Abstract
Pseudomonas aeruginosa and species of Acinetobacter calcoaceticus-baumanii complex are multiresistant intrahospital opportunistic pathogens, able to acquire carbapenemases and produce outbreaks with high morbidity and mortality. Pseudomonas putida has also emerged with similar characteristics. The aim of this research was to characterize the Metallo-β-lactamases (MBLs) detected by surveillance in Paraguay in the first 5 years of their circulation in hospitals. The coexistence of KPC and OXA-type carbapenemases was also investigated. 70 MBL-producing strains from inpatients were detected from clinical samples and rectal swab from 11 hospitals. The strains were identified by manual, automated, and molecular methods. Antimicrobial susceptibility was studied by Kirby-Bauer and automated methods, while colistin susceptibility was determined by broth macrodilution. MBLs were investigated by synergy with EDTA against carbapenems and PCR, and their variants by sequencing. KPC and OXA-carbapenemases were investigated by PCR. Clonality was studied by pulsed-field gel electrophoresis (PFGE). The results demonstrated the circulation of blaVIM-2 (60%), blaNDM-1 (36%), and blaIMP-18 (4%). The MBL-producing species were P. putida (45.7%), P. aeruginosa (17.2%), A. baumannii (24.3%), A. pittii (5.7%), A. nosocomialis, (4.3%) A. haemolyticus (1.4%), and A. bereziniae (1.4%). PFGE analysis showed one dominant clone for A. baumannii, a predominant clone for half of the strains of P. aeruginosa, and a polyclonal spread for P. putida. In the first 5 years of circulation in Paraguay, MBLs were disseminated as unique variants per genotype, appeared only in Pseudomonas spp. and Acinetobacter spp., probably through horizontal transmission between species and vertical by some successful clones.
Keywords: MBL, Pseudomonas aeruginosa, Pseudomonas putida, Acinetobacter spp., PFGE
Introduction
Gram-negative bacteria resistant to carbapenems, such as Acinetobacter baumannii and Pseudomonas aeruginosa, are emerging causes of health care-associated infections and of global public health concerns [1]. Pseudomonas putida has also emerged as a multiresistant nosocomial opportunistic pathogen [2–4]. The most widespread mechanism of carbapenem resistance is the production of carbapenemases [5]. These enzymes can be classified according to their molecular characteristics into classes A, B, and D. The classes A (ex. KPC) and D (the OXAs) carbapenemases include enzymes that hydrolyze their substrates forming an acyl-enzyme through a serine of the active site, while the class B β-lactamases are metalloenzymes or metallo-β-lactamases (MBLs) that use at least one zinc ion from the active site to facilitate the hydrolysis of beta-lactam. The most important types of MBLs due to epidemiological spread and clinical relevance are IMP, VIM [6], and NDM [7]. SPM is characteristic of P. aeruginosa in Brazil [8].
The surveillance of MBLs dissemination in Paraguay began in 2009, with 21 laboratories members of the survaillance of the Antimicrobial Resistance Network dependent on the Central Laboratory of Public Health—Ministry of Public Health and Social Welfare. The aim of this study was to characterize the MBLs detected among Pseudomonas spp. and A. calcoaceticus-baumannii complex isolates in Paraguay in the first 5 years of their circulation in hospitals. The characterization involved identification of species and clonality of MBL-producing strains, determination of genotype and subtype of MBL, and coexistence of KPC and OXA-type carbapenemases in the strains.
Materials and methods
From November 2009 to December 2015, clinical carbapenem-resistant isolates of Pseudomonas spp. and A. calcoaceticus-baumannii complex collected from hospitalized patients from 11 Paraguayan hospitals were sent to the Central Laboratory of Public Health in Paraguay and submitted to phenotypic detection of MBL by inhibition with EDTA [9]. Only one isolate for each patient was included for further studies, except in one case where two isolates with different resistance phenotypes, recovered from tracheal secretion within 4 months of difference were included.
Species identification was performed by classical and automated methods (Vitek 2 Compact–Biomerieux, France). For A. calcoaceticus-baumannii complex, in addition to this a multiplex PCR was performed to detect the presence of blaOXA-51 according to the protocol described in the literature, which allows the detection of blaOXA-23; blaOXA-24 and blaOXA-58 [10]. The blaOXA-51 negative strains were subjected to species-specific PCR for Acinetobacter pittii and Acinetobacter nosocomialis according to published protocol [11]. Isolates of Acinetobacter spp. that were negative for blaOXA-51 and species-specific PCR were subjected to rpoβ amplification PCR [12] and sequencing.
Antimicrobial susceptibility tests were performed by automated method (Vitek 2 Compact-Biomerieux, France). Susceptibility to colistin was determined by broth macrodilution [13]. The results were interpreted according to the CLSI manual [13].
On surveillance, routine genotyping of MBLs was performed by PCR for blaIMP, blaVIM, and blaNDM according to the protocol of the regional reference laboratory [14], however, for this work, the genotypes were verified according to multiplex PCR [15]. The blaSPM—for P. aeruginosa strains—was investigated according to the published protocol [16]. The detection of the Serino-carbapenemase gene blaKPC was performed for P. aeruginosa isolates [17].
To determine the variant of each MBL genotype, PCR and subsequent sequencing were made according to the published protocol for IMP and VIM [18]. To determine NDM variants, primers designed by the Laboratório de Pesquisa em Infecção Hospitalar (LAPIH) from the Oswaldo Cruz Institute-Fiocruz, Brazil [19] were used (Table 1).
Table 1.
Primers used in this work
| Primer | Gene | Sequence 5′ → 3′ | Amplicon size (pb) | Ref | |
|---|---|---|---|---|---|
| A. pittii | Apit-F | rpoβ | TGGGCAGTTACCAGATTGACCTA | 147 | [11] |
| Apit-R | AACCAGCAGCTTCCATTTGACG | ||||
| A. nosocomialis | Anos-F | rpoβ | GCCGCTCGTGAACGTGTAATC | 394 | [11] |
| Anos-R | CATCGTGTGGCATATCTTCAAC | ||||
| Amplification of rpoB | Ac696- F | rpoβ | TAYCGYAAAGAYTTGAAAGAAG | 350 | [12] |
| Ac1093-R | CMACACCYTTGTTMCCRTGA | ||||
| MBL genotyping | VIM-F | blaVIM | AGTGGTGAGTATCCGACAG | 261 | [15] |
| VIM-R | ATGAAAGTGCGTGGAGAC | ||||
| IMP-UF | blaIMP | GGYGTTTWTGTTCATACWTCKTTYGA | 404 | [15] | |
| IMP-UR | GGYARCCAAACCACTASGTTATCT | ||||
| NDM-F | blaNDM | AGCACACTTCCTATCTCGAC | 512 | [15] | |
| NDM-R | GGCGTAGTGCTCAGTGTC | ||||
| SPM-F | blaSPM | AGACCGCGATTTCTATTCTT | 505 | [16] | |
| SPM-R | AGTTCCTTCGGCTTTATCAT | ||||
| OXA genotyping | OXA-51 F | blaOXA- 51 | TAATGCTTTGATCGGCCTTG | 353 | [10] |
| OXA-51 R | TGGATTGCACTTCATCTTGG | ||||
| OXA-23 F | blaOXA- 23 | GATCGGATTGGAGAACCAGA | 501 | [10] | |
| OXA-23 R | ATTTCTGACCGCATTTCCAT | ||||
| OXA-24 F | blaOXA- 24 | GGTTAGTTGGCCCCCTTAAA | 246 | [10] | |
| OXA-24 R | AGTTGAGCGAAAAGGGGATT | ||||
| OXA 58-F | blaOXA- 58 | AAGTATTGGGGCTTGTGCTG | 599 | [10] | |
| OXA 58-R | CCCCTCTGCGCTCTACATAC | ||||
| KPC screening | KPC-F | blaKPC | AACAAGGAATATCGTTGATG | 916 | [17] |
| KPC-R | AGATGATTTTCAGAGCCTTA | ||||
| MBL variants | VIM1-F | blaVIM-1 | TGTTAAAAGTTATTAGTAGTTTATTG | 801 | [18] |
| VIM1-R | CTACTCGGCGACTGAGC | ||||
| VIM2-F | blaVIM-2 | ATGTTCAAACTTTTGAGTAAG | 801 | [18] | |
| VIM2-R | CTACTCAACGACTGAGCG | ||||
| IMP1-F | blaIMP-1 | ATGAGCAAGTTATCTGTATTC | 741 | [18] | |
| IMP1-R | TTAGTTGCTTGGTTTTGATGG | ||||
| IMP2-F | blaIMP-2 | ATGAAGAAATTATTTGTTTTATG | 741 | [18] | |
| IMP2-R | TTAGTTACTTGGCTGTGATG | ||||
| NDM-F | blaNDM | CGAAGCTGAGCACCGCATTA | 764 | [19] | |
| NDM-R | TCAGCGCAGCTTGTCGGC |
Macrogen Inc.-Korea performed sequencing. Using the Genbank database through the BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi) the sequences were analyzed, and further studied by multiple alignments with BioEdit software comparing them with reference sequences available from NCBI.
To perform PFGE, the PulseNet protocol was used [20]. Restriction enzymes (Invitrogen by Thermo Fisher Scientific) and PFGE running conditions for P. aeruginosa, P. putida and Acinetobacter spp. (protocols not included in PulseNet Network) were taken from the scientific literature [21, 22] with adjusted running times, 18.8 h for Acinetobacter spp. and 18.7 h for Pseudomonas spp. Salmonella serovar Braenderup H9812 (restricted with Xba I) was used as the standard strain. The images of gels were analyzed (Gel Doc EZ Imager-BioRad) using Gel Compare II Software (Applied Maths) to obtain the relationship between patterns. We determine clusters using the Unweighted Pair Group Mean (UPGMA) method, with 90% Dice’s similarity coefficients among patterns to define each clonal group.
Results
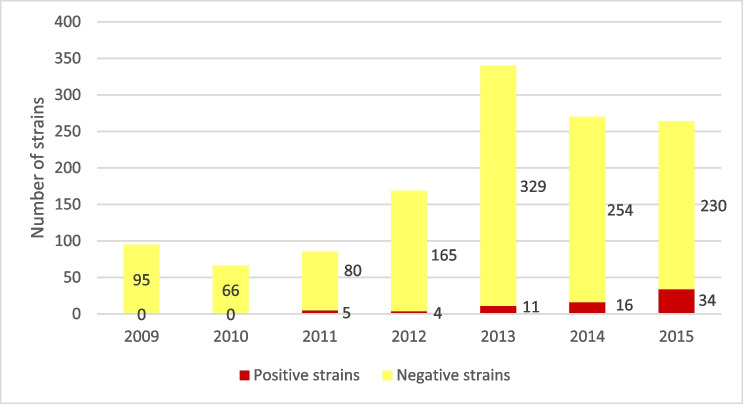
Out of 1289 clinical carbapenem-resistant isolates of Pseudomonas spp. and A. calcoaceticus-baumannii complex studied between 2009 and 2015, 70 strains were MBL positive (5.4%). The first MBL-positive strain appeared in 2011 and the prevalence was 5.8% in 2011, 2.4% in 2012, 3.2% in 2013, 5.9% in 2014, and 12.8% in 2015 (see Fig. 1).
Fig. 1.
Number of MBL-positive isolates out of the total strains studied per year
The blaVIM (n = 60%) was the most frequently MBL gene detected, followed by blaNDM (n = 36%) and blaIMP (n = 4%). No blaSPM or blaKPC genes were detected. Only the blaOXA-51 was detected in 65% of Acinetobacter spp. isolates (17 strains), since in A. pittii (4 strains), A. nosocomialis (3 strains), A. bereziniae (1 strain), and A. haemolyticus (1 strain) OXA-type carbapenemases were not detected.
Regarding geographical origin, all the positive strains came from the Capital and Central Department of Paraguay. Concerning the source, 67% came from several clinical samples, and 33% from rectal swabs. blaNDM was detected mainly in rectal swabs (64%), blaVIM was mainly associated with clinical samples (83.3%) and the only three blaIMP were isolated from blood culture, cerebrospinal fluid, and pleural fluid, respectively.
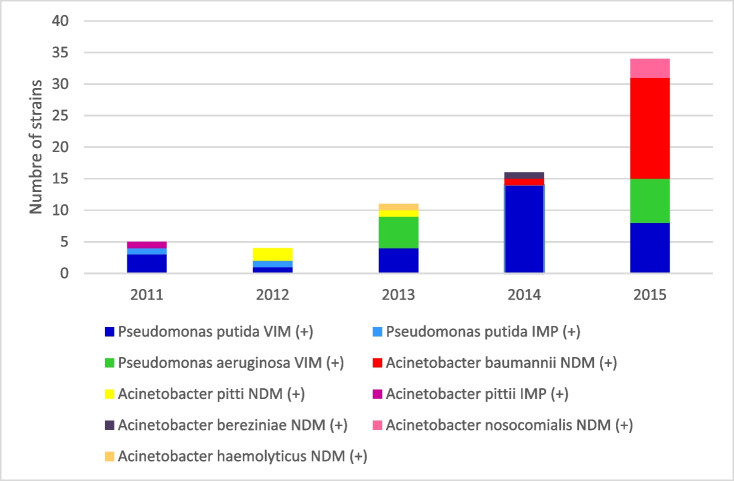
About the association between MBL genotype and bacterial species, blaVIM was detected in P. aeruginosa (n = 12) and P. putida (n = 30). blaIMP was detected in two P. putida strains and one A. pittii strain, while blaNDM appeared only in species of Acinetobacter spp.: A. baumannii (n = 17), A. nosocomialis (n = 3), A. pittii (n = 3), A. haemolyticus (n = 1) and A. bereziniae (n = 1). Figure 2 shows the proportion of detection of each genotype with respect to the carrier species and the time. blaVIM was the most prevalent genotype until 2014, mainly associated with P. putida.
Fig. 2.
Number of MBL-positive isolates per year of each bacterial species according to MBL genotype
Regarding the MBL variant genes, all blaNDM sequences aligned 100% with the reference sequence blaNDM-1 (GenBank FN396876.1), blaVIM, 38/42 strains aligned 99 to 100% with the reference sequence blaVIM-2 (GenBank FN396876.1) (it was not possible to determine the blaVIM allelic variant from four isolates, because they were not viable at the time of sequencing). Concerning to blaIMP, the sequence of one P. putida isolate and A. pittii corresponded to blaIMP-18. The blaIMP allelic variant of one P. putida was not sequenced due to lack of viability too.
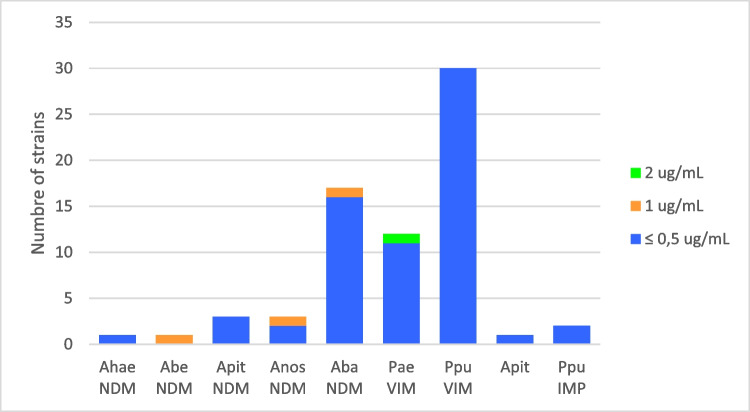
Concerning resistance profile (antibiotype), it was variable, and Table 2 shows the summary. The definition of resistance classification was according to Magiorakos et al. [23]. Only colistin maintained 100% susceptibility in all isolates (MIC range ≤ 0.5 to 1 μg/mL, MIC50 and MIC90 = 0.5 μg/mL) (Fig. 3). Regarding P. aeruginosa, less than 50% were also susceptible to amikacin (5 strains), aztreonam (4 strains), and/or ciprofloxacin (3 strains). Concerning to P. putida, most of the isolates remained susceptible to amikacin (97%). All Acinetobacter spp. remained susceptible to tigecycline; some strains were also susceptible to amikacin (46%), gentamicin (42%), ciprofloxacin (38.5%), trimethoprim-sulfamethoxazole (35%), and ampicillin-sulbactam (11.6%). P. putida has a higher proportion of XDR (78%) and VIM antibiotypes II and III coincide with some of the IMP. Several of the isolates share the same pattern. The antibiotype mentioned in Table 2 is the same as the dendrograms that appear later (Figs. 4, 5, 6, 7, and 8).
Table 2.
Antimicrobial resistance characteristics of the MBL-positive strains
| Species | Genotype | Nr. of strains | Resistance pattern (not-sensitive) | Sensitive pattern | AT | RP |
|---|---|---|---|---|---|---|
| P. aeruginosa | VIM | |||||
| 7 | CAZ, IPM, MPM, PIT, CPM, ATM, GEN, CIP,AMI | COL | I | XDR | ||
| 2 | CAZ, IPM, MPM, PIT, CPM, GEN, CIP | ATM,AMI,COL | II | MDR | ||
| 1 | CAZ, IPM, MPM, PIT, CPM, ATM, GEN | CIP,AMI,COL | III | MDR | ||
| 2 | CAZ, IPM, MPM, PIT, CPM, GEN | ATM,CIP,AMI,COL | IV | MDR | ||
| P. putida | VIM | 1 | CAZ, IPM, MPM, PIT, CPM, ATM, GEN, CIP,AMI | COL | I | XDR |
|
VIM IMP |
13 1 |
CAZ, IPM, MPM, PIT, CPM, ATM, GEN, CIP CAZ, IPM, MPM, PIT, CPM, ATM, GEN, CIP |
AMI,COL AMI,COL |
II II |
XDR XDR | |
|
VIM IMP |
9 1 |
CAZ, IPM, MPM, PIT, CPM, ATM, CIP CAZ, IPM, MPM, PIT, CPM, ATM, CIP |
GEN,AMI,COL GEN,AMI,COL |
III III |
XDR XDR |
|
| VIM | 4 | CAZ, IPM, MPM, PIT, CPM, ATM, GEN | CIP,AMI,COL | IV | MDR | |
| VIM | 2 | CAZ, IPM, MPM, PIT, CPM, GEN, CIP | ATM, AMI,COL | V | MDR | |
| VIM | 1 | CAZ, IPM, MPM, PIT, CPM, GEN | ATM,CIP,AMI,COL | VI | MDR | |
|
Acinetobacter spp. |
NDM | 13 Ab | CAZ,IPM, MPM, PIT, CPM, GEN, AMI, CIP, SUT,AMS | COL,TGC | I | XDR |
| 2 Ab | CAZ,IPM, MPM, PIT, CPM, GEN, CIP, SUT, AMS | COL,AMI,TGC | II | MDR | ||
| 1 An | CAZ,IPM, MPM, PIT, CPM, SUT, AMS | COL,GEN,AMI,CIP, TGC | III | MDR | ||
| 1 Ab | CAZ,IPM, MPM, PIT, CPM, CIP, SUT | COL,GEN,AMI,AMS,TGC | IV | MDR | ||
| 7 Ap, An, Ab, Abe * | CAZ,IPM, MPM, PIT, CPM | COL,GEN,AMI,CIP, SUT,TGC | V | MDR | ||
| 1 Ah | CAZ,IPM, MPM, PIT, CPM, AMI | COL,GEN,CIP,SUT, AMS,TGC | VI | MDR | ||
| IMP | 1 Ap | CAZ,IPM, PIT, CPM, AMS | MPM,COL,GEN,AMI,CIP,SUT, AMS,TGC | VII | MDR | |
AbA. baumannii, AnA. nosocomialis, AbeA. bereziniae, ApA. pittii, AhA. haemolyticus, *7: Ap n = 3, An n = 2, Ab n = 1, Abe n = 1. AT antibiotype, RP resistance profile, CAZ ceftazidime, CPM cefepime, IPM imipenem, MPM meropenem, PIT piperacillin/tazobactam, ATM aztreonam, GEN gentamicin, AMI amikacin, CIP ciprofloxacin, COL colistin, SUT trimethoprim/sulphametoxazole, AMS ampicillin/sulbactam, TGC tigecycline
Fig. 3.
Minimal inhibitory concentrations of colistin. Abbreviations: Ahae: A. haemolyticus, Abe: A. bereziniae, Apit: Acinetobacter pittii, Anos: A. nosocomialis, Aba: A. baumannii, Pae: P. aeruginosa, Pput: P. putida
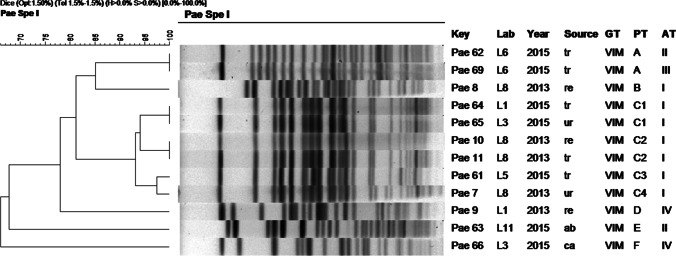
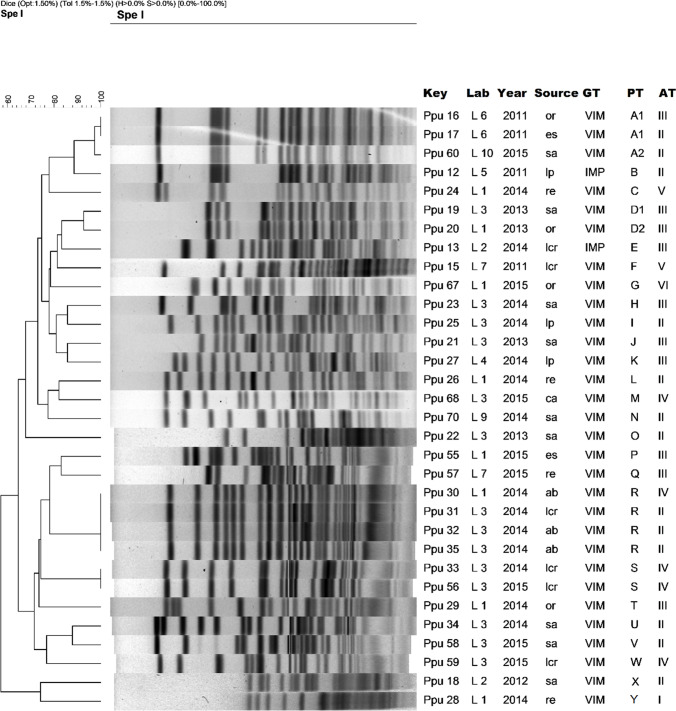
Fig. 4.
Dendrogram of molecular typing of P. aeruginosa isolates. Abbreviations: tr: traqueal, re: rectal swab, ur: urine, ab: abdominal fluid, ca: catheter, Lab: origin (hospital laboratory) GT: genotype, PT: pulsotype, AT: antibiotype
Fig. 5.
Dendrogram of molecular typing of P. putida strains. Abbreviations: ur: urine, sp: sputum, bl: blood, pl: pleural fluid, re: rectal swab, tr: traqueal, cf: cerebrospinal fluid, ca: catheter, ab: abdominal fluid, GT: genotype, PT: pulsotype, AT: antibiotype
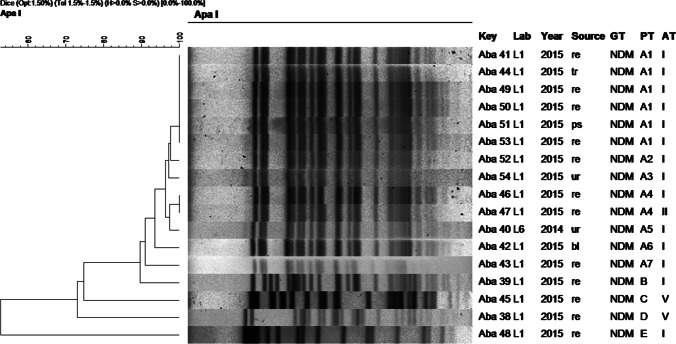
Fig. 6.
Dendrogram of molecular typing of A. baumannii isolates. Abbreviations: ur: urine, re: rectal swab, ps: ps: secretion, tr: traqueal, bl: blood, GT: genotype, PT: pulsotype, AT: antibiotype
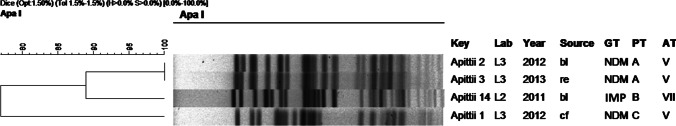
Fig. 7.
Dendrogram of molecular typing of A. pittii strains. Abbreviations: bl: blood, re: rectal swab, cf: cerebrospinal fluid, GT: genotype, PT: pulsotype, AT: antibiotype. Obs: The strains A. pittii 1 and A. pittii 2 were studied in a previous publication [17]
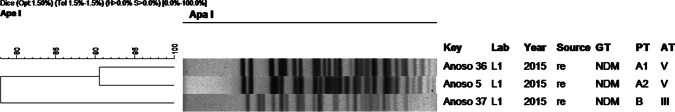
Fig. 8.
Dendrogram of molecular typing of A. nosocomialis isolates. Abreviations: re: rectal swab, GT: genotype, PT: pulsotype, AT: antibiotype
About molecular typing with PFGE, VIM-2-producing P. aeruginosa (Fig. 4) revealed six different patterns or pulsotypes and four antibiotypes. Six isolates (50%) belong to dominant pulsotype C, including four subtypes. These strains were isolated in 2013 and 2015, came from four different hospitals and had the same antibiotype. The strains were isolated from both clinical samples and rectal swabs. Despite presenting different antibiotypes, the two isolates from the same patient (Pae 62 and Pae 69 isolates) belonged to the same pulsotype (pattern A). Figure 4 shows the dendrogram and complementary data of the strains studied.
PFGE analysis for P. putida (Fig. 5) showed 25 different pulsotypes in the VIM-producing strains with only six antibiotypes, and no large clonal group was observed. However, there are isolates from different hospitals belonging to the same pulsotype. The IMP-producing strains belonged to different clones than VIM-producing isolates but share some of the antibiotypes. The strains came from both clinical samples and rectal swabs.
The dendrogram for NDM-producing A. baumannii isolates (Fig. 6) revealed five different pulsotypes (A-E) and three antibiotypes (I, II, and V). Pulsotype A including seven subtypes (A1-A7) was found in 13 strains, 12 of them from the same hospital (L1), and all except one with the same antibiotype. The first NDM-producing A. baumannii isolate detected in 2014 recovered from a different hospital (L6) also belonged to the pulsotype A. The four remaining strains presented different patterns. In this group of strains, 12/17 came from rectal swabs.
Molecular typing with PFGE for A. pittii (Fig. 7) revealed three different pulsotypes (A–C) for two antibiotypes, one of them also present in A. baumannii (V, see Fig. 6). The first two NDM-producing A. pittii isolates, recovered from the same hospital, belonged to different pulsotypes (pulsotype A and C), already investigated in a previous publication [24]. Pulsotype A has with two closely related strains from the same hospital, isolated in different years, and the B and C pulsotypes are observed in one strain each. All NDM-positive strains showed the same antibiotypes, and they came from the same hospital. The IMP positive strain shows its own pulsotypes and antibiotype.
A. nosocomialis strains revealed two different pulsotypes (Fig. 8) and two antibiotypes, one of them (V) also present in A. baumannii and A. pittii. Two strains were grouped as A1 and A2 subtypes, while the third strain have a different pulsotype. All strains were isolated from the rectal swab.
Two strains were not included in the previous dendrograms, since they were typified as A. haemolyticus (antibiotype VI present in this single strain) and A. bereziniae (antibiotype V also present in A. baumannii and A. pittii).
Discussion
There are few studies published about intrahospital carbapenemases in Paraguayan non-fermenting gram-negative rods using molecular methods. One of them is from Pasteran et al. [24] which includes our strains A. piitii 1 and A. pitti 2 (see Fig. 7). Pasteran et al. identified those two strains as A. pittii by MALDI-TOF and were not clonally related by PFGE. These findings are consistent with our results. The study published by Rodriguez et al. [25] including A. baumannii paraguayan strains among others, found only OXA-23 carbapenemase and no MBL. None of these strains is part of the present work. In 2021, Melgarejo et al. [26] studying fermenting and non-fermenting gram-negative bacilli in Paraguay, found strains of A. baumannii with OXA 51, OXA 23, NDM, and NDM + OXA 58, and P. aeruginosa with NDM, but all those strains were isolated in 2021. Our work shows the beginning and the first years of MBLs dissemination in non-fermenting gram-negative bacilli and their behavior over time. Since then, MBL-producing strains have not ceased to be detected and as expected, the isolates increased gradually. It is possible to affirm that in Paraguay the MBL began to appear in BGNNF to spread years later in fermenting gram-negative bacilli also since the first publication in an Enterobacterales is from the year 2016 [27].
Regarding the MBL-producing species, P. putida, P. aeruginosa and A. baumannii, were the most prevalent in this investigation, as in another studies [8]. As expected, A. baumannii turned out to be the most prevalent of its genus (65%), followed by A. pittii and A. nosocomialis. This last two species, as well as A. haemolyticus and A. bereziniae, less prevalent, have been sporadically described as species involved in hospital infections [28, 29]. The most frequent association between bacterial species and genotype was observed with P. putida and VIM (43%), followed by A. baumannii associated with NDM (24%). It is interesting to note that NDM was detected in Acinetobacter spp. from 2011 to 2014 in “non-baumannii” species (A. pittii, A. haemolyticus, A. nosocomialis, A. bereziniae). However, appeared in A. baumannii in 2015 with practically absolute predominance.
Although IMP, VIM, and NDM genotypes are the most frequent MBLs in the world, [7, 30], the frequency is variable, characteristic of each country, region, or hospital, consequently, studies published, show a variable prevalence [31, 32]. Before the description of NDM, the most frequently detected MBLs in the world were IMP and VIM type, being VIM the most predominant [5, 30]. In our study, the distribution percentage found was 60% VIM, 36% NDM, and 4% IMP, but the prevalence varied over to the years. In fact, the VIM-producing, the most frequently isolated until 2014, was surpassed in 2015 by NDM associated to A. baumannii evidencing the great dissemination capacity of the mechanism in this genomespecies. In 2015 by the way, NDM was considered to becoming the most commonly and distributed carbapenemase worldwide [30].
Regarding the allelic variants described, VIM-1 and VIM-2 are the most frequent, reported in Europe [7, 33] and Asia [7]. In Latin American countries VIM-2 has been mostly detected [7] in Chile and Venezuela [34], Argentina [35], Brazil [36], Uruguay [37], and other countries, especially in P. aeruginosa followed by P. fluorescens. In this study, we demonstrate the circulation of VIM-2, associated with P. putida and P. aeruginosa. The allelic variant NDM-1, found in the present study, has been reported around the world, including Latin American countries such as Colombia, where it was detected in A. baumannii and A. nosocomialis [38], NDM-1 was also confirmed in A. baumannii [39] and in A. pittii [40] in Brazil. The IMP has been described with a lower prevalence than others. IMP-1 was the first variant described, followed later by IMP-2 [41], subsequently more variants were described. In this study, the IMP-18 subtype was detected in a lower prevalence. This variant has been detected in P. aeruginosa in Brazil [42].
In this work, only OXA-51, intrinsic to A. baumannii, was detected. None other OXA were found associated with MBL. In fact, the coexistence of OXA-type carbapenemase with NDM is unusual but has been documented [43].
Multiple clonalities of P. putida has been reported [44]. Some studies reported several P. putida clones with the same VIM-2 transposon in plasmids, together with a high proportion of MBL-producing strains compared to P. aeruginosa, suggesting that P. putida is a reservoir of these elements MDR transferable [44]. This situation could have been observed in this study, where in 2014 P. aeruginosa was not isolated and 43% of the VIM-2-producing strains were P. putida versus 17% of P. aeruginosa.
European reports suggest intrahospital transmission of VIM associated with P. aeruginosa [45]. Publications from Argentina [35], Spain [45], Colombia [46], and Brazil [47] show polyclonal propagation as well as spread by successful clones. In our study, we observed intra and out-off-hospital spread of the predominant clonal group. An outbreak would have started in one hospital, lasted 2 years and spread at two other hospitals.
Clonal diversity as well as dissemination of successful clonal groups of A. baumannii has been demonstrated in several studies [48, 49]. All this has been verified in the present work. NDM-producing A. baumannii presented a group of strains with a common clonal origin (13/17 isolates), all epidemiologically related, since 12 were from the same hospital (L1) and from the same year (2015). This could describe an outbreak in a hospital (L1) that would have originated in another hospital (L6), which had only one closely related isolate (95% similarity).
With this study, we ascertain the intra and interhospital dissemination of MBL-type carbapenemases, through polyclonal and successful clones capable of generating outbreaks, evidenced with the pulsotype A of A. baumannii, pulsotypes C of P. aeruginosa and R of P. putida. We also observed evidence of dissemination by mobile genetic elements due the presence of the same MBL variant in different species like VIM-2 in both P. aeruginosa and P. putida, IMP-18 in both P. putida and A. pitti, and NDM-1 in different Acinetobacter species. The presence of the same genotype variant and the same antibiotype in clonally unrelated strains suggests the spread of resistance by mobile genetic elements. It will be very interesting to compare this work with the subsequent evolution of carbapenemases in non-fermenting gram-negative rods in Paraguay, and the study of mobile genetic elements in more recent times.
Obviously, it is necessary to maintain and strengthen the surveillance of MBLs, as well as in-hospital containment measures to avoid the dissemination of these resistance mechanisms and others that could be successfully transferred through mobile genetic elements sometimes associated with successful clones, both within the same hospital, as between hospitals in Paraguay. The transfer of patients from one hospital to another or the fact that in Paraguay health personnel work at the same time in different hospitals, could be part of the problem of the spread of colonizing bacteria of intrahospital origin.
Acknowledgements
We gratefully thank all professionals who are members of the Resistance Surveillance Network of the Ministry of Public Health and Social Welfare of Paraguay for their interest and participation in science and surveillance, and for sending strains for study by their own means. Our sincere thanks to Claudia Candia Ibarra, former Technical Coordinator, and Norma Colucci, former General Coordinator of Unidad Coordinadora de Proyectos del Laboratorio Central de Salud Pública (Project Coordination Unit of the Central Public Health Laboratory) for their invaluable support.
Funding
This work was supported by grants from the IEBAS-FOCEM Project “Research, Education and Biotechnologies applied to Health” (agreement COF 03/11).
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethical approval
Ethical approval was received from the Institutional Ethics Committee (IEC) of the Central Laboratory of Public Health, (approval no. CEI / LCSP 41/030314).
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
Responsible Editor: Luis Nero
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Rossana Franco, Email: rossanafranco@gmail.com.
Ivson Cassiano de Oliveira Santos, Email: ivsoncass@gmail.com.
Mario Fabián Martinez Mora, Email: mfmarmora@gmail.com.
Patricia Violeta Araújo López, Email: patriciamicologiaparaguay@gmail.com.
Vivian Estela Takahasi Alvarez, Email: vivitakahasi@gmail.com.
Flavia Helena Ortiz Arce, Email: flavia.arce@gmail.com.
Graciela Lird, Email: gracielalird@hotmail.com.
Marlene Silvagni, Email: mar.sil@live.com.
Anibal Kawabata, Email: bhatayazz2204@hotmail.com.
María Carolina Rojas Fariña, Email: carolinarojasfar@gmail.com.
Mirna Fabiola Agüero Fernández, Email: fabiolaguero@gmail.com.
Thamirys R. Tavares e Oliveira, Email: thamirysrachel@gmail.com.
Claudio M. Rocha-de-Souza, Email: claudio.rocha@ioc.fiocruz.br
Ana Paula D’ Alincourt Carvalho Assef, Email: anapdca@ioc.fiocruz.br.
References
- 1.World Health Organization. Guidelines for the prevention and control of carbapenem-resistant Enterobacteriaceae, Acinetobacter baumannii, and Pseudomonas aeruginosa in health care facilities. [Internet]. 2017 [cited 2020 Mar 16]. http://www.ncbi.nlm.nih.gov/books/NBK493061/ [PubMed]
- 2.Kim SE, Park SH, Park HB, Park KH, Kim SH, Jung SI, et al. Nosocomial Pseudomonas putida Bacteremia: high rates of carbapenem resistance and mortality. Chonnam Med J. 2012;48(2):91–95. doi: 10.4068/cmj.2012.48.2.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan G, Xi Y, Yuan P, Sun Z, Yang D. Risk factors and antimicrobial resistance profiles of Pseudomonas putida infection in Central China, 2010–2017. Med (Baltimore) 2019;98(44):e17812. doi: 10.1097/MD.0000000000017812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fernández M, Porcel M, de la Torre J, Molina-Henares MA, Daddaoua A, Llamas MA, et al. Analysis of the pathogenic potential of nosocomial Pseudomonas putida strains. Front Microbiol [Internet]. 2015 [cited 2022 Jul 12];6. https://www.frontiersin.org/articles/10.3389/fmicb.2015.00871 [DOI] [PMC free article] [PubMed]
- 5.Bush K, Jacoby GA. Updated functional classification of β-lactamases. Antimicrob Agents Chemother. 2010;54(3):969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Molecular diversity of extended-spectrum β-lactamases and carbapenemases, and antimicrobial resistance | Journal of Intensive Care | Full Text [Internet]. [cited 2020 Jul 28]. https://jintensivecare.biomedcentral.com/articles/10.1186/s40560-020-0429-6 [DOI] [PMC free article] [PubMed]
- 7.Kazmierczak KM, Rabine S, Hackel M, McLaughlin RE, Biedenbach DJ, Bouchillon SK, et al. Multiyear, multinational survey of the incidence and global distribution of metallo-β-lactamase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2016;60(2):1067–1078. doi: 10.1128/AAC.02379-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Labarca JA, Salles MJC, Seas C, Guzmán-Blanco M. Carbapenem resistance in Pseudomonas aeruginosa and Acinetobacter baumannii in the nosocomial setting in Latin America. Crit Rev Microbiol. 2014;27:1–17. doi: 10.3109/1040841X.2014.940494. [DOI] [PubMed] [Google Scholar]
- 9.Protocolo-WHONET-XIII-Taller-Mar-del-Plata1.pdf [Internet]. [cited 2020 Apr 27]. http://antimicrobianos.com.ar/ATB/wp-content/uploads/2011/01/Protocolo-WHONET-XIII-Taller-Mar-del-Plata1.pdf
- 10.Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27(4):351–353. doi: 10.1016/j.ijantimicag.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Li XM, Choi JA, Choi IS, Kook JK, Chang YH, Park G, et al. Development and evaluation of species-specific PCR for detection of nine Acinetobacter species. Ann Clin Lab Sci. 2016;46(3):270–278. [PubMed] [Google Scholar]
- 12.Gundi VAKB, Dijkshoorn L, Burignat S, Raoult D, La Scola B. Validation of partial rpoB gene sequence analysis for the identification of clinically important and emerging Acinetobacter species. Microbiol. 2009;155(7):2333–2341. doi: 10.1099/mic.0.026054-0. [DOI] [PubMed] [Google Scholar]
- 13.Weinstein MP (2019) Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing
- 14.Protocolo de PCR para la detección de carbapenemasas | antimicrobianos.com.arantimicrobianos.com.ar [Internet]. [cited 2022 Jul 13]. http://antimicrobianos.com.ar/2013/?cat=52
- 15.Detección MBL-Multiplex | antimicrobianos.com.arantimicrobianos.com.ar [Internet]. [cited 2020 Apr 27]. http://antimicrobianos.com.ar/2017/01/3708/
- 16.Pasterán F, Rapoport M, Petroni A, Faccone D, Corso A, Galas M, et al. Emergence of PER-2 and VEB-1a in Acinetobacter baumannii strains in the Americas. Antimicrob Agents Chemother. 2006;50(9):3222–3224. doi: 10.1128/AAC.00284-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pasteran FG, Otaegui L, Guerriero L, Radice G, Maggiora R, Rapoport M, et al. Klebsiella pneumoniae Carbapenemase–2, Buenos Aires. Argentina Emerg Infect Dis. 2008;14(7):1178–1180. doi: 10.3201/eid1407.070826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Characterization of the new metallo-β-lactamase VIM-13 and its integron-borne gene from a Pseudomonas aeruginosa clinical isolate in Spain | Antimicrobial Agents and Chemotherapy [Internet]. [cited 2020 Apr 27]. https://aac.asm.org/content/52/10/3589.long [DOI] [PMC free article] [PubMed]
- 19.Da Silva IR, Aires CAM, Conceição-Neto OC, de Oliveira Santos IC, Ferreira Pereira N, Moreno Senna JP, et al. Distribution of clinical NDM-1-producing gram-negative bacteria in Brazil. Microb Drug Resist. 2019;25(3):394–399. doi: 10.1089/mdr.2018.0240. [DOI] [PubMed] [Google Scholar]
- 20.Standard operating procedure for pulsenet pfge of escherichia coli O157:H7, Escherichia coli non-O157 (STEC), Salmonella serotypes, shigella sonnei and shigella flexneri :16
- 21.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutiérrez O, Juan C, Cercenado E, Navarro F, Bouza E, Coll P, et al. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa isolates from Spanish hospitals. Antimicrob Agents Chemother. 2007;51(12):4329–4335. doi: 10.1128/AAC.00810-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 24.Pasteran F, Mora MM, Albornoz E, Faccone D, Franco R, Ortellado J, et al. Emergence of genetically unrelated NDM-1-producing Acinetobacter pittii strains in Paraguay. J Antimicrob Chemother. 2014;69(9):2575–2578. doi: 10.1093/jac/dku139. [DOI] [PubMed] [Google Scholar]
- 25.Rodríguez CH, Balderrama Yarhui N, Nastro M, Nuñez Quezada T, Castro Cañarte G, Magne Ventura R, et al. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii in South America. J Med Microbiol. 2016;65(10):1088–1091. doi: 10.1099/jmm.0.000328. [DOI] [PubMed] [Google Scholar]
- 26.Melgarejo-Touchet N, Brítez CM, Busignani S, Falcón M, López E, Laconich M, et al. Caracterización molecular de carbapenemasas en bacilos gramnegativos circulantes en hospitales de Paraguay Primer cuatrimestre 2021. Mem Inst Investig En Cienc Salud. 2021;19(2):49–58. doi: 10.18004/mem.iics/1812-9528/2021.019.02.49. [DOI] [Google Scholar]
- 27.Kiese MR, Ortiz H, Almada P, Arguello R, Melgarejo N, Martínez C, et al. Escherichia coli metalobetalactamasa en un hospital de alta complejidad en Paraguay. Rev Virtual Soc Paraguaya Med Interna. 2016;1:120–123. doi: 10.18004/rvspmi/2312-3893/2016.03(02)120-123. [DOI] [Google Scholar]
- 28.Turton JF, Shah J, Ozongwu C, Pike R. Incidence of Acinetobacter Species Other than A baumannii among Clinical Isolates of Acinetobacter: Evidence for Emerging Species. J Clin Microbiol. 2010;48(4):1445–9. doi: 10.1128/JCM.02467-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Álvarez-Buylla A, Culebras E, Picazo JJ. Identification of Acinetobacter species: is Bruker biotyper MALDI-TOF mass spectrometry a good alternative to molecular techniques? Infect Genet Evol. 2012;12(2):345–349. doi: 10.1016/j.meegid.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 30.Patel G, Bonomo RA. “Stormy waters ahead”: global emergence of carbapenemases. Front Microbiol [Internet]. 2013 Mar 14 [cited 2020 Apr 27];4. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3596785/ [DOI] [PMC free article] [PubMed]
- 31.Rossi Gonçalves I, Dantas RCC, Ferreira ML, da Batistão DWF, Gontijo-Filho PP, Ribas RM. Carbapenem-resistant Pseudomonas aeruginosa: association with virulence genes and biofilm formation. Braz J Microbiol. 2016;48(2):211–7. doi: 10.1016/j.bjm.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reddy RM, Kumar S, Kumar M. Characterization of VIM and IMP Metallo-β-lactamases (MBL) in Pseudomonas aeruginosa isolated in a tertiary care hospital. Int J Curr Microbiol Appl Sci. 2017;6(3):1461–1467. doi: 10.20546/ijcmas.2017.603.167. [DOI] [Google Scholar]
- 33.Fritsche TR, Sader HS, Toleman MA, Walsh TR, Jones RN. Emerging metallo-b-lactamase–mediated resistances: a summary report from the Worldwide SENTRY Antimicrobial Surveillance Program :3 [DOI] [PubMed]
- 34.Mendes RE, Castanheira M, Garcia P, Guzman M, Toleman MA, Walsh TR, et al. First isolation of blaVIM-2 in Latin America: report from the SENTRY Antimicrobial Surveillance Program. Antimicrob Agents Chemother. 2004;48(4):1433–1434. doi: 10.1128/AAC.48.4.1433-1434.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pagniez G, Radice M, Cuirolo A, Rodríguez O, Rodríguez H, Vay C, et al (2006) Prevalencia de metalo-β-lactamasas en Pseudomonas aeruginosa resistentes a carbapenemes en un Hospital Universitario de Buenos Aires. Rev Argent Microbiol 6 [PubMed]
- 36.Sader HS, Castanheira M, Mendes RE, Toleman M, Walsh TR, Jones RN. Dissemination and diversity of metallo-β-lactamases in Latin America: report from the SENTRY Antimicrobial Surveillance Program. Int J Antimicrob Agents. 2005;25(1):57–61. doi: 10.1016/j.ijantimicag.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 37.Ingold AJ, Castro M, Nabón A, Borthagaray G, Márquez C (2011) Detección del gen codificante de la metalo-β-lactamasa VIM-2 en un integrón de clase 1 asociado con el gen blaCTX-M-2 en un aislamiento clínico de Pseudomonas aeruginosa en el Uruguay: primera comunicación. Rev Argent Microbiol 5 [DOI] [PubMed]
- 38.Rojas LJ, Wright MS, De La Cadena E, Motoa G, Hujer KM, Villegas MV, et al. Initial assessment of the molecular epidemiology of blaNDM-1 in Colombia. Antimicrob Agents Chemother. 2016;60(7):4346–4350. doi: 10.1128/AAC.03072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pillonetto M, Arend L, Vespero EC, Pelisson M, Chagas TPG, Carvalho-Assef APD, et al. First report of NDM-1-producing Acinetobacter baumannii sequence type 25 in Brazil. Antimicrob Agents Chemother. 2014;58(12):7592–7594. doi: 10.1128/AAC.03444-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pagano M, Poirel L, Martins AF, Rozales FP, Zavascki AP, Barth AL, et al. Emergence of NDM-1-producing Acinetobacter pittii in Brazil. Int J Antimicrob Agents. 2015;45(4):444–445. doi: 10.1016/j.ijantimicag.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 41.Zhao WH, Hu ZQ. IMP-type metallo-β-lactamases in Gram-negative bacilli: distribution, phylogeny, and association with integrons. Crit Rev Microbiol. 2011;37(3):214–226. doi: 10.3109/1040841X.2011.559944. [DOI] [PubMed] [Google Scholar]
- 42.Picão RC, Andrade SS, Nicoletti AG, Campana EH, Moraes GC, Mendes RE, et al. Metallo-β-lactamase detection: comparative evaluation of double-disk synergy versus combined disk tests for IMP-, GIM-, SIM-, SPM-, or VIM-producing isolates. J Clin Microbiol. 2008;46(6):2028–2037. doi: 10.1128/JCM.00818-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mathlouthi N, El Salabi AA, Ben Jomàa-Jemili M, Bakour S, Al-Bayssari C, Zorgani AA, et al. Early detection of metallo-β-lactamase NDM-1- and OXA-23 carbapenemase-producing Acinetobacter baumannii in Libyan hospitals. Int J Antimicrob Agents. 2016;48(1):46–50. doi: 10.1016/j.ijantimicag.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 44.Juan C, Zamorano L, Mena A, Alberti S, Perez JL, Oliver A. Metallo- -lactamase-producing Pseudomonas putida as a reservoir of multidrug resistance elements that can be transferred to successful Pseudomonas aeruginosa clones. J Antimicrob Chemother. 2010;65(3):474–478. doi: 10.1093/jac/dkp491. [DOI] [PubMed] [Google Scholar]
- 45.(2011) Detección de un brote epidémico por Pseudomonas aeruginosa multirresistente productora de metalo-beta-lactamasa. Rev Clínica Esp 211(4):187–91 [DOI] [PubMed]
- 46.Crespo MP, Woodford N, Sinclair A, Kaufmann ME, Turton J, Glover J, et al. Outbreak of carbapenem-resistant Pseudomonas aeruginosa producing VIM-8, a novel metallo-β-lactamase, in a tertiary care center in Cali. Colombia J Clin Microbiol. 2004;42(11):5094–5101. doi: 10.1128/JCM.42.11.5094-5101.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dantas RCC, Silva RT e, Ferreira ML, Gonçalves IR, Araújo BF, de Campos PA, et al. Molecular epidemiological survey of bacteremia by multidrug resistant Pseudomonas aeruginosa: the relevance of intrinsic resistance mechanisms. PLoS ONE [Internet]. 2017 May 8 [cited 2020 Apr 29];12(5). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5421754/ [DOI] [PMC free article] [PubMed]
- 48.Villalon P, Valdezate S, Medina-Pascual MJ, Rubio V, Vindel A, Saez-Nieto JA. Clonal diversity of nosocomial epidemic Acinetobacter baumannii strains isolated in Spain. J Clin Microbiol. 2011;49(3):875–882. doi: 10.1128/JCM.01026-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Villalón P, Valdezate S, Cabezas T, Ortega M, Garrido N, Vindel A, et al. Endemic and epidemic Acinetobacter baumannii clones: a twelve-year study in a tertiary care hospital. BMC Microbiol [Internet]. 2015 Feb 25 [cited 2020 Apr 29];15. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4352537/ [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.