Abstract
The MYB/MYBL1::QKI fusion induces the protooncogene, MYB, and deletes the tumor suppressor gene, QKI. MYB/MYBL1::QKI rearrangement was previously reported only in angiocentric glioma (AG) and diffuse low-grade glioma. This report compares 2 tumors containing the MYB/MYBL1::QKI fusion: a diffuse pediatric-type high-grade glioma (DPedHGG) in an 11-year-old boy and an AG in a 46-year-old woman. We used immunohistochemistry, next-generation sequencing, and methylation profiling to characterize each tumor and compare our findings to the literature on AG and tumors with the MYB/MYBL1::QKI rearrangement. Both tumors were astrocytic with angiocentric patterns. The MYB::QKI fusion-positive DPedHGG, which recurred once, was accompanied by TP53 mutation and amplification of CDK6 and KRAS, suggesting malignant transformation secondary to additional genetic aberrations. The second case was the adult AG with MYBL1::QKI fusion, which mimicked ependymoma based on histopathology and its dot- and ring-like epithelial membrane antigen positivity. Combined with a literature review, our results suggest that MYB/MYBL1 alterations are not limited to low-grade gliomas, including AG. AG is most common in the cerebra of children and adolescents but exceptional cases occur in adults and the acquisition of additional genetic mutations may contribute to high-grade glioma. These cases further demonstrate that molecular characteristics, morphologic features, and clinical context are essential for diagnosis.
Keywords: Angiocentric glioma, Methylation profile, MYB/MYBL1-alteration, Next-generation sequencing, Pediatric-type high-grade glioma
INTRODUCTION
Angiocentric gliomas (AGs) are rare, diffusely infiltrating pediatric-type low-grade gliomas (LGGs) characterized by an angiocentric pattern and elongated astrocytic morphology with monomorphic bipolar cells. These cells possess ependymoma-like characteristics, such as dot- and ring-like epithelial membrane antigen (EMA)-positivity. AG was first described in the fourth edition of the World Health Organization (WHO) Classification of Tumors of the Central Nervous System. Zhang et al (1) described the MYB rearrangement in AGs, followed by Bandopadhayay et al (2), who identified the complete MYB::QKI rearrangement in AG.
There is debate about whether AGs are intracortical ependymomas or a new type of tumor. MYB/MYBL1::QKI fusion has been demonstrated in AG; however, we are unaware of reports of this fusion in ependymomas. To date, no MYB/MYBL1::QKI fusion has been reported in CNS or non-CNS tumors—except for 1 case of ganglioglioma, which may have demonstrated an indirect MYB::QKI fusion due to a 6q23.3-q26 deletion (3); therefore, it is unclear whether this ganglioglioma has an MYB-QKI fusion. This fusion is generally reported in diffuse LGGs (previous isomorphic glioma) (4) and AGs (2, 4–8). Here, we present an MYB::QKI fusion-positive diffuse pediatric-type high-grade glioma (DPedHGG) in a pediatric patient and an MYBL1::QKI fusion-positive AG in an adult patient. The first case suggests that tumors with MYB/MYBL1::QKI fusions should only be considered indolent when the fusion is the only driver of alteration in LGG. However, it may become a high-grade tumor if other genetic mutations accompany the fusion.
MATERIALS AND METHODS
We describe 2 cases: an 11-year-old boy and a 46-year-old woman. We additionally reviewed all previously published reports of molecular genetically confirmed MYB/MYBL1 fusion-positive brain tumors.
The Seoul National University Hospital’s Institutional Review Board provided regulatory oversight of this study (1905-108-1035 and 1906-020-1037). This study was a retrospective review of pathologic reports, virtual images, electronic medical records, and anonymized and curated next-generation sequencing (NGS). No patients were examined directly, and the Board waived the requirement for informed consent.
Histopathologic and immunohistochemical examinations
Neutral formalin-fixed paraffin-embedded (FFPE) tissues were cut into 3-μm slices for Hematoxylin and eosin (H&E) staining and immunohistochemical (IHC) examination. IHC staining was performed using the standard avidin-biotin-peroxidase method and the BenchMark ULTRA system (Roche Diagnostics, Indianapolis, US). The tissue sections were stained with anti-GFAP (C6F2 monoclonal, 1:200, DAKO, Glostrup, Denmark), anti-IDH-1 (H09, monoclonal, 1:100, Dianova, Hamburg, Germany), anti-p53 (DO-7, monoclonal, 1:100, DAKO), anti-c-MYB (Ab117635, polyclonal, 1:200, Abcam, Cambridge, UK), anti-H3K27me3 (C36B11, monoclonal, 1:100, Cell Signaling, Boston, MA), anti-synaptophysin (27G12, monoclonal, 1:200, Novocastra, Newcastle, UK), Anti-Histone H3, K27M (ABE419, polyclonal, 1:700, Millipore, Temecula, CA, US), and anti-EZHIP (HPA004003, polyclonal, 1:300, Atlas, Stockholm, Sweden). The mitotic rate was assisted by phosphorylated histone H3 (pHH3) (369A-15, polyclonal, 1:100, Cell Marque, Rocklin, CA, US). The Ki-67 labeling index (MIB-1 monoclonal, 1:100, DAKO) was calculated using the Sectra Ki-67 morphometric analyzer on virtual Leica Biosystems slides. We used known positive tissues or internal positive controls for IHC-positive controls; primary antibodies were omitted when IHC was performed for the negative control.
The samples were reviewed by 2 pathologists (Y.Y.S. and S.H.P.) according to histopathological criteria established by the fifth edition of the WHO Classification of CNS Tumors (9).
DNA and RNA extraction for NGS, O6 methylguanine methyltransferase promoter methylation-specific PCR, and microsatellite instability test
Representative areas of the tumor from FFPE tissue on H&E-stained sections with at least 90% tumor cell content were outlined for macrodissection. DNA/RNA extraction was performed using the Maxwell RSC DNA/RNA FFPE Kit (Promega, Madison, WI) according to the manufacturer’s instructions.
We used the methylation-specific polymerase chain reaction (MSP) technique to analyze the methylation of the methylguanine methyltransferase promoter (MGMTp). The prepared DNA was modified via sodium bisulfite treatment using the EZ DNA Methylation-Gold Kit (D5005; Zymo Research, Orange, CA). The primer sequences used for MGMTp were: methylated forward, 5′-TTT CGA CGT TCG TAG GTT TTC GC-3′; methylated reverse, 5′-GCA CTC TTC CGA AAA CGA AAC G-3′; unmethylated forward, 5′-TTT GTG TTT TGA TGT TTG TAG GTT TTT GT-3′; unmethylated reverse, 5′-AAC TCC ACA CTC TTC CAA AAA CAA AAC A-3′. The annealing temperature was set to 64°C. The obtained polymerase chain reaction products were electrophoresed on 2% agarose gels and visualized under ultraviolet illumination after staining with ethidium bromide.
NGS studies with customized brain tumor-targeted gene panels
NGS studies were performed with tumor DNA and RNA extracted from FFPE tumor tissue and NEXTSeq Dx505 using a customized brain tumor gene panel. The FiRST brain tumor panel (BTP) was established by the Department of Pathology, Seoul National University Hospital (SNUH), and approved by the Korea Food and Drug Administration. FiRST BTP assesses 224 brain tumor-associated genes and 151 fusion genes. The fusion genes were sequenced using RNA. The NGS data were analyzed as previously reported (10). Briefly, somatic mutations were detected using the Genome Analysis Toolkit (GATK) Mutect2 v4.1.4.1. with default parameters (11). To avoid germline variant contamination, we used the gnomad.hg19.vcf Genome Aggregation Database (gnomAD) (12) and 1000 g_pon.hg19.vcf files, which include a standard panel for 1000 genomes. The files were provided by the GATK resource bundle. After calling somatic mutations, all variants were annotated by ANNOVAR (https://doc-openbio.readthedocs.io/projects/annovar/en/latest/) (13).
Methylation study using an EPIC 850K BeadChip microarray
DNA methylation array analysis was performed using the Infinium MethylationEPIC 850K BeadChip microarray. DNA methylation data analysis was performed using the MethylationArrayAnalysis package (version 1.14.0) for R programming (R 4.0.3). Unsupervised nonlinear dimension reduction was performed by selecting the 10 000 most variably methylated probes. The resulting distance matrix was used as input for t-distributed stochastic neighbor embedding analysis ([t-SNE]; Rtsne package version 0.15). The nondefault parameters were distance = TRUE, perplexity = 20, and theta = 0.5. Cases were colored on the t-SNE plot for effective visualization using the ggplot2 package (version 3.3.3). T-SNE analysis with DNA methylation data of this case and the previously reported methylation class of the brain tumors (14–16). IDAT files were uploaded to either version 11b4 or 12.5 of the online CNS tumor methylation classifier (MC) (https://www.molecularneuropathology.org).
RESULTS
Case 1: A diffuse MYB::QKI fusion-positive high-grade glioma in a child
An 11-year-old boy presented with generalized tonic seizures, without clonic movements, that lasted for approximately 1 minute and were accompanied by drooling, urination, and about 10-minute period of postictal drowsiness. The patient was previously healthy, with an unremarkable developmental history and no family history of brain tumors.
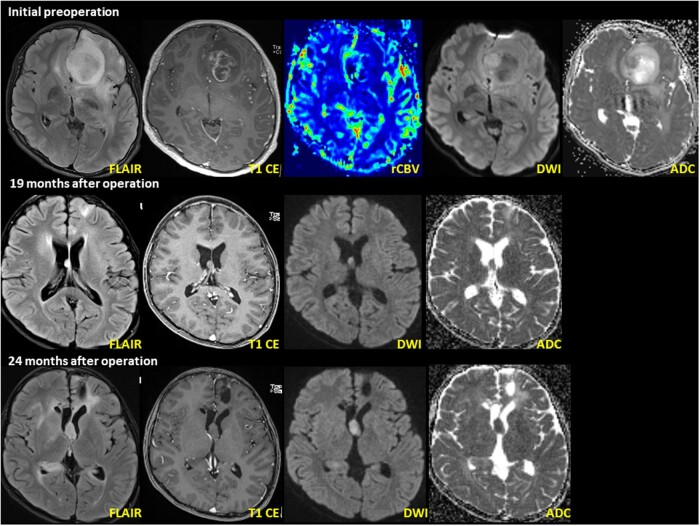
Brain magnetic resonance imaging (MRI) showed a 3.6 cm × 3.0 cm × 2.4 cm heterogeneously enhancing mass in the left frontal lobe with T2 high signal intensity, suggesting HGG (Fig. 1). Axial T2 FLAIR images showed high intensity with necrosis. Axial T1-weighted images with gadolinium contrast enhancement showed peripheral enhancement, while axial dynamic susceptibility contrast images showed increased cerebral blood volume in the solid portion of the tumor. There was also extensive peritumoral edema, left lateral ventricular compression, and midline shifting.
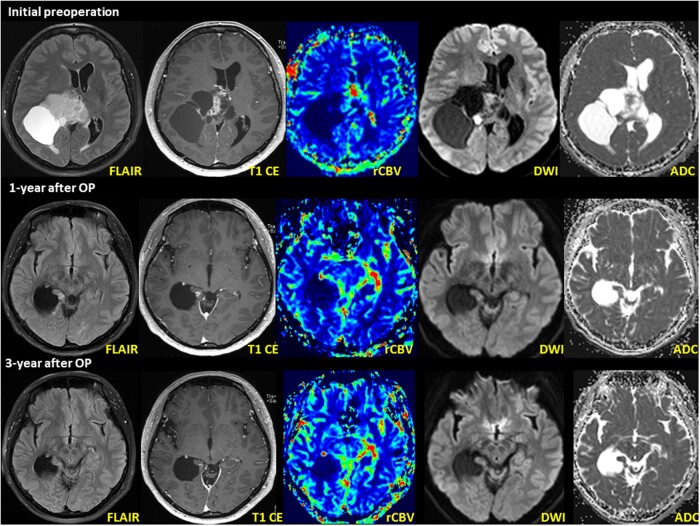
Figure 1.
DPedHGG with MYB::QKI fusion-positivity in a 11-year-old boy presenting with recurrent seizure. Initial MRI shows a well-defined, 3.6 cm × 3.0 cm × 2.4 cm (anteroposterior × transverse × craniocaudal) tumor in the left frontal lobe. The axial T2 FLAIR image shows high intensity with necrosis and restricted diffusion within the enhanced area. Follow-up MRI (pictures in the second layer) 19 months after the initial operation shows a newly developed nodule at the septum pellucidum. The nodule shows T2 hyperintensity on the axial T2 FLAIR image. A second operation was performed to resect the mass in the septum pellucidum. Postoperative MRI (pictures in the third layer) 3 months after the second operation (24 months after initial operation) shows recurrent tumors at the septum pellucidum and right lateral ventricular wall. The recurrent tumor’s imaging characteristics resemble the previous tumor at the septum pellucidum.
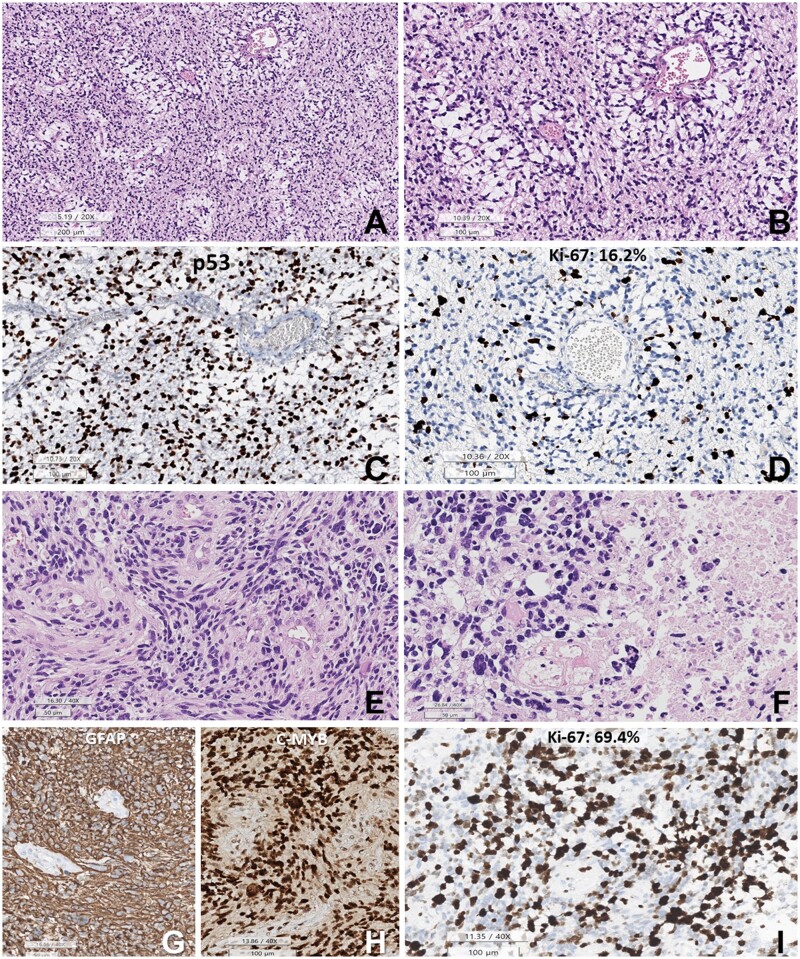
Craniotomy and subtotal tumor removal were performed. The initial tumor displayed ill-demarcated high-grade astrocytic and angiocentric patterns, increased cellularity, mild nuclear pleomorphism, microvascular proliferation, and no necrosis (Fig. 2).
Figure 2.
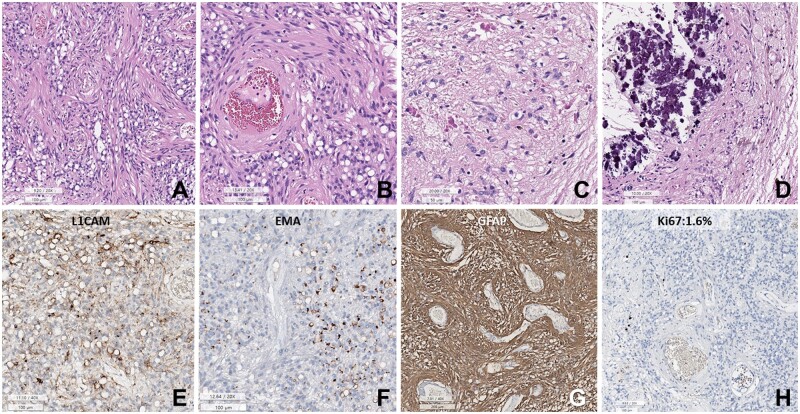
Histopathology of the initial DPedHGG, MYB::QKI fusion-positive tumor. (A, B) The tumor is composed of angiocentric astrocytic cells. The angiocentric areas are myxoid. (C) p53 is strongly positive in the tumor cell nuclei. (D) Ki-67 labeling index is 16.2%. (E) Histopathology of the recurrent tumor shows sheet-like growth of elongated glial cells with angiocentric whirling. (F) Pleomorphic nuclei of tumor cells and necrosis are observed. (G) GFAP is positive in the tumor cells (negative area: blood vessels). (H) c-MYB immunostain is positive in the tumor cell nuclei. (I) The Ki-67 (MIB-1) labeling index is high (69.4%) (A, B, E, F: HE, C: p53, D, I: Ki-67 immunohistochemistry, G: GFAP, H: c-MYB. Scale bars: A: 200 µm, B–D: 200 µm, E–G: 50 µm, H, I: 100 µm).
Immunohistochemistry (IHC) results revealed that the tumor was diffusely and robustly positive for GFAP and negative for IDH-1. The mitotic rate was 11/10 in the high-power field (HPF), assisted by pHH3. The Ki-67 labeling index was 16.2%. The tumor cells were robustly positive for p53, c-MYB, and H3K27 me3 but negative for synaptophysin, K27M, and EZHIP. There were rare EMA-positive dots in the tumor cells. The NGS study revealed a TP53 mutation (R273C) without other mutations. In addition, there was no MGMTp methylation in the MSP.
Postoperative brain MRI showed a nonenhanced, ill-defined T2 high-signal intensity lesion involving the left external capsule and coursing to the left temporal lobe along the corpus callosum genu, suggesting a residual lesion. The nodule showed T2 hyperintensity on axial T2 FLAIR images and subtle homogeneous enhancement on axial T1-weighted images with gadolinium contrast enhancement. Three weeks after the initial surgery, the patient’s brain MRI revealed a nonenhanced diffusion-restrictive lesion at the deep margin of the left frontal lobe, suggesting a recurrent or residual tumor.
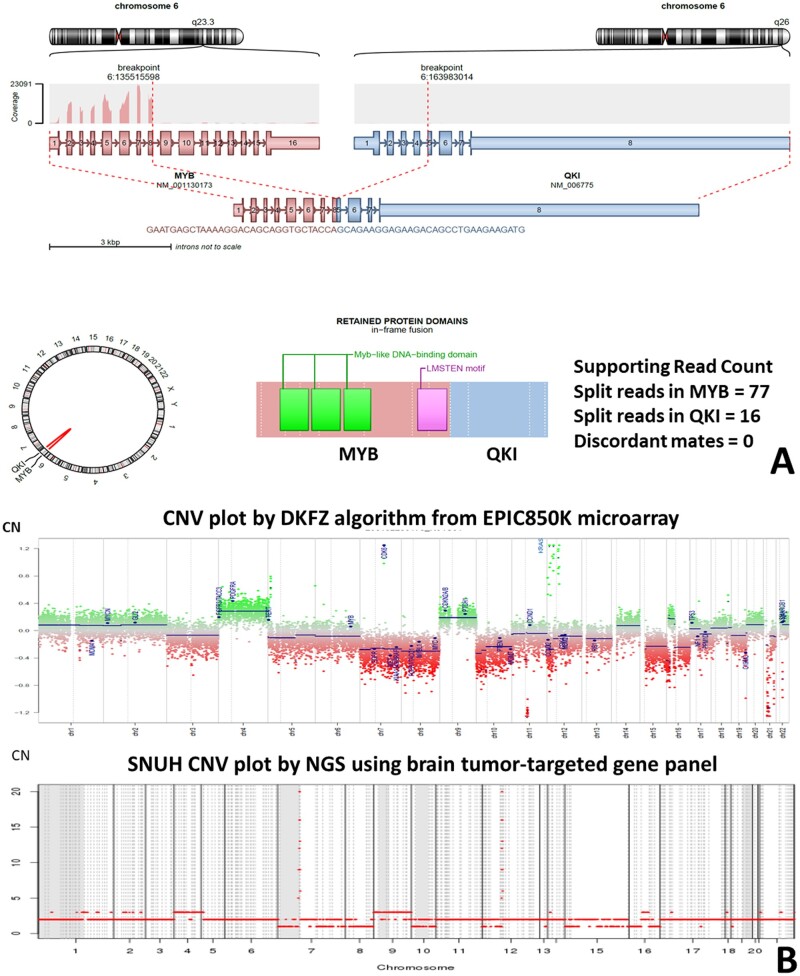
He underwent concurrent chemoradiation therapy (temozolomide + 50.4 + 10.8 Gy/34 fractionated). Brain MRI studies performed at 6, 9, and 12 months postoperatively revealed a continuous reduction in tumor size. However, MRI at 19 months postoperatively revealed a T2-hyperintense new nodule in the septum pellucidum (Fig. 1). The patient underwent a second operation 20 months after the first operation for a recurrent brain tumor. The tumor showed higher cellularity, increased nuclear polymorphism, and more microvascular proliferation than the initial tumor. However, there was no necrosis (Fig. 2). The IHC profile was similar to that of the first tumor, diffusely positive for GFAP, P53, and c-MYB. The mitotic rate and Ki-67 labeling index were significantly higher than the previous tumor, indicating 54/10 HPF (mitotic rate determination assisted by the use of pHH3) and 69.4% of the Ki-67 labeling index. NGS using the FiRST BTP revealed MYB::QKI fusion, TP53-mutation (R273C), BRIP1-mutation (L974fs), and amplification of CDK6 (copy number: 19, normal copy number: 0) and KRAS (copy number: 9) (Fig. 3). MGMTp methylation and microsatellite instability were absent.
Figure 3.
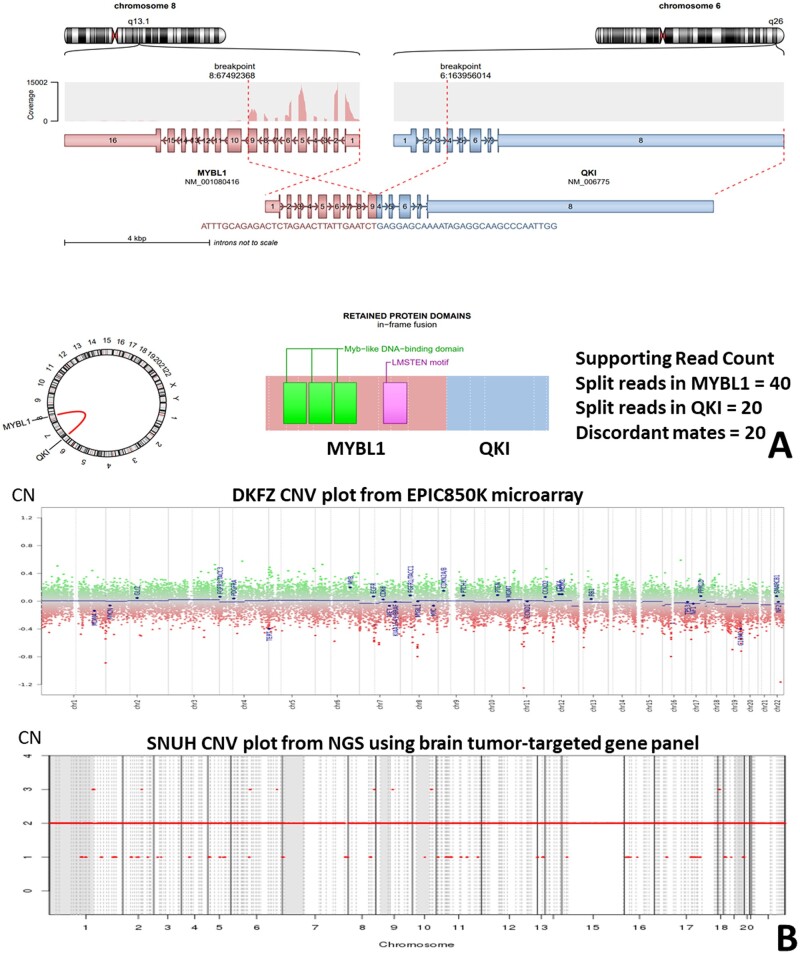
(A) Arriba plot of HGG with MYB::QKI fusion. (B) The copy number aberration of the high-grade glioma with MYB::QKI fusion. There is CDK6 and KRAS amplification. The upper plot depicts data obtained from Illumina methylationEPIC 850K array data by DKFZ CNV algorithm (normal copy number: 0). The lower plot depicts the NGS study using a customized FiRST brain tumor-targeted gene panel of Seoul National University Hospital (SNUH) (normal copy number: 2).
The Deutsches Krebsforschungzentrum (DKFZ) methylation class has been updated several times to improve the classification of brain tumors with methylation profiles. The methylation class of this tumor matched ‘glioblastoma, IDH-wildtype (matching score: 0.99920)’ with DKFZ version v11b4 MC. However, the updated DKFZ v12.5 MC matched this tumor to a ‘DpedHGG, H3-wildtype and IDH-wildtype’ (matching score: 0.68791).
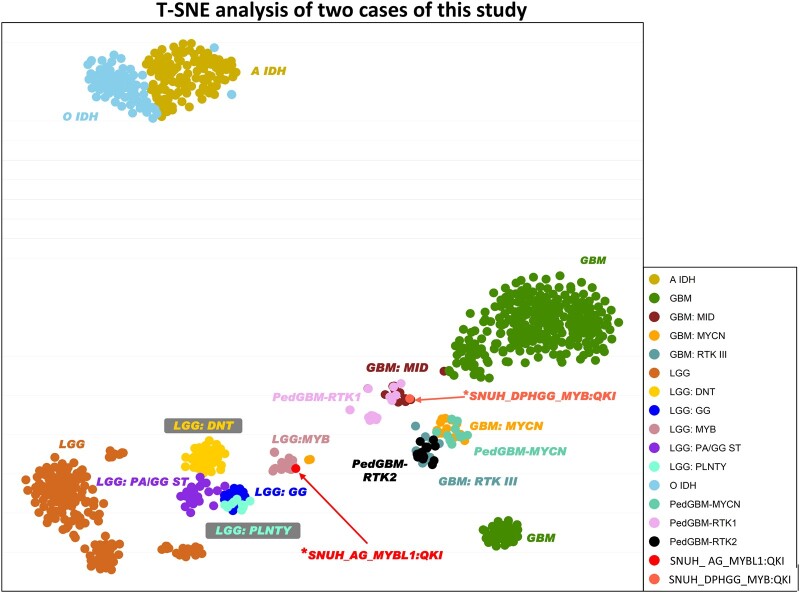
On t-SNE analysis with DNA methylation data by v11b4, this case clustered with previously reported pedGBM RTK1 and GBM-MID (Fig. 4) (14–16). We could not display t-SNE graphs in v12.5 because v12.5 methylation clustering data could not be obtained from published papers or authors.
Figure 4.
T-SNE analysis of the study cases. Using DKFZ v11b4, case 1 (*SNUH DPHGG, MYB::QKI fusion-positive) is clustered with PedGBM-RTK1 and GMB-MD. Case 2 (*SNUH_AG, MYBL1::QKI fusion-positive) is clustered with LGG-MYB.
Case 2: AG with MYB::QKI fusion-positive in an adult
A 46-year-old previously healthy woman presented with vision impairment. Brain MRI showed a large, well-demarcated, solid, and cystic mass in the right cerebral hemisphere involving the splenium of the corpus callosum and thalamus (Fig. 5). The solid portion showed internal heterogeneous enhancement, mildly increased cerebral blood volume, and focal nodular diffusion restrictions accompanied by petechial hemorrhage and calcifications, a leftward midline shift, and tight subfalcine and uncal herniations. However, there was no apparent peritumoral edema. The radiological differential diagnoses included pleomorphic xanthoastrocytoma, ganglioglioma, oligodendroglioma, and pilocytic astrocytoma. Gross total resection was performed; however, the cyst wall tumor was retained. The residual tumor was treated with radiotherapy (50.4 + 9 Gy).
Figure 5.
MRI of AG, MYBL1::QKI fusion-positive, WHO Grade 1 in a 46-year-old woman (case 2). Initial MRI shows an 8.1 cm × 6.8 cm × 7.6 cm large solid and cystic tumor in the right frontoparietal region involving the splenium of the corpus callosum and right thalamus. An MRI obtained 1-year after surgery (second row) and postoperative radiation treatment showed a new, small nodule within the right medial temporal lobe adjacent to the surgical bed margin. The nodule shows T2 hyperintensity on the axial T2 FLAIR image. MRI revealed a small, enhancing nodule that decreased in size between 2 and 3 years postoperatively (third row). The axial T1 weighted image shows less-prominent gadolinium contrast enhancement than the initial postoperative MRI.
The tumor was comprised sheets of rounded cells, with prominent cytoplasmic vacuolation and blood vessels (Fig. 6). Perivascular fascicular arrangement of slender bipolar tumor cells was seen. Microvascular proliferation and necrosis were not observed. Mitoses were not found on pHH3 IHC. The EMA was positive, with a dot- and ring-like appearance (Fig. 6). Tumor cells were diffusely positive for GFAP and focally positive for L1CAM but negative for p53. C-MYB was negative because this tumor had an MYBL1 rearrangement, not an MYB rearrangement. The Ki-67 labeling index was 1.6%.
Figure 6.
Histopathology of AG with MYBL1::QKI fusion-positive in a 46-year-old woman. (A) Sheets of small round cells with prominent perivascular slender cytoplasmic processes and numerous cytoplasmic vacuoles. (B) Spindle-shaped tumor cells are accentuated in the perivascular area. (C) The Rosenthal fibers are rich in a focal area of the tumor. (D) Massive calcifications are focally present. (E) Tumor cells are focally positive for L1CAM. (F) Dot- or ring-like positivity for EMA. (G) The tumor cells are positive for GFAP. (H) Ki-67 labeling index is 1.6% (A–F: H&E, E: L1CAM, F: EMA, G: GFAP, H: Ki-67 immunohistochemistry. Scale bars: A, B, D–F, H: 100 µm, C: 50 µm, G: 200 µm).
An NGS study using FiRST BTP revealed an MYBL1::QKI fusion with no other pathogenic alterations (Fig. 7A). The methylation class was matched to AG via DKFZ version 11b4 (score: 0.99514) and 12.5 (score: 0.99978). Copy number aberrations were not found in NGS data using the methylationEPIC 850K array data analyzed by the DKFZ algorithm and our customized brain tumor panel (Fig. 7B). The t-SNE analysis showed clustering with LGG-MYB of previously reported cases (Fig. 4) (14–16). There was no evidence of recurrence during the 52-month follow-up period.
Figure 7.
(A) Arriba plot of AG with MYBL1::QKI fusion. (B) This tumor (case 2) shows a balanced copy number. The upper plot depicts the methylationEPIC 850K microarray data obtained by the DKFZ CNV algorithm (normal copy number: 0). The lower plot depicts the results of the NGS study using a customized FiRST brain tumor-targeted SNUH gene panel (normal copy number: 2).
DISCUSSION
Only 6 papers on MYB/MYBL1 fusion-positive AG have been published in the English literature to date (Table) (1, 2, 7, 8, 17–19). AG is a rare tumor that commonly occurs in children and young adults (median age: 15–17 years, range: 2–46 years) (Table) (5, 20); 72% were younger than 11 and 90% were younger than 21 years. Unusually, the MYBL1::QKI fusion-positive AG in this study developed in a 46-year-old woman. No apparent sexual predilection has been reported; however, confirmed cases are disproportionately male predominant (2:1). AGs occur in the supratentorial cortex and frequently involve leptomeninges (5); less commonly, however; they are found in the thalamus and brainstem (8, 17). The clinical manifestations reflect the areas of involvement. Patients mostly present with seizures; however, neurological deficits, such as cranial nerve palsy or developmental delay, have also been reported in patients with brainstem involvement (7, 8, 17, 20). Case 2 in our study presented with visual impairment.
TABLE.
Epidemiology of the previously reported genetically confirmed angiocentric gliomas
| References | No. | Age | Sex | Clinical features | DX | Site | Genetics | Op and adjuvant Tx | Outcome |
|---|---|---|---|---|---|---|---|---|---|
| Zhang et al, 2013 (1) | 1 | 3–8 | M | NM | GG | Cerebral cortex | MYB rearrangement | NM | NM |
| 2 | >8 | M | NM | AG | Cerebral cortex | MYB rearrangement | NM | NM | |
| Ramkissoon et al, 2013 (19) | 3 | <2 | F | NM | AG | NM | focal 6q23.3/MYB del | NM | NM |
| 4 | 2–10 | F | NM | AG | NM | focal 6q23.3/MYB del | NM | NM | |
| 5 | 2–10 | M | NM | AG | NM | focal 6q23.3/MYB del | NM | NM | |
| Qaddoumi et al, 2016 (18) | 6 | 3 | M | NM | AG | Parietal | MYB::QKI fusion | NM | NM |
| 7 | 11 | M | NM | AG | Parietal | MYB::QKI fusion | NM | NM | |
| 8 | 5 | F | NM | AG | Frontal | MYB::QKI fusion | NM | NM | |
| 9 | 11 | M | NM | AG | Temporal | MYB::QKI fusion | NM | NM | |
| 10 | 37 | M | NM | AG | Temporal | MYB::QKI+BRAF V600E | NM | NM | |
| 11 | 15 | F | NM | AG | Temporal | MYB::QKI fusion | NM | NM | |
| 12 | 3 | M | NM | AG | Frontoparietal | MYB::QKI fusion | NM | NM | |
| 13 | 5 | M | NM | AG | Temporal | MYB::QKI fusion | NM | NM | |
| 14 | 7 | M | NM | AG | Parietal | MYB::QKI fusion | NM | NM | |
| 15 | 10 | F | NM | AG | Temporal | MYB::QKI fusion | NM | NM | |
| 16 | 17 | M | NM | AG | Occipital | MYB::QKI+BRAF V600E | NM | NM | |
| 17 | 19 | F | NM | AG | Frontal | MYB::QKI fusion | NM | NM | |
| 18 | 41 | F | NM | AG | Temporal | MYB::QKI fusion | NM | NM | |
| 19 | 7 | M | NM | AG | Temporal | MYB::QKI fusion | NM | NM | |
| 20 | 3 | F | NM | AG | Temporal | QKI | NM | NM | |
| Bandopadhayay et al, 2016 (2) | 21 | NM | NM | NM | AG | NM | MYB::QKI fusion | NM | NM |
| 22 | NM | NM | NM | AG | NM | MYB::QKI fusion | NM | NM | |
| 23 | NM | NM | NM | AG | NM | MYB::QKI fusion | NM | NM | |
| 24 | NM | NM | NM | AG | NM | MYB::QKI fusion | NM | NM | |
| 25 | NM | NM | NM | AG | NM | MYB::QKI fusion | NM | NM | |
| 26 | NM | NM | NM | AG | NM | MYB::QKI fusion | NM | NM | |
| 27 | NM | NM | NM | AG | NM | Other MYB mutation | NM | NM | |
| Chan et al, 2017 (7) | 28 | 7 | M | Sixth nerve palsy | AG | Inferior Pons | MYB::QKI fusion | Biopsy only | NM |
| D’Aronco et al, 2017 (8) | 29 | 7 | M | Developmental delay | AG | Pons and medulla | MYB::QKI fusion | Biopsy only, Unresectable, Carboplatin, and Vincristine+Bevacizumab | Initially progressed but stable size in 12 months |
| 30 | 3 | F | Seventh nerve palsy | AG | Brainstem | MYB::QKI fusion | Biopsy only, Unresectable, carboplatin, and vincristine+Bevacizumab, mTOR inhibitor | Initially progressed but stable in 4 years | |
| Lake et al, 2020 (17) | 31 | 5 | M | NM | AG | Thalamus | MYB::QKI fusion | Radiotherapy | NM |
| 32 | 2 | M | NF1 patient | AG | Lt frontal | MYB::QKI fusion | Chemotherapy | NM | |
| Suh et al, Present case | 33 | 11 | M | Tonic seizure | HGG | Lt frontal | MYB::QKI fusion | CCRT+trial | One recurs 20 months after surgery and no further recurs 40 months after the initial surgery |
| Suh et al, Present case | 34 | 46 | F | Visual impairment | AG | Rt hemisphere CC and Thalamus | MYBL1::QKI fusion | GTR only | No recur for 52 months F-U |
GG, gangliogloma; AG, angiocentric glioma, CNS WHO grade 1; NM, not mentioned; CC, corpus callosum; CCRT, concurrent chemo-radiotherapy; GTR, gross total resection; F-U, follow-up.
AGs have characteristic histopathologic features, appearing as angiogenic astrocytic tumors composed of bipolar fibrillary cells with slender and elongated nuclei and cytoplasm. The spindling effect is more pronounced in perivascular areas. As with case 2 of this study, caution is needed, given that spindling can be confused with the anuclear fibrillary zone of the perivascular pseudorosettes observed in ependymomas. The mitotic rate and Ki-67 labeling index have been low in reported cases. These tumors were diffusely positive for GFAP and S-100 and dot- or ring-like EMA positivity. EMA positivity is similar to ependymoma, which may cause confusion between the 2 tumor types (6).
The MYB/MYBL1 rearrangement is a defining genetic event in AG, which usually carries no further pathogenic mutations (2, 7, 18). According to Qaddoumi et al. (18), all AGs harbored an MYB::QKI fusion (13/15; 87%), MYB::ESR1 fusion (1/15), or QKI rearrangement (1/15). However, 2 AGs with an MYB::QKI fusion also harbored a BRAF p.V600E mutation (18). The QKI rearranged case requires verification as it was studied with FISH and only split signals of the QKI gene could be detected using FISH break-apart probes without apparent involvement of MYB or MYBL1. Therefore, the authors could not completely rule out a cryptic MYB::QKI aberration or QKI fusion with another gene partner. Given the results of-our 2 cases, gene fusion may not be the only driver of AGs.
MYB encodes a transcription factor and may be a protooncogene that may be activated into an oncogene (21). It contains a negative regulatory domain at the C-terminus of the MYB gene. QKI encodes Quaking, an RNA-binding protein expressed in the nervous system. Moreover, it is considered a tumor suppressor gene (21).
The MYB::QKI fusion is an intrachromosomal rearrangement in chromosome 6 that is considered oncogenic. The MYB::QKI fusion loci (78%) were mainly between MYB exon 15 and QKI exon 5, and rarely between MYB exon 9 and QKI exon 5 (18). MYB is a protooncogene with increased expression in LGG and HGGs (22). These fusions promote tumor cell proliferation in vitro and formed HGG in vivo studies (2). The oncogenic function of defective MYBL1 was confirmed by in vitro studies, where truncating MYBL1 transformed NIH3T3 cells which formed malignant tumors; however, wildtype MYBL1 transfected cells did not (19). These fusion proteins functioned as transcription factors and drove tumorigenesis via multiple mechanisms (2). There are 3 hypothetical mechanisms of oncogenesis: MYB or MYBL1 activation by truncation of the negative regulatory domain in the C-terminus, disruption of the tumor suppressor QKI, or enhancer translocations that drive constitutively active MYB::QKI or MYBL1::QKI expression (2).
MYB/MYL1 alterations have not been reported in HGG (1–3, 7, 8, 17–19, 21). In this study, MYB::QKI fusion-positive HGG harbored CDK6 and KRAS amplifications, and TP53 and BRIP1 mutation. KIT and CDK6 were reportedly associated with MYB activation (2). However, the HGG in this study demonstrated gene-level amplification of CDK6 and KRAS. These additional mutations appear to cause tumors with MYB/MYBL1 alterations to act more aggressively from a histopathological and biological standpoint. Case 1 showed marked nuclear pleomorphism, high mitotic rate, microvascular proliferation, and necrosis with a high Ki-67 index.
In most cases of AG, the prognosis has been good with surgical resection alone. However, in some clinically aggressive cases, the tumor may enlarge to where resection is impossible due to the location (8). Two boys (5 and 2 years old) with thalamic and brainstem MYB::QKI fusion-positive AGs presented with neurological deficits (Table). The former underwent subtotal tumor resection with radiation therapy and multiple kinase inhibitors. In contrast, the latter underwent gross total resection with chemotherapy. Despite adjuvant chemoradiation therapy, continuous tumor progression was observed at the original or distant spinal sites.
CONCLUSION
The novel findings of this report are (1) the age of the patient with the AG (46 years old) with MYBL1::QKI fusion and (2) the discovery of the MYB::QKI fusion in a HGG. Both tumors demonstrated angiocentric histopathologic features. The MYB-altered HGG exhibited additional mutations, including a TP53 and BRIP1 mutation and amplification of CDK6 and KRAS. These observations confirmed that MYB::QKI or MYBL1::QKI fusions were not limited to LGG, supratentorial loci, and children. Instead, the tumors with these fusions can have additional genomic events, potentially leading to HGG, and can occur anywhere within the brains and brainstem of children and adults. We suggest that molecular findings, morphologic characteristics, and clinical information must be combined for accurate diagnosis in the molecular era.
Contributor Information
Ye Yoon Suh, Department of Pathology, Seoul National University College of Medicine, Seoul, Republic of Korea.
Kwanghoon Lee, Department of Pathology, Seoul National University College of Medicine, Seoul, Republic of Korea.
Yu-Mi Shim, Department of Pathology, Seoul National University College of Medicine, Seoul, Republic of Korea.
Ji Hoon Phi, Department of Neurosurgery, Seoul National University College of Medicine, Seoul, Republic of Korea.
Chul-Kee Park, Department of Neurosurgery, Seoul National University College of Medicine, Seoul, Republic of Korea.
Seung-Ki Kim, Department of Neurosurgery, Seoul National University College of Medicine, Seoul, Republic of Korea.
Seung Hong Choi, Department of Radiology, Seoul National University College of Medicine, Seoul, Republic of Korea.
Hongseok Yun, Department of Genomic Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea.
Sung-Hye Park, Department of Pathology, Seoul National University College of Medicine, Seoul, Republic of Korea; N euroscience Research Institute, Seoul National University College of Medicine, Seoul, Republic of Korea.
FUNDING
This study was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute, funded by the Ministry of Health & Welfare, Republic of Korea (Grant No. HI14C1277); and the Institute for Information & Communications Technology Promotion grant funded by the Korean government (MSIP) (No. 2019-0567).
CONFLICT OF INTEREST
The authors have no duality or conflicts of interest to declare.
DATA AVAILABILITY
The corresponding author’s datasets for this study are available on reasonable request.
REFERENCES
- 1. Zhang J, Wu G, Miller CP, et al. ; St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet 2013;45:602–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bandopadhayay P, Ramkissoon LA, Jain P, et al. MYB-QKI rearrangements in angiocentric glioma drive tumorigenicity through a tripartite mechanism. Nat Genet 2016;48:273–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Roth JJ, Santi M, Rorke-Adams LB, et al. Diagnostic application of high resolution single nucleotide polymorphism array analysis for children with brain tumors. Cancer Genet 2014;207:111–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wefers AK, Stichel D, Schrimpf D, et al. Isomorphic diffuse glioma is a morphologically and molecularly distinct tumour entity with recurrent gene fusions of MYBL1 or MYB and a benign disease course. Acta Neuropathol 2020;139:193–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lian F, Wang LM, Qi XL, et al. MYB-QKI rearrangement in angiocentric glioma. Clin Neuropathol 2020;39:263–70 [DOI] [PubMed] [Google Scholar]
- 6. Lum DJ, Halliday W, Watson M, et al. Cortical ependymoma or monomorphous angiocentric glioma? Neuropathology 2008;28:81–6 [DOI] [PubMed] [Google Scholar]
- 7. Chan E, Bollen AW, Sirohi D, et al. Angiocentric glioma with MYB-QKI fusion located in the brainstem, rather than cerebral cortex. Acta Neuropathol 2017;134:671–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. D'Aronco L, Rouleau C, Gayden T, et al. Brainstem angiocentric gliomas with MYB-QKI rearrangements. Acta Neuropathol 2017;134:667–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Louis DN, Perry A, Wesseling P, et al. The 2021 WHO classification of tumors of the central nervous system: A summary. Neuro Oncol 2021;23:1231–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim H, Lim KY, Park JW, et al. Sporadic and Lynch syndrome-associated mismatch repair-deficient brain tumors. Lab Invest 2022;102:160–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Benjamin D, Sato T, Cibulskis K, et al. Calling somatic snvs and indels with mutect2. BioRxiv 2019;861054
- 12. Karczewski KJ, Francioli LC, Tiao G, et al. ; Genome Aggregation Database Consortium. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020;581:434–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Capper D, Jones DTW, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature 2018;555:469–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huse JT, Snuderl M, Jones DT, et al. Polymorphous low-grade neuroepithelial tumor of the young (PLNTY): An epileptogenic neoplasm with oligodendroglioma-like components, aberrant CD34 expression, and genetic alterations involving the MAP kinase pathway. Acta Neuropathol 2017;133:417–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Korshunov A, Schrimpf D, Ryzhova M, et al. H3-/IDH-wild type pediatric glioblastoma is comprised of molecularly and prognostically distinct subtypes with associated oncogenic drivers. Acta Neuropathol 2017;134:507–16 [DOI] [PubMed] [Google Scholar]
- 17. Lake JA, Donson AM, Prince E, et al. Targeted fusion analysis can aid in the classification and treatment of pediatric glioma, ependymoma, and glioneuronal tumors. Pediatr Blood Cancer 2020;67:e28028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Qaddoumi I, Orisme W, Wen J, et al. Genetic alterations in uncommon low-grade neuroepithelial tumors: BRAF, FGFR1, and MYB mutations occur at high frequency and align with morphology. Acta Neuropathol 2016;131:833–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ramkissoon LA, Horowitz PM, Craig JM, et al. Genomic analysis of diffuse pediatric low-grade gliomas identifies recurrent oncogenic truncating rearrangements in the transcription factor MYBL1. Proc Natl Acad Sci USA 2013;110:8188–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fuller LD, Prayson RA. Molecular immunohistochemical profile of angiocentric glioma. J Epilepsy Res 2020;10:79–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jain P, Resnick AC. MYB-QKI drives childhood brain tumors via tripartite mechanism. Cell Cycle 2017;16:390–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tatevossian RG, Tang B, Dalton J, et al. MYB upregulation and genetic aberrations in a subset of pediatric low-grade gliomas. Acta Neuropathol 2010;120:731–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The corresponding author’s datasets for this study are available on reasonable request.