Abstract
Super-resolution imaging techniques have provided unprecedentedly detailed information by surpassing the diffraction-limited resolution of light microscopy. However, in order to derive high quality spatial resolution, many of these techniques require high laser power, extended imaging time, dedicated sample preparation, or some combination of the three. These constraints are particularly evident when considering three-dimensional (3D) super-resolution imaging. As a result, high-speed capture of 3D super-resolution information of structures and dynamic processes within live cells remains both desirable and challenging. Recently, a highly effective approach to obtain 3D super-resolution information was developed that can be employed in commonly available laboratory microscopes. This development makes it both scientifically possible and financially feasible to obtain super-resolution 3D information under certain conditions. This is accomplished by converting 2D single-molecule localization data captured at high speed within subcellular structures and rotationally symmetric organelles. Here, a high-speed 2D single-molecule tracking and post-localization technique, known as single-point edge-excitation sub-diffraction (SPEED) microcopy, along with its 2D-to-3D transformation algorithm is detailed with special emphasis on the mathematical principles and Monte Carlo simulation validation of the technique.
Keywords: SPEED microscopy, Super-resolution microscopy, Single-molecule microscopy, STORM
1. Introduction
Limited by the two-dimensional constraints of the detector, imaging three-dimensional structures and molecules in their native state has been a long-standing goal of light microscopists. Although several microscopy advancements have overcome this limitation through a mixture of optical and computational methodologies, such methods are both challenging and costly to implement. However, alternative techniques may be utilized to resolve these issues. Methods such as optical sectioning, which enable the derivation of axial (z-dimension) information by combining several lateral (XY) images at different axial positions, and post-imaging mathematical approaches of transforming two-dimensional data into virtual three-dimensional images by taking advantage of the physical properties of symmetrical structures. Here, we briefly review techniques available for deriving 3D data from 2D information, describe the key concepts for localizing single-molecules, and review the 2D-to-3D transformation algorithm application in different biological samples.
2. Approaches for 2D-to-3D image reconstruction
2.1. Optical sectioning
Optical sectioning is the process of serial imaging a sample at different axial positions then combining the images to create a 3D image. This methodology has provided significant insight into both clinical and foundational research problems. For example, 3D reconstruction via optical sectioning has been employed in clinical pathology to reconstruct 3D images of breast tissue. This enabled a detailed and comprehensive understanding of the nuclear structures for both benign and malignant regions of human breast tissue[1]. These techniques have also been widely employed in foundational research in a variety of models, such as studying the inner ear[2] and lens[3] of mice as well as muscle degradation in Drosophila melanogaster[4].
While many techniques utilize some form of optical sectioning, the most prominent techniques enabling the reconstruction of 3D information from 2D images are maximum intensity projection, orthographic projection, volume rendering, and surface reconstructions[5]. Maximum intensity projection is the most commonly employed of these techniques and utilizes raw 2D data to produce a single 3D image stack that consists of the maximum pixel value found at each discrete pixel location within the dataset. The maximum value pixels are then combined in a pointillistic manner to generate a 3D volumetric impression[6]. While these images provide valuable information, a common criticism is that images generated using this methodology lack a sense of depth; however, this can be corrected through the use of animations. The illusion of depth is achieved via animations where the sample can be rotated in space a small number of degrees, at which point the computer then recalculates the maximum pixel values at the new orientation. The perception of depth is thus achieved through a clever application of orientation based animation and recalculation[6]. Notably, axial resolution is reduced when the lateral plane rotates beyond 30–45° in the Z-dimension [6]. However, by orthographic projection, the Z-axis resolution may be conserved. Differing from maximum intensity projection, orthographic projection exploits the curvature of a spheroidal or semi- spheroidal sample to gain equatorial data by comparing it to the anterior region of the sample. As demonstrated in 2010 by S. Bassnett and Y. Shi in 2010, the authors implement orthographic projection microscopy to accurately quantify the cell density of a mouse’s lens epithelium[3]. A process that typically miscalculates the equatorial cell density when applying maximum intensity projection.
Another technique relying upon optical sectioning is volume rendering. This technique consists of assembling multiple 2D slices that undergo 3D reconstruction to form what is termed as a ‘volume model.’ In this system the X, Y, and Z points are labeled with the fluorescent intensity and are rendered as an image that visualizes the internal structure[7]. Volume renders are a relatively accurate representation of the underlying data and lead to fewer incorrect conclusions regarding localization[8]. Lastly, surface reconstruction is an optical sectioning method in which virtual 3D models are rendered from 2D sets of images[7]. The voxels of interest are assigned a range of intensity values where a marching cube algorithm is utilized to provide the 3D structure[6]. This methodology has been utilized to make several novel and impactful observations. As was previously mentioned, dorsal oblique deterioration in Drosophila melanogaster was observed in real time. This was accomplished via a 3D timelapse reconstruction of 2D images of the nucleus that enabled the visualization of the real time breaking down of the muscle[4].
The generation of a 3D reconstruction enables excellent analysis of samples, enabling a more granular understanding of cellular structures. Information derived from a 2D analysis alone is often insufficient to enable an in-depth analysis of either a cellular structure nor its dynamic processes. Despite the obvious advantages, this methodology is not without its challenges. Chiefly, the quality of a 3D reconstruction via optical sectioning is largely dependent on the axial sampling. Undersampling excludes or dampens pertinent information and produces aliasing artifacts[9], where oversampling unnecessarily extends the image acquisition time, increasing the probability of photobleaching. However, following the Nyquist-Shannon sampling theorem [10], [11] with regards to the correct number of images needed in the axial dimension considering the objective’s depth-of-field (DOF), the illumination and emission wavelengths, and the lens immersion and specimen medium refractive indices [12], will greatly improve the reconstruction’s likeness to the original sample. Additionally, image segmentation, or selecting a structure or region of interest (ROI) within a sample, whose lateral position varies in the axial dimension, results in poor image quality [2]. However, automatization has greatly reduced the time spent on this process and has improved the 3D reconstruction of these images[13], [14].
2.2. 2D-to-3D transformation algorithm
Although several techniques implement some sort of post-imaging analysis of the data, the 2D-to-3D transformation algorithm quantifies the 3D information from 2D data without the need for complex optical setups[15]. This is accomplished by taking advantage of the physical properties of radially symmetric structures and assumes that the location of molecule will remain the same if the properties of the structure do not change. For example, the nuclear pore complex (NPC) exhibits 8-fold rotational symmetry with its scaffold proteins and thus the distance from the radial center of one scaffold protein would be the same for a scaffold protein of the same type[16]. To resolve the radial distance, an area matrix is used to accurately determine the radial position of a molecule. This process is described further in Section 4.2.
3. Single particle tracking and localization microscopy
3.1. General principles of single-molecule microscopy
The 2D-to-3D transformation algorithm relies inherently upon high quality 2D single-molecule localizations. Capturing the requisite 2D data may be achieved through a variety of different techniques, but ultimately relies upon several key concepts. First, single molecules must be sufficiently spatially or temporally separated to permit the mathematical localization of the emitter[17]. Second, the sensitivity of the optical system, which is most influenced by the numerical aperture (NA) of the objective and the detector quality[18]. Lastly, the quality of the emission source, meaning the object being imaged via fluorescent microscopy. With optimized laser power, target selection, and labeling efficiency, most fluorescent proteins and dyes can provide a stable number of photons prior to quenching and are therefore capable of being localized at the single-molecule level.
3.1.1. Optical system sensitivity
Numerical aperture was originally defined by Ernst Abbe as , with representing one-half angular aperture of the objective and denoting the refractive index of the medium between the objective and the sample. NA may independently improve the overall spatial resolution of an image as the NA effects both the photon collection efficiency of the emission source and the area of illumination. This concept is illustrated by the point spread function (PSF) shape and size. Two PSFs exist within an image plane, an illumination PSF (iPSF) and an emission PSF (ePSF). With respect to the objective NA the iPSF may be adjusted to reduce background noise from emitters in the lateral and axial planes. However, for moving particles, reducing the width of the ePSF, will be partially dependent upon the sensitivity and speed of the detector. A good detector will reduce the width of the ePSF by increasing the amount of localizations between the start and finish of the molecule’s trajectory.
Regarding the sensitivity of the detector, this will have a role in not only the spatial resolution but the temporal resolution as well. For single-molecule detection, two major types of detectors are typically used, electron multiplying charge-coupled device (EMCCD) and scientific complementary metal oxide semiconductor (sCMOS) cameras[19], [20]. Both camera types effectively convert photons into electrons with a typical quantum efficiency (QE) above 90% and demonstrate very low camera noise[19], [20]. This differs from background noise as camera noise results from both the current that is thermally generated by the detector and the detector’s conversion of pixel photocurrent into signal voltage; referred to as dark noise and read noise, respectively. Interestingly, sCMOS cameras with parallel charge-voltage conversion for each pixel induces pixel-based readout and pattern noise compared to EMCCD sensors. However, by noise correction algorithm (NCS)[21] and automatic correction of sCMOS-related noise (ACsN)[20], sCMOS cameras preserve the characteristics of the fluorescence signal. Improving QE and reducing camera noise play significant roles when accurately localizing single molecules.
3.1.2. Quality of the emission source
In addition to the optical setup, the quality of the emission source must meet some minimum requirements to effectively localize single molecules. For fluorescent molecules, this is generally measured by the brightness of a molecule. Brightness is quantified by multiplying how strongly a fluorescent molecule absorbs light at a particular wavelength, defined as extinction coefficient (ε), by how efficiently the absorbed light is converted into emitted light, defined as quantum yield (Φ)[22], [23]. Although, having a fluorescent molecule that is very good at absorbing light but inefficient at converting that absorbed energy into emitted light, may limit its usefulness in single-molecule microcopy studies. This is best illustrated by the organic dye Cyanine-3 (Cy3), which has an extinction coefficient of 150,000 and quantum yield of 0.15[23]. The inverse is exemplified by mBanana, a fluorescent protein with an extinction coefficient of 6000 and a quantum yield of 0.70[23].
Furthermore, photobleaching, or the time associated with irreversibly modifying a fluorescent molecule so that the molecule cannot be excited again, is both a positive and negative quality that may be exploited in single-molecule microscopy environments[24]. As a positive, photobleaching may reduce the quantity of fluorescent molecules within the imaging area prior to image capture, allowing new fluorescently active molecules to enter. By reducing the concentration of active fluorescent molecules results in a direct decrease of overall background noise and a providing a more precise localization of a single molecule. Negatively speaking, when an emission source has a short photobleaching time, signal photons captured within a given time may be insufficient for distinguishing the single molecule from the background. In the case for moving particles, a trajectory may end prematurely and, as a result, providing insufficient information regarding the molecular functions or properties of the moving particle. For example, if an eGFP were passively diffusing through the nuclear pore complex (NPC) and underwent photobleaching halfway through the complex, then the trajectory would be incomplete and thus unusable for determining the transport efficiency of eGFP through the NPC.
In addition, fluorescence intermittency (or blinking), a phenomenon of random switching between bright and dark states of the emitter under a continuous excitation, also has negative and positive aspects. This phenomenon was typically found in some molecular fluorophores and colloidal quantum dots, which is believed to be caused by the competition between the radiative and non-radiative relaxation pathways [25]. Negatively speaking, for example, a repeated cycle of fluorescence blinking behavior, observed for single nanoparticles, green fluorescent protein (GFP) molecules, or Alexa Fluor 647 dyes, can significantly challenge the determination of Fluorescence Resonance Energy Transfer (FRET) efficiency in single molecule FRET measurements using these fluorophores. While, on the other hand, fluorescence blinking of these fluorophores or reversibly photoswitchable fluorescent proteins can form the fundamental underpinning for super-resolution microscopy imaging such as stochastic optical reconstruction microscopy (STORM)[26] and photoactivated localization microscopy (PALM)[27].
Alternatives to overcome issues regarding fluorophore stability could be increasing a sample’s labeling efficiency and the number of fluorophores on a sample, or substituting the fluorophore with a more robust inorganic equivalent, such as a quantum dot[28] or plasmonic nanoparticle[29]. However, both approaches have major experimental design limitations, including loss-of-function or aberrant behavior of the labeled molecule and limited labeling sites due to structural properties of the conjugate or target molecule.
Similar to the detector, the emission source and environment will produce noise that will distort the true location of a single molecule. This noise arises from off-target illumination of neighboring emission sources, which may come from fluorescently labeled molecules or from endogenous molecules with fluorescent properties typically denoted as autofluorescence[30]. However, off-target illumination can be reduced by applying different angles of excitation light. For example, total internal reflection fluorescence (TIRF) microscopy excites only the bottom 100–200 nm layer of a cell sample, which drastically limits the background noise that would come from above that bottom layer. The major drawback of this approach is that it severely limits the axial range of imaging beyond the excited layer. When selecting 2D localizations to utilize for the 2D-to-3D transformation, it is recommended to compare the signal to noise as a signal-to-noise ratio (SNR). This is a useful estimator for determining the quality of your 2D-localizaitions.
3.1.3. Localizing the single-molecule
After the above parameters are optimized, and single molecule images are taken with sufficient SNR, it is then necessary to determine from where in Euclidean space the target molecule is emitting. The raw image will not provide sufficiently precise location data regarding the target molecule’s location. In order to localize the molecule at the nm scale, overcoming the diffraction limit is required. Defined by Ernst Abbe in the late 1800 s, the resolution limit as in the lateral plane (x, y) and in the axial plane (z), where d is the diffraction limit, is the wavelength, and is the numerical aperture. Meaning, for conventional light microscopy, spatial resolution is limited to approximately 200 nm laterally and 500 nm axially[31], [32]. As a result of these physical limitations, it is impossible to obtain a high enough resolution for single particle localization, so algorithms are used to predict where exactly the molecule is in 2D space. These fitting algorithms use statistical analysis to delineate the signal of the target molecule from noisy, pixelated images of ROIs. Typically, these algorithms assume that within the ROI, there is only a single target molecule emitting light, which means that it is necessary during image collection to have either spatially or temporally separated fluorescent signals.
There are many different types of fitting algorithms that could be applied to the images, and while some of them are more precise and sophisticated than others, that sophistication generally comes with more difficult implementation, and may be unnecessary depending on the desired level of precision and accuracy. One relatively imprecise and inaccurate method is to use a max value estimator, which assumes that the target molecule is located in the center of the pixel with the greatest intensity value, which makes this method highly constrained by pixel size. This concept can be further improved by instead using a weighted average of all of the pixels in the ROI, creating a centroid estimator. While the centroid estimator is slightly more precise than the max value estimator, it will always bias results towards the center of the ROI as a result of the inclusion of background pixels. If the target molecule is emitting light isotropically, and the PSF is assumed to have radial symmetry, fitting the signal to a Gaussian function for localization has been shown to be quite accurate, and is widely used for single molecule localization[33].
3.2. SPEED microscopy
SPEED microscopy is designed to localize and track either mobile or immobile fluorescent single molecules as they occupy space within or peripheral to biological channels and cavities at a spatiotemporal resolution of up to ≤ 10 – 20 nm and 0.4 ms[34], [35]. One of the features of SPEED that allows for such high temporal and spatial resolutions is the utilization of an inclined single-point illumination in the focal plane. To achieve the inclined single-point illumination volume, we first require a vertical single-point illumination. The laser beam is aligned to pass through the central axis of the objective by adjusting the reflection mirror, so that we get center excitation beam or vertical single-point illumination. Next, the center excitation beam is shifted a specific distance off the central axis of objective by a micrometer stage, so as to generate an inclined illumination volume at 45° in the vertical direction (Fig. 1A). The shift distance is determined by several parameters: the focal length of objective, the refractive indexes of the different mediums on the optical path, and the inclined angle required. The inclined single-point illumination volume of SPEED microscopy has better axial resolution and acceptable lateral resolution (∼320–230 nm in the x, y, and z directions with the tilted angle between 350 and 550 for a 488-nm excitation laser). This incline provides a couple of major benefits that improve 2D localization. For example, it reduces out-of-focus background fluorescence, and generates a smaller effective illumination volume in the axial direction. When studying nuclear pore complexes (NPCs), for example, it is possible to only excite a single NPC in all three dimensions. In addition, because it is possible to directly focus the inclined beam onto the focal plane, a higher laser power density can be applied onto the target without having to increase laser power, resulting in a higher number of excited photons in the targeted region. This also reduces the impact of photobleaching and phototoxic effects on the cell, permitting more accurate imaging of live cells. An optical chopper has been set up to further reduce the probability of the negative effects of applying a laser to a living cell, allowing the beam to only be interacting with the cell for 1/10 of the frames captured during imaging. Finally, the detector utilized in SPEED microscopy is an on-chip multiplication gain CCD camera rather than a photomultiplier tube (PMT), which allows for direct recording of spatial information and a larger area of photon collection. The CCD camera paired with such a small illumination volume results in a detection speed of around 5000 frames per second, which minimizes localization error. All of these aspects of SPEED microscopy allow for a greater SNR. To be clarified, the design and application of SPEED microscopy is different from highly inclined and laminated optical sheet (HILO) microscopy that has been well used for single-molecule imaging inside cells. As aforementioned, SPEED uses a focused laser beam while HiLo has a parallel laser beam through the focal plane.
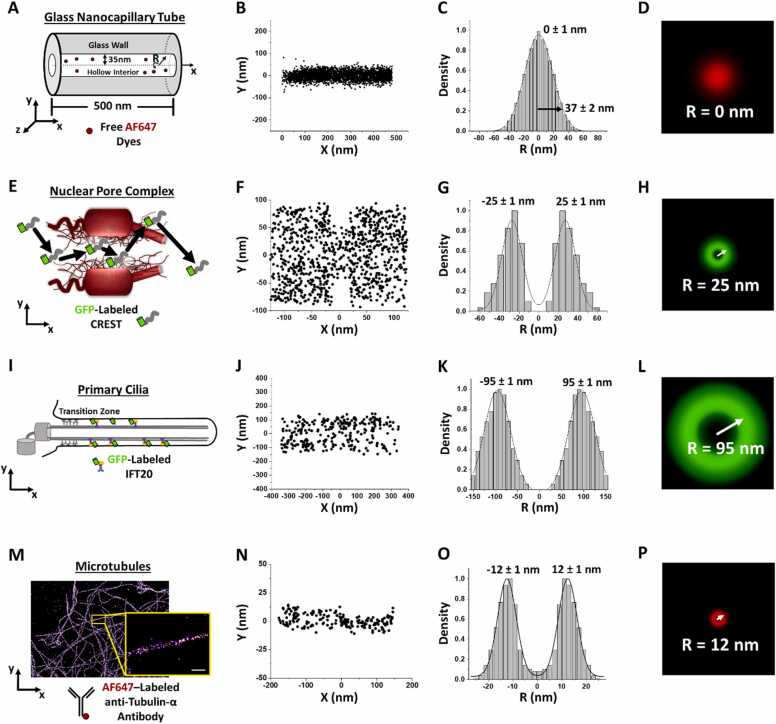
Fig. 1.
2D-to-3D Transformation of different fluorophore localizations within various structures. (A) Illustration of a glass nanocapillary tube with a length of 500 nm and a hollow interior radius of 35 ± 0.25 nm with freely diffusing Alexa Fluor 647 (AF647) dyes. (B) X and y 2D localizations from previously published AF647 single-particle tracking events inside the glass nanocapillary tube[42]. (C) Density plot of 2D localizations from B with a 37 ± 2 nm at two standard deviations from the mean, 0 ± 1 nm. (D) Radial density map from C with the R peak noted in white. (E) Illustration of the nuclear pore complex (NPC) with a model trajectory for GFP-labeled intrinsically disordered protein, calcium-responsive transactivator (CREST). (F) X and y 2D localizations from previously published GFP-CREST single-particle tracking events inside the NPC[43]. (G) Density plot of 2D localizations between x-dimension − 20–20 nm from F with peak fittings at − 25 ± 1 nm and 25 ± 1 nm, showing CREST’s NPC transport route as ∼25 nm. (H) Radial density map from G. (I) Illustration of GFP-labeled intraflagellar transport (IFT)20 protein in the shaft of a primary cilium. (J) X and y 2D localizations from previously published GFP-IFT20 single-particle tracking events inside a primary cilium[42]. (K) Density plot of 2D localizations from J with peak fittings at − 95 ± 1 nm and 95 ± 1 nm, showing IFT20 active transport route as ∼95 nm. (L) Radial density map from K. (M) 2D STORM data showing the reconstructed super-resolution image of AF647-labeled anti-tubulin-α antibody localized to tubulin. Scale bar = 50 nm. (N) X and y 2D localizations of AF647-anti-tubulin-α antibody within COS-7 cells. (O) Density plot of 2D localizations from N with peak fittings at − 12 ± 1 nm and 12 ± 1 nm, showing the radius of the microtubule structure, ∼12 nm. (P) Radial density map from part O.
Figure has been reproduced with permission [15].
As described previously, Gaussian distribution fitting functions are excellent tools with which to accurately and precisely determine single molecule location beyond the diffraction limit. SPEED utilizes the ImageJ plugin GDSC-SMLM to perform a series of tasks: fitting the single molecules and determining spatial location, determining fluorescence intensity of the molecules, determining intensity of the background, and solving for the Gaussian width of the single molecule spots[36]. By determining the Gaussian width of a single molecule, it is possible to filter out frames that display wider or narrower signals, suggesting that there are either multiple fluorescent molecules in a single ROI, or the signal is merely background noise. After calculating the SNR, it is possible to also filter out frames that have an SNR ≤ 10, maintaining only the images that will yield the best localization precisions. The localization precisions of stationary molecules are determined by the standard deviation of multiple measurements of the central point, while for mobile molecules the localization precision can be solved with the following equation, , where F is equal to 2, N is the number of collected photons, a is the effective pixel size of the detector, b is the standard deviation of the background in photons per pixel, and , where s0 is the standard deviation of the PSF in the focal plane, D is the diffusion coefficient of the substrate, and Δt is the image acquisition time[18].
There are two major types of information that can be collected using SPEED after analysis of the 2D single-molecule localizations. The first of which is dynamics information, which includes diffusion coefficients, transport efficiencies, and interaction or dwell time, and is resolved with a temporal resolution of 0.4 – 2 ms. The second type is structural information, which requires the superposition of hundreds of single-molecule trajectories, which is achieved with a temporal resolution of 1 – 3 s[18]. With longer acquisitions, the sample may shift from its original position within the illumination area producing incoherent information. Even with a short time acquisition, sample drift may occur as well. To identify sample drift, a marker image is taken before and after data collection and then compared. For example, when collecting single-molecule mCherry-labeled messenger ribonucleoproteins (mRNPs) interacting with a the GFP-labeled NPC by SPEED microscopy, a widefield marker image of the entire GFP-labeled nuclear envelope (NE) was taken before and after imaging the mRNPs. Then, when comparing the two images, if the NE shifted, the shift can be recorded and corrected in localizing single-molecule trajectories and in the determination of overall localization precision of the experiments. The acquisition could be even excluded from the final dataset if the shift is significant and hard to be corrected [37], [38]. There are several approaches to reduce and correct sample drift, including imaging software, optical hardware, and environmental hardware, however, a combination of the three is required for high-speed single-molecule microscopy.
3.3. 3D SMLM techniques
There are several single-molecule approaches to gain Z-dimensional data by capturing and contorting the emission PSF [17], [39], [40], [41]. One such technique captures the emission PSFs at multiple focal planes by multiple detectors, known multifocal or bi-plane imaging. Typically, out-of-focus signal weakens the ability to obtain high-resolution images and is considered only as a parameter to avoid. However, 3D SMLM techniques implementing multifocal/bi-plane imaging can simultaneously capture multiple focal planes, rendering different Z-dimensional profiles of an emitter’s PSF. Comparing the off-focal plane image to the focal plane image resolves the distance of an emitter in the z-dimension. Another technique to gain axial dimensional data is through PSF engineering, in which the emission PSF is contorted by back focal plane modulation to create a contorted PSF indicative of the emitter’s axial position. Techniques using PSF engineering include astigmatism, phase-ramp, double-helix, accelerating beam, corkscrew, and tetrapod. For a more robust explanation of these different 3D SMLM approaches, we recommend L. von Diezmann et al. [17].
4. Application of 2D-to-3D transformation algorithm
4.1. 2D-to-3D transformation applied to different structures
Several structures, both non-biological and biological, possess radially symmetric qualities that may utilize the 2D-to-3D transformation algorithm. As demonstrated in Fig. 1, we transformed 2D localizations collected within a glass nanocapillary (GNC) tube[42], nuclear pore complex (NPC)[43], primary cilia[42], and from the exterior of a microtubule into 3D density maps.
Within the GNC, the 2D-to-3D transformation algorithm offered an alternative to helium scanning transmission ion microscopy (HeSTIM) for determining the internal diameter of the GNC (Fig. 1A). To accomplish this high quality 2D localizations of Alexa Fluor 647 dyes as they freely diffuse through the GNC hollow interior (Fig. 1B). Then transforming the 2D localizations into a 3D radial density plot (Fig. 1C) and map (Fig. 1D) using the 2D-to-3D transformation algorithm. With an estimated diameter of 37 ± 2 nm, the 2D-to-3D transformation calculation coincides with the HeSTIM observation of 35 ± 0.25 nm[42]. Although HeSTIM provides better accuracy, the reduced in experimental time and complexity the 2D-to-3D transformation algorithm offers may be a worthy substitution.
In addition to GNC tube experiments, the 2D-to-3D transformation algorithm has been applied to several sub-cellular molecules and structures, including intrinsically disordered proteins (IDPs) transport through the NPC[43] (Fig. 1E – H), diffusion of intraflagellar transport (IFT) protein complexes through the primary cilia[42] (Fig. 1I – 1 L), and resolving the diameter of tubulin by antibody labeling (Fig. 1M – P). The sensitivity of the 2D-to-3D transformation algorithm was further demonstrated in three different environments. For the NPC and primary cilia environments, the 3D transport routes of GFP-labeled transiting particles within live cells was resolved using the 2D-to-3D transformation algorithm. 2D-to-3D transformation algorithm also enabled us to reveal the nucleocytoplasmic transport routes for calcium-responsive transactivator (CREST) and other IDPs, which challenged an established rule for passive diffusion through the NPC, known as the size exclusion barrier [44], [45], [46]. The virtual 3D method further suggested that the active and passive diffusion of molecules in primary cilium use the peripheral and central luminal regions respectively[42]. In both instances, the 3D virtual images of these transport routes were necessary in revealing novel information regarding the sub-cellular structure and protein’s function. Moreover, to demonstrate the 2D-to-3D transformation algorithm adaptability to 2D localizations collected by other microscopy techniques, the diameter of tubulin was resolved using stochastic optical reconstruction microscopy (STORM) with fluorescently-labeled anti-tubulin antibody in fixed wt COS7 cells (Fig. 1M – P). The diameter of 24 ± 1 nm as determined by the 2D-to-3D transformation algorithm coincides with previously published results[47].
4.2. Mathematical concept of the 2D-to-3D transformation algorithm
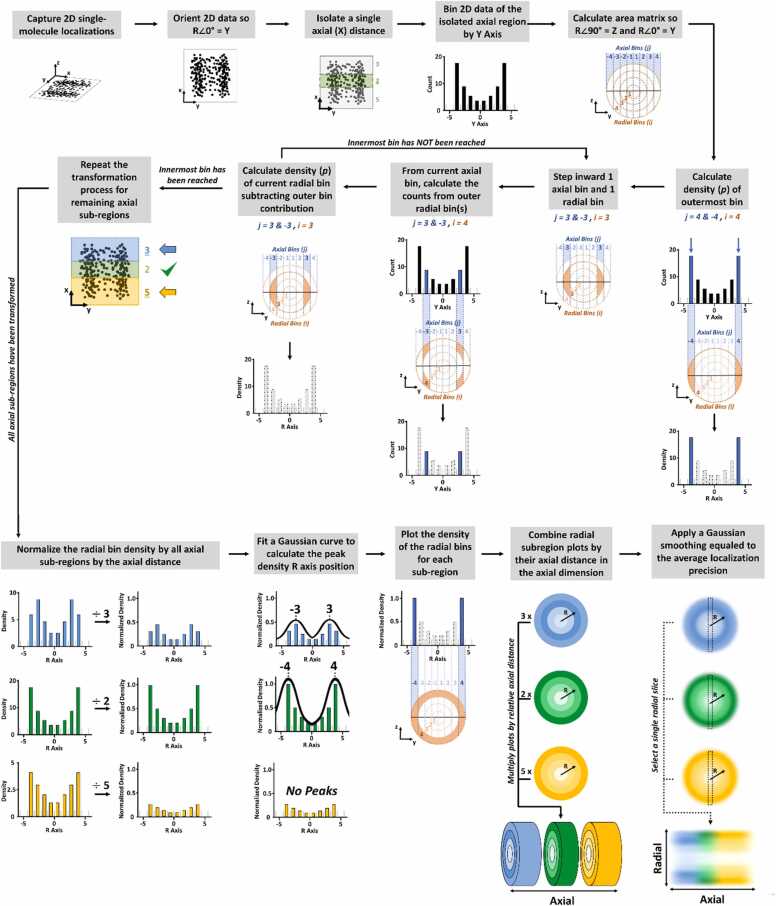
Assuming high quality single-molecule localizations within a rotationally symmetric structure are obtained, then the 2D-to-3D transformation algorithm may be applied[15]. The Fig. 2 flowchart demonstrates the transformation of 2D localizations into a radial density plot. After capturing 2D single-molecule localizations, the localizations are oriented such that the radial (r) angle of 0° equals the y-dimension. Then the 2D localizations are binned in the y-axis by the optimal bin size (Δr). The optimal bin size is determined by testing the smallest possible bin size that yields no statistical difference by a Chi square analysis of the y-axis histogram and a back-calculated histogram of the density plot. The 2D localizations within the x-dimension may all undergo y-axis binning, as shown in GNC tube from Fig. 1C & D, or a x-dimension segment may be isolated based on the biological properties exhibited by the target molecule, as shown in the NPC for Fig. 1G & H for the hydrophobic properties exhibited by the proteins found within the NPC central scaffold region (−20 to 20 nm). Once the 2D localizations are binned by the y-axis, the area matrix may be calculated by setting r angle of 90° equaled to the z-dimension. This calculated area matrix consists of both axial bins (i) and radial bins (j), which allows each sub-region area (A(i,j)) to be calculated. To begin transforming the binned y-axis data into radial density information, the spatial probability density () of the outermost bin, where i equals j, is calculated by measuring the area (S(i,j)) as shown in equation 1. Then stepping inward 1 axial and 1 radial bin, such that i and j are still equal, the area is calculated using the same equation (Eq. 1). Fig. 2.
Fig. 2.
Flowchart illustrating the 2D-to-3D density transformation algorithm and subsequent normalization of multiple axial subregions. The 2D-to-3D transformation algorithm converts the y-dimensional histogram into r-dimensional density information. This process is based on the area matrix that was developed for the given structure from which the y-dimensional information was obtained.
Figure has been reproduced with permission [15].
From the same axial bin, the outer radial bin (i ≠ j, i > j) areas (A(i,j)) are calculated using equation 2.
However, if i ≠ j and i < j than the area (A(i,j)) is 0 since the radial bin can only be equal or greater than the axial bin. Once the innermost bin is reached and the areas of all the subregions are calculated than the number of events in each axial bin (Nj) can be accurately determined using equation 3.
Now the radial density results may be combined with other radial density results from different x-axis segments to create a full virtual 3D image.
4.3. Experimental validation by simulation
To evaluate the robustness and reliability of this technique, a variety of simulations were devised. These simulations operate on the principles of Monte Carlo Simulation, which is also referred to as a multiple probability simulation. In essence, such a simulation takes into account the possibility of multiple outcomes from an event, thereby predicting the most probable outcome for the inputted variables[48], [49]. The first simulation was devised specifically to interrogate the reproducibility of the 2D-to-3D transformation algorithm. This simulation found that there are two critical parameters required for an accurate 3D density map reconstruction; single-molecule localization error and the number of single-molecule locations[27], [50]. The degree of single-molecule localization error tolerable within the algorithm is directly correlated to the radius of the symmetrical structure being evaluated. For instance, a structure with a radius of 25 nm, the approximate radius of the central channel of the nuclear pore complex[51], [52], [53], [54], is only reproducible if the localization error remains below 21 nm. It is notable that the number of 21 nm is slightly smaller than the theoretical single-molecule localization error predicted by the Rayleigh criterion[55]. This slight discrepancy is most probably due to the sensitivity of the inner bins of the area matrix that are more sensitive to noise due to the slightly smaller area of the inner bins when compared to the outer bins. Further, researchers found the relationship between localization error and radius could best be demonstrated as the ratio of radius over precision (R/P), where it was found that the minimal R/P ratio was equal to 1.19, while the optimal ratio was ≥ 2.0. This is noteworthy as SPEED microscopy is capable of an average of 10 nm localization precision, or 2.5 R/P.
Regarding the number of points required to accurately reconstruct a 3D density distribution, it was found that 100 or 350 single-molecule localizations would provide a reproducibility of 90% and 99%, respectively. It is important to note that while this number is seemingly small, it is higher than the minimum Nyquist sampling theorem estimation of 38 single-molecule localizations[56]. Interestingly, it was found that 1000 single-molecule localizations with an appropriately small localization error resulted in a 3D mean location accuracy of 1 nm, indicating that the accuracy of the 3D reconstruction could extend to a boundlessly small measure provided sufficient single-molecule data with a sufficiently small localization error[18], [57].
The 2D-to-3D transformation algorithm was next interrogated for the accurate reproduction of multiple transport routes through the symmetrical structure. It is possible that multiple transport routes through a structure are available to a macromolecule of interest, therefore it was critical to determine the viability of the 2D-to-3D transformation algorithm in differentiating between these multiple possible routes. The simulation demonstrated an ability to differentiate between multiple routes. However, it was shown that at the minimal R/P of 1.19 that ∼200 single-molecule localizations were required to achieve 90% reproducibility when compared to the ∼100 localizations required for a single transport route. Interestingly, the ability of the algorithm to distinguish between multiple transport routes increased as the R/P increased, with only ∼100 localizations required to differentiate between multiple transport routes with an R/P of 2.5[57].
Lastly, the robustness of this algorithm was interrogated in light of the physical properties of the symmetrical structure. While Monte Carlo simulations and the 2D-to-3D transformation algorithm assume a perfect system, the realities of biology are rarely so precise. Further simulations were devised to evaluate the efficacy of this algorithm under conditions where the structure is compressed, not perfectly symmetrical, exhibits varying labeling efficiency, as well as the impacts of fold symmetry and rotation. These simulations found that compression of the structure, labeling efficiency, and the rotation of the structure do not overly impact the accuracy and reproducibility of the 2D-to-3D transformation algorithm so long as multiple datasets are averaged together and appropriately binned. Notably, the labeling efficiency did not change significantly so long as a minimum of half the objects are labeled[57]. Taken together, these simulations demonstrate that the 2D-to-3D localization algorithm is both accurate and robust with a relatively low number of single-molecule localizations required to accurately and reproducibly generate a 3D probability density map.
4.4. Limitations of the 2D-to-3D transformation algorithm
The single largest limitation associated with the 2D-to-3D transformation algorithm is the inability of this algorithm to utilized to determine the symmetry of an object. The algorithm inherently functions based upon the principle that there is a constant number of single molecules along the single radial bin within the structure. As a result, this algorithm is capable of deriving virtual 3D information from 2D localizations, but not capable of determining the relative symmetry of a biological structure. Further, as the algorithm currently exists, it is only possible to derive 3D information from rotationally symmetrical structures. As many biological structures are symmetrical, but not radially symmetrical, or irregularly shaped; this method is limited to only a few biological structures.
5. Summary and outlook
Obtaining three-dimensional data for super-resolution single-molecule microscopy remains both challenging and desirable. The development of the 2D-to-3D transformation algorithm for SPEED microscopy has somewhat addressed this need by providing a method of deriving 3D information from 2D images without the addition of costly microscope modifications or software. As a result, this virtual 3D technique for single molecules remains one of the most accessible methods currently available to researchers. This methodology, as it currently exists, provides the spatial probability density maps for single molecules as they transfer through rotationally symmetric structures. At the time of writing, relatively few structures have been studied with this technique. Specifically, the nuclear pore complex, glass nanopores, primary cilium, and microtubules have been interrogated. The results of these studies have provided significant and novel insight into the dynamics of transiting macromolecules in a variety of cellular processes. However, this technique can be applied to any rotationally symmetric structure providing several more potential targets yet to be interrogated. Further, as this methodology is further refined, it is the intention of the authors to further develop this algorithm to derive spatial information from cellular structures that are symmetric, but not rotationally symmetric, such as endosomes and mitochondria.
Funding
The project is supported by grants from the US NIH (GM122552 to W.Y.).
CRediT authorship contribution statement
Samuel L. Junod: Conceptualization, Data curation, Software, Writing – original draft. Mark Tingey: Writing – original draft, Writing – review & editing. Coby Rush: Writing – original draft. Abdullah Alkurdi: Writing – original draft. Khushi Bajoria: Writing – original draft. Weidong Yang: Supervision, Conceptualization, Methodology, Writing – review & editing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Liu S., Weaver D.L., Taatjes D.J. Three-dimensional reconstruction by confocal laser scanning microscopy in routine pathologic specimens of benign and malignant lesions of the human breast. Histochem Cell Biol. 1997;107(4):267–278. doi: 10.1007/s004180050112. [DOI] [PubMed] [Google Scholar]
- 2.Kopecky B.J., et al. Three-dimensional reconstructions from optical sections of thick mouse inner ears using confocal microscopy. J Microsc. 2012;248(3):292–298. doi: 10.1111/j.1365-2818.2012.03673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassnett S., Shi Y. A method for determining cell number in the undisturbed epithelium of the mouse lens. Mol Vis. 2010;16:2294–2300. [PMC free article] [PubMed] [Google Scholar]
- 4.Du T., Wasser M. 3D image stack reconstruction in live cell microscopy of Drosophila muscles and its validation. Cytom Part A. 2009;75A(4):329–343. doi: 10.1002/cyto.a.20701. [DOI] [PubMed] [Google Scholar]
- 5.Elliott A.D. Confocal microscopy: principles and modern practices. Curr Protoc Cytom. 2020;92(1) doi: 10.1002/cpcy.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trusk T.C. In: Basic Confocal Microscopy. Jerome W.G., Price R.L., editors. Springer International Publishing; Cham: 2018. 3D Reconstruction of confocal image data; pp. 279–307. [Google Scholar]
- 7.Zhang Q., Eagleson R., Peters T.M. Volume visualization: a technical overview with a focus on medical applications. J Digit Imaging. 2011;24(4):640–664. doi: 10.1007/s10278-010-9321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shihavuddin A., et al. Smooth 2D manifold extraction from 3D image stack. Nat Commun. 2017;8(1):15554. doi: 10.1038/ncomms15554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stefanoiu A., et al. Artifact-free deconvolution in light field microscopy. Opt Express. 2019;27(22):31644–31666. doi: 10.1364/OE.27.031644. [DOI] [PubMed] [Google Scholar]
- 10.Nyquist H. Certain topics in telegraph transmission theory. Trans Am Inst Electr Eng. 1928;47(2):617–644. [Google Scholar]
- 11.Shannon C.E. Communication in the presence of noise. Proc IRE. 1949;37(1):10–21. [Google Scholar]
- 12.Wilson T., Tan J.B. Three dimensional image reconstruction in conventional and confocal microscopy. Bioimaging. 1993;1(3):176–184. [Google Scholar]
- 13.D’Antuono R., Pisignano G. ZELDA: A 3D image segmentation and parent-child relation plugin for microscopy image analysis in napari. Front Comput Sci. 2022:3. [Google Scholar]
- 14.Wang A., et al. A novel deep learning-based 3D cell segmentation framework for future image-based disease detection. Sci Rep. 2022;12(1):342. doi: 10.1038/s41598-021-04048-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruba A., et al. 3D tracking-free approach for obtaining 3D super-resolution information in rotationally symmetric biostructures. J Phys Chem B. 2019;123(24):5107–5120. doi: 10.1021/acs.jpcb.9b02979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma J., et al. Super-resolution mapping of scaffold nucleoporins in the nuclear pore complex. J Cell Sci. 2017;130(7):1299–1306. doi: 10.1242/jcs.193912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Diezmann L., Shechtman Y., Moerner W.E. Three-dimensional localization of single molecules for super-resolution imaging and single-particle tracking. Chem Rev. 2017;117(11):7244–7275. doi: 10.1021/acs.chemrev.6b00629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y., et al. High-speed super-resolution imaging of rotationally symmetric structures using SPEED microscopy and 2D-to-3D transformation. Nat Protoc. 2021;16(1):532–560. doi: 10.1038/s41596-020-00440-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van den Eynde R., et al. Quantitative comparison of camera technologies for cost-effective super-resolution optical fluctuation imaging (SOFI) J Phys: Photonics. 2019;1(4) [Google Scholar]
- 20.Mandracchia B., et al. Fast and accurate sCMOS noise correction for fluorescence microscopy. Nat Commun. 2020;11(1):94. doi: 10.1038/s41467-019-13841-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu S., et al. sCMOS noise-correction algorithm for microscopy images. Nat Methods. 2017;14(8):760–761. doi: 10.1038/nmeth.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bielec K., et al. Analysis of brightness of a single fluorophore for quantitative characterization of biochemical reactions. J Phys Chem B. 2020;124(10):1941–1948. doi: 10.1021/acs.jpcb.0c00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Day R.N., Davidson M.W. The fluorescent protein palette: tools for cellular imaging. Chem Soc Rev. 2009;38(10):2887–2921. doi: 10.1039/b901966a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demchenko A.P. Photobleaching of organic fluorophores: quantitative characterization, mechanisms, protection. Methods Appl Fluoresc. 2020;8(2) doi: 10.1088/2050-6120/ab7365. [DOI] [PubMed] [Google Scholar]
- 25.Baddeley D., et al. Light-induced dark states of organic fluochromes enable 30 nm resolution imaging in standard media. Biophys J. 2009;96(2):L22–L24. doi: 10.1016/j.bpj.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rust M.J., Bates M., Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3(10):793–796. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Betzig E., et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313(5793):1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 28.He X., Gao L., Ma N. One-step instant synthesis of protein-conjugated quantum dots at room temperature. Sci Rep. 2013;3(1):2825. doi: 10.1038/srep02825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y., van Asdonk K., Zijlstra P. A robust and general approach to quantitatively conjugate enzymes to plasmonic nanoparticles. Langmuir. 2019;35(41):13356–13363. doi: 10.1021/acs.langmuir.9b01879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Croce A.C., Bottiroli G. Autofluorescence spectroscopy and imaging: a tool for biomedical research and diagnosis. Eur J Histochem. 2014;58(4):2461. doi: 10.4081/ejh.2014.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leung B.O., Chou K.C. Review of super-resolution fluorescence microscopy for biology. Appl Spectrosc. 2011;65(9):967–980. doi: 10.1366/11-06398. [DOI] [PubMed] [Google Scholar]
- 32.Huang B., Bates M., Zhuang X. Super-resolution fluorescence microscopy. Annu Rev Biochem. 2009;78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von Diezmann A., Shechtman Y., Moerner W.E. Three-dimensional localization of single molecules for super-resolution imaging and single-particle tracking. Chem Rev. 2017;117(11):7244–7275. doi: 10.1021/acs.chemrev.6b00629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma J., Yang W. Three-dimensional distribution of transient interactions in the nuclear pore complex obtained from single-molecule snapshots. Proc Natl Acad Sci. 2010;107(16):7305. doi: 10.1073/pnas.0908269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma J., et al. Self-regulated viscous channel in the nuclear pore complex. Proc Natl Acad Sci. 2012;109(19):7326. doi: 10.1073/pnas.1201724109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herbert, A., Single Molecule Light Microscopy ImageJ Plugins,. 2014.
- 37.Li Y., et al. Nuclear export of mRNA molecules studied by SPEED microscopy. Methods. 2019;153:46–62. doi: 10.1016/j.ymeth.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma J., et al. High-resolution three-dimensional mapping of mRNA export through the nuclear pore. Nat Commun. 2013;4:2414. doi: 10.1038/ncomms3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Y., et al. Advances in 3D single particle localization microscopy. APL Photonics. 2019;4(6) [Google Scholar]
- 40.Song K.-H., et al. Three-dimensional biplane spectroscopic single-molecule localization microscopy. Optica. 2019;6(6):709–715. doi: 10.1364/optica.6.000709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y., et al. Global fitting for high-accuracy multi-channel single-molecule localization. Nat Commun. 2022;13(1):3133. doi: 10.1038/s41467-022-30719-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo W., et al. Axonemal lumen dominates cytosolic protein diffusion inside the primary cilium. Sci Rep. 2017;7(1):15793. doi: 10.1038/s41598-017-16103-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Junod, S.L., et al., Nucleocytoplasmic transport of intrinsically disordered proteins studied by high-speed super-resolution microscopy. Protein Science, 2020. n/a(n/a). [DOI] [PMC free article] [PubMed]
- 44.Patel S.S., et al. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell. 2007;129(1):83–96. doi: 10.1016/j.cell.2007.01.044. [DOI] [PubMed] [Google Scholar]
- 45.Sakiyama Y., et al. Spatiotemporal dynamics of the nuclear pore complex transport barrier resolved by high-speed atomic force microscopy. Nat Nanotechnol. 2016;11:719. doi: 10.1038/nnano.2016.62. [DOI] [PubMed] [Google Scholar]
- 46.Ma J., Goryaynov A., Yang W. Super-resolution 3D tomography of interactions and competition in the nuclear pore complex. Nat Struct Mol Biol. 2016;23(3):239–247. doi: 10.1038/nsmb.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boca M., et al. Probing protein interactions in living mammalian cells on a microtubule bench. Sci Rep. 2015;5(1):17304. doi: 10.1038/srep17304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mooney, C.Z., Monte carlo simulation. 1997: Sage.
- 49.Mahadevan, S., Monte Carlo Simulation. 1997, New York and Basel Switzerland: Marcel Dekker.
- 50.Thompson R.E., Larson D.R., Webb W.W. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82(5):2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tingey M., et al. Spelling out the roles of individual nucleoporins in nuclear export of mRNA. Nucleus. 2022;13(1):170–193. doi: 10.1080/19491034.2022.2076965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zimmerli C.E., et al. Nuclear pores dilate and constrict in cellulo. Science. 2021;374(6573):eabd9776. doi: 10.1126/science.abd9776. [DOI] [PubMed] [Google Scholar]
- 53.Stoffler D., et al. Cryo-electron tomography provides novel insights into nuclear pore architecture: implications for nucleocytoplasmic transport. J Mol Biol. 2003;328(1):119–130. doi: 10.1016/s0022-2836(03)00266-3. [DOI] [PubMed] [Google Scholar]
- 54.Schuller A.P., et al. The cellular environment shapes the nuclear pore complex architecture. Nature. 2021;598(7882):667–671. doi: 10.1038/s41586-021-03985-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ram S., Ward E.S., Ober R.J. Beyond Rayleigh's criterion: a resolution measure with application to single-molecule microscopy. Proc Natl Acad Sci. 2006;103(12):4457–4462. doi: 10.1073/pnas.0508047103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson M.A., et al. Methods in enzymology. Elsevier; 2010. Molecules and methods for super-resolution imaging; pp. 27–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruba A., et al. 3D tracking-free approach for obtaining 3D super-resolution information in rotationally symmetric biostructures. J Phys Chem B. 2019;123(24):5107–5120. doi: 10.1021/acs.jpcb.9b02979. [DOI] [PMC free article] [PubMed] [Google Scholar]