Abstract
This perspective draws on the record of ancient pathogen genomes and microbiomes illuminating patterns of infectious disease over the course of the Holocene in order to address the following question. How did major changes in living circumstances involving the transition to and intensification of farming alter pathogens and their distributions? Answers to this question via ancient DNA research provide a rapidly expanding picture of pathogen evolution and in concert with archaeological and historical data, give a temporal and behavioral context for heath in the past that is relevant for challenges facing the world today, including the rise of novel pathogens.
Keywords: ancient DNA, pathogens, disease, health
The process of domestication, which largely started during the Holocene in multiple locations around the globe, altered human subsistence, culture, and health. This agricultural transition is also linked to a demographic transition, where fertility rates increased, and an epidemiological transition (often called the first epidemiological transition), where the parasitic and infectious disease burden increased as a result of poor sanitary conditions associated with sedentism and increased close contact with domesticated animals (1–3). However, the agricultural transition, like the changes in population density and disease ecology, was a process rather than a rapid shift (4–6), occurring over hundreds if not thousands of years and to varying degrees in different places. It also affected the built environment and created conditions that allowed for eventual urbanization and more recently, industrialization. Below, we offer perspectives about using crucial information from modern genomics, bioarchaeology, epidemiology, and biomedical and veterinary sciences in tandem with ancient DNA data to address questions about how changing human behaviors and environments during the Holocene have fostered pathogen and parasite successes.
What Are Pathogens?
Anthropologically oriented studies of ancient biomolecules are uniquely situated to characterize our ancestral relationship with microbes, including pathogenic relationships. However, in relation to human health, microbes cannot be classified into a simple binary of pathogens or probiotics. The relationship between humans and microbes is ecological and evolutionary, which suggests that it may be time to question the concept of pathogens altogether (7), at least as a general label. Traditionally, a pathogen is an infectious agent that causes disease, and many of these agents are obligate pathogens in that their evolutionary strategy inevitably results in harm to their host; in contrast, some pathogens are opportunistic and typically do not cause disease to hosts, but they can when that host is compromised. Alternatively, the microbe–host relationship may be described using terms such as commensalism, where one organism benefits and the other is neither harmed nor benefited, and mutualism, where there is a mutually beneficial relationship. These labels are often attached to microbes, without ecological consideration. For example, ebolaviruses are typically considered pathogens. Although current investigations of ebolaviruses do not assume they specifically evolved to harm humans, if the host is human, the outcome is often dire. If our goal is to understand the biology of ebolaviruses more generally, the term “pathogen” may be a poor fit. Efforts to find the natural host of ebolaviruses do so by seeking organisms that exhibit little consequence from the viral “infection” (8), a more commensal relationship. The ecosystem, the host, the evolutionary strategies, and the relationships matter.
Increased population growth and environmental changes resulting from the sociocultural behavioral modifications enabled or necessitated by intensive plant cultivation and animal domestication likely contributed to the emergence and reemergence of infectious diseases. Over 60% of emerging infectious diseases have zoonotic origins, and a subset of pathogens persists in the environment as zoonotic infections to date (9–11). Zoonoses are defined by the World Health Organization as diseases and infections that are naturally transmitted between vertebrate animals and people (https://www.who.int/news-room/fact-sheets/detail/zoonoses). Emerging infectious diseases still cause global public health and socioeconomic threats as evidenced by the ongoing COVID-19 pandemic and other recent epidemics (12, 13). Mechanisms by which pathogens emerge and reemerge are yet to be understood fully because they are determined by many complex factors operating at the level of the host, the pathogen, and the environment (14). Identifying the processes that act at different levels to influence pathogen emergence, maintenance, and transmission requires a One Health approach, which recognizes that there is a connection between people, animals, plants, and their shared environment (15). For pathogens with an ancient history in humans, a One Health approach involves integrating data from such disciplines as archaeology, history, epidemiology, biology, and traditions of healing (16). New genomic tools provide novel opportunities to retrieve ancient pathogen DNA from humans, animals, and the environment to add additional rich sources of data (17, 18). Generally, the term ancient DNA refers to damaged and short DNA fragments obtained from subfossil material, including bones and preserved soft tissues, and this can also apply to DNA damaged by formalin fixation in historical samples, such as biopsy specimens (18). Together, these fields enable identification and understanding of the effects of ancient diseases, including the sociocultural contexts in which these diseases occurred. Here, we discuss some factors related to dietary shifts, sedentary lifestyles, and animal husbandry as well as climate changes that may have influenced the success of pathogens after the agricultural transition.
Ancient Environment as a Driver of Pathogens.
Environmental and ecological factors, such as rainfall, temperature, humidity, land use change, deforestation, agriculture biodiversity loss, and latitude, influence zoonotic disease emergence and successful transmission and can alter the distribution of existing pathogens (9). The Holocene is characterized by climate stability and warm temperatures that provide opportunities for growth of infective pathogen stages as well the introduction of vectors and vector-borne pathogens (19, 20). DNA recovered from ancient people shows the presence of pathogens typically transmitted by insect vectors, such as ticks, fleas, lice, and tsetse flies (ref. 21; e.g., refs. 22 and 23) and could be used to infer vector distributions and examine pathogen diversity through time. In addition, environmental changes can be highly dependent on human behavior and cultural factors. Ancient environmental DNA has been retrieved from soil, sediments, ice, and water and has provided valuable information on biodiversity, including changes in vegetation cover, animal species, and parasites (24–27). There are mixed results relating the effects of biodiversity to the emergence of infectious diseases. In some instances, increased biodiversity is associated with reduced infectious disease, possibly due to the dilution effect, which postulates that the presence of multiple hosts reduces the likelihood of disease occurrence by reducing parasite density in hosts and interfering with transmission dynamics within and between hosts (28, 29). For instance, Sokolow et al. (30) reported reduced prevalence of human schistosomiasis with the reintroduction of a snail (vector) predator in a community in Senegal. However, this is possibly true for pathogens that infect multiple hosts and have complex life cycles, but it is unlikely to affect those that are host specific and have highly infective parasite stages that are resistant to environmental changes (31). The alternate hypothesis is that increased biodiversity may lead to increased emergence of zoonoses because of the many parasites that might spill over to humans (32). Studies that support this hypothesis report the increased burden of disease (e.g., cutaneous leishmaniasis, simian retroviruses) in intact forests as opposed to deforested areas (33, 34). Because ancient data on both biodiversity and pathogens exist, studies should focus on characterizing the associations between the two, and this could possibly inform conservation management and disease prevention and control strategies today.
Sedentary Lifestyles and Animal Husbandry Practices.
The domestication of animals and cultivation of plants led to sedentary lifestyles for both humans and animals (35). This resulted in increased population sizes and densities; thus, it facilitated transmissibility of infectious pathogens, especially those transmitted through contact and respiratory droplets, and eventually promoted the maintenance of so-called “crowd diseases.” This new lifestyle resulted in sustained interactions between humans and domesticated animals and hence, facilitated zoonotic pathogen transfer within and between these hosts, where the latter probably served as reservoirs or conduits from wildlife with the potential for incubating new pathogens (36). Pathogens that can infect a broad range of hosts (also referred to as generalist species), such as helminths and other soil-transmitted pathogens, must have thrived in this new environment (37, 38). Archeological data on animal husbandry practices are important for understanding the contexts of disease transmission and/or emergence. These data can include feeds used, grazing techniques, disease management, manure disposal methods, housing structures, storage of produce, culling of animals, and population breeding techniques. Ancient data on interactions between domesticated and nondomesticated animals (wildlife) and humans are equally important for studies of zoonoses. Because wildlife is not exposed to husbandry practices, their infectious disease dynamics are different.
Dietary Shifts for Humans.
The Neolithic “revolution” clearly resulted in changes in human diet. Ancient human diets have been inferred from analyses of a range of data, including stable isotopes, dental wear, dental calculus, coprolites, and zooarchaeological remains (39–42), and these data show a diet shift during the Holocene from one with a sole reliance on hunting and gathering to one largely dominated by the consumption of domesticated animals and plants (35). Arguably, at least in some cases, this entailed a loss of dietary breadth (such as reduced consumption of meat and greater reliance on domesticated grains), reduced quality of diets (e.g., fewer micronutrients, such as iron), and increased vulnerability to food shortages, all of which could have resulted in declines in health (43). Domestication of animals, such as goats and sheep, led to the acquisition of diseases, such as Brucellosis, through ingestion of animal products, such as milk and cheese (44, 45). Diets also influence the gut microbiome of both animals and humans and hence, play an important role in the general well-being of hosts by influencing nutrition, immune responses, and chronic diseases (46–48). Plants cultivated and consumed by humans during epidemics may also have medicinal uses that could result in selection pressure on pathogens (49).
Given that nutritional status is a crucial determinant of immune competence and thus, disease susceptibility (50), combining dietary data with paleodemographic, paleopathological, and pathogen paleogenetic data to examine the associations between diet (breadth, quality, and/or severe shortages), infection, and disease outcomes during and after the transition to agriculture is important. Previous work has examined some (but not all) of these intersections in other contexts. For example, Fellows Yates et al. (51) examined dental biofilms of humans and Neanderthals, identifying core bacterial genera and microbial coadaptation with our lineage’s diet. Richards and Montgomery (52) summarized studies to date that link stable isotopes and paleopathology (ref. 53 is a recent example). The Beaumont and Montgomery (54) application of incremental dentine analysis of dietary isotopes can produce high-resolution data regarding the timing of severe nutritional stress during development, which can be integrated into the developmental origins of health and disease framework to assess the effects of early life stress on later health outcomes. Some studies have demonstrated the potential for integrating demographic data (mortality or conversely, survivorship) as a reflection of underlying health that is affected by diet (e.g., refs. 55 and 56). These and similar studies provide models for research that addresses the effects of agricultural diets on health and how those effects might have varied across time and space. However, the simultaneous analysis of all these lines of evidence—linked at the individual level to enable clear inferences about the nature of interactions—remains an untapped resource for richly detailed reconstructions of the intersections of diet, disease, and subsistence transitions.
Our Microbial Ecosystems.
The classic concept of “diseases of civilization” has fed academic pursuits for decades, inspiring debates about thrifty genes (57) and hygiene hypotheses (58, 59) as well as about the postulate that the transition to urban (and much later, industrialized) lifestyles had a varied but high impact on health. Strachan (58, 59) is credited with posing the hygiene hypothesis, the concept that our changed relationship with microbes, then attributed to family size, household amenities, and perceptions of cleanliness, impacted asthma and hay fever incidence. The desire and effort to build off, modify, and improve the scope and precision of the hypothesis continue and include the disappearing microbiota (60) and old friends (61) variants, each with their own compelling arguments about how the changed relationships of microbial commensals, symbionts, and pathogens impact health, including in particular, inflammation and autoimmunity. In such efforts, a fair argument to abandon the hygiene hypothesis is made (62); the hypothesis fosters the public perception that “cleanliness” leads to allergy risk—it does not, at least not exactly. Despite the calls to abandon it, the phrase perpetuates (63–65), and the essence and rigidity of the hypothesis give structure to explore similar concepts, even when the original lacks nuance or misleads the public. Replacing such cornerstone hypotheses when they well leverage investigations may be less productive than simply updating them. In that spirit, given the patterns in nature currently observed, what would be the “hygiene hypothesis 2.0?” How do pathogens fit into this picture, and how can ancient DNA and the archaeological record add to the discussion of this hypothesis?
Lifestyle and Microbial Relationships.
Without question, our lifestyles impact our relationships with microbes. The most studied human microbiome, the gut, demonstrates a clear pattern where subsistence practices impact microbiome composition (66). Specifically, gut microbiomes in individuals practicing traditional lifestyles show more diversity and a different composition than those from individuals with more urban–industrial–metropolitan lifestyles. For example, Yatsunenko et al. (67) found that the subsistence farmer lifestyle had differences from a metropolitan lifestyle, with the microbiome taxonomic composition of traditional populations clustering together and more diverse when compared with metropolitan populations, even if these more traditional farmers were continents apart: for example, in Africa and South America. Moreover, traditional hunter-gatherer populations have microbiome features different from other lifestyles (68, 69), likely attributed to fiber digestion. In addition, the taxonomic and functional composition of hunter-gatherer gut microbiomes from Africa and South America clusters more tightly together (68) within the broader cluster of microbiomes from people with traditional subsistence practices (including subsistence farming), and it is distinct from urban–industrial–metropolitan-derived gut microbiomes. Gut microbiome data from ancient contexts also shared these more traditional features (70). The pattern observed by these cornerstone studies has been observed repeatedly (ref. 71 has a review), and yet, a most fundamental question remains. Does the loss of microbiome diversity or a change in microbiome composition matter to health?
The human microbiome, its composition shaped by lifestyle and behavior, also associates with a broad range of complex diseases (72). However, “hygiene,” even broadly defined, remains insufficient to predict microbiome composition. For example, with respect to the gut, much of the microbiome composition is most strongly associated with diet, exercise, and body composition (72, 73). The evolutionary relationship between the microbiome and host is intimate and dynamic and affects resistance to pathogens. The microbiome forces putative pathogens to compete with commensals, and hosts have evolved mechanisms to foster a more cooperative relationship with microbes (74), creating a coimmunity (75). One of many emerging discoveries is that the gut microbiome impacts risks for active tuberculosis (TB) infection (76). In contrast to aiding pathogen resistance, a broken relationship (a dysbiosis) can turn commensal relationships into pathogenic ones. In essence, an “opportunistic pathogen” often reveals a tragic story of a failed relationship between the host and their microbiota, where a microecological event occurs that changes the relationship of a microbe from mutualistic or commensalistic to one that is pathogenic. For example, Helicobacter pylori is linked to the majority of stomach cancer cases and a wide range of other gastric symptoms and disease. Yet, most of the human population harbors H. pylori, implying a predominately commensal relationship, and H. pylori infection appears to bestow some benefits, such as a reduced risks of asthma, atopic disease, and other inflammatory diseases (77).
The gut microbiome is also a reservoir of antibiotic-resistant genes that have been associated with the emergence of drug-resistant pathogens (78, 79). Antibiotic resistance is ancient and widely spread in the environment (80). The ancient resistome (antibiotic-resistant genes together with their precursors in pathogenic and nonpathogenic bacteria) can provide insight into the evolutionary history of these regions (81). Metagenomic analysis of ancient samples, such as soil, water, plants, and coprolites, is thus integral in understanding the drivers of ancient gut microbiome composition.
Microbes, microbiomes, and their role in disease are increasingly clearly a microecological phenomenon, a relationship among the microbes and their host. Revealing these ecosystem dynamics is essential to our next stage of microbiome understanding (82). For example, with respect to the impact on the gut microbiome, it is not simply the loss of diversity observed in the microbiomes of those with industrialize lifestyles compared with those with traditional lifestyles but metabolic resilience potential for key aspects of microbiome functions, such as short-chain fatty acid production (83). Important efforts and questions remain in characterizing keystone taxa and functional diversity and applying ecological theory to predict and ideally, foster healthy and resilient human microbiomes. Efforts to bring such ecological studies to prehistoric context remain challenging, but they are feasible (84) and essential for determining the extent to which the Neolithic transition and later, industrialization have impacted our biology. Given that the urban–industrial lifestyle has indeed affected our relationship with microbes for the better and sometimes, for the worse, such ancient data could contribute critical wisdom.
Bioarchaeological Context
A hallmark of bioarchaeology is careful attention to context, and ancient pathogen DNA data promise to improve our understanding of cultural, environmental, and biological contexts further. Pathogen paleogenetic data may provide a means to examine the widely held perspective that the transition from hunting and gathering to agriculture (and subsequent urbanization) was accompanied by a decline in human health (85). This perspective typically does not account for variation within groups characterized as hunter-gatherers and agriculturalists; it assumes that changes in factors, such as diet, group size, sedentism, and inequality, that accompany the transition to agriculture occurred simultaneously, uniformly and unidirectionally, and it ignores the potential that these factors might have exerted differential effects on, among other things, exposure to pathogens (85).
The integration of ancient pathogen DNA might disrupt such potentially oversimplified perspectives and foster a view of transitioning populations as heterogeneous. Paleogenetic data informative about pathogens and disease ecologies can be combined with data on age, sex, stature, social status or wealth inequality, residence location (e.g., urban vs. rural), and other variables to clarify within- and between-population variation in pathogen loads, exposures to zoonotic infection, and outcomes of infection. For example, some diseases (crowd diseases) require certain threshold population sizes to be maintained in human populations. Analysis of pathogens in light of paleodemographic and settlement pattern data would clarify the population-dynamic tipping points for maintenance of these diseases in the past. Further, many infectious diseases disproportionately affect infants and young children. Resulting high rates of infant and childhood mortality produce relatively low life expectancies at birth, but typically, people who survive this vulnerable early period can expect to live relatively long lives on average (86). Targeted analysis of pathogens in the skeletons of young children would clarify whether observed reductions in the mean age at death in agricultural populations reflect general declines in health or a combination of higher fertility and increased burdens of infant deaths from infectious diseases. Paleogenetic data can also provide a means for expanding beyond focusing on a single disease at a time and on those diseases with pathognomonic skeletal pathologies to consider also parasite burdens and other coinfections that are not visible skeletally or in archaeological contexts. This may allow for the discernment of otherwise hidden heterogeneity in frailty, as described by Wood and colleagues (87), as well as enable bioarchaeological examination of syndemics [the co-occurrence of multiple conditions that produce outcomes worse than their individual effects (88, 89)]. For example, recent studies, although not focused on the transition to agriculture, demonstrate the rich potential for paleogenetic data (combined with or independent of skeletal signs of infection or other health burdens) to reveal comorbidities or coinfections in past populations that might have shaped health outcomes beyond the individual effects of each etiology. Studies have found evidence of coinfection with Mycobacterium tuberculosis and Mycobacterium leprae in sites from Egypt, Hungary, Israel, and Sweden dating from the first to thirteenth centuries (90), Yersinia pestis (plague) and Treponema pallidum pertenue (yaws) in postmedieval Lithuania (91), and Y. pestis and Haemophilus influenzae (meningitis) in early medieval England (92). Further, Guellil et al. (92) raise the possibility of Y. pestis infection enhancing preexisting or opportunistic bacterial infections via its immunosuppressive effects.
Pathogen paleogenetic data may help clarify whether the introduction of novel pathogens to human populations, as enabled and facilitated by settled agriculture, by itself resulted in declines in health. Alternatively, did this process simply increase the number of potential competing causes of death without substantially altering individual risks of death or population-level patterns of general health? That is, paleogenetics, in combination with skeletal data on mortality or survival (reflective of general patterns of health), might make it possible to discern whether a shift in disease scapes that accompanied agriculture simply entailed a greater variety of possible morbid conditions but not an increase in morbidity and mortality overall. If findings indeed suggest that exposure to increased varieties of pathogens was associated with increased mortality overall, bioarchaeological data can clarify the syndemic effects at play within the specific context (i.e., the biosocial factors that interacted with newly introduced pathogens to produce declines in health).
Studies of the transition to agriculture have just recently begun to include paleogenetic data, although not yet pathogen DNA (at the time of writing). Recently, for example, Marciniak et al. (93) assessed the relationship between stature (determined via DNA and a sign of developmental stress) and farming, yielding both genomic evidence and skeletal evidence of a decline in health for early European farmers. Although this study focuses on ancient people rather than pathogens, it points a way forward for integrating pathogen DNA further into studies of this subsistence transition.
Pathogen Genomics
Using ancient and modern genetic and genomic data has provided insights into the relationships and geographic distributions of pathogens today and in the past. However, the question about what is a pathogen gains added complexity when trying to discern the cause(s) of disease in ancient individuals. For example, today we know of many cases where the majority of strains of a particular microorganism do not cause disease, but a subset does (e.g., Escherichia coli, Yersinia enterocolitica), often depending on the presence of virulence factors. These virulence factors matter, but it seems that in many cases, the human ecology (here, including such things as the current state of the immune system, competition with commensals, and coinfections as noted above) likely matters more. For example, pathogen coinfections are ubiquitous in nature, and clinical outcomes are dependent on their interaction with each other and with the host. The outcomes might be beneficial, detrimental, or inconsequential to hosts. In particular, helminth infections were common in the past, and yet, there are scarce ancient data on their coinfections with other pathogens. Helminths are known to influence outcomes of some infectious diseases, including malaria and tuberculosis (94–96). Untangling these relationships requires modern functional and epidemiological data from both veterinary and human biomedical sciences. Reconstructing this picture in ancient microbes that no longer exist adds to this challenge.
Ancient DNA analyses provide a window into past patterns of pathogen distributions and evolutionary changes as well as timing. An excellent example is that of Y. pestis, a bacterial pathogen for which we currently have the most genome data from ancient individuals. These data show that in addition to being the cause of the historically known plague pandemics, Y. pestis also affected people as early as the Late Neolithic and Early Bronze Age (e.g., refs. 23 and 97–101). Significantly, analyses of the earliest cases point to the presence of strains that may not have been flea transmitted (inferred from genetic analyses), leaving the mode(s) of transmission and potential reservoirs unclear, as well as cases that point to an extended period of overlap with strains that do show the genomic changes (both chromosomal and plasmid) that signal flea transmission or at least better flea transmission (102–105). Functional studies will be important to assess how the presence/absence of specific changes affects transmission and virulence. Phylogenetic analyses show diversification during the Late Neolithic/Early Bronze Age (∼5,000 to 3,000 y ago) of these lineages along with those leading to extant Y. pestis lineages (including 0.PE2, 0.PE4, 0.PE5, and 0.PE7 primarily found in rodents in central Eurasia). This leads to questions about the ecology and spread of these lineages within and among species during this period in particular and underscores the importance of understanding the extent and history of Y. pestis from a One Health perspective.
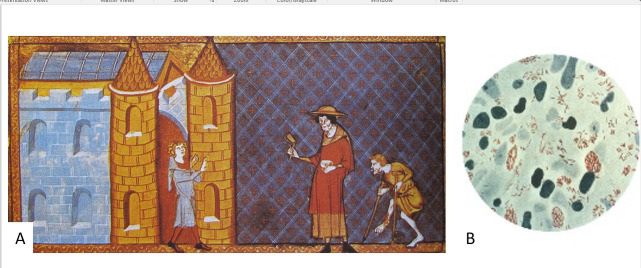
Ancient genomic analyses of mycobacteria, including M. leprae (Fig. 1) and M. tuberculosis complex (MTBC), have also resulted in surprises. For example, unlike the findings for Y. pestis, both M. leprae and the MTBC were found to have younger than expected most recent common ancestors (both roughly 5,000 y ago), suggesting that these are not the “heirlooms” that we thought (106–109). Studies have also revealed significant diversity of strains through time and coupled with studies of modern strains in both humans and animals, the clear interchange of these pathogens from humans to other species and back again (e.g., refs. 107 and 109–111). These mycobacteria are also among the first where we can see evidence for genetic susceptibility of particular subsets of ancient populations. For example, Krause-Kyora et al. (112) extracted DNA from 85 individuals with paleopathological indications of lepromatous leprosy buried at St. Jørgen’s leprosarium (dated between AD 1270 and AD 1550) in Denmark, and they were able to genotype an allele characteristic of major histocompatibility complex, class II, DR beta 1 (HLA DRB1*15:01, a susceptibility marker in modern populations) in 69 individuals who also tested positive for M. leprae in their DNA analyses. After comparing these results with those from control samples, they found that the individuals with leprosy were more likely to have the susceptibility allele at this locus. Susceptibility to severe TB was linked to a variant of the tyrosine kinase 2 gene in homozygotes that causes impairment of the Interleukin 23 responsive pathway (113). Ancient genome analyses in Europeans show that the frequency of this allele has declined since the Bronze Age, likely as morbidity and mortality from TB increased, perhaps as strains of TB were introduced (113–115). Such analyses are also beginning to see the impact of other pathogens on human variation. In particular, recent evidence suggests that the very high mortality produced by the fourteenth-century pandemic (caused by Y. pestis and commonly known as the Black Death) had a selective effect on human immune loci (116).
Fig. 1.
(A) A fourteenth-century miniature of two lepers being denied entry to the city. Reproduced from Speculum historiale of Vincent de Beauvais (circa 1184 to 1264) Bibliothèque de l’Arsenal, folio 373r (fourteenth-century manuscript). (B) A photomicrograph of M. leprae (in red) taken from a leprosy skin lesion. Reproduced from the Centers for Disease Control Public Health Image Library (1979, US Government public domain).
Ancient DNA has also begun to provide some insights into coinfections as noted above. Coinfections are likely influenced by factors including geographic overlap, similar transmission routes, and pathogen influences on host immunity (e.g., opportunistic infections brought about by weakened immune responses). It is also likely that most people had helminths given the numerous archeoparasitological reports on helminths, such as Trichuris trichiuria, Enterobius vermicularis, Ascaris species, and Schistosoma species (117). Data on ancient helminth coinfections with other parasites are scarce. DNA analyses of helminths from ancient coprolites can add to this record; however, most analyses to date are from isolated coprolites or cesspits (e.g., refs. 115 and 118) and thus, are not linked to specific individuals.
Ancient pathogen studies have important limitations. Most ancient pathogen DNA data (like most modern genomic data) are heavily biased geographically, and this bias is often true of the comparative modern data. Also, what ancient pathogen DNA datasets we have are influenced by the pathogen load at the time of death, the part of the body that remains, the climate, and in the case of viruses, likely whether it is composed of double-stranded DNA (single-stranded DNA or RNA is less likely to preserve). Other limitations include our ability to discern whether an extinct microbe was in fact a pathogen. Ancient pathogen studies to date focus on those with modern representatives. Ancient pathogen data availability is also affected by the archaeological record, with more data from complete adult burials from formal cemeteries. Children, individuals who were cremated, single burials (often characteristic of hunter-gatherer populations), and parts of the world where paleopathological analyses are limited are often poorly represented.
Conclusions: How Should We Move Forward?
Ancient DNA analyses of pathogens offer often unexpected insights into the origin/timing of some infectious diseases in human populations as well as exchanges back and forth between humans and other animals. Ancient DNA research has also enhanced our understanding of how the microbiome has changed over time and in different contexts. However, there are many unanswered questions about when and where specific microbes/parasites/pathogens became endemic in humans, their distributions in the past, and how these might be linked to agricultural transitions and subsequent urbanization. Were there crowd diseases in urban centers in the Americas precontact? If so, what were they, what was their zoonotic origin (e.g., from camelids? From turkeys or wildfowl?), and how common were they? How do we assess ancient animal diseases as well considering that most pathogens have zoonotic origins and considering that epidemics in domesticated animals (and plants) ultimately affect the health of the populations that rely on them for food? Where and when did animals acquire such pathogens, and what enabled them to maintain them?
During most of human evolution, population sizes and densities were low, and the assumption has been that the pathogens and parasites primarily affecting us were heirloom pathogens afflicting us since (and prior to) our divergence with other hominids (1, 3, 119, 120). In addition, ancient hunter-gatherer populations were likely affected by pathogens that are found in the environment, including those encountered in food or water, or that could be transmitted via animal reservoirs and vectors. Such exposures were not static given the ecological and climatic changes over time and space as humans moved around the landscape … and out of Africa. Some new pathogens that might “spill over” (e.g., coronaviruses, Ebola virus, bird flu, and Nipah virus) likely burned out quickly in these typically low-density populations due to rapid depletion of susceptible hosts. Today, the world population is growing, surpassing 8 billion, and this growth has been associated with anthropogenic activities, such as encroachment into wildlife habitats, logging and deforestation, and rapid long-distance travel, which increase the likelihood of infectious disease emergence and spread (9, 121, 122). It is inevitable that humans, animals, and wildlife will continue to interact and that zoonotic spillovers will continue to occur. A clear grasp of the processes that influence these spillovers will provide insight for prediction of disease emergence and development of strategies for disease prevention and response. Ancient DNA analyses help us understand when (and hopefully, also aspects of how/why) past jumps occurred, although it is important to keep in mind that much of our data may reflect the most recent successful jump and radiation of a particular pathogen rather than the first “jump.”
The pathogenic relationships that burden society today are a product of ancient events. Ancient genes, exposed to over a billion years of natural, microbial, and chemical warfare, may be repurposed to resist antibiotic medications. Ancient host-commensal relationships can become unstable by exposure to new poor-fitting human hosts. Ancient human microbiome relationships can be disrupted and become dysbiotic, resulting in an overreaction of host immunity. While we see the evidence of these events, “top down,” through health and disease today, we lack the details about the precise precipitating ancient events and contexts that ultimately gave rise to contemporary observable patterns. We are thus left with important questions. What ancient and widespread genes are poised for novel antimicrobial resistance? Why is one host relationship commensal and another pathogenic? How do the ecosystems in and surrounding our bodies change? Addressing these questions is a matter of finding the coveted biological “mechanism,” the molecular agents. The molecular characterization informs biological evolution, forms the bases for medications, and lays the most basal of foundations, wisdom, for building healthier relationships to microbes. There is no more direct line of research into these ancient events than to study ancient microbes directly via ancient biomolecules.
Acknowledgments
Author contributions
C.M.L., M.Y.A., S.N.D., and A.C.S. wrote the paper.
Competing interest
C.L. is an editor of an upcoming book for which A.C.S. has written a chapter. They are also coauthors on a review published in 2022.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
There are no data underlying this work.
References
- 1.Cohen M. N., Health and the Rise of Civilization (Yale University Press, 1989). [Google Scholar]
- 2.Bocquet-Appel J.-P., Bar-Yosef O., Eds., The Neolithic Demographic Transition and Its Consequences (Springer, 2008). [Google Scholar]
- 3.Zuckerman M. K., Harper K. N., Barrett R., Armelagos G. J., The evolution of disease: Anthropological perspectives on epidemiologic transitions. Glob. Health Action 7, 23303 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larson G., et al. , Current perspectives and the future of domestication studies. Proc. Natl. Acad. Sci. U.S.A. 111, 6139–6146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeder M. A., The origins of agriculture in the Near East. Curr. Anthropol. 52, S221–S235 (2011). [Google Scholar]
- 6.Stock J. T., Pinhasi R., Eds., Human Bioarchaeology of the Transition to Agriculture (John Wiley & Sons, Ltd., 2011). [Google Scholar]
- 7.Casadevall A., Pirofski L. A., Microbiology: Ditch the term pathogen. Nature 516, 165–166 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Peterson A. T., Carroll D. S., Mills J. N., Johnson K. M., Potential mammalian filovirus reservoirs. Emerg. Infect. Dis. 10, 2073–2081 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones K. E., et al. , Global trends in emerging infectious diseases. Nature 451, 990–993 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolfe N. D., Dunavan C. P., Diamond J., Origins of major human infectious diseases. Nature 447, 279–283 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor L. H., Latham S. M., Woolhouse M. E., Risk factors for human disease emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356, 983–989 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bloom D. E., Cadarette D., Infectious disease threats in the twenty-first century: Strengthening the global response. Front. Immunol. 10, 549 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keni R., Alexander A., Nayak P. G., Mudgal J., Nandakumar K., COVID-19: Emergence, spread, possible treatments, and global burden. Front. Public Health 8, 216 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gebreyes W. A., et al. , The global one health paradigm: Challenges and opportunities for tackling infectious diseases at the human, animal, and environment interface in low-resource settings. PLoS Negl. Trop. Dis. 8, e3257 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atlas S., One Health: Its Origins and Future One Health: The Human-Animal-Environment Interfaces in Emerging Infectious Diseases, Mackenzie J., Jeggo M., Daszak P., Richt J., Eds. (Current Topics in Microbiology and Immunology, Springer, Berlin, Germany, 2012), vol. 365. [Google Scholar]
- 16.Marciniak S., Poinar H., “Ancient pathogens through human history: A paleogenomic perspective” in Paleogenomics, Lindqvist C., Rajora O., Eds. (Springer, Berlin, Germany, 2018), pp. 115–138. [Google Scholar]
- 17.Stone A. C., Ozga A. T., “Ancient DNA in the study of ancient disease” in Ortner’s Identification of Pathological Conditions in Human Skeletal Remains, Buikstra J. E., Ed. (Academic Press, London, United Kingdom, ed. 3, 2019), pp. 183–210. [Google Scholar]
- 18.Orlando L., et al. , Ancient DNA analysis. Nat. Rev. Methods Primers 1, 14 (2021). [Google Scholar]
- 19.Gowdy J., Our hunter-gatherer future: Climate change, agriculture and uncivilization. Futures 115, 102488 (2020). [Google Scholar]
- 20.Panagiotakopulu E., Buckland P. C., A thousand bites: Insect introductions and late Holocene environments. Quat. Sci. Rev. 156, 23–35 (2017). [Google Scholar]
- 21.Lima V. S., et al. , Chagas disease in ancient hunter-gatherer population, Brazil. Emerg. Infect. Dis. 14, 1001–1002 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barbieri R., et al. , A 2,000-year-old specimen with intraerythrocytic Bartonella quintana. Sci. Rep. 10, 10069 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spyrou M. A., et al. , The source of the Black Death in fourteenth-century central Eurasia. Nature 606, 718–724 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pedersen M. W., et al. , Ancient and modern environmental DNA. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20130383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hahn A., Mid-to Late Holocene climatic and anthropogenic influences in Mpondoland, South Africa. Quat. Sci. Rev. 261, 106938 (2021). [Google Scholar]
- 26.Wang Y., et al. , Late Quaternary dynamics of Arctic biota from ancient environmental genomics. Nature 600, 86–92 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seersholm F. V., et al. , DNA evidence of bowhead whale exploitation by Greenlandic Paleo-Inuit 4,000 years ago. Nat. Commun. 7, 13389 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood C. L., et al. , Does biodiversity protect humans against infectious disease? Ecology 95, 817–832 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Civitello D. J., et al. , Biodiversity inhibits parasites: Broad evidence for the dilution effect. Proc. Natl. Acad. Sci. U.S.A. 112, 8667–8671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sokolow S. H., et al. , Reduced transmission of human schistosomiasis after restoration of a native river prawn that preys on the snail intermediate host. Proc. Natl. Acad. Sci. U.S.A. 112, 9650–9655 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohr J. R., et al. , Towards common ground in the biodiversity-disease debate. Nat. Ecol. Evol. 4, 24–33 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keesing F., et al. , Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature 468, 647–652 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolfe N. D., et al. , Naturally acquired simian retrovirus infections in central African hunters. Lancet 363, 932–937 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Walsh J. F., Molyneux D. H., Birley M. H., Deforestation: Effects on vector-borne disease. Parasitology 106 (suppl.), S55–S75 (1993). [DOI] [PubMed] [Google Scholar]
- 35.Larsen C. S., The agricultural revolution as environmental catastrophe: Implications for health and lifestyle in the Holocene. Quat. Int. 150, 12–20 (2006). [Google Scholar]
- 36.Pearce-Duvet J. M., The origin of human pathogens: Evaluating the role of agriculture and domestic animals in the evolution of human disease. Biol. Rev. Camb. Philos. Soc. 81, 369–382 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Woolhouse M. E., Gowtage-Sequeria S., Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 11, 1842–1847 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wells K., Morand S., Wardeh M., Baylis M., Distinct spread of DNA and RNA viruses among mammals amid prominent role of domestic species. Glob. Ecol. Biogeogr. 29, 470–481 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macko S. A., et al. , Documenting the diet in ancient human populations through stable isotope analysis of hair. Philos. Trans. R. Soc. Lond. B Biol. Sci. 354, 65–75 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warinner C., et al. , Direct evidence of milk consumption from ancient human dental calculus. Sci. Rep. 4, 7104 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sistiaga A., Berna F., Laursen R., Goldberg P., Steroidal biomarker analysis of a 14,000 years old putative human coprolite from Paisley Cave, Oregon. J. Archaeol. Sci. 41, 813–817 (2014). [Google Scholar]
- 42.Warinner C., Speller C., Collins M. J., A new era in palaeomicrobiology: Prospects for ancient dental calculus as a long-term record of the human oral microbiome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20130376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larsen C. S., Animal source foods and human health during evolution. J. Nutr. 133 (11 suppl. 2), 3893S–3897S (2003). [DOI] [PubMed] [Google Scholar]
- 44.Bendrey R., et al. , Approaching ancient disease from a One Health perspective: Interdisciplinary review for the investigation of zoonotic brucellosis. Int. J. Osteoarchaeol. 30, 99–108 (2020). [Google Scholar]
- 45.D’Anastasio R., Staniscia T., Milia M. L., Manzoli L., Capasso L., Origin, evolution and paleoepidemiology of brucellosis. Epidemiol. Infect. 139, 149–156 (2011). [DOI] [PubMed] [Google Scholar]
- 46.David L. A., et al. , Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shreiner A. B., Kao J. Y., Young V. B., The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 31, 69–75 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kau A. L., Ahern P. P., Griffin N. W., Goodman A. L., Gordon J. I., Human nutrition, the gut microbiome and the immune system. Nature 474, 327–336 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Esteban I., et al. , Coastal palaeoenvironments and hunter-gatherer plant-use at Waterfall Bluff rock shelter in Mpondoland (South Africa) from MIS 3 to the Early Holocene. Quat. Sci. Rev. 250, 106664 (2020). [Google Scholar]
- 50.Scrimshaw N. S., Taylor C. E., Gordon J. E., Interactions of nutrition and infection. Monogr. Ser. World Health Organ. 57, 3–329 (1968). [PubMed] [Google Scholar]
- 51.Fellows Yates J. A., et al. , The evolution and changing ecology of the African hominid oral microbiome. Proc. Natl. Acad. Sci. U.S.A. 118, e2021655118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richards M., Montgomery J., “Isotope analysis and paleopathology: A short review and future developments” in The Global History of Paleopathology: Pioneers and Prospects, Buikstra J. E., Roberts C. A., Eds. (Oxford University Press, Oxford, United Kingdom, 2012), pp. 718–731. [Google Scholar]
- 53.Curto A., et al. , Diet and disease in Tomar, Portugal: Comparing stable carbon and nitrogen isotope ratios between skeletons with and without signs of infectious disease. J. Archaeol. Sci. 105, 59–69 (2019). [Google Scholar]
- 54.Beaumont J., Montgomery J., Oral histories: A simple method of assigning chronological age to isotopic values from human dentine collagen. Ann. Hum. Biol. 42, 407–414 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Reitsema L. J., Vercellotti G., Boano R., Subadult dietary variation at Trino Vercellese, Italy, and its relationship to adult diet and mortality. Am. J. Phys. Anthropol. 160, 653–664 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Redfern R. C., DeWitte S. N., Beaumont J., Millard A. R., Hamlin C., A new method for investigating the relationship between diet and mortality: Hazard analysis using dietary isotopes. Ann. Hum. Biol. 46, 378–387 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Neel J. V., Diabetes mellitus: A “thrifty” genotype rendered detrimental by “progress?” Am. J. Hum. Genet. 14, 353–362 (1962). [PMC free article] [PubMed] [Google Scholar]
- 58.Strachan D. P., Hay fever, hygiene, and household size. BMJ 299, 1259–1260 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strachan D. P., Family size, infection and atopy: The first decade of the “hygiene hypothesis.” Thorax 55 (suppl. 1), S2–S10 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blaser M. J., Falkow S., What are the consequences of the disappearing human microbiota? Nat. Rev. Microbiol. 7, 887–894 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rook G. A., Lowry C. A., Raison C. L., Microbial ‘Old Friends,’ immunoregulation and stress resilience. Evol. Med. Public Health 2013, 46–64 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bloomfield S. F., et al. , Time to abandon the hygiene hypothesis: New perspectives on allergic disease, the human microbiome, infectious disease prevention and the role of targeted hygiene. Perspect. Public Health 136, 213–224 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bach J. F., The hygiene hypothesis in autoimmunity: The role of pathogens and commensals. Nat. Rev. Immunol. 18, 105–120 (2018). [DOI] [PubMed] [Google Scholar]
- 64.Finlay B. B., et al. , The hygiene hypothesis, the COVID pandemic, and consequences for the human microbiome. Proc. Natl. Acad. Sci. U.S.A. 118, e2010217118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scudellari M., News feature: Cleaning up the hygiene hypothesis. Proc. Natl. Acad. Sci. U.S.A. 114, 1433–1436 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.De Filippo C., et al. , Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. U.S.A. 107, 14691–14696 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yatsunenko T., et al. , Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Obregon-Tito A. J., et al. , Subsistence strategies in traditional societies distinguish gut microbiomes. Nat. Commun. 6, 6505 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schnorr S. L., et al. , Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 5, 3654 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tito R. Y., et al. , Insights from characterizing extinct human gut microbiomes. PLoS One 7, e51146 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sonnenburg E. D., Sonnenburg J. L., The ancestral and industrialized gut microbiota and implications for human health. Nat. Rev. Microbiol. 17, 383–390 (2019). [DOI] [PubMed] [Google Scholar]
- 72.Manor O., et al. , Health and disease markers correlate with gut microbiome composition across thousands of people. Nat. Commun. 11, 5206 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.De Luca F., Shoenfeld Y., The microbiome in autoimmune diseases. Clin. Exp. Immunol. 195, 74–85 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McLaren M. R., Callahan B. J., Pathogen resistance may be the principal evolutionary advantage provided by the microbiome. Philos. Trans. R. Soc. London. B, Biol. Sci. 375, 20190592 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chiu L., et al. , Protective microbiota: From localized to long-reaching co-immunity. Front. Immunol. 8, 1678 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Khan N., et al. , Alteration in the gut microbiota provokes susceptibility to tuberculosis. Front. Immunol. 7, 529 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reshetnyak V. I., Burmistrov A. I., Maev I. V., Helicobacter pylori: Commensal, symbiont or pathogen? World J. Gastroenterol. 27, 545–560 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van Schaik W., The human gut resistome. Philos. Trans. R. Soc. London. B, Biol. Sci. 370, 20140087 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Anthony W. E., Burnham C. D., Dantas G., Kwon J. H., The gut microbiome as a reservoir for antimicrobial resistance. J. Infect. Dis. 223 (12 suppl. 2), S209–S213 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.D’Costa V. M., et al. , Antibiotic resistance is ancient. Nature 477, 457–461 (2011). [DOI] [PubMed] [Google Scholar]
- 81.Wright G. D., Poinar H., Antibiotic resistance is ancient: Implications for drug discovery. Trends Microbiol. 20, 157–159 (2012). [DOI] [PubMed] [Google Scholar]
- 82.Relman D. A., The human microbiome: Ecosystem resilience and health. Nutr. Rev. 70 (suppl. 1), S2–S9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jacobson D. K., et al. , Analysis of global human gut metagenomes shows that metabolic resilience potential for short-chain fatty acid production is strongly influenced by lifestyle. Sci. Rep. 11, 1724 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jacobson D. K., et al. , Functional diversity of microbial ecologies estimated from ancient human coprolites and dental calculus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 375, 20190586 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Milner G. R., Wood J. W., Boldsen J. L., “Paleodemography” in Biological Anthropology of the Human Skeleton, Katzenberg M. A., Saunders S. R., Eds. (Wiley Blackwell, 2019), pp. 593–633. [Google Scholar]
- 86.Gage T. B., DeWitte S., Wood J. W., “Demography. Part 1. Mortality and migration” in Human Biology: An Evolutionary and Biocultural Perspective, Stinson S., Bogin B., O’Rourke D. H., Eds. (Wiley-Blackwell, 2012), pp. 695–756. [Google Scholar]
- 87.Wood J. W., Milner G. R., Harpending H. C., Weiss K. M., The osteological paradox: Problems of inferring prehistoric health from skeletal samples. Curr. Anthropol. 33, 343–370 (1992). [Google Scholar]
- 88.Singer M., Clair S., Syndemics and public health: Reconceptualizing disease in bio-social context. Med. Anthropol. Q. 17, 423–441 (2003). [DOI] [PubMed] [Google Scholar]
- 89.Larsen C. S., Crespo F., Paleosyndemics: A bioarchaeological and biosocial approach to study infectious diseases in the past. Centaurus 64, 181–196 (2022). [Google Scholar]
- 90.Donoghue H. D., et al. , Co-infection of Mycobacterium tuberculosis and Mycobacterium leprae in human archaeological samples: A possible explanation for the historical decline of leprosy. Proc. Biol. Sci. 272, 389–394 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Giffin K., et al. , A treponemal genome from an historic plague victim supports a recent emergence of yaws and its presence in 15th century Europe. Sci. Rep. 10, 9499 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guellil M., et al. , An invasive Haemophilus influenzae serotype b infection in an Anglo-Saxon plague victim. Genome Biol. 23, 22 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Marciniak S., et al. , An integrative skeletal and paleogenomic analysis of stature variation suggests relatively reduced health for early European farmers. Proc. Natl. Acad. Sci. U.S.A. 119, e2106743119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cox F. E., Concomitant infections, parasites and immune responses. Parasitology 122 (suppl.), S23–S38 (2001). [DOI] [PubMed] [Google Scholar]
- 95.Hartgers F. C., Yazdanbakhsh M., Co-infection of helminths and malaria: Modulation of the immune responses to malaria. Parasite Immunol. 28, 497–506 (2006). [DOI] [PubMed] [Google Scholar]
- 96.Karo-Atar D., Khan N., Divangahi M., King I. L., Helminth-mediated disease tolerance in TB: A role for microbiota? PLoS Pathog. 17, e1009690 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schuenemann V. J., et al. , Targeted enrichment of ancient pathogens yielding the pPCP1 plasmid of Yersinia pestis from victims of the Black Death. Proc. Natl. Acad. Sci. U.S.A. 108, E746–E752 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wagner D. M., et al. , Yersinia pestis and the plague of Justinian 541-543 AD: A genomic analysis. Lancet Infect. Dis. 14, 319–326 (2014). [DOI] [PubMed] [Google Scholar]
- 99.Rasmussen S., et al. , Early divergent strains of Yersinia pestis in Eurasia 5,000 years ago. Cell 163, 571–582 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rascovan N., et al. , Emergence and spread of basal lineages of Yersinia pestis during the Neolithic decline. Cell 176, 295–305.e10 (2019). [DOI] [PubMed] [Google Scholar]
- 101.Yu H., et al. , Paleolithic to Bronze Age Siberians reveal connections with first Americans and across Eurasia. Cell 181, 1232–1245.e20 (2020). [DOI] [PubMed] [Google Scholar]
- 102.Andrades Valtueña A., et al. , Stone Age Yersinia pestis genomes shed light on the early evolution, diversity, and ecology of plague. Proc. Natl. Acad. Sci. U.S.A. 119, e2116722119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Demeure C. E., et al. , Yersinia pestis and plague: An updated view on evolution, virulence determinants, immune subversion, vaccination, and diagnostics. Genes Immun. 20, 357–370 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hinnebusch B. J., Jarrett C. O., Bland D. M., “Fleaing” the plague: Adaptations of Yersinia pestis to its insect vector that lead to transmission. Annu. Rev. Microbiol. 71, 215–232 (2017). [DOI] [PubMed] [Google Scholar]
- 105.Bland D. M., Miarinjara A., Bosio C. F., Calarco J., Hinnebusch B. J., Acquisition of Yersinia murine toxin enabled Yersinia pestis to expand the range of mammalian hosts that sustain flea-borne plague. PLoS Pathog. 17, e1009995 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vågene Å., et al. , Geographically dispersed zoonotic tuberculosis in pre-contact New World human populations. Nat. Commun. 13, 1195 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bos K. I., et al. , Pre-Columbian mycobacterial genomes reveal seals as a source of New World human tuberculosis. Nature 514, 494–497 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schuenemann V. J., et al. , Genome-wide comparison of medieval and modern Mycobacterium leprae. Science 341, 179–183 (2013). [DOI] [PubMed] [Google Scholar]
- 109.Pfrengle S., et al. , Mycobacterium leprae diversity and population dynamics in medieval Europe from novel ancient genomes. BMC Biol. 19, 220 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Urban C., et al. , One health approaches to trace Mycobacterium leprae's zoonotic potential through time. Front. Microbiol. 12, 762263 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Inlamea O. F., et al. , Evolutionary analysis of Mycobacterium bovis genotypes across Africa suggests co-evolution with livestock and humans. PLoS Negl. Trop. Dis. 14, e0008081 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Krause-Kyora B., et al. , Ancient DNA study reveals HLA susceptibility locus for leprosy in medieval Europeans. Nat. Commun. 9, 1569 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Boisson-Dupuis S., et al. , Tuberculosis and impaired IL-23-dependent IFN-γ immunity in humans homozygous for a common TYK2 missense variant. Sci. Immunol. 3, eaau8714 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kerner G., et al. , Human ancient DNA analyses reveal the high burden of tuberculosis in Europeans over the last 2,000 years. Am. J. Hum. Genet. 108, 517–524 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sabin S., et al. , A seventeenth-century Mycobacterium tuberculosis genome supports a Neolithic emergence of the Mycobacterium tuberculosis complex. Genome Biol. 21, 201 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Klunk J., et al. , Evolution of immune genes is associated with the Black Death. Nature, 10.1038/s41586-022-05349-x (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Le Bailly M., Maicher C., Roche K., Dufour B., Accessing ancient population lifeways through the study of gastrointestinal parasites: Paleoparasitology. Appl. Sci. (Basel) 11, 4868 (2021). [Google Scholar]
- 118.Søe M. J., et al. , Ancient DNA from latrines in Northern Europe and the Middle East (500 BC-1700 AD) reveals past parasites and diet. PLoS One 13, e0195481 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sprent J. F. A., “Parasitism, immunity and evolution” in The Evolution of Living Organisms, Leeper G. S., Ed. (Melbourne University Press, Melbourne, Australia, 1962), pp. 149–165. [Google Scholar]
- 120.Armelagos G. J., Brown P. J., Turner B., Evolutionary, historical and political economic perspectives on health and disease. Soc. Sci. Med. 61, 755–765 (2005). [DOI] [PubMed] [Google Scholar]
- 121.McMichael A. J., Environmental and social influences on emerging infectious diseases: Past, present and future. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359, 1049–1058 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Allen T., et al. , Global hotspots and correlates of emerging zoonotic diseases. Nat. Commun. 8, 1124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There are no data underlying this work.