Summary
Background
Tongue images (the colour, size and shape of the tongue and the colour, thickness and moisture content of the tongue coating), reflecting the health state of the whole body according to the theory of traditional Chinese medicine (TCM), have been widely used in China for thousands of years. Herein, we investigated the value of tongue images and the tongue coating microbiome in the diagnosis of gastric cancer (GC).
Methods
From May 2020 to January 2021, we simultaneously collected tongue images and tongue coating samples from 328 patients with GC (all newly diagnosed with GC) and 304 non-gastric cancer (NGC) participants in China, and 16 S rDNA was used to characterize the microbiome of the tongue coating samples. Then, artificial intelligence (AI) deep learning models were established to evaluate the value of tongue images and the tongue coating microbiome in the diagnosis of GC. Considering that tongue imaging is more convenient and economical as a diagnostic tool, we further conducted a prospective multicentre clinical study from May 2020 to March 2022 in China and recruited 937 patients with GC and 1911 participants with NGC from 10 centres across China to further evaluate the role of tongue images in the diagnosis of GC. Moreover, we verified this approach in another independent external validation cohort that included 294 patients with GC and 521 participants with NGC from 7 centres. This study is registered at ClinicalTrials.gov, NCT01090362.
Findings
For the first time, we found that both tongue images and the tongue coating microbiome can be used as tools for the diagnosis of GC, and the area under the curve (AUC) value of the tongue image-based diagnostic model was 0.89. The AUC values of the tongue coating microbiome-based model reached 0.94 using genus data and 0.95 using species data. The results of the prospective multicentre clinical study showed that the AUC values of the three tongue image-based models for GCs reached 0.88–0.92 in the internal verification and 0.83–0.88 in the independent external verification, which were significantly superior to the combination of eight blood biomarkers.
Interpretation
Our results suggest that tongue images can be used as a stable method for GC diagnosis and are significantly superior to conventional blood biomarkers. The three kinds of tongue image-based AI deep learning diagnostic models that we developed can be used to adequately distinguish patients with GC from participants with NGC, even early GC and precancerous lesions, such as atrophic gastritis (AG).
Funding
The National Key R&D Program of China (2021YFA0910100), Program of Zhejiang Provincial TCM Sci-tech Plan (2018ZY006), Medical Science and Technology Project of Zhejiang Province (2022KY114, WKJ-ZJ-2104), Zhejiang Provincial Research Center for Upper Gastrointestinal Tract Cancer (JBZX-202006), Natural Science Foundation of Zhejiang Province (HDMY22H160008), Science and Technology Projects of Zhejiang Province (2019C03049), National Natural Science Foundation of China (82074245, 81973634, 82204828), and Chinese Postdoctoral Science Foundation (2022M713203).
Abbreviations: AFP, alpha fetoprotein; AG, atrophic gastritis; AI, artificial intelligence; APINet, attentive pairwise interaction neural network; AUC, area under the curve; BC, breast cancer; CA, carbohydrate antigen; CEA, carcinoembryonic antigen; CRC, colorectal cancer; DT, decision tree learning; EC, esophageal cancer; GC, gastric cancer; HBPC, hepatobiliary pancreatic carcinoma; HC, healthy control; KNN, K-nearest neighbours; LC, lung cancer; NGC, non-gastric cancers; PCoA, principal coordinates analysis; SG, superficial gastritis; SVM, support vector machine; TCM, traditional Chinese medicine; TransFG, transformer architecture for fine-grained recognition
Keywords: Gastric cancer, Tongue images, Artificial intelligence, Traditional Chinese medicine, Tongue coating microbiome
Research in context.
Evidence before this study
Gastric cancer (GC) is one of the most lethal types of cancer across all countries and ethnicities. The diagnosis and screening of GC still rely on gastroscopy, but its application is greatly limited because of its invasiveness, high cost and the need for professional endoscopists. In addition, liquid biopsy techniques, such as the analysis of circulating tumour cells (CTCs), circulating tumour DNA (ctDNA) and exosomes, are difficult to be widely use in the diagnosis and screening of GC due to insufficient accuracy and high price. Tongue images, which reflect the health state of the whole body according to the theory of traditional Chinese medicine (TCM), have been widely used in China for thousands of years. For the first time, we applied artificial intelligence (AI) deep learning to evaluate the diagnostic value of tongue images in the context of GC. PubMed searches for those terms related to this approach performed up to September, 2022, with no language restrictions, did not reveal any publications in this area.
Added value of this study
We conducted a prospective multicentre clinical study and recruited participants from 10 centres across China to further evaluate the role of tongue images in the diagnosis of GC. Moreover, we verified this approach in another independent external validation cohort from 7 centres. In addition, we applied three completely different deep learning models to evaluate the value of tongue images for GC diagnosis and screening to reduce the deviation of conclusions caused by the model differences. The results show that tongue images can be used as a stable tool for the diagnosis of GC, and the results were independent of diet, region, or the type of AI deep learning model.
Implications of all the available evidence
For the first time, we explored the role of tongue images in the diagnosis of cancer, especially GC. We found that tongue images can be used as a stable means of GC diagnosis and are significantly superior to conventional blood biomarkers. Because of the convenience of tongue image collection, we believe that tongue images can be an effective, noninvasive method to perform an auxiliary diagnosis anywhere. Considering the substantial burden of GCs in China and across the globe, we believe that tongue images, in combination with the widespread use of AI deep learning approaches, might be the most cost-efficient, noninvasive and acceptable approach for the screening and early detection of GCs, which will also have considerable socioeconomic effects.
Introduction
According to the latest data, gastric cancer (GC) is the fourth leading cause of cancer-related death worldwide.1 In 2020, there were an estimated 1.09 million new GC cases and 0.77 million GC-related deaths. Among them, an estimated 0.48 million new cases and 0.37 million deaths have occurred in China, accounting for approximately half of the world's cases.1 The diagnosis and screening of GC still rely on gastroscopy, but its application is greatly limited because of its invasiveness, high cost and the need for professional endoscopists. In addition, due to the lack of specific symptoms during the early stage and the poor specificity and sensitivity of clinical disease markers, more than 60% of patients were found to exhibit local or distant metastasis at the time of diagnosis.2 The 5-year survival rate of patients with localized early-stage GC was more than 60%, while that of patients with local and distant metastasis decreased significantly to 30% and 5%, respectively.2 Therefore, there is an urgent need for new GC diagnosis or screening methods to improve the rate of early-stage diagnosis and improve the prognosis of this population.
Traditional Chinese medicine (TCM) is a medical science and cultural heritage empirically applied and reserved by Chinese people for thousands of years.3 Tongue image diagnosis is one of the most important components of TCM in the diagnosis of diseases. The theory of TCM suggests that changes detected using tongue images (the colour, size and shape of the tongue and the colour, thickness and moisture content of the tongue coating) can reflect the health state of the human body, which is especially closely related to gastric diseases.4, 5, 6 Recent studies have shown that changes in tongue images and tongue coatings are closely related to the oral/tongue coating microbiome.7 Additionally, many studies have confirmed that the oral/tongue coating microbial group has good diagnostic value for pancreatic cancer,8 liver carcinoma,9 colorectal cancer10 and other tumours, as well as gastritis,11 rheumatoid arthritis,12 chronic hepatitis B13 and other diseases. However, the relationship between the changes in tongue images or the tongue coating microbiome and GC and the value of tongue images or the tongue coating microbiome in the diagnosis and screening of GC have not been studied.
Artificial intelligence (AI) may be useful in screening, diagnosing, and treating various diseases by enabling the accurate analysis of diagnostic clinical images, the identification of therapeutic targets, and the processing large of datasets.14,15 Cheung et al. developed a deep learning system to evaluate the risk of cardiovascular disease by measuring the calibre of retinal vessels, which can be used to effectively predict the risk of cardiovascular disease.16 Takenaka et al. developed a deep neural network for the evaluation of endoscopic images from patients with ulcerative colitis that identified those in endoscopic remission with 90.1% accuracy and those in histologic remission with 92.9% accuracy.17 At present, the application of AI in tongue image diagnosis of TCM mainly focuses on the standardization of tongue feature extraction to eliminate the differences caused by human interpretation.18, 19, 20, 21 For the first time, we applied AI deep learning to establish diagnostic models of GC based on tongue images or the tongue coating microbiome and evaluated their value in GC diagnosis, which could provide a scientific basis for the tongue image diagnosis theory of TCM.
In this study, we simultaneously collected tongue images and tongue coating samples from 328 patients with GC and 304 non-gastric cancer (NGC) participants, and 16 S rDNA was used to characterize the microbiome of these tongue coating samples. Then, AI deep learning models were established to evaluate the value of tongue images and the tongue coating microbiome in the diagnosis of GC. In addition, considering that tongue imaging is more convenient and cost-efficient as a diagnostic tool, we conducted a prospective multicentre clinical study and recruited 937 patients with GC and 1911 participants with NGC from 10 centres across China to further evaluate the role of tongue images in the diagnosis of GC. Moreover, 294 patients with GC and 521 participants with NGC from 7 centres were recruited for independent external validation. Finally, we recruited patients with oesophageal cancer, liver cancer, colorectal cancer and lung cancer to evaluate the differential diagnostic value of the model.
Methods
Clinical specimens
From May 2020 to January 2021, tongue images and tongue coating samples were simultaneously collected from 328 patients with GC and 304 participants with NGC, including 155 healthy controls (HCs) and 149 with atrophic gastritis (AG) in the Zhejiang Cancer Hospital and the First Affiliated Hospital of Zhejiang Chinese Medical University. In addition, from May 2020 to March 2022, we conducted a prospective multicentre clinical study and recruited 937 patients with GC and 1911 participants with NGC from 10 centres across China to further evaluate the role of tongue images in the diagnosis of GC. Moreover, we verified this approach in another independent external validation cohort, including 294 patients with GC and 521 participants with NGC from 7 centres. The inclusion and exclusion criteria used for all patients with GC and participants with NGC were the same as previously described. HCs, superficial gastritis (SG), and AG were confirmed by gastroscopy and pathology. Finally, we recruited 104 patients with oesophageal cancer (EC), 134 patients with hepatobiliary pancreatic carcinoma (HBPC), 106 patients with colorectal cancer (CRC) and 184 patients with lung cancer (LC) from Zhejiang Cancer Hospital to evaluate the differential diagnostic value of the model. The inclusion and exclusion criteria are detailed in the Supplementary Materials.
The study was approved by the centralized ethics board used by 17 of the participating centres (IRB-2019-56) and all patients provided written informed consent. The informed consent form clearly informs the patient that all clinical information such as tongue images, age, sex, TNM staging and so on will be used for publication by the investigator, and the participant agrees and is approved by the Ethics Committee.
Sample size calculation
Assuming a prevalence of 0.3 and a sample sensitivity of 0.8, the sample size needed for a two-sided 95% sensitivity confidence interval with a width of at most 0.07, is 1764. Assuming a prevalence of 0.3 and a sample specificity of 0.8, the sample size needed for a two-sided 95% specificity confidence interval with a width of at most 0.07, is 756. The whole table sample size required so that both confidence intervals have widths less than 0.07, is 1764, the larger of the two sample sizes.22,23 Considering that the larger the sample size is, the more accurate the results will be. In order to make our results more accurate, we actually recruited 3663 samples, which is larger than the calculated sample size (1764).
DNA extraction, library construction and sequencing
Microbial DNA was extracted using the E.Z.N.A. Tissue DNA Kit (D3396-01; Omega, Norcross, Georgia, USA) following the manufacturer's instructions as described previously. The AxyPrep PCR Clean-up Kit (AP-PCR-500G; Corning, NY, USA) was used to separate, extract and purify the PCR products, and the products were quantified using Quant-iT PicoGreen dsDNA Reagent (P7581, Thermo Scientific, Waltham, MA, USA). After quality determination, libraries that passed quality control were sequenced with a NovaSeq sequencer for 2 × two terminal sequencing of 250 bp at LC-Bio Co., Ltd.
Three tongue image-based AI deep learning models
Three different AI deep learning models, including the attentive pairwise interaction neural network (APINet) model, transformer architecture for fine-grained recognition (TransFG) model and DeepLabV3+ model, were established to evaluate the value of tongue images in the diagnosis of GC. APINet24 consists of three parts: the feature extraction module, the feature selection module and the classification module. In practical applications, only by fully comparing a pair of samples can we discover the commonalities and characteristics. Following guidelines for the use of TransFG,25 we split the tongue images into a sequence of n small flattened patches. We cut the images into nonoverlapping square regions. In this way, the deep learning model could be used to choose the key patches that facilitate classification. The input patches were mapped to a latent embedding space after the linear projection module. We could use semantic segmentation for image classification because each image contains only one tongue. Another deep learning model, DeepLabV3+, was used for tongue image classification.26 As with most segmentation models, the segmentation results were generated by downsampling the input images and upsampling the deep features. The three tongue image-based models all had a 50% probability as a cut-off value. If the sample probability was greater than 50%, the result was considered positive; otherwise, it was considered negative. The deep learning environments contained cuda-11.1 and pytorch-1.9.0, and we used Python 3 to implement the models and train them on NVIDIA-A100. The detailed construction steps used to establish the three models are shown in the supplementary materials.
Three blood tumour marker-based machine learning models and two fusion models
We used three typical methods in machine learning, including decision tree learning (DT), support vector machines (SVMs) and k-nearest neighbours (KNN) models, for the recognition of GC according to blood indicators. Then, based on the APINet model and the TransFG model, we designed two fusion models using blood indicators and tongue images called API fusion and transfusion fusion, respectively. The prediction of each case depended on both the characteristics of the tongue images and the levels of blood indicators. The detailed construction steps used to generate the three blood tumour marker-based and two fusion models are also shown in the supplementary materials.
Tongue microbiome evaluation (TME) model
We also designed an TME model based on the tongue coating microbiome to evaluate the value of the tongue microbiome in the diagnosis of GC. The detailed construction steps used to establish the TME model are also shown in the supplementary materials.
Statistical analysis
All statistical analyses were performed using SPSS 23.0 software (SPSS Inc., Chicago, IL, USA). Independent sample t-test was used to analyze the measurement data such as age, BMI and blood tumor indicators of the population cohort. And the counting data such as sex, smoking and alcohol consumption were tested by Chi-square test. The count data were analysed using the chi-square test. Results with P < 0.05 were considered statistically significant.
This study is registered at ClinicalTrials.gov, NCT01090362.
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
Both tongue images and the tongue coating microbiome can be used as diagnostic tools for GC
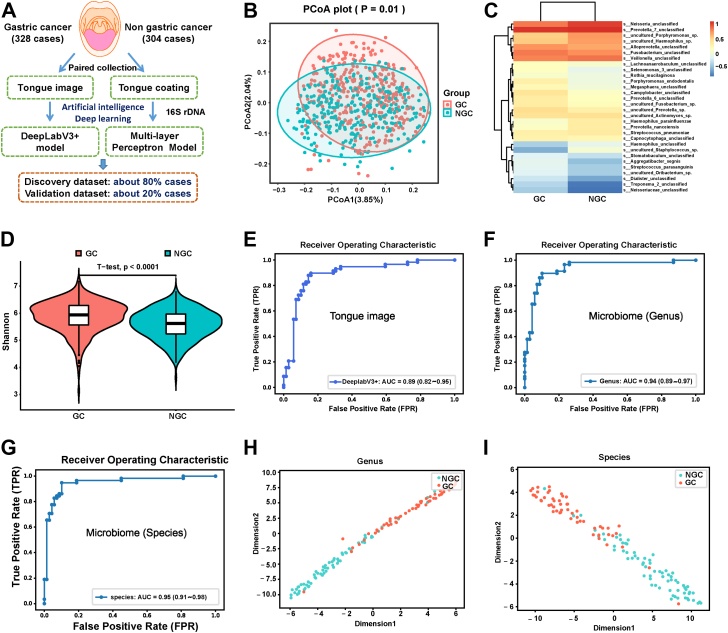
As shown in Fig. 1A, we established AI diagnostic models based on tongue images and the tongue coating microbiome to evaluate the value of tongue images and the tongue coating microbiome in the diagnosis of GC. The general clinical information, such as age, sex, smoking status, and drinking status, was well matched between the two groups (Table S1), and more detailed information is shown in Tables S2 and S3.
Fig. 1.
Both tongue images and the tongue coating microbiome can be used as diagnostic tools for gastric cancer (GC). (A) Flow diagram of the study design. We simultaneously collected tongue images and tongue coating samples from 328 patients with GC and 304 non-gastric cancer (NGC) participants, and 16 S rDNA was used to characterize the microbiome of these tongue coating samples. Then, artificial intelligence (AI) deep learning models were established to evaluate the value of tongue images and the tongue coating microbiome in the diagnosis of GC. Approximately 80% of the cases were used as the training dataset, and approximately 20% of the cases were used as the validation dataset. (B) Principal coordinate analysis (PCoA) showed that there were significant differences in the microbiome between GCs and NGCs. (C) Heatmap of the top 30 species that differed between patients with GC and participants with NGC. (D) The species richness in patients with GC was significantly higher than that in participants with NGC according to Simpson index analysis. (E) The ROC curve and AUC of the internal validation of the DeepLabV3+ model based on the tongue images. (F–G) The ROC curve and AUC of the internal validation of the tongue microbiome evaluation (TME) model based on the tongue coating microbiome at the genus and species levels. (H–I) The distribution of patients with GC and participants with NGC is displayed at the genus and species levels in the internal verification.
As shown in Fig. 1B, principal coordinate analysis (PCoA),27 PCoA 1 and PCoA 2 are two principal coordinate components. Each point in the figure represents a sample, the red points represent the sample of GC, and the green points represent the sample of NGC, which showed that there were significant differences in the microbiome between GCs and NGCs. The heatmap between patients with GC and participants with NGC also indicated a significant difference between them (Fig. 1C). Regarding the alpha diversity, the species richness in patients with GC was significantly higher than that in participants with NGC (Fig. 1D, p < 0.0001, estimated by the Shannon index), indicating a higher number of species in the tongue coating of patients with GC. We observed that 28 of the top 50 operational taxonomic units (OTUs) (28/50, 56%) showed significant abundance differences between patients with GC and participants with NGC. Among them, 21 OTUs (42%) were upregulated and 7 OTUs (14%) were downregulated in patients with GC compared with participants with NGC (Fig. S1A and Table S4). For example, the abundance of s__Alloprevotella_unclassified increased gradually, while the abundance of s__Veillonella_unclassified decreased gradually during the progression from normal, precancerous, early-stage GC, and late-stage GC. Furthermore, 7 OTUs showed significant alterations across GC stage I to GC stage IV; for example, the abundance of s__Alloprevotella_unclassified increased gradually, while the abundance of s__Veillonella_unclassified decreased gradually during the progression from stage 1 to stage IV in GC (Fig. S1B). These results suggested that the changes in tongue coating microorganisms are closely related to the occurrence and development of GC and may be used as promising markers for the diagnosis of GC.
In addition, the area under the curve (AUC) value of tongue image-based model for the diagnosis of GC was 0.89 (Fig. 1E). The sensitivity, specificity and accuracy were shown in Table S5. Moreover, we established an TME model based on the tongue coating microbiome, and the observed the AUC values were 0.94 at the genus level (Fig. 1F) and 0.95 at the species level (Fig. 1G). The sensitivity, specificity and accuracy were also shown in Table S5. The output diagram of the TME model showed that patients with GC and participants with NGC could be clearly distinguished at the genus and species levels (Fig. 1H and I). In addition, the TME model can well distinguish patients with GC from participants with NGC at the levels of family and order levels (Fig. S2 and Table S5).
Tongue imaging is a stable noninvasive diagnostic tool for GC
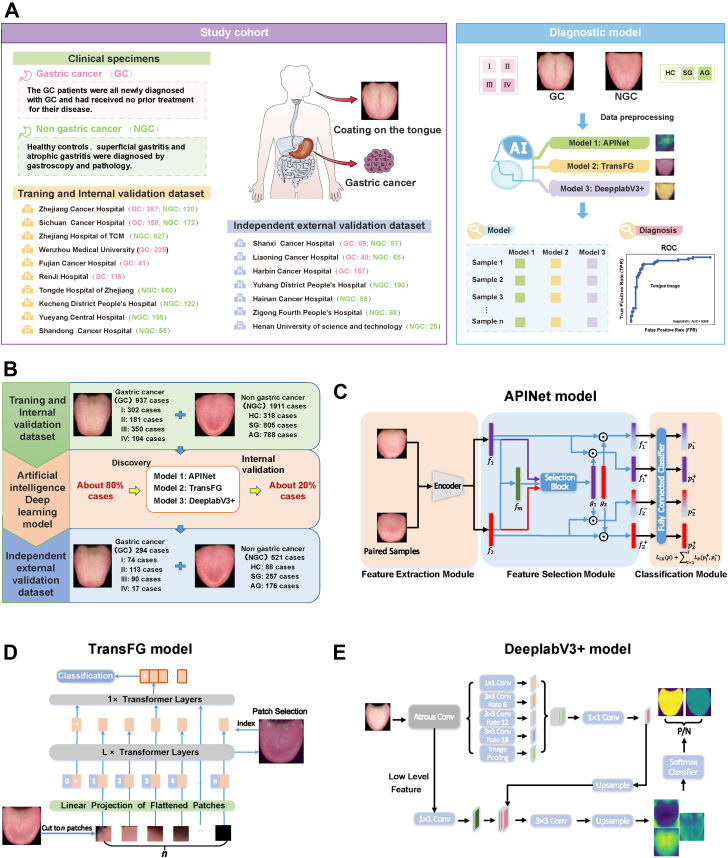
Considering that tongue imaging as a diagnostic tool is more convenient and cost-efficient than the tongue coating microbiome, we conducted a prospective multicentre clinical study to further evaluate the stability of tongue imaging in the diagnosis of GC. To eliminate the influence of regional, dietary, and centre differences on the study, we conducted a nationwide multicentre clinical study that included 17 centres. As shown in Fig. 2A and B, we recruited 937 patients with GC and 1911 participants with NGC from 10 centres to establish 3 tongue image-based AI diagnostic models. Approximately 80% of the participants were used as the training dataset, and approximately 20% of the participants were used as the internal validation dataset. In addition, 294 patients with GC and 521 participants with NGC from 7 centres were recruited as an independent external validation dataset to verify the value of the three diagnostic models.
Fig. 2.
This was a prospective multicentre clinical study. (A–B) Flow diagram of study design: We recruited 937 patients with GC and 1911 participants with NGC from 10 centres to establish 3 tongue image-based artificial intelligence diagnostic models. Approximately 80% of the cases were used as the training dataset, and approximately 20% of the cases were used as the internal validation dataset. In addition, 294 patients with GC and 521 participants with NGC from 7 centres were recruited as the independent external validation dataset to verify the value of the three diagnostic models. (C) The method used to establish model 1 (APINet model) and its process. (D) The method used to establish model 2 (TransFG model) and its process. (E) The method used to establish model 3 (DeepLabV3+ model) and its process.
In addition to collecting tongue images obtained from all the participants, we collected all clinical information, including age, sex, height, weight, family history, smoking status, drinking status, TNM staging, blood tumour markers and so on. As shown in Table 1, there were 937 patients with GC and 1911 participants with NGC in the training and internal validation datasets, including 288 patients with TNM stage I GC, 180 patients with TNM stage II GC, 359 patients with TNM stage III GC, 110 patients with TNM stage IV GC, 448 HCs, 701 patients with SG, and 762 patients with AG. More detailed information is shown in Tables S6 and S7. As shown in Table 2, the independent external validation dataset included 74 patients with TNM stage I GC, 113 patients with TNM stage II GC, 90 patients with TNM stage III GC, 17 patients with TNM stage IV GC, 88 HCs, 257 patients with SG, and 176 patients with AG. More detailed information is shown in Tables S8 and S9. The general patient information, such as age, sex, body mass index (BMI), smoking and drinking status, was well matched between the GC and NGC groups in the training, internal validation and independent external validation datasets (Table 1, Table 2). In addition, the levels of blood markers, such as carcinoembryonic antigen (CEA), carbohydrate antigen 424 (CA424), CA724, CA125, CA19-9, CA50, alpha-fetoprotein (AFP) and ferritin were significantly higher in patients with GC than in those with NGC (Table 1, Table 2).
Table 1.
Clinical characteristics of the GC and NGC participants in the training and internal validation datasets.
| Clinical indexes | Training and internal validation dataset |
||
|---|---|---|---|
| GC (n = 937) | NGC (n = 1911) | P value | |
| Subgroup information | TNM stage I = 288; II = 180; III = 359; IV = 110 | HC = 448; SG = 701; AG = 762 | N/A |
| Age (Years) | 63.33 ± 11.19 | 62.80 ± 8.45 | 0.21 |
| Sex (Female/Male) | 299/638 | 653/1258 | 0.23 |
| BMI (kg/m2) | 22.53 ± 3.19 | 22.67 ± 3.10 | 0.26 |
| Smoking (Yes/No/Unknown) | 244/652/41 | 526/1385/0 | 0.87 |
| Drinking (Yes/No/Unknown) | 314/582/41 | 614/1297/0 | 0.13 |
| HP (Positive/Negative/Unknown) | 167/158/612 | 256/235/1420 | 0.83 |
| Family history (Yes/No/Unknown) | 89/741/107 | N/A | N/A |
| Pathological type (Adenocarcinoma/Others/Unknown) | 860/57/20 | N/A | N/A |
| Tumour size (≤5 cm/>5 cm/Unknown) | 492/274/171 | N/A | N/A |
| Tumour location (U/M/L/W/Unknown) | 149/261/453/21/53 | N/A | N/A |
| Grade of differentiation (High/Medium + Medium-low/Low/Undifferentiated/Unknown) | 43/319/507/4/64 | N/A | N/A |
| TNM type (I/II/III/IV) | 288/180/359/110 | N/A | N/A |
| HER2 (0/1/2/3/Unknown) | 326/114/135/49/313 | N/A | N/A |
| MMR (dMMR/pMMR/Unknown) | 46/514/377 | N/A | N/A |
| Lauren type (Intestinal/Diffuse/Mixed/Unknown) | 93/86/60/698 | N/A | N/A |
| Nerve invasion (Yes/No/Unknown) | 200/205/532 | N/A | N/A |
| Vascular tumour thrombus (Yes/No/Unknown) | 206/202/529 | N/A | N/A |
| CEA (ng/ml) | (n = 904) 11.86 ± 78.78 |
(n = 1096) 2.00 ± 1.82 |
<0.0001 |
| CA242 (U/ml) | (n = 605) 24.01 ± 94.83 |
(n = 814) 6.75 ± 7.53 |
<0.0001 |
| CA724 (U/ml) | (n = 651) 13.24 ± 44.16 |
(n = 775) 2.59 ± 1.97 |
<0.0001 |
| AFP (ng/ml) | (n = 901) 14.95 ± 149.92 |
(n = 923) 2.73 ± 1.89 |
0.012 |
| CA125 (U/ml) | (n = 694) 21.12 ± 47.72 |
(n = 912) 12.77 ± 8.55 |
<0.0001 |
| CA19-9 (U/ml) | (n = 906) 151.80 ± 1152.65 |
(n = 1071) 12.83 ± 9.51 |
<0.0001 |
| CA50 (U/ml) | (n = 581) 29.39 ± 138.17 |
(n = 555) 10.52 ± 17.91 |
0.001 |
| Ferritin (ng/ml) | (n = 588) 124.71 ± 144.03 |
(n = 531) 57.72 ± 74.91 |
<0.0001 |
CA, Carbohydrate antigen; CEA, Carcinoembryonic antigen; GC, Gastric cancer; HP, helicobacter pylori; NGC, Non-gastric cancer.
Table 2.
Clinical characteristics of the GC and NGC participants in the independent external validation dataset.
| Clinical indexes | Independent external validation dataset |
||
|---|---|---|---|
| GC (n = 294) | NGC (n = 521) | P value | |
| Subgroup information | TNM stage I = 74; II = 113; III = 90; IV = 17 | HC = 88; SG = 257; AG = 176 | N/A |
| Age (Years) | 62.28 ± 9.43 | 61.55 ± 8.87 | 0.27 |
| Sex (Female/Male) | 87/207 | 176/345 | 0.22 |
| BMI (kg/m2) | 22.98 ± 3.41 | 22.66 ± 3.06 | 0.16 |
| Smoking (Yes/No) | 92/202 | 139/382 | 0.16 |
| Drinking (Yes/No) | 90/204 | 184/337 | 0.35 |
| HP (Positive/Negative/Unknown) | 53/47/194 | 88/90/343 | 0.57 |
| Family history (Yes/No) | 39/255 | N/A | N/A |
| Pathological type (Adenocarcinoma/Others/Unknown) | 281/9/4 | N/A | N/A |
| Tumour size (≤5 cm/>5 cm/Unknown) | 174/111/9 | N/A | N/A |
| Tumour location (U/M/L/W/Unknown) | 69/63/151/7/4 | N/A | N/A |
| Grade of differentiation (High/Medium + Medium-low/Low/Unknown) | 23/120/147/4 | N/A | N/A |
| TNM type (I/II/III/IV) | 74/113/90/17 | N/A | N/A |
| HER2 (0/1/2/3/Unknown) | 87/74/34/24/75 | N/A | N/A |
| MMR (dMMR/pMMR/Unknown) | 17/166/111 | N/A | N/A |
| Lauren type (Intestinal/Diffuse/Mixed/Unknown) | 36/36/43/179 | N/A | N/A |
| Nerve invasion (Yes/No/Unknown) | 92/52/150 | N/A | N/A |
| Vascular tumour thrombus (Yes/No/Unknown) | 68/78/148 | N/A | N/A |
| CEA (ng/ml) | (n = 263) 5.40 ± 23.06 |
(n = 297) 1.63 ± 1.27 |
0.009 |
| CA242 (U/ml) | (n = 230) 29.13 ± 89.52 |
(n = 171) 4.51 ± 3.82 |
<0.0001 |
| CA724 (U/ml) | (n = 252) 11.15 ± 30.09 |
(n = 167) 2.79 ± 2.07 |
<0.0001 |
| AFP (ng/ml) | (n = 264) 12.97 ± 94.21 |
(n = 288) 2.54 ± 1.70 |
0.073 |
| CA125 (U/ml) | (n = 226) 17.19 ± 35.82 |
(n = 283) 11.98 ± 8.09 |
0.033 |
| CA19-9 (U/ml) | (n = 239) 41.59 ± 127.84 |
(n = 286) 14.42 ± 10.08 |
0.001 |
| CA50 (U/ml) | (n = 144) 28.98 ± 73.52 |
(n = 167) 8.56 ± 6.38 |
0.001 |
| Ferritin (ng/ml) | (n = 129) 102.85 ± 136.16 |
(n = 182) 42.79 ± 73.26 |
<0.0001 |
CA, Carbohydrate antigen; CEA, Carcinoembryonic antigen; GC, Gastric cancer; HP, helicobacter pylori; NGC, Non-gastric cancer.
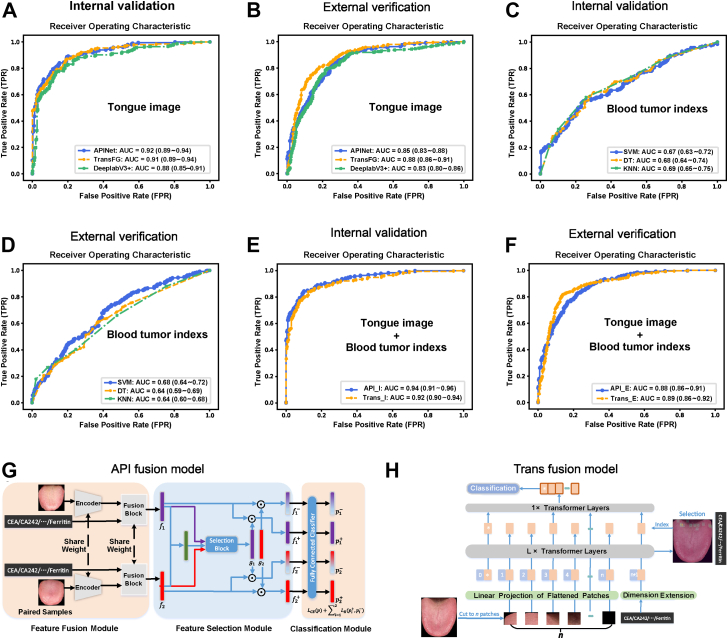
We applied three completely different deep learning models to evaluate the value of tongue images for GC diagnosis and screening to reduce the deviations in the conclusions caused by the model differences. Two completely different models, the APINet model (Fig. 2C) and TransFG model (Fig. 2D), were used in addition to the previous DeepLabV3+ model (Fig. 2E). The internal validation results showed that the AUC values of the three tongue imaging models in diagnosing GC ranged from 0.88 to 0.92 (Fig. 3A), the sensitivity ranged from 0.83 to 0.88, the specificity ranged from 0.78 to 0.81, and the accuracy ranged from 0.81 to 0.83 (Table S10). Interestingly, the independent external validation also showed that the tongue images could be used to easily distinguish patients with GC from participants with NGC; the AUC values were 0.83–0.88 (Fig. 3B), the sensitivity values were 0.84–0.90, the specificity values were 0.70–0.70, and the accuracy values were 0.75–0.77 (Table S10). Thus, tongue images can be used as a stable tool for the diagnosis of GC, and this approach is not affected by diet, region or the type of AI deep learning model used.
Fig. 3.
The AI diagnostic model based on tongue images performed significantly better than the combined analysis of the levels of 8 tumour indicators in blood in the diagnosis of GC, and combined analysis of tongue images and the levels of 8 tumour indicators in blood can further improve the diagnosis of GC. (A) The ROC curves and AUCs obtained during the internal validation of the three models based on tongue images. (B) The ROC curves and AUCs obtained during the external validation of the three models based on tongue images. (C) The ROC curves and AUCs obtained during the internal validation of the three models (SVM, DT, and KNN) based on the levels of 8 tumour indexes in blood. (D) The ROC curves and AUCs obtained during the external validation of the three models based on the levels of 8 tumour indexes in blood. (E) The ROC curves and AUCs obtained during the internal validation of the two fusion models. (F) The ROC curves and AUCs obtained during the external validation of the two fusion models. (G) The method used to establish the API fusion diagnostic model and its process. (H) The method used to establish the transfusion diagnostic model and its process.
To further evaluate the value of tongue imaging in diagnosing and screening GC, we compared the analysis of tongue images with the analysis of blood tumour indexes that have clinical application value. We collected data regarding the blood levels of classic tumour markers, including CEA, CA242, CA724, CA125, CA19-9, CA50, AFP, and ferritin. We established three machine learning models (SVM, DT and KNN models) integrating the levels of the eight blood indexes, in which the training, internal verification and external verification datasets of the models were consistent with those used in the tongue image model (excluding the cases with missing blood indexes). The AUC values ranged from 0.67 to 0.69 in the internal validation and from 0.64 to 0.68 in the external verification (Fig. 3C and D). The sensitivity, specificity and accuracy in the internal validation and the external verification are shown in Table S10. The results show that the value of the AI diagnostic model based on tongue images was significantly better than that of the combined analysis of the levels of 8 blood tumour indicators for the diagnosis of GC.
Based on the APINet model and TransFG model, we designed two fusion models of blood indicators and tongue images called API fusion and transfusion (Fig. 3G and H). The AUC values ranged from 0.92 to 0.94 in the internal validation and from 0.88 to 0.89 in the external verification (Fig. 3E and F), which were slightly higher than those obtained using the tongue image diagnosis model alone. The sensitivity, specificity and accuracy in the internal validation and the external verification are shown in Table S10. Therefore, the analysis of tongue images combined with the levels of 8 blood tumour indicators can further improve the diagnosis value of GC.
Correlation between the tongue image-based model and clinical information
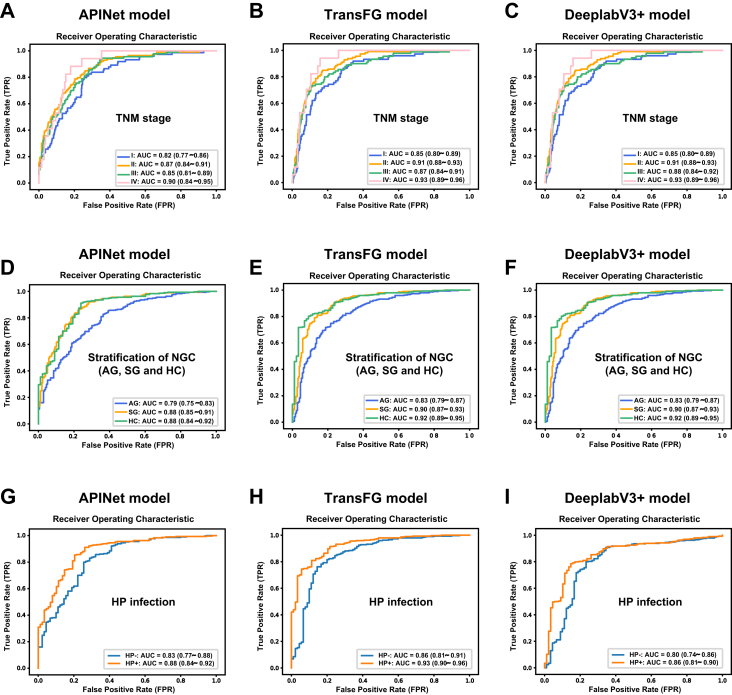
We further analysed the differences in the diagnostic value of the tongue image-based model in patients with different TNM stages of GC, as well as in participants with NGC, such as HCs and SG and patients with AG. As shown in Fig. 4A–C, the AUC values of the three tongue image-based models for patients with TNM stage I were 0.82–0.85, those for patients with TNM stage II were 0.87–0.91, those for patients with TNM stage III were 0.85–0.88, and those for patients with TNM stage IV were 0.90–0.93 in the external verification. Tongue imaging analysis had good diagnostic value for predicting early-stage GC, and the diagnostic value for predicting advanced GC was slightly higher than that for predicting early-stage GC. As shown in Fig. 4D–F, the AUC values of the three tongue image-based models for predicting the HCs were 0.88–0.92, those for predicting the SG patients were 0.88–0.90, and those for predicting the patients with AG were 0.79–0.83, indicating that tongue images can not only be used to distinguish HCs and SG patients from patients with GC but also to distinguish patients with AG from patients with GC. In addition, the value of the analysis of tongue images in distinguishing HCs and SG patients from patients with GC was higher than that of distinguishing patients with AG from patients with GC.
Fig. 4.
Tongue image-based models can be used to well distinguish patients with GC from participants with NGC, even those with early-stage GC and precancerous lesions, such as AG, and the diagnostic value of the tongue image-based model for GC has some relationship with tumour size. The ROC curves and AUCs obtained using the APINet model (A), TransFG model (B) and DeepLabV3+ model (C) for GC with different TNM stages. The ROC curves and AUCs obtained using the APINet model (D), TransFG model (E) and DeepLabV3+ model (F) for different participants with NGC (HCs, SG and AG). The ROC curves and AUCs obtained using the APINet model (G), TransFG model (H) and DeepLabV3+ model (I) for HP infection.
In addition, we analysed the correlation between clinical information, such as family history, tumour size, grade of differentiation, tumour location, and expression of HER2, and the diagnostic value of the tongue image-based model. As shown in Fig. 4G–I, the diagnostic value of the tongue image-based model for GC has some relationship with HP infection. The AUC value was 0.86–0.93 for HP-positive patients, while that of HP negative patients decreased slightly to 0.80–0.86. However, due to the small number of patients with HP infection information collected in this study, larger sample verification is needed in the future. In addition, we found that there was no significant correlation between clinical information, such as family history, smoking, drinking, tumour size, degree of differentiation, tumour location and HER2 expression, and the diagnostic value of the tongue image-based model (Figs. S3 and S4).
Characteristics of GC tongue images and traceability of the tongue image models
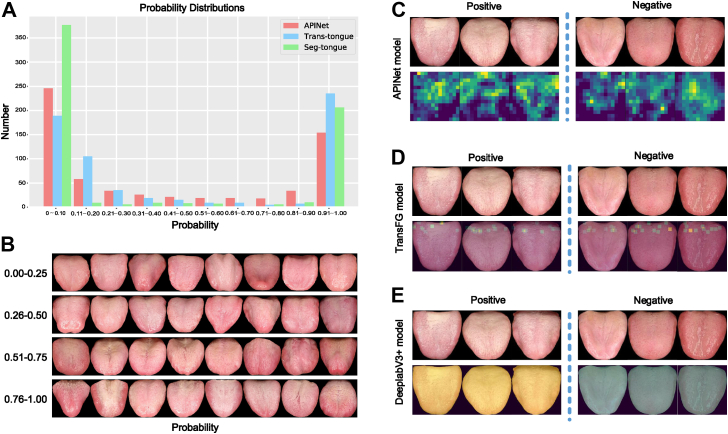
We evaluated the performance of the model by observing the probability distribution of the model. As shown in Fig. 5A, in the analysis of the probability distribution in the external verification of the three tongue image models, most of the cases were distributed on both sides; that is, there were fewer cases with a vague diagnosis in the middle of 0.41–0.60. This result suggested that the models could distinguish patients with GC from participants with NGC very well. In addition, it can be seen from the tongue images with different probabilities of less than 50% that they were judged as NGC since they had the characteristics of ruddy and thin coating on the tongue. In addition, as the probability increased, the thickness of the tongue coating gradually increased, and the water level of the tongue surface decreased (Fig. 5B).
Fig. 5.
Probability distribution and traceability of the three tongue image models. (A) In the probability distribution performed during the external verification of the three tongue image models, most of the cases were distributed on both sides; that is, there were fewer cases with a vague diagnosis in the middle of 0.41–0.60. (B) Representative tongue images with different probabilities (the intersection of the three models). (C) Output diagram of the APINet model. (D) Traceability of the TransFG model. (E) Traceability of the DeeplabV3+ model.
In addition, we intend to visualized the results and aimed to explain the basis of the classification. We incorporated six pairs of images that contained all the samples in subfigure an into APINet. Then, we found the top-activated channel of the gate vector corresponding to the displayed images in the input pairs. Subsequently, the relevant feature maps (14 × 14) before global pooling are shown in Fig. 5C. The activated features indicate the most important components of the classification. Obviously, all the areas of interest were concentrated on the tongue without being affected by the background. To illustrate the most discriminative patches in the tongue classification used for the TransFG model, we took advantage of its innate multihead attention mechanism. The classification patches most likely to be used were selected based on the attention weights in the first L-1 transformer layers. K (K = 12) patches were incorporated into the last layer for further classification, which are highlighted in the original input images, as shown in Fig. 5D. All of the regions of interest were in the area of the tongue, and the background area was useless for the prediction of the model. For the DeepLabV3+ model, we display the semantic segmentation results for some samples in Fig. 5E. The pixels belonging to the yellow area were divided into the positive category, while the pixels belonging to the green area were divided into the negative category. The pixels classified into the positive and negative categories were both on the tongue area. The probability that the input images are predicted to be positive was determined by the ratio of the number of positive pixels to the total number of positive and negative pixels, so the prediction of the image category was not affected by the background area.
Differential diagnostic value of the tongue image-based model for other cancers
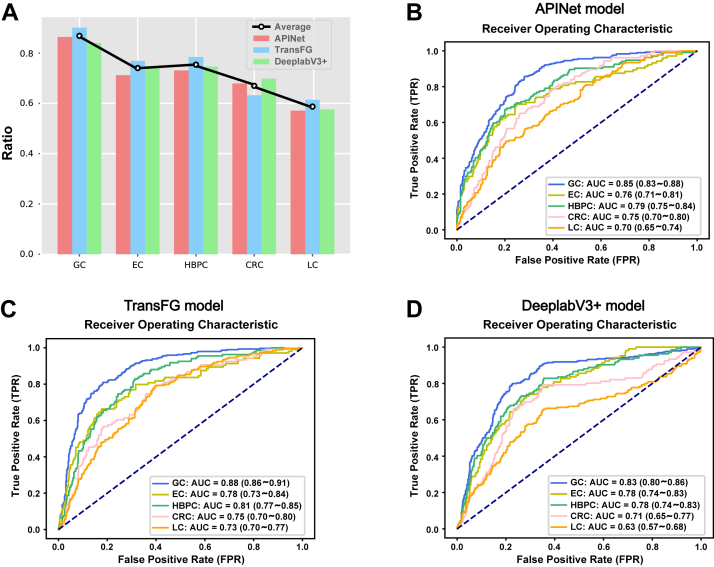
To assess the specificity and effectiveness of the diagnostic model based on the tongue images for GC, we also recruited 104 patients with EC, 134 patients with HBPC, 106 patients with CRC and 184 patients with LC to evaluate the diagnostic value of the models for other cancers. The general information was well matched, such as age, sex, BMI, smoking status, drinking status and TNM stage, was well matched between patients with GC and patients with other cancers (Table S11); more detailed information is shown in Tables S12–S15. The results showed that the tongue image models had the highest diagnostic value for GC (the specificity was 0.84–0.90, and the AUC was 0.83–0.88) and had some diagnostic effect in the prediction of other tumours, such as EC (the specificity was 0.71–0.77, and the AUC was 0.76–0.78), HBPC (the specificity was 0.73–0.78, and the AUC was 0.78–0.81) and CRC (the specificity was 0.63–0.70, and the AUC was 0.71–0.75), but the diagnostic value was further reduced in the prediction of other nondigestive tract tumours, such as LC (the specificity was 0.57–0.61, and the AUC was 0.63–0.73) (Fig. 6 and Table S16). The results showed that the tongue image models were the most useful for GC diagnosis and had a certain effect in the diagnosis of digestive tract tumours such as EC, HBPC and CRC, but these results need to be verified by more samples.
Fig. 6.
The diagnostic value of the tongue image-based diagnostic models for other tumours (EC, HBPC, CRC, LC). (A) The specificity of the tongue-based diagnostic models for GC and other tumours. (B) The ROC and AUC obtained using the APINet model for GC and other tumours. (C) The ROC and AUC obtained using the TransFG model for GC and other tumours. (D) The ROC and AUC obtained using the DeepLabV3+ model for GC and other tumours. CRC, colorectal cancer; EC, oesophageal cancer; HBPC, hepatobiliary pancreatic carcinoma; LC, lung cancer.
Discussion
Due to occult clinical symptoms and the dependence of diagnosis and screening on gastrointestinal endoscopy, the diagnosis rate of early-stage gastrointestinal tumours is low, and the prognosis is poor, which places a heavy burden on society and the economy.28 There is an urgent need to develop noninvasive and effective screening and diagnostic methods to improve the rate of the detection of early-stage digestive system tumours. AI has illuminated a clear path towards an evolving healthcare system replete with enhanced precision and computing capabilities, which play an increasingly important role in cancer screening and diagnosis.29 Wu et al. found that the use of an AI system during upper gastrointestinal endoscopy significantly reduced the gastric neoplasm miss rate in a single-centre, tandem randomized controlled trial.30 Dong et al. previously showed that a deep learning radiomic nomogram can be used to predict the number of lymph node metastases in locally advanced patients with GC in an international multicentre study.31 In our study, we conducted an observational, prospective, multicentre clinical study to evaluate the value of tongue images and the tongue coating microbiome in the screening and diagnosis of GC and other gastrointestinal tumours.
The colour, thickness and moisture content of the tongue coating are important components of tongue image diagnosis, and the formation of the tongue coating is closely related to the microbiome.32 Therefore, we simultaneously investigated the diagnostic value of tongue images and the tongue coating microbiome in GC. Previous studies have shown that the tongue coating microbiome can be used as a diagnostic tool for malignant tumours, including EC,33 GC34 and LC.35 Our study once again confirmed the diagnostic value of the tongue coating microbiome in GC. Considering that tongue imaging analysis as a diagnostic tool is more convenient and cost-efficient than the analysis of the tongue coating microbiome, our research focused on tongue imaging data. Our large sample multicentre clinical study showed that the AUC values of the three tongue image-based models for GC reached 0.88–0.92 in the internal verification and reached 0.83–0.88 in the independent external verification, which were significantly superior to the combined analysis of the levels of eight blood biomarkers. In addition, the fusion model of tongue images and tumour blood indicators further improved the diagnostic value. Further stratified analysis showed that the tongue image-based models could be used to well distinguish patients with GC from participants with NGC, even those with early-stage GC, and precancerous lesions, such as AG.
Tongue image diagnosis can be an effective, noninvasive method to perform an auxiliary diagnosis anywhere, which can support the global needs of the primary healthcare systems.36 However, the recognition of the use of tongue images by TCM doctors is subjective and challenging.37 At present, many efforts have been made in AI in terms of the standardization of tongue image diagnosis, namely, preprocessing, tongue detection, segmentation, feature extraction, and tongue analysis, especially in TCM.38 Xu et al. developed a multitask joint learning model for segmenting and classifying tongue images using a deep neural network, which could optimally extract the tongue image features.39 Meng et al. proposed a novel feature extraction framework called constrained high dispersal neural networks to extract unbiased features and to reduce human labour for tongue image diagnosis in TCM.40 Previous research on AI in tongue diagnosis mainly focused on the standardization of tongue diagnosis to reduce human error. Our study is the first to apply AI deep learning to explore the value of tongue images in tumour diagnosis, and provides a powerful basis for the use of tongue images and the tongue coating microbiome as diagnostic methods for GC. Considering the instability of AI models, multiple AI models are used separately to verify the clinical value of tongue images in GC. The three models independently used the method of two classifications for learning, and there should be no multiplicity problem. And all the three models demonstrate the discriminative validity over tongue images, which showed that tongue image can be used as a stable tool for GC diagnosis, and it is not affected by AI depth learning model type.
According to the theory of TCM, the tongue is a mirror for the internal organs, reflecting the body's physiological and clinicopathological condition.37 The colour, size and form, motion, substances, coating, and geometric shape of the tongue, as well as changes in the tongue body, are just a few qualities that must be evaluated that are related to the health state of the patient.41 In our study, the traceability of the three tongue image models showed that all the models focused on the tongue and were not affected by the background. The tongue images that were determined by the models to be from participants with NGC had the characteristics of a ruddy and thin tongue coating, while the tongue images determined to be from patients with GC had the characteristics of a significantly increased tongue coating thickness and decreased water level on the tongue surface. This finding shows that evaluating the state of human health according to the physical characteristics of tongue images in TCM theory has a scientific basis. In addition, an increasing number of studies have shown that the oral/tongue coating microbiome is closely related to digestive tract diseases,42,43 and the oral/tongue coating microbiome is significantly different among different tongue images.44,45 This finding provides a scientific basis for tongue imaging to be used as a diagnostic tool for GC.
However, there are still some shortcomings to our research. This study only included a Chinese population, and the applicability of the findings to other ethnic groups needs to be further verified. In addition, the methods based on deep learning were all data-driven. The robustness of these deep-learning models is closely related to the amount and diversity of training data. However, collecting all the data associated with a specific task is not realistic. Moreover, our NGC cohort only includes HC, AG and SG, more lesions such as erosion, ulcers, intestinal metaplasia, and high grade intraepithelial neoplasia were not included in our study design, which may reduce the clinical significance of this method. In addition, to observe the specificity of tongue images in the diagnosis of GC, the tongue images obtained from patients with other tumours were collected for differential diagnosis. The results showed that the tongue image models were the most useful for GC diagnosis and had some diagnostic effects in evaluating EC, HBPC and CRC, but the diagnostic value was further reduced in the prediction of other nondigestive tract tumours, such as LC. However, due to the small number of samples obtained from patients with gastrointestinal tumours, such as EC, HBPC and CRC, the role of tongue imaging in the diagnosis of gastrointestinal tumours (except GC) needs further multicentre and large-sample clinical research.
In short, considering the substantial burden of GC and other gastrointestinal tumours in China and across the globe, we believe that tongue images, in combination with the widespread use of AI deep learning approaches, might be the most cost-efficient, noninvasive and acceptable approach for the screening and early detection of GC, which will also have considerable socioeconomic effects.
In conclusion, tongue image analysis conducted as a means of noninvasive diagnosis and screening of GC performed significantly better than the combined analysis of the levels of 8 blood indicators. The three kinds of tongue image-based AI deep learning diagnostic models that we developed can be used to adequately distinguish patients with GC from participants with NGC, even those with early-stage GC and precancerous lesions, such as AG. However, additional studies are needed to determine its applicability to other populations, such as those in different parts of the world and those with different races. In addition, the diagnostic value of tongue image analysis for other gastrointestinal tumours, such as EC, HBPC and CRC, needs further multicentre and large-sample clinical research. We need to further promote the application of tongue image analysis in the screening and diagnosis of GC to improve the diagnosis rate of early-stage GC. Moreover, this study provides strong scientific support for the theory of tongue image diagnosis in TCM.
Contributors
X-DC conceived the study and acquired the funding. LY, Z-YX and J-JQ carried out clinical research, collected clinical samples and analysed clinical data, and wrote the paper. LY and S-CZ established the artificial intelligence diagnosis model and wrote the paper. Y-FS, P-CY, YW, Z-HB, Y-HX, J-CS, W-YH, T-HC, X-LC, CH, PZ, C-WD, Y-NW, NJ, BL, Y-WX, B-PJ, H-YG, K-QC, JL, HW, X-BW, X-QG, XL, GZ, Z-CZ, JY, H-YY, L-CC, Z-SY, H-QY, YB, XC, P-ZZ, LW, W-TZ, Y-FT, MC, YY, G-HY, HR, X-LG, J-LX, H-DX, Y-LZ, L-BD, S-JZ, and H-YF participated in clinical sample collection. All authors have read and approved the final manuscript. X-DC, LY, Z-YX, J-JQ, LY and S-CZ had access to dataset and had final responsibility for the decision to submit for publication.
Data sharing statement
The raw reads of 16 S rDNA were deposited into the NCBI SRA database (Accession Number: Bioproject PRJNA752841). The code used in this study and all supporting data are available upon request.
Declaration of interests
All authors declare no competing interests.
Acknowledgments
This study was supported by the National Key R&D Program of China (2021YFA0910100), Program of Zhejiang Provincial TCM Sci-tech Plan (2018ZY006), Medical Science and Technology Project of Zhejiang Province (2022KY114, WKJ-ZJ-2104), Zhejiang Provincial Research Center for Upper Gastrointestinal Tract Cancer (JBZX-202006), Natural Science Foundation of Zhejiang Province (HDMY22H160008), Science and Technology Projects of Zhejiang Province (2019C03049), National Natural Science Foundation of China (82074245, 81973634, 82204828), and Chinese Postdoctoral Science Foundation (2022M713203).
Footnotes
Translation For the Chinese translation of the abstract see Supplementary Materials section.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.eclinm.2023.101834.
Appendix A. Supplementary data
References
- 1.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Thrift A.P., El-Serag H.B. Burden of gastric cancer. Clin Gastroenterol Hepatol. 2020;18:534–542. doi: 10.1016/j.cgh.2019.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang W.Y., Zhou H., Wang Y.F., et al. Current policies and measures on the development of traditional Chinese medicine in China. Pharmacol Res. 2021;163 doi: 10.1016/j.phrs.2020.105187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui J., Cui H., Yang M., et al. Tongue coating microbiome as a potential biomarker for gastritis including precancerous cascade. Protein Cell. 2019;10:496–509. doi: 10.1007/s13238-018-0596-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanawong R., Obafemi-Ajayi T., Liu D., et al. Tongue image analysis and its mobile app development for health diagnosis. Adv Exp Med Biol. 2017;1005:99–121. doi: 10.1007/978-981-10-5717-5_5. [DOI] [PubMed] [Google Scholar]
- 6.Zhou J., Li S., Wang X., et al. Weakly supervised deep learning for tooth-marked tongue recognition. Front Physiol. 2022;13 doi: 10.3389/fphys.2022.847267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han S., Yang X., Qi Q., et al. Potential screening and early diagnosis method for cancer: tongue diagnosis. Int J Oncol. 2016;48:2257–2264. doi: 10.3892/ijo.2016.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu H., Ren Z., Li A., et al. Tongue coating microbiome data distinguish patients with pancreatic head cancer from healthy controls. J Oral Microbiol. 2019;11 doi: 10.1080/20002297.2018.1563409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu H., Ren Z., Li A., et al. Deep sequencing reveals microbiota dysbiosis of tongue coat in patients with liver carcinoma. Sci Rep. 2016;6 doi: 10.1038/srep33142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han S., Chen Y., Hu J., et al. Tongue images and tongue coating microbiome in patients with colorectal cancer. Microb Pathog. 2014;77:1–6. doi: 10.1016/j.micpath.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Wu Z.F., Zou K., Xiang C.J., et al. Helicobacter pylori infection is associated with the co-occurrence of bacteria in the oral cavity and the gastric mucosa. Helicobacter. 2021;26 doi: 10.1111/hel.12786. [DOI] [PubMed] [Google Scholar]
- 12.Kroese J.M., Brandt B.W., Buijs M.J., et al. The oral microbiome in early rheumatoid arthritis patients and individuals at risk differs from healthy controls. Arthritis Rheumatol. 2021;73(11):1986–1993. doi: 10.1002/art.41780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y., Mao Y.F., Tang Y.S., et al. Altered oral microbiota in chronic hepatitis B patients with different tongue coatings. World J Gastroenterol. 2018;24:3448–3461. doi: 10.3748/wjg.v24.i30.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goyal H., Sherazi S.A.A., Mann R., et al. Scope of artificial intelligence in gastrointestinal oncology. Cancers (Basel) 2021;13:5494. doi: 10.3390/cancers13215494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu G., Sun K., Xu C., et al. Accurate recognition of colorectal cancer with semi-supervised deep learning on pathological images. Nat Commun. 2021;12:6311. doi: 10.1038/s41467-021-26643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheung C.Y., Xu D., Cheng C.Y., et al. A deep-learning system for the assessment of cardiovascular disease risk via the measurement of retinal-vessel calibre. Nat Biomed Eng. 2021;5:498–508. doi: 10.1038/s41551-020-00626-4. [DOI] [PubMed] [Google Scholar]
- 17.Takenaka K., Ohtsuka K., Fujii T., et al. Development and validation of a deep neural network for accurate evaluation of endoscopic images from patients with ulcerative colitis. Gastroenterology. 2020;158:2150–2157. doi: 10.1053/j.gastro.2020.02.012. [DOI] [PubMed] [Google Scholar]
- 18.Hu Y., Wen G., Luo M., et al. Fully-channel regional attention network for disease-location recognition with tongue images. Artif Intell Med. 2021;118 doi: 10.1016/j.artmed.2021.102110. [DOI] [PubMed] [Google Scholar]
- 19.Hu Y., Wen G., Liao H., et al. Automatic construction of Chinese herbal prescriptions from tongue images using CNNs and auxiliary latent therapy topics. IEEE Trans Cybern. 2021;51:708–721. doi: 10.1109/TCYB.2019.2909925. [DOI] [PubMed] [Google Scholar]
- 20.Pang B., Zhang D., Wang K. The bi-elliptical deformable contour and its application to automated tongue segmentation in Chinese medicine. IEEE Trans Med Imaging. 2005;24:946–956. doi: 10.1109/TMI.2005.850552. [DOI] [PubMed] [Google Scholar]
- 21.Wang X., Zhang B., Yang Z., et al. Statistical analysis of tongue images for feature extraction and diagnostics. IEEE Trans Image Process. 2013;22:5336–5347. doi: 10.1109/TIP.2013.2284070. [DOI] [PubMed] [Google Scholar]
- 22.Hajian-Tilaki K. Sample size estimation in diagnostic test studies of biomedical informatics. J Biomed Inform. 2014;48:193–204. doi: 10.1016/j.jbi.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Newcombe R.G. Two-sided confidence intervals for the single proportion: comparison of seven methods. Stat Med. 1998;17:857–872. doi: 10.1002/(sici)1097-0258(19980430)17:8<857::aid-sim777>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 24.Wu L., Wang Y., Li X., et al. Deep attention-based spatially recursive networks for fine-grained visual recognition. IEEE Trans Cybern. 2019;49:1791–1802. doi: 10.1109/TCYB.2018.2813971. [DOI] [PubMed] [Google Scholar]
- 25.He J., Chen J., Liu S., et al. A transformer architecture for fine-grained recognition. arXiv. 2022 preprint arXiv:2103.07976 2021. [Google Scholar]
- 26.Liang-Chieh Chen Y.Z., George P., Schroff F., Adam H. Proceedings of the European Conference on Computer Vision (ECCV) 2018. Encoder-decoder with atrous separable convolution for semantic image segmentation; pp. 801–818. [Google Scholar]
- 27.Gower J.C. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika. 1966;53(3–4):325–338. [Google Scholar]
- 28.Peery A.F., Crockett S.D., Murphy C.C., et al. Burden and cost of gastrointestinal, liver, and pancreatic diseases in the United States: update 2021. Gastroenterology. 2021;162(2):621–644. doi: 10.1053/j.gastro.2021.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomasian N.M., Kamel I.R., Bai H.X. Machine intelligence in non-invasive endocrine cancer diagnostics. Nat Rev Endocrinol. 2021;18:1–15. doi: 10.1038/s41574-021-00543-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu L., Shang R., Sharma P., et al. Effect of a deep learning-based system on the miss rate of gastric neoplasms during upper gastrointestinal endoscopy: a single-centre, tandem, randomised controlled trial. Lancet Gastroenterol Hepatol. 2021;6(9):700–708. doi: 10.1016/S2468-1253(21)00216-8. [DOI] [PubMed] [Google Scholar]
- 31.Dong D., Fang M.J., Tang L., et al. Deep learning radiomic nomogram can predict the number of lymph node metastasis in locally advanced gastric cancer: an international multicenter study. Ann Oncol. 2020;31:912–920. doi: 10.1016/j.annonc.2020.04.003. [DOI] [PubMed] [Google Scholar]
- 32.Ren Z., Wang H., Cui G., et al. Alterations in the human oral and gut microbiomes and lipidomics in COVID-19. Gut. 2021;70:1253–1265. doi: 10.1136/gutjnl-2020-323826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang X., Lu B., Xiao P., et al. Microbial characteristics of common tongue coatings in patients with precancerous lesions of the upper gastrointestinal tract. J Healthc Eng. 2022;2022 doi: 10.1155/2022/7598427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu S., Xiang C., Wu J., et al. Tongue coating bacteria as a potential stable biomarker for gastric cancer independent of lifestyle. Dig Dis Sci. 2021;66:2964–2980. doi: 10.1007/s10620-020-06637-0. [DOI] [PubMed] [Google Scholar]
- 35.Vogtmann E., Hua X., Yu G., et al. The oral microbiome and lung cancer risk: an analysis of 3 prospective cohort studies. J Natl Cancer Inst. 2022;114(11):1501–1510. doi: 10.1093/jnci/djac149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wen G., Ma J., Hu Y., et al. Grouping attributes zero-shot learning for tongue constitution recognition. Artif Intell Med. 2020;109 doi: 10.1016/j.artmed.2020.101951. [DOI] [PubMed] [Google Scholar]
- 37.Matos L.C., Machado J.P., Monteiro F.J., et al. Can traditional Chinese medicine diagnosis Be parameterized and standardized? A narrative review. Healthcare (Basel) 2021;9:177. doi: 10.3390/healthcare9020177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tania M.H., Lwin K., Hossain M.A. Advances in automated tongue diagnosis techniques. Integr Med Res. 2019;8:42–56. doi: 10.1016/j.imr.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Q., Zeng Y., Tang W., et al. Multi-task joint learning model for segmenting and classifying tongue images using a deep neural network. IEEE J Biomed Health Inform. 2020;24:2481–2489. doi: 10.1109/JBHI.2020.2986376. [DOI] [PubMed] [Google Scholar]
- 40.Meng D., Cao G., Duan Y., et al. Tongue images classification based on constrained high dispersal network. Evid Based Complement Alternat Med. 2017;2017 doi: 10.1155/2017/7452427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z.C., Zhang S.P., Yuen P.C., et al. Intra-rater and inter-rater reliability of tongue coating diagnosis in traditional Chinese medicine using smartphones: quasi-delphi study. JMIR Mhealth Uhealth. 2020;8 doi: 10.2196/16018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kitamoto S., Nagao-Kitamoto H., Jiao Y., et al. The intermucosal connection between the mouth and gut in commensal pathobiont-driven colitis. Cell. 2020;182:447–462.e14. doi: 10.1016/j.cell.2020.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y., Cui J., Liu Y., et al. Oral, tongue-coating microbiota, and metabolic disorders: a novel area of interactive research. Front Cardiovasc Med. 2021;8 doi: 10.3389/fcvm.2021.730203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu J., Xiang C., Zhang C., et al. Microbial biomarkers of common tongue coatings in patients with gastric cancer. Microb Pathog. 2019;127:97–105. doi: 10.1016/j.micpath.2018.11.051. [DOI] [PubMed] [Google Scholar]
- 45.Ye J., Cai X., Yang J., et al. Bacillus as a potential diagnostic marker for yellow tongue coating. Sci Rep. 2016;6 doi: 10.1038/srep32496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.