ABSTRACT
Several models were developed to study the pathogenicity of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as well as the in vivo efficacy of vaccines and therapeutics. Since wild-type mice are naturally resistant to infection by ancestral SARS-CoV-2 strains, several transgenic mouse models expressing human angiotensin-converting enzyme 2 (hACE2) were developed. An alternative approach has been to develop mouse-adapted SARS-CoV-2 strains. Here, we compared the clinical progression, viral replication kinetics and dissemination, pulmonary tropism, and host innate immune response dynamics between the mouse-adapted MA10 strain and its parental strain (USA-WA1/2020) following intranasal inoculation of K18-hACE2 mice, a widely used model. Compared to its parental counterpart, the MA10 strain induced earlier clinical decline with significantly higher viral replication and earlier neurodissemination. Importantly, the MA10 strain also showed a wider tropism, with infection of bronchiolar epithelia. While both SARS-CoV-2 strains induced comparable pulmonary cytokine/chemokine responses, many proinflammatory and monocyte-recruitment chemokines, such as interleukin-6 (IL-6), tumor necrosis factor alpha (TNF-α), IP-10/CXCL10, and MCP-1/CCL2, showed an earlier peak in MA10-infected mice. Furthermore, both strains induced a similar downregulation of murine Ace2, with only a transient downregulation of Tmprss2 and no alterations in hACE2 expression. Overall, these data demonstrate that in K18-hACE2 mice, the MA10 strain has a pulmonary tropism that more closely resembles SARS-CoV-2 tropism in humans (airways and pneumocytes) than its parental strain. Its rapid replication and neurodissemination and early host pulmonary responses can have a significant impact on the clinical outcomes of infection and are, therefore, critical features to consider for study designs using these strains and mouse model.
IMPORTANCE The COVID-19 pandemic, caused by SARS-CoV-2, is still significantly impacting health care systems around the globe. Refined animal models are needed to study SARS-CoV-2 pathogenicity as well as efficacy of vaccines and therapeutics. In line with this, thorough evaluation of animal models and virus strains/variants are paramount for standardization and meaningful comparisons. Here, we demonstrated differences in replication dynamics between the Wuhan-like USA-WA1/2020 strain and the derivative mouse-adapted MA10 strain in K18-hACE2 mice. The MA10 strain showed accelerated viral replication and neurodissemination, differential pulmonary tropism, and earlier pulmonary innate immune responses. The observed differences allow us to better refine experimental designs when considering the use of the MA10 strain in the widely utilized K18-hACE2 murine model.
KEYWORDS: ACE2-expressing mice, COVID-19, K18-hACE2 mice, MA10, mouse-adapted virus, respiratory infection, SARS-CoV-2, WA1, mACE2, mouse model, viral replication
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an enveloped, positive-sense, single-stranded RNA virus belonging to the Betacoronavirus genus (order Nidovirales, family Coronaviridae) (1, 2). Since the initial case reported at a seafood market in Wuhan, Hubei Province, China in December 2019 (3–5), SARS-CoV-2 has rapidly become a pandemic (6) with 660 million confirmed cases and nearly 6.7 million deaths (1.01%) as of 31 December 2022 (7). To infect the host, the viral spike glycoprotein (S) of SARS-CoV-2 utilizes the human angiotensin-converting enzyme 2 (hACE2) as the major entry receptor (8). After virus binding, priming and cleavage of the S glycoprotein by the host transmembrane serine protease 2 (TMPRSS2), conformational changes in S trimers allow for subsequent fusion of the cellular membrane and viral envelope, facilitating virus entry (8, 9). CD147, neuropilin 1, and the neutral amino acid transporter B0AT1 were also identified as host receptors/factors that may have a potential role in SARS-CoV-2 infection (10–14).
SARS-CoV-2 is the causative agent of coronavirus disease 2019 (COVID-19), causing respiratory disease of variable severity that may lead to the development of acute respiratory distress syndrome (ARDS) (15–17). Although the exact mechanisms by which SARS-CoV-2 induces ARDS are not fully understood, the induction of a cytokine storm, characterized by increased levels of inflammatory cytokines and chemokines, such as interleukin-2 (IL-2), IL-6, IL-10, tumor necrosis factor alpha (TNF-α), granulocyte-macrophage colony-stimulating factor (GM-CSF), monocyte chemoattractant protein-1 (MCP-1/CCL-2), macrophage inflammatory protein 1 alpha (MIP-1α/CCL3), and CXC-chemokine ligand 10 (CXCL10), is considered to be one of the driving factors (18–22).
The low binding affinity between the viral S protein of the ancestral Wuhan-like (USA-WA1/2020) SARS-CoV-2 strains and murine ACE2 (mACE2) renders conventional mouse strains as naturally resistant to infection, which has posed a significant challenge for the development of murine models of COVID-19 (23, 24). This challenge was in part countered with the emergence of variants of concern (VOCs) possessing the N501Y mutation on the S glycoprotein, which increases the S-mACE2 interaction and also allows for infection of airway epithelia (25). Several strategies have also been developed to increase the susceptibility of laboratory mice to SARS-CoV-2 infection such as using transgenic mice expressing hACE2 under the K18 promoter (K18-hACE2) (26–29), transduction of hACE2 using an adenoviral delivery system (30), or using mouse-adapted SARS-CoV-2 strains (31–34). The K18-hACE2 mouse model expresses the hACE2 transgene along with the mACE2 receptor (27) and is a common in vivo model used to study SARS-CoV-2 pathogenesis and efficacy of therapeutics and vaccines. The main limitation of this model is its high lethality associated with SARS-CoV-2 neuroinvasion and subsequent clinical decline following intranasal challenge (35–38), limiting long-term studies. The mouse-adapted SARS-CoV-2 MA strain was developed by Dinnon et al. (33) via engineering amino acid residues Q498Y/P499T of the receptor binding domain (RBD) on the S protein involved in the S-mACE2 interface to allow binding of the S glycoprotein to mACE2. To increase the virulence of this engineered virus, Leist et al. (34) developed the mouse-adapted MA10 strain by 10 subsequent passages of the SARS-CoV-2 MA strain in BALB/cAnNHsd mice, resulting in increased virulence in both young and aged BALB/cAnNHsd mice. Importantly, both the MA and MA10 strains demonstrated acquired bronchiolar epithelial tropism, a feature not identified in K18-hACE2 mice infected with the USA-WA1/2020 strain of SARS-CoV-2 (35).
Refined models to study SARS-CoV-2 pathogenicity are still needed, and a potential alternative approach is to use the MA10 strain, which has an expanded cellular tropism, in the transgenic K18-hACE2 mouse model. Although it was recently demonstrated that this mouse-adapted strain induces high lethality in K18-hACE2 mice (39), an extensive pathogenicity study to comparatively identify differences with its parental strain and better inform its use in this humanized mouse model has not yet been conducted. In this study, a time course investigation following intranasal challenge of K18-hACE2 mice with the mouse-adapted MA10 strain was conducted and compared to its parental strain USA-WA1/2020. Clinical outcomes, viral replication kinetics, dissemination, tropism, and histomorphological changes in the lung and brain, pulmonary cytokine/chemokine responses, and pulmonary mAce2, hACE2, and Tmprss2 expression dynamics were quantitated and compared. This study highlights the importance of thoroughly characterizing SARS-CoV-2 animal models for preclinical trials to better define relevant experimental observation time points/data collection points and assist in drawing adequate interpretations.
RESULTS
SARS-CoV-2 MA10 strain induces earlier and more severe weight loss with comparable lethality to the USA-WA1/2020 parental strain in intranasally infected K18-hACE2 mice.
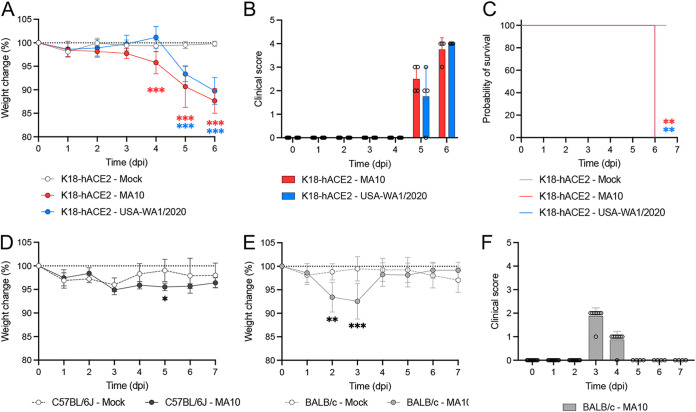
K18-hACE2 mice were intranasally infected with 2 × 104 PFU of either the SARS-CoV-2 MA10 strain or its parental strain USA-WA1/2020 and monitored for 6 to 7 days, at which point euthanasia criteria was met in all animals. Significant weight loss was first observed in the MA10-infected mice at 4 days postinfection (dpi) (4.22 ± 2.37%; P ≤ 0.001), and body weight continued to decrease throughout the remainder of the experiment (Fig. 1A). Significant weight loss in mice infected with the same dose of SARS-CoV-2 USA-WA1/2020 strain was first identified at 5 dpi instead (6.63 ± 1.61%; P ≤ 0.001). Peak in weight loss occurred at 6 dpi for both MA10- and USA-WA1/2020-infected mice with 12.34 ± 2.64% (P ≤ 0.001) and 10.23 ± 2.86% (P ≤ 0.001) loss, respectively. Besides weight loss, clinical signs (increased respiratory rate/effort, reluctance to move, tremors, and/or poor responsiveness) were initially noted at 5 dpi for both groups, with a maximum score reached at 6 dpi (Fig. 1B), time at which humane euthanasia endpoint was reached (see Fig. 6C). Altogether, these data demonstrate that the MA10 strain of SARS-CoV-2 induces clinical decline at least 24 h earlier than the ancestral USA-WA1/2020 strain in K18-hACE2 mice.
FIG 1.
SARS-CoV-2 MA10 and USA-WA1/2020 strains induced weight loss and lethality in K18-hACE2 mice. Mice were intranasally infected with 2 × 104 PFU of SARS-CoV-2 MA10 (red; n = 12), USA-WA1/2020 strain (blue; n = 12), or mock infected with 50 μL of MEM (gray; n = 4). Body weight (A), clinical signs (B), and survival (C) of K18-hACE2 mice were monitored daily. Body weight of C57BL/6J (D) and BALB/c (E) mice intranasally infected with 2 × 104 PFU of SARS-CoV-2 MA10. Clinical disease developed in infected BALB/c (F) but not in C57BL/6J mice. Infection of BALB/c and C57BL/6J was nonlethal at this dose, and animals recovered following infection. Points and bars represent the mean ± standard deviation. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
FIG 6.
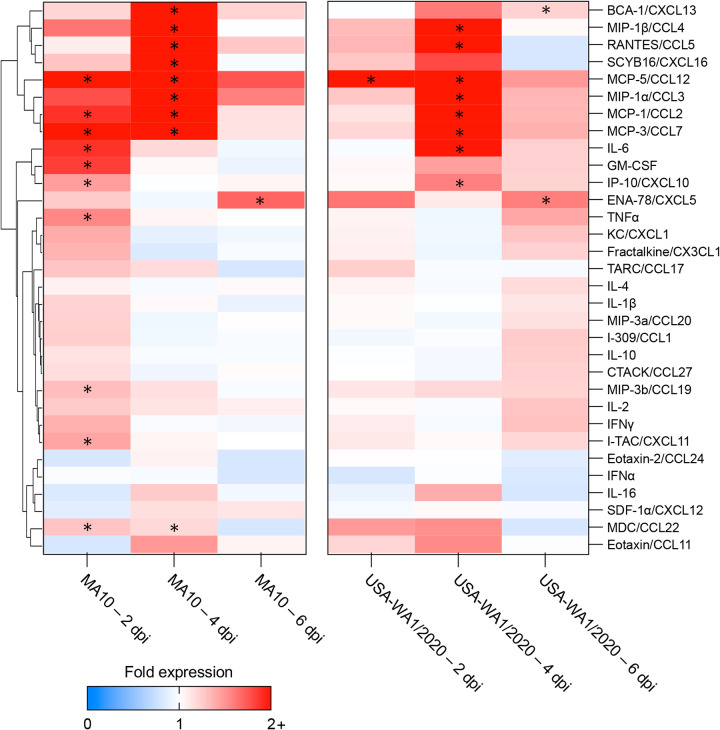
Cytokine and chemokine profiling in the lungs of MA10- and USA-WA1/2020-infected K18-hACE2 mice. The proinflammatory cytokine and chemokine response occurs more rapidly in the lung of K18-hACE2 mice infected with the MA10 strain compared to those infected with USA-WA1/2020 strain. Heatmap depicting the fold change of protein expression in the lung of infected mice at 2, 4, and 6 dpi compared to mock infected K18-hACE2 mice. Color scale depicts differences in fold expression. Cytokines and chemokines that are significantly upregulated compared to the mock are marked with an asterisk.
Comparatively, MA10-infected C57BL/6J mice, the background mouse strain of the K18-hACE2 mice, and BALB/c mice showed significant and transient weight loss at 5 dpi (2.58 ± 0.81%; P = 0.029) or as early as 2 dpi (5.41 ± 3.17%; P ≤ 0.001), respectively (Fig. 1D and E). Clinical signs (ruffled fur and lethargy) were observed at 3 and 4 dpi (Fig. 1F) in infected BALB/c mice but not in infected C57BL/6J mice (data not shown). All BALB/c infected mice recovered by 5 dpi (Fig. 1E and F), and no lethality was recorded in either infected BALB/c or in C57BL/6J mice by 7 dpi (data not shown).
SARS-CoV-2 MA10 strain induces rapid replication and an earlier peak in viral titers in the lung of K18-hACE2 mice compared to that of the SARS-CoV-2 USA-WA1/2020 parental strain.
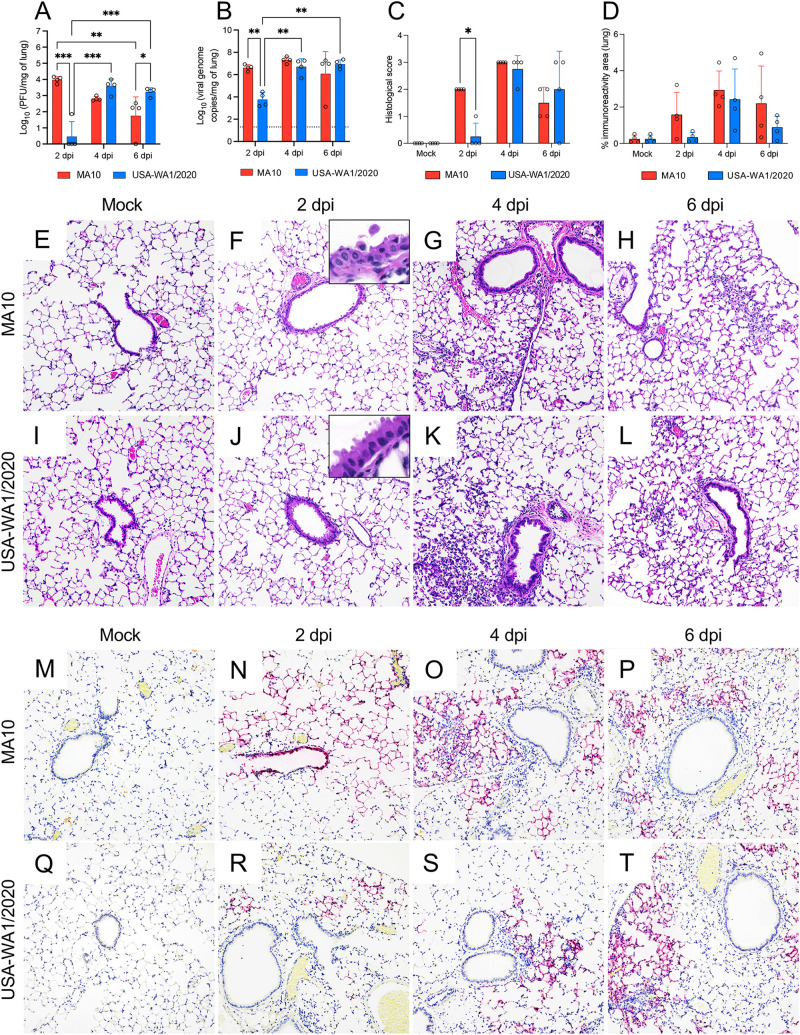
To evaluate viral replication kinetics, four infected K18-hACE2 mice per group were euthanized at 2, 4, and 6 dpi, and the level of infectious virus and viral RNA in the lung were quantitated. At 2 dpi, infectious virus was detectable in the lungs of all MA10-infected K18-hACE2 mice, whereas only one-quarter of mice infected with USA-WA1/2020 had detectable virus (Fig. 2A). Comparatively, viral titers were significantly higher in the lung of MA10-infected K18-hACE2 mice (P ≤ 0.001), with a >3 log difference compared to the USA-WA1/2020-infected K18-hACE2 mice (Fig. 2A). Viral titers for USA-WA1/2020-infected K18-hACE2 mice reached comparable values to those in the MA10-infected group (2 dpi) only at 4 dpi and thereafter. At 6 dpi, a significantly higher viral titer was observed in the lung of mice infected with the USA-WA1/2020 parental strain compared with the MA10 strain (P = 0.047). Viral genomic RNA showed similar dynamics during infection, with a rapid spike at 2 dpi for the MA10-infected K18-hACE2 mice, which displayed an ~2.8 log increase compared to the USA-WA1/2020-infected mice (P = 0.004) (Fig. 2B). These results illustrate accelerated replication dynamics of the MA10 strain in the lungs of K18-hACE2 infected mice compared to the WA1/2020 parental strain.
FIG 2.
Comparative temporal analysis of SARS-CoV-2 MA10 and USA-WA1/2020 replication and pathological alterations in the lung of K18-hACE2 mice. Infectious viral particles (A) and viral RNA (B) were quantified in the lung of infected mice at 2, 4, and 6 dpi; the MA10-infected mice showed significantly higher viral titers and viral RNA in the lung by 2 dpi compared to the parental strain. Histological scores (C) and percentage of SARS-CoV-2 immunoreactivity measured by quantitative IHC (D) in the lung of K18-hACE2 mice. Histopathological alterations in the lung appear earlier in the MA10-infected mice compared to the parental strain. Bars represent the mean ± standard deviation. Dotted line represents the limit of detection. Temporal histologic lesions (E to L) and viral antigen abundance and distribution (M to T) in the lung of MA10- and USA-WA1/2020-infected K18-hACE2 mice. Bronchiolar epithelial degeneration and necrosis with viral antigen located within bronchiolar epithelium were evident in the MA10-infected group (F and N, and inset) at 2 dpi but not in the USA-WA1/2020-infected group (J and R, and inset). Mild-to-moderate pneumonia with a predominantly mononuclear cell infiltrate were evident at 4 dpi and persisted at 6 dpi in both infected groups (G, H, K, and L), with abundant viral antigen within AT1 and AT2 cells (M to T). H&E (E to L) and Fast Red (viral antigen) (M to T); ×200 total magnification. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
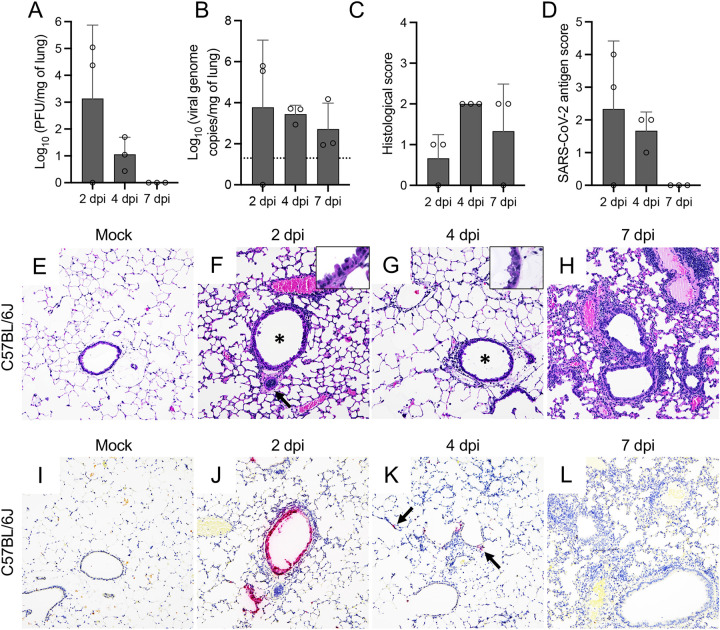
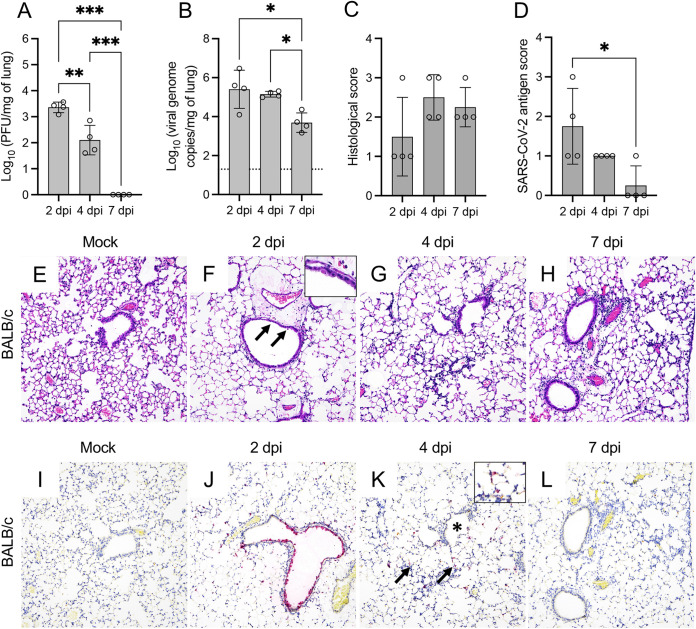
Infectious virus/viral RNA were detectable in the lungs of two out of three of the MA10-infected C57BL/6J mice at 2 dpi and in the lungs of all C57BL/6J mice at 4 dpi (Fig. 3A and B). Even though MA10 viral RNA was detected in the lung at 7 dpi, no infectious virus was recovered. Infectious virus was detectable in the lungs of all BALB/c infected mice at 2 and 4 dpi but not at 7 dpi (Fig. 4A). In contrast, viral RNA was detected in all infected BALB/c mice at all time points (Fig. 4B). Peak in viral titers and viral RNA in these mice occurred at 2 dpi.
FIG 3.
SARS-CoV-2 MA10 infection of C57BL/6J mice. Infectious viral particles (A) and viral RNA (B) were quantified in the lung of infected C57BL/6J mice at 2, 4, and 7 dpi. Histological scores (C) and scoring for SARS-CoV-2 antigen abundance (D) in the lung of C57BL/6J mice are shown. Bars represent the mean ± standard deviation. Dotted line represents the limit of detection. Temporal histologic lesions (E to H) and viral antigen abundance and distribution (I to L) in the lung of MA10-infected C57BL/6J mice. At 2 dpi, there was evidence of bronchiolar (F, asterisk) epithelial degeneration and necrosis (F, inset), with abundant viral antigen (J), mild peribronchiolar inflammation, and blood vessels with hypertrophied (activated) endothelium (F, arrow). Similar alterations were noted at 4 dpi (G, inset), but these resolved by 7 dpi (H). Mild interstitial pneumonia was first identified at 4 dpi (G) and persisted until the end of the study (7 dpi) (H). At 4 dpi, viral antigen was rare and only identified within the cytoplasm of scattered pneumocytes (K, arrows). No viral antigen was noted at 7 dpi (L). H&E (E to H) and Fast Red (viral antigen) (I to L); ×200 total magnification. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
FIG 4.
SARS-CoV-2 MA10 induces injury in the lung of BALB/c mice. Infectious viral particles (A) and viral RNA (B) were quantified in the lung of infected BALB/c mice at 2, 4, and 7 dpi. Histological scores (C) and scoring of SARS-CoV-2 antigen abundance (D) in the lung of BALB/c mice are presented. Bars represent the mean ± standard deviation. Dotted line represents the limit of detection. Temporal histologic lesions (E to H) and viral antigen abundance and distribution (I to L) in the lung of MA10-infected BALB/c mice. There is evidence of bronchiolar degeneration/necrosis at 2 dpi (F, arrows and inset), with abundant viral antigen (J) and mild peribronchiolar inflammation. At 4 dpi, there is evidence of interstitial pneumonia with alveolar septa expanded by mononuclear inflammatory cells, and alveolar spaces contain similar inflammatory cells and karyorrhectic debris (G). Similar histologic changes persist at 7 dpi (H). At 4 dpi, viral antigen is mostly frequent in the cytoplasm of AT2 cells within areas of pneumonia (K, arrows and inset) but spare the bronchiolar epithelium (K, asterisk). Viral antigen is rarely detected at 7 dpi (L). H&E (E to H) and Fast Red (viral antigen) (I to L); ×200 total magnification. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Differential tropism of the SARS-CoV-2 MA10 strain compared to that of the USA-WA1/2020 parental strain is associated with bronchiolitis and subsequent interstitial pneumonia.
To comprehensively characterize differences between these strains in the lung of K18-hACE2 mice, we evaluated the histologic alterations in both infected groups as well as viral tropism, abundance, and spread of viral antigen via immunohistochemistry (IHC) and quantitative image analysis throughout the course of infection (Fig. 2C and Fig. 2E to T). In the MA10-infected K18-hACE2 mice, histologic alterations were evident at 2 dpi and were mainly characterized by multifocal necrotizing bronchiolitis featuring bronchiolar epithelial vacuolation, rounding and detachment, attenuation, and loss of polarity and cilia accompanied by a minimal infiltrate of peribronchiolar lymphocytes and histiocytes (Fig. 2F). In comparison, only sporadic perivascular cuffs composed of minimal lymphocytes were noted in the USA-WA1/2020-infected mice at this time point, with no evidence of airway damage (Fig. 2J). At 4 dpi, all MA10 and USA-WA1/2020-infected K18-hACE2 mice showed evidence of comparable mild to moderate multifocal interstitial pneumonia with predominantly mononuclear cells (lymphocytes and histiocytes) (Fig. 2G and K). In the MA10-infected K18-hACE2 mice, the bronchiolar epithelium was fully regenerated, and no changes were noted in the airways of USA-WA1/2020-infected K18-hACE2 mice as also noted in previous reports (35). Interstitial pneumonia variably persisted until the end of the experiment (6 dpi) (Fig. 2H and L). Histologic lesions were semiquantitatively scored based on their severity (Fig. 2C); the significant difference identified at 2 dpi between MA10- and USA-WA1/2020-infected K18-hACE2 mice is attributable to the bronchiolitis that developed exclusively in the former group at 2 dpi (P = 0.029) (Fig. 2F).
We subsequently compared viral tropism of these two strains and quantitatively determined the abundance of SARS-CoV-2 N antigen in the lungs via IHC (Fig. 2D and Fig. 2M to T). Correlating with the histologic changes noted at 2 dpi and accelerated elevation of viral titers in the MA10-infected mice, intracytoplasmic viral antigen was abundant within the affected bronchiolar epithelium as well as within neighboring alveolar type 1 and 2 (AT1 and AT2) cells (Fig. 2N). On subsequent time points, viral antigen was limited to the cytoplasm of only AT1 and AT2 cells (Fig. 2O and P). Comparatively and as previously described, USA-WA1/2020 showed exclusive tropism for AT1 and AT2 cells, with no viral antigen detected within bronchiolar epithelium at any time point (Fig. 2R to T). To determine differences in viral spread/abundance between these two strains, we quantitated the immunoreactive area as a percentage of the entire pulmonary parenchyma evaluated (Fig. 2D). Although not statistically significant, a trend similar to viral load data was noted, with a higher immunoreactive area for SARS-CoV-2 N antigen in MA10-infected K18-hACE2 mice at 2 dpi than in USA-WA1/2020-infected animals that subsequently becomes similar at 4 and 6 dpi.
Similar to MA10-infected K18-hACE2 mice, histologic lesions in MA10-infected C57BL/6J mice were initially evident at 2 dpi and were characterized by necrotizing bronchiolitis with abundant epithelial viral antigen and mild peribronchiolar/perivascular mononuclear cell infiltration (Fig. 3C, F, and J). This was subsequently followed by an increase in peribronchiolar and perivascular inflammatory cells at 4 dpi and mild interstitial pneumonia noted both at 4 and 7 dpi (Fig. 3G and H). By 7 dpi, bronchiolar epithelial histomorphological changes were resolved, and viral antigen was no longer detected (Fig. 3L). Histologic changes in the lungs of MA10-infected BALB/c mice were similar (Fig. 4C and Fig. 4E to H), with more frequent foci of interstitial pneumonia and intra-alveolar debris than C57BL/6J mice (Fig. 4G and H). At 2 dpi, viral antigen was detected in the bronchiolar epithelium and was more frequently within alveolar epithelial cells than in MA10-infected C57BL/6J but not as widespread as in infected K18-hACE2 mice (Fig. 4J). At 4 dpi, no viral antigen was detected in the bronchiolar epithelium but persisted within AT2 cells (Fig. 4K to L).
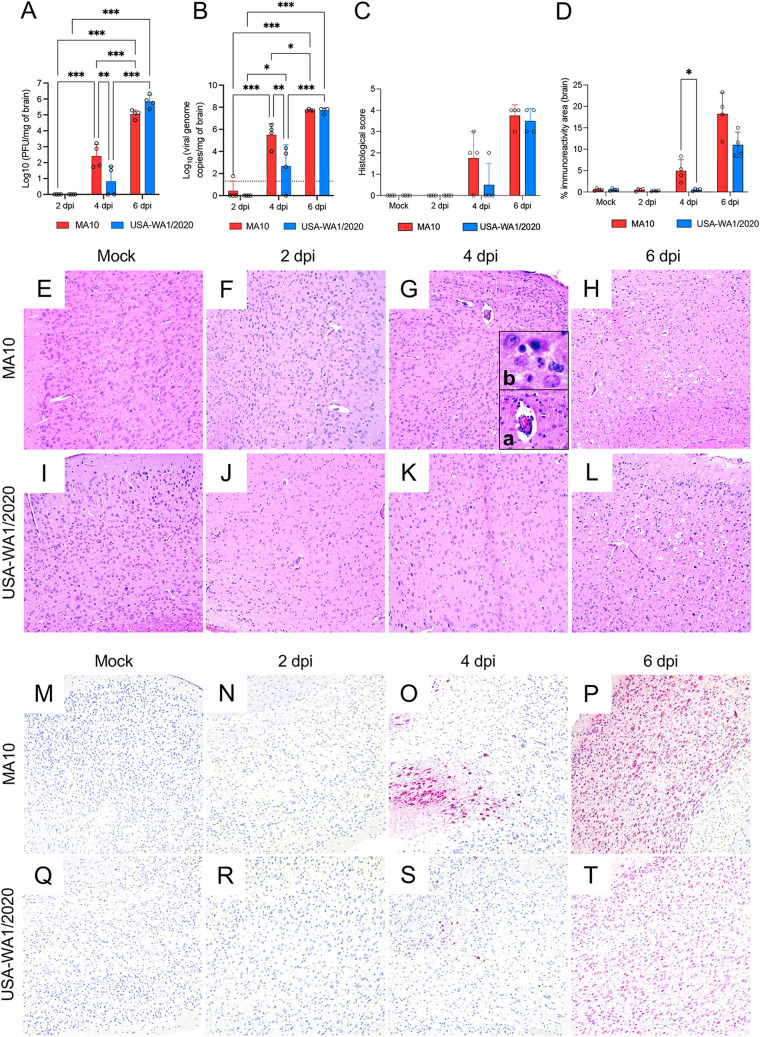
SARS-CoV-2 MA10 strain exhibits similar neurotropism but accelerated neurodissemination compared to that of the USA-WA1/2020 parental strain in K18-hACE2 mice.
The temporal infection dynamics in the brain of both MA10- and USA-WA1/2020-infected K18-hACE2 mice were comparatively evaluated. At 2 dpi, no detectable infectious virus was identified in the brain of either group (Fig. 5A), with a marginal amount of viral RNA detected in the brain of a single MA10-infected mouse (Fig. 5B). At 4 dpi, infectious virus and viral RNA were detected in the brain of all MA10-infected mice but only in 2 of 4 and 3 of 4 K18-hACE2 mice infected with the USA-WA1/2020 strain, respectively. At 4 dpi, viral titers and viral RNA copies per milligram of brain were significantly higher in the MA10-infected K18-hACE2 mice than in the USA-WA1/2020-infected animals (~1.6 log, P = 0.006; ~2.8 log, P = 0.007, respectively). Viral titers and viral RNA copies per milligram of brain reached their peak at 6 dpi, with comparable levels between both groups.
FIG 5.
Comparative temporal analysis of SARS-CoV-2 MA10 and USA-WA1/2020 replication and pathological alterations in the brain (cerebral cortex) of K18-hACE2 mice. Infectious viral particles (A) and viral RNA (B) were quantified in the brain of infected mice at 2, 4, and 6 dpi. Histological scores (C) and percentage of SARS-CoV-2 immunoreactivity (D) in the brain of K18-hACE2 mice is shown. Bars represent the mean ± standard deviation. Dotted line represents the limit of detection. Temporal histologic lesions (E to L) and viral antigen abundance and distribution (M to T) in the brain of K18-hACE2 mice. Histologic changes are noted as early as 4 dpi and are more pronounced on the MA10-infected group (G) with delicate perivascular lymphocytic cuffs (G, inset a), sporadic pyknotic nuclei, and increased microglial cells and scattered neutrophils within the neuroparenchyma (G, inset b). At this time point, viral antigen is significantly more abundant in the MA10-infected group compared to that of the group infected with the parental USA-WA1/2020 strain. As previously reported, neuronal vacuolation/necrosis are prominent at 6 dpi (H and L) with diffuse and abundant expression of viral antigen within neuronal bodies and processes (P and T) with cerebellar sparing as previously reported. H&E (E to H) and Fast Red (viral antigen) (I to L); ×200 total magnification. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
In addition, we analyzed morphological changes and viral tropism and spread as described for the lung. No morphological changes or viral antigen were observed in the brain of either group at 2 dpi (Fig. 5C, D, F, J, N, and R). At 4 dpi, histologic alterations were initially seen in both groups of mice, characterized by sporadic neuronal necrosis and microgliosis with occasional lymphocytic perivascular cuffs and few neutrophils within the parenchyma (Fig. 5G and K); these alterations were more pronounced in the MA10-infected animals with identifiable alterations in 3 of 4 animals compared to only 1 of 4 animals in the USA-WA1/2020 group. Viral antigen was detected within clusters of neuronal bodies and neuronal processes within the anterior olfactory nucleus and orbital area of the cerebral cortex as well as the mitral layer of the olfactory bulb in all MA10-infected K18-hACE2 mice and only 1 of 4 USA-WA1/2020-infected mice (Fig. 5O and S). Immunoreactive area quantification revealed a significantly higher SARS-CoV-2 N immunoreactive area in the MA10-infected group than in USA-WA1/2020-infected K18-hACE2 mice (P = 0.029) (Fig. 5D), indicating more rapid neurodissemination. At 6 dpi, abundant viral antigen was diffusely detected in neurons (with sparing of the cerebellum) in both groups (Fig. 5P and T), accompanied by neuronal vacuolation and necrosis (Fig. 5H and L) as previously described (35). The results show that the MA10 strain retains neurotropism and neuro disseminates occurs more rapidly compared to the USA-WA1/2020 parental strain.
No infectious virus, viral RNA, viral antigen, or histological alterations were detected in the brain of MA10-infected C57BL/6J and BALB/c mice throughout study time points (data not shown), confirming the absence of neurodissemination in these mouse models.
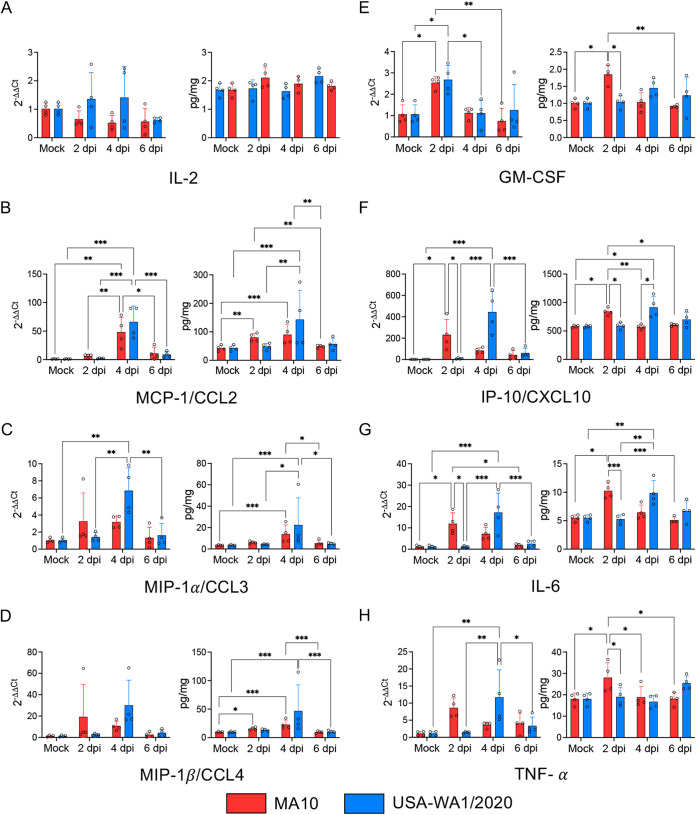
Pulmonary host cytokine and chemokine responses are temporally delayed in USA-WA1/2020-infected K18-hACE2 mice.
Severe SARS-CoV-2 infections are associated with a cytokine storm in the lung of patients, mainly characterized by increased expression of proinflammatory cytokines (18–20). The protein expression levels of 32 cytokines were evaluated in the lung of K18-hACE2 mice infected with SARS-CoV-2 MA10 and USA-WA1/2020 strains (see Fig. S1 in the supplemental material). Even though 16 cytokines were significantly upregulated in the lung of MA10-infected K18-hACE2 mice compared to those in mock-infected animals, only 10 were significantly upregulated in the lung of USA-WA1/2020-infected mice (Fig. 6). In addition, the expression of five of the upregulated cytokines/chemokines (BCA-1/CXCL13, MCP-1/CCL2, MCP-3/CCL7, IL-6, and IP-10/CXCL10) was significantly delayed in the lung of USA-WA1/2020-infected K18-hACE2 mice (Fig. 6; see also Fig. S1). No change in alpha interferon (IFN-α) expression was observed in either group, suggestive of inhibition of type I interferons by SARS-CoV-2.
SARS-CoV-2 MA10 strain induces an early expression of proinflammatory cytokines and chemokines in the lungs of K18-hACE2 mice compared to that of its parental strain USA-WA1/2020. Protein expression of 32 cytokines/chemokines was measured in the lung of mock-, MA10-, and USA-WA1/2020-infected K18-hACE2 mice at 2 dpi, 4 dpi, and 6 dpi (n = 4). Data analyzed by nonparametric Wilcoxon tests. Download FIG S1, TIF file, 1.0 MB (1MB, tif) .
Copyright © 2023 Thieulent et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gene expression of a subset of eight cytokines and chemokines reported to be related to SARS-CoV-2 pathogenesis in human patients (19, 40) was also evaluated to correlate with protein expression results. Results were consistent with protein measurements (Fig. 7). No change in IL-2 expression was observed (Fig. 7A). Overexpression of MCP-1/CCL2 (Fig. 7B), MIP-1α/CCL3 (Fig. 7C), MIP-1β/CCL4 (Fig. 7D), and GM-CSF (Fig. 7E) was noted, all of which are associated with monocyte/macrophage recruitment and responses. Interestingly, IP-10/CXCL10, IL-6 and TNF-α showed a delayed upregulation in the lung of USA-WA1/2020-infected K18-hACE2 mice (Fig. 7F to H). Overall, these data show that both strains induced an increase of proinflammatory cytokines/chemokines involved in macrophage recruitment, which are temporally delayed in the lung of USA-WA1/2020-infected K18-hACE2 mice compared to those in the MA10-infected animals.
FIG 7.
Cytokine and chemokine dynamics (protein and gene expression) in the lungs of MA10- and USA-WA1/2020-infected K18-hACE2 mice. The proinflammatory cytokine and chemokine response occurs earlier in the course of infection in the MA10-infected group compared to the parental strain. Relative gene expression and protein expression of IL-2 (A), MCP-1/CCL2 (B), MIP-1α/CCL3 (C), MIP-1β/CCL4 (D), GM-CSF (E), IP-10/CXCL10 (F), IL-6 (G), and TNF-α (H) were evaluated in the lungs of mock-, MA10-, and WA1-infected mice at 2, 4, and 6 dpi. Bars represent the mean ± standard deviation. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
The cytokine expression levels were also evaluated in the lung of MA10-infected C57BL/6J and BALB/c mice. In infected C57BL/6J mice, cytokine and chemokine fluctuations were mostly marginal. Among the relevant upregulated cytokines, chemokines of chemoattractant nature to monocytes were identified (i.e., MCP-5/CCL12, MIP-1α/CCL3, MIP-1β/CCL4) (see Fig. S2 in the supplemental material). These results could be associated with the overall lower susceptibility of this mouse strain to SARS-CoV-2, but also the small sample size could have contributed. Among the 10 overexpressed cytokines/chemokines detected in the infected BALB/c mice (see Fig. S3 in the supplemental material), seven of them were similar to those upregulated in the lung of MA10-infected K18-hACE2 mice (BCA-1/CXCL13, IL-6, IL-10/CXCL10, MCP-5/CCL12, MDC/CCL22, RANTES/CCL5, and SCYB16/CXCL16).
Expression of proinflammatory cytokines and chemokines in the lungs of MA10-infected C57BL/6J mice. Protein expression of 31 cytokines/chemokines was measured in the lung of mock- and MA10-infected C57BL/6J mice at 2 dpi, 4 dpi, and 7 dpi (n = 3). Data analyzed by nonparametric Wilcoxon tests. Download FIG S2, TIF file, 0.3 MB (340.9KB, tif) .
Copyright © 2023 Thieulent et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of proinflammatory cytokines and chemokines in the lungs of MA10-infected BALB/c mice. Protein expression of 31 cytokines/chemokines was measured in the lung of mock- and MA10-infected BALB/c mice at 2 dpi, 4 dpi, and 7 dpi (n = 4). Data analyzed by nonparametric Wilcoxon tests. Download FIG S3, TIF file, 0.4 MB (371.8KB, tif) .
Copyright © 2023 Thieulent et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
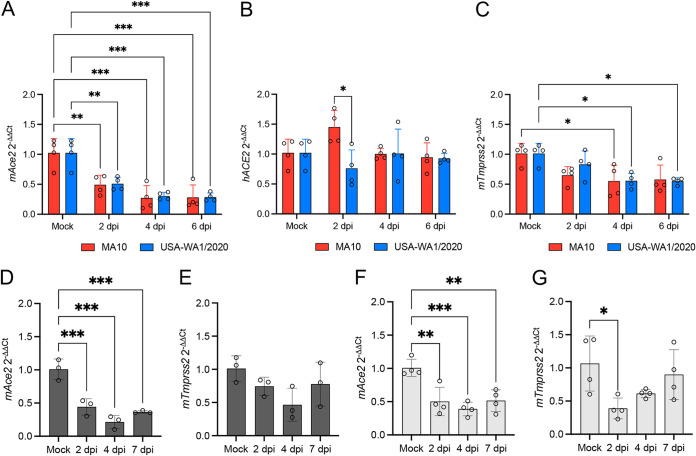
SARS-CoV-2 induces downregulation of mouse Ace2 expression but not human ACE2 in the lungs of K18-hACE2 mice.
The expression of human ACE2 (hACE2), mouse Ace2 (mAce2), and mouse Tmprss2 (mTmprss2) were evaluated in the lung of infected mice. The specificity (inclusivity/exclusivity) of the primers targeting hACE2 and mAce2 was first validated using mRNA derived from the lungs of uninfected C57BL/6J mice, which do not express hACE2, and mRNA isolated from human Caco-2 cells, known to express hACE2 (41–45) (see Fig. S4A and B in the supplemental material). Additionally, both primer sets targeting mAce2 and hACE2 successfully amplified mAce2 and hACE2 transcripts from the lung of K18-hACE2 mice (Fig. S4C).
Validation of the specificity of the mAce2 and hACE2 primer pairs. Primers used for mAce2 and hACE2 expression measurements were tested in mRNA extracted from lung lysates of C57BL/6J mice (A), in mRNA derived from human colorectal adenocarcinoma (Caco-2) cell pellets (B) and in mRNA extracted from lungs lysates of K18-hACE2 mice (C). Only mAce2 or hACE2 transcripts are detected in the lung of C57BL/6J mice or Caco-2 cells, respectively (A and B), while both mAce2 and hACE2 transcripts are expressed in the lung of K18-hACE2 mice (C). ΔCT values represent the difference of CT between the gene of interest and housekeeping genes. N.D., not detected. Download FIG S4, TIF file, 0.1 MB (88.2KB, tif) .
Copyright © 2023 Thieulent et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A significant reduction of mAce2 expression was observed at 2 dpi in the lung of MA10- (P = 0.005) and USA-WA1/2020-infected (P = 0.007) K18-hACE2 mice and remained low over the course of infection (P ≤ 0.001 for MA10- and USA-WA1/2020-infected mice) (Fig. 8A). No change in relative hACE2 expression was observed at any stage of infection compared to that of the mock-infected mice (Fig. 8B). The relative mTmprss2 expression was significantly reduced at 4 dpi in the lung of MA10-infected K18-hACE2 mice (P = 0.034) and USA-WA1/2020-infected mice at 4 dpi (P = 0.037) and 6 dpi (P = 0.034) (Fig. 8C).
FIG 8.
Mouse Ace2 (mAce2) expression, but not hACE2, is downregulated in the lung of SARS-CoV-2-infected K18-hACE2, C57BL/6J, and BABL/c mice. The relative expression of mAce2 (A), hACE2 (B), and mTmprss2 (C) was evaluated in the lung of K18-hACE2 mice at 2, 4, and 6 dpi following infection with SARS-CoV-2 MA10 and USA-WA1/2020 strains. The relative expression of mAce2 (D and F) and mTmprss2 (E and G) expression were evaluated at 2, 4, and 7 dpi in the lung of mock and MA10-infected C57BL/6J and BALB/c mice, respectively. Bars represent the mean ± standard deviation. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
To evaluate if the downregulation of mAce2 and mTmprss2 occurs specifically in the lung of infected K18-hACE2 mice, we also determined the expression levels in the lungs of infected C57BL/6J and BALB/c mice. A similar trend in mAce2 downregulation was noted in the lung of infected C57BL/6J (P ≤ 0.001) (Fig. 8D) and BALB/c mice (P = 0.002) (Fig. 8F). As anticipated, no hACE2 expression was detected in either C57BL/6J or BALB/c mice (data not shown). A significant downregulation of mTmprss2 expression was observed at 2 dpi in the lung of BALB/c mice (P = 0.017) (Fig. 8G) but not in C57BL/6J mice (Fig. 8E).
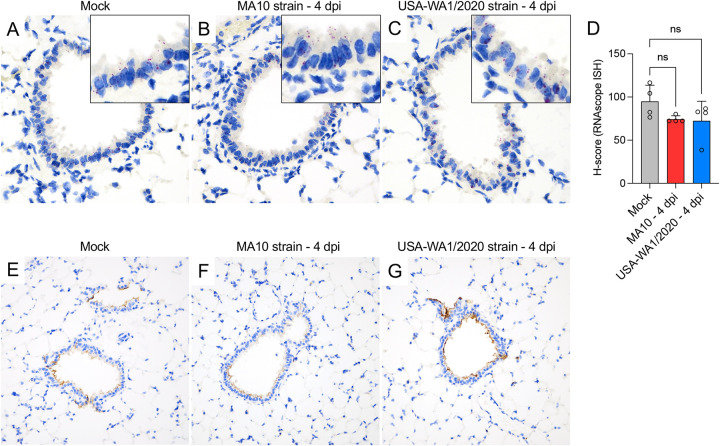
The mAce2 expression and distribution was also analyzed by RNAscope in situ hybridization (ISH) using a mouse-specific probe and by IHC in K18-hACE2 mice (Fig. 9). Expression of mAce2/ACE2 mainly involved bronchiolar epithelial cells with apical protein expression, and sporadic expression in AT2 cells (Fig. 9A to C and Fig. 9E to G). Tissue-based quantification of mAce2 noted a reduction trend of mAce2 expression at 4 dpi in both infected groups; statistical significance was not reached as observed via quantitative PCR (qPCR) analysis, likely due to differences in assay’s sensitivity (Fig. 9D, MA10 versus Mock, P = 0.078; USA-WA1/2020 versus Mock, P = 0.753). No differences in the distribution and abundance of ACE2 expression were identified between mock-infected and both infected groups at 4 dpi (Fig. 9E to G).
FIG 9.
Expression and distribution of mAce2 mRNA and ACE2 protein in the lungs of K18-hACE2 mice. mAce2 mRNA expression and distribution was evaluated by RNAscope ISH in the lung of mock-, MA10-, and USA-WA1/2020-infected mice at 4 dpi (A to C). Expression was predominantly observed within the bronchiolar epithelium, with expression in only rare pneumocytes (A to C, insets). mAce2 mRNA expression was semiquantitated in the bronchiolar epithelium by H-score calculation; while a reduction trend in expression level was noted, this was not statistically significant (D). Distribution of ACE2 protein in the lung of mock-, MA10-, and USA-WA1/2020-infected mice at 4 dpi (E to G). ACE2 has an apical distribution predominantly within bronchiolar epithelium and rarely in AT2 cells. Fast Red (mAce2 mRNA) (A to C) and DAB (brown, ACE2) (E to G). ×200 total magnification.
DISCUSSION
Since the discovery of SARS-CoV-2, several animal models have been used to understand its pathophysiology (46–48). Based on the low affinity interaction between murine ACE2 and the RBD of the ancestral, Wuhan-like SARS-CoV-2 S glycoprotein, the transgenic mouse expressing hACE2 under the K18 promoter (K18-hACE2) has become, among others, an extensively used model to study SARS-CoV-2 pathogenesis and preclinically assess therapeutic and vaccine efficacy (29). The Syrian hamster is another popular model for such studies with some advantageous features compared to the K18-hACE2 model, including clinical recovery following infection (i.e., nonlethal model), lack of neuroinvasion/neurodissemination, and development of pronounced bronchointerstitial pneumonia with subsequent intense airway reparative response. Such features have allowed better parallelism with disease in humans (49–51). However, not all animal biosafety level 3 (ABSL-3) facilities have the necessary infrastructure to handle this rodent model and also the lack of immunological reagents available limits their utilization (52). Currently, there is no animal model that perfectly recapitulates all of the features of COVID-19, and, therefore, careful selection and continued refinement of appropriate rodent models to test hypotheses, vaccine efficacy, and treatments related to understanding the mechanisms of SARS-CoV-2 in humans is extremely important. As an alternative approach, Leist et al. developed a mouse-adapted SARS-CoV-2 strain (namely, MA10) engineered to bind mACE2 and to which conventional mouse strains are permissive (34). To date, the vast majority of the studies in the literature have evaluated the pathogenicity of Wuhan-like SARS-CoV-2 and VOCs (i.e., Alpha, Beta, Delta, and Omicron variants) in K18-hACE2 mice (35, 53–55), with only a single study evaluating the replication of the mouse-adapted MA10 strain in this specific mouse model (39). Thus, far, no in-depth characterization of the differences in infection dynamics and pathophysiology between the mouse-adapted MA10 strain and the counterpart USA-WA1/2020 in K18-hACE2 mice has been undertaken. Therefore, the main objective of this study was to compare the dynamics and differential pathogenesis of both MA10 and its parental strain, USA-WA1/2020, on the widely used K18-hACE2 mouse model. Overall, our study demonstrated that the mouse-adapted MA10 strain induces earlier clinical decline, accelerated viral replication, earlier shift in pulmonary host responses, and displays differential tropism compared to its parental counterpart in K18-hACE2 mice. This comparison is highly relevant, as it provides critical comparative data on viral tropism and temporal dynamics that would better inform study design when utilizing these strains in this widely used mouse model.
Based on the analysis of lungs from autopsied patients, SARS-CoV-2 antigen is mainly detected in the majority of bronchiolar epithelial cell types and occasional AT1 and AT2 cells (56–58). In this study, we demonstrated that the MA10 strain can infect both bronchiolar and alveolar (AT1 and AT2) epithelia in K18-hACE2 mice, whereas the tropism of the USA-WA1/2020 strain is restricted only to alveolar (AT1 and AT2) cells. Since differences in tropism could be related to cell-specific expression of ACE2, the main host receptor for SARS-CoV-2, we sought to analyze its expression in the lung of mice. We demonstrated that mAce2 is mainly expressed in the bronchiolar epithelium and in scattered AT2 cells (59). Similar localization of hACE2, when driven by the K18 promoter, was recently demonstrated (35), but surprisingly, no viral antigen was detected within bronchiolar epithelia after USA-WA1/2020 infection. Interestingly, when comparing viral tropism of the MA10 strain in alveolar cells of K18-hACE2 and BALB/c mice, we observed that, while viral antigen is diffusely expressed in both AT1 and AT2 cells in K18-hACE2, viral antigen seems mostly restricted to AT2 cells in BALB/c mice. Results from our study suggest that the MA10 strain exploits both mACE2 and hACE2 receptor, and utilization of mACE2 is likely responsible for the increased cellular tropism and infection of bronchiolar epithelium. Such observation is also supported by the earlier neuroinvasion noted in MA10-infected K18-hACE2 mice. While the bronchiolar epithelial infection is transient, the use of the MA10 strain in the K18-hACE2 mice seems to be an alternative model with a more comparable tropism to that seen in COVID-19 patients than that of the USA-WA1/2020 strain.
Interestingly, our results demonstrate a pronounced and specific downregulation of mAce2 expression after SARS-CoV-2 infection, in the absence of similar changes in hACE2 transcripts, regardless of the virus strain used. This is in contrast to a previous study (29), where hACE2 expression was reported to be decreased in the lung of K18-hACE2 mice after infection with a similar dose of USA-WA1/2020. The hACE2 expression in this previous study was measured in copy per mg of tissue (i.e., absolute quantification), which cannot be compared head-to-head with relative quantification (29). However, the specificity of the primer-probe combination used and the absence of mAce2 amplification were not reported. The specificity of our assays was thoroughly evaluated in wild-type and hACE2 transgenic mice, and the Caco-2 human-derived cell line known for expressing ACE2 (41–45). The reduction of mAce2 expression was also observed in the lung of infected C57BL/6J and BALB/c mice, confirming that it is indeed mAce2, not hACE2, that is downregulated in the lung of infected K18-hACE2 mice. The inability to detect a discernible decline of ACE2 protein expression by IHC is explained by the cross-reactive nature of the anti-ACE2 antibody used for both hACE2 and mACE2. Unfortunately, there were no monospecific antisera to either hACE2 or mACE2 to make a clear distinction between the expression of these two receptors in mouse tissues using IHC. It was previously demonstrated that mAce2 expression is downregulated in the lung of mice infected with SARS-CoV by Western blotting (60). Similar results were obtained with influenza A H1N1 infections (61) and influenza A H5N1 (62). The lack of downregulation of hACE2 could possibly be associated with the high number of transgene copies in K18-hACE2 mice as previously reported (63). Further studies are needed to understand the pathogenic relevance of this change and its potential impact on pulmonary responses. To our knowledge, no previous study reports a change in Tmprss2 expression after SARS-CoV-2 infection. Here, we identified a reduction of Tmprss2 expression in the lung of K18-hACE2 mice regardless of the SARS-CoV-2 strain used and a transient reduction in the lung of BALB/c mice infected with the MA10 strain. Interestingly, only a trend of Tmprss2 downregulation was observed in the lungs of infected C57BL/6J mice, which is the least susceptible mouse strain to MA10 infection (34). Epithelial turnover after SARS-CoV-2 infection may be an explanation for the transient Tmprss2 downregulation, since Tmprss2 is rich within bronchiolar epithelial cells (see Fig. S5 in the supplemental material).
Expression of mTmprss2 mRNA in the lung of mice using RNAscope ISH. Tmprss2 is rich within bronchiolar epithelia (inset 1) and sporadic in AT1 and AT2 cells (inset 2). Fast Red (mTmprss2 mRNA); ×200 total magnification. Download FIG S5, TIF file, 3.9 MB (4MB, tif) .
Copyright © 2023 Thieulent et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
An additional important feature of MA10 infection in K18-hACE2 mice is related to the earlier pulmonary innate immune responses and differences in the cytokine/chemokine inflammatory signature compared to the parental strain. This, along with the overexpression of a higher number of proinflammatory cytokines/chemokines in the lung of MA10-infected mice, is related to the accelerated viral replication with rapid increase in viral titers and with the differential pulmonary tropism compared to its parental strain. The lack of IL-2 overexpression, coupled with the increase of cytokines/chemokines involved in macrophage and monocyte recruitment (i.e., IL-6, MCP-1/CCL2, and GM-CSF) reflects the fact that the local cellular response in the infected lung is mediated primarily by macrophages rather than lymphocytes (35). Of note, the absence of a pronounced IFN-α-mediated response in the lung of K18-hACE2 mice after infection with both strains suggests inhibition of type I interferons, which is consistent with previous reports in infected human patients (64).
The lack of SARS-CoV-2 neuroinvasion in conventional laboratory mice (i.e., BALB/c and C57BL/6J) showed in this as well as other studies (31, 65, 66) further highlights that hACE2 is a key but likely not the sole factor associated with the neuropathogenesis in the K18-hACE2 model (35). As previously reported, lethality in this mouse model is associated with viral neuroinvasion and neurodissemination via transport to the olfactory bulb originating from axonal processes traversing the olfactory neuroepithelium (35). Based on the rapid neurodissemination following intranasal challenge with the MA10 strain, we hypothesize that the presence of both mACE2 and hACE2 facilitates entry and rapid dissemination, explaining the higher level of viral titers/viral RNA and antigen in the brain by 4 dpi and the earlier weight loss. This difference occurs within a narrow time frame following infection as both MA10- and USA-WA1/2020-infected K18-hACE2 mice reach similar levels of viral replication, antigen abundance, histologic alterations, and clinical decline/mortality by 6 dpi.
Based on all of these observations, the use of the MA10 strain in the K18-hACE2 model provides a better representation of human COVID-19 in terms of viral tropism and progression of bronchointerstitial pneumonia, yet it is still imperfect and with important limitations that overall preclude the mouse from being the most faithful or relevant model of COVID-19. Major limitations in the utilization of both wild-type and K18-hACE2 mice have been identified (here and elsewhere) and mainly include marginal disease phenotypes (in wild-type mice) and absence of interstitial pneumonia uniquely depicting the features of diffuse alveolar damage (DAD) and subsequent reparative fibrosis that characterize pneumonia in human patients (absent in both wild-type and transgenic mice). Additionally, SARS-CoV-2 neuroinvasion and neurodissemination specifically makes the K18-hACE2 model invariably and prematurely fatal and, thus, limits its use for studying virus-specific immune responses, pulmonary repair mechanisms, and viral long-term effects (i.e., post-acute sequela of COVID-19 [long COVID-19]). Another important consideration relates to the virus strain used, and the utilization of the mouse-adapted MA10 strain may not necessarily reflect the pathogenicity of VOCs currently circulating in the population. Therefore, the mouse model cannot be appraised as the sole model for comprehensive understanding of SARS-CoV-2 pathogenesis, and multiple alternative animal models are needed to understand different aspects of COVID-19 in a complementary manner. Consequently, thorough characterization of virus strains in specific animal models and identification of their limitations is necessary to assess translational potential and further refine model systems.
To conclude, this study provides further insight into the comparative dynamics of SARS-CoV-2 MA10 and USA-WA1/2020 strains in the K18-hACE2 transgenic mouse model along with temporal pulmonary mAce2, hACE2, and mTmprss2 expression analysis. This study provides extensive evidence that the MA10 strain replicates and disseminates more rapidly in the lungs and brain of K18-hACE2, exhibits differences in pulmonary tropism, and induces an earlier proinflammatory and monocyte chemoattractant chemokine response compared to the parental USA-WA1/2020 strain; usage of both mACE2 and hACE2 as host receptors by MA10 is likely the underlying mechanism driving these dynamics. These differences could be of significant impact to experimental conditions, and, therefore, this study provides critical information that should be considered for study design based on strain selection, strain differences in cell tropism, and the experimental time points selected for downstream analysis. Using the MA10 strain along with the K18-hACE2 mouse model represents an additional model for understanding the mechanisms governing SARS-CoV-2 pathogenesis and evaluating the efficacy of therapeutics and vaccines.
MATERIALS AND METHODS
Biosafety.
All aspects of this study were approved by the Institutional Biosafety and Recombinant DNA Safety Committee, Office of Environmental Health and Safety at Louisiana State University (LSU) prior to study initiation. Work with SARS-CoV-2 was performed in a biosafety level-3 (BSL-3 and ABSL-3) laboratory at the School of Veterinary Medicine, LSU by personnel equipped with powered air-purifying respirators.
Ethical statement.
All experimental procedures were approved by the Institutional Animal Care and Use Committee at Louisiana State University (IACUC protocol number 20-091). Euthanasia was performed via isoflurane overdose and subsequent cervical dislocation following the American Veterinary Medical Association (AVMA) Guidelines for the Euthanasia of Animals.
Cells and viruses.
African green monkey VERO C1008 cells (Vero 76, clone E6, Vero E6) (ATCC CRL-1587, Manassas, VA) were maintained in minimum essential medium (MEM) with Earl’s salts and l-glutamine (Corning, Corning, NY) supplemented with 10% fetal bovine serum (FBS) (HyClone Laboratories, Inc., Logan, UT), 100 U/mL of penicillin and 100 μg/mL streptomycin (Gibco, Carlsbad, CA), and 0.25 μg/mL of amphotericin B (Gibco). Caco-2 colorectal adenocarcinoma cells (ATCC HTB-37) were maintained in MEM as described above but contained 20% FBS. Cells were incubated at 37°C and 5% CO2 in a humidified incubator.
The SARS-CoV-2 mouse-adapted strain, MA10 variant (NR-55329), and the SARS-CoV-2 isolate USA-WA1/2020 (NR-52281) were obtained from BEI Resources (Manassas, VA, USA). Virus stocks (passage 3) were prepared in Vero E6 cells, titrated by plaque assay, and stored at −80°C for experimental infections. The genomes of both SARS-CoV-2 MA10 and USA-WA1/2020 strains at passage 3 were fully sequenced at the Iowa State University Veterinary Diagnostic Laboratory, Ames, IA. Consensus genome sequences were analyzed using Geneious R7 (BioMatters, Auckland, New Zealand) and demonstrated to possess 99.9% and 100% homology with the reference genome sequences, respectively (MA10 strain [GenBank accession number MT952602.1]; USA-WA1/2020 strain [GenBank accession number MW811435.1]).
Mice.
Twelve-week-old male B6.Cg-Tg(K18-ACE2)2Prlmn/J (strain number 034860, K18-hACE2; n = 28) and 14-week-old male C57BL/6J mice (strain no. 000664; n = 12) were obtained from The Jackson Laboratory (JAX, Bar Harbor, ME). Additionally, 10-week-old male BALB/cAnNHsd mice (strain number 047, BALB/c; n = 16) were obtained from Envigo (Indianapolis, IN). Mice were housed in groups of 3 to 4 per cage under a 12-h light/12-h dark cycle with ad libitum access to water and a standard chow diet.
Experimental design and intranasal inoculation.
Since K18-hACE2 mice typically reach humane euthanasia criteria (associated with viral neurodissemination) within the first week following SARS-CoV-2 infection (28, 29, 35, 67), all experiments were performed for a maximum duration of 7 dpi. K18-hACE2 mice were randomly assigned to the MA10-infected group (n = 12), WA1-infected group (n = 12), or the mock-infected group (n = 4). C57BL/6J and BALB/c mice were randomly assigned to the MA10-infected group (n = 9 and n = 12, respectively) or the mock-infected group (n = 3 and n = 4, respectively). For intranasal inoculation, mice were anesthetized with 4% isoflurane (Fluriso; VetOne, Boise, ID) (oxygen, 3 to 4 L/min) in an induction chamber until deeply anesthetized. Mice were intranasally inoculated with either 50 μL (25 μL per nostril) of MEM containing 2 × 104 PFU of the SARS-CoV-2 MA10 strain, SARS-CoV-2 USA-WA1/2020 strain, or mock inoculated with 50 μL of plain MEM. Mice were then transferred to their cages and monitored until completely recovered (approximately 10 min). Three (C57BL/6J) and four (K18-hACE2 and BALB/c) infected mice per group were euthanized at 2 dpi, 4 dpi, and 7 dpi or when mice reached predetermined humane euthanasia criteria. All mock-infected mice were euthanized at 6 dpi (K18-hACE2) or 7 dpi (C57BL/6J and BALB/c).
Clinical monitoring.
An IACUC-approved clinical scoring system was utilized to monitor disease progression as previously described (35). Clinical signs and body weight were monitored once a day for the duration of the experiment (7 dpi). A score of 0 or 1 was given for each of the following parameters: body weight loss greater than 15%; rapid/shallow respiration or increased respiratory effort; appearance (ruffled fur, hunched posture); responsiveness; and neurologic signs (e.g., tremors). Cumulative scores of 4 or greater for two consecutive observation periods, weight loss greater than or equal to 15%, severe respiratory distress, or lack of responsiveness were the criteria considered for electing humane euthanasia.
Tissue collection and processing.
The main left bronchus was clamped using Kelly forceps, and the right lung was insufflated with instillation of ~750 μL of 1% low-melting-point agarose (Thermo Fisher Scientific, Waltham, MA) diluted in 1× phosphate-buffered saline using a 24-gauge catheter placed into the trachea. The left lung was divided into ~30 mg samples and stored at −80°C either as fresh frozen or in RNAlater solution (Invitrogen, Waltham, MA) in 1.5 mL microfuge tubes containing 2.4 mm metal grinding beads (Omni International, Kennesaw, GA). Subsequently, the skull cap was removed, and ~30 mg of left frontal cortex was collected and stored at −80°C in 1.5 mL microfuge tubes containing 2.4 mm metal grinding beads. The insufflated right lung and head were fixed in 10% neutral buffered formalin at a 20:1 fixative to tissue ratio for a minimum of 72 h before removal from BSL-3 in accordance with an approved institutional standard operating procedure.
Viral RNA isolation and quantitation by SARS-CoV-2 ORF1ab-specific real-time RT-qPCR.
Approximately 30 mg of fresh frozen lung and brain were homogenized using a Qiagen TissueLyser II (Qiagen, Valencia, CA) in MEM (1:10 volume; 10% tissue homogenates) by two dissociation cycles of 2 min at 30 Hz. Samples were clarified by centrifugation at 3,000 × g for 10 min at 4°C. A total of 100 μL of clarified 10% tissue homogenate were collected, and viral RNA isolation was subsequently performed using the Taco nucleic acid automatic extraction system (GeneReach Biotechnology Corporation, Taichung, Taiwan) as previously described (68) and eluted in 100 μL of elution buffer. Reverse transcription-quantitative PCR (RT-qPCR) targeting ORF1ab of SARS-CoV-2 (69, 70) was performed using a 7500 fast real-time PCR system (Applied Biosystems). Sequences of primers, probe, and thermal cycling are shown in Table 1. RT-qPCRs were performed in a total volume of 20 μL containing 5 μL of 4× TaqPath 1-step RT-qPCR master mix (Applied Biosystems), 1 μL of 20× primers (500 nM)/probe (250 nM) mix, 6 μL of nuclease-free water, and 5 μL of extracted RNA. For absolute quantitation of viral RNA, a standard curve using a synthesized in vitro transcribed RNA containing a 132-bp fragment from the SARS-CoV-2 ORF1ab gene was used. Briefly, the target sequence was chemically synthesized and cloned into the pGEM-3Z vector (Promega, Madison, WI), and in vitro RNA transcription was performed as previously described (71, 72). Synthesized RNA was purified using the MEGAclear transcription clean-up kit (Thermo Fisher Scientific) and quantitated using a Qubit 4 fluorometer (Invitrogen). The analytical sensitivity of the assay was determined to be 1 genome copy/μL.
TABLE 1.
Primers and probe sequences used for SARS-CoV-2 RNA quantitation
| Target | Primers and probe sequences (5′–3′)a | Cycling temp and time | Reference |
|---|---|---|---|
| SARS-CoV-2 | HKU-ORF1-F: TGGGGYTTTACRGGTAACCT | 25°C, 2 min | 69 |
| ORF1ab | HKU-ORF1-R: AACRCGCTTAACAAAGCACTC | 50°C, 15 min | |
| HKU-ORF1-P: FAM-TAGTTGTGATGCWATCATGACTAG-BHQ1 | 95°C, 2 minb | ||
| 95°C, 3 secb | |||
| 60°C, 30 secb |
F, forward; R, reverse; P, probe.
40 cycles of amplification.
Virus titer determination by plaque assay.
Clarified (10%) lung and brain homogenates were subjected to 10-fold serial dilutions in MEM and inoculated in Vero E6 cells preseeded in 12-well plates. After 1 h of adsorption at 37°C, cells were overlaid with complete MEM containing 0.75% carboxymethylcellulose (Sigma-Aldrich, Saint-Louis, MO) and incubated for 72 h at 37°C and 5% CO2 in a humidified incubator. Media was finally removed, and cells were fixed and stained with 0.2% crystal violet solution (Sigma-Aldrich) in 10% neutral buffered formalin for 4 h and subsequently rinsed with tap water. Plaques were counted, and viral titers were determined as PFU per milligram of tissue.
Chemokine and cytokine analysis.
Chemokine and cytokine protein expression analysis was performed on collected lung tissue using the Bio-Plex Pro mouse chemokine panel 31-plex (Bio-Rad, Hercules, CA). Approximately 30 mg of lung was homogenized in 500 μL of Bio-Plex cell lysing solution (Bio-Rad) according to the manufacturer’s recommendations using a Qiagen TissueLyser II as described above, followed by one freeze-thaw cycle at −80°C. Samples were centrifuged at 4,500 × g for 10 min at 4°C and subsequently diluted 1:10 in sample diluent, processed following the manufacturer’s recommendations, and read on a Bio-Plex 200 system (Bio-Rad). Each sample was analyzed in duplicate wells. The concentration of interferon-alpha (IFN-α) was measured using the mouse IFN-α enzyme-linked immunosorbent assay (ELISA) kit (Invitrogen) according to the manufacturer’s recommendations. Each sample was analyzed in duplicate wells, and absorbance was read at 450 nm using a BOLT automated ELISA system (Gold Standard Diagnostics, Davis, CA). Cytokine/chemokine concentrations were calculated as picograms per milligram of tissue.
Total cellular RNA isolation and gene expression analysis for selected cytokines/chemokines.
Gene expression analysis of cellular genes involved in SARS-CoV-2 binding/entry (mAce2, hACE2, and mTmprss2) as well as a subset of cytokines/chemokines, including Mcp1/Ccl2, Mip1a/Ccl3, Mip1b/Ccl4, Gm-csf, Il2, Il6, Ip10/Cxcl10, and Tnfa, were measured as previously described (73). Approximately 30 mg of frozen lung tissue was homogenized in 600 μL of Buffer RLT Plus (Qiagen) containing 1% of β-mercaptoethanol using a Qiagen TissueLyser II (Qiagen) as described above. Total RNA was extracted using the RNeasy Plus minikit (Qiagen) according to the manufacturer’s recommendations, and then 2 μg of total RNA was reverse transcribed using the high-capacity cDNA reverse transcription kit (Applied Biosystems) according to the manufacturer’s recommendations. The 2× PowerUp SYBR green master mix (Applied Biosystems) was used for qPCRs, and these were performed using a QuantStudio 5 real-time PCR system (Applied Biosystems) as previously described (73). Melt curve analysis was performed to confirm nonspecific amplifications along with the inclusion of nontemplate controls. Specific forward and reverse primers for genes of interest were designed using the Primer-BLAST website (NCBI, NIH) and are shown in Table 2. Specificity of the mAce2 and hACE2 primers were confirmed using RNA extracted from lungs of C57BL/6J mice and Caco-2 (Caco2) (ATCC HTB-37) cell pellets. Quantitative PCR efficiencies and cycle threshold (CT) values were determined using LinRegPCR version 2021.2 (74). Reactions were performed in duplicate and expression of genes of interest were normalized to three stable housekeeping genes (Actb, Gusb, and Rpl13b) as previously described (75).
TABLE 2.
Primer sequences used for gene expression analysis
| Target (GenBank accession no.)a | Forward primer (5′–3′) | Reverse primer (5′–3′) | Product length (bp) |
|---|---|---|---|
| Human ACE2 (NM_001371415.1) | AGAAAGCAGTCTGCCATCCC | GCTGTCAGGAAGTCGTCCAT | 97 |
| Mouse Ace2 (NM_001130513.1) | GTCATGGATGCGCTTTGGAT | CTTGGGTTGGGCACTGCTTA | 88 |
| Tmprss2 (NM_015775.2) | TGACGGGGTAGCACATTGTC | CACACGGGATACCAGGCTTT | 117 |
| Ccl2 (NM_011333.3) | TGACCCCAAGAAGGAATGGG | ACCTTAGGGCAGATGCAGTT | 104 |
| Ccl3 (NM_011337.2) | ACATTCCTGCCACCTGCATA | GAAGAGTCCCTCGATGTGGC | 74 |
| Ccl4 (NM_013652.2) | CTAACCCCGAGCAACACCAT | CCATTGGTGCTGAGAACCCT | 97 |
| Cxcl10 (NM_021274.2) | CTGAGTCCTCGCTCAAGTGG | GTCGCACCTCCACATAGCTT | 69 |
| Gm-csf (NM_009969.4) | GCCAGTTCTTGGAAGGGCTTAT | ATCTCCTGGCCCTTATCAGCTA | 99 |
| Il-2 (NM_008366.3) | GAAACTCCCCAGGATGCTCA | CGCAGAGGTCCAAGTTCATCT | 99 |
| Il-6 (NM_031168.2) | GGGACTGATGCTGGTGACAA | ACAGGTCTGTTGGGAGTGGT | 90 |
| Tnfα (NM_013693.3) | AGCCGATGGGTTGTACCTTG | ATAGCAAATCGGCTGACGGT | 99 |
| Actb (NM_007393.5) | CACTGTCGAGTCGCGTCC | TCATCCATGGCGAACTGGTG | 89 |
| Gusb (NM_001357025.1) | GGCGATGGACCCAAGATACC | ACCCTTGGGATACCACAACT | 84 |
| Rpl13a (NM_001357025.1) | CCCACAAGACCAAGAGAGGC | CACCATCCGCTTTTTCTTGTCA | 92 |
Ace2/ACE2, angiotensin converting enzyme 2; Tmprss2, transmembrane serine protease 2; Ccl, C-C motif chemokine ligand; Cxcl10, C-X-C motif chemokine ligand 10; Gm-csf, granulocyte-macrophage colony-stimulating factor; Il, interleukin; Tnfα, tumor necrosis factor α; Actb, actin, beta; Gusb, glucuronidase beta; Rpl13a, ribosomal protein L13a.
Histopathology.
Following fixation, the whole head was decalcified in Immunocal Decalcifier (StatLab, McKinney, TX) for 5 days followed by thorough rinsing in tap water per manufacturer’s recommendations before performing a midsagittal section. Tissues (lung and heads) were subsequently processed, embedded in paraffin, and 4 μm sections stained with hematoxylin and eosin following standard histological procedures. Histological alterations were semiquantitatively scored as follows: 0, no histologic alterations; 1, minimal; 2, mild; 3, moderate; and 4, severe.
SARS-CoV-2-specific immunohistochemistry.
For SARS-CoV-2 IHC, 4 μm sections of formalin-fixed paraffin-embedded tissue were mounted on positively charged Superfrost Plus slides and subjected to IHC using antinucleocapsid rabbit polyclonal antibody (3A) as previously described (76–82) on the automated BOND-RXm platform (Leica Biosystems, Nussloch, Germany). SARS-CoV-2 N antigen abundance was semiquantitatively scored as follows: 0, no viral protein observed; 1, up to 5% positive target cells per 400× field examined; 2, 5 to 25% positive target cells per 400× field examined; 3, up to 50% positive target cells per 400× field examined; and 4, >50% positive target cells per 400× field examined. Additionally, the immunoreactive area was quantitated as indicated below.
For ACE2, tissue sections were prepared as described above and epitope retrieval performed using a ready-to-use EDTA-based solution (pH 9.0; Leica Biosystems) at 100°C for 20 min. Sections were then incubated with a ready-to-use hydrogen peroxide solution (Leica Biosystems) for 5 min and subsequently a rabbit monoclonal anti-ACE2 antibody (clone E5O6J [Cell Signaling Technologies, Danvers, MA, USA] diluted at 1:100 in primary antibody diluent [Leica Biosystems]) for 30 min at room temperature. Slides were finally incubated with a polymer-labeled goat anti-rabbit IgG coupled with horseradish peroxidase (8 min). 3′,3′ diaminobenzidine (DAB) was used as the chromogen (10 min), and counterstaining was performed with hematoxylin for 5 min. Slides were dried and mounted as indicated above.
RNAscope in situ hybridization for mouse Ace2.
For mouse Ace2 mRNA RNAscope ISH, an antisense probe targeting mouse Ace2 (GenBank accession number NM_001130513.1; catalog number 863818) with no cross-reactivity to hACE2 was used. For mouse Tmprss2 mRNA RNAscope ISH, an anti-sense probe targeting mouse Tmprss2 (GenBank accession number NM_015775.2; catalog number 496728) was used. The RNAscope ISH assay was performed using the RNAscope 2.5 LSx reagent kit (Advanced Cell Diagnostics, Newark, CA) on the automated BOND RXm platform (Leica Biosystems, Buffalo Grove, IL) as described previously (35).
Quantitative image analysis of SARS-CoV-2 antigen distribution and abundance.
Digitized whole-slide scans were analyzed using the image analysis software HALO (Indica Labs, Inc., Corrales, NM). Slides were manually annotated to include only the brain and/or lung parenchyma. Visualization threshold values were adjusted in viewer settings to reduce background signal. Area quantification (AQ) was performed to determine percentages of SARS-CoV-2 N antigen immunoreactivity in lungs and brains.
Quantitative image analysis of mouse Ace2 abundance.
Quantitative analysis for RNAscope ISH to mouse Ace2 was performed in QuPath 0.3.1 digital pathology image analysis software. Guidelines established by ACD were followed with some modifications. Briefly, stain vectors were adjusted for each slide, respectively, before pursuing further analysis. Since mouse Ace2 is most abundant in the bronchiolar epithelium, five approximately 300-μm-diameter bronchioles were randomly selected for analysis. The cell detection algorithm was performed with default values, and after cell segmentation was performed, the subcellular detection algorithm was applied for detection of Fast Red (Ace2) spots using default values except that minimum spot size was set to 0.1 and thresholds were fine-tuned for detection accuracy. The estimated number of spots were determined per cell per region and exported into an Excel file. Spots/cell (excluding clusters) for each tissue compartment were used to compute an H-score (range of 0 to 400) by binning cells with different levels of expression into separate bins (bin 0 [0 dots/cell], bin 1 [1 to 3 dots/cell], bin 2 [4 to 9 dots/cell], bin 3 [10 to 15 dots/cell], and bin 4 [greater than or equal to 15 dots/cell]). The H-score was computed using the weighted formula as follows: H-score = ∑ (bin number × % cells per bin). H-scores were subsequently used for statistical analysis.
Statistical analysis.
Statistical analysis was performed using GraphPad Prism v9.3.1 statistical analysis software (GraphPad, San Diego, CA) and JMP16 Pro (Cary, NC, USA). Kaplan-Meier curves were used for survival analysis. Weight changes were analyzed using two-way analysis of variance (ANOVA) with Sidak’s as the post hoc test. Viral loads (genomic copies/mg and PFU/mg) and relative gene expression levels were analyzed using two-way ANOVA with Tukey’s as the post hoc test. Clinical, histopathology, and viral antigen abundance scores and RNAscope ISH H-scores were analyzed using the nonparametric Kruskal-Wallis test followed by a Dunn’s post hoc test using the mock group as control. Chemokine and cytokine protein concentrations and percentages of immunoreactivity area were analyzed using the nonparametric Kruskal-Wallis test and a Dunn’s post hoc test for joint ranks. Fold change of cytokine and chemokine protein concentrations was calculated by dividing the protein concentrations of infected mice by the average protein concentrations of mock-infected mice, and a heatmap was build using package “pheatmap” in R. Data are expressed as mean ± standard deviation (SD). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
ACKNOWLEDGMENTS
The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: SARS-related coronavirus 2, isolate USA-WA1/2020, NR-52281. The following reagent was obtained through BEI Resources, NIAID, NIH: SARS-related coronavirus 2, mouse-adapted, MA10 variant (in isolate USA-WA1/2020 backbone), infectious clone (ic2019-nCoV MA10) in Calu-3 cells, NR-55329, contributed by Ralph S. Baric.
The authors would like to kindly acknowledge the Histology and Immunohistochemistry Laboratory at the Louisiana Animal Disease Diagnostic Laboratory (LADDL) and Abbie Fish, LSU Environmental Health and Safety. We also thank the personnel of the Comparative Pathology Laboratory at the National Emerging Infectious Diseases Laboratories (NEIDL), Boston University.
We declare that we have no competing interests.
Conceptualization, M.C.; Methodology, C.J.T., W.D., N.A.C., M.C., and U.B.R.B.; Protocol validation, C.J.T. and M.C.; Formal analysis, C.J.T., X.W., N.A.C., and M.C.; Writing—original draft preparation, C.J.T. and M.C.; Writing—review and editing, C.J.T., W.D., N.A.C., U.B.R.B., J.A.R., and M.C.; Supervision, M.C.; Project administration, M.C.; Funding acquisition, M.C.
All authors have read and agreed to the published version of the manuscript.
This study was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM130555 in addition to start-up funds (PG009641) from the School of Veterinary Medicine, Louisiana State University (to M.C.). We also acknowledge support from the interagency partnership program between NIH and the U.S. Department of Agriculture’s (USDA) National Institute of Food and Agriculture (NIFA) Dual Purpose with Dual Benefit: Research in Biomedicine and Agriculture Using Agriculturally Important Domestic Animal Species grant number 2019-67016-29102 (award number AWD-47990-1; to U.B.R.B.), fee-based funds (PG008671 to U.B.R.B. and M.C.), and the shared instrumentation grant S10-OD030269 (to N.A.C.).
J.A.R. was funded through grants from the Center of Emerging and Zoonotic Infectious Diseases (CEZID) from National Institute of General Medical Sciences (NIGMS) under award number P20GM130448, and the NIAID supported Center of Excellence for Influenza Research and Response (CEIRR, contract number 75N93021C00016).
Contributor Information
Mariano Carossino, Email: mcarossino1@lsu.edu.
Michael J. Imperiale, University of Michigan—Ann Arbor
REFERENCES
- 1.Hu B, Guo H, Zhou P, Shi Z-L. 2021. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol 19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. 2020. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. 2020. The proximal origin of SARS-CoV-2. 4. Nat Med 26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, Yuan M-L, Zhang Y-L, Dai F-H, Liu Y, Wang Q-M, Zheng J-J, Xu L, Holmes EC, Zhang Y-Z. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cucinotta D, Vanelli M. 2020. WHO declares COVID-19 a pandemic. Acta Biomed 91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johns Hopkins University of Medicine. 2022. Johns Hopkins University & Medicine: Coronavirus Resource Center. Johns Hopkins Coronavirus Resource Center. https://coronavirus.jhu.edu/. Retrieved 27 October 2022. [Google Scholar]
- 8.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A, Müller MA, Drosten C, Pöhlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuyama S, Nagata N, Shirato K, Kawase M, Takeda M, Taguchi F. 2010. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol 84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang K, Chen W, Zhang Z, Deng Y, Lian J-Q, Du P, Wei D, Zhang Y, Sun X-X, Gong L, Yang X, He L, Zhang L, Yang Z, Geng J-J, Chen R, Zhang H, Wang B, Zhu Y-M, Nan G, Jiang J-L, Li L, Wu J, Lin P, Huang W, Xie L, Zheng Z-H, Zhang K, Miao J-L, Cui H-Y, Huang M, Zhang J, Fu L, Yang X-M, Zhao Z, Sun S, Gu H, Wang Z, Wang C-F, Lu Y, Liu Y-Y, Wang Q-Y, Bian H, Zhu P, Chen Z-N. 2020. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Sig Transduct Target Ther 5:283. doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cantuti-Castelvetri L, Ojha R, Pedro LD, Djannatian M, Franz J, Kuivanen S, van der Meer F, Kallio K, Kaya T, Anastasina M, Smura T, Levanov L, Szirovicza L, Tobi A, Kallio-Kokko H, Österlund P, Joensuu M, Meunier FA, Butcher SJ, Winkler MS, Mollenhauer B, Helenius A, Gokce O, Teesalu T, Hepojoki J, Vapalahti O, Stadelmann C, Balistreri G, Simons M. 2020. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daly JL, Simonetti B, Klein K, Chen K-E, Williamson MK, Antón-Plágaro C, Shoemark DK, Simón-Gracia L, Bauer M, Hollandi R, Greber UF, Horvath P, Sessions RB, Helenius A, Hiscox JA, Teesalu T, Matthews DA, Davidson AD, Collins BM, Cullen PJ, Yamauchi Y. 2020. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 370:861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson CB, Farzan M, Chen B, Choe H. 2022. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol 23:3–20. doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. 2020. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang F, Kream RM, Stefano GB. 2020. Long-term respiratory and neurological sequelae of COVID-19. Med Sci Monit 26:e928996. doi: 10.12659/MSM.928996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goyal P, Choi JJ, Pinheiro LC, Schenck EJ, Chen R, Jabri A, Satlin MJ, Campion TR, Nahid M, Ringel JB, Hoffman KL, Alshak MN, Li HA, Wehmeyer GT, Rajan M, Reshetnyak E, Hupert N, Horn EM, Martinez FJ, Gulick RM, Safford MM. 2020. Clinical characteristics of Covid-19 in New York City. N Engl J Med 382:2372–2374. doi: 10.1056/NEJMc2010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tenforde MW. 2020. Symptom duration and risk factors for delayed return to usual health among outpatients with COVID-19 in a multistate health care systems network — United States, March–June 2020. MMWR Morb Mortal Wkly Rep 69:993–998. doi: 10.15585/mmwr.mm6930e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hojyo S, Uchida M, Tanaka K, Hasebe R, Tanaka Y, Murakami M, Hirano T. 2020. How COVID-19 induces cytokine storm with high mortality. Inflamm Regen 40:37. doi: 10.1186/s41232-020-00146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu B, Huang S, Yin L. 2021. The cytokine storm and COVID-19. J Med Virol 93:250–256. doi: 10.1002/jmv.26232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ragab D, Salah Eldin H, Taeimah M, Khattab R, Salem R. 2020. The COVID-19 cytokine storm; what we know so far. Front Immunol 11:1446. doi: 10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neufeldt CJ, Cerikan B, Cortese M, Frankish J, Lee J-Y, Plociennikowska A, Heigwer F, Prasad V, Joecks S, Burkart SS, Zander DY, Subramanian B, Gimi R, Padmanabhan S, Iyer R, Gendarme M, El Debs B, Halama N, Merle U, Boutros M, Binder M, Bartenschlag R. 2022. SARS-CoV-2 infection induces a proinflammatory cytokine response through cGAS-STING and NF-κB. Commun Biol 5:45. doi: 10.1038/s42003-021-02983-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costela-Ruiz VJ, Illescas-Montes R, Puerta-Puerta JM, Ruiz C, Melguizo-Rodríguez L. 2020. SARS-CoV-2 infection: the role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev 54:62–75. doi: 10.1016/j.cytogfr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Damas J, Hughes GM, Keough KC, Painter CA, Persky NS, Corbo M, Hiller M, Koepfli K-P, Pfenning AR, Zhao H, Genereux DP, Swofford R, Pollard KS, Ryder OA, Nweeia MT, Lindblad-Toh K, Teeling EC, Karlsson EK, Lewin HA. 2020. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc Natl Acad Sci USA 117:22311–22322. doi: 10.1073/pnas.2010146117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conceicao C, Thakur N, Human S, Kelly JT, Logan L, Bialy D, Bhat S, Stevenson-Leggett P, Zagrajek AK, Hollinghurst P, Varga M, Tsirigoti C, Tully M, Chiu C, Moffat K, Silesian AP, Hammond JA, Maier HJ, Bickerton E, Shelton H, Dietrich I, Graham SC, Bailey D. 2020. The SARS-CoV-2 spike protein has a broad tropism for mammalian ACE2 proteins. PLoS Biol 18:e3001016. doi: 10.1371/journal.pbio.3001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Liu J, Plante KS, Plante JA, Xie X, Zhang X, Ku Z, An Z, Scharton D, Schindewolf C, Widen SG, Menachery VD, Shi P-Y, Weaver SC. 2022. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature 602:294–299. doi: 10.1038/s41586-021-04245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bao L, Deng W, Huang B, Gao H, Liu J, Ren L, Wei Q, Yu P, Xu Y, Qi F, Qu Y, Li F, Lv Q, Wang W, Xue J, Gong S, Liu M, Wang G, Wang S, Song Z, Zhao L, Liu P, Zhao L, Ye F, Wang H, Zhou W, Zhu N, Zhen W, Yu H, Zhang X, Guo L, Chen L, Wang C, Wang Y, Wang X, Xiao Y, Sun Q, Liu H, Zhu F, Ma C, Yan L, Yang M, Han J, Xu W, Tan W, Peng X, Jin Q, Wu G, Qin C. 2020. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 27.McCray PB, Pewe L, Wohlford-Lenane C, Hickey M, Manzel L, Shi L, Netland J, Jia HP, Halabi C, Sigmund CD, Meyerholz DK, Kirby P, Look DC, Perlman S. 2007. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J Virol 81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yinda CK, Port JR, Bushmaker T, Owusu IO, Purushotham JN, Avanzato VA, Fischer RJ, Schulz JE, Holbrook MG, Hebner MJ, Rosenke R, Thomas T, Marzi A, Best SM, de Wit E, Shaia C, van Doremalen N, Munster VJ. 2021. K18-hACE2 mice develop respiratory disease resembling severe COVID-19. PLoS Pathog 17:e1009195. doi: 10.1371/journal.ppat.1009195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winkler ES, Bailey AL, Kafai NM, Nair S, McCune BT, Yu J, Fox JM, Chen RE, Earnest JT, Keeler SP, Ritter JH, Kang L-I, Dort S, Robichaud A, Head R, Holtzman MJ, Diamond MS. 2020. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat Immunol 21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hassan AO, Case JB, Winkler ES, Thackray LB, Kafai NM, Bailey AL, McCune BT, Fox JM, Chen RE, Alsoussi WB, Turner JS, Schmitz AJ, Lei T, Shrihari S, Keeler SP, Fremont DH, Greco S, McCray PB, Perlman S, Holtzman MJ, Ellebedy AH, Diamond MS. 2020. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell 182:744–753. doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu H, Chen Q, Yang G, He L, Fan H, Deng Y-Q, Wang Y, Teng Y, Zhao Z, Cui Y, Li Y, Li X-F, Li J, Zhang N-N, Yang X, Chen S, Guo Y, Zhao G, Wang X, Luo D-Y, Wang H, Yang X, Li Y, Han G, He Y, Zhou X, Geng S, Sheng X, Jiang S, Sun S, Qin C-F, Zhou Y. 2020. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science 369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan F, Li E, Wang T, Li Y, Liu J, Wang W, Qin T, Su R, Pei H, Wang S, Feng N, Zhao Y, Yang S, Xia X, Gao Y. 2022. Characterization of two heterogeneous lethal mouse-adapted SARS-CoV-2 variants recapitulating representative aspects of human COVID-19. Front Immunol 13:821664. doi: 10.3389/fimmu.2022.821664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dinnon KH, Leist SR, Schäfer A, Edwards CE, Martinez DR, Montgomery SA, West A, Yount BL, Hou YJ, Adams LE, Gully KL, Brown AJ, Huang E, Bryant MD, Choong IC, Glenn JS, Gralinski LE, Sheahan TP, Baric RS. 2020. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leist SR, Dinnon KH, Schäfer A, Tse LV, Okuda K, Hou YJ, West A, Edwards CE, Sanders W, Fritch EJ, Gully KL, Scobey T, Brown AJ, Sheahan TP, Moorman NJ, Boucher RC, Gralinski LE, Montgomery SA, Baric RS. 2020. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell 183:1070–1085. doi: 10.1016/j.cell.2020.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carossino M, Kenney D, O’Connell AK, Montanaro P, Tseng AE, Gertje HP, Grosz KA, Ericsson M, Huber BR, Kurnick SA, Subramaniam S, Kirkland TA, Walker JR, Francis KP, Klose AD, Paragas N, Bosmann M, Saeed M, Balasuriya UBR, Douam F, Crossland NA. 2022. Fatal neurodissemination and SARS-CoV-2 tropism in K18-hACE2 mice is only partially dependent on hACE2 expression. Viruses 14:535. doi: 10.3390/v14030535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vidal E, López-Figueroa C, Rodon J, Pérez M, Brustolin M, Cantero G, Guallar V, Izquierdo-Useros N, Carrillo J, Blanco J, Clotet B, Vergara-Alert J, Segalés J. 2022. Chronological brain lesions after SARS-CoV-2 infection in hACE2-transgenic mice. Vet Pathol 59:613–626. doi: 10.1177/03009858211066841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song E, Zhang C, Israelow B, Lu-Culligan A, Prado AV, Skriabine S, Lu P, Weizman O-E, Liu F, Dai Y, Szigeti-Buck K, Yasumoto Y, Wang G, Castaldi C, Heltke J, Ng E, Wheeler J, Alfajaro MM, Levavasseur E, Fontes B, Ravindra NG, Van Dijk D, Mane S, Gunel M, Ring A, Kazmi SAJ, Zhang K, Wilen CB, Horvath TL, Plu I, Haik S, Thomas J-L, Louvi A, Farhadian SF, Huttner A, Seilhean D, Renier N, Bilguvar K, Iwasaki A. 2021. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J Exp Med 218:e20202135. doi: 10.1084/jem.20202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumari P, Rothan HA, Natekar JP, Stone S, Pathak H, Strate PG, Arora K, Brinton MA, Kumar M. 2021. Neuroinvasion and encephalitis following intranasal inoculation of SARS-CoV-2 in K18-hACE2 mice. Viruses 13:132. doi: 10.3390/v13010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Natekar JP, Pathak H, Stone S, Kumari P, Sharma S, Auroni TT, Arora K, Rothan HA, Kumar M. 2022. Differential pathogenesis of SARS-CoV-2 variants of concern in human ACE2-expressing mice. Viruses 14:1139. doi: 10.3390/v14061139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thwaites RS, Sanchez Sevilla Uruchurtu A, Siggins MK, Liew F, Russell CD, Moore SC, Fairfield C, Carter E, Abrams S, Short C-E, Thaventhiran T, Bergstrom E, Gardener Z, Ascough S, Chiu C, Docherty AB, Hunt D, Crow YJ, Solomon T, Taylor GP, Turtle L, Harrison EM, Dunning J, Semple MG, Baillie JK, Openshaw PJ, >Baillie JK, Semple MG, Openshaw PJ, Alex B, Bach B, Barclay WS, Bogaert D, Chand M, Cooke GS, Docherty AB, Dunning J, da Silva Filipe A, Fletcher T, Green CA, Gupta RK, Harrison EM, Hiscox JA, Ho AYW, Horby PW, Ijaz S, Khoo S, Klenerman P, Law A, Lim WS, On Behalf of the Isaric4c Investigators, et al. 2021. Inflammatory profiles across the spectrum of disease reveal a distinct role for GM-CSF in severe COVID-19. Sci Immunol 6:eabg9873. doi: 10.1126/sciimmunol.abg9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sherman EJ, Emmer BT. 2021. ACE2 protein expression within isogenic cell lines is heterogeneous and associated with distinct transcriptomes. Sci Rep 11:15900. doi: 10.1038/s41598-021-95308-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ren X, Glende J, Al-Falah M, de Vries V, Schwegmann-Wessels C, Qu X, Tan L, Tschernig T, Deng H, Naim HY, Herrler G. 2006. Analysis of ACE2 in polarized epithelial cells: surface expression and function as receptor for severe acute respiratory syndrome-associated coronavirus. J Gen Virol 87:1691–1695. doi: 10.1099/vir.0.81749-0. [DOI] [PubMed] [Google Scholar]
- 43.Corpetti C, Del Re A, Seguella L, Palenca I, Rurgo S, De Conno B, Pesce M, Sarnelli G, Esposito G. 2021. Cannabidiol inhibits SARS-Cov-2 spike (S) protein-induced cytotoxicity and inflammation through a PPARγ-dependent TLR4/NLRP3/Caspase-1 signaling suppression in Caco-2 cell line. Phytother Res 35:6893–6903. doi: 10.1002/ptr.7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bojkova D, Klann K, Koch B, Widera M, Krause D, Ciesek S, Cinatl J, Münch C. 2020. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature 583:469–472. doi: 10.1038/s41586-020-2332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reus P, Schneider A-K, Ulshöfer T, Henke M, Bojkova D, Cinatl J, Ciesek S, Geisslinger G, Laux V, Grättinger M, Gribbon P, Schiffmann S. 2021. Characterization of ACE inhibitors and AT1R antagonists with regard to their effect on ACE2 expression and infection with SARS-CoV-2 using a Caco-2 cell model. Life (Basel) 11:810. doi: 10.3390/life11080810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muñoz-Fontela C, Dowling WE, Funnell SGP, Gsell P-S, Riveros-Balta AX, Albrecht RA, Andersen H, Baric RS, Carroll MW, Cavaleri M, Qin C, Crozier I, Dallmeier K, de Waal L, de Wit E, Delang L, Dohm E, Duprex WP, Falzarano D, Finch CL, Frieman MB, Graham BS, Gralinski LE, Guilfoyle K, Haagmans BL, Hamilton GA, Hartman AL, Herfst S, Kaptein SJF, Klimstra WB, Knezevic I, Krause PR, Kuhn JH, Le Grand R, Lewis MG, Liu W-C, Maisonnasse P, McElroy AK, Munster V, Oreshkova N, Rasmussen AL, Rocha-Pereira J, Rockx B, Rodríguez E, Rogers TF, Salguero FJ, Schotsaert M, Stittelaar KJ, Thibaut HJ, Tseng C-T, et al. 2020. Animal models for COVID-19. Nature 586:509–515. doi: 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muñoz-Fontela C, Widerspick L, Albrecht RA, Beer M, Carroll MW, de Wit E, Diamond MS, Dowling WE, Funnell SGP, García-Sastre A, Gerhards NM, de Jong R, Munster VJ, Neyts J, Perlman S, Reed DS, Richt JA, Riveros-Balta X, Roy CJ, Salguero FJ, Schotsaert M, Schwartz LM, Seder RA, Segalés J, Vasan SS, Henao-Restrepo AM, Barouch DH. 2022. Advances and gaps in SARS-CoV-2 infection models. PLoS Pathog 18:e1010161. doi: 10.1371/journal.ppat.1010161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Renn M, Bartok E, Zillinger T, Hartmann G, Behrendt R. 2021. Animal models of SARS-CoV-2 and COVID-19 for the development of prophylactic and therapeutic interventions. Pharmacol Ther 228:107931. doi: 10.1016/j.pharmthera.2021.107931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Imai M, Iwatsuki-Horimoto K, Hatta M, Loeber S, Halfmann PJ, Nakajima N, Watanabe T, Ujie M, Takahashi K, Ito M, Yamada S, Fan S, Chiba S, Kuroda M, Guan L, Takada K, Armbrust T, Balogh A, Furusawa Y, Okuda M, Ueki H, Yasuhara A, Sakai-Tagawa Y, Lopes TJS, Kiso M, Yamayoshi S, Kinoshita N, Ohmagari N, Hattori S-I, Takeda M, Mitsuya H, Krammer F, Suzuki T, Kawaoka Y. 2020. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci USA 117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bednash JS, Kagan VE, Englert JA, Farkas D, Tyurina YY, Tyurin VA, Samovich SN, Farkas L, Elhance A, Johns F, Lee H, Cheng L, Majumdar A, Jones D, Mejia OR, Ruane-Foster M, Londino JD, Mallampalli RK, Robinson RT. 2022. Syrian hamsters as a model of lung injury with SARS-CoV-2 infection: pathologic, physiologic, and detailed molecular profiling. Translational Res 240:1–16. doi: 10.1016/j.trsl.2021.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gruber AD, Firsching TC, Trimpert J, Dietert K. 2022. Hamster models of COVID-19 pneumonia reviewed: how human can they be? Vet Pathol 59:528–545. doi: 10.1177/03009858211057197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warner BM, Safronetz D, Kobinger GP. 2017. Syrian hamsters as a small animal model for emerging infectious diseases: advances in immunologic methods. Adv Exp Med Biol 972:87–101. doi: 10.1007/5584_2016_135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Radvak P, Kwon H-J, Kosikova M, Ortega-Rodriguez U, Xiang R, Phue J-N, Shen R-F, Rozzelle J, Kapoor N, Rabara T, Fairman J, Xie H. 2021. SARS-CoV-2 B.1.1.7 (alpha) and B.1.351 (beta) variants induce pathogenic patterns in K18-hACE2 transgenic mice distinct from early strains. Nat Commun 12:6559. doi: 10.1038/s41467-021-26803-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tarrés-Freixas F, Trinité B, Pons-Grífols A, Romero-Durana M, Riveira-Muñoz E, Ávila-Nieto C, Pérez M, Garcia-Vidal E, Perez-Zsolt D, Muñoz-Basagoiti J, Raïch-Regué D, Izquierdo-Useros N, Andrés C, Antón A, Pumarola T, Blanco I, Noguera-Julián M, Guallar V, Lepore R, Valencia A, Urrea V, Vergara-Alert J, Clotet B, Ballana E, Carrillo J, Segalés J, Blanco J. 2022. Heterogeneous infectivity and pathogenesis of SARS-CoV-2 variants Beta, Delta and Omicron in transgenic K18-hACE2 and wildtype mice. Front Microbiol 13:840757. doi: 10.3389/fmicb.2022.840757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee KS, Wong TY, Russ BP, Horspool AM, Miller OA, Rader NA, Givi JP, Winters MT, Wong ZYA, Cyphert HA, Denvir J, Stoilov P, Barbier M, Roan NR, Amin MS, Martinez I, Bevere JR, Damron FH. 2022. SARS-CoV-2 Delta variant induces enhanced pathology and inflammatory responses in K18-hACE2 mice. PLoS One 17:e0273430. doi: 10.1371/journal.pone.0273430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hui KPY, Cheung M-C, Perera RAPM, Ng K-C, Bui CHT, Ho JCW, Ng MMT, Kuok DIT, Shih KC, Tsao S-W, Poon LLM, Peiris M, Nicholls JM, Chan MCW. 2020. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. Lancet Respir Med 8:687–695. doi: 10.1016/S2213-2600(20)30193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu J, Li Y, Liu Q, Yao Q, Wang X, Zhang H, Chen R, Ren L, Min J, Deng F, Yan B, Liu L, Hu Z, Wang M, Zhou Y. 2021. SARS-CoV-2 cell tropism and multiorgan infection. Cell Discov 7:17. doi: 10.1038/s41421-021-00249-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bhatnagar J, Gary J, Reagan-Steiner S, Estetter LB, Tong S, Tao Y, Denison AM, Lee E, DeLeon-Carnes M, Li Y, Uehara A, Paden CR, Leitgeb B, Uyeki TM, Martines RB, Ritter JM, Paddock CD, Shieh W-J, Zaki SR. 2021. Evidence of severe acute respiratory syndrome coronavirus 2 replication and tropism in the lungs, airways, and vascular endothelium of patients with fatal coronavirus disease 2019: an autopsy case series. J Infect Dis 223:752–764. doi: 10.1093/infdis/jiab039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muhl L, He L, Sun Y, Mäe MA, Pietilä R, Liu J, Genové G, Zhang L, Xie Y, Leptidis S, Mocci G, Stritt S, Osman A, Anisimov A, Hemanthakumar KA, Räsänen M, Hansson EM, Björkegren J, Vanlandewijck M, Blomgren K, Mäkinen T, Peng X-R, Hu Y, Ernfors P, Arnold TD, Alitalo K, Lendahl U, Betsholtz C. 2022. The SARS-CoV-2 receptor ACE2 is expressed in mouse pericytes but not endothelial cells: implications for COVID-19 vascular research. Stem Cell Rep 17:1089–1104. doi: 10.1016/j.stemcr.2022.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, Huan Y, Yang P, Zhang Y, Deng W, Bao L, Zhang B, Liu G, Wang Z, Chappell M, Liu Y, Zheng D, Leibbrandt A, Wada T, Slutsky AS, Liu D, Qin C, Jiang C, Penninger JM. 2005. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med 11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu X, Yang N, Tang J, Liu S, Luo D, Duan Q, Wang X. 2014. Downregulation of angiotensin-converting enzyme 2 by the neuraminidase protein of influenza A (H1N1) virus. Virus Res 185:64–71. doi: 10.1016/j.virusres.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zou Z, Yan Y, Shu Y, Gao R, Sun Y, Li X, Ju X, Liang Z, Liu Q, Zhao Y, Guo F, Bai T, Han Z, Zhu J, Zhou H, Huang F, Li C, Lu H, Li N, Li D, Jin N, Penninger JM, Jiang C. 2014. Angiotensin-converting enzyme 2 protects from lethal avian influenza A H5N1 infections. Nat Commun 5:3594. doi: 10.1038/ncomms4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Low BE, Hosur V, Lesbirel S, Wiles MV. 2022. Efficient targeted transgenesis of large donor DNA into multiple mouse genetic backgrounds using bacteriophage Bxb1 integrase. Sci Rep 12:5424. doi: 10.1038/s41598-022-09445-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, Péré H, Charbit B, Bondet V, Chenevier-Gobeaux C, Breillat P, Carlier N, Gauzit R, Morbieu C, Pène F, Marin N, Roche N, Szwebel T-A, Merkling SH, Treluyer J-M, Veyer D, Mouthon L, Blanc C, Tharaux P-L, Rozenberg F, Fischer A, Duffy D, Rieux-Laucat F, Kernéis S, Terrier B. 2020. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Currey JM, Rabito F, Maness NJ, Blair RV, Rappaport J, Qin X, Kolls JK, Srivastava AK. 2022. C57BL/6J mice are not suitable for modeling severe SARS-CoV-2 Beta and Gamma variant infection. Viruses 14:966. doi: 10.3390/v14050966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pan T, Chen R, He X, Yuan Y, Deng X, Li R, Yan H, Yan S, Liu J, Zhang Y, Zhang X, Yu F, Zhou M, Ke C, Ma X, Zhang H. 2021. Infection of wild-type mice by SARS-CoV-2 B.1.351 variant indicates a possible novel cross-species transmission route. Sig Transduct Target Ther 6:420. doi: 10.1038/s41392-021-00848-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oladunni FS, Park J-G, Pino PA, Gonzalez O, Akhter A, Allué-Guardia A, Olmo-Fontánez A, Gautam S, Garcia-Vilanova A, Ye C, Chiem K, Headley C, Dwivedi V, Parodi LM, Alfson KJ, Staples HM, Schami A, Garcia JI, Whigham A, Platt RN, Gazi M, Martinez J, Chuba C, Earley S, Rodriguez OH, Mdaki SD, Kavelish KN, Escalona R, Hallam CRA, Christie C, Patterson JL, Anderson TJC, Carrion R, Dick EJ, Hall-Ursone S, Schlesinger LS, Alvarez X, Kaushal D, Giavedoni LD, Turner J, Martinez-Sobrido L, Torrelles JB. 2020. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat Commun 11:6122. doi: 10.1038/s41467-020-19891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carossino M, Lee P-YA, Nam B, Skillman A, Shuck KM, Timoney PJ, Tsai Y-L, Ma L-J, Chang H-FG, Wang H-TT, Balasuriya UBR. 2016. Development and evaluation of a reverse transcription-insulated isothermal polymerase chain reaction (RT-iiPCR) assay for detection of equine arteritis virus in equine semen and tissue samples using the POCKIT system. J Virol Methods 234:7–15. doi: 10.1016/j.jviromet.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 69.Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, Liu Y, Ng DYM, Wan CKC, Yang P, Wang Q, Peiris M, Poon LLM. 2020. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem 66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vogels CBF, Brito AF, Wyllie AL, Fauver JR, Ott IM, Kalinich CC, Petrone ME, Casanovas-Massana A, Catherine Muenker M, Moore AJ, Klein J, Lu P, Lu-Culligan A, Jiang X, Kim DJ, Kudo E, Mao T, Moriyama M, Oh JE, Park A, Silva J, Song E, Takahashi T, Taura M, Tokuyama M, Venkataraman A, Weizman O-E, Wong P, Yang Y, Cheemarla NR, White EB, Lapidus S, Earnest R, Geng B, Vijayakumar P, Odio C, Fournier J, Bermejo S, Farhadian S, Dela Cruz CS, Iwasaki A, Ko AI, Landry ML, Foxman EF, Grubaugh ND. 2020. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat Microbiol 5:1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carossino M, Li Y, Lee P-YA, Tsai C-F, Chou P-H, Williams D, Skillman A, Frank Cook R, Brown G, Chang H-FG, Wang H-TT, Balasuriya UBR. 2017. Evaluation of a field-deployable reverse transcription-insulated isothermal PCR for rapid and sensitive on-site detection of Zika virus. BMC Infect Dis 17:778. doi: 10.1186/s12879-017-2852-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carossino M, Barrandeguy ME, Erol E, Li Y, Balasuriya UBR. 2019. Development and evaluation of a one-step multiplex real-time TaqMan RT-qPCR assay for the detection and genotyping of equine G3 and G14 rotaviruses in fecal samples. Virol J 16:49. doi: 10.1186/s12985-019-1149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carossino M, Dini P, Kalbfleisch TS, Loynachan AT, Canisso IF, Cook RF, Timoney PJ, Balasuriya UBR. 2019. Equine arteritis virus long-term persistence is orchestrated by CD8+ T lymphocyte transcription factors, inhibitory receptors, and the CXCL16/CXCR6 axis. PLoS Pathog 15:e1007950. doi: 10.1371/journal.ppat.1007950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, van den Hoff MJB, Moorman AFM. 2009. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37:e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yin R, Tian F, Frankenberger B, de Angelis MH, Stoeger T. 2010. Selection and evaluation of stable housekeeping genes for gene expression normalization in carbon nanoparticle-induced acute pulmonary inflammation in mice. Biochem Biophys Res Commun 399:531–536. doi: 10.1016/j.bbrc.2010.07.104. [DOI] [PubMed] [Google Scholar]
- 76.Hall JS, Knowles S, Nashold SW, Ip HS, Leon AE, Rocke T, Keller S, Carossino M, Balasuriya U, Hofmeister E. 2021. Experimental challenge of a North American bat species, big brown bat (Eptesicus fuscus), with SARS-CoV-2. Transbound Emerg Dis 68:3443–3452. doi: 10.1111/tbed.13949. [DOI] [PubMed] [Google Scholar]
- 77.Cool K, Gaudreault NN, Morozov I, Trujillo JD, Meekins DA, McDowell C, Carossino M, Bold D, Mitzel D, Kwon T, Balaraman V, Madden DW, Artiaga BL, Pogranichniy RM, Roman-Sosa G, Henningson J, Wilson WC, Balasuriya UBR, García-Sastre A, Richt JA. 2022. Infection and transmission of ancestral SARS-CoV-2 and its alpha variant in pregnant white-tailed deer. Emerg Microbes Infect 11:95–112. doi: 10.1080/22221751.2021.2012528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gaudreault NN, Trujillo JD, Carossino M, Meekins DA, Morozov I, Madden DW, Indran SV, Bold D, Balaraman V, Kwon T, Artiaga BL, Cool K, García-Sastre A, Ma W, Wilson WC, Henningson J, Balasuriya UBR, Richt JA. 2020. SARS-CoV-2 infection, disease and transmission in domestic cats. Emerg Microbes Infect 9:2322–2332. doi: 10.1080/22221751.2020.1833687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gaudreault NN, Carossino M, Morozov I, Trujillo JD, Meekins DA, Madden DW, Cool K, Artiaga BL, McDowell C, Bold D, Balaraman V, Kwon T, Ma W, Henningson J, Wilson DW, Wilson WC, Balasuriya UBR, García-Sastre A, Richt JA. 2021. Experimental re-infected cats do not transmit SARS-CoV-2. Emerg Microbes Infect 10:638–650. doi: 10.1080/22221751.2021.1902753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gaudreault NN, Cool K, Trujillo JD, Morozov I, Meekins DA, McDowell C, Bold D, Carossino M, Balaraman V, Mitzel D, Kwon T, Madden DW, Artiaga BL, Pogranichniy RM, Roman-Sosa G, Wilson WC, Balasuriya UBR, García-Sastre A, Richt JA. 2022. Susceptibility of sheep to experimental co-infection with the ancestral lineage of SARS-CoV-2 and its alpha variant. Emerg Microbes Infect 11:662–675. doi: 10.1080/22221751.2022.2037397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meekins DA, Morozov I, Trujillo JD, Gaudreault NN, Bold D, Carossino M, Artiaga BL, Indran SV, Kwon T, Balaraman V, Madden DW, Feldmann H, Henningson J, Ma W, Balasuriya UBR, Richt JA. 2020. Susceptibility of swine cells and domestic pigs to SARS-CoV-2. Emerg Microbes Infect 9:2278–2288. doi: 10.1080/22221751.2020.1831405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Carossino M, Ip HS, Richt JA, Shultz K, Harper K, Loynachan AT, Del Piero F, Balasuriya UBR. 2020. Detection of SARS-CoV-2 by RNAscope in situ hybridization and immunohistochemistry techniques. Arch Virol 165:2373–2377. doi: 10.1007/s00705-020-04737-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SARS-CoV-2 MA10 strain induces an early expression of proinflammatory cytokines and chemokines in the lungs of K18-hACE2 mice compared to that of its parental strain USA-WA1/2020. Protein expression of 32 cytokines/chemokines was measured in the lung of mock-, MA10-, and USA-WA1/2020-infected K18-hACE2 mice at 2 dpi, 4 dpi, and 6 dpi (n = 4). Data analyzed by nonparametric Wilcoxon tests. Download FIG S1, TIF file, 1.0 MB (1MB, tif) .
Copyright © 2023 Thieulent et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of proinflammatory cytokines and chemokines in the lungs of MA10-infected C57BL/6J mice. Protein expression of 31 cytokines/chemokines was measured in the lung of mock- and MA10-infected C57BL/6J mice at 2 dpi, 4 dpi, and 7 dpi (n = 3). Data analyzed by nonparametric Wilcoxon tests. Download FIG S2, TIF file, 0.3 MB (340.9KB, tif) .
Copyright © 2023 Thieulent et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of proinflammatory cytokines and chemokines in the lungs of MA10-infected BALB/c mice. Protein expression of 31 cytokines/chemokines was measured in the lung of mock- and MA10-infected BALB/c mice at 2 dpi, 4 dpi, and 7 dpi (n = 4). Data analyzed by nonparametric Wilcoxon tests. Download FIG S3, TIF file, 0.4 MB (371.8KB, tif) .
Copyright © 2023 Thieulent et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Validation of the specificity of the mAce2 and hACE2 primer pairs. Primers used for mAce2 and hACE2 expression measurements were tested in mRNA extracted from lung lysates of C57BL/6J mice (A), in mRNA derived from human colorectal adenocarcinoma (Caco-2) cell pellets (B) and in mRNA extracted from lungs lysates of K18-hACE2 mice (C). Only mAce2 or hACE2 transcripts are detected in the lung of C57BL/6J mice or Caco-2 cells, respectively (A and B), while both mAce2 and hACE2 transcripts are expressed in the lung of K18-hACE2 mice (C). ΔCT values represent the difference of CT between the gene of interest and housekeeping genes. N.D., not detected. Download FIG S4, TIF file, 0.1 MB (88.2KB, tif) .
Copyright © 2023 Thieulent et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of mTmprss2 mRNA in the lung of mice using RNAscope ISH. Tmprss2 is rich within bronchiolar epithelia (inset 1) and sporadic in AT1 and AT2 cells (inset 2). Fast Red (mTmprss2 mRNA); ×200 total magnification. Download FIG S5, TIF file, 3.9 MB (4MB, tif) .
Copyright © 2023 Thieulent et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.