Significance
tRNA-derived small RNAs (tDRs) regulate gene expression at multiple levels. Some tDRs are abundant in human biofluids, where they are mostly present outside extracellular vesicles. This poses the intriguing question about how extracellular tDRs resist degradation by extracellular RNases. In this work, we solved this mystery by finding out that some of the most frequently detected extracellular tDRs are nicked forms of full-length tRNAs and are therefore dsRNA molecules. Nicked tRNAs cannot be studied by standard molecular biology and/or sequencing techniques because they are discontinuous. However, we developed several methods for the analysis of nicked tRNAs under native conditions and showed their presence in cells and biofluids. We also uncovered a potential intercellular communication pathway mediated by stable nonvesicular RNAs.
Keywords: extracellular RNA, liquid biopsies, tRNA halves, RNA stability, tRF
Abstract
Nonvesicular extracellular RNAs (nv-exRNAs) constitute the majority of the extracellular RNAome, but little is known about their stability, function, and potential use as disease biomarkers. Herein, we measured the stability of several naked RNAs when incubated in human serum, urine, and cerebrospinal fluid (CSF). We identified extracellularly produced tRNA-derived small RNAs (tDRs) with half-lives of several hours in CSF. Contrary to widespread assumptions, these intrinsically stable small RNAs are full-length tRNAs containing broken phosphodiester bonds (i.e., nicked tRNAs). Standard molecular biology protocols, including phenol-based RNA extraction and heat, induce the artifactual denaturation of nicked tRNAs and the consequent in vitro production of tDRs. Broken bonds are roadblocks for reverse transcriptases, preventing amplification and/or sequencing of nicked tRNAs in their native state. To solve this, we performed enzymatic repair of nicked tRNAs purified under native conditions, harnessing the intrinsic activity of phage and bacterial tRNA repair systems. Enzymatic repair regenerated an RNase R-resistant tRNA-sized band in northern blot and enabled RT-PCR amplification of full-length tRNAs. We also separated nicked tRNAs from tDRs by chromatographic methods under native conditions, identifying nicked tRNAs inside stressed cells and in vesicle-depleted human biofluids. Dissociation of nicked tRNAs produces single-stranded tDRs that can be spontaneously taken up by human epithelial cells, positioning stable nv-exRNAs as potentially relevant players in intercellular communication pathways.
Extracellular RNAs (exRNAs) circulating in human bodily fluids can enable disease diagnosis before the onset of clinical symptoms (1). Beyond applications as disease biomarkers in liquid biopsies (2), exRNAs are also involved in intercellular communication pathways between cells in different tissues (3) and in host–pathogen interactions (4–6).
One key aspect governing both exRNA functionality and utility as biomarkers is their stability against ubiquitous extracellular RNases (7). This can be achieved by RNA encapsulation inside extracellular vesicles (EVs) such as exosomes and microvesicles (8, 9). However, despite the functional relevance (10) and biotechnological applications (11) of EV-encapsulated RNA, the majority of exRNAs in cell culture (12–15) and in human plasma (15–18) are not transported as part of EV cargo.
Transfer RNA-derived RNAs (tDRs) are among the most abundant nonvesicular small RNAs (nv-exRNAs) in cell culture (12, 13). Inside cells, tRNA cleavage and the consequent generation of specific tDRs is a conserved response to stress in all kingdoms of life (19–21). In humans, RNase A superfamily members, such as RNase 5 (Angiogenin), are responsible for stress-induced tRNA cleavage at the anticodon, generating stress-induced tRNA halves (tiRNAs) (22, 23). tiRNAs can regulate gene expression at various levels, including global inhibition of translation initiation (24, 25). Other, shorter tDRs can bind to mRNAs and regulate their translation (26) or silence genes by a miRNA-like mechanism (27).
Extracellular tDRs were first reported in EVs from murine immune cells (28) but were later shown to be present mainly outside vesicles in mouse serum (29, 30). In human breast cancer cell lines, tDRs can be detected in vesicular fractions but are overwhelmingly more abundant in EV-depleted ultracentrifugation supernatants (12). While a heterogeneous population of tDRs is detectable inside cells, extracellular nonvesicular tDRs are mainly 5′ tRNA halves of 30 or 31 nucleotides, derived from tRNAGly and tRNAGlu. These specific fragments are also ubiquitous in human biofluids (31). A possible explanation for the extracellular enrichment of these fragments is their enhanced stability against degradation, because 5′ tRNAGlyGCC halves of 30 to 31 nt can form RNase-resistant homodimers in vitro (32).
To study exRNA processing in more detail, we added RNase inhibitors (RI) to cell-conditioned media and uncovered a population composed of nonvesicular ribosomes and full-length tRNAs (33). Knock-out of RNase 1 in K562 cells also shaped exRNA profiles from tRNA halves into full-length tRNAs (34). Thus, most nonvesicular tRNA halves are generated directly in the extracellular space by endonucleolytic cleavage of extracellular full-length tRNAs (33–35).
We have found that extracellular ribosomes can induce dendritic cell activation in vitro in an exRNA-dependent manner (33). However, the involvement of nonvesicular exRNAs (nv-exRNAs) in intercellular communication pathways faces an important conceptual challenge. As a consequence of the strong ribonuclease activity that characterizes the extracellular space (36), nv-exRNAs are expected to be rapidly degraded unless protected by RNA-binding proteins. How, then, do these RNAs resist degradation, diffuse to recipient cells, and trigger downstream effects, or even remain measurable and thus serve as potential disease biomarkers?
In this study, we incubated protein-free RNA or purified ribonucleoprotein complexes in human biofluids and measured their decay kinetics by northern blot. As expected, the protective effect of RNA-binding proteins was confirmed. Interestingly, though, we also found that certain specific naked tRNAs are intrinsically stable. Furthermore, the stability of 5′ tRNA halves generated from the endonucleolytic cleavage of unstable tRNAs was extremely high, with half-lives of hours in certain human biofluids. Also surprisingly, these ultra-stable RNAs are not single stranded. Developing and employing different strategies of enzymatic repair and electrophoretic or chromatographic separations under native conditions, we show that these RNAs natively exist as full-length tRNAs containing broken phosphodiester bonds. Commonly used RNA purification methods, sequencing, and denaturing northern blotting do not detect these forms, forcing them to melt into single-stranded tRNA halves. Nevertheless, at low rates, nicked tRNAs also seem to disassemble spontaneously into tRNA halves in the extracellular space. These extracellular nonvesicular tDRs are unstable, but they can be efficiently internalized by recipient cells.
Results
Identification of Naked RNAs that Are Stable in the Presence of Serum.
To search for intrinsically stable RNAs in extracellular samples, we first screened the most abundant cellular transcripts to see if, in the absence of their protein counterparts, they can resist degradation in serum-containing media (Fig. 1A). Unsurprisingly, naked rRNAs were degraded in less than one minute as judged by lack of northern blot signals in the absence of added RI. In contrast, and consistent with our previous observations (33), full-length tRNALysUUU was present at input levels after 1.5 h, suggesting that this tRNA is not efficiently targeted by serum RNases. This was not observed for other tRNAs like tRNAGlyGCC, which, like rRNAs, were undetectable after one minute (Fig. 1A).
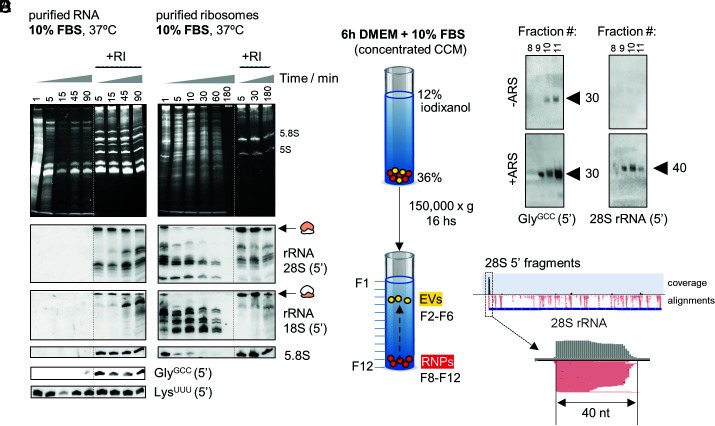
Fig. 1.
Identification of stable nonvesicular RNAs. (A and B) Northern blot of different rRNAs and tRNAs after incubating purified RNA (A) or ribosomes (B) from human cells in 10% FBS. (C and D) Northern blot of 5’ tRNAGlyGCC (D, Left) or 5′ 28S rRNA-derived fragments (D, Right) in extracellular nonvesicular fractions purified by density gradients (C). U2-OS cells were treated or not with NaAsO2 (ARS) before collecting the CCM. (E) Read coverage in small RNA-seq data of extracellular ribosomes [from Tosar et al. (33)], revealing enrichment of 40-nt 5′-derived small RNAs among all other 28S rRNA-derived fragments.
Ribosomal Proteins Stabilize rRNAs and Protect rRNA-Derived Fragments.
To assess the effect of RNA-binding proteins on the stability of nv-exRNAs, we purified whole ribosomes from cells (SI Appendix, Fig. S1) and studied rRNA decay kinetics in 10% FBS (Fig. 1B). In ribosomes, full-length rRNAs and rRNA-derived fragments were detectable for at least 10 and 60 min at 37 °C, respectively, even in the absence of RI. Thus, ribosomes are more stable than naked rRNAs and ribosomal proteins greatly stabilize extracellular rRNA-derived fragments.
Based on these results, we searched for nonvesicular rRNA-derived fragments in a cell-conditioned medium (CCM) obtained in the presence of 10% FBS. We separated EVs from RNP complexes by flotation on iodixanol density gradients (Fig. 1C). Extensive characterization of this methodology has shown that the densest fractions (F8-F12) are devoid of EVs (33, 37). We reproduced our previous findings (33) showing the presence of 5′ tRNAGlyGCC halves of 30 to 31 nt and the absence of full-length tRNAs in these nonvesicular fractions (Fig. 1D). Interestingly, northern blot detection of these fragments was enhanced when cells were treated with the cytotoxic agent sodium arsenite (Fig. 1D, +ARS). Additionally, using a probe complementary to the first 20 nucleotides of the 28S rRNA resulted in a single band of 40 nt in the conditioned medium of arsenite-treated cells. Purified extracellular ribosomes analyzed by small RNA-seq (33) also showed enrichment of these 40-nt 5′ fragments (Fig. 1E). Detection of these fragments in the presence of serum suggests that at least some stable RNA-containing RNPs are produced after the release and subsequent fragmentation of extracellular ribosomes, in agreement with recent findings (38).
Naked Full-Length tRNALysUUU Is Intrinsically Stable in Biofluids.
To precisely measure tRNA half-lives, we repeated the previous assay with better temporal resolution (Fig. 2A), confirming complete degradation of rRNAs and the 7SL RNA within seconds, while tRNALysUUU was still detectable after 1 h at 37 °C. The full-length tRNAGlyGCC was also rapidly degraded, generating fragments that lasted as much as the full-length tRNALysUUU. In fact, tRNALysUUU and tRNAGlyGCC constitute two extreme cases in a wide range of extracellular stabilities shown by different tRNAs (SI Appendix, Fig. S2A). In vitro incubation of RNA with recombinant human RNase 1 (r-RNase1), representing the most common RNase in human blood, resulted in virtually identical results (Fig. 2A). This observation argues against the possibility of tRNALysUUU being stabilized by the interaction with FBS-derived proteins or metabolites. Therefore, we conclude that naked tRNALysUUU is intrinsically resistant to the action of RNase A family members.
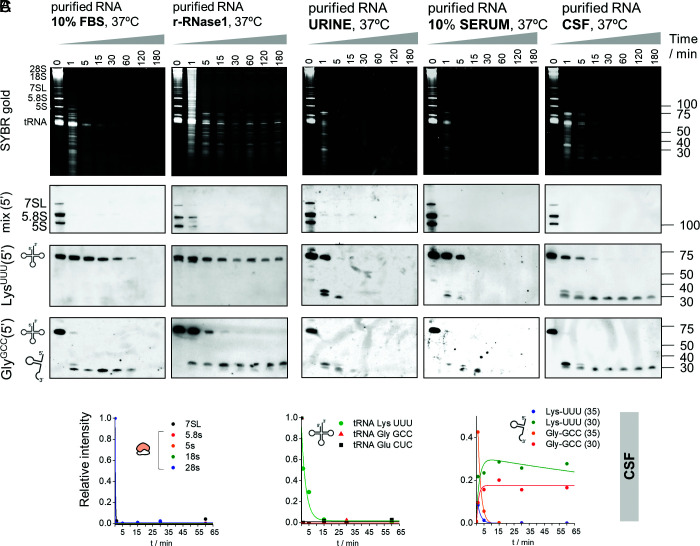
Fig. 2.
Naked tRNA halves are extremely stable in human biofluids. (A) Northern blot of several RNAs after incubating purified total RNA from human cells in 10% FBS or with recombinant human RNase 1 for different periods. (B) Samples were also incubated in human urine, 10% serum, and CSF. (C) Data were fitted to a kinetic decay model, as explained in SI Appendix, Supplementary Methods. Half-lives in each biofluid, calculated after inclusion of experimental replicates, are presented in SI Appendix, Fig. S3 B and C. r-RNase1: recombinant human RNase 1.
We then repeated these assays in human biofluids including urine, diluted and undiluted serum, and cerebrospinal fluid (CSF) (Fig. 2B and SI Appendix, Figs. S2B and S3A). Strikingly, the stability of full-length tRNALysUUU was always higher than the stability of any other tested RNA, irrespective of the sample type. RNA decay kinetics were highly consistent among independent replicates (SI Appendix, Fig. S2B).
Glycine tRNA Halves Produced in Biofluids Are Extremely Stable.
Full-length tRNAGlyGCC was almost completely degraded in less than one minute in 10% FBS (Fig. 2A) and in human biofluids (Fig. 2B). However, its cleavage resulted in the formation of 5′ halves that showed remarkable long half-lives, even in undiluted human sera (Fig. 2 A and B and SI Appendix, Fig. S3A). For instance, the half-life (t1/2) of 5′ tRNAGlyGCC halves of 30 nt in CSF was >2 h (Fig. 2C; 2 ± 1 h when replicates were combined, SI Appendix, Fig. S3 B and C), meaning that these RNAs are highly stable once they are generated and will therefore persist in biofluids.
A closer inspection of the data shows strong differences in the stability of fragments derived from the same parental tRNAs but with slightly different lengths. Consistent with our previous report (33), tRNAGlyGCC is first cleaved at the anticodon loop, generating 34 to 35-nt 5′ halves that rapidly disappear. These fragments are subsequently replaced by highly stable shorter fragments of approximately 30 to 31 nt (t1/2 = 6 ± 1, 8 ± 2, and 126 ± 65 min in urine, diluted serum and CSF, respectively, Fig. 2B), with a cleavage site at the start of the anticodon loop. An additional cleavage site in the TΨC loop (position 54) could be identified and exposed by using lower r-RNase1 concentrations (SI Appendix, Fig. S4A).
The sequential production of first longer (34 to 35 nt) and then shorter (30–31) 5′ halves from tRNAGlyGCC was also observed with recombinant human RNase 5 or Angiogenin (r-Ang; SI Appendix, Fig. S5). Thus, cleavage sites and tRNA processing dynamics seem to be conserved among different RNase A family members.
Overall, tRNAs are more resistant to degradation than other longer noncoding RNAs, but differences in stability among tRNAs (e.g., tRNALysUUU vs. tRNAGlyGCC) are also substantial (>30-fold in CSF). Consistent with our previous reports (32, 33), naked 5′ tRNAGlyGCC halves of 30 to 31 nt are accumulated at higher RNase concentrations or after longer incubations in biofluids and are extremely stable.
Nicked tRNAs Are a Source of 5’ and 3’ tRNA Halves.
Next, we compared the relative stabilities of 5′ and 3′ tRNAGlyGCC-derived fragments (SI Appendix, Fig. S6). Surprisingly, 30 to 35-nt 3′ fragments were observed, and their rate of decay was comparable to their 5′ counterparts. In certain biofluids, such as FBS (SI Appendix, Fig. S6A) and urine (SI Appendix, Fig. S6B), 3′ tRNA-derived fragments <20 nt were observed after short incubations (1 min; black asterisks). However, these fragments were rapidly degraded, in sharp contrast with the 5′ and 3′ halves.
We considered the possibility that 5′ and 3′ tRNAGlyGCC halves could remain physically associated with each other after RNase cleavage, representing a full-length tRNA bearing a cleaved phosphodiester bond at the anticodon loop (i.e., in the form of “nicked tRNAs”). This would explain the similar decay kinetics of each half among different biofluids. The introduction of any irreversible denaturation steps in analytical determinations, such as those used in standard molecular biology approaches, would induce dissociation of nicked tRNAs into single-stranded tRNA halves. Therefore, testing this hypothesis required the generation of new assays capable of interrogating oligomeric RNA complexes under native conditions.
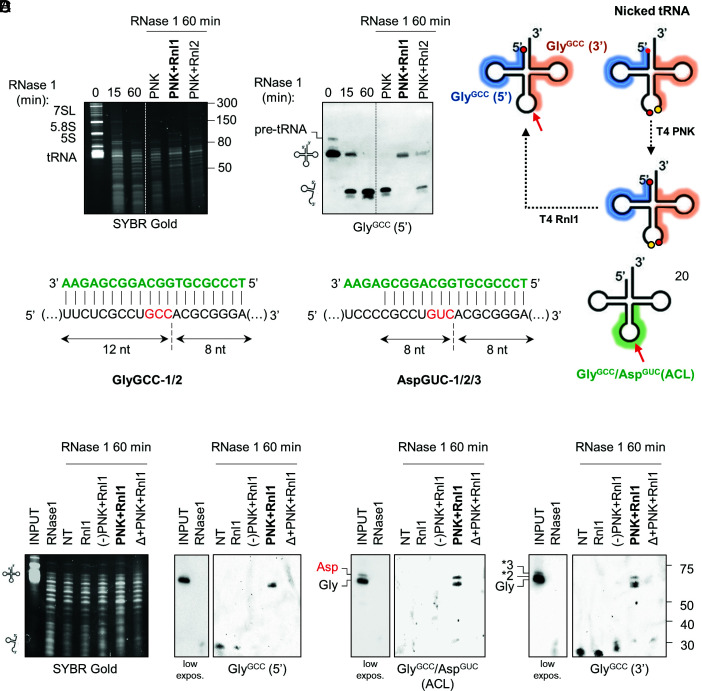
Nicked tRNAs are the natural substrates of T4 polynucleotide kinase (PNK) and T4 RNA ligase 1 (Rnl1) (39). When Escherichia coli is infected by the T4 Phage, a bacterial anticodon nuclease is activated and cleaves the host’s tRNALys in an attempt to prevent the translation of viral proteins (40). This results in a nicked tRNA bearing a 3′ cyclic phosphate (3′ cP) and a 5′-OH adjacent to the cleavage site. However, the phage evolved two enzymes capable of performing end-healing (PNK) and tRNA repair (Rnl1). We harnessed the intrinsic activity of these enzymes and used them to investigate the native structure of human tRNA halves in extracellular samples.
To optimize the system, we purified total RNA from cells and incubated the RNA with r-RNase1 for 0, 15, or 60 min (Fig. 3A). As expected, by 60 min, full-length tRNAGlyGCC was completely degraded and converted to a collection of tDRs (Fig. 3A). RNase 1 degradation products were purified by phenol-free silica-based solid phase extraction (SPE) columns and treated with either PNK alone, PNK followed by Rnl1, or PNK followed by Rnl2 (a dsRNA-specific ligase). Surprisingly, treatment with PNK and Rnl1 regenerated a single band of approximately the size of the cognate full-length tRNA (Fig. 3 A and B). Ligation with Rnl2 was less efficient.
Fig. 3.
Glycine tRNA halves identified by northern blot are nicked tRNAs. (A) RNase 1-treated RNA (60 min) was incubated with the indicated enzymatic combinations. Repair of tRNAGlyGCC was analyzed by northern blot using a 5′-targeting probe. (B) schematic representation of the nicked tRNA repair strategy and 5′ (blue) and 3′ (orange) probe binding sites in tRNAGlyGCC. (C) Design of a third probe targeting the anticodon loop (ACL) of tRNAGlyGCC and tRNAAspGUC. (D) enzymatic repair of tRNAGlyGCC evidenced with either the 5′, ACL or 3′ probes. NT: fragmented RNA purified by SPE but without treatment with the enzymatic repair cocktail. (-)PNK: mutant PNK lacking phosphatase activity. Δ: heat.
To demonstrate that the tRNA-sized religated products are indeed repaired tRNAs, we designed a third probe (termed ACL, for anticodon loop) bridging both sides of the anticodon of tRNAGlyGCC (Fig. 3C). This probe was designed so that the Tm of its pairing with either the 5′ or the 3′ half was below the hybridization temperature of the assay (42 °C). Thus, the ACL probe would fail to detect 5′ or 3′ tDRs but should be able to hybridize with full-length or repaired tRNAGlyGCC. Due to sequence similarities among tRNAs, ACL also recognizes the anticodon loop of tRNAAspGUC, but fortunately, these tRNAs migrate slightly differently in denaturing urea gels, making this assay multiplex. Strikingly, treatment of RNase 1-treated RNA with PNK and Rnl1 regenerated tRNA-sized products observable with the 5′, the 3′ or the ACL probes (Fig. 3D). Furthermore, heating (Δ) and then cooling the RNase-treated RNA before the addition of the enzymatic cocktail (Δ + PNK + Rnl1) prevented the generation of a tRNA-sized band, demonstrating that ligation occurs in cis under the assay conditions. Rnl1 alone or following incubation with a mutant version of T4 PNK lacking its 3′ phosphatase activity [(-)PNK] also failed to reconstitute a full-length tRNA.
Next, we studied whether tRNA halves generated in human biofluids (Fig. 2B) are in fact nicked tRNAs. Indeed, a short incubation in CSF (1 min) produced 5′ tRNAAspGUC halves that could be enzymatically repaired back into a full-length tRNA (SI Appendix, Fig. S7A). Enzymatic repair of tRNAGlyGCC produced after longer (1 h) incubations in CSF was also highly efficient (SI Appendix, Fig. S7B). However, the main religated product was approximately 11 nt shorter than the parental full-length tRNA, consistent with an irreversible loss of the entire anticodon loop (7 nt) plus the single-stranded NCCA 3′ overhang.
In summary, nicked tRNAs are produced in vitro and in biofluids using different tRNAs as substrates. These nicked tRNAs can be repaired enzymatically to regenerate an almost full-length tRNA, presumably lacking the 3′ NCCA overhang in in vitro settings (34, 41). Additional bases can be trimmed after prolonged exposure to extracellular RNases, but these shorter forms are still reparable using our assay.
Nicked tRNAs Are Irreversibly Melted by Popular RNA Extraction Methods.
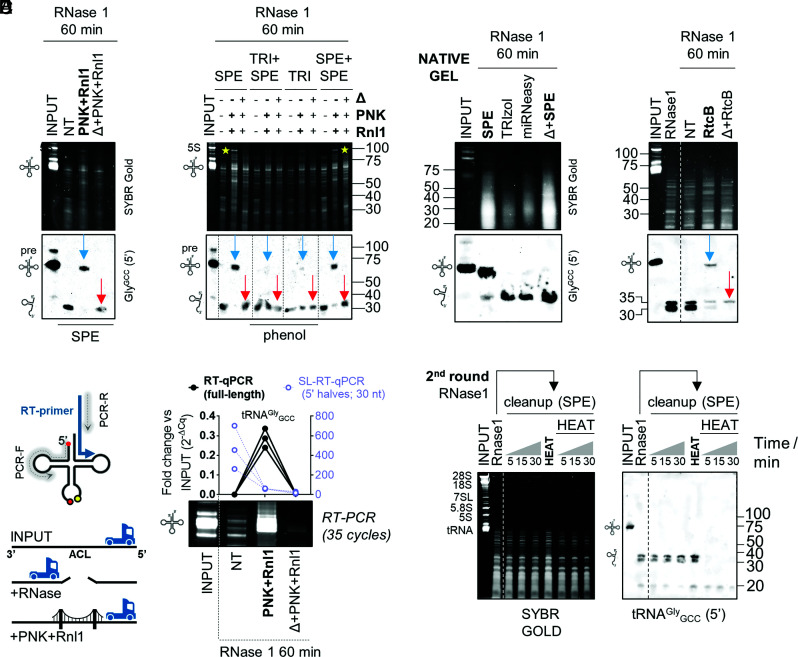
Whereas heating RNase-treated RNA before tandem incubation with PNK and Rnl1 prevented the formation of a tRNA-sized band, this also affected the detection of monomeric 5′ and 3′ tRNA halves (Fig. 3D; Δ + PNK + Rnl1 lane). However, when we repeated SPE purification under conditions optimized for small RNA recovery (SI Appendix, Fig. S7C), monomeric tRNA halves were now detectable in the control (i.e., heat) reaction (Fig. 4A; red arrows).
Fig. 4.
Nicked tRNAs are stable, dissociated by phenol, can be repaired by PNK+Rnl1 or by RtcB, and cannot be reverse-transcribed otherwise. (A) Enzymatic nicked tRNA repair under optimized SPE conditions. Blue arrow: repaired product (ligation in cis). Red arrow: single-stranded tRNA halves generated by heat before enzymatic treatment. (B) RNA purification by TRIzol (TRI) impairs enzymatic nicked tRNA repair. Yellow stars: other RNAs are also repaired (ligated in cis) by our protocol. (C) Northern blot of RNAse1-treated RNA purified by SPE, TRIzol, miRNeasy or heated before SPE purification, after separation in native gels. (D) Nicked tRNA repair with RtCB from E. coli. (E) RNase treatment generates a roadblock for thermostable reverse transcriptases (RT) at the anticodon loop (ACL), inhibiting RT-PCR amplification of nicked tRNAs. Treated samples were analyzed by end-point RT-PCR (SYBR gold-stained gel) or RT-qPCR for the full-length tRNAGlyGCC (black, Left axis) or by SL-RT-qPCR for 5′ tRNAGlyGCC halves of 30 nt (purple, Right axis). (F) Purification of RNase1-treated RNA by SPE and reexposure to RNase 1, with or without previous heat denaturation. Δ: heat.
Given that heat irreversibly affects nicked tRNAs, we studied the effects of phenol-based RNA extraction methods, because this chemical agent, like heat, can disrupt base-pairing interactions. A recent study of different RNA purification methods for liquid biopsies found that the miRNeasy kit (Qiagen) recovered a broad spectrum of exRNAs associated with different carrier subclasses (31). However, when we purified RNase1-treated RNA with this kit following the manufacturer’s instructions, tRNA halves were no longer amenable to enzymatic repair (SI Appendix, Fig. S7D). Similar results were obtained when comparing phenol-free SPE-based purification vs. TRIzol (Fig. 4B). Interestingly, adding a second SPE-based purification round recovered reparable tRNA halves with a yield close to 100%. Thus, guanidine salts included in the SPE binding buffer do not affect nicked tRNAs, while heat and phenol induce their irreversible separation. As previously observed, the length of the repaired tRNA was slightly shorter (4 to 5 nt) than that of the parental full-length tRNA (SI Appendix, Fig. S7E).
To confirm the effects of phenol and heat by an orthogonal method, we studied RNase-1-treated RNAs by native northern blot (run on TB + Mg2+ gels) (Fig. 4C). Under native conditions, the electrophoretic migration of RNase1-treated tRNAGlyGCC was almost identical to that of untreated full-length tRNAs. Only <10% of the total signal in the treated lane corresponded to bona-fide tDRs. This confirms that the rapid disappearance of the full-length tRNA band observed in Fig. 2A is in fact an artifact caused by denaturing conditions, with >90% of tRNAs remaining as nicked tRNAs after 1 h of enzymatic digestion. Interestingly, the migration of nicked tRNAs in native gels was slightly faster than that of full-length tRNAs, consistent with the irreversible loss of the NCCA 3′ overhang inferred from previous assays (SI Appendix, Fig. S7E). Additionally, heat or standard RNA purification methods such as TRIzol or miRNeasy, unlike phenol-free RNA cleanup columns (SPE), induced the irreversible dissociation of nicked tRNAs (Fig. 4C).
The 3′–5′ RNA Ligase RtcB Can also Ligate Nicked tRNAs.
RtcB is a ligase involved in tRNA splicing and RNA repair in all domains of life (42–44). Unlike the 5′–3′ T4 Rnl1, RtcB seals broken RNAs with 2′,3′ cyclic phosphate (2,3-cP) and 5′-OH ends (45). Thus, RtcB should be sufficient to repair RNase1-treated RNAs without prior end-healing by T4 PNK. Indeed, RtcB regenerated a tRNA-sized band when incubated with RNase1-treated RNAs, and generation of this ligation product was inhibited by heat (Fig. 4D). Once again, the repaired tRNA-sized band was 4 to 5 nt shorter than the parental full-length tRNA (SI Appendix, Fig. S7F).
Nicked tRNAs Are Reverse-Transcribed Inefficiently unless Repaired.
Broken phosphodiester bonds could act as roadblocks inhibiting reverse transcription (RT) of nicked tRNAs in their native state, preventing the analysis of nicked tRNAs by RT-PCR or sequencing. To evaluate this possibility, we used a thermostable retroviral reverse transcriptase primed by a gene-specific RT primer aligning to the 3′ end of tRNAGlyGCC (Fig. 4E). Forward and reverse PCR primers were placed closed to the 5′ and 3′ ends of tRNAGlyGCC, respectively. Surprisingly, RNase-1-treated RNA was not amplified unless enzymatic repair (T4 PNK + T4 Rnl1) was performed before RT. Consistent with northern blot results (Figs. 3D and 4 A–D), heating the samples before enzymatic repair also inhibited RT-PCR amplification (Fig. 4E). Conversely, 5′ tRNAGlyGCC halves of 30 nt increased 200 to 700-fold in RNase1-treated samples vs. input, illustrating the limitations of small RNA expression analysis in samples containing nicked forms of their parental RNAs.
Nicked tRNAs Are Resistant to RNase 1 Cleavage In Vitro.
Having observed that nicked tRNAs can be purified efficiently by SPE, we generated and purified nicked tRNAs and treated them with r-RNase 1 (Fig. 4F). Nicked tRNAs were not degraded by this treatment (up to 30 min at 37 °C). However, heating and then cooling the nicked tRNAs before exposure to r-RNase1 induced complete degradation of the tRNA halves, now in their single-stranded form. This effect was not induced by heat itself. In conclusion, nicked tRNAs are stable reservoirs of 5′ tRNA halves, which are degradation prone once dissociated from their 3′ counterparts.
Nicked tRNAs as a Source of Intracellular Stress-Induced tRNA Halves.
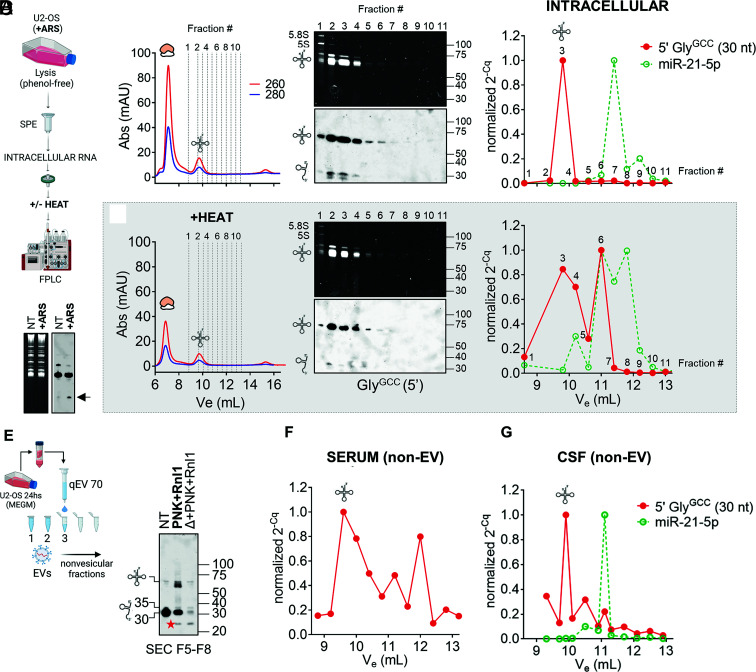
Our enzymatic repair assays were not useful to interrogate the existence of nicked tRNAs inside cells due to the large excess of full-length tRNAs in intracellular samples. We thus resorted to a different strategy based on intracellular RNA fractionation under nondenaturing conditions by size-exclusion chromatography (SEC) (Fig. 5 A–D).
Fig. 5.
Detection and analysis of nicked tRNAs in intracellular (A–D) and extracellular (E–G) samples. (A) Schematic representation of SEC-based (native) intracellular RNA fractionation as performed in (C) and (D). (B) Northern blot of intracellular RNAs showing the presence of 5′ tRNA halves in arsenite (ARS)-treated samples (i.e., tiRNAs). (C) Purified RNA from ARS-treated U2-OS cells was separated on a Superdex 75 column using an FPLC system. Selected fractions were analyzed by northern blot (Center) or by stem-loop RT-qPCR (Right). Cq values were normalized to the fraction containing the highest signal. (D) Same as in (C), but the RNA was heat-denatured before injection. (E) Cell-conditioned, RI-treated, serum-free medium from U2-OS cells was fractionated with an Izon 70-nm qEVoriginal column to prepare EV-depleted fractions. Northern blot analysis of tRNAs & tDRs in pooled nonvesicular extracellular fractions (F5 to F8) before and after enzymatic repair with T4 PNK and T4 Rnl1. Red star: a ~24-nt band that was only observed in the presence of Rnl1. (F and G) Separation by SEC (as done in C) of purified RNA from Proteinase K-treated ultracentrifugation supernatants of human serum (E) or CSF (F). Selected eluted fractions were analyzed by SL-RT-qPCR using primers specific for 5′ tRNAGlyGCC halves of 30 nt (red) and miR-21-5p (green). A tRNA icon in this figure indicates fractions where full-length tRNAs are expected to elute (if present).
To stimulate the formation of stress-induced tRNA halves inside cells (tiRNAs), U2-OS cells were treated with sodium arsenite (23). Upregulation of tRNA halves in stressed cells was confirmed by northern blot (Fig. 5B). We then lysed tiRNA-containing stressed cells by phenol-free methods, purified intracellular RNA by SPE to avoid potential nicked RNA denaturation, and size-fractionated the purified RNA by SEC in RNase-free PBS as running buffer. Two peaks with ABS 260 nm > ABS 280 nm were observed, corresponding to the elution of rRNAs and tRNAs (Fig. 5C, Left). The elution of full-length tRNAs at elution volume (Ve) = 9.80 mL was confirmed by northern blot (Fig. 5C, Center; fractions 2 to 4). Surprisingly, intracellular 5′ tRNA halves (of 35 and 30 nt) coeluted with full-length tRNAs, suggesting that tiRNAs are not single-stranded molecules inside cells. We considered the possibility that the sensitivity of our DIG-based northern blot assay was not sufficient to detect monomeric tiRNAs, which would be expected at Ve ≥ 11 mL (fraction ≥ #6) based on the injection of synthetic single-stranded RNAs of 30 nt (32). To study this, a highly sensitive stem-loop RT-qPCR (SL-RT-qPCR) assay was used (Fig. 5C, Right). This assay is specific for mature miRNAs or tRNA halves and does not amplify their precursors (pre-miRNAs or full-length tRNAs, respectively) (32, 46). miR-21-5p eluted at Ve = 11.4 mL (fraction #7), 5′ tRNAGlyGCC halves of 30 nt (Fig. 5C) or 35 nt (SI Appendix, Fig. S8A) were again undetectable, except in fraction #3, containing most full-length tRNAs. Heating the RNA before injection (Fig. 5D) shifted the amplification of 5′ tRNAGlyGCC halves toward fraction #6 (Ve = 11 mL), consistent with the expected denaturation of nicked tRNAs and the release of single-stranded tRNA halves, which now coeluted with miR-21-5p.
Overall, while we do not discard that bona fide single-stranded intracellular tiRNAs could be degraded upon cell lysis, our results strongly suggest that at least some (if not most) intracellular tiRNAs are predominantly present in the form of nicked tRNAs.
Detection of Nonvesicular Nicked tRNAs in Cell-Conditioned Media.
To explore the presence of nicked tRNAs in extracellular samples, we collected cell-conditioned media (CCM, t = 24 h) from U2-OS cells grown under-serum-free conditions and separated EVs from nonvesicular proteins and RNAs by SEC, using commercial qEV columns (Fig. 5E). To preserve both nicked tRNAs and single-stranded tDRs, we added RI to the media. As expected in RI-treated serum-free CCM (33), we observed full-length tRNAs in nonvesicular fractions by northern blot (Fig. 5E; NT lane), albeit most of the signal corresponded to ~30-nt fragments. In this case, sequential treatment with PNK and Rnl1 generated at least three bands with sizes close to, but slightly shorter than, the size of the parental full-length tRNA. Once again, heating and cooling the samples before ligation abrogated generation of these products, arguing against nonspecific ligation in trans. The lowest and most intense band had a size of 61 nt, consistent with the main ligation product observed when incubating RNAs in CSF for long periods of time (SI Appendix, Fig. S7B). As previously discussed, this suggests the presence of trimmed forms of nicked tRNAs, lacking both the 3′ NCCA tail and the entire anticodon loop. Once formed, these ligation products could be heated and remained resistant to the processive 3′ to 5′ exonuclease RNase R (SI Appendix, Fig. S8B), suggesting the ligated products are highly structured. In contrast, the remaining 30 to 35-nt band after enzymatic repair was completely degraded by RNase R.
Interestingly, a band of approximately 24 nt was generated by the combined action of PNK and Rnl1 in both heated and nonheated samples (Fig. 5E; red stars). Because this band was resistant to RNase R (SI Appendix, Fig. S8B), we speculate that it corresponds to a circularized RNA generated by head-to-tail ligation of 5′ tRNA halves. Formation of this product in nonheated samples implies that at least some extracellular tRNA halves are present in fragment form in RI-treated CCM. This suggests that nicked tRNAs are spontaneously separating and releasing single-stranded tRNA halves into the extracellular space. Their detection is facilitated by the low RNase activity in the conditions of this assay.
Nonvesicular Nicked tRNAs Circulate in Human Biofluids.
Having validated a method capable of separating nicked tRNAs from single-stranded tRNA halves (Fig. 5 C and D), we implemented this methodology to address the question of whether nicked tRNAs circulate in human biofluids. To do so, small volumes of serum or CSF were diluted in PBS, and EVs were pelleted by ultracentrifugation. RNA was then isolated from Proteinase K-treated supernatants and fractionated by SEC. Surprisingly, 30-nt 5′ tRNAGlyGCC halves could be amplified by SL-RT-qPCR (Fig. 5 F and G), mostly in the size range corresponding to full-length tRNAs, which are expected to be absent in these samples (Fig. 2B).
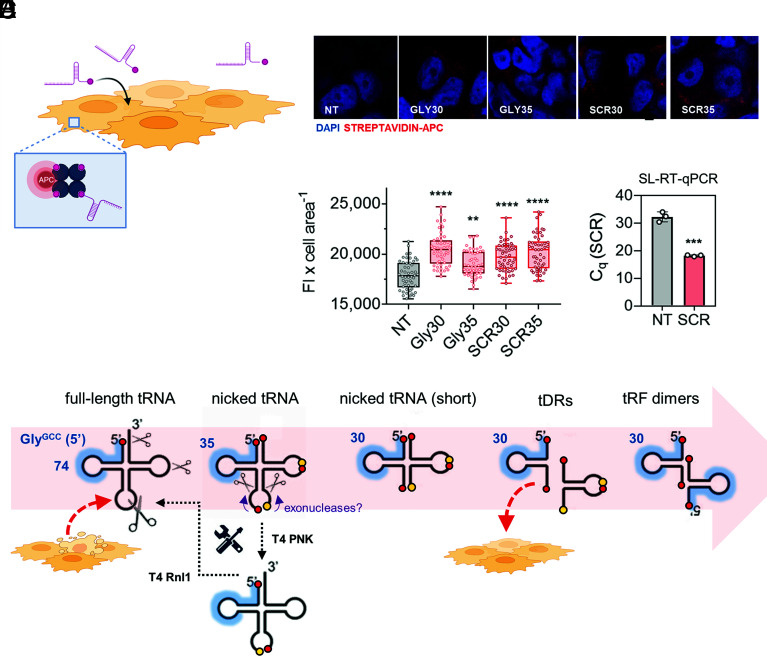
Spontaneous Uptake of Nonvesicular tRNA Halves by Human Epithelial Cells.
Unless stabilized by dimerization or RNP complex formation, we would expect single-stranded tRNA halves to be degraded in a matter of seconds in extracellular samples with high RNase activity (Fig. 4F). However, if stable nicked tRNAs diffuse from the sites in which they are generated and they can reach farther cellular populations, their spontaneous dissociation could release single-stranded tRNA halves close to the surface of potentially recipient cells. Could cells then internalize these single-stranded tDRs?
We incubated synthetic and biotinylated 5′ tRNAGlyGCC halves of 30 and 35 nt, or scrambled (SCR) versions of these oligonucleotides, with MCF-7 cells grown in serum-free media (Fig. 6A). No lipids or transfection reagents were used. After 30 min, cells were thoroughly washed, fixed, stained with APC-coupled streptavidin and observed under a confocal microscope. Surprisingly, the red signal of the fluorophore’s emission was observed within cellular boundaries in all conditions where labeled RNAs were included (Fig. 6 B and C). As an orthogonal approach, we incubated cells with nonbiotinylated SCR oligonucleotides (which do not exist in recipient cells), washed cells thoroughly, and observed a drop of almost 15 PCR cycles when comparing with nontreated cells, based on a SCR-specific SL-RT-qPCR assay (Fig. 6D). Taken together, these results show that human epithelial cells can spontaneously incorporate single-stranded small RNAs present in the media. Although this uptake route seems to be sequence- independent, most naked exRNAs will degrade before having the opportunity to interact with recipient cells. However, tRNA halves are extremely stable in extracellular samples (Fig. 2), presumably due to their transport in the form of nicked tRNAs. For this reason, and because their extracellular concentration is increased in the presence of cell stress (Fig. 1D), we foresee the involvement of extracellular tRNA halves in intercellular communication pathways mediated by naked, nonvesicular RNAs (Fig. 6E).
Fig. 6.
Nicked tRNAs are a source of nonvesicular single-stranded tRNA halves that can be taken up by recipient cells. (A) Synthetic 5′ biotinylated RNAs were added to the media of MCF-7 cells, and oligonucleotide uptake was confirmed by confocal microscopy using APC-coupled streptavidin (B and C) or by SL-RT-qPCR (D). (E) Proposed model.Red circles: phosphate groups. Yellow circles: OH groups.
Discussion
This work challenges the widespread belief that all RNAs are intrinsically unstable and cannot circulate in extracellular samples unless in the context of RNPs, lipoproteins and/or EVs. Although this holds true for rRNA-derived fragments (Fig. 1), full-length or nicked tRNAs showed surprisingly long half-lives in human biofluids, even when incubated in their naked forms. While these RNAs might also be present in RNP complexes in the extracellular space, our results demonstrate that protein complexation is not a prerequisite for their remarkable extracellular stability.
An aspect of this study that certainly deserves more attention is the differences in extracellular stabilities observed among tRNAs. We are tempted to speculate that posttranscriptionally modified bases at the anticodon are at least partially responsible for these different behaviors. However, the difference in stability between tRNAs is less pronounced when considering that nicked tRNAs, and not tDRs, are the stable degradation intermediates dictating the abundance of nonvesicular glycine tRNA halves.
Is the full-length tRNAGlyGCC unstable in human biofluids? It is efficiently cleaved at several positions by extracellular RNases. So the immediate answer would be “yes”. However, the result of these cleavage events is a molecule that probably still resembles a tRNA, although it bears some broken phosphodiester bonds. Thus, although tRNALysUUU and tRNAGlyGCC show completely different behaviors when analyzed by northern blot after being exposed to RNases, this difference is exaggerated because standard northern blotting forces nicked tRNAs to denature. This is also the case when purifying RNA using phenol and when RNA is heated at any stage in a protocol (e.g., as done during small RNA-seq library preparation). The important conclusion is that we do not always apprehend the true, native form of RNA because most available analytical techniques contain denaturation steps at some point. Additionally, nicked tRNAs cannot be reverse-transcribed and amplified unless repaired or denatured into fragments.
The case of tRNALysUUU is also interesting. Although it is highly resistant to degradation, once cleaved by r-RNase 1 or by FBS-derived RNase A, it typically does not survive as a nicked tRNA (Fig. 2A). This implicates extracellular RNases in the degradation of tDRs (47). In contrast, tRNAGlyGCC is sensitive to initial cleavage events, but nicked tRNAs produced therefrom are intrinsically stable. Differential stabilities among distinct tRNA sequences could be considered a new example of noncanonical “moonlighting” functions of tRNAs, what has been related to the diversity of the tRNA isodecoder pool (48).
In bacteria, phage infection (40) or DNA-damage (49) induces regulated tRNA cleavage at the anticodon loop. This process can be reversed by host-encoded tRNA ligases such as RtcB (44) or by phage antitoxins like T4 PNK and T4 Rnl1. Thus, the existence of nicked tRNAs that can be enzymatically repaired back into full-length tRNAs is well established in prokaryotes.
In plants, the phloem sap (PS) has been shown to contain 70 to 80-nt RNAs, which resemble tRNAs, that are capable of inhibiting translation. Importantly, this activity was lost when purifying PS RNA by TRIzol (50). Thus, plants also seem to generate nicked tRNAs in their vasculature. Nonvesicular tRNA halves were also recently identified in the extracellular apoplastic fluid (51). The theoretical capacity of nicked tRNAs to act as regulators of the translation machinery was recognized early (52). However, nicked tRNAs were not considered as stable degradation intermediates in the subsequent literature (53).
Stress-induced tRNA cleavage at the anticodon loop is thought to be an irreversible process. Nevertheless, it should be considered that most of the enzymatic activities used in this work to repair anticodon-cleaved nicked tRNAs are present in human cells (54, 55). Reversibility of stress-induced nicked tRNA formation in cellulo arises as an exciting possibility (56).
In the clinical setting, double-stranded RNAs usually require encapsulation in lipid nanoparticles or GalNAc conjugation for efficient uptake, but single-stranded oligonucleotides are spontaneously endocytosed (57). Thus, our proposed model where nonvesicular nicked tRNAs are carriers of tRNA halves that can transfer information to recipient cells is at least feasible (Fig. 6E).
On the applied side, we took advantage of phage and bacterial enzymes involved in damaged tRNA repair to develop an analytical technique that can be used to detect and measure stable nonvesicular RNAs circulating in biofluids. Although we focused mostly on tRNAs, close inspection of gels shows additional bands that could be restored by combined treatment with PNK and Rnl1 and that were lost in heated samples (Fig. 4B, yellow stars). This strongly suggests that the nonvesicular RNAome is much more complex than previously thought, in accordance with recent findings (33). Many stable nonvesicular RNAs are single-stranded molecules tightly bound and protected by exRNA-binding proteins (15, 16, 38). However, intrinsically stable RNAs could also circulate freely in biofluids (58). These molecules are predicted to be both highly structured and nicked. There are sequencing methods that can efficiently handle highly structured RNAs (59, 60). However, nicked RNAs are more challenging because they contain roadblocks for reverse transcriptases (i.e., the broken phosphodiester bonds) and because nicked RNAs are dissociated by phenol or heat. Our enzymatic repair protocol therefore increases the number and types of RNA molecules that can be analyzed and used as disease biomarkers.
Materials and Methods
Detailed versions of these protocols and additional methods are included in SI Appendix, Supplementary Materials and Methods.
Biofluids and Cell Culture.
Biofluids samples were obtained at UC San Diego (CSF) or at the Pasteur Institute of Montevideo (serum, urine), and deidentified prior to use. Samples were spun down at 2,000 × g before storage at ≤ −20 °C. Fetal bovine serum (FBS) was from Gibco. Cell-conditioned media (CCM) were obtained from either U2-OS cells grown under serum-free (MEGM, Lonza) or serum-containing conditions (10% FBS), with or without addition of murine RI (NEB). Intracellular tRNA-derived fragments were generated by exposing cells to 500 µM sodium arsenite (Sigma) for 2 h.
RNA Purification by SPE.
RNA was purified by silica-based SPE using Monarch RNA Cleanup Kits (for in vitro assays) or the Total RNA Miniprep Kit for intracellular samples (NEB).
RNA Decay Assays.
For determination of RNA half-lives in biofluids, 1 μg heated and refolded U2-OS total RNA was incubated at 37 °C for variable periods with 50 μL undiluted or PBS-diluted biofluid samples. Digested RNAs were purified by SPE and analyzed by northern blot.
In Vitro RNA Digestion.
For in vitro generation of nicked tRNAs and/or tDRs, 1 μg heated and refolded U2-OS total RNA was incubated with recombinant human RNase 1 (Bon Opus Bio; 0.0625 μg/mL) for 15, 30, or 60 min at 37 °C, and purified by SPE.
RNA Ligation Assays.
In vitro-digested or CCM-derived nonvesicular RNA was incubated for 1 h at 37 °C in a 10 μL reaction containing 20 U RI, 1 mM ATP, 1X T4 PNK reaction buffer, 10 U T4 RNA ligase 1 (Rnl1, NEB), and/or 10 U of T4 PNK (wild type or 3′ phosphatase minus, NEB). For RtcB ligation, reaction mixtures contained 5 μL in vitro-digested digested RNA, 20 U RI, 1X RtcB ligase buffer, 1 mM GTP, 1 mM Mn2+, and 1 μM RtcB ligase from E. coli (NEB).
Preparation of Nonvesicular RNAs from Cell-Conditioned Media.
Nonvesicular RNAs from U2-OS cell-conditioned media were separated from EVs by either iodixanol 12 to 36% density gradients (33, 37) or by size-exclusion chromatography using 70-nm qEVoriginal columns (IZON).
Chromatographic Fractionation of Nonvesicular RNAs from Biofluids.
EVs were depleted from human serum or CSF samples by ultracentrifugation (256,000 × g and 4 °C for 1 h). The supernatants were concentrated by ultrafiltration (10 kDa MWCO), treated with phenol-free RNA Binding Buffer (included in RNA Cleanup Kits from NEB) and Proteinase K, and the RNA was purified by SPE. Purified RNAs were then injected in a Superdex 75 10/300 column (GE), using an Äkta Pure FPLC system. Fractions of 0.2 mL were collected and analyzed by stem-loop RT-qPCR.
Northern Blot, RT-qPCR for Full-Length tRNAs and SL-RT-qPCR for tDRs.
DIG-based northern blot and stem-loop RT-qPCR were done as described in ref. 33 and 32, respectively. RT-PCR for full-length tRNAs was done using Superscript IV at 50 °C. Probes, primers, and assay conditions are provided in SI Appendix, Supplementary Materials and Methods.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Sergio Bianchi and other members of the Functional Genomics Laboratory (IPMon) and the Analytical Biochemistry Unit (UdelaR). This work was supported in part by the NIH (R01GM126150, R01GM146997), the National Cancer Institute (NCI) and NIH Office of the Director (UG3/UH3CA241694), and Universidad de la República, Uruguay (CSIC I+D_2020_433). B.C. received a fellowship from the National Agency of Research and Innovation (ANII, Uruguay; POS_NAC_M_2020_1_163868). J.P.T., E.C., and A.C. are members of the National System of Researchers (ANII, Uruguay) and the Program for the Development of Basic Science (PEDECIBA, Uruguay).
Author contributions
J.P.T. designed research; B.C., M.L.C., M.C., V.B., and J.P.T. performed research; I.L., P.I., K.W., A.C., and J.P.T. contributed new reagents/analytic tools; B.C., E.C., and J.P.T. analyzed data; B.C., M.L.C., M.C., V.B., E.C., I.L., P.I., K.W., and A.C. edited the manuscript; and J.P.T. wrote the paper.
Competing interest
The authors have patent filings to disclose, J.P.T., B.C., A.C. and K.W. have filled a provisional patent in the US related to this work.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Moufarrej M. N., et al. , Early prediction of preeclampsia in pregnancy with cell-free RNA. Nature 602, 689–694 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heitzer E., Haque I. S., Roberts C. E. S., Speicher M. R., Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 20, 71–88 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Thomou T., et al. , Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 542, 450–455 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia-Silva M. R., et al. , Extracellular vesicles shed by Trypanosoma cruzi are linked to small RNA pathways, life cycle regulation, and susceptibility to infection of mammalian cells. Parasitol. Res. 113, 285–304 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Cai Q., et al. , Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360, 1126–1129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buck A. H., et al. , Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat. Commun. 5, 5488 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tosar J. P., Witwer K., Cayota A., Revisiting extracellular RNA release, processing, and function. Trends Biochem. Sci. 46, 438–445 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skog J., et al. , Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valadi H., et al. , Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Mateescu B., et al. , Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - An ISEV position paper. J. Extracell. Vesicles 6, 1286095 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Brien K., Breyne K., Ughetto S., Laurent L. C., Breakefield X. O., RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 21, 585–606 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tosar J. P., et al. , Assessment of small RNA sorting into different extracellular fractions revealed by high-throughput sequencing of breast cell lines. Nucleic Acids Res. 43, 5601–5616 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei Z., et al. , Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat. Commun. 8, 1145 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Q., et al. , Supermeres are functional extracellular nanoparticles replete with disease biomarkers and therapeutic targets. Nat. Cell Biol. 23, 1240–1254 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turchinovich A., Weiz L., Langheinz A., Burwinkel B., Characterization of extracellular circulating microRNA. Nucleic Acids Res. 39, 7223–7233 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arroyo J. D., et al. , Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U.S.A. 108, 5003–5008 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geekiyanage H., Rayatpisheh S., Wohlschlegel J. A., Brown R., Ambros V., Extracellular microRNAs in human circulation are associated with miRISC complexes that are accessible to anti-AGO2 antibody and can bind target mimic oligonucleotides. Proc. Natl. Acad. Sci. U.S.A. 117, 24213–24223 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vickers K. C., Palmisano B. T., Shoucri B. M., Shamburek R. D., Remaley A. T., MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 13, 423–435 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson D. M., Lu C., Green P. J., Parker R., tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14, 2095–2103 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y., et al. , Stress-induced tRNA-derived RNAs: A novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 36, 6048–6055 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.David M., Borasio G. D., Kaufmann G., Bacteriophage T4-induced anticodon-loop nuclease detected in a host strain restrictive to RNA ligase mutants. Proc. Natl. Acad. Sci. U.S.A. 79, 7097–7101 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu H., et al. , Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 583, 437–442 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Yamasaki S., Ivanov P., Hu G.-F., Anderson P., Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 185, 35–42 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivanov P., Emara M. M., Villen J., Gygi S. P., Anderson P., Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 43, 613–623 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyons S. M., et al. , eIF4G has intrinsic G-quadruplex binding activity that is required for tiRNA function. Nucleic Acids Res. 48, 6223–6233 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim H. K., et al. , A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature 552, 57–62 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuscu C., et al. , tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA 24, 1093–1105 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nolte'T Hoen E. N. M., et al. , Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 40, 9272–9285 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dhahbi J. M., et al. , 5’ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genomics 14, 298 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y., et al. , Identification and characterization of an ancient class of small RNAs enriched in serum associating with active infection. J. Mol. Cell Biol. 6, 172–174 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Srinivasan S., et al. , Small RNA sequencing across diverse biofluids identifies optimal methods for exRNA isolation. Cell 177, 446–462 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tosar J. P., et al. , Dimerization confers increased stability to nucleases in 5’ halves from glycine and glutamic acid tRNAs. Nucleic Acids Res. 46, 9081–9093 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tosar J. P., et al. , Fragmentation of extracellular ribosomes and tRNAs shapes the extracellular RNAome. Nucleic Acids Res. 48, 12874–12888 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nechooshtan G., Yunusov D., Chang K., Gingeras T. R., Processing by RNase 1 forms tRNA halves and distinct Y RNA fragments in the extracellular environment. Nucleic Acids Res. 48, 8035–8049 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanadgol N., König L., Drino A., Jovic M., Schaefer M. R., Experimental paradigms revisited: oxidative stress-induced tRNA fragmentation does not correlate with stress granule formation but is associated with delayed cell death. Nucleic Acids Res. 50, 6919–6937 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sorrentino S., The eight human “canonical” ribonucleases: Molecular diversity, catalytic properties, and special biological actions of the enzyme proteins. FEBS Lett. 584, 2194–2200 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Jeppesen D. K., et al. , Reassessment of exosome composition. Cell 177, 428–445 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tosar J. P., Cayota A., Witwer K., Exomeres and supermeres: Monolithic or diverse? J. Extracell. Biol. 1, e45 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwer B., Sawaya R., Ho C. K., Shuman S., Portability and fidelity of RNA-repair systems. Proc. Natl. Acad. Sci. U.S.A. 101, 2788–2793 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaufmann G., Anticodon nucleases. Trends Biochem. Sci. 25, 70–74 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Akiyama Y., et al. , Selective cleavage at CCA ends and anticodon loops of tRNAs by stress-induced RNases. Front. Mol. Biosci. 9, 791094 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Englert M., Sheppard K., Aslanian A., Yates J. R., Söll D., Archaeal 3′-phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proc. Natl. Acad. Sci. U.S.A. 108, 1290–1295 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Popow J., et al. , HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science 331, 760–764 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Tanaka N., Shuman S., RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J. Biol. Chem. 286, 7727–7731 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chakravarty A. K., Subbotin R., Chait B. T., Shuman S., RNA ligase RtcB splices 3′-phosphate and 5′-OH ends via covalent RtcB-(histidinyl)-GMP and polynucleotide-(3′)pp(5′)G intermediates. Proc. Natl. Acad. Sci. U.S.A. 109, 6072–6077 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen C., et al. , Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33, e179 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li G., et al. , Distinct stress-dependent signatures of cellular and extracellular tRNA-derived small RNAs. Adv. Sci. 9, e2200829 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Avcilar-Kucukgoze I., Kashina A., Hijacking tRNAs from translation: Regulatory functions of tRNAs in mammalian cell physiology. Front. Mol. Biosci. 7, 610617 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hughes K. J., Chen X., Burroughs A. M., Aravind L., Wolin S. L., An RNA repair operon regulated by damaged tRNAs. Cell Rep. 33, 108527 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang S., Sun L., Kragler F., The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol. 150, 378–387 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zand Karimi H., et al. , Arabidopsis apoplastic fluid contains sRNA- and circular RNA-protein complexes that are located outside extracellular vesicles. Plant Cell 34, 1863–1881 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thompson D. M., Parker R., Stressing out over tRNA cleavage. Cell 138, 215–219 (2009). [DOI] [PubMed] [Google Scholar]
- 53.Chen Q., Zhang X., Shi J., Yan M., Zhou T., Origins and evolving functionalities of tRNA-derived small RNAs. Trends Biochem. Sci. 46, 790–804 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kroupova A., et al. , Molecular architecture of the human tRNA ligase complex. Elife 10, e71656 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pinto P. H., et al. , ANGEL2 is a member of the CCR4 family of deadenylases with 2’,3’-cyclic phosphatase activity. Science 369, 524–530 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Akiyama Y., et al. , RTCB complex regulates stress-induced tRNA cleavage. Int. J. Mol. Sci. 23, 13100 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Levin A. A., Treating disease at the RNA Level with oligonucleotides. N. Engl. J. Med. 380, 57–70 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Tosar J. P., Die hard: Resilient RNAs in the blood. Nat. Rev. Mol. Cell Biol. 22, 373–373 (2021). [DOI] [PubMed] [Google Scholar]
- 59.Qin Y., et al. , High-throughput sequencing of human plasma RNA by using thermostable group II intron reverse transcriptases. RNA 22, 111–128 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Behrens A., Rodschinka G., Nedialkova D. D., High-resolution quantitative profiling of tRNA abundance and modification status in eukaryotes by mim-tRNAseq. Mol. Cell 81, 1802–1815.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.