Abstract
Clinical genomic testing is becoming routine in prostate cancer, as biomarker-driven therapies such as poly-ADP ribose polymerase (PARP) inhibitors and anti-PD1 immunotherapy are now approved for select men with castration resistant prostate cancer harboring alterations in DNA repair genes. Challenges for precision medicine in prostate cancer include an overall low prevalence of actionable genomic alterations, and a still limited understanding of the impact of tumor heterogeneity and co-occurring alterations on treatment response and outcomes across diverse patient populations. Expanded tissue-based technologies such as whole genome sequencing, transcriptome analysis, epigenetic analysis, and single-cell RNA sequencing have not yet entered the clinical realm and could potentially improve upon our understanding of how molecular features of tumors, intra-tumoral heterogeneity, and the tumor microenvironment impact therapy response and resistance. Blood-based technologies including cell-free DNA, circulating tumor cells, and extracellular vesicles are less invasive molecular profiling resources that could also help capture intra-individual tumor heterogeneity and track dynamic changes that occur in the context of specific therapies. Furthermore, molecular imaging is an important biomarker tool within the framework of prostate cancer precision medicine with a capability to detect heterogeneity across metastases and potential therapeutic targets less invasively. Here, we review recent technological advances that may help promote the future implementation and value of precision oncology testing for patients with advanced prostate cancer.
Keywords: prostate cancer, precision oncology, molecular profiling, molecular imaging, integrative analysis
Introduction
Advances in sequencing technologies over the last decade have allowed for a comprehensive view of the prostate cancer genome, epigenome, and transcriptome, identifying drivers of prostate cancer initiation and progression and diverse mechanisms of treatment resistance.1 Based on the field’s growing understanding of the prostate cancer genome, predictive biomarkers have translated to clinic practice with two classes of genomically-driven therapies (poly-ADP ribose polymerase (PARP) inhibitors and anti-program cell death protein 1 (PD1) immunotherapy) now FDA-approved for the treatment of select men with metastatic castration resistant prostate cancer (CRPC) harboring specific aberrations in DNA repair genes.
Despite these exciting advances, there are still several barriers that limit the widespread clinical implementation of genomic sequencing, including cost, access, and feasibility based on often limited tissue availability or quality. Further, only a subset of prostate cancers harbor actionable genomic aberrations, and even then not all patients respond durably to genomically-selected therapy.2–4 This may be due to a number of factors including the presence of co-occurring genomic and non-genomic alterations, intra-patient tumor heterogeneity, and the development of acquired resistance. Integrative analyses combining genomics with other features such as transcriptome and epigenome, as well characteristics of the tumor microenvironment, may provide additional insights into identifying patients most likely to benefit from our current therapies and inform the development of novel biomarker-driven and precision therapy approaches for patients. These types of integrative analyses are still maturing and are not yet applied clinically. Moreover, methods for linking large emerging datasets with clinical information will be essential for the more accurate prediction of treatment response.
The study of advanced prostate cancer and tumor evolution in the context of therapy is particularly challenged compared to other tumor types due to the challenges of accessing metastatic tissue given the predominance of sclerotic bone metastases that are often difficult to biopsy and yield limited tumor for molecular studies. In addition, the analysis of a tumor from one region might fail to capture intra-individual tumor heterogeneity. Furthermore, the genomic profiles of bulk tumors may not capture intra-tumoral heterogeneity, which may contribute to drug sensitivity and influence resistance patterns.5–7 Another major barrier in the broad implementation of precision medicine in prostate cancer is our currently limited understanding of the impact of molecular, genetic, and environmental factors across diverse racial and ethnic populations. This should be understood because the incidence of prostate cancer and its mortality rates can vary substantially.8,9 Barriers also exist that that limit the availability of testing and access to targeted therapies worldwide.
In this review, we discuss recent technological advances that may help promote the future implementation and value of precision oncology testing for patients with advanced prostate cancer.
Tissue-based technologies
Since metastatic tissue acquisition in prostate cancer is a challenge, primary archival specimens are often used to assess for actionable alterations involving DNA repair genes, as these alterations tend to be early events in prostate cancer and therefore detectable through primary tumor sequencing analysis.10,11 Mateo et al.10 reported a high concordance of DNA repair alterations when comparing primary prostate cancer and metastatic CRPC tissues. All alterations involving CDK12, BRCA2, ATM, PALB2, and MSH6 found in metastatic CRPC biopsies of 9 of 61 patients were detectable in patient-matched, diagnostic, treatment-naive primary tissues. Conversely, increased AR, TP53, RB1, and PI3K/AKT pathway alterations were enriched in CRPC compared to same-patient primary samples, indicating that these likely emerged later. Schweizer et al.11 reported 84% concordance of DNA repair gene mutations between primary prostate cancer samples and paired metastatic tissue or cell-free DNA (cfDNA) from 51 patients. These data further support DNA repair alterations being early events, and the majority (90%) of patients enrolled on the Phase 3 PROfound trial of olaparib were screened for DNA repair aberrations using targeted exome sequencing of primary archival prostate tissues with an overall sequencing success rate of approximately 70%.12 Therefore, primary tumor targeted sequencing is a viable approach to evaluate for somatic DNA repair aberrations, particularly when metastatic tumor biopsies are not feasible. Circulating tumor DNA (ctDNA) may also be considered a tissue alternative to assess for DNA repair mutations. Tukachinsky et al.13 evaluated ctDNA from 3,334 patients with mCRPC, including 1,674 screening samples from rucaparib trials (TRITON 2 and TRITON 3) using the Foundation Medicine ctDNA targeted assay; 94% had detectable ctDNA (median ctDNA fraction 7.5%). Overall, 72/837 patients had BRCA1/2 mutations detected in tissue, 67 (93%) of which were also identified using ctDNA. Limitations of using ctDNA include decreased sensitivity for detection of copy number alterations (e.g., BRCA2 deletions) and the presence of confounding factors in blood such as clonal hematopoiesis alterations that may also involve DNA repair genes (e.g., ATM). When looking for acquired phenotypic changes in prostate cancer (e.g., histologic transformation) and for studying non-genomic treatment resistance pathways, metastatic biopsy is preferred. Tissue-based sequencing is of particular value when considering expanded molecular profiling for research using emerging technologies such as whole genome sequencing, transcriptome analysis, in situ analyses, or integrative epigenetic studies.
Whole genome sequencing (WGS)
Along with technological advances and an overall decline in sequencing costs, WGS is positioned as a possible future tool to identify more detailed genomic aberrations in prostate cancer including those involving non-coding regions and structural variations. Prostate cancer WGS studies have revealed important alterations not detectable by targeted exome approaches. In CRPC, amplification of an upstream enhancer of the androgen receptor (AR) gene leads to resistance to AR pathway inhibitor (ARPI) therapy.14–17 A comprehensive analysis of nearly 200 patients with metastatic CRPC defined distinct genomic subgroups by WGS, including a cluster with a tandem duplication phenotype that correlated with biallelic CDK12 inactivation and another cluster showing homologous recombination deficiency features with numerous deletions and BRCAness-associated gene alterations.17 In addition, a mutational signature for homologous recombination deficiency may be informative for PARP inhibitor or platinum response even in those that lack DNA repair mutations,18 supporting a possible utility of using expanded genomic analysis over targeted exome profiling in the clinic. Studies in other diseases have shown the feasibility of using WGS in the clinic, including WGS of pediatric cancers.19
Transcriptome analysis
Commercial gene expression classifiers such as Decipher® 20–22 and the Oncotype DX® Genomic Prostate Score23 have demonstrated prognostic value in patients with primary prostate cancer. Transcriptome sequencing is not currently used clinically in patients with advanced metastatic disease. Bolis et al.24 integrated the transcriptional profiles of prostate cancers at various disease stages ranging from normal prostate tissue to primary prostate cancer and metastatic CRPC. Principal component analysis showed transcriptional changes related to different disease stages. They performed trajectory analysis to characterize disease progression and found that most prostate cancers evolve from normal tissue by continuously increasing AR signaling and increasing pseudotime resulted in a gradual upregulation of cell cycle-related genes and concomitant downregulation of androgen-responsive genes. Recently, the PAM50 clustering model based on gene expression data of primary prostate cancers classified prostate cancer patients into three molecular subtypes (luminal A, luminal B, basal)25; the basal subtype (detected in primary prostate tissue) has been associated with inferior outcomes in patient with advanced disease treated with ARPI or docetaxel chemotherapy versus luminal. Ongoing studies are underway to better understand the predictive and prognostic value of these gene expression signatures. Additionally, an integrated neuroendocrine prostate cancer (NEPC) score, based on the expression of a set of 70 genes,26 may identify NEPC tumors or those patients at high risk for the development of NEPC progression after AR-targeted therapies. Treatment-emergent NEPC shares similar genomics as castration resistant adenocarcinoma, indicating a possible utility of transcriptomics over genomics as a prognostic or diagnostic assay in the context of advanced prostate cancer and NEPC. Other gene signatures and mRNA expression-based analyses may complement genomic testing in the future, and biomarker-driven clinical trials incorporating mRNA profiling such as the Alliance CRPC umbrella trial A032102 (PREDICT) are now being launched.
Transcriptome analysis does not always detect cell type composition within the tissue to distinguish tumor cell populations, immune cells, and other cells of the tumor microenvironment. For the analysis of cell type-specific expression profiles in heterogenous samples, computational deconvolution of bulk transcriptome data is required.27 Wu et al.28 examined gene expression data of primary prostate cancer tissue and normal prostate tissue using CIBERSORT, a tool for deconvolution, and analyzed the proportion of 22 immune cells infiltrating in the microenvironment and their prognostic effects in prostate cancer. They found that the detection of M1 macrophages and neutrophils in prostate cancer tissue was associated with poor prognosis. In metastatic CRPC, transcriptome analysis using CIBERSORT has revealed substantial variation in overall immune infiltrate–related transcripts among tumor biopsy sites, as well as heterogeneity in inferred immune cell populations.29 Single-cell RNA sequencing (scRNA-seq) has demonstrated utility in assessing intratumoral heterogeneity at single cell resolution as well as the complexity of the microenvironment.30–32 In a recent study by Chen et al.,33 scRNA-seq revealed that prostate cancer cells modulate infiltrating T cells to express KLK3, which establishes a pre-metastatic niche in lymph nodes. They also showed cancer-associated fibroblast-marker-expressing endothelial cells were enriched in CRPC and promoted cancer cell invasion. These results indicate that the tumor heterogeneity and the microenvironment may play important roles in disease progression and metastasis through close cell-cell communication. Another scRNA-seq analysis of metastatic CRPC revealed that resistance to enzalutamide was associated with cancer cell–intrinsic epithelial–mesenchymal transition and transforming growth factor-β (TGF-β) signaling, suggesting the clinical utility of inhibiting TGF-β.34 In addition, NEPC cells showed divergent expression programs driven by HOXB5, HOXB6 and NR1D2 as well as transcriptional regulators promoting lineage plasticity, which might help inform therapeutic strategies for NEPC. Furthermore, scRNA-seq and single-cell assays for transposase-accessible chromatin (ATAC) sequencing of prostate cancer resistance models revealed pre-existing and treatment-persistent cell subpopulations, which may lead to the prediction of the risk of recurrence and disease progression.35 Spatial transcriptome sequencing is a newly emerged technology, allowing for the profiling and visualization of cells while they remain in their tissues. Understanding cell-cell interactions in a spatial context between tumor cells and within the tumor microenvironment may inform resistance subtypes and therapeutic strategies.36 Brady et al.7 performed digital spatial profiling of metastatic prostate tissues to assess and quantify transcript and protein abundance in spatially-distinct regions tissue specimens. Although there was high intratumor concordance for the status of particular gene expression signatures including AR activity and NEPC-associated genes, they showed the juxtaposition of AR+/NE− and AR−/NE+ tumor phenotypes within the same metastasis. Furthermore, digital spatial profiling revealed that most metastatic tumors are devoid of significant inflammatory infiltrates and express low-to-absent immune checkpoint proteins CTLA4, PD1, and PD-L1, supporting the very low response rates of immune checkpoint inhibitors observed in the majority of prostate cancer patients. Therefore, scRNA-seq and spatial transcriptome analyses could contribute to future precision oncology in prostate cancer by characterizing transcriptional diversity in the tumor and its microenvironment.
Epigenetic analysis
Epigenetic alterations, including changes in DNA methylation and histone modifications, influence gene expression and are key factors driving prostate cancer initiation, progression, and treatment resistance.37 There are several techniques for epigenetic analysis that may be applied to clinical specimens, including bisulfite sequencing, 5-hydroxymethylcytosine (5hmC) sequencing, ATAC sequencing, and chromatin immunoprecipitation (ChIP) sequencing.
Bisulfite sequencing is commonly used technology for profiling DNA methylation with single-base resolution. This method is based on the finding that treatment with sodium bisulfite leads to deamination of unmethylated cytosines into uracils, while methylated cytosines (both 5-methylcytosine [5mC] and 5hmC) remain unchanged.38 As early events in prostate cancer, hypermethylation of the promoter regions of several genes have been reported such as APC, CCND2, GSTP1, RARB2, and RASSF1.39–41 ConfirmMDx is a multiplex epigenetic assay42,43 that combines three methylation regions including GAS6, GSTP1, and HAPLN3 as a classifier for distinguishing prostate cancer from benign tissues.44 In CRPC, unsupervised hierarchical clustering of recurrent hypomethylated regions (rHMRs) identified a novel cluster with significantly higher methylation levels at rHMRs as well as a cluster of treatment-emergent NEPC.45 The novel hypermethylated cluster was enriched for mutations involving TET2, IDH1, BRAF, and DNMT3B. Although DNA methylation changes are usually associated with poor clinical outcomes, methylation of the promoter region of SRD5A2 gene has been correlated with better prognosis in CRPC.46 Further studies exploring the role of DNA methylation in prostate cancer progression could help develop novel strategies for precision oncology biomarkers and therapies in prostate cancer.
5hmC is the first oxidative product of 5mC catalyzed by ten-eleven translocation enzymes and another important epigenetic regulator of transcription.47 Oxidative bisulfite sequencing48 and Tet-assisted bisulfite sequencing49 are both single-base resolution sequencing strategies which distinguish between 5mC and 5hmC. Additionally, several bisulfite-free methods to detect 5hmC at base resolution have recently been developed.50,51 Sjöström et al.52 reported that 5hmC levels in metastatic CRPC associate with gene expression to a greater degree than promoter methylation or copy number, especially in androgen response genes, and 5hmC has the ability to track disease-specific gene activation.
ATAC sequencing is a method for detecting chromatin accessibility across the genome. It uses Tn5 transposase to cut and tag adapters to regions of accessible chromatin which correspond to transcription factor binding sites and nucleosome positioning.53 For instance, integrative analysis of gene expression and ATAC sequencing of CHD1 loss prostate cancer models revealed substantial changes in open and closed chromatin with associated transcriptomic changes, which resulted in the emergence of plasticity via upregulation of transcription factors that promote non-luminal lineage programs.54 ChIP sequencing for histone modification marks such as H3K27ac, H3K4me3, and H3K27me3 might be helpful for detecting distinct prostate cancer subtypes55 and/or other epigenetic events associated with prostate cancer progression.56
Blood-based technologies
In order to overcome some of the current limitations associated with tumor biopsies, liquid biopsies including analysis of ctDNA, circulating tumor cells (CTCs), and extracellular vesicles (EVs) in the blood are emerging as noninvasive tools for precision oncology.
Circulating tumor DNA
ctDNA is well recognized as an important biomarker tool in several cancer types.57–59 In prostate cancer, ctDNA tumor fraction in cfDNA is prognostic60 and ctDNA is capable of detecting common recurrent prostate cancer aberrations in metastatic CRPC including those involving DNA repair genes, with high concordance with matched tissue biopsies.13,61–66 Additionally, rapid autopsy studies have shown that ctDNA can capture more driver alterations than multiple randomly selected tissue samples, indicating its utility in detecting intra-patient tumor heterogeneity.67 In practice, several commercial targeted ctDNA platforms are available to assess for targetable aberrations in CRPC, particularly in cases when tumor tissue is not available and a new biopsy is not feasible to obtain. While ctDNA is convenient and noninvasive, there are limitations. ctDNA analysis may detect aberrations involving both the tumor as well as other cells in the circulation, such as white blood cells. Normal leukocytes harboring clonal hematopoiesis of indeterminate potential (CHIP) variants may confound ctDNA test interpretation, which is especially relevant if they involve BRCA or ATM or other genes linked to PARP inhibitor approval.66,68,69 Additionally, ctDNA is diluted by cfDNA from non-cancer cells and declines with therapy response such that not all patients will have detectable ctDNA, leading to difficulties with the identification of mutations and copy number aberrations.66,70 Further studies exploring baseline and dynamic changes in ctDNA are needed to validate ctDNA as prognostic and response biomarker and will provide additional insights into the evolution of specific clonal and subclonal lesions that occur during prostate cancer disease progression and in the context of therapies.
cfDNA methylation analysis of plasma is not currently used clinically in prostate cancer, but is being developed across cancer types for early detection strategies and tumor classification.71,72 In prostate cancer, dynamics in cytosine modification profiles of cfDNA has been shown to be a predictive biomarker for abiraterone treatment response.73 Additionally, Mahon et al.73 showed that undetectable methylated GSTP1 is a favorable prognostic biomarker in metastatic CRPC. More recently, whole genome bisulfite sequencing analysis of cfDNA from NEPC patients revealed that the methylation patterns detected in cfDNA reflected those observed in biopsy tissues,74 which include NEPC-associated epigenetic changes such as hypermethylation of ASXL3 and SPDEF and hypomethylation of INSM1 and CDH2.26 In a recent study by Berchuck et al,75 a NEPC Risk Score was developed using methylated DNA immunoprecipitation coupled with next-generation sequencing (MeDIP-seq) to predict the presence of NEPC using differentially methylated regions detected from NEPC and CRPC tumor samples. MeDIP-seq was then applied to cfDNA which showed that this tissue-informed analysis resulted in high sensitivity and specificity for detecting NEPC. Overall, these results support the possible utility of using cfDNA methylation as a monitoring tool which may be particularly relevant when detecting epigenetically driven subtypes of advanced disease such as NEPC.
Recently, it has been reported that cfDNA fragment characteristics can also help infer nucleosome positioning and transcription factor binding sites.76 Ulz et al.77 developed an accessibility score to estimate transcription factor activity based on cfDNA sequencing and nucleosome footprint analysis. They analyzed two cfDNA samples from a prostate cancer patient collected in a 12-month interval, during which the adenocarcinoma transdifferentiated to a treatment-emergent NEPC and showed reduced accessibilities of the binding sites of AR, HOXB13, NKX3–1, and REST, indicating that the accessibility score can distinguish NEPC from prostate adenocarcinoma. cfDNA fragment analysis has also been feasible in other cancer types such as early-stage colorectal cancer.77 The analysis of nucleosome positioning in cfDNA could overcome some of the limitations of mutation-based ctDNA analysis with a potentially higher detection sensitivity.59
Circulating tumor cells
CTCs offer not only quantitative information but also the ability to isolate heterogenous cell populations, quantify gene expression, detect splice variants, and measure specific protein expression. In prostate cancer, the enumeration of CTCs has been shown to be a prognostic biomarker,78,79 and the detection of AR splice variant 7 (AR-V7) in CTCs may predict resistance to abiraterone and enzalutamide.80,81 The PROPHECY study, a multicenter, prospective-blinded clinical trial, investigated the impact of CTC AR-V7 detection in men with metastatic CRPC starting ARPI treatment on progression-free survival (PFS) and OS.82 Detection of AR-V7 in CTCs by two assays was significantly associated with shorter PFS (median PFS 3.1 vs 6.9 months and 3.1 vs 6.1 months, respectively) and OS (median OS 10.8 vs 27.2 months and 8.4 vs 25.5 months, respectively). On the other hand, a recent study detected the transcriptional profile of CTCs from metastatic prostate cancer patients using a multiplex gene expression biomarker panel including AR splice variants, AR targets, and NEPC markers.83 The result showed that increased expression of AR-regulated genes was independently associated with shorter OS on multivariate analysis, while AR splice variant status was not significant. Additionally, Scher et al.84 quantified digital pathology features of CTCs from 179 metastatic CRPC patients. They classified individual CTCs into 15 phenotypic subtypes and revealed that low CTC phenotypic heterogeneity was associated with better OS in patients treated with ARPIs (median OS 28.1 vs. 8.8 months), whereas patients with an increasing heterogeneity score had a higher risk of death on ARPIs relative to taxane chemotherapy. In recent years, several single-cell analyses of CTC have revealed prostate tumor heterogeneity that could contribute to patients’ prognosis.85–87 Miyamoto et al.85 conducted scRNA-seq of 77 CTCs from 13 CRPC patients and showed the activation of noncanonical WNT signaling was associated with resistance to ARPIs. Conteduca et al.87 reported a patient with both primary prostate adenocarcinoma and liver metastasis with NEPC morphology at the time of initial presentation, whose CTCs reflected the state of intra-patient tumor heterogeneity. This supports the promise of CTCs in representing the molecular profiles of metastases, though much still remain to be learned about the origin of CTCs and how well they reflect the molecular landscape across heterogeneous tumors. Conteduca et al. performed single-cell copy-number variation analysis of CTCs and found copy-number heterogeneity involving tumor suppressor genes, such as RB1, TP53, and PTEN, associated with differential detection of AR and NEPC marker protein expression in CTCs87. These states of heterogeneity were highly similar to those observed in tumor biopsies, indicating the feasibility of extending CTC analysis at the single-cell level to integrate genomics with protein expression. Furthermore, drug sensitivity testing of ex vivo cultured CTCs could contribute to future precision oncology efforts.88
Extracellular vesicles (EVs)
EVs are secreted by cells and detected in almost every biological fluid, especially blood. There has been growing interest in cancer EVs due to their unique functions as intercellular messengers and their diagnostic and therapeutic potential.89 EVs may serve as biomarkers for the early diagnosis of prostate cancer90 and for the detection of advanced disease.91 Additionally, exosomal specific microRNAs and exosomal AR-V7 have been shown to be potential prognostic biomarkers and for prediction of ARPI response in CRPC patients.92,93 The commonly used techniques for isolation of EVs include ultracentrifugation, size-exclusion chromatography, ultrafiltration, and immunoaffinity capture.94 However, due to their small diameter and the co-existence of different types of vesicles, their isolation is challenging. Furthermore, since the current isolation technologies are usually time-intensive, the development of reliable and efficient isolation procedures would be mandatory for the clinical applications of EVs.
Molecular imaging
Molecular imaging allows for the visualization and quantification of specific markers or biological processes across anatomic disease sites.95 18F-Fluciclovine positron emission tomography (PET) with computed tomography (CT) imaging detects amino acid transporter upregulation in prostate cancer versus surrounding tissues, and was FDA approved in 2016 for the detection of recurrent prostate cancer.96 Prostate specific membrane antigen (PSMA) PET imaging is now recognized for its even higher sensitivity and specificity for identifying prostate cancer recurrence and metastasis97 and is rapidly replacing 18F-Fluciclovine PET. In 2020, the FDA approved Ga-PSMA-11 PET/CT for the initial diagnosis and staging of prostate cancer patients with suspected metastases and the imaging of patients with biochemical recurrence after prostatectomy or radiation therapy. Piflufolastat F 18 was also approved as the second PSMA-targeted PET imaging agent in 2021 for the same prostate cancer imaging indications as Ga-PSMA-11.
In metastatic CRPC, PSMA/PET imaging is also useful to identify candidates for the PSMA-directed radionuclide therapy Lu-PSMA-617. The Phase III VISION trial comparing Lu-PSMA-617 plus standard of care versus standard of care alone improved progression free survival and overall survival for men with metastatic CRPC previously treated with ARPI and taxane chemotherapy. All patients had PSMA-positive disease identified by Ga-PSMA-11 PET/CT. In the VISION trial, PSMA positivity was defined as at least one PSMA-positive metastatic lesion with PSMA uptake greater than liver, and no PSMA negative soft tissue or visceral lesions ≥1cm or lymph nodes ≥2.5cm. Lu-PSMA-617 was approved by the FDA in March 2022 for patients with metastatic CRPC after progression on ARPI and docetaxel, and Ga-PSMA-11 was also approved as a companion diagnostic imaging test. This will expand the number of patients with advanced disease receiving PSMA PET/CT scans. Understanding how baseline PSMA-PET correlates with clinical features, PSMA dynamics on therapy, and PSMA PET characteristics at progression may help refine PSMA PET as biomarker in the context of Lu-PSMA-617. Patterns at progression may influence future sequencing of other PSMA-targeted drugs in development. Beyond targeting PSMA, PSMA PET/CT may also be useful in assessing tumor dynamics and response in the context of other prostate cancer therapies such as ARPI and chemotherapy. PSMA expression is indirectly regulated by the AR, and a subset of CRPC tumors may lose PSMA expression in later stages of the disease in conjunction with loss of AR. In the VISION trial, 12.6% did not meet inclusion criteria based on PSMA-imaging. Several studies incorporating dual-tracer PET/CT have reported that metastatic CRPC patients with low PSMA expression or PSMA-negative fluorodeoxyglucose (FDG)-positive discordant lesions have poor prognosis.98,99 This could be due to NEPC transformation or AR-negative prostate cancer, which is supported by a recent study showing the positive correlation between levels of FDG uptake-associated genes with NEPC gene signatures in PSMA-suppressed tumors.100 In addition, Wang et al.101 also demonstrated that 24% of PSMA-negative, FDG-positive disease was found in patients with an early PSA progression during castration. These results suggest that dual-tracer PET/CT might enable the earlier diagnosis of metabolically active PSMA-suppressed disease for earlier or more aggressive management. Fluorodihydrotestosterone F18 ([18F]-FDHT) PET/CT directly images AR-expressing tissues.102 A recent study analyzing 133 metastatic CRPC patients using molecular imaging with FDHT and FDG PET/CT also showed that 49% of patients had at least one FDHT-negative, FDG-positive lesion, which was the most potent imaging phenotype with respect to adverse prognosis.103 Therefore, dual-tracer with FDHT and FDG imaging could also be a future diagnostic or prognostic biomarker.
The presence of PSMA-negative or AR-negative CRPC lesions may lead to the suspicion of treatment emergent NEPC differentiation, but it is not a strategy to uniquely identify NEPC. Recently, Puca et al.104 found that delta-like protein 3 (DLL3), which is an inhibitory ligand of the Notch signaling pathway,105 is aberrantly expressed on the cell surface of the majority of NEPC.104 DLL3 is also aberrantly expressed in small cell lung cancer (SCLC). ImmunoPET imaging with 89Zr-labeled SC16 antibody is capable of detecting DLL3 positive SCLC and NEPC in preclinical models.106,107 Korsen et al.107 performed in vivo 89Zr-SC16 PET imaging and biodistribution studies using xenograft models of NCI-H660, which is a DLL3-positive NEPC cell line, and DU145, which is a DLL3-negative AR independent prostate cancer cell line. They showed 89Zr-SC16 PET imaging can uniquely detect NEPC lesions, indicating that this technology might be useful for the early detection of NEPC in the future and for selection for DLL3-targeted therapies such as T cell engagers.
Although there are several barriers for translating novel molecular imaging tools into daily clinical practice including expense and time to validate novel tracers, a lack of established framework for multicenter trials, and variable quality of imaging acquisition and analysis,108 molecular imaging may play an important role in future precision oncology in prostate cancer and has great potential to guide more effective and less invasive target detection.
Data integration
Most clinical data as well as molecular information are not well integrated, which provides cumbersome datasets. Since genes, transcripts, proteins, metabolites, and other molecules interact with each other to regulate cellular processes, integrative analysis of multi-omics data is needed for better disease classification, prediction of biomarkers, and understanding of disease biology.109 Ramazzotti et al.110 established a new cancer subtyping method integrating multi-omics data, called Cancer Integration via Multikernel Learning (CIMLR). They applied CIMLR to multi-omics data from 36 cancer types including 490 primary prostate tumors. CIMLR found three clusters in prostate cancer, and one cluster showed significantly worse outcomes compared to the other clusters, characterized by loss of TRIM35, reduced expression of RHOBTB2, high promoter methylation, and high prevalence of TP53 mutation and/or loss.
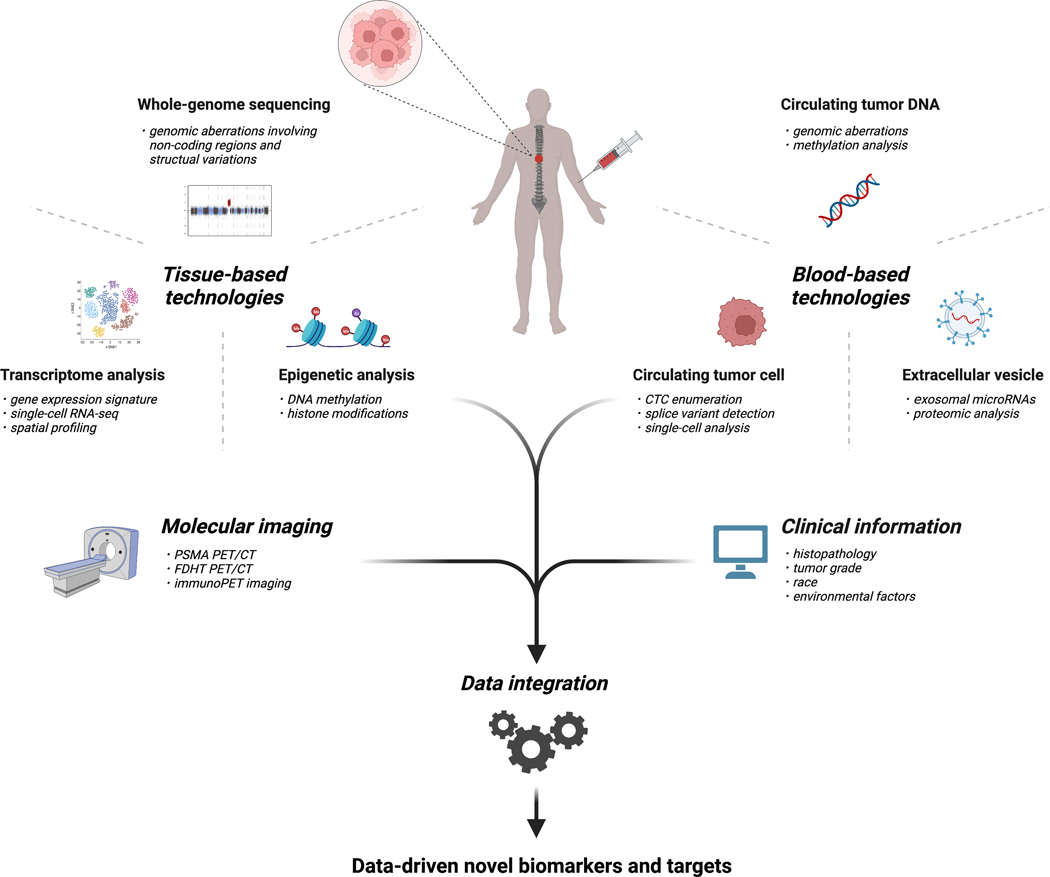
Deep learning-based multi-omics data integration has also been developed.111,112 In prostate cancer, a recent study investigated the Cancer Genome Atlas (TCGA) prostate adenocarcinoma dataset using deep learning and similarity network fusion.113 From the two models, six multi-omics biomarkers, TELO2, ZMYND19, miR-143, miR-378a, and methylation status of two CpG loci, were selected for multi-omics panel construction. This panel was shown to be a potential biomarker for the early detection of prostate cancer patients at high recurrence risk. Elmarakeby et al.114 developed a deep-learning predictive model named P-NET to predict cancer state in prostate cancer patients on the basis of biological information including mutations, copy number alterations, methylation, and gene expression. P-NET accurately classified metastatic CRPC versus primary prostate cancers. Moreover, this visible neural network model revealed novel alterations which strongly contributed to predictive performance in genes, such as MDM4 and FGFR1. Recently, other novel computational methods have been applied to multi-omics integrative methods,115 including models incorporating histopathology imaging,116 although there are few multimodal studies to date incorporating radiology. Deep learning systems have demonstrated high proficiency at Gleason grading of prostate biopsy specimens117 and have provided support for computational three-dimensional histology analysis.118 Advances in computational methods will further enable the integration of multimodal data including molecular data, histopathology, radiology, and clinical data such as race/ethnicity, tumor grade, recurrence, treatment response, and long term outcomes. This could lead to the development of data-driven novel biomarkers for prostate cancer and a better understanding of its complex nature (Figure).
Figure.
Potential workflow for the future implementation of precision oncology testing for patients with advanced prostate cancer. Tools for implementing precision oncology in advanced prostate cancer include tissue-based technologies, blood-based technologies, and molecular imaging. Clinical information as well as molecular features obtained from the novel technologies are integrated for the detection and application of novel biomarkers and targets.
Conclusions
Precision oncology in prostate cancer is a rapidly evolving field. However, there remain substantial challenges for implementing precision oncology more effectively and more broadly. We have focused on novel technologies and findings that could be used to overcome certain barriers in the advanced prostate cancer setting. Multiple different layers of information including genome, transcriptome, and epigenome could help refine predictive biomarkers and define subclasses that will better predict patient outcomes. The integration of these multi-omics data and clinical information with computational methods including deep learning will also be important. Additionally, integrative analysis might help understand the clinical impact of co-occurring alterations and rare molecular aberrations, leading to broader application of precision oncology for prostate cancer patients. Liquid biopsies and molecular imaging are less invasive technologies that can capture intra-individual heterogeneity, which could affect the treatment response and prognosis. Several biomarker-driven targets are emerging in prostate cancer and clinical trials evaluating the agents against these targets are ongoing. The tremendous progress in the field has only been possible because of large multidisciplinary collaboration and patient engagement.
Acknowledgements.
The figure was created with BioRender.com.
Conflict of Interest
H.B. has served as consultant/advisory board member for Janssen, Astellas, Astra Zeneca, Merck, Pfizer, Foundation Medicine, Blue Earth Diagnostics, Amgen, Oncorus, LOXO, Daicchi Sankyo and has received research funding from Janssen, AbbVie/Stemcentrx, Eli Lilly, Millennium Pharmaceuticals, Bristol Myers Squibb.
Funding
K.M. is supported by The Uehara Memorial Foundation Fellowship. H.B. is supported by the Prostate Cancer Foundation, National Cancer Institute R37 CA241486-01A1, and Department of Defense PCRP W81XWH-17-1-0653.
REFERENCES
- 1.Koboldt DC, Steinberg KM, Larson DE, Wilson RK, Mardis ER. The next-generation sequencing revolution and its impact on genomics. Cell. Sep 26 2013;155(1):27–38. doi: 10.1016/j.cell.2013.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stockley TL, Oza AM, Berman HK, et al. Molecular profiling of advanced solid tumors and patient outcomes with genotype-matched clinical trials: the Princess Margaret IMPACT/COMPACT trial. Genome Medicine. 2016;8(1)doi: 10.1186/s13073-016-0364-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Massard C, Michiels S, Ferté C, et al. High-Throughput Genomics and Clinical Outcome in Hard-to-Treat Advanced Cancers: Results of the MOSCATO 01 Trial. Cancer Discovery. 2017;7(6):586–595. doi: 10.1158/2159-8290.cd-16-1396 [DOI] [PubMed] [Google Scholar]
- 4.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nature Medicine. 2017;23(6):703–713. doi: 10.1038/nm.4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nature Reviews Clinical Oncology. 2018;15(2):81–94. doi: 10.1038/nrclinonc.2017.166 [DOI] [PubMed] [Google Scholar]
- 6.Marusyk A, Janiszewska M, Polyak K. Intratumor Heterogeneity: The Rosetta Stone of Therapy Resistance. Cancer Cell. Apr 13 2020;37(4):471–484. doi: 10.1016/j.ccell.2020.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brady L, Kriner M, Coleman I, et al. Inter- and intra-tumor heterogeneity of metastatic prostate cancer determined by digital spatial gene expression profiling. Nature Communications. 2021;12(1)doi: 10.1038/s41467-021-21615-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsing AW, Yeboah E, Biritwum R, et al. High prevalence of screen detected prostate cancer in West Africans: implications for racial disparity of prostate cancer. J Urol. Sep 2014;192(3):730–5. doi: 10.1016/j.juro.2014.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dess RT, Hartman HE, Mahal BA, et al. Association of Black Race With Prostate Cancer–Specific and Other-Cause Mortality. JAMA Oncology. 2019;5(7):975. doi: 10.1001/jamaoncol.2019.0826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mateo J, Seed G, Bertan C, et al. Genomics of lethal prostate cancer at diagnosis and castration resistance. Journal of Clinical Investigation. 2020;130(4):1743–1751. doi: 10.1172/jci132031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schweizer MT, Sivakumar S, Tukachinsky H, et al. Concordance of DNA Repair Gene Mutations in Paired Primary Prostate Cancer Samples and Metastatic Tissue or Cell-Free DNA. JAMA Oncology. 2021;7(9):1378. doi: 10.1001/jamaoncol.2021.2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hussain M, Mateo J, Fizazi K, et al. Survival with Olaparib in Metastatic Castration-Resistant Prostate Cancer. New England Journal of Medicine. 2020;383(24):2345–2357. doi: 10.1056/nejmoa2022485 [DOI] [PubMed] [Google Scholar]
- 13.Tukachinsky H, Madison RW, Chung JH, et al. Genomic Analysis of Circulating Tumor DNA in 3,334 Patients with Advanced Prostate Cancer Identifies Targetable BRCA Alterations and AR Resistance Mechanisms. Clin Cancer Res. Jun 1 2021;27(11):3094–3105. doi: 10.1158/1078-0432.CCR-20-4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Viswanathan SR, Ha G, Hoff AM, et al. Structural Alterations Driving Castration-Resistant Prostate Cancer Revealed by Linked-Read Genome Sequencing. Cell. Jul 12 2018;174(2):433–447 e19. doi: 10.1016/j.cell.2018.05.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quigley DA, Dang HX, Zhao SG, et al. Genomic Hallmarks and Structural Variation in Metastatic Prostate Cancer. Cell. Jul 26 2018;174(3):758–769 e9. doi: 10.1016/j.cell.2018.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeda DY, Spisak S, Seo JH, et al. A Somatically Acquired Enhancer of the Androgen Receptor Is a Noncoding Driver in Advanced Prostate Cancer. Cell. Jul 12 2018;174(2):422–432 e13. doi: 10.1016/j.cell.2018.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Dessel LF, Van Riet J, Smits M, et al. The genomic landscape of metastatic castration-resistant prostate cancers reveals multiple distinct genotypes with potential clinical impact. Nature Communications. 2019;10(1)doi: 10.1038/s41467-019-13084-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Sarkar N, Dasgupta S, Chatterjee P, et al. Genomic attributes of homology-directed DNA repair deficiency in metastatic prostate cancer. JCI Insight. 2021;6(23)doi: 10.1172/jci.insight.152789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rusch M, Nakitandwe J, Shurtleff S, et al. Clinical cancer genomic profiling by three-platform sequencing of whole genome, whole exome and transcriptome. Nature Communications. 2018;9(1)doi: 10.1038/s41467-018-06485-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erho N, Crisan A, Vergara IA, et al. Discovery and Validation of a Prostate Cancer Genomic Classifier that Predicts Early Metastasis Following Radical Prostatectomy. PLoS ONE. 2013;8(6):e66855. doi: 10.1371/journal.pone.0066855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jairath NK, Dal Pra A, Vince R Jr., et al. A Systematic Review of the Evidence for the Decipher Genomic Classifier in Prostate Cancer. Eur Urol. Mar 2021;79(3):374–383. doi: 10.1016/j.eururo.2020.11.021 [DOI] [PubMed] [Google Scholar]
- 22.Feng FY, Huang HC, Spratt DE, et al. Validation of a 22-Gene Genomic Classifier in Patients With Recurrent Prostate Cancer: An Ancillary Study of the NRG/RTOG 9601 Randomized Clinical Trial. JAMA Oncol. Apr 1 2021;7(4):544–552. doi: 10.1001/jamaoncol.2020.7671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brooks MA, Thomas L, Magi-Galluzzi C, et al. GPS Assay Association With Long-Term Cancer Outcomes: Twenty-Year Risk of Distant Metastasis and Prostate Cancer–Specific Mortality. JCO Precision Oncology. 2021;(5):442–449. doi: 10.1200/po.20.00325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolis M, Bossi D, Vallerga A, et al. Dynamic prostate cancer transcriptome analysis delineates the trajectory to disease progression. Nature Communications. 2021;12(1)doi: 10.1038/s41467-021-26840-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao SG, Chang SL, Erho N, et al. Associations of Luminal and Basal Subtyping of Prostate Cancer With Prognosis and Response to Androgen Deprivation Therapy. JAMA Oncology. 2017;3(12):1663. doi: 10.1001/jamaoncol.2017.0751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beltran H, Prandi D, Mosquera JM, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. Mar 2016;22(3):298–305. doi: 10.1038/nm.4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Avila Cobos F, Vandesompele J, Mestdagh P, De Preter K. Computational deconvolution of transcriptomics data from mixed cell populations. Bioinformatics. Jun 1 2018;34(11):1969–1979. doi: 10.1093/bioinformatics/bty019 [DOI] [PubMed] [Google Scholar]
- 28.Wu Z, Chen H, Luo W, et al. The Landscape of Immune Cells Infiltrating in Prostate Cancer. Front Oncol. 2020;10:517637. doi: 10.3389/fonc.2020.517637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nava Rodrigues D, Rescigno P, Liu D, et al. Immunogenomic analyses associate immunological alterations with mismatch repair defects in prostate cancer. Journal of Clinical Investigation. 2018;128(10):4441–4453. doi: 10.1172/jci121924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tirosh I, Izar B, Prakadan SM, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352(6282):189–196. doi: 10.1126/science.aad0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puram SV, Tirosh I, Parikh AS, et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell. Dec 14 2017;171(7):1611–1624 e24. doi: 10.1016/j.cell.2017.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roerink SF, Sasaki N, Lee-Six H, et al. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature. 2018;556(7702):457–462. doi: 10.1038/s41586-018-0024-3 [DOI] [PubMed] [Google Scholar]
- 33.Chen S, Zhu G, Yang Y, et al. Single-cell analysis reveals transcriptomic remodellings in distinct cell types that contribute to human prostate cancer progression. Nature Cell Biology. 2021;23(1):87–98. doi: 10.1038/s41556-020-00613-6 [DOI] [PubMed] [Google Scholar]
- 34.He MX, Cuoco MS, Crowdis J, et al. Transcriptional mediators of treatment resistance in lethal prostate cancer. Nature Medicine. 2021;27(3):426–433. doi: 10.1038/s41591-021-01244-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taavitsainen S, Engedal N, Cao S, et al. Single-cell ATAC and RNA sequencing reveal pre-existing and persistent cells associated with prostate cancer relapse. Nature Communications. 2021;12(1)doi: 10.1038/s41467-021-25624-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grauel AL, Nguyen B, Ruddy D, et al. TGFβ-blockade uncovers stromal plasticity in tumors by revealing the existence of a subset of interferon-licensed fibroblasts. Nature Communications. 2020;11(1)doi: 10.1038/s41467-020-19920-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conteduca V, Hess J, Yamada Y, Ku S-Y, Beltran H. Epigenetics in prostate cancer: clinical implications. Translational Andrology and Urology. 2021;10(7):3104–3116. doi: 10.21037/tau-20-1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frommer M, McDonald LE, Millar DS, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. Mar 1 1992;89(5):1827–31. doi: 10.1073/pnas.89.5.1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bastian PJ, Yegnasubramanian S, Palapattu GS, et al. Molecular Biomarker in Prostate Cancer: The Role of CpG Island Hypermethylation. European Urology. 2004;46(6):698–708. doi: 10.1016/j.eururo.2004.07.022 [DOI] [PubMed] [Google Scholar]
- 40.Kang GH, Lee S, Lee HJ, Hwang KS. Aberrant CpG island hypermethylation of multiple genes in prostate cancer and prostatic intraepithelial neoplasia. The Journal of Pathology. 2004;202(2):233–240. doi: 10.1002/path.1503 [DOI] [PubMed] [Google Scholar]
- 41.Henrique R, Jerónimo C, Teixeira MR, et al. Epigenetic Heterogeneity of High-Grade Prostatic Intraepithelial Neoplasia: Clues for Clonal Progression in Prostate Carcinogenesis. Molecular Cancer Research. 2006;4(1):1–8. doi: 10.1158/1541-7786.mcr-05-0113 [DOI] [PubMed] [Google Scholar]
- 42.Stewart GD, Van Neste L, Delvenne P, et al. Clinical Utility of an Epigenetic Assay to Detect Occult Prostate Cancer in Histopathologically Negative Biopsies: Results of the MATLOC Study. Journal of Urology. 2013;189(3):1110–1116. doi: 10.1016/j.juro.2012.08.219 [DOI] [PubMed] [Google Scholar]
- 43.Partin AW, Van Neste L, Klein EA, et al. Clinical Validation of an Epigenetic Assay to Predict Negative Histopathological Results in Repeat Prostate Biopsies. Journal of Urology. 2014;192(4):1081–1087. doi: 10.1016/j.juro.2014.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patel PG, Wessel T, Kawashima A, et al. A three-gene DNA methylation biomarker accurately classifies early stage prostate cancer. Prostate. Oct 2019;79(14):1705–1714. doi: 10.1002/pros.23895 [DOI] [PubMed] [Google Scholar]
- 45.Zhao SG, Chen WS, Li H, et al. The DNA methylation landscape of advanced prostate cancer. Nature Genetics. 2020;52(8):778–789. doi: 10.1038/s41588-020-0648-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ge R, Wang Z, Montironi R, et al. Epigenetic modulations and lineage plasticity in advanced prostate cancer. Ann Oncol. Apr 2020;31(4):470–479. doi: 10.1016/j.annonc.2020.02.002 [DOI] [PubMed] [Google Scholar]
- 47.Song CX, Szulwach KE, Fu Y, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. Jan 2011;29(1):68–72. doi: 10.1038/nbt.1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Booth MJ, Ost TW, Beraldi D, et al. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nat Protoc. Oct 2013;8(10):1841–51. doi: 10.1038/nprot.2013.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu M, Hon GC, Szulwach KE, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. Jun 8 2012;149(6):1368–80. doi: 10.1016/j.cell.2012.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y, Siejka-Zielińska P, Velikova G, et al. Bisulfite-free direct detection of 5-methylcytosine and 5-hydroxymethylcytosine at base resolution. Nat Biotechnol. Apr 2019;37(4):424–429. doi: 10.1038/s41587-019-0041-2 [DOI] [PubMed] [Google Scholar]
- 51.Sun Z, Vaisvila R, Hussong LM, et al. Nondestructive enzymatic deamination enables single-molecule long-read amplicon sequencing for the determination of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Genome Res. Jan 19 2021;31(2):291–300. doi: 10.1101/gr.265306.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sjöström M, Zhao S, Small EJ, et al. 5-hydroxymethylcytosine as a liquid biopsy biomarker in mCRPC. Journal of Clinical Oncology. 2021/02/20 2021;39(6_suppl):148–148. doi: 10.1200/JCO.2021.39.6_suppl.148 [DOI] [Google Scholar]
- 53.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. Dec 2013;10(12):1213–8. doi: 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Z, Zhou C, Li X, et al. Loss of CHD1 Promotes Heterogeneous Mechanisms of Resistance to AR-Targeted Therapy via Chromatin Dysregulation. Cancer Cell. 2020;37(4):584–598.e11. doi: 10.1016/j.ccell.2020.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stelloo S, Nevedomskaya E, Kim Y, et al. Integrative epigenetic taxonomy of primary prostate cancer. Nature Communications. 2018;9(1)doi: 10.1038/s41467-018-07270-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pomerantz MM, Qiu X, Zhu Y, et al. Prostate cancer reactivates developmental epigenomic programs during metastatic progression. Nature Genetics. 2020;52(8):790–799. doi: 10.1038/s41588-020-0664-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwarzenbach H, Hoon DSB, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nature Reviews Cancer. 2011;11(6):426–437. doi: 10.1038/nrc3066 [DOI] [PubMed] [Google Scholar]
- 58.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. Aug 2013;10(8):472–84. doi: 10.1038/nrclinonc.2013.110 [DOI] [PubMed] [Google Scholar]
- 59.Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nature Reviews Cancer. 2017;17(4):223–238. doi: 10.1038/nrc.2017.7 [DOI] [PubMed] [Google Scholar]
- 60.Choudhury AD, Werner L, Francini E, et al. Tumor fraction in cell-free DNA as a biomarker in prostate cancer. JCI Insight. Nov 2 2018;3(21)doi: 10.1172/jci.insight.122109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Romanel A, Gasi Tandefelt D, Conteduca V, et al. Plasma AR and abiraterone-resistant prostate cancer. Sci Transl Med. Nov 4 2015;7(312):312re10. doi: 10.1126/scitranslmed.aac9511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Azad AA, Volik SV, Wyatt AW, et al. Androgen Receptor Gene Aberrations in Circulating Cell-Free DNA: Biomarkers of Therapeutic Resistance in Castration-Resistant Prostate Cancer. Clinical Cancer Research. 2015;21(10):2315–2324. doi: 10.1158/1078-0432.ccr-14-2666 [DOI] [PubMed] [Google Scholar]
- 63.Wyatt AW, Azad AA, Volik SV, et al. Genomic Alterations in Cell-Free DNA and Enzalutamide Resistance in Castration-Resistant Prostate Cancer. JAMA Oncol. Dec 1 2016;2(12):1598–1606. doi: 10.1001/jamaoncol.2016.0494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Annala M, Vandekerkhove G, Khalaf D, et al. Circulating Tumor DNA Genomics Correlate with Resistance to Abiraterone and Enzalutamide in Prostate Cancer. Cancer Discov. Apr 2018;8(4):444–457. doi: 10.1158/2159-8290.CD-17-0937 [DOI] [PubMed] [Google Scholar]
- 65.Sumiyoshi T, Mizuno K, Yamasaki T, et al. Clinical utility of androgen receptor gene aberrations in circulating cell-free DNA as a biomarker for treatment of castration-resistant prostate cancer. Sci Rep. Mar 11 2019;9(1):4030. doi: 10.1038/s41598-019-40719-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mizuno K, Sumiyoshi T, Okegawa T, et al. Clinical Impact of Detecting Low-Frequency Variants in Cell-Free DNA on Treatment of Castration-Resistant Prostate Cancer. Clinical Cancer Research. 2021;27(22):6164–6173. doi: 10.1158/1078-0432.ccr-21-2328 [DOI] [PubMed] [Google Scholar]
- 67.Pereira B, Chen CT, Goyal L, et al. Cell-free DNA captures tumor heterogeneity and driver alterations in rapid autopsies with pre-treated metastatic cancer. Nature Communications. 2021;12(1)doi: 10.1038/s41467-021-23394-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu Y, Ulrich BC, Supplee J, et al. False-Positive Plasma Genotyping Due to Clonal Hematopoiesis. Clinical Cancer Research. 2018;24(18):4437–4443. doi: 10.1158/1078-0432.ccr-18-0143 [DOI] [PubMed] [Google Scholar]
- 69.Jensen K, Konnick EQ, Schweizer MT, et al. Association of Clonal Hematopoiesis in DNA Repair Genes With Prostate Cancer Plasma Cell-free DNA Testing Interference. JAMA Oncol. Jan 1 2021;7(1):107–110. doi: 10.1001/jamaoncol.2020.5161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bettegowda C, Sausen M, Leary RJ, et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Science Translational Medicine. 2014;6(224):224ra24–224ra24. doi: 10.1126/scitranslmed.3007094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu R-H, Wei W, Krawczyk M, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nature Materials. 2017;16(11):1155–1161. doi: 10.1038/nmat4997 [DOI] [PubMed] [Google Scholar]
- 72.Luo H, Zhao Q, Wei W, et al. Circulating tumor DNA methylation profiles enable early diagnosis, prognosis prediction, and screening for colorectal cancer. Sci Transl Med. Jan 1 2020;12(524)doi: 10.1126/scitranslmed.aax7533 [DOI] [PubMed] [Google Scholar]
- 73.Gordevičius J, Kriščiūnas A, Groot DE, et al. Cell-Free DNA Modification Dynamics in Abiraterone Acetate-Treated Prostate Cancer Patients. Clin Cancer Res. Jul 15 2018;24(14):3317–3324. doi: 10.1158/1078-0432.Ccr-18-0101 [DOI] [PubMed] [Google Scholar]
- 74.Beltran H, Romanel A, Conteduca V, et al. Circulating tumor DNA profile recognizes transformation to castration-resistant neuroendocrine prostate cancer. J Clin Invest. Apr 1 2020;130(4):1653–1668. doi: 10.1172/JCI131041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Berchuck JE, Baca SC, McClure HM, et al. Detecting Neuroendocrine Prostate Cancer Through Tissue-Informed Cell-Free DNA Methylation Analysis. Clinical Cancer Research. 2021:clincanres.CCR. doi: 10.1158/1078-0432.ccr-21-3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Cell. Jan 14 2016;164(1–2):57–68. doi: 10.1016/j.cell.2015.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ulz P, Perakis S, Zhou Q, et al. Inference of transcription factor binding from cell-free DNA enables tumor subtype prediction and early detection. Nature Communications. 2019;10(1)doi: 10.1038/s41467-019-12714-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.De Bono JS, Scher HI, Montgomery RB, et al. Circulating Tumor Cells Predict Survival Benefit from Treatment in Metastatic Castration-Resistant Prostate Cancer. Clinical Cancer Research. 2008;14(19):6302–6309. doi: 10.1158/1078-0432.ccr-08-0872 [DOI] [PubMed] [Google Scholar]
- 79.Goldkorn A, Ely B, Quinn DI, et al. Circulating Tumor Cell Counts Are Prognostic of Overall Survival in SWOG S0421: A Phase III Trial of Docetaxel With or Without Atrasentan for Metastatic Castration-Resistant Prostate Cancer. Journal of Clinical Oncology. 2014;32(11):1136–1142. doi: 10.1200/jco.2013.51.7417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Antonarakis ES, Lu C, Wang H, et al. AR-V7 and Resistance to Enzalutamide and Abiraterone in Prostate Cancer. New England Journal of Medicine. 2014;371(11):1028–1038. doi: 10.1056/nejmoa1315815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Antonarakis ES, Lu C, Luber B, et al. Androgen Receptor Splice Variant 7 and Efficacy of Taxane Chemotherapy in Patients With Metastatic Castration-Resistant Prostate Cancer. JAMA Oncology. 2015;1(5):582. doi: 10.1001/jamaoncol.2015.1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Armstrong AJ, Halabi S, Luo J, et al. Prospective Multicenter Validation of Androgen Receptor Splice Variant 7 and Hormone Therapy Resistance in High-Risk Castration-Resistant Prostate Cancer: The PROPHECY Study. Journal of Clinical Oncology. 2019;37(13):1120–1129. doi: 10.1200/jco.18.01731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sperger JM, Emamekhoo H, McKay RR, et al. Prospective Evaluation of Clinical Outcomes Using a Multiplex Liquid Biopsy Targeting Diverse Resistance Mechanisms in Metastatic Prostate Cancer. Journal of Clinical Oncology. 2021;39(26):2926–2937. doi: 10.1200/jco.21.00169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Scher HI, Graf RP, Schreiber NA, et al. Phenotypic Heterogeneity of Circulating Tumor Cells Informs Clinical Decisions between AR Signaling Inhibitors and Taxanes in Metastatic Prostate Cancer. Cancer Research. 2017;77(20):5687–5698. doi: 10.1158/0008-5472.can-17-1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miyamoto DT, Zheng Y, Wittner BS, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349(6254):1351–1356. doi: 10.1126/science.aab0917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Malihi PD, Graf RP, Rodriguez A, et al. Single-Cell Circulating Tumor Cell Analysis Reveals Genomic Instability as a Distinctive Feature of Aggressive Prostate Cancer. Clinical Cancer Research. 2020;26(15):4143–4153. doi: 10.1158/1078-0432.ccr-19-4100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Conteduca V, Ku S-Y, Fernandez L, et al. Circulating tumor cell heterogeneity in neuroendocrine prostate cancer by single cell copy number analysis. npj Precision Oncology. 2021;5(1)doi: 10.1038/s41698-021-00211-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yu M, Bardia A, Aceto N, et al. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345(6193):216–220. doi: 10.1126/science.1253533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nature Reviews Molecular Cell Biology. 2018;19(4):213–228. doi: 10.1038/nrm.2017.125 [DOI] [PubMed] [Google Scholar]
- 90.Tavoosidana G, Ronquist G, Darmanis S, et al. Multiple recognition assay reveals prostasomes as promising plasma biomarkers for prostate cancer. Proc Natl Acad Sci U S A. May 24 2011;108(21):8809–14. doi: 10.1073/pnas.1019330108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Soekmadji C, Corcoran NM, Oleinikova I, et al. Extracellular vesicles for personalized therapy decision support in advanced metastatic cancers and its potential impact for prostate cancer. The Prostate. 2017;77(14):1416–1423. doi: 10.1002/pros.23403 [DOI] [PubMed] [Google Scholar]
- 92.Huang X, Yuan T, Liang M, et al. Exosomal miR-1290 and miR-375 as Prognostic Markers in Castration-resistant Prostate Cancer. European Urology. 2015;67(1):33–41. doi: 10.1016/j.eururo.2014.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Del Re M, Biasco E, Crucitta S, et al. The Detection of Androgen Receptor Splice Variant 7 in Plasma-derived Exosomal RNA Strongly Predicts Resistance to Hormonal Therapy in Metastatic Prostate Cancer Patients. European Urology. 2017;71(4):680–687. doi: 10.1016/j.eururo.2016.08.012 [DOI] [PubMed] [Google Scholar]
- 94.Tang Y-T, Huang Y-Y, Zheng L, et al. Comparison of isolation methods of exosomes and exosomal RNA from cell culture medium and serum. International Journal of Molecular Medicine. 2017;40(3):834–844. doi: 10.3892/ijmm.2017.3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Weissleder R.Molecular imaging in cancer. Science. May 26 2006;312(5777):1168–71. doi: 10.1126/science.1125949 [DOI] [PubMed] [Google Scholar]
- 96.Parent EE, Schuster DM. Update on (18)F-Fluciclovine PET for Prostate Cancer Imaging. J Nucl Med. May 2018;59(5):733–739. doi: 10.2967/jnumed.117.204032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Killock D.PSMA PET–CT improves staging. Nature Reviews Clinical Oncology. 2020/06/01 2020;17(6):337–337. doi: 10.1038/s41571-020-0364-4 [DOI] [PubMed] [Google Scholar]
- 98.Hofman MS, Violet J, Hicks RJ, et al. [ 177 Lu]-PSMA-617 radionuclide treatment in patients with metastatic castration-resistant prostate cancer (LuPSMA trial): a single-centre, single-arm, phase 2 study. The Lancet Oncology. 2018;19(6):825–833. doi: 10.1016/s1470-2045(18)30198-0 [DOI] [PubMed] [Google Scholar]
- 99.Thang SP, Violet J, Sandhu S, et al. Poor Outcomes for Patients with Metastatic Castration-resistant Prostate Cancer with Low Prostate-specific Membrane Antigen (PSMA) Expression Deemed Ineligible for (177)Lu-labelled PSMA Radioligand Therapy. Eur Urol Oncol. Nov 2019;2(6):670–676. doi: 10.1016/j.euo.2018.11.007 [DOI] [PubMed] [Google Scholar]
- 100.Bakht MK, Lovnicki JM, Tubman J, et al. Differential Expression of Glucose Transporters and Hexokinases in Prostate Cancer with a Neuroendocrine Gene Signature: A Mechanistic Perspective for 18F-FDG Imaging of PSMA-Suppressed Tumors. Journal of Nuclear Medicine. 2020;61(6):904–910. doi: 10.2967/jnumed.119.231068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang B, Liu C, Wei Y, et al. A Prospective Trial of 68Ga-PSMA and 18F-FDG PET/CT in Nonmetastatic Prostate Cancer Patients with an Early PSA Progression During Castration. Clinical Cancer Research. 2020;26(17):4551–4558. doi: 10.1158/1078-0432.ccr-20-0587 [DOI] [PubMed] [Google Scholar]
- 102.Beattie BJ, Smith-Jones PM, Jhanwar YS, et al. Pharmacokinetic Assessment of the Uptake of 16β−18F-Fluoro-5α-Dihydrotestosterone (FDHT) in Prostate Tumors as Measured by PET. Journal of Nuclear Medicine. 2010;51(2):183–192. doi: 10.2967/jnumed.109.066159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fox JJ, Gavane SC, Blanc-Autran E, et al. Positron Emission Tomography/Computed Tomography–Based Assessments of Androgen Receptor Expression and Glycolytic Activity as a Prognostic Biomarker for Metastatic Castration-Resistant Prostate Cancer. JAMA Oncology. 2018;4(2):217. doi: 10.1001/jamaoncol.2017.3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Puca L, Gavyert K, Sailer V, et al. Delta-like protein 3 expression and therapeutic targeting in neuroendocrine prostate cancer. Sci Transl Med. Mar 20 2019;11(484)doi: 10.1126/scitranslmed.aav0891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Geffers I, Serth K, Chapman G, et al. Divergent functions and distinct localization of the Notch ligands DLL1 and DLL3 in vivo. J Cell Biol. Jul 30 2007;178(3):465–76. doi: 10.1083/jcb.200702009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sharma SK, Pourat J, Abdel-Atti D, et al. Noninvasive Interrogation of DLL3 Expression in Metastatic Small Cell Lung Cancer. Cancer Research. 2017;77(14):3931–3941. doi: 10.1158/0008-5472.can-17-0299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Korsen J, Kalidindi T, Khitrov S, et al. Delta-like Ligand 3 (DLL3) is a novel target for molecular imaging of Neuroendocrine Prostate Cancer. Journal of Nuclear Medicine. 2020;61(supplement 1):133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mankoff DA, Farwell MD, Clark AS, Pryma DA. Making Molecular Imaging a Clinical Tool for Precision Oncology. JAMA Oncology. 2017;3(5):695. doi: 10.1001/jamaoncol.2016.5084 [DOI] [PubMed] [Google Scholar]
- 109.Subramanian I, Verma S, Kumar S, Jere A, Anamika K. Multi-omics Data Integration, Interpretation, and Its Application. Bioinform Biol Insights. 2020;14:1177932219899051. doi: 10.1177/1177932219899051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ramazzotti D, Lal A, Wang B, Batzoglou S, Sidow A. Multi-omic tumor data reveal diversity of molecular mechanisms that correlate with survival. Nature Communications. 2018;9(1)doi: 10.1038/s41467-018-06921-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Poirion OB, Chaudhary K, Garmire LX. Deep Learning data integration for better risk stratification models of bladder cancer. AMIA Jt Summits Transl Sci Proc. 2018;2017:197–206. [PMC free article] [PubMed] [Google Scholar]
- 112.Chaudhary K, Poirion OB, Lu L, Garmire LX. Deep Learning–Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clinical Cancer Research. 2018;24(6):1248–1259. doi: 10.1158/1078-0432.ccr-17-0853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang T-H, Lee C-Y, Lee T-Y, Huang H-D, Hsu JB-K, Chang T-H. Biomarker Identification through Multiomics Data Analysis of Prostate Cancer Prognostication Using a Deep Learning Model and Similarity Network Fusion. Cancers. 2021;13(11):2528. doi: 10.3390/cancers13112528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Elmarakeby HA, Hwang J, Arafeh R, et al. Biologically informed deep neural network for prostate cancer discovery. Nature. 2021;598(7880):348–352. doi: 10.1038/s41586-021-03922-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang T, Shao W, Huang Z, et al. MOGONET integrates multi-omics data using graph convolutional networks allowing patient classification and biomarker identification. Nature Communications. 2021;12(1)doi: 10.1038/s41467-021-23774-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Popovici V, Budinská E, Čápková L, et al. Joint analysis of histopathology image features and gene expression in breast cancer. BMC Bioinformatics. May 11 2016;17(1):209. doi: 10.1186/s12859-016-1072-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nagpal K, Foote D, Tan F, et al. Development and Validation of a Deep Learning Algorithm for Gleason Grading of Prostate Cancer From Biopsy Specimens. JAMA Oncology. 2020;6(9):1372. doi: 10.1001/jamaoncol.2020.2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xie W, Reder NP, Koyuncu C, et al. Prostate Cancer Risk Stratification via Nondestructive 3D Pathology with Deep Learning–Assisted Gland Analysis. Cancer Research. 2022;82(2):334–345. doi: 10.1158/0008-5472.can-21-2843 [DOI] [PMC free article] [PubMed] [Google Scholar]