Abstract
Five versions of the Chlamydomonas reinhardtii reference genome have been produced over the last two decades. Here we present version 6, bringing significant advances in assembly quality and structural annotations. PacBio-based chromosome-level assemblies for two laboratory strains, CC-503 and CC-4532, provide resources for the plus and minus mating-type alleles. We corrected major misassemblies in previous versions and validated our assemblies via linkage analyses. Contiguity increased over ten-fold and >80% of filled gaps are within genes. We used Iso-Seq and deep RNA-seq datasets to improve structural annotations, and updated gene symbols and textual annotation of functionally characterized genes via extensive manual curation. We discovered that the cell wall-less classical reference strain CC-503 exhibits genomic instability potentially caused by deletion of the helicase RECQ3, with major structural mutations identified that affect >100 genes. We therefore present the CC-4532 assembly as the primary reference, although this strain also carries unique structural mutations and is experiencing rapid proliferation of a Gypsy retrotransposon. We expect all laboratory strains to harbor gene-disrupting mutations, which should be considered when interpreting and comparing experimental results. Collectively, the resources presented here herald a new era of Chlamydomonas genomics and will provide the foundation for continued research in this important reference organism.
A major update to the Chlamydomonas reference genome and structural annotations is presented, revealing large genomic changes that have occurred in laboratory culture.
IN A NUTSHELL.
Background: Chlamydomonas reinhardtii (Chlamydomonas) is an important reference organism. The first draft genome for the species was sequenced 20 years ago. The current assembly, version 5, contains many gaps and some misassemblies. Although the structural annotations are generally of high quality, some gene models are missing, and many genes have misleading names. Finally, the reference genome has always been based on CC-503, a mating-type plus strain that was mutagenized to achieve a cell wall-less phenotype.
Question: We aimed to update the Chlamydomonas reference genome and annotations using long-read sequencing. We also sequenced a second strain, the mating-type minus CC-4532. Via comparison, we tested whether the mutagenesis of CC-503 resulted in any gene-disrupting structural mutations.
Findings: We produced highly contiguous genome assemblies for CC-503 and CC-4532 that were validated by linkage analyses. Most of the filled gaps were in genic regions, leading to substantial improvements in the gene models. Gene symbols were manually overhauled, providing a reliable nomenclature that can serve as a template for green algal genome projects. We discovered that the CC-503 genome harbors many large mutations and is unstable, making it an unsuitable reference. Genomic instability may stem from the deletion of the helicase RECQ3. We also fnd that the CC-4532 genome is experiencing an ongoing proliferation of transposable elements. We expect that all strains carry some large laboratory mutations.
Next steps: The CC-4532 genome is presented as the new Chlamydomonas reference. The assembly and annotation bring great advancements and will serve as a foundation for future updates. However, no single strain can provide a perfect reference, and we anticipate a truly representative Chlamydomonas pan-genome.
Introduction
The unicellular green alga Chlamydomonas (Chlamydomonas reinhardtii) is one of the primary model organisms in plant and cell biology. Chlamydomonas has been instrumental to discoveries in photosynthesis, chloroplast biology, and cilia structure and function, facilitated by its experimental tractability and amenability to classical genetics (Salomé and Merchant 2019). More recently, the species has been used as a powerful model for investigating the eukaryotic cell cycle (Cross and Umen 2015) and conserved mechanisms of sexual reproduction (Ning et al. 2013; Fédry et al. 2017), for discovery of optogenetic tools (Deisseroth and Hegemann 2017), and for in situ structural analyses by cryo-electron microscopy (Engel et al. 2015; Freeman Rosenzweig et al. 2017). Genome-wide mutant libraries form part of a growing suite of tools for exploiting high-throughput functional genomics approaches (Li et al. 2019; Fauser et al. 2022). As the most thoroughly studied green alga, Chlamydomonas also serves as an integral reference for the rapidly expanding fields of algal biology and biotechnology (Crozet et al. 2018; Blaby-Haas and Merchant 2019). The Chlamydomonas Genome Project was initiated two decades ago (Grossman et al. 2003; Merchant et al. 2007), and its continued development has kept the species at the forefront of plant and algal genomics (Blaby et al. 2014). Maintained at Phytozome (Goodstein et al. 2012), the genome assembly and structural annotations are a fundamental resource for contemporary Chlamydomonas research.
The Chlamydomonas genome is ∼111 Mb in length, GC-rich (∼64% genome-wide) and consists of 17 chromosomes. Preceded by two preliminary versions (Grossman et al. 2003), the initial draft genome (v3) was assembled from ∼13 × coverage of Sanger-sequenced reads (Merchant et al. 2007). Utilizing targeted sequencing of assembly gaps and molecular mapping data (Kathir et al. 2003; Rymarquis et al. 2005), the first chromosome-level assembly (v4) quickly followed in 2008 (Table 1). With the onset of next-generation sequencing, the v5 assembly was released in 2012 and applied both 454 and further Sanger sequencing to target all remaining gaps, successfully filling approximately half of those in v4 (Blaby et al. 2014). At 111.1 Mb, with 1,441 gaps (∼3.7% of the genome) and 37 unplaced scaffolds (∼2.0% of the genome), v5 has been the most long-standing release to date.
Table 1.
Comparison of assembly metrics between v6 assemblies, previous reference genome versions, and the CC-1690 assembly
| Assembly strain/version | CC-503 v4 | CC-503 v5 | CC-503 v6 | CC-4532 v6 | CC-1690 |
|---|---|---|---|---|---|
| Year | 2008 | 2012 | 2022 | 2022 | 2020 |
| Technology | Sanger | Sanger + 454 | PacBio + Illumina | PacBio + Illumina | Nanopore + Illumina |
| Total length (Mb) | 112.3 | 111.1 | 111.5 | 114.0 | 111.1 |
| Unplaced scaffolds/contigs | 71 | 37 | 42 | 40 | 1 |
| Unplaced length (Mb) | 9.68 | 2.20 | 1.45 | 1.72 | 1.65 |
| Total contigs | 2,739 | 1,495 | 145 | 120 | 21 |
| Contig N50 (Mb) | 0.09 | 0.22 | 2.92 | 2.65 | 3.58 |
| GC (%) | 64.1 | 64.1 | 64.1 | 64.1 | 64.1 |
| Gaps/Ns (%) | 7.54 | 3.65 | 1.66 | 0.81 | <0.01 |
| Transposable elements (%) | 9.84 | 10.61 | 10.80 | 12.42 | 11.24 |
| Microsatellites (%) | 1.32 | 1.43 | 1.72 | 1.76 | 1.65 |
| Satellite DNA (%) | 3.33 | 3.68 | 4.79 | 5.25 | 5.09 |
Unplaced sequence was assembled as scaffolds in v4 and v5, and contigs in all other assemblies. The single unplaced contig in the released version of the CC-1690 assembly was later assembled to the right arm of chromosome 15 (Chaux-Jukic et al. 2021).
Although the assembly metrics of v5 represented a considerable achievement, there remained substantial room for improvement relative to the highest quality Sanger-sequenced contemporaries. A decade earlier, near-complete assemblies featuring just tens of gaps in the most repetitive regions had been produced for Arabidopsis (Arabidopsis thaliana) (Arabidopsis Genome Initiative 2000) and rice (Oryza sativa) (Goff et al. 2002). Recently, long-read sequencing technologies have provided a platform to achieve similar contiguity, and even complete telomere-to-telomere assemblies, for far more complex genomes such as maize (Zea mays) (Jiao et al. 2017; Liu et al. 2020). Pacific Biosciences (PacBio) sequencing has been applied to close relatives of Chlamydomonas, yielding assemblies more contiguous than v5 for multiple unicellular and multicellular volvocine algae (Hamaji et al. 2018; Craig et al. 2021a; Yamamoto et al. 2021). Most recently, O'Donnell et al. (2020) used ultra-long Nanopore sequencing (Liu et al. 2019) to produce an unannotated assembly of Chlamydomonas strain CC-1690 (classically named 21gr) featuring only four gaps. It is worth noting that many of the gaps in the v5 assembly are expected to be in genic regions (Tulin and Cross 2016), and improvements to contiguity should therefore advance biological discovery via improved structural and functional annotation.
Perhaps of greater significance than contiguity, recent studies have highlighted inconsistencies between genetic mapping and the v5 assembly, potentially indicating misassemblies. Salomé and Merchant (2019) reported that the phytoene synthase gene (PSY1) is presently located on chromosome 2, although its corresponding white mutant lts1 was mapped to chromosome 11 (McCarthy et al. 2004). Likewise, Ozawa et al. (2020) characterized MTHI1, which encodes an octotricopeptide repeat protein and is mutated in the non-photosynthetic strain ac46, and observed that the gene is located on chromosome 17 despite having been mapped to chromosome 15 (Dutcher et al. 1991). Notably, both inconsistencies were introduced during the transition from v4 to v5, raising the possibility that past assembly improvements may have come at the expense of new errors.
There is also a potential issue with the classical reference strain, the cell wall-less CC-503 (cw92), which was chosen to meet the high DNA yield requirements of the early genome project. The cw phenotype was induced by mutagenesis of the mating-type plus (mt+) “wild-type” strain 137c+ (later deposited as CC-125) with the methylating agent N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) (Hyams and Davies 1972). MNNG primarily induces G:C to A:T transitions, although it can also induce double-strand breaks (DSBs) and chromosomal aberrations in high doses (Kaina 2004; Wyatt and Pittman 2006). For CC-503, the cw phenotype shows aberrant segregation in crosses, suggesting that there may be more than one causal mutation (Davies 1972; Hyams and Davies 1972). However, no causal mutations have been identified, and the potential genome-wide effects of mutagenesis in CC-503 have not been analyzed. More broadly, little is known about the extent of structural mutations, such as transposable element (TE) insertions and large duplications and deletions, during routine laboratory culture, which have the potential to introduce substantial genomic heterogeneity among strains.
Finally, a single strain does not represent the genomic diversity present among Chlamydomonas laboratory strains, which are interrelated but not isogenic. This fact is most obvious for the mating-type locus (MT) located on the left arm of chromosome 6. The plus (MT+) and minus (MT−) alleles, which, respectively, control the sexual differentiation of plus or minus gametes, feature a small number of mating-type-specific genes and several rearrangements that suppress crossover recombination (Ferris et al. 2010; De Hoff et al. 2013). While the CC-503 reference harbors the MT+ sequence, an MT− assembly is only available for the divergent field isolate CC-2290 (S1D2) (Ferris et al. 2010). Furthermore, all previous assembly versions have only included sequence and structural annotations for the nuclear genome, despite the relevance of organelle biology in the Chlamydomonas literature and the long availability of resources for the organelle genomes (Vahrenholz et al. 1993; Maul et al. 2002; Smith and Lee 2009; Gallaher et al. 2018).
Beyond the assembly itself, the structural annotations, which define the genomic coordinates of genes and the proteins they encode, are the foundation of omics analyses, most notably high-throughput transcriptomics and proteomics. The Chlamydomonas structural annotations have also been subject to several rounds of improvement (see Blaby et al. 2014; Blaby and Blaby-Haas 2017). Previous versions incorporated evidence from expressed sequence tags (ESTs) and assembled cDNAs, with protein homology support from Volvox carteri genes (Prochnik et al. 2010). The annotations performed for v5 incorporated over one billion RNA-seq reads, resulting in several major changes to gene models (Blaby and Blaby-Haas 2017). The most recent v5 annotation (v5.6) features 17,741 protein-coding genes with 1,785 alternative transcripts. Recent advances in sequencing again provide substantial opportunities to update structural annotations. For example, Gallaher et al. (2021) used PacBio Iso-Seq (long-read sequencing of cDNA) to discover more than 100 polycistronic loci in Chlamydomonas (i.e. genes producing a single transcript that encodes more than one protein), although these data have not yet been used to systematically improve structural annotations.
Here we present the first major update to the Chlamydomonas Genome Project in nearly a decade. We present PacBio-based assemblies for the classical mt+ reference strain CC-503 and for the mt− laboratory strain CC-4532, bringing extensive improvements to both assembly and annotation quality. Using comparative analyses, we specifically tested whether the mutagenesis of CC-503 has resulted in genomic aberrations and explored the wider influence of TE insertions in the genomes of Chlamydomonas laboratory strains. We found that the CC-503 genome carries many large structural mutations predicted to affect ∼100 genes, while the genomes of all laboratory strains are likely to harbor a non-negligible and potentially highly variable number of TE insertions. We therefore present the CC-4532 assembly as the primary v6 reference genome and discuss the implications of mutation in the laboratory. These updates mark the start of an exciting new era for Chlamydomonas genomics, with developing opportunities to produce high-quality assemblies and annotations for several strains and divergent isolates of the species.
Results and discussion
CC-4532 version 6: a long-read Chlamydomonas reference assembly
As the first step in updating the reference genome, we produced de novo contig-level assemblies from high-coverage (>120×) PacBio Sequel datasets for the mt+ CC-503 and mt− CC-4532 strains. In line with the reported inconsistencies with mapping data, we detected multiple contradictions between the prior v5 assembly and the newly assembled contigs of both CC-503 and CC-4532. We thus reassembled all well-supported contigs to chromosomes without reference to previous versions, which we primarily achieved by mapping the contigs to the near-complete Nanopore-based CC-1690 assembly (O'Donnell et al. 2020). This approach not only allowed contigs to be placed on chromosomes in a manner consistent across all three assemblies, but also enabled the estimation of gap lengths between remaining contig breaks in the PacBio assemblies relative to CC-1690. We refer to these assemblies as CC-503 v6 and CC-4532 v6, respectively, to highlight that they are both the product of version 6 of the genome project. We validated all structural changes by reanalyzing previously published linkage data (Kathir et al. 2003; Liu et al. 2018). In addition, recent knowledge of centromeric (Lin et al. 2018; Craig et al. 2021a) and subtelomeric (Chaux-Jukic et al. 2021) repeats provided extrinsic validation. While the CC-4532 v6 and CC-1690 assemblies are entirely consistent relative to each other and all supporting evidence, we identified remaining inconsistencies in the CC-503 v6 assembly, indicative of genomic rearrangements unique to this strain. We describe these structural mutations further below, while the following text focuses on CC-4532 v6 as the primary reference assembly.
CC-4532 v6 is considerably more contiguous than previous versions (Table 1). The number of contigs decreased by an order of magnitude relative to v5, from 1,495 to 120, with a corresponding increase in the contig-level N50 from 0.22 to 2.65 Mb (i.e. contigs ≥2.65 Mb represent >50% of the assembly length). Although unplaced sequence only fell from 2.20 to 1.72 Mb, the 40 highly repetitive unplaced contigs in CC-4532 v6 mostly represent newly assembled sequences that are unrelated to the 37 unplaced scaffolds in v5, all but three of which are now at least partially placed on chromosomes. With a genome size of 114.0 Mb, CC-4532 v6 is ∼3 Mb larger than v5 and the CC-1690 assembly. This discrepancy can be explained in part by redundancy between the unplaced contigs and the gaps to which they presumably correspond, since gap lengths (represented by unknown bases, i.e. Ns) were estimated relative to CC-1690. However, we attribute most of the biological increase in genome size to TE activity in the laboratory. In the following sections, we present a thorough assessment of the assembly and annotation improvements.
A note on CC-4532 and laboratory strain haplotypes
CC-4532 has been widely used in transcriptomics analyses and was initially selected for genome sequencing to obtain an assembly of the MT− allele. While its promotion to the new reference over other widely used strains may raise concerns, we note that there is no optimum or authoritative reference strain for Chlamydomonas. Laboratory strains are thought to be derived from the haploid progeny of a diploid zygospore isolated by G. M. Smith in 1945. Their genomes are thus comprised of two haplotypes, although their frequencies are unbalanced; one haplotype covers only a maximum of 25% of the genome, but generally much less (Gallaher et al. 2015). The two haplotypes differ at ∼2% of sites and many between-haplotype variants are expected to be functionally important. Gallaher et al. (2015) arbitrarily defined haplotype 1 as being that of the classical reference CC-503, with haplotype 2 referring to any region featuring the alternative haplotype in other strains. Laboratories use a variety of strains, including the oldest “wild types” (e.g. 137c+/CC-125 and 21gr/CC-1690) and those derived from subsequent crosses. Therefore, most strains in use differ genetically from the reference genome in multiple genomic regions, introducing variants in hundreds of genes.
CC-4532 is a putative subclone of CC-621 (NO−) and is partly descended from 137c+ (the progenitor of CC-503), although the exact crosses that produced the strain are unknown. It carries haplotype 1 at more than 95% of the genome and will thus provide a similar user experience as a reference strain. We later discuss remaining issues with a CC-4532 reference and solutions to producing a fully representative reference assembly for Chlamydomonas laboratory strains.
The version 6 assembly corrects misassemblies of version 5
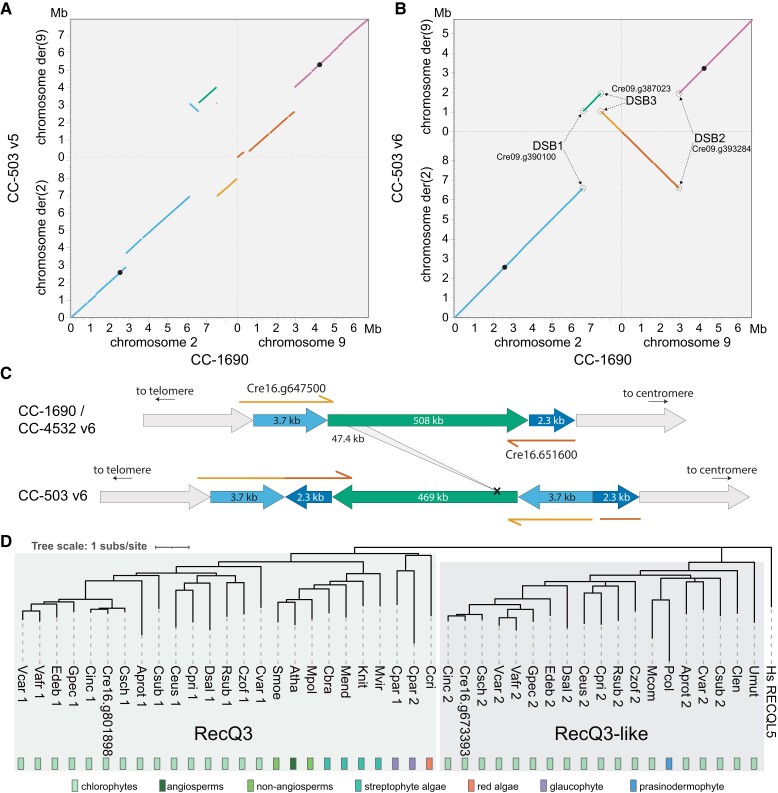
The CC-4532 v6 assembly has major structural differences relative to v5, affecting the ordering and orientation of sequence both within and between chromosomes. Only six chromosomes (1, 4, 6, 7, 13, and 14) remained consistent with respect to the ordering of scaffolds in v5. The extent of the changes to the remaining 11 chromosomes ranged from minor intra-chromosomal reordering of short contigs to major inter-chromosomal rearrangements affecting megabases of sequence. An overview of the between-chromosome changes is presented in Figure 1A.
Figure 1.
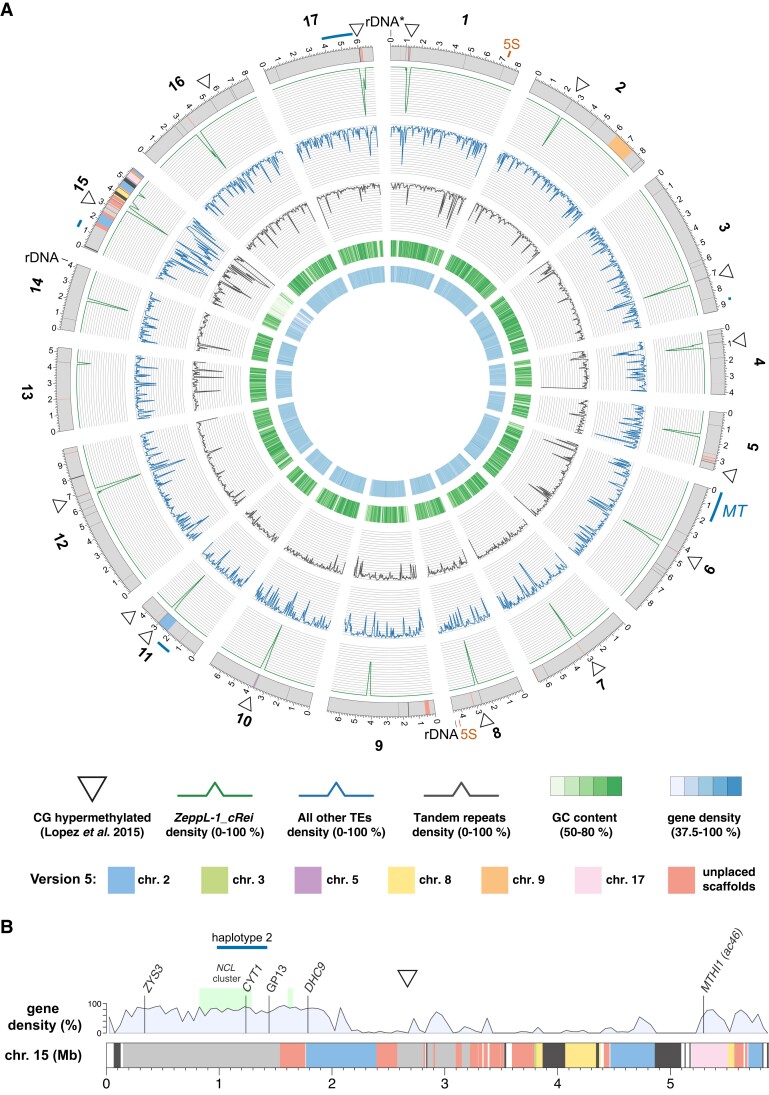
The CC-4532 version 6 assembly. A, Circos plot (Krzywinski et al. 2009) representation of the CC-4532 v6 genome. Gray outer bands represent chromosomes, with colors highlighting genomic regions that were assembled on other chromosomes or unplaced scaffolds in v5. Dark gray regions represent gaps between contigs, with any gaps <10 kb increased to 10 kb to aid visualization. Outer lines in dark blue represent haplotype 2 regions, including the mating-type locus (MT) and flanking regions on chromosome 6. All metrics were calculated for 50-kb windows. Tandem repeats combine microsatellite and satellite annotations. CG-hypermethylated regions were taken from Lopez et al. (2015) and mapped from v5 to v6 coordinates, with some neighboring regions merged to a single marker in the plot (see Supplemental Figure 2 for all regions). B, Linear representation of chromosome 15. Colors are as in (A), with dark gray representing assembly gaps. Light gray regions were present on chromosome 15 in v5, while white regions are newly assembled in v6. See Supplemental Dataset 2 for coordinates linking v5 and v6 assembly regions. Molecular markers are from Kathir et al. (2003) and the light green boxes represent the NCL gene clusters described by Boulouis et al. (2015). CYT1 was previously recorded as CYTC1. GP13 is a marker derived from cloned genomic DNA.
Many of the changes occurred in proximity to the most repetitive genomic regions, particularly the putative centromeres and the subtelomeres, as well as regions corresponding to unplaced scaffolds in v5. Although approximate centromeric locations were predicted from molecular mapping (Preuss and Mets 2002), genomic coordinates and sequence characteristics have only recently been reported. Lin et al. (2018) identified 200–800 kb regions tightly linked to the centromeres that featured multiple open reading frames (ORFs) encoding proteins with reverse transcriptase domains. Craig et al. (2021a) linked these ORFs to an L1 LINE retrotransposon homologous to Zepp, the centromeric component of the trebouxiophyte alga Coccomyxa subellipsoidea (Blanc et al. 2012). Termed Zepp-like (ZeppL) elements in Chlamydomonas, this TE forms highly localized clusters at the putative centromeres, although in v5, chromosomes 2, 3, 5, and 8 featured two clusters, and chromosomes 11 and 15 lacked clusters (Lin et al. 2018; Craig et al. 2021a). Chlamydomonas subtelomeres were recently shown to feature large satellite arrays termed Sultans, with other complex repeats present at specific chromosome termini (Chaux-Jukic et al. 2021). Subtelomeres are capped by the telomeric repeat (TTTTAGGG)n (Petracek et al. 1990). Due to their complexity, subtelomeres were previously poorly assembled, and only half of chromosome termini featured a scaffold terminating in telomeric repeats in v5.
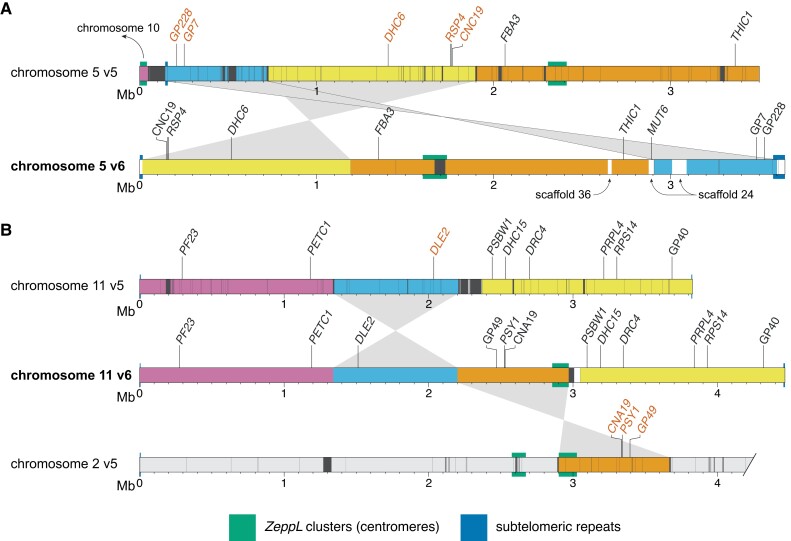
Comparisons of chromosomes 5 (Figure 2A) and 11 (Figure 2B) between v5 and v6 illustrate the types of misassemblies that affected these regions. In v5, the left arm of chromosome 5 terminated in a 47-kb contig featuring a ZeppL cluster (purple, Figure 2A), which in v6 is assembled within the putative centromere of chromosome 10 (Supplemental Figure 1D). The remaining regions of chromosome 5, consisting of three blocks of ∼0.7, 1.2, and 1.7 Mb (light blue, yellow, and orange, respectively; Figure 2A), are now rearranged and reorientated. The misassembly of the light blue and yellow regions featured a large gap corresponding to part of scaffold 24 (containing MUT6), while the misassembly of the yellow and orange regions featured subtelomeric repeats that are now correctly placed at the left arm terminus in v6. Thus, the reassembled chromosome 5 features a single internal centromere, subtelomeric repeats at both termini, and is congruent with the molecular map (Kathir et al. 2003). On chromosome 11, the movement of an ∼750-kb region (orange) from chromosome 2 simultaneously resolved the absence of a putative centromere on chromosome 11 and the presence of two ZeppL clusters on chromosome 2 (Figure 2B). This region includes PSY1, which was mapped genetically to chromosome 11 (McCarthy et al. 2004; Salomé and Merchant 2019). Independently, an ∼860-kb region (light blue) was inverted, consistent with the tight linkage of PETC1 and DLE2 (full gene names provided in Supplemental Dataset 23; Kathir et al. 2003). Misassemblies affecting other chromosomes are shown in Supplemental Figure 1.
Figure 2.
Version 5 misassemblies and their resolution in version 6. Chromosome segments are colored to show the reordering and reorientation of specific regions, and dark gray regions represent assembly gaps. Markers inconsistent with the molecular map of Kathir et al. (2003) are shown in vermillion text. Gene symbols (in italics) were updated where applicable. “GP” markers were derived from genomic DNA, while “CAN” and “CNC” markers were derived from cDNA. Note that the plot was made using CC-503 v6 to simplify mapping between versions. CC-503 v6 and CC-4532 v6 are entirely syntenic for chromosomes 5 and 11. A, Reassembly of chromosome 5. The purple region was reassigned to chromosome 10. White regions on the v6 chromosome correspond to sequence not assembled on the v5 chromosome (e.g. the region containing MUT6 corresponds to part of scaffold 24 in v5). In the original map RSP4 corresponded to the pf1 marker (and the neighboring RSP6 to pf26; not shown; Dutcher 2014). Updated gene symbols: FBA3 was ALD, THIC1 was THI8. B, Reassembly of chromosome 11; only the first 4.2 Mb of chromosome 2 is shown. Genes that originally corresponded to genetic markers are: PSY1, lts1; PF23, pf23 (Yamamoto et al. 2017); DRC4, pf2 (Dutcher 2014); PRPL4, ery1; RPS14, cry1. Updated gene symbols: PETC1 was PETC, DLE2 was VFL2, DHC15 was ODA2, PSBW1 was PSBW.
By far the most substantial changes affected chromosome 15, which approximately tripled in length from 1.92 Mb in v5 (the shortest chromosome) to 5.87 Mb in CC-4532 v6, acquiring sequence previously assigned to chromosomes 2, 3, 8, and 17, as well as 15 unplaced scaffolds (Figure 1B). The sequence reassembled from chromosomes 2 (∼1.2 Mb) and 17 (∼0.3 Mb) each featured a marker gene previously mapped to chromosome 15: DHC9 (Porter et al. 1996; Kathir et al. 2003) and the aforementioned MTHI1 (Dutcher et al. 1991; Ozawa et al. 2020), respectively. Some of the sequence reassembled from chromosome 8 (∼0.4 Mb) and unplaced scaffolds (∼1.1 Mb total) featured ZeppL elements, explaining the absence of centromeric repeats on chromosome 15 in v5. We attribute the degree of past misassembly to the unique sequence characteristics of chromosome 15. Its repeat content (47.2%) is substantially higher, and its gene density lower (36.7%), than the remaining 16 chromosomes (mean 17.7% and 79.0%, respectively; Supplemental Dataset 1). Furthermore, this pattern is not uniform: the gene density of the chromosome arms (67.1%, ∼2.1 Mb left and ∼0.6 Mb right) approaches that of other chromosomes, while the internal region is massively repetitive (66.7%) and gene-poor (10.9%). As a result, chromosome 15 remains the most fragmented in CC-4532 v6, featuring 10 gaps spanning 9.2% of the chromosome length, relative to a mean of three gaps and 0.4% for the remaining chromosomes. We expect that many of the unplaced contigs belong to chromosome 15, although their extreme repeat content (69.8%) hinders efforts to place them without longer reads.
The unusual features of chromosome 15 raise questions about its evolutionary origins, gene content, and chromosomal environment. Except for MTHI1, all marker genes (ZYS3, CYT1, and DHC9) are located within the relatively gene-rich left arm of the chromosome. This region is also notable for containing almost all the NCL (NUCLEAR CONTROL OF CHLOROPLAST GENE EXPRESSION-LIKE) genes, encoding a family of RNA-binding proteins that is experiencing ongoing diversification (Boulouis et al. 2015). All but one of the 49 NCL genes are on chromosome 15, with 43 present in a cluster spanning ∼460 kb, and three forming a shorter upstream cluster that was assembled on scaffold 19 in v5 (Figure 1B). The mutation responsible for the yellow-in-the-dark mutant y1 was also mapped to the left arm of chromosome 15 and is linked to DHC9 (Porter et al. 1996). The unknown Y1 gene might thus have been assigned to either chromosome 2 or an unplaced scaffold in v5. The remainder of chromosome 15 contains only 145 genes, 80 of which are in the highly repetitive internal region. Although most of these genes are not functionally annotated, we expect at least some to be essential (e.g. the plastid 50S ribosomal protein gene PRPL3). It would be interesting to determine if much of chromosome 15 is heterochromatic, and if so, whether genes are expressed from heterochromatic environments (as is the case for many genes on the repeat-rich dot chromosome in Drosophila melanogaster; Riddle and Elgin 2018). Similarly, it would be interesting to explore whether the high repeat content results in an atypical recombination landscape on chromosome 15, and whether similarly high repeat contents are found on homologous chromosomal regions in closely related species.
Assembly improvements reveal novel genic sequence and hypermethylated centromeres
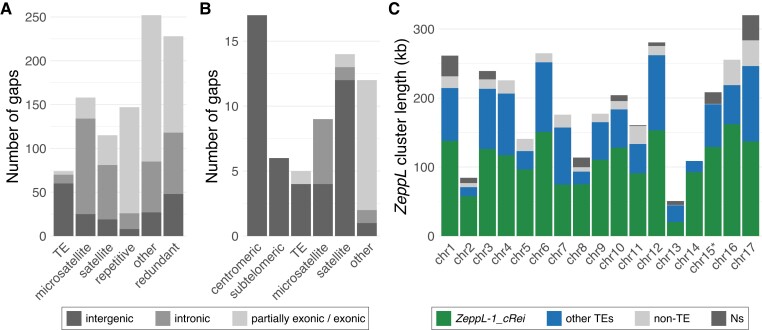
To assess the functional effect of assembly improvements in CC-4532 v6, we next analyzed the filled and remaining assembly gaps relative to the gene and repeat landscape of the Chlamydomonas genome. We annotated almost 1,000 filled v5 gaps based on their sequence context in CC-4532 v6, either as “TE” (∼8% of the gaps), “microsatellite” (16%) or “satellite” (12%) if the novel sequence featured >50% of the corresponding repeat class, “repetitive” (15%) if the sequence otherwise had >25% repeat content, and “other” (26%) for less repetitive sequences (Figure 3A). We further classified gaps relative to genic features annotated de novo in CC-4532 v6 (described below), as either entirely intergenic (∼19% of the gaps), entirely intronic (34%) or at least partially exonic (47%) (i.e. the filled sequence featured some novel exonic sequence). Tandem repeats were associated with nearly four times as many gaps as TEs, despite covering almost half as much of the genome (Table 1). Furthermore, while 81% of TE-associated gaps were intergenic, 84% of gaps associated with tandem repeats were within genes (Figure 3A). These results are consistent with the underrepresentation of TEs (Philippsen et al. 2016) and overrepresentation of tandem repeats (Zhao et al. 2014) in introns, and are consistent with our own annotation of repeats by site class (Supplemental Dataset 3). The high proportion of genic gaps supports the study of Tulin and Cross (2016), which identified more than 100 “hidden” exons by comparing a de novo-assembled transcriptome to the v5 assembly. Overall, our results suggest that prior targeted gap filling was largely successful in assembling intergenic TEs, while the higher density of intronic tandem repeats precluded the more complete assembly of genic regions by Sanger and short-read technologies. Finally, 23% of gaps were not filled in v6, but instead lost redundant sequence from one or both flanks (class “redundant”, Figure 3A). Approximately half of these cases resulted in the removal of redundant exonic sequence, providing further potential to improve structural annotation.
Figure 3.
Filled gaps and the remaining assembly challenges in CC-4532 version 6. A, Repeat classification of v5 gaps filled in CC-4532 v6. Bars are split into entirely intergenic gaps, entirely intronic gaps and gaps with at least partial exonic overlap. See main text for details of gap definitions by repeat class. B, Classification of the remaining gaps in CC-4532 v6, shading follows (A). “Other” gaps were associated with other repeat types (e.g. large duplications) or were not clearly associated with repeats. C, Summary of the length of putative centromeric ZeppL clusters. Colors represent the number of bases annotated as ZeppL-1_cRei (the only ZeppL family in Chlamydomonas), any other TE, non-TE sequence, and assembly gaps (Ns). Note that chromosome 15 contains two short ZeppL clusters downstream of the main cluster (Supplemental Dataset 5), which are not shown.
The CC-4532 v6 chromosomes still contain 63 gaps that generally coincide with the most repetitive genomic regions. Approximately one-third fall within the putative centromeres and subtelomeres, with another third accounted for by tandem repeats, especially large satellites (Figure 3B). Despite the complexity of the repeats present at subtelomeres, 26 of the 34 chromosome termini are capped with telomeric repeats. Among the incomplete termini are the two ribosomal DNA (rDNA) arrays on the right arms of chromosome 8 and 14 (Figure 1A; note that the chromosome 1 rDNA array is truncated and likely non-functional in laboratory strains, but potentially not so in field isolates (Chaux-Jukic et al. 2021)). One gap corresponds to the 5S rDNA array on chromosome 1, while the second 5S rDNA array on chromosome 8 is putatively complete (Figure 1A). Although approximately half of the microsatellite-associated gaps are intronic, almost all the remaining repeat-associated gaps are intergenic. Unfortunately, 12 gaps contain exonic sequence, potentially affecting 18 genes based on comparison to de novo annotation of CC-503 v6 (Supplemental Dataset 4). Most of these gaps are not obviously repetitive (“other” class, Figure 3B) and will be prime targets for future manual finishing.
Following the misassembly corrections, each v6 chromosome features a single localized cluster of ZeppL elements (Figure 1A), except for chromosome 15, where we identified two minor clusters (∼30 and 9 kb) downstream of the major cluster. Although most putative centromeres feature at least one gap, they are not particularly long; by comparison to the CC-1690 assembly, we estimate that more than 95% of putatively centromeric sequence is assembled in CC-4532 v6 (Figure 3C; Supplemental Dataset 5). Based on the span of ZeppL elements, the putative centromeres range from 51 to 320 kb, with a mean of 192 kb. Approximately 60% of the sequence is composed of the ZeppL element itself, with most of the remaining sequence contributed by other TEs (Figures 1, A and 3C; see also Supplemental Figure 2 for CC-1690), especially Dualen LINEs (Craig et al. 2021a). Satellite DNA does not appear to be a major component of the clusters (except chromosome 16; Supplemental Dataset 5), although we observed satellites immediately flanking the clusters on some chromosomes (e.g. 4 and 5; Supplemental Figure 2). The structure of these regions warrants further study, as does the localization of centromeric histone H3, which may be encoded by two paralogous genes in Chlamydomonas (Cui et al. 2015).
Finally, we revisited the genomic landscape of CG methylation (C5-methylcytosine, 5mC) in Chlamydomonas. Lopez et al. (2015) identified 23 hypermethylated loci relative to a genomic background of very low methylation (<1%). We determined that 19 of the hypermethylated regions coincide with the putative centromeres on 11 chromosomes, with a further two localizing to subtelomeres (Figure 1A; Supplemental Figure 2). Chaux-Jukic et al. (2021) called CG methylation directly from Nanopore reads, which facilitates mapping to highly repetitive regions, revealing ubiquitous hypermethylation of subtelomeres. Using the same Nanopore dataset (Liu et al. 2019), we extended this analysis to the entire CC-1690 assembly and established that all putative centromeres are hypermethylated (Supplemental Figure 2). Alongside subtelomeres, a few other highly repetitive regions were hypermethylated (e.g. a ∼200-kb region on the left arm of chromosome 12), while we observed many more localized methylation peaks of smaller magnitude. Presumably, these regions were previously overlooked due to the limitations of mapping short-read bisulfite sequencing data to repeats and the incompleteness of the most repetitive regions in v5. Strenkert et al. (2022) reported an atypical chromatin architecture for the previously identified hypermethylated regions, suggesting that the hypermethylated centromeres, subtelomeres, and potentially some other repeat-rich islands, may constitute heterochromatin in Chlamydomonas.
Linkage data validates the CC-1690 and version 6 assemblies
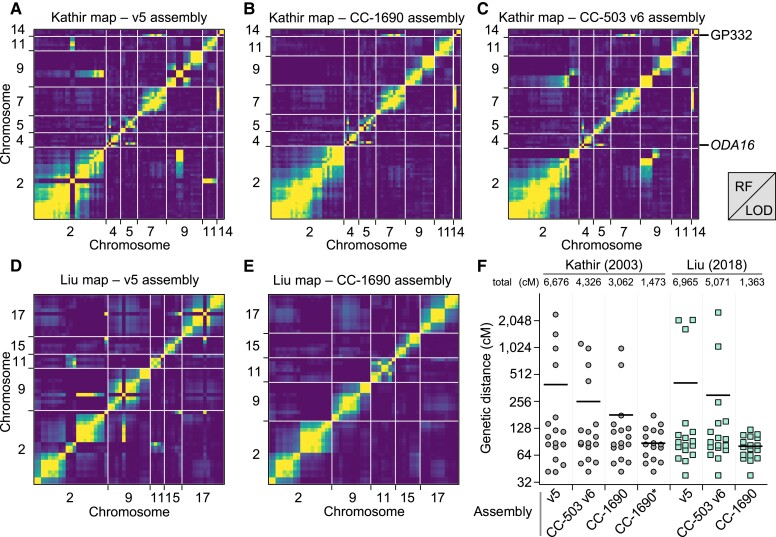
To systematically validate the improvements between v5 and v6, we turned to two independent genetic recombination datasets. We primarily compared v5 to the CC-1690 assembly, since CC-1690 was used as a reference to scaffold the v6 assemblies, and the CC-1690 and CC-4532 v6 assemblies are entirely syntenic. We repeated these analyses using CC-503 v6 following the discovery of outstanding inconsistencies in this assembly. We first identified the v5 chromosomal coordinates of 239 molecular markers described by Kathir et al. (2003) (Supplemental Dataset 6). We then ordered the genotype data used to generate the genetic map based on the v5 coordinates before estimating a new genetic map with the R/QTL package (Broman et al. 2003). To assess the concordance between assigned and true genomic positions, we visualized recombination frequencies between marker pairs: two unlinked markers should exhibit random segregation and appear as dark blue squares (low log of the odds score), whereas linked markers should appear in yellow. While most markers agreed with their v5 chromosomal locations, we identified 10 misplaced markers, eight of which mapped to chromosomes 2 or 9 (Figure 4A; Supplemental Figure 3). Markers CNA19 and GP49 were located on chromosome 2 in v5, but showed strong linkage with chromosome 11. Satisfyingly, both markers relocated to chromosome 11 in CC-1690 (Figure 4B) and subsequently in both v6 assemblies (Figure 2B). We also resolved the genomic location of most other mismapped markers when using CC-1690 coordinates. Conversely, inconsistencies remained between chromosomes 2 and 9 when using the CC-503 v6 coordinates (Figure 4C), which as detailed below stems from a putative chromosomal rearrangement unique to CC-503. The two further misplaced markers remained apparently wrongly assigned when using CC-1690 or CC-503 v6 coordinates: GP332 and ODA16, which were assigned to the top of chromosome 14 and 4, respectively, in both assemblies. The genetic mapping data indicated strong linkage between GP332 and chromosome 7 markers (CNC43, CHL27A and GLTR1), and between ODA16 and chromosome 5 markers (DHC6, CNC19 and RSP6; see Figure 2A). In both cases, the chance of the regions corresponding to these sequences being misassembled in the exact same location on independent contigs in CC-1690, CC-4532 v6, and CC-503 v6 is negligible, and their previous mapping locations or associated sequences are presumably incorrect.
Figure 4.
Validation of the Chlamydomonas genome reassemblies by recombination maps. A, Partial plot of recombination frequencies between molecular markers from Kathir et al. (2003). Strong linkage is indicated by a yellow color; absence of linkage is shown as dark blue. B and C, Partial recombination frequency plots between the same molecular markers with updated genomic coordinates according to the CC-1690 (B) or CC-503 v6 (C) assembly. Note that the markers GP332 and ODA16 are consistently mismapped. D and E, Partial recombination frequency plots between informative single-nucleotide polymorphisms (SNPs) extracted from Liu et al. (2018), using the genomic coordinates from the v5 (D) or CC-1690 (E) assemblies. RF, recombination fraction; LOD, logarithm of the odds. F, Gradual improvement of the estimation of genetic map length, from v5, to CC-503 v6, to CC-1690. Chromosome lengths are plotted in centimorgans (cM) for each increment of the genetic maps. CC-1690* denotes the use of CC-1690 genomic coordinates with the removal or the GP332 and ODA16 molecular markers from the analysis. Total map length, in cM, is listed above each dot plot. Horizontal bar, mean.
We followed the same steps to generate a genetic map from whole-genome resequencing data of tetrads derived from crosses between two Quebec field isolates (Liu et al. 2018). We reduced the data to keep only single-nucleotide polymorphisms (SNPs) that were informative of haplotype transitions (164 SNPs). Again, the deduced recombination map largely agreed with v5 chromosomal positions, except for 14 SNPs, eight of which had been wrongly assigned to chromosome 2 (Figure 4D). The CC-1690 genomic coordinates corrected all mismapping (Figure 4E) and greatly reduced the overall length of the genetic map, from over 6,000 cM using v5 coordinates to ∼1,400 cM with CC-1690 coordinates (Figure 4F). As with the molecular markers, any discordance between CC-1690 and CC-503 v6 mapped to the putative rearrangement affecting chromosomes 2 and 9. We therefore conclude that CC-1690, and thus the v6 assemblies, receives strong recombination support from two independent mapping datasets, which were derived from a laboratory strain (CC-1690) and diverse field isolates (CC-1952 in one case, CC-2935 and CC-2936 in the other). It is now expected that the order and orientation of chromosomal sequence in the CC-1690, CC-4532 v6 and CC-503 v6 assemblies represents the biological reality for these strains.
The CC-503 genome is unstable and harbors major structural mutations
Following the discovery of remaining inconsistencies between CC-503 v6 and the CC-4532 v6 and CC-1690 assemblies, we set out to characterize structural mutations in the CC-503 genome. This endeavor was possible since the three assemblies feature the same ancestral haplotype over most of their genomes, meaning that any variant segregating uniquely in CC-503 could be attributed to mutation arising in the laboratory, potentially as a result of historic mutagenesis.
The most conspicuous mutation affected chromosomes 2 and 9. Indeed, these chromosomes were misassembled in all past versions, and changes that occurred between v4 and v5 were noted previously (Lin et al. 2013). In v5, the aberration was misassembled as a complex translocation that would have involved at least five DSBs (Figure 5A). This mistake presumably occurred due to conflicting evidence between contig assembly, based on mutant-state CC-503 sequencing reads, and longer range scaffolding based on wild-type linkage data from other laboratory strains and field isolates. Via manual inspection of the CC-503 v6 contigs, we inferred that chromosomes 2 and 9 have instead experienced a putative reciprocal translocation, with an inversion affecting part of the fragment translocated from chromosome 2 to 9 (Figure 5B). This model posits three DSBs, one on chromosome 9 (DSB2 between purple and vermilion, Figure 5B) and two on chromosome 2 (DSB1 between blue and green, and DSB3 green and orange). The 0.9-Mb inversion shares DSB1 with the translocation event, suggesting that all three DSBs occurred, and were subsequently misrepaired, simultaneously. Notably, all DSBs and their repair events were associated with insertions and deletions (InDels), ranging from a few bp to 1,950 bp, and all were predicted to disrupt coding sequence (CDS) relative to the CC-4532 de novo structural annotations (Supplemental Figure 4). For example, the deletion at DSB2 entirely removed the second exon of a gene (Cre09.g390100) encoding a 318-amino acid (aa) protein with an S-adenosylmethioine-dependent methyltransferase domain, with the remaining (and presumably pseudogenized) exons now split between the derived chromosomes 2 and 9 in CC-503 v6 (Supplemental Figure 5). Illumina resequencing data from CC-125 (the progenitor of CC-503) mapped across the deletions at each DSB (Supplemental Figure 6), confirming that the mutation is unique to CC-503.
Figure 5.
Structural mutations in the CC-503 version 6 genome. A, B, Dotplot representation of chromosomes 2 and 9 between CC-503 v5 and CC-1690 (A), and CC-503 v6 and CC-1690 (B). Colors link fragments between panels (A) and (B). Black dots represent putative centromeres. CC-503 chromosomes are named as derivatives (der) based on their centromeres. Genes disrupted by DSBs are labeled. C, Schematic diagram of the duplication–inversion–duplication (dupINVdup) and deletion double mutation. The duplicated flanks (light and dark blue) are shown 50 × the scale of the main inverted fragment (green). Disrupted and partly duplicated genes are labeled. The left flank is predicted to have formed a gene fusion in CC-503 v6.1, although this is entirely based on ab initio prediction. The 47.4-kb internal deletion is represented by the gray ribbon. D, Protein-based phylogeny of the RecQ3 and RecQ3-like subfamilies of RecQ helicases in Archaeplastida. Branches with bootstrap values <50% were removed. Full species names and protein IDs can be found in Supplemental Dataset 9.
Remarkably, we identified 71 additional structural mutations (i.e. >50 bp) present in CC-503 v6 and absent in CC-4532 v6 and CC-1690, putatively affecting 103 genes (Supplemental Dataset 7). This number excludes TEs, which are presented separately below. In full, we called 63 deletions (cumulatively 302.1 kb and including events >10 kb), six duplications, one insertion and one inversion. Many of the mutations were complex; for example, the duplications were often associated with InDels and inversions. One of the most striking mutations was a ∼508-kb inversion between 0.81 and 1.32 Mb on chromosome 16 (Figure 5C). Inspection of the two DSBs and their subsequent repair revealed that this event is an unusual dupINVdup (duplication–inversion–duplication) mutation (Brand et al. 2015), in which both flanks (3.7 kb to the left and 2.3 kb to the right) of the unique inverted sequence are duplicated and themselves inverted. Genic sequence was disrupted and partially duplicated at both flanks (Figure 5C). Surprisingly, the inverted region itself harbored a 47-kb deletion that partially or fully deleted 10 genes (Supplemental Dataset 7).
Although it is tempting to directly attribute the exceptional number of structural mutations in CC-503 to its past mutagenesis with MNNG (Hyams and Davies 1972), we unexpectedly observed that 46 of the 72 structural mutations were not present in past assembly versions (Supplemental Dataset 7), including the chromosome 16 dupINVdup/deletion. Previous assemblies were primarily based on Sanger sequencing from the initial genome project, while the v6 PacBio sequencing was performed on a CC-503 culture obtained from the Chlamydomonas Resource Center by Gallaher et al. (2015). Given that many of the mutations are shared between past versions and CC-503 v6, some of which are very distinctive (e.g. the reciprocal translocation described above), the more recently acquired culture undoubtedly shares a clonal common ancestor with that used in the original genome project. It therefore appears that approximately two thirds of the structural mutations have occurred over the past two decades, and that the CC-503 genome may be unstable. Two main lines of evidence support this hypothesis. First, in a reciprocal analysis we discovered only 10 structural mutations unique to CC-4532 v6 (Supplemental Dataset 8, see below) and no large rearrangements in CC-1690, suggesting an elevated rate of chromosomal aberrations in CC-503. Second, many of the mutations were complex and featured large InDels or duplications at their repair points, potentially indicating a deficiency in DSB repair. High rates of deletions, duplications, and rearrangements have recently been documented in the Chlamydomonas field isolate CC-2931, however this was partly attributed to TE activity and similar patterns of mutational complexity at repair points were not observed (López-Cortegano et al. 2022).
We attempted to find candidate loci for genomic instability by examining each gene affected by a mutation that was common to CC-503 v6 and all past assembly versions, under the assumption that these mutations could have originated during mutagenesis, or at least prior to the initial genome project. We identified a RecQ helicase gene (Cre16.g801898) as a possible candidate, which was fully deleted in CC-503 as part of a 48-kb deletion on chromosome 16 that partially or fully deleted at least six genes (note that this is unrelated to the chromosome 16 deletion described above, see Supplemental Dataset 7). RecQ helicases have been referred to as “guardians of the genome” and play key roles in genome maintenance and all DSB repair pathways in humans (Croteau et al. 2014; Lu and Davis 2021). Many eukaryotes possess multiple RecQ helicase genes that belong to ancient gene families, with five genes in human and seven in Arabidopsis (Dorn and Puchta 2019). We performed a phylogenetic analysis including the protein encoded by the deleted gene Cre16.g801898 and homologous proteins in algae and plants, which demonstrated that Cre16.g801898 encodes a putative ortholog of the plant RecQ3 subfamily (Figure 5D), which is homologous to human RECQ-LIKE HELICASE 5 (RECQL5; Wiedemann et al. 2018). Furthermore, the RecQ3 subfamily is present across Archaeplastida (the green lineage plus red algae and glaucophytes). Interestingly, our analysis also revealed a green algal-specific subfamily, RecQ3-like, which formed a clade with the canonical RecQ3 subfamily (Figure 5D). All analyzed species from the Chlorophyceae and Trebouxiophyceae had both RecQ3 and RecQ3-like subfamily genes, indicating strong conservation. However, the RecQ3 subfamily appeared to be absent in prasinophytes (e.g. Micromonas spp.) and ulvophytes (Caulerpa lentillifera and Ulva mutabilis). Such a deep evolutionary division between the RecQ3 and RecQ3-like subfamilies is roughly analogous to the plant-specific RecQsim subfamily, which forms a clade with the eukaryotic RecQ6/WRN group (Wiedemann et al. 2018).
The specific functions of RecQ helicases have not been studied in green algae and it is difficult to draw parallels with other species, since the evolution of RecQ helicases is dynamic in many lineages. Certain plants have lost specific subfamilies and duplicated others; for instance, the moss Physcomitrium patens lacks RecQ1 or RecQ3 genes but has two RecQsim paralogs, and Arabidopsis lacks a RecQ6 gene but has two RecQ4 paralogs. All subfamilies appear to be represented in Chlamydomonas, although only a mutant of the RecQ5 subfamily gene (Cre15.g634701; homologous to human RECQL4), which is unable to undergo cell division, has been described (Tulin and Cross 2014). These findings suggest that neo- and subfunctionalization may occur in RecQ helicase evolution and that orthologous proteins may not have identical functions in different species. In humans, RECQL5 downregulation results in genomic instability and chromosomal rearrangements, and recql5 mutants are associated with tumorigenesis (Lu and Davis 2021). However, Arabidopsis recq3 mutants were viable and had no growth abnormalities, although this observation does not rule out longer term genomic instability (Röhrig et al. 2018). It remains to be seen if the deletion of RECQ3 in Chlamydomonas can explain the genomic instability of CC-503, and it will likely never be known if this specific deletion was caused by mutagenesis or arose later in culture.
Finally, we also identified a candidate mutation for the cell wall-less phenotype of CC-503. A 6.0-kb deletion on chromosome 1 almost entirely removed a putative prolyl 4-hydroxylase (P4H) gene (Cre01.g800047; Supplemental Figure 7). P4Hs catalyze the formation of 4-hydroxyproline (Gorres and Raines 2010), a major post-translational modification of the hydroxyproline-rich glycoproteins that comprise the Chlamydomonas cell wall (Woessner and Goodenough 1994; Sumper and Hallmann 1998). The Chlamydomonas genome encodes more than 20 putative P4Hs, and although their specific roles are generally unknown, P4Hs have different expression patterns and are unlikely to be redundant. Keskiaho et al. (2007) showed that the knockdown of P4H-1 (now annotated as PFH12; Cre03.g160200), was sufficient to induce abnormal cell wall assembly. Notably, the deleted gene in CC-503 has one paralog, PFH5 (Cre01.g014650; encoding a protein sharing 76% aa identity with Cre01.g800047). This paralog is immediately downstream of the deleted gene and appears to be intact, although its regulation may be affected by the deletion. It is therefore unclear whether the loss of Cre01.g800047 alone is responsible for the cw phenotype. Indeed, as introduced, more than one mutation may underlie the loss of the cell wall (Davies 1972; Hyams and Davies 1972).
Major duplications and insertions in the CC-4532 genome
We also identified 10 non-TE structural mutations unique to CC-4532 v6 and absent in CC-503 v6 and CC-1690. These mutations are predicted to disrupt eight genes (Supplemental Dataset 8, Supplemental Figure 8). The largest mutations were both duplications: 24.5 kb on chromosome 3 and 89.1 kb on chromosome 12, which together caused the duplication of 17 complete genes. Using a coverage-based approach, Flowers et al. (2015) inferred the presence of several large duplications among various laboratory strains, hinting that duplications may be an important source of laboratory mutation. Interestingly, three-gene-disrupting insertions in CC-4532 v6 consisted entirely of a satellite, MSAT-11_cRei, ranging from ∼8 kb to >19 kb (two caused assembly gaps and their full length is unknown). For example, one insertion interrupted the first exon of a gene possibly encoding nicotinate phosphoribosyltransferase (Cre03.g188800), which catalyzes the first step of the nicotinamide adenine dinucleotide salvage pathway. MSAT-11_cRei arrays consist of a 1.9-kb tandemly repeated monomer and are present on chromosomes 7 and 12 in all three available genomes. We also detected two additional unique insertions in CC-1690. Similarly, MSAT-11_cRei de novo insertions have been observed in experimental lines of the field isolate CC-2931 (López-Cortegano et al. 2022). There are very few observations of de novo satellite dissemination and its mechanisms are generally unclear (Ruiz-Ruano et al. 2016), although rolling circle replication and reinsertion via extrachromosomal circular DNA intermediates has been proposed (Navrátilová et al. 2008). Collectively, these results suggest that all laboratory strains may harbor at least a small number of gene-disrupting structural mutations relative to the ancestral wild-type.
TE proliferation in the laboratory and the strain history of 137c
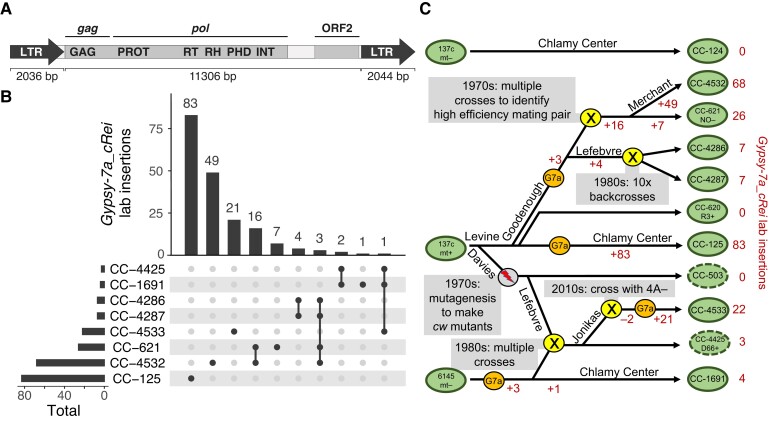
We next aimed to characterize the extent of TE activity in the CC-503 v6 and CC-4532 v6 genomes. We identified 26 TE insertions unique to CC-503 v6 (nine of which were absent in v5, suggesting recent activity; Supplemental Dataset 10) and 109 insertions unique to CC-4532 v6 (Supplemental Dataset 11, Supplemental Figure 8), which collectively involved 14 different TE families. Remarkably, 86 of the 109 CC-4532 v6 insertions were of the same 15.4-kb Gypsy long-terminal repeat (LTR) retrotransposon (Gypsy-7a_cRei, Figure 6A), adding ∼1.3 Mb of unique sequence (all TE insertions ∼1.4 Mb). Together with the large duplications and insertions described above, these TE insertions were responsible for the expanded length of the CC-4532 v6 assembly, which is more than 1% longer than CC-1690 (Table 1). Gypsy-7a_cRei has not previously been reported as active, and we identified no insertions in CC-503 v6, where the element is present as only one partial and two full-length ancestral copies. Only 10 of the 86 insertions were predicted to disrupt CDS (in some cases breaking the annotated gene model; Supplemental Dataset 11), and we observed intergenic insertions 2.6 times more frequently than expected by chance. Gypsy-7a_cRei may have a mechanism of targeted insertion, or genic insertions may have been selected against in the laboratory. The Gypsy-7a_cRei Gag-Pol polyprotein contains a plant homeodomain finger, an accessory domain found in several Chlamydomonas TEs (Perez-Alegre et al. 2005; Craig 2021) that may be involved in chromatin remodeling to minimize deleterious insertions (Kapitonov and Jurka 2003). Nonetheless, intergenic insertions may still affect gene expression, and we observed 10 insertions into introns and 25 into untranslated regions (UTRs), including the 3′-UTR of TUB2, the gene encoding beta-tubulin.
Figure 6.
Gypsy-7a_cRei insertions and the strain history of 137c+. A, Structure of the 15.4-kb Gypsy LTR retrotransposon. LTR subparts are shown as block arrows; note that the left LTR is missing the final 8-bp of the right LTR. The two ORFs are highlighted within the 11.3-kb internal section, and the gag and pol sections of the polyprotein are indicated. Protein domains: GAG, group-specific antigen; PROT, pepsin-like aspartate protease; RT, reverse transcriptase; RH, RNAse H; PHD, plant homeodomain finger; INT, integrase. B, Upset plot (Lex et al. 2014) showing the number of shared and strain-specific laboratory insertions of Gypsy-7a_cRei in select laboratory strains. Ancestral copies of Gypsy-7a_cRei are excluded. C, Schematic diagram representing a putative strain history of several interrelated laboratory strains. Presented is the most parsimonious interpretation of the shared and independent insertions (B) coupled with known strain histories. Green ovals represent strains as indicated, with a dashed line indicating cell wall defective strains. A gray circle indicates the MNNG mutagenesis that produced cw mutants. Yellow circles indicate crosses as labeled. Orange circles indicate likely activation of Gypsy-7a_cRei (“G7a”). Changes in the net number of Gypsy-7a_cRei loci due to addition by retrotransposition (+) or loss during crossings (−) are indicated in red. The names of several key Chlamydomonas researchers (R.P. Levine, D.R. Davies, U.W. Goodenough, P.A. Lefebvre, S.S. Merchant) are indicated where relevant.
We next used whole-genome resequencing data (Gallaher et al. 2015) to test whether Gypsy-7a_cRei is active in any other laboratory strains. We analyzed 14 laboratory strains, including the oldest extant strains (CC-124, CC-125, CC-1009, CC-1010, CC-1690, and CC-1691) that are parental to all laboratory strains. Insertions were identified by extracting read pairs where one read mapped uniquely to a non-repetitive genomic region and the other mapped to Gypsy-7a_cRei (see Supplemental Dataset 12 for insertion coordinates). This approach retrieved 68 of the 86 Gypsy-7a_cRei insertions in CC-4532 v6, the difference being attributable to insertions occurring in the ∼8 years between the Illumina and PacBio sequencing, or the inability to call insertions in repetitive regions (e.g. centromeres, see Supplemental Figure 8). All strains carry two to four ancestral Gypsy-7a_cRei copies, depending on their proportions of haplotype 1 and 2 (collectively three copies in haplotype 1 and one in haplotype 2). Six of the 14 strains (CC-124, CC-503, CC-620, CC-1690, CC-1009, CC-1010) had only these ancestral loci, despite being propagated for over seven decades, suggesting that Gypsy-7a_cRei is largely quiescent. However, in a few strains, particularly those descended from 137c+, we observed massive expansions of Gypsy-7a_cRei, like that in CC-4532. Indeed, CC-125, the linear descendant of 137c+, had the most novel insertions of any strain (83, Figure 6B). This result was unexpected, since there are no new insertions in CC-503, which was derived from 137c+ by mutagenesis, and no insertions in CC-620, another direct descendant of 137c+. CC-4532 shared 19 of its 68 laboratory insertions with CC-621 (Figure 6B), which corroborates our understanding that CC-4532 and CC-621 are both subclones of NO− from Ursula Goodenough that have been separated by at least three decades. Strains CC-4286 and CC-4287 also had some shared and unique insertions relative to CC-4532 and CC-621, indicating shared ancestry.
We attempted to reconcile the distribution of the Gypsy-7a_cRei insertions with described strain histories (Pröschold et al. 2005; Gallaher et al. 2015), which is presented as the proposed strain history in Figure 6C. Since all insertions were unique to CC-125, we hypothesize that Gypsy-7a_cRei became active in the 137c+ culture that became CC-125 after being separated from the cultures that became CC-503 and CC-620, which occurred several decades ago. Gypsy-7a_cRei became active independently in a strain from the laboratory of Ursula Goodenough (NO−/CC-621) that was produced by crossing 137c+ and unknown strains, and it remains active and continues to expand in strains derived from NO−, e.g. CC-4286 and CC-4287 from Paul Lefebvre and CC-4532 from Sabeeha Merchant. A third reactivation of Gypsy-7a_cRei likely occurred in Ruth Sager's 6145 strain, which eventually became CC-1691. This event contributed novel insertions to strain D66+ (CC-4425), which in turn contributed a single laboratory insertion to Martin Jonikas' strain, CC-4533. This last strain, the parental strain of the Chlamydomonas Library Project (CLiP), may represent a fourth reactivation of Gypsy-7a_cRei (or an increase in transposition frequency) since it carries 21 private insertions despite being separate from CC-4425 by approximately a decade.
Aside from Gypsy-7a_cRei, the most active TE family was MRC1, with 17 insertions in CC-503 v6 and 16 insertions in CC-4532 v6 (Supplemental Datasets 10 and 11). MRC1 was originally described as a non-autonomous LTR element (Kim et al. 2006), however we recently reclassified it as a non-autonomous Chlamys Penelope-like element (Craig et al. 2021b). Gallaher et al. (2015) and Neupert et al. (2020) reported activity of MRC1, and it may generally be one of the most active TEs in the laboratory. We identified four active DNA transposons that have been described previously, namely one insertion each of Gulliver (Ferris 1989), Tcr1 (Schnell and Lefebvre 1993), and Tcr3 (Wang et al. 1998) (hAT, Kyakuja, and EnSpm superfamilies, respectively), and three insertions of the non-autonomous hAT family Bill (Kim et al. 2006). The eight remaining TEs have only been described in Repbase (Bao et al. 2015) or the more recent Chlamydomonas TE library (Craig 2021).
Collectively, these results suggest that TE activity between laboratory strains can be highly heterogenous, with the potential for rapid TE proliferation to cause significant increases in genome size and to disrupt genic sequence. Indeed, serendipitous or screened-for TE insertions have caused several informative Chlamydomonas mutants (e.g. Moseley et al. (2002); Helliwell et al. (2015)) and have led to the discovery of many of the TEs active in laboratory strains. It is presently unclear why suppression of Gypsy-7a_cRei is unstable in certain strains, and why this family exhibits a far higher transposition frequency than other active TEs upon activation. Similar copy number variation among laboratory strains has been reported for the non-autonomous DIRS retrotransposon TOC1 (Day et al. 1988), although curiously we did not find any de novo insertions of this element in CC-503 v6 nor CC-4532 v6. Given the wealth of transcriptomics data available, it would be interesting to explore the expression patterns of Gypsy-7a_cRei and other TE genes under various stress and culture conditions. It is possible that certain avoidable conditions induce transposition, as has been documented elsewhere [e.g. temperature-sensitive TEs in V. carteri (Ueki and Nishii 2008) and Arabidopsis (Ito et al. 2011)].
Version 6 structural annotations
We annotated both the CC-4532 v6 and CC-503 v6 assemblies de novo, incorporating Iso-Seq data, more than 500 Gb of RNA-seq data, and protein homology from the growing number of green algal structural annotations. Notably, more than 1.6 billion strand-specific 150-bp RNA-seq read pairs were introduced from the JGI Gene Atlas (https://phytozome-next.jgi.doe.gov/geneatlas/), which assessed gene expression under 25 conditions. We predicted gene models using several annotation tools, with the model receiving the best support from transcriptomic and protein homology evidence retained in cases of redundancy. Focusing on CC-4532 v6, we then made several further improvements (see below) to the de novo gene models to arrive at the final CC-4532 v6 annotation, named CC-4532 v6.1, featuring 16,801 protein-coding genes (Table 2). The number of predicted alternative transcripts also increased more than eight-fold relative to v5.6. Dedicated analyses will be required to validate these new isoforms (see Labadorf et al. 2010; Raj-Kumar et al. 2017). One highlight of the annotations was that the longest transcripts overlap for 29% of adjacent genes, 64% of which are on opposite strands (see examples in Figure 7). While the longest transcripts may not always be the most abundant, this result nevertheless speaks to the compactness of the genome. Overlapping models were essentially absent from v5.6 (1% of neighboring genes) and were made possible by Iso-Seq support, and the present count may be an underestimate since these data do not cover all genes. Although poorly characterized, overlapping genes are a feature of many eukaryotes (Wright et al. 2022) and can be widespread in the most compact genomes (Williams et al. 2005). This result may have important implications for understanding gene regulation in Chlamydomonas.
Table 2.
Comparison of structural annotations between reference genome versions
| Annotation | CC-503 v4.3 | CC-503 v5.6 | CC-503 v6.1 | CC-4532 v6.1 |
|---|---|---|---|---|
| Nuclear genes | 17,114 | 17,741 | 16,795 | 16,801a |
| Alternative transcripts | / | 1,789 | 14,874 | 14,979 |
| TE genes | / | / | 647 | 810 |
| Low coding potential genes | / | / | 1,435 | 1,417 |
| Plastome genes | / | / | 74b | 74b |
| Mitogenome genes | / | / | 8 | 8 |
| BUSCO (chlorophyta_odb10, N = 1,519) | C:96.7% S:96.0%,D:0.7%] F:1.3%,M:2.0% |
C:98.9% [S:98.2%,D:0.7%] F:0.3%,M:0.8% |
C:100.0% [S:99.3%,D:0.7%] F:0.1%,M:0.0% |
C:99.8% [S:98.8%,D:1.0%] F:0.1%,M:0.1% |
Abbreviations: C, complete; S, single-copy; D, duplicated; F, fragmented; M, missing.
CC-4532 v6.1 contains 16 MT+ specific genes (see below).
The three trans-spliced exons of psaA are here counted as individual models.
Figure 7.
Browser views of example gene models improved between v5.6 and CC-4532 v6.1. A, PF20, CC-4532 v6 coordinates: chromosome 4, 3,483,590–3,493,250. B, NRAMP2, CC-4532 v6 coordinates: chromosome 7, 1,258,513–1,267,855. Note that the redundant sequences (boxed) are not included in the gene model converted from v5.6, since these duplicated sequences do not exist in CC-4532 v6. No peptides were identified for NRAMP2. H3K4me3 ChIP-seq (dark blue peaks) marks promoters. The v5 assembly track shows an alignment of v5 contigs to CC-4532 v6, with assembly gaps appearing as unmapped regions and redundant sequence as overlapping regions. Peptides are from mass spectrometry analysis of the proteome. Coordinates for v5.6 gene models (orange) were converted to CC-4532 v6. Thick blocks represent CDS, thin blocks UTRs, and joining lines introns. Forward strand mappings are shown in pink and reverse in blue. Red and green mismatches at the end of Iso-Seq reads correspond to poly(A) tails.
Since so many of the v5 assembly gaps were within genes, the assembly improvements provided considerable potential to improve gene models. Highlighted by Tulin and Cross (2016) as a gene featuring “hidden exons”, PARALYZED FLAGELLA 20 (PF20) encodes a 606-aa protein important for cilia function (Smith and Lefebvre 1997). The filling of a v5 assembly gap in PF20 resulted in the correction of the gene model in CC-4532 v6.1, adding three new exons (exons 9, 10, and 11 in CC-4532 v6.1) and shifting the 3′ splice site of exon 8 (Figure 7A). A second example is the putative metal ion transporter NATURAL RESISTANCE-ASSOCIATED MACROPHAGE PROTEIN 2 (NRAMP2), which featured two gaps in v5 that were both classified as “redundant” in our prior analysis. While one “gap” duplicated only 26 bp of intronic sequence, the second duplicated exons 10 and 11, fortuitously maintaining the reading frame and resulting in the erroneous repetition of 63 aa in the v5 protein (Figure 7B). Finally, while PF20 and NRAMP2 were annotated as single genes in v5, some genes were incorrectly split into separate models by gaps (Supplemental Figure 9). We chose these examples from hundreds of affected genes, demonstrating the scale of improvement made possible by assembly improvements.
We further focused on specific issues that have been previously highlighted. Cross (2016) showed that more than 4,000 v5 gene models have in-frame upstream ORFs, many of which likely correspond to genuine N-terminal protein extensions based on comparison to V. carteri orthologs. To address this issue, we generally annotated the first in-frame start codon for each predicted mRNA as the start codon in the v6 annotations. NRAMP2 also exemplifies this change, with the CC-4532 v6.1 protein extended by 126 aa at its N terminus (Figure 7B). Second, two studies (Blaby and Blaby-Haas 2017; Craig et al. 2021a) reported more than 100 strongly supported gene models that are absent from the v5 annotations. Many of these genes were present in the v4 annotations (e.g. PSBW1), and 25 are part of polycistronic transcripts (Gallaher et al. 2021). We attempted to transfer any strongly supported gene model from the v4.3, v5.6 or preliminary CC-503 v6 annotations to CC-4532 v6.1 if they were absent in the preliminary de novo annotation. Third, we manually curated a modest number of genes of interest, including 12 encoding selenoproteins (Novoselov et al. 2002) that were all previously misannotated due to their use of the canonical stop codon “TGA” to encode selenocysteine. Finally, as detailed below, the CC-4532 v6.1 annotation was supplemented with MT+-specific genes and genes found in the organelle genomes.
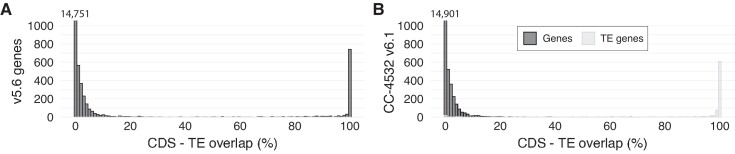
Two further changes caused the nuclear gene count to fall by 940 between v5.6 and CC-4532 v6.1. First, we previously found that several hundred v5.6 genes have low coding potential and are unlikely to represent protein-coding genes (Craig et al. 2021a). This designation was reached by combining evidence from functional annotation, comparative genomics, population genetics, and intrinsic features of Chlamydomonas genes and CDS (codon usage bias and the strength of translation initiation sites, i.e. Kozak-like sequences). We repeated these analyses on the preliminary v6 annotations, conservatively calling 1,417 “low coding potential” gene models in CC-4532 v6.1 (Table 2; Supplemental Figures 10 and 11). Validating these analyses, we found no peptide support for these models in our proteomics analysis (see below). We did not include these models in the main annotations, but they are available as Supplemental Datasets 13 and 14. Many of these loci may be long noncoding RNA (lncRNA) genes that contain spurious short ORFs, or short ORFs located within the UTRs of other genes. Second, we previously identified ∼1,000 genes in v5.6 that are likely part of TEs (Craig et al. 2021a). There are ∼220 TE families in the Chlamydomonas genome, and although only a fraction is active in laboratory strains, many TEs are likely active in the wider species (Craig 2021). Since most TE copies are not degraded, their genes can be readily identified by gene prediction algorithms. Unknowingly including TE genes within annotations can confound analyses, such as analyses of methylation, chromatin states, or small RNA targeting, where substantial differences may be expected between non-TE and TE genes. Genome projects therefore generally aim to exclude TE genes, while highly curated annotations of model organisms may include TEs as defined entities.
When comparing v5.6 genes and TE coordinates, the distribution of their intersect is highly bimodal; 1,023 genes have a >30% overlap between their CDS and TEs, and 908 genes have >80% overlap (Figure 8A). We obtained similar distributions in the preliminary v6 annotations, indicating that most genes can be cleanly divided into TE and non-TE subsets. To designate high-confidence TE genes, we required a gene with a high CDS-TE overlap to have either sequence similarity to a known TE-encoded protein or a functional domain. This analysis resulted in the inclusion of 810 TE genes in CC-4532 v6.1 (Figure 8B, Table 2) and 647 in CC-503 v6.1 (Supplemental Figure 12), which are integrated in the associated GFF3 files under the field “transposable_element_gene”. Users should be aware that these TE gene sets are not exhaustive, and projects requiring TE coordinates in general should use annotations derived from the dedicated repeat library (Supplemental Dataset 15).
Figure 8.
TE genes in v5.6 and CC-4532 v6.1. A, Overlap between gene CDS and TEs) in v5.6. The number of genes with 0% overlap is indicated above the first bar. B, Overlap between gene CDS and TEs in CC-4532 v6.1. Genes were split into non-TE and TE genes.
The mating-type locus and haplotype 2
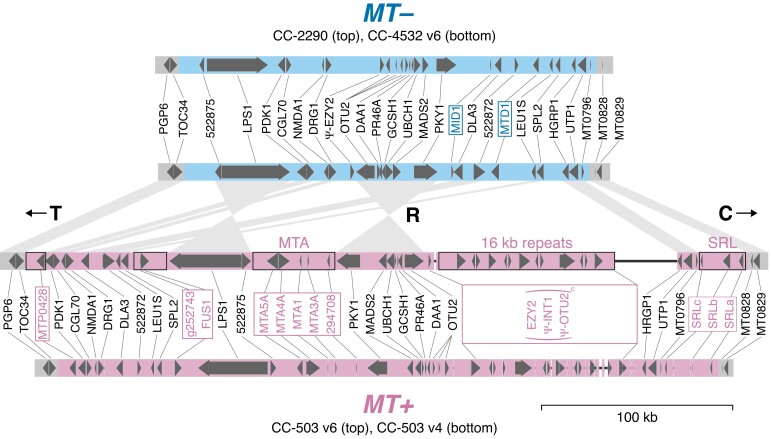
The mating-type locus (MT) on the left arm of chromosome 6 is naturally within a region where strains carry different haplotypes: mt+ strains carry haplotype 1, and mt− strains carry haplotype 2. Except for genes unique to either allele, MT genes have homologs present on both alleles (i.e. gametologs), although those within the rearranged (R) domain are generally not syntenic between MT+ and MT− (Ferris and Goodenough 1994; Ferris et al. 2002). Since CC-503 is mt+, past assembly versions have lacked the two MT− specific genes, MINUS DOMINANCE 1 (MID1) and MATING TYPE REGION D-1 (MTD1). With the reference now based on the mt− CC-4532, the situation is reversed; however, this is a greater issue since there are at least 16 MT+ specific genes in five MT+-specific regions, three of which originated from autosomal insertion (MTP0428, the MTA region and the SRL region; De Hoff et al. 2013). To address this issue, we appended a 375-kb MT+ R domain contig extracted from CC-503 v6 to the reference CC-4532 v6 assembly. To avoid potential mismapping of omics data, we hardmasked (i.e. replaced with Ns) any gametologous regions on the appended contig so that only sequences corresponding to MT+-specific regions and genes were included. Finally, we manually curated all R domain gene models and appended MT+-specific genes to the CC-4532 v6.1 annotation. CC-4532 v6 should thus be suitable for analyses of data from both mt+ and mt− strains, and we expect that the availability of highly contiguous and well-annotated assemblies of both alleles will be a major resource for the Chlamydomonas community.
We compared our resources for CC-503 v6 and CC-4532 v6 to the existing curated MT+ (CC-503 v4) and MT− (CC-2290) annotations of De Hoff et al. (2013) (Figure 9). The gapless CC-4532 MT− R domain (∼211 kb) was entirely syntenic with that of CC-2290 (∼218 kb), although intergenic regions were often unalignable due to polymorphic repeats. The only major change in both MT− and MT+ affected OTUBAIN PROTEIN 2 (OTU2), which was extended to incorporate the genes 155027 and MT0618 into a single-gene model (i.e. the correct OTU2 was split across three-gene models in CC-2290 and CC-503 v4). The MT+ allele of OTU2 was recently shown to function in the uniparental inheritance of the plastome (Joo et al. 2022). In MT+, OTU2 is located immediately upstream of an MT+-specific region termed the “16-kb repeats” (Ferris et al. 2002), consisting of a 17.2-kb tandemly repeated region containing multiple copies of EARLY ZYGOTE 2 (EZY2), INTEGRASE 1 (INT1) and what was previously annotated as OTU2 (i.e. the repeats contain duplicates of only a 3′ fragment of the full OTU2 gene, which may be pseudogenized). INT1 shares strong sequence similarity with the proteins of DIRS retrotransposons from Chlamydomonas (e.g. TOC3; Goodwin and Poulter 2004) and is likely derived from a TE family that is no longer present elsewhere in the genome. Although the reverse transcriptase domain is missing, INT1 does contain sequence encoding the RNAse H and methyltransferase domains of a DIRS element in addition to the “integrase” (actually a tyrosine recombinase). Assuming INT1 has not been co-opted, the multiple copies of EZY2, which produce zygote-specific transcripts (Ferris et al. 2002), may be the only functional genes in the repeat. The MT+-specific regions are collectively responsible for the larger size of the MT+ allele. However, the assembly of the 16-kb repeats remains incomplete in CC-503 v6, with two gaps relative to CC-1690 (which is also mt+). We detected no structural variants indicative of mutations between CC-503 v6 and CC-1690 in the R domain, suggesting that CC-503 v6 provides a typical representation of all mt+ laboratory strains across this region. Notably, there were two full-length copies of OTU2 annotated in v5 (Joo et al. 2022), however we found no evidence for this state in either CC-503 v6 or CC-1690, and this was likely a misassembly of the regions flanking the 16-kb repeats.
Figure 9.
Assembly and annotation comparisons of the plus (MT+) and minus (MT−) alleles of the mating-type locus rearranged (R) domain. Block arrows represent protein-coding genes. Mating-type-specific gene symbols are boxed. CC-503 v6 MT+-specific regions that were not hardmasked in the MT+ contig appended to CC-4532 v6 are outlined in black. Synteny between CC-4532 v6 MT− and CC-503 v6 MT+ genes is represented by wedges. T and C refer to the telomere-proximal and centromere-proximal domains, respectively. Only copies of EZY2 within the 16-kb repeats are included in the MT+ gene annotation. Copies of OTU2 within the 16-kb repeats are truncated and are marked as putative pseudogenes (as are INT1 copies, see main text). Thin black lines represent assembly gaps. The CC-2290 and CC-503 v4 annotations are from De Hoff et al. (2013). Gene symbols are from De Hoff et al. (2013), except for symbols updated herein (Supplemental Dataset 16).
More broadly, CC-4532 contains five haplotype 2 regions spanning 4.6% of the genome (Figure 1A; Supplemental Figure 8) and featuring 818 genes (Supplemental Dataset 17). Unlike our analysis of structural mutations above, we did not perform a systematic analysis of structural variation present between the two haplotypes; the CC-503–CC-4532 comparison captures less than one fifth of the total haplotype variation among laboratory strains (which can affect up to ∼25% of the genome), and this question would be best addressed by assembling and comparing genomes of additional strains. Furthermore, without additional genomes, it is currently impossible to distinguish ancestral structural variants from derived laboratory mutations in these regions. We did, however, revise the coordinates of the haplotype 2 blocks reported by Gallaher et al. (2015) relative to CC-4532 v6 (Supplemental Dataset 18), since some were affected by assembly corrections. The distribution of haplotype blocks among many of the most widely used laboratory strains is shown in Supplemental Figure 13.
Organelle genomes and structural annotations
The genomes of the plastid and mitochondria, the plastome and mitogenome, respectively, encode abundant cellular proteins and contribute disproportionately to the transcriptome: 46% of all mRNA in the cell is transcribed from the plastome, and just eight mitochondrial genes contribute 1.4% to the total mRNA pool (Gallaher et al. 2018). We recently produced high-quality assemblies and annotations of the plastome and mitogenome (Gallaher et al. 2018), which are now included in the v6 releases (Table 2). Importantly, there are no genetic variants to distinguish the organelle genomes of CC-4532 and CC-503, since the laboratory strains are putatively descended from one zygote and the multicopy organelle genomes are inherited uniparentally.
The circular 205.6-kb plastome carries 72 protein-coding genes, with two (psbA and I-CreI) duplicated in the inverted repeat regions. Many of the genes are expressed from polycistronic transcripts. Cavaiuolo et al. (2017) used small RNA profiling to accurately map the plastid genes, and we incorporated their improvements to the v6.1 annotations. The psaA gene, which encodes photosystem I chlorophyll a binding apoprotein A1, is expressed as three separate transcripts that are trans-spliced to generate the mature mRNA molecule (Kück et al. 1987). The three separate genes that contribute to the mature transcript are out of order and in different orientations, and we therefore assigned three separate, but sequential, gene IDs (CreCp.g802280, CreCp.g802281, and CreCp.g802282) to the three psaA exons.
The linear 15.8-kb mitogenome carries eight protein-coding genes, which are expressed from a single bidirectional promoter. Seven of these genes encode components of the respiratory complex, while the eighth, reverse transcriptase-like (rtl), is likely required for mitogenome replication (Smith and Craig 2021). We incorporated the more accurate annotations of Salinas-Giegé et al. (2017), who demonstrated that the 5′-end of each mature mitochondrial transcript begins immediately at the start codon (i.e. there are no 5′-UTRs).
Gene model validation
To validate the CC-4532 v6.1 annotation, we first queried all predicted proteins against the BUSCO (Benchmarking Universal Single-Copy Orthologs) chlorophyte dataset (Manni et al. 2021), with the number of fragmented and missing genes dropping from five and eleven, respectively, in v5.6, to only one and two in CC-4532 v6.1 (Table 2). Notably, CC-503 v6.1 had no missing genes, and upon inspection, the two missing genes in CC-4532 v6 were found within the small number of remaining genic gaps (see above). Nevertheless, we consider the CC-4532 v6.1 annotation to be superior to that of CC-503: many more genes are affected by major loss-of-function mutations in CC-503, although none are genes in the BUSCO dataset (many of which may be essential).
We next turned to chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) data of trimethylated histone H3 lysine 4 (H3K4me3), which reliably mark promoter regions (Ngan et al. 2015; Strenkert et al. 2022; see Figure 7). We queried 1,224 H3K4me3 peaks that had been called as intergenic relative to the v5 genome and v5.6 annotation, assigning 244 peaks to gene TSSs in CC-4532 v6.1. Approximately 30% of the genes newly associated with H3K4me3 peaks did not have gene IDs mapped forward from v5.6, suggesting that the improvements can be attributed to both the inclusion of new genes and changes to the TSSs of existing genes. It is not surprising that almost 1,000 H3K4me3 peaks remain unannotated, since they are expected to be associated with features other than protein-coding genes, such as lncRNAs (Strenkert et al. 2022). Furthermore, ∼40% of the remaining peaks coincided with TEs, which may be an underappreciated source of active promoters in Chlamydomonas.
Finally, we queried the v5.6 and CC-4532 v6.1 predicted proteins against a pool of proteomics data. We identified at least one unambiguously assigned peptide for 14,339 v5.6 proteins and 14,841 v6.1 proteins, an increase of 3.5% (Supplemental Dataset 19). The v6.1 total included 14,770 proteins encoded by the nuclear genome (including TE proteins), 65 from the plastome, and six from the mitogenome. We noted a 7.2% increase in the total number of unique peptides assigned to CC-4532 v6.1 relative to v5.6, and a 7.0% increase in the total number of peptides. These increases can be attributed to several improvements in v6.1, including the incorporation of entirely new nuclear genes, the inclusion of new exons within previous assembly gaps, and the N-terminal ORF extensions. For example, we identified three unique peptides assigned to the previously “hidden” exons of PF20 (Figure 7A). The addition of the organelle annotations also contributed substantially. This was especially true for the total number of peptides, since the 65 plastome-encoded proteins with identified peptides accounted for 5.0% of all peptides assigned to CC-4532 v6.1, and the six mitogenome-encoded proteins accounted for 0.018%. Notably, these estimates are far lower than the total mRNA contribution from the organelles to the cell mentioned above.
Gene IDs
Starting with the v4 annotations and becoming standard for all genes in v5.5, Chlamydomonas locus IDs have taken the form CreYY.gNNNNNN, where YY is the chromosome number and NNNNNN is a unique number that nominally increases along the chromosome (Blaby et al. 2014). We successfully mapped existing “Cre” IDs from v5.6 to 15,224 nuclear genes in the CC-4532 v6.1 annotation (90.6%, Supplemental Dataset 20). The remaining gene models were either novel or had changed considerably relative to their v5.6 counterparts (e.g. due to gene model mergers or splits). For these 2,277 CC-4532 v6.1 genes with no v5 equivalent (including most TE and all organelle genes), new NNNNNN numbers were introduced, ranging from 800,000 to 802,251 and increasing with genomic coordinates. Plastome and mitogenome genes were assigned locus identifiers from CreCp.g802263 to CreCp.g802335, and from CreMt.g802337 to CreMt.g802344, respectively. Since we also annotated the CC-503 v6 assembly (and many more genomes may follow), it was necessary to distinguish between orthologous gene models annotated in each assembly. We therefore included a four-digit strain-specific suffix to the IDs: CreYY.gNNNNNN_4532 for CC-4532 v6.1 and CreYY.gNNNNNN_0503 for CC-503 v6.1. With CC-4532 becoming the reference, gene models in other assemblies (including CC-503 v6) will be attributed IDs based on their mapping to this annotation.
It is also imperative to note that the misassembly corrections and the CC-503 structural rearrangements resulted in many genes having CC-4532 v6.1 gene IDs that refer to the wrong chromosome (i.e. YY number). Similarly, the NNNNNN numbers may not be contiguous. In fact, this was already an issue for some IDs in v5 due to assembly changes relative to v4. Unfortunately, both the YY and NNNNNN numbers are now meaningless (and potentially misleading), and users are cautioned that no spatial information should be extracted from the IDs alone. To counter any confusion, we devised a spatially correct and strain-specific “associated locus ID” for each gene. They follow the format XXXX_YY_NNNNN, where XXXX is the strain identifier from the Chlamydomonas Resource Center, YY is the chromosome number, and NNNNN is a unique gene number that increases along the chromosome, with odd numbers for forward strand genes and even numbers for reverse strand genes. Successive IDs feature NNNNN numbers separated by 3 or 4 unused numbers depending on relative strandedness (rising to 53 or 54 for genes on either side of an assembly gap), serving as placeholders for possible new gene models. As an example, in CC-4532 v6.1, PSY1 has the primary ID Cre02.g095092_4532 and the associated locus ID 4532_11_52343, with the latter providing the correct chromosomal location (Figure 2B). These IDs also carry additional information as optional suffixes; for instance, all TE genes feature the suffix “_TE”, making them instantly recognizable. The associated locus IDs have a one-to-one relationship with the existing “Cre” IDs (Supplemental Dataset 19) and we envision that they will be used in parallel (e.g. to simultaneously assess spatial information).
Expert annotation and gene symbols
Over decades of research, Chlamydomonas genes have been assigned a gene symbol, designed to uniquely identify and succinctly characterize a given locus. In v5.6, 5,130 genes (28.9%) were annotated with a gene symbol (Supplemental Dataset 21). These symbols have been derived from several sources, including protein function, mutant phenotypes, and orthology (Supplemental Note 1). The gene symbols are a powerful tool for interpreting, analyzing, and communicating research in Chlamydomonas, especially for large-scale and systems biology research. Unfortunately, automated annotation has driven the proliferation of uninformative gene symbols. For example, the root “ANK” was used to assign gene symbols to 20 genes in v5.6 due solely to the presence of a predicted ankyrin repeat domain. Similarly, there are 51 HEL genes (encoding proteins with a DEAD/DEAH box helicase domain) and 35 DNJ genes (encoding proteins with a DnaJ domain) in v5.6. The presence of a gene symbol may imply that the gene has been at least partially characterized and perhaps has a validated function corresponding to the name, while these examples provide no information beyond their automated domain annotations. Furthermore, some symbols rely on erroneous predictions. For example, NIK1 (Cre14.g629650) was named from homology to a nickel (Ni) transporter; however, Chlamydomonas has no known Ni-requiring genes and no nutritional requirement for Ni (Blaby-Haas et al. 2016).
The Chlamydomonas annotations are frequently used to guide the annotation of newly sequenced Chlorophyte genomes (Roth et al. 2017), which propagates the low information or misinformation throughout the Chlorophyte lineage. Therefore, we sought to improve and update the gene symbols, which consisted of three phases: (1) the addition of new gene symbols wherever those annotations were based on expert analysis or empirical data, (2) transfer of a primary gene symbol to “previous identifiers” for uninformative and misleading gene symbols, and (3) reformatting or changing existing gene symbols to conform to a uniform style.
We added 610 new gene symbols to the CC-4532 v6.1 annotation. The majority of these were assigned in collaboration with the authors of individual chapters in the forthcoming third edition of the Chlamydomonas Sourcebook. Still others were based on recent publications. We reclassified 1,332 v5.6 gene symbols as “previous identifiers”, preserving connections to historical research that may have used those symbols (Supplemental Dataset 22). As a result, there are now 4,408 out of 16,801 (26.2%) genes with a gene symbol in v6.1 (excluding TE genes). An additional 549 genes had their gene symbol replaced, altered, or reformatted to improve clarity, highlight orthologies, and unify formatting. This effort was guided by several rules, updated and expanded from our previous work (Blaby et al. 2014), which are documented in Supplemental Note 1. We recommend that they be applied for the naming of all Chlamydomonas genes going forward.
Beyond symbols, many genes have a definition line (defline) and associated comments. These may include a description of the gene function, relevant expression data, paralogy and orthology information, and links to related peer-reviewed literature. This last feature, in the form of PMID accession numbers, has also been expanded and updated from 1,852 genes supported by one or more PMIDs (2,626 total PMIDs) in v5.6, to 3,042 genes (4,697 total PMIDs) in CC-4532 v6.1 (Supplemental Dataset 21).
Finally, the rate at which genes are expertly annotated in the literature outpaces that of updates to the Chlamydomonas genome and structural annotations. We have therefore created a dedicated email account, chlamy.updates@gmail.com, to receive and store user updates. We encourage users to send curated annotation updates. This may include gene symbol suggestions, textual annotation, PMIDs, expression data, functional validation, among other information. We also welcome manually curated gene models (preferably in GFF3 format), either for entirely new genes or for evidence-based corrections of existing models. We are committed to collating this information so that future updates are both efficient and representative of recent advances in Chlamydomonas research.
The present and future of the Chlamydomonas Genome Project
For almost two decades, the Chlamydomonas Genome Project has been based on the mt+ strain CC-503. In version 6, we have presented near-complete assemblies for both CC-503 and the mt− strain CC-4532. Following the discovery of numerous structural mutations affecting CC-503, CC-4532 v6 was chosen to serve as the reference genome. Despite its replacement, CC-503 v6 remains a valuable resource, especially for the MT+ allele and the existing organelle genomes that were appended to CC-4532 v6.
It is now clear that laboratory strains can differ extensively from each other, both genetically and phenotypically. Most of this variation stems from the mosaic of two haplotypes that comprise the genome of each strain (Gallaher et al. 2015). These developments have led to the “know thy strain” maxim: researchers are encouraged to consider the genetic differences that exist between the reference genome and the strains used in experimental work (Salomé and Merchant 2019). Our results suggest that these differences should not only be considered with respect to ancestral variation between the haplotypes, but also to derived variation arising by laboratory mutation. Although CC-503 may be an extreme case, the CC-4532 genome harbors 10 structural mutations and more than 100 TE insertions. Indeed, analyses by Gallaher et al. (2015) and Flowers et al. (2015) previously inferred the presence of many derived structural variants among strains, including several large duplications. It has also been estimated that ∼5−10% of all de novo mutations in Chlamydomonas experimental lines are structural (i.e. >50 bp; López-Cortegano et al. 2022), supporting a prominent role for structural evolution in the laboratory. While many of the most characteristic laboratory phenotypes were caused by mutations (e.g. nit1 and nit2), it is likely that all strains have experienced unique structural mutations (including TE proliferation at various rates), many of which disrupt genes. It is also possible that independently maintained cultures of the same strain differ due to independent mutations. Laboratory strains have been maintained clonally for as many as 75 years and mutations are an unavoidable consequence, especially if strains are evolving under relaxed selection. The implications of “laboratory domestication” have been considered in other model systems such as Caenorhabditis elegans (Sterken et al. 2015), and laboratory mutations should be carefully considered when evaluating experimental results. This may be particularly relevant in strains that have been selected for, and often actively mutagenized to achieve specific traits (e.g. cell wall-less strains with increased transformation efficiency).
With the continuous developments in long-read sequencing, we are entering an exciting era of Chlamydomonas genomics. A pan-genome project has been initiated, targeting genome assemblies for multiple laboratory strains and field isolates. Indeed, concurrent with the publication of this work, chromosome-level assemblies for the field isolates CC-1952 and CC-2931 have been released (López-Cortegano et al. 2022), as has the first essentially complete genome for a laboratory strain (Payne et al. 2022). As demonstrated herein, many insights can only be gleaned by comparing the genomes of different strains, and we can expect substantial benefits from sequencing additional strains and isolates moving forward. With respect to the two laboratory haplotypes, a “laboratory pan-genome” could be produced where all haplotype 1 and 2 regions are represented, capturing all ancestral variation present among laboratory strains. This dataset could potentially take the form of consensus assemblies for each haplotype, with genomes from several strains used to infer the ancestral state at the time of isolation. Such an ancestral reconstruction would arguably be the most representative and strain-agnostic Chlamydomonas reference genome possible, since differences between any laboratory strain and the reference would easily be recognized as a mutation. Furthermore, similar to resources developed for several important plants (Bayer et al. 2020), the species-level pan-genome aims to incorporate the far greater diversity present among Chlamydomonas field isolates (Flowers et al. 2015; Craig et al. 2019). There also remains substantial scope to further enhance structural annotations, especially with the continued growth in the availability of omics data for Chlamydomonas and related species. Such prospects are expected to reveal novel aspects of Chlamydomonas biology, continuing the development of the species as an integral model in plant and algal biology.
Materials and methods
Strains and DNA sequencing
CC-503 was obtained from the Chlamydomonas Resource Center in 2012. CC-4532 has been propagated in Sabeeha Merchant's group since the late 1990s (see Gallaher et al. 2015), when it was received from Ursula Goodenough. Cultures were grown as described previously (Gallaher et al. 2015).
Genomic DNA was extracted from frozen cell pellets and used for library preparation and sequencing at the U.S. Department of Energy Joint Genome Institute. Libraries were constructed using a SMRTbell Template Prep Kit 1.0 and size-selected to 10–50 kb on a SAGE Blue Pippin instrument. Sequencing was performed on a PacBio Sequel platform in continuous long reads mode using a 10-hour movie time, generating ∼127 × and 176 × coverage for CC-503 and CC-4532, respectively (CC-503 mean read length was 3.58 kb; CC-4532 mean read length was 9.88 kb). Additional Illumina sequencing was performed on a HiSeq2000 platform (150-bp paired-end reads, ∼400-bp insert) to ∼50 × (CC-503) and 55 × (CC-4532) coverage, as reported in Gallaher et al. (2015).
Assembly of CC-4532 v6 and CC-503 v6 genomes
Preliminary contig-level assemblies were produced from the PacBio datasets. CC-503 was assembled using MECAT v1.1 (genomeSize = 130,000,000 ErrorRate = 0.02 Overlapper = mecat2asmpw; Xiao et al. 2017) and CC-4532 using Canu v1.8 (genomeSize = 120,000,000; Koren et al. 2017). Reads were mapped to the raw assembly using BLASR, and error correction was performed using a single pass of Arrow correction from the GenomicConsensus toolkit. Remaining consensus errors were corrected using the strain-appropriate Illumina data. Illumina reads were aligned using bwa mem (Li 2013) and SNPs and InDels to be corrected were identified using GATK UnifiedGenotyper (McKenna et al. 2010). The corrections were verified by mapping the Illumina reads to the corrected consensus sequence.
The CC-1690 assembly (O'Donnell et al. 2020) was used to scaffold the preliminary contigs of each assembly to chromosomes. Contigs were mapped to the CC-1690 assembly using minimap2 v2.17 (-ax asm5; Li 2018) to produce PAF (Pairwise mApping Format) files. These mapping data were used to manually order and orientate uniquely mapping contigs (i.e. the majority of the contig received a mapping quality of 60) relative to each CC-1690 chromosome. Any inconsistencies between the contigs and CC-1690 chromosomes were inspected against the raw PacBio reads from the relevant strain (CC-503 or CC-4532) using IGV v2.7.2 (Robinson et al. 2011). In a small number of cases, a misassembled contig was split, while for CC-503 some genuine inconsistencies caused by structural mutations were supported by the raw reads and maintained. Several short contigs that mostly featured satellite DNA were manually removed since they appeared to duplicate a region already assembled on a larger contig. Other short contigs entirely consisting of subtelomeric repeats, which generally did not map uniquely, were assigned to chromosome termini by specific alignment and phylogenetic analysis (see Chaux-Jukic et al. 2021).
Gap lengths between contigs were estimated relative to CC-1690 and the appropriate number of Ns were inserted between contigs. Occasionally the estimated “gap” length was negative, suggesting redundant sequence at the termini of neighboring contigs. These contig termini were compared, trimmed to remove redundant sequence, and subsequently merged where possible. Arbitrary gaps of 100 Ns were inserted between contigs that could not be successfully merged.
Repeat annotation
TE sequence was identified in each assembly by providing the latest Chlamydomonas repeat library to RepeatMasker v4.0.9 (Smit et al. 2013–2015). This library features updated consensus models for all Chlamydomonas repeats available in Repbase (https://www.girinst.org/repbase/) together with >100 newly curated repeats (Craig 2021). Any putative TE copy divergent by >20% from its consensus sequence was removed. ZeppL clusters were identified as the span from the first two consecutive ZeppL-1_cRei copies to the final two consecutive ZeppL-1_cRei copies on each chromosome (except for chromosome 15, where three distinct clusters were observed; see Results and Discussion).
Microsatellites and satellite DNA were primarily identified using Tandem Repeats Finder (Benson 1999) with parameters “2 7 7 80 10 50 1000” (i.e. a minimum alignment score of 50 and a maximum monomer size of 1000 bp). Tandem repeats consisting of ≥2 monomers were split into microsatellites (monomers <10 bp) and satellite DNA (monomers ≥10 bp). If a region was called as both, priority was given to satellite DNA since shorter monomers are frequently nested within larger ones. Satellite DNA annotations were supplemented with curated satellites identified by RepeatMasker from the repeat library, several of which have monomers longer than the detection limit of Tandem Repeats Finder.
Genome-wide CG methylation was quantified for the CC-1690 assembly following Chaux-Jukic et al. (2021). Briefly, the raw signal of the CC-1690 Nanopore reads (i.e. fast5 files) generated by Liu et al. (2019) were mapped to the CC-1690 assembly using Tombo (https://nanoporetech.github.io/tombo/) and CG methylation was called using DeepSignal (Ni et al. 2019).
Validation of assembly improvements
Misassemblies in the v5 assembly were identified by mapping the v5 contigs to the chromosomal CC-503 v6 and CC-4532 v6 assemblies using minimap2 as described above. Genomic coordinates of intra- and inter-chromosomal inconsistencies were assessed manually from the PAF files and converted to input files for Circos (Krzywinski et al. 2009) and karyoploteR (Gel and Serra 2017) to produce Figures 1 and 2 and Supplemental Figure 1.
To enable convenient liftover of coordinates between assemblies, a five-way Cactus whole-genome alignment (WGA) (Armstrong et al. 2020) was produced including the v4, v5, CC-503 v6, CC-4532 v6, and CC-1690 assemblies. Each assembly was soft-masked for repeats by providing coordinates of TEs and tandem repeats (see above) to BEDtools v2.26.0 maskfasta (-soft) (Quinlan and Hall 2010). An arbitrary guide tree for Cactus was provided as “(CC-4532_v6:0.001, (CC-1690:0.001, (CC-503_v4:0.001, (CC-503_v5:0.001, CC-503_v6:0.001):0.001):0.001):0.001)”, and all assemblies were set to reference quality. Liftover of genomic coordinates in BED (Browser Extensible Data) format could then be performed between any two assemblies in the HAL (Hierarchical ALignment) format WGA using the HAL tools command halLiftover (Hickey et al. 2013). This approach was used to convert v5 coordinates of hypermethylated regions (Lopez et al. 2015) to CC-4532 v6 (Figure 1) and CC-1690 (Supplemental Figure 2) coordinates. Coordinates of v5 assembly gaps were also converted to CC-4532 v6 coordinates to investigate the sequence properties of filled gaps in the updated assembly (see Figure 3).
The genotyping data from Kathir et al. (2003) were kindly provided by Paul Lefebvre. The genomic coordinates (v5 assembly, as chromosome and position, in bp) were determined for all markers by BLAST search in Phytozome using the sequence deposited for each marker (https://www.chlamycollection.org/BAC/MARKER_index.htm), or by keyword search using the gene name in Phytozome. The markers were then ordered based on their assigned v5 chromosome and position. All genotyping data were assembled into a tab-delimited file and used as input for R/QTL (Broman et al. 2003) with the functions read.cross, est.rf, plotMap, plotRF, and summaryMap. The genotyping data for the CC-2935 × CC-2936 progeny (12 full tetrads) were obtained from Liu et al. (2018). Since the genotypes were encoded as either 1 or 2, a matrix (of the same size as the genotype matrix) was calculated whereby each n + 1 position received the difference between the genotype at the n + 1 position and the genotype at position n. Any SNP with a value not equal to zero was retained to estimate the genetic map, as described above. The genomic coordinates of mismapped markers or SNPs were manually corrected based on the CC-503 v6 or CC-1690 assemblies before re-running the genetic map construction, as above. The quality of the assemblies was assessed by plotting the recombination frequencies across the entire genome (plotRF) and by calculating the total length of the genetic map (summaryMap).
Structural annotations
Protein-coding genes for the CC-4532 v6 and CC-503 v6 assemblies were annotated using several sources of evidence. Input data were ∼1.6 billion 150-bp paired-end RNA-seq reads from the JGI Gene Atlas (strain CC-1690), ∼6.4 million 454-sequenced ESTs generated by previous versions of the genome project (CC-1690), and ∼1.6 million PacBio Iso-Seq reads (pooled samples from CC-4532, CC-5390, CC-4348, CC-4349, CC-4565, CC-4566, and CC-4567, see Gallaher et al. 2021). The Gene Atlas samples are described by Sreedasyam et al. (2022) and can be browsed at https://phytozome-next.jgi.doe.gov/geneatlas/. Specifically for the CC-4532 v6 annotation, ∼520 million unpaired 50-bp RNA-seq reads were included that were generated from CC-4532 grown under a range of conditions including heterotrophic and photoautotrophic growth, and in iron (Fe)-replete and Fe-limited media (NCBI SRA accessions PRJNA842032 and PRJNA717804). The RNA-seq and 454 reads were first assembled using PERTRAN (Shu et al. 2013), which conducts genome-guided transcriptome short-read assembly via GSNAP (Wu and Nacu 2010) and builds splice alignment graphs after alignment validation, realignment and correction. Iso-Seq circular consensus sequencing (CCS) reads were corrected and collapsed using a pipeline that aligns CCS reads to the genome with GMAP (Wu and Watanabe 2005), performs intron correction for small InDels in splice junctions (if any), and clusters alignments where all introns are shared for multi-exonic transcripts, or have 95% overlap for single-exon transcripts. A combined assembly of all transcriptomic data was then produced using PASA (Haas et al. 2003), yielding 287,891 assembled transcripts for CC-4532 v6 and 293,991 for CC-503 v6.
Preliminary loci were then identified using a combination of several tools and the relevant transcriptome assembly or splice alignments. This complex pipeline involved extensive post-processing, including the transfer of “missing” genes from previous assemblies, the identification of low coding potential and TE genes, and the manual curation of several gene models. These steps are described in detail in Supplemental Note 2.
ChIP-seq and proteomics
Intergenic H3K4me3 chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) peaks called against the v5 assembly were retrieved from Strenkert et al. (2022). Peak coordinates from the three time points in their experiment were merged and subsequently converted to CC-4532 v6 coordinates using halLiftover (see above; a peak was defined as successfully lifted over if ≥90% of sites were converted). Following Strenkert et al. (2022), distance from the midpoint of each peak to the nearest transcription start site (TSS) was calculated, and a peak was assigned to a TSS if it was within 750 bp. Peaks that were still classified as intergenic after this analysis were compared to the TE annotation and conservatively called TE-associated if ≥80% of sites within the peak overlapped a single TE copy.
The proteomics analysis was performed as in Gallaher et al. (2018), using datasets generated in that study. Briefly, peptides were identified by mass spectrometry and compared to the v5.6 and CC-4532 v6.1 predicted proteins. The total number of gene models encoding proteins with at least one assigned peptide was estimated, as was the total number of unique peptides assigned to each annotation, and the total number of peptides assigned overall.
Identification of structural mutations and TE insertions
Structural variants (i.e. >50 bp) were called between the CC-503 v6 and CC-4532 v6 assemblies using MUM&Co (O'Donnell and Fischer 2020), which identifies putative variants from MUMmer alignments (Kurtz et al. 2004). MUM&Co was run on each chromosome individually, and for chromosomes 2 and 9, the CC-503 v6 chromosomes were split at the translocation breakpoints and the relevant parts of each chromosome were included. All variant calls were then visualized and curated in IGV by comparing the CC-503 v6, CC-4532 v6 and CC-1690 assemblies (using alignments produced by minimap2, as performed above) and raw PacBio reads. Variants called within tandem repeats or within regions where CC-4532 carried haplotype 2 were not considered. Confirmed variants were polarized as mutations by comparing the three assemblies, i.e. the allele present in two assemblies (one of the v6 assemblies and CC-1690) was assumed to be ancestral. Structural mutations unique to CC-1690 were not called.
Structural mutations identified in CC-503 v6 were subsequently compared to past assembly versions and were called as consistent (present) or inconsistent (absent). Genes putatively affected by structural mutations were identified from the assembly and annotation featuring the ancestral state, i.e. genes from the CC-4532 v6.1 annotation were identified at the regions overlapping CC-503 v6 structural mutations (see Supplemental Datasets 7 and 8 and 10 and 11).
TE insertions were called as specific cases of insertion variants called by MUM&Co. All insertions were compared against the annotations derived from the Chlamydomonas repeat library (see above) and called as TE insertions where genomic coordinates of a TE perfectly intersected those of the insertion. Similarly, a small number of “deletions” were called as excision events of cut-and-paste DNA transposons (e.g. Gulliver).
To identify insertions of Gypsy-7a_cRei in laboratory strains, whole-genome resequencing data from 14 strains (Gallaher et al. 2015) were aligned using bwa mem (Li 2013) to a version of CC-4532 v6 that had been hardmasked for TEs and had the Chlamydomonas repeat library appended (causing all reads derived from Gypsy-7a_cRei copies to map to the single consensus sequence of this TE in the repeat library). Read pairs with at least one read mapped to the Gypsy-7a_cRei sequence and a mapping quality score >10 were extracted with samtools view (v1.15) (“-b -h -P -q 10”) (Danecek et al. 2021). The resulting BAM files were used to generate bedgraph files of read coverage using bedtools genomecov v2.30 (“-bg -split”) (Quinlan and Hall 2010). Peaks with <5 reads were filtered out. The resulting tracks were visualized in IGV v2.9.4 (Robinson et al. 2011). Peaks of coverage were manually identified for each strain.
Phylogenetic analysis of RecQ3 helicases
Peptide sequences were collected by searching for similar proteins to Cre16.g801898, Cre16.g673393, and At4g35740 using the Phycocosm (Grigoriev et al. 2021), Phytozome (Goodstein et al. 2012) and NCBI databases. Sequences were aligned with MAFFT (v7.305) (Katoh and Standley 2013) through the CIPRES web portal (Miller et al. 2010) and phylogenetic reconstruction was performed using W-IQ-TREE with default parameters (substitution model auto and ultrafast bootstrapping; Trifinopoulos et al. 2016). The consensus tree was visualized in iTOL (Letunic and Bork 2019), and the sequences from the subtree representing the RecQ3 subfamily were extracted, realigned, and used to build a RecQ3 phylogeny.
Accession numbers
CC-4532 v6 is available at Phytozome (https://phytozome-next.jgi.doe.gov). CC-4532 PacBio reads and the CC-4532 v6 assembly and annotation are deposited at NCBI under the BioProject PRJNA887768. CC-503 PacBio reads and the CC-503 v6 assembly and annotation are deposited at NCBI under the BioProject PRJNA887764.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Note 1 . Gene symbol naming rules.
Supplemental Note 2 . Detailed gene annotation methods.
Supplemental Figure 1 . Misassemblies in version 5 and their resolution in version 6.
Supplemental Figure 2 . CG methylation and repeat landscape of the CC-1690 assembly.
Supplemental Figure 3 . Full recombination frequency plots for the estimation of the genetic maps.
Supplemental Figure 4 . Summary of InDels present at CC-503 reciprocal translocation/inversion double-strand breaks and repair points.
Supplemental Figure 5 . Browser views of genes at double-strand breaks associated with the CC-503 reciprocal translocation/inversion mutation.
Supplemental Figure 6 . Browser views of whole-genome resequencing data at double-strand breaks associated with the CC-503 reciprocal translocation/inversion mutation.
Supplemental Figure 7 . Browser view of the CC-503 specific deletion of a prolyl 4-hydroxylase gene.
Supplemental Figure 8 . CC-4532 v6 haplotype 2 regions and unique structural mutations.
Supplemental Figure 9 . Browser view of a v5.6 split gene model merged in CC-4532 v6.1.
Supplemental Figure 10 . Analyses of coding potential for CC-4532 v6.1.
Supplemental Figure 11 . Analyses of coding potential for CC-503 v6.1.
Supplemental Figure 12 . Intersect between coding sequence of CC-503 v6.1 gene models and transposable elements.
Supplemental Figure 13 . Genomic distribution of haplotypes 1 and 2 among laboratory strains.
Supplemental Dataset 1 . Summary statistics, gene density and repeat content of the CC-4532 v6 chromosomes.
Supplemental Dataset 2 . Coordinate map between CC-4532 v6 chromosome 15 and the v5 assembly.
Supplemental Dataset 3 . Summary statistics of the CC-4532 v6 genome split by site class with respect to the CC-4532 v6.1 annotation.
Supplemental Dataset 4 . Metrics and sequence context of all CC-4532 v6 assembly gaps.
Supplemental Dataset 5 . Putative centromere metrics of the CC-1690 and CC-4532 v6 assemblies.
Supplemental Dataset 6 . Approximate genomic coordinates from the markers of Kathir et al. (2003).
Supplemental Dataset 7 . Curated structural mutations in the CC-503 v6 assembly.
Supplemental Dataset 8 . Curated structural mutations in the CC-4532 v6 assembly.
Supplemental Dataset 9 . Proteins, alignment and phylogeny for RECQ3 analysis.
Supplemental Dataset 10 . Curated TE insertions/excisions in the CC-503 v6 assembly.
Supplemental Dataset 11 . Curated TE insertions/excisions in the CC-4532 v6 assembly.
Supplemental Dataset 12 . Approximate coordinates of Gypsy-7a_cRei copies among laboratory strains.
Supplemental Dataset 13 . CC-4532 low coding potential genes.
Supplemental Dataset 14 . CC-503 low coding potential genes.
Supplemental Dataset 15 . Latest Chlamydomonas repeat library (v3.4).
Supplemental Dataset 16 . Mating-type locus R domain genes in CC-4532 v6.1 and CC-503 v6.1.
Supplemental Dataset 17 . Haplotype 2 regions in CC-4532 v6.
Supplemental Dataset 18 . Haplotype 2 coordinates in v5 and CC-4532 v6, and changes between the assembly versions.
Supplemental Dataset 19 . Comparison of proteomic validation of v5.6 and CC-4532 v6.1 proteins.
Supplemental Dataset 20 . Master annotation table of CC-4532 v6.1.
Supplemental Dataset 21 . Automated and expert annotations of v5.6 and CC-4532 v6.1 structural annotations.
Supplemental Dataset 22 . List of genes with gene symbols or previous identifiers in v5.6 and v6.1.
Supplemental Dataset 23 . Full names of gene symbols present in the main text.
Supplementary Material
Acknowledgments
We thank four reviewers for their comments that substantially improved an earlier version. We would like to thank the following for their expert advice in assigning descriptions and gene symbols to the v6.1 annotations: Jean Alric, Marius Arend, Ariane Atteia, Olga Baidukova, Steven G. Ball, Matteo Ballotari, Gabriella Benko, Christoph Benning, Robert Bloodgood, Alexandre-Viola Bohne, Pierre Cardol, Yen Peng Chew, Yves Choquet, José L. Crespo, David Dauvillée, Dion Dunford, Susan K. Dutcher, Emilio Fernández-Reyes, Aurora Galván, Michel Goldschmidt-Clermont, Arthur R. Grossman, Patrice P. Hamel, Thomas Happe, Peter Hegemann, Michael Hippler, Martin Jonikas, Steve King, J. Clark Lagarias, Stéphane D. Lemaire, Younghua Li-Beisson, Takuya Matsuo, David Mitchell, Aurora Nedelcu, Jöerg Nickelsen, Adrian Nievergeld, Krishna K. Niyogi, Junmin Pan, Dhruv Patel, Matthew C. Posewitz, Claire Remacle, Nicolas Rouhier, Emanuel Sanz-Luque, Michael Schroda, James Umen, Setsuko Wakao, Florent Waltz, Robert Willows, Felix Willmund, George B. Witman, Francis-André Wollman, Katia Wostrikoff, and William Zerges. We would like to thank Julianne Oshiro and Jordan L. Chastain for assistance tracking down PMID accessions for incorporation into Phytozome.
Contributor Information
Rory J Craig, California Institute for Quantitative Biosciences, University of California, Berkeley, California 94720, USA; Institute of Ecology and Evolution, School of Biological Sciences, University of Edinburgh, Edinburgh EH9 3FL, UK.
Sean D Gallaher, California Institute for Quantitative Biosciences, University of California, Berkeley, California 94720, USA.
Shengqiang Shu, United States Department of Energy, Joint Genome Institute, Berkeley, California 94720, USA.
Patrice A Salomé, Department of Chemistry and Biochemistry, University of California, Los Angeles, California 90095, USA; Institute for Genomics and Proteomics, University of California, Los Angeles, California 90095, USA.
Jerry W Jenkins, HudsonAlpha Genome Sequencing Center, HudsonAlpha Institute for Biotechnology, Huntsville, Alabama 35806, USA.
Crysten E Blaby-Haas, The Molecular Foundry, Lawrence Berkeley National Laboratory, Berkeley, California 94720, USA.
Samuel O Purvine, Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, Richland, Washington 99354, USA.
Samuel O’Donnell, Laboratory of Computational and Quantitative Biology, UMR 7238, CNRS, Institut de Biologie Paris-Seine, Sorbonne Université, Paris 75005, France.
Kerrie Barry, United States Department of Energy, Joint Genome Institute, Berkeley, California 94720, USA.
Jane Grimwood, HudsonAlpha Genome Sequencing Center, HudsonAlpha Institute for Biotechnology, Huntsville, Alabama 35806, USA.
Daniela Strenkert, California Institute for Quantitative Biosciences, University of California, Berkeley, California 94720, USA.
Janette Kropat, Department of Chemistry and Biochemistry, University of California, Los Angeles, California 90095, USA.
Chris Daum, United States Department of Energy, Joint Genome Institute, Berkeley, California 94720, USA.
Yuko Yoshinaga, United States Department of Energy, Joint Genome Institute, Berkeley, California 94720, USA.
David M Goodstein, United States Department of Energy, Joint Genome Institute, Berkeley, California 94720, USA.
Olivier Vallon, Unité Mixte de Recherche 7141, CNRS, Institut de Biologie Physico-Chimique, Sorbonne Université, Paris 75005, France.
Jeremy Schmutz, United States Department of Energy, Joint Genome Institute, Berkeley, California 94720, USA; HudsonAlpha Genome Sequencing Center, HudsonAlpha Institute for Biotechnology, Huntsville, Alabama 35806, USA.
Sabeeha S Merchant, California Institute for Quantitative Biosciences, University of California, Berkeley, California 94720, USA; Department of Molecular and Cell Biology, University of California, Berkeley, California 94720, USA; Department of Plant and Microbial Biology, University of California, Berkeley, California 94720, USA; Division of Environmental Genomics and Systems Biology, Lawrence Berkeley National Laboratory, Berkeley, California 94720, USA.
Funding
The work (proposals: 10.46936/10.25585/60007932, 10.46936/10.25585/60001051, and 10.46936/10.25585/60000843) conducted by the U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337), a DOE Office of Science User Facility, is supported by the Office of Science of The U.S. Department of Energy operated under Contract No. DE-AC02-05CH11231. Proteomics analyses were performed under the Facilities Integrating Collaborations for User Science Program Proposals 49262, 49840, 49960, and 50797 and used resources at the US DOE Joint Genome Institute and the Environmental Molecular Sciences Laboratory (EMSL; grid.436923.9), which are DOE Office of Science User Facilities.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408(6814): 796–815 [DOI] [PubMed] [Google Scholar]
- Armstrong J, Hickey G, Diekhans M, Fiddes IT, Novak AM, Deran A, Fang Q, Xie D, Feng S, Stiller J, et al. (2020) Progressive Cactus is a multiple-genome aligner for the thousand-genome era. Nature 587(7833): 246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O (2015) Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6(1): 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer PE, Golicz AA, Scheben A, Batley J, Edwards D (2020) Plant pan-genomes are the new reference. Nat Plants 6(8): 914–920 [DOI] [PubMed] [Google Scholar]
- Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27(2): 573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaby-Haas CE, Castruita M, Fitz-Gibbon ST, Kropat J, Merchant SS (2016) Ni induces the CRR1-dependent regulon revealing overlap and distinction between hypoxia and cu deficiency responses in Chlamydomonas reinhardtii. Metallomics 8(7): 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaby-Haas CE, Merchant SS (2019) Comparative and functional algal genomics. Annu Rev Plant Biol 70(1): 605–638 [DOI] [PubMed] [Google Scholar]
- Blaby IK, Blaby-Haas CE (2017) Genomics and functional genomics in Chlamydomonas reinhardtii. InHippler M, eds, Chlamydomonas: Molecular Genetics and Physiology. Springer, Berlin, pp 1–26 [Google Scholar]
- Blaby IK, Blaby-Haas CE, Tourasse N, Hom EF, Lopez D, Aksoy M, Grossman A, Umen J, Dutcher S, Porter M, et al. (2014) The Chlamydomonas genome project: a decade on. Trends Plant Sci 19(10): 672–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Agarkova I, Grimwood J, Kuo A, Brueggeman A, Dunigan DD, Gurnon J, Ladunga I, Lindquist E, Lucas S, et al. (2012) The genome of the polar eukaryotic microalga Coccomyxa subellipsoidea reveals traits of cold adaptation. Genome Biol 13(5): R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulouis A, Drapier D, Razafimanantsoa H, Wostrikoff K, Tourasse NJ, Pascal K, Girard-Bascou J, Vallon O, Wollman FA, Choquet Y (2015) Spontaneous dominant mutations in Chlamydomonas highlight ongoing evolution by gene diversification. Plant Cell 27(4): 984–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand H, Collins RL, Hanscom C, Rosenfeld JA, Pillalamarri V, Stone MR, Kelley F, Mason T, Margolin L, Eggert S, et al. (2015) Paired-duplication signatures mark cryptic inversions and other complex structural variation. Am J Hum Genet 97(1): 170–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA (2003) R/qtl: qTL mapping in experimental crosses. Bioinformatics 19(7): 889–890 [DOI] [PubMed] [Google Scholar]
- Cavaiuolo M, Kuras R, Wollman FA, Choquet Y, Vallon O (2017) Small RNA profiling in Chlamydomonas: insights into chloroplast RNA metabolism. Nucleic Acids Res 45(18): 10783–10799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaux-Jukic F, O'Donnell S, Craig RJ, Eberhard S, Vallon O, Xu Z (2021) Architecture and evolution of subtelomeres in the unicellular green alga Chlamydomonas reinhardtii. Nucleic Acids Res 49(13): 7571–7587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig RJ (2021) The evolutionary genomics of Chlamydomonas. PhD thesis. University of Edinburgh, Edinburgh, UK. 10.7488/era/1603 [DOI]
- Craig RJ, Böndel KB, Arakawa K, Nakada T, Ito T, Bell G, Colegrave N, Keightley PD, Ness RW (2019) Patterns of population structure and complex haplotype sharing among field isolates of the green alga Chlamydomonas reinhardtii. Mol Ecol 28(17): 3977–3993 [DOI] [PubMed] [Google Scholar]
- Craig RJ, Hasan AR, Ness RW, Keightley PD (2021a) Comparative genomics of Chlamydomonas. Plant Cell 33(4): 1016–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig RJ, Yushenova IA, Rodriguez F, Arkhipova IR (2021b) An ancient clade of Penelope-like retroelements with permuted domains is present in the green lineage and protists, and dominates many invertebrate genomes. Mol Biol Evol 38(11): 5005–5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross FR (2016) Tying down loose ends in the Chlamydomonas genome: functional significance of abundant upstream open reading frames. G3 (Bethesda) 6(2): 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross FR, Umen JG (2015) The Chlamydomonas cell cycle. Plant J 82(3): 370–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau DL, Popuri V, Opresko PL, Bohr VA (2014) Human RecQ helicases in DNA repair, recombination, and replication. Annu Rev Biochem 83(1): 519–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozet P, Navarro FJ, Willmund F, Mehrshahi P, Bakowski K, Lauersen KJ, Perez-Perez ME, Auroy P, Gorchs Rovira A, Sauret-Gueto S, et al. (2018) Birth of a photosynthetic chassis: a MoClo toolkit enabling synthetic biology in the microalga Chlamydomonas reinhardtii. ACS Synth Biol 7(9): 2074–2086 [DOI] [PubMed] [Google Scholar]
- Cui J, Zhang Z, Shao Y, Zhang K, Leng P, Liang Z (2015) Genome-wide identification, evolutionary, and expression analyses of histone H3 variants in plants. Biomed Res Int 2015: 341598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. (2021) Twelve years of SAMtools and BCFtools. Gigascience 10(2): giab008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DR (1972) Cell wall organisation in Chlamydomonas reinhardi. The role of extra-nuclear systems. Mol Gen Genet 115(4): 334–348 [DOI] [PubMed] [Google Scholar]
- Day A, Schirmer-Rahire M, Kuchka MR, Mayfield SP, Rochaix JD (1988) A transposon with an unusual arrangement of long terminal repeats in the green alga Chlamydomonas reinhardtii. EMBO J 7(7): 1917–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Hoff PL, Ferris P, Olson BJSC, Miyagi A, Geng S, Umen JG (2013) Species and population level molecular profiling reveals cryptic recombination and emergent asymmetry in the dimorphic mating locus of C. reinhardtii. PLoS Genet 9(8): e1003724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K, Hegemann P (2017) The form and function of channelrhodopsin. Science 357(6356): eaan5544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn A, Puchta H (2019) DNA Helicases as safekeepers of genome stability in plants. Genes (Basel) 10(12): 1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher SK (2014) The awesome power of dikaryons for studying flagella and basal bodies in Chlamydomonas reinhardtii. Cytoskeleton 71(2): 79–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher SK, Power J, Galloway RE, Porter ME (1991) Reappraisal of the genetic map of Chlamydomonas reinhardtii. J Hered 82(4): 295–301 [DOI] [PubMed] [Google Scholar]
- Engel BD, Schaffer M, Kuhn Cuellar L, Villa E, Plitzko JM, Baumeister W (2015) Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. eLife 4: e04889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauser F, Vilarrasa-Blasi J, Onishi M, Ramundo S, Patena W, Millican M, Osaki J, Philp C, Nemeth M, Salome PA, et al. (2022) Systematic characterization of gene function in the photosynthetic alga Chlamydomonas reinhardtii. Nat Genet 54(5): 705–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fédry J, Liu Y, Pehau-Arnaudet G, Pei J, Li W, Tortorici MA, Traincard F, Meola A, Bricogne G, Grishin NV, et al. (2017) The ancient gamete fusogen HAP2 is a eukaryotic class II fusion protein. Cell 168(5): 904–915.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ (1989) Characterization of a Chlamydomonas transposon, Gulliver, resembling those in higher-plants. Genetics 122(2): 363–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ, Armbrust EV, Goodenough UW (2002) Genetic structure of the mating-type locus of Chlamydomonas reinhardtii. Genetics 160(1): 181–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ, Goodenough UW (1994) The mating-type locus of Chlamydomonas reinhardtii contains highly rearranged DNA sequences. Cell 76(6): 1135–1145 [DOI] [PubMed] [Google Scholar]
- Ferris P, Olson BJ, De Hoff PL, Douglass S, Casero D, Prochnik S, Geng S, Rai R, Grimwood J, Schmutz J, et al. (2010) Evolution of an expanded sex-determining locus in Volvox. Science 328(5976): 351–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers JM, Hazzouri KM, Pham GM, Rosas U, Bahmani T, Khraiwesh B, Nelson DR, Jijakli K, Abdrabu R, Harris EH, et al. (2015) Whole-genome resequencing reveals extensive natural variation in the model green alga Chlamydomonas reinhardtii. Plant Cell 27(9): 2353–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman Rosenzweig ES, Xu B, Kuhn Cuellar L, Martinez-Sanchez A, Schaffer M, Strauss M, Cartwright HN, Ronceray P, Plitzko JM, Forster F, et al. (2017) The eukaryotic CO2-concentrating organelle is liquid-like and exhibits dynamic reorganization. Cell 171(1): 148–162.e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallaher SD, Craig RJ, Ganesan I, Purvine SO, McCorkle S, Grimwood J, Strenkert D, Davidi L, Roth MS, Jeffers TL, et al. (2021) Widespread polycistronic gene expression in green algae. Proc Natl Acad Sci U S A 118(7): e2017714118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallaher SD, Fitz-Gibbon ST, Glaesener AG, Pellegrini M, Merchant SS (2015) Chlamydomonas genome resource for laboratory strains reveals a mosaic of sequence variation, identifies true strain histories, and enables strain-specific studies. Plant Cell 27(9): 2335–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallaher SD, Fitz-Gibbon ST, Strenkert D, Purvine SO, Pellegrini M, Merchant SS (2018) High-throughput sequencing of the chloroplast and mitochondrion of Chlamydomonas reinhardtii to generate improved de novo assemblies, analyze expression patterns and transcript speciation, and evaluate diversity among laboratory strains and wild isolates. Plant J 93(3): 545–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gel B, Serra E (2017) Karyoploter: an R/bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics 33(19): 3088–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al. (2002) A draft sequence of the rice genome (Oryza sativa L. Ssp. japonica). Science 296(5565): 92–100 [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40(D1): D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin TJ, Poulter RT (2004) A new group of tyrosine recombinase-encoding retrotransposons. Mol Biol Evol 21(4): 746–759 [DOI] [PubMed] [Google Scholar]
- Gorres KL, Raines RT (2010) Prolyl 4-hydroxylase. Crit Rev Biochem Mol Biol 45(2): 106–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev IV, Hayes RD, Calhoun S, Kamel B, Wang A, Ahrendt S, Dusheyko S, Nikitin R, Mondo SJ, Salamov A, et al. (2021) Phycocosm, a comparative algal genomics resource. Nucleic Acids Res 49(D1): D1004–D1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman AR, Harris EE, Hauser C, Lefebvre PA, Martinez D, Rokhsar D, Shrager J, Silflow CD, Stern D, Vallon O, et al. (2003) Chlamydomonas reinhardtii at the crossroads of genomics. Eukaryot Cell 2(6): 1137–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK Jr, Hannick LI, Maiti R, Ronning CM, Rusch DB, Town CD, et al. (2003) Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res 31(19): 5654–5666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaji T, Kawai-Toyooka H, Uchimura H, Suzuki M, Noguchi H, Minakuchi Y, Toyoda A, Fujiyama A, Miyagishima S, Umen JG, et al. (2018) Anisogamy evolved with a reduced sex-determining region in volvocine green algae. Commun Biol 1(1): 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell KE, Collins S, Kazamia E, Purton S, Wheeler GL, Smith AG (2015) Fundamental shift in vitamin B12 eco-physiology of a model alga demonstrated by experimental evolution. ISME J 9(6): 1446–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey G, Paten B, Earl D, Zerbino D, Haussler D (2013) HAL: a hierarchical format for storing and analyzing multiple genome alignments. Bioinformatics 29(10): 1341–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyams J, Davies DR (1972) Induction and characterization of cell-wall mutants of Chlamydomonas reinhardi. Mutat Res 14(4): 381–389 [Google Scholar]
- Ito H, Gaubert H, Bucher E, Mirouze M, Vaillant I, Paszkowski J (2011) An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472(7341): 115–119 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Peluso P, Shi J, Liang T, Stitzer MC, Wang B, Campbell MS, Stein JC, Wei X, Chin CS, et al. (2017) Improved maize reference genome with single-molecule technologies. Nature 546(7659): 524–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo S, Kariyawasam T, Kim M, Jin E, Goodenough U, Lee JH (2022) Sex-linked deubiquitinase establishes uniparental transmission of chloroplast DNA. Nat Commun 13(1): 1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaina B (2004) Mechanisms and consequences of methylating agent-induced SCEs and chromosomal aberrations: a long road traveled and still a far way to go. Cytogenet Genome Res 104(1–4): 77–86 [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J (2003) The esterase and PHD domains in CR1-like non-LTR retrotransposons. Mol Biol Evol 20(1): 38–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathir P, LaVoie M, Brazelton WJ, Haas NA, Lefebvre PA, Silflow CD (2003) Molecular map of the Chlamydomonas reinhardtii nuclear genome. Eukaryot Cell 2(2): 362–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM (2013) MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30(4): 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskiaho K, Hieta R, Sormunen R, Myllyharju J (2007) Chlamydomonas reinhardtii has multiple prolyl 4-hydroxylases, one of which is essential for proper cell wall assembly. Plant Cell 19(1): 256–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KS, Kustu S, Inwood W (2006) Natural history of transposition in the green alga Chlamydomonas reinhardtii: use of the AMT4 locus as an experimental system. Genetics 173(4): 2005–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM (2017) Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27(5): 722–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19(9): 1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kück U, Choquet Y, Schneider M, Dron M, Bennoun P (1987) Structural and transcription analysis of two homologous genes for the P700 chlorophyll a-apoproteins in Chlamydomonas reinhardii: evidence for in vivo trans-splicing. EMBO J 6(8): 2185–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL (2004) Versatile and open software for comparing large genomes. Genome Biol 5(2): R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labadorf A, Link A, Rogers MF, Thomas J, Reddy AS, Ben-Hur A (2010) Genome-wide analysis of alternative splicing in chlamydomonas reinhardtii. Bmc Genomics 11(1): 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P (2019) Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47(W1): W256–W259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lex A, Gehlenborg N, Strobelt H, Vuillemot R, Pfister H (2014) Upset: visualization of intersecting sets. IEEE Trans Vis Comput Graph 20(12): 1983–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997
- Li H (2018) Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34(18): 3094–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Patena W, Fauser F, Jinkerson RE, Saroussi S, Meyer MT, Ivanova N, Robertson JM, Yue R, Zhang R, et al. (2019) A genome-wide algal mutant library and functional screen identifies genes required for eukaryotic photosynthesis. Nat Genet 51(4): 627–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Cliften PF, Dutcher SK (2018) MAPINS, a highly efficient detection method that identifies insertional mutations and complex DNA rearrangements. Plant Physiol 178(4): 1436–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Miller ML, Granas DM, Dutcher SK (2013) Whole genome sequencing identifies a deletion in protein phosphatase 2A that affects its stability and localization in Chlamydomonas reinhardtii. PLoS Genet 9(9): e1003841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Fang L, Yu G, Wang D, Xiao CL, Wang K (2019) Detection of DNA base modifications by deep recurrent neural network on Oxford nanopore sequencing data. Nat Commun 10(1): 2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Huang J, Sun X, Li J, Hu Y, Yu L, Liti G, Tian D, Hurst LD, Yang S (2018) Tetrad analysis in plants and fungi finds large differences in gene conversion rates but no GC bias. Nat Ecol Evol 2(1): 164–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Seetharam AS, Chougule K, Ou S, Swentowsky KW, Gent JI, Llaca V, Woodhouse MR, Manchanda N, Presting GG, et al. (2020) Gapless assembly of maize chromosomes using long-read technologies. Genome Biol 21(1): 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Cortegano E, Craig RJ, Chebib J, Balogun EJ, Keightley PD (2022) Rates and spectra of de novo structural mutation in Chlamydomonas reinhardtii. Genome Res. 10.1101/gr.276957.122 [DOI] [PMC free article] [PubMed]
- Lopez D, Hamaji T, Kropat J, De Hoff P, Morselli M, Rubbi L, Fitz-Gibbon S, Gallaher SD, Merchant SS, Umen J, et al. (2015) Dynamic changes in the transcriptome and methylome of Chlamydomonas reinhardtii throughout its life cycle. Plant Physiol 169(4): 2730–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Davis AJ (2021) Human RecQ helicases in DNA double-strand break repair. Front Cell Dev Biol 9: 640755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM (2021) BUSCO Update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol 38(10): 4647–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul JE, Lilly JW, Cui L, dePamphilis CW, Miller W, Harris EH, Stern DB (2002) The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. Plant Cell 14(11): 2659–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy SS, Kobayashi MC, Niyogi KK (2004) White mutants of Chlamydomonas reinhardtii are defective in phytoene synthase. Genetics 168(3): 1249–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20(9): 1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318(5848): 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RJ, Pfeiffer W, Schwartz T (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. In2010 Gateway Computing Environments Workshop (GCE). IEEE, pp 1–8. 10.1109/GCE.2010.5676129. [DOI] [Google Scholar]
- Moseley JL, Page MD, Alder NP, Eriksson M, Quinn J, Soto F, Theg SM, Hippler M, Merchant S (2002) Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14(3): 673–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navrátilová A, Koblížková A, Macas J (2008) Survey of extrachromosomal circular DNA derived from plant satellite repeats. BMC Plant Biol 8(1): 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert J, Gallaher SD, Lu Y, Strenkert D, Segal N, Barahimipour R, Fitz-Gibbon ST, Schroda M, Merchant SS, Bock R (2020) An epigenetic gene silencing pathway selectively acting on transgenic DNA in the green alga Chlamydomonas. Nat Commun 11(1): 6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngan CY, Wong CH, Choi C, Yoshinaga Y, Louie K, Jia J, Chen C, Bowen B, Cheng H, Leonelli L, et al. (2015) Lineage-specific chromatin signatures reveal a regulator of lipid metabolism in microalgae. Nat Plants 1(8): 15107. [DOI] [PubMed] [Google Scholar]
- Ni P, Huang N, Zhang Z, Wang DP, Liang F, Miao Y, Xiao CL, Luo F, Wang J (2019) Deepsignal: detecting DNA methylation state from nanopore sequencing reads using deep-learning. Bioinformatics 35(22): 4586–4595 [DOI] [PubMed] [Google Scholar]
- Ning J, Otto TD, Pfander C, Schwach F, Brochet M, Bushell E, Goulding D, Sanders M, Lefebvre PA, Pei J, et al. (2013) Comparative genomics in Chlamydomonas and Plasmodium identifies an ancient nuclear envelope protein family essential for sexual reproduction in protists, fungi, plants, and vertebrates. Genes Dev 27(10): 1198–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoselov SV, Rao M, Onoshko NV, Zhi H, Kryukov GV, Xiang Y, Weeks DP, Hatfield DL, Gladyshev VN (2002) Selenoproteins and selenocysteine insertion system in the model plant cell system, Chlamydomonas reinhardtii. EMBO J 21(14): 3681–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell S, Chaux F, Fischer G (2020) Highly contiguous nanopore genome assembly of Chlamydomonas reinhardtii CC-1690. Microbiol Resour Announc 9(37): e00726–00720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell S, Fischer G (2020) MUM&Co: accurate detection of all SV types through whole-genome alignment. Bioinformatics 36(10): 3242–3243 [DOI] [PubMed] [Google Scholar]
- Ozawa SI, Cavaiuolo M, Jarrige D, Kuras R, Rutgers M, Eberhard S, Drapier D, Wollman FA, Choquet Y (2020) The OPR protein MTHI1 controls the expression of two different subunits of ATP synthase CFo in Chlamydomonas reinhardtii. Plant Cell 32(4): 1179–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne ZL, Penny GM, Turner TN, Dutcher SK (2022) A gap-free genome assembly of Chlamydomonas reinhardtii and detection of translocations induced by CRISPR-mediated mutagenesis. Plant Commun. 10.1016/j.xplc.2022.100493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Alegre M, Dubus A, Fernandez E (2005) REM1, A new type of long terminal repeat retrotransposon in Chlamydomonas reinhardtii. Mol Cell Biol 25(23): 10628–10638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petracek ME, Lefebvre PA, Silflow CD, Berman J (1990) Chlamydomonas telomere sequences are A + T-rich but contain three consecutive G–C base pairs. Proc Natl Acad Sci U S A 87(21): 8222–8226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippsen GS, Avaca-Crusca JS, Araujo APU, DeMarco R (2016) Distribution patterns and impact of transposable elements in genes of green algae. Gene 594(1): 151–159 [DOI] [PubMed] [Google Scholar]
- Porter ME, Knott JA, Myster SH, Farlow SJ (1996) The dynein gene family in Chlamydomonas reinhardtii. Genetics 144(2): 569–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D, Mets L (2002) Plant centromere functions defined by tetrad analysis and artificial chromosomes. Plant Physiol 129(2): 421–422 [Google Scholar]
- Prochnik SE, Umen J, Nedelcu AM, Hallmann A, Miller SM, Nishii I, Ferris P, Kuo A, Mitros T, Fritz-Laylin LK, et al. (2010) Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 329(5988): 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pröschold T, Harris EH, Coleman AW (2005) Portrait of a species: Chlamydomonas reinhardtii. Genetics 170(4): 1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6): 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj-Kumar PK, Vallon O, Liang C (2017) In silico analysis of the sequence features responsible for alternatively spliced introns in the model green alga Chlamydomonas reinhardtii. Plant Mol Biol 94(3): 253–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle NC, Elgin SCR (2018) The Drosophila dot chromosome: where genes flourish amidst repeats. Genetics 210(3): 757–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29(1): 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhrig S, Dorn A, Enderle J, Schindele A, Herrmann NJ, Knoll A, Puchta H (2018) The RecQ-like helicase HRQ1 is involved in DNA crosslink repair in Arabidopsis in a common pathway with the Fanconi anemia-associated nuclease FAN1 and the postreplicative repair ATPase RAD5A. New Phytol 218(4): 1478–1490 [DOI] [PubMed] [Google Scholar]
- Roth MS, Cokus SJ, Gallaher SD, Walter A, Lopez D, Erickson E, Endelman B, Westcott D, Larabell CA, Merchant SS, et al. (2017) Chromosome-level genome assembly and transcriptome of the green alga Chromochloris zofingiensis illuminates astaxanthin production. Proc Natl Acad Sci U S A 114(21): E4296–E4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Ruano FJ, López-León MD, Cabrero J, Camacho JPM (2016) High-throughput analysis of the satellitome illuminates satellite DNA evolution. Sci Rep 6(1): 28333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymarquis LA, Handley JM, Thomas M, Stern DB (2005) Beyond complementation. Map-based cloning in Chlamydomonas reinhardtii. Plant Physiol 137(2): 557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas-Giegé T, Cavaiuolo M, Cognat V, Ubrig E, Remacle C, Duchene AM, Vallon O, Marechal-Drouard L (2017) Polycytidylation of mitochondrial mRNAs in Chlamydomonas reinhardtii. Nucleic Acids Res 45(22): 12963–12973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé PA, Merchant SS (2019) A series of fortunate events: introducing Chlamydomonas as a reference organism. Plant Cell 31(8): 1682–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell RA, Lefebvre PA (1993) Isolation of the Chlamydomonas regulatory gene NIT2 by transposon tagging. Genetics 134(3): 737–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu S, Goodstein D, Rokhsar D (2013) PERTRAN: genome-guided RNA-seq read assembler. OSTIgov: US Department of Energy—Office of Scientific and Technical Information [Google Scholar]
- Smit AFA, Hubley R, Green P (2013–2015) RepeatMasker Open-4.0. http://www.repeatmasker.org
- Smith DR, Craig RJ (2021) Does mitochondrial DNA replication in Chlamydomonas require a reverse transcriptase? New Phytol 229(3): 1192–1195 [DOI] [PubMed] [Google Scholar]
- Smith DR, Lee RW (2009) Nucleotide diversity of the Chlamydomonas reinhardtii plastid genome: addressing the mutational-hazard hypothesis. BMC Evol Biol 9(1): 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EF, Lefebvre PA (1997) PF20 Gene product contains WD repeats and localizes to the intermicrotubule bridges in Chlamydomonas flagella. Mol Biol Cell 8(3): 455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedasyam A, Plott C, Shadkhawat Hossain M, Lovell JT, Grimwood J, Jenkins JW, Daum C, Barry K, Carlson J, Shu Set al. (2022) JGI Plant Gene Atlas: an updateable transcriptome resource to improve structural annotations and functional gene descriptions across the plant kingdom. Biorxiv. 10.1101/2022.09.30.510380 [DOI]
- Sterken MG, Snoek LB, Kammenga JE, Andersen EC (2015) The laboratory domestication of Caenorhabditis elegans. Trends Genet 31(5): 224–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strenkert D, Yildirim A, Yan J, Yoshinaga Y, Pellegrini M, O'Malley RC, Merchant SS, Umen JG (2022) The landscape of Chlamydomonas histone H3 lysine 4 methylation reveals both constant features and dynamic changes during the diurnal cycle. Plant J 112(2): 352–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumper M, Hallmann A (1998) Biochemistry of the extracellular matrix of Volvox. Int Rev Cytol 180: 51–85 [DOI] [PubMed] [Google Scholar]
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ (2016) W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 44(W1): W232–W235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin F, Cross FR (2014) A microbial avenue to cell cycle control in the plant superkingdom. Plant Cell 26(10): 4019–4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin F, Cross FR (2016) Patching holes in the Chlamydomonas genome. G3 (Bethesda) 6(7): 1899–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki N, Nishii I (2008) Idaten is a new cold-inducible transposon of Volvox carteri that can be used for tagging developmentally important genes. Genetics 180(3): 1343–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahrenholz C, Riemen G, Pratje E, Dujon B, Michaelis G (1993) Mitochondrial DNA of Chlamydomonas reinhardtii: the structure of the ends of the linear 15.8-kb genome suggests mechanisms for DNA replication. Curr Genet 24(3): 241–247 [DOI] [PubMed] [Google Scholar]
- Wang SC, Schnell RA, Lefebvre PA (1998) Isolation and characterization of a new transposable element in Chlamydomonas reinhardtii. Plant Mol Biol 38(5): 681–687 [DOI] [PubMed] [Google Scholar]
- Wiedemann G, van Gessel N, Kochl F, Hunn L, Schulze K, Maloukh L, Nogue F, Decker EL, Hartung F, Reski R (2018) Recq helicases function in development, DNA repair, and gene targeting in Physcomitrella patens. Plant Cell 30(3): 717–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BA, Slamovits CH, Patron NJ, Fast NM, Keeling PJ (2005) A high frequency of overlapping gene expression in compacted eukaryotic genomes. Proc Natl Acad Sci U S A 102(31): 10936–10941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woessner JP, Goodenough UW (1994) Volvocine cell walls and their constituent glycoproteins: an evolutionary perspective. Protoplasma 181(1–4): 245–258 [Google Scholar]
- Wright BW, Molloy MP, Jaschke PR (2022) Overlapping genes in natural and engineered genomes. Nat Rev Genet 23(3): 154–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Nacu S (2010) Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26(7): 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK (2005) GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21(9): 1859–1875 [DOI] [PubMed] [Google Scholar]
- Wyatt MD, Pittman DL (2006) Methylating agents and DNA repair responses: methylated bases and sources of strand breaks. Chem Res Toxicol 19(12): 1580–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao CL, Chen Y, Xie SQ, Chen KN, Wang Y, Han Y, Luo F, Xie Z (2017) MECAT: fast mapping, error correction, and de novo assembly for single-molecule sequencing reads. Nat Methods 14(11): 1072–1074 [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Hamaji T, Kawai-Toyooka H, Matsuzaki R, Takahashi F, Nishimura Y, Kawachi M, Noguchi H, Minakuchi Y, Umen JG, et al. (2021) Three genomes in the algal genus Volvox reveal the fate of a haploid sex-determining region after a transition to homothallism. Proc Natl Acad Sci U S A 118(21): e2100712118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R, Obbineni JM, Alford LM, Ide T, Owa M, Hwang J, Kon T, Inaba K, James N, King SM, et al. (2017) Chlamydomonas DYX1C1/PF23 is essential for axonemal assembly and proper morphology of inner dynein arms. PLoS Genet 13(9): e1006996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Guo C, Sutharzan S, Li P, Echt CS, Zhang J, Liang C (2014) Genome-wide analysis of tandem repeats in plants and green algae. G3 (Bethesda) 4(1): 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.