Significance
The brassinosteroid (BR) receptor BRI1 provides a paradigm for understanding receptor-mediated signaling in plants and contribution of post-translational modifications. Here, we show that BRI carries SUMO modifications in planta on two intracellular lysine residues and that temperature elevation triggers BRI1 deSUMOylation mediated by the Desi3a SUMO protease. Importantly, BRI1 deSUMOylation leads to downregulation of BR signaling via increased BRI1 interaction with the BIK1 negative regulator and increased BRI1 endocytosis. Loss of BRI1 deSUMOylation in desi3a mutants boosts plant responses to elevated ambient temperature, indicating that BRI1 deSUMOylation acts as a brake to keep temperature responses in check. Our study uncovers a new post-translational modification targeting BRI1 and sheds light on its functional outcome for environmentally controlled plant growth.
Keywords: Arabidopsis, brassinosteroids, BRI1, SUMO, temperature
Abstract
Brassinosteroids (BRs) are a class of steroid molecules perceived at the cell surface that act as plant hormones. The BR receptor BRASSINOSTEROID INSENSITIVE1 (BRI1) offers a model to understand receptor-mediated signaling in plants and the role of post-translational modifications. Here we identify SUMOylation as a new modification targeting BRI1 to regulate its activity. BRI1 is SUMOylated in planta on two lysine residues, and the levels of BRI1 SUMO conjugates are controlled by the Desi3a SUMO protease. Loss of Desi3a leads to hypersensitivity to BRs, indicating that Desi3a acts as a negative regulator of BR signaling. Besides, we demonstrate that BRI1 is deSUMOylated at elevated temperature by Desi3a, leading to increased BRI1 interaction with the negative regulator of BR signaling BIK1 and to enhanced BRI1 endocytosis. Loss of Desi3a or BIK1 results in increased response to temperature elevation, indicating that BRI1 deSUMOylation acts as a safety mechanism necessary to keep temperature responses in check. Altogether, our work establishes BRI1 deSUMOylation as a molecular crosstalk mechanism between temperature and BR signaling, allowing plants to translate environmental inputs into growth response.
Brassinosteroids (BRs) are a class of plant hormones controlling various aspects of plant development and stress responses (1). Genetic, biochemical, and structural biology studies have revealed that BRs are perceived at the cell surface by the BRASSINOSTEROID INSENSITIVE1 (BRI1) leucine-rich-repeat receptor-like kinase (LRR-RLK) (2–7). BR binding to BRI1 promotes its heterodimerization with the LRR-RLK BRI1-ASSOCIATED RECEPTOR KINASE (BAK1) (8, 9) to form a competent receptor complex. cis- and trans-phosphorylation of BRI1 and BAK1 fully activates the receptor complex and initiates a protein phosphorylation-mediated signaling cascade, which ultimately regulates the activity of the BRASSINAZOLE RESISTANT(BZR)-family of transcription factors (10, 11). Among these, BZR1 and BZR2 (BES1) were shown to control the expression of thousands of BR-responsive genes important for plant growth and stress response (12, 13).
Inability to properly produce, sense, or transduce the BR signal results in characteristic BR-deficient/insensitive phenotypes that include short hypocotyls in the dark, dwarf stature in the light, altered vascular development, prolonged vegetative phase, and reduced male fertility (5, 14–16). In contrast, BR overproduction or enhanced signaling activities are associated with increased growth (7). The precise control of BR perception at the cell surface is therefore crucial to ensure proper plant development and completion of the plant life cycle in an ever-changing environment. As the major BR receptor and a long-lived protein, BRI1 is regulated by several mechanisms to keep its basal activity in check and to desensitize BRI1 after BR signaling. The BRI1 kinase is first kept in its basal state by an autoinhibitory C-terminal tail (17). Phosphorylation of the C-terminal tail upon BR binding likely releases autoinhibition for the full activation of BRI1. BRI1 also interacts with an inhibitory protein named BRI1 KINASE INHIBITOR1 (BKI1) that prevents interaction between BRI1 and BAK1 in resting cells (18). BR binding to BRI1 triggers BKI1 tyrosine phosphorylation and release in the cytosol, allowing the formation of an active BRI1-BAK1 receptor complex (18, 19). Additional mechanisms were coopted to stop BRI1 from firing after perception of BRs and allow plant cells to go back to the resting state. Autophosphorylation of residue S891 in the G-loop occurs late after BR perception and deactivates BRI1 via inhibiting its ATP binding (20). BRI1 is also subjected to internalization from the cell surface and vacuolar degradation using several mechanisms. First, the KINASE-ASSOCIATED PROTEIN PHOSPHATASE (KAPP) was proposed to regulate BRI1 through interaction between its forkhead-associated domain and BRI1's cytoplasmic domain (21). KAPP also colocalizes with the BAK1-related coreceptor SOMATIC EMBRYOGENESIS RECEPTOR KINASE1 (SERK1) at the plasma membrane (PM) and interacts with SERK1 in endosomes suggesting that KAPP-mediated dephosphorylation of BRI1 and SERK1 down-regulates BR signaling (22). Second, BRI1 degradation was shown to require PROTEIN PHOSPHATASE2A (PP2A)-mediated dephosphorylation triggered by methylation of the PP2A using a leucine carboxyl methyltransferase (23). Most importantly, BRI1 undergoes endocytosis and degradation in the vacuole (24). This is controlled by lysine(K)-63 linked polyubiquitin chain conjugation to BRI1 intracellular domain driven by the PLANT U-BOX12 (PUB12) and PLANT U-BOX13 (PUB13) E3 ligases (25, 26). BRI1 ubiquitination promotes BRI1 internalization from the cell surface and is essential for proper sorting in endosomes and vacuolar targeting (25). BRI1 endocytosis was initially thought to be independent of ligand binding (24). However, the fact that BRI1 ubiquitination is dependent on both BRI1 kinase activity and ligand perception suggests that BRI1 internalization and vacuolar degradation may be regulated by BRs (25, 26). BRI1 endocytosis is also under the control of environmental signals that impinge on growth via BR responses. Elevation of ambient growth temperature decreases BRI1 protein accumulation to boost root elongation (27). The crosstalk between BR signaling and temperature responses likely uses BRI1 ubiquitination, as the expression of an ubiquitination-defective BRI1 variant lacking 25 lysine residues in BRI1 intracellular domain abolishes BRI1 degradation upon warmer temperature (27).
BRI1 serves as the archetypal plant receptor protein in the study of post-translational modifications (PTMs) and their interplay and their role in signaling/signal integration. Here show that BRI1 is SUMOylated and that BRI1 SUMO conjugates are regulated by the DESUMOYLATING ISOPEPTIDASE 3a (Desi3a) small ubiquitin-like modifier (SUMO) protease. Most importantly, we uncover that BRI1 SUMOylation drops following growth at heightened temperatures, leading to increased BRI1 interaction with the negative regulator BOTRYTIS-INDUCED KINASE 1 (BIK1) and increased BRI1 endocytosis to attenuate BR-dependent growth. Finally, we demonstrate that such downregulation in BR signaling is required to restrict elevated temperature-induced growth responses. Overall, our work shed light on a new PTM targeting BRI1 and highlights its interplay with BRI1 ubiquitin-mediated endocytosis in the control of environmentally regulated plant growth.
Results
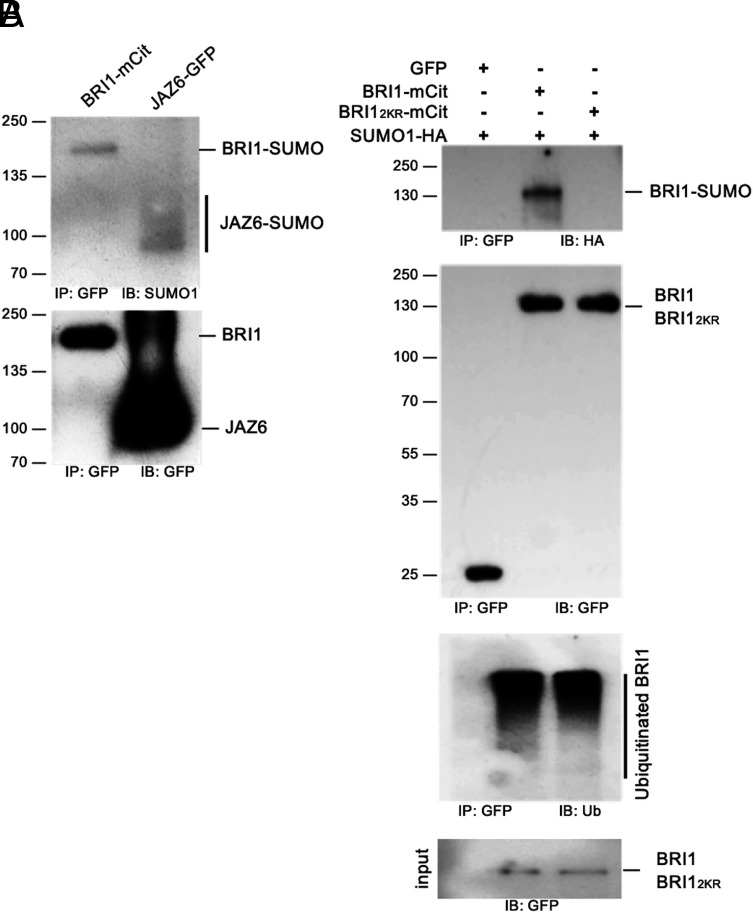
BRI1 Is Decorated with SUMO Modifications In Planta.
BRI1 was previously demonstrated to be modified by ubiquitination on intracellular lysine residues (25). Ubiquitin is the founding member of a class of PTMs named ubiquitin-like modifiers that share a core β-grasp fold of approximately 70 amino acids and that can be reversibly attached to proteins or other cellular constituents such as lipids to regulate their activity (28). To evaluate if BRI1 is modified by other ubiquitin-like modifications, we first sought to monitor if BRI1 is SUMOylated in planta. Transgenic plants expressing a functional BRI1 fusion to the mCitrine yellow fluorescent protein (mCit) under the control of BRI1 promoter were used to immunoprecipitate BRI1-mCit protein with micromagnetic beads coupled to anti-GFP antibodies. Probing BRI1-mCit immunoprecipitates with anti-SUMO1 (AtSUMO1) revealed a SUMO-specific signal at the size of BRI1-mCit, similar to what is observed for the positive controls JASMONATE ZIM DOMAIN PROTEIN 6 (JAZ6) fused to Green Fluorescent Protein (GFP) (29) (Fig. 1A). This indicates that BRI1 is conjugated with SUMO1 under standard growth conditions. In contrast to the smear-like signals obtained for BRI1 with anti-Ub antibodies (25, 26), the SUMO1-specific signal associated to BRI1 migrated as a sharp band close to the molecular weight of BRI1 indicating that BRI1 carries a very limited number of SUMO modifications.
Fig. 1.
BRI1 is SUMOylated in vivo on two intracellular lysine residues. A. In vivo SUMOylation analyses of BRI1. Immunoprecipitation was carried out using anti-GFP antibodies on solubilized protein extracts from mono-insertional homozygous BRI1-mCitrine plants or the JAZ6-GFP positive control plants. Detection of immunoprecipitated proteins used the anti-GFP (Bottom) and AtSUMO1 (Top) antibodies. B. In vivo SUMOylation analyses of BRI1 and BRI12KR. BRI1-mCit and the BRI12KR-mCit variant mutated for residues K1066,1118R were transiently expressed in N. benthamiana leaves prior to immunoprecipitation using anti-GFP antibodies on solubilized protein extracts. GFP alone was used as negative control. All constructs were co-expressed with SUMO1-HA. Detection of immunoprecipitated proteins used the AtSUMO1, anti-GFP, and anti-ubiquitin antibodies.
To gain further insight into BRI1 SUMOylation, we searched for possible SUMO sites in BRI1 as previously done with FLS2 (30). We identified lysine residues K1066 and K1118 in the BRI1 kinase domain as putative SUMO sites. Residue K1066 is strictly conserved in Arabidopsis BRI1-like proteins and more generally in plant BRI1 homologs (SI Appendix, Fig. S1A). A conserved lysine is also found in a close context to K1118 in Arabidopsis BRI1-Like proteins (BRLs) and plant BRI1 homologs. To decipher if both lysine residues are actual targets of SUMO in planta, we generated the K1066R and K1118R BRI1 variants where the corresponding lysine has been substituted to arginine to maintain the positive charge while preventing SUMOylation. Mutation of each of the two lysine residues in BRI1 decreased SUMO conjugation compared to wild-type (WT) BRI1 when transiently expressed in Nicotiana benthamiana (SI Appendix, Fig. S1B), suggesting that these are bona fide SUMO sites. Mutation of both lysine residues completely abolished BRI1 SUMOylation (Fig. 1B), indicating that these are the only SUMO sites in BRI1. Mutation of K1066 and K1118 however did not alter significantly the overall ubiquitination pattern of BRI1 (Fig. 1B). This observation is consistent with the fact that BRI1 is heavily ubiquitinated in planta and that mutation of 25 lysine residues in BRI1 intracellular domain, including K1066 and K1118, is required to completely abolish BRI1 ubiquitination (25). Altogether, our work reveals that BRI1 can be post-translationally modified at residues K1066 and K1118 by either ubiquitin or SUMO.
BRI1 SUMOylation Is Regulated by the SUMO Protease Desi3a.
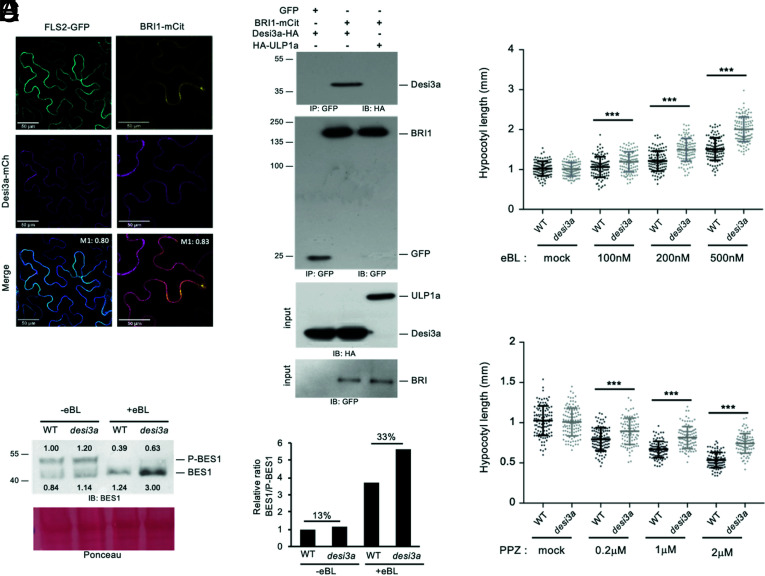
The levels of SUMO conjugates of another plant PM protein, FLS2, are controlled by the balance between SUMO E2-CONJUGATING ENZYME 1, which is capable of directly transferring SUMO onto target residues (31), and the Desi3a SUMO protease that deSUMOylates FLS2 (30). Desi3a has been shown to be degraded upon flagellin perception, allowing the accumulation of SUMO-FLS2 conjugates and triggering intracellular immune signaling. To examine if Desi3a also controls the levels of SUMO-BRI1, we first determined whether Desi3a is found in overlapping expression territories with BRI1. Publicly available genome-wide expression data reveals that Desi3a has a broad expression profile overlapping with BRI1. We confirmed these observations by qRT-PCR using RNA extracted from various Arabidopsis tissues (SI Appendix, Fig. S2). We next addressed whether Desi3a protein colocalizes with BRI1 at the PM. Transient expression of a Desi3a-mCherry (Desi3a-mCh) fusion protein in N. benthamiana confirmed the presence of Desi3a at the cell surface (Fig. 2A). A similar pattern was observed with the BRI1-mCit or FLS2-GFP functional fusions. In addition, a clear colocalization at the cell surface is observed between Desi3a and BRI1 or FLS2 (Manders coefficient MBRI1 = 0.83 and MFLS2 = 0.80), to the resolution of the confocal microscope. Besides, the fluorescence profile of Desi3a-mCh clearly overlapped with BRI1-mCit at the PM, similarly to what is observed for FLS2-GFP (SI Appendix, Fig. S3) (30). We next tested whether the Desi3a SUMO protease had the ability to interact with BRI1. To this purpose, we took advantage of the transient expression in N. benthamiana since BRI1 is also SUMOylated in this experimental system. BRI1-mCit was transiently expressed together with Desi3a-haemaglutenin (HA) and subjected to immunoprecipitation using GFP beads. Probing BRI1-mCit immunoprecipitates with anti-HA antibodies revealed the presence of Desi3a-HA (Fig. 2B). Expression of GFP alone with Desi3a was used as a negative control and failed to capture any interaction (Fig. 2B), indicating that BRI1 interacts in vivo with Desi3a. Furthermore, we used the ULP1a SUMO protease that deSUMOylates the BZR1 transcription factor (32) as another negative control and observed no interaction with BRI1 (Fig. 2B).
Fig. 2.
SUMO protease Desi3a interacts with BRI1 and negatively regulates BR signaling. A. Confocal microscopy and colocalization analyses between BRI1 and Desi3a. BRI1-mCit and Desi3a-mcherry (Desi3a-mCh) were transiently expressed in N. benthamiana leaves. The Manders colocalization coefficient is shown in the overlay channel. (Scale bars, 50 μm.) B. In vivo interaction between BRI1 and Desi3a. BRI1-mCit and Desi3a-HA were transiently expressed in N. benthamiana leaves prior to immunoprecipitation using anti-GFP antibodies on solubilized protein extracts. GFP alone and the ULP1a SUMO protease were used as negative controls. Detection of immunoprecipitated proteins used the anti-HA and anti-GFP antibodies. C. Hypocotyl length of 6-d-old WT and desi3a mutant plants grown on plates containing mock or increasing concentrations of eBL. Experiments were carried out in triplicates. Error bars represent SD (n = 40). The asterisks indicate a statistically significant difference between WT and desi3a (two-way ANOVA with Sidak’s multiple comparison test). D. Hypocotyl length of 6-d-old WT and desi3a mutant plants on plates containing mock or increasing concentrations of PPZ. Experiments were carried out in triplicates. Error bars represent SD (n = 40). The asterisks indicate a statistically significant difference between WT and desi3a (two-way ANOVA with Sidak’s multiple comparison test). E. Phosphorylation state of the BES1 transcription factor in WT or desi3a plants treated with mock or 1 μM eBL for 20 min. Detection of BES1 was performed with anti-BES1 antibodies. Signal intensities of BES1 and P-BES1 are indicated. F. Quantification of the ratio between BES1 and phosphorylated BES1 (P-BES1) shown in E. Percentage increase compared to mock is shown.
Altogether, this indicates that BRI1 specifically interacts with the PM-localized Desi3a SUMO protease and that the SUMOylation status of BRI1 is controlled by the Desi3a SUMO protease.
Desi3a-Mediated deSUMOylation of BRI1 Controls BR Signaling.
The fact that Desi3a interacts with BRI1 prompted us to investigate the role of Desi3a in BR signaling. To this purpose, we measured hypocotyl length of WT and desi3a knockout plants grown in light and treated with mock, epibrassinolide (eBL), or the BR biosynthetic inhibitor propiconazole (PPZ). Consistent with previous observations, WT plants exposed to eBL showed longer hypocotyls (Fig. 2C). desi3a mutant plants however displayed a stronger sensitivity to exogenous eBL, as attested by the longer hypocotyl length for eBL-treated plants relative to mock. Such hypersensitivity to BRs is also highlighted by the increased resistance to the dwarfing effect of PPZ (Fig. 2D). The desi3a/Desi3a-HA line, which is complemented for the desi3a immune phenotypes (30), also failed to show such BR-related phenotypes when grown on eBL or PPZ (SI Appendix, Fig. S4 A–D). These observations provide genetic proof for the role of Desi3a in BR signaling and indicate that Desi3a acts as a negative regulator of the BR pathway.
To better characterize the role of Desi3a in plant BR responses, we next investigated the phosphorylation status of the BR pathway downstream transcription factor BES1 that serves as readout of BR signaling. BES1 indeed exists under a phosphorylated inactive form (P-BES1) and dephosphorylated active form (BES1), and exogenous BR application promotes the conversion of P-BES1 into its dephosphorylated active BES1 form (11). WT and desi3a plants were therefore treated with mock or eBL and BES1 phosphorylation status revealed by immunoblotting using anti-BES1 antibodies. desi3a displayed a greater sensitivity to BL compared to WT plants, as attested by the enhanced conversion of the BES1 pool into its unphosphorylated fast migrating form (Fig. 2 E and F), consistent with the results from the eBL/PPZ dose-response. Altogether, these observations clearly argue for a role of Desi3a in negatively regulating BR signaling through its control of BRI1 SUMOylation.
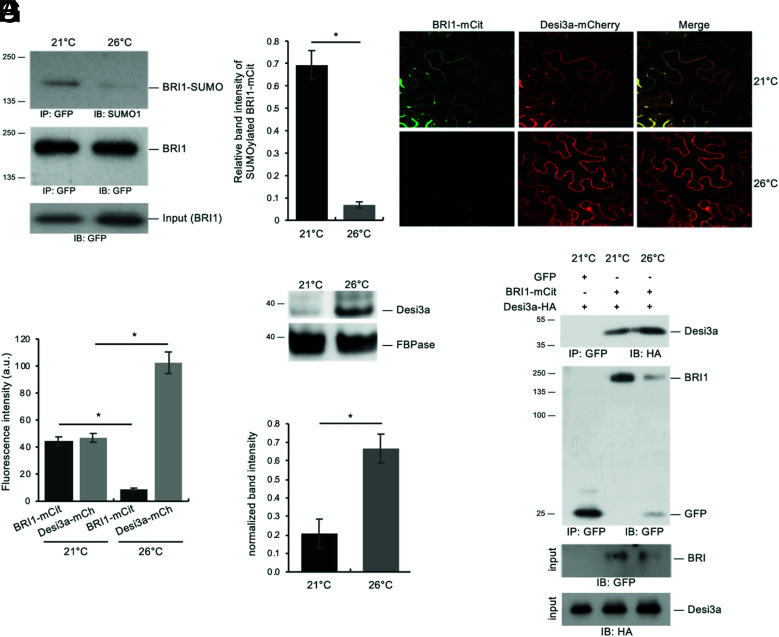
Temperature Elevation Regulates BRI1 deSUMOylation.
To shed light on the biological relevance of Desi3a and BRI1 SUMOylation, we first searched for possible stimuli/conditions increasing or decreasing the accumulation of BRI1 SUMO conjugates. Among conditions tested, we focused our attention on the role of ambient temperature elevation since previously reported to post-translationally regulate BRI1 (27). We first evaluated if BRI1 protein levels were affected by growth temperature elevation. Seedlings from plants expressing BRI1-mCit were grown at 21 °C and 26 °C as previously done (27), and protein levels determined by immunoblotting. Plants subjected to elevated temperature displayed lower BRI1 levels (SI Appendix, Fig. S5A), similarly to what has been described in roots only (27). To determine the impact of temperature on BRI1 SUMOylation, BRI1-mCit was immunoprecipitated from plants grown at 21 °C or 26 °C. More tissues were harvested for 26 °C-grown plants to compensate for the lower BRI1 levels and immunoprecipitates normalized to show equivalent BRI1-mCit signals, as visualized using anti-GFP antibodies (Fig. 3A), before being probed with anti-SUMO1 antibodies to detect the SUMOylated pool of BRI1. Plants grown at 21 °C or 26 °C clearly showed different accumulation of BRI1 SUMO conjugates. Notably, temperature elevation was accompanied with lower SUMOylated BRI1 (Fig. 3A). Quantification of the BRI1 SUMO signals obtained, relative to the levels of immunoprecipitated BRI1, revealed a sevenfold decrease at elevated temperature (Fig. 3B). Such a drop in BRI1 SUMO levels at higher temperature may be a direct consequence of increased Desi3a levels. To test this, we first investigated the influence of temperature on the accumulation of BRI1 and Desi3a proteins using transient expression in N. benthamiana. Plants agroinfiltrated with BRI1-mCit and Desi3a-mCh were exposed to either 21 °C or 26 °C for 2 d and imaged at the confocal microscope. Several independent experiments were carried out and multiple regions of interest analyzed to overcome the variability in transformation efficiency. Overall, temperature elevation reproducibly decreased BRI1-mCit accumulation when co-expressed with Desi3a-mCh (Fig. 3 C and D), reminiscent of the effect of warmer temperature on BRI1 accumulation previously observed in Arabidopsis roots (27). This was also accompanied by an increase in the fluorescence associated with Desi3a (Fig. 3 C and D). The impact of temperature elevation on Desi3a accumulation was confirmed using the desi3a/Desi3a-HA transgenic Arabidopsis plants constitutively expressing the Desi3a-HA fusion protein. Temperature elevation promoted the accumulation of Desi3a-HA protein (Fig. 3 E and F), pointing to a post-transcriptional regulation of Desi3a by temperature. We then addressed how temperature affects the ability of BRI1 and Desi3a to interact in vivo. Agroinfiltrated plants exposed to 26 °C reproducibly harbored lower BRI1-mCit accumulation (Fig. 3G), consistent with our confocal microscopy observations. BRI1 immunoprecipitates however showed increased Desi3a-HA proteins levels at 26 °C, pointing to the increased interaction between BRI1 and Desi3a when plant experiences heightened temperature (Fig. 3G). Taken together, these observations indicate that the increased accumulation of Desi3a and stronger interaction with BRI1 likely explains the drop in BRI1 SUMOylation observed at 26 °C.
Fig. 3.
Desi3a deSUMOylates BRI1 upon changes in ambient temperature. A. In vivo SUMOylation analyses of BRI1 at 21 °C or 26 °C. Immunoprecipitation was carried out using anti-GFP antibodies on solubilized protein extracts from mono-insertional homozygous BRI1-mCitrine plants grown at 21 °C or 26 °C. Detection of immunoprecipitated proteins used the anti-SUMO1 and anti-GFP antibodies. B. Quantification of BRI1 SUMO at 21 °C and 26 °C relative to immunoprecipitated BRI1-mCit levels. Error bars represent SD (n = 3). The asterisks indicate a statistically significant difference between 21 °C and 26 °C (Mann–Whitney). C. Confocal microscopy analyses of BRI1 and Desi3a at 21 °C and 26 °C. BRI1-mCit and Desi3a-mch were transiently expressed in N. benthamiana leaves and incubated at 21 °C or 26 °C for 2 d. Similar confocal detection settings were used to compare the effect of temperature on BRI1 and Desi3a proteins levels. Several independent experiments were carried out and multiple regions of interest analyzed to overcome the variability in transformation efficiency. (Scale bars, 20 μm.) D. Quantification of BRI1 and Desi3a fluorescence levels in experiments carried out as in C. Multiple regions of interest analyzed for to overcome the variability in transformation efficiency. Error bars represent SD (n = 15). The asterisk indicates a statistically significant difference between BRI1-mCit or Desi3a-mCh at 21 °C and 26 °C (Mann–Whitney). E. Western blot analyses monitoring the accumulation of Desi3a-HA protein in Arabidopsis plants grown at 21 °C or 26 °C. Detection of Desi3a-HA was performed with anti-HA antibodies. The membrane was stripped and probed with anti-FBPase antibodies as loading control. F. Quantification of Desi3a protein at 21 °C and 26 °C relative to FBPase levels. Error bars represent SD (n = 3). The asterisks indicate a statistically significant difference in Desi3a protein level at 21 °C and 26 °C (Mann–Whitney). G. In vivo interaction between BRI1 and Desi3a at 21 °C and 26 °C. BRI1-mCit and Desi3a-HA were transiently expressed in N. benthamiana and incubated at 21 °C or 26 °C for 2 d prior to immunoprecipitation using anti-GFP antibodies on solubilized protein extracts. GFP alone was used as negative control. Detection of immunoprecipitated proteins used the anti-HA and anti-GFP antibodies.
Desi3a-Mediated deSUMOylation of BRI1 Controls Temperature Responses.
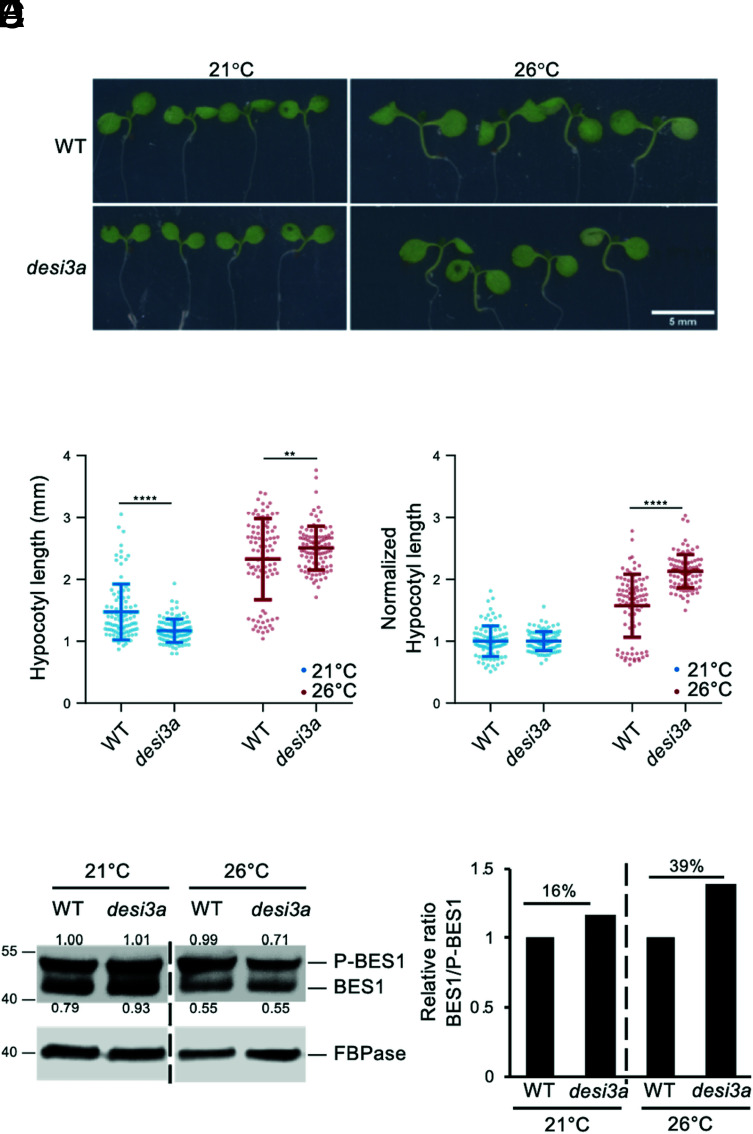
Plant responses to temperature are highly dependent on plant hormones and other environmental factors such as light (27, 33). We therefore assessed the genetic contribution of BR signaling to plant responses to elevated temperature in our own conditions by scoring hypocotyl length of WT, bri1, and the bes1-D constitutive BR response mutant at 21 °C or 26 °C. Consistent with previous observations, WT seedlings grown at elevated temperature elongated their hypocotyls (SI Appendix, Fig. S5 B–D). bes1-D showed much greater responses to increased temperature than WT, while bri1 failed to respond (SI Appendix, Fig. S5 B–D), suggesting that BR signaling positively impinges on plant temperature responses in hypocotyls. We next investigated the role of SUMO/deSUMOylation upon temperature elevation by comparing hypocotyl length of WT and desi3a at 21 °C or 26 °C. desi3a mutants showed slightly shorter hypocotyls at 21 °C compared to WT seedlings but elongated more at 26 °C (Fig. 4 A and B). The increased response to temperature of desi3a is also highlighted by the elevated ratio of hypocotyl length at 26 °C/21 °C for desi3a (Fig. 4C). In contrast to desi3a, the desi3a/DESI3a-HA line did not show hypersensitivity to temperature elevation (SI Appendix, Fig. S4 E and F), attesting that the loss of Desi3a is responsible for the observed effect. This suggests that Desi3a is a negative regulator of temperature responses or that conversely, SUMOylation has a positive role on hypocotyl elongation during temperature elevation. We next monitored the phosphorylation status of BES1 as a readout of BR pathway activation upon increased temperature. Compared to WT plants, desi3a reproducibly showed a mild increase in dephosphorylated BES1:phosphorylated BES1 ratio when exposed to a small change in ambient temperature (Fig. 4 D and E and SI Appendix, Fig. S5 E–H). Although exogenous application of saturating concentrations of BRs triggers full conversion of P-BES1 in BES1, the mild response observed to a subtle change in environmental conditions confirms that loss of Desi3a yields enhanced BR signaling and BR-dependent growth at 26 °C. Furthermore, this is consistent with bes1-D greatly over-responding to higher temperature.
Fig. 4.
Desi3a is a negative regulator of plant responses to temperature elevation. A. Phenotype of 6-d-old WT and desi3a mutant plants grown at 21 °C or 26 °C. Representative pictures are shown. B. Hypocotyl length of 6-d-old WT and desi3a mutant plants grown at 21 °C or 26 °C. Experiments were carried out in triplicates. Error bars represent SD (n = 40). The asterisks indicate a statistically significant difference between WT and desi3a (two-way ANOVA with Sidak’s multiple comparison test). C. Normalized hypocotyl length from WT and desi3a mutant plants grown at 21 °C and 26 °C for 6 d, relative to 21 °C. Experiments were carried out in triplicates. Error bars represent SD (n = 40). The asterisks indicate a statistically significant difference between WT and desi3a (two-way ANOVA with Sidak’s multiple comparison test). D. Phosphorylation state of the BES1 transcription factor in WT or desi3a plants grown at 21 °C or 26 °C. Detection of BES1 was performed with anti-BES1 antibodies. Signal intensities of BES1 and P-BES1 are indicated. E. Normalized BES1 to phosphorylated BES1 (P-BES1) ratios from western blot shown in D, relative to WT. Replicate experiments are shown in SI Appendix, Fig. S5 E–H.
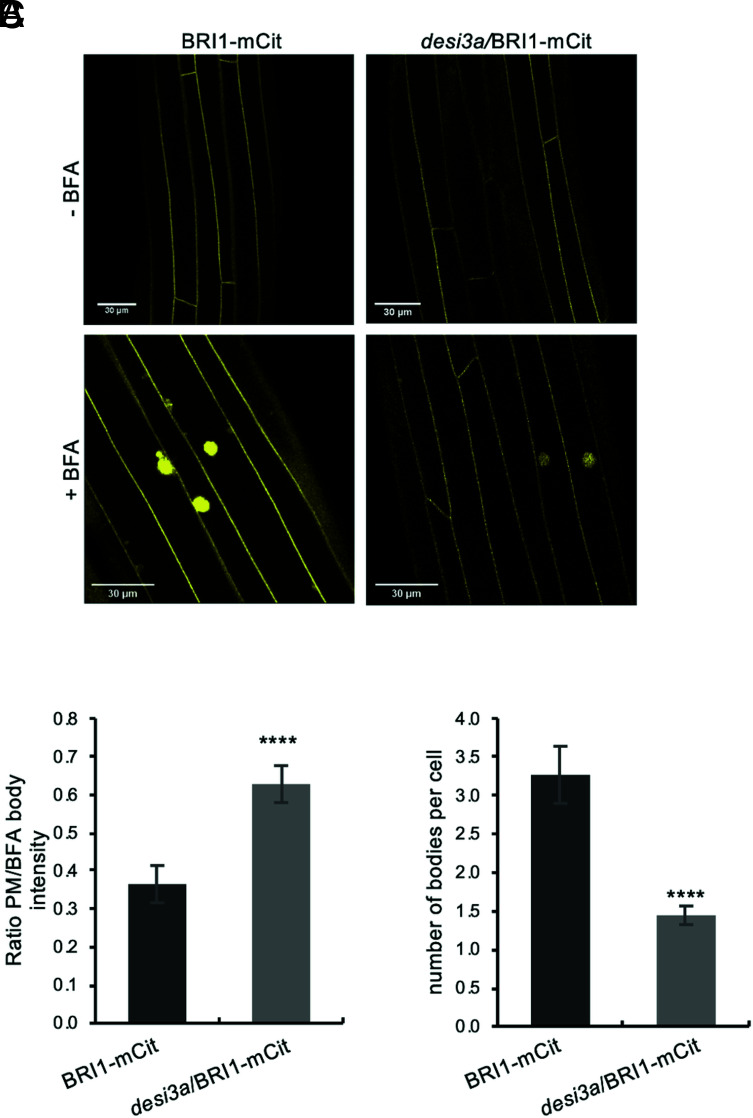
Next, we sought to decipher the specific role of BRI1 SUMO/deSUMOylation in plant responses to elevated temperature and the underlying mechanism(s) by genetically impacting on BRI1 SUMOylation. We reasoned that generating transgenic plants expressing the non-SUMOylated BRI12KR version would prevent us from reaching any solid conclusion on the role of BRI1 SUMO/deSUMOylation since mutation of K1066 and K1118 would also impact on BRI1 ubiquitination. We decided instead to characterize deeper the impact of altered BRI1 SUMO levels at residues K1066 and K1118 using the desi3a mutant background. This offers the great advantage to grasp the interplay between both post-translational modifications at K1066 and K1118. We therefore crossed desi3a with the BRI1-mCit reporter line and isolated desi3a/BRI1-mCit double homozygous plants. These plants were imaged at the confocal microscope to observe any possible change in BRI1 distribution in the cell. No obvious change of BRI1 distribution between the PM and endosomes could be observed for BRI1-mCit or desi3a/BRI1-mCit lines. To better investigate the possible effect of BRI1 SUMO on BRI1 dynamics, we then took advantage of the fungal toxin Brefeldin A (BFA) that inhibits endosomal trafficking in Arabidopsis roots and hypocotyls (34, 35) and thus creates large aggregates of trans-Golgi network/early endosomal compartments. Endocytosed BRI1-mCit was found in BFA bodies when plants were challenged with BFA in the presence of the translation inhibitor cycloheximide (CHX) (Fig. 5A), similar to what has been reported in roots (24, 25, 36). However, quantification of several parameters pointed to a reduction in BRI1 endocytosis in the desi3a mutant compared to WT plants. First, the ratio of PM over intracellular BFA-trapped BRI1-mCit fluorescence is increased in desi3a compared to the WT background (Fig. 5B). Second, loss of Desi3a is accompanied with a reduction in the number of BFA bodies per cell (Fig. 5C). Taken together, these observations indicate the loss of Desi3a and SUMOylation at residues K1066 and K1118 directly or indirectly decreased the endocytic flux of BRI1.
Fig. 5.
Desi3a-dependent deSUMOylation regulates BRI1 endocytosis and interaction with BIK1. A. Confocal microscopy analyses of BRI1-mCit and desi3a/BRI1-mCit dark-grown hypocotyls. Similar confocal detection settings were used to compare the fluorescence intensity in the two transgenic lines. (Scale bars, 30 μm.) B. Quantification of the ratio between PM and BFA body fluorescence signal intensities of BRI1-mCit and desi3a/BRI1-mCit. C. Quantification of the number of BFA bodies per cell in BRI1-mCit and desi3a/BRI1-mCit. Experiments were carried out in triplicates. Error bars represent SD (n = 9). The asterisks indicate a statistically significant difference between BRI1-mCit and desi3a/BRI1-mCit (Mann–Whitney).
BIK1 Is Recruited to deSUMOylated BRI1 to Dampen Temperature Responses.
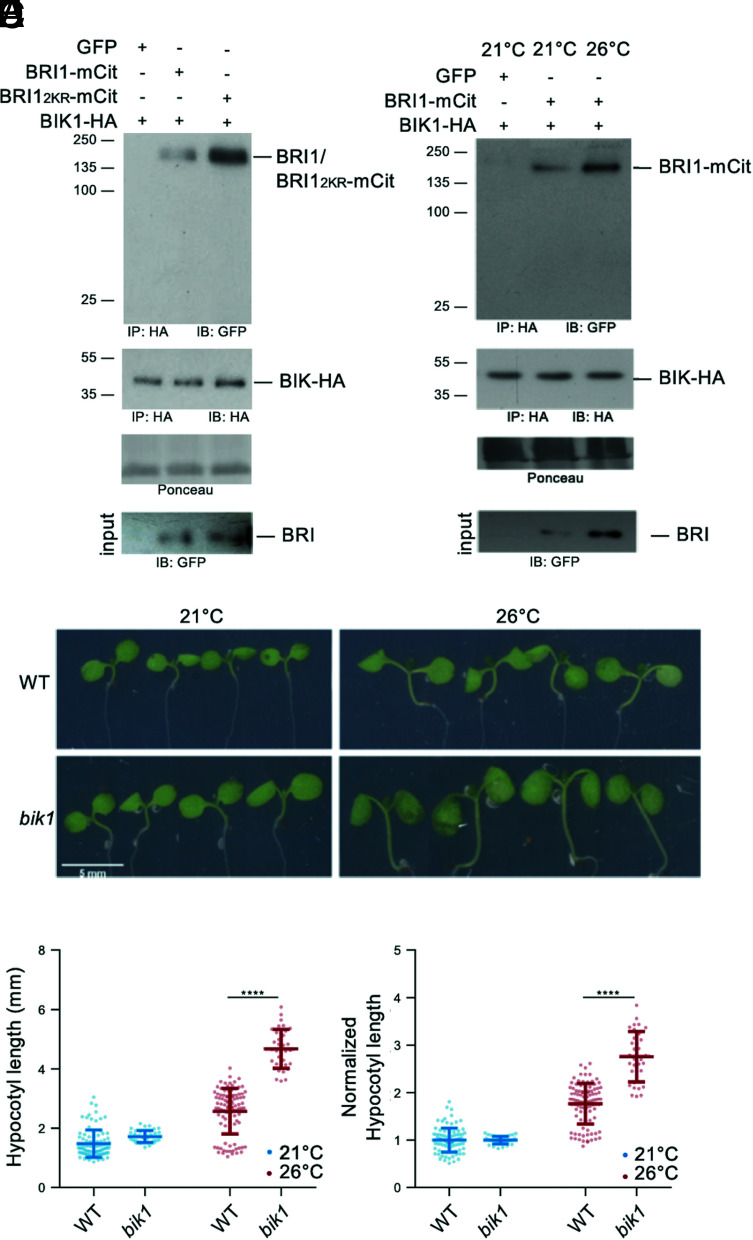
Lack of SUMOylation renders FLS2 unable to mount proper immune responses due to the increased interaction with the downstream receptor-like cytoplasmic kinase (RLCK) BIK1 (30), which acts as a positive regulator of FLS2-mediated signaling (37). BIK1 is also known to participate to BR signaling, although as negative regulator, where it is associated with BRI1 in resting cells and is released from the BRI1-BAK1 receptor complex upon BR perception (38). To shed light on the direct functional consequences of BRI1 SUMO/deSUMOylation, we therefore investigated if BRI1 SUMOylation affects the BRI1-BIK1 interaction using transient expression in N. benthamiana. Immunoprecipitation of BIK1-HA using HA beads followed by probing with anti-GFP antibodies failed to detect any interaction with free GFP (Fig. 6A). In contrast, BIK1-HA was able to interact with BRI1-mCit (Fig. 6A), consistent with previous reports (38). Strikingly, BIK1-HA showed a much stronger interaction with the SUMO-defective BRI12KR (Fig. 6A). Considering that BRI1 SUMO levels are lower at 26 °C, we sought to determine the influence of temperature elevation on the BRI1-BIK1 interaction. BRI1 immunoprecipitates recovered higher BIK1 protein levels at 26 °C compared to 21 °C (Fig. 6B), consistent with the fact that BIK1 shows stronger interaction with the non-SUMOylatable BRI12KR variant. This evidence indicates that, similarly to what was observed for FLS2 (30), SUMOylation reduces the interaction of BRI1 with the downstream kinase BIK1. Genetically, BIK1 negatively regulates BR signaling, with bik1 mutant showing increased BR responses even upon high concentration of the BRZ BR biosynthetic inhibitor (38). To further illustrate the contribution of BRI1 SUMOylation to temperature responses and determine the possible role of BIK1 in this process, we phenotyped the previously published bik1 loss-of-function mutant at 21 °C and 26 °C. In contrast to the mild increase in hypocotyl length observed in WT seedlings, bik1 hypocotyls dramatically elongated in response to heightened temperature (Fig. 6 C–E), illustrating that BIK1 is a negative regulator of plant responses to warmth. A bik1/BIK1::BIK1-HA line, which complements the bik1 BR phenotypes (38), did not show such hypersensitivity to increased temperature (SI Appendix, Fig. S6 A and B). Altogether, these observations indicate that BIK1 is recruited to deSUMOylated BRI1 when plants experience increased temperature to limit growth responses.
Fig. 6.
Desi3a-dependent deSUMOylation regulates BRI1 interaction with BIK1. A. In vivo interaction between BIK1 and WT BRI1 or BRI12KR. BIK1-HA was co-expressed with BRI1-mCit or BRI12KR-mCit in N. benthamiana leaves prior to immunoprecipitation using anti-HA antibodies on solubilized protein extracts. GFP alone was used as negative control. Detection of immunoprecipitated proteins used the anti-GFP and anti-HA antibodies. B. In vivo interaction between BIK1 and BRI1 at 21 °C or 26 °C. BIK1-HA and BRI1-mCit were transiently expressed in N. benthamiana and incubated at 21 °C or 26 °C for 2 d prior to immunoprecipitation using anti-HA antibodies. GFP alone was used as negative control. Detection of immunoprecipitated proteins used the anti-GFP and anti-HA antibodies. C. Phenotype of 6-d-old WT and bik1 mutant plants grown at 21 °C or 26 °C. Representative pictures are shown. D. Hypocotyl length of 6-d-old WT and bik1 mutant plants grown at 21 °C or 26 °C. Experiments were carried out in triplicates. Error bars represent SD (n = 40). The asterisks indicate a statistically significant difference between WT and bik1 (two-way ANOVA with Sidak’s multiple comparison test). E. Normalized hypocotyl length from WT and bik1 mutant plants grown at 21 °C and 26 °C for 6 d, relative to 21 °C. Experiments were carried out in triplicates. Error bars represent SD (n = 40). The asterisks indicate a statistically significant difference between WT and bik1 (two-way ANOVA with Sidak’s multiple comparison test).
Discussion
The BR receptor BRI1 serves as a model to grasp the intricate mechanisms of RLK-mediated signaling in plants (39). The activity of BRI1 is regulated by post-translational modifications required to initiate, amplify, or dampen BR signaling. Phosphorylation between BRI1, the SERK co-receptors, and different downstream receptor-like cytoplasmic kinases mostly activates BR signaling (39), while ubiquitination leads to BRI1 degradation and signal attenuation (25, 26). Several endogenous and exogenous cues also converge at the level of BRI1 protein to influence BR-dependent growth. For example, glucose was shown to increase BRI1 endocytosis to control, in part, changes in root architecture associated with fluctuations in light intensity and photosynthetic activity (40). Similarly, elevation of ambient temperature impinges on BR-dependent root growth by triggering BRI1 destabilization (27). We show here that BRI1 is subjected to SUMO/deSUMOylation and that this is also required to control plant responses to increased temperature. BRI1 is decorated with SUMO under standard conditions and deSUMOylated by the Desi3a SUMO protease when plants are grown at warmer temperature (SI Appendix, Fig. S7). Desi3a-mediated BRI1 deSUMOylation favors BRI1 interaction with the negative regulator of BR signaling BIK1 and also promotes BRI1 endocytosis. desi3a and bik1 mutants both overrespond to heightened temperature, indicating that both Desi3a and BIK1 act as negative regulators of temperature responses. Thus, our work revealed a new role for SUMO in the regulation of BR signaling, in addition to the SUMO/deSUMOylation of the BZR1, BES1, and CESTA (CES) BR transcription factors (32, 41, 42), and also highlights an original mechanism to integrate temperature input into BR-dependent growth responses.
Heat stress triggers growth repression in plants and has been associated with an increase in global levels of SUMO conjugates (43). In contrast, exposure to elevated ambient temperature boosts plant growth, and here we show that this is accompanied with reduced BRI1 SUMOylation. The major driver of the elongation observed when plants face warmth is the phytohormone auxin (44). Temperature elevation reduces PHYTOCHROME B activity and induces PHYTOCHROME INTERACTING FACTOR4 (PIF4) expression to increase auxin biosynthesis (45–52). PIF4, as well as other PIFs, directly binds to the promoters of auxin biosynthesis genes, such as YUCCA8 (YUC8) and YUC9, TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1, and CYTOCHROME P450 FAMILY79B to increase auxin-responsive gene expression and tissue elongation (45, 46, 48, 50–53). BRs were also reported to participate to warmer temperature response in both aerial and underground tissues, although their contribution is opposite in both organs where BRs are known to differentially regulate growth (14, 54). In roots, temperature elevation promotes BRI1 destabilization to down-regulate BR signaling and to stimulate root elongation (27). In aerial parts, PIF4 boosts BR biosynthesis and BZR1 binds to the promoter of PIF4 at elevated temperature to increase its expression (55, 56), thus amplifying shoot transcriptional responses to heightened temperature and promoting hypocotyl elongation. Our findings now shed light on another level of control of BR signaling by temperature in aerial parts, with increased temperature promoting BIK1 recruitment to the BR receptor complex and decreasing BRI1 levels through Desi3a-mediated BRI1 deSUMOylation (SI Appendix, Fig. S7). This new layer of integration between BR signaling and temperature elevation negatively regulates temperature-dependent growth responses, as loss of Desi3a or BIK1 yields increased hypocotyl elongation upon heightened temperature. Auxin and BRs are well known to act synergistically to promote the transcription of growth-promoting genes and cell elongation (57). The SUMO-dependent regulation of BRI1 we have uncovered likely allows plants to dampen BR signaling to balance the synergistic effect between auxin and BRs and to attenuate the BR-dependent feedforward loop activating PIF4, thus preventing plants from over-elongating upon warm temperature.
The precise molecular mechanisms underlying this new regulatory level are still unclear. SUMOylation may have a direct inhibitory role on BRI1 endocytosis so that Desi3a-triggered BRI1 deSUMOylation at 26 °C increases internalization of BRI1. There are few reports describing a role for SUMO in inhibiting endocytosis. For example, SUMOylation of the TRANSIENT RECEPTOR POTENTIAL CATION CHANNEL SUBFAMILLY M4 (TRPM4) Ca2+-activated nonselective cation channel impairs TRPM4 endocytosis and leads to elevated TRPM4 density at the cell surface (58). Alternatively, SUMO/deSUMOylation of BRI1 may indirectly impact on BRI1 dynamics via the interplay between SUMO and Ub in the control of BRI1 endocytosis. BRI1 internalization from the cell surface is driven by massive ubiquitination decorating many cytosol-exposed lysine residues through the PUB12 and PUB13 E3 Ub ligases (25, 26). Additionally, elevated ambient temperature was shown in roots to destabilize BRI1 in an ubiquitin-dependent manner (27). Similarly to Ub, SUMO is covalently attached to proteins using lysine indicating that SUMO modifications potentially compete with Ub for the same sites to alter protein functions (59). During NF-κB activation for example, IκBα is ubiquitinated and degraded to release its inhibition of NF-κB (60). In contrast, SUMOylation at the same site prevents ubiquitination and turnover of IκBα (61), therefore inhibiting NF-κB activation. BRI1 SUMOylation at residues K1066 and K1118 may therefore limit its ubiquitination at standard growth temperature. The deSUMOylation of BRI1 observed at elevated temperature and driven by Desi3a would free additional lysine residues for ubiquitination, thus boosting Ub-mediated endocytosis of BRI1. No significant difference in the Ub profile of BRI1 could however be observed in the SUMO-defective BRI12KR form. The presence of many target lysines for Ub in BRI1 yields a large high molecular weight BRI1 Ub smear that likely masks the effect of lack of ubiquitination at K1066 and K1118 in BRI12KR. Consistently, the mutation of a single Ub target lysine in BRI1 identified by proteomics had no significant impact of the overall Ub profile of BRI1 (25). Considering the prominent role of Ub in endocytosis of PM proteins, we propose that temperature-regulated SUMO/deSUMOylation of BRI1 allows plant to fine-tune BRI1 Ub-mediated endocytosis and BR signaling (SI Appendix, Fig. S7). Whether PUB12 and PUB13 are responsible for the linkage of additional Ub chains to deSUMOylated BRI1 upon increased temperature or whether yet to be characterized temperature-regulated E3 Ub ligase is involved will have to be tackled in the future.
The FLS2 LRR-RLK flagellin receptor shows increased SUMOylation upon flagellin perception mediated by Desi3a degradation, thus releasing BIK1 from FLS2 (30). BIK1 being a positive regulator of FLS2 signaling, FLS2-SUMO conjugates positively regulate downstream innate immune signaling (30, 37). BRI1 and FLS2 are often compared since both represent model for the large plant LRR-RLK family. Despite their radically different biological outputs, BRI1- and FLS2-mediated signaling pathways share striking parallels. Both receptors predominantly localize at the PM where they bind their respective ligands and fire (62, 63). BRI1 and FLS2 both use the same subset of co-receptors to initiate signaling (8, 9, 64, 65). Structural and biochemical studies revealed that BRs and the flg22 flagellin peptide both act as molecular glue to form or stabilize signaling-competent receptor complexes (3, 6, 66). In both cases, ligand binding triggers cis- and trans-phosphorylation events within the receptor complexes to reach full activation (67, 68). Downstream of the receptor complexes is found RLCKs that are direct substrates of the receptor complexes (69). In particular, the RLCK BIK1 is shared between both pathways but is a positive regulator for immune responses and a negative regulator for BR signaling (37, 38, 70). BIK1 has been shown to negatively regulate BR signaling through direct association with BRI1 (38). After BR perception, BIK1 is phosphorylated by BRI1, causing its dissociation from the receptor. The precise function of BIK1 in BR signaling is therefore still unclear. Regardless, the Desi3a- and SUMO-regulated interaction of BIK1 with BRI1 and FLS2 now emerges as a possible common regulatory mechanisms of ligand-binding LRR-RLKs and corresponding signaling pathways. Although BRI1 and FLS2 have been shown to be confined to different nanodomains at the PM and the corresponding pathways to use different phosphocode in early phases (71), how signaling specificity is maintained along both pathways that rely on several shared components will deserve more attention in the future.
Material and Methods
Plant Material and Growth Conditions.
The genotypes used in this study are WT (Col0), bri1 T-DNA knockout (GABI_134E10) (19), BRI1::BRI1-mCit (19), desi3a-1 T-DNA knockout (SALK_151016C) (30), desi3a/Desi3a-HA (30), bik1 T-DNA knockout (SALK_005291) (37), bik1/BIK1::BIK1-HA (38), bes1-D (11), and 35S::JAZ6-GFP (29). BRI1K1066R, BRI1K1118R, and the BRI12KR variant carrying the K1066R and K1118R substitutions were generated by site-directed mutagenesis of the pDONR221-BRI1 (19) using the primers listed (SI Appendix, Table S1). Final destination vectors are obtained by recombination using the pB7m34GW destination vectors (72), the entry vectors pDONRP4P1r-BRI1prom (19), pDONR221-BRI1 or mutated BRI1 versions, and pDONRP2rP3-mCitrine (19).
After seed sterilization and stratification, seeds were placed for germination on solid agar plates containing half-strength Linsmaier and Skoog medium without sucrose. They were grown vertically in growth chambers under long-day conditions (16 h light/8 h dark, 90 μE m−2 s−1) at 21 °C or 26 °C. For the specific growth conditions, refer to figure legends.
Infiltration of N. benthamiana leaves was performed using standard procedures and used the binary vectors carrying BRI1::BRI1-mCit (19), 35S::Desi3a-mCh and FLS2::FLS2-GFP (30).
Chemical Treatments.
The final concentrations of the chemicals are indicated in the figure legends. The chemical stock solutions are in the following concentrations: 6 mM eBL (Sigma) in DMSO, 1 mM PPZ (Sigma), in water, 100 mM CHX (Sigma) in EtOH, and 10 mM BFA (Sigma) in DMSO.
Hypocotyl Measurements.
Seeds were sterilized and stratified for 4 d. For dark-grown hypocotyl measurements, seeds were exposed to the light for 6 h and then placed in dark at 21 °C or 26 °C for 3 d. For light-grown hypocotyl measurements, seeds were directly exposed to light at 21 °C or 26 °C for 6 d. Plates were scanned and hypocotyls measured using Fiji ImageJ software. The mean and SD were calculated by combining the three replicates.
Immunoprecipitation and Western Blot Analysis.
For detection of proteins from crude extracts, total proteins were extracted from ~50 mg plant material using Laemmli extraction buffer, using a 1:3 w/v ratio between tissue powder and extraction buffer. After debris elimination, proteins were separated by SDS-PAGE. Protein detection was carried out using peroxidase-coupled anti-HA-Peroxidase antibodies (Roche, dilution 1/4,000), peroxidase-coupled anti-GFP antibodies (Milteneyi, dilution 1/5,000), anti-SUMO1 antibodies (73), anti-BES1 antibodies (11), and anti-FBPase (Agrisera, dilution 1/5,000). To quantify the ratio between BES1 and P-BES1, signal intensity obtained with anti-BES1 antibodies and corresponding to BES1 and P-BES1 was determined using Image J. Western blot analyses were performed in triplicates. Representative blots are shown in figures. For the loading control using anti-FBPase antibodies, the same membranes were stripped and used.
Immunoprecipitation experiments were carried out as previously described (25), using the μMACS GFP and HA isolation kits (Miltenyi Biotec). Input and immunoprecipitated fractions were separated by SDS-PAGE and subjected to western blot analyses as described above.
Confocal Microscopy.
Dark-grown hypocotyls were treated with 100 µM CHX and 50 µM BFA for 15 min under vacuum before transfer to the corresponding temperatures prior to imaging. Hypocotyls were mounted in the same solution and imaged on a Leica SP8 confocal laser scanning microscopes (www.leica-microsystems.com). The 514-nm laser line was used to image BRI1-mCit. Laser intensity and detection settings were kept constant in individual sets of experiments to allow the direct comparison of fluorescence levels. To image entire hypocotyl cells, the TileScan and Z-stack option was used. Due to the length of hypocotyl cells, the fluorescence intensity of the total PM was not possible. Rather, the intensity of different region of interest of equivalent size and corresponding to the PM was measured. The PM/BFA body ratio corresponds to the mean fluorescence of the PM portions and the mean fluorescence of BFA bodies.
For localization and colocalization of transiently expressed proteins in N. benthamiana, the 488, 514, 561 nm laser line were used to image FLS2-GFP, BRI1-mCit, and Desi3a-mCh, respectively. Colocalization analyses and determination of the Manders’ coefficient, highlighting the fraction of GFP/mCit signals colocalizing with mCh, were carried out using the ImageJ plugin JACoP (74).
Statistical Analyses.
Data are shown as the average of three individual biological replicates, unless stated otherwise. Statistical analyses were performed with the software GraphPad Prism 7 software. Statistical significance of hypocotyl length between genotypes and/or conditions was assessed using one-way or two-way ANOVA with post hoc Sidak or Tukey post hoc tests. Experiments had at least n = 40 seedlings in each biological replicate. Quantification of western blots used the non-parametric Mann–Whitney (two genotypes/conditions) or Kruskal–Wallis (from three genotypes or conditions) tests. Statistical significance is defined as follows: *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001, and ****P ≤ 0.0001. Different letters indicate means that were statistically different by Tukey’s multiple testing method (P ≤ 0.05).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Cyril Zipfel, Libo Shan, Yanhai Yin, and Eugenia Russinova for sharing the bik1 mutant and bik1/BIK1::BIK1-HA complemented lines and anti-BES1 antibodies. We would also like to acknowledge the imaging facility from the Fédération de Recherche Agrobiosciences Interactions et Biodiversité of Toulouse (FRAIB). This work was supported by research grants from the French National Research Agency (ANR-17-CE20-0026-01 to G.V.) and the French Laboratory of Excellence (project “TULIP” grant nos. ANR–10–LABX–41 and ANR–11–IDEX–0002–02 to G.V.).
Author contributions
A.S. and G.V. designed research; M.N.-A., M.S., F.D., P.K.B., and M.M. performed research; M.N.-A., M.S., and A.S. analyzed data; and G.V. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. J.C. is a guest editor invited by the Editorial Board.
Contributor Information
Ari Sadanandom, Email: ari.sadanandom@durham.ac.uk.
Grégory Vert, Email: Gregory.Vert@univ-tlse3.fr.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Clouse S. D., Brassinosteroid signal transduction: From receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23, 1219–1230 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He Z., et al. , Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 288, 2360–2363 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Hothorn M., et al. , Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474, 467–471 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kinoshita T., et al. , Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433, 167–171 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Li J., Chory J., A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938 (1997). [DOI] [PubMed] [Google Scholar]
- 6.She J., et al. , Structural insight into brassinosteroid perception by BRI1. Nature 474, 472–476 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z. Y., Seto H., Fujioka S., Yoshida S., Chory J., BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410, 380–383 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Li J., et al. , BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Nam K. H., Li J., BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Wang Z. Y., et al. , Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2, 505–513 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Yin Y., et al. , BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109, 181–191 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Sun Y., et al. , Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19, 765–777 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu X., et al. , A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65, 634–646 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Clouse S. D., Langford M., McMorris T. C., A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111, 671–678 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J., Nagpal P., Vitart V., McMorris T. C., Chory J., A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272, 398–401 (1996). [DOI] [PubMed] [Google Scholar]
- 16.Szekeres M., et al. , Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171–182 (1996). [DOI] [PubMed] [Google Scholar]
- 17.Wang X., et al. , Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev. Cell 8, 855–865 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Wang X., Chory J., Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313, 1118–1122 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Jaillais Y., et al. , Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 25, 232–237 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oh M. H., Wang X., Clouse S. D., Huber S. C., Deactivation of the Arabidopsis BRASSINOSTEROID INSENSITIVE 1 (BRI1) receptor kinase by autophosphorylation within the glycine-rich loop. Proc. Natl. Acad. Sci. U.S.A. 109, 327–332 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stone J. M., Trotochaud A. E., Walker J. C., Clark S. E., Control of meristem development by CLAVATA1 receptor kinase and kinase-associated protein phosphatase interactions. Plant Physiol. 117, 1217–1225 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shah K., Russinova E., Gadella T. W. Jr., Willemse J., De Vries S. C., The Arabidopsis kinase-associated protein phosphatase controls internalization of the somatic embryogenesis receptor kinase 1. Genes Dev. 16, 1707–1720 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu G., et al. , Methylation of a phosphatase specifies dephosphorylation and degradation of activated brassinosteroid receptors. Sci. Signal 4, ra29 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geldner N., Hyman D. L., Wang X., Schumacher K., Chory J., Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 21, 1598–1602 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martins S., et al. , Internalization and vacuolar targeting of the brassinosteroid hormone receptor BRI1 are regulated by ubiquitination. Nat. Commun. 6, 6151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou J., et al. , Regulation of Arabidopsis brassinosteroid receptor BRI1 endocytosis and degradation by plant U-box PUB12/PUB13-mediated ubiquitination. Proc. Natl. Acad. Sci. U.S.A. 115, E1906–E1915 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martins S., et al. , Brassinosteroid signaling-dependent root responses to prolonged elevated ambient temperature. Nat. Commun. 8, 309 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vierstra R. D., The expanding universe of ubiquitin and ubiquitin-like modifiers. Plant Physiol. 160, 2–14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srivastava A. K., et al. , SUMO suppresses the activity of the Jasmonic acid receptor CORONATINE INSENSITIVE1. Plant Cell 30, 2099–2115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orosa B., et al. , SUMO conjugation to the pattern recognition receptor FLS2 triggers intracellular signalling in plant innate immunity. Nat. Commun. 9, 5185 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tozluoglu M., Karaca E., Nussinov R., Haliloglu T., A mechanistic view of the role of E3 in sumoylation. PLoS Comput. Biol. 6, e1000913 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Srivastava M., et al. , SUMO conjugation to BZR1 enables Brassinosteroid signallint to integrate environmental cues to shape plant growth. Curr. Biol. 30, 1410–1423 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fei Q., et al. , Effects of auxin and ethylene on root growth adaptation to different ambient temperatures in Arabidopsis. Plant Sci. 281, 159–172 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Rakusova H., et al. , Termination of shoot gravitropic responses by auxin feedback on PIN3 polarity. Curr. Biol. 26, 3026–3032 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Robinson D. G., Jiang L., Schumacher K., The endosomal system of plants: Charting new and familiar territories. Plant Physiol. 147, 1482–1492 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Di Rubbo S., et al. , The clathrin adaptor complex AP-2 mediates endocytosis of brassinosteroid insensitive1 in Arabidopsis. Plant Cell 25, 2986–2997 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu D., et al. , A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. U.S.A. 107, 496–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin W., et al. , Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor-like cytoplasmic kinase BIK1. Proc. Natl. Acad. Sci. U.S.A. 110, 12114–12119 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Belkhadir Y., Jaillais Y., The molecular circuitry of brassinosteroid signaling. New Phytol. 206, 522–540 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Singh M., Gupta A., Laxmi A., Glucose control of root growth direction in Arabidopsis thaliana. J. Exp. Bot. 65, 2981–2993 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khan M., et al. , Interplay between phosphorylation and SUMOylation events determines CESTA protein fate in brassinosteroid signalling. Nat. Commun. 5, 4687 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang L., et al. , Sumoylation of BRI1-EMS-SUPPRESSOR 1 (BES1) by the SUMO E3 ligase SIZ1 negatively regulates Brassinosteroids signaling in Arabidopsis thaliana. Plant Cell Physiol. 60, 2282–2292 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Kurepa J., et al. , The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J. Biol. Chem. 278, 6862–6872 (2003). [DOI] [PubMed] [Google Scholar]
- 44.de Lima C. F. F., Kleine-Vehn J., De Smet I., Feraru E., Getting to the root of belowground high temperature responses in plants. J. Exp. Bot. 10, 202 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Fiorucci A. S., et al. , PHYTOCHROME INTERACTING FACTOR 7 is important for early responses to elevated temperature in Arabidopsis seedlings. New Phytol. 226, 50–58 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gray W. M., Ostin A., Sandberg G., Romano C. P., Estelle M., High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 95, 7197–7202 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung J. H., et al. , Phytochromes function as thermosensors in Arabidopsis. Science 354, 886–889 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Koini M. A., et al. , High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 19, 408–413 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Legris M., et al. , Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354, 897–900 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Stavang J. A., et al. , Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J. 60, 589–601 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Sun J., Qi L., Li Y., Chu J., Li C., PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS Genet. 8, e1002594 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Franklin K. A., et al. , Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. U.S.A. 108, 20231–20235 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chung B. Y. W., et al. , An RNA thermoswitch regulates daytime growth in Arabidopsis. Nat. Plants 6, 522–532 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gonzalez-Garcia M. P., et al. , Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138, 849–859 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Ibanez C., et al. , Brassinosteroids dominate hormonal regulation of plant thermomorphogenesis via BZR1. Curr. Biol. 28, 303–310.e303 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Martinez C., et al. , PIF4-induced BR synthesis is critical to diurnal and thermomorphogenic growth. EMBO J. 37, e99552 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nemhauser J. L., Mockler T. C., Chory J., Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2, E258 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kruse M., et al. , Impaired endocytosis of the ion channel TRPM4 is associated with human progressive familial heart block type I. J. Clin. Invest. 119, 2737–2744 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cuijpers S. A. G., Willemstein E., Vertegaal A. C. O., Converging Small Ubiquitin-like Modifier (SUMO) and ubiquitin signaling: Improved methodology identifies co-modified target proteins. Mol. Cell Proteomics 16, 2281–2295 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen J., Chen Z. J., Regulation of NF-kappaB by ubiquitination. Curr. Opin. Immunol. 25, 4–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Desterro J. M., Rodriguez M. S., Hay R. T., SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol. Cell 2, 233–239 (1998). [DOI] [PubMed] [Google Scholar]
- 62.Irani N. G., et al. , Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat. Chem. Biol. 8, 583–589 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Robatzek S., Chinchilla D., Boller T., Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20, 537–542 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chinchilla D., et al. , A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Heese A., et al. , The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. U.S.A. 104, 12217–12222 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun Y., et al. , Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342, 624–628 (2013). [DOI] [PubMed] [Google Scholar]
- 67.Schulze B., et al. , Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 285, 9444–9451 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang X., et al. , Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev. Cell 15, 220–235 (2008). [DOI] [PubMed] [Google Scholar]
- 69.Ortiz-Morea F. A., He P., Shan L., Russinova E., It takes two to tango - molecular links between plant immunity and brassinosteroid signalling. J. Cell Sci. 133, 246728 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang J., et al. , Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7, 290–301 (2010). [DOI] [PubMed] [Google Scholar]
- 71.Perraki A., et al. , Phosphocode-dependent functional dichotomy of a common co-receptor in plant signalling. Nature 561, 248–252 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Karimi M., Depicker A., Hilson P., Recombinational cloning with plant gateway vectors. Plant Physiol. 145, 1144–1154 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Conti L., et al. , Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell 20, 2894–2908 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bolte S., Cordelieres F. P., A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.