Abstract
Severe fever with thrombocytopenia syndrome virus (SFTSV) is a phenuivirus that has rapidly become endemic in several East Asian countries. The large (L) protein of SFTSV, which includes the RNA-dependent RNA polymerase (RdRp), is responsible for catalysing viral genome replication and transcription. Here, we present 5 cryo-electron microscopy (cryo-EM) structures of the L protein in several states of the genome replication process, from pre-initiation to late-stage elongation, at a resolution of up to 2.6 Å. We identify how the L protein binds the 5′ viral RNA in a hook-like conformation and show how the distal 5′ and 3′ RNA ends form a duplex positioning the 3′ RNA terminus in the RdRp active site ready for initiation. We also observe the L protein stalled in the early and late stages of elongation with the RdRp core accommodating a 10-bp product-template duplex. This duplex ultimately splits with the template binding to a designated 3′ secondary binding site. The structural data and observations are complemented by in vitro biochemical and cell-based mini-replicon assays. Altogether, our data provide novel key insights into the mechanism of viral genome replication by the SFTSV L protein and will aid drug development against segmented negative-strand RNA viruses.
INTRODUCTION
Severe fever with thrombocytopenia syndrome virus (SFTSV) is a segmented negative-strand RNA virus (sNSV) that belongs to the Phenuiviridae family in the order Bunyavirales (1). SFTSV was first isolated in China in 2009 (2). Since then, SFTSV has been identified in several other East Asian countries, including Japan (3), South Korea (4), Vietnam (5) and Taiwan (6). In addition to humans, SFTSV is known to infect a range of other mammalian hosts, including domesticated animals, such as cats and dogs, as well as wild animals like rodents, boar, and deer (7–9). SFTSV is thought to be predominantly transmitted via tick bite. However, there is some evidence of human-to-human transmission (2,10,11). For these reasons, SFTSV is considered a significant threat to global public health but antivirals or other therapeutic approaches and countermeasures are lacking.
A key target for the development of antiviral strategies is the bunyaviral large (L) protein, which includes the viral RNA-dependent RNA polymerase (RdRp) (12). The SFTSV L protein is ∼240 kDa in size and responsible for catalysing viral genome replication and transcription. SFTSV particles contain three genome segments of single-stranded RNA with negative polarity, denoted S, M and L segment. During genome replication, negative-sense viral RNA (vRNA) is copied initially into positive-sense complementary RNA (cRNA), which is then used as a template to produce progeny vRNA (12). Several bunyaviruses are known to initiate genome replication de novo by internally producing a short primer, which is subsequently realigned to the 3′ template extremity via a prime-and-realign mechanism. For example, in the case of La Crosse virus (LACV), belonging to the Peribunyaviridae family, cRNA synthesis is initiated internally at position 4 of the 3′ vRNA end and the produced short primer is then realigned to the 3′ terminus (13,14). In contrast, for Lassa virus (LASV), which is a member of the Arenaviridae family, cRNA synthesis is also initiated internally but primer realignment results in an extra G nucleotide at the 5′ end of the vRNA and cRNA (15). An alternative mechanism involves initiating genome replication de novo terminally without the need for realignment in a similar fashion to that seen for influenza virus (16). For SFTSV, biochemical data explain terminal initiation and internal initiation equally well and so further studies are needed before concluding whether the SFTSV L protein uses either or both modes to initiate genome replication (17). Bunyavirus transcription is initiated via a process known as cap-snatching, where the bunyavirus L protein binds to and cleaves off capped fragments of cellular mRNA and uses these as capped primers. This process relies on the endonuclease and a cap-binding activity of the L protein in addition to the RdRp core (18).
Individual endonuclease and cap-binding domains of different bunyaviral L proteins have been investigated by X-ray crystallography (12,17,19,20,21,22,23,24,25,26,27). Recently, cryo-electron microscopy (cryo-EM) data from single-particle analysis became available describing relevant functional states of Machupo virus (MACV), LACV and LASV L proteins in complex with vRNA, emphasizing how these complex molecular machines undergo major conformational changes during viral genome replication and transcription (13,14,28,29). However, to date, structures of the SFTSV and Rift valley fever virus (RVFV) L proteins, as models for phenuiviruses, are only available in an apo state, i.e. without viral RNA (17,30,31,32,33,34). These structures demonstrated that the overall architecture of the phenuivirus L protein was similar to the structure of the heterotrimeric influenza virus polymerase complex and the peribunyavirus LACV L protein but also identified some notable differences between bunyavirus and influenza virus polymerases, the most apparent being differences in the fingertip's insertion (SFTSV L protein residues 913–920). The fingertip's insertion is mostly conserved among bunyaviruses (35), presumably because it contains part of the RdRp active site motif F. However, it curiously adopts an alternative conformation in SFTSV compared to LACV L. While representing important first steps, these apo structures of the SFTSV and RVFV L proteins have not yet allowed for an in-depth analysis of protein-RNA interactions: like the structure of the 5′ hook or the mechanism of prime and realign (17). Such information is however crucial for our understanding of the structure-function relationships underlying viral genome replication and transcription, which is important for antiviral drug development against emerging bunyaviruses.
To address this gap, we took a stepwise approach using a combination of biochemical in vitro assays and cryo-EM to visualize and further understand how the structure of the SFTSV L protein changes upon RNA binding and how the L protein catalyses viral genome replication. Thus, here we present 5 cryo-EM structures of the SFTSV L protein in different biologically relevant states from pre-initiation to late-stage elongation, which are refined to resolutions between 2.6 and 3.5 Å. This structural data is validated by testing selected mutants in a cell-based mini-replicon system. Ultimately, this data provides detailed insights into the mechanism of genome replication by the SFTSV L protein as well as the basis for comparison with other negative-strand RNA viruses like influenza virus and other bunyaviruses.
MATERIALS AND METHODS
Expression and purification of SFTSV L protein
The L gene of SFTSV AH 12 (accession no. HQ116417), which contains a C-terminal StrepII-tag, was chemically synthesized (Centic Biotech, Germany) and cloned into an altered pFastBacHT B vector (17). A point mutation (D112A) was introduced by mutagenic PCR to reduce the SFTSV L proteins intrinsic endonuclease activity (24). DH10EMBacY Escherichia coli cells were used to generate recombinant baculoviruses, which were subsequently used for protein expression in Hi5 insect cells. The harvested Hi5 insect cells were resuspended in Buffer A (50 mM HEPES[NaOH] pH 7.0, 1 M NaCl, 10% [w/v] Glycerol and 2 mM DTT), supplemented with 0.05% (v/v) Tween20 and protease inhibitors (Roche, cOmplete mini), lysed by sonication, and centrifuged twice (20 000 × g for 30 min at 4°C). Soluble protein was loaded on Strep-TactinXT beads (IBA Lifesciences) and eluted with 50 mM Biotin (Applichem) in Buffer B (50 mM HEPES[NaOH] pH 7.0, 500 mM NaCl, 10% [w/v] Glycerol and 2 mM DTT). L protein-containing fractions were pooled and diluted 1:1 with Buffer C (20 mM HEPES[NaOH] pH 7.0) before loading on a heparin column (HiTrap Heparin HP, Cytiva). Proteins were eluted with Buffer A and concentrated using centrifugal filter units (Amicon Ultra, 30 kDa MWCO). The proteins were further purified by size-exclusion chromatography using a Superdex 200 column (Cytiva, formerly GE Life Sciences) in buffer B. Purified L proteins were concentrated as described above, flash frozen and stored at −80 °C.
Polymerase assay
For the standard polymerase assay, 0.9 μM of the SFTSV L protein was incubated sequentially with 1.8 μM of single-stranded 5′ promoter RNA and 1.8 μM of single-stranded 3′ promoter RNA in assay buffer (100 mM HEPES, 50 mM KCl, 2.5 mM MgCl2 and 2 mM DTT, pH 7) on ice for 20 min (Supplementary Table S1). For primer-dependent reactions, 18 μM of the respective primer was added (making for a 10-fold molar excess to promoter RNA) to SFTSV L protein bound to promoter RNA and the mix incubated for an additional 20 min on ice. The polymerase reaction was started by the addition of NTPs (0.2 mM ATP/CTP/UTP and 0.1 mM GTP spiked with 166 nM, 5 μCi[α]32P-GTP, unless stated otherwise) in a final reaction volume of 10 μl. After incubation at 30°C for 2 h, the reaction was stopped by adding an equivalent volume of RNA loading buffer (98% formamide, 18 mM EDTA, 0.025 mM SDS, xylene cyanol and bromophenol blue) and heating the samples at 95°C for 5 min. Products were then separated by denaturing gel electrophoresis using 25% polyacrylamide 7 M urea gels and 0.5-fold Tris–borate–EDTA running buffer. Signals were visualized by phosphor screen autoradiography using a Typhoon FLA-7000 phosphorimager (Fujifilm) operated with the FLA-7000 software.
Rift valley fever virus mini-replicon system
The experiments were performed using the RVFV mini-replicon system described previously (36). L genes were amplified using mutagenic PCR from a pCITE2a-L template to produce either wild-type or mutated L gene expression cassettes. PCR products were gel purified and quantified spectrophotometrically. All mutations were confirmed by sequencing of the PCR products. BSR-T7 cells were transfected per well of a 24 well plate with 250 ng of L gene PCR product, 500 ng of pCITE expressing NP, 750 ng of pCITE expressing the mini-genome RNA encoding Renilla luciferase (Ren-Luc), and 10 ng of pCITE expressing the firefly luciferase as an internal control, all under control of a T7 promoter. For transfection we used Lipofectamine 2000 according to the manufacturer's instructions. At 24 h post-transfection, either total cellular RNA was extracted for Northern blot analysis using an RNeasy Mini kit (Qiagen) or cells were lysed in 100 μl of passive lysis buffer (Promega) per well, and firefly luciferase and Ren-Luc activity quantified using the dual-luciferase reporter assay system (Promega). Ren-Luc levels were corrected with the firefly luciferase levels (resulting in standardized relative light units [sRLU]) to compensate for differences in transfection efficiency or cell density. Data were evaluated in Prism and are always presented as the mean of a given amount (n) of biological replicates as well as the respective standard deviation (SD). To confirm general expressibility of the L protein mutants in BSR-T7 cells, the cells were transfected with 500 ng L gene PCR product expressing C-terminally 3xFLAG-tagged L protein mutants per well. Cells were additionally infected with Modified Vaccinia virus Ankara expressing a T7 RNA polymerase (MVA-T7) to boost the expression levels and, thereby facilitate detection by immunoblotting. At 24 h post-transfection, cells were lysed in 50 μl passive lysis buffer (Promega) per well. After cell lysis and separation in a 3–8% Tris-acetate polyacrylamide gel (Invitrogen), proteins were transferred to a nitrocellulose membrane (Cytiva). FLAG-tagged L protein mutants were detected using horseradish peroxidase-conjugated anti-FLAG M2 antibody (1:9000) (A8592; Sigma-Aldrich) and bands were visualized by chemiluminescence using Clarity Max Western ECL Substrate (Bio-Rad) and a FUSION SL image acquisition system (Vilber Lourmat).
Northern blot analysis
For the northern blot analysis, 600 ng of total cellular RNA was separated in a 1.5% agarose-formaldehyde gel and transferred onto a Roti-Nylon plus membrane (pore size 0.45 μM, Carl Roth). After UV cross-linking and methylene blue staining to visualize 28 S rRNA, the blots were hybridized with a 32P-labelled riboprobe targeting the Ren-Luc gene. Transcripts of the Ren-Luc gene and complementary replication intermediate RNA of the minigenome were visualized by autoradiography using a Typhoon FLA-7000 phosphorimager (Fujifilm) operated with the FLA-7000 software. Quantification of signals for antigenomic RNA and mRNA was done in Fiji.
Sample preparation for cryo-EM 5′ HOOK structure
The SFTSV L protein at a concentration of 3 μM in assay buffer (100 mM HEPES, 50 mM KCl, 2.5 mM MgCl2, and 2 mM DTT, pH 7) was mixed with single-stranded L 5′ (1–20) cRNA in a 2-fold molar excess. The mixture was incubated on ice for 30 min and then centrifuged at 15 000 × g for 10 min at 4°C. Aliquots of 3 μl were applied to Quantifoil R 2/1 Au G200F4 grids, immediately blotted for 2 s, and plunge frozen into liquid ethane/propane cooled to liquid nitrogen temperature using a FEI Vitrobot Mark IV (4°C, 100% humidity, blotting force –10).
Early-elongation, early-elongation-endo and late-elongation structures
The SFTSV L protein at a concentration of 3 μM in assay buffer (100 mM HEPES, 50 mM KCl, 2.5 mM MgCl2, and 2 mM DTT, pH 7) was mixed sequentially with single-stranded L 5′ (1–20) cRNA and single-stranded L 3′ (1–20) cRNA modified with 6 additional A bases, in a 2-fold molar excess. After 30 min of incubation on ice, the reaction was initiated by addition of NTPs (0.63 mM ATP/CTP/GTP/nhUTP). After incubation at 30°C for 2 h, the sample was centrifuged at 15 000 × g for 10 min at 4°C. Aliquots of 3 μl were applied to Quantifoil R 2/1 Au G200F4 grids, immediately blotted for 2 s, and plunge frozen into liquid ethane/propane cooled to liquid nitrogen temperature using a FEI Vitrobot Mark IV (4°C, 100% humidity, blotting force –10).
Resting structure
The SFTSV L protein at a concentration of 3 μM in assay buffer (100 mM HEPES, 50 mM KCl, 2.5 mM MgCl2, and 2 mM DTT, pH 7) was mixed with single-stranded L 5′ (1–20) cRNA and single-stranded L 3′ (1–20) cRNA in a 2-fold molar excess. After 30 min of incubation on ice, the reaction was supplemented with a short 2 nt primer (AC), which was at a 1:1 molar ratio with the RNA species. After incubation at 30°C for 2 h, the sample was then centrifuged at 15 000 × g for 10 min at 4°C. Aliquots of 3 μl were applied to Quantifoil R 2/1 Au G200F4 grids, immediately blotted for 2 s, and plunge frozen into liquid ethane/propane cooled to liquid nitrogen temperature using a FEI Vitrobot Mark IV (4°C, 100% humidity, blotting force –10).
Cryo-EM single-particle analysis
Data collection
Grids were loaded into a 300-kV Titan Krios transmission electron microscope (Thermo Scientific) equipped with a K3 direct electron detector and a post-column GIF BioQuantum energy filter (Gatan). Micrograph movies were typically collected using the EPU software (Thermo Scientific) at a nominal magnification of 105 000× with a pixel size of 0.85 Å using a defocus range of –0.8 to –2.6 μm, unless stated otherwise. The sample was exposed with a dose of 15 electrons/physical pixel per second (estimated over vacuum) for 2.5 s and thus, a total dose of 50 electrons/Å2. Recorded movie frames were fractionated into 50 frames. A summary of the key data collection statistics for each cryo-EM structure is provided in Supplementary Table S2.
Image processing
All movie frames were imported in RELION 3.1 (37) or RELION 4.0 (38) and motion corrected using its own implementation of MotionCor2 (39). Particles were picked automatically using the pre-trained BoxNet in Warp (40) and the calculation of contrast transfer function (CTF) estimation and correction was done in RELION with CTFFIND4 (41). Picked particles were extracted and processed in RELION 3.1/4.0 with visual inspection done with UCSF Chimera (42). First, particles were subjected to 2D classification at 4× binned pixel size typically as an initial check of the quality of the dataset. Then, all particles were used for 3D classification in an iterative manner using a 50 Å low pass filtered map from apo SFTSV L protein (17). Incomplete, low resolution or 3D classes containing damaged particles were excluded from further data analyses. The remaining particles were re-extracted with the original pixel size value for further 3D classification and refinement as described for the 5′ hook structure (Supplementary Figures S1 and S2); early-elongation, early-elongation-endo and late-elongation structures (Supplementary Figures S3 and S4); and, resting structure (Supplementary Figures S5 and S6). All cryo-EM maps were CTF refined and Bayesian polished before final refinement in relion except the resting structure which, after 3D classification, was imported as a particle stack in cryoSPARC (43) where final refinement was done with non-uniform refinement (44).
All final cryo-EM density maps were generated by the post-processing feature in RELION and sharpened or blurred into MTZ format using CCP-EM (45) except the early-elongation-endo structure where the final map was sharpened using DeepEMhancer (46) and the resting structure, which was sharpened using the local sharpening function of cryoSPARC. The resolutions of the cryo-EM density maps were estimated at the 0.143 gold standard Fourier Shell Correlation (FSC) cut-off. A local resolution was calculated using relion 3.1/4.0 except the resting structure which was calculated in cryoSPARC and displayed with Chimera visualization tools. A summary of the key processing statistics for each cryo-EM structure is provided in Supplementary Table S2.
Model building, refinement and validation
The published corrected apo structure of the SFTSV L protein (30) was used as the starting point for the RNA-bound structures published in this study which then underwent iterative rounds of model building in Coot (47) and real-space refinement in Phenix (48). The comprehensive validation tool in Phenix was used to assess model quality (49,50). Coordinates and EM maps are deposited in the EMDB and PDB databases as indicated in Table 1. Figures have been generated using CCP4mg (51), PyMOL (Schrödinger), and UCSF ChimeraX (52). A summary of the key refinement and validation statistics for each cryo-EM structure is provided in Supplementary Table S2.
Table 1.
Overview of cryo-EM structures. An overview of the cryo-EM structures is provided with the reported resolution (Å), any RNA ligands present, PDB and EMDB accession codes, as well as supporting comments highlighting the key features of each structure
| Name | Res (Å) | RNA Ligands | PDB Code | EMDB Code | Comment |
|---|---|---|---|---|---|
| 5′ Hook | 3.4 | L 5′ (1–20) | 8AS6 | EMD-15607 | Only 5′ cRNA is bound to the L protein in the hook-like motif. The L protein core is clear and motif F is unstructured. |
| Early-elongation | 2.6 | L 5′ (1–20) L 3′ + 6A (1–26) | 8AS7 | EMD-15608 | The 5′ and 3′ cRNA form a distal duplex structure with the 3′ cRNA shown feeding into the L protein core. Short 3 nt product in L protein core. Stalling nhUTP clearly bound at + 1 position. |
| Early-elongation-endo | 3.6 | L 5′ (1–20) L 3′ + 6A (1–26) | 8ASB | EMD-15610 | The 5′ and 3′ cRNA form a distal duplex structure with the 3′ cRNA shown feeding into the L protein core. There is also a short 3 nt-long product in L protein core. However, in this structure, we see the endonuclease in a raised conformation unlike that seen elsewhere. Stalling nhUTP clearly bound at +1 position. |
| Late-elongation | 2.6 | L 5′ (1–20) L 3′ + 6A (1–26) | 8ASD | EMD-15614 | The distal duplex structure is disrupted and there is a long 10-bp product-template duplex in the L protein core. Stalling nhUTP clearly bound at +1 position. The CBD can also be fit with the putative cap-mRNA binding site occupied by the Arginine blocking motif. |
| Resting | 3.2 | L 5′ (1–20) L 3′ (1–20) Primer AC (1–2) | 8ASG | EMD-15615 | The 5′ cRNA is bound as in the 5′ HOOK structure. Excess of 3′ cRNA is bound to the 3′ secondary binding site. A small number of 3′ cRNA nts can be fit in the L protein core but the region of map for the AC primer is not resolved. |
RESULTS
Overview
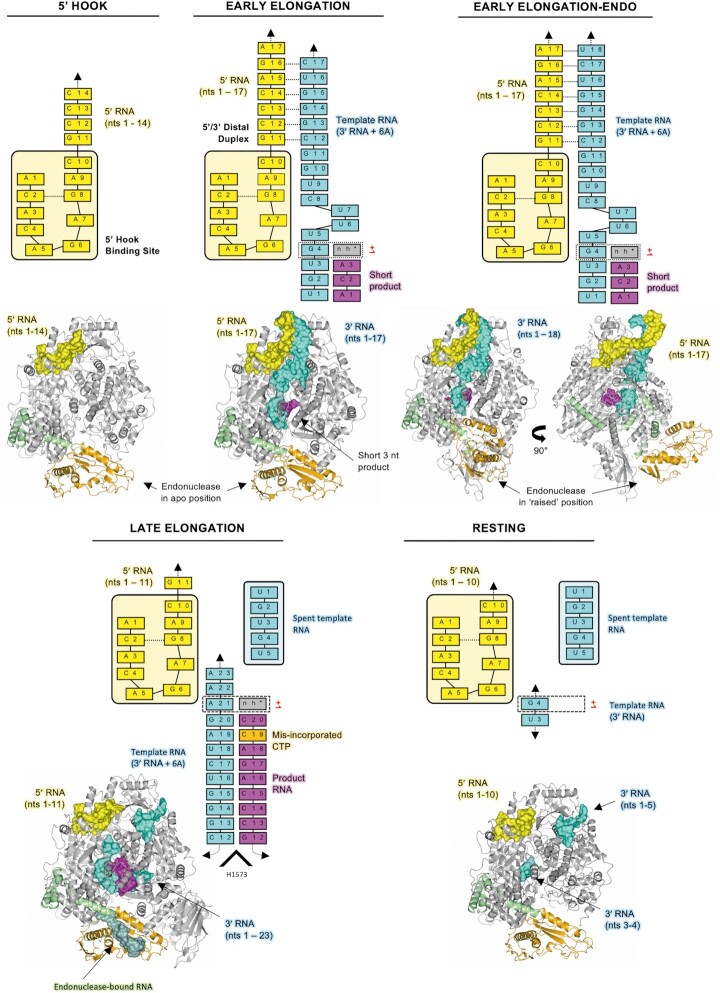
We have determined five structures of the SFTSV L protein in several biologically relevant states ranging from pre-initiation to late-stage elongation. An overview of all structures including their key features is provided in Table 1. First, we incubated the L protein with nt 1–20 of the L segment 5′ cRNA (5′ hook, 3.4 Å) (Supplementary Table S1; Supplementary Figures S1 and S2). This initial structure enabled us to identify the pocket in which the L protein coordinates the 5′ RNA terminus in a so-called hook-like conformation, which is known to be important for activation of the polymerase function (17).
To stall the L protein in an active genome replication mode, we incubated the L protein with nts 1–20 of the L segment 5′ cRNA and a modified 3′ cRNA template with 6 additional A’s (Supplementary Table S1). These were incubated together in the presence of ATP, CTP and GTP spiked with non-hydrolysable UTP (nhUTP) (Supplementary Figures S3 and S4). With this, we observed three elongation-state structures: two in which the L protein is stalled at position G4 of the modified 3′ cRNA template (EARLY-ELONGATION, 2.6 Å and EARLY-ELONGATION-ENDO, 3.6 Å) and another in which the L protein is stalled at position A21 of the modified 3′ cRNA template (LATE-ELONGATION, 2.6 Å). While the binding of viral RNAs in the two early-elongation structures appear similar with the distal 5′ cRNA region forming a duplex with the 3′ cRNA template, there is a key difference in the overall conformation of the L protein. In the EARLY-ELONGATION-ENDO structure, the endonuclease domain adopts an entirely different ‘raised’ conformation by comparison to the endonuclease domain in the early-elongation structure, which is broadly similar to that seen in the apo and late-elongation structures. In the latter, the CBD can also be fitted and the putative cap-binding site appears blocked by the arginine blocking motif previously described (30–33). Therefore, in total, we obtained 3 structures of the L protein stalled in active genome replication mode: two stalled in the early stages of elongation, and a final structure stalled in a late stage of elongation.
To identify the structural basis for replication priming, we incubated the L protein with nts 1–20 of the L segment 5′ cRNA and 3′ cRNA termini and added a short 2 nt (AC) primer (Supplementary Table S1; Supplementary Figures S5 and S6). Processing of data collected from this grid led to a single structure (RESTING, 3.2 Å), in which the 5′ RNA hook binds in the same way as that seen in our other structures. However, rather than formation of the distal duplex and the 3′ cRNA template entering the active site, we observe the 3′ cRNA terminus binding to an alternative region, denoted the 3′ secondary binding site in analogy to similar binding sites identified in LASV (28) and LACV L proteins (13) and also the influenza virus polymerase complex (53). The distal duplex is not formed in this structure and we observe a weak signal in this region of the map that likely corresponds to an average of the RNA strands in multiple different conformations. Nevertheless, this structure demonstrates that both the 3′ and 5′ RNA termini can be bound simultaneously to the hook-binding and secondary binding sites, respectively. We propose that this might represent a resting conformation after finishing one round of genome replication and just before another pre-initiation complex is formed with the distal duplex stabilizing the 3′ RNA and guiding it into the RdRp active site for initiation. However, in this structure, we can also confidently fit 2 nts of the 3′ cRNA in the active site, which indicates that the observed map represents a mixture of resting and pre-initiation states, which would explain the relative lack of definition in some regions of the map, making it challenging to interpret.
The L protein during pre-initiation of viral genome replication
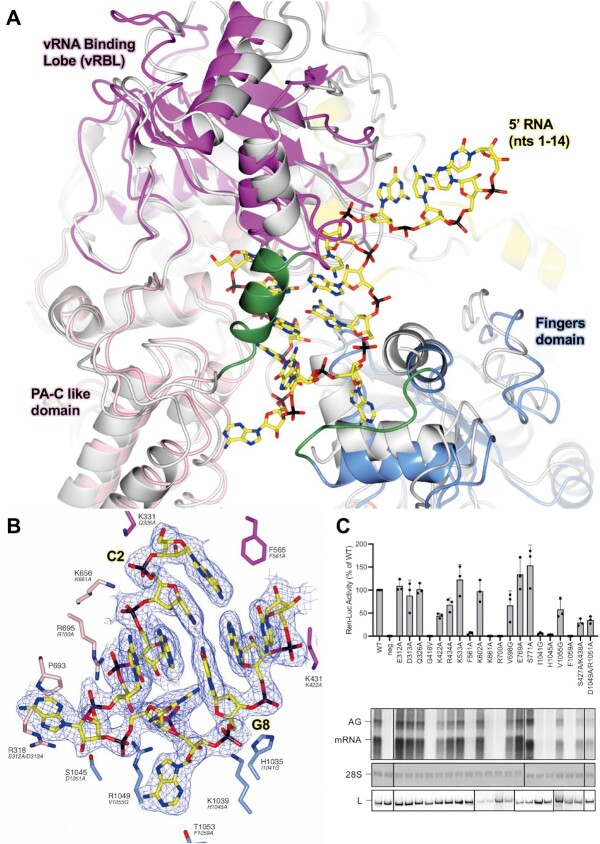
The 5′ HOOK structure demonstrates that the SFTSV L protein undergoes several conformational changes as a result of binding to the terminal 20 nts of the 5′ cRNA in comparison to the apo structure (PDB: 6L42, 6Y6K and 7ALP) (30–33). The most apparent changes being an opening up of the vRNA binding lobe (vRBL), the neighbouring fingers domain, and the PA-C like core lobe, allowing the 5′ RNA to bind in a positively charged cleft (Figure 1A). This cleft is covered by a short 3-turn α-helix (M437–H447) and supported on the periphery by a short loop (H1038–D1046) (Figure 1A). Of note, both of these structural motifs were missing in the published apo structures, suggesting here that the 5′ RNA has a role in stabilizing this region of the L protein.
Figure 1.
Conformational changes following the binding of the 5′ cRNA hook. (A) The SFTSV L protein (5′ HOOK) is shown with the published apo structure of the SFTSV L protein (PDB: 7ALP, backbone coloured grey) superposed by SSM in CCP4mg (51). A short 3-turn α-helix (M437–H447) and loop (H1038–D1046), which are both stabilized by the binding of the 5′ cRNA nts, are shown in green. (B) A close-up of the SFTSV L protein hook-binding site is shown with the early-elongation map overlaid. Bound 5′ cRNA is shown and coloured yellow with the exception of C2 and G8, which together form the only cognate base-base interaction and are labelled. Residues from the SFTSV L protein are labelled and coloured according to the assigned domain (pink for the PA-C like-domain, blue for the fingers, and magenta for the vRBL) with the corresponding RVFV L protein residue shown in black italicized text underneath. Not all hook-interacting residues are shown for clarity. (C) RVFV mini-replicon data for L protein with mutations in the proposed 5′ hook binding site presenting luciferase reporter activity (in standardized relative light units relative to the wild-type L protein (WT)). Data were presented as mean values ± SD of three biological replicates (n = 3). All biological replicates are shown as black dots (top panel). Middle panels present Northern blotting results with signals for antigenomic viral RNA (AG, equal to cRNA), viral mRNA (mRNA) and 28 S ribosomal RNA (28 S) as a loading control, and the bottom panel shows Western blot detection of FLAG-tagged L proteins (L) to demonstrate general expressibility of the mutants. Uncropped original blots/gels are provided in the Supplementary Data.
Structure of the 5′ cRNA hook
We find the 5′ cRNA bound in each of our L protein structures and, depending on the structure, we can fit 11–17 nts into the corresponding map region. From this data, we found that the first 10 nts of the SFTSV 5′ RNA form the characteristic hook-like motif (Figure 1B). In addition, in our structures, the 5′ RNA hook only appears to be stabilized by one canonical base-base interaction between C2 and G8. The SFTSV hook structure hence appears to be closer to that of LASV, which is also stabilized by one canonical base-base interaction (28), in comparison to that of LACV, which is stabilized by two canonical base–base interactions (14) (Supplementary Figure S7). In addition to the C2/G8 base-pairing interaction, amino acid sidechains from the fingers, finger node, and the core lobe contribute towards 5′ RNA hook coordination.
For example, the K656 sidechain is shown to interact with the backbone phosphates of nts C2 and A3 (Figure 1B). The N765 sidechain also interacts with nt A3, with the sidechain amino group coordinating one of the A3 base nitrogens, while the carboxyl group interacts with the A3 ribose O2′ hydroxyl. There are two sidechain interactions stabilizing nt C4: R1049 and H447. These coordinate the C4 ribose O2′ hydroxyl and C4 base oxygen, respectively. The R695 sidechain also stacks against the C4 base providing additional stability. The A5 base is sandwiched between the R318 and P693 sidechains. Similarly, there are not many interactions visible stabilizing the G6 base, only the H447 sidechain coordinating the G6 ribose oxygen (Supplementary Figure S8A). In addition to this interaction, in our early-elongation structure, we noticed an interesting ring-shaped blob of density between the G6 base and the adjacent R1043 sidechain (Supplementary Figure S8B). After exploring several different options, including individual NTPs bound non-specifically at this position, we concluded that the best fit was a single HEPES molecule, which is the buffer used during the L protein/cRNA incubation step. Here, the HEPES ring is shown sandwiched in between the G6 base and the R1043 sidechain (Supplementary Figure S8B). The base A7 is shown to sit comfortably in a relatively tight pocket delineated by the sidechains of F1032, L1036, F1048, R1049, and M1052 (Supplementary Figure S8C). Here, the R1049 sidechain also coordinates the A7 backbone phosphate. A9 is coordinated by the S562 and H563 sidechains, which interact with the A9 O2′ hydroxyl and a base nitrogen atom, respectively (Supplementary Figure S8D). Finally, the C10 base is sandwiched between the A9 base and the K431 sidechain. There is then a sharp turn between nt 10 and 11 of the 5′ RNA marking the end of the hook motif.
Mutational analysis of 5′ RNA hook-binding residues
To investigate the importance of the key 5′ RNA hook-binding residues, we mutated 21 different amino acids of the L protein (18 single, 2 double-mutants) and tested for genome replication and transcription activity in a viral mini-replicon system. In this system, genomic viral RNA is produced by the T7 polymerase from a plasmid and then serves as a template for viral genome replication and transcription, producing Ren-Luc mRNA, antigenomic viral RNA and subsequently also further genomic viral RNA copies, all catalyzed by L and N proteins (36). As we had a RVFV mini-replicon system already established, we mapped the identified SFTSV residues onto the closely-related RVFV L protein (Supplementary Figure S9), and then tested the corresponding positions in this RVFV mini-replicon system (36). Of the 21 mutants tested, 11 showed wild type-like activity (i.e. Ren-Luc activity, >50%) and 3 mutants showed a slightly reduced activity (i.e. Ren-Luc activity, 8–49%) (Supplementary Table S3, Figure 1C). However, mutations of 7 residues led to a strong decrease in RVFV L protein activity (i.e. Ren-Luc activity, <7%), including: G418V (SFTSV: G427), F561A (SFTSV: F565), K661A (SFTSV: K656), R700A (SFTSV: R695), I1041G (SFTSV: H1035), H1045A (SFTSV: K1039) and F1059A (SFTSV: T1053) (Supplementary Table S3, Figure 1C). Of those residues critical for L protein function in the RVFV mini-replicon system, the corresponding K656, R695, and K1039 directly interact with A3, C4/A5, and G8 of the 5′ cRNA in our structure of the SFTSV L protein. In addition, H1035 appears to line the hook binding cleft, whereas T1053 has a long interaction (>4 Å) with the A7 base of the 5′ cRNA, while G427 and F565 are adjacent to the A1 and C10 nts of the 5′ cRNA (Figure 1B). Overall, this analysis suggests that the 5′ RNA binding site is important for both genome replication and transcription as no selective defect in RNA synthesis could be detected. This matches previous observations for LASV L protein (28).
The transition from pre-initiation to elongation
To capture the SFTSV L protein in an actively replicating state, we incubated the L protein with nts 1–20 of the SFTSV L 5′ cRNA and a 3′ cRNA template modified with 6 additional A’s at the 5′ end together with ATP, GTP, CTP and nhUTP. The SFTSV L 3′ cRNA has only one A in the first 20 nts and this is at position 19. If we had stalled the L protein using nhUTP with this un-modified 3′ cRNA then the difference between the stalled and un-stalled products would have been only 1 nt and this would have been difficult to visualize biochemically. Thus, we decided to add six additional A’s to the 5′ end of the 3′ cRNA boosting the overall size of the template from 20 nts to 26 nts. If the L protein stalled at the first A in the 3′ cRNA template, this would have produced a stalling product at 19 nts, whereas the un-stalled product would now be 26 nts. This greater difference in product size allowed us to visualize the stalling reaction clearly by biochemical means.
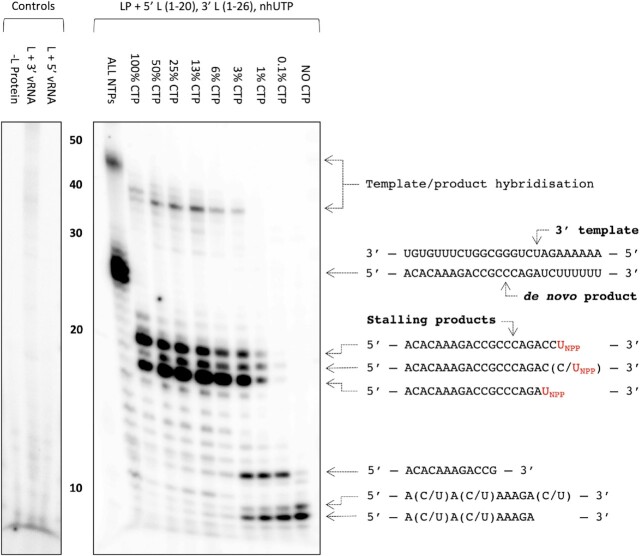
After cryo-EM data collection and processing, we identified a major class in which the L protein appeared to be stalled in the later stages of elongation with a 10-bp product-template duplex in the L protein core (LATE-ELONGATION, 2.6 Å; Supplementary Figures S3 and S4). Upon further inspection, we discovered that the L protein had seemingly stalled at A21 of the 3′ cRNA. Using similar conditions, biochemical analysis showed that incorporation of nhUTP into the NTP mix leads to two major products: one at 18 nts, which was the expected stalling product length given that the first A in the template sequence is at position 19, and a second, 20 nt-long product. As it is known that viral RNA polymerases are relatively low-fidelity enzymes (54), we hypothesized that the second major stalling product at 20 nts was probably the result of the L protein misincorporating CTP at A19 in the template before stalling at A21. To test this hypothesis, we titrated CTP out of the NTP mix spiked with nhUTP and found that as we reduced the concentration of CTP, the intensity of the larger, 20 nt band, decreased in a corresponding manner (Figure 2). At the same time, the intensity of the smaller 18 nt band increased (Figure 2). Thus, we could demonstrate both in vitro and structurally that the SFTSV L protein can misincorporate CTP in place of UTP. There is also a minor product at ∼19 nts (Figure 2). We think this could either be termination after a second misincorporated CTP or possibly the result of misincorporating nhUTP at this position. The second of these two options seems less likely but that may explain why this product has a much weaker intensity. It is also noteworthy that as we titrate CTP out of the NTP mix, the intensity of two smaller bands in the 9–15 nt range increases (Figure 2). We think these represent early termination events where the L protein reaches a poly-G sequence in the 3′ cRNA strand and then stalls due to lack of CTP. This would explain why the intensity of these products increases as CTP is titrated out of the NTP mix.
Figure 2.
Stalling of the SFTSV L protein and misincorporation of CTP for UTP. The effect of removing CTP from the NTP mix in the presence of nhUTP (UNPP) on the polymerase activity of purified SFTSV L protein (LP) was tested in vitro. The reactions were carried out with the L 5′ cRNA (nts 1–20) and L 3′ cRNA (nts 1–26) present under standard polymerase assay conditions (see Materials and Methods), with the exception of the NTPs used in the CTP titration conditions, which were diluted 1:10 resulting in a final concentration of 20 μM ATP, UTP and 10 μM GTP spiked with 166 nM, 5 μCi[α]32P-GTP. The final concentration of CTP in the NTP mix varied from 20 to 0 μM. Products were separated by denaturing gel electrophoresis and visualized by autoradiography. Uncropped original blots/gels are provided in the Supplementary Data.
Processing of the cryo-EM data identified two other major classes with the L protein stalled at a much earlier point in the 3′ cRNA template and with significant differences regarding the endonuclease domain (Supplementary Figures S3 and S4). Whereas in the EARLY-ELONGATION structure the endonuclease adopts a similar conformation to that seen in the published apo structure (30–33), in the EARLY-ELONGATION-ENDO structure, the endonuclease adopts a ‘raised’ conformation (Supplementary Figures S3 and S4). After inspecting the initial maps, we found that in both early elongation structures the 3′ RNA template was positioned with 4 nts into the L protein core (3′-UGUG…-5′), with a short 3 nt product (5′-ACA-3′) and the nhUTP at the +1 position, where we would actually expect CTP. Given that we knew the L protein could misincorporate CTP in place of UTP, we considered it possible the L protein could also misincorporate UTP in place of CTP. If this was true, the L protein could presumably also misincorporate nhUTP leading to the observed early stalling product.
To test this hypothesis, we selected the SFTSV S9U segment vRNA as this genome end has a naturally occurring poly-G sequence. If the L protein can incorporate UTP in place of CTP, then in the absence of CTP, the L protein should be able to read-through the poly-G sequence (nts 11–15) by incorporating UTP. If not, then we should observe a stalling product around 11 nts. We additionally used a 16 nt primer (Primer B-OH, Supplementary Table S1) to increase any stalling product size by 13 nts, accounting for the 3 bp overlap with the template. In our in vitro polymerase assay setup, when we remove CTP from the NTP mix but retain UTP, we still see the full-length product (Supplementary Figure S10). When we then also titrate UTP out of the NTP mix, we see the emergence of a product at the approximate size of the expected primed stalling product (Supplementary Figure S10). Together, this strongly suggests that the L protein can also incorporate UTP in place of CTP supporting our hypothesis that the early-stalling structures we observed are likely the result of nhUTP being misincorporated in place of CTP. To conclude, we obtained 3 structures of the SFTSV L protein stalled in actively replicating states: two structures stalled at position G4 of the 3′ cRNA (EARLY-ELONGATION and EARLY-ELONGATION-ENDO), and one structure stalled at position U21 of the 3′ cRNA (LATE-ELONGATION), which we refer to as early- and late-stage elongation states, respectively.
The L protein early elongation structures
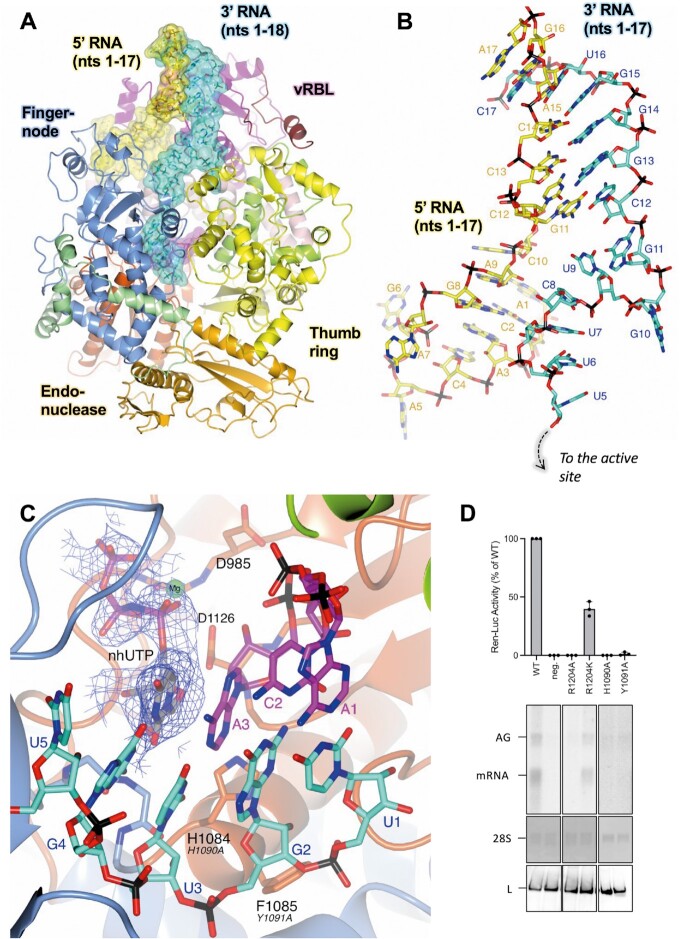
The two early elongation structures, denoted EARLY-ELONGATION and EARLY-ELONGATION-ENDO, demonstrate that when both 5′ and 3′ 1–20 nts cRNAs are present, the SFTSV L protein undergoes further remodelling, mostly involving residues in the thumb and thumb ring domains. These structural changes allow the L protein to accommodate the 5′/3′ distal duplex region, which is shown to curl around the vRBL, adopting a similar conformation to the full promoters observed for LACV (14) and LASV (28) (Figure 3A). However, there is a key difference in the number of canonical base-pair interactions between the 5′ and 3′ RNA strands. Unlike in other related bunyaviruses, the conserved 5′ and 3′ RNA termini of the SFTSV L segment are not particularly complementary with theoretically only 11 matches within the first 20 nts, including only 2 exact matches (nts 13 and 14) in the distal duplex region (nts 11–18) (Supplementary Figure S11). However, in the EARLY-ELONGATION and EARLY-ELONGATION-ENDO structures, we observe 7 bp interactions in the distal duplex. This is possible because the 5′ and 3′ RNA strands are shifted by 1 nt in respect to each other, meaning that G11 of the 5′ cRNA base-pairs with C12 of the 3′ cRNA, C12 of the 5′ cRNA base-pairs with G13 of the 3′ cRNA and so on (Figure 3B). In LACV, the 5′/3′ distal duplex is stabilized by 6 base-pair interactions (13), whereas, in LASV there are 8 base-pair interactions (28). This shift between the 3′ and 5′ RNA strands presumably enables the SFTSV L protein to overcome the general lack of complementarity in the 3′/5′ duplex region.
Figure 3.
The L protein at early-elongation. (A) The SFTSV L protein (EARLY-ELONGATION) is shown with key L protein domains coloured according to the following colour scheme: endonuclease (orange), endonuclease linker (light green), PA-C like domain (light pink), vRBL (magenta), fingers and finger node (blue), thumb (green), and thumb ring (yellow). RNAs are shown as sticks with surface overlaid (50% transparency) and coloured either yellow (5′ cRNA), cyan (3′ cRNA), or magenta (progeny RNA). (B) A close-up of the 5′/3′ distal duplex in the EARLY-ELONGATION structure is shown with RNA nts labelled starting from either the 5′ end for the 5′ cRNA or from the 3′ end for the 3′ cRNA. (C) The active site in the EARLY-ELONGATION structure is shown with the respective map overlaid. The map is clipped to the nhUTP and contoured to 4 σ. Residues from the SFTSV L protein are labelled and coloured according to the assigned domain (blue for the fingers domain, and dark orange for the palm domain) with the corresponding RVFV L protein residue shown in black italicized text underneath. (D) RVFV mini-replicon data for L protein with mutations in the vicinity of the active site presenting luciferase reporter activity (in standardized relative light units relative to the wild-type L protein (WT)). Data are presented as mean values ± SD of three biological replicates (n = 3). All biological replicates are shown as black dots (top panel). Middle panels present northern blotting results with signals for antigenomic viral RNA (AG), viral mRNA (mRNA) and 28 S ribosomal RNA (28 S) as a loading control, and the bottom panel shows Western blot detection of FLAG-tagged L proteins (L) to demonstrate general expressibility of the mutants. Uncropped original blots/gels are provided in the Supplementary Data.
In both the EARLY-ELONGATION and EARLY-ELONGATION-ENDO structures, we can reliably fit the 3′ cRNA residues found in the space between the distal duplex and the template entry channel. Notably, the G11 base stacks against the K533 sidechain whilst also interacting directly with the H535 sidechain (Supplementary Figure S12). There is a sharp turn which allows the G10 base to sit in a pocket delineated by W1342–K1347 and L1399–S1400 (Supplementary Figure S12). While, the bases U9, C8, and U7 can be fit, this region is by comparison to the rest of the 3′ cRNA less defined in both maps, which is most likely due to a low number of interactions with protein residues. The C8 base interacts with the S561 sidechain and the U7 backbone phosphate is coordinated by the K1401 sidechain (Supplementary Figure S12). The remaining template RNA nts 1–6 of the 3′ cRNA, are overall much better defined and can be fit with high confidence in both structures. Bases U6 and U5 are found in the template entrance channel and G4 at the +1 position in the active site (Supplementary Figure S12). The only close interaction between the L protein and U6 and U5 is between the R871 sidechain and the U5 phosphate. In addition to the stalling nhUTP molecule, the G4 nt is coordinated by the Y923 and Q1080 carbonyls. There is only one close interaction between the remaining 3′ cRNA and the L protein and this is between the G1081 carbonyl and the U3 ribose.
The stalling nhUTP contributes towards the coordination of a single Mg2+ ion, also found at the +1 position, with each of the nhUTP phospho-oxygens contributing to one coordinating bond (Figure 3C). The A986 carbonyl and sidechains of D985 and D1126 provide the three remaining coordination bonds required to complete the bipyramidal Mg2+ coordination shell. The volume for the 3 nt product strand in both early elongation structures is noisy but nevertheless good enough to fit the cognate bases for the template U1, G2 and U3 bases, which are A1, C2 and A3, respectively (Supplementary Figure S13). There are some interactions between the L protein and product RNA nts but these are limited. A3 is stabilized by interactions from the S1125 and D1126 sidechains, which interact with the ribose O2′ and O3′ hydroxyls. There is also a relatively long interaction between the A3 phosphate and the S1183 sidechain. G2 is sandwiched in between A1 and A3 while the G2 phosphate interacts with the R1197 sidechain of L. There are no close protein interactions stabilizing A1, which likely contributes to the poor map quality around this particular nt.
The L protein undergoes significant remodelling as elongation progresses
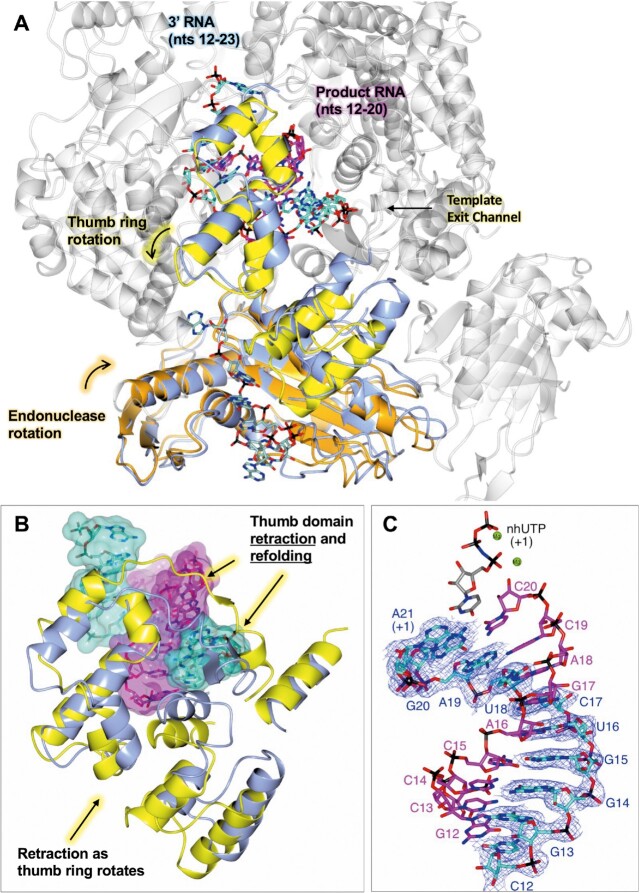
There are several noticeable changes to the L protein comparing pre-initiation to early-stage and then late-stage elongation. The most apparent being an expansion of the L protein core, which leads to an increase in the internal diameter of the L protein by ∼6 Å (Supplementary Figure S14). This opening up allows the L protein core to accommodate not just the small 3 nt product we observe in the EARLY-ELONGATION and EARLY-ELONGATION-ENDO structures but also the 10-bp product-template duplex we find in the LATE-ELONGATION structure. Additionally, in comparison to the apo structure there is a rotation of the endonuclease domain by ∼30° along the Z axis towards the L protein core (Figure 4A). This movement buries α4 of the endonuclease (residues 134–159, (24)) deeper within the L protein and, in doing so, partially opens the putative product exit channel in preparation for the later stages of elongation. It also provides space for the latter half of the thumb ring domain (roughly composed of L1509–K1577) to rotate away from the L protein core, backfilling the space left free by the movement of the endonuclease domain (Figure 4A and B). Finally, a beta-hairpin of the thumb domain (T1333–V1340) refolds and moves away from the L protein core (Figure 4B) unblocking the template exit channel.
Figure 4.
Conformational changes as the L protein proceeds from pre-initiation to late-stage elongation. (A) The SFTSV L protein (LATE-ELONGATION) is shown with the 5′ HOOK structure of the SFTSV L protein superposed by SSM in CCP4mg (51). Parts in grey are very similar in both structures and only the LATE-ELONGATION structure is depicted. Domains of the LATE-ELONGATION structure that were shown to either rotate or remodel as the L protein moves from pre-initiation, i.e. hook binding, to late elongation are coloured (orange for the endonuclease and yellow for the thumb ring domain). The corresponding 5′ HOOK structure domains are shown in blue. RNAs are shown as sticks with surface overlaid (50% transparency) and coloured either cyan (3′ cRNA), magenta (progeny RNA), or pale green (endonuclease-bound RNA). Movements to the endonuclease and thumb ring domains precipitated by the progression to late elongation are indicated. (B) A close-up of the remodelling which leads to the opening of the template exit channel. As in A, the thumb domain and RNA of the LATE-ELONGATION structure is coloured yellow and cyan/magenta, respectively. The apo structure is shown in blue. (C) The 10-bp product-template duplex including the non-hydrolysable UTP (nhUTP) from the LATE-ELONGATION structure are shown as sticks with the respective map overlaid. The map is clipped to the 3′ cRNA strand and contoured to 4 σ. Catalytically important magnesium ions (Mg) are shown as green spheres.
State of RNA at late-stage elongation
While in our early-stage elongation structures the 5′/3′ RNA distal duplex is maintained, in the later-stage LATE-ELONGATION structure, the distal duplex is disrupted completely. The SFTSV L protein is stalled through the incorporation of nhUTP at position A21 in the 3′ RNA template and the L protein inner cavity contains a 10-bp product-template duplex (Figure 4C). For related polymerases, such as LASV and LACV L proteins, the product-template duplex in the RdRp active site cavity was found to be 8 bp and 9–10 bp, respectively (13,28). The late-elongation map is of sufficient quality and resolution (2.6 Å) to unambiguously identify each base in the growing strand. This led to the interesting finding that instead of stalling at the first A in the 3′ cRNA sequence (position 19), the L protein instead stalls at position 21 on the template, which is the second A in the overall sequence and the first A in the 6A tail of the modified template used.
Product exit channel, template exit channel, and the 3′ secondary binding site
At the base of the SFTSV L protein core, the product-template duplex splits. This seems to be enabled partly by thumb-ring residue H1573, the sidechain of which protrudes directly into the path of the growing duplex. Unfortunately, the bottom of the SFTSV L protein core does not overlay nicely with the same space in the RVFV L protein. It was not possible therefore to explore the impact of mutating the equivalent of H1573 in the RVFV mini-replicon system. Additionally, recombinant expression of a D112A/H1573A SFTSV L protein mutant did not yield sufficient amounts of protein for in vitro testing. The growing strand then exits via the product exit channel, while the now used template exits via the template exit channel in the opposite direction. The putative product exit channel is open under apo/resting conditions and is formed of residues from the fingers (T839 and R835) and thumb ring domains (R1374, R1375 and R1526), as well as the endonuclease linker (E219 and E220) (30–33). The sidechains of these residues create a pore through which the product RNA can leave the L protein core (Supplementary Figure S15).
The template exit channel is formed by residues from the thumb ring domain. In the published apo structures (30–33), this template exit channel is blocked. In the EARLY-ELONGATION and EARLY-ELONGATION-ENDO structures, we observe that the blocking motifs either rotate (L1509–K1577) or refold (T1333–V1340) and thereby, partially clear the channel aperture in preparation for template exit. As the L protein moves from early to late-stage elongation, the L protein core opens up and the associated global movement creates sufficient space for the template 3′ RNA to exit the L protein core via the template exit channel.
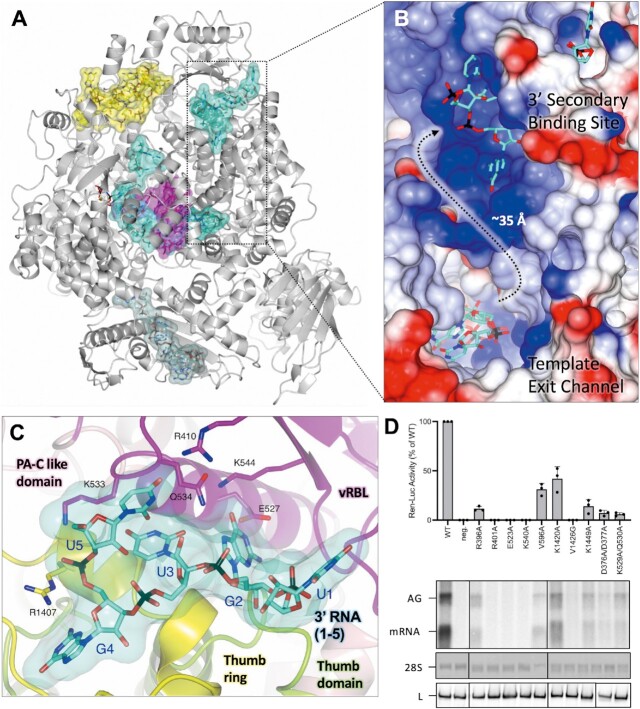
Several bunyaviruses and influenza viruses are known to coordinate 3′ RNA at secondary binding sites within the viral polymerase (13,28,29,53,55,56,57). In the LATE-STAGE ELONGATION structure reported here, we find nts 1–5 of the 3′ cRNA bound in a tunnel immediately adjacent to the vRBL (Figure 5A). This area broadly corresponds to the 3′ secondary binding sites observed in LASV (28) and LACV (13) L proteins. Unfortunately, in the polished late-elongation map, it is not possible to fit an unbroken chain of RNA between the 3′ cRNA in the L protein core and the RNA bound to the 3′ secondary binding site. However, analysis of the surface electrostatics in this area reveals a positively charged cleft connecting the template exit channel to the 3′ secondary binding site (Figure 5B). We hypothesized that the lack of volume connecting the 3′ RNA in the L protein core and the RNA bound to the 3′ secondary binding site might be due to over-sharpening of the map in this area.
Figure 5.
Interaction of 3′ cRNA at the 3′ secondary binding site. (A) The SFTSV L protein (LATE-ELONGATION) is shown with the protein depicted as ribbon in grey. RNAs are shown as sticks with surface overlaid (50% transparency) and coloured either yellow (5′ cRNA), cyan (3′ cRNA), magenta (progeny RNA), or pale green (endonuclease-bound RNA). (B) A close-up of the indicated area in A is shown as surface coloured by electrostatic potential demonstrating that the path connecting the template exit channel and 3′ secondary binding site has a largely positive charge. (C) The 3′ secondary binding site is shown with the L protein vRBL in magenta, the PA-C like domain in pink, thumb in green and thumb ring in yellow. The 3′ cRNA nts bound to the 3′ secondary binding site are shown as sticks with surface overlaid (50% transparency). Side chains of key interacting amino acids, coloured according to the protein domain, are shown as sticks and labelled accordingly. (D) RVFV mini-replicon data for L protein with mutations in the 3′ secondary binding site presenting luciferase reporter activity (in standardized relative light units relative to the wild-type L protein (WT)). Data are presented as mean values ± SD of three biological replicates (n = 3). All biological replicates are shown as black dots (top panel). Middle panels present Northern blotting results with signals for antigenomic viral RNA (AG), viral mRNA (mRNA) and 28 S ribosomal RNA (28 S) as a loading control, and the bottom panel shows Western blot detection of FLAG-tagged L proteins (L) to demonstrate general expressibility of the mutants. Uncropped original blots/gels are provided in the Supplementary Data.
To assess this, we produced a blurred late-elongation map and then re-examined this area of the L protein. In this blurred map, although the volume between the template exit channel and the 3′ secondary binding site is still rather amorphous, we clearly see the volume connecting the 3′ cRNA in the L protein core with the RNA bound to the 3′ secondary binding site (Supplementary Figure S16). Considering that the sequence of the RNA in the 3′ secondary binding site matches the first 5 nts of the 3′ cRNA used for this structure, and that the distance from the template exit channel to the secondary binding site (∼35 Å) fits with the estimated length of 6 nts of RNA that are missing in the 3′ cRNA chain, we concluded that the RNA bound to the 3′ secondary binding site is likely the tail-end of the 3′ cRNA stalled in the L protein core. This follows similar observations in other sNSV polymerases, including influenza virus polymerase complex, where after being copied, the template 3′ end rebinds to the polymerase in a secondary binding site analogous to the secondary binding site described here (53).
There are extensive protein–RNA interactions at the 3′ secondary binding site which stabilize the bound RNA (Figure 5C). The first nt fit into the map at this site corresponds to U1 in the 3′ RNA sequence and is found on the extremity of the L protein surrounded by several flexible sidechains (D383, K386 and E387), however, none of these form close interactions with the U1 base. This is not too surprising as we observed something similar for the LASV L protein (28). Nucleotide G2 sits in a pocket delineated by sidechains of Y408, E527, K544, L1254, P1256, I1344 and V1413. The E527, K544 and Q534 sidechains contribute to the coordination bonds and further stabilize the G2 base. The base of the third nt, U3, interacts with the Q534 sidechain while the U3 phosphate interacts with the R1418 sidechain. The base of G4 is coordinated by the carbonyl of Y1341 and the Y1414 sidechain. The base of the fifth and final fit nt, U5, is sandwiched between the K405 sidechain on one side and the K533-Q534 sidechains on the other. There is also a direct interaction between the R410 sidechain and the base O4′ carbonyl.
Mutational analysis of residues at the 3′ secondary binding site
To test for the functional relevance of the secondary 3′ RNA binding site, we first compared the LATE-ELONGATION structure to the RVFV L and found that several of the key interacting residues are highly conserved (Supplementary Figure S17). We then mutated these key residues and tested 8 single and 2 double mutants in the RVFV mini-replicon system (Supplementary Table S3, Figure 5D). The majority of the mutations resulted in a loss of L protein activity of more than 90%, including: R401A (SFTSV: R410), E523A (SFTSV: E527), K540A (SFTSV: K544), V1426G (SFTSV: V1413), as well as the double-mutants D376A/D377A (SFTSV: S382/D383) and K529A/Q530A (SFTSV: K533/Q534). The remaining mutations resulted in a significantly reduced but not totally abolished L protein activity (between 10 and 42% activity remaining), including R396A (SFTSV: K405), V596A (SFTSV: K599), K1420A (SFTSV: R1407) and K1449A (SFTSV: K1435). This demonstrates that 3′ RNA binding to the secondary binding site is essential for both genome replication and transcription, as also demonstrated for LASV L protein (28).
Initiation and priming of genome replication by the SFTSV L protein
To test how the SFTSV L protein could initiate replication, we incubated the L protein with 5′ cRNA, 3′ cRNA and a short 2 nt primer (AC) in the absence of any NTPs. Given that the AC primer could only base-pair with either one of the two UG repeats at the 3′ end of the 3′ cRNA, we had hoped this might allow us to visualize a priming loop (as seen in influenza virus) (58) or prime-and-realign loop (as reported for LACV) (13). Processing of this dataset gave one major class, which we have named a RESTING state structure. Here, the 5′ cRNA hook is bound in the normal manner nestled betwixt the vRBL, PA-C like domain and fingers, while the 3′ cRNA is bound to the 3′ secondary binding site. Whereas in the early-elongation structures in which the 3′ RNA clearly feeds into the L protein core, in the RESTING structure, the distal duplex is absent. The RESTING structure is reminiscent therefore of the LASV 3END-CORE (PDB: 7OEA) (28) structure in which excess 3′ vRNA binds to the 3′ secondary binding site instead of entering the L protein core. Although we clearly see 3′ cRNA binding to the 3′ secondary binding site, we also found that we could fit 4 nts of the template 3′ cRNA in the L protein core, however, only 2 of these (U3 and G4) could be fit with confidence. The region of the map where these 3′ RNA nts are found is rather noisy but we think that similar to our early-elongation structures, the nts in the L protein core are likely the start of the 3′ cRNA. Although we could not visualize initiation, given that the 3′ cRNA repeatedly appears to reach at least 4 nts into the L protein core, and that we detect an RNA product corresponding exactly to the terminus of the 3′ template without additional or missing nts in our biochemical assays (17), we think that for vRNA synthesis SFTSV primes internally and then realigns this primer to the 3′ template extremity before proceeding with replication. How this works for transcription remains unclear and needs further investigation.
Insights into the role and activity of the SFTSV L protein endonuclease
Inspection of our LATE-ELONGATION structure revealed, quite unexpectedly, RNA bound across the SFTSV L protein endonuclease (Supplementary Figure S18A). Analysis of the surface electrostatics in this area demonstrates that the RNA is bound across a positively charged cleft that includes the metal-coordination site and active site (Supplementary Figure S18B). Here, we can fit a ∼7 nt RNA bound across the SFTSV L protein endonuclease domain. It is important to note that, in these experiments, the L protein used should be endonuclease inactive due to the mutation D112A in the active site (17,24). This is presumably why we observe uncleaved RNA. The volume for the endonuclease-bound RNA is of variable quality and it is difficult to determine base identity. However, we could fit the following sequence: 5′-ACACAGA-3′ and propose that the RNA bound to the endonuclease is likely an excess of 5′ cRNA. Indeed, in our cryo-EM experiments the 5′ cRNA is used at a 2-fold molar excess compared to the L protein.
There are no close interactions between the first two nts, A1 and C2, and the endonuclease or other parts of the L protein. The A3 phosphate forms an interaction with the R1532 sidechain of the thumb ring domain. The next two nts, C4 and A5, sit across two pockets which form the endonuclease metal-coordination site. In influenza virus endonuclease, these are known as the P1 and P0 pockets, respectively (59). Interestingly, despite mutation of one of the metal-coordinators (D112A), the map suggests there is a single Mg2+ ion coordinated by E126 and the A5 phosphate. While there are no close interactions between the C4 and A5 bases, the C4 phosphate interacts with the sidechain of Y149, as well as the main chain carbonyl, amide, and sidechain of T110. In addition to coordinating the bound Mg2+ ion, the A5 phosphate forms a long interaction with the E126 sidechain. The next nt, G6, sits in a pocket equivalent of the P1 pocket in influenza viruses (59). The G6 base is then sandwiched between the K1558 sidechain and adjacent A5 base. The G6 phosphate meanwhile interacts with the amide and sidechain of T129. There is then a sharp turn which sees the base of the next nt A7 sandwiched between the sidechains of R131 and R832. There is also an interaction between the A7 phosphate and the R131 sidechain and between the A7 ribose ring-oxygen and the amide and sidechain of S132. At this point, the quality of the map degrades to such an extent that the fitting of further nts to determine the RNA sequence is no longer possible.
DISCUSSION
The structural and functional data presented here provide insights into how the SFTSV L protein replicates the viral genome (Figure 6). First, in the pre-initiation state, compared to the apo conformation there is a relaxation of the vRBL and elements of the PA-C like domain revealing a positively charged cleft into which the 5′ RNA binds in a hook-like conformation. This is followed by the formation of a distal duplex structure between the 5′ and 3′ RNA which orients and positions the template RNA 3′ terminus such that it can feed into the L protein core and be positioned in the RdRp active site in preparation for initiation. Previously, we had proposed two alternative mechanisms for de novo initiation of genome replication by the SFTSV L protein (17). Either the L protein initiates terminally at nt 1 of the template and then directly proceeds to elongation or the L protein initiates internally at position 3 of the template generating either an AC or ACA primer, which is then realigned to positions 1 and 2 or 1–3 of the template RNA before elongation (17). In both cases, the end result is a blunt-end product and so it is difficult to discriminate between the two mechanisms biochemically. Furthermore, a prime-and-realign scenario has already been detected for several other sNSVs such as influenza viruses, peribunyaviruses and arenaviruses (13,15,60,61,62,63,64).
Figure 6.
Overview of the SFTSV L protein at resting, pre-initiation, early-elongation, and late-elongation stages. A cartoon representation of each SFTSV L protein structure is shown in grey with the endonuclease and endonuclease-linker highlighted in orange and green, respectively. RNAs are shown as surface and coloured either yellow (5′ cRNA), cyan (3′ cRNA), magenta (progeny RNA), or pale green (endonuclease-bound RNA). In addition, the RNAs present in each structure are shown schematically and labelled.
Notably, in the early elongation structures, we found the 3′ cRNA positioned at least 4 nts into the RdRp active site suggesting an internal initiation scenario requiring subsequent realignment. Given the lack of an initiation stage structure, it is however not possible to reliably identify a potential priming or prime-and-realign loop which, in related RdRps, is stabilizing the first nucleotide and directing realignment. The prime-and-realign loop in LACV was suggested to work initially by stabilizing the 3′ vRNA in a position which allows an AGU triplet to form at positions 4–6 on the 3′ vRNA (13). By releasing the tension built up upon template progression, the prime-and-realign loop then supposedly pushes the 3′ vRNA backwards whilst the AGU triplet remains in the active site, thereby repositioning the AGU triplet to positions 1 to 3 on the 3′ vRNA (13). Despite the lack of direct structural evidence, we made several observations that could be important for the SFTSV de novo initiation mechanism of genome replication. Firstly, we note that in our early-stage elongation structures, there is a bunching of the 3′ RNA at the template entrance channel with nts U6, U7 and C8 forming a kink. Secondly, G10 of the 3′ RNA is anchored in a pocket delineated by residues W1342 to K1347 as well as L1399 to S1400. Thirdly, the distal duplex stays intact even as the template reaches 4 nts into the active site. Finally, while the vast majority of amino acid residues in the L protein core overlay nearly perfectly in each of our structures (accounting for the general opening up of the inner cavity itself), we note that the sidechain of R1197 adopts one of two conformations. In fact, it is pointing either upward into the L protein core (pre-initiation) or laying flush with the growing RNA duplex (elongation). Although this difference is very subtle, the R1197 is of particular interest as it sits around 4–5 nts into the L protein core. Moreover, we know from our mutational analysis that mutating the equivalent of SFTSV L R1197 in the RVFV L protein (R1204) to a K reduces the L protein activity by over half and mutating it to an A completely abolishes L protein activity.
Based on the above, it seems possible that, at initiation, the R1197 sidechain hinders progression of the internally initiated product-template duplex while the distal duplex region of the promoter is still intact. Tension created in the template strand by the attempted template progression could lead to the template slipping backwards in the active site leading to the compressed RNA formation (nts 6–8) we see in our early elongation structures and thereby realignment of the primer to the 3′ terminus–in effect, a ‘spring-loaded’ prime-and-realign mechanism. This would result in the blunt-end product that we detect in our biochemical assays and also explain the lack of an obvious priming loop or prime-and-realign loop as seen in other polymerases. The blunt-ended template-product duplex might then be able to push the R1197 side chain aside upon strand progression without any further re-alignment. Of course, this proposed mechanism is speculative and the possibility remains that a potentially very flexible priming or prime-and-realign loop exists in a similar fashion to that seen in either influenza virus or LACV (13,58).
As the L protein moves from early-elongation to late-stage elongation we find that the 5′/3′ distal duplex structure dissolves. At late-stage elongation, progeny RNA is released via a channel adjacent to the endonuclease domain, whereas spent template RNA is fed across the surface of the L protein in the direction of a 3′ secondary binding site. In the late-elongation structure, the RNA fit in the 3′ secondary binding site is 5′-UGUGU-3′, which corresponds to the first 5 nts of the 3′ RNA. We expect that the 3′ RNA terminus binds to the 3′ secondary binding site with the remaining spent 3′ RNA then free to bulge out from the L protein in a similar fashion to that proposed for influenza virus (53). In the cell, this RNA would presumably then be encapsidated by the viral N protein maintaining the viral ribonucleoprotein complex. However, structural data on this process is currently missing. As observed for LASV L protein (28), binding of the 3′ vRNA to the secondary binding site and 5′ hook binding is not mutually exclusive. Likewise, we could not detect any major changes in the 5′ hook binding site upon 3′ cRNA binding to the secondary binding site, leaving it unclear what triggers the release of the 5′ terminus at late elongation to achieve complete genome replication. Looking at the resting state structure it is conceivable that binding of the 3′ end to the secondary binding site and re-binding of the 5′ hook after passing through the RdRp active site supports the distal duplex formation, which then serves to guide the 3′ template terminus into the active site leading to the re-formation of a pre-initiation complex. However, further work looking at this recycling episode of the genome replication process is needed before any solid conclusions can be made.
Endonuclease domains have now been identified in many different bunyavirus L proteins and in each of these, the endonuclease is located at the N-terminus of the L protein (12,19,21,22,23,25,26,27,65,66). While we can fit the SFTSV endonuclease into each of our structures published here, in the late-elongation structure we can also visualize RNA bound across the endonuclease active site making it the first structure of a bunyaviral endonuclease with bound RNA substrate. This allows us to explore how the SFTSV endonuclease engages with substrate RNA. It has been hypothesized from biochemical studies that the phenuivirus endonuclease prefers purine-rich RNA species as substrates (21). However, we could observe only one base-specific interaction between the hydroxyl group of Y76 and a C base right in the centre of the bound RNA. At this position the protein and RNA seem too close to accommodate a G base, whereas an A could in theory fit (Supplementary Figure S18C). In addition, Y76 appears to be stabilized in its position by contacts to neighbouring side chains which form part of the endonuclease-linker arm. All other protein-RNA contacts in the late-elongation structure were solely formed between the protein and the RNA backbone. Therefore, while we cannot find structural evidence for a preference of purine-rich substrates, we do think that the late-elongation structure suggests a disadvantage for G-rich RNA substrates. In the endonuclease assays described by Jones et al., the GA-rich RNA substrate mainly consisted of A bases, which would not be contradictory to our observations (21). Furthermore, as the length of the bound RNA in our structure is rather short (only 7 nts are fit), we cannot exclude that there may be further protein-RNA contacts with longer RNAs leading to the detected lower cleavage efficiency of pyrimidine-rich RNA substrates compared to purine-rich RNA substrates.
In addition to sequence specificity, regulation of the SFTSV endonuclease is also of particular interest not least because the endonuclease domain is thought to be central to the bunyaviruses ability to snatch capped-RNA fragments from host cells. For LASV, we found that the endonuclease is autoinhibited by two alternative mechanisms: either an inhibitory peptide or the C-terminal helix of the endonuclease domain blocks the endonuclease active site (28). It has been proposed that the C-terminal α-helix of the SFTSV endonuclease (residues 211–233) could be responsible for controlling substrate access to the endonuclease active site (Supplementary Figure S19A) (24). However, in each of our cryo-EM structures, we find that at around 2-turns into this particular α-helix (∼L214), the helical arrangement unfolds, the protein chain bends by ∼90° and subsequently forms the EN linker (Supplementary Figure S19B). The exception to this observation is the early-elongation-endo structure. In this structure the helix still unfolds around the same place after around 2-turns, but instead of veering away from the endonuclease active site, the strand proceeds for an additional 4–5 amino acids (Supplementary Figure S19C), which sit on top of the endonuclease active site. In a superposition of the endonuclease-bound RNA in the late-elongation structure and the early-elongation-endo structure, the described 4–5 additional residues clash with the endonuclease-bound RNA (Supplementary Figure S19D). Thus, it would seem that in the raised endonuclease conformation access to the endonuclease active site is precluded. The early-elongation-endo structure therefore likely demonstrates one mechanism of endonuclease regulation in SFTSV, which is similar to what has been observed for LASV (28) and compatible with the mechanism proposed by Wang et al. (24). However, as in LASV, there may be further mechanisms of inhibition in SFTSV L we have yet to discover.
Another interesting observation is the apparent ability of the SFTSV RdRp to misincorporate nucleotides when access to particular NTPs was restricted. A relatively low fidelity is a common feature of sNSV RNA polymerases but we also recognize that in vitro assay conditions may well produce artifacts. In cells, levels of all NTPs are not equal. For example, CTP and GTP are present at relatively low concentrations (≤0.5 mM) when compared to UTP (∼1 mM) (67,68). The level of ATP is coupled to the metabolic status of the cell and is normally >2 mM (67,68). The intracellular NTP concentrations are therefore even higher than those used in our in vitro polymerase assay (∼0.2 mM) and may positively influence L protein fidelity. The mononegavirus respiratory syncytial virus (RSV) polymerase is capable of producing new RNA either de novo or via primer extension (69). However, in in vitro biochemical assays, it was found that the ratio of de novo vs primer extension RNA synthesis is influenced by NTP concentration reinforcing this idea that both the concentration of NTPs and the ratio of individual NTPs are important for RNA synthesis (69). Indeed, one suggested mechanism of action of the antiviral ribavirin is described to work by competitively inhibiting the cellular enzyme inosine 5′-monophosphate dehydrogenase, which ultimately leads to GTP depletion (70). In addition, at high concentrations (≥100 μM) ribavirin has been shown to be incorporated as a GTP analogue into nascent viral RNA acting as a mutagen in following rounds of genome replication and transcription (71,72). Further factors known to influence the fidelity of polymerases are the availability and concentration of divalent metal ions with Mn2+ leading to higher rates of misincorporation compared to Mg2+ (73). Our results on the misincorporation of CTP for UTP and vice versa under our assay conditions emphasize that NTP concentrations and relative ratios are important to consider for in vitro drug testing against NSV RdRp targets. Although out of scope for this study, we think it would be particularly interesting to investigate whether bunyaviral infections have an influence on the intracellular NTP pools as significant changes in dNTP levels have recently been observed in bacteria in response to bacteriophage infection (74). Related to this topic, the low fidelity of the RdRp somewhat limits the confidence in the RNA product modelled in our late-elongation structure. In the corresponding in vitro reaction, we detect two products, one most likely with a misincorporated CTP instead of UTP at position 19 of the product as we used nhUTP (>95% purity). Therefore, although we modelled the 20mer product with the described misincorporation, the cryo-EM data are most likely a mixture of both the 18mer and the 20mer. Interestingly, the mismatch does not result in a distortion of the RNA duplex, a scenario which has been described in the literature both for DNA and RNA suggesting that the rare tautomers giving rise to these mismatches are in fact relatively widespread in nucleic acids (75,76).
To conclude, the biochemical and structural data presented here has provided us with key insights into how genome replication is catalysed by the SFTSV L protein. We have shown that there are specific changes to the L protein that occur upon 5′ RNA hook binding and that several L protein domains adapt further upon addition of 3′ RNA to accommodate the 5′/3′ RNA distal duplex. Our data show that as the L protein moves from pre-initiation to late-stage elongation, the L protein undergoes further remodelling with the inner cavity of the L protein opening up to accommodate the 10-bp product-template duplex. Similar to other sNSV’s, we demonstrate that the SFTSV L protein has a functional 3′ secondary binding site, which is occupied under both resting conditions and also during the later stages of elongation. This structural data is accompanied by a comprehensive mutational analysis allowing us to identify key amino acid residues involved in binding RNA at the 5′ RNA hook binding site and 3′ secondary binding site. Altogether, this data provides an excellent foundation up on which further questions can be addressed including how the SFTSV L protein catalyses viral transcription.
DATA AVAILABILITY
Coordinates and structure factors or maps included in this paper have been deposited in the Worldwide Protein Data Bank (wwPDB) and the Electron Microscopy Data Bank (EMDB) with the following accession codes: SFTSV L protein bound to 5′ promoter RNA [5′ HOOK] EMD-15607 PDB-8AS6; SFTSV L protein early elongation structure [EARLY-ELONGATION] EMD-15608 PDB-8AS7; SFTSV L protein early elongation raised endonuclease structure [EARLY-ELONGATION-ENDO] EMD-15610 PDB-8ASB; SFTSV L protein late elongation structure [LATE-ELONGATION] EMD-15614 PDB-8ASD; SFTSV L protein resting structure [RESTING] EMD-15615 PDB-8ASG.
Supplementary Material
ACKNOWLEDGEMENTS
We want to thank Stephan Günther and Tomas Kouba for support and helpful discussions throughout the project. We thank Imre Berger for providing the DH10EMBaY E. coli and the team of the Eukaryotic Expression Facility (EEF) at European Molecular Biology Laboratory (EMBL) Grenoble for support and advice. We also acknowledge support by Carolin Seuring, Cornelia Cazey and Ulrike Laugks for access to the Cryo-EM multi-user facility at CSSB and providing time for sample preparation, screening, and data collection; Wolfgang Lugmayr for help and support in using the CSSB partition on the DESY computer cluster for cryo-EM data processing. We also want to thank Benoît Arragain for feedback on the manuscript.
Notes
Present address: Emmanuelle R.J. Quemin. Department of Virology, Institute for Integrative Biology of the Cell (I2BC), Centre National de la Recherche Scientifique (CNRS) UMR9198, Gif-sur-Yvette, France.
Contributor Information
Harry M Williams, Bernhard Nocht Institute for Tropical Medicine (BNITM), Hamburg, Germany.
Sigurdur R Thorkelsson, Centre for Structural Systems Biology, Leibniz Institute of Virology, Hamburg, Germany.
Dominik Vogel, Bernhard Nocht Institute for Tropical Medicine (BNITM), Hamburg, Germany.
Morlin Milewski, Bernhard Nocht Institute for Tropical Medicine (BNITM), Hamburg, Germany.
Carola Busch, Bernhard Nocht Institute for Tropical Medicine (BNITM), Hamburg, Germany.
Stephen Cusack, European Molecular Biology Laboratory, Grenoble, France.
Kay Grünewald, Centre for Structural Systems Biology, Leibniz Institute of Virology, Hamburg, Germany; University of Hamburg, Hamburg, Germany.
Emmanuelle R J Quemin, Centre for Structural Systems Biology, Leibniz Institute of Virology, Hamburg, Germany.
Maria Rosenthal, Bernhard Nocht Institute for Tropical Medicine (BNITM), Hamburg, Germany; Fraunhofer Institute for Translational Medicine and Pharmacology (ITMP), Discovery Research ScreeningPort, Hamburg, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Leibniz Association's Leibniz competition programme [K72/2017]; part of this work was performed at the Cryo-EM multi-user Facility at CSSB, headed by K.G. and supported by the UHH and DFG [INST 152/772-1, 774-1, 775-1, 776-1]; S.T. benefited from a travel grant from the Leibniz Institute for Virology; E.Q. was supported by an individual fellowship from the Alexander von Humboldt Foundation and a Klaus Tschira Boost Fund; M.R. received funding from the German Federal Ministry of Education and Research [01KI2019]. Funding for open access charge: Leibniz Association, institutional funding.
Conflict of interest statement. None declared.
REFERENCES
- 1. Casel M.A., Park S.J., Choi Y.K.. Severe fever with thrombocytopenia syndrome virus: emerging novel phlebovirus and their control strategy. Exp. Mol. Med. 2021; 53:713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yu X.J., Liang M.F., Zhang S.Y., Liu Y., Li J.D., Sun Y.L., Zhang L., Zhang Q.F., Popov V.L., Li C.et al.. Fever with thrombocytopenia associated with a novel bunyavirus in China. N. Engl. J. Med. 2011; 364:1523–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Takahashi T., Maeda K., Suzuki T., Ishido A., Shigeoka T., Tominaga T., Kamei T., Honda M., Ninomiya D., Sakai T.et al.. The first identification and retrospective study of Severe Fever with Thrombocytopenia Syndrome in Japan. J. Infect. Dis. 2014; 209:816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim K.H., Yi J., Kim G., Choi S.J., Jun K.I., Kim N.H., Choe P.G., Kim N.J., Lee J.K., Oh M.D.. Severe fever with thrombocytopenia syndrome, South Korea, 2012. Emerg. Infect. Dis. 2013; 19:1892–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tran X.C., Yun Y., Van An L., Kim S.H., Thao N.T.P., Man P.K.C., Yoo J.R., Heo S.T., Cho N.H., Lee K.H.. Endemic severe fever with thrombocytopenia Syndrome, Vietnam. Emerg. Infect. Dis. 2019; 25:1029–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lin T.L., Ou S.C., Maeda K., Shimoda H., Chan J.P., Tu W.C., Hsu W.L., Chou C.C.. The first discovery of severe fever with thrombocytopenia syndrome virus in Taiwan. Emerg. Microbes Infect. 2020; 9:148–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen C., Li P., Li K.F., Wang H.L., Dai Y.X., Cheng X., Yan J.B.. Animals as amplification hosts in the spread of severe fever with thrombocytopenia syndrome virus: a systematic review and meta-analysis. Int. J. Infect. Dis. 2019; 79:77–84. [DOI] [PubMed] [Google Scholar]
- 8. Jin C., Jiang H., Liang M., Han Y., Gu W., Zhang F., Zhu H., Wu W., Chen T., Li C.et al.. SFTS virus infection in nonhuman primates. J. Infect. Dis. 2015; 211:915–925. [DOI] [PubMed] [Google Scholar]
- 9. Niu G., Li J., Liang M., Jiang X., Jiang M., Yin H., Wang Z., Li C., Zhang Q., Jin C.et al.. Severe fever with thrombocytopenia syndrome virus among domesticated animals, China. Emerg. Infect. Dis. 2013; 19:756–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu Y., Li Q., Hu W., Wu J., Wang Y., Mei L., Walker D.H., Ren J., Wang Y., Yu X.J.. Person-to-person transmission of severe fever with thrombocytopenia syndrome virus. Vector Borne Zoonotic Dis. 2012; 12:156–160. [DOI] [PubMed] [Google Scholar]
- 11. Koga S., Takazono T., Ando T., Hayasaka D., Tashiro M., Saijo T., Kurihara S., Sekino M., Yamamoto K., Imamura Y.et al.. Severe fever with Thrombocytopenia Syndrome Virus RNA in Semen, Japan. Emerg. Infect. Dis. 2019; 25:2127–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reguera J., Gerlach P., Rosenthal M., Gaudon S., Coscia F., Gunther S., Cusack S.. Comparative structural and functional analysis of bunyavirus and arenavirus cap-snatching endonucleases. PLoS Pathog. 2016; 12:e1005636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arragain B., Durieux Trouilleton Q., Baudin F., Provaznik J., Azevedo N., Cusack S., Schoehn G., Malet H.. Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription. Nat. Commun. 2022; 13:902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arragain B., Effantin G., Gerlach P., Reguera J., Schoehn G., Cusack S., Malet H.. Pre-initiation and elongation structures of full-length La Crosse virus polymerase reveal functionally important conformational changes. Nat. Commun. 2020; 11:3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vogel D., Rosenthal M., Gogrefe N., Reindl S., Gunther S.. Biochemical characterization of the Lassa virus L protein. J. Biol. Chem. 2019; 294:8088–8100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oymans J., Te Velthuis A.J.W.. A mechanism for priming and realignment during influenza A virus replication. J. Virol. 2018; 92:e01773-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vogel D., Thorkelsson S.R., Quemin E.R.J., Meier K., Kouba T., Gogrefe N., Busch C., Reindl S., Gunther S., Cusack S.et al.. Structural and functional characterization of the severe fever with thrombocytopenia syndrome virus L protein. Nucleic Acids Res. 2020; 48:5749–5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Olschewski S., Cusack S., Rosenthal M.. The cap-Snatching mechanism of Bunyaviruses. Trends Microbiol. 2020; 28:293–303. [DOI] [PubMed] [Google Scholar]
- 19. Fernandez-Garcia Y., Reguera J., Busch C., Witte G., Sanchez-Ramos O., Betzel C., Cusack S., Gunther S., Reindl S.. Atomic structure and biochemical characterization of an RNA endonuclease in the N terminus of Andes virus L protein. PLoS Pathog. 2016; 12:e1005635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gogrefe N., Reindl S., Gunther S., Rosenthal M.. Structure of a functional cap-binding domain in Rift Valley fever virus L protein. PLoS Pathog. 2019; 15:e1007829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jones R., Lessoued S., Meier K., Devignot S., Barata-Garcia S., Mate M., Bragagnolo G., Weber F., Rosenthal M., Reguera J.. Structure and function of the Toscana virus cap-snatching endonuclease. Nucleic Acids Res. 2019; 47:10914–10930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morin B., Coutard B., Lelke M., Ferron F., Kerber R., Jamal S., Frangeul A., Baronti C., Charrel R., de Lamballerie X.et al.. The N-terminal domain of the arenavirus L protein is an RNA endonuclease essential in mRNA transcription. PLoS Pathog. 2010; 6:e1001038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wallat G.D., Huang Q., Wang W., Dong H., Ly H., Liang Y., Dong C.. High-resolution structure of the N-terminal endonuclease domain of the Lassa virus L polymerase in complex with magnesium ions. PLoS One. 2014; 9:e87577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang W., Shin W.J., Zhang B., Choi Y., Yoo J.S., Zimmerman M.I., Frederick T.E., Bowman G.R., Gross M.L., Leung D.W.et al.. The cap-Snatching SFTSV endonuclease domain is an antiviral target. Cell Rep. 2020; 30:153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao S., Xu G., He G., Peng Y., Liang C.. Characterization of an endonuclease in rice stripe tenuivirus Pc1 in vitro. Virus Res. 2019; 260:33–37. [DOI] [PubMed] [Google Scholar]
- 26. Reguera J., Weber F., Cusack S.. Bunyaviridae RNA polymerases (L-Protein) have an N-Terminal, influenza-Like endonuclease domain, essential for viral cap-Dependent transcription. PLoS Pathog. 2010; 6:e1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rosenthal M., Gogrefe N., Vogel D., Reguera J., Rauschenberger B., Cusack S., Gunther S., Reindl S.. Structural insights into reptarenavirus cap-snatching machinery. PLoS Pathog. 2017; 13:e1006400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kouba T., Vogel D., Thorkelsson S.R., Quemin E.R.J., Williams H.M., Milewski M., Busch C., Gunther S., Grunewald K., Rosenthal M.et al.. Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity. Nat. Commun. 2021; 12:7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peng R.C., Xu X., Jing J.M., Wang M., Peng Q., Liu S., Wu Y., Bao X.C., Wang P.Y., Qi J.X.et al.. Structural insight into arenavirus replication machinery. Nature. 2020; 579:615–619. [DOI] [PubMed] [Google Scholar]
- 30. Cusack S., Rosenthal M.. Errors in the deposited SFTSV L protein structure. Nat Microbiol. 2021; 6:549–550. [DOI] [PubMed] [Google Scholar]
- 31. Wang P., Liu L., Liu A., Lou Z.. Reply to: errors in the deposited SFTSV L protein structure. Nat Microbiol. 2021; 6:551–552. [DOI] [PubMed] [Google Scholar]
- 32. Wang P., Liu L., Liu A., Yan L., He Y., Shen S., Hu M., Guo Y., Liu H., Liu C.et al.. Author correction: structure of severe fever with thrombocytopenia syndrome virus L protein elucidates the mechanisms of viral transcription initiation. Nat Microbiol. 2021; 6:697–698. [DOI] [PubMed] [Google Scholar]
- 33. Wang P., Liu L., Liu A., Yan L., He Y., Shen S., Hu M., Guo Y., Liu H., Liu C.et al.. Structure of severe fever with thrombocytopenia syndrome virus L protein elucidates the mechanisms of viral transcription initiation. Nat Microbiol. 2020; 5:864–871. [DOI] [PubMed] [Google Scholar]
- 34. Wang X., Hu C., Ye W., Wang J., Dong X., Xu J., Li X., Zhang M., Lu H., Zhang F.et al.. Structure of rift valley fever virus RNA-dependent RNA polymerase. J. Virol. 2022; 96:e0171321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Amroun A., Priet S., de Lamballerie X., Querat G.. Bunyaviridae RdRps: structure, motifs, and RNA synthesis machinery. Crit. Rev. Microbiol. 2017; 43:753–778. [DOI] [PubMed] [Google Scholar]
- 36. Jerome H., Rudolf M., Lelke M., Pahlmann M., Busch C., Bockholt S., Wurr S., Gunther S., Rosenthal M., Kerber R.. Rift Valley fever virus minigenome system for investigating the role of L protein residues in viral transcription and replication. J. Gen. Virol. 2019; 100:1093–1098. [DOI] [PubMed] [Google Scholar]
- 37. Zivanov J., Nakane T., Scheres S.H.W.. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ. 2020; 7:253–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kimanius D., Dong L., Sharov G., Nakane T., Scheres S.H.W.. New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochem. J. 2021; 478:4169–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A.. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017; 14:331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tegunov D., Cramer P.. Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods. 2019; 16:1146–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mindell J.A., Grigorieff N.. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 2003; 142:334–347. [DOI] [PubMed] [Google Scholar]
- 42. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 43. Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A.. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017; 14:290–296. [DOI] [PubMed] [Google Scholar]
- 44. Punjani A., Zhang H.W., Fleet D.J.. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods. 2020; 17:1214–1221. [DOI] [PubMed] [Google Scholar]
- 45. Burnley T., Palmer C.M., Winn M.. Recent developments in the CCP-EM software suite. Acta Crystallogr. D Struct. Biol. 2017; 73:469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sanchez-Garcia R., Gomez-Blanco J., Cuervo A., Carazo J.M., Sorzano C.O.S., Vargas J.. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 2021; 4:874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta. Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Afonine P.V., Poon B.K., Read R.J., Sobolev O.V., Terwilliger T.C., Urzhumtsev A., Adams P.D.. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 2018; 74:531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Afonine P.V., Klaholz B.P., Moriarty N.W., Poon B.K., Sobolev O.V., Terwilliger T.C., Adams P.D., Urzhumtsev A.. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D Struct. Biol. 2018; 74:814–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Williams C.J., Headd J.J., Moriarty N.W., Prisant M.G., Videau L.L., Deis L.N., Verma V., Keedy D.A., Hintze B.J., Chen V.B.et al.. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 2018; 27:293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McNicholas S., Potterton E., Wilson K.S., Noble M.E.. Presenting your structures: the CCP4mg molecular-graphics software. Acta. Crystallogr. D Biol. Crystallogr. 2011; 67:386–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pettersen E.F., Goddard T.D., Huang C.R.C., Meng E.E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E.. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021; 30:70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wandzik J.M., Kouba T., Karuppasamy M., Pflug A., Drncova P., Provaznik J., Azevedo N., Cusack S.. A structure-Based model for the complete transcription cycle of influenza polymerase. Cell. 2020; 181:877–893. [DOI] [PubMed] [Google Scholar]
- 54. Campagnola G., McDonald S., Beaucourt S., Vignuzzi M., Peersen O.B.. Structure-function relationships underlying the replication fidelity of viral RNA-dependent RNA polymerases. J. Virol. 2015; 89:275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fan H., Walker A.P., Carrique L., Keown J.R., Serna Martin I., Karia D., Sharps J., Hengrung N., Pardon E., Steyaert J.et al.. Structures of influenza A virus RNA polymerase offer insight into viral genome replication. Nature. 2019; 573:287–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gerlach P., Malet H., Cusack S., Reguera J.. Structural insights into bunyavirus replication and its regulation by the vRNA promoter. Cell. 2015; 161:1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Peng Q., Liu Y.Q., Peng R.C., Wang M., Yang W., Song H., Chen Y.H., Liu S., Han M., Zhang X.Z.et al.. Structural insight into RNA synthesis by influenza D polymerase. Nat. Microbiol. 2019; 4:1750–1759. [DOI] [PubMed] [Google Scholar]
- 58. te Velthuis A.J.W., Robb N.C., Kapanidis A.N., Fodor E.. The role of the priming loop in influenza A virus RNA synthesis. Nat Microbiol. 2016; 1:16029. [DOI] [PubMed] [Google Scholar]
- 59. Kumar G., Cuypers M., Webby R.R., Webb T.R., White S.W.. Structural insights into the substrate specificity of the endonuclease activity of the influenza virus cap-snatching mechanism. Nucleic Acids Res. 2021; 49:1609–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Deng T., Vreede F.T., Brownlee G.G.. Different de novo initiation strategies are used by influenza virus RNA polymerase on its cRNA and viral RNA promoters during viral RNA replication. J. Virol. 2006; 80:2337–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Garcin D., Kolakofsky D.. Tacaribe arenavirus RNA synthesis in vitro is primer dependent and suggests an unusual model for the initiation of genome replication. J. Virol. 1992; 66:1370–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Garcin D., Lezzi M., Dobbs M., Elliott R.M., Schmaljohn C., Kang C.Y., Kolakofsky D.. The 5′ ends of Hantaan virus (Bunyaviridae) rnas suggest a prime-and-realign mechanism for the initiation of RNA synthesis. J. Virol. 1995; 69:5754–5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Polyak S.J., Zheng S., Harnish D.G.. 5' Termini of Pichinde Arenavirus S-Rnas and Messenger-Rnas contain nontemplated nucleotides. J. Virol. 1995; 69:3211–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Prehaud C., Lopez N., Blok M.J., Obry V., Bouloy M.. Analysis of the 3' terminal sequence recognized by the Rift Valley fever virus transcription complex in its ambisense S segment. Virology. 1997; 227:189–197. [DOI] [PubMed] [Google Scholar]
- 65. Holm T., Kopicki J.D., Busch C., Olschewski S., Rosenthal M., Uetrecht C., Gunther S., Reindl S.. Biochemical and structural studies reveal differences and commonalities among cap-snatching endonucleases from segmented negative-strand RNA viruses. J. Biol. Chem. 2018; 293:19686–19698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kuang W., Zhang H., Cai Y., Zhang G., Deng F., Li H., Hu Z., Guo Y., Wang M., Zhou Y.et al.. Insights into two-Metal-Ion catalytic mechanism of cap-Snatching endonuclease of Ebinur Lake virus in Bunyavirales. J. Virol. 2022; 96:e0208521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kong Z., Jia S., Chabes A.L., Appelblad P., Lundmark R., Moritz T., Chabes A.. Simultaneous determination of ribonucleoside and deoxyribonucleoside triphosphates in biological samples by hydrophilic interaction liquid chromatography coupled with tandem mass spectrometry. Nucleic Acids Res. 2018; 46:e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Traut T.W. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994; 140:1–22. [DOI] [PubMed] [Google Scholar]
- 69. Noton S.L., Deflube L.R., Tremaglio C.Z., Fearns R.. The respiratory syncytial virus polymerase has multiple RNA synthesis activities at the promoter. PLoS Pathog. 2012; 8:e1002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Streeter D.G., Witkowski J.T., Khare G.P., Sidwell R.W., Bauer R.J., Robins R.K., Simon L.N.. Mechanism of action of 1- -D-ribofuranosyl-1,2,4-triazole-3-carboxamide (Virazole), a new broad-spectrum antiviral agent. Proc. Natl. Acad. Sci. U.S.A. 1973; 70:1174–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Crotty S., Maag D., Arnold J.J., Zhong W., Lau J.Y., Hong Z., Andino R., Cameron C.E.. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 2000; 6:1375–1379. [DOI] [PubMed] [Google Scholar]
- 72. Nystrom K., Waldenstrom J., Tang K.W., Lagging M.. Ribavirin: pharmacology, multiple modes of action and possible future perspectives. Future Virol. 2019; 14:153–160. [Google Scholar]
- 73. Dominguez O., Ruiz J.F., Lain de Lera T., Garcia-Diaz M., Gonzalez M.A., Kirchhoff T., Martinez A.C., Bernad A., Blanco L.. DNA polymerase mu (Pol mu), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J. 2000; 19:1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tal N., Millman A., Stokar-Avihail A., Fedorenko T., Leavitt A., Melamed S., Yirmiya E., Avraham C., Brandis A., Mehlman T.et al.. Bacteria deplete deoxynucleotides to defend against bacteriophage infection. Nat. Microbiol. 2022; 7:1200–1209. [DOI] [PubMed] [Google Scholar]
- 75. Kimsey I.J., Petzold K., Sathyamoorthy B., Stein Z.W., Al-Hashimi H.M.. Visualizing transient Watson-Crick-like mispairs in DNA and RNA duplexes. Nature. 2015; 519:315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bebenek K., Pedersen L.C., Kunkel T.A.. Replication infidelity via a mismatch with Watson-Crick geometry. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:1862–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors or maps included in this paper have been deposited in the Worldwide Protein Data Bank (wwPDB) and the Electron Microscopy Data Bank (EMDB) with the following accession codes: SFTSV L protein bound to 5′ promoter RNA [5′ HOOK] EMD-15607 PDB-8AS6; SFTSV L protein early elongation structure [EARLY-ELONGATION] EMD-15608 PDB-8AS7; SFTSV L protein early elongation raised endonuclease structure [EARLY-ELONGATION-ENDO] EMD-15610 PDB-8ASB; SFTSV L protein late elongation structure [LATE-ELONGATION] EMD-15614 PDB-8ASD; SFTSV L protein resting structure [RESTING] EMD-15615 PDB-8ASG.