ABSTRACT
Accumulating evidence indicates a potential role for bacterial lipopolysaccharide (LPS) in the overactivation of the immune response during SARS-CoV-2 infection. LPS is recognized by Toll-like receptor 4, mediating proinflammatory effects. We previously reported that LPS directly interacts with SARS-CoV-2 spike (S) protein and enhances proinflammatory activities. Using native gel electrophoresis and hydrogen–deuterium exchange mass spectrometry, we showed that LPS binds to multiple hydrophobic pockets spanning both the S1 and S2 subunits of the S protein. Molecular simulations validated by a microscale thermophoresis binding assay revealed that LPS binds to the S2 pocket with a lower affinity compared to S1, suggesting a role as an intermediate in LPS transfer. Congruently, nuclear factor-kappa B (NF-κB) activation in monocytic THP-1 cells is strongly boosted by S2. Using NF-κB reporter mice followed by bioimaging, a boosting effect was observed for both S1 and S2, with the former potentially facilitated by proteolysis. The Omicron S variant binds to LPS, but with reduced affinity and LPS boosting in vitro and in vivo. Taken together, the data provide a molecular mechanism by which S protein augments LPS-mediated hyperinflammation.
Keywords: COVID-19, SARS-CoV-2, spike protein, lipopolysaccharide, TLR4, hyperinflammation
Introduction
Pulmonary and systemic hyperinflammation are some of the prominent hallmarks of severe COVID-19 disease. These dysregulated inflammatory reactions trigger the onset of sepsis and acute respiratory distress syndrome (ARDS), both of which have been documented in nearly all deceased patients (Guan et al., 2020; Zhou et al., 2020). Sepsis and ARDS are initiated by a full-blown activation of the immune response, typically via pattern recognition receptors such as the Toll-like receptors, leading to a cytokine storm that causes excessive and damaging inflammatory effects (Rittirsch et al., 2008). Patients with metabolic syndrome are at a higher risk to develop severe COVID-19 disease involving sepsis and ARDS (Cai et al., 2020a; Zhou et al., 2020). Interestingly, metabolic syndrome is associated with a high blood level of bacterial lipopolysaccharide (LPS) due to gut dysbiosis and translocation of bacterial components into the systemic circulation (Awoyemi et al., 2018). LPS is the main component of the outer membrane of Gram-negative bacteria and is recognized by Toll-like receptor 4 (TLR4) (Poltorak et al., 1998) in complex with its lipid-binding co-receptor MD-2 (Kim et al., 2007). LPS contains lipid A, which consists of six acyl tails connected to a phosphorylated diglucosamine headgroup covalently connected to a heterogeneous polysaccharide chain. LPS also binds to an intermediary receptor, CD14 (Hailman et al., 1996). LPS transfer through this series of proteins is a potent trigger of the inflammatory response in sepsis (Medzhitov and Janeway, 2000). A clinical report on severely ill COVID-19 patients with pneumonia demonstrated significantly elevated levels of bacterial LPS (Sirivongrangson et al., 2020). Non-survivor COVID-19 patients also presented increased systemic levels of LPS and soluble CD14 during hospitalization compared to survivors, indicating a link between elevated LPS level and death (Teixeira et al., 2021). We previously showed that SARS-CoV-2 spike (S) protein can bind to LPS, with a similar affinity to CD14, and boost proinflammatory activity (Petruk et al., 2020), thus providing a potential link between high-risk COVID-19 patients and observed symptoms like ARDS and sepsis. Nevertheless, the detailed molecular mechanisms of interaction between the S protein and LPS are still unclear.
The S protein is a class I viral fusion protein trimer on the surface of SARS-CoV-2 (Ke et al., 2020). The protein is made of two subunits: the S1 subunit that is involved in receptor binding and the S2 subunit that facilitates membrane fusion. The structure of S protein ectodomain (ECD) has been resolved in both open and closed conformations using cryogenic electron microscopy (cryo-EM) (Cai et al., 2020b; Walls et al., 2020). Importantly, accumulating structural evidence from cryo-EM has revealed that S protein is able to bind to various non-polar molecules such as linoleic acid (Toelzer et al., 2020), polysorbate (Bangaru et al., 2020), and haem metabolites (Rosa et al., 2021) by employing hydrophobic cryptic pockets in the N-terminal domain (NTD) and the receptor binding domain (RBD). A neutron reflectometry study showed that the SARS-CoV-2 S protein significantly degrades lipid bilayers by extracting lipids from the membrane, suggesting a strong association of the protein with lipid molecules (Luchini et al., 2021). Using computational docking and molecular dynamics (MD) simulations, we predicted that LPS may bind to an inter-protomeric groove near the S1/S2 cleavage site (Petruk et al., 2020). This groove has a group of partially exposed hydrophobic residues that could accommodate the LPS lipid tails, primarily found on the S2 subunit (hence designated as the ‘S2 pocket’). It is not known, however, whether LPS can bind to the NTD and RBD pockets.
In this study, we established LPS interactions with both S1 and S2 subunits by blue native polyacrylamide gel electrophoresis (BN-PAGE) and mapped LPS binding to the S2 pocket as well as the NTD and RBD pockets by amide hydrogen–deuterium exchange mass spectrometry (HDXMS). Calculation of potentials of mean force (PMFs) within a MD simulation framework, supported by microscale thermophoresis (MST) binding experiments, indicates that LPS binds with strong affinities to the RBD and NTD pockets on the S1 subunit, while the S2 pocket represents a weaker binding site compared to CD14, suggesting its potential role as an intermediate in the LPS receptor transfer cascade (Huber et al., 2018). Furthermore, monocytic THP-1 cell assays show boosting of nuclear factor-kappa B (NF-κB) activation by LPS in the presence of the S2 subunit, but not with the S1 subunit. NF-κB reporter mice show enhanced inflammatory response with both S1 and S2 subunits of the S protein. Finally, the boosting effect on LPS is conserved in Omicron S protein in vitro and in vivo, albeit at a moderately reduced level. Moreover, the Omicron variant shows a reduced affinity to the LPS-binding site on the RBD, as shown by MST, tryptophan fluorescence quenching assays, and MD simulations. Collectively, a molecular mechanism of how the SARS-CoV-2 S protein augments LPS-mediated hyperinflammation emerges, whereby the S protein acts as an additional LPS delivery system to its receptors.
Results
LPS and lipid A bind to SARS-CoV-2 S1 and S2
To test binding of LPS or lipid A with the S protein subunits S1 and S2, we used BN-PAGE and western botting, using His-tag-specific antibodies (Petruk et al., 2020). Intact S protein migrated at a molecular mass of ∼480 kDa (Figure 1A, left; Petruk et al., 2020). The S1 and S2 subunits migrated at molecular masses of around 242 and 146 kDa, respectively. The S1 and S2 band intensity decreased with increasing doses of LPS, denoting an interaction between the proteins and LPS, as illustrated by histograms (Figure 1A, right). We observed that changes in migration were more pronounced for S2 compared to S1. Next, we evaluated the binding of S1 and S2 to lipid A. S protein was included for comparison (Figure 1B). We found that both subunits bound to lipid A, as shown by a decrease of band intensities corresponding to S1 and S2 at 242 and 146 kDa, respectively. Simultaneously, we observed formation of a high-molecular-weight complex in the stacking gel. Interestingly, lipid A binding to S2 was already significant at 100 µg/ml of lipid A. Sodium dodecylsulfate–PAGE analysis of S, S1, and S2 under denaturing conditions showed homogenous bands (Supplementary Figure S1A). BN-PAGE analysis showed that lipid A binding to S, S1, and S2 resulted in formation of higher molecular mass bands compared with the monomer. This is not unexpected, particularly for the S2 subunit, which has been reported to assume a post-fusion trimer structure that is more stable and rigid, both in the absence or presence of ACE2 (Cai et al., 2020b). To explore whether interactions between S2 and LPS were dependent on the pre- and/or post-fusion conformations of S2, we used a mutant-S2 subunit having targeted amino acids substituted with proline residues (F817P, A892P, A899P, A942P, K986P, and V987P) to block fusogenic structural rearrangements in S2 (Zhang et al., 2021). After incubation with increasing doses of LPS, the proline-substituted S2 mutant (hereafter referred to as S2 P-mutant) was analysed on BN-PAGE (Supplementary Figure S1B) followed by western blotting analysis. As with S2 (Figure 1A), we observed bands of different molecular weights in addition to a more pronounced monomer band of 146 kDa. Furthermore, in the presence of LPS, almost no visible S2 protein was observed, even at lower concentrations (e.g. 100 µg/ml), suggesting a stronger binding of LPS to S2 in its pre-fusion conformation. In addition, we did not observe any bands at higher molecular weights in the top stacking gel, as seen with S2:lipid A (Figure 1B).
Figure 1.
Analysis of binding of S1 and S2 subunits to LPS and lipid A. (A and B) SARS-CoV-2 S protein or its subunits, S1 and S2, was incubated with 0–500 µg/ml LPS (A) or lipid A (B), separated using BN-PAGE, and detected by western blotting. One representative image of three independent experiments is shown (n = 3). The intensity of the bands at 480, 242, and 146 kDa, corresponding to molecular mass of monomeric SARS-CoV-2 S protein, S1, and S2 subunits, respectively, was quantified using Image Lab software 6.1 from Bio-Rad.
HDXMS reveals multiple lipid A/LPS-binding sites on SARS-CoV-2 S protein
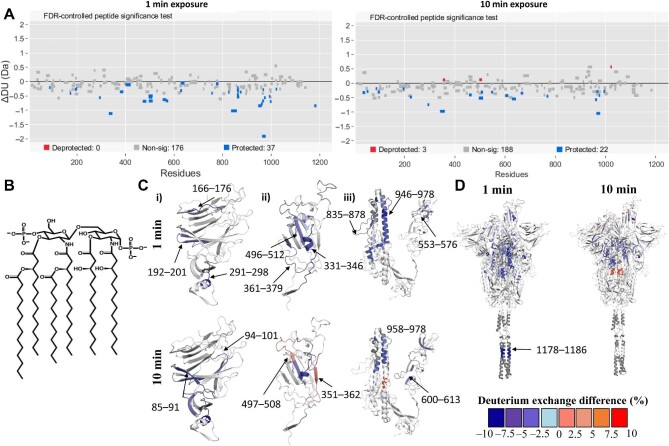
To map the binding sites of lipid A and LPS on S protein, we compared HDXMS of S protein saturated with either lipid molecules. Our HDXMS analysis included pepsin fragment peptides (217 peptides in free S protein state) covering ∼82% of the S protein sequence in the free and bound states (Supplementary Figure S2). HDXMS has previously been used to probe dynamics of the S protein (Raghuvamsi et al., 2021; Costello et al., 2022) and LPS (Fiorentino et al., 2021). Previous studies have shown that deuterium exchange in the fast time scales, i.e. <60 min, predominantly provides a readout of solvent accessibility compared to hydrogen-bonding propensity (Ramirez-Sarmiento and Komives, 2018; Markwick et al., 2019). To map the binding sites of lipid A and LPS with S protein, we carried out deuterium exchange (labelling times t = 1 min and 10 min). Comparative deuterium exchange analysis and statistical significance tests on the differences in deuterium exchange values revealed the binding hotspots of LPS/lipid A on S protein and the respective effect on dynamics. Peptides showing significant differences (confidence intervals (CI) > 99%, Supplementary Table S1) were mapped onto the structure of S protein. HDXMS analysis revealed multiple non-contiguous surface loci spanning several domains—RBD, NTD, and S2 domain (Figure 2A–D; Supplementary Figure S3)—showing decreased deuterium exchange in the lipid A/LPS bound states. Interestingly, protected peptides spanning the NTD (residues 94–101, 192–199, and 221–229) and RBD (residues 331–346 and 495–512) overlay pockets were predicted to be optimal for binding to lipid A (Figure 2Ci and ii). These lipid-binding pockets were reported in previous cryo-EM studies wherein densities corresponding to a polysorbate detergent or lipid tails were identified (Bangaru et al., 2020; Toelzer et al., 2020). Antiparallel β-strands form a sandwich structure, a convenient binding site for aliphatic tails of lipid A. In the case of the RBD, the lipid A-binding pocket consists of a short α-helical region called the ‘gating helix’ along with antiparallel β-strands. Furthermore, several peptides spanning the NTD (residues 291–298), subdomain (residues 553–576), S2 subunit (residues 835–878), and HR1 domain (residues 947–978) showed protection from deuterium exchange in the presence of lipid A (Figure 2Ciii). These peptides are at the inter-protomer interface of the trimeric S protein, forming a hydrophobic pocket, which coincides with our previously computationally predicted LPS interaction site (Petruk et al., 2020). Interestingly, this region is located close to the functionally significant S1/S2 cleavage site, which plays a critical role in viral entry (Figure 2D).
Figure 2.
Lipid A binding hotspots on S protein revealed by HDXMS. (A) Woods plots showing differences in deuterium exchange (∆DU, Y-axis) between S protein:lipid A state and free S protein state at deuterium labelling times of 1 and 10 min. The length of the lines represents the length of each pepsin-proteolyzed peptide listed along the X-axis from N- to C-termini. Peptides showing significant differences (CI > 99%) are highlighted as per colour key. (B) Chemical structure of E. coli lipid A molecule. (C and D) Differences in deuterium exchange between S protein:lipid A and free S protein states at labelling times of 1 min and 10 min are mapped onto the respective S protein domain structures NTD (i), RBD (ii), and S2 domain (iii) and full-length S protein structure (residues 14–1208). Only the peptides showing significant deuterium exchange differences are mapped onto the structure. Deuterium exchange difference (%) values are mapped onto the structures as per the colour key. White and grey regions on the structure represent those with no significant change or sequence coverage in the presence of lipid A.
Overall, lipid A- and LPS-bound S proteins showed comparable deuterium exchange profiles with equivalent peptides binding to lipid A and LPS (Figure 2; Supplementary Figure S3). Protection from deuterium exchange was observed in peptides spanning NTD (residues 94–101, 170–176, 200–212, and 221–229) at difference in deuterium exchange of 1 or 10 min in the presence of LPS (Supplementary Figure S3A, Bi and ii). Peptides spanning the RBD (residues 331–346 and 496–512) and S2-binding pockets (residues 859–869 and 946–978) showed protection at 1 and 10 min labelling times (Supplementary Figure S3Bii and iii), similar to the lipid A:S protein state. Peptide 1178–1187 spanning the HR2 region also showed protection in the presence of both lipid A and LPS at 1 and 10 min labelling times, which is consistent with recent computational predictions and an NMR study (Chiliveri et al., 2021; Zuzic et al., 2022), suggesting the potential for binding of hydrophobic molecules such as lipids. Increases in deuterium exchange observed in peptides spanning the NTD (residues 170–176), RBD (residues 453–470), and S2 domain (residues 970–985, 1019–1025, and 1051–1063) indicate local destabilization of binding pockets to accommodate the considerably larger and more heterogeneous LPS molecule compared to lipid A. An interesting similarity between lipid A- and LPS-bound S protein is seen with the protection from deuterium exchange at the receptor-binding motif of the RBD, a crucial component involved in ACE2 receptor interactions. In both lipid–S protein complexes, no peptides showed EX1 deuterium exchange kinetics (Supplementary Figures S2C and S5).
HDXMS at a longer labelling time (t = 100 min) showed increases in deuterium exchange throughout the S protein in the presence of lipid A or LPS (Supplementary Table S1 and Figure S4). Increases in deuterium exchange at the NTD (Supplementary Table S1) and RBD (Supplementary Table S1) pockets may result from widening or destabilization of the local secondary structure to accommodate more of the bulky, multiply-acylated lipid A component of LPS, in accordance with our simulation data described below. In the S2-binding pocket, increases in deuterium exchange observed at the central helical bundle and flanking peptides (residues 600–613, 929–981, and 853–859) in lipid A- and LPS-bound S protein were indicative of disruption of inter-protomer contacts of the S protein (Supplementary Table S1 and Figure S3), potentially exacerbated by accommodation of more than one lipid molecule, also supported by our simulations below. Long-range effects of binding to these two pockets have also been characterized by cryo-EM studies, whereby the binding of haem metabolites to the NTD results in allosteric changes of antibody epitopes (Rosa et al., 2021), while linoleic acid binding to the RBD shifts the conformational equilibrium of the S protein to favour the down state (Toelzer et al., 2020).
S protein binding sites have different affinities for LPS
Our previous study showed that LPS binding to the S2 pocket involved residues from both S1 and S2 subunits (Petruk et al., 2020). To determine whether LPS binding to this site requires both S1 and S2, we performed MD simulations of LPS bound to this site without S1. LPS remained stably bound to the isolated S2, as assessed by measurement of the root mean square deviation (RMSD) of the lipid with respect to its initial conformation, which remained low and comparable to the RMSD observed during simulations of the whole S ECD (Supplementary Figure S8). This suggests that the key residues for LPS binding in this pocket are those found on S2. For comparison, simulations of LPS bound to this binding site in a truncated system containing only S1 showed LPS detachment from the binding site, and consequently, significantly higher RMSD values. This indicates that LPS binding to S1 observed in our BN-PAGE experiment does not involve the S2 pocket, but rather involves the pockets in the NTD and RBD only.
To characterize the dynamics of LPS binding to the NTD and RBD, we performed unbiased MD simulations whereby a lipid A molecule was placed nearby the NTD and RBD facing the hydrophobic pockets. In all simulations, the lipid tails spontaneously inserted into the binding sites. The hydrophobic pockets expanded to accommodate the lipid tails, especially on the RBD in which the pocket volume became significantly larger in simulations upon complexation with lipid A (Supplementary Figure S9). Solvent accessible surface area (SASA) of individual lipid tails showed that the NTD pocket can accommodate up to five tails, whereas the flexible RBD site can accommodate as many as all six lipid tails of lipid A (Figure 3A and B) upon pocket expansion. Congruently, the binding of lipid A to RBD resulted in a lower percentage of its lipid tail atoms being exposed to solvent compared to binding to NTD, suggesting the potential for stronger binding (Figure 3C and D). Structural clustering analysis suggested that NTD can accommodate a limited length of each of the lipid tails (∼5 carbon atoms distal from the glucosamine units), whereas the RBD can accommodate the entirety of the tails (Figure 3E and F).
Figure 3.
Spontaneous binding of lipid A to NTD and RBD pockets. (A and B) One lipid A molecule was placed nearby the polysorbate-binding site on NTD or the linoleic acid-binding site on RBD and three independent replicas of 1000 ns MD simulations were performed. The number of lipid tails buried was calculated based on the SASA of individual lipid tails. A cut-off of 0.5 nm2 was used to categorise the lipid tail as buried. (C and D) Percentage of lipid tail atoms exposed to solvent defined as those found within 0.4 nm of any water molecule. Thick lines show average over repeat simulations and shaded areas indicate SD. (E and F) Cluster analysis was performed with an RMSD cut-off of 0.4 nm. The central structures of the top clusters are shown. Lipid A in orange, stick representation; NTD and RBD in cyan, cartoon representation. Enlarged images show hydrophobic residues on NTD and RBD that interact with the lipid tails.
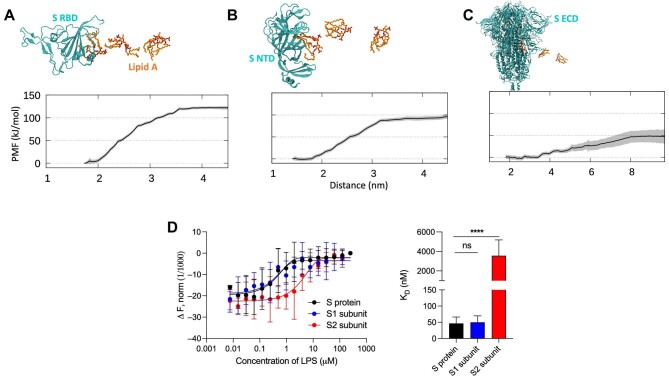
To estimate lipid A affinities to all three identified binding sites, we performed PMF calculations. Adequate sampling was achieved as indicated by Supplementary Figure S10. The PMFs suggested that lipid A binds most strongly to the RBD pocket, with a free energy estimated to be ∼125 kJ/mol (Figure 4A). Intriguingly, this is comparable to the value previously calculated for CD14 (Huber et al., 2018), which serves as a conduit for LPS in the TLR4 pathway. This result is thus consistent with our previous MST experiment showing similar KD values for LPS interaction with S protein and CD14 (Petruk et al., 2020). The NTD pocket exhibited a slightly lower binding affinity with an estimated free energy of ∼95 kJ/mol (Figure 4B). This agrees with our unbiased simulations showing that lipid A binding to NTD involves a reduced portion of the lipid tails compared to the RBD (Figure 3E and F). Finally, the S2 pocket represented the weakest lipid A-binding site with a free energy value of ∼50 kJ/mol (Figure 4C). The percentage of lipid tail exposure to water in each of the umbrella sampling windows showed a less dramatic increase in solvent exposure during lipid dissociation for the S2 pocket in contrast with the RBD (Supplementary Figure S11). Nevertheless, the free energy value is still significant, indicating that a free lipid A molecule would favourably bind to this site. However, in the presence of proteins with higher affinities to lipid A, such as CD14, the lipid A molecule-bound equilibrium would presumably shift, suggesting that the S2 pocket could play a role as an intermediary in the TLR4 cascade.
Figure 4.
Quantification of lipid A binding to S protein. (A) The calculated PMF for lipid A (un)binding to the RBD pocket is shown (bottom), with snapshots along the pathway indicated (top), based on umbrella sampling simulations. Protein in cyan, cartoon representation; lipid A in orange, stick representation. The reaction coordinate is the distance between the centers of mass of lipid A and the protein. (B and C) Similar PMF calculations for the NTD and S2 pocket, respectively. (D) The binding affinity of LPS to S1 and S2 was quantified by MST. Representative binding curves are shown on the left, while KD constants obtained from the curves are summarized with the histogram on the right. KD for S protein is 46.7 ± 19.7 nM, for S1 subunit 50.0 ± 20.0 nM, and for S2 3.6 ± 1.6 µM. Data are shown as mean ± SD (n = 6).
To verify the predicted affinities, we performed MST and determined the KD values for LPS binding to S1 and S2 independently (Figure 4D). The KD obtained for S1 was in the same range as for the S protein, 50.0 ± 20.0 and 46.7 ± 19.7 nM, respectively. The low KD value is in good agreement with our PMF calculations showing the presence of high-affinity binding pockets in the NTD and RBD. Conversely, the KD obtained for S2 was 3.6 ± 1.6 µM, which supports the weaker-affinity binding site predicted by our PMF calculation.
Our BN-PAGE experiment showed that LPS alters the migration of S2 more prominently compared to S1 (Figure 3), particularly for the prefusion stabilized construct (Supplementary Figure S7B), suggesting a more pronounced interaction with S2. It must be stressed, however, that the concentration of LPS used in these experiments was well above the KD values determined by MST (10–50 µM), and therefore, the BN-PAGE analysis reported binding to LPS in a saturated system. Due to the large size of the S2 pocket and the partially exposed lipid tails of bound LPS, we hypothesized that more than one LPS molecule could occupy the binding site through lipid–lipid interactions, which would explain the marked changes in migration pattern of S2 in the presence of LPS. To test this hypothesis, we performed an unbiased MD simulation with three additional lipid A molecules placed outside the S2 pocket. Within 300 ns, all three lipid A molecules spontaneously entered the S2 pocket and formed a small lipid A aggregate within the binding site and remained stably bound for the remainder of the simulation (Supplementary Figure S12). The S2 pocket could therefore act as a pool for small LPS aggregates and may be able to serve a role in forward transfer along the TLR4 relay.
LPS binding to S protein subunits boosts proinflammatory responses
We have previously shown that S protein, when combined with ultralow-threshold levels of LPS, boosts NF-κB activation in monocytic THP-1 cells and proinflammatory cytokine release in human blood (Petruk et al., 2020). Since both S1 and S2 subunits bind to LPS, we next explored whether these S protein subunits could enhance proinflammatory responses as well. Therefore, we first incubated THP-1 cells with increasing concentrations of LPS (0–1 ng/ml) and a constant amount (5 nM) of S1 or S2. After 20 h, the levels of NF-κB were measured. The results reported in Figure 5A (left) showed that the S2 alone stimulates NF-κB production. This observation prompted us to investigate whether the preparation could be contaminated by LPS, which has been reported for various commercial S protein preparations (Ouyang et al., 2021). Indeed, we found that the endotoxin levels were 3.5 and 1401.1 fg/µg in S1 and S2, respectively. It is of note that the level of LPS observed in the S2 preparation did not yield any detectable proinflammatory response in THP-1 cells per se; 1 and 10 pg/ml LPS yielded no NF-κB activation (n = 3, P = 0.9982 and 0.9998, respectively). Nevertheless, taking this confounding endogenous LPS contamination into account, the results demonstrated that exogenously added LPS at ultra-low levels indeed induced a boosting of NF-κB. Surprisingly, no boosting effect was observed for S1, either alone or in combination with LPS. Nevertheless, the proinflammatory effect of LPS was not even supressed by the addition of S1. No tested condition was cytotoxic, excluding low cell response to stimuli due to death (Figure 5A, right). To verify that S2 had a stronger proinflammatory effect because of LPS contamination, we performed the experiments on THP-1 cells in the presence of polymyxin B, a well-established neutralizer of LPS (Avedissian et al., 2019). Figure 5B shows that the addition of polymyxin B suppresses the NF-κB activation by either S2 alone or S2 mixed with LPS, indicating an LPS-mediated effect by the S2 preparation. As expected, no visible alteration in NF-κB levels was observed in the case of THP-1 cells treated with the S1 subunit alone or with LPS and/or polymyxin B. The results with the S2 preparation prompted us to investigate the behaviour of the prefusion stabilized construct, i.e. S2 P-mutant. The LPS level in this variant was 5.7 fg/µg protein, comparable to the levels observed in S protein and S1 subunit. Correspondingly, the prefusion stabilized S2 construct showed less intrinsic proinflammatory activity per se but retained a significant ability to boost the responses to ultra-low levels of LPS (Supplementary Figure S13). Hence, the stabilized S2 retains the inflammation-boosting activity previously demonstrated for the whole S protein (Petruk et al., 2020).
Figure 5.
S2 subunit boosts the proinflammatory response to LPS. (A) NF-κB activation (left) and cell viability (right) were measured in THP-1-XBlue-CD14 cells stimulated with increasing doses (0–1 ng/ml) of LPS and a constant amount (5 nM) of S1 and S2. Lysed cells were used as negative control for cell viability. Data are presented as mean ± SD of four independent experiments performed in triplicate (n = 4). ****P ≤ 0.0001, determined using two-way analysis of variance (ANOVA) with Tukey's multiple comparisons test. (B) THP-1-XBlue-CD14 cells were treated with 5 nM S1 or S2, alone or in combination with 0.25 ng/ml LPS and/or 100 µg/ml polymyxin B. After 20 h incubation, NF-κB activation (left) and cell viability (right) were measured. Lysed cells were used as negative control for cell viability. Data are presented as mean ± SD of four independent experiments all performed in triplicate (n = 4). ****P ≤ 0.0001, determined using ordinary one-way ANOVA with Tukey's multiple comparisons test.
Using mice reporting NF-κB activation, we previously showed that neither 2 µg LPS nor 5 µg SARS-CoV-2 S protein alone could induce a significant proinflammatory response; however, when combined together, a marked boosting of inflammation was observed (Petruk et al., 2020; Puthia et al., 2022). Using a similar setup, we investigated the proinflammatory responses to S1 and S2 and the subcutaneously administrated LPS. S1 alone did not yield any measurable NF-κB activation (Figure 6A). S1 combined with 2 µg LPS resulted in a significant proinflammatory response. S2 alone yielded a proinflammatory response per se, which was not unexpected given the detected LPS content in this preparation (Supplementary Figure S14). Like the in vitro results (Figure 5), addition of polymyxin B to the S2 preparation reduced the proinflammatory response, indicating that LPS was causing the NF-κB activation (Supplementary Figure S14). Analogously to the in vitro data, the prefusion stabilized S2 form showed a significant boosting effect on LPS responses (Figure 6B). Taken together, the results showed that S1 has a boosting effect on LPS-induced inflammation. Indirectly, the results also showed that S2’s inherent proinflammatory effect was due to minute LPS-contaminants, resulting in a boosted NF-κB response to the combination. The proline-stabilized S2 construct, which contained less contaminating LPS, exhibited a low proinflammatory activity alone, but significantly boosted the response to 2 µg LPS.
Figure 6.
LPS binding to S protein subunits boosts in vivo proinflammatory responses in NF-κB reporter mice. (A and B) LPS alone or in combination with S1 (A) or S2 (B) was subcutaneously deposited on the left and right sides on the back of transgenic BALB/c Tg(NF-κB-RE-luc)-Xen reporter mice. In vivo imaging of NF-κB reporter gene expression was performed using the IVIS Spectrum system. Representative images show bioluminescence at 1, 3, and 6 h after subcutaneous deposition and bar charts show bioluminescence emitted from these reporter mice. Areas of subcutaneous deposition and region of interest for data analysis are depicted as dotted circles. Data are presented as mean ± SEM (n = 4). P-values were determined using one-way ANOVA with Dunnett posttest. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001; ns, not significant.
Intriguingly, S1 displays a boosting effect on LPS-induced inflammation in vivo but not in vitro, suggesting additional mechanisms in the former. We hypothesize that one potential reason for this in vitro–in vivo difference is the interactions with proteases secreted in the tissue during inflammation, such as neutrophil elastase (NE) that could modify the structure of the S1 subunit and alter its affinity to LPS. To test this hypothesis, we incubated THP-1 cells with the digested or undigested S1 subunit in the absence and presence of LPS. Indeed, S1 mixed with LPS and then digested with NE for 15 min was able to increase NF-κB production compared to the mixtures without NE and pre-digested S1 (Supplementary Figure S15). This supports our hypothesis that LPS binding to the high-affinity sites in S1 in vivo is compromised by proteolysis during inflammation resulting in an onward transfer to LPS receptors. The proteolytic sites of these enzymes and how they weaken LPS binding, however, would require further studies.
Omicron S protein binds to LPS and boosts proinflammatory responses
The Omicron S protein has 37 mutations, which lead to considerable escape from antibody neutralization (Cao et al., 2021). None of these mutations occur in the putative LPS-binding sites described in this study. To determine whether LPS binding is preserved in Omicron S protein, we first performed BN-PAGE experiments. Similar to the Wuhan strain, the S protein band intensity decreased with increasing doses of LPS (Supplementary Figure S16A), indicating interactions between Omicron S protein and LPS. We then investigated the boosting effects of Omicron S protein on LPS-mediated proinflammatory responses in THP-1 cells. Both the Wuhan and Omicron S proteins enhanced NF-κB production compared to LPS alone (Supplementary Figure S16B). At all LPS concentrations, the degree of boosting by Omicron S protein was lower than that of the original S protein. We then performed in vivo bioimaging of NF-κB reporter mice injected subcutaneously with Omicron and Wuhan S proteins on a background of the ultra-low LPS levels as used above and in previous publications (Petruk et al., 2020; Puthia et al., 2022). The proinflammatory response of LPS was boosted by Omicron S protein compared to that in the mice injected with LPS alone after an incubation period of 3 h (Supplementary Figure S16C). Congruent with the in vitro results, the boosting effect was reduced compared to that of the Wuhan strain (Supplementary Figure S16D).
To understand the underlying molecular mechanism of the reduced proinflammatory boosting effect, we first performed MST and determined the KD values of LPS binding to Omicron S protein (Supplementary Figure S17A). Intriguingly, the KD value was much higher than that for the Wuhan S protein and comparable to that for the original S2 (Figure 4D), suggesting the loss of the high-affinity binding sites. As S1 contains the high-affinity binding sites, we then performed MST for the Omicron S1, and indeed, the KD value obtained was much higher than that for the Wuhan counterpart (Supplementary Figure S17C). Tryptophan fluorescence quenching assay showed compatible results with MST data, i.e. higher doses of LPS are needed to reach the quenching of intrinsic fluorescence in Omicron S protein than in Wuhan S protein. Fluorescence data of S1 were also compatible with the MST data (Supplementary Figure S17D), as indicated by a lower KD for the Wuhan variant, but the maximum intrinsic fluorescence intensities for the two subunits were similar, suggesting the same effect of LPS upon the three-dimensional structure of the proteins. MST analysis also showed that the KD value for lipid A binding to Omicron S1 protein was significantly higher than that for Wuhan S1 protein (Supplementary Figure S17E).
The RBD pocket on S1 represents the highest-affinity LPS-binding site (Figure 4A); we thus performed unbiased MD simulations of lipid A bound to Omicron RBD. Lipid A remained stably bound and interacted with equivalent hydrophobic residues as found in Wuhan S protein. However, the gating helix—a short α-helix that shifts to allow entry of the lipid molecule—spontaneously unfolded in simulations of Omicron RBD (Supplementary Figure S17F). While the sequence of the helix itself is conserved in the Omicron variant, three serine residues immediately downstream of the helix (S371, S373, and S375) are mutated to non-polar residues. These serine residues interacted with the hydroxyl moieties of the lipid headgroup in the original RBD simulations. In the Omicron RBD simulations, these interactions were lost, and the mutated residues moved away from the lipid headgroup, resulting in an unfolding of the nearby gating helix. To investigate the effect of this helix unfolding on the free energy of binding, we performed PMF calculations. We obtained a free energy value of ∼50 kJ/mol (Supplementary Figure S17G), which was much lower than that for the Wuhan RBD and comparable to that for the S2 pocket, in agreement with the MST experiments. The reduced free energy is caused by an increase in lipid tail exposure to solvent due to the unfolded gating helix. Taken together, the Omicron S protein retains its LPS-binding capability and the associated proinflammatory boosting effect, albeit at a reduced level, possibly due to the loss of the high-affinity binding site on the RBD.
Discussion
There is growing evidence that bacterial LPS plays a central role in severe COVID-19 complications (Kruglikov et al., 2020; Petruk et al., 2020; Sirivongrangson et al., 2020; Camell et al., 2021; Hoel et al., 2021; Kruglikov and Scherer, 2021; Teixeira et al., 2021). Overstimulation of the TLR4 pathway by LPS triggers a hyperinflammatory state that can lead to sepsis and ARDS. In this study, we showed that LPS binds to SARS-CoV-2 S protein at various sites resulting in amplified proinflammatory responses. In the TLR4 pathway, LPS is transferred from the outer membrane of Gram-negative bacteria through a series of receptors to the TLR4:MD-2 complex, which initiates downstream signal activation (Ryu et al., 2017). An affinity gradient exists between LPS and these receptors to favour a one-way transfer (Huber et al., 2018). Here, we characterize the LPS interaction sites on S protein and their respective affinities to LPS and propose how the S protein fits into the LPS transfer cascade by acting as an additional delivery mediator to the host LPS receptors (Figure 7).
Figure 7.
Proposed model for S protein boosting effect on LPS-mediated proinflammatory response. In the presence of S protein, LPS micelles disaggregate due to binding to sites on the S1 and S2 subunits, facilitating downstream LPS transfer to CD14. Subsequently, LPS is transferred to the TLR4:MD-2 complex, which triggers proinflammatory reactions. Thus, the S protein acts as an intermediate in the LPS receptor transfer cascade. Proteases, lipids, and metabolites may modulate the LPS interaction with binding sites in S1.
Previous studies suggested that SARS-CoV-2 S protein binds to TLR4 directly and activates related immune responses (Aboudounya et al., 2021; Zhao et al., 2021). Our experiment with polymyxin B, an LPS-depleting agent, however, suggests that those results could be caused by LPS contamination. In the presence of polymyxin B, the inflammatory-boosting effect by S2 was eliminated, indicating that LPS is categorically essential and the S protein alone is not able to activate the TLR4 pathway. Additionally, a recent study showed that specific blocking of LPS by the peptide TCP-25 completely abrogates S protein–LPS-induced lung inflammation in an experimental mouse model (Puthia et al., 2022). Moreover, our findings that the intrinsic proinflammatory activity of the two different S2 preparations is correlated to the presence of minute LPS contaminants elegantly confirmed the LPS dependence of the observed inflammation. Indeed, two recent studies showed that commercially produced S protein preparations contain varying concentrations of LPS, which correlate with their abilities to induce cytokine expression in blood cells (Ouyang et al., 2021; Cinquegrani et al., 2022). Similar to our results, the cytokine production by these S protein reagents was abrogated by polymyxin B. This further corroborates our proposed mechanism whereby the S protein is an intermediary rather than a direct causality of hyperinflammation.
Around 8 million sequences of SARS-CoV-2 genome have been deposited on the Global Initiative on Sharing All Influenza Data (GISAID) platform (Elbe and Buckland-Merrett, 2017) and almost all of the residues on the S protein have been detected to mutate at least once since its discovery. We calculated the percentage of occurrence of amino acid changes from hydrophobic to non-hydrophobic residues for the LPS-binding residues and found a value of well below 0.01% for all positions in the three binding pockets (Supplementary Figure S18A). The LPS-binding residues are therefore highly conserved in emerging SARS-CoV-2 variants and unlikely to mutate in the future. Hence, these sites could be attractive targets for development of therapeutics. When compared to sequences from other related beta-coronaviruses, most of these residues are also conserved (Supplementary Figure S18B), suggesting a potentially universal LPS-binding capability. We previously showed that SARS-CoV S protein interacts with LPS similarly to SARS-CoV-2 (Petruk et al., 2020); further studies would be required to determine LPS interactions with S proteins from other coronaviruses.
LPS binding to the NTD and RBD is interesting due to their instrumental role in immune evasion of SARS-CoV-2. Haem metabolites binding to the NTD yields profound conformational changes that inhibit neutralizing antibodies (Rosa et al., 2021). As LPS binds to the same pocket, it is possible that it could also alter the conformation of the antigenic supersite (McCallum et al., 2021b). Linoleic acid binding to the RBD bridges two adjacent RBDs (Toelzer et al., 2020), hence favouring the down state, which reduces accessibility to antibody epitopes. Similarly, the charged headgroup of LPS protrudes out of the binding pocket and could potentially interact with a neighbouring RBD and shift the open–closed equilibrium of the S protein. Various mutations found on the NTD and RBD of SARS-CoV-2 variants accelerate community transmission and vaccine breakthrough (McCallum et al., 2021a). None of these mutations appear in any of the LPS-binding pockets, implying that the S proteins from these variants are competent to bind to LPS. Indeed, analyses of the Omicron S protein showed that the boosting of LPS responses is preserved (Supplementary Figure S16). The overall LPS binding, as determined using BN-PAGE under conditions where S protein is saturated with LPS, is retained in the Omicron variant. However, the affinity of Omicron S protein to LPS is reduced. The KD obtained by MST for Omicron S protein is similar to the KD for the Wuhan S2 subunit, a finding compatible with the S2 site's role in LPS binding and proinflammatory boosting. We also observed a moderate decrease in the LPS boosting effect compared to the Wuhan strain in vitro and in vivo. This observation is indeed interesting as the Omicron variant produces less severe COVID-19 symptoms (Shuai et al., 2022). Intriguingly, a differential effect of LPS on maximal intrinsic fluorescence was particularly observed between the Wuhan and Omicron S proteins, while the difference is less marked between the corresponding S1 subunits, suggesting different exposure patterns of tryptophan residues in S and S1, respectively.
The evolutionary origin of LPS-binding capacity by the SARS-CoV-2 S protein remains an enigma. From a structural standpoint, the presence of partially exposed hydrophobic patches that allow for LPS binding is counterintuitive. Some of the residues in the S2 pocket that interact with LPS acyl tails form the central core of the S protein post-fusion structure (Cai et al., 2020b). These residues become more exposed to solvent upon S1 shedding, suggesting that they may be a part of the ‘spring-loaded’ S2 core that drives the pre- to post-fusion conformational transition. The LPS-binding site could therefore arise as a side effect of the inclusion of these metastable features required for viral infection.
The LPS-binding sites could also have emerged to promote direct bacteria–virus interaction. Interkingdom synergy between bacteria and viruses at the host–pathogen interface plays monumental roles in various co-infections (Neu and Mainou, 2020). It is possible that SARS-CoV-2 has evolved to bind to Gram-negative bacteria via S protein–LPS interactions to modulate certain aspects of its viral life cycle. If so, it may be speculated that the occurrence of the Omicron variant having a preserved, albeit reduced interaction with LPS and possibly bacteria reflects an evolutionary adaptation of SARS-CoV-2 to the human system, dominated by interactions with receptors such as ACE2. However, the exact role of these interactions would require further studies.
In conclusion, we report in detail the molecular mechanism of interaction between the SARS-CoV-2 S protein and bacterial LPS, suggestive of how this interaction leads to enhanced TLR4-mediated inflammatory responses. Our study also provides novel therapeutic avenues for drug development against hyperinflammation observed in severe COVID-19 cases.
Materials and methods
S protein expression and purification
S protein expression and purification for use in HDXMS experiments were carried out as described previously (Raghuvamsi et al., 2021). For BN-PAGE, MST, tryptophan fluorescence quenching, THP-1 cell assay, and in vivo experiments, SARS-CoV-2 S, S1, S2 (wild-type and P-mutant), as well as Omicron SARS-CoV-2 S and S1 produced by ACROBiosystems (USA) were used. Further details are included in Supplementary Methods.
BN-PAGE
SARS-CoV-2 S1, S2, S2 P-mutant, as well as Omicron SARS-CoV-2 S (2 µg) were incubated with increasing doses of Escherichia coli LPS or lipid A (0.1–0.5 mg/ml) for 30 min at 37°C in 20 µl as final volume. Samples were subsequently mixed with loading buffer and analysed as described in Supplementary Methods. All experiments were performed at least 3 times.
HDXMS
Deuterium labelling was performed on free S protein as well as S protein incubated with E. coli LPS or lipid A molecules solubilized in PBS at pH 7.4. All reactions were performed at 37°C in PBS buffer reconstituted in 99% D2O attaining at final concentration of 90% for 1, 10, or 100 min before quenching as described in Supplementary Methods. Approximately 100 pmol of quenched reaction mixture was then subjected to pepsin digestion and subsequent mass spectrometry as described in Supplementary Methods. Deuteros 2.0 software was used to perform statistical analysis to identify CI and generate Woods plots (Lau et al., 2021).
MST
SARS-CoV-2 S, S1, S2 as well as Omicron S and S1 (40 µg) were labelled and 5 µl of 20 nM labelled proteins were incubated with 5 µl of increasing concentrations of LPS or lipid A (0.007–250 µM) in 10 mM Tris at pH 7.4. MST analysis was then performed as described in Supplementary Methods. Results shown are mean values ± standard deviation (SD) of six measurements.
Quenching of intrinsic fluorescence
The intrinsic fluorescence of 0.15 µM SARS-CoV-2 S protein (Wuhan and Omicron) and 0.30 µM S1 (Wuhan and Omicron), titrated with increasing concentrations of LPS (0–4 µM), was analyzed by recording protein emission fluorescence spectra at 300–450 nm following excitation at 280 nm. Fluorescence spectra were recorded as described in Supplementary Methods and the results are shown as mean values ± SD of three measurements.
MD simulations
E. coli lipid A was placed 3 nm away from the NTD, RBD (Wuhan and Omicron), and S2 pockets of the S protein and allowed to spontaneously bind to the pockets in unbiased all-atom MD simulations. Three replicates of 1 µs (NTD and RBD) or 500 ns (S2) simulations were performed using parameters described in Supplementary Methods. To estimate the binding affinity of lipid A to each binding pocket, we performed PMF calculations. Cluster analysis was performed on unbiased simulations and the central structure of the top cluster was used as the starting coordinates for a steered MD simulation. Umbrella sampling MD simulations were subsequently run using protocols described in Supplementary Methods.
NF-κB activation assay
THP-1-XBlue-CD14 reporter cells were treated with 5 nM SARS-CoV-2 S, S1, S2, S2 P-mutant, as well as Omicron SARS-CoV-2 S in the presence of increasing doses of LPS (0–1 ng/ml). The cells were incubated at 37°C for 20 h and subsequently the NF-κB activation was analysed as described in Supplementary Methods. Data shown are mean values ± SD from at least three independent experiments performed in triplicate.
Mouse inflammation model and in vivo imaging
BALB/c Tg(NF-κB-RE-luc)-Xen reporter mice were subcutaneously injected with a preparation of 5 µg SARS-CoV-2 S, S1, S2 as well Omicron SARS-CoV-2 S and S1 mixed with 2 µg LPS. Mice were subsequently imaged at 1, 3, and 6 h after the subcutaneous deposition as described in Supplementary Methods.
Supplementary Material
Acknowledgements
Simulations were performed on resources of the National Supercomputing Centre, Singapore (https://www.nscc.sg), the A*STAR Computational Resource Centre (A*CRC), and the supercomputer Fugaku provided by RIKEN through the HPCI System Research Project (Project ID: hp200303). The authors thank Ann-Charlotte Strömdahl (Lund University) for excellent technical assistance with THP-1 cells.
Contributor Information
Firdaus Samsudin, Bioinformatics Institute (BII), Agency for Science, Technology and Research (A*STAR), Singapore 138671, Singapore.
Palur Raghuvamsi, Bioinformatics Institute (BII), Agency for Science, Technology and Research (A*STAR), Singapore 138671, Singapore; Department of Biological Sciences, National University of Singapore, Singapore 117543, Singapore.
Ganna Petruk, Division of Dermatology and Venereology, Department of Clinical Sciences, Lund University, SE-22184 Lund, Sweden.
Manoj Puthia, Division of Dermatology and Venereology, Department of Clinical Sciences, Lund University, SE-22184 Lund, Sweden.
Jitka Petrlova, Division of Dermatology and Venereology, Department of Clinical Sciences, Lund University, SE-22184 Lund, Sweden.
Paul MacAry, Life Sciences Institute, Centre for Life Sciences, National University of Singapore, Singapore 117546, Singapore.
Ganesh S Anand, Department of Biological Sciences, National University of Singapore, Singapore 117543, Singapore; Department of Chemistry, The Pennsylvania State University, PA 16801, USA.
Peter J Bond, Bioinformatics Institute (BII), Agency for Science, Technology and Research (A*STAR), Singapore 138671, Singapore; Department of Biological Sciences, National University of Singapore, Singapore 117543, Singapore.
Artur Schmidtchen, Division of Dermatology and Venereology, Department of Clinical Sciences, Lund University, SE-22184 Lund, Sweden; Copenhagen Wound Healing Center, Bispebjerg Hospital, Department of Biomedical Sciences, University of Copenhagen, DK-2400 Copenhagen, Denmark.
Funding
This work was supported by BII (A*STAR) core funds, grants FY21_CF_HTPO SEED_ID_BII_C211418001 funded by A*STAR, the Swedish Research Council (projects 2017-02341 and 2020-02016), Edvard Welanders Stiftelse and Finsenstiftelsen (Hudfonden), the Torsten Söderberg, Crafoord, and Österlund Foundations, Stiftelsen Lars Hiertas Minne, the Royal Physiographic Society of Lund, and the Swedish Government Funds for Clinical Research (ALF).
Conflict of interest: A.S. is a founder and shareholder of in2cure AB, a company developing therapies based on host defense peptides. A patent application related to the present work, with A.S., G.P. and M.P. listed as inventors, has been filed.
Author contributions: P.J.B. and A.S. designed the study. F.S. performed molecular modelling and simulations. P.R. conducted HDXMS experiments. G.P. conducted BN-PAGE, in vitro NF-κB cell activation, and tryptophan fluorescence quenching assays. M.P. performed mouse inflammation model and in vivo bioimaging. J.P. performed MST binding assay. P.J.B., A.S. and G.S.A. supervised the experiments and simulations. All authors drafted and finalised the manuscript.
References
- Aboudounya M.M., Holt M.R., Heads R.J. (2021). SARS-CoV-2 Spike S1 glycoprotein is a TLR4 agonist, upregulates ACE2 expression and induces pro-inflammatory M1 macrophage polarisation. bioRxiv, 10.1101/2021.08.11.455921 [DOI]
- Avedissian S.N., Liu J., Rhodes N.J.et al. (2019). A review of the clinical pharmacokinetics of polymyxin B. Antibiotics 8, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awoyemi A., Trøseid M., Arnesen H.et al. (2018). Markers of metabolic endotoxemia as related to metabolic syndrome in an elderly male population at high cardiovascular risk: a cross-sectional study. Diabetol. Metab. Syndr. 10, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangaru S., Ozorowski G., Turner H.L.et al. (2020). Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate. Science 370, 1089–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q., Chen F., Wang T.et al. (2020a). Obesity and COVID-19 severity in a designated hospital in Shenzhen, China. Diabetes Care. 43, 1392–1398. [DOI] [PubMed] [Google Scholar]
- Cai Y., Zhang J., Xiao T.et al. (2020b). Distinct conformational states of SARS-CoV-2 spike protein. Science 369, 1586–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camell C.D., Yousefzadeh M.J., Zhu Y.et al. (2021). Senolytics reduce coronavirus-related mortality in old mice. Science 373, eabe4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Wang J., Jian F.et al. (2021). Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 602, 657–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiliveri S.C., Louis J.M., Ghirlando R.et al. (2021). Transient lipid-bound states of spike protein heptad repeats provide insights into SARS-CoV-2 membrane fusion. Sci. Adv. 7, eabk2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinquegrani G., Spigoni V., Iannozzi N.T.et al. (2022). SARS-CoV-2 Spike protein is not pro-inflammatory in human primary macrophages: endotoxin contamination and lack of protein glycosylation as possible confounders. Cell Biol. Toxicol. 38, 667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello S.M., Shoemaker S.R., Hobbs H.T.et al. (2022). The SARS-CoV-2 spike reversibly samples an open-trimer conformation exposing novel epitopes. Nat. Struct. Mol. Biol. 29, 229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S., Buckland-Merrett G. (2017). Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob. Chall. 1, 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorentino F., Sauer J.B., Qiu X.et al. (2021). Dynamics of an LPS translocon induced by substrate and an antimicrobial peptide. Nat. Chem. Biol. 17, 187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W., Ni Z., Hu Y.et al. (2020). Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 382, 1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailman E., Vasselon T., Kelley M.et al. (1996). Stimulation of macrophages and neutrophils by complexes of lipopolysaccharide and soluble CD14. J. Immunol. 156, 4384–4390. [PubMed] [Google Scholar]
- Hoel H., Heggelund L., Reikvam D.H.et al. (2021). Elevated markers of gut leakage and inflammasome activation in COVID-19 patients with cardiac involvement. J. Intern. Med. 289, 523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber R.G., Berglund N.A., Kargas V.et al. (2018). A thermodynamic funnel drives bacterial lipopolysaccharide transfer in the TLR4 pathway. Structure 26, 1151–1161.e4. [DOI] [PubMed] [Google Scholar]
- Ke Z., Oton J., Qu K.et al. (2020). Structures, conformations, and distributions of SARS-CoV-2 spike protein trimers on intact virions. Nature 588, 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.M., Park B.S., Kim J.I.et al. (2007). Crystal structure of the TLR4–MD-2 complex with bound endotoxin antagonist Eritoran. Cell 130, 906–917. [DOI] [PubMed] [Google Scholar]
- Kruglikov I.L., Scherer P.E. (2021). Preexisting and inducible endotoxemia as crucial contributors to the severity of COVID-19 outcomes. PLoS Pathog. 17, e1009306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglikov I.L., Shah M., Scherer P.E. (2020). Obesity and diabetes as comorbidities for COVID-19: underlying mechanisms and the role of viral–bacterial interactions. eLife 9, e61330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A.M., Claesen J., Hansen K.et al. (2021). Deuteros 2.0: peptide-level significance testing of data from hydrogen deuterium exchange mass spectrometry. Bioinformatics 37, 270–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchini A., Micciulla S., Corucci G.et al. (2021). Lipid bilayer degradation induced by SARS-CoV-2 spike protein as revealed by neutron reflectometry. Sci. Rep. 11, 14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwick P.R.L., Peacock R.B., Komives E.A. (2019). Accurate prediction of amide exchange in the fast limit reveals thrombin allostery. Biophys. J. 116, 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., Bassi J., De Marco A.et al. (2021a). SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science 373, 648–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., De Marco A., Lempp F.A.et al. (2021b). N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 184, 2332–2347.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R., Janeway C.J. (2000). Innate immunity. N. Engl. J. Med. 343, 338–344. [DOI] [PubMed] [Google Scholar]
- Neu U., Mainou B.A. (2020). Virus interactions with bacteria: partners in the infectious dance. PLoS Pathog. 16, e1008234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W., Xie T., Fang H.et al. (2021). Variable Induction of pro-inflammatory cytokines by commercial SARS-CoV-2 spike protein reagents: potential impacts of LPS on in vitro modeling and pathogenic mechanisms in vivo. Int. J. Mol. Sci. 22, 7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk G., Puthia M., Petrlova J.et al. (2020). SARS-CoV-2 spike protein binds to bacterial lipopolysaccharide and boosts proinflammatory activity. J. Mol. Cell Biol. 12, 916–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poltorak A., He X., Smirnova I.et al. (1998). Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282, 2085–2088. [DOI] [PubMed] [Google Scholar]
- Puthia M., Tanner L., Petruk G.et al. (2022). Experimental model of pulmonary inflammation induced by SARS-CoV-2 spike protein and endotoxin. ACS Pharmacol. Transl. Sci. 5, 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuvamsi P., Tulsian N., Samsudin F.et al. (2021). SARS-CoV-2 S protein:ACE2 interaction reveals novel allosteric targets. eLife 10, e63646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Sarmiento C.A., Komives E.A. (2018). Hydrogen–deuterium exchange mass spectrometry reveals folding and allostery in protein–protein interactions. Methods 144, 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittirsch D., Flierl M.A., Ward P.A. (2008). Harmful molecular mechanisms in sepsis. Nat. Rev. Immunol. 8, 776–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A., Pye V.E., Graham C.et al. (2021). SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity. Sci. Adv. 7, eabg7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J.K., Kim S.J., Rah S.H.et al. (2017). Reconstruction of LPS transfer cascade reveals structural determinants within LBP, CD14, and TLR4–MD2 for efficient LPS recognition and transfer. Immunity 46, 38–50. [DOI] [PubMed] [Google Scholar]
- Shuai H., Chan J.F.-W., Hu B.et al. (2022). Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature 603, 693–699. [DOI] [PubMed] [Google Scholar]
- Sirivongrangson P., Kulvichit W., Payungporn S.et al. (2020). Endotoxemia and circulating bacteriome in severe COVID-19 patients. Intensive Care Med. Exp. 8, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira P.C., Dorneles G.P., Filho P.C.S.et al. (2021). Increased LPS levels coexist with systemic inflammation and result in monocyte activation in severe COVID-19 patients. Int. Immunopharmacol. 100, 108125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toelzer C., Gupta K., Yadav S.K.N.et al. (2020). Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein. Science 370, 725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A.et al. (2020). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 180, 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.Y., Wang J., Dokholyan N.V. (2021). Prefusion spike protein stabilization through computational mutagenesis. Proteins Struct. Funct. Bioinf. 89, 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Kuang M., Li J.et al. (2021). SARS-CoV-2 spike protein interacts with and activates TLR4. Cell Res. 31, 818–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R.et al. (2020). Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet North Am. Ed. 395, 1054–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuzic L., Samsudin F., Shivgan A.T.et al. (2022). Uncovering cryptic pockets in the SARS-CoV-2 spike glycoprotein. Structure 30, 1062–1074.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.