Summary
This study sought to examine the association between DNA methylation and body mass index (BMI) and the potential of BMI-associated cytosine-phosphate-guanine (CpG) sites to provide information about metabolic health. We pooled summary statistics from six trans-ethnic epigenome-wide association studies (EWASs) of BMI representing nine cohorts (n = 17,034), replicated these findings in the Women’s Health Initiative (WHI, n = 4,822), and developed an epigenetic prediction score of BMI. In the pooled EWASs, 1,265 CpG sites were associated with BMI (p < 1E−7) and 1,238 replicated in the WHI (FDR < 0.05). We performed several stratified analyses to examine whether these associations differed between individuals of European and African descent, as defined by self-reported race/ethnicity. We found that five CpG sites had a significant interaction with BMI by race/ethnicity. To examine the utility of the significant CpG sites in predicting BMI, we used elastic net regression to predict log-normalized BMI in the WHI (80% training/20% testing). This model found that 397 sites could explain 32% of the variance in BMI in the WHI test set. Individuals whose methylome-predicted BMI overestimated their BMI (high epigenetic BMI) had significantly higher glucose and triglycerides and lower HDL cholesterol and LDL cholesterol compared to accurately predicted BMI. Individuals whose methylome-predicted BMI underestimated their BMI (low epigenetic BMI) had significantly higher HDL cholesterol and lower glucose and triglycerides. This study confirmed 553 and identified 685 CpG sites associated with BMI. Participants with high epigenetic BMI had poorer metabolic health, suggesting that the overestimation may be driven in part by cardiometabolic derangements characteristic of metabolic syndrome.
Keywords: obesity, DNA methylation, epigenomics, epigenome-wide association study, BMI, metabolic disease, prediction, adiposity
DNA methylation is increasingly used as a biomarker of health and associates with BMI. Do et al. constructed an epigenetic BMI score and observed differential metabolic health among individuals whose epigenetically predicted BMI deviates from their actual BMI. These findings suggest that epigenetic BMI prediction could provide information relevant to metabolic health.
Introduction
Globally, the prevalence of obesity is rising with an estimated 650 million adults obese, representing 19.5% of the adult population.7,8 Obesity has been found to accompany a multitude of molecular and metabolic perturbations including impaired cell signaling, insulin resistance, hyperlipidemia, and hypertension.9,10,11 Ultimately these perturbations can lead to the early onset of chronic diseases; an individual living with obesity has a 37% increased risk of type 2 diabetes12 and 67%–85% increased risk of cardiovascular disease compared to individuals living without obesity.13 With a growing population of individuals living with obesity, it is increasingly important to understand the molecular mechanisms dysregulated by obesity to further elucidate both early markers of disease progression and novel therapeutic targets.
Epigenetic mechanisms are molecularly mediated changes in gene function which do not change the DNA sequence. DNA methylation, the most widely characterized epigenetic mechanism, occurs when a methyl group attaches to the cytosine in a cytosine-guanine nucleotide (CpG) pair.14 DNA methylation has been shown to influence gene expression by blocking transcription factor binding and recruiting chromatin remodelers.15 As a functional mechanism influencing gene expression, DNA methylation may be on a disease pathway and could provide insight into important therapeutic targets. DNA methylation has also become an important biomarker of health, for example with the development of epigenetic clocks, which can provide accurate estimates of individual age based on the methylation status of a representative set of CpG sites.16 Individuals whose DNA methylation deviates from their actual chronological age, such that their epigenetically predicted age is higher than their actual age, have been shown to have higher rates of cancer, cardiovascular disease, diabetes, and mortality.17 We hypothesized that DNA methylation-based prediction of obesity measures could provide another useful metric of health, particularly metabolic health.
Several studies have examined the relationship between DNA methylation and body mass index (BMI), a commonly used measure of obesity.1,2,3,4,5,6,18,19,20,21 Obesity has been significantly associated with differential DNA methylation, and Mendelian randomization analyses have suggested that while this differential methylation appears to be a consequence of the state of obesity at many CpG sites, some CpGs show evidence consistent with causal roles in obesity.18,3 While several large-scale studies have identified sites associated with obesity, it is likely that additional sites will be detectable only with large sample sizes, because observed DNA methylation differences are often subtle.22 Thus, a goal of this study is to conduct a well-powered epigenome-wide association study (EWAS) meta-analysis of BMI in nine population-based cohort studies to detect previously unidentified sites associated with obesity. The identification of novel sites can reveal unique molecular signatures of various BMI phenotypes (including metabolically healthy/unhealthy BMI) and may enable improved prediction of BMI. Previous studies have reported that a collection of methylation-based predictors can explain between 4.7% and 18% of the variance in BMI.18,6,23,24 In conducting this EWAS of BMI, we may have better predictive capacity by incorporating the novel CpG sites identified in the EWAS meta-analysis. As such, a secondary aim of this study is to examine whether BMI-associated CpG sites can predict BMI. As with epigenetic age, deviations from epigenetically predicted BMI may be associated with several relevant health outcomes and could be used as an informative metric of overall health and/or a predictor of future cardiovascular disease. Thus, we examined whether individuals whose BMI was poorly predicted by DNA methylation (DNA methylation overpredicts their actual BMI or DNA methylation underpredicts their actual BMI) have differential metabolic health status.
Subjects and methods
Participants
Our discovery analysis used data from 17,034 participants from six published EWASs of individuals of European descent (n = 11,220), African descent (n = 3,134), and South Asian descent (n = 2,680, Table 1). The six studies were based on nine cohorts: Atherosclerosis Risk in Communities study (ARIC),25 Melbourne Collaborative Cohort Study (MCCS),26 Lifelines DEEP,27 Lothian Birth Cohort (LBC) 1921 and 1936,28 Bogalusa Heart Study (BHS),29 the Research on Obesity and Diabetes among African Migrants (RODAM),30 the Kooperative Gesundheitsforschung in der Region Augsburg (KORA),31 the London Life Sciences Prospective Population Study (LOLIPOP),32 and Italian cardiovascular component of the European Prospective Investigation into Cancer and Nutrition (EPICOR).33 Replication analyses were conducted in three ancillary studies from the Women’s Health Initiative (WHI)34: Epigenetic Mechanisms of Particulate Matter-Mediated Cardiovascular Disease (EMPC, a.k.a. AS315), the Integrative Genomics for Risk of Coronary Heart Disease and Related Phenotypes in WHI cohort (BAA23), and Bladder Cancer and Leukocyte Methylation (AS311). All studies used procedures in accordance with ethical standards of the responsible committee on human experimentation and obtained informed consent from all participants. More information can be found in the individual study references. In the WHI, individuals were excluded if BMI and blood samples for DNA methylation were not measured within the same year. Extreme levels of BMI (<17 kg/m2 and >75 kg/m2) were excluded. Further description of the discovery and replication cohorts is described in supplemental methods.
Table 1.
Study characteristics of discovery analyses
| Reference | Study population | n | Exposure | Outcome | Sample | Covariates |
|---|---|---|---|---|---|---|
| Demerath et al.1 | Atherosclerosis Risk in Communities (ARIC) | 2,097 | BMI | DNA methylation β-value | leukocytes | age, sex, study center, total white blood cell differentials, education, household income, cigarette smoking, current alcohol use, leisure physical activity, cell composition,35 top 10 PCs of genetic relatedness and batch effects (row, plate number and chip number) |
| Geurts et al.5 | Melbourne Collaborative Cohort Study (MCCS) | 5,361 | BMI Z score | DNA methylation M-values | dried blood spot, mononuclear cells, buffy coats | age, sex, smoking status, country of birth, sample type, cell composition,35 and study, plate and chip included as random effects |
| Meeks et al.2 | Research on Obesity and Diabetes among African Migrants (RODAM) study | 547 | BMI | DNA methylation M-values | whole blood | age, sex, recruitment site, cell composition,35 hybridization batch, array position and first PC of genetic relatedness |
| Shah et al.6 | Lothian Birth Cohort (LBC) and Lifelines DEEP | 2,116 | DNA methylation M-values | BMI Z score | whole blood | age, sex, batch effects, complete blood cell count adjusted for in sensitivity analyses |
| Sun et al.4 | Bogalusa Heart Study (BHS) | 1,485 | BMI | DNA methylation β-value | whole blood | age, sex, current smoking status, cell composition35 included as fixed effects with batch array as a random effect |
| Wahl et al.3 | KORA, LOLIPOP, EPICOR | 5,458 | DNA methylation β-value | BMI | whole blood | top 20 PCs of control probes, cell composition,35 age, sex, smoking status, physical activity index and alcohol consumption |
BMI, DNA methylation, and covariates
BMI was defined as weight in kg/(height in m)2. Methodologies obtaining weight and height differed among the studies, but all used standard methods. One study transformed BMI values to obtain a normal distribution.5 Relevant variables in our replication analysis included race/ethnicity, age, physical activity, and smoking status, which were all based on self-report. Smoking status was defined as current, former, or never.
DNA methylation was measured in several cell types including CD4+ T cells, mononuclear cells, and whole blood. DNA methylation in all studies was measured using the Illumina 450K Infinium Methylation BeadChip. DNA methylation was estimated as the proportion of methylated signal relative to combined unmethylated and methylated signal for a specific CpG site, defined as the β value. Quality control procedures of the previous studies have been reported in detail and they did not differ substantially across studies. In the WHI, all methylation data were quality controlled and normalized using beta-mixture quantile normalization. In replication analyses, chip and row on chip were included as technical covariates in all models to adjust for batch effects. Cell composition was estimated using methods derived by Houseman et al.35
Statistical analysis
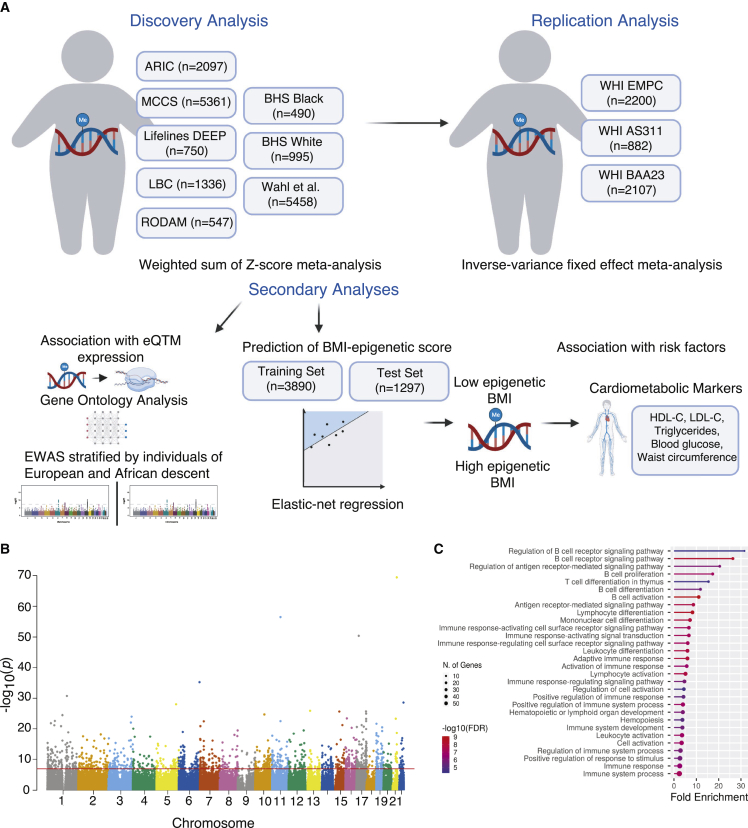
A summary of our analyses is included in Figure 1A. Our primary method was weighted sum of Z score meta-analysis.36 This method utilizes Z scores from individual study summary statistics computed from inverse-normal p values and the direction of effect to determine significant sites. This was chosen as the primary method for meta-analysis since the studies did not all have equivalent exposure-outcome definition (DNA methylation was defined as exposure in two studies and outcome in four studies) and BMI was transformed in one study. The EWAS was adjusted for genomic inflation via the method of genomic control37 (λ = 1.89) and significance was defined as p < 1 × 10−7. To annotate CpG sites, we used the Illumina HumanMethylation450K annotation file.
Figure 1.
BMI associates with differential methylation
(A) Description of study analyses, created in BioRender.com.
(B) Manhattan plot of the association between DNA methylation and BMI. Each dot represents a CpG site, with genomic location on the x axis and –log10(p value) on the y axis.
(C) Top pathways identified in gene ontology analysis.
The significant sites were examined for replication within WHI. Models were stratified by ancillary study. Covariates in this analysis included age, race/ethnicity, cell composition, the top three principal components of genetic relatedness, row on chip, smoking status, clinical trial arm, and case-control status (BAA23 and AS311). To account for potential chip-to-chip differences in measurement and to adjust for batch effects, chip was included as a random effect for each BeadChip in our model. Stratified analyses were combined using inverse-variance weighted (IVW) meta-analysis.38 Significance was defined by false discovery rate (FDR) q value < 0.05.
BMI prediction score
To examine the degree to which methylation can predict BMI and the secondary cardiometabolic outcomes associated with BMI, we used elastic net regression models with the significant sites to predict log-normalized BMI. The WHI cohorts were randomly divided into a training and test set (80% and 20%, respectively) with an equal BMI distribution. We used elastic net regression on the training set with 10-fold cross validation to select a predictive model, which we subsequently tested in the test set. Using the significant sites and coefficients selected by the model, a DNA methylation prediction score was developed by multiplying the coefficient by the individual β-value and summing over all the sites for each individual. We then evaluated the performance of the DNA methylation score in the test set, both in terms of how accurately it predicted BMI (metrics: R2 and median absolute deviation) and how well it predicted obesity status (BMI ≥ 30 kg/m2) (metrics: sensitivity and specificity).
Using the predicted BMI values, we examined the patterns among outliers in the prediction model. Individuals were split into categories based on regressing the predicted BMI on the actual BMI. Accurately predicted individuals were defined as those with residuals between −0.04 and 0.04 (accurate epigenetic BMI). Individuals outside of this range were split into two groups: residual below −0.04 (low epigenetic BMI or individuals whose methylome-predicted BMI underestimated their BMI) and residual above 0.04 (high epigenetic BMI or individuals whose methylome-predicted BMI overestimated their BMI). These thresholds were defined based on the 10% and the 90% distribution of the residuals. Using these categories, we examined cardiometabolic differences including waist circumference, triglycerides, HDL cholesterol, LDL cholesterol, and blood glucose among these categories using linear regression models regressing log-normalized cardiometabolic markers on DNA methylation prediction category adjusted for age, race/ethnicity, smoking status, and physical activity. To aid interpretability, results were reported based on the change in average value in the text. In sensitivity analyses, we further examined results using thresholds defined by the 20% and 80% distribution of residuals and using continuous residuals.
Functional annotation
To examine how individual CpG sites associate with gene expression, we used summary statistics from two EWASs of gene expression in the Grady Trauma Project (GTP) and Multi-Ethnic Study of Atherosclerosis (MESA) studies.39 Summary statistics on the CpG-transcript associations were extracted for relevant sites identified in primary and secondary analyses. We conducted gene ontology (GO) analysis of the genes identified in the CpG-transcript associations using the PANTHER database (www.pantherdb.org). Significant pathways were defined as those with FDR < 0.05.
Sensitivity analyses
We conducted several sensitivity analyses. In the discovery meta-analysis, we examined the influence of specific studies on the results using leave-one-out analyses to examine the degree that each study is influencing the results. We also compared results from the weighted sum of Z score meta-analysis with results that would be obtained using an IVW meta-analysis in studies with the same exposure-outcome definition. We examined CpG sites which were exclusively found to be significant in models including individuals of African descent versus European descent (defined by self-reported race/ethnicity) for interaction by self-reported race/ethnicity and BMI. We used linear mixed-effect models adjusting for age, cell composition, smoking status, WHI study randomization arm, case-control status, and row with a random effect for chip. In our replication analysis in WHI, models were additionally adjusted for diet quality, physical activity level, and socioeconomic status.
Results
Our discovery analysis included 17,034 participants from six EWASs (Figure 1A, Tables 1 and S1). The definition of BMI and DNA methylation differed, with several studies transforming these values in the models (Table 1). The covariates in the model also differed with all studies adjusting for age and sex and the majority adjusting for cell composition and smoking status. Most studies were conducted in ancestrally homogeneous populations. However, among studies including mixed ethnicities, analyses were stratified by self-reported race/ethnicity. When pooling results from all studies, 1,265 CpG sites were associated with BMI (Figure 1B, Table S2, p < 1E−7) with 498 of the sites having a consistent direction of effect in all the cohorts meta-analyzed. More than half of the significant sites (726 CpG sites) were positively associated with BMI.
In the WHI, 367 women were excluded due to missing BMI, extreme levels of BMI, or overlap, leaving 4,822 women included in the replication cohort (Table S3). Of the 1,265 sites identified in the discovery analysis, 1,254 were analyzed after QC. In the WHI, 1,238 CpG sites were significantly associated with BMI (Table S4, FDR q value < 0.05). These 1,238 CpG sites annotated to 742 unique genes. Additionally, 147 of these genes were annotated to more than one BMI-associated CpG site, with 382 CpG sites annotated to these 147 genes. With the large sample size, our meta-analysis confirmed 553 CpG sites previously reported in the literature40 and discovered 685 novel CpG sites (defined as not previously identified in EWASs of BMI). We examined how the replicated sites associated with differential gene expression based on previously published analyses of the GTP and MESA cohort.39 Of the 1,238 CpG sites, 317 sites associated with 35 genes in MESA (1,103 CpG-transcript associations; Table S5) and 35 sites associated with 45 genes in GTP (79 CpG-transcript associations; Table S6). One site associated with the same mRNA transcript in both cohorts, cg25653947, which was positively associated with expression of TOP1MT. We performed a GO analysis of the differentially expressed genes and found enrichment in pathways related to the adaptive immune system with regulation in B and T cell pathways (Figure 1C, Table S7).
We next re-performed our discovery EWAS stratified by European vs. African descent (defined by self-reported race/ethnicity). We found 936 and 130 CpG sites that were associated with BMI in the analyses restricted to individuals from European (n = 11,220) and African (n = 2,587) descent, respectively. Of the 130 significant CpG sites in the analysis of individuals of African descent, 43 unique sites were significant only in that population (Tables S8 and S9). We examined these sites for interaction in the WHI non-Hispanic white and African American individuals. We found that five CpG sites had a nominally significant interaction with BMI by race/ethnicity (Table 2, Figure S1). In three CpG sites (cg15391590, cg27113059, cg25652701), we excluded one outlier and tested whether the interaction remained. Results remained significant after the exclusion of individual outliers (cg15391590 pinteraction = 0.032, cg27113059 pinteraction = 0.011, cg25652701 pinteraction = 0.020), though none were significant after Bonferroni adjustment for 43 tests. Two of these sites were quantitative trait methylation loci in the GTP cohort: cg25212453 negatively associated with expression of TNFRSF13B and COCH and cg08122652 negatively associated with expression of LGALS3BP and OTOF (Table S10).
Table 2.
Interaction between BMI and race/ethnicity in WHI
| CpG Site | Main effect estimate (BMI) | Interaction effect estimate | Interaction SE | Interaction Z score |
Interaction p value |
|---|---|---|---|---|---|
| cg25652701 | −8.00E−5 | −5.8E−04 | 1.4E−04 | −4.29 | 1.8E−05 |
| cg25212453 | −1.91E−4 | 5.01E−04 | 1.6E−04 | 3.05 | 0.0022 |
| cg08122652 | −2.30E−2 | 8.9E−04 | 3.6E−04 | 2.47 | 0.014 |
| cg27113059 | 8.87E−5 | −2.3E−04 | 9.59E−05 | −2.36 | 0.018 |
| cg15391590 | −1.29E−4 | −2.4E−04 | 1.1E−04 | −2.18 | 0.029 |
Interaction term reflects increase or decrease in slope for African Americans compared to non-Hispanic White individuals.
We next explored the potential of DNA methylation to predict BMI using the 1,238 CpG sites from the replication analysis. After model tuning using elastic-net regression in a training set (n = 3,858), 398 sites were selected for the model (Table S11). These sites accounted for 32% of the variance in BMI in the test set (median absolute deviation = 0.040, n = 964). The addition of age, race/ethnicity, physical activity, and cell composition as predictors only marginally improved the adjusted R2 (Table 3). In the combined training and test set (n = 4,822), these sites accounted for 36% of the variance in BMI. For comparison, we constructed a similar predictor in our WHI training set using the 83 CpG sites identified by Mendelson et al.18 and examined its performance. In the combined training and test set, a prediction score based on these 83 CpG sites accounted for 29% of the variance in BMI.
Table 3.
Predicting BMI from DNA methylation using elastic net regression
| Predictors | Root-mean-square error (RMSE) | Adjusted R2 | Median absolute deviation |
|---|---|---|---|
| 398 CpG sites | 0.0702 | 0.3169 | 0.0401 |
| 398 CpG sites + age | 0.0699 | 0.3229 | 0.0355 |
| 398 CpG sites + age + ethnicity | 0.0699 | 0.3474 | 0.0354 |
| 398 CpG sites + age + ethnicity + cell composition | 0.0699 | 0.3473 | 0.0356 |
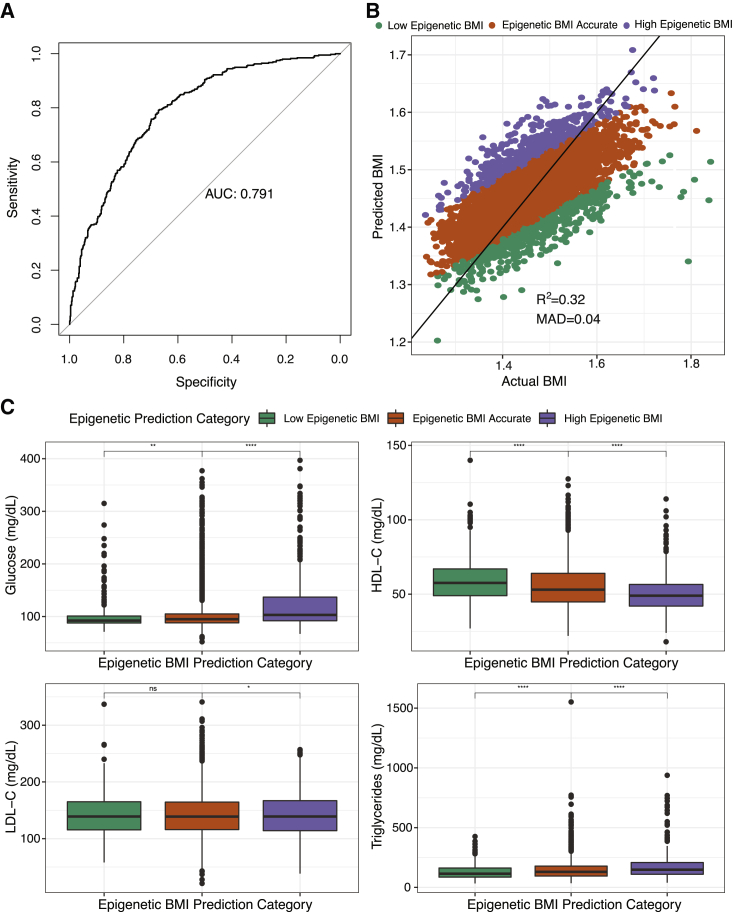
We next assessed the potential of this DNA methylation-based BMI score to predict obesity, defined as BMI > 30. In our test set (n = 964), the area under the curve was 0.79, and using a cutoff of 30 for our BMI score, the sensitivity was 0.82 and the specificity was 0.62 (Figure 2A). Individuals were then categorized based on how well methylation predicted BMI. On average, DNA methylation tended to underpredict BMI in the test set (Figure 2B). Individuals with high epigenetic BMI had 20.5 mg/dL higher blood glucose (SE = 2.0, p = 2.7E−24), 31 mg/dL higher triglycerides (SE = 4.3, p = 6.9E−13), 4.3 mg/dL lower HDL cholesterol (SE: 0.68, p = 1.4E−10), and 3.3 mg/dL lower LDL cholesterol (SE = 2.0, p = 0.047) compared with accurate predicted BMI. In contrast, individuals with low epigenetic BMI had 5.2 mg/dL lower blood glucose (SE = 2.2, p = 0.02), 23.7 mg/dL lower triglycerides (SE = 4.8, p = 1.4E−08), and 3.0 mg/dL higher HDL cholesterol (Figure 2C, Table 4, SE = 0.8, p = 0.0004) compared to accurate predicted BMI. We found consistent results in two sensitivity analyses. Significant results remained when using the 80% and 20% distribution of the epigenetic score to define groups. We also examined whether the residual between predicted BMI and actual BMI associated with cardiometabolic markers and found consistent results with two exceptions: LDL cholesterol did not associate with the residual value and waist circumference positively associated with the residual value (Table S12).
Figure 2.
Epigenetic BMI accurately predicts BMI
(A) Receiver operating characteristic curve showing the performance of the DNA methylation prediction score identifying obesity. AUC denotes area under the curve. Y axis is the sensitivity (true positive rate) and the x axis is specificity (1-false positive rate).
(B) Scatterplot of predicted BMI from elastic net regression of 398 CpG sites by actual BMI. Individuals categorized based on the residual of predicted BMI regressed on actual BMI. MAD denotes median absolute deviation between predicted and actual BMI.
(C) Boxplot of the association between epigenetic prediction category and blood glucose (mg/dL), high-density lipoprotein (HDL-C, mg/dL), low-density lipoprotein (LDL-C, mg/dL), and triglycerides (mg/dL). Significance defined as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Table 4.
Association between epigenetic BMI groups and log-normalized cardiometabolic risk factors
| Estimate | SE | p value | |
|---|---|---|---|
| Waist circumference (cm, n = 4,356) | |||
| High epigenetic BMI | 5.88E−03 | 4.07E−03 | 0.14 |
| Low epigenetic BMI | 9.04E−04 | 3.98E−03 | 0.82 |
| Blood glucose (mg/dL, n = 3,823) | |||
| High epigenetic BMI | 6.00E−02 | 5.80E−03 | 2.7E−24 |
| Low epigenetic BMI | −1.86E−02 | 6.48E−03 | 0.002 |
| Blood triglycerides (mg/dL, n = 3,829) | |||
| High epigenetic BMI | 7.63E−02 | 1.04E−02 | 6.9E−13 |
| Low epigenetic BMI | −6.34E−02 | 1.15E−02 | 1.4E−08 |
| HDL cholesterol (mg/dL, n = 3,832) | |||
| High epigenetic BMI | −3.75E−02 | 5.39E−03 | 1.4E−12 |
| Low epigenetic BMI | 2.46E−02 | 5.99E−03 | 4.0E−04 |
| LDL cholesterol (mg/dL, n = 3,740) | |||
| High epigenetic BMI | −1.19E−02 | 6.05E−03 | 0.050 |
| Low epigenetic BMI | −239E−03 | 6.71E−03 | 0.72 |
Model adjusted for race/ethnicity, smoking status, age, and physical activity.
We conducted several sensitivity analyses. We first examined how the results changed in a leave-one-out meta-analysis (Table S13). Excluding the results from Wahl et al.3 led to the largest reduction in significant sites resulting in 536 significant CpG sites, likely due to a reduction in power. We next compared results obtained using Z score vs. IVW meta-analysis in cohorts with the same exposure-outcome relationship (ARIC, RODAM, BHS White, and BHS Black). In the IVW and Z score meta-analysis of these four studies, 1,939 CpG sites and 1,433 CpG sites, respectively, were significantly associated with BMI (p < 1E−7) with 935 overlapping sites among methods. Among the sites identified significant in either analysis, the correlation between the test statistics obtained using Z score vs. IVW meta-analysis was 0.98. The meta test statistics tend to be smaller when identified using weighted sum of Z score meta-analysis, suggesting our main results may be more conservative than what would be obtained using IVW meta-analysis. Finally, in the main EWAS, we examined how the results changed when adjusted for diet, physical activity, and income status. Overall, 1,161, 1,167 and 1,160 CpG sites remained associated with BMI when additional covariates were included to adjust for diet quality, physical activity, and income, respectively.
Discussion
We performed a large meta-EWAS of BMI, identifying a unique methylomic signature of BMI. The majority of the sites associated with BMI in the discovery cohort (99%) were replicated in the WHI and were found to associate with genes enriched for several metabolic and inflammatory pathways. Moreover, we found five CpG sites that are differentially associated with BMI between non-Hispanic White individuals and African Americans, two of which may play a role in gene expression. Finally, we constructed a score based on 398 CpG sites that had reasonable predictive ability for BMI and was associated with several cardiometabolic risk factors. Individuals whose measured BMI was higher than predicted by their methylome were found to have poorer metabolic health including higher blood glucose and triglycerides and lower HDL cholesterol compared to individuals whose BMI was accurately predicted.
In this study we identified 1,238 CpG sites that were significantly associated with BMI in several race/ethnicity groups. 329 of the 1,238 CpG sites were associated with differential gene expression in Kennedy et al.,39 and the differentially expressed transcripts were highly enriched for immune response pathways, particularly the adaptive immune response, with the top pathways regulating B and T cell signaling. This is consistent with previous studies reporting that genes near BMI-associated CpG sites are enriched for immune pathways.18,41 Low-grade inflammation is a hallmark of obesity and can lead to significant metabolic dysregulation.42 Mechanistic studies have identified DNA methylation as a key player in promoting macrophage polarization in response to obesity, with more M1 macrophages associated with obesity.43
We also examined how associations between DNA methylation and BMI differed when stratified by race/ethnicity. Racial and ethnic differences in adiposity have been well established. While African Americans have been found to have higher risk for cardiovascular diseases compared to non-Hispanic White individuals, they have consistently been shown to have lower visceral adipose tissue and lower body fat percentage compared to non-Hispanic White individuals.44,45 Of the five CpG sites with a significant interaction between BMI and race/ethnicity, two sites were associated with differential expression in four mRNA transcripts related to inflammatory pathways and hearing. Two of these genes, TNFRSF13B and LGALS3BP, have been found to regulate NF-κB signaling and to be upregulated with obesity,46,47,48 and methylation of CpG sites near LGALS3BP was highly associated with increased BMI both in our study and another.18 Our study found a positive and less negative association between BMI and methylation in cg25212453 and cg08122652, respectively (in WHI African Americans compared to non-Hispanic White individuals) and a negative association between methylation in these two sites and expression in LGALS3BP and TNFRSF13B (in GTP). This could be consistent with a scenario where the upregulation of these genes typically accompanying obesity is not as severe in African Americans compared to non-Hispanic White individuals, suggesting a potentially advantageous effect on inflammatory profiles in African Americans. Low-grade inflammation in obesity leads to significant metabolic dysregulation.42 However, there is some epidemiological data that suggests individuals of African descent may not be as prone to increased inflammatory profiles when living with obesity.49,50 Noting that our interaction analysis was limited in sample size, our study may help provide some mechanistic explanation to these differences in the relationship between inflammation and adiposity in individuals of African descent. Investigation in larger samples is warranted to further explore this idea.
We also found that DNA methylation was predictive of BMI, with the score we developed based on 398 CpG sites explaining 32% of the variance in BMI in an independent test set. Previous studies constructing scores based on smaller samples have been able to explain between 4.7% and 18% of the variance in BMI.6,18,23,24 DNA methylation has been found to be an accurate predictor of current BMI and a poor predictor of future BMI.24 Outliers in the epigenetic BMI model predicted a unique phenotype. Individuals with high epigenetic BMI or whose BMI was overpredicted by the epigenetic markers had poorer cardiometabolic markers compared to accurately predicted BMI. Because this occurs across the full range of BMI, this may suggest that epigenetic BMI prediction may be identifying individuals with poor health regardless of their BMI and that these sites may be useful biomarkers to examine further. Our findings related to LDL cholesterol were inconsistent with other cardiometabolic markers. We found that individuals with high epigenetic BMI had lower LDL cholesterol as compared to individuals with accurate epigenetic BMI; however, these results were only nominally significant (p = 0.0497) and so should be interpreted with caution.
Our study had several important limitations. We focused on BMI as a measure of adiposity due to its availability in many large datasets. However, examining more sensitive measures of adiposity such as body fat percentage may provide greater insight into relevant pathways and could ameliorate our analysis of outliers since these results may be due to limitations of BMI. Our discovery analysis was stratified based on race/ethnicity based on self-report. Thus, it is unclear whether we are identifying molecular differences due to ancestry or social construct. Additionally, while we were able to explore stratified analyses based on self-reported race/ethnicity, our analyses in individuals of African descent (n = 3,134) were substantially smaller compared to those of European descent (n = 11,220), further highlighting a well-known issue of genomic research under-representing African populations.51 Moreover, these populations, which include African Americans, Ghanaians, and European-residing Ghanaians, are not homogeneous in genetic ancestry, living environment, lifestyles, and other factors. Our interaction and expression analyses were conducted in African American populations from the WHI and GTP, so these results may be generalizable only for this population. In particular, the racial disparities in the US may be an underlying cause of these results, as opposed to differences in ancestry. For example, in the US, African Americans are much more likely to live in poverty compared to non-Hispanic White individuals.52 In our results, we may be identifying compensatory mechanisms of structural racism which may be driven by environmental exposures, for example, ambient particulate matter exposure, stress, and lack of access to health care, as well as obesity. Another potential limitation is that the training and test set in our prediction analyses come from the same population (WHI). Future research efforts could test this model in another population to examine the reproducibility of these findings.
Overall, this study yields several important discoveries. We identified novel sites associated with BMI and found suggestive evidence for a unique molecular profile associated with obesity in individuals of African descent. We additionally found that epigenetic markers can predict BMI well and may be able to distinguish individuals whose metabolic health does not align with their BMI. Future studies should examine whether BMI-associated methylation is differential by metabolic health status.
Acknowledgments
We would like to acknowledge and thank Drs. Sonia Shah and Peter Visscher for sharing summary statistics which have been included in the discovery portion of this analysis. The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through 75N92021D00001, 75N92021D00002, 75N92021D00003, 75N92021D00004, and 75N92021D00005. WHI EMPC (AS315) was supported by NIEHS grant R01-ES020836 (L.H., A.A.B., E.A.W.). WHI AS311 was supported by American Cancer Society award 125299-RSG-13–100-01-CCE (P.B.). WHI-BAA23 was supported by NHLBI Broad Agency Announcement contract HHSN268201300006C. The RODAM study was supported by the Intramural Research Program of the National Human Genome Research Institute of the National Institutes of Health (NIH) through the Center for Research on Genomics and Global Health (CRGGH) and by the European Commission under the Framework Programme (grant number 278901). The CRGGH is also supported by the National Institute of Diabetes and Digestive and Kidney Diseases and the Office of the Director at the NIH (Z01HG200362). The Atherosclerosis Risk in Communities study has been funded in whole or in part with federal funds from the National Heart, Lung, and Blood Institute (NHLBI), National Institutes of Health, Department of Health and Human Services (contract numbers HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700004I, and HHSN268201700005I). The authors thank the staff and participants of the ARIC study for their important contributions. Funding was also supported by NIH 5RC2HL102419 and R01NS087541. Additional acknowledgments can be found in the supplemental information.
Author contributions
W.L.D., K.M.V.N., and K.N.C. conceived of the study. W.L.D., K.M.V.N., L.R.S., A.D.S., and A.K.S. developed the methods. E.D., W.G., S.L., W.C., D.S., R.M., P.-A.D., K.M., A.A., and C.A. computed and shared the summary statistics from the participating studies in the discovery analysis. L.H., S.H., T.L.A., P.B., K.M.J., E.A.W., and A.A.B. curated and processed the data used in the replication analyses. A.H.S., J.E.M., and R.N. provided cohort expertise for the replication data. W.L.D. performed all statistical analyses with support from K.N.C. W.L.D. drafted the manuscript with content expertise and review from all authors. All authors read and approved of the final manuscript.
Declaration of interests
K.M.J. is an employee of Bristol Myers Squibb.
Published: January 16, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.12.014.
Web resources
European Genome-Phenome Archive, https://ega-archive.org/
Supplemental information
Data and code availability
Summary statistics analyzed in our discovery meta-analysis were previously generated in six previously published EWASs.1,2,3,4,5,6 Summary statistics from Wahl et al.3 were accessed from the European Genome-Phenome Archive (accession number EGAS00001001922). For the other five EWASs, summary statistics for top sites in each study are available as supplemental data in the relevant publications,1,2,4,5,6 and complete summary statistics were obtained from the corresponding author of each study. For the three WHI ancillary datasets (AS311, AS315, and BA23) analyzed in our replication study, data are available through WHI via paper proposals (https://www.whi.org/md/working-with-whi-data), and data from BA23 are also available via dbGaP (accession code phs001335.v2.p3). The code used to generate these findings is available from the corresponding author upon reasonable request.
References
- 1.Demerath E.W., Guan W., Grove M.L., Aslibekyan S., Mendelson M., Zhou Y.-H., Hedman Å.K., Sandling J.K., Li L.-A., Irvin M.R., et al. Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Hum. Mol. Genet. 2015;24:4464–4479. doi: 10.1093/hmg/ddv161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meeks K.A.C., Henneman P., Venema A., Burr T., Galbete C., Danquah I., Schulze M.B., Mockenhaupt F.P., Owusu-Dabo E., Rotimi C.N., et al. An epigenome-wide association study in whole blood of measures of adiposity among Ghanaians: the RODAM study. Clin. Epigenetics. 2017;9:103. doi: 10.1186/s13148-017-0403-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wahl S., Drong A., Lehne B., Loh M., Scott W.R., Kunze S., Tsai P.-C., Ried J.S., Zhang W., Yang Y., et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541:81–86. doi: 10.1038/nature20784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun D., Zhang T., Su S., Hao G., Chen T., Li Q.-Z., Bazzano L., He J., Wang X., Li S., Chen W. Body Mass Index Drives Changes in DNA Methylation. Circ. Res. 2019;125:824–833. doi: 10.1161/CIRCRESAHA.119.315397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geurts Y.M., Dugué P.A., Joo J.E., Makalic E., Jung C.H., Guan W., Nguyen S., Grove M.L., Wong E.M., Hodge A.M., et al. Novel associations between blood DNA methylation and body mass index in middle-aged and older adults. Int. J. Obes. 2018;42:887–896. doi: 10.1038/ijo.2017.269. [DOI] [PubMed] [Google Scholar]
- 6.Shah S., Bonder M.J., Marioni R.E., Zhu Z., McRae A.F., Zhernakova A., Harris S.E., Liewald D., Henders A.K., Mendelson M.M., et al. Improving Phenotypic Prediction by Combining Genetic and Epigenetic Associations. Am. J. Hum. Genet. 2015;97:75–85. doi: 10.1016/j.ajhg.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Organisation for Economic Co-operation and Development . 2017. Obesity Update 2017.https://www.oecd.org/health/obesity-update.htm [Google Scholar]
- 8.World Health Organization . WHO; 2016. Obesity and Overweight.https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight [Google Scholar]
- 9.Asghar A., Sheikh N. Role of immune cells in obesity induced low grade inflammation and insulin resistance. Cell. Immunol. 2017;315:18–26. doi: 10.1016/j.cellimm.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 10.O'Flanagan C.H., Bowers L.W., Allot E.H., Hursting S.D. Vol. 10. IARC Working Group; 2017. Molecular and metabolic mechanisms underlying the obesity–cancer link. (Energy Balance and Obesity). [Google Scholar]
- 11.Hall J.E., do Carmo J.M., da Silva A.A., Wang Z., Hall M.E. Obesity-Induced Hypertension. Circ. Res. 2015;116:991–1006. doi: 10.1161/CIRCRESAHA.116.305697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narayan K.M.V., Boyle J.P., Thompson T.J., Gregg E.W., Williamson D.F. Effect of BMI on Lifetime Risk for Diabetes in the U.S. Diabetes Care. 2007;30:1562–1566. doi: 10.2337/dc06-2544. [DOI] [PubMed] [Google Scholar]
- 13.Khan S.S., Ning H., Wilkins J.T., Allen N., Carnethon M., Berry J.D., Sweis R.N., Lloyd-Jones D.M. Association of Body Mass Index With Lifetime Risk of Cardiovascular Disease and Compression of Morbidity. JAMA Cardiol. 2018;3:280–287. doi: 10.1001/jamacardio.2018.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith Z.D., Meissner A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 15.Greenberg M.V.C., Bourc’his D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019;20:590–607. doi: 10.1038/s41580-019-0159-6. [DOI] [PubMed] [Google Scholar]
- 16.Horvath S., Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 2018;19:371–384. doi: 10.1038/s41576-018-0004-3. [DOI] [PubMed] [Google Scholar]
- 17.Fransquet P.D., Wrigglesworth J., Woods R.L., Ernst M.E., Ryan J. The epigenetic clock as a predictor of disease and mortality risk: a systematic review and meta-analysis. Clin. Epigenetics. 2019;11:62. doi: 10.1186/s13148-019-0656-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mendelson M.M., Marioni R.E., Joehanes R., Liu C., Hedman Å.K., Aslibekyan S., Demerath E.W., Guan W., Zhi D., Yao C., et al. Association of Body Mass Index with DNA Methylation and Gene Expression in Blood Cells and Relations to Cardiometabolic Disease: A Mendelian Randomization Approach. PLoS Med. 2017;14:e1002215. doi: 10.1371/journal.pmed.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al Muftah W.A., Al-Shafai M., Zaghlool S.B., Visconti A., Tsai P.C., Kumar P., Spector T., Bell J., Falchi M., Suhre K. Epigenetic associations of type 2 diabetes and BMI in an Arab population. Clin. Epigenetics. 2016;8:13. doi: 10.1186/s13148-016-0177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aslibekyan S., Demerath E.W., Mendelson M., Zhi D., Guan W., Liang L., Sha J., Pankow J.S., Liu C., Irvin M.R., et al. Epigenome-wide study identifies novel methylation loci associated with body mass index and waist circumference. Obesity. 2015;23:1493–1501. doi: 10.1002/oby.21111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campanella G., Gunter M.J., Polidoro S., Krogh V., Palli D., Panico S., Sacerdote C., Tumino R., Fiorito G., Guarrera S., et al. Epigenome-wide association study of adiposity and future risk of obesity-related diseases. Int. J. Obes. 2005;42:2022–2035. doi: 10.1038/s41366-018-0064-7. [DOI] [PubMed] [Google Scholar]
- 22.Breton C.V., Marsit C.J., Faustman E., Nadeau K., Goodrich J.M., Dolinoy D.C., Herbstman J., Holland N., LaSalle J.M., Schmidt R., et al. Small-Magnitude Effect Sizes in Epigenetic End Points are Important in Children's Environmental Health Studies: The Children's Environmental Health and Disease Prevention Research Center's Epigenetics Working Group. Environ. Health Perspect. 2017;125:511–526. doi: 10.1289/EHP595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCartney D.L., Hillary R.F., Stevenson A.J., Ritchie S.J., Walker R.M., Zhang Q., Morris S.W., Bermingham M.L., Campbell A., Murray A.D., et al. Epigenetic prediction of complex traits and death. Genome Biol. 2018;19:136. doi: 10.1186/s13059-018-1514-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reed Z.E., Suderman M.J., Relton C.L., Davis O.S.P., Hemani G. The association of DNA methylation with body mass index: distinguishing between predictors and biomarkers. Clin. Epigenetics. 2020;12:50. doi: 10.1186/s13148-020-00841-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am. J. Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 26.Milne R.L., Fletcher A.S., MacInnis R.J., Hodge A.M., Hopkins A.H., Bassett J.K., Bruinsma F.J., Lynch B.M., Dugué P.A., Jayasekara H., et al. Cohort Profile: The Melbourne Collaborative Cohort Study (Health 2020) Int. J. Epidemiol. 2017;46 doi: 10.1093/ije/dyx085. 1757–1757i. [DOI] [PubMed] [Google Scholar]
- 27.Tigchelaar E.F., Zhernakova A., Dekens J.A.M., Hermes G., Baranska A., Mujagic Z., Swertz M.A., Muñoz A.M., Deelen P., Cénit M.C., et al. Cohort profile: LifeLines DEEP, a prospective, general population cohort study in the northern Netherlands: study design and baseline characteristics. BMJ Open. 2015;5:e006772. doi: 10.1136/bmjopen-2014-006772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deary I.J., Gow A.J., Taylor M.D., Corley J., Brett C., Wilson V., Campbell H., Whalley L.J., Visscher P.M., Porteous D.J., Starr J.M. The Lothian Birth Cohort 1936: a study to examine influences on cognitive ageing from age 11 to age 70 and beyond. BMC Geriatr. 2007;7:28. doi: 10.1186/1471-2318-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berenson G.S., Bogalusa Heart Study Investigators Bogalusa Heart Study: a long-term community study of a rural biracial (Black/White) population. Am. J. Med. Sci. 2001;322:293–300. [PubMed] [Google Scholar]
- 30.Agyemang C., Beune E., Meeks K., Owusu-Dabo E., Agyei-Baffour P., Aikins A.d.G., Dodoo F., Smeeth L., Addo J., Mockenhaupt F.P., et al. Rationale and cross-sectional study design of the Research on Obesity and type 2 Diabetes among African Migrants: the RODAM study. BMJ Open. 2014;4:e004877. doi: 10.1136/bmjopen-2014-004877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holle R., Happich M., Löwel H., Wichmann H.E., MONICA/KORA Study Group KORA--a research platform for population based health research. Gesundheitswesen. 2005;67(Suppl 1):S19–S25. doi: 10.1055/s-2005-858235. [DOI] [PubMed] [Google Scholar]
- 32.Chambers J.C., Loh M., Lehne B., Drong A., Kriebel J., Motta V., Wahl S., Elliott H.R., Rota F., Scott W.R., et al. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. Lancet Diabetes Endocrinol. 2015;3:526–534. doi: 10.1016/S2213-8587(15)00127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bendinelli B., Masala G., Saieva C., Salvini S., Calonico C., Sacerdote C., Agnoli C., Grioni S., Frasca G., Mattiello A., et al. Fruit, vegetables, and olive oil and risk of coronary heart disease in Italian women: the EPICOR Study. Am. J. Clin. Nutr. 2011;93:275–283. doi: 10.3945/ajcn.110.000521. [DOI] [PubMed] [Google Scholar]
- 34.The Women’s Health Initiative Study Group Design of the Women’s Health Initiative clinical trial and observational study. Controlled Clin. Trials. 1998;19:61–109. doi: 10.1016/s0197-2456(97)00078-0. [DOI] [PubMed] [Google Scholar]
- 35.Houseman E.A., Accomando W.P., Koestler D.C., Christensen B.C., Marsit C.J., Nelson H.H., et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee C.H., Cook S., Lee J.S., Han B. Comparison of Two Meta-Analysis Methods: Inverse-Variance-Weighted Average and Weighted Sum of Z-Scores. Genomics Inform. 2016;14:173–180. doi: 10.5808/GI.2016.14.4.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 38.Woolf B. On estimating the relation between blood group and disease. Ann. Hum. Genet. 1955;19:251–253. doi: 10.1111/j.1469-1809.1955.tb01348.x. [DOI] [PubMed] [Google Scholar]
- 39.Kennedy E.M., Goehring G.N., Nichols M.H., Robins C., Mehta D., Klengel T., Eskin E., Smith A.K., Conneely K.N. An integrated -omics analysis of the epigenetic landscape of gene expression in human blood cells. BMC Genom. 2018;19:476. doi: 10.1186/s12864-018-4842-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Do W.L., Gohar J., McCullough L.E., Galaviz K.I., Conneely K.N., Narayan K.M.V. Examining the association between adiposity and DNA methylation: A systematic review and meta-analysis. Obes. Rev. 2021;22:e13319. doi: 10.1111/obr.13319. [DOI] [PubMed] [Google Scholar]
- 41.Do W.L., Nguyen S., Yao J., Guo X., Whitsel E.A., Demerath E., Rotter J.I., Rich S.S., Lange L., Ding J., et al. Associations between DNA methylation and BMI vary by metabolic health status: a potential link to disparate cardiovascular outcomes. Clin. Epigenetics. 2021;13:230. doi: 10.1186/s13148-021-01194-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reilly S.M., Saltiel A.R. Adapting to obesity with adipose tissue inflammation. Nat. Rev. Endocrinol. 2017;13:633–643. doi: 10.1038/nrendo.2017.90. [DOI] [PubMed] [Google Scholar]
- 43.Wang X., Cao Q., Yu L., Shi H., Xue B., Shi H. Epigenetic regulation of macrophage polarization and inflammation by DNA methylation in obesity. JCI Insight. 2016;1:e87748. doi: 10.1172/jci.insight.87748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Després J.P. Body Fat Distribution and Risk of Cardiovascular Disease. Circulation. 2012;126:1301–1313. doi: 10.1161/CIRCULATIONAHA.111.067264. [DOI] [PubMed] [Google Scholar]
- 45.Dugas L.R., Cao G., Luke A.H., Durazo-Arvizu R.A. Adiposity is not equal in a multi-race/ethnic adolescent population: NHANES 1999-2004. Obesity. 2011;19:2099–2101. doi: 10.1038/oby.2011.52. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y., Kent J.W., Jr., Olivier M., Ali O., Broeckel U., Abdou R.M., Dyer T.D., Comuzzie A., Curran J.E., Carless M.A., et al. QTL-based association analyses reveal novel genes influencing pleiotropy of metabolic syndrome (MetS) Obesity. 2013;21:2099–2111. doi: 10.1002/oby.20324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu G., Xia Z., Deng F., Liu L., Wang Q., Yu Y., Wang F., Zhu C., Liu W., Cheng Z., et al. Inducible LGALS3BP/90K activates antiviral innate immune responses by targeting TRAF6 and TRAF3 complex. PLoS Pathog. 2019;15:e1008002. doi: 10.1371/journal.ppat.1008002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Challa T.D., Straub L.G., Balaz M., Kiehlmann E., Donze O., Rudofsky G., Ukropec J., Ukropcova B., Wolfrum C. Regulation of De Novo Adipocyte Differentiation Through Cross Talk Between Adipocytes and Preadipocytes. Diabetes. 2015;64:4075–4087. doi: 10.2337/db14-1932. [DOI] [PubMed] [Google Scholar]
- 49.Doumatey A.P., Lashley K.S., Huang H., Zhou J., Chen G., Amoah A., Agyenim-Boateng K., Oli J., Fasanmade O., Adebamowo C.A., et al. Relationships Among Obesity, Inflammation, and Insulin Resistance in African Americans and West Africans. Obesity. 2010;18:598–603. doi: 10.1038/oby.2009.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hakim O., Bello O., Ladwa M., Peacock J.L., Umpleby A.M., Charles-Edwards G., Amiel S.A., Goff L.M. The Link between Obesity and Inflammatory Markers in the Development of Type 2 Diabetes in Men of Black African and White European Ethnicity. Nutrients. 2020;12:3796. doi: 10.3390/nu12123796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fatumo S., Chikowore T., Choudhury A., Ayub M., Martin A.R., Kuchenbaecker K. A roadmap to increase diversity in genomic studies. Nat. Med. 2022;28:243–250. doi: 10.1038/s41591-021-01672-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carnethon M.R., Pu J., Howard G., Albert M.A., Anderson C.A.M., Bertoni A.G., Mujahid M.S., Palaniappan L., Taylor H.A., Willis M., et al. Cardiovascular Health in African Americans: A Scientific Statement From the American Heart Association. Circulation. 2017;136:e393–e423. doi: 10.1161/CIR.0000000000000534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Summary statistics analyzed in our discovery meta-analysis were previously generated in six previously published EWASs.1,2,3,4,5,6 Summary statistics from Wahl et al.3 were accessed from the European Genome-Phenome Archive (accession number EGAS00001001922). For the other five EWASs, summary statistics for top sites in each study are available as supplemental data in the relevant publications,1,2,4,5,6 and complete summary statistics were obtained from the corresponding author of each study. For the three WHI ancillary datasets (AS311, AS315, and BA23) analyzed in our replication study, data are available through WHI via paper proposals (https://www.whi.org/md/working-with-whi-data), and data from BA23 are also available via dbGaP (accession code phs001335.v2.p3). The code used to generate these findings is available from the corresponding author upon reasonable request.