Abstract
World is in the middle of the pandemic (COVID-19), caused by SARS-COV-2 virus, which is a significant global health crisis after Spanish influenza in the beginning of 20th century. Progressive drastic steps have been enforced to minimize the transmission of the disease. Likewise, in the current years, antimicrobial resistance (AMR) has been referred as one of the potential perils to the global economy and health; however, it is now veiled under the present pandemic. During the current pandemic, AMR to available frontline antibiotics may prove fatal and life threatening to bacterial and fungal infections during routine procedures like elective surgery, C-sections, etc. Currently, a swift elevation in multidrug-resistant organisms (MDROs), like carbapenem-resistant New Delhi metallo-β-lactamase (NDM)-producing Acinetobacter baumannii, Enterobacterales, extended-spectrum β-lactamase (ESBL)-producing Klebsiella pneumoniae, methicillin-resistant Staphylococcus aureus (MRSA), multi-triazole-resistant Aspergillus fumigatus and pan-echinocandin-resistant Candida glabrata has been seen. Thereupon, the global outbreak of COVID-19 also offers some important ramification for developing antimicrobial drug resistance. This article aims to highlights episodes and aspects of AMR prevalence, impact of management and mismanagement of COVID-19 crisis, hospital settings, community, environment, and travel on the AMR during the current pandemic.
Keywords: Antimicrobial resistance (AMR), Covid-19, Bacteria, Fungi, Disinfectants, Travel
Introduction
The ongoing pandemic has overwhelmed healthcare systems globally and at the same time, increase in AMR and MDR (multi-drug resistant) pathogens maintains to risk the world health through momentous morbidity, mortality, and economic loss [1]. AMR can be defined as an increasing resistance to frontline lifesaving antibiotics that undermines the ability to treat common and serious infectious diseases. A recent study reports, owing to the (COVID-19) pandemic, higher number of patients have been admitted in hospitals for pragmatic antimicrobial treatment that might not be applicable [2], [3], potentially leading to resistant infections worldwide. Although the COVID-19 treatment with antimicrobial therapy is non effective, there are other various reasons for prescribing the antimicrobials: the presence of clinical manifestation that are similar to bacterial or other pneumonias, the possibility of secondary infections, and protocols of healthcare functioning, that might recommend the antimicrobial usage [4]. Though, the antimicrobial treatment in COVID-19 patients could be justifiable in case of bacterial or fungal secondary infection, while for AMR, optimal therapies and appropriate reduction of dose or withdrawal of antimicrobial treatment. and antimicrobial stewardship directed on promoting the line of empirical treatment when a co-infection is present would be of paramount importance [4].
An increasing number of studies indicates that AMR has been growing, after the antibiotic treatment for COVID-19 patients, however, the analysis of existing AMR and comparative range of the related pathogens is still unclear. Studying the occurrence of AMR in COVID-19 patients is of paramount importance. There is a clear indication that over consumption of antimicrobial in humans has led to resistant pathogens that adversely affects the human health, perhaps, among the top ten list, AMR is considered as the potential global threats to human health, food security, and development [5], [6], [7]. A crucial information divergent exists about the occurrence, and characteristics of secondary infections with bacteria and fungi, including AMR among the COVID-19 patients.
As per the World health organization, as on 10th November 2022, 6,583,163 lives are lost due to COVID-19 globally since the outbreak and AMR is currently estimated to kill approximately 700,000 people, annually. Assuming that the given data for both are accurate, a rough comparison evaluates that death rates for COVID-19 will stay fixed for the remaining year, however, AMR will kill over 130,000 people annually alone [8]. Moreover, AMR deaths are expected to rise to 10 million deaths every year by 2050, whereas COVID-19 may hopefully be controlled in a much briefer period [1]. This article discusses the impact of various factors on AMR prevalence during the current pandemic, for example, prevalence of bacterial and fungal secondary infections, incoherent use of antimicrobial in covid-19 patients, impact of the management of COVID-19 in hospital settings, community, environment, and travel ( Fig. 1) [9].
Fig. 1.
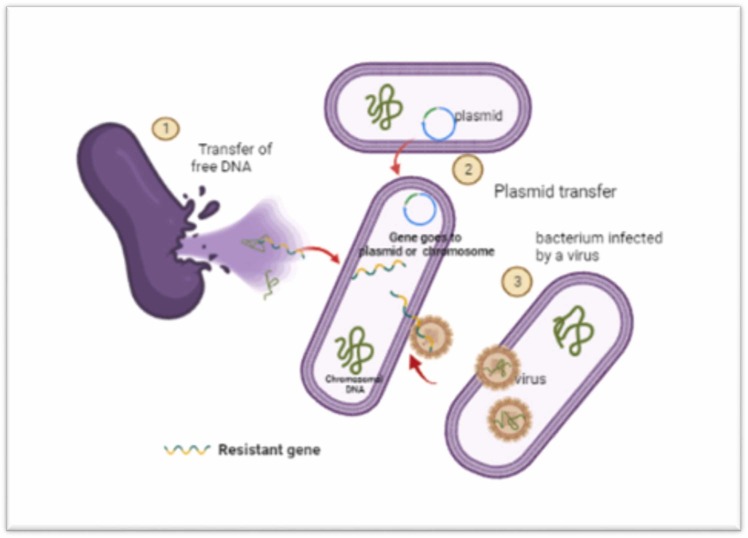
AMR by bacteria is acquired through mutated “resistance genes” via vertical transfer; gene intake from lysed bacteria, that can also occur between different strains; plasmids as carrier of genes as a horizontal transfer between one bacterium to the other and viruses transporting the genes.
Secondary infections during the pandemic
Bacterial infections
One of the important issues is the secondary bacterial infections among COVID-19 patients. Until now 1–10 % of cases have been recorded to have secondary infections. However, regardless of the proportionality of low recognition of secondary infections, relatively more usage of antibiotics has been recorded during the treatment of COVID-19 patients. This is being practiced even after the WHO recommendation, that is contrary to the consumption of antimicrobials for COVID-19 patients. There are various studies which reports an elevation in MDR bugs during the ongoing pandemic ( Table 1). One of the retrospective studies reported, that the colonization of carbapenem-resistant Enterobacterales has raised from 6.7 % to 50 % from 2019 to 2020. In China, Li et al. presented the study reported the isolation of 159 bacterial strains from 102 COVID-19 patients having secondary infections, among them, the most common was Acinetobacter baumannii (35.8 %; n = 57), that was followed by Klebsiella pneumoniae and Stenotrophomonas maltophilia with (30.8 %; n = 49) (6.3 %; n = 10), respectively. Additionally, the rate of carbapenem resistance for A. baumannii was 91.2 % and K. pneumoniae 75.5 %. Another study in France, found that 26 COVID-19 patients in ICU with severe respiratory disease were having a bacterial co-infection, among which five isolates were resistant to third generation cephalosporins and two for amoxicillin/clavulanate. Likewise, another study of 19 COVID-19 patients who were admitted to ICU had a coinfection with 17 MDR- A. baumannii and a MRSA. Fu et al. reported five cases of ICU COVID-19 patients with co- infections of MDRs like, extended-spectrum β-lactamase (ESBL)-producing K. pneumoniae, Burkholderia cepacian, P. aeruginosa and S. maltophilia. Additionally, New Delhi metallo-β-lactamase (NDM)-producing Enterobacter cloacae was reported to be a causative agent for secondary infection in five COVID-19 patients in New York (USA). A retrospective study reported 1959 rare isolates with 29 % (569) resistant pathogens while as secondary infection with resistant pathogens ranged from 0.2 % to 100 %. Multi-drug resistant organisms, MRSA, carbapenem-resistant Acinetobacter baumannii, Pseudomonas aeruginosa, Klebsiella pneumoniae and Candida auris were among the commonly isolated organisms. This study also reported higher prevalence of AMR outside of Europe and in ICU settings [10], [11], [12], [13], [14], [15], [16], [17], [18].
Table 1.
Some of the AMR studies from different nations during the COVID-19 pandemic.
| Secondary Infection | Country of study | Year | Settings | Number of patients | Resistance | Ref. |
|---|---|---|---|---|---|---|
| E. coli, Klebsiella, S. aureus (MSSA), S. aureus (MRSA), P. aeruginosa. and Enterobacter species | Iran | 2020 | Outpatients/inpatients | 340 | Cotrimoxazole, piperacillin, ceftazidime, and cefepime | [9] |
| A. baumannii, K. pneumoniae, and S. maltophilia. | China | 2020 | ICU | 102 | Carbapenem- and Methicillin | [19] |
| K. pneumoniae, A. baumannii, S. maltophilia, C. albicans, and Pseudomonas spp. | China | 2020 | ICU | 190 | Carbapenem | [20] |
| A. baumannii. | Iran | 2020 | ICU | 19 | colistin | [14] |
|
K. pneumoniae and 4 Enterobacter cloacae |
USA | 2020 | ICU | 13 | ceftazidime/avibactam | [21] |
| A. baumannii, and K. pneumonia. | India | 2021 | ICU | 750 | MDR, Carbapenem 47 % | [22] |
| Acinetobacter spp. | Serbia | 2021 | ICU | 611 | imipenem, meropenem, and ciprofloxacin | [23] |
| K. pneumoniae, and A. baumannii. | Egypt | 2021 | ICU | 197 | (PDR)K. pneumoniae, and (MDR) A. baumannii | [24] |
| E. coli and K. pneumonia | Pakistan | 2021 | ICU | 856 | E. coli to ciprofloxacin and ampicillin. K. pneumoniae to ampicillin and amoxycillin | [25] |
| S. aureus, and P. aeruginosa | Italy | 2021 | Outpatients/inpatients | 255 | oxacillin, vancomycin, carbapenems, colistin, third- and fourth-generation cephalosporins. | [26] |
| A. baumannii and E. coli | India | 2021 | ICU | 7309 | MDR | [27] |
| E. coli | Indonesia | 2021 | Outpatients/inpatients | 148 | ofloxacin, aztreonam, Fosfomycin, piperacillin, amoxicillin, cefadroxil, and ampicillin |
[28] |
|
E. coli, K. pneumoniae, and P. aeruginosa. |
Turkey | 2021 | Outpatients/inpatients | 3532 | SBL producing Enterobacterales MDR |
[29] |
|
E. coli, A. baumannii,K. pneumoniae, S. aureus and E. faecalis. |
Italy | 2021 | ICU | 89 | MDR | [30] |
|
A. baumannii, K. pneumoniae. E. coli and P. aeruginosa |
Saudi Arabia |
2022 | Outpatients/inpatients | 108 | piperacillin/tazobactam and trimethoprim/sulfamethoxazole. |
[31] |
|
K. pneumonia and A. baumannii |
Iran | 2022 | ICU | 553 | carbapenem-resistant | [32] |
| Acinetobacter spa | Iran | 2022 | ICU | 38 | amikacin, gentamycin, imipenem, and cefixime. |
[33] |
|
A. baumannii, K. pneumoniae, P. aeruginosa, and E. faecium |
Turkey | 2022 | ICU | 119 | tigecycline. | [34] |
|
K. pneumoniae and A. baumannii. |
India | 2022 | inpatients | 122 | Carbapenem | [35] |
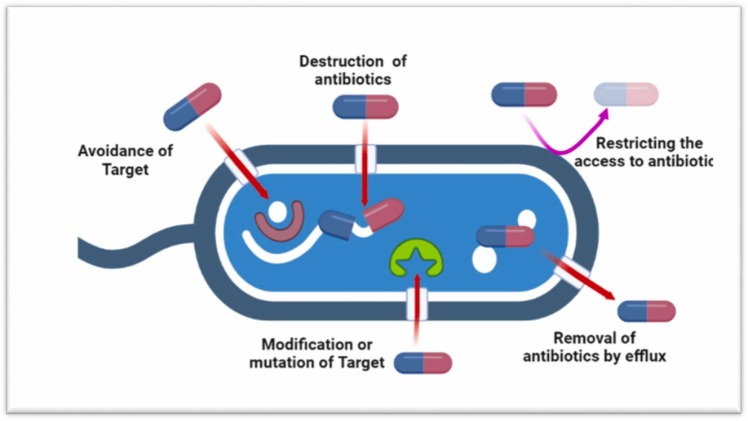
A bacterium is considered as resistant to any specific antimicrobial agent when the drug substance – after a recommended dose – does not effectively kill or inhibit the multiplication of the bacterium. Even though, many bacteria are characteristically resistant to antimicrobials, however, others can develop resistance via gene mutations (for example insertions and deletions) and horizontal gene transfer through conjugation, transformation, or transduction by temperate phages ( Fig. 2) [36], [37]. The mechanism of antimicrobial resistance in bacteria can be summarized into different categories: (1) restrictive uptake of a drug; (2) altering a drug target; (3) drug inactivation; (4) efflux pump Fig. 2). The restrictive drug uptake, inactivation of drug, and active drug efflux are considered as the mechanism of intrinsic resistance while acquired resistance may be modification of the drug target of the microbial cell, inactivation of drug molecule, as well as drug efflux pump [38]. Gram negative and gram-positive bacteria varies in the mode of mechanism due to the differences in their cellular structure. Gram negative bacteria may opt all the four main mechanisms, however gram-positive bacteria mostly adopt the restrictive drug uptake as it lacks an LPS outer membrane, hence having limited capacity for specific drug efflux mechanisms [39], [40]. In gram negative bacteria, the LPS layer offers a barrier to various types of drug molecules imparting them with innate resistance to several groups of antimicrobial drugs [41]. Fig. 2 demonstrates the general antimicrobial resistance mechanisms.
Fig. 2.
Antimicrobial resistance in bacterial co-infections. Bacteria can develop the mechanism that avoid the target either by producing enzymes capable of destroying the drug or modifying their structure. Bacteria use efflux pumps to remove the antimicrobial or reduce the affinity by mutating the target.
Fungal infections
Though not recognized well, but fungal infections constitute greatly to human mortality. The unwarranted usage of antifungals has raised the prevalence of invasive mycoses, that have resistance to many antifungals [42] (Fig. 2). In China and worldwide a study on SARS and influenza virus suggests an increased threat of invasive mycoses in COVID-19 immuno-compromised patients. Life-saving surgeries and treatments needs appropriate diagnosis and appropriate treatment for the co-fungal infections [43]. In a study by Posteraro et al., 53-day clinical course of a COVID-19 patient having type 2 diabetes mellitus, was found to have bloodstream infection by three different organisms, Morganella morganii, MRSA, and Candida glabrata [44]. After the 13 days treatment with caspofungin, C. glabrata was found to have FKS-associated pan-echinocandin resistance. In New Delhi (India), candidaemia was found in 15 COVID-19 critical patients, among them 10 cases were MDR Candida auris which accounted for 6 deaths [45]. Mohamed et al. observed a severe COVID-19 pneumonia with a co-infection of Aspergillus fumigatus, a multi-triazole-resistant strain [46]. A cohort of 31 subjects found, 19.4 % were having aspergillosis and other studies done in Indian hospitals during the COVID-19 treatment, were detected with candidemia in 2.5 % critical patients, among which 53 % died. Moreover, 66 % of them were suffering from a persistent fungemia despite the treatment with antifungal therapy [47], [48]. For many years, azole group has been established as the gold standard treatment against fungal infections. However, Denning et al. in a study demonstrated a greater occurrence of resistance to triazole by Aspergillus fumigatus among the patients with chronic fungal diseases. Therefore, fungal infections among the COVID-19 patients bear a constant treatment challenge that demands examining the non-traditional solutions to combat the drug resistance menace post COVID-19.
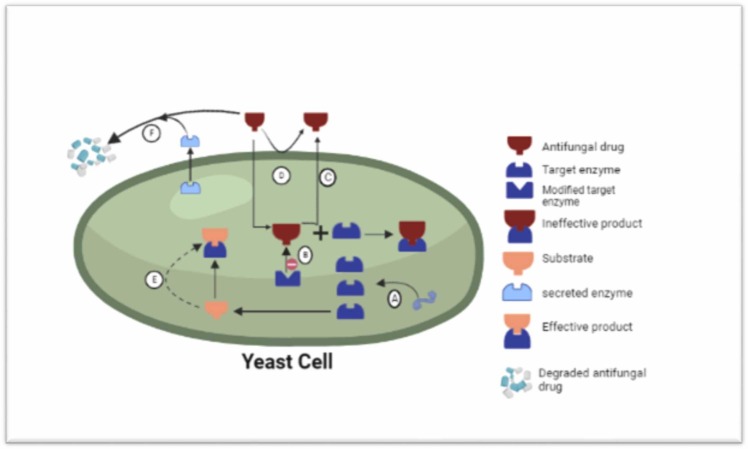
One of the mechanisms for the resistance is fungi tend to produce elevated amount of enzymes that are the main target for azole group and other antifungal drugs, hence preventing the suppression of metabolic reactions. Another mechanism used is the modification of the arrangement of targeted enzyme that minimize the attachment efficacy of azole significantly [49]. Additionally fungal cells can actively pump out the antifungal drugs by using the efflux pumps. Moreover, cell can also bypass the metabolic cascades that the drug is known to target. The fungi have the ability to produce extracellular enzymes that can dissociate the antifungal compounds [50], [51], [52] ( Fig. 3).
Fig. 3.
Fungal resistance against antifungal compounds. (A) Fungi likely increase the production of enzymes that are main targets for the drugs. This counters the inhibition of metabolic activities. (B) Modifying the spatial structure of target enzyme minimizes the drug binding efficiency. (C) Efflux pumps efficiently pumps out the antifungal drugs out of the cell. (D) fungal cell wall/membrane blocks the penetration of antifungals. (E) The fungal cell escapes the routine pathway that the drug recognizes to target. (E) The secretion of extracellular enzymes having the ability to degrade the antifungal drug.
Perspective
Instead, great discretion is needed for the fact that improper use or abuse of antimicrobials is recognized to be one of a potential operator for the occurrence of AMR. Significant approach on AMR focusses on minimizing the inappropriate utilization of antimicrobials by clinicians and health practitioners. Several nations, which have relatively made advancement in this field, may easily confront reduced AMR infections compared to the nations who have achieved little or no success in minimizing the antibiotic consumption. The antibiotics by clinicians should be considered very critically and carefully, as frequent antibiotic prescription of antibiotics makes public believe that all antibiotics are effective for the treatment of viral infections in general and SARS-COV-2 in particular.
Incoherent application of disinfectants
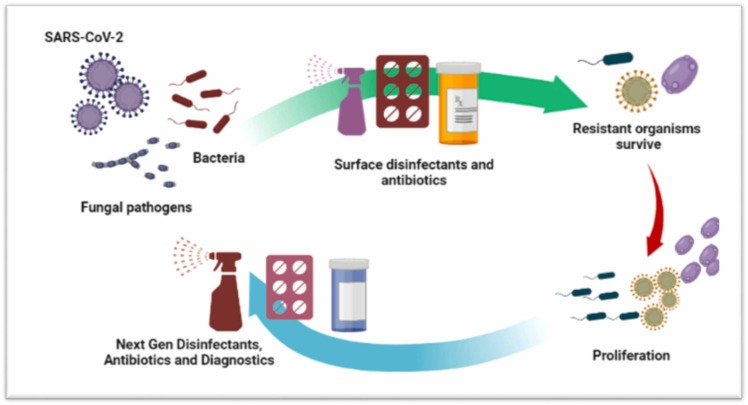
Beyond the clinics, almost all the nations have been exercising measures, intended at reducing the spread of SARS-CoV-2, that varied from physical distancing to complete lock down. However, the constant instructions from the beginning have been regular hand washing with soaps and/or the use of sanitizers Nevertheless, the use of disinfectants and antimicrobial soaps has exponentially increased over the period of this pandemic and is likely to continue [1], [53]. Undoubtedly such practices may help in combating the spread of virus; however, it has an adverse effect that could emerge from increased usage of products that are mainly microbiocidal (Fig. 3). Ultimately, these microbiocidal found in most of the disinfectants and surface cleaners may also lead to development of AMR, as these increased concentrations of biocides usually find their way into wastewater treatments plants and other water bodies [54], [55]. Most importantly if the concentration of biocide is very high, possibly the maximum inhibition of bacteria will take place, however, if the concentration increases but does not reach the minimum inhibitory concentration required for varying microbial flora, this would in turn offer an opportunity to the emergence of AMR. Moreover, preventive treatment for bacterial secondary infection among COVID-19 patients could be also responsible to elevated levels of antibiotics in the water bodies and the receiving environments which may result in the evolution of AMR [56], [57] ( Fig. 4).
Fig. 4.
Incoherent usage of disinfectants leading to proliferation of resistant microbes that would be needing next generation antibiotics.
Hospital settings
The health management system throughout the globe has been under immense burden which led towards several modifications in clinical practice that may have an impact on AMR. Regarding the COVID-19 management in health care settings, most of the world governments have recommended some measures for the prevention and control of infection through droplets, aerosols and direct contact with potentially contaminated surfaces. These strategies like extra vigilance of hygiene, sterilization techniques may lower down the circulation of AMR bacteria within clinical settings. However, it would be of paramount importance to collect the information on the prevalence of prior and post AMR infections during the current pandemic, to evaluate and contain the AMR menace at local and thereby at global level. Clinicians and researchers need to work hand in hand for the study and correlation of whole genome sequences of clinical isolates, prior, during and post COVID-19 pandemic, which is one of the promising technique to interpret the changes in the transition of AMR mechanisms [58], [59], [60], [61].
Travel and AMR
Travel has been also seen to have impact on the transmission of AMR. Transfer of important AMR genes between nations is inarguable. Among the primary genes responsible for resistance to last option antibiotic carbapenem was initially identified in India [62], and thereafter isolated throughout the world [63]. Moreover, inception of the mcr1 gene responsible for conferring resistance to colistin an important antibiotic, was initially isolated in China, however now found to exist worldwide [64], [65]. First time, the emergence of tigecycline resistance gene tet (X4), has recently found in China. One of the “pandemic” labeled gene i.e., CTX-M genes emerged among environmental bacteria is found worldwide Nevertheless, a viral pandemic has even more rapid effect of infection, usually with symptoms, hence transmission of AMR may lead to colonization and shedding [66], [67], [68], [69]. Restrictions of travel during the pandemic on a massive scale may have a huge impact on slowing the spread of AMR.
Conclusion and outlook
The current pandemic will be remembered as a momentous event in the history of humankind. Eventually, COVID-19 would be an important comparison for defining the development of AMR and illustrating the challenges to contain it, once it has evolved. Unfortunately, a widespread misconception seen among the common masses is that antibiotics like azithromycin, amoxicillin, amoxiclav etc., are effective in treating COVID-19. Additionally, suspected cases that are self-isolating and confirmed asymptomatic COVID-19 cases are reported to use antibiotics. Therefore, it is pertinent to create greater awareness in the public about the consequences of overuse and misuse of antibiotics. The reasonable understanding of appropriate usage of antibiotic would be instrumental in preventing the occurrence of AMR. However, among the emerging infectious diseases, AMR will persist over and above the COVID-19 pandemic. Likewise, the current and other pandemics, AMR too are a problem that “knows no borders’ and is considered as more intriguing problem. Undoubtedly, AMR presents lesser immediate impact on everyday life, though potentially greater and far-reaching adverse effects on humankind. Therefore, management strategies to reduce the emergence of AMR should be investigated and implemented at local and global level.
Remarks
Apart from current pandemic, the clinicians in developing countries have been heavily prescribing antibiotics even for the common cold where they are least or not effective at all. Furthermore, other factors which are considered as a criminal offense in the developed parts of the world, for example, over the counter usage of antibiotics and their prophylactic consumption, are a normal practice in developing regions. This abuse of antibiotics is an additional challenge that needs action from policy makers at national and international institutional level.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Rossen J.W.A., Tedim A.P., Murray A.K. The novel coronavirus COVID-19 outbreak: global implications for antimicrobial resistance. Front Microbiol. 2020;1:1020. doi: 10.3389/fmicb.2020.01020. 〈www.frontiersin.org〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nori P., et al. Bacterial and fungal coinfections in COVID-19 patients hospitalized during the New York City pandemic surge. Infect Control Hosp Epidemiol. 2021;42(1) doi: 10.1017/ice.2020.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rawson T.M., Wilson R.C., Holmes A. Understanding the role of bacterial and fungal infection in COVID-19. Clin Microbiol Infect. 2021;27(1) doi: 10.1016/j.cmi.2020.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Langford B.J., et al. Antibiotic prescribing in patients with COVID-19: rapid review and meta-analysis. Clin Microbiol Infect. 2021;27(4) doi: 10.1016/j.cmi.2020.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Razazi K., et al. Risks of ventilator-associated pneumonia and invasive pulmonary aspergillosis in patients with viral acute respiratory distress syndrome related or not to Coronavirus 19 disease. Crit Care. 2020;24(1) doi: 10.1186/s13054-020-03417-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Council of Canadian Academies. When antibiotics fail. The Expert Panel on the Potential Socio-Economic Impacts of Antimicrobial Resistance in Canada; 2019.
- 7.Talebi Bezmin Abadi A., Rizvanov A.A., Haertlé T., Blatt N.L. World Health Organization report: current crisis of antibiotic resistance. BioNanoScience. 2019;9(4) doi: 10.1007/s12668-019-00658-4. [DOI] [Google Scholar]
- 8.IACG (2019). No Time to Wait: Securing the Future From Drug-Resistant Infections. Report to the Secretary-General of the United Nations. (IACG).
- 9.Mahmoudi H. Bacterial co-infections and antibiotic resistance in patients with COVID-19. GMS Hyg Infect Control. 2020;15 doi: 10.3205/dgkh000370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang B., et al. Quadas-c: a tool for assessing risk of bias in comparative diagnostic accuracy studies. Ann Intern Med. 2021;174(11) doi: 10.7326/M21-2234. [DOI] [PubMed] [Google Scholar]
- 11.Guisado-Gil A.B., et al. Impact of the COVID-19 pandemic on antimicrobial consumption and hospital-acquired candidemia and multidrug-resistant bloodstream infections. Antibiotics. 2020;9(11) doi: 10.3390/antibiotics9110816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Contou D., et al. Bacterial and viral co-infections in patients with severe SARS-CoV-2 pneumonia admitted to a French ICU. Ann Intensive Care. 2020;10(1) doi: 10.1186/s13613-020-00736-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez S., et al. Increase in hospital-acquired carbapenem-resistant Acinetobacter baumannii infection and colonization in an acute care hospital during a surge in COVID-19 admissions — New Jersey, February–July 2020. MMWR Morb Mortal Wkly Rep. 2020;69(48) doi: 10.15585/mmwr.mm6948e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharifipour E., et al. Evaluation of bacterial co-infections of the respiratory tract in COVID-19 patients admitted to ICU. BMC Infect Dis. 2020;20(1) doi: 10.1186/s12879-020-05374-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calderon M., et al. Evaluation of procalcitonin-guided antimicrobial stewardship in patients admitted to hospital with COVID-19 pneumonia. JAC Antimicrob Resist. 2021;3(3) doi: 10.1093/jacamr/dlab133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerra D., et al. Bacterial co-infection and COVID-19 in ICU. Intensive Care Med Exp. 2021;9(SUPPL 1) [Google Scholar]
- 17.Salehi M., et al. Oropharyngeal candidiasis in hospitalised COVID-19 patients from Iran: species identification and antifungal susceptibility pattern. Mycoses. 2020;63(8) doi: 10.1111/myc.13137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J., et al. Etiology and antimicrobial resistance of secondary bacterial infections in patients hospitalized with COVID-19 in Wuhan, China: a retrospective analysis. Antimicrob Resist Infect Control. 2020;9(1) doi: 10.1186/s13756-020-00819-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J., et al. Etiology and antimicrobial resistance of secondary bacterial infections in patients hospitalized with COVID-19 in Wuhan, China: a retrospective analysis. Antimicrob Resist Infect Control. 2020;9(1) doi: 10.1186/s13756-020-00819-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sang L., et al. Secondary infection in severe and critical covid-19 patients in China: a multicenter retrospective study. Ann Palliat Med. 2021;10(8) doi: 10.21037/apm-21-833. [DOI] [PubMed] [Google Scholar]
- 21.Gomez-Simmonds A., et al. Carbapenemase-producing Enterobacterales causing secondary infections during the COVID-19 crisis at a New York City hospital. J Antimicrob Chemother. 2021;76(2) doi: 10.1093/jac/dkaa466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palanisamy N., et al. Clinical profile of bloodstream infections in COVID-19 patients: a retrospective cohort study. BMC Infect Dis. 2021;21(1) doi: 10.1186/s12879-021-06647-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Despotovic A., et al. The impact of covid-19 on the profile of hospital-acquired infections in adult intensive care units. Antibiotics. 2021;10(10) doi: 10.3390/antibiotics10101146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meawed T.E., Ahmed S.M., Mowafy S.M.S., Samir G.M., Anis R.H. Bacterial and fungal ventilator associated pneumonia in critically ill COVID-19 patients during the second wave. J Infect Public Health. 2021;14(10) doi: 10.1016/j.jiph.2021.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeshan B., et al. The usage of antibiotics by COVID-19 patients with comorbidities: the risk of increased antimicrobial resistance. Antibiotics. 2022;11(1) doi: 10.3390/antibiotics11010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caruso P., et al. Antibiotic resistance in diabetic foot infection: how it changed with COVID-19 pandemic in a tertiary care center. Diabetes Res Clin Pract. 2021;175 doi: 10.1016/j.diabres.2021.108797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saini V., et al. Paradigm shift in antimicrobial resistance pattern of bacterial isolates during the covid-19 pandemic. Antibiotics. 2021;10(8) doi: 10.3390/antibiotics10080954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wardoyo E.H., Suardana I.W., Yasa I.W.P.S., Sukrama I.D.M. Antibiotics susceptibility of Escherichia coli isolates from clinical specimens before and during covid-19 pandemic. Iran J Microbiol. 2021;13(2) doi: 10.18502/ijm.v13i2.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karataş M., Yaşar-Duman M., Tünger A., Çilli F., Aydemir Ş., Özenci V. Secondary bacterial infections and antimicrobial resistance in COVID-19: comparative evaluation of pre-pandemic and pandemic-era, a retrospective single center study. Ann Clin Microbiol Antimicrob. 2021;20(1) doi: 10.1186/s12941-021-00454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Temperoni C., Caiazzo L., Barchiesi F. High prevalence of antibiotic resistance among opportunistic pathogens isolated from patients with covid-19 under mechanical ventilation: results of a single-center study. Antibiotics. 2021;10(9) doi: 10.3390/antibiotics10091080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bazaid A.S., et al. Bacterial coinfection and antibiotic resistance profiles among hospitalised COVID-19 patients. Microorganisms. 2022;10(3) doi: 10.3390/microorganisms10030495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pourajam S., et al. Secondary bacterial infection and clinical characteristics in patients with COVID-19 admitted to two intensive care units of an academic hospital in Iran during the first wave of the pandemic. Front Cell Infect Microbiol. 2022;12 doi: 10.3389/fcimb.2022.784130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jamnani A.N., Montazeri M., Mirzakhani M., Moosazadeh M., Haghighi M. Evaluation of bacterial coinfection and antibiotic resistance in patients with COVID-19 under mechanical ventilation. SN Compr Clin Med. 2022;4(1) doi: 10.1007/s42399-021-01114-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bahçe Y.G., Acer Ö., Özüdoğru O. Evaluation of bacterial agents isolated from endotracheal aspirate cultures of Covid-19 general intensive care patients and their antibiotic resistance profiles compared to pre-pandemic conditions. Microb Pathog. 2022;164 doi: 10.1016/j.micpath.2022.105409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boorgula S.Y., Yelamanchili S., Kottapalli P., Naga M.D. An update on secondary bacterial and fungal infections and their antimicrobial resistance pattern (AMR) in COVID-19 confirmed patients. J Lab Phys. 2022;14(03) doi: 10.1055/s-0041-1741438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blair J.M.A., Webber M.A., Baylay A.J., Ogbolu D.O., Piddock L.J.V. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13(1) doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 37.Loh B., Chen J., Manohar P., Yu Y., Hua X., Leptihn S. A biological inventory of prophages in A. baumannii genomes reveal distinct distributions in classes, length, and genomic positions. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.579802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blair J.M.A., Richmond G.E., Piddock L.J.V. Multidrug efflux pumps in Gram-negative bacteria and their role in antibiotic resistance. Future Microbiol. 2014;9(10) doi: 10.2217/FMB.14.66. [DOI] [PubMed] [Google Scholar]
- 39.Chancey S.T., Zähner D., Stephens D.S. Acquired inducible antimicrobial resistance in Gram-positive bacteria. Future Microbiol. 2012;7(8) doi: 10.2217/fmb.12.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abushaheen M.A., et al. Antimicrobial resistance, mechanisms and its clinical significance. Disease-a-Month. 2020;66(6) doi: 10.1016/j.disamonth.2020.100971. [DOI] [PubMed] [Google Scholar]
- 41.Martinez J.L. General principles of antibiotic resistance in bacteria. Drug Discov Today: Technol. 2014;11(1) doi: 10.1016/j.ddtec.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 42.Brown G.D., Denning D.W., Gow N.A.R., Levitz S.M., Netea M.G., White T.C. Hidden killers: human fungal infections. Sci Transl Med. 2012;4(165) doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 43.Song G., Liang G., Liu W. Fungal co-infections associated with global COVID-19 pandemic: a clinical and diagnostic perspective from China. Mycopathologia. 2020;185(4) doi: 10.1007/s11046-020-00462-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Posteraro B., et al. Pan-echinocandin-resistant candida glabrata bloodstream infection complicating covid-19: a fatal case report. J Fungi. 2020;6(3) doi: 10.3390/jof6030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chowdhary A., Tarai B., Singh A., Sharma A. Multidrug-resistant candida auris infections in critically Ill Coronavirus disease patients, India, April–July 2020. Emerg Infect Dis. 2020;26(11) doi: 10.3201/eid2611.203504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mohamed A., et al. Multi-triazole-resistant Aspergillus fumigatus and SARS-CoV-2 co-infection: a lethal combination. Med Mycol Case Rep. 2021;31 doi: 10.1016/j.mmcr.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang D., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA - J Am Med Assoc. 2020;323(11) doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guan W., et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18) doi: 10.1056/nejmoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Denning D.W., et al. High-frequency triazole resistance found in nonculturable Aspergillus fumigatus from lungs of patients with chronic fungal disease. Clin Infect Dis. 2011;52(9) doi: 10.1093/cid/cir179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown G.D., Denning D.W., Gow N.A.R., Levitz S.M., Netea M.G., White T.C. Hidden killers: human fungal infections. Sci Transl Med. 2012;4(165) doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 51.Kim J.Y. Human fungal pathogens: why should we learn. J Microbiol. 2016;54(3) doi: 10.1007/s12275-016-0647-8. [DOI] [PubMed] [Google Scholar]
- 52.Stappers M.H.T., Brown G.D. Candida albicans: cellular and molecular biology: second edition. 2017. Host immune responses during infections with Candida albicans. [DOI] [Google Scholar]
- 53.Najib S.Y., et al. Utilization of physical and chemical microbial load reduction agents for SARS-CoV-2: toxicity and development of drug resistance implications. J Appl Pharm Sci. 2022;2(1) doi: 10.7324/JAPS.2021.120101. [DOI] [Google Scholar]
- 54.Mahoney A.R., Safaee M.M., Wuest W.M., Furst A.L. The silent pandemic: emergent antibiotic resistances following the global response to SARS-CoV-2. iScience. 2021;24(4) doi: 10.1016/j.isci.2021.102304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruan Z., Feng Y. BacWGSTdb, a database for genotyping and source tracking bacterial pathogens. Nucleic Acids Res. 2016;44(D1) doi: 10.1093/nar/gkv1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;55(3) doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Levy S.B. Active efflux, a common mechanism for biocide and antibiotic resistance. J Appl Microbiol Symp Suppl. 2002;92(1) doi: 10.1046/j.1365-2672.92.5s1.4.x. [DOI] [PubMed] [Google Scholar]
- 58.World Health Organization. Including outreach and campaigns, in the context of the COVID-19 pandemic, Who. no. May; 2020.
- 59.Estrich C.G., et al. Estimating COVID-19 prevalence and infection control practices among US dentists. J Am Dent Assoc. 2020;151(11) doi: 10.1016/j.adaj.2020.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.WHO. Including outreach and campaigns, in the context of the COVID-19 pandemic. Who. no. May; 2020.
- 61.WHO. COVID-19: occupational health and safety for health workers. COVID-19: occupational health and safety for health workers, no. February; 2021.
- 62.Liang Z., et al. Molecular basis of NDM-1, a new antibiotic resistance determinant. PLoS One. 2011;6(8) doi: 10.1371/journal.pone.0023606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nordmann P., Poirel L., Walsh T.R., Livermore D.M. The emerging NDM carbapenemases. Trends Microbiol. 2011;19(12) doi: 10.1016/j.tim.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 64.Castanheira M., Griffin M.A., Deshpande L.M., Mendes R.E., Jones R.N., Flamm R.K. Detection of mcr-1 among Escherichia coli clinical isolates collected worldwide as part of the SENTRY Antimicrobial Surveillance Program in 2014 and 2015. Antimicrob Agents Chemother. 2016;60(9) doi: 10.1128/AAC.01267-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Y.Y., et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2) doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 66.Ball T.A., Fedorka-Cray P.J., Horovitz J., Thakur S. Molecular characterization of Salmonella spp. from cattle and chicken farms in Uganda. Online J Public Health Inf. 2018;10(1) doi: 10.5210/ojphi.v10i1.8934. [DOI] [Google Scholar]
- 67.Cantón R., Coque T.M. The CTX-M beta-lactamase pandemic. Curr Opin Microbiol. 2006;9(5) doi: 10.1016/j.mib.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 68.Cantón R., González-Alba J.M., Galán J.C. CTX-M enzymes: origin and diffusion. Front Microbiol. 2012;3(APR) doi: 10.3389/fmicb.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.ÖstholmBalkhed Å., Tärnberg M., Nilsson M., Nilsson L.E., Hanberger H., Hällgren A. Duration of travel-associated faecal colonisation with ESBL-producing enterobacteriaceae – a one year follow-up study. PLoS One. 2018;13(10) doi: 10.1371/journal.pone.0205504. [DOI] [PMC free article] [PubMed] [Google Scholar]