Abstract
Inflammatory bowel disease (IBD) is an immune-mediated chronic intestinal disorder with major phenotypes: ulcerative colitis (UC) and Crohn’s disease (CD). Multiple studies have identified over 240 IBD susceptibility loci. However, most studies have centered on European (EUR) and East Asian (EAS) populations. The prevalence of IBD in non-EUR, including African Americans (AAs), has risen in recent years. Here we present the first attempt to identify loci in AAs using a trans-ancestry Bayesian approach (MANTRA) accounting for heterogeneity between diverse ancestries while allowing for the similarity between closely related populations. We meta-analyzed genome-wide association studies (GWAS) and Immunochip data from a 2015 EUR meta-analysis of 38 155 IBD cases and 48 485 controls and EAS Immunochip study of 2824 IBD cases and 3719 controls, and our recent AA IBD GWAS of 2345 cases and 5002 controls. Across the major IBD phenotypes, we found significant evidence for 92% of 205 loci lead SNPs from the 2015 meta-analysis, but also for three IBD loci only established in latter studies. We detected 20 novel loci, all containing immunity-related genes or genes with other evidence for IBD or immune-mediated disease relevance: PLEKHG5;TNFSFR25 (encoding death receptor 3, receptor for TNFSF15 gene product TL1A), XKR6, ELMO1, BC021024;PI4KB;PSMD4 and APLP1 for IBD; AUTS2, XKR6, OSER1, TET2;AK094561, BCAP29 and APLP1 for CD; and GABBR1;MOG, DQ570892, SPDEF;ILRUN, SMARCE1;CCR7;KRT222;KRT24;KRT25, ANKS1A;TCP11, IL7, LRRC18;WDFY4, XKR6 and TNFSF4 for UC. Our study highlights the value of combining low-powered genomic studies from understudied populations of diverse ancestral backgrounds together with a high-powered study to enable novel locus discovery, including potentially important therapeutic IBD gene targets.
Introduction
Inflammatory bowel disease (IBD) is a chronic intestinal disorder of the gastrointestinal tract with two major and genetically related phenotypes, ulcerative colitis (UC) and Crohn’s disease (CD). UC entails continuous inflammation restricted to mucosal layers of the rectum and colon. In contrast, CD involves transmural, discontinuous inflammation primarily of the small or large intestine but can affect any portion of the gastrointestinal tract. IBD pathogenesis is incompletely understood but has been attributed to dysregulated intestinal immunity, especially in response to intestinal microbiota, and primarily in genetically susceptible individuals.
Studies have identified over 240 IBD genetic susceptibility loci in subjects of European ancestry (EUR), primarily via genome-wide association studies (GWAS) and, in particular, GWAS meta-analyses (1–8). Approximately 35 IBD loci have also been established in East Asian (EAS) populations via a handful of individual GWAS and several focused studies to replicate loci identified in whites (5,9–12). A few loci appear Asian specific (13). We performed a GWAS on African American (AA) subjects with IBD and detected significant replication evidence for 13 loci established in EUR, including NOD2 and PTGER4 (2). We also detected at genome-wide significance universal risk alleles at established loci at HLA-DRB1 for UC and near USP25 for IBD, as well as novel African-specific single-nucleotide polymorphism (SNPs) for UC at ZNF649 and LSAMP and African-specific variants with locus-specific replication at five additional loci (P < 1.5 × 10−6).
A major objective of our 2017 IBD GWAS in AAs was to identify novel loci, given that the 80% admixed West African ancestral genome of AAs is known to have a higher concentration of unique SNPs as well as lower linkage disequilibrium. However, our GWAS was limited in power by modest sample size. In the present study, the primary purpose is to identify novel loci for IBD, loci not observed whole-genome significant in any prior study, via the increased power from a meta-analysis of GWAS data from multiple ethnicities made possible by the Bayesian partition model using the software MANTRA. This Bayesian approach takes advantage of existing differences in LD architecture between the study populations to better detect the association signal at the causal variant. Compared with traditional meta-analysis methods, MANTRA produces significant improvements in performance by considering the degree of relatedness between ancestries.
Results
We first compared SNPs with log10 Bayes’ factor (log10BF) ≥ 6.0 detected in our three-population meta-analysis for replication of the 200 loci defined as significant in the Liu et al. (5) parent study. We found significant association evidence (log10BF ≥ 6.0), for SNPs at 177 of these 200 loci (88.5%) with requiring the SNPs to be within 250 Kb of a lead Liu et al. significant SNP (per their definition of locus width). Among 205 of the 232 lead Liu et al. SNPs (10 excluded from analysis from homologous pairings, and the three NOD2 mutations were excluded with only EUR data as well as 15 additional SNPs without genotypes in EAS or AA data sets), 189 (92%) likewise met significance in our meta-analysis. Interestingly, among 24 lead SNPs that the Liu et al. MANTRA meta-analysis reported no variants with log10BF ≥6.0 in the three phenotypes tested (and all being the sole SNP for their locus that met criteria for significance), in our meta-analysis 10 SNPs either showed log10BF ≥6.0 (four SNPs) or we detected an SNP within 100 Kb (142–72 238 bp from the Liu et al. SNP) that met significance.
We then examined if our analysis detected significant loci at the 25 novel IBD loci detected in the de Lange et al. (14) British GWAS meta-analysis, which combined an additional 12 160 EUR IBD cases and 13 145 population controls. We detected three loci (at SLC19A3-CCL20, AKAP11/TNFSF11 and NCF4) with one or more significant SNPs, log10BF ≥ 6.0, within 10 Kb of a novel de Lange et al. locus lead SNP.
Among the three major IBD phenotypic classes (CD, UC and all IBD), we detected 20 novel loci with log10BF ≥ 6.0 (Table 2). Association results for the AA–EUR–EAS trans-ancestry meta-analyses for all SNPs with log10BF ≥ 6 are found in Supplementary Material, Tables S1–S3 for IBD, CD and UC, respectively. SupplementaryMaterial, Tables S4–S6 contain the AA–EUR GWAS trans-ancestry meta-analyses results for IBD, CD and UC. Our results for the lead Liu et al. and de Lange et al. SNPs we observed as significant are noted in Supplementary Material, Tables S8 and S9.
Table 2.
Top novel associations for IBD, CD and UC
| Phenotype | Leading SNP | Chr | Position | Annotation | A1 | A2 | Gene | log10BF | PPAHet |
Total
sample size |
Direction |
Effect
allele Freq. AA |
Effect
allele Freq. EUR |
Effect
allele Freq. EAS |
PMAE-AA | PMAE-EUR | PMAE-EAS |
Number of
significant SNPs in locus |
Significant SNPs in
locus |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IBD | rs2986751 | 1 | 6 534 781 | UTR5 | A | G |
PLEKHG5; TNFSFR25 |
6.21 | 0.07 | 41 999 | − | 0.291 | 0.065 | -0.174 | -0.171 | 1 | |||
| IBD | rs3811406 | 1 | 151 254 041 | ncRNA_ Exonic |
A | G |
BC021024; PI4KB; PSMD4 |
8.06 | 0.073 | 100 530 | − | 0.200 | 0.230 | 0.370 | -0.072 | -0.071 | -0.070 | 2 | rs3811406*, rs2031797 |
| IBD | rs12532822 | 7 | 37 393 177 | UTR5 | A | C | ELMO1 | 8.24 | 1 | 100 530 | − + − | 0.053 | 0.171 | 0.048 | -0.011 | 0.049 | -0.473 | 1 | |
| IBD | rs79315643 | 8 | 10 773 414 | Intronic | A | G | XKR6 | 23.38 | 1 | 100 530 | +++ | 0.869 | 0.780 | 0.905 | 0.056 | 0.012 | 0.732 | 1 | |
| IBD | rs230261 | 19 | 36 363 470 | Exonic | A | G | APLP1 | 6.14 | 0.186 | 100 530 | ++− | 0.115 | 0.030 | 0.002 | 0.138 | 0.141 | 0.123 | 1 | |
| CD | rs974801 | 4 | 106 071 064 | Intronic | A | G |
TET2; AK094561 |
7.73 | 0.074 | 74 249 | +++ | 0.707 | 0.620 | 0.385 | 0.069 | 0.069 | 0.068 | 3 | rs17035289, rs974801 rs10010325 |
| CD | rs2293503 | 7 | 70 250 125 | Intronic | T | C | AUTS2 | 6.05 | 1 | 68 840 | +− | 0.463 | 0.439 | 0.187 | -0.090 | 1 | |||
| CD | rs3801944 | 7 | 107 255 548 | Intronic | A | G | BCAP29 | 7.05 | 0.074 | 74 249 | +++ | 0.256 | 0.280 | 0.289 | 0.073 | 0.072 | 0.073 | 3 | rs3801944*, rs10273733, rs2808 |
| CD | rs79315643 | 8 | 10 773 414 | Intronic | A | G | XKR6 | 18.50 | 1 | 74 249 | +++ | 0.869 | 0.780 | 0.905 | 0.123 | 0.010 | 0.811 | 1 | |
| CD | rs230261 | 19 | 36 363 470 | Exonic | A | G | APLP1 | 6.13 | 0.211 | 74 249 | ++− | 0.115 | 0.030 | 0.002 | 0.164 | 0.169 | 0.152 | 1 | |
| CD | rs2143606 | 20 | 42 838 550 | Intronic | A | G | OSER1 | 8.80 | 0.066 | 74 249 | +++ | 0.274 | 0.531 | 0.436 | 0.076 | 0.076 | 0.076 | 1 | |
| UC | rs10465507 | 1 | 173 162 439 | Intronic | A | C | TNFSF4 | 6.18 | 1 | 75 264 | − + − | 0.496 | 0.245 | 0.050 | 0.000 | 0.001 | -0.770 | 1 | |
| UC | rs115484865 | 6 | 29 604 124 | Intergenic | A | G |
GABBR1; MOG |
6.51 | 0.19 | 33 017 | −+ | 0.115 | 0.206 | 0.132 | 0.159 | 1 | |||
| UC | rs16869677 | 6 | 33 876 082 | ncRNA_ Exonic |
A | G | DQ570892 | 11.59 | 1 | 75 264 | + − + | 0.095 | 0.081 | 0.141 | 0.046 | -0.001 | 0.470 | 1 | |
| UC | rs73407795 | 6 | 34 518 271 | Intronic | A | G |
SPDEF; ILRUN |
8.04 | 1 | 75 264 | +++ | 0.141 | 0.064 | 0.061 | 0.053 | 0.051 | 0.526 | 1 | |
| UC | rs3822921 | 6 | 35 057 331 | UTR3 | A | G |
ANKS1A; TCP11; AY927475 |
7.93 | 1 | 75 264 | - − + | 0.024 | 0.127 | 0.058 | -0.038 | -0.030 | 0.539 | 2 | rs3822921*, rs11755266 |
| UC | rs79315643 | 8 | 10 773 414 | Intronic | A | G | XKR6 | 7.23 | 1 | 75 264 | + − + | 0.865 | 0.780 | 0.905 | 0.003 | -0.003 | 0.572 | 1 | |
| UC | rs72661359 | 8 | 79 677 725 | Intronic | A | C | IL7 | 7.88 | 0.993 | 75 264 | −++ | 0.071 | 0.058 | 0.000 | 0.110 | 0.133 | 2.733 | 1 | |
| UC | rs2940716 | 10 | 50 124 724 | Intronic | A | G |
LRRC18; WDFY4 |
7.61 | 1 | 75 264 | −++ | 0.277 | 0.269 | 0.000 | 0.005 | 0.006 | 2.934 | 1 | |
| UC | rs9911533 | 17 | 38 775 476 | Intergenic | A | G |
CCR7; SMARCE1; KRT222; KRT24; KRT25 |
7.98 | 0.099 | 75 264 | +++ | 0.783 | 0.616 | 0.723 | 0.080 | 0.078 | 0.077 | 11 | rs7221109, rs757411, rs9911533*, rs1013971, rs4890093, rs9906785, rs7217237, rs2315020, rs2159430, rs2462963, rs726848 |
Notes: Trans-ancestry association analysis results for all novel SNPs are shown in this table for IBD, CD and UC. The significance threshold for the trans-ancestry meta-analysis was set at log10BF ≥ 6. Summary statistics of GWAS and immunochip data sets were analyzed using MANTRA to identify novel risk loci for each of the three phenotypes. Phenotype, traits of interest (IBD, CD, UC); SNP, single-nucleotide polymorphism; Chr, chromosome; Position, base pair position (hg19); A1, effect allele; A2, other allele; Annotation, the functional classification of the variant; Genes, names of most proximal candidate genes ; log10BF, log 10 Bayes factor in favor of association, PPAHet, posterior probability of association showing evidence of heterogeneity; Total sample size, lists the sample sizes used in the analysis per trait; Effect direction, denotes direction of effect with + for positive allelic effect for effect allele, and − for negative allelic effect for effect allele; Effect Allele Freq., allele frequency of the effect allele for the population; PMAE-AA, posterior mean allelic effect for AA cohort; PMAE-EUR, posterior mean allelic effect for EUR cohort; PMAE-EAS, posterior mean allelic effect for EA cohort; Number of significant SNPs in locus, shows the number of SNPs that met the threshold for significance (log10BF ≥ 6); Significant SNPs in locus, lists the SNPs that met the threshold for significance (log10BF ≥ 6), * denotes the SNP that had the highest Bayes factor in the locus.
Inflammatory Bowel Disease phenotype
We detected the strongest signal in our study (log10BF 23.38) at 8p23.1 within an intronic variant at Kell blood group complex subunit-related family, Member 6 (XKR6). This locus on chromosome (Chr) 8 was detected in all three phenotypes (CD, UC and IBD). The SNP rs79315643 is at least 16 Mb away from any established IBD locus. The direction of effect was the same in all three populations. However, the size of the effects was heterogenous, with the strongest posterior mean allelic effect in the EAS cohort. In addition to XKR6, four other loci reached the evidence threshold, located proximal to genes BC021024;PI4KB;PSMD4, engulfment and cell motility protein 1 (ELMO1) and APLP1 in the IBD phenotype analysis. Among these, SNP rs12532822, located in the 5′ UTR of ELMO1, showed the most substantial evidence (log10BF 8.24) with similar evidence observed for SNP rs3811406 (log10BF 8.06) that maps to non-coding RNA BC021024. The signals in BC021024;PI4KB;PSMD4 (both rs3811406 and PI4KB SNP rs2031797, log10BF 7.63), and APLP1 showed homogenous effects across the three populations, whereas ELMO1, like XKR6, showed heterogeneous effects. The AA versus EUR GWAS trans-ancestry analysis for IBD showed a significant signal at rs2986751 (log10BF 6.21). This SNP is a 5′ UTR variant in PLEKHG5 and <9 Kb from TNFSFR25. Analysis of whole-blood and Genotype-Tissue Expression (GTEx) expression quantitative loci analysis (eQTL) databases shows that rs2986751 is a cis-eQTL for TNFSFR25 (P = 2.28 × 10−46).
Crohn’s disease
We identified nine novel SNP associations for CD (Table 2). Similar to that observed for IBD overall, the strongest novel CD signal (log10BF 18.50) was at rs79315643 (XKR6). SNP rs230261 (APLP1) likewise showed evidence in both IBD and CD (log10BF 6.13). SNP rs230261 at 19q13.12 is not in LD with the lead SNP rs587259 (log10BF 6.81 near LSM14A) in the known CD locus 1.7 Mb away (15). We also found three novel SNPs rs3801944, rs10273733, and rs2808 in Chr 7 near BCAP29 (log10BF 6.41–7.05) associated with CD. All three SNPs showed evidence of homogeneity across the three studied populations. Although our CD-associated SNPs in BCAP29 are proximal to a known risk locus for UC (SLC26A3; DLD), this UC locus has not, to date, been implicated in CD (3,14). Additionally, SNPs we observed significant in UC at this known UC locus 189 Kb from rs2808 (e.g. rs78058114; log10BF 19.56 in UC) showed no evidence for CD, suggesting that BCAP29 may be an entirely separate and novel IBD locus. We also report three SNPs mapping to an intronic region near ten-eleven translocation enzyme 2 (TET2). In the GWAS trans-ancestry meta-analysis between AA and EUR, we found a novel signal at rs2293503 (log10BF 6.05). The SNP at 7q11.22 is an intronic variant within autism susceptibility candidate 2 (AUTS2).
Ulcerative colitis
In the UC analysis, we found 20 novel SNP associations that reached our threshold for significance (Table 2). The most significant novel SNP association for the UC analysis is rs16869677 (log10BF 11.59) mapping to DQ570892 (in hg38 referred to as RP3-468B3.2) and located just outside the human leukocyte antigen (HLA) region. We also detected three SNP associations (log10BF 7.55–8.04) at 6p21.31 near genes SPDEF, ANKS1A and TCP11. Given the proximity of these SNPs to the HLA region, we checked for independence and did not find any significant LD between our SNPs compared with the known HLA associations in the EUR, AA and EAS cohorts. Eleven SNPs spanning a 100 Kb region in 17q21.2 (log10BF 6.12–7.62) mapped to a region near genes CCR7;SMARCE1;KRT24. We also report the first evidence of associations near TNFSF4 (log10BF 6.18) on Chr 1 and near genes LRRC18 and WDFY4 (log10BF 7.61) on Chr 10. Finally, we found SNP associations in Chr 8 at rs72661359 (interleukin 7 [IL7]) and rs79315643 (XKR6). The UC novel associations identified on Chr 6, 8, and 10 all depict strong evidence of deviation from homogeneity in allelic effects across the three populations. Their posterior mean allelic effects indicate that the associations may be more specific to the EAS ancestry. In contrast, the variants on Chr 17 appear to show homogeneity across the three ethnicities. In the UC GWAS trans-ancestry meta-analysis of AA and EUR, we found a novel and homogeneous signal at rs115484865 (log10BF 6.51) also on Chr 6. This SNP is an intergenic variant between GABBR1 and MOG.
Discussion
This study is the first meta-analysis in IBD to combine an AA GWAS data set with genome-wide data sets of other ethnicities, notably a EUR ancestry GWAS data set and an EAS ancestry Immunochip data set. We evaluated genotypes from 100 530 individuals, 43 324 IBD cases and 57 206 controls. The ancestry proportions within the total data set were 86.2% EUR, 6.5% EAS and 7.3% AA. In total, we detected 20 novel loci that met the criteria of log10BF ≥ 6.0 for genome-wide significance. Importantly, the value of combining the GWAS genotype data of the 7347 AA study subjects with the 93 183 subjects of EUR or EAS ancestry and utilizing a Bayesian analysis is patent when one considers that only two novel loci were detected from the AA GWAS subjects alone with using a standard association analysis. Additionally, these 20 loci were detected above that of the Liu study, which evaluated the same EUR and EAS data sets but with an additional 3303 Indian or Iranian study subjects.
The internal validity of our study was established by our replicating 92% of SNPs that were evaluated from the Liu et al. (5) study. Complete replication was not expected, given that to limit heterogeneity, our study did not include summary data from 4481 Liu et al. subjects from India and Iran, and conversely, lack of evidence for some loci within the AA data set may have detracted from replication. However, we did observe significant evidence for 10 loci that Liu et al. reported no variants with log10BF > 6.0, likely gaining association evidence from our meta-analyzed AA data set. Further integrity of our study is demonstrated by our meta-analysis detecting significant evidence for three loci detected in the de Lange et al. study (14), a significantly more powerful meta-analysis that combined a new EUR GWAS of 23 305 cases and controls meta-analyzed with the Liu et al. data set.
Of the 20 newly identified loci, 10 had immunologic associations. One of the more provocative findings in our study is the IBD association of SNP rs2986751 that is within PLEKHG5 and is cis-eQTL with TNFSFR25. The protein from the PLEKHG5 activates the nuclear factor kappa B (NFkB) signaling pathway. PLEKHG5 mutations have been found in both autosomal recessive Distal Spinal Muscular Atrophy 4 and Charcot–Marie–Tooth disease. Dubinsky et al. (16) identified a SNP in the PLEKHG5 region with suggestive evidence for association with CD surgery. Perhaps a more relevant candidate gene is TNFRSF25, <9 Kb centromeric from rs2986751. The gene product of TNFRSF25 is death receptor 3 (DR3), one of the two major receptors for TNFSF15 gene product TL1A. TL1A is currently recognized as a promising new drug target for IBD (17–19). TNFSF15 was the first gene identified by GWAS in IBD (specifically CD phenotype in a Japanese population study) (20), and is the major CD risk gene in EAS populations with no risk for UC (Liu et al. (5) maximal EAS associated SNP rs13300483 odds ratio for CD 1.70, P = 1.2 × 10−36, for UC P = 0.85). Interestingly, TNFSF15 has risk for both CD and UC in EUR with similar association evidence for both phenotypes although much lower odds ratios (1.18 for CD [P = 8.2 × 10−43] and 1.14 for UC [P = 4 × 10−26]) than observed in EAS CD, consistent with our finding of the TNFRSF25 SNP observed association with IBD and no heterogeneity in posterior mean allelic effect (PMAE) between EUR and AA. DR3 is expressed in lymphocytes and found in lymphocyte-rich tissues, especially the small intestine. DR3 regulates lymphocyte homeostasis, apoptosis and activation of NFkB. DR3 expressing T cells were found increased in IBD (21). Our associated SNP is also just 22 Kb from the lead SNP (rs2986736, an SNP just 8 Kb p-telomeric of TNFRSF25), which showed genome-wide significant association (P < 10−16) with multiple sclerosis, a phenotype highly associated with co-existing IBD (22).
Among the other novel risk loci that we found in our study, which have immunologic functions, are IL7 and chemokine receptor 7 (CCR7) in UC, and ELMO1 in IBD. These candidate genes within three novel loci provide internal validation for our approach, given their strong evidence for roles in IBD. IL7, a pro-inflammatory cytokine that causes expansion of B and T cells, has long been established as having a role in UC. Early studies showed that IL7 expressed in colonic epithelial cells and goblet cells act on the IL7 receptor (IL7R) in intestinal mucosal lymphocytes, and IL7 transgenic mice develop chronic colitis (23,24). IL7 expression may also have important therapeutic consequences for IBD. In circulating T cells, IL-7 signaling was enriched in IBD patients with a more aggressive course of UC and a more refractory course of CD (25). Multi-tissue eQTL analysis of the SNP in this locus, rs72661359, reveals that it is in cis-eQTL with IL7. In a separate study, IL7 and IL7R were found to be increased in colon biopsies of IBD patients non-responsive to anti-TNF therapy, with IL7R also increased in non-responders to anti-α4β7 integrin IBD biologic therapy (vedolizumab). In human T-lymphocyte cultures, IL-7 induced upregulation and activation of the α4β7 heterodimer (26). Our study is the first report of a genetic association in IL7 with UC. IL7R was significantly associated with UC in the first UC GWAS meta-analysis, and it has also been significantly associated with primary biliary cirrhosis and multiple sclerosis (1).
The UC Chr 17 locus at 17q21.2 also showed the broadest association with 11 SNPs having log10BF above 6.1, the SNPs spanning a region of 100 000 bp with the peak SNP, rs9911533 (log10BF 8.0) in an intergenic region 53 Kb p-terminal of the CCR7 gene with the locus extending through SMARCE1 and keratin gene, KRT24. Interestingly, we previously reported significant admixture association evidence that included this region in our AA immunochip study for both IBD and CD (27). The top SNP in Chr 17, rs9911533, was in cis-eQTL with both SMARCE1 and CCR7. CCR7 is a major regulator of leukocyte trafficking present on T cells and dendritic cells. Such mechanisms are established as important in IBD as reduction of leukocyte trafficking to the intestinal epithelium is a major therapy for IBD. CCR7 knockout caused exacerbation of the TNFΔARE model of ileitis, and the knockout results in effector-memory T cells in inflamed ileal tissue and decreased trafficking of T cells into mesenteric lymph nodes (28,29). A CCR7 M7V start-loss variant, rs2228015, was recently found significantly associated with CD in a large-scale EUR exome sequencing association analysis (30). The variant was also observed to be associated with the trait, lymphocyte count (31). However, rs2228015 is not in LD with our rs9911533 UC association. The other proximal candidate genes do not have overt functional roles related to IBD, although KRT24 is expressed in the colon and showed evidence of association in early onset colorectal cancer (32).
In IBD, the association of a SNP in the 5′ UTR of ELMO1, a gene widely expressed in immune and epithelial cells with important functions in phagocytosis of apoptotic cells and bacteria, and with expression associated with degree of reactive oxidase species production, also provides a level of internal validation to our study and demonstrates the potential importance of our findings (33). Blood and small intestine expression quantitative trait loci analysis of rs12532822 shows that it is in cis-eQTL with ELMO1. ELMO1 depletion in enteroids was found to inhibit bacterial internalization and recruitment of monocytes with a decrease in pro-inflammatory cytokine production. Its knockout was shown protective of dextran sodium sulfate model of colitis (34). The gene has been significantly associated with diabetic nephropathy but also showed association evidence in pediatric CD in a Scandinavian study (35,36).
Our strongest association signal was log10BF 23.4 in IBD for rs79315643 located within the second intron of XKR6 on 8p23.1, the SNP also significantly associated with CD (log10BF 18.50) and UC (log10BF 7.23). It maps to the second intron of XKR6. XKR6 has been associated with systemic lupus erythematosus (SLE) and eosinophilic esophagitis (37–39). XKR6 is expressed in red blood cells, thyroid, colon, duodenum, esophagus, small intestine and stomach and codes for one of the transmembrane proteins of the Kell blood group of antigens. Since very little is known about its function, its key role in IBD pathogenesis is uncertain. However, its expression suggests that it may be immune related (39). The SNP rs79315643 is in cis-eQTL with FAM167A, BLK and FDFT1. FAM167A and BLK are part of a susceptibility locus associated with multiple autoimmune diseases, whereas FDFT1 has been identified as a potential blood-based biomarker to predict disease activity in UC (40–43).
For IBD and CD, we detected a synonymous exonic APLP1 association at 19q13.12. APLP1 encodes a member of the highly conserved amyloid precursor protein gene family. The encoded protein is a membrane-associated glycoprotein cleaved by secretases in a manner similar to amyloid beta A4 precursor protein cleavage. This cleavage liberates an intracellular cytoplasmic fragment that may act as a transcriptional activator (44,45). It may also play a role in synaptic maturation during cortical development. APLP1 has also been shown to be upregulated in the uninvolved colon sample of a CD patient (46). It is highly expressed in neuroendocrine tumors of the gastrointestinal tract with enhanced expression in metastatic lesions, which indicates that APLP1 may be upregulated during tumor dissemination (47). GTEx multi-tissue eQTL analysis reveals that the SNP in this locus, rs230261, is also in cis-eQTL with APLP1.
For the IBD phenotype, we detected an association with rs2031797 mapping to an intronic region of PI4KB. PI4KB is responsible for the synthesis and maintenance of the Golgi and trans-Golgi network phosphatidylinositol 4-phosphate (PI4P) pools. PI4P plays an essential role in cell signaling, lipid transport and as a precursor for higher phosphoinositides. It is also shown that mutations in PI4KB can also be found in cancers affecting the large intestine (48). Single and multi-tissue eQTL analysis shows that rs2031797 is in cis-eQTL with PSMD4. PSMD4 encodes part of the 19-s regulator base that forms the 26S proteasome complex, which is important in protein homeostasis (49). Researchers studying cancer development evaluated IBD patients and found upregulated expression of Nrf2 and proteasome subunit proteins, including PSMD4 at inflammatory sites of IBD tissues, leading to enhanced proteasome activity and apoptosis protection of human colonocytes (50).
In CD, we detected SNP associations pointing to a region in 4q24 near the TET2 gene. The SNP in this region is in cis-eQTL with VPS53 and TET2. The TET2 gene is in a known UC locus (5), but GWA studies have yet to implicate the gene in CD. This gene may have a role in pro-inflammatory cytokine IL-6 expression in mice (51). We also implicate OSER1, also known as C20orf111, in the region of 20q13.12. Evaluation of SNP rs2143606 in blood and multi-tissue eQTL databases revealed that it is in cis-eQTL with OSER1. Little is known about the function of this gene. It is thought to have an increase in expression in cells undergoing hydrogen peroxide-induced apoptosis. We uncovered a fourth locus on Chr 7 associated with CD pointing to BCAP29. This gene may play a role in anterograde transport of membrane proteins from the endoplasmic reticulum to the Golgi and may be involved in CASP8-mediated apoptosis. Currently, no studies have reported the association of BCAP29 variants (rs3801944, rs10273733 and rs2808) with CD. Our genome-wide trans-ancestry meta-analyses of the AA and EUR CD samples revealed a signal in an intronic variant within AUTS2. A study on genome-wide gene expression differences in CD revealed that the AUTS2 gene is downregulated in CD samples compared with healthy controls (52).
For UC, several associations were found in the region 6p21.31 including rs73407795 (SPDEF), rs3822921(ANKS1A) and rs11755266 (TCP11;AY927475). SPDEF is the major regulator of Paneth and goblet cells, the first line of defense against gut pathogens that secretes antimicrobial peptides and mucus (53). SPDEF has been recently identified as a novel target for the enrichment of intestinal epithelial stem cells and mucosal healing (54). ANKS1A, also known as ODIN, regulates the epidermal growth factor receptor and EphA receptor signaling pathways (55). As a target of Src family kinases, which are implicated in the development of some colorectal cancers, ODIN may play a role in cancer cell signaling mechanisms (56). These three UC-associated SNPs (rs73407795, rs3822921 and rs11755266) were found to be in cis-eQTL with the inflammation and lipid regulator with UBA-like and NBR1-like domains (ILRUN). ILRUN, formerly known as C6orf106, regulates inflammation and antiviral responses (57). This gene was identified as having suggestive evidence of gene-smoking interaction in UC (58). In our trans-ancestry meta-analyses of the AA and EUR UC GWAS samples, the new signal in rs115484865 is found between GABBR1 and MOG. MOG is a target antigen in autoimmune diseases like multiple sclerosis (59).
Our study also discovered a unique UC association in the region 1q25.1 mapping to TNFSF4. The associated SNP in the region, rs10465507, is in cis-eQTL with TNFSF4 in both single and multi-tissue eQTL databases. TNFSF4 encodes a protein that plays a role in T-cell antigen-presenting cell interactions and mediates the binding of activated T cells to vascular endothelial cells (60). Polymorphisms in this gene are associated with susceptibility to autoimmune diseases such as SLE and primary Sjogren’s syndrome (60,61).
The other remaining novel UC locus detected was within introns of two genes: WD repeat-and FYVE domain-containing protein 4 (WDFY4) and leucine-rich repeat containing protein 18 (LRRC18). This locus has shown genome-wide association evidence with SLE in EAS and EUR populations, and an Immunochip-wide analysis found suggestive evidence of WDFY4 and smoking for UC and IBD but not for CD (58). Analysis of eQTL database in blood revealed that rs2940716 was strongly associated with the expression level of WDFY4. WDFY4 lymphocyte conditional knockout mice showed a decrease in subpopulations of B cells in the periphery and impaired B-cell antibody response to antigen stimulation (62). LRRC18 is thought to have a role in the regulation of spermatogenesis and sperm maturation.
In summary, we have performed one of the largest trans-ancestry analyses in IBD and involving three disparate ancestries: AA, EUR and EAS populations. We have demonstrated that leveraging the use of multiethnic groups can help identify additional novel loci in IBD. The results of this study highlight the value of utilizing prior association evidence from much larger studies in other populations to enable novel discovery by combining data with much smaller cohorts from understudied and diverse ancestral populations.
Materials and Methods
Data
We used aggregate summary-level statistics derived from EUR, EAS and AA descent. The EUR and EAS data were generated by the combined genome-wide or Immunochip genotype data, as reported by Liu et al. (5). The EUR data were generated on 73 076 independent IBD cases and controls of EUR ancestry from 15 countries in Europe, North America and Oceania. The EAS data were generated on 6598 IBD cases and controls recruited from Japan, South Korea and Hong Kong.
The AA data were generated by our GWAS meta-analysis (2). Briefly, the study was a meta-analysis of two high-density, genome-wide scans on AAs with IBD and population controls. In GWAS 1, AA samples with IBD (1258 cases) were recruited by the Johns Hopkins Multicenter AA IBD Study, Cedars-Sinai Medical Center and other Genetics Research Centers of the National Institute of Diabetes and Digestive and Kidney Diseases IBD Genetics Consortium, whereas data from 1678 AA controls were from the Health and Retirement Study made available via dbGAP. In GWAS 2, 1087 IBD cases were obtained by Emory University from the GENESIS study, and 3324 controls were obtained from the Kaiser Research Program on Genes, Environment, and Health study. Samples were genotyped for GWAS 1 on the Illumina Omni 2.5 (~2.3 million SNPs) or Omni2.5 Exome (~2.6 million SNPs) arrays and for GWAS 2 on the Affymetrix Axiom Genome-wide AFR1 Array world Array 3 (~894 000 SNPs). GWAS 1 and GWAS 2 cohorts were then combined and meta-analyzed using METAL using an inverse-variance, fixed-effects model (2).
Summary-level statistics in each population were evaluated separately for the major phenotypes of CD and UC and all IBD (CD, UC and IBD-undifferentiated). SNPs with risk for ambiguity (i.e. homologous pairings A-T or C-G pairings) were removed prior to analysis to remove variants that correct allele assignment between populations cannot be matched with certainty. The remaining variants were then used for the trans-ancestry meta-analysis.
Ethical approval
Approval for this study was obtained from the Institutional Review Boards of all individual participating centers. Written informed consent was obtained from all the participants in each of the studies (2,5).
All authors had access to the study data and reviewed and approved the final manuscript.
Trans-ancestry meta-analysis
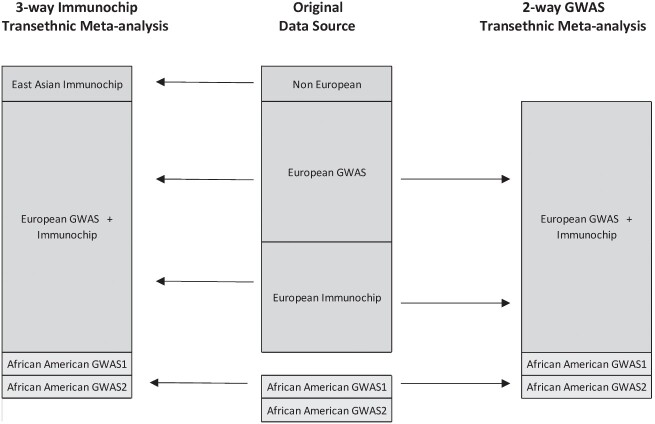
We performed a trans-ancestry meta-analysis for each of the major IBD phenotypes using the GWAS and Immunochip data from the three population data sets, as illustrated in Fig. 1 (2,5). Our meta-analysis was limited to the common SNPs between the GWAS and Immunochip data sets. Meta-analyses of all three study population SNPs (i.e. EUR, EAS and AA) were limited to those within the EAS Immunochip data set, given that only Immunochip data were available for EASs in the Liu et al. study. Trans-ancestry meta-analyses limited to the AA and EUR populations were conducted separately using the larger GWAS data sets available from these two populations to cover the whole genome and rare variants not otherwise on the Immunochip. To account for heterogeneity in allelic effects between diverse ancestry groups while allowing for similarity in allelic effects between closely related populations, we adopted a Bayesian model as implemented in meta-analysis of Trans-ethnic association (MANTRA) algorithm (6). Compared with a purely random effects analysis, the advantage of this approach is the modeling of allelic heterogeneity between ethnic groups. Populations are assigned to clusters based on a prior model of relatedness and observed effect sizes by means of the Bayesian partition model. Groups within the same ethnic cluster are assumed to have the same underlying allelic effects, and different clusters are assumed to have different underlying allelic effects. Using this approach allows for the expected heterogeneity between populations. Although MANTRA partitions study populations based on allele frequency similarity, as a method based on summary statistics, it does not incorporate local ancestry information (63).
Figure 1.
Diagram of source population data sets with cases and controls. This is an illustration of the source data sets used in the current trans-ancestry association analysis. The top box in the center of the figure represents the breakdown of cases and controls by population used in the 2015 meta-analysis (5). The bottom box in the center of the figure depicts the breakdown of cases and controls used in the AA GWAS meta-analysis (1).
MANTRA estimates the strength of association using the log10BF computed for each SNP. We employed a log10BF threshold equal to or ≥6.0 to determine strong evidence for genome-wide significance in AAs utilizing significance criteria of association evidence of loci established by GWAS in the other populations and in the 2015 Liu et al. parent study (5,6). Functional annotation of genetic variants was carried out using the web-based annotation engine, wANNOVAR (64).
In total, our study included 38 155 IBD cases and 48 485 controls from the EUR GWAS and Immunochip cohort, 2824 IBD cases and 3719 controls from the EAS Immunochip data and 2345 IBD cases and 5002 controls as part of the AA cohort (Table 1).
Table 1.
Cohort sample sizes for GWAS and Immunochip trans-ancestry meta-analysis
| Population | Cohort | Cases | Controls | Total sample size |
|---|---|---|---|---|
| EUR | EUR CD | 20 550 | 41 642 | 62 192 |
| EUR UC | 17 647 | 47 179 | 64 826 | |
| EUR IBD | 38 155 | 48 485 | 86 640 | |
| EA | EAS CD | 1690 | 3719 | 5409 |
| EAS UC | 1134 | 3719 | 4853 | |
| EAS IBD | 2824 | 3719 | 6543 | |
| AA | AA CD | 1646 | 5002 | 6648 |
| AA UC | 583 | 5002 | 5585 | |
| AA IBD | 2345 | 5002 | 7347 |
Notes: Listed in the table are the sample sizes for the cases and controls per phenotype used in the IBD trans-ancestry meta-analysis.
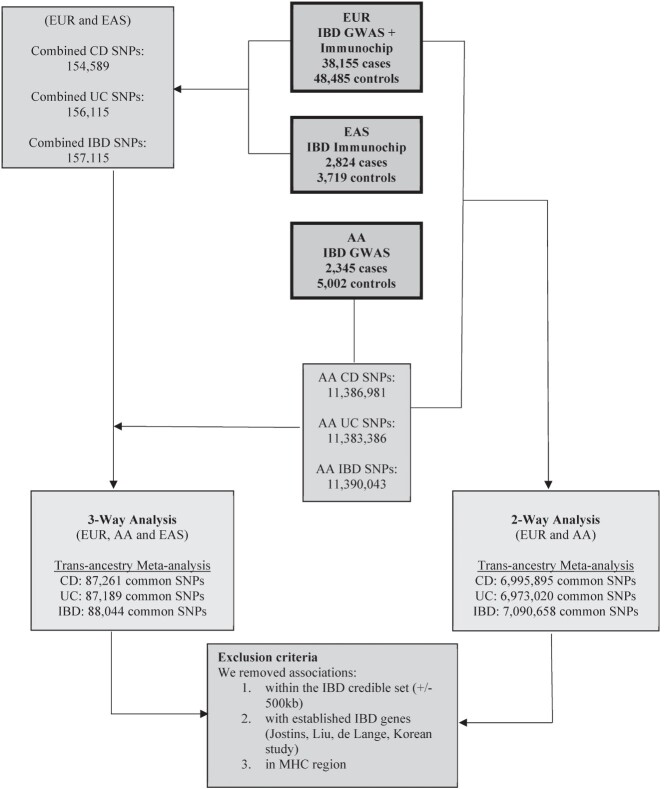
Among the SNPs that achieved significance at log10BF ≥ 6, we focused on the associations outside of known IBD credible sets for the phenotype of interest and established IBD genes. We also excluded SNPs inside the HLA region on Chr 6 (Fig. 2). All SNPs in the filtered subset were not in linkage disequilibrium with SNPs reported associated within established IBD loci.
Figure 2.
Workflow to identify novel SNP associations with CD, UC and IBD. This is an overview of the workflow used in determining new risk loci in the IBD trans-ancestry meta-analysis. Summary statistics from EUR, EA and AA cohorts were combined using MANTRA for each of the phenotypes of interest (UC, CD and IBD). The left-hand side of the figure depicts the common SNPs carried over in the downstream analysis for the EUR, EAS and AA trans-ancestry meta-analysis. The right-hand side of the figure displays the common SNPs carried over in the downstream analysis for the EUR and AA trans-ancestry meta-analysis. SNPs with a log 10 Bayes Factor ≥ 6 were considered to be genome-wide significant. Variants within 500 Kb of established risk loci for the IBD phenotype of interest or in the MHC region were removed from the analysis.
Heterogeneity analysis
Across the three ancestries, deviation from homogeneity was assessed using the posterior probability of association. A posterior probability of heterogeneity of >0.95 would provide strong evidence of a deviation from homogeneity in allelic effects across the ethnic groups (6).
eQTL analysis
We used several eQTL repositories to interrogate SNPs meeting log10BF ≥ 6 criteria for association with mRNA gene expression. Single-tissue eQTL analysis examined the results of NESDA NTR Conditional eQTL Catalog, eQTL summary data from the eQTLGen consortium and Blood eQTL browser data set (Supplementary Material, Table S7). We also looked at Multi-tissue eQTLs in the GTEx consortium database (65) as it can improve power for eQTL discovery by modeling patterns of sharing across all available tissues in the database (66–68) (Supplementary Material, Fig. S1).
Conflict of Interest statement. None declared.
Funding
National Institutes of Health (DK062431 to S.R.B. and C.L.S., DK087694 to S.K., DK062413 to D.P.B.M).
Data Availability
The full set of summary statistics for all SNPs (in addition to those with Bayes Factor ≥ 6 shared in Supplementary Tables) will be made available via the NIDDK IBD Genetics Consortium, the sponsor of the study, and accessible at https://ibdgc.datacommons.io/.
Supplementary Material
Contributor Information
Roberto Y Cordero, Department of Genetics, Genomics, and Informatics, University of Tennessee Health Science Center, Memphis, TN 38163, USA.
Jennifer B Cordero, Department of Genetics, Genomics, and Informatics, University of Tennessee Health Science Center, Memphis, TN 38163, USA.
Andrew B Stiemke, Department of Genetics, Genomics, and Informatics, University of Tennessee Health Science Center, Memphis, TN 38163, USA.
Lisa W Datta, Meyerhoff Inflammatory Bowel Disease Center, Department of Medicine, School of Medicine, Johns Hopkins University, Baltimore, MD 21231, USA.
Steven Buyske, Department of Statistics and Biostatistics, Rutgers University, Piscataway, NJ 08854, USA.
Subra Kugathasan, Department of Pediatrics and Department of Human Genetics, Emory University School of Medicine, Atlanta, GA 30322, USA.
Dermot P B McGovern, F. Widjaja Foundation Inflammatory Bowel and Immunobiology Research Institute, Cedars Sinai Medical Center, Los Angeles, CA 90048, USA.
Steven R Brant, Meyerhoff Inflammatory Bowel Disease Center, Department of Medicine, School of Medicine, Johns Hopkins University, Baltimore, MD 21231, USA; Rutgers Crohn’s and Colitis Center of New Jersey, Department of Medicine, Rutgers Robert Wood Johnson Medical School, Rutgers, The State University of New Jersey, New Brunswick, NJ 08901, USA; Human Genetics Institute of New Jersey and Department of Genetics, School of Arts and Sciences, Rutgers, The State University of New Jersey, Piscataway, NJ 08854, USA.
Claire L Simpson, Department of Genetics, Genomics, and Informatics, University of Tennessee Health Science Center, Memphis, TN 38163, USA.
References
- 1. Anderson, C.A., Boucher, G., Lees, C.W., Franke, A., D’Amato, M., Taylor, K.D., Lee, J.C., Goyette, P., Imielinski, M., Latiano, A. et al. (2011) Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet., 43, 246–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brant, S.R., Okou, D.T., Simpson, C.L., Cutler, D.J., Haritunians, T., Bradfield, J.P., Chopra, P., Prince, J., Begum, F., Kumar, A. et al. (2017) Genome-wide association study identifies African-specific susceptibility loci in African Americans with inflammatory bowel disease. Gastroenterology, 152, 206–217.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jostins, L., Ripke, S., Weersma, R.K., Duerr, R.H., McGovern, D.P., Hui, K.Y., Lee, J.C., Schumm, L.P., Sharma, Y., Anderson, C.A. et al. (2012) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature, 491, 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kenny, E.E., Pe'er, I., Karban, A., Ozelius, L., Mitchell, A.A., Ng, S.M., Erazo, M., Ostrer, H., Abraham, C., Abreu, M.T. et al. (2012) A Genome-wide scan of Ashkenazi Jewish Crohn’s disease suggests novel susceptibility loci. PLoS Genet., 8, e1002559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu, J.Z., van Sommeren, S., Huang, H., Ng, S.C., Alberts, R., Takahashi, A., Ripke, S., Lee, J.C., Jostins, L., Shah, T. et al. (2015) Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet., 47, 979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morris, A.P. (2011) Transethnic meta-analysis of genomewide association studies. Genet. Epidemiol., 35, 809–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Parkes, M., Barrett, J.C., Prescott, N., Tremelling, M., Anderson, C.A., Fisher, S.A., Roberts, R.G., Nimmo, E.R., Cummings, F.R., Soars, D. et al. (2007) Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn disease susceptibility. Nat. Genet., 39, 830–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yamazaki, K., Umeno, J., Takahashi, A., Hirano, A., Johnson, T.A., Kumasaka, N., Morizono, T., Hosono, N., Kawaguchi, T., Takazoe, M. et al. (2013) A genome-wide association study identifies 2 susceptibility loci for Crohn’s disease in a Japanese population. Gastroenterology, 144, 781–788. [DOI] [PubMed] [Google Scholar]
- 9. Juyal, G., Negi, S., Sood, A., Gupta, A., Prasad, P., Senapati, S., Zaneveld, J., Singh, S., Midha, V., van Sommeren, S. et al. (2015) Genome-wide association scan in north Indians reveals three novel HLA-independent risk loci for ulcerative colitis. Gut, 64, 571–579. [DOI] [PubMed] [Google Scholar]
- 10. Ye, B.D., Choi, H., Hong, M., Yun, W.J., Low, H.Q., Haritunians, T., Kim, K.J., Park, S.H., Lee, I., Bang, S.Y. et al. (2016) Identification of ten additional susceptibility loci for ulcerative colitis through Immunochip analysis in Koreans. Inflamm. Bowel Dis., 22, 13–19. [DOI] [PubMed] [Google Scholar]
- 11. Yang, S.K., Hong, M., Choi, H., Zhao, W., Jung, Y., Haritunians, T., Ye, B.D., Kim, K.J., Park, S.H., Lee, I. et al. (2015) Immunochip analysis identification of 6 additional susceptibility loci for Crohn’s disease in Koreans. Inflamm. Bowel Dis., 21, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hong, M., Ye, B.D., Yang, S.K., Jung, S., Lee, H.S., Kim, B.M., Lee, S.B., Hong, J., Baek, J., Park, S.H. et al. (2018) Immunochip meta-analysis of inflammatory bowel disease identifies three novel loci and four novel associations in previously reported loci. J. Crohns Colitis, 12, 730–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ng, S.C., Tsoi, K.K., Kamm, M.A., Xia, B., Wu, J., Chan, F.K. and Sung, J.J. (2012) Genetics of inflammatory bowel disease in Asia: systematic review and meta-analysis. Inflamm. Bowel Dis., 18, 1164–1176. [DOI] [PubMed] [Google Scholar]
- 14. de Lange, K.M., Moutsianas, L., Lee, J.C., Lamb, C.A., Luo, Y., Kennedy, N.A., Jostins, L., Rice, D.L., Gutierrez-Achury, J., Ji, S.G. et al. (2017) Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet., 49, 256–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ellinghaus, D., Jostins, L., Spain, S.L., Cortes, A., Bethune, J., Han, B., Park, Y.R., Raychaudhuri, S., Pouget, J.G., Hubenthal, M. et al. (2016) Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat. Genet., 48, 510–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dubinsky, M.C., Kugathasan, S., Kwon, S., Haritunians, T., Wrobel, I., Wahbeh, G., Quiros, A., Bahar, R., Silber, G., Farrior, S. et al. (2013) Multidimensional prognostic risk assessment identifies association between IL12B variation and surgery in Crohn’s disease. Inflamm. Bowel Dis., 19, 1662–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Banfield, C., Rudin, D., Bhattacharya, I., Goteti, K., Li, G., Hassan-Zahraee, M., Brown, L.S., Hung, K.E., Pawlak, S. and Lepsy, C. (2020) First-in-human, randomized dose-escalation study of the safety, tolerability, pharmacokinetics, pharmacodynamics and immunogenicity of PF-06480605 in healthy subjects. Br. J. Clin. Pharmacol., 86, 812–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Danese, S., Klopocka, M., Scherl, E.J., Romatowski, J., Allegretti, J.R., Peeva, E., Vincent, M.S., Schoenbeck, U., Ye, Z., Hassan-Zahraee, M. et al. (2021) Anti-TL1A antibody PF-06480605 safety and efficacy for ulcerative colitis: a phase 2a single-arm study. Clin. Gastroenterol. Hepatol., 19, 2324–2332.e2326. [DOI] [PubMed] [Google Scholar]
- 19. Furfaro, F., Alfarone, L., Gilardi, D., Correale, C., Allocca, M., Fiorino, G., Argollo, M., Zilli, A., Zacharopoulou, E., Loy, L. et al. (2021) TL1A: a new potential target in the treatment of inflammatory bowel disease. Curr. Drug Targets, 22, 760–769. [DOI] [PubMed] [Google Scholar]
- 20. Yamazaki, K., McGovern, D., Ragoussis, J., Paolucci, M., Butler, H., Jewell, D., Cardon, L., Takazoe, M., Tanaka, T., Ichimori, T. et al. (2005) Single nucleotide polymorphisms in TNFSF15 confer susceptibility to Crohn’s disease. Hum. Mol. Genet., 14, 3499–3506. [DOI] [PubMed] [Google Scholar]
- 21. Bamias, G., Martin, C., 3rd, Marini, M., Hoang, S., Mishina, M., Ross, W.G., Sachedina, M.A., Friel, C.M., Mize, J., Bickston, S.J. et al. (2003) Expression, localization, and functional activity of TL1A, a novel Th1-polarizing cytokine in inflammatory bowel disease. J. Immunol., 171, 4868–4874. [DOI] [PubMed] [Google Scholar]
- 22. International Multiple Sclerosis Genetics Consortium (2019) Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science, 365, eaav7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Watanabe, M., Ueno, Y., Yajima, T., Iwao, Y., Tsuchiya, M., Ishikawa, H., Aiso, S., Hibi, T. and Ishii, H. (1995) Interleukin 7 is produced by human intestinal epithelial cells and regulates the proliferation of intestinal mucosal lymphocytes. J. Clin. Invest., 95, 2945–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Watanabe, M., Ueno, Y., Yajima, T., Okamoto, S., Hayashi, T., Yamazaki, M., Iwao, Y., Ishii, H., Habu, S., Uehira, M. et al. (1998) Interleukin 7 transgenic mice develop chronic colitis with decreased interleukin 7 protein accumulation in the colonic mucosa. J. Exp. Med., 187, 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee, J.C., Lyons, P.A., McKinney, E.F., Sowerby, J.M., Carr, E.J., Bredin, F., Rickman, H.M., Ratlamwala, H., Hatton, A., Rayner, T.F. et al. (2011) Gene expression profiling of CD8+ T cells predicts prognosis in patients with Crohn disease and ulcerative colitis. J. Clin. Invest., 121, 4170–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Belarif, L., Danger, R., Kermarrec, L., Nerrière-Daguin, V., Pengam, S., Durand, T., Mary, C., Kerdreux, E., Gauttier, V., Kucik, A. et al. (2019) IL-7 receptor influences anti-TNF responsiveness and T cell gut homing in inflammatory bowel disease. J. Clin. Invest., 129, 1910–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang, C., Haritunians, T., Okou, D.T., Cutler, D.J., Zwick, M.E., Taylor, K.D., Datta, L.W., Maranville, J.C., Liu, Z., Ellis, S. et al. (2015) Characterization of genetic loci that affect susceptibility to inflammatory bowel diseases in African Americans. Gastroenterology, 149, 1575–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McNamee, E.N., Masterson, J.C., Veny, M., Collins, C.B., Jedlicka, P., Byrne, F.R., Ng, G.Y. and Rivera-Nieves, J. (2015) Chemokine receptor CCR7 regulates the intestinal TH1/TH17/Treg balance during Crohn’s-like murine ileitis. J. Leukoc. Biol., 97, 1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Genua, M., Sgambato, A. and Danese, S. (2015) Editorial: CCR7 is required for leukocyte egression in an experimental model of Crohn’s disease-like ileitis. J. Leukoc. Biol., 97, 1000–1002. [DOI] [PubMed] [Google Scholar]
- 30. Sazonovs, A., Stevens, C.R., Venkataraman, G.R., Yuan, K., Avila, B., Abreu, M.T., Ahmad, T., Allez, M., Ananthakrishnan, A.N., Atzmon, G. et al. (2022) Large-scale sequencing identifies multiple genes and rare variants associated with Crohn’s disease susceptibility. Nat. Genet., 54, 1275–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. John, S.E., Antony, D., Eaaswarkhanth, M., Hebbar, P., Channanath, A.M., Thomas, D., Devarajan, S., Tuomilehto, J., Al-Mulla, F., Alsmadi, O. et al. (2018) Assessment of coding region variants in Kuwaiti population: implications for medical genetics and population genomics. Sci. Rep., 8, 16583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hong, Y., Ho, K.S., Eu, K.W. and Cheah, P.Y. (2007) A susceptibility gene set for early onset colorectal cancer that integrates diverse signaling pathways: implication for tumorigenesis. Clin. Cancer Res., 13, 1107. [DOI] [PubMed] [Google Scholar]
- 33. Kakoki, M., Bahnson, E.M., Hagaman, J.R., Siletzky, R.M., Grant, R., Kayashima, Y., Li, F., Lee, E.Y., Sun, M.T., Taylor, J.M. et al. (2019) Engulfment and cell motility protein 1 potentiates diabetic cardiomyopathy via Rac-dependent and Rac-independent ROS production. JCI Insight, 4, e127660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sayed, I.M., Suarez, K., Lim, E., Singh, S., Pereira, M., Ibeawuchi, S.R., Katkar, G., Dunkel, Y., Mittal, Y., Chattopadhyay, R. et al. (2020) Host engulfment pathway controls inflammation in inflammatory bowel disease. FEBS J., 287, 3967–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shimazaki, A., Kawamura, Y., Kanazawa, A., Sekine, A., Saito, S., Tsunoda, T., Koya, D., Babazono, T., Tanaka, Y., Matsuda, M. et al. (2005) Genetic variations in the gene encoding ELMO1 are associated with susceptibility to diabetic nephropathy. Diabetes, 54, 1171–1178. [DOI] [PubMed] [Google Scholar]
- 36. Parmar, A.S., Lappalainen, M., Paavola-Sakki, P., Halme, L., Färkkilä, M., Turunen, U., Kontula, K., Aromaa, A., Salomaa, V., Peltonen, L. et al. (2012) Association of celiac disease genes with inflammatory bowel disease in Finnish and Swedish patients. Genes Immun., 13, 474–480. [DOI] [PubMed] [Google Scholar]
- 37. Alonso-Perez, E., Suarez-Gestal, M., Calaza, M., Ordi-Ros, J., Balada, E., Bijl, M., Papasteriades, C., Carreira, P., Skopouli, F.N., Witte, T. et al. (2012) Further evidence of subphenotype association with systemic lupus erythematosus susceptibility loci: a European cases only study. PLoS One, 7, e45356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Joo, Y.B., Lim, J., Tsao, B.P., Nath, S.K., Kim, K. and Bae, S.-C. (2018) Genetic variants in systemic lupus erythematosus susceptibility loci, XKR6 and GLT1D1 are associated with childhood-onset SLE in a Korean cohort. Sci. Rep., 8, 9962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kottyan, L.C., Davis, B.P., Sherrill, J.D., Liu, K., Rochman, M., Kaufman, K., Weirauch, M.T., Vaughn, S., Lazaro, S., Rupert, A.M. et al. (2014) Genome-wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nat. Genet., 46, 895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hom, G., Graham, R.R., Modrek, B., Taylor, K.E., Ortmann, W., Garnier, S., Lee, A.T., Chung, S.A., Ferreira, R.C., Pant, P.V. et al. (2008) Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N. Engl. J. Med., 358, 900–909. [DOI] [PubMed] [Google Scholar]
- 41. Gregersen, P.K., Amos, C.I., Lee, A.T., Lu, Y., Remmers, E.F., Kastner, D.L., Seldin, M.F., Criswell, L.A., Plenge, R.M., Holers, V.M. et al. (2009) REL, encoding a member of the NF-kappaB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat. Genet., 41, 820–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lessard, C.J., Li, H., Adrianto, I., Ice, J.A., Rasmussen, A., Grundahl, K.M., Kelly, J.A., Dozmorov, M.G., Miceli-Richard, C., Bowman, S. et al. (2013) Variants at multiple loci implicated in both innate and adaptive immune responses are associated with Sjögren’s syndrome. Nat. Genet., 45, 1284–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Burakoff, R., Pabby, V., Onyewadume, L., Odze, R., Adackapara, C., Wang, W., Friedman, S., Hamilton, M., Korzenik, J., Levine, J. et al. (2015) Blood-based biomarkers used to predict disease activity in Crohn’s disease and ulcerative colitis. Inflamm. Bowel Dis., 21, 1132–1140. [DOI] [PubMed] [Google Scholar]
- 44. Cao, X. and Südhof, T.C. (2001) A transcriptionally (correction of transcriptively) active complex of APP with Fe65 and histone acetyltransferase Tip60. Science, 293, 115–120. [DOI] [PubMed] [Google Scholar]
- 45. Gao, Y. and Pimplikar, S.W. (2001) The gamma-secretase-cleaved C-terminal fragment of amyloid precursor protein mediates signaling to the nucleus. Proc. Natl. Acad. Sci. U. S. A., 98, 14979–14984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dooley, T.P., Curto, E.V., Reddy, S.P., Davis, R.L., Lambert, G.W., Wilborn, T.W. and Elson, C.O. (2004) Regulation of gene expression in inflammatory bowel disease and correlation with IBD drugs: screening by DNA microarrays. Inflamm. Bowel Dis., 10, 1–14. [DOI] [PubMed] [Google Scholar]
- 47. Arvidsson, Y., Andersson, E., Bergström, A., Andersson, M.K., Altiparmak, G., Illerskog, A.C., Ahlman, H., Lamazhapova, D. and Nilsson, O. (2008) Amyloid precursor-like protein 1 is differentially upregulated in neuroendocrine tumours of the gastrointestinal tract. Endocr. Relat. Cancer, 15, 569–581. [DOI] [PubMed] [Google Scholar]
- 48. Waugh, M.G. (2014) Amplification of chromosome 1q genes encoding the phosphoinositide signalling enzymes PI4KB, AKT3, PIP5K1A and PI3KC2B in breast cancer. J. Cancer, 5, 790–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Collins, G.A. and Goldberg, A.L. (2017) The logic of the 26S proteasome. Cell, 169, 792–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kruse, M.-L., Friedrich, M., Arlt, A., Röcken, C., Egberts, J.-H., Sebens, S. and Schäfer, H. (2016) Colonic lamina propria inflammatory cells from patients with IBD Induce the nuclear factor-E2 related factor-2 thereby leading to greater proteasome activity and apoptosis protection in human colonocytes. Inflamm. Bowel Dis., 22, 2593–2606. [DOI] [PubMed] [Google Scholar]
- 51. Zhang, Q., Zhao, K., Shen, Q., Han, Y., Gu, Y., Li, X., Zhao, D., Liu, Y., Wang, C., Zhang, X. et al. (2015) Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature, 525, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wu, F., Dassopoulos, T., Cope, L., Maitra, A., Brant, S.R., Harris, M.L., Bayless, T.M., Parmigiani, G. and Chakravarti, S. (2007) Genome-wide gene expression differences in Crohn’s disease and ulcerative colitis from endoscopic pinch biopsies: insights into distinctive pathogenesis. Inflamm. Bowel Dis., 13, 807–821. [DOI] [PubMed] [Google Scholar]
- 53. Noah, T.K., Kazanjian, A., Whitsett, J. and Shroyer, N.F. (2010) SAM pointed domain ETS factor (SPDEF) regulates terminal differentiation and maturation of intestinal goblet cells. Exp. Cell Res., 316, 452–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nishimura, R., Shirasaki, T., Tsuchiya, K., Miyake, Y., Watanabe, Y., Hibiya, S., Watanabe, S., Nakamura, T. and Watanabe, M. (2019) Establishment of a system to evaluate the therapeutic effect and the dynamics of an investigational drug on ulcerative colitis using human colonic organoids. J. Gastroenterol., 54, 608–620. [DOI] [PubMed] [Google Scholar]
- 55. Lee, H., Noh, H., Mun, J., Gu, C., Sever, S. and Park, S. (2016) Anks1a regulates COPII-mediated anterograde transport of receptor tyrosine kinases critical for tumorigenesis. Nat. Commun., 7, 12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Emaduddin, M., Edelmann, M.J., Kessler, B.M. and Feller, S.M. (2008) Odin (ANKS1A) is a Src family kinase target in colorectal cancer cells. Cell Commun. Signal., 6, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ambrose, R.L., Brice, A.M., Caputo, A.T., Alexander, M.R., Tribolet, L., Liu, Y.C., Adams, T.E., Bean, A.G.D. and Stewart, C.R. (2020) Molecular characterisation of ILRUN, a novel inhibitor of proinflammatory and antimicrobial cytokines. Heliyon, 6, e04115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yadav, P., Ellinghaus, D., Rémy, G., Freitag-Wolf, S., Cesaro, A., Degenhardt, F., Boucher, G., Delacre, M., International IBD Genetics Consortium, Peyrin-Biroulet, L. et al. (2017) Genetic factors interact with tobacco smoke to modify risk for inflammatory bowel disease in humans and mice. Gastroenterology, 153, 550–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. von Büdingen, H.C., Tanuma, N., Villoslada, P., Ouallet, J.C., Hauser, S.L. and Genain, C.P. (2001) Immune responses against the myelin/oligodendrocyte glycoprotein in experimental autoimmune demyelination. J. Clin. Immunol., 21, 155–170. [DOI] [PubMed] [Google Scholar]
- 60. Cunninghame Graham, D.S., Graham, R.R., Manku, H., Wong, A.K., Whittaker, J.C., Gaffney, P.M., Moser, K.L., Rioux, J.D., Altshuler, D., Behrens, T.W. et al. (2008) Polymorphism at the TNF superfamily gene TNFSF4 confers susceptibility to systemic lupus erythematosus. Nat. Genet., 40, 83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nordmark, G., Kristjansdottir, G., Theander, E., Appel, S., Eriksson, P., Vasaitis, L., Kvarnström, M., Delaleu, N., Lundmark, P., Lundmark, A. et al. (2011) Association of EBF1, FAM167A(C8orf13)-BLK and TNFSF4 gene variants with primary Sjögren’s syndrome. Genes Immun., 12, 100–109. [DOI] [PubMed] [Google Scholar]
- 62. Yuan, Q., Li, Y., Li, J., Bian, X., Long, F., Duan, R., Ma, X., Gao, F., Gao, S., Wei, S. et al. (2018) WDFY4 is involved in symptoms of systemic lupus erythematosus by modulating B cell fate via noncanonical autophagy. J. Immunol., 201, 2570–2578. [DOI] [PubMed] [Google Scholar]
- 63. Martin, E.R., Tunc, I., Liu, Z., Slifer, S.H., Beecham, A.H. and Beecham, G.W. (2018) Properties of global- and local-ancestry adjustments in genetic association tests in admixed populations. Genet. Epidemiol., 42, 214–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chang, X. and Wang, K. (2012) wANNOVAR: annotating genetic variants for personal genomes via the web. J. Med. Genet., 49, 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Aguet, F., Brown, A.A., Castel, S.E., Davis, J.R., He, Y., Jo, B., Mohammadi, P., Park, Y., Parsana, P., Segrè, A.V. et al. (2017) Genetic effects on gene expression across human tissues. Nature, 550, 204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li, G., Shabalin, A.A., Rusyn, I., Wright, F.A. and Nobel, A.B. (2018) An empirical Bayes approach for multiple tissue eQTL analysis. Biostatistics, 19, 391–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Flutre, T., Wen, X., Pritchard, J. and Stephens, M. (2013) A statistical framework for joint eQTL analysis in multiple tissues. PLoS Genet., 9, e1003486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sul, J.H., Han, B., Ye, C., Choi, T. and Eskin, E. (2013) Effectively identifying eQTLs from multiple tissues by combining mixed model and meta-analytic approaches. PLoS Genet., 9, e1003491. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The full set of summary statistics for all SNPs (in addition to those with Bayes Factor ≥ 6 shared in Supplementary Tables) will be made available via the NIDDK IBD Genetics Consortium, the sponsor of the study, and accessible at https://ibdgc.datacommons.io/.