Abstract
NF-E2-related factor 2 (NRF2) plays a crucial role in the maintenance of cellular homeostasis by regulating various enzymes and proteins that are involved in the redox reactions utilizing sulfur. While substantial impacts of NRF2 on mitochondrial activity have been described, the precise mechanism by which NRF2 regulates mitochondrial function is still not fully understood. Here, we demonstrated that NRF2 increased intracellular persulfides by upregulating the cystine transporter xCT encoded by Slc7a11, a well-known NRF2 target gene. Persulfides have been shown to play an important role in mitochondrial function. Supplementation with glutathione trisulfide (GSSSG), which is a form of persulfide, elevated the mitochondrial membrane potential (MMP), increased the oxygen consumption rate (OCR) and promoted ATP production. Persulfide-mediated mitochondrial activation was shown to require the mitochondrial sulfur oxidation pathway, especially sulfide quinone oxidoreductase (SQOR). Consistently, NRF2-mediated mitochondrial activation was also dependent on SQOR activity. This study clarified that the facilitation of persulfide production and sulfur metabolism in mitochondria by increasing cysteine availability is one of the mechanisms for NRF2-dependent mitochondrial activation.
Keywords: xCT, Mitochondria, Cystine, Cysteine, Persulfide, Mitochondrial membrane potential
Graphical abstract
Highlights
-
•
Cystine restriction reduces persulfides and inhibits mitochondrial activity.
-
•
Persulfide supplementation promotes mitochondrial activity.
-
•
NRF2 activation increases persulfides and promotes mitochondrial activity.
-
•
Upregulation of xCT induced by NRF2 activation results in perusulfide increase.
-
•
NRF2-mediated mitochondrial activation relies on persulfide metabolism.
1. Introduction
The KEAP1 (Kelch-like ECH-associated protein 1)-NRF2 (nuclear factor-erythroid 2-related factor 2) system is an important cytoprotective mechanism against oxidative and electrophilic stresses [1]. Under basal conditions, NRF2 is constitutively subjected to ubiquitination and proteasomal degradation through binding to KEAP1, a substrate adaptor protein of E3 ubiquitin ligase. In the presence of oxidative and electrophilic stresses, KEAP1 is inactivated, and NRF2 is stabilized. Consequently, NRF2 translocates into the nucleus, binds to antioxidant response elements (AREs), recruits coactivators for enhancer formation and activates the transcription of a battery of cytoprotective genes, including SLC7A11, encoding the cystine transporter xCT [[2], [3], [4]]. In addition to its antioxidant function [5], NRF2 exerts a potent anti-inflammatory function [6,7]. Recent studies have also clarified that appropriate activation of NRF2 has antiaging effects by limiting the accumulation of oxidative damage in tissues [[8], [9], [10]]. Moreover, NRF2 has been reported to confer resistance against mitochondrial toxins [11,12], protect against mitochondria-related disorders such as Parkinson's disease and Alzheimer's disease [13,14] and improve skeletal muscle performance and exercise endurance with increased oxygen consumption [15,16]. These reports suggest that NRF2 plays a crucial role in the regulation of mitochondrial function. Indeed, NRF2 has been shown to impact mitochondrial function as a master regulator of cellular redox homeostasis [17]. However, the precise mechanism by which NRF2 enhances mitochondrial activity is still not fully understood.
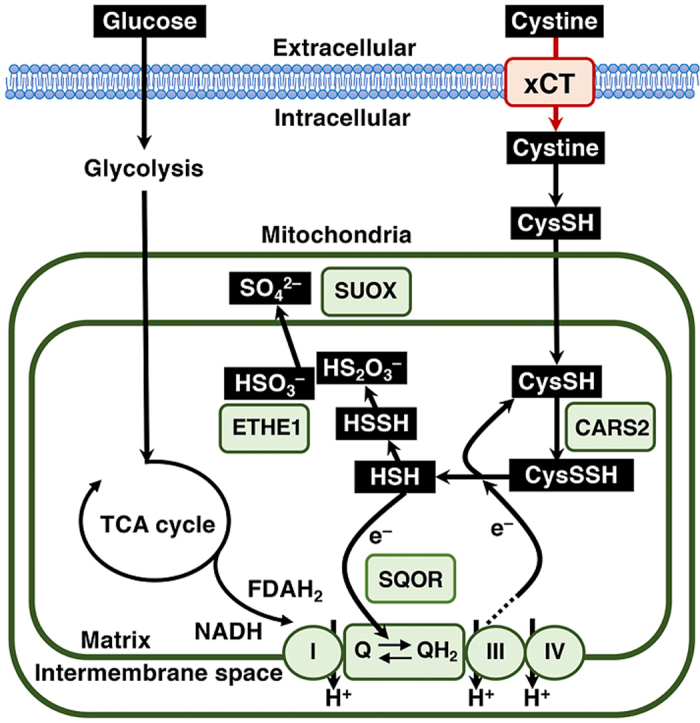
Mitochondria are key organelles functioning as the “energy powerhouse” in eukaryotic cells, which principally rely on redox reactions. The physiological importance of mitochondria has been widely appreciated, and mitochondrial dysfunction is associated with various diseases [[18], [19], [20]]. Recently, we found that mitochondrial sulfur metabolism makes an important contribution to cellular energy production [21]. Cysteinyl tRNA synthetase (CARS) has been identified as a dual enzyme possessing two independent catalytic activities: cysteinyl tRNA synthesizing activity and cysteine persulfide synthesizing activity. Cysteine persulfide production in mitochondria mediated by CARS2, which is a mitochondrial isoform of CARS, is required for the maintenance of the mitochondrial membrane potential (MMP) [21], indicating a critical contribution of persulfides to MMP generation. Cysteine persulfide in mitochondria was shown to be reductively metabolized to sulfide depending on the activity of the electron transport chain (ETC) [21], implying that ETC-derived electrons are accepted by cysteine persulfide. Sulfide is oxidized by sulfide quinone oxidoreductase (SQOR), which is followed by subsequent sulfur oxidation by a persulfide-oxidizing enzyme, ethylmalonic encephalopathy 1 protein (ETHE1), and sulfite oxidase (SUOX), resulting in the excretion of sulfate and thiosulfate [22]. Because sulfide accumulation inhibits ETC activity [23,24] and because SQOR transfers electrons from sulfide to ubiquinone of the ETC [25], the sulfur oxidation pathway is thought to protect cells from sulfide toxicity and to support electron transport efficiency by bringing ETC-derived electrons back to ubiquinone of the ETC.
Because NRF2 promotes cystine uptake by upregulating SLC7A11, which encodes the cystine transporter xCT [26], we hypothesized that NRF2 enhances mitochondrial activity by increasing cellular cysteine availability to facilitate mitochondrial sulfur metabolism. In this study, we found that cystine availability is one of the major determinants of intracellular persulfides and that persulfide supplementation promotes mitochondrial function. Mitochondrial activation induced by NRF2 activation due to KEAP1 inhibition relies on cystine uptake, followed by persulfide production and sulfur oxidation.
2. Materials and methods
2.1. Reagents
Dimethyl sulfoxide (DMSO) was purchased from Wako Pure Chemicals (Osaka, Japan). Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) was purchased from Abcam (Cambridge, UK). Antimycin A, rotenone and sulfasalazine (SASP) were obtained from Santa Cruz Biotechnology (Dallas, TX, USA). Oligomycin was purchased from Alomone Labs (Jerusalem, Israel). Glutathione trisulfide (GSSSG) was synthesized as previously described [27,28]. GSSSG was dissolved in distilled water to prepare a 1 mM stock solution until analysis. Sodium acetate, 30 mM, was added to adjust the stock solution to a pH of 5.0. The stock solution was diluted with culture medium to the indicated final concentration.
2.2. Cell culture
Hepa1c1c7, HeLa and H1650 cells were cultured and maintained in low glucose Dulbecco's Modified Eagle Medium (DMEM; Wako Pure Chemicals, Osaka, Japan) containing 10% fetal bovine serum (FBS; Biosera, Kansas, MO, USA) and 1% penicillin/streptomycin (Nacalai Tesque, Kyoto, Japan) under 5% CO2 at 37 °C.
2.3. Cell viability and growth assays
Hepa1c1c7, HeLa and H1650 cells (5.0 × 104 cells/well in 24-well plates) were cultured in low glucose DMEM containing 10% FBS for 24 h. The culture medium was removed, and the cells were washed twice with PBS. The cells were further cultured in cystine-depleted low glucose DMEM supplemented with 10% FBS and cystine (20 or 200 μM) under 5% CO2 at 37 °C for designated periods of time (0, 12 and 24 h). Viability and growth were measured by staining with 0.4% trypan blue and manual counting with a hemocytometer. The percentage of viability was defined as the percent of live cells per total cells.
2.4. Mitochondrial membrane potential assay
The mitochondrial membrane potential (MMP) was measured using a Cell Meter JC-10 Mitochondrion Membrane Potential Assay Kit (AAT Bioquest, Sunnyvale, CA, USA) according to the manufacturer's instructions. Cells were seeded in 4-chamber 35-mm culture dishes at a density of 1 × 105 cells/well for confocal microscopy and in 96-well black-walled clear-bottom plates at a density of 2 × 104 cells/well and incubated for designated periods of time. JC-10 dye loading solutions were added to cells and incubated at 37 °C and 5% CO2 for 1 h. Then, assay buffer was added. For the microplate reader assay, the fluorescence intensities were measured at Ex/Em = 490/525 nm and 540/590 nm with a microplate reader (SpectraMax M2e, Molecular Devices, San Jose, CA, USA). For imaging, the cells were observed at Ex/Em = 488/515-535 nm and 552/570-610 nm under confocal microscopy (TCS SP8, Leica, Wetzlar, Germany). The fluorescence intensity was measured by ImageJ (National Institute of Health).
2.5. Persulfide detection
Intracellular persulfides were detected using Sulfane Sulfur Probe 4 (SSP4, Dojindo, Kumamoto, Japan) according to the manufacturer's instructions. The cells were seeded in 4-chamber 35 mm culture dishes at a density of 1 × 105 cells/well and incubated for designated periods of time. After two washes with serum-free low glucose DMEM, 20 μM SSP4 working solution containing 0.5 mM cetyltrimethylammonium bromide was added to the cells and incubated at 37 °C and 5% CO2 for 15 min. Then, the cells were washed twice with PBS and observed at Ex/Em = 488/515-535 nm with confocal microscopy (TCS SP8, Leica). The fluorescence intensity was measured by ImageJ (National Institute of Health).
2.6. Immunoblot analysis
The cells were seeded in 35 mm culture dishes at a density of 2 × 105 cells/well and incubated for designated periods of time. After the designated treatment, whole cells were lysed in 100 μl of sodium dodecyl sulfate buffer. The samples (10 μl) were subjected to electrophoresis on 7.5% or 10% sodium dodecyl sulfate-polyacrylamide gels for 90–120 min at 10 mA and then transferred onto polyvinylidene difluoride membranes for 100 min at 200 mA. The membranes were blocked in 5% skim milk in Tris-buffered saline with Tween 20 (TBS-T) for 1 h and incubated overnight at 4 °C in the presence of primary antibodies at dilutions of 1:1000–1:3000 in 5% skim milk in TBS-T or Signal Enhancer HIKARI for Western Blotting and ELISA Solution A (Nacalai Tesque). After four washes with TBS-T, the membranes were incubated with the corresponding species-appropriate secondary antibodies at dilutions of 1:1000–1:3000 in 5% skim milk in TBS-T or TBS-T for 1 h. Then the immunoreactive bands on the membrane were visualized with ECL Prime Western Blotting Detection Reagents (GE Healthcare, Chicago, IL, USA) or Chemi-Lumi One L (Nacalai Tesque), and the images were captured using the LAS-4000 mini (Fujifilm, Tokyo, Japan). The antibodies used were as follows: anti-NRF2 (sc-13032, Santa Cruz Biotechnology) and anti-α-tubulin (T9026, Sigma‒Aldrich, St. Louis, MO, USA). The antibody against KEAP1 was described previously [29].
2.7. RNA purification and quantitative real-time PCR (RT‒PCR)
Total RNA samples were prepared from cells using ISOGEN (Wako Pure Chemicals). cDNAs were synthesized from 500 ng of total RNA using ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo, Osaka, Japan) in three independent experiments. Quantitative real-time PCR was performed with a QuantStudio real-time PCR system (Thermo Fisher Scientific, Waltham, MA, USA) using KAPA SYBR FAST qPCR Master Mix (Kapa Biosystems, Wilmington, MA, USA). β-Actin and Hprt were used as internal controls for normalization. The primer sequences used in the quantitative real-time PCR are listed in Supplementary Table 1.
2.8. Oxygen consumption rate (OCR) measurements
Hepa1c1c7 cells were seeded in Agilent Seahorse XFp cell culture miniplates (Agilent Technologies, Santa Clara, CA, USA) at a density of 1.2 × 104 cells/well and cultured with low glucose DMEM that was phenol red-free and cystine-deficient (Cell Science & Technology Inst., Inc) containing 10% FBS in the presence of 200 μM cystine and/or 10 μM GSSSG under 5% CO2 at 37 °C for 24 h. Then, the culture medium was changed to XF DMEM (Cat# 103575-100, Agilent Technologies) supplemented with 2 mM glutamine, 1 mM sodium pyruvate, 10 mM glucose and/or 10 μM GSSSG and cultured for 1 h in a CO2-free 37 °C incubator before performing a Mito Stress Test (Agilent Technologies). In the GSSSG and GSSG supplementation study, 10 μM GSSSG or GSSG was added to XF DMEM containing 2 mM glutamine, 1 mM sodium pyruvate and 10 mM glucose, and the pH was adjusted to 7.40. The oxygen consumption rate (OCR) was measured with an XFp flux analyzer (Agilent Technologies) using the manufacturer's protocols. For the Mito Stress Test, the following drug concentrations were used: 1 μM oligomycin, 150 nM FCCP, and 1 μM rotenone +1 μM antimycin A (Agilent Technologies).
H1650 cells were seeded in XF24 cell culture microplates (Agilent Technologies, Santa Clara, CA, USA) at a density of 4 × 104 cells/well and cultured with low glucose DMEM containing 10% FBS for 24 h. Then, the culture medium was changed to XF DMEM (Agilent Technologies) supplemented with 2 mM glutamine, 1 mM sodium pyruvate and 25 mM glucose and cultured for 1 h in a CO2-free 37 °C incubator before performing the Mito Stress Test (Agilent Technologies). The oxygen consumption rate (OCR) was measured with an XF24 flux analyzer (Agilent Technologies) using the manufacturer's protocols. For the Mito Stress Test, the following drug concentrations were used: 1 μM oligomycin, 100 nM FCCP, and 1 μM rotenone +1 μM antimycin A (Agilent Technologies).
2.9. Small interfering RNA (siRNA) transfection
Mouse Nfe2l2 (Nrf2) (cat. # L-040766-00-0005), mouse Keap1 (cat. # M-041104-01-0005), mouse Cars2 (cat. # L-060862-01-0005), mouse Sqor (cat. # M-063889-00-0005), mouse Ethe1 (cat. # L-056791-01-0005), mouse Suox (cat. # L-053903-01-0005), human KEAP1 (cat. # L-012453-00-0020), human SLC7A11 (cat. # M-007612-01-0010), human SQOR (cat. # M-008271-01-0010), human ETHE1 (cat. # M-012508-00-0005), and human SUOX (cat. # L-008310-00-0005) siRNAs were purchased from Dharmacon (Lafayette, CO, USA). Human NFE2L2 (Nrf2) siRNAs were purchased from Invitrogen (Waltham, MA, USA, cat. # HSS107128). Human CARS2 siRNAs were purchased from Ambion (Austin, TX, USA, cat. # 4392420). MISSION siRNA Universal Negative Controls (Sigma‒Aldrich) were used as controls. Hepa1c1c7, HeLa and H1650 cells were transfected with 100 nM siRNA using RNA iMAX reagent (Invitrogen) according to the manufacturer's protocol. At 24 h after transfection, the medium was changed to fresh medium containing the indicated reagents.
2.10. ATP measurements
Hepa1c1c7 cells were seeded in 96-well flat clear-bottom plates (View PlateTM-96-F TC, PerkinElmer, cat. # 6005182, USA) at a density of 5 × 103 cells/well and cultured with low glucose DMEM that was phenol red-free and cystine-deficient (Cell Science & Technology Inst., Inc.) containing 10% FBS in the presence of cystine (200 μM or 20 μM) under 5% CO2 at 37 °C for 24 h. For the GSSSG and GSSG supplementation study, 10 μM GSSSG or 10 μM GSSG was added to medium containing 200 μM cystine. The ADP/ATP ratio measurement was performed according to the manufacturer's instructions (ADP/ATP Ratio Assay Kit, cat. # MAK135, Sigma‒Aldrich, USA).
2.11. Statistical analysis
Experiments were repeated independently at least three times. All data are expressed as the mean ± S.D. The confidence interval was calculated for the evaluation of fold changes. α < 0.05 was considered to be statistically significant. An unpaired Student's t-test was used to analyze differences between two groups. One-way analysis of variance (ANOVA) followed by Tukey's post hoc test was used to analyze differences among three groups. P < 0.05 was considered to be statistically significant. These analyses were performed using GraphPad Prism 9.5 (Graph Pad Software, San Diego, CA, USA) and JMP pro 15 (SAS Institute, Cary, NC, USA).
3. Results
3.1. Cystine restriction reduces intracellular persulfides and inhibits mitochondrial activity
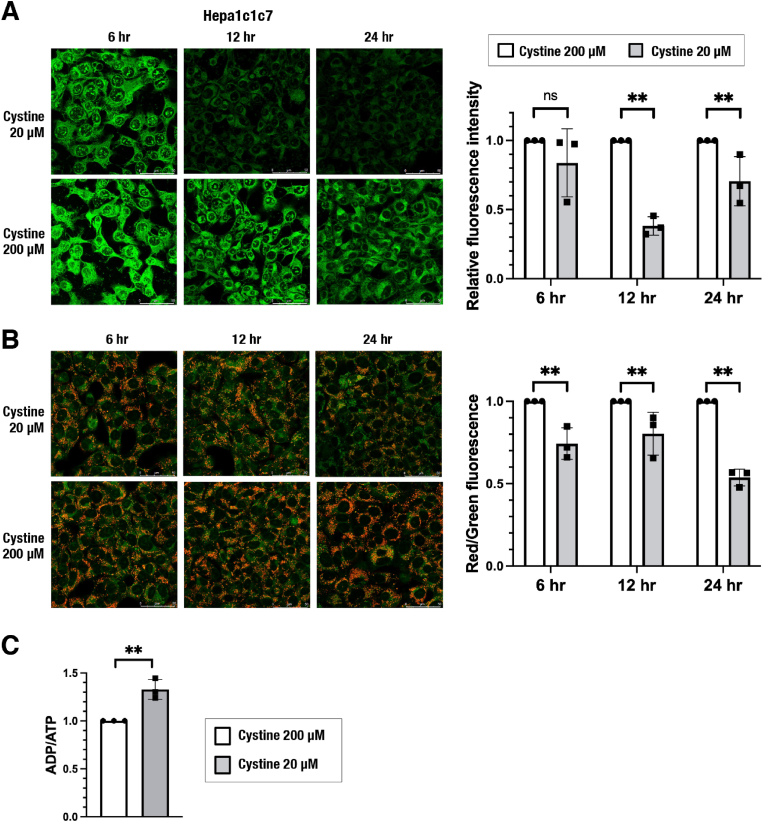
Based on our previous finding that cysteine is converted to cysteine persulfide by CARS2 in mitochondria and that CARS2-dependent mitochondrial persulfide production contributes to the maintenance of mitochondrial activity [21], we first examined how cysteine availability impacted intracellular persulfide levels and mitochondrial function. Because one of the major cysteine sources is extracellular cystine, cells were challenged with cystine restriction for up to 24 h Hepa1c1c7 cells exhibited a reduction in intracellular persulfides, as evaluated by the fluorescent probe SSP4 [30] (Fig. 1A), and a decrease in mitochondrial membrane potential (MMP), as evaluated by the fluorescent probe JC-10 (Fig. 1B), after 12 h of exposure to 20 μM cystine compared with 200 μM cystine. Similar effects were observed in HeLa and H1650 cells after 24 h of exposure to 20 μM cystine (Supplementary Figs. 1A and 1B). Cystine restriction slowed the cell number increase but did not affect cell viability during the 24 h after initiation of cystine restriction (Supplementary Figs. 1C and 1D), suggesting that the MMP decrease in cystine-restricted cells was not due to cell death. We next examined ATP production in Hepa1c1c7 cells exposed to 20 μM cystine for 24 h and found that cystine restriction elevated the ADP/ATP ratio (Fig. 1C). These results indicated that cystine restriction reduces intracellular persulfides and suppresses mitochondrial energy metabolism, which is consistent with our previous observation that mitochondrial persulfide production is necessary for the maintenance of mitochondrial activity.
Fig. 1.
Cystine restriction decreases intracellular persulfides and inhibits mitochondrial activity in Hepa1c1c7 cells.
(A) Detection of intracellular persulfides by the SSP4 fluorescence probe. Representative images from 4 to 5 images of randomly selected areas per experiment from three independent experiments (left) and relative fluorescence intensities (right) are shown. Scale bars correspond to 50 μm (left). (B) Mitochondrial membrane potential (MMP) analyzed by using JC-10 fluorescence imaging. Representative images from 5 to 7 images of randomly selected areas per experiment from three independent experiments (left) and relative red and green fluorescence ratios (right) are shown. Scale bars correspond to 50 μm (left). (C) ADP/ATP ratio measurement. Relative ratios are shown.
At 24 h after cell seeding, the culture medium was replaced with fresh medium containing 20 μM or 200 μM cystine. SSP4 staining (A) and JC-10 staining (B) were conducted 6 h, 12 h and 24 h after the medium change. ADP/ATP ratio measurement (C) were conducted 24 h after the medium change. The data are presented as the mean ± S.D. of three independent experiments. **α < 0.01 compared with the cystine 200 μM group, determined using confidence interval estimation. ns: not significant. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Glutathione trisulfide (GSSSG) supplementation promotes mitochondrial activity
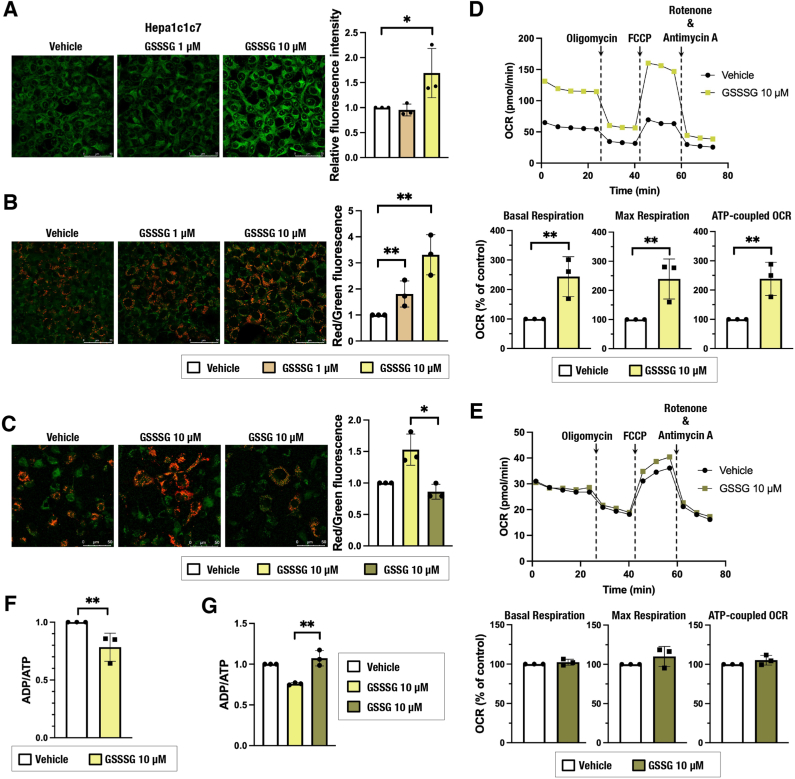
We next asked whether exogenous persulfide supplementation activates mitochondrial function. Glutathione trisulfide (GSSSG) was added to the culture medium of Hepa1c1c7 cells as a source of persulfides, and intracellular persulfides and mitochondrial function were evaluated after 24 h. Supplementation with GSSSG increased persulfides (Fig. 2A) and elevated MMP (Fig. 2B). The increase of MMP was not induced by GSSG supplementation (Fig. 2C). OCR was increased by GSSSG (Fig. 2D) but not by GSSG (Fig. 2E). Similarly, the ADP/ATP ratio was lowered by GSSSG (Fig. 2F) but not changed by GSSG (Fig. 2G). These results suggest that increased persulfides contributed to the activation of mitochondrial function. A persulfide increase and MMP elevation were also observed in GSSSG-treated HeLa cells (Supplementary Fig. 1E).
Fig. 2.
Glutathione trisulfide (GSSSG) supplementation increases intracellular persulfides and promotes mitochondrial function in Hepa1c1c7 cells. (A) Detection of intracellular persulfides by the SSP4 fluorescence probe. Representative images from 4 to 5 images of randomly selected areas per experiment from three independent experiments (left) and relative fluorescence intensities (right) are shown. Scale bars correspond to 50 μm. (B, C) MMP analyzed by using JC-10 fluorescence imaging. Representative images from 5 to 7 images of randomly selected areas per experiment from three independent experiments (left) and relative red and green fluorescence ratios (right) are shown. Scale bars correspond to 50 μm. (D, E) Oxygen consumption rate (OCR) analysis. A representative OCR respirogram from three independent experiments is shown (top). Respiration parameters were calculated from the results of three independent experiments (bottom). Basal respiration is an average of the OCR values before the addition of oligomycin. Maximum respiration is the average OCR value after the addition of FCCP. ATP-coupled OCR is the difference between basal respiration and the average OCR value after oligomycin addition. Oligomycin (1 μM), FCCP (0.15 μM) and antimycin A + rotenone (1 μM each) were used. (F, G) ADP/ATP ratio measurement. Relative ratios are shown.
At 24 h after cell seeding, the culture medium was replaced with 200 μM cystine-containing fresh medium with vehicle (water) or GSSSG (1 μM or 10 μM). Analyses were conducted 24 h after the medium change. The data are presented as the mean ± S.D. of three independent experiments. *α < 0.05 and **α < 0.01 compared with the vehicle (water) group, determined using confidence interval estimation (A, B, D, E and F). An unpaired Student's t-test was conducted for comparison of GSSSG-treated cells and GSSG-treated cells (C and G). *p < 0.05, **p < 0.01. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. The sulfur oxidation pathway is required for persulfide-mediated mitochondrial activation
Mitochondria are equipped with a sulfur oxidation pathway that consists of sulfide:quinone oxidoreductase (SQOR), ethylmalonic encephalopathy protein 1 (ETHE1) and sulfite oxidase (SUOX), which are responsible for oxidizing sulfide, persulfide and sulfite, respectively [22,[31], [32], [33]]. Persulfides generated by CARS2 in mitochondria are reduced to sulfide in an ETC-dependent manner [21], and the resultant sulfide together with persulfides are metabolized to thiosulfate and sulfate via sequential and concurrent oxidation processes.
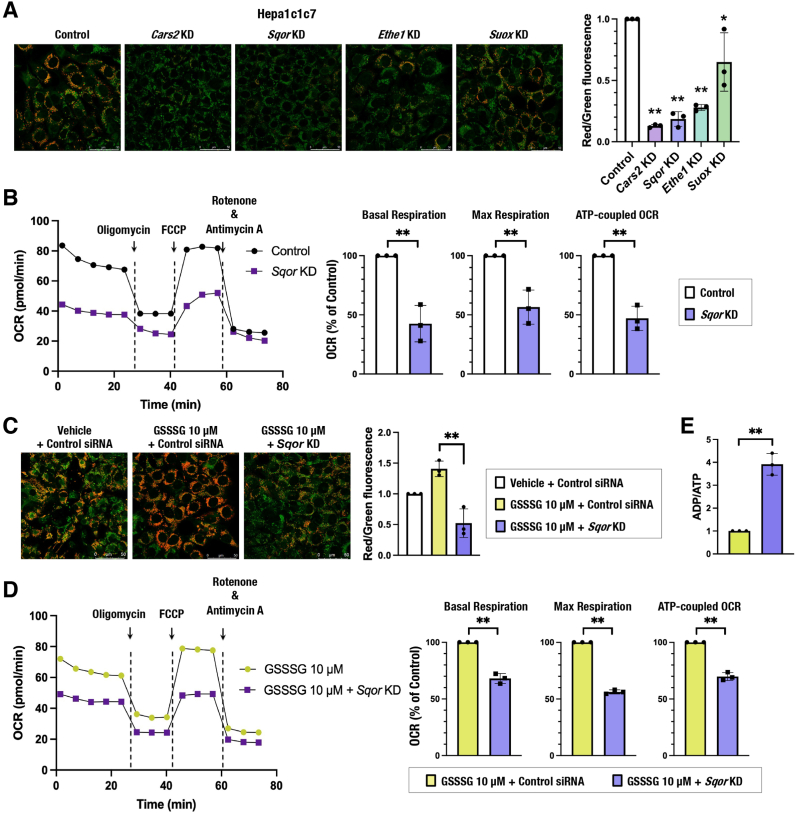
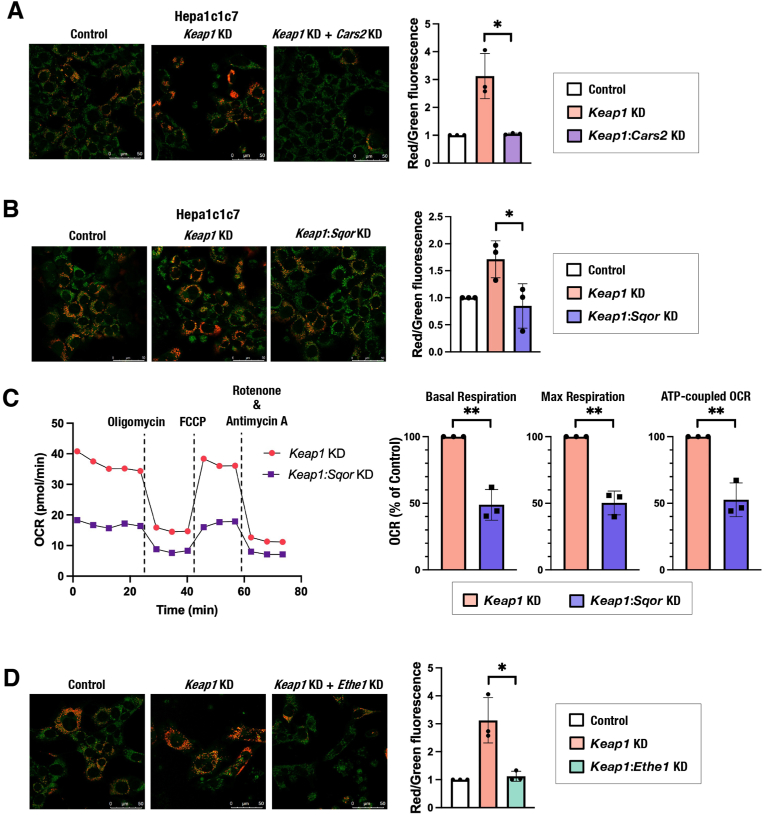
To clarify the significance of the sulfur oxidation pathway in mitochondrial function, we inhibited the sulfur oxidation pathway by knocking down genes encoding SQOR, ETHE1 and SUOX and examined their effects on MMP in Hepa1c1c7, HeLa and H1650 cells (Supplementary Figs. 2A and 2B). Cars2/CARS2 knockdown was used as a positive control. The MMP was decreased by the reduction of these enzymes, except for SUOX (Fig. 3A and Supplementary Fig. 2C), and the OCR was reduced by SQOR inhibition (Fig. 3B and Supplementary Fig. 2D). These results suggested that CARS2-mediated persulfide production and SQOR- and ETHE1-mediated sulfur oxidation are required for mitochondrial activity, which is consistent with previous reports [21,34]. Importantly, GSSSG-induced mitochondrial activation, i.e., elevation of MMP, OCR and relative ATP amount vs. ADP, was not observed in Sqor-knockdown Hepa1c1c7 cells (Fig. 3C–E), indicating that the sulfur oxidation pathway, especially SQOR, is required for GSSSG-induced mitochondrial activation.
Fig. 3.
The sulfur oxidation pathway is required for persulfide-mediated mitochondrial activation in Hepa1c1c7 cells.
(A and C) MMP was analyzed by using JC-10 fluorescence imaging for Hepa1c1c7 cells with Cars2, Sqor, Ethe1 and Suox knockdown (A) and GSSSG treatment and/or Sqor knockdown (C). Representative images from 5 to 7 images of randomly selected areas per experiment from three independent experiments (left) and relative red and green fluorescence ratios (right) are shown. Scale bars correspond to 50 μm. (B and D) OCR analysis. A representative OCR respirogram from three independent experiments is shown (left). Respiration parameters were calculated from the results of three independent experiments (right). (E) ADP/ATP ratio measurement. Relative ratios are shown.
At 24 h after siRNA transfection, the culture medium was replaced with 200 μM cystine-containing fresh medium (A and B) and 200 μM cystine-containing fresh medium with vehicle (water) or 10 μM GSSSG (C and D). Analyses were conducted 24 h after the medium change. The data are presented as the mean ± S.D. of three independent experiments. *α < 0.05 and **α < 0.01 compared with the control cells (A and B) and with GSSSG-treated control cells (D and E), determined using confidence interval estimation. An unpaired Student's t-test was conducted for comparison of GSSSG-treated cells with and without Sqor knockdown (C). *p < 0.01. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.4. NRF2 activation increases intracellular persulfides and promotes mitochondrial activity
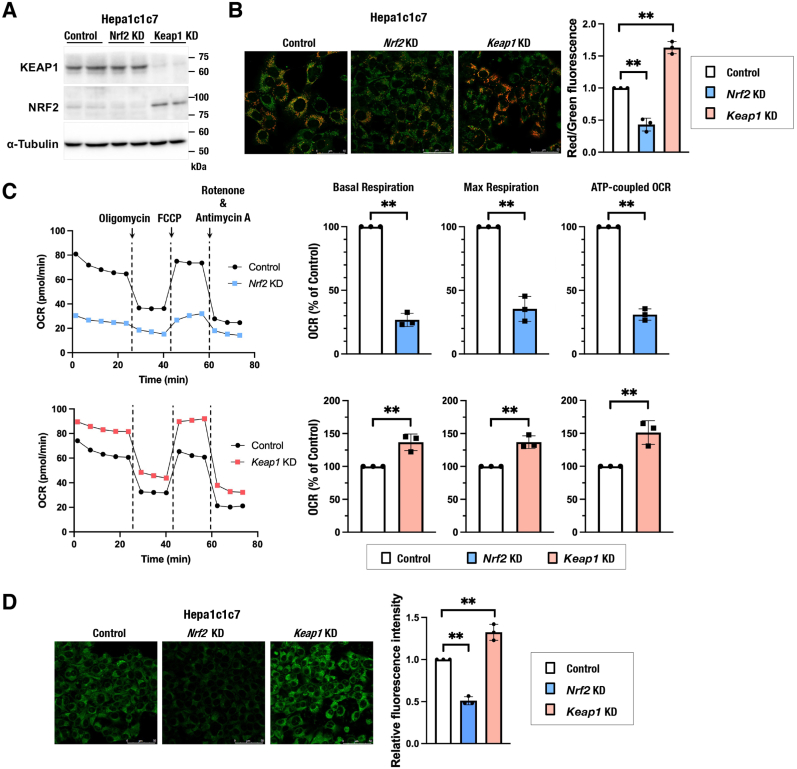
To investigate the impacts of NRF2 activity on mitochondrial function in this experimental setting, MMP and OCR were examined in Hepa1c1c7, HeLa and H1650 cells with different NRF2 activities. Hepa1c1c7 cells were treated with siRNAs against Nrf2 and Keap1 (Fig. 4A) and used for MMP and OCR analyses. NRF2 inhibition decreased MMP and OCR, whereas NRF2 activation by KEAP1 inhibition increased MMP and OCR (Fig. 4B and C). Similar effects of NRF2 inhibition and NRF2 activation were observed in HeLa and H1650 cells (Supplementary Figs. 3A–3C). Consistent with a positive correlation between persulfide levels and mitochondrial activities, intracellular persulfides were decreased and increased in Nrf2-knockdown and Keap1-knockdown Hepa1c1c7 cells, respectively (Fig. 4D). These results suggested that NRF2 activation promotes mitochondrial function by increasing intracellular persulfides.
Fig. 4.
NRF2 promotes mitochondrial activity and increases intracellular persulfides in Hepa1c1c7 cells.
(A) Verification of the knockdown efficiency of Nrf2 and Keap1 in Hepa1c1c7 cells. Representative immunoblot analysis of NRF2 and KEAP1 proteins. α-Tubulin was used as a loading control. (B) MMP was analyzed using JC-10 fluorescence imaging for Hepa1c1c7 cells with Nrf2 and Keap1 knockdown. Representative images from 5 to 7 images of randomly selected areas per experiment from three independent experiments (left) and relative red and green fluorescence ratios (right) are shown. Scale bars correspond to 50 μm. (C) OCR analysis. Representative OCR respirograms from three independent experiments are shown (left). Respiration parameters were calculated from the results of three independent experiments (right). (D) Detection of intracellular persulfides by the SSP4 fluorescence probe. Representative images from 4 to 5 images of randomly selected areas per experiment from three independent experiments (left) and relative fluorescence intensities (right) are shown. Scale bars correspond to 50 μm.
At 24 h after siRNA transfection, the culture medium was replaced with 200 μM cystine-containing fresh medium. Analyses were conducted 24 h after the medium change. The data are presented as the mean ± S.D. of three independent experiments. **α < 0.01 compared with the control cells, determined using confidence interval estimation. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.5. NRF2-mediated activation of mitochondrial function requires cystine uptake via xCT
We wanted to know how NRF2 increases intracellular persulfides. Because Slc7a11, which encodes a cystine transporter xCT, is a well-established NRF2 target gene [26] and because persulfide-synthesizing enzymes, including CARS2, did not seem to be directly regulated by NRF2, we hypothesized that NRF2 increases persulfides by promoting cystine uptake through xCT upregulation.
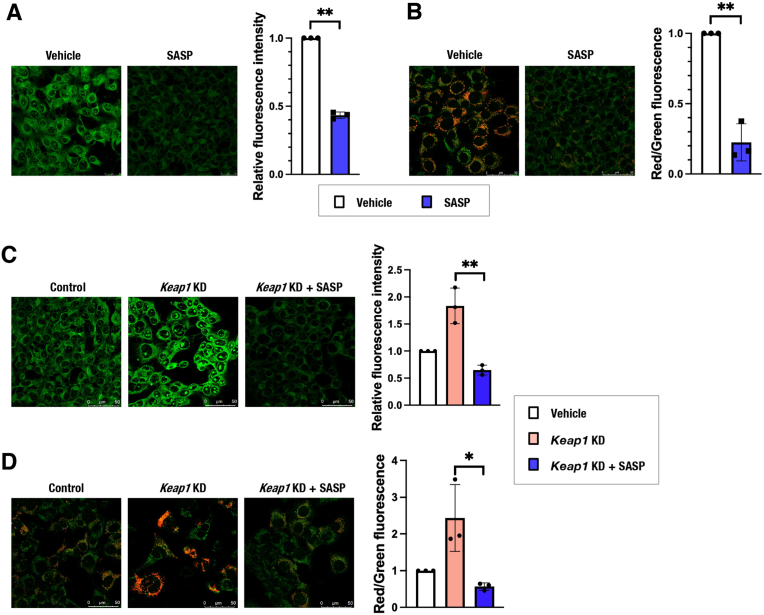
Sulfasalazine (SASP), an inhibitor of xCT, decreased intracellular persulfides and MMP in Hepa1c1c7 cells (Fig. 5A and B) as well as in HeLa and H1650 cells (Supplementary Figs. 4A and 4B), consistent with the decreased cystine availability resulting in reduced persulfides and mitochondrial inhibition. Although loss-of-function experiments of Slc7a11 in Hepa1c1c7 cells were not successful due to poor knockdown efficiency, SLC7A11 knockdown in HeLa and H1650 cells was efficient enough to show a significant decrease in MMP (Supplementary Figs. 4C and 4D). Increased persulfides and elevated MMP by Keap1 knockdown were reversed by SASP treatment in Hepa1c1c7 cells (Fig. 5C and D), suggesting that the NRF2-mediated persulfide increase and mitochondrial activation depend on increased cystine uptake.
Fig. 5.
The cystine transporter xCT is responsible for NRF2-mediated mitochondrial activation in Hepa1c1c7 cells.
(A and C) Detection of intracellular persulfides by the SSP4 fluorescence probe in Hepa1c1c7 cells treated with SASP. Representative images from 4 to 5 images of randomly selected areas per experiment from three independent experiments (left) and relative fluorescence intensities (right) are shown. Scale bars correspond to 50 μm. (B and D) MMP was analyzed by using JC-10 fluorescence imaging in Hepa1c1c7 cells treated with SASP. Representative images from 5 to 7 images of randomly selected areas per experiment from three independent experiments (left) and relative red and green fluorescence ratios (right) are shown. Scale bars correspond to 50 μm.
At 24 h after cell seeding (A and B) and 24 h of control siRNA or Keap1 siRNA transfection (C and D), the culture medium was replaced with 200 μM cystine-containing fresh medium with vehicle (DMSO) or 500 μM SASP. Analyses were conducted 24 h after the medium change. The data are presented as the mean ± S.D. of three independent experiments. **α < 0.01 compared with the vehicle (DMSO) group, determined using confidence interval estimation (A and B). An unpaired Student's t-test was conducted for comparison of Keap1-knockdown cells with and without SASP treatment (C and D). *p < 0.05, *p < 0.01. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.6. NRF2-mediated activation of mitochondrial function requires the mitochondrial sulfur oxidation pathway
To further examine whether NRF2-mediated mitochondrial activation requires persulfide production and sulfur oxidation pathway activity, we conducted simultaneous inhibition of KEAP1 and mitochondrial sulfur-metabolizing enzymes. The simultaneous knockdown of Keap1 and Cars2 reversed the MMP elevation induced by Keap1 knockdown in Hepa1c1c7 cells (Fig. 6A), suggesting that NRF2-mediated mitochondrial activation depends on CARS2-mediated persulfide production. The simultaneous knockdown of Keap1 and Sqor reversed the MMP elevation and OCR increase induced by Keap1 knockdown (Supplementary Fig. 5A, Fig. 6B and C). Similar effects on MMP were observed in HeLa and H1650 cells (Supplementary Figs. 5B and 5C). The simultaneous knockdown of Keap1 and Ethe1 reversed the MMP elevation induced by Keap1 knockdown (Fig. 6D). These results suggested that NRF2-mediated mitochondrial activation depends on the sulfur oxidation pathway in mitochondria. Thus, NRF2 activation caused by Keap1 inhibition promotes mitochondrial function by increasing cystine uptake and subsequent persulfide production and sulfur oxidation.
Fig. 6.
Persulfide production and the sulfur oxidation pathway are required for NRF2-mediated mitochondrial activation.
(A, B and D) MMP was analyzed by using JC-10 fluorescence imaging in Hepa1c1c7 cells with Keap1 and/or Cars2 knockdown (A), Sqor knockdown (B) and Ethe1 knockdown (D). Representative images from 5 to 7 images of randomly selected areas per experiment from three independent experiments (left) and relative red and green fluorescence ratios (right) are shown. Scale bars correspond to 50 μm. (C) OCR analysis. A representative OCR respirogram from three independent experiments is shown (left). Respiration parameters were calculated from the results of three independent experiments (right).
At 24 h after siRNA transfection, the culture medium was replaced with 200 μM cystine-containing fresh medium. Analyses were conducted 24 h after the medium change. The data are presented as the mean ± S.D. of three independent experiments. **α < 0.01 compared with the control cells, determined using confidence interval estimation (C). An unpaired Student's t-test was conducted for comparison of Keap1 single knockdown cells and double knockdown cells (A, B and D). *p < 0.05. Panels A and D share the same control samples and Keap1 knockdown samples. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Accumulating evidence has suggested that cysteine persulfide and related persulfides are found in abundant quantities in both prokaryotic and eukaryotic cells and play important roles in antioxidant and anti-inflammatory responses [27,[35], [36], [37], [38]]. Among the enzymes that have been reported to possess persulfide-synthesizing activity, including cystathionine β-synthetase (CBS), cystathionine γ-lyase (CSE), 3-mercaptopyruvate sulfurtransferase (3MST) and CARS [21,27,39,40], CARS2, a mitochondrial isoform of CARS, is regarded as the major contributor to the production of persulfide-containing metabolites judging from the results of loss-of-function experiments [21]. This study reproduced the result showing the necessity of CARS2 for persulfide generation and mitochondrial function and demonstrated important roles of the sulfur oxidation pathway catalyzed by SQOR and ETHE1 in mitochondrial activity. Based on these results, we identified xCT-dependent cystine uptake as one of the major activities of NRF2 in promoting mitochondrial function. Considering that cystine is also a substrate of CBS and CSE for direct generation of cysteine persulfide [27,41], cystine availability is most likely to be an important determinant of intracellular persulfide levels. NRF2-mediated mitochondrial activation depends on persulfide production and the sulfur oxidation pathway, verifying the significance of sulfur metabolism for mitochondrial function.
How persulfides support mitochondrial function is possibly explained by the following mechanisms that are tightly intertwined with one another. Considering their antioxidant function [27], persulfides are likely to quench reactive oxygen species generated in mitochondria and prevent oxidative damage to mitochondrial proteins. Considering a previous result that cysteine persulfide is converted to sulfide in an ETC-dependent manner [21], persulfides are likely to serve as electron acceptors for electrons from the ETC and may prevent aberrant electron acceptance by dioxygen and consequently avoid generation of excessive reactive oxygen species. Considering their possible roles in the synthesis of iron-sulfur clusters [42], persulfides may serve as a decoy to protect persulfide-based sulfur transfer for the incorporation of sulfur into iron-sulfur clusters, which is essential for ETC function. The necessity of sulfide oxidation by SQOR for persulfide-mediated mitochondrial activation implies three aspects of sulfide metabolism: protection from sulfide toxicity [23,24], recovery of electrons leaked from the ETC via persulfides and regeneration of persulfides from sulfide. Sulfide accumulation needs to be avoided for mitochondrial integrity, whereas electrons transferred from sulfide to ubiquinone participate in the ETC for mitochondrial energy production. And sulfide should become hydropersulfide after being oxidized by SQOR, resulting in the regeneration of mitochondrial persulfides.
Regarding the relationship between NRF2 activity and mitochondrial function, a previous study described that MMP was reduced and elevated in mouse embryonic fibroblasts (MEFs) and primary cortical neural cells derived from Nrf2-and Keap1-knockout mice, respectively [43], which is quite similar to our current results. Another report described that NRF2 activation facilitates fatty acid oxidation in the mitochondria of MEFs as well as those isolated from the brain, liver and heart [44]. These studies concluded that NRF2 promotes mitochondrial function by increasing the availability of substrates, NADH and FADH2, for mitochondrial respiration. A fatty acid-metabolizing enzyme in peroxisomes, ACOX2, has been shown to be one of the target genes of NRF2 [45], which may contribute to the increased supply of NADH and FADH2 by NRF2 for mitochondrial respiration. Because increased NADH/NAD+ ratio could limit metabolic processes that require NAD+ such as those catalyzed by dehydrogenases, metabolic impacts of NRF2 activation may be more complex. Nevertheless, our study indicated that elevated mitochondrial activity is primarily attributable to the NRF2-mediated increase in cysteine availability, which is expected to secure the efficiency of ETC function. Therefore, taken together, the results suggest that NRF2 increases proton-donating substrates and improves electron transport efficiency in mitochondria.
A previous study showed that sulfide activates NRF2 by inactivating KEAP1 through the formation of a disulfide bond between two cysteine residues, Cys226 and Cys613 [46], which appears to be an indirect effect of sulfide considering that sulfide is not an oxidant. Indeed, this study suggested the involvement of H2O2 production in the upregulation of NRF2 by sulfide [46]. We surmise that reactive oxygen species generated from mitochondria due to inhibitory action of excessive sulfide might induce the disulfide bond formation between two cysteine residues, Cys226 and Cys613, resulting in the NRF2 activation. This study also showed that NRF2 induces SQOR expression and proposed a feedback loop between NRF2 and sulfide [46]. In our experimental setting, in Hepa1c1c7 cells, NRF2 binding was detected in the first intron of the Sqor gene in a chromatin-immunoprecipitation assay (unpublished observation). However, the NRF2 contribution to SQOR protein level was limited, implying that the feedback loop between NRF2 and sulfide is likely to be context dependent.
Intriguingly, glutathione synthesis was shown to be enhanced in Dictyostelium discoideum under starvation, which resulted in cysteine sequestration to limit the mitochondrial use of cysteine, especially the use of cysteine sulfur atoms for iron-sulfur cluster synthesis by desulfuration [47]. This previous study suggested the competitive utilization of cysteine by glutathione synthesis in the cytoplasm and desulfuration-coupled persulfide production in mitochondria. While NRF2 directly promotes anabolic processes utilizing cysteine, such as glutathione synthesis by inducing the rate limiting enzyme γ-glutamylcysteine synthetase [1] and protein translation by maintaining the reduced states of ribosomal subunits [48], the promotion of mitochondrial sulfur metabolism, i.e., persulfide synthesis and subsequent sulfur oxidation, are an indirect and passive effect of NRF2 activation through the increasing cysteine supply. When NRF2 strongly and persistently promotes glutathione synthesis, which often occurs in NRF2-activated cancer cells with KEAP1 mutations [49], mitochondrial cysteine demand may not be satisfied, resulting in mitochondrial suppression. In addition, NRF2-activated cancer cells are subjected to a glutamate shortage and, consequently, insufficient anaplerosis for TCA cycle intermediates, which decreases substrates for electron donation to the ETC and inhibits mitochondrial activity [50,51]. Therefore, we surmise that the sufficient availability of NADH and FADH2 for electron donation to the ETC is an important prerequisite for NRF2-mediated mitochondrial activation, which is dependent on persulfide production and sulfur oxidation.
Author contributions
M.M.A. and K.A. designed the study, conducted the experiments, analyzed the data and wrote an early draft of the paper. E.S. conducted the experiments and analyzed the data. S.M. and H.S. analyzed and interpreted the data and wrote the paper. T. Abe provided an essential experimental system and analyzed the data. T. Akaike supervised the research, analyzed the data and wrote the paper. H.M. designed the study, supervised the research, analyzed the data and wrote the paper.
Declaration of competing interest
The authors declare no competing financial or nonfinancial interests.
Acknowledgments
We thank the Biomedical Research Core of the Institute of Development, Aging and Cancer for their technical support.
This work was supported by the Tokyo Biochemical Research Foundation (M.M.A.), by JSPS grant numbers 19K18747 (A.K.), 20K07320 (H.S.), 21K15358 (S.M.), 18H05277 (T. Akaike), 21H05258 (T. Akaike, H.M.), 21H05263 (T. Akaike), 21H05264 (H.M.), and 21H04799 (H.M.), and by AMED JP21zf0127001 (T. Abe). The funders had no role in the study design, data collection and analysis, decision to publish or manuscript preparation.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102624.
Contributor Information
Takaaki Akaike, Email: takaike@med.tohoku.ac.jp.
Hozumi Motohashi, Email: hozumim@med.tohoku.ac.jp.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
No data was used for the research described in the article.
References
- 1.Yamamoto M., Kensler T.W., Motohashi H. The KEAP1-NRF2 system: a thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol. Rev. 2018;98:1169–1203. doi: 10.1152/physrev.00023.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sekine H., Okazaki K., Ota N., Shima H., Katoh Y., Suzuki N., Igarashi K., Ito M., Motohashi H., Yamamoto M. The mediator subunit MED16 transduces NRF2-activating signals into antioxidant gene expression. Mol. Cell Biol. 2015;36:407–420. doi: 10.1128/MCB.00785-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okazaki K., Papagiannakopoulos T., Motohashi H. Metabolic features of cancer cells in NRF2 addiction status. Biophys Rev. 2020;12:435–441. doi: 10.1007/s12551-020-00659-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okazaki K., Anzawa H., Liu Z., Ota N., Kitamura H., Onodera Y., Alam M.M., Matsumaru D., Suzuki T., Katsuoka F., et al. Enhancer remodeling promotes tumor-initiating activity in NRF2-activated non-small cell lung cancers. Nat. Commun. 2020;11:5911. doi: 10.1038/s41467-020-19593-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Honkura Y., Matsuo H., Murakami S., Sakiyama M., Mizutari K., Shiotani A., Yamamoto M., Morita I., Shinomiya N., Kawase T., et al. NRF2 is a key target for prevention of noise-induced hearing loss by reducing oxidative damage of cochlea. Sci. Rep. 2016;6 doi: 10.1038/srep19329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi E.H., Suzuki T., Funayama R., Nagashima T., Hayashi M., Sekine H., Tanaka N., Moriguchi T., Motohashi H., Nakayama K., et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 2016;7 doi: 10.1038/ncomms11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki T., Murakami S., Biswal S.S., Sakaguchi S., Harigae H., Yamamoto M., Motohashi H. Systemic activation of NRF2 alleviates lethal autoimmune inflammation in scurfy mice. Mol. Cell Biol. 2017;37 doi: 10.1128/MCB.00063-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oishi T., Matsumaru D., Ota N., Kitamura H., Zhang T., Honkura Y., Katori Y., Motohashi H. Activation of the NRF2 pathway in Keap1-knockdown mice attenuates progression of age-related hearing loss. NPJ Aging Mech Dis. 2020;6:14. doi: 10.1038/s41514-020-00053-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wati S.M., Matsumaru D., Motohashi H. NRF2 pathway activation by KEAP1 inhibition attenuates the manifestation of aging phenotypes in salivary glands. Redox Biol. 2020;36 doi: 10.1016/j.redox.2020.101603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao M., Murakami S., Matsumaru D., Kawauchi T., Nabeshima Y.I., Motohashi H. NRF2 pathway activation attenuates ageing-related renal phenotypes due to α-klotho deficiency. J. Biochem. 2022;171:579–589. doi: 10.1093/jb/mvac014. [DOI] [PubMed] [Google Scholar]
- 11.Shih A.Y., Imbeault S., Barakauskas V., Erb H., Jiang L., Li P., Murphy T.H. Induction of the Nrf2-driven antioxidant response confers neuroprotection during mitochondrial stress in vivo. J. Biol. Chem. 2005;280:22925–22936. doi: 10.1074/jbc.M414635200. [DOI] [PubMed] [Google Scholar]
- 12.Calkins M.J., Jakel R.J., Johnson D.A., Chan K., Kan Y.W., Johnson J.A. Protection from mitochondrial complex II inhibition in vitro and in vivo by Nrf2-mediated transcription. Proc. Natl. Acad. Sci. U.S.A. 2005;102:244–249. doi: 10.1073/pnas.0408487101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cook A.L., Vitale A.M., Ravishankar S., Matigian N., Sutherland G.T., Shan J., Sutharsan R., Perry C., Silburn P.A., Mellick G.D., et al. NRF2 activation restores disease related metabolic deficiencies in olfactory neurosphere-derived cells from patients with sporadic Parkinson's disease. PLoS One. 2011;6 doi: 10.1371/journal.pone.0021907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uruno A., Matsumaru D., Ryoke R., Saito R., Kadoguchi S., Saigusa D., Saito T., Saido T.C., Kawashima R., Yamamoto M. Nrf2 suppresses oxidative stress and inflammation in app knock-in alzheimer's disease model mice. Mol. Cell Biol. 2020;40 doi: 10.1128/MCB.00467-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uruno A., Yagishita Y., Katsuoka F., Kitajima Y., Nunomiya A., Nagatomi R., Pi J., Biswal S.S., Yamamoto M. Nrf2-Mediated regulation of skeletal muscle glycogen metabolism. Mol. Cell Biol. 2016;36:1655–1672. doi: 10.1128/MCB.01095-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Onoki T., Izumi Y., Takahashi M., Murakami S., Matsumaru D., Ohta N., Wati S.M., Hatanaka N., Katsuoka F., Okutsu M., et al. Skeletal muscle-specific Keap1 disruption modulates fatty acid utilization and enhances exercise capacity in female mice. Redox Biol. 2021;43 doi: 10.1016/j.redox.2021.101966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinkova-Kostova A.T., Abramov A.Y. The emerging role of Nrf2 in mitochondrial function. Free Radic. Biol. Med. 2015;88(Pt B):179–188. doi: 10.1016/j.freeradbiomed.2015.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McBride H.M., Neuspiel M., Wasiak S. Mitochondria: more than just a powerhouse. Curr. Biol. 2006;16:R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 19.Chandel N.S. Evolution of mitochondria as signaling organelles. Cell Metabol. 2015;22:204–206. doi: 10.1016/j.cmet.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 20.Suomalainen A., Battersby B.J. Mitochondrial diseases: the contribution of organelle stress responses to pathology. Nat. Rev. Mol. Cell Biol. 2018;19:77–92. doi: 10.1038/nrm.2017.66. [DOI] [PubMed] [Google Scholar]
- 21.Akaike T., Ida T., Wei F.Y., Nishida M., Kumagai Y., Alam M.M., Ihara H., Sawa T., Matsunaga T., Kasamatsu S., et al. Cysteinyl-tRNA synthetase governs cysteine polysulfidation and mitochondrial bioenergetics. Nat. Commun. 2017;8:1177. doi: 10.1038/s41467-017-01311-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olson K.R. H2S and polysulfide metabolism: conventional and unconventional pathways. Biochem. Pharmacol. 2018;149:77–90. doi: 10.1016/j.bcp.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 23.Nicholls P., Kim J.K. Oxidation of sulphide by cytochrome aa3. Biochim. Biophys. Acta. 1981;637:312–320. doi: 10.1016/0005-2728(81)90170-5. [DOI] [PubMed] [Google Scholar]
- 24.Nicholls P., Kim J.K. Sulphide as an inhibitor and electron donor for the cytochrome c oxidase system. Can. J. Biochem. 1982;60:613–623. doi: 10.1139/o82-076. [DOI] [PubMed] [Google Scholar]
- 25.Ziosi M., Di Meo I., Kleiner G., Gao X.H., Barca E., Sanchez-Quintero M.J., Tadesse S., Jiang H., Qiao C., Rodenburg R.J., et al. Coenzyme Q deficiency causes impairment of the sulfide oxidation pathway. EMBO Mol. Med. 2017;9:96–111. doi: 10.15252/emmm.201606356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasaki H., Sato H., Kuriyama-Matsumura K., Sato K., Maebara K., Wang H., Tamba M., Itoh K., Yamamoto M., Bannai S. Electrophile response element-mediated induction of the cystine/glutamate exchange transporter gene expression. J. Biol. Chem. 2002;277:44765–44771. doi: 10.1074/jbc.M208704200. [DOI] [PubMed] [Google Scholar]
- 27.Ida T., Sawa T., Ihara H., Tsuchiya Y., Watanabe Y., Kumagai Y., Suematsu M., Motohashi H., Fujii S., Matsunaga T., et al. Reactive cysteine persulfides and S-polythionation regulate oxidative stress and redox signaling. Proc. Natl. Acad. Sci. U.S.A. 2014;111:7606–7611. doi: 10.1073/pnas.1321232111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamid H.A., Tanaka A., Ida T., Nishimura A., Matsunaga T., Fujii S., Morita M., Sawa T., Fukuto J.M., Nagy P., et al. Polysulfide stabilization by tyrosine and hydroxyphenyl-containing derivatives that is important for a reactive sulfur metabolomics analysis. Redox Biol. 2019;21 doi: 10.1016/j.redox.2019.101096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watai Y., Kobayashi A., Nagase H., Mizukami M., McEvoy J., Singer J.D., Itoh K., Yamamoto M. Subcellular localization and cytoplasmic complex status of endogenous Keap1. Gene Cell. 2007;12:1163–1178. doi: 10.1111/j.1365-2443.2007.01118.x. [DOI] [PubMed] [Google Scholar]
- 30.Shieh M., Ni X., Xu S., Lindahl S.P., Yang M., Matsunaga T., Flaumenhaft R., Akaike T., Xian M. Shining a light on SSP4: a comprehensive analysis and biological applications for the detection of sulfane sulfurs. Redox Biol. 2022;56 doi: 10.1016/j.redox.2022.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rudolph M.J., Johnson J.L., Rajagopalan K.V., Kisker C. The 1.2 A structure of the human sulfite oxidase cytochrome b(5) domain. Acta Crystallogr D Biol Crystallogr. 2003;59:1183–1191. doi: 10.1107/s0907444903009934. [DOI] [PubMed] [Google Scholar]
- 32.Brito J.A., Sousa F.L., Stelter M., Bandeiras T.M., Vonrhein C., Teixeira M., Pereira M.M., Archer M. Structural and functional insights into sulfide:quinone oxidoreductase. Biochemistry. 2009;48:5613–5622. doi: 10.1021/bi9003827. [DOI] [PubMed] [Google Scholar]
- 33.Kabil O., Banerjee R. Characterization of patient mutations in human persulfide dioxygenase (ETHE1) involved in H2S catabolism. J. Biol. Chem. 2012;287:44561–44567. doi: 10.1074/jbc.M112.407411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vitvitsky V., Kumar R., Libiad M., Maebius A., Landry A.P., Banerjee R. The mitochondrial NADH pool is involved in hydrogen sulfide signaling and stimulation of aerobic glycolysis. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toohey J.I. Sulfur signaling: is the agent sulfide or sulfane? Anal. Biochem. 2011;413:1–7. doi: 10.1016/j.ab.2011.01.044. [DOI] [PubMed] [Google Scholar]
- 36.Fukuto J.M., Carrington S.J., Tantillo D.J., Harrion J.G., Ignarro L.J., Freeman B.A., Chen A., Wink D.A. Small molecule signaling agents: the integrated chemistry and biochemistry of nitrogen oxide, oxides of carbon, dioxygen, hydrogen sulfide, and their derived species. Chem. Res. Toxicol. 2012;25:769–793. doi: 10.1021/tx2005234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimizu T., Shen J., Fang M., Zhang Y., Hori K., Trinidad J.C., Bauer C.E., Giedroc D.P., Masuda S. Sulfide-responsive transcriptional repressor SqrR functions as a master regulator of sulfide-dependent photosynthesis. Proc. Natl. Acad. Sci. U.S.A. 2017;114:2355–2360. doi: 10.1073/pnas.1614133114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang T., Ono K., Tsutsuki H., Islam W., Akaike T., Sawa T. Enhanced cellular polysulfides negatively regulate TLR4 signaling and mitigate lethal endotoxin shock. Cell Chem Biol. 2019;26:686–698.e4. doi: 10.1016/j.chembiol.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 39.Nishida M., Sawa T., Kitajima N., Ono K., Inoue H., Ihara H., Motohashi H., Yamamoto M., Suematsu M., Kurose H., et al. Hydrogen sulfide anion regulates redox signaling via electrophile sulfhydration. Nat. Chem. Biol. 2012;8:714–724. doi: 10.1038/nchembio.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanaoka K., Sasakura K., Suwanai Y., Toma-Fukai S., Shimamoto K., Takano Y., Shibuya N., Terai T., Komatsu T., Ueno T., et al. Discovery and mechanistic characterization of selective inhibitors of H2S-producing enzyme: 3-mercaptopyruvate sulfurtransferase (3MST) targeting active-site cysteine persulfide. Sci. Rep. 2017;7 doi: 10.1038/srep40227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yadav P.K., Martinov M., Vitvitsky V., Seravalli J., Wedmann R., Filipovic M.R., Banerjee R. Biosynthesis and reactivity of cysteine persulfides in signaling. J. Am. Chem. Soc. 2016 Jan 13;138(1):289–299. doi: 10.1021/jacs.5b10494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parent A., Elduque X., Cornu D., Belot L., Le Caer J.P., Grandas A., Toledano M.B., D'Autréaux B. Mammalian frataxin directly enhances sulfur transfer of NFS1 persulfide to both ISCU and free thiols. Nat. Commun. 2015;6:5686. doi: 10.1038/ncomms6686. [DOI] [PubMed] [Google Scholar]
- 43.Holmström K.M., Baird L., Zhang Y., Hargreaves I., Chalasani A., Land J.M., Stanyer L., Yamamoto M., Dinkova-Kostova A.T., Abramov A.Y. Nrf2 impacts cellular bioenergetics by controlling substrate availability for mitochondrial respiration. Biol Open. 2013;2:761–770. doi: 10.1242/bio.20134853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ludtmann M.H., Angelova P.R., Zhang Y., Abramov A.Y., Dinkova-Kostova A.T. Nrf2 affects the efficiency of mitochondrial fatty acid oxidation. Biochem. J. 2014;457:415–424. doi: 10.1042/BJ20130863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knatko E.V., Tatham M.H., Zhang Y., Castro C., Higgins M., Dayalan Naidu S., Leonardi C., de la Vega L., Honda T., Griffin J.L., et al. Downregulation of Keap1 confers features of a fasted metabolic state. iScience. 2020;23 doi: 10.1016/j.isci.2020.101638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hourihan J.M., Kenna J.G., Hayes J.D. The gasotransmitter hydrogen sulfide induces nrf2-target genes by inactivating the keap1 ubiquitin ligase substrate adaptor through formation of a disulfide bond between cys-226 and cys-613. Antioxidants Redox Signal. 2013;19:465–481. doi: 10.1089/ars.2012.4944. [DOI] [PubMed] [Google Scholar]
- 47.Kelly B., Carrizo G.E., Edwards-Hicks J., Sanin D.E., Stanczak M.A., Priesnitz C., Flachsmann L.J., Curtis J.D., Mittler G., Musa Y., et al. Sulfur sequestration promotes multicellularity during nutrient limitation. Nature. 2021;591:471–476. doi: 10.1038/s41586-021-03270-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chio I.I.C., Jafarnejad S.M., Ponz-Sarvise M., Park Y., Rivera K., Palm W., Wilson J., Sangar V., Hao Y., Öhlund D., et al. NRF2 promotes tumor maintenance by modulating mRNA translation in pancreatic cancer. Cell. 2016;166:963–976. doi: 10.1016/j.cell.2016.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mitsuishi Y., Taguchi K., Kawatani Y., Shibata T., Nukiwa T., Aburatani H., Yamamoto M., Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 50.Sayin V.I., LeBoeuf S.E., Singh S.X., Davidson S.M., Biancur D., Guzelhan B.S., Alvarez S.W., Wu W.L., Karakousi T.R., Zavitsanou A.M., et al. Activation of the NRF2 antioxidant program generates an imbalance in central carbon metabolism in cancer. Elife. 2017;6 doi: 10.7554/eLife.28083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Romero R., Sayin V.I., Davidson S.M., Bauer M.R., Singh S.X., LeBoeuf S.E., Karakousi T.R., Ellis D.C., Bhutkar A., Sánchez-Rivera F.J., et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat. Med. 2017;23:1362–1368. doi: 10.1038/nm.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.