Abstract
Purpose
Circulating cell-free single-stranded DNA (ccf-ssDNA) is extracellular DNA and it is a useful biomarker for the diagnosis of tumors and predicting the prognosis of tumors. However, the clinical usefulness of ccf-ssDNA in colorectal cancer (CRC) is not well known. Thus, the purpose of this study was to investigate the clinical usefulness of ccf-ssDNA in CRC.
Methods
The study was conducted on 44 patients who had undergone surgery for CRC, and ccf-ssDNA level was measured before surgery and statistical analysis was performed on clinical factors.
Results
The association between ccf-ssDNA level and clinicopathological factors was analyzed and compared, and these factors included age, sex, body mass index, diabetes mellitus, hypertension, tumor markers (carcinoembryonic antigen and carbohydrate antigen 19-9), tumor location, size, stage (TNM), recurrence, and death. The group with a ccf-ssDNA level of ≥7.5 ng/μL had a lower age (P=0.010), and was associated with diabetes mellitus (P=0.037) and lymph node metastasis (P=0.049). Multivariate analysis of disease-free survival showed that lymph node metastasis and ccf-ssDNA level (hazard ratio, 10.011; 95% confidence interval, 2.269–44.175; P=0.002) were independent prognostic factors for recurrence. In terms of overall survival, there were no statistically significant results except for vascular invasion.
Conclusion
This study showed that ccf-ssDNA level in plasma in CRC patients was an independent prognostic factor that could predict recurrence non-invasively. In this regard, further evaluation with a prospective, large sample size study will be needed to obtain additional results.
Keywords: Colon cancer, Colorectal cancer recurrence, Circulating cell-free single-stranded DNA, Circulating cell-free DNA, Liquid biopsy
INTRODUCTION
Colorectal cancer (CRC) is the most common cancer in the world after breast cancer and lung cancer in terms of incidence, with >1,800,000 new patients in 2020 and has the second highest mortality rate [1]. In Korea, according to the National Cancer Registration Program, it is the fourth most common cancer with the third highest mortality rate after lung cancer and liver cancer [2].
In recent years, many technological advances have been made in the diagnosis and treatment of CRC (e.g., surgery, radiation therapy, chemotherapy, and targeted therapy), and through these advances, the patient’s survival rate has been significantly improved to 65% [3]. Nevertheless, in patients with advanced CRC, recurrence including distant metastasis still occurs in many patients, leading to poor prognosis with a significant impact on their quality of life [4,5]. Therefore, to have a customized treatment strategy for CRC patients, the most important issue is to select patients with a high risk of recurrence and select an appropriate treatment. To date, the standard for evaluating the risk of recurrence in CRC patients is the clinicopathological stage. However, the prognosis varies from patient to patient despite individuals have the same clinicopathological risk factors. Thus, a new biomarker that can predict the risk of recurrence is needed.
Circulating cell-free DNA (ccfDNA) exists in serum or plasma as extracellular DNA and it has been attracting attention as a useful biomarker for tumor selection, treatment effect determination, and prognosis through various studies to date [6,7]. Leon et al. [8] have reported that cell-free DNA (cfDNA) level is higher in tumor patients than in normal healthy individuals and other studies have shown the same results. These cfDNAs are known to arise from activated metabolism of tumors or necrosis of tumor cells. Thus, researchers mostly agree that there is an association between the primary tumor burden and the cfDNA level in plasma. Therefore, it has been reported that cfDNA is useful as an early diagnostic test in various tumors including kidney cancer, lung cancer, testicular tumor, and CRC, showing the possibility of non-invasive liquid biopsy in tumor patients [9,10]. In addition, cfDNA level in the serum is higher in bladder cancer patients than in benign prostatic hyperplasia patients, and it has been reported that cfDNA level plays an important role in predicting the prognosis of bladder cancer patients [11]. A recent meta-analysis has shown that the higher the cfDNA level in breast cancer patients, has the lower the disease-free survival rate and overall survival rate in the presence of circulation tumor DNA (ctDNA) [12].
Studies on CRC patients have also reported that cfDNA level is increased in CRC patients along with other tumors than in healthy individuals, and Hao et al. have shown significant results in predicting the prognosis in metastatic CRC when ALU115 and segmental index (ALU247/115) are analyzed through reverse transcription polymerase chain reaction of cfDNA [13,14].
Despite the clinical usefulness of cfDNA, it is still difficult to know how cfDNA is secreted from tumor cells into the blood, and various studies have reported their origin, structure, and types. cfDNA is composed of double-stranded DNA (dsDNA) and single-stranded DNA (ssDNA) and it is known to be secreted mainly due to cell apoptosis or necrosis, and recently, it has been reported that active secretion of cells through microvesicles such as exosomes is important for their composition [15]. In addition, with the development of current technology, the composition and structure of cfDNA according to the length are being studied, and clinical association has been reported accordingly [16,17]. Although clinical studies of cfDNA and CRC with various characteristics have been reported, ssDNA and dsDNA levels and their clinical association have not been reported yet.
Therefore, the aim of this study was to evaluate clinical usefulness and investigate role of prognostic factor of ssDNA and dsDNA levels in CRC patients.
METHODS
Patients and samples
This study protocol was approved by the Institutional Review Board of Soonchunhyang University Cheonan Hospital, and this study was conducted after written consent was obtained from patients included in the study. From January 2015 to December 2015, 69 patients diagnosed with CRC and underwent surgery at the Department of Surgery at Soonchunhyang University Cheonan Hospital were enrolled in this study. Postoperative stage of these patients was I–IV, and surgically resectable patients were included in this study. Patients under 18 years of age or over 75 years of age were excluded, and patients with concurrent cancer at the time of surgery and those who received chemotherapy or radiotherapy before surgery were also excluded. In addition, the study was not conducted according to the exclusion criteria for patients with poor blood collection condition or ethical issues. According to the exclusion criteria, 25 patients were excluded and finally 44 patients were enrolled in this study. Patients’ clinicopathological records were collected retrospectively and analyzed through computerized records. For survival analysis, outpatient visit records were checked or patient’s survival period was checked over the phone. Recurrence was defined as cases with evidence of tumor or diagnosis on abdominal computed tomography, chest computed tomography, PET-CT, and colonoscopy. The median follow-up period was 57 months. Tumor stage was defined according to Tumor-Node-Metastasis (TNM) in accordance with the American Joint Committee on Cancer (AJCC)/International Union 7th Edition.
For patients’ blood sampling, 10 mL of whole blood was collected into a tube containing K3-EDTA before surgery. After separating plasma using a centrifuge (1,600 g, 10 minutes) within 4 hours, sample was placed into a DNA tube and then stored in a cryogenic freezer at −80°C until analysis.
Isolation of ccfDNA from plasma
ccfDNA was isolated from plasma (100 μL) using a Maxwell RSC Instrument (AS4500; Promega, Madison, WI, USA) and a Maxwell RSC ccfDNA Plasma Kit (AS1480, Promega) according to the user manual. In the last elution step, 50 μL X TE buffer was used according to the user manual.
Quantification of ccfDNA levels from plasma
Quantitative analysis of dsDNA and ssDNA levels was performed by fluorescence analysis using a Quantus Fluorometer (E6150, Promega). dsDNA level was measured using a QuantiFluor dsDNA System (E2670, Promega), and dsDNA level was measured using a QuantiFluor ssDNA System (E3190, Promega).
Statistical analysis
Statistical analysis was performed using PASW Statistics version 18.0 (SPSS Inc., Chicago, IL, USA). For categorical clinicopathological factors, the chi-square test or Fisher exact test was used, and for continuous variables, independent-sample t-test or Mann- Whitney test was used. Overall survival rate and disease-free survival rate were analyzed and compared using the Kaplan-Meier curve method and log-rank test. For univariate and multivariate analyses, Cox proportional hazards model was used. P<0.05 was defined as a statistically significant result. The optimal cutoff value for ccfDNA level for predicting recurrence was determined by analyzing the receiver operating characteristic (ROC) curve.
RESULTS
Baseline clinicopathological data
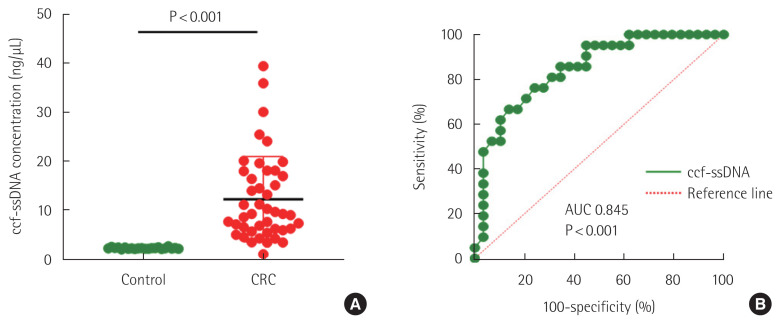
This study was conducted on 44 patients who underwent surgery for CRC. The mean age of these patients was 63.98±11.96 (mean±standard deviation [SD]) years and there were 15 female patients and 29 male patients. The median survival period was 3.9 years (range, 0.8–5.8 years) and the median disease-free survival period was 2.5 years (range, 0.5–5.8 years). Mean ssDNA and dsDNA levels of these patients were 12.15±8.74 ng/μL and 1.01±0.79 ng/μL, respectively. The optimal circulating cell-free single-stranded DNA (ccf-ssDNA) cutoff value for recurrence was 7.5 ng/μL (area under curve, 0.845, 95% confidence interval [CI], 0.7380–0.9533, P< 0.001) (Fig. 1). Therefore, there were 19 (43.1%) group I patients with ccf-ssDNA level less than 7.5 ng/μL and 25 (56.9%) group II patients with ccf-ssDNA level higher than 7.5 ng/μL. There were 14 (31.8%) patients with right sided colon cancer and 30 (69.2%) patients with left sided colon cancer including rectum. In terms of stage, there were seven (15.9%) patients in stage I, 17 (38.6%) patients in stage II, 16 (36.4%) patients in stage III, and four (9.1%) patients in stage IV. Recurrence occurred in 14 (31.8%) of 44 patients, and eight (18.1%) patients died.
Fig. 1.
Comparison of the ccf-ssDNA levels in plasma samples from healthy individuals and colorectal cancer (CRC) patients. (A) ccf-ssDNA level in CRC patients and control were compared. (B) Receiver operating characteristic curves for recurrence in CRC patients. ccf-ssDNA, circulating cell-free single-stranded DNA; AUC, area under curve.
Association between ccf-ssDNA level and clinicopathological characteristics of patients with CRCs
Associations between ccf-ssDNA level and clinicopathological factors were analyzed and compared, and these clinicopathological factors included age, sex, body mass index, diabetes mellitus, hypertension, tumor markers (carcinoembryonic antigen [CEA], carbohydrate antigen 19-9 [CA19-9]), tumor location, size, stage (TNM), recurrence, and death. As a result, when comparing between the two groups (group I <7.5 ng/μL vs. group II ≥7.5 ng/μL) according to ccf-ssDNA level, it was confirmed that the age was lower in group II (P<0.01). In addition, the group with ccf-ssDNA level higher than 7.5 ng/μL showed an association with diabetes mellitus (P=0.037). ccf-ssDNA was not associated with T stage in comparison with pathological factors, but it was associated with N stage which was lymph node metastasis (P=0.049). ccf-ssDNA showed no statistically significant association with other risk factors related to the prognosis of CRC, such as lymph node invasion, vascular invasion, or perineural invasion. In terms of recurrence, three (15.7%) patients in group I and 11 (44.0%) patients in group II showed a statistically significant association with recurrence as ccf-ssDNA level increased (P=0.047) (Table 1).
Table 1.
Association between ccf-ssDNA concentration and clinicopathological factors in colorectal cancer patients
| Characteristics | ccf-ssDNA | ||
|---|---|---|---|
|
| |||
| <7.5 ng/μL (n=19) | ≥7.5 ng/μL (n=25) | P-value | |
| Age (yr), mean±SD | 68.30±8.87 | 59.24±13.26 | 0.010 |
|
| |||
| Age group | 0.317 | ||
| <65 yr | 7 (36.84) | 13 (52.00) | |
| ≥65 yr | 12 (63.16) | 12 (48.00) | |
|
| |||
| Sex | 0.737 | ||
| Male | 12 (63.16) | 17 (68.00) | |
| Female | 7 (36.84) | 8 (32.00) | |
|
| |||
| Body mass index (kg/m2), mean±SD | 23.21±3.22 | 23.42±4.42 | 0.859 |
|
| |||
| Diabetes mellitus | 0.037 | ||
| No | 15 (78.95) | 12 (48.00) | |
| Yes | 4 (21.05) | 13 (52.00) | |
|
| |||
| Hypertension | 0.967 | ||
| No | 9 (47.37) | 12 (48.00) | |
| Yes | 10 (52.63) | 13 (52.00) | |
|
| |||
| Neutrophil (/μL), mean±SD | 4,213.16±1,806.28 | 5,276.00±2,307.94 | 0.105 |
|
| |||
| CEA | 0.530 | ||
| <5 ng/μL | 8 (42.11) | 13 (52.00) | |
| ≥5 ng/μL | 11 (57.89) | 12 (48.00) | |
|
| |||
| CA19-9 (U/mL), median (IQR) | 15.5 (8.2–31.1) | 8.6 (5.0–18.1) | 0.115 |
|
| |||
| Tumor location | 0.533 | ||
| Left | 7 (36.84) | 7 (28.00) | |
| Right | 12 (63.16) | 18 (72.00) | |
|
| |||
| Tumor size (cm), mean±SD | 5.49±2.30 | 4.51±2.62 | 0.167 |
|
| |||
| pT | 0.059 | ||
| 1 | 1 (5.26) | 4 (16.00) | |
| 2 | 0 | 5 (25.00) | |
| 3 | 13 (68.42) | 11 (44.00) | |
| 4 | 5 (26.32) | 2 (8.00) | |
|
| |||
| pN | 0.049 | ||
| 0 | 14 (73.68) | 1 (44.00) | |
| 1, 2 | 5 (26.32) | 14 (56.00) | |
|
| |||
| Harvest LN, mean±SD | 21.47±6.71 | 21.18±9.29 | 0.910 |
|
| |||
| pM | 0.122 | ||
| 0 | 19 (100.00) | 21 (84.00) | |
| 1 | 0 | 4 (16.00) | |
|
| |||
| Overall stage | 0.026 | ||
| I&II | 14 (73.68) | 10 (40.00) | |
| III&IV | 5 (26.32) | 15 (60.00) | |
|
| |||
| Lymphatic invasion | 1.000 | ||
| No | 14 (73.68) | 16 (76.19) | |
| Yes | 5 (26.32) | 5 (23.81) | |
|
| |||
| Venous invasion | 0.698 | ||
| No | 16 (84.21) | 16 (76.19) | |
| Yes | 3 (15.79) | 5 (23.81) | |
|
| |||
| Perineural invasion | 0.473 | ||
| No | 13 (68.42) | 17 (80.95) | |
| Yes | 6 (31.58) | 4 (19.05) | |
|
| |||
| Chemotherapy | 0.632 | ||
| No | 7 (36.84) | 11 (44.00) | |
| Yes | 12 (63.16) | 14 (56.00) | |
|
| |||
| Recurrence | 0.047 | ||
| No | 16 (84.21) | 14 (56.00) | |
| Yes | 3 (15.79) | 11 (44.00) | |
|
| |||
| Mortality | 0.667 | ||
| No | 15 (78.95) | 21 (84.00) | |
| Yes | 4 (21.05) | 4 (16.00) | |
Values are presented as number (%) unless otherwise indicated.
ccf-ssDNA, circulating cell-free single-stranded DNA; SD, standard deviation; CEA, carcinoembryonic antigen; CA19-9, carbohydrate antigen 19-9; IQR, interquartile range; pT, pathologic T stage; pN, pathologic N stage; pM, pathologic M stage, LN, lymph node.
Association between lymph node metastasis and ccf-ssDNA level in CRC patients
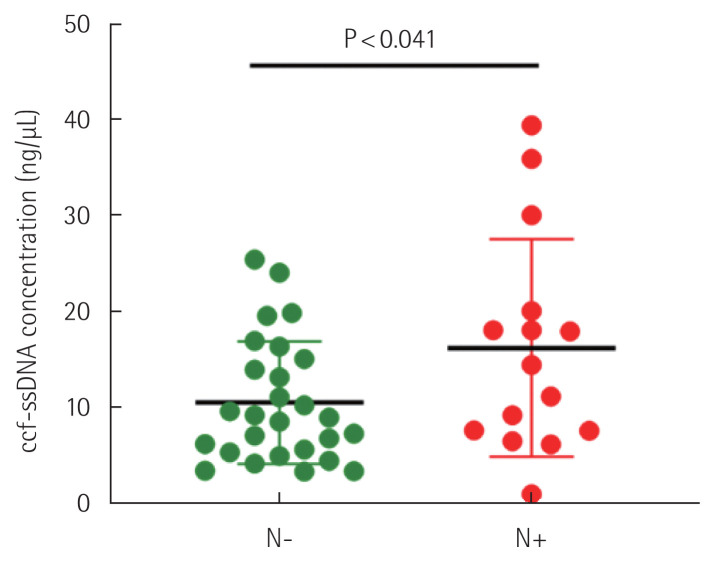
Among 44 patients, 19 patients had lymph node metastasis, and of these 19 patients, five patients were in group I and 14 patients were in group II. ccf-ssDNA level was 10.46±6.40 ng/μL (mean±SD) in patients without lymph node metastasis, and 16.49±10.65 ng/μL (mean±SD) in patients with lymph node metastasis. When statistical analysis of ccf-ssDNA levels in these two groups was performed, it was confirmed that level was significantly higher in patients with lymph node metastasis (P<0.041) (Fig. 2).
Fig. 2.
Comparison of the circulating cell-free single-stranded DNA (ccf-ssDNA) plasma levels in patients with (N+) and without (N−) lymph node metastasis.
Univariate and multivariate survival analyses of survival
Independent factors affecting disease-free survival and overall survival in CRC were identified using the Cox proportional hazard model. When univariate analysis of disease-free survival rate was performed, diabetes mellitus (hazard ratio [HR], 2.921; 95% CI, 1.004–8.503; P=0.049), lymph node metastasis (HR, 3.482; 95% CI, 1.195–10.143; P=0.022), and ccf-ssDNA level (group II vs. I) (HR, 4.383; 95% CI, 1.232–15.598; P=0.023) showed statistically significant results. When multivariate analysis of these factors was performed, AJCC stage (III, IV vs. I, II) (HR, 0.589; 95% CI, 0.067–5.195; P=0.634) did not show any statistically significant results. However, lymph node metastasis (HR, 7.211; 95% CI, 2.064–25.193; P=0.002) and ccf-ssDNA level (HR, 10.011; 95% CI, 2.269–44.175; P=0.002) were shown to be independent prognostic factors for recurrence (Table 2).
Table 2.
Univariate and multivariate Cox proportional hazard regression analysis of disease-free survival in patients with colorectal cancer
| Variable | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
|
|
|
|||
| Hazard ratio (95% CI) | P-value | Hazard ratio (95% CI) | P-value | |
| Age (<65 yr vs. ≥65 yr) | 0.856 (0.299–2.446) | 0.771 | ||
|
| ||||
| Sex (male vs. female) | 0.588 (0.184–1.881) | 0.365 | ||
|
| ||||
| DM (yes vs. no) | 2.921 (1.004–8.503) | 0.049 | 1.715 (0.582–5.053) | 0.328 |
|
| ||||
| Tumor location (right vs. left) | 1.883 (0.523–6.783) | 0.333 | ||
|
| ||||
| Vascular invasion (yes vs. no) | 1.577 (0.483–5.143) | 0.450 | ||
|
| ||||
| Lymphatic invasion (yes vs. no) | 1.436 (0.441–4.673) | 0.450 | ||
|
| ||||
| Perineural invasion (yes vs. no) | 1.586 (0.488–5.161) | 0.443 | ||
|
| ||||
| Lymph node metastasis (+ vs. −) | 3.482 (1.195–10.143) | 0.022 | 7.211 (2.064–25.193) | 0.002 |
|
| ||||
| AJCC stage (I, II vs. III, IV) | 2.905 (0.989–8.535) | 0.052 | 0.589 (0.067–5.195) | 0.634 |
|
| ||||
| ccf-ssDNA concentration (<7.5 ng/μL vs. ≥7.5 ng/μL) | 4.383 (1.232–15.598) | 0.023 | 10.011 (2.269–44.175) | 0.002 |
CI, confidence interval; DM, diabetes mellitus; AJCC, American Joint Committee on Cancer; ccf-ssDNA, circulating cell-free single-stranded DNA.
When univariate analysis of overall survival rate was performed, age (HR, 7.991; 95% CI, 1.010–63.235; P=0.049) and vascular invasion (HR, 4.334; 95% CI, 1.067–17.604; P=0.040) showed statistically significant association with prognosis. However, ccf-ssDNA level (HR, 0.593; 95% CI, 0.167–2.104; P=0.419) did not show statistically significant results. When multivariate analysis was performed, age, AJCC stage, and ccf-ssDNA level did not show significant results as prognostic factors, although vascular invasion (HR, 6.413; 95% CI, 1.556–26.430; P=0.010) was shown to be an independent prognostic factor (Table 3).
Table 3.
Univariate and multivariate Cox proportional hazard regression analysis of overall survival in patients with colorectal cancer
| Variable | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
|
|
|
|||
| Hazard ratio (95% CI) | P-value | Hazard ratio (95% CI) | P-value | |
| Age (<65 yr vs. ≥65 yr) | 7.991 (1.010–63.235) | 0.049 | 4.983 (1.556–26–430) | 0.144 |
|
| ||||
| Sex (male vs. female) | 0.750 (0.194–2.904) | 0.678 | ||
|
| ||||
| DM (yes vs. no) | 1.689 (0.489–5.841) | 0.407 | ||
|
| ||||
| Tumor location (right vs. left) | 0.695 (0.196–2.468) | 0.574 | ||
|
| ||||
| Vascular invasion (yes vs. no) | 4.334 (1.067–17.604) | 0.040 | 6.413 (1.556–26.430) | 0.010 |
|
| ||||
| Lymphatic invasion (yes vs. no) | 1.243 (0.239–6.449) | 0.796 | ||
|
| ||||
| Perineural invasion (yes vs. no) | 1.993 (0.473–8.396) | 0.347 | ||
|
| ||||
| Lymph node metastasis (+ vs. −) | 0.581 (0.117–2.877) | 0.505 | ||
|
| ||||
| AJCC stage (I, II vs. III, IV) | 2.829 (0.868–9.215) | 0.084 | 0.685 (0.187–3.015) | 0.685 |
|
| ||||
| ccf-ssDNAconcentration (<7.5 ng/μL vs. ≥7.5 ng/μL) | 0.593 (0.167–2.104) | 0.419 | 1.468 (0.389–5.544) | 0.571 |
CI, confidence interval; DM, diabetes mellitus; AJCC, American Joint Committee on Cancer; ccf-ssDNA, circulating cell-free single-stranded DNA.
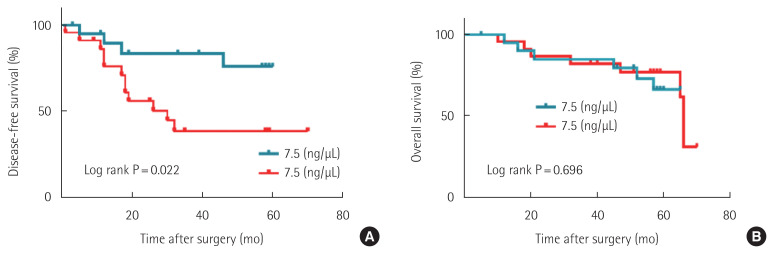
To analyze the survival rate for these patients, Kaplan-Meier curve analysis and log-rank test were performed. As a result, it was confirmed that the 5-year disease-free survival rate was lower in group II patients than in group I patients (P=0.022). However, there was no statistical difference in the overall survival rate between the two groups (P=0.696) (Fig. 3).
Fig. 3.
Kaplan-Meier survival curves for disease-free survival (A) and overall survival (B) according to circulating cell-free single-stranded DNA (ccf-ssDNA) level in patients with colorectal cancer.
DISCUSSION
CRC is one of the cancers with the highest mortality rate worldwide, and predicting its recurrence is the most important issue along with technological advances in diagnosis and treatment. To date, the most important factor in predicting the prognosis of CRC patients is the pathology of the tumor. However, these pathological risk factors and molecular biomarkers alone are not accurate in predicting recurrence, and therefore other reliable non-invasive prognostic factors are needed.
This study showed that ccf-ssDNA level measured in plasma collected from CRC patients before surgery was an independent prognostic factor that could predict recurrence. This is the first study to measure ccf-ssDNA before surgery in CRC patients and to investigate its association with recurrence along with long-term follow-up. In conclusion, for disease-free survival rate, when ccf-ssDNA level was higher than 7.5 ng/μL, which was the cutoff, the HR and statistical significance were higher than those of AJCC stage and vascular invasion. On the other hand, CEA measured before surgery did not show any association with recurrence, and early CRC patients showed normal findings. Czeiger et al. [14] reported that ccf-DNA level measured before surgery was high in CRC patients who died or recurred within 5 years, and as a result, it was an effective prognostic factor. Also, in this study, when ccf-ssDNA level of CRC patients was compared with that of healthy individuals, confirming that the ccf-ssDNA level was high as in other studies [14,16,18]. ccf-dsDNA level was also higher in CRC patients than in normal subjects. However, in this study, unlike ccf-ssDNA, ccf-dsDNA could not confirm a significant cutoff level in recurrence and it did not show any association with other clinicopathological factors.
There are two ways to conduct research using cfDNA, a quantitative analysis of cfDNA and a method to identify tumor-specific markers by analyzing circulating tumor DNA through sequence analysis using next-generation sequencing or polymerase chain reaction. All these methods are effective methods for analyzing cfDNA, but measuring tumor-specific ctDNA showed better results in predicting the prognosis [18]. However, ctDNA is known to account for 0.01% to 50% of total cfDNA, making it difficult to represent the increased cfDNA due to necrosis and apoptosis in the tumor microenvironment [19]. Recently, significant results have been reported for the diagnosis, treatment, and prediction of the prognosis of tumors through technological advances in cfDNA quantitative analysis [20,21]. The most important part in this study was to separate and measure ccf-ssDNA and ccf-dsDNA, respectively, and their values vary depending on the extraction method for cfDNA, and some studies have shown that ccf-ssDNA is significantly lower than its actual level [22]. Through previously reported studies, it was possible to confirm a significantly higher DNA extraction result in ccf-ssDNA measurement using the magnetic beam method rather than other column methods, therefore, in this study, the accuracy of ccf-ssDNA level could be improved through these methods [23].
In this study, there was a statistically significant association between ccf-ssDNA level and lymph node metastasis, and ccf-ssDNA level was higher in patients with lymph node metastasis. These results showed the possibility that ccf-ssDNA could predict lymph node metastasis, thus it is considered to be an important factor in the decision of surgical treatment. In patients with early CRC, extensive resection, including lymph node dissection, remains controversial. In CRC population screening, when additional surgery was performed for colorectal polyp adenocarcinoma, lymph node metastasis was found in 8% of cases. Therefore, it is thought that overtreatment and complications can be reduced by predicting lymph node metastasis through ccf-ssDNA measurement.
According to a recent meta-analysis, increased cfDNA in breast cancer patients showed a significant association with metastasis to the axillary lymph node, a sentinel lymph node [24]. Some studies have reported the association between high cfDNA level or ctDNA methylation with lymph node metastasis and advanced stage [25,26]. Lymph node metastasis is an important pathway for local and distant metastasis in CRC, and the association between lymph node metastasis and ccf-ssDNA is believed to play a role in recurrence. Recently, it has been reported that cfDNA can enter healthy cells and bind to their genome, causing DNA damage. This bound cfDNA can cause DNA rearrangement, translocation, and deletion, eventually leading to chemical resistance [27]. A series of genometastasis in which cfDNA in plasma contributes to tumorigenesis and induces metastasis through transfection of nucleic acids in susceptible cells in tissues has recently been reported [28]. However, which cfDNA causes this genometastasis remains unclear. Based on results of this study, it is believed that ccf-ssDNA related to lymph node metastasis and recurrence might play a role in genometastasis, and additional verification is required.
Huang et al. [17] have reported that the ssDNA/dsDNA ratio is significantly associated with the diagnostic usefulness and tumor stage in gastric cancer patients, but the ssDNA/dsDNA ratio did not show any association with other clinicopathological factors or prognosis in this study. Also, in these studies, levels of ssDNA and dsDNA were significantly higher than those of healthy individuals as in this study, but they showed no association with prognosis.
Based on literature review this is the first study to confirm that ccf-ssDNA is an independent prognostic factor for recurrence in CRC, but this study has several limitations. First, the cutoff value of ccf-ssDNA was selected through ROC analysis, thus it is necessary to verify it in an independent cohort. Second, due to the single center and small sample size of this study, a large number of patients and multicenter studies are needed to apply results to predict the prognosis in CRC. Third, since cfDNA level is non-specific and affected by other diseases (acute infection, chronic disease, inflammation), there are limitations of a retrospective study and bias in not excluding chronic diseases.
In conclusion, we confirmed that ccf-ssDNA level is a simple and cost-effective biomarker for predicting recurrence in CRC patients. In addition, this study provides more accurate information to evaluate the risk of recurrence in CRC patients with high ccf-ssDNA level before surgery, and it is thought to be able to help make more accurate decisions along with other risk factors for patients who need chemotherapy after surgery. Although it is difficult to directly apply clinically based on the results of this study, it is believed that this study will help suggest potential as a novel cfDNA biomarker. Therefore, in order to become a clinically applicable biomarker in CRC, a prospective, multicenter study with a large number of samples and additional verification on ccf-ssDNA will be required.
ACKNOWLEDGMENTS
This study was supported by Soonchunhyang University Research Fund and was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (grant nos. 2019R1I1A3A01061911 and 2021R1G1A1094891).
This researcher would like to thank PhD. Jong Seok Moon of Soonchunhyang Institute of Medi-Bio Science for his help.
Footnotes
Moo-Jun Baek is an editorial board member of the journal but was not involved in the peer reviewer selection, evaluation, or decision process of this article. No other potential conflicts of interest relevant to this article were reported.
REFERENCES
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Jung KW, Won YJ, Hong S, Kong HJ, Lee ES. Prediction of cancer incidence and mortality in Korea, 2020. Cancer Res Treat. 2020;52:351–8. doi: 10.4143/crt.2020.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–41. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 4.Andre N, Schmiegel W. Chemoradiotherapy for colorectal cancer. Gut. 2005;54:1194–202. doi: 10.1136/gut.2004.062745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lurje G, Zhang W, Lenz HJ. Molecular prognostic markers in locally advanced colon cancer. Clin Colorectal Cancer. 2007;6:683–90. doi: 10.3816/CCC.2007.n.037. [DOI] [PubMed] [Google Scholar]
- 6.Taback B, Hoon DS. Circulating nucleic acids and proteomics of plasma/serum: clinical utility. Ann N Y Acad Sci. 2004;1022:1–8. doi: 10.1196/annals.1318.002. [DOI] [PubMed] [Google Scholar]
- 7.Taback B, Hoon DS. Circulating nucleic acids in plasma and serum: past, present and future. Curr Opin Mol Ther. 2004;6:273–8. [PubMed] [Google Scholar]
- 8.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646–50. [PubMed] [Google Scholar]
- 9.Banki F, Mason RJ, Oh D, Hagen JA, DeMeester SR, Lipham JC, et al. Plasma DNA as a molecular marker for completeness of resection and recurrent disease in patients with esophageal cancer. Arch Surg. 2007;142:533–9. doi: 10.1001/archsurg.142.6.533. [DOI] [PubMed] [Google Scholar]
- 10.Ellinger J, Wittkamp V, Albers P, Perabo FG, Mueller SC, von Ruecker A, et al. Cell-free circulating DNA: diagnostic value in patients with testicular germ cell cancer. J Urol. 2009;181:363–71. doi: 10.1016/j.juro.2008.08.118. [DOI] [PubMed] [Google Scholar]
- 11.Ellinger J, Bastian PJ, Ellinger N, Kahl P, Perabo FG, Buttner R, et al. Apoptotic DNA fragments in serum of patients with muscle invasive bladder cancer: a prognostic entity. Cancer Lett. 2008;264:274–80. doi: 10.1016/j.canlet.2008.01.038. [DOI] [PubMed] [Google Scholar]
- 12.Tan G, Chu C, Gui X, Li J, Chen Q. The prognostic value of circulating cell-free DNA in breast cancer: a meta-analysis. Medicine (Baltimore) 2018;97:e0197. doi: 10.1097/MD.0000000000010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hao TB, Shi W, Shen XJ, Qi J, Wu XH, Wu Y, et al. Circulating cell-free DNA in serum as a biomarker for diagnosis and prognostic prediction of colorectal cancer. Br J Cancer. 2014;111:1482–9. doi: 10.1038/bjc.2014.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Czeiger D, Shaked G, Sebbag G, Vakhrushev A, Flomboym A, Lior Y, et al. Elevated cell-free DNA measured by a simple assay is associated with increased rate of colorectal cancer relapse. Am J Clin Pathol. 2016;145:852–7. doi: 10.1093/ajcp/aqw068. [DOI] [PubMed] [Google Scholar]
- 15.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–65. [PubMed] [Google Scholar]
- 16.Qi J, Qian C, Shi W, Wu X, Jing R, Zhang L, et al. Alu-based cell-free DNA: a potential complementary biomarker for diagnosis of colorectal cancer. Clin Biochem. 2013;46:64–9. doi: 10.1016/j.clinbiochem.2012.08.026. [DOI] [PubMed] [Google Scholar]
- 17.Huang X, Zhao Q, An X, Pan J, Zhao L, Shen L, et al. The ratio of ssDNA to dsDNA in circulating cell-free DNA extract is a stable indicator for diagnosis of gastric cancer. Pathol Oncol Res. 2020;26:2621–32. doi: 10.1007/s12253-020-00869-1. [DOI] [PubMed] [Google Scholar]
- 18.Basnet S, Zhang ZY, Liao WQ, Li SH, Li PS, Ge HY. The prognostic value of circulating cell-free DNA in colorectal cancer: a meta-analysis. J Cancer. 2016;7:1105–13. doi: 10.7150/jca.14801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han X, Wang J, Sun Y. Circulating tumor DNA as biomarkers for cancer detection. Genomics Proteomics Bioinformatics. 2017;15:59–72. doi: 10.1016/j.gpb.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee TH, Montalvo L, Chrebtow V, Busch MP. Quantitation of genomic DNA in plasma and serum samples: higher concentrations of genomic DNA found in serum than in plasma. Transfusion. 2001;41:276–82. doi: 10.1046/j.1537-2995.2001.41020276.x. [DOI] [PubMed] [Google Scholar]
- 21.Ponti G, Maccaferri M, Manfredini M, Kaleci S, Mandrioli M, Pellacani G, et al. The value of fluorimetry (Qubit) and spectrophotometry (NanoDrop) in the quantification of cell-free DNA (cfDNA) in malignant melanoma and prostate cancer patients. Clin Chim Acta. 2018;479:14–9. doi: 10.1016/j.cca.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 22.Sorber L, Zwaenepoel K, Deschoolmeester V, Roeyen G, Lardon F, Rolfo C, et al. A comparison of cell-free DNA isolation kits: isolation and quantification of cell-free DNA in plasma. J Mol Diagn. 2017;19:162–8. doi: 10.1016/j.jmoldx.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 23.Volckmar AL, Sultmann H, Riediger A, Fioretos T, Schirmacher P, Endris V, et al. A field guide for cancer diagnostics using cell-free DNA: from principles to practice and clinical applications. Genes Chromosomes Cancer. 2018;57:123–39. doi: 10.1002/gcc.22517. [DOI] [PubMed] [Google Scholar]
- 24.Lee JH, Jeong H, Choi JW, Oh HE, Kim YS. Liquid biopsy prediction of axillary lymph node metastasis, cancer recurrence, and patient survival in breast cancer: a meta-analysis. Medicine (Baltimore) 2018;97:e12862. doi: 10.1097/MD.0000000000012862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Payne RE, Hava NL, Page K, Blighe K, Ward B, Slade M, et al. The presence of disseminated tumour cells in the bone marrow is inversely related to circulating free DNA in plasma in breast cancer dormancy. Br J Cancer. 2012;106:375–82. doi: 10.1038/bjc.2011.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roth C, Pantel K, Muller V, Rack B, Kasimir-Bauer S, Janni W, et al. Apoptosis-related deregulation of proteolytic activities and high serum levels of circulating nucleosomes and DNA in blood correlate with breast cancer progression. BMC Cancer. 2011;11:4. doi: 10.1186/1471-2407-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mittra I, Khare NK, Raghuram GV, Chaubal R, Khambatti F, Gupta D, et al. Circulating nucleic acids damage DNA of healthy cells by integrating into their genomes. J Biosci. 2015;40:91–111. doi: 10.1007/s12038-015-9508-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Olmo DC, Dominguez C, Garcia-Arranz M, Anker P, Stroun M, Garcia-Verdugo JM, et al. Cell-free nucleic acids circulating in the plasma of colorectal cancer patients induce the oncogenic transformation of susceptible cultured cells. Cancer Res. 2010;70:560–7. doi: 10.1158/0008-5472.CAN-09-3513. [DOI] [PubMed] [Google Scholar]