Abstract
Environmental toxins and toxicants can damage proteins and threaten cellular proteostasis. Most current methodologies to identify misfolded proteins in cells survey the entire proteome for sites of changed reactivity. We describe and apply a quantitative proteomics methodology to identify destabilized proteins based on their binding to the human Hsp40 chaperone DNAJB8. These protein targets are validated by an orthogonal limited proteolysis assay using parallel reaction monitoring. We find that brief exposure of HEK293T cells to meta-arsenite increases the affinity of two dozen proteins to DNAJB8, including known arsenite-sensitive proteins. In particular, arsenite treatment destabilizes both the pyruvate dehydrogenase complex E1 subunit and several RNA-binding proteins. This platform can be used to explore how environmental toxins impact cellular proteostasis, and to identify the susceptible proteome.
INTRODUCTION
Exposure to environmental toxins threatens the structural integrity of proteins.1 Structural changes due to oxidation, covalent modification, or non-covalent binding can cause proteins to misfold, leading to aggregation or loss of function.2 For example, the heavy metal arsenic (As) binds to protein sulfhydryl groups and generates reactive oxygen species (ROS).3 Consequent accumulation of misfolded proteins and oxidative stress activates the Heat Shock Response (HSR) to induce chaperones and restore protein homeostasis.4 Activation of HSR or other similar misfolded protein stress responses is a common response to heavy metals, electrophilic pesticides/herbicides, and other environmental toxins. While measuring these responses indicates whether a given toxin is likely to be inducing protein misfolding, it does not indicate which proteins are misfolding, and hence which cellular pathways are being affected by the exposure.
Most current approaches to identify misfolded proteins measure proteome-wide solvent accessibility by mass spectrometry to infer conformational changes.5,6 Stability of Proteins from Rates of Oxidation (SPROX) analyzes protein methionine oxidation in cellular lysates, with varying chaotrope concentrations to measure proteins’ ΔGunfolding.7,8 Fast Photochemical Oxidation of Proteins (FPOP) measures the exposure of protein sites in cells or organisms to in situ generated hydroxide radicals.9 Limited Proteolysis (LiP) measures proteome-wide susceptibility to proteolytic cleavage.10 Covalent protein painting measures differences in protein folding based on accessible lysine ε-amines after proteins are exposed to electrophilic reagents.11 Cellular Thermal Shift Assay (CETSA) measures proteome-wide susceptibility to aggregation with increasing temperature.12 Each technique offers a unique approach using quantitative proteomics to assess protein stability in a cell.
Alternatively, the cell identifies misfolded proteins through recognition by chaperones. A highly promiscuous chaperone is Hsp70, which relies upon members of the Hsp40 family to identify and recruit misfolded protein clients for refolding; one third of the proteome relies on this cycle under basal conditions13–16. Release of clients from Hsp40 to Hsp70 can be blocked by an H-to-Q mutation in the Hsp40 J-domain, stabilizing misfolded protein binding.17
DNAJB8 is notable among human Hsp40s for its dual nuclear and cytosolic localization, formation of oligomers, and slow client release kinetics.18–21 We previously used affinity purification and quantitative proteomics to identify hundreds of cellular protein clients of overexpressed human Hsp40 DNAJB8H31Q with high reproducibility and statistical confidence.22 Herein we exploit the ability of DNAJB8H31Q to recognize misfolded protein clients to develop a platform for identifying proteins that are destabilized in response to exogenous stress (Figure 1). We demonstrate this approach in HEK293T cells treated with trivalent arsenic, a toxic metal that causes widespread damage to nucleic acids and proteins, leading to genomic and metabolic instability.23
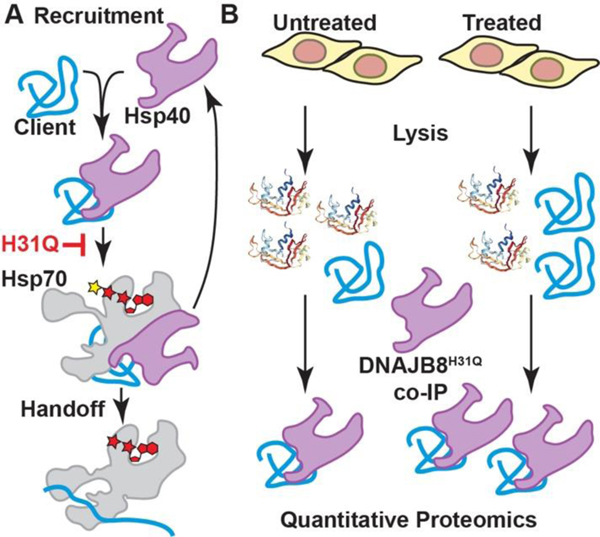
Figure 1.
Design of the Hsp40 affinity assay for misfolded proteins. A) DNAJB8 binding and handoff to Hsp70 is interrupted by an H31Q mutation in the J-domain. B) If cellular treatment increases the misfolded population of a DNAJB8 client protein, then the apparent affinity of that protein for DNAJB8H31Q will increase.
MATERIALS AND METHODS
Materials:
Bovine Serum Albumin (BSA), Dulbecco’s Modified Eagle Media (DMEM), Dulbecco’s phosphate-buffered saline (DPBS), 10 cm plates, and 6 well plates were from VWR. Roche Protease Inhibitor cocktail w/o EDTA (PIC), 1,4-dithiothreitol (DTT), HEPES, sodium meta arsenite (NaAsO2), Cd(NO3)2•(H2O)4 were from Sigma Aldrich. Sodium chloride (NaCl), Tris-Hydrochloride (Tris-HCl), Triton X-100, sodium deoxycholate, KCl, MgCl2, CaCl2, Ag(NO3)2, Na2S2O3, urea, Ca(O2C2H3)2), glycerol, sodium dodecyl sulfate (SDS), poly D-lysine, and sequencing grade trypsin were from Thermo Fisher Scientific. Proteinase K (PK) was from Promega. Nanopure water was purified using a Millipore Milli-Q Laboratory lab 4 Chassis Reagent Water System. 5 μm and 3 μm Aqua C18 resins were from Phenomenex. Sepharose-4B beads, anti-M2 Flag magnetic beads, tris (2-carboxyethyl)phosphine hydrochloride (TCEP), and iodoacetamide were from Millipore Sigma. 250 μm diameter fused silica columns were from Agilent. 100 μm diameter fused silica columns were from Polymicro. Strong cation exchange resin was from Partisphere, GE Healthcare. Rapigest was from Aobious (Gloucester, MA). TMT-6plex isotopic labels were from Pierce. Bradford reagent was purchased from Bio-rad.
AP-TMT-MudPIT:
TMT-AP-MS experiments were performed as previously described22. For each sixplex TMT-AP-MS, six 10 cm plates of HEK293T cells were transfected by the calcium phosphate method with 5 μg of plasmid DNA encoding FlagDNAJB8H31Q in the pFLAG backbone. Plates were treated with heavy metal salts or vehicle at 40–46 hours post transfection. Cells were harvested by scraping in DPBS and lysed in 9 parts RIPA Buffer (150 mM NaCl, 50 mM Tris pH 7.5, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS) and 1 part 10x PIC for 30 min on ice. Lysate was separated from cell debris by centrifugation at 21,000 x g for 15 minutes at 4 °C. Protein in the lysate was quantified by Bradford. Lysates were pre-cleared with 15 μL Sepharose-4B beads for 30 min at 4 °C, then centrifuged at 1,500 x g for 1 min to pellet beads. Lysate was then separated and incubated with 15 μL of M2 anti-Flag Magnetic Beads and rotated overnight at 4 °C. The anti-Flag beads were washed the next day four times with RIPA buffer. Each wash included rotation for 10 minutes at ambient temperature. Proteins bound to the anti-Flag beads were eluted by boiling for 5 min at 100 °C in 30 μL of Laemmli concentrate (120 mM Tris pH 6.8, 60% glycerol, 12% SDS, brilliant phenol blue to color). 5 μL of the elutes were saved for silver stain analysis and the remainder was prepped for mass spectrometry and TMT-labeled.
Only MS quality organic solvents were used during sample preparation. The composition for buffer A is 5% acetonitrile, 0.1% formic acid in water. The composition for Buffer B is 80% acetonitrile, 0.1% formic acid. The composition for Buffer C is 500 mM ammonium acetate in Buffer A. The first arsenic exposure TMT-MS run was performed as one-dimensional LC/MS/MS on an Orbitrap Fusion Tribrid mass spectrometer (Thermo) interfaced with a nanoAquity UPLC (Waters) system. All other MS runs were performed with a two-dimensional LC/MS/MS setup on an LTQ Orbitrap Velos Pro hybrid mass spectrometer (Thermo) interfaced with an Easy-nLC 1000 (Thermo) according to standard MuDPIT protocols24. For each run, MS/MS spectra were extracted using MSConvert (version 3.0.21144) with Peak Picking Filtering. MS/MS spectra was then searched by FragPipe25 against a Uniprot human proteome database (06/11/2021 release) containing 40858 human sequences (longest entry for each protein). MS/MS spectra were also searched against 20429 select decoys (e.g albumen, porcine trypsin, contaminants etc.). FragPipe searches allowed for static modification of cysteine residues (57.02146 Da, acetylation), and N-termini and lysine residues (229.1629 Da, TMT-tagging), half tryptic peptidolysis specificity, and mass tolerance of 20 ppm for precursor mass and 20 ppm for product ion masses. Spectra matches were assembled and filtered by MSFragger (version 3.2). Decoy proteins, common contaminants, immunoglobulins and keratins were filtered from the final protein list. Quantitation in FragPipe was performed by averaging TMT reporter ion intensities for all spectra associated with an individual peptide.
Statistical Analysis:
Protein-level intensities were normalized to the intensity of bait (DNAJB8) in each TMT channel. To combine the multiple TMT runs, we used a version of the scaled reference approach26. A scaling factor was obtained from averaging the bait-normalized integrated TMT reporter ion intensities for each protein across the 3 control conditions in each AP-MS run. Each bait-normalized protein intensity was then divided by this scaling factor. Unadjusted p-values were converted to q-values (local false discovery rates) using Storey’s modification to the method of Benjamini and Hochberg27,28. Unadjusted p-values were ranked in increasing order and the q-value for the ith protein determined from:
Storey’s modification is performed by determining the overrepresentation of low p-values to infer a global false discovery rate, and then scaling local false discovery rates accordingly. The π-factor for this scaling was 0.84 for the arsenic treatment TMT-AP-MS data set and 0.79 for the cadmium treatment.
Limited Proteolysis:
The limited proteolysis procedure was optimized from standard protocols29. 1 mg/ml stocks were made from 25 mg of lyophilized Proteinase K (PK) dissolved in a storage buffer (50 mM Tris-HCl, 2 mM calcium acetate, pH 8.0) suitable for PK and stored at –70 °C. The following concentrations of PK were prepared from serial dilutions from 1mg/ml aliquot: 0.5 mg/ml, 0.2 mg/ml, 0.1 mg/ml, 0.05 mg/ml, 0.02 mg/ml, and added to lysate to yield 1:200, 1:500, 1:1000, 1:2000, and 1:5000 wt/wt protease: substrate protein ratios respectively. For each digestion, 2 μl PK was added to a 200 μg aliquot of protein lysate and incubated for 1 min at 25.0 °C. Samples were then boiled for 5 min to quench PK activity. Three separate digestions were performed for the no PK condition for each lysate sample. The sample set-up and the calculation of fraction remaining for each peptide is shown in Figure S1. Samples were prepared for mass spectrometry, spiked with an internal standard peptide NH2-VFFAEDVGSNK-CO2H to 83 nM and analyzed using LC-MS/MS and parallel reaction monitoring (PRM).
FlagPDHA1 co-IP:
Immunoprecipitations were performed similarly to those of FlagDNJAB8H31Q with the exception that cells were crosslinked prior to lysis with 1 mM DSP/1% DMSO/DPBS for 30 mins while rotating at ambient temperature. DSP was quenched with 100 mM Tris pH 8 (final concentration) with rotating for 15 minutes at room temperature
RESULTS AND DISCUSSION
Protein misfolding stresses leads to extensive transcriptional, translational, and post-translational remodeling of the cell.[30] To isolate stress-induced protein misfolding from these pleiotropic effects, we limited cellular exposure to brief 15 min treatments. We validated that 15 min. 500 μM sodium meta-arsenite (NaAsO2) induces expression of the HSR target HSPA1A in HEK293T cells (Figure S2). Because HSR is activated by misfolded protein accumulation, this suggests that 500 μM arsenite treatment causes protein misfolding in only 15 min. This response is not suppressed by overexpression of FlagDNAJB8H31Q (Figure S2), indicating that our recognition element for misfolded protein does not prevent protein destabilization.
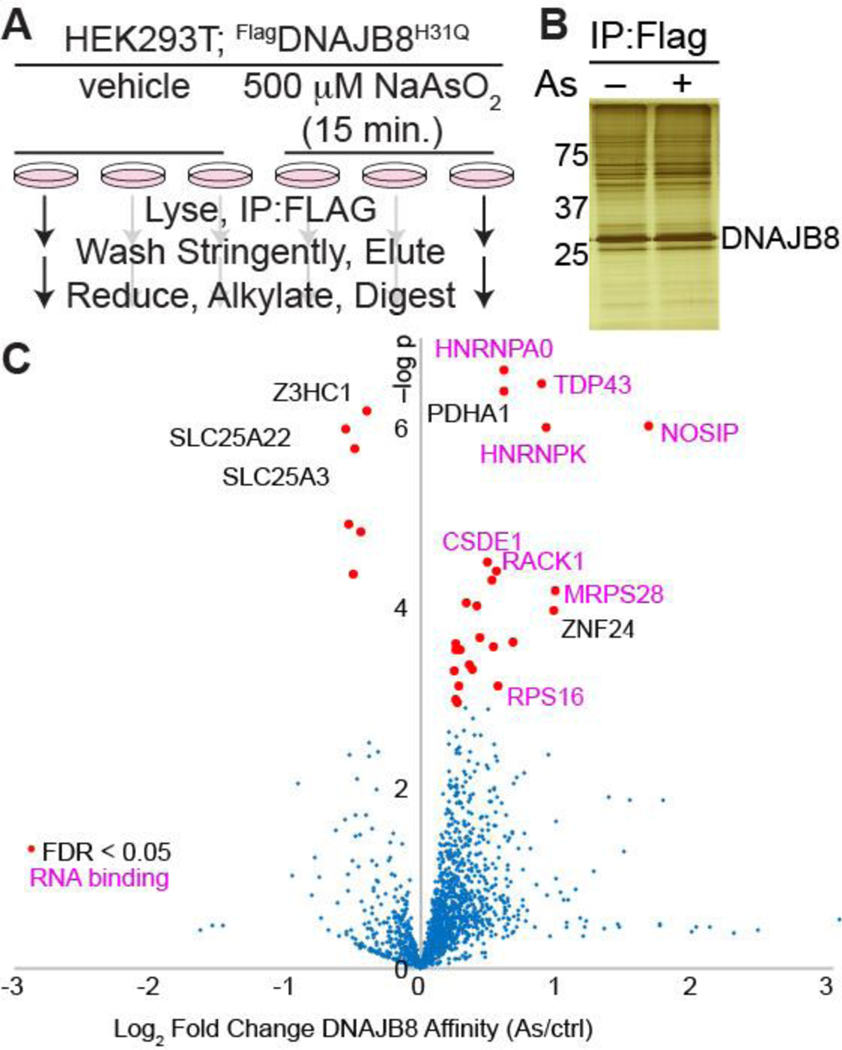
We used the experimental approach illustrated in Figure 2A to determine proteins that misfold in response to arsenite exposure. FlagDNAJB8H31Q was transiently overexpressed in HEK293T cells, followed by 15 min. NaAsO2 treatment and immediate Flag immunoprecipitation from cellular lysate. Co-immunoprecipitated proteins were identified and quantified by LC/LC-MS/MS in concert with TMT isobaric tagging.31 Overall, 24 biological replicates (12 treated and 12 controls) were analyzed through four 6-plex TMT runs. Most observed protein clients slightly increase affinity to DNAJB8 following arsenite treatment, likely because peptides from proteins with increased DNAJB8 binding following arsenite treatment are more represented in the pooled peptide mixture, and thus have higher chances of identification during shotgun proteomics. However, the bulk DNAJB8 associated proteome does not change (Figure 2B). 24 proteins have significantly greater affinity for FlagDNAJB8H31Q in response to the arsenite treatment. These proteins include PDHA1 and 17 ribosomal RNA-binding proteins, including HNRNPA0, TDP-43, RACK1, and RPS16 (Figure 2C and Table S1).
Figure 2.
Arsenite treatment increases the affinity of a select subset of proteins with DNAJB8H31Q. A) Experimental protocol. B) Representative silver stain for proteins co-immunoprecipitated with DNAJB8H31Q. C) Volcano plot illustrating the effect of cellular As treatment on protein interactions with DNAJB8H31Q. Red dots represent proteins with significantly increased interaction with DNAJB8H31Q, using a false discovery rate threshold (FDR) of 5% (n = 12 biological replicates in 4 TMT-AP-MS runs). Protein names in purple are RNA-binding proteins.
Arsenite generally induces misfolding and aggregation of RNA-binding proteins into stress granules.32,33 However, canonical stress granule markers34 G3BP1 and eIF4G1 are not enriched in DNAJB8H31Q pull-downs from arsenite-treated cells, indicating that DNAJB8H31Q is not co-precipitating intact stress granules (Table S1). TDP-43 is a ribonuclear protein that forms aggregates in ALS and other proteinopathies.35 It accumulates in cytoplasmic and nuclear condensates in response to arsenite treatment, due to displacement from RNA and post-translational modification.36 Yeast RACK1 migrates to stress granules in response to arsenite.37 It is particularly interesting that PDHA1, the alpha subunit of the pyruvate dehydrogenase complex (PDC),38 is destabilized by arsenite. Inhibition of PDC is a major contributor to arsenite-induced metabolic disfunction.39 We are encouraging that our assay primarily finds proteins that are known to be arsenite-sensitive.
The Hsp40 Affinity mass spectrometry experiments are comparisons between two sets of twelve AP-MS preparations, analyzed as four individual multiplexed injections of six samples each. To indicate the robustness of hits from this assay, the Strictly Standardized Mean Differences (SSMD) were compared between each multiplexed injection (Figure S3). SSMDs represent the mean differences between the treated and untreated populations, normalized by root-mean-square standard deviations. The most affected proteins reproduce well across each replicate. It is worth noting that arsenite treatment does not affect the bulk proteome of eluted DNAJB8H31Q co-IPs by silver stain of the eluate (Figure S4). The arsenite pulldowns collectively identified and quantified 1696 unique proteins, not including 28 keratin/immunoglobulin proteins that were excluded from the set, and 24 proteins that were identified but whose TMT reporter ions could not be quantified. We previously characterized 562 high confidence DNAJB8H31Q interactors from AP-MS in unstressed HEK293T cells22. Those included 379 of the proteins found in this study, and 21/30 of our high confidence arsenite-sensitive proteins. It is possible that stress conditions, by increasing the affinity and thus recovery of a select proteome, also increase the likelihood of identifying those proteins from data-dependent analysis. Piette et al recently40 reported comprehensive interactomes of human Hsp40 and Hsp70 family proteins (unlike our work, these all had active J-domains), finding 37 high confidence interactors of DNAJB8 and another 479 proteins that could not be reliably distinguished from background. We find 22/37 of their high-confidence interactors in our data set, with most of the missing proteins being either Hsp70s (which bind through the J-domain) or Hsp70 binding proteins (Hsp40s, CHIP, FKBP8 etc.), consistent with expected differences in recovery between the wild-type protein and a J-inactive variant. Conversely, 11/31 of our high confidence arsenite-sensitive proteins are found in their study, and none are among their high confidence interactors.
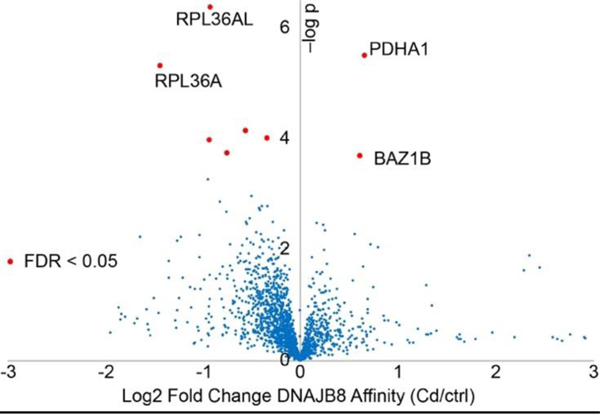
To determine whether the arsenite response is specific, we treated cells with Cd2+, another heavy metal that induces cellular apoptosis through generation of ROS.[41] 15 min. treatment with 200 μM Cd(NO3)2 induces HSR in HEK293T cells (Figure S5). Cells were treated with Cd or vehicle for 15 min. and then immediately lysed, and the DNAJB8H31Q interacting proteome quantified by TMT-AP-MS (Figure 3 and Table S2). As with arsenite-treated cells, the bulk DNAB8H31Q interacting proteome is unaffected (Figure S6). Unlike with arsenite, the only proteins destabilized by acute Cd treatment are PDHA1 and BAZ1B (Figure 3 and Figure S7). Rather, many proteins slightly lose affinity for DNAJB8 following treatment, perhaps due to direct binding to Cd. As with arsenite, Cd exposure inhibits pyruvate dehydrogenase activity42. TDP-43 association with DNAJB8 is unchanged, despite reports that Cd2+ treatment (100 μM, 2 h) promotes TDP-43 aggregation in Cos7 cells.35 The difference in proteome destabilization between the two heavy metals suggests that ROS generation is not adequate to explain their effects on protein stability following acute arsenite exposure.
Figure 3.
Cadmium treatment (200 μM Cd(NO3)2 for 15 min.) only affects the DNAJB8H31Q affinity of a few proteins. The experimental protocol is similar to Figure 1A. n = 12 biological replicates in 4 TMT-AP-MS runs.
Protein sequences are optimized for a native conformational landscape, and hence damage that modulates charge, sterics, or binding partners has the potential to induce misfolding. However, binding to the native state will generally stabilize that state, decreasing the propensity to access misfolded states. This is the basis of ligand target discovery through stabilization as widely employed in CETSA12. Because our assay enriches Hsp40 clients, it will enrich proteins with enhanced destabilization in response to treatment and should be inherently disadvantaged for discovery of stabilized protein targets. However, we would still expect to see proteins that are stabilized, such as are observed on the left in Figure 2C and Figure 3 if, for example, direct metal binding increases ΔGunfolding.
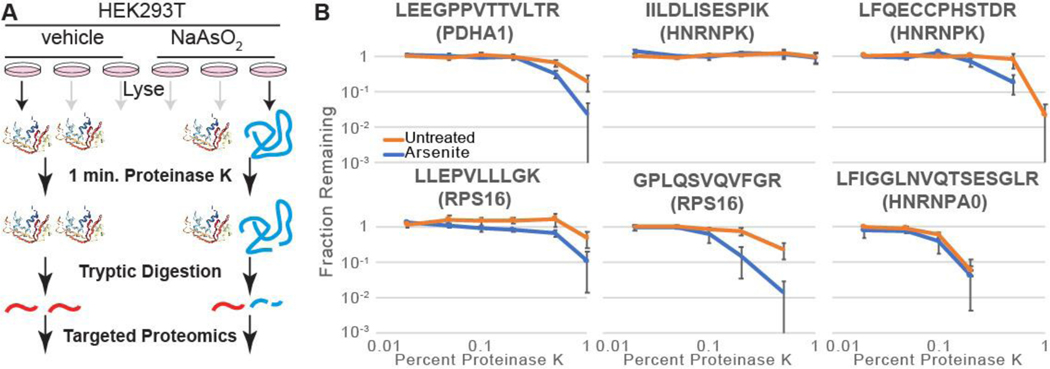
Although our short treatments and immediate processing avoid many pleiotropic effects (e.g. altered transcription, translation, trafficking, and degradation, etc.), unforeseen variables besides protein stability could impact DNAJB8-client affinity. Hence we applied an orthogonal assay, LiP, to validate and prioritize proteins with arsenite-induced binding to DNAJB8 (Figure 4A). LiP as a discovery technology is challenged by the need to identify and quantify peptides in a proteome that has been rendered complex by the use of two orthogonal proteases; for example, hit overlap with SPROX from the same samples is about ~20%43,44. We targeted select peptides from our hit proteins to alleviate this challenge. The PK concentration gradient was optimized to bracket the full range of changes in observable protein on a Coomassie-stained SDS-PAGE gel (Figure S8). Bulk protein band intensity decreases at a PK:protein ratio of 1:1000 and little intact protein is observable at 1:100 PK:protein. These values are somewhat lower than commonly reported conditions, which range from 1:33 to 1:10045,46. Peptides for monitoring were chosen in the Picky software47, based on chemical properties amenable to PRM and to maximize distribution across the chromatographic gradient. The PRM method had a median CV of of 20%, with all but one peptide below 30% (Table S3). Biological replicates showed similar CV values (Table S4). Precision was unaffected by choice of MS2 resolving power (Table S5), consistent with a recent report on the dispensability of high resolution for PRM48, so 7500 was used.
Figure 4.
A) Schematic illustrating how limited proteolysis differentiates between different conformations of a protein. B) Proteinase K susceptibility curves for four peptides as monitored by LiP-PRM. Error bars represent standard error (n = 3).
We treated cells with arsenite or vehicle for 15 min., followed by immediate lysis. Lysates were treated with varying concentration of proteinase K for 1 min, heat-quenched, tryptically digested, and peptides from candidate proteins quantified by Parallel Reaction Monitoring (PRM).49 If a cellular treatment increases the PK proteolysis yield of a peptide, that implies that the protein is destabilized in the vicinity of that sequence45. Limited proteolysis is a local measure of protein conformation and thus is subject to false negatives at the protein level. Hence, a given peptide being equally sensitive to proteolysis with or without cellular treatment does not imply that the entire protein is unaffected.
We determined the proteolytic susceptibility with and without 15 min. cellular arsenite treatment for 13 peptides from PDHA1, RSP16, RPS3, TDP-43, HNRNPK, RACK1, and HNRNPA0. Although RPS3 was a lower significance protein from our screen, we included it in these targeted LiP experiments because arsenite causes release of RPS3 from RNA50, which could affect protein stability. Peptides from PDHA1 and RPS16 are more susceptible to proteolysis upon arsenite treatment, implying that arsenite destabilizes these proteins (Figures 4B and S9A). The peptides chosen for TDP-43, HNRNPA0, RACK1 and RPS3 do not show a clear increase in proteolytic sensitivity; those proteins might still be destabilized by arsenite in unsampled regions of the protein. HNRNPK increases sensitivity at two peptides, but not at three others. No peptide became less sensitive to PK following cellular arsenite treatment. We expanded the TDP-43 evaluation to four additional peptides, of which three showed arsenic sensitivity (Figure S9B and Table S6). Surprisingly, all three peptides come from structured regions within the RNA-binding domains of the protein.51 The two peptides that do not have increased proteolytic susceptibility in response to cellular arsenic exposure are in intrinsically disordered regions; one is in the linker between the RNA binding domains and the other is in the unstructured C-terminus. The general validation of protein destabilization by an orthogonal method demonstrates that DNAJB8H31Q affinity successfully identified proteins that are destabilized by arsenite treatment.
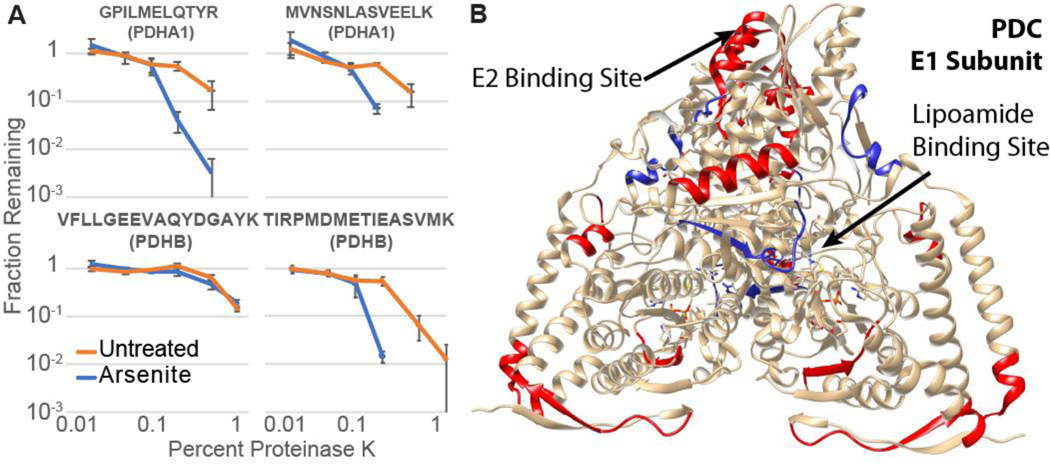
The sensitivity of PDHA1 to arsenite is surprising. PDC inhibition by arsenite is generally ascribed to arsenite coordination to vicinal thiols in the lipoamide cofactor3, though other evidence strongly points to lipoamide binding being unnecessary for inhibition by arsenic.52 PDC is composed of three subunits, including an E1 heterotetramer containing two PDHB and two PDHA1 proteins.38 Lipoamide is anchored covalently to DLAT in the E2 subunit, but reacts in the groove of PDHB in the E1 subunit. We considered the hypothesis that arsenite destabilizes the rest of the E1 subunit. PDHB did show increased affinity to DNAJB8H31Q in our initial screen, but with low significance (FC = 1.3 ± 0.3, q = 0.06). The binding of DLAT to DNAJB8H31Q was not meaningfully unaffected by arsenite exposure (FC = 0.93 ± 0.48, q = 0.69). Because LiP measures local proteolytic sensitivity,[41] we used targeted LiP to determine whether PDHA1 and PDHB are globally destabilized. Eleven peptides from PDHA1 and PDHB were selected, covering most of the protein sequences (Figure 5A, Figure S9C, and Table S7). Most locations on both proteins are destabilized by cellular arsenite treatment (Figure 5B), indicating that the proteins undergo an extensive conformational change with arsenite treatment. In particular, cellular arsenite increases proteolytic susceptibility at the E1-E2 interface (IMEGPAFNFLDAPAVR and TIRPMDMETIEASVMK peptides). The peptide at the lipoamide binding site (VFLLGEEVAQYDGAYK), however, is not affected. That lead us to explore the hypothesis that perhaps cellular inactivation of PDC is caused by complex destabilization and disassembly of E1. We cloned affinity tagged FlagPDHA1 and expressed by transfecting in HEK293T cells. As expected for a dynamic complex, FlagPDHA1 requires intracellular crosslinking to co-immunoprecipitate PDHB and DLAT (Figure S10). However, neither 15 min. nor 4 h cellular arsenite treatment (500 μM) decreases the yield of PDHB and DLAT recovery with FlagPDHA1. Hence, despite complex destabilization by arsenite, the complex does not significantly dissociate (Figures S11–S14 and Supplemental Discussion).
Figure 5.
A) Proteinase K susceptibility curves for four peptides from the E1 subunit of PDC as monitored by LiP-PRM. Error bars represent standard error (n = 3). B) Crystal structure of the dimeric PDC E1 subunit (PDB: 1NI4)55,56 with As destabilized peptides are colored red, and the two peptides that are not As-sensitive are blue. The arrow indicates the location of the lipoamide co-factor.
In conclusion, we have developed a platform to identify proteins that are destabilized in response to cellular stress. The DNAJB8H31Q immunoprecipitation assay identifies destabilized proteins using the same criterion as the cell: increased binding to a chaperone. Identification of likely destabilized proteins in the screening step enables targeted LiP-PRM as a mechanistically orthogonal, and technically straightforward, validation step. Using this technology, we identified proteins that are destabilized by just a brief 15 min. cellular arsenite exposure. Most of these proteins are known to be functionally perturbed by arsenite, with changes in post-translational modifications and even aggregation. However, it has not previously been demonstrated that these proteins are destabilized, nor that arsenite can induce conformational changes inside living cells within 15 min. of treatment. Furthermore, we have found that arsenite destabilizes both members of the E1 subunit of PDC. This platform will be useful in further understanding how environmental toxins and toxicants perturb proteome integrity.
DNAJB8 is localized to the nucleus and cytosol53, consistent with the lack of genetically encoded localization sequences, suggesting that this approach might be limited to those two environments. Nevertheless, 13% and 1% of high-confidence DNAJB8H31Q interactors are mitochondrial and secretory proteins respectively22. Given the high affinity of DNAJB8H31Q for its clients, it is possible that the protein can engage substrates after lysis. If this is the case, then DNAJB8H31Q could be used to profile across cellular environments, and suggests that cellular expression of DNAJB8H31Q may be dispensable for the assay. Alternatively, another way to assay for protein misfolding in the ER may be to append the distinctive C-terminus of DNJAB8 onto the native ER Hsp40 DNAJB11, with the H53Q mutation to block handoff to the ER Hsp70 BiP. Similarly to DNAJB8, secreted DNAJB11 has irreversible client binding in the absence of Hsp7054. Mammalian mitochondria do not have Type II (DNAJB) Hsp40 chaperones13. For that environment, it would be necessary to evaluate whether DNAJB8H31Q with mitochondrial matrix or IMS localization sequences a) perturb mitochondrial proteostasis and b) are able to reach, fold, and function in those environments. Further investigation will establish what fraction of the proteome can be assayed by this approach.
Supplementary Material
ACKNOWLEDGMENT
We thank H.T. Wu for assistance with Skyline, and the NIH for S10 OD010669. This work was supported by the University of California, Riverside and a Society of Analytical Chemists of Pittsburgh Starter Grant.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
The authors declare no competing financial interest.
REFERENCES
- [1].Tamás M; Sharma S; Jacobsen T; Christen P Heavy Metals and Metalloids As a Cause for Protein Misfolding and Aggregation. Biomolecules. 2014, 4, 252–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Petrov D; Zagrovic B; Microscopic Analysis of Protein Oxidative Damage: Effect of Carbonylation on Structure, Dynamics, and Aggregability of Villin Headpiece. J. Am. Chem. Soc 2011, 133, 7016–7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Shen S; Li X; Cullen WR; Weinfield M; Lee XC Arsenic Binding to Proteins. Chem. Rev 2013, 113, 7769–7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Alford BD; Brandman O Quantification of Hsp90 availability reveals differential coupling to the heat shock response. J. Cell. Biol 2018, 217, 3809–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kaur U; Meng H; Lui F; Ma R; Ogburn RN; Johnson JHR; Fitzgerald MC; Jones ML Proteome-Wide Structural Biology: An Emerging Field for the Structural Analysis of Proteins on the Proteomic Scale. J. Proteome Res 2018, 17, 3614–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Genereux JC Mass spectrometric approaches for profiling protein folding and stability. Adv Protein Chem Struct Biol., 2019, 118, 111–144. [DOI] [PubMed] [Google Scholar]
- [7].Adhikari J; West GM; Fitzgerald MC Global Analysis of Protein Folding Thermodynamics for Disease State Characterization. J. Proteome Res 2015, 14, 2287–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhang T; Wolfe C; Pierle A; Welle KA; Hryhoernko JR; Ghaemmaghami S. Proteome-wide modulation of degradation dynamics in response to growth arrest PNAS, 2017, 114, DOI: 10.1073/pnas.1710238114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chea EE; Jones LM Modifications generated by fast photochemical oxidation of proteins reflect the native conformations of proteins. Protein Sci, 2018, 27, 1047–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Leuenberger P; Ganscha S; Kahraman A; Cappelletti V; Boersema PJ; Von Mering C; Classen M; Picotti P. Cell-wide analysis of protein thermal unfolding reveals determinants of thermostability. Science 2017, 355. [DOI] [PubMed] [Google Scholar]
- [11].Bamberger C; Pankow S; Martínez-Bartolomé S; Ma M; Diedrich J; Rissman RA; Yates JR; Protein Footprinting via Covalent Protein Painting Reveals Structural Changes of the Proteome in Alzheimer’s Disease. J. Proteome Res 2021, DOI 10.1021/acs.jproteome.0c00912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Savitski MM; Reinhard FBM; Franken H; Werner T; Savitski MF; Eberhard D; Molina DM; afari R; Dovega RB; Klaeger S; Kuster B; Nordlund P; Bantscheff M; Drewes G. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science 2014, 346, 1255784 DOI: 10.1126/science.1255784. [DOI] [PubMed] [Google Scholar]
- [13].Kampinga HH; Craig EA The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. 2010, 11, 570–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bukau B; Weissman J; Horwich A. Molecular Chaperones and Protein Quality Control. Cell. 2006, 125, 443–451. [DOI] [PubMed] [Google Scholar]
- [15].Kim YE; Hipp MS; Bracher A; Hayer-Hartl M; Hartl FU Molecular chaperone functions in protein folding and proteostasis Annu. Rev. Biochem. 2013, 82, 323–355. [DOI] [PubMed] [Google Scholar]
- [16].Labbadia J; Morimoto RI The Biology of Proteostasis in Aging and Disease. Annu. Rev. Biohem 2015, 84, 435–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Otero JH; Lizak B; Feige MJ; Hendershot LM Dissection of Structural and Functional Requirements That Underlie the Interaction of ERdj3 Protein with Substrates in the Endoplasmic Reticulum. J. Biol. Chem 2014, 289, 27504–27512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yamashita M; Hirohashi Y; Torigoe T; Kusumoto H; Murai A; Imagawa T; Sato N. Dnajb8, a Member of the Heat Shock Protein 40 Family Has a Role in the Tumor Initiation and Resistance to Docetaxel but Is Dispensable for Stress Response. PLoS One. 2016, DOI: 10.1371/journal.pone.0146501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hageman J; Rujano MA; van Waarde MAWH; Kakkar V; Dirks RP; Govorukhina N; Oosterveld-Hut HMJ; Lubsen NH; Kampinga HH A DNAJB Chaperone Subfamily with HDAC-Dependent Activities Suppresses Toxic Protein Aggregation. Mol. Cell. 2010, 37, 355–369. [DOI] [PubMed] [Google Scholar]
- [20].Kakkar V; Månsson C; de Mattos EP; Bergink S; van der Zwaag M; van Waarde M, M. A. W. H.; Kloosterhuis NJ; Melki R; van Cruchten RTP; Al-Karadaghi S; Arosio P; Dobson CM; Knowles TPJ; Bates GP; van Deursen JM; Linse S; van de Sluis B; Emanuelsson C; Kampinga HH The S/T-Rich Motif in the DNAJB6 Chaperone Delays Polyglutamine Aggregation and the Onset of Disease in a Mouse Model. . Mol. Cell 2016, 62, 272–283. [DOI] [PubMed] [Google Scholar]
- [21].Gillis J; Schipper-Krom S; Juenemann K; Gruber A; Coolen S; van den Nieuwendijk R; van Veen H; Overkleeft H; Goedhart J; Kampinga HH; Reits EA The DNAJB6 and DNAJB8 Protein Chaperones Prevent Intracellular Aggregation of Polyglutamine Peptides. J. Biol. 2013, 288, 17225–17237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Mei L; Montoya MR; Quanrud GM; Tran M; Villa-Sharma A; Huang H; Genereux JC Bait Correlation Improves Interactor Identification by Tandem Mass Tag-Affinity Purification-Mass Spectrometry. J. Proteome. Res 2020, 19, 1565–1573. [DOI] [PubMed] [Google Scholar]
- [23].Naujokas MF; Anderson B; Ahsan H; Aposhian HV; Graziano JH; Thompson C; Suk WA The broad scope of health effects from chronic arsenic exposure: update on a worldwide public health problem Environ. Health. Perspect 2013, 121, 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Washburn MP; Wolters D; Yates JR Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol 2001, 19, 242–247. [DOI] [PubMed] [Google Scholar]
- [25].Kong AT; Leprevost FV; Avtonomov DM; Mellacheruvu D; Nesvizhskii AI MSFragger: ultrafast and comprehensive peptide identification in shotgun proteomics. Nat. Meth. 2017, 14, 513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Plubell DL; Wilmarth PA; Zhao Y; Fenton AM; Minnier J; Reddy AP; Klimek J; Yang X; David LL; Pamir N. Extended Multiplexing of Tandem Mass Tags (TMT) Labeling Reveals Age and High Fat Diet Specific Proteome Changes in Mouse Epididymal Adipose Tissue. Mol. Cell. Proteomics 2017, 16, 873–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yekutieli D; Benjamini Y; Resampling-based false discovery rate controlling multiple test procedures for correlated test statistics. J. Stat. Plan. Inference 1999, 82, 171–196. [Google Scholar]
- [28].Storey JD; Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Schopper S; Kahraman A; Leunberger P; Feng Y; Piazza I; Muller O; Boersema PJ; Picotti P Measuring protein structural changes on a proteome-wide scale using limited proteolysis-coupled mass spectrometry. Nat. Protocol. 2017, 1, 2391–2410. [DOI] [PubMed] [Google Scholar]
- [30].Cao SS; Kaufman RJ Endoplasmic reticulum stress and oxidative stress in cell fate decision and human disease. Antioxid. Redox Signal 2014, 21, 396–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Rauniyar N; Yates JR; Isobaric labeling-based relative quantification in shotgun proteomics. J. Proteome Res 2014, 13, 5293–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Turakhiya A; Meyer SR; Marincola G; Schlosser A; Hofmann K. ZFAND1 Recruits p97 and the 26S Proteasome to Promote the Clearance of Arsenite-Induced Stress Granules. Mol. Cell 2018, 906–919. [DOI] [PubMed] [Google Scholar]
- [33].Tam S; Geller R; Spiess C; Frydman J. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat. Cell Biol 2006, 8, DOI 10.1038/ncb1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wheeler JR; Matheny T; Jain S; Abrisch R; Parker R. Distinct stages in stress granule assembly and disassembly. Elife 2016, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hergesheimer RC; Chami AA; De Assis DR; Vourc P; Andres CR; Corcia P. A role for SUMOylation in the Formation and Cellular Localization of TDP-43 Aggregates in Amyotrophic Lateral Sclerosis. Brain 2019, 1176–1194.30938443 [Google Scholar]
- [36].Cohen TJ; Hwang AW; Restrepo CR; Yuan C; Trojanowski JQ; Lee YMY An acetylation switch controls TDP-43 function and aggregation propensity. Nat. Commun 2015, DOI: 10.1038/ncomms6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Satoh R; Tanaka A; Kita A; Morita T; Matsumura Y; Umeda N; Takada M; Hayashi S; Tani T; Shinmyozu K; Sugiura R. Role of the RNA-Binding Protein Nrd1 in Stress Granule Formation and Its Implication in the Stress Response in Fission Yeast.PLoS One. 2012, 7, DOI: 10.1371/journal.pone.0029683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Patel MS; Nemeria NS; Furey W; Jordan F. The pyruvate dehydrogenase complexes: structure-based function and regulation. J. Biol. Chem. 2014, 289, 16615–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Petrick JS; Jagadish B; Mash EA; Aposhian HV Monomethylarsonous acid (MMA(III)) and arsenite: LD(50) in hamsters and in vitro inhibition of pyruvate dehydrogenase. Chem. Res. Toxicol 2001, 14, 651–656. [DOI] [PubMed] [Google Scholar]
- [40].Piette BL; Alerasool N; Lin Z; Lacoste J; Hiu M; Lam Y; Qian W. Tran S; Larsen B; Campos E; Peng J; Gingras AC; Taipale M. Comprehensive interactome profiling of the human Hsp70 network highlights functional differentiation of J domains. Mol. Cell 2021, 81, 2549–2565. [DOI] [PubMed] [Google Scholar]
- [41].Gobe G; Crane D; Mitochondria, reactive oxygen species and cadmium toxicity in the kidney. Toxicology. Letters 2010, 198, 49–55. [DOI] [PubMed] [Google Scholar]
- [42].Tynecka Z;Malm A. Substrate-dependent cadmium toxicity affecting energy-linked K+/86Rb transport in Staphylococcus aureus. J. Basic Microbiol 1998, 36, 447–452. [DOI] [PubMed] [Google Scholar]
- [43].Meng H; Fitzgerald MC Proteome-Wide Characterization of Phosphorylation-Induced Conformational Changes in Breast Cancer. J. Proteome Res 2018, 17, 1129–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Liu F; Fitzgerald MC Large-Scale Analysis of Breast Cancer-Related Conformational Changes in Proteins using Limited Proteolysis. J. Proteome Res 2016, 15, 4666–4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Feng Y; De Franceschi G; Kahraman A; Soste M; Melnik A; Boersema PJ; Polverino de Laureto P; Nikolaev Y; Oliveira AP; Picotti P. Global analysis of protein structural changes in complex proteomes. Nat. Biotech 2014, 32, 1036–1044. [DOI] [PubMed] [Google Scholar]
- [46].To P; Whitehead B; Tarbox HE; Fried SD Nonrefoldability is Pervasive Across the E. coli Proteome. J. Am. Chem. Soc 2021, 143, 11435–11448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zauber H; Kirchner M; Selbach M. Picky: a simple online PRM and SRM method designer for targeted proteomics Nat. Meth 2018, 15, 156–157. [DOI] [PubMed] [Google Scholar]
- [48].Heil LR; Remes PM; Maccoss MJ Comparison of unit resolution versus high-resolution accurate mass for parallel reaction monitoring. BioRxiv 2021, 2021.05.04.442680. [DOI] [PubMed] [Google Scholar]
- [49].Peterson AC; Russell JD; Bailey DJ; Westphall MS; Coon JJ Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol. Cell. Proteomics 2012, 11, 1475–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Trendel J; Schwarzl T; Horos R; Prakash A; Bateman A; Hentze MW; Krigsveld J. The Human RNA-Binding Proteome and Its Dynamics during Translational Arrest. Cell. 2019, 176, 391–403. [DOI] [PubMed] [Google Scholar]
- [51].Johnson BS; McCaffery JM; Lindquist S; Gitler AD A yeast TDP-43 proteinopathy model: Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. PNAS. 2008, 105, 6439–6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Samikkannu T; Chen C;Yih L; Wang ASS; Lin S; Chen T; Jan K. Reactive oxygen species are involved in arsenic trioxide inhibition of pyruvate dehydrogenase activity. Chem. Res. Toxicol 2003, 409–414. [DOI] [PubMed] [Google Scholar]
- [53].Hageman J; Van Waarde MAWH; Zylicz A; Walerych D; Kampinga HH The diverse members of the mammalian HSP70 machine show distinct chaperone-like activities. Biochem. J 2011, 435, 127–142. [DOI] [PubMed] [Google Scholar]
- [54].Genereux JC; Qu S; Zhou M; Ryno LM; Wang S; Shoulders MD; Kaufman RJ; Lasmézas CI; Kelly JW; Wiseman RL Unfolded protein response-induced ERdj3 secretion links ER stress to extracellular proteostasis. EMBO J. 2015, 34, 4–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ciszak EM; Korotchkina LG; Dominiak PM; Sidhu S; Patel MS; Structural Basis for Flip-Flop Action of Thiamin Pyrophosphate-dependent Enzymes Revealed by Human Pyruvate Dehydrogenase. J. Biol. Chem 2003, 278, 21240–21246. [DOI] [PubMed] [Google Scholar]
- [56].Pettersen EF; Goddard TD; Huang CC; Couch GS; Greenblatt DM; Meng EC; Ferrin TE; UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004, 25, 1605–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.