Abstract
Purpose
The objective of this study was to analyze the microbial profile of individuals with peri-implantitis (PI) compared to those of periodontally healthy (PH) subjects and periodontitis (PT) subjects using Illumina sequencing.
Methods
Buccal, supragingival, and subgingival plaque samples were collected from 109 subjects (PH: 30, PT: 49, and PI: 30). The V3-V4 region of 16S rRNA was sequenced and analyzed to profile the plaque microbiota.
Results
Microbial community diversity in the PI group was higher than in the other groups, and the 3 groups showed significantly separated clusters in the buccal samples. The PI group showed different patterns of relative abundance from those in the PH and PT groups depending on the sampling site at both genus and phylum levels. In all samples, some bacterial species presented considerably higher relative abundances in the PI group than in the PH and PT groups, including Anaerotignum lactatifermentans, Bacteroides vulgatus, Faecalibacterium prausnitzii, Olsenella uli, Parasutterella excrementihominis, Prevotella buccae, Pseudoramibacter alactolyticus, Treponema parvum, and Slackia exigua. Network analysis identified that several well-known periodontal pathogens and newly recognized bacteria were closely correlated with each other.
Conclusions
The composition of the microbiota was considerably different in PI subjects compared to PH and PT subjects, and these results could shed light on the mechanisms involved in the development of PI.
Keywords: Dental plaque, High-throughput nucleotide sequencing, Microbiota, Peri-implantitis, Periodontitis
Graphical Abstract
INTRODUCTION
Advances in the surgical and prosthodontic techniques of dental implants have led to a survival rate of more than 96% in implants that function for a long period [1]. However, as many implants are placed in patients every year, the prevalence of peri-implant disease has gradually increased and has recently been reported to be 45% at the patient level [2]. Peri-implantitis (PI) is a progressive peri-implant disease that involves an inflammatory lesion in the peri-implant mucosa with additional loss of the supporting bone after initial bone remodeling [3].
Poor oral hygiene, a history of severe periodontitis (PT), and no regular maintenance care have been suggested as risk indicators for PI [4]. In particular, bacteria in the biofilm of teeth or implants produce toxins that cause tissue damage and further intensify the host’s inflammatory response, eventually leading to the destruction of the surrounding tissue [5,6]. Both PT and PI are inflammatory diseases associated with oral polymicrobial infection [7]. However, the characteristics of the tissues surrounding implants present distinct differences compared to that of the tooth, including an absence of the periodontal ligament, a poor vascular system, and the arrangement of the connective tissues [8]. Due to these differences, PI is likely to worsen and easily spread to supporting bone [8,9]. Understanding the characteristics of the microbiota associated with PI may help to develop therapeutic methods and effective prevention specific to PI.
The microbiota associated with biofilms surrounding healthy implants have been reported to be similar to those surrounding healthy teeth [10,11]. Studies published in the early 2000s found that the microbial composition associated with peri-implant disease was similar to that of chronic PT [10,12]. Most of these studies used conventional methods such as culture techniques, microscopy, and the Sanger method. Recently, next-generation sequencing (NGS) has been introduced. NGS can perform multiple reactions at the same time, carry out sequencing with a small amount of samples, and detect non-culturable organisms; it has also revolutionized sequencing in terms of the read length, accuracy, time, and cost [13]. NGS is used in various fields of dentistry, such as research on periodontal disease-related microorganisms [14], the analysis of mutations of cancer-related genes in oral squamous cell carcinoma [15], and the analysis of salivary microbiota related to Sjögren syndrome [16]. In particular, Illumina sequencing has been a widely used NGS platform. The advantages of this platform are a lower cost per sequence, a lower per-base error rate, and a greater number of reads with a higher average quality than that of other platforms [13].
The objective of this study was to analyze the microbial profile of subjects with PI compared to that of periodontally healthy (PH) subjects and subjects with PT using Illumina sequencing.
MATERIALS AND METHODS
Study population and clinical examination
Plaque samples were obtained from PH subjects (n=30), PT subjects (n=49), and PI subjects (n=30) at the Department of Periodontology, Pusan National University Dental Hospital, Yangsan, Republic of Korea. The sampling period was from March 2018 to August 2018. The inclusion criteria were as follows: (a) the presence of 20 or more teeth, (b) the absence of systemic diseases that may affect periodontal status as evidenced by the patient’s medical history, (c) no periodontal treatment within the last 3 months, and (d) no history of systemic antibiotic or anti-inflammatory drug use within the past 6 months. The criteria for exclusion were as follows: (a) pregnant or breastfeeding, (b) acute infection or chronic mucosal lesion of the oral cavity, and (c) heavy smoking (>20 cigarettes/day).
Full-mouth clinical examinations were carried out by 1 periodontist to characterize the periodontal and peri-implant status. These examinations evaluated the probing pocket depth (PPD), clinical attachment level (CAL), plaque index, and gingival index (GI). The diagnosis of diseases followed guidelines presented by the 2017 World Workshop on the Classification of Periodontal and Peri-implant Diseases and Conditions [17]. Periodontal health was diagnosed when subjects exhibited PPD ≤3 mm, bleeding on probing ≤10%, no clinical attachment loss, and no radiological bone loss. PT was diagnosed as follows: 1) interdental clinical attachment loss is detectable at ≥2 non-adjacent teeth, or 2) buccal of oral clinical attachment loss ≥3 mm with PPD >3 mm is detectable at ≥2 teeth. PI was diagnosed in subjects who had an implant with radiographic evidence of marginal bone loss ≥3 mm and/or PPD ≥6 mm in conjunction with profuse bleeding. The CAL of the implant was calculated as the distance from a fixed landmark (implant-abutment junction) to the bottom of the implant sulcus/pocket [18]. Each subject was assigned to only 1 group, and there were no diseased implants in either the PH or PT group. Written informed consent was obtained from all participants prior to conducting the study, and the experimental protocol was approved by the Institutional Review Board (IRB) of Pusan National University Dental Hospital (IRB No. PNUDH-2017-023).
Plaque sample collection
Plaque samples were obtained during the full-mouth periodontal examination. Buccal and supragingival samples were collected from the PH, PT, and PI subjects, while subgingival samples were only collected from the PT and PI subjects. The buccal samples were collected from the mucosa of both cheeks using a sterile micro-brush and were placed in a separate sterile 1.5-mL microcentrifuge tube. Before collecting the supragingival samples, the site was dried with a cotton roll to isolate the sample from any saliva or blood. The subgingival samples were acquired from the deepest periodontal pocket and peri-implant site using a sterile Gracey curette, after which the samples were placed in the tube mentioned above. All the samples were collected and stored at −80°C for subsequent processing.
Extraction of total genomic DNA
Total DNA extraction from the samples was performed using a Gram-positive DNA purification kit (Lucigen, Middleton, WI, USA) following the manufacturer’s instructions. The final concentration measurements were conducted with a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and the samples were stored at −80°C until use.
Polymerase chain reaction amplification and sequencing analysis
The V3-V4 region of the 16S ribosomal RNA gene was amplified by polymerase chain reaction, and the purified amplicons were pooled in an equimolar and paired-end sequence (2×300) on a MiSeq platform (Illumina, San Diego, CA, USA). The raw FastQ files were demultiplexed and processed using the tools available in QIIME2 (version 2019.7) [19]. The taxonomy classification was performed with a naïve Bayes machine-learning classifier using representative sequence sets for each dada2 sequence variant. The Human Oral Microbiome Database (HOMD, v15.2) [20] was used as the reference database.
Bioinformatic analysis, statistical analysis, and visualization
Several normality tests were performed for each set of data. The mean clinical parameters were compared using 1-way analysis of variance (ANOVA) followed by the post hoc Games-Howell test with P<0.05 as the statistical significance level. The operational taxonomic unit table was rarefied, and then alpha diversity was evaluated by 2 metrics: the species richness was estimated using the Chao1 index, and the Shannon index was used to estimate the evenness of the sample microbiota. The Mann-Whitney U test was performed to evaluate the significance of differences in the alpha diversity indices among the groups (P<0.05). The unweighted UniFrac distance was used to evaluate the similarities of microbial community composition among all samples, and the overall structure was visualized with a principal coordinate analysis (PCoA) plot. The non-parametric permutational ANOVA test was used to analyze group differences with 1,000 permutations [21]. The linear discriminant analysis effect size with default settings was used to identify taxa whose abundances were significantly different among the groups [22]. All the PCoA analyses, scatter diagrams, and box plots were conducted using R (R Foundation, Vienna, Austria).
Network analysis
To visualize interactions and characterize the microbial community, Sparse Correlations for Compositional Data (SparCC) [23] was used to calculate the Spearman correlation coefficient with the corresponding P value between each pair of genera. The network was then visualized using Cytoscape [24].
RESULTS
The clinical characteristics of the PH, PT, and PI subjects are listed in Table 1. Compared with the PH group, the mean values of PPD, CAL, GI, and the plaque index in the PT and PI groups showed significant differences (P<0.01). The mean values of both GI and the plaque index were significantly higher in the PT group than in the PI group (P<0.05).
Table 1. Characteristics of the subjects.
| Parameters | PH (n=30) | PT (n=49) | PI (n=30) | P value | |||
|---|---|---|---|---|---|---|---|
| PH-PT | PH-PI | PT-PI | |||||
| Patient age (yr) | 26.9±5.7 | 50.7±9.8 | 55.8±10.1 | ||||
| Sex | |||||||
| Male | 15 (50.0) | 36 (73.5) | 15 (50.0) | ||||
| Female | 15 (50.0) | 13 (26.5) | 15 (50.0) | ||||
| PPD (mm; average of all sites) | 2.4±0.04 | 3.5±0.8 | 3.2±0.4 | <0.01 | <0.01 | 0.19 | |
| PPD (mm; subgingival sampled site) | N/A | 7.7±3.1 | 7.8±2.7 | N/A | N/A | 0.98 | |
| CAL (mm) | 2.4±0.04 | 3.9±1.2 | 3.7±0.7 | <0.01 | <0.01 | 0.54 | |
| GI | 0.1±0.01 | 0.9±0.3 | 0.6±0.2 | <0.01 | <0.01 | <0.05 | |
| Plaque index | 0.1±0.01 | 0.5±0.06 | 0.4±0.03 | <0.01 | <0.01 | <0.01 | |
Values are presented as mean±standard deviation or number (%). P<0.05, analysis of variance with the post hoc Games-Howell test.
PPD: periodontal pocket depth, CAL: clinical attachment level, GI: gingival index, PH: periodontally healthy, PT: periodontitis, PI: peri-implantitis, N/A: not available.
Microbial community diversity
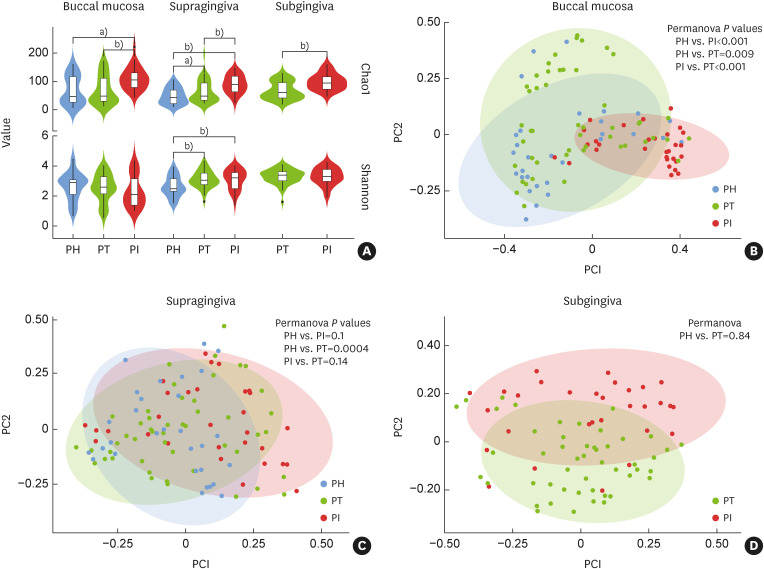
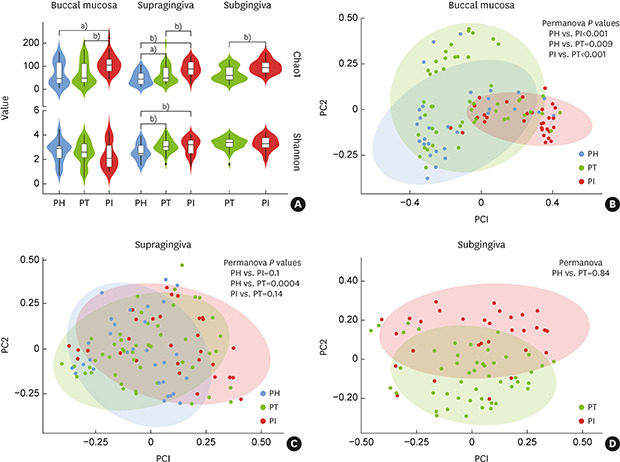
The Chao1 index showed that the species richness in all samples was significantly higher in the PI group than in the other groups (P<0.01). The Shannon index showed that the evenness of the sample microbiota in both the PI and PT groups was significantly higher than that in the PH groups in the supragingival samples (P<0.01), but no significant difference was found in the other samples (Figure 1A).
Figure 1. Alpha and beta diversities of microbial communities grouped by disease status and sampling sites. (A) Alpha diversity values in the buccal, supragingival, and subgingival samples were calculated by the Chao1 index and Shannon index. (B-D) Beta diversity values in the buccal, supragingival, and subgingival samples, respectively.
PH: periodontally healthy, PI: peri-implantitis, PT: periodontitis.
a)P<0.05; b)P<0.01.
Figure 1B showed a clear distinction and significant difference among the microbial clusters of the PI, PT, and PH groups in the buccal samples (P<0.01). In the supragingival and subgingival samples, the microbial clustering was not significant (Figure 1C and D).
Relative abundance of microbial composition
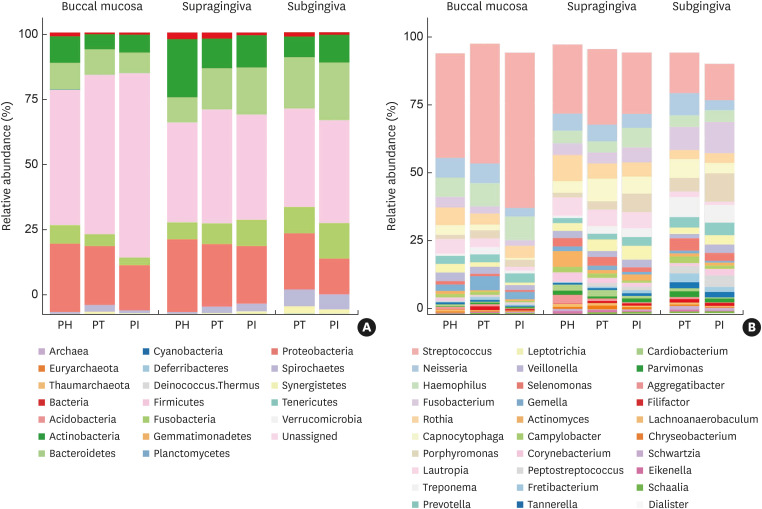
At the phylum level, the 7 most abundant phyla were Firmicutes, Proteobacteria, Actinobacteria, Bacteroidetes, Fusobacteria, Spirochaetes, and Synergistetes (Figure 2A). The relative abundances of Firmicutes, Spirochaetes, and Synergistetes in the diseased groups (PT and PI) were higher than those in the healthy group (PH) for both the buccal and supragingival samples. When comparing the diseased groups, Firmicutes and Actinobacteria showed higher relative abundances in the PI group than in the PT group in the buccal samples. In the subgingival samples, the relative abundances of Firmicutes, Actinobacteria, Bacteroidetes, and Fusobacteria in the PI group were higher than those in the PT group.
Figure 2. Relative abundance of the microbial composition at the (A) phylum level and (B) genus level according to disease status and sampling sites. Graphs show the proportions of mean relative abundances of each phylum or genus, which were calculated from the samples within the respective subjects.
PH: periodontally healthy, PI: peri-implantitis, PT: periodontitis.
At the genus level, 10 genera (Streptococcus, Neisseria, Haemophilus, Fusobacterium, Rothia, Capnocytophaga, Porphyromonas, Lautropia, Treponema, and Prevotella) comprised more than 50% of the total sequences in each group (Figure 2B). Compared with those of the PH and PT subjects, the bacterial communities in the PI subjects had higher levels of Haemophilus, Fusobacterium, Porphyromonas, and Leptotrichia in the supragingival samples; and Fusobacterium, Porphyromonas, Leptotrichia, Veillonella, Corynebacterium, and Peptostreptococcus in the subgingival samples.
Comparison of significantly different relative abundances
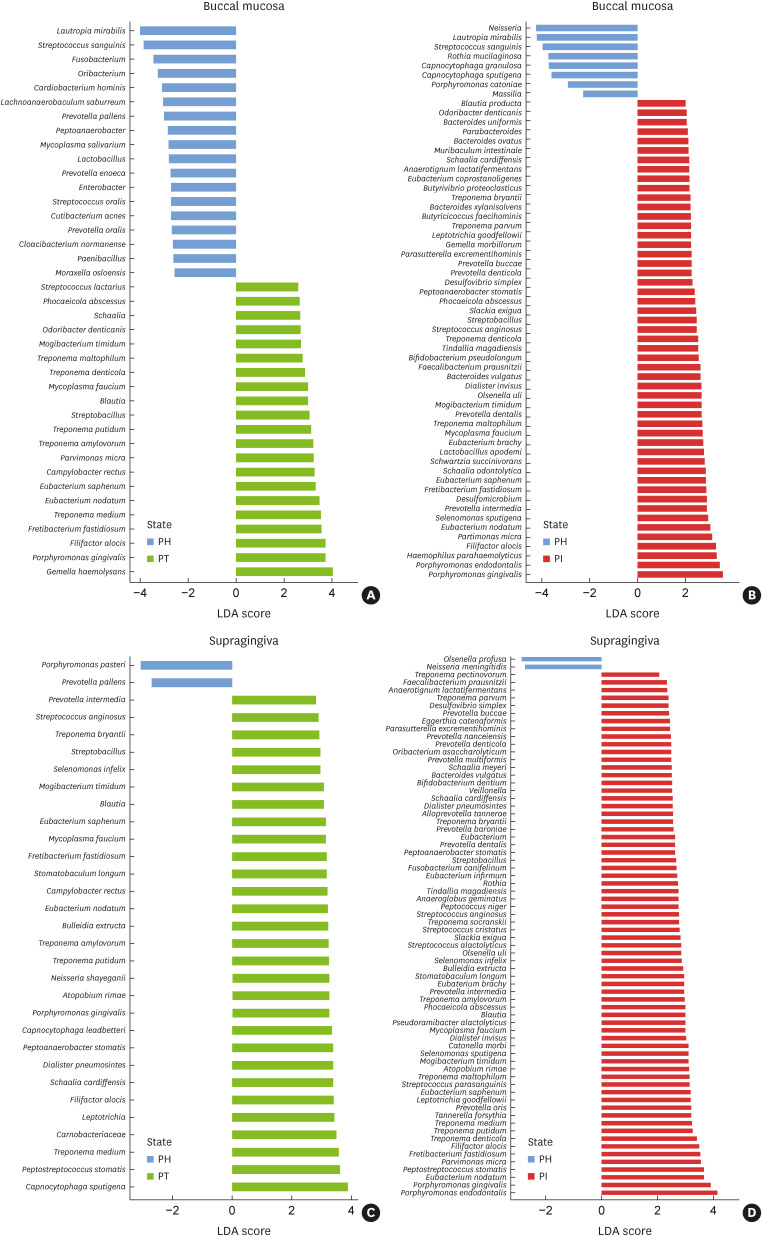
Several species in the diseased groups presented significantly different relative abundances compared to those in the PH group in the buccal (PT: 21 species, PI: 51 species) (Figure 3A and B) and supragingival samples (PT: 29 species, PI: 68 species) (Figure 3C and D). In the buccal samples, the relative abundances of Eubacterium spp., Treponema spp., Porphyromonas gingivalis, Filifactor alocis, Mogibacterium timidum, Parvimonas micra, Fretibacterium fastidiosum, and Mycoplasma faucium were high in both the PT and PI groups. In the supragingival samples, the relative abundances of Eubacterium nodatum, Eubacterium saphenum, Treponema spp., Prevotella intermedia, P. gingivalis, F. alocis, M. timidum, F. fastidiosum, and M. faucium were high in both diseased groups.
Figure 3. Significantly different relative abundances of microbiota according to (A) the disease status, (B) buccal mucosa, (C) supragingival plaque, and (D) supragingival plaque. These were identified by LDA coupled with effect size measurements. Only taxa that met the LDA significance threshold of 2.0 are shown.
LDA: linear discriminant analysis, PH: periodontally healthy, PI: peri-implantitis, PT: periodontitis.
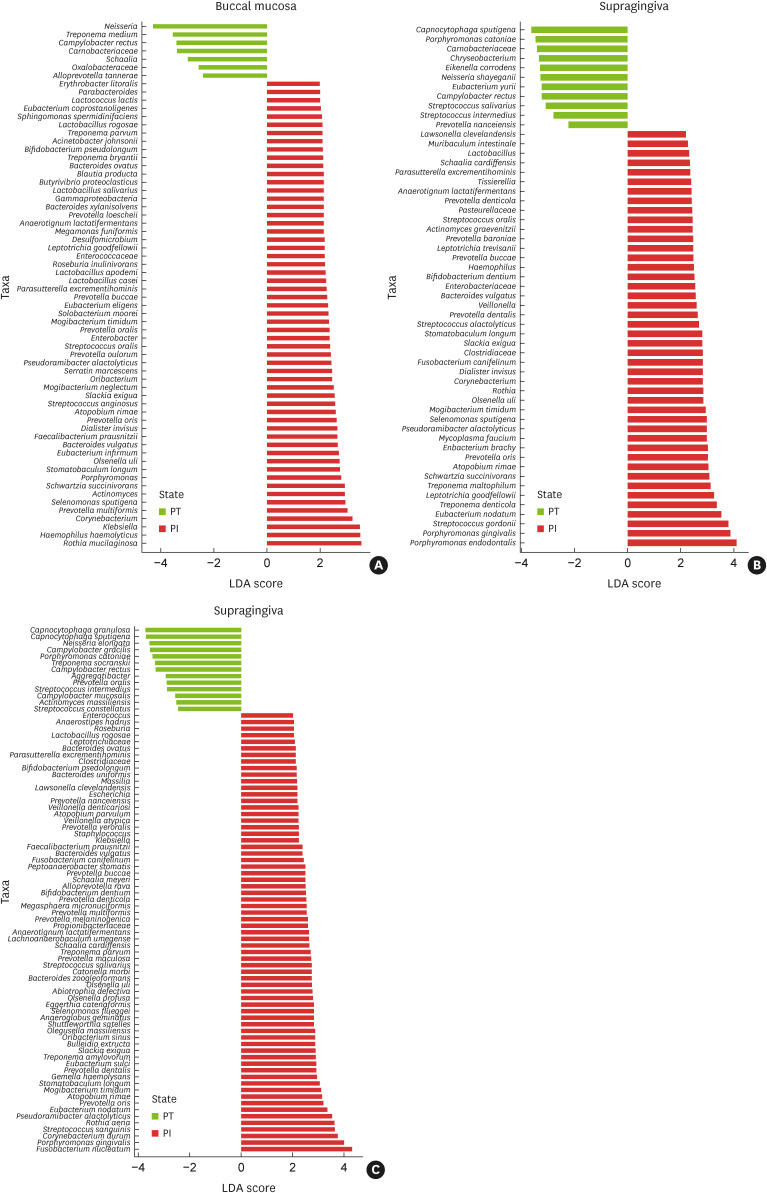
When comparing the diseased groups, 57, 44, and 67 species in the buccal, supragingival, and subgingival samples, respectively, exhibited significantly higher relative abundances in the PI group than in the PT group (Figure 4A-C). These include P. gingivalis, E. nodatum, Pseudoramibacter alactolyticus, Solobacterium moorei, and Porphyromonas endodontalis. In contrast, considerably fewer species (7, 11, and 13 in the buccal, supragingival, and subgingival samples, respectively) presented significantly higher relative abundances in the PT group than in the PI group, including Capnocytophaga spp., Campylobacter spp., and Porphyromonas catoniae.
Figure 4. Significantly different relative abundances of microbiota between subjects with PT and subjects with PI in (A) buccal mucosa, (B) supragingival plaque, and (C) subgingival plaque. The LDA effect size was used to identify taxa that had an abundance significantly different between groups. Only taxa that met the LDA significance threshold of 2.0 are shown.
LDA: linear discriminant analysis, PT: periodontitis, PI: peri-implantitis.
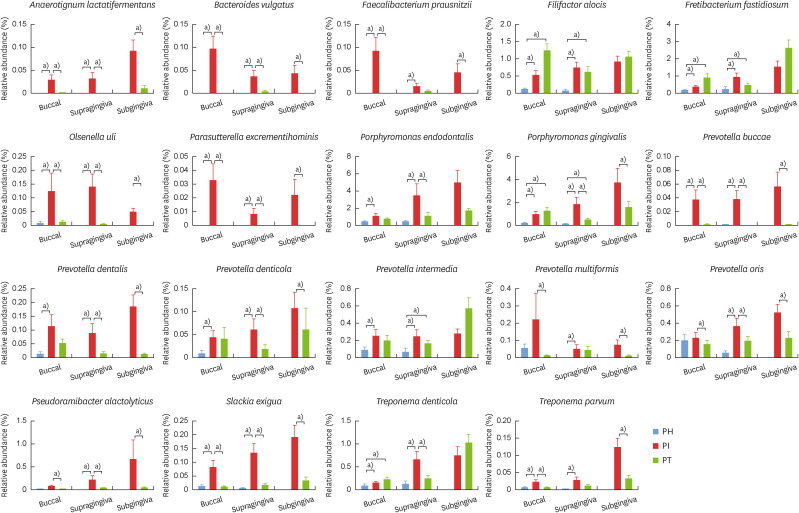
Relative abundances of significant bacterial species
Well-known periodontal pathogens, such as P. gingivalis, Prevotella spp., Treponema spp., F. alocis, and F. fastidiosum were prevalent in both the PT and PI groups, and most showed high relative abundances (Figure 5). In contrast, several bacteria previously unrecognized in periodontal tissue, such as Anaerotignum lactatifermentans, Bacteroides vulgatus, Faecalibacterium prausnitzii, Olsenella uli, Parasutterella excrementihominis, Prevotella buccae, P. alactolyticus, and Slackia exigua, were considerably more significantly abundant in the PI group than in the PH and PT groups.
Figure 5. The relative abundances of significant bacterial species. Graphs show the proportion of mean relative abundance for well-known periodontal pathogens and newly identified bacteria within PH, PT, and PI subjects in buccal mucosa, supragingival plaque, and subgingival plaque.
PH: periodontally healthy, PI: peri-implantitis, PT: periodontitis.
a)Above significant linear discriminant analysis threshold of 2.0.
Bacterial correlation networks
The bacterial networks of the supragingival and subgingival samples established more complex and larger clusters than those of the buccal samples. In both the supragingival and subgingival samples, a cluster of bacterial species in the diseased groups (PI and PT) presented cooperative interactions. In the supragingival samples, dense connections were noted between Treponema spp., Eubacterium spp., Tannerella forsythia, P. gingivalis, F. fastidiosum, and F. alocis in both the PT and PI subjects (Figure 6B). In the subgingival samples, Treponema spp., T. forsythia, M. timidum, M. faucium, P. endodontalis, and F. alocis formed a dense network (Figure 6C). In the buccal samples, the nodes of E. nodatum and F. alocis were highly connected with other bacteria in the network (Figure 6A).
Figure 6. Bacterial correlation networks among PH, PT, and PI subjects in (A) buccal mucosa, (B) supragingival plaque, and (C) subgingival plaque. To visualize interactions and characterize the microbial community, the Sparse Correlations for Compositional Data (SparCC) method and Cytoscape were used. The nodes denote the species and the connections represent the existence of correlations.
PH: periodontally healthy, PI: peri-implantitis, PT: periodontitis.
DISCUSSION
In the shift from symbiosis to dysbiosis, keystone pathogens interfere with host immune reactions and contribute to homeostatic destruction. Moreover, in dysbiosis, pathobionts show increased relative abundance and cause destructive inflammation and bone loss [25]. In implants, this dysbiosis could cause PI and, if untreated, result in destruction of osseointegration, leading to implant loss [26]. Because the microbiome is related to oral disease, the present study analyzed the microbiome in relation to PI. In this study, the clinical periodontal parameters were similar in the PT and PI subjects. However, many more bacterial species showed significantly higher relative abundances in the PI group than in the PT group in both the supragingival and subgingival samples. Given these results, it could be inferred that, compared to PT, PI is a more complex inflammatory response that involves more bacteria.
Both the PT and PI groups presented higher alpha diversity of the microbiota than was observed in the PH group. Specifically, the PI group showed more enrichment of the microbiota than the PT group, which was consistent with the results of previous studies [27,28]. Since inflammation damages the periodontal tissue, thereby supplying an important source of nutrients, more diverse and abundant microbiota could be observed in an inflammatory state than in a healthy state [29]. In this regard, the microbiota may have been more diverse in the PI group than in the PT group because the peri-implant invasion was more vulnerable to the development of inflammation and exhibited a more aggressive response than the periodontal tissue [9]. A previous study also reported that severe inflammation was more likely to occur in peri-implant tissue than in periodontal tissue, leading to differences in the core microbiota between PI and PT [28]. The beta diversity analysis showed significant separation of the microbial clusters of the 3 groups (P<0.05) in the buccal samples. Therefore, as microbiome analysis from buccal samples is easy and noninvasive, it could be useful for screening these diseases [14,30]. In the subgingival samples, although there was no significant difference, the microbial clusters of PT and PI showed distinct tendencies. A distinctive separation between the PT and PI microbiota was also observed in subgingival samples in a previous study [31].
The relative abundances of specific phyla and genera were higher in the diseased groups than those in the PH group. As the relative abundances of Firmicutes, Spirochaetes, and Synergistetes were higher in both the PT and PI groups than in the PH group, these phyla could be considered to be highly associated with the inflammatory response. When comparing only the diseased groups, some phyla and genera were more abundant in the PI subjects than in the PT subjects. Since PI is a more severe inflammatory response than PT [9] and the subgingival pocket is directly related to the disease condition, the higher abundances of Firmicutes, Actinobacteria, Bacteroidetes, and Fusobacteria in the PI group than in the PT group in the subgingival samples of this study may reflect the aggressive progression of PI.
This study identified microbiota with significantly higher relative abundances in both the PT and PI groups than in the PH group. In the buccal and supragingival samples, these bacteria included M. timidum and M. faucium, as well as several well-known periodontal pathogens (including P. gingivalis, F. alocis, F. fastidiosum Treponema spp., Eubacterium spp., P. intermedia, and P. micra). A previous study reported that Porphyromonas spp. and Prevotella spp. were more abundant in diseased implants than in healthy implants [32]. Eubacterium spp., T. denicola, and P. intermedia were also reported to be more prevalent in diseased implants [33]. Since the relative abundances of these bacteria are significantly higher in inflammatory conditions, they can be considered as keystone pathogens contributing to the progression of PT or PI, or pathobionts accelerating disease progression by adapting well to the altered environment caused by the disease.
Several bacterial species had significantly different relative abundance levels between the PT and PI subjects. The abundance of P. gingivalis, E. nodatum, P. alactolyticus, S. moorei, and P. endodontalis was higher in the PI subjects. Most of these bacteria have not been identified in periodontal tissue by conventional methods, but recent studies using NGS also reported the existence of P. endodontalis, F. alocis, F. fastidiosum, P. alactolyticus, S. moorei, and Peptostreptococcus stomatis in samples from patients with PI [31]. P. endodontalis is considered to be abundant in infected root canals [34], capable of inducing osteoclastogenesis [35], and a core bacterium of PT [36]. A previous study also found P. stomatis and S. moorei in samples from patients with PI, but their properties are unknown [31]. In particular, the genus Peptostreptococcus is known to cause abscesses and necrotic soft tissue infection [37].
Additionally, this study found, at low levels of relative abundance, the presence of bacterial species that have rarely been mentioned in other studies. Compared to those in the PH and PT groups, these bacteria had considerably higher proportions in the PI group, which presented significant relative abundances at all sites. These bacteria included A. lactatifermentans, B. vulgatus, O. uli, P. excrementihominis, P. buccae, and S. exigua. However, there is little information, to our knowledge, on the function of these bacteria in periodontal tissue. S. exigua has been reported to be prevalent in periapical infections and has been identified in deep PI pockets [38]. O. uli has been found to be abundant in infected root canals [34], but its role in PI is unknown. Moreover, F. prausnitzii and P. alactolyticus also presented similar tendencies to these bacteria. P. alactolyticus is known to be common within canals with irreversible pulpitis [39]. In addition, it has recently been found in diseased implants [40]. Taken together, although most of these bacteria had a substantially low relative abundance and little is known about their role in PI, they might play an important role in the dysbiosis of PI.
In the bacterial community interaction network, some microbiota interacted with other bacteria in both diseased groups. In the network, well-known periodontal pathogens, including the red complex, Treponema spp., Prevotella spp., E. nodatum, F. alocis, and F. fastidiosum were also found in this study. T. maltophilum, M. timidum, and P. endodontalis were found in this study, but few have been reported in periodontal tissues in the past. Most of these bacteria exhibited dense connectivity with other species in both PT and PI subjects. Moreover, more complex and larger clusters were established in both the supragingival and subgingival samples than in the buccal samples. Therefore, it could be assumed that some of these microbes, as common strains associated with both inflammatory diseases, function like pathobionts.
The mean ages in each subject group (PH, PT, and PI) were 26.9, 50.7, and 55.8 years, respectively. One of the limitations of the present study was that the mean age of PH differed considerably from that of the other groups. Furthermore, in this study, the PH subjects were employed as a control group for the diseased groups. A more systematic comparison would have been possible if subjects with healthy implants were additionally recruited to the control group. Since PT and PI are diseases involving multifactorial risk factors, it is necessary to conduct sampling from the same subjects in order to control for subject-dependent factors in these groups more clearly. Therefore, future research will need to address these limitations.
In conclusion, compared with the PH and PT groups, the composition of the microbiota showed considerable differences in the PI group, and this result could be associated with the aggressive and complicated nature of PI. The microbial profile specific to PI, as identified by NGS, could provide significant evidence relevant for the treatment of this disease. Further research into the role of unique bacteria found in this study in regard to PI will be helpful for establishing an effective and optimal treatment protocol for this disease.
Footnotes
Funding: This research was supported by the National Research Foundation of Korea (NRF) funded by the Ministry of Science & ICT (MEST; NRF-2017M3A9B6062021 and NRF-2017M3A9B6062026).
Conflict of Interest: No potential conflict of interest relevant to this article was reported.
- Conceptualization: Hyun-Joo Kim, Hee Sam Na, Ju-Youn Lee.
- Data curation: Hyun-Joo Kim, Dae-Hee Ahn, Yeuni Yu, Hyejung Han, Si Yeong Kim, Ji-Young Joo.
- Formal analysis: Yeuni Yu, Hyejung Han, Si Yeong Kim, Ji-Young Joo, Jin Chung.
- Funding acquisition: Jin Chung, Ju-Youn Lee.
- Investigation: Hyun-Joo Kim, Dae-Hee Ahn, Yeuni Yu, Hyejung Han, Si Yeong Kim, Ji-Young Joo.
- Methodology: Jin Chung, Hee Sam Na, Ju-Youn Lee.
- Project administration: Jin Chung, Hee Sam Na, Ju-Youn Lee.
- Writing-original draft: Hyun-Joo Kim, Dae-Hee Ahn.
- Writing-review & editing: Hyun-Joo Kim, Dae-Hee Ahn, Yeuni Yu, Hyejung Han, Si Yeong Kim, Ji-Young Joo, Jin Chung, Hee Sam Na, Ju-Youn Lee.
References
- 1.Hasegawa T, Kawabata S, Takeda D, Iwata E, Saito I, Arimoto S, et al. Survival of Brånemark System Mk III implants and analysis of risk factors associated with implant failure. Int J Oral Maxillofac Implants. 2017;46:267–273. doi: 10.1016/j.ijom.2016.10.014. [DOI] [PubMed] [Google Scholar]
- 2.Derks J, Schaller D, Håkansson J, Wennström JL, Tomasi C, Berglundh T. Effectiveness of implant therapy analyzed in a Swedish population: prevalence of peri-implantitis. J Dent Res. 2016;95:43–49. doi: 10.1177/0022034515608832. [DOI] [PubMed] [Google Scholar]
- 3.Renvert S, Persson GR, Pirih FQ, Camargo PM. Peri-implant health, peri-implant mucositis, and peri-implantitis: case definitions and diagnostic considerations. J Clin Periodontol. 2018;45(Suppl 20):S278–S285. doi: 10.1111/jcpe.12956. [DOI] [PubMed] [Google Scholar]
- 4.Berglundh T, Armitage G, Araujo MG, Avila-Ortiz G, Blanco J, Camargo PM, et al. Peri-implant diseases and conditions: consensus report of workgroup 4 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J Periodontol. 2018;89(Suppl 1):S313–S318. doi: 10.1002/JPER.17-0739. [DOI] [PubMed] [Google Scholar]
- 5.Esposito M, Hirsch JM, Lekholm U, Thomsen P. Biological factors contributing to failures of osseointegrated oral implants. (II). Etiopathogenesis. Eur J Oral Sci. 1998;106:721–764. doi: 10.1046/j.0909-8836..t01-6-.x. [DOI] [PubMed] [Google Scholar]
- 6.Page RC, Kornman KS. The pathogenesis of human periodontitis: an introduction. Periodontol 2000. 1997;14:9–11. doi: 10.1111/j.1600-0757.1997.tb00189.x. [DOI] [PubMed] [Google Scholar]
- 7.Charalampakis G, Leonhardt Å, Rabe P, Dahlén G. Clinical and microbiological characteristics of peri-implantitis cases: a retrospective multicentre study. Clin Oral Implants Res. 2012;23:1045–1054. doi: 10.1111/j.1600-0501.2011.02258.x. [DOI] [PubMed] [Google Scholar]
- 8.Belibasakis GN, Charalampakis G, Bostanci N, Stadlinger B. Peri-implant infections of oral biofilm etiology. Adv Exp Med Biol. 2015;830:69–84. doi: 10.1007/978-3-319-11038-7_4. [DOI] [PubMed] [Google Scholar]
- 9.Carcuac O, Berglundh T. Composition of human peri-implantitis and periodontitis lesions. J Dent Res. 2014;93:1083–1088. doi: 10.1177/0022034514551754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shibli JA, Melo L, Ferrari DS, Figueiredo LC, Faveri M, Feres M. Composition of supra- and subgingival biofilm of subjects with healthy and diseased implants. Clin Oral Implants Res. 2008;19:975–982. doi: 10.1111/j.1600-0501.2008.01566.x. [DOI] [PubMed] [Google Scholar]
- 11.Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol 2000. 2005;38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- 12.Botero JE, González AM, Mercado RA, Olave G, Contreras A. Subgingival microbiota in peri-implant mucosa lesions and adjacent teeth in partially edentulous patients. J Periodontol. 2005;76:1490–1495. doi: 10.1902/jop.2005.76.9.1490. [DOI] [PubMed] [Google Scholar]
- 13.Frey KG, Herrera-Galeano JE, Redden CL, Luu TV, Servetas SL, Mateczun AJ, et al. Comparison of three next-generation sequencing platforms for metagenomic sequencing and identification of pathogens in blood. BMC Genomics. 2014;15:96. doi: 10.1186/1471-2164-15-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei Y, Shi M, Zhen M, Wang C, Hu W, Nie Y, et al. Comparison of subgingival and buccal mucosa microbiome in chronic and aggressive periodontitis: a pilot study. Front Cell Infect Microbiol. 2019;9:53. doi: 10.3389/fcimb.2019.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakagaki T, Tamura M, Kobashi K, Omori A, Koyama R, Idogawa M, et al. Targeted next-generation sequencing of 50 cancer-related genes in Japanese patients with oral squamous cell carcinoma. Tumour Biol. 2018;40:1010428318800180. doi: 10.1177/1010428318800180. [DOI] [PubMed] [Google Scholar]
- 16.Sembler-Møller ML, Belstrøm D, Locht H, Enevold C, Pedersen AM. Next-generation sequencing of whole saliva from patients with primary Sjögren’s syndrome and non-Sjögren’s sicca reveals comparable salivary microbiota. J Oral Microbiol. 2019;11:1660566. doi: 10.1080/20002297.2019.1660566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caton JG, Armitage G, Berglundh T, Chapple IL, Jepsen S, Kornman KS, et al. A new classification scheme for periodontal and peri-implant diseases and conditions - Introduction and key changes from the 1999 classification. J Periodontol. 2018;89(Suppl 1):S1–S8. doi: 10.1002/JPER.18-0157. [DOI] [PubMed] [Google Scholar]
- 18.Duarte PM, de Mendonça AC, Máximo MB, Santos VR, Bastos MF, Nociti FH., Jr Effect of anti-infective mechanical therapy on clinical parameters and cytokine levels in human peri-implant diseases. J Periodontol. 2009;80:234–243. doi: 10.1902/jop.2009.070672. [DOI] [PubMed] [Google Scholar]
- 19.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010;2010:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- 22.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedman J, Alm EJ. Inferring correlation networks from genomic survey data. PLOS Comput Biol. 2012;8:e1002687. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hajishengallis G. Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol. 2014;35:3–11. doi: 10.1016/j.it.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leonhardt A, Dahlén G, Renvert S. Five-year clinical, microbiological, and radiological outcome following treatment of peri-implantitis in man. J Periodontol. 2003;74:1415–1422. doi: 10.1902/jop.2003.74.10.1415. [DOI] [PubMed] [Google Scholar]
- 27.Yu XL, Chan Y, Zhuang L, Lai HC, Lang NP, Keung Leung W, et al. Intra-oral single-site comparisons of periodontal and peri-implant microbiota in health and disease. Clin Oral Implants Res. 2019;30:760–776. doi: 10.1111/clr.13459. [DOI] [PubMed] [Google Scholar]
- 28.Maruyama N, Maruyama F, Takeuchi Y, Aikawa C, Izumi Y, Nakagawa I. Intraindividual variation in core microbiota in peri-implantitis and periodontitis. Sci Rep. 2014;4:6602. doi: 10.1038/srep06602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012;10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Na HS, Kim SY, Han H, Kim HJ, Lee JY, Lee JH, et al. Identification of potential oral microbial biomarkers for the diagnosis of periodontitis. J Clin Med. 2020;9:1549. doi: 10.3390/jcm9051549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koyanagi T, Sakamoto M, Takeuchi Y, Maruyama N, Ohkuma M, Izumi Y. Comprehensive microbiological findings in peri-implantitis and periodontitis. J Clin Periodontol. 2013;40:218–226. doi: 10.1111/jcpe.12047. [DOI] [PubMed] [Google Scholar]
- 32.Neilands J, Wickström C, Kinnby B, Davies JR, Hall J, Friberg B, et al. Bacterial profiles and proteolytic activity in peri-implantitis versus healthy sites. Anaerobe. 2015;35:28–34. doi: 10.1016/j.anaerobe.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 33.Zheng H, Xu L, Wang Z, Li L, Zhang J, Zhang Q, et al. Subgingival microbiome in patients with healthy and ailing dental implants. Sci Rep. 2015;5:10948. doi: 10.1038/srep10948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rôças IN, Alves FR, Santos AL, Rosado AS, Siqueira JF., Jr Apical root canal microbiota as determined by reverse-capture checkerboard analysis of cryogenically ground root samples from teeth with apical periodontitis. J Endod. 2010;36:1617–1621. doi: 10.1016/j.joen.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Ma N, Yang D, Okamura H, Teramachi J, Hasegawa T, Qiu L, et al. Involvement of interleukin\xe2\x80\x9123 induced by Porphyromonas endodontalis lipopolysaccharide in osteoclastogenesis. Mol Med Rep. 2017;15:559–566. doi: 10.3892/mmr.2016.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 2013;7:1016–1025. doi: 10.1038/ismej.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mombelli A, Décaillet F. The characteristics of biofilms in peri-implant disease. J Clin Periodontol. 2011;38(Suppl 11):203–213. doi: 10.1111/j.1600-051X.2010.01666.x. [DOI] [PubMed] [Google Scholar]
- 38.Kim KS, Rowlinson MC, Bennion R, Liu C, Talan D, Summanen P, et al. Characterization of Slackia exigua isolated from human wound infections, including abscesses of intestinal origin. J Clin Microbiol. 2010;48:1070–1075. doi: 10.1128/JCM.01576-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irani S. Orofacial bacterial infectious diseases: an update. J Int Soc Prev Community Dent. 2017;7:S61–S67. doi: 10.4103/jispcd.JISPCD_290_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pokrowiecki R, Mielczarek A, Zaręba T, Tyski S. Oral microbiome and peri-implant diseases: where are we now? Ther Clin Risk Manag. 2017;13:1529–1542. doi: 10.2147/TCRM.S139795. [DOI] [PMC free article] [PubMed] [Google Scholar]