Significance
The entomopathogenic fungus Metarhizium robertsii offers an environmentally friendly alternative to chemical insecticides for biocontrol of insect pests. Upon insect cuticle induction, fungal pathogens produce appressoria that develop enormous turgor pressure to breach the host cuticle. A better understanding of the regulatory mechanism underlying appressorium-mediated cuticle penetration is critical for improvement of its efficacy. Here, we reveal an important regulatory role of the histone lysine methyltransferase ASH1 in appressorium turgor generation. ASH1 is significantly up-regulated upon cuticle exposure. Furthermore, ASH1-mediated H3K36me2 activates the peroxin gene pex16. PEX16 is essential for the biogenesis of peroxisomes that promote lipid hydrolysis to produce large amounts of glycerol for turgor generation. The ASH1–PEX16 regulatory pathway may lead to strategies for improvement of fungal biopesticides.
Keywords: entomopathogenic fungi, host–microbe interactions, histone methylation, appressorium turgor generation, mosquitoes
Abstract
Entomopathogenic fungi infect insects by penetrating through the cuticle into the host body. To breach the host cuticle, some fungal pathogens produce specialized infection cells called appressoria, which develop enormous turgor pressure to allow cuticle penetration. However, regulatory mechanisms underlying appressorium turgor generation are poorly understood. Here, we show that the histone lysine methyltransferase ASH1 in the insecticidal fungus Metarhizium robertsii, which is strongly induced during infection of the mosquito cuticle, regulates appressorium turgor generation and cuticle penetration by activating the peroxin gene Mrpex16 via H3K36 dimethylation. MrPEX16 is required for the biogenesis of peroxisomes that participate in lipid catabolism and further promotes the hydrolysis of triacylglycerols stored in lipid droplets to produce glycerol for turgor generation, facilitating appressorium-mediated insect infection. Together, the ASH1–PEX16 pathway plays a pivotal role in regulating peroxisome biogenesis to promote lipolysis for appressorium turgor generation, providing insights into the molecular mechanisms underlying fungal pathogenesis.
Entomopathogenic fungi such as Metarhizium robertsii are considered environmentally friendly alternatives to chemical pesticides; these fungi can naturally cause epidemics among host insects and thus be used to control pest populations. Importantly, entomopathogenic fungi are the only insect pathogens able to infect their hosts by penetration through the cuticle and are especially suitable for controlling insects with piercing and sucking mouthparts, such as adult mosquitoes (1, 2). But fungal bioinsecticides tend to be less effective than chemical insecticides, limiting their widespread application (3). Therefore, a deeper understanding of the molecular mechanism underlying fungal pathogenicity is required to improve the efficacy of fungal biopesticides.
During the course of infection in insects, penetration into the host cuticle is a prerequisite for successful fungal infection. After adhering to the cuticle, the fungal conidium germinates and differentiates into a specialized infection structure called an appressorium (4, 5). Appressorium-mediated cuticle penetration is accompanied by the combined action of cuticle-degrading enzymes and turgor pressure (6, 7). As an appressorium develops, lipid droplets, which are dynamic intracellular lipid storage organelles, gradually transport from the conidium to the appressorium, where the triacylglycerols (TAGs) stored within the lipid droplets are catabolized into free fatty acids and glycerol via a process known as lipolysis (8, 9). The accumulating glycerol generates enormous internal turgor pressure, which is required for appressorium-mediated penetration through the insect cuticle (10–12). Although it has been suggested that the glycerol derived from lipid droplets serves as an osmolyte for turgor pressure in entomopathogenic fungi (7), the regulatory mechanism underlying appressorium turgor generation remains elusive.
Appressorium differentiation, lipid mobilization, and turgor generation involve various cellular biological processes, which require rapid response and finely tuned regulation of gene expression programs (13, 14). Epigenetic regulatory mechanisms, especially histone methylation, which regulates gene expression at the transcriptional level, have been shown to play an important role in the infection-related morphogenesis of pathogenic fungi (15, 16). We previously revealed that appressorium differentiation in M. robertsii is regulated by the KMT2 (histone lysine (K) methyltransferase 2, catalyzing H3K4 methylation)-Cre1-Hyd4 coordinated regulatory pathway (15). Whether histone lysine methylation also regulates the expression of genes involved in appressorium-mediated penetration, especially in appressorium turgor generation, is unknown. By examining the transcriptomic profiles of the entomopathogenic fungus over the course of mosquito infection, we discovered that a putative histone methyltransferase, a homologue of the ASH1 (absent, small, or homeotic discs 1) protein in Drosophila melanogaster (17), was significantly up-regulated during cuticle penetration (14). ASH1 specifically catalyzes H3K36 methylation in other fungi and has a major impact on genome stability (18, 19). This indicates the potential role of ASH1-mediated histone methylation in entomopathogenic fungi for the regulation of cuticle penetration.
In this study, we reveal that MrASH1 (ASH1 from M. robertsii, EXU96232.1) plays a crucial role in the coordinated regulation of appressorium turgor generation by directly activating the peroxin gene Mrpex16 via H3K36 dimethylation (H3K36me2). PEX16 is required for the biogenesis of peroxisomes where fatty acids are efficiently degraded, which further promotes the hydrolysis of lipid droplets to release glycerol for enormous appressorial turgor pressure, allowing the fungus to breach the insect cuticle and cause infection. These findings provide insights into the epigenetic mechanism of peroxisome biogenesis and appressorium-mediated insect infection by a fungal pathogen.
Results
MrASH1, an H3K36me2 Methyltransferase in M. robertsii, Positively Regulates Fungal Pathogenicity.
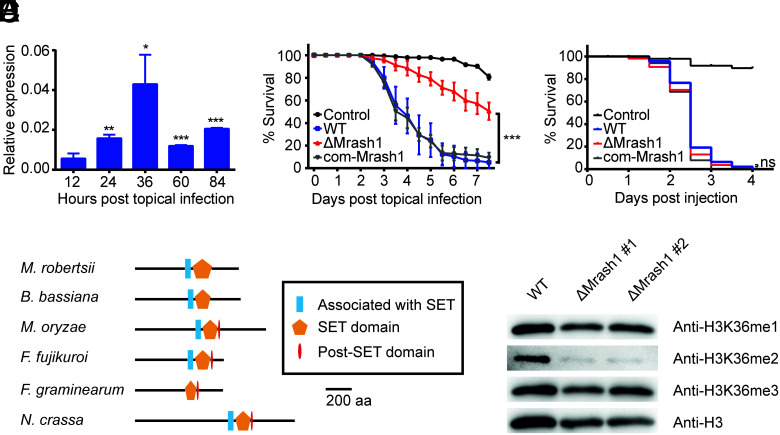
A transcription level analysis of M. robertsii wild type (WT) over the course of topical infection in female adult Anopheles stephensi mosquitoes demonstrated that Mrash1 expression was markedly up-regulated after topical infection and peaked at 36 h after infection when the fungus was penetrating the insect cuticle to enter the hemocoel (Fig. 1A). To determine the function of Mrash1 in the entomopathogenic fungus, we constructed an Mrash1 knockout strain (ΔMrash1) and the complemented strain com-Mrash1 (SI Appendix, Fig. S1). Conidial germination was not affected by Mrash1 deletion (SI Appendix, Fig. S2). A topical infection bioassay of A. stephensi mosquitoes showed that the virulence of ΔMrash1 was significantly reduced compared with that of WT (P < 0.001), while the complemented strain com-Mrash1 showed recovery of virulence (Fig. 1B). However, an injection bioassay in which fungal conidia were directly injected into the mosquito hemocoel showed no significant differences in virulence between WT and ΔMrash1 (Fig. 1C). These results indicated that Mrash1 is required for topical fungal infection of the host cuticle rather than for subsequent infection inside the insect hemocoel.
Fig. 1.
MrASH1, an H3K36me2 methyltransferase in M. robertsii, affects fungal pathogenicity. (A) qPCR analysis of Mrash1 transcription during topical infection of A. stephensi by the WT strain. Data are shown as the mean ± SD of three technical replicates. Significant differences compared with that at 12 h after topical infection were determined by Student's t test. *P < 0.05, **P < 0.01, and ***P < 0.001. gpd was used as a reference gene. The experiments were repeated twice with similar results. (B) Survival of female adult A. stephensi mosquitoes following topical application of conidial suspension (6 × 106 conidia/ml) of the WT, ΔMrash1, and com-Mrash1 strains. The control mosquitoes were treated with 0.01% Triton X-100. Each treatment was replicated three times, with 50 mosquitoes per replicate. Significant differences compared with those in WT were determined by the log-rank (Mantel–Cox) test. ***P < 0.001. The experiments were repeated twice with similar results. (C) Survival of female adult A. stephensi mosquitoes after injection of conidial suspension (138 nL of 1 × 106 conidia/mL) of the WT, ΔMrash1, and com-Mrash1 strains. The control mosquitoes were injected with 0.01% Triton X-100 in PBS. Fifty mosquitoes were used in each treatment. Significant differences compared with those in WT were determined by the log-rank (Mantel–Cox) test. ns, not significant. (D) Protein domain structures of MrASH1 and its homologues in other fungal species. aa, amino acid. (E) Western blotting analysis of histone modifications in the WT and two ΔMrash1 mutants. Histones extracted from M. robertsii mycelia grown in MM2 medium were resolved by 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and probed with specific antibodies against H3K36me1, H3K36me2, H3K36me3, and a C-terminal peptide of histone H3.
By protein sequence alignment, we found that MrASH1 homologues were present in pathogenic fungi, fruit fly (D. melanogaster), and mammals (SI Appendix, Fig. S3). Furthermore, MrASH1 was evolutionally closer to homologues from another entomopathogenic fungus, Beauveria bassiana (identity rate 59.64%), and shared high similarity in protein structure with ASH1 homologues of several filamentous fungi, including plant pathogens, such as Magnaporthe oryzae and Fusarium graminearum, and the model fungus Neurospora crassa (Fig. 1D). All of these homologues possessed a SET (Su(var)3-9, Enhancer-of-zeste, Trithorax) domain, and most of them had an AWS (Associated with SET) domain and a Post-SET domain. Since ASH1 in Fusarium fujikuroi and Neurospora crassa has been reported to catalyze H3K36 methylation (18, 19), MrASH1 likely has similar catalytic activity. Thus, we extracted the fungal histones and determined the histone methylation levels in different strains by western blotting. We found that H3K36 dimethylation (H3K36me2) levels were drastically reduced in ΔMrash1 compared with WT (Fig. 1E), but H3K36 mono- and trimethylation (H3K36me1 and H3K36me3) signals were not significantly altered after Mrash1 deletion. These observations indicated that MrASH1 is specifically responsible for H3K36me2 modification in M. robertsii.
MrASH1 Is Required for Cuticle Penetration, Appressorium Turgor Generation, and Fungal Lipolysis.
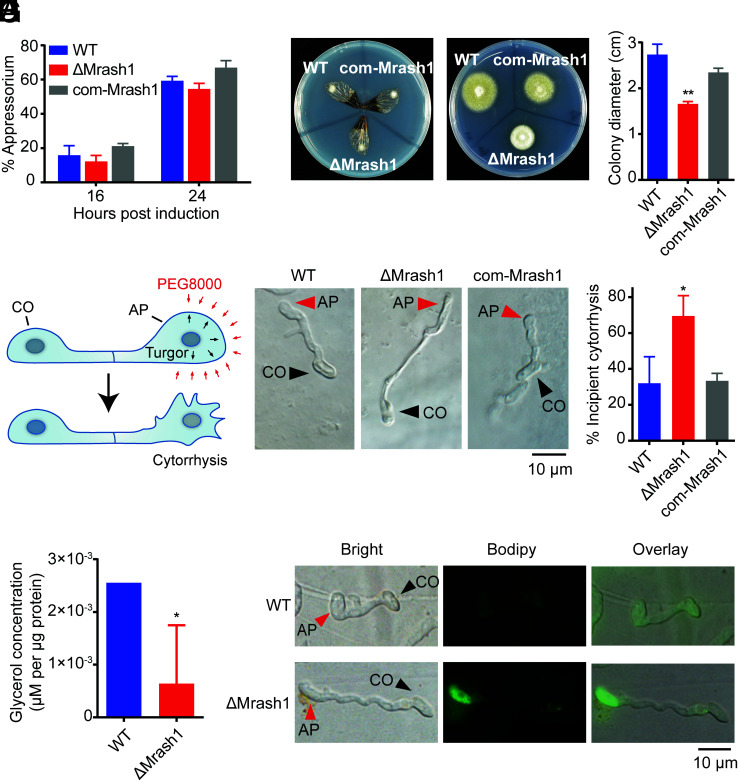
To determine whether MrASH1 modulates appressorium differentiation, we inoculated conidia of WT, ΔMrash1, and com-Mrash1 on hydrophobic plates to induce appressorium differentiation (15). We found that deletion of Mrash1 did not impact the rate of appressorium formation (Fig. 2A), suggesting that the decreased virulence of ΔMrash1 was not due to appressorium differentiation.
Fig. 2.
ΔMrash1 exhibits defects in cuticle penetration, appressorium turgor generation, glycerol accumulation, and lipid droplet degradation in M. robertsii. (A) The percentage of germinated conidia with appressoria relative to the total germinated conidia at 16 h and 24 h after induction in MM-Gly medium on hydrophobic plates. Data are shown as the mean ± SD of three biological replicates. The experiments were repeated twice with similar results. (B) The penetration capacity of WT, ΔMrash1, and com-Mrash1 was assayed using cicada wings. Mycelial blocks inoculated on the cicada wings were incubated for 2 d (Left). Then, the cicada wings were removed, and the plates were incubated for an additional 5 d (Right). (C) Colony diameters of WT, ΔMrash1, and com-Mrash1 at 5 d after removing the cicada wings in the cuticle penetration assay. Data are shown as the mean ± SD of three biological replicates. Significant differences compared with those in WT were determined by Student's t test. **P < 0.01. The experiments were repeated twice with similar results. (D) A schematic model of the incipient cytorrhysis assay. Black and red arrows in the upper image indicate appressorium turgor pressure and exogenous pressure exerted on the appressorium cell wall by PEG8000, respectively. The appressorium collapsed when the pressure of PEG8000 exceeded the appressorium turgor pressure, as shown in the lower image. (E) Microscopic images of WT, ΔMrash1, and com-Mrash1 appressoria induced on cicada wings after immersion in PEG8000 for 10 min in an incipient cytorrhysis assay. (F) Percentage of appressoria induced on cicada wings that underwent incipient cytorrhysis after immersion in PEG8000 for 10 min. Data are shown as the mean ± SD of three biological replicates. Significant differences compared with those in WT were determined by Student's t test. *P < 0.05. The experiments were repeated twice with similar results. (G) Intracellular glycerol levels in WT and ΔMrash1 appressoria induced on hydrophobic plates. Data are shown as the mean ± SD of three biological replicates. Significant differences compared with those in WT were determined by Student's t test. *P < 0.05. (H) Distribution of lipid droplets in appressoria of WT and ΔMrash1 induced on cicada wings. Lipid droplets were stained with BODIPY. CO, conidium; AP, appressorium.
We next examined the insect cuticle penetration ability of the WT, ΔMrash1, and com-Mrash1 appressoria on cicada wings. The cicada wing bioassay showed that the penetration capability of ΔMrash1 was dramatically decreased compared with that of WT (Fig. 2 B and C), while the com-Mrash1 strain rescued the penetration defect, indicating that Mrash1 plays a crucial role in appressorium-mediated penetration of the insect cuticle.
During topical infection, entomopathogenic fungi secrete proteases and chitinases to degrade the insect cuticle. To test whether Mrash1 regulates insect cuticle penetration by affecting the expression of the main cuticle-degrading enzymes (20), we determined the gene transcription levels of two proteases and five chitinases involved in cuticle degradation by qPCR. The transcription levels of these genes were not down-regulated in ΔMrash1 compared with those in WT, and some genes were even up-regulated (SI Appendix, Fig. S4), indicating that Mrash1 did not affect cuticle penetration capability by regulating cuticle-degrading enzymes.
Turgor pressure is required for appressorium-mediated penetration by M. robertsii (7). We used an incipient cytorrhysis assay to test whether Mrash1 affected appressorium turgor pressure (Fig. 2D). Cytorrhysis provides a measure of internal cellular turgor based on the external solute (such as PEG8000 solution) concentration necessary to collapse the appressorium (21). The ratio of incipient cytorrhysis occurring in the appressorium is negatively related to the turgor pressure, i.e., appressoria with high turgor pressure result in a low ratio of incipient cytorrhysis. We inoculated fungal conidia on cicada wings for 18 h and then immersed them in PEG8000 solution. The results showed that only a small percentage (32.0% ± 12.1%) of appressoria in WT were not intact because of the external permeation pressure, whereas a large percentage (69.5% ± 9.2%) of appressoria in ΔMrash1 showed obvious cytorrhysis (Fig. 2 E and F). This phenotypic defect was restored in the complemented strain com-Mrash1. Collectively, these observations indicated that deletion of Mrash1 led to decreased turgor pressure in the appressorium, suggesting that Mrash1 is required for appressorium turgor generation.
A previous study has indicated that glycerol, a product of lipid degradation in fungi, is the main osmolyte for turgor generation (10). We thereby examined the glycerol concentration in appressoria of WT, ΔMrash1, and com-Mrash1 inoculated on hydrophobic plates (15). In comparison with WT, the appressoria of which accumulated glycerol to a concentration of 2.56 × 10−3 μM/μg protein, the ΔMrash1 appressoria generated only 6.40 × 10−4 μM glycerol/μg protein (Fig. 2G) at 20 h after inoculation, the time point at which glycerol was produced for turgor generation (SI Appendix, Fig. S5). Glycerol is produced from TAGs stored within lipid droplets via lipolysis (11). Lipid droplets, serving as energy storage organelles composed of neutral lipids in conidia, translocate to the appressorium and vanish during appressorium development due to the high rate of lipid degradation. Therefore, we further examined the changes in lipid droplets in the appressoria of WT and ΔMrash1 by staining with BODIPY. We found that the lipid droplets were almost completely degraded in the WT appressorium, whereas abundant lipid droplets were still distributed in the ΔMrash1 appressorium, indicating that MrASH1 plays an important role in lipolysis of TAGs within lipid droplets during appressorium development (Fig. 2H). These results indicated that the severely decreased virulence of the ΔMrash1 mutant was due to the reduced appressorium turgor pressure required to breach the host cuticle, which resulted from defective lipolysis of TAGs and decreased glycerol production in the appressorium.
MrASH1-Mediated H3K36me2 Modification Directly Regulates the Transcription of Lipid Metabolism–Related Genes.
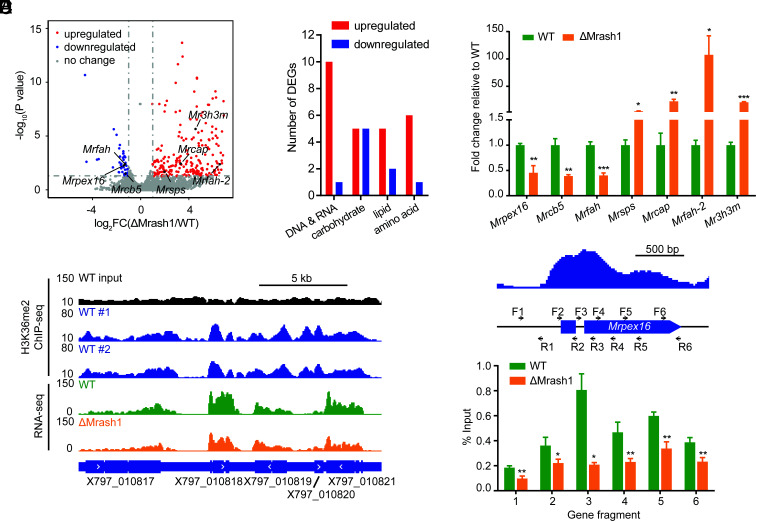
Given that MrASH1 is a histone methyltransferase responsible for H3K36me2 that affects gene transcription, we reasoned that MrASH1 might modulate glycerol production in the appressorium by regulating genes involved in lipid metabolism. To test this hypothesis and identify genes regulated by MrASH1, we conducted RNA-seq to analyze changes in global gene expression in WT and ΔMrash1 strains upon exposure to insect cuticles. In total, 281 genes showed significant differences (≥2-fold change, P < 0.05) between the two strains, with 235 genes up-regulated and 46 genes down-regulated in ΔMrash1 compared with WT (Fig. 3A). These differentially expressed genes (DEGs) were overrepresented in the functional GO categories of catalytic activity, binding, metabolic process, and cellular process (SI Appendix, Fig. S6). Notably, three down-regulated genes in ΔMrash1, including the peroxin gene Mrpex16 (X797_007679) and two fatty acid hydroxylase genes Mrcb5 (X797_008851) and Mrfah (X797_002674), were involved in lipid catabolism. Moreover, four up-regulated genes, including the squalene/phytoene synthase gene Mrsps (X797_006414), CDP-alcohol phosphatidyltransferase gene Mrcap (X797_002723), fatty acid hydroxylase gene Mrfah-2 (X797_006379), and 3-hydroxy-3-methylglutaryl-CoA-synthase gene Mr3h3m (X797_006417), were also involved in lipid anabolism (Fig. 3 A and B). RT–qPCR confirmed that these seven genes had transcriptional changes consistent with the RNA-seq data (Fig. 3C). Collectively, the transcription of these seven lipid metabolism–related genes is regulated by MrASH1 upon cuticle exposure.
Fig. 3.
MrASH1 positively regulates the peroxin gene Mrpex16 in M. robertsii. The mycelia of WT and ΔMrash1 incubated in MM2 medium supplemented with adult locust cuticle for 8 h were analyzed by a combined RNA-seq and H3K36me2 ChIP-seq analysis. (A) Volcano plot of DEGs (fold change ≥ 2, P < 0.05) between WT and ΔMrash1 identified by transcriptome analysis. Significantly up-regulated, down-regulated, and unchanged genes in ΔMrash1 compared with those in WT are marked in red, blue, and gray, respectively. Lipid metabolism–related genes are marked in black. (B) DEGs by gene ontology category (primary metabolism). Red and blue columns represent the number of genes up-regulated and down-regulated in ΔMrash1 compared with that in WT, respectively. (C) qPCR analysis of lipid metabolism–related genes in the mycelium of WT and ΔMrash1 cultured in MM2 medium supplemented with adult locust cuticle. Data are shown as the mean ± SD of three technical replicates. Statistical significance was determined with Student's t test. *P < 0.05, **P < 0.01, and ***P < 0.001. gpd was used as a reference gene. The experiments were repeated twice with similar results. (D) Representative genome browser view of the enrichment of H3K36me2 in two WT replicates and mRNA signals in WT and ΔMrash1. (E) H3K36me2 levels at the Mrpex16 gene locus in WT and ΔMrash1. The upper image shows H3K36me2 enrichment at the Mrpex16 gene locus in WT, as determined by ChIP-seq analysis, and the black arrows indicate the position and polarity (5′-3′) of the primers used for ChIP–qPCR. The lower image shows ChIP–qPCR validation of ChIP-seq shown as the percentage of the signal from immunoprecipitation over the input in WT and ΔMrash1. Data are shown as the mean ± SD of three technical replicates. Statistical significance was determined with Student's t test. *P < 0.05 and **P < 0.01. The experiments were repeated twice with similar results.
Next, to examine whether these DEGs were directly regulated by MrASH1-mediated H3K36me2 modification, we performed chromatin immunoprecipitation (ChIP) using an H3K36me2 antibody followed by DNA sequencing (ChIP-seq) and profiled the chromatin landscape of H3K36me2 in the WT strain upon cuticle exposure. Genome-wide analysis revealed that H3K36me2 was predominantly distributed across the coding region of the genes, with more accumulation at the transcription start site (Fig. 3D). Among all the DEGs identified by RNA-seq data analysis, the coding regions of 27 down-regulated and 171 up-regulated genes in ΔMrash1 were modified with H3K36me2, indicating that these genes were directly regulated by MrASH1-mediated H3K36me2; thus, these genes were identified as MrASH1 targets. Notably, all seven DEGs involved in lipid metabolism identified by RNA-seq were modified by H3K26me2 in WT as they lost the H3K36me2 modification in ΔMrash1, which was confirmed by ChIP quantitative PCR (ChIP–qPCR) (Fig. 3E and SI Appendix, Fig. S7). These results indicated that these seven lipid metabolism–related genes were directly regulated by MrASH1 via H3K36me2. Among them, three genes, including Mrpex16, were activated, while the other four lipid metabolism–related genes were repressed by MrASH1-mediated H3K36me2.
Mrpex16, the Key Target of MrASH1, Is Involved in Regulating Appressorium Turgor Generation and Cuticle Penetration.
To assess whether these lipid metabolism–related genes, regulated by MrASH1-mediated H3K36me2, were involved in cuticle penetration, we constructed individual deletion mutants of these genes and performed a cuticle penetration assay. Of these, deletion of only Mrpex16, but not the other six genes, attenuated cuticle penetration on cicada wings in comparison with WT (Fig. 4 A and B and SI Appendix, Fig. S8). This finding indicated that Mrpex16 might function as the key target of MrASH1 to regulate appressorium-mediated cuticle penetration.
Fig. 4.
Mrpex16 affects cuticle penetration, appressorium turgor generation, glycerol accumulation, lipid droplet degradation, and fungal pathogenicity in M. robertsii. (A) The penetration capacity of the WT and two ΔMrpex16 strains was assayed using cicada wings. Mycelial blocks inoculated on the cicada wings were incubated for 2 d (Left). Then, the cicada wings were removed, and the plates were incubated for an additional 5 d (Right). (B) Colony diameters of the WT and two ΔMrpex16 strains at 5 d after removing the cicada wings in the cuticle penetration assay. Data are shown as the mean ± SD of three biological replicates. Significant differences compared with those in WT were determined by Student's t test. **P < 0.01. The experiments were repeated twice with similar results. (C) Percentage of germinated conidia with appressoria relative to the total germinated conidia at 16 h and 24 h after induction in MM-Gly medium on hydrophobic plates. Data are shown as the mean ± SD of three biological replicates. The experiments were repeated twice with similar results. (D) Microscopic images of the WT and two ΔMrpex16 strains appressoria induced on cicada wings after immersion in PEG8000 for 10 min in an incipient cytorrhysis assay. (E) Percentage of appressoria induced on cicada wings that underwent incipient cytorrhysis after immersion in PEG8000 for 10 min. Data are shown as the mean ± SD of three biological replicates. Significant differences compared with those in WT were determined by Student's t test. **P < 0.01 and ***P < 0.001. (F) Intracellular glycerol levels in WT and ΔMrpex16 appressoria induced on hydrophobic plates. Data are shown as the mean ± SD of three biological replicates. Significant differences compared with those in WT were determined by Student's t test. *P < 0.05. The experiments were repeated twice with similar results. (G) Distribution of lipid droplets in appressoria of WT and ΔMrpex16 induced on cicada wings. Lipid droplets were stained with BODIPY. CO, conidium; AP, appressorium. (H) Survival of female adult A. stephensi mosquitoes following topical application of conidial suspensions (6 × 106 conidia/mL) of the WT and two ΔMrpex16 strains. The control mosquitoes were treated with 0.01% Triton X-100. Each treatment was replicated three times, with 50 mosquitoes per replicate. Significant differences compared with those in WT were determined by the log-rank (Mantel–Cox) test. *P < 0.05 and ***P < 0.001. The experiments were repeated twice with similar results. (I) Survival of female adult A. stephensi mosquitoes after injection of conidial suspensions (138 nL of 1 × 106 conidia/mL) of the WT and two ΔMrpex16 strains. The control mosquitoes were injected with 0.01% Triton X-100 in PBS. Fifty mosquitoes were used in each treatment. Significant differences compared with those in WT were determined by the log-rank (Mantel–Cox) test. ***P < 0.001. The experiments were repeated twice with similar results.
By protein sequence alignment, we found that MrPEX16 (EXU99250.1) was homologous to the peroxin PEX16 in the yeast Yarrowia lipolytica (33.75% identity) and the filamentous fungus Aspergillus kawachii (51.43% identity) with a conserved PEX16 domain (SI Appendix, Fig. S9). We found that deletion of Mrpex16 did not affect conidial germination or appressorium formation (SI Appendix, Fig. S10 and Fig. 4C) but markedly impaired cuticle penetration in M. robertsii (Fig. 4 A and B). Turgor generation in appressoria was significantly reduced in the ΔMrpex16 mutant compared with that in WT, as indicated by a dramatically higher rate of cell collapse at the same osmolarity in the incipient cytorrhysis assay (Fig. 4 D and E). Moreover, glycerol accumulation was significantly reduced in ΔMrpex16 appressoria (Fig. 4F). Accordingly, the lipid droplets were not utilized in ΔMrpex16 during appressorium development (Fig. 4G). Due to the defect in cuticle penetration, deletion of Mrpex16 markedly reduced fungal virulence against the A. stephensi mosquito in topical bioassays (P < 0.05) compared with that of the WT strain (Fig. 4H). However, the virulence of the ΔMrpex16 strain was not attenuated when the cuticle was bypassed by direct infection of conidia into the mosquito hemocoel (Fig. 4I), suggesting that Mrpex16 is specifically required for appressorium-mediated cuticle infection. These results showed that the phenotypic defects of the ΔMrpex16 strain resembled those of the ΔMrash1 strain. Therefore, as the key target of MrASH1-mediated H3K36me2, Mrpex16 plays a crucial role in lipid metabolism and appressorium-mediated cuticle penetration.
MrPEX16 Regulates Peroxisome Biogenesis to Promote Lipolysis for Appressorium Turgor Generation.
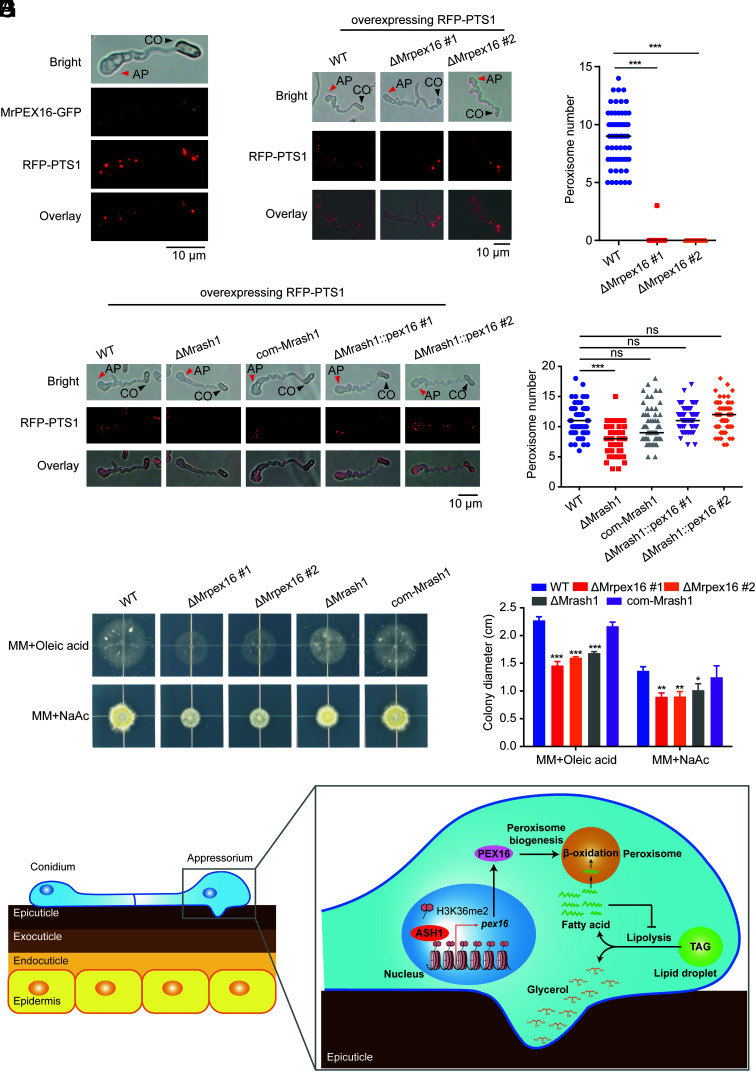
As a peroxin family gene, Mrpex16 was recently shown to be indispensable for peroxisome formation in the white koji fungus, A. kawachii (22). To determine whether Mrpex16 is also involved in peroxisome formation in M. robertsii, we first investigated the localization of MrPEX16. We constructed transgenic strains that simultaneously overexpressed MrPEX16 fused with GFP (MrPEX16–GFP) and RFP fused with peroxisome-targeting signal 1 (RFP–PTS1). PTS1 is a peptide in the C terminus of peroxisome-targeting proteins that mediates the transport of these proteins into peroxisomes. Therefore, fluorescent protein–fused PTS1 can be used as a peroxisome marker for visualization of peroxisomes in fungal cells (23). As shown in Fig. 5A, peroxisomes marked with RFP–PTS1 (red) were distributed in fungal cells and especially enriched in the appressorium. MrPEX16–GFP (green) colocalized exclusively with RFP–PTS1 (red) in the fungal conidia and appressorium, indicating that MrPEX16 localizes in peroxisomes of M. robertsii.
Fig. 5.
MrASH1 activates Mrpex16 to regulate peroxisome biogenesis and fatty acid utilization in M. robertsii. (A) Subcellular locations of PEX16 fused with GFP and peroxisomes marked with RFP–PTS1 in the appressorium of WT induced in MM-Gly medium on hydrophobic plates for 20 h. CO, conidium; AP, appressorium. (B) Subcellular locations of peroxisomes marked with RFP–PTS1 in the appressoria of the WT and two ΔMrpex16 strains induced in MM-Gly medium on hydrophobic plates for 20 h. CO, conidium; AP, appressorium. (C) Quantification of peroxisomes in the appressoria of the WT and two ΔMrpex16 strains induced in MM-Gly medium on hydrophobic plates for 20 h. In total, 50 to 100 appressoria were detected for each strain. Horizontal lines represent the medians. Significant differences in the median compared with those in WT were determined by Student's t test. ***P < 0.001. The experiments were repeated twice with similar results. (D) Subcellular locations of peroxisomes marked with RFP–PTS1 in the appressoria of the WT, ΔMrash1, com-Mrash1, and two ΔMrash1::pex16 strains induced in MM-Gly medium on hydrophobic plates for 20 h. CO, conidium; AP, appressorium. (E) Quantification of peroxisomes in the appressoria of the WT, ΔMrash1, com-Mrash1, and two ΔMrash1::pex16 strains induced in MM-Gly medium on hydrophobic plates for 20 h. In total, 50 to 100 appressoria were detected for each strain. Horizontal lines represent the medians. Significant differences in the median compared with those in WT were determined by Student's t test. ***P < 0.001. ns, not significant. The experiments were repeated twice with similar results. (F) Fungal colonies of the WT, two ΔMrpex16 strains, ΔMrash1, and com-Mrash1 strains grown on MM plates supplied with 2.5 mM oleic acid (a long-chain fatty acid) or 50 mM NaAc (sodium acetate, a short-chain fatty acid) for 7 d. (G) Colony diameters of the WT, two ΔMrpex16 strains, ΔMrash1, and com-Mrash1 strains on fatty acid or acetate plates. Data are shown as the mean ± SD of three biological replicates. Significant differences compared with those in WT were determined by Student's t test. *P < 0.05, **P < 0.01, and ***P < 0.001. The experiments were repeated twice with similar results. (H) Schematic model showing the regulatory mechanism of the ASH1–PEX16 regulatory pathway in appressorium turgor generation and cuticle penetration in the insect pathogenic fungus. Upon host cuticle exposure, the up-regulated epigenetic regulator MrASH1 activates the target gene Mrpex16 via H3K36me2 modification on chromatin to control peroxisome biogenesis in the appressorium. Efficient degradation of fatty acids in the peroxisome via β-oxidation prevents their accumulation from inhibiting lipolysis and in turn promotes the hydrolysis of TAGs within lipid droplets to produce large amounts of glycerol for appressorium turgor pressure generation.
To observe the peroxisomes in the appressorium, we overexpressed RFP–PTS1 in the WT and ΔMrpex16 strains. We found that numerous peroxisomes (dot-like structures, red fluorescence) were present in the appressorium of the WT strain (Fig. 5B), but no peroxisome was observed in the ΔMrpex16 appressorium (Fig. 5C), indicating that Mrpex16 is required for peroxisome biogenesis in M. robertsii. Furthermore, the number of peroxisomes was dramatically reduced in the ΔMrash1 strain compared with that in the WT strain, while the complemented strain showed recovery of the defect (Fig. 5 D and E). Notably, overexpression of Mrpex16 rescued the defect in the biogenesis of peroxisomes in the ΔMrash1 strain (Fig. 5 D and E), indicating that MrASH1 regulates peroxisome formation via MrPEX16 at a genetic level. These results showed that the histone methyltransferase ASH1 orchestrates peroxisomal gene pex16 transcription during appressorium development, revealing a role of histone methylation in peroxisome biogenesis.
During the course of fungal appressorium formation, TAGs stored within lipid droplets are converted to free fatty acids and glycerol via a process known as lipolysis (11). Subsequently, glycerol accumulates to generate turgor pressure, while fatty acids enter the peroxisome for further catabolism via peroxisomal β-oxidation (24). Fungal cells with defective peroxisomes may exhibit aberrant accumulation of fatty acids, which would inhibit TAG mobilization from lipid droplets (25, 26). To test whether MrPEX16 and MrASH1 affected peroxisomal fatty acid degradation, we compared the fatty acid utilization ability of ΔMrpex16 and ΔMrash1 with that of WT by inoculating these strains on minimal medium (MM) plates supplemented with different carbon sources. On medium containing either NaAc (sodium acetate, a short-chain fatty acid) or oleic acid (a long-chain fatty acid) as the sole carbon source (23, 27), the growth rate of the ΔMrpex16 strain was significantly lower than that of WT cultured on both media (Fig. 5 F and G). Similar to the ΔMrpex16 strain, the ΔMrash1 mutant also grew slower than the WT strain on both plates, but the growth defect was restored in the com-Mrash1 strain (Fig. 5 F and G). These data indicated that MrASH1 and MrPEX16 regulate fatty acid utilization. Together, these results revealed that MrASH1 regulates peroxisome biogenesis via activation of Mrpex16 and subsequently manipulates lipid droplet–derived fatty acid utilization, which facilitates TAG mobilization and breakdown from lipid droplets and ultimately increases the consequent glycerol production in the appressorium.
Discussion
In this research, we revealed a regulatory role of the histone methyltransferase MrASH1 in appressorium turgor generation and cuticle penetration in the entomopathogenic fungus M. robertsii. As shown in Fig. 5H, upon host cuticle exposure, the up-regulated epigenetic regulator MrASH1 activates the target gene Mrpex16 via H3K36me2 modification to control peroxisome biogenesis in the appressorium. Peroxisomes provide sites for efficient degradation of fatty acids via peroxisomal β-oxidation, which promotes the upstream hydrolysis of TAGs within lipid droplets to produce large amounts of glycerol for appressorium turgor pressure. Together, our findings reveal that the ASH1–PEX16 regulatory pathway plays a crucial role in regulating the biogenesis of peroxisomes, which promotes the lipolysis of lipid droplets for appressorium turgor generation, thus enabling the fungus to breach the insect cuticle and cause insect infection. This newly discovered mechanism by which epigenetic factors regulate appressorium-mediated insect infection provides insights into the mechanism of fungal pathogenesis in insects.
ASH1-mediated H3K36 methylation plays a vital role in gene regulation. Based on our combined RNA-seq and ChIP-seq analyses, 27 and 171 genes were activated and repressed by ASH1-mediated H3K36me2 upon cuticle exposure, respectively. In the phytopathogenic fungus F. fujikuroi, ASH1 activates target genes by antagonizing the PRC2 complex, which catalyzes H3K27 methylation for gene silencing (19). However, in the model fungus N. crassa, gene transcription in the chromosomal region with H3K36me modification catalyzed by ASH1 is down-regulated (18). These results indicate that ASH1 can either positively or negatively regulate gene transcription. Several studies have shown that ASH1 has pleiotropic effects on fungal pathogenicity. In the phytopathogenic fungus F. fujikuroi, ASH1 affects fungal pathogenicity by regulating the expression of secondary metabolism gene clusters for toxin synthesis (19). In the rice blast fungus M. oryzae, deletion of the ash1 homologue gene Mokmt2h resulted in a reduction in appressorium formation and virulence toward wheat (16). Here, we report that ASH1 in the entomopathogenic fungus M. robertsii does not influence appressorium formation but regulates appressorium-mediated turgor generation during cuticle penetration. This study provides insights into the mechanisms by which ASH1 regulates fungal pathogenicity.
Peroxisomes are formed by two distinct pathways: de novo synthesis at the endoplasmic reticulum (ER) and the growth and fission of mature peroxisomes (28). The peroxin PEX16 is one of the key regulators of peroxisome biogenesis. However, its relative contribution to peroxisome formation varies depending on the organism. In Y. lipolytica, PEX16 participates in peroxisomal fission (29), while in humans and plants (e.g., Arabidopsis thaliana), PEX16 is involved in the de novo biogenesis of peroxisomes (30, 31). In this study, we revealed that PEX16 is required for the de novo biogenesis of peroxisomes in an insect pathogenic fungus. These findings indicate that PEX16 is a multifaceted regulator of peroxisome biogenesis and functions in remarkably diverse ways in different organisms. The peroxisome, which is a metabolic organelle, is central to fatty acid metabolism, and excessive levels of fatty acids result in lipotoxicity and limited lipolysis (32). During appressorium development, lipid droplets are mobilized, in which the stored TAGs are catabolized into glycerol and fatty acids (8). Fatty acids are transported into peroxisomes and degraded via the β-oxidation cycle, which provides acetyl-CoA for energy mobilization (24). Thereby, the efficient degradation of fatty acids via β-oxidation in peroxisomes prevents their accumulation from inhibiting lipolysis and in turn promotes the upstream hydrolysis of TAGs, which ensures that large amounts of glycerol can be successfully produced and accumulated for the generation of enormous turgor pressure. In M. robertsii, we discovered that deletion of Mrpex16 completely abolished peroxisome formation and thus inhibited the mobilization and breakdown of lipid droplets in the appressorium, leading to decreased glycerol production and turgor generation and finally impairing appressorium-mediated cuticle infection. These results indicate that the lipolysis of lipid droplets requires the coordinated metabolic function of peroxisomes and that the peroxisomal fatty acid degradation-mediated feedback mechanism couples the functions of peroxisomes and lipid droplets. Together, our study links the role of peroxisomes in lipid metabolism with appressorium turgor generation and cuticle penetration.
Appressorium differentiation and turgor generation are complex morphogenetic processes that are tightly linked to finely coordinated gene regulation. Although an array of genes and signaling pathways have been characterized (33, 34), epigenetic regulatory mechanisms involved in appressorium formation and function, especially in appressorium turgor generation, are poorly understood. Our previous study revealed that the KMT2 (for H3K4me3)-Cre1-Hyd4 pathway performs coordinated regulation of appressorium formation (15). In this study, we discovered the role of histone modification in the biogenesis of peroxisomes and uncovered the regulation of the ASH1 (for H3K36me2)-PEX16 regulatory pathway in appressorium-mediated turgor generation and cuticle penetration. These findings provide comprehensive insights into infection-related morphogenesis and appressorium-mediated infection in an insect pathogenic fungus. Collectively, our findings show that multiple layers of epigenetic mechanisms are being used to orchestrate gene expression programs and cellular processes to precisely and concordantly regulate appressorium-mediated infection by insect pathogenic fungi and inform strategies to improve fungal pathogenicity against mosquitoes, leading to better control of mosquito-borne diseases.
Materials and Methods
Fungal Culture and Mosquito Rearing.
The fungus M. robertsii ARSEF 2575 was routinely grown on PDA (BD Difco) at 27 °C. A. stephensi (Dutch strain) mosquitoes were maintained based on a previous study (35). Experimental details can be found in SI Appendix, Materials and Methods.
Construction of M. robertsii Gene Deletion, Complementation, and Overexpression Strains.
For gene deletion, complementation, and overexpression, each gene fragment was amplified by PCR from the ARSEF 2575 genomic DNA as a template and then subcloned into the respective binary vector. All the gene deletion, complementation, and overexpression strains were constructed using Agrobacterium tumefaciens–mediated transformation. Experimental details can be found in SI Appendix, Materials and Methods.
Insect Bioassays.
To assess fungal pathogenicity and virulence, bioassays by topical infection and direct injection using conidial suspension were performed with female adult A. stephensi mosquitoes (36). Experimental details can be found in SI Appendix, Materials and Methods.
Appressorium Induction.
Conidia of the WT and mutant strains were harvested in 0.01% (vol/vol) Triton X-100 from 14-d-old fungal cultures on PDA. Appressorium induction was performed on hydrophobic surface and cicada wings. Experimental details can be found in SI Appendix, Materials and Methods.
Cuticle Penetration Assay.
Autoclaved cicada wings were lined on MM+1% glucose agar plates. Mycelium blocks of the WT and mutant strains were picked from 2-d-old fungal cultures on PDA and inoculated on the wings for 2 d. The wings with cultures were then removed, and the plates were incubated for an additional 5 d. The colony sizes were measured and compared between WT and mutants.
Incipient Cytorrhysis Assay.
Appressorium turgor was measured via an incipient cytorrhysis assay (21). Appressoria of the WT and mutant strains were induced on cicada wings. Individual wings were dipped in PEG8000 solution (6.5 g in 10 mL distilled water) for 10 min, and the percentage of appressoria undergoing incipient cytorrhysis was determined from 100 appressoria for each strain.
Glycerol Assay.
Appressoria of the WT and mutant strains induced on hydrophobic plastic surfaces via the methods described above were harvested by scraping with a plastic spatula and then homogenized with liquid nitrogen using a mortar and pestle. The glycerol content of the debris was then measured using a commercial glycerol assay kit (Applygen E1012-50) according to the manufacturer's instructions. Each strain was assayed in triplicate.
Lipid Droplet Observation.
Appressoria of the WT and mutant strains induced on cicada wings were stained with BODIPY (Invitrogen D3922) to detect neutral lipids (9). The stained lipid droplets in the appressoria were observed and photographed with a microscope (Olympus BX53). Experimental details can be found in SI Appendix, Materials and Methods.
Microscopic Observation of MrPEX16–GFP and RFP–PTS1 Localization.
To observe the localization of MrPEX16–GFP and RFP–PTS1, appressoria of the M. robertsii strains on hydrophobic surfaces were induced. Peroxisomes marked with dot-like RFP signals in appressoria were further observed and photographed with a microscope (Olympus BX53). Experimental details can be found in SI Appendix, Materials and Methods.
Western Blotting Analysis.
Histones were acid-extracted as previously described (15). Proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and then transferred to polyvinylidene fluoride membranes and blotted using standard procedures. Primary antibodies specific to H3K36me1 (Abcam ab9048), H3K36me2 (active motif 39255), H3K36me3 (Abcam ab9050), and H3 (Abmart P30266S) were used for western blotting. The secondary antibody used was Peroxidase AffiniPure Goat Anti-Rabbit IgG (H+L) (Jackson 111-035-003). Experimental details can be found in SI Appendix, Materials and Methods.
ChIP-Seq and ChIP–qPCR.
ChIP was performed as described previously with some modifications (16, 37). Two separate samples of the WT strain were prepared. Briefly, mycelia were generated by the insect cuticle exposure method. Chromatin of the fungal cultures was fixed and sheared into 150- to 300-bp fragments. The soluble chromatin fraction was immunoprecipitated using an antibody specific for H3K36me2 (active motif 39255). DNA fragments were recovered from immunoprecipitated chromatin or total chromatin (input DNA) by treatment with proteinase K and used for ChIP-seq or ChIP–qPCR. Experimental details can be found in SI Appendix, Materials and Methods.
RNA Extraction, RT–qPCR, and RNA-Seq.
Total RNA was extracted using an RNAiso Plus kit (Takara D9108A), and cDNA was synthesized using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara DRR047A) according to the manufacturer's instructions. Then, qPCR was performed using a Hieff qPCR SYBR Green Master Mix kit (Yeason 11201ES08) and a PikoReal instrument (Thermo N11471). Experimental details can be found in SI Appendix, Materials and Methods.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This study was supported by grants from the National Key R&D Program of China (grants nos. 2020YFC1200100 and 2018YFA0900502), the National Natural Science Foundation of China (grant nos. 31970470, 32021001, 32230015, 82261128007 and 31772534), the Strategic Priority Research Program of Chinese Academy of Sciences (grant no. XDPB16), Youth Innovation Promotion Association CAS (grant no. 2021272), Shanghai Talent Development Funding (grant no. 2020121). The author gratefully acknowledges the support of SANOFI Scholarship Program.
Author contributions
L.W., Y.L., and S.W. designed research; L.W., Y.L., J.C., W.Z., L.D., Y.Z., and F.L. performed research; G.W. contributed new reagents/analytic tools; L.W., Y.L., X.C., and G.W. analyzed data; and L.W., Y.L., and S.W. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
MrASH1 ChIP-seq and RNA-seq data have been deposited in the National Center for Biotechnology Information Sequence Read Archive with accession number PRJNA839849 (38). This study did not generate any unique code.
Supporting Information
References
- 1.Lovett B., et al. , Transgenic Metarhizium rapidly kills mosquitoes in a malaria-endemic region of Burkina Faso. Science 364, 894–897 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Scholte E. J., et al. , An entomopathogenic fungus for control of adult African malaria mosquitoes. Science 308, 1641–1642 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Lovett B., Bilgo E., Diabate A., St Leger R., A review of progress toward field application of transgenic mosquitocidal entomopathogenic fungi. Pest. Manag. Sci. 75, 2316–2324 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Ortiz-Urquiza A., Keyhani N. O., Action on the surface: Entomopathogenic fungi versus the insect cuticle. Insects 4, 357–374 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryder L. S., Talbot N. J., Regulation of appressorium development in pathogenic fungi. Curr. Opin. Plant Biol. 26, 8–13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keyhani N. O., Lipid biology in fungal stress and virulence: Entomopathogenic fungi. Fungal Biol. 122, 420–429 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Wang C., Wang S., Insect pathogenic fungi: Genomics, molecular interactions, and genetic improvements. Annu. Rev. Entomol. 62, 73–90 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Thines E., Weber R. W., Talbot N. J., MAP kinase and protein kinase A-dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea. Plant Cell 12, 1703–1718 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang C., St Leger R. J., The Metarhizium anisopliae perilipin homolog MPL1 regulates lipid metabolism, appressorial turgor pressure, and virulence. J. Biol. Chem. 282, 21110–21115 (2007). [DOI] [PubMed] [Google Scholar]
- 10.de Jong J. C., McCormack B. J., Smirnoff N., Talbot N. J., Glycerol generates turgor in rice blast. Nature 389, 244–244 (1997). [Google Scholar]
- 11.Foster A. J., Ryder L. S., Kershaw M. J., Talbot N. J., The role of glycerol in the pathogenic lifestyle of the rice blast fungus Magnaporthe oryzae. Environ. Microbiol. 19, 1008–1016 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Gao Q., Shang Y., Huang W., Wang C., Glycerol-3-phosphate Acyltransferase contributes to triacylglycerol biosynthesis, lipid droplet formation, and host invasion in Metarhizium robertsii. Appl. Environ. Microbiol. 79, 7646–7653 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gómez-Díaz E., Jordà M., Peinado M. A., Rivero A., Epigenetics of host-pathogen interactions: The road ahead and the road behind. PLoS Pathog. 8, e1003007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai Y., et al. , In vivo gene expression profiling of the entomopathogenic fungus Beauveria bassiana elucidates its infection stratagems in Anopheles mosquito. Sci. China Life Sci. 60, 839–851 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Lai Y., et al. , Coordinated regulation of infection-related morphogenesis by the KMT2-Cre1-Hyd4 regulatory pathway to facilitate fungal infection. Sci. Adv. 6, eaaz1659 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pham K. T., et al. , MoSET1 (histone H3K4 methyltransferase in Magnaporthe oryzae) regulates global gene expression during Infection-related morphogenesis. PLoS Genet. 11, e1005385 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindehell H., Glotov A., Dorafshan E., Schwartz Y. B., Larsson J., The role of H3K36 methylation and associated methyltransferases in chromosome-specific gene regulation. Sci. Adv. 7, eabh4390 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bicocca V. T., Ormsby T., Adhvaryu K. K., Honda S., Selker E. U., ASH1-catalyzed H3K36 methylation drives gene repression and marks H3K27me2/3-competent chromatin. eLife 7, e41497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janevska S., et al. , Elucidation of the two H3K36me3 histone methyltransferases Set2 and Ash1 in Fusarium fujikuroi unravels their different chromosomal targets and a major impact of Ash1 on genome stability. Genetics 208, 153–171 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang W., Shang Y., Chen P., Gao Q., Wang C., MrpacC regulates sporulation, insect cuticle penetration and immune evasion in Metarhizium robertsii. Environ. Microbiol. 17, 994–1008 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Oses-Ruiz M., Sakulkoo W., Littlejohn G. R., Martin-Urdiroz M., Talbot N. J., Two independent S-phase checkpoints regulate appressorium-mediated plant infection by the rice blast fungus Magnaporthe oryzae. Proc. Natl. Acad. Sci. U.S.A. 114, E237–E244 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimoto D., et al. , Pex16 is involved in peroxisome and woronin body formation in the white koji fungus, Aspergillus luchuensis mut. kawachii. J. Biosci. Bioeng. 127, 85–92 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Pang M. Y., Lin H. Y., Hou J., Feng M. G., Ying S. H., Different contributions of the peroxisomal import protein Pex5 and Pex7 to development, stress response and virulence of insect fungal pathogen Beauveria bassiana. J. Appl. Microbiol. 132, 509–519 (2022). [DOI] [PubMed] [Google Scholar]
- 24.Falter C., Reumann S., The essential role of fungal peroxisomes in plant infection. Mol. Plant Pathol. 23, 781–794 (2022), 10.1111/mpp.13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujihara N., Sakaguchi A., Tanaka S., Peroxisome biogenesis factor PEX13 is required for appressorium-mediated plant infection by the anthracnose fungus Colletotrichum orbiculare. Mol. Plant Microbe Interact. 23, 436–445 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Wang Z. Y., Soanes D. M., Kershaw M. J., Talbot N. J., Functional analysis of lipid metabolism in Magnaporthe grisea reveals a requirement for peroxisomal fatty acid beta-oxidation during appressorium-mediated plant infection. Mol. Plant Microbe Interact. 20, 475–491 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Asakura M., Okuno T., Takano Y., Multiple contributions of peroxisomal metabolic function to fungal pathogenicity in Colletotrichum lagenarium. Appl. Environ. Microbiol. 72, 6345–6354 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim P. K., Mullen R. T., PEX16: A multifaceted regulator of peroxisome biogenesis. Front. Physiol. 4, 241 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo T., et al. , A signal from inside the peroxisome initiates its division by promoting the remodeling of the peroxisomal membrane. J. Cell Biol. 177, 289–303 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim P. K., Mullen R. T., Schumann U., Lippincott-Schwartz J., The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J. Cell Biol. 173, 521–532 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin Y., Cluette-Brown J. E., Goodman H. M., The peroxisome deficient Arabidopsis mutant sse1 exhibits impaired fatty acid synthesis. Plant Physiol. 135, 814–827 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding L., et al. , Peroxisomal β-oxidation acts as a sensor for intracellular fatty acids and regulates lipolysis. Nat. Metab. 3, 1648–1661 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson R. A., Talbot N. J., Under pressure: Investigating the biology of plant infection by Magnaporthe oryzae. Nat. Rev. Microbiol. 7, 185–195 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Ryder L. S., et al. , A sensor kinase controls turgor-driven plant infection by the rice blast fungus. Nature 574, 423–427 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Wang S., et al. , Fighting malaria with engineered symbiotic bacteria from vector mosquitoes. Proc. Natl. Acad. Sci. U.S.A. 109, 12734–12739 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei G., et al. , Insect pathogenic fungus interacts with the gut microbiota to accelerate mosquito mortality. Proc. Natl. Acad. Sci. U.S.A. 114, 5994–5999 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamaru H., et al. , Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat. Genet. 34, 75–79 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Wang L., et al. , Regulatory mechanism of histone lysine methyltransferase ASH1 (MrASH1)in Metarhizium robertsii. SRAdatabase. https://www.ncbi.nlm.nih.gov/sra/PRJNA839849.2022-05-21.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
MrASH1 ChIP-seq and RNA-seq data have been deposited in the National Center for Biotechnology Information Sequence Read Archive with accession number PRJNA839849 (38). This study did not generate any unique code.