Significance
Multidrug-resistant pathogens, especially those that form polymicrobial biofilms, pose an emerging threat to human health. Here, we designed and tested short tryptophan-based, self-assembling antimicrobial peptides. The lead candidate, Peptide K6, showed bactericidal and antibiofilm activity in vitro and cleared a mixed biofilm-based infection in mice. This peptide warrants further development as an antibiotic candidate.
Keywords: antimicrobial peptides, nanostructures, oligopeptide antibiotics
Abstract
The emergence of multidrug-resistant bacterial pathogens is a growing threat to global public health. Here, we report the development and characterization of a panel of nine–amino acid residue synthetic peptides that display potent antibacterial activity and the ability to disrupt preestablished microbial biofilms. The lead peptide (Peptide K6) showed bactericidal activity against Pseudomonas aeruginosa and Staphylococcus aureus in culture and in monocultures and mixed biofilms in vitro. Biophysical analysis revealed that Peptide K6 self-assembled into nanostructured micelles that correlated with its strong antibiofilm activity. When surface displayed on the outer membrane protein LamB, two copies of the Peptide K6 were highly bactericidal to Escherichia coli. Peptide K6 rapidly increased the permeability of bacterial cells, and resistance to this toxic peptide occurred less quickly than that to the potent antibiotic gentamicin. Furthermore, we found that Peptide K6 was safe and effective in clearing mixed P. aeruginosa–S. aureus biofilms in a mouse model of persistent infection. Taken together, the properties of Peptide K6 suggest that it is a promising antibiotic candidate and that design of additional short peptides that form micelles represents a worthwhile approach for the development of antimicrobial agents.
Multidrug resistance (MDR) bacterial infection is a global public health problem driven by the increasing number of microorganisms that show intrinsic and acquired resistance to multiple different types of antibiotics. Many MDR pathogens form biofilms, surface-attached structures formed by communities of bacterial cells that are embedded in the heterogeneous extracellular matrix. These complex structures exacerbate antibiotic resistance in many ways that include altering the microenvironment where cells replicate, impairing the penetration of antibiotics into cells, and induction of highly drug-resistant phenotypic states (1). Polymicrobial biofilms further promote the interspecies transfer of antibiotic resistance genes and can have synergic effects on drug resistance properties. For example, Pseudomonas aeruginosa and Staphylococcus aureus are frequently found together in chronic wounds and in the lungs of cystic fibrosis patients; these mixed infections are associated with increased disease severity and healthcare costs (2–4). S. aureus increases the resistance of P. aeruginosa to tobramycin when these two organisms are grown in mixed biofilms (5). In turn, P. aeruginosa can produce 2-n-heptyl-4-hydroxyquinoline N-oxide, a compound that protects S. aureus from the antimicrobial activity of vancomycin (6). Thus, new antimicrobials should ideally show activity not only against problematic antibiotic-resistant species but also against polymicrobial biofilms such as those formed by P. aeruginosa and S. aureus.
Antimicrobial peptides (AMPs) are an exciting category of therapeutic agents that show great promise in treating infections due to MDR organisms. However, the high cost of manufacturing synthetic peptides has slowed the development of clinically useful AMPs. AMPs generally contain 10 to 100 amino acid residues (7). Although shorter AMPs can be made at a lower cost, these are generally considered to have much weaker antimicrobial efficacy (8). In the APD3 Antimicrobial Peptide Database (9), 72 of 3,324 AMPs also display antibiofilm activity, but only 2 of these are less than 10 residues in length (10). Natural products such as polymyxins are modified peptides of 10 residues in length that are potent antibacterials but are now considered antibiotics of last resort because of their narrow spectrum of activity and relatively high toxicity (11). Thus, enhancing the potency, spectrum, and safety of synthetic AMPs is a major unmet challenge in the field.
AMPs that self-assemble into nanostructures can improve antimicrobial properties by extending the in vivo half-life of the peptide and increasing its local concentration (12–16). Recent studies reported that tryptophan (W) residues in short peptides promote the formation of self-assembled nanostructures (17, 18). Meanwhile, triple tryptophan (WWW) has been reported to be a potent motif that targets bacterial membranes (19, 20). In this study, we aimed to develop tryptophan-based short AMPs that self-assemble into nanostructures that show inhibitory activity against biofilm-forming P. aeruginosa and S. aureus either alone or in mixed infection models.
Here, we report the characterization of AMPs that were based on six center-symmetric, 9–amino acid peptides that contain WWW or double tryptophan (WW) motifs. Among the six peptides tested, one—Peptide K6—showed the most potent antibacterial activity against P. aeruginosa and S. aureus. Peptide K6 self-assembled into nanostructured micelles and exhibited potent antibiofilm activity against a P. aeruginosa–S. aureus polymicrobial biofilm formed in vitro. Moreover, Peptide K6 was safe and effective in clearing P. aeruginosa–S. aureus polymicrobial biofilm in a mouse infection model. The strong antibiotic and antibiofilm activities of Peptide K6 warrant its further development as a therapeutic.
Results
Design and Synthesis of Center-Symmetric, 9–Amino Acid Short Peptides with WWW or WW Motifs.
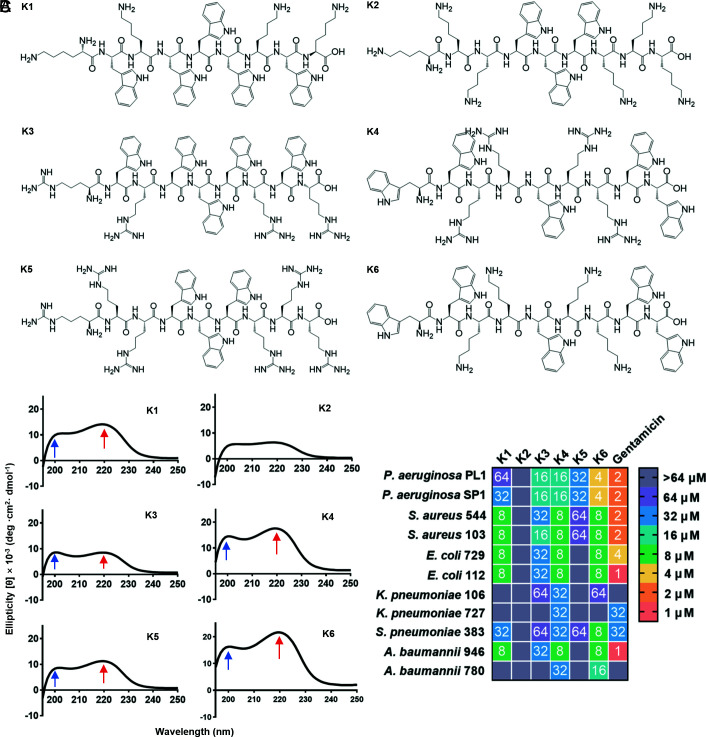
We designed peptides based on known structure–function relationships and principles of self-assembly and the following principles: 1) peptide length was set as short as 9 amino acids to keep synthetic costs low, and the amino acids were placed in a center-symmetric manner to keep the cytotoxicity against mammalian cells low (21); 2) hydrophobic residue percentage was set to 30% to 60% (which conforms to the characteristics of antibiofilm peptides in the Antimicrobial Peptide Database) to ensure that the peptides were amphiphilic when combined with polar residues; 3) WWW or WW motifs were placed in the middle or at the ends of the peptide to provide hydrophobicity and potentiate self-assembly; and 4) arginine or lysine residues were included to introduce positive net charges of +4 or +6. Based on this design rationale, six peptides, designated as Peptides K1–K6, were synthesized (Fig. 1A and SI Appendix, Table S1). All of the peptides were carboxyamidated to increase their stability and net charge. Their molecular weights were confirmed via matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (MALDI–TOF MS). The peptides were determined to be more than 95% pure by analytical reverse-phase (RP)-HPLC, and their measured molecular weights were in agreement with the predicted values (SI Appendix, Table S1).
Fig. 1.
Characteristics of peptides with nine residues. (A) Scheme chemical structural formula and sequence of the designed peptides. (B) Circular dichroism spectrum of Peptides K1–K6. (C) Minimal inhibitory concentration (MIC) values of peptides and gentamicin against bacteria isolated from clinical infection were determined as described in Materials and Methods and shown in a color code for the dilution of a standard stock of the indicated peptide (1 to 64 μM) that showed complete growth inhibition.
Circular Dichroism (CD) Spectra, Antibacterial Potency, and Antibacterial Spectrum of Peptides K1–K6.
Self-assembled short peptides were reported to have unique CD spectrum patterns (18, 22–24). Thus, we obtained the CD spectra of Peptides K1–K6. Briefly, 64 μM in 10 mM PBS of each peptide was treated by sonication to promote possible uniform self-assembled nanostructures. As shown in Fig. 1B, Peptides K1 and K3–K6 exhibited two positive bands with maxima at 200 nm and 220 nm, respectively (arrowed). This was not the case for Peptide K2. The spectra of Peptides K1 and K3–K6 were very similar to that observed by other researchers on a short peptide that self-assembled into nanostructures (22), suggesting that Peptides K1 and K3–K6 likely have self-assembly activities, while K2 is deficient in this property.
We next assessed the antibacterial activity of these peptides against bacterial species that cause MDR infections worldwide including P. aeruginosa, S. aureus, Escherichia coli, Klebsiella pneumoniae, Streptococcus pneumoniae, and Acinetobacter baumannii (25); some of these clinical isolates were multidrug resistant (SI Appendix, Table S2). Peptides K3, K4, and K6 exhibited the broadest antimicrobial spectra (Fig. 1C). There was some variability in the sensitivity of different clinical isolates of the same bacterial species. For example, K. pneumoniae strain 727 was resistant to Peptides K3 and K6, while strain 106 showed mild sensitivity to these two peptides. We speculate that changes in the composition of extracellular polysaccharides such as capsule polysaccharides or lipopolysaccharide O-antigen might account for such differences in sensitivity. Peptide K2 showed no antibacterial activity against any tested strains. These results suggest that the ability to form self-assembled nanostructures is important to the antibacterial potency and spectrum of these synthetic peptides (Fig. 1B) (26).
Peptide K6 Shows Bactericidal Activity against P. aeruginosa and S. aureus in Monoculture or Mixed Coculture Formats.
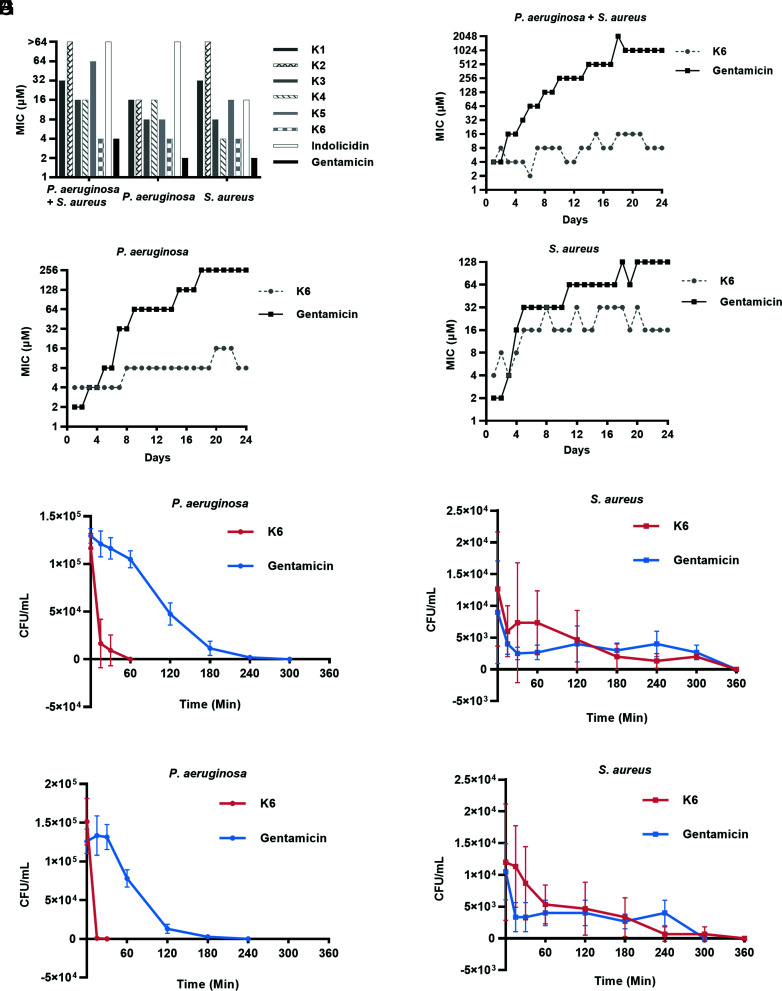
Given reports that interactions between P. aeruginosa and S. aureus promote antibiotic resistance (27, 28), we next tested the antimicrobial activity of the six peptides against P. aeruginosa PAO-1, S. aureus SP1, and a mixture of the strains (P. aeruginosa + S. aureus) along with two positive controls, indolicidin, and gentamicin. Indolicidin is a 13–amino acid, tryptophan-rich AMP, which shares similarities with Peptides K1–K6 (SI Appendix, Fig. S1), and was reported to show potent inhibitory activity against both P. aeruginosa and S. aureus (29, 30). However, indolicidin did not have a high antimicrobial activity (Minimal inhibitory concentration (MIC) > 64 μM) against P. aeruginosa (Fig. 2A), which may be explained by strain variations and differences in experimental conditions. Among all peptides, Peptide K6 exhibited the highest antimicrobial activity, which was close to that of gentamicin. As such, we decided to focus on Peptide K6 in the follow-up studies.
Fig. 2.
Peptide K6 exhibited high antimicrobial activity and low potential to develop resistance. (A) Minimal inhibitory concentration (MIC) of peptides and antibiotics against single bacterium/mixed bacteria. (B–D) Resistance development of mixed bacteria (B), P. aeruginosa (C), and S. aureus (D) to Peptide K6 and the antibiotic gentamicin. (E–H) Time course of Peptide K6 killing at its MIC against P. aeruginosa (E) and S. aureus (F) in mixed cultures; Peptide K6 at 2× its MIC against P. aeruginosa (G) and S. aureus (H) in mixed cultures.
AMPs have a unique advantage over antibiotics because they are less likely to induce resistance (31). We used a standard end point dilution assay monitoring growth for 24 d to determine whether P. aeruginosa, S. aureus, and P. aeruginosa + S. aureus develop resistance against Peptide K6. Starting from day 2, bacteria used in the MIC assay were subcultured from the culture grown at the highest sub-MIC of antimicrobial agents on the previous day. As shown in Fig. 2B, P. aeruginosa + S. aureus mixed cultures developed higher resistance (MIC = 1,024 μM at day 24) against gentamicin compared with the monocultures of either organism (MIC ≤ 256 μM) during the entire 24-d time course (Fig. 2 C and D); this result is consistent with the observed synergistic interactions between P. aeruginosa and S. aureus that apparently enhance antibiotic resistance in polymicrobial infections (28). Notably, all of P. aeruginosa, S. aureus, and P. aeruginosa + S. aureus remained sensitive (MIC ≤ 32 μM) to Peptide K6 during the entire 24-d period (Fig. 2 B–D), demonstrating a significant advantage of Peptide K6 over gentamicin regarding bacterial resistance development.
We carried out bactericidal killing assays to determine how rapidly Peptide K6 kills P. aeruginosa and S. aureus cells. At 1x MIC (4 μM), Peptide K6 killed all P. aeruginosa and S. aureus cells within 60 min and 360 min, respectively (Fig. 2 E and F). At 2× MIC (8 μM), Peptide K6 killed all P. aeruginosa and S. aureus cells within 30 min and 360 min, respectively (Fig. 2 G and H). Notably, Peptide K6 killed P. aeruginosa much more rapidly than S. aureus, suggesting that either the peptidoglycan layer of Gram-positive S. aureus may represent a barrier to penetration of Peptide K6 or alternatively, Peptide K6 attacks a uniquely Gram-negative target in P. aeruginosa such as its outer membrane or its lipopolysaccharide in addition to a target present in both bacterial species such as the inner membrane. Thus, Peptide K6 has bactericidal activity against P. aeruginosa and S. aureus individually or in mixed culture, and development of resistance to Peptide K6 occurred less rapidly than it did to the potent conventional antibiotic gentamicin.
Peptide K6 Self-Assembles into Nanostructured Micelles.
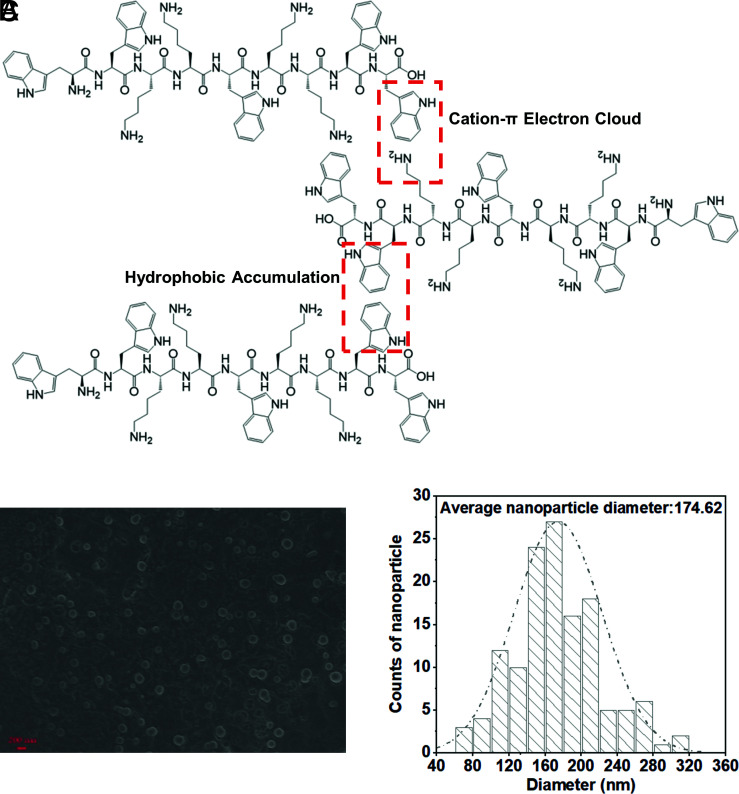
Recent studies reported that Trp residues in short peptides promote the formation of self-assembling nanostructures (17, 18, 32). Peptide K6 contains two WW motifs and two positively charged KK motifs at both ends that flank a central W residue. This configuration was predicted to form cation–π bonds and hydrophobic interactions between the W aromatic rings (Fig. 3A) that could lead to the self-assembly of a micelle-like structure through amphipathic interactions. Scanning electron microscopy (SEM) was used to test whether self-assembled Peptide K6 in PBS at pH 7.4 was detectable. Indeed, nanostructured micelles, with an average diameter of 174 ± 48.82 nm, were observed (Fig. 3 B and C). These observations are with the idea that the WW and KK motifs in Peptide K6 promote the self-assembly of micelle-like structures as predicted. In contrast, Peptide K2 was predicted to have the lowest level of self-assembly based on the CD spectrum (Fig. 1B), suggesting that the formation of nanostructured micelles correlates with bactericidal activity.
Fig. 3.
Peptide K6 can form nanoparticles. (A) Conceptual graphs with the main self-assembled driving force of Peptide K6. (B) SEM images of Peptide K6 were taken at its MIC for P. aeruginosa (4 μM). (C) The average size of Peptide K6.
Cell Surface Display of Peptide K6 Causes Disruption of the Gram-Negative Cell Envelope.
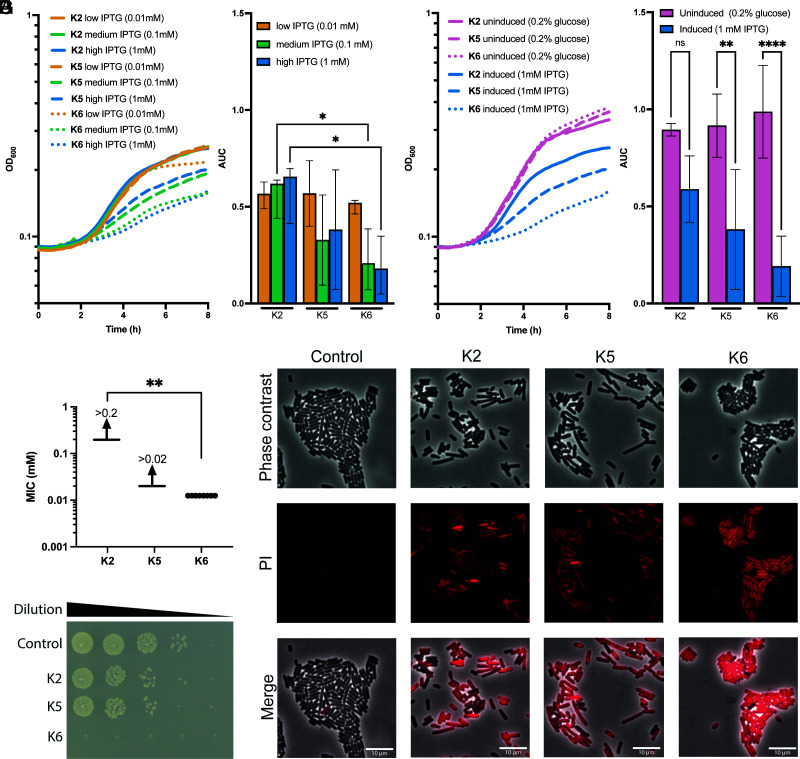
We sought an orthogonal approach to confirm the relative potency of our synthetic peptides by tethering them to a bacterial cell surface protein. For these experiments, we used a system developed by Brown (33) that allows display of tandem peptides on a surface-exposed loop of the LamB outer membrane protein of E. coli. LamB is a highly abundant protein in E. coli, with an estimated 30,000 molecules of this protein exposed on the cell surface (34). We displayed the Peptides K2, K5, and K6 in an extracellular surface–exposed loop of LamB by creating a genetic fusion where the respective fusion proteins are expressed under the control of the lac promoter and LacI system, which is IPTG inducible in E. coli. The three fusions contained 2 to 3 identical repeats of the Peptides K2, K5, or K6 displayed in tandem with identical spacer sequences. The LamB fusion protein displaying the Peptide K6 showed statistically more growth inhibition in an inducer-dependent fashion than the Peptide K2, while the Peptide K5 showed some inhibition that did not reach statistical significance (Fig. 4 A and B). These differences were further enhanced by comparing IPTG-induced cultures with those grown in glucose in order to repress the lac promoter (Fig. 4 C and D). We also performed assays to determine whether LamB fusions displaying Peptides K2, K5, and K6 were bactericidal after inducing their expression for 2 h (SI Appendix, Fig. S2) and found that the LamB–K6 fusion protein was approximately 100-fold more bactericidal than the LamB–K2 fusion protein. Furthermore, when tested as synthetic peptides, Peptide K6 was approximately 20-fold more toxic than Peptide K2 for E. coli in MIC assays (Fig. 4 E and F). To confirm that Peptide K6 was more toxic to E. coli than the Peptides K2 and K5, we also used imaging of cells stained with the dye propidium iodide (PI), which monitors the integrity of the cell membrane. Virtually, all E. coli cells exposed to Peptide K6 showed a profound loss of membrane integrity compared with the untreated cells or those exposed to the less potent Peptides K2 and K5 (Fig. 4G). Given these findings confirming that Peptide K6 had the most potent antimicrobial activity, we carried out further studies of this peptide as described below.
Fig. 4.
Peptide K6 showed bactericidal activity for E. coli when surface displayed on the outer membrane protein LamB or as a synthetic peptide. (A and B) Elevated expression of Peptide K6 reduces cell viability. Growth curves of E. coli OMNIMAX harboring an IPTG-inducible plasmid expressing K2, K5, or K6. Peptides are expressed as repeated sequences: K2 and K5 have 3 repeats, and K6 has 2 repeats. Linker sequences between repeats are LQTQATS. Cells were grown in LB medium supplemented with 25 µg/mL chloramphenicol and 0.01 mM, 0.1 mM, or 1 mM IPTG (A). Mean area under the curve (AUC) values ± SD retrieved from growth curves. Significant differences are indicated by an asterisk (∗P < 0.05; n = 4) (B). (C and D) Growth curves of E. coli OMNIMAX harboring an IPTG-inducible plasmid expressing K2, K5, and K6. Peptides are expressed as repeated sequences: K2 and K5 have 3 repeats, and K6 has 2 repeats. Linker sequences between repeats are LQTQATS. Cells were grown in LB medium supplemented with 25 µg/mL chloramphenicol 0.2% glucose (uninduced) or 1 mM IPTG (induced) (C). AUC values ± SD retrieved from growth curves. Significant differences are indicated by asterisks (∗P < 0.05; n = 4) (D). (E) Minimal inhibitory concentrations of the Peptides K2, K5, and K6 against E. coli OMNIMAX were determined. About 150 µL LB with 2-fold dilutions of peptides (starting at 0.02 mM for K5 and 0.2 mM for the others) were incubated with OD600 = 0.02 of E. coli OMNIMAX. Plates were incubated for 16 h at 37 °C with no agitation. No growth was determined as less than 50% OD600 of the control without peptides. Significant differences are calculated to a theoretical value for K2 of 0.21 (∗P < 0.05; n = 4). (F) Peptide K6 rapidly kills. 10-fold dilutions of E. coli OMNIMAX were plated on LB subsequent to 30 min of treatment with the respective peptides or PBS in PBS at 37 °C. (G) Synthetic Peptide K6 rapidly kills E. coli. Phase-contrast and propidium iodide (PI)–stained cells and a merged image are shown for E. coli cells treated with control (PBS) or 0.1 mM of Peptide K2 or K6 in PBS for 10 min. PI-positive cells lost membrane integrity and are considered nonviable.
Peptide K6 Has Antibiofilm Activity against Mixed P. aeruginosa–S. aureus Biofilms.
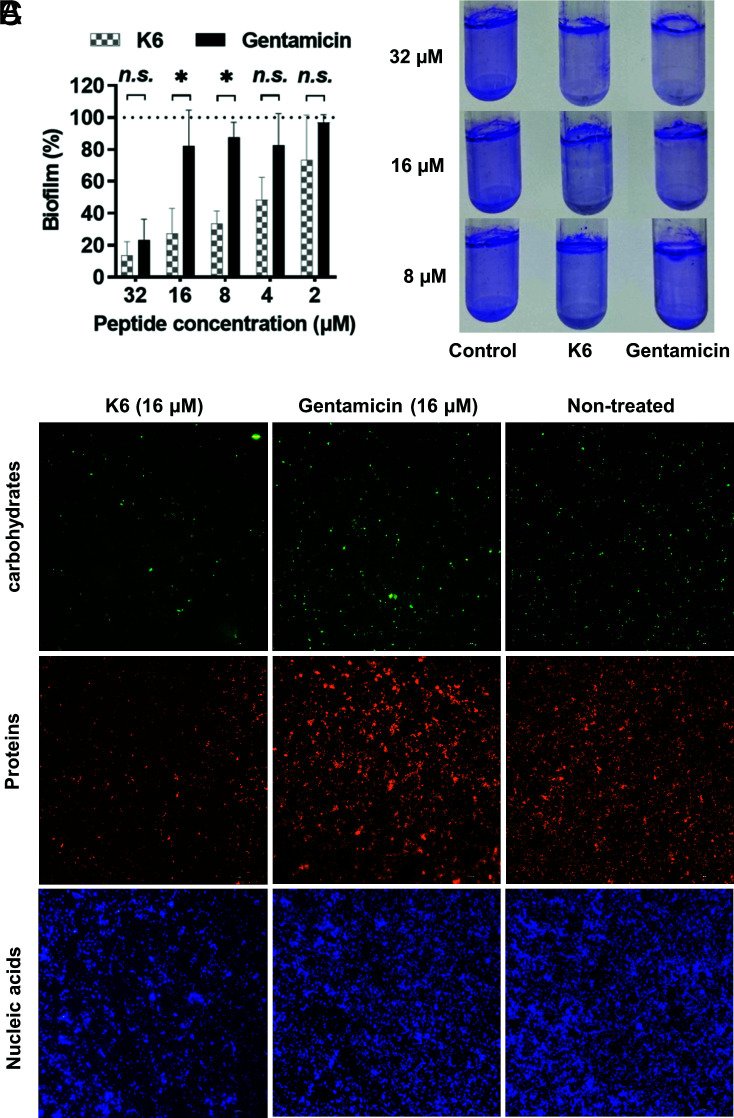
Some AMPs have a direct antibiofilm activity (35, 36). A biofilm clearance assay was used to test whether Peptide K6 has a direct antibiofilm activity. Although gentamicin shows antimicrobial activity against a mixture of planktonic P. aeruginosa–S. aureus (Fig. 2A), it does not have a direct antibiofilm activity and thus was useful as a control to assess the antibiofilm properties of Peptide K6. At concentrations <32 μM, gentamicin had little impact on preestablished mixed P. aeruginosa–S. aureus biofilms. In contrast, at concentrations of 8 to 16 μM, Peptide K6 largely removed the biofilm (Fig. 5 A and B), demonstrating that this micelle-forming peptide has biofilm-disrupting properties.
Fig. 5.
Peptide K6 exhibited higher antibiofilm activity than gentamicin with disruption of an extracellular matrix with nanoparticle structure. (A) Elimination of biofilm formation by Peptide K6 or gentamicin using polystyrene 96-well round-bottom plates. Results were expressed as the biofilm percentage, measured using crystal violet staining. (B) Elimination of biofilm formation by Peptide K6 or gentamicin determined by crystal violet staining using polystyrene round-bottom tubes. (C) Content of an extracellular matrix of established mixed biofilms treated with Peptide K6 or gentamicin. Results were measured using a fluorescence microscope.
Microbial biofilms consist of bacterial cells and an extracellular matrix composed of nucleic acids, carbohydrates, and proteins (37). We next sought to visualize these components in biofilms either untreated or treated with Peptide K6 or gentamicin. Specifically, Hoechst 33342, fluorescein isothiocyanate–labeled concanavalin A (FITC-ConA), and SYPRO orange were used to stain the nucleic acids, carbohydrates, and proteins, respectively. As shown in Fig. 5C, Peptide K6 removed a major portion of extracellular carbohydrates and proteins as well as nucleic acids compared with the untreated group, while gentamicin had little observable effect on their concentration in the biofilm. These results suggest that Peptide K6 was able to disperse biofilms more efficiently than gentamicin through its more potent ability to attack an extracellular biofilm matrix. Although Peptide K6 and gentamicin exhibited a similar MIC level against planktonic P. aeruginosa–S. aureus mixture, Peptide K6 was superior to gentamicin in its ability to disperse these two bacterial species, polymicrobial antibiofilm activity.
We also found that Peptide K6 was superior to gentamicin in its ability to inhibit de novo formation of a P. aeruginosa–S. aureus polymicrobial biofilm (SI Appendix, Fig. S3A). To provide additional insights into the antibiofilm activity of Peptide K6, we performed plate-based assays for swimming, swarming and twitching motility behaviors because these have previously been implicated in biofilm formation (38, 39). Peptide K6 significantly decreased all motility behaviors (SI Appendix, Fig. S3B), suggesting that the peptide might inhibit biofilm formation at sub-MIC concentrations by disrupting flagella and type IV pilus-dependent movement of P. aeruginosa cells.
Peptide K6 Disrupted the Integrity of Cell Membrane of P. aeruginosa and S. aureus.
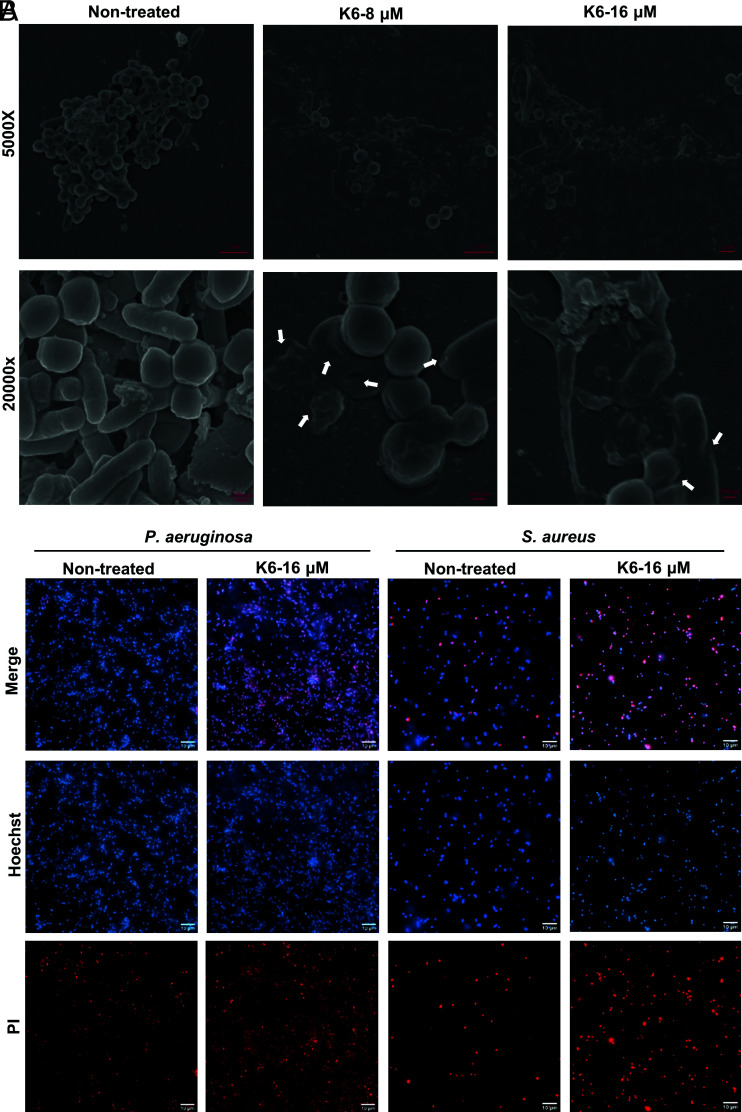
To investigate the cell biologic effects of Peptide K6, we used SEM to compare the appearance of bacterial cells in untreated biofilm samples to those treated with Peptide K6. In untreated samples, the bacilli (P. aeruginosa) and cocci (S. aureus) had intact cell structures. In contrast, Peptide K6 treatment led P. aeruginosa and S. aureus cells to appear shrunken (Fig. 6A), suggesting that the peptide disrupts the cell envelop integrity of both Gram-negative and Gram-positive bacterial cells.
Fig. 6.
Peptide K6 can permeabilize bacterial cell membranes in biofilms. (A) SEM images of established mixed biofilm treated with Peptide K6. (B) Live/dead fluorescence images of established P. aeruginosa or S. aureus biofilm treated with Peptide K6. Hoechst can stain live and dead cells and emits blue fluorescence, whereas PI only binds to DNA from dead cells and emits red fluorescence.
To test whether Peptide K6 causes an increase in the permeability of the bacterial cell membrane, we imaged live and dead cells in biofilm samples that were treated with Peptide K6 or PBS. All bacteria should stain with the cell membrane–permeable dye (Hoechst 33342, blue), while only dead bacteria should be stained with the nonpermeable fluorescent red dye PI. After 1 h, a larger proportion of both P. aeruginosa and S. aureus showed red fluorescence compared with the PBS group, demonstrating that Peptide K6 was able to disrupt the integrity of the cell membrane of both Gram-negative and Gram-positive bacteria (Fig. 6B).
Determination of Mammalian Cell Lytic and Cytotoxic Activity of the Peptide K6.
Low mammalian cell toxicity is a prerequisite for AMPs that are intended for clinical use. We carried out a hemolytic assay on mouse erythrocytes and a cytotoxicity assay on RAW264.7 mouse macrophages to assess the toxicity of Peptides K1–K6. In the hemolytic assay, at the concentrations tested (4 to 64 μM), Peptides K1–K6 exhibited less than 10% lysis of red blood cells (SI Appendix, Fig. S4), which is generally considered an acceptable level of hemolytic activity (32). In the cytotoxicity assay, at the concentrations tested (4 to 64 μM), all peptides except Peptide K5 exhibited less than 20% cytotoxicity against RAW264.7 macrophages, which is generally an acceptable level for this cell line (32). Given the properties of the Peptide K6 in our in vitro potency and biofilm disruption assays, this AMP was prioritized for testing in a mouse model of biofilm-based infection.
Peptide K6 Displayed Low Systemic Toxicity but Cleared a Mixed P. aeruginosa–S. aureus Infection in a Mouse Model of a Biofilm-Associated Infection.
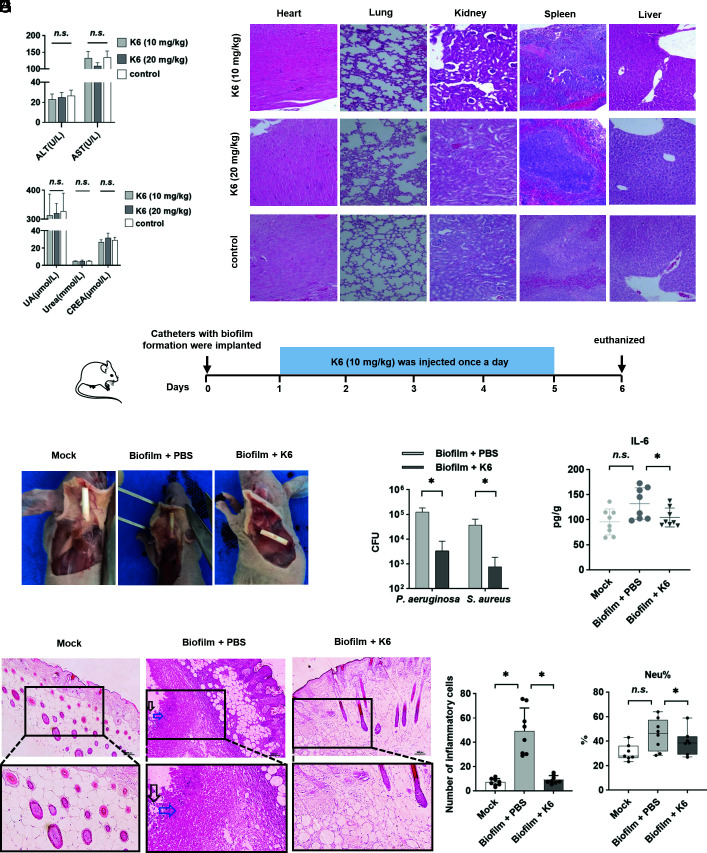
In order to test the in vivo safety of Peptide K6, 10 mg/kg or 20 mg/kg was subcutaneously injected into the skin on the back of mice. Serum levels of alanine aminotransferase (ALT), aspartate aminotransferase (AST), uric acid (UA), urea, and creatinine (CREA) from both doses were within normal range and did not differ from the control group, indicating that the tested concentrations of Peptide K6 did not cause acute damage to the liver or kidney (Fig. 7A). Furthermore, histological examination revealed no abnormalities in the heart, liver, spleen, kidneys, or lungs in all test groups (Fig. 7B), suggesting the Peptide K6 has little toxicity.
Fig. 7.
Peptide K6 was safe and efficient in clearing P. aeruginosa–S. aureus biofilm in mice. (A) Renal and liver function-related indexes reflecting nephrotoxic and hepatotoxic injury after intraperitoneal injection of 10 or 20 mg/kg Peptide K6, respectively. Error bars represented the SD from the mean of 3 biological replicates. (B) Histological analysis of the heart, lung, kidney, spleen, and liver tissues by H&E staining of the treatment groups that received different concentrations of Peptide K6. (C) Schematic of the experimental protocol for a biofilm-associated catheter mouse model. (D) Effects of Peptide K6 on the dorsal skin of mouse infected with the biofilm-associated catheter. (E) Bactericidal activity of Peptide K6 against persister cells derived from catheter biofilms. (F) Effects of Peptide K6 on the IL-6 level in the skin tissue. Data were shown as the mean ± SD (n = 8). (G) Histopathological H&E staining of the skin tissue. Black arrows indicated the inflammatory cell infiltrates, and blue arrows indicated necrotic cells. (H) Numbers of inflammatory cells in histopathological H&E staining of the skin tissue. Data were shown as the mean ± SD (n = 8). (I) Effects of Peptide K6 on the Neu% in the mouse serum. Data were shown as the mean ± SD (n = 8).
To assess the in vivo efficacy of Peptide K6 in the therapeutic treatment of an existing polymicrobial biofilm, a subcutaneous catheter mouse model was employed. In this infection model, 1-cm catheters were preincubated in a mixture of P. aeruginosa–S. aureus (or PBS as mock) for 24 h to allow the formation of P. aeruginosa–S. aureus polymicrobial biofilm on the catheters. The biofilm-coated catheters were then subcutaneously implanted into the back of nude mice, followed by injection of Peptide K6 or PBS once a day for 5 d (Fig. 7C). At day 6, Fig. 7D, the skin tissue of the Peptide K6 treatment group (biofilm + Peptide K6) recovered similarly to the noninfected control catheters (mock), while mice in the control group (treated only with PBS) displayed copious amounts of pus and tissue damage, consistent with ongoing infection. The bacteria adherent to each catheter were enumerated by determining CFU released after mild sonication of the implanted devices. As shown in Fig. 7E, Peptide K6 significantly reduced the number of both P. aeruginosa and S. aureus that colonized the catheters. Furthermore, the skin tissue adjacent to the catheters was collected and evaluated by histological staining or homogenized and analyzed for IL-6, a marker of acute inflammation. Compared with PBS controls, Peptide K6 treatment reduced the levels of IL-6 (Fig. 7F) and the numbers of infiltrated inflammatory cells (black arrows in Fig. 7 G and H) in the skin tissue. Moreover, in mice exposed to biofilm-coated catheters, Peptide K6 treatment reduced neutrophil levels in blood compared with untreated controls (Fig. 7I), suggesting that Peptide K6 treatment reduced systemic inflammation compared with the untreated group. Taken together, these findings demonstrate the in vivo efficacy of Peptide K6 in disrupting an established P. aeruginosa–S. aureus biofilm implanted on a foreign device in a murine model of polymicrobial infection.
Discussion
P. aeruginosa and S. aureus are often MDR pathogens associated with nosocomial and community-acquired infections. Both are particularly troublesome agents that can be deadly in patients with chronic obstructive pulmonary disease or cystic fibrosis (40). Immunosuppressed individuals and burn patients are also prone to lethal infections due to these organisms. Moreover, polymicrobial infections that include biofilms populated by P. aeruginosa and S. aureus frequently display enhanced drug resistance against many antibiotics (6, 28, 41). However, polymicrobial models are often overlooked for the development of antimicrobial agents (42). For example, the novel designed self-assembled AMPs mentioned in this paper all focused on the action on a single strain without considering the antimicrobial activity of multibacterial coexistence (32). Although activity of colistin–AMP combinations against P. aeruginosa and Staphylococcus double-species biofilms has been performed in vitro, in vivo activity studies are still lacking (43). Here, we employed both in vitro and in vivo polymicrobial models to screen for synthetic, AMPs that can target these two challenging MDR pathogens. It is known that changes in sequence permutation and amino acid composition in antimicrobial peptides can substantially alter their activity spectrum (44). Thus, peptide libraries can be extended and optimized to contain very potent antimicrobial compounds. In this study, we particularly focused on peptides that contained multiple tryptophan (W) motifs in that these have been reported to have an affinity for bacterial cell membranes and assemble into nanostructures (12–16) and designed a panel of short peptides with potential nanostructures and anti-polymicrobial biofilm activity. To obtain the ideal peptide, we synthesized peptides that had a high hydrophobic ratio (>50%) with a net charge of +4 in order to keep the charge and hydrophobicity in balance (45, 46) or a higher net positive charge of +6 with only 33.3% of its residues being hydrophobic. This composition of residues is similar to the peptide composition of polymyxin B, which is a 10-residue, cyclic natural product lipopeptide present in the library of 72 antibiofilm peptides.
During the screening of a panel of newly designed center-symmetric, 9–amino acid short peptides with WWW or WW motifs, we discovered that Peptides K3, K4, and K6 exhibited broad-spectrum antibacterial activity against all six leading bacterial species that are associated with worldwide MDR problem (Fig. 1C). While our charge and hydrophobic composition principles were successful for these 4 AMPs, others AMPs that had similar predicted properties were far less active. For example, Peptides K1, K2, and K5 showed low antimicrobial potency, and this suggested that the sequence of the AMP was also contributing to its antimicrobial potency perhaps by affecting higher-order secondary/tertiary conformations or oligomeric nanostructures.
Peptide K6 exhibited that the lowest MIC against P. aeruginosa and S. aureus was found to self-assemble into nanostructured micelles (Fig. 3C). Like Peptide K6, other octapeptides rich in tryptophan have recently been reported to form nanostructured micelles (32); however, the average diameter of Peptide K6 micelles we observed was much larger (174 nm vs. <40 nm) when compared with these previously described octapeptides. Furthermore, Peptide K6 was able to disrupt the integrity of bacterial cell membrane in as early as 1 h at a lower MIC concentration (Fig. 6B) than the previously described nanostructure peptides (47).
We also showed that a dimer of Peptide K6 was strongly bactericidal when expressed as an externally exposed loop on the LamB outer membrane protein of E. coli. This suggests that if properly localized, even two units of Peptide K6 might have potent bactericidal activity. We appreciate that this fusion protein may display toxicity during its transport through the inner membrane or during its insertion in the outer membrane as well. Nonetheless, the toxicity displayed by the LamB–Peptide K6 fusion protein may allow both the selection of modestly resistant mutants of E. coli and biochemical interaction studies that could together provide a better understanding of the mechanism of action and targets of the Peptide K6 AMP antibiotic for this bacterial species. Synthetic Peptide K6 was also highly toxic for E. coli compared with the Peptide K2. Further work will be needed to establish whether the self-assembly of the Peptide K6 into micelles is a key feature of the antimicrobial activity of Peptide K6 and its low toxicity toward mammalian cell lines.
Our studies also showed that it is less likely for P. aeruginosa and S. aureus to develop resistance against Peptide K6 compared with gentamicin (Fig. 2 B–D). This property is a potential advantage of Peptide K6 over the aminoglycoside antibiotic gentamicin, which can induce resistance in P. aeruginosa through the upregulation of drug efflux pumps. If Peptide K6 indeed works at the level of the bacterial cell envelop, then resistance due to drug efflux may be less of a worry in the clinical development of Peptide K6. Although cationic aminoglycoside antibiotics are potent, these broad-spectrum antibiotics suffer from side effects including nephrotoxicity when used in patients systemically. Thus, aerosolized formulations of gentamicin have been adopted because this drug is not absorbed through pulmonary mucosal surfaces. Despite the mildly cationic nature of Peptide K6, our preliminary analyses suggest that subcutaneous administration of Peptide K6 did not display either local (i.e., injection site) or systemic toxicity in the form of kidney or liver damage (Fig. 7 A and B). Furthermore, unlike gentamicin, Peptide K6 exhibited a direct antibiofilm activity (Fig. 5) in vitro at relatively low concentrations. This feature of Peptide K6 might provide another rationale for its further development as a clinically useful antibiotic particularly for MDR organisms.
Insertion of a peripheral venous catheter is one of the most common medical invasive procedures used in hospitals and is frequently associated with nosocomial infections as a result of bacterial colonization of these foreign bodies within patients (48, 49). We adopted a P. aeruginosa and S. aureus mixed biofilm-coated catheter model in mice (50) that allowed us to evaluate Peptide K6 for treatment efficacy of a polymicrobial infection under in vivo conditions. Our data showed that a dose of Peptide K6 at 10 mg/kg per day for 5 d significantly reduced the inflammation and bacterial load in the host, while no acute toxicity was observed (Fig. 7), demonstrating both in vivo safety and effectiveness of Peptide K6. Future studies will focus on determining the in vivo safety of the use of Peptide K6 for an extended time and investigating whether Peptide K6 composed of D-amino acids enhances its in vivo efficacy. Taken together, Peptide K6 showed strong antibiotic and antibiofilm activity against a model polymicrobial infection. Thus, Peptide K6 is a promising candidate for further evaluation and improvement as an antibiotic for MDR pathogens of clinical significance.
Materials and Methods
Bacterial Strains and Growth Conditions.
Bacterial strains (listed in SI Appendix, Table S2) were provided by the First Affiliated Hospital of Sun Yat-sen University, and the growth conditions were shown in SI Appendix, Materials and Methods.
AMP Synthesis.
Peptides were synthesized by GL Biochem Ltd. (Shanghai, China) using a solid-phase peptide synthesis strategy. The crude peptide was purified by RP-HPLC; the final purities of the peptide were >95%, and the molecular weights were analyzed by MALDI–TOF MS (Linear Scientific Inc., USA). Additional details are provided in SI Appendix, Materials and Methods.
SEM Characterization of Peptides.
For the preparation of SEM samples, 10 μL Peptide K6 concentration of 4 μM in PBS was dropped on tinfoil after sonicating for 30 min and sprayed with gold after drying. All samples were observed using a Gimini SEM 500.
Clearance of Established Biofilm Detection.
The clearing effect of Peptide K6 on a biofilm formed by mixed bacteria was detected by crystal violet staining, and solubilized crystal violet was detected spectrophotometrically at its absorbance maximum (595 nm). The change of extracellular polymeric substances present in preformed biofilm was determined by staining extracellular polymeric substances using Hoechst (0.5 μg/mL) for nucleic acids, SYPRO orange (diluted in 5,000 times) for proteins, and FITC-ConA (5 μg/mL) for carbohydrates and visualized by a fluorescence microscope (Eclipse Ti2-E; Nikon). Additional details are provided in SI Appendix, Materials and Methods.
Growth Curve Measurement of LamB Peptide Expressing E. coli.
Growth kinetics were essentially performed as previously described (51, 52). Briefly, E. coli OMNIMAX expressing peptides of interest as part of LamB were grown in a preculture for ~16 h in LB, 0.2% glucose with aeration, and shaking at 37 °C. Precultures were adjusted to OD600 = 0.05 in LB with 0.2% glucose or IPTG (0.01 mM, 0.1 mM, and 1 mM). The OD600 was monitored every 10 min in a microplate reader at 37 °C with shaking.
In Vivo Safety Assay.
Twelve six-week-old female athymic NU/NU nude mice (Crl:NU-Foxn1nu) purchased from Charles Rivers Laboratories (Beijing, China) were randomly divided into three groups (4 mice in each group). Blood was collected to detect the levels of ALT, AST, urea, CREA, and UA in the serum. Mouse organs (heart, liver, spleen, lung, and kidney) were stained with hematoxylin and eosin (H&E) for histological analysis. Additional details are provided in SI Appendix, Materials and Methods.
Subcutaneously Implanted Catheter Infection Model.
A model for biofilm-contaminated catheters was developed based on the observations of the catheter biofilm model infected by S. aureus (50). This animal study was performed according to protocols and guidelines approved by the Laboratory Animal Welfare and Ethics Committee of the Guangzhou Institute of Biomedicine and Health, Chinese Academy of Sciences (N2022002). A 1-cm central venous catheter with preformed biofilm was implanted into the skin on the back of the mice, and the wounds were sutured. After 24 h, PBS or Peptide K6 (10 mg/kg) was injected at the site of the implanted catheter in the back of the mice for five consecutive days. Mice were killed on the sixth day, and the catheter, EDTA anticoagulant blood, and skin tissue from the infection site were collected. Samples were processed in four parallel pipelines as shown in SI Appendix, Materials and Methods.
Detailed information on all other methods, including CD spectroscopy characterization, MIC measurement, bactericidal assay, inhibition of biofilm formation measurement, motility assay, SEM characterization of biofilms, fluorescence microscopy, hemolysis/cytotoxicity assay, and statistical analysis, is available in SI Appendix, Materials and Methods.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work has been supported by the National Key Research and Development Program of China (grant no. 2020YFA0907800) awarded to W.Z., the National Natural Science Foundation of China (grant no. 31900056 awarded to W.Z. and 32000096 awarded to X.M.), the Shenzhen Science and Technology Innovation Program (KQTD20200820145822023) awarded to W.Z. and X.M., and the Foshan Science and Technology Innovation Program (2120001010795) awarded to S.C. Work in the laboratory of M.K.W was supported by the HHMI. Work in the laboratory of J.J.M. was also supported by grant number AI-018045 from the National Institute of Allergy and Infectious Diseases (NIAID) to J.J.M. The contents of the manuscript describing the results of the study are solely the responsibility of the authors and do not necessarily represent the official views of the NIH and NIAID.
Author contributions
S.C., W.Z., J.J.M., and X.M. designed research; S.C., H.G., F.G.Z., S.Z., and J.T. performed research; B.X., Y.C., and P.C. contributed new reagents/analytic tools; S.C., H.G., F.G.Z., S.Z., and J.T. analyzed data; and S.C., H.G., F.G.Z., S.Z., J.T., M.K.W., W.Z., J.J.M., and X.M. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
Reviewers: B.W.D., The University of Texas at Austin; and W.S., The Forsyth Institute.
Contributor Information
Wenjing Zhao, Email: zhaowj29@ms.sysu.edu.cn.
John J. Mekalanos, Email: john_mekalanos@hms.harvard.edu.
Xiangyu Mou, Email: mouxy5@ms.sysu.edu.cn.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Stewart P. S., Costerton J. W., Antibiotic resistance of bacteria in biofilms. Lancet (London, England) 358, 135–138 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Yung D. B. Y., Sircombe K. J., Pletzer D., Friends or enemies? The complicated relationship between Pseudomonas aeruginosa and Staphylococcus aureus Mol. Microbiol. 116, 1–15 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Fischer A. J., et al. , Sustained coinfections with Staphylococcus aureus and pseudomonas aeruginosa in cystic fibrosis. Am. J. Respir. Crit. Care. Med. 203, 328–338 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ibberson C. B., et al. , Co-infecting microorganisms dramatically alter pathogen gene essentiality during polymicrobial infection. Nat. Microbiol. 2, 17079 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dehbashi S., Alikhani M. Y., Tahmasebi H., Arabestani M. R., The inhibitory effects of Staphylococcus aureus on the antibiotic susceptibility and virulence factors of Pseudomonas aeruginosa: A549 cell line model. AMB Express 11, 50 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giulia Orazi G. A. O. T., Pseudomonas aeruginosa alters Staphylococcus aureus sensitivity to vancomycin in a biofilm model of cystic fibrosis infection. mBio 8, e00873-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jhong J. H., et al. , dbAMP: An integrated resource for exploring antimicrobial peptides with functional activities and physicochemical properties on transcriptome and proteome data. Nucleic Acids Res. 47, D285–D297 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong N., et al. , Strand length-dependent antimicrobial activity and membrane-active mechanism of arginine- and valine-rich beta-hairpin-like antimicrobial peptides. Antimicrob. Agents Chemother 56, 2994–3003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang G., Li X., Wang Z., APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 44, D1087–1093 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lakshmaiah Narayana J., et al. , Two distinct amphipathic peptide antibiotics with systemic efficacy. Proc. Natl. Acad. Sci. U.S.A. 117, 19446–19454 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Satlin M. J., Jenkins S. G., "151–Polymyxins" in Infectious Diseases, Cohen J., Powderly W. G., Opal S. M., Eds. (Elsevier, ed. 4, 2017), pp. 1285–1288.e1282. [Google Scholar]
- 12.Xie X., et al. , Host-guest interaction driven peptide assembly into photoresponsive two-dimensional nanosheets with switchable antibacterial activity. CCS Chem. 3, 1949–1962 (2021). [Google Scholar]
- 13.Nilsson D. M. R. B. L., Multicomponent peptide assemblies. Chem. Soc. Rev. 47, 3659–3720 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang Y., Chen Y., Song Z., Tan Z., Cheng J., Recent advances in design of antimicrobial peptides and polypeptides toward clinical translation. Adv. Drug. Deliv. Rev. 170, 261–280 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Tan P., Fu H., Ma X., Design, optimization, and nanotechnology of antimicrobial peptides: From exploration to applications. Nano. Today 39, 101229 (2021). [Google Scholar]
- 16.Engelberg Y., Landau M., The Human LL-37(17–29) antimicrobial peptide reveals a functional supramolecular structure. Nat. Commun. 11, 3894 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhardwaj I., Jha D., Admane P., Panda A. K., Haridas V., Self-assembling tryptophan-based designer peptides as intracellular delivery vehicles. Bioorg. Med. Chem. Lett. 26, 672–676 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Zhang J., Liu S., Li H., Tian X., Li X., Tryptophan-based self-assembling peptides with bacterial flocculation and antimicrobial properties. Langmuir 36, 11316–11323 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Chou S., et al. , Peptides with triplet-tryptophan-pivot promoted pathogenic bacteria membrane defects. Front. Microbiol. 11, 537 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zarena D., Mishra B., Lushnikova T., Wang F., Wang G., The pi configuration of the WWW motif of a short Trp-rich peptide is critical for targeting bacterial membranes, disrupting preformed biofilms, and killing methicillin-resistant Staphylococcus aureus. Biochemistry 56, 4039–4043 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee H., Yang S., Shin S. Y., Improved cell selectivity of symmetric α-helical peptides derived from Trp-rich antimicrobial peptides. Bull. Korean Chem. Soc. 41, 930–936 (2020). [Google Scholar]
- 22.Gupta M., et al. , Self-assembly of a dipeptide- containing conformationally restricted dehydrophenylalanine residue to form ordered nanotubes. Adv. Mater. 19, 858–861 (2007). [Google Scholar]
- 23.Maity S., Das P., Reches M., Inversion of supramolecular chirality by sonication-induced organogelation. Sci. Rep. 5, 16365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo Z., et al. , Helix self-assembly behavior of amino acid-modified camptothecin prodrugs and its antitumor effect. ACS Appl. Mater. Interfaces 12, 7466–7476 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Anonymous, Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 399, 629–655 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu L., et al. , Antimicrobial activity and membrane-active mechanism of tryptophan zipper-like beta-hairpin antimicrobial peptides. Amino. Acids 47, 2385–2397 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Radlinski L., et al. , Pseudomonas aeruginosa exoproducts determine antibiotic efficacy against Staphylococcus aureus. PLoS Biol 15, e2003981 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fugère A., et al. , Interspecific small molecule interactions between clinical isolates of Pseudomonas aeruginosa and Staphylococcus aureus from adult cystic fibrosis patients. PLoS One 9, e86705 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Batista Araujo J., Sastre de Souza G., Lorenzon E. N., Indolicidin revisited: Biological activity, potential applications and perspectives of an antimicrobial peptide not yet fully explored. World J. Microbiol. Biotechnol. 38, 39 (2022). [DOI] [PubMed] [Google Scholar]
- 30.Mishra R., et al. , Natural anti-biofilm agents: Strategies to control biofilm-forming pathogens. Front. Microbiol. 11, 566325 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan H., et al. , Influence of molecular structure on the antimicrobial function of phenylenevinylene conjugated oligoelectrolytes. Chem. Sci. 7, 5714–5722 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fang Y., et al. , Biomaterial-interrelated bacterial sweeper: Simplified self-assembled octapeptides with double-layered trp zipper induces membrane destabilization and bacterial apoptosis-like death. Small Methods 5, e2101304 (2021). [DOI] [PubMed] [Google Scholar]
- 33.Brown S., Metal-recognition by repeating polypeptides. Nat. Biotechnol. 15, 269–272 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Gibbs K. A., et al. , Complex spatial distribution and dynamics of an abundant Escherichia coli outer membrane protein, LamB. Mol. Microbiol. 53, 1771–1783 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Dostert M., Trimble M. J., Hancock R. E. W., Antibiofilm peptides: Overcoming biofilm-related treatment failure. RSC Adv. 11, 2718–2728 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yasir M., Willcox M. D. P., Dutta D., Action of antimicrobial peptides against bacterial biofilms. Materials (Basel, Switzerland) 11, 2468 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park J. S., Choi H. Y., Kim W. G., The nitrite transporter facilitates biofilm formation via suppression of nitrite reductase and is a new antibiofilm target in Pseudomonas aeruginosa. mBio 11, e00878-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Felise H. B., et al. , An inhibitor of gram-negative bacterial virulence protein secretion. Cell Host. Microbe. 4, 325–336 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Treuner-Lange A., et al. , PilY1 and minor pilins form a complex priming the type IVa pilus in Myxococcus xanthus. Nat. Commun. 11, 5054 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pang Z., Raudonis R., Glick B. R., Lin T. J., Cheng Z., Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 37, 177–192 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Beaudoin T., et al. , Staphylococcus aureus interaction with Pseudomonas aeruginosa biofilm enhances tobramycin resistance. NPJ. Biofilms Microbiomes 3, 25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bottery M. J., Pitchford J. W., Friman V. P., Ecology and evolution of antimicrobial resistance in bacterial communities. ISME J. 15, 939–948 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jorge P., Grzywacz D., Kamysz W., Lourenco A., Pereira M. O., Searching for new strategies against biofilm infections: Colistin-AMP combinations against Pseudomonas aeruginosa and Staphylococcus aureus single- and double-species biofilms. PLoS One 12, e0174654 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mishra B., et al. , Sequence permutation generates peptides with different antimicrobial and antibiofilm activities. Pharmaceuticals (Basel, Switzerland) 13, 271 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu M., et al. , Molecular design and anti-melanoma activity of a novel bullfrog antibacterial peptide RGD-chimera. Oncol. Lett. 21, 115 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gunasekaran P., et al. , Synthesis of Fmoc-Triazine amino acids and its application in the synthesis of short antibacterial peptidomimetics. Int. J. Mol. Sci. 21, 3602 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Q., et al. , De novo design of a pH-triggered self-assembled beta-hairpin nanopeptide with the dual biological functions for antibacterial and entrapment. J. Nanobiotechnol. 19, 183 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Collet C., et al. , Effect of motor imagery and actual practice on learning professional medical skills. BMC Med. Educ. 21, 59 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skulec R., Callerova J., Vojtisek P., Cerny V., Two different techniques of ultrasound-guided peripheral venous catheter placement versus the traditional approach in the pre-hospital emergency setting: A randomized study. Int. Emergency Med. 15, 303–310 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Park S. C., et al. , Anti-biofilm effects of synthetic antimicrobial peptides against drug-resistant Pseudomonas aeruginosa and staphylococcus aureus planktonic cells and biofilm. Molecules 24, 4560 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seper A., et al. , Extracellular nucleases and extracellular DNA play important roles in Vibrio cholerae biofilm formation. Mol. Microbiol. 82, 1015–1037 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moisi M., et al. , Characterizing the hexose-6-phosphate transport system of Vibrio cholerae, a utilization system for carbon and phosphate sources. J. Bacteriol. 195, 1800–1808 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.