Abstract
Recent advances in fluorescence imaging permit large-scale recording of neural activity and dynamics of neurochemical release with unprecedented resolution in behaving animals. Calcium imaging with highly optimized genetically encoded indicators provides mesoscopic view of neural activity from genetic defined populations at cellular and subcellular resolution. Rigorously improved voltage sensors and microscopy allow for robust spike imaging of populational neurons in various brain regions. In addition, recent protein engineering efforts in the past few years have led to development of sensors for neurotransmitters and neuromodulators. Here, we discuss the development and applications of these genetically encoded fluorescent indicators in reporting neural activity in response to various behavior in different biological systems, as well as in drug discovery. We also report a simple model to guide sensor selection and optimization.
Keywords: Fluorescence imaging, genetically encoded indicators, biosensors, drug discovery, neuromodulation, neurocircuit imaging
Introduction
To study neural circuitry, the action of one cell within a network of others, one would precisely measure and perturb specific neuronal populations and molecules in behaving animals which are engaged in performing the computation or function of interest. The discovery and heterogeneous expression of green fluorescent protein (GFP) promoted the development of a vast array of genetically encoded sensors that have been created to monitor neurotransmission, synaptic spillover, excitable membrane potential, calcium dynamics, vesicle trafficking, receptor mobilization, and other biochemical events. For example, the development of genetically-encoded calcium indicators (GECIs) such as GCaMP and its color variant jRGECO, combined with advanced imaging modalities, has revolutionized systems neuroscience (Chen et al 2013, Dana et al 2016, Tian et al 2009) by permitting mesoscopic recording of intracellular calcium as a proxy for electrical activity (Grienberger & Konnerth 2012). Genetically-encoded or hybrid voltage indicators (GEVIs) have also been engineered and significantly improved to directly report spiking patterns and sub-threshold voltage activity. In addition, genetically-encoded sensors for specific neurochemicals (GENIs) report release dynamics with high spatiotemporal resolutions and molecular specificity. Collectively, application of these sensors provides new opportunities for in vivo dissection of neural circuits underlying behavior across various species. In this review, we outline a workflow for sensor development, provide a simple model to guide sensor selection and optimization, and discuss a broad range of applications in neuroscience and pharmacology.
Imaging neuronal activity in genetically defined population.
The GECIs can be categorized into two types, based on Förster resonance energy transfer (FRET) between two fluorescent proteins (FPs) or intensity changes of single FPs (Figure 1a). Miyawaki et al. engineered the very first FRET-based Ca2+ sensor, Cameleon, composed of a Ca2+-sensitive calmodulin (CaM) and a calmodulin-binding peptide M13, sandwiched by two GFP variants (BFP-GFP or CFP-YFP) (Miyawaki et al 1997). Ca2+ binding induces CaM-M13 interaction to increase the FRET efficiency, reflected by changes in the dual-emission of donor and acceptor. Further improvement on such FRET based sensors include chromophore orientation tuning with Venus variants to improve the response dynamic range (Nagai et al 2004), and CaM-M13 replacement with Troponin C (TnC) or structure-guided redesign of calmodulin-M13 interface to minimize cellular perturbation(Mank et al 2008, Palmer et al 2006). The ratiometric measurement of such FRET sensors confers correction of motion and blood flow artifacts and allows quantification of intracellular Ca2+ concentration. However, FRET sensors often show less sensitivity compared with single FP-based Ca2+ sensors.
Figure 1: A schematic of genetically encoded sensors for calcium (GECIs), voltage (GEVIs) and neurochemicals (GENIs).
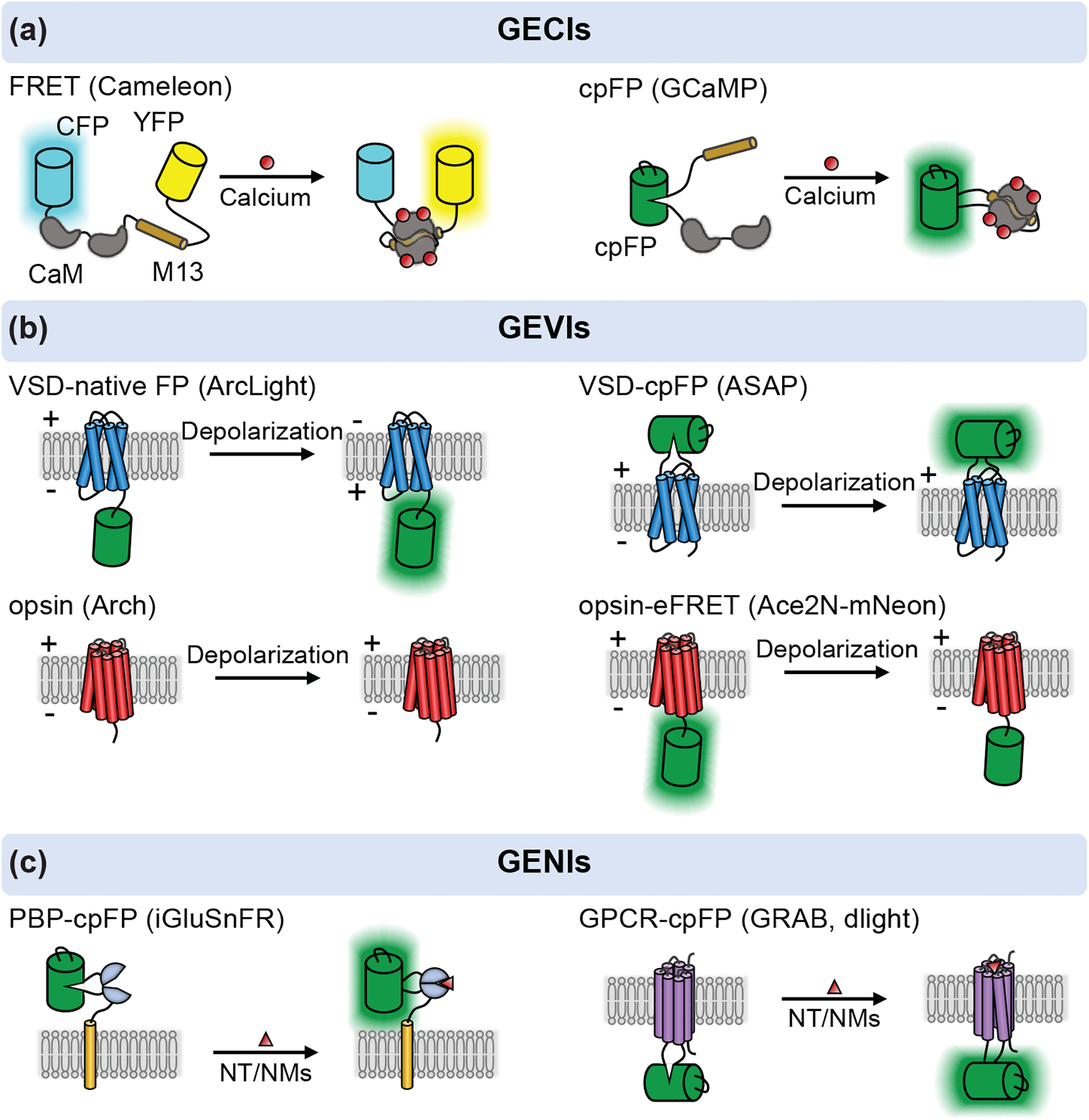
(a) GECIs based on FRET and cpFP. The FERT based calcium sensor is developed by inserting the CaM-M13 in between a donor and acceptor fluorophores, such as CFP-YFP or BFP-GFP (e.g. Cameleon). The cpFP based sensor is intensiometric, in which a cpFP is inserted in between CaM and M13/RS20 (e.g. GCaMP). (b) GEVIs utilize the voltage-sensitive domain (VSD) or opsin as scaffold. The VSD based sensors are developed by attaching a native FP to the C-terminus of VSD (e.g. ArcLight) or inserting a cpGFP into the extracellular S3-S4 loop of VSD (e.g. ASAP series). The light-driven proton pumps (opsin) are functionally reversed to action as a voltage sensitive optical element (e.g. Arch). A bright FP is attached to the opsin to address the dimness of opsin based GEVIs via electrochromic FRET (eFRET), as shown in Ace2N-mNeon. The FP is replaced with HaloTag - Janelia fluro dyes to develop a hybrid voltage sensor, namely Voltron. (c) Two class of ligand-binding scaffolds, PBPs and GPCRs, are used to developed GENIs. In both cases, a cpFP is inserted into the hinge region of PBPs (e.g. iGluSnFR) or intracellular loop 3 of GPCRs (e.g. GRAB, dLight). Examples named in the figure are as follows: Cameleon (Miyawaki et al 1997), GCaMP (Nagai et al 2001), ArcLight (Jin et al 2012), ASAP series (St-Pierre et al 2014, Villette et al 2019, Yang et al 2016), Arch (Kralj et al 2011), Ace2N-mNeon (Gong et al 2015), Voltron (Abdelfattah et al 2019), iGluSnFR (Marvin et al 2013), GRAB (Sun et al 2018) and dLight (Patriarchi et al 2018).
The emergence of single FP-based Ca2+ sensors originates from the creation of circularly-permuted FP (cpFP) (Baird et al 1999). The environment-sensitive cpFP can be modulated by the Ca2+ binding induced conformational changes of CaM-M13/RS20. Early versions of the cpFP-based Ca2+ sensors include Pericam and GCaMP, both using the CaM and M13, with a cpYFP or cpGFP inserted (Nagai et al 2001, Nakai et al 2001). Iterative protein evolution has greatly improved the sensors’ sensitivity and pushed them into practical use in vivo. In particular, GCaMP6, a big breakthrough in this field, has been widely used in the neuroscience community for neural activity imaging due to superior signal-to-noise ratio (SNR) when probing calcium in individual neuron as well as ensembles in vivo (Chen et al 2013). Further optimization of GCaMP6 has led to jGCaMP7 and jGCaMP8 series. The jGCaMP7 shows further improved sensitivity and bright basal fluorescence, allowing high-quality Ca2+ imaging in spines and somas (Dana et al 2019). The latest jGCaMP8 was engineered by swapping the M13 peptide with an endothelial nitric oxide (NO) synthase peptide. The rational design confers faster kinetics to the sensor, which benefits the tracking of action potentials. A similar strategy was also applied in developing the XCaMP series, in which the M13 was replaced by a ckkap peptide from a neuronal protein CaMKK, which is shown to respond to calcium linearly (Inoue et al 2019).
To improve signal-noise ratio of calcium imaging, subcellularly-targeted GECIs have also been developed. Axon-GCaMP6 is a specialized GECI enriching axonal structures by fusing growth-associated protein-43 (GAP43) (El-Husseini Ael et al 2001) at the N-terminus of GCaMP6m. Axon-GCaMP enabled the in vivo recordings of orientation and direction tuning of axons projecting from L4 V1 neurons across cortical layers without somatodendritic Ca2+ signal contamination (Broussard et al 2018). The expression of GECIs can also be restricted to neuronal soma to reduce overlapping fluorescence from surrounding neuropil to substantially improve SNR and specificity of imaging.
In addition, red-shifted, far-red and near-infrared GECIs have been developed to enable multiplex imaging and to potentially enhance imaging depth, allowing researchers to explore previously unreachable brain regions. R-GECO1 and RCaMP1 were the first red-shifted GECIs designed by replacing cpGFP in GCaMPs with cpmApple and cpmRuby, respectively (Akerboom et al 2013, Zhao et al 2011). Further optimization has generated the broadly used jRGECO1a, jRCaMP1a, and jRCaMP1b (Dana et al 2016). The jRGECO1a is more sensitive but suffers from blue-light induced photoactivation and lysosomal accumulation. To tackle this issue, a new variant called KGECO1, based on cpFusionRed, was engineered while retaining high response (Shen et al 2018). The color spectrum was further shifted to far-red in FR-GECO using the monomeric far-red fluorescent protein mKelly to enable sensitive detection of single action potentials in neurons (Dalangin et al 2020).
Recent breakthroughs in protein engineering efforts have also led to the development of non-GFP based NIR GECIs (Shcherbakova et al 2015). Taking advantage of biliverdin-binding NIR FPs, the first NIR GECI, NIR-GECO1, was designed by inserting the CaM-RS20 module into the split NIR mIFP(Qian et al 2020, Qian et al 2019). A brighter, FRET-based NIR GECI, named iGECI, utilized two bright NIR FPs miRFP670 and miRFP720, flanking the CaM-RS20 for FRET (Shemetov et al 2021). Further optimization in the brightness and photostability of these sensors would significantly improve SNR for in vivo imaging.
Genetically encoded voltage indicators (GEVIs), based on voltage-sensing domain (VSD) of voltage-sensitive phosphatases or light-driven proton pumps (opsin), have been developed to directly resolve firing patterns and coding properties of targeted neurons, such as rapid sequential firing, hyperpolarizing, and subthreshold depolarizing (Knopfel & Song 2019). These GEVIs are engineered to measure membrane potential (Vm) changes (Figure 1b). Though voltage imaging remains challenging compared to calcium imaging, recent rigorous sensor engineering efforts have enabled spiking imaging in genetically defined neuronal population in behaving animals.
The development and applications of GEVIs have been reviewed extensively and in a great depth elsewhere (Pal & Tian 2020, Panzera & Hoppa 2019, Wang et al 2019). The current GEVIs are still far from optimal. Inadequate sensitivity, along with the requirement of kHz acquisition and very limited imaging duration, have limited their utilization to a few laboratories. We expect a practically useful GEVI, in conjunction with GECIs, to provide more precise dissection of information processing in the brain. Nevertheless, GECIs and GEVIs have paved the way for genetically-encoded indicators, providing the foundation for the engineering and application of other sensors, including the neurochemical sensors as reviewed below.
Imaging beyond spikes with neurotransmitter and neuromodulator sensors
Neurotransmitters (NT) and neuromodulators (NM) are essential signaling molecules for information processing in the brain. There are more than 100 known NT and NM molecules (Kovacs 2004), classified as amino acids, monoamines, neuropeptides, purines and lipids, based on their structures. Fast-acting NTs, namely glutamate and gamma-aminobutyric acid (GABA), act through ligand-gated ion channels and G-protein coupled receptors (GPCRs) to modulate firing rate and excitability of post-synaptic cells. In contrast to fast-acting NTs, NMs almost exclusively bind to GPCRs to initiate molecular signaling cascades that modulate synaptic strength, neuronal excitability and circuit dynamics on timescales of sub-seconds to hours (Guillaumin & Burdakov 2021, Nadim & Bucher 2014). Altered NT and NM release is central to pathogenesis of neurological and psychiatric disorders. Although the anatomical characterizations of NT and NM projections and their functional significance are understood at a moderate level, many outstanding questions remain in regard to the structural and molecular basis and key computations underlying their release. There is a pressing need to increase experimental capacity to precisely measure the dynamics of these molecules with sub-second and single-cellular resolution, high molecular specificity and cell-type specific ideally, across the full course of a behavioral paradigm.
Inspired by the design of GECIs, we and others have developed a toolkit of genetically-encoded single-FP based indicators to probe various NT and NM systems (Figure 1c). To date, two main categories of single-FP sensors have been developed, classified by their ligand-binding scaffolds: bacterial periplasmic binding proteins (PBPs) and GPCRs.
PBP scaffolds are attractive for sensor engineering due to the conserved and large conformational change upon ligand binding. These proteins usually consist of two domains linked by a hinge region; this structure is conserved across other PBPs (Quiocho & Ledvina 1996). The ligand binding site is located in between the two domains, and the protein typically adopts two distinct conformations: a ligand-free (Apo) and a ligand-bound (Sat) state. These two conformations can interconvert via the hinge region upon ligand binding and releasing. By connecting these PBPs with a circularly-permuted FP, we can achieve more versatile and intensiometric measurements of the ligand transients in real-time. Using this design platform, highly sensitive sensors have been developed for glutamate – iGluSnFR (Helassa et al 2018, Marvin et al 2013, Marvin et al 2018), acetylcholine - iAChSnFR (Borden et al 2020), gamma-aminobutyric acid - iGABASnFR (GABA) (Marvin et al 2019), nicotine – iNicSnFR (Shivange et al 2019), ATP- iATPSnFR (Lobas et al 2019), glucose (Hu et al 2018, Keller et al 2021, Mita et al 2019) , and more recently serotonin-iSeroSnFR (Unger et al 2020).
The development of iGluSnFR pioneered the field of NT sensor development, paving the way for developing other NT/NM sensors. For example, the acetylcholine sensor, iAChSnFR, consists of a hyperthermophilic homologue of B. subtilis OpuBC from Thermoanaerobacter sp.X513 and a circularly permuted superfolder GFP (cpsfGFP) (Borden et al 2020). A critical challenge for developing iAChSnFR is the specificity tuning. The X513-OpuBC binds to both choline and acetylecholine, and the affinity for choline (kd=8uM) is tighter than acetylcholine (kd = 95uM). With the guidance of crystal structure modeling and modifications on the binding pockets, hinge region, protein interface between X513 and cpSFGFP, and junctions between the binding protein and cpSFGFP, the specificity and affinity of iAChSnFR has shifted from choline towards acetylcholine, despite the molecular similarity between the two. Expressing iAChSnFR in HEK293T cells yields an apparent of 2.9+−1.6uM, and maximum dynamic range of 10+−1.4. Furthermore, iAChSnFR has been utilized for in vivo recordings in mice, fish, flies, and worms (Borden et al 2020).
The recent development of the PBP-based serotonin sensor iSeroSnFR furthered the process of altering specificity and selectivity of the binding protein. By applying a machine learning strategy and computational modeling, the binding pocket of iAChSnFR was radically redesigned to bind serotonin while ablating acetylcholine and choline binding. The finalized iSeroSnFR conveys 19 mutations with a ~5,000-fold increase in serotonin binding specificity compared to iAChSnFR. The large dynamic range of this sensor permits in vivo detection of distinct serotonin transients across various brain regions and behaviors, including fear conditioning and sleep-wakefulness cycles (Unger et al 2020).
To further expand the toolbox, endeavors to create genetically-encoded sensors based on GPCRs as a binding module have started to rise (Andreoni et al 2019, Sabatini & Tian 2020, Wang et al 2018). Not only are GPCRs the largest and most diverse group of membrane receptors in eukaryotes, but they also are native targets of NTs and NMs in the brain. The readily available binding module for endogenous NTs and NMs of interest, as well as their highly conserved structures led to the development of a universal design approach for NM sensors. The existing GPCR structures combined with computational simulation approaches suggest that the largest conformational change upon ligand binding is in the intracellular loop 3 (IL3), which is the domain crucial for recruiting G-proteins, that lies in between transmembrane domains TM5 and TM6 (Tikhonova & Costanzi 2009) (for solved GPCR structures see (Zhang et al 2015). Utilizing this conformational change, two major single FP intensiometric sensors, the Light and GRAB families, have been developed. The general design strategies for GPCR-based sensors are similar to the PBP-based sensors, except they use the binding moiety from GPCRs. For the dLight1 series, based on three dopamine receptors (DRD1, 2 and 4), IL3 of various DA receptors was completely removed and the cpGFP was inserted directly onto the TMs with short linkers (Patriarchi et al 2018), while the GRABDA series, based on the DRD2 receptor used different linkers and IL3 insertion sites (Sun et al 2018). By systematic site saturation mutagenesis (SSM) on the linker region, GFP, and on the receptors, dLight1 achieved an affinity range from 4nM to 2.3uM, dynamic range from 170% to 930%. Similarly, GRABDA has affinity of 7–130nM, and dynamic range from 90% to 340%. The high affinity and high specificity of the DA sensors demonstrated excellent fluorescent sensitivity and brightness in response to ligand binding, making them ideally suited for in vivo imaging. This strategy has been successfully expanded to generate more GPCRs-based biosensors for detecting other neuromodulators, including acetylcholine (GRABACh), NE (nLight, GRABNE), serotonin (psychLight, GRAB5-HT), adenosine (GRABAdo), ATP (GRABATP), endocannabinoid (GRABeCB) (Dong et al 2020, Dong et al 2021, Feng et al 2019, Jing et al 2020, Jing et al 2018, Peng et al 2020, Wan et al 2021, Wu et al 2021).
Workflow for development and optimization of NT/NM sensors
Despite the considerable progress in NT/NM sensors that has been made in recent years, the existing NTs with applicable sensors are still far from enough. Here, we summarize the workflow of how to develop and optimize NT/NM sensors with GPCR-Activation Based dopamine sensors (GRABDA) as an example, aiming to facilitate the expansion of sensor families (Figure 2).
Figure 2: A workflow for the development and optimization of GPCR based neurochemical sensors.
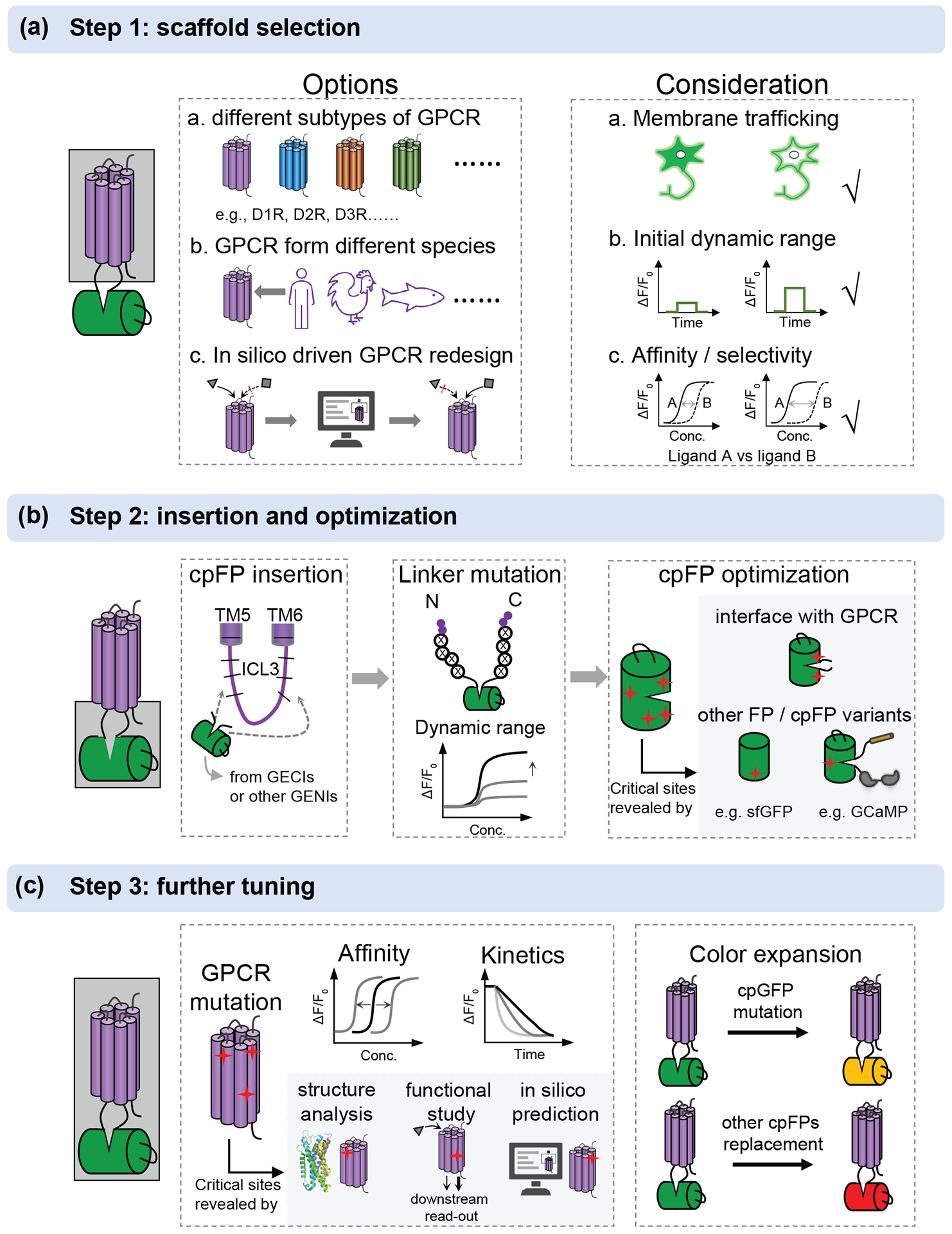
(a) The options and consideration for selection of an appropriate scaffold. GPCRs can be from different subtypes, different species and redesign in silicon. A good scaffold should have good membrane trafficking, high initial dynamic range after cpGFP insertion, appropriate affinity and high selectivity for ligand of interest. (b) After choosing a good scaffold, cpFP insertion, linker optimization and cpFP optimization can be performed sequentially. The critical sites in cpFP optimization are mainly on the interface with GPCR or learned from other FP or cpFP variants, as highlighted in gray. (c) Further tuning can be performed by mutating GPCR to tune the affinity and kinetics. The potential sites in GPCR can be obtained from the reported GPCR structures, previous function studies by downstream signaling detection and in silico prediction, as highlighted in gray. The color can be expanded by introducing mutation to cpGFP or replacing with other cpFPs, including commonly used cpmApple.
The first step is choosing a good sensing domain which will determine the overall performance of the sensor (Figure 2a). GPCRs are more desirable due to the lack of the corresponding PBPs for most NT/NMs, like dopamine. Moreover, diverse subtypes of GPCR offer more choices. A potential candidate should be evaluated according to a set of criteria including membrane trafficking, affinity and selectivity, and the initial dynamic range. During the development of GRABDA sensors, D1R and D2R showed excellent membrane trafficking when cpGFP was inserted in the preliminary screening of five human dopamine receptor subtypes. D2R was chosen for GRABDA sensors, because of its higher affinity than D1R. While D1R showed higher selectivity of DA over NE, as shown in the dLight1.3b sensor. GPCRs from different species or GPCR redesign by computational approach also provide additional options.
To increase the coupling of fluorescence changes to conformational changes induced by ligand binding, systematic truncation of the ICL3 is required to determine the appropriate position of cpGFP insertion into the ICL3 of GPCRs, followed by SSM on the linker region (Figure 2b). After that, GRABDA1m sensor with maximal ΔF/F0 about 90% was generated. Further optimization of GRABDA can firstly focus on the cpGFP, especially on the interface with GPCR. Other potential sites on cpGFP can be obtained from the differential sites in alignment with brighter GFP variants, like sfGFP and mClover3, or cpGFP variants from other sensors including GECIs and GEVIs. A 2 to 3-fold improvement in response was observed in this step for the next-generation GRABDA sensors. These steps are not always sequential. Sometimes, individual optimization of linker region and cpGFP followed by a combination of beneficial mutations may help to maximize the sensitivity, as presented in the development of GRABACh3.0.
Dynamic range and brightness are the priority for the previous steps. Additional parameters should be considered, including affinity, kinetics and color expansion, as they also determine the in vivo performance (Figure 2c). The affinity can be tuned by rational design on the ligand-binding pocket to match the concentration of NT release in physiological conditions in vivo in which the resolved structures are of great help. In addition, previous downstream functional analysis also provides potential sites for affinity tuning, especially as some of them are outside of the ligand-binding pocket. For example, in the GRABDA sensor, the T205M mutation on D2R that has been previously identified by evolution-based computational approaches and validated by downstream signaling detection assay was introduced to generate high affinity versions. Fast on and off kinetics are always expected for sensors, but there usually exists a compromise between kinetics and affinity. For example, the high-affinity GRABDA1h and GRABDA2h show slower off rate than low-affinity GRABDA1m and GRABDA2m, respectively. The color of sensors can be slightly yellow-shifted by introducing mutation on cpGFP or further red-shifted by replacing with other cpFPs, such as cpmApple used to develop red GRABDA sensors.
Theoretic considerations to choose and optimize sensors
No tool is without its limitations. GPCR-based sensors have similar ligand affinity to endogenous receptors, which may reduce effective dynamic range and interfere with endogenous signaling pathways. PBP-based sensors, on the other hand, have large dynamic range but may have compromised sensitivity at lower concentrations of release. The appropriate combination of intrinsic parameters of a sensor including brightness, expression, dynamic range, apparent affinity and kinetics must be matched to the extrinsic properties of the system which includes the size, shape, and the frequency and concentration of release. To guide the selection of the sensor that is most appropriate to applications and future improvement, especially for NM sensors, we provided a simple model based on kinetics, and dynamic ranges of GRAB5-HT1.0 and iSeroSnFR to illustrate the relationship between these parameters (Figure 3).
Figure 3: Computational modeling to guide sensor optimizations.
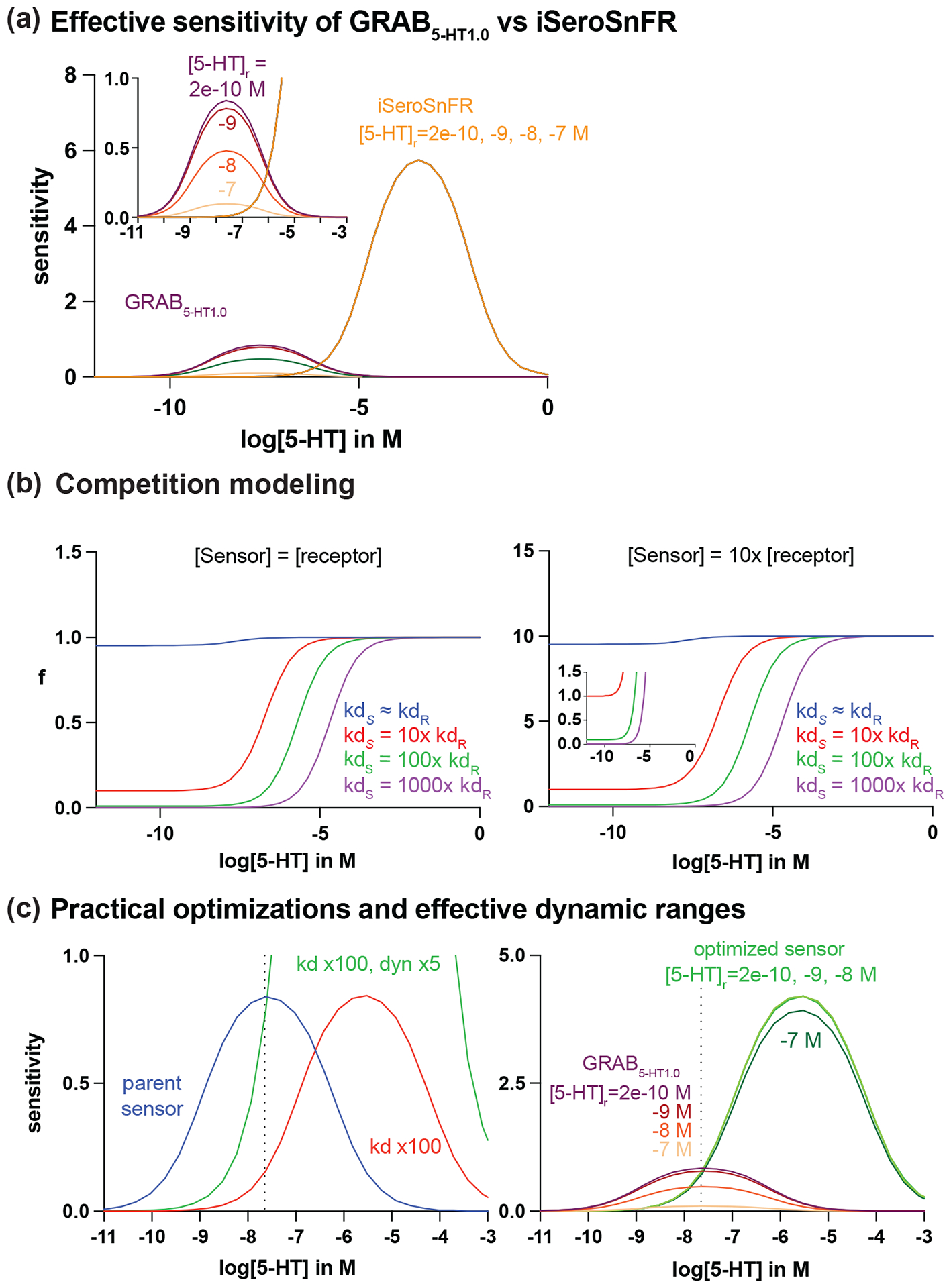
(a) Modeling of effective sensitivity of GRAB5-HT1.0 and iSeroSnFR at resting 5-HT concentration ([5-HT]r) of 200nM, 20nM, 2nM, and 200pM. (b) A 100-fold (green) or greater increase in sensor kd relative to that of the native receptor minimizes ligand buffering effect when sensor and receptor expression is equal (left) and when sensor expression is 10 times higher than receptor expression (right). (c) Left panel: a practical optimization for GRAB5-HT1.0 to maintain or enhance sensitivity while minimizing competition is to increase both the kd and the maximum dynamic range of the parent sensor (blue). According to the model, this can be achieved with a 100-fold increase in kd and a 5-fold increase in maximum dynamic range (green). Right panel: the optimized sensor is more tolerant to changes in [5-HT]r compared to the parent GRAB5-HT1.0.
We define the dissociation and association rates of receptor or sensor, and yield kd as a rate constant at half of the maximum dynamic range.
| [1] |
| [2] |
When kd of the sensor is similar to the endogenous receptor, we need to factor in the competition of ligand binding. We thus define f as the ratio of the fractional occupancy of sensor and native receptor with respective to ligand concentration, in which fractional occupancy (Θ) is defined as the fraction of receptors that is in a bound state (Motulsky & Neubig 2010).
| [3] |
We next define as the sensor’s fluorescence change relative to its baseline fluorescence based upon ligand binding. According to the specific binding equation, we can obtain a function of with respective to ligand concentration:
| [4] |
We assume both PBP-based sensors and GPCR-based sensors have Hill coefficient of one. F([Ligend]) is a function of fluorescence intensity respective to ligand concentration. is a constant calculated from function F([Ligend]) when ligand concentration is zero. is the maximum dynamic range of the sensor at its saturation ligand concentration.
This leads to the definition of the sensitivity of the sensor as the first derivative of function :
| [5] |
When resting ligand concentration is above zero, the effective sensitivity is affected. We finally defined the effective sensitivity as,
| [6] |
where [L]r is resting NM releasing concentration. GRAB5-HT1.0 (magenta) showed much higher sensitivity at the range of 1nM to 1μM compared to iSeroSnFR (yellow), while the sensitivity starts to decrease when the release concentration is beyond 10nM. (Figure 3a). However, the effective sensitivity of GRAB5-HT1.0 significantly decreases with the increase of extracellular ligand concentration at equilibrium, whereas the effective sensitivity of iSeroSnFR remains unchanged (Figure 3a). Therefore, it is concerning that GRAB5-HT1.0 may have compromised sensitivity when extracellular 5-HT concentration is higher at resting state. In contrast, iSeroSnFR would theoretically be less affected by resting concentration. Both serotonin sensors have shown capabilities of recording in vivo (Unger et al 2020, Wan et al 2021); iSeroSnFR may not fully utilize its dynamic range at nM release range, whereas GRAB5-HT1.0 could potentially encounter saturation. In other words, the intrinsic properties of both sensors can be further optimized.
As the dissociation rate of GRAB5-HT1.0 is similar to the endogenous 5-HT2 receptor (kd of 5-HT2R = 20nM (Kelly & Sharif 2006), the potential competition with endogenous receptor can be predominant especially for long-term expression. Based on the model, if a sensor’s dissociation rate can be increased about 100-fold higher than the endogenous receptor, the competition at the physiological range of release is minimal, which will significantly reduce the potential buffering effect (Figure 3b left). However, by simply increasing the dissociation rate, the sensitivity of the sensor will be reduced (Figure 3c left). To compensate for the loss of sensitivity, we also need to increase the dynamic range. Therefore, to maximize the effective sensitivity with minimal buffering, a practical optimization goal (green) for GRAB5-HT1.0 is to increase the by 100-fold while increasing the dynamic range by 5-fold as predicted by the model (Figure 3c right). On the other hand, to optimize iSeroSnFR, the model predicted to decrease its kd by 100-fold while maintaining the dynamic range. Theoretically, if the dynamic range of iSeroSnFR can be further increased by 5-fold, the sensitivity would be able to detect the release between 100nM and 1mM without comprised effective sensitivity or buffering effect. However, this ideal sensor may be practically difficult to engineer.
The other important factor affecting the SNR of imaging is the expression level of the sensor. We assume that the arbitrary sensor concentration is 10× higher than native receptors to achieve sufficient SNR. Thus, the fraction will up shift and more favorable to sensors than endogenous receptors. However, a 100× higher kd is still able to minimize the competition even sensor expression level is 10× higher than the endogenous receptors (Figure 3b right). Again, to avoid competition with endogenous receptors, we need to design a sensor with a higher kd than native receptors. By increasing maximum dynamic range and basal fluorescence, we can boost effective sensitivity at the lower release concentration. This modeling did not account for diffusion rate, distance of diffusion, ligand removal from transporter or enzyme, and assuming equilibrium is reached when binding. Together, our model suggests that a sensor with higher dissociation rate compared to endogenous receptors and higher dynamic range is preferred to offer both great sensitivity and fast kinetics with a possibly reduced buffering effect.
Imaging NM release in behaving animals
The fast-growing toolkit of genetically-encoded sensors has allowed researchers to study complex neural systems and circuitry across a range of animal models with flexible experimental design. These tools are optimally tuned to answer biological questions about the functionality, pharmacology, and interactions of different molecules involved in chemical neurotransmission. One of the primary advantages is the sub-second temporal resolution. This will allow real-time alignment of neural activity across the full course of the behaviors. Additionally, the use of spectrally-separated sensors permit a lot of flexibility in experimental design, as they can be multiplexed to record and compare transients from different neurochemicals simultaneously (Patriarchi et al 2020, Sun et al 2020). For example, researchers can use calcium indicators in conjunction with NM and NT sensors to interrogate the interplay of cellular activity and different neurochemical release phenomena.
Genetically-encoded sensors offer the unique advantage of cell-type specific viral strategies for targeting distinct neuronal populations. Researchers can use a host of approaches to narrow the scope of sensor expression. For broad distinction of neurons, promoters such as CaMKII or hSyn are often utilized (Kügler et al 2003, Nieuwenhuis et al 2021, Wang et al 2013). Viral constructs can also be expressed under promoters designed to localize expression to subclasses of neurons, e.g., the GAD67 promoter selectively expresses in GABAergic neurons (Rasmussen et al 2007). For additional specificity, researchers can employ transgenic animal models to limit sensor expression. Gene recombination systems, such as Cre/lox, are widely used to constrain sensor expression to specific cell types (Bouabe & Okkenhaug 2013, Kim et al 2018). These approaches are generally combined with local or systematic viral injections to selectively label and image from region- or projection-specific cell types at scale.
The array of existing sensors combined with various imaging modalities provides an expanded technical arsenal for the interrogation of neural circuits, systems, and behaviors. Fiber photometry combined with genetically-encoded sensors enables sub-second recording of calcium and neurochemical dynamics in freely-moving behavior in rodents elucidating neurobiological phenomena underlying innate and learned behaviors albeit lack of the single-cell resolution (Sabatini & Tian 2020). Additionally, photometric recording is suitable for targeting deep subcortical nuclei and densely fasciculated projections where difficult to access with either one-photon or multi-photon microscopy. For example, an adenosine sensor (GRABAdo) and serotonin sensors (GRAB5-HT and iSeroSnFR) have revealed fluctuating dynamics of extracellular adenosine and serotonin, respectively, over the entire course of the sleep-wake cycle in mouse cortical and subcortical regions (Unger et al 2020) (Figure 4a, from (Peng et al 2020)). With fiber photometry, dLight has been used to great effect to understand downstream effects of dopaminergic neuromodulation. Recent work in the field of neural reinforcement learning illustrated a dissociation between dopaminergic VTA spiking and DA release in the NAc using dLight. The authors showed that DA release in the NAc core covaried with reward history and expectations, independently of dopaminergic VTA neuron spiking (Mohebi et al 2019). Other research has revealed positive and negative DA modulation leads to cell-type specific, asynchronous fluctuations in spiny projection neuron PKA levels (Lee et al 2020).
Figure 4: Behavioral and pharmacological applications of NT/NM sensors.
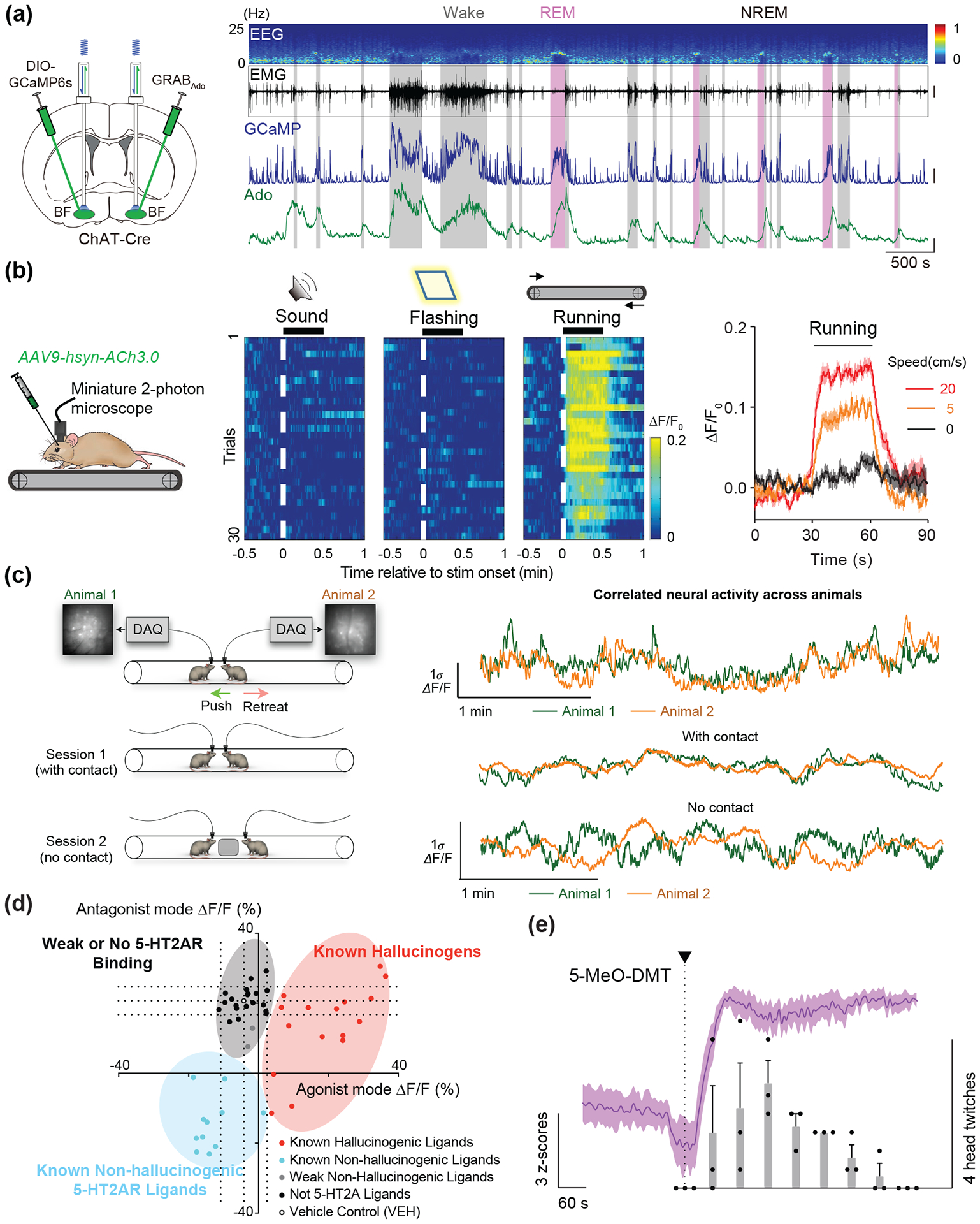
(a) Long-term recording of adenosine release in cholinergic neurons using the GRABAdo sensor. Changes in adenosine-dependent fluorescence can be compared to cholinergic calcium activity and EEG/EMG signal across the full time-course of the sleep/wake cycle (Peng et al 2020). (b) Imaging GRABAch3.0 sensor using miniature 2-photon microscopy in a treadmill task in mice. Single-cell changes in ΔF/F0 response are tractable during different stages of the task, and across running speeds (Jing et al 2020). (c) Simultaneous calcium imaging in dmPFC of two mice during a social interaction test reveals correlations in neural activity during contact vs. no contact sessions (Kingsbury et al 2019). (d) psychLight based characterization of compounds based on 5-HT2AR binding and hallucinogenic potential (Dong, et al., 2021). (e) psychLight tracks in vivo action of 5-MeO-DMT administration over the course of the head-twitch response (Dong et al 2021).
One-photon and multi-photon microscopy combined with NM sensors, on the other hand, provide the cellular or subcellular map of distinct release in response to electrical stimuli or behavior. For example, two-photon imaging of dopamine with a fast dopamine sensor dLight1.3b revealed the rise and fall of local synaptic release within 200ms before the post-synaptic current reaches plateau (Condon et al 2021). Wide-field mesoscopic recordings of GRABACh sensors in the S1 cortex have led to insights on single-cell cholinergic activity within cortical neuron populations in response to systemic pharmacological manipulation (Jing et al 2020). Additionally, single- and multi-photon microendoscopy techniques have been adapted and optimized for freely-moving behavior (Ziv & Ghosh 2015). These adaptations also permit high-resolution imaging in deep brain regions (Zhang et al 2019). Miniature two-photon microscopy has been used to demonstrate visual cortex cholinergic response during a treadmill task in mice using the ACh3.0 sensor (Figure 4b, from (Jing et al 2020)). It is worth mentioning that the intrinsic properties of current NM sensors need to be further optimized to be applied to broad application with all microscopies.
What is particularly exciting about the multitude of ways these sensors can be implemented in vivo is the broad range of novel questions and experimental designs to which they can be applied. A prominent topic in neuro-behavioral research that will benefit from the discussed in vivo imaging methods is social behavior. Social behaviors represent a range of interactions where two or more animals are engaging with each other and relate to a wide array of important and translatable lines of research including mental health topics (Lim & Young 2006). Researchers have previously encountered roadblocks in measuring neural activity during social behaviors due to the freedom of movement required in many social behavior tests. Recent work has begun to take advantage of recording setups that allow unrestrained movement to investigate circuit specific activity during behaviors such as neutral interaction (Gunaydin et al 2014), social defeat (Muir et al 2018) , social isolation (Matthews et al 2016, Mita et al 2019), and social reward (Hung et al 2017). It is even possible to record from multiple animals who are actively interacting and correlate changes in neural activity between animals (Figure 4c, from (Kingsbury et al 2019)). All the examples noted thus far utilize calcium imaging with transgenic Cre lines to ensure cell type specificity. However, the advent of NM and NT sensors allows for specific and meaningful observations of dynamic changes in neurocircuitry to be conducted in model species that do not have transgenic options.
Genetically-encoded sensors can be modified and used across an array of different species, both vertebrate and invertebrate. GRABACh sensors have been validated in Drosophila and mice, and GRABDA has been shown to track dopamine fluctuations in Drosophila, zebrafish, and mice (Jing et al 2020, Sun et al 2018, Sun et al 2020). There are also advantages to using these sensors in wild-type animals, namely that they can serve to better model certain ethology. For example, researchers interested in understanding the neurobiology of paternal behavior may prefer to use mandarin voles as a model, as mandarin voles are one of the few rodent species to show a high level of paternal infant-directed care (Tai & Wang 2001, Tai et al 2001). Mandarin vole researchers used the dLight1 sensor to record dopamine release into nucleus accumbens (NAc) during infant-directed paternal behaviors, allowing for a specificity of real-time, unrestrained, region-specific NT recording without a need for transgenics (He et al 2021). Similarly, the GRABDA was used in zebra finches to monitor dopamine dynamics in the dopaminergic mesocortical circuit during cultural transmission of vocal behavior (Tanaka et al 2018). The use of neurochemical sensors in non-traditional models is still in its infancy but we predict will see a large increase in popularity. Future directions may see a large increase in the diversity of animal models to allow for the examination of behaviors, conditions, or characteristics not available in inbred lab mice or rats. The California mouse, for example, exhibits high levels of female-female aggression (Trainor et al 2011) and sex-specific changes in anxiety behavior (Wright et al 2020), allowing for unique insights into sex differences in anxiety-driven neuromodulation. Forays into sensor application in non-human primates have already begun (Sadakane et al 2015, Seidemann et al 2016), and represent a future direction of research where sensor neuroimaging could be done in complex cortical regions that do not present in rodents. Neurochemical sensors can be applied in cases like these to probe important questions using the most translatable animal model and ethological experimental design.
Spectrally shifted sensors further push the boundaries of in vivo imaging possibilities. Spatiotemporal overlap in neurochemical release, either from synaptic co-release or from converging inputs, is a common phenomenon but one that has been historically difficult to visualize. By using spectrally distinct sensors, researchers can monitor and correlate activity of multiple neurochemicals without sacrificing spatiotemporal resolution. Red-shifted iGluSnFR (Wu et al 2018) and red and yellow-shifted dLight (Patriarchi et al 2020), and red GRABDA (Sun et al 2020) have already been engineered. We recognize the importance of continued efforts to produce an expanded palette of neurochemical sensors and the wide-reaching implications these developments have on the ability to measure interactions and complexities of in vivo neurochemical systems. Additionally, the universally applied sensor engineering methods can also be used to expand the scope of neurochemical sensors. For example, neurolipid indicators have permitted exploration of clinically relevant neurophysiology of endogenous lipid activity. Namely, the GRABeCB2.0 sensor has revealed in vivo endocannabinoid transients across multiple brain regions in response to foot-shock, locomotion, and seizure (Dong et al 2020, Farrell et al 2021).
Application of NM sensors in drug discovery
Besides imaging endogenous NM release dynamics, GPCR-based NM sensors, such as psychLight, have been recently applied to probe specific ligand-induced conformational changes of the 5-HT2A receptor to predict behavioral outcomes. The cell-based high-throughput assays using 5-HT2A now enable early identification of abusive drugs and the development of 5-HT2AR dependent non-hallucinogenic therapeutics at scale.
35% of all Food and Drug Administration (FDA) approved medications bind to GPCRs (Hauser et al 2017). The size, shape, and amino acid composition of the orthosteric binding site is very well suited to designing small synthetic molecules (Shoichet & Kobilka 2012). The ligand-binding specific conformation of the receptor dictates its function and diverse behavior via different conformation-dependent downstream pathways. It is known that there are multiple signaling pathways for GPCRs, and it is possible to bias the signaling of a given GPCR through either a specific G protein or through β-arrestin induced by designer compounds (Smith et al 2018), and the biased agonism on GPCRs has been utilized in pharmacology to reduce the side effects of some designer compounds (Whalen et al 2011). However, the mechanistic action of designer drugs at both molecular and cellular levels is not known. There is a pressing need to develop technologies that can access drugs with addictive potentials at scale at the early stage of drug discovery.
Recently, an open source of 14 optimized bioluminescence resonance energy transfer (BRET) based sensors, TRUPATH, has facilitated the understanding of drug actions on GPCRs (Olsen et al 2020). By attaching a RLuc8 on the optimized location of 14 different Ga unit and GFP2 on Gg units, and BRET can happen only when two fluorophores in the correct proximation and orientation, TRUPATH can specifically indicate the physical association of combinations of Gα, Gβ, Gγ. Before binding of ligands, the G-proteins are in a heterotrimer state. RLuc8 and GFP2 are in a proximity so a BRET signal can be observed. Upon binding, Gα subunit exchanges bound GDP with GTP. The disassociation of Gα unit from the βγ subunit will result in a loss of BRET signal, which indicates when the specific G-proteins that are tagged with FP are being engaged. Thus, one can identify specific transducer complexes that are activated by a specific drug in response to a GPCR of interest. Furthermore, TRUPATH verified that both in wt κOR and chemogenetic κOR Ro1 activated canonical Gai3 and novel GaGustducin transducers to a similar extent, which was the pathway that was previously reported from in vivo experiments (Mueller et al 2005).
On the other hand, the use of GPCR-based sensor is a promising strategy to directly monitor the specific receptor conformational changes induced by ligand instead of secondary signaling molecules. 5-HT2A receptor is the major target of psychedelics which have shown promising antidepressant effect albeit severe hallucinogenic effect (Chi & Gold 2020). Recent research has shown that the hallucination effect is not necessary for treating depression (Cameron et al 2021). Thus, it is imperative to screen novel compounds for 5-HT2AR that do not trigger hallucination but still retain therapeutic effects. To do so, psychLight was engineered by coupling the fluorescence changes of a cpGFP to ligand-binding induced conformational changes of the 5-HT2A receptor (Dong et al 2021). psychLight can categorize compounds into weak or no 5-HT2AR binding, hallucinogens, and non-hallucinogens by real-time fluorescent read out (Figure 4d). One of the novel compounds AAZ-A-154 was discovered from the assay as a non-hallucinogen. The non-hallucinogenic property of AAZ-A-154 was then validated in vivo with head-twitch response (HTR) experiment in mice. With forced swim test and glucose preference test in genetic model of depression, AAZ-A-154 was shown to be a non-hallucinogenic psychedelic analog that exhibits anti-depressant properties.
The genetically encoded approach to design sensors also has advantages of studying the action of drug on receptor in vivo. Together with fiber photometry recording, psychLight signals are able to correlate 5-MeO-DMT action in mPFC during the HTR experiment. An intraperitoneal (IP) injection of 5-MeO-DMT takes about a minute to reach mPFC and initiate head twitching (Figure 4e). With GPCR-based sensors directly monitoring drug actions in vivo, PBP-based sensors can have a major role in neuropharmacology as well. For example, monitoring NM release induced by exogeneous drugs.
Summary and outlook
The development of genetically-encoded sensors has opened doors in neuroscience research that were previously untenable, particularly in the context of understanding neural circuit bases of complex, naturalistic behavior. By reducing restrictions on animal model and experimental design, these sensors allow for the broadening of circuits- and systems-level neuroscience to encompass a larger array of questions and permit unprecedented exploration of neurobehavioral systems in real-time.
To date, there are a vast amount of NT/NMs lacking precise detection tools, including neuropeptides and lipid transmitters. By taking advantage of naturally designed GPCRs, a similar strategy to GRAB sensor and Light sensor development pipeline can be generalized to cover more neurochemicals, which will greatly facilitate studies revealing their emergent roles under physiological and pathological conditions. In addition, the intrinsic properties of existing sensors demand further iterative optimization to be broadly applied with all microscopy techniques. The model we reported here provides theoretical guidance for future optimization. To achieve these goals, precise rational design aided by computational approach is expected for broad use, including machine leaning, as applied in iSeroSnFR (Unger et al 2020) or the recently developed AlphaFold 2.0 for protein structure prediction with unprecedented accuracy (Jumper et al 2021, Tunyasuvunakool et al 2021). A high-throughput screening system is crucial to maximize sensor optimization efficiency, since GPCR-based sensors can currently only be screened in mammalian cells. Fluorescence-activating cell sorting (FACS) combined with image-based screening or the well-known CRISPR/Cas9 system that has been applied in evolution of fluorescent proteins and voltage sensors (Erdogan et al 2020, Lee et al 2020, Piatkevich et al 2018) hold great promise for screening of NT/NM sensors.
Extending the sensitivity, specificity, and color palette of neuromodulator sensors will continue to create rich opportunities for minimally invasive, multiplex imaging of neurochemical signaling dynamics. Together with advanced microscopy, opsin-based optogenetics and cell atlas, fluorescent imaging with next generation genetically encoded indicators will bring our experimental capabilities to previously impossible level, thus uncovering brain mechanisms, and potentially transform our understanding of how neurochemical inputs influence cell and circuit function in health and disease.
Acknowledgements
The authors would like to thank Alessio Andreoni for his critiques and helpful comments on modeling.
Footnotes
Disclosure statements
LT is a co-founder of Seven Biosciences. All other authors have nothing to declare.
Literature cited
- Abdelfattah AS, Kawashima T, Singh A, Novak O, Liu H, et al. 2019. Bright and photostable chemigenetic indicators for extended in vivo voltage imaging. Science 365: 699–704 [DOI] [PubMed] [Google Scholar]
- Akerboom J, Carreras Calderon N, Tian L, Wabnig S, Prigge M, et al. 2013. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front Mol Neurosci 6: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreoni A, Davis CMO, Tian L. 2019. Measuring brain chemistry using genetically encoded fluorescent sensors. Current Opinion in Biomedical Engineering 12: 59–67 [Google Scholar]
- Baird GS, Zacharias DA, Tsien RY. 1999. Circular permutation and receptor insertion within green fluorescent proteins. Proc Natl Acad Sci U S A 96: 11241–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden PM, Zhang P, Shivange AV, Marvin JS, Cichon J, et al. 2020. A fast genetically encoded fluorescent sensor for faithful in vivo acetylcholine detection in mice, fish, worms and flies. bioRxiv: 2020.02.07.939504 [Google Scholar]
- Bouabe H, Okkenhaug K. 2013. Gene Targeting in Mice: a Review. Methods Mol Biol 1064: 315–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broussard GJ, Liang Y, Fridman M, Unger EK, Meng G, et al. 2018. In vivo measurement of afferent activity with axon-specific calcium imaging. Nat Neurosci 21: 1272–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron LP, Tombari RJ, Lu J, Pell AJ, Hurley ZQ, et al. 2021. A non-hallucinogenic psychedelic analogue with therapeutic potential. Nature 589: 474–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, et al. 2013. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499: 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi T, Gold JA. 2020. A review of emerging therapeutic potential of psychedelic drugs in the treatment of psychiatric illnesses. J Neurol Sci 411: 116715. [DOI] [PubMed] [Google Scholar]
- Condon AF, Robinson BG, Asad N, Dore TM, Tian L, Williams JT. 2021. The residence of synaptically released dopamine on D2 autoreceptors. Cell Reports 36: 109465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalangin R, Drobizhev M, Molina RS, Aggarwal A, Patel R, et al. 2020. Far-red fluorescent genetically encoded calcium ion indicators. bioRxiv: 2020.11.12.380089 [Google Scholar]
- Dana H, Mohar B, Sun Y, Narayan S, Gordus A, et al. 2016. Sensitive red protein calcium indicators for imaging neural activity. eLife 5: e12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H, Sun Y, Mohar B, Hulse BK, Kerlin AM, et al. 2019. High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat Methods 16: 649–57 [DOI] [PubMed] [Google Scholar]
- Dong A, He K, Dudok B, Farrell JS, Guan W, et al. 2020. A fluorescent sensor for spatiotemporally resolved endocannabinoid dynamics in vitro and in vivo. preprint Rep., Neuroscience [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Ly C, Dunlap LE, Vargas MV, Sun J, et al. 2021. Psychedelic-inspired drug discovery using an engineered biosensor. Cell 184: 2779–92.e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Husseini Ael D, Craven SE, Brock SC, Bredt DS. 2001. Polarized targeting of peripheral membrane proteins in neurons. J Biol Chem 276: 44984–92 [DOI] [PubMed] [Google Scholar]
- Erdogan M, Fabritius A, Basquin J, Griesbeck O. 2020. Targeted In Situ Protein Diversification and Intra-organelle Validation in Mammalian Cells. Cell Chem Biol 27: 610–21 e5 [DOI] [PubMed] [Google Scholar]
- Farrell JS, Colangeli R, Dong A, George AG, Addo-Osafo K, et al. 2021. In vivo endocannabinoid dynamics at the timescale of physiological and pathological neural activity. Neuron 109: 2398–403.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Zhang C, Lischinsky JE, Jing M, Zhou J, et al. 2019. A Genetically Encoded Fluorescent Sensor for Rapid and Specific In Vivo Detection of Norepinephrine. Neuron 102: 745–61.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Huang C, Li JZ, Grewe BF, Zhang Y, et al. 2015. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science 350: 1361–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienberger C, Konnerth A. 2012. Imaging Calcium in Neurons. Neuron 73: 862–85 [DOI] [PubMed] [Google Scholar]
- Guillaumin MCC, Burdakov D. 2021. Neuropeptides as Primary Mediators of Brain Circuit Connectivity. Front. Neurosci 0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaydin LA, Grosenick L, Finkelstein JC, Kauvar IV, Fenno LE, et al. 2014. Natural neural projection dynamics underlying social behavior. Cell 157: 1535–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser AS, Attwood MM, Rask-Andersen M, Schioth HB, Gloriam DE. 2017. Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov 16: 829–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Zhang L, Hou W, Zhang X, Young LJ, et al. 2021. Paraventricular Nucleus Oxytocin Subsystems Promote Active Paternal Behaviors in Mandarin Voles. J. Neurosci 41: 6699–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helassa N, Durst CD, Coates C, Kerruth S, Arif U, et al. 2018. Ultrafast glutamate sensors resolve high-frequency release at Schaffer collateral synapses. Proc Natl Acad Sci U S A 115: 5594–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Wei Y, Wang D, Su N, Chen X, et al. 2018. Glucose monitoring in living cells with single fluorescent protein-based sensors. RSC Advances 8: 2485–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung LW, Neuner S, Polepalli JS, Beier KT, Wright M, et al. 2017. Gating of social reward by oxytocin in the ventral tegmental area. Science 357: 1406–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M, Takeuchi A, Manita S, Horigane SI, Sakamoto M, et al. 2019. Rational Engineering of XCaMPs, a Multicolor GECI Suite for In Vivo Imaging of Complex Brain Circuit Dynamics. Cell 177: 1346–60 e24 [DOI] [PubMed] [Google Scholar]
- Jin L, Han Z, Platisa J, Wooltorton JRA, Cohen LB, Pieribone VA. 2012. Single action potentials and subthreshold electrical events imaged in neurons with a fluorescent protein voltage probe. Neuron 75: 779–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing M, Li Y, Zeng J, Huang P, Skirzewski M, et al. 2020. An optimized acetylcholine sensor for monitoring in vivo cholinergic activity. Nature Methods 17: 1139–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing M, Zhang P, Wang G, Feng J, Mesik L, et al. 2018. A genetically encoded fluorescent acetylcholine indicator for in vitro and in vivo studies. Nat Biotechnol 36: 726–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, et al. 2021. Highly accurate protein structure prediction with AlphaFold. Nature [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller JP, Marvin JS, Lacin H, Lemon WC, Shea J, et al. 2021. In vivo glucose imaging in multiple model organisms with an engineered single-wavelength sensor. Cell Rep 35: 109284. [DOI] [PubMed] [Google Scholar]
- Kelly CR, Sharif NA. 2006. Pharmacological evidence for a functional serotonin-2B receptor in a human uterine smooth muscle cell line. J Pharmacol Exp Ther 317: 1254–61 [DOI] [PubMed] [Google Scholar]
- Kim H, Kim M, Im S-K, Fang S. 2018. Mouse Cre-LoxP system: general principles to determine tissue-specific roles of target genes. Lab Anim Res 34: 147–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury L, Huang S, Wang J, Gu K, Golshani P, et al. 2019. Correlated Neural Activity and Encoding of Behavior across Brains of Socially Interacting Animals. Cell 178: 429–46.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopfel T, Song C. 2019. Optical voltage imaging in neurons: moving from technology development to practical tool. Nat Rev Neurosci 20: 719–27 [DOI] [PubMed] [Google Scholar]
- Kovacs GL. 2004. The Endocrine Brain: Pathophysiological Role of Neuropeptide-Neurotransmitter Interactions. EJIFCC 15: 107–12 [PMC free article] [PubMed] [Google Scholar]
- Kralj JM, Douglass AD, Hochbaum DR, Maclaurin D, Cohen AE. 2011. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat Methods 9: 90–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kügler S, Kilic E, Bähr M. 2003. Human synapsin 1 gene promoter confers highly neuron-specific long-term transgene expression from an adenoviral vector in the adult rat brain depending on the transduced area. Gene Ther 10: 337–47 [DOI] [PubMed] [Google Scholar]
- Lee J, Liu Z, Suzuki PH, Ahrens JF, Lai S, et al. 2020. Versatile phenotype-activated cell sorting. Science Advances 6: eabb7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim MM, Young LJ. 2006. Neuropeptidergic regulation of affiliative behavior and social bonding in animals. Horm Behav 50: 506–17 [DOI] [PubMed] [Google Scholar]
- Lobas MA, Tao R, Nagai J, Kronschlager MT, Borden PM, et al. 2019. A genetically encoded single-wavelength sensor for imaging cytosolic and cell surface ATP. Nat Commun 10: 711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank M, Santos AF, Direnberger S, Mrsic-Flogel TD, Hofer SB, et al. 2008. A genetically encoded calcium indicator for chronic in vivo two-photon imaging. Nat Methods 5: 805–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin JS, Borghuis BG, Tian L, Cichon J, Harnett MT, et al. 2013. An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat Methods 10: 162–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin JS, Scholl B, Wilson DE, Podgorski K, Kazemipour A, et al. 2018. Stability, affinity, and chromatic variants of the glutamate sensor iGluSnFR. Nat Methods 15: 936–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin JS, Shimoda Y, Magloire V, Leite M, Kawashima T, et al. 2019. A genetically encoded fluorescent sensor for in vivo imaging of GABA. Nat Methods 16: 763–70 [DOI] [PubMed] [Google Scholar]
- Matthews T, Danese A, Wertz J, Odgers CL, Ambler A, et al. 2016. Social isolation, loneliness and depression in young adulthood: a behavioural genetic analysis. Soc Psychiatry Psychiatr Epidemiol 51: 339–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita M, Ito M, Harada K, Sugawara I, Ueda H, et al. 2019. Green Fluorescent Protein-Based Glucose Indicators Report Glucose Dynamics in Living Cells. Anal Chem 91: 4821–30 [DOI] [PubMed] [Google Scholar]
- Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, et al. 1997. Fluorescent indicators for Ca2+based on green fluorescent proteins and calmodulin. Nature 388: 882–87 [DOI] [PubMed] [Google Scholar]
- Mohebi A, Pettibone JR, Hamid AA, Wong J-MT, Vinson LT, et al. 2019. Dissociable dopamine dynamics for learning and motivation. Nature 570: 65–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motulsky HJ, Neubig RR. 2010. Analyzing Binding Data. Current Protocols in Neuroscience 52: 7.5.1–7.5.65 [DOI] [PubMed] [Google Scholar]
- Mueller KL, Hoon MA, Erlenbach I, Chandrashekar J, Zuker CS, Ryba NJP. 2005. The receptors and coding logic for bitter taste. Nature 434: 225–29 [DOI] [PubMed] [Google Scholar]
- Muir J, Lorsch ZS, Ramakrishnan C, Deisseroth K, Nestler EJ, et al. 2018. In Vivo Fiber Photometry Reveals Signature of Future Stress Susceptibility in Nucleus Accumbens. Neuropsychopharmacology 43: 255–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadim F, Bucher D. 2014. Neuromodulation of Neurons and Synapses. Current Opinion in Neurobiology 0: 48–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Sawano A, Park ES, Miyawaki A. 2001. Circularly permuted green fluorescent proteins engineered to sense Ca2+. Proc Natl Acad Sci U S A 98: 3197–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. 2004. Expanded dynamic range of fluorescent indicators for Ca(2+) by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci U S A 101: 10554–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai J, Ohkura M, Imoto K. 2001. A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nat Biotechnol 19: 137–41 [DOI] [PubMed] [Google Scholar]
- Nieuwenhuis B, Haenzi B, Hilton S, Carnicer-Lombarte A, Hobo B, et al. 2021. Optimization of adeno-associated viral vector-mediated transduction of the corticospinal tract: comparison of four promoters. Gene Ther 28: 56–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen RHJ, DiBerto JF, English JG, Glaudin AM, Krumm BE, et al. 2020. TRUPATH, an open-source biosensor platform for interrogating the GPCR transducerome. Nat Chem Biol 16: 841–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal A, Tian L. 2020. Imaging voltage and brain chemistry with genetically encoded sensors and modulators. Curr Opin Chem Biol 57: 166–76 [DOI] [PubMed] [Google Scholar]
- Palmer AE, Giacomello M, Kortemme T, Hires SA, Lev-Ram V, et al. 2006. Ca2+ indicators based on computationally redesigned calmodulin-peptide pairs. Chem Biol 13: 521–30 [DOI] [PubMed] [Google Scholar]
- Panzera LC, Hoppa MB. 2019. Genetically Encoded Voltage Indicators Are Illuminating Subcellular Physiology of the Axon. Front Cell Neurosci 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patriarchi T, Cho JR, Merten K, Howe MW, Marley A, et al. 2018. Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science 360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patriarchi T, Mohebi A, Sun J, Marley A, Liang R, et al. 2020. An expanded palette of dopamine sensors for multiplex imaging in vivo. Nature Methods 17: 1147–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W, Wu Z, Song K, Zhang S, Li Y, Xu M. 2020. Regulation of sleep homeostasis mediator adenosine by basal forebrain glutamatergic neurons. Science 369 [DOI] [PubMed] [Google Scholar]
- Piatkevich KD, Jung EE, Straub C, Linghu C, Park D, et al. 2018. A robotic multidimensional directed evolution approach applied to fluorescent voltage reporters. Nature Chemical Biology 14: 352–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Cosio DMO, Piatkevich KD, Aufmkolk S, Su WC, et al. 2020. Improved genetically encoded near-infrared fluorescent calcium ion indicators for in vivo imaging. PLoS Biol 18: e3000965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Piatkevich KD, Mc Larney B, Abdelfattah AS, Mehta S, et al. 2019. A genetically encoded near-infrared fluorescent calcium ion indicator. Nat Methods 16: 171–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiocho FA, Ledvina PS. 1996. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol Microbiol 20: 17–25 [DOI] [PubMed] [Google Scholar]
- Rasmussen M, Kong L, Zhang G-r, Liu M, Wang X, et al. 2007. Glutamatergic or GABAergic neuron-specific, long-term expression in neocortical neurons from helper virus-free HSV-1 vectors containing the phosphate-activated glutaminase, vesicular glutamate transporter-1, or glutamic acid decarboxylase promoter. Brain Research 1144: 19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini BL, Tian L. 2020. Imaging Neurotransmitter and Neuromodulator Dynamics In Vivo with Genetically Encoded Indicators. Neuron 108: 17–32 [DOI] [PubMed] [Google Scholar]
- Sadakane O, Masamizu Y, Watakabe A, Terada S-I, Ohtsuka M, et al. 2015. Long-Term Two-Photon Calcium Imaging of Neuronal Populations with Subcellular Resolution in Adult Non-human Primates. Cell Reports 13: 1989–99 [DOI] [PubMed] [Google Scholar]
- Seidemann E, Chen Y, Bai Y, Chen SC, Mehta P, et al. 2016. Calcium imaging with genetically encoded indicators in behaving primates. eLife 5: e16178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova DM, Shemetov AA, Kaberniuk AA, Verkhusha VV. 2015. Natural photoreceptors as a source of fluorescent proteins, biosensors, and optogenetic tools. Annu Rev Biochem 84: 519–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemetov AA, Monakhov MV, Zhang Q, Canton-Josh JE, Kumar M, et al. 2021. A near-infrared genetically encoded calcium indicator for in vivo imaging. Nat Biotechnol 39: 368–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Dana H, Abdelfattah AS, Patel R, Shea J, et al. 2018. A genetically encoded Ca(2+) indicator based on circularly permutated sea anemone red fluorescent protein eqFP578. BMC Biol 16: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivange AV, Borden PM, Muthusamy AK, Nichols AL, Bera K, et al. 2019. Determining the pharmacokinetics of nicotinic drugs in the endoplasmic reticulum using biosensors. Journal of General Physiology 151: 738–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoichet BK, Kobilka BK. 2012. Structure-based drug screening for G-protein-coupled receptors. Trends Pharmacol Sci 33: 268–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Lefkowitz RJ, Rajagopal S. 2018. Biased signalling: from simple switches to allosteric microprocessors. Nat Rev Drug Discov 17: 243–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre F, Marshall JD, Yang Y, Gong Y, Schnitzer MJ, Lin MZ. 2014. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat Neurosci 17: 884–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Zeng J, Jing M, Zhou J, Feng J, et al. 2018. A Genetically Encoded Fluorescent Sensor Enables Rapid and Specific Detection of Dopamine in Flies, Fish, and Mice. Cell 174: 481–96 e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Zhou J, Dai B, Qian T, Zeng J, et al. 2020. Next-generation GRAB sensors for monitoring dopaminergic activity in vivo. Nature Methods 17: 1156–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai F, Wang TZ. 2001. Social organization of mandarin voles in burrow system. Acta Theriol Sin 21: 50–56 [Google Scholar]
- Tai F, Wang TZ, Zhao YJ. 2001. Mating system of mandarin vole (Microtus mandarinus). Acta Zoologica Sinica 47: 260–67 [Google Scholar]
- Tanaka M, Sun F, Li Y, Mooney R. 2018. A mesocortical dopamine circuit enables the cultural transmission of vocal behaviour. Nature 563: 117–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Hires SA, Mao T, Huber D, Chiappe ME, et al. 2009. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nature Methods 6: 875–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonova IG, Costanzi S. 2009. Unraveling the structure and function of G protein-coupled receptors through NMR spectroscopy. Curr Pharm Des 15: 4003–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor BC, Pride MC, Landeros RV, Knoblauch NW, Takahashi EY, et al. 2011. Sex Differences in Social Interaction Behavior Following Social Defeat Stress in the Monogamous California Mouse (Peromyscus californicus). PLOS ONE 6: e17405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunyasuvunakool K, Adler J, Wu Z, Green T, Zielinski M, et al. 2021. Highly accurate protein structure prediction for the human proteome. Nature [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger EK, Keller JP, Altermatt M, Liang R, Matsui A, et al. 2020. Directed Evolution of a Selective and Sensitive Serotonin Sensor via Machine Learning. Cell 183: 1986–2002.e26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villette V, Chavarha M, Dimov IK, Bradley J, Pradhan L, et al. 2019. Ultrafast Two-Photon Imaging of a High-Gain Voltage Indicator in Awake Behaving Mice. Cell 179: 1590–608 e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Peng W, Li X, Qian T, Song K, et al. 2021. A genetically encoded sensor for measuring serotonin dynamics. Nat Neurosci 24: 746–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Jing M, Li Y. 2018. Lighting up the brain: genetically encoded fluorescent sensors for imaging neurotransmitters and neuromodulators. Curr Opin Neurobiol 50: 171–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Kim CK, Ting AY. 2019. Molecular tools for imaging and recording neuronal activity. Nature Chemical Biology 15: 101–10 [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang C, Szábo G, Sun Q-Q. 2013. Distribution of CaMKIIα expression in the brain in vivo, studied by CaMKIIα-GFP mice. Brain Research 1518: 9–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen EJ, Rajagopal S, Lefkowitz RJ. 2011. Therapeutic potential of beta-arrestin- and G protein-biased agonists. Trends Mol Med 17: 126–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright EC, Hostinar CE, Trainor BC. 2020. Anxious to see you: Neuroendocrine mechanisms of social vigilance and anxiety during adolescence. The European journal of neuroscience 52: 2516–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Abdelfattah AS, Zhou H, Ruangkittisakul A, Qian Y, et al. 2018. Genetically Encoded Glutamate Indicators with Altered Color and Topology. ACS Chem Biol 13: 1832–37 [DOI] [PubMed] [Google Scholar]
- Wu Z, He K, Chen Y, Li H, Pan S, et al. 2021. An ultrasensitive GRAB sensor for detecting extracellular ATP in vitro and in vivo. bioRxiv: 2021.02.24.432680 [Google Scholar]
- Yang HH, St-Pierre F, Sun X, Ding X, Lin MZ, Clandinin TR. 2016. Subcellular Imaging of Voltage and Calcium Signals Reveals Neural Processing In Vivo. Cell 166: 245–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Zhao Q, Wu B. 2015. Structural Studies of G Protein-Coupled Receptors. Mol Cells 38: 836–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Liang B, Barbera G, Hawes S, Zhang Y, et al. 2019. Miniscope GRIN Lens System for Calcium Imaging of Neuronal Activity from Deep Brain Structures in Behaving Animals. Current Protocols in Neuroscience 86: e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Araki S, Wu J, Teramoto T, Chang YF, et al. 2011. An expanded palette of genetically encoded Ca(2)(+) indicators. Science 333: 1888–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Ghosh KK. 2015. Miniature microscopes for large-scale imaging of neuronal activity in freely behaving rodents. Current Opinion in Neurobiology 32: 141–47 [DOI] [PubMed] [Google Scholar]


