Graphical abstract
Keywords: Makgeolli, Rice wine, Lactic acid bacteria, Microbial community, Metabolite, Lactobacillus
Highlights
-
•
Commercial non-pasteurized makgeolli generally contained lactic acid bacteria (LAB).
-
•
The most abundant and frequently detected genus in makgeolli was Lactobacillus.
-
•
LAB maintained about 5 log CFU/mL of viable cell number until 30 days of cold storage.
-
•
LAB did not affect the quality of makgeolli during low-temperature storage.
Abstract
Makgeolli, the traditional Korean rice wine, is generally considered to contain lactic acid bacteria (LAB) despite its bacterial inoculation-free brewing process. The existence of LAB in makgeolli often presents inconsistent trends in microbial profiles and cell numbers. Therefore, to establish LAB-related insights, 94 commercial non-pasteurized products were collected and microbial communities and metabolites were analyzed using 16S rRNA amplicon sequencing and GC–MS, respectively. All samples contained various LAB genera and species, with an average viable cell number of 5.61 log CFU/mL. Overall, 10 LAB genera and 25 LAB species were detected; the most abundant and frequent LAB genus was Lactobacillus. There was no significant change in the LAB composition profile or lactic acid content during low-temperature storage, indicating the presence of LAB did not significantly affect the quality of makgeolli under low-temperature storage conditions. Overall, this study contributes to understand the microbial profile and role of LAB in makgeolli.
1. Introduction
Makgeolli is a traditional Korean rice-based, fermented alcoholic beverage. Unlike other fermented liquors like wine and beer, various microorganisms, including molds, yeasts, and bacteria, coexist in makgeolli. Additionally, it has a unique feature wherein saccharification by molds and alcoholic fermentation by yeasts occur simultaneously. The microbiome and mycobiome of makgeolli remain viable in non-pasteurized raw products, which thus have a shorter shelf life, usually 30 days at refrigerated temperatures, compared to that of other fermented liquors.
Lactic acid bacteria (LAB) are generally considered one of the existing microorganisms in makgeolli. In addition to probiotic strain isolation (Lee et al., 2014, Moon et al., 2014, Park et al., 2015), the microbiome of makgeolli and its shifting dynamics over time (Chai et al., 2015, Min et al., 2012) have been reported previously. However, unlike yeasts and molds, which are intentionally inoculated to obtain the intended results, makgeolli contains LAB despite not being inoculated; therefore, the origin, variety, and contribution of LAB to makgeolli remain unclear. There has been little research on which genera or species exist and dominate in makgeolli. To date, the reported viable cell numbers, dominant species, and microbiome composition have been inconsistent, resulting in a lack of standardized trends regarding LAB in makgeolli (Jin et al., 2008, Jung et al., 2012).
LAB are generally considered as contaminants and need to be controlled in alcoholic beverage products (García-Ruiz et al., 2008, Garofalo et al., 2015, Suzuki et al., 2008). However, in makgeolli, existence of LAB is usually considered an advantage and is advertised as a probiotic property, even though the influence and contribution of LAB to the liquor remain unclear. In fact, the microbiome is unstable in response to external stimuli, such as temperature. If the liquor is not stored at low temperatures, the makgeolli microbiome and its metabolism shift toward a phase that proceeds to spoilage (Kim et al., 2011). Therefore, fundamental insights regarding the role of LAB in the quality of makgeolli are needed to establish quality control principles based on their contributions.
To discover the role of LAB in makgeolli, an intrinsic understanding of the existence of LAB including viable cell numbers, microbial profiles, and impact on the quality of makgeolli during fermentation and storage needs to be investigated. Therefore, in this study, we determined the existing LAB in makgeolli, their numbers, genera and species, and their frequencies by monitoring various commercially available non-pasteurized makgeolli products. Microbial profiles and alteration of metabolites in samples during low-temperature storage were monitored using 16S rRNA gene amplicon sequencing and GC–MS-based metabolomic approaches, respectively.
2. Materials and methods
2.1. Sample collection
In total, 94 kinds of commercially available non-pasteurized raw makgeolli products were purchased from local markets in Korea. Samples were collected within 10 days of manufacture to minimize changes during cold storage. All samples were kept in an ice box maintaining low temperature and transported to the laboratory within an hour for analysis. All available information marked on the product labels was recorded, including the product name, alcohol content (%), ingredients (including raw material and type of inoculated starter), and date of manufacture and/or shelf life. At days 10 and 30 from the date of manufacture, samples were collected for viable cell number determination as well as physicochemical, microbial community, and metabolic profile analyses. Except the samples for viable cell count, all collected samples were stored at −80 °C for further analyses.
2.2. Viable cell count of LAB and physiochemical analysis
The pH of each makgeolli sample was measured using a pH meter (pH-250L, ISTEK, Seoul, Korea). Titratable acidity (TA) as lactic acid was measured by titrating 5 mL of each commercial makgeolli sample with 0.1 N NaOH solution to pH 8.3. To determine the viable LAB cell numbers, samples were serially diluted in peptone water and inoculated on 3MTM Petrifilm LAB count plates (3M™ Microbiology, Saint Paul, Minnesota, USA). The colony forming units on each plate were counted after incubation at 30 °C for 48 h. The results are presented as the mean values of three replicate measurements.
2.3. Microbial profiling
For analyzing the microbial community and metabolome, makgeolli samples were centrifuged at 12,000 rpm for 5 min at 4 °C. The precipitates and supernatants were used for 16S rRNA gene amplicon sequencing and GC–MS analysis, respectively. To isolate DNA from the samples, an AccuFAST automated system (AccuGene, Incheon, Korea) was used according to the manufacturer’s instructions. The V3-V4 region of the 16S rRNA gene was amplified as previously described (Yoon et al., 2021) using KAPA HiFi HosStart ReadyMix (Kapa Biosystems, Wilmington, MA, USA) and primers 341f/805r (King et al., 2019) containing Nextera adaptors. The PCR products (∼428 bp) were purified using HiAccuBeads (AccuGene, South Korea). Amplicon libraries pooled at an equimolar ratio were sequenced on an Illumina MiSeq system using the MiSeq Reagent Kit v3 for 600 cycles (Illumina, San Diego, CA, USA).
All raw datasets were denoised by correcting amplicon errors and the exact amplicon sequence variants (ASVs) were inferred using DADA2 v1.16 (Callahan et al., 2016). The SILVA release 138 rRNA reference database (Glöckner et al., 2017) was used to create a naïve Bayes classifier to classify the ASVs obtained from DADA2. Downstream analyses of quality-filtered and chimera-filtered reads were performed using the QIIME2-2022.2 software package (Bolyen et al., 2019). Each sequence obtained from the DADA2 datasets was assigned a taxonomy with a threshold of 99% pairwise identity using QIIME2 workflow scripts and the SILVA release 138 rRNA reference database classifier.
2.4. Metabolic profiling
The protocol for sample derivatization and GC–MS analysis conditions were similar to those of previous study with some modifications (Seo et al., 2016). In brief, 20 μL of sample supernatant, 90 μL of distilled water, and 20 μL of ribitol solution (internal standard, ribitol in water at 0.5 mg/mL) were blended and lyophilized. After preprocessing, 100 μL of methoxyamine hydrochloride (20 mg/mL in pyridine) was added to the dried sample, the mixture was ultrasonicated for 20 min, gently vortexed, and incubated with shaking for 90 min at 30 °C. For derivatization, 50 μL of N-methyl-N-trimethylsilyl-trifluoroacetamide (MSTFA) was added and the solution was vortexed and incubated for 30 min at 37 °C. After centrifugation at 12,000 rpm and 4 °C for 5 min, the supernatant was used for GC–MS analysis on a GC–MS-QP2020 instrument (Shimadzu, Kyoto, Japan) equipped with an RTX-5MS column (Restek, Bellefonte, PA, USA). The temperature program was designed as follows: the initial GC oven temperature was set at 80 °C, held for 2 min, increased to 330 °C at a ramp rate of 15 °C/min, and held for 6 min at the final temperature. The carrier gas (helium) flow rate was 1 mL/min, the m/z range was set at 85–500, and the electron impact ionization was set at 70 eV. The raw data were converted to netCDF format files using the GC–MS post-run analysis software (Shimadzu, Kyoto, Japan), followed by processing for peak detection and alignment using MetAlign software (https://www.metAlign.nl). The resulting csv files were analyzed using AIoutput software to identify peaks based on retention time and retention index. The final data were further processed for principal component analysis (PCA) using SIMCA version 16 (Umetrics, Umea, Sweden).
2.5. Statistical analysis
Statistical analysis was conducted using IBM SPSS statistics version 26 (SPSS Inc., Chicago, IL, USA). After testing for normality, non-parametric data were analyzed using the Mann-Whitney U test, with significance set at p less than 0.05.
3. Results
3.1. Properties and characteristics of commercial makgeolli
Various makgeolli products with different combinations of properties, such as alcohol content, raw material, saccharification agents, and food additives, were collected (Table S1). The average alcohol content was 7.30 ± 2.30%; the alcohol content of 58 products was below 6%, whereas that of 36 products was above 6%; the highest alcohol content was 15%. The density and alcohol contents of makgeolli differ based on whether water was added to the product and the amount of water added after fermentation. Thirteen samples used wheat, 89 contained non-glutinous rice, and 17 contained glutinous rice. Sixty-two products were brewed by inoculation of the traditional Korean saccharification agent, nuruk. Food additives, such as sweeteners and acid regulators, were added to most products, whereas 29 products were free of food additives.
Makgeolli environment is generally acidic. Average pH value and titratable acidity was 4.29 ± 0.29 and 0.51 ± 0.21%, respectively (Fig. 1a& 1b). The pH value and titratable acidity showed no significant changes during low-temperature storage. Fig. 1c shows the number of viable LAB cells at 10 and 30 days from the date of manufacture. The average viable cell numbers of LAB on day 10 and 30 post-manufacture were 5.61 ± 1.92 log CFU/mL and 5.39 ± 1.79 log CFU/mL, respectively. The cell counts between days 10 and 30 were not significantly different. Fifteen samples showed a difference of more than 1 log CFU/mL between days 10 and 30, of which 14 samples showed a decrease and one showed an increase in number. These results indicate that, on average, approximately 5 log CFU/mL of LAB exist in commercial makgeolli products and that a large number of viable LAB cells remain until 30 days of low-temperature storage.
Fig. 1.
Physicochemical properties of makgeolli samples at day 10 and day 30 from manufacture. (a) pH value, (b) titratable acidity (TA), and (c) viable cell numbers of LAB were shown.
3.2. Microbial characteristics
The bacteria in the commercial makgeolli samples were classified into 11 phyla, mainly belonging to Firmicutes, Proteobacteria, Bacteroidetes, and Actinobacteria. In 94.95% of the samples, the phylum Firmicutes was dominant, representing 75.27 ± 18.49% (maximum 99.74%, minimum 18.68%) of the population on average. The average relative abundance of the overall LAB genera in commercial makgeolli was 58.49 ± 23.07%. Among LAB, the genera Aerococcus, Carnobacterium, Enterococcus, Tetragenococcus, Lactobacillus, Pediococcus, Leuconostoc, Weissella, Lactococcus, and Streptococcus were detected in the commercial makgeolli samples. At the species level, the following 25 species were detected in makgeolli: Carnobacterium viridans, Enterococcus casseliflavus, Tetragenococcus halophilus, Lactobacillus acidipiscis, Lactobacillus brevis, Lactobacillus delbrueckii, Lactobacillus hamsteri, Lactobacillus helveticus, Lactobacillus anihotivorans, Lactobacillus paralimentarius, Lactobacillus paraplantarum, Lactobacillus plantarum, Lactobacillus pontis, Lactobacillus zeae, Pediococcus acidilactici, Pediococcus ethanolidurans, Leuconostoc fallax, Leuconostoc mesenteroides, Weissella beninensis, Weissella ghanensis, Weissella hellenica, Weissella paramesenteroides, Weissella viridescens, Lactococcus garvieae, and Streptococcus luteciae.
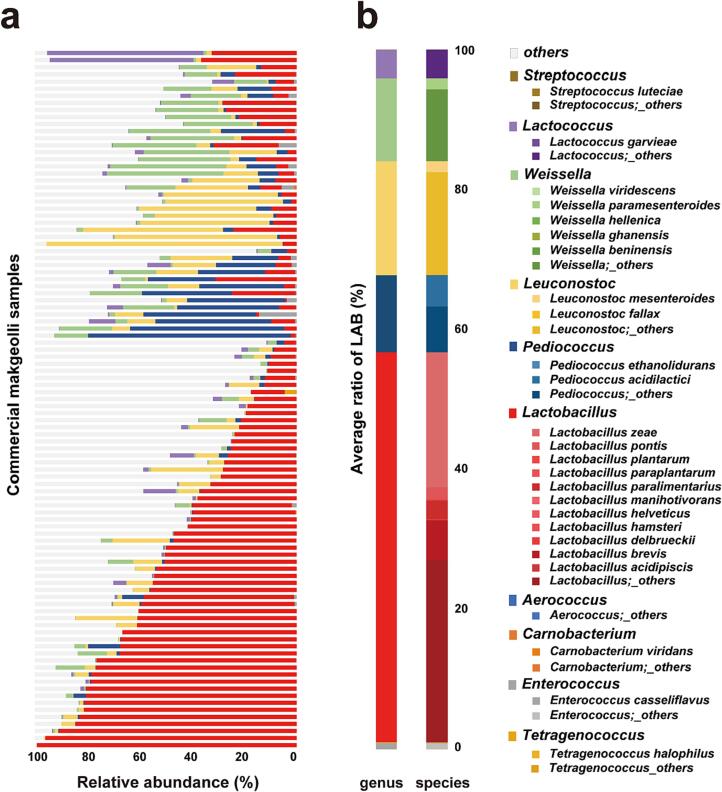
Five LAB genera were identified as the prevailing genera in makgeolli (Fig. 2a & 2b). Commercial makgeolli samples were aligned based on the most abundant among the LAB genera, indicating that Lactobacillus was the dominant genus in 58 samples (Fig. 2a). In the other samples, the genera Weissella, Pediococcus, Leuconostoc, and Lactococcus were the most abundant. To compare the abundance of the LAB genera themselves, the ratio of LAB genera and species was calculated by averaging the relative abundance in all collected samples (Fig. 2b). Among all LAB genera, the genus Lactobacillus was dominant, accounting for 55.76% of the LAB genera, followed by Leuconostoc (16.32%), Weissella (11.86%), Pediococcus (11.04%), and Lactococcus (4.02%).
Fig. 2.
Overall profile of LAB in makgeolli. (a) Composition of microbial community in makgeolli at the genus level. Samples were aligned based on the most abundant genus among the LAB genera. (b) Average ratio of LAB genera and species in makgeolli. The proportion of each LAB genus and species when the sum of relative abundance in all LAB was considered as 100.
Table 1 shows the average relative abundance and detection rate for each LAB genus and species in commercial makgeolli samples. Lactobacillus, the most abundant genus, accounted for an average of 32.61 ± 27.31% of the relative abundance in the makgeolli microbiome, followed by Leuconostoc, Weissella, and Pediococcus. Similarly, the most frequent LAB genus detected in all samples was Lactobacillus, followed by Leuconostoc, Weissella, Lactococcus, and Pediococcus. Thus, Lactobacillus was the most abundant and frequent genus in makgeolli. Among the identified species, Lactobacillus zeae had the highest relative abundance and detection rate as it was detected in 92% of the samples, accounting for an average relative abundance of 11.26 ± 18.41% (max 95.77%). Likewise, Lactobacillus brevis accounted for an average of 3.32 ± 7.47% (max 45.45%) of the makgeolli microbiome and was found in 80% of the samples. One of the most abundant species on average was Pediococcus acidilactici, with an average relative abundance of 2.62 ± 10.16% (max 76.30%). Lactobacillus paralimentarius detected in 71% of the samples, with an average of 1.6 ± 6.52% (max 58.65%). Leuconostoc mesenteroides and Weissella paramesenteroides were detected in 34% and 39% of the samples, respectively, representing an average relative abundance of 0.9%. Lactobacillus pontis accounted for an average of 1 ± 5.63% (max 51.95%) of the makgeolli microbiome.
Table 1.
Average proportions and frequencies of LAB in various commercial makgeolli during low-temperature storage.
| Genus | Species | Average (%) |
Frequency (%) |
||
|---|---|---|---|---|---|
| Day 10 | Day 30 | Day 10 | Day 30 | ||
| Aerococcus | 0.00 | 0.00 | 1.01 | 0.00 | |
| Aerococcus;_others | 0.00 | 0.00 | 1.01 | 0.00 | |
| Carnobacterium | 0.01 | 0.01 | 2.02 | 3.03 | |
| Carnobacterium viridans | 0.00 | 0.00 | 0.00 | 1.01 | |
| Carnobacterium;_others | 0.01 | 0.01 | 2.02 | 2.02 | |
| Enterococcus | 0.52 | 0.46 | 44.44 | 49.49 | |
| Enterococcus casseliflavus | 0.03 | 0.03 | 5.05 | 10.10 | |
| Enterococcus;_others | 0.48 | 0.44 | 41.41 | 44.44 | |
| Tetragenococcus | 0.05 | 0.00 | 4.04 | 2.02 | |
| Tetragenococcus halophilus | 0.01 | 0.00 | 2.02 | 1.01 | |
| Tetragenococcus_others | 0.05 | 0.00 | 3.03 | 1.01 | |
| Lactobacillus | 32.61 | 31.45 | 100.00 | 100.00 | |
| Lactobacillus acidipiscis | 0.00 | 0.00 | 6.06 | 5.05 | |
| Lactobacillus brevis | 3.32 | 2.83 | 80.81 | 78.79 | |
| Lactobacillus delbrueckii | 0.04 | 0.03 | 11.11 | 12.12 | |
| Lactobacillus hamsteri | 0.00 | 0.00 | 1.01 | 0.00 | |
| Lactobacillus helveticus | 0.01 | 0.01 | 4.04 | 4.04 | |
| Lactobacillus manihotivorans | 0.01 | 0.00 | 3.03 | 3.03 | |
| Lactobacillus paralimentarius | 1.60 | 1.57 | 70.71 | 66.67 | |
| Lactobacillus paraplantarum | 0.00 | 0.00 | 2.02 | 4.04 | |
| Lactobacillus plantarum | 0.10 | 0.07 | 14.14 | 12.12 | |
| Lactobacillus pontis | 1.00 | 0.73 | 23.23 | 17.17 | |
| Lactobacillus zeae | 11.26 | 13.15 | 91.92 | 92.93 | |
| Lactobacillus;_others | 15.28 | 13.06 | 100.00 | 100.00 | |
| Pediococcus | 6.46 | 5.39 | 79.80 | 77.78 | |
| Pediococcus acidilactici | 2.62 | 2.01 | 35.35 | 32.32 | |
| Pediococcus ethanolidurans | 0.00 | 0.00 | 1.01 | 1.01 | |
| Pediococcus;_others | 3.83 | 3.38 | 66.67 | 68.69 | |
| Leuconostoc | 9.54 | 10.46 | 96.97 | 96.97 | |
| Leuconostoc fallax | 0.00 | 0.00 | 1.01 | 0.00 | |
| Leuconostoc mesenteroides | 0.91 | 0.97 | 34.34 | 36.36 | |
| Leuconostoc;_others | 8.63 | 9.49 | 96.97 | 96.97 | |
| Weissella | 6.94 | 8.03 | 92.93 | 90.91 | |
| Weissella beninensis | 0.00 | 0.00 | 1.01 | 0.00 | |
| Weissella ghanensis | 0.01 | 0.01 | 6.06 | 7.07 | |
| Weissella hellenica | 0.00 | 0.00 | 1.01 | 0.00 | |
| Weissella paramesenteroides | 0.92 | 1.03 | 39.39 | 40.40 | |
| Weissella viridescens | 0.00 | 0.00 | 2.02 | 2.02 | |
| Weissella;_others | 6.01 | 6.99 | 89.90 | 87.88 | |
| Lactococcus | 2.35 | 2.26 | 80.81 | 80.81 | |
| Lactococcus garvieae | 0.01 | 0.01 | 5.05 | 5.05 | |
| Lactococcus;_others | 2.34 | 2.25 | 80.81 | 80.81 | |
| Streptococcus | 0.01 | 0.01 | 14.14 | 15.15 | |
| Streptococcus luteciae | 0.00 | 0.00 | 3.03 | 5.05 | |
| Streptococcus;_others | 0.00 | 0.00 | 11.11 | 10.10 | |
| Others | 41.51 | 41.94 | 100.00 | 100.00 | |
To determine whether the microbiome profile was altered during storage, changes in the microbial taxonomic distribution, alpha diversity, and beta diversity were investigated. There was no significant difference in alpha and beta diversity between days 10 and 30 after manufacture, indicating that there were no significant changes in the microbial profile during low-temperature storage (Fig. S1). Additionally, comparing the microbial proportion and frequency at days 10 and 30 revealed no significant alterations in the average relative abundance and frequency of each LAB genus during low-temperature storage (Table 1). Thus, there was no significant alteration in LAB profile during low-temperature storage.
3.3. Factors contributing to LAB genus abundance
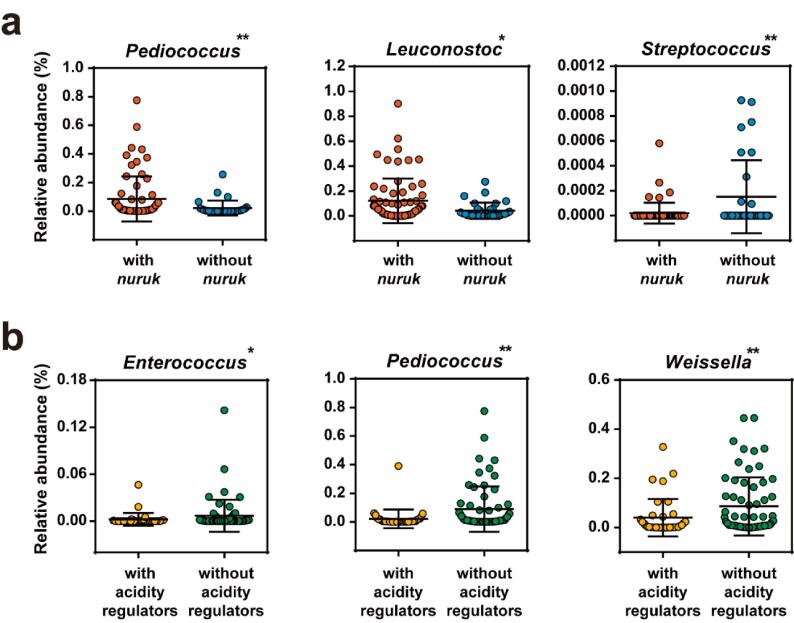
Based on each characteristic factor of makgeolli samples, including alcohol content, inoculated starter type, and addition of an acidity regulator, factors that could affect the LAB genus were verified. Two factors, nuruk usage and acidity regulator usage, were relevant to the relative abundance of some LAB genera. Fig. 3a shows that the commercial makgeolli products prepared using nuruk demonstrated a higher relative abundance of the genera Pediococcus and Leuconostoc and a lower relative abundance of the genus Streptococcus compared to that in the samples without nuruk. Likewise, commercial products supplemented with acidity regulators, such as citric acid and lactic acid, showed relatively lower relative abundances of the genera Enterococcus, Pediococcus, and Weissella (Fig. 3b). The other characteristics of each commercial product, including alcohol content and raw starch material type, showed no significant effect on LAB genera.
Fig. 3.
Comparison of significantly altered relative abundance of LAB genera based on the addition of (a) nuruk and (b) acidity regulators.
3.4. Metabolite changes during low-temperature storage
PCA was performed to investigate the metabolite changes in makgeolli during low-temperature storage (Fig. 4a). Makgeolli samples stored for 10 and 30 days at refrigeration temperature were not clearly separated on the PCA score plot, indicating that the metabolite profile was hardly altered during the storage period. Of the 162 peaks detected using GC–MS, 63 metabolites were identified. Although some metabolites were significantly different, there was no significant change in the contents of lactic acid, the major metabolite of LAB, indicating that LAB presence did not significantly affect the quality of makgeolli under low-temperature storage conditions (Fig. 4b).
Fig. 4.
Metabolite profile during low-temperature storage. (a) Principal component analysis score plot derived from the GC–MS dataset of day 10 and day 30 samples (R2X = 0.351, Q2 = 0.296). (b) Comparison of lactic acid content between day 10 and day 30 samples.
4. Discussion
Despite differences in manufacturing methods, raw materials, and saccharification agents, various types of LAB were detected in all analyzed commercial makgeolli samples. The number of viable cells in LAB was determined to be above 5 log CFU/mL on average, which was slightly lower than the results of previous studies reporting viable cell numbers of LAB as 7–8 log CFU/mL on average (Kim et al., 2018, Park et al., 2017, Yoon et al., 2012). These differences may be attributed to differences in the makgeolli manufacturing methods or storage temperatures. The presence of a large number of LAB in makgeolli without bacterial inoculation during manufacture suggests that LAB proliferated actively during the fermentation period.
To date, the previously reported dominating and existing species in makgeolli have been widely varied, leading to lack of information on the commonly existing LAB genera. Overall, the LAB genera presenting in makgeolli or dominating the system remain to be established. For example, in some previous studies, L. plantarum, W. cibaria, and P. pentosaseus were reported as the dominant species in makgeolli, along with the detection of various species belonging to Lactobacillus including L. paracasei, L. arizonensis, L. brevis, L. fermentum, and several species belonging to Weissella, Leuconostoc, and Pediococcus (Jin et al., 2008, Jung et al., 2012, Park et al., 2017, Yoon et al., 2012). In the current study, we revealed that the most abundant and frequent LAB genus in commercial makgeolli was Lactobacillus, which could be considered as the dominant genus in the liquor. Moreover, the most abundant and frequent LAB species was Lactobacillus zeae, which is known to be isolated from raw milk and is closely related to L. casei, L. paracasei, and L. rhamnoses (Desai et al., 2006, Lim et al., 2008). In addition, L. brevis, P. acidilactici, Leuconostoc mesenteroides, and Weissella paramesenteroides also showed a high relative abundance and frequency, which could be expected to exist in the liquor. Additionally, among 25 LAB species, most belonged to the genus Lactobacillus, followed by the genera Weissella, Pediococcus, and Leuconostoc. Notably, 18 species were reported for the first time in rice wine, including Carnobacterium viridans, Enterococcus casseliflavus, Tetragenococcus halophilus, Lactobacillus hamsteri, Lactobacillus helveticus, Lactobacillus manihotivorans, Lactobacillus paralimentarius, Lactobacillus paraplantarum, Lactobacillus pontis, Lactobacillus zeae, Pediococcus ethanolidurans, Leuconostoc fallax, Weissella beninensis, Weissella ghanensis, Weissella hellenica, Weissella viridescens, Lactococcus garvieae, and Streptococcus luteciae. Meanwhile, a considerable number of previously reported LAB species were not detected in this study, even though we analyzed 94 commercial makgeolli samples. This could be attributed to the bacterial inoculation-free manufacture of makgeolli and the use of various kinds of saccharification agents. Collectively, we confirmed that makgeolli contains various genera and species of LAB in its microbiome, and that the most abundant LAB genus is Lactobacillus.
The origin of LAB in makgeolli remains unclear. All samples in the current study contained LAB, which limited the investigation of their origin. Although factors such as use of a saccharification agent or acidity regulator were found to affect the abundance of LAB genera in makgeolli, identifying the origin of the bacterial community remains challenging. The saccharification agent, which has been reported to possess its own bacterial community, is a potential source of the LAB in makgeolli (Bal et al., 2015, Song et al., 2013). Another possible source is the brewing environment (Hu et al., 2021). However, further studies on the presence of LAB in the materials used and changes during the manufacture period are required to elucidate the origin of LAB in makgeolli.
In the current study, during low-temperature storage, the viable LAB cell number and relative abundance of each LAB species were not altered significantly. Further, lactic acid, the noteworthy metabolite of LAB, did not show significant differences in content between days 10 and 30, indicating that although LAB exist in the makgeolli system, they have a minimal impact on metabolite changes in makgeolli during low-temperature storage. However, as no specific LAB inoculation was performed during brewing, viable LAB cell numbers exceeding 5 log CFU/mL could be considered to indicate cell proliferation in the makgeolli system at relatively high temperatures (22–28 °C) during the fermentation period. If LAB proliferate in makgeolli during fermentation, they may participate in altering the metabolic profile and quality of the liquor. Dynamic alterations in LAB cell numbers and profiles during the fermentation period have been reported, although the results vary slightly among studies. According to Chai et al. (Chai et al., 2015), LAB genera dominated the makgeolli system from day 2, with the population shifting complementarily between Pediococcus and Weissella. Jung et al. (Jung et al., 2012) reported complicated results showing that some species of Lactobacillus and Weissella vanished, whereas some species of Lactobacillus increased in proportion with time. Overall, the role of LAB in makgeolli needs to be understood for quality control, quality enhancement, and the development of inoculation strains. Similar to wine, where specific LAB species are inoculated to induce malolactic fermentation, specific species could be inoculated if LAB are found to be necessary for makgeolli brewing. If the presence of LAB or certain species has beneficial effects on makgeolli quality, a mixture of LAB species could be developed for inoculation. However, if LAB existence is found to have a poor impact on liquor quality, strategies to control their microbial proportion in the system would need to be formulated.
Although LAB possibly might have small impact on the metabolic profile of makgeolli during low-temperature storage, they present and maintain their viable cell number until at least the shelf life of the product, indicating the potential of makgeolli to be considered as a probiotic bacteria-containing alcoholic beverage. The notified probiotic species in Korea, Lactobacillus helveticus and Lactobacillus plantarum, were detected in makgeolli samples in the present study. In addition, some strains of Enterococcus casseliflavus, Tetragenococcus halophilus, Lactobacillus delbrueckii, Lactobacillus paraplantarum, Lactobacillus zeae, Pediococcus acidilactici, Leuconostoc mesenteroides, Weissella hellenica, Weissella paramesenteroides, and Lactococcus garvieae, which were detected in this study, have been reported to possess probiotic potential (Ayyash et al., 2020, Barigela and Bhukya, 2021, Huang et al., 2021, Inayah et al., 2022, Kim et al., 2022, Pabari et al., 2020, Panthee et al., 2019, Safari et al., 2016, Son et al., 2018). These species collectively accounted for 4.65% of the relative abundance of the makgeolli microbiome, and a total cell number of 7.15 log CFU per bottle of makgeolli. Although this number is less than 8–9 log CFU, which the usually considered a standard probiotic cell number, its potential to alter the host microbiome needs to be studied further.
5. Conclusion
To our knowledge, this is the first study to provide an overview of the average LAB cell number in non-pasteurized commercial makgeolli products along with their LAB profiles. It is confirmed that LAB exist in makgeolli, displaying various species with an average viable cell number of 5.61 log CFU/mL. Overall, 10 LAB genera and 25 LAB species were detected; the most abundant and frequent LAB genus was Lactobacillus. The microbial profile and cell number were minimally altered under low-temperature storage and had minimal impact on the metabolite profile of makgeolli. Overall, a bottle of makgeolli may contain LAB species with probiotic properties and considerable cell numbers until shelf life, which might provide potential probiotic benefits. Although further studies examining the contribution of LAB to makgeolli and on their role and origin are needed, the present study provides useful insights into the microbial diversity of LAB in makgeolli. Collectively, the results of this study could contribute to enhance the understanding of basic microbial properties, especially those related to LAB, in the traditional Korean hazy liquor makgeolli.
CRediT authorship contribution statement
Jeongmin Cha: Conceptualization, Methodology, Data curation, Software, Formal analysis, Writing – original draft. Kwang-Moon Cho: Software, Formal analysis. Sun Jae Kwon: Software. Seong-Eun Park: Visualization, Investigation. Eun-Ju Kim: Investigation. Seung-Ho Seo: Formal analysis, Investigation. Hong-Seok Son: Supervision, Project administration, Writing – review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) (No. 2019R1C1C1002208 and 2020R1I1A1A0106992012), a Korea University Grant, and the Institute of Biomedical Science and Food Safety, CJ-Korea University Food Safety Hall at Korea University, Republic of Korea.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochx.2022.100552.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Ayyash M., Abu-Jdayil B., Itsaranuwat P., Almazrouei N., Galiwango E., Esposito G., Hunashal Y., Hamed F., Najjar Z. Exopolysaccharide produced by the potential probiotic Lactococcus garvieae C47: Structural characteristics, rheological properties, bioactivities and impact on fermented camel milk. Food Chemistry. 2020;333 doi: 10.1016/j.foodchem.2020.127418. [DOI] [PubMed] [Google Scholar]
- Bal J., Yun S.-H., Choi M.-S., Yeo S.-H., Kim J.-M., Kim D.-H. Pyrosequencing reveals bacterial diversity in Korean traditional wheat-based nuruk. Journal of Microbiology. 2015;53(12):812–819. doi: 10.1007/s12275-015-5516-3. [DOI] [PubMed] [Google Scholar]
- Barigela A., Bhukya B. Probiotic Pediococcus acidilactici strain from tomato pickle displays anti-cancer activity and alleviates gut inflammation in-vitro. 3. Biotech. 2021;11(1):1–11. doi: 10.1007/s13205-020-02570-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A.…Asnicar F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature biotechnology. 2019;37(8):852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nature methods. 2016;13(7):581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai C., Lim G.S., Kim Y.J., Oh S.W. Microbial community changes in Makgeolli during brewing. Journal of the Institute of Brewing. 2015;121(2):304–308. [Google Scholar]
- Desai A., Shah N.P., Powell I. Discrimination of dairy industry isolates of the Lactobacillus casei group. Journal of dairy science. 2006;89(9):3345–3351. doi: 10.3168/jds.S0022-0302(06)72371-2. [DOI] [PubMed] [Google Scholar]
- García-Ruiz A., Bartolomé B., Martínez-Rodríguez A.J., Pueyo E., Martín-Álvarez P.J., Moreno-Arribas M. Potential of phenolic compounds for controlling lactic acid bacteria growth in wine. Food Control. 2008;19(9):835–841. [Google Scholar]
- Garofalo C., Osimani A., Milanović V., Taccari M., Aquilanti L., Clementi F. The occurrence of beer spoilage lactic acid bacteria in craft beer production. Journal of food science. 2015;80(12):M2845–M2852. doi: 10.1111/1750-3841.13112. [DOI] [PubMed] [Google Scholar]
- Glöckner F.O., Yilmaz P., Quast C., Gerken J., Beccati A., Ciuprina A., Bruns G., Yarza P., Peplies J., Westram R., Ludwig W. 25 years of serving the community with ribosomal RNA gene reference databases and tools. Journal of biotechnology. 2017;261:169–176. doi: 10.1016/j.jbiotec.2017.06.1198. [DOI] [PubMed] [Google Scholar]
- Hu Y., Lei X., Zhang X., Guan T., Wang L., Zhang Z., Yu X., Tu J., Peng N., Liang Y., Zhao S. Characteristics of the Microbial Community in the Production of Chinese Rice-Flavor Baijiu and Comparisons With the Microflora of Other Flavors of Baijiu. Frontiers in microbiology. 2021;12 doi: 10.3389/fmicb.2021.673670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.-H., Chen Y.-H., Chen J.-H., Hsu P.-S., Wu T.-H., Lin C.-F., Peng C.-C., Wu M.-C. A potential probiotic Leuconostoc mesenteroides TBE-8 for honey bee. Scientific reports. 2021;11(1):1–13. doi: 10.1038/s41598-021-97950-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inayah I., Wibowo M.S., Julianti E., Suciati T. Characterization of Lactobacillus zeae as probiotic and starter culture for tamarillo fermented product. Food Science and Technology. 2022;42 [Google Scholar]
- Jin J., Kim S.-Y., Jin Q., Eom H.-J., Han N.-S. Diversity analysis of lactic acid bacteria in Takju, Korean rice wine. Journal of microbiology and biotechnology. 2008;18(10):1678–1682. [PubMed] [Google Scholar]
- Jung M.-J., Nam Y.-D., Roh S.W., Bae J.-W. Unexpected convergence of fungal and bacterial communities during fermentation of traditional Korean alcoholic beverages inoculated with various natural starters. Food microbiology. 2012;30(1):112–123. doi: 10.1016/j.fm.2011.09.008. [DOI] [PubMed] [Google Scholar]
- Kim D.H., Kim S.-A., Jo Y.M., Seo H., Kim G.Y., Cheon S.W., Yang S.H., Jeon C.O., Han N.S. Probiotic potential of Tetragenococcus halophilus EFEL7002 isolated from Korean soy Meju. BMC microbiology. 2022;22(1):1–17. doi: 10.1186/s12866-022-02561-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.Y., Kim D., Park P., Kang H.-I., Ryu E.K., Kim S.M. Effects of storage temperature and time on the biogenic amine content and microflora in Korean turbid rice wine. Makgeolli. Food Chemistry. 2011;128(1):87–92. doi: 10.1016/j.foodchem.2011.02.081. [DOI] [PubMed] [Google Scholar]
- Kim N.H., Jun S.H., Lee S.H., Hwang I.G., Rhee M.S. Microbial diversities and potential hazards of Korean turbid rice wines (makgeolli): Multivariate analyses. Food microbiology. 2018;76:466–472. doi: 10.1016/j.fm.2018.07.008. [DOI] [PubMed] [Google Scholar]
- King W.L., Siboni N., Williams N.L., Kahlke T., Nguyen K.V., Jenkins C., Dove M., O’Connor W., Seymour J.R., Labbate M. Variability in the composition of Pacific oyster microbiomes across oyster families exhibiting different levels of susceptibility to OsHV-1 μvar disease. Frontiers in microbiology. 2019;10:473. doi: 10.3389/fmicb.2019.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.R., Kim H.R., Kim J.-H., Ahn B.-H., Lee J.-E. Isolation of lactic acid bacteria and its application for synbiotics makgeolli fermentation. Current Topics in Lactic Acid Bacteria and Probiotics. 2014;2(1):27–33. [Google Scholar]
- Lim S.-D., Kim K.-S., Do J.-R. Physiological characteristics and ACE inhibitory activity of Lactobacillus zeae RMK354 isolated from raw milk. Food Science of Animal Resources. 2008;28(5):587–595. [Google Scholar]
- Min J.-H., Kim Y.-H., Kim J.-H., Choi S.-Y., Lee J.-S., Kim H.-K. Comparison of microbial diversity of Korean commercial Makgeolli showing high β-glucan content and high antihypertensive activity, respectively. Mycobiology. 2012;40(2):138–141. doi: 10.5941/MYCO.2012.40.2.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon Y.-J., Baik S.-H., Cha Y.-S. Lipid-lowering effects of Pediococcus acidilactici M76 isolated from Korean traditional makgeolli in high fat diet-induced obese mice. Nutrients. 2014;6(3):1016–1028. doi: 10.3390/nu6031016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabari K., Pithva S., Kothari C., Purama R.K., Kondepudi K.K., Vyas B.R.M., Kothari R., Ambalam P. Evaluation of probiotic properties and prebiotic utilization potential of Weissella paramesenteroides isolated from fruits. Probiotics and Antimicrobial Proteins. 2020;12(3):1126–1138. doi: 10.1007/s12602-019-09630-w. [DOI] [PubMed] [Google Scholar]
- Panthee S., Paudel A., Blom J., Hamamoto H., Sekimizu K. Complete genome sequence of Weissella hellenica 0916-4-2 and its comparative genomic analysis. Frontiers in Microbiology. 2019:1619. doi: 10.3389/fmicb.2019.01619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Seo J.S., Kim S.-A., Shin S.-Y., Park J.-H., Han N.S. Microbial diversity of commercial makgeolli and its influence on the organoleptic characteristics of Korean rice sourdough, jeung-pyun. Journal of Microbiology and Biotechnology. 2017;27(10):1736–1743. doi: 10.4014/jmb.1708.08003. [DOI] [PubMed] [Google Scholar]
- Park Y.-U., Kim M.-D., Jung D.-H., Seo D.-H., Jung J.-H., Park J.-G., Hong S.-Y., Cho J.-Y., Park S.-Y., Park J.-W., Shin W.-C., Park J.-W. Probiotic properties of lactic acid bacteria isolated from Korean rice wine Makgeolli. Food Science and Biotechnology. 2015;24(5):1761–1766. [Google Scholar]
- Safari R., Adel M., Lazado C.C., Caipang C.M.A., Dadar M. Host-derived probiotics Enterococcus casseliflavus improves resistance against Streptococcus iniae infection in rainbow trout (Oncorhynchus mykiss) via immunomodulation. Fish & Shellfish Immunology. 2016;52:198–205. doi: 10.1016/j.fsi.2016.03.020. [DOI] [PubMed] [Google Scholar]
- Seo S.-H., Park S.-E., Yoo S.-A., Lee K.I., Na C.-S., Son H.-S. Metabolite profiling of Makgeolli for the understanding of yeast fermentation characteristics during fermentation and aging. Process Biochemistry. 2016;51(10):1363–1373. [Google Scholar]
- Son S.-H., Yang S.-J., Jeon H.-L., Yu H.-S., Lee N.-K., Park Y.-S., Paik H.-D. Antioxidant and immunostimulatory effect of potential probiotic Lactobacillus paraplantarum SC61 isolated from Korean traditional fermented food, jangajji. Microbial Pathogenesis. 2018;125:486–492. doi: 10.1016/j.micpath.2018.10.018. [DOI] [PubMed] [Google Scholar]
- Song S.H., Lee C., Lee S., Park J.M., Lee H.-J., Bai D.-H., Yoon S.-S., Choi J.B., Park Y.-S. Analysis of microflora profile in Korean traditional nuruk. Journal of microbiology and biotechnology. 2013;23(1):40–46. doi: 10.4014/jmb.1210.10001. [DOI] [PubMed] [Google Scholar]
- Suzuki K., Asano S., Iijima K., Kitamoto K. Sake and beer spoilage lactic acid bacteria—a review. Journal of the Institute of Brewing. 2008;114(3):209–223. [Google Scholar]
- Yoon S.-S., Choi J.-A., Kim K.-H., Song T.-S., Park Y.-S. Populations and potential association of Saccharomyces cerevisiae with lactic acid bacteria in naturally fermented Korean rice wine. Food science and biotechnology. 2012;21(2):419–424. [Google Scholar]
- Yoon W., Park S.H., Lee J.S., Byeon J.H., Kim S.H., Lim J., Yoo Y. Probiotic mixture reduces gut inflammation and microbial dysbiosis in children with atopic dermatitis. Australasian Journal of Dermatology. 2021;62(3):e386–e392. doi: 10.1111/ajd.13644. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.