Abstract
The heart develops in a synchronized sequence of proliferation and differentiation of cardiac progenitor cells (CPCs) from two anatomically distinct pools of cells, the first heart field (FHF) and second heart field (SHF). Congenital heart defects arise upon dysregulation of these processes, many of which are restricted to derivatives of the FHF or SHF. Of the conserved set of signaling pathways that regulate development, the Wnt signaling pathway has long been known for its importance in SHF development. The source of such Wnts has remained elusive, though it has been postulated that these Wnts are secreted from ectodermal or endodermal sources. The central question remains unanswered: Where do these Wnts come from? Here, we show that CPCs autoregulate SHF development via Wnt through genetic manipulation of a key Wnt export protein (Wls), scRNA-seq analysis of CPCs, and use of our precardiac organoid system. Through this, we identify dysregulated developmental trajectories of anterior SHF cell fate, leading to a striking single ventricle phenotype in knockout embryos. We then applied our findings to our precardiac organoid model and found that Wnt2 is sufficient to restore SHF cell fate in our model of disrupted endogenous Wnt signaling. In this study, we provide a basis for SHF cell fate decision—proliferation vs. differentiation—autoregulated by CPCs through Wnt.
Keywords: heart development, second heart field, Wnt, Wls, organoid
Wnt signaling is required for second heart field (SHF) development (1). Some have thought that the source of these Wnts is from ectodermal or endodermal sources (2, 3). However, we found that Wnts are enriched in first heart field (FHF) progenitors while Wnt activity is higher in SHF progenitors (4). This led us to test whether the FHF is influencing SHF development. While others have perturbed Wnt signaling in Wnt-receiving cells through manipulation of β-catenin levels (1), the obligatory transcriptional effector of Wnt signaling, we instead focused on eliminating the ability of cells to secrete Wnts. To suppress Wnt secretion, we used a mouse strain harboring loxp sites flanking the gene encoding Wls (5), a protein required for export of Wnt ligands (6, 7). This allowed us to study which populations of cells that secrete Wnt affect heart development (Fig. 1A).
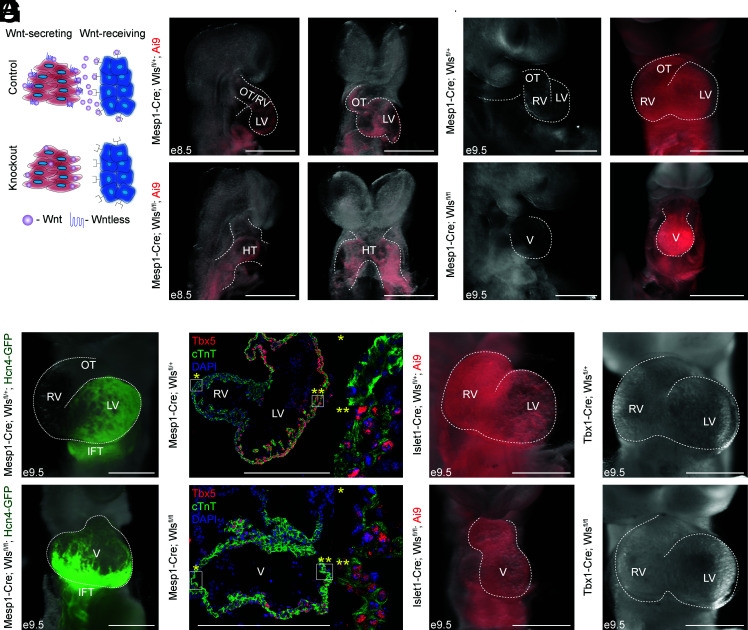
Fig. 1.
Knockout of Wntless in CPCs leads to FHF-derived single chamber phenotype. (A) Schematic detailing conditional knockout of the gene Wls. (B) e8.5 Mesp1-Cre; Wlsfl/+ (control), and Mesp1-Cre; Wlsfl/fl (knockout) embryos. (C ) e9.5 control and knockout embryos. (D) Fluorescent overlay images of Hcn4-GFP reporter in control and knockout embryos. (E ) Immunostaining of control and knockout sections of embryo hearts for cTnt (green), a cardiomyocyte marker, and Tbx5 (red), an LV marker with emphases on the right (*) and left (**) sides of the resulting chambers. (F ) Front images of Islet1-Cre control and knockout embryos. Fifty percent shows significant heart defects. (G) Front images of Tbx1-Cre control and knockout embryos (OT, outflow tract; IFT, inflow tract; RV, right ventricle; LV, left ventricle; V, ventricle). White scale bars indicate 500 μM.
As there are no validated Cre drivers that induce recombination with high efficiency in the early FHF, we first deleted Wls in precardiac mesoderm using Mesp1-Cre (8). Interestingly, upon knockout of Wls, we found a heart defect starting from embryonic day (e) 8.5 (Fig. 1B). Control embryos formed the primitive outflow tract/right ventricle (OT/RV) and left ventricles (LVs), expected at this developmental stage. However, a structure reminiscent of the cardiac crescent—which is transiently present from e7.75 to e8.25—and early heart tube persisted in knockout embryos at e8.5. The heart defect became more pronounced by e9.5, where, in comparison to expected LV, RV, and OT formation in control embryos, knockout embryos exhibit a single ventricle-like structure (Fig. 1C ).
The presence of a cardiac crescent-like/early heart tube structure (FHF) in knockout embryos suggests that the single ventricle phenotype may be due to dysregulation of SHF development. To test this, we utilized an Hcn4-Green Fluorescent Protein (GFP) reporter mouse strain to visualize FHF progenitors. As expected, GFP+ cells were localized in the LV of control embryos. However, knockout embryos showed an expansion of the GFP-expressing domain across the single ventricle (Fig. 1D). Additionally, immunostaining showed that the entire ventricle was positive for the LV marker Tbx5 in knockout embryos, while only the LV was positive for Tbx5 in control embryos (Fig. 1E ), consistent with a decrease in SHF derivatives. These results suggest that mesodermal Wnts are required for SHF development.
To determine the source of mesodermal Wnts more specifically, we leveraged multiple precardiac Cre drivers with shared expression domains to conditionally knockout Wls. We first used Islet1-Cre mice, as their Cre expression begins shortly after gastrulation in FHF progenitors before marking the SHF (4, 9, 10). We chose this particular Cre driver as it is activated in both heart fields and expressed early in heart development (11), which we confirmed by observing Red Fluorescent Protein (RFP) expression in both LV and RV in Cre-recombined control embryos (Fig. 1F ). Interestingly, we observed a similar single ventricle phenotype in Islet1-Cre; Wls-knockout embryos (Fig. 1F ), albeit with variable penetrance, perhaps due to differing domains of recombination previously observed in this particular Islet1-Cre driver. These results suggest that the source of Wnts is from a common population of cells derived from both Mesp1+ and Islet1+ cells—the derivatives of the FHF and SHF. To test which heart field is responsible for the phenotype, we blocked Wnt secretion in the Tbx1+ domain of the anterior SHF (aSHF). No heart defect was observed in Tbx1-Cre; Wls-knockout embryos (Fig. 1G ). This indicates that Wnts from the Tbx1 lineage are dispensable for SHF development. Together, these results support that cardiac progenitor cells (CPCs), likely the FHF, provide Wnts for SHF development.
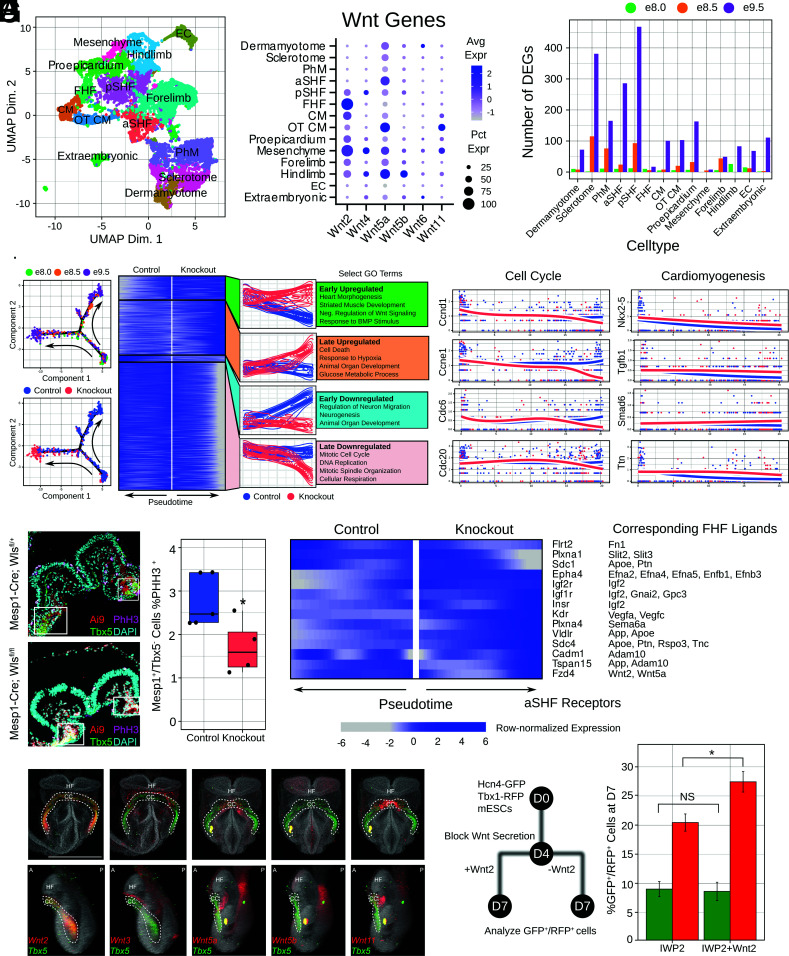
To understand the observed defect at the transcriptional level, we isolated Mesp1+ cells from Mesp1-Cre; Ai9; Wlsfl/+ and Wlsfl/fl mice at three developmental time points (e8.0, e8.5, and e9.5) for scRNA-seq. We performed fluorescence-activated cell sorting to isolate RFP+ cells and generated sequencing libraries. We performed dimensionality reduction through UMAP and annotated clusters through known marker genes (Fig. 2A). Our full cluster identification and quality control strategy is detailed in SI Appendix, Online Methods. We then analyzed Wnts in each population of mesoderm. Based on levels of expression of relevant Wnts, and our Wls loss of function studies, we focused on three populations as putative sources of instructive Wnts—posterior SHF (pSHF), FHF, and OT cardiomyocytes—though the OT cardiomyocytes are known to derive from Tbx1+ cells. The FHF cluster highly expressed Wnt2, while the pSHF cluster expressed low/undetectable levels of Wnts (Fig. 2B). These data further support our hypothesis that the FHF may be a source of instructive Wnts.
Fig. 2.
SHF development is dysregulated through decreased proliferation without Mesp1+ Wnts. (A) UMAP clustering based on cell type. (B) Expression of Wnts across cell populations. (C ) Quantification of differentially expressed genes (DEGs) across time points and cell populations. (D) aSHF developmental trajectories from control and knockout embryos and heatmaps identifying differential gene trends with expression dynamics of cell cycle and cardiomyogenesis genes. (E ) Quantification of percentage of Mesp1+/Tbx5− cells undergoing proliferation. White boxes highlight the Tbx5-positive regions which were excluded from quantification. Control average = 2.77%. Knockout average = 1.71%. *P = 0.039. (F ) Differentially expressed ligand–receptor pairs in knockout embryos in the aSHF. (G) Frontal (Top) and lateral (Bottom) views of whole-mount in situ hybridization of candidate Wnts with Tbx5 cohybridization in 4ss embryos (HF, head fold; CC, cardiac crescent; A, anterior; P, posterior). The white scale bar indicates 500 μM. Auto fluorescent debris is noted in Wnt5a/5b/11 embryos. (H ) Flow cytometry quantifications of Hcn4-GFP; Tbx1-RFP-positive cells at day 7 of differentiation after IWP-2 and IWP-2+Wnt2 treatment. IWP-2 %GFP+ average = 9%; %RFP+ average = 20.44%. IWP-2+Wnt2 %GFP+ average = 8.61%; %RFP+ average = 27.43%. GFP+ cells P = 0.86 (NS = not significant); RFP+ cells *P = 0.016.
We then asked how Wls deletion affected mesodermal cells outside a generalized response due to secondary effects of having a defective heart. To do this, we explored genes that were differentially expressed in unique clusters of cells. Notably, this pool of genes was enriched for Gene Ontology terms related to cell cycle, Wnt signaling, and mesodermal organ tissue and development. This suggested that, outside the nonspecific stress response in all tissues, Wls deficiency leads to perturbation of tissue development in specific mesodermal processes. The majority of these differentially expressed genes were identified in aSHF, pSHF, and sclerotome (Fig. 2C ), suggesting that these tissues are most perturbed by loss of Wnt signaling.
Given the perturbation of aSHF cells, we performed trajectory reconstruction of control and knockout cells in the aSHF. We reconstructed a branched trajectory in which control and knockout cells are indistinguishable early at e8.0, before reaching disparate states at later time points (Fig. 2D). We were interested in genes differentially expressed across the control and knockout paths at the branch point, focusing on genes with differential expression at the end states. We classified four clusters of genes based on expression dynamics (Fig. 2D). In general, GO terms relevant to heart development were enriched in upregulated gene clusters while GO terms related to cell cycle were enriched in downregulated clusters (Fig. 2D), pointing to potential impaired proliferation. Given that proper SHF development requires maintenance of proliferation and delayed differentiation, the upregulation of aSHF genes and downregulation of proliferation genes suggests dysregulation of cell identity after premature differentiation. We verified this in vivo by examining proliferation of Mesp1+/Tbx5− cells to quantify proliferation of the aSHF and found consistent decreased proliferation of SHF progenitors based on this staining (Fig. 2E ). Taken together, these results point to a decreased cell proliferation in the aSHF as a key mechanism for the knockout heart phenotype.
We then performed a basic ligand–receptor analysis (Fig. 2F ) to identify potential candidate signaling pairs affected by Wls knockout. Interestingly, we found that genes do not change significantly in the FHF as a result of Wls knockout; thus, there are no receptors or ligands whose expression is changing in the FHF. We instead focused on receptors whose expression is changing in the aSHF (as in Fig. 2E ), with the corresponding ligand expressed in at least 25% of cells in the FHF. As expected, many receptors are increased in expression level in the knockout, potentially indicative of a compensatory response to loss of Wnt signaling. In particular, the Wnt receptor Fzd4, which has been shown to interact with Wnt2, increases in level in the knockout.
To determine the specificity of Wnt expression to the FHF, we performed whole-mount in situ hybridization (12) of candidate Wnts with cohybridization of Tbx5, and found that, among our candidate genes identified from scRNA-seq, only Wnt2 was localized to the FHF (Fig. 2G ). Based on this, we used our precardiac organoid system, which recapitulates FHF/SHF development at both transcriptomic and functional levels, to determine whether Wnt2 affects formation of the SHF in the setting of blocked Wnt secretion (Fig. 2H ). We blocked Wnt secretion at day 4 of differentiation using the small molecule IWP-2 as previously described (4) and treated organoids with recombinant Wnt2 protein. Remarkably, upon cotreatment with Wnt2 protein, we observed an increase in SHF formation in IWP-2 treated cells, with no observable impact on the FHF. This demonstrates that Wnt2, which is expressed by the FHF, is sufficient to promote SHF development.
In summary, we show that Wls-deficient CPCs dysregulate developmental trajectories of aSHF cells, leading to impaired proliferation. These results demonstrate a critical role of precardiac (likely FHF) Wnts in SHF fate decisions, highlighting the importance of coordinated Wnt signals in heart field development and chamber formation. Further research into the instructive role of Wnt signaling in human models will help elucidate these complex cell–cell morphogenic signaling relationships and better understand the etiology of congenital heart disease.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported by the NIH, the Saving tiny Hearts Society, the American Heart Association, and the Department of Defense.
Author contributions
M.M., S.K., P.A., H.U., and C.K. designed research; M.M., S.K., M.J.A., X.L., D.S., M.H., B.L., T.K., S.M., E.T., M.L., P.A., and H.U. performed research; M.M., S.K., M.J.A., X.L., D.S., M.H., B.L., S.M., E.T., M.L., P.A., H.U., and C.K. analyzed data; and M.M. and C.K. wrote the paper.
Competing interest
The authors declare no competing interest.
Data, Materials, and Software Availability
scRNA-seq data have been deposited in Synapse; GitHub (https://www.synapse.org/#!Synapse:syn24200678/files/; https://github.com/skannan4/wls).
Supporting Information
References
- 1.Klaus A., Saga Y., Taketo M. M., Tzahor E., Birchmeier W., Distinct roles of Wnt/ β-catenin and Bmp signaling during early cardiogenesis. Proc. Natl. Acad. Sci. U.S.A. 104, 18531–18536 (2007), 10.1073pnas.0703113104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kemp C., Willems E., Abdo S., Lambiv L., Leyns L., Expression of all Wnt genes and their secreted antagonists during mouse blastocyst and postimplantation development. Dev. Dyn. 233, 1064–1075 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Tzahor E., Lassar A. B., Wnt signals from the neural tube block ectopic cardiogenesis. Genes Dev. 15, 255–260 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen P., et al. , Precardiac organoids form two heart fields via Bmp/Wnt signaling. Nat. Commun. 9, 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carpenter A. C., Rao S., Wells J. M., Campbell K., Lang R. A., Generation of mice with a conditional null allele for Wntless. Genesis 48, 554–558 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartscherer K., Pelte N., Ingelfinger D., Boutros M., Secretion of Wnt ligands requires Evi, a conserved transmembrane protein. Cell 125, 523–533 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Bänziger C., et al. , Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell 125, 509–522 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Saga Y., et al. , MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development 126, 3437–3447 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Ma Q., Zhou B., Pu W. T., Reassessment of Isl1 and Nkx2-5 cardiac fate maps using a Gata4-based reporter of Cre activity. Dev. Biol. 323, 98–104 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Y., et al. , Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev. Biol. 304, 286–296 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang L., et al. , Isl1 Cre reveals a common Bmp pathway in heart and limb development. Development 133, 1575–1585 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson M. J., Magidson V., Kageyama R., Lewandoski M., Fgf4 maintains Hes7 levels critical for normal somite segmentation clock function. Elife 9, 1–22 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
scRNA-seq data have been deposited in Synapse; GitHub (https://www.synapse.org/#!Synapse:syn24200678/files/; https://github.com/skannan4/wls).