Abstract
Computational machine learning (ML)-based frameworks could be advantageous for scalable analyses in neuropathology. A recent deep learning (DL) framework has shown promise in automating the processes of visualizing and quantifying different types of amyloid-β deposits as well as segmenting white matter (WM) from gray matter (GM) on digitized immunohistochemically stained slides. However, this framework has only been trained and evaluated on amyloid-β-stained slides with minimal changes in preanalytic variables. In this study, we evaluated select preanalytical variables including magnification, compression rate, and storage format using three digital slides scanners (Zeiss Axioscan Z1, Leica Aperio AT2, and Leica Aperio GT 450), on over 60 whole slide images, in a cohort of 14 cases having a spectrum of amyloid-β deposits. We conducted statistical comparisons of preanalytic variables with repeated measures analysis of variance evaluating the outputs of two DL frameworks for segmentation and object classification tasks. For both WM/GM segmentation and amyloid-β plaque classification tasks, there were statistical differences with respect to scanner types (p < 0.05) and magnifications (p < 0.05). Although small numbers of cases were analyzed, this pilot study highlights the significance of preanalytic variables that may alter the performance of ML algorithms.
Keywords: Alzheimer disease, Amyloid-β, Deep learning, Digital pathology, Machine learning, Slide scanner, Whole slide imaging
INTRODUCTION
Whole-slide imaging has become an increasingly popular modality to assess brain tissues. With the help of a digital slide scanner, ultra high-resolution whole slide images (WSIs) are generated to aid in the preservation of tissue details (1). WSIs can be viewed and annotated through computer software such as Aperio ImageScope, ZEN, and QuPath (2). The digitization of tissue information allows for the application of computational approaches, which include but are not limited to machine learning (ML) and image processing that can aid with automated analyses of tissues.
Many types of pathologies within the brain define the neuropathologic classification of many neurodegenerative diseases (3). For example, amyloid-β deposits, in the form of plaques, are a hallmark pathological feature of Alzheimer disease (AD) (3). It is becoming advantageous to have more quantitative assessments of these pathologies for deeper phenotyping that is paving the way for precision medicine approaches for these devastating diseases (4–6). The manual quantification of pathologies, such as amyloid-β plaques, can be a time-consuming task that has been automated through Convolutional Neural Networks (CNN) (7), a type of ML framework. Other deep learning (DL) studies in pathology have applied similar techniques for WSI analysis (7–9). However, the performance of the aforementioned CNN models has not been fully demonstrated for WSIs scanned under variable conditions (i.e., magnifications, compression rates, etc.).
Many promising DL-based studies in neuropathology utilize WSIs from a single scanner, with single WSI formatting settings (10–13). Such design choices lead to a study with little or no variation in preanalytical variables such as image format, image compression rate, and scanner types. Despite displaying competitive performance, studies may not adequately assess generalizability; this lack of preanalytical variable diversity could be a concern for the reported performance metrics (10–13). Research on organ tissues other than brain has revealed a model trained with data from diverse settings, such as different scanners, outperform models trained with single-source WSIs (14). Although 2 WSIs from different scanners may look identical to a human evaluator, they may look distinct to the DL model due to the scanner, formatting applied to the scan, and/or the digital watermark left by preprocessing software. Different formatting settings or scanners introduce variables including but are not limited to compression standard, compression rate, storage format, and magnification. The pixel values such as µm/pixel, in each WSI, may differ due to compression or other variables and yet display an identical image to the evaluator. Since DL models learn through backpropagation (15) and thus “see” the pixel values, not the overall picture like an expert, they may be affected by the change in these preanalytic variables.
Concerning the performance of DL frameworks when presented data with different preanalytical variables, few studies have tested and observed degradation in performance with different tissue areas and quantities (16, 17), different scanners (18), and different class distributions (19). Another study has experimented with a single framework facing changes in storage format and architecture used (20). In real-world model deployment, data from different scanners or generated with different scanning formatting will likely be evaluated by the model, leading to a concern about the performance metrics displayed in studies that demonstrate no variance in preanalytical variables. Studies that present the same preanalytical variables in training and testing data do not adequately test a model’s generalizability despite good reported performance metrics. Therefore, there may be many published DL models with good reported performance that can only replicate good performance when fed data similar to the training set.
Our study seeks to provide a proof of concept in the neuropathology realm examining the potential prediction effects of different preanalytic variables of DL models. We aim to test the generalizability capacity of 2 DL models, one for segmentation and the other for classification, trained using WSIs from a single scanner by testing them on the same slides scanned with different scanners and scanner settings. By displaying and comparing the qualitative and quantitative outputs of 2 different DL tasks when applied to data with different preanalytical variables, we report and highlight effects by such variable changes.
MATERIALS AND METHODS
Datasets
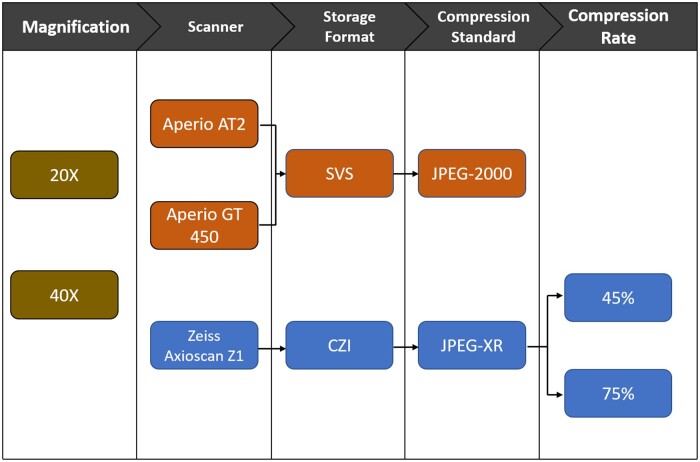
We utilized WSI from a total of 14 cases (see Supplementary Data Table S2 for demographic details) from slides of formalin-fixed paraffin-embedded 5 µm sections of postmortem human brain temporal cortex immunohistochemically stained with an antibody against amyloid-β diluted 1:1600 (4G8, BioLegend, formally Covance, San Diego, CA, USA); all sections were subjected to standard procedures on automated machines, pretreatment included 10 minutes in 87% formic acid; endogenous peroxidases were blocked with 3% hydrogen peroxide. All antibody staining was conducted on an autostainer (DAKO AutostainerLink48, Agilent, Santa Clara, CA, USA) utilizing proper positive and negative control for the antibody. All staining was conducted using proper controls by the University of California Davis Histology Core, which is a Clinical Laboratory Improvement Amendments (CLIA) and College of American Pathologists (CAP) accredited laboratory that also operates under the best laboratory practices standards and meets all Federal, State of California and UC Davis guidelines and regulations. The stained slides were digitized to create 6 different WSIs datasets having different preanalytic variables (see Fig. 1 for schematic). In the following sections, we describe each of our datasets according to its preanalytical variables. The names of the datasets reflect a formatting of scanner-magnification-compression. To ensure fair performance through similar processing times for each WSI, all 40× WSIs were resized to 20× through PyVips package. Other than resizing, no preprocessing was done in any of the WSIs. No preprocessing was applied prior to feeding the WSIs to the model to avoid any digital watermarks generated by the image software such as ImageScope or ZEN. All scanners undergo routine servicing once a year. The AT2 scanner was purchased in 2016, the Zeiss Axio Z1 Scanner was purchased in 2019, and the GT450 was purchased in 2021. We selected the standard processing method for all scanners, which included but were not limited to standard automatic color profile and tissue detection. Tissue detection automatically crops the WSI to ensure reduced background. Color profile normalizes the pixel values for optimal monitor display. The effects of cropping and color profile can be observed in Supplementary Data Figure S1.
Figure 1.
Schematic representation of preanalytical variables evaluated. Variables linked by arrows are nested, for example, all Zeiss Axioscan Z1 data employed are in CZI storage format.
AT2-20×
A total of 14 slides from our cohort were scanned into JPEG-2000 compressed .svs files. The WSIs were digitized by Leica Aperio AT2 at 20× magnification. This dataset contains the same preanalytical variables as the WSIs used for training of both amyloid-β deposit detection and white matter (WM)/gray matter (GM) segmentation.
AT2-40×
All 14 slides from our cohort scanned into JPEG-2000 compressed .svs files. The WSIs were digitized by Leica Aperio AT2 at 40× magnification. This dataset presents only 1 preanalytical variable change, that is, magnitude change, as the WSIs used for training of both evaluated DL frameworks. Case 11 displayed cover slip deadherence not seen in other datasets.
GT450-40×
A total of 14 slides from our cohort were scanned into JPEG-2000 compressed .svs files. The WSIs were digitized by Leica Aperio GT450 at 40× magnification. Despite having the same storage format, and compression standard, these WSIs came from a different Leica scan than the AT2.
Axio-Z1-40×-45
A total of 13 slides from our cohort were scanned into JPEG-XR compressed .czi files. The WSIs were digitized by Zeiss Axio Z1 scanner at 40× magnification. The JPEG-XR compression reduced the size of the file by 55%.
Axio-Z1-40×-75
A total of 13 slides from our cohort were scanned into JPEG-XR compressed .czi files. The WSIs were digitized by Zeiss Axio Z1 scanner at 40× magnification. The JPEG-XR compression reduced the size of the file by 25%.
Evaluated pipelines
This study evaluated 2 pretrained models (overall workflow is depicted in Fig. 2). The first model, aimed at WM/GM segmentation (8), was trained on 20× JPEG-2000 compressed .svs slides digitized from Leica Aperio AT2. The second model, aimed at detecting amyloid-β plaques (7), was trained on 20× JPEG-2000 compressed .svs slides. We performed no tuning or additional training on any of the 2 models. The preanalytic variables for the data used in the 2 pretrained models displayed constant scanner (Aperio AT2), constant magnification (20×), consistent storage format (SVS), and constant compression standard (JPEG-2000), matching the preanalytic variables from the AT2-20× dataset.
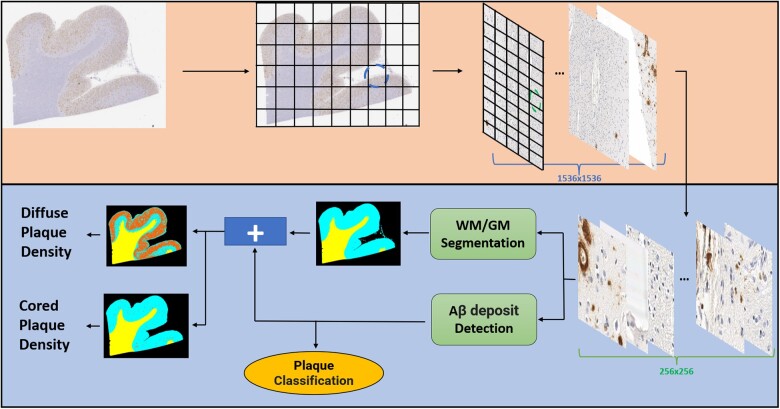
Figure 2.
Convolutional Neural Network-based deep learning (DL) pipeline employed in this study. The approach for prediction is patch-based; therefore, whole slide image must be patched prior to analysis. The different blue and green segmented circled on the gridded images in the top figure (light orange box) panel refer to the different resolutions (1536 × 1536 and 256 × 256, respectively) patched by the framework. There are 2 DL modules responsible for predictions (lower figure panel—light blue box), a white matter (WM)/gray matter (GM) segmentation and an amyloid-β deposit detection with subsequent classification module which operate on the 256 × 256 pixel resolution. For heatmaps in the bottom left corner of the figure, WM is represented in yellow, GM as cyan, background as black, and plaques as orange (figure adapted from [23]).
Both models employed CNN-based DL. The amyloid-β deposit detection was originally trained on a version of VGG (21). The WM/GM segmentation model was trained on a version of ResNet-18 (22). Both ResNet and VGG are commonly used CNN-based DL architectures. The pipeline used to generate both models’ predictions was similar to the one described in (23). We patched each WSI in 256 × 256 segments and those patches were the input to both classification and segmentation models simultaneously as depicted in Figure 2. Although the input is the same, each model performs different tasks, while the ResNet performs patch-based segmentation, the VGG model performs classification and detection of amyloid-β present in each patch.
The ResNet-based WM/GM segmentation module outputs a heatmap with yellow, cyan, and black representing WM, GM, and background, respectively (Supplementary Data Fig. S2). The model also outputs WM and GM size in µm/pixel, which we use to calculate the WM/GM ratio. The VGG-based amyloid-β deposit detection module outputs separate heatmaps based on each plaque classification (cored and diffuse) colored in red. Counts/area of each plaque classification were also generated by incorporating the WM/GM predictions. All codes related to these processes are located in this GitHub (https://github.com/ucdrubinet/BrainSec, last accessed January 10, 2023).
Registration
Due to the distinct field-of-view and automatic tissue detection present in each scanner, the output WSI files from different scanners (see Supplementary Data Table S1 for additional details on each WSIs parameters) are not aligned and present different tissue sizes and aspect ratios despite being generated from the same slide. Automatic cropping caused loss of tissue area for select Zeiss scans (Supplementary Data Fig. S1).
Hence, to register the WSIs and ensure as much alignment and as little loss of tissue as possible, we employed a technique for restained histological WSI coregistration (24). Aligning restained WSIs is similar to our task since the tissue borders are similar between restained slides. However, due to the difference in magnification in some WSIs, we also needed to resize the 40× files into 20× to ensure similar tissue size. We achieved this by calculating the resizing factor that allowed for the height and width difference to be minimal when compared to the original AT2-20× WSI. Due to distinct aspect ratios from different scanners, the height and width from resized WSIs were not able to match the ones from the original AT2-20× WSI. This required an additional manual tuning step to the registration technique employed. All codes related to these processes are located in this GitHub (https://github.com/smujiang/Re-stained_WSIs_Registration, last accessed January 10, 2023).
Statistical analysis
Because all slides were scanned using each of the scanners, repeated measures analysis of variance (ANOVA) was used to compare differences in WM/GM segmentation and amyloid-β core or diffuse plaque counts derived from the ML models across preanalytic variables. Key factors of interest included scanner, magnification, and compression rate. Not all combinations of factors were considered, so separate analyses were conducted for each comparison of interest, including all relevant data. For example, when considering compression rate, only outcomes from the slides on the Zeiss Axioscan were included. All analyses were conducted using Python and a p value of less than 0.05 was considered statistically significant.
RESULTS
Effects of preanalytic variables on the amyloid-β deposit detection/classification model
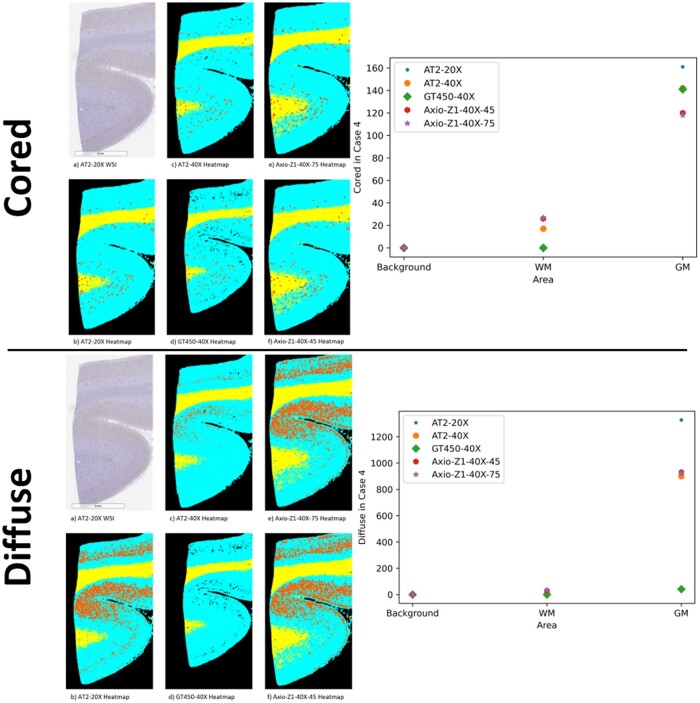
The amyloid-β deposit detection with subsequent plaque classification outputs counts for cored and diffuse amyloid-β plaques. The module acquires these counts by detecting and then classifying all deposits located in the WSI. A comparison of these predictions for a single case can be seen in Figure 3. We observed some disagreement in prediction between the different datasets for both diffuse and cored plaques. Figure 3 shows an example with heatmaps and accompanying quantitative results for plaque counts in background, GM, and WM. Figure 4 is a graphical representation of the quantitative results for cored and diffuse plaque counts and GM/WM ratios across all cases based on the preanalytic variable.
Figure 3.
Schematic of heatmaps of white matter (WM)/gray matter (GM) segmentation and plaques counts for case 4. Top panel—GM/WM cored plaque heatmap (left) and counts by area based on select preanalytic variables (right). Bottom panel—GM/WM diffuse plaque heatmap (left) and counts by area based on select preanalytic variables (right). A zoomed-in area (not the whole slide image) of case 4 was chosen to aid in visualization. For heatmaps, plaques are depicted in orange, background as black, WM as yellow, and GM as cyan.
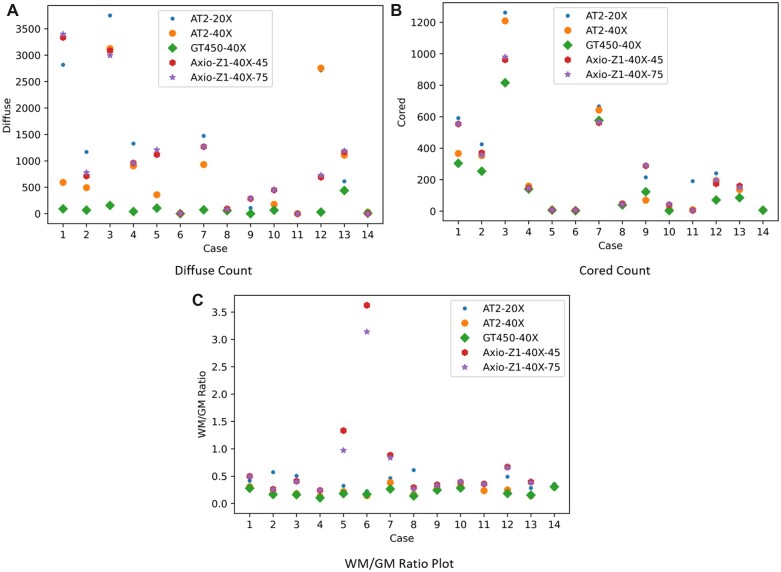
Figure 4.
White matter (WM)/gray matter (GM) ratio, cored, and diffuse plaque (A), cored plaque (B) counts, and WM/GM ratio (C) for each case by preanalytic variable. Cases with none/low likelihood Alzheimer disease (AD) [5, 6, 8, 10, 11, 14] typically had low numbers of core plaques, while cases with high likelihood AD [1, 3, 4, 12, 13] had higher counts. More information on the demographics of cases located in Supplementary Data Table S2. Further details on case 6 for GM/WM ratio is located within Supplementary Data Fig. S4.
By acquiring the quantitative results for cored/diffuse plaque counts and applying ANOVA, we can test whether the preanalytical variables affect the target outcome (deposit counts). Table 1 shows that magnification and scanner type are 2 preanalytical variables with the most effect on our DL predictions. Results from Table 1 show similar effect observed in the case of the segmentation model, where magnification and scanner (GT450) are the preanalytical variables with the most effect on DL predictions.
Table 1.
p-Values for ANOVA Tests
| Statistical analysis values |
||||
|---|---|---|---|---|
| Axio-Z1-40×-45 versus Axio-Z1-40×-75 | AT2-20× versus Axio-Z1-40×-75 | AT2-20× versus GT450-40× | AT2-20× versus AT2-40× | |
| Cored count | 0.4024 | 0.0738 | 0.0078 | 0.0160 |
| Diffuse count | 0.2272 | 0.4290 | 0.0073 | 0.0705 |
| WM/GM ratio | 0.0853 | 0.2475 | 0.0005 | 0.0013 |
ANOVA, analysis of variance; GM, gray matter; WM, white matter.
Effects of preanalytic variables on the WM/GM segmentation model
The WM/GM segmentation module yields WM/GM ratio as a quantitative measure that can be used in statistical analysis. We plotted the WM/GM ratios for the different datasets evaluated in Figure 4. When applying ANOVA to those values, we can check whether the preanalytical variables affect the target (WM/GM ratio). Table 1 summarizes our results; both magnification and scanner type (GT450) have significant effects on DL prediction outcomes.
The WM/GM segmentation map also outputs a heatmap of the segmented WSI, in this heatmap, we have GM predictions denoted as cyan, WM as yellow, and background as black (Fig. 3 and Supplementary Data Figs. S2 and S4). This method of visualizing the results is a better indicator of stable performance, as that is the final product to be analyzed by the expert, as well as the map to be used for the calculation of densities of deposits and structures seen in the WM/GM. As seen in Figure 4, changing scanners and magnification has an effect on our model’s predictions. For case 7, when comparing the results from AT2-20×, Axio-Z1-40×-75, and Axio-Z1-40×-45, there are prediction disagreements between GM and WM close to the boundaries of GM and background (Supplementary Data Fig. S2).
Saliency maps
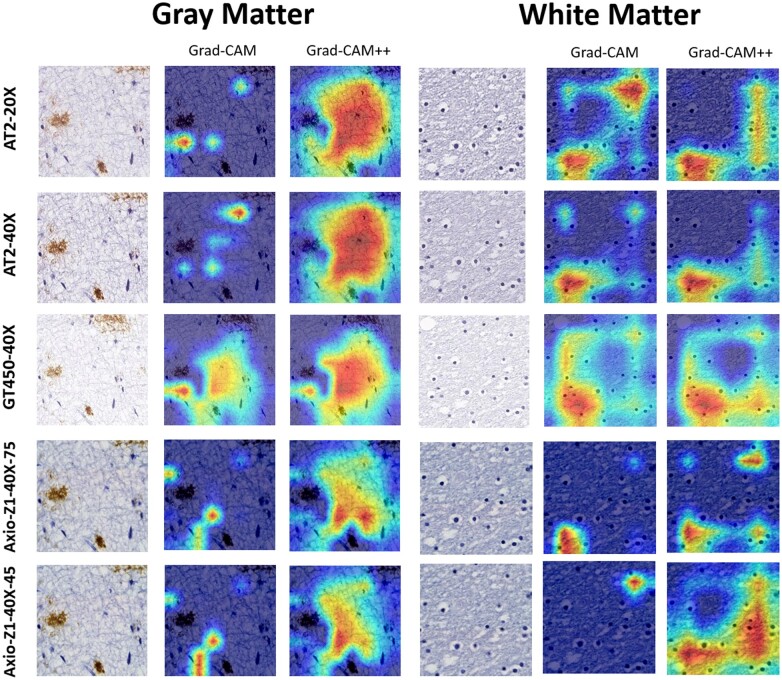
When analyzing heatmaps and quantitative scores acquired from the 2 DL frameworks, we can assess how the preanalytical variables affect the outputs. However, this information only tells us how the final output was affected, but the effect on the prediction process of the DL frameworks is still unknown. Saliency maps allow us to tap into the black box nature of DL models and learn a bit about their prediction process, more specifically, how much the different locations in each image contributed to the final output. We employed Class Activation Mapping (CAM) (25), Grad-CAM (26), and Grad-CAM++ (27) as methods to acquire the saliency maps. By analyzing the saliency maps generated from each 256 × 256 patch, we can observe which areas of each patch contributed most to the final model output and how these areas may differ according to the preanalytical variables. That is especially relevant for the WM/GM segmentation DL model, as there is no obvious single structure linked to the predictions such as an amyloid-β plaque classification, as it relies on features such as texture of the tissue, as shown previously (23).
The Grad-CAM presented in Figure 5 shows that despite the prediction outcome remaining constant as GM in all the cases shown, the areas that led the WM/GM frameworks to reach that conclusion were different. The Grad-CAM++ displays less differences, pointing toward a higher level of agreement that occurs when taking in consideration a more complex interpretability framework. The same effect is observed when the agreed predictions are WM, as seen in Figure 5.
Figure 5.
Grad-CAM and Grad-CAM++ of agreed predictions of gray matter and white matter. All datasets agreed on the tile’s prediction and got the correct prediction.
We are also able to see in Supplementary Data Figure S3 the difference remains when the final output disagrees. This patch is taken from the patch with high WM-GM prediction disagreement observed between AT2-20× and Axio-Z1 datasets in case 7 (Supplementary Data Fig. S2). Despite a level of overlap in the saliency map, the AT2-20× CAM covers a wider area than its Axio-Z1 counterparts.
DISCUSSION
Recent studies utilizing ML/DL in pathology have been successful in displaying high prediction performance (7–9, 23). However, due to the black box nature of trained DL models, rigorous testing is needed since there is no guarantee a model trained without domain adaptation (DA) techniques (28–30) will have the same performance when applied to data with different preanalytic variables. Furthermore, as other studies have shown feeding WSIs with different preanalytical variables may degrade performance (17), we must extend the rigorous testing to account for such differences. Studies including generalizability testing are limited (17–20).
Our study’s AT2-20× dataset constitutes a fair baseline: it shares the same scanner, magnification, brain region, storage format, stain, and compression standard as the training data. Therefore, when performing tests on this data subset, we expected the model to achieve the most accurate performance in both DL modules as there is no variance in preanalytical variables. Here, we utilized the AT2-20× in the training set, so it was the gold standard of performance for all other datasets. By evaluating the DL frameworks on AT2-20× and then evaluating on other datasets with different preanalytical variables, we investigated the effect preanalytical variables on DL generalizability in WSIs and observed some level disagreement in both DL frameworks.
We demonstrated the detection of amyloid-β plaques in brain WSI trained on 20× AT2 slides is affected by WSI magnification (40×) and GT450 scanner. We saw a similar effect for both diffuse and cored amyloid-β plaques. In addition to the statistical analysis result, we can reliably identify an overall effect of preanalytical variables on amyloid-β plaque counts when observing heatmaps and counts per area.
We also observed performance differences when our generalizability test was applied to a WM/GM segmentation task. Our results revealed WSI magnification (40×) and scanner type (GT450) have an impact on predicted WM/GM ratio. Our observation of WM/GM segmentation heatmaps and saliency maps also displays unstable WM/GM predictions when applied to WSIs from GT450 scanner (Fig. 5). Some outlier WM/GM predicted heatmaps can be observed in Zeiss scanners having stark differences when compared to the AT2-20× heatmap (Supplementary Data Fig. S4).
The outlier performance from GT450 expanded to all cases employed in this study. When analyzing the overall scan from the GT450 in comparison to other scanners, subjective observations denoted an increase in brightness and white tones. We hypothesized the different standard color profile applied to the scan (i.e., ICC profile) is responsible for the difference observed. Since scanning was done with default parameters, the difference in color profile may extend to the software version employed at the time of the scan. Studies have argued a normalization step is required to match performance between scanners or different scanning protocols (31). In future works, we will examine how color normalization may alter results within the preanalytic variable realm (32–34). Preliminary experiments revealed Reinhard normalization (32) to be a suitable intervention to address GT450 performance differences (Supplementary Data Fig. S5).
Although this study contributes to the field, there are some limitations to consider. First, there is the misalignment of digitalized tissue caused by the different scanners’ field of view. Due to such misalignment, we could not perfectly overlap the heatmap predictions. Such limitation prevented us from using our WM/GM annotated ground truth to reliably calculate Intersection over Union (IoU) or DICE coefficient between our WSIs digitized from the same slides. Both DICE coefficient and IoU have been used to compare ground truth and predictions on a pixel-by-pixel basis (35, 36). This misalignment prevented automated tile comparison, as a human observer was required to fine tune registration for each individual area compared. Second, there was also the use of only 20× and 40× magnification; additional works with other magnifications such as 10× and 5× may be advantageous as file sizes may be smaller and easier to process. Third, our study examined only a limited number of cases from a single brain bank. Due to the large file size of WSIs, especially at 40× magnification, it becomes a time-consuming task to generate predictions for both WM/GM segmentation and amyloid-β deposit assessment, approximately 6 hours per 20× slide when employing an NVIDIA Tesla T4 GPU. We processed a total of 65 slides, which account for almost 400 hours of GPU use. Lastly, we utilized the AT2-20× as the gold standard to conduct comparisons. To our knowledge, although checklists for ML algorithms in medical imaging have been proposed (34), there are no gold standards for preanalytic variables for digital pathology when conducting ML algorithms. The choice of using AT2-20× as gold standard is due to data of same preanalytical variables being employed in training. This choice best matches the recommendations of item 7, regarding data sources, in previous works (37) as test data from AT2-20× match the trained model best. Unlike other medical imaging fields such as Radiology that have standard file formats, there have been no proposed standards in digital pathology and there exist many options given the vast array of available slide scanners and associated settings in the WSI realm. This study highlights the importance of denoting scanner types, magnifications, as well as compression rates when conducting such workflows.
Generalizability is a crucial challenge for deploying DL in real-life pathology problems. Currently, in the field, there are studies seeking to perform DA techniques to address generalizability from diverse preanalytical variables in ML frameworks (28–30). These efforts are important to advance the generalizability of frameworks in the field and address the unwanted effects we observe when varying preanalytical variables. Unlike normalization, DA does not need any additional preprocessing steps for generalization to many different scanners. The application of these DA techniques has also been shown in the WSI domain (30) and would be a great candidate to address the performance difference we observed.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank the families and participants of the University of California Davis Alzheimer’s Disease Research Centers (ADRC) for their generous donations as well as ADRC staff and faculty for their contributions. The views and opinions expressed in this article are those of the author and do not necessarily reflect the official policy or position of any public health agency of California or of the US government. We also thank the UC Davis Health Department of Pathology and Laboratory Medicine as well as the laboratory of Dr. Alexander “Sandy” Borowsky for the use of digital slide scanners.
Contributor Information
Luca Cerny Oliveira, Department of Electrical and Computer Engineering, University of California Davis, Davis, California, USA.
Zhengfeng Lai, Department of Electrical and Computer Engineering, University of California Davis, Davis, California, USA.
Danielle Harvey, Department of Public Health Sciences, University of California, Davis, Davis, California, USA.
Kevin Nzenkue, Department of Pathology and Laboratory Medicine, University of California Davis, Sacramento, California, USA.
Lee-Way Jin, Department of Pathology and Laboratory Medicine, University of California Davis, Sacramento, California, USA.
Charles Decarli, Department of Neurology, University of California, Davis, Sacramento, California, USA.
Chen-Nee Chuah, Department of Electrical and Computer Engineering, University of California Davis, Davis, California, USA.
Brittany N Dugger, Department of Pathology and Laboratory Medicine, University of California Davis, Sacramento, California, USA.
FUNDING
Resources for this study were funded in part by grants from the National Institute on Aging of the National Institutes of Health under Award Numbers R01AG062517, P30AG072972, and R01AG056519, and a research grant from the California Department Of Public Health (19-10611) with partial funding from the 2019 California Budget Act.
CONFLICTS OF INTEREST
Authors have no conflicts of interest to disclose related to this work.
Supplementary Data
Supplementary Data can be found at academic.oup.com/jnen.
REFERENCES
- 1. Al-Janabi S, Huisman A, Van Diest PJ. Digital pathology: Current status and future perspectives. Histopathology 2012;61:1–9 [DOI] [PubMed] [Google Scholar]
- 2. Bankhead P, Loughrey MB, Fernandez JA, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep 2017;7:16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dugger BN, Dickson DW. Pathology of neurodegenerative diseases. Cold Spring Harb Perspect Biol 2017;9:a028035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shakir MN, Dugger BN. Advances in deep neuropathological phenotyping of Alzheimer disease: Past, present, and future. J Neuropathol Exp Neurol 2022;81:2–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McKenzie AT, Marx GA, Koenigsberg D, et al. ; PART working group. Interpretable deep learning of myelin histopathology in age-related cognitive impairment. Acta Neuropathol Commun 2022;10:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vizcarra JC, Gearing M, Keiser MJ, et al. Validation of machine learning models to detect amyloid pathologies across institutions. Acta Neuropathol Commun 2020;8:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tang Z, Chuang KV, DeCarli C, et al. Interpretable classification of Alzheimer's disease pathologies with a convolutional neural network pipeline. Nat Commun 2019;10:2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lai Z, Guo R, Xu W, et al. Automated grey and white matter segmentation in digitized aβ human brain tissue slide images. In: 2020 IEEE International Conference on Multimedia & Expo Workshops (ICMEW). IEEE; 2020
- 9. Litjens G, Kooi T, Bejnordi B, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60–88 [DOI] [PubMed] [Google Scholar]
- 10. Hekler A, Utikal JS, Enk AH, et al. Deep learning outperformed 11 pathologists in the classification of histopathological melanoma images. Eur J Cancer 2019;118:91–96 [DOI] [PubMed] [Google Scholar]
- 11. Aresta G, Araujo T, Kwok S, et al. Bach: Grand challenge on breast cancer histology images. Med Image Anal 2019;56:122–139 [DOI] [PubMed] [Google Scholar]
- 12. Hsu WW, Guo JM, Pei L, et al. A weakly supervised deep learning-based method for glioma subtype classification using WSI and mpMRIs. Sci Rep 2022;12:6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kiani A, Uyumazturk B, Rajpurkar P, et al. Impact of a deep learning assistant on the histopathologic classification of liver cancer. NPJ Digit Med 2020;3:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Balkenhol MC, Tellez D, Vreuls W, et al. Deep learning assisted mitotic counting for breast cancer. Lab Invest 2019;99:1596–1606 [DOI] [PubMed] [Google Scholar]
- 15. LeCun Y, Boser B, Denker J, et al. Handwritten digit recognition with a back-propagation network. Adv Neural Inf Process Syst 1989:2 [Google Scholar]
- 16. Vali-Betts E, Krause KJ, Dubrovsky A, et al. Effects of image quantity and image source variation on machine learning histology differential diagnosis models. J Pathol Inform 2021;12:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jang HJ, Song IH, Lee SH. Generalizability of deep learning system for the pathologic diagnosis of various cancers. Appl Sci 2021;11:808. 10.3390/app11020808. Accessed January 10, 2023 [DOI] [Google Scholar]
- 18. Yan W, Huang L, Xia L, et al. MRI manufacturer shift and adaptation: Increasing the generalizability of deep learning segmentation for MR images acquired with different scanners. Radiol Artif Intell 2020;2:e190195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sathitratanacheewin S, Sunanta P, Pongpirul K. Deep learning for automated classification of tuberculosis-related chest X-Ray: dataset distribution shift limits diagnostic performance generalizability. Heliyon 2020;6:e04614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jones AD, Graff JP, Darrow M, et al. Impact of pre‐analytical variables on deep learning accuracy in histopathology. Histopathology 2019;75:39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556, 2014. Available at: https://arxiv.org/abs/1409.1556. Accessed January 10, 2023
- 22. He K, Zhang X, Ren S, et al. Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2016. Available at: https://arxiv.org/abs/1512.03385v1. Accessed January 10, 2023
- 23. Lai Z, Oliveira LC, Guo R, et al. BrainSec: Automated brain tissue segmentation pipeline for scalable neuropathological analysis. IEEE Access 2022;10:49064–49079. Available at: https://ieeexplore.ieee.org/document/9766171. Accessed January 10, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jiang J, Larson NB, Prodduturi N, et al. Robust hierarchical density estimation and regression for re-stained histological whole slide image co-registration. PLoS One 2019;14:e0220074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou B, Khosla A, Lapedriza A, et al. Learning deep features for discriminative localization. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2016. Available at: https://arxiv.org/abs/1512.04150. Accessed January 10, 2023
- 26. Selvaraju RR, Cogswell M, Das A, et al. Grad-cam: Visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE International Conference on Computer Vision; 2017. Available at: https://arxiv.org/abs/1610.02391v4. Accessed January 10, 2023
- 27. Chattopadhay A, Sarkar A, Howlader P, et al. Grad-cam++: Generalized gradient-based visual explanations for deep convolutional networks. In: 2018 IEEE Winter Conference on Applications of Computer Vision (WACV). IEEE; 2018. Available at: https://arxiv.org/abs/1710.11063v3. Accessed January 10, 2023
- 28. Breen J, Zucker K, Orsi NM, et al. Assessing domain adaptation techniques for mitosis detection in multi-scanner breast cancer histopathology images. In: MICCAI 2021: Biomedical Image Registration, Domain Generalisation and out-of-Distribution Analysis, Strasbourg, France, September 27–October 01, 2021. Cham: Springer; 2021: 14–22. ISBN: 9783030972806
- 29. Aviles J, Talou GD, Camara O, et al. Domain adaptation for automatic aorta segmentation of 4D flow magnetic resonance imaging data from multiple vendor scanners. In: International Conference on Functional Imaging and Modeling of the Heart 2021:112–21 [Google Scholar]
- 30. Panfilov E, Tiulpin A, Klein S, et al. Improving robustness of deep learning based knee MRI segmentation: Mixup and adversarial domain adaptation. In: Proceedings of the IEEE/CVF International Conference on Computer Vision Workshops; 2019:450–9
- 31. Khan AM, Rajpoot N, Treanor D, et al. A nonlinear mapping approach to stain normalization in digital histopathology images using image-specific color deconvolution. IEEE Trans Biomed Eng 2014;61:1729–38 [DOI] [PubMed] [Google Scholar]
- 32. Reinhard E, Adhikhmin M, Gooch B, et al. Color transfer between images. IEEE Comput Grap Appl 2001;21:34–41 [Google Scholar]
- 33. Roy S, Panda S, Jangid M. Modified reinhard algorithm for color normalization of colorectal cancer histopathology images. In: 29th European Signal Processing Conference (EUSIPCO). IEEE; 2021:1231–5
- 34. Vahadane A, Peng T, Sethi A, et al. Structure-preserving color normalization and sparse stain separation for histological images. IEEE Trans Med Imaging 2016;35:1962–71 [DOI] [PubMed] [Google Scholar]
- 35. Rahman MA, Wang Y. Optimizing intersection-over-union in deep neural networks for image segmentation. In: International Symposium on Visual Computing International Symposium on Visual Computing. Cham: Springer; 2016:234–44
- 36. Zou KH, Warfield SK, Bharatha A, et al. Statistical validation of image segmentation quality based on a spatial overlap index1: Scientific reports. Acad Radiol 2004;11:178–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mongan J, Moy L, Kahn CE. Jr Checklist for artificial intelligence in medical imaging (CLAIM): A guide for authors and reviewers. Radiol Artif Intell 2020;2:e200029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.