Abstract
Background
Long non-coding RNA (lncRNA) is one of the most essential forms of transcripts, playing crucial regulatory roles in the development of cancers and diseases without protein-coding ability. It was assumed that short ORFs (sORFs) in lncRNA were weak to translate proteins. However, recent research has shown that sORFs can encode peptides, which increases the difficulty to identify lncRNA. Therefore, identifying lncRNAs with sORFs facilitates finding novel regulatory factors.
Results
In this paper, we propose LncCat for identifying lncRNA based on category boosting (CatBoost) and ORF-attention features. LncCat combines five types of features to encode transcript sequences and employs CatBoost to build a prediction model. In addition, the visualization comparison reveals that the ORF-attention features between lncRNAs and protein-coding transcripts are significantly distinct. The comparison results show that LncCat outperforms competing methods on several benchmark datasets. For Matthew’s Correlation Coefficient (MCC), LncCat achieves 0.9503, 0.9219, 0.8591, 0.8672, and 0.9047 on the human, mouse, zebrafish, wheat, and chicken datasets, with improvements ranging from 1.90% to 7.82%, 1.49–17.63%, 6.11–21.50%, 3.02–51.64% and 5.35–26.90%, respectively. Moreover, LncCat dramatically improves the MCC by at least 11.90%, 12.96% and 42.61% on sORF test datasets of human, mouse, and zebrafish, respectively.
Conclusions
Experiments indicate that LncCat performs better both on long ORF and sORF datasets, and ORF-attention features show positive effects on predicting lncRNA. In brief, LncCat is a reliable method for identifying lncRNA. Additionally, a user-friendly web server is developed for academics at http://cczubio.top/lnccat.
Keywords: LncRNAs identification, Ensemble learning, ORF-attention features, Small ORF
Graphical Abstract

1. Introduction
According to the Encyclopedia of DNA Elements (ENCODE) project, 80% of the human genome have biochemical functions. Less than 2% of the genome can be translated into protein, and the remaining 98% is non-coding [1], [2]. Non-coding RNAs (ncRNAs) are divided into two categories based on their length: lncRNAs (Long non-coding RNAs, length above 200 nucleotides) and small ncRNAs [3]. LncRNA is an essential component of ncRNA, and approximately 70% of non-coding sequences are transcribed into lncRNAs [4]. LncRNAs were once considered as “noise” of transcription because of their lower expression level and lower sequence conservation compared to message RNAs (mRNAs) [5]. However, the growing evidence indicates that lncRNAs are a vital part of the transcripts and widely exist in eukaryotes [6].
Researchers have attached much attention to lncRNAs because of their significant regulatory functions [7]. According to [8], [9], [10], [11], [12], lncRNAs play crucial roles in metabolic processes, chromosome dynamics, and cell differentiation. Studies revealed that lncRNAs are relevant to a variety of complex human diseases, such as lung cancer [13], Alzheimer’s disease [14], and cardiovascular diseases [15], which indicates a substantial linkage between lncRNAs and disease [16], [17]. According to Wang and Chekanova [18], lncRNAs play crucial roles in various biological processes in plants, including flowering time control, organogenesis in roots, gene silencing, photomorphogenesis in seedlings, and plant reproduction. These lncRNAs regulate gene expression through different mechanisms and play a significant role in plant growth and development. Overall, lncRNAs play crucial roles in most organisms and significantly impact life activities [19], [20]. An open reading frame (ORF) is the nucleotide sequence between the start codon and the nearest stop codon. Small ORFs (sORFs) are defined as ORFs that are less than 300 nt in length. RNAs with sORFs are usually considered non-coding RNA because their small size does not typically allow for the production of a full-length protein. However, recent research has shown that sORFs can produce small peptides with significant biological functions [21]. Consequently, several sequences containing sORFs were previously assumed to be lncRNAs, but subsequent investigation revealed their coding potential.
With the latest genome-wide studies, biotechnologies such as high-throughput technology have provided thousands of unclassified transcripts. Identifying lncRNAs is a fundamental step to reveal their functions and mechanisms. Based on machine learning technology, numerous approaches for differentiating lncRNAs from protein-coding transcripts (PCTs) have been developed. CPC [22] (Coding-Potential Calculator) is a method for evaluating the coding potential of nucleotide sequences by comparing the sequences with the protein database. However, the alignment process is extremely time-consuming and limited by the quality of the database. Hence, researchers have developed several alignment-free methods to avoid the disadvantages caused by alignment. CPAT [23] (Coding-Potential Assessment Tool) builds a logistic regression model with the Fickett TESTCODE score [24] and hexamer score of open reading frame (ORF) regions [25] to assess the differences in nucleotide positions and codon usage between non-coding transcripts and PCTs. CNCI [26] (Coding-Noncoding Index) is developed by using a support vector machine (SVM) with adjoining nucleotide triplets (ANTs) matrices and codon bias. PLEK [27] (predictor of long non-coding RNAs and messenger RNAs based on an improved k-mer scheme) utilizes an improved k-mer scheme and selects SVM as its model to classify the sequences. CPC2 [28] is an upgrade of CPC, an alignment-free method based on the sequence intrinsic features. CPC2 employs SVM to learn the different patterns between lncRNA and PCTs. LncFinder [29] combines SVM with sequence intrinsic features, secondary structure features and EIIP-derived physicochemical features [30] to identify lncRNAs. mRNN [31] encodes RNA sequences with one-hot and adopts the deep learning model called Recursive Neural Network (RNN) to recognize lncRNA. And RNAsamba [32] is a neural network model to detect lncRNA by whole sequence and ORF information. In addition, RNAsamba can predict transcripts with small ORFs.
On long ORF datasets, several methods have achieved promising results. However, the properties of sORFs make it challenging to distinguish lncRNAs and PCTs, some of these approaches perform poorly on sORF datasets. It is necessary to develop a more accurate and effective model to discover new lncRNAs, especially those with sORFs. The discovery of new lncRNAs and their functions can help to better understand the novel functions of transcripts and improve our knowledge of gene regulation and cellular processes.
Ensemble learning is a branch of machine learning that employs and combines multiple learners to improve accuracy [33], [34]. It can be divided into the Bagging algorithm [35] and Boosting algorithm. Bagging algorithm increases the generalizability of a model by reducing its variance. The Boosting algorithm transforms weak learners into strong learners to improve the model’s accuracy. Boosting algorithm includes adaptive boosting (AdaBoost) [36], gradient boosting decision tree (GBDT), extreme gradient boosting (XGBoost) [37], light gradient boosting machine (LightGBM) [38], and categorical boosting (CatBoost) [39]. GBDT and AdaBoost are the most popular boosting algorithms. XGBoost and CatBoost are the improvements of GBDT and show promising performance in biology and medicine [40], [41], [42]. Although many studies have attempted to apply these methods for bioinformatics, CatBoost is faster, more flexible, and more sensitive.
In this article, we propose a new method named LncCat, which fuses multiple sequence features and employs the CatBoost to build the prediction model to distinguish lncRNAs from PCTs. First, four types of sequence-derived features are collected, including codons-related, GC-related, sequence-related, and peptide-related features. Additionally, LncCat introduces ORF-attention features derived from ORFs. Second, the prediction model is constructed by using the CatBoost algorithm and the above sequence-based and ORF-attention features. And then, to validate the model, LncCat is compared with eight state-of-the-art methods (CPAT, CNCI, PLEK, CPC2, LncFinder, CPPred, mRNN, and RNAsamba) on five species datasets. In addition, LncCat is validated on three datasets with sORFs. In terms of Matthew’s Correlation Coefficient (MCC), the compared results show that LncCat outperforms other methods on five species datasets and achieves improving MCC by at least 11.90%, 12.96%, and 42.61% on the human-sORF dataset, mouse-sORF dataset, and zebrafish-sORF dataset, respectively. Finally, a web server is developed and deployed on http://cczubio.top/lnccat/. The source code and datasets are accessible at https://github.com/a525076133/LncCat.
2. Materials and methods
2.1. Datasets
Previous research suggests that a rigorous dataset is essential for building a reliable prediction model. The datasets of this study are sourced from Han’s [29] and Tong’s [43] studies. In Tong’s study, human coding sequences are collected from NCBI RefSeq [44], and human ncRNAs, mouse, and zebrafish datasets are downloaded from Ensembl [45]. In Han’s study, human and mouse datasets are obtained from GENCODE [46], and zebrafish and wheat datasets are obtained from Ensembl. Both databases contain ncRNAs and PCTs annotated manually. Since datasets between or within two databases may overlap, the open-source program CD-HIT [47] is used to reduce the redundancy of the datasets by a threshold of 80% to get a rigorous dataset. After that, ncRNAs with the length shorter than 200 nt are eliminated. Finally, the dataset is divided into training, validation, and test datasets, which account for 70%, 10%, and 20% of the total, respectively. All datasets are summarized in Table 1.
Table 1.
Summary of the five species datasets.
| Species | Database | Coding sequences |
LncRNAs |
||||
|---|---|---|---|---|---|---|---|
| Training | Validation | Test | Training | Validation | Test | ||
| Human | RefSeq, Ensembl, GENCODE |
16, 072 | 2, 296 | 4, 592 | 14, 756 | 2, 108 | 4, 217 |
| Mouse | Ensembl, GENCODE |
14, 494 | 2, 071 | 4, 142 | 7, 494 | 1, 071 | 2, 142 |
| Zebrafish | Ensembl | 11, 123 | 1, 589 | 3, 179 | 3, 066 | 439 | 877 |
| Wheat | Ensembl | 3, 284 | 470 | 939 | 3, 763 | 538 | 1, 076 |
| Chicken | Ensembl | 4, 706 | 673 | 1, 345 | 2, 727 | 390 | 780 |
2.2. Sequence encoding methods
In this experiment, five types of features are introduced to construct the prediction model. First, codon-related, guanine-cytosine (GC)-related, and sequence-related features are extracted, which can be calculated directly from the sequence. Second, the peptide-related features are extracted from PCT- or lncRNA-encoded peptides or putative peptides. Third, because of the significance of ORF in coding sequences, ORF-related features are extracted, such as ORF length and ORF coverage. Finally, to investigate the efficacy of ORF-attention features, 1) the codon-related, GC-related, sequence-related, and peptide-related features of the top three longest ORFs are extracted, and 2) pre-training model based on Bidirectional Encoder Representations from Transformers (BERT) is used to enhance ORF information by encoding whose translating peptide sequences.
2.2.1. Codon-related features
In molecular biology, a codon is a nucleotide triplet in RNA. Panwar B et al. [48] have revealed that codons of lncRNAs and PCTs are typically distinct, and several codon-related features have been utilized to predict lncRNA. A stop codon is a codon that indicates the end of protein translation. Stop codon count is the number of stop codons in the transcript. Stop codon frequency is calculated by dividing the number of stop codons in the transcript by the length of the transcript. Another frequently used codon-related feature is the Fickett TESTCODE score [24], also known as the Fickett score. The Fickett score for a particular transcript is derived from the weighted nucleotide frequency of the entire transcript.
2.2.2. GC-related features
GC content is the proportion of guanine (G) or cytosine (C) nitrogenous base in an RNA or DNA sequence. A previous study has revealed that the GC content of coding regions is typically higher than non-coding sequences [49]. GC1, GC2, and GC3 can be calculated as the proportion of G and C in the first, second, and third codon positions, respectively. GC frame score refers to the variance of the number of GCs content in the three ORFs. The same method can get the GC1 frame score, GC2 frame score, and GC3 frame score.
2.2.3. Sequence-related features
Recent research has shown the feasibility of transcript-derived features for lncRNA identification [50]. The sequence length is calculated directly from the sum of the nucleotides in the sequence. K-mer refers to a specific subsequence comprising k nucleotides, which is one of the most frequently used features in lncRNA identification. Composition, Transition, and Distribution (CTD) refer to the descriptors of the entire transcript sequence based on the nucleotides’ composition, transition, and distribution. Hexamer-based features are variants of the k-mer features. Hexamer-based features measure the hexamer usage bias between coding and non-coding sequences, including hexamer usage bias or hexamer score, distance to PCTs, and distance ratio.
2.2.4. Peptide-related features
Peptide-related features refer to the properties of the peptide or putative peptide encoded by the ORF or putative ORF of the RNA sequence. Peptide-related features are important indicators because there are numerous different properties in terms of the sequence structure between lncRNAs and PCTs. For instance, Kang et al. [30] assume that the chemical properties of peptides encoded by coding sequences differ from those of putative peptides generated by non-coding sequences. Therefore, Some of these features, including molecular weight (Mw), theoretical isopotential point (PI), and measures of hydrophilicity (Gravy) and stability (Instability index), are introduced to identify lncRNA.
2.2.5. ORF-attention features
ORF is a reading frame that has translated potential. ORF-related features are one of the most important features to identify lncRNA because the PCT’s ORFs are generally longer than lncRNA’s. And many ORF-related features are explored and employed for lncRNA identification. ORF length is an essential feature for distinguishing lncRNAs and PCTs since long putative ORFs are few in lncRNA sequences. ORF coverage is the ratio between the length of the longest ORF and the transcript length. Besides the above features, ORF-attention features are also introduced. The ORF-attention features are derived from the codon-related, GC-related, sequence-related, and peptide-related features of the longest ORFs or putative ORFs. Furthermore, a recently proposed ORF-dominance feature [51] has been proven effective for classifying LncRNAs and PCTs, which is used to build models on small ORF datasets.
Orfipy [52] is a tool written in Python programming language that can extract ORF more efficiently. Orfipy implements a core ORF search algorithm implemented by Cython technology. The entire sequence is input to extract ORFs by the default parameters. Generally, the translation of eukaryotes starts from the ATG. In rare cases, translation in eukaryotes can be initiated from codons other than ATG. A well-documented case is the GTG start of a ribosomal P protein of the fungus [53]. Other examples, such as [54], [55], [56], [57], can start from ATG and TTG. Orfipy utilized Standard Code (transl table=1), which included the start codons (ATG, TTG, and CTG) and the stop codons (TAA, TAG, and TGA). All translation tables provided by Orfipy are accessible at https://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi?chapter=cgencodes.
2.2.6. Pre-trained ORF BERT
BERT is a machine learning technique based on a transformer for natural language processing (NLP), which has achieved state-of-the-art performance in various fields [58]. In addition, BERT comprises stacked transformer encode layers. BERT can accurately capture the bidirectional contextual information of texts to encode raw sequences. However, an ORF may be longer than 1000 nt, and BERT is not good at processing long sequences because of the increased memory and time consumption. The peptide sequences translated by the longest ORF in lncRNAs or PCTs sequences are used, which reduces the sequence length to one-third so that BERT can effectively process it. The pre-trained BERT model can get the vector representation of the sequence by contextual information, which can help capture the potential information of ORF and improve the accuracy of the prediction.
2.3. Constructing LncCat model
CatBoost is a new gradient-boosting decision tree (GBDT) algorithm that can handle the features in the training phase. CatBoost employs unbiased gradient estimation to solve the over-fitting issue. In addition, CatBoost can work with categorical features with minimal information loss. Moreover, CatBoost can be executed on GPU to accelerate the training process. Because of the above advances, CatBoost is implemented as a classifier for lncRNA identification. In this experiment, CatBoost is used to build the classification model using the five types of features listed above. The framework of this study is displayed in Fig. 1. First, RNA sequences of five different species are collected from NCBI RefSeq, GENCODE, and Ensembl. Second, five types of features are calculated from the sequences, including codon-related, GC-related, sequence-related, peptide-related features, and ORF-attention features. Next, LncCat is built by CatBoost with the above features to identify lncRNAs. And then, LncCat is compared with eight different methods and evaluated by seven standard metrics. Finally, a user-friendly web server is developed and freely available for academics.
Fig. 1.
The framework of LncCat. First, lncRNAs and PCTs of five species are collected from GENCODE, Ensembl, and RefSeq. And then, sequences with similarity> 80% are removed by CD-HIT; Second, transcripts are encoded by codon-related, GC-related, sequence-related, peptide-related, and ORF-attention features; Third, build a CatBoost classifier with the above five types of features; Finally, LncCat is compared with eight state-of-the-art methods on seven main metrics; A user-friendly web server is developed and freely available for academics.
2.4. Evaluation metrics
To comprehensively evaluate the performance of LncCat, seven commonly used evaluation metrics are employed, including accuracy (ACC), sensitivity (SEN/Recall), specificity (SPE), F-measure (F1), precision (PRE), Matthew’s correlation coefficient (MCC), and area under the ROC curve (AUC). And the metrics are calculated by the following equations:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
Where TP, TN, FP, and FN represent the number of true positives, true negatives, false positives, and false negatives, respectively. In our evaluation, lncRNAs are labeled as the positive class, and PCTs are labeled as the negative class. ACC is used to evaluate the overall predictive capability of the prediction model. And MCC is a more reliable indicator [59] that produces a high score only when the prediction performs well in all four confusion matrix categories (true positive, false negative, true negative, and false positive), which is proportional to the size of both the positive and negative elements in the dataset. The receiver operating characteristic (ROC) curves are generated by plotting the false positive rate (1-SP) versus the number of false positives (SN) for different cutoff thresholds. The AUC score is the area under the ROC curve. The ROC curve serves as a visual representation of the overall performance.
2.5. The methods used in experiments
The methods involved in the comparison include CPAT [23], CNCI [26], CPC2 [28], LncFinder [29], PLEK [27], CPPred [43], mRNN [31], and RNAsamba [32]. SVM is used by the CNCI, PLEK, CPC2, LncFinder, and CPPred algorithms. CPAT employs logistic regression. RNAsamba and mRNN are based on deep learning methods. According to their respective manuals, CPPred and RNAsamba can process the sORF dataset. Table 2 lists the version and website.
Table 2.
The version and website of eight compared methods.
| Methods | Version | Website |
|---|---|---|
| CPAT | v3.0.0 | http://cpat.readthedocs.io/en/latest |
| CNCI | version 2 | http://www.bioinfo.org/software/cnci |
| CPC2 | v1.0.1 | http://cpc2.cbi.pku.edu.cn |
| LncFinder | v1.1.5 | http://github.com/HAN-Siyu/LncFinder |
| PLEK | v1.2 | http://sourceforge.net/projects/plek |
| CPPred | - | http://www.rnabinding.com/CPPred |
| mRNN | - | http://github.com/hendrixlab/mRNN |
| RNAsamba | - | http://github.com/apcamargo/RNAsamba |
3. Experimental result
3.1. Feature visualization on five species datasets with and without ORF-attention
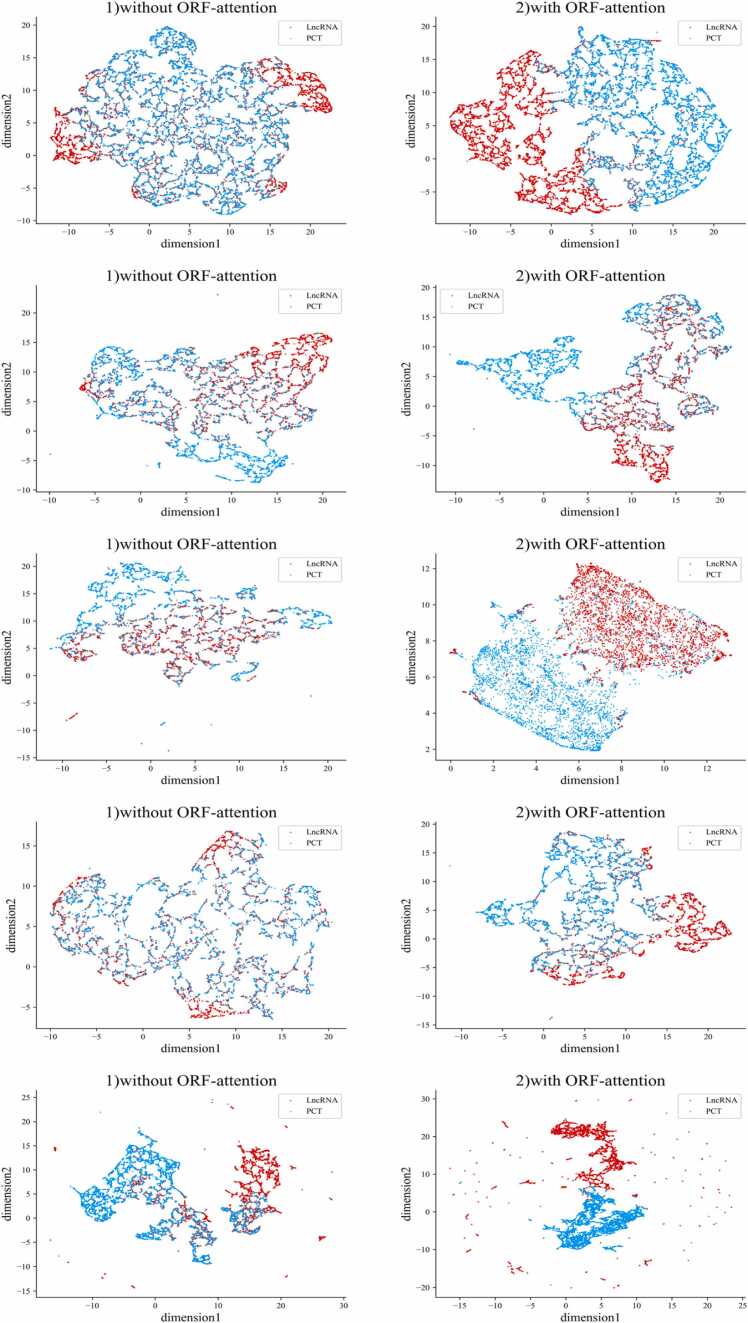
The features used by LncCat have been described previously. To better demonstrate the impact of ORF-attention features on the model, Uniform Manifold Approximation and Projection (UMAP) is used to visualize the distribution of lncRNAs and PCTs to illustrate the effect of ORF-attention features. The features are mapped into two-dimensional spaces. The distributions of five species sequences without ORF-attention features are shown in Fig. 2-(1)s. The distributions of five species sequences with ORF-attention features are shown in Fig. 2-(2)s. It can be observed that sequences without ORF-attention features are not adequately divided into two clusters. With the addition of ORF-attention features, LncCat can effectively differentiate between lncRNAs and PCTs, which demonstrates the effectiveness of ORF-attention features. In addition, Supplementary Table S1–1 provides a comprehensive description of all features of this experiment. Figs. S1–1 to Figs. S1–S5 show the feature importance of each feature.
Fig. 2.
Distribution of lncRNAs and PCTs on five datasets. (1)s represent the distribution of RNA sequences without ORF-attention on human, mouse, zebrafish, wheat, and chicken datasets. (2)s represent the distribution of RNA sequences with ORF-attention on human, mouse, zebrafish, wheat, and chicken datasets.
3.2. Evaluations by comparison with state-of-the-art methods on five datasets
In this section, LncCat is compared with eight methods, including CPAT, CNCI, CPC2, LncFinder, PLEK, CPPred, mRNN and RNAsamba on five species datasets, including human (Homo sapiens), mouse (Mus musculus), zebrafish (Danio rerio), wheat (Triticum aestivum), and chicken (Gallus gallus). For CPAT, CNCI, LncFinder and PLEK, the re-trained models are constructed by the same datasets as LncCat for a comprehensive and fair comparison.
The first comparison is on the human dataset. Table 3 displays the main metrics for each method.
Table 3.
Comparison of LncCat and eight methods on the human dataset.
| Methods | PRE | SEN | SPE | ACC | F1 | MCC | AUC |
|---|---|---|---|---|---|---|---|
| CPAT | 0.9330 | 0.9405 | 0.9379 | 0.9392 | 0.9367 | 0.8782 | 0.9835 |
| CNCI | 0.8967 | 0.9753 | 0.8968 | 0.9344 | 0.9343 | 0.8721 | 0.9271 |
| CPC2 | 0.9391 | 0.9476 | 0.9436 | 0.9455 | 0.9433 | 0.8909 | 0.9865 |
| LncFinder | 0.9515 | 0.9675 | 0.9547 | 0.9608 | 0.9594 | 0.9217 | 0.9873 |
| PLEK | 0.9472 | 0.9825 | 0.9497 | 0.9654 | 0.9645 | 0.9313 | 0.9918 |
| CPPred | 0.9389 | 0.9545 | 0.9429 | 0.9485 | 0.9466 | 0.8969 | 0.9872 |
| mRNN | 0.9592 | 0.9488 | 0.9630 | 0.9562 | 0.9540 | 0.9122 | 0.9774 |
| RNAsamba | 0.9837 | 0.9429 | 0.9856 | 0.9651 | 0.9628 | 0.9308 | 0.9943 |
| LncCat | 0.9666 | 0.9819 | 0.9688 | 0.9751 | 0.9742 | 0.9503 | 0.9966 |
From Table 3, LncCat outperforms other methods with F1 of 0.9742 and MCC of 0.9503, followed by PLEK (F1: 0.9645; MCC: 0.9313). In addition, LncCat achieves the highest ACC and AUC (ACC: 0.9751; AUC: 0.9966). CNCI achieves an MCC of 0.8721, which is slightly inferior to CPAT. PLEK reaches the highest SEN of 0.9825, indicating its strong ability to predict positive classes. CPPred has a 0.33% advantage over CPC2 in F1 and a 0.06% advantage in MCC. In terms of deep learning methods, RNAsamba achieves greater MCC and F1 than mRNN (RNAsamba: F1, 0.9628, MCC, 0.9308; mRNN: F1, 0. 9540, MCC, 0. 9122).
In addition, nine methods are evaluated on the mouse dataset, as it is one of the most important species. CPC2 is a species-neutral classification method that can be applied to the transcriptions of non-model organisms. Table 4 displays the performance of the different methods on the mouse dataset. For the mouse dataset, LncCat achieves the best performance among these methods with F1 of 0.9487 and MCC of 0.9219. In addition, ACC and AUC are the highest as well. PLEK performs poorly on the mouse dataset, obtaining only an MCC of 0.7456. The MCC of CPC2 and CNCI is comparable. The MCC of CPC2 is 0.8661, while the MCC of CNCI is 0.8687. LncFinder is inferior to LncCat but superior to other methods, with F1 of 0.9391 and MCC of 0.9070. Neither mRNN nor RNAsamba is satisfactory, with MCC values of 0.5971 and 0.6664, respectively.
Table 4.
Comparison of LncCat and eight methods on the mouse dataset.
| Methods | PRE | SEN | SPE | ACC | F1 | MCC | AUC |
|---|---|---|---|---|---|---|---|
| CPAT | 0.9138 | 0.9099 | 0.9556 | 0.9400 | 0.9119 | 0.8664 | 0.9796 |
| CNCI | 0.8687 | 0.9641 | 0.9247 | 0.9381 | 0.9139 | 0.8687 | 0.9473 |
| CPC2 | 0.8871 | 0.9393 | 0.9382 | 0.9386 | 0.9125 | 0.8661 | 0.9805 |
| LncFinder | 0.9183 | 0.9608 | 0.9558 | 0.9575 | 0.9391 | 0.9070 | 0.9822 |
| PLEK | 0.7754 | 0.9057 | 0.8643 | 0.8784 | 0.8355 | 0.7456 | 0.9532 |
| CPPred | 0.8953 | 0.9580 | 0.9421 | 0.9475 | 0.9256 | 0.8863 | 0.9843 |
| mRNN | 0.7384 | 0.9466 | 0.6064 | 0.7901 | 0.8297 | 0.5971 | 0.8460 |
| RNAsamba | 0.8081 | 0.9074 | 0.7470 | 0.8336 | 0.8548 | 0.6676 | 0.9021 |
| LncCat | 0.9296 | 0.9687 | 0.9620 | 0.9643 | 0.9487 | 0.9219 | 0.9920 |
To visually demonstrate the performance of each method, the ROC curves of the nine methods on the human and mouse datasets are shown in Fig. 3. LncCat achieves the highest AUC on human and mouse datasets, with an AUC score of 0.9966 and 0.9920, respectively. On the human dataset, PLEK achieves an AUC of 0.9918, which is the second-best method; however, on the mouse dataset, it only achieves 0.9532. CPPred achieves an AUC of 0.9843, which is 0.0210 greater than LncFinder. But in MCC, LncFinder is 0.0207 higher than CPPred. RNAsamba gets the second highest AUC (AUC: 0.9943) on the human dataset but only gets an AUC of 0.9021 on the mouse dataset. MCC and F1 are more reliable metrics for unbalanced datasets. Therefore, F1 and MCC bar charts of human and mouse datasets are plotted and displayed in Fig. S1–6 and Figs. S1–S7.
Fig. 3.
Comparison of CPAT, CNCI, CPC2, LncFinder, PLEK, CPPred, mRNN, RNAsamba and LncCat by ROC curves. (A) ROC curves on the human dataset. (B) ROC curves on the mouse dataset.
The comparison methods are evaluated on the other three species: zebrafish, wheat, and chicken. The MCC and F1 of nine methods on three datasets are shown in Fig. 4 (A, B). On zebrafish, wheat, and chicken datasets, the MCC of LncCat achieves 0.8858, 0.7971, and 0.8735, showing an improvement ranging from 6.11% to 21.50%, 3.02–60.60%, and 5.34–26.91%, respectively. On wheat datasets, LncFinder achieves the highest SEN with 0.9582, while CPPred achieves the highest SPE with 0.9286. However, LncCat is more comprehensive, with the highest MCC and F1. RNAsamba does not perform well on wheat, possibly because it is unsuitable for the plant. As shown in Fig. 4 (C, D, and E), LncCat outperforms the other eight methods with an AUC of 0.9827 for zebrafish, 0.9780 for wheat, and 0.9882 for chicken, which is 3.16%, 2.16%, and 1.64% higher than the second-best method, respectively. More detailed metrics for each method are shown in Table S1–2 to Tables S1–S5.
Fig. 4.
(A) MCC of CPAT, CNCI, CPC2, LncFinder, PLEK, CPPred, mRNN, RNAsamba, and LncCat on zebrafish, wheat, and chicken datasets. (B) F1 of nine methods on zebrafish, wheat, and chicken datasets. ROC curves of nine methods on (C) zebrafish, (D) wheat, and (E) chicken datasets.
3.3. Distribution of the predicting score
The predicting score indicates the possibility that a transcript is a lncRNA or PCT. A heatmap of prediction scores provides a visual representation of each method’s classification capability. As shown in Fig. 5, the darker color indicates a higher probability that the transcript is lncRNA, whereas a lighter color means a higher probability that the transcript is PCT. LncCat is in the last column with a clear distinction between lncRNAs and PCTs.
Fig. 5.
The distribution of predicting score heatmaps on (A) human, (B) mouse, (C) zebrafish, (D) wheat, and (E) chicken datasets.
3.4. The comparison of different cutoff values
The optimal cutoff values for predicting scores may vary in different species, and utilizing inappropriate cutoff values can cause poor results. The MCC and F1 corresponding to different cutoff values for five species datasets are presented in Tables 5 and 6. When MCC and F1 achieve optimum, the cutoff varies between 0.4 and 0.6. In this experiment, the cutoff value is set at 0.5, which means that the prediction score is greater than or equal to 0.5 for lncRNA and less than 0.5 for PCT. Detailed evaluation results of cutoff values are shown in Table S1–7 to Tables S1–S11.
Table 5.
MCC corresponding to different cutoff values on five species datasets.
| 0.1 | 0.2 | 0.3 | 0.4 | 0.5 | 0.6 | 0.7 | 0.8 | 0.9 | |
|---|---|---|---|---|---|---|---|---|---|
| Human | 0.9393 | 0.9462 | 0.9486 | 0.9507 | 0.9503 | 0.9493 | 0.9479 | 0.9457 | 0.9368 |
| Mouse | 0.9081 | 0.9171 | 0.9203 | 0.9209 | 0.9219 | 0.9250 | 0.9228 | 0.9213 | 0.9103 |
| Zebrafish | 0.8368 | 0.8421 | 0.8462 | 0.8535 | 0.8521 | 0.8450 | 0.8466 | 0.8465 | 0.8435 |
| Wheat | 0.8447 | 0.8536 | 0.8613 | 0.8629 | 0.8672 | 0.8649 | 0.8634 | 0.8624 | 0.8465 |
| Chicken | 0.8789 | 0.8950 | 0.9012 | 0.9051 | 0.9047 | 0.9043 | 0.9009 | 0.8994 | 0.8851 |
Table 6.
F1 corresponding to different cutoff values on five species datasets.
| 0.1 | 0.2 | 0.3 | 0.4 | 0.5 | 0.6 | 0.7 | 0.8 | 0.9 | |
|---|---|---|---|---|---|---|---|---|---|
| Human | 0.9685 | 0.9721 | 0.9734 | 0.9744 | 0.9742 | 0.9737 | 0.9728 | 0.9715 | 0.9664 |
| Mouse | 0.9393 | 0.9454 | 0.9476 | 0.9481 | 0.9488 | 0.9508 | 0.9492 | 0.9480 | 0.9402 |
| Zebrafish | 0.8716 | 0.8763 | 0.8798 | 0.8856 | 0.8846 | 0.8789 | 0.8796 | 0.8786 | 0.8744 |
| Wheat | 0.9291 | 0.9333 | 0.9368 | 0.9375 | 0.9393 | 0.9381 | 0.9371 | 0.9360 | 0.9266 |
| Chicken | 0.9236 | 0.9340 | 0.9379 | 0.9404 | 0.9401 | 0.9397 | 0.9373 | 0.9358 | 0.9255 |
3.5. The performance of LncCat on cross-species datasets
The cross-species experiments are conducted to validate the generalization of LncCat. Fig. 6 shows the MCC of five species models validated on five species datasets. The vertical coordinate refers to the different species models, and the horizontal coordinate indicates different species datasets. The intersection indicates the MCC of a species model on a species dataset. From Fig. 6, the model performs best when the dataset is the same species as the model. The human model is validated on mouse and zebrafish datasets, achieving an MCC of 0.93 and 0.90, respectively. Human, mouse, and zebrafish models perform well on each other’s datasets but poorly on wheat, with MCC of 0.67, 0.63, and 0.66. One hypothesis is that the plant and animal RNAs are far from homology. When species differ significantly, re-training with new datasets is necessary. When species differ significantly, it is necessary to re-training the model with new datasets.
Fig. 6.
The MCC of LncCat on cross-species datasets.
3.6. The comparison on the small ORFs datasets
LncCat outperforms other methods on the above five datasets, and other methods also achieve acceptable results. But most of them are not designed for the datasets with sORFs. In addition, LncCat and eight methods were validated on three sORF datasets. The sORF test datasets of human, mouse, and zebrafish are collected from [43], which are derived from Ensembl. Human-sORF test dataset contains 639 lncRNAs and 641 PCTs. The mouse-sORF test dataset contains 993 lncRNAs and 846 PCTs. The zebrafish-sORF test dataset contains 497 lncRNAs and 387 PCTs. The sORF test datasets do not overlap with the preceding long ORFs datasets and can be used as independent test datasets. The summary of the sORF test datasets is shown in Table 7.
Table 7.
Summary of the sORF datasets.
| Datasets | LncRNAs | PCTs |
|---|---|---|
| Human-sORF | 639 | 641 |
| Mouse-sORF | 993 | 846 |
| Zebrafish-sORF | 497 | 387 |
The MCC and F1 of nine methods on three sORF datasets are shown in Tables 8 and 9. Two bar charts are plotted in Fig. 7 (A, B). LncCat exhibits significant advantages on three sORF datasets, with MCC leading by at least 11.90%, 12.95%, and 42.61% on human-sORF, mouse-sORF, and zebrafish-sORF datasets, respectively. CPC2 performs poorly on sORF datasets and even achieves a negative MCC on the zebrafish-sORF dataset. PLEK performs adequately on the human-sORF dataset but poorly on the other two datasets. RNAsamba achieves the second-best MCC on the mouse-sORF dataset, but the MCC is still 12.95% lower than LncCat. On the zebrafish-sORF dataset, the MCC between LncCat and the second-best method comes to 42.61%. The F1 of LncCat on the human-sORF dataset is 11.90% higher than PLEK. The F1 of LncCat improves by 5.49% and 20.14% for the mouse-sORF dataset and zebrafish-sORF dataset, respectively. Detailed metrics are displayed in Tables S12 to S14.
Table 8.
MCC of nine methods on three small ORF datasets.
| Species | CPAT | CNCI | CPC2 | LncFinder | PLEK | CPPred | mRNN | RNAsamba | LncCat |
|---|---|---|---|---|---|---|---|---|---|
| Human | 0.2866 | 0.3982 | 0.1344 | 0.2390 | 0.7668 | 0.4484 | 0.6044 | 0.7404 | 0.8858 |
| Mouse | 0.2884 | 0.4101 | 0.1404 | 0.4754 | 0.4019 | 0.4868 | 0.5971 | 0.6676 | 0.7971 |
| Zebrafish | 0.1965 | 0.1968 | -0.0869 | 0.3824 | 0.3549 | 0.2783 | 0.2812 | 0.4473 | 0.8735 |
Table 9.
F1 of nine methods on three small ORF datasets.
| Species | CPAT | CNCI | CPC2 | LncFinder | PLEK | CPPred | mRNN | RNAsamba | LncCat |
|---|---|---|---|---|---|---|---|---|---|
| Human | 0.7049 | 0.7362 | 0.6722 | 0.6934 | 0.8871 | 0.7543 | 0.8153 | 0.8751 | 0.9431 |
| Mouse | 0.7302 | 0.7665 | 0.7056 | 0.7868 | 0.7626 | 0.7907 | 0.8297 | 0.8548 | 0.9098 |
| Zebrafish | 0.6874 | 0.7033 | 0.6544 | 0.7435 | 0.7423 | 0.7120 | 0.6715 | 0.7043 | 0.9449 |
Fig. 7.
MCC (A) and F1 (B) of CPAT, CNCI, CPC2, LncFinder, PLEK, CPPred, mRNN, RNAsamba and LncCat on three small ORF datasets. ROC curves of nine methods on (C) human-sORF, (D) mouse-sORF and (E) zebrafish-sORF datasets.
Fig. 7 (C, D, E) shows the ROC curves of nine methods on three sORF datasets. LncCat acquires the best AUC. On the human-sORF dataset, PLEK is 3.00% inferior to LncCat and outperforms the other methods with an AUC of 0.9553. The ROC curve of PLEK is steeper than LncCat’s initially, but the inflection point appears earlier, and the AUC score is lower than LncCat’s. On the mouse-sORF dataset, LncCat achieves a 7.56% advantage over the second-best method (LncCat: 0.9777; RNAsamba: 0.9021). CNCI performs poorly on three sORFs datasets, achieving 0.5847, 0.6221, and 0.5876 for the human-sORF, mouse-sORF, and zebrafish-sORF datasets, respectively. On the mouse-sORF dataset, the ROC curve of CPAT is nearly a straight line. Consequently, LncCat is a stable and efficient method and performs well on long ORFs and sORF datasets.
The heat maps of predicting scores on three datasets are shown in Fig. 8. It can be observed that LncCat can effectively distinguish lncRNAs and PCTs. The method with poor performance does not show a clear boundary between lncRNAs and PCTs.
Fig. 8.
The distribution of predicting score heatmaps on (A) human-sORF dataset, (B) mouse-sORF dataset, and (C) zebrafish-sORF dataset.
3.7. Comparison Ribo-seq datasets
In some public datasets, the annotation of LncRNAs is simple, and transcripts are considered LncRNAs because of lacking sufficient long ORFs. Many of these transcripts are subsequently discovered to have coding potential or even to contain functional sORFs. Some peptides translated by sORFs are much shorter than most known proteins but play vital functional roles in various organisms. The emergence of ribosome profiling (also known as ribosome sequencing, Ribo-seq) [60] has significantly facilitated the comprehension of the translational dynamics within the cell. Ribo-seq can systematically monitor the cell translation process, identify which regions of a message are being translated, and help to determine the proteome of complex organisms. Therefore, using the data validated by Ribo-seq is more persuasive.
Study [61] provides 7264 Ribo-seq ORFs, involving a consensus set of Ribo-seq ORFs identified by seven recent experimental publications mapped to GENCODE version 35 annotations. The transcripts (LncRNAs and PCTs) are obtained by mapping the transcript IDs from the annotation files to the GENCODE version 35. The transcripts are divided into the training dataset, test dataset, and validation dataset in the proportion of 7:2:1. Table 10 provides the details of the datasets.
Table 10.
The details of the Ribo-seq dataset.
| Datasets | LncRNAs | PCTs |
|---|---|---|
| Training dataset | 3668 | 10,783 |
| Test dataset | 1049 | 3082 |
| Validation dataset | 525 | 1541 |
From Table 11 and Fig. 9, LncCat obtains an MCC of 0.8004, which is 6.34% higher than the second-best method mRNN. The deep learning methods mRNN achieves an MCC of 0.7370, and RNAsamba achieves an MCC of 0.7254. But CPAT misclassifies many coding sequences into non-coding sequences. Although CPPred obtains the highest SEN, it misclassifies many coding sequences, which leads to poor performance. LncFinder achieves an ACC of 0.8877, which is higher than mRNN and RNAsamba (mRNN: 0.8816; RNAsamba: 0.8840), but its MCC is only 0.6983.
Table 11.
Comparison of LncCat and CPAT, CNCI, CPC2, LncFinder, PLEK, CPPred, mRNN, RNAsamba on Ribo-seq dataset.
| PRE | SEN | SPE | ACC | F1 | MCC | AUC | |
|---|---|---|---|---|---|---|---|
| CPAT | 0.7637 | 0.4120 | 0.9565 | 0.8180 | 0.5352 | 0.4664 | 0.9035 |
| CNCI | 0.5303 | 0.9828 | 0.7038 | 0.7746 | 0.6889 | 0.5987 | 0.8325 |
| CPC2 | 0.4747 | 0.9285 | 0.6502 | 0.7209 | 0.6282 | 0.5038 | 0.8382 |
| LncFinder | 0.8000 | 0.7436 | 0.9367 | 0.8877 | 0.7708 | 0.6973 | 0.9362 |
| PLEK | 0.4815 | 0.9066 | 0.6674 | 0.7282 | 0.6290 | 0.5003 | 0.8469 |
| CPPred | 0.5104 | 0.9590 | 0.6869 | 0.7560 | 0.6662 | 0.5629 | 0.8755 |
| mRNN | 0.6972 | 0.9438 | 0.8605 | 0.8816 | 0.8019 | 0.7370 | 0.9531 |
| RNAsamba | 0.7182 | 0.8942 | 0.8806 | 0.8840 | 0.7966 | 0.7253 | 0.9513 |
| LncCat | 0.8716 | 0.8284 | 0.9585 | 0.9254 | 0.8495 | 0.8004 | 0.9739 |
Fig. 9.
ROC curves of CPAT, CNCI, CPC2, LncFinder, PLEK, CPPred, mRNN, RNAsamba, and LncCat on the Ribo-seq dataset.
K-fold cross-validation can ensure that all points are considered at least once. We validate LncCat with 5-fold cross-validation and 10-fold cross-validation on the Ribo-seq dataset. The performance of each fold is shown in Tables 12 and 13. Overall, LncCat can achieve a stable and satisfactory performance for each fold.
Table 12.
Metrics of LncCat on Ribo-seq dataset by 5-fold cross-validation.
| PRE | SEN | SPE | ACC | F1 | MCC | AUC | |
|---|---|---|---|---|---|---|---|
| Fold1 | 0.8690 | 0.8194 | 0.9578 | 0.9225 | 0.8434 | 0.7926 | 0.9717 |
| Fold2 | 0.8787 | 0.8170 | 0.9600 | 0.9225 | 0.8467 | 0.7959 | 0.9728 |
| Fold3 | 0.8601 | 0.8074 | 0.9565 | 0.9194 | 0.8329 | 0.7805 | 0.9705 |
| Fold4 | 0.8761 | 0.8508 | 0.9595 | 0.9322 | 0.8633 | 0.8184 | 0.9786 |
| Fold5 | 0.8800 | 0.8098 | 0.9628 | 0.9242 | 0.8434 | 0.7947 | 0.9720 |
Table 13.
Metrics of LncCat on Ribo-seq dataset by 10-fold cross-validation.
| PRE | SEN | SPE | ACC | F1 | MCC | AUC | |
|---|---|---|---|---|---|---|---|
| Fold1 | 0.8780 | 0.8362 | 0.9604 | 0.9288 | 0.8566 | 0.8097 | 0.9755 |
| Fold2 | 0.8672 | 0.8178 | 0.9571 | 0.9215 | 0.8418 | 0.7903 | 0.9693 |
| Fold3 | 0.8770 | 0.8086 | 0.9601 | 0.9206 | 0.8414 | 0.7897 | 0.9754 |
| Fold4 | 0.8797 | 0.8199 | 0.9599 | 0.9230 | 0.8487 | 0.7981 | 0.9722 |
| Fold5 | 0.8526 | 0.8143 | 0.9562 | 0.9225 | 0.8330 | 0.7829 | 0.9735 |
| Fold6 | 0.8706 | 0.8253 | 0.9568 | 0.9225 | 0.8473 | 0.7960 | 0.9709 |
| Fold7 | 0.8765 | 0.8629 | 0.9593 | 0.9351 | 0.8696 | 0.8265 | 0.9783 |
| Fold8 | 0.8743 | 0.8407 | 0.9592 | 0.9293 | 0.8571 | 0.8105 | 0.9788 |
| Fold9 | 0.8926 | 0.8060 | 0.9660 | 0.9244 | 0.8471 | 0.7989 | 0.9739 |
| Fold10 | 0.8560 | 0.8238 | 0.9551 | 0.9230 | 0.8396 | 0.7891 | 0.9714 |
3.8. Integration model
There is insufficient data to construct a prediction model for unexplored and unannotated species. A multi-species integrated model is built for these underexplored species as a reference. The training dataset combines human, mouse, zebrafish, wheat, and chicken training datasets. And LncCat is compared with CPAT, CNCI, CPC2, LncFinder, PLEK, CPPred, mRNN, and RNAsamba on integrated test datasets and sORF integrated test datasets. Table 14 and Table 15 show the main metrics of the nine methods.
Table 14.
Comparison on the integrated dataset.
| Methods | PRE | SEN | SPE | ACC | F1 | MCC | AUC |
|---|---|---|---|---|---|---|---|
| CPAT | 0.8857 | 0.8917 | 0.9262 | 0.9127 | 0.8887 | 0.8169 | 0.9627 |
| CNCI | 0.8747 | 0.9402 | 0.9137 | 0.9240 | 0.9062 | 0.8441 | 0.9285 |
| CPC2 | 0.9054 | 0.8856 | 0.9408 | 0.9192 | 0.8954 | 0.8298 | 0.9709 |
| LncFinder | 0.9234 | 0.9280 | 0.9507 | 0.9418 | 0.9257 | 0.8779 | 0.9741 |
| PLEK | 0.8272 | 0.8946 | 0.8803 | 0.8859 | 0.8596 | 0.7654 | 0.9510 |
| CPPred | 0.9166 | 0.8969 | 0.9477 | 0.9279 | 0.9067 | 0.8481 | 0.9751 |
| mRNN | 0.9410 | 0.8492 | 0.9659 | 0.9203 | 0.8928 | 0.8324 | 0.9574 |
| RNAsamba | 0.9704 | 0.7907 | 0.9846 | 0.9089 | 0.8714 | 0.8121 | 0.9810 |
| LncCat | 0.9425 | 0.9651 | 0.9623 | 0.9634 | 0.9537 | 0.9236 | 0.9915 |
Table 15.
Comparison on the sORF integrated dataset.
| Methods | PRE | SEN | SPE | ACC | F1 | MCC | AUC |
|---|---|---|---|---|---|---|---|
| CPAT | 0.5755 | 0.9202 | 0.2289 | 0.5966 | 0.7081 | 0.2085 | 0.7482 |
| CNCI | 0.6171 | 0.9347 | 0.3410 | 0.6568 | 0.7434 | 0.3477 | 0.6050 |
| CPC2 | 0.5445 | 0.9192 | 0.1265 | 0.5481 | 0.6839 | 0.0753 | 0.7617 |
| LncFinder | 0.6186 | 0.9347 | 0.3453 | 0.6588 | 0.7445 | 0.3517 | 0.7640 |
| PLEK | 0.7091 | 0.9023 | 0.5784 | 0.7509 | 0.7941 | 0.5131 | 0.8412 |
| CPPred | 0.6537 | 0.9150 | 0.4493 | 0.6970 | 0.7626 | 0.4167 | 0.8385 |
| mRNN | 0.7219 | 0.8793 | 0.6153 | 0.7557 | 0.7929 | 0.5166 | 0.7961 |
| RNAsamba | 0.8146 | 0.8483 | 0.7807 | 0.8166 | 0.8311 | 0.6314 | 0.8740 |
| LncCat | 0.8786 | 0.9756 | 0.8469 | 0.9153 | 0.9245 | 0.8346 | 0.9717 |
As shown in Table 14, LncCat achieves MCC of 0.9236 and F1 of 0.9537, which improves the MCC and F1 from 4.58% to 15.82% and 2.80–9.41%, respectively. Table 15 shows that the performance of LncCat on the sORF-integrated dataset is also satisfactory.
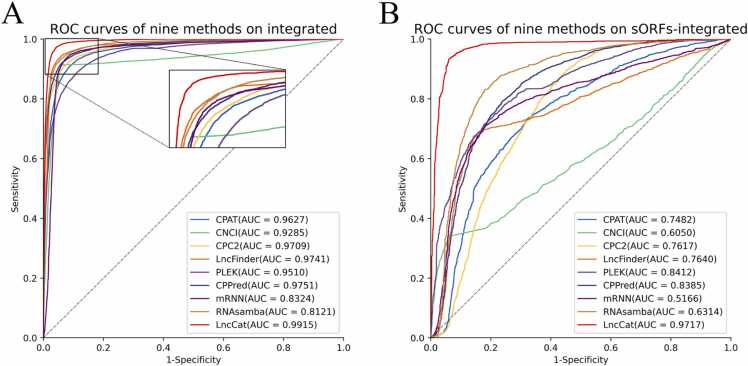
Fig. 10 shows the ROC curves for the integrated and sORF-integrated datasets. LncCat achieves an AUC of 0.9915, which is higher than other methods on the integrated dataset ranging from 1.64% to 6.30%. On the sORF-integrated dataset, The AUC of LncCat outperforms the second-best method by 13.05%.
Fig. 10.
ROC curves of LncCat and CPAT, CNCI, CPC2, LncFinder, PLEK, CPPred, mRNN, RNAsamba on (A) integrated dataset and (B) sORF integrated dataset.
The comparison results demonstrate that LncCat can provide favorable overall performance, and we expect LncCat will provide a reliable reference for underexplored species. The detailed results are shown in Tables S1–15 and S1-S16. The MCC and F1 of nine methods on integrated and sORF-integrated datasets are shown in Figs. S1-S8 and S1–S9.
4. Discussion
Many new lncRNAs and PCT have been generated since the advance of sequencing technology. Previous research has shown that lncRNAs are linked to human diseases and play roles in various cancers. The identification of lncRNAs is an important first step in exploring their functions and mechanisms. Previous methods have produced satisfactory results on long ORF datasets, but identifying sequences with sORFs remains challenging.
In previous studies, ORF length and ORF coverage have shown strong classification ability and are wildly used to predict lncRNA with long ORFs. Based on the clues, the ORF-attention features are constructed from the top three longest ORFs, including codons-related, GC-related, sequence-related, and peptide-related features. Additionally, the pre-trained BERT model has shown effectiveness in representing protein sequences. The attention mechanism in BERT allows it to capture the dependencies between the amino acids in a peptide sequence. Therefore, LncCat employs the BERT model to represent peptide sequences encoded by ORFs as a part of ORF-attention features. Furthermore, to enhance the feature abundance, ORF-attention features source from the top three longest ORFs. The comparison results indicate ORF-attention features can deeply excavate potential information in the ORFs. Moreover, to cover the non-ORF region, widely used lncRNA features are merged to generate the final feature spaces. Finally, a 599-dimensional feature vector is obtained to identify lncRNA.
CatBoost is a high-performance gradient boosting implementation. LncCat employs CatBoost to construct the prediction model with the above features to identify lncRNA on five species datasets and the Ribo-seq dataset. According to comparison experimental results, LncCat improves MCC by at least 1.90%, 1.49%, 6.11%, 3.02% and 5.34% on the human, mouse, zebrafish, wheat and chicken datasets. For sORF datasets, LncCat achieves a considerable improvement and increases the MCC by at least 11.90%, 12.96%, and 42.61% on the human-sORF dataset, mouse-sORF dataset, zebrafish-sORF dataset. The stable performance of LncCat on the long ORFs datasets and the sORF datasets indicates that ORF-attention features are effective and reliable.
LncCat still gets the preferable MCC in cross-species experiments. The human model and mouse model perform tightly on the other three species datasets because of similar homology. Chicken and zebrafish are oviparous, and their models perform similarly on the other three species datasets. Animal models, such as human, mouse, zebrafish and chicken, show slightly lower performances on the plant dataset because of the poor genomic homology between animals and plants. The cross-species experiments between animal and plant species reflect that LncCat performs better on homologous species and achieves relatively satisfactory results for non-homologous species. Besides, for the other species models, LncCat also achieves favorable results, which shows the higher generalization on cross-species datasets rather than one species dataset alone.
Species may lack annotations, and it is impossible to build models for all species. We build a multi-species integrated model by the training dataset of five species. The MCC of LncCat is 0.9236 on the long ORF integration dataset, which is 4.58∼15.82% higher than the compared models. The MCC on the sORF dataset is 20.32% higher than the second-best model. Additionally, LncCat has been deployed on the web server and can provide a reference for under-explored species.
The source code and datasets of this experiment are available for academics at https://github.com/a525076133/LncCat. The model can be re-trained by the user’s data. Moreover, a user-friendly web server is developed with free accessibility at http://cczubio.top/lnccat/. The web server provides the identification of lncRNA for multiple species with FASTA format sequences as input. Users can upload a FASTA file or input sequences in the text area. Currently, five species and integrated species are available on this web server, including human, mouse, wheat, chicken and zebrafish. The results will be replied to the user’s email.
sORFs may translate peptides, and peptides have been one of the most promising cures for cancer in recent years [62]. In the following work, we will collect the sORF-translated peptide sequences and annotate the functions of peptides on a large scale by protein family’s alignment and modeling to drive the process of peptides in cancer therapy.
In summary, the experimental results show that ORF-attention features are crucial and effective in identifying lncRNA on both long ORF and sORF datasets. In all, LncCat is an effective method for lncRNA identification.
5. Conclusion
LncRNA identification is a very significant task in bioinformatics. Although researchers have achieved satisfactory results on long ORFs datasets, there is still considerable space for improvement on the sORF datasets. In this study, we propose a method called LncCat, based on ensemble learning called CatBoost. Five types of features are used to encode transcripts into vectors, and CatBoost is employed to construct the prediction model. The extensive experimental results on five species show that LncCat performs well on five datasets and three sORF datasets. In terms of MCC, LncCat achieved 0.9503, 0.9219, 0.8591, 0.8672, and 0.9047 on human, mouse, zebrafish, wheat, and chicken datasets, with improvement ranging from 1.90% to 7.82%, 1.49–17.63%, 6.11–21.50%, 3.02–51.64% and 5.35–26.90%, respectively. In addition, LncCat increases MCC by at least 6.88%, 22.98%, and 39.43 on the human-sORF dataset, mouse-sORF dataset, and zebrafish-sORF dataset compared with the other eight methods. Moreover, the feature comparing results show that the ORF-attention features effectively differentiate lncRNAs from PCTs. In all, the ORF-attention feature we proposed can help distinguish LncRNAs from PCTs, especially for the sORF dataset. LncCat is a reliable and robust machine learning model for identifying lncRNAs. A user-friendly web server is available at http://cczubio.top/lnccat/.
Funding
This research was funded by the National Natural Science Foundation of China (No. 62072212).
CRediT authorship contribution statement
Shaocong Wang conceived the algorithm, developed the program, and wrote the manuscript. Sen Yang, Hongqi Feng, Yan Wang and Xinye Ni helped with manuscript editing and design. Zixi Yang and Sen Yang prepared the datasets. All authors have read and agreed to the published version of the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.02.012.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Data Availability Statement
Publicly available datasets were analyzed in this study. Codes and data are available at http://cczubio.top/lnccat/ and https://github.com/a525076133/LncCat.
References
- 1.Pennisi E. ENCODE project writes eulogy for junk DNA. Science. 2012;337:1159–1161. doi: 10.1126/science.337.6099.1159. [DOI] [PubMed] [Google Scholar]
- 2.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., et al. The transcriptional landscape of the mammalian genome. Sci, N Ser. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 4.The ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 6.Lee J.T. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 7.Li R., ZHu H., Luo Y. Understanding the long non-coding RNA biological function through its structure conservation. Int J Mol Sci. 2016;17:702. doi: 10.3390/ijms17050702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhartiya D., Kapoor S., Jalali S., Sati S., Kaushik K., Sachidanandan C., et al. Conceptual approaches for lncRNA drug discovery and future strategies. Expert Opin Drug Discov. 2012;7:503–513. doi: 10.1517/17460441.2012.682055. [DOI] [PubMed] [Google Scholar]
- 9.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.da Rocha S.T., Boeva V., Escamilla-Del-Arenal M., Ancelin K., Granier C., Matias N.R., et al. Jarid2 Is Implicated in the Initial Xist-Induced Targeting of PRC2 to the Inactive X Chromosome. Mol Cell. 2014;53:301–316. doi: 10.1016/j.molcel.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y., Tao Y., Liao Q. Long noncoding RNA: a crosslink in biological regulatory network. Brief Bioinforma. 2018;19:930–945. doi: 10.1093/bib/bbx042. [DOI] [PubMed] [Google Scholar]
- 12.O’Leary V.B., Ovsepian S.V., Carrascosa L.G., Buske F.A., Radulovic V., Niyazi M., et al. PARTICLE, a Triplex-Forming Long ncRNA, Regulates Locus-Specific Methylation in Response to Low-Dose Irradiation. Cell Rep. 2015;11:474–485. doi: 10.1016/j.celrep.2015.03.043. [DOI] [PubMed] [Google Scholar]
- 13.Shi X., Sun M., Liu H., Yao Y., Kong R., Chen F., et al. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer: GAS5 REGULATES PROLIFERATION AND APOPTOSIS OF NSCLC. Mol Carcinog. 2015;54:E1–E12. doi: 10.1002/mc.22120. [DOI] [PubMed] [Google Scholar]
- 14.Ng S.-Y., Lin L., Soh B.S., Stanton L.W. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 2013;29:461–468. doi: 10.1016/j.tig.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Congrains A., Kamide K., Oguro R., Yasuda O., Miyata K., Yamamoto E., et al. Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis. 2012;220:449–455. doi: 10.1016/j.atherosclerosis.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 16.Ning S., Zhang J., Wang P., Zhi H., Wang J., Liu Y., et al. Lnc2Cancer: a manually curated database of experimentally supported lncRNAs associated with various human cancers. Nucleic Acids Res. 2016;44:D980–D985. doi: 10.1093/nar/gkv1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen G., Wang Z., Wang D., Qiu C., Liu M., Chen X., et al. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2012;41:D983–D986. doi: 10.1093/nar/gks1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H.-L.V., Chekanova J.A. In: Rao M.R.S., editor. vol. 1008. Springer Singapore; Singapore: 2017. Long Noncoding RNAs in Plants; pp. 133–154. (Long Non Coding RNA Biology). [DOI] [Google Scholar]
- 19.Hu R., Sun X. lncRNATargets: A platform for lncRNA target prediction based on nucleic acid thermodynamics. J Bioinform Comput Biol. 2016;14 doi: 10.1142/S0219720016500165. [DOI] [PubMed] [Google Scholar]
- 20.Ma L., Bajic V.B., Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:924–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Röhrig H., Schmidt J., Miklashevichs E., Schell J., John M. Soybean ENOD40 encodes two peptides that bind to sucrose synthase. Proc Natl Acad Sci USA. 2002;99:1915–1920. doi: 10.1073/pnas.022664799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong L., Zhang Y., Ye Z.-Q., Liu X.-Q., Zhao S.-Q., Wei L., et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L., Park H.J., Dasari S., Wang S., Kocher J.-P., Li W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gkt006. e74–e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fickett J.W. Recognition of protein coding regions in DNA sequences. Nucleic Acids Res. 1982;10:5303–5318. doi: 10.1093/nar/10.17.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fickett J.W., Tung C.-S. Assessment of protein coding measures. Nucleic Acids Res. 1992;20:6441–6450. doi: 10.1093/nar/20.24.6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun L., Luo H., Bu D., Zhao G., Yu K., Zhang C., et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gkt646. e166–e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li A., Zhang J., Zhou Z. PLEK: a tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinforma. 2014;15:311. doi: 10.1186/1471-2105-15-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang Y.-J., Yang D.-C., Kong L., Hou M., Meng Y.-Q., Wei L., et al. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017;45:W12–W16. doi: 10.1093/nar/gkx428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han S., Liang Y., Ma Q., Xu Y., Zhang Y., Du W., et al. LncFinder: an integrated platform for long non-coding RNA identification utilizing sequence intrinsic composition, structural information and physicochemical property. Brief Bioinforma. 2019;20:2009–2027. doi: 10.1093/bib/bby065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nair A.S., Sreenadhan S.P. A coding measure scheme employing electron-ion interaction pseudopotential (EIIP) Bioinformation. 2006;1:197–202. [PMC free article] [PubMed] [Google Scholar]
- 31.Hill S.T., Kuintzle R., Teegarden A., Merrill E., Danaee P., Hendrix D.A. A deep recurrent neural network discovers complex biological rules to decipher RNA protein-coding potential. Nucleic Acids Res. 2018;46:8105–8113. doi: 10.1093/nar/gky567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Camargo A.P., Sourkov V., Pereira G.A.G., Carazzolle M.F. RNAsamba: neural network-based assessment of the protein-coding potential of RNA sequences. NAR Genom Bioinforma. 2020;2:lqz024. doi: 10.1093/nargab/lqz024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griffiths P., Nendel C., Pickert J., Hostert P. Towards national-scale characterization of grassland use intensity from integrated Sentinel-2 and Landsat time series. Remote Sens Environ. 2020;238 doi: 10.1016/j.rse.2019.03.017. [DOI] [Google Scholar]
- 34.Gomez C., Mangeas M., Petit M., Corbane C., Hamon P., Hamon S., et al. Use of high-resolution satellite imagery in an integrated model to predict the distribution of shade coffee tree hybrid zones. Remote Sens Environ. 2010;114:2731–2744. doi: 10.1016/j.rse.2010.06.007. [DOI] [Google Scholar]
- 35.Chrysafis I., Mallinis G., Gitas I., Tsakiri-Strati M. Estimating Mediterranean forest parameters using multi seasonal Landsat 8 OLI imagery and an ensemble learning method. Remote Sens Environ. 2017;199:154–166. doi: 10.1016/j.rse.2017.07.018. [DOI] [Google Scholar]
- 36.Freund Y., Schapire R.E. A decision-theoretic generalization of on-line learning and an application to boosting. J Comput Syst Sci. 1997;55:119–139. doi: 10.1006/jcss.1997.1504. [DOI] [Google Scholar]
- 37.Chen T., Guestrin C. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. ACM,; San Francisco California USA: 2016. XGBoost: a scalable tree boosting system; pp. 785–794. [DOI] [Google Scholar]
- 38.Sun X., Liu M., Sima Z. A novel cryptocurrency price trend forecasting model based on LightGBM. Financ Res Lett. 2020;32 doi: 10.1016/j.frl.2018.12.032. [DOI] [Google Scholar]
- 39.Prokhorenkova L., Gusev G., Vorobev A., Dorogush A.V., Gulin A. CatBoost: unbiased boosting with categorical features. ArXiv. 2019 170609516 [Cs] [Google Scholar]
- 40.Huang G., Wu L., Ma X., Zhang W., Fan J., Yu X., et al. Evaluation of CatBoost method for prediction of reference evapotranspiration in humid regions. J Hydrol. 2019;574:1029–1041. doi: 10.1016/j.jhydrol.2019.04.085. [DOI] [Google Scholar]
- 41.Fan J., Wang X., Zhang F., Ma X., Wu L. Predicting daily diffuse horizontal solar radiation in various climatic regions of China using support vector machine and tree-based soft computing models with local and extrinsic climatic data. J Clean Prod. 2020;248 doi: 10.1016/j.jclepro.2019.119264. [DOI] [Google Scholar]
- 42.Waqas Khan P., Byun Y.-C., Lee S.-J., Park N. Machine learning based hybrid system for imputation and efficient energy demand forecasting. Energies. 2020;13:2681. doi: 10.3390/en13112681. [DOI] [Google Scholar]
- 43.Tong X., Liu S. CPPred: coding potential prediction based on the global description of RNA sequence. Nucleic Acids Res. 2019;47 doi: 10.1093/nar/gkz087. e43–e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zerbino D.R., Achuthan P., Akanni W., Amode M.R., Barrell D., Bhai J., et al. Ensembl 2018. Nucleic Acids Res. 2018;46:D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frankish A., Diekhans M., Ferreira A.-M., Johnson R., Jungreis I., Loveland J., et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47:D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 48.Panwar B., Arora A., Raghava G.P. Prediction and classification of ncRNAs using structural information. BMC Genom. 2014;15:127. doi: 10.1186/1471-2164-15-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pozzoli U., Menozzi G., Fumagalli M., Cereda M., Comi G.P., Cagliani R., et al. Both selective and neutral processes drive GC content evolution in the human genome. BMC Evolut Biol. 2008;8:99. doi: 10.1186/1471-2148-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang C., Yang L., Zhou M., Xie H., Zhang C., Wang M.D., et al. LncADeep: an ab initio lncRNA identification and functional annotation tool based on deep learning. Bioinformatics. 2018;34:3825–3834. doi: 10.1093/bioinformatics/bty428. [DOI] [PubMed] [Google Scholar]
- 51.Suenaga Y., Kato M., Nagai M., Nakatani K., Kogashi H., Kobatake M., et al. Open reading frame dominance indicates protein‐coding potential of RNAs. EMBO Rep. 2022:23. doi: 10.15252/embr.202154321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh U., Wurtele E.S. orfipy: a fast and flexible tool for extracting ORFs. Bioinformatics. 2021;37:3019–3020. doi: 10.1093/bioinformatics/btab090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abramczyk D., Tchórzewski M., Grankowski N. Non-AUG translation initiation of mRNA encoding acidic ribosomal P2A protein in Candida albicans: Alternative start codon of P-protein gene from Candida albicans. Yeast. 2003;20:1045–1052. doi: 10.1002/yea.1020. [DOI] [PubMed] [Google Scholar]
- 54.Sugihara H., Andrisani V., Salvaterra P.M. Drosophila choline acetyltransferase uses a non-AUG initiation codon and full length RNA is inefficiently translated. J Biol Chem. 1990;265:21714–21719. [PubMed] [Google Scholar]
- 55.Prats H., Kaghad M., Prats A.C., Klagsbrun M., Lélias J.M., Liauzun P., et al. High molecular mass forms of basic fibroblast growth factor are initiated by alternative CUG codons. Proc Natl Acad Sci USA. 1989;86:1836–1840. doi: 10.1073/pnas.86.6.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takahashi K., Maruyama M., Tokuzawa Y., Murakami M., Oda Y., Yoshikane N., et al. Evolutionarily conserved non-AUG translation initiation in NAT1/p97/DAP5 (EIF4G2) Genomics. 2005;85:360–371. doi: 10.1016/j.ygeno.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 57.Hann S.R., King M.W., Bentley D.L., Anderson C.W., Eisenman R.N. A non-AUG translational initiation in c-myc exon 1 generates an N-terminally distinct protein whose synthesis is disrupted in Burkitt’s lymphomas. Cell. 1988;52:185–195. doi: 10.1016/0092-8674(88)90507-7. [DOI] [PubMed] [Google Scholar]
- 58.Devlin J., Chang M.-W., Lee K., Toutanova K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding, 2018. https://doi.org/10.48550/ARXIV.1810.04805.
- 59.Chicco D., Jurman G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020;21:6. doi: 10.1186/s12864-019-6413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ingolia N.T., Ghaemmaghami S., Newman J.R.S., Weissman J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mudge J.M., Ruiz-Orera J., Prensner J.R., Brunet M.A., Calvet F., Jungreis I., et al. Standardized annotation of translated open reading frames. Nat Biotechnol. 2022;40:994–999. doi: 10.1038/s41587-022-01369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu L., Ye C., Hu X., Yang S., Zhu C. ACP-check: An anticancer peptide prediction model based on bidirectional long short-term memory and multi-features fusion strategy. Comput Biol Med. 2022 doi: 10.1016/j.compbiomed.2022.105868. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Data Availability Statement
Publicly available datasets were analyzed in this study. Codes and data are available at http://cczubio.top/lnccat/ and https://github.com/a525076133/LncCat.