Abstract
Microbial utilization and conversion of organic one-carbon compounds, such as formate and methanol that can be easily produced from CO2, has emerged as an attractive approach for biorefinery. In this study, we discovered Clostridium beijerinckii NCIMB 8052, a typical solventogenic Clostridium strain, to be a native formate-utilizing bacterium. 13C isotope analysis showed that formate could be metabolized via both assimilation and dissimilation pathways in C. beijerinckii NCIMB 8052. Notably, the use of formate as the supplementary substrate by this strain could significantly enhance its glucose consumption and ABE (acetone-butanol-ethanol) production, largely due to the up-regulation of genes responsible for glycolysis and glucose transport under formate stress. Based on these findings, we further improved formate tolerance of C. beijerinckii NCIMB 8052 by adaptive laboratory evolution, generating an evolved strain Cbei-FA01. The Cbei-FA01 strain could produce 23.0 g/L of ABE solvents using glucose and formate as dual substrates, ∼50% higher than that of the wild-type strain under the same condition. Moreover, such a promotion effect of formate on ABE production by Cbei-FA01 was also observed in fermenting a glucose-xylose mixture. This work reveals a previously unreported role of formate in biological ABE production, providing a new approach to utilize this one-carbon source.
Keywords: Formate utilization, Clostridium beijerinckii, Metabolic pathways, Adaptive laboratory evolution, ABE solvents
1. Introduction
The use of one-carbon (C1) gases (e.g., CO2, CO, and CH4) to produce value-added biomass and chemicals has emerged as a potential route to reduce greenhouse gas emission and broaden the carbon sources for biorefinery [[1], [2], [3], [4]]. To overcome the difficulties of handling C1 gases directly, chemical transformation of these gases into liquids such as formate as the feedstock for the following biological conversion has offered an alternative approach to utilize these C1 sources. Notably, formate is considered as a CO2-equivalent carbon source because it can be easily obtained from CO2 via electrochemical catalysis at high specificity and efficiency [[5], [6], [7]].
Natural formatotrophs are able to grow on formate as the sole carbon source [[8], [9], [10]]. However, they are generally less suitable for industrial use due to their relatively poor growth [5]. Thus, adapting model microorganisms, such as Escherichia coli [[11], [12], [13], [14]] and yeasts [15], for formate utilization has been exerted in the last decade. Although some progresses have been made, the highest cell density and formate consumption achieved by these artificial cells were significantly lower than those based on conventional carbon sources such as glucose [5]. Thus, the use of mixed substrates consisting of sugars and formate, in which sugars are consumed as the primary carbon sources for biomass and product synthesis and formate acts as a supplementary substrate, has emerged as an industrially feasible approach for mix-carbon biorefinery [16,17].
As a large and heterogenous group of bacteria, the genus Clostridium has great potential for industrial application. Clostridial ABE (Acetone, Butanol and Ethanol) fermentation is a traditional industrial-scale biological process with renewed interests recently. The major Clostridium species include Clostridium acetobutylicum, Clostridium beijerinckii, Clostridium saccaroperbutylacetonicum, and Clostridium saccharoacetobutylicum [18], normally producing 20–25 g/L ABE solvents using different substrates such as hexose and pentose sugars, wheat straw, potato, and corn fiber [[19], [20], [21]]. Of these clostridia, C. beijerinckii is one of the best-studied solventogenic species, capable of using hexose and pentose sugars simultaneously [22]. This enables C. beijerinckii to be used for ABE production from different lignocellulosic hydrolysates [23]. Of note, C. beijerinckii has been found to re-assimilate CO2 and H2 in fermenting sugars, which was presumed to be attributed to an incomplete Wood-Ljungdahl (WL) pathway and a complete reversed pyruvate ferredoxin oxidoreductase/pyruvate-formate-lyase (rPFOR/PFL)-dependent pathway in this anaerobe [24]. Both the WL and rPFOR/PFL pathways are known to be associated with formate assimilation during their operation (KEGG Pathway Database); thus, formate may be utilized through these pathways in C. beijerinckii during its ABE fermentation.
In this study, we investigated the formate metabolism of C. beijerinckii NCIMB 8052 by employing both the 13C isotopomer tracer and genetic analyses. An interesting finding was that the formate metabolism by C. beijerinckii NCIMB 8052 could significantly increase its glucose consumption, consequently enhancing ABE production. The underlying mechanism for this phenomenon was also analyzed. Based on these findings, we further enhanced the formate tolerance and consumption of C. beijerinckii NCIMB 8052 through adaptive laboratory evolution approach. The evolved strain showcased much improved performance in ABE fermentation using glucose and formate as dual substrates compared to the wild-type strain. The results from this work will be useful for developing a more economically viable process for biological ABE production.
2. Materials and methods
2.1. Strains, media, and reagents
All the strains and plasmids used in this study are listed in Supplementary Table S1. The E. coli strain DH5α was adopted as the host for plasmid cloning. The E. coli cells were grown in lysogeny broth (LB) medium (containing 10 g/L tryptone, 5 g/L yeast extract, and 10 g/L sodium chloride) [25] at 37 °C supplemented with ampicillin (100 μg/mL) when needed. The C. beijerinckii NCIMB 8052 strain and other mutants were grown anaerobically at 37 °C in CGM medium [26] or the modified MP2 medium [27] supplemented with erythromycin (40 μg/mL) when needed. The modified MP2 contained the following compounds per liter: various amounts of glucose; MgSO4·7H2O, 0.2 g; MnSO4·H2O, 0.01 g; FeSO4·7H2O, 0.01 g; NaCl, 0.01 g; p-aminobenzoic acid, 1.0 mg; biotin, 0.01 mg; thiamine, 1.0 mg; KH2PO4, 0.5 g; K2HPO4, 0.6 g; (NH4)2SO4, 2.0 g; and CH3COONa·3H2O, 10.9 g. All solid plates were supplemented with 2% (wt/vol) agar. Resazurin was used as an oxygen indicator, and final concentration was 0.5 mg/L. All the anaerobic operations were performed in an anaerobic chamber (Coy Laboratory Products, Inc., USA) at 37 °C.
All primers were synthesized by Tsingke Biotechnology Co., Ltd. (Shanghai, China). The primer sequences are listed in Supplementary Table S2. Restriction enzymes (Thermo Fisher Scientific, Shanghai, China) and T4 DNA ligase (Takara Bio, Shanghai, China) were used to construct plasmids. Plasmids were extracted by plasmid extraction kit (Axygen, Shanghai, China). DNA sequencing was conducted by Tsingke Biotechnology Co., Ltd. (Shanghai, China). KOD plus Neo and KOD FX high-fidelity DNA polymerase (Toyobo, Osaka, Japan) were applied to PCR amplification reactions. Common reagents (Sinopharm Chemical Reagent Co., Ltd, Shanghai, China) were purchased to prepare media. 13C-labeled sodium formate (Sigma-Aldrich, Shanghai, China) was applied to isotope tracer experiments.
2.2. Construction of plasmids
We used the ClosTron website (http://clostron.com/) to design the intron site and primers. Here, all the gene numbers were under GenBank accession CP000721.1 [28]. The plasmid pWJ1-fdh for the disruption of fdh (Cbei_3801) was constructed as follows: the plasmid pWJ1 [29] was digested with Bsp1407I/XhoI, yielding the liner plasmid. Next, two DNA fragments (250 and 121 bp) were obtained by PCR amplification with template pWJ1 using primers 3801-IBS/EBS universal and 3801-EBS2/3801-EBS1d, respectively. Then the two DNA fragments were assembled by overlapping PCR using the primers 3801-IBS/3801-EBS1d. The resulting DNA fragment was digested with Bsp1407I/XhoI and then ligated to the abovementioned liner plasmid pWJ1 by T4 DNA ligase, generating the target plasmid pWJ1-fdh.
2.3. Electro-transformation and gene inactivation
The disruption of fdh was achieved by the group II intron (TargeTron) technology [30]. Electrocompetent cells were prepared as previously described [31] with modification. Briefly, cells were incubated in 50 mL CGM medium and harvested at the optical density (OD600) ∼ 0.8, then washed twice by ETM buffer (92.4 g/L sucrose, 0.2 g/L Na2HPO4·12H2O, 0.7 g/L NaH2PO4·2H2O, 1.8 g/L MgCl2·6H2O) at 4 °C, and resuspended by 1 mL ET buffer (92.4 g/L sucrose, 0.2 g/L Na2HPO4·12H2O, 0.7 g/L NaH2PO4·2H2O) making the electrocompetent cells. Per 200 μL electrocompetent cells were thawed on ice and then mixed with 2 μg of plasmid DNA for 10 min. The mixture was placed in a 2 mm-gap electroporation cuvette (Harvard Apparatus, MA, USA) and immediately pulsed using a Gene Pulser Xcell microbial electroporation system (Bio-Rad, USA) with parameters set at 1.8 kV, 200 Ω and 25 μF. Next, the cells were recovered in 0.5 mL CGM medium for 6 h, plated onto CGM agar plates (supplemented with 40 μg/mL of erythromycin), and then incubated at 37 °C for 24 h. When the colonies were visible on the agar plate, the colony-PCR-based screening was performed to identify the desired insertion events. Finally, the obtained DNA fragments by PCR amplification were sequenced to further confirm intron insertions.
2.4. Fermentation
Inoculum preparation and fermentations of C. beijerinckii NCIMB 8052 were carried out anaerobically in CGM and the modified MP2, respectively. Briefly, 50 μl of frozen stock was transferred into 5 mL of CGM medium and then incubated anaerobically at 37 °C for 12–24 h. When the seed cells reached OD600 of ∼1.0, 5% (vol/vol) of cells were transferred into the modified MP2 medium (30 mL) for fermentation in 50 mL round-bottom tubes. Sodium formate was added into media at the beginning of fermentation when needed.
2.5. Analytical methods
Cell density (OD600) was determined by spectrophotometer (DU730, Beckman Coulter). Assays of products (butanol, acetone and ethanol) were performed as previously described [32]. In brief, samples were taken at appropriate time intervals and then centrifuged at 7, 000 g for 20 min at 4 °C. The products in supernatant were then analyzed using a gas chromatograph (7890 A, Agilent, Wilmington, DE, USA) equipped with a flame ionization detector (Agilent) and a capillary column (EC-Wax; Alltech), details were described previously [32].
Glucose and formate were determined by HPLC (model 1200, Agilent, USA) equipped with an Aminex HPX-87H ion-exclusion column (Bio-Rad) and a differential refractive index detector (RID). 5 mM of H2SO4 was used as mobile phase at 0.6 mL/min flow rate. The column temperature was set at 30 °C.
2.6. GC-MS and isotopomer analysis
The C. beijerinckii NCIMB 8052 strain was grown anaerobically at 37 °C in the modified MP2 medium containing 5 g/L of glucose and 1 g/L of 13C-labeled sodium formate as the carbon sources. The 2 mL grown cells were harvested when they reached OD600 of 2.5 and then hydrolyzed with 6 M HCl for 12 h at 105 °C. The samples were dried in a vacuum centrifuge and then derivatized with 100 μL of pyridine and 50 μL of N-methyl-N-[tert-butyldimethylsilyl]trifluoroacetamide (MTBSTFA, M − 108) for 1 h at 85 °C. Following filtration (0.22-μm pore size; Millipore), 1 μL of samples were injected into the GC-MS system (Agilent 7890B–5977B, Agilent, USA) equipped with a HP-5MS column (19091S–433UI, 30 m × 0.25 mm × 0.25 μm), the carrier gas was Helium. The mass spectrometer (Agilent MS-5977B) was operated in the electron impact (EI) mode at 70 eV. GC-MS data were analyzed as described previously [33].
2.7. Quantitative real-time PCR (qRT-PCR)
The C. beijerinckii NCIMB 8052 strain was grown anaerobically in the modified MP2 medium (60 g/L glucose) with and without the supplementation of formate (4 g/L) at 37 °C. Cells were harvested when reaching OD600 of 4.5. Total RNA was extracted using the ultrapure RNA kit (CW0581, CWbio, China) according to the manufacturer's instructions. cDNA was obtained by TransScript® One-Step gDNA Removal and cDNA Synthesis SuperMix kit (AT311-03, Trans, China). The RNA and cDNA concentrations were determined by using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA). The manipulation of qRT-PCR was the same as described previously [34]. The primers used for qRT-PCR are listed in Supplementary Table S2. Here, the gene (Cbei_R0001) coding for 16S ribosomal RNA was used as the internal control [22].
2.8. Adaptive laboratory evolution experiment
The experiment started with the C. beijerinckii NCIMB 8052 strain. The initial cultivation was performed anaerobically at 37 °C in 6 mL of the modified MP2 medium containing 10 g/L glucose as the carbon source. The grown cells were then transferred into the modified MP2 medium containing 10 g/L glucose and 4 g/L sodium formate for the first round of adaptive evolution. Cells were passed on every 24 h. Once cells exhibited obviously increased growth rate after multiple rounds of subculturing, sodium formate added into the medium was increased. Samples were taken every passage and then stored at −80 °C with 20% (vol/vol) glycerol.
3. Results
3.1. Formate metabolism promotes growth and glucose utilization of C. beijerinckii NCIMB 8052
Formate is known to be toxic to microorganisms [35,36]. To assess the ability of C. beijerinckii NCIMB 8052 to use formate, we examined its growth with glucose as the primary carbon source and formate as the supplementary substrate. The experiment was carried out with 60 g/L glucose and gradually enhanced concentrations of formate (HCOONa) (2, 4, and 6 g/L). As expected, we observed formate consumption by C. beijerinckii NCIMB 8052, which enhanced with increasing initial formate concentrations in the medium (Fig. 1A). No obvious inhibition on the cell growth occurred in the presence of 2 g/L formate, while the growth was slightly and heavily impaired with the supplementation of 4 and 6 g/L formate, respectively (Fig. 1B). Interestingly, the supplementation of 2 and 4 g/L formate significantly promoted glucose consumption by C. beijerinckii NCIMB 8052, approximately 25% and 23% higher, respectively, than those in the absence of formate (Fig. 1C). Consequently, 17.0 and 19.1 g/L ABE solvents were produced by C. beijerinckii NCIMB 8052 with the supplementation of 2 g/L and 4 g/L formate, respectively, while the ABE titer reached only 13.9 g/L without formate (Fig. 1D). However, the ABE production of this strain was only 3.9 g/L in the presence of 6 g/L formate (Fig. 1D), which should be attributed to the poor cell growth under high formate concentration (Fig. 1B). According to these results, we demonstrate that C. beijerinckii NCIMB 8052 is a native formate-utilizing bacterium.
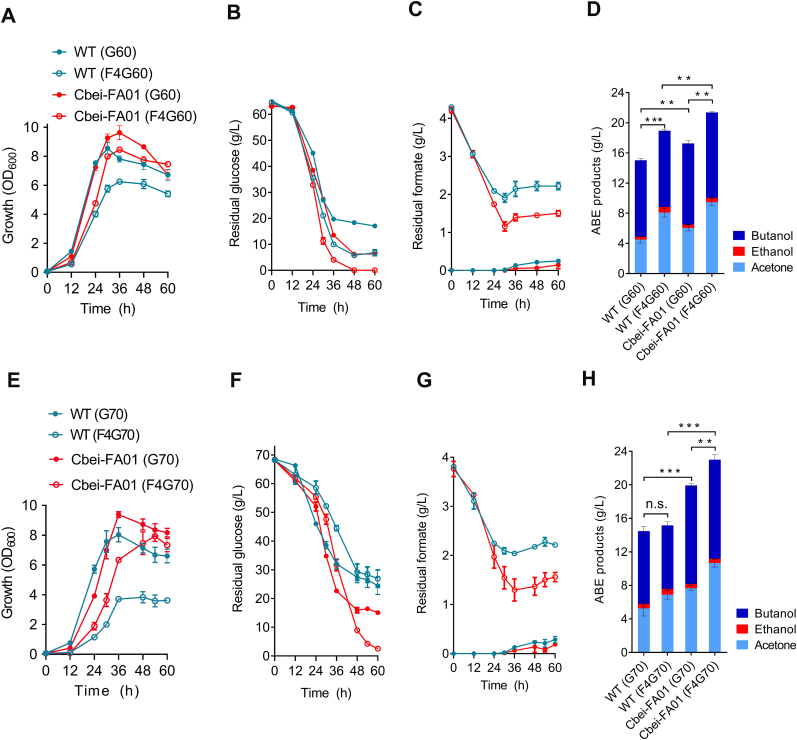
Fig. 1.
Formate tolerance and utilization of C. beijerinckii NCIMB 8052 as well as the promotion of formate on cellular performance. (A, B) Formate consumption and influence of formate on the growth of C. beijerinckii NCIMB 8052. The modified MP2 medium contained 60 g/L glucose supplemented with different amounts of sodium formate (0, 2, 4, and 6 g/L). F0G60: 0 g/L formate and 60 g/L glucose; F2G60: 2 g/L sodium formate and 60 g/L glucose; F4G60: 4 g/L sodium formate and 60 g/L glucose; F6G60: 6 g/L sodium formate and 60 g/L glucose. (C) Glucose consumption of C. beijerinckii NCIMB 8052 in the presence of different formate concentrations. (D) ABE production at 60 h of C. beijerinckii NCIMB 8052 in the presence of different formate concentrations. (E) Glucose transport and glycolysis pathways. The former was shadowed in blue and the latter was in orange. Genes in C. beijerinckii NCIMB 8052 were predicted in Table S3. PTS, phosphotransferase system; ABC, ATP-dependent ATP-binding cassette transporter; SGLT, sodium/glucose transporter; Glck, glucokinase; Pgi, phosphoglucose isomerase; Pfk, phosphofructokinase; Fba, fructose-1,6-bisphosphate aldolase; Tpi, triose-phosphate isomerase; Gap, glyceraldehyde-3-phosphate dehydrogenase; Pgk, phosphoglycerate kinase; Pgm, phosphoglycerate mutase; Eno, enolase; Pyk, pyruvate kinase. (F, G) The transcription changes of glucose transport and glycolysis genes after the supplementation of 4 g/L formate. Data were presented as mean ± standard deviation (error bars; n = 3). Statistical analysis was performed using a two-tailed Student's t-test. ***p < 0.001; **p < 0.01; *p < 0.05.
To better understand the promotion effect of formate on the glucose consumption of C. beijerinckii NCIMB 8052, we compared the transcriptional changes of the crucial genes responsible for glycolysis and glucose transportation in the presence and absence of 4 g/L formate (Fig. 1E, Supplementary Table S3). The samples for the analysis were collected at the logarithmic growth stage of cells. The results showed that all the examined glucose transporter genes, including the phosphotransferase system (PTS), ATP-binding cassette (ABC) transporters and sodium-dependent glucose transporters (SGLTs), were significantly up-regulated with formate (Fig. 1F); meanwhile, multiple genes for the glycolysis pathway were also up-regulated with the addition of formate (Fig. 1G). In the contrary, when the concentration of formate was increased to 6 g/L, the majority of the genes for the glycolysis pathway were significantly down-regulated (Supplementary Fig. S1), which may partially explain the poor cell growth and glucose consumption under this formate concentration. All these results indicate the promotion effect on the glucose transportation and metabolism of C. beijerinckii NCIMB 8052 by an appropriate concentration of formate, thereby enhancing the glucose consumption of this bacterium for ABE production.
3.2. Putative functional formate metabolic pathways in C. beijerinckii NCIMB 8052 and 13C-tracer analysis
As mentioned above, an incomplete WL pathway and a complete rPFOR/PFL pathway responsible for CO2 re-assimilation have been proposed in C. beijerinckii [24]. The results from sequence alignment showed that most of the enzymes in these pathways in C. beijerinckii shared >30% amino acid sequence identity with the counterparts in Acatobacterium woodii (Supplementary Table S4), an anaerobic bacterium capable of growing on formate as the sole carbon source [10]. Considering that formate can be integrated into the WL and rPFOR/Pfl pathways during their operation (Fig. 2A), these pathways may play roles in the formate metabolism of C. beijerinckii.
Fig. 2.
13C isotope analysis of formate metabolic flux in C. beijerinckii NCIMB 8052. (A) Potential formate metabolic pathways in C. beijerinckii NCIMB 8052. FTL, formate-tetrahydrofolate ligase; FOLD, bifunctional protein: methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase; METF, methylenetetrahydrofolate reductase; METR, 5-methyltetrahydrofolate corrinoid/iron sulfur protein methyltransferase; GLYA, glycine hydroxymethyltransferase; SDAA, l-serine dehydratase; METH, methionine synthase; FDH, formate dehydrogenase; CODH, carbon monoxide dehydrogenase; PFL, pyruvate formate-lyase; PFOR, pyruvic-ferredoxin oxidoreductase. CODH/ACS, carbon monoxide dehydrogenase/acetyl-CoA synthase, gray shading indicates no homologous proteins in the genome. The green lines represent the incomplete WLP pathway, the red represents the THF cycle, the yellow shows the direct formation of formate into pyruvate, and the blue shows the reaction in which formate is oxidized and fixed again. (B) The proportion of 13C-labeled amino acids in C. beijerinckii NCIMB 8052 after adding 13C-labeled sodium formate into the medium. Ala, alanine; gly, glycine; val, valine; leu, leucine; pro, proline; thr, threonine; phe, phenylalanine; asp, aspartic acid; glu, glutamic acid; lys, lysine; his, histidine; tyr, tyrosine; ser, serine; met, methionine. (C) The labeling pattern of metabolites in C. beijerinckii NCIMB 8052 with 13C-labeled sodium formate. Red circles denote the 13C atoms arising from 13C-labeled sodium formate. (D) The fractions of 13C-labeled methionine and serine in C. beijerinckii NCIMB 8052 after adding 13C-labeled sodium formate into the medium. M + 0, unlabeled; M + (1–5), one to five labeled carbons. (E) The ratios of 13C-labeled C1-3 (the first to third carbon) and C2-3 (the second to third carbon) fragments in proteinogenic serine and alanine. Data were representative of three biological replicates. Data were presented as mean ± standard deviation (error bars; n = 3). Statistical analysis was performed using a two-tailed Student's t-test. ***p < 0.001; **p < 0.01; *p < 0.05.
To confirm this possibility, we performed isotopomer analysis using 13C-labeled formate as the tracer. The exponentially grown cells in the medium (containing 5 g/L glucose supplemented with 1 g/L 13C-labeled sodium formate) were collected for measuring mass isotopomer patterns of proteinogenic amino acids using gas chromatography-mass spectrometry (GC-MS). The results showed that all the detected amino acids were labeled by 13C (Fig. 2B), demonstrating that formate was really utilized by C. beijerinckii NCIMB 8052, although the contribution degree of dissimilation and assimilation pathways to formate metabolism remained unknown.
We further examined the 13C-labeling profile of alanine, serine, and methionine synthesized from formate. The results showed that methionine and serine contained significant proportions of 13C-labeled amino acids (35.5% and 11.6%, respectively, mostly m+1), suggesting the incorporation of exogenous formate into the synthesis of methionine (labeled at the fifth carbon) and serine (labeled at the third carbon) via the methyl branch of the WL pathway (Fig. 2C and D). In addition, higher ratios of 13C-labeled C1‒3 against 13C-labeled C2‒3 (m+1) in serine and alanine were observed (Fig. 2E), indicating that quite a lot of formate was incorporated into these two amino acids via the rPFL or rPFOR enzymes and the intermediate pyruvate (Fig. 2C).
3.3. Enhanced formate tolerance and utilization of C. beijerinckii NCIMB 8052 via adaptive laboratory evolution
Since formate as a supplementary substrate could promote glucose consumption and ABE production in C. beijerinckii NCIMB 8052 (Fig. 1C and D), whether improving formate tolerance and utilization can further reinforce this phenotype? To explore this possibility, C. beijerinckii NCIMB 8052 was subjected to the adaptive evolution strategy with gradually increased formate levels.
As depicted in Fig. 3A, the strain was first cultivated in the medium containing 10 g/L glucose (without formate) for sufficient growth. Then, the grown cells were sub-cultured every 24 h in the medium containing 10 g/L glucose and 4 g/L sodium formate, during which the initial optical density of cells (OD600) after each passage was controlled at 0.1. When an obviously increased growth rate occurred, the concentration of formate was enhanced for the next round of adaptive evolution. Through this approach, the tolerance of C. beijerinckii NCIMB 8052 to formate was gradually enhanced, generating an evolved strain (named as Cbei-FA01) that grew much better than the wild-type strain in the presence of 10 g/L formate (Fig. 3B). Simultaneously, the Cbei-FA01 strain could consume 4 g/L formate and all the glucose (10 g/L) within 48 h, while the wild-type strain only consumed 3.0 g/L formate and 4.3 g/L glucose within the same period (Fig. 3C and D). These results demonstrate the enhanced formate tolerance of C. beijerinckii NCIMB 8052 after the adaptive laboratory evolution, which endowed the strain with improved ability in using formate and glucose as dual substrates.
Fig. 3.
Adaptive laboratory evolution facilitates formate tolerance and consumption by C. beijerinckii NCIMB 8052. (A) The laboratory evolution strategy. The growth of the C. beijerinckii NCIMB 8052 strain and the evolved strain Cbei-FA01 were indicated with green and red line, respectively. (B–D) The cell growth and consumption of formate and glucose of the Cbei-FA01 stain and the wild-type strain. The initial concentrations of both glucose and sodium formate in the medium were 10 g/L. Data were representative of three biological replicates. Data were presented as mean ± standard deviation (error bars; n = 3). Statistical analysis was performed using a two-tailed Student's t-test. ***p < 0.001; **p < 0.01.
3.4. Characterization of the evolved strain Cbei-FA01 in ABE production
To further examine the performance of the Cbei-FA01 strain in ABE production with adequate supply of carbon sources, the strain was cultivated in the medium containing approximate 60 g/L glucose. The results showed that the Cbei-FA01 strain exhibited significantly improved growth, glucose and formate consumption, and ABE production compared to the wild-type strain whether in the presence or absence of formate (Fig. 4A‒D). In addition, the promotion of formate on cellular performance was still observed for both the wild-type and Cbei-FA01 strains under this glucose concentration (Fig. 4A‒D). Encouragingly, the advantage of the Cbei-FA01 strain over the wild-type strain in the presence of formate was further strengthened when the glucose concentration was increased to about 70 g/L. Under this condition, Cbei-FA01 grew better than the wild-type strain (Fig. 4E); simultaneously, Cbei-FA01 could consume 65.8 g/L glucose and 2.2 g/L formate and produce 23.0 g/L ABE solvents within 60 h, 59.5%, 37.4%, and 51.7%, respectively, higher than those of the wild-type strain (Fig. 4F‒H).
Fig. 4.
The performance of the Cbei-FA01 strain in ABE production using glucose and formate as dual substrates. (A–D) Improved growth, consumption of glucose and formate, and ABE production by Cbei-FA01 against the wild-type strain with the supplementation of around 60 g/L glucose and 4 g/L sodium formate. (E–H) Improved growth, consumption of glucose and formate, and ABE production by Cbei-FA01 against the wild-type strain under higher glucose concentration (with around 70 g/L glucose and 4 g/L sodium formate). Data were presented as mean ± standard deviation (error bars; n = 3). Statistical analysis was performed using a two-tailed Student's t-test. ***p < 0.001; **p < 0.01; n.s., no significant.
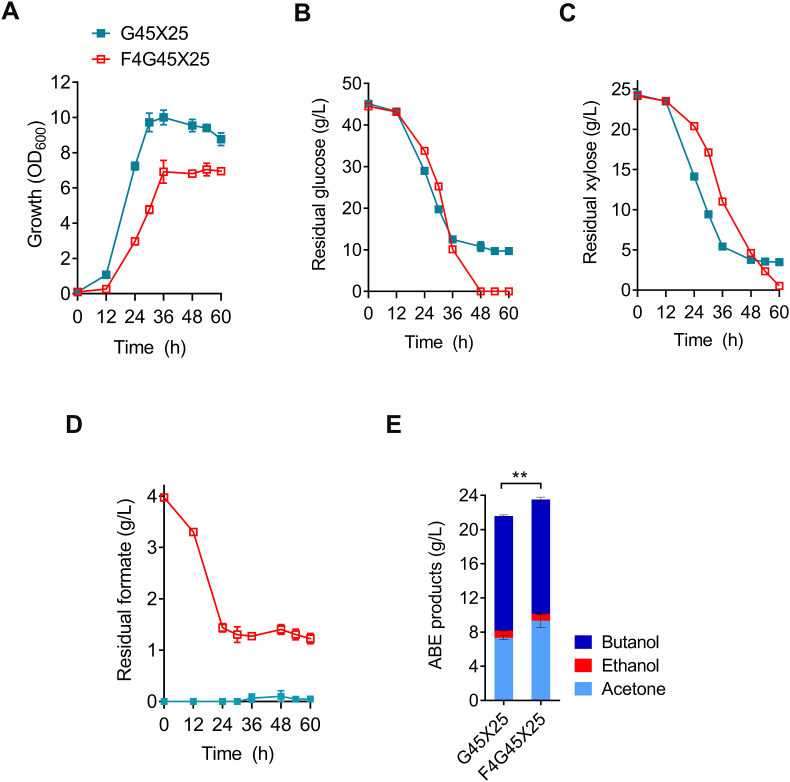
Next, we sought to test if the addition of formate could also improve the use of other sugars by the Cbei-FA01 strain. To this end, Cbei-FA01 was cultivated in the medium containing a glucose-xylose sugar mixture (simulating the carbon sources of lignocellulosic hydrolysate). As expected, enhanced consumption of both glucose and xylose was observed for Cbei-FA01 after the supplementation of formate. Although the cell growth was slightly impaired by formate, Cbei-FA01 exhausted glucose and xylose after 60 h of fermentation with the supplementation of formate, while 9.7 g/L glucose and 3.5 g/L xylose were left in the absence of formate (Fig. 5A‒C). Simultaneously, 2.8 g/L formate was consumed by Cbei-FA01 (Fig. 5D). Consequently, the final ABE solvents produced by Cbei-FA01 reached 23.5 g/L with the supplementation of formate, 9.0% higher than that in the absence of formate (Fig. 5E).
Fig. 5.
The promotion effect of formate on the fermentation of mixed hexose and pentose sugars by Cbei-FA01. (A–C) Improved growth and consumption of glucose (45 g/L) and xylose (25 g/L) of Cbei-FA01 with the supplementation of 4 g/L formate. (D) Formate consumption by Cbei-FA01. (E) Improved ABE production of Cbei-FA01 with the supplementation of formate. Data were presented as mean ± standard deviation (error bars; n = 3). Statistical analysis was performed using a two-tailed Student's t-test. **p < 0.01.
4. Discussion
Formate is an attractive CO2-equivalent resource for biorefinery. This work shows that the major ABE-producing strain C. beijerinckii NCIMB 8052 has an innate capability to metabolize this C1 carbon source. Although native formate assimilation pathways have been reported in some typical microbial hosts such as Methylobacterium extorquens, Acatobacterium woodii, and Butyribacterium methylotrophicum [8,10,37], the potential of solventogenic Clostridium species in formate metabolism remains hardly explored. Here, the possible formate assimilation and dissimilation pathways were identified using 13C isotope analysis and an unreported promotion effect of formate on glucose and xylose consumption was discovered in C. beijerinckii NCIMB 8052. Furthermore, the tolerance and metabolism of this strain to formate was enhanced by adaptive laboratory evolution approach, showcasing the potential of this bacterium in formate utilization.
Lignocellulosic biomass is a huge carbon source that can be converted to chemicals and fuels through chemical and biological routes [38,39]. C. beijerinckii is capable of using various feedstocks [40]. This characteristic enabled it to efficiently use hexose and pentose sugars in lignocellulosic biomass for ABE production [41,42]. Formate as a chemical component present in lignocellulosic hydrolysates, due to the chemical pretreatment of raw materials, has been known to inhibit microbial growth [43,44]. Thus, the presence of formate metabolic routes may allow C. beijerinckii NCIMB 8052 to overcome formate inhibition and grow at relatively higher formate concentrations, which is helpful for the application of this bacterium in ABE production from lignocellulosic hydrolysates.
Although C. beijerinckii NCIMB 8052 has been proven to be a native formate-utilizing bacterium in this study, the exact relative contributions of the reverse PFL/PFOR pathways and the THF cycle to formate metabolism remains unclear (Fig. 2A), because we could not shut down the THF cycle which has direct influence on microbial basic metabolism [45]. Here, it can be concluded that FDH-mediated formate oxidation reaction (generating CO2) is crucial for the formate metabolism in C. beijerinckii NCIMB 8052 based on the findings that the disruption of fdh (encoding formate dehydrogenase) greatly impaired formate consumption as well as the transcription of fdh was significantly up-regulated (30.3-fold) under formate stress (Supplementary Fig. S2 and Fig. S3). The use of formate to produce CO2 through FDH-mediated oxidation pathway can provide extra reducing power, probably beneficial for the formation of ethanol and butanol (two major compounds in ABE solvents) in C. beijerinckii NCIMB 8052. The generated CO2 may be further assimilated by C. beijerinckii NCIMB 8052 via the reverse PFOR reaction or other carbon fixation reactions such as phosphoenolpyruvate carboxylation. In addition, microbial growth on formate requires a high rate of reducing power regeneration. C. beijerinckii NCIMB 8052 is also a potent hydrogen producer [46,47]; thus, the consumption of formate may affect the formation of hydrogen, besides ABE solvents, in this Clostridium strain, which remains to be determined.
Adaptive laboratory evolution has unique advantages over classical genetic engineering in improving microbes to efficiently utilize unfavorable substrates [[48], [49], [50], [51]]. Herein, this strategy finds utility in promoting formate tolerance and utilization of C. beijerinckii NCIMB 8052. However, we noticed that the improvement of formate consumption by Cbei-FA01 was not as significant as that of formate tolerance (Fig. 3), indicating no direct causal relationship between these two phenotypes. A possible reason for this phenomenon is that the rapid metabolism of glucose as the preferred substrate may cause CCR (carbon catabolite repression)-like inhibition on formate metabolism. This limitation may be overcome by rational strain design or a modified adaptive laboratory evolution process to reduce the dependence of cell growth on glucose.
The commonly used substrates in clostridial ABE fermentation are sugars or starchy materials. A complete substitution of these traditional substrates with cheaper feedstocks such as lignocellulose in ABE production is economically unviable so far. Partial replacement of sugars or starchy materials with other cheaper feedstocks is an alternative approach. The formate metabolism as well as its promotion on glucose and xylose utilization of C. beijerinckii NCIMB 8052 makes formate a cheap material worth considering. However, it should be noted that C. beijerinckii has a large number of strains with different phenotypes during fermentation; thus, whether the promotion of formate on the ABE production of C. beijerinckii NCIMB 8052 is applicable to other C. beijerinckii strains remains unknown, which needs further exploration.
In summary, the results in this study suggest the potential of C. beijerinckii NCIMB 8052 in using formate as a secondary substrate for ABE production. Formate metabolism will provide additional carbon source and reducing power for cell growth and product formation. Our future work will continue to enhance formate assimilation in C. beijerinckii through multiple strategies, which will allow more substitution of sugars with formate in ABE production.
5. Conclusions
This study reveals the potential of C. beijerinckii NCIMB 8052 in the utilization of formate as well as the promotion effect of formate on glucose and xylose consumption in this bacterium. The pathways and the associated genes that are potentially responsible for the formate metabolism in C. beijerinckii NCIMB 8052 were analyzed. The possibility of developing a C. beijerinckii strain capable of efficiently consuming both sugars and formate was also demonstrated through adaptive laboratory evolution. The findings and strain improvement strategies in this study will be useful for formate biorefinery.
Declaration of competing interest
The authors declare that they have no competing interests.
CRediT authorship contribution statement
Ran Zhao: Investigation, Writing – original draft. Wenyue Dong: Formal analysis. Chen Yang: Resources. Weihong Jiang: Conceptualization, Supervision, Writing – review & editing, Project administration. Jinzhong Tian: Conceptualization, Methodology. Yang Gu: Conceptualization, Writing – review & editing, Supervision, Project administration.
Acknowledgements
This work was supported by the National Key R&D Program of China (2018YFA0901500 and 2021YFC2103500), Science and Technology Commission of Shanghai Municipality (21DZ1209100), DNL Cooperation Fund, CAS (DNL202013), and Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project (TSBICIP-KJGG-016).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2023.01.005.
Contributor Information
Jinzhong Tian, Email: tianjinzhong@cemps.ac.cn.
Yang Gu, Email: ygu@cemps.ac.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Sun X., Atiyeh H.K., Kumar A., Zhang H., Tanner R.S. Biochar enhanced ethanol and butanol production by Clostridium carboxidivorans from syngas. Bioresour Technol. 2018;265:128–138. doi: 10.1016/j.biortech.2018.05.106. [DOI] [PubMed] [Google Scholar]
- 2.Charubin K., Papoutsakis E.T. Direct cell-to-cell exchange of matter in a synthetic Clostridium syntrophy enables CO2 fixation, superior metabolite yields, and an expanded metabolic space. Metab Eng. 2019;52:9–19. doi: 10.1016/j.ymben.2018.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Lemgruber R. de SP., Valgepea K., Tappel R., Behrendorff J.B., Palfreyman R.W., Plan M., et al. Systems-level engineering and characterisation of Clostridium autoethanogenum through heterologous production of poly-3-hydroxybutyrate (PHB) Metab Eng. 2019;53:14–23. doi: 10.1016/j.ymben.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Roy M., Yadav R., Chiranjeevi P., Patil S.A. Direct utilization of industrial carbon dioxide with low impurities for acetate production via microbial electrosynthesis. Bioresour Technol. 2021;320 doi: 10.1016/j.biortech.2020.124289. [DOI] [PubMed] [Google Scholar]
- 5.Bang J., Ahn J.H., Lee J.A., Hwang C.H., Kim G.B., Lee J., et al. Synthetic formatotrophs for one‐carbon biorefinery. Adv Sci. 2021;8 doi: 10.1002/advs.202100199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng T., Liu C., Guo C., Zhang M., Li X., Jiang Q., et al. Copper-catalysed exclusive CO2 to pure formic acid conversion via single-atom alloying. Nat Nanotechnol. 2021;16:1386–1393. doi: 10.1038/s41565-021-00974-5. [DOI] [PubMed] [Google Scholar]
- 7.Zheng T., Zhang M., Wu L., Guo S., Liu X., Zhao J., et al. Upcycling CO2 into energy-rich long-chain compounds via electrochemical and metabolic engineering. Nat Catal. 2022;5:388–396. doi: 10.1038/s41929-022-00775-6. [DOI] [Google Scholar]
- 8.Crowther G.J., Kosály G., Lidstrom M.E. Formate as the main branch point for methylotrophic metabolism in Methylobacterium extorquens AM1. J Bacteriol. 2008;190:5057–5062. doi: 10.1128/JB.00228-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandra T.S., Shethna Y.I. Oxalate, formate, formamide, and methanol metabolism in Thiobacillus novellus. J Bacteriol. 1977;131:389–398. doi: 10.1128/jb.131.2.389-398.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moon J., Dönig J., Kramer S., Poehlein A., Daniel R., Müller V. Formate metabolism in the acetogenic bacterium Acetobacterium woodii. Environ Microbiol. 2021;23:4214–4227. doi: 10.1111/1462-2920.15598. [DOI] [PubMed] [Google Scholar]
- 11.Bang J., Lee S.Y. Assimilation of formic acid and CO2 by engineered Escherichia coli equipped with reconstructed one-carbon assimilation pathways. Proc Natl Acad Sci USA. 2018;115:E9271–E9279. doi: 10.1073/pnas.1810386115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu H., Liao J.C. A modified serine cycle in Escherichia coli coverts methanol and CO2 to two-carbon compounds. Nat Commun. 2018;9:3992. doi: 10.1038/s41467-018-06496-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bang J., Hwang C.H., Ahn J.H., Lee J.A., Lee S.Y. Escherichia coli is engineered to grow on CO2 and formic acid. Nat Microbiol. 2020;5:1459–1463. doi: 10.1038/s41564-020-00793-9. [DOI] [PubMed] [Google Scholar]
- 14.Kim S., Lindner S.N., Aslan S., Yishai O., Wenk S., Schann K., et al. Growth of E. coli on formate and methanol via the reductive glycine pathway. Nat Chem Biol. 2020;16:538–545. doi: 10.1038/s41589-020-0473-5. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez de la C.J., Machens F., Messerschmidt K., Bar-Even A. Core catalysis of the reductive glycine pathway demonstrated in yeast. ACS Synth Biol. 2019;8:911–917. doi: 10.1021/acssynbio.8b00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahn J.H., Bang J., Kim W.J., Lee S.Y. Formic acid as a secondary substrate for succinic acid production by metabolically engineered Mannheimia succiniciproducens. Biotechnol Bioeng. 2017;114:2837–2847. doi: 10.1002/bit.26435. [DOI] [PubMed] [Google Scholar]
- 17.Harris D.M., Van der Krogt Z.A., Van Gulik W.M., Van Dijken J.P., Pronk J.T. Formate as an auxiliary substrate for glucose-limited cultivation of penicillium chrysogenum: impact on penicillin G production and biomass yield. Appl Environ Microbiol. 2007;73:5020–5025. doi: 10.1128/AEM.00093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Millat T., Winzer K. Mathematical modelling of clostridial acetone-butanol-ethanol fermentation. Appl Microbiol Biotechnol. 2017;101:2251–2271. doi: 10.1007/s00253-017-8137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarangi P.K., Nanda S. In: Recent advancements in biofuels and bioenergy utilization. Sarangi P.K., Nanda S., Mohanty P., editors. Springer; Singapore: 2018. Recent developments and challenges of acetone-butanol-ethanol fermentation; pp. 111–123. [DOI] [Google Scholar]
- 20.Li Y., Tang W., Chen Y., Liu J., Lee C.F. Potential of acetone-butanol-ethanol (ABE) as a biofuel. Fuel. 2019;242:673–686. doi: 10.1016/j.fuel.2019.01.063. [DOI] [Google Scholar]
- 21.Li S., Huang L., Ke C., Pang Z., Liu L. Pathway dissection, regulation, engineering and application: lessons learned from biobutanol production by solventogenic clostridia. Biotechnol Biofuels. 2020;13:39. doi: 10.1186/s13068-020-01674-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao H., Li Z., Jiang Y., Yang Y., Jiang W., Gu Y., et al. Metabolic engineering of D-xylose pathway in Clostridium beijerinckii to optimize solvent production from xylose mother liquid. Metab Eng. 2012;14:569–578. doi: 10.1016/j.ymben.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 23.Cho D.H., Lee Y.J., Um Y., Sang B.-I., Kim Y.H. Detoxification of model phenolic compounds in lignocellulosic hydrolysates with peroxidase for butanol production from Clostridium beijerinckii. Appl Microbiol Biotechnol. 2009;83:1035–1043. doi: 10.1007/s00253-009-1925-8. [DOI] [PubMed] [Google Scholar]
- 24.Sandoval-Espinola W.J., Chinn M.S., Thon M.R., Bruno-Bárcena J.M. Evidence of mixotrophic carbon-capture by n-butanol-producer Clostridium beijerinckii. Sci Rep. 2017;7 doi: 10.1038/s41598-017-12962-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertani G. Studies on lysogenesis I: the mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiesenborn D.P., Rudolph F.B., Papoutsakis E.T. Thiolase from Clostridium acetobutylicum ATCC 824 and its role in the synthesis of acids and solvents. Appl Environ Microbiol. 1988;54:2717–2722. doi: 10.1128/aem.54.11.2717-2722.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C.-K., Blaschek H.P. Effect of acetate on molecular and physiological aspects of Clostridium beijerinckii NCIMB 8052 solvent production and strain degeneration. Appl Environ Microbiol. 1999;65:499–505. doi: 10.1128/AEM.65.2.499-505.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y., Li X., Mao Y., Blaschek H.P. Single-nucleotide resolution analysis of the transcriptome structure of Clostridium beijerinckii NCIMB 8052 using RNA-Seq. BMC Genom. 2011;12:479. doi: 10.1186/1471-2164-12-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun Z., Chen Y., Yang C., Yang S., Gu Y., Jiang W. A novel three-component system-based regulatory model for D-xylose sensing and transport in Clostridium beijerinckii. Mol Microbiol. 2015;95:576–589. doi: 10.1111/mmi.12894. [DOI] [PubMed] [Google Scholar]
- 30.Heap J.T., Pennington O.J., Cartman S.T., Carter G.P., Minton N.P. The Clostron: a universal gene knock-out system for the genus Clostridium. J Microbiol Methods. 2007;70:452–464. doi: 10.1016/j.mimet.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 31.Mermelstein L.D., Welker N.E., Bennett G.N., Papoutsakis E.T. Expression of cloned homologous fermentative genes in Clostridium acetobutylicum ATCC 824. Nat Biotechnol. 1992;10:190–195. doi: 10.1038/nbt0292-190. [DOI] [PubMed] [Google Scholar]
- 32.Ren C., Gu Y., Hu S., Wu Y., Wang P., Yang Y., et al. Identification and inactivation of pleiotropic regulator CcpA to eliminate glucose repression of xylose utilization in Clostridium acetobutylicum. Metab Eng. 2010;12:446–454. doi: 10.1016/j.ymben.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 33.Xiong W., Lin P.P., Magnusson L., Warner L., Liao J.C., Maness P.-C., et al. CO2 -fixing one-carbon metabolism in a cellulose-degrading bacterium Clostridium thermocellum. Proc Natl Acad Sci USA. 2016;113:13180–13185. doi: 10.1073/pnas.1605482113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L., Liu Y., Yang Y., Jiang W., Gu Y. A novel dual-cre motif enables two-way autoregulation of CcpA in Clostridium acetobutylicum. Appl Environ Microbiol. 2018;84:e00114–e00118. doi: 10.1128/AEM.00114-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warnecke T., Gill R.T. Organic acid toxicity, tolerance, and production in Escherichia coli biorefining applications. Microb Cell Factories. 2005;4:25. doi: 10.1186/1475-2859-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicholls P. Formate as an inhibitor of cytochrome c oxidase. Biochem Biophys Res Commun. 1975;67:610–616. doi: 10.1016/0006-291X(75)90856-6. [DOI] [PubMed] [Google Scholar]
- 37.Humphreys J.R., Hebdon S.D., Rohrer H., Magnusson L., Urban C., Chen Y.-P., et al. Establishing Butyribacterium methylotrophicum as a platform organism for the production of biocommodities from liquid C1 metabolites. Appl Environ Microbiol. 2022;88 doi: 10.1128/aem.02393-21. e02393-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen G., Yuan X., Chen S., Liu S., Jin M. High titer cellulosic ethanol production from sugarcane bagasse via DLCA pretreatment and process development without washing/detoxifying pretreated biomass. Renew Energy. 2022;186:904–913. doi: 10.1016/j.renene.2022.01.062. [DOI] [Google Scholar]
- 39.Yuan X., Chen X., Shen G., Chen S., Yu J., Zhai R., et al. Densifying lignocellulosic biomass with sulfuric acid provides a durable feedstock with high digestibility and high fermentability for cellulosic ethanol production. Renew Energy. 2022;182:377–389. doi: 10.1016/j.renene.2021.10.015. [DOI] [Google Scholar]
- 40.Yang Y., Nie X., Jiang Y., Yang C., Gu Y., Jiang W. Metabolic regulation in solventogenic clostridia: regulators, mechanisms and engineering. Biotechnol Adv. 2018;36:905–914. doi: 10.1016/j.biotechadv.2018.02.012. [DOI] [PubMed] [Google Scholar]
- 41.Guo T., Tang Y., Zhang Q., Du T., Liang D., Jiang M., et al. Clostridium beijerinckii mutant with high inhibitor tolerance obtained by low-energy ion implantation. J Ind Microbiol Biotechnol. 2012;39:401–407. doi: 10.1007/s10295-011-1017-5. [DOI] [PubMed] [Google Scholar]
- 42.Liu Z.-Y., Yao X.-Q., Zhang Q., Liu Z., Wang Z.-J., Zhang Y.-Y., et al. Modulation of the acetone/butanol ratio during fermentation of corn stover-derived hydrolysate by Clostridium beijerinckii strain NCIMB 8052. Appl Environ Microbiol. 2017;83 doi: 10.1128/AEM.03386-16. e03386-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Danon B., van der Aa L., de Jong W. Furfural degradation in a dilute acidic and saline solution in the presence of glucose. Carbohydr Res. 2013;375:145–152. doi: 10.1016/j.carres.2013.04.030. [DOI] [PubMed] [Google Scholar]
- 44.Jönsson L.J., Martín C. Pretreatment of lignocellulose: formation of inhibitory by-products and strategies for minimizing their effects. Bioresour Technol. 2016;199:103–112. doi: 10.1016/j.biortech.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 45.Hong Y., Ren J., Zhang X., Wang W., Zeng A.-P. Quantitative analysis of glycine related metabolic pathways for one-carbon synthetic biology. Curr Opin Biotechnol. 2020;64:70–78. doi: 10.1016/j.copbio.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y., Li J., Meng J., Sun K., Yan H. A neutral red mediated electro-fermentation system of Clostridium beijerinckii for effective co-production of butanol and hydrogen. Bioresour Technol. 2021;332 doi: 10.1016/j.biortech.2021.125097. [DOI] [PubMed] [Google Scholar]
- 47.Li J., Zhang Y., Sun K., Liu W., Yan H., Meng J. Optimization of a cathodic electro-fermentation process for enhancing co-production of butanol and hydrogen via acetone-butanol-ethanol fermentation of Clostridium beijerinckii. Energy Convers Manag. 2022;251 doi: 10.1016/j.enconman.2021.114987. [DOI] [Google Scholar]
- 48.Chen F.Y.-H., Jung H.-W., Tsuei C.-Y., Liao J.C. Converting Escherichia coli to a synthetic methylotroph growing solely on methanol. Cell. 2020;182:933–946. doi: 10.1016/j.cell.2020.07.010. [DOI] [PubMed] [Google Scholar]
- 49.Espinosa M.I., Gonzalez-Garcia R.A., Valgepea K., Plan M.R., Scott C., Pretorius I.S., et al. Adaptive laboratory evolution of native methanol assimilation in Saccharomyces cerevisiae. Nat Commun. 2020;11:5564. doi: 10.1038/s41467-020-19390-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bennett R.K., Gregory G.J., Gonzalez J.E., Har J.R.G., Antoniewicz M.R., Papoutsakis E.T. Improving the methanol tolerance of an Escherichia coli methylotroph via adaptive laboratory evolution enhances synthetic methanol utilization. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.638426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan Z., Zhang J., Bao J. Increasing cellulosic ethanol production by enhancing phenolic tolerance of Zymomonas mobilis in adaptive evolution. Bioresour Technol. 2021;329 doi: 10.1016/j.biortech.2021.124926. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.