Significance
During the interphase of the cell cycle, chromatin is accessible to the transcription machinery, which allows gene expression. When cells enter mitosis, chromosomes strongly condense, which facilitates their symmetrical segregation at the end of mitosis. However, condensed mitotic chromosomes prevent interactions between the DNA and many transcription factors, which correlates with a reduction in gene expression. This transcriptional silencing is generally regarded as a passive consequence of mitotic chromatin condensation. In this study, we show that mitotic chromosome condensation has an unexpected biological significance beyond mitosis: It resets the transcriptome to protect cells from uncontrolled transcriptional drift during the following interphase. Therefore, by resetting the chromatin, mitotic chromosome condensation contributes to the homeostatic control of transcription and gene expression regulation.
Keywords: transcription, mitosis, chromatin, chromosomes, cell cycle
Abstract
Mitotic entry correlates with the condensation of the chromosomes, changes in histone modifications, exclusion of transcription factors from DNA, and the broad downregulation of transcription. However, whether mitotic condensation influences transcription in the subsequent interphase is unknown. Here, we show that preventing one chromosome to condense during mitosis causes it to fail resetting of transcription. Rather, in the following interphase, the affected chromosome contains unusually high levels of the transcription machinery, resulting in abnormally high expression levels of genes in cis, including various transcription factors. This subsequently causes the activation of inducible transcriptional programs in trans, such as the GAL genes, even in the absence of the relevant stimuli. Thus, mitotic chromosome condensation exerts stringent control on interphase gene expression to ensure the maintenance of basic cellular functions and cell identity across cell divisions. Together, our study identifies the maintenance of transcriptional homeostasis during interphase as an unexpected function of mitosis and mitotic chromosome condensation.
During mitotic entry, eukaryotic chromosomes undergo condensation, a drastic compaction process that makes them manageable units for the mitotic spindle to segregate them between daughter cells (1). In addition, mitotic chromosome compaction remodels genome architecture, preventing the association of many transcription and chromatin regulators to cis regulatory elements and shutting down many interphase genes (2, 3). At the same time, it has been shown in yeast but also in mammals that active RNA polymerase II is present on chromatin during mitosis to maintain cell cycle regulation and basic cellular functions (3–6). At the end of mitosis, interphase chromatin structures and gene expression programs are reestablished in a precise and timely coordinated manner (7, 8). First, cells enhance the expression of housekeeping genes, whereas genes involved in the maintenance of cellular identity and functions are reactivated later (4, 9). This correlates with the timely reestablishment of interphase chromatin architecture and reloading of the transcription machinery. Although mitotic promoter bookmarking and histone modifications participate, the mechanisms enforcing controlled genome reactivation at mitotic exit remain unclear (7, 9, 10). While the relative transcriptional silencing of mitotic chromosomes has been observed and accepted a long time ago, it is typically regarded as a passive consequence of mitotic chromatin condensation and it remains unclear whether it has any biological significance beyond the M phase. Here, we investigated the effect of mitotic chromosome condensation on regulation of gene expression and discovered that it is important for transcriptional homeostasis during interphase.
Results
To investigate the possible consequences of mitotic chromosome condensation on gene expression in the next cell cycle, we took advantage of a recently developed approach that allows one to interfere with the condensation of a single chromosome at a time. Indeed, beyond preventing kinetochore assembly excision of a single yeast centromere prevents the condensation of the entire host chromosome (11, 12). This observation established that at least in budding yeast, condensation is a chromosome-autonomous process, is initiated at centromeres, and proceeds through the sequential mobilization of the kinase Ipl1, the protein shugoshin (Sgo1), and the sirtuin Hst2. These proteins mediated histone deacetylation, chromatin compaction, and condensin-dependent contraction of the chromosome arms (11–13). In the present study, we investigated the effect of preventing the condensation of a single chromosome (chr IV) on its transcription by precisely deleting the 444 nucleotides forming its centromere (CEN4). CEN4, flanked by lox recombination sites (CEN4*; Fig. 1A) (12, 14), was excised on demand through activation of a chimeric Cre protein fused to an estradiol-binding domain. We confirmed that CEN4* was quickly and efficiently excised from the chromosome in the majority of the cell population 30 minutes after estradiol treatment (SI Appendix, Fig. S1A).
Fig. 1.
Failure to achieve mitotic chromosome condensation results in general upregulation of gene expression in cis. (A) The centromere excision assay. (B) RNAseq assessment of gene expression at each chromosome and at different time points after CEN4* excision. (C) RNAseq assessment of gene expression at each chromosome 180 min after CEN4* excision in CEN4* and CEN4* H3S10D cells.
RNA-sequencing (RNAseq) at several time points after CEN4* excision (see SI Appendix, Fig. S1B for principal component analysis, PCA) enabled us to analyze the effect of CEN4* excision on gene expression of all yeast chromosomes, using increasingly stringent P-adj cutoffs. Strikingly, CEN4* excision was followed by a specific, significant, and progressive upregulation of transcription of genes located on chromosome IV. The amplitude (Log2 FC) of this increase and its significance were much more pronounced than at any other chromosome (Fig. 1B and SI Appendix, Fig. S1 C–E and Tables S1 and S2). These transcriptional changes became apparent 60 min after excision of CEN4*, although at present, we cannot exclude that at earlier time points changes in the transcriptome occurred but were obfuscated by the experimental approach. The enrichment of upregulated genes at chromosome IV was confirmed by unbiased hierarchical clustering. These analyses identified a large cluster of upregulated genes that were significantly enriched for genes located on chromosome IV (SI Appendix, Fig. S2 A–D). In contrast, other clusters contained genes that were either upregulated, downregulated, or unaffected upon CEN4* excision and were not enriched for genes belonging to chromosome IV (SI Appendix, Fig. S2 A–D). Consistent with the notion that upregulation of the genes on chromosome IV upon CEN4* excision resulted simply from their localization, Gene Ontology (GO) analysis could not classify them into any specific pathway (SI Appendix, Fig. S3 A and B). As a formal control, we also excised a random patch of noncentromeric DNA on one arm of chromosome IV using the same cre/lox system (SI Appendix, Fig. S4 A and B). However, this had no significant effect on gene expression (SI Appendix, Fig. S4 C and D), demonstrating that transcriptional deregulation is specific to the excision of CEN4*.
Beyond affecting its condensation cycle, deleting the centromere of a chromosome prevents it to segregate to the bud at mitosis, possibly causing changes in chromosome copy number in the population over time. Although the time frame of our experiment is probably too short for having a strong impact on chromosome IV copy number in the population (the cells undergo in average 1.5 divisions during the 180 min of the time course), we still wanted to know next what is the contribution of chromosome condensation, specifically, on the expression of chromosome IV upon excision of its centromere. Yeast centromeres gate the condensation of host chromosomes through recruitment of aurora B/Ipl1, which in turn initiates the condensation of the chromosome arms through phosphorylating Serine 10 of histone H3 (H3S10). As a consequence, the condensation defect resulting from the loss of the centromere is largely suppressed in cells expressing phospho-mimicking H3S10D (12). To test whether the increased gene expression observed on chromosome IV upon CEN4* excision was specifically caused by the failure of this chromosome to condense at mitosis, we asked whether this effect was suppressed in H3S10D-expressing cells. As expected, gene expression at chromosome IV was upregulated in CEN4* cells 180 min after CEN4 excision, while other chromosomes were not affected. However, in CEN4* H3S10D cells, in which chromatin condensation is constitutive throughout the cell cycle (12), the effect of centromere excision on gene expression at chromosome IV was substantially and significantly reduced (Fig. 1C). Since H3S10D suppresses specifically the condensation defects of cen-less chromosomes and not kinetochore assembly, we conclude that the centromere limits gene expression in cis through its function in licensing mitotic chromosome condensation and not through its role in chromosome copy number, at least in the time frame of our experiment.
Since gene transcription on chromosome IV was by far most significantly affected, we wondered how CEN4* excision affected genes on other chromosomes. GO analysis indicated an enrichment of a number of terms, such as carbohydrate metabolic process (SI Appendix, Fig. S3A). This included several genes involved in galactose metabolism like GAL1, GAL2, GAL7, and GAL10 (SI Appendix, Fig. S5A). These genes operate downstream of GAL3, which is a galactose-inducible transcription activator (15). Strikingly, GAL3 is located on chromosome IV, and therefore, we hypothesized that activation of GAL genes might be the consequence of increased expression of GAL3. Indeed, even in absence of galactose, GAL3 was progressively activated over time upon CEN4* excision (SI Appendix, Fig. S5A), suggesting that unscheduled activation of GAL genes located outside chromosome IV is caused by aberrant expression of GAL3 (SI Appendix, Fig. S5A). A similar effect was observed for the INO2 gene, which encodes an activator of phospholipid synthesis genes on chromosome IV as well (16) (SI Appendix, Fig. S5B). INO2 expression increased significantly upon excision of CEN4*, which was followed by induction of its target genes INO1, CHO1, ACS2, and ACC1 (SI Appendix, Fig. S5B), all of which are on other chromosomes. Thus, loss of CEN4* unleashes the expression of chromosome IV genes, even in absence of cognate signals, subsequently impacting the expression of downstream genes located on other chromosomes (SI Appendix, Fig. S5C).
Interestingly, in addition to causing aberrant expression of specific transcriptional programs, such as the GAL or INO systems, excision of CEN4* also affected several genes located outside chromosome IV that are involved in the regulation of translation, ribosome biogenesis, and metabolism. These pathways are often downregulated in response to stress and partially overlapped with the environmental stress response (ESR) program (17, 18) (SI Appendix, Figs. S3 A and B and S5D). We are not aware of the presence of any ESR transcription factors on chromosome IV, but we did notice upregulation of the two main activators of the ESR, MSN2 and MSN4 (Dataset S1), which are located on chromosome XIII and XI, respectively. Thus, although the mechanism remains to be identified, the loss of a single centromere may be sensed by the cell, leading to activation of the ESR (SI Appendix, Fig. S5C). Possibly, this response was due to an imbalance in gene expression, a process known to compromise proteostasis and cause ESR activation, such as for example in aneuploid cells or cells subjected to replication stress (12, 19–21). Taken together, these data show that centromeres suppress unscheduled activation of transcription in cis and that loss of a single centromere induces the ESR.
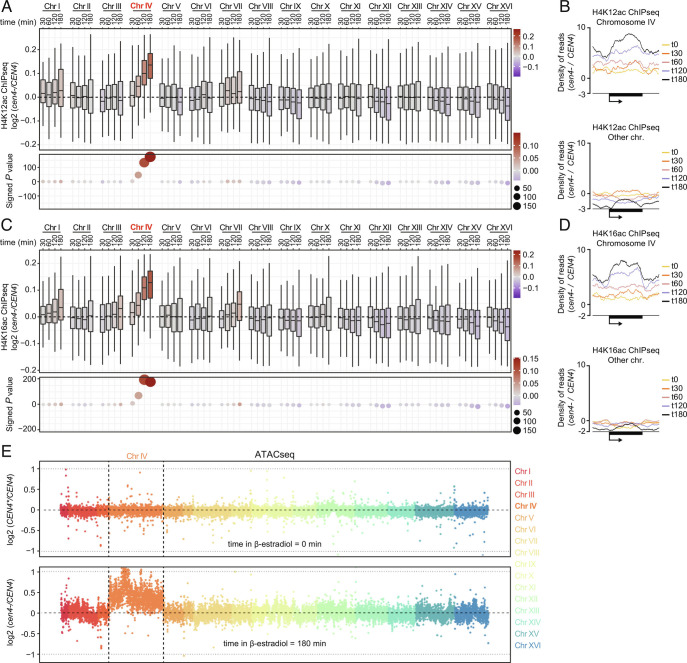
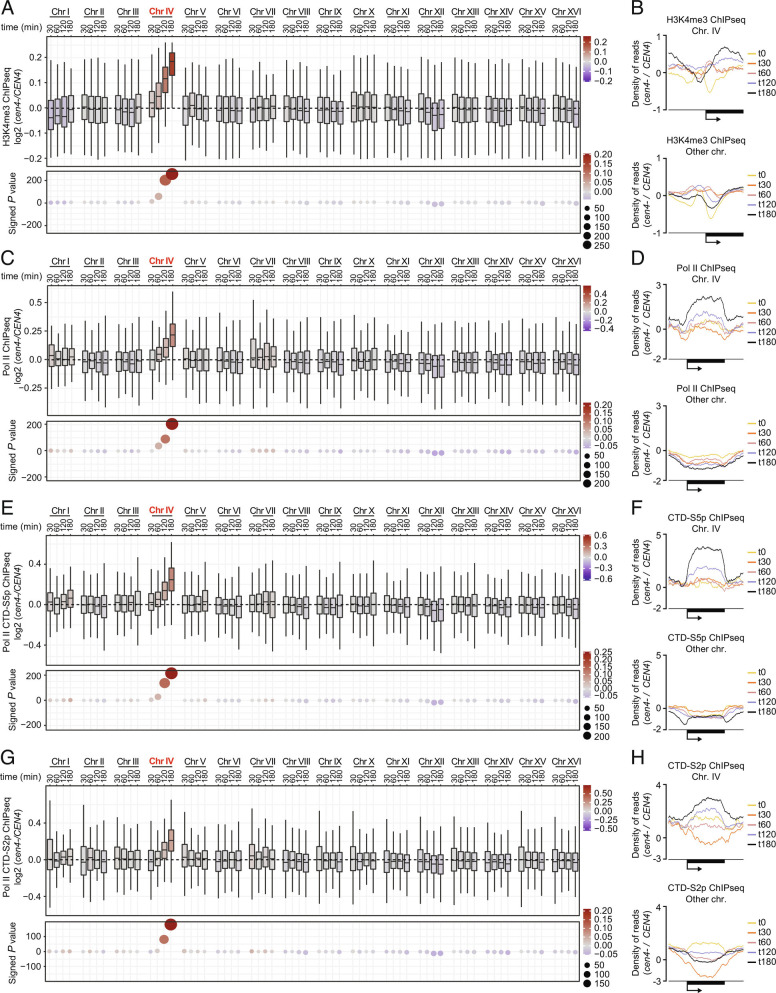
Given that the histone deacetylase Hst2 mediates mitotic chromatin condensation downstream of centromeres (11, 12), we wondered whether upregulation of gene expression at chr IV correlated with corresponding chromosome-specific chromatin alterations. Supporting this idea, the occurrence of H4K12ac and H4K16ac on chromosome IV was substantially increased over time after CEN4* excision, as determined by ChIP-sequencing (ChIPseq) (see SI Appendix, Fig. S6 A and B for PCA), whereas the other chromosomes remained largely unaffected (Fig. 2 A–D and SI Appendix, Figs. S7 A and B and S8 A and B). Consistently, ATACseq experiments revealed a strong increase in chromatin accessibility on chromosome IV specifically (Fig. 2E and SI Appendix, Fig. S8C), showing that centromere inactivation causes chromatin opening, or prevents chromatin compaction, relative to the other chromosomes. Supporting this interpretation, the occurrence of the active promoter mark H3K4me3 was significantly increased at promoters on chromosome IV after β-estradiol treatment (Fig. 3 A and B and SI Appendix, Figs. S6C, S7 A and B, and S9A for PCA). This was associated with a clear increase in total RNA polymerase II (Pol II) levels at chromosome IV, as well as the transcriptionally active forms of Pol II, CTD-S5p and CTD-S2p (Fig. 3 C–H and SI Appendix, Figs. S6 D–F, S7 A and B. and S9 B–D for PCA). In contrast, Pol II levels at other chromosomes declined upon CEN4* excision (Fig. 3 D, F, and H and SI Appendix, Fig. S9 B–D), suggesting that upregulation of gene expression at chromosome IV titrates Pol II. These data would suggest that yeast genes compete for the pool of Pol II and are therefore consistent with the notion that Pol II is present in limiting amounts (22). Importantly, acetylation of histones on chromosome IV peaked 120 min after CEN4* excision (Fig. 2 A and C) while the occurrence of H3K4me3, Pol II, Pol II CTD-S5p, and Pol II CTD-S2p was maximal at 180 min (Fig. 3 A, C, E, and G). Note that we have observed a mild increase of the number of mapped reads at chromosome IV in ChIPseq input samples over time and that precautions were taken to prevent bias during our above differential analysis (see Materials and Methods and SI Appendix, Fig. S10). The fact that this normalization does not affect the conclusion of our experiment supports our conclusion that the increased recruitment of Pol II on centromere-less chromosome IV is more merely due to an increased copy number of that chromosome. Together, these data indicate that the failure to recruit Hst2 and condense the chromosome in cen4- cells results first in increased histone acetylation and chromatin opening and subsequently in increased H3K4 methylation at promoters and transcription initiation.
Fig. 2.
Centromere loss triggers chromatin relaxation in cis. H4K12ac (A) and H4K16ac (C) occurrence at each chromosome and at different time points after CEN4* excision. Metagene analysis of the top 20% ChIP signals showing increasing presence of H4K12ac (B) and H4K16ac (D) at chromosome IV genes and decreasing presence at genes on other chromosomes after centromere excision (see SI Appendix, Fig. S9 for metagenes of 100% of ChIP signals). (E) ATACseq experiments showing chromatin accessibility at time points 0 min (Upper) and 180 min (Lower) at each chromosome after CEN4* excision in CEN4* and in CEN4 cells. Each chromosome is represented using a dedicated color. y axis: chromosome coordinates. Each dot corresponds to bin of 10 Kb of chromosomal DNA.
Fig. 3.
Lack of mitotic condensation triggers spontaneous transcriptional initiation. Assessment of H3K4me3 (A), Pol II (C), Pol II CTD-S5p (E), and Pol II CTD-S2p (G) occurrence using ChIPseq at each chromosome and at different time points after CEN4* excision. Metagene analysis of the top 20% ChIP signals shows increasing presence of H3K4me3 (B), Pol II (D), Pol II CTD-S5p (F), and Pol II CTD-S2p (H) at chromosome IV genes and a decrease at other chromosomes after centromere excision.
Importantly, the condensation defect provoked by CEN4* excision emerged as the most likely mechanism for the deregulation of individual genes: Unbiased hierarchical clustering analysis highlighted a cluster of genes at which an increase in positive histone marks and active Pol II clearly correlated with increased gene expression. This cluster was highly significantly enriched for chromosome IV genes (SI Appendix, Fig. S11A). While some of the other clusters were enriched for certain GO terms, this “chromosome IV” cluster was not enriched for any particular pathway (SI Appendix, Fig. S11B). Thus, the lack of histone deacetylation, Pol II removal, and demethylation of chromatin on active genes was indeed the reason for the overactivation of genes on chromosome IV upon CEN4* excision (SI Appendix, Fig. S11C).
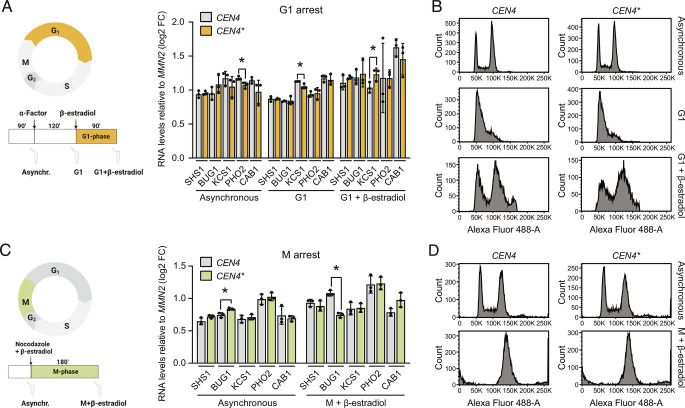
Given the fact that in budding yeast mitotic condensation has only a modest effect on gene expression during the M phase (5, 9), we hypothesized that centromere inactivation rather affected gene expression in the next interphase. In agreement with this idea, cells arrested in their cell cycle prior to chromosome condensation using hydroxyurea (HU), which arrests yeast cells in the S phase right before anaphase onset (S phase partially overlaps with early phases of mitosis in budding yeast) (23–25), did not upregulate chromosome IV genes upon CEN4* excision, as determined using a panel of strongly upregulated genes as reporters (Fig. 4 A and B). The expression of these genes was normalized to control gene MNN2 (SI Appendix, Fig. S11D). Similar results were obtained with cells arrested in G1 using alpha-factor or in prometaphase using nocodazole (Fig. 5 A–D). Thus, excision of CEN4* does not affect gene expression when cells are arrested prior to chromosome condensation, which takes place at the metaphase-to-anaphase transition in yeast. In stark contrast, when cells were released from HU arrest into mitosis CEN4* excision had a strong effect on transcriptional activation within 90 min, i.e., after the cells had traversed and completed mitosis, and remained high afterward (150 min time point; Fig. 4 B and C). These data establish that i) progression through mitosis is required for centromere excision to cause aberrant gene upregulation and ii) expression of genes located on chromosome IV remains abnormally high even after the cell has exited from mitosis. We conclude that chromosome condensation is important for setting correct transcriptional output levels during interphase. Thus, centromeres help maintain transcriptional homeostasis through their role in mitotic chromosome condensation.
Fig. 4.
Failure to condense chromatin at mitotic entry results in general upregulation of gene expression during interphase. (A) Layout of CEN4* excision in S-phase arrest and release experiments. (B) RT-qPCR analysis of the expression of five selected genes from chromosome IV at different time points before and after S-phase arrest and release. *P values < 0.05; **P values < 0.01. (C) Analysis of the cell cycle using flow cytometry performed with the cells indicated in B.
Fig. 5.
Centromere excision has no effect on gene expression in cells arrested in G1 and M phases. (A) Layout of CEN4* excision in G1-phase arrest experiments (Left). RT-qPCR analysis of the expression of six selected genes from chromosome IV at different time points before and after G1-phase arrest (Right). *P values < 0.05. (B) Analysis of the cell cycle using flow cytometry performed with the cells indicated in A. (C) Layout of CEN4* excision in M-phase arrest experiments (Left). RT-qPCR analysis of the expression of six selected genes from chromosome IV at different time points before and after M-phase arrest (Right). *P values < 0.05. (D) Analysis of the cell cycle using flow cytometry performed with the cells indicated in C.
Discussion.
Together, our data demonstrate that chromosome condensation has an important effect on regulation of gene expression in the following interphase. Failure of a mitotic chromosome to condense in mitosis results in spontaneous recruitment of the transcription machinery to an excessively relaxed chromatin, triggering deregulated transcription of the entire chromosome in the next interphase, and liberating genes from transcriptional control by upstream regulatory pathways (SI Appendix, Fig. S11E). Since yeast and mammalian chromatin remain significantly active during mitosis (4, 5), we propose that mitotic condensation, rather than passively silencing chromatin by physically excluding transcription factors in the M phase, serves as a mechanism for the cell to maintain transcriptional homeostasis in the following interphase by preventing unlicensed transcription. In this regard, it is interesting that a single locus, the centromere, instructs histone deacetylation and limits whole-chromosome gene expression in cis. Whether centromeres only contribute to regulating transcription via spreading of histone deacetylation or if they support additional regulatory mechanisms like histone methylation remains to be investigated (11, 12, 26–29). Our results also indicate that preventing chromatin condensation is sufficient to uncouple inducible genes, like the GAL genes, from their cognate signals. This stresses an underestimated prominence of chromatin regulation during adaptive activation and repression of transcription.
The effect of chromosome condensation on epigenetic markers and on gene expression in the ensuing interphase could also be relevant for asymmetrically dividing cells, such as stem cells, in which gene expression programs need to be reset to allow for maintenance of pluripotency, cellular identity, and determining cell fate (4, 10, 26, 27, 30, 31). Our study reveals an unexpected mechanism by which cells prevent postmitotic transcriptional drifting, providing new inroads for the exploration of mechanisms that promote the timely reactivation of gene expression at mitosis exit and thus safeguard cell identity and homeostasis (4). Our results may also contribute to a better understanding of the etiology of diseases involving malfunctioning centromeres (32, 33).
Materials and Methods
Strains and Culture Conditions.
All strains used in this study (SI Appendix, Table S3) are derived from the Winston S288c genetic background and were grown at 30 °C in a YPD medium [1% (w/v) yeast extract, 2% (w/v) peptone, and 2% (w/v) dextrose].
CEN4* and mCh* Excision Assays.
Excision assays and assessment of excision efficiency were performed as in ref. 12 with minor modifications (SI Appendix, SI Materials and Methods).
RNA Sequencing.
Total RNA purification was performed as in ref. 34. Library preparation, sequencing, and data analysis are described in SI Appendix, SI Materials and Methods.
Cell Synchronization.
Cells were arrested in G1, S, and M phases using a-factor, HU, and nocodazole, respectively. For detailed procedures and analysis of cell cycle by flow cytometry, see SI Appendix, SI Materials and Methods.
Real-Time qPCR (RT-qPCR).
Total RNA purification and reverse transcriptions were performed as in ref. 35 (SI Appendix, SI Materials and Methods). A complete list of primers used for RT-qPCR can be found in SI Appendix, Table S4.
Chromatin Immunoprecipitation Sequencing (ChIPseq).
Our ChIPseq procedure was adapted from ref. 12. For details about sample and library preparation, sequencing, and data analysis, see SI Appendix, SI Materials and Methods.
ATAC Sequencing.
Library preparation of ATAC-seq samples was performed as described in ref. 36 (SI Appendix, SI Materials and Methods). Details about sequencing of libraries and data analysis are available in SI Appendix, SI Materials and Methods.
Statistical Analysis.
Data processing, data visualization, and statistical methods are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Appendix 01 (PDF)
Dataset S1 (XLSX)
Dataset S2 (XLSX)
Dataset S3 (XLSX)
Acknowledgments
Sequencing was performed by the GenomEast platform, a member of the “France Genomique” consortium ANR-10-INBS-0009 and the Genomics Core Facility (GCF), Norwegian University of Science and Technology (NTNU). We thank Dr. Ignacio Garcia for biostatistical insights. Funding: The Faculty of Medicine and Health Sciences at NTNU and Central Norway Regional Health Authority (GCF). Norwegian Health Authority South-East grants 2017064, 2018012, and 2019096 (J.M.E., P.C.); The Norwegian Cancer Society grants 182524 and 208012 (J.M.E.); Research Council of Norway grants 261936, 294916, 301268, 314811, and 287911 (M.B., J.M.E., P.C.); Research Council of Norway Centers of Excellence funding scheme 262652 (J.M.E.); French National Research Agency ANR-10-INBS-0009 (S.L., B.J.); and Swiss National Science Foundation grant 31003-A-105904 (Y.B.).
Author contributions
P.C. designed research; L.R.-A. and P.C. performed research; L.R.-A., P.H., S.L.G., X.Z., B.J., and P.C. analyzed data; B.J. supervision; M.B. and J.M.E. resources; P.C. resources, supervision; and L.R.-A., P.H., S.L.G., X.Z., M.B., Y.B., J.M.E., and P.C. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Yves Barral, Email: yves.barral@bc.biol.ethz.ch.
Jorrit M. Enserink, Email: j.m.enserink@ibv.uio.no.
Pierre Chymkowitch, Email: pierre.chymkowitch@ibv.uio.no.
Data, Materials, and Software Availability
The RNAseq, ATACseq and H4K12ac, H4K16ac, H3K4me3, Pol II, Pol II CTD-S2p, and Pol II CTD-S5p ChIPseq data have been deposited to the Gene Expression Omnibus with accession number GSE20225. All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Antonin W., Neumann H., Chromosome condensation and decondensation during mitosis. Curr. Opin. Cell Biol. 40, 15–22 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Gottesfeld J. M., Forbes D. J., Mitotic repression of the transcriptional machinery. Trends Biochem Sci 22, 197–202 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Naumova N., et al. , Organization of the mitotic chromosome. Science 342, 948–953 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palozola K. C., et al. , Mitotic transcription and waves of gene reactivation during mitotic exit. Science 358, 119–122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Granovskaia M. V., et al. , High-resolution transcription atlas of the mitotic cell cycle in budding yeast. Genome Biol. 11, R24 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raccaud M., Suter D. M., Transcription factor retention on mitotic chromosomes: Regulatory mechanisms and impact on cell fate decisions. FEBS Lett. 592, 878–887 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Abramo K., et al. , A chromosome folding intermediate at the condensin-to-cohesin transition during telophase. Nat. Cell Biol. 21, 1393–1402 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H., et al. , Chromatin structure dynamics during the mitosis-to-G1 phase transition. Nature 576, 158–162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palozola K. C., Liu H., Nicetto D., Zaret K. S., Low-level, global transcription during mitosis and dynamic gene reactivation during mitotic exit. Cold Spring Harb. Symp. Quant Biol. 82, 197–205 (2018), 10.1101/sqb.2017.82.034280. [DOI] [PubMed] [Google Scholar]
- 10.Timmers H. T. M., Verrijzer C. P., Mitotic chromosomes: Not so silent after all. Dev. Cell 43, 119–121 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Wilkins B. J., et al. , A cascade of histone modifications induces chromatin condensation in mitosis. Science 343, 77–80 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Kruitwagen T., Chymkowitch P., Denoth-Lippuner A., Enserink J., Barral Y., Centromeres license the mitotic condensation of yeast chromosome arms. Cell 175, 780–795.e715 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kruitwagen T., Denoth-Lippuner A., Wilkins B. J., Neumann H., Barral Y., Axial contraction and short-range compaction of chromatin synergistically promote mitotic chromosome condensation. Elife 4, e1039 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warsi T. H., Navarro M. S., Bachant J., DNA topoisomerase II is a determinant of the tensile properties of yeast centromeric chromatin and the tension checkpoint. Mol. Biol. Cell 19, 4421–4433 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phenix H., et al. , Quantitative epistasis analysis and pathway inference from genetic interaction data. PLoS Comput. Biol. 7, e1002048 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heyken W. T., Repenning A., Kumme J., Schuller H. J., Constitutive expression of yeast phospholipid biosynthetic genes by variants of Ino2 activator defective for interaction with Opi1 repressor. Mol. Microbiol. 56, 696–707 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Shore D., Zencir S., Albert B., Transcriptional control of ribosome biogenesis in yeast: Links to growth and stress signals. Biochem. Soc. Trans. 49, 1589–1599 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gasch A. P., et al. , Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241–4257 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shor E., Fox C. A., Broach J. R., The yeast environmental stress response regulates mutagenesis induced by proteotoxic stress. Plos Genet. 9, e1003680 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang Y.-L., et al. , Yeast Cip1 is activated by environmental stress to inhibit Cdk1–G1 cyclins via Mcm1 and Msn2/4. Nat. Commun. 8, 56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terhorst A., et al. , The environmental stress response causes ribosome loss in aneuploid yeast cells. Proc. Natl. Acad. Sci. U.S.A. 117, 17031–17040 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swaffer M. P., et al. , RNA polymerase II dynamics and mRNA stability feedback scale mRNA in proportion to cell size. bioRxiv [Preprint] (2022). 10.1101/2021.09.20.461005 Accessed 20 December 2022. [DOI]
- 23.Slater M. L., Effect of reversible inhibition of deoxyribonucleic acid synthesis on the yeast cell cycle. J. Bacteriol. 113, 263–270 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivanova T., et al. , Budding yeast complete DNA synthesis after chromosome segregation begins. Nat. Commun. 11, 2267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishnan V., Nirantar S., Crasta K., Cheng A. Y., Surana U., DNA replication checkpoint prevents precocious chromosome segregation by regulating spindle behavior. Mol. Cell 16, 687–700 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Palozola K. C., Lerner J., Zaret K. S., A changing paradigm of transcriptional memory propagation through mitosis. Nat. Rev. Mol. Cell Biol. 20, 55–64 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y., et al. , Widespread mitotic bookmarking by histone marks and transcription factors in pluripotent stem cells. Cell Rep. 19, 1283–1293 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teves S. S., et al. , A stable mode of bookmarking by TBP recruits RNA polymerase II to mitotic chromosomes. Elife 7, e35621 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu J. Y., et al. , Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102, 279–291 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Neurohr G., et al. , A midzone-based ruler adjusts chromosome compaction to anaphase spindle length. Science 332, 465–468 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Roubinet C., White I. J., Baum B., Asymmetric nuclear division in neural stem cells generates sibling nuclei that differ in size, envelope composition, and chromatin organization. Curr. Biol. 31, 3973–3983.e3974 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barra V., Fachinetti D., The dark side of centromeres: Types, causes and consequences of structural abnormalities implicating centromeric DNA. Nat. Commun. 9, 4340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smurova K., De Wulf P., Centromere and pericentromere transcription: Roles and regulation … in sickness and in health. Front. Genet. 9, 674 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chymkowitch P., et al. , TORC1-dependent sumoylation of Rpc82 promotes RNA polymerase III assembly and activity. Proc. Natl. Acad. Sci. U.S.A. 114, 1039–1044 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chymkowitch P., et al. , Cdc28 kinase activity regulates the basal transcription machinery at a subset of genes. Proc. Natl. Acad. Sci. U.S.A. 109, 10450–10455 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hendrickson D. G., Soifer I., Wranik B. J., Botstein D., Scott McIsaac R., Simultaneous profiling of DNA accessibility and gene expression dynamics with ATAC-Seq and RNA-Seq. Methods Mol. Biol. 1819, 317–333 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S1 (XLSX)
Dataset S2 (XLSX)
Dataset S3 (XLSX)
Data Availability Statement
The RNAseq, ATACseq and H4K12ac, H4K16ac, H3K4me3, Pol II, Pol II CTD-S2p, and Pol II CTD-S5p ChIPseq data have been deposited to the Gene Expression Omnibus with accession number GSE20225. All study data are included in the article and/or SI Appendix.