Abstract
Cancer is a global public health concern. Alterations in epigenetic processes are among the earliest genomic aberrations occurring during cancer development and are closely related to progression. Unlike genetic mutations, aberrations in epigenetic processes are reversible, which opens the possibility for novel pharmacological treatments. Non-coding RNAs (ncRNAs) represent an essential epigenetic mechanism, and emerging evidence links ncRNAs to carcinogenesis. Epigenetic drugs (epidrugs) are a group of promising target therapies for cancer treatment acting as coadjuvants to reverse drug resistance in cancer. The present review describes central epigenetic aberrations during malignant transformation and explains how epidrugs target DNA methylation, histone modifications and ncRNAs. Furthermore, clinical trials focused on evaluating the effect of these epidrugs alone or in combination with other anticancer therapies and other ncRNA-based therapies are discussed. The use of epidrugs promises to be an effective tool for reversing drug resistance in some patients with cancer.
Keywords: cancer, therapy, epigenetic mechanisms, non-coding RNA, epigenetic drugs
1. Introduction
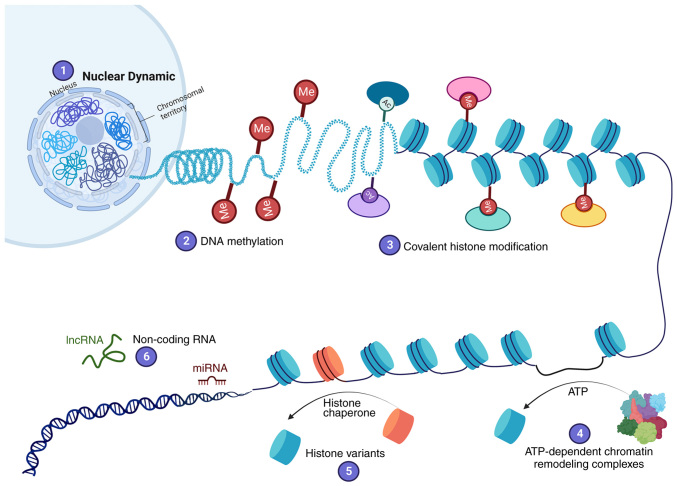
Cancer is one of the leading cause of death worldwide; according to estimates from the World Health Organization, ~10 million cancer deaths occurred in 2020 (1). Cancer results from DNA aberrations that cause the deregulation of pathways controlling cellular processes involved in proliferation, cell survival, apoptosis and DNA repair. Mutations in tumor suppressor genes and oncogenes are responsible for cancer initiation, promotion and progression (2). However, carcinogenesis cannot be explained solely by genetic alterations, as it also involves epigenetic processes. Epigenetic changes are defined as inheritable modifications in gene expression that do not result from changes in DNA sequences (3). Epigenetic mechanisms were discovered as the knowledge regarding DNA structure increased. DNA is packaged in the nucleus by histones into a structure called chromatin. The basic unit of chromatin is the nucleosome; each nucleosome comprises one histone octamer composed of H3, H4, H2A and H2B subunits, with 147 bp of DNA coiling around them. Chromatin is classified as either heterochromatin or euchromatin. Heterochromatin is a region of DNA that is highly condensed and transcriptionally inactive, whereas euchromatin is less condensed and is actively transcribed (4). The chromatin structure creates a barrier to DNA replication, damage repair or access of transcription machinery to DNA. Chromatin is highly dynamic to allow different regulators or transcriptional factors to access DNA (5). Various epigenetic mechanisms regulate the states of euchromatin or heterochromatin. Six epigenetic mechanisms altering chromatin structure have been described (Fig. 1): i) Nuclear dynamic; ii) DNA methylation; iii) covalent histone modification; iv) ATP-dependent chromatin remodeling complexes; v) histone variants; and vi) non-coding RNA (ncRNA), including microRNA (miRNA/miR) and long ncRNA (lncRNA) (5–7).
Figure 1.
Six epigenetic mechanisms involved in the regulation of gene expression. (1) Nuclear dynamics: The structural and three-dimensional organization of the genome in the nucleus, and short- and long-range interconnected transcriptional regulation, which impacts gene expression. (2) DNA methylation: The addition of a methyl group to cytosine nucleotides by DNA methyltransferases, which induces transcriptional repression. (3) Covalent histone modification: The addition of chemical groups to the amino terminus of histones, which can increase or decrease DNA compaction. (4) ATP-dependent chromatin remodeling complexes: Insertion, removal and displacement of nucleosomes along the DNA through ATP hydrolysis to regulate DNA accessibility. (5) Histone variants: Histone chaperones exchange canonical histones for histone variants (H2A.Z, H2A.X, H3.3 and CENP-A). (6) Non-coding RNA: Non-coding RNA expression promotes transcriptional silencing. Overall, this enables chromatin structure modification to regulate gene expression. Ac, acetyl group; lncRNA, long non-coding RNA; Me, methyl group; miRNA, microRNA.
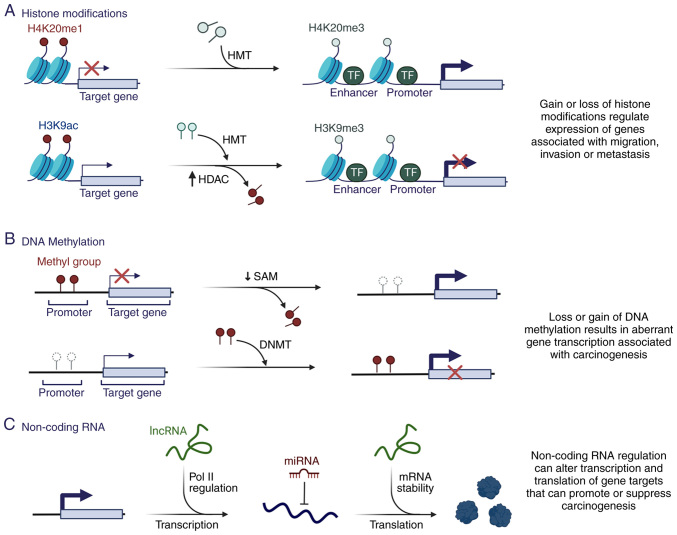
A number of studies have demonstrated that dysregulation of epigenetic mechanisms is a critical contributor to carcinogenesis (8–10) (Fig. 2), mainly due to regulation of DNA accessibility and the compactness of chromatin structure, favoring the regulation of different genes necessary for carcinogenesis (Fig. 2A and B).
Figure 2.
Epigenetic alterations and their contribution to carcinogenesis. (A) Loss or gain of different histone modifications can induce aberrant gene expression and promote carcinogenesis in different tumors. The gain of H3K4me3 by HMT and the loss of H3K9ac by HDAC contributes to the expression of genes associated with carcinogenesis. (B) Alterations in the pattern of DNA methylation and the expression of enzymes, such as DNMTs, are found in several tumors. DNA hypomethylation in various regions increases genomic instability and activates proto-oncogenes, whereas DNA hypermethylation favors the silencing of genes, such as tumor suppressors, which contributes to carcinogenesis. (C) Changes in non-coding RNA expression patterns serve an important role in regulating the initiation and progression of various tumors because they can inhibit or increase gene expression through binding to various target genes. Ac, acetylation; DNMT, DNA methyltransferase; H, histone; HDAC, histone deacetylase; K, lysine; HMT, histone methyltransferase; lncRNA, long non-coding RNA; me1, monomethylation; me3, trimethylation; miRNA, microRNA; Pol II, RNA polymerase II; SAM, S-adenosyl methionine; TF, transcription factor.
Unlike genetic mutations, the cancer epigenomic landscape can be reprogrammed. It represents one of the most promising therapeutic targets for treatment and reversal of drug resistance. Epigenetic alterations in cancer development and progression may be the basis for individual variations in drug response (11,12). The present review focuses on epigenetic reprogramming during tumorigenesis and recent progress in developing clinical strategies to target epigenetic codes.
2. Epigenetic mechanisms in cancer, roles in carcinogenesis and therapeutic approaches
DNA methylation in cancer
DNA methylation is the most well-studied epigenetic modification. It is a stable gene-silencing mechanism that regulates gene expression and chromatin architecture through the covalent addition of a methyl group to the C5 cytosines by DNA methyltransferases (DNMTs), producing 5-methylcytosine (5mC) (Fig. 1) (12). A total of five members of the DNMT family have been identified in mammals: DNMT1, DNMT2, DNMT3A, DNMT3B and DNMT3-like (DNMT3L) (12). However, only DNMT1, DNMT3A and DNMT3B exhibit catalytic activity for DNA methylation (13). The de novo methyltransferases DNMT3A and DNMT3B establish DNA methylation during embryonic development (14,15), whereas DNMT1 is a housekeeping methyltransferase that methylates preexisting hemi-methylated DNA and preserves DNA methylation during DNA replication (14). DNMT3L has sequence homology with DNMT3A/3B but has no catalytic domain; however, it is an essential accessory protein for de novo methylation (16). Although the mechanism remains unclear, DNMT3L modulates the activity of other DNMTs, including DNMT3A (17) and DNMT2, which act as RNA methyltransferases (18).
DNA methylation is reversible and is mediated by three members of the ten-eleven translocation (TET) family: TET1, TET2 and TET3 (19). TET enzymes function in the demethylation of 5mC to 5-hydroxymethylcytosine (5hmC), which is converted to unmethylated cytosine through sequential oxidation to produce 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) intermediates (20). 5fC and 5caC are subsequently removed by DNA glycosylases and the base excision repair machinery to generate unmethylated cytosines at TET-targeted sites, or they are passively demethylated by the loss of 5hmC during cell division (20). Therefore, TET enzymes regulate the chromatin structure and gene expression (21).
DNA methylation serves a fundamental role in different processes, including embryonic development (22), X chromosome inactivation (23), genomic imprinting (24) and transcriptional silencing (25). CpG islands are often methylated in genome sequences rich in cytosine-guanine nucleotide pairs in normal tissues (16). By contrast, promoter regions are generally unmethylated, except for the inactive X chromosome, silenced alleles of imprinted genes and tissue-specific genes (21). DNA methylation can also be detected in repetitive sequences, either in tandem (satellite DNA), or short- or long-interspersed nuclear elements; these modifications mainly help maintain genomic integrity (26).
Approximately 60% of all gene promoters are enriched in CpG islands; therefore, these genes are potentially epigenetically regulated (26). In cancer cells, alterations in DNA methylation are the first epigenetic marker associated with cancer development (Fig. 2B) (27). Cancer cell genomes are hypomethylated relative to normal counterparts, whereas a paradoxical increment of site-specific CpG methylation is observed in the gene promoter regions (28). Loss of methylation in repetitive regions of the genome is the main cause of DNA hypomethylation (29). It is associated with genomic instability, changes in gene imprinting and increased cancer risk (30). In addition, DNA methylation is substantially more frequent than gene inactivation by genetic mutations (31). It can induce gene silencing directly or indirectly by inhibiting the binding of specific transcription factors or by recruiting methyl-CpG-binding domain proteins, respectively (27). The most crucial tumor-suppressor gene silencing occurs through epigenetic mechanisms (32). This includes p14, p16/inhibitors of CDK4 (33), retinoblastoma, cyclin-dependent kinase inhibitor 2 and methylguanine-DNA-methyltransferase, which favor uncontrolled cellular proliferation (34). In addition, alterations in DNA methylation patterns detected in hematological malignancies are frequently associated with the aberrant activity of enzymes regulating DNA methylation, such as DNMT3A (35).
Alterations in the epigenome and deregulated epigenetic mechanisms cannot entirely explain the process of multistage carcinogenesis. However, epigenetic changes are generally of a clonal nature, and occur in the early generation of cancer cells; this initiates genetic instability resulting in the acquisition of genetic mutations in tumor suppressor genes and the activation of genetic mutations in oncogenes that facilitate cell transformation (28). In addition, epigenetic alterations can lead to the development of drug-resistant phenotypes (Fig. 2). Epigenetic inactivation can directly determine tumor chemosensitivity and influence drug resistance and post-therapy clinical outcomes (36–38). These findings and the reversible nature of the underlying changes have made epigenetic alterations potential rational targets for therapeutic approaches. Classes of chromatin-modifying drugs have been approved by the US Food and Drug Administration (FDA), including two main DNMT inhibitors (DNMTi) and histone deacetylase (HDAC) inhibitors (HDACi; Fig. 3A) (39). Their application can reactivate tumor suppressor genes and synergistically suppress the proliferation of cancer cell lines in vitro and in vivo (40,41). In addition, DNMTi are involved in responses to radiotherapy and chemotherapy treatment, sensitizing several chemo- and/or radiation-resistant cancer types in the clinical setting (Fig. 3D) (42,43). For example, treatment with 5-Aza-2′-deoxycytidine (5-aza-D) has been shown to be effective in reversing cisplatin resistance in bladder cancer cells, which is mainly associated with promoter demethylation of the HOXA9 gene (44).
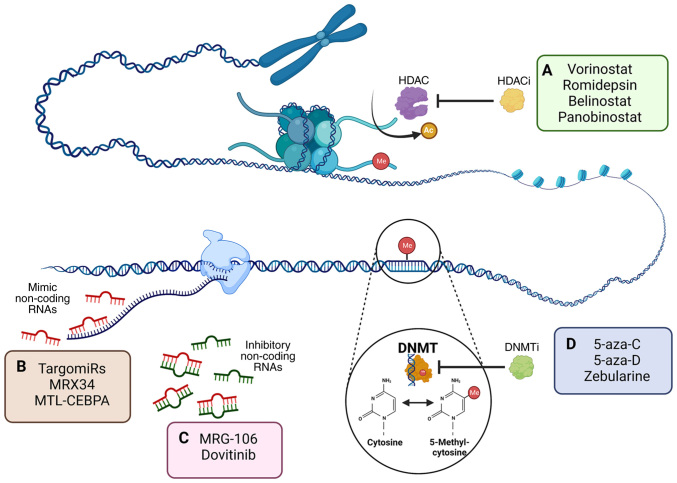
Figure 3.
Inhibitor epigenetic modifications and action in cancer. (A) DNA methylation is catalyzed by DNMTs and involves the addition of a methyl group to cytosine, producing 5-methylcytosine. DNMTi, such as 5-aza-C and 5-aza-D, prevent DNA methylation and restore the function of aberrantly silenced genes in cancer. Therapeutic approaches targeting non-coding RNAs in cancer may be directed to one of two aims: (B) Restoring tumor suppressor activity through mimicking molecules (TargomiRs and MRX34) and synthetic RNAs (MTL-CEBPA) or (C) inhibiting oncogene expression through the employment of molecules that promote the degradation of target mRNAs (MRG-106 and Dovitinib). (D) Histone tails in the nucleosome can be post-translationally modified by the covalent attachment of acetyl groups. Histone deacetylation catalyzed by HDAC modifies the histone code. Various HDACi have been approved by the US Food and Drug Administration for their use as antineoplastic drugs, including vorinostat, romidepsin, belinostat and panobinostat. 5-aza-C, 5-azacytidine; 5-aza-D, 5-aza-2′-deoxycytidine; Ac, acetyl group; DNMT, DNA methyltransferase; DNMTi, DNMT inhibitors; HDAC, histone deacetylase; HDACi, HDAC inhibitors; Me, methyl group; MTL-CEBPA, RNA duplex acting of c/ebpa gen transcription; CEBPA, CCAAT/enhancer-binding protein alpha; MRX34, mimic of miR-34a.
DNMTi for cancer treatment
Drugs that inhibit DNA cytosine methylation are divided into two groups: i) Nucleoside analogs, including 5-azacytidine (5-aza-C; Vidaza) and 5-aza-D (Dacogen® or decitabine) and zebularine (Fig. 3A) (45); and ii) non-nucleoside drugs, including procainamide, hydralazine, epigallocatechin-3-gallate (EGCG), N-phthalyl-L-tryptophan and MG98 (43,46). Nucleoside analogs are characterized by different modifications of the cytosine ring, such as replacement of carbon with nitrogen at position 5 of the pyrimidine ring (Fig. 4) (47). Cellular uptake of nucleotide analogs is dependent on human equilibrative nucleoside transporter (hENT)-1 and hENT2 (48). Following absorption, they are successively phosphorylated by intracellular deoxycytidine kinase to produce the active tri-phosphorylated metabolite 5-aza-2′-deoxycytidine 5′-triphosphate (49). This subsequently contributes to the loss of methylation through its incorporation into newly synthesized DNA and covalent bond formation with DNMTs (50). Thus, the enzyme remains bound to DNA and its methyltransferase function is blocked (51). Furthermore, the covalent bond compromises DNA functionality by triggering DNA damage signaling, leading to the degradation of these enzymes and the passive demethylation of the cellular genome during DNA replication (45). 5-aza-C and 5-aza-D are the most studied demethylating agents commonly used as chemotherapeutic agents that produce numerous biological effects (Table I) (52,53).
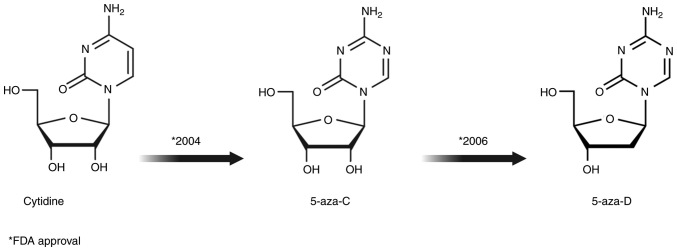
Figure 4.
Chemical structures of DNA demethylating agents (cytidine analogs 5-aza-C and 5-aza-D) approved by the United States Food and Drug Administration as antineoplastic drugs. 5-aza-C, 5-azacytidine; 5-aza-D, 5-aza-2′-deoxycytidine.
Table I.
Different effects of 5-aza-C and 5-aza-D in cancer cells.
| First author/s, year | Feature | 5-aza-C | 5-aza-D | (Refs.) |
|---|---|---|---|---|
| Stresemann and Lyko, 2008 | RNA incorporation | 80-90% | No | (45) |
| Stresemann and Lyko, 2008 | DNA incorporation | 20-40% | 100% | (45) |
| Stresemann and Lyko, 2008 | DNA strand breaks | Weaker DNA damage compared with 5-aza-D | Increased DNA damage compared with 5-aza-C | (45) |
| Stresemann and Lyko, 2008 | Demethylating capacity | Weaker DNA demethylating activity than 5-aza-D | 90% more than 5-aza-C | (45) |
| Nguyen et al, 2010 | Cell cycle | Arrest at phase G1 | Increases the number of cells arrested in G2 phase and mitosis | (77) |
| Hollenbach et al, 2010 | Gene expression | Decreasing the expression of genes related to the cell cycle and metabolic processes | Regulates genes related to cell differentiation | (62) |
| Zheng et al, 2012; Soengas et al, 2001 | Mechanism of action | Transcriptional regulation and induced hypomethylation | Induced demethylation, removal of methylation, histone modifications or a combination of both | (52,222) |
5-aza-C, 5-azacytidine; 5-aza-D, 5-aza-2′-deoxycytidine; n/a, not applicable.
5-aza-C
5-aza-C was first synthesized in 1960 for use as a classical cytostatic agent and was later shown to inhibit DNA methylation in human cell lines (54–56). It is the first epigenetic drug (epidrug) proposed for use in cancer therapeutics, specifically in patients with acute myelodysplastic syndrome (MDS). In May 2004, the US FDA approved it as a first line treatment for MDS (57). It is also used to treat other hematological malignancies, such as chronic myeloid leukemia (58).
As 5-aza-C contains a ribose in its structure (Fig. 4), it can be incorporated into RNA and DNA (59). There are two main mechanisms of action for 5-aza-C. The first mechanism of epigenetic effects is attributed to its incorporation into DNA, where it induces DNA hypomethylation after several rounds of replication and DNA damage due to adduct formation; therefore, it has been hypothesized that 5-aza-C leads to the activation of repressed tumor suppressor genes (60). On the other hand, most of 5-aza-C (80–90%) is incorporated into RNA (60), which can modify the synthesis of certain proteins by altering the activity of posttranscriptional mechanisms and inducing apoptosis (61–63). Its mechanism of action is concentration-dependent, with high concentrations regulating gene transcription, whereas lower doses induce hypomethylation and inhibit DNMTs (61). Although the main mechanism associated with clinical response observed in patients treated with 5-aza-C remains controversial, the rationale behind the currently approved low-dose therapy for MDS is based on the theory that the induction of hypomethylation with subsequent re-expression of methylated genes will modify gene expression patterns and lead to cell differentiation, apoptosis or senescence of the malignant clone (63).
In preclinical studies, 5-aza-C inhibited the antiapoptotic transcription factor NimA related kinase by decreasing the phosphorylation of the upstream regulator IKKα/β, which induces upregulation of the proapoptotic protein phorbol-12-myristate-13-acetate-induced protein 1, resulting in increased cell death (64,65). Furthermore, 5-aza-C has been proposed to inhibit Wnt signaling, a pathway involved in oncogene expression in acute myeloid leukemia (AML) and other cancer types, such as colon cancer (66,67).
5-aza-D
5-aza-D is a potent DNMTi included in the group of analogs of the nucleoside cytidine. Because it contains a deoxyribose, it can be incorporated into DNA (Fig. 4) (59,62), unlike 5-aza-C, which predominantly incorporates into RNA (Table I). 5-aza-D covalently binds to DNA during the S-phase of the cell cycle leading to rapid loss of methylated cytosine (66). In 2006, the FDA approved its use in MDS, and it is currently used to treat chronic myelomonocytic leukemia and AML (68).
5-aza-D has direct and indirect effects on gene expression (52). The effect is direct when its incorporation into DNA alters its methylation (69). This is more evident at low concentrations, in which the formation of 5-aza-D/DNMT complexes is limited and does not prevent DNA synthesis (51). High concentrations lead to growth arrest and cell death through increased adduct formation and impaired DNA polymerase function (70). Indirect effects on gene expression are mainly mediated by histone modifications, such as acetylation and methylation (71–73). Treatment with 5-aza-D abolished transcription factor RUNX3 silencing through decreased H3K9 methylation, which favored gene expression, independent of changes in promoter methylation (74). Similar effects of 5-aza-D at the p14/p16 locus have been reported in bladder and gastric cancer cells (73,75).
5-aza-D and 5-aza-C are used to treat various hematological cancer types, including AML and MDS, because of their ability to reactivate silenced tumor suppressor genes and DNA damage-induced cell cycle arrest and apoptosis (76). Both drugs induce complete responses and hematologic improvement; however, prolonged overall survival has only been shown for 5-aza-C. Although the drugs only slightly differ in their molecular structures, there are essential differences in their mechanisms of action (Table I) (77). There are >135 epigenetics-related clinical completed studies in different phases (49 in phase 1; 67 in phase 2; 22 in phase 3; and 1 in phase 4); Of which, 17 phase 3 interventional clinical trials using 5-aza-D and 5-aza-C in combination with chemotherapy drugs or immunotherapy in cancer have completed the recruitment stage (Table II) (78).
Table II.
Interventional clinical trials of DNA demethylating agents 5-aza-C and 5-aza-D in cancer.
| Drug | Cancer type | Therapeutic approach | ClinicalTrials.gov identifier |
|---|---|---|---|
| 5-aza-C | Patients newly diagnosed with AML | Talirine combined with 5-aza-C or 5-aza-D | NCT02785900 |
| Adults with AML | Pracinostat in combination with 5-aza-C | NCT03151408 | |
| MDS | Comparing the efficacy and safety of Eltrombopag in combination with 5-aza-C vs. placebo plus 5-aza-C | NCT02158936 | |
| Relapse or refractory DLBL | Avelumab in combination regimens that include an immune agonist, 5-aza-C, a CD20 antagonist and/or conventional chemotherapy | NCT02951156 | |
| AML | Venetoclax in combination with 5-aza-C vs. 5-aza-C alone | NCT02993523 | |
| AML with flt3 gene mutation | Gilteritinib plus 5-aza-C or 5-aza-C alone | NCT02752035 | |
| AML with idh2 gene mutation | AG-221 (a small molecule inhibitor of the IDH2 enzyme) compared with BSC only, 5-aza-C plus BSC, low-dose cytarabine plus BSC, or intermediate-dose cytarabine plus BSC | NCT02577406 | |
| AML with idh1 gene mutation | Ivosidenib vs. placebo in combination with 5-aza-C | NCT03173248 | |
| MDS | MBG453 or placebo added to 5-aza-C | NCT04266301 | |
| AML | 5-aza-C with or without nivolumab or midostaurin, or 5-aza-D and cytarabine alone | NCT03092674 | |
| AML | Vadastuximab talirine vs. placebo in combination with 5-aza-C or 5-aza-D | NCT02785900 | |
| 5-aza-D | Metastatic TNBC | 5-aza-D plus carboplatin | NCT03295552 |
| Ovary cancer | Lower dose 5-aza-D plus carboplatin-paclitaxel regimen | NCT02159820 | |
| MDS with P53 mutation | 5-aza-D and arsenic trioxide | NCT03377725 | |
| AML | Standard combination chemotherapy plus 5-aza-D | NCT02172872 | |
| AML | Clofarabine or daunorubicin hydrochloride and cytarabine followed by 5-aza-D | NCT02085408 | |
| AML | 5-aza-D with or without nivolumab or midostaurin, or cytarabine and 5-aza-D alone | NCT03092674 |
5-aza-C, 5-azacytidine; 5-aza-D, 5-aza-2′-deoxycytidine; AML, acute myeloid leukemia; MDS, myelodysplastic syndrome; TNBC, triple-negative breast cancer; DLBL, diffuse large B-cell lymphoma; flt3, fms related receptor tyrosine kinase 3; IDH2, isocitrate dehydrogenase; BSC, best supportive care; MBG453, sabatolimab.
Zebularine
In the early 1990s, second-generation nucleoside analogs were synthesized, such as zebularine [1-(β-D-ribofuranosyl)-2(1H)-pyrimidinone], a cytidine analog that lacks the amino (N) group at position four of the pyrimidine ring (79). It has high stability under physiological conditions and inhibits DNA methylation through its action on DNMTs and cytidine deaminase, an enzyme responsible for resistance to nucleoside DNA methylation analogs (80). In addition, zebularine has low cytotoxicity, which could translate into longer treatments with low doses to maintain a demethylated state at an acidic or a neutral pH (80). By comparison, 5-aza-C and 5-aza-D exhibit significant toxicity, low instability under physiological conditions and short half-lives (59), which may complicate their use in clinical settings.
Non-nucleoside small-molecule compounds
Furthermore, a chemically diverse group of non-nucleoside small-molecule compounds, such as hydralazine and procainamide have been reported as DNMTi (81–83). These drugs that are not incorporated into DNA or RNA but inhibit DNMT activity alone or in combination with chemo/radiotherapy, have shown promising results in various cancer types (82). Other small-molecule compounds, such as guadecitabine, epigallocatechin-3-gallate (EGCG), RG-108 and MG98, exhibit a DNA hypomethylating effect either through the competitive inhibition or through the degradation of DNMT (46,84). In addition, the inhibition of histone acetylation and generation of reactive oxygen species are effects associated with the treatment with biologically active non-nucleoside compounds (85,86).
Covalent histone modifications in cancer
In eukaryotic cells, 145–147 bp DNA is wrapped around histone octamers, comprising a core of histone dimers 2A, H2B, H3 and H4, to form nucleosomes, the basic units of chromatin. Histone tails extending from the core of the nucleosome can be post-translationally modified by methylation, acetylation, phosphorylation, SUMOylation and ubiquitination (Fig. 1) (87). Histone modifications serve essential roles in various cellular processes, including DNA repair, replication, stemness and changes in cell state (88). Aberrant histone post-translation modifications occur during the initiation and progression of cancer (89).
Histone acetylation and deacetylation
Two families of antagonistic enzymes control histone acetylation: Histone acetyltransferases (HATs), which acetylate lysine residues at the N-terminus of histone proteins, and HDACs, which remove acetyl groups from histones. Histone acetylation reduces attraction between negative and positive charges of DNA and histone tails, respectively (90,91). This structural change favors the decompaction of chromatin, allowing access to the gene sequence for its transcription. The altered activity of HATs observed in several types of cancer, including esophageal, lung and liver cancer, has been associated with the aberrant activation of genes whose expression is necessary for maintaining tumor growth (92–94). Chemical modifications to histones form the basis of histone codes and may be associated with gene expression or silencing (88). Based on their sequence similarities to yeast HDACs, the 18 identified human HDACs are divided into four classes: i) Class I isoenzyme HDACs, which include HDAC1, HDAC2, HDAC3 and HDAC8; ii) class II HDACs, which are homologous to yeast protein lysine deacetylase and are further divided into class IIa (including HDAC4, HDAC5, HDAC7 and HDAC9) and class IIb (HDAC6 and HDAC10) subgroups; iii) class III HDACs are similar to yeast sirtuins (SIRTs), and include SIRT1-SIRT7; and iv) class IV HDACs, which comprise only HDAC11 (95). SIRTs are NAD+-dependent enzymes, whereas the other three classes are zinc cation (Zn2+)-dependent HDACs (96).
HDACs serve a role in primary biological functions, such as transcription, metastasis (97), autophagy, cell cycle (98), DNA damage repair, angiogenesis (99), stress responses (100) and senescence (101), through histone deacetylation and through non-histone proteins. However, aberrant expression of HDACs can contribute to carcinogenesis by altering gene expression through the deacetylation of different histone residues (Table III) (84). In addition, HDACs can deacetylate non-histone proteins involved in apoptosis, differentiation, cell proliferation control and transcription (102). HDAC upregulation in solid and hematologic tumors promotes tumor suppressor gene silencing or alters gene expression of oncogenic pathways (99). Furthermore, HDAC upregulation is useful for the diagnosis and prognosis of gastric and breast cancer (103).
Table III.
Effects of HDACs on carcinogenesis.
| HDAC | Cancer types | Substrate (histone and non-histone) | Cellular processes affected |
|---|---|---|---|
| HDAC1 | Lung, gastric, liver, breast, ovarian, prostate, renal, bladder and hematological | H4K16 and H3K56 | Differentiation, proliferation, metastasis, apoptosis and cell cycle |
| HDAC2 | Medulloblastoma, gastric, pancreatic, colorectal, breast, ovarian, prostate, renal, bladder and hematological | H4K16 and H3K56 | Differentiation, proliferation, metastasis, apoptosis and cell cycle |
| HDAC3 | Lung, gastric, breast, ovarian, prostate, bladder, melanoma and hematological | H3K9, H3K14, H4K5 and H4K12 | Differentiation, proliferation, metastasis, apoptosis, cell cycle and inflammation |
| HDAC4 | Hematological (lymphoblastic leukemia) and colorectal tumors | Not defined | Angiogenesis |
| HDAC5 | Hematological, bladder, renal, breast, medulloblastoma | p53 | Proliferation, angiogenesis apoptosis and cell cycle |
| HDAC6 | Hematological (lymphoblastic leukemia) | α-tubulin, HSP90 and cortactin | Inflammation, metastasis, angiogenesis, cell cycle and apoptosis |
| HDAC7 | Hematological (lymphoblastic leukemia) | Not defined | Angiogenesis and proliferation |
| HDAC8 | Neuroblastoma, melanoma and hematological | ERRα | Differentiation, apoptosis and cell cycle |
| HDAC9 | Hematological | Not defined | Proliferation and apoptosis |
| HDAC10 | Cervical tumors and chronic lymphocytic leukemia | Not defined | Angiogenesis and cell cycle |
| HDAC11 | Lung, mixed lobular and ductal breast carcinoma, mantle cell lymphoma and myeloproliferative neoplasms | Not defined | Proliferation and survival |
| SIRT1, SIRT2, SIRT3, SIRT6 and SIRT7 | Breast, head and neck, non-small cell lung, colon, prostate, ovarian, gastric, melanoma and hepatocellular | H4K16, H3K9, H3K18, H3K56, H4K16, p53, p73, PTEN, FOXO1, FOXO3a, FOXO4, NICD, MEF2, HIF-1α, HIF-2α, TAF(I)68, SREBP-1c, β-catenin, RelA/p65, PCAF, TIP60, p300, SUV39H1, H4K16, H3K56, α-tubulin and ATP-citrate lyase SETD8 | Differentiation, proliferation, metastasis, apoptosis, cell cycle and inflammation |
H3K/H4K, histone 2/4 lysine; HIF, hypoxia-inducible factor; HDAC, histone deacetylase; MEF2, myocyte-specific enhancer factor 2; NICD, Notch 1 intracellular domain; PCAF, P300/CBP-associated factor; SIRT, sirtuin; SUV39H1, suppressor of variegation 3–9 homolog 1; ERRα, estrogen-related receptor α; FOXO, forkhead box transcription factors; TAF(I)68, TBP-associated factor; SREBP-1c, sterol regulatory element-binding protein-1c; SETD8, SET domain-containing protein 8.
Histone methyltransferase (HMT)
Histone methylation is an essential post-transcriptional modification generally occurring at the lysine and arginine residues of histones H3 and H4 through the addition of methyl groups (104). Mono-, di- or trimethylated states may occur in both lysine and arginine, which subsequently affects transcriptional regulation (88). Histone methylation is mediated by HMTs, of which there are six major classes of histone lysine methyltransferase complexes (KMT1-KMT6) (97,105). The identification of lysine demethylases revealed the reversibility of lysine methylation (106). H3K4-trimethylation (me3), H3K36me3 and H3K79me3 are associated with transcriptional activation, whereas methylation of H3K9me3, H4K20me3 and H3K27me3 is associated with a repressive chromatin state. The interaction and level of methylation of these histone marks serve an important role in transcriptional regulation (103). Furthermore, mutations detected in different cancer types (bladder, melanoma and lung cancer) have been associated with altered activity of methylating enzymes (107). Mutations in enhancer of zeste homolog 2 (EZH2) are found in follicular and diffuse large B-cell lymphomas, gliomas and bone cancers affecting H3K27 (108). The HMT SET domain containing 1A is upregulated in breast cancer, favoring the increase in H3K4me3 in genes associated with migration and metastasis (109). In addition, H3K36me3 is altered in various cancers, such as chondroblastoma, and colon, and head and neck cancer (110,111). These alterations in histone modifications and methylation enzymes contribute to changes in gene expression that may promote carcinogenesis. Therefore, HMT inhibitors have been developed for use as antineoplastic drugs. Clinical trials of protein arginine N-methyltransferase 1, EZH2 and lysine-specific histone demethylase 1 inhibitors have been summarized by Yang et al (112).
HDACi for cancer therapy
The increasing understanding of the reversibility of epigenetic processes has led to the identification of drugs that inhibit HDAC activation and prevent tumor development. Four HDACi affecting different cellular processes are FDA-approved (105,113), including vorinostat, romidepsin, belinostat and panobinostat (Fig. 3A; Table IV). Additionally, interventional clinical trials using HDACi in combination with therapeutic drugs for cancer are summarized in Table V.
Table IV.
HDACi approved by the US FDA for cancer treatment and their effects on cellular processes.
| First author/s, year | HDACi | Target HDAC | Target cellular processes | Type of cancer | Year approved by the FDA | (Refs.) |
|---|---|---|---|---|---|---|
| Bubna, 2015; Mann et al, 2007 | Vorinostat (SAHA) | Class I and II | Induces cell cycle arrest and apoptosis; inhibits angiogenesis, and downregulates immunosuppressive interleukins | CTCL | 2006 | (115, 223) |
| Grant et al, 2010 | Romidepsin (FK288) | Class I | Induces differentiation, cell cycle arrest and apoptosis | CTCL | 2009 | (224) |
| Poole, 2014 | Belinostat (PXD101) | Class I, II and IV | Induces apoptosis through the reactivation of suppressor genes | Peripheral T-cell lymphoma | 2014 | (124) |
| Moore, 2016 | Panobinostat (LBH589) | Pan | Induces cell cycle arrest and apoptosis | CTCL and multiple myeloma | 2015 | (225) |
CTCL, cutaneous T-cell lymphoma; HDAC, histone deacetylase; HDACi, HDAC inhibitors; SAHA, suberoylanilide hydroxamic acid; US FDA, United States Food and Drug Administration.
Table V.
Interventional clinical studies of HDACi agents in cancer.
| HDACi | Cancer type | Treatment | ClinicalTrials.gov identifier |
|---|---|---|---|
| Vorinostat (SAHA) | Small cell lung | Carboplatin and etoposide in combination with vorinostat | NCT00702962 |
| Non-small cell lung | Maximum tolerated dose of the combination of vorinostat, cisplatin, pemetrexed and radiation therapy | NCT01059552 | |
| Pancreatic | Maximum tolerated dose of vorinostat plus radiation therapy | NCT00831493 | |
| Breast | Combination of lapatinib and vorinostat | NCT01118975 | |
| Metastatic or recurrent gastric | Vorinostat plus capecitabine and cisplatin | NCT01045538 | |
| Romidepsin (FK288) | Non-small cell lung | Romidepsin in combination with erlotinib hydrochloride | NCT01302808 |
| Pancreatic | Romidepsin (depsipeptide) in combination with gemcitabine | NCT00379639 | |
| HPV+ nasopharyngeal, HPV+ cervical and liposarcoma | 5-aza-D in combination with romidepsin | NCT01537744 | |
| Belinostat (PXD101) | Solid tumors | PXD101 alone and in combination with 5-fluorouracil | NCT00413322 |
| Ovarian | Belinostat plus carboplatin or paclitaxel or both | NCT00421889 | |
| Non-small cell lung | PXD101 in combination with paclitaxel plus carboplatin in chemotherapy-naïve patients | NCT01310244 | |
| Neuroendocrine | Belinostat in combination with cisplatin and etoposide | NCT00926640 | |
| Small cell lung | |||
| Malignant epithelial | |||
| Panobinostat | Soft tissue sarcoma | Panobinostat alone | NCT01136499 |
| (LBH589) | Breast | Combination of panobinostat and letrozole | NCT01105312 |
| Small cell lung | Combination of panobinostat, etoposide and cisplatin | NCT01160731 | |
| Renal | Combination of panobinostat and sorafenib | NCT01005797 | |
| Prostate | Panobinostat in combination with docetaxel and prednisone | NCT00663832 | |
| Pancreatic | Panobinostat in combination with bortezomib | NCT01056601 | |
| Thyroid | Panobinostat alone | NCT01013597 | |
| Brain | Panobinostat alone | NCT00848523 | |
| Hodgkin Lymphoma | Panobinostat alone | NCT00742027 | |
| Non-Hodgkin Lymphoma | Combination of panobinostat and ketoconazole | NCT00503451 |
HDACi, histone deacetylase inhibitors; HPV, human papillomavirus; SAHA, suberoylanilide hydroxamic acid.
Vorinostat
Vorinostat (also known as suberoylanilide hydroxamic acid) is a class I- and class II HDACi structurally belonging to the hydroxamate group; it does not inhibit class III HDACs (114). Vorinostat binds to zinc in the catalytic site of HDAC, which blocks the access of target proteins and inhibits its HDAC activity (115). Therefore, this HDACi can modify expression of proteins through structural changes on chromatin, but also by modifying the activity of non-histones proteins (p53, EKLF, E2F1, HIF-1 and GATA) (115,116). Clinical studies have demonstrated that vorinostat treatment reduced solid tumors when administered alone or in combination with alkylating agents, proteasome inhibitors, anthracyclines and antiangiogenic and/or antimetabolite agents (78,113,117,118).
Romidepsin (Istodax®)
Romidepsin is a bicyclic peptide derived from Chromobacterium violaceum and is a class I HDACi (particularly against HDAC1, HDAC2, HDAC3 and HDAC8) with only weak activity against class II HDACs (119). Its mechanism of action is based on binding to zinc in the pocket domain of HDACs, inhibiting the interaction with target proteins (120). Treatment with romidepsin shows promising effects in hematological tumors, such as cutaneous T-cell lymphoma and other peripheral T-cell lymphomas (PTCLs), with clinical studies demonstrating a reduction in solid tumors, including colon, sarcoma, breast and renal cancer (121). Furthermore, romidepsin has been used in combination with other cancer treatments, including cisplatin, paclitaxel, CC-486 and 5-aza-D, which are safe and effective for solid and hematological tumors (122).
Belinostat
Belinostat has been used as a monotherapy to treat relapsed or refractory PTCL (123). Belinostat is a hydroxamate-type inhibitor of class I, II and IV HDACs; however, its mechanism of action has not yet been elucidated (124). Nevertheless, clinical studies have used belinostat to treat solid tumors, including colon, breast, pancreas, prostate and bladder cancer (78). Furthermore, belinostat has been reported to act synergistically with the proteasome inhibitor bortezomib and with the alkylating agents cisplatin and carboplatin against solid tumors in vitro (78,125,126).
Panobinostat
Panobinostat is a pan-HDACi belonging to the hydroxamate family. Some clinical studies have tested its use alone or in combination with other drugs to particularly treat solid tumors, such as sarcoma, breast, lung, renal, prostate, pancreas, thyroid and malignant brain cancer, including hematological cancer types such as Hodgkin and non-Hodgkin lymphoma; however, the mechanism of action is uncharacterized (78,127).
3. ncRNAs in epigenetic control
In addition to DNA methylation and histone modifications, ncRNAs also serve a key role in epigenetic control (Fig. 1). ncRNAs are divided into two main groups: Housekeeping and regulatory ncRNA. Housekeeping ncRNAs include transfer RNAs, ribosomal RNAs, small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs), which enable the flow of genetic information from DNA to proteins (128,129). snRNAs and snoRNAs participate in the modulation of alternative splicing, which drives mRNA maturation (130). In some instances, these splicing-devoted ncRNAs have epigenetic regulatory functions, as is the case with snRNAs U1 and 7SK, which modulate transcription (130). U1 is associated with the general transcription initiation factor transcription factor IIH (131), and 7SK interacts with the chromatin factor and transcriptional regulator high mobility group AT-hook 1 (132).
By contrast, regulatory ncRNAs are non-constitutively expressed RNAs that serve roles in epigenetic control by modulating chromatin structure, RNA polymerase activity and mRNA stability. Regulatory ncRNAs are divided according to length: Small [small non-coding RNA (sncRNA); ≤200 nucleotides] and long [long non-coding RNA (lncRNA); >200 nucleotides] (128). The best-understood sncRNAs are miRNAs/miRs and small interfering RNAs (siRNAs). Other small ncRNAs include trans-acting siRNAs, small-scan siRNAs, repeated-associated siRNAs and piwi-interacting RNAs (piRNAs) (133,134). Classification of lncRNAs is even more complex, and efforts have been made to obtain more comprehensive annotations. Thus, one classification system for lncRNAs involves four subdivisions: i) Long intergenic non-coding RNAs (lincRNAs) and intronic lncRNAs; ii) sense and antisense lncRNAs; iii) cis- or trans-acting lncRNAs; and iv) transcriptional or post-transcriptional lncRNAs (135). Notably, the function of several lncRNAs remains unknown; therefore, they are denoted as non-functional RNAs and considered as transcriptional noise (136). Circular RNAs (circRNAs) can be consider as lncRNAs in terms of size (mean length ~50 nt); however, due to their structure, some authors consider them as a third group of ncRNAs (137). They comprise between one and five exons and flanking introns, but differ from the previously mentioned lncRNAs since they are single-stranded RNA that form a loop by covalently joining their 5′ and 3′ ends (135).
sncRNAs in cancer
siRNAs and miRNAs are the most understood sncRNAs. They are double-stranded RNAs (dsRNAs) whose genesis and function partially overlap. Both can promote RNA interference (RNAi), a post-translational regulatory mechanism that provokes gene silencing through mRNA degradation (138). The genesis of siRNAs and miRNAs relies on Dicer, an RNase III enzyme that cleaves dsRNAs (139). A capped and polyadenylated RNA with a stem-loop structure is cleaved by Drosha to form genetically coded precursor miRNAs (pre-miRNAs) (140). miRNA genes are transcribed in the cell nucleus by RNA polymerase II giving rise to primary miRNAs (pri-miRNA) (141). Pre-miRNAs and dsRNAs are processed by Dicer in the cytoplasm and interact with the RNA-induced silencing complex (RISC) (142). The siRNA or miRNA sequence guides RISC to the target mRNA (139). After binding to its target by base pairing, argonaute 2 endonuclease (a part of RISC) cleaves the mRNA, causing gene silencing (138,143).
There are essential differences between siRNAs and miRNAs: siRNAs are molecules 21–23 nucleotides long that can modulate more than one mRNA because they are partially complementary to their targets; whereas miRNAs are 19–25 nucleotides long and highly specific to one mRNA sequence, but they can potentially regulate more than one target gene (138). The partial complementarity of siRNAs with the gene sequence suggests that inhibitory activity is not solely due to the direct degradation of target mRNA by the RISC complex but can also increase the mRNA decay through the rapid deadenylation of target transcripts (144,145). In addition, siRNAs can recruit chromatin-targeted RNAi silencing components to heterochromatin (146).
piRNAs are likely miRNAs that are genetically coded in the piRNA cluster of genes and are highly expressed in gonadal tissue to regulate transposable elements in the germline (147). Precursor piRNAs (pre-piRNAs) are transcribed by RNA polymerase II. Once the newly transcribed RNA leaves the nucleus, pre-piRNAs interact with the germline-specific subclade or argonaute proteins known as PIWI proteins (148). Transgenerational inheritance of piRNAs, such as maternal inheritance through oocytes, alters the progeny phenotype, suggesting that it may contribute to diverse cellular processes over generations (149).
There is strong evidence showing the association between miRNAs and cancer development (150–153). miRNA expression is commonly dysregulated in various tumors and deletion of miRNA genes is also observed in other oncogenic genes (Fig. 2C). For example, miR-15a and miR-16-1 are commonly deleted in B-cell chronic lymphocytic leukemia (154). These miRNAs are tumor suppressors that inhibit the expression of the antiapoptotic factor Bcl-1 (155).
By contrast, miRNAs with oncogenic potential are known oncogenes. The best-studied oncomiR is the miR-17-92 cluster (known as oncomiR-1); it is part of chromosome 13 and includes miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1 and miR-92a-1, which are often dysregulated in hematopoietic and solid cancers (156). Transcriptional regulation of miRNAs is also common in cancer. For instance, transcription factors and the tumor suppressor protein p53 are the most commonly mutated genes in several human cancer types (e.g., prostate, ovary, skin, lung and breast cancer), and thus, a high number of dysregulated miRNAs are associated with carcinogenesis (157,158). Notably, the highly mutagenic landscape seen in the p53 gene gives rise to different variants, each with a particular behavior, with specific miRNA targets (159). Notably, several oncoviruses express proteins that inactivate p53 and consequently alter host miRNAs (160,161).
Therapeutic oligonucleotides are an emerging alternative treatment
The therapeutic potential of miRNAs is delayed through problems associated with the toxicity of the administration methods and biological factors (clearance, non-specific distribution and degradation in circulation) that may affect the efficacy of the molecules. In addition to side effects unwanted related mainly with the possible activation immune response and off-target silencing of RNAs that play essential roles in cell mechanisms. Synthetic RNA molecules improve biostability and reduce toxicity through chemical modifications, such as 20-fluoro-35 and 20-O-methyl, indicating that they may solve some of the problems associated with miRNAs (161).
lncRNAs in cancer
lncRNAs are the most abundant RNA, with 16,000-100,000 known human lncRNA genes (162). They share similarities with mRNAs because they exhibit transcriptional regulation, and a considerable number of them are 3′ polyadenylated and 5′ capped and undergo lncRNA splicing (159).
lncRNAs modulate gene transcription and epigenetics in the cell nucleus and regulate mRNA stability, protein translation and competing endogenous RNA (ceRNA) in the cytoplasm (163). These RNAs have various mechanisms, such as transmitting transcriptional signals, functioning as decoys, scaffolds, RNA enhancers or coding for short peptides (163,164).
Signal lncRNAs regulate transcription in response to several stimuli such as inflammatory response, metabolic changes and oxidative stress (163). Their transcription occurs at specific times and locations, thus serving as molecular signals. For example, X-chromosome inactivation occurs during female development (165). The lncRNA Xist is transcribed from the X chromosome that will be inactivated, and the sole expression of Xist indicates active silencing at the genomic locus. Signal lncRNAs can be classified as decoys or scaffolds (166), and a single lncRNA can have more than one mode of action. For instance, lincRNA-p21 has essential functions in cancer development by modulating the transcription of p21 downstream of p53 activity (167). lincRNA-p21 expression is a marker of DNA damage and apoptosis (168). However, it is also a scaffold molecule for the transcriptional complex assembled over the p21 promoter by recruiting heterogeneous nuclear ribonucleoprotein (hnRNP)-K, which mediates p53 binding to such promoters (169). Furthermore, lincRNA-p21 interacts with MDM2 proto-oncogene (MDM2) to release p53 from repression (166).
Decoy lncRNAs trap transcriptional regulatory factors by presenting decoy binding sites, thus lowering the availability of elements needed for mRNA transcription (170). Scaffold lncRNAs allow the formation of multiple complexes that either enhance or repress transcription (164). For instance, paraspeckles are mammalian-specific protein-rich nuclear organelles assembled on top of the lncRNA NEAT1. These organelles can trap RNAs and nucleoplasmic proteins, thereby reducing their availability (171). Gastric cancer lncRNA serves as a modular scaffold for the histone-modified components WD repeat-containing protein 5 and lysine acetyltransferase 2A in gastric cancer. It guides these components to target genes to specify histone modification patterns (172). The guiding function of lncRNAs helps direct ribonucleoproteins to target genes, and enhancer lncRNAs influence the three-dimensional organization of DNA (164).
It has been demonstrated that some ncRNAs possess open reading frames encoding peptides (173). pri-miRNAs may encode proteins if transported to the cytosol without processing (174). circRNAs can also be translated into protein through the internal ribosome entry site (IRES) (175). Several lncRNAs have been identified in a number cancer types. For instance, lncRNA HOXB-AS3 encodes a conserved 53-amino-acid-long peptide that interacts with hnRNPA1 to repress glucose metabolism reprogramming, thus suppressing colon cancer growth (176). Notably, the lncRNA peptidome has been established for colorectal adenocarcinoma (177). Five universally expressed lncRNA peptides show greater expression in tumors than normal tissues, suggesting that they are promising biomarkers for this type of cancer (154). Furthermore, the peptide products of these lncRNAs are more stable than RNA molecules in circulation; therefore, patient plasma can be a reliable sample for diagnosis (178).
circRNAs in cancer
circRNAs are ncRNAs that are similar in length to lncRNAs but they lack 5′ and 3′ modifications and are covalently linked, forming a closed circular structure (129). These circular structures are usually formed between the 5′ and 3′ spliced sites of exons, although intron circularization also exists (179). Intronic circRNAs (ciRNAs) are distinguished from exonic circRNAs (180). Exonic circRNAs are transported to the cytoplasm by ATP-dependent RNA helicases, whereas ciRNAs remain in the nucleus and modulate transcription (129).
Cytoplasmic circRNAs act as miRNA sponges that regulate downstream targets or bind proteins (181). For instance, numerous circRNAs are sponges of miR-7, a crucial tumor suppressor in several cancer types. ciRS-7 expression is upregulated in colorectal and gastric cancer, and blocks the suppressive function of miR-7 (181). ciRS-7 contains ~70 conserved miR-7 target sites, and downregulation of this miRNA promotes retinoblastoma and astrocytoma development (182). circZFR binds to different miRNAs and exerts opposing functions; it inhibits gastric cancer proliferation through PTEN upregulation by targeting the miR-130a/107 (183). Conversely, circFZR promotes hepatocellular carcinoma proliferation by modulating miR-1261/4302/3619 (184).
In addition, circRNAs interact with RNA-binding proteins, which regulate RNA transcription, splicing, stability, localization and translation (185). The interaction of circRNAs with proteins acts as either a decoy or a scaffold (186). For instance, circ-DNMT1, originating from the splicing of DNMT1, is upregulated in breast cancer and contributes to cell proliferation (187). It interacts with p53 and ribonucleoprotein hnRNPD and mediates the nuclear translocation of both proteins (187). The binding of circ-DNMT1 to hnRNPD stabilizes DNMT1 mRNA and increases its translation (188). circ-forkhead box O3 (FOXO3) expression serves a determining role in the control of cell cycle progression. The formation of a ternary complex with CDK2 and p21 proteins inhibits the G1/S transition, mainly by blocking the interaction of CDK2 with cyclin E (189). circ-FOXO3 expression is dysregulated in tumor cells (breast and renal cancer), while, under oxidative stress conditions, its upregulation is associated with cell cycle arrest and apoptosis (190). The interaction of Circ-FOXO3 with MDM2 facilitates p53 ubiquitination in the cytosol, which reduces FOXO3 degradation by MDM2 and upregulates the expression of the p53 upregulated modulator of apoptosis gene (187).
Some circRNAs can be translated into peptides due to IRESs or N6-methyladenosine modifications (184). For example, circ-FBXW7 encodes a 21 kDa protein that inhibits cancer cell proliferation and reduces c-Myc half-life; its expression is positively associated with the overall survival of patients with glioblastoma (191).
ncRNAs as therapeutic tools in cancer
RNA-based therapy is an expanding drug category with rapid success found through the development of an mRNA-based vaccine against severe acute respiratory syndrome coronavirus-2 to tackle the coronavirus disease 2019 pandemic (192). ncRNA-based therapies are aimed at modulating the transcriptional or post-transcriptional expression of endogenous mRNAs to control the expression of specific proteins involved in cancer development (193). Most ncRNA therapies are based on miRNAs because they potentially target several genes simultaneously, which is useful in multigenetic diseases (137). However, this multi-target action may also have off-target drawbacks, and thus, siRNAs are preferred for target identification and drug discovery (138). Similarly, lncRNAs may be used to directly target or modulate other molecules, such as miRNAs, with various molecules reaching the clinical trial phase (194). To the best of our knowledge, there are no current preclinical reports using circRNAs alone as targets or therapeutic vectors for cancer treatment; however, this will likely change in the future (195).
Targeting ncRNAs in cancer serves one of two aims: To restore tumor suppressor activity or to inhibit oncogene expression (Fig. 3C). The ncRNA may directly related with activation of genes controlling growth and differentiation cellular, or promote the transformation through indirect inhibition of genes of downstream pathways (196). In the second case, ncRNAs modulate gene expression through RNAi or through transcriptional gene silencing, which can result in long-term stable, inheritable epigenetic changes (197). This second mechanism is more commonly described for lncRNAs and RNAi than for sncRNAs, although both types of ncRNAs can exert each mode of action (144).
ncRNA replacement therapy (mimics)
Replacement therapy involves delivering mimicking molecules, synthetic RNA (particularly sncRNA), to restore downregulated targets (Fig. 3B) (198). Exogenous inhibitory RNAs mimic tiers in the RNAi pathway; synthetic RNAs can be delivered to cells in three ways. The first is artificial miRNAs (amiRNAs), which are equivalent to endogenous pri-miRNAs. amiRNAs are double-stranded stems flanked by a single-stranded basal segment and an apical loop containing a recombinant pri-miRNA scaffold and siRNA insert (199). These synthetic RNAs enter the cell nucleus, undergo Drosha processing and are exported to the cytoplasm to be loaded into the Dicer complex. Therapeutically, amiRNAs can exert a long-lasting effect due to their stable expression, which implies treatment regimens with few doses, thus reducing the risk of toxicity. In addition, they do not saturate miRNA machinery (198). Second, vector-based short hairpin RNAs are the most widely used molecules for silencing in mammalian cells. They mimic pre-miRNAs, which are processed double-stranded stem-loop molecules that undergo Dicer processing. Third, synthetic siRNAs are molecules of dsRNA that can be directly processed by Dicer in a similar manner to miRNAs (198,199). This last group includes small activating RNAs (saRNAs), which are dsRNAs that specifically target and activate promoters. saRNAs are processed by argonaute proteins, which translocate as a complex to the nucleus, bind to the promoter to open the chromatin, and recruit positive transcriptional regulators to the complex (200). Tables VI and VII present examples of ncRNAs associated with cancer development and therapeutic approaches applied in clinical trials.
Table VI.
Examples of replacement therapies for non-coding RNAs.
| First author/s, year | Target | Role in cancer | Therapeutic approach | Effect | (Refs.) |
|---|---|---|---|---|---|
| Gilles and Slack, 2018 | let-7 miRNA | Low let-7 expression is associated with inferior survival in different types of cancer | Synthetic let-7b is delivered directly by adenovirus and lentivirus | Reduces NSCLC xenograft tumor growth by targeting HGMA2, which is commonly upregulated in NSCLC | (226) |
| Segal et al, 2020 | Synthetic let-7b miRNA conjugated with lipophilic moieties | (227) | |||
| Wu et al, 2015; Pan et al, 2021; Li et al, 2021 | let-7 agomirs, which are chemically modified and end-labeled miRNAs that mimic endogenous miRNAs but with increased stability | Its use decreases invasion and migration of cholangiocarcinoma, sensitizes epithelial ovarian cancer to cisplatin treatment, and inhibits hepatocellular carcinoma | (228–230) | ||
| Hong et al, 2020 | miR-34a | Commonly lost or downmodulated in various solid tumors | MRX34 is a liposomal formulation for restoring miR-34a | Reduction of Bcl-2, DNAJB1, CTNNB1, FOXP1 and HDAC1 expression levels | (231) |
| van Zandwijk et al, 2017; Winata et al, 2017 | miR-15/16 | Downregulated in chronic lymphocytic leukemia, melanoma, colorectal and bladder cancer, and other solid tumors | TargomiRs comprising miRNA mimics based on the miR-15/16 consensus sequence, packaged in EDVs (non-living bacterial minicells) that allow for efficient packaging of nucleic acids and targeting moiety. | In various cancer types, increased levels of the miR-15/16 family result in tumoral growth inhibition | (232,233) |
| The EDVs are targeted to anti-EGFR-specific antibodies | |||||
| Filippova et al, 2021; Williams and Pickard, 2016; Liu et al, 2021 | GAS5 lncRNA | Tumor suppressor in multiple solid tumors | This is a stem-loop structure within the GAS5 hormone response element mimic | Cell lines exhibit increased apoptosis and reduced cell proliferation | (234–236) |
| Smaldone and Davies, 2010 | H19 lncRNA | Highly expressed in malignant tissue but low or undetectable in adult tissue | BC-819 is a double-stranded DNA plasmid that codes for diphtheria toxin A driven by the H19 promoter and is selectively expressed in tumor cells | Selective toxin expression results in selective tumor cell destruction through the inhibition of protein synthesis in the tumor cells | (237) |
| Zhao et al, 2019; Reebye et al, 2014 | CEBPA-saRNA | A transcriptional regulator that serves a key role in maintaining liver function; its expression is lost in HCC | MTL-CEBPA comprises a double-stranded RNA formulated into a SMARTICLES® liposomal nanoparticle | CEBPA-saRNA reduces tumor burden and simultaneously improves liver function in HCC | (238,239) |
HCC, hepatocellular carcinoma; HDAC, histone deacetylase; lncRNA, long non-coding RNA; miR/miRNA, microRNA; NSCLC, non-small cell lung cancer; saRNA, small activating RNA; DNAJB1, DnaJ heat shock protein family (Hsp40) member B1; HMGA2, high mobility group AT-hook 2; CTNNB1, catenin β1 gene; FOXP1, forkhead box protein P1; TargomiRs, miRNA mimics based on the miR-15/16 consensus sequence; EDVs, extracellular directed vesicles; MTL-CEBPA, RNA duplex activating of the c/ebpa gen transcription; CEBPA, CCAAT/enhancer-binding protein alpha; CEBPA-saRNA, CCAAT/enhancer-binding protein α-small activating RNA.
Table VII.
Interventional clinical trials of replacement therapies for non-coding RNAs.
| Therapeutic oligonucleotide | Cancer type | Therapeutic approach | ClinicalTrials.gov identifier |
|---|---|---|---|
| MRX34 | Primary liver cancer or other selected solid tumors and hematologic malignancies | Monotherapy with MRX34 to investigate the safety and tolerability of escalating doses | NCT01829971 |
| TargomiRs | Recurrent malignant pleural mesothelioma and non-small cell lung cancer | Single-drug TargomiR | NCT02369198 |
| DTA-H19 | Unresectable, locally advanced pancreatic cancer and advanced-stage ovarian cancer | Monotherapy intratumoral with DTA-H19 | NCT00826150 |
| Non-muscle invasive bladder cancer | Monotherapy BC-819 instilled intravesical into the bladder | NCT03719300 | |
| Superficial transitional cell bladder carcinoma and locally advanced pancreatic adenocarcinoma | BC-819 and bacillus Calmette-Guérin vaccine | NCT01878188 | |
| BC-819 | Advanced HCC | BC-819 and gemcitabine | NCT01413087 |
| (inodiftagene | |||
| vixteplasmid) | Lung, ovarian, pancreatic, gall bladder, HCC, neuroendocrine, and cholangiocarcinoma | Monotherapy or in combination with sorafenib | NCT02716012 |
| MTL-CEBPA | Solid tumor in adult population | MTL-CEBPA in combination with a PD-1 inhibitor pembrolizumab | NCT04105335 |
| Advanced HCC | MTL-CEBPA plus sorafenib or sorafenib alone | NCT04710641 | |
| Advanced HCC | MTL-CEBPA, atezolizumab and bevacizumab | NCT05097911 |
HCC, hepatocellular carcinoma; PD-1, programmed cell death protein 1; TargomiR, miRNA mimics based on the miR-15/16 consensus sequence; MTL-CEBPA, RNA duplex activating of the c/ebpa gen transcription; CEBPA, CCAAT/enhancer-binding protein α; MRX34, mimic of miR-34a; DTA, DNA plasmid that carries the gene for the diphtheria toxin A.
ncRNA inhibition therapy
Inhibition therapy depletes oncogenic ncRNA expression. Antisense oligonucleotides (ASOs) are 18–30 bp in length and bind to target mRNAs and promote their degradation by RNAse H, which recognizes the DNA:RNA hybrid and cleaves the RNA in the complex (201). Alternatively, ASOs can bind to miRNAs [anti-miRNA oligonucleotide (AMOs) also known as antagomirs] (202). ASOs and AMOs are subjected to nuclease degradation and weakly bind to proteins; thus, their stability and uptake must be improved by specific chemical modifications (201). One such modification is locked nucleic acids (LNAs), a methylene linkage connecting the 2′ and 4′ carbons of the RNA backbone ribose (203). BlockmiRs are another class of ASOs that specifically target the miRNA binding site in mRNAs, thus blocking miRNA binding (204).
Aptamers are a class of synthetic DNA or RNA oligonucleotides that target proteins. They recognize the tertiary or quaternary structure of proteins rather than the linear sequence, as is the case for ASOs; thus, aptamers are also known as chemical antibodies (205). Gapmers are ASOs containing a central DNA flanked by modified oligonucleotides. These molecules are considered powerful options for targeting nuclear lncRNAs due to the high level of RNAse H activity observed during application (206).
Small molecules can also inhibit ncRNAs. These molecules target either RNA-interacting molecules or ncRNAs. For example, bisphenol A and diethylstilbestrol can modify chromatin structure and gene activation via the increased expression of the lncRNA HOTAIR (207). Their advantage over oligonucleotides is that they are easily delivered, soluble, bioavailable, stable and can be designed on a structure-based approach. In addition, some small-molecule inhibitors approved by the FDA can manipulate ncRNAs; this would shorten the time taken to translate experiments to clinical therapy (208).
Endogenously expressed lncRNAs and circRNAs function as ceRNAs by sequestering miRNAs (209). This inherent ability of lncRNAs to block the activity of miRNAs was exploited to engineer decoys, also referred to as target mimics or sponges (177). Both lncRNAs and artificial sponges harbor short sequences that are homologous to miRNA binding sites on their endogenous targets (210). Therefore, decoys sequester miRNAs and prevent their binding to targets (211). Engineered sponges typically contain 10 binding sites separated by a few nucleotides (212). Sponges with imperfect binding sites provoking a bulge are more effective in attracting miRNAs and are more stable (213). Tough decoys are 60-bp-long harpin sponges with an internal bulge containing two miRNA sites (214).
The CRISPR-CRISPR associated protein 9 (CRISPR-Cas9) toolbox enables the manipulation of genes based on RNA-guided DNA recognition (193). A single-guide RNA is used to mediate DNA endonuclease Cas-9 to introduce gaps into the DNA; thus, CRISPR-Cas9 is designed to remove DNA fragments from the genome (215). The addition of donor DNA with the CRISPR-Cas9 system may cause DNA breakage through homology-directed repair, which uses and introduces the donor DNA into the genome. Although several studies have confirmed the potential therapeutic value of CRISPR-Cas9, there are still concerns regarding off-target effects and ethical concerns raised by this technique in germline cells (216–219). Tables VIII and IX present examples of ncRNA-inhibitory therapies and clinical trials for cancer, respectively.
Table VIII.
Inhibitory non-coding RNAs in cancer therapy.
| First author/s, year | Type | Target | Role of target in cancer | Mechanism of action | Effects | (Refs.) |
|---|---|---|---|---|---|---|
| Seto et al, 2018 | ASO | miR-155 | miR-155 is associated with poor prognosis in lymphoma and leukemia | Cobomarsen (MRG-106) is a locked nucleic acid-modified ASO inhibitor of miR-155 | Inhibition of miR-155 reverses malignant tumor phenotype by inducing apoptosis | (240) |
| Ijaz et al, 2018; Ohanian et al, 2018 | ASO | Grb2 involved in the progression and development of different cancer types | Grb2 is (BP1001) comprises an antisense oligodeoxynucleotide targeted against the translation initiation site of the Grb2 transcript | L-Grb2 ASO inhibition results in reduced proliferation of cancer cells and enhanced survival of mice bearing leukemia xenografts | Grb2 gene | (241,242) |
| Golan et al, 2015; Timar and Kashofer, 2020 | siRNA | KRAS mRNA | KRAS is mutated in ~30% of all tumor cases | System LODER® (Local Drug EluteR), a miniature biodegradable polymeric matrix for local drug release of KRAS (G12D) siRNA within the tumor | KRAS inhibition induces inhibition of tumor growth and induction of apoptosis in tumor cells In mice, survival rates are improved | (243,244) |
| Zhang et al, 2021; Shortridge et al, 2022 | Small molecule inhibitor | miR-21 | miR-21 is an antiapoptotic factor in cancer cells and is upregulated in breast, ovarian and lung cancer, as well as in glioblastomas | ITX-0052 is a small molecule that exhibits strong binding activity to pre-miR-21, inhibiting its processing | These compounds reduce cellular proliferation and miR-21 expression levels in cancer cell lines and increase various tumor suppressor protein expression levels | (245,246) |
| Dovitinib (TKI258) is a tyrosine kinase small-molecule inhibitor that selectively targets the precursor of-miR-21 | ||||||
| Donlic et al, 2018; Goyal et al, 2021 | MALAT1 lncRNA | MALAT1 is upregulated in multiple cancer types | Diphenylfuran analogs targeted at the triple helix structure of MALAT1 lncRNA | In non-small cell lung cancer, knockdown of MALAT1 inhibits tumor growth and metastasis by downregulating PI3K/AKT signaling | (247,248) |
ASO, antisense oligonucleotide; FGFR, fibroblast growth factor receptor; lncRNA, long non-coding RNA; miR, microRNA; siRNA, small interfering RNA; Grb2, growth factor receptor-bound protein 2; L-Grb2, liposomal antisense oligodeoxynucleotide directed to Grb2; MALAT1, metastasis associated lung adenocarcinoma transcript 1.
Table IX.
Interventional clinical trials of inhibitory non-coding RNAs.
| Therapeutic oligonucleotide | Cancer type | Treatment | ClinicalTrials.gov identifier |
|---|---|---|---|
| Cobomarsen | CTCL | MRG-106 monotherapy | NCT03837457 |
| (MRG-106) | CTCL | MRG-106 or vorinostat | NCT03713320 |
| CTCL, CLL, ATLL, and DLBCL | MRG-106 monotherapy | NCT02580552 | |
| RAML and ALL | Combination of BP1001 and concurrent low-dose Ara-C | NCT01159028 | |
| Dovitinib | mRCC | Dovitinib vs. sorafenib | NCT01223027 |
| (TK1258) | Solid tumors | Dovitinib monotherapy | NCT02116803 |
ALL, acute lymphoblastic leukemia; Ara-C, cytarabine; CLL, chronic lymphocytic leukemia; CTCL, cutaneous T-cell lymphoma; BP1001, L-Grb-2 antisense oligonucleotide; RAML, recurrent acute myeloid leukemia; mRCC, metastatic renal cell carcinoma; DLBC, diffuse large B-cell lymphoma; ATLL, adult T-cell leukemia/lymphoma.
4. Conclusions
Although carcinogenesis has a complex and multifactorial origin, alterations in epigenetic processes are considered among the first genomic aberrations that occur during cancer development and are closely related to its progression. The dynamic and reversible nature of epigenetic modifications has allowed the use of epidrug agents that improve cancer treatment, including reducing chemoresistance. Although the use of epidrugs in combination with chemotherapeutic agents may have clinical benefits, several issues must be considered. The reversible nature of methylation patterns persists after drug treatment, and re-methylation and silencing are major unresolved issues. The toxicity and instability of 5-aza-C and 5-aza-D may complicate their use in clinical settings under physiological conditions (59,62). Furthermore, the lack of specificity of the methylating agent may result in the activation of normally silenced genes and contribute to tumorigenesis (62); this poses a significant obstacle in epigenetic therapy.
ncRNAs also represent an essential epigenetic mechanism. Thousands of ncRNAs have been identified and their roles in carcinogenesis are defined. This enables their clinical application through the development of ncRNA-based diagnostic kits for the early detection of cancer and for anticancer therapy. However, this field is not as successful as epidrugs because there is still a large gap between the identification of functional ncRNAs and their clinical applications. In addition, delivery-associated toxicity, poor transfection efficiency, systemic clearance, non-specific biodistribution, degradation in circulation and rapid renal clearance, have delayed the therapeutic potential of ncRNAs (220,221).
The use of epidrugs promises to be an effective tool to help overcome drug resistance in some patients with cancer, particularly for hematological cancer types. However, multidisciplinary work is needed to optimize drug engineering and to design clinical trials to assess the safety and scope of these drugs in cancer therapy for solid tumors.
Acknowledgements
Not applicable.
Funding Statement
This work was supported by the National Council on Science and Technology (CONACYT), Instituto Nacional de Cancerología, Mexico (grant no. CF-2019-263979).
Availability of data and materials
Not applicable.
Authors' contributions
LJCM, EVU, CS, ML, EDLCH and ACP performed the bibliographic review and wrote and critically revised the manuscript. ACP conceived and directed the manuscript. Data authentication is not applicable. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Mantovani F, Collavin L, Del Sal G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2019;26:199–212. doi: 10.1038/s41418-018-0246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deans C, Maggert KA. What do you mean, ‘epigenetic’? Genetics. 2015;199:887–896. doi: 10.1534/genetics.114.173492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bashyam MD, Animireddy S, Bala P, Naz A, George SA. The Yin and Yang of cancer genes. Gene. 2019;704:121–133. doi: 10.1016/j.gene.2019.04.025. [DOI] [PubMed] [Google Scholar]
- 5.Chen P, Li W, Li G. Structures and functions of chromatin fibers. Annu Rev Biophys. 2021;50:95–116. doi: 10.1146/annurev-biophys-062920-063639. [DOI] [PubMed] [Google Scholar]
- 6.Ferreira HJ, Esteller M. Non-coding RNAs, epigenetics, and cancer: Tying it all together. Cancer Metastasis Rev. 2018;37:55–73. doi: 10.1007/s10555-017-9715-8. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L, Lu Q, Chang C. Epigenetics in health and disease. Adv Exp Med Biol. 2020;1253:3–55. doi: 10.1007/978-981-15-3449-2_1. [DOI] [PubMed] [Google Scholar]
- 8.Lu Y, Chan YT, Tan HY, Li S, Wang N, Feng Y. Epigenetic regulation in human cancer: The potential role of epi-drug in cancer therapy. Mol Cancer. 2020;19:79. doi: 10.1186/s12943-020-01197-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding X, He M, Chan AWH, Song QX, Sze SC, Chen H, Man MKH, Man K, Chan SL, Lai PBS, et al. Genomic and epigenomic features of primary and recurrent hepatocellular carcinomas. Gastroenterology. 2019;157:1630–1645.e6. doi: 10.1053/j.gastro.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Malouf GG, Taube JH, Lu Y, Roysarkar T, Panjarian S, Estecio MR, Jelinek J, Yamazaki J, Raynal NJ, Long H, et al. Architecture of epigenetic reprogramming following Twist1-mediated epithelial-mesenchymal transition. Genome Biol. 2013;14:R144. doi: 10.1186/gb-2013-14-12-r144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miranda Furtado CL, Dos Santos Luciano MC, Silva Santos RD, Furtado GP, Moraes MO, Pessoa C. Epidrugs: targeting epigenetic marks in cancer treatment. Epigenetics. 2019;14:1164–1176. doi: 10.1080/15592294.2019.1640546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet. 2018;19:81–92. doi: 10.1038/nrg.2017.80. [DOI] [PubMed] [Google Scholar]
- 13.Ren W, Gao L, Song J. Structural basis of DNMT1 and DNMT3A-mediated DNA methylation. Genes (Basel) 2018;9:620. doi: 10.3390/genes9120620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Z, Zhang Y. Role of mammalian DNA methyltransferases in development. Annu Rev Biochem. 2020;89:135–158. doi: 10.1146/annurev-biochem-103019-102815. [DOI] [PubMed] [Google Scholar]
- 15.Smith ZD, Meissner A. DNA methylation: Roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 16.Zhang ZM, Lu R, Wang P, Yu Y, Chen D, Gao L, Liu S, Ji D, Rothbart SB, Wang Y, et al. Structural basis for DNMT3A-mediated de novo DNA methylation. Nature. 2018;554:387–391. doi: 10.1038/nature25477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Veland N, Lu Y, Hardikar S, Gaddis S, Zeng Y, Liu B, Estecio MR, Takata Y, Lin K, Tomida MW, et al. DNMT3L facilitates DNA methylation partly by maintaining DNMT3A stability in mouse embryonic stem cells. Nucleic Acids Res. 2019;47:152–167. doi: 10.1093/nar/gky947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loaeza-Loaeza J, Beltran AS, Hernández-Sotelo D. DNMTs and impact of CpG content, transcription factors, consensus motifs, lncRNAs, and histone marks on DNA methylation. Genes (Basel) 2020;11:1336. doi: 10.3390/genes11111336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Onodera A, González-Avalos E, Lio CWJ, Georges RO, Bellacosa A, Nakayama T, Rao A. Roles of TET and TDG in DNA demethylation in proliferating and non-proliferating immune cells. Genome Biol. 2021;22:186. doi: 10.1186/s13059-021-02384-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016;30:733–750. doi: 10.1101/gad.276568.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sabino JC, de Almeida MR, Abreu PL, Ferreira AM, Caldas P, Domingues MM, Santos NC, Azzalin CM, Grosso AR, de Almeida SF. Epigenetic reprogramming by TET enzymes impacts co-transcriptional R-loops. Elife. 2022;11:e69476. doi: 10.7554/eLife.69476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C, Fan Y, Li G, Xu X, Duan J, Li R, Kang X, Ma X, Chen X, Ke Y, et al. DNA methylation reprogramming of functional elements during mammalian embryonic development. Cell Discov. 2018;4:41. doi: 10.1038/s41421-018-0039-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dossin F, Pinheiro I, Żylicz JJ, Roensch J, Collombet S, Le Saux A, Chelmicki T, Attia M, Kapoor V, Zhan Y, et al. SPEN integrates transcriptional and epigenetic control of X-inactivation. Nature. 2020;578:455–460. doi: 10.1038/s41586-020-1974-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tucci V, Isles AR, Kelsey G, Ferguson-Smith AC, Erice Imprinting Group Genomic imprinting and physiological processes in mammals. Cell. 2019;176:952–965. doi: 10.1016/j.cell.2019.01.043. [DOI] [PubMed] [Google Scholar]
- 25.Zhang M, Wu J, Zhong W, Zhao Z, He W. DNA-methylation-induced silencing of DIO3OS drives non-small cell lung cancer progression via activating hnRNPK-MYC-CDC25A axis. Mol Ther Oncolytics. 2021;23:205–219. doi: 10.1016/j.omto.2021.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kennedy EM, Goehring GN, Nichols MH, Robins C, Mehta D, Klengel T, Eskin E, Smith AK, Conneely KN. An integrated-omics analysis of the epigenetic landscape of gene expression in human blood cells. BMC Genomics. 2018;19:476. doi: 10.1186/s12864-018-4842-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanwal R, Gupta K, Gupta S. Cancer epigenetics: An introduction. Methods Mol Biol. 2015;1238:3–25. doi: 10.1007/978-1-4939-1804-1_1. [DOI] [PubMed] [Google Scholar]
- 28.Klutstein M, Nejman D, Greenfield R, Cedar H. DNA methylation in cancer and aging. Cancer Res. 2016;76:3446–3450. doi: 10.1158/0008-5472.CAN-15-3278. [DOI] [PubMed] [Google Scholar]
- 29.Sheaffer KL, Elliott EN, Kaestner KH. DNA hypomethylation contributes to genomic instability and intestinal cancer initiation. Cancer Prev Res (Phila) 2016;9:534–546. doi: 10.1158/1940-6207.CAPR-15-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hatziapostolou M, Iliopoulos D. Epigenetic aberrations during oncogenesis. Cell Mol Life Sci. 2011;68:1681–1702. doi: 10.1007/s00018-010-0624-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guerrero-Preston R, Michailidi C, Marchionni L, Pickering CR, Frederick MJ, Myers JN, Yegnasubramanian S, Hadar T, Noordhuis MG, Zizkova V, et al. Key tumor suppressor genes inactivated by ‘greater promoter’ methylation and somatic mutations in head and neck cancer. Epigenetics. 2014;9:1031–1046. doi: 10.4161/epi.29025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faam B, Ghaffari MA, Khorsandi L, Ghadiri AA, Totonchi M, Amouzegar A, Fanaei SA, Azizi F, Shahbazian HB, Hashemi Tabar M. RAP1GAP functions as a tumor suppressor gene and is regulated by DNA methylation in differentiated thyroid cancer. Cytogenet Genome Res. 2021;161:227–235. doi: 10.1159/000516122. [DOI] [PubMed] [Google Scholar]
- 33.Chantre-Justino M, Gonçalves da Veiga Pires I, Cardoso Figueiredo M, Dos Santos Moreira A, Alves G, Faria Ornellas MH. Genetic and methylation status of CDKN2A (p14ARF/p16INK4A) and TP53 genes in recurrent respiratory papillomatosis. Hum Pathol. 2022;119:94–104. doi: 10.1016/j.humpath.2021.11.008. [DOI] [PubMed] [Google Scholar]
- 34.Han B, Yang X, Zhang P, Zhang Y, Tu Y, He Z, Li Y, Yuan J, Dong Y, Hosseini DK, et al. DNA methylation biomarkers for nasopharyngeal carcinoma. PLoS One. 2020;15:e0230524. doi: 10.1371/journal.pone.0230524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoang NM, Rui L. DNA methyltransferases in hematological malignancies. J Genet Genomics. 2020;47:361–372. doi: 10.1016/j.jgg.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steele N, Finn P, Brown R, Plumb JA. Combined inhibition of DNA methylation and histone acetylation enhances gene re-expression and drug sensitivity in vivo. Br J Cancer. 2009;100:758–763. doi: 10.1038/sj.bjc.6604932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bao Y, Oguz G, Lee WC, Lee PL, Ghosh K, Li J, Wang P, Lobie PE, Ehmsen S, Ditzel HJ, et al. EZH2-mediated PP2A inactivation confers resistance to HER2-targeted breast cancer therapy. Nat Commun. 2020;11:5878. doi: 10.1038/s41467-020-19704-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vijayaraghavalu S, Labhasetwar V. Nanogel-mediated delivery of a cocktail of epigenetic drugs plus doxorubicin overcomes drug resistance in breast cancer cells. Drug Deliv Transl Res. 2018;8:1289–1299. doi: 10.1007/s13346-018-0556-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Z, Li HQ, Liu F. DNA methyltransferase inhibitors and their therapeutic potential. Curr Top Med Chem. 2018;18:2448–2457. doi: 10.2174/1568026619666181120150122. [DOI] [PubMed] [Google Scholar]
- 40.Kedhari Sundaram M, Hussain A, Haque S, Raina R, Afroze N. Quercetin modifies 5′CpG promoter methylation and reactivates various tumor suppressor genes by modulating epigenetic marks in human cervical cancer cells. J Cell Biochem. 2019;120:18357–18369. doi: 10.1002/jcb.29147. [DOI] [PubMed] [Google Scholar]
- 41.Sanaei M, Kavoosi F, Behjoo H. Effect of valproic acid and zebularine on SOCS-1 and SOCS-3 gene expression in colon carcinoma SW48 cell line. Exp Oncol. 2020;42:183–187. doi: 10.32471/exp-oncology.2312-8852.vol-42-no-3.15113. [DOI] [PubMed] [Google Scholar]
- 42.Capdevila J, Arqués O, Hernández Mora JR, Matito J, Caratù G, Mancuso FM, Landolfi S, Barriuso J, Jimenez-Fonseca P, Lopez Lopez C, et al. Epigenetic EGFR gene repression confers sensitivity to therapeutic BRAFV600E blockade in colon neuroendocrine carcinomas. Clin Cancer Res. 2020;26:902–909. doi: 10.1158/1078-0432.CCR-19-1266. [DOI] [PubMed] [Google Scholar]
- 43.Marques-Magalhães Â, Graça I, Henrique R, Jerónimo C. Targeting DNA methyltranferases in urological tumors. Front Pharmacol. 2018;9:366. doi: 10.3389/fphar.2018.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xylinas E, Hassler MR, Zhuang D, Krzywinski M, Erdem Z, Robinson BD, Elemento O, Clozel T, Shariat SF. An epigenomic approach to improving response to neoadjuvant cisplatin chemotherapy in bladder cancer. Biomolecules. 2016;6:37. doi: 10.3390/biom6030037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stresemann C, Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int J Cancer. 2008;123:8–13. doi: 10.1002/ijc.23607. [DOI] [PubMed] [Google Scholar]
- 46.Chakrawarti L, Agrawal R, Dang S, Gupta S, Gabrani R. Therapeutic effects of EGCG: A patent review. Expert Opin Ther Pat. 2016;26:907–916. doi: 10.1080/13543776.2016.1203419. [DOI] [PubMed] [Google Scholar]
- 47.Beisler JA. Isolation, characterization, and properties of a labile hydrolysis product of the antitumor nucleoside, 5-azacytidine. J Med Chem. 1978;21:204–208. doi: 10.1021/jm00200a012. [DOI] [PubMed] [Google Scholar]
- 48.Rahman MF, Raj R, Govindarajan R. Identification of structural and molecular features involved in the transport of 3′-Deoxy-nucleoside analogs by human equilibrative nucleoside transporter 3. Drug Metab Dispos. 2018;46:600–609. doi: 10.1124/dmd.117.079400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Momparler RL, Derse D. Kinetics of phosphorylation of 5-aza-2′-deoxyycytidine by deoxycytidine kinase. Biochem Pharmacol. 1979;28:1443–1444. doi: 10.1016/0006-2952(79)90454-4. [DOI] [PubMed] [Google Scholar]
- 50.Santi DV, Garrett CE, Barr PJ. On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell. 1983;33:9–10. doi: 10.1016/0092-8674(83)90327-6. [DOI] [PubMed] [Google Scholar]
- 51.Seelan RS, Mukhopadhyay P, Pisano MM, Greene RM. Effects of 5-Aza-2′-deoxycytidine (decitabine) on gene expression. Drug Metab Rev. 2018;50:193–207. doi: 10.1080/03602532.2018.1437446. [DOI] [PubMed] [Google Scholar]
- 52.Zheng Z, Li L, Liu X, Wang D, Tu B, Wang L, Wang H, Zhu WG. 5-Aza-2′-deoxycytidine reactivates gene expression via degradation of pRb pocket proteins. FASEB J. 2012;26:449–459. doi: 10.1096/fj.11-190025. [DOI] [PubMed] [Google Scholar]
- 53.Sorm F, Pískala A, Cihák A, Veselý J. 5-Azacytidine, a new, highly effective cancerostatic. Experientia. 1964;20:202–203. doi: 10.1007/BF02135399. [DOI] [PubMed] [Google Scholar]
- 54.Sorm F, Vesely J. The activity of a new antimetabolite, 5-azacytidine, against lymphoid leukaemia in AK mice. Neoplasma. 1964;11:123–130. [PubMed] [Google Scholar]
- 55.Case DC., Jr 5-azacytidine in refractory acute leukemia. Oncology. 1982;39:218–221. doi: 10.1159/000225641. [DOI] [PubMed] [Google Scholar]
- 56.Tanaka K, Appella E, Jay G. Developmental activation of the H-2K gene is correlated with an increase in DNA methylation. Cell. 1983;35:457–465. doi: 10.1016/0092-8674(83)90179-4. [DOI] [PubMed] [Google Scholar]
- 57.Vogler WR, Miller DS, Keller JW. 5-Azacytidine (NSC 102816): A new drug for the treatment of myeloblastic leukemia. Blood. 1976;48:331–337. doi: 10.1182/blood.V48.3.331.331. [DOI] [PubMed] [Google Scholar]
- 58.Kaminskas E, Farrell A, Abraham S, Baird A, Hsieh LS, Lee SL, Leighton JK, Patel H, Rahman A, Sridhara R, et al. Approval summary: Azacitidine for treatment of myelodysplastic syndrome subtypes. Clin Cancer Res. 2005;11:3604–3608. doi: 10.1158/1078-0432.CCR-04-2135. [DOI] [PubMed] [Google Scholar]
- 59.Ganesan A, Arimondo PB, Rots MG, Jeronimo C, Berdasco M. The timeline of epigenetic drug discovery: From reality to dreams. Clin Epigenetics. 2019;11:174. doi: 10.1186/s13148-019-0776-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Duchmann M, Itzykson R. Clinical update on hypomethylating agents. Int J Hematol. 2019;110:161–169. doi: 10.1007/s12185-019-02651-9. [DOI] [PubMed] [Google Scholar]
- 61.Schaefer M, Hagemann S, Hanna K, Lyko F. Azacytidine inhibits RNA methylation at DNMT2 target sites in human cancer cell lines. Cancer Res. 2009;69:8127–8132. doi: 10.1158/0008-5472.CAN-09-0458. [DOI] [PubMed] [Google Scholar]
- 62.Hollenbach PW, Nguyen AN, Brady H, Williams M, Ning Y, Richard N, Krushel L, Aukerman SL, Heise C, MacBeth KJ. A comparison of azacitidine and decitabine activities in acute myeloid leukemia cell lines. PLoS One. 2010;5:e9001. doi: 10.1371/journal.pone.0009001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Venturelli S, Berger A, Weiland T, Essmann F, Waibel M, Nuebling T, Häcker S, Schenk M, Schulze-Osthoff K, Salih HR, et al. Differential induction of apoptosis and senescence by the DNA methyltransferase inhibitors 5-azacytidine and 5-aza-2′-deoxycytidine in solid tumor cells. Mol Cancer Ther. 2013;12:2226–2236. doi: 10.1158/1535-7163.MCT-13-0137. [DOI] [PubMed] [Google Scholar]
- 64.Jin S, Cojocari D, Purkal JJ, Popovic R, Talaty NN, Xiao Y, Solomon LR, Boghaert ER, Leverson JD, Phillips DC. 5-Azacitidine induces NOXA to prime AML cells for venetoclax-mediated apoptosis. Clin Cancer Res. 2020;26:3371–3383. doi: 10.1158/1078-0432.CCR-19-1900. [DOI] [PubMed] [Google Scholar]
- 65.Fabre C, Grosjean J, Tailler M, Boehrer S, Adès L, Perfettini JL, de Botton S, Fenaux P, Kroemer G. A novel effect of DNA methyltransferase and histone deacetylase inhibitors: NFkappaB inhibition in malignant myeloblasts. Cell Cycle. 2008;7:2139–2145. doi: 10.4161/cc.7.14.6268. [DOI] [PubMed] [Google Scholar]
- 66.Carbajo-García MC, Corachán A, Segura-Benitez M, Monleón J, Escrig J, Faus A, Pellicer A, Cervelló I, Ferrero H. 5-aza-2′-deoxycitidine inhibits cell proliferation, extracellular matrix formation and Wnt/β-catenin pathway in human uterine leiomyomas. Reprod Biol Endocrinol. 2021;19:106. doi: 10.1186/s12958-021-00790-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Linnekamp JF, Kandimalla R, Fessler E, de Jong JH, Rodermond HM, van Bochove GGW, The FO, Punt CJA, Bemelman WA, van de Ven AWH, et al. Pre-operative decitabine in colon cancer patients: Analyses on WNT target methylation and expression. Cancers (Basel) 2021;13:2357. doi: 10.3390/cancers13102357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Santini V. How I treat MDS after hypomethylating agent failure. Blood. 2019;133:521–529. doi: 10.1182/blood-2018-03-785915. [DOI] [PubMed] [Google Scholar]
- 69.Mabaera R, Greene MR, Richardson CA, Conine SJ, Kozul CD, Lowrey CH. Neither DNA hypomethylation nor changes in the kinetics of erythroid differentiation explain 5-azacytidine's ability to induce human fetal hemoglobin. Blood. 2008;111:411–420. doi: 10.1182/blood-2007-06-093948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Susanto JM, Colvin EK, Pinese M, Chang DK, Pajic M, Mawson A, Caldon CE, Musgrove EA, Henshall SM, Sutherland RL, et al. The epigenetic agents suberoylanilide hydroxamic acid and 5-AZA-2′ deoxycytidine decrease cell proliferation, induce cell death and delay the growth of MiaPaCa2 pancreatic cancer cells in vivo. Int J Oncol. 2015;46:2223–2230. doi: 10.3892/ijo.2015.2894. [DOI] [PubMed] [Google Scholar]
- 71.Evans IC, Barnes JL, Garner IM, Pearce DR, Maher TM, Shiwen X, Renzoni EA, Wells AU, Denton CP, Laurent GJ, et al. Epigenetic regulation of cyclooxygenase-2 by methylation of c8orf4 in pulmonary fibrosis. Clin Sci (Lond) 2016;130:575–586. doi: 10.1042/CS20150697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 73.Meng CF, Zhu XJ, Peng G, Dai DQ. Promoter histone H3 lysine 9 di-methylation is associated with DNA methylation and aberrant expression of p16 in gastric cancer cells. Oncol Rep. 2009;22:1221–1227. doi: 10.3892/or_00000558. [DOI] [PubMed] [Google Scholar]
- 74.Lee SH, Kim J, Kim WH, Lee YM. Hypoxic silencing of tumor suppressor RUNX3 by histone modification in gastric cancer cells. Oncogene. 2009;28:184–194. doi: 10.1038/onc.2008.377. [DOI] [PubMed] [Google Scholar]
- 75.Nguyen CT, Weisenberger DJ, Velicescu M, Gonzales FA, Lin JCY, Liang G, Jones PA. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res. 2002;62:6456–6461. [PubMed] [Google Scholar]
- 76.Griffiths EA, Gore SD. Epigenetic therapies in MDS and AML. Adv Exp Med Biol. 2013;754:253–283. doi: 10.1007/978-1-4419-9967-2_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nguyen AN, Hollenbach PW, Richard N, Luna-Moran A, Brady H, Heise C, MacBeth KJ. Azacitidine and decitabine have different mechanisms of action in non-small cell lung cancer cell lines. Lung Cancer (Auckl) 2010;1:119–140. doi: 10.2147/LCTT.S11726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Home-ClinicalTrials.gov. [ November 22; 2022 ];https://clinicaltrials.gov/ [Google Scholar]
- 79.Hurd PJ, Whitmarsh AJ, Baldwin GS, Kelly SM, Waltho JP, Price NC, Connolly BA, Hornby DP. Mechanism-based inhibition of C5-cytosine DNA methyltransferases by 2-H pyrimidinone. J Mol Biol. 1999;286:389–401. doi: 10.1006/jmbi.1998.2491. [DOI] [PubMed] [Google Scholar]
- 80.Yoo CB, Cheng JC, Jones PA. Zebularine: A new drug for epigenetic therapy. Biochem Soc Trans. 2004;32:910–912. doi: 10.1042/BST0320910. [DOI] [PubMed] [Google Scholar]
- 81.Ferguson LR, Tatham AL, Lin Z, Denny WA. Epigenetic regulation of gene expression as an anticancer drug target. Curr Cancer Drug Targets. 2011;11:199–212. doi: 10.2174/156800911794328510. [DOI] [PubMed] [Google Scholar]
- 82.Dueñas-Gonzalez A, Coronel J, Cetina L, González-Fierro A, Chavez-Blanco A, Taja-Chayeb L. Hydralazine-valproate: A repositioned drug combination for the epigenetic therapy of cancer. Expert Opin Drug Metab Toxicol. 2014;10:1433–1444. doi: 10.1517/17425255.2014.947263. [DOI] [PubMed] [Google Scholar]
- 83.Singh N, Dueñas-González A, Lyko F, Medina-Franco JL. Molecular modeling and molecular dynamics studies of hydralazine with human DNA methyltransferase 1. ChemMedChem. 2009;4:792–799. doi: 10.1002/cmdc.200900017. [DOI] [PubMed] [Google Scholar]
- 84.Daher-Reyes GS, Merchan BM, Yee KWL. Guadecitabine (SGI-110): An investigational drug for the treatment of myelodysplastic syndrome and acute myeloid leukemia. Expert Opin Investig Drugs. 2019;28:835–849. doi: 10.1080/13543784.2019.1667331. [DOI] [PubMed] [Google Scholar]
- 85.Salvador LA, Luesch H. Discovery and mechanism of natural products as modulators of histone acetylation. Curr Drug Targets. 2012;13:1029–1047. doi: 10.2174/138945012802008973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang Y, Wang X, Han L, Zhou Y, Sun S. Green tea polyphenol EGCG reverse cisplatin resistance of A549/DDP cell line through candidate genes demethylation. Biomed Pharmacother. 2015;69:285–290. doi: 10.1016/j.biopha.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 87.Alaskhar Alhamwe B, Khalaila R, Wolf J, von Bülow V, Harb H, Alhamdan F, Hii CS, Prescott SL, Ferrante A, Renz H, et al. Histone modifications and their role in epigenetics of atopy and allergic diseases. Allergy Asthma Clin Immunol. 2018;14:39. doi: 10.1186/s13223-018-0259-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lawrence M, Daujat S, Schneider R. Lateral thinking: How histone modifications regulate gene expression. Trends Genet. 2016;32:42–56. doi: 10.1016/j.tig.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 89.Lakshmaiah KC, Jacob LA, Aparna S, Lokanatha D, Saldanha SC. Epigenetic therapy of cancer with histone deacetylase inhibitors. J Cancer Res Ther. 2014;10:469–478. doi: 10.4103/0973-1482.137937. [DOI] [PubMed] [Google Scholar]
- 90.Albaugh BN, Arnold KM, Denu JM. KAT(ching) metabolism by the tail: Insight into the links between lysine acetyltransferases and metabolism. Chembiochem. 2011;12:290–298. doi: 10.1002/cbic.201000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang J, Song C, Zhan X. The role of protein acetylation in carcinogenesis and targeted drug discovery. Front Endocrinol (Lausanne) 2022;13:972312. doi: 10.3389/fendo.2022.972312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cohen I, Poręba E, Kamieniarz K, Schneider R. Histone modifiers in cancer: Friends or foes? Genes Cancer. 2011;2:631–647. doi: 10.1177/1947601911417176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fan P, Zhao J, Meng Z, Wu H, Wang B, Wu H, Jin X. Overexpressed histone acetyltransferase 1 regulates cancer immunity by increasing programmed death-ligand 1 expression in pancreatic cancer. J Exp Clin Cancer Res. 2019;38:47. doi: 10.1186/s13046-019-1044-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sun XJ, Man N, Tan Y, Nimer SD, Wang L. The role of histone acetyltransferases in normal and malignant hematopoiesis. Front Oncol. 2015;5:108. doi: 10.3389/fonc.2015.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang P, Wang Z, Liu J. Role of HDACs in normal and malignant hematopoiesis. Mol Cancer. 2020;19:5. doi: 10.1186/s12943-020-01182-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Carraway HE, Malkaram SA, Cen Y, Shatnawi A, Fan J, Ali HEA, Abd Elmageed ZY, Buttolph T, Denvir J, Primerano DA, Fandy TE. Activation of SIRT6 by DNA hypomethylating agents and clinical consequences on combination therapy in leukemia. Sci Rep. 2020;10:10325. doi: 10.1038/s41598-020-70622-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hu XT, Xing W, Zhao RS, Tan Y, Wu XF, Ao LQ, Li Z, Yao MW, Yuan M, Guo W, et al. HDAC2 inhibits EMT-mediated cancer metastasis by downregulating the long noncoding RNA H19 in colorectal cancer. J Exp Clin Cancer Res. 2020;39:270. doi: 10.1186/s13046-020-01783-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Körholz K, Ridinger J, Krunic D, Najafi S, Gerloff XF, Frese K, Meder B, Peterziel H, Vega-Rubin-de-Celis S, Witt O, Oehme I. Broad-spectrum HDAC inhibitors promote autophagy through FOXO transcription factors in neuroblastoma. Cells. 2021;10:1001. doi: 10.3390/cells10051001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chun P. Histone deacetylase inhibitors in hematological malignancies and solid tumors. Arch Pharm Res. 2015;38:933–949. doi: 10.1007/s12272-015-0571-1. [DOI] [PubMed] [Google Scholar]
- 100.Jęśko H, Wencel P, Strosznajder RP, Strosznajder JB. Sirtuins and their roles in brain aging and neurodegenerative disorders. Neurochem Res. 2017;42:876–890. doi: 10.1007/s11064-016-2110-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Willis-Martinez D, Richards HW, Timchenko NA, Medrano EE. Role of HDAC1 in senescence, aging, and cancer. Exp Gerontol. 2010;45:279–285. doi: 10.1016/j.exger.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26:5420–5432. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- 103.Audia JE, Campbell RM. Histone modifications and cancer. Cold Spring Harb Perspect Biol. 2016;8:a019521. doi: 10.1101/cshperspect.a019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bergamin E, Sarvan S, Malette J, Eram MS, Yeung S, Mongeon V, Joshi M, Brunzelle JS, Michaels SD, Blais A, et al. Molecular basis for the methylation specificity of ATXR5 for histone H3. Nucleic Acids Res. 2017;45:6375–6387. doi: 10.1093/nar/gkx224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mohan M, Herz HM, Shilatifard A. SnapShot: Histone lysine methylase complexes. Cell. 2012;149:498–498.e1. doi: 10.1016/j.cell.2012.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.From the American Association of Neurological Surgeons (AANS), American Society of Neuroradiology (ASNR), Cardiovascular and Interventional Radiology Society of Europe (CIRSE), Canadian Interventional Radiology Association (CIRA), Congress of Neurological Surgeons (CNS), European Society of Minimally Invasive Neurological Therapy (ESMINT), European Society of Neuroradiology (ESNR), European Stroke Organization (ESO), Society for Cardiovascular Angiography and Interventions (SCAI), Society of Interventional Radiology (SIR) et al. Multisociety consensus quality improvement revised consensus statement for endovascular therapy of acute ischemic stroke. Int J Stroke. 2018;13:612–632. doi: 10.1016/j.jvir.2017.11.026. [DOI] [PubMed] [Google Scholar]
- 107.McGrath J, Trojer P. Targeting histone lysine methylation in cancer. Pharmacol Ther. 2015;150:1–22. doi: 10.1016/j.pharmthera.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 108.Wan YCE, Liu J, Chan KM. Histone H3 mutations in cancer. Curr Pharmacol Rep. 2018;4:292–300. doi: 10.1007/s40495-018-0141-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Salz T, Deng C, Pampo C, Siemann D, Qiu Y, Brown K, Huang S. Histone methyltransferase hSETD1A is a novel regulator of metastasis in breast cancer. Mol Cancer Res. 2015;13:461–469. doi: 10.1158/1541-7786.MCR-14-0389. [DOI] [PubMed] [Google Scholar]
- 110.Behjati S, Tarpey PS, Presneau N, Scheipl S, Pillay N, Van Loo P, Wedge DC, Cooke SL, Gundem G, Davies H, et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet. 2013;45:1479–1482. doi: 10.1038/ng.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Papillon-Cavanagh S, Lu C, Gayden T, Mikael LG, Bechet D, Karamboulas C, Ailles L, Karamchandani J, Marchione DM, Garcia BA, et al. Impaired H3K36 methylation defines a subset of head and neck squamous cell carcinomas. Nat Genet. 2017;49:180–185. doi: 10.1038/ng.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yang T, Yang Y, Wang Y. Predictive biomarkers and potential drug combinations of epi-drugs in cancer therapy. Clin Epigenetics. 2021;13:113. doi: 10.1186/s13148-021-01098-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bates SE. Epigenetic therapies for cancer. N Engl J Med. 2020;383:650–663. doi: 10.1056/NEJMra1805035. [DOI] [PubMed] [Google Scholar]
- 114.Yaseen A, Chen S, Hock S, Rosato R, Dent P, Dai Y, Grant S. Resveratrol sensitizes acute myelogenous leukemia cells to histone deacetylase inhibitors through reactive oxygen species-mediated activation of the extrinsic apoptotic pathway. Mol Pharmacol. 2012;82:1030–1041. doi: 10.1124/mol.112.079624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bubna AK. Vorinostat-an overview. Indian J Dermatol. 2015;60:419. doi: 10.4103/0019-5154.160511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Richon VM. Cancer biology: Mechanism of antitumour action of vorinostat (suberoylanilide hydroxamic acid), a novel histone deacetylase inhibitor. Br J Cancer. 2006;95((Suppl 1)):S2–S6. doi: 10.1038/sj.bjc.6603463. [DOI] [Google Scholar]
- 117.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 118.Siegel D, Hussein M, Belani C, Robert F, Galanis E, Richon VM, Garcia-Vargas J, Sanz-Rodriguez C, Rizvi S. Vorinostat in solid and hematologic malignancies. J Hematol Oncol. 2009;2:31. doi: 10.1186/1756-8722-2-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nakajima H, Kim YB, Terano H, Yoshida M, Horinouchi S. FR901228, a potent antitumor antibiotic, is a novel histone deacetylase inhibitor. Exp Cell Res. 1998;241:126–133. doi: 10.1006/excr.1998.4027. [DOI] [PubMed] [Google Scholar]
- 120.VanderMolen KM, McCulloch W, Pearce CJ, Oberlies NH. Romidepsin (Istodax, NSC 630176, FR901228, FK228, depsipeptide): A natural product recently approved for cutaneous T-cell lymphoma. J Antibiot (Tokyo) 2011;64:525–531. doi: 10.1038/ja.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Marshall JL, Rizvi N, Kauh J, Dahut W, Figuera M, Kang MH, Figg WD, Wainer I, Chaissang C, Li MZ, Hawkins MJ. A phase I trial of depsipeptide (FR901228) in patients with advanced cancer. J Exp Ther Oncol. 2002;2:325–332. doi: 10.1046/j.1359-4117.2002.01039.x. [DOI] [PubMed] [Google Scholar]
- 122.Petrich A, Nabhan C. Use of class I histone deacetylase inhibitor romidepsin in combination regimens. Leuk Lymphoma. 2016;57:1755–1765. doi: 10.3109/10428194.2016.1160082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.O'Connor OA, Horwitz S, Masszi T, Van Hoof A, Brown P, Doorduijn J, Hess G, Jurczak W, Knoblauch P, Chawla S, et al. Belinostat in patients with relapsed or refractory peripheral T-cell lymphoma: Results of the pivotal phase II BELIEF (CLN-19) study. J Clin Oncol. 2015;33:2492–2499. doi: 10.1200/JCO.2014.59.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Poole RM. Belinostat: First global approval. Drugs. 2014;74:1543–1554. doi: 10.1007/s40265-014-0270-0. [DOI] [PubMed] [Google Scholar]
- 125.Autin P, Blanquart C, Fradin D. Epigenetic drugs for cancer and microRNAs: A focus on histone deacetylase inhibitors. Cancers (Basel) 2019;11:1530. doi: 10.3390/cancers11101530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Spratlin JL, Pitts TM, Kulikowski GN, Morelli MP, Tentler JJ, Serkova NJ, Eckhardt SG. Synergistic activity of histone deacetylase and proteasome inhibition against pancreatic and hepatocellular cancer cell lines. Anticancer Res. 2011;31:1093–1103. [PMC free article] [PubMed] [Google Scholar]
- 127.Lee MJ, Kim YS, Kummar S, Giaccone G, Trepel JB. Histone deacetylase inhibitors in cancer therapy. Curr Opin Oncol. 2008;20:639–649. doi: 10.1097/CCO.0b013e3283127095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gibney ER, Nolan CM. Epigenetics and gene expression. Heredity (Edinb) 2010;105:4–13. doi: 10.1038/hdy.2010.54. [DOI] [PubMed] [Google Scholar]
- 129.He J, Xie Q, Xu H, Li J, Li Y. Circular RNAs and cancer. Cancer Lett. 2017;396:138–144. doi: 10.1016/j.canlet.2017.03.027. [DOI] [PubMed] [Google Scholar]
- 130.Khanna A, Stamm S. Regulation of alternative splicing by short non-coding nuclear RNAs. RNA Biol. 2010;7:480–485. doi: 10.4161/rna.7.4.12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kwek KY, Murphy S, Furger A, Thomas B, O'Gorman W, Kimura H, Proudfoot NJ, Akoulitchev A. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat Struct Mol Biol. 2002;9:800–805. doi: 10.1038/nsb862. [DOI] [PubMed] [Google Scholar]
- 132.Eilebrecht S, Brysbaert G, Wegert T, Urlaub H, Benecke BJ, Benecke A. 7SK small nuclear RNA directly affects HMGA1 function in transcription regulation. Nucleic Acids Res. 2011;39:2057–2072. doi: 10.1093/nar/gkq1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Szymanski M, Erdmann VA, Barciszewski J. Noncoding RNAs database (ncRNAdb) Nucleic Acids Res. 2007;35:D162–D164. doi: 10.1093/nar/gkl994. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chu CY, Rana TM. Small RNAs: Regulators and guardians of the genome. J Cell Physiol. 2007;213:412–419. doi: 10.1002/jcp.21230. [DOI] [PubMed] [Google Scholar]
- 135.Abi A, Farahani N, Molavi G, Gheibi Hayat SM. Circular RNAs: Epigenetic regulators in cancerous and noncancerous skin diseases. Cancer Gene Ther. 2020;27:280–293. doi: 10.1038/s41417-019-0130-x. [DOI] [PubMed] [Google Scholar]
- 136.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 137.Ding X, Zhang S, Li X, Feng C, Huang Q, Wang S, Wang S, Xia W, Yang F, Yin R, et al. Profiling expression of coding genes, long noncoding RNA, and circular RNA in lung adenocarcinoma by ribosomal RNA-depleted RNA sequencing. FEBS Open Bio. 2018;8:544–555. doi: 10.1002/2211-5463.12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lam JKW, Chow MYT, Zhang Y, Leung SWS. siRNA versus miRNA as therapeutics for gene silencing. Mol Ther Nucleic Acids. 2015;4:e252. doi: 10.1038/mtna.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 140.Kim VN. MicroRNA biogenesis: Coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 141.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tomari Y, Zamore PD. Perspective: Machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 143.Paturi S, Deshmukh MV. A glimpse of ‘dicer biology’ through the structural and functional perspective. Front Mol Biosci. 2021;8:643657. doi: 10.3389/fmolb.2021.643657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Alagia A, Jorge AF, Aviñó A, Cova TFGG, Crehuet R, Grijalvo S, Pais AACC, Eritja R. Exploring PAZ/3′-overhang interaction to improve siRNA specificity. A combined experimental and modeling study. Chem Sci. 2018;9:2074–2086. doi: 10.1039/C8SC00010G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chen L, Dahlstrom JE, Lee SH, Rangasamy D. Naturally occurring endo-siRNA silences LINE-1 retrotransposons in human cells through DNA methylation. Epigenetics. 2012;7:758–771. doi: 10.4161/epi.20706. [DOI] [PubMed] [Google Scholar]
- 147.Guo B, Li D, Du L, Zhu X. piRNAs: Biogenesis and their potential roles in cancer. Cancer Metastasis Rev. 2020;39:567–575. doi: 10.1007/s10555-020-09863-0. [DOI] [PubMed] [Google Scholar]
- 148.Sparmann A. piRNAs-guardians of the germline. Nat Res. 2019 [Google Scholar]
- 149.Iwasaki YW, Siomi MC, Siomi H. PIWI-interacting RNA: Its biogenesis and functions. Annu Rev Biochem. 2015;84:405–433. doi: 10.1146/annurev-biochem-060614-034258. [DOI] [PubMed] [Google Scholar]
- 150.Caramuta S, Egyházi S, Rodolfo M, Witten D, Hansson J, Larsson C, Lui WO. MicroRNA expression profiles associated with mutational status and survival in malignant melanoma. J Invest Dermatol. 2010;130:2062–2070. doi: 10.1038/jid.2010.63. [DOI] [PubMed] [Google Scholar]
- 151.Chi J, Ballabio E, Chen XH, Kušec R, Taylor S, Hay D, Tramonti D, Saunders NJ, Littlewood T, Pezzella F, et al. MicroRNA expression in multiple myeloma is associated with genetic subtype, isotype and survival. Biol Direct. 2011;6:23. doi: 10.1186/1745-6150-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Garzon R, Garofalo M, Martelli MP, Briesewitz R, Wang L, Fernandez-Cymering C, Volinia S, Liu CG, Schnittger S, Haferlach T, et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci USA. 2008;105:3945–3950. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 154.Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, Taccioli C, Zanesi N, Garzon R, Aqeilan RI, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci USA. 2008;105:5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603–1614. doi: 10.1038/cdd.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: Lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 158.Liao JM, Cao B, Zhou X, Lu H. New insights into p53 functions through its target microRNAs. J Mol Cell Biol. 2014;6:206–213. doi: 10.1093/jmcb/mju018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Madrigal T, Hernández-Monge J, Herrera LA, González-De la Rosa CH, Domínguez-Gómez G, Candelaria M, Luna-Maldonado F, Calderón González KG, Díaz-Chávez J. Regulation of miRNAs expression by mutant p53 gain of function in cancer. Front Cell Dev Biol. 2021;9:695723. doi: 10.3389/fcell.2021.695723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tornesello ML, Annunziata C, Tornesello AL, Buonaguro L, Buonaguro FM. Human oncoviruses and p53 tumor suppressor pathway deregulation at the origin of human cancers. Cancers (Basel) 2018;10:213. doi: 10.3390/cancers10070213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hassler MR, Turanov AA, Alterman JF, Haraszti RA, Coles AH, Osborn MF, Echeverria D, Nikan M, Salomon WE, Roux L, et al. Comparison of partially and fully chemically-modified siRNA in conjugate-mediated delivery in vivo. Nucleic Acids Res. 2018;46:2185–2196. doi: 10.1093/nar/gky037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22:96–118. doi: 10.1038/s41580-021-00330-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gao N, Li Y, Li J, Gao Z, Yang Z, Li Y, Liu H, Fan T. Long non-coding RNAs: The regulatory mechanisms, research strategies, and future directions in cancers. Front Oncol. 2020;10:598817. doi: 10.3389/fonc.2020.598817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics. 2016;14:42–54. doi: 10.1016/j.gpb.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mathy NW, Chen XM. Long non-coding RNAs (lncRNAs) and their transcriptional control of inflammatory responses. J Biol Chem. 2017;292:12375–12382. doi: 10.1074/jbc.R116.760884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Amirinejad R, Rezaei M, Shirvani-Farsani Z. An update on long intergenic noncoding RNA p21: A regulatory molecule with various significant functions in cancer. Cell Biosci. 2020;10:82. doi: 10.1186/s13578-020-00445-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hall JR, Messenger ZJ, Tam HW, Phillips SL, Recio L, Smart RC. Long noncoding RNA lincRNA-p21 is the major mediator of UVB-induced and p53-dependent apoptosis in keratinocytes. Cell Death Dis. 2015;6:e1700. doi: 10.1038/cddis.2015.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Dai X, Kaushik AC, Zhang J. The emerging role of major regulatory RNAs in cancer control. Front Oncol. 2019;9:920. doi: 10.3389/fonc.2019.00920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fox AH, Nakagawa S, Hirose T, Bond CS. Paraspeckles: Where long noncoding RNA meets phase separation. Trends Biochem Sci. 2018;43:124–135. doi: 10.1016/j.tibs.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 172.Sun TT, He J, Liang Q, Ren LL, Yan TT, Yu TC, Tang JY, Bao YJ, Hu Y, Lin Y, et al. LncRNA GClnc1 promotes gastric carcinogenesis and may Act as a modular scaffold of WDR5 and KAT2A complexes to specify the histone modification pattern. Cancer Discov. 2016;6:784–801. doi: 10.1158/2159-8290.CD-15-0921. [DOI] [PubMed] [Google Scholar]
- 173.Wang J, Zhu S, Meng N, He Y, Lu R, Yan GR. ncRNA-encoded peptides or proteins and cancer. Mol Ther. 2019;27:1718–1725. doi: 10.1016/j.ymthe.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Xing J, Liu H, Jiang W, Wang L. LncRNA-encoded peptide: Functions and predicting methods. Front Oncol. 2021;10:622294. doi: 10.3389/fonc.2020.622294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kong S, Tao M, Shen X, Ju S. Translatable circRNAs and lncRNAs: Driving mechanisms and functions of their translation products. Cancer Lett. 2020;483:59–65. doi: 10.1016/j.canlet.2020.04.006. [DOI] [PubMed] [Google Scholar]
- 176.Huang JZ, Chen M, Chen D, Gao XC, Zhu S, Huang H, Hu M, Zhu H, Yan GR. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol Cell. 2017;68:171–184.e6. doi: 10.1016/j.molcel.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 177.Chen Y, Long W, Yang L, Zhao Y, Wu X, Li M, Du F, Chen Y, Yang Z, Wen Q, et al. Functional peptides encoded by long non-coding RNAs in gastrointestinal cancer. Front Oncol. 2021;11:777374. doi: 10.3389/fonc.2021.777374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chakraborty S, Andrieux G, Hasan AMM, Ahmed M, Hosen MI, Rahman T, Hossain MA, Boerries M. Harnessing the tissue and plasma lncRNA-peptidome to discover peptide-based cancer biomarkers. Sci Rep. 2019;9:12322. doi: 10.1038/s41598-019-48774-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Welden JR, Stamm S. Pre-mRNA structures forming circular RNAs. Biochim Biophys Acta Gene Regul Mech. 2019;1862:194410. doi: 10.1016/j.bbagrm.2019.194410. [DOI] [PubMed] [Google Scholar]
- 180.Eger N, Schoppe L, Schuster S, Laufs U, Boeckel JN. Circular RNA splicing. In: Xiao J, editor. Circular RNAs. Advances in Experimental Medicine and Biology. Vol. 1087. Springer; Singapore: 2018. pp. 41–52. [DOI] [PubMed] [Google Scholar]
- 181.Nisar S, Bhat AA, Singh M, Karedath T, Rizwan A, Hashem S, Bagga P, Reddy R, Jamal F, Uddin S, et al. Insights into the role of CircRNAs: Biogenesis, characterization, functional, and clinical impact in human malignancies. Front Cell Dev Biol. 2021;9:617281. doi: 10.3389/fcell.2021.617281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhao J, Tao Y, Zhou Y, Qin N, Chen C, Tian D, Xu L. MicroRNA-7: A promising new target in cancer therapy. Cancer Cell Int. 2015;15:103. doi: 10.1186/s12935-015-0259-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Liu T, Liu S, Xu Y, Shu R, Wang F, Chen C, Zeng Y, Luo H. Circular RNA-ZFR inhibited cell proliferation and promoted apoptosis in gastric cancer by sponging miR-130a/miR-107 and modulating PTEN. Cancer Res Treat. 2018;50:1396–1417. doi: 10.4143/crt.2017.537. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 184.Yu CY, Kuo HC. The emerging roles and functions of circular RNAs and their generation. J Biomed Sci. 2019;26:29. doi: 10.1186/s12929-019-0523-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Jiang MP, Xu WX, Hou JC, Xu Q, Wang DD, Tang JH. The emerging role of the interactions between circular RNAs and RNA-binding proteins in common human cancers. J Cancer. 2021;12:5206–5219. doi: 10.7150/jca.58182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular RNA-protein interactions: Functions, mechanisms, and identification. Theranostics. 2020;10:3503–3517. doi: 10.7150/thno.42174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Du WW, Yang W, Li X, Awan FM, Yang Z, Fang L, Lyu J, Li F, Peng C, Krylov SN, et al. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene. 2018;37:5829–5842. doi: 10.1038/s41388-018-0369-y. [DOI] [PubMed] [Google Scholar]
- 188.Chen S, Thorne RF, Zhang XD, Wu M, Liu L. Non-coding RNAs, guardians of the p53 galaxy. Semin Cancer Biol. 2021;75:72–83. doi: 10.1016/j.semcancer.2020.09.002. [DOI] [PubMed] [Google Scholar]
- 189.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wang C, Tao W, Ni S, Chen Q. Circular RNA circ-Foxo3 induced cell apoptosis in urothelial carcinoma via interaction with miR-191-5p. Onco Targets Ther. 2019;12:8085–8094. doi: 10.2147/OTT.S215823. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 191.Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, Huang N, Yang X, Zhao K, Zhou H, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110:304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Park JW, Lagniton PNP, Liu Y, Xu RH. mRNA vaccines for COVID-19: What, why and how. Int J Biol Sci. 2021;17:1446–1460. doi: 10.7150/ijbs.59233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Damase TR, Sukhovershin R, Boada C, Taraballi F, Pettigrew RI, Cooke JP. The limitless future of RNA therapeutics. Front Bioeng Biotechnol. 2021;9:628137. doi: 10.3389/fbioe.2021.628137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Qian Y, Shi L, Luo Z. Long non-coding RNAs in cancer: Implications for diagnosis, prognosis, and therapy. Front Med (Lausanne) 2020;7:612393. doi: 10.3389/fmed.2020.612393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: Opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ling H. Non-coding RNAs: Therapeutic strategies and delivery systems. Adv Exp Med Biol. 2016;937:229–237. doi: 10.1007/978-3-319-42059-2_12. [DOI] [PubMed] [Google Scholar]
- 197.Weinberg MS, Morris KV. Transcriptional gene silencing in humans. Nucleic Acids Res. 2016;44:6505–6517. doi: 10.1093/nar/gkw139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kotowska-Zimmer A, Pewinska M, Olejniczak M. Artificial miRNAs as therapeutic tools: Challenges and opportunities. Wiley Interdiscip Rev RNA. 2021;12:e1640. doi: 10.1002/wrna.1640. [DOI] [PubMed] [Google Scholar]
- 199.Turner AMW, Morris KV. Controlling transcription with noncoding RNAs in mammalian cells. Biotechniques. 2010;48:9–16. doi: 10.2144/000113442. [DOI] [PubMed] [Google Scholar]
- 200.Yoon S, Rossi JJ. Therapeutic potential of small activating RNAs (saRNAs) in human cancers. Curr Pharm Biotechnol. 2018;19:604–610. doi: 10.2174/1389201019666180528084059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Scoles DR, Minikel EV, Pulst SM. Antisense oligonucleotides: A primer. Neurol Genet. 2019;5:e323. doi: 10.1212/NXG.0000000000000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Raghavendra P, Pullaiah T. Advances in Cell and Molecular Diagnostics. Elsevier; 2018. RNA-based applications in diagnostic and therapeutics for cancer; pp. 33–55. [DOI] [Google Scholar]
- 203.Ratti M, Lampis A, Ghidini M, Salati M, Mirchev MB, Valeri N, Hahne JC. MicroRNAs (miRNAs) and long non-coding RNAs (lncRNAs) as new tools for cancer therapy: First steps from bench to bedside. Targ Oncol. 2020;15:261–278. doi: 10.1007/s11523-020-00717-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Vicentini C, Galuppini F, Corbo V, Fassan M. Current role of non-coding RNAs in the clinical setting. Noncoding RNA Res. 2019;4:82–85. doi: 10.1016/j.ncrna.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Giudice V, Mensitieri F, Izzo V, Filippelli A, Selleri C. Aptamers and antisense oligonucleotides for diagnosis and treatment of hematological diseases. Int J Mol Sci. 2020;21:3252. doi: 10.3390/ijms21093252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Maruyama R, Yokota T. Knocking down long noncoding RNAs using antisense oligonucleotide gapmers. Methods Mol Biol. 2020;2176:49–56. doi: 10.1007/978-1-0716-0771-8_3. [DOI] [PubMed] [Google Scholar]
- 207.Bhan A, Hussain I, Ansari KI, Bobzean SAM, Perrotti LI, Mandal SS. Bisphenol-A and diethylstilbestrol exposure induces the expression of breast cancer associated long noncoding RNA HOTAIR in vitro and in vivo. J Steroid Biochem Mol Biol. 2014;141:160–170. doi: 10.1016/j.jsbmb.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Simonson B, Das S. MicroRNA therapeutics: The next magic bullet? Mini Rev Med Chem. 2015;15:467–474. doi: 10.2174/1389557515666150324123208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, Ren D, Ye X, Li C, Wang Y, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79. doi: 10.1186/s12943-018-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Banks IR, Zhang Y, Wiggins BE, Heck GR, Ivashuta S. RNA decoys: An emerging component of plant regulatory networks? Plant Signal Behav. 2012;7:1188–1193. doi: 10.4161/psb.21299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Ebert MS, Sharp PA. MicroRNA sponges: Progress and possibilities. RNA. 2010;16:2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Bernardo BC, Ooi JY, Lin RC, McMullen JR. miRNA therapeutics: A new class of drugs with potential therapeutic applications in the heart. Future Med Chem. 2015;7:1771–1792. doi: 10.4155/fmc.15.107. [DOI] [PubMed] [Google Scholar]
- 214.Yoo J, Hajjar R, Jeong D. Generation of efficient miRNA inhibitors using tough decoy constructs. In: Ishikawa K, editor. Cardiac Gene Therapy. Methods in Molecular Biology. Vol. 1521. Humana Press; New York, NY: 2017. pp. 41–53. [DOI] [PubMed] [Google Scholar]
- 215.Feng R, Patil S, Zhao X, Miao Z, Qian A. RNA therapeutics-research and clinical advancements. Front Mol Biosci. 2021;8:710738. doi: 10.3389/fmolb.2021.710738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Aimo A, Castiglione V, Rapezzi C, Franzini M, Panichella G, Vergaro G, Gillmore J, Fontana M, Passino C, Emdin M. RNA-targeting and gene editing therapies for transthyretin amyloidosis. Nat Rev Cardiol. 2022;19:655–667. doi: 10.1038/s41569-022-00683-z. [DOI] [PubMed] [Google Scholar]
- 217.Dever DP, Bak RO, Reinisch A, Camarena J, Washington G, Nicolas CE, Pavel-Dinu M, Saxena N, Wilkens AB, Mantri S, et al. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature. 2016;539:384–389. doi: 10.1038/nature20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby R, Olson EN. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Lee SWL, Paoletti C, Campisi M, Osaki T, Adriani G, Kamm RD, Mattu C, Chiono V. MicroRNA delivery through nanoparticles. J Control Release. 2019;313:80–95. doi: 10.1016/j.jconrel.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Kawakami S, Hashida M. Targeted delivery systems of small interfering RNA by systemic administration. Drug Metab Pharmacokinet. 2007;22:142–151. doi: 10.2133/dmpk.22.142. [DOI] [PubMed] [Google Scholar]
- 222.Soengas MS, Capodieci P, Polsky D, Mora J, Esteller M, Opitz-Araya X, McCombie R, Herman JG, Gerald WL, Lazebnik YA, et al. Inactivation of the apoptosis effector Apaf-1 in malignant melanoma. Nature. 2001;409:207–211. doi: 10.1038/35051606. [DOI] [PubMed] [Google Scholar]
- 223.Mann BS, Johnson JR, Cohen MH, Justice R, Pazdur R. FDA approval summary: Vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist. 2007;12:1247–1252. doi: 10.1634/theoncologist.12-10-1247. [DOI] [PubMed] [Google Scholar]
- 224.Grant C, Rahman F, Piekarz R, Peer C, Frye R, Robey RW, Gardner ER, Figg WD, Bates SE. Romidepsin: A new therapy for cutaneous T-cell lymphoma and a potential therapy for solid tumors. Expert Rev Anticancer Ther. 2010;10:997–1008. doi: 10.1586/era.10.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Moore D. Panobinostat (Farydak): A novel option for the treatment of relapsed or relapsed and refractory multiple myeloma. P T. 2016;41:296–300. [PMC free article] [PubMed] [Google Scholar]
- 226.Gilles ME, Slack FJ. Let-7 microRNA as a potential therapeutic target with implications for immunotherapy. Expert Opin Ther Targets. 2018;22:929–939. doi: 10.1080/14728222.2018.1535594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Segal M, Biscans A, Gilles ME, Anastasiadou E, De Luca R, Lim J, Khvorova A, Slack FJ. Hydrophobically modified let-7b miRNA enhances biodistribution to NSCLC and downregulates HMGA2 in vivo. Mol Ther Nucleic Acids. 2020;19:267–277. doi: 10.1016/j.omtn.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Wu L, Wang Q, Yao J, Jiang H, Xiao C, Wu F. MicroRNA let-7g and let-7i inhibit hepatoma cell growth concurrently via downregulation of the anti-apoptotic protein B-cell lymphoma-extra large. Oncol Lett. 2015;9:213–218. doi: 10.3892/ol.2014.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Pan X, Wang G, Wang B. MicroRNA-1182 and let-7a exert synergistic inhibition on invasion, migration and autophagy of cholangiocarcinoma cells through down-regulation of NUAK1. Cancer Cell Int. 2021;21:161. doi: 10.1186/s12935-021-01797-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Li M, Dou J, Pan M, Xu H, Xu Z. MicroRNA-7 agomir is a potential bioactive material for breast cancer therapy by inhibiting breast cancer stem cell tumorigenicity. Mater Express. 2021;11:824–831. doi: 10.1166/mex.2021.2001. [DOI] [Google Scholar]
- 231.Hong DS, Kang YK, Borad M, Sachdev J, Ejadi S, Lim HY, Brenner AJ, Park K, Lee JL, Kim TY, et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br J Cancer. 2020;122:1630–1637. doi: 10.1038/s41416-020-0802-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.van Zandwijk N, Pavlakis N, Kao SC, Linton A, Boyer MJ, Clarke S, Huynh Y, Chrzanowska A, Fulham MJ, Bailey DL, et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: A first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017;18:1386–1396. doi: 10.1016/S1470-2045(17)30621-6. [DOI] [PubMed] [Google Scholar]
- 233.Winata P, Williams M, McGowan E, Nassif N, van Zandwijk N, Reid G. The analysis of novel microRNA mimic sequences in cancer cells reveals lack of specificity in stem-loop RT-qPCR-based microRNA detection. BMC Res Notes. 2017;10:600. doi: 10.1186/s13104-017-2930-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Filippova EA, Fridman MV, Burdennyy AM, Loginov VI, Pronina IV, Lukina SS, Dmitriev AA, Braga EA. Long Noncoding RNA GAS5 in breast cancer: Epigenetic mechanisms and biological functions. Int J Mol Sci. 2021;22:6810. doi: 10.3390/ijms22136810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Williams GT, Pickard MR. Long non-coding RNAs: New opportunities and old challenges in cancer therapy. Transl Cancer Res. 2016;5((Suppl 3)):S564–S565. doi: 10.21037/tcr.2016.09.04. [DOI] [Google Scholar]
- 236.Liu W, Zhan J, Zhong R, Li R, Sheng X, Xu M, Lu Z, Zhang S. Upregulation of long noncoding RNA_GAS5 suppresses cell proliferation and metastasis in laryngeal cancer via regulating PI3K/AKT/mTOR signaling pathway. Technol Cancer Res Treat. 2021;20:1533033821990074. doi: 10.1177/1533033821990074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Smaldone MC, Davies BJ. BC-819, a plasmid comprising the H19 gene regulatory sequences and diphtheria toxin A, for the potential targeted therapy of cancers. Curr Opin Mol Ther. 2010;12:607–616. [PubMed] [Google Scholar]
- 238.Zhao X, Reebye V, Hitchen P, Fan J, Jiang H, Sætrom P, Rossi J, Habib NA, Huang KW. Mechanisms involved in the activation of C/EBPα by small activating RNA in hepatocellular carcinoma. Oncogene. 2019;38:3446–3457. doi: 10.1038/s41388-018-0665-6. [DOI] [PubMed] [Google Scholar]
- 239.Reebye V, Sætrom P, Mintz PJ, Huang KW, Swiderski P, Peng L, Liu C, Liu X, Lindkaer-Jensen S, Zacharoulis D, et al. Novel RNA oligonucleotide improves liver function and inhibits liver carcinogenesis in vivo. Hepatology. 2014;59:216–227. doi: 10.1002/hep.26669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Seto AG, Beatty X, Lynch JM, Hermreck M, Tetzlaff M, Duvic M, Jackson AL. Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. Br J Haematol. 2018;183:428–444. doi: 10.1111/bjh.15547. [DOI] [PubMed] [Google Scholar]
- 241.Ijaz M, Wang F, Shahbaz M, Jiang W, Fathy AH, Nesa EU. The role of Grb2 in cancer and peptides as Grb2 antagonists. Protein Pept Lett. 2018;24:1084–1095. doi: 10.2174/0929866525666171123213148. [DOI] [PubMed] [Google Scholar]
- 242.Ohanian M, Tari Ashizawa A, Garcia-Manero G, Pemmaraju N, Kadia T, Jabbour E, Ravandi F, Borthakur G, Andreeff M, Konopleva M, et al. Liposomal Grb2 antisense oligodeoxynucleotide (BP1001) in patients with refractory or relapsed haematological malignancies: A single-centre, open-label, dose-escalation, phase 1/1b trial. Lancet Haematol. 2018;5:e136–e146. doi: 10.1016/S2352-3026(18)30021-8. [DOI] [PubMed] [Google Scholar]
- 243.Golan T, Khvalevsky EZ, Hubert A, Gabai RM, Hen N, Segal A, Domb A, Harari G, David EB, Raskin S, et al. RNAi therapy targeting KRAS in combination with chemotherapy for locally advanced pancreatic cancer patients. Oncotarget. 2015;6:24560–24570. doi: 10.18632/oncotarget.4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Timar J, Kashofer K. Molecular epidemiology and diagnostics of KRAS mutations in human cancer. Cancer Metastasis Rev. 2020;39:1029–1038. doi: 10.1007/s10555-020-09915-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Zhang P, Liu X, Abegg D, Tanaka T, Tong Y, Benhamou RI, Baisden J, Crynen G, Meyer SM, Cameron MD, et al. Repro-gramming of protein-targeted small-molecule medicines to RNA by ribonuclease recruitment. J Am Chem Soc. 2021;143:13044–13055. doi: 10.1021/jacs.1c02248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Shortridge MD, Chaubey B, Zhang HJ, Pavelitz T, Olsen GL, Calin GA, Varani G. Drug-like small molecules that inhibit expression of the oncogenic microRNA-21. bioRxiv. doi: 10.1101/2022.04.30.490150. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Donlic A, Morgan BS, Xu JL, Liu A, Roble C, Jr, Hargrove AE. Discovery of small molecule ligands for MALAT1 by tuning an RNA-binding scaffold. Angew Chem Int Ed Engl. 2018;57:13242–13247. doi: 10.1002/anie.201808823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Goyal B, Yadav SRM, Awasthee N, Gupta S, Kunnumakkara AB, Gupta SC. Diagnostic, prognostic, and therapeutic significance of long non-coding RNA MALAT1 in cancer. Biochim Biophys Acta Rev Cancer. 2021;1875:188502. doi: 10.1016/j.bbcan.2021.188502. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.