Abstract
Light is a major environmental factor for seed germination. Red light-activated phytochrome B (phyB) promotes seed germination by modulating the dynamic balance of two phytohormones, gibberellic acid (GA) and abscisic acid (ABA). How phyB modulates ABA biosynthesis after perceiving a light signal is not yet well understood. Here, we identified the noncoding RNA HIDDEN TREASURE 1 (HID1) as a repressor of ABA biosynthesis acting downstream of phyB during Arabidopsis thaliana seed germination. Loss of HID1 function led to delayed phyB-dependent seed germination. Photoactivated phyB promoted the accumulation of HID1 in the radicle within 48 h of imbibition. Our transcriptomics analysis showed that HID1 and phyB co-regulate the transcription of a common set of genes involved in ABA and GA metabolism. Through a forward genetic screen, we identified three ABA biosynthesis genes, ABA DEFICIENT 1 (ABA1), ABA2, and ABA3, as suppressors of HID1. We further demonstrated that HID1 directly inhibits the transcription of 9-CIS-EPOXYCAROTENOID DIOXYGENASE (NCED9), a gene encoding a key rate-limiting enzyme of ABA biosynthesis. HID1 interacts with ARABIDOPSIS TRITHORAX-RELATED7 (ATXR7), an H3K4me3 methyltransferase, inhibiting its occupancy and H3K4me3 modification at the NCED9 locus. Our study reveals a nuclear mechanism of phyB signaling transmitted through HID1 to control the internal homeostasis of ABA and GA, which gradually optimizes the transcriptional network during seed germination.
The Arabidopsis noncoding RNA HID1 positively regulates phyB-mediated seed germination by modulating abscisic acid biosynthesis.
Introduction
Seeds monitor the environment in order to germinate at the optimal time, which is a life-or-death decision, and selecting the right moment is crucial for subsequent robust seedling establishment. Seed germination is affected by numerous environmental factors including light, temperature, humidity, and nutrition (Weitbrecht et al., 2011). The phytohormones gibberellic acid (GA) and abscisic acid (ABA) are major internal signaling molecules that play central roles in the regulation of seed germination (Seo et al., 2009). Seed germination is promoted by decreased ABA levels and increased GA levels (Seo et al., 2006); therefore, the balance between these two antagonistic hormones determines the capacity of a seed for germination (de Wit et al., 2016).
Light is a critical factor during seed germination (Borthwick et al., 1952). It is well acknowledged that the phytochrome photoreceptors play important roles in light-initiated seed germination. In Arabidopsis thaliana, five genes (phyA to phyE) encode phytochromes, among which phyA and phyB play dominant roles in the regulation of seed germination (Shinomura et al., 1994; 1996). phyB mediates red light-induced seed germination, whereas phyA triggers canopy light-induced seed germination (Lee et al., 2012). Upon red light irradiation, photoactivated phyB directly interacts with PHYTOCHROME INTERACTING FACTOR 1 (PIF1), a negative regulator of seed germination, leading to the rapid phosphorylation and degradation of PIF1 via the 26S proteasome (Oh et al., 2004; 2006). The phyB–PIF1 de-repression module ultimately promotes increased levels of GA and reduced levels of ABA, facilitating seed germination (Oh et al., 2007; Kim et al., 2008; Cho et al., 2012; Shi et al., 2013; 2015). Subsequently, the phyB–FHY3 (FAR-RED ELONGATED HYPOCOTYL 3) interaction further increases the levels of bioactive GA to promote germination (Jiang et al., 2016; Yang et al., 2020; Liu et al., 2021). It is well-established that ABA levels in imbibed seeds decrease dramatically after exposure to red light; however, little is known regarding the link between phyB and the genes involved in ABA biosynthesis, through which phyB activation promotes seed germination during prolonged imbibition.
Recent genome-wide profiling data sets have shown that noncoding RNAs (ncRNAs) occupy a significant portion of the transcriptome in eukaryotes (Berretta and Morillon, 2009; Mattick et al., 2010). Increasing evidence indicates that ncRNAs regulate gene expression at both the transcriptional and post-transcriptional levels. Long noncoding RNAs (lncRNAs), which are longer than 200 nucleotides (nts), comprise a sizable set of ncRNAs that perform diverse functions in vivo (Statello et al., 2021). In plants, thousands of lncRNAs have been identified or predicted, but only a few have been fully functionally characterized (Wang and Chekanova, 2017). Recent studies have begun to uncover the regulatory role of nucleus-localized lncRNAs that associate with chromatin and regulate gene transcription at the nexus of environmental responses and plant development (Yu et al., 2019). Several studies suggest that certain microRNAs, including miR163 (Chung et al., 2016), miR390 (Sarkar Das et al., 2018), and miR408 (Jiang et al., 2021), play a role in the control of seed germination; however, the functions of lncRNAs in seed germination remain largely elusive.
Our previous studies have revealed that HIDDEN TREASURE 1 (HID1), a conserved 236-nt noncoding RNA in the plant kingdom, acts as a chromatin-bound lncRNA and directs transcriptional regulation underlying light-dependent seedling development (Wang et al., 2014a, 2014b; 2018a, 2018b). In the present study, we identified HID1 as a positive regulator of phyB-dependent seed germination. HID1 acts genetically downstream of phyB. The expression of HID1 is gradually enriched in the radicle during seed germination in a phyB-dependent manner. The phyB-HID1 axis modulates the expression of genes involved in ABA and GA metabolism and signaling after 24 h of imbibition. The results of our molecular and forward genetic analyses suggest that HID1 promotes phyB-mediated seed germination predominantly by repressing ABA biosynthesis. HID1 directly represses the transcription of NCED9, a gene encoding a key ABA biosynthesis enzyme, and inhibits the occupancy of ATXR7 at NCED9. Therefore, our study uncovers another layer of phyB-dependent regulation of seed germination mediated by a lncRNA, which converts external light signals into internal phytohormone signaling to promote seed germination.
Results
HID1 acts as a positive regulator of phyB-dependent seed germination
To identify ncRNAs that function during red light-induced seed germination in Arabidopsis thaliana, we utilized the phyB-dependent germination protocol (Shi et al., 2013) and screened a collection of Agrobacterium tumefaciens-transferred DNA (T-DNA) insertion mutants that were compromised in the expression of specific ncRNAs for delayed or accelerated seed germination phenotypes. In principle, seeds were exposed to far-red light (5 μmol m−2 s−1) followed by red light (20 μmol m−2 s−1), before incubation in the dark for five days (phyB-on conditions) (Figure 1A).
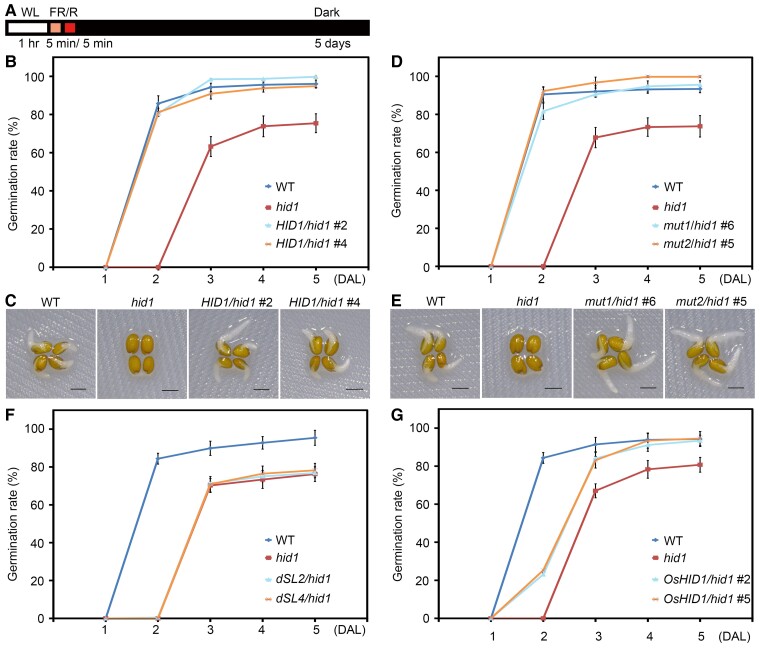
Figure 1.
The ncRNA mutant hid1 exhibits delayed germination under phyB-on conditions. A, Schematic illustration of phyB-on conditions. Seeds were exposed to continuous WL (50 μmol m−2 s−1) for 1 h before exposure to far-red light (FR) (5 μmol m−2 s−1) for 5 min (min) to inactivate phyB. The seeds were then irradiated with red light (R) (20 μmol m−2 s−1) for 5 min to activate phyB before incubation in the dark for the indicated time (5 days). B, Germination rates of WT, hid1, and pHID1:HID1/hid1 (HID1/hid1) seeds under phyB-on conditions. Germination rates were recorded every 24 h after light treatment. Mean ± standard deviation (SD), n = 3. C, Representative images of WT, hid1, and HID1/hid1 seeds at 48 h under phyB-on conditions. Scale bar, 1 mm. D, Germination rates of WT, hid1, mut1/hid1, and mut2/hid1 seeds under phyB-on conditions. Mean ± SD, n = 3. E, Representative images of WT, hid1, mut1/hid1, and mut2/hid1 seeds at 48 h under phyB-on conditions. Scale bar, 1 mm. F, Germination rates of WT, hid1, dSL2/hid1, and dSL4/hid1 seeds under phyB-on conditions. Mean ± SD, n = 3. G, Germination rates of WT, hid1, and OsHID1/hid1 seeds under phyB-on conditions. Mean ± SD, n = 3.
In the screening, the ncRNA mutant hidden treasure 1 (hid1) exhibited a reduced germination rate during phyB-dependent seed germination (Figure 1B). The germination rate of wild-type (WT) seeds was roughly 80% following 48 h dark incubation after light treatment (DAL), and this rate gradually increased to almost 100% during prolonged dark incubation. However, the hid1 mutants did not germinate following 48 h DAL. After 5 days of DAL, only ∼70% of the hid1 mutant seeds had germinated, while WT seeds had a germination frequency of almost 100%. Under phyB-off conditions, during which phyB is inactive, neither WT nor hid1 seeds germinated (Supplemental Figure S1, A and B).
In a previous study, we showed that the T-DNA insertion of the hid1 mutant used in these experiments occurs upstream of a polycistronic ncRNA cluster including HID1, causing a dramatic reduction in the expression of all four ncRNAs (Wang et al., 2014a, 2014b). Expression of HID1 under the control of its endogenous promoter in the hid1 background (pHID1:HID1/hid1) rescued the elongated hypocotyl phenotype of hid1 to that of WT (Wang et al., 2014a, 2014b). In the present study, we observed that the germination rate of pHID1:HID1/hid1 seeds was comparable to that of WT seeds (Figure 1, B, and C). Considering that ∼60% of hid1 seeds germinated at 48 h under white light (WL) after 3 days of cold stratification (Supplemental Figure S1, C and D), but hid1 seeds had not germinated at all following 48 h DAL under phyB-on conditions, these results indicate that HID1 acts as a positive regulator of phyB-mediated seed germination. We also tested whether HID1 is involved in phyA-mediated seed germination. Following 96 h and 120 h of DAL under phyA-on conditions, during which phyA is activated by a second far-red irradiation after 48 h dark treatment, the hid1 mutants had a lower seed germination rate than WT (Supplemental Figure S1, E and F), suggesting that HID1 plays a role in phyA-dependent germination.
Next, to determine whether HID1 acts directly as a bona fide ncRNA in seed germination, we took advantage of our previously generated transgenic lines expressing mutated HID1 to predict the HID1 open-reading frame in the hid1 mutant: (1) Mut1, mutation of ATG to ATA; and (2) Mut2, deletion of ATG to AG (Wang et al., 2014a, 2014b). As expected, the two transgenic lines phenocopied the WT under phyB-on conditions (Figure 1, D and E), suggesting that HID1 functions as a noncoding RNA to promote red light-induced seed germination.
Our previous study showed that two stem loops of HID1, SL2, and SL4, are essential for HID1-mediated hypocotyl elongation under continuous red light; therefore, we evaluated their importance in HID1-regulated seed germination. We examined the germination rate of WT, hid1, and transgenic lines expressing HID1 mutants that lack SL2 or SL4 in the hid1 mutant background under phyB-on conditions. These mutants had a germination rate similar to that of hid1 mutants, indicating that SL2 and SL4 are essential for HID1-mediated seed germination (Figure 1F).
To assess whether the function of HID1 in phyB-dependent seed germination is conserved among monocots and dicots, we examined the seed germination rate of WT, hid1, and transgenic lines expressing the rice (Oryza sativa) HID1 homolog, OsHID1, driven by the OsHID1 endogenous promoter in the hid1 background (Wang et al., 2014a, 2014b). Following 24 h DAL, OsHID1 transgenic seeds had a consistently higher germination rate than that of the hid1 mutants under phyB-on conditions (Figure 1G), suggesting that the functions of AtHID1 and OsHID1 may be conserved in phyB-dependent seed germination. Taken together, these results suggest that being a noncoding RNA, HID1 promotes light-dependent seed germination.
phyB promotes the accumulation of HID1 in radicles
To examine the spatial expression pattern of HID1 during seed germination under phyB-on conditions, we used a pHID1:GUS reporter construct to study HID1 promoter activity in stable transgenic plants. Interestingly, the HID1 promoter is primarily active in the embryo and is highly active in the radicle following 48 h DAL (Figure 2A). Under phyB-off conditions, during which phyB is inactivated by illumination with far-red light, the HID1 promoter was almost inactive following 48 h DAL. These results suggest that expression of the HID1 promoter in the radicle is modulated by phyB during seed germination.
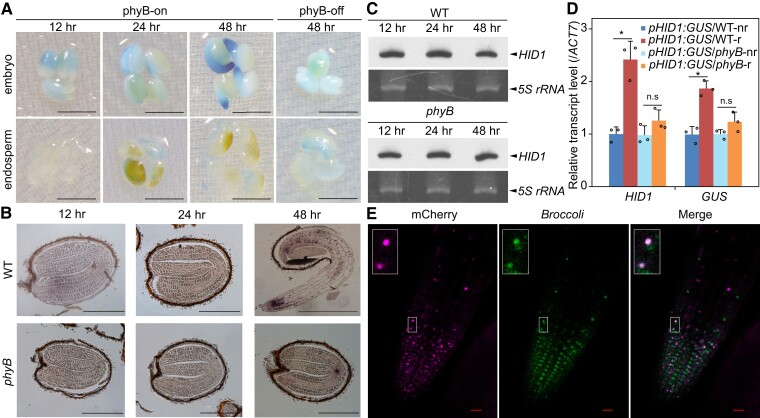
Figure 2.
PhyB promotes nuclear accumulation of HID1 in the radicle. A, GUS staining showing the expression pattern of pHID1:GUS/WT transgenic seeds under phyB-on and phyB-off conditions. Scale bar, 1 mm. B, In situ hybridization of WT and phyB seeds under phyB-on conditions with the HID1 antisense probe. Scale bar, 0.5 mm. C, RNA gel blots showing the expression levels of HID1 in WT and phyB seeds under phyB-on conditions. 5S rRNA was used as the loading control. D, RT-qPCR showing the mRNA levels of HID1 and GUS in the non-radicle (nr) and radicle (r) parts of pHID1:GUS/WT and pHID1:GUS/phyB transgenic seeds at 48 h under phyB-on conditions. Mean ± SD, n = 3. n.s, no significance. * P ≤ 0.05 (two-tailed Student's t-test). E, RNA imaging of Broccoli-HID1 in Broccoli-HID1/hid1 seeds possessing the 35S:mCherry-SV40-NLS cassette at 48 h after DAL under phyB-on conditions. Scale bar, 20 μm. An enlarged view of selected Broccoli-HID1 and mCherry-SV40-NLS fluorescence signals are shown in the top left corner.
Next, we performed in situ hybridization to investigate the expression of HID1 in imbibed seeds of the WT and phyB lines following 12, 24, and 48 h DAL under phyB-on conditions. In embryos from both WT and phyB mutant seeds, the expression of HID1 displayed a dispersed pattern at 12 h and 24 h under phyB-on conditions (Figure 2B). Similar to our observation of HID1 promoter activity (pHID1:GUS), HID1 was mainly expressed in the radicle following 48 h DAL (Figure 2B); however, loss of phyB abolished the expression of HID1 in the radicle following 48 h DAL (Figure 2B), suggesting that phyB promotes the accumulation of HID1 in the radicle during seed germination. Furthermore, RNA gel blotting showed that HID1 was constantly expressed in imbibed WT seeds during seed germination, and its expression was not obviously affected in phyB (Figure 2C). To precisely examine the changes in HID1 expression in the radicle of imbibed seeds, we generated a transgenic line expressing pHID1:GUS in the phyB mutant. We assessed the expression levels of β-glucuronidase (GUS) and endogenous HID1 in the radicle and non-radicle cells of pHID1:GUS/WT and pHID1:GUS/phyB seeds following 48 h DAL. RT-qPCR demonstrated that both GUS and HID1 had significantly elevated expression levels in the radicle of the pHID1:GUS/WT line as compared with non-radicle cells; however, in pHID1:GUS/phyB seeds, GUS and HID1 had similar expression levels in radicle and non-radicle cells (Figure 2D). Taken together, these data suggest that phyB promotes the accumulation of HID1 in the radicle during seed germination.
To further examine the subcellular localization of HID1 in vivo, we applied the RNA aptamer Broccoli to the RNA imaging of HID1 (Filonov et al., 2014). It has been reported that Broccoli gives rise to a fluorescence signal at 507 nm following excitation in the appropriate working solution supplemented with the chemical compound DFHBI-1 T. We generated stable transgenic lines expressing pHID1:Broccoli-HID1 in hid1 (Supplemental Figure S2A). Our genetic and phenotypic analyses showed that a high expression level of Broccoli-HID1 fully rescued the delayed germination phenotype of hid1 to that of WT under phyB-on conditions (Supplemental Figure S2B).
To better assess the nuclear localization of HID1, we introduced the nuclear marker 35S:mCherry-SV40-NLS into the pHID1:Broccoli-HID1 line. In vivo imaging of HID1 in the root tip of WL-grown seedlings (initiated with their roots in DFHBI-1 T solution) showed that the fluorescence signal of Broccoli-HID1 clearly overlapped with that of mCherry-SV40-NLS (Supplemental Figure S2, C and D), supporting our previous finding that HID1 is primarily nucleus-localized. Taking advantage of this Broccoli-tagged HID1 transgenic line, we found that HID1 was also enriched in the nuclei of radicle cells following 48 h DAL (Figure 2E), although the nuclear marker mCherry-SV40-NLS was expressed at low levels at this stage. Taken together, these results suggest that HID1 is gradually enriched in the radicle and nucleus-localized under phyB-on conditions.
HID1 and phyB co-regulate the expression of a large set of genes at 48 h under phyB-on conditions
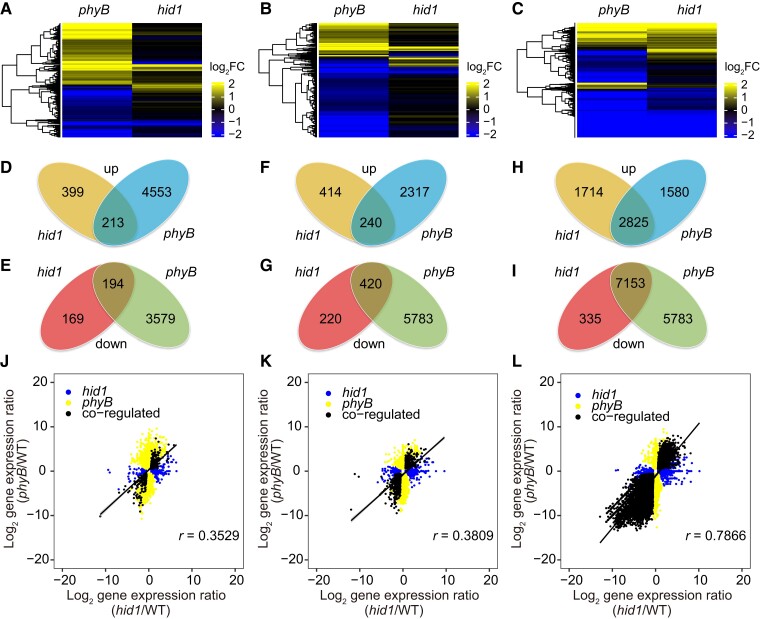
To further explore the function of HID1 in phyB-dependent seed germination, we incubated imbibed WT, hid1, and phyB seeds under phyB-on conditions and then sampled each group following 12, 24, and 48 h DAL for deep mRNA sequencing. Differentially expressed genes (DEGs; phyB/WT, hid1/WT, q value <0.05, fold change >1.5) were selected for further analysis (Supplemental Data Set 1). From the comprehensive transcriptomics analysis, we observed that a significant portion of the transcriptome was regulated by phyB during the three selected time points (Figure 3, A–C). In total, the phyB/WT comparison yielded 8,539 DEGs following 12 h DAL and 17,341 DEGs following 48 h DAL. However, the hid1/WT comparison yielded only 975 DEGs following 12 h DAL and 12,027 DEGs following 48 h DAL (Figure 3, D–G). Interestingly, the differences in the transcriptome revealed by the hid1/WT and phyB/WT comparisons were similar following 48 h DAL under phyB-on conditions (Figure 3, H and I). Two sets of DEGs, comprising 2,825 co-upregulated DEGs and 7,153 co-downregulated DEGs, accounted for more than half of the phyB-regulated genes and more than 80% of the hid1-regulated genes, indicating that phyB and HID1 co-regulate a large set of genes following 48 h DAL. Similarly, the expression levels of most of the hid1- and phyB DEGs were not highly correlated following 12 h or 24 h DAL (Figure 3, J–K), while their directions were highly correlated following 48 h DAL (R = 0.7866) (Figure 3L). This genome-wide analysis revealed that HID1 regulates a subset of genes following 24 h DAL but mainly regulates gene expression following 48 h DAL, which is different from the rapid response mediated by PIF1.
Figure 3.
HID1 and phyB co-regulate a large set of genes at the transcriptional level at 48 h under phyB-on conditions. A–C, Cluster analysis of hid1- and phyB-regulated genes following 12, 24, and 48 h DAL under phyB-on conditions. The scale bar shows the log2 transformation of the ratio. D, F, and H, Venn diagrams showing the overlap between hid1- and phyB-upregulated genes following 12, 24, and 48 h DAL under phyB-on conditions. E, G, and I, Venn diagrams showing the overlap between hid1- and phyB-downregulated genes following 12, 24, and 48 h DAL under phyB-on conditions. J, K, and L, Correlation scatterplots of the log2 fold change values between hid1- and phyB-regulated genes at 12, 24, and 48 h under phyB-on conditions.
It is well-established that the balance between signaling pathways activated by GA and ABA is an important regulator of seed germination, and it has been reported that phyB represses PIF1 following 12 h DAL to regulate the expression of many genes in the GA and ABA pathways. We chose several phyB–PIF1-regulated GA and ABA biosynthesis and metabolism genes as examples and measured their expression levels by RT-qPCR using total RNA samples extracted from the WT, hid1, PIF1OX (transgenic line overexpressing PIF1), and phyB lines following 12 h DAL. Consistent with previous studies, the levels of four genes, 9-CIS-EPOXYCAROTENOID DIOXYGENASE (NCED9), ABA INSENSITIVE 5 (ABI5), SOMNUS (SOM), and REPRESSOR OF GA1–3 (RGA), were upregulated in the phyB mutant and PIF1OX lines as compared with those in the WT line following 12 h DAL (Supplemental Figure S3, A–C). Following 12 h DAL, the levels of CYP707A2, EXPANSIN 8 (EXP8), and EXP10 were downregulated in the phyB and PIF1OX lines (Supplemental Figure S3, A–C), but the expression levels of these genes were either unaffected or regulated in the opposite direction in the hid1 line. Following 48 h DAL, the above-mentioned genes in ABA and GA metabolism were co-regulated by HID1 and phyB (Supplemental Figure S3D). Taken together, these results indicate that HID1 may not participate in phyB–PIF1-mediated rapid transcriptional regulation occurring within 12 h of seed imbibition.
The phyB-HID1 axis regulates multiple phytohormones and cellular metabolism genes following 48 h DAL
To further decode the phyB-HID1 regulatory network functioning during seed germination, we performed gene ontology (GO) analysis using the AgriGO software. Following 48 h DAL, the upregulated genes in the phyB and hid1 lines were enriched in several GO terms: response to ABA stimulus, ABA metabolic processes, seed development, and response to water. In contrast, the downregulated genes were primarily enriched in two GO terms: response to hormone stimuli and cellular metabolism (Figure 4A).
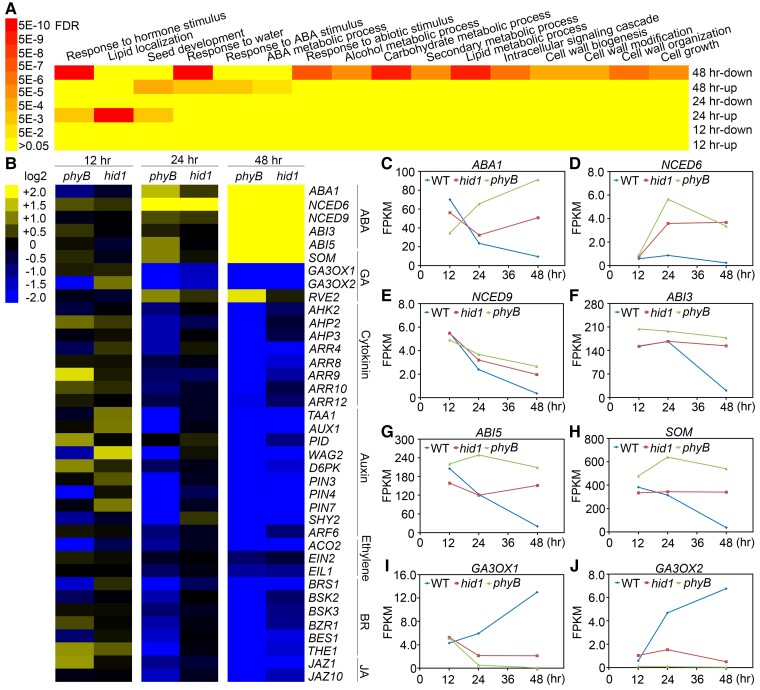
Figure 4.
HID1 promotes phyB-dependent seed germination by regulating the expression of metabolic and signaling genes related to ABA and GA. A, GO term analysis of differentially expressed genes in hid1 mutants in comparison with WT following 12, 24, and 48 h DAL. “up” or “down” indicates upregulated or downregulated genes, respectively, in hid1 mutants in comparison with WT. B, Cluster analysis of hid1- and phyB-regulated genes. The scale bar shows the log2 transformation of the ratio. C–J, FPKM (fragments per kilobase per million mapped reads) values (from the RNA-seq data) of the indicated genes in WT, hid1, and phyB following 12, 24, and 48 h DAL under phyB-on conditions.
Based on our time-course transcriptomics analysis, we further analyzed DEGs related to plant hormone regulation in the hid1 and phyB mutants. In accordance with the results mentioned above, the expression levels of phytohormone metabolic and signaling genes were mostly unaffected in the hid1 line following 12 h DAL, whereas they were mildly affected following 24 h DAL, suggesting that HID1 plays a minor role in seed germination at these timepoints (Figure 4B). Following 48 h DAL, the expression levels of ABA biosynthesis genes, including ABA1, NCED6, and NCED9, as well as positive regulators of the ABA signaling pathway, including ABI3 and ABI5, were co-upregulated in the hid1 and phyB lines (Figure 4, C–H). The expression levels of GA biosynthesis genes, including GA3OX1 and GA3OX2, as well as many other plant hormone-related genes, were downregulated in the hid1 and phyB lines (Figure 4, I and J). In addition to ABA and GA, other plant hormones including cytokinin, auxin, brassinosteroids (BRs), ethylene, and jasmonic acid, have also been found to regulate seed germination. The temporal dynamics of the transcriptomics datasets of the WT line showed that the expression levels of ABA biosynthesis and signaling genes gradually decreased during seed germination under phyB-on conditions, while the expression levels of biosynthesis and signaling genes related to GA, BR, auxin, and cytokinin gradually increased (Figure 4B; and Supplemental Figure S4). Taken together, these results suggest that loss of HID1 causes transcriptional reprogramming of the phytohormone network, promoting ABA signaling and inhibiting signaling downstream of GA and other germination-promoting phytohormones.
To further explore the genetic relationship between HID1 and phyB, we crossed our previously generated 35S:HID1/hid1 line with the phyB single mutant to obtain a homozygous 35S:HID1/phyB transgenic line. Overexpression of HID1 alleviated the seed germination defect of phyB mutants under phyB-on conditions (Supplemental Figure S5A). We also generated hid1 phyB double mutants. From the seed germination analysis, we found that the hid1 phyB double-mutant failed to germinate under phyB-on conditions, which phenocopied the phyB single mutant (Supplemental Figure S5, A and B). RT-qPCR demonstrated that the mRNA levels of selected genes involved in ABA biosynthesis and signaling were significantly upregulated in the hid1 line compared to the WT. The expression levels of these genes in the phyB and hid1 phyB double mutants were ∼20-fold greater than in the WT (Supplemental Figure S5C). By contrast, the expression levels of GA biosynthesis genes were downregulated in the hid1, phyB, and hid1 phyB double-mutant lines (Supplemental Figure S5D). Taken together, these results suggest that HID1 acts genetically downstream of phyB to mediate transcriptome changes that promote seed germination.
HID1 promotes phyB-dependent seed germination predominantly by suppressing ABA biosynthesis
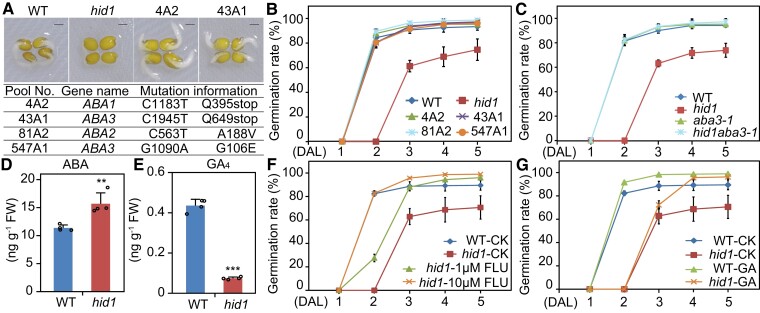
To further explore the mechanism by which HID1 regulates phyB-dependent seed germination, we performed forward genetic screening to identify key downstream components of the phyB-HID1 pathway. The screening process identified four independent recessive alleles in the M2 population of the ethylmethanesulfonate (EMS)-mutagenized hid1 mutant, and mutations in each of these alleles completely suppressed the delayed germination phenotype of hid1 under phyB-on conditions (Figure 5, A and B). Map-based cloning enabled the identification of three ABA biosynthesis genes, ABA1, ABA2, and ABA3, as candidate suppressors of HID1 (Figure 5A). The mutants 4A2 and 43A1 contained a single C to T nucleotide change in the coding region of ABA1 and ABA3, respectively, resulting in an amino acid change to a stop codon. Mutants 81A2 and 547A1 contained a nucleotide change in the coding region of ABA2 and ABA3, resulting in a missense mutation in their amino acid sequence (Figure 5A). We generated a hid1 aba3-1 double-mutant to validate the genetic relationship between HID1 and ABA3. From the phenotypic analysis, we found that the germination rates of aba3 and hid1 aba3 were similar to that of the WT line (Figure 5C), indicating that ABA3 acts downstream of HID1 during phyB-dependent seed germination. Next, we examined the endogenous levels of ABA and GA in imbibed WT and hid1 seeds following 48 h DAL under phyB-on conditions and found that ABA levels in the hid1 seeds were significantly higher than those in the WT seeds (Figure 5D). By contrast, the content of GA4, the major bioactive GA, was much lower in the hid1 seeds than in the WT seeds, while the concentrations of precursor GA9 and deactivated GA51 were increased in the hid1 seeds than in the WT seeds (Figure 5E; and Supplemental Figure S6). These results indicate that HID1 promotes low levels of ABA and high levels of GA following 48 h DAL.
Figure 5.
HID1 promotes phyB-dependent seed germination by repressing ABA biosynthesis. A, Representative images of WT, hid1, and two genetic suppressor lines of hid1 (4A2 and 43A1) following 48 h DAL under phyB-on conditions. Scale bar, 1 mm. The table underneath shows mutational information for the HID1 suppressor genes. B, Germination rates of WT, hid1, and four genetic suppressor lines of hid1 (4A2, 43A1, 81A2, and 547A1) following 48 h DAL under phyB-on conditions. Germination frequencies were recorded every 24 h after light treatment. Mean ± SD, n = 3. C, Germination rates of WT, hid1, aba3-1, and the hid1 aba3-1 double-mutant following 48 h DAL under phyB-on conditions. Mean ± SD, n = 3. D and E, Quantitation of endogenous ABA (D) and GA4 (E) levels in WT and hid1 mutant seeds following 48 h DAL under phyB-on conditions. FW, fresh weight. Mean ± SD, n = 4. ** P ≤ 0.01, *** P ≤ 0.001 (two-tailed Student's t-test). F and G, Germination rates of WT and hid1 mutant seeds exposed to different chemical treatments. CK, no chemical treatment; FLU, Fluridone; GA, GA3. Mean ± SD, n = 3.
Subsequently, to determine whether the ABA level in the hid1 mutant line is the primary contributor to HID1-mediated seed germination, we treated WT and hid1 mutant seeds with the ABA synthesis inhibitor fluridone (FLU). Exposure to 10 µM FLU fully rescued the delayed germination phenotype of the hid1 mutants to that of the WT (Figure 5F). In addition, the delayed germination phenotype of the hid1 mutants was only slightly rescued following the application of 5 μM bioactive GA3 to the MS media (Figure 5G). Collectively, these results demonstrate that activation of the phyB-HID1 axis promotes seed germination following 48 h DAL predominantly by repressing ABA biosynthesis.
HID1 binds to NCED9 chromatin and inhibits it transcription
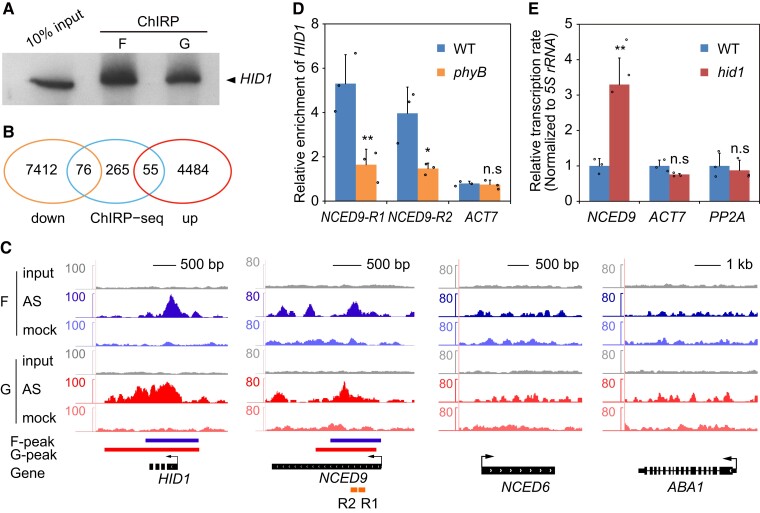
The results of our previous study suggested that HID1 binds chromatin and regulates transcription (Wang et al., 2014a, 2014b). To gain further insight into the molecular mechanisms underlying HID1 function, we investigated the genomic binding sites of HID1 using chromatin isolation by RNA purification (ChIRP) and subsequent high-throughput sequencing (Chu et al., 2011). We compared two chemical crosslinking strategies for fixing HID1:DNA:protein interactions with formaldehyde and glutaraldehyde. Using probes complementary to HID1 RNA, 15% and 12% of the total HID1 RNA were specifically retrieved by 1% formaldehyde and 1% glutaraldehyde crosslinking, respectively (Figure 6A). Strong HID1 peaks were identified using each of the crosslinkers, suggesting that the HID1 ChIRP-seq procedure obtained a reliable signal.
Figure 6.
HID1 associates with the NCED9 locus and represses its transcription. A, RNA gel blots of HID1. F indicates 1% (v/v) formaldehyde crosslinking, and G indicates 1% (v/v) glutaraldehyde crosslinking. B, Venn diagram of HID1-regulated and HID1-bound genes identified by ChIRP-seq. The overlapping ChIRP peaks were identified by both formaldehyde- and glutaraldehyde-crosslinked ChIRP-seq. C, HID1 association with the NCED9 locus by ChIRP-seq. F indicates formaldehyde crosslinking and G indicates glutaraldehyde crosslinking. D, ChIRP-qPCR showing the differences in HID1 occupancy on chromatin at NCED9 in WT and phyB. Mean ± SD, n = 3. n.s, no significance. * P ≤ 0.05, ** P ≤ 0.01 (two-tailed Student's t-test). E, The nascent RNA levels of NCED9 in WT and hid1 seeds following 48 h DAL under phyB-on conditions were determined by the nuclear run-on assay. 5S rRNA was used as an internal control. Mean ± SD, n = 4. n.s, no significance. ** P ≤ 0.01 (two-tailed Student's t-test).
In total, 396 genes that gave peaks in the ChIRP-seq experiments with both crosslinkers were considered HID1-bound gene candidates (Supplemental Data Set 2). Among these genes, 55 were upregulated and 76 were downregulated in the hid1 mutants at 48 h under phyB-on conditions (Figure 6B). Interestingly, we found strong peaks at the genomic locus of NCED9, a key enzyme in ABA biosynthesis (Figure 6C). Subsequent ChIRP-qPCR experiments confirmed the binding of HID1 on the chromatin of the NCED9 gene body (Figure 6D). Notably, the occupancy of HID1 at NCED9 was greatly reduced in the phyB mutants, suggesting that phyB is required for the binding of HID1 at NCED9 during seed germination (Figure 6D). We next examined the transcription rate of NCED9 by performing nuclear run-on (NRO) assays in WT and hid1 mutant lines. Loss of HID1 significantly increased the NCED9 transcription rate, while the transcription rates of ACT7 and PP2A were unaffected (Figure 6E), suggesting that HID1 inhibits NCED9 expression at the transcriptional level.
HID1 inhibits the enrichment of H3K4me3 on its repressed target genes
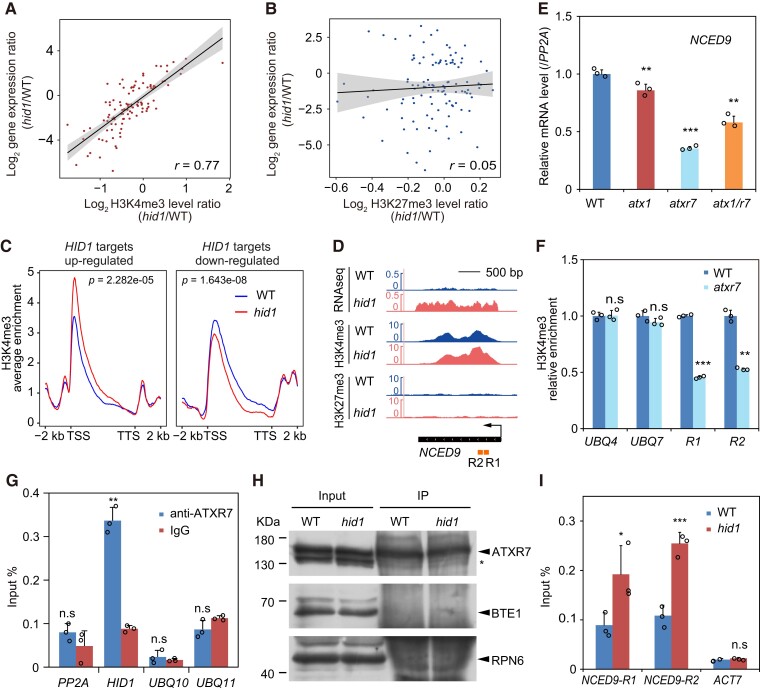
LncRNAs have been shown to be key regulators of histone modification on chromatin at specific genomic loci to modulate gene expression in cis and/or in trans (Statello et al., 2021). To assess whether HID1 mediates changes in histone modification on its target genes during phyB-dependent seed germination, we examined the genomic profiles of the active gene expression mark H3K4me3 and the repressive gene expression mark H3K27me3 in imbibed WT and hid1 seeds following 48 h DAL under phyB-on conditions. To this end, antibodies against H3K4me3 and H3K27me3 were used in chromatin-immunoprecipitation (ChIP)-seq experiments (Supplemental Data Sets 3 and 4). The 131 differentially expressed and HID1-bound genes were selected for subsequent analysis. In hid1 (hid1/WT), a change in the H3K4me3 level (R = 0.77), but not in the H3K27me3 level (R = 0.05), was positively correlated with changes in the expression of the 131 HID1 target genes (Figure 7, A and B). These results suggest that HID1-directed transcriptional changes may be coupled to H3K4me3 changes. We next created a meta-plot of H3K4me3 modification on HID1 target genes. The density of H3K4me3 was markedly higher on the 5′-end of HID1-repressed target genes in hid1 compared with WT, while H3K4me3 density was reduced on HID1-activated genes (Figure 7C). These results indicate that HID1 may modulate H3K4me3 occupancy and regulate target gene expression.
Figure 7.
HID1 antagonizes the accumulation of H3K4me3 and ATXR7 at the NCED9 locus. The correlation scatterplots show the log2 fold change values of the mRNA and H3K4me3 levels (A) and the H3K27me3 levels (B) in hid1 as compared with WT at 48 h under phyB-on conditions. r, Pearson's correlation coefficient. C, Meta-plots of the ChIP-seq signal of H3K4me3 for hid1-upregulated genes and hid1-downregulated genes in WT (blue curve) and hid1 (red curve). P value as indicated by the Wilcoxon rank-sum test. D, Integrative Genomics Viewer (IGV) screenshots of the RNA-seq, H3K4me3 ChIP-seq, and H3K27me3 ChIP-seq signals at NCED9. The gene model is shown at the bottom by the black bar. The arrow indicates the direction of transcription. E, RT-qPCR showing the mRNA levels of NCED9 in WT, atx1, atxr7, and atx1 atxr7 seeds following 48 h DAL under phyB-on conditions. Mean ± SD, n = 3. *** P ≤ 0.001 (two-tailed Student's t-test). F, ChIP-qPCR showing the relative enrichment of H3K4me3 on the indicated genes in WT and atxr7. Mean ± SD, n = 3. n.s, no significance. ** P ≤ 0.01, *** P ≤ 0.001 (two-tailed Student's t-test). UBQ4 and UBQ7 were used as controls. G, RIP-qPCR showing the association between ATXR7 and HID1. Mean ± SD, n = 3. n.s, no significance. ** P ≤ 0.01 (two-tailed Student's t-test). H, Immunoblots showing the protein expression levels of ATXR7 in the input and immunoprecipitates (IP) from WT and hid1. BTE1 and RPN6 were used as the loading controls. I, ChIP-qPCR showing the enrichment of ATXR7 at the indicated DNA regions of NCED9 in WT and hid1. ACT7 was used as the control. Mean ± SD, n = 3. n.s, no significance. *** P ≤ 0.001 (two-tailed Student's t-test).
HID1 disrupts the binding of ATXR7 at NCED9
In Arabidopsis, H3K4me3 is catalyzed by methyltransferases embedded in conserved COMPASS-like complexes (Hu and Du, 2022). Seven methyltransferases catalyze H3K4 methylation in Arabidopsis, including Trithorax group proteins ARABIDOPSIS TRITHORAX 1–5 (ATX1–5) and ATX-related (ATXR) proteins 3 and 7 (Hu and Du, 2022). Among these methyltransferases, ATX1 mediates the establishment of H3K4me3 and activates the expression of genes regulating plant meristem homeostasis, flowering, and ABA-dependent stress responses (He et al., 2021). ATXR7, an Arabidopsis homolog of yeast SET1, is also a methyltransferase for H3K4me3 (Tamada et al., 2009) and is known to play a role in seed dormancy (Liu et al., 2011). Recalling that the level of H3K4me3 at the NCED9 locus was increased in the hid1 mutant compared with WT (Figure 7D) and that HID1 binding coincided with the H3K4me3 peak at the 5′-end of NCED9 (Figure 6C), we speculated that HID1 mediates the transcriptional repression of NCED9 by regulating H3K4me3 modification. We initially measured the expression of NCED9 in atx1 and atxr7 single mutants and found that the mRNA level of NCED9 was strongly downregulated in the atxr7 mutant (more than two-fold) but only slightly reduced in atx1 (Figure 7E). Moreover, the expression of NCED9 in the atx1 atxr7 double-mutant closely resembled that of the atxr7 single mutant (Figure 7E). These results raised the possibility that NCED9 is a direct target of ATXR7; therefore, we used the atxr7 single mutant to determine whether ATXR7 is essential for H3K4me3 deposition at NCED9. We found that depletion of ATXR7 resulted in a marked reduction in the H3K4me3 level at NCED9, but not at the control genes UBQ4 and UBQ7 (Figure 7F), suggesting that ATXR7 is required for H3K4me3 deposition and activation of NCED9.
We next examined whether HID1 inhibits the enrichment of ATXR7 at NCED9. By generating an antibody against endogenous ATXR7 in Arabidopsis (Supplemental Figure 7), we assessed the interaction between HID1 and ATXR7 in vivo by RNA immunoprecipitation (RIP) using total protein lysates extracted from imbibed WT seeds following 48 h DAL under phyB-on conditions. Our results showed that ATXR7 co-immunoprecipitated with HID1 in vivo (Figure 7, G and H). To further determine the role of HID1 in the targeting of NCED9 by ATXR7, we compared the enrichment of ATXR7 at NCED9 in the WT and hid1 mutant lines. ChIP-qPCR data clearly demonstrated that the occupancy of ATXR7 on NCED9 chromatin within ATXR7 peak regions was greatly enhanced in the hid1 mutant compared with the WT, suggesting that HID1 inhibits the binding of ATXR7 at NCED9 (Figure 7I).
Discussion
Plants have evolved ingenious mechanisms to regulate seed germination. In general, from imbibition to radicle protrusion, it takes ∼2 days for Arabidopsis seeds to germinate after perceiving a light signal via photoreceptors (Seo et al., 2009; Shi et al., 2013). A better understanding of how photoreceptors transduce external light signals into internal phytohormone signaling to induce genome-wide changes in transcription during seed germination will provide important insight into seed biology. In the present study, we explored the molecular framework underlying a transcriptional cascade through which phyB-promoted radicle accumulation of the ncRNA HID1 inhibits ABA biosynthesis to promote seed germination during prolonged imbibition following 24 h DAL (Figure 8). It is likely that red light-activated phyB mediates short- and long-term effects to decrease the transcription of ABA biosynthesis genes. It is well accepted that for short-term effects, phyB promotes the degradation of PIF1 via the 26S proteosome. For long-term effects, at least in part, phyB promotes the reduction of ABA biosynthesis and signaling genes transcription via HID1. The results of this study demonstrate that activation of the phyB-HID1 genetic axis promotes red light-initiated seed germination. Our work establishes a new link between phyB and ABA biosynthesis genes via HID1, revealing another layer of light-dependent regulation of seed germination mediated by noncoding RNA.
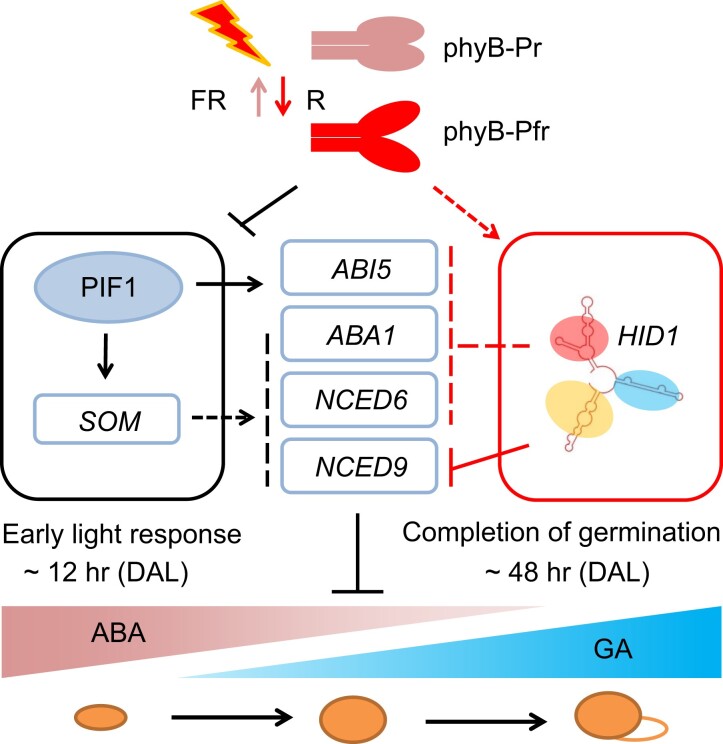
Figure 8.
A simplified working model of phyB-dependent seed germination mediated by HID1. Previous studies have shown that upon activation by red light, the Pfr form of phyB mediates the rapid degradation of PIF1 as an early light response. This inhibits PIF1-induced transcription of ABA biosynthesis genes via a SOMNUS-dependent pathway. Following 48 h DAL, the expression of the noncoding RNA HID1 in the radicles is significantly increased in a phyB-dependent manner. HID1 directly and indirectly inhibits the expression of ABA biosynthesis genes to promote phyB-dependent seed germination. Solid arrows, direct positive regulation; Dashed arrows, indirect positive regulation; Solid bars, direct negative regulation; Dashed bars, indirect negative regulation.
It has been reported that ABA content decreases rapidly in imbibed seeds. The decrease in ABA content at the beginning of seed imbibition is not dependent on light quality; however, the continuous decrease in ABA levels that occurs during prolonged imbibition (seeds imbibed for more than 24 h) is regulated by light (Seo et al., 2009). The manner by which light signaling regulates ABA biosynthesis and maintains the germination signal in imbibed seeds is unclear. Our genetic and transcriptomics analyses suggest that HID1 acts downstream of phyB to regulate ABA biosynthesis (Figure 3; and Supplemental Figure S4). Forward genetic screening identified three ABA biosynthesis genes, ABA1, ABA2, and ABA3, as candidate targets of HID1 suppression, suggesting that HID1 inhibits the biosynthesis of ABA in imbibed seeds (Figure 5). Indeed, the ABA levels in the hid1 mutants were higher than those in the WT line. ChIRP-seq and nuclear run-on experiments demonstrated that HID1 directly binds to the chromatin of the ABA biosynthesis gene NCED9 and inhibits its transcription. Hence, these results indicate that HID1 inhibits ABA biosynthesis to promote seed germination primarily by inhibiting the expression of ABA biosynthesis genes, both directly and indirectly. Interestingly, exogenous application of the ABA biosynthesis inhibitor FLU completely rescued the hid1 mutant phenotype to that of the WT, yet the exogenous application of GA hardly improved the germination rate of hid1 seeds (Figure 6). These results suggest that, although the ABA content in the imbibed seeds following 48 h DAL under phyB-on conditions was already low, modulation of ABA levels at this stage may have a stronger effect on seed germination in comparison with an increase in GA levels. Thus, the phyB-HID1-ABA1/NCED6/NCED9 transcriptional cascade may play a regulatory role in the maintenance of the signal-promoting seed germination.
Previous studies have demonstrated the presence of a decision center in the embryonic radicle of dormant seeds, which plays an important role in the decision to germinate in diverse environments. In the radicle, the responses to ABA and GA occur in a distinct spatial configuration, with the ABA response enriched in the tip of the radicle and the GA response enriched in the hypocotyl of the embryo, including the vascular tissue (Oh et al., 2006; Topham et al., 2017). The pronounced expression of HID1 in radicle cells following 48 h DAL can be related to its function in modulating ABA biosynthesis. Investigation into the manner by which phyB mediates the spatiotemporal expression of HID1 is underway. Photoactivated phyB may promote the expression and activity of HID1 during seed germination as a long-term effect. We speculate that there may exist a general repression mechanism or low positive effect immediately after the activation of phyB by red light in order to maintain HID1 accumulation at a certain level in the radicle following 12 and 24 h DAL. After 24 h DAL, the significant increase in HID1 expression in the radicle may be achieved by the gradual activation or deactivation of positive or negative regulators of HID1 transcription, respectively. Future studies are required to delineate the manner by which phyB precisely regulates the expression of HID1 in the decision center of the radicle.
As a large transcript reservoir, chromatin-associated lncRNAs can contribute to transcriptional regulation in cis or in trans by interacting with chromatin modifiers, mediating the recruitment or decoying of chromatin modifiers at specific genomic loci to modulate transcription (Long et al., 2017; Statello et al., 2021). Several research works in plants have revealed that chromatin-associated lncRNAs can regulate gene transcription by modulating histone H3K4me3 modification. For instance, COLDAIR (Liu et al., 2020) and MAS (Zhao et al., 2018) in Arabidopsis, as well as LAIR (Wang et al., 2018a, 2018b) in rice, mediate the recruitment of H3K4me3 to activate gene expression and affect Arabidopsis flowering and rice yield. However, the manner by which lncRNAs function in trans to regulate H3K4me3 and gene transcription is poorly understood. In this study, we found that ectopic expression of HID1 in the hid1 mutant rescued the delayed seed germination phenotype under phyB-on conditions, suggesting that HID1 functions in trans to regulate gene transcription during seed germination. We further explored the repressive role of HID1 on H3K4me3 deposition and gene transcription at NCED9 in trans. We demonstrated that HID1 inhibits the occupancy of ATXR7, a methyltransferase for H3K4me3, at NCED9 and reduces the levels of H3K4me3 modification and NCED9 transcription. Our results suggest a likely role for HID1 as a molecular decoy that interfaces with epigenetic machinery to regulate the chromatin environment at specific gene loci during the completion of seed germination. It is also possible that HID1 may function by affecting the interactions among components in the ATXR7-containing methyltransferase complexes and/or impairing ATXR7 H3K4 methyltransferase activity. The mechanisms by which chromatin-associated lncRNAs, including HID1, interact with ATXR7 in terrestrial plants to regulate H3K4me3 levels and gene transcription merit further investigation.
Our series of work has identified HID1, a plant-specific and conserved noncoding RNA, as an essential player in the control of multiple light-dependent biological processes including seed germination, seedling greening during de-etiolation, and photomorphogenesis. Interestingly, in all scenarios, HID1 acts as a negative regulator on chromatin to repress the transcription of different target genes in trans beyond Polycomb Repressive Complex 2 (PRC2)-mediated gene repression. We speculate that HID1 may modulate chromatin dynamics at specific genomic loci through various mechanisms, including, but not limited to, interfacing with epigenetic machinery, transcription factors, and transcriptional co-regulators in a light-dependent manner. Moreover, future studies should investigate ways in which the HID1–chromatin interaction can be modulated to regulate the transcription of specific target genes in light-mediated plant growth and development.
Materials and methods
Plant materials and growth conditions
The ecotype of all WT Arabidopsis thaliana used in this study was Columbia-0 (Col-0). The hid1 mutant (SALK_017318), the transgenic lines used in this study (HID1/hid1#2, HID1/hid1 #4, mut1/hid1#6, mut2/hid1#5, dSL2/hid1, dSL4/hid1, OsHID1/hid1#2, OsHID1/hid1#5, and 35S:HID1/WT), and phyB-9 have been reported previously (Wang et al., 2014a, 2014b). 35S:HID1/phyB was obtained by crossing 35S:HID1/WT with phyB-9. aba3-1 (CS157), atx1 (SALK_149002), and atxr7 (SALK_149692) were ordered from the Arabidopsis Biological Resource Center. hid1 aba3-1 double mutants were obtained by crossing hid1 with aba3-1, and atx1 atxr7 double mutants were obtained by crossing atx1 with atxr7. Arabidopsis seed sterilization, stratification, and standard seedling growth experiments were performed as previously described (Zhu et al., 2008). For seed germination-related experiments, all plants were grown side-by-side, and the seeds were harvested on the same day. The seeds were dried at room temperature for 2–6 months prior to conducting experiments, unless otherwise stated. Before performing any experiments, the germination rates of all the seeds were checked to ensure almost 100% germination at Day 3 after planting under continuous WL.
Germination assays
Including sterilization and plating, seeds were exposed to WL (50 μmol m−2 s−1) for 1 h. Under phyB-off conditions, seeds were exposed to far-red light (5 μmol m−2 s−1) for 5 min to inactivate phyB. Under phyB-on conditions, seeds were then irradiated with red light (20 μmol m−2 s−1) for 5 min to activate phyB. Under phyA-on conditions, seeds were irradiated with far-red light (FR) (5 μmol m−2 s−1) for 4 h, to activate phyA at 48 h and then incubated in the dark for 3 days. Under WL conditions, seeds were exposed to continuous WL for 1 h, stratified at 4°C for 3 days in the dark before being planted, and then incubated under continuous WL for a further 5 days. Germination rates were recorded every 24 h after planting. The seeds were then incubated in the dark for the indicated time, and the germination rates were determined in a dark room. At least 120 seeds were used for each experimental treatment, and three biological replicates were performed for each statistical analysis.
Plasmid construction and generation of transgenic plants
To generate the pHID1:GUS construct, the 1.5 kb upstream regulatory sequence of HID1 was cloned into the Gateway vector pDONR221 (Invitrogen) using BP Clonase and transferred into the binary vector pBGWFS7 by the LR reaction (Invitrogen). To generate the pHID1:Broccoli-HID1 construct, the Broccoli sequence was inserted into HID1 by PCR-based mutagenesis using a pHID1:HID1 plasmid as the template and then cloned into the BamHI site of pCAMBIA1300 . To generate the 35S:mCherry-SV40-NLS construct, the SV40-NLS sequence was fused with the mCherry coding sequence at the 3′end by PCR and then cloned into the BamHI/XbaI sites of pCAMBIA3300.
Agrobacterium tumefaciens strain GV3101 was used to transform Arabidopsis plants of the indicated backgrounds using the floral dip method (Clough and Bent, 1998). All transgenic plants were obtained by antibiotic selection and verified by immunoblotting and/or RNA gel blotting. pHID1:GUS/phyB was obtained by crossing pHID1:GUS/WT with phyB-9.
All cloning primers are listed in Supplemental Table S1.
GUS staining
GUS staining was performed as previously reported (Zhang et al., 2008a, 2008b).
In situ hybridization
In situ hybridization was performed as previously described (Wu and Wagner, 2012). Digoxigenin (DIG)-labeled HID1 antisense and sense probes were transcribed using the AmpliScribe T7-Flash Transcription Kit (Illumina).
RNA gel blotting
RNA gel blotting was performed as previously reported (Liu et al., 2013). The sequence used to probe HID1 is listed in Supplemental Table S1.
RNA imaging
Plant materials were incubated in RNA Imaging Buffer [20 mM HEPES-KOH (pH 7.5), 0.3 mM MgCl2, 100 mM KCl, 200 μM DFHBI-1 T] for 5 min at room temperature. Images were taken using a Zeiss LSM800 confocal microscope. For the Broccoli channel, the excitation wavelength was 488 nm, and the emission wavelength range was 490–520 nm. For the mCherry channel, the excitation wavelength was 561 nm, and the emission wavelength range was 600–670 nm.
Quantitative RT-PCR and RNA-seq
Total RNA was isolated using the Spectrum Plant Total RNA Kit (STRN50, Sigma). For quantitative RT-PCR (RT-qPCR), 1 μg total RNA was used to synthesize cDNA with SSRIII transcriptase (Invitrogen). Real-time PCR was performed using the SYBR Premix Ex Taq Kit (Takara) on a 7500 Fast Real-Time PCR Amplifier (Applied Biosystems). ACTIN7, 5S rRNA, and PP2A were used as the internal controls as indicated in different figures. The primers used for RT-qPCR are listed in Supplemental Table S1.
For RNA-seq, three biological replicates for each sample were used for library generation. Poly(A) RNA from 1 μg total RNA was used to generate each cDNA library according to the TruSeq RNA Sample Prep Kit (Illumina) protocol. All libraries were sequenced on a HiSeq 2500 System (Illumina) with 150 bp paired-end reads. RNA-seq reads were mapped to TAIR10 using HISAT2 (version 2.1.0). Differentially expressed genes were defined as those that differed by more than 1.5-fold with a false discovery rate <0.01 using the edgeR software (version 2.0.0) (Robinson et al., 2010).
ChIP and ChIP-seq library preparation
ChIP was carried out as outlined previously, with minor modifications (Gendrel et al., 2005). Briefly, 0.3 g seedlings grown under phyB-on conditions for 48 h were collected, crosslinked with 1% formaldehyde for 20 min under a vacuum, and ground into a fine powder in liquid nitrogen. Chromatin was isolated and sheared into fragments (200–500 bp) by sonication and incubated with antibody-bound Sera-Mag SpeedBeads Protein A/G Magnetic Particles (17152104010150, GE Healthcare) overnight at 4°C. Precipitated DNA was reverse-crosslinked, digested with RNase A and Proteinase K, and purified using the ChIP DNA Clean & Concentrator Kit (D5205, Zymo Research) according to the manufacturer's instructions. Three biological replicates for each sample were subjected to ChIP-seq. All ChIP-seq libraries were generated with the NEBNext® Ultra II DNA Library Prep Kit for Illumina® (E7645S, NEB) according to the manufacturer's instructions. All libraries were sequenced on a HiSeq2500 System with 150 bp paired-end reads. The antibodies used for ChIP were anti-H3K4me3 (04-745, Millipore, 1:200 dilution), anti-H3K27me3 (07-449, Millipore, 1:100 dilution), and anti-ATXR7 (raised in rabbit using the synthetic peptide LIRDGENKLRPITLC as the antigen, Beijing Protein Innovation, 1:200 dilution). The primers used for ChIP-qPCR are listed in Supplemental Table S1.
ChIP-seq data analysis
For all ChIP-seq data sets, reads were aligned to the Arabidopsis genome (TAIR10) (Lamesch et al., 2012) using bowtie2 (version 2.3.4) (Langmead and Salzberg, 2012) with default parameters. Only unique alignments of high mapping quality (MAPQ > 30) were retained by SAMtools (Li et al., 2009). The alignments were converted to bigWig files using the bamCoverage tool from deepTools2 (version 3.1.3) (Ramirez et al., 2016) with parameters “–binSize 5 –normalizeUsing CPM”. Meta-gene-averaged ChIP-seq profiles were generated in deepTools2 using the computeMatrix tool and plotted with the ggplot2 (version 3.2.1) package.
Suppressor screening and gene mapping
The hid1 mutant seeds were soaked in water overnight, treated with 0.4% (v/v) EMS solution for 8 h, washed extensively with sterile water, and dried. Plants (5,000 M1) were grown in groups of 1,000. M2 plants were screened for the WT phenotype as suppressor candidates. Four independent M3 candidate suppressors that were no longer segregated for the hid1 phenotype were identified and crossed with Landsberg erecta to generate the mapping population. Rough mapping and fine mapping were carried out as previously described (Qu and Qin, 2014). The markers used for map-based cloning were designed according to the Monsanto Arabidopsis polymorphism and Landsberg sequence collections.
Quantitation of endogenous ABA and GA levels
For each genotype, 0.3 g seeds grown under phyB-on conditions for 48 h were collected, frozen in liquid nitrogen, and ground into a fine powder in a dark room. Endogenous ABA and GA were purified and measured as previously described (Jiang et al., 2021). Statistical analysis was performed on four experiments using individual batches of seeds.
Immunoprecipitation and immunoblotting
For immunoprecipitation (IP) assays, seed samples (0.5 g) were frozen in liquid nitrogen, ground into a powder, and placed in lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM EDTA, 10% (v/v) glycerol, 0.1% (v/v) Tween®20, 1 mM PMSF, and 1× protease inhibitors). Samples were centrifuged at 16,000 × g for 10 min, and the total protein concentration of the supernatants was determined by the Bradford method. A 50 μg aliquot of total protein was taken as input, and 500 μg total protein was incubated with 5 μl purified anti-ATXR7 antibody in 500 μl lysis buffer for 4 h at 4°C. Subsequently, 20 μl well-washed Protein A/G magnetic beads (17152104010150, GE Healthcare) were added to each sample, and the tube was rotated for 2 h at 4°C. The beads were washed three times with lysis buffer, and proteins were eluted from the beads by boiling in 1× SDS loading buffer. Immunoblotting was performed as previously reported (Zhu et al., 2008). The anti-BTE1 (1:1000 dilution) and anti-RPN6 (1:2000 dilution) antibodies used in this study have been reported previously (Wang et al., 2022).
Chromatin isolation by RNA purification
ChIRP was performed as previously described, with some modifications (Chu et al., 2011). A total of 0.3 g seeds were used for each assay. The materials were fixed using two methods in the dark room. For reversible chemical crosslinking, fixation was performed with 1% (v/v) formaldehyde in PBS at room temperature for 20 min under a vacuum. For nonreversible crosslinking, fixation was performed with 1% (v/v) glutaraldehyde in PBS at room temperature for 20 min under a vacuum. For both crosslinking methods, the reactions were quenched with glycine (0.125 M final concentration) for 5 min under a vacuum. Antisense DNA probes were designed against the full-length HID1 sequence and biotinylated at the 3′ end (Invitrogen). Unlabeled probes were used as a control. DNA was purified using the ChIP DNA Clean & Concentrator Kit (D5205, Zymo Research) according to the manufacturer's instructions, and purified DNA was subjected to high-throughput sequencing. All ChIRP-seq libraries were generated using the NEBNext® Ultra II DNA Library Prep Kit for Illumina (E7645S, NEB) according to the manufacturer's instructions. All libraries were sequenced on a HiSeq 2500 System with 150 bp paired-end reads. The primers used for ChIRP-qPCR are listed in Supplemental Table S1.
ChIRP-seq data analysis
Raw ChIRP sequence reads were mapped to the TAIR10 genome assembly (Lamesch et al., 2012) of Arabidopsis via bowtie2 (version 2.1.0) (Langmead and Salzberg, 2012) with default parameters. Only uniquely mapped reads of high MAPQ > 30 were retained for further analysis. Peak calling was performed using MACS (version 1.4.2) (Zhang et al., 2008a, 2008b) with the following parameters: –keep-dup 1 –gsize 1.2e8 –Pvalue 1e5 –keep-dup = auto –to-large –nomodel –shiftsize 100. TAIR10 genome release was used for genomic region annotation of mapped ChIRP peaks. We defined ChIRP-associated genes as a ChIRP peak located in its gene body, 1500 bp upstream, or 500 bp downstream. Multiple gene-localized sites were retained for further verification.
Nuclear run-on assay
The nuclear run-on assay was performed as previously described (Zhu et al., 2016). A total of 0.3 g seeds were used for each assay. Four independent biological replicates were performed and analyzed. 5S rRNA was used as the internal control. The primers used for the nuclear run-on assay are listed in Supplemental Table S1.
RNA immunoprecipitation
RIP was performed as previously described (Wierzbicki et al., 2008). A total of 0.3 g seeds were used for each assay. Three independent biological replicates were performed and analyzed. For immunoblotting, proteins were eluted from the beads by boiling in 1× SDS loading buffer. The antibodies used in this assay were ani-ATXR7 (raised in rabbit using the synthetic peptide LIRDGENKLRPITLC as the antigen, Beijing Protein Innovation, 1:200 dilution) and Rabbit IgG (PP64, Millipore, 1:200 dilution). The primers used for RIP are listed in Supplemental Table S1.
Statistical analysis
The germination rates, all of the RT-qPCR, ChIP-qPCR, ChIRP-qPCR, RIP-qPCR results, and endogenous ABA and GA levels obtained in this study are presented as means ± standard deviation (SD). The numbers of biological replicates in each experiment are noted in the figure legends. Statistical analysis was performed by a two-tailed Student's t-test. Wilcoxon rank-sum tests for the ChIP-seq signal of H3K4me3 levels in each sample were performed using R (https://www.R-project.org/). Details about the statistical analysis are provided in Supplemental Data Set 5.
Accession numbers
Accession numbers obtained from the Arabidopsis Information Resource (TAIR) for the genes mentioned in this article are as follows: HID1, AT2G35747; PHYB, AT2G18790; ABA1, AT5G67030; NCED6, AT3G24220; NCED9, AT1G78390; ABI3, AT3G24650; ABI5, AT2G36270; SOM, AT1G03790; GA3OX1, AT1G15550; GA3OX2, AT1G80340; RVE2, AT5G37260; AHK2, AT5G35750; AHP2, AT3G29350; AHP3, AT5G39340; ARR4, AT1G10470; ARR8, AT2G41310; ARR9, AT3G57040; ARR10, AT4G31920; ARR12, AT2G25180; TAA1, AT1G70560; AUX1, AT2G38120; PID, AT2G34650; WAG2, AT3G14370; D6PK, AT5G55910; PIN3, AT1G70940; PIN4, AT2G01420; PIN7, AT1G23080; SHY2, AT1G04240; ARF6, AT1G30330; ACO2, AT1G62380; EIN2, AT5G03280; EIL1, AT2G27050; BRS1, AT4G30610; BSK2, AT5G46570; BSK3, AT4G00710; BZR1, AT1G75080; BES1, AT1G19350; THE1, AT5G54380; JAZ1, AT1G19180; JAZ10, AT5G13220; PIF1, AT2G20180; ACT7, AT5G09810; PP2A, AT1G13320; CYP707A2, AT2G29090; RGA, AT2G01570; RGL2, AT3G03450; EXP8, AT2G40610; EXP10, AT1G26770; ATX1, AT2G31650; ATXR7, AT5G42400; UBQ4, AT5G20620; UBQ7, AT2G35635; UBQ10, AT4G05320; UBQ11, AT4G05050; BTE1, AT2G40630; RPN6, AT1G29150.
Supplementary Material
Acknowledgments
We are grateful to Drs. Peiyong Xin and Jinfang Chu (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for detecting the ABA and GA levels. We thank Prof. Giltsu Choi (Korea Advanced Institute of Science and Technology, Korea) for providing PIF1OX seeds and Dr. Xuncheng Wang for bioinformatics assistance.
Contributor Information
Yuqiu Wang, State Key Laboratory of Protein and Plant Gene Research, School of Advanced Agricultural Sciences and School of Life Sciences, Peking-Tsinghua Center for Life Sciences, Peking University, Beijing 100871, China.
Yangyang Fan, State Key Laboratory of Protein and Plant Gene Research, School of Advanced Agricultural Sciences and School of Life Sciences, Peking-Tsinghua Center for Life Sciences, Peking University, Beijing 100871, China.
De Fan, State Key Laboratory of Protein and Plant Gene Research, School of Advanced Agricultural Sciences and School of Life Sciences, Peking-Tsinghua Center for Life Sciences, Peking University, Beijing 100871, China; Peking University Institute of Advanced Agricultural Sciences, Shandong Laboratory of Advanced Agricultural Sciences in Weifang, Weifang, Shandong 261325, China.
Xiaoli Zhou, State Key Laboratory of Protein and Plant Gene Research, School of Advanced Agricultural Sciences and School of Life Sciences, Peking-Tsinghua Center for Life Sciences, Peking University, Beijing 100871, China.
Yuntong Jiao, State Key Laboratory of Protein and Plant Gene Research, School of Advanced Agricultural Sciences and School of Life Sciences, Peking-Tsinghua Center for Life Sciences, Peking University, Beijing 100871, China.
Xing Wang Deng, State Key Laboratory of Protein and Plant Gene Research, School of Advanced Agricultural Sciences and School of Life Sciences, Peking-Tsinghua Center for Life Sciences, Peking University, Beijing 100871, China; Peking University Institute of Advanced Agricultural Sciences, Shandong Laboratory of Advanced Agricultural Sciences in Weifang, Weifang, Shandong 261325, China.
Danmeng Zhu, State Key Laboratory of Protein and Plant Gene Research, School of Advanced Agricultural Sciences and School of Life Sciences, Peking-Tsinghua Center for Life Sciences, Peking University, Beijing 100871, China.
Data availability
RNA-seq, ChIP-seq, and ChIRP-seq data sets generated in this study have been deposited in the Gene Expression Omnibus (GEO) under accession GSE195748. All other study data are included in the article and/or supporting information.
Supplemental data
Supplemental Figure S1. ncRNA hid1 mutant seeds show a delayed germination phenotype under white light and phyA-on conditions.
Supplemental Figure S2. Broccoli-HID1 is biologically functional.
Supplemental Figure S3. phyB and PIF1, but not HID1, co-regulate the expression levels of a set of genes following 12 h DAL under phyB-on conditions.
Supplemental Figure S4. Expression patterns of genes involved in phytohormone signaling, cell wall loosening, water transport, and oil body formation over time during phyB-dependent seed germination.
Supplemental Figure S5. HID1 and phyB co-regulate metabolic and signaling genes related to ABA and GA.
Supplemental Figure S6. HID1 is essential for GA homeostasis following 48 h DAL under phyB-on conditions.
Supplemental Figure S7. Specificity test of the Anit-ATXR7 antibody in Arabidopsis.
Supplemental Figure S8. Unprocessed immunoblots in this study.
Supplemental Table S1. Primers and probes used in this study.
Supplemental Data Set 1. Differentially expressed genes in the WT and mutants over time during phyB-dependent seed germination obtained from the RNA-seq analysis.
Supplemental Data Set 2. The list of HID1-bound genes identified by ChIRP-seq.
Supplemental Data Set 3. Gene lists summarizing ChIP-seq analysis of H3K4me3 levels in WT, hid1 and phyB seeds.
Supplemental Data Set 4. Gene lists summarizing ChIP-seq analysis of H3K27me3 levels in WT, hid1 and phyB seeds.
Supplemental Data Set 5. t-test results.
Supplemental Data Set 6. Summary of mapped reads for RNA-seq, ChIP-seq, and ChIRP-seq in each of the samples.
Funding
This work was supported by the National Natural Science Foundation of China (grant 91940301 to D.Z.), the National Key R&D Program of China (grant 2016YFA0500800 to D.Z.), the National Natural Science Foundation of China grants (31621001 to X.W.D., 31970259 to Y.W.).
References
- Berretta J, Morillon A (2009) Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep 10(9): 973–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borthwick HA, Hendricks SB, Parker MW, Toole EH, Toole VK (1952) A reversible photoreaction controlling seed germination. Proc Natl Acad Sci U S A 38(8): 662–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JN, Ryu JY, Jeong YM, Park J, Song JJ, Amasino RM, Noh B, Noh YS (2012) Control of seed germination by light-induced histone arginine demethylation activity. Dev Cell 22(4):736–748 [DOI] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY (2011) Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 44(4): 667–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung PJ, Park BS, Wang H, Liu J, Jang IC, Chua NH (2016) Light-Inducible MiR163 targets PXMT1 transcripts to promote seed germination and primary root elongation in arabidopsis. Plant Physiol 170(3): 1772–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16(6): 735–743 [DOI] [PubMed] [Google Scholar]
- de Wit M, Galvao VC, Fankhauser C (2016) Light-mediated hormonal regulation of plant growth and development. Annu Rev Plant Biol 67(1): 513–537 [DOI] [PubMed] [Google Scholar]
- Filonov GS, Moon JD, Svensen N, Jaffrey SR (2014) Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J Am Chem Soc 136(46): 16299–16308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Martienssen R, Colot V (2005) Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods 2(3): 213–218 [DOI] [PubMed] [Google Scholar]
- He K, Cao X, Deng X (2021) Histone methylation in epigenetic regulation and temperature responses. Curr Opin Plant Biol 61: 102001. [DOI] [PubMed] [Google Scholar]
- Hu H, Du J (2022) Structure and mechanism of histone methylation dynamics in Arabidopsis. Curr Opin Plant Biol 67: 102211. [DOI] [PubMed] [Google Scholar]
- Jiang A, Guo Z, Pan J, Yang Y, Zhuang Y, Zuo D, Hao C, Gao Z, Xin P, Chu J, et al. (2021) The PIF1-miR408-PLANTACYANIN repression cascade regulates light-dependent seed germination. Plant Cell 33(5): 1506–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Xu G, Jing Y, Tang W, Lin R (2016) Phytochrome B and REVEILLE1/2-mediated signalling controls seed dormancy and germination in Arabidopsis. Nat Commun 7: 12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Yamaguchi S, Lim S, Oh E, Park J, Hanada A, Kamiya Y, Choi G (2008) SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 20(5): 1260–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. (2012) The Arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res 40(D1): D1202–D1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with bowtie 2. Nat Methods 9(4): 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Piskurewicz U, Tureckova V, Carat S, Chappuis R, Strnad M, Fankhauser C, Lopez-Molina L (2012) Spatially and genetically distinct control of seed germination by phytochromes A and B. Genes Dev 26(17): 1984–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25(16): 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Yang L, Li J, Tang W, Li J, Lin R (2021) FHY3 Interacts with phytochrome B and regulates seed dormancy and germination. Plant Physiol 187(1): 289–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TT, Zhu D, Chen W, Deng W, He H, He G, Bai B, Qi Y, Chen R, Deng XW (2013) A global identification and analysis of small nucleolar RNAs and possible intermediate-sized non-coding RNAs in Oryza sativa. Mol Plant 6(3): 830–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Geyer R, van Zanten M, Carles A, Li Y, Horold A, van Nocker S, Soppe WJ (2011) Identification of the Arabidopsis REDUCED DORMANCY 2 gene uncovers a role for the polymerase associated factor 1 complex in seed dormancy. PLoS One 6(7): e22241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZW Liu, Zhao N, YN Su, SS Chen, XJ He (2020) Exogenously overexpressed intronic long noncoding RNAs activate host gene expression by affecting histone modification in Arabidopsis. Sci Rep 10(1): 3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Y, Wang X, Youmans DT, Cech TR (2017) How do lncRNAs regulate transcription? Sci Adv 3(9): eaao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, Taft RJ, Faulkner GJ (2010) A global view of genomic information–moving beyond the gene and the master regulator. Trends Genet 26(1): 21–28 [DOI] [PubMed] [Google Scholar]
- Oh E, Kim J, Park E, Kim JI, Kang C, Choi G (2004) PIL5, A phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16(11): 3045–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Hu J, Yusuke J, Jung B, Paik I, Lee HS, Sun TP, Kamiya Y, Choi G (2007) PIL5, A phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19(4): 1192–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Kamiya Y, Bae G, Chung WI, Choi G (2006) Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J 47(1): 124–139 [DOI] [PubMed] [Google Scholar]
- Qu LJ, Qin G (2014) Generation and identification of Arabidopsis EMS mutants. Methods Mol Biol 1062: 225–239 [DOI] [PubMed] [Google Scholar]
- Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, Manke T (2016) Deeptools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44(W1): W160–W165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) Edger: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1): 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar Das S, Yadav S, Singh A, Gautam V, Sarkar AK, Nandi AK, Karmakar P, Majee M, Sanan-Mishra N (2018) Expression dynamics of miRNAs and their targets in seed germination conditions reveals miRNA-ta-siRNA crosstalk as regulator of seed germination. Sci Rep 8(1): 1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Hanada A, Kuwahara A, Endo A, Okamoto M, Yamauchi Y, North H, Marion-Poll A, Sun TP, Koshiba T, et al. (2006) Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J 48(3): 354–366 [DOI] [PubMed] [Google Scholar]
- Seo M, Nambara E, Choi G, Yamaguchi S (2009) Interaction of light and hormone signals in germinating seeds. Plant Mol Biol 69(4): 463–472 [DOI] [PubMed] [Google Scholar]
- Shi H, Wang X, Mo X, Tang C, Zhong S, Deng XW (2015) Arabidopsis DET1 degrades HFR1 but stabilizes PIF1 to precisely regulate seed germination. Proc Natl Acad Sci U S A 112(12): 3817–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Zhong S, Mo X, Liu N, Nezames CD, Deng XW (2013) HFR1 Sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell 25(10): 3770–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura T, Nagatani A, Chory J, Furuya M (1994) The induction of seed germination in Arabidopsis thaliana is regulated principally by phytochrome B and secondarily by phytochrome A. Plant Physiol 104(2): 363–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura T, Nagatani A, Hanzawa H, Kubota M, Watanabe M, Furuya M (1996) Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc Natl Acad Sci U S A 93(15): 8129–8133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statello L, Guo CJ, Chen LL, Huarte M (2021) Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 22(2): 96–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada Y, Yun JY, Woo SC, Amasino RM (2009) ARABIDOPSIS TRITHORAX-RELATED7 is required for methylation of lysine 4 of histone H3 and for transcriptional activation of FLOWERING LOCUS C. Plant Cell 21(10): 3257–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topham AT, Taylor RE, Yan D, Nambara E, Johnston IG, Bassel GW (2017) Temperature variability is integrated by a spatially embedded decision-making center to break dormancy in Arabidopsis seeds. Proc Natl Acad Sci U S A 114(25): 6629–6634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HV, Chekanova JA (2017) Long noncoding RNAs in plants. Adv Exp Med Biol 1008: 133–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Fan X, Lin F, He G, Terzaghi W, Zhu D, Deng XW (2014a) Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc Natl Acad Sci U S A 111(28): 10359–10364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Fan Y, Fan Z, Zhou Y, Zhang X, Wang R, Sun Y, Zhang Y, He W, Deng Y, et al. (2022) The Arabidopsis DREAM complex antagonizes WDR5A to modulate histone H3K4me2/3 deposition for a subset of genome repression. Proc Natl Acad Sci U S A 119(27): e2206075119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li J, Deng XW, Zhu D (2018a) Arabidopsis noncoding RNA modulates seedling greening during deetiolation. Sci China Life Sci 61(2): 199–203 [DOI] [PubMed] [Google Scholar]
- Wang Y, Luo X, Sun F, Hu J, Zha X, Su W, Yang J (2018b) Overexpressing lncRNA LAIR increases grain yield and regulates neighbouring gene cluster expression in rice. Nat Commun 9(1): 3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang X, Deng W, Fan X, Liu TT, He G, Chen R, Terzaghi W, Zhu D, Deng XW (2014b) Genomic features and regulatory roles of intermediate-sized non-coding RNAs in Arabidopsis. Mol Plant 7(3): 514–527 [DOI] [PubMed] [Google Scholar]
- Weitbrecht K, Muller K, Leubner-Metzger G (2011) First off the mark: early seed germination. J Exp Bot 62(10): 3289–3309 [DOI] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS (2008) Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135(4): 635–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MF, Wagner D (2012) RNA In situ hybridization in Arabidopsis. Methods Mol Biol 883: 75–86 [DOI] [PubMed] [Google Scholar]
- Yang L, Jiang Z, Jing Y, Lin R (2020) PIF1 And RVE1 form a transcriptional feedback loop to control light-mediated seed germination in Arabidopsis. J Integr Plant Biol 62(9): 1372–1384 [DOI] [PubMed] [Google Scholar]
- Yu Y, Zhang Y, Chen X, Chen Y (2019) Plant noncoding RNAs: hidden players in development and stress responses. Annu Rev Cell Dev Biol 35(1): 407–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Feng S, Chen F, Chen H, Wang J, McCall C, Xiong Y, Deng XW (2008a) Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. Plant Cell 20(6): 1437–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008b) Model-based analysis of ChIP-seq (MACS). Genome Biol 9(9): R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Li J, Lian B, Gu H, Li Y, Qi Y (2018) Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA. Nat Commun 9(1): 5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Maier A, Lee JH, Laubinger S, Saijo Y, Wang H, Qu LJ, Hoecker U, Deng XW (2008) Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell 20(9): 2307–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Wang Y, Qin N, Wang F, Wang J, Deng XW, Zhu D (2016) Arabidopsis small nucleolar RNA monitors the efficient pre-rRNA processing during ribosome biogenesis. Proc Natl Acad Sci U S A 113(42): 11967–11972 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq, ChIP-seq, and ChIRP-seq data sets generated in this study have been deposited in the Gene Expression Omnibus (GEO) under accession GSE195748. All other study data are included in the article and/or supporting information.