Significance
Mutations in the SORL1 gene confer exceptionally high risk of Alzheimer’s disease (AD)—on par with the risk associated with mutations in APPPSEN1, and PSEN2—bringing it into the top 4 of genes considered causal of AD. SORLA, encoded by the SORL1 gene, is a sorting receptor acting together with the protein complex retromer to ensure endosomal recycling of cell-surface proteins, of great importance in maintaining neuronal integrity and function by guiding various neuroreceptors to synaptic membranes. It is important to understand the role of SORLA in this trafficking pathway and how this relates to AD. Here, we show that SORLA dimerizes and that the mode of dimerization reinforces retromer assembly and function, presenting strategies for AD therapeutics.
Keywords: Alzheimer’s disease, SORL1, dimerization, minigene, pathogenicity
Abstract
SORL1, the gene encoding the large multidomain SORLA protein, has emerged as only the fourth gene that when mutated can by itself cause Alzheimer’s disease (AD), and as a gene reliably linked to both the early- and late-onset forms of the disease. SORLA is known to interact with the endosomal trafficking regulatory complex called retromer in regulating the recycling of endosomal cargo, including the amyloid precursor protein (APP) and the glutamate receptor GluA1. Nevertheless, SORLA’s precise structural–functional relationship in endosomal recycling tubules remains unknown. Here, we address these outstanding questions by relying on crystallographic and artificial-intelligence evidence to generate a structural model for how SORLA folds and fits into retromer-positive endosomal tubules, where it is found to dimerize via both SORLA’s fibronectin-type-III (3Fn)- and VPS10p-domains. Moreover, we identify a SORLA fragment comprising the 3Fn-, transmembrane, and cytoplasmic domains that has the capacity to form a dimer, and to enhance retromer-dependent recycling of APP by decreasing its amyloidogenic processing. Collectively, these observations generate a model for how SORLA dimer (and possibly polymer) formation can function in stabilizing and enhancing retromer function at endosome tubules. These findings can inform investigation of the many AD-associated SORL1 variants for evidence of pathogenicity and can guide discovery of novel drugs for the disease.
The sortilin-related receptor 1 (SORL1) gene is strongly associated with Alzheimer’s disease: while common SORL1 single-nucleotide polymorphisms are associated with a modest but GWAS-significant risk of the late-onset form of AD (1–3), recent exome sequencing studies have provided evidence that rare loss-of-function genetic variants can be considered causal for AD (4, 5). Moreover, besides the loss-of-function variants, a huge number of potentially deleterious SORL1 missense mutations are also uncovered by exome sequencing studies, providing evidence that SORL1 is probably one of the genes harboring the most pathogenic variants that lead to AD in the human genome (4, 6, 7). However, the pathogenicity of most SORL1 variants is unclear, in part due to lack of detailed knowledge on physiologically relevant binding partners, as well as missing overall structural information about the protein encoded by SORL1.
This protein, termed SORLA (sorting-related receptor with type A repeats, but also sometimes called SORL1 like its gene) is a 250-kDa type-1 transmembrane receptor, containing many different domains that, from the N-terminal end, begins with a VPS10p-domain, followed by a YWTD-repeated β-propeller together with an EGF-domain, 11 complement-type repeat (CR)-domains, as well as 6 fibronectin-type-III (3Fn)-domains within the luminal (or ectodomain) region, which is continued by a single-pass transmembrane alpha-helix and a short C-terminal cytoplasmic domain (Fig. 1A).
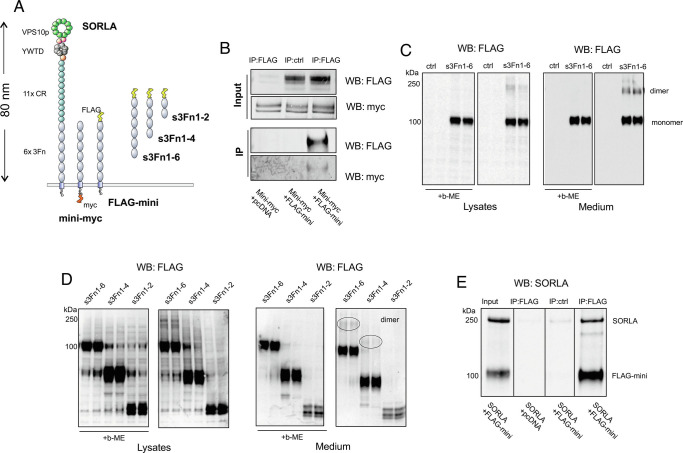
Fig. 1.
SORLA dimerization by its 3Fn domains. (A) Schematic representation showing domain architectures of SORLA, mini-myc, FLAG-mini, s3Fn1-6, s3Fn1-4, and s3Fn1-2. The overall length of the SORLA extracellular region was estimated based on its homology model. (B) Coimmunoprecipitation analysis of lysates from HEK293 cells transfected with mini-myc together with either an empty vector (pcDNA3.1) or FLAG-mini using FLAG as bait. Precipitated proteins were detected by WB using myc and FLAG antibodies. Representative blots from two independent experiments. (C) WB analysis of lysate and conditioned medium from CHO cells transfected with s3Fn1-6 treated with either 5% or 0% β-mercaptoethanol (β-ME) detected with FLAG antibody. Representative blots from three independent experiments. (D) WB analysis of lysate and conditioned medium from CHO cells transfected with s3Fn1-6, s3Fn1-4, and s3Fn1-2 treated with either 5% or 0% β-ME detected with FLAG antibody. Representative blots from three independent experiments. (E) Coimmunoprecipitation analysis of lysates from HEK293 cells transfected with FLAG-mini together with either an empty vector (pcDNA3.1) or SORLA using FLAG as bait. Precipitated proteins were detected by WB using SORLA antibody. Representative blots from four independent experiments.
The cytoplasmic domain, often referred to as the tail of the protein, contains binding sites for the retromer multiprotein complex (8, 9) that forms an oligomer that coats endosome tubules (10, 11). This interaction is key for SORLA to exert its known role within endosomal recycling pathways, by facilitating the recycling of certain plasma membrane proteins such as the amyloid precursor protein (APP) and peptides such as the amyloid beta (Aβ) peptide (12–15). Impaired binding between SORLA and retromer also lowers the level of SORLA maturation (8, 14, 16), which can report on the level of functional receptor activity, although the process is not yet completely understood (17). These findings all add to the emerging picture of how the endosomal recycling pathway is of high importance for a number of (patho)physiological processes (18) and is currently seen as a therapeutic target for AD therapies (19–21).
VPS10p-domains are also found in extracellular regions of members of the VPS10p-domain receptor family, which all participate in intracellular trafficking of multiple and diverse cargo molecules (22, 23). The YWTD- and CR-domains, known from members of the low-density lipoprotein receptor family, are also engaged in direct ligand binding activities, whereas the function of the six 3Fn-domains is largely unknown as they have not yet been assigned any role in ligand binding for SORLA (24), despite a vast number of pathogenic mutations that map to this region of the protein, which hints toward an important function (6, 25). It is interesting to note that 3Fn-domains are found in a number of proteins that form dimers, and that protein dimerization often occurs by direct homodimerization between 3Fn-domains.
The other members of the VPS10p-domain receptor family have all been shown to form dimers. They can dimerize by interactions formed by their VPS10p-domains (e.g., Sortilin) (26–28), as well as by interactions between their polycystic kidney disease domains (e.g., SorCS1 and SorCS2), which have 3Fn domain-like topology (29, 30). The capacity of SORLA to form dimers has only very recently surfaced, but it suggests that several parts of the protein can be involved in this process and that SORLA may form dimers with different conformations depending on its cellular localization (31).
Here, we first show that sites for SORLA dimerization reside in both its 3Fn-domains as well as in its VPS10p-domain and identify that SORLA dimers are predominantly located together with retromer in endosomes. Next, we identify a SORLA fragment that comprises the 3Fn-, transmembrane, and cytoplasmic domains (termed SORLA minireceptor) that has the capacity to form a dimer. These observations generate a model for how SORLA dimerization, which may potentially form polymers, may have an effect on stabilizing both retromer and the endosome tubule. In cells, the SORLA minireceptor can shift APP away from endosomal amyloidogenic processing by increasing its recycling out of the endosome, showing how SORLA dimerization is associated with this receptor’s activity in the retromer-dependent endosomal recycling pathway.
Results
SORLA Forms a Dimer via Its 3Fn-Domains.
To start delineating the dimer capacity of the SORLA 3Fn-domains, we generated two SORLA minireceptor constructs comprising the six 3Fn-domains, the transmembrane region, and the cytoplasmic tail with either an N-terminal FLAG (corresponding to amino acids DYKDDDDK)- or C-terminal myc-tag (named FLAG-mini and mini-myc, respectively; Fig. 1A). Additionally, we also produced soluble forms of the minireceptor without the transmembrane region and cytoplasmic tail, containing two, four, or six 3Fn-domains (s3Fn1-2, s3Fn1-4, and s3Fn1-6, respectively), to investigate the 3Fn-domains in their isolated form.
First, we tested the ability of the two membrane-bound minireceptors to form a complex using coimmunoprecipitation analysis of cells transfected with FLAG-mini and mini-myc. We could clearly precipitate mini-myc using an anti-FLAG antibody when both minireceptors were coexpressed, while cells that were only transfected with mini-myc but not expressing FLAG-mini showed no precipitation of mini-myc (Fig. 1B). This demonstrates that two minireceptors form a complex but does not exclude whether this could be mediated by the cytoplasmic tails that could be part of larger cytosolic complexes.
In a second set of experiments, when testing the expression of the s3Fn1-6 construct, we observed a clear signal corresponding to a dimer by western blot (WB) analysis after normal SDS–PAGE separation of proteins in both lysates and medium from transfected Chinese hamster ovary (CHO) cells (Fig. 1C). This dimer formation was sensitive to treatment of the samples with β-mercaptoethanol (β-ME), reducing intradomain disulfide-bridges present in the first and sixth 3Fn-domains, indicating that the dimer formation depends on the structural integrity of correctly folded 3Fn-domains, and is independent of the SORLA transmembrane and tail domains.
Next, we sought to determine if any of the six 3Fn-domains are indispensable for this dimer formation, by analyzing the dimerization of s3Fn1-4 and s3Fn1-2 in comparison to the s3Fn1-6 fragment (Fig. 1D). The fragment lacking the two C-terminal 3Fn-domains (s3Fn1-4) produced a signal representing dimer formation to a similar extent as the construct containing all six 3Fn-domains. The shortest fragment we tested, containing only the two most N-terminal 3Fn-domains (s3Fn1-2), did not show any dimer formation under these experimental conditions. This result suggests that dimerization is independent of the fifth and sixth 3Fn-domains, but we did not pursue further dissection of the binding site in the current study. We note that there is a general lower detection and/or secretion of the shortest s3Fn1-2 fragment, but even upon very long exposure times of the WB membrane we were unable to detect signs of dimer formation for this short fragment.
Finally, we also tested whether the FLAG-minireceptor could interact with the full-length SORLA protein to form heterodimers. We cotransfected HEK293 cells with FLAG-mini and untagged full-length SORLA, prepared cell lysates, and precipitated proteins using anti-FLAG antibody. By WB analysis using a polyclonal serum against the SORLA extracellular fragment, we could clearly show that FLAG-mini also precipitates full-length (~250 kDa) SORLA in cells (Fig. 1E). Lysates from cells cotransfected with FLAG-mini and an empty vector (pcDNA3.1) or double transfected cells treated in parallel but without the anti-FLAG antibody in the IP reaction served as negative control experiments.
Combined, these data provide evidence of SORLA dimer formation via its 3Fn-domains, in vitro and in cells.
Cellular Analysis of SORLA Dimerization in Endosomes.
We next turned our attention to the cellular localization of SORLA dimer formation. We used constructs for full-length SORLA proteins with C-terminal FLAG- or GFP-tags (Fig. 2A) and applied the proximity ligation assay (PLA) to study where in cells SORLA forms dimers. This method relies on immunostaining using secondary antibodies tagged with short oligonucleotides able to hybridize when the two antibody-bound target molecules come into close proximity (i.e., form dimers) allowing for rolling-circle amplification. For visualization by confocal microscopy, fluorescently labeled oligos are bound to complementary sequences on the amplicon, represented by red dots.
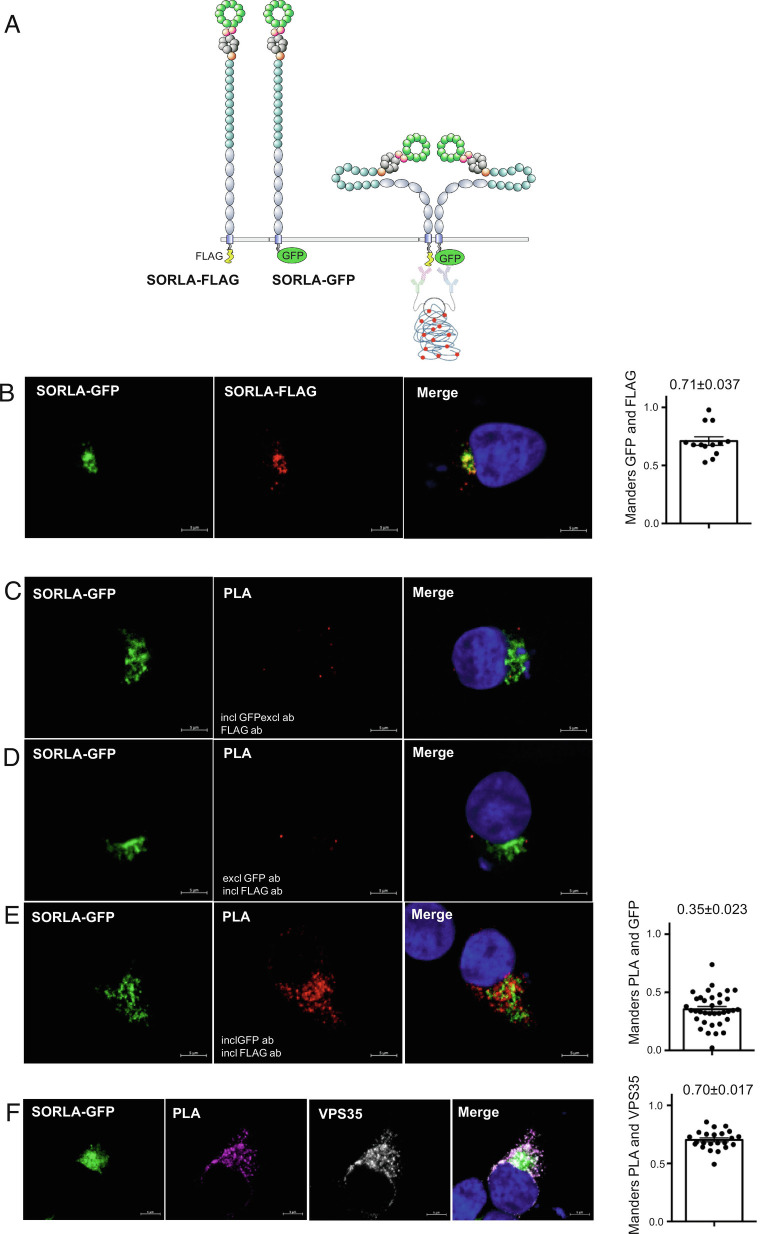
Fig. 2.
SORLA dimers are located within endosomes. (A) Schematic representation showing SORLA proteins with C-terminal FLAG-tag (SORLA-FLAG) or GFP-tag (SORLA-GFP). A red PLA signal will be formed when the two antibody-bound SORLA constructs get into close contact representing dimerization. (B) Representative immunofluorescence images of HEK293 cells transfected with SORLA-FLAG and SORLA-GFP and immunostained for FLAG and GFP. Bar graph in the Right panel illustrates quantifications of colocalization between FLAG and GFP signals using Mander’s (n = 13 from two independent experiments). (C–E) Representative immunofluorescence images after PLA (red) of HEK293 cells transfected with SORLA-FLAG and SORLA-GFP (green) treated with GFP (C), FLAG (D) or both antibodies (E). Bar graph in the Right panel illustrates quantifications of colocalization between GFP (green) and PLA (red) signals using Mander’s (n = 36 from two independent experiments). (F) Representative immunofluorescence images after PLA (purple) of HEK293 cells transfected with SORLA-FLAG and SORLA-GFP (green) counterstained with VPS35 (white). Bar graph in the Right panel illustrates quantifications of colocalization between PLA (purple) and VPS35 (white) signals using Mander’s (n = 23). (Scale bars, 5 µm.) Error bars indicate SEM.
First, we tested whether either of the two C-terminal tags affected the localization of the tagged receptor forms using double-transfected HEK293 cells and determination of their degree of colocalization. From this experiment, we observed no indication that the tags lead to differences in cellular localization as they showed a strong overlap and Mander’s coefficient of 0.71 (Fig. 2B). Next, we performed control experiments for the PLA setup excluding either the anti-FLAG (Fig. 2C) or anti-GFP (Fig. 2D) primary antibodies that in both settings abolished formation of the PLA signal. However, when including both antibodies, we observed a strong PLA signal as detected by confocal microscopy (Fig. 2E), confirming that two full-length SORLA proteins can form a dimer within cells. However, when we determined the degree of colocalization between the PLA signal (representing SORLA dimers) and SORLA-GFP (representing both monomers and dimers), we noticed a clear lack of colocalization for several PLA signals, and Mander’s coefficient confirmed an overlap of only 0.35 (Fig. 2E), suggesting that only a fraction of the total cellular pool of SORLA protein exists as dimers. This may mean that monomers or dimers are formed depending on where in the cell SORLA is localized.
We have previously demonstrated how SORLA associates with retromer, the multiprotein master regulator of endosomal protein trafficking, in endosomes (8, 32), and it has been shown how the related receptor Sortilin forms a dimer specifically in acidic compartments (26–28). We therefore hypothesized that SORLA dimers are also predominantly formed in the endosome. Costaining for the PLA signal representing SORLA dimers and the VPS35 subunit of the retromer core representing endosomal compartments showed a high degree of colocalization with Mander’s coefficient of 0.70, confirming that the majority of SORLA dimers are located together with retromer (Fig. 2F).
Endosomal Retromer Binding to a SORLA Dimer.
We next wanted to confirm that the cellular SORLA pool is represented by a mixture of both monomers and dimers using the bimolecular fluorescence complementation (BiFC) assay as an independent approach. This technique is based upon structural complementation between non-fluorescent N-terminal and C-terminal fragments of a split fluorescent protein (such as Venus, a derivative of GFP) fused to bait and prey proteins, respectively. Upon interaction between the bait and prey proteins, the split Venus domains come into close proximity, enabling them to refold into a functional β-barrel structure containing the fluorophore (Fig. 3A). Thereby, dimerization of the two proteins (in our case SORLA) drives the formation of a fluorescent protein that can be visualized by fluorescent microscopy.
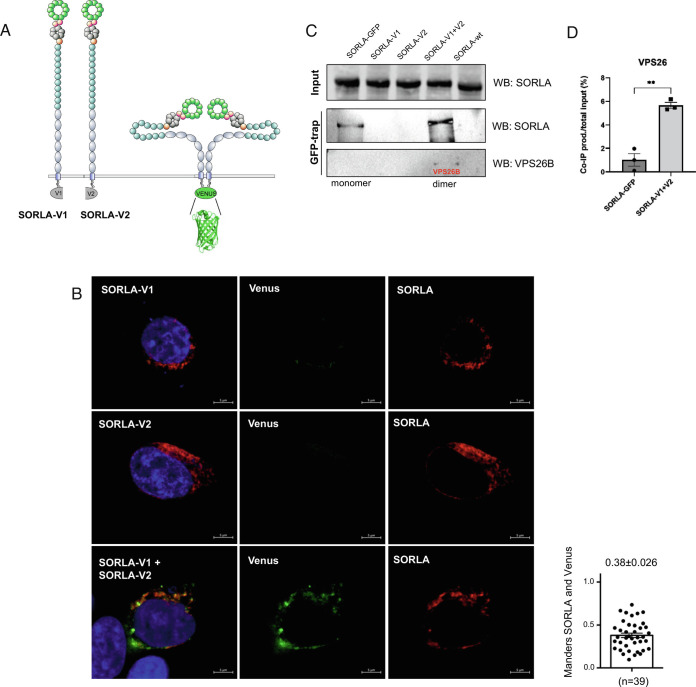
Fig. 3.
Retromer binds to the SORLA dimer. (A) Schematic depiction showing SORLA proteins with C-terminal fusions of N-terminal (SORLA-V1) and C-terminal (SORLA-V2) fragments of the Venus fluorescent protein. Upon interaction between two SORLA proteins, the Venus fragments associate and refold to produce a fluorescent molecule. (B) Representative immunofluorescence images of HEK293 cells transfected with SORLA-V1 (Top), SORLA-V2 (Middle) or both (Bottom) followed by immunostaining with SORLA antibody. The bar graph shows colocalization between reconstituted Venus (green) and SORLA (red) signals using Mander’s (n = 39 from three independent experiments). Scale bars, 5 µm (C) GFP-trap immunoprecipitation analysis of lysates from HEK293 cells transfected with SORLA-GFP, SORLA-V1, SORLA-V2, both SORLA-V1 and SORLA-V2, or SORLA. Precipitated proteins were detected by WB using SORLA and VPS26b antibodies. Error bars indicate SEM. Representative blots from three independent experiments. (D) Densiometric quantification of co-IP product (VPS26) relative to total input (100%) from three independent experiments.
Accordingly, two SORLA constructs were engineered to contain C-terminal fusions of either the N-terminal (Met1-Gln157) or C-terminal (Lys158-Lys238) fragment of the Venus fluorescent protein (33). These two SORLA constructs were termed SORLA-V1 and SORLA-V2, containing either the N-terminal or C-terminal part of the Venus fluorescent protein, respectively.
Transfection of HEK293 cells with each of the two SORLA BiFC constructs individually did not produce any green fluorescent signals (Fig. 3B). In contrast, cotransfection with both constructs revealed specific green cytoplasmic signals, indicating that SORLA dimerization had occurred (Fig. 3 B, Bottom). Simultaneous staining with a SORLA antibody showed a predominant perinuclear localization of the SORLA molecules (red) in agreement with previous studies demonstrating that SORLA is mainly localized to the perinuclear region of transfected HEK293 cells (34, 35). Quantification of the overlap between the green Venus signal specific for SORLA dimers and the total SORLA pool identified by the anti-SORLA staining confirmed that only a fraction of SORLA exists as dimers (Mander’s coefficient 0.38). Notably, this value is similar to the degree of overlap between monomer and dimer as estimated by the PLA experiment (0.35, Fig. 2E). Moreover, this finding is in agreement with a model suggesting that SORLA is a dimer mainly within the retromer-coated endosomal structures, whereas SORLA monomers reside in different perinuclear compartments such as the TGN.
We next wanted to test if retromer binds preferentially to the SORLA monomer or dimer. For that purpose, we applied the GFP-trap technique that relies on beads conjugated with a nanobody that interacts with the Venus protein (as well as GFP) only in their folded state. However, the nanobody used for the GFP-trap does not bind either the V1- or the V2-fragments with sufficient affinity to allow precipitation of these isolated parts of the Venus protein (36).
We transfected HEK293 cells with SORLA-GFP, SORLA-V1, SORLA-V2, combined SORLA-V1 and SORLA-V2, and full-length SORLA without tags, and confirmed that SORLA forms a dimer that can be precipitated with the GFP-trap (Fig. 3C). As anticipated SORLA-GFP is also precipitated using the GFP-trap, representing both monomer and dimer SORLA, whereas the precipitate from the reconstituted Venus protein only contains the SORLA dimer fraction. By re-probing the membrane with an antibody against the VPS26b subunit of retromer, we could demonstrate that the endosomal SORLA dimer fraction indeed exists almost entirely in complex with retromer (Fig. 3C). Quantification of the co-IP product (VPS26) relative to total input (100%) was carried out by densiometric scanning of membranes, showing that the level of bound VPS26 is significantly increased for the SORLA dimer (6%) compared with the monomer (1%) (Fig. 3D). However, as only a fraction corresponding to ~6% of the VPS26 (i.e., retromer) pool is precipitated under our experimental conditions, it suggests that the SORLA-retromer units are part of higher order complexes not easily precipitated. This tool will allow for future analysis of the interactome of SORLA monomer and dimers, but has thus far confirmed that SORLA forms a dimer in endosomes, where it also forms a complex with retromer.
The SORLA 3Fn-Domain Minireceptor Decreases Endosomal Processing of APP.
We and others have previously shown how full-length SORLA protein decreases APP trafficking in the secretory pathway as well as assisting in APP recycling out of the endosome, resulting in a decrease in both sAPPα and sAPPβ/Aβ-peptide products (13, 37, 38). As the minireceptor does not harbor the CR-domains required for the direct binding of the APP protein (39), nor the VPS10p-domain suggested to bind the Aβ peptide (12, 40), it is not known whether this remaining part of SORLA that constitutes the minireceptor has an impact on APP biology. However, it has previously been shown that pharmacological chaperones that stabilize the retromer complex, and increase APP recycling from the endosome to the cell surface, change APP processing away from amyloidogenic cleavage by β-secretase (which occurs in the endosome) toward the nonamyloidogenic processing by α-secretase (which occurs at the plasma membrane) (41). We therefore hypothesized that the SORLA minireceptor, forming a dimer and binding to retromer [which is also a dimer on the tubular endosome membrane (10)] at the same time, could exert similar biological effects as small molecule chaperones that stabilize the retromer complex.
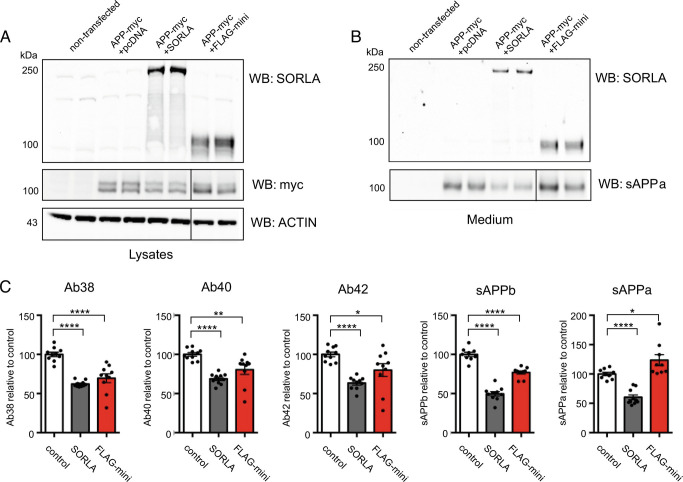
To investigate this, we transfected murine-derived N2a cells with an APP-myc construct for expression of the human APP protein either in the absence of any exogenous SORLA (+pcDNA3.1), together with the full-length SORLA protein (+SORLA), or together with the SORLA minireceptor (+FLAG-mini), and analyzed APP proteolysis using a WB approach (Fig. 4 A and B). Additionally, we quantified the levels of different APP metabolites by mesoscale discovery (MSD) assays specific for human-sequence APP-derived fragments (Fig. 4C). In line with previous findings, the presence of SORLA decreased both nonamyloidogenic and amyloidogenic processing of APP, as evidenced by significantly reduced levels of sAPPα, sAPPβ, and Aβ-peptides of 38, 40, and 42 amino acids in length (Fig. 4 B and C). To our surprise, we also observed a significant decrease of APP amyloidogenic processing in the presence of FLAG-mini, with significantly decreased production of sAPPβ and all three Aβ peptides (Aβ38/Aβ40/Aβ42). However, whereas the full-length SORLA protein also decreased sAPPα production, FLAG-mini significantly increased this nonamyloidogenic fragment (Fig. 4 B and C), representing a cellular and endosomal phenocopy of chemical chaperones that stabilize retromer (41) (c.f. Fig. 4 of that paper).
Fig. 4.
SORLA 3Fn minireceptor reduces the pathogenic APP processing pathway. (A, B) Representative immunoblots of lysates and conditioned medium from nontransfected (ctrl) N2a cells or cotransfected with APP-myc together with either an empty vector (pcDNA3.1), SORLA, or FLAG-mini using antibodies against SORLA, myc and actin for lysates (A) or WO2 specifically binding to human sAPPα in medium samples (B). β-actin levels are used as loading controls (n = 10 from five independent experiments) (C) Quantifications of APP processing products using MSD immunoassays to monitor levels of human Aβ38, Aβ40, Aβ42, sAPPβ, and sAPPα in conditioned medium from transfected N2a cells (n = 10 from five independent experiments). Solid vertical lines in the blots for myc, ACTIN and sAPPα indicate deletion of lanes from the membranes. Error bars indicate SEM.
Possible SORLA Multimerization by VPS10p-Domain and 3Fn-Domains.
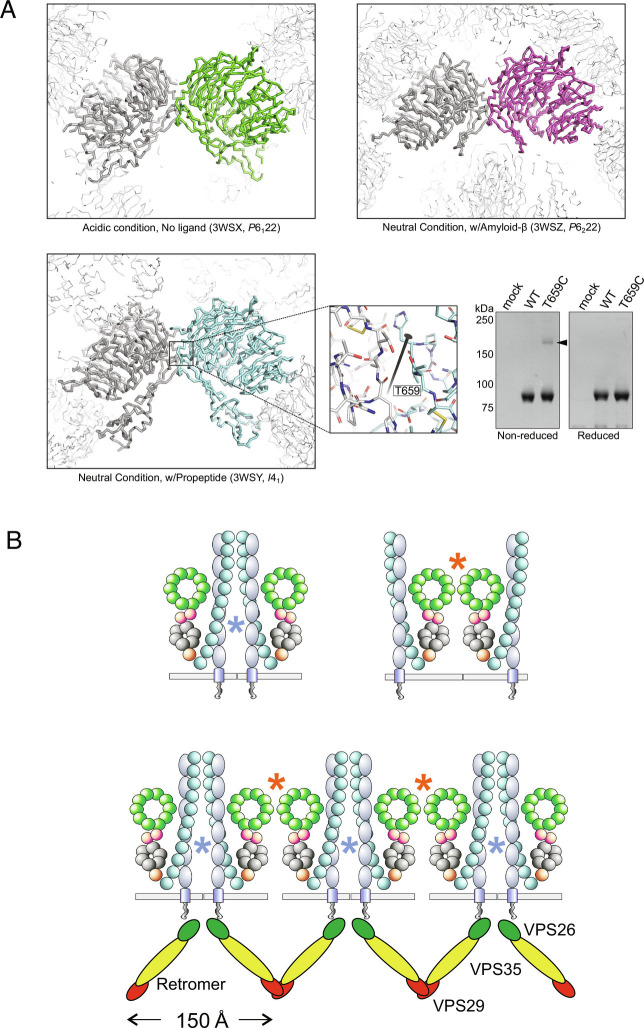
As described above, SORLA potentially forms a homodimer via its VPS10p-domain (31). Therefore, we also tested whether the VPS10p-domain can form dimers on its own by pull-down assay using an engineered tagged SORLA VPS10p construct. All three crystal structures of the human SORLA VPS10p-domain reported so far show the presence of a dimer formed in the exact same manner: related to a crystallographic neighbor by twofold crystallographic symmetry despite their different space groups (Fig. 5A). Examining these structures, we found that Thr659 is located at a suitable position to form a disulfide bond across the twofold symmetric axis if it is mutated to Cysteine (Fig. 5A). Based on that concept, we created the T659C mutant for the SORLA VPS10p-domain, and PA-tagged SORLA VPS10p-domain (1 to 753) for both wild-type and the T659C mutant was pulled-down from culture medium by NZ-1 (42) immobilized Sepharose-resin. Following electrophoresis, gel analysis showed a fraction of T659C mutant forms a covalently bonded dimer, whereas wild-type showed no dimer formation under these conditions (Fig. 5A), indicating that the SORLA VPS10p-domain can form a dimer in a specific manner.
Fig. 5.
SORLA dimerization by the VPS10p-domain. (A) Three crystal structures of the SORLA VPS10p-domain reported so far (PDBID: 3WSX, 3WSY, 3WSZ: a monomer in the asymmetric unit is colored as green, cyan, and magenta, respectively) with crystallographic neighbors. Their space groups are also described in their captions. One specific neighbor forming a dimer by twofold rotation is described as thick gray ribbon, and other neighbors are described as thin white ribbons. The close-up view of 3WSY shows the surroundings of Thr659over the crystallographic twofold rotation axis depicted as a black bar with the twofold symbol. The gel panel shows the result of a pull-down assay followed by SDS-PAGE for the wild-type and the T659C mutant of the human SORLA VPS10p-domain. indicating that only the T659C mutant can form a covalently linked dimer (black arrowhead). (B) Schematics of two different local dimers by either 3Fn- or VPS10p-domain sites, as well as a suggestive model showing how SORLA if forming polymers could engage with the polymeric retromer coat on endosome tubules. The two putative dimer interfaces, one involving the Vps10 domains and one involving the 3Fn domains, are depicted as red and blue asterisks, respectively. The lower figure illustrates how one possible arrangement of these two interfaces, consistent with their relative orientations as predicted by the AlphaFold model shown in Fig. 6, could lead to polymerization of SORLA in a way that would both fit with and possibly stabilize the polymer of retromer known to exist on the tubular membrane.
Based on the presence of two independent sites for dimer formation; the VPS10p-domain and the 3Fn-domains, we envision at least two different conformational models of SORLA: one where two receptors form a local dimer that can be held together by either the 3Fn-domain site, the VPS10p-domain site, or by both, as depicted schematically on the right sides of Figs. 2A and 3A, and one where each of the two different dimerization sites originate from different receptor molecules that may lead to a large polymer of SORLA that coats the endosome tubule (10) (Fig. 5B).
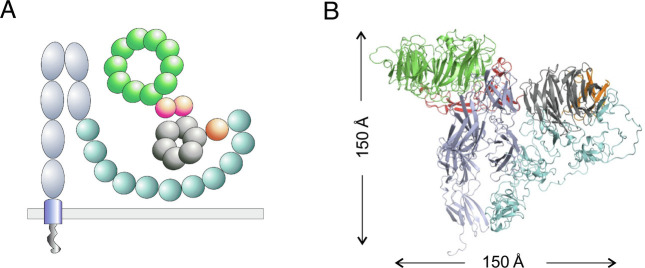
To gain insight into which of these models may be more plausible, we constructed a three-dimensional model of the full SORLA ectodomain using the tools of the deep learning program AlphaFold (43, 44). Although AlphaFold has been used to generate predicted structures for the entire human proteome, including SORLA (43, 44), we found its original modeling of SORLA (and other type 1 transmembrane proteins) to be fundamentally flawed. All type 1 transmembrane proteins have parts of their structures in three distinct topological compartments (one on each side of the membrane plus a transmembrane helix); however, in the human proteome modeling, the AlphaFold algorithm was applied to the entire protein sequence for every protein, including type 1 transmembrane proteins, producing in the case of SORLA a model in which the transmembrane helix was implausibly embedded in the middle of the ectodomain structure, with the cytoplasmic tail wrapped around it.
Since the creators of AlphaFold generously made their algorithm available to the scientific community, we were able to implement it locally and apply it only to the ectodomain portion of SORLA, producing the model shown schematically (Fig. 6A) and as a complete backbone fold (Fig. 6B); the color coding in the two is the same. Examination of this model shows that the likely sites of dimerization in the VPS10p- and 3Fn-domain regions are oriented with respect to each other in a way that is more consistent with the polymer model in Fig. 5B than with a local, closed dimer depicted in Figs. 2A and 3A. If correct, this model, in which SORLA polymerization both fits with and enhances the polymerization of retromer on the tubular endosome membrane, may explain why SORLA is unique for endosome sorting compared with other VPS10p-domain containing receptors and other retromer-binding receptors. We note that the model also highly depends on the assembly with many other factors (18), but it is consistent with our earlier observation that SORLA dimerization is independent of the fifth and sixth 3Fn-domains—these domains project away from the largely globular rest of the ectodomain model, as they presumably need to do in order to attach to the transmembrane helix (Fig. 6).
Fig. 6.
Putative structural organization of the SORLA luminal region. (A) Schematics representing domain architectures and predicted folding of the full SORLA luminal domain. (B) AlphaFold model showing secondary and tertiary structures of the compact SORLA ectodomain depicted by the molecular graphics program PyMOL. Structures in A and B are presented with the same color code: VPS10p (green), 10CC (red), YWTD (gray), EGF (orange), CR (turquoise), and 3Fn (light blue). Binding of APP to SORLA occurs by the CR-domains, which are not involved in the dimer formation and likely accessible for ligand interactions.
Discussion
SORLA is an important protein for understanding the major neurodegenerative disorder Alzheimer’s disease, as genetic linkage studies continuously find SORL1 to be highly associated with AD. Therefore, elucidating its structure–function relationship is vital to identify which of the hundreds of mutations that have been found in this protein are truly pathogenic, and why. In this work, we have focused on the questions of whether SORLA dimerizes in the cell, whether that dimer is the form that binds to the core retromer heterotrimer, and what the consequences of both might be.
We have shown by a variety of approaches that SORLA indeed forms dimers, both in vitro and in vivo, and have dissected which of the many structural domains of this large protein are involved in dimer formation. We found that the 3Fn-domains, but not all of them, can form dimers on their own and mediate dimerization of the whole protein. We find the same is true of the VPS10p-domain. We have shown that in the cell dimerization primarily occurs on the endosomal membrane, where SORLA colocalizes with retromer through the association of its C-terminal cytosolic tail with VPS26 (illustrated in Fig. 5B). Using the AlphaFold algorithm, we have generated a full structural model of the entire SORLA ectodomain, comprising most of the protein and the entire part that is located inside the endosome tubule (Fig. 6).
By addressing these motivating questions, our findings clarify a number of issues regarding SORLA’s structure and, more importantly, its function. Structurally, our findings clarify how the large holoprotein folds to fit inside the approximately 50 to 100-nm diameter of the endosome tubule (45). Moreover, the proposed folding model suggests how and where the folded SORLA monomer can dimerize, and potentially may polymerize, inside the tubule.
The functional consequences of how SORLA is suggested to structurally fold and to dimerize are twofold. Most important are the consequences for the retromer and the endosomal recycling pathway it serves. Retromer is a molecular machine that has been found to require the interaction of multiple modules, either proteins or protein complexes, in regulating the formation and stabilization of endosomal tubules and the sorting and trafficking of cargo out of the endosome via these tubules. Retromer’s central module is the heterotrimeric core comprising VPS29, VPS35, and VPS26 (a or b) that forms an arch-shaped dimer on the cytosolic side of the endosomal tubular membrane. This dimer is proposed to further self-associate via interactions between the VPS26 proteins at the bottom of the arch to form a polymeric network that wraps around the outside of the tubule, thereby acting to both construct and stabilize the tubule (10, 11, 46) (i.e., Fig. 5B of this paper). All the other of retromer’s key modules interact with this heterotrimer core to abet retromer’s stabilization and function, and to date all these modules bind the heterotrimer on the cytosolic side of the endosomal tubule. Our findings implicate the SORLA dimer as an intratubular module that also aids in retromer stabilization and function, by binding the heterotrimer core through the tubular membrane. The interpretation that SORLA functions to stabilize retromer is supported by findings showing that a primary depletion of SORLA leads to a secondary reduction in VPS26b and vice versa (13).
Our findings’ second functional consequence is, at this point, more inferential. Many of retromer’s cargo proteins that are recycled through the endosome, particularly those linked to AD, turn out to be neuronal transmembrane proteins recycled as dimers, in particular glutamate receptors and APP (15). Disrupting their retromer-dependent recycling, either by depleting SORLA or VPS26, leads to accelerated Aβ production, a triggering event in AD’s ‘amyloid pathology’ or synaptic dysfunction and the first step in AD’s neurodegenerative processes. SORLA dimerization appears to form an ideal intratubular platform onto which dimeric glutamate receptors and APP can bind and then successfully recycle. Future studies will be able to rely on our findings to identify the specific manner in which either glutamate receptor or APP dimers bind the SORLA dimeric platform inside endosomal tubules.
Finally, our SORLA structure–function findings offer genetic and even therapeutic insight. SORL1 pathogenic variants are spread across the entire protein coding region, including a high number leading to missense mutations in the 3Fn-domains as well as a number in the VPS10p-domain (6). Our work suggests that characterization and assessment of pathogenicity of 3Fn-domain and VPS10p-domain variants should include functional testing of their impact on dimerization. Therapeutically, it is noteworthy that the SORLA minireceptor effectively functions similarly to retromer pharmacological chaperones, which by stabilizing retromer enhance retromer’s overall activity. While we have developed and used the SORLA minireceptor as an experimental tool to clarify SORLA’s structural–functional relationship, this minireceptor can inform AD drug discovery, e.g., as the minireceptor has a size compatible with viral packaging and delivery for possible use in gene therapy. The therapeutic promise of such a discovery program would certainly be relevant to the subset of AD patients who carry loss-of-function pathogenic SORL1 variants; but since SORLA and VPS26b are also found reliably deficient in many ‘sporadic’ AD brains, these drug discovery efforts promise to extend to all forms of AD more broadly.
Materials and Methods
Plasmids and Constructs.
The SORLA coding sequence was inserted into pDEST-ORF-V1 (#73637, Addgene) and pDEST-ORF-V2 (#73638, Addgene) vectors already containing the two Venus fragments (V1 and V2) obtaining the SORLA-V1 and SORLA-V2 constructs as previously described (33).
The SORLA expression constructs including untagged SORLA (from human cDNA), SORLA-GFP, and SORLA-FLAG were cloned into the pcDNA3.1 vector as previously described in detail (47). The APP-myc construct was generated from human cDNA and introduced in the pcDNA3.1 vector followed by insertion of a C-terminal myc-tag as explained previously (48). For the recombinant expression of SORLA VPS10p-domain, the coding sequence corresponding to 1 to 753 residues of human SORLA was fused with PA-tag coding sequence on its C terminus and inserted into pcDNA3.1 in a same way previously described (40).
Cloning.
The boundaries between the six SORLA 3Fn domains were determined in-silico using online software including PsiPred (49), Scratch SSpro (50), and Yaspin (51) to predict secondary structures within the amino acid sequence. Subsequently, an alignment of the six SORLA 3Fn domains was generated based on their highly conserved pattern of secondary structures and few conserved amino acids using Clustal Omega software.
The FLAG-tagged 3Fn minireceptor (FLAG-mini) was generated by a single-step PCR reaction (Herculase II Fusion, Agilent) using the primer pair (fwd: 5′-CAGGATCCGACTACAAGGACCACGACGGCGACTACAAGGACCACGACATCGACTACAAGGACGACGACGACAAGGAGTTGACTGTGTACAAAGTACAG-3′ and rev: 5′-GTGAATTCTCAGGCTATCACCATGG-3′) amplifying a human SORL1 fragment downstream of Glu1552 (the predicted beginning of 3Fn cassette) to the final protein stop codon. Additionally, the forward primer adds a sequence encoding a 3× FLAG tag to the 5′ end of the amplicon. The PCR product was cloned into a Psectag2B expression vector (#V90020, Invitrogen) using BamHI and EcoRI restriction enzymes.
The myc-tagged 3Fn minireceptor (mini-myc) was generated similarly using the primer pair (fwd: 5′-CAGGATCCGAGTTGACTGTGTACAAAGvTAC-3′ and rev: 5′-GTGAATvTCTCACAGATCCTCTTCTGAGATGAGTTTTTGTTCGGCTATCACCATGGG-3′) adding a myc-tag to the C-terminal end of the minireceptor.
Site-Directed Mutagenesis.
Soluble forms of the 3Fn minireceptor containing two, four, or six 3Fn-domains (s3Fn1-2, s3Fn1-4, and s3Fn1-6) was generated by site-directed mutagenesis introducing stop codons in the predicted 3Fn-domain boundaries. Primers were designed following instructions from the manufacturer: s3Fn1-2: fwd; 5′-CATTACCACCATAAAAGGATGAGTGATCCCACCACCAGATATC-3′, rev; 5′-GATATCTGGTGGTGGGATCACTCATCCTTTTATGGTGGTAATG-3′, s3Fn1-4: fwd; 5′-GTTGTAGTGAAGATGATCTGAGACAGCAGGCTTCCACCC-3′, rev; 5′-GGGTGGAAGCCTGCTGTCTCAGATCATCTTCACTACAAC-3′ and s3Fn1-6: fwd: 5’-CCTGCTGTACGATGAGTGAGGGTCTGGTGCAGATGC-3′, rev: 5′-GCATCTGCACCAGAC CCTCACTCATCGTACAGCAGG-3’). Site-directed mutagenesis was performed with the FLAG-mini construct as template and using the Quickchange XL site-directed mutagenesis kit (#20052, Agilent) following the protocol from the manufacturer. For the production of the SORLA VPS10p-domain T659C mutant, the corresponding sequence was amplified with the wild-type plasmid as template and the primer pair (fwd: 5′-TGCTTCAATGGAGAGGACTTTGACAGGCCG-3′, rev: 5′- ACAGGCATGGGGGGTCCGCC -3′), and circularized by the ligation reaction with T4 PNK (New England Biolabs Inc. M0201).
Cell Culture.
HEK293 and N2a cells were cultured in Dulbecco’s Modified Eagles Medium (Sigma) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (P/S) in a humidified 5% CO2 incubator at 37 °C. CHO cells were grown in F12 Kaighn’s modification medium (Cytvia) supplemented with 10% FBS and 1% P/S in a humidified 5% CO2 incubator at 37 °C. Transient transfections of cells were performed using Fugene 6 Transfection Reagent kit (Promega) with a Fugene:DNA ratio of 3:1 for HEK293 and CHO cells and 4:1 for N2a cells.
APP Processing.
Approximately 5 × 105 N2a cells were seeded per well in a 6-well plate and grown for 24 h. Cells were then transiently cotransfected with plasmids encoding APP-myc together with either SORLA, FLAG-mini or an empty pcDNA3.1 vector. The medium was changed to serum-free medium 48 h post transfection and incubated for another 48 h before harvesting cell lysates and conditioned medium.
Immunoblotting.
Cell supernatant was collected, and cell pellets were lysed on ice for 1 h in lysis buffer (1 M Tris HCl pH 8.1, 0.5 M EDTA, 1% Triton-X-100, 1% NP40) supplemented with Complete™ protease inhibitor (Sigma). Total protein concentration in lysates was measured using the BCA assay kit (Sigma).
Lysate and media samples were mixed with SDS and 5% β-ME (unless otherwise noted) followed by heating at 95 °C for 5min. Proteins were separated by SDS–PAGE gel electrophoresis using precast 4 to 12% Bis-Tris gradient gels (Thermo) and transferred onto nitrocellulose membranes by either wet blotting (Thermo) or dry blotting (Thermo). The membranes were blocked for 1 h at room-temperature in blocking buffer (Tris-Base 0.25M, NaCl 2.5M, skimmed milk 2%, tween-20 2%) followed by incubation in primary antibodies overnight. at 4 °C. Membranes were incubated in HRP-conjugated secondary antibodies (DAKO) diluted 1:1,500 for 1 h at room temperature, and finally the proteins were detected using either ECL prime detection reagent (Sigma) or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo) with the iBright imaging system (Thermo).
Antibodies: anti-FLAG (#F1804, Sigma), WO2 (#MABN10, Merck), anti-myc (#R950-25, Thermo), anti-LR11 (#612633, BD Transduction Laboratories), anti-sol-SORLA (5387, C. M. Pedersen Aarhus University), anti-VPS26b (#92575, Novus Biologicals), and anti-β-actin (#A5441, Sigma).
Immunoassays.
Levels of sAPPα and sAPPβ were measured using the sAPPα/sAPPβ kit (#K15120E, MSD) whereas levels of Aβ38, Aβ40 and Aβ42 were determined using the V-plex Aβ panel 1 (4G8) kit (#K15200E, MSD). A fourfold serial dilution of the calibrators was prepared following instructions from the manufacturer. The precoated 4-spot plates were incubated at room temperature in either 3% blocker A solution or Diluent 35 for 1 h followed by washing in either Tris wash buffer or PBS + 0.05% Tween-20. Calibrators and undiluted cell supernatants (25 μL) were incubated in the plate for 1 h at room temperature. Subsequently the plates were washed, followed by incubation in Read Buffer T for 10 min at room temperature followed by detection of electrochemilumiscent signals using a SECTOR® Imager (MSD). Concentrations of sAPPα, sAPPβ, Aβ38, Aβ40, and Aβ42 were calculated on Microsoft Excel by fitting to a standard curve generated from the calibrators.
Coimmunoprecipitation.
For co-IP between FLAG-mini and mini-myc: Cells were collected 48 h after transfection and lysed on ice for 10 min in CHAPS lysis buffer (1 M Tris HCl pH 8.0, 0.5 M EDTA, 0.5% CHAPS) supplemented with Complete™ protease inhibitor (Sigma). Protein G dynabeads (Thermo) were incubated with 2.5 µg FLAG antibody (#F1804, Sigma) for 10 min at room temperature and washed in PBS pH 7.4. Beads were incubated with 200 µg cell lysate overnight at 4 °C followed by washing in PBS pH 7.4. Bound proteins were eluted from the beads with SDS sample buffer supplemented with 5% β-ME for analysis by WB.
For co-IP between FLAG-mini and SORLA: HEK293 cells were transfected followed by 48 h of incubation before harvesting cell lysates (as described for immunoblotting). Gammabind G sepharose beads (GE-Healthcare) were coated with 2.5 μg anti-FLAG antibody (#F1804, Sigma) for 2 h at room temperature. Beads were washed and incubated in 70 μg of total protein from cell lysates overnight at 4 °C. Finally, beads were washed and bound proteins were eluted from the beads using SDS sample buffer supplemented with 5% β-ME. All washing steps were performed in PBS + 0.05% tween-20.
Immunocytochemistry (ICC) and Confocal Microscopy.
Approximately 5 × 104 HEK293 cells were seeded on glass coverslips coated with poly-L-lysine 0.1% (Sigma) followed by 24 h of incubation before transfection. After another 24 h cells were fixed in 4% PFA for 10 min at room temperature with subsequent washing in PBS pH 7.4. Coverslips were washed in PBS with 0.1% triton-x-100 and blocked in PBS supplemented with 10% FBS for 30 min at room temperature. After blocking, the cells were incubated overnight. at 4 °C with primary antibodies Next, cells were washed in PBS with 0.1% triton-x-100 and incubated in Alexa fluor secondary antibodies (Invitrogen, 1:500) for 1 h at room temperature. Cells were then washed in PBS, and nuclei were counterstained with Höechst (Abcam, 1:50,000) for 10 min at room temperature. Finally, the coverslips were mounted on glass slides with DAKO fluorescence mounting medium (Agilent), and after drying stained cells were imaged using a Zeiss LSM780 confocal microscope. Antibodies: anti-FLAG 1:1,000 (#F1804, Sigma), anti-GFP 1:2,000 (#ab6556, abcam), anti-VPS35 1:200 (#EB06268, Everest Biotech) and anti-sol-SORLA 1:200 (5387, Aarhus University).
BiFC.
Approximately 5 × 104 HEK293 cells were seeded on glass coverslips coated with poly-L-lysine 0.1% (Sigma) and grown for 24 h before transfection. Cells were transfected with 500 ng of either SORL1-V1, SORL1-V2, SORL1-GFP, or both SORL1-V1 and SORL1-V2. Transfected cells were incubated for another 24 h and fixed with 4% PFA for 10 min followed by washing in PBS. Counter-staining with SORLA antibody (sol-SORLA, 5387) diluted 1:200 and Höechst was performed as described for ICC above. The coverslips were mounted on glass slides using DAKO mounting medium (Agilent), and cells were visualized on a Zeiss LSM780 confocal microscope.
In Situ PLA.
HEK293 cells were seeded on 0.1% poly-L-lysine (Sigma)-coated glass coverslips and incubated for 24 h before transfection. The cells were cotransfected with 500-ng plasmids encoding either SORLA-FLAG or SORLA-GFP and incubated for 24 h. The transfected cells were fixed in 4% PFA for 10 min and washed in PBS followed by incubation in primary antibodies. Cells were incubated overnight at 4 °C in either anti-FLAG antibody (F1804 M2, Sigma) or anti-GFP antibody (ab5665, Abcam) or both diluted 1:1,000 and 1:2,000 respectively. Additionally, costaining with anti-VPS35 antibody (#EB06268, Everest Biotech) diluted 1:200 overnight was performed for visualization of endosomes. Next, the cells were washed in PBS with 0.1% triton-x-100 and incubated in PLA probes (Sigma) for 1 h at 37 °C. Cells were then washed in wash buffer A (Sigma) and incubated in ligase (Sigma) for 30 min at 37 °C. The coverslips were washed in wash buffer A followed by incubation in polymerase (Sigma) for 100 min at 37 °C. Subsequently, the cells were first washed in wash buffer B followed by washing in 0.01× wash buffer B for 1min. After the PLA procedure the cells were stained with an Alexa-fluor secondary antibody (Invitrogen, 1:500) recognizing VPS35 as described for ICC. Finally, coverslips were mounted on glass slides with Duolink® In Situ Mounting Medium with DAPI (Sigma) and imaged by confocal microscopy (LSM780, Zeiss). Colocalization was presented by Mander’s correlation coefficient (52) quantified using the ImageJ (53) Plugin JaCOP (54) and the same settings were applied to all images.
GFP-Trap Immunoprecipitation.
SORLA dimers formed by the interaction between C-terminal venus fragments (and SORLA-GFP as control) were immunoprecipitated using GFP-Trap Magnetic Particles (#M-270, Chromotek) following the manufacturer’s instructions. Then, 500 ng of total lysate from transfected HEK293 cells was incubated with 25 μL magnetic bead slurry for 1 h at 4 °C with end-over-end rotation. Subsequently, beads were washed in wash buffer (10 mM Tris/HCl pH 7.5, 150 mM NaCl, 0.05% Nonidet-P40, 0.5 mM EDTA) and eluated using SDS sample buffer.
Pull-Down Assay.
The plasmids encoding recombinant human SORLA VPS10p-domain with C-terminal PA-tag for wild-type and T659C mutant were transfected to HEK293T cell with polyethyleneimine, and secreted tagged protein was captured from its culture medium by NZ-1 immobilized Sepharose-resin (42). After washing three times with 20 mM Tris-HCl pH 8.0 and 150 mM NaCl (Tris-buffered saline), protein was eluted by SDS sample buffer. The sample was run on the SDS-PAGE gel (Invitrogen Novex WedgeWell 4 to 12% Tris-Glycine Gel) either with or without 200 mM DTT, and the stained gel by Coomassie Blue was visualized by iBright 1500 (Thermo).
Structure Modeling with AlphaFold.
The structural modeling for the whole extracellular domain of human SORLA was performed using the locally installed AlphaFold v.2.2.0 (43) with the data preset of reduced database, due to hardware limitations. The sequence of the extracellular portion without its signal sequence and propeptide (corresponding to amino acids 82 to 2,126) derived from human SORLA (NP_003096.2) was used as a query. Subsequent energy minimization was achieved by NAMD (55) (http://www.ks.uiuc.edu/Research/namd/) with the Generalized Born Implicit Solvent mode (56, 57).
All figures of 3D structures were prepared using PyMOL (The PyMOL Molecular Graphics System, Version 2.5 Schrödinger, LLC.).
Statistical Analysis.
The paired Student t test was used to compare and analyze statistical significance between samples using Graphpad Prism 8.0 (La Jolla, CA, USA) to generate graphs and to calculate P-values. P-values above 0.05 were considered nonsignificant (ns), while P < 0.05 (*), P < 0.01 (**), P < 0.001 (***), and P < 0.0001 (****) were considered significantly different.
Acknowledgments
We thank Sandra Bonnesen for excellent technical assistance. The Not Another Molecular Dynamics (NAMD) program was developed by the Theoretical and Computational Biophysics Group in the Beckman Institute for Advanced Science and Technology at the University of Illinois at Urbana-Champaign. Funding: The part of the study performed in the Andersen laboratory is funded by grants from The Alzheimer’s Association, The Danish Alzheimer’s Research Foundation, and The Novo Nordic Foundation.
Author contributions
Y.K., S.A.S., G.A.P., and O.M.A. designed research; A.M.G.J., Y.K., E.F., C.B.V., and O.M.A. performed research; A.M.G.J., Y.K., E.F., S.A.S., G.A.P., and O.M.A. analyzed data; and Y.K., C.B.V., S.A.S., G.A.P., and O.M.A. wrote the paper.
Competing interest
The authors have organizational affiliations, stock ownership, and patent filings to disclose. G.A.P., S.A.S., and O.M.A. have financial interest in Retromer Therapeutics, Inc., but none of the research was funded by or influenced by the company. Aarhus University (A.M.G.J. and O.M.A.) holds a patent to the use of the SORLA minereceptor for AD gene-therapy.
Footnotes
Reviewers: P.J.C., University of Bristol; and T.E.W., Max-Delbrueck-Center for Molecular Medicine.
Contributor Information
Gregory A. Petsko, Email: gpetsko@bwh.harvard.edu.
Olav M. Andersen, Email: o.andersen@biomed.au.dk.
Data, Materials, and Software Availability
Atomic coordinates for the predicted SORL1 ectodomain model are available upon request (58). All study data are included in the main text.
References
- 1.Jansen I. E., et al. , Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 51, 404–413 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wightman D. P., et al. , A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer’s disease. Nat. Genet. 53, 1276–1282 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellenguez C., et al. , New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet. 54, 412–436 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holstege H., et al. , Exome sequencing identifies rare damaging variants in ATP8B4 and ABCA1 as risk factors for Alzheimer’s disease. Nat. Genet. 54, 1786–1794 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheltens P., et al. , Alzheimer’s disease. Lancet 397, 1577–1590 (2021), 10.1016/S0140-6736(20)32205-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holstege H., et al. , Characterization of pathogenic SORL1 genetic variants for association with Alzheimer’s disease: A clinical interpretation strategy. Eur. J. Hum. Genet. 25, 973–981 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raghavan N. S., et al. , Whole-exome sequencing in 20,197 persons for rare variants in Alzheimer’s disease. Ann. Clin. Transl. Neurol. 5, 832–842 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fjorback A. W., et al. , Retromer binds the FANSHY sorting motif in sorLA to regulate amyloid precursor protein sorting and processing. J. Neurosci. 32, 1467–1480 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang T. Y., et al. , SNX27 and SORLA interact to reduce amyloidogenic subcellular distribution and processing of amyloid precursor protein. J. Neurosci. 36, 7996–8011 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kovtun O., et al. , Structure of the membrane-assembled retromer coat determined by cryo-electron tomography. Nature 561, 561–564 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leneva N., Kovtun O., Morado D. R., Briggs J. A. G., Owen D. J., Architecture and mechanism of metazoan retromer:SNX3 tubular coat assembly. Sci. Adv. 7, eabf8598 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caglayan S., et al. , Lysosomal sorting of amyloid-beta by the SORLA receptor is impaired by a familial Alzheimer’s disease mutation. Sci. Transl. Med. 6, 223ra220 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Andersen O. M., et al. , A genetically modified minipig model for Alzheimer’s disease with SORL1 haploinsufficiency Cell Rep. Med. 3, 100740, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simoes S., et al. , Alzheimer’s vulnerable brain region relies on a distinct retromer core dedicated to endosomal recycling. Cell Rep. 37, 110182 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mishra S., et al. , The Alzheimer’s gene SORL1 is a regulator of endosomal traffic and recycling in human neurons. Cell. Mol. Life Sci. 79, 162 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christensen S. K., et al. , Endosomal trafficking is required for glycosylation and normal maturation of the Alzheimer’s-associated protein sorLA. bioRxiv [Preprint] (2020), 10.1101/2020.07.12.199885 Accessed 1 August 2022. [DOI]
- 17.Rovelet-Lecrux A., et al. , Impaired SorLA maturation and trafficking as a new mechanism for SORL1 missense variants in Alzheimer disease. Acta Neuropathol. Commun. 9, 196 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cullen P. J., Steinberg F., To degrade or not to degrade: Mechanisms and significance of endocytic recycling. Nat. Rev. Mol. Cell Biol. 19, 679–696 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Small S. A., Petsko G. A., Retromer in Alzheimer disease, Parkinson disease and other neurological disorders. Nat. Rev. Neurosci. 16, 126–132 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Small S. A., Simoes-Spassov S., Mayeux R., Petsko G. A., Endosomal traffic jams represent a pathogenic hub and therapeutic target in Alzheimer’s disease. Trends Neurosci. 40, 592–602 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Small S. A., Petsko G. A., Endosomal recycling reconciles the Alzheimer’s disease paradox. Sci. Transl. Med. 12, eabb1717 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willnow T. E., Petersen C. M., Nykjaer A., VPS10P-domain receptors–Regulators of neuronal viability and function. Nat. Rev. Neurosci. 9, 899–909 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Hermey G., The Vps10p-domain receptor family. Cell. Mol. Life Sci. 66, 2677–2689 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monti G., Andersen O. M., 20 years anniversary for SORLA/SORL1 (1996–2016). Recept. Clin. Invest. 4, e1611 (2017). [Google Scholar]
- 25.Verheijen J., et al. , A comprehensive study of the genetic impact of rare variants in SORL1 in European early-onset Alzheimer’s disease. Acta Neuropathol. 132, 213–224 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leloup N., et al. , Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release. Nat. Commun. 8, 1708 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Januliene D., et al. , Acidic environment induces dimerization and ligand binding site collapse in the Vps10p domain of sortilin. Structure 25, 1809–1819.e3 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Yabe-Wada T., Matsuba S., Unno M., Onai N., Crystal structure of the ligand-free form of the Vps10 ectodomain of dimerized Sortilin at acidic pH. FEBS Lett. 592, 2647–2657 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Januliene D., et al. , Hidden twins: SorCS neuroreceptors form stable dimers. J. Mol. Biol. 429, 2907–2917 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Leloup N., Chataigner L. M. P., Janssen B. J. C., Structural insights into SorCS2-nerve growth factor complex formation. Nat. Commun. 9, 2979 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X., et al. , Cryo-EM structures reveal distinct apo conformations of sortilin-related receptor SORLA. Biochem. Biophys. Res. Commun. 600, 75–79 (2022). [DOI] [PubMed] [Google Scholar]
- 32.Bhalla A., et al. , The location and trafficking routes of the neuronal retromer and its role in amyloid precursor protein transport. Neurobiol. Dis. 47, 126–134 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Al-Akhrass H., et al. , A feed-forward loop between SorLA and HER3 determines heregulin response and neratinib resistance. Oncogene 40, 1300–1317 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herskowitz J. H., et al. , GGA1-mediated endocytic traffic of LR11/SorLA alters APP intracellular distribution and amyloid-beta production. Mol. Biol. Cell 23, 2645–2657 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burgert T., et al. , SORLA-dependent and -independent functions for PACS1 in control of amyloidogenic processes. Mol. Cell. Biol. 33, 4308–4320 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Croucher D. R., et al. , Bimolecular complementation affinity purification (BiCAP) reveals dimer-specific protein interactions for ERBB2 dimers. Sci. Signal. 9, ra69 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Andersen O. M., et al. , SorLA/LR11, a neuronal sorting receptor that regulates processing of the amyloid precursor protein. Proc. Natl. Acad. Sci. U.S.A. 102, 13461–13466 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dodson S. E., et al. , Loss of LR11/SORLA enhances early pathology in a mouse model of amyloidosis: Evidence for a proximal role in Alzheimer’s disease. J. Neurosci. 28, 12877–12886 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mehmedbasic A., et al. , SorLA complement-type repeat domains protect the amyloid precursor protein against processing. J. Biol. Chem. 290, 3359–3376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kitago Y., et al. , Structural basis for amyloidogenic peptide recognition by sorLA. Nat. Struct. Mol. Biol. 22, 199–206 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Mecozzi V. J., et al. , Pharmacological chaperones stabilize retromer to limit APP processing. Nat. Chem. Biol. 10, 443–449 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujii Y., et al. , PA tag: A versatile protein tagging system using a super high affinity antibody against a dodecapeptide derived from human podoplanin. Protein Expr. Purif. 95, 240–247 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tunyasuvunakool K., et al. , Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willingham M. C., Hanover J. A., Dickson R. B., Pastan I., Morphologic characterization of the pathway of transferrin endocytosis and recycling in human KB cells. Proc. Natl. Acad. Sci. U.S.A. 81, 175–179 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hierro A., et al. , Functional architecture of the retromer cargo-recognition complex. Nature 449, 1063–1067 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spoelgen R., et al. , Interaction of the cytosolic domains of sorLA/LR11 with the amyloid precursor protein (APP) and b-secretase b-site APP-cleaving enzyme. J. Neurosci. 26, 418–428 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kinoshita A., et al. , Demonstration by fluorescence resonance energy transfer of two sites of interaction between the low-density lipoprotein receptor-related protein and the amyloid precursor protein: Role of the intracellular adapter protein Fe65. J. Neurosci. 21, 8354–8361 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones D. T., Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 (1999). [DOI] [PubMed] [Google Scholar]
- 50.Cheng J., Randall A. Z., Sweredoski M. J., Baldi P., SCRATCH: A protein structure and structural feature prediction server. Nucleic Acids Res. 33, W72–76 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin K., Simossis V. A., Taylor W. R., Heringa J., A simple and fast secondary structure prediction method using hidden neural networks. Bioinformatics 21, 152–159 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Manders E. M. M., Verbeek F. J., Aten J. A., Measurement of co-localization of objects in dual-colour confocal images. J. Microsc. 169, 375–382 (1993). [DOI] [PubMed] [Google Scholar]
- 53.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bolte S., Cordelieres F. P., A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Phillips J. C., et al. , Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 153, 044130 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanner D. E., Chan K. Y., Phillips J. C., Schulten K., Parallel generalized born implicit solvent calculations with NAMD. J. Chem. Theory Comput. 7, 3635–3642 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanner D. E., Phillips J. C., Schulten K., GPU/CPU algorithm for generalized born/solvent-accessible surface area implicit solvent calculations. J. Chem. Theory Comput. 8, 2521–2530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kitago Y., Andersen O. M., Petsko G. A., AlphaFold2prediction of hSORLA extracelluar domain. ModelArchive. https://modelarchive.org/doi/10.5452/ma-zgbg4. Deposited 30 December 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Atomic coordinates for the predicted SORL1 ectodomain model are available upon request (58). All study data are included in the main text.