Abstract
Antibody “ultralong” third heavy chain complementarity-determining regions (CDR H3) appear unique to bovine antibodies and may enable binding to difficult epitopes that shorter CDR H3 regions cannot easily access. Diversity is concentrated in the “knob” domain of the CDR H3, which is encoded by the DH gene segment and sits atop a β-ribbon “stalk” that protrudes far from the antibody surface. Knob region cysteine content is quite diverse in terms of total number of cysteines, sequence position, and disulfide bond pattern formation. We investigated the role of germline cysteines in production of a diverse CDR H3 structural repertoire. The relationship between DH polymorphisms and deletions relative to germline at the nucleotide level, as well as diversity in cysteine and disulfide bond content at the structural level, were ascertained. Structural diversity is formed through (i) DH polymorphisms with altered cysteine positions, (ii) DH deletions and (iii) new cysteines that arise through somatic hypermutation (SH) that form new, unique disulfide bonds to alter the knob structure. Thus, a combination of mechanisms at both the germline and somatic immunogenetic levels result in diversity in knob region cysteine content, contributing to remarkable complexity in knob region disulfide patterns, loops, and antigen binding surface.
Introduction
The structural diversity of antibodies, which for nearly all species occurs within the immunoglobulin (Ig) scaffold, enables the vertebrate adaptive immune system to neutralize a myriad of foreign antigens. The heavy chain third heavy chain complementarity-determining region (CDR H3) is encoded by rearranged variable (VH), diversity (DH), and joining (JH) gene segments and is the most diverse part of the antibody molecule (1–5). The length of the CDR H3 loop varies in different species and is usually important in antigen binding (6). The antigen binding site (paratope) of most human antibodies is flat or undulating, with CDR H3s typically ranging from 8–16 amino acids in length (5, 7, 8). The ability to bind certain classes of antigens, including viral spike and other glycoproteins, has been attributed to longer and protruding CDR H3 structures on human antibodies (9–14). Bovine antibodies have much longer CDR H3 regions than any other species examined, with a typical average length of over 23 amino acids. Remarkably, approximately 10% of Bos taurus antibodies, in all isotypes, have exceptionally long CDR H3 regions that are 40 to 70 amino acids in length (1, 5, 15–18). These ultralong CDR H3s may enable binding to concave or cryptic epitopes that antigen binding sites with shorter CDR H3s cannot easily access. Cows are the only vertebrates studied that can mount a rapid broadly neutralizing response against the engineered HIV gp140 SOSIP Env timer, and several monoclonal antibodies with broadly neutralizing activity had ultralong CDR H3s (19). The ultralong CDR H3 of one such antibody, NC-Cow1, has a unique ability to navigate through the glycan shield of the HIV spike to reach the cryptic CD4 binding site (16, 19, 20). Cows, therefore, have an unusual humoral immune system characterized by antibodies with ultralong CDR H3 regions that may have unique protective properties against certain antigens.
Compared to other species, the bovine antibody repertoire is limited in terms of the number of germline genetic components, and thus has a lower potential for combinatorial diversity (5, 15, 21–23). The functional gene segments of the bovine antibody repertoire appear to include only 12 VH, 23 DH, and 2 JH at the heavy chain locus, 25 Vλ and 3 Jλ at the λ locus and 8 Vκ, and 3 Jκ at the κ locus (5, 21, 24, 25). There is even less potential for combinatorial diversity in bovine heavy chains with ultralong CDR H3s, as all antibodies in this subset appear to use the same germline VH, DH, and JH gene segments: IGHV1–7, IGHD8–2, IGHJ2–4 (15, 16, 21–24). The single very long DH gene segment has two polymorphic alleles: IGHD8–2*01 and IGHD8–2*02 (22, 25, 26). The frequencies of these alleles in the bovine population and their relevance to ultralong CDR H3 diversity has not been explored. Thus, there are severe limitations in germline potential diversity for ultralong CDR H3 antibodies, however polymorphic DH regions could contribute to diversity across the Bos taurus population or specifically in heterozygote animals.
Conventionally, antibody repertoire diversity is generated through V(D)J recombination (including junctional insertions and/ or deletions) prior to antigen exposure, and somatic hypermutation (SH) after antigen exposure (3, 4, 15, 27). The limited potential for combinatorial diversity of bovine antibodies, particularly the subset with ultralong CDR H3s, suggests that diversity of the bovine antibody repertoire is primarily achieved through SH and possibly additional unknown mechanisms. Several species, including cows, however, activate SH prior to antigen exposure, thus allowing for further diversification of the primary repertoire (28, 29). Unlike antibodies from other species (and bovine antibodies with shorter CDR H3s), mature ultralong CDR H3 antibodies have low amino acid variability in the CDR H1 and CDR H2 regions (15, 21). Diversity in the bovine ultralong antibody repertoire is concentrated in the CDR H3 region which is largely encoded by the DH gene (15, 21). Studies of the knob domain have suggested that the CDR H3 may be the only CDR used for antigen recognition by bovine antibodies with ultralong CDR H3s (19, 20, 30–32). This suggests that the CDR H1, H2, L1, L2, and L3 of ultralong antibodies may not interact with antigen and function primarily to support and stabilize the CDR H3. Indeed, CDR H3s can be transplanted between antibodies and retain function (19), and knobs can be cleaved from the antibody and still bind antigen (32). Thus, both the binding and diversity properties of ultralong CDR H3 antibodies appear to reside within the ultralong CDR H3, as opposed to the rest of the molecule.
Crystal structures have revealed a conserved structural paradigm for bovine ultralong CDR H3s; they are comprised of a β-ribbon “stalk” upon which sits a disulfide-bonded “knob” mini-domain (1, 5, 16, 33). The extended “stalk” protrudes from the typical antibody surface and is formed by two antiparallel β-strands which can vary in length. The “knob” domain contains disulfide bonded cysteine residues and three short anti-parallel β-strands at its core (1, 5, 16). A CTTVHQ motif encoded by the 3’ end of IGHV1–7 initiates the ascending β-strand of the stalk, with junctional diversity residues comprising the rest of the ascending β-strand (5, 16, 21). The DH gene segment encodes the knob region and a portion of the stalk’s descending β-strand, which usually contains alternating stacking aromatic residues. The C terminal end of the descending β-strand is encoded by IGHJ2–4 (5, 16, 21). The underlying genetic features encoding the structural architecture are established, but diversity generating mechanisms that alter cysteine diversity and therefore the disulfide bond patterns of the knob have yet to be deciphered in detail.
The CDR H3 knob regions are quite diverse in amino acid sequence, shape, orientation, and disulfide patterns (1, 5, 16). While the presence of 3 very short anti-parallel β-strands appears conserved in knob regions based on available crystal structures, there is dramatic variability in amino acid sequence as well as overall sequence length (1, 5, 16). Knob regions typically have an even number of cysteines, and the sequence position of the first cysteine is nearly universally conserved (1, 5, 16). Otherwise, knob region cysteines are diverse in terms of their positions and participation in disulfide bond patterns (1, 5, 16). The IGHD8–2*01 sequence contains 19 RGYW/WRCY SH hotspots, recognized by activation-induced cytidine deaminase (AID), and 38 out of 48 amino acid codons can mutate to a cysteine with a single nucleotide point mutation (15). IGHD8–2*01 was observed to have a high frequency of internal deletions that altered cysteine content, with in-frame deletions surviving clonal selection (21). With the high density of RGYW/WRCY motifs in the DH region, over 96% of these deletions overlapped an AID hotspot (21). This data suggests that the ability to genetically alter cysteine content and positions would have significant impact on the knob structure by altering disulfide-bonded loops.
A key mechanism for generating knob region structural diversity appears to involve changes in cysteine position and disulfide bond pattern that arise through SH by means of point mutations to form cysteine codons, and internal deletions that alter cysteine content or position (1, 16, 21). In this regard, the relationship between cysteine location, relative to the germline at the sequence level, and disulfide bond pattern on the structural level, has not been fully investigated. Additionally, differences in knob structure resulting from the usage of each DH allele have not been examined. Furthermore, the impact of nucleotide deletions on ultralong CDR H3 structures, and particularly cysteine position, is unknown. A more thorough understanding of how changes in ultralong DH region cysteine content at the sequence level result in certain disulfide bonding patterns will be useful for understanding repertoire generation and development as well as for antibody engineering applications, such as knob region rational design or knob peptide molecular evolution. We analyzed the sequences and crystal structures of seven ultralong CDR H3s to determine germline DH allele identity, location of nucleotide deletions (where applicable) and compared disulfide bond patterns and DH region cysteine locations relative to germline cysteines. We sought to identify conserved cysteines and disulfide bond patterns on both sequence and structural levels. Our analysis revealed that the DH germline allele of five of the antibodies is IGHD8–2*02, including three antibodies with DH deletions, and IGHD8–2*01 for two of the antibodies. We found that allele usage and DH deletions, in conjunction with SH, contribute to diversity in DH region cysteine distribution and disulfide bond connectivity. Further we have analyzed heavy chain deep sequence data to determine germline cysteine conservation, and somatically generated cysteine frequency. While several germline cysteines are typically conserved on each antibody, new cysteines arise through SH and form unique disulfide bond connectivity patterns.
Materials and Methods
Data Sources.
The variable regions encoding seven ultralong CDR H3 antibodies that have published crystal structures were obtained from the Protein Data Bank: A01 (PDB: 5ILT), B11 (PDB: 5IHU), BLV1H12 (PDB: 4K3D), BLV5B8 (PDB: 4K3E), E03 (PDB: 5IJV), Bov6 (PDB: 6E9Q) and NC-Cow1 (PDB: 6OO0). The germline gene segments IGHV1–7, IGHD8–2*01 or IGHD8–2*02, and IGHJ2–4 were from IMGT (https://www.imgt.org/, accession KT723008).
Alignments and DH analysis.
The CLUSTALW tool (https://www.genome.jp/tools-bin/clustalw) was used to align an in silico rearranged germline VDJ sequence pairwise with the DNA sequences. For the in silico rearranged sequences, coding and in-frame nucleotide sequences of IGHV1–7, IGHD8–2*01 or IGHD8–2*02, and IGHJ2–4 were assembled to create two electronic VDJ recombined germline sequences, one for each DH allele. Since V-D and D-J junctional regions do not exist in the germline, we added the VH-DH junctional nucleotides of each mature antibody gene to produce an identical VH-DH junction for the in silico rearranged germline sequence for each pairwise alignment. The slow/accurate alignment parameter and CLUSTAL output format was applied for all alignments. The weight matrix used was IUB for nucleotide alignments and BLOSUM for amino acid alignments.
The nucleotide and amino acid sequences of the two DH alleles, IGHD8–2*01 and IGHD8–2*02, were aligned to identify differences that may be reflected in the sequences of antibodies that use each allele. This analysis included mapping RGYW and WRCY SH hotspots recognized by AID and nucleotides that can form a cysteine codon with a single mutation. The most probable germline DH allele used by each antibody was assigned based on comparison of the alignment score and number of gaps in pairwise nucleotide alignments with each electronic germline sequence at each set of gap penalty values tested. Gap open penalty values ranged from 15–50 and gap extension penalty values ranged from 1–30 (Supplemental Table 1). The electronic germline DH allele with a consistently higher alignment score and lower number of gaps, at each set of gap penalty values, was designated as the most probable DH allele used by the aligned antibody gene. The most probable location of DH region deletions on each antibody, relative to the germline DH of each, was assessed. We assumed a single deletion event was more biologically probable than multiple, so the deletion location was determined based on alignments with gap penalty values that resulted in a single gap in the alignment.
Cysteine positions and disulfide bonding patterns within the DH region of each antibody, with respect to its corresponding germline DH allele, were analyzed at the sequence and structural levels. First, antibodies with DH regions equivalent in length to each germline DH allele were grouped by allele usage and the two groups were aligned by amino acid sequence to each electronically rearranged germline sequence separately. In another analysis, amino acid sequences of antibodies with DH region deletions were separately aligned with the corresponding germline DH sequence and with the DH sequence of another antibody that served as a germline surrogate. The germline surrogate served to compare changes in cysteine positions and disulfide bonding patterns on the structural level since structural data is not available for any germline ultralong CDR H3 antibodies. Antibodies with available crystal structure data that are most homologous in sequence to each DH allele were selected as structural surrogates for each germline DH allele. The germline structural surrogates selected were BLV1H12 for IGHD8–2*01 and B11 for IGHD8–2*02. For each structural analysis, cysteines in the mature antibody that align with germline cysteines were indicated on the amino acid alignment, primary and secondary structure topology diagram, and crystal structure. Primary and secondary structure topology diagrams illustrate the β-strands and connecting loops [as defined by (Stanfield et al. 2016)]. DH region deletion locations, where applicable, are indicated on each germline surrogate and defined as deletion locations relative to the germline DH at the corresponding sequence positions on the surrogate. Variables evaluated in the structural analyses include cysteine position and distribution on primary and secondary structures, disulfide bonding pattern, and spatial distribution of cysteines on crystal structures.
NGS analysis and allele determination.
Next generation sequence analysis of 204 cow VH regions was performed according to Safonova et.al. (34). Only ultralong CDR H3s were used for the NGS analysis. Ultralong CDR H3s were defined as CDR H3s exceeding 150 nt and derived from VDJ sequences with IGHV1–7 as the best V gene match. The percentage of ultralong CDR H3s varies from 0.3% to 5.4% across 204 samples with the average percentage 1.9%. We define the score of an alignment between two sequences as the number of differences (excluding starting and ending gaps) normalized by the alignment length. The average score of alignments between an IGHD8–2 allele and an ultralong CDR H3 varies from 0.38 to 0.45 across all subjects thus indicating an extremely high mutation rate. To minimize the impact of poorly aligned ultralong CDR H3s, we apply two filters. First, we say that an ultralong CDR H3 is assigned to sequence s1 rather than sequence s2 if score(s2, CDR H3) – score(s1, CDR H3) > diffmin, where diffmin is the score(s1, s2). If we replace s1 with IGHD8–2*01 and s2 with IGHD8–2*02, then |score(s1, CDR H3) – score(s2, CDR H3)| is expected to be close to score(IGHD8–2*01, IGHD8–2*02)=0.046. However, as Supplemental Figure 1A shows, only ~3.69% of ultralong CDRH3s (8 CDR H3s in absolute numbers) per sample have diff above 0.04. Similarly, only ~5.85% of ultralong CDRH3s (39 CDR H3s in absolute numbers) have diff above 0.03. To recruit more CDR H3s and avoid possible impacts of clonal expansion, we decreased the value of diffmin to 0.02. The average percentage of used ultralong CDR H3s per sample is 14.83% (Supplemental Figure 1A). This filter allows us to discard ambiguous ultralong CDR H3s that are aligned to IGHD8–2*01 and IGHD8–2*02 with similar scores. Since a high mutation rate makes alignments less accurate, we additionally discard ultralong CDR H3s with alignment scores above 0.3 because they correspond to alignments with less than 70% identity. Supplemental Figure 1B shows a subject that is likely homozygous for allele IGHD8–2*01. While all ultralong CDR H3s with low mutation rates (scores below 0.3) have lower alignment scores for allele IGHD8–2*01 compared to IGHD8–2*02 (and thus are assigned to IGHD8–2*01), higher mutation rates create confusion making many ultralong CDR H3s with scores above 0.3 equally distant from both alleles. Figure S1C shows that the latter observation holds true for a subject that is likely homozygous by IGHD8–2*02. To classify the state of IGHD8–2 for a single subject, we compute the fraction of non-discarded ultralong CDRH3s assigned to IGHD8–2*01. If the fraction is above 0.9, we classify the subject as homozygous for IGHD8–2*01. If the fraction is below 0.1, we classify the subject as homozygous for IGHD8–2*02. Otherwise, we classify the subject as heterozygous. Supplemental Figure 1E shows the distribution of the fractions of ultralong CDR H3s assigned to IGHD8–2*01 across 204 subjects.
Results
Germline DH allele length and cysteine position diversity
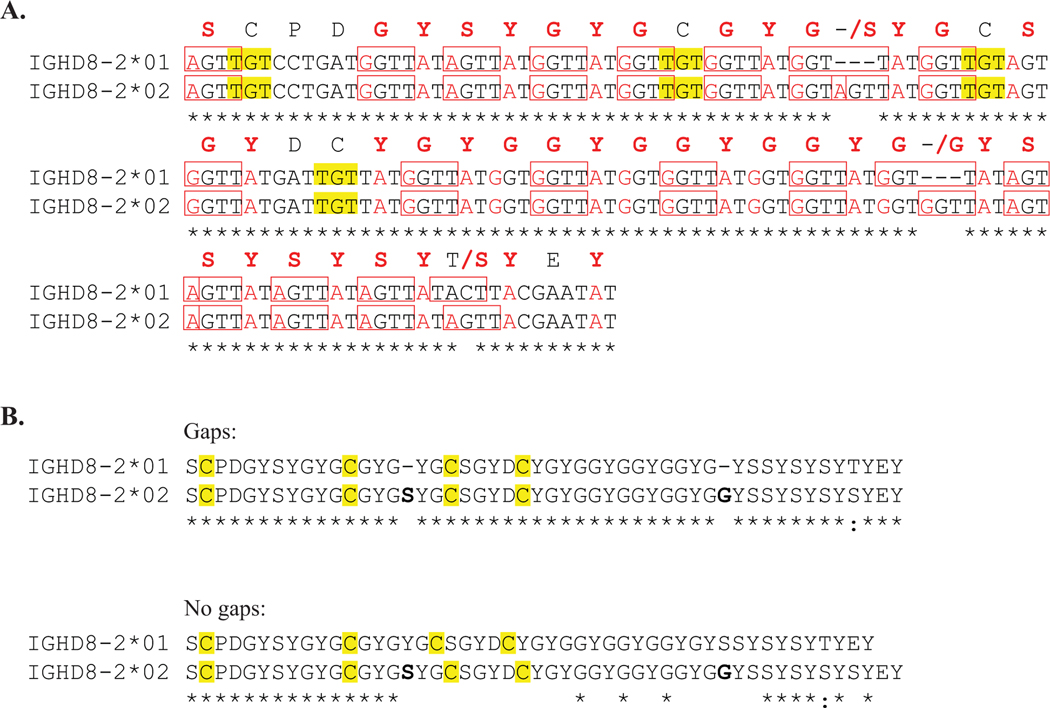
The two known DH alleles used to encode antibodies with ultralong CDR H3 regions, IGHD8–2*01 and IGHD8–2*02, were aligned at the nucleotide and amino acid sequence levels (Figure 1). The DH alleles are very similar with 95% identity at the nucleotide level, with several repeating units that encode G-Y-G. Both have a high density of RGYW SH hotspots (Figure 1, A). Notably, the sequence length of IGHD8–2*02 is six nucleotides (two amino acids) longer than IGHD8–2*01. The DH alleles are identical except at three positions: one position has a difference in nucleotide and amino acid identity (Thr in *01 and Ser in *02) and the other two positions consist of gaps in the IGHD8–2*01 sequence where an additional amino acid is inserted in the IGHD8–2*02 sequence. The sequence location of the IGHD8–2*02 serine insertion is between cysteines 2 and 3, resulting in a single amino acid position shift of cysteines 3 and 4 relative to the locations of cysteines 1 and 2 in IGHD8–2*01 (Figure 1, B). Cysteine 1 at position D2 and cysteine 2 at position D12 are conserved between both germline genes. The conserved sequence in both alleles encode knob structural features; the CPDG motif at the N-terminal end forms turn 1 of the knob with the cysteine participating in a conserved disulfide bond, and the alternating aromatic residues at the C-terminal end forms the descending strand of the stalk. Thus, the two DH alleles are identical in encoding key structural determinants (turn, disulfide bond, and descending stalk) but are divergent in their length and in two cysteine positions.
Figure 1. Two ultralong DH alleles are highly homologous but differ in length and cysteine position.
Alignment of IGHD8–2*01 and IGHD8–2*02 sequences. Asterisks below alignments indicate positions of homology between the aligned sequences. A. Nucleotide sequences are aligned with the corresponding amino acid residue above the center nucleotide of each codon. Positions with nonhomologous amino acids between the two sequences show the amino acid symbol or gap (marked by a hyphen) of IGHD8–2*01 followed by a slash and then the amino acid symbol corresponding to the IGHD8–2*02 sequence. Nucleotide sequences of RGYW and WRCY hot spots for SH are boxed. Individual nucleotides that can form a cysteine codon with a single mutation are in red text. Cysteine codons are highlighted in yellow. B. Amino acid alignment with cysteines highlighted in yellow and inserted amino acids in the IGHD8–2*02 sequence, relative to IGHD8–2*01, in bold. The upper alignment (Gaps) indicates positions of IGHD8–2*02 insertions, with a hyphen, at the location on the IGHD8–2*01 sequence. The lower alignment (No Gaps) does not contain any gaps and illustrates the shift in position of the third and fourth cysteines caused by the difference in DH allele sequence length.
Structural conservation of germline cysteines
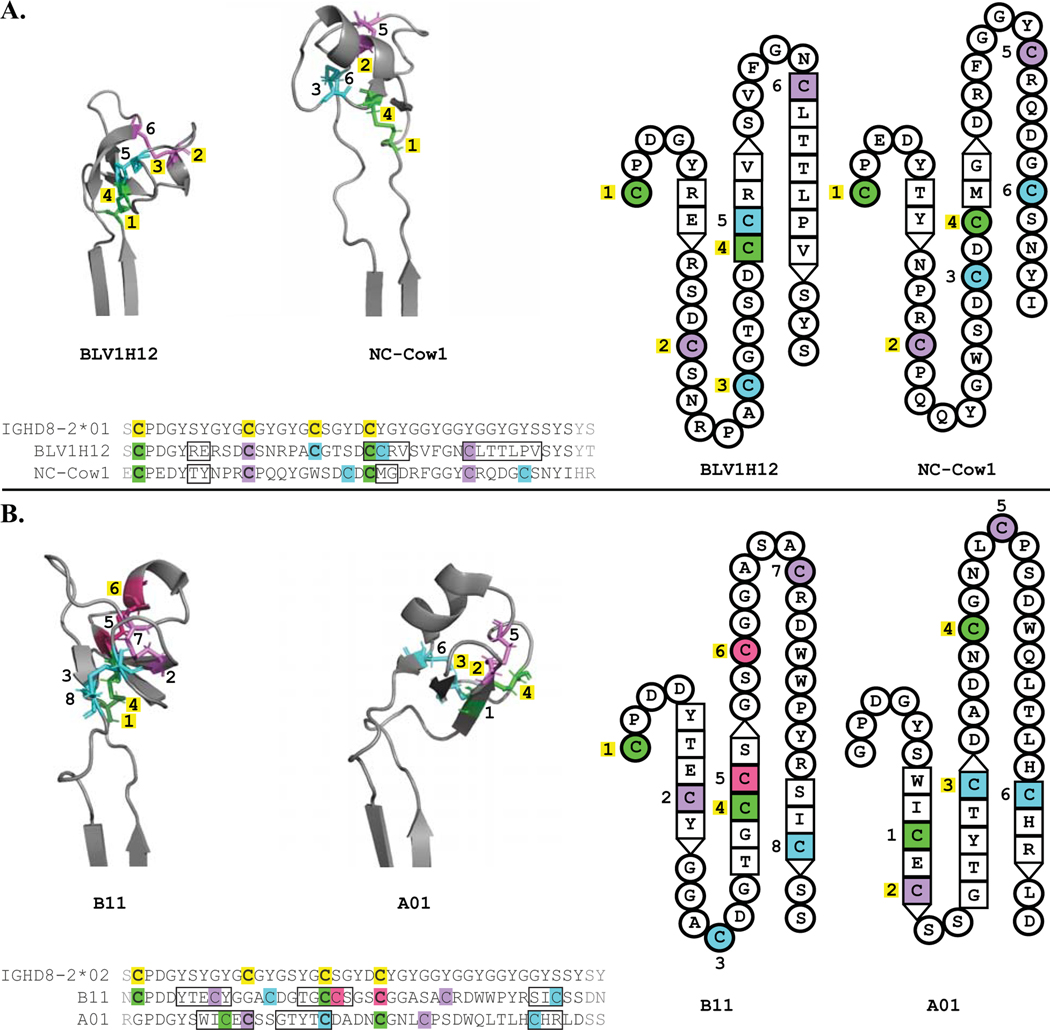
To determine the usage and structural positions of germline-encoded cysteines in affinity matured antibodies, a DH allele was assigned to each of seven antibodies with known structures based on nucleotide alignment scores (Supplemental Table 1). The alignment scores of BLV1H12 (PDB: 4K3D) and NC-Cow1 (PDB: 6OO0) were higher with IGHD8–2*01. Alignment scores of A01 (PDB: 5ILT), B11 (PDB: 5IHU), BLV5B8 (PDB: 4K3E), E03 (PDB: 5IJV) and Bov6 (PDB: 6E9Q) were higher with IGHD8–2*02. Four antibodies were encoded by full length DH regions in the absence of deletions; BLV1H12 and NC-Cow1 with IGHD8–2*01 and B11 and A01 with IGHD8–2*02 (Figure 2A and B, left). Although IGHD8–2*02 is longer than IGHD8–2*01, all three antibodies with deletions (BLV5B8, E03, Bov6) consistently had higher alignment scores with IGHD8–2*02 and each had fewer DH region nucleotide mismatches with IGHD8–2*02 than IGHD8–2*01. Of the seven antibodies, two use IGHD8–2*01 (BLV1H12 and NC-Cow1) and neither contain SH generated deletions, and five use IGHD8–2*02 with two being full length (B11 and A01) and three containing deletions (BLV5B8, E03, and Bov6).
Figure 2. Most germline cysteines are conserved on each antibody CDR H3 structure.
Three (left) and two (right) dimensional structural representations of the CDR H3 knob regions, with amino acid alignments below, grouped by DH allele used: BLV1H12 and NC-Cow1 use IGHD8–2*01 (A.), B11 and A01 use IGHD8–2*02 (B.). Cysteines which form disulfide bonds have the same color, with the first conserved cysteine green, with each subsequent cysteine participating in a unique disulfide bond colored purple, cyan, and pink. Cysteine color coding is consistent between the left and right sequence and structural representations. The number of each sequential cysteine is indicated, with those that correspond to germline encoded cysteines highlighted in yellow. Residues that comprise beta strands are represented as squares on the topology diagrams, with interacting beta sheet residues adjacent to one another. Amino acid sequence alignment features include cysteines highlighted in colors consistent with each structural representation, germline cysteines are in bold text and highlighted in yellow, cysteines that align with germline cysteines are also in bold text, beta sheet residues are in boxes, and amino acids not depicted on topology diagrams (e.g. residues outside of the DH region) are in gray text.
Conserved disulfide bonds using germline cysteines could have integral roles in forming the knob structural scaffold while new disulfide bonds and cysteine positions can arise through SH to generate diversity in knob surface shape. Therefore, we determined (i) the conservation of germline cysteines and (ii) the conservation of germline encoded disulfide bonds (Figure 2). In order to assess disulfide bond and deletion structural positions, we used comparator antibody structures that encoded the full-length region of either IGHD8–2*01 (BLV1H12) or IGHD8–01*02 (B11). B11 was chosen as a comparator over A01 because it maintains the highly conserved first cysteine position. At least three out of four germline DH cysteine positions, including germline cysteine position 4, are conserved on all four of the antibodies encoded by full length DH regions (e.g. without deletions). The fourth germline encoded cysteine, located on strand 2 of the β-sheet core, or its adjacent loops, forms a disulfide bond with the first DH cysteine. Germline cysteine position 1 is conserved on BLV1H12, NC-Cow1, and B11. A01 is a rare exception in bovine heavy chains, as this cysteine is highly conserved in ultralong CDR H3 sequences. A01 has a glycine at this location; DH cysteine 1 in A01 is instead located on strand 1 and forms a disulfide bond with the fourth DH cysteine (located 5 amino acids downstream from strand 2). The other conserved disulfide bond is between germline cysteine 2 and an SH-generated cysteine on or within 2 amino acids of the third loop turn. Germline cysteine position 2 is located on loop 2 of both IGHD8–2*01 antibodies (Figure 2, A) and on strand 1 of A01, the IGHD8–2*02 antibody with germline cysteine 2 conserved (Figure 2, B). Thus, the second germline cysteine is somewhat structurally conserved between IGHD8–2*01 and IGHD8–2*02 antibodies but can be located on either a loop (IGHD8–2*01) or strand (IGHD8–2*02). The relative sequence and knob structural position of the cysteines participating in these bonds are conserved on antibodies using each germline DH. Other DH cysteine positions and disulfide bonds are diverse within the conserved scaffold of three anti-parallel beta-strands connected by loops of varying length. For example, a disulfide bond between germline cysteine 3 (on loop 2) and SH cysteine 5 (on strand 2) is distinct to BLV1H12 (Figure 2A) and a disulfide bond between SH cysteine 5 (on strand 2) and SH cysteine 6 (on loop 3) is unique to B11 (Figure 2B). These examples illustrate how changes in additional cysteine positions and disulfide bond patterns through SH serve as mechanisms for generating diversity in knob domain shape.
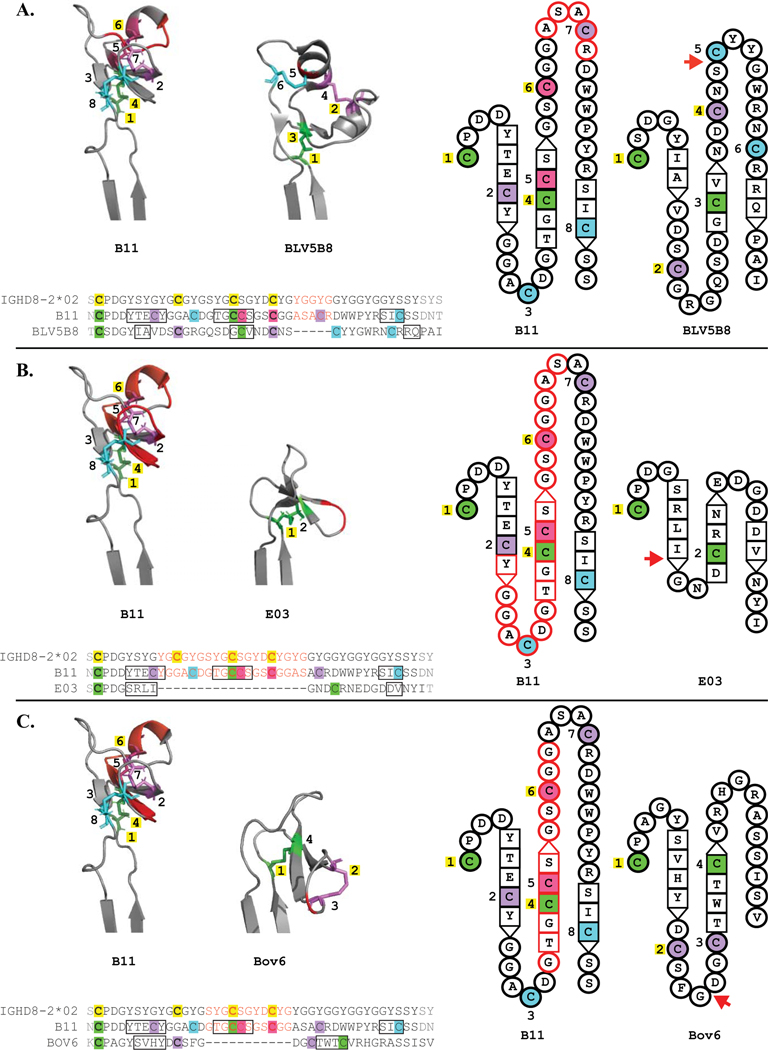
Structural locations of deletions
To determine the structural location of DH region deletions, we evaluated deletion positions relative to the structure of an antibody with a full-length DH (Figure 3). All antibodies in this analysis use germline IGHD8–2*02. In BLV5B8, three of four germline cysteines are conserved, and a deletion occurs C-terminal to the last germline cysteine (Figure 3A). Two SH-generated cysteines arose downstream of the last germline cysteine and these two new cysteines form a disulfide bond with each other. E03 only has two cysteines, which form a single disulfide bond, and three germline DH cysteines are deleted (Figure 3B). For Bov6, cysteine 2 forms a disulfide bond with cysteine 3, located on loop 2. Two germline DH cysteines are deleted on Bov6, however SH-generated cysteines have also formed outside of the deletion locations (Figure 3C). The disulfide bond between the first cysteine and a cysteine located on strand 2 is conserved on all the antibodies. Thus, because of the deletion, the conserved disulfide between germline cysteines 1 and 4 was disrupted where a new cysteine replaces germline 4 in strand 2. Germline cysteines 2, 3 and 4 were deleted on E03 (Figure 3B) and germline cysteines 3 and 4 were deleted on Bov6 (Figure 3C), however an SH-generated cysteine arose on strand 2 that forms a disulfide bond to cysteine 1 on both antibodies. To summarize, deletion events both delete and alter the positions of germline encoded cysteines, however the conserved disulfide bond between germline cysteine 1 and an SH cysteine on strand 2 are maintained.
Figure 3. Deletions within DH genes impact knob region structure and cysteine distribution.
Structural representations and amino acid alignments of three ultralong CDRH3 antibodies with DH region deletions, BLV5B8 (A.), E03 (B.) and Bov6 (C.). Each is analyzed in comparison to B11, which is the same length as IGHD8–2*02. Cysteine color coding, cysteine numbering and representation of beta sheets follow the same convention as in Figure 2, apart from added features pertaining to deletions within DH regions. Deletion location is indicated with a red arrow adjacent to each topology diagram, red hyphens in each sequence alignment, and a red backbone on each B11 crystal structure. Amino acids on B11 that occupy positions corresponding to the locations of deletions are in red text on the sequence alignments, outlined in red on each B11 topology diagram, and represented with a red backbone on each B11 crystal structure.
DH allele usage and germline cysteine conservation
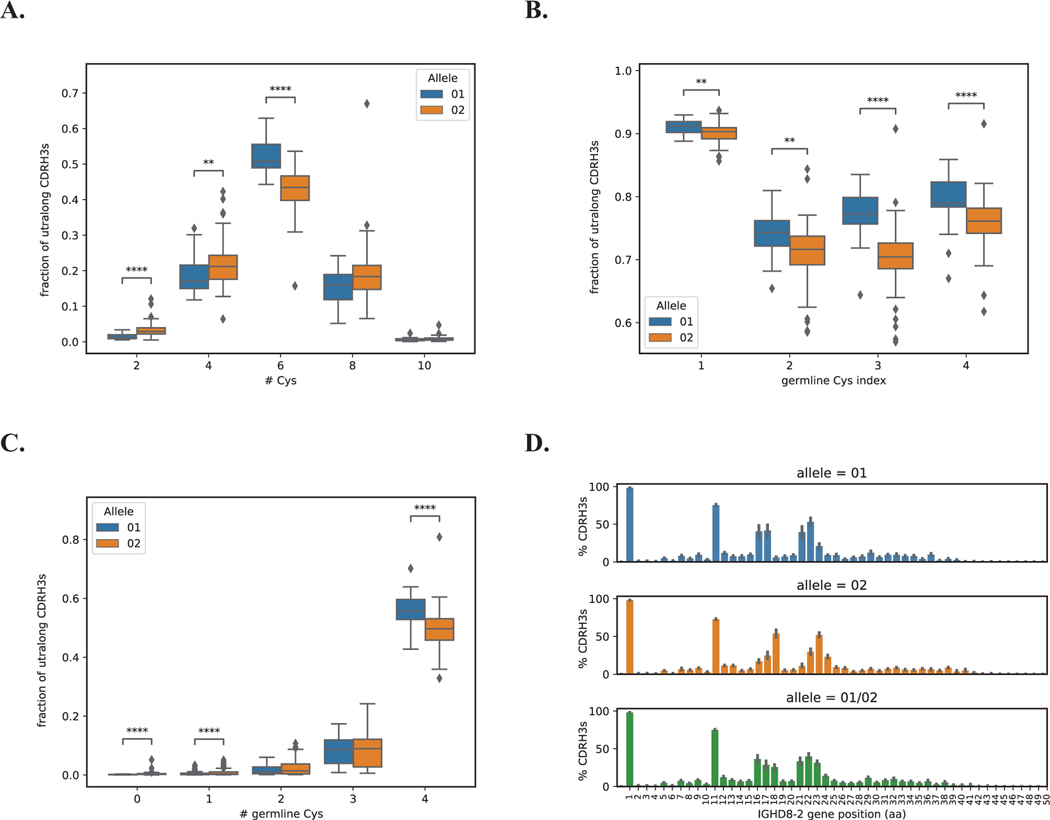
To analyze patterns of germline cysteine conservation, we processed repertoire-sequencing (Rep-Seq) data from IgG antibody repertoires of 204 cows (34). For each Rep-Seq dataset, we extracted ultralong CDR H3 regions and aligned them to both alleles of IGHD8–2. The average percent identities of alignments to the closest allele of IGHD8–2 vary from 54.55% to 62.09% across 204 subjects thus indicating that ultralong CDRH3s have extremely high mutation rates. Since a high mutation rate can make allele assignment ambiguous, we used ultralong CDR H3s with percent identities of alignments above 70% only. To assign alleles of IGHD8–2 within a single subject, we computed the fraction of ultralong CDR H3s that have higher percent identities of alignments to IGHD8–2*01 compared to IGHD8–2*02. If the fraction is close to 1, we classified the subject as homozygous for IGHD8–2*01. If the fraction is close to 0, we classified the subject as homozygous for IGHD8–2*02. Otherwise, we classified the subject as heterozygous for alleles IGHD8–2*01 and IGHD8–2*02. Details of the allele assignment procedure are described in Supplemental Note “Assignment of IGHD8–2 alleles”. As a result, 35 (17%), 84 (41%) and 85 (42%) subjects were classified as homozygous for allele IGHD8–2*01, homozygous for allele IGHD8–2*02, and heterozygous for both alleles, respectively. Thus, IGHD8–2*02 was used at over twice the frequency of IGHD8–2*01.
To determine whether germline DH alleles impact the cysteine content of the repertoire, we analyzed the cysteine number and germline conservation in homozygotes. Figure 4A shows that subjects homozygous for IGHD8–2*01 have slightly higher fractions of ultralong CDR H3s with six cysteines (the average fraction is 0.52) compared to subjects homozygous for IGHD8–2*02 (the average fraction is 0.43), P-value=1.41×10−11. Here and further, we used the Kruskal-Wallis test (35). A germline cysteine is conserved in an ultralong CDR H3 if positions corresponding to it in the alignment represent the germline codon TGT or the mutated codon TGC. Figure 4B illustrates that germline cysteines are more conserved in ultralong CDR H3s derived from IGHD8–2*01, P-values for germline cysteines ordered according to their appearance in IGHD8–2 are 6.04×10−3, 1.31×10−3, 5.29×10−12, and 1.34×10−5. Of the germline cysteines, cysteine 1 is the most conserved at over 90%, followed by cysteine 4. Notably these two cysteines often form a disulfide bond with one another as described above. IGHD8–2*01 is also characterized by a higher fraction of ultralong CDR H3s that have all four conserved germline cysteines compared to allele IGHD8–2*02, P-value=1.77×10−5 (Figure 4C).
Figure 4. Allele usage and germline cysteine conservation and diversity in ultralong CDR H3 knobs.
Homozygous alleles IGHD8–2*01 and IGHD8–2*02 are shown in blue and orange, respectively. Heterozygous alleles (IGHD8–2*01/02) are shown in green. A. Fractions of ultralong CDRH3s with 2, 4, 6, 8, and 10 cysteines derived from homozygous alleles IGHD8–2*01 and IGHD8–2*02. B. Fractions of ultralong CDR H3s with conserved germline cysteines in homozygous alleles IGHD8–2*01 and IGHD8–2*02. C. Fractions of ultralong CDR H3s derived from homozygous alleles IGHD8–2*01 and IGHD8–2*02 with 0, 1, 2, 3, and 4 germline cysteines. D. Percentages of ultralong CDR H3 derived from the homozygous allele IGHD8–2*01 (top), the homozygous allele IGHD8–2*02 (middle), and heterozygous alleles IGHD8–2*01/02 (bottom) that have cysteines at positions corresponding to positions 1–50 of the germline D gene. CDR H3s were cropped and aligned by first position of the DH gene. P-values were computed using the Kruskal-Wallis test and denoted as follows: **<0.01, ****<0.0001. Non-significant P-values are not shown. Each box in Figure 4 shows the quartiles of the distribution. The whiskers show the rest of the distribution, except for outliers found using a function of the interquartile range implemented by the Seaborn package in Python.
To determine whether heterozygotes have greater cysteine positional diversity, we analyzed the proportion of cysteine at each position (Figure 4D). While heterozygous alleles of IGHD8–2 contain 8 amino acid positions where at least 20% of CDRH3s have cysteines, homozygous alleles IGHD8–2*01 and IGHD8–2*02 contain 7 and 6 such positions, respectively. Thus, subjects with two different ultralong CDR H3 germline DH alleles that differ in cysteine location have a repertoire that contains cysteine at more locations than homozygotes.
Discussion
The antibody repertoire of cows is unusual in having few VH, DH, and JH regions to contribute to the combinatorial diversity process, but appears to broadly use cysteine content and disulfide bond diversity created through germline cysteines and somatic hypermutation to create novel antigen binding structures (15, 36). As an extreme example of this, ultralong CDR H3 regions of cows appear to use the knob mini-domain to bind antigen (20, 32). Immunological evolution in the knob region of cow CDR H3s appears to function as a disulfide mediated mini-fold structure generator, as several crystal structures show remarkably diverse sequence content and disulfide patterns in knob regions. The knob region is exclusively encoded by a single DH in homozygous animals, or either IGHD8–2*01 or IGHD8–2*02 in heterozygotes. As ultralong CDR H3 antibodies appear to use single VH, DH, and JH regions, and a limited number of VL regions, there is essentially very little combinatorial diversity that can occur within this subclass of antibodies. Despite this limitation, massive somatic hypermutation can diversify cysteine content and position, providing new disulfide loops as structural scaffolds to bind antigen (5, 15). Other mechansisms, such as gene replacement or gene conversion may also play a role in diversity generation in cattle, but these have not been investigated in depth. Here we identify polymorphisms in the DH region as an additional mechanism to enhance the diversity of the repertoire, in terms of its cysteine content and position.
In characterizing available crystal structures of ultralong CDR H3 antibodies, we found representatives that use each germline DH allele. Detailed analysis allowed us to map germline cysteines onto the crystal structures. Notably, germline cysteine 1 forms a conserved disulfide bond, often with germline cysteine 4. Consistent with the structural analysis, deep sequencing showed that germline cysteine 1 is nearly universally conserved in ultralong CDR H3 regions (e.g. >90%, Figure 4), whereas the second most conserved germline cysteine is cysteine 4, however at a much lower frequency. Taken together, these results suggest that the naïve precursor (germline) CDR H3 region may often contain a 1–4 disulfide bond. As there are only two remaining cysteines, 3 and 4, they may also pair with one another. Alternatively, ‘free’ cysteines may exist in the germline structure, awaiting SH to position a new cysteine nearby to form a new disulfide bond. As these analyses used a limited number of crystal structures (two for IGHD8–2*01 and five for IGHD8–2*02), further expansion of the structural dataset will determine whether these conclusions are generalizeable for ultralong CDR H3 antibodies. The cow ultralong CDR H3 repertoire is unique in that a single precursor VDJ rearrangement (albeit with junctional diversity outside the knob region) appears to account for generation of the entire repertoire. No structural studies have yet been described on this unique germline CDR H3, and the indirect evidence presented here on the possible disulfide pattern is a first attempt to understand its structural dynamics and early evolution. Further, as more structures are solved it may be possible to use predictive algorithms to classify sequences into different structural templates based on cysteine content and position.
The two DH alleles were present in significant frequencies in mature antibody sequences from a population of 204 cows. In this cow population, 17% of cows were IGHD8–2*01 homozygotes and 41% were IGHD8–2*02 homozygotes. Thus, these alleles appear to be utilized in natural immune responses. Of the available crystal structures, we could identify examples of antibodies encoded by each of the two IGHD8–2 variants. The two DH alleles are nearly identical except for their cysteine positions (Figure 1). Heterozygous cows therefore have two germline starting points, with different loop lengths, from which novel disulfide-based loop structures may evolve. Furthermore, deletion events can further alter the cysteine positions encoded by either DH variant. Whether such heterozygous cows have advantages in immunity as a result of this added potential structural diversity has yet to be determined. Polymorphisms in V, D, and J regions are common in all vertebrate species. When amino acid changes occur in polymorphic alleles, they are often single residue changes. The IGHD8–2 region, however, is unusual in having nearly identical variants which only differ in length, and as a consequence, cysteine position. It is unknown whether the germline VDJ recombined ultralong CDR H3 encodes a knob domain that adopts a structure with a single disulfide pattern, or whether structural plasticity occurs that allows different disulfide patterns to form. In the latter scenario, a single gene sequence may produce more than one structure. In other vertebrates, alternative folding of germline CDR H3 residues can indeed produce different paratope structures, as a deviation from the “one gene one protein” paradigm (37–40). Whether such a mechanism extends toward multiple cysteine patterns in ultralong CDR H3 antibodies is currently unknown, however, the covalent nature of disulfide bonds could stabilize germline precursors enabling further SH processes to ‘lock’ certain disulfide patterns favorable to binding a given antigen in place during immunological evolution. The different alleles for IGHD8–2 may encode alternative different precursor structures, enabling alternative evolutionary paths during SH. Thus, in addition to combinatorial and junctional diversity as key mechanisms to provide for a diverse immunoglobulin repertoire, cysteine positional polymorphisms and heterozygosity can additionally add to immune receptor diversity.
Supplementary Material
Key Points.
Bovine ultralong CDR H3s use cysteine polymorphisms to generate structural diversity.
Different disulfides form between germline and mutated cysteines within CDR H3.
Acknowledgements
We thank Duncan McGregor, Pavel Pevzner, Ruiqi Huang, Jeremy Haakenson, and Abigail Kelley for helpful conversations during the course of this work.
Grant Support
This work was supported by grants R01GM105826 and R01HD088400 to V.V.S.
Abbreviations
- CDR H3
third complementary determining region of the heavy chain
- SH
somatic hypermutation
References
- 1.Stanfield RL, Wilson IA, and Smider VV. 2016. Conservation and diversity in the ultralong third heavy-chain complementarity-determining region of bovine antibodies. Sci Immunol 1: aaf7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fugmann SD, Lee AI, Shockett PE, Villey IJ, and Schatz DG. 2000. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol 18: 495–527. [DOI] [PubMed] [Google Scholar]
- 3.Kato L, Stanlie A, Begum NA, Kobayashi M, Aida M, and Honjo T. 2012. An evolutionary view of the mechanism for immune and genome diversity. J Immunol 188: 3559–3566. [DOI] [PubMed] [Google Scholar]
- 4.Smider V, and Chu G. 1997. The end-joining reaction in V(D)J recombination. Semin Immunol 9: 189–197. [DOI] [PubMed] [Google Scholar]
- 5.Wang F, Ekiert DC, Ahmad I, Yu W, Zhang Y, Bazirgan O, Torkamani A, Raudsepp T, Mwangi W, Criscitiello MF, Wilson IA, Schultz PG, and Smider VV. 2013. Reshaping antibody diversity. Cell 153: 1379–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrios Y, Jirholt P, and Ohlin M. 2004. Length of the antibody heavy chain complementarity determining region 3 as a specificity-determining factor. J Mol Recognit 17: 332–338. [DOI] [PubMed] [Google Scholar]
- 7.Johnson G, and Wu TT. 1998. Preferred CDRH3 lengths for antibodies with defined specificities. Int Immunol 10: 1801–1805. [DOI] [PubMed] [Google Scholar]
- 8.Zemlin M, Klinger M, Link J, Zemlin C, Bauer K, Engler JA, Schroeder HW Jr., and Kirkham PM. 2003. Expressed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. J Mol Biol 334: 733–749. [DOI] [PubMed] [Google Scholar]
- 9.Collis AVJ, Brouwer AP, and Martin ACR. 2003. Analysis of the antigen combining site: correlations between length and sequence composition of the hypervariable loops and the nature of the antigen. J Mol Biol 325: 337–354. [DOI] [PubMed] [Google Scholar]
- 10.Irimia A, Sarkar A, Stanfield RL, and Wilson IA. 2016. Crystallographic identification of lipid as an integral component of the epitope of HIV broadly neutralizing antibody 4E10. Immunity 44: 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jardine JG, Sok D, Julien JP, Briney B, Sarkar A, Liang CH, Scherer EA, Henry Dunand CJ, Adachi Y, Diwanji D, Hsueh J, Jones M, Kalyuzhniy O, Kubitz M, Spencer S, Pauthner M, Saye-Francisco KL, Sesterhenn F, Wilson PC, Galloway DM, Stanfield RL, Wilson IA, Burton DR, and Schief WR. 2016. Minimally mutated HIV-1 broadly neutralizing antibodies to guide reductionist vaccine design. PLoS Pathog 12: e1005815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steichen JM, Kulp DW, Tokatlian T, Escolano A, Dosenovic P, Stanfield RL, McCoy LE, Ozorowski G, Hu X, Kalyuzhniy O, Briney B, Schiffner T, Garces F, Freund NT, Gitlin AD, Menis S, Georgeson E, Kubitz M, Adachi Y, Jones M, Mutafyan AA, Yun DS, Mayer CT, Ward AB, Burton DR, Wilson IA, Irvine DJ, Nussenzweig MC, and Schief WR. 2016. HIV vaccine design to target germline precursors of glycan-dependent broadly neutralizing antibodies. Immunity 45: 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Protocol GPI, Koff WC, Wilson IA, Burton DR, and Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477: 466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwong PD, and Wilson IA. 2009. HIV-1 and influenza antibodies: seeing antigens in new ways. Nat Immunol 10: 573–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haakenson JK, Huang R, and Smider VV. 2018. Diversity in the cow ultralong CDR H3 antibody repertoire. Front Immunol 9: 1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanfield RL, Haakenson J, Deiss TC, Criscitiello MF, Wilson IA, and Smider VV. 2018. The unusual genetics and biochemistry of bovine immunoglobulins. Adv Immunol 137: 135–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berens SJ, Wylie DE, and Lopez OJ. 1997. Use of a single VH family and long CDR3s in the variable region of cattle Ig heavy chains. Int Immunol 9: 189–199. [DOI] [PubMed] [Google Scholar]
- 18.Shojaei F, Saini SS, and Kaushik AK. 2003. Unusually long germline DH genes contribute to large sized CDR3H in bovine antibodies. Mol Immunol 40: 61–67. [DOI] [PubMed] [Google Scholar]
- 19.Sok D, Le KM, Vadnais M, Saye-Francisco KL, Jardine JG, Torres JL, Berndsen ZT, Kong L, Stanfield R, Ruiz J, Ramos A, Liang CH, Chen PL, Criscitiello MF, Mwangi W, Wilson IA, Ward AB, Smider VV, and Burton DR. 2017. Rapid elicitation of broadly neutralizing antibodies to HIV by immunization in cows. Nature 548: 108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stanfield RL, Berndsen ZT, Huang R, Sok D, Warner G, Torres JL, Burton DR, Ward AB, Wilson IA, and Smider VV. 2020. Structural basis of broad HIV neutralization by a vaccine-induced cow antibody. Sci Adv 6: eaba0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deiss TC, Vadnais M, Wang F, Chen PL, Torkamani A, Mwangi W, Lefranc MP, Criscitiello MF, and Smider VV. 2019. Immunogenetic factors driving formation of ultralong VH CDR3 in Bos taurus antibodies. Cell Mol Immunol 16: 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koti M, Kataeva G, and Kaushik AK. 2008. Organization of D(H)-gene locus is distinct in cattle. Dev Biol (Basel) 132: 307–313. [DOI] [PubMed] [Google Scholar]
- 23.Koti M, Kataeva G, and Kaushik AK. 2010. Novel atypical nucleotide insertions specifically at VH-DH junction generate exceptionally long CDR3H in cattle antibodies. Mol Immunol 47: 2119–2128. [DOI] [PubMed] [Google Scholar]
- 24.Ma L, Qin T, Chu D, Cheng X, Wang J, Wang X, Wang P, Han H, Ren L, Aitken R, Hammarstrom L, Li N, and Zhao Y. 2016. Internal duplications of DH, JH, and C region genes create an unusual IgH gene locus in cattle. J Immunol 196: 4358–4366. [DOI] [PubMed] [Google Scholar]
- 25.Liljavirta J, Niku M, Pessa-Morikawa T, Ekman A, and Iivanainen A. 2014. Expansion of the preimmune antibody repertoire by junctional diversity in Bos taurus. PLoS One 9: e99808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smider BA, and Smider VV. 2020. Formation of ultralong DH regions through genomic rearrangement. BMC Immunol 21: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kocks C, and Rajewsky K. 1988. Stepwise intraclonal maturation of antibody affinity through somatic hypermutation. Proc Natl Acad Sci U S A 85: 8206–8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liljavirta J, Ekman A, Knight JS, Pernthaner A, Iivanainen A, and Niku M. 2013. Activation-induced cytidine deaminase (AID) is strongly expressed in the fetal bovine ileal Peyer’s patch and spleen and is associated with expansion of the primary antibody repertoire in the absence of exogenous antigens. Mucosal Immunol 6: 942–949. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Y, Jackson SM, and Aitken R. 2006. The bovine antibody repertoire. Dev Comp Immunol 30: 175–186. [DOI] [PubMed] [Google Scholar]
- 30.Macpherson A, Birtley JR, Broadbridge RJ, Brady K, Schulze MED, Tang Y, Joyce C, Saunders K, Bogle G, Horton J, Kelm S, Taylor RD, Franklin RJ, Selby MD, Laabei M, Wonfor T, Hold A, Stanley P, Vadysirisack D, Shi J, van den Elsen J, and Lawson ADG. 2021. The chemical synthesis of knob domain antibody fragments. ACS Chem Biol 16: 1757–1769. [DOI] [PubMed] [Google Scholar]
- 31.Macpherson A, Laabei M, Ahdash Z, Graewert MA, Birtley JR, Schulze ME, Crennell S, Robinson SA, Holmes B, Oleinikovas V, Nilsson PH, Snowden J, Ellis V, Mollnes TE, Deane CM, Svergun D, Lawson AD, and van den Elsen JM. 2021. The allosteric modulation of complement C5 by knob domain peptides. eLife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macpherson A, Scott-Tucker A, Spiliotopoulos A, Simpson C, Staniforth J, Hold A, Snowden J, Manning L, van den Elsen J, and Lawson ADG. 2020. Isolation of antigen-specific, disulphide-rich knob domain peptides from bovine antibodies. PLoS Biol 18: e3000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong J, Finn JA, Larsen PA, Smith TPL, and Crowe JE Jr. 2019. Structural diversity of ultralong CDRH3s in seven bovine antibody heavy chains. Front Immunol 10: 558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Safonova Y, Shin SB, Kramer L, Reecy J, Watson CT, Smith TPL, and Pevzner PA. 2022. Variations in antibody repertoires correlate with vaccine responses. Genome Res 32: 791–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kruskal WH, and Wallis WW. 1952. Use of Ranks in One-Criterion Variance Analysis. J Am Stat Assoc 47: 583–621. [Google Scholar]
- 36.Haakenson JK, Deiss TC, Warner GF, Mwangi W, Criscitiello MF, and Smider VV. 2019. A broad role for cysteines in bovine antibody diversity. Immunohorizons 3: 478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin J, A. E. t. Beuscher, S. E. Andryski, R. C. Stevens, and P. G. Schultz. 2003. Structural plasticity and the evolution of antibody affinity and specificity. J Mol Biol 330: 651–656. [DOI] [PubMed] [Google Scholar]
- 38.Adhikary R, Yu W, Oda M, Walker RC, Chen T, Stanfield RL, Wilson IA, Zimmermann J, and Romesberg FE. 2015. Adaptive mutations alter antibody structure and dynamics during affinity maturation. Biochemistry 54: 2085–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goel M, Krishnan L, Kaur S, Kaur KJ, and Salunke DM. 2004. Plasticity within the antigen-combining site may manifest as molecular mimicry in the humoral immune response. J Immunol 173: 7358–7367. [DOI] [PubMed] [Google Scholar]
- 40.Manivel V, Bayiroglu F, Siddiqui Z, Salunke DM, and Rao KV. 2002. The primary antibody repertoire represents a linked network of degenerate antigen specificities. J Immunol 169: 888–897. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.