 0.2 μm confocal slices. Images were captured at 1 frame per 1 min. Time min:sec.
Video S3, related to Figure 3. Single filopodium growth from a HeLa cell expressing Myo10MD-FRB and CDHR2TM-FKBP. Live cell movie of maximum intensity projections of HeLa cell expressing Myo10MD-FRB (green) and CDHR2TM-FKBP (magenta) treated with rapalog. 3 μm maximum intensity projections are composed of 3 × 0.2 μm confocal slices. Images were captured at 1 frame per ~2 sec. Time min:sec. Scale bar = 5 μm.
Video S4, related to Figure 3. Docking the motor-dead construct, Myo10MDx, to the membrane fails to elongate filopodia. Live cell movie of maximum intensity projections of HeLa cell expressing Myo10MDx-FRB (green) and CDHR2TM-FKBP (magenta) treated with rapalog. 3 μm maximum intensity projections are composed of 17 × 0.2 μm confocal slices. Images were captured at 1 frame per 15 sec. Time min:sec.
Video S5, related to Figure 3. Docking the pointed-end directed motor, Myo6MD, to the membrane fails to elongate filopodia. Live cell movie of maximum intensity projections of HeLa cell expressing Myo6MD-FRB (green) and CDHR2TM-FKBP (magenta) treated with rapalog. 3 μm maximum intensity projections are composed of 11 × 0.3 μm confocal slices. Images were captured at 1 frame per 15 sec. Time min:sec.
Video S6, related to Figure 4. Filopodia elongation in HeLa cells treated with Lat A. Live cell movie of maximum intensity projections of HeLa cell expressing Myo10MD-FRB (green) and CDHR2TM-FKBP (magenta) treated with rapalog after a 7-day incubation with Latrunculin A (LatA) for 75 min. 3 μm maximum intensity projections are composed of 16 × 0.2 μm confocal slices.
Video S7, related to Figure 4. Filopodia length sum increases following a Lat A washout. Live cell movie of maximum intensity projections of HeLa cell expressing Myo10MD-FRB (green) and CDHR2TM-FKBP (magenta) treated with rapalog after a 7-day incubation with Latrunculin A (LatA) and then with a LatA washout. Cells were imaged for 7–5 min with the Lat A washout at 35 min. 3 μm maximum intensity projections are composed of 18 × 0.18 μm confocal slices. Images were captured at 1 frame per 5 min. Time min:sec.
Video S8, related to Figure 4. Membrane-bound Myo10MD drives filopodia elongation without CellMask treatment. Live cell movie of maximum intensity projections of HeLa cell expressing Myo10MD-FRB (green) and CDHR2TM-FKBP (magenta) treated with rapalog for 140 min. 3 μm maximum intensity projections are composed of 18 × 0.18 μm confocal slices. Time hr:min:sec.
Video S9, related to Figure 4. CellMask addition after rapalog treatment drives a second stage of filopodia elongation. Live cell movie of maximum intensity projections of HeLa cell expressing Myo10MD-FRB (green) and CDHR2TM-FKBP (magenta) treated with CellMask 35 min after rapalog induction, and then imaged for a total of 140 min. 3 μm maximum intensity projections are composed of 18 × 0.18 μm confocal slices. Images were captured at 1 frame per 5 min. Time hr:min:sec.
Video S10, related to Figure 5. Membrane bound Myo3aΔKMD (top) and Myo15aMD (bottom) drive filopodia elongation. Live cell movie of maximum intensity projections of HeLa cell expressing Myo3aΔKMD-FRB (top) or Myo15aMD (bottom) (green) and CDHR2TM-FKBP (magenta) treated with rapalog. 3 μm maximum intensity projections are composed of 16 × 0.2 μm (Myo3aΔKMD) and 18 × 0.18 μm (Myo15aMD) confocal slices. Images were captured at 1 frame per 5 min. Time min:sec.
Figure 7: Model for the contribution of myosin-generated force in protrusion growth.
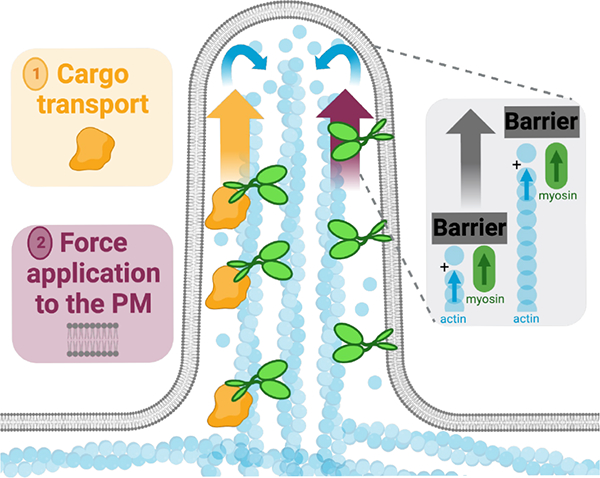
Our studies reveal that, in addition to (1) delivering cargoes that promote protrusion growth or stability, barbed end directed force exerted by membrane bound myosins can also promote protrusion growth. Force application by myosin likely serves in parallel with force generated by the barbed-ends of elongating core bundle filaments, to displace the physical barrier posed by the plasma membrane.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Anti-GFP (chicken) | Aves Labs | Cat# GFP-1020; RRID: AB_10000240 |
| Anti-Vasp (rabbit) | Cell Signaling Technologies | Cat# 3132S; RRID: AB_2213392 |
| Anti-mCherry (rat) | Invitrogen | Cat# M11217; RRID: AB_2536611 |
| Anti-Fascin (mouse) | Agilent | Cat# M356701-8; RRID: AB_2278596 |
| Anti-DIAPH1 (rabbit) | Bethyl Laboratories | Cat# A300-078A; RRID: AB_2092939 |
| Anti-GAPDH (rabbit) | Cell Signaling Technologies | Cat# 2118L; RRID: AB_561053 |
| IRDye 800CW Donkey Anti-Rabbit IgG | LI-COR | Cat# 926-32213; RRID: AB_621848 |
| Donkey Anti-Mouse Alexa Fluor 647 IgG (H+L) | Thermo Fischer Scientific | Cat# A31571; RRID: AB_162542 |
| Goat Anti-Chicken Alexa Fluor 488 IgY (H+L) | Thermo Fischer Scientific | Cat# A11039; RRID: AB_2534096 |
| Donkey anti-Rabbit Alexa Fluor 647 IgG (H+L) | Thermo Fischer Scientific | Cat# A31573; RRID: AB_2536183 |
| Goat Anti-Rat Alexa Fluor 568 IgG (H+L) | Thermo Fischer Scientific | Cat# A11077; RRID: AB_2534121 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| E. coli DH5-Alpha competent cells | Molecular Cell Biology Resource Core, Vanderbilt Medical Center | Item# DH5 Alpha |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Alexa Fluor 568 Phalloidin | Invitrogen | Cat# A12380 |
| Alexa Fluor 647 Phalloidin | Invitrogen | Cat# A22287 |
| ProLong Gold Antifade Reagent | Invitrogen | Cat# P36930 |
| 16% Paraformaldehyde solution | Electron Microscopy Sciences | Cat# 15710 |
| Triton X-100 | Sigma | Cat# T8787 |
| Q5 High-Fidelity DNA Polymerase | NEB | Cat# M0491S |
| PfuUltra High-Fidelity DNA Polymerase | Agilent | Cat# 200523 |
| Lipofectamine 2000 | Thermo Fischer Scientific | Cat# 11668019 |
| A/C Heterodimerizer | Takara | Cat# 635057 |
| Cytochalasin D | Sigma | Cat# C2618 |
| Latrunculin A | Molecular Probes | Cat# 12370 |
| CellMask Orange plasma membrane dye | Thermo Fischer Scientific | Cat# C10046 |
| Celllytic M lysis buffer | Sigma | Cat# C2978 |
| Protease Inhibitor | Roche | Cat# 04906845001 |
| Laminin | Corning | Cat# 354232 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| LLC-PK1-CL4 | Gift from Dr. Carolyn Slayman (Yale University) | N/A |
| HeLa | Gift from Dr. Ryoma Ohi (University of Michigan) | N/A |
| HEK293FT | ATCC | CRL-3216 |
| B16-F1 Melanoma Cells | ATCC | CRL-6323 |
|
| ||
| Oligonucleotides | ||
|
| ||
| siRNA targeting sequence: human DIAPH1: Accell SMARTPool | Horizon Discovery | Cat# E-010347-00-0005 |
| siRNA targeting sequence: non-targeting control pool: Accell | Horizon Discovery | Cat# D-001910-10-05 |
|
| ||
| Recombinant DNA | ||
|
| ||
| FKBP-myc | Addgene | Cat# 20211 |
| FRB-myc | Addgene | Cat# 20228 |
| mCherry-FKBP-myc | This paper | N/A |
| EGFP-FRB-myc | This paper | N/A |
| CDHR2TM-mCherry-FKBP-myc | This paper | N/A |
| EGFP-Myo10MD-FRB-myc | This paper | N/A |
| EGFP-Myo10FL | Gift from Dr. Richard Cheney (University of North Carolina) | N/A |
| EGFP-Myo10MD-R220AswitchIdead-FRB-myc | This paper | N/A |
| EGFP-Myo10MD-G437AswitchIIdead-FRB-myc | This paper | N/A |
| EGFP-Myo6MD-FRB-myc | This paper | N/A |
| BTKPH-mCherry-FKBP-myc | This paper | N/A |
| FKBP-myc-mCherry-TH1 | This paper | N/A |
| EGFP-Myo3aΔKMD-FRB-myc | This paper | N/A |
| EGFP-Myo15aMD-FRB-myc | This paper | N/A |
| EGFP-Myo5bMD-FRB-myc | This paper | N/A |
| EGFP-Myo1aMD-FRB-myc | This paper | N/A |
| EGFP-Myo5b3IQMD-FRB-myc | This paper | N/A |
| EGFP-Myo7bMD-FRB-myc | This paper | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| NIS Elements AR Analysis | https://www.nikoninstruments.com/Products/Software | N/A |
| Prism9 | GraphPad | N/A |
| FIJI/ImageJ | http://imagej.net/Fiji/Downloads | N/A |
|
| ||
| Other | ||
|
| ||
| 35 mm glass bottom dishes | CellVis | D35-20-1.5-N |
Highlights.
Force generated by membrane-bound myosin motor domains promotes filopodial elongation
Protrusion elongation is partially limited by availability of actin and membrane
Motors domains of myosin-10, -3a and -15a elongate protrusions when membrane bound
Motor domains elongate protrusions independent of the mode of membrane binding
ACKNOWLEDGMENTS
The authors would like to thank all members of the M.J.T. laboratory, members of the Vanderbilt Microtubules and Motors Club, L.M.M., and Dr. Tatsuki Koyama (Dept. of Biostatistics at Vanderbilt) for their feedback and guidance. We thank Dr. Richard Cheney, Dr. Uri Manor, and Dr. Christopher Yengo for their gift of template DNA used for the myosin-10, -15a and -3a motor constructs, respectively. Light microscopy was performed in part through use of the VUMC Cell Imaging Shared Resource. This work was supported by the NIH NIDDK National Research Service Award F31-DK130599 (G.N.F.), and NIH grants R01-DK125546, R01-DK095811, and R01-DK111949 (M.J.T.). We also acknowledge the Lacks family and are grateful for the use of HeLa cells, which heavily contributed to the discoveries in this work.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
DECLARATION OF INTEREST
The authors declare they have no competing financial interest in this work. declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Tilney LG, Derosier DJ, and Mulroy MJ (1980). The organization of actin filaments in the stereocilia of cochlear hair cells. J Cell Biol 86, 244–259. 10.1083/jcb.86.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mooseker MS, and Tilney LG (1975). Organization of an actin filament-membrane complex: Filament polarity and membrane attachment in the microvilli of intestinal epithelial cells. J Cell Biol 67, 725–743. 10.1083/jcb.67.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Small JV (1988). The actin cytoskeleton. Electron Microsc Rev 1, 155–174. 10.1016/s0892-0354(98)90010-7. [DOI] [PubMed] [Google Scholar]
- 4.Houdusse A, and Titus MA (2021). The many roles of myosins in filopodia, microvilli and stereocilia. Curr Biol 31, R586–R602. 10.1016/j.cub.2021.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kovar DR, and Pollard TD (2004). Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci U S A 101, 14725 LP–14730. 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mogilner A, and Oster G (1996). Cell motility driven by actin polymerization. Biophys J 71, 3030–3045. 10.1016/S0006-3495(96)79496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Footer MJ, Kerssemakers JWJ, Theriot JA, and Dogterom M (2007). Direct measurement of force generation by actin filament polymerization using an optical trap. Proc Natl Acad Sci U S A 104, 2181–2186. 10.1073/pnas.0607052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peskin CS, Odell GM, and Oster GF (1993). Cellular motions and thermal fluctuations: the Brownian ratchet. Biophys J 65, 316–324. 10.1016/S0006-3495(93)81035-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mogilner A, and Rubinstein B (2005). The physics of filopodial protrusion. Biophys J 89, 782–795. 10.1529/biophysj.104.056515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Theriot JA (2000). The polymerization motor. Traffic 1, 19–28. 10.1034/j.1600-0854.2000.010104.x. [DOI] [PubMed] [Google Scholar]
- 11.Mogilner A, and Oster G (1996). Cell motility driven by actin polymerization. Biophysical Journal 71, 3030–3045. 10.1016/S0006-3495(96)79496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill TL, and Kirschner MW (1982). Bioenergetics and kinetics of microtubule and actin filament assembly-disassembly. Int Rev Cytol 78, 1–125. [PubMed] [Google Scholar]
- 13.Hill TL, and Kirschner MW (1982). Subunit treadmilling of microtubules or actin in the presence of cellular barriers: possible conversion of chemical free energy into mechanical work. Proc Natl Acad Sci U S A 79, 490–494. 10.1073/pnas.79.2.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higashida C, Miyoshi T, Fujita A, Oceguera-Yanez F, Monypenny J, Andou Y, Narumiya S, and Watanabe N (2004). Actin Polymerization-Driven Molecular Movement of mDia1 in Living Cells. Science 303, 2007 LP–2010. 10.1126/science.1093923. [DOI] [PubMed] [Google Scholar]
- 15.Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, and Gertler FB (2002). Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell 109, 509–521. 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- 16.Dobramysl U, Jarsch IK, Inoue Y, Shimo H, Richier B, Gadsby JR, Mason J, Szałapak A, Ioannou PS, Correia GP, et al. (2021). Stochastic combinations of actin regulatory proteins are sufficient to drive filopodia formation. J Cell Biol 220. 10.1083/jcb.202003052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McConnell RE, Benesh AE, Mao S, Tabb DL, and Tyska MJ (2011). Proteomic analysis of the enterocyte brush border. Am J Physiol - GLP 300, 6914–6926. 10.1152/ajpgi.00005.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacquemet G, Stubb A, Saup R, Miihkinen M, Kremneva E, Hamidi H, and Ivaska J (2019). Filopodome Mapping Identifies p130Cas as a Mechanosensitive Regulator of Filopodia Stability. Curr Biol 29, 202–216 e207. 10.1016/j.cub.2018.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krey JF, Wilmarth PA, David LL, and Barr-Gillespie PG (2017). Analysis of the Proteome of Hair-Cell Stereocilia by Mass Spectrometry. Methods Enzymol 585, 329–354. 10.1016/bs.mie.2016.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warshaw DM, Guilford WH, Freyzon Y, Krementsova E, Palmiter KA, Tyska MJ, Baker JE, and Trybus KM (2000). The light chain binding domain of expressed smooth muscle heavy meromyosin acts as a mechanical lever. J Biol Chem 275, 37167–37172. 10.1074/jbc.M006438200. [DOI] [PubMed] [Google Scholar]
- 21.Uyeda TQP, Abramson PD, and Spudich JA (1996). The neck region of the myosin motor domain acts as a lever arm to generate movement. Proc Natl Acad Sci USA 93, 4459–4464. 10.1073/pnas.93.9.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rayment I, Rypniewski WR, Schmidt-Bäse K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, and Holden HM (1993). Three-dimensional structure of myosin subfragment-1: A molecular motor. Science 261, 50–58. 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 23.Sellers JR (2000). Myosins: A diverse superfamily. Biochim Biophys Acta 1496, 3–22. 10.1016/S0167-4889(00)00005-7. [DOI] [PubMed] [Google Scholar]
- 24.Chen ZY, Hasson T, Zhang DS, Schwender BJ, Derfler BH, Mooseker MS, and Corey DP (2001). Myosin-VIIb, a novel unconventional myosin, is a constituent of microvilli in transporting epithelia. Genomics 72, 285–296. 10.1006/geno.2000.6456. [DOI] [PubMed] [Google Scholar]
- 25.Conzelman KA, and Mooseker MS (1987). The 110-kD protein-calmodulin complex of the intestinal microvillus is an actin-activated MgATPase. J Cell Biol 105, 313–324. 10.1083/jcb.105.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benesh AE, Nambiar R, McConnell RE, Mao S, Tabb DL, and Tyska MJ (2010). Differential localization and dynamics of class I myosins in the enterocyte microvillus. Mol Biol Cell 21, 970–978. 10.1091/mbc.E09-07-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider ME, Dosé AC, Salles FT, Chang W, Erickson FL, Burnside B, and Kachar B (2006). A new compartment at stereocilia tips defined by spatial and temporal patterns of myosin IIIa expression. J Neurosci 26, 10243–10252. 10.1523/JNEUROSCI.2812-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belyantseva IA, Boger ET, and Friedman TB (2003). Myosin XVa localizes to the tips of inner ear sensory cell stereocilia and is essential for staircase formation of the hair bundle. Proc Natl Acad Sci U S A 100, 13958–13963. 10.1073/pnas.2334417100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hasson T, Heintzelman MB, Santos-Sacchi J, Corey DP, and Mooseker MS (1995). Expression in cochlea and retina of myosin VIIa, the gene product defective in Usher syndrome type 1B. Proc Natl Acad Sci U S A 92, 9815–9819. 10.1073/pnas.92.21.9815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petersen KJ, Goodson HV, Arthur AL, Luxton GW, Houdusse A, and Titus MA (2016). MyTH4-FERM myosins have an ancient and conserved role in filopod formation. Proc Natl Acad Sci U S A 113, E8059–E8068. 10.1073/pnas.1615392113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berg JS, Derfler BH, Pennisi CM, Corey DP, and Cheney RE (2000). Myosin-X, a novel myosin with pleckstrin homology domains, associates with regions of dynamic actin. J Cell Sci 113 Pt 19, 3439–3451. [DOI] [PubMed] [Google Scholar]
- 32.Bohil AB, Robertson BW, and Cheney RE (2006). Myosin-X is a molecular motor that functions in filopodia formation. Proc Natl Acad Sci U S A 103, 12411–12416. 10.1073/pnas.0602443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weck ML, Crawley SW, Stone CR, and Tyska MJ (2016). Myosin-7b Promotes Distal Tip Localization of the Intermicrovillar Adhesion Complex. Curr Biol 26, 2717–2728. 10.1016/j.cub.2016.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feeser EA, Ignacio CM, Krendel M, and Ostap EM (2010). Myo1e binds anionic phospholipids with high affinity. Biochemistry 49, 9353–9360. 10.1021/bi1012657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazerik JN, and Tyska MJ (2012). Myosin-1A targets to microvilli using multiple membrane binding motifs in the tail homology 1 (TH1) domain. J Biol Chem 287, 13104–13115. 10.1074/jbc.M111.336313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hokanson DE, and Ostap EM (2006). Myo1c binds tightly and specifically to phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate. Proc Natl Acad Sci U S A 103, 3118–3123. 10.1073/pnas.0505685103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plantard L, Arjonen A, Lock JG, Nurani G, Ivaska J, and Strömblad S (2010). PtdIns(3,4,5)P3 is a regulator of myosin-X localization and filopodia formation. J Cell Sci 123, 3524–3534. 10.1242/jcs.069609. [DOI] [PubMed] [Google Scholar]
- 38.Zhang H, Berg JS, Wang Y, Lång P, Sousa AD, Bhaskar A, Cheney RE, and Strömblad S (2004). Myosin-X provides a motor-based link between integrins and the cytoskeleton. Nat Cell Biol 6, 523–531. 10.1038/ncb1136. [DOI] [PubMed] [Google Scholar]
- 39.Yu IM, Planelles-Herrero VJ, Sourigues Y, Moussaoui D, Sirkia H, Kikuti C, Stroebel D, Titus MA, and Houdusse A (2017). Myosin 7 and its adaptors link cadherins to actin. Nat Commun 8, 15864. 10.1038/ncomms15864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crawley SW, Shifrin DA, Grega-Larson NE, McConnell RE, Benesh AE, Mao S, Zheng Y, Zheng QY, Nam KT, Millis BA, et al. (2014). Intestinal brush border assembly driven by protocadherin-based intermicrovillar adhesion. Cell 157, 433–446. 10.1016/j.cell.2014.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berg JS, and Cheney RE (2002). Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat Cell Biol 4, 246–250. 10.1038/ncb762. [DOI] [PubMed] [Google Scholar]
- 42.Tokuo H, and Ikebe M (2004). Myosin X transports Mena/VASP to the tip of filopodia. Biochem and Biophys Res Comm 319, 214–220. 10.1016/j.bbrc.2004.04.167. [DOI] [PubMed] [Google Scholar]
- 43.Inobe T, and Nukina N (2016). Rapamycin-induced oligomer formation system of FRB-FKBP fusion proteins. J Biosc Bioeng 122, 40–46. 10.1016/j.jbiosc.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 44.Banaszynski LA, Liu CW, and Wandless TJ (2005). Characterization of the FKBP.rapamycin.FRB ternary complex. J Am Chem Soc 127, 4715–4721. 10.1021/ja043277y. [DOI] [PubMed] [Google Scholar]
- 45.Vandekerckhove J, Deboben A, Nassal M, and Wieland T (1985). The phalloidin binding site of F-actin. EMBO J 4, 2815–2818. 10.1002/j.1460-2075.1985.tb04008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vignjevic D, Kojima SI, Aratyn Y, Danciu O, Svitkina T, and Borisy GG (2006). Role of fascin in filopodial protrusion. J Cell Biol. 10.1083/jcb.200603013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schäfer C, Faust U, Kirchgeßner N, Merkel R, and Hoffmann B (2011). The filopodium. Cell Adh Migr 5, 431–438. 10.4161/cam.5.5.17400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. (2015). Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 49.Sasaki N, and Sutoh K (1998). Structure-mutation analysis of the ATPase site of Dictyostelium discoideum myosin II. Adv Biophys 35, 1–24. [PubMed] [Google Scholar]
- 50.Spector I, Shochet NR, Kashman Y, and Groweiss A (1983). Latrunculins: novel marine toxins that disrupt microfilament organization in cultured cells. Science 219, 493–495. 10.1126/science.6681676. [DOI] [PubMed] [Google Scholar]
- 51.Raucher D, and Sheetz MP (1999). Characteristics of a membrane reservoir buffering membrane tension. Biophys J 77, 1992–2002. 10.1016/S0006-3495(99)77040-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raucher D, and Sheetz MP (1999). Membrane expansion increases endocytosis rate during mitosis. J Cell Biol 144, 497–506. 10.1083/jcb.144.3.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raucher D, and Sheetz MP (2000). Cell spreading and lamellipodial extension rate is regulated by membrane tension. Journal of Cell Biology 148. 10.1083/jcb.148.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salim K, Bottomley MJ, Querfurth E, Zvelebil MJ, Gout I, Scaife R, Margolis RL, Gigg R, Smith CI, Driscoll PC, et al. (1996). Distinct specificity in the recognition of phosphoinositides by the pleckstrin homology domains of dynamin and Bruton’s tyrosine kinase. EMBO J 15, 6241–6250. 10.1002/j.1460-2075.1996.tb01014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fukuda M, Kojima T, Kabayama H, and Mikoshiba K (1996). Mutation of the pleckstrin homology domain of Bruton’s tyrosine kinase in immunodeficiency impaired inositol 1,3,4,5-tetrakisphosphate binding capacity. J Biol Chem 271, 30303–30306. 10.1074/jbc.271.48.30303. [DOI] [PubMed] [Google Scholar]
- 56.Revenu C, Athman R, Robine S, and Louvard D (2004). The co-workers of actin filaments: from cell structures to signals. Nat Rev Mol Cell Biol 5, 635–646. 10.1038/nrm1437. [DOI] [PubMed] [Google Scholar]
- 57.Dai J, and Sheetz MP (1995). Mechanical properties of neuronal growth cone membranes studied by tether formation with laser optical tweezers. Biophys J 68, 988–996. 10.1016/S0006-3495(95)80274-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sheetz MP (2001). Cell control by membrane-cytoskeleton adhesion. Nat Rev Mol Cell Biol 2, 392–396. 10.1038/35073095. [DOI] [PubMed] [Google Scholar]
- 59.Condeelis J (1993). Life at the leading edge: the formation of cell protrusions. Annu Rev Cell Biol 9, 411–444. 10.1146/annurev.cb.09.110193.002211. [DOI] [PubMed] [Google Scholar]
- 60.Pollard TD, and Borisy GG (2003). Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465. 10.1016/S0092-8674(03)00120-X. [DOI] [PubMed] [Google Scholar]
- 61.Crawley SW, Weck ML, Grega-Larson NE, Shifrin DA Jr., and Tyska MJ (2016). ANKS4B Is Essential for Intermicrovillar Adhesion Complex Formation. Dev Cell 36, 190–200. 10.1016/j.devcel.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li J, He Y, Weck ML, Lu Q, Tyska MJ, and Zhang M (2017). Structure of Myo7b/USH1C complex suggests a general PDZ domain binding mode by MyTH4-FERM myosins. Proc Natl Acad Sci U S A 114, E3776–E3785. 10.1073/pnas.1702251114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grati M, and Kachar B (2011). Myosin VIIa and sans localization at stereocilia upper tip-link density implicates these Usher syndrome proteins in mechanotransduction. Proc Natl Acad Sci U S A 108, 11476–11481. 10.1073/pnas.1104161108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Manor U, Disanza A, Grati M, Andrade L, Lin H, Di Fiore PP, Scita G, and Kachar B (2011). Regulation of stereocilia length by myosin XVa and whirlin depends on the actin-regulatory protein Eps8. Curr Biol 21, 167–172. 10.1016/j.cub.2010.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ebrahim S, Avenarius MR, Grati M, Krey JF, Windsor AM, Sousa AD, Ballesteros A, Cui R, Millis BA, Salles FT, et al. (2016). Stereocilia-staircase spacing is influenced by myosin III motors and their cargos espin-1 and espin-like. Nat Commun 7, 10833. 10.1038/ncomms10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arthur AL, Crawford A, Houdusse A, and Titus MA (2021). VASP-mediated actin dynamics activate and recruit a filopodia myosin. Elife 10. 10.7554/eLife.68082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Raucher D, and Sheetz MP (2000). Cell spreading and lamellipodial extension rate is regulated by membrane tension. J Cell Biol 148, 127–136. 10.1083/jcb.148.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Orly G, Naoz M, and Gov NS (2014). Physical model for the geometry of actin-based cellular protrusions. Biophys J 107, 576–587. 10.1016/j.bpj.2014.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bornschlogl T, and Bassereau P (2013). The sense is in the fingertips: The distal end controls filopodial mechanics and dynamics in response to external stimuli. Commun Integr Biol 6, e27341. 10.4161/cib.27341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chinowsky CR, Pinette JA, Meenderink LM, Lau KS, and Tyska MJ (2020). Nonmuscle myosin-2 contractility-dependent actin turnover limits the length of epithelial microvilli. Mol Biol Cell 31, 2803–2815. 10.1091/mbc.E20-09-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Medeiros NA, Burnette DT, and Forscher P (2006). Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat Cell Biol 8, 215–226. 10.1038/ncb1367. [DOI] [PubMed] [Google Scholar]
- 72.Gong R, Jiang F, Moreland ZG, Reynolds MJ, de Los Reyes SE, Gurel P, Shams A, Heidings JB, Bowl MR, Bird JE, and Alushin GM (2022). Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia. Sci Adv 8, eabl4733. 10.1126/sciadv.abl4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moreland ZG, Jiang F, Aguilar C, Barzik M, Gong R, Shams A, Faaborg-Andersen C, Werth JC, Harley R, Sutton DC, et al. (2021). Myosin-driven Nucleation of Actin Filaments Drives Stereocilia Development Critical for Hearing. bioRxiv, 2021.2007.2009.451618. 10.1101/2021.07.09.451618. [DOI] [Google Scholar]
- 74.Sanderson JM (2012). Resolving the kinetics of lipid, protein and peptide diffusion in membranes. Mol Membr Biol 29, 118–143. 10.3109/09687688.2012.678018. [DOI] [PubMed] [Google Scholar]
- 75.Cicuta P, Keller SL, and Veatch SL (2007). Diffusion of liquid domains in lipid bilayer membranes. J Phys Chem B 111, 3328–3331. 10.1021/jp0702088. [DOI] [PubMed] [Google Scholar]
- 76.Takagi Y, Farrow RE, Billington N, Nagy A, Batters C, Yang Y, Sellers JR, and Molloy JE (2014). Myosin-10 produces its power-stroke in two phases and moves processively along a single actin filament under low load. Proc Natl Acad Sci U S A 111, E1833–1842. 10.1073/pnas.1320122111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ropars V, Yang Z, Isabet T, Blanc F, Zhou K, Lin T, Liu X, Hissier P, Samazan F, Amigues B, et al. (2016). The myosin X motor is optimized for movement on actin bundles. Nat Commun 7, 12456. 10.1038/ncomms12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Krey JF, Krystofiak ES, Dumont RA, Vijayakumar S, Choi D, Rivero F, Kachar B, Jones SM, and Barr-Gillespie PG (2016). Plastin 1 widens stereocilia by transforming actin filament packing from hexagonal to liquid. J Cell Biol 215, 467–482. 10.1083/jcb.201606036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Perrin BJ, Strandjord DM, Narayanan P, Henderson DM, Johnson KR, and Ervasti JM (2013). beta-Actin and fascin-2 cooperate to maintain stereocilia length. J Neurosci 33, 8114–8121. 10.1523/JNEUROSCI.0238-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sekerkova G, Zheng L, Mugnaini E, and Bartles JR (2006). Differential expression of espin isoforms during epithelial morphogenesis, stereociliogenesis and postnatal maturation in the developing inner ear. Dev Biol 291, 83–95. 10.1016/j.ydbio.2005.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morales EA, Arnaiz C, Krystofiak ES, Zanic M, and Tyska MJ (2022). Mitotic Spindle Positioning (MISP) is an actin bundler that selectively stabilizes the rootlets of epithelial microvilli. Cell Rep 39, 110692. 10.1016/j.celrep.2022.110692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bartles JR, Zheng L, Li A, Wierda A, and Chen B (1998). Small espin: a third actin-bundling protein and potential forked protein ortholog in brush border microvilli. J Cell Biol 143, 107–119. 10.1083/jcb.143.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bretscher A, and Weber K (1979). Villin: the major microfilament-associated protein of the intestinal microvillus. Proc Natl Acad Sci U S A 76, 2321–2325. 10.1073/pnas.76.5.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bretscher A, and Weber K (1980). Fimbrin, a new microfilament-associated protein present in microvilli and other cell surface structures. J Cell Biol 86, 335–340. 10.1083/jcb.86.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.O’Connell CB, Tyska MJ, and Mooseker MS (2007). Myosin at work: Motor adaptations for a variety of cellular functions. Biochim Biophys Acta 11773, 615–630. 10.1016/j.bbamcr.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 86.Komaba S, Inoue A, Maruta S, Hosoya H, and Ikebe M (2003). Determination of human myosin III as a motor protein having a protein kinase activity. J Biol Chem 278, 21352–21360. 10.1074/jbc.M300757200. [DOI] [PubMed] [Google Scholar]
- 87.Bird JE, Takagi Y, Billington N, Strub MP, Sellers JR, and Friedman TB (2014). Chaperone-enhanced purification of unconventional myosin 15, a molecular motor specialized for stereocilia protein trafficking. Proc Natl Acad Sci U S A 111, 12390–12395. 10.1073/pnas.1409459111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bornschlogl T, Romero S, Vestergaard CL, Joanny JF, Van Nhieu GT, and Bassereau P (2013). Filopodial retraction force is generated by cortical actin dynamics and controlled by reversible tethering at the tip. Proc Natl Acad Sci U S A 110, 18928–18933. 10.1073/pnas.1316572110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dose AC, Ananthanarayanan S, Moore JE, Burnside B, and Yengo CM (2007). Kinetic mechanism of human myosin IIIA. J Biol Chem 282, 216–231. 10.1074/jbc.M605964200. [DOI] [PubMed] [Google Scholar]
- 90.Homma K, and Ikebe M (2005). Myosin X is a high duty ratio motor. J Biol Chem 280, 29381–29391. 10.1074/jbc.M504779200. [DOI] [PubMed] [Google Scholar]
- 91.Hebron KE, Li EY, Arnold Egloff SA, von Lersner AK, Taylor C, Houkes J, Flaherty DK, Eskaros A, Stricker TP, and Zijlstra A (2018). Alternative splicing of ALCAM enables tunable regulation of cell-cell adhesion through differential proteolysis. Sci Rep 8, 3208. 10.1038/s41598-018-21467-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lord SJ, Velle KB, Mullins RD, and Fritz-Laylin LK (2020). SuperPlots: Communicating reproducibility and variability in cell biology. J Cell Biol 219. 10.1083/jcb.202001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1, related to Figure 1. Membrane bound Myo10MD drives robust elongation of filopodia. (Top) Myo10MD and CDHR2TM: no rapalog. Live cell movie of maximum intensity projections of HeLa cell expressing Myo10MD-FRB (green) and CDHR2TM-FKBP (magenta) not treated with rapalog. 3 μm maximum intensity projections are composed of 16 × 0.2 μm confocal slices. (Bottom) Myo10MD and CDHR2TM: rapalog treatment. Live cell movie of maximum intensity projections of HeLa cell expressing Myo10MD-FRB (green) and CDHR2TM-FKBP (magenta) treated with rapalog. 3 μm maximum intensity projections are composed of 21 × 0.15 μm confocal slices. Images were captured at 1 frame per 15 sec. Time min:sec.
Video S2, related to Figure 1. HeLa cells expressing Myo10MD, without CDHR2TM, and treated with rapalog do not elongate filopodia. Live cell movie of maximum intensity projections of HeLa cell expressing Myo10Md-FRB (green) treated with rapalog. 3 μm maximum intensity projections are composed of 16 × 0.2 μm confocal slices. Images were captured at 1 frame per 1 min. Time min:sec.
Video S3, related to Figure 3. Single filopodium growth from a HeLa cell expressing Myo10MD-FRB and CDHR2TM-FKBP. Live cell movie of maximum intensity projections of HeLa cell expressing Myo10MD-FRB (green) and CDHR2TM-FKBP (magenta) treated with rapalog. 3 μm maximum intensity projections are composed of 3 × 0.2 μm confocal slices. Images were captured at 1 frame per ~2 sec. Time min:sec. Scale bar = 5 μm.
Video S4, related to Figure 3. Docking the motor-dead construct, Myo10MDx, to the membrane fails to elongate filopodia. Live cell movie of maximum intensity projections of HeLa cell expressing Myo10MDx-FRB (green) and CDHR2TM-FKBP (magenta) treated with rapalog. 3 μm maximum intensity projections are composed of 17 × 0.2 μm confocal slices. Images were captured at 1 frame per 15 sec. Time min:sec.
Video S5, related to Figure 3. Docking the pointed-end directed motor, Myo6MD, to the membrane fails to elongate filopodia. Live cell movie of maximum intensity projections of HeLa cell expressing Myo6MD-FRB (green) and CDHR2TM-FKBP (magenta) treated with rapalog. 3 μm maximum intensity projections are composed of 11 × 0.3 μm confocal slices. Images were captured at 1 frame per 15 sec. Time min:sec.
Video S6, related to Figure 4. Filopodia elongation in HeLa cells treated with Lat A. Live cell movie of maximum intensity projections of HeLa cell expressing Myo10MD-FRB (green) and CDHR2TM-FKBP (magenta) treated with rapalog after a 7-day incubation with Latrunculin A (LatA) for 75 min. 3 μm maximum intensity projections are composed of 16 × 0.2 μm confocal slices.
Video S7, related to Figure 4. Filopodia length sum increases following a Lat A washout. Live cell movie of maximum intensity projections of HeLa cell expressing Myo10MD-FRB (green) and CDHR2TM-FKBP (magenta) treated with rapalog after a 7-day incubation with Latrunculin A (LatA) and then with a LatA washout. Cells were imaged for 7–5 min with the Lat A washout at 35 min. 3 μm maximum intensity projections are composed of 18 × 0.18 μm confocal slices. Images were captured at 1 frame per 5 min. Time min:sec.
Video S8, related to Figure 4. Membrane-bound Myo10MD drives filopodia elongation without CellMask treatment. Live cell movie of maximum intensity projections of HeLa cell expressing Myo10MD-FRB (green) and CDHR2TM-FKBP (magenta) treated with rapalog for 140 min. 3 μm maximum intensity projections are composed of 18 × 0.18 μm confocal slices. Time hr:min:sec.
Video S9, related to Figure 4. CellMask addition after rapalog treatment drives a second stage of filopodia elongation. Live cell movie of maximum intensity projections of HeLa cell expressing Myo10MD-FRB (green) and CDHR2TM-FKBP (magenta) treated with CellMask 35 min after rapalog induction, and then imaged for a total of 140 min. 3 μm maximum intensity projections are composed of 18 × 0.18 μm confocal slices. Images were captured at 1 frame per 5 min. Time hr:min:sec.
Video S10, related to Figure 5. Membrane bound Myo3aΔKMD (top) and Myo15aMD (bottom) drive filopodia elongation. Live cell movie of maximum intensity projections of HeLa cell expressing Myo3aΔKMD-FRB (top) or Myo15aMD (bottom) (green) and CDHR2TM-FKBP (magenta) treated with rapalog. 3 μm maximum intensity projections are composed of 16 × 0.2 μm (Myo3aΔKMD) and 18 × 0.18 μm (Myo15aMD) confocal slices. Images were captured at 1 frame per 5 min. Time min:sec.
Data Availability Statement
Microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


