Significance
Human coronavirus 229E(HCoV-229E) and NL63 (HCoV-NL63) are endemic worldwide and cause mild upper respiratory infections or occasionally, more severe lower respiratory tract infections. Mice are not permissive to these virus infections primarily because they lack receptors for these viruses. The recent emergence of COVID-19 emphasizes the need to develop animal models for these CoVs. Here, we generated mouse models for 229E and NL63 by exogenous delivery of their receptors, hAPN and hACE2, respectively, using adenoviruses. We show that these mouse models are useful for analyzing immune responses and for evaluating vaccines and potential therapeutic drugs against HCoV-229E and HCoV-NL63. Moreover, CCCoV-infected mice were partially protected from SARS-CoV-2 infection.
Keywords: HCoV-229E, HCoV-NL63, SARS-CoV-2, mouse model, therapeutics, vaccine
Abstract
Human coronavirus 229E (HCoV-229E) and NL63 (HCoV-NL63) are endemic causes of upper respiratory infections such as the “common cold” but may occasionally cause severe lower respiratory tract disease in the elderly and immunocompromised patients. There are no approved antiviral drugs or vaccines for these common cold coronaviruses (CCCoV). The recent emergence of COVID-19 and the possible cross-reactive antibody and T cell responses between these CCCoV and SARS-CoV-2 emphasize the need to develop experimental animal models for CCCoV. Mice are an ideal experimental animal model for such studies, but are resistant to HCoV-229E and HCoV-NL63 infections. Here, we generated 229E and NL63 mouse models by exogenous delivery of their receptors, human hAPN and hACE2 using replication-deficient adenoviruses (Ad5-hAPN and Ad5-hACE2), respectively. Ad5-hAPN- and Ad5-hACE2-sensitized IFNAR−/− and STAT1−/− mice developed pneumonia characterized by inflammatory cell infiltration with virus clearance occurring 7 d post infection. Ad5-hAPN- and Ad5-hACE2-sensitized mice generated virus-specific T cells and neutralizing antibodies after 229E or NL63 infection, respectively. Remdesivir and a vaccine candidate targeting spike protein of 229E and NL63 accelerated viral clearance of virus in these mice. 229E- and NL63-infected mice were partially protected from SARS-CoV-2 infection, likely mediated by cross-reactive T cell responses. Ad5-hAPN- and Ad5-hACE2-transduced mice are useful for studying pathogenesis and immune responses induced by HCoV-229E and HCoV-NL63 infections and for validation of broadly protective vaccines, antibodies, and therapeutics against human respiratory coronaviruses including SARS-CoV-2.
Coronaviruses cause respiratory, neuronal, and intestinal infections in animals and humans (1, 2). To date, seven coronaviruses have been known to infect human and cause respiratory diseases of varying severity, ranging from common cold to severe pneumonia, including severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The latter three are zoonotic, highly pathogenic, and have or are causing outbreaks, epidemics, or pandemics. The other four coronaviruses, HCoV-229E, HCoV-NL63, HCoV-OC43, and HCoV-HKU1, circulate globally in the human population and cause approximately one-third of common cold infections in humans (3, 4). In the elderly, children, and immunocompromised patients, these four HCoVs occasionally can cause life-threatening pneumonia and bronchiolitis (5–8). HCoV-229E is typically associated with the common cold (9), although a few healthy adults infected with 229E developed acute respiratory distress syndrome (10). Our recent study also showed that HCoV-NL63 readily mutates and has the potential to cause severe respiratory disease in humans (11).
Coronaviruses are taxonomically classified into four genera, α, β, γ, and δ coronaviruses. SARS-CoV, SARS-CoV-2, MERS-CoV, OC43, and HKU-1 are β coronaviruses, while HCoV-229E and NL63 belong to the α coronavirus genus. Receptor-mediated entry is the first step of virus infection of host cells. HCoV-229E uses human aminopeptidase N (hAPN, also known as CD13) as its receptor (12), while HCoV-NL63 uses human angiotensin-converting enzyme 2 (hACE2) as its receptor, which is the same receptor used by SARS-CoV and SARS-CoV-2 (13–17). hAPN and hACE2 are both ectopeptidase that are abundantly expressed in the respiratory tract (18–21).
Animal models are important for assessing virus pathogenicity, immunity, and for development and preclinical evaluation of vaccines or antiviral agents. Unfortunately, there are no robust animal models available for 229E and NL63 infections. Mice are the most widely used experimental animal, offering the convenience of small size, ease of genetic manipulation, and wide availability. Lassnig and colleagues demonstrated that susceptibility to HCoV-229E was only possible in immunocompromised hAPN transgenic mice (hAPN+/+STAT1−/−), using a 229E strain that had been adapted to grow in cells from hAPN+/+STAT1−/− mice (22). These mice developed only mild disease. There is no mouse model reported for NL63.
We previously developed mouse models for MERS and COVID-19 by transducing mice with a recombinant, replication-deficient adenovirus expressing hDPP4 and hACE2, respectively (23, 24), which were useful for MERS-CoV and SARS-CoV-2 studies.
Here, using similar technology, we generated mouse models for HCoV-229E and HCoV-NL63 infections by exogenous delivery of hAPN and hACE2 using adenovirus. Ad5-hAPN- and Ad5-hACE2-sensitized IFNAR−/− and STAT1−/− mice developed pneumonia characterized by inflammatory cell infiltration with virus clearance occurring 7 d after infection. Ad5-hAPN- and Ad5-hACE2-sensitized mice elicited virus-specific T cells and neutralizing antibody responses after 229E or NL63 infection. Remdesivir and vaccine candidates targeting spike proteins of 229E and NL63 were each able to accelerate viral clearance of each virus in Ad5-hAPN- and Ad5-hACE2-transduced/infected mice, respectively. CCCoV-infected mice were partially protected from SARS-CoV-2 infection, which was likely mediated by cross-reactive T cells. These two mouse models will be useful for experimental studies on the pathogenesis and immunity of 229E and NL63 viruses, understanding the consequences of cross-reactive immune responses between CoVs and for the development of multiple interventions against multiple human respiratory coronaviruses, including SARS-CoV-2. This strategy would be also useful in the evaluation of pancoronavirus vaccines.
Results
Development of Mice Sensitized for HCoV-229E Infection.
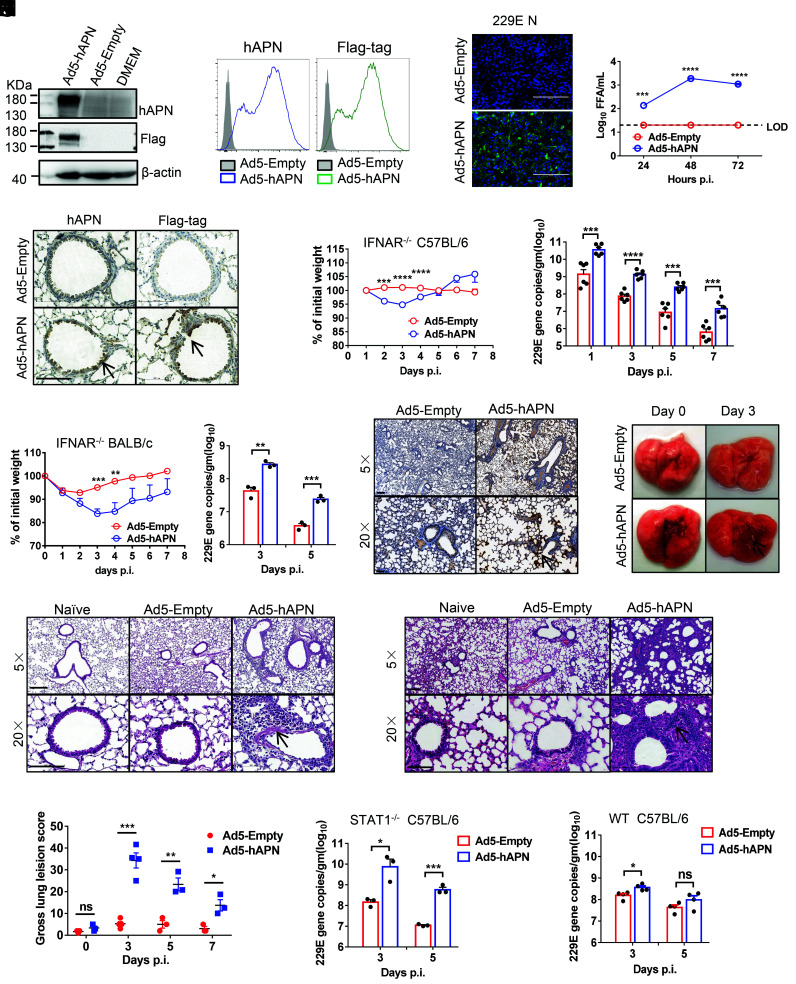
HCoV-229E uses the human APN (hAPN) molecule as its cellular receptor (12). The adenoviral vector expressing hAPN under the control of the CMV promoter was generated as previously described (25–27). hAPN expression was detected by immunoblot and flow cytometry on the cell surface, when we transduced mouse 17CL-1 cells with Ad5-hAPN, but not Ad5-empty (an adenoviral vector with no expression cassette) [multiplicity of infection (MOI) = 20), (Fig. 1 A and B). To determine whether hAPN rendered cells susceptible to 229E infection, we infected Ad5-Empty and Ad5-hAPN-transduced cells with 229E. The 229E antigen (N protein) was detected by immunofluorescence assays in Ad5-hAPN-transduced cells (Fig. 1C). Control cells were resistant to 229E infection, whereas 229E replicated to high titers in Ad5-hAPN-transduced cells (Fig. 1D). We transduced IFNAR−/− C57BL/6 mice with 2.5 × 108 focus forming unit (FFU) of Ad5-hAPN or Ad5-Empty and detected hAPN with an antibody that recognized human hAPN as previously described, (23). hAPN expression in the airway and alveolar epithelium was observed (Fig. 1E). Five days later, mice were infected with 1.5 × 105 TCID50 of 229E and were monitored over a 7-d time course. Ad5-hAPN-transduced IFNAR−/− C57BL/6 mice infected with 229E lost up to 5 to 10% of their body weight in the first 4 d of infection, virus grew to a high viral load in lung tissue and gradually declined over the course of the infection (Fig. 1F). Ad5-hAPN-transduced IFNAR−/− BALB/c mice lost up to 15% of their body weight and virus also replicated to a high viral load in the lung tissue after 229E infection, indicating that mice of BALB/c genetic background were more susceptible to 229E infection (Fig. 1G). As expected, following virus challenge, viral antigen (N protein) was detected in the lungs of mice previously transduced with Ad5-hAPN but not Ad5-empty control (Fig. 1H). Lung tissues from IFNAR−/− C57BL/6 and BALB/c mice had evidence of interstitial pneumonia with perivascular and interstitial inflammatory cell infiltrates (Fig. 1 I and J ). Consistent with the histological findings, examination of gross lung specimens from infected Ad5-hAPN-transduced mice revealed increased vascular congestion and hemorrhage (Fig. 1K), and the gross lung pathology was scored (Fig. 1L). We found that Ad5-hAPN-transduced STAT1−/− mice (Fig. 1M ) support viral replication in their lungs. Of note, 229E only minimally replicated in Ad5-hAPN-transduced WT C57BL/6 mice (Fig. 1N ).
Fig. 1.
Development of mice sensitized to 229E infection. (A and B) To assess hAPN expression and surface localization, 17CL-1 cells were transduced with Ad5-hAPN or Ad5-Empty with MOI of 20 at 37 °C for 4 h. The hAPN expression was monitored by western blot assay (A) or flow cytometry (B). (C and D) Ad5-hAPN-transduced 17CL-1 cells were infected with 229E at MOI of 0.04 at 48 h post transduction, and viral antigen was determined by immunofluorescence assays at 48 h.p.i. (C), and viral titers were determined by focus forming assay (FFA) at 24, 48, and 72 h.p.i. (D). (Scale bars = 200 µm.) (E) Five days after transduction with 2.5 × 108 FFU of Ad5-hAPN or Ad5-Empty in 75 μL DMEM intranasally, lungs were harvested from mice, fixed in zinc formalin, and embedded in paraffin. Sections were stained with an anti-hAPN antibody and anti-Flag antibody (brown color). Arrowheads indicate regions with hAPN expression. (Scale bars = 100 µm.) (F and G) Ad5-hAPN or Ad5-Empty-transduced IFNAR−/− C57BL/6 and BALB/c mice were intranasally infected with 1.5 × 105 TCID50 of 229E in 75 μL DMEM. Weight changes in 6 to 8-wk-old IFNAR−/− C57BL/6 (F) and BALB/c mice (G) were monitored daily (n ≥ 3 mice per group). To obtain viral replication kinetics in IFNAR−/− C57BL/6 (F) and BALB/c mice (G), lungs were harvested and homogenized at the indicated time points, and the viral load was detected by RT-qPCR. Viral loads are expressed as gene copies /g lung tissue (n = 3 mice per group per time point). Data are representative of two independent experiments. (H) Three days post infection, lungs were harvested from IFNAR−/− C57BL/6 mice, fixed in zinc formalin, and embedded in paraffin. Sections were stained with a rabbit anti-229E nucleocapsid protein polyclonal antibody. Arrowheads indicate regions with 229E antigen expression. (Bars = 200 and 50 μm, Top and Bottom, respectively). (I and J) Representative HE staining of lungs from IFNAR−/− C57BL/6(I) and BALB/c (J) mice harvested at 3 d.p.i. Arrowheads indicate regions with interstitial pneumonia with perivascular and interstitial inflammatory cell infiltrates. (Bars = 200 and 100 μm, Top and Bottom, respectively). (K) Photographs of lung specimens isolated from infected IFNAR−/− C57BL/6 mice at indicated time points are shown. Arrowheads indicate regions with vascular congestion and hemorrhage. (L) Gross lung lesion scores are mean ± SE (error bars) and were graded based on the percentage of lung area affected (n = 3 or 4 mice per group per time point). (M and N) Ad5-hAPN or Ad5-Empty-transduced STAT1−/− C57BL/6(L) and WT C57BL/6 (M) mice were intranasally infected with 1.5 × 105 TCID50 of 229E in 75 μL DMEM. Lungs were harvested and homogenized at the indicated time points, and the viral load was detected by RT-qPCR. Viral loads are expressed as gene copies /g lung tissue (n = 3 mice per group per time point). Data are representative of two independent experiments. (*P ≤ 0.05, **P ≤ 0.005, ***P ≤ 0.0005, ****P ≤ 0.0001).
Development of Mice Sensitized for NL63 Infection.
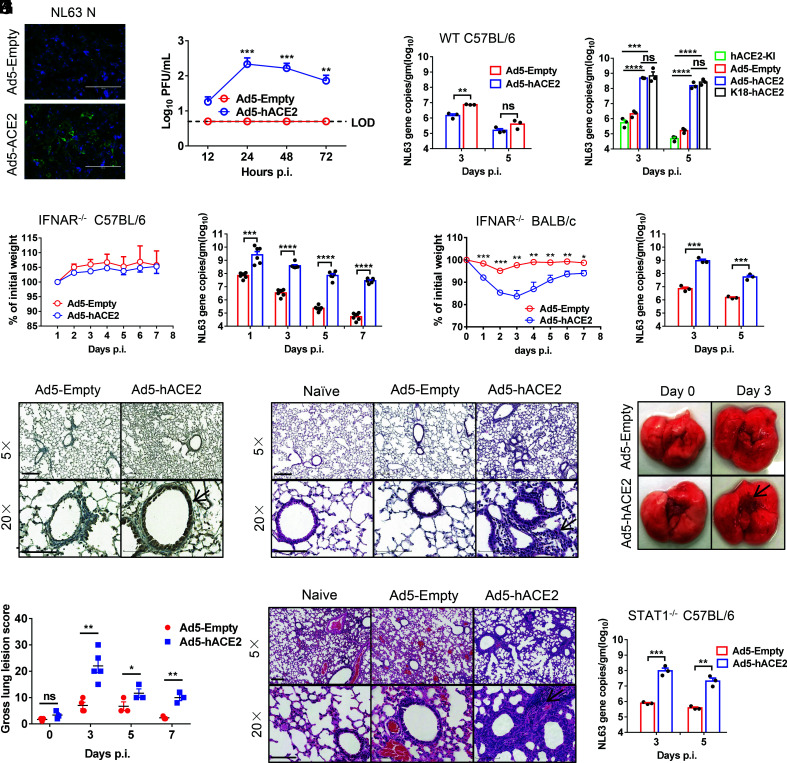
HCoV-NL63 uses human ACE2 to enter cells, same receptor used by SARS-CoV and SARS-CoV-2. The adenoviral vector expressing hACE2 was generated as previously described (25–27). We transduced mouse 17CL-1 cells with Ad5-hACE2 or Ad5-empty (MOI = 100) and found that hACE2 was expressed on the cell surface as previously described (24). To determine whether hACE2 rendered cells susceptible to NL63 infection, Ad5-Empty and Ad5-hACE2-transduced cells were infected with NL63. HCoV-NL63 antigen (N protein) was detected by immunofluorescence assays in Ad5-hACE2-transduced cells (Fig. 2A). Control cells were resistant to NL63 infection, whereas NL63 virus replication was observed in Ad5-hACE2-transduced cells (Fig. 2B). We transduced mice with 2.5 × 108 FFU of Ad5-hACE2 or Ad5-Empty and observed hACE2 expression in the airway and alveolar epithelium as previously described (24). Ad5-hACE2-transduced WT C57BL/6 mice were not susceptible to NL63 infection (Fig. 2C). Similarly, hACE2 knockin mice also did not support NL63 infection. K18-hACE2 mice is a useful model for studying COVID-19 pathogenesis and for assessing therapeutic interventions (28). We found that K18-hACE2 mice also supported NL63 infection and the viral load in lung tissue was similar to Ad5-hACE2-sensitized IFNAR−/− C57BL/6 mice (Fig. 2D), indicating that innate immunity and the expression level of entry receptor are both key factors restricting NL63 infection in vivo. Next, we transduced mice lacking IFN-I signaling (IFNAR−/−) with 2.5 × 108 FFU of Ad5-hACE2 or Ad5-Empty. Five days later, mice were infected with 1 × 104 plaque forming unit(PFU) of HCoV-NL63 and were monitored over a 7-d time course. Ad5-hACE2-transduced IFNAR−/− C57BL/6 mice infected with NL63 showed a slight, but not a significant trend of slower weight gain comparing with the control group. The virus grew to a relatively high viral load in lung tissue of Ad5-hACE2-transduced IFNAR−/− C57BL/6, and the viral load gradually declined over the course of the infection (Fig. 2E). In contrast, Ad5-hACE2-transduced IFNAR−/− BALB/c mice infected with NL63 lost up to 15% of their body weight in the first 1 to 5 d of infection, and viral loads in lung were significantly higher in Ad5-hACE2-transduced mice, indicating mice on BALB/c background were more susceptible to NL63 infection (Fig. 2F). As expected, viral antigen (N protein) was detected in the lungs of mice transduced with Ad5-hACE2 but not Ad5-empty control (Fig. 2G). Histological examination of lungs showed interstitial pneumonia in infected IFNAR−/− C57BL/6 and BALB/c mice (Fig. 2 H and I). Examination of the lungs revealed vascular congestion and hemorrhage (Fig. 2J), and the gross lung pathology was scored (Fig. 2K). We found that Ad5-hACE2-transduced STAT1−/− C57BL/6 (Fig. 2L) mice supported viral infection.
Fig. 2.
Development of mice sensitized to NL63 infection. (A and B) Ad5-hACE2-transduced 17CL-1 cells were infected with NL63 at MOI of 0.02 at 48 h post transduction, and virus antigen was determined by immunofluorescence assays at 48 h.p.i. (A), and virus titers were determined by plaque assay at 12,24, and 48 h.p.i. (B). (Scale bars = 200 µm.) (C) Five days after WT C57BL/6 mice were transduced with 2.5 × 108 FFU of Ad5-hACE2 or Ad5-Empty in 75 μL DMEM intranasally, mice were intranasally infected with 1 × 104 PFU of NL63 in 75 μL DMEM, lungs were harvested and homogenized at the indicated time points, and the viral load was detected by RT-qPCR. Viral loads are expressed as gene copies /g lung tissue (n = 3 mice per group per time point). Data are representative of two independent experiments. (D) Ad5-hACE2-transduced IFNAR−/− C57BL/6 mice, hACE2 C57BL/6 knockin mice, and K18-hACE2 mice were intranasally infected with 1 × 104 PFU of NL63 in 75 μL DMEM, lungs were harvested and homogenized at the indicated time points, and the viral load was detected by RT-qPCR. Viral loads are expressed as gene copies/g lung tissue (n = 3 mice per group per time point). Data are representative of two independent experiments. (E and F) Ad5-hACE2- or Ad5-Empty-transduced IFNAR−/− C57BL/6 and BALB/c mice were intranasally infected with 1.0 × 104 PFU of NL63 in 75 μL DMEM. Weight changes in 6 to 8-wk-old IFNAR−/− C57BL/6 (E) and BALB/c (F) mice were monitored daily (n = 3 mice per group). To obtain virus kinetics in IFNAR−/− C57BL/6 (E) and BALB/c mice (F), lungs were harvested and homogenized at the indicated time points, and the viral load was detected by RT-qPCR. Viral loads are expressed as gene copies /g lung tissue (n = 3 mice per group per time point). Data are representative of two independent experiments. (G) Three days post infection, lungs were harvested from IFNAR−/− C57BL/6 mice, fixed in zinc formalin, and embedded in paraffin. Sections were stained with mice sera from VRP-NL63-N-immunized mice. Arrowheads indicate regions with NL63 antigen expression. (Bars = 200 and 100 μm, Top and Bottom, respectively). (H and I) Representative HE staining of lungs from IFNAR−/− C57BL/6 (G) and BALB/c (J) mice harvested at 3 d.p.i. Arrowheads indicate regions with interstitial pneumonia with perivascular and interstitial inflammatory cell infiltrates. (Bars = 200 and 100 μm, Top and Bottom, respectively). (J) Photographs of lung specimens isolated from infected IFNAR−/− C57BL/6 mice at indicated time points are shown. Arrowheads indicate regions with vascular congestion and hemorrhage. (K) Gross lung lesion scores are mean ± SE (error bars) and were graded based on the percentage of lung area affected (n = 3 or 4 mice per group per time point). (L) Ad5-hACE2- or Ad5-Empty-transduced STAT1−/− C57BL/6 mice were intranasally infected with 1.0 × 104 PFU of NL63 in 75 μL DMEM. Lungs were harvested and homogenized at the indicated time points, and the viral load was detected by RT-qPCR. Viral loads are expressed as gene copies/g lung tissue (n = 3 mice per group per time point). Data are representative of two independent experiments. (*P ≤ 0.05, **P ≤ 0.005, ***P ≤ 0.0005, ****P ≤ 0.0001).
HCoV-229E Infection Induced Viral-Specific T Cell Response and Neutralizing Antibodies in Mice.
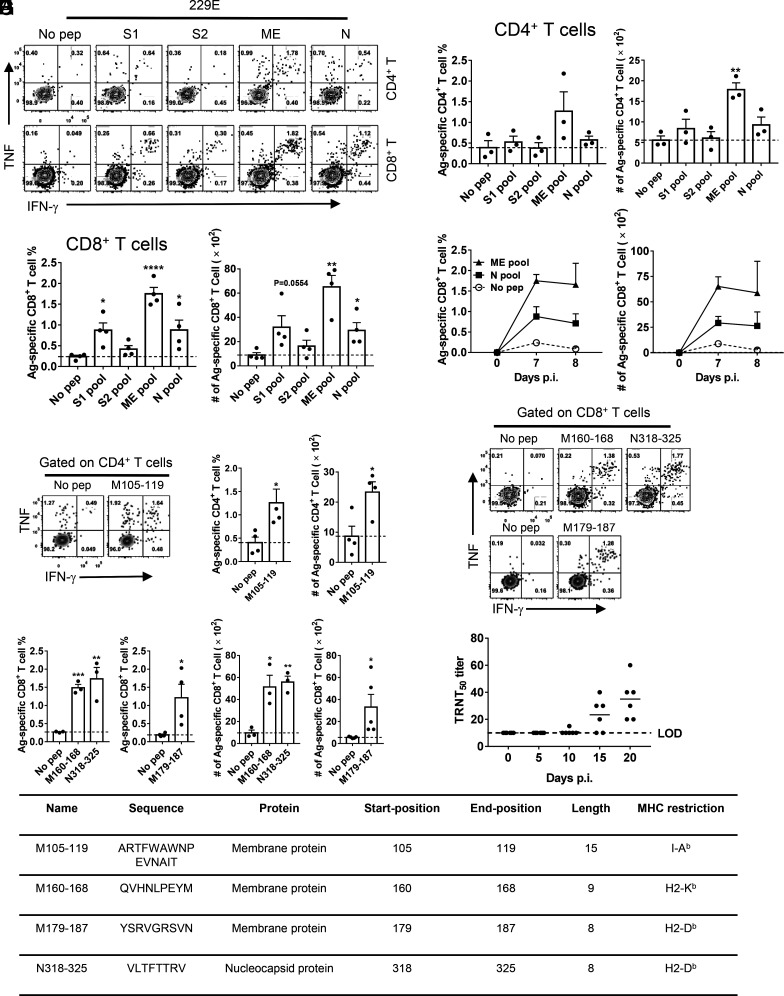
Virus-specific T cells are critical for highly pathogenic CoV clearance in mice (29, 30). To further characterize the T cell response in 229E-infected mice, peptide pools encompassing 229E-structural proteins were synthesized and used to stimulate cells harvested from bronchoalveolar lavage fluids (BALF) of infected mice. In HCoV-229E-infected IFNAR−/− C57BL/6 mice, we found that CD4+ and CD8+ T cell epitopes were predominantly found in the membrane (M) and envelope (E) and N proteins (Fig. 3 A–C). In contrast, CD4+ and CD8+ T cell epitopes in HCoV-229E-infected IFNAR−/− BALB/c mice were predominantly found in the S1 region of the S protein, M, E, and N proteins (SI Appendix, Fig. S1 A–C).The virus-specific CD8+ T cell responses peaked at day 7 p.i. and 8 p.i. in IFNAR−/− C57BL/6 and BALB/c mice, respectively (Fig. 3D and SI Appendix, Fig. S1 D and E). We further screened predicted immunodominant CD4+ and CD8+ T cell epitopes from M/E or N peptide pools using T cell epitope consensus servers [Rankpep, Immune Epitope Database and Analysis Resource (IEDB) and SYFPEITHI] (31 to 33). M105–119 was the immunodominant CD4+ T cell epitope (Fig. 3E) and M160–168, M179–187, and N318–325 were the dominant CD8+ T cell epitopes (Fig. 3 F and G) recognized in HCoV-229E-infected IFNAR−/− C57BL/6 mice. The sequence and MHC restriction of dominant HCoV-229E-CD4+ and -CD8+ T cell epitopes are shown in Fig. 3H. Ad5-hAPN-sensitized mice produced neutralizing antibodies in their sera after infection as determined by TCID50 reduction neutralizing test (TRNT50) (Fig. 3I).
Fig. 3.
229E infection induces virus-specific T cell response and neutralizing antibodies in mice. (A–C) Ad5-hAPN-transduced IFNAR−/− C57BL/6 mice were infected with 1.5 × 105 TCID50 of 229E. To identify 229E T cell responses, single-cell suspensions were prepared from the BALF of transduced/ infected IFNAR−/− C57BL/6 mice and stimulated with 2 μM structural protein peptide pools for 5 to 6 h in the presence of brefeldin A. Flow plots [(A), 7 d.p.i.], and summary of frequencies and cell numbers of 229E-specific CD4+ (B) and CD8+ T cells (C) (determined by IFN-γ intracellular staining) are shown (n = 3 or 4 mice per group per time point). (D) To determine the kinetics of virus-specific T cell responses in 229E -infected IFNAR−/− C57BL/6 mice, single-cell suspensions were prepared from the BALF of transduced/infected IFNAR−/− C57BL/6 mice at indicated time points and stimulated with 2 μM ME or N protein peptide pools for 5 to 6 h in the presence of brefeldin A. The frequencies (Left) and cell numbers (Right) of 229E-specific T cells from BALF are shown (n = 3 mice per time point). (E) Confirmation of 229E-specific-CD4+ T cell epitopes in IFNAR−/− C57BL/6 mice (7 d.p.i.). Flow plots (Left) and summary panels (Right) are shown. (F and G) Confirmation of 229E-specific-CD8+ T cell epitopes in IFNAR−/− C57BL/6 mice (7 d.p.i.). Flow plots (F) and summary panels (G) are shown. (H) Characteristics of 229E-derived CD4+ and CD8+ T cell epitopes in IFNAR−/− C57BL/6 mice. (I) TRNT50 titers of the sera of hAPN-transduced/229E-infected IFNAR−/− C57BL/6 mice were determined at the indicated time points. (n = 6 mice). (*P ≤ 0.05, **P ≤ 0.005, ***P ≤ 0.0005, ****P ≤ 0.0001).
NL63 Infection-Induced Viral-Specific T Cell Response and Neutralizing Antibodies in Mice.
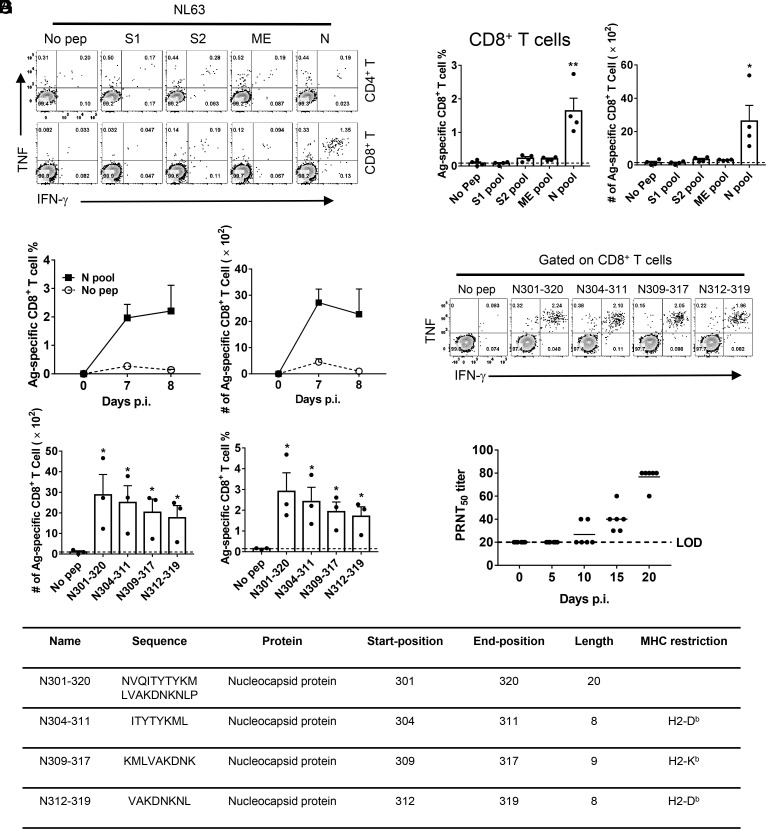
Similarly, to further characterize the T cell response in NL63-infected mice, peptide pools encompassing NL63-structural proteins were synthesized and used to stimulate cells harvested from infected mouse BALFs. In NL63-infected IFNAR−/− C57BL/6 and BALB/c mice, we found that CD8+ T cell epitopes were predominantly located in the N protein (Fig. 4 A and B and SI Appendix, Fig. S2 A and B). The virus-specific CD8+ T cell responses peaked at day 7 p.i. and 8 p.i. in IFNAR−/− C57BL/6 and BALB/c mice, respectively [Fig. 4C and SI Appendix, Fig. S2C). We further screened for immunodominant CD8+ T cell epitopes within N peptide pools, which were predicted using T cell epitope consensus servers (Rankpep, IEDB and SYFPEITHI)] (31–33), and identified the N301–320 region containing multiple CD8+ T cell epitopes (N304–311, N309–317, N312–319) (Fig. 4 D and E). The sequence and MHC restriction of dominant NL63-CD8+ T cell epitopes are shown in Fig. 4F. Ad5-hACE2-sensitized mice also produced neutralizing antibodies in the sera after infection as determined by plaque reduction neutralizing test (PRNT50) (Fig. 4G).
Fig. 4.
NL63 infection-induced virus-specific T cell response and neutralizing antibodies in mice. (A and B) Ad5-hACE2-transduced IFNAR−/− C57BL/6 mice were infected with 1.0 × 104 PFU of NL63. To identify NL63 T cell responses, single-cell suspensions were prepared from the BALF of transduced/infected IFNAR−/− C57BL/6 mice and stimulated with 2 μM structural protein peptide pools for 5 to 6 h in the presence of brefeldin A. Flow plots [(A), 7 d.p.i.], and summary of frequencies and cell numbers of NL63-specific CD8+ T cells (B) (determined by IFN-γ intracellular staining) are shown (n = 4 mice per time point). (C) To determine the kinetics of virus-specific T cell responses in IFNAR−/− C57BL/6 mice, single-cell suspensions were prepared from the BALF of transduced/ infected IFNAR−/− C57BL/6 mice at indicated time points and stimulated with 2 μM N protein peptide pools for 5 to 6 h in the presence of brefeldin A. The frequencies (Left) and cell numbers (Right) of NL63-specific CD8+ T cells from BALF are shown (n = 3 mice at 7 d.p.i. and n = 4 mice at 8 d.p.i.). (D and E) Confirmation of NL63-specific-CD8+ T cell epitopes in IFNAR−/− C57BL/6 mice (8 d.p.i.). Flow plots (D) and summary panels (E) are shown. (F) Characteristics of NL63-derived CD8+ T cell epitopes in IFNAR−/− C57BL/6 mice. (G) PRNT50 titers of the sera of hACE2-transduced/NL63-infected IFNAR−/− C57BL/6 mice were determined at indicated time points p.i. (n = 6 mice). (*P ≤ 0.05, **P ≤ 0.005, ***P ≤ 0.0005, ****P ≤ 0.0001).
HCoV-229E and NL63 Mouse Models Are Useful for Evaluating Antiviral Therapies and Vaccines.
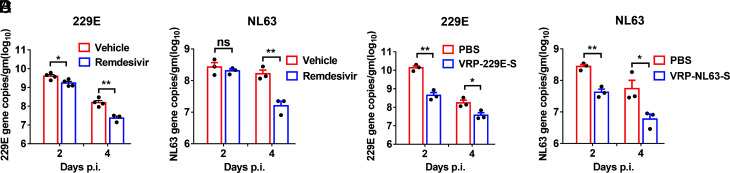
Remdesivir (RDV, GS-5734), is a monophosphoramidate prodrug of an adenosine analog with potent activity against an array of RNA virus through the targeting of the viral RNA-dependent RNA polymerase (RdRp) (34). It can inhibit 229E, OC43, SARS-CoV, and MERS-CoV replication in vitro, and treatment with remdesivir ameliorates SARS and COVID-19 (35–37). Therefore, remdesivir was granted emergency use approval by the US Food and Drug Administration for COVID-19 treatment, based on recently published results (38–40). However, whether remdesivir reduces 229E and NL63 replication in vivo remains unclear. Treatment with remdesivir administered 1 d before infection and for 4 d after infection continued at a dose of 50 mg/kg twice daily resulted in significantly accelerated HCoV-229E and HCoV-NL63 clearance at 4 d post infection (Fig. 5 A and B), indicating it has broad efficacy against CoV infections in vivo.
Fig. 5.
Remdesivir and VRP-S vaccinations protected mice from 229E and NL63 infections. (A and B) For remdesivir treatment, Ad5-hAPN-transduced and Ad5-hACE2-transduced IFNAR−/− C57BL/6 mice were pretreated with remdesivir (50 mg/kg, bid s.c.) or equivalent vehicle at day -1 p.i., respectively. 229E and NL63 viral loads were determined by RT-qPCR. Viral loads are expressed as gene copies/g lung tissue [n = 3 (A) or n = 4 (B) mice per group per time point]. Data are representative of two independent experiments. (C and D) IFNAR−/− C57BL/6 mice were immunized with 2 × 105 infectious unit(IU) of 229E- VRP-S in the footpad in 50 μL PBS or immunized with 2 × 105 IU of NL63-VRP-S intranasally in 50 μL DMEM, respectively. Mice were transduced and infected with 1.5 × 105 TCID50 of 229E or 1.0 × 104 PFU of NL63 3 wk after VRP booster. 229E (C) or NL63 (D) viral loads in the lungs were measured at the indicated time points. Viral loads are expressed as gene copies/g lung tissue (n = 3 or 4 mice per group per time point). (*P ≤ 0.05, **P ≤ 0.005, ***P ≤ 0.0005, ****P ≤ 0.0001).
To evaluate the utility of Ad5-hAPN/Ad5-hACE2-sensitized mice in vaccine evaluation, we developed Venezuelan equine encephalitis replicon particles (VRPs) expressing the 229E or NL63 spike protein (VRP-S). Intranasal immunization with HCoV-229E VRP-S or HCoV-NL63 VRP-S reduced HCoV-229E or HCoV-NL63 replication, respectively, in the lungs of IFNAR−/− C57BL/6 mice (Fig. 5 C and D), indicating these models are generally useful for immune response study and vaccine evaluation.
HCoV-229E- and NL63-Infected Mice were Partially Protected from SARS-CoV-2 Infection.
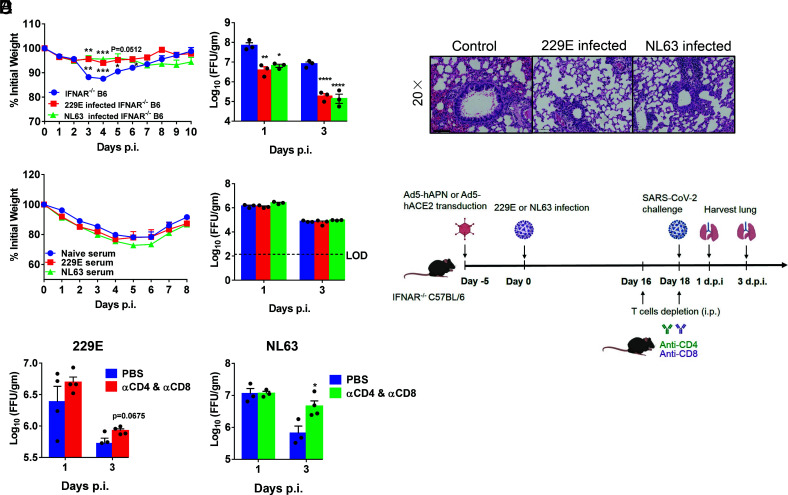
It has hitherto been unclear whether prior infection with CCCoV has cross-protective immune responses to a pathogenic virus such as SARS-CoV-2. To address this question experimentally, initially, Ad5-hAPN- and Ad5-hACE2-transduced mice were infected with 229E and NL63. Twenty-one days later, mice were challenged with SARS-CoV-2 Beta (B.1.351) variant and monitored over a 10-d time course postchallenge. Compared with the control group, 229E- and NL63-infected mice challenged with SARS-CoV-2 showed reduced weight loss and accelerated virus clearance at day 1 and day 3. (Fig. 6A). Examination of HE-stained lung tissues from 229E- and NL63-infected mice challenged with SARS-CoV-2 showed slightly decreased vascular congestion and hemorrhage and interstitial pneumonia compared with control mice (Fig. 6B).
Fig. 6.
HCoV-229E- and NL63-infected mice were partially protected from SARS-CoV-2 infection. (A and B) Ad5-hAPN- and Ad5-hACE2-transduced IFNAR−/− C57BL/6 mice were intranasally infected with 1.5 × 105 TCID50 of 229E and 1.0 × 104 PFU of NL63 in 75 μL DMEM, respectively. Twenty one days postinfection, mice were infected with 5 × 104 PFU of SARS-CoV-2 Beta (B.1.351) variant. Weight changes (n = 4 or 5 mice) were monitored daily, and viral titers in lungs were measured at the indicated time points (A), and hematoxylin/eosin staining of sections of paraffin-embedded lungs is shown at 4 d.p.i. (B). (n = 3 mice per group per time point). Scale bar, 100 μm. (C) For adoptive transfer of serum, Ad5-hAPN- and Ad5-hACE2-transduced IFNAR−/− C57BL/6 mice were intranasally infected with 1.5 × 105 TCID50 of 229E and 1.0 × 104 PFU of NL63 in 75 μL DMEM, respectively. The mouse immune sera were obtained 4 wk postinfection. Then, 150 μL serum was transferred into Ad5-hACE2-transduced mice intravenously (i.v.) 1 d before SARS-CoV-2 infection. Weight changes (n = 5 mice) were monitored daily, and viral titers in lungs were measured at the indicated time points (n = 3 mice per group per time point). (D) The schematic diagrams of T cell depletion from 229E or NL63 infected mice and SARS-CoV-2 challenge. (E) For systemic depletion of CD4+ or CD8+ T cells, mice were infected with 229E (Left) or NL63 (Right). Eighteen days later, mice were injected intraperitoneally (i.p.) with 0.5-mg anti-CD4 antibody (clone GK1.5) and 0.1-mg anti-CD8 antibody (clone 2.43) 2 d before and on the day of SARS-CoV-2 (beta variant) infection. Virus titers in the lungs were measured at the indicated time points. Titers are expressed as FFU/g tissue (n = 3 or 4 mice per group per time point). (*P ≤ 0.05, **P ≤ 0.005, ***P ≤ 0.0005, ****P ≤ 0.0001).
To explore whether the protective effect was mediated by cross-reactive antibody, 150 μL immune sera from 229E- (TRNT50 titer against 229E = 1:160) and NL63- (FRNT50 titer against NL63 = 1:320) infected mice were adoptively transferred to naïve mice one day prior to SARS-CoV-2 infection. This passive transfer of antibody had no effect on weight loss and viral clearance (virus load changes) (Fig. 6C), which suggested that cross-reacting antibodies were not the main mechanism for the observed protection. The contribution of 229E or NL63 infection-elicited cross-protective T cell responses in controlling SARS-CoV-2 infection was then investigated. Sixteen days after receptor-transduced IFNAR−/− C57BL/6 mice were infected with 229E or NL63, we used anti-CD4 and anti-CD8 antibody to deplete both CD4+ and CD8+ T cells 2 d prior to SARS-CoV-2 challenge (Fig. 6D). For this experiment, we used beta variant of SARS-CoV-2 as challenge virus, since this variant can directly infect WT mice without ACE2 receptor transduction (41–43). The efficacy of T cell depletion is shown in SI Appendix, Fig. S3. Our results showed that the virus load in the lung was higher in NL63-infected mice with T cell depletion at 3 d postinfection, indicating that cross-reactive T cell responses induced by CCCoV infections might provide a protective role against SARS-CoV-2 infection. 229E-infected mice showed a similar trend although the difference was not statistically significant (Fig. 6E).
Discussion
HCoV-229E and HCoV-NL63 are endemic human coronaviruses that typically cause mild upper respiratory disease. However, in the immunodeficient population, they could cause severe pneumonia. Compared with other HCoVs, HCoV-NL63 is more frequently associated with croup (44). So far, there is no mouse model reported for NL63. The only report for the 229E mouse model was using a mouse-adapted 229E virus with immunocompromised hAPN transgenic mice (hAPN+/+STAT1−/−) reported by Lassnig and colleagues (22). Development of adenovirus vectors expressing the 229E and NL63 receptors in mouse lungs is an efficient and rapid strategy to allow sensitization of all mouse strains and all genetically modified mice to 229E or NL63 infection. Previously, utilizing this strategy, we successfully constructed mouse models for MERS-CoV and SARS-CoV-2, respectively (23, 24). In this study, we described two mouse models that are useful for investigating 229E and NL63 immunity, pathogenesis, and for evaluating antiviral therapies.
Our results revealed that HCoV-229E replicated more effectively than HCoV-NL63 in vitro and in vivo. Ad5-hAPN-transduced IFNAR−/− C57BL/6 mice infected with 229E lost up to 5 to 10% of their body weight in the first 1 to 4 d of infection. In contrast, Ad5-hACE2-transduced IFNAR−/− C57BL/6 mice infected with NL63 did not show significant weight loss. HCoV-229E and HCoV-NL63 replicated similarly in IFNAR−/− BALB/c mice with more significant weight loss. Lassnig and colleagues demonstrated that susceptibility to HCoV-229E was only possible in immunocompromised hAPN transgenic mice (hAPN+/+STAT1−/−), using a 229E strain that had been adapted to grow in cells from hAPN+/+STAT1−/− mice. Our model potentially could be used to evaluate all HCoV-229E clinical strain infections without prior mouse-adaptation (22).
Significant viral RNA were detected in the lungs of HCoV-229E- and HCoV-NL63-infected Ad5-Empty-transduced IFNAR−/− mice. Based on our results in Fig. 1 C and D, HCoV-229E does not replicate in the absence of hAPN. Lassnig and colleagues also demonstrated that hAPN is required but not sufficient to confer susceptibility in vivo (22). We speculate that 229E virus RNA detected in Ad5-Empty-transduced IFNAR−/− mice was derived from the remaining viruses of primary infection, although there was no viral replication, which could be also be true for the HCoV-NL63 mouse model.
Ad5-hAPN- and Ad5-hACE2-transduced IFNAR−/− and STAT1−/− mice supported 229E and NL63 replication, respectively. The levels of replication were comparable in these two strains of mice, although Ad5-hACE2-transduced IFNAR−/− mice infected with NL63 showed a slightly higher viral load postinfection. IFNAR and STAT1 are both key molecules in the innate immune system. STAT1 is downstream of IFNAR. IFN-I binds to IFNAR and then activate STAT1 to produce numerous interferon-stimulated genes (ISG). ISGs will serve to establish a cellular antiviral state. IFNAR deficiency only interrupts type I interferon signaling, while STAT1 deficiency will block all the type I/II/III interferon pathways, as well as other biological processes related to STAT1 signaling. Thus, we speculated that IFNAR−/− and STAT1−/− double knockout will be redundant, and these mice will behave similar to IFNAR−/− or STAT1−/− mice infected with 229E. Of note, although HCoV-229E and HCoV-NL63 infections could be established in IFNAR−/− mice and STAT1−/− mice, they have limitations when used for studying immunopathogenesis, since they are deficient in the IFN response. In one of our previous studies, the type I interferon pathway was proved to be critical for generating robust T cell responses against SARS-CoV-2 infection. Ad5-hAPN- and Ad5-hACE2-sensitized mice generated virus-specific T cells and neutralizing antibodies after 229E or NL63 infection. The dominant specific T cell epitopes vary across different human coronaviruses. In a C57BL/6 background, the dominant 229E-specific CD8+ T cell epitopes were in M and N protein, whereas the dominant NL63-specific CD8+ T cell epitopes were only found in N protein. Our previous study showed that in MERS-CoV-infected C57BL/6 mice, the dominant virus-specific CD8+ T cell epitopes were in S and M protein (23). SARS-CoV-2-specific CD4+ and CD8+ T cell responses were higher in Ad5-ACE2-transduced mice than those observed in 229E- and NL63-infected mice (24) (45). This could be either because of more robust SARS-CoV-2 replication in mouse respiratory tract or inefficient activation of virus-specific T cell in 229E- and NL63-infected mice deficient of type I interferon signaling.
Remdesivir, a broad-spectrum antiviral agent, was initially developed for the treatment of Ebola virus infection (46). It has been proposed as a promising option for treating COVID-19 (47, 48). Remdesivir exhibits effective antiviral activity in animal models including SARS-CoV (35), MERS-CoV (49), and SARS-CoV-2 (24). In our study, treatment with remdesivir 1 d before infection and twice daily thereafter resulted in significantly accelerated 229E and NL63 clearance by 4 but not 2 d postinfection. In SARS-CoV- and SARS-CoV-2-infected mice, remdesivir significantly cleared viruses in the lung starting at 2 d postinfection (24, 35), indicating that remdesivir might be less efficient against 229E and NL63 infection.
Sager et al. (50) reported that hospitalized patients previously infected with CCCoV had lower rates of intensive care unit (ICU) admissions and higher rates of survival than hospitalized CCCoV-uninfected patients. Several other studies demonstrated that CCCoV-specific T cells cross-react with SARS-CoV-2 (51–53). However, hitherto, there has been no direct evidence suggesting that previous CCCoV infections were protective against subsequent SARS-CoV-2 infection in vivo. Here, we found that 229E and NL63 partially protected against SARS-CoV-2 infection, and this protection was likely mediated by cross-reactive T cells in vivo. The detailed mechanisms for such protection in vivo require further studies.
In summary, we successfully generated 229E and NL63 mouse models using Ad5-hAPN- and Ad5-hACE2, respectively. These two mouse models will be useful for expediting studies of 229E and NL63 pathogenesis and the development of vaccines and therapeutics with broadly protective effects against multiple human respiratory coronaviruses including SARS-CoV-2. In particular, they would help understand immune interactions and cross protection between common cold coronaviruses and SARS-CoV-2 infection and for the development of pancoronavirus vaccines.
Materials and Methods
Mice, Viruses, and Cells.
Specific pathogen-free 6 to 8 wk-old C57BL/6 mice were purchased from Hunan SJA Laboratory Animal Co. and maintained in the Animal Care Facilities at the Guangzhou Medical University. STAT1−/− and IFNAR−/− C57BL/6 mice were purchased from Jackson Laboratories. The IFNAR−/− BALB/c mice were generated by backcrossing IFNAR−/− C57BL/6 with WT BALB/c mice for at least 10 generations. hACE2-KI and K18-hACE2 mice were purchased from Gempharmatech Co. All protocols were approved by the Institutional Animal Care and Use Committees of the Guangzhou Medical University. The 229E strain (ATCC VR-740) was cultured on MRC-5 cells. NL63 strain (NR-470) was obtained from BEI, and cultured on LLC-MK2 cells. The SARS-CoV-2 Beta (B.1.351) variant (Accession number: MT123290) used in this research was isolated from a COVID-19 patient in Guangzhou and passaged on Vero E6 and Calu 3 2B4 cells. Experiments related to authentic SARS-CoV-2 were conducted in Guangzhou Customs District Technology Center BSL-3 Laboratory. African Green monkey kidney-derived Vero E6 cells, 17CL-1 cells, MRC-5 cells, and LLC-MK2 cells were grown in Dulbecco's modified Eagle's medium (DMEM, GIBCO, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS). Calu-3 2B4 cells were grown in MEM (GIBCO, Grand Island, NY) supplemented with 20% FBS. The human serotype 5 adenoviral vector expressing hAPN or hACE2 under the control of the CMV promoter was previously described (27, 54).
Chemicals and Peptides.
Remdesivir (Cat. No. HY-104077) and sulfobutylether-β-cyclodextrin (Cat. No. HY-17031) were purchased from MedChemExpress. A set of 20-mer peptides overlapping by 10 amino acids encompassing the four 229E and NL63 structural [S1, S2, N, and ME encompassing the N- and C-terminal portions of the spike (S) glycoprotein, the nucleocapsid (N) protein, and the transmembrane (M) and envelope (E) proteins] were synthesized by GL Biochem Ltd., and used for stimulation of T cells.
Transduction and Infection of 17Cl-1 Cells and Western Blot Analysis.
Recombinant adenoviral vectors expressing hAPN with Flag tag (Ad5-hAPN) and recombinant adenoviral vectors expressing hACE2 with C9 tag (Ad5-hACE2) were prepared and used as previously described (27, 54). 17Cl-1 cells were transduced with Ad5-hAPN and Ad5-hACE2 at a MOI of 20 and at a MOI of 100, respectively for 4 h at 37 °C, with Ad5-Empty as control. Extracts were prepared 48 h posttransduction. Identical amounts of protein were separated on an 8% SDS/PAGE gel and transferred to PVDF membranes. For hAPN detection, membranes were stained with a mouse anti-human hAPN antibody (clone OTI3F8, Origene, Rockville, MD), a mouse anti-Flag antibody (Cat: A01429, Genscript, Nanjing), or a mouse anti–β-actin (Cat: A01546, Genscript, Nanjing). For hACE2 detection, membranes were stained with a mouse anti-human hACE2 antibody (clone OTI2G7, Origene, Rockville, MD), a mouse anti-C9 antibody (clone Rho 1D4, EMD Millipore, Temecula, CA) or a mouse anti–β-actin (Cat: A01546, Genscript, Nanjing). Proteins were detected using a SuperSignal West Pico Trial Kit (Thermo Scientific). For 229E infection, 17Cl-1 cells were transduced with Ad5- hAPN or Ad5-Empty for 48 h with MOI of 20. The transduced cells were infected with 229E with MOI of 0.04. For NL63 infection, 17Cl-1 cells were transduced with Ad5-hACE2 or Ad5-Empty for 48 h with MOI of 100. The transduced cells were infected with NL63 at MOI = 0.02. Culture supernatants and cells were collected at the indicated time points and analyzed for infectious virus by TCID50 or plaque forming assay (see below).
Immunofluorescence Assays.
The transduced cells were infected with 229E with MOI of 0.04 and infected with NL63 at MOI = 0.02. After 48 h, cells were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. Cells were then incubated with mouse sera harvested from VRP-229E/NL63-N-immunized mice (1: 100 dilution), followed by an Alexa Fluor 488-labeled goat anti-mouse secondary antibody (Cat. No.: 115-545-062, Jackson ImmunoResearch Laboratories, Inc. West Grove, PA). Images of the cells were taken using a EVOS FL inverted microscope at ×20.
Transduction and Infection of Mice.
Mice were lightly anesthetized with isoflurane and transduced intranasally with 2.5 × 108 FFU of Ad5-hAPN, Ad5-hACE2, or Ad5-Empty in 75 μL DMEM. Five days posttransduction, mice were infected intranasally with 229E (1.5 × 105 TCID50), or NL63 (1 × 104 PFU) in a total volume of 75 μL DMEM. Mice were monitored and weighted daily. For cross infection experiments, 21 d postinfection, 229E- or NL63-infected mice were infected with 5 × 104 PFU of SARS-CoV-2 Beta (B.1.351) variant. Mice were monitored and weighed daily.
NL63 Plaque Assay.
Viruses were serially diluted in DMEM. LLC-MK2 cells in 12-well plates were infected with viruses and inoculated at 37 °C in 5% CO2 for 1 h with gentle rocking every 15 min. After removing the inocula, plates were overlaid with 0.9% carboxymethylcellulose containing 2% FBS warmed to 37 °C per well. After further incubation for 5 d, cells were fixed with 10% formaldehyde for 2 h, then overlays were removed, and plaques were visualized by staining with 0.1% crystal violet. Viral titers were calculated as PFU per mL.
229E TCID50 Test.
MRC-5 cells were seeded in 96-well plates one day before infection. Virus cultures were serially diluted with virus growth media (DMEM supplemented with 2% FBS) and used to inoculate MRC-5 cells at 37 °C for 1 h. Inocula were then removed before adding 200 μL virus growth media warmed to 37 °C per well. After 5 to 7 d, TCID50 was calculated by the Reed-Muench method.
229E Focus Forming Assay (FFA) Test.
Virus was titered using an FFA. Huh7 cells were seeded in 96-well plates one day before infection. Virus cultures or lung homogenate was serially diluted and used to inoculate Huh7 cells at 34 °C for 1 h. Inocula were then removed before adding 150 μL 1.2% carboxymethylcellulose warmed to 34 °C per well. After 48 h, cells were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. Cells were then incubated with a rabbit anti-229E nucleocapsid protein polyclonal antibody (Cat. No.: 40640-T62, Sino Biological, Inc. Beijing), followed by an HRP-labeled goat anti-rabbit secondary antibody (Cat. No.: 109-035-088, Jackson ImmunoResearch Laboratories, Inc. West Grove, PA). The foci were visualized by TrueBlue Peroxidase Substrate (KPL, Gaithersburg, MD), and counted with an ELISPOT reader (Cellular Technology Ltd. Cleveland, OH). Viral titers were calculated as FFU per mL.
SARS-CoV-2 FFA Test.
Virus was titered using an FFA. Vero E6 cells were seeded in 96-well plates one day before infection. Virus cultures or lung homogenate was serially diluted and used to inoculate Vero E6 cells at 37 °C for 1 h. Inocula were then removed before adding 125 μL 1.6% carboxymethylcellulose warmed to 37 °C per well. After 24 h, cells were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. Cells were then incubated with a rabbit anti-SARS-CoV-2 nucleocapsid protein polyclonal antibody (Cat. No.: 40143-T62, Sino Biological, Inc. Beijing), followed by an HRP-labeled goat anti-rabbit secondary antibody (Cat. No.: 109-035-088, Jackson ImmunoResearch Laboratories, Inc. West Grove, PA). The foci were visualized by TrueBlue Peroxidase Substrate (KPL, Gaithersburg, MD), and counted with an ELISPOT reader (Cellular Technology Ltd. Cleveland, OH). Viral titers were calculated as FFU per mL or per gram tissue.
229E TCID50 Reduction Neutralization Test (TRNT50).
MRC-5 cells were seeded in 96-well plates one day before infection. Serum samples were serially diluted in DMEM and mixed with an equal volume of 229E containing 100 TCID50. After incubation at 37 °C for 1 h, aliquots were added to MRC-5 cells in 96-well plates and incubated at 37 °C in 5% CO2 for 1 h, following steps were the same as TCID50. TRNT50 was the half-maximum neutralizing titer.
NL63 Focus Reduction Neutralization Test (FRNT50).
LLC-MK2 cells were seeded in 96-well plates one day before infection. Serum samples were serially diluted in DMEM and mixed with an equal volume of NL63 containing 80 to 100 PFU. After incubation at 37 °C for 1 h, aliquots were added to LLC-MK2 cells in 96-well plates and incubated at 37 °C in 5% CO2 for 1 h. Inocula were then removed before adding 150 μL 0.9% carboxymethylcellulose warmed to 37 °C per well. After 5 d, cells were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. Cells were then incubated with mouse sera harvested from VRP-NL63-N-immunized mice (1: 200 dilution), followed by an HRP-labeled goat anti-mouse secondary antibody (Cat. No.: 115-035-062, Jackson ImmunoResearch Laboratories, Inc. West Grove, PA). The foci were visualized by TrueBlue™ Peroxidase Substrate (KPL, Gaithersburg, MD). FRNT50 were half-maximum neutralizing titer.
Remdesivir Treatment.
Remdesivir was dissolved in DMSO in 100 mg/mL and diluted with 12% Sulfobutylether-β-Cyclodextrin (pH = 5). Mice were injected subcutaneously (s.c.) with 25 mg/kg remdesivir twice daily and the control group subcutaneously (s.c.) with a solvent.
VRPs and Mouse Immunization.
VRPs expressing 229E and NL63 spike proteins (S/N) were constructed as previously described (55). Mice were primed and boosted two times (3 wk after priming) with 2 × 105 infectious units (IU) of VRP-S/N in the left footpad in 50 μL PBS or intranasally (i.n.) in 50 μL DMEM after light anesthesia with isoflurane. Sera were collected 1 to 2 wk after second time booster. For challenge experiment, mice were infected with 229E or NL63 viruses three weeks post VRP booster.
Preparation of Cells from BALF.
Mice were sacrificed at the indicated time points. BALF was acquired by inflating lungs with 1-mL complete RPMI 1640 medium via cannulation of the trachea followed by lavaging four times. Cells in the BALF were collected by centrifugation.
Flow Cytometry.
The following monoclonal antibodies were used: rat anti-mouse CD8α-Alexa 488 (clone 53-6.7, Cat. No. 100723, Biolegend, San Diego, CA), anti-mouse CD16/32-PerCP-Cy5.5 (clone 93, Cat. No. 45-0161-82, eBioscience, San Diego, CA), anti-mouse CD4-eFluor 450 (cloneRM4-5, Cat. No. 48-0042-82 eBioscience, San Diego, CA), anti-mouse TNF-PE (cloneMP6-XT22, Cat. No. 12-7321-82Bioscience, San Diego, CA), anti-mouse IFN-γ-APC (cloneXMG1.2, Cat. No. 17-7311-82, eBioscience, San Diego, CA), mouse anti-Flag antibody (Cat: A01429, Genscript, Nanjing), mouse anti-C9 antibody (clone Rho 1D4, Cat. No. MAB5356, EMD Millipore), anti-mouse CD3-APC (clone 145-2C11, Cat. No. 100312, eBioscience, San Diego, CA), anti-mouse CD45-PE (clone 30-F11, Cat. No. 103105, eBioscience, San Diego, CA), live-dead-BV510 (Cat. No. L23105, Thermo Scientific, Massachusetts, CA).
For intracellular cytokine staining (ICS), lymphocytes were cultured in 96-well dishes at 37 °C for 5 to 6 h in the presence of 2 μM peptide pool and brefeldin A (BD Biosciences). Cells were then labeled for cell-surface markers, fixed/permeabilized with Cytofix/Cytoperm Solution (BD Biosciences), and labeled with anti-IFN-γ and anti-TNF antibodies. All flow cytometry data were acquired on a BD FACSVerse and were analyzed using FlowJo software.
Histology and Immunohistochemistry.
Animals were anesthetized and transcardially perfused with PBS followed by zinc formalin. Lungs were fixed in zinc formalin. For routine histology, tissue sections (~4 µm each) were stained with hematoxylin and eosin. To detect hAPN and hACE2 expression, sections were incubated with a blocking reagent (Rodent Block-M, Biocare Medical, Pacheco, CA) incubated with a mouse anti-human hAPN antibody (1:150 dilution, clone OTI3F8, Origene, Rockville, MD), a mouse anti-Flag antibody (1: 500 dilution, Cat: A01429, Genscript, Nanjing). To detect hACE2 expression, sections were incubated with a blocking reagent (Rodent Block-M, Biocare Medical, Pacheco, CA) incubated with a mouse anti-human hACE2 antibody (1:100 dilution, Cat: MAB933, R&D Systems), a mouse anti-C9 antibody (1:200 dilution, clone Rho 1D4, EMD Millipore, Temecula, CA), then incubated with a secondary polymer (Mouse Envision, Dako, Carpinteria, CA), followed by incubation with DAB+ (Dako). To detect 229E and NL63 virus antigen expression, sections were incubated with a blocking reagent (Rodent Block-M, Biocare Medical, Pacheco, CA) incubated with a rabbit anti-229E nucleocapsid protein polyclonal antibody (1:200 dilution, Cat. No.: 40640-T62, Sino Biological, Inc. Beijing) or mouse sera harvested from VRP-NL63-N-immunized mice (1: 100 dilution), then incubated with a secondary polymer (Mouse Envision, Dako, Carpinteria, CA), followed by incubation with DAB (Dako).
RNA Extraction and RT-qPCR.
Total RNA was extracted from infected lungs using Trizol (Invitrogen) according to the manufacturer’s protocol. RT-PCR were performed using the One Step PrimeScript™ RT-PCR Kit (Takara, Japan), in which samples were processed in duplicate using the following cycling protocol: 42 °C for 5 min, 95 °C for 10 s, followed by 45 cycles at 95 °C for 5 s and 60 °C for 34 s. The primer and probe sequences used for RT-PCR are targeted against the nucleocapsid (N) genes of 229E and NL63 are as follows: 229E-Forward: 5′-CGCAAGAATTCAGAACCAGAG--3′, 229E-Reverse: 5′-GGGAGTCAGGTTCTTCAACAA-3′, 229E-N-FAM probe: 5′-CCACAC TTCAATCAAAAGCTCCCAAATG-3′; NL63-Forward:5′-AGGACCTTAAATTCAGACAACGTTCT -3′, NL63-Reverse: 5′-GATTACGTTTGCGATTAC CAAGACT-3′, NL63-N-FAM probe: 5′-TAACA GTTTTAGCACCTTCCTTAGCAACCCAAACA-3′.
Statistical Analysis.
ANOVA and Student’s t tests were used to analyze differences in mean values between groups using GraphPad Prism 7. All results are expressed as mean ± SEM and were corrected for multiple comparisons. P values of <0.05 were considered statistically significant. (*P ≤ 0.05, **P ≤ 0.005, ***P ≤ 0.0005, ****P ≤ 0.0001).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work is supported by the grants National Natural Science Foundation of China (81861168032, 82100117, 82025001, 8181101118, and 91842106); N_HKU737/18 from the Research Grants Council of Hong Kong, Ministries of Science and Technology, Education of Guangdong province (2020B1111330001, 2020A111128008, 2020B1111320003, 2020KZDZX1158, and B195001248); Zhongnanshan Medical Foundation of Guangdong Province (ZNSA-2020013); State Key Laboratory of Respiratory Disease, Guangdong-Hong Kong–Macao Joint Laboratory of Respiratory Infectious Disease (GHMJLRID-Z-202101); National Key R&D Program of China (2021YFC2301700); Guangzhou Institute of Respiratory Health Open Project (Funds provided by China Evergrande Group, 2020GIRHHMS24); Guangdong Natural Science Foundation General Program (2018A030313965); PhD Startup Foundation from Guangzhou Women and Children's Medical Center (2018-2020); Guangzhou Science and Technology Planning Program Municipal School/Institute-Joint Funded Program (202102010143); and Guangdong Medical Technology Research Foundation (B2022233). We thank Dr. Ralph Baric for providing VRP vaccine vector.
Author contributions
D.L., Jingxian Zhao, M.P., and Jincun Zhao designed research; D.L., C.C., D.C., A.Z., F.L., Z.Z., C.K.P.M., J.D., X.L., Y.J., J.S., Y.W., Y.L., Y.Z., L.W., Z.Z., J.Zhuo, J.W., W.R., D.W., S.Z., Y.T., S.L., X.L., P.W., J.Y., F.C., S.H., F.S., Z.Q., W.T., and Jingxian Zhao performed research; Z.C. contributed new reagents/analytic tools; D.L. and M.P. analyzed data; and D.L., Jingxian Zhao, M.P., and Jincun Zhao wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
Reviewers: R.B., University of North Carolina (UNC) at Chapel Hill; L.E., Fundacion General Consejo Superior de Investigaciones Científicas (CSIC); and E.d.W., National Institute of Allergy and Infectious Diseases
Contributor Information
Jingxian Zhao, Email: zhaojingxian@gird.cn.
Malik Peiris, Email: malik@hku.hk.
Jincun Zhao, Email: zhaojincun@gird.cn.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Fields B. N., Fields Virology, Knipe D. M., Howley P. M., Eds. (Lippincott Williams & Wilkins, 2013), vol. 2, pp. 825–858. [Google Scholar]
- 2.Wu Y., et al. , Nervous system involvement after infection with COVID-19 and other coronaviruses. Brain. Behav. Immun. 87, 18–22 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Hoek L., Human coronaviruses: What do they cause? Antivir. Ther. 12, 651–658 (2007). [PubMed] [Google Scholar]
- 4.Davis B. M., et al. , Human coronaviruses and other respiratory infections in young adults on a university campus: Prevalence, symptoms, and shedding. Influenza Other Respir. Viruses 12, 582–590 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pene F., et al. , Coronavirus 229E-related pneumonia in immunocompromised patients. Clin. Infect. Dis. 37, 929–932 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh E. E., Shin J. H., Falsey A. R., Clinical impact of human coronaviruses 229E and OC43 infection in diverse adult populations. J. Infect. Dis. 208, 1634–1642 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorse G. J., et al. , Human coronavirus and acute respiratory illness in older adults with chronic obstructive pulmonary disease. J. Infect. Dis. 199, 847–857 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui J., Li F., Shi Z. L., Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 17, 181–192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradburne A. F., Bynoe M. L., Tyrrell D. A., Effects of a “new” human respiratory virus in volunteers. Br. Med. J. 3, 767–769 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vassilara F., et al. , A rare case of human coronavirus 229E associated with acute respiratory distress syndrome in a healthy adult. Case Rep. Infect. Dis. 2018, 6796839 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y., et al. , Discovery of a subgenotype of human coronavirus NL63 associated with severe lower respiratory tract infection in China, 2018. Emerg. Microbes Infect. 9, 246–255 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeager C. L., et al. , Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357, 420–422 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W., et al. , The S proteins of human coronavirus NL63 and severe acute respiratory syndrome coronavirus bind overlapping regions of ACE2. Virology 367, 367–374 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou P., et al. , A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W., et al. , Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann M., et al. , SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofmann H., et al. , Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. U.S.A. 102, 7988–7993 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamming I., et al. , Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 203, 631–637 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolb A. F., et al. , Characterization of functional domains in the human coronavirus HCV 229E receptor. J. Gen. Virol. 77, 2515–2521 (1996). [DOI] [PubMed] [Google Scholar]
- 20.Mina-Osorio P., The moonlighting enzyme CD13: Old and new functions to target. Trends Mol. Med. 14, 361–371 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang G., et al. , Human coronavirus 229E infects polarized airway epithelia from the apical surface. J. Virol. 74, 9234–9239 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lassnig C., et al. , Development of a transgenic mouse model susceptible to human coronavirus 229E. Proc. Natl. Acad. Sci. U.S.A. 102, 8275–8280 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao J., et al. , Rapid generation of a mouse model for Middle East respiratory syndrome. Proc. Natl. Acad. Sci. U.S.A. 111, 4970–4975 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun J., et al. , Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell 182, 734–743.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao J., et al. , Rapid generation of a mouse model for Middle East respiratory syndrome. Proc. Natl. Acad. Sci. U.S.A. 111, 4970–4975 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCray P. B. Jr., et al. , Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 81, 813–821 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia H. P., et al. , ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 79, 14614–14621 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng J., et al. , COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature 589, 603–607 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao J., Zhao J., Perlman S., T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J. Virol. 84, 9318–9325 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao J., et al. , Airway memory CD4(+) T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity 44, 1379–1391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reche P. A., et al. , Enhancement to the RANKPEP resource for the prediction of peptide binding to MHC molecules using profiles. Immunogenetics 56, 405–419 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Rammensee H. G., Chemistry of peptides associated with MHC class I and class II molecules. Curr. Opin. Immunol. 7, 85–96 (1995). [DOI] [PubMed] [Google Scholar]
- 33.Dhanda S. K., et al. , IEDB-AR: Immune epitope database-analysis resource in 2019. Nucleic Acids Res. 47, W502–W506 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siegel D., et al. , Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f][triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of Ebola and emerging viruses. J. Med. Chem. 60, 1648–1661 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Sheahan T. P., et al. , Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 9, eaal3653 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown A. J., et al. , Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antiviral Res. 169, 104541 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun J., et al. , Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell 182, 734–743.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin W., et al. , Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 368, 1499–1504 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang G. X., et al. , Minimally invasive procedure for diagnosis and treatment of vallecular cysts in children: Review of 156 cases. Eur. Arch. Otorhinolaryngol. 277, 3407–3414 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Grein J., et al. , Compassionate use of remdesivir for patients with severe Covid-19. N. Engl. J. Med. 382, 2327–2336 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan T., et al. , Infection of wild-type mice by SARS-CoV-2 B.1.351 variant indicates a possible novel cross-species transmission route. Signal Transduct. Target. Ther. 6, 420 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kant R., et al. , Common laboratory mice are susceptible to infection with the SARS-CoV-2 beta variant. Viruses 13, 2263 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shuai H., et al. , Emerging SARS-CoV-2 variants expand species tropism to murines. EBio Med. 73, 103643 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Hoek L., et al. , Croup is associated with the novel coronavirus NL63. PLoS Med 2, e240 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhuang Z., et al. , Correction: Mapping and role of T cell response in SARS-CoV-2-infected mice. J. Exp. Med. 218, e2020218710052021c (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pardo J., et al. , The journey of remdesivir: From Ebola to COVID-19. Drugs Context 9, 2020–2024 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beigel J. H., et al. , Remdesivir for the treatment of COVID-19 - final report. N. Engl. J. Med. 383, 1813–1826 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh A. K., et al. , Remdesivir in COVID-19: A critical review of pharmacology, pre-clinical and clinical studies. Diabetes Metab. Syndr. 14, 641–648 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Wit E., et al. , Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. U.S.A. 117, 6771–6776 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sagar M., et al. , Recent endemic coronavirus infection is associated with less-severe COVID-19. J. Clin. Invest. 131, e143380 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mateus J., et al. , Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science 370, 89–94 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woldemeskel B. A., et al. , Healthy donor T cell responses to common cold coronaviruses and SARS-CoV-2. J. Clin. Invest. 130, 6631–6638 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woldemeskel B. A., Garliss C. C., Blankson J. N., SARS-CoV-2 mRNA vaccines induce broad CD4+ T cell responses that recognize SARS-CoV-2 variants and HCoV-NL63. J. Clin. Invest. 131, e149335 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anderson R. D., et al. , A simple method for the rapid generation of recombinant adenovirus vectors. Gene. Ther. 7, 1034–1038 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Deming D., et al. , Vaccine efficacy in senescent mice challenged with recombinant SARS-CoV bearing epidemic and zoonotic spike variants. PLoS Med. 3, e525 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.