Abstract
The nucleic acid therapeutics field has made tremendous progress in the past decades. Continuous advances in chemistry and design have led to many successful clinical applications, eliciting even more interest from researchers including both academic groups and drug development companies. Many preclinical studies in the field focus on improving the delivery of antisense oligonucleotide drugs (ONDs) and/or assessing their efficacy in target tissues, often neglecting the evaluation of toxicity, at least in early phases of development. A series of consensus recommendations regarding regulatory considerations and expectations have been generated by the Oligonucleotide Safety Working Group and the Japanese Research Working Group for the International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use S6 and Related Issues (WGS6) in several white papers. However, safety aspects should also be kept in sight in earlier phases while screening and designing OND to avoid subsequent failure in the development phase. Experts and members of the network “DARTER,” a COST Action funded by the Cooperation in Science and Technology of the EU, have utilized their collective experience working with OND, as well as their insights into OND-mediated toxicities, to generate a series of consensus recommendations to assess OND toxicity in early stages of preclinical research. In the past few years, several publications have described predictive assays, which can be used to assess OND-mediated toxicity in vitro or ex vivo to filter out potential toxic candidates before moving to in vivo phases of preclinical development, that is, animal toxicity studies. These assays also have the potential to provide translational insight since they allow a safety evaluation in human in vitro systems. Yet, small preliminary in vivo studies should also be considered to complement this early assessment. In this study, we summarize the state of the art and provide guidelines and recommendations on the different tests available for these early stage preclinical assessments.
Keywords: antisense oligonucleotides, early preclinical safety assessment, toxicity, predictive assays
Introduction
Nucleic acid therapeutics (NATs) based on the use of antisense oligonucleotide drugs (ONDs) was, for many years, a niche for research in rare disorders, as traditional treatments were not available. However, considering that only a small fraction of the human genome has been successfully drugged to date [1], the approval of a dozen ONDs, including eight since 2018, has opened the doors to their application not only to a wider range of rare diseases but also to many common disorders. Because the mechanism of action of ONDs is related to the well-understood Watson–Crick–Franklin base-pairing rules, ONDs are straightforward to design, and this has increased the number of academic laboratories and small companies evaluating their potential to treat a myriad of disorders.
However, while many reference publications advise on how to design and evaluate the efficacy of these drugs, fewer address the evaluation of their safety and toxicological aspects. While many basic researchers may consider they are not likely to bring these molecules to a stage where formal safety assessment is required, there are several safety considerations that should be taken into account as early as possible during the discovery process. This would ensure correct interpretation of experiments and increase probability of success in the eventual future transition of a candidate molecule to clinical trials. Aspects such as the sequence, the chemistry of the backbone or the use of delivery systems to ensure efficient access to target tissues will influence the safety of these drugs and should be considered already at project initiation.
Broadly speaking, ONDs can be used to inhibit or restore the expression of some target genes by diverse mechanisms, including degrading messenger RNA (mRNA) transcripts causing gene silencing/knockdown or altering the splicing of pre-mRNAs [2]. For example, transcript degradation of mRNA can be achieved by antisense oligonucleotides (ASOs) recruiting RNase H1 cleaving DNA-RNA hybrids; by small interfering RNAs (siRNAs) that mediate target RNA degradation through RNA-induced silencing complex (RISC); or by splice-switching OND, which may be designed to skip regular exons in the pre-mRNA and thus create mRNA isoforms that encode nonfunctional proteins or trigger degradation of the mRNA by nonsense-mediated decay [3,4].
However, to restore functional protein expression in the treatment of disorders caused by splicing alterations, splice switching OND may be designed to promote exon inclusion, pseudo-exon exclusion, exon skipping, or splicing redirection in pre-mRNA [5]. More recently, ONDs have also been shown to upregulate gene expression by targeting nonproductive splicing events [6] or through the binding to regulatory elements, for example, upstream open reading frames [7,8].
As part of the endeavors of the network “DARTER,” a COST Action funded by the Cooperation in Science and Technology of the EU (www.antisenserna.eu), we aim to provide researchers with some guidelines on safety considerations during preclinical research, also called “discovery phase,” when designing these versatile drugs. A summary of the different assays that may be included from early on in the discovery phase is shown in Table 1 and expanded in the text of our article. In this article, we will focus on single-stranded ASOs, in particular phosphorothioate (PS)-ASOs because most of the published assays are described for that design, but most concepts also apply to other chemistries and mechanisms of action and we will provide examples for siRNAs when available.
Table 1.
Summary of the Main Recommended Predictive Assays and the Associated References for Detailed Protocols
| OND toxicity | Predictive assays | In vitro/in vivo | References for protocols |
|---|---|---|---|
| Off-target effects | In silico evaluation/in vitro validation | In silico and in vitro | Guidelines published by the OSWG in 2012 (Lindow et al. [16]; Michel et al. [146]; Yoshida et al. [147]) |
| Immunostimulatory effects | • Quantification of cytokines/chemokines release in human PBMC or WBA by ELISA • Quantification of cytokines as well as CCL22 mRNA levels in BJAB cells by qRT-PCR |
In vitro | Coch et al. [81]; Lankveld et al. [82]; Sewing et al. [104]; Anderson et al. [83] |
| Toxicities in high exposure organs | • Cytotoxicity by caspase assay Predictive assays for hepatotoxicity: • Quantification of LDH and ATP in primary hepatocytes • Caspase assay in transfected mouse 3T3 fibroblasts or human HepG2 cells • Evaluation of liver enzymes in mice/NHP Predictive assays for nephrotoxicity: • Quantification of EGF in human kidney tubule epithelial cells • Quantification of kidney injury biomarkers using chip-cultured HRPTEC • Quantification of urinary biomarkers (eg, β2-microglobulin and KIM-1) by ELISA |
In vitro
In vitro In vitro In vivo In vitro In vitro In vivo |
Anderson et al. [83] Sewing et al. [104,105] Dieckmann et al. [94] Burel et al. [95] Moisan et al. [114] Nieskens et al. [115] Echevarría and Goyenvalle [117] |
| Thrombocytopenia | • Evaluation of platelet activation in human or NHP platelet-rich plasma or whole blood by flow cytometry (activation of CD62P and PAC-1) | In vitro and in vivo | Narayanan et al. [90]; Sewing et al. [71]; Slingsby et al. [85] |
| Inhibition of coagulation | • Quantification of PT and aPTT in vitro in human/mouse/NHP citrated plasma | In vitro | Echevarría et al. [122]; Relizani et al. [125] |
| Complement activation | • Quantification of split products of the APC (C3a, Bb, and C5a) in vitro in human/NHP/mouse serum | In vitro | Aupy et al. [123]; Henry et al. [129]; Sewing et al. [71] |
| CNS-specific toxicities | • Prediction of neurotoxicity from sequence features • Quantification of spontaneous calcium oscillations in primary cortical neuronal cultures |
In silico
In vitro |
Hagedorn et al. [27] Hagedorn et al. [27] |
APC, alternative pathway of the complement; aPTT, activated partial thromboplastin time; ATP, adenosine triphosphate; BJAB, EBV-negative Burkitt-like lymphoma cell line; CCL22, C-C Motif Chemokine Ligand 22; CNS, central nervous system; EGF, epidermal growth factor; ELISA, enzyme-linked immunosorbent assay; HRPTEC, human renal proximal tubule epithelial cells; KIM-1, kidney injury molecule-1; LDH, lactate dehydrogenase; mRNA, messenger RNA; NHP, non human primate; OND, oligonucleotide drug; OSWG, Oligonucleotide Safety Working Group; PBMC, peripheral blood mononuclear cell; PT, prothrombin time; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; WBA, whole blood assay.
The target tissue and delivery route should also be carefully considered since high exposure organs will be different depending on local or systemic delivery. In this review, we will mostly discuss systemically delivered OND, which may accumulate in the liver or kidney, but we will also separately discuss the safety aspect of direct delivery to the central nervous system (CNS), which has gained increased interest over the past few years, especially since the clinical success of nusinersen.
Overall Strategy and Considerations for OND Safety Testing
For OND projects aiming to reach the clinic, it is useful to understand the expectations of health authorities approving clinical trials. In 2020, the Japanese Pharmaceuticals and Medical Devices Agency issued a guideline for preclinical safety assessment of oligonucleotide therapeutics. In 2021 the U.S. Food and Drug Administration (FDA) issued a draft guidance, “Nonclinical Testing of Individualized Antisense Oligonucleotide Drug Products for Severely Debilitating or Life-Threatening Diseases,” providing a high-level overview of the key issues they will consider in the special situation of developing an OND to treat extremely rare mutations present on a single or very small number of patients (not for commercial development).
No specific formal guidelines for preclinical testing of ONDs to treat more common conditions have been issued to date by the International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use (ICH), FDA, or European Medical Agency (EMA), and so the testing principles normally follow the overall ICH M3(r2) guideline for small molecules. However, with great support from both sponsors and regulators, a series of consensus recommendations regarding regulatory considerations and expectations for OND have been generated by the Oligonucleotide Safety Working Group (OSWG) and other working groups in several white papers [9–20].
As briefly discussed in the Introduction, many academic researchers may not aim for clinical trials but “only” design and use ONDs to modulate targets to answer some specific biological questions. However, ONDs are not inert molecules and unless safety properties are understood and ideally selected for during potency screening of such research tools, scientists run the risk of incorrect interpretation of results due to undesired effects confounding the results. Although the requirements may be more rigorous for potential clinical candidates, the same principles and considerations apply for early safety assessment of tool compounds.
The main determinants of OND properties are chemistry, sequence, and design. For optimal performance, the choice of OND chemistry and design should be matched with intended use. Depending on ribose modification pattern, single-stranded ASOs with a PS backbone with a DNA gap can be used to reduce target transcripts via RNase H mechanism or without a DNA gap as steric blockers to modify splicing. With neutral backbone, for example, phosphorodiamidate morpholino oligomer (PMO) single-stranded ASOs can be used for splice modulation but not for RNase H mechanism.
RISC-dependent mechanisms such as target transcript degradation or microRNA mimics generally require a double-stranded design. In addition to mechanism, the chemistry and design also influence distribution and uptake. In general, single-stranded ASOs with PS backbone show productive uptake (leading to a pharmacodynamic effect) in several cell types on their own [21], whereas neutral backbone ASOs and double-stranded OND mostly need a delivery system or high doses for sufficient activity.
After having decided on the chemistry and design, the next step is screening for sequences with sufficient desired activity. Several guidelines have been published for OND sequence selection in silico [22–24]. Sequence motifs known to be associated with undesired safety effects should ideally be avoided already during the in silico design stage. Published examples of such motifs include cytosine guanine motifs (CpG) motifs activating Toll-like receptor (TLR)9 (immunostimulatory effects), polyG motifs, polypyrimidine, and other sequence motifs associated with toxicity in the liver [25,26] or neuronal toxicity flags [27]. Despite perfect homology, many sequences show poor activity so potency must be confirmed in vitro [28–30] using quantitative polymerase chain reaction for knockdown mechanisms followed by selection of the most potent sequences for further testing of for example, safety properties.
There are several points to consider when establishing assays and models for safety assessment of ONDs:
-
1.
Is the assay intended for screening/filtering out undesired properties or for thorough characterization of for example, a clinical candidate?
-
2.
Which reference ONDs should be used to validate the assay?
-
3.
What should the study design look like?
-
4.
Which are the best readouts?
Most established in vitro cellular models developed for safety screening of small molecules are either not relevant due to toxicity mechanism(s), not applicable for ONDs, or are not sensitive enough to pick up OND-induced toxicities. More complex in vitro models such as microphysiological systems (MPS) may be able to bridge this gap [31]. MPS are likely more reflecting the in vivo situation, but the cost and limited throughput will often prevent larger scale screening. At present, these models are likely best suited for characterization of a smaller number of really promising candidates.
Selection of in vivo models also needs careful consideration. Some safety concerns, such as proinflammatory manifestations, show relatively poor translation between species but may still be relevant to assess and select against to increase probability of success in regulatory toxicology studies. Moreover, it is not uncommon that ONDs showing good tolerability in naive wild-type animals trigger toxicity in disease models used for target validation and pharamokinetic/pharmacodynamic studies. To avoid confounding (toxicity) factors in such studies, safety characterization in the model of choice may be warranted before running larger pivotal studies.
The right reference compounds should be selected to validate and optimize the assay. These reference compounds should ideally be of the same chemistry and design as the OND aimed to be identified, for example, 3-10-3 locked nucleic acid (LNA)-DNA-LNA gapmer with full PS backbone modification or a GalNAc-conjugated fully 2′OMethyl modified 20mer PS backbone steric blocker. Smaller companies or academic research groups normally do not have a historic library of such reference compounds to use, but careful search of published articles can often identify some examples of safe and toxic reference compounds for a specific readout.
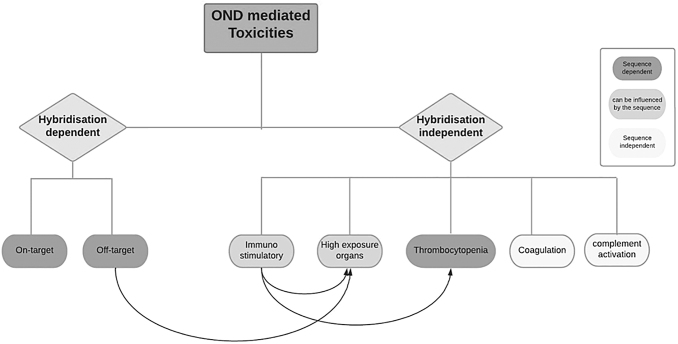
With a relatively slow onset to showing toxicity, OND safety in vitro and in vivo assays may require adapted study design, for example, longer duration than for other modalities. The safety considerations with ONDs can be categorized as: (1) hybridization and sequence dependent, (2) hybridization independent but sequence dependent, and (3) hybridization and sequence independent (“backbone effect”) (Fig. 1).
FIG. 1.
OND-associated toxicities: schematic representation of the most common OND-mediated toxicities. Some of these effects are strictly chemistry and design specific (sequence independent, white boxes), some are mostly class specific but can be influenced by the sequence (light gray boxes), and others are sequence dependent (dark gray boxes). The arrows represent the impact of a specific effect on another, for example, immunostimulatory effects play a role on thrombocytopenia and toxicities observed in high exposure organs. OND, oligonucleotide drug.
Hybridization- and Sequence-Dependent Effects: On- and Off-Target Safety
Potential on-target toxicities
Before a therapeutic candidate can move forward in the preclinical pipeline, its safety profile needs to be considered and optimized. This should start already during the selection of the intended target, by assessing the potential for so-called on-target toxicity or exaggerated pharmacology. On-target toxicity can result in too strong intended effect (eg, hypoglycemia induced by a diabetes treatment) or an adverse response to the drug in a tissue, which is not the intended target tissue. Although no clear examples of on-target safety issues have been published, ONDs can show a much longer effect duration compared with many other drug modalities: the washout period may be weeks or even months should adverse on-target effects be observed. For targets with clear on-target risk, strategies of oligo-based antidotes have been described [32,33], although those would require a dedicated safety assessment package before being applied in the clinic.
Until better tools are available, the assessment of on-target toxicity mostly relies on compiling available information on the target with regard to its biological function, tissue expression pattern, and from published information of other drugs for the same target to assess potential risks for patients. In vitro assessments ideally should be performed in biologically relevant cell lines or preferably, in primary human cells or MPS when possible, and in vivo testing is generally required. In vivo assessment requires a species cross-reactive OND or a species active surrogate molecule that is sufficiently potent and safe and should be of the same chemistry and design as the candidate OND. Detailed considerations for assessing on-target toxicities for OND can be found in the study by Kornbrust et al. [15].
Potential off-target toxicities
However, hybridization-dependent off-target toxicities are those where ONDs act on transcripts other than the intended one via Watson–Crick–Franklin base pairing. These off-target effects may be severe, such as acute hepatotoxicity, and will be discussed further in the Toxicities in high exposure organs section. Several studies have described ways to assess hybridization-dependent off-target effects [34–38]. Some straightforward steps that should be followed when assessing off-target toxicities of gapmer ASOs using sequence alignment tools and sequencing data repositories (ie, transcriptomics) as summarized by the OSWG [16]:
-
1.
Perform an in silico analysis (eg, Basic Local Alignment Search Tool search optimized for short sequences or other tools such as RNAhybrid [39]) of the entire pre-mRNA transcriptome (including both introns and exons) to search for potential complementary sequences (off-target candidates) using defined criteria. From the predictions, prioritize the sequences with no or as few predicted hybridization off-targets as possible. This should be the obvious approach for gapmer ASOs or siRNA where numerous sequence options exist but may be more challenging for steric blockers where choice of sequence is significantly restricted. For OND intended for clinical development, it may be a good idea to also assess and minimize potential off-targets for the species that will be used for regulatory toxicity studies. However, it should be kept in mind that sequence-based affinity predictors do not consider secondary structures nor RNA binding proteins that can influence the activity of ASO [40].
-
2.
Reduce the list of potential off-target transcripts: for example, those expressed exclusively in organs with insufficient exposure, for example, CNS in the case of systemically administered PS backbone ASOs.
-
3.
Perform in vitro concentration-response activity margin assessment in cells expressing both the primary target and the off-targets identified by in silico approaches using, for example, revese transcription-quantitative polymerase chain reaction or RNA-seq. The ASO exposure is ideally done by free (or “gymnotic”) uptake, but since all cell models may not support this, other delivery methods such as lipid transfection and electroporation can be considered, in which case the nonspecific impact of the delivery method should be taken into account.
-
4.
The size of an acceptable in vitro margin depends on the potential consequence of meaningful modulation of the off-target at the intended dose levels. For off-targets lacking clear in vitro margins to primary target, follow the same principles as for on-target safety assessment, that is, assessing tissue expression profile, described biological role, pattern of developmental expression, effect of other compounds hitting the same target, and phenotype of genetically modified animals and human polymorphisms. Also, the potential of the off-target to trigger an adverse effect could be evaluated in relevant in vitro system (eg, MPS) and even in vivo using appropriate surrogate OND of the same chemistry and design targeting the animal version of the off-target.
In contrast to gapmer ASOs, ONDs that modulate pre-mRNA splicing by blocking access of splicing factors (general called steric-blocking oligonucleotides or SBO) may result in off-target events by causing unwanted splicing effects at near-complementary sites, or by causing changes in transcript levels (expression changes). A recent study analyzed the off-target splicing effects of 81 SBO and the off-target expression changes of a subset of 46 of those [41]. They found that off-target splicing effects are more predictable and predominantly hybridization dependent, whereas the off-target expression effects are less reproducible, although more frequent, and are probably driven by other mechanisms beyond hybridization such as transcriptional events and experimental variation.
Therefore, off-target expression effects of SBOs need to be investigated and validated in vitro in cells (and not only in silico) before being considered as bona fide off-target effects. In silico models to predict SBO off-target effects are often of limited use because they miss the majority of unwanted events. Therefore, conducting in vitro experiments such as RNA sequencing are preferable. Off-target hybridization-dependent splicing changes may be reduced by combining two SBOs at lower doses each, introducing mismatches and using shorter sequences [41]. The mode of delivery is also an important factor. Off-target mis-splicing events appear to be a greater problem in vitro when delivered to the cell by lipid transfection than in vivo, perhaps because in vivo there is a much lower amount of ASO available to reach the target [40].
When assessing potential hybridization-dependent on- and off-target toxicities for oligonucleotides, sequence differences may result in insufficient activity of the clinical candidate in animal test species. In such cases, surrogate ONDs designed for the test species (often rodent) may be developed and used in parallel to the human candidate to assess potential on-target safety in regulatory toxicity studies.
In addition, chemical modifications that protect against metabolizing nucleases result in slow tissue elimination and therefore in a longer duration of the effect of the ONDs. For example, the GalNAc-conjugated siRNA inclisiran shows maintained reduction of the target proprotein convertase subtilisin/kexin type 9 serine protease 6 months after a single subcutaneous administration. [42]. The potential mechanism for this observation has been described as a depot effect in lysosomes of the hepatocytes [43]. Although ASO gapmers show a shorter effect duration than siRNAs, several weeks are significantly longer than observed for most small-molecule therapeutics [44–47]. While this represents an advantage for patients, because less frequent administration is required, the sustained effect may require longer periods of time (weeks or months) for a potential adverse effect to resolve. Since long-term effects may be difficult to pick up in vitro in two-dimensional culture, this highlights the need for more complex system such as MPS, which can be kept in culture for several weeks [31].
Finally, the limited productive uptake of ONDs in certain cell types needs to be addressed when assessing hybridization-dependent on- and off-target safety risks. The limited productive uptake distribution often observed means that the real risk of any on- or off-target effect would be restricted to cell types with good productive uptake. For example, naked ASOs generally do not pass across the blood–brain barrier so the risk for on- and off-target toxicities in the CNS is rather negligible after systemic administration [21,45]. The tissue distribution of therapeutic oligonucleotides will vary depending on the administration routes and delivery systems such as conjugates or formulations, so understanding the productive uptake distribution for such new conditions is critical for proper risk assessment of potential on- and off-target toxicities.
Hybridization Independent But Sequence-Dependent Toxicities
In contrast with on- and off-target effects, some toxicities are hybridization independent but still depend on the nucleic acid sequence. These include proinflammatory manifestations and effects in high exposure organs, such as the liver and kidney.
Immunostimulatory effects
ONDs have long been recognized for their immune-stimulatory effects, which greatly depend on their design, chemistry, and nucleotide sequence [48,49]. ONDs can activate the innate immune system through binding to pattern recognition receptors (PRRs) such as the TLRs. While these immunostimulatory properties may be deliberately chosen for vaccine adjuvants or for cancer and autoimmune disease therapies [50–52], they are mostly undesired for other OND types.
The oligonucleotide sequence has been shown to be a key feature defining the immunomodulatory effects of ONDs. One of the best known example is the DNA CpG motif, which strongly activates TLR9 [53,54], particularly in the presence of optimal flanking sequences [55,56]. Examples of other proinflammatory sequences include RNA rich in guanine uracile motif or adenine uracil motifs, which trigger TLR7 and TLR8 responses [57,58] and U-rich sequences in siRNA [59–62]. The cytoplasmic PRRs such as retinoic acid-inducible gene I and protein kinase R are not activated by a specific nucleobase sequence but are instead triggered by structural features typical for viral RNA, such as the presence of uncapped 5′triphosphate in single- or double-stranded RNA, or blunt-ends in double-stranded RNA [63]. In contrast with proinflammatory sequences, some motifs have been associated with an immunosuppressant activity in particular for PS-containing ONDs [64–66].
To modify the immunostimulatory potential of a given sequence, chemical modifications have been introduced over the years. While it is commonly accepted that the PS modification has immunostimulatory effects [67,68], such effects have never been reported for OND with neutral backbones such as PMOs (at least when they are unconjugated) [69]. OND modifications frequently include 5′-methylation of cytosine residues to suppress the immune stimulatory effect of CpG DNA sequences [56,70]. Although ribose modifications such as 2′F, 2′methoxyethyl (MOE) and LNA were primarily introduced to increase affinity and further nuclease protection, their presence can also at least partly reduce the inflammatory stimuli [62,70,71].
Despite these efforts in design and chemical modifications, some of these modified ONDs still induce proinflammatory effects that can manifest in different ways in the clinic, including injection site reactions, flu-like symptoms, and thrombocytopenia [72–75]. Therefore, specific screening for immunostimulatory adverse effects is highly recommended in early preclinical stages to filter out potential proinflammatory candidates and prevent unexpected harmful effects in clinical development.
Rodents have been widely used for in vivo screening and they are considered particularly sensitive to immune stimulation, which does not always translate to humans. Treatment of mice with high doses of PS-ONDs has shown increased levels of circulating cytokines and chemokines [76,77] and resulted in a dose-dependent lymphoid hyperplasia with enlargement of spleen and lymph nodes as well as lymphohistiocytic cell infiltration often seen in multiple tissues [56,60,61,78,79]. In addition to in vivo assessment, in vitro tests have been designed to predict cytokine release using either isolated peripheral blood mononuclear cell or whole blood assay [71,80–82].
These offer the advantage of being species specific (human or non human primate [NHP]), which may be more relevant. Following incubation with various doses of ONDs, cytokines such as interleukin-6, interferon (IFN)-alpha, and tumor necrosis factor-alpha are generally measured in cell culture supernatants by enzyme-linked immunosorbent assay (ELISA) allowing fast, easy, and reliable predictions. An additional useful predictive assay was recently described using BJAB cells, an Epstein-Barr virus (EBV)-negative Burkitt-like lymphoma cell line, which is highly sensitive to stimulation by CpG and non-CpG oligonucleotides in a sequence and TLR9-dependent manner [83]. The induction of the cytokine CCl22 in OND-treated BJAB cells, measured by qRT-PCR, was shown to reliably predict the proinflammatory profile of gapmer ASOs.
Thrombocytopenia
Thrombocytopenia, that is, decrease in platelet counts, has been occasionally reported in preclinical models (rodents and NHPs) and in three recent clinical trials following treatment with ONDs, in particular PS-ASO (volanesorsen, inotersen, and drisapersen) [84]. Two phenotypes of platelet count decrease have been distinguished: phenotype 1 characterized by a moderate but not clinically severe drop in platelet count and the rarer phenotype 2 of severe thrombocytopenia [85]. In contrast with PS-ASO, severe thrombocytopenia has not been reported for siRNA drugs, neither in preclinical studies nor in clinical trials, but encapsulation of siRNA in lipid nanoparticles has been shown to cause decrease in platelet counts in rats, presumably induced by the cationic lipid molecules themselves [86,87].
Following the observed results in humans with PS-ASO, retrospective analysis of NHP and human data performed by the company Ionis revealed that thrombocytopenia is sequence dependent and consistent across species (dose–response and time of onset). Approximately 40% of the 102 evaluated sequences induce a phenotype 1 platelet decline in NHP compared with 20% of 16 sequences observed in humans [88].
The underlying mechanisms of thrombocytopenia are still being investigated, and several immune- and non-immune-mediated hypotheses have been proposed. The presence of PS backbone has been shown to increase the risk of platelet activation [71,89]. Mechanistic investigations using non-CpG ASOs with therapeutically common modifications indicate no effect on the bone marrow or thrombopoiesis and have largely ruled out a platelet factor-4 (PF4) mechanism [71,90]. Instead, Narayanan et al. showed that thrombocytopenia is associated with increases in total immunoglobulin M (IgM), antiplatelet IgM, and/or anti-PF4 IgM and that monocyte activation contributes to increased platelet sequestration in the spleen and liver, leading to decreased platelets in peripheral blood [90].
More recently, the sequence-specific binding of OND (PS-ASO specifically) to platelet glycoprotein VI was shown to activate human platelets triggering formation of platelet–leukocyte aggregates [85]. These findings also highlight the possibility of a genetic susceptibility component given that donors with higher platelet glycoprotein VI levels had greater OND-induced platelet activation.
Overall, published studies and publically available data suggest that diverse risk factors are relevant for thrombocytopenic events observed in the clinic. Importantly, in vitro tools have been implemented to address some of the risk factors originating from the nature of the OND molecule early on at design and screening stage. Platelet activation can indeed be determined by measuring the activation of marker P-selectin (CD62P) and PAC-1 (activated GPIIb/IIIa) in platelet-rich plasma by flow cytometry [71]. Since in vitro platelet activation represents only a potential risk with no full validation of translation to thrombocytopenia in the clinic, these readouts should only serve as prioritization criteria for choosing the best OND from a pool of molecules with different behavior. This precaution is particularly meaningful in projects in which the route of administration and treatment duration entail a theoretical risk, that is, typically chronic systemic applications.
Toxicities in high exposure organs
The highest concentrations of ONDs are generally found in the liver and kidney after systemic administration. The accumulation of ONDs per se in these organs is not necessarily associated with toxicities, which rather depend on the combination of OND sequence, chemistry, and designs. For ONDs of lower affinity chemistry (eg, 2′OMe, MOE), high doses leading to very high tissue concentrations are normally required to trigger kidney toxicity in preclinical species, but a different pattern is observed with ONDs of higher affinity chemistry [78], where there are examples of acute kidney injury in early clinical studies [91,92]. Importantly, gapmers using both moderate-affinity MOE and high-affinity LNA have caused unexpected renal toxicity in humans that had not been detected or predicted from the in vitro and in vivo screening assays run at the time [92,93], illustrating the importance of improved screening models for these effects, and the importance of monitoring relevant biomarkers in clinical studies.
As a first step to identify potential accumulation-driven cytotoxicity liabilities of ONDs, a caspase assay has been described using transient transfection or electroporation of ONDs in HepG2, 3T3, or Hepa1–6 cell lines and measuring caspase 3/7 activation [83,94]. This assay allows to deselect sequences with a high cytotoxicity liability early on before moving to more complex safety relevant in vitro models or in vivo studies.
Hepatotoxicity
Sequence-dependent toxicity of high-affinity ONDs has been observed in the liver relatively often during the discovery phase. In mice dosed only a single or a few times with some LNA or cEt gapmers, acute and subacute toxicities characterized by single cell necrosis, pronounced liver enzymes elevation, morbidity, and mortality have been reported [26,95,96]. Several studies have investigated these subacute toxicities and revealed different underlying mechanisms and identified sequence-specific motifs associated with hepatotoxicity [25,26].
A suggested mechanism involves PS backbone-dependent binding to key intracellular proteins in a sequence- and chemistry-dependent manner. Given the higher hydrophobic nature of high-affinity modifications, they also show higher affinity to a number of intracellular proteins compared with the same sequence with, for example, MOE chemistry [97–99]. Other molecular mechanisms have been proposed, including cell death as a cellular consequence to increased RNA degradation resulting from nonselective hybridization [71,94,95,100]. Indeed, hepatotoxicity was shown to be attenuated by RNase H1 knockdown [95].
In the case of siRNA, the hepatotoxicity observed in rodents has also been largely attributed to RNAi-mediated off-target effects [87], but not to the perturbation of RNAi pathways. This highlights the need to screen for hybridization-dependent off-target effects, as previously mentioned in the Potential off-target toxicities section.
Interestingly, it was recently shown that controlling PS stereochemistry in LNA gapmers or replacing PS in the DNA gap with methoxypropyl phosphonate or mesylphopsphoramidate can significantly improve their therapeutic index by improving safety without compromising activity [101–103], although this has not been tested in humans yet. In a similar detoxification effort, it was shown that the off-target effects observed with specific siRNA can be mitigated by modulating seed-pairing using a destabilizing chemical modification [36], analogous to placing an OMe in position 2 of the DNA part of gapmers [99].
In parallel to these design modifications reducing OND hepatotoxicity, predictive in vitro models have been established to filter out liver toxic candidates. The hepatotoxic potential of LNA gapmers can be evaluated in primary hepatocytes by measurement of extracellular lactate dehydrogenase and adenosine triphosphate levels following gymnotic delivery, as well as measurement of miR-122 expression in cell culture supernatants [104,105]. These studies showed that similar cytotoxicity readouts could be measured in mouse and human hepatocytes and correlated with the observed in vivo hepatotoxicity in mice, thus confirming the relevance of rodent hepatotoxicity to predict human hepatotoxicity.
Kidney toxicity
Following systemic administration, a large proportion of the administered dose of ONDs also end up accumulating in the kidneys, including the charge neutral backbones such as PMO [106,107], sometimes with kidney toxicity occurring. Renal lesions are generally restricted to the proximal tubules, in which the highest uptake is observed in contrast with kidney medulla [21,108]. Nonetheless, glomerulopathies were previously reported in mouse and monkey studies with the 2′OMe PS steric blocker drisapersen developed for the treatment of Duchenne muscular dystrophy [109], but these may have been linked to the chronic complement activation and inflammatory effects of the ASO. Rats develop a spontaneous lesion called chronic progressive nephropathy [110,111] that can be enhanced by kidney accumulation of PS backbone ASOs [78].
Renal toxicity was mostly regarded as an accumulation-related toxicity and primarily sequence unspecific until more acute tubular lesions were reported with high-affinity ONDs, such as LNA [112]. It has been suggested that these effects may be related to excessive accumulation of RNase H-dependent off-target transcripts and/or specific protein binding as described above for hepatotoxicity [113]. It is possible that the adverse sequence- and chemistry-dependent ASO:protein interactions proposed for liver toxicity also underlie the kidney toxicity observed for higher affinity modifications.
To screen out potential nephrotoxic candidates, several studies have reported useful predictive assays in the past few years. Moisan et al. identified the elevation of extracellular epidermal growth factor (EGF) as a robust and sensitive in vitro biomarker of LNA-induced cytotoxicity in human kidney proximal tubular epithelial cells [114]. More recently, another group explored the utility of in vitro systems to predict acute kidney injury and demonstrated cytotoxicity and induction in kidney injury biomarkers using chip-cultured human renal proximal tubule epithelial cells [115].
Besides these in vitro predictive assays, several specific early biomarkers of toxicity can be evaluated in vivo in mice (treated with high doses of ASO) to predict toxicity in preclinical development and exclude nephrotoxic candidates before moving forward to larger safety studies [109,116]. Urinary biomarkers of kidney toxicity can be quantified in urine collected from treated mice either shortly after ONDs injection to evaluate the potential acute kidney toxicity or after several weeks of repeated treatment to assess the potential long-term renal toxicity induced by the accumulation of ONDs in the kidneys.
These biomarkers include general ones such as total protein, albumin, or creatinine as well as specific kidney injury biomarkers such as β2-microglobulin, renin, kidney injury molecule-1, IFN-gamma-induced protein 10, Cystatin C, EGF, Lipocalin-2-NGAL, clusterin, and osteopontin as described in the studies by Echevarría and Goyenvalle [117] and Sandelius et al. [118]. Finally, more general biomarkers of renal toxicity can also be measured in the serum or plasma of mice treated with high doses of ONDs, such as urea, albumin, creatinine, and total protein.
Importantly, the development of predictive in vitro models of kidney toxicity and the improvement in OND design have allowed identification of potent ONDs with significant reduction in toxicity as illustrated by the development of GalNAc conjugates, which conferred a more favorable safety profile at the cellular level [119]. However, one should keep in mind that translation of such findings may be limited due to breakdown of conjugates in vivo resulting in the unmodified sequence of the OND with all its potential renal safety liabilities.
Sequence- and Hybridization-Independent Effects
Besides these hybridization-independent toxicities that are impacted by the sequence, other toxicities are independent of both hybridization and sequence. These toxicities are generally driven by the plasma concentration reaching above a threshold level and include prolongation of coagulation time and activation of the alternative complement system. These findings can occasionally be observed in clinical studies but can be reduced by adapting dosing regimen and are in most cases of low magnitude with limited impact of clinical safety.
Inhibition of coagulation
Prolongation of coagulation time often observed with PS-ASO results from the PS backbone interacting with the Tenase complex in the coagulation cascade [120,121]. It is considered a class effect, modulated by interactions of the OND with plasma proteins in a sequence-independent way. At low plasma concentrations, the PS modification selectively prolongs the partial thromboplastin time. However, at high plasma concentrations, both the intrinsic and extrinsic pathways are affected, suggesting additional inhibitory effects [120]. Prolongation of clotting times can be screened relatively easily both in vivo and in vitro in mouse, NHP, and human serum [88,122]. Following incubation of the OND in citrated serum, both the prothrombin time and the activated partial thromboplastin time can be measured as described in the studies by Echevarría et al. [122], Aupy et al. [123], Henry et al. [124], and Relizani et al. [125].
Complement activation
Systemic administration of PS containing ONDs has been reported to elicit the complement alternative pathway due to plasma protein binding [126,127]. Although this hybridization-independent effect is mainly related to the PS backbone of single-stranded ASOs, unexpected complement activation has been observed with some specific sequences in the case of tricyclo-DNA for example [123]. Single-stranded PS backbone ASOs interact directly with plasma factor H (a negative regulator of the complement cascade). This interaction has been shown to reduce the free levels of inhibitor, leading to an uncontrolled amplification of the cascade and a release of split products such as Bb and anaphylatoxins C3a and C5a [126]. Repeated activation of the complement induced by chronic dosing of PS-ONDs can result in C3 depletion, eventually leading to altered complement function, secondary inflammation, and vasculitis [112,113,128].
While humans appear less sensitive to complement activation than NHP [14], it is recommended to routinely assess complement activation in preclinical safety studies of new OND candidates in NHPs to minimize complement-driven toxicities in NHP of longer duration. Early preclinical safety assessment can also be performed in mice to screen out particular toxic candidates as it was previously shown with tricyclo-DNA sequences [123]. More importantly, protocols to assess complement activation in vitro have been described in several studies, allowing a relevant assessment directly in human serum. Increasing concentrations of the candidate OND are generally incubated in serum, plasma, or whole blood from mice, NHP, or human, and the different split products of the alternative complement pathway (Bb, C3a, and C5a) are then measured by ELISA [71,123,129].
Since these effects on complement and coagulation are driven by reaching above a threshold level in plasma, they are generally transient in nature and disappear when the OND is cleared from plasma by tissue uptake, within hours from systemic administration [44–47]. Moreover, the dosing regimens are often adapted in the clinic (favoring slow intravenous infusion to bolus administration, for example) leading to reduced plasma concentrations and therefore rarely exceeding activation thresholds [128,130,131]. However, these effects should be carefully considered in toxicity studies in which much higher doses are administered, in particular in Cynomolgus monkeys, which tend to present higher sensitivity [112].
Toxicities Associated with CNS Local Delivery
ASOs are generally charged and have a molecular weight of ∼5,000 to 10,000 Da: too large to cross the blood–brain barrier by simple diffusion and reach an effective concentration in the brain or spinal cord. Currently, the most frequently used CNS administration route in humans is intrathecal (IT) administration, and in rodent models intracerebroventricular delivery (ICV). This results in an immediate high and long-lasting ASO concentration in the cerebral spinal fluid and brain and significant pharmacodynamic effects up to 6 months after the last dosing [132–134]. Although immediately after both IT and ICV administration, there is a peak in systemic exposure, this peak lowers rapidly after dosing.
Moreover, the overall doses administered are lower compared with systemic delivery, together making the risks for peripheral toxicity lower for CNS applications [45]. However, some acute neurotoxic effects have been reported after ASO administration in rodent brain, and at least some of the potential on- and off-target toxicities associated with systemic delivery also appear to occur in the CNS [135]. This means that most of the previously mentioned assays are also useful to predict the safety profile of ONDs aimed for CNS delivery (eg, assay predicting proinflammatory profile); however, some specific toxicities have been observed and reported in rodents following CNS direct delivery.
ICV administration of ASOs has been shown to induce an immune response in rodent brain [136,137] that could persist up to 2 months after the last administration. This suggests a prolonged immune response to the treatment, and as described in the previous paragraph, these immune effects are dependent on sequence and chemical modifications of the OND.
The most overt neurotoxicity in preclinical rodent studies is the occurrence of slight tremors and seizure-like activities immediately after OND administration. In this context, sequence-specific toxicity was studied following CNS delivery by Hagedorn et al. where acute tolerability behavior was assessed 1 h after ICV bolus injection in the mouse brain of 148 different LNA-modified ASOs with full PS backbone. They found that in particular the number and position of guanine nucleotides in the 3′-end of the ASO increased toxicity, while there was a decrease when adenine nucleotides were substituted [27]. Recent studies demonstrated that reducing the PS content of ONDs leads to increased tolerability in the CNS [138,139].
Despite these advances in safety assessment, it remains difficult to predict human toxicity from animal models. This has again be highlighted for instance by the recently stopped large multinational phase 3 study—GENERATION HD1 trial (NCT03761849) investigating a gapmer ASO lowering both wild-type and mutant huntingtin protein [140]. In March 2021, dosing in this trial was stopped because the high-dose patient group performed worse on clinical rating scales and had higher frequencies of serious adverse events. Ongoing studies are aiming to better predict neurotoxicity with human-based cell models, in line with the European Union directives on animal use in science to advance the development of alternative model systems to replace animal studies [141].
For an OND-based approach that targets (pre) mRNA, the use of a human model is essential since there are considerable differences in expressed RNA between mouse and human [142] as well as in neuronal versus non-neuronal cells. In a study using human fibroblasts and nusinersen, the only FDA- and EMA-approved OND for a CNS disorder, the off-target effects of the splice modulating therapeutic OND nusinersen caused widespread alterations in gene expression (including innate immunity) and aberrant splicing [143]. However, the interpretation of RNA sequencing studies after OND delivery in cell and animal models is still challenging, distinguishing between off- and on-target effects as well as acute and long-term effects.
Primary cortical neuronal cultures were used to evaluate more than 1,600 LNA-modified PS ASO, using spontaneous calcium oscillations as a readout. This in vitro assay was found to accurately predict acute neurotoxicity found in mice [27]. Using human induced pluripotent stem cell-derived neuronal models for the in vitro assessment of seizure liability is promising for ONDs [144,145]. More studies where neurotoxicity is assessed both in animals and in human-based cell models will be needed to better predict OND neurotoxicity.
Conclusions
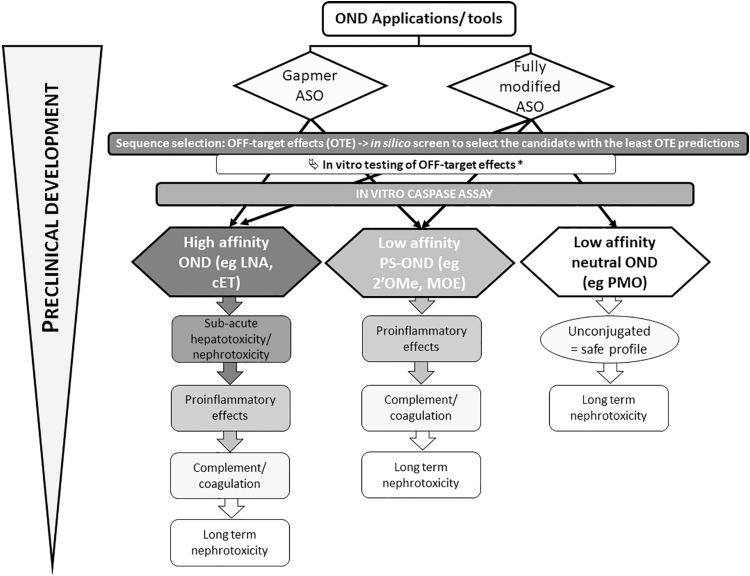
OND-associated toxicities depend on sequence, chemistry, design, dose, duration, and the delivery route. We mostly focused here on systemic administration and direct delivery to the CNS and summarized the main type of toxicities than can be observed in preclinical research. More importantly, we have provided a list of predictive assays available to filter out potential toxic candidates during this preclinical research (Table 1). We also aimed to recapitulate the most commonly encountered types of toxicity depending on the mode of action (mostly described for RNase H-dependent ASO gapmer or steric blocking ASO) and the chemistry (high, medium/low affinity) to help scientists prioritize the type of assay for their preclinical assessment (Fig. 2).
FIG. 2.
Simplified recommendation path to assess OND-associated toxicities depending on the design and chemistry: for all types of ONDs, it is highly recommended to perform in silico analysis during sequence selection to select the candidates with the least predicted OTE, followed by in vitro testing of the predictive OTE at a suitable time in the screening cascade. *Depending on the resources available, the in vitro screening may be run on a smaller set of candidates, for example, after proinflammatory assessment in the cascade. The overall cytotoxicity of OND can then be assessed in vitro using caspase assays, which offer good throughput to select safe compounds. Depending on the application and chemistry selected, the priority for predictive assay is then different since high-affinity ONDs present higher probabilities of inducing subacute hepatotoxicity or nephrotoxicity than low-affinity PS-ONDs or even ASO such as PMO, which have never been reported to induce such toxicities (when unconjugated). ASO, antisense oligonucleotide; cET, constrained ethyl; LNA, locked nucleic acid; MOE, methoxyethyl; OTE, off-target effects; PMO, phosphorodiamidate morpholino oligomer; PS, phosphorothioate.
High-affinity chemistries on PS backbone are indeed more likely to trigger subacute hepatotoxicity or nephrotoxicity, compared with chemistries with more moderate affinity (such as MOE and Me modifications). In contrast, charge neutral PMO has never been reported to induce such effects, but this may not hold true when PMO is conjugated or formulated to enhance their uptake. The last few years have witnessed tremendous progress in delivery systems aiming at improving the distribution of ONDs to target tissues.
These include development of lipidic ligands, peptide and antibody conjugates, or lipid-based nanoparticles. In this context, early preclinical assessment of safety aspects is crucial to ensure their future transition to the clinic. Far too often, preclinical studies focus on assessing only the improved delivery and efficacy, neglecting the importance of thorough early toxicity evaluation. With these guidelines, we hope to ultimately improve the success rate in the development phase and avoid safety signals in regulatory toxicity studies and ultimately in clinical studies that could have been identified earlier on.
So far, most ONDs have been developed to treat rare genetic disorders without existing therapeutic options. This has allowed some degree of acceptability of safety findings in terms of risk–benefit, but this assessment will likely be different in the coming years given the growing interest for ONDs and their development for much more common diseases for which therapeutic alternatives are available.
However, in parallel, our increasing understanding of the mechanisms underlying these various toxicities and the development of more complex in vitro tissue models will likely lead to more predictive and more sensitive test systems in the coming years. This will further improve the process to identify potent and safe ONDs and facilitate NAT drugs to reach their full potential.
Author Disclosure Statement
None of this funding represents a conflict of interest with the content of this review.
Funding Information
This work was supported by funding from Cooperation of Science and Technology (COST) Action CA17103 (networking grant to V.A.-G.). A.G. acknowledges funding from the Institut National de la santé et la recherche médicale (INSERM) and the Université Paris-Saclay (France). V.A.-G. acknowledges funding from Ikerbasque (Basque Foundation for Science). C.J.-M. acknowledges funding the Instituto de Salud Carlos III and FEDER “una manera de hacer Europa” (PI19/0122 grant to C.J.-M.) and the Spanish Center for Biomedical Network Research on Rare Diseases (ACCI2018-02 grant to C.J.-M.). S.S. is an employee of F. Hoffmann La-Roche, Switzerland. P.A. is an employee of AstraZeneca R&D, Sweden.
References
- 1. Warner KD, Hajdin CE and Weeks KM. (2018). Principles for targeting RNA with drug-like small molecules. Nat Rev Drug Discov 17:547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arechavala-Gomeza V and Garanto A. (2022). Antisense RNA therapeutics: a brief overview. Methods Mol Biol 2434:33–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maquat LE. (2004). Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 5:89–99. [DOI] [PubMed] [Google Scholar]
- 4. Nomakuchi TT, Rigo F, Aznarez I and Krainer AR. (2016). Antisense oligonucleotide-directed inhibition of nonsense-mediated mRNA decay. Nat Biotechnol 34:164–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arechavala-Gomeza V, Khoo B and Aartsma-Rus A. (2014). Splicing modulation therapy in the treatment of genetic diseases. Appl Clin Genet 7:245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lim KH, Han Z, Jeon HY, Kach J, Jing E, Weyn-Vanhentenryck S, Downs M, Corrionero A, Oh R, et al. (2020). Antisense oligonucleotide modulation of non-productive alternative splicing upregulates gene expression. Nat Commun 11:3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liang X-H, Shen W, Sun H, Migawa MT, Vickers TA and Crooke ST. (2016). Translation efficiency of mRNAs is increased by antisense oligonucleotides targeting upstream open reading frames. Nat Biotechnol 34:875–880. [DOI] [PubMed] [Google Scholar]
- 8. Liang X-H, Sun H, Shen W, Wang S, Yao J, Migawa MT, Bui H-H, Damle SS, Riney S, et al. (2017). Antisense oligonucleotides targeting translation inhibitory elements in 5’ UTRs can selectively increase protein levels. Nucleic Acids Res 45:9528–9546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alton EW, Boushey HA, Garn H, FH, Green, Hodges M, Martin RJ, Murdoch RD, Renz H, Shrewsbury SB, et al. (2012). Clinical expert panel on monitoring potential lung toxicity of inhaled oligonucleotides: consensus points and recommendations. Nucleic Acid Ther 22:246–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berman CL, Cannon K, Cui Y, Kornbrust DJ, Lagrutta A, Sun SZ, Tepper J, Waldron G and Younis HS. (2014). Recommendations for safety pharmacology evaluations of oligonucleotide-based therapeutics. Nucleic Acid Ther 24:291–301. [DOI] [PubMed] [Google Scholar]
- 11. Berman CL, Barros SA, Galloway SM, Kasper P, Oleson FB, Priestley CC, Sweder KS, Schlosser MJ and Sobol Z. (2016). OSWG recommendations for genotoxicity testing of novel oligonucleotide-based therapeutics. Nucleic Acid Ther 26:73–85. [DOI] [PubMed] [Google Scholar]
- 12. Capaldi D, Teasdale A, Henry S, Akhtar N, den Besten C, Gao-Sheridan S, Kretschmer M, Sharpe N, Andrews B, et al. (2017). Impurities in oligonucleotide drug substances and drug products. Nucleic Acid Ther 27:309–322. [DOI] [PubMed] [Google Scholar]
- 13. Cavagnaro J, Berman C, Kornbrust D, White T, Campion S and Henry S. (2014). Considerations for assessment of reproductive and developmental toxicity of oligonucleotide-based therapeutics. Nucleic Acid Ther 24:313–325. [DOI] [PubMed] [Google Scholar]
- 14. Henry SP, Seguin R, Cavagnaro J, Berman C, Tepper J and Kornbrust D. (2016). Considerations for the characterization and interpretation of results related to alternative complement activation in monkeys associated with oligonucleotide-based therapeutics. Nucleic Acid Ther 26:210–215. [DOI] [PubMed] [Google Scholar]
- 15. Kornbrust D, Cavagnaro J, Levin A, Foy J, Pavco P, Gamba-Vitalo C and Guimond A. (2013). Oligo safety working group exaggerated pharmacology subcommittee consensus document. Nucleic Acid Ther 23:21–28. [DOI] [PubMed] [Google Scholar]
- 16. Lindow M, Vornlocher HP, Riley D, Kornbrust DJ, Burchard J, Whiteley LO, Kamens J, Thompson JD, Nochur S, et al. (2012). Assessing unintended hybridization-induced biological effects of oligonucleotides. Nat Biotechnol 30:920–923. [DOI] [PubMed] [Google Scholar]
- 17. Marlowe JL, Akopian V, Karmali P, Kornbrust D, Lockridge J and Semple S. (2017). Recommendations of the oligonucleotide safety working group's formulated oligonucleotide subcommittee for the safety assessment of formulated oligonucleotide-based therapeutics. Nucleic Acid Ther 27:183–196. [DOI] [PubMed] [Google Scholar]
- 18. Schubert D, Levin AA, Kornbrust D, Berman CL, Cavagnaro J, Henry S, Seguin R, Ferrari N and Shrewsbury SB. (2012). The oligonucleotide safety working group (OSWG). Nucleic Acid Ther 22:211–212. [DOI] [PubMed] [Google Scholar]
- 19. Tessier Y, Achanzar W, Mihalcik L, Amuzie C, Andersson P, Parry JD, Moggs J, and Whiteley LO. (2021). Outcomes of the European Federation of Pharmaceutical Industries and Associations Oligonucleotide Working Group survey on nonclinical practices and regulatory expectations for therapeutic oligonucleotide safety assessment. Nucleic Acid Ther 31:7–20. [DOI] [PubMed] [Google Scholar]
- 20. Hirabayashi Y, Maki K, Kinoshita K, Nakazawa T, Obika S, Naota M, Watanabe K, Suzuki M, Arato T, et al. (2021). Considerations of the Japanese Research Working Group for the ICH S6 & related issues regarding nonclinical safety assessments of oligonucleotide therapeutics: comparison with those of biopharmaceuticals. Nucleic Acid Ther 31:114–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hung G, Xiao X, Peralta R, Bhattacharjee G, Murray S, Norris D, Guo S and Monia BP. (2013). Characterization of target mRNA reduction through in situ RNA hybridization in multiple organ systems following systemic antisense treatment in animals. Nucleic Acid Ther 23:369–378. [DOI] [PubMed] [Google Scholar]
- 22. Aartsma-Rus A. (2012). Overview on AON design. Methods Mol Biol 867:117–129. [DOI] [PubMed] [Google Scholar]
- 23. Chiba S, Lim KRQ, Sheri N, Anwar S, Erkut E, Shah MNA, Aslesh T, Woo S, Sheikh O, et al. (2021). eSkip-Finder: a machine learning-based web application and database to identify the optimal sequences of antisense oligonucleotides for exon skipping. Nucleic Acids Res 49:W193–W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lennox KA and Behlke MA. (2020). Tips for Successful lncRNA Knockdown Using Gapmers. Methods Mol Biol 2176:121–140. [DOI] [PubMed] [Google Scholar]
- 25. Burdick AD, Sciabola S, Mantena SR, Hollingshead BD, Stanton R, Warneke JA, Zeng M, Martsen E, Medvedev A, et al. (2014). Sequence motifs associated with hepatotoxicity of locked nucleic acid—modified antisense oligonucleotides. Nucleic Acids Res 42:4882–4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hagedorn PH, Yakimov V, Ottosen S, Kammler S, Nielsen NF, Høg AM, Hedtjärn M, Meldgaard M, Møller MR, et al. (2013). Hepatotoxic potential of therapeutic oligonucleotides can be predicted from their sequence and modification pattern. Nucleic Acid Ther 23:302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hagedorn PH, Brown JM, Easton A, Pierdomenico M, Jones K, Olson RE, Mercer SE, Li D, Loy J, et al. (2022). Acute neurotoxicity of antisense oligonucleotides after intracerebroventricular injection into mouse brain can be predicted from sequence features. Nucleic Acid Ther 32:151–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. López-Martínez A, Soblechero-Martín P and Arechavala-Gomeza V. (2022). Evaluation of exon skipping and dystrophin restoration in in vitro models of duchenne muscular dystrophy. Methods Mol Biol 2434:217–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tomkiewicz TZ, Suárez-Herrera N, Cremers FPM, Collin RWJ and Garanto. A (2021). Antisense oligonucleotide-based rescue of aberrant splicing defects caused by 15 pathogenic variants in ABCA4. Int J Mol Sci 22:4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Toonen LJA, Rigo F, van Attikum and Hvan Roon-Mom WMC. (2017). Antisense oligonucleotide-mediated removal of the polyglutamine repeat in spinocerebellar ataxia type 3 mice. Mol Ther Nucleic Acids 8:232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ramsden D, Belair DG, Agarwal S, Andersson P, Humphreys S, Dalmas DA, Stahl SH, Maclauchlin C and Cichocki JA. (2022). Leveraging microphysiological systems to address challenges encountered during development of oligonucleotide therapeutics. ALTEX 39:273–296. [DOI] [PubMed] [Google Scholar]
- 32. Crosby JR, Zhao C, Zhang H, MacLeod AR, Guo S and Monia BP. (2015). Reversing antisense oligonucleotide activity with a sense oligonucleotide antidote: proof of concept targeting prothrombin. Nucleic Acid Ther 25:297–305. [DOI] [PubMed] [Google Scholar]
- 33. Zlatev I, Castoreno A, Brown CR, Qin J, Waldron S, Schlegel MK, Degaonkar R, Shulga-Morskaya S, Xu H, et al. (2018). Reversal of siRNA-mediated gene silencing in vivo. Nat Biotechnol 36:509–511. [DOI] [PubMed] [Google Scholar]
- 34. Hagedorn PH, Hansen BR, Koch T and Lindow M. (2017). Managing the sequence-specificity of antisense oligonucleotides in drug discovery. Nucleic Acids Res 45:2262–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jackson AL and Linsley PS. (2010). Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Discov 9:57–67. [DOI] [PubMed] [Google Scholar]
- 36. Janas MM, Schlegel MK, Harbison CE, Yilmaz VO, Jiang Y, Parmar R, Zlatev I, Castoreno A, Xu H, et al. (2018). Selection of GalNAc-conjugated siRNAs with limited off-target-driven rat hepatotoxicity. Nat Commun 9:723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kamola PJ, Kitson JD, Turner G, Maratou K, Eriksson S, Panjwani A, Warnock LC, Douillard Guilloux GA, Moores K, et al. (2015). In silico and in vitro evaluation of exonic and intronic off-target effects form a critical element of therapeutic ASO gapmer optimization. Nucleic Acids Res 43:8638–8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Watt AT, Swayze G, Swayze EE and Freier SM. (2020). Likelihood of nonspecific activity of gapmer antisense oligonucleotides is associated with relative hybridization free energy. Nucleic Acid Ther 30:215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rehmsmeier M, Steffen P, Hochsmann M and Giegerich R. (2004). Fast and effective prediction of microRNA/target duplexes. RNA 10:1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Scharner J, Ma WK, Zhang Q, Lin K-T, Rigo F, Bennett CF, and Krainer AR. (2020). Hybridization-mediated off-target effects of splice-switching antisense oligonucleotides. Nucleic Acids Res 48:802–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Holgersen EM, Gandhi S, Zhou Y, Kim J, Vaz B, Bogojeski J, Bugno M, Shalev Z, Cheung-Ong K, et al. (2021). Transcriptome-wide off-target effects of steric-blocking oligonucleotides. Nucleic Acid Ther 31:392–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ray KK, Landmesser U, Leiter LA, Kallend D, Dufour R, Karakas M, Hall T, Troquay RPT, Turner T, et al. (2017). Inclisiran in patients at high cardiovascular risk with elevated LDL cholesterol. N Engl J Med 376:1430–1440. [DOI] [PubMed] [Google Scholar]
- 43. Brown CR, Gupta S, Qin J, Racie T, He G, Lentini S, Malone R, Yu M, Matsuda S, Shulga-Morskaya S, et al. (2020). Investigating the pharmacodynamic durability of GalNAc-siRNA conjugates. Nucleic Acids Res 48:11827–11844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bosgra S, Sipkens J, de Kimpe S, den Besten C, Datson N and van Deutekom J. (2019). The pharmacokinetics of 2’-O-methyl phosphorothioate antisense oligonucleotides: experiences from developing exon skipping therapies for duchenne muscular dystrophy. Nucleic Acid Ther 29:305–322. [DOI] [PubMed] [Google Scholar]
- 45. Geary RS, Norris D, Yu R and Bennett CF. (2015). Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv Drug Deliv Rev 87:46–51. [DOI] [PubMed] [Google Scholar]
- 46. Yu RZ, Kim T-W, Hong A, Watanabe TA, Gaus HJ, and Geary RS. (2007). Cross-species pharmacokinetic comparison from mouse to man of a second-generation antisense oligonucleotide, ISIS 301012, targeting human apolipoprotein B-100. Drug Metab Dispos 35:460–468. [DOI] [PubMed] [Google Scholar]
- 47. Yu RZ, Lemonidis KM, Graham MJ, Matson JE, Crooke RM, Tribble DL, Wedel MK, Levin AA, and Geary RS. (2009). Cross-species comparison of in vivo PK/PD relationships for second-generation antisense oligonucleotides targeting apolipoprotein B-100. Biochem Pharmacol 77:910–919. [DOI] [PubMed] [Google Scholar]
- 48. Agrawal S and Kandimalla ER. (2004). Role of Toll-like receptors in antisense and siRNA [corrected]. Nat Biotechnol 22:1533–1537. [DOI] [PubMed] [Google Scholar]
- 49. Krieg AM. (1998). The CpG motif: implications for clinical immunology. BioDrugs 10:341–346. [DOI] [PubMed] [Google Scholar]
- 50. Kline JN and Krieg AM. (2008). Toll-like receptor 9 activation with CpG oligodeoxynucleotides for asthma therapy. Drug News Perspect 21:434–439. [DOI] [PubMed] [Google Scholar]
- 51. Krieg AM. (2006). Therapeutic potential of Toll-like receptor 9 activation. Nat Rev Drug Discov 5:471–484. [DOI] [PubMed] [Google Scholar]
- 52. Krieg AM and Davis HL. (2001). Enhancing vaccines with immune stimulatory CpG DNA. Curr Opin Mol Ther 3:15–24. [PubMed] [Google Scholar]
- 53. Bauer S, Kirschning CJ, Häcker H, Redecke V, Hausmann S, Akira S, Wagner H and Lipford GB. (2001). Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc Natl Acad Sci U S A 98:9237–9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Krieg AM, Yi AK, Matson S, Waldschmidt TJ, Bishop GA, Teasdale R, Koretzky GA and Klinman DM. (1995). CpG motifs in bacterial DNA trigger direct B-cell activation. Nature 374:546–549. [DOI] [PubMed] [Google Scholar]
- 55. Hartmann G, Weeratna RD, Ballas ZK, Payette P, Blackwell S, Suparto I, Rasmussen WL, Waldschmidt M, Sajuthi D, et al. (2000). Delineation of a CpG phosphorothioate oligodeoxynucleotide for activating primate immune responses in vitro and in vivo. J Immunol 164:1617–1624. [DOI] [PubMed] [Google Scholar]
- 56. Krieg AM. (2002). CpG motifs in bacterial DNA and their immune effects. Annu Rev Immunol 20:709–760. [DOI] [PubMed] [Google Scholar]
- 57. Forsbach A, Nemorin J-G, Montino C, Müller C, Samulowitz U, Vicari AP, Jurk M, Mutwiri GK, Krieg AM, et al. (2008). Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J Immunol 180:3729–3738. [DOI] [PubMed] [Google Scholar]
- 58. Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H and Bauer S. (2004). Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303:1526–1529. [DOI] [PubMed] [Google Scholar]
- 59. Diebold SS, Massacrier C, Akira S, Paturel C, Morel Y and Reis e Sousa C. (2006). Nucleic acid agonists for Toll-like receptor 7 are defined by the presence of uridine ribonucleotides. Eur J Immunol 36:3256–3267. [DOI] [PubMed] [Google Scholar]
- 60. Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, Noronha A, Manoharan M, Akira S, et al. (2005). Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med 11:263–270. [DOI] [PubMed] [Google Scholar]
- 61. Judge AD, Bola G, Lee ACH and MacLachlan I. (2006). Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther 13:494–505. [DOI] [PubMed] [Google Scholar]
- 62. Sioud M. (2006). Single-stranded small interfering RNA are more immunostimulatory than their double-stranded counterparts: a central role for 2’-hydroxyl uridines in immune responses. Eur J Immunol 36:1222–1230. [DOI] [PubMed] [Google Scholar]
- 63. Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann K-K, et al. (2006). 5’-Triphosphate RNA is the ligand for RIG-I. Science 314:994–997. [DOI] [PubMed] [Google Scholar]
- 64. Alharbi AS, Garcin AJ, Lennox KA, Pradeloux S, Wong C, Straub S, Valentin R, Pépin G, Li H-M, et al. (2020). Rational design of antisense oligonucleotides modulating the activity of TLR7/8 agonists. Nucleic Acids Res 48:7052–7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bayik D, Gursel I and Klinman DM. (2016). Structure, mechanism and therapeutic utility of immunosuppressive oligonucleotides. Pharmacol Res 105:216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Valentin R, Wong C, Alharbi AS, Pradeloux S, Morros MP, Lennox KA, Ellyard JI, Garcin AJ, Ullah TR, et al. (2021). Sequence-dependent inhibition of cGAS and TLR9 DNA sensing by 2’-O-methyl gapmer oligonucleotides. Nucleic Acids Res 49:6082–6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Liang H, Nishioka Y, Reich CF, Pisetsky DS and Lipsky PE. (1996). Activation of human B cells by phosphorothioate oligodeoxynucleotides. J Clin Invest 98:1119–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhao Q, Temsamani J, Iadarola PL, Jiang Z and Agrawal S. (1996). Effect of different chemically modified oligodeoxynucleotides on immune stimulation. Biochem Pharmacol 51:173–182. [DOI] [PubMed] [Google Scholar]
- 69. Zhang A, Uaesoontrachoon K, Shaughnessy C, Das JR, Rayavarapu S, Brown KJ, Ray PE, Nagaraju K, van den Anker JN, et al. (2015). The use of urinary and kidney SILAM proteomics to monitor kidney response to high dose morpholino oligonucleotides in the mdx mouse. Toxicol Rep 2:838–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Henry S, Stecker K, Brooks D, Monteith D, Conklin B and Bennett CF. (2000). Chemically modified oligonucleotides exhibit decreased immune stimulation in mice. J Pharmacol Exp Ther 292:468–479. [PubMed] [Google Scholar]
- 71. Sewing S, Roth AB, Winter M, Dieckmann A, Bertinetti-Lapatki C, Tessier Y, McGinnis C, Huber S, Koller E, et al. (2017). Assessing single-stranded oligonucleotide drug-induced effects in vitro reveals key risk factors for thrombocytopenia. PLoS One 12:e0187574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Crooke ST, Baker BF, Witztum JL, Kwoh TJ, Pham NC, Salgado N, McEvoy BW, Cheng W, Hughes SG, et al. (2017). The effects of 2’-O-methoxyethyl containing antisense oligonucleotides on platelets in human clinical trials. Nucleic Acid Ther 27:121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rudin CM, Holmlund J, Fleming GF, Mani S, Stadler WM, Schumm P, Monia BP, Johnston JF, Geary R, et al. (2001). Phase I Trial of ISIS 5132, an antisense oligonucleotide inhibitor of c-raf-1, administered by 24-hour weekly infusion to patients with advanced cancer. Clin Cancer Res 7:1214–1220. [PubMed] [Google Scholar]
- 74. Thomas GS, Cromwell WC, Ali S, Chin W, Flaim JD and Davidson M. (2013). Mipomersen, an apolipoprotein B synthesis inhibitor, reduces atherogenic lipoproteins in patients with severe hypercholesterolemia at high cardiovascular risk: a randomized, double-blind, placebo-controlled trial. J Am Coll Cardiol 62:2178–2184. [DOI] [PubMed] [Google Scholar]
- 75. Voit T, Topaloglu H, Straub V, Muntoni F, Deconinck N, Campion G, De Kimpe SJ, Eagle M, Guglieri M, et al. (2014). Safety and efficacy of drisapersen for the treatment of Duchenne muscular dystrophy (DEMAND II): an exploratory, randomised, placebo-controlled phase 2 study. Lancet Neurol 13:987–996. [DOI] [PubMed] [Google Scholar]
- 76. Monteith DK, Henry SP, Howard RB, Flournoy S, Levin AA, Bennett CF and Crooke ST. (1997). Immune stimulation—a class effect of phosphorothioate oligodeoxynucleotides in rodents. Anticancer Drug Des 12:421–432. [PubMed] [Google Scholar]
- 77. Paz S, Hsiao J, Cauntay P, Soriano A, Bai L, Machemer T, Xiao X, Guo S, Hung G, et al. (2017). The distinct and cooperative roles of Toll-like receptor 9 and receptor for advanced glycation end products in modulating in vivo inflammatory responses to select CpG and non-CpG oligonucleotides. Nucleic Acid Ther 27:272–284. [DOI] [PubMed] [Google Scholar]
- 78. Frazier KS. (2015). Antisense oligonucleotide therapies: the promise and the challenges from a toxicologic pathologist's perspective. Toxicol Pathol 43:78–89. [DOI] [PubMed] [Google Scholar]
- 79. Monteith DK and Levin AA. (1999). Synthetic oligonucleotides: the development of antisense therapeutics. Toxicol Pathol 27:8–13. [DOI] [PubMed] [Google Scholar]
- 80. Burel SA, Machemer T, Baker BF, Kwoh TJ, Paz S, Younis H and Henry SP. (2022). Early-stage identification and avoidance of antisense oligonucleotides causing species-specific inflammatory responses in human volunteer peripheral blood mononuclear cells. Nucleic Acid Ther 32:457–472. [DOI] [PubMed] [Google Scholar]
- 81. Coch C, Lück C, Schwickart A, Putschli B, Renn M, Höller T, Barchet W, Hartmann G and Schlee. M (2013). A human in vitro whole blood assay to predict the systemic cytokine response to therapeutic oligonucleotides including siRNA. PLoS One 8:e71057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lankveld DPK, Van Loveren H, Baken KA and Vandebriel RJ. (2010). In vitro testing for direct immunotoxicity: state of the art. Methods Mol Biol 598:401–423. [DOI] [PubMed] [Google Scholar]
- 83. Anderson BA, Freestone GC, Low A, De-Hoyos CL, Iii WJD, ME Østergaard, Migawa MT, Fazio M, Wan WB, et al. (2021). Towards next generation antisense oligonucleotides: mesylphosphoramidate modification improves therapeutic index and duration of effect of gapmer antisense oligonucleotides. Nucleic Acids Res 49:9026–9041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Andersson P. (2022). Preclinical safety assessment of therapeutic oligonucleotides. Methods Mol Biol 2434:355–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Slingsby MHL, Vijey P, Tsai I-T, Roweth H, Couldwell G, Wilkie AR, Gaus H, Goolsby JM, Okazaki R, et al. (2022). Sequence-specific 2’-O-methoxyethyl antisense oligonucleotides activate human platelets through glycoprotein VI, triggering formation of platelet-leukocyte aggregates. Haematologica 107:519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Chi X, Gatti P and Papoian T. (2017). Safety of antisense oligonucleotide and siRNA-based therapeutics. Drug Discov Today 22:823–833. [DOI] [PubMed] [Google Scholar]
- 87. Janas MM, Harbison CE, Perry VK, Carito B, Sutherland JE, Vaishnaw AK, Keirstead ND and Warner G. (2018). The nonclinical safety profile of GalNAc-conjugated RNAi therapeutics in subacute studies. Toxicol Pathol 46:735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Henry SP, Narayanan P, Shen L, Bhanot S, Younis HS and Burel SA. (2017). Assessment of the effects of 2’-methoxyethyl antisense oligonucleotides on platelet count in cynomolgus nonhuman primates. Nucleic Acid Ther 27:197–208. [DOI] [PubMed] [Google Scholar]
- 89. Flierl U, Nero TL, Lim B, Arthur JF, Yao Y, Jung SM, Gitz E, Pollitt AY, Zaldivia MTK, et al. (2015). Phosphorothioate backbone modifications of nucleotide-based drugs are potent platelet activators. J Exp Med 212:129–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Narayanan P, Shen L, Curtis BR, Bourdon MA, Nolan JP, Gupta S, Hoffmaster C, Zhou F, Christian B, et al. (2018). Investigation into the mechanism(s) that leads to platelet decreases in cynomolgus monkeys during administration of ISIS 104838, a 2’-MOE-modified antisense oligonucleotide. Toxicol Sci 164:613–626. [DOI] [PubMed] [Google Scholar]
- 91. van Poelgeest EP, Swart RM, Betjes MGH, Moerland M, Weening JJ, Tessier Y, Hodges MR, Levin AA and Burggraaf J. (2013). Acute kidney injury during therapy with an antisense oligonucleotide directed against PCSK9. Am J Kidney Dis 62:796–800. [DOI] [PubMed] [Google Scholar]
- 92. van Poelgeest EP, Hodges MR, Moerland M, Tessier Y, Levin AA, Persson R, Lindholm MW, Dumong Erichsen K, H Ørum, et al. (2015). Antisense-mediated reduction of proprotein convertase subtilisin/kexin type 9 (PCSK9): a first-in-human randomized, placebo-controlled trial. Br J Clin Pharmacol 80:1350–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. van Meer L, van Dongen M, Moerland M, de Kam M, Cohen A and Burggraaf. J (2017). Novel SGLT2 inhibitor: first-in-man studies of antisense compound is associated with unexpected renal effects. Pharmacol Res Perspect 5:e00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Dieckmann A, Hagedorn PH, Burki Y, Brügmann C, Berrera M, Ebeling M, Singer T and Schuler F. (2018). A sensitive in vitro approach to assess the hybridization-dependent toxic potential of high affinity gapmer oligonucleotides. Mol Ther Nucleic Acids 10:45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Burel SA, Hart CE, Cauntay P, Hsiao J, Machemer T, Katz M, Watt A, Bui H-H, Younis H, et al. (2016). Hepatotoxicity of high affinity gapmer antisense oligonucleotides is mediated by RNase H1 dependent promiscuous reduction of very long pre-mRNA transcripts. Nucleic Acids Res 44:2093–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Swayze EE, Siwkowski AM, Wancewicz EV, Migawa MT, Wyrzykiewicz TK, Hung G, Monia BP and Bennett CF. (2007). Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res 35:687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Liang X, Sun H, Shen W and Crooke ST. (2015). Identification and characterization of intracellular proteins that bind oligonucleotides with phosphorothioate linkages. Nucleic Acids Res 43:2927–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Shen W, De Hoyos CL, Sun H, Vickers TA, Liang X-H and Crooke ST. (2018). Acute hepatotoxicity of 2’ fluoro-modified 5-10-5 gapmer phosphorothioate oligonucleotides in mice correlates with intracellular protein binding and the loss of DBHS proteins. Nucleic Acids Res 46:2204–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Shen W, De Hoyos CL, Migawa MT, Vickers TA, Sun H, Low A, Bell TA, Rahdar M, Mukhopadhyay S, et al. (2019). Chemical modification of PS-ASO therapeutics reduces cellular protein-binding and improves the therapeutic index. Nat Biotechnol 37:640–650. [DOI] [PubMed] [Google Scholar]
- 100. Kamola PJ, Maratou K, Wilson PA, Rush K, Mullaney T, McKevitt T, Evans P, Ridings J, Chowdhury P, et al. (2017). Strategies for in vivo screening and mitigation of hepatotoxicity associated with antisense drugs. Mol Ther Nucleic Acids 8:383–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Funder ED, Albæk N, Moisan A, Sewing S and Koch. T (2020). Refining LNA safety profile by controlling phosphorothioate stereochemistry. PLoS One 15:e0232603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Vasquez G, Freestone GC, Wan WB, Low A, De Hoyos CL, Yu J, Prakash TP, ME Ǿstergaard, Liang X-H, et al. (2021). Site-specific incorporation of 5’-methyl DNA enhances the therapeutic profile of gapmer ASOs. Nucleic Acids Res 49:1828–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Vasquez G, Migawa MT, Wan WB, Low A, Tanowitz M, Swayze EE and Seth PP. (2022). Evaluation of phosphorus and non-phosphorus neutral oligonucleotide backbones for enhancing therapeutic index of gapmer antisense oligonucleotides. Nucleic Acid Ther 32:40–50. [DOI] [PubMed] [Google Scholar]
- 104. Sewing S, Boess F, Moisan A, Bertinetti-Lapatki C, Minz T, Hedtjaern M, Tessier Y, Schuler F, Singer T and Roth. AB (2016). Establishment of a predictive in vitro assay for assessment of the hepatotoxic potential of oligonucleotide drugs. PLoS One 11:e0159431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Sewing S, Minz T and Boess F. (2019). In vitro assessment of the hepatotoxicity potential of therapeutic oligonucleotides. Methods Mol Biol 2036:249–259. [DOI] [PubMed] [Google Scholar]
- 106. Carver MP, Charleston JS, Shanks C, Zhang J, Mense M, Sharma AK, Kaur H and Sazani P. (2016). Toxicological characterization of exon skipping phosphorodiamidate morpholino oligomers (PMOs) in non-human primates. J Neuromuscul Dis 3:381–393. [DOI] [PubMed] [Google Scholar]
- 107. Sazani P, Ness KPV, Weller DL, Poage DW, Palyada K and Shrewsbury SB. (2011). Repeat-dose toxicology evaluation in cynomolgus monkeys of AVI-4658, a phosphorodiamidate morpholino oligomer (PMO) drug for the treatment of duchenne muscular dystrophy. Int J Toxicol 30:313–321. [DOI] [PubMed] [Google Scholar]
- 108. Butler M, Stecker K and Bennett CF. (1997). Cellular distribution of phosphorothioate oligodeoxynucleotides in normal rodent tissues. Lab Invest 77:379–388. [PubMed] [Google Scholar]
- 109. Frazier KS, Sobry C, Derr V, Adams MJ, Besten CD, De Kimpe S, Francis I, Gales TL, Haworth R, et al. (2014). Species-specific inflammatory responses as a primary component for the development of glomerular lesions in mice and monkeys following chronic administration of a second-generation antisense oligonucleotide. Toxicol Pathol 42:923–935. [DOI] [PubMed] [Google Scholar]
- 110. Obert LA and Frazier KS. (2019). Intrarenal renin-angiotensin system involvement in the pathogenesis of chronic progressive nephropathy-bridging the informational gap between disciplines. Toxicol Pathol 47:799–816. [DOI] [PubMed] [Google Scholar]
- 111. Hard GC, Banton MI, Bretzlaff RS, Dekant W, Fowles JR, Mallett AK, McGregor DB, Roberts KM, Sielken RL, et al. (2013). Consideration of rat chronic progressive nephropathy in regulatory evaluations for carcinogenicity. Toxicol Sci 132:268–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Engelhardt JA, Fant P, Guionaud S, Henry SP, Leach MW, Louden C, Scicchitano MS, Weaver JL, Zabka TS, et al. (2015). Scientific and regulatory policy committee points-to-consider paper*: drug-induced vascular injury associated with nonsmall molecule therapeutics in preclinical development: Part 2. Antisense oligonucleotides. Toxicol Pathol 43:935–944. [DOI] [PubMed] [Google Scholar]
- 113. Andersson P and den Besten C. (2019). CHAPTER 20 preclinical and clinical drug-metabolism, pharmacokinetics and safety of therapeutic oligonucleotides. In Advances in Nucleic Acid Therapeutics. The Royal Society of Chemistry, pp. 474–531. [Google Scholar]
- 114. Moisan A, Gubler M, Zhang JD, Tessier Y, Dumong Erichsen K, Sewing S, Gérard R, Avignon B, Huber S, et al. (2017). Inhibition of EGF uptake by nephrotoxic antisense drugs in vitro and implications for preclinical safety profiling. Mol Ther Nucleic Acids 6:89–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Nieskens TTG, Magnusson O, Andersson P, Söderberg M, Persson M and Sjögren A-K. (2021). Nephrotoxic antisense oligonucleotide SPC5001 induces kidney injury biomarkers in a proximal tubule-on-a-chip. Arch Toxicol 95:2123–2136. [DOI] [PubMed] [Google Scholar]
- 116. Relizani K, Griffith G, Echevarría L, Zarrouki F, Facchinetti P, Vaillend C, Leumann C, Garcia L and Goyenvalle A. (2017). Efficacy and safety profile of tricyclo-DNA antisense oligonucleotides in duchenne muscular dystrophy mouse model. Mol Ther Nucleic Acids 8:144–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Echevarría L and Goyenvalle A. (2022). Preclinical evaluation of the renal toxicity of oligonucleotide therapeutics in mice. Methods Mol Biol 2434:371–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Sandelius Å, J Basak, Hölttä M, Sultana S, Hyberg G, Wilson A, Andersson P and Söderberg M. (2020). Urinary kidney biomarker panel detects preclinical antisense oligonucleotide-induced tubular toxicity. Toxicol Pathol 48:981–993. [DOI] [PubMed] [Google Scholar]
- 119. Sewing S, Gubler M, Gérard R, Avignon B, Mueller Y, Braendli-Baiocco A, Odin M and Moisan A. (2019). GalNAc conjugation attenuates the cytotoxicity of antisense oligonucleotide drugs in renal tubular cells. Mol Ther Nucleic Acids 14:67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Sheehan JP and Lan HC. (1998). Phosphorothioate oligonucleotides inhibit the intrinsic tenase complex. Blood 92:1617–1625. [PubMed] [Google Scholar]
- 121. Sheehan JP and Phan TM. (2001). Phosphorothioate oligonucleotides inhibit the intrinsic tenase complex by an allosteric mechanism. Biochemistry 40:4980–4989. [DOI] [PubMed] [Google Scholar]
- 122. Echevarría L, Aupy P, Relizani K, Bestetti T, Griffith G, Blandel F, Komisarski M, Haeberli A, Svinartchouk F, et al. (2019). Evaluating the impact of variable phosphorothioate content in tricyclo-DNA antisense oligonucleotides in a duchenne muscular dystrophy mouse model. Nucleic Acid Ther 29:148–160. [DOI] [PubMed] [Google Scholar]
- 123. Aupy P, Echevarría L, Relizani K, Zarrouki F, Haeberli A, Komisarski M, Tensorer T, Jouvion G, Svinartchouk F, et al. (2020). Identifying and avoiding tcDNA-ASO sequence-specific toxicity for the development of DMD Exon 51 skipping therapy. Mol Ther Nucleic Acids 19:371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Henry SP, Novotny W, Leeds J, Auletta C, and Kornbrust DJ. (1997). Inhibition of coagulation by a phosphorothioate oligonucleotide. Antisense Nucleic Acid Drug Dev 7:503–510. [DOI] [PubMed] [Google Scholar]
- 125. Relizani K, Echevarría L, Zarrouki F, Gastaldi C, Dambrune C, Aupy P, Haeberli A, Komisarski M, Tensorer T, et al. (2021). Palmitic acid conjugation enhances potency of tricyclo-DNA splice switching oligonucleotides. Nucleic Acids Res 11:17–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Henry SP, Giclas PC, Leeds J, Pangburn M, Auletta C, Levin AA and Kornbrust DJ. (1997). Activation of the alternative pathway of complement by a phosphorothioate oligonucleotide: potential mechanism of action. J Pharmacol Exp Ther 281:810–816. [PubMed] [Google Scholar]
- 127. Henry SP, Beattie G, Yeh G, Chappel A, Giclas P, Mortari A, Jagels MA, Kornbrust DJ and Levin AA. (2002). Complement activation is responsible for acute toxicities in rhesus monkeys treated with a phosphorothioate oligodeoxynucleotide. Int Immunopharmacol 2:1657–1666. [DOI] [PubMed] [Google Scholar]
- 128. Shen L, Engelhardt JA, Hung G, Yee J, Kikkawa R, Matson J, Tayefeh B, Machemer T, Giclas PC and Henry SP. (2016). Effects of repeated complement activation associated with chronic treatment of cynomolgus monkeys with 2’-O-methoxyethyl modified antisense oligonucleotide. Nucleic Acid Ther 26:236–249. [DOI] [PubMed] [Google Scholar]
- 129. Henry SP, Jagels MA, Hugli TE, Manalili S, Geary RS, Giclas PC and Levin AA. (2014). Mechanism of alternative complement pathway dysregulation by a phosphorothioate oligonucleotide in monkey and human serum. Nucleic Acid Ther 24:326–335. [DOI] [PubMed] [Google Scholar]
- 130. Crooke ST, Baker BF, Kwoh TJ, Cheng W, Schulz DJ, Xia S, Salgado N, Bui H-H, Hart CE, et al. (2016). Integrated safety assessment of 2’-O-methoxyethyl chimeric antisense oligonucleotides in nonhuman primates and healthy human volunteers. Mol Ther 24:1771–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Shen L, Frazer-Abel A, Reynolds PR, Giclas PC, Chappell A, Pangburn MK, Younis H, and Henry SP. (2014). Mechanistic understanding for the greater sensitivity of monkeys to antisense oligonucleotide-mediated complement activation compared with humans. J Pharmacol Exp Ther 351:709–717. [DOI] [PubMed] [Google Scholar]
- 132. Jafar-Nejad P, Powers B, Soriano A, Zhao H, Norris DA, Matson J, DeBrosse-Serra B, Watson J, Narayanan P, et al. (2021). The atlas of RNase H antisense oligonucleotide distribution and activity in the CNS of rodents and non-human primates following central administration. Nucleic Acids Res 49:657–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Kordasiewicz HB, Stanek LM, Wancewicz EV, Mazur C, McAlonis MM, Pytel KA, Artates JW, Weiss A, Cheng SH, et al. (2012). Sustained therapeutic reversal of huntington's disease by transient repression of huntingtin synthesis. Neuron 74:1031–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Rigo F, Chun SJ, Norris DA, Hung G, Lee S, Matson J, Fey RA, Gaus H, Hua Y, et al. (2014). Pharmacology of a central nervous system delivered 2’-O-methoxyethyl-modified survival of motor neuron splicing oligonucleotide in mice and nonhuman primates. J Pharmacol Exp Ther 350:46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Peng Ho S, Livanov V, Zhang W, Li J, and Lesher T. (1998). Modification of phosphorothioate oligonucleotides yields potent analogs with minimal toxicity for antisense experiments in the CNS. Brain Res Mol Brain Res 62:1–11. [DOI] [PubMed] [Google Scholar]
- 136. Elepfandt P, Rupprecht S, Schöning-Burkhardt B, Volk H-D, and Woiciechowsky C. (2002). Oligodeoxynucleotides induce brain inflammation in rats when infused intracerebroventricularly. Neurosci Lett 322:107–110. [DOI] [PubMed] [Google Scholar]
- 137. Toonen LJA, Casaca-Carreira J, Pellisé-Tintoré M, Mei H, Temel Y, Jahanshahi and Avan Roon-Mom WMC. (2018). Intracerebroventricular administration of a 2’-O-methyl phosphorothioate antisense oligonucleotide results in activation of the innate immune system in mouse brain. Nucleic Acid Ther 28:63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Moazami M, Rembestsy-Brown J, Wang F, Krishnamurthy PM, Brown R and Watts. JK (2021). Quantifying and mitigating motor phenotypes induced by antisense oligonucleotides in the central nervous system. BioRxiv, BioRxiv. doi: 10.1101/2021.02.14.431096. [DOI]
- 139. Zarrouki F, Relizani K, Bizot F, Tensorer T, Garcia L, Vaillend C and Goyenvalle A. (2022). Partial restoration of brain dystrophin and behavioral deficits by exon skipping in the muscular dystrophy X-linked (mdx) mouse. Ann Neurol 92:213–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Tabrizi SJ, Estevez-Fraga C, van Roon-Mom WMC, Flower MD, Scahill RI, Wild EJ, Muñoz-Sanjuan I, Sampaio C, Rosser AE and Leavitt BR. (2022). Potential disease-modifying therapies for Huntington's disease: lessons learned and future opportunities. Lancet Neurol 21:645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Marshall LJ, Constantino H and Seidle. T (2022). Phase-in to phase-out-targeted, inclusive strategies are needed to enable full replacement of animal use in the European Union. Animals (Basel) 12:863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Lin S, Lin Y, Nery JR, Urich MA, Breschi A, Davis CA, Dobin A, Zaleski C, Beer MA, et al. (2014). Comparison of the transcriptional landscapes between human and mouse tissues. Proc Natl Acad Sci U S A 111:17224–17229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Ottesen EW, Luo D, Singh NN and Singh. RN (2021). High concentration of an ISS-N1-targeting antisense oligonucleotide causes massive perturbation of the transcriptome. Int J Mol Sci 22:8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Schmidt BZ, Lehmann M, Gutbier S, Nembo E, Noel S, Smirnova L, Forsby A, Hescheler J, Avci HX, et al. (2017). In vitro acute and developmental neurotoxicity screening: an overview of cellular platforms and high-throughput technical possibilities. Arch Toxicol 91:1–33. [DOI] [PubMed] [Google Scholar]
- 145. Tukker AM, Wijnolts FMJ, de Groot A and Westerink RHS. (2020). Applicability of hiPSC-derived neuronal cocultures and rodent primary cortical cultures for in vitro seizure liability assessment. Toxicol Sci 178:71–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Michel S, Schirduan K, Shen Y, Klar R, Tost J and Jaschinski F. (2021). Using RNA-seq to assess off-target effects of antisense oligonucleotides in human cell lines. Mol Diagn Ther 25:77–85. [DOI] [PubMed] [Google Scholar]
- 147. Yoshida T, Naito Y, Yasuhara H, Sasaki K, Kawaji H, Kawai J, Naito M, Okuda H, Obika S and Inoue T. (2019). Evaluation of off-target effects of gapmer antisense oligonucleotides using human cells. Genes Cells 24:827–835. [DOI] [PMC free article] [PubMed] [Google Scholar]