Summary
To understand the fine differential elements that can lead to or prevent acute respiratory distress syndrome (ARDS) in COVID-19 patients, it is crucial to investigate the immune response architecture. We herein dissected the multiple layers of B cell responses by flow cytometry and Ig repertoire analysis from acute phase to recovery. Flow cytometry with FlowSOM analysis showed major changes associated with COVID-19 inflammation such as an increase of double-negative B-cells and ongoing plasma cell differentiation. This paralleled COVID-19-driven expansion of two disconnected B-cell repertoires. Demultiplexing successive DNA and RNA Ig repertoire patterns characterized an early expansion of IgG1 clonotypes with atypically long and uncharged CDR3, the abundance of this inflammatory repertoire being correlated with ARDS and likely pejorative. A superimposed convergent response included convergent anti-SARS-CoV-2 clonotypes. It featured progressively increasing somatic hypermutation together with normal-length or short CDR3 and it persisted until a quiescent memory B-cell stage after recovery.
Subject areas: Immunology, Respiratory medicine
Graphical abstract

Highlights
-
•
Severe COVID-19 expands double-negative B cells, plasma cells, and B10 cells
-
•
Parallel DNA and RNA repertoires dissociate high Ig expression from cell expansion
-
•
COVID-19 memory repertoire reveals short CDR-H3 and progressive affinity maturation
-
•
Early increase of IgG1 clonotypes with long uncharged CDR3 correlates with ARDS
Immunology; Respiratory medicine
Introduction
The SARS-Coronavirus-2 (SARS-CoV-2) virus has currently affected about 680 million people worldwide, eventually requiring admission to Intensive Care Unit (ICU) and killing about 6.9 million patients (Johns Hopkins Coronavirus Resource Center: https://coronavirus.jhu.edu/map.html). From the beginning, clinical presentation of COVID-19 has been highly variable, from asymptomatic or mild to severe, with acute respiratory distress syndrome (ARDS) as the major and eventually fatal complication,1,2 requiring prolonged mechanical ventilation and yielding complications hereby.3 Immune responses diversely modulate COVID-19 onset and evolution. Notably, vaccination has massively demonstrated that protective specific immunity can prevent COVID-19 or ensure its benign evolution with faster clearance of the virus and reduced need for hospitalization, even for vaccinees facing the delta or omicron variants in situations where cellular immunity persists but specific antibodies are less efficient.4,5,6,7 Rather than protective immunity, responses to SARS-CoV-2 infection can also yield uncontrolled inflammation and then participate to ARDS development, as notably seen in the elderly or in patients with type I interferon defects.8,9 The fine balance between these two sides of immune responses is thus of major importance for the evolution and prognosis of the disease. Notably, ICU COVID-19 patients rapidly develop anti-SARS-CoV-2 adaptive immune responses which, although reaching much higher levels than in mild patients, fail to protect them.10,11 Accordingly, patients at this stage of strong viral burden and inflammation become unresponsive to passive immunotherapy with specific mAbs or convalescent plasma.12,13 Immune dysfunction is clearly multifactorial in ICU patients, with atypical myeloid cell expansion and increased levels of pro-inflammatory cytokines correlating with T cell lymphocytopenia and a broad T cell activation including Th1, Th2, Th17, and MAIT cells.14,15,16,17,18,19 Cellular anomalies are also present on the B-cell side, and differ between patients with mild COVID-19 and ICU patients, the latter showing an increase of blood plasma cells and of double-negative (DN) [IgDneg, CD27neg] B-cells suggesting extra-follicular B-cell activation.10,11 Single-cell analysis of SARS-CoV2-responding B cells has also shown that they include both cells induced into early extra-follicular activation and germinal center B-cells later differentiating into memory B cells.20 Many aspects of the B-cell responses, however, remain to be understood, notably regarding the specific role of a given B-cell sub-population or a given component of the Ig repertoire for either protection, pathologic inflammation or even immunosuppression, in conditions where severe COVID-19 triggers biased immune activation pathways. Notably for COVID-19 patients entering the ICU with a highly variable prognosis, it is unknown whether B-cell parameters correlate with evolution in the ICU and notably with the development of ARDS needing mechanical ventilation, with all complications hereby.
To fill these gaps, we undertook a high-resolution profiling of blood B-cells by superimposing Ig repertoires explored both at the DNA level (evaluating cell abundance with regard to clonotypic features) and the mRNA level (then exploring the relative expression of Ig clonotypes in both membrane-type and secretedtype IgH transcripts). Although plasma cells highly transcribing IgH genes would have an increased weight in global RNA-based repertoires, the split analysis of BCR transcripts and secreted-type transcripts for each Ig class more specifically appreciates the repertoires from B-lymphocytes and plasma cells, respectively. In parallel to repertoire analyses, we also finely analyzed B-cell sub-populations by flow cytometry.
Finally, and by contrast to previous studies which compared ICU patients with patients affected by mild COVID-19, our study aimed at evaluating among ICU patients, two cohorts either responsive to oxygen therapy or affected with ARDS. We then looked for correlation between the molecular determinants of the B-cell response and the evolution of the disease among those critically ill patients.
Results
Characteristics of COVID-19+ individuals
We recruited 35 patients with severe COVID-19, 22 (62%) with ARDS, according to the consensus definition.21 Blood samples were analyzed by flow cytometry (30 patients), DNA repertoire (28 patients) and RNA repertoire (19 patients). The median ages in the ARDSpos and ARDSneg COVID-19+ groups, 64 and 57 years old, respectively, did not significantly differ (Student’s t test, p = 0.14). Comparison of clinical features, age-and sex-ratio ARDSpos/neg COVID-19+ and control groups is presented in Table S1. ICU samples were drawn at D0-3 (D0) and D7-10 (D7) after admission and at M4 in recovered patients. PBMCs isolated from whole blood using Ficoll were cryopreserved. Information about gender, age, Body Mass Index (BMI), outcome (alive or dead) and preexisting chronic disease was collected. Patients’ characteristics are reported in Table S2.
Immunophenotypic B-cell differential in COVID19 patients with and without ARDS
B-cell subsets were analyzed by flow cytometry for 53 samples from 30 severe COVID19 patients (19 with and 11 without ARDS) and 9 healthy controls. CD45 and CD19 were used as gating markers for B-cells. Phenotypic markers included IgD, CD10, CD24, CD27, CD38 to follow most B-cell subsets as reported by Sanz et al.11 Moreover, we added the PD-L1 (CD274) marker to identify B-cells with putative immune regulatory activities.22 All samples were globally analyzed for these markers using the FlowSOM R package.23 Based on these analyses, 81 phenotypic clusters were generated used a 9 × 9 grid (Figures 1A, 1B andS1). Examination of clusters found them associated into 8 metaclusters, the phenotypic analysis of which by biparametric histograms confirmed correspondence with the expected phenotype of 8 major B-cell subsets (Figure S1). Further dissociation of metaclusters by FlowSOM only discerned small clusters at the periphery of the minimum spanning tree (MST) (not shown), lacking functional relevance. We thus rather defined the regulatory B-cell subsets of interest in a supervised manner, adding 4 regulatory metaclusters close to the center of the MST: metacluster 9 corresponds the so-called “double-negative” (DN) IgDneg/CD27neg B-cells, whereas metaclusters 10, 11 and 12 contains CD24high/CD27+ B-cells (so-called B10 regulatory B-cells24). These subsets have indeed been implicated in immune regulatory functions.11,25 DN B-cells included clusters 45 and 36 (Figures 1 and S1). CD24high/CD27+ B-cells were found in three small branches of the MST corresponding to clusters 8 and 18, 9 and 17 as well as 35 and 27, and differences between these 3 metaclusters were mostly related to IgD and CD38 expression (Figures 1A and S1).
Figure 1.
Flowsom analysis of the different B cell subsets
(A) Minimum spanning tree of the 81 clusters and 12 metaclusters identified after FlowSOM analysis.
(B) Color codes and correspondences of the 12 metaclusters with their B-cell subset counterpart (cardinal phenotypic features are indicated).
(C) tSNE analysis of the 12 FlowSOM metaclusters. The color code is the same than in 1B. Each B-cell subset is pointed by an arrow.
(D) Minimum spanning tree of the 81 clusters and the 12 metaclusters according to the COVID-19 course. Diameter of each cluster ball is proportional to the relative abundance of its cell content.
(E) tSNE overlay of the 12 flow metaclusters for B-cells from controls versus patients at D7 and M4.
Altogether, this flow cytometry mapping captures all expected functional B-cells subsets, and includes not only the main circulating subsets such as naive or memory B-cells, but also rarer compartments such as transitional B-cells, marginal zone (MZ) B-cells as well as CD24neg/CD38high plasmablasts (Figures 1A and S1). PD-L1 expression by plasmablasts and MZ B-cells additionally suggests that these two subsets could exert some immunoregulatory functions. tSNE (t-distributed stochastic neighbor embedding) analysis consolidated the validity of all above-described metaclusters, by showing that each metacluster could be ascribed to a specific tSNE territory (Figure 1C and S2). Although distant on the MST, tSNE territories of cluster 10, 11 and 12 were logically close together, because of high CD24 expression (Figure 1C and S2).
B-cell subsets from healthy controls were dominated by the IgD+/CD27- B-cell compartment, with less abundant IgD-/CD27+ memory and hardly detectable transitional, DN B-cells, B10 cells or plasmablasts (Figures 1D and S3 and S4). In contrast, severe SARS-CoV-2 infection was associated with an initial increase of transitional B-cells mainly in ARDSneg patients (see Metacluster 1, Figure S4), indicating increased production or circulation of recently differentiated B-cells. COVID19 was also characterized by a shift from naive to memory B-cells as early as D0 for ARDSpos patients, while delayed at D7 for ARDSneg patients, with maximum increase of the memory B-cell compartment at M4 (Figure 1D and see Metaclusters 2 and 4, Figure S4). These phenotypic changes were confirmed by tSNE analysis (Figure 1E).
We observed in the acute phase of severe COVID-19, major alterations of B-cell sub-populations typical of the extra-follicular response reported by Woodruff et al., noticeably involving the population of DN cells (Metacluster 9, Figure S4), previously associated with auto-immune conditions and inflammation.11,24 In addition to a memory B-cell shift, we noticed an expansion of CD27+/IgD-/CD24high B cells (metacluster 10 on Figure S4, carrying markers of B10 Breg compartment) whereas levels of IgD+/CD24high B-cells remained stable (Metaclusters 11 and 12, Figure S4). An expansion of CD27+/IgD+/PDL1+ B-cells (Metacluster 3, Figure S4), which reached significance at M4. These cells, that evoke marginal zone Bregs, were those with the highest PD-L1 expression (Figure S1). Some plasmablasts expansion (as Metaclusters 5, 6 and 8, Figure S4) was also visible and associated with PDL1 expression, again evoking regulatory cells. Alterations of B-cell compartments partly persisted after recovery and patients at M4 showed a persistent increase of transitional, DN, and regulatory B-cells, mainly CD24high/CD27+ B-cells but also PD-L1 expressing marginal zone B-cells and plasmablasts.
Analysis of circulating B-cells thus shows that, beyond the inflammatory phase and the memory B-cell expansion, several cell compartments were durably impacted after COVID-19 recovery, including for an increased ongoing plasma cell differentiation (even if heterogeneous), and for cells known with regulatory functions and/or polyreactivity. This suggests the prolonged superimposition of a classical memory B-cell response with a parallel expansion of B-cells with extra-follicular features, the expansion of which was previously noticed in severe compared to mild COVID-19 patients.11,26,27 We also note that at M4, PD-L1+ B-cells were less abundant in ARDSpos than in ARDSneg patients, suggesting their clinically relevant participation to a better control of inflammation in the latter.
Oligoclonal humoral response and expansion of circulating IgG1+ B-cells during the acute phase of SARS-CoV-2 infection
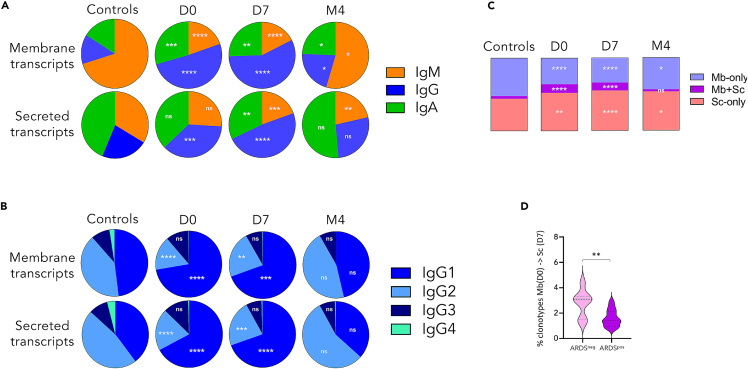
To in-depth analyze the anti-SARS-CoV-2 global response, circulating B-cell repertoires were characterized in a cohort of 35 patients hospitalized in ICU for severe COVID-19 disease (22 with and 13 without ARDS). As stated above, D0, D7 and M4 blood samples were collected. Overall, both on RNA and DNA templates, a significant decrease in the diversity of circulating Ig repertoires was observed during the acute phase (D0/D7) (Figure 2). Following Ig diversity by analyzing the frequency distribution curve of the CDR3 length, readily showed oligoclonal expansions in the acute phase repertoires: over the polyclonal background, small peaks corresponding to transient clonotypic expansions were detected in most ICU patients whereas controls showed smooth distribution curves of highly diversified clonotypes (Figure 2A). Accordingly, the elevated Gini diversity index and relative abundance of the most abundant (“top 100”) Ig clonotypes on DNA and RNA templates scored a strongly decreased diversity during the acute phase (Figures 2B–2E). Noticeably the long-term kinetics of this alteration differed in RNA- or DNA-based repertoires: Although the RNA-repertoire diversity was back to normal after recovery at M4 patients, the DNA-based analysis eventually showed a persistent oligoclonal pattern (Figures 2B and 2C). Such uncoupled aspects suggest that at M4, amplified clonotypes were more represented among circulating memory B-cells, than within those cells the most actively transcribing Ig genes, i.e. circulating plasma cells. The transient loss of diversity manifested in all of our severe COVID-19 patients, without any significant difference related to the ARDS status (Figures 2 and S5).
Figure 2.
Oligoclonal response during SarsCov2 infection
Total size distribution frequency of the Ig repertoire sequenced reads of one healthy volunteer and 3 representatives severe COVID-19 patients on DNA template (A). Gini diversity index of circulating Ig repertoires on DNA (B), membrane and secreted Ig transcripts on RNA templates (C). Relative abundance of the top 100 circulating clonotypes on DNA (D), membrane and secreted Ig transcripts on RNA templates (E).
To analyze the predominantly selected IGHV-JH gene rearrangements during COVID-19, we quantified and clustered the differentially expressed IGHV-JH pairs (VJ pairs) at both DNA and RNA levels in COVID-19 patients when compared to controls, with a p value of 0.05 and a false discovery rate (FDR) of 0.1 after the Benjamini-Hochberg correction using the EdgeR package. Twenty-one VH-JH pairs were increased for DNA and RNA but with an overlap of 3 VJ pairs, IGHV2-5_IGHJ4, IGHV2-70_IGHJ4, IGHV1-24_IGHJ5 (Table S3). Although associated with various JH segments, several VH genes also appeared as overexpressed at both the DNA and RNA level, such as IGHV1-24, IGHV2-4, IGHV3-9, IGHV3-11, IGHV3-23, IGHV3-30 and IGHV4-34. By contrast seventeen and four VH-JH pairs were down regulated for DNA and RNA, respectively (Table S3). Heatmaps of differentially expressed VH-JH pairs reveal that COVID-19 patients had an early and homogeneous bias of Ig repertoires at both DNA and RNA levels by comparison to controls (Figure 3), with no significant difference between ARDSpos and ARDSneg patients in this approach. At M4, repertoire biases clearly persisted at the DNA level for about 2/3 of the patients.
Figure 3.
Immunoglobulin repertoire clusters by heavy chain gene segment usage
Heatmaps of variable heavy (IGHV) and joining (IGHJ) gene usage on DNA (A) and RNA templates (B) differentially expressed between samples collected at D0 and D7 from patients with or without ARDS versus healthy subjects. Each heatmap displays normalized IGHV-IGHJ gene expression across the groups. Groups (upper line) have the following color code: healthy subjects (red), patients without ARDS (yellow), patients with ARDS (red). Time points (second line) indicate sampling day for healthy subjects (red) and for patients, Day 0 (D0, green), Day 7 (D7, dark green), Day 14 (D14, light blue), Month 4 (M4, dark blue).
Additional major changes of Ig expression at the RNA level were found during the acute phase (D0/D7) and related to class/subclass usage in both ARDSpos and ARDSneg patients (Figures 4 and S6A). This included a strong and early (starting from D0) increase of the [class-switched-Ig/IgM] ratio (Figure 4A), mainly seen for membrane transcripts and partially normalized in recovered patients. The same loss of IgM predominance affected secreted transcripts in a delayed manner, only significant at D7. Both IgG and IgA expression were increased but IgG predominated among switched transcripts, mostly with a strong increase of IgG1 and a relative decrease of IgG2/IgG3 (Figure 4B). This transient increase was equivalently observed in patients with or without ARDS (Figure S6A). In addition, comparing the membrane and the secreted repertoires from the same patients showed their increased overlap during the acute period (D0 and D7), i.e., with more abundant shared clonotypes, as well as an increased proportion of secreted transcripts at the expense of membrane transcripts (Figure 4C). These elements indicate ongoing plasma cell differentiation at this stage. When analyzing sequences from successive time points in the same patient, an overlap also appeared between those clonotypes expressed at D0 in the membrane form and at Day7 in the secreted form, although less abundantly in ARDSpos patients who might develop less efficient humoral responses (Figure 4D).
Figure 4.
Changes of class/subclass usage, according to the membrane or secreted nature of Ig transcripts, during the SARS-CoV-2 infection
(A and B) Ig class (A) and IgG subclass (B) distribution.
(C) Average distribution of circulating clonotypes between membrane and secretory repertoires within each repertoire.
(D) Frequency of transcripts from the D0 membrane repertoire have shifted to the secreted repertoire at D7 in the same patient, according to ARDS status.
COVID-19 repertoire CDR3 features in ICU patients
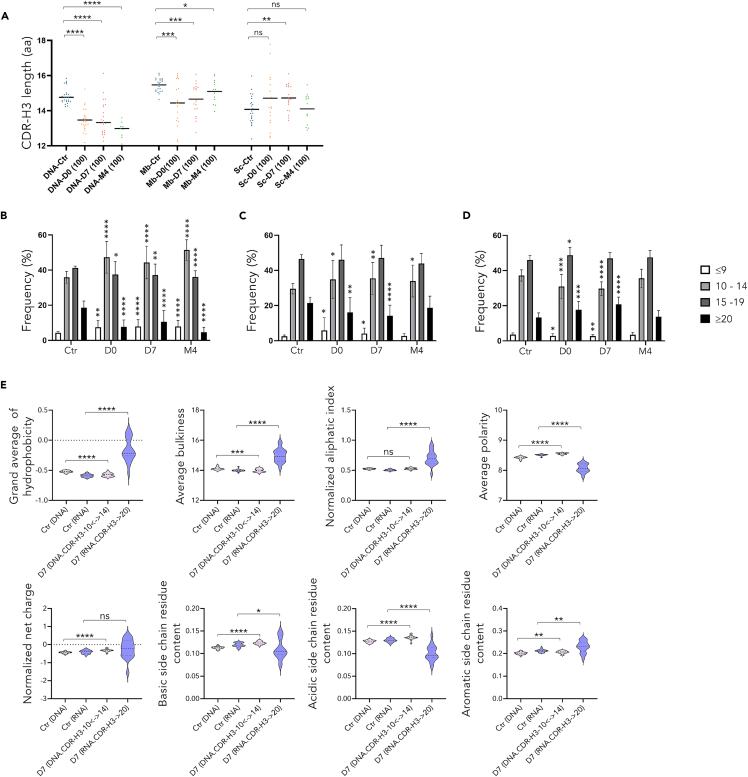
DNA and RNA repertoires showed differential evolution (Figures 5A–5F and S7). In contrast with the repertoires arising from DNA templates (Figure 5A), examination of CDR-H3 from RNA repertoires showed that mean CDR-H3 length was significantly increased in the Ig repertoires of COVID-19 patients during the acute phase, compared with controls, but this did not persist at M4 (Figure 5B). Analyzing with more details this transient increase, it in fact appeared the most significant for IgG (Figure 5E), with most notably an amplification of IgG1 transcripts with long CDR-H3 (Figure 5F).
Figure 5.
IgG1 expansion associated with few mutations but significantly increased CDR3 length in the Ig repertoire
Average length of CDR3 for each group from DNA repertoire (A) membrane and secreted transcripts (B) and according to class (E) or IgG subclass distribution (F) from RNA template. V region somatic hypermutation frequency for DNA repertoire (C) and according to membrane or secreted-type transcripts from each group (D) Ig class (G) or IgG subclass (H).
Both cohorts of ICU patients in fact slightly differed in the kinetics of this expansion. Although not statistically significant, mean CDR3-H3 length of Ig transcripts appeared temporarily higher in ARDSpos than ARDSneg patients at Day 0. Relative expansion of this long CDR3 subset in fact occurred later and persisted at M4 in ARDSneg patients (Figure S7A). Analysis focused on specific Ig classes and subclasses showed for each switched class of transcripts the same trend for longer CDR3-H length at Day0 contrasting with shorter length at distance from the acute period, reaching significant difference for IgG and IgA switched transcripts at M4 in ARDSpos patients (Figures S7B and S7C).
COVID-19 repertoire SHM features
Assessing somatic hypermutation (SHM) on the total repertoire only showed a minor global increase during the acute phase, before reaching a significantly higher SHM rate at M4 (Figures 5C, 5F–5H). Under more precise scrutiny of the various Ig classes, SHM in fact showed a differential impact in ICU patients compared to controls, initially increasing for IgM while apparently decreasing for IgG1 (Figures 5E and 5F). This acute phase kinetics likely corresponds to the transient invasion of the switched B-cell compartments by recent migrants, newly switched to IgG (particularly IgG1 and IgG3). For both γ1 membrane and secreted transcripts, such recently switched cells first exhibited a lower SHM rate than controls (in whom γ1 transcript by contrast likely originated from memory cells). In all cases, SHM rate in γ1 transcript however proved higher than for μ transcripts. In recovered patients at M4, all classes of transcripts, either switched or unswitched, had reached a higher rate of SHM than measured in naive controls, especially for the γ1 and γ3 membrane transcripts and for both membrane and secreted α transcripts.
ARDSpos patients also ended at M4 with the highest SHM load on γ1 secreted transcripts, higher than in ARDSneg patients. By contrast, ARDSneg patients showed a trend toward a higher rate of SHM on γ3 secreted transcripts than ARDSpos patients (Figure S7E).
Convergent anti-SARS-CoV-2 clonotypic response in severe COVID-19 patients
To characterize the COVID-19 response signature in the circulating B-cell repertoires of our patients, we first searched for the presence of specific anti-SARS-CoV-2 clonotypes (SC for specific clonotype) listed in the CovAbDab database (The Oxford Protein Informatics Group public database: http://opig.stats.ox.ac.uk/webapps/covabdab/) which now documents thousands of published or patented human SARS-CoV-2-specific binding antibodies and nanobodies.28 As reported in the literature,29,30 we also searched and found, convergent clonotypes (CCs) shared by different individuals of our cohorts in response to COVID-19.
Although low, the relative representation of such “public” COVID-19 clonotypes in DNA and RNA repertoires from blood cells was significantly higher in severe COVID-19 patients than in controls, as a footprint of the public human antiSARS-CoV-2 humoral response on individual repertoires (Figure 6A). As expected, these public clonotypes were often associated with the IgM class at D0 but underwent a class switch at D7, largely toward IgG1 and IgA (Figure 6B), whereas their low SHM at D7 suggested that they were carried by naive cells (Figure 6C). With regard to the clinical status, a noticeable difference appeared between ARDSneg patients in whom public clonotypes were earlier associated with switched isotypes at D0 and ARDSpos patients, in whom they first mostly associated with IgM, possibly reflecting a delay in class switching of the most specific clonotypes in those patients developing ARDS (Figure S5B). Of interest, the CDR-H3 length of the SC clonotypes expressed in our ICU patients and homologous to specific anti-SARS-CoV-2 CoV-AbDab clonotypes, which intrinsically relate to the Ag-specific response, was significantly shorter than the mean length of human control repertoires (Figure 6D). CCs identified among patients from our study also showed no increase of CDR-H3 length by comparison to controls.
Figure 6.
Characteristic of CoV-AbDab and convergent SARS-CoV-2 clonotypes, present in the circulating Ig repertoires of severe COVID-19 patients
Relative abundance of CoV-AbDab specific SARS-CoV-2 clonotypes (SC) and convergent clonotypes (CC, shared by several patients of this study) in circulating Ig repertoires (A). Class and IgG subclass distribution (B) V region somatic hypermutation frequency (C) and average length of CDR3 (D) of SC and CC clonotypes, on RNA template.
IgH repertoires characterized by CDR-H3 and SHM features, thus appear to include two superimposed repertoires: one with short mean CDR3 length including public clonotypes representative of the SARS-CoV-2-specific response, shared between COVID-19 patients from this study or from previous studies, and the other, preponderant in the expressed RNA analysis, characterized by enlarged hydrophobic CDR-H3 but for which the precise CDR3 sequence remains private to each patient.
Layering of 2 clonally distinct responses identified at the DNA and RNA levels
RNA repertoires evaluate relative expression of each clonotype and show transient increase of Ig sequences with long CDR3 in COVID-19 patients. By contrast, DNA repertoires yielding a single functional VDJ copy for each cell and thus evaluating relative cell numbers, failed to show any change in CDR3 length. Analysis to the 100 most amplified clonotypes at the time of infection within the DNA repertoire even showed a significant decrease in mean CDR-H3 length (Figure 7A), contrasting to the increased mean CDR-H3 length characterizing the 100 most highly expressed clonotypes within RNA secretory repertoires (Figure 7A). Of interest, considering these dominant repertoires, the membrane Ig transcripts showed a similar profile as the DNA repertoire with clonotypes carrying significantly shorter CDR-H3. Thus, the distinct CDR-H3 length distribution of the most abundant cells (dominant DNA clonotypes) on one side, and the most expressed Ig (dominant RNA clonotypes, originating from activated or differentiated cells) on the other side, suggests that increased numbers of cells with short CDR3 (notably in the 10–14 aa range) proliferate and durably persist at 4 months, while in parallel, cells with long CDR3 (above 20 codons), get actively but transiently differentiated into plasma cells (in the D0-D7 period) (Figures 7B–7D).
Figure 7.
Most expressed clonotypes of the DNA
(A and B) and RNA (A, C, D) repertoires capture 2 aspects of humoral responses with distinct CDR-H3 features. Average CDR-H3length for the 100 most represented clonotypes (100) at D0, D7 and M4, on DNA and RNA repertoires according to membrane or secreted transcripts (A). Average length distribution of CDR-H3 (with ≤9, 10 to 14, 15 to 19, and ≥20 amino acids) of the D7 and M4-100 main clonotypes in DNA repertoires (B) membrane (C) and secreted transcripts (D). Physico-chemical properties of the D7-100 most represented DNA 10 to 14 aa-CDR-H3 and RNA ≥20 aa CDR-H3 clonotypes compared with those of the control DNA and RNA repertoires, respectively (E).
Additional and noticeable CDR-H3 physicochemical properties differentially characterize these clonotypes. Predominant DNA clonotypes (with encoded CDR-H3 in the 10 to 14aa range) showed, on average, decreased hydrophobicity and increased polarity, with normal charge and basic/acidic side chain residue content. By contrast, the specifically abundant RNA clonotypes ([long-CDR-H3] in the >20 aa range) showed increased hydrophobicity and aliphatic index, together with decreased polarity and decreased basic/acidic side chain residue content (Figure 7E). Noticeably, the latter features did not appear to be specific to SARS-CoV-2 infection but intrinsically linked to a subset of [long-CDR-H3] clonotypes which, although less abundantly expressed, are already detectable in control naive repertoires (Figure S8).
Focusing on the top 100 clonotypes, these data demonstrate the superimposition among PBMC of two different humoral responses, reflecting the amplification of two different sets of clonotypes. The [long-CDR-H3] set represents increased expression (likely impacted by plasmablast and plasma cell expansion which biases the RNA repertoire), and probably witnesses an early first line of defense which rapidly generates circulating Ig at the acute phase. The second set associates with the amplification of B cell clones with a short CDR3, as captured by DNA repertoire changes persisting over time, and likely represents the constitution of the memory B-cell pool.
Discussion
Evolution of severe COVID-19 is an exemplary demonstration that protective immunity is not just a matter of quantity of immune cells and mediators, but rather a matter of kinetics, with on-time and rapidly coordinated innate and adaptive T and B-cell responses eventually able to control the viral burden and then limit pathogenic dysregulated inflammation.31,32 Interpreting immune responses in severe ICU patients is not unequivocal, because not only hyperinflammation, but also hypoinflammation correlate with the occurrence of ARDS, the most severe and potentially lethal complication of COVID-19.33 Beside inflammation, various alterations of circulating immune cells were indeed found associated with a poor prognosis and the eventual occurrence of ARDS. This included the early observation of lymphopenia, the expansion of various myeloid suppressive cells, the increased circulation of TREG cells with specific phenotypes, together with metabolic anomalies responsible for lymphoid dysfunction.17,31,34,35,36,37 Although B lymphocytes are major contributors to protective adaptive immune responses, they also participate to early innate responses and carry immunoregulatory functions, but their role in the early evolution of severe COVID-19 remains unclear. In this study of ICU COVID-19patients evolving or not toward ARDS, we tried to obtain high resolution snapshots of the B cell landscape combining flow cytometry and molecular analysis of Ig repertoires, to differentially analyze the architecture of the B-cell response and to search correlations with early clinical evolution.
Increase in DN B-cells and plasma cells has been previously reported in COVID-19 patients.11,27,38 Postmortem studies of lymph nodes and spleens from patients who died from severe COVID-19, noticed the local abundance of extra-follicular DN B-cells, similar to those found in peripheral blood and including virus-specific B-cells, in situations where the deregulated immune response featured a defect of germinal center B cells and T follicular helper cells.38 Here, flow cytometry analysis of blood lymphocytes identified 12 B-cell metaclusters, not only including DN B-cells and CD24neg/CD38high plasmablasts, but also with the phenotype of naive, memory, transitional as well as MZ B-cells. Some of these subsets have been suggested to exert immunoregulatory functions, as indicated by PD-L1 expression on some plasmablasts and MZ B-cells, whereas tSNE analysis additionally identifies DN B-cells and cells with the classical BREG CD24high/CD27+ phenotype. Acute phase ICU COVID-19 patients, but also recovered patients, showed increased recently differentiated transitional B-cells, memory B-cells, DN cells (IgDneg/CD27neg), plasmablasts, and CD24high/CD27+ BREG cells, expansion of such populations appearing earlier in ARDSpos patients than in ARDSneg patients. By contrast, PD-L1 expression was higher in ARDSneg patients and might participate to a better control of inflammation.
Regarding repertoire changes, oligoclonal expansion with clonotypes enriched both in cell number and expression level was a prominent anomaly in all ICU patients and durably impacted the memory cell compartment, with decreased diversity of the DNA repertoire at M4. The minor age differences between COVID-19 patients (ARDSneg: mean 57-year-old and ARDSpos mean 64-year-old) and healthy controls (mean 45-year-old) in our study makes it legitimate to assume that the M4 loss of diversity is a signature of the recent disease. An age bias indeed appears unlikely in such groups of adults, because in prior scientific literature, variations of diversity attributed to immune senescence were only reported in very old adults compared to young individuals (mean 82.7 versus 23.8 years old, respectively).39
Broad B-cell activation during the acute phase also translated into increased SHM of IgM-associated clonotypes and a major shift from IgM to class-switched Ig expression involving both IgA and IgG but the strongest for IgG1 (sparing IgG2 and IgG3) and for secreted-type transcripts, in agreement with plasmacytosis. ARDSpos patients then showed a delay in the class switching of convergent clonotypes by comparison to ARDSneg patients. In the acute phase, ARDSneg patients also showed a more efficient shift of some clonotypes from an early expression as membrane-type transcripts to a late expression as secreted Ig, suggesting earlier secretion of the corresponding antibodies.
In addition to the ARDS-associated delay in CSR of SARS-CoV2 clones, SHM of IgG3 secreted transcripts was lower in ARDSpos than in ARDSneg patients, suggesting less efficient maturation of the Ag-specific response. By reference to the altered germinal centers documented postmortem in COVID patients with fatal evolution,38,40 it is also possible that alterations of lymphoid structures differentially contribute to evolution of ARDSneg and ARDSpos patients, with increased extra-follicular commitment eventually resulting in decreased GC entry. In agreement with other studies,29,30,38 IgH transcripts from the IgG class (most notably γ1 and γ3) globally carried a lower SHM load in our patients at D7 than in controls. Although this low SHM rate of IgG1 was previously interpreted as indicating that COVID-19 does not initiate robust SHM, analysis of successive timepoints however suggests that this could simply mark the transient expansion and circulation of newly switched cells. SHM indeed clearly occurs for μ transcripts in these patients, and the mean SHM level in D7 γ transcripts, although lower than in control memory cells, is however already higher than for D7 μm transcripts, before finally reaching at M4 the same level as for γ transcripts from control memory cells. These data also fit with single cell analysis which previously documented near germline SARS-CoV-2-specific Ig sequences in the initial stage of the disease whereas they were lately found modified by SHM in memory cells several months after recovery.20 That SHM occurs after CSR does not stand as a COVID-related specific feature but perfectly fits with the now acknowledged order of events, with CSR demonstrated to mostly occur before the entry into germinal centers and to SHM during normal immune responses.41
Regarding VDJ associations and CDR3 assembly, the immune response of patients is marked by a COVID-19 footprint, with increased occurrence of specific VH-JH pairs and of clonotypes abundantly represented within the CovAbDab database of known anti-spike Abs. Such COVID-related clonotypes first associate with IgM expressed at D0, but later associate with class-switched Ig, the latter association being significantly higher in ARDSneg patients. Another striking aspect of the early class-switched response is the specific expansion of a [long CDR-H3] subset of abundantly transcribed clonotypes, which was higher in ARDSpos patients. CDR3 features in fact highlighted two parallel variations of the Ig response in patients. A standard Ag-specific Ig response including SARS-CoV-2- specific and convergent clonotypes with normal CDR3 length develops and is the most durable, persisting after recovery. Its long-term impact on the DNA repertoire rather than on the RNA repertoire indicates that it is supported by memory B cells rather than plasma cells. In parallel but more transiently, occurs an early increase of expressed Ig with long, hydrophobic and aliphatic CDR3. It associates with an early class-switched response and is probably of lower specificity, as a first line of defense rapidly yielding circulating Ig at the acute phase in a context of exacerbated inflammation. Fast kinetics of this response correlates with the occurrence of ARDS. The prognostic implication of this inflammatory B-cell response among ICU patients, is reminiscent of the extra-follicular B cell response evidenced by cell cytometry, the amount of which correlates with ARDS in our study, and which was previously reported to be stronger in severe COVID-19 patients than in those with mild disease.11 This element also argues for a fate decision of immune cells according which a bias toward stronger engagement into the extra-follicular rather than the germinal center pathway would be correlated with a bad prognosis.
CDR3 length is a clonally stable feature, unaffected by late VDJ remodeling in activated B-cells. The two B cell compartments identified by specific Ig repertoire and CDR3 features are thus clearly disconnected and cannot correspond to successive activation stages of the same B-cell clones. Rather, they likely correspond to cells with differential activation requirements and differential functions. Although such a study in patients obviously remains descriptive, our data point not only on two different B-cell activation pathways, either extra-follicular or GC-dependent, but also the involvement of B cell subsets with disconnected repertoire structure and dynamics. Repertoire features indeed define an early and transient inflammatory B-cell response in parallel to the conventional Ag-specific response.
In conclusion, our results characterize in details two parallel aspects of the B-cell response in ICU COVID-19 patients. They also suggest the existence of an optimal ratio between these two B-cell arms in severe COVID-19, because the expansion of inflammatory repertoire (featuring long, hydrophobic and uncharged CDR3 regions) correlates with ARDS. Whether the inflammatory features of the B cell response either simply witness pathologic inflammation, or early participate to deleterious inflammation or immunosuppression will remain to be determined in other models.
Limitations of the study
By separately analyzing DNA templates which can characterize B cell diversity independently from their gene expression level, and RNA templates expected to vary in abundance, class-switch and splicing, this study intends to highlight those B-cell compartments which are specifically mobilized by the response to COVID-19. These molecular analyses might however have limitations regarding their translation into cellular and physiological aspects of the immune response. Notably, plasma cells undergo a strong (about 7-fold up-regulation) but still incomplete switch from membrane-type to secreted-type transcripts,42 and this strong trend still do not provide an absolute marker of plasma cell Ig transcripts vs lymphoid transcripts. Beyond the plasma cell case, RNA repertoires evaluate B-cell transcriptional activity but their connection with B-cell activation remains to be fully described. Their overlay with the DNA repertoire, reflecting B cell numbers, will need to be more extensively incorporated to future studies, whereas characterization of sorted B-cell subsets will also help elucidate the functional sub-structure of Ig repertoires. Finally, and as for any study in the complex situation of the ICU, we should acknowledge that preexisting baseline differences between the groups of patients included could participate to some of the variations observed in the kinetics of their immune response to COVID-19.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| FITC Mouse Anti-Human IgD (IA6-2) - Dil = 1/40 | BD Biosciences | Cat# 555778; RRID: AB_396113 |
| PE Mouse Anti-Human PDL1 (29E,2A3) - Dil = 1/10 | Biolegend | Cat# 329706; RRID: AB_940368 |
| PC5.5 Mouse Anti-Human CD27 (1A4CD27) - Dil = 1/10 | Beckman Coulter | Cat# B21444; RRID: AB_2934286 |
| KO Mouse Anti-Human CD45 (J33) - Dil = 1/10 | Beckman Coulter | Cat# B36294; RRID: AB_2833027 |
| PC7 Mouse Anti-Human CD38 (LS198-4-3) - Dil = 1/10 | Beckman Coulter | Cat# B49198; RRID: AB_2934287 |
| APC Mouse Anti-Human CD24 (ALB9) - Dil = 1/10 | Beckman Coulter | Cat# A87785; RRID: AB_2934288 |
| APC-AF700 Mouse Anti-Human CD10 (ALB1) - Dil = 1/10 | Beckman Coulter | Cat# B49223; RRID: AB_2934289 |
| APC-AF750 Mouse Anti-Human CD19 (J3-119) - Dil = 1/10 | Beckman Coulter | Cat# A94681; RRID:AB_2833030 |
| Biological samples | ||
| Blood samples from 35 ICU patients infected with SARS-CoV-2, see Table S2 | University Hospital Centers from Rennes (22), Limoges (9) and Lyon. | N/A |
| Blood samples from 50 healthy volunteers | University Hospital Centers from Limoges. | N/A |
| Blood samples from 18 ICU patients infected with SARS-CoV-2, see Table S2 | University Hospital Centers from Limoges. | N/A |
| Blood samples from 6 ICU patients infected with SARS-CoV-2, see Table S2 | University Hospital Centers from Lyon. | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Protoscript II RT (Race RepSeq) | Biolabs | Cat# M0368L |
| Deoxynucleotide Solutions, Mix | Biolabs | Cat# N0447S |
| Uracil-DNA glycosylase (Race RepSeq) | Biolabs | Cat# M0280S |
| Phusion high-fidelity DNA Pol (Race RepSeq) | Biolabs | Cat# M0530S |
| Ampure XP 5 mL | Beckman Coulter | Cat# A63880 |
| MiSeq Reagent Kit v3 (600-cycle) | Illumina | Cat# MS-102-3003 |
| AmpliTaq Gold DNA Polymerases | Thermofisher | Cat# 4398823 |
| HighPrep™ PCR Clean Up | Magbio | Cat# AC-60250 |
| Critical commercial assays | ||
| Tempus Spin RNA Isolation Kit | Thermofischer | Cat# 4380204 |
| EDTA K2 | BD Vacutainer | N/A |
| Deposited data | ||
| Raw sequencing data | This paper | Gene Expression Omnibus (GEO) accession number: GSE222431 |
| Covabdab database | Raybould et al28 | https://opig.stats.ox.ac.uk/webapps/covabdab/ |
| Oligonucleotides | ||
| Primers for DNA IGH library, see Table S4 | This paper | N/A |
| Primers for RACE-RepSeq IGH library, see Table S4 | This paper | N/A |
| Software and algorithms | ||
| Python 3.9 | Python Software Foundation | https://www.python.org/ |
| R 4.0.1 | The R Foundation | https://www.r-project.org/ |
| Immcantation Package | Gupta et al.43 | https://immcantation.readthedocs.io/ |
| Migec 1.2.9 | Shugay et al.44 | https://github.com/mikessh/migec |
| IMGT HighV-Quest | the international ImMunoGeneTics information system. Brochet et al.45 | https://www.imgt.org/IMGTindex/IMGTHighV-QUEST.php |
| FlowSOM | Saeys Unit - Data Mining and Modeling for Biomedicine, University of Gent | Van Gassen et al.23 |
| FlowCore | Life Sciences Department, Computational Biology Program, Division of Public Health Sciences, Fred Hutchinson Cancer Research Center, Seattle, Washington 98,109-1024, USA. | Ellis B, Haaland P, Hahne F, Le Meur N, Gopalakrishnan N, Spidlen J, Jiang M, Finak G (2022). _flowCore: flowCore: Basic structures for flow cytometry data_. R package version 2.10.0. |
| Kaluza | Beckman Life Science | https://www.beckman.fr/en/flow-cytometry/software |
| Adobe Photoshop CS5 version 12.0 x64 | Adobe Systems | https://www.adobe.com/fr/photoshop/ |
| Alakazam | Interdepartmental Program in Computational Biology and Bioinformatics, Yale University, New Haven, CT 06511 | Gupta et al.43 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Virginie Pascal (virginie.pascal@unilim.fr).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Patient recruitment and sampling
Blood samples from 35 ICU patients (30 for flow cytometry, 28 for DNA repertoire, 19 for RNA repertoire) infected with SARS-CoV-2 and admitted at University Hospital Centers from Rennes (22), Limoges (9) and Lyon (4), were included in this study. Patients with preexisting malignant or immune disorders or under immunosuppressive therapy were excluded. Individuals were tested for SARS-CoV-2 by real-time reverse-transcriptase–polymerase-chain-reaction (RT-PCR) using nasopharyngeal swabs. Patients tested positive were enrolled in the study upon ICU admission, due to severe symptoms of COVID-19 and were followed over multiple time points (D0, D7 and 4 months after admission). Among the 35 patients with severe COVID-19, 22 with ARDS (63%) and 13 without ARDS (37%). Samples from healthy volunteers, recruited between the ages of 18 and 75 and free from any pathology (COVID-19 negative, cell blood count, blood creatinine and serum immunoglobulin level without particularity), were used as controls for flow cytometry (9 samples) and DNA and RNA repertoires (26 and 24 samples, respectively). Comparison of clinical features of controls and patients were provided in Tables S1 and S2.
Ethics statement
All enrolled participants gave written, informed consent in accordance with the Declaration of Helsinki. The study design was approved by our ethical committee (CHU Rennes, n°35RC20_9795_HARMONICOV, ClinicalTrials.gov Identifier: NCT04373200).
Method details
Blood samples processing and storage
For Flow cytometry, Peripheral Blood Mononuclear Cells (PBMCs) were obtained by gradient centrifugation from EDTA samples and stored in DMSO at −80°C. For characterization of circulating repertoires, the Paxgene kit (Qiagen, Germantown, USA) was used to extract genomic DNA and total RNA from whole blood. Nucleic acids were stored at −80°C until experiments were performed.
Flow cytometry
Multiparametric flow cytometry (FCM) analysis was performed on 62 thawed PBMC samples. Monoclonal antibody (mAb) combination used for this study was IgD-FITC/PDL1-PE/CD27-PC5.5/CD45-KO/CD38-PC7/CD24-APC/CD10-APC-AF700/CD19-APC-AF750 (detailed in key resources table). Data acquisition was performed with a Navios flow cytometer and list mode data files (LMD) were re-analyzed with Kaluza software (Beckman-Coulter, Miami, FL, USA). LMD files from controls, ARDSpos and ARDSneg patients at Day 0 (D0) Day 7 (D7) and Month 4 (M4), were separately merged with Kaluza. Bi-exponential transformation was performed using “estimateLogicle” and “transform” functions of the flowCore R package (https://git.bioconductor.org/packages/flowCore). Self-Organizing Map (SOM) for flow cytometry and t-distributed stochastic neighbor embedding (t-SNE) analyzes were performed on CD45high, SS low CD19 positive gated B-cells using the FlowSOM R package.23 Overlay of tSNE graphs was done with the Adobe Photoshop∗ software (version 12.0 x64, Adobe Systems Incorporated, San Jose, CA).
DNA IGH library preparation
IGH CDR3 regions were amplified and sequenced from 750 ng of total DNA. Two steps were performed to polymerase chain reaction (PCR) amplification of IGH gene rearrangements. First, we amplified genomic DNA with primers BIOMED-2.46 We incorporated barcodes with a second PCR (nested PCR). Sequences are provided in Table S4.
Sequencing was performed using a Miseq apparatus and MiSeq Reagent Kit v3 (600-cycles) (Illumina, San Diego, CA, USA). Library preparation began with PCR amplification on 750ng of blood DNA with AmpliTaq Gold (Thermofisher) using BIOMED-2 primers. Cycling conditions were 30 sat 98°C, 34 cycles of 10 sat 98°C, 30 sat 62°C, 30 sat 72°C and final elongation 5 minat 72°C. PCR products were purified using 1.8x volume of purification beads (HighPrep™ PCR Clean-up System – Gaithersburg, USA) and resuspended in 40μL of water. Thereafter, Illumina adapter and tag sequences were added by primer extension. For this, PCR product (2μL) was reamplified with Taq Phusion (NewEngland Biolabs®). Cycling conditions were 30 seconds at 98°C, 12 cycles of 30 seconds at 98°C, 30 seconds at 62°C, 30 seconds at 72°C, and final elongation 5 minutes at 72°C. The PCR products were, once again, purified using 1.8 x volume of HighPrep™ purification beads and resuspended in 40μl of water. Finally, libraries were quantified by Qubit dosage (Thermofischer).
RACE-RepSeq IGH library preparation
For each sample, 500 ng of total RNA was reverse-transcribed to cDNA by a 5’RACE PCR using a ProtoScript II (New England Biolabs) and a cap race primer carrying unique molecular identifiers (UMIs) associated with consensus reverse primers for IgH Cμ, Cγ and Cα constant region. For each of the μ, γ, and α mRNA, the initial cDNA synthesis relied on two types of reverse primers, hC(n)-Mb-race or hC(n)-Sc-race, respectively targeting either the membrane-type exons (M1 or M2), or the specific secreted-type segments of the last CH exon (CHS). This RACE-PCR strategy separately evaluated the repertoire of either membrane-type (Mb) (mostly relevant to lymphocytes), or secreted-type (Sc) (most relevant to plasma cells) IgH transcripts (primer sequences listed in Table S4).
For preparing libraries, 5 μl of cDNA were amplified with Taq Phusion (New England Biolabs), using a universal forward primer and a nested reverse primer selected within the CH1 exon of the Cμ, Cγ and Cα genes. For Cγ transcripts, consensus CH1-anchored primers were chosen identical to all γ genes, but with amplicons including an initial CH1 segment as a unique identifier of each of the γ1, γ2, γ3 and γ4 isotypes. Cycling conditions were 2 minat 98°C, 32 cycles of 10 secat 98°C, 30 secat 62°C, 45 secat 72°C and final elongation 5 minat 72°C. PCR products were purified using (0.6 x reaction volume) AMPure XP beads (Beckman Coulter), and resuspended in 20μl of NEB elution buffer (Qiagen). Thereafter, 150ng of amplified product was barcoded and tagged with Illumina adapters through 12 cycles of primer extension with Taq Phusion (30 seconds at 98°C, 30 seconds at 65°C, 30 seconds at 72) with a final elongation 5 minutes at 72°C. Tagged PCR products were purified using (0.6 x reaction volume) AMPure XP beads and mixed with a stoichiometric ratio. Resulting libraries were gel purified (QIAGEN), quantified by qPCR (NEBNext Library Quant Kit for Illumina - New England Biolabs) before sequencing.
Quantification and statistical analysis
Sequencing and data analysis
Libraries from DNA or RNA templates were sequenced with an Illumina MiSeq, standard Illumina sequencing primers and MiSeq Reagent Kit v3 (600 cycles). For DNA libraries, paired-end reads were merged using the flash software47 and concatenating non-overlapping pairs. For RNA libraries, the migec script44 was used for UMI-demultiplexing, sequence correction and consensus assembly. The IGHV, IGHD, and IGHJ gene segments and the CDR-H3 junctions were identified using the IMGT/HighV-QUEST sequence alignment software.45 Quality filtering of sequences only kept reads including a productive CDR-H3 region. For cDNA-templated IGH reads, isotypes and subclasses were called by matching to the constant region gene segment. Sequences were finally assembled into clonal groups with the Change-o toolkit43 based on common IGHV gene, IGHJ gene and junction length and a maximal Hamming distance between junction amino acid sequences. Each clonal group is identified by a unique set of IGHV gene, IGHJ gene and a junctional coding sequence, corresponding to the most frequent CDR3 sequence of this clonal group.
The isotypes were determined by searching for the sequences of the isotype-specific primers IGHM, IGHG and IGHA described in Table S4. IGHG subclasses were defined by comparing 5 nucleotide bases in the IGHG sequences distinguishing IGHG1, IGHG2, IGHG3 and IGHG4 genes:
-
•
chr14:105743011>G (IGHG1, IGHG3); chr14:105743011>A (IGHG2, IGHG4)
-
•
chr14:105743013>G (IGHG1, IGHG3); chr14:105743013>A (IGHG2, IGHG4)
-
•
chr14:105743025>A (IGHG1); chr14:105743025>G (IGHG2, IGHG3, IGHG4)
-
•
chr14:105743036>A (IGHG1); chr14:105743036>G (IGHG2, IGHG3, IGHG4)
-
•
chr14:105743051>G (IGHG1, IGHG2, IGHG3); chr14:105743051>G (IGHG4)
For each repertoire, the gini diversity index and the proportion of membrane and secretory clonotypes were calculated and repertoires restricted to the 100 most represented circulating clonotypes were generated. Subclass fractions were determined for each subject by dividing the number of clones for a given subclass by the total number of clones for that isotype category. In parallel, the average length of CDR-H3 and the median somatic mutation frequency of reads were calculated for each clone. Physico-chemical properties of clonotypes were calculated by using R package alakazam.43
Differentially expressed (DE) IGHV-IGHJ germinal pairs were identified between controls, and patients with and without ARDS at D0 and D7 after admission for adjusted p value <0.05 (Benjamini-Hochberg multiple testing correction) (edgeR R package). Hierarchical clustering heatmaps of the DE IGHV-IGHJ pairs were represented for all samples collected at D0, D7, D14 and Month 4 (M4) after admission including controls, using the euclidean distance for IGHV_IGHJ, the Pearson distance for samples, and the Ward.D2 linkage method for IGHV-IGHJ pairs and samples (complexHeatmap R package). DE IGHV-IGHJ pairs were annotated by their abundance in the CoV-AbDab database.28 The CoV-AbDab database used for the heatmaps consisted in 2696 distinct COVID-19 Human antibodies (Ab) published between May 4th, 2020 to Nov 1st, 2021, corresponding to 209 distinct Human IGHV-IGHJ pairs. While many of these pairs feature private clonotypes, with single or rare (<5) occurrences in the database, the 25% most represented of these pairs accounted for more than 80% of all the Abs included in the CoV-AbDab (culminating with 179 occurrences for the IGHV3-30_IGHJ4 pairs). To identify convergent rearranged IGH and CoV-AbDab clonotypes among patients with SARS-CoV-2 infection, we retrieved hits on the CoV-AbDab database for each clonal group, based on the same IGHV gene, IGHJ gene and a minimum of 90% CDR-H3 amino acid sequence similarity.
Statistical analysis
Statistical tests were performed in GraphPad Prism 8.4.3 with unpaired t test. In all analyses where statistical significance was tested, significance was defined as: ∗∗∗p value <0.001; ∗∗p value <0.01; ∗p value <0.05; not significant (NS): p value >0.05. Analysis of variance and non-parametric Wilcoxon tests were performed with R.
Acknowledgments
We thank all the donors, the staff in charge of patients care, clinical data collection, biological sample collection and the staff in charge of ethical and administrative issues. We thank Dr. Mehdi Alizadeh for help with sequencing. This work was supported by the Fondation pour la Recherche Médicale (FRM) and the Agence Nationale de la Recherche(ANR), Flash Covid-19 joint grant (HARMONICOV to MC).
Author contributions
Conceptualization, M.Cog., J.Feu. and K.T.; Methodology, M.Cog., J.Feu., D.Ri., and V.P.; Investigation, V.P., M.D., D.Ri., R.J., S.G., M.Cor., J.Fer., and M.R.; Formal Analysis, P.d.R. and D.Ro.; Writing – Original Draft, M.Cog., J.Feu., and V.P.; Writing – Review and Editing, M.Cog., J.Feu., K.T., J.M.T., and V.P.; Funding Acquisition, M.Cog., J.M.T and K.T.; Resources, J.M.T., F.R., M.L.S., B.F., T.D., G.M., and F.V.; Supervision, M.Cog., J.Feu., and K.T.
Declaration of interests
The authors declare no competing interest.
Inclusion and diversity
We support inclusive, diverse and equitable conduct of research. One or more of the authors of this paper self-identifies as a member of the LGBTQIA+ community.
Published: February 21, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106260.
Supplemental information
Data and code availability
RACE-RepSeq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. DNA repertoire and FCM data reported in this paper will be shared by the lead contact upon request.
All original code is available in this paper’s supplemental information.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Guan W.-J., Ni Z.-Y., Hu Y., Liang W.-H., Ou C.-Q., He J.-X., Liu L., Shan H., Lei C.-L., Hui D.S.C., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helms J., Tacquard C., Severac F., Leonard-Lorant I., Ohana M., Delabranche X., Merdji H., Clere-Jehl R., Schenck M., Fagot Gandet F., et al. High risk of thrombosis in patients with severe SARS-CoV-2 infection: a multicenter prospective cohort study. Intensive Care Med. 2020;46:1089–1098. doi: 10.1007/s00134-020-06062-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collie S., Champion J., Moultrie H., Bekker L.-G., Gray G. Effectiveness of BNT162b2 vaccine against omicron variant in South Africa. N. Engl. J. Med. 2022;386:494–496. doi: 10.1056/NEJMc2119270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J., Chandrashekar A., Sellers D., Barrett J., Jacob-Dolan C., Lifton M., McMahan K., Sciacca M., VanWyk H., Wu C., et al. Vaccines elicit highly conserved cellular immunity to SARS-CoV-2 omicron. Nature. 2022;603:493–496. doi: 10.1038/s41586-022-04465-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pouwels K.B., Pritchard E., Matthews P.C., Stoesser N., Eyre D.W., Vihta K.-D., House T., Hay J., Bell J.I., Newton J.N., et al. Effect of Delta variant on viral burden and vaccine effectiveness against new SARS-CoV-2 infections in the UK. Nat. Med. 2021;27:2127–2135. doi: 10.1038/s41591-021-01548-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.VanBlargan L.A., Errico J.M., Halfmann P.J., Zost S.J., Crowe J.E., Purcell L.A., Kawaoka Y., Corti D., Fremont D.H., Diamond M.S. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. 2022;28:490–495. doi: 10.1038/s41591-021-01678-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Q., Bastard P., COVID Human Genetic Effort. Cobat A., Casanova J.-L. Human genetic and immunological determinants of critical COVID-19 pneumonia. Nature. 2022;603:587–598. doi: 10.1038/s41586-022-04447-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun B., Feng Y., Mo X., Zheng P., Wang Q., Li P., Peng P., Liu X., Chen Z., Huang H., et al. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg. Microbes Infect. 2020;9:940–948. doi: 10.1080/22221751.2020.1762515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woodruff M.C., Ramonell R.P., Nguyen D.C., Cashman K.S., Saini A.S., Haddad N.S., Ley A.M., Kyu S., Howell J.C., Ozturk T., et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat. Immunol. 2020;21:1506–1516. doi: 10.1038/s41590-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Libster R., Pérez Marc G., Wappner D., Coviello S., Bianchi A., Braem V., Esteban I., Caballero M.T., Wood C., Berrueta M., et al. Early high-titer plasma therapy to prevent severe Covid-19 in older adults. N. Engl. J. Med. 2021;384:610–618. doi: 10.1056/NEJMoa2033700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.So-Osman C., Valk S.J. High-dose immunoglobulins from convalescent donors for patients hospitalised with COVID-19. Lancet. 2022;399:497–499. doi: 10.1016/S0140-6736(22)00112-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agrati C., Sacchi A., Bordoni V., Cimini E., Notari S., Grassi G., Casetti R., Tartaglia E., Lalle E., D’Abramo A., et al. Expansion of myeloid-derived suppressor cells in patients with severe coronavirus disease (COVID-19) Cell Death Differ. 2020;27:3196–3207. doi: 10.1038/s41418-020-0572-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flament H., Rouland M., Beaudoin L., Toubal A., Bertrand L., Lebourgeois S., Rousseau C., Soulard P., Gouda Z., Cagninacci L., et al. Outcome of SARS-CoV-2 infection is linked to MAIT cell activation and cytotoxicity. Nat. Immunol. 2021;22:322–335. doi: 10.1038/s41590-021-00870-z. [DOI] [PubMed] [Google Scholar]
- 16.Kuri-Cervantes L., Pampena M.B., Meng W., Rosenfeld A.M., Ittner C.A.G., Weisman A.R., Agyekum R.S., Mathew D., Baxter A.E., Vella L.A., et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020;5:eabd7114. doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roussel M., Ferrant J., Reizine F., Le Gallou S., Dulong J., Carl S., Lesouhaitier M., Gregoire M., Bescher N., Verdy C., et al. Comparative immune profiling of acute respiratory distress syndrome patients with or without SARS-CoV-2 infection. Cell Rep. Med. 2021;2:100291. doi: 10.1016/j.xcrm.2021.100291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., Levantovsky R., Malle L., Moreira A., Park M.D., et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T., et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sokal A., Chappert P., Barba-Spaeth G., Roeser A., Fourati S., Azzaoui I., Vandenberghe A., Fernandez I., Meola A., Bouvier-Alias M., et al. Maturation and persistence of the anti-SARS-CoV-2 memory B cell response. Cell. 2021;184:1201–1213.e14. doi: 10.1016/j.cell.2021.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.ARDS Definition Task Force. Ranieri V.M., Rubenfeld G.D., Thompson B.T., Ferguson N.D., Caldwell E., Fan E., Camporota L., Slutsky A.S. Acute respiratory distress syndrome: the Berlin Definition. JAMA. 2012;307:2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 22.Catalán D., Mansilla M.A., Ferrier A., Soto L., Oleinika K., Aguillón J.C., Aravena O. Immunosuppressive mechanisms of regulatory B cells. Front. Immunol. 2021;12:611795. doi: 10.3389/fimmu.2021.611795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Gassen S., Callebaut B., Van Helden M.J., Lambrecht B.N., Demeester P., Dhaene T., Saeys Y. FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A. 2015;87:636–645. doi: 10.1002/cyto.a.22625. [DOI] [PubMed] [Google Scholar]
- 24.Jenks S.A., Cashman K.S., Zumaquero E., Marigorta U.M., Patel A.V., Wang X., Tomar D., Woodruff M.C., Simon Z., Bugrovsky R., et al. Distinct effector B cells induced by unregulated toll-like receptor 7 contribute to pathogenic responses in systemic lupus erythematosus. Immunity. 2018;49:725–739.e6. doi: 10.1016/j.immuni.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosser E.C., Mauri C. Regulatory B cells: origin, phenotype, and function. Immunity. 2015;42:607–612. doi: 10.1016/j.immuni.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Sosa-Hernández V.A., Torres-Ruíz J., Cervantes-Díaz R., Romero-Ramírez S., Páez-Franco J.C., Meza-Sánchez D.E., Juárez-Vega G., Pérez-Fragoso A., Ortiz-Navarrete V., Ponce-de-León A., et al. B cell subsets as severity-associated signatures in COVID-19 patients. Front. Immunol. 2020;11:611004. doi: 10.3389/fimmu.2020.611004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cervantes-Díaz R., Sosa-Hernández V.A., Torres-Ruíz J., Romero-Ramírez S., Cañez-Hernández M., Pérez-Fragoso A., Páez-Franco J.C., Meza-Sánchez D.E., Pescador-Rojas M., Sosa-Hernández V.A., et al. Severity of SARS-CoV-2 infection is linked to double-negative (CD27- IgD-) B cell subset numbers. Inflamm. Res. 2022;71:131–140. doi: 10.1007/s00011-021-01525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raybould M.I.J., Kovaltsuk A., Marks C., Deane C.M. CoV-AbDab: the coronavirus antibody database. Bioinformatics. 2021;37:734–735. doi: 10.1093/bioinformatics/btaa739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen S.C.A., Yang F., Jackson K.J.L., Hoh R.A., Röltgen K., Jean G.H., Stevens B.A., Lee J.-Y., Rustagi A., Rogers A.J., et al. Human B cell clonal expansion and convergent antibody responses to SARS-CoV-2. Cell Host Microbe. 2020;28:516–525.e5. doi: 10.1016/j.chom.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.-E., Katsaounou P., et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27:992–1000.e3. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sinha P., Calfee C.S., Cherian S., Brealey D., Cutler S., King C., Killick C., Richards O., Cheema Y., Bailey C., et al. Prevalence of phenotypes of acute respiratory distress syndrome in critically ill patients with COVID-19: a prospective observational study. Lancet Respir. Med. 2020;8:1209–1218. doi: 10.1016/S2213-2600(20)30366-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395 doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galván-Peña S., Leon J., Chowdhary K., Michelson D.A., Vijaykumar B., Yang L., Magnuson A.M., Chen F., Manickas-Hill Z., Piechocka-Trocha A., et al. Profound Treg perturbations correlate with COVID-19 severity. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2111315118. e2111315118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harb H., Benamar M., Lai P.S., Contini P., Griffith J.W., Crestani E., Schmitz-Abe K., Chen Q., Fong J., Marri L., et al. Notch4 signaling limits regulatory T-cell-mediated tissue repair and promotes severe lung inflammation in viral infections. Immunity. 2021;54:1186–1199.e7. doi: 10.1016/j.immuni.2021.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reizine F., Lesouhaitier M., Gregoire M., Pinceaux K., Gacouin A., Maamar A., Painvin B., Camus C., Le Tulzo Y., Tattevin P., et al. SARS-CoV-2-Induced ARDS associates with MDSC expansion, lymphocyte dysfunction, and arginine shortage. J. Clin. Immunol. 2021;41:515–525. doi: 10.1007/s10875-020-00920-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaneko N., Kuo H.-H., Boucau J., Farmer J.R., Allard-Chamard H., Mahajan V.S., Piechocka-Trocha A., Lefteri K., Osborn M., Bals J., et al. Loss of Bcl-6-expressing T follicular helper cells and germinal centers in COVID-19. Cell. 2020;183:143–157.e13. doi: 10.1016/j.cell.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Bourcy C.F.A., Angel C.J.L., Vollmers C., Dekker C.L., Davis M.M., Quake S.R. Phylogenetic analysis of the human antibody repertoire reveals quantitative signatures of immune senescence and aging. Proc. Natl. Acad. Sci. USA. 2017;114:1105–1110. doi: 10.1073/pnas.1617959114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Röltgen K., Nielsen S.C.A., Silva O., Younes S.F., Zaslavsky M., Costales C., Yang F., Wirz O.F., Solis D., Hoh R.A., et al. Immune imprinting, breadth of variant recognition, and germinal center response in human SARS-CoV-2 infection and vaccination. Cell. 2022;185:1025–1040.e14. doi: 10.1016/j.cell.2022.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roco J.A., Mesin L., Binder S.C., Nefzger C., Gonzalez-Figueroa P., Canete P.F., Ellyard J., Shen Q., Robert P.A., Cappello J., et al. Class-switch recombination occurs infrequently in germinal centers. Immunity. 2019;51:337–350.e7. doi: 10.1016/j.immuni.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peterson M.L., Perry R.P. The regulated production of mu m and mu s mRNA is dependent on the relative efficiencies of mu s poly(A) site usage and the c mu 4-to-M1 splice. Mol. Cell Biol. 1989;9:726–738. doi: 10.1128/mcb.9.2.726-738.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gupta N.T., Vander Heiden J.A., Uduman M., Gadala-Maria D., Yaari G., Kleinstein S.H. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics. 2015;31:3356–3358. doi: 10.1093/bioinformatics/btv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shugay M., Britanova O.V., Merzlyak E.M., Turchaninova M.A., Mamedov I.Z., Tuganbaev T.R., Bolotin D.A., Staroverov D.B., Putintseva E.V., Plevova K., et al. Towards error-free profiling of immune repertoires. Nat. Methods. 2014;11:653–655. doi: 10.1038/nmeth.2960. [DOI] [PubMed] [Google Scholar]
- 45.Brochet X., Lefranc M.-P., Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Dongen J.J.M., Langerak A.W., Brüggemann M., Evans P.a.S., Hummel M., Lavender F.L., Delabesse E., Davi F., Schuuring E., García-Sanz R., et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 47.Magoč T., Salzberg S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RACE-RepSeq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. DNA repertoire and FCM data reported in this paper will be shared by the lead contact upon request.
All original code is available in this paper’s supplemental information.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.