Abstract
Identifying the potential associations between microbes and diseases is the first step for revealing the pathological mechanisms of microbe-associated diseases. However, traditional culture-based microbial experiments are expensive and time-consuming. Thus, it is critical to prioritize disease-associated microbes by computational methods for further experimental validation. In this study, we proposed a novel method called MNNMDA, to predict microbe-disease associations (MDAs) by applying a Matrix Nuclear Norm method into known microbe and disease data. Specifically, we first calculated Gaussian interaction profile kernel similarity and functional similarity for diseases and microbes. Then we constructed a heterogeneous information network by combining the integrated disease similarity network, the integrated microbe similarity network and the known microbe-disease bipartite network. Finally, we formulated the microbe-disease association prediction problem as a low-rank matrix completion problem, which was solved by minimizing the nuclear norm of a matrix with a few regularization terms. We tested the performances of MNNMDA in three datasets including HMDAD, Disbiome, and Combined Data with small, medium and large sizes respectively. We also compared MNNMDA with 5 state-of-the-art methods including KATZHMDA, LRLSHMDA, NTSHMDA, GATMDA, and KGNMDA, respectively. MNNMDA achieved area under the ROC curves (AUROC) of 0.9536 and 0.9364 respectively on HDMAD and Disbiome, better than the AUCs of compared methods under the 5-fold cross-validation for all microbe-disease associations. It also obtained a relatively good performance with AUROC 0.8858 in the combined data. In addition, MNNMDA was also better than other methods in area under precision and recall curve (AUPR) under the 5-fold cross-validation for all associations, and in both AUROC and AUPR under the 5-fold cross-validation for diseases and the 5-fold cross-validation for microbes. Finally, the case studies on colon cancer and inflammatory bowel disease (IBD) also validated the effectiveness of MNNMDA. In conclusion, MNNMDA is an effective method in predicting microbe-disease associations.
Availability
The codes and data for this paper are freely available at Github https://github.com/Haiyan-Liu666/MNNMDA.
Keywords: Microbe-disease association; Matrix nuclear norm; Gaussian interaction profile kernel similarity; Functional similarity, heterogeneous information network
Graphical Abstract
In this study, we proposed a novel method called MNNMDA, to predict microbe-disease associations (MDAs) by applying a minimizing Matrix Nuclear Norm method into known microbe and disease data. Specifically, we explored literatures and related databases to obtain high-quality known relationship between microbes and diseases. By incorporating multiple similarity networks between microbes and diseases, we formulated the microbe-disease association prediction problem as a low-rank matrix completion problem, which was solved by minimizing the nuclear norm of a matrix with a few regularization terms.
1. Introduction
Small in size and simple in structure, human microbe or microorganism is a category of tiny living organisms reproducing quickly and distributing widely [1], [2]. Human microbes are distributed in and on various organs of the human body, such as oral cavity, skin, gut, lung, gastrointestinal tract, and so on. Gut is called the largest microecosystem in the human body, since 80% human microbes are distributed in it [3], [4].
More and more studies have proved that microbes play important roles in the development, treatment and prognosis of many diseases, such as neuronal diseases and cancers [5], [6], [7]. Identifying disease-related microbes not only help reveal the pathological mechanism of human diseases, but also provide potential biomarkers for disease diagnosis and prognosis. Although biological experiment is the gold standard to confirm disease-microbe associations, it is a long process with a high cost. Computational models can prioritize confident disease-associated microbes for further experimental validation.
Recently, machine learning technologies have been widely used and achieved remarkable performances in association predictions, such as lncRNA-disease association prediction [8], [9], [10], [11], [12], [13], drug repositioning [14], [15], [16], [17], [18], [19], miRNA-disease association prediction [20], [21], [22], [23] and so on. Simultaneously, many computational methods have been developed to help identify the relationship between microbes and diseases. For instance, Chen et al. proposed the first KATZ-based computation method called KATZHMDA for predicting microbe-disease associations in 2016 [14]. Wang et al. proposed a LRLSHMDA method based on Laplacian regularized least squares framework in 2017, which used semi-supervised computational models to predict microbial-disease associations without negative samples, and achieved good results [24]. Shi et al. proposed a method called BMCMDA based on binary matrix completion to predict potential MDAs in 2018 [25]. Li et al. proposed a KATZBNRA method based on the KATZ model and bipartite network recommendation algorithm to discover potential associations between microbes and diseases in 2019 [26]. Yan et al. proposed a prediction method BRWMDA based on bi-random walk to predict potential MDAs in 2020 [27]. Liu et al. developed a multi-component Graph Attention Network based framework (MGATMDA) for predicting microbe-disease associations in 2021 [28]. Xu et al. developed a novel computational method (MDAKRLS) to discover potential MDAs based on the Kronecker regularized least squares in 2021 [29]. Hua et al. proposed a multi-view graph convolutional network (MVGCNMDA) to reveal disease-associated microbes using specific data augmentation and multi-view attention blocks in 2022 [30]. Chen et al. proposed a method based on heterogeneous network and metapath aggregated graph neural network (MATHNMDA) to predict MDAs in 2022 [31]. The above computational methods mainly utilized a basic assumption that microbes with similar functions will share similar non-interaction or interaction patterns with phenotypical diseases [32], [33].
Matrix factorization and matrix completion techniques are useful tools and have been widely applied to association prediction [34], [35], [36], [37], [38]. Yang et al. proposed to use a bounded nuclear norm regularization (BNNR) method to complete the drug-disease matrix under the low-rank assumption [39]. Liang et al. proposed a novel antiviral drug repositioning DRMNN method, which formulated the drug repositioning problem as a low-rank matrix completion problem solved by minimizing the nuclear norm of a matrix with a few regularization terms [40].
In this study, we proposed a novel matrix decomposition-based method to predict the association between microbes and diseases. Specifically, we first collected and downloaded data about microbes and diseases from the literature. Second, in order to mine more valuable similarity information, we employed different methods to explore the similarity of microbes and diseases. Third, we constructed a heterogeneous microbe-disease network, which integrates the microbe similarity network, disease similarity network and the microbe-disease association data. Fourth, we used the nuclear norm minimization method to obtain the microbes most likely to be involved in the pathogenesis of a disease. Finally, experimental results and case studies showed that MNNMDA is significantly better than the baseline methods on the HMDAD, Disbiome and Combined Dataset (Peryton and MicroPhenoDB).
2. Materials and methods
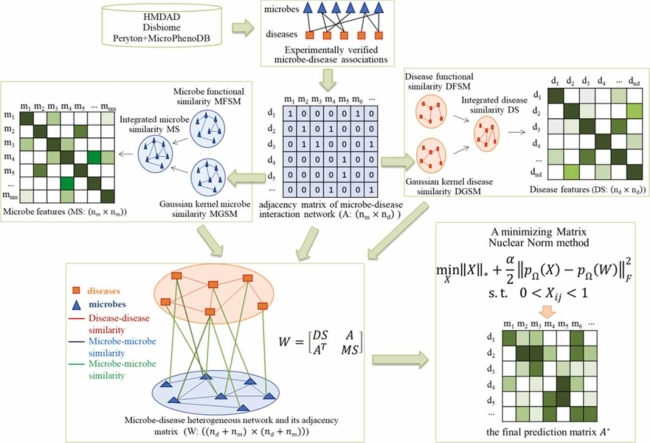
The workflow of MNNMDA for inferring novel MDAs was illustrated in Fig. 1. First, we explored literatures and related databases to obtain high-quality known relationship between microbes and diseases. Second, we calculated multiple similarity networks between microbes and diseases. Third, we built a heterogeneous network based on multi-view features of microbes and diseases. Finally, we applied the minimizing matrix nuclear norm method on the heterogeneous network to calculate the final possibility score of each microbe-disease pair.
Fig. 1.
The overall workflow of MNNMDA.
2.1. Materials
To establish the human microbe-disease interaction network, we retrieve known microbe-disease associations from the Human microbe-disease Association Database (HMDAD) (http://www.cuilab.cn/hmdad), which included 483 experimentally confirmed microbe-disease associations between 39 diseases and 292 microbes [41]. In HMDAD, a microbe-disease pair may include multiply entries from different evidence. Here we consider the same microbe-disease associations from different evidence as a pair. Subsequently, we obtain 450 various associations after removing the superfluous associations. In addition, Janssens et al. released a new microbe-disease association database named Disbiome (https://disbiome.ugent.be/home), where 5573 experimentally confirmed human microbe-disease associations were collected from previously published literature and different databases, including 240 diseases and 1098 microbes [42]. In Disbiome, a microbe-disease pair may be recorded more than one time according to different detection methods. Here we neglect the information of detection methods. After filtering out repetitive data, we finally download 4351 associations between 218 diseases and 1052 microbes. In addition, to evaluate the ability of MNNMDA in handling comprehensive data from multiple datasets, we referred as “Combined data” the dataset compiled by Liu et al. [28], using two different datasets Peryton [43] and MicroPhenoDB [44]. Peryton has 43 diseases and 1396 microbes with 7900 microbe-disease associations; MicroPhenoDB is a collection of microbe-disease association datasets such as HMDAD, Disbiome, VFDB [45], and CARD [46]. Among them, MicroPhenoDB contains 5511 associations between 1774 microbes and 500 diseases. After removing redundant data, 9600 associations were obtained, including 2456 microbes and 537 diseases. Overall, the specific statistics of the three microbe-disease association datasets were shown in Table 1.
Table 1.
The statistics for three microbe–disease association datasets used in this study.
| Dataset | # Microbes | # Diseases | # Associations |
|---|---|---|---|
| HMDAD | 292 | 39 | 450 |
| Disbiome | 1052 | 218 | 4351 |
| Combined Data | 2546 | 537 | 9660 |
We formatted an adjacency matrix of the known human microbe-disease interaction network as , that is, if there exists the experimentally verified relationship between the disease and the microbe , equals to 1, otherwise 0. The association between diseases and microbes are represented as a binary matrix where:
| (1) |
The elements with the values of 1 in are microbe-disease association data and taken as positive samples. The zero entities in are unknown microbe-disease pairs and taken as unlabeled samples.
2.2. Functional similarity of microbes
In this study, we calculated microbe functional similarity based on the method proposed by Kamneva et al. [47]. In order to accurately calculate the functional similarity for a given pair of microbes, we first retrieved protein-protein functional association network from STRING v11 database (https://string-db.org.). More details about the calculation of microbe functional similarity could be found in [47]. We adopt the similarity scores to obtain a × microbe functional similarity matrix MFSM, where MFSM () represents the similarity between microbe and microbe .
2.3. Functional similarity of diseases
Based on the assumption that similar diseases tend to interact with similar genes [48], [49], we calculated disease functional similarity based on the functional associations between disease-related genes. The latest released HumanNet v2.0 database (https://www.inetbio.org/humannet/download.php) is available for effectively accessing gene interactions [50], [51], where each interaction has an associated log-likelihood score that evaluates the probability of a functional linkage between genes. For a disease pair and , we first derived their related gene sets and respectively, where is the number of genes in and is the number of genes in . We define the functional association between gene and gene set as follows:
| (2) |
Where represents the functional similarity score between genes, which is defined as follows:
| (3) |
Where is the normalized of genes, which is defined as follows:
| (4) |
Where and denote the maximum and minimum in HumanNet, respectively.
Finally, we formulated the disease functional similarity as:
| (5) |
2.4. Gaussian interaction profile kernel similarity for microbes
We calculate the microbe Gaussian interaction profile kernel similarity via known experimentally confirmed human microbe-disease association network, which based on the assumption that functionally similar diseases generally tend to present interaction or non-interaction patterns with similar microbes. Specifically, the Gaussian interaction profile kernel similarity of microbe and can be defined as follows [52]:
| (6) |
Where represents the interaction between the microbe and each disease, represents the normalized kernel bandwidth and is defined as follows:
| (7) |
Where is the original bandwidth that affects . For the sake of simplicity, we set according to previous relevant study [14]. represents the total number of microbes. at the row and column denotes the similarity between microbe and .
2.5. Gaussian interaction profile kernel similarity for diseases
Similarly, the Gaussian interaction profile kernel similarity of diseases and can be computed by (8), (9):
| (8) |
| (9) |
Where represents the interaction between the disease and each microbe, represents the normalized kernel bandwidths, and represents the total number of diseases. Again, we set the original bandwidth to 1 based on previous research experience [14]. at the row and column denotes the similarity between disease and . It should be noted that in each cross-validation experiment, the similarities of diseases and microbes will be recalculated.
2.6. Integrated similarities for diseases and microbes
It is worth noticing that not all diseases have known associated genes. If a given disease have no related genes, we cannot calculate the functional similarity scores between the disease and other diseases. Therefore, we established a new similarity network for diseases by integrating multiple disease similarity networks calculated from different perspectives, namely the disease Gaussian interaction profile kernel similarity and the disease functional similarity. Specifically, the integrated disease similarity can be defined as follows:
| (10) |
Similarly, the integrated microbe similarity was calculated as follows:
| (11) |
2.7. Building a heterogeneous network
We constructed a human microbe-disease interaction network by using human microbes and diseases interaction associations, a microbe similarity network and a diseases similarity network. Then, the heterogeneous human microbe-disease network can be expressed as a bipartite graph , where M represented all of microbes, D represents all of diseases, and E represents the interaction of microbes and microbes, diseases and diseases, and microbes and diseases. The heterogeneous network is represented by adjacency matrix . By the similarity between the microbes (MS) and the similarity between the diseases (DS), the coefficients of similarity can construct a heterogeneity network.
| (12) |
The submatrix represents the relationship network between the microbe and the disease, is the transposition of , the adjacency matrix of the and are the disease network and the microbe network, and their weights are set to the similarity of the paired microbe and disease, and the range is [0,1].
2.8. MNNMDA for predicting microbe-disease associations
The nuclear norm is the sum of the singular values of the matrix, which is used to constrain the low rank of the matrix. For sparse data, the matrix has a low rank and contains a lot of redundant information, which can be used to recover data and extract features. The nuclear norm has been widely used in various fields and has achieved good results [39]. Generally, when a matrix has a low rank, the kernel norm minimization problem can be expressed as:
| (13) |
Where represents the kernel norm of , which is defined as the sum of all singular values of . The kernel norm minimization model is a convex optimization problem.
In order to predict the microbe-disease association, the elements in the microbe similarity matrix and the disease similarity matrix are in the interval [0,1]. The elements in the correlation matrix A are 0 or 1. The predicted value of the unknown entry is expected to be in the range of [0,1], where a predicted value close to 1 suggests that it may be indicative of an association and vice versa. However, in the above matrix completion model (12), the entries in the completed matrix can be any real values in (, ). However, it has no practical significance for values greater than 1 and less than 0. Therefore, it is important to add a constraint to the matrix completion model to ensure that the missing elements that are not found are in the interval [0,1]. In addition, because there may be a lot of “noise” data in the microbe and disease data, the microbe-disease relocation model should tolerate the potential noise as much as possible. The noise-tolerant matrix completion model is:
| (14) |
Where is the measurement noise level, is a set of index pairs () containing all known entries in , and is the projection operator on .
| (15) |
However, for this model with inequality constraints, there are many difficulties in solving. For example: how to choose the appropriate model parameters and how to choose an effective solution algorithm. Therefore, we usually replace the inequality constraint model with a regularized model. The introduction of soft regularization can not only tolerate unknown noise, but also provide a lot of convenience for us to solve. Then the model can be rewritten as the following:
| (16) |
Where denotes the Frobenius norm, is the parameter that balances the nuclear specification and the error term. To solve the optimization problem in Eq. (16), we chose the more classic alternating direction multiplier method (ADMM) [53]. It is worth noting that our objective function is convex. With the introduction of the auxiliary matrix , the ADMM framework can be optimized in the following equivalent form.
| (17) |
Therefore, the enhanced Lagrange function becomes the following:
| (18) |
where is the Lagrange multiplier and is the penalty parameter. The solution process of MNNMDA belongs to iterative solution. Therefore, when we iterate times, we need to calculate the value of iterations , and according to the result of the iteration.
Update: Repeat the following steps until convergence or reaching a predetermined number of iterations. Fix and and calculate a matrix to minimize Eq. (18).
| (19) |
Here, is the optimal solution of , if and only if holds,
| (20) |
where represents the adjoint operator of . Then, the closed solution becomes
| (21) |
where is the identity operator. denotes the inverse operator of and is equal to . It’s worth noting that . Considering the interval [0,1] constraint, we limit the range of the elements of to [0,1] such that
| (22) |
Fix and and calculate a matrix to minimize Eq. (18).
| (23) |
where is the singular value shrinkage operator defined as:
| (24) |
Where is the singular values of which is larger than , while and are the left and right singular vectors corresponding to , respectively.
Fix and and calculate a matrix .
| (25) |
where is the learning rate, which is set to 1 in this study for simplicity. Iterate according to the above iteration rules until convergence, and finally, we obtain the matrix after convergence. Therefore, after supplying the adjacency matrix of the microbe-disease heterogeneous network to MNNMDA, we can obtain an updated microbe-disease association matrix , where the unknown entries in are filled up, the final prediction matrix is:
| (26) |
The entries in with prediction scores close to 1 indicate the potential microbe-disease associations.
2.9. Evaluation methods
The performances of computational methods were evaluated by standard fivefold cross-validation (5-fold CV) under the following three different settings:
(i) 5-fold CVS1 (overall testing): CV on microbe-disease pairs-random known entries in A (i.e., microbe-disease pairs) are selected for testing.
(ii) 5-fold CVS2 (horizontal testing for diseases): CV on diseases-random rows in A (i.e., diseases) are blinded for testing.
(iii) 5-fold CVS3 (vertical testing for microbes): CV on microbes-random columns in A (i.e., microbes) are blinded for testing.
For 5-fold CVS1, we randomly divide all known microbe-disease associations into five equal and uncrossed groups. For each round, one group of microbe-disease associations (i.e., positive samples) with an equal-ratio set of unknown randomly sampled microbe-disease pairs (i.e., negative samples) are selected as test samples in turn. And the remaining four groups of microbe-disease associations together with the rest of unknown microbe-disease pairs are utilized to train the model. Specifically, all test samples would first obtain their prediction scores in each round and then be prioritized according to their scores. For a positive (or negative) test sample (microbe-disease pair), we consider that the model successfully predicts the microbe-disease pairs if its ranking is higher (or lower) than a specific threshold. As such, we could obtain the corresponding precision, true positive rates (TPR, sensitivity/recall) and false positive rates (FPR, 1-specificity) by setting different thresholds. Here, precision measures the percentage of the positive test samples among all samples that are predicted as positive with the given threshold. Sensitivity/recall is defined as the percentage of the positive test samples whose rankings are higher than the given threshold. Specificity means the percentage of the negative test samples that are ranked lower than the given threshold. Subsequently, the receiver operating characteristics (ROCs) curves and precision-recall (PR) curves could be drawn by plotting TPR versus FPR and precision versus recall at different thresholds, respectively. The performance is measured by area under ROC curve (AUC) and area under PR curve (AUPR). To reduce the influence of random division, each experiment is repeatedly conducted for 10 times. And the final AUC and AUPR scores are calculated over the average of 10 repetitions. Similarly, for 5-fold CVS2 and 5-fold CVS3, we randomly sample 20 % rows and columns in the adjacent matrix A as test samples, while the rest of the rows and columns are considered as training samples, respectively. It should be noted that 5-fold CVS2 and 5-fold CVS3 are set to predict novel microbe-disease associations for new diseases and new microbes, respectively.
3. Results
We demonstrated the performance of MNNMDA by comparing it with five methods from previous studies on the three data sets (i.e., HMDAD, Disbiome and Combined Data) under three different 5-fold CV settings (i.e., 5-fold CVS1, 5-fold CVS2, and 5-fold CVS3). In addition, we use AUPR and AUC values as indicators to assess the performance of MNNMDA. We also implemented case studies on two common diseases to further confirm the effectiveness of our method in predicting potential disease-related microbes.
3.1. Parameter tuning
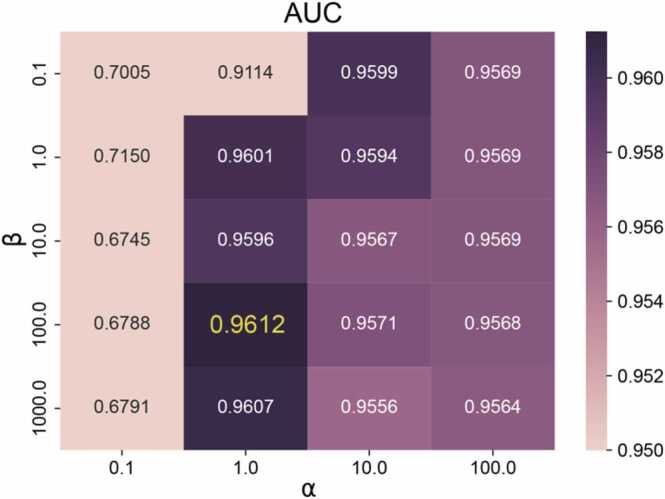
In this section, we investigated the influence of two important parameters in Formula (18), the balances the nuclear specification and the error term α and the penalty parameter β. We conducted two parameter combination experiments on the HMDAD dataset to select the optimal parameter combination of the MNNMDA method. First, we tuned from the list and tuned from the list , then searched the best values on the 4 × 5 grid expanded by both and . We recorded the performance of MNNMDA for each pairwise value of (, ) under 5-fold CVS1 in terms of AUC (Fig. 2). Finally, we picked up the pair of (, ) = (1, 100), which achieves the highest one among 20 values of AUC, as the best value of (, ), and further applied them in all the following experiments.
Fig. 2.
The AUC values on different and for the 5-fold CVS1 on HMDAD dataset.
3.2. MNNMDA is robust to fake associations
We assessed the robustness of our method on the HMDAD dataset by manually adding a few fake associations. Specifically, we randomly selected a predefined percentage of unknown associations and converted their labels to known associations. The percentage varies from 1 % to 5 %. The 5-fold CVS1 is then implemented on the new known microbe-disease associations to evaluate the performance of our method. The results were shown in Table 2, from which we can see that the AUCs and the AUPRs are stable with 5 % fake associations. It indicates that our method is robust to a small percentage of fake associations.
Table 2.
The robustness of MNNMDA.
| Percentage | AUC | AUPR |
|---|---|---|
|
0% 1% 2% 3% 4% 5% |
0.958 0.939 0.940 0.926 0.918 0.916 |
0.662 0.630 0.674 0.685 0.686 0.681 |
3.3. Comparison with previous methods
To evaluate the performance of our proposed method, we compare MNNMDA with the following state-of-the-art methods on the same data set.
KATZHMDA [14] is a KATZ-based computational method, which calculated based on the number and length of paths between two nodes in microbe-disease heterogeneous network.
LRLSHMDA [24] is a semi-supervised learning calculation model based on Laplacian regularized least squares classification.
NTSHMDA [54] is a computational model based on random walk and network topology similarity to predict the associations between microbes and diseases.
GATMDA [51] is a computational framework, which combines inductive matrix completion and graph attention networks to complete the prediction task.
KGNMDA [55] is a knowledge graph neural network method for predicting microbe-disease associations.
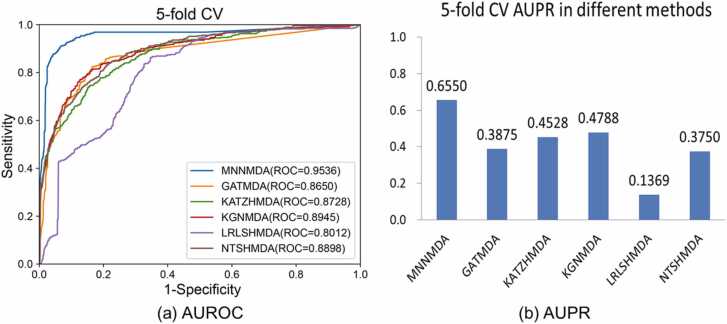
Our method was compared with five baselines under 5-fold CVS1 on the HMDAD dataset. We used the AUC value and AUPR value as indicators to evaluate each method. For better visual comparison, the corresponding ROC curves and AUPR values of MNNMDA, KATZHMDA, LRLSHMDA, NTSHMDA, GATMDA, and KGNMDA were shown in Fig. 3 Obviously, MNNMDA achieves the best performance compared to the other five methods, with an average AUC value of 0.9536 and an average AUPR is 0.6550. The results indicated that our method is effective in predicting novel microbe-disease associations.
Fig. 3.
Comparing the ROC curves and AUPR values of MNNMDA and other five methods based on 5-fold CVS1 on the HMDAD dataset.
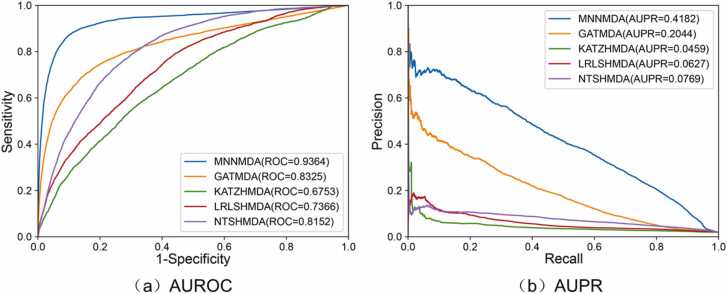
To further evaluate the effectiveness of our model, we conduct experiments under 5-fold CV on the Disbiome dataset and the Combined dataset (Peryton and MicroPhenoDB). We compare our method with four baseline methods on the data set Disbiome. The results under the 5-fold CVS1 were shown in Fig. 4, which demonstrated that our method consistently outperforms the four baseline methods with an average AUC of 0.9364 and an average AUPR of 0.4182.
Fig. 4.
Comparison of the ROC curves and PR curves of MNNMDA and four other methods under the 5-fold CVS1 on the Disbiome dataset.
The performances of MNNMDA under the 5-fold CVS1 on HMDAD, Disbiome, and Combined Data were summarized in Table 3. It can be seen that our method has the best AUC and AUPR on the HMDAD dataset. The main reason may be that the Disbiome and Combined Data are sparser than HMDAD. The density of HMDAD is 3.95 %, the density of the Disbiome is 1.90 %, and the density of combined data is 0.71 %. So the method MNNMDA can achieve relatively better training on HMDAD than Disbiome and Combined Data.
Table 3.
Performance of the MNNMDA method under 5-fold CVS1 on three datasets.
| Dataset | AUC | AUPR |
|---|---|---|
|
HMDAD Disbiome Combined Data |
0.9536 0.9364 0.8858 |
0.6550 0.4182 0.3014 |
To further verify the performance of our method, we made 5-fold CVS2 and 5-fold CVS3 on the HMDAD dataset. In addition, comparing our method with five baseline methods, and the results were shown in Table 4, it could be significantly discovered from Table 4 that our method is slightly superior to other baseline methods in terms of AUC and AUPR under the settings 5-fold CVS3. Overall, our method is able to identify novel microbe-disease associations under different scenarios.
Table 4.
Performance comparison between five baseline methods and our method under the 5-fold CVS2 and 5-fold CVS3 on HMDAD data set.
| Methods | 5-fold CVS2 |
5-fold CVS3 |
||
|---|---|---|---|---|
| AUC | AUPR | AUC | AUPR | |
|
KATZHMDA LRLSHMDA NTSHMDA GATMDA KGNMDA MNNMDA |
0.6573 0.6568 0.7231 0.6000 0.7541 0.7250 |
0.1579 0.1637 0.0861 0.1702 0.1534 0.4214 |
0.9079 0.9094 0.9111 0.8683 0.9093 0.9737 |
0.6579 0.6588 0.6568 0.6442 0.6582 0.9575 |
From all CV results, we observe that the AUC value of MNNMDA method under 5-fold CVS3 setting is significantly better than that under 5-fold CVS1and 5-fold CVS2 settings. For new diseases and new microbes, we have no known association pairs for them to train the model, which results in lower AUC value under 5-fold CVS1 and 5-fold CVS2. Furthermore, various methods achieve generally better performance under 5-fold CVS3 than under 5-fold CVS2 on HMDAD dataset. As the number of microbes (292) is much more than that of diseases (39), the microbe similarity matrix (292 × 292) is thus more informative than the disease similarity matrix (39 × 39). Therefore, new microbes can obtain more abundant and accurate information from neighbors than new diseases.
3.4. Case study
In this section, we select two common diseases of colon cancer and inflammatory bowel disease (IBD) for case studies on the HMDAD dataset to further verify the predictive ability of the MNNMDA method. For each disease, microbes that have known associations with the disease are first removed. Then the predicted scores of candidate microbes are sorted in descending order according to the MNNMDA method. Finally, we verify whether the top 10 microbes associated with the disease are confirmed by the previous publications.
Colon cancer, a common malignant digestive tract tumor, has a high incidence ranking second on the list of digestive tract tumor, greatly threatening human life and health [29], [56], [57], [58]. More and more evidences show that the occurrence and development of colon cancer is closely associated with the imbalance of microbial community [59]. We applied MNNMDA to the case study of colon cancer. Of the top 10 predicted microbes, 8 have been validated based on existing biological literature (see Table 5). For example, Oxalobacter formigenes might stand for a pathogenic factor in colon cancer, when antibiotics are prescribed generously [60]. Helicobacter pylori infection was found to be considered as a risk increase factor of left-sided colon cancer [61], [62]. Typically, it is reported that colon cancer patients aged 60 years or older who have inserted a metal stent preoperatively are identified as a risk factor for Clostridium difficile infection [63], [64] Staphylococcus aureus is a kind of tannase-producing bacteria, its activity may be related to the development of colon cancer [65]. The high prediction accuracy rates demonstrate that our model could be used in real-life applications.
Table 5.
The top 10 potential microbes related to colon cancer identified by MNNMDA.
| Disease | Microbes | Rank | Evidence |
|---|---|---|---|
| Colon cancer | Helicobacter pylori | 1 | PMID: 22294430 PMID: 30636955 |
| Oxalobacter formigenes | 2 | PMID: 32880090 | |
| Tropheryma whipplei | 3 | Unconfirmed | |
| Prevotella copri | 4 | PMID: 35682578 | |
| Corynebacterium | 5 | PMID: 31609493 | |
| Staphylococcus aureus | 6 | PMID: 26198124 | |
| Clostridium difficile | 7 | PMID: 29245331 PMID: 21152135 |
|
| Desulfovibrio | 8 | PMID: 19496818 | |
| Helicobacter pylori | 9 | PMID: 30636955 PMID: 22294430 |
|
| Dietzia maris | 10 | Unconfirmed |
In addition, the main types of inflammatory bowel disease (IBD) include Crohn's disease and ulcerative colitis, which is caused in part by bacteria that may activate the patient's immune system to attack foreign substances. Once the patient's immune system is activated, it is difficult to regulate and destroy the gastrointestinal tract, resulting in IBD symptoms [66]. Recent researches have shown that a wide range of microbes are closely associated with IBD. MNNMDA method was used to calculate the top 10 microorganisms with the highest correlation row scores and analyze whether they were related to IBD. In Table 6, 8 of them have been verified to be related to IBD in the studies. Clostridium coccoides are less represented in A-IBD patients [67]. Lachnospiraceae and other taxa identified as translocating bacteria or targets of systemic immunity in IBD concomitantly exhibited heightened transcriptional activity and growth rates in IBD patient gut microbiomes [68]. The changes in the abundance of Proteobacteria of IBD patients are not only related to current activity but also to the course of the disease [69]. Research shows a significant relationship between Staphylococcus and IBD [70]. In a word, these two sets of case studies validate the powerful capability of MNNMDA in inferring new possible microbes for diseases again.
Table 6.
The top 10 potential microbes related to inflammatory bowel disease (IBD) identified by MNNMDA.
| Disease | Microbes | Rank | Evidence |
|---|---|---|---|
| IBD | Actinobacteria | 1 | Unconfirmed |
| Proteobacteria | 2 | PMID: 31530835 | |
| Lachnospiraceae | 3 | PMID: 35976997 | |
| Propionibacterium | 4 | PMID: 19847949 | |
| Propionibacterium acnes | 5 | PMID: 28630242 | |
| Pseudomonas | 6 | PMID: 31662859 | |
| Staphylococcus | 7 | PMID: 27239107 PMID:23679203 |
|
| Clostridium coccoides | 8 | PMID: 27687331 | |
| Verrucomicrobiaceae | 9 | PMID: 22572638 | |
| Oxalobacteraceae | 10 | Unconfirmed |
4. Discussion
Identifying microbe-disease associations is a hot topic, which can not only provide great insight into understanding the complex pathogenic mechanism of human diseases, but also reduces the cost and time of biological experiments. For example, systematic identification of potential pathological microorganisms can help doctors or biologists to identify biomarkers for diagnosis and treatment clinically or experimentally [7], [71], [72], [73], especially for complex human diseases. In addition, computational prediction of pathogenic microorganisms can help pharmacologists or biologists effectively narrow down the field of candidate compounds [74], [75]. This may further guide them in planning experiments and thus reducing costs. Therefore, a series of related information databases have been established. Based on the data in these information databases, more and more computational models have been proposed and applied to the field of microbe-disease association prediction, and these prediction models have achieved remarkable results. However, previous computational methods suffer from two major challenges. On the one hand, few methods did not suffer from decrease of accuracy owing to the high rate of false positive and false negative samples in the microbe-disease association database. On the other hand, most of them did not systematically integrate the biological knowledge information of microbes and diseases. That may be not made reasonable predictions about new diseases or microbes.
The reliable performance of MNNMDA results from several major factors as follows: to begin with, the observed experimentally confirmed human microbe-disease associations are reliable. In addition, the integration of multiple prior biological informations about microbes and diseases complements and improves the microbe similarity and disease similarity, respectively, which potentially enhances the prediction capability of our method.
Although the good prediction performance of MNNMDA method, there are some limitations that are expected to be further improved in the future. For example, our method cannot be applied to make predictions for all new microbes and new diseases. It is because for a new disease without any known associated genes, and new microbes that are lack of protein-protein interaction information, it fails to obtain its similarity between other diseases and it that is essential for new disease prediction. But this limitation could be overcome by collecting more prior information, such as microbe sequence similarity, disease gene-based similarity network and disease symptom similarity network or developing other effective similarity calculation method. In addition, there are too few known microbe-disease associations in the database. The data is imbalanced, leading to prediction deviation. Finally, though our model could predict potential disease-associated microbes, it still cannot determine how the microbial abundances influence disease status. We could further handle this problem by incorporating the microbial abundance information into the heterogeneous network.
5. Conclusion
An in-depth study of the microbe-disease relations will not only help doctors understand the complex pathogenic mechanism of human diseases, but also provide new insight for microbe-oriented medicine. In this work, we proposed a method based on the minimum nuclear specification, named MNNMDA, for human microbe-disease association prediction. We fully exploit multiply sources of biological data to construct similarity features for diseases and microbes. MNNMDA not only can restrict all predicted matrix entry values within a specific interval, but also exhibit robustness to tolerate potentially noise similarity calculations. We compared our method with five state-of-the-art methods based on the database HMDAD. MNNMDA achieved the average AUC value of 0.9536 based on the 5-fold CV. In addition, MNNMDA achieved good results in two common human diseases: colon cancer and IBD. Among the predicted top 10 microbe candidates, 8 microbes were confirmed by the previous research literature, respectively. Overall, MNNMDA is effective and promising in identifying potential microbe-disease associations.
Funding
This study was supported by Hunan Key Laboratory Cultivation Base of the Research and Development of Novel Pharmaceutical Preparations (No.2016TP1029), Hunan Provincial Innovation Platform and Talents Program (No. 2018RS3105), the Foundation of Hunan Educational Committee (Grant No. 19A060), the Provincial Key R & D Projects of Hunan Provincial Science and Technology Department (No. 2022SK2074), and the Training Plan for Young Backbone Teachers in Hunan Province.
CRediT authorship contribution statement
Haiyan Liu (First Author): Methodology, Formal analysis, and Writing – Original Draft; Pingping Bing: Methodology and Writing – Review & Editing; Meijun Zhang: Data Curation, Writing - Original Draft; Geng Tian: Writing – Review & Editing; Jun Ma: Visualization, Investigation; Haigang Li: Software, Validation; Meihua Bao: Visualization, Writing - Review & Editing Kunhui He: Writing – Review & Editing; Jianjun He (Corresponding Author): Conceptualization, Funding Acquisition, Resources, Writing - Review & Editing; Binsheng He (Corresponding Author): Supervision, Writing - Review & Editing; Jialiang Yang (Corresponding Author): Conceptualization, Resources, Writing – Review & Editing and Funding acquisition.
Declaration of Competing Interest
All authors disclosed no relevant relationships.
Contributor Information
Jianjun He, Email: hejianjun@csmu.edu.cn.
Binsheng He, Email: hbscsmu@163.com.
Jialiang Yang, Email: yangjl@geneis.cn.
References
- 1.Wu C., et al. PRWHMDA: human microbe-disease association prediction by random walk on the heterogeneous network with PSO. Int J Biol Sci. 2018;14(8):849–857. doi: 10.7150/ijbs.24539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng L., et al. gutMDisorder: a comprehensive database for dysbiosis of the gut microbiota in disorders and interventions. Nucleic Acids Res. 2020;48(D1):D554–D560. doi: 10.1093/nar/gkz843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gill S.R., et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312(5778) doi: 10.1126/science.1124234. 1355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng L., et al. gutMGene: a comprehensive database for target genes of gut microbes and microbial metabolites. Nucleic Acids Res. 2021 doi: 10.1093/nar/gkab786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zou S., Zhang J., Zhang Z. Novel human microbe-disease associations inference based on network consistency projection. Sci Rep. 2018;8(1):8034. doi: 10.1038/s41598-018-26448-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang H., et al. Prioritizing disease-related microbes based on the topological properties of a comprehensive network. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.685549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang M., et al. A multi-omics machine learning framework in predicting the survival of colorectal cancer patients. Comput Biol Med. 2022;146 doi: 10.1016/j.compbiomed.2022.105516. [DOI] [PubMed] [Google Scholar]
- 8.Yang X., et al. A network based method for analysis of lncRNA-disease associations and prediction of lncRNAs implicated in diseases. PLOS One. 2014;9(1) doi: 10.1371/journal.pone.0087797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng L., et al. DincRNA: a comprehensive web-based bioinformatics toolkit for exploring disease associations and ncRNA function. Bioinformatics. 2018;34(11):1953–1956. doi: 10.1093/bioinformatics/bty002. [DOI] [PubMed] [Google Scholar]
- 10.Du B., et al. Predicting LncRNA-disease association based on generative adversarial network. Current Gene Ther. 2021 doi: 10.2174/1566523221666210506131055. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J., Sun Q., Liang C. Prediction of lncRNA-disease associations based on robust multi-label learning. Curr Bioinform. 2021;16(9):1179–1189. [Google Scholar]
- 12.Xiao X., et al. BPLLDA: predicting lncRNA-disease associations based on simple paths with limited lengths in a heterogeneous network. Front Genet. 2018;9:411. doi: 10.3389/fgene.2018.00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W., et al. Inferring latent disease-lncRNA associations by faster matrix completion on a heterogeneous network. Front Genet. 2019;10:769. doi: 10.3389/fgene.2019.00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang W., et al. Predicting potential drug-drug interactions by integrating chemical, biological, phenotypic and network data. BMC Bioinform. 2017;18(1):18. doi: 10.1186/s12859-016-1415-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meng Y., et al. A weighted bilinear neural collaborative filtering approach for drug repositioning. Brief Bioinform. 2022 doi: 10.1093/bib/bbab581. [DOI] [PubMed] [Google Scholar]
- 16.Yang J., et al. Human geroprotector discovery by targeting the converging subnetworks of aging and age-related diseases. Geroscience. 2020;42(1):353–372. doi: 10.1007/s11357-019-00106-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu C., et al. An improved anticancer drug-response prediction based on an ensemble method integrating matrix completion and ridge regression. Mol Ther Nucleic Acids. 2020;21:676–686. doi: 10.1016/j.omtn.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang X., et al. Indicator regularized non-negative matrix factorization method-based drug repurposing for COVID-19. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.603615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai L., et al. Drug repositioning based on the heterogeneous information fusion graph convolutional network. Brief Bioinform. 2021;22(6) doi: 10.1093/bib/bbab319. [DOI] [PubMed] [Google Scholar]
- 20.Chen H., Zhang Z. Similarity-based methods for potential human microRNA-disease association prediction. BMC Med Genom. 2013;6:12. doi: 10.1186/1755-8794-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong J., et al. A five-gene signature for predicting the prognosis of colorectal cancer. Current Gene Ther. 2021;21(4):280–289. doi: 10.2174/1566523220666201012151803. [DOI] [PubMed] [Google Scholar]
- 22.Xu J., et al. LRMCMDA: predicting miRNA-disease association by integrating low-rank matrix completion with miRNA and disease similarity information. IEEE Access. 2020;8:80728–80738. [Google Scholar]
- 23.He B., et al. DGHNE: network enhancement-based method in identifying disease-causing genes through a heterogeneous biomedical network. Brief Bioinform. 2022;23(6) doi: 10.1093/bib/bbac405. [DOI] [PubMed] [Google Scholar]
- 24.Wang F., et al. LRLSHMDA: laplacian regularized least squares for human microbe-disease association prediction. Sci Rep. 2017;7(1):7601. doi: 10.1038/s41598-017-08127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi J.Y., et al. BMCMDA: a novel model for predicting human microbe-disease associations via binary matrix completion. BMC Bioinform. 2018;19(Suppl 9):281. doi: 10.1186/s12859-018-2274-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li S., Xie M., Liu X. A novel approach based on bipartite network recommendation and KATZ model to predict potential micro-disease associations. Front Genet. 2019;10:1147. doi: 10.3389/fgene.2019.01147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan C., et al. BRWMDA: predicting microbe-disease associations based on similarities and bi-random walk on disease and microbe networks. IEEE/ACM Trans Comput Biol Bioinform. 2020;17(5):1595–1604. doi: 10.1109/TCBB.2019.2907626. [DOI] [PubMed] [Google Scholar]
- 28.Dayun L., et al. MGATMDA: predicting microbe-disease associations via multi-component graph attention network. IEEE/ACM Trans Comput Biol Bioinform. 2021 doi: 10.1109/TCBB.2021.3116318. [DOI] [PubMed] [Google Scholar]
- 29.Xu D., et al. MDAKRLS: predicting human microbe-disease association based on Kronecker regularized least squares and similarities. J Transl Med. 2021;19(1):66. doi: 10.1186/s12967-021-02732-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hua M., et al. MVGCNMDA: multi-view graph augmentation convolutional network for uncovering disease-related microbes. Interdiscip Sci. 2022;14(3):669–682. doi: 10.1007/s12539-022-00514-2. [DOI] [PubMed] [Google Scholar]
- 31.Chen Y., Lei X. Metapath aggregated graph neural network and tripartite heterogeneous networks for microbe-disease prediction. Front Microbiol. 2022;13 doi: 10.3389/fmicb.2022.919380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bao W., Jiang Z., Huang D.-S. Novel human microbe-disease association prediction using network consistency projection. BMC Bioinform. 2017;18(S16) doi: 10.1186/s12859-017-1968-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L., et al. A bidirectional label propagation based computational model for potential microbe-disease association prediction. Front Microbiol. 2019;10:684. doi: 10.3389/fmicb.2019.00684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hao Y.-J., et al. Application of a deep matrix factorization model on integrated gene expression data. Current Bioinform. 2020;15(4):359–367. [Google Scholar]
- 35.Qiu Y., Ching W.-K., Zou Q. Matrix factorization-based data fusion for the prediction of RNA-binding protein and alternative splicing event associations during epithelial-mesenchymal transition. Brief Bioinform. 2021;22(6) doi: 10.1093/bib/bbab332. [DOI] [PubMed] [Google Scholar]
- 36.Ding Y., et al. Identification of drug-target interactions via multiple kernel-based triple collaborative matrix factorization. Brief Bioinform. 2022;23(2) doi: 10.1093/bib/bbab582. [DOI] [PubMed] [Google Scholar]
- 37.Xu J., et al. CMF-Impute: an accurate imputation tool for single-cell RNA-seq data. Bioinformatics. 2020;36(10):3139–3147. doi: 10.1093/bioinformatics/btaa109. [DOI] [PubMed] [Google Scholar]
- 38.Huang L., et al. Matrix completion with side information and its applications in predicting the antigenicity of influenza viruses. Bioinformatics. 2017;33(20):3195–3201. doi: 10.1093/bioinformatics/btx390. [DOI] [PubMed] [Google Scholar]
- 39.Yang M., et al. Drug repositioning based on bounded nuclear norm regularization. Bioinformatics. 2019;35(14):i455–i463. doi: 10.1093/bioinformatics/btz331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang H., et al. Repositioning drugs on human influenza a viruses based on a novel nuclear norm minimization method. Front Physiol. 2020;11 doi: 10.3389/fphys.2020.597494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma W., et al. An analysis of human microbe-disease associations. Brief Bioinform. 2017;18(1):85–97. doi: 10.1093/bib/bbw005. [DOI] [PubMed] [Google Scholar]
- 42.Janssens Y., et al. Disbiome database: linking the microbiome to disease. BMC Microbiol. 2018;18(1):50. doi: 10.1186/s12866-018-1197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skoufos G., et al. Peryton: a manual collection of experimentally supported microbe-disease associations. Nucleic Acids Res. 2021;49(D1):D1328–D1333. doi: 10.1093/nar/gkaa902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao G., et al. MicroPhenoDB associates metagenomic data with pathogenic microbes, microbial core genes, and human disease phenotypes. Genom Proteom Bioinform. 2020;18(6):760–772. doi: 10.1016/j.gpb.2020.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen L., et al. VFDB 2016: hierarchical and refined dataset for big data analysis--10 years on. Nucleic Acids Res. 2016;44(D1):D694–7. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jia B., et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45(D1):D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kamneva O.K. Genome composition and phylogeny of microbes predict their co-occurrence in the environment. PLOS Comput Biol. 2017;13(2) doi: 10.1371/journal.pcbi.1005366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei H., Liu B. iCircDA-MF: identification of circRNA-disease associations based on matrix factorization. Brief Bioinform. 2020;21(4):1356–1367. doi: 10.1093/bib/bbz057. [DOI] [PubMed] [Google Scholar]
- 49.Xu J., Li Y. Discovering disease-genes by topological features in human protein-protein interaction network. Bioinformatics. 2006;22(22) doi: 10.1093/bioinformatics/btl467. 2800-5. [DOI] [PubMed] [Google Scholar]
- 50.Hwang S., et al. HumanNet v2: human gene networks for disease research. Nucleic Acids Res. 2019;47(D1):D573–D580. doi: 10.1093/nar/gky1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Long Y., et al. Predicting human microbe-disease associations via graph attention networks with inductive matrix completion. Brief Bioinform. 2021;22(3) doi: 10.1093/bib/bbaa146. [DOI] [PubMed] [Google Scholar]
- 52.van Laarhoven T., Nabuurs S.B., Marchiori E. Gaussian interaction profile kernels for predicting drug-target interaction. Bioinformatics. 2011;27(21) doi: 10.1093/bioinformatics/btr500. 3036-43. [DOI] [PubMed] [Google Scholar]
- 53.Candès E., Recht B. Simple bounds for recovering low-complexity models. Math Program. 2012;141(1–2):577–589. [Google Scholar]
- 54.Luo J., Long Y. NTSHMDA: prediction of human microbe-disease association based on random walk by integrating network topological similarity. IEEE/ACM Trans Comput Biol Bioinform. 2020;17(4):1341–1351. doi: 10.1109/TCBB.2018.2883041. [DOI] [PubMed] [Google Scholar]
- 55.Jiang C., et al. KGNMDA: a knowledge graph neural network method for predicting microbe-disease associations. IEEE/ACM Trans Comput Biol Bioinform. 2022 doi: 10.1109/TCBB.2022.3184362. [DOI] [PubMed] [Google Scholar]
- 56.Zhang L., et al. Analysis on regulatory network linked to Hpa gene in invasion and metastasis of colon cancer. Saudi J Biol Sci. 2017;24(3):504–507. doi: 10.1016/j.sjbs.2017.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu H., et al. Evaluating DNA methylation, gene expression, somatic mutation, and their combinations in inferring tumor tissue-of-origin. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.619330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He B., et al. TOOme: a novel computational framework to infer cancer tissue-of-origin by integrating both gene mutation and expression. Front Bioeng Biotechnol. 2020;8:394. doi: 10.3389/fbioe.2020.00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moore W.E.C., Moore L.H. Intestinal floras of populations that have a high risk of colon cancer. App Environ Microbiol. 1995;61(9):3202–3207. doi: 10.1128/aem.61.9.3202-3207.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ravikumar Y., Begum R.F., Velmurugan R. Oxalobacter formigenes reduce the risk of kidney stones in patients exposed to oral antibiotics: a case-control study. Int Urol Nephrol. 2021;53(1):13–20. doi: 10.1007/s11255-020-02627-3. [DOI] [PubMed] [Google Scholar]
- 61.Teimoorian F., et al. Association of helicobacter pylori infection with colon cancer and adenomatous polyps. Iran J Pathol. 2018;13(3):325–332. [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y., et al. Helicobacter pylori infection and colorectal cancer risk: evidence from a large population-based case-control study in Germany. Am J Epidemiol. 2012;175(5) doi: 10.1093/aje/kwr331. 441-50. [DOI] [PubMed] [Google Scholar]
- 63.Li B., et al. When omeprazole met with asymptomatic clostridium difficile colonization in a postoperative colon cancer patient: a case report. Medicine. 2017;96(49) doi: 10.1097/MD.0000000000009089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yeom C.H., et al. Risk factors for the development of clostridium difficile-associated colitis after colorectal cancer surgery. J Korean Soc Coloproctol. 2010;26(5) doi: 10.3393/jksc.2010.26.5.329. (329-33) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim H., et al. Inhibitory effect of lactobacillus plantarum extracts on HT-29 colon cancer cell apoptosis induced by staphylococcus aureus and its alpha-toxin. J Microbiol Biotechnol. 2015;25(11) doi: 10.4014/jmb.1504.04047. 1849-55. [DOI] [PubMed] [Google Scholar]
- 66.Lomax A.E., et al. Effects of gastrointestinal inflammation on enteroendocrine cells and enteric neural reflex circuits. Auton Neurosci. 2006;126–127 doi: 10.1016/j.autneu.2006.02.015. (250-7) [DOI] [PubMed] [Google Scholar]
- 67.Prosberg M., et al. The association between the gut microbiota and the inflammatory bowel disease activity: a systematic review and meta-analysis. Scand J Gastroenterol. 2016;51(12):1407–1415. doi: 10.1080/00365521.2016.1216587. [DOI] [PubMed] [Google Scholar]
- 68.Vujkovic-Cvijin I., et al. The systemic anti-microbiota IgG repertoire can identify gut bacteria that translocate across gut barrier surfaces. Sci Transl Med. 2022;14(658):eabl3927. doi: 10.1126/scitranslmed.abl3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vester-Andersen M.K., et al. Increased abundance of proteobacteria in aggressive Crohn's disease seven years after diagnosis. Sci Rep. 2019;9(1):13473. doi: 10.1038/s41598-019-49833-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kojima A., et al. Aggravation of inflammatory bowel diseases by oral streptococci. Oral Dis. 2014;20(4) doi: 10.1111/odi.12125. 359-66. [DOI] [PubMed] [Google Scholar]
- 71.Takahashi M.K., et al. A low-cost paper-based synthetic biology platform for analyzing gut microbiota and host biomarkers. Nat Commun. 2018;9(1):3347. doi: 10.1038/s41467-018-05864-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou Y., et al. Gut microbiota offers universal biomarkers across ethnicity in inflammatory bowel disease diagnosis and infliximab response prediction. mSystems. 2018;3(1) doi: 10.1128/mSystems.00188-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu T., Dai Q., He P.-A. Identification of potential immune-related biomarkers in gastrointestinal cancers. Curr Bioinform. 2021;16(9):1203–1213. [Google Scholar]
- 74.Barrows N.J., et al. A screen of FDA-approved drugs for inhibitors of zika virus infection. Cell Host Microbe. 2016;20(2) doi: 10.1016/j.chom.2016.07.004. (259-70) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou T., et al. High-content screening in hPSC-neural progenitors identifies drug candidates that inhibit zika virus infection in fetal-like organoids and adult brain. Cell Stem Cell. 2017;21(2):274–283. doi: 10.1016/j.stem.2017.06.017. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]